- 1College of Forestry, Co-Innovation Center for the Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2Foreign Disease-Weed Science Research Unit, USDA, Agricultural Research Service, Fort Detrick, MD, United States
- 3ARS Research Participation Program, Oak Ridge Institute for Science and Education, Oak Ridge, TN, United States
- 4Technical Center for Animal, Plant and Food Inspection and Quarantine of Shanghai Customs, Shanghai, China
- 5Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China
Early and accurate detection of the causal pathogen Phytophthora sojae is crucial for effective prevention and control of root and stem rot and seedling damping-off of soybean. In the present study, a novel isothermal amplification assay was developed for detecting P. sojae. This 25 min assay included a two-step approach. First, a pair of novel primers, PSYPT-F and PSYPT-R were used to amplify a specific fragment of the Ypt1 gene of P. sojae in a 20 min recombinase polymerase amplification (RPA) step. Second, lateral flow dipsticks (LFD) were used to detect and visualize RPA amplicons of P. sojae within 5 min. This RPA-LFD assay was specific to P. sojae. It yielded negative detection results against 24 other Phytophthora, one Globisporangium, and 14 fungal species. It was also found to be sensitive, detecting as low as 10 pg of P. sojae genomic DNA in a 50-μL reaction. Furthermore, P. sojae was detected from artificially inoculated hypocotyls of soybean seedlings using this novel assay. In a comparative evaluation using 130 soybean rhizosphere samples, this novel assay consistently detected P. sojae in 55.4% of samples, higher than other three methods, including loop-mediated isothermal amplification (54.6%), conventional PCR (46.9%), and leaf-disc baiting (38.5–40.0%). Results in this study indicated that this rapid, specific, and sensitive RPA-LFD assay has potentially significant applications to diagnosing Phytophthora root and stem rot and damp-off of soybean, especially under time- and resource-limited conditions.
Introduction
Phytophthora sojae is one of the most devastating pathogens of soybean crops (Glycine max), causing damping-off on seedlings and root and stem rot on older plants. Areas that receive heavy rain may suffer plant mortality and yield losses up to 100% (Tyler, 2007; Dorrance, 2018). An estimated annual worldwide loss of 1–2 billion U.S. dollar has been caused by this pathogen (Wrather and Koenning, 2006; Tyler, 2007). P. sojae was first reported as a novel causal pathogen of soybean root and stem rot in Indiana and Ohio, United States (Kaufmann and Gerdemann, 1958). Thereafter, it has become widespread in many soybean-producing countries (Schmitthenner, 1985; Erwin and Ribeiro, 1996). After assessing its potential risks to agricultural and economic security, the Ministry of Agriculture of the People’s Republic of China identified P. sojae as a quarantine pest in 20071, whereas it was discovered in Jilin and Heilongjiang Provinces in 1989 (Su and Shen, 1993). Spread of this pathogen has been accelerated by China’s increasing international and interprovincial trade and transportation of soybean seeds and plants (Cui et al., 2010; Wu et al., 2017). To date, the pathogen has been found in the Inner Mongolia Autonomous Region, Xinjiang Uygur Autonomous Region, Huanghe-Huaihe River Basin and Yangtze River Basin (Chen and Wang, 2017), as well as Jilin, Heilongjiang (Su and Shen, 1993), Fujian (Cui et al., 2010; Wu et al., 2017), and Anhui (Dai Y.L. et al., 2015) Provinces.
Rapid detection of P. sojae is a crucial step toward effective management of soybean root and stem rot and seedling damping-off. Traditionally, detection methods for P. sojae include isolation from symptomatic plant tissues and baiting from soil (Erwin and Ribeiro, 1996). Subsequent pathogen identification based on morphological characters and DNA sequence data is usually time-consuming and requires trained expertise. A variety of molecular detection methods including conventional PCR (Wang et al., 2006; Bienapfl et al., 2011; Xiong et al., 2019), quantitative PCR (Wang et al., 2006; Bienapfl et al., 2011; Haudenshield et al., 2017), LAMP assays (Dai et al., 2012, Dai Y.L. et al., 2015), and a recombinase polymerase amplification (RPA) assay targeting the atp9–nad9 region of the mitochondrial genome (Rojas et al., 2017) have been developed for P. sojae. However, field application of PCR-based methods is limited due to their long time span and requirement for thermocyclers and gel electrophoresis. Furthermore, specificity to P. sojae of previously developed methods has been challenged by newly emerging pathogens (Rojas et al., 2017; Xiong et al., 2019), such as P. sansomeana (Hansen et al., 2009), also a pathogen of soybean, and P. melonis and P. vignae, two sister species phylogenetically related to P. sojae (Yang et al., 2017). Thus, a rapid and P. sojae-specific method that can be performed under time- and resource-limited conditions is warranted.
In the present study, a novel RPA assay targeting the Ypt1 gene of P. sojae was developed. The RPA amplicons were designed to be detected using lateral flow dipsticks (LFD) in real-time. Additionally, specificity to P. sojae of this assay was validated by testing against P. sansomeana, P. melonis, P. vignae, and other oomycete and fungal species.
Materials and Methods
Isolate Selection of Phytophthora Species
Twenty-nine isolates of P. sojae were tested in this study (Table 1). The 11 isolates with determined pathotypes (races) including R2, R3, R6, R8, R12, R14, R17, R19, R20, R28, and R31 were provided by Dr. Brett Tyler at Oregon State University, United States and Dr. Jinhuo Peng at Dalian Animal and Plant Quarantine Bureau, China (Table 1). The remaining 18 P. sojae isolates were recovered from root and stem tissues of diseased soybean crops in Jiangsu, Fujian, and Yunnan Provinces, China. Isolates belonging to 24 other Phytophthora, one Globisporangium, and 14 fungal species were used for specificity evaluation. All isolates were maintained in collections at Department of Plant Pathology at Nanjing Agricultural University and Department of Forest Protection at Nanjing Forestry University in Nanjing, China.
