- 1Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
- 2Department of Infectious Diseases, Wenzhou Sixth People's Hospital, Wenzhou Central Hospital Medical Group, Wenzhou, China
Background: Whole-genome sequencing (WGS) is a viable and financially feasible tool for timely and comprehensive diagnosis of drug resistance in developed countries. With the increase in the incidence of multidrug-resistant tuberculosis (MDR-TB), second-line anti-TB drugs are gaining importance. However, genetic resistance to second-line anti-TB drugs based on WGS has not been fully studied.
Methods: We randomly selected 100 MDR-TB and 10 non-MDR-TB isolates from a hospital in Zhejiang Province, China. Drug susceptibility tests against 13 anti-TB drugs were performed, and 34 drug resistance-related genes were analyzed using WGS in all isolates. For each drug, the accuracy, sensitivity, specificity, and positive and negative predictive values of WGS were compared with those of the conventional drug susceptibility test.
Results: The overall sensitivity and specificity for WGS were respectively, 99.0 and 100.0% for isoniazid (INH), 99.0 and 100.0% for rifampicin (RIF), 94.8 and 65.3% for ethambutol (EMB), 86.2 and 84.4% for pyrazinamide (PZA), 95.6 and 95.6% for levofloxacin (LFX), 89.5 and 65.3% for moxifloxacin (MFX), 91.3 and 95.1% for streptomycin (SM), 90.9 and 99.0% for kanamycin, 90.9 and 100.0% for amikacin, 88.9 and 98.0% for capreomycin, 87.0 and 85.1% for prothionamide (PTO), 85.7 and 99.0% for para-aminosalicylic acid (PAS), and 66.7 and 95.9% for clofazimine (CLO).
Conclusions: WGS is a promising approach to predict resistance to INH, RIF, PZA, LFX, SM, second-line injectable drugs (SLIDs), and PTO with satisfactory accuracy, sensitivity, and specificity of over 85.0%. The specificity of WGS in diagnosing resistance to EMB, and high-level resistance to MFX (2.0 mg/L) needs to be improved.
Introduction
In 2017, there were ~558,000 new cases of rifampicin-resistant tuberculosis (RR-TB) worldwide, including 82% multidrug-resistant (MDR-TB) cases with resistance to isoniazid (INH), and rifampicin (RIF). Only 28% of MDR/RR-TB cases have been detected worldwide (World Health Organization, 2018a). GeneXpert MTB/RIF as a sensitive and quick diagnostic tool is the recommended initial diagnostic test to detect pulmonary TB and RIF resistance (World Health Organization, 2013; Denkinger et al., 2014; Steingart et al., 2014). Line probe assays (LPAs) enable rapid drug-susceptibility testing for RIF and INH resistance and Mycobacterium tuberculosis detection. However, conventional culture and drug susceptibility tests (DSTs) for second-line agents are currently still indispensable in countries with documented or suspected cases of extensive drug resistant TB (XDR-TB) to ensure quality testing of second-line agents, following the World Health Organization (WHO) policy guidance (Denkinger et al., 2014; Steingart et al., 2014). The cumbersome technology, high infrastructure requirements, and long cycle has made accessibility to DST is low. In China, only 18% cases of DR-TB were documented (World Health Organization, 2018a).
Studies have confirmed the viability and financial feasibility of whole-genome sequencing (WGS) in the diagnosis of clinical resistance to first-line anti-TB drugs, especially in developed countries (Walker et al., 2015; Farhat et al., 2016; Pankhurst et al., 2016; Miotto et al., 2017; Allix-Béguec et al., 2018; Zignol et al., 2018). With the rise in the incidence of DR-TB in China, South Africa, Russia, and other developing countries, second-line anti-TB drugs are becoming increasingly important. WGS can quickly and comprehensively detect all possible drug resistance genes simultaneously, which could provide more information for clinical treatment, especially MDR-TB. However, there are only a few studies on the accuracy of WGS in predicting resistance to second-line anti-TB agents. As the country with the second highest burden of DR-TB worldwide, it would be more meaningful and useful if WGS could accurately diagnose drug resistance to second- and first-line drugs.
Therefore, in this study, we evaluated the performance of a WGS approach for diagnosing drug susceptibility and resistance to 100 MDR-TB, and 10 non-MDR-TB clinical isolates in China. The results of the WGS were compared with phenotypic DST results of 13 anti-TB drugs including drugs less studied but receiving increasing attention [para-aminosalicylic acid (PAS), prothionamide (PTO), and clofazimine (CLO)].
Materials and Methods
Clinical Isolates
In total, 100 clinical MDR Mycobacterium tuberculosis (MTB) isolates were randomly selected from preserved MDR-TB strains between January 1, 2014 and June 30, 2017 at the Sixth People's Hospital of Wenzhou city, Zhejiang Province, China. To better evaluate test specificities, 10 clinical non-MDR-TB isolates were also selected randomly. This was a retrospective study and the strains included were all stocked strains. All isolates and the reference strain MTB H37Rv were thawed and cultured on Löwenstein–Jensen medium. A colloidal gold assay (Genesis Biodetection and Biocontrol Ltd., Hangzhou, Zhejiang Province, China) to detect MPB64 antigen was used routinely to differentiate MTB from NTM. All NTM isolates were excluded from the study. After extracting the DNA, we confirmed TB further using polymerase chain reaction (PCR) amplification, and Sanger sequencing of 16s rRNA. If a patient had multiple isolates, the earlier isolates were excluded.
Drug Susceptibility Test
The drug susceptibility tests of all clinical isolates and the reference strain MTB H37Rv to 13 anti-TB drugs were carried out according to the Clinical and Laboratory Standards Institute (CLSI) and World Health Organization (WHO) guidelines. All antibiotics, except pyrazinamide (PZA) and PTO, were tested using the proportion method on commercial Löwenstein–Jensen medium with antibiotics (Baso, Zhuhai, Guangzhou Province, China). The critical concentrations were 0.2 mg/L for INH, 40.0 mg/L for RIF, 2.0 mg/L for ethambutol (EMB), 2.0 mg/L for levofloxacin (LFX), 2.0 mg/L for moxifloxacin (MFX), 4.0 mg/L for streptomycin (SM), 30.0 mg/L for amikacin (AM), 30.0 mg/L for kanamycin (KM), 40.0 mg/L for capreomycin (CM), 1.0 mg/L for PAS, and 1.0 mg/L for CLO, respectively, according to the CLSI guidelines, and WHO guidelines (Institute., CALS, 2011; World Health Organization, 2018b). The results were determined after 3 weeks incubation at 37°C. The susceptibility of MTB to PZA and PTO was evaluated using an automated Mycobacterial Growth Indicator Tube 960 system (Becton Dickinson Diagnostic Systems, Franklin Lakes, NJ, USA) according to the manufacturer's instructions at critical concentrations of 100.0 and 2.5 mg/L, respectively. The well-known limitation of technical feasibility and reproducibility of the phenotypic DST of PZA, EMB, and PTO, required the DST to be performed at least twice for these three drugs. If the two results were inconsistent, a third test was performed. All experiments using live MTB were performed in a biosafety level 2 plus laboratory.
