- Laboratory of Molecular Plant Pathology, College of Plant Science, Jilin University, Changchun, China
The tea plant [Camellia sinensis (L.) O. Kuntze] is one of the most important leaf crops, and it is widely used for the production of non-alcoholic beverages worldwide. Tea also has a long history of medicinal use. Colletotrichum camelliae Massee is one of the dominant fungal pathogens that infects tea leaves and causes severe tea anthracnose disease. To analyze the molecular biology of C. camelliae, the quantification of pathogen gene expression by the RT-qPCR method is necessary. Reliable RT-qPCR results require the use of stable reference genes for data normalization. However, suitable reference genes have not been reported in C. camelliae thus far. In this study, 12 candidate genes (i.e., CcSPAC6B12.04c, CcWDR83, Cchp11, Ccnew1, CcHplo, CcRNF5, CcHpcob, CcfaeB-2, CcYER010C, CcRNM1, CcUP18, and CcACT) were isolated from C. camelliae and assessed as potential reference genes. The expression stability of these genes in C. camelliae during spore germination and mycelial growth and interaction with host plants was first evaluated using several statistical algorithms, such as geNorm, NormFinder, and Bestkeeper. A web-based analysis program, Refinder, was then used to find the most suitable reference genes. Our results indicated that Cenew1, CcHplo, and CcSPAC6B12.04c were the most stable reference genes in C. camelliae under all conditions. Our work provided the most suitable reference genes for future studies performed to quantify the target gene expression levels of C. camelliae.
Introduction
Colletotrichum includes a wide range of fungal pathogens that cause serious diseases in various plants in tropical, subtropical, and temperate regions (Kubo, 2012; Yan et al., 2018). Their economic impacts have led to extensive studies on diverse aspects of fungal biology, including fungi–plant interactions, genomics and genetics, the cell biology of pathogen infection and colonization, and fungal virulence factors (De Silva et al., 2017; Villa-Rivera et al., 2017; Yan et al., 2018). Some species have been used as models for studying infection strategies and host-parasite interactions (De Silva et al., 2017; Yan et al., 2018). Colletotrichum camelliae is one of the dominant fungal pathogens that infects tea plants (Wang Y. et al., 2016; Lu et al., 2018). C. camelliae can damage tea leaves and cause several tea diseases, such as tea anthracnose, tea leaf blight, and tea brown blight (Chen and Chen, 1990; Liu F. et al., 2015; Wang L. et al., 2016; Wang Y. et al., 2016; Lu et al., 2018).
The quantification of functional gene expression levels is one of the most important aspects in the systematic study of gene transcription and regulation (Marcial-Quino et al., 2016). The reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) method is frequently used to quantify target gene expression levels (Derveaux et al., 2010; Borowski et al., 2014; Mcintosh et al., 2016). The RT-qPCR method is simple, reproducible, highly sensitive, and practical in detecting gene transcription (Klein, 2002; Hao et al., 2014). However, reliable RT-qPCR results require suitable reference genes for data analysis. The use of inadequate reference genes may result in incorrect expression data (Amil-Ruiz et al., 2013; Galli et al., 2015). Thus, the suitable reference genes in C. camelliae should be constantly expressed among the samples, and their expression is assumed to be unaffected by different experimental conditions (Bustin, 2002; Galli et al., 2015).
ACT, TUB, GAPDH, and 18SRNA are often used as reference genes because of their functions in basic cellular processes, cell structure maintenance, or primary metabolism (Galli et al., 2015). Reports have shown that several traditional reference genes were used in Colletotrichum spp.; for example, the ACT gene was used in C. higginsianum not only to quantify fungal growth but also to normalize the expression of the MFS transporter gene ChMFS1 (Narusaka et al., 2009; Liu et al., 2017). In C. acutatum and C. gloeosporioides, the β-TUB gene was used to normalize the expression of two ABC genes, CaABC1 and CgABCF2 (Kim et al., 2014; Zhou et al., 2017). In C. lindemuthianum and C. coccodes, two conserved genes, GAPDH and 18SRNA, were also used as reference genes (Ben-Daniel et al., 2012; Pereira et al., 2013; Galli et al., 2015). However, no suitable reference genes have been reported in C. camelliae for RT-qPCR analysis, although stably expressed referenced genes are believed to occur in C. camelliae, even under different experimental conditions. Thus, suitable reference genes in C. camelliae should be identified.
In the present study, we evaluated the stability of 12 candidate genes to identify the most suitable reference genes for transcript normalization in C. camelliae during spore germination and mycelial growth and interaction with hosts. To evaluate the efficacy of selected reference genes, we investigated the expression of a target gene involved with an ABC transporter during C. camelliae spore germination and mycelial growth and its interaction with tea plants. To our knowledge, this is the first analysis of the expression stability of suitable reference genes in C. camelliae.
Materials and Methods
Plants, Pathogen Materials, and Treatments
Tea plant Camellia sinensis cultivar Longjing 43 (LJ43) was used for all assays. Two-year-old tea were grown in a microbe-free climate chamber under 12-h light/12-h dark conditions at 25°C and 60–80% relative humidity before inoculation. For fungal inoculation, mature leaves of 2-year-old LJ43 were collected randomly. The C. camelliae isolate CCA was isolated from a diseased garden in Fancun, which is located in Hangzhou, China. The fungal isolate was cultivated on PDA plates at 22°C in a climate chamber (12-h light/12-h dark) for 10 days. C. camelliae spores were then collected, washed, and frozen at −80°C in 0.8% NaCl with a concentration of 108 spores mL–1 as previously indicated (Liu S. et al., 2015).
For the inoculation of tea plants, spores were diluted in ddH2O with a final concentration of 106 spore mL–1. Six to eight droplets (20 μL for one droplet) of diluted spores were applied to each single detached tea leaf. The leaves were wounded with a narrow razor blade before inoculation. In each treatment, at least 40 mature leaves were randomly selected from more than 20 tea plants. For the control, spores were incubated in ddH2O. Infection was carried out on a bench at room temperature. After infection for different times (e.g., 12, 14, or 24 h), the fungi were recovered from tea leaves and then frozen at −80°C for RNA assays. Three independent biological replicates were performed.
For the effects of tea catechins (Aladdin, China) on C. camelliae gene expression, the fungus was incubated on PDA solid media (Lu et al., 2018). The compound was dissolved in ddH2O and then mixed with sterile melted PDA medium to obtain a final concentration of 0.25 mg mL–1. The PDA medium was then poured into 9.0 cm diameter Petri plates for the inoculation with 0.8 cm disks of C. camelliae CCA. Each treatment was performed in 3 replicates. The PDA plates containing ddH2O (without any tea catechins) were used as the control. Fresh spores of C. camelliae CCA were also used as a 0-h control. The mycelia were harvested at 3 and 6 days, respectively. The fresh spores and mycelia were frozen at −80°C and used for RNA assays.
Total RNA Extraction and Reverse Transcription
Approximately 0.1 mg fungal fresh mycelia or 106 spores were used for RNA extraction. The samples were first frozen in liquid nitrogen and homogenized using a Tissue Lyser (Qiagen, Hilden, Germany) for 2 × 30 sec at 30 strokes/sec. RNA was then extracted using 1.0 mL TRIzol® Reagent (Life Technologies, Foster City, CA, United States) according to the manufacturer’s instructions. Finally, the total RNA was dissolved in nuclease-free water. The purity and concentration of the isolated RNA were estimated by a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, United States). RNA with an A260/A280 ratio of 1.8–2.0 was used for cDNA synthesis. The quality and integrity of the purified RNA templates was further confirmed by agarose gel electrophoresis. cDNA was synthesized from 1.0 μg of total RNA using the PrimeScriptTM RT reagent kit with gDNA Eraser (Takara, Dalian, China) to remove the genomic DNA contamination.