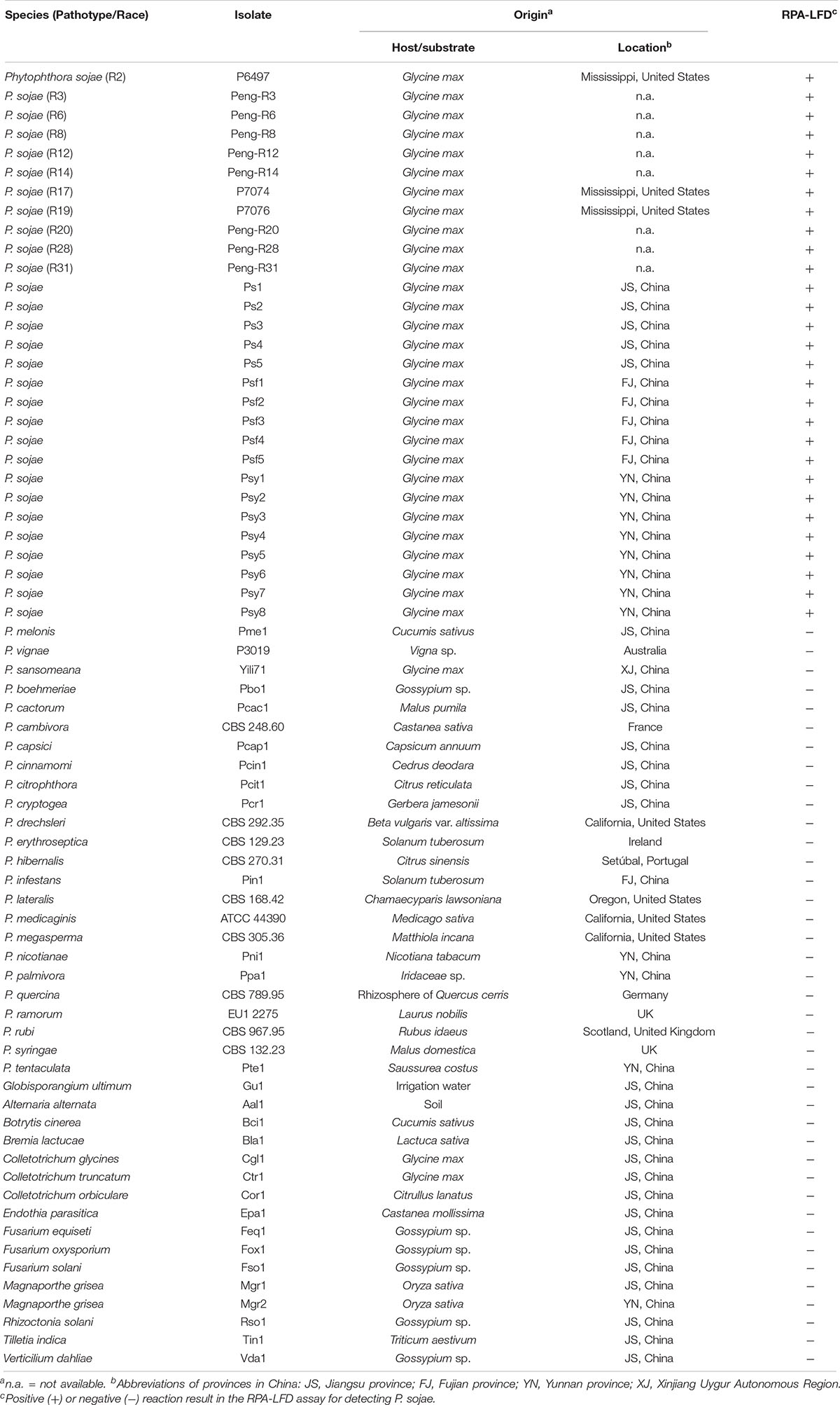
Table 1. List of oomycete and fungal isolates used in this study and their detection results in the recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay.
Culture Conditions and DNA Extraction
Phytophthora and Globisporangium isolates were cultured in 10% clarified V8 juice agar at 25°C in the dark. Fungal isolates were maintained in potato dextrose agar at 25°C in the dark.
For extracting genomic DNAs (gDNAs), each oomycete or fungal isolate was grown in 10% clarified V8 juice or potato dextrose broth, respectively, at 25°C for 4–5 days, harvested, and freeze dried. gDNAs were extracted using a DNAsecure Plant Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Total DNAs were extracted from artificially inoculated soybean tissues using an NaOH lysis method (Wang et al., 1993). Environmental DNAs (eDNAs) from rhizosphere samples were extracted using a FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, Ohio, United States). Specifically, 400 mg of each soil sample was placed in a lysing matrix E 2-mL tube, followed by the addition of 978 mL of phosphate buffer and 122 mL of MT buffer (MP Biomedicals, Solon, OH, United States). Mixtures in lysing tubes were homogenized using a FastPrep FP120 instrument (MP Biomedicals, Solon, OH, United States) at speed 6 for 40 s. Extraction of eDNAs was completed following manufacturer’s instructions.
DNA concentrations were quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). All DNA extractions were stored at −20°C until use.
Primers and Probe Design
Sequences of the Ypt1 gene of P. sojae (GenBank accession No. DQ162958) and its phylogenetically close species were downloaded from GenBank (Benson et al., 2018). Multiple sequence alignment by Clustal W (Larkin et al., 2007) was carried out using BioEdit version 7.0.5 (Hall, 1999). Several combinations of RPA primers and probe targeting the P. sojae-specific fragment within the 478-nt sequence (Figure 1) were designed according to RPA guidelines and manufacturer’s instructions for Twist Amp® DNA amplification kit (TwistDx Ltd., Cambridge, United Kingdom), followed by testing in RPA to identify the optimal primer set. A pair of forward primer and a 5′-biotin-labeled reverse primer (Table 2) met the requirement for the specific detection of P. sojae Ypt1 gene according to the TwistAmp® nfo kit (TwistDx Ltd., Cambridge, United Kingdom). Thereafter, a nfo DNA probe (Table 2) used for the LFD visualization (Milenia Biotec, Giessen, Germany) was designed based on the sequences of RPA primers. This nfo probe was labeled with a fluorescein amidite (FAM) at the 5′ end, a base analog tetrahydrofuran (THF) inserted between the 30th and 31st bases, and a C3 spacer at the 3′ end (Table 2). The primers and probe (Table 2) were synthesized by GenScript (Nanjing, China).