WGS and Phylogenetic Tree Construction
MTB colonies were scraped from the Löwenstein–Jensen medium and genomic DNA was extracted using the Dneasy Blood & Tissue Kit (QIAGEN, Valencia, CA, USA), following the manufacturer's protocol. All sequencing libraries were prepared using the Nextera XT Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer's protocol and sequenced on Illumina Miseq, Illumina Hiseq, and BGISEQ-500 platforms, with depths of at least 50-fold coverage. After filtering the low-quality reads, the reads were aligned to the MTB H37Rv (GenBank NC000962.3) reference sequence using Bowtie2 (version 2.3.3.1) with default parameters. Single nucleotide polymorphisms (SNPs) were detected using SAMtools (version 1.6) with a minimum sequencing depth of 10 reads without strand bias and a frequency of no <10%.
Since duplicative clones or closely clustered strains would decrease mutation variety and effective statistical sample numbers, considering all 100 MDR strains were collected in one hospital, we built a phylogenetic tree using reliable SNPs of the whole genome to exclude possible genetic simplicity of the isolates. The phylogenetic tree was constructed using the maximum likelihood method using MEGA (version 7.0) software based on the reliable SNPs, with H37Rv as the root. If the mutation ratio of a certain site was above 80%, it was considered a fixed mutation. SNPs in highly repetitive regions such as PE and PPE, GC-enriched sequences, and drug resistance-related genes were removed from the analysis. The strains with a genetic distance of no more than 12 SNPs were considered recent transmission (Walker et al., 2013; Yang et al., 2017).
Interpretation of Mutations Related to Resistance
The genetic resistance diagnosis of INH (katG, inhA, and ahpC-promoter), RIF (rpoB), PZA (pncA), EMB (embB and embA), LFX (gyrA and gyrB), MFX (gyrA), AM (rrs), KM (eis and rrs), CM (tlyA and rrs), SM (rrs and rpsL), PTO (inhA and ethA), PAS (folC, thyA, dfrA, and ribD), and CLO (rv0678) were based on resistance mutation sites reported in previous studies (Table S1) (Chakraborty et al., 2013; Walker et al., 2015; Zhang et al., 2015a,b; Farhat et al., 2016; Miotto et al., 2017; Allix-Béguec et al., 2018; Liu et al., 2018). Isolates containing resistance mutations were diagnosed as genetically resistant.
In order to find new possible resistance-related mutations, we further analyzed all mutations in genes above and another 15 resistance-related genes for INH (nat, ndh, iniA, iniB, and iniC), PZA (rpsA and panD), EMB (embC, embR, and ubiA), SM (gidB), and CLO resistance (rv1979c, rv2535c, mmpL3, and mmpL5) (Table S1) (Wong et al., 2011; Zhang et al., 2013, 2015a; Hartkoorn et al., 2014; Shi et al., 2014; Vilchèze and Jacobs, 2014; Almeida et al., 2016; Li et al., 2017; Ismail et al., 2018; Sun et al., 2018; Khosravi et al., 2019; Tulyaprawat et al., 2019).
Statistical Analyses
The phenotypic DST was used as the gold standard to calculate the accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), and their 95% confidence intervals of WGS by Stata 13.1. The sensitivity and specificity are the percentages of true positives and negatives, respectively. The sensitivity and specificity are also the proportion of strains that were genetically resistant and sensitive among all phenotypic resistant and sensitive strains, respectively. PPV and NPV were calculated using the relevant formula.
Results
Basic and Clustering Information of Strains
In total, 110 strains were reconfirmed as MTB by aligning the sequencing reads to the reference genome of MTB H37Rv. After constructing the phylogenetic tree of the MDR strains, five pairs of isolates were found to be closely related, suggesting recent transmission events (Figure S1). The clustering rate of MDR isolates was 5.0%. The low clustering rate of the isolates excluded the possibility of low genetic variety of the isolates, albeit they were collected in one hospital and, therefore, indicated low risks of over-estimating the weight of specific rare mutations carried by the strains. Among them, 79 MDR and 8 non-MDR-TB strains belonged to lineage 2, and 21 MDR and 2 non-MDR TB strains belonged to lineage 4 (Coll et al., 2014).
DST
The susceptibility test for all drugs, except CLO, was performed on all clinical isolates; CLO susceptibility test was performed using only 80 isolates because of the low resistance rate. The antibiotic phenotypic resistance rate of all the clinical strains was as follows: INH, 91.8%; RIF, 90.9%; SM, 62.7%; LFX, 59.1%; PZA, 59.1%; EMB, 52.7%; MFX, 34.5%; PTO, 20.9%; AM, 10.0%; KM, 10.0%; CM, 8.2%; PAS, 6.4%; and CLO, 5.0%.
Comparison of WGS and Phenotypic DST Results
Drugs With High Accuracy
Inh
Among the 110 strains, 101, and 9 strains were INH-resistant and INH-sensitive, respectively. The most common INH-resistant mutations were KatG S315N/T, inhA c-15t, and KatG S315T+inhA c-15t, which accounted for 66.3, 11.9, and 5.0% of the phenotypically resistant strains, respectively (Table S2). Four INH-resistant strains had early termination or frameshift mutations in the katG gene (Q88*, W198*, 334 g-in + 713 gcg-in, and 2044 t-in). Six resistant strains carried resistance-associated mutations in the promoter of ahpC. According to previous criteria, the accuracy, sensitivity, specificity, PPV, and NPV, and their 95% confidence intervals of the WGS diagnosis for INH were 93.6%, 93.1% (85.6–96.9%), 100.0% (62.9–100.0%), 100.0% (95.1–100.0%), and 56.3% (30.6–79.2%), respectively (Table 1). Seven phenotypically resistant but genetically sensitive strains showed inconsistent results between the DST and WGS according to previous diagnosis criteria. Six of these seven strains carried mutations in katG, a well-defined INH resistance-related gene, namely Q88P+M257V, W91R, A122D, Q127P, A312E, and D419Y. After including the new mutations in the diagnosis criteria, the accuracy, and sensitivity improved to 99.1 and 99.0% (93.8–99.9%), respectively. No mutations in iniB and iniC were detected. Mutations in Ndh (Q52R, A154V, and M138V) and IniA (R479P and D198G) found in six INH-resistant isolates were all present with KatG S315T, except in one phenotypically resistant isolate with only IniA K526R.
