- Institute of Animal Husbandry Veterinary, Jinzhou Medical University, Jinzhou, China
Chinese sacbrood virus (CSBV) is the major cause and lead to the collapse of Apis cerana colonies. VP1, the structural protein of CSBV, shows the highest variation in the amino acid sequences among proteins from different CSBV strains as well as exhibits excellent immunogenicity. However, its function with host protein still remains unclear. To clarify its function with host protein, we screened out host cellular proteins that interact with VP1 using the membrane protein yeast two-hybrid system. In addition, we verified interactions between heat shock protein 70 cognate 5 (Hsp70-c5) and VP1 using glutathione S-transferase (GST) pull-down and co-immunoprecipitation assays. VP1 and Hsp70-c5 were colocalized in the cytoplasm and nucleus. Using western blot and real-time polymerase chain reaction (PCR), Hsp70-c5 expression in CSBV-infected larvae was upregulated compared with that in healthy larvae. We observed that when we silenced Hsp70-c5, VP1 expression was significantly downregulated. These results demonstrate that Hsp70-c5 is involved in at least one stage(s) of the viral life cycle.
Introduction
Apis mellifera (Am) and Apis cerana (Ac) are major honey bee species in the global beekeeping industry (Zhang et al., 2014). They are heavily infected by different vital viruses (Chen et al., 2004). Among honeybee viruses, Chinese sacbrood virus (CSBV) is the most serious threat to bee health and has caused wide spread concern among beekeepers and researchers (Sun et al., 2018). CSBV usually infects 2-day-old larvae and prevents them from pupating, ultimately leading to their death. The first CSBV inflection was reported in 1972 in A. cerana in Fogang and Conghua, Guangdong province, China (Zhang et al., 2002; Sun et al., 2017). In 2008, epidemic outbreaks of CSD caused the death of individual bees and decimated entire bee colonies in Liaoning Province (Mingxiao et al., 2011). Currently, the virus has been frequently reported in numerous regions in China and has had devastating impacts on apiculture.
Chinese sacbrood virus is a typical positive-stranded small RNA virus with a large open reading frame encoding a polypeptide consisting of structural proteins (VP1, VP2, VP3, and VP4) and non-structural proteins (Fei et al., 2015). The VP1 gene has the highest amino acid sequence variation among different CSBV strains and contains the common and major epitope of CSBV (Cheng et al., 2011). Fei et al. (2015) reported that the structural proteins can induce high antibody levels and enhance lymphocyte proliferation, of which VP1 has good immunogenicity. CSBV is classified into four subgroups based on the VP1 sequences (Mingxiao et al., 2013). The existence of different CSBV subgroups could be associated with regional variations (Fiil et al., 2008; Hu et al., 2016). Despite the classification of CSBV into four subgroups based on the VP1 sequences, these subgroups do not exhibit substantial differences with respect to physical and chemical properties, pathogenicity, and immunogenicity (Sato et al., 1994). However, host–cell interactions with partner proteins and the functions of VP1 remain unknown.
Currently, the analytical methods for evaluation of protein–protein interactions mainly include assays such as co-immunoprecipitation (Co-IP), yeast two-hybridization (Y2H), molecular fluorescence complementation, and glutathione S-transferase (GST) pull-down as well as phage display technology (Vaughan et al., 1996; Stagljar et al., 1998; Ozalp et al., 2005; Rajagopala and Uetz, 2011; Luo et al., 2014). Among these, the Y2H system is highly efficient and is used in in vivo screening for protein–protein interactions (Johnsson and Varshavsky, 1994) However, it has some key limitations. For example, the traditional Y2H system is unable to analyze interactions of proteins that cannot access the nucleus such as transmembrane proteins. To address the limitations of existing analytical methods used for the evaluation of transmembrane protein interactions, a Y2H system based on ubiquitin reconstitution has been proposed (Schaafhausen et al., 2011). The advantage of a split-ubiquitin Y2H system is that both bait and prey plasmids adopt strong promoter and high copy number plasmids, which increase protein expression and facilitate the screening of proteins with weak interactions (Wu et al., 2018).
To study the functions of VP1, we have screened out host cellular proteins that interact with VP1 using the membrane protein Y2H system. The results indicated that host heat shock protein 70 cognate 5 (Hsp70-c5) interacts with VP1. GST pull-down and Co-IP assays were used to verify interactions between Hsp70-c5 and VP1, and they were colocalized mainly in the nucleus and cytoplasm. We report here that Hsp70-c5 expression in CSBV-infected larvae was significantly upregulated compared with that in healthy larvae. Han et al. (2013) also found Hsp70 was significantly up-regulated in honeybee larvae challenged by the CSBV. When we silenced Hsp70-c5, VP1 expression was significantly downregulated, demonstrating that Hsp70-c5 is involved in at least one stage(s) of the viral life cycle. The results provide insights that could facilitate further investigations on CSBV pathogenesis.
Materials and Methods
Plasmids, Virus, cDNA Library, and Main Reagents
Plasmids pBT3STE, pET32a, pTT5, pTT5-VP1-His, pEGFP-C2, and pGEX-6P-1-VP1 were provided by the Laboratory of Life Sciences Research Institute of Jinzhou Medical University. The CSBV VP1 gene, 2–3-day-old A. cerana larvae, NMY32 yeast, and A. cerana larvae yeast cDNA libraries were constructed in our laboratory. We purchased Escherichia coli DH5α and BL21 from TransGen Biotech (Beijing, China) and GST-tagged Protein Purification Kit from Sangon Biotech (Shanghai, China).
Construction and Evaluation of the CDNA Library
Total RNA from bee larvae was isolated using Trizol reagent (Invitrogen, CA, United States) according to the manufacturer’s instructions. The integrity of the total RNA was analyzed via 1% agarose gel electrophoresis. The concentration and purity of the total RNA were determined by measuring the absorbance at 260 and 280 nm using a spectrophotometer (Eppendorf AG, Hamburg, Germany). mRNA was then isolated from the purified samples using the Oligotex mRNA Kit (Qiagen), and used for constructing the Y2H cDNA library. Briefly, the purified mRNA sample was used for double-stranded cDNA (dscDNA) synthesis. The purified dscDNAs were ligated into a pPR3-N vector (Clontech Mountain View, CA, United States) and then electrotransformed into E. coli. The quality of the library was calculated as follows: CFU (colony-forming units)/mL = number of colonies on the plate/10 μL × dilution factor × 1 × 10–3 μL. Therefore, total CFU of the library = CFU/mL × total volume of the library liquid (mL). Single bacterial colonies were picked for PCR, and the PCR products were analyzed using 1% agarose gel electrophoresis.
Construction of pBT3STE-VP1 Bait Plasmid and Plasmid
Total RNA was extracted from purified CSBV and 2–3-day-old A. cerana larvae using Trizol reagent according to the manufacturer’s instructions (Invitrogen, CA, United States). RNA was reverse-transcribed into cDNA using a first-strand cDNA synthesis kit (TransGen, Beijing, China). Three pairs of primers were designed for pBT3STE-VP1, pET32a-Hsp70-c5, pEGFP-C2-Hsp70-c5-Flag, and pTT5-Hsp70-c5-Flag. Restriction enzyme sites were inserted based on the CSBV VP1 sequence (GenBank access No. HM237361.1) and the Hsp70 gene (GenBank access No. MH122655.1) (Table 1). PCR cycling conditions were as follows: initial denaturation at 94°C for 2 min followed by 30 cycles at 94°C for 45 s, 58°C for 45 s (pBT3STE-VP1) or 55°C for 45 s (pET32a-Hsp70-c5, pEGFP-C2-Hsp70-c5-Flag, and pTT5-Hsp70-c5-Flag), and 72°C for 45 s, and a final extension step at 72°C for 10 min. The PCR product was purified and double digested, following which, the plasmid was extracted and inserted into the vector using the designed restriction site. Finally, the plasmid was transformed into E. coli DH5α and identified by endonuclease cleavage and sequencing (Synbio Technologies, Suzhou, China).

Table 1. Synthetic oligonucleotides for plasmid construction of pET32a-Hsp70-c5, pBT3STE-VP1, pEGFP-C2-Hsp70-c5, pTT5-Hsp70-c5 -Flag and siRNA.
Function and Self-Activation Detection of Bait Plasmid
To test bait plasmid with respect to its self-activation and function, it was co-transformed with the control plasmids into the reporter NMY32 yeast strain. Transformants were grown on SD/-Trp/-Leu, SD/-Trp/-Leu/-His, and SD/-Trp/-Leu/-His/-Ala agar plates for 3–5 days (Table 2). The self-activation and function of the bait protein expression plasmid pBT3STE-VP1 was detected based on the growth of the colonies on the plates.
Apis cerana Larvae cDNA Library Screening Using pBT3STE-VP1 as the Bait
After ensuring that our bait was functional in the membrane protein Y2H system by function and self-activation detection, we initiated the screening of host cellular proteins that interacted with VP1. We transformed the cDNA library into the NMY32 cells, which contained the appropriate pBT3STE-VP1 decoy plasmid, at 30°C for 3 days. We selected all the potentially positive colonies on SD/-Trp/-Leu and 60 mM 3-amino-1, 2, 4-triazole (3-AT) SD/-Trp/-Leu/-Ade/-His selective media for 1–2 days at 30°C. Yeast colonies cultured on the selective medium could potentially interact with bait and prey proteins. The proteins were recovered from yeast and retransformed into E. coli DH5α for subsequent analysis. The proteins were sequenced by Synbio Technologies, Co., Ltd. (Jiangsu, China). The sequences were analyzed using DNSTART biological software (V7.10) and aligned using BLAST in the NCBI/GenBank databases to obtain information on host protein interactions.
His and Hsp70-c5 Fusion Protein Expression and Purification From E. coli
The plasmid pET32a-Hsp70-c5 was transformed into E. coli BL21 (DE3). The single selected bacterial colonies were inoculated into 10 mL of Luria-Bertani (LB) medium and incubated at 37°C for 12 h. The cultures (1000 μL) were inoculated into 100 mL LB (100 μg/mL Amp) and cultured at 37°C until the absorbance at 600 nm reached 0.6–0.8. Protein expression was induced by the addition of 0.5 mM isopropyl β-D-thiogalactoside at 28°C for 8 h. The cell precipitates were collected to detect Hsp70-c5 expression using 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and His-fusion proteins were purified using a His-tagged Proteins Purification Kit (Bio-Works, Sweden).