Selection of Candidate Reference Genes and Primer Design
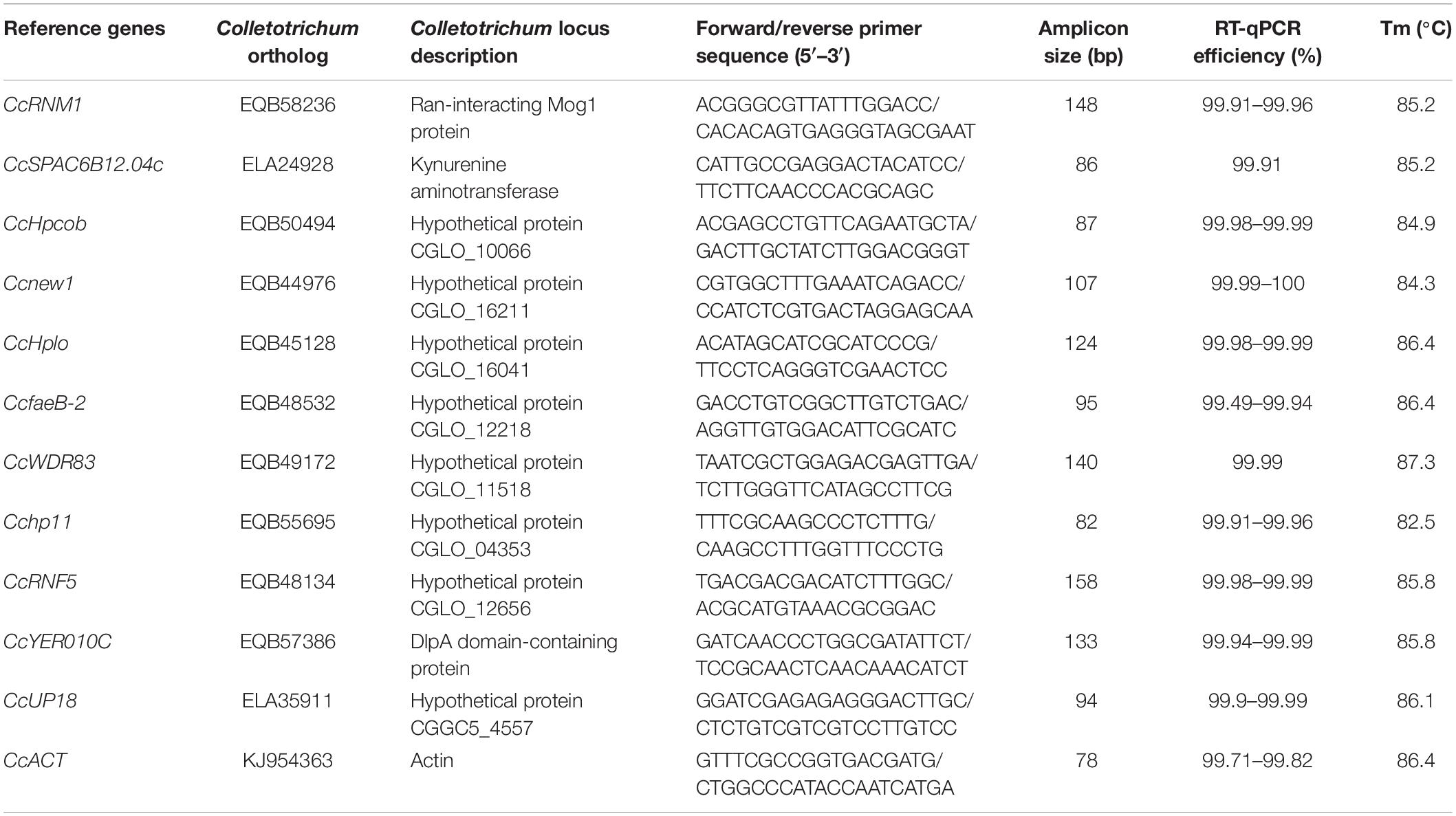
CcACT was chosen as a PCR reference gene in the present study according to previous reports (Wang Y. et al., 2016; Ning et al., 2018). Eleven candidate reference genes were chosen from the transcriptomic data of C. camelliae during the infection of tea plants (Supplementary Table S1). The open reading frame (ORF) sequences of 11 candidate reference genes from C. camelliae were first cloned using 2x Primer Star mix (Takara, Dalian, China) as the polymerase. The purified PCR products were ligated into the pEASY®-Blunt simple cloning vector (TranGen, Beijing, China) and then transformed into Escherichia coli. The bacterial liquids were sequenced by Comate Bioscience (Comate, Changchun, China). A bioinformatics analysis of the reference gene was performed by BLAST1. The RT-qPCR primers for all candidate genes were designed by Primer-BLAST2 and are presented in Table 1.
Quantitative Real-Time PCR
For qPCR analysis, approximately 20 ng of cDNA was mixed with 0.2 mM gene-specific primers and SYBR Green Supermix in a total volume of 10 μL. The qPCR was performed using a LightCycler® 480 system (Roche) according to the manufacturer’s instructions. The PCR program consisted of a preliminary step of 1 min at 95°C followed by 40 cycles at 95°C for 15 s and at 60°C for 34 s. No-template and no-RT controls for each primer pair were included. Each qPCR was performed in triplicate, and each experiment was independently repeated for three times. Standard curves were drawn to determine the amplification efficiency (E) and correlation coefficient (R2) of the diluted series on the basis of the 10-fold diluted cDNA series (Wu et al., 2016). We used the following equation to calculate the qPCR efficiency: E = (10{−1/slope}−1) × 100%.
Validation of Reference Genes
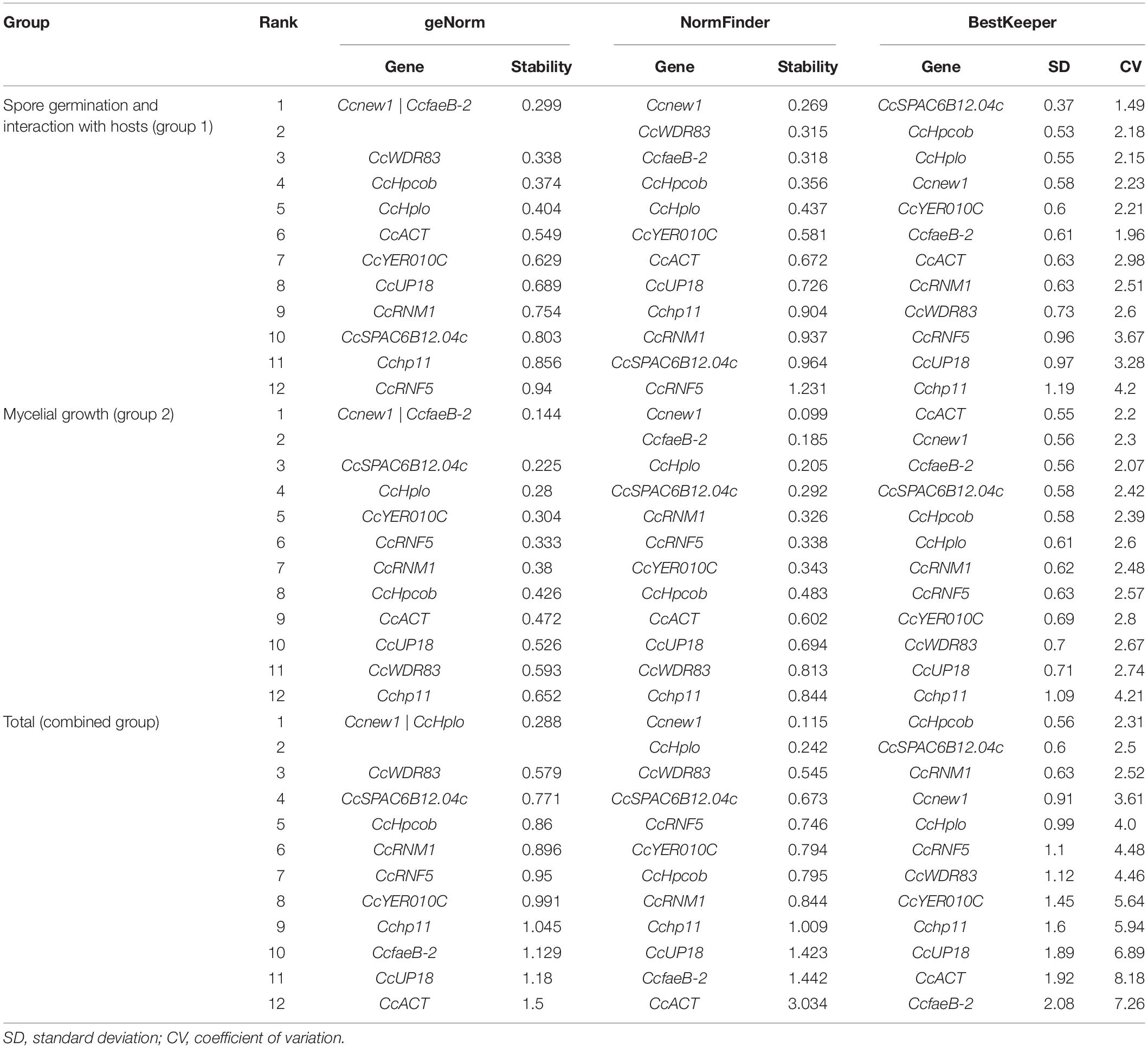
The expression of the 12 candidate genes was first evaluated according to the quantification cycle (Cq) value. Determination of the expression stability of the genes was then performed with three statistical algorithms (Bestkeeper, NormFinder, and geNorm) for the evaluation and selection of reference genes (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004; Marcial-Quino et al., 2016). The Cq values obtained for each of the analyzed genes were used to monitor the stability of the genes with these three methods, thus identifying the best reference genes for the normalization of data in RT-qPCR analyses (Marcial-Quino et al., 2016). Finally, Refinder was used to comprehensively evaluate and rank the reference genes from experimental data (Xie et al., 2011).