Figure 1. Sequence of the Ypt1 gene of Phytophthora sojae (GenBank accession No. DQ162958). Nucleotides targeted by forward primers PSYPT-F, PSYPT-Probe, and reverse primer PSYPT-R of the novel recombinase polymerase amplification assay are below respective arrows. Arrows indicate the direction of amplification. Primer sequences are provided in Table 2.

Table 2. Oligonucleotide primers and probe designed for the recombinase polymerase amplification-lateral flow dipstick assay in this study.
RPA-LFD Assay
Recombinase polymerase amplification-Lateral flow dipsticks assay was performed according to the quick guide of TwistAmp® nfo kit (TwistDx Ltd., Cambridge, United Kingdom). Briefly, each 50 μL reaction contained 29.5 μL of rehydration buffer (supplied in the kit), 2.1 μL of each of forward and reverse primers (10 μM), 0.6 μL of probe (10 μM), 12.2 μL of nuclease-free water (nfH2O; Thermo Fisher Scientific, Wilmington, DE, United States), and 1 μL of DNA template. After mixing by vortex, 2.5 μL of 280-mM magnesium acetate was added to each reaction for initiating amplification. RPA was performed at 39°C in a SimpliAmpTM thermal cycler instrument (Model A24812, Thermo Fisher Scientific, Wilmington, DE, United States) for 20 min with non-heated lid and a vortex and spin step after the first 4 min. To detect RPA amplicons, 10 μL of RPA product was mixed with 90 μL of phosphate buffered saline with Tween 20 (PBST) running buffer. Then 10 μL of the mixture was added to the sample pad of a HybriDetect 1 LFD (Milenia Biotec GmbH, Giessen, Germany) using a pipettor. The LFD was dipped into a tube containing 200 μL of PBST and incubated at room temperature (aver. 22°C) for up to 5 min until a control line was visible. When test and control lines were simultaneously visible, it was a positive detection. If only the control line was visible, it was a negative detection. All LFDs were then air-dried and photographed using a Canon PowerShot SX730 HS camera.
RPA-LFD Assay Specificity and Sensitivity
Specificity of the RPA-LFD assay was evaluated against all isolates listed in Table 1. Each RPA reaction included 10 ng of purified gDNA. RPA-LFD assay was performed in triplicate against each isolate.
To determine sensitivity, 10-fold dilutions of P. sojae gDNA (isolate P6497) ranging from 100 to 0.001 ng per μL were used as DNA templates in the RPA-LFD assay. nfH2O was used in no-template control (NTC) reactions. This RPA-LFD assay was repeated in triplicate for each concentration of gDNA template under the same conditions described above.
Detection of P. sojae in Artificially Inoculated Soybean Seedlings Using RPA-LFD
Seedlings of soybean cultivar Hefeng 47 were grown in a glasshouse at a day/night temperature of 25/20°C and a 16 h photoperiod. P. sojae isolate P6497 was cultured in rye grains mixed with 10% clarified V8 juice at 25°C in the dark for 3 days. Hypocotyls of 4-day-old soybean seedlings were wounded using a sterile inoculation needle. A P. sojae-colonized rye grain was placed on the wound site of each of three seedlings. A sterile grain was used for each of three non-inoculated seedlings. Hypocotyl tissues were then covered using parafilm to keep rye grains attached and maintain humidity. Development of symptoms was recorded daily. At approximately 72 h after inoculation, total gDNAs at the wounded site of hypocotyls were extracted as described above. Concentrations of gDNA extractions were measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) and adjusted to 10 ng per μL by adding nfH2O. The RPA-LFD assay was performed as described above using the hypocotyl total DNA extractions as templates. This experiment was repeated once. Purified gDNA (10 ng per μL) of P. sojae isolate P6497 and nfH2O were included in each repeat as a positive control and NTC, respectively.
Comparative Evaluation of Detection Assays Using Soybean Rhizosphere Samples
One hundred and thirty rhizosphere samples (0- to 10-cm depth) were collected from soybean fields in seven cities of the Heilongjiang Province in China, namely Daqing, Haerbin, Jiamusi, Jixi, Mudanjiang, Qiqihaer, and Yichun, from 2008 to 2014 (Table 3). After sampling, they were stored in 1-gallon Ziploc bags and transported in ice boxes to laboratories at Nanjing Agricultural University and Nanjing Forestry University. eDNAs were extracted from all samples and quantified as described above.
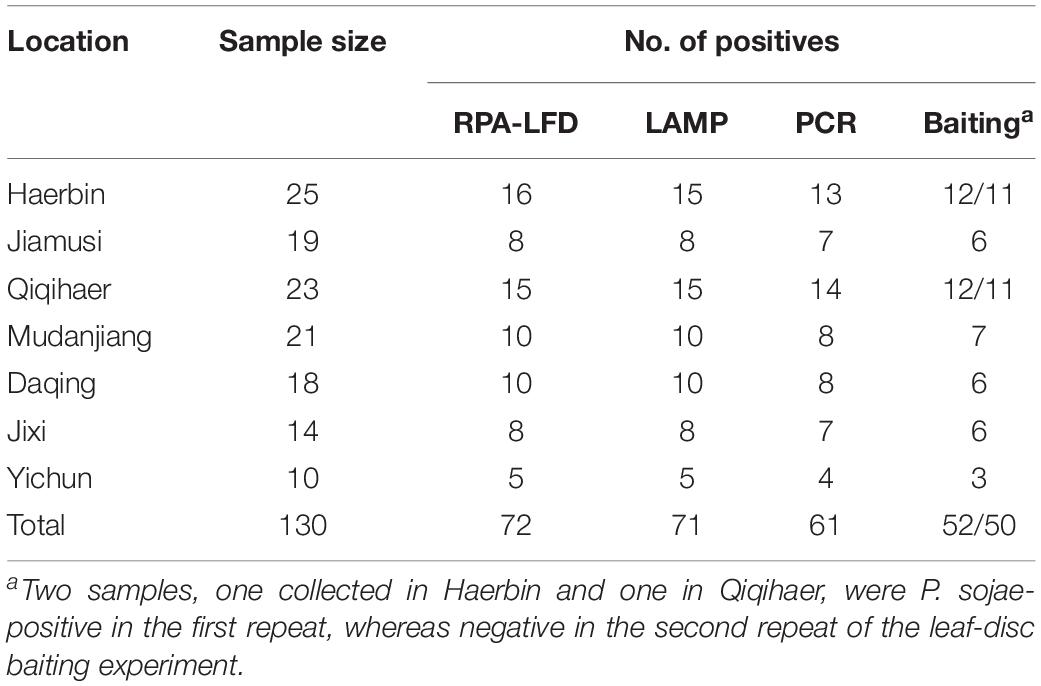
Table 3. Detection of Phytophthora sojae using a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay developed in this study, and three previously used methods including Loop-mediated isothermal amplification (LAMP), conventional PCR, and leaf-disc baiting on 130 rhizosphere samples collected from soybean fields in Heilongjiang Province, China.