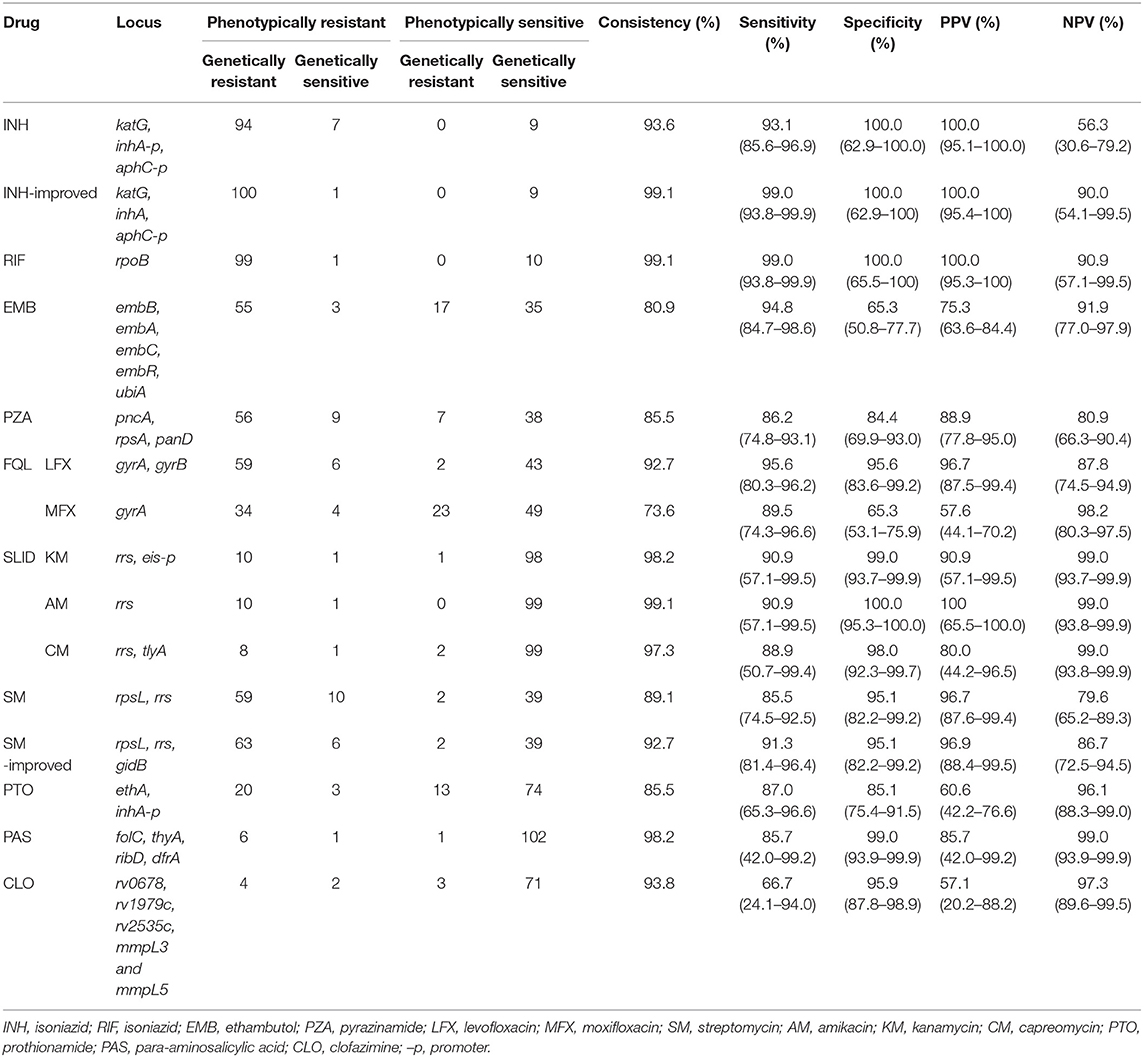
Table 1. Whole-genome sequencing (WGS) compared with phenotypic drug sensitivity test (DST) in drug resistance diagnosis of Mycobacterium tuberculosis (MT).
Rif
Genetic resistance against RIF was mostly diagnosed as mutations in the 81-bp rpoB RIF resistance-determining region (RRDR). The most prevalent drug-resistant mutations were S450L/F/W, D435V, H445D/L/Y, L452P, and Q432L, which accounted for 64.0, 13.0, 7.0, 1.0, and 2.0%, respectively, among 100 phenotypically resistant strains. There were also six strains with double mutations, namely V170F+H445Y, L430P+D435G, L430P+D435N, L430P+H445N, D435A+H445D, D435A+L452P, and H445Q+L452S. One isolate carried rpoB H674Y+1291 gcc-in causing frameshift in RRDR and was considered RIF-resistance. Only one isolate with inconsistent results between the WGS diagnosis and phenotypic DST had no mutation in the rpoB gene (Table S3). The accuracy, sensitivity, specificity, PPV, and NPV of RIF WGS diagnosis were 999.1%, 99.0% (93.8–99.9%), 100.0% (65.5–100.0%), 100.0% (95.3–100.0%), and 90.9% (57.1–99.5%), respectively (Table 1).
pza
PZA resistance is associated with full-length mutations in pncA (Yadon et al., 2017; Liu et al., 2018). Among the 65 resistant strains, 86.2% (56/65) strains carried mutation in pncA (Table S5). We also screened mutations in rpsA and panD, which are associated with resistance to PZA (Zhang et al., 2013; Shi et al., 2014). Mutations in rpsA were found in seven resistant strains, which all had a pncA mutation; no mutation was detected in panD. We included all mutations that cause amino acid changes in pyrazinamidase (encoded by pncA) as the resistance diagnosis criteria, and the accuracy, sensitivity, specificity, PPV, and NPV of the WGS diagnosis were 85.5%, 86.2% (74.8–93.1%), 84.4% (69.9–93.0%), 88.9% (77.8–95.0%), and 80.9% (66.3–90.4%) (Table 1).
Six new mutations in pncA were identified including PncA A28D, D136G, M175R, T177P, and S59Y+T153P. The same position but different amino acid mutations, D136Y and M175I/V, were previously detected in PZA-R strains, which further supported the notion that these new mutations result in PZA resistance. Totally, nine phenotypically resistant but genetically sensitive isolates had no mutations in pncA, panD, and rpsA, indicating undiscovered mechanisms of resistance. For seven phenotypically sensitive but genetically resistant strains, four strains carried PncA L35R, T47I, T47P, and F106C respectively, among which L35R, T47P, and F106C have not been found in PZA-R isolates before.
lfx
For LFX, the most common drug-resistant mutations occurred in the quinolone resistance determining region (QRDR) of gyrA (Mayer and Takiff, 2014). The most frequent drug-resistant mutations were D94A/G/H/N/Y, A90V, and S91P, which accounted for 49.2, 24.6, and 4.6%, respectively. Additionally, nine (13.8%) resistant isolates, namely G88A+D94A, A90V+D94A, A90V+D94H, A90V+D94G, A90V+D94Y, two each with A90V+D94N and S91P+D94A, contained two common drug-resistant mutations in QRDR. Among the other six phenotypically resistant strains, three were genetic wildtype and the other three strains carried mutations of QRDR, namely GyrA G158V+GyrB D461N, GyrB V636L, and GyrA I287T. Two phenotypically sensitive strains carried GyrA A90V and D94A mutations (Table S6).
The resistance-associated region in GyrB was codons 500–540. Nine strains carrying the T500A, E501V, E501D, A504V, and G512R mutations were found, but all occurred simultaneously with GyrA A90V, or D94G/N. The accuracy, sensitivity, specificity, PPV, and NPV of LFX WGS diagnosis were 92.7%, 95.6% (80.3–96.2%), 95.6% (83.6–99.2%), 96.7% (87.6–99.4%), and 87.8% (74.5–94.9%), respectively (Table 1).