For the validation and quantification of the target gene (CcABC8) expression after pathogen infection, the 2–ΔΔCT method was used (Livak and Schmittgen, 2001). Statistical analyses were performed using a Student’s homoscedastic two-tailed t-test. Statistical significance is considered at ∗P < 0.05 and ∗∗P < 0.01. Three replicates of three independent experiments were performed. The qPCR primers for CcABC8 included CcABC8S (5′-TCCCTCCTCCTGACTCTCCT-3′) and CcABC8A (5′-TGGATCAATGTTGTCACGGA-3′).
Results
Isolation and Characterization of Candidate Reference Genes
The candidate reference genes were identified from the C. camelliae transcriptome by a homology analysis with Colletotrichum spp. (Supplementary Table S1). The transcripts of these genes were slightly changed during C. camelliae spore germination and during its interaction with host plants, thereby we chose them for further analysis. The ORF sequence of each gene was cloned from C. camelliae CCA based on the transcriptomic data (Supplementary Figure S1 and Supplementary Table S2). Protein-protein BLAST analysis indicated that two genes had the highest homology ( 89.5 and 98.9%) with C. fructicola Nara gc5, while others had the highest homology with C. gloeosporioides Cg-14 (from 82.3 to 99%) (Alkan et al., 2013; Gan et al., 2013). Among these candidate reference genes, one was similar to the ran-interacting Mog1 protein and named CcRNM1, a second gene was similar to kynurenine aminotransferase and named CcSPAC6B12.04c, a third gene was a DlpA domain-containing protein and referred to as CcYER010, and other genes were annotated as hypothetical proteins and named CcHpcob, Ccnew1, CcHplo, CcfaeB-2, CcWDR83, Cchp11, CcRNF5, and CcUP18.
The bioinformatics results for the amino acid sequence information are shown in Supplementary Table S2. The total amino acid number for the candidate reference gene was from 78 for Ccnew1 to 876 for CcHpcob; therefore, the lowest predicted molecular weight (MW) was 8.6 kDa for Ccnew1, and the highest MW was 100.5 kDa for CcHpcob. Most of the proteins had MWs ranging from 20 to 65 kDa. The predicted isoelectric point (pI) was from 4.3 for CcRNM1 to 11.0 for CcUP18, and many of proteins had pI values from 4.5 to 7.5.
Determination of Primer Specificity and Efficiency
Based on the candidate reference genes observed, 12 pairs of primers were designed (Table 1). A traditional reference gene, CcACT, was also isolated and included. The cDNA was synthesized from RNA obtained from organisms isolated at 12 h after infection of the plant cultivar Longjing 43. C. camelliae spores inoculated with ddH2O were used as a non-plant inoculation control (CK).
The melting curves observed from the PCR amplification products for each of the genes showed a single distinct sharp peak (Figure 1A). The specificity of the primers for all genes was further tested in 2.0% agarose gels. For all the candidate reference genes, a single PCR amplification band of the expected size was observed (Figure 1B), indicating that primer dimers and non-specific amplified products were not generated (Marcial-Quino et al., 2016).
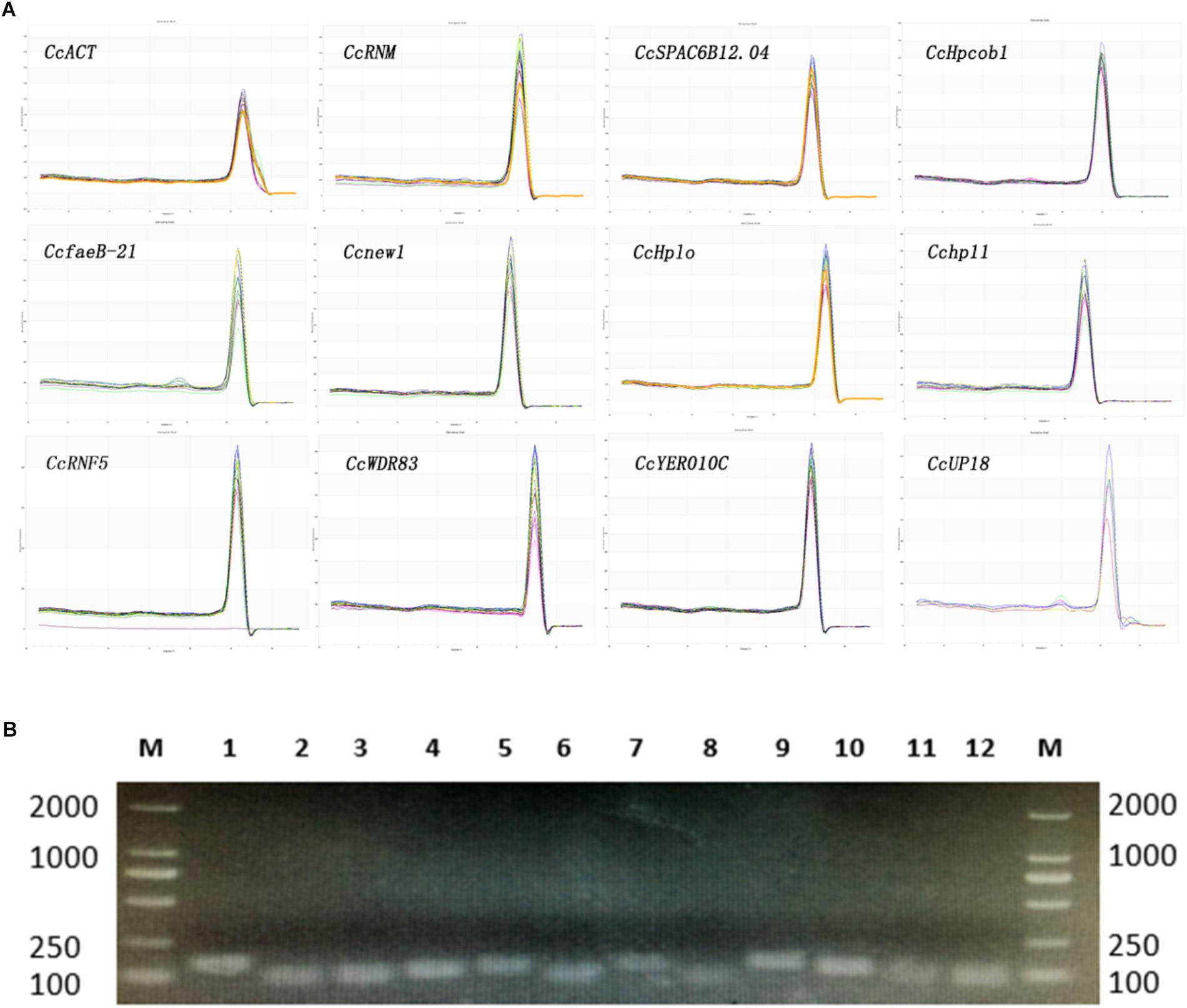
Figure 1. Confirmation of the primer specificity and amplicon size. (A) Melting curve analysis of 12 candidate reference genes. All RT-qPCR products had a single melting curve indicating the breakdown of only one PCR product. (B) Amplification results for 12 candidate genes using a C. camelliae cDNA template. M: DL2000 DNA marker. 1–12. CcRNM1, CcSPAC6B12.04c, CcHpcob, Ccnew1, CcHplo, CcfaeB-2, CcWDR83, Cchp11, CcRNF5, CcYER010C, CcUP18, and CcACT.