GST Pull-Down Assay
The glutathione S-transferase (GST)–VP1 fusion protein was incubated with prepared glutathione agarose beads (Bio-Works, Sweden) at 4°C for 3 h. The beads were incubated overnight at 4°C with the 0.1 mg/mL of input protein His-Hsp70-c5. The agarose complex was eluted, separated, collected, and centrifuged. Finally, the collected protein was analyzed using 8% SDS-PAGE electrophoresis and western blot. As negative control, GST was incubated alone on beads with the E. coli lysates.
Co-immunoprecipitation Assay
The plasmids pTT5-VP1-His and pTT5-Hsp70-c5-Flag were co-transfected into HEK293T cells using Lipofectamine 2000 (GeneCopoeiaTM, United States). After 24 h of transfection, the cells were collected, treated with radio-immunoprecipitation assay buffer (NaCl, 0.0584 g; Tris–HCL, 100 μL; NP40 25 μL, 0.5%; add water to make up volume to 5 mL) on ice for 30 min. Cellular debris was pelleted by centrifugation (Thermo Scientific Fisher, Waltham, MA, United States) at 12000 rpm at 4°C for 10 min. In all, 500 μL of the cell lysate above was transferred to a 1.5-mL microcentrifuge tube and the cell lysate was incubated with 4 μg of 6∗His, His-tagged antibody (Proteintech, Wuhan, China) at 4°C for 1 h. Further, 20 μL of resuspended protein A/G PLUS-Agarose (Santa Cruz Biotechnology, CA, United States) volume was also added. Subsequently, the tubes were capped and incubated at 4°C for 1 h to overnight. Immunoprecipitates were collected by centrifugation at 1500 rpm at 4°C for 3 min. The supernatant was carefully aspirated and discarded. Co-IP products were eluted with 5×SDS loading buffer (Beyotime, Shanghai, China), boiled for 5 min, and identified by western blotting.
Immunofluorescence Staining and Confocal Microscopy
The plasmids pTT5-VP1-His and pEGFP-C2-Hsp70-c5-Flag were co-transfected into HEK293T cells using Lipofectamine 2000 (GeneCopoeiaTM, United States). After 24 h of transfection, the cells grown on glass coverslips (Biosorfa, Zhejiang, China) were fixed in 4% paraformaldehyde (Solarbio, Beijing, China) in phosphate-buffered saline (PBS) for 20 min at room temperature. The cells were then washed three times in PBS. The cells were permeabilized with 0.5% Triton X-100 (Sangon Biotech, Shanghai, China) in PBS for 30 min at room temperature and blocked in 2% bovine serum albumin (Solarbio, Beijing, China) in TBST (Tris–HCl, 10 mM; pH, 8.0; NaCl, 150 mM; Tween 20, 0.05%) for 1 h at room temperature. The coverslips were then incubated with 6∗His, His-tagged antibody (1:400 dilution) (Proteintech, Wuhan, China) overnight at 4°C. Subsequently, the cells were washed three times in PBS for 5 min. The coverslips were then incubated with secondary antibody and protected from light for 1 h. The secondary antibody was goat anti-mouse antibody (1:2000 dilution) (Abcam, Cambridge, United States). We used 49,69-diamidino-2-phenylindole dihydrochloride (DAPI) (Coolaber, Beijing, China) (1:750 dilution) to stain the cell nuclei and protected from light for 20 min. Finally, the cells were examined using a laser scanning confocal microscope (LEICA, Germany).
Hsp70-c5 Level After Larvae Infected With CSBV
Three-days-old A. cerana larvae were used to this experiment. A total of 20 three-days-old larvae were retrieved from the same colony, following the method of Hu et al. (2016), and randomly distributed into 2 groups, with each group containing 10 larvae. Group I were fed with CSBV at 1.25 × 107 copies/larva (Hu et al., 2016), group II were fed with basic larval diet (BLD) and placed in constant temperature and humidity conditions (T = 34°C, RH = 95%). Each larva was fed 20 μL of a virus suspension mixed with an equal amount of BLD. BLD was used subsequently for daily feeding. The clinical signs in each group of larvae were examined and recorded every day until death of larvae. The dead larvae were detected by RT-PCR for the following viruses during the experiments: BQCV, ABPV, CBPV, DWV, KBV, IAPV, and CSBV. Hsp70-c5 level after larvae infected with CSBV in this study were analyzed by RT-PCR, the copy number of Hsp70-c5 is calculated according to the method of Zhou et al. (2017). Drawing a histogram using GraphPad Prism 7.0. Primers for real-time PCR are listed in Table 1. The above assay was performed in triplicate.
We also conducted western blot analysis. Equal amounts of protein samples (12 μg/lane) were separated by 12% SDS-PAGE and then transferred to polyvinylidene fluoride membranes. After blocking, the membranes were incubated overnight at room temperature with anti-Hsp70 (1:1000 dilution). Following three washes, the membranes were further incubated with goat anti-mouse antibody for 1 h. Protein bands were detected using the ECL Western Blotting Substrate (United States). The above assay was performed in triplicate.
Effect of Silencing Hsp70-c5 on VP1 Expression
We performed silencing experiments to investigate the function of Hsp70-c5 during viral infection. Three pairs of siRNAs of Hsp70-c5 were designed and synthesized by Guangzhou RiboBio, Co., Ltd. (Table 1). A total of 40 3-days-old larvae were retrieved from the same colony, following the method of Hu et al. (2016), and randomly distributed into 4 groups (groupI-IV), with each group containing 10 larvae. Feeding in constant temperature and humidity conditions (T = 34°C, RH = 95%). After 24 h, group I was fed with BLD containing 1 μg siRNA1, group II was fed with BLD containing 1 μg siRNA2, group III was fed with BLD containing 1 μg siRNA3, group IV was fed BLD alone. 24 h after feeding the siRNA, the diet was replaced with BLD without siRNA and was supplemented daily until the larvae entered the defecation period. After 72 h, three larvae were randomly selected from each group for real-time PCR analysis.
After screening the best siRNA for inhibiting Hsp70-c5. A total of 30 three-days-old larvae were retrieved from the same colony, following the method of Hu et al. (2016), and randomly distributed into 3 groups (groupV–VII), with each group containing 10 larvae. Group V larvae were fed with BLD containing 1 μg siRNA, while group VI and group VII were fed with BLD alone. After 24 h, group V was fed BLD containing 1 μg siRNA3 and 1 × 107 copies/μL CSBV (Hu et al., 2016), while group VI was fed BLD with only the same copy number of CSBV. Group VII was fed BLD alone. After 24 h, the diet was switched to BLD without siRNA3 or virus, and was supplemented daily until the larvae entered the defecation period. After 72 h, three larvae were randomly selected from each group for real-time PCR analysis. VP1 expression in larvae was detected using the TransStart Tip Green qPCR SuperMix (TransGen, Beijing, China). The copy number was calculated according to the method of Zhou et al. (2017).
Results
Construction of the Y2H cDNA Library
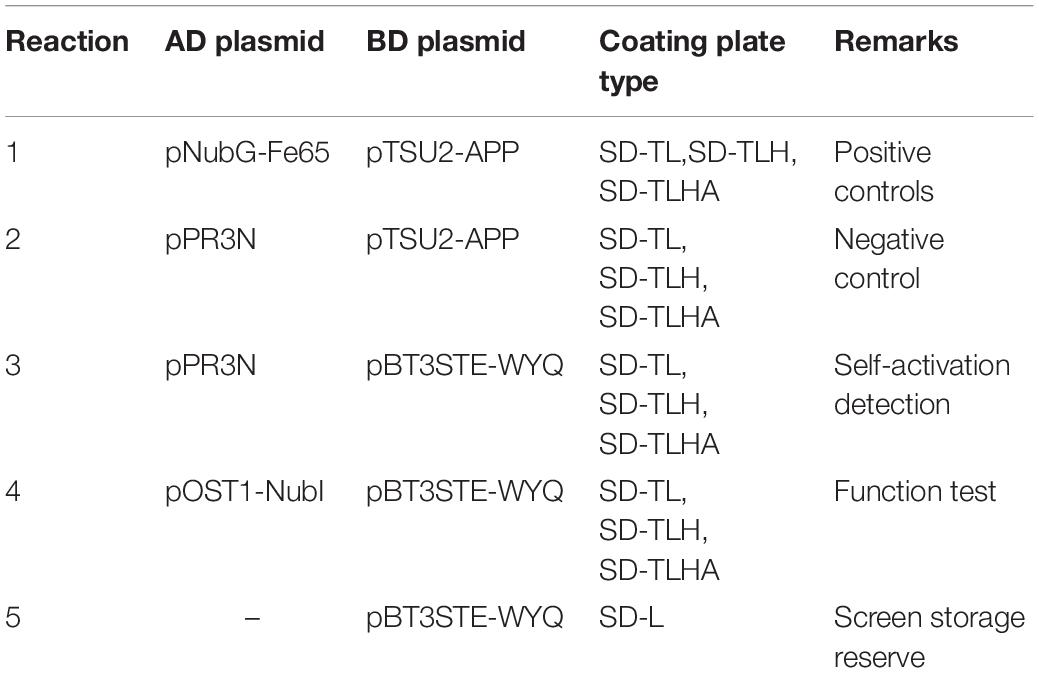
To identify cellular factors that potentially interact with CSBV VP1, we needed to construct and evaluate a cDNA library. The integrity of the total RNA extracted using Trizol reagent was assessed via 1% agarose gel electrophoresis, which revealed three bands corresponding to ribosomal 28S, 18S, and 5S RNA; there was no visible degradation (Figure 1A). The OD260/OD280 ratio of total RNA was 1.95, with a concentration of 139.4 ng/μL, indicating that the total RNA had sufficient purity and quality to construct the cDNA library. After mRNA isolation, the total amount of mRNA was determined to be 5.6 μg. The results of electrophoretic detection are shown in Figure 1B; the mRNA bands were clear, distributed in a diffuse manner, and the bands were evenly distributed, fully meeting the needs for library construction. The library titer was 3 × 106 CFU/mL. The total storage capacity of the original bacterial solution after conversion of 5 mL was 1.5 × 107 CFU. The results of PCR revealed that most inserts were greater than 1200 bp, and the average length was about 1500 bp. None of the electrophoresis lanes running the 24 monoclonal PCR products were blank; therefore the recombination rate of the library was 100%, and the library index was qualified (Figure 2).