Second-line-injectable-drugs-slid
A total of 10 strains carried rrs-a1401g, the most prevalent resistance-related mutations of second-line injectable drugs (SLIDs). All 10 strains with rrs-a1401g were phenotypically resistant to AM and KM, whereas 8 out of the 10 were phenotypically resistant to CM. One strain with a resistant phenotype did not have any known resistance-related mutation (Tables S8–S10). One isolate with eis (c-10t), which was reported to cause KM resistance, was phenotypically susceptible to KM. For AM, the accuracy, sensitivity, specificity, PPV, and NPV of the WGS diagnosis were 99.1%, 90.9% (57.1–99.5%), 100.0% (95.3–100.0%), 100.0% (65.5–100.0%), and 99.0% (93.8–99.9%), respectively. For KM, the accuracy, sensitivity, specificity, PPV, and NPV of WGS diagnosis were 98.2%, 90.9% (57.1–99.5%), 99.0% (93.7–99.9%), 90.9% (57.1–99.5%), and 99.0% (93.7–99.9%), respectively. For CM, the accuracy, sensitivity, specificity, PPV, and NPV of WGS diagnosis were 97.3%, 88.9% (50.7–99.4%), 98.0% (92.3–99.7%), 80.0% (44.2–96.5%), and 99.0% (93.8–99.9%), respectively (Table 1).
Among the 69 SM-resistant strains, the most common drug-resistant mutations were RpsL K43R, K88R, rrs a514c, and RpsL K43T+rrs a514c, which accounted for 71.0, 7.2, 5.8, and 1.4%, respectively. Two other strains carrying RpsL K43R or rrs a514c mutation were phenotypically sensitive to SM (Table S11). Mutations in the gidB gene were diverse and were reported to result in low-level resistance to SM (Tudó et al., 2010; Wong et al., 2011). Except for frameshift mutations, few SNP mutations have been identified to cause SM resistance. We found five new frameshift mutations of gidB including 102g_del, 351g_del, 386g_in, 115c_del, and 114c_del (together with RpsL K43R) in 5 SM-R isolates, respectively. After including frameshift mutations of gidB in the diagnosis criteria, the accuracy and sensitivity of the SM WGS diagnosis increased from 89.1 and 85.5% (74.5–92.5%) to 92.7 and 91.3% (81.4–96.4%), respectively (Table 1).
Pto
PTO resistance is mainly associated with mutations in ethA and the promoter of inhA (Morlock et al., 2003; Brossier et al., 2011; Vilchèze and Jacobs, 2014). In this study, 23 and 87 strains were PTO phenotype resistant and sensitive, respectively. The mutation of inhA (c-15t) alone without mutation in ethA was found in 13.0% PTO-resistant and 4.6% PTO-susceptible strains. Double mutation of the inhA (c-15t) and ethA genes (ethA t-11c, 1114t-del, EthA W45R, R54H, W116C, G184D, and L190P) was detected in 34.8% of the PTO-resistant and 1.1% PTO-susceptible strains (EthA T342A, Table S12). The previous drug resistance diagnostic criteria only included inhA (Miotto et al., 2017). EthA is the activator of PTO; thus, from the perspective of mechanism, mutations in ethA that result in loss of EthA function could lead to drug resistance (Vilchèze and Jacobs, 2014). Therefore, pooled frameshifts and premature stop codons were included in our diagnostic criteria (Table S1). The accuracy, sensitivity, specificity, PPV, and NPV of the PTO WGS diagnosis were 85.5%, 87.0% (65.3–96.6%), 85.1% (75.4–91.5%), 60.6% (42.2–76.6%), and 96.1% (88.3–99.0%), respectively (Table 1).
Drugs With Moderate Accuracy
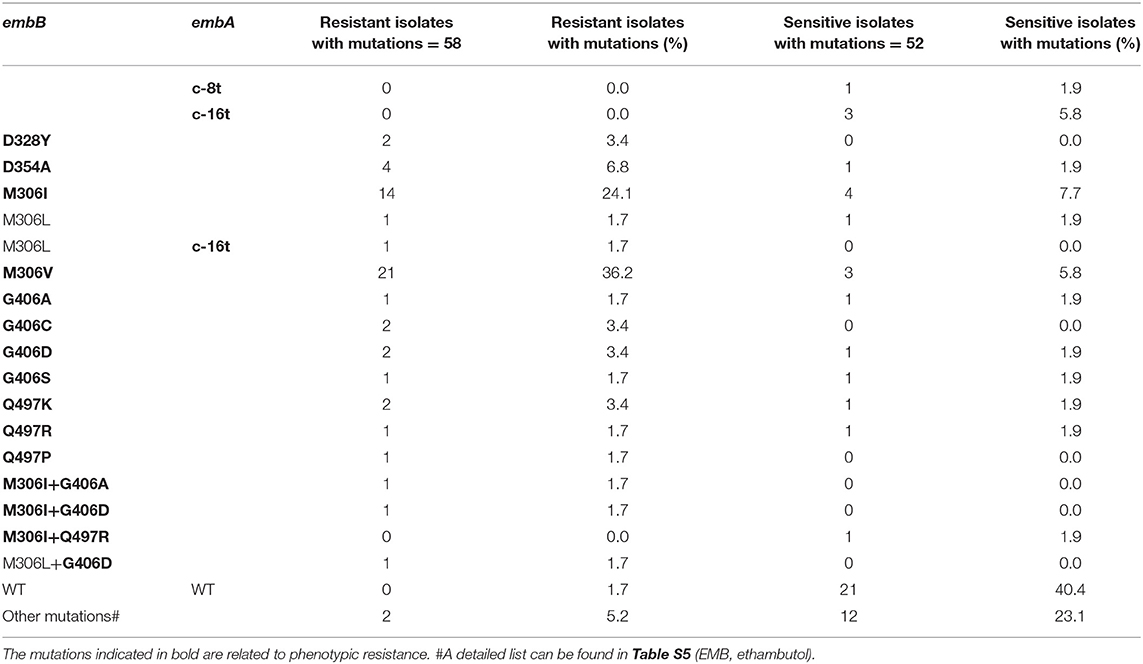
Emb
There were 58 and 52 strains with EMB phenotype resistance and sensitivity, respectively. The most common drug-resistant mutations were EmbB M306V and M306I, which were found in 21 and 16 of 58 resistant isolates, respectively. However, these two mutations were also detected in 3 and 5 of 52 sensitive isolates. Table 2 and Table S4 show the distribution of these mutations in EMB-resistant and -susceptible strains. The accuracy, sensitivity, specificity, PPV, and NPV of EMB WGS diagnosis were 80.9%, 94.8% (84.7–98.7%), 65.3% (50.8–77.7%), 75.3% (63.6–84.4%), and 91.9% (77.0–97.9%), respectively (Table 1). Most mutations in embA, embC, embR, and ubiA found in EMB resistance isolates were present together with mutations in embB (Table S4). Only two out of three strains with phenotypic resistance to EMB but genetic sensitivity carried EmbA V206M+A542V, and embC g-43c mutations, respectively. The other phenotypically resistant isolate carried EmbB M306L.