In a second experiment, the efficiency of the primers was detected using a 10-fold dilution series of cDNA from C. camelliae CCA (CK and 12 h). For all of the above cDNA samples, the PCR amplification efficiencies for the candidate genes varied from 99.49% for CcfaeB-2 to 100.00% for Ccnew1 (Table 1). The primers yielded linear amplification on a range of cDNA concentrations and the correlation coefficient R2 were from 93.2% for Ccnew1 to 99.97% for CcSPAC6B12.04c (Figure 2). For the gene primers, such as CcSPAC6B12.04c, Cchp11, CcRNM1, and CcfaeB-2, R2 was over 99%, indicating that these primer pairs were well-suited for amplification of the gene even when using very low cDNA input.
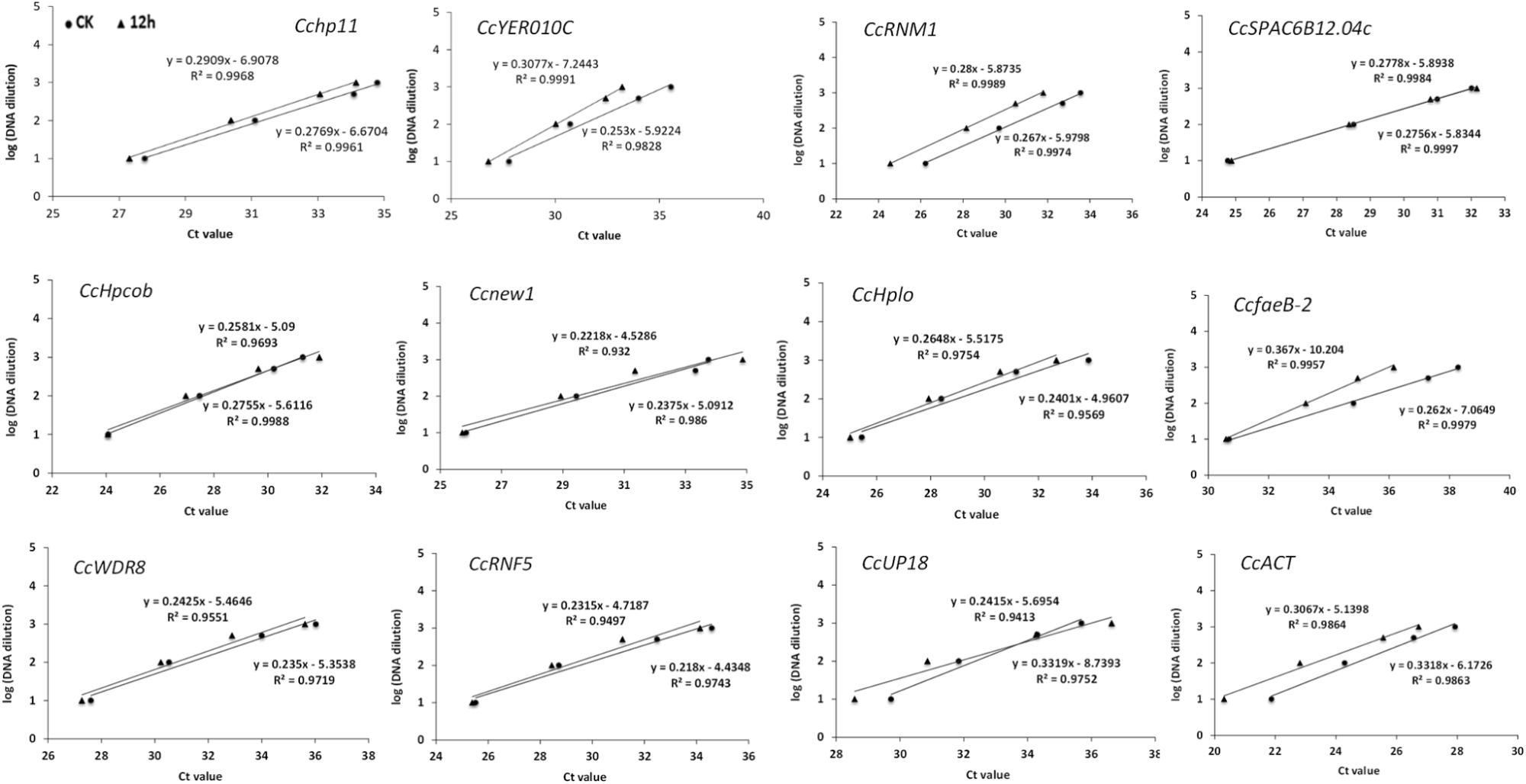
Figure 2. Validation of primers for RT-qPCR quantification of the tea pathogen. The primer efficiency for the RT-qPCR quantification of the gene was determined using a serial dilution of cDNA templates from C. camelliae. The respective correlation coefficients (R2) are indicated.
Expression Profiles of Candidate Reference Genes
A reliable reference gene should present a constant expression level among different samples or different conditions (Galli et al., 2015); thus, we next checked the Cq values to evaluate the expression of these candidate reference genes. Total RNA was extracted from C. camelliae CCA fresh spores (un-germinated spores, CcFS0h), C. camelliae grown on PDA plates for 3 days (mycelium, CcPM3d) and 6 days (mycelium, CcPM6d), C. camelliae grown on PDA plates with tea catechins for 3 days (mycelium, CcPCM3d) and 6 days (mycelium, CcMPC6d), C. camelliae incubated on tea plant LJ43 for 14 h (CcTP14h) and C. camelliae spores incubated with ddH2O for 14 h (control, spores germinated in ddH2O, CcGe14h). Total RNA was reverse transcribed into cDNA and then used for RT-qPCR.
As shown in Figure 3, the 12 candidate reference genes showed a narrow Cq range among all experimental series. The Cq values ranged from 23.5 to 28.7. Transcription of CcACT showed the most abundant level, while the gene CcfaeB-2 was the least abundant transcript. It indicated that each reference gene had varied expression ranges in all studied samples (Hao et al., 2014). CcHpcob had the lowest variation in expression across the studied reference genes, while CcfaeB-2 showed the highest variation in expression (Figure 3).
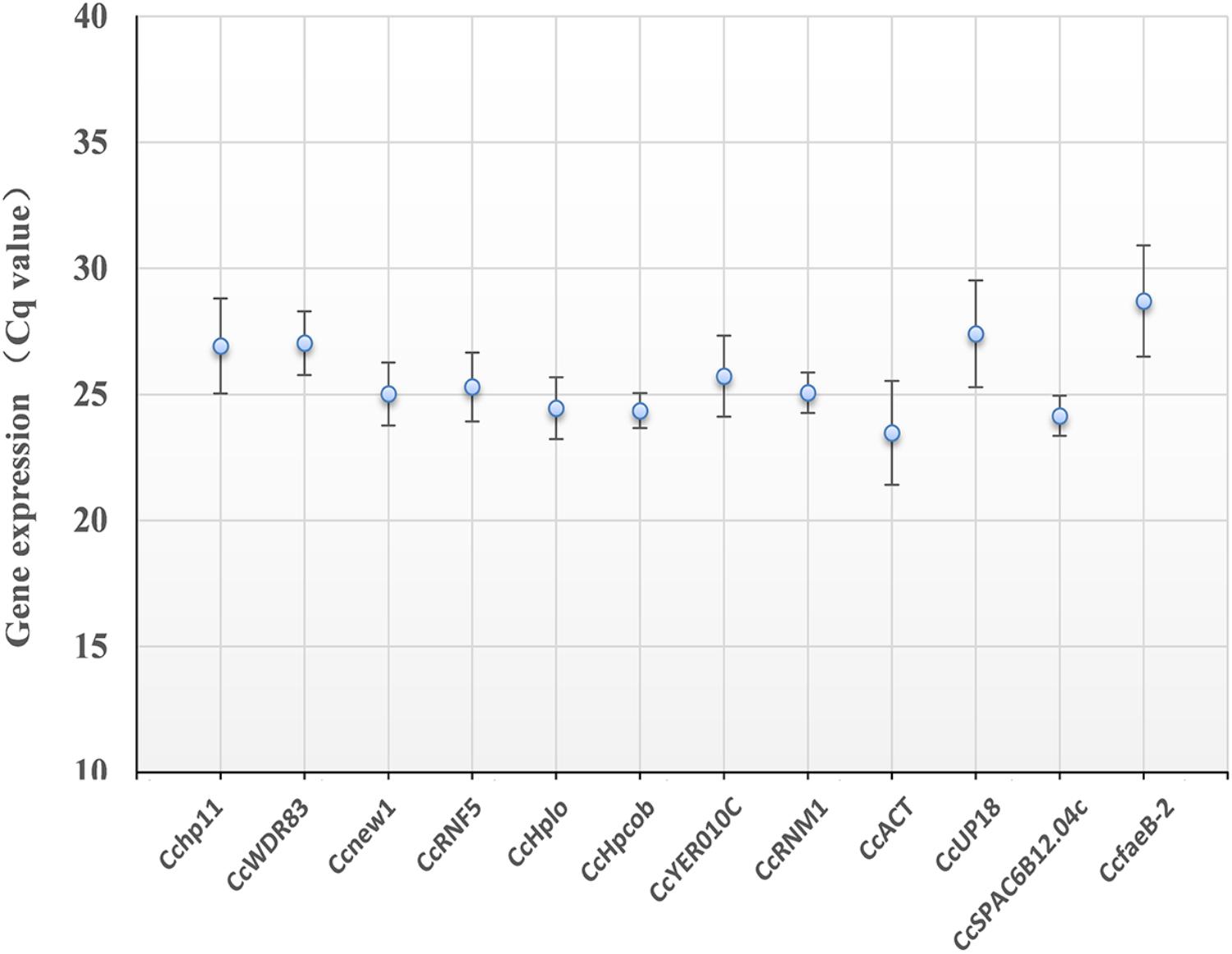
Figure 3. Average cycle threshold (Cq) values for 12 candidate reference genes. The filled dot symbol indicates the mean Cq values. The bars indicate standard deviation.