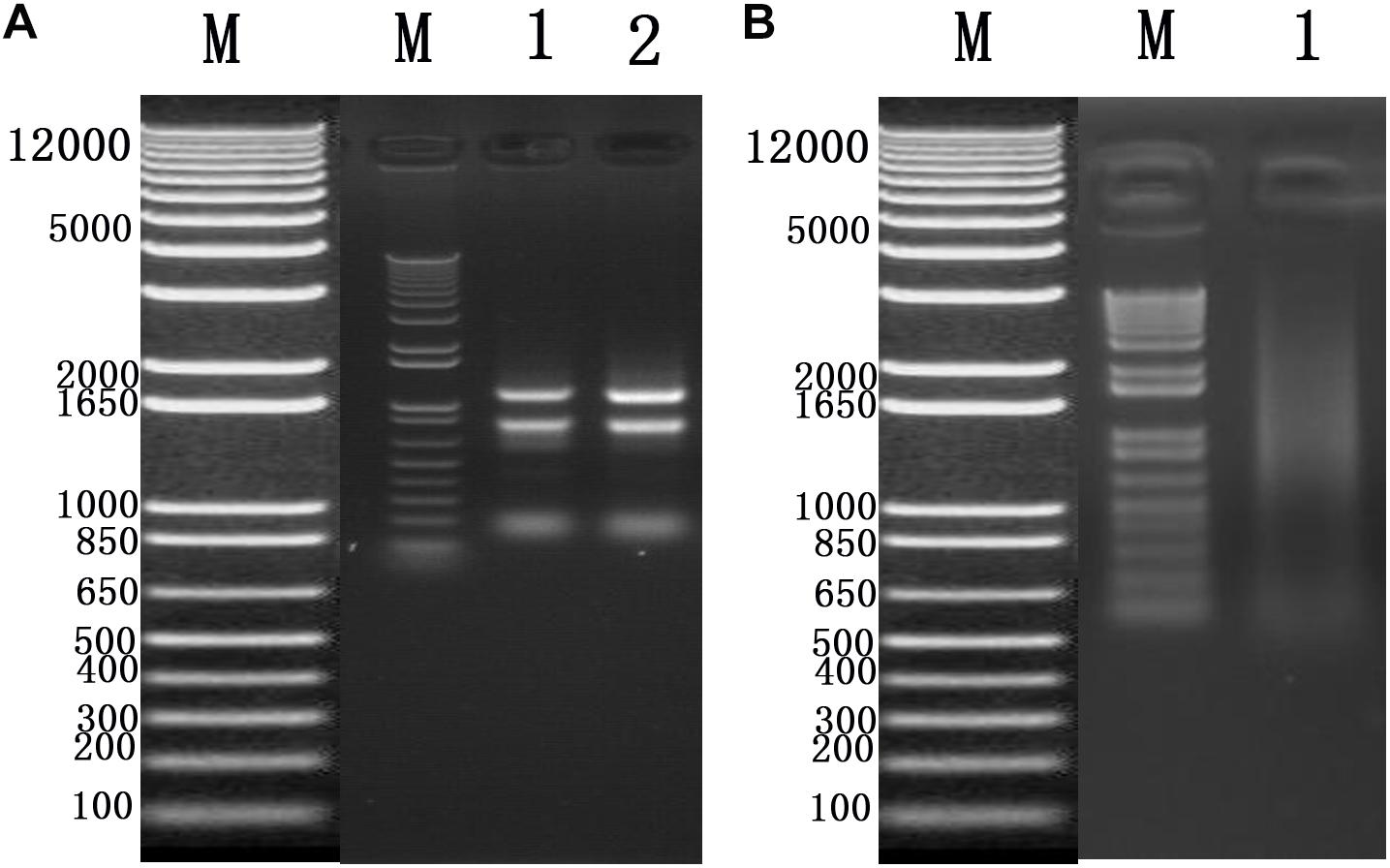
Figure 1. (A) Total RNA of Chinese bee larvae: M:DNA Marker DL12000, lane1-2:total RNA of Chinese bee larvae. (B) Chinese bee larval mRNA isolation results: M:DNA Marker DL12000, lane1:Chinese bee larval mRNA.
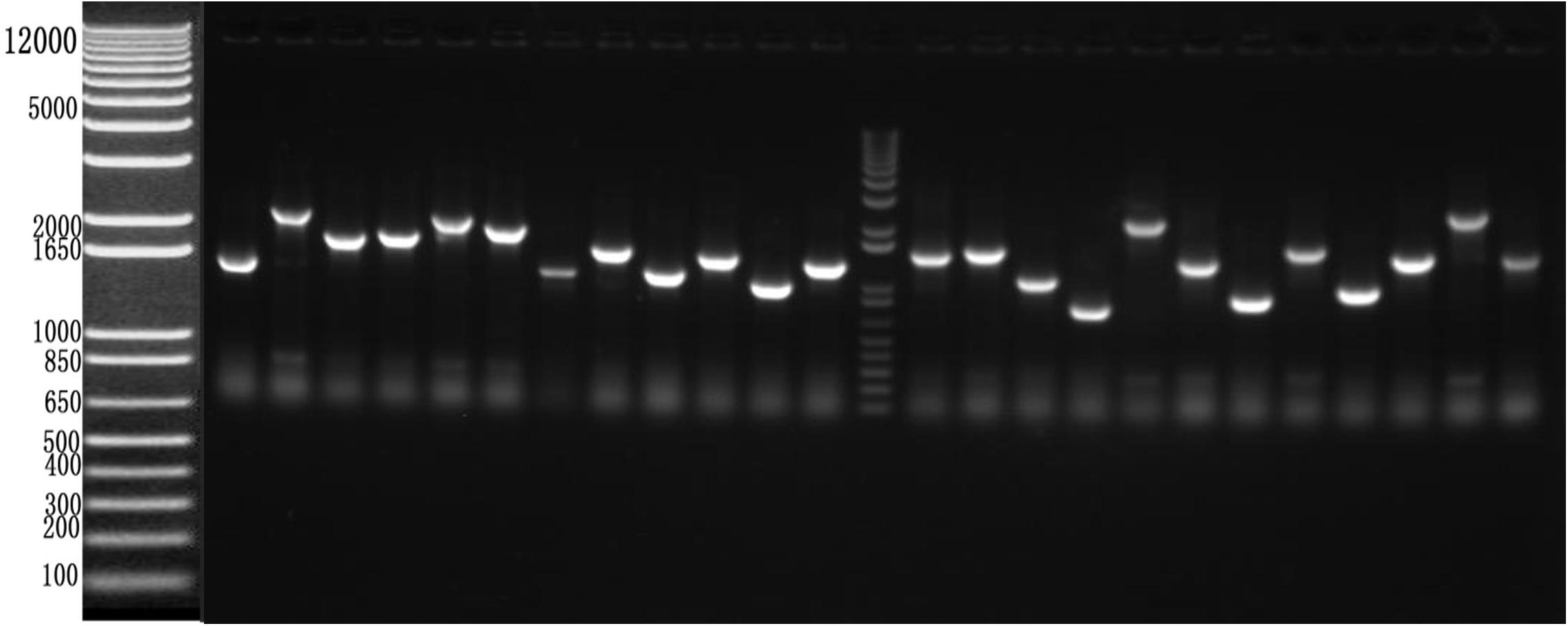
Figure 2. Recombination rate and insert fragment identification of membrane protein yeast cDNA library of Apis cerana cerana.
Construction of pBT3STE-VP1 Bait Plasmid
After RT-PCR using a specific primer, a fragment of approximately 945 bp was generated (Figure 3A). Sequencing and alignment results revealed that the nucleotide sequence identity exceeded 99% similarity with the reference strain. Then, the VP1 gene was cloned into the plasmid pBT3STE using the Sif I restriction site. Restriction digestion and sequencing analysis indicated that pBT3STE-VP1 bait plasmid was successfully constructed.
Figure 3. Construction of pBT3STE-VP1, pET32a-Hsp70-C5 and pTT5-Hsp70-c5 -Flag plasmid. (A) Digestion by Sif I, lane 1: DNA Marker DL2000, lane 2: pBT3STE-VP1 plasmid digested by restriction endonuclease. (B) Digestion by NcoI and HindIII, lane 1: DNA marker DL2000, lane 2:pET32a-Hsp70-c5 plasmid digested by restriction endonuclease. (C) Digestion by EcorI and HindIII, lane 1: DNA marker DL2000, lane 2:pTT5-Hsp70-c5 plasmid digested by restriction endonuclease. (D) Digestion by EcorI and KpnI, lane 1: DNA marker DL2000, lane 2:pEGFP-C2-Hsp70-c5 plasmid digested by restriction endonuclease.
Function and Self-Activation Detection of pBT3STE-VP1 Plasmid
To analyze the self-activation level of the pBT3STE-VP1 bait plasmid in yeast cells, pBT3STE-VP1 and various plasmids were co-transfected into NMY32 cells and assayed on selective plates. Analysis of the function and self-activation of VP1 revealed that white clones grew on all the SD/-Trp/-Leu plates. White clones were present on the SD/-Trp/-Leu/-His and SD/-Trp/-Leu/-Ade/-His solid plates in the positive control and functional test groups but were absent in the negative control and self-activation detection groups (Figure 4). Therefore, the pBT3STE-VP1 bait plasmid had functions and exhibited no self-activation in the assay.