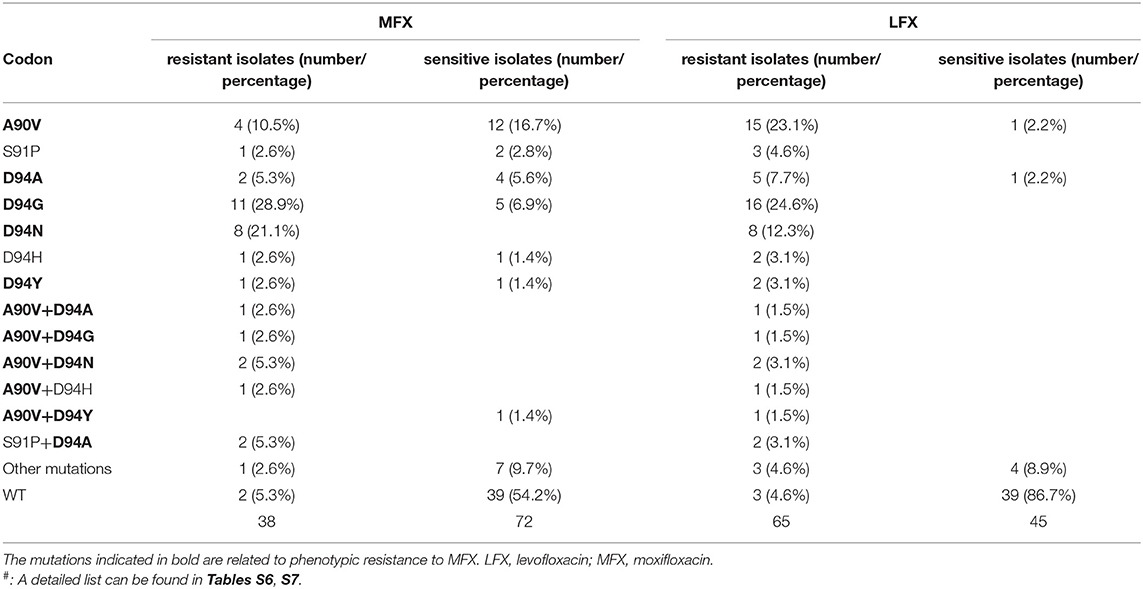
Mfx
Among the 65 strains resistant to LFX, 38 were also resistant to MFX. For MFX, the most common resistance mutation sites were GyrA A90V, D94G, D94H, D94N, and D94Y. The distribution of the above sites in resistant and susceptible strains is shown in Table 3. Outside the QRDR, the phenotype of the one isolate carrying GyrA I287T was resistant (Table S7). The accuracy, sensitivity, specificity, PPV, and NPV of the WGS diagnosis were 73.6%, 89.5% (74.3–96.6%), 65.3% (53.1–75.9%), 57.6% (44.1–70.2%), and 98.2% (80.3–97.5%), respectively (Table 1).
Drugs With Insufficient Evaluation
Pas
PAS resistance is mainly related to the folate metabolism pathway, and the main genes involved include thyA, folC, ribD, and dfrA (Mathys et al., 2009; Chakraborty et al., 2013; Zhao et al., 2014; Zhang et al., 2015b; Cheng et al., 2016). In this study, the seven resistant strains carried the following mutations: two strains with FolC I43T, one strain with FolC I43T+thyA t-31c, two strains with FolC S150G, one strain with FolC S150G+ThyA H75N, and one wildtype strain. Of the 103 sensitive strains, 10 carried mutations in the above-mentioned genes, namely DfrA Q28L, DfrA E90A, FolC F374L (three strains), FolC G283D, thyA t-31c, ThyA S160G, thyA 290g_in, and FolC S150G+ThyA A262V (Table S13). Resistance-related mutations reported previously were included in the diagnostic criteria (Table S1), and the accuracy, sensitivity, and specificity of the WGS diagnosis were 98.2%, 85.7% (42.0–99.2%), and 99.0% (93.9–99.9%), respectively.
Clo
Resistance to CLO is mainly related to rv0678, rv1979c, rv2535c, mmpL3, and mmpL5, of which rv0678 is the most reported gene in which mutations occur (Hartkoorn et al., 2014; Zhang et al., 2015a; Almeida et al., 2016; Li et al., 2017). The distribution of rv0678 mutations in drug-resistant strains was as follows: among six phenotypically resistant isolates, two carried L32S, one each carried R50Q and Q115P mutations, and the other two isolates were genetic wildtypes. There were 74 phenotypically sensitive isolates, consisting of 71 wild type, and 1 each carrying R50Q, E104G, and V1L (Table S14). According to a previous study, CLO resistance-related mutations are dispersed throughout the rv0678 gene (Zhang et al., 2015a). Therefore, we suggest that as long as a mutation occurs in rv0678, it could be judged as resistant. The accuracy, sensitivity, and specificity of the WGS diagnosis were 93.8%, 66.7% (24.1–94.0%), and 95.9% (87.8–98.9%), respectively. However, the positive predictive value was only 57.1% (20.1–88.2%) due to the low resistance rate and unclear mechanism of resistance. After analysis of rv1979c, rv2535c, mmpL3, and mmpL5, we found Rv1979c T265A in 1 CLO-resistant strain, which occurred simultaneously with Rv0678 Q115P and 10 mutations in 13 CLO-sensitive strains, namely MmpL3 W731G, Q435E, and D152H, MmpL5 Q666H, L367P, G246S, and H77Y, Rv1979c R348Q, and V221G, Rv2535c G306E (Table S14).
WGS Analysis of Non-MDR
We analyzed 34 resistance-related genes among 10 non-MDR isolates and identified mutations in four isolates, among which one was INH mono-resistant, one was resistant to SM and PZA and the other 8 were pan-susceptible. The INH mono-resistant isolate carried a mutation in inhA c-15t, which caused cross-resistance to both PTO and INH. One isolate resistant to SM and PZA carried only one mutation in RpsL K43R. The other two isolates were phenotypically sensitive to all drugs, but they carried mutations in resistance-related genes, namely pepQ c-3t and UbiA P122R, respectively (Table S15).
Discussion
In this study, we compared the results of phenotypic DST and WGS for 13 anti-TB drugs. Among the 13 drugs, DST profiles of LFX, MFX, AM, KM, CM, SM, PTO, CLO, and PAS have not been extensively studied using the WGS method. The data showed that the accuracy, sensitivity, and specificity of WGS in diagnosing the resistance against INH, RIF, PZA, LFX, AM, KM, CM, SM, and PTO were more than 85.0%, suggesting that WGS is a promising tool to predict resistance to these drugs. The highly consistent results of the WGS and DST for first-line anti-TB drugs, namely INH, RIF, and PZA and common second-line anti-TB drugs, namely LFX, AM, KM, CM, SM, were consistent with previously reported resistance profiles (Walker et al., 2015; Allix-Béguec et al., 2018).
WGS has several advantages over traditional DST (Miotto et al., 2017; Allix-Béguec et al., 2018). Firstly, WGS can quickly and comprehensively detect all possible drug resistance genes simultaneously. Therefore, for MDR-TB patient, DST results can be obtained for more drugs to provide an optimal treatment regimen and monitor therapy, or adjust the regimen based on WGS after a side effect. Furthermore, WGS can identify new resistance-related mutations. Secondly, it is easier to deliver strain DNA or inactivated strains to a center laboratory with WGS capabilities than to deliver live TB strains to perform DST, considering the current low accessibility to second-line drugs DST in China and other developing countries because of its difficulty and high infrastructure requirements (World Health Organization, 2018a). Thirdly, it has potential to be time-conserving and economical in the near future, if technically sound. WGS can decrease diagnostic time to 5–15 days in practice compared to the 2–6 weeks for traditional DST. Furthermore, the current commercial sequencing service for MTB WGS is $80–100 per isolate in China, as it is comparable to or slightly more economical than DST for MDR patients who need to undergo all second-line drug tests.