The RPA-LFD assay along with three previously described detection methods for P. sojae were comparatively evaluated using the same set of 130 samples. eDNAs were used as templates in the novel RPA-LFD assay, as well as LAMP (Dai et al., 2012) and conventional PCR (Wang et al., 2006) assays. In a modified leaf-disc baiting assay (Erwin and Ribeiro, 1996; Malvick and Grunden, 2004), rhizosphere samples were dried at room temperature for 3 days. Approximately 300 g of each sample was saturated by adding distilled water and maintained under the saturated condition at room temperature for 5 days. Thirty leaf discs of soybean cultivar Hefeng 47 (2 cm in diameter) were pressed onto the surface of each saturated rhizosphere sample and incubated at room temperature in the dark for 2 to 3 days. After incubation, leaf discs were placed onto PARP selective media (containing pimarcin, ampicillin, rifampicin, and pentachloronitrobenzene) to recover isolates. Each isolate of recovered Phytophthora species was examined for characteristic oospores of P. sojae. Representative isolates were identified by sequencing the internal transcribed spacer region (Cooke et al., 2000). Each detection method was repeated once against all 130 rhizosphere samples.
Results
Specificity and Sensitivity of the RPA-LFD Aassay
In the evaluation of specificity, identical results were observed among three repeats of the experiments. All dipsticks had a visible control line, indicating valid tests. Test lines were visible on dipsticks using gDNAs of P. sojae isolates. No test lines were observed on dipsticks of other species or NTC (Figures 2, 3).
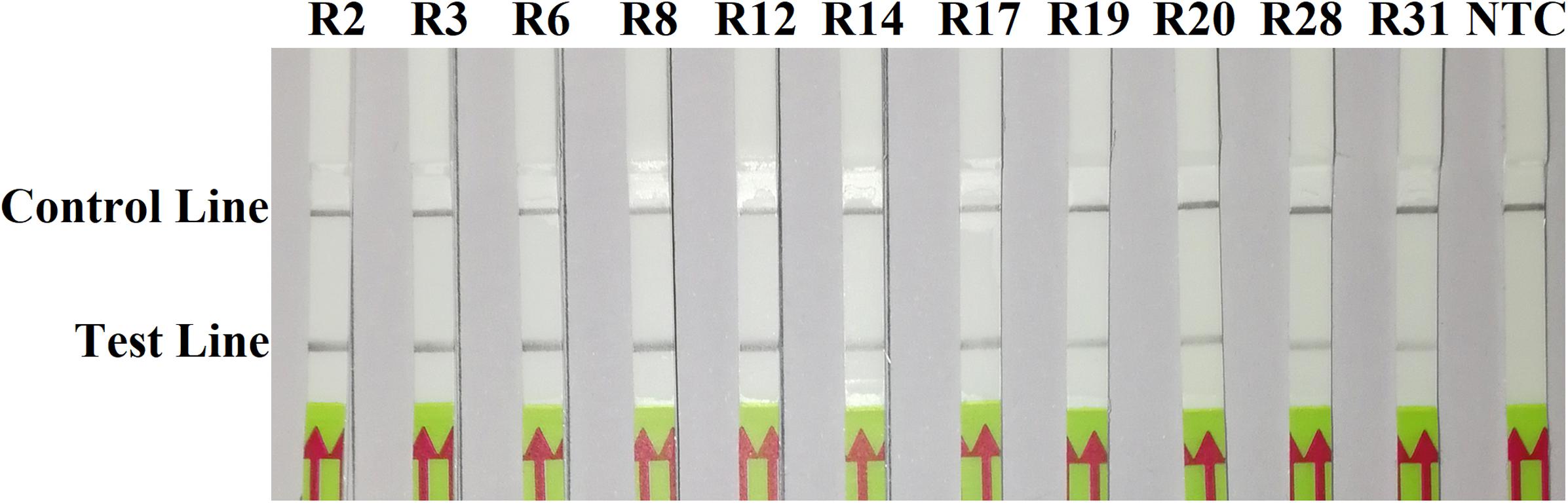
Figure 2. Evaluation of specificity of the novel recombinase polymerase amplification-lateral flow dipstick assay using 11 isolates belonging to different pathotypes of Phytophthora sojae. Nuclease-free water was used in place of DNA templates in a no-template control (NTC). Dipsticks of the first repeat are shown, as results were identical among three repeats of the experiment.