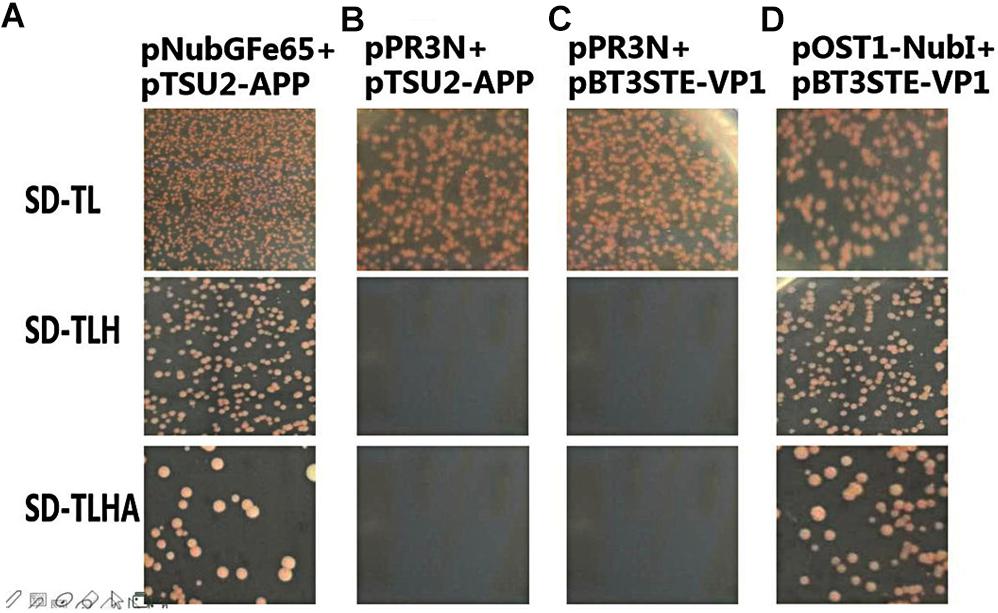
Figure 4. Function and self-activation detection of pBT3STE-VP1 plasmid. Panel (A) is the positive control group: pNubGFe65 and pTSU2-APP. Panel (B) is the Negative control group: pPR3N and pTSU2-APP. Panel (C) is the self-activation assay of the bait plasmid pBT3STE-VP1: pPR3N and pBT3STE-VP1. Panel (D) is the functional test group: pOST1-NubI and pBT3STE-VP1.
Apis cerana Larvae cDNA Library Screening Using pBT3STE-VP1 as the Bait
To screen host proteins that interplay with the pBT3STE-VP1 bait using the Y2H system, the bait plasmid pBT3STE-VP1 was hybridized using the cDNA library of A. cerana. The clones were screened, yielding 40 positive bait–prey interacting yeast from the membrane protein Y2H system, and passed the detection of reporter genes (Supplementary Figure S1). In addition, sequence analysis of the positive plasmids indicated that these 40 plasmids represented 23 cDNA of potential protein genes interacting with VP1 (Supplementary Table S1). The gene screened was Hsp70 gene (GenBank access No. MH122655.1, named Hsp70-c5) which identity with the A. cerana heat shock 70 kDa protein cognate 5 (GenBank access No. XM017048256.1) is 83.35%.
His and Hsp70-c5 Fusion Protein Expression and Purification From E. coli
Apis cerana Hsp70-c5 complete cDNA had an overall length of 1833 bp. Recombinant plasmids pET32a-Hsp70-c5 were constructed using NcoI and HindIII (Figure 3B). The expression plasmids were transformed into BL21 (DE3). After induction, His-Hsp70-c5 recombinant protein could be purified effectively using the His-tagged Protein Purification Kit. Protein expression levels were analyzed using SDS-PAGE. The results indicated that the recombinant proteins had molecular weights of 87 kDa. However, the HIS protein in the negative control had a molecular weight of approximately 20 kDa (Figure 5).
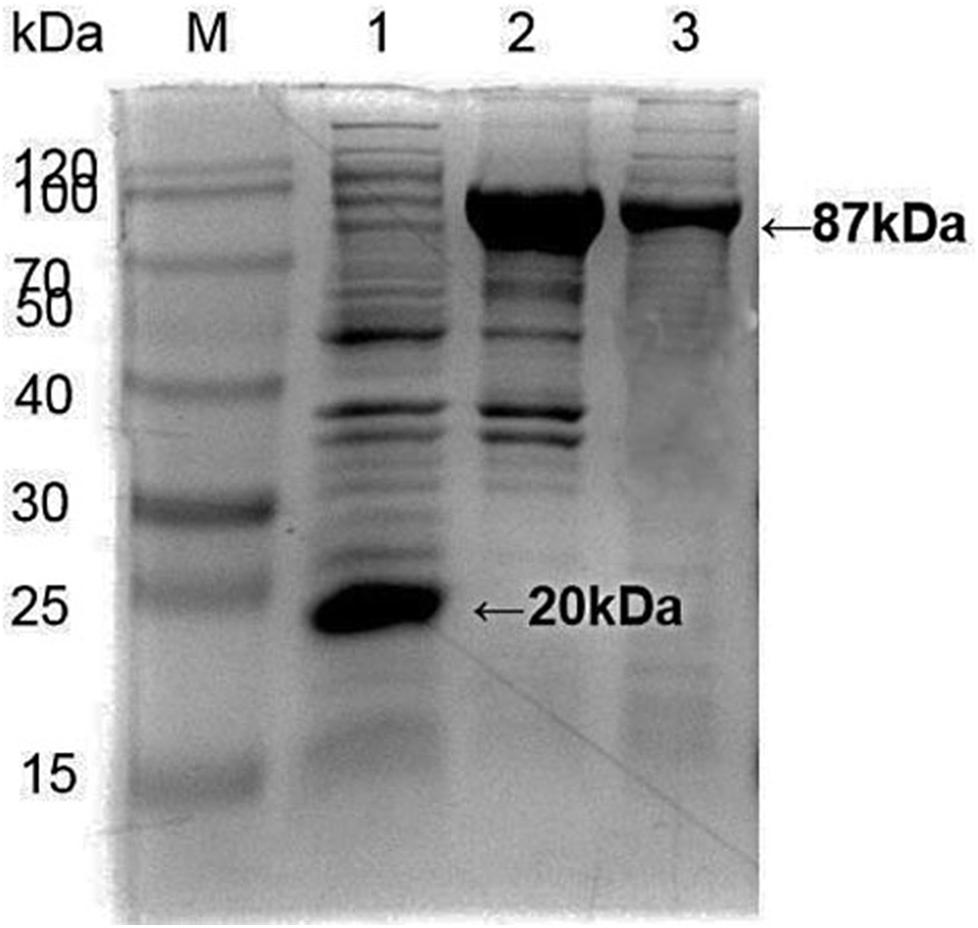
Figure 5. Expression and purification of E. coli HIS and Hsp70-c5 Fusion protein. Purification of Recombinant protein pET-32a-Hsp70-c5, M: the low molecular weight standard protein Marker, lane 1: Expression of pET-32a, lane 2: Expression of HIS-Hsp70-c5, lane 3: pET-32a-Hsp70-c5 purification.
GST Pull-Down Assay
To detect the binding ability between VP1 and host Hsp70-c5 in vitro, GST pull-down assay was performed with GST, GST-VP1, and Hsp70-c5. The experimental results were analyzed using Coomassie blue staining and western blotting (Figure 6). Compared with the negative control group, the experimental and positive control groups exhibited a protein band with a molecular weight of approximately 87 kDa, consistent with the expected Hsp70-c5 protein sizes (Upper), indicating that Hsp70-c5 and GST-VP1 may have formed a complex. Western blotting was performed with anti-His antibody. His-Hsp70-c5 protein appeared at a molecular mass of approximately 87 kDa (Lower). The results indicate that there is a strong interaction between VP1 and Hsp70-c5.
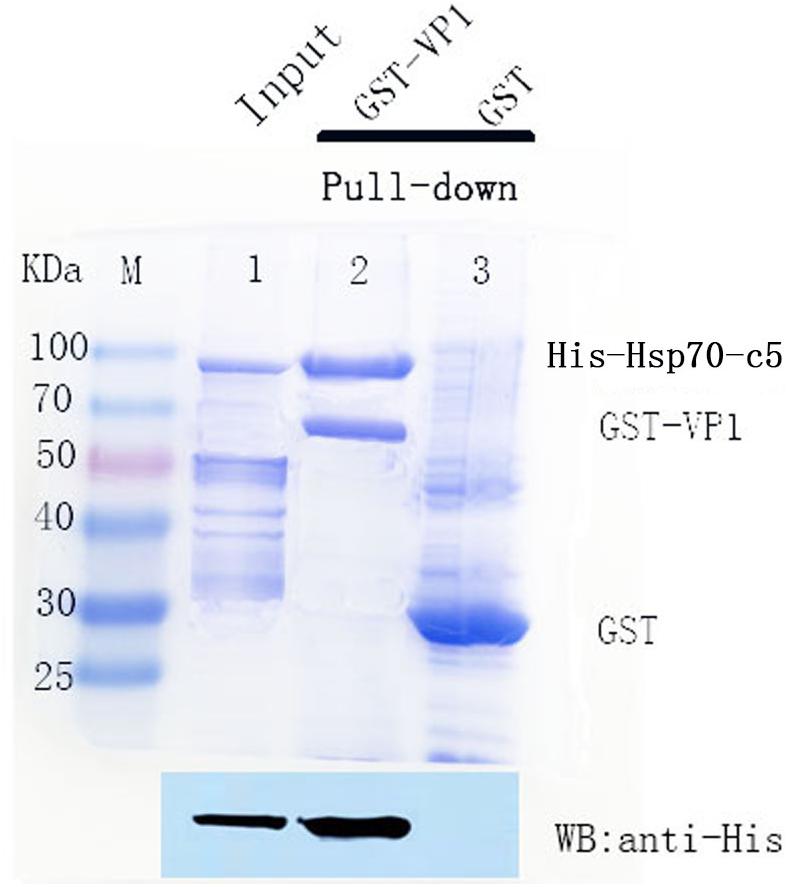
Figure 6. SDS-PAGE (8%, Coomassie blue stained) and western blotting analysis of glutathione S-transferase (GST) pull-down samples. Compared with the negative control group, the experimental and positive control groups exhibited a protein band with a molecular weight of approximately 87 kDa, consistent with the expected Hsp70-c5 protein sizes (Upper), indicating that Hsp70-c5 and GST-VP1 may have formed a complex. Western blotting was performed with anti-His antibody. His-Hsp70-c5 protein appeared at a molecular mass of approximately 87 kDa (Lower). GST-VP1 pull down Hsp70-c5-His. SDS-PAGE analysis (Coomassie blue-stained) of GST-VP1 pull down samples (Upper), and western blotting was performed using anti-His antibody (Lower).