Expression Stability of the Candidate Genes
First, to calculate the reference gene expression stability, geNorm software was used (Vandesompele et al., 2002). According to geNorm, the candidate gene that had the lowest value was considered the most stable gene (Zhao et al., 2019). As shown in Table 2, Ccnew1, CcfaeB-2, CcWDR83, CcHpcob, and CcHplo were the top five stable candidate reference genes during C. camelliae spore germination and its interaction with hosts (group 1: CcFS, CcGe, and CcTP), while Ccnew1, CcfaeB-2, CcSPAC6B12.04c, CcHplo, and CcYER010C were the top five genes during mycelial growth (group 2: CcM). Three genes, Ccnew1, CcfaeB-2 and CcHplo, were detected in both groups, indicating that these genes were stable under each condition. Furthermore, in a combined group analyzed by geNorm software, the top five stable candidate reference genes were Ccnew1, CcHplo, CcWDR83, CcSPAC6B12.04c, and CcHpcob. This finding indicates that these genes are relatively stable during spore germination and mycelial growth and infection processes.
NormFinder software is based on a mathematical model of separate analyses of sample subgroups and the estimation of both intra- and intergroup expression variations (Andersen et al., 2004; Wu et al., 2016). Genes with stable expression were indicated by low average expression stability values (Wu et al., 2016). Based on NormFinder analysis, Ccnew1, CcWDR83, CcfaeB-2, CcHpcob, and CcHplo were the top five stable candidate reference genes in group 1, which were the same as the genes identified by geNorm software (Table 2). In group 2, the top five stable genes were Ccnew1, CcfaeB-2, CcHplo, CcSPAC6B12.04c, and CcRNM1. Interestingly, Ccnew1, CcfaeB-2, and CcHplo were also observed in both groups. This result was the same as that found by the geNorm analysis and further confirmed that these genes were stable under each condition. In the combined group, the top five stable genes were Ccnew1, CcHplo, CcWDR83, CcSPAC6B12.04c, and CcRNF5. Based on NormFinder, these genes are relatively stable under all conditions.
Bestkeeper, which calculates the CP standard deviation (SD) and the coefficient of variance (CV) for each gene, was additionally used (Pfaffl et al., 2004; Marcial-Quino et al., 2016). Stable reference genes have a relatively low coefficient of variance and standard deviation (CV ± SD). Genes with SD values < 1 are considered stable and thus are suitable as reference genes (Marcial-Quino et al., 2016). The results of analysis for the 12 reference genes showed markedly stable expression in both group 1 and group 2 samples except Cchp11 (Table 2). However, under the combined condition, the suitable reference genes observed by Bestkeeper were CcHpcob, CcSPAC6B12.04c, CcRNM1, Ccnew1, and CcHplo.
To identify the most suitable reference genes, RefFinder was used to analyze stability of the candidate reference genes. The outcome of four programs (ΔCt, Bestkeeper, geNorm, and NormFinder) were integrated by RefFinder (Xie et al., 2011), and Ccnew1, CcfaeB-2, CcHpcob, and CcWDR83 appeared to be the most stable reference genes during C. camelliae spore germination and its interaction with hosts (group 1), while Ccnew1, CcfaeB-2, CcSPAC6B12.04c, and CcHplo were the most stable reference genes during mycelial growth (group 2) (Figures 4A,B). Ccnew1 and CcfaeB-2 seemed stable in each group when target gene expression was analyzed separately. When target gene expression was analyzed under both conditions, Ccnew1, CcHplo, CcSPAC6B12.04c, and CcWDR83 were the most stable reference genes (Figure 4C). However, CcfaeB-2, CcHpcob, and CcWDR83 were observed as the least stable reference genes under at least one condition (Figures 4A–C). In conclusion, we considered Ccnew1, CcHplo, and CcSPAC6B12.04c to be the most stable reference genes that could be used to compare and analyze the target gene expression in C. camelliae during spore germination, mycelial growth and its interaction with host plants.
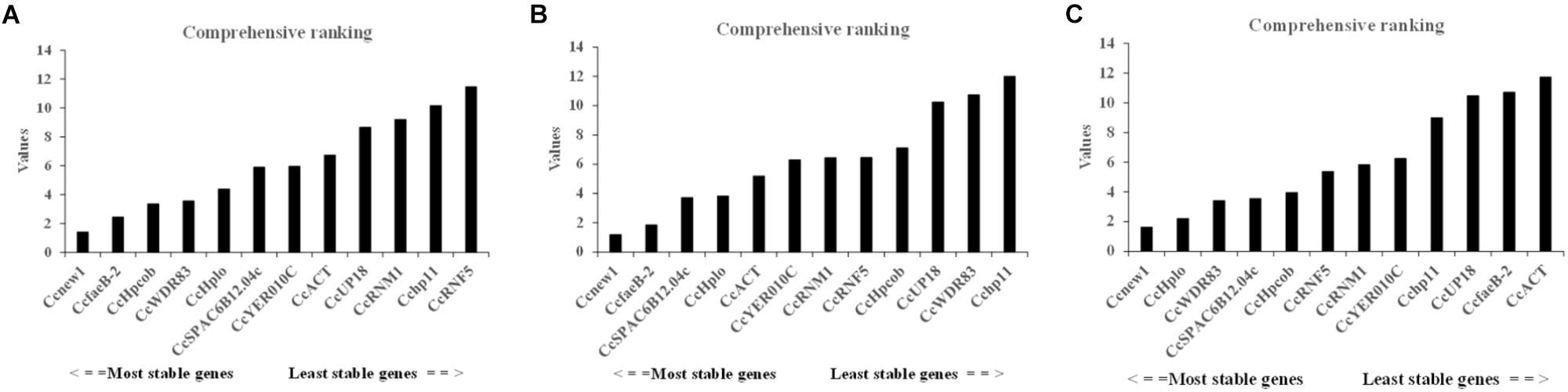
Figure 4. Expression stability of the candidate reference genes in C. camelliae as calculated by RefFinder. (A) Expression stability of the candidate reference genes during C. camelliae spore germination and interaction with tea plants. (B) Expression stability of the candidate reference genes during C. camelliae mycelial growth. (C) Expression stability of the candidate reference genes in C. camelliae under the combined condition.
Evaluation of Reference Genes
To evaluate and compare the functional gene expression in C. camelliae, the Cenew1, CcSPAC6B12.04c, CcHplo, Cchp11, CcACT, and CcUP18 genes were selected as reference genes for RT-qPCR. Cenew1 was the most stable reference gene under all conditions. CcSPAC6B12.04c was the most suitable candidate reference gene even when using a low cDNA input. CcHplo was also very stable during mycelial growth and under the combined condition. CcACT was the least stable reference gene under the combined condition, while the Cchp11 and CcUP18 genes were the least stable reference genes under all conditions. We chose one target gene that may be involved with the ABC transporter (CcABC8) to test its expression.