In our study, new or rarely reported mutations in known resistance-related genes were identified. In detail, mutations of INH-resistant isolates containing KatG A122D, and M257V have not been reported and mutations such as Q88P, M257I/T, W91R, Q127P, A312E, and D419Y have been rarely reported (Vilchèze and Jacobs, 2014; Allix-Béguec et al., 2018). One phenotypic resistant but genetic sensitive strain carried IniA K526R. Although iniA overexpression may lead to lower susceptibility to INH (Alland et al., 2000; Colangeli et al., 2005), the consequence of mutations in IniA K526R to INH resistance require further validation. Another mutation in Nat G207R was found in both susceptible and resistant strains in previous studies (Upton et al., 2001; Ramaswamy et al., 2003), whereas in our study, it was found in all 14 strains belonging to lineage 4.4, indicating it could be a lineage marker instead of resistance-related mutation.
For EMB, two phenotypically resistant but genetically sensitive isolates carried EmbA V206M+A542V and embC g-43c, respectively. Since EmbA V206M was reported as a phylogenetic SNP (Walker et al., 2015; Allix-Béguec et al., 2018), EmbA A542V might be a new EMB resistance-related mutation. EmbC g-43c mutation was found previously in both EMB-R and EMB-S isolates (Walker et al., 2015; Allix-Béguec et al., 2018); therefore, the role of this mutation in EMB resistance is complex and requires further study. New PncA mutations including A28D, D136G, M175R, T177P, and S59Y+T153P and frameshift mutations of gidB including 102g, 351g, 386g, 115c, and 114c deletion or insertion were identified as PZA or SM resistance-related mutations.
For PTO, since the inhA c-15t mutation is well-known to cause PTO resistance as reported in a series of published studies (Morlock et al., 2003; Tan et al., 2017), the previous diagnostic criteria only included inhA c-15t (Miotto et al., 2017). EthA activates PTO by acting as a monooxygenase (Wang et al., 2007); therefore, a mechanism involving loss of function of EthA could also result in resistance to PTO (Brossier et al., 2011). After we included pooled frameshifts and premature stop codons of EthA in the drug resistance diagnosis list, the accuracy, sensitivity, and specificity of the WGS reached above 85%. In addition to frameshift and premature stop codon mutations, point mutations with amino acid changes in EthA were found in 24.1% (21/87) of sensitive and 17.4% (4/23) of resistant isolates. Interestingly, we found that a frameshift mutation or early termination of ethA did not necessarily lead to phenotypic resistance (Table S11). Furthermore, 30 of the 87 (34.5%) strains with ethA gene mutations were still phenotypically sensitive to PTO. This phenomenon may have been due to the presence of multiple monooxygenases in MTB, which likely compensated for the inactivation of EthA (Morlock et al., 2003).
For EMB and MFX (2.0 mg/L), our results suggested that the WGS approach would not adequately replace DST but might have potential after clearer interpretation of gene mutations are achieved in affecting DST. For EMB, the specificity is lower than expected (65.3%, 50.8%−77.7%), as mutations like EmbB M306I, M306V, G406, and Q497 were detected in both sensitive and resistant strains. The phenomenon was also reported in previous studies (Lee et al., 2004; Engström et al., 2012; Cheng et al., 2014). Mutations like M306I only causing a slight increase in MIC, which is sometimes not enough to be diagnosed as resistance in phenotypic DST, might explain the inconsistency (Starks et al., 2009; Plinke et al., 2011). For isolates with contradictory result between WGS and DST, the DST was repeated at least twice to confirm. The unsatisfactory consistency of results for EMB may also be due to inappropriate critical concentrations and poor repeatability of phenotypic DST.
For MFX, the inconsistency between WGS and DST was mainly due to the mutation of GyrA A90V, which was found in 4 (10.5%) resistant and 12 (16.7%) sensitive isolates. GyrA A90V mutation was reported to cause a high level of LFX resistance but low level of MFX resistance (Sirgel et al., 2012; Mayer and Takiff, 2014). Other mutations in QRDR, including D94A, D94G, and D94Y were also found in both MFX-resistant and -sensitive isolates, which lowers the specificity of the diagnosis of MFX resistance.
Because of the low resistance rate and limited number of strains, only six CLO- and seven PAS-resistant strains were included in the study. Therefore, although the sensitivities and specificities were rather high, the result of the WGS diagnosis of CLO and PAS resistance was not convincing. To accurately evaluate the performance of WGS in predicting the DST of CLO and PAS, more resistant clinical strains, especially from patients who used these two drugs, are needed in the future studies. For CLO, apart from rv0678, mutations in rv1979c, rv2535c, mmpL3, and mmpL5 were less reported and vague in contribution to CLO resistance. In our study, 11 mutations were detected in CLO-sensitive isolates, which have not been reported. A possible explanation for these phenomena could be that these mutations increased the resistance slightly and were not detected in our study.
The main reasons for the discrepancy between WGS and DST are as follows in our study. Firstly, the mechanism of drug resistance is not clear, and there are still several unknown resistance-related mutations related to PAS and CLO. Secondly, some drugs such as PZA and EMB have poor DST consistency and repeatability. DST of these drugs was performed more than once to mitigate these limitations. For PZA, the PncA V9A, or V139A mutation were detected in both phenotypically resistant and sensitive isolates. According to previous studies, PncA V9A resulted in susceptibility in vitro but resistance in vivo and PncA V139G/L was found in PZA-R strains (Yadon et al., 2017; Allix-Béguec et al., 2018). Instability of PZA DST and inconsistent results of the in vivo and in vitro DST may explain the discrepancy. Thirdly, it is recognized that the resistance of MTB to PTO and PZA is related to the ethA and pncA genes, but the resistance-related mutations are scattered throughout the gene without a hot spot, which makes it difficult to interpret resistance through the mutation site. Finally, heterogeneous resistance has frequently been found in the gyrA and ethA genes. Resistance-related mutations and wild-type alleles occur simultaneously, making it difficult for phenotypic DST to reflect true susceptibility results, which underscores the value of genetic sequencing over DST (van Rie et al., 2005; Sun et al., 2012).
Our study has some limitations. First, China has a vast territory, and this was a single-center study with a limited number of strains. Thus, the strains selected in this study are not wholly representative of China. Second, because of the low resistance rate, the results are underpowered for some drugs. For example, the resistance rate of AK, KM, PAS, and CLO was <10%, leading to a high false measurement index; therefore, the high specificity of these drugs might not be representative. We have to admit that the study is particularly unconvincing for the drugs with low resistance. On the other hand, for INH, and RIF, the small number of non-resistant strains affected the ranges of the CI and NPV. Thirdly, some mutations, such as EmbB M306I, only cause low-grade drug resistance (Plinke et al., 2011), and the use of a critical concentration method for DST may lead to the misdiagnosis of these resistances, which could also be one of the reasons for the inconsistency between phenotype and genotype in this study. Minimum inhibitory concentration (MIC) test could be better for studying drug resistance methods. However, commercial microplates were not available when the study was carried out. Considering the amount of drugs, numbers of isolates, higher contamination rates, and low quality control of in-house microliter plates, we determined to perform DST mostly using commercial L-J media.