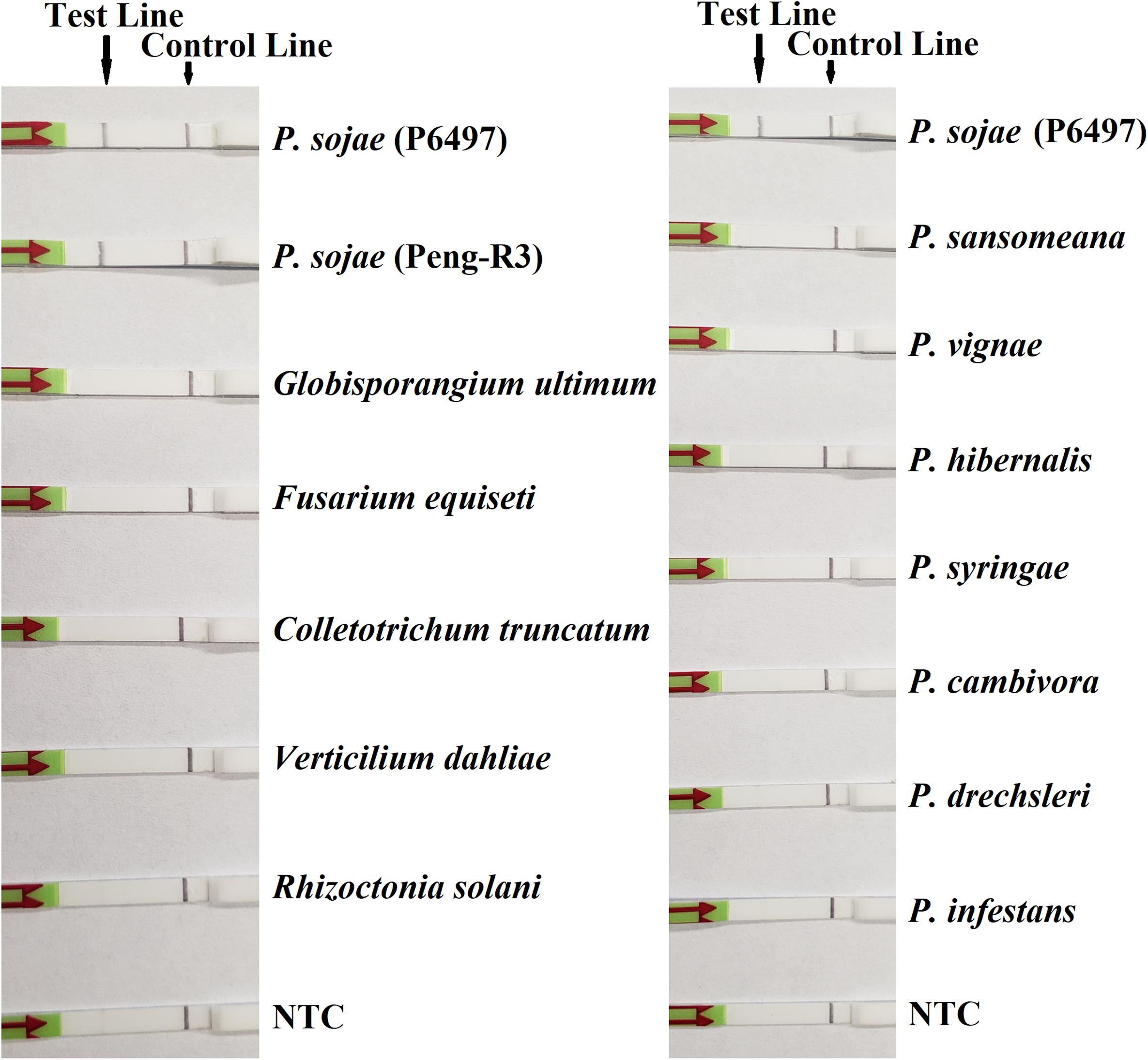
Figure 3. Evaluation of specificity of the novel recombinase polymerase amplification-lateral flow dipstick assay using Phytophthora sojae isolates and other oomycete and fungal species. Nuclease-free water was used in place of DNA templates in no-template controls (NTC). Dipsticks of selected isolates are shown. All results are listed in Table 1.
In sensitivity evaluation, all dipsticks had visible control lines. Test lines were visible on dipsticks correlating with 100, 10, 1, 0.1, or 0.01 ng of P. sojae gDNA template used per each RPA reaction. No test lines were observed on those with 0.001 or 0.0001 ng of gDNA, or NTC (Figure 4). The results at all gDNA concentrations were consistent among three repeats of the experiment.
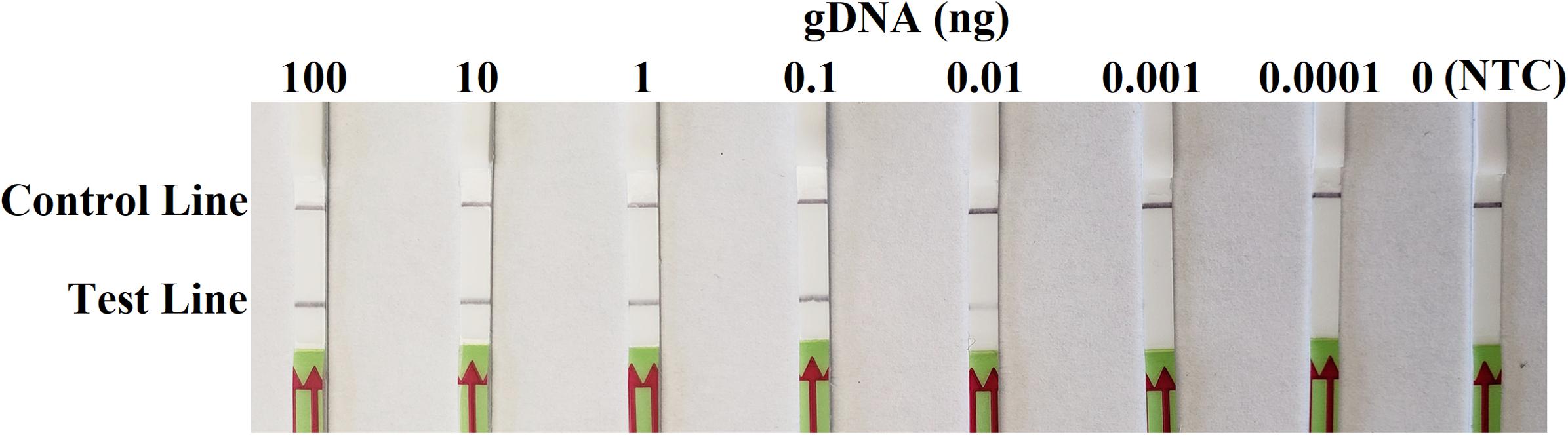
Figure 4. Evaluation of sensitivity of the novel recombinase polymerase amplification-lateral flow dipstick assay using 10-fold dilutions of genomic DNA (gDNA) of Phytophthora sojae isolate P6497 as templates. Nuclease-free water was used in place of DNA templates in a no-template control (NTC). Dipsticks of the first repeat are shown, as results were identical among three repeats of the experiment.
Detection of P. sojae in Artificially Inoculated Soybean Seedlings Using RPA-LFD
On the third day after inoculation, three inoculated seedlings had severe wilting with discoloration at wound sites. There was no discoloration on three wounded, non-inoculated hypocotyls, although a slight wilting might be observed. In the RPA-LFD assay, all dipsticks had visible control lines. Test lines were visible on three dipsticks with total DNAs extracted from inoculated hypocotyls, whereas no test lines were observed on those from three non-inoculated hypocotyls (Figure 5). Results were identical between two repeats of the experiment.