As shown in Figures 5D–F, the expression level of CcABC8 showed no significant differences between C. camelliae spore germination (CcGe) and interaction with the tea plant (CcTP) when we used CcACT, Cchp11, and CcUP18 as the reference genes. When we used Cenew1, CcSPAC6B12.04c, or CcHplo as the reference genes, we detected significant CcABC8 gene induction during pathogen interaction with the tea plant (Figures 5A–C).
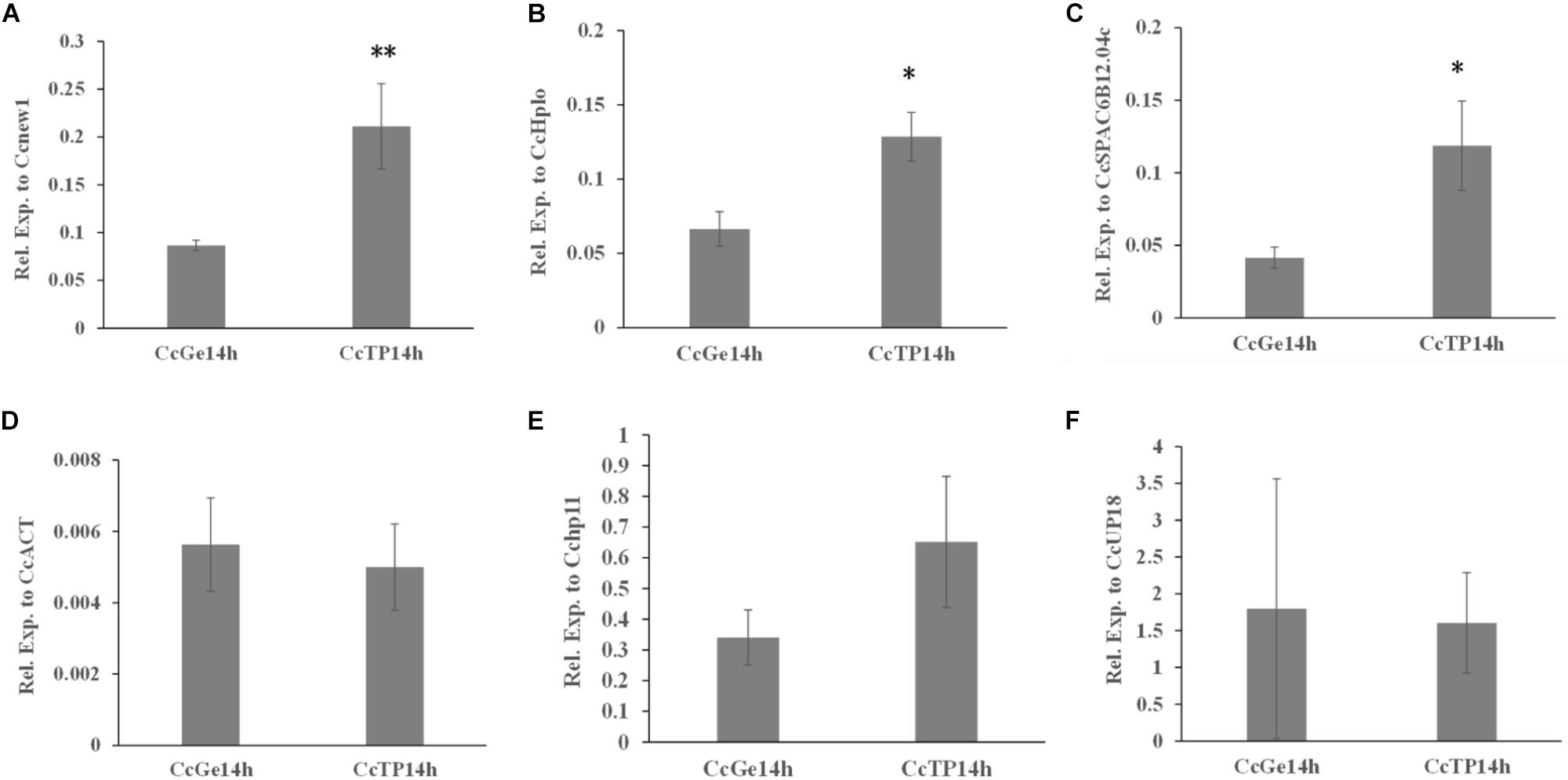
Figure 5. Expression profile of CcABC8 in C. camelliae during spore germination and its interaction with tea plants. The most stable reference genes (Cenew1, CcHplo, and CcSPAC6B12.04c) and the least stable reference genes (CcACT, Cchp11, and CcUP18) were used to normalize the expression data. (A) Cenew1; (B) CcHplo; (C) CcSPAC6B12.04c; (D) CcACT; (E) Cchp11; and (F) CcUP18. The data show the mean expression ± standard deviation calculated from three biological replicates. ∗P < 0.05; ∗∗P < 0.01.
In addition, the expression of CcABC8 during C. camelliae mycelial growth was also tested. Since the C. camelliae spores were incubated on PDA plates at the beginning (CcFS0h) of the experiment, we used CcFS0h as the control. As shown in Figures 6A–D, the expression of CcABC8 increased during C. camelliae growth on PDA plates (CcPM) or PDA plates with catechins (CcPCM) for 3 and 6 days when using Cenew1, CcSPAC6B12.04c, CcHplo, or CcACT as reference genes, respectively. This indicates that the gene was induced during mycelial growth. The expression of CcABC8 was higher after growing on PDA plates for 6 days than 3 days (Figures 6A–D). However, the expression of CcABC8 was not significantly increased in fungal mycelium compared with the control when using Cchp11 and CcUP18 as reference genes (Figures 6E,F).
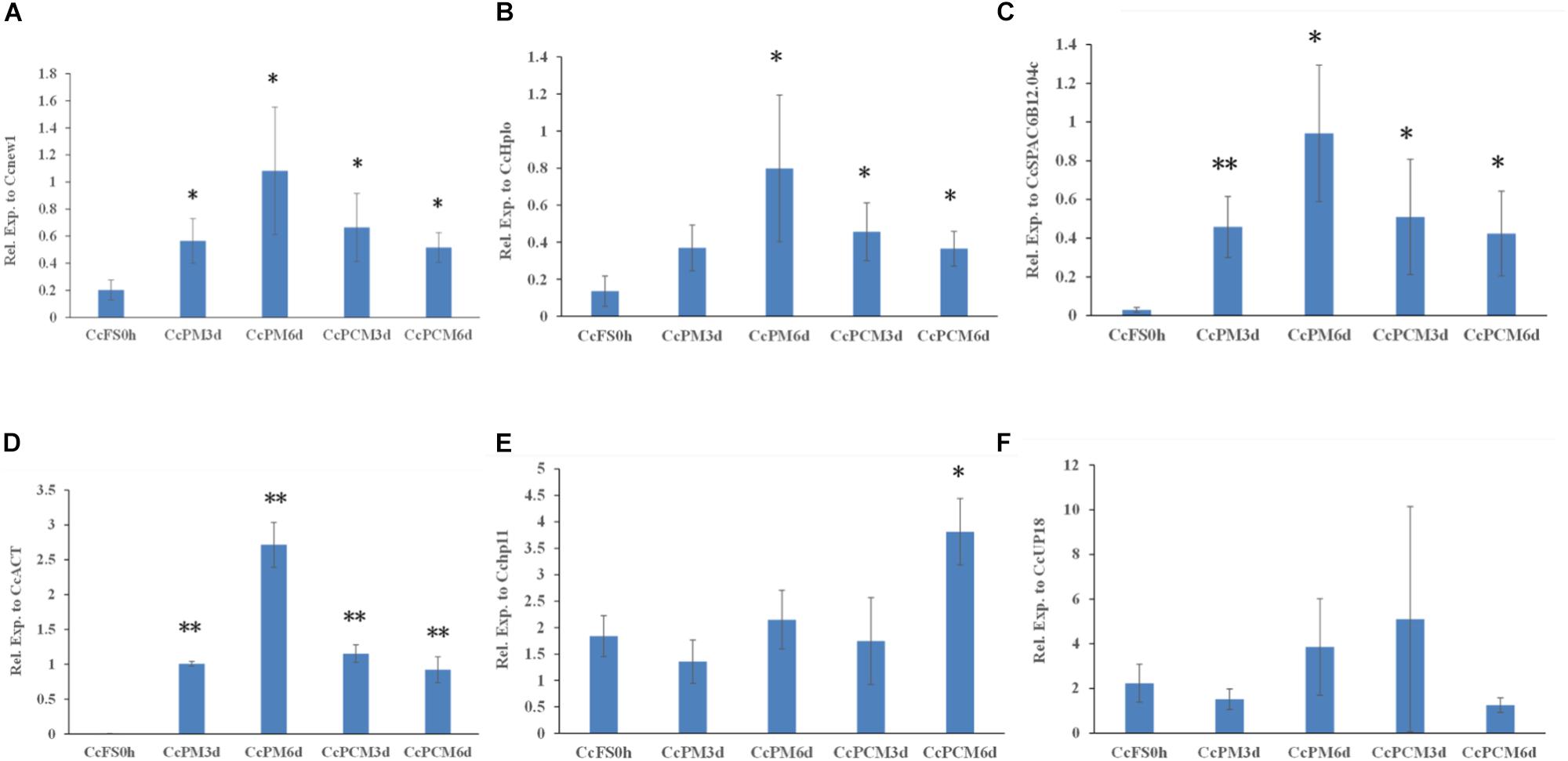
Figure 6. Expression profile of CcABC8 in C. camelliae during mycelial growth. The reference genes (Cenew1, CcHplo, CcSPAC6B12.04c, CcACT, Cchp11, and CcUP18) were used to normalize the expression data. (A) Cenew1; (B) CcHplo; (C) CcSPAC6B12.04c; (D) CcACT; (E) Cchp11; and (F) CcUP18. The data show the mean expression ± standard deviation calculated from three biological replicates. ∗P < 0.05; ∗∗P < 0.01.