In conclusion, WGS is a promising approach to predict resistance to drugs such as INH, RIF, PZA, LFX, AM, KM, SM, SM, and PTO with a satisfactory accuracy, sensitivity, and specificity of over 85.0%. The specificity of WGS diagnosis for EMB and MFX (2.0 mg/L) was not high enough and needs to be improved. For other drugs such as PAS and CLO, inclusion of more strains, especially resistant strains, is needed to improve data interpretation before widespread clinical adoption.
Data Availability
The raw data of WGS has already been submitted to the SRA database and can be accessed by the public at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA522942. The BioProject accession number is PRJNA522942. The SRA accession numbers for 110 isolates are from SAMN10961303 to 10961412.
Author Contributions
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. JC and WZ: conception and design of study, revising the manuscript critically for important intellectual content. XC, GH, SW, and SL: acquisition of data, analysis and interpretation of data. XC and JC: drafting the manuscript.
Funding
This work was supported by the National Science and Technology Major Project of China (2017ZX10302301-001) and National Natural Science Foundation of China (81761148027).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01741/full#supplementary-material
References
Alland, D., Steyn, A. J., Weisbrod, T., Aldrich, K., and Jacobs, W. R. (2000). Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 182, 1802–1811. doi: 10.1128/JB.182.7.1802-1811.2000
Allix-Béguec, C., Arandjelovic, I., Bi, L., Beckert, P., Bonnet, M., Bradley, P., et al. (2018). Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415. doi: 10.1056/NEJMoa1800474
Almeida, D., Ioerger, T., Tyagi, S., Li, S. Y., Mdluli, K., Andries, K., et al. (2016). Mutations in pepQ confer Low-Level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 60, 4590–4599. doi: 10.1128/AAC.00753-16
Brossier, F., Veziris, N., Truffot-Pernot, C., Jarlier, V., and Sougakoff, W. (2011). Molecular investigation of resistance to the antituberculous drug ethionamide in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55, 355–360. doi: 10.1128/AAC.01030-10
Chakraborty, S., Gruber, T., Barry, C. E., Boshoff, H. I., and Rhee, K. Y. (2013). Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339, 88–91. doi: 10.1126/science.1228980
Cheng, S., Cui, Z., Li, Y., and Hu, Z. (2014). Diagnostic accuracy of a molecular drug susceptibility testing method for the antituberculosis drug ethambutol: a systematic review and meta-analysis. J. Clin. Microbiol. 52, 2913–2924. doi: 10.1128/JCM.00560-14
Cheng, V. W., Leung, K. S., Kwok, J. S., Leung, R. K., Yang, K. Y., Chan, R. C., et al. (2016). Phylogenetic and structural significance of dihydrofolate synthase (folC) mutations in drug-resistant Mycobacterium tuberculosis. Microb. Drug Resist. 22, 545–551. doi: 10.1089/mdr.2015.0193
Colangeli, R., Helb, D., Sridharan, S., Sun, J., Varma-Basil, M., Hazbón, M. H., et al. (2005). The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55, 1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x
Coll, F., McNerney, R., Guerra-Assunção, J. A., Glynn, J. R., Perdigão, J., Viveiros, M., et al. (2014). A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5:4812. doi: 10.1038/ncomms5812
Denkinger, C. M., Schumacher, S. G., Boehme, C. C., Dendukuri, N., Pai, M., and Steingart, K. R. (2014). Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 44, 435–446. doi: 10.1183/09031936.00007814
Engström, A., Morcillo, N., Imperiale, B., Hoffner, S. E., and Juréen, P. (2012). Detection of first- and second-line drug resistance in Mycobacterium tuberculosis clinical isolates by pyrosequencing. J. Clin. Microbiol. 50, 2026–2033. doi: 10.1128/JCM.06664-11
Farhat, M. R., Sultana, R., Iartchouk, O., Bozeman, S., Galagan, J., Sisk, P., et al. (2016). Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am. J. Respir. Crit. Care Med. 194, 621–630. doi: 10.1164/rccm.201510-2091OC
Hartkoorn, R. C., Uplekar, S., and Cole, S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrobial Agents Chemother. 58, 2979–2981. doi: 10.1128/AAC.00037-14
Institute., CALS. (2011). “Susceptibility testing of Mycobacteria, Nocardia, and other aerobic actinomycetes; Approved Standard–Second edition. CLSI document M24-A2, CLSI, Wayne, PA, USA, 2011.”.).
Ismail, N., Omar, S. V., Ismail, N. A., and Peters, R. P. H. (2018). Collated data of mutation frequencies and associated genetic variants of bedaquiline, clofazimine and linezolid resistance in Mycobacterium tuberculosis. Data Brief 20, 1975–1983. doi: 10.1016/j.dib.2018.09.057
Khosravi, A. D., Sirous, M., Abdi, M., and Ahmadkhosravi, N. (2019). Characterization of the most common embCAB gene mutations associated with ethambutol resistance in Mycobacterium tuberculosis isolates from Iran. Infect. Drug Resist. 12, 579–584. doi: 10.2147/IDR.S196800
Lee, A. S., Othman, S. N., Ho, Y. M., and Wong, S. Y. (2004). Novel mutations within the embB gene in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 4447–4449. doi: 10.1128/AAC.48.11.4447-4449.2004
Li, W., Sanchez-Hidalgo, A., Jones, V., de Moura, V. C., North, E. J., and Jackson, M. (2017). Synergistic interactions of MmpL3 inhibitors with antitubercular compounds in vitro. Antimicrob. Agents Chemother. 61:e02399-16. doi: 10.1128/AAC.02399-16
Liu, W., Chen, J., Shen, Y., Jin, J., Wu, J., Sun, F., et al. (2018). Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Hangzhou, China. Clin. Microbiol. Infect. 24, 1011–1016. doi: 10.1016/j.cmi.2017.12.012
Mathys, V., Wintjens, R., Lefevre, P., Bertout, J., Singhal, A., Kiass, M., et al. (2009). Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 53, 2100–2109. doi: 10.1128/AAC.01197-08
Mayer, C., and Takiff, H. (2014). The molecular genetics of fluoroquinolone resistance in Mycobacterium tuberculosis. Microbiol. Spectr. 2, M2–M9. doi: 10.1128/microbiolspec.MGM2-0009-2013
Miotto, P., Tessema, B., Tagliani, E., Chindelevitch, L., Starks, A. M., Emerson, C., et al. (2017). A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Euro. Respirat. J. 50:1701354. doi: 10.1183/13993003.01354-2017
Morlock, G. P., Metchock, B., Sikes, D., Crawford, J. T., and Cooksey, R. C. (2003). EthA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 47, 3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003
Pankhurst, L. J., Del Ojo Elias, C., Votintseva, A. A., Walker, T. M., Cole, K., Davies, J., et al. (2016). Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med. 4, 49–58. doi: 10.1016/S2213-2600(15)00466-X
Plinke, C., Walter, K., Aly, S., Ehlers, S., and Niemann, S. (2011). Mycobacterium tuberculosis embB codon 306 mutations confer moderately increased resistance to ethambutol in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2891–2896. doi: 10.1128/AAC.00007-10