Co-IP Assay
To further determine whether the interaction between VP1 and Hsp70-c5 occurs in vitro, Co-IP was performed in HEK293T cells co-expressing Hsp70-c5-FLAG and VP1-His. Recombinant plasmid pTT5-Hsp70-c5-Flag was constructed using EcoRI and HindIII (Figure 3C). Co-IP assay was performed in HEK293T cells co-expressing Hsp70-c5-Flag and VP1-His. The results show that Hsp70-c5-Flag could be co-immunoprecipitated with anti-His antibodies or anti-Flag when co-expressed with VP1-His, demonstrating an interaction between Hsp70-c5-Flag and VP1-His in the cells (Figure 7).
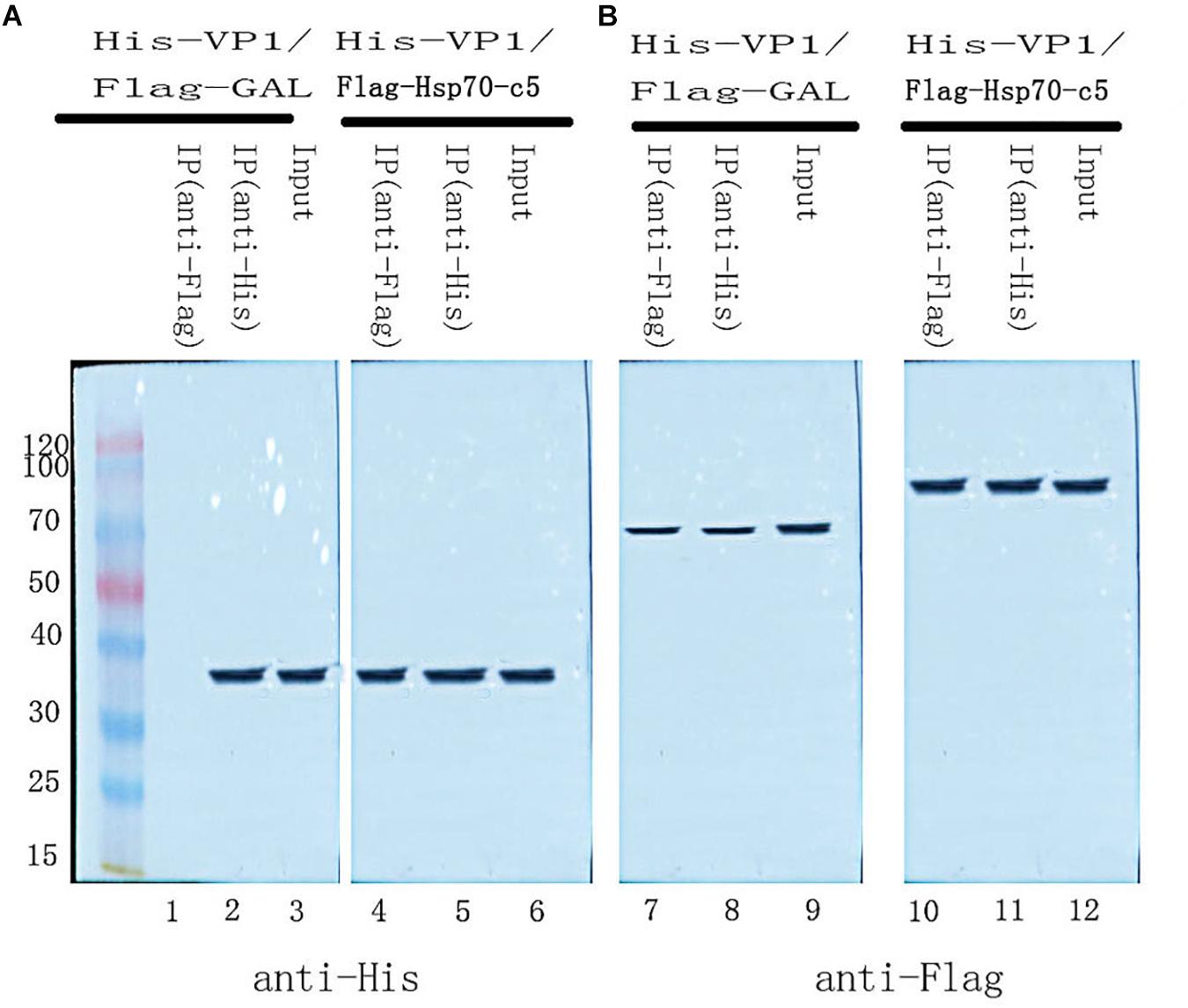
Figure 7. Used Co-IP assay analysis interaction between VP1 and Hsp70-c5. Immunoprecipitation of VP1 and Hsp70-c5. HEK293T cells were co-transfected with the expression plasmids for His-tagged VP1 and FLAG-tagged Hsp70-c5. The resulting Input and IP samples (anti-His or anti-Flag) were subjected to SDS-page electrophoresis, and protein expression was detected using His and Flag antibodies. (A) The resulting Input and IP samples (anti-His or anti-Flag) were subjected to SDS-page electrophoresis, and protein expression was detected using His antibodies. (B) The resulting Input and IP samples (anti-His or anti-Flag) were subjected to SDS-page electrophoresis, and protein expression was detected using Flag antibodies.
Immunofluorescence Staining and Confocal Microscopy
Following the determination of interactions between Hsp70-c5 and VP1, we investigated whether these two proteins colocalized in the cells. To determine whether VP1 and Hsp70-c5 proteins localized within the same cellular compartment, recombinant pEGFP-C2-Hsp70-c5-Flag plasmid was constructed using EcoRI and KpnI restriction sites (Figure 3D). The recombinant plasmids pTT5-VP1-His and pEGFP-C2-Hsp70-c5-Flag were co-transfected into HEK293T cells. As shown by the green fluorescent dots in Figure 8, Flag-Hsp70-c5 localization in the cytoplasm and nucleus, His-VP1 also localization in the cytoplasm and nucleus. VP1 and Hsp70-c5 colocalized in an ER-like pattern, which was particularly evident in the cytoplasm and nucleus. Colocalization results showed that co-expressed VP1 and Hsp70-c5 could completely overlap in cells. Our data illustrate that VP1 and Hsp70-c5 were colocalized primarily in cytoplasm (Figure 8).
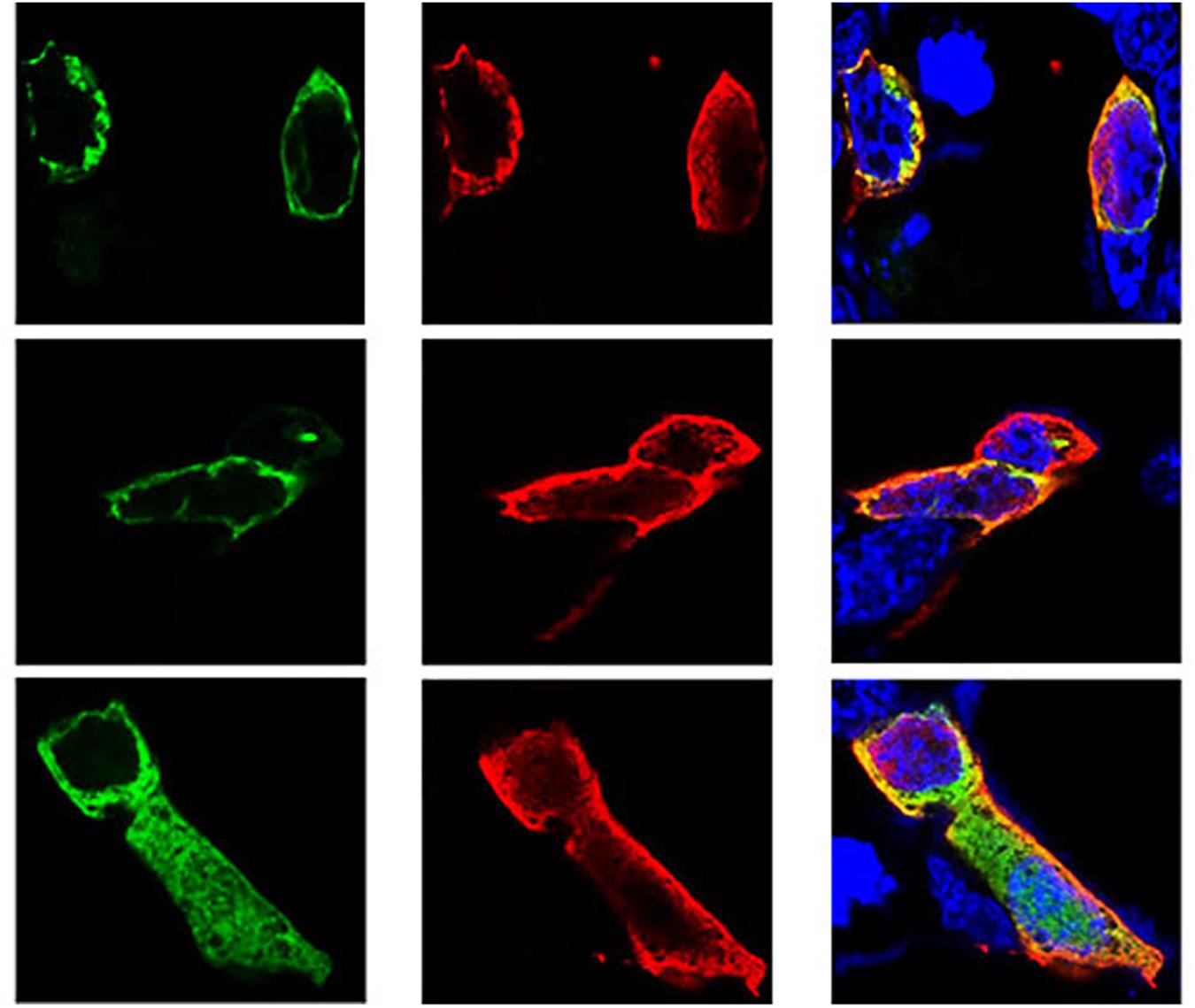
Figure 8. Colocalization of VP1 and Hsp70-c5 proteins using immunofluorescence confocal. Green represents Flag-Hsp70-c5, red represents His-VP1, blue DAPI represents nucleus, co-localization shows that red is almost completely coincident with green. Colocalization of VP1 and Hsp70-c5 is indicated in yellow in this merged.