These results indicated that (i) the use of unstable reference genes will lead to differences in the relative transcript profile and (ii) the use of different suitable references could have diverse significant results during qPCR.
Discussion
To analyze the gene functions of the tea plant pathogen C. camelliae, gene expression differences might be important to analyze (Lu et al., 2018). RT-qPCR has become an important technique for studying gene transcript profiles as its sensitivity, accuracy, and reproducibility (Klein, 2002; Bustin et al., 2005; Hao et al., 2014; Wang et al., 2017). The reliability of the results of gene expression in RT-qPCR studies is dependent on the use of suitable reference genes for the microbe and the condition under study (Galli et al., 2015; Huang et al., 2018). The expression of the reference genes should not change with time or under different experimental conditions (Bustin, 2002; Melgar-Rojas Pedro et al., 2015). However, the stability among traditionally used reference genes is relative, and there is no single gene that has a constant stable expression under all experimental conditions (Radonic et al., 2004; Czechowski et al., 2005). Under specific experimental conditions, it might be misleading to use previously identified reference genes for the normalization of target gene expression in C. camelliae without first investigating their stability (Amil-Ruiz et al., 2013; Galli et al., 2015). Therefore, CcSPAC6B12.04c, CcWDR83, Cchp11, Ccnew1, CcRNF5, CcHpcob, CcfaeB-2, CcYER010C, CcRNM1, CcUP18, and CcACT were selected here for validation under the experimental conditions.
The expression of candidate reference genes were first evaluated with the Cq value in RT-qPCR. The Cq values for most of the tested samples were approximately 25. Even after 1000-fold dilution, the Cq value for nine candidate genes was still lower than 35.0. All candidate reference genes had very good linear amplification, and four of them had R2 values greater than 99%. Notably, the R2 value of CcSPAC6B12.04 was 100%. The Cq value comparison provided an approximation of the stability of gene expression.
The programs geNorm, NormFinder, and Bestkeeper were then used to determine which reference gene was most suitable for transcript normalization during C. camelliae spore germination, mycelial growth, and fungal interaction with the tea plants. Among the 12 candidate reference genes, Ccnew1 was ranked first in both the geNorm and NormFinder analyses under all conditions. The Cq SD value of Ccnew1 was < 1 based on the Bestkeeper program, which was consistent with reference genes with SD values < 1 that are considered stable (Pfaffl et al., 2004; Marcial-Quino et al., 2016). Taken together, Ccnew1 was the most stable reference gene for the detection of target gene expression not only during C. camelliae spore germination and its interaction with hosts, but also during mycelial growth. Here, the result indicates that Ccnew1 is a universal reference gene that is stably expressed under different experimental conditions in this study.
We further used Refinder analysis to reduce bias or avoid contradictory results caused by the use of individual methods, ΔCt, Bestkeeper, geNorm, and NormFinder (Xie et al., 2011; Marcial-Quino et al., 2016). Based on this, the most stable reference genes were Ccnew1, CcHplo and CcSPAC6B12.04c, while the least stable genes were Cchp11 and CcUP18. Interestingly, CcfaeB-2 was ranked the second most stable gene not only in C. camelliae spore germination but also in mycelial growth (Figures 4A,B), whereas it ranked the second least stable reference gene under all conditions (Figure 4C). One reason that explains the difference may be the highest variation in expression of CcfaeB-2 (Figure 3).
During previous studies of gene expression in Colletotrichum spp., ACT was often used to normalize qPCR because it was stably expressed in many other microbes (Narusaka et al., 2009; Liu et al., 2017). Similarly, CcACT seems stably expressed during C. camelliae mycelial growth (Figure 4B). However, under the combined conditions, CcACT was ranked as the least stably expressed reference gene (Figure 4C). Nevertheless, if only the expression of target genes was detected during mycelial growth, then CcACT could be a choice for reference gene (Figure 6D). In conclusion, the commonly used reference genes need to be reconfirmed according to specific experimental conditions.
To validate the suitability of potential reference genes, the expression profile of a target gene was assessed in C. camelliae, with Cenew1, CcHplo, CcSPAC6B12.04c, CcACT, Cchp11, and CcUP18 as internal reference genes. The gene expression patterns were highly similar but the expression levels were significantly different from that of the treatments when the most stably expressed reference genes were used, while the transcript levels could be inaccurate or present no significant differences when the least stably expressed reference genes were used (Figures 5, 6). Thus, using a reliable reference gene is a prerequisite for accurate RT-qPCR data analyses of C. camelliae.
Conclusion
To our knowledge, this is the first report describing the identification of suitable reference genes for RT-qPCR analyses in C. camelliae. We evaluated 12 candidate reference genes for the normalization of gene expression in C. camelliae. Common statistical algorithms and a web-based analysis program were used and indicated that Cenew1, CcHplo, and CcSPAC6B12.04c were the most stable reference genes. In addition, Cchp11 and CcUP18 seem to be unsuitable as internal controls under the experimental conditions we tested. Additionally, the analysis of the CcABC8 expression level confirmed the importance of selecting suitable reference genes for the normalization of RT-qPCR data. The reference genes selected here provide important choices for target gene expression and functional studies in C. camelliae.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
SL designed the experiments. SH, TA, and RA performed the experiments. SL, SH, and RA analyzed the data. SH and SL wrote the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (31370689 and 30972405).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Huchen Chen for excellent experimental assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02055/full#supplementary-material
Footnotes
References
Alkan, N., Meng, X., Friedlander, G., Reuveni, E., Sukno, S., Sherman, A., et al. (2013). Global aspects of pacC regulation of pathogenicity genes in Colletotrichum gloeosporioides as revealed by transcriptome analysis. Mol. Plant Microbe Int. 26, 1345–1358. doi: 10.1094/MPMI-03-13-0080-R
Amil-Ruiz, F., Garrido-Gala, J., Blanco-Portales, R., Folta, K. M., Muñoz-Blanco, J., and Caballero, J. L. (2013). Identification and validation of reference genes for transcript normalization in strawberry (Fragaria× ananassa) defense responses. PLoS One 8:e70603. doi: 10.1371/journal.pone.0070603
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.can-04-0496
Ben-Daniel, B.-H., Bar-Zvi, D., and Tsror Lahkim, L. (2012). Pectate lyase affects pathogenicity in natural isolates of Colletotrichum coccodes and in pelA gene-disrupted and gene-overexpressing mutant lines. Mol. Plant Pathol. 13, 187–197. doi: 10.1111/j.1364-3703.2011.00740.x
Borowski, J. M., Galli, V., Silva Messias, R., Perin, E. C., Buss, J. H., and Anjos e Silva, S. D. (2014). Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 239, 1187–1200. doi: 10.1007/s00425-014-2041-2
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–39. doi: 10.1677/jme.0.0290023
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR-A perspective. J. Mol. Endocrinol. 34, 597–601.
Chen, Z., and Chen, X. (1990). Diagnosis and Treatment of Tea Tree Diseases. China: Shanghai Scientific and Technical Publishers.
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
De Silva, D., Crous, P., Ades, P., Hyde, K., and Taylor, P. (2017). Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol. Rev. 31, 155–168. doi: 10.1016/j.fbr.2017.05.001
Derveaux, S., Vandesompele, J., and Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. doi: 10.1016/j.ymeth.2009.11.001
Galli, V., Borowski, J. M., Perin, E. C., Messias, R., da, S., Labonde, J., et al. (2015). Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 554, 205–214. doi: 10.1016/j.gene.2014.10.049
Gan, P., Ikeda, K., Irieda, H., Narusaka, M., O’Connell, R. J., Narusaka, Y., et al. (2013). Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197, 1236–1249. doi: 10.1111/nph.12085
Hao, X., Horvath, D. P., Chao, W. S., Yang, Y., Wang, X., and Xiao, B. (2014). Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L.) O. Kuntze). Int. J Mol. Sci. 15, 22155–22172. doi: 10.3390/ijms151222155
Huang, N., Ling, H., Liu, F., Su, Y., Mao, H., Zhang, X., et al. (2018). Identification and evaluation of PCR reference genes for host and pathogen in sugarcane-Sporisorium scitamineum interaction system. BMC Genomics 19:479. doi: 10.1186/s12864-018-4854-z
Kim, S., Park, S. Y., Kim, H., Kim, D., Lee, S. W., Kim, H. T., et al. (2014). Isolation and Characterization of the Colletotrichum acutatutn ABC Transporter CaABC1. Plant Pathol. J. 30, 375–383. doi: 10.5423/PPJ.OA.08.2014.0077
Klein, D. (2002). Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8, 257–260. doi: 10.1016/s1471-4914(02)02355-9
Kubo, Y. (2012). Appressorium function in Colletotrichum orbiculare and prospect for genome based analysis. Morphogenesis Pathogenicity Fungi 22, 115–131. doi: 10.1007/978-3-642-22916-9_7
Liu, F., Weir, B. S., Damm, U., Crous, P. W., Wang, Y., Liu, B., et al. (2015). Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35, 63–86. doi: 10.3767/003158515X687597
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., and Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4:e07295. doi: 10.7554/eLife.07295
Liu, L., Yan, Y., Huang, J., Hsiang, T., Wei, Y., Li, Y., et al. (2017). A Novel MFS transporter gene ChMfs1 is important for hyphal morphology, conidiation, and pathogenicity in Colletotrichum higginsianum. Front. Microbiol. 8:1953. doi: 10.3389/fmicb.2017.01953
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Q., Wang, Y., Li, N., Ni, D., Yang, Y., and Wang, X. (2018). Differences in the characteristics and pathogenicity of Colletotrichum camelliae and C. fructicola isolated from the tea plant (Camellia sinensis (L.) O. Kuntze). Front. Microbiol. 9:3060. doi: 10.3389/fmicb.2018.03060
Marcial-Quino, J., Fierro, F., De la Mora-De la Mora, I., Enríquez-Flores, S., Gómez-Manzo, S., Vanoye-Carlo, A., et al. (2016). Validation of housekeeping genes as an internal control for gene expression studies in Giardia lamblia using quantitative real-time PCR. Gene 581, 21–30. doi: 10.1016/j.gene.2016.01.018
Mcintosh, C. H., Baird, J., Zinser, E., Woods, D. J., Campbell, E. M., and Bowman, A. S. (2016). Reference gene selection and RNA preservation protocol in the cat flea, Ctenocephalides felis, for gene expression studies. Parasitology 143, 1532–1542. doi: 10.1017/S0031182016001025
Melgar-Rojas Pedro, C. A. J., Fuentes-Santamaría, V., Gabaldón-Ull María, C., Juiz José, M., and Bernd, S. (2015). Validation of reference genes for RT-qPCR analysis in noise-induced hearing loss: a study in wistar rat. PLoS One 10:e0138027. doi: 10.1371/journal.pone.0138027
Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M., et al. (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x
Ning, H., Hui, L., Feng, L., Yachun, S., Weihua, S., Huaying, M., et al. (2018). Identification and evaluation of PCR reference genes for host and pathogen in sugarcane-Sporisorium scitamineum interaction system. BMC Genomics 19:479. doi: 10.1186/s12864-018-4854-z
Pereira, M. F., de Araújo Dos Santos, C. M., de Araújo, E. F., de Queiroz, M. V., and Bazzolli, D. M. S. (2013). Beginning to understand the role of sugar carriers in Colletotrichum lindemuthianum: the function of the gene mfs1. J. Microbiol. 51, 70–81. doi: 10.1007/s12275-013-2393-5
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Radonic, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1.
Villa-Rivera, M. G., Conejo-Saucedo, U., Lara-Marquez, A., Cano-Camacho, H., Lopez-Romero, E., and Zavala-Paramo, M. G. (2017). The role of virulence factors in the pathogenicity of Colletotrichum sp. Curr. Protein Pept. Sci. 18, 1005–1018. doi: 10.2174/1389203717666160813160727
Wang, L., Wang, Y., Cao, H., Hao, X., Zeng, J., Yang, Y., et al. (2016). Transcriptome analysis of an anthracnose-resistant tea plant cultivar reveals genes associated with resistance to Colletotrichum camelliae. PLoS One 11:e0148535. doi: 10.1371/journal.pone.0148535
Wang, Y., Hao, X., Wang, L., Bin, X., Wang, X., and Yang, Y. (2016). Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Sci. Rep. 6:35287. doi: 10.1038/srep35287
Wang, M., Li, Q., Xin, H., Chen, X., Zhu, X., and Li, X. (2017). Reliable reference genes for normalization of gene expression data in tea plants (Camellia sinensis) exposed to metal stresses. PLoS One 12:e0175863. doi: 10.1371/journal.pone.0175863
Wu, Z., Tian, C., Jiang, Q., Li, X., and Zhuang, J. (2016). Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 6:19748. doi: 10.1038/srep19748
Xie, F., Sun, G., Stiller, J. W., and Zhang, B. (2011). Genome-Wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS One 6:1–12. doi: 10.1371/journal.pone.0026980
Yan, Y., Yuan, Q., Tang, J., Huang, J., Zheng, L., Hsiang, T., et al. (2018). Colletotrichum higginsianum as a model for understanding host–pathogen interactions: a Review. Int. J Mol. Sci. 19:2142. doi: 10.3390/ijms19072142
Zhao, Z., Wang, L., Yue, D., Ye, B., Li, P., Zhang, B., et al. (2019). Evaluation of reference genes for normalization of RT-qPCR gene expression data for Trichoplusia ni cells during Antheraea pernyi (Lepidoptera: Saturniidae) multicapsid nucleopolyhedrovirus (AnpeNPV) infection. J. Insect Sci. 19:4. doi: 10.1093/jisesa/iey133
Keywords: reference genes, tea plant, Colletotrichum camelliae, RT-qPCR, gene expression
Citation: He S, An T, AR and Liu S (2019) Validation of Reliable Reference Genes for RT-qPCR Studies of Target Gene Expression in Colletotrichum camelliae During Spore Germination and Mycelial Growth and Interaction With Host Plants. Front. Microbiol. 10:2055. doi: 10.3389/fmicb.2019.02055
Received: 29 April 2019; Accepted: 20 August 2019;
Published: 04 September 2019.
Edited by:
Raffaella Balestrini, Institute for Sustainable Plant Protection, Italian National Research Council (IPSP-CNR), ItalyReviewed by:
Taurai Tasara, University of Zurich, SwitzerlandArunachalam Ramaiah, University of California, Irvine, United States
Matteo Caser, University of Turin, Italy
Copyright © 2019 He, An, A and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouan Liu, c2hvdWFuQGpsdS5lZHUuY24=
†These authors have contributed equally to this work
 Shengnan He
Shengnan He Tai An†
Tai An† Shouan Liu
Shouan Liu