- 1Cell Biology of Bacterial Pathogenicity Team, Laboratory of Molecular Microbiology and Structural Biochemistry, CNRS UMR 5086, University of Lyon, Lyon, France
- 2Cell to Cell DNA Transfer Team, Laboratory of Molecular Microbiology and Structural Biochemistry, CNRS UMR 5086, University of Lyon, Lyon, France
Acinetobacter baumannii is a multidrug-resistant nosocomial opportunistic pathogen that is becoming a major health threat worldwide. In this study, we have focused on the A. baumannii DSM30011 strain, an environmental isolate that retains many virulence-associated traits. We found that its genome contains two loci encoding for contact-dependent growth inhibition (CDI) systems. These systems serve to kill or inhibit the growth of non-sibling bacteria by delivering toxins into the cytoplasm of target cells, thereby conferring the host strain a significant competitive advantage. We show that one of the two toxins functions as a DNA-damaging enzyme, capable of inducing DNA double-stranded breaks to the chromosome of Escherichia coli strain. The second toxin has unknown catalytic activity but stops the growth of E. coli without bactericidal effect. In our conditions, only one of the CDI systems was highly expressed in the A. baumannii DSM30011 strain and was found to mediate interbacterial competition. Surprisingly, the absence of this CDI system promotes adhesion of A. baumannii DSM30011 to both abiotic and biotic surfaces, a phenotype that differs from previously described CDI systems. Our results suggest that a specific regulation mediated by this A. baumannii DSM30011 CDI system may result in changes in bacterial physiology that repress host cell adhesion and biofilm formation.
Introduction
In Gram-negative bacteria, the two-partner secretion (TPS) pathway, also known as type Vb secretion system (T5bSS), mediates the translocation across the outer membrane of large, mostly virulence-related, TpsA proteins (Guérin et al., 2017). Functions of the TpsA secreted through the TPS pathway are diverse ranging from cytolysis, adhesion, and iron acquisition to contact-dependent growth inhibition (CDI) (van Ulsen et al., 2014). CDI system was the first secretion system identified to deliver toxin into neighboring cells, arming bacteria with a killing mechanism for outcompeting non-kin cells and establishment of self-communities (Aoki et al., 2005). Growth inhibition involves direct physical contact between bacteria and depends on the production of toxin–antitoxin pairs (Willett et al., 2015). This mechanism exploits the CdiA/CdiB subfamily of TPS systems to export CdiA to the surface through the cognate CdiB transporter and deliver into the cytosol of the target bacterium the last ∼300 C-terminal toxic residues of the CdiA proteins, called CdiA-CT. The C-terminal domain is delimited by conserved motifs of unknown function such as (Q/E)LYN in Burkholderia, VENN in most bacteria, or yet other motifs in Pseudomonas (Aoki et al., 2005; Anderson et al., 2012; Mercy et al., 2016). The presence of the cytoplasmic immunity protein CdiI protects CDI+ bacteria by interacting with the cognate CdiA-CT toxin and neutralizing its toxic activity. CdiA-CT is highly variable and shows various folds and activities (tRNase, DNase, and pore forming), allowing for a wide diversity of distinct toxins to be deployed to target bacteria (Zhang et al., 2011).
Contact-dependent growth inhibition systems are widespread among Gram-negative bacteria, as cdi gene clusters are found in several α-, β-, and γ-proteobacteria. They have been extensively studied in Enterobacteria and Burkholderia species, and recent work investigated their role in Acinetobacter species (Harding et al., 2017). Acinetobacter baumannii can be found associated with severe infections in humans, exhibiting multidrug resistance and causing fatal infections in susceptible hosts, such as patients in intensive care units. A. baumannii resists desiccation and forms biofilms that may contribute to its persistence in the clinical devices, causing acute infections. The molecular mechanisms implicated in infection by A. baumannii and the virulence factors associated with this process are still unclear. Recent studies investigated the potential implication of TPS systems in A. baumannii pathogenesis. The TpsA proteins characterized in strains A. baumannii ATCC 19606(T) and clinical AbH12O-A2 are both adhesins that mediate adherence to eukaryotic cells (Darvish Alipour Astaneh et al., 2014; Pérez et al., 2016), and TpsA of A. baumannii AbH12O-A2 was shown to contribute to virulence in models of mouse systemic infection and Caenorhabditis elegans (Pérez et al., 2016). Interestingly, our in silico analysis revealed that these two adhesins associated with their respective CdiB and CdiI partners constitute putative CDI systems, suggesting a potential involvement of these systems in the virulence of A. baumannii. This is in line with studies in other organisms suggesting a role for CDI systems beyond bacterial competition. Indeed, several CdiA promotes bacterial auto-aggregation and biofilm formation in Escherichia coli, Pseudomonas aeruginosa, and Burkholderia thailandensis (Garcia et al., 2013; Ruhe et al., 2015; Mercy et al., 2016), as well as intracellular escape and immune evasion of Neisseria meningitidis (Talà et al., 2008), functions that are required for the virulence of several pathogens (Gallagher and Manoil, 2001; Rojas et al., 2002; Guilhabert and Kirkpatrick, 2005; Gottig et al., 2009). Recently, in silico analysis revealed the identification of more than 40 different CDI systems in pathogenic Acinetobacter genomes that have been sorted into type I and II groups (De Gregorio et al., 2019). While sequencing the genome from A. baumannii DSM30011 strain (Repizo et al., 2017), we have also identified two cdiBAI loci potentially encoding type I and II CDI systems. A. baumannii DSM30011, an environmental strain isolated in 1944 from resin-producing guayule plants, has many of the characteristics of clinical strains and was shown to use a type 6 secretion system (T6SS) for bacterial competition and colonization in the model organism Galleria mellonella (Repizo et al., 2015). In this study, we used live-cell microscopy to characterize the function of CdiA-CT toxins when produced in E. coli cells. Using transcriptional fusions, we show that only one CDI system is expressed in A. baumannii DSM30011 and promotes interbacterial competition but is surprisingly a limiting factor for the adhesion process.
Results
The Acinetobacter baumannii DSM30011 Genome Contains Two Predicted CDI Systems
In the course of this study, we performed a bioinformatic search to obtain the global repartition and representation of TPS systems among A. baumannii species. Each subset of TpsA was used to blast against the A. baumannii sequence database. Based on their sequence, TpsA proteins can be phylogenetically classified into at least five subfamilies with distinct functions: (i) the CDI CdiA proteins (Aoki et al., 2005), (ii) the hemolysins/cytolysins such as ShlA of Serratia marcescens (Braun et al., 1992), (iii) the adhesins such as filamentous hemagglutinin (FHA) of Bordetella pertussis (Relman et al., 1989), (iv) HxuA-type proteins involved in iron acquisition (Fournier et al., 2011), and (v) TpsA with unknown specific activities (Faure et al., 2014). The blast search revealed that the CDI system subfamily is predominantly represented within A. baumannii strains with the exception of some genomes comprising Hxu system homologues. We detected two loci in our A. baumannii DSM30011 laboratory model strain containing gene organization related to the “Escherichia coli”-type CDI systems (Figure 1A). We renamed them as cdi1Ab30011 (encoding proteins PNH15603.1, PNH15604.1, and PNH15605.1) and cdi2Ab30011 (encoding proteins PNH14818.1, PNH14817.1, and PNH14816.1). The CdiA proteins of these systems share only 9.6% identity overall, but both contain the highly conserved VENN motif (PF04829) that delimits the N- and C-terminal (CT) domains. The sequence analysis of CdiA1 revealed that the N-terminal domain of this very large protein (532 kDa) harbors long stretches of imperfect repeats predicted to form β-helix folds, that is, β-strand structure organized in fibrous (Kajava et al., 2001), which classifies it as a type II CdiA protein (De Gregorio et al., 2019). CdiA1-CT does not contain any conserved domain. The smaller CdiA2 protein (204 kDa) belongs to the type I CdiA and its CdiA2-CT preceded by the small-helical DUF637 domain found in many CdiA proteins (Iyer et al., 2011) contains a Tox-REase7 nuclease domain. Three potential orphan cdiI genes encoding the PNH15606.1, PNH15607.1, and PNH15608.1 proteins are located downstream of the cdi1 locus. Indeed, PNH15606.1 and PNH15607.1 proteins contain an Imm23 domain, and PNH15608.1 protein has a Smi1/Knr4 superfamily domain typically found in immunity proteins present in bacterial polymorphic toxin systems (Iyer et al., 2011; Zhang et al., 2011).