Hsp70-c5 Level After Larvae Infected With CSBV
These experiments were performed to further investigate the changes in Hsp70-c5 expression in larvae after CSBV infection. The expression levels of Hsp70-c5 in each group of larvae were detected via SYBR Green RT-PCR. Hsp70-c5 expression in larvae was detected using TransStart Tip Green qPCR SuperMix (TransGen, Beijing, China), and the copy number was calculated according to the method of Zhou et al. (2017). The results indicated that Hsp70-c5 was expressed in honeybee larvae. Hsp70-c5 expression levels were low in healthy larvae, whereas it was high in the CSBV-infected larvae (Figure 9). All dead larvae found during the experiments were analyzed by RT-PCR, and the results showed that all of the other honeybee viruses were undetectable, whereas CSBV was detectable.
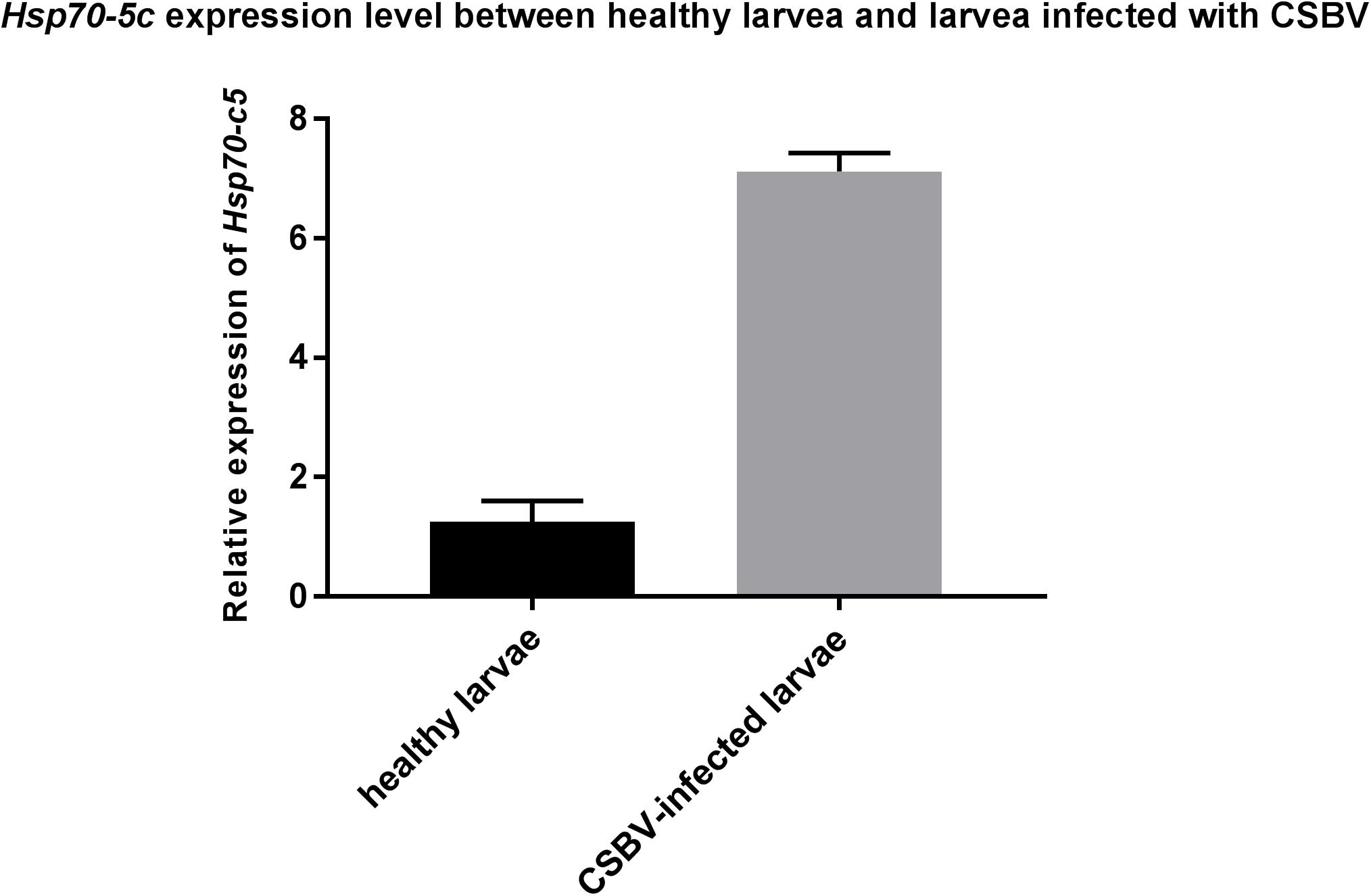
Figure 9. Hsp70-c5 level after larvae infected with CSBV. Hsp70-c5 level after larvae infected with CSBV. The bars represent relative fold change of Hsp70-c5 expression. The black bars indicates expression of Hsp70-c5 in the larvae, and gray bars denote higher expression of Hsp70-c5 in the infected larvae. Error bar is standard deviation.
Western blot results indicated that Hsp70-c5 was expressed in honeybee larvae and its molecular weight was approximately 70 kDa. Hsp70-c5 gene expression levels were low in healthy larvae, whereas they were high in the CSBV-infected larvae (Figure 10). The results indicated that Hsp70-c5 expression considerably increased in CSBV-infected larvae. Therefore, we conclude that Hsp70-c5 upregulation can affect viral assembly processes or enhance host antiviral effects.
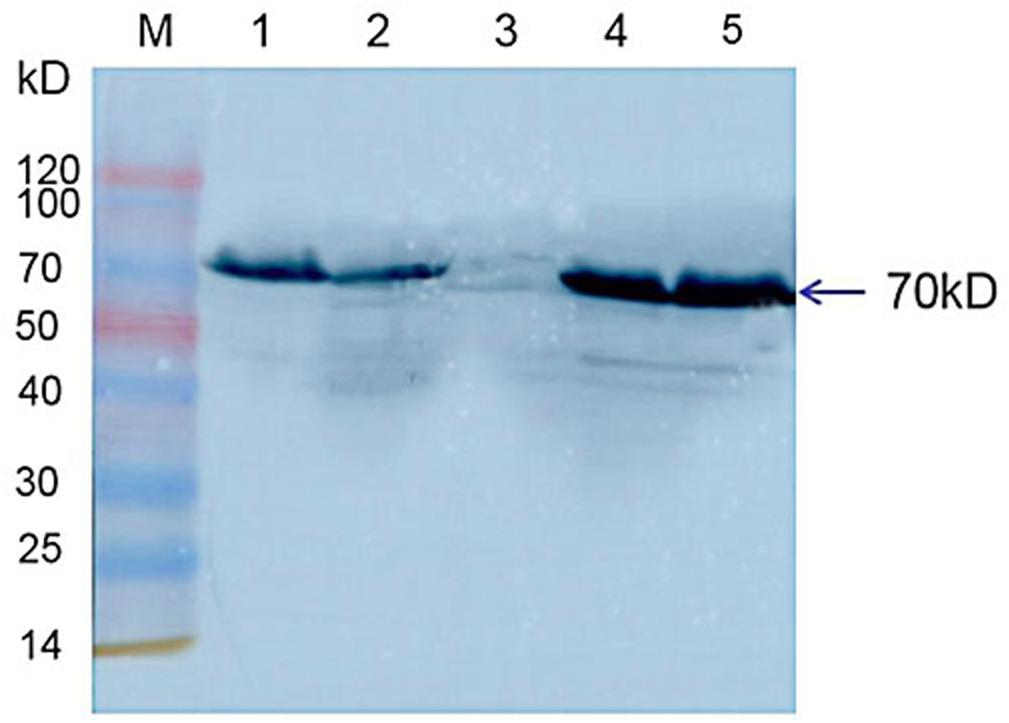
Figure 10. Detection of Hsp70-c5 expression in three groups of healthy and diseased larvae. M: Low molecular weight standard protein Marker; 1, 2: The expression of Hsp70-c5 in healthy larvae; 4, 5: Expression of Hsp70-c5 in the infected larva of CSBV.
Effect of Silencing Hsp70-c5 on VP1 Expression
We have founded that Hsp70-c5 expression was upregulated in vivo when the larvae were infected with CSBV. We performed silencing experiments to investigate the function of Hsp70-c5 during CSBV infection more specifically. Compared with siRNA1 and siRNA2, siRNA3 had the highest inhibitory effect on Hsp70-c5. Therefore, siRNA3 was chosen for the subsequent experiment. Comparing the expression levels of Hsp70-c5 in groups V and VII, we found that Hsp70-c5 expression was downregulated in groups V. Comparing the expression levels of VP1 in groups V and VI, we found that when we silenced Hsp70-c5, VP1 expression was significantly downregulated, indicating that Hsp70-c5 can promote the replication of CSBV in larvae.
Discussion
The honeybee is an important insect species for crop pollination and plays a crucial role in agricultural production (Potts et al., 2011). CSBV is the pathogen that majorly infects A. cerana, resulting in severe and lethal infections in colonies and eventual losses of entire colonies (Feng et al., 2013). VP1 reportedly has the highest variation in amino acid sequences among different CSBV strains. In addition, it harbors the major CSBV epitope (Mingxiao et al., 2011). Cheng et al. (2011) demonstrated that VP1 is an excellent immunogen in CSBV and structural proteins are a major component of the virus, although the role of VP1 in viral infection and its interaction with host proteins remains unclear. Studies on the interaction between VP1 and host proteins are important and could improve our understanding of the molecular mechanisms underlying viral proliferation and pathogenesis and enable the identification of checkpoints for the development of appropriate therapeutics.