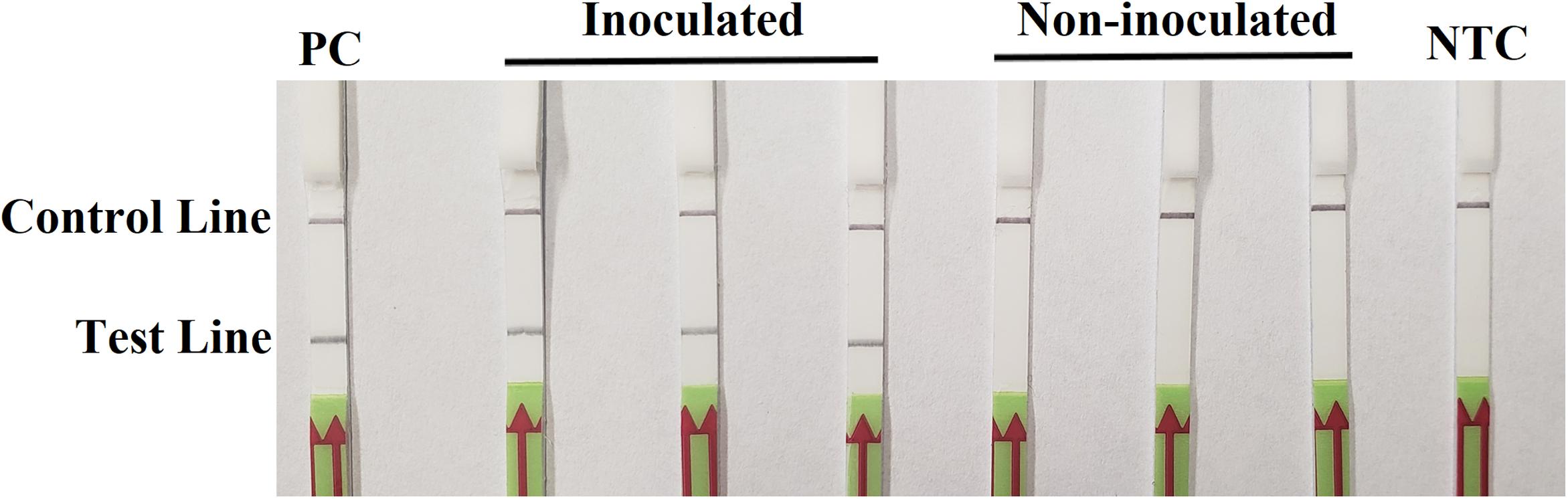
Figure 5. Detection of Phytophthora sojae in artificially inoculated soybean seedlings using the recombinase polymerase amplification-lateral flow dipstick assay. Genomic DNA (10 ng) of P. sojae isolate P6497 was used as the template in a positive control (PC). Nuclease-free water was used in a no-template control (NTC). Dipsticks of the first repeat are shown, as results were identical between two repeats of the experiment.
Comparative Evaluation of Detection Assays Using Rhizosphere Samples
Detection results were identical between two repeats of the RPA-LFD, LAMP, and PCR assays. P. sojae was detected in 72 of 130 (55.4%) samples (Table 3) using the novel RPA-LFD assay. These 72 positive samples were collected from Haerbin (16 of 25), Jiamusi (8 of 19), Qiqihaer (15 of 23), Mudanjiang (10 of 21), Daqing (10 of 18), Jixi (8 of 14), and Yichun (5 of 10). Using the LAMP assay, 71 samples (54.6%) were detected as positive (Table 3). All 71 positive samples in the LAMP assay were also detected as positive in the RPA-LFD assay (Table 3). P. sojae was detected in one sample collected from Haerbin using the RPA-LFD assay, but not detectable using the LAMP assay. Using the conventional PCR assay, 61 samples (46.9%) were determined as positive. They were also positive in both RPA-LFD and LAMP assays (Table 3). The positive detection rate using PCR was lower than those of both isothermal amplification assays in each city (Table 3).
Phytophthora sojae was recovered from soybean leaf-disc baits deployed in 52 (40%) and 50 (38.5%) samples in two repeats of the experiment, at lower detection rates than other methods (Table 3). One sample collected from Haerbin and one from Qiqihaer were positive in the first repeat, whereas P. sojae was not recovered from these two samples in the second repeat of the experiment using the baiting method (Table 3).
Discussion
Accurate and rapid detection of P. sojae in plants and soil is a critical step toward effective prevention and management of soybean root and crown rot and seedling damping-off. In this study, a novel method was developed to detect P. sojae using the RPA-LFD assay. Evaluations in the study determined this assay as specific to P. sojae. It was also found to be sensitive, detecting as low as 10 pg per μL of gDNA, and P. sojae in soil samples at a higher rate than three previously developed methods, namely LAMP (Dai et al., 2012), conventional PCR (Wang et al., 2006), and leaf-disc baiting. High sensitivity and specificity, and several other advantages make this novel RPA-LFD assay a potentially useful method in high-throughput testing under time- and resource-limited conditions.
Recombinase polymerase amplification assay in combination with LFD for the diagnosis of P. sojae shows a high degree of specificity. Although many previous methods were believed as P. sojae-specific when they were developed (Wang et al., 2006; Bienapfl et al., 2011; Haudenshield et al., 2017), their accuracy has been challenged by newly emerging pathogens (Rojas et al., 2017; Xiong et al., 2019), such as P. sansomeana (Hansen et al., 2009), another species pathogenic to soybeans, and phylogenetic sister taxa of P. sojae, such as P. melonis and P. vignae. Rojas et al. (2017) reported that an RPA assay targeting the mitochondrial atp9–nad9 region was specific to the genus Phytophthora and several species including P. sojae. This high specificity has also been found in the novel RPA-LFD assay targeting Ypt1 gene in this study. As demonstrated in the specificity evaluation, this novel RPA-LFD assay detected DNAs of P. sojae, while had no positive reactions to those of 24 other Phytophthora species, including P. sansomeana, P. melonis, and P. vignae (Table 1).