Ramaswamy, S. V., Reich, R., Dou, S. J., Jasperse, L., Pan, X., Wanger, A., et al. (2003). Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003
Shi, W., Chen, J., Feng, J., Cui, P., Zhang, S., Weng, X., et al. (2014). Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg. Microbes Infect. 3:e58. doi: 10.1038/emi.2014.61
Sirgel, F. A., Warren, R. M., Streicher, E. M., Victor, T. C., van Helden, P. D., and Böttger, E. C. (2012). GyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 67, 1088–1093. doi: 10.1093/jac/dks033
Starks, A. M., Gumusboga, A., Plikaytis, B. B., Shinnick, T. M., and Posey, J. E. (2009). Mutations at embB codon 306 are an important molecular indicator of ethambutol resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 53, 1061–1066. doi: 10.1128/AAC.01357-08
Steingart, K. R., Schiller, I., Horne, D. J., Pai, M., Boehme, C. C., and Dendukuri, N. (2014). Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane. Database Syst. Rev. 1:D9593. doi: 10.1002/14651858.CD009593.pub3
Sun, G., Luo, T., Yang, C., Dong, X., Li, J., Zhu, Y., et al. (2012). Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 206, 1724–1733. doi: 10.1093/infdis/jis601
Sun, Q., Xiao, T. Y., Liu, H. C., Zhao, X. Q., Liu, Z. G., Li, Y. N., et al. (2018). Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrobial Agents Chemother 62:e01279-17. doi: 10.1128/AAC.01279-17
Tan, Y., Su, B., Zheng, H., Song, Y., Wang, Y., and Pang, Y. (2017). Molecular characterization of prothionamide-resistant Mycobacterium tuberculosis isolates in southern china. Front. Microbiol. 8:2358. doi: 10.3389/fmicb.2017.02358
Tudó, G., Rey, E., Borrell, S., Alcaide, F., Codina, G., Coll, P., et al. (2010). Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis clinical isolates in the area of Barcelona. J. Antimicrob. Chemother. 65, 2341–2346. doi: 10.1093/jac/dkq322
Tulyaprawat, O., Chaiprasert, A., Chongtrakool, P., Suwannakarn, K., and Ngamskulrungroj, P. (2019). Association of ubiA mutations and high-level of ethambutol resistance among Mycobacterium tuberculosis Thai clinical isolates. Tuberculosis 114, 42–46. doi: 10.1016/j.tube.2018.11.006
Upton, A. M., Mushtaq, A., Victor, T. C., Sampson, S. L., Sandy, J., Smith, D. M., et al. (2001). Arylamine N-acetyltransferase of Mycobacterium tuberculosis is a polymorphic enzyme and a site of isoniazid metabolism. Mol. Microbiol. 42, 309–317. doi: 10.1046/j.1365-2958.2001.02648.x
van Rie, A., Victor, T. C., Richardson, M., Johnson, R., van der Spuy, G. D., Murray, E. J., et al. (2005). Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 172, 636–642. doi: 10.1164/rccm.200503-449OC
Vilchèze, C., and Jacobs, W. R. (2014). Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol. Spectr. 2, M2–M14. doi: 10.1128/microbiolspec.MGM2-0014-2013
Walker, T. M., Ip, C. L., Harrell, R. H., Evans, J. T., Kapatai, G., Dedicoat, M. J., et al. (2013). Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146. doi: 10.1016/S1473-3099(12)70277-3
Walker, T. M., Kohl, T. A., Omar, S. V., Hedge, J., Del Ojo Elias, C., Bradley, P., et al. (2015). Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis. 15, 1193–1202. doi: 10.1016/S1473-3099(15)00062-6
Wang, F., Langley, R., Gulten, G., Dover, L. G., Besra, G. S., Jacobs, W. R., et al. (2007). Mechanism of thioamide drug action against tuberculosis and leprosy. J. Exp. Med. 204, 73–78. doi: 10.1084/jem.20062100
Wong, S. Y., Lee, J. S., Kwak, H. K., Via, L. E., Boshoff, H. I., and Barry, C. E. (2011). Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55, 2515–2522. doi: 10.1128/AAC.01814-10
World Health Organization [WHO] (2013). Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children. Geneva: WHO. Available online at: https://www.who.int/tb/publications/xpert-mtb-rif-assay-diagnosis-policy-update/en/
World Health Organization [WHO] (2018a). Global Tuberculosis Report 2018. Geneva: WHO. Available online at: http://www.who.int/tb/publications/global~report/en/
World Health Organization [WHO] (2018b). Technical Report on Critical Concentrations for TB Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant TB. Geneva: WHO. Available online at: https://www.who.int/tb/publications/2018/WHO_technical_report_concentrations_TB_drug_susceptibility/en/
Yadon, A. N., Maharaj, K., Adamson, J. H., Lai, Y. P., Sacchettini, J. C., Ioerger, T. R., et al. (2017). A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat. Commun. 8:588. doi: 10.1038/s41467-017-00721-2
Yang, C., Luo, T., Shen, X., Wu, J., Gan, M., Xu, P., et al. (2017). Transmission of multidrug-resistant Mycobacterium tuberculosis in Shanghai, China: a retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect. Dis. 17, 275–284. doi: 10.1016/S1473-3099(16)30418-2
Zhang, S., Chen, J., Cui, P., Shi, W., Zhang, W., and Zhang, Y. (2015a). Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 70, 2507–2510. doi: 10.1093/jac/dkv150
Zhang, S., Chen, J., Shi, W., Liu, W., Zhang, W., and Zhang, Y. (2013). Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2:e34. doi: 10.1038/emi.2013.38
Zhang, X., Liu, L., Zhang, Y., Dai, G., Huang, H., and Jin, Q. (2015b). Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob. Agents Chemother. 59, 1320–1324. doi: 10.1128/AAC.03695-14
Zhao, F., Wang, X. D., Erber, L. N., Luo, M., Guo, A. Z., Yang, S. S., et al. (2014). Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 1479–1487. doi: 10.1128/AAC.01775-13
Zignol, M., Cabibbe, A. M., Dean, A. S., Glaziou, P., Alikhanova, N., Ama, C., et al. (2018). Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect. Dis. 18, 675–683. doi: 10.1016/S1473-3099(18)30073-2
Keywords: drug resistance, drug susceptibility test, whole-genome sequencing, multidrug-resistant tuberculosis, mutation
Citation: Chen X, He G, Wang S, Lin S, Chen J and Zhang W (2019) Evaluation of Whole-Genome Sequence Method to Diagnose Resistance of 13 Anti-tuberculosis Drugs and Characterize Resistance Genes in Clinical Multi-Drug Resistance Mycobacterium tuberculosis Isolates From China. Front. Microbiol. 10:1741. doi: 10.3389/fmicb.2019.01741
Received: 27 April 2019; Accepted: 15 July 2019;
Published: 31 July 2019.
Edited by:
Thomas Dick, Hackensack Meridian Health Center for Discovery and Innovation, United StatesReviewed by:
Amit Kaushik, Johns Hopkins University, United StatesMaria Rosalia Pasca, University of Pavia, Italy
Copyright © 2019 Chen, He, Wang, Lin, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiazhen Chen, amlhemhlbl9jaGVuQDE2My5jb20=
†These authors have contributed equally to this work
 Xinchang Chen
Xinchang Chen Guiqing He2†
Guiqing He2† Jiazhen Chen
Jiazhen Chen Wenhong Zhang
Wenhong Zhang