Chinese sacbrood virus is a typical positive single-stranded RNA virus. RNA viruses often rely on host factors to proliferate in cells (Snijder, 2008; Tang et al., 2012; Reid et al., 2015); therefore, we screened out the host proteins interacting with CSBV VP1 using the membrane protein Y2H system in the present study. According to the results, 23 host proteins that interacted with VP1 were identified. The interacting proteins were associated with critical biological functions, such as stress response, protein phosphorylation, signal transduction, protein targeting, cytoskeleton organization, DNA repair, protein regulation, ribosome metabolism. Among these interacting proteins, Hsp70 is majorly significant in research owing to its roles in viral proliferation and immunity (Kantengwa et al., 1995; Carsillo et al., 2009).
HSPs are widely expressed in prokaryotes and eukaryotes as stress-inducible molecular chaperons (Shukla and Pitha, 2012). Based on the molecular weights of HSPs, they can be classified as Hsp100, Hsp90, Hsp70, Hsp60, Hsp40, small Hsp family, and ubiquitin, with Hsp70 being the most widely studied (Venetianer et al., 1994). Zhang (2013) reported that there are at least five Hsp70s in honeybee, such as Hsp70Ab, Hsc70-4, Hsc70-3, Hsc70-5, Hsp70-4L. Hsp70 is involved in immune responses and participates in the life of a virus through folding, transporting, positioning, assembling, or degrading activities (Risler et al., 2012). Increasing evidence suggests that Hsp70 plays key roles in viral replication and assembly in several RNA viruses (Nagy et al., 2011; Lahaye et al., 2012; Katoh et al., 2017), such as those of dengue (Taguwa et al., 2015), avian infectious bronchitis (Shan et al., 2018), and influenza A (Manzoor et al., 2014). Alzhanova et al. (2014) observed that Hsp70 could bind to the structural proteins of beet yellows virus to form a complex and participate in viral assembly. In addition, Jiang et al. (2015) demonstrated that Hsp70 probably participates in rice stripe viral replication by interacting with viral RNA-dependent RNA polymerase (RdRp). In the present study, utilizing the GST pull-down and Co-IP assays, we have revealed an interaction that exists between structural protein VP1 and Hsp70-c5. In addition, we speculate that the interaction between VP1 and Hsp70-c5 in A. cerana plays a role in viral assembly.
Confocal microscopy was used to investigate the colocalization of Hsp70-c5 and VP1 in HEK293T cells. VP1 and Hsp70-c5 were mainly colocalized in the nucleus and cytoplasm. Ren et al. (2012) demonstrated that Hsp70 migrated into the raft fraction after Japanese encephalitis virus infection, whereas Ye et al. (2001) observed that in Hantaan virus infection, Hsp70 forms a complex with nuclear proteins, which translocates from the cytoplasm to nucleoli, a process associated with viral assembly. Hsp70 influences viral assembly and enhances host antiviral immunity in the course of viral infection (Kim et al., 2013b). Ohgitani et al. (1999) analyzed the movement of Hsp70 from the cytoplasm to the nucleus and then back to the cytoplasm at different stages of infection and observed that in the early stages of infection, due to the presence of stress factors, Hsp70 aggregates in the nucleus and may be involved in the transcription processes of viruses and hosts, viral DNA replication, and assembly of nucleocapsid proteins. However, Hsp70 aggregates in the cytoplasm and is involved in the protection of infected cells from host immunity in the later stages of infection. Therefore, we speculated that Hsp70-c5 migrates during the course of CSBV infection, a process that potentially influences viral assembly and immune processes.
Hsp70 upregulation after viral infection has been widely reported (Padwad et al., 2010). Lahaye et al. (2012) observed that Hsp70 upregulation was correlated with excess amounts of viral proteins, suggesting that Hsp70 plays an active role in the rabies viral life cycle. Vesicular stomatitis viral infection results in Hsp70 upregulation, which ultimately induces innate immune responses that facilitate viral clearance (Kim et al., 2013a). In the present study, Hsp70-c5 expression in CSBV-infected larvae was significantly upregulated when compared with healthy larvae. Thus, we conclude that Hsp70-c5 upregulation can affect viral assembly processes or enhance host antiviral effects.
However, further studies are required to determine whether the interactions between Hsp70-c5 and VP1 play a role in CSBV infection. De-la-Re-Vega et al. (2017) observed that Hsp70 is involved in the process of herpes virus infection, thereby promoting herpes virus replication. Taguwa et al. (2015) found that when dengue virus infects the body, Hsp70 is involved in every step of viral infection, i.e., from replication to assembly to release, to help the virus complete the infection. Han et al. (2013) also found Hsc70 and Hsp90 participate in the facilitation of viral production in host cells during the infection cycle. In the larval experiment, we found that HSP70 expression was upregulated in vivo when the larvae were infected with CSBV. We performed silencing experiments to investigate the function of Hsp70-c5 during CSBV infection more specifically and found that when we silenced Hsp70-c5, VP1 expression was significantly downregulated, indicating that Hsp70-c5 can promote the replication of CSBV in larvae. These results demonstrate that Hsp70-c5 is involved in at least one stage of the viral life cycle.
In summary, we screened out the host protein, Hsp70-c5, as an interacting partner with VP1 of CSBV using the membrane protein Y2H system and observed the interaction between VP1 and Hsp70-c5 using the GST pull-down and Co-IP assays. VP1 and Hsp70-c5 were colocalized mainly in the nucleus and cytoplasm and Hsp70-c5 expression in CSBV-infected larvae was significantly upregulated. Our study supports a positive regulatory role for Hsp70-c5 in the CSBV virus infection cycle. Our findings could facilitate the study of the mechanisms underlying CSBV infection and host antiviral defense. The host–pathogen interaction reported in the present study could serve as a foundation for the development of novel therapeutics and prevention strategies against CSBV infections.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The use of the experimental animals involved in the manuscript is in compliance with the relevant provisions of the Animal Welfare and Ethics of Experimental Animals of the Experimental Animal Center of Jinzhou Medical University, China.
Author Contributions
XZ and DF designed the study and wrote the manuscript. LS, ML, DF, SH, CW, YM, and MM performed the experiments and analyzed the data. All authors approved the final version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31772760), award for “Liaoning Distinguished Professor,” and the Natural Science Foundation of Liaoning Province (No. 20180550289).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02192/full#supplementary-material
FIGURE S1 | Screening of proteins interacting with pBT3STE-VP1 plasmid and yeast Library Gel electrophoresis of PCR products amplified from putatively positive prey plasmids.
TABLE S1 | Analysis of sequencing results.
References
Alzhanova, D. V., Napuli, A. J., Creamer, R., and Dolja, V. V. (2014). Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. Embo J. 20, 6997–7007. doi: 10.1093/emboj/20.24.6997
Carsillo, T., Carsillo, M., Traylor, Z., Rajala-Schultz, P., Popovich, P., Niewiesk, S., et al. (2009). Major histocompatibility complex haplotype determines Hsp70 -dependent protection against measles virus neurovirulence. J. Virol. 83, 5544–5555. doi: 10.1128/JVI.02673-08
Chen, Y., Zhao, Y., Hammond, J., Hsu, H. T., Evans, J., and Feldlaufer, M. (2004). Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Invert Pathol. 87, 84–93. doi: 10.1016/j.jip.2004.07.005
Cheng, J., Zhang, P., Ma, M. X., Li, M., and Yang, S. (2011). Predication of spatial structure and b cell epitope of vp1 protein of chinese sacbrood virus ln-qy strain. Chin. J. Biol. 24, 280–284.
De-la-Re-Vega, E., Sánchez-Paz, A., Gallardo-Ybarra, C., Lastra-Encinas, M. A., Castro-Longoria, R., Grijalva-Chon, J. M., et al. (2017). The Pacific oyster (Crassostrea gigas) Hsp70 modulates the Ostreid herpes virus 1 infectivity. Fish Shellfish Immunol. 71, 127–135. doi: 10.1016/j.fsi.2017.09.079
Fei, D., Haochun, Z., Qingyun, D., Lili, J., Qiang, W., Yi, Z., et al. (2015). Correction: codon optimization, expression in escherichia coli, and immunogenicity of recombinant Chinese sacbrood virus (csbv) structural proteins vp1, vp2, and vp3. PLoS One 10:e0134423. doi: 10.1371/journal.pone.0134423
Feng, M., Fang, Y., Han, B., Zhang, L., Lu, X., and Li, J. (2013). Novel aspects of understanding molecular working mechanisms of salivary glands of worker honeybees (Apis mellifera) investigated by proteomics and phosphoproteomics. J Proteom. 87, 1–15. doi: 10.1016/j.jprot.2013.05.021
Fiil, B. K., Qiu, J. L., Petersen, K., Petersen, M., and Mundy, J. (2008). Coimmunoprecipitation (co-ip) of nuclear proteins and chromatin immunoprecipitation (chip) from arabidopsis. Csh Protoc. 3, 1–9. doi: 10.1101/pdb.prot5049
Han, B., Zhang, L., Feng, M., Fang, Y., and Li, J. (2013). An integrated proteomics reveals pathological mechanism of honeybee (Apis cerena) sacbrood disease. J. Proteome Res. 12, 1881–1897. doi: 10.1021/pr301226d
Hu, Y., Fei, D., Jiang, L., Wei, D., Li, F., Diao, Q., et al. (2016). A comparison of biological characteristics of three strains of chinese sacbrood virus in Apis cerana. Sci. Rep. 6:37424. doi: 10.1038/srep37424
Jiang, S., Lu, Y., Li, K., Lin, L., Zheng, H., Yan, F., et al. (2015). Heat shock protein 70 is necessary for rice stripe virus infection in plants. Mol. Plant Pathol. 15, 907–917. doi: 10.1111/mpp.12153
Johnsson, N., and Varshavsky, A. (1994). Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A. 91, 10340–10344. doi: 10.1073/pnas.91.22.10340
Kantengwa, S., Müller, I., Louis, J., and Polla, B. S. (1995). Infection of human and murine macrophages with leishmania major is associated with early parasite heat shock protein synthesis but fails to induce a host cell stress response. Immunol. Cell Biol. 73, 73–80. doi: 10.1038/icb.1995.12
Katoh, H., Kubota, T., Nakatsu, Y., Tahara, M., Kidokoro, M., and Takeda, M. (2017). Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J. Virol. 91:e02220-16. doi: 10.1128/JVI.02220-16
Kim, M. Y., Ma, Y., Zhang, Y., Li, J., Shu, Y., and Oglesbee, M. (2013a). Hsp70 -dependent antiviral immunity against cytopathic neuronal infection by vesicular stomatitis virus. J. Virol. 87, 10668–10678. doi: 10.1128/JVI.00872-13
Kim, M. Y., Shu, Y., Carsillo, T., Zhang, J., Yu, L., Peterson, C., et al. (2013b). Hsp70 and a novel axis of type i interferon-dependent antiviral immunity in the measles virus-infected brain. J. Virol. 87, 998–1009. doi: 10.1128/JVI.02710-12
Lahaye, X., Vidy, A., Fouquet, B., and Blondel, D. (2012). Hsp70 protein positively regulates rabies virus infection. J. Virol. 86, 4743–4751. doi: 10.1128/JVI.06501-11
Luo, L., King, N. P., Yeo, J. C., Jones, A., and Stow, J. L. (2014). Single-step protease cleavage elution for identification of protein-protein interactions from gst pull-down and mass spectrometry. Proteomics 14, 19–23. doi: 10.1002/pmic.201300315
Manzoor, R., Kuroda, K., Yoshida, R., Tsuda, Y., Fujikura, D., Miyamoto, H., et al. (2014). Heat shock protein 70 modulates influenza a virus polymerase activity. J. Biol. Chem. 289, 7599–7614. doi: 10.1074/jbc.M113.507798
Mingxiao, M., Ming, L., Jian, C., Song, Y., Shude, W., and Pengfei, L. (2011). Molecular and biological characterization of Chinese sacbrood virus LN isolate. Comp. Funct. Genom. 2011:409386. doi: 10.1155/2011/409386
Mingxiao, M., Yanna, Y., Xiaoli, X., Lin, Z., Yongfei, L., and Zhidong, L. (2013). Genetic characterization of VP1 gene of seven Sacbrood virus isolated from three provinces in northern China during the years 2008–2012. Virus Res. 176, 78–82. doi: 10.1016/j.virusres.2013.04.018
Nagy, P. D., Wang, R. Y., Pogany, J., Hafren, A., and Makinen, K. (2011). Emerging picture of host chaperone and cyclophilin roles in rna virus replication. Virology 411, 374–382. doi: 10.1016/j.virol.2010.12.061
Ohgitani, E., Kobayashi, K., Takeshita, K., and Imanishi, J. (1999). Biphasic translocation of a 70 kDa heat shock protein in human cytomegalovirus-infected cells. J. Gen. Virol. 80, 63–68. doi: 10.1099/0022-1317-80-1-63
Ozalp, C., Szczesna-Skorupa, E., and Kemper, B. (2005). Bimolecular fluorescence complementation analysis of cytochrome p450 2c2, 2e1, and NADPH-cytochrome p450 reductase molecular interactions in living cells. Drug Metab. Dispos. 33, 1382–1390. doi: 10.1124/dmd.105.005538
Padwad, Y. S., Mishra, K. P., Jain, M., Chanda, S., and Ganju, L. (2010). Dengue virus infection activates cellular chaperone Hsp70 in THP-1 cells: downregulation of Hsp70 by siRNA revealed decreased viral replication. Viral Immunol. 23, 557–565. doi: 10.1089/vim.2010.0052
Potts, S. G., Breeze, T. D., Bailey, A. P., and Balcombe, K. G. (2011). Pollination services in the uk: how important are honeybees? Agric. Ecosyst. Environ. 142, 137–143. doi: 10.1098/rsbl.2018.0001
Rajagopala, S. V., and Uetz, P. (2011). “Analysis of protein–protein interactions using high-throughput yeast two-hybrid screens,” in Network Biology Methods in Molecular Biology, eds G. Cagney, and A. Emili, (Totowa, NJ: Humana Press), 1–29. doi: 10.1007/978-1-61779-276-2_1
Reid, C., Airo, A., and Hobman, T. (2015). The virus-host interplay: biogenesis of+ RNA replication complexes. Viruses 7, 4385–4413. doi: 10.3390/v7082825
Ren, H., Qi, H., Cao, M., et al. (2012). Association of heat-shock protein 70 with lipid rafts is required for Japanese encephalitis virus infection in Huh7 cells. J. Gen. Virol. 93, 61–71. doi: 10.1099/vir.0.034637-0
Risler, J. K., Kenny, A. E., Palumbo, R. J., Gamache, E. R., and Curcio, M. J. (2012). Host co-factors of the retrovirus-like transposon Ty1. Mobile DNA 3:12. doi: 10.1186/1759-8753-3-12
Sato, T., Hanada, M., Bodrug, S., Irie, S., Iwama, N., Boise, L. H., et al. (1994). Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. U.S.A. 91, 9238–9242. doi: 10.1073/pnas.91.20.9238
Schaafhausen, A., Rost, S., Oldenburg, J., and Müller, C. R. (2011). Identification of VKORC1 interaction partners by split-ubiquitin system and coimmunoprecipitation. Thromb. Haemost. 105, 285–294. doi: 10.1160/TH10-07-0483
Shan, L. P., Chen, X. H., Ling, F., Zhu, B., and Wang, G. X. (2018). Targeting heat shock protein 70 as an antiviral strategy against grass carp reovirus infection. Virus Res. 247, 1–9. doi: 10.1016/j.virusres.2018.01.005
Shukla, H. D., and Pitha, P. M. (2012). Role of hsp90 in systemic lupus erythematosus and its clinical relevance. Autoimmune Dis. 2012:728605. doi: 10.1155/2012/728605
Snijder, D. E. J. (2008). The +Rna Virus Replication and Transcription Complex: Hijacking Membranes to Accommodate Viral Rna Synthesis in the Cytoplasm. Leiden: Universiteit Leiden.
Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 95, 5187–5192. doi: 10.1073/pnas.95.9.5187
Sun, L., Li, M., Fei, D., Hu, Y., and Ma, M. (2017). Chinese sacbrood virus infection in apis mellifera, Shandong, China, 2016. Virus Res. 242, 96–99. doi: 10.1016/j.virusres.2017.09.014
Sun, L., Li, M., Fei, D., Wang, J., Li, L., and Mingxiao, M. (2018). Preparation and application of egg yolk antibodies against chinese sacbrood virus infection. Front. Microbiol. 9:1814. doi: 10.3389/fmicb.2018.01814
Taguwa, S., Maringer, K., Li, X., Bernal-Rubio, D., Rauch, J. N., Gestwicki, J. E., et al. (2015). Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163, 1108–1123. doi: 10.1016/j.cell.2015.10.046
Tang, T., Wu, C., Li, J., Ren, G., Huang, D., and Liu, F. (2012). Stress-induced Hsp70 from musca domestica plays a functionally significant role in the immune system. J. Insect Physiol. 58, 1226–1234. doi: 10.1016/j.jinsphys.2012.06.007
Vaughan, T. J., Williams, A. J., Pritchard, K., Osbourn, J. K., Pope, A. R., Earnshaw, J. C., et al. (1996). Isolation of human antibodies with sub-nanomolar affinities directly from a large non-immunized phage display library. Nat. Biotechnol. 14, 309–314. doi: 10.1038/nbt0396-309
Venetianer, A., Pirity, M., and Hever-Szabo, A. (1994). The function of heat-shock proteins in stress tolerance. Cell Biol. Int. 18, 605–616. doi: 10.1006/cbir.1994.1087
Wu, P., Yu, H., Xu, J., Wu, J., Getachew, A., Tu, Y., et al. (2018). Purification of Chinese sacbrood virus (CSBV), gene cloning and prokaryotic expression of its structural protein VP1. Mol. Biotechnol. 60, 901–911. doi: 10.1007/s12033-018-0121-4
Ye, L., Liu, Y., Yang, S., Liao, W., and Wang, C. (2001). Increased expression of Hsp70 and co-localization with nuclear protein in cells infected with the hantaan virus. Chin. Med. J. 114, 535–539.
Zhang, Q. (2013). Characterization and Inducible Expression of Hsp70 Family in the Silkworm. Chongqing: Institute of Biological Sciences, Chongqing University, 1–65. doi: 10.7666/d.D355941
Zhang, Q., Yang, Y., Liang, Y., Lu, X., and Zhang, J. (2002). Study on structure of the Chinese sacbrood virus^ nucleic acid. J. Chin. Electron Microsc. Soc. 21, 331–334.
Zhang, Y., Zhang, G., Huang, X., and Han, R. (2014). Proteomic analysis of Apis cerana and apis mellifera larvae fed with heterospecific royal jelly and by csbv challenge. PLoS One 9:e0102663. doi: 10.1371/journal.pone.0102663
Keywords: Chinese sacbrood virus, VP1, heat shock protein 70 cognate 5, yeast two-hybrid screening, glutathione S-transferase pull-down, co-immunoprecipitation
Citation: Zhang X, Fei D, Sun L, Li M, Ma Y, Wang C, Huang S and Ma M (2019) Identification of the Novel Host Protein Interacting With the Structural Protein VP1 of Chinese Sacbrood Virus by Yeast Two-Hybrid Screening. Front. Microbiol. 10:2192. doi: 10.3389/fmicb.2019.02192
Received: 27 May 2019; Accepted: 06 September 2019;
Published: 26 September 2019.
Edited by:
Douglas Paul Gladue, United States Department of Agriculture, United StatesReviewed by:
Daniel C. Pevear, VenatoRx Pharmaceuticals, United StatesXingqi Guo, Shandong Agricultural University, China
Jianke Li, Institute of Apiculture Research (CAAS), China
Copyright © 2019 Zhang, Fei, Sun, Li, Ma, Wang, Huang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingxiao Ma, bG5qem1teEAxNjMuY29t
†These authors have contributed equally to this work
 Xiyan Zhang
Xiyan Zhang Dongliang Fei
Dongliang Fei Li Sun
Li Sun Ming Li
Ming Li YueYu Ma
YueYu Ma Chen Wang
Chen Wang Sichao Huang
Sichao Huang Mingxiao Ma
Mingxiao Ma