Sensitivity of RPA-LFD assay reported here is adequate if not higher than most previously developed methods. In the sensitivity evaluation using gDNA, the detection lower limit for this RPA-LFD assay was 0.01 ng (10 pg) in a 50 μL RPA reaction (Figure 4). It was at least 100 and 10 times more sensitive than a conventional PCR assay (Wang et al., 2006) and Ypt1-based LAMP assay (Dai T.T. et al., 2015), respectively, and equally sensitive as an A3aPro-specific LAMP assay (Dai et al., 2012). In the comparative evaluation using field soil samples, the RPA-LFD assay resulted in the highest detection rate of P. sojae among four evaluated methods (Table 3). The only higher sensitivity reported so far was 100 fg in a PCR-based method using a set of four SCAR primers (Xiong et al., 2019). However, RPA has the advantage in using fewer primers and special equipment, as well as its significantly shorter amplification time.
Several advantages make this RPA-LFD assay useful under time- and resource-limited conditions. First, RPA reaction does not require specialized equipment such as LAMP devices, PCR thermal cyclers, or electrophoresis systems. Second, RPA reactions could be performed within a wider temperature range between 25 and 45°C (James and Macdonald, 2015; Daher et al., 2016). In contrast, PCR-based methods require stringent control of various temperatures, while LAMP assays require a consistently high temperature for amplification, approximately 64°C. Third, the RPA-LFD assay is a time-saving diagnostic tool. This two-step assay only requires 20 min for RPA and less than 5 min for LFD detection. The reaction durations usually double for LAMP assays and are at least 90 min for PCR. Fourth, the RPA-LFD assay does not require a fluorometer to monitor the fluorescent signal. RPA results can be directly visualized on the dipsticks, making this method much simpler to operate than any other methods. Due to the rapid disease development and field-to-field spread of P. sojae (Erwin and Ribeiro, 1996), simplicity and time-saving are important merits of diagnostic tools, especially when disease prevent and pathogen eradication are urgent and a large quantity of samples are required to be processed. Fifth, RPA assays are more resistant to inhibitors such as host DNA as compared to other isothermal detection methods, such as LAMP, although false negative results can also occur (Rosser et al., 2015; Moore and Jaykus, 2017; Ahmed et al., 2018). In this study, total DNAs containing both pathogen and host gDNAs were extracted from P. sojae-inoculated soybean hypocotyls, while no false negative result was yielded (Figure 5). This finding indicated that inhibitory effects of soybean gDNA was unlikely involved in the present RPA assay.
A pipeline framework of developing a novel RPA-LFD assay for a specific Phytophthora species has been demonstrated in this study, including designing specific RPA primers, optimizing reaction conditions of RPA and LFD visualization, and evaluating the assay’s sensitivity and specificity. The unique sequence of the Ypt1 gene of P. sojae was targeted here, while other genetic markers could be utilized for developing isothermal amplification assays for P. sojae (Dai et al., 2012) and other plant pathogens. With the increasing availability of genome sequences, identification of species-specific markers has become easier and more affordable. For example, a comparative genomics approach has been applied for designing LAMP primers specific to Phytophthora cinnamomi (Dai et al., 2019). A similar approach has been used for developing a Pectobacterium species-specific RPA-LFD assay (Ahmed et al., 2018). It is not unexpected that additional RPA assays using a diverse of genetic markers will be developed for detecting an array of important Phytophthora species in the future.
Conclusion
A novel RPA-LFD assay was developed for the accurate, simple and rapid detection of P. sojae. The specific primers combination was determined by targeting the Ypt1 gene. The RPA-LFD assay could perform at the temperature range of 25–45°C within 25 min. This assay has several notable advantages. Only a primer pair plus a probe are required to detect trace amounts of DNA. Meanwhile, the amplicons could generate visible lines on LFD, while no gel electrophoresis is required. Additionally, sensitivity evaluation revealed that RPA-LFD assay could detect as low as 10 pg gDNA of P. sojae. Furthermore, the RPA-LFD assay successfully detected P. sojae in inoculated plant tissues and infested soil samples at higher rates than LAMP, PCR, and leaf-disc baiting methods. Based on the above findings, this RPA-LFD assay has great potential to be adapted as a routine test for detecting P. sojae, especially under time- and resource-limited conditions.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
TD, XY, XZ, and DS conceived and designed the experiments, contributed the reagents, materials and analysis tools, and wrote the manuscript. TH, BJ and YX performed the experiments and analyzed the data.
Funding
This work was supported by the National Natural Science Foundation of China (31500526), China Postdoctoral Science Foundation (2016T790467), Overseas Research and Study Project of Excellent Young and Middle-aged Teachers and Principals in Colleges and Universities of Jiangsu Province of 2018, the Priority Academic Program Development of Jiangsu Higher Education Institutions, the General Program of Jiangsu Science and Technology Department (SBK 2019021775), Research Project of China Customs (2019HK044), and Special Fund for Agro-scientific Research in the Public Interest (201503112).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Brett Tyler at Oregon State University, United States and Dr. Jinhuo Peng at Dalian Animal and Plant Quarantine Bureau for providing isolates and DNAs of Phytophthora species used in this study. USDA is an equal opportunity provider and employer.
Footnotes
- ^ http://www.aqsiq.gov.cn/; June 22, 2016
References
Ahmed, F. A., Larrea-Sarmiento, A., Alvarez, A. M., and Arif, M. (2018). Genome-informed diagnostics for specific and rapid detection of Pectobacterium species using recombinase polymerase amplification coupled with a lateral flow device. Sci. Rep. 8: 15972
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., et al. (2018). GenBank. Nucleic Acids Res. 46, D41–D47. doi: 10.1093/nar/gkx1094
Bienapfl, J. C., Malvick, D. K., and Percich, J. A. (2011). Specific molecular detection of Phytophthora sojae using conventional and real-time PCR. Fungal Biol. 115, 733–740. doi: 10.1016/j.funbio.2011.05.007
Chen, X. R., and Wang, Y. C. (2017). “Phytophthora sojae,” in Biological Invasions and Its Management in China, Vol. 2, eds F. H. Wan, M. X. Jiang, and A. B. Zhan (Singapore: Springer), 199–223.