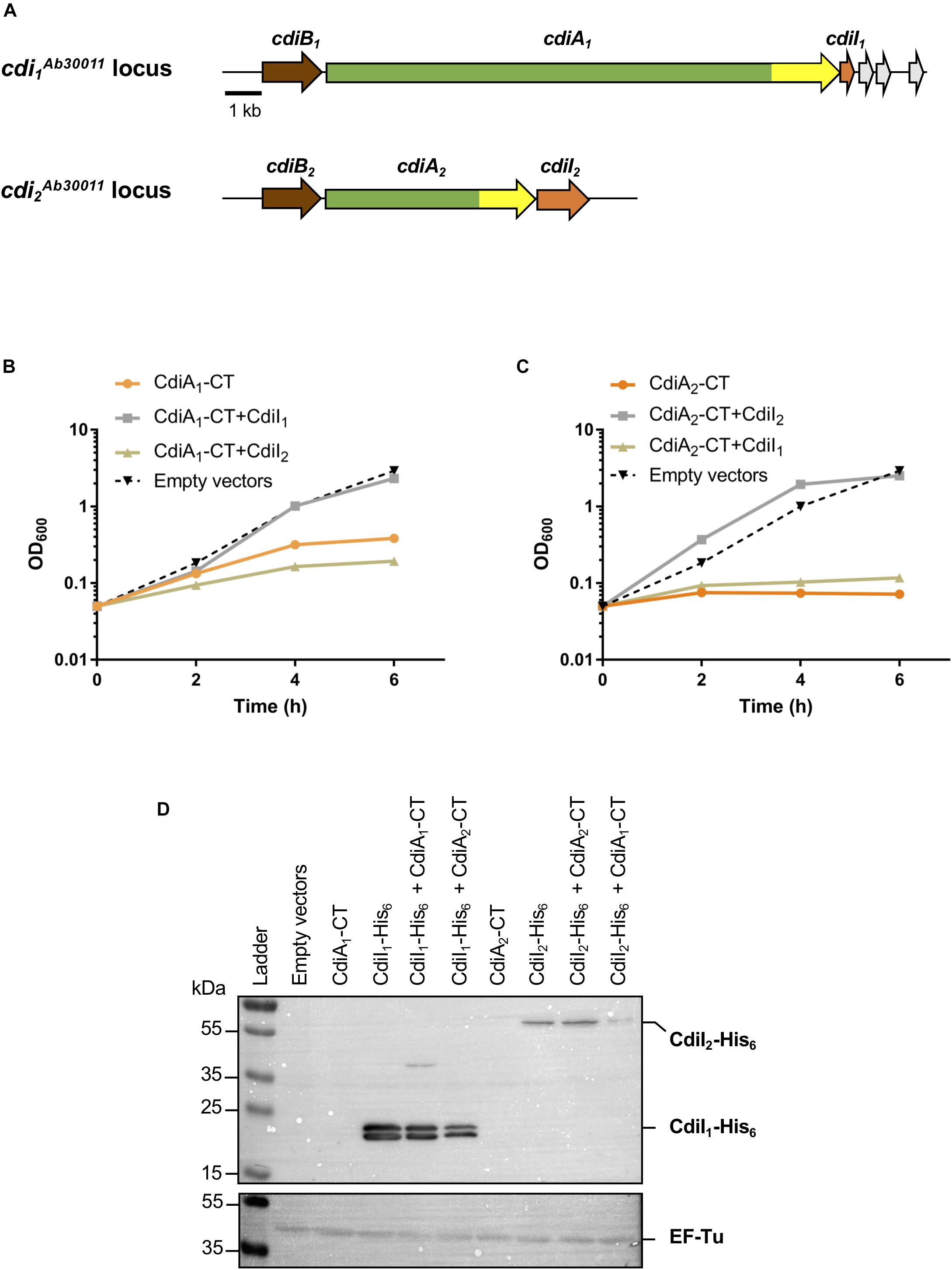
Figure 1. The production of CT domains of CdiA1 or CdiA2 in Escherichia coli is toxic, and cdiI genes encode immunity proteins. (A) Schematic of the cdi1Ab30011 and cdi2Ab30011 loci. Genes encoding putative CdiB transporters, CdiA exoproteins, and CdiI immunity proteins are colored, respectively, in brown, green, and orange. Gray arrows represent putative orphan immunity genes. The highly variable nucleotide regions encoding CdiA-CT domains are highlighted in yellow. Black bar represents the 1-kb scale. Based on sequenced Acinetobacter baumannii DSM30011 genome (Repizo et al., 2017), CdiB1, CdiA1, and CdiI1 correspond, respectively, to PNH15603.1, PNH15604.1, and PNH15605.1 proteins and CdiB2, CdiA2, and CdiI2 to PNH14818.1, PNH14817.1, and PNH14816.1 proteins. (B,C) Effect of CdiA1-CT (B) or CdiA2-CT (C) production in E. coli DH5α strain. CdiA-CT production was induced at 0 min with or without CdiI, and growth was monitored by measuring the optical density at 600 nm (OD600). (D) After 6 h of culture, E. coli cell extracts containing an indicated set of produced proteins were analyzed to probe the production of CdiI-His6 and the cytoplasmic EF-Tu control using the anti-pentaHis (top panel) and anti-EF-Tu monoclonal antibodies (bottom panel). Molecular weight marker (kDa) is indicated on the left.
CdiA1-CT/CdiI1 and CdiA2-CT/CdiI2 Are Two Non-interchangeable Toxin–Antitoxin Pairs
To address the toxicity of CT domains, we generated pBAD33 plasmid derivatives producing each CdiA-CT (from the VENN motif to the stop codon) in the presence of arabinose. To assess the CdiI immunity property, nucleotide sequences encoding CdiI fused to 6xHis tag were introduced in pTrc99a plasmid and induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG). The production of CdiA1-CT alone stops the growth of the E. coli DH5α strain after 4 h of induction (Figure 1B). Unlike CdiA1-CT, CdiA2-CT is highly toxic in E. coli where its induction inhibits the growth (Figure 1C). Cells coproducing CdiI1 and CdiA1-CT or CdiI2 and CdiA2-CT are protected from the toxic effect of the toxins and exhibited growth equivalent to that of cells containing the empty vector (Figures 1B,C). In contrast, the production of CdiI1 or CdiI2 with CdiA2-CT or CdiA1-CT, respectively, does not suppress toxicity. Both CdiI-6xHis were detected in E. coli cells in the presence or absence of CdiA-CT using Western blot experiment (Figure 1D) showing that the inability to rescue the growth defect caused by a non-cognate CdiA-CT is not due to a lack of CdiI production. CdiA1-CT/CdiI1 and CdiA2-CT/CdiI2 therefore function as pairs of toxin–antitoxin, and these systems are not interchangeable.
CdiA2-CTAb30011 Toxin Induces DNA Damage in E. coli
CdiA2-CT contains a restriction endonuclease-like domain belonging to the Tox-REase7 family (Pfam PF15649) mostly found in CdiA of Pseudomonas and Acinetobacter species (Zhang et al., 2012; Mercy et al., 2016). To determine whether CdiA2-CT displays nuclease activity when expressed in E. coli cells, we performed real-time microscopy visualization of the nucleoid-associated HU protein after induction of the toxin. HU is a widely conserved histone-like protein very abundant in the bacterial cytoplasm that binds to DNA in a non-specific manner. Owing to its nucleoid association, HU localization reveals the global organization of the chromosome and potential alterations. We grew a CdiA2-CT-producing E. coli MG1655 strain in which the hupA gene encoding the α-subunit of HU is fused to the mcherry gene at the endogenous locus. Twenty minutes after CdiA2-CT induction, the majority of the cells contain organized and well-segregated nucleoids (Figure 2 and Supplementary Movie S1). However, after 60 min, the nucleoids condense as a dense mass at midcell of the bacteria or show a diffused localization pattern. This global nucleoid disorganization is followed by cell filamentation indicative of cell division inhibition. These chromosome alterations are associated with cell death, as CdiA2-CT-producing cells are not able to form viable colony units (Supplementary Figure S1A). No filamentation is observed after induction of the CdiA2-CT in recA- cells showing that cell division arrest depends on the homologous recombination RecA protein (Supplementary Figure S1B). In contrast to bacteria producing only CdiA2, cells coproducing the CdiI2 immunity protein suffer no loss of viability and retain normal chromosome organization and cell division (Figure 2, Supplementary Movie S2, and Supplementary Figure S1A). The perturbation of DNA organization together with the RecA-dependent filamentation observed in the presence of CdiA2-CT suggests that this toxin induces DNA damage to the chromosomal DNA. To test this hypothesis, we examined the localization pattern of the RecA protein, which has been reported to polymerize into large intracellular structures in response to DNA lesions (Lesterlin et al., 2014). To do so, we used an E. coli MG1655 strain expressing a carboxy-terminal fusion of RecA to green fluorescent protein (GFP) and wild-type (wt) RecA, both expressed from wt chromosome recA promoters. Before induction of CdiA2-CT, RecA-GFP fluorescence appears uniformly distributed in most cells (Figure 3; t = 0 h and Supplementary Movie S3). From 20 min after production of CdiA2-CT, fluorescent RecA-GFP structures form in the majority of cells, and we observed the formation of RecA bundles reported to promote homologous recombination repair between distant regions of homology (Lesterlin et al., 2014). As expected, RecA-GFP fluorescence is diffuse when CdiA2-CT is co-expressed with CdiI2, reflecting the absence of DNA damage (Figure 3 and Supplementary Movie S4). Altogether, these results demonstrate that CdiA2-CT creates multiple DNA breaks that cannot be repaired by the cells leading to growth inhibition, loss of chromosome organization, and eventual cell death.