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., and Brasier, C. M. (2000). A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. doi: 10.1006/fgbi.2000.1202
Cui, L. K., Yin, W. X., Tang, Q. H., Dong, S. M., Zheng, X. B., Zhang, Z. G., et al. (2010). Distribution, pathotypes, and metalaxyl sensitivity of Phytophthora sojae from Heilongjiang and Fujian Provinces in China. Plant Dis. 94, 881–884. doi: 10.1094/pdis-94-7-0881
Daher, R. K., Stewart, G., Boissinot, M., and Bergeron, M. G. (2016). Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 62, 947–958. doi: 10.1373/clinchem.2015.245829
Dai, T. T., Lu, C. C., Lu, J., Dong, S. M., Ye, W. W., Wang, Y. C., et al. (2012). Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 334, 27–34. doi: 10.1111/j.1574-6968.2012.02619.x
Dai, T. T., Yang, X., Hu, T., Li, Z. Y., Xu, Y., and Lu, C. C. (2019). A novel LAMP assay for the detection of Phytophthora cinnamomi utilizing a new target gene identified from genome sequences. Plant Dis. doi: 10.1094/PDIS-04-19-0781-RE
Dai, T. T., Zheng, X. B., and Wu, X. Q. (2015). LAMP assay for rapid detection of Phytophthora sojae based on its Ypt1 gene. Acta Phytopathologica Sinica 6, 576–584. doi: 10.3389/fmicb.2017.01920
Dai, Y. L., Liu, T. F., Zhang, L. F., Li, Z. H., Guo, W. W., Cao, S., et al. (2015). First report of Phytophthora root and stem rot caused by Phytophthora sojae on soybean in Taihe. China. Plant Dis. 99, 1861–1862. doi: 10.1094/pdis-04-15-0450-pdn
Dorrance, A. E. (2018). Management of Phytophthora sojae of soybean: a review and future perspectives. Can. J. Plant Pathol. 40, 210–219. doi: 10.1080/07060661.2018.1445127
Erwin, D. C., and Ribeiro, O. K. (1996). Phytophthora diseases worldwide. St. Paul, Minn: APS Press.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hansen, E. M., Wilcox, W. F., Reeser, P. W., and Sutton, W. (2009). Phytophthora rosacearum and P. sansomeana, new species segregated from the Phytophthora megasperma "complex". Mycologia 101, 129–135. doi: 10.3852/07-203
Haudenshield, J. S., Song, J. Y., and Hartman, G. L. (2017). A novel, multiplexed, probe-based quantitative PCR assay for the soybean root-and stem-rot pathogen, Phytophthora sojae, utilizes its transposable element. PLoS One 12:e0176567. doi: 10.1371/journal.pone.0176567
James, A., and Macdonald, J. (2015). Recombinase polymerase amplification: emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev. Mol. Diagn. 15, 1475–1489. doi: 10.1586/14737159.2015.1090877
Kaufmann, M. J., and Gerdemann, J. W. (1958). Root and stem rot of soybean caused by Phytophthora sojae n. sp. Phytopathology 48, 201–208.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Malvick, D. K., and Grunden, E. (2004). Traits of soybean-infecting Phytophthora populations from Illinois agricultural fields. Plant Dis. 88, 1139–1145. doi: 10.1094/pdis.2004.88.10.1139
Moore, M. D., and Jaykus, L. A. (2017). Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Sci. Rep. 7:40244. doi: 10.1038/srep40244
Rojas, J. A., Miles, T. D., Coffey, M. D., Martin, F. N., and Chilvers, M. I. (2017). Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 101, 1171–1181. doi: 10.1094/pdis-09-16-1225-re
Rosser, A., Rollinson, D., Forrest, M., and Webster, B. L. (2015). Isothermal recombinase polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasites Vectors 8:446. doi: 10.1186/s13071-015-1055-3
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368.
Su, Y., and Shen, C. (1993). The discovery and biological characteristic studies of Phytophthora megasperma f. sp. glycinea on soyabean in China. Acta Phytopathologica Sinica 23, 341–347.
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Wang, H., Qi, M., and Cutler, A. J. (1993). A simple method of preparing plant samples for PCR. Nucleic Acids Res. 21, 4153–4154. doi: 10.1093/nar/21.17.4153
Wang, Y. C., Zhang, W. L., Wang, Y., and Zheng, X. B. (2006). Rapid and sensitive detection of Phytophthora sojae in soil and infected soybeans by species-specific polymerase chain reaction assays. Phytopathology 96, 1315–1321. doi: 10.1094/PHYTO-96-1315
Wrather, J. A., and Koenning, S. R. (2006). Estimates of disease effects on soybean yields in the United States 2003 to 2005. J. Nematol. 38, 173–180.
Wu, M., Li, B., Liu, P., Weng, Q., Zhan, J., and Chen, Q. (2017). Genetic analysis of Phytophthora sojae populations in Fujian. China. Plant Pathol. 66, 1182–1190. doi: 10.1111/ppa.12666
Xiong, Q., Xu, J., Zheng, X. Y., Zhu, Y., Zhang, C., Wang, X. L., et al. (2019). Development of seven novel specific SCAR markers for rapid identification of Phytophthora sojae: the cause of root- and stem-rot disease of soybean. Eur. J. Plant Pathol. 153, 517–531. doi: 10.1007/s10658-018-1579-4
Keywords: oomycetes, plant destroyers, field diagnosis, Phytophthora sansomeana, Phytophthora melonis, Phytophthora vignae
Citation: Dai T, Yang X, Hu T, Jiao B, Xu Y, Zheng X and Shen D (2019) Comparative Evaluation of a Novel Recombinase Polymerase Amplification-Lateral Flow Dipstick (RPA-LFD) Assay, LAMP, Conventional PCR, and Leaf-Disc Baiting Methods for Detection of Phytophthora sojae. Front. Microbiol. 10:1884. doi: 10.3389/fmicb.2019.01884
Received: 15 May 2019; Accepted: 30 July 2019;
Published: 09 August 2019.
Edited by:
Christopher Rensing, Fujian Agriculture and Forestry University, ChinaReviewed by:
Liliana M. Cano, University of Florida, United StatesStefania Tegli, University of Florence, Italy
Copyright © 2019 Dai, Yang, Hu, Jiao, Xu, Zheng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danyu Shen, c2hlbmRhbnl1QG5qYXUuZWR1LmNu
 Tingting Dai
Tingting Dai Xiao Yang
Xiao Yang Tao Hu1
Tao Hu1 Danyu Shen
Danyu Shen