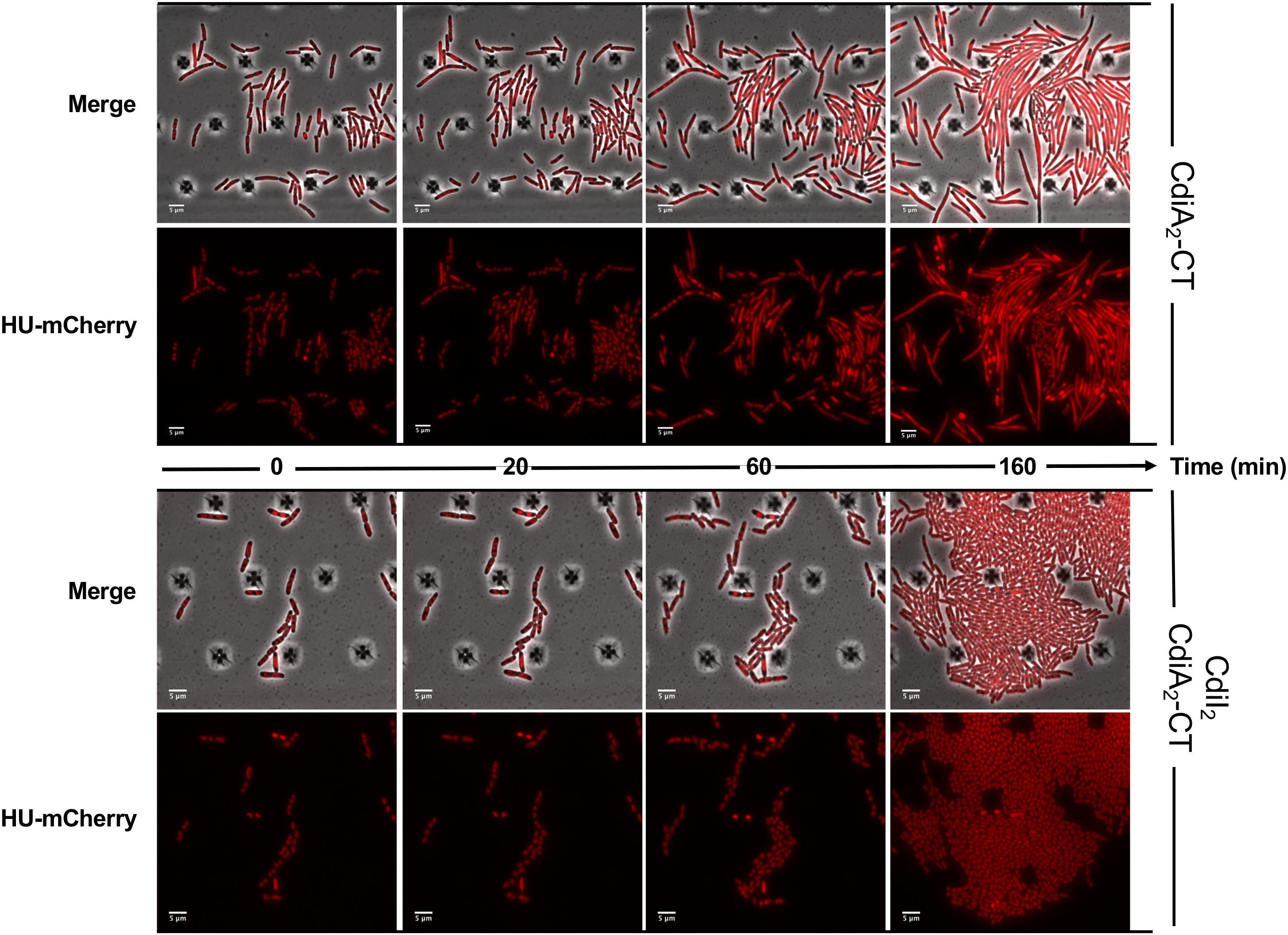
Figure 2. The production of CdiA2-CT in Escherichia coli disorganizes the nucleoid localization. Time-lapse fluorescence microscopy of the recombinant HU-mCherry protein produced by E. coli MG1655 strain after induction of the CdiA2-CT in the absence or in presence of its cognate CdiI2 immunity protein. The scale bar corresponds to 5 μm.
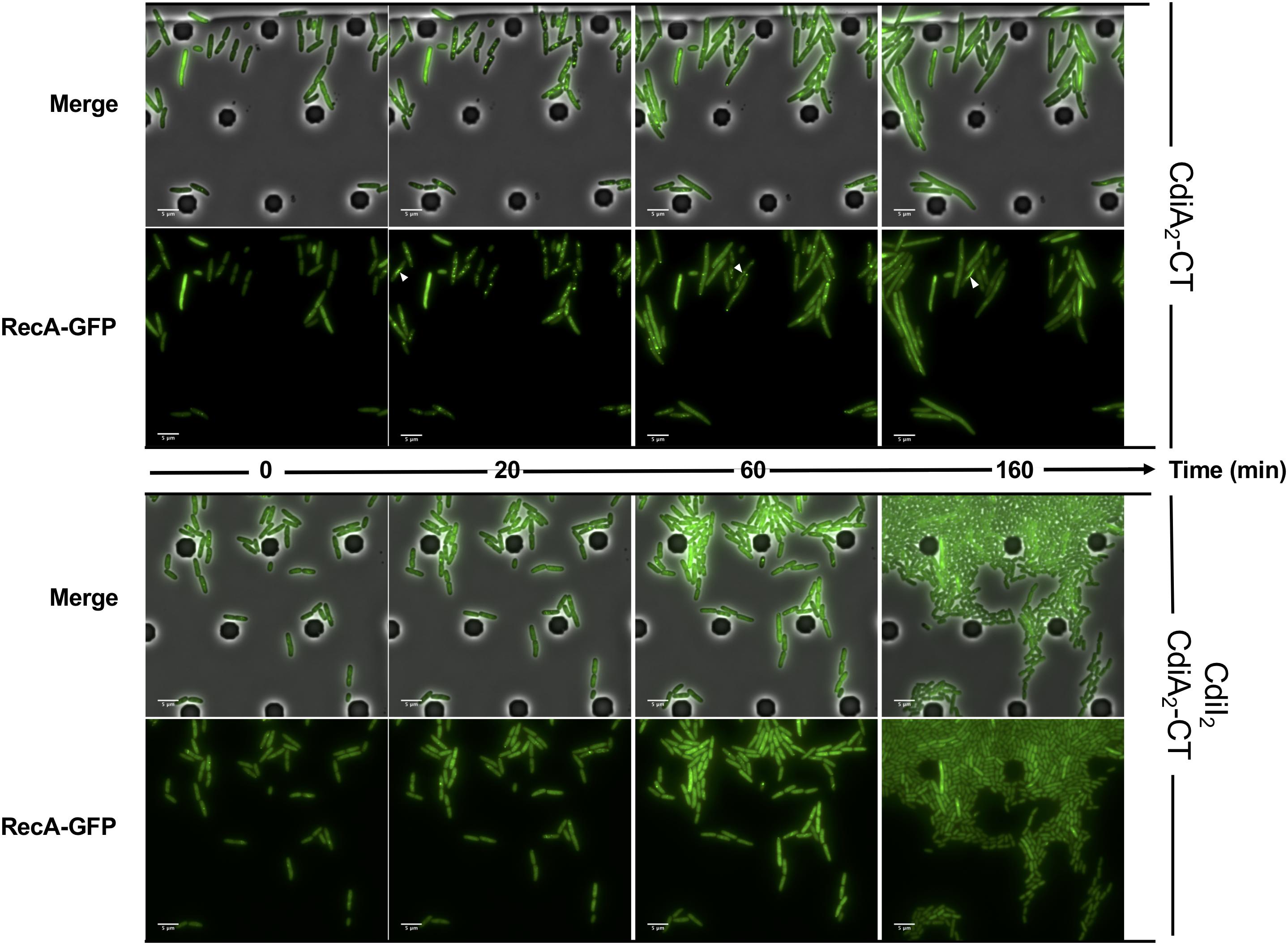
Figure 3. The production of CdiA2-CT in Escherichia coli induces DNA damages. Time-lapse fluorescence microscopy of the recombinant RecA-GFP protein produced by E. coli MG1655 strain after induction of the CdiA2-CT in the absence or in the presence of its cognate CdiI2 immunity protein. White arrows indicate examples of bundles of RecA-GFP. The scale bar corresponds to 5 μm. GFP, green fluorescent protein.
CdiA1-CT Toxin Inhibits the Growth of E. coli
In order to get insight into the mechanism of growth inhibition generated by the production of CdiA1-CT in E. coli (Figure 1B), we analyzed in real-time microscopy the localization pattern of the recombinant HU-mCherry and RecA-GFP proteins after induction of CdiA1-CT in the presence or absence of its cognate CdiI1 immunity protein. Microscopy analysis showed no diffusion of the HU-mCherry in CdiA1-CT-producing cells and RecA-GFP formed no spot or bundle, indicating that the toxin does not induce nucleoid disruption nor DNA damage (Figure 4A and Supplementary Movies S5–S8). However, we noticed CdiA1-CT production led to increased cell size (Figure 4B) and division arrest in more than half of the cell population (Figure 4C). These results are consistent with the observed cell viability decrease 2 h post production of the CdiA1-CT followed by a plateau of the number of colony-forming unit (CFU)/ml indicating a cell growth defect (Supplementary Figure S2A). To assess if these arrested cells are alive, we used the Live/Dead assay on the basis of the green fluorescent SYTO9 entering all cells to stain nucleic acid and the propidium iodide entering only dead cells with damaged cytoplasmic membranes. Our results showed the CdiA1-CT-producing cells are not dying once they have stopped growing and that the percentage of cell death was similar to that of cells producing CdiA1-CT in the presence of CdiI1 (Supplementary Figure S2B).

Figure 4. The CdiA1-CT toxin stops the growth of Escherichia coli. (A) Fluorescence microscopy of recombinant HU-mCherry or RecA-GFP produced by E. coli MG1655 strains 130 min after induction of CdiA1-CT in the absence or in the presence of its cognate CdiI1 immunity protein. The scale bar is indicated on the lower left. (B) Cell length analysis of RecA-GFP producing E. coli MG1655 strains 3 h after induction of CdiA1-CT in the absence or in the presence of its cognate CdiI1 immunity protein. n indicates the number of analyzed cells. (C) HU-mCherry producing E. coli MG1655 strains were grown in M9-casa after induction of CdiA1-CT with or without CdiI1, and real-time microscopy was performed in order to calculate the number of division over 45 single bacteria. GFP, green fluorescent protein.
Unlike the cdiBAI2 Locus, the cdiBAI1 Genes Are Expressed in A. baumannii DSM30011
Most of the cdi genes are not expressed in laboratory growth conditions (Cope et al., 1994; Rojas et al., 2002; Mercy et al., 2016). In order to get insight into the expression profile of cdi genes present in A. baumannii DSM30011, we constructed plasmids containing transcriptional fusions between DNA region upstream of these genes and the gfp reporter. A. baumannii DSM30011 reporter strains were grown in liquid Luria–Bertani (LB)-rich medium at 37°C with agitation and static conditions, and the GFP fluorescence intensity was measured at 6 and 8 h, respectively. Equivalent results were obtained for each growth condition (Figure 5A). The level of GFP fluorescence of PcdiB2, PcdiA2, and PcdiI2 fusions is identical to that of the promoterless fusion, indicating that DNA regions upstream of these genes do not have any promoter activity and that the cdi2 locus might not be expressed under the growth conditions tested (Figure 5A). In contrast, the PcdiB1 fusion produces a really high level of GFP compared to the regions upstream the cdiA1 and cdiI1 genes that do not exhibit any promoter activity (Figure 5A). In addition, the GFP fluorescence is quite homogeneous within the population, as all individual bacteria produce high levels of GFP fluorescence (Figure 5B). In order to determine whether CdiA1 is produced in the tested conditions, we tried to detect this protein using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Overnight cultures were fractionated to analyze the protein profile of the whole cell lysate and secreted fraction. A band with a slower mobility than 250-kDa molecular weight marker was detected in the secreted fraction of the wt strain, which was identified as CdiA1 protein by mass spectrometry (Figure 5C). For controls, we confirmed that this band disappeared in the ΔcdiA1 mutant and that the CdiA1 transport required CdiB1 because no CdiA1 is secreted in a ΔcdiB1 mutant probably owing to the instability of the protein that is trapped in the periplasm, which is consistent with previous reports on FHA undergoing a rapid proteolytic degradation in the periplasm in the absence of its transporter FhaC (Jacob-Dubuisson et al., 1997; Guédin et al., 1998). Furthermore, the secretion of CdiA1 is not due to cell lysis, as the cytoplasmic EF-Tu protein is only found in the whole cell fraction and Hcp, identified by mass spectrometry, the main component of the T6SS, is detected in the secreted fraction (Figure 5C). Knowing that CdiA1 is highly produced and can be visualized by Coomassie blue staining, this suggests that the three cdiBAI1 genes are transcribed as a polycistronic mRNA from the same promoter upstream cdiB1.
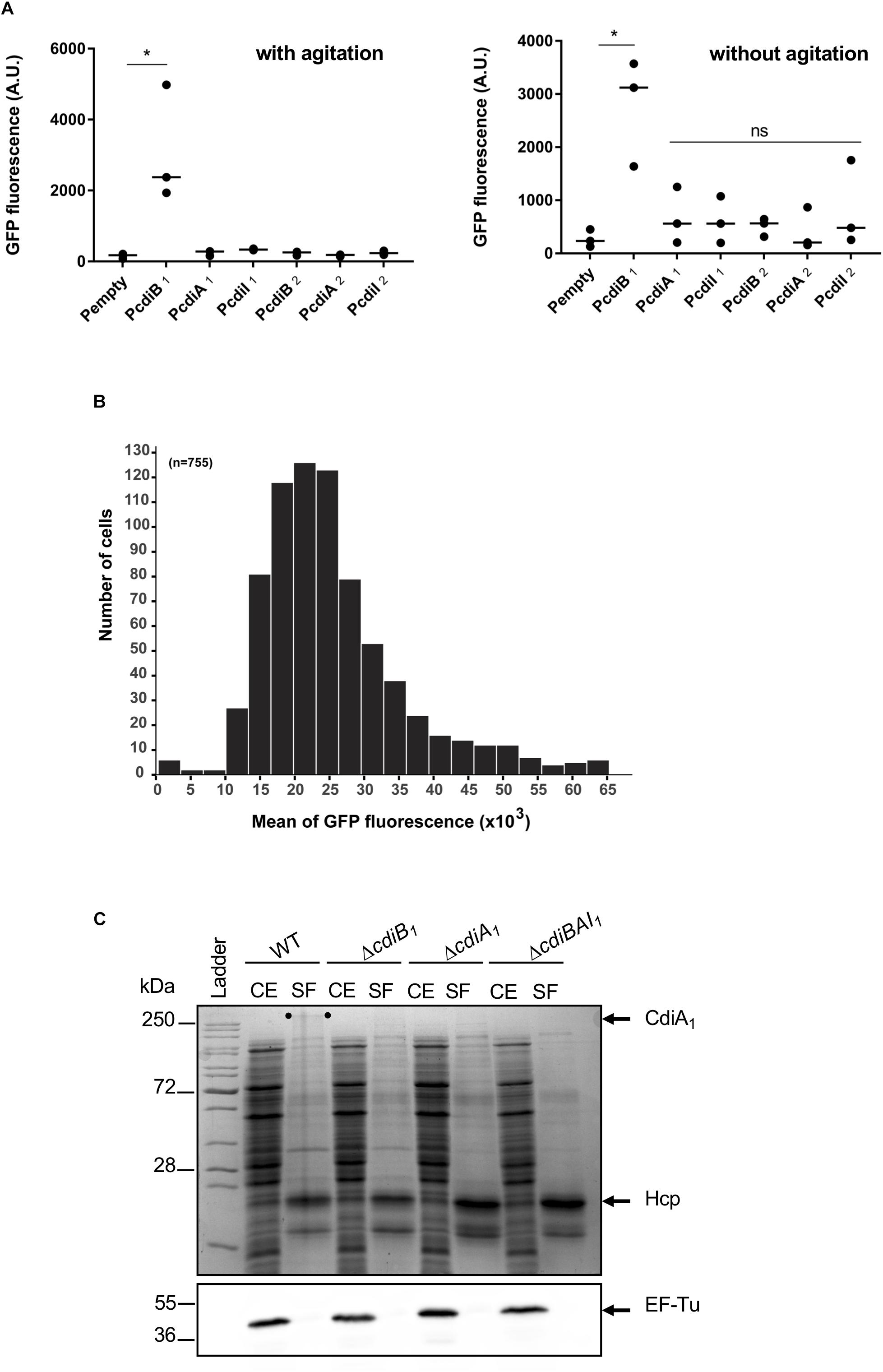
Figure 5. Only the cdiBAI1 genes are expressed in Acinetobacter baumannii DSM3011. (A) GFP fluorescence measurements for potential promoter regions of cdi genes. A strain harboring plasmid with a gfp reporter gene without a promoter (Pempty) served as a negative control. For statistical analyses, a non-parametric one-way ANOVA with Dunn’s multiple comparison test was performed. ∗p = 0.0184 (with agitation) and p = 0.028 (without agitation); ns: not significant. (B) Repartition of the GFP fluorescence intensities within a sibling population harboring the PcdiB1 fusion. n = number of analyzed cells. (C) Cell extract (CE) and secreted fraction (SF) of wild-type and mutant strains grown overnight were analyzed by Coomassie blue staining and Western blot analysis using anti-EF-Tu antibody. The migration position of CdiA1 protein is indicated by two dots. Molecular marker (kDa) is indicated on the left. GFP, green fluorescent protein.
The cdiBAI1 Locus Mediates Bacterial Competition
As only the CDI1 system is turned on under the tested conditions and we have not yet determined the regulatory pathway of the cdi2 locus, we pursued in A. baumannii the characterization of the cdiBAI1 system. To determine whether this system functions as a CDI system, the entire locus was replaced by a kanamycin cassette or deleted generating the ΔcdiBAI1:kn and ΔcdiBAI1 mutants. Next, we performed competition experiments by mixing the wt or ΔcdiBAI1 attacker strains with the ΔcdiBAI1:kn target strain and measure the CFU/ml of the target over time. As seen in Figure 6, wt strain inhibits the growth of the isogenic ΔcdiBAI1:kn target strain by ∼1 log after 6 h of growth competition, whereas no growth inhibition is observed with the ΔcdiBAI1 attacker strain. No significant viability defect between the wt and mutant strains was observed, indicating that the ability of wt strain to outcompete the ΔcdiBAI1 is not due to a growth rate difference (Supplementary Figure S3).
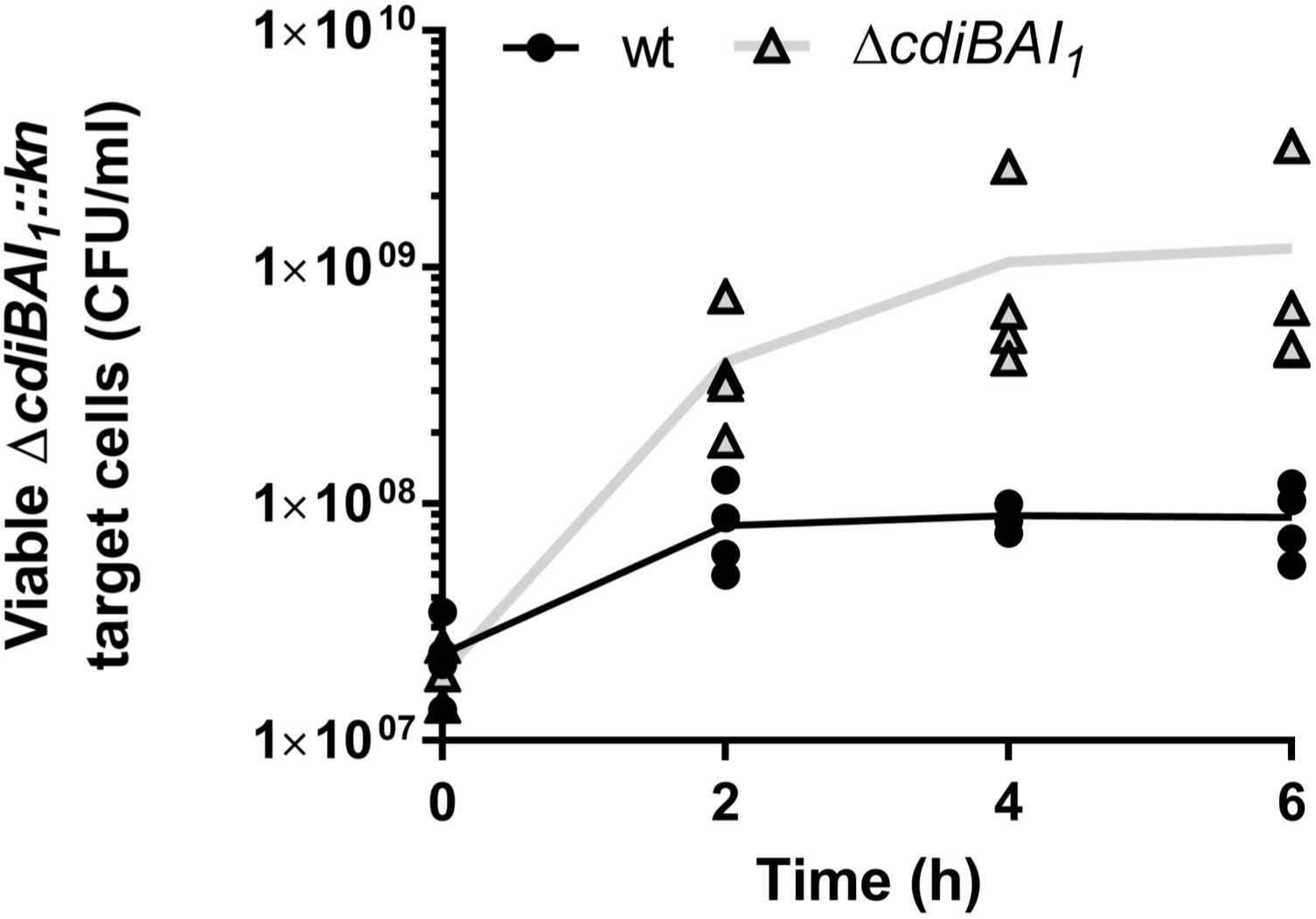
Figure 6. Growth inhibition mediated by the CDI1 system of strain Acinetobacter baumannii DSM30011. Both attacker and target strains were grown in LB separately to OD600 = 0.35 and mixed to a 4:1 (attacker:target) ratio. At various times of co-incubation, the cdiBAI1:kn target was spread on LB agar supplemented with kanamycin to measure the colony-forming unit (CFU)/ml. For statistical analyses, a non-parametric Mann–Whitney test was performed. ∗p = 0.0286 at 4 and 6 h. GFP, green fluorescent protein; LB, Luria–Bertani.
cdi1 Locus Decreases A. baumannii DSM30011 Biofilm Formation and Adhesion to Epithelial Cells
CdiA proteins can promote biofilm formation and/or attachment to eukaryotic cells (Rojas et al., 2002; Plamondon et al., 2007; Schmitt et al., 2007; Gottig et al., 2009; Neil and Apicella, 2009; Garcia et al., 2013; Ruhe et al., 2015; Mercy et al., 2016). To investigate whether cdi1 locus contributes to biofilm formation, we quantified in 96-well polystyrene plates over a 24-h time period the biofilm biomass formed by the wt and cdiBAI1 mutant strains. After 3 h, the capacity to form biofilm of the mutant was lower than that of the wt (Figure 7A). Surprisingly, cdiBAI1 mutant generated twice as much biofilm mass as the wt after 5 and 24 h (Figure 7A). Analysis of the depth bacterial growth on glass-bottom slides by confocal microscopy confirmed that the cdiBAI1 mutant exhibited the highest ability to form a bacterial 3D structured layer especially after 5 h (Figure 7B and Supplementary Figure S4). To determine whether the cdi1 locus is also implicated in the adhesion of A. baumannii DSM30011 to biotic surfaces, we compared the adhesiveness of wt and isogenic cdiBAI1 mutant strains to A549 epithelial cells by CFU measurement and confocal microscopy analysis to directly visualize A. baumannii strains and confirm CFU counts. No difference between strains was observed after 2 h of infection with exponential cultures grown in LB liquid medium (Figure 7C; exponential). The biofilm increase is a late phenotype (Figure 7A), and we noticed that on agar plate the ΔcdiBAI1 colonies were slightly smaller than those of the wt, reflecting a potential change in physiology depending on the growth condition. For these reasons, we directly analyzed the capacity of bacteria scratched from the agar plate to bind to host cells. As seen in Figure 7C (solid), the deletion of the cdi1 locus led to a significant 2.5-fold increase in the proportion of cell-associated bacteria 2-h post infection. Furthermore, the number of cell-attached bacteria detected by immunofluorescence using an anti-Acinetobacter antibody was also higher for the cdiBAI1 mutant (Figure 7D). Z-stack reconstruction confirmed that the bacteria are well attached to the cell surface and not endocyted by the host cells (Supplementary Figure S5).
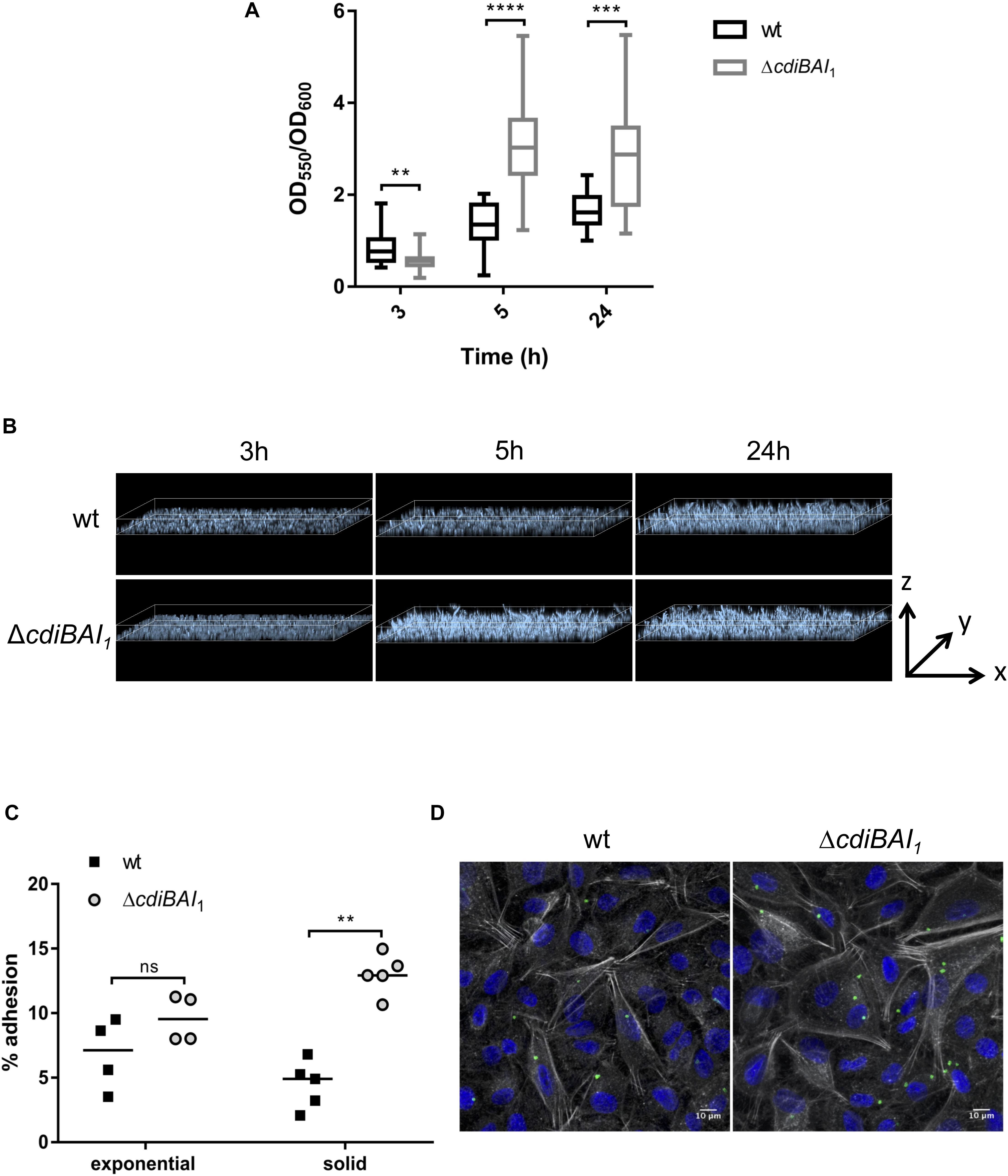
Figure 7. The adhesive properties of Acinetobacter baumannii DSM30011 is enhanced in the absence of cdiBAI1 locus. (A) Biofilm formation assayed by crystal violet staining. A. baumannii DSM30011 wild-type (wt) and ΔcdiBAI1 strains were statically grown at 37°C for 3, 5, and 24 h. The biofilm biomass was estimated by calculating the OD550/OD600 ratio from three independent experimental replicates. For statistical analyses, a non-parametric Mann–Whitney test was performed. ∗∗p = 0.0045, ∗∗∗p = 0.0004, ∗∗∗∗p < 0.0001. (B) Confocal microscopy of biofilm formed by the wild-type and ΔcdiBAI1 strains grown on glass-bottom slides in static conditions. Images correspond to the depth analysis of a 3D reconstruction of z-stacks obtained for the different strains at 3, 5, and 24 h. The height of the z axis is between 0 and 10 μm. Bacteria were labeled with DAPI. (C) Adhesion assay in which A549 cells were infected with A. baumannii DSM30011 wt or ΔcdiBAI1 strains at a MOI of 100 for 2 h. Data are expressed as a percentage of A. baumannii adhesion obtained from the ratio between CFU/ml before and after infection. For statistical analyses, a non-parametric Mann–Whitney test was performed. ∗∗p = 0.0079, ns: not significant. (D) Representative immunofluorescence images obtained by confocal microscopy and analyzed with ImageJ of A549 cells infected with bacteria grown in solid. Images correspond to Z-projections at average intensity. A. baumannii strains were detected with an anti-A. baumannii antibody (green). The actin cytoskeleton and nucleus were labeled with phalloidin (white) and DAPI (blue), respectively. MOI, multiplicity of infection.
Discussion
The knowledge on virulence mechanisms and factors contributing to the pathogenic potential of Acinetobacter baumannii is limited, and a deeper understanding of its infection mechanisms may shed light on new strategies for drug development. On the basis of recent studies that characterized the potential implication of secretion systems including the TPS pathway, we performed a blast search using TpsA of diverse functions to evaluate their distribution within A. baumannii. The basic pattern reflected in this search was that several cdi loci could be identified in a large number of A. baumannii species and confirmed a previous study (De Gregorio et al., 2019). Strikingly, other subfamilies of TPS systems were absent with the exception of HxuA homologues found in some A. baumannii strains, which are TpsA involved in iron acquisition (Cope et al., 1994, 1995, 1998). These findings suggest that CDI systems may be significant players in A. baumannii. In this work, we used the environmental A. baumannii DSM30011 strain with two CDI systems.
Most of the cdi genes appear to be under tight regulatory control (Mercy et al., 2016) or only expressed during infection (Rojas et al., 2002; Aoki et al., 2010). Our results show that A. baumannii DSM30011 cells differentially produce its CDI systems. Indeed, cdi2 locus is not expressed under the rich medium and growth conditions used in this study and undergoes negative regulation. Interestingly, we noticed the presence of a putative pho box in the cdiB2 promoter region, suggesting a potential regulation by the transcriptional regulator PhoB through a differential phosphate level, as it has already been characterized for the Pseudomonas aeruginosa tps genes (Faure et al., 2013). Although additional experiments will be necessary to identify the regulatory circuits controlling the expression of this cdiA gene and its role in A. baumannii, we were able to show that CdiA2-CT, produced intracellularly in Escherichia coli, induces multiple DNA damages that the target cell cannot repair, leading to its death. This finding is consistent with the presence of a Tox-REase-7-fold domain and strongly suggests that CdiA2-CT functions as a cytoplasmic DNase to degrade nucleic acids like several other CdiA toxins (Willett et al., 2015). In contrast, the CdiA1 protein is highly secreted in A. baumannii, and the expression of cdiBAI1 genes is quite homogeneous within the population, in comparison with Burkholderia thailandensis, which expresses a high level of cdi genes in a stochastic manner (Anderson et al., 2012). In addition, in the A. baumannii SDF strain, we also observed the presence of two CDI systems, one of which is not repressed (Supplementary Figures S6, S7), and other studies have shown the constitutive activation of CDI systems within several Acinetobacter species in growth laboratory conditions (Perez et al., 2007; Darvish Alipour Astaneh et al., 2014; Harding et al., 2017), suggesting that Acinetobacter might not keep CDI system in an inactive state. Interestingly, we and others have shown that several Acinetobacter species often coproduce the CDI and the T6SS (Carruthers et al., 2013; Repizo et al., 2015; Harding et al., 2017; unpublished data), and in Pseudomonas aeruginosa, they are both regulated by the post-transcriptional RsmA regulator (Mercy et al., 2016; Allsopp et al., 2017). These observations might reflect that a co-regulation exists between these two systems, but additional work is needed to understand the real link between CDI and T6SS co-regulation in Acinetobacter.
Although we established that the CDI1 system is functional and arrests the growth of neighboring A. baumannii bacteria that do not contain the cdi1 locus, we have not yet investigated the mechanisms involved. However, we have shown that the production of CdiA1-CT in the cytoplasm of E. coli stops bacterial growth by inhibiting cell division rather than cell death, indicating that this domain might be responsible for the A. baumannii growth arrest during bacterial competition. CdiA1-CT does not contain any known conserved domains, but the use of fluorescent proteins reporting chromosome compaction state and DNA damage allowed us to exclude that this toxin functions as a DNase. The induction of growth arrest by CdiA1-CT can arise from different mechanisms of action. Many CdiA toxins act as nucleases that degrade tRNA or rRNA arresting growth by blocking the translation (Willett et al., 2015), whereas CdiAEC93 from E. coli EC93, a ionophore that dissipates the proton motive force, inhibits the growth by depleting ATP levels (Aoki et al., 2009). Further investigation will be necessary to identify the functional activity of CdiA1 toxin of A. baumannii DSM30011.
CdiA proteins might be multifunctional, which would be quite conceivable given their large sizes and might therefore have a broader role than bacterial competition. Indeed, studies in A. baumannii but also in other bacteria show that CdiA functions as adhesins mediating adhesion to epithelial cells or structuring a biofilm through bacteria–bacteria interactions (Balder et al., 2007; Plamondon et al., 2007; Darvish Alipour Astaneh et al., 2014; Ruhe et al., 2015; Pérez et al., 2016). Interestingly, CdiA1 of A. baumannii DSM30011 is a very large protein whose amino acid sequence is mostly constituted of β-helical and FHA repeats found in a number of TpsA adhesins (Jacob-Dubuisson et al., 2001). However, we found that CDI1 system does not promote biofilm formation nor adhesion to eukaryotic cells, but instead, its absence increases the adhesiveness of A. baumannii. The mechanism enabling the limitation of cell–cell or surface adhesion by the CDI1 system is not yet understood and remains to be discovered. Recent studies suggest a role for CDI beyond bacterial competition in collective behavior between sibling immune cells. In P. aeruginosa, the deletion of cdi locus increases the production of cyanide and swarming motility potentially via post-transcriptional regulation (Melvin et al., 2017). Burkholderia uses the contact-dependent signaling (CDS) mechanism to modulate gene expression in immune recipient bacteria that are dependent on the activity of the CdiA-CT toxin but independent of CDI growth inhibitory function (Garcia et al., 2016); and in E. coli, CDI modulates the cellular (p)ppGpp levels to increase the number of persister cells (Ghosh et al., 2018). It is therefore possible that the A. baumannii CDI1 system could also impact overall bacterial physiology by fine-tuning cellular responses.
Experimental Procedures
Growth Conditions, Strains, Plasmids, and Primers
Strains/plasmids and primers used in this study are listed in Supplementary Tables S1, S2, respectively. Acinetobacter baumannii and Escherichia coli strains were grown in LB, EZ-Rich Defined Medium (RDM), or M9 medium containing 0.2% glucose and 0.4% casamino acid (M9-casa) supplemented with appropriate antibiotics: 30 μg/ml of chloramphenicol (Cm), 50 μg/ml of ampicillin (Amp), 50 μg/ml of kanamycin (Kn), 15 μg/ml of gentamycin (Gm), and 50 (liquid) or 200 μg/ml (solid) of carbenicillin (Cb).
Strain Construction
Gene insertion in the E. coli chromosome was performed by λRed recombination (Datsenko and Wanner, 2000; Yu et al., 2000). Mutant alleles were transferred by phage P1 transduction to generate the final strains. The kan and cat genes were removed using site-specific recombination induced by expression of the Flp recombinase from plasmid pCP20 (Datsenko and Wanner, 2000).
cdiBAI1 mutants were constructed following the Tucker et al. (2014) protocol. Briefly, the FRT (Flippase Recognition Target) site-flanked kanamycin resistance cassette was amplified from the pKD4 plasmid with primers containing 100-nt extension with homology to the flanking regions of cdiBAI locus. After the PCR product was electroporated into A. baumannii competent cells carrying pAT02 plasmid, which produces the RecAb recombinase, mutants were selected on Kn 50 μg/ml, and the presence of integrated kanamycin cassette was verified by PCR. The kan gene was removed using site-specific recombination induced by expression of the Flp recombinase from plasmid pFLP2 (Hoang et al., 2000).
cdiA1 and cdiB1 mutants were constructed by amplifying 2 kb with homology to the flanking regions of the genes. PCR products were combined by overlapping extension PCR and cloned into pUC18T-mini-Tn7-Ap SacI/BamHI. The apramycin (Apr) resistance cassette was then amplified from pMHL2-2 and cloned between the 2-kb-flanking regions with NcoI/SacI restriction site. Then, the 2-kb-flanked apramycin cassette was amplified by PCR and electroporated into A. baumannii competent cells, and mutants were selected on Apr 50 μg/ml. The presence of integrated cassette was verified by PCR.
Toxicity Assays in E. coli
CdiA-CT domains were cloned with the artificial Shine-Dalgarno sequence into the pBAD33 plasmid using SacI and SalI. The cdiI genes were PCR amplified with a reverse primer encoding a 6xHis C-terminal tag and cloned into the pTrc99a plasmid using NcoI and BamHI. Plasmid cloning was verified by Sanger sequencing (GATC Biotech). To perform toxicity assay, an overnight culture in LB with 0.5% glucose was washed in LB and diluted to an OD600 ∼ 0.05 in LB with 100 μM of IPTG and 1% arabinose to produce the immunity protein and the CdiA-CT domain, respectively. At indicated time of culture at 37°C, cells were washed in LB, and CFU/ml was calculated by plating onto LB agar plates containing Cm, Amp, and glucose 0.5%. The production of immunity proteins was verified by Western blot analyses. Briefly, the cell extract was separated on 12% SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes, and probed with primary mouse anti-pentaHis (Qiagen) or anti EF-Tu.
Live-Cell Microscopy Experiment
Live and Dead Assay
Overnight cultures in RDM supplemented with 0.5% glucose were washed and diluted to OD600 of 0.05 with 100 μM of IPTG and 1% arabinose and grown further at 37°C for 3 h 30 min before treatment with the LIVE/DEAD® BacLightTM Bacterial Viability Kit. To control the functionality of the Live/Dead assay, we treated the samples with ethanol to kill the bacteria, and as expected, the percentage of cell death reaches ∼100% in this condition (Supplementary Figure S2B). Cells were stained for 20 min, washed, resuspended in RDM, and spread over a RDM or M9-casa 1% agarose pad.
Time-Lapse Experiments
Overnight cultures in RDM or M9-casa media supplemented with 0.5% glucose were diluted to OD600 of 0.05 with 0.5% glucose and grown further to OD600 = 0.1. Cultures were loaded to B04A microfluidic chamber (ONIX, CellASIC®) with 5 psi for 1 min. Medium supplemented with 1% arabinose was maintained at 1 psi with a constant temperature of 37°C. Cells were imaged every 10 min for 6 h.
Image Acquisition and Analysis
Conventional wide-field fluorescence microscopy imaging was carried out on an Eclipse Ti-E microscope (Nikon), equipped with ×100/1.45 oil Plan Apo Lambda phase objective, FLash4 V2 CMOS camera (Hamamatsu), and using NIS software for image acquisition. Acquisition setting was 10 ms for Syto9, 100 ms for GFP, 10 ms for propidium iodide, and 100 ms for mCherry, using 50% power of a fluo LED Spectra X light source at 488- and 560-nm excitation wavelengths, respectively. Cell counting and length analysis were performed using MicrobeJ (Ducret et al., 2016).
Quantification of Promoter Activities
The Pempty transcriptional fusion was constructed by cloning the gfpmut2 gene from pUA66 plasmid as an EcoRI/BglII PCR fragment cloned into pWH1266. To construct transcriptional fusions, 500 bp corresponding to the putative promoter regions was amplified as a BamHI/SacI PCR fragment and cloned into pWH1266-Pempty-gfp. To quantify promoter activities, overnight cultures in LB were diluted to an OD600 of 0.05 and grown at 37°C for 6 h. Two hundred microliters of cells was transferred into well of a black 96-well plate (Greiner), and the absorbance at 600 nm and fluorescence (excitation 485 nm, emission 530 nm) were measured using TECAN Spark multimode plate reader. The relative fluorescence was expressed as the intensity of fluorescence divided by the absorbance at 600 nm after subtracting the values of blank sample. Fluorescence repartition of the PcdiB1-gfp was performed by diluting an overnight culture in RDM to an OD600 of 0.05 and grown further at 37°C to an OD600 ∼ 0.8 before spreading the cells over a RDM 1% agarose pad. The analysis was performed using MicrobeJ (Ducret et al., 2016).
Detection of CdiA1
Cell extract and secreted fractions were prepared as follows. Bacterial cells from overnight culture grown in LB were harvested by centrifugation at 4,000 × g for 20 min, and pellets (cell extracts) were resuspended in loading buffer. The supernatant fraction was centrifuged at 13,000 × g for 20 min at 4°C. Proteins from the supernatant (secreted fraction) were then precipitated with 12% (w/v) trichloroacetic acid (TCA), washed with acetone, air-dried, and resuspended in loading buffer. The protein samples were then heated to 95°C for 10 min, separated by SDS-PAGE, and revealed by Coomassie blue staining or immunoblotting.
Growth Competition Assays
The attackers and target strains were grown overnight separately in LB medium without antibiotic at 37°C. Overnight cultures were diluted to an OD600 of 0.05 in LB medium without antibiotic and grown at 37°C with shaking to an OD600 ∼ 0.35. Strains were then mixed at an attacker to target cell ratio of 4:1, and the mixed bacteria were grown at 37°C with agitation at 160 rpm. At indicated times, the mix was serially diluted and plated onto kanamycin-containing LB agar plates to determine the CFU/ml of the target strain.
Biofilm Assays
Biofilm assays were performed following the O’Toole (2011) protocol. Briefly, overnight cultures were inoculated into LB at a final OD600 of 0.1. One hundred microliters of culture was added to 96-well polystyrene plates and incubated over a 5-h period without shaking. At the indicated times, cells in suspension were removed, and the wells washed twice with distilled water. One hundred twenty-five microliters of 0.1% crystal violet was then added to the wells, incubated for 10 min, and washed three times with 125 μl of distilled water. To solubilize the crystal violet, 125 μl of 30% acetic acid was added and the absorbance measured at 550 nm. The ratio OD550/OD600 was calculated to normalize the biofilm formation due to variation in bacterial growth.
For biofilm imaging, bacteria were grown in BM2 minimal medium (Repizo et al., 2015) supplemented with 10 mM of potassium glutamate (BM2G) as carbon source in a glass-bottom 24-well μ-plate. Cultures were inoculated at an initial OD600 of 0.1 from an overnight culture grown in LB and then incubated at 37°C under static conditions. At indicated times, the biofilm was labeled with DAPI, and images were taken with a confocal Zeiss LSM800 Airyscan microscope and analyzed with ImageJ and Imaris software.
A. baumannii Infection of A549 Cells
A549 cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% of fetal calf serum and 1% L-glutamine at 37°C with 5% CO2 atmosphere. For adhesion assays and microscopy analysis, A549 cells were first seeded into 24-well tissue culture plates at 5 × 105 cells/well to obtain a monolayer. A. baumannii strains were grown on LB agar plate for 24 h at 37°C to form a “lawn” covering. Bacteria were scraped from the agar surface and resuspended in 1 ml of LB to form a homogeneous suspension of ∼108 CFU/ml. Cells were then infected at a multiplicity of infection (MOI) of 100 of A. baumannii in 500 μl of complete medium per well. Plates were centrifuged at 400 × g for 5 min and then incubated for 2 h at 37°C with 5% CO2 atmosphere. Cells were then washed five times with phosphate-buffered saline (PBS) 1 × and either lysed with 0.1% sodium deoxycholate (5-min incubation) or fixed with 3% paraformaldehyde (15 min incubation). The number of CFU/ml before and after infection was determined to calculate the percentage of bacterial adhesion. The number of CFU/ml of the inocula was also used to verify the MOI and ensure equivalent numbers of bacteria were used for wt and mutant strains.
Immunofluorescence Labeling and A549 Cell Microscopy
After fixation, cells were incubated at room temperature for 1 h in a PBS 1 × 0.1% saponin and 10% horse serum solution for permeabilization and blocking. Cells were then labeled at room temperature with primary rabbit anti-Acinetobacter antibody mix diluted at 1/20,000 in the same solution for 1 h followed by two washes in PBS 1 × 0.1% saponin. Secondary antibodies (anti-rabbit Alexa-488 [1/1,000], phalloidin-568 [1/250], and DAPI nuclear dye [1/1,000] to label A. baumannii, actin cytoskeleton, and host cell nucleus, respectively) were then mixed and incubated for 45 min, followed by two washes in PBS 1 × 0.1% saponin, one wash in PBS 1×, and one wash in distilled water before mounting with ProLong Gold. Coverslips were examined on a Zeiss LSM800 laser scanning confocal microscopes and analyzed with ImageJ software (Schindelin et al., 2012). Z-stack tool and Z-projection for maximum intensity were used for image presentation.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MR performed the majority of the experiments. SR, LC, JC, and CL also conducted the experiments. MR, SS, and CL participated in the experimental design and data analysis. SB conceived the project, designed and undertook experiments, interpreted data, and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by a FINOVI young principal investigator grant awarded to SS. MR has a Ph.D. fellowship from the French Ministry of Research. JC was funded by the FINOVI Foundation in partnership to the ATIP-Avenir grant to CL. SS and CL was supported by an INSERM staff scientist contract. SB was supported by the CNRS staff scientist contract.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank E. Gueguen at the MAP, University of Lyon, for helpful discussions and support and for providing pUA66 plasmid. We are grateful to B. Davies at the University of Texas at Austin for providing the pAT02 plasmid, to X. Charpentier at the CIRI (INSERM, University of Lyon, CNRS, ENS) for the pMHL2-2 plasmid, and to R. Voulhoux at the CNRS-Aix-Marseille University for the anti EF-Tu antibody. We thank the proteomic facility of the Protein Science Facility of the SFR Biosciences for the mass spectrometry.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02450/full#supplementary-material
References
Allsopp, L. P., Wood, T. E., Howard, S. A., Maggiorelli, F., Nolan, L. M., Wettstadt, S., et al. (2017). RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, 7707–7712. doi: 10.1073/pnas.1700286114
Anderson, M. S., Garcia, E. C., and Cotter, P. A. (2012). The burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 8:e1002877. doi: 10.1371/journal.pgen.1002877
Aoki, S. K., Diner, E. J., de Roodenbeke, C., Burgess, B. R., Poole, S. J., Braaten, B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442. doi: 10.1038/nature09490
Aoki, S. K., Pamma, R., Hernday, A. D., Bickham, J. E., Braaten, B. A., and Low, D. A. (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. doi: 10.1126/science.1115109
Aoki, S. K., Webb, J. S., Braaten, B. A., and Low, D. A. (2009). Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J. Bacteriol. 191, 1777–1786. doi: 10.1128/JB.01437-1438
Balder, R., Hassel, J., Lipski, S., and Lafontaine, E. R. (2007). Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect. Immun. 75, 2765–2775. doi: 10.1128/IAI.00079-77
Braun, V., Hobbie, S., and Ondraczek, R. (1992). Serratia marcescens forms a new type of cytolysin. FEMS Microbiol. Lett. 100, 299–305. doi: 10.1016/0378-1097(92)90225-d
Carruthers, M. D., Nicholson, P. A., Tracy, E. N., and Munson, R. S. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388
Cope, L. D., Thomas, S. E., Hrkal, Z., and Hansen, E. J. (1998). Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66, 4511–4516.
Cope, L. D., Thomas, S. E., Latimer, J. L., Slaughter, C. A., Müller-Eberhard, U., and Hansen, E. J. (1994). The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13, 863–873. doi: 10.1111/j.1365-2958.1994.tb00478.x
Cope, L. D., Yogev, R., Muller-Eberhard, U., and Hansen, E. J. (1995). A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177, 2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995
Darvish Alipour Astaneh, S., Rasooli, I., and Mousavi Gargari, S. L. (2014). The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606(T). Microb. Pathog. 74, 42–49. doi: 10.1016/j.micpath.2014.07.007
Datsenko, K., and Wanner, B. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Gregorio, E., Zarrilli, R., and Di Nocera, P. P. (2019). Contact-dependent growth inhibition systems in acinetobacter. Sci. Rep. 9:154. doi: 10.1038/s41598-018-36427-36428
Ducret, A., Quardokus, E. M., and Brun, Y. V. (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1:16077. doi: 10.1038/nmicrobiol.2016.77
Faure, L. M., Garvis, S., de Bentzmann, S., and Bigot, S. (2014). Characterization of a novel two-partner secretion system implicated in the virulence of Pseudomonas aeruginosa. Microbiol. Read. Engl. 160, 1940–1952. doi: 10.1099/mic.0.079616-79610
Faure, L. M., Llamas, M. A., Bastiaansen, K. C., de Bentzmann, S., and Bigot, S. (2013). Phosphate starvation relayed by PhoB activates the expression of the Pseudomonas aeruginosa σvreI ECF factor and its target genes. Microbiol. Read. Engl. 159, 1315–1327. doi: 10.1099/mic.0.067645-67640
Fournier, C., Smith, A., and Delepelaire, P. (2011). Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity: HxuA releases haem from haemopexin. Mol. Microbiol. 80, 133–148. doi: 10.1111/j.1365-2958.2011.07562.x
Gallagher, L. A., and Manoil, C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001
Garcia, E. C., Anderson, M. S., Hagar, J. A., and Cotter, P. A. (2013). BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition: CDI system protein-mediated biofilm. Mol. Microbiol. 89, 1213–1225. doi: 10.1111/mmi.12339
Garcia, E. C., Perault, A. I., Marlatt, S. A., and Cotter, P. A. (2016). Interbacterial signaling via burkholderia contact-dependent growth inhibition system proteins. Proc. Natl. Acad. Sci. U.S.A. 113, 8296–8301. doi: 10.1073/pnas.1606323113
Ghosh, A., Baltekin, Ö, Wäneskog, M., Elkhalifa, D., Hammarlöf, D. L., Elf, J., et al. (2018). Contact-dependent growth inhibition induces high levels of antibiotic-tolerant persister cells in clonal bacterial populations. EMBO J. 37:e98026. doi: 10.15252/embj.201798026
Gottig, N., Garavaglia, B. S., Garofalo, C. G., Orellano, E. G., and Ottado, J. (2009). A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PloS One 4:e4358. doi: 10.1371/journal.pone.0004358
Guédin, S., Willery, E., Locht, C., and Jacob-Dubuisson, F. (1998). Evidence that a globular conformation is not compatible with FhaC-mediated secretion of the Bordetella pertussis filamentous haemagglutinin. Mol. Microbiol. 29, 763–774. doi: 10.1046/j.1365-2958.1998.00970.x
Guérin, J., Bigot, S., Schneider, R., Buchanan, S. K., and Jacob-Dubuisson, F. (2017). Two-partner secretion: combining efficiency and simplicity in the secretion of large proteins for bacteria-host and bacteria-bacteria interactions. Front. Cell. Infect. Microbiol. 7:148. doi: 10.3389/fcimb.2017.00148
Guilhabert, M. R., and Kirkpatrick, B. C. (2005). Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute a biofilm maturation to X. fastidios and colonization and attenuate virulence. Mol. Plant Microb. Interact. MPMI 18, 856–868. doi: 10.1094/MPMI-18-0856
Harding, C. M., Pulido, M. R., Di Venanzio, G., Kinsella, R. L., Webb, A. I., Scott, N. E., et al. (2017). Pathogenic acinetobacter species have a functional type i secretion system and contact-dependent inhibition systems. J. Biol. Chem. 292, 9075–9087. doi: 10.1074/jbc.M117.781575
Hoang, T. T., Kutchma, A. J., Becher, A., and Schweizer, H. P. (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72. doi: 10.1006/plas.1999.1441
Iyer, L. M., Zhang, D., Rogozin, I. B., and Aravind, L. (2011). Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 39, 9473–9497. doi: 10.1093/nar/gkr691
Jacob-Dubuisson, F., Buisine, C., Willery, E., Renauld-Mongénie, G., and Locht, C. (1997). Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J. Bacteriol. 179, 775–783. doi: 10.1128/jb.179.3.775-783.1997
Jacob-Dubuisson, F., Locht, C., and Antoine, R. (2001). Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40, 306–313. doi: 10.1046/j.1365-2958.2001.02278.x
Kajava, A. V., Cheng, N., Cleaver, R., Kessel, M., Simon, M. N., Willery, E., et al. (2001). Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42, 279–292. doi: 10.1046/j.1365-2958.2001.02598.x
Lesterlin, C., Ball, G., Schermelleh, L., and Sherratt, D. J. (2014). RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature 506, 249–253. doi: 10.1038/nature12868
Melvin, J. A., Gaston, J. R., Phillips, S. N., Springer, M. J., Marshall, C. W., Shanks, R. M. Q., et al. (2017). Pseudomonas aeruginosa contact-dependent growth inhibition plays dual role in host-pathogen interactions. mSphere 2, e336–e317. doi: 10.1128/mSphere.00336-317
Mercy, C., Ize, B., Salcedo, S. P., de Bentzmann, S., and Bigot, S. (2016). Functional characterization of Pseudomonas contact dependent growth inhibition (CDI) systems. PLoS One 11:e0147435. doi: 10.1371/journal.pone.0147435
Neil, R. B., and Apicella, M. A. (2009). Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect. Immun. 77, 2285–2293. doi: 10.1128/IAI.01502-1508
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47, e2437. doi: 10.3791/2437
Pérez, A., Merino, M., Rumbo-Feal, S., Álvarez-Fraga, L., Vallejo, J. A., Beceiro, A., et al. (2016). The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 8, 959–974. doi: 10.1080/21505594.2016.1262313
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/AAC.01464-1466
Plamondon, P., Luke, N. R., and Campagnari, A. A. (2007). Identification of a novel two-partner secretion locus in Moraxella catarrhalis. Infect. Immun. 75, 2929–2936. doi: 10.1128/IAI.00396-397
Relman, D. A., Domenighini, M., Tuomanen, E., Rappuoli, R., and Falkow, S. (1989). Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. U.S.A. 86, 2637–2641. doi: 10.1073/pnas.86.8.2637
Repizo, G. D., Gagné, S., Foucault-Grunenwald, M.-L., Borges, V., Charpentier, X., Limansky, A. S., et al. (2015). Differential Role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265
Repizo, G. D., Viale, A. M., Borges, V., Cameranesi, M. M., Taib, N., Espariz, M., et al. (2017). The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol. Evol. 9, 2292–2307. doi: 10.1093/gbe/evx162
Rojas, C. M., Ham, J. H., Deng, W.-L., Doyle, J. J., and Collmer, A. (2002). HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. U.S.A. 99, 13142–13147. doi: 10.1073/pnas.202358699
Ruhe, Z. C., Townsley, L., Wallace, A. B., King, A., Van der Woude, M. W., Low, D. A., et al. (2015). CdiA promotes receptor-independent intercellular adhesion. Mol. Microbiol. 98, 175–192. doi: 10.1111/mmi.13114
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schmitt, C., Turner, D., Boesl, M., Abele, M., Frosch, M., and Kurzai, O. (2007). A functional two-partner secretion system contributes to adhesion of Neisseria meningitidis to epithelial cells. J. Bacteriol. 189, 7968–7976. doi: 10.1128/JB.00851-857
Talà, A., Progida, C., De Stefano, M., Cogli, L., Spinosa, M. R., Bucci, C., et al. (2008). The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell. Microbiol. 10, 2461–2482. doi: 10.1111/j.1462-5822.2008.01222.x
Tucker, A. T., Nowicki, E. M., Boll, J. M., Knauf, G. A., Burdis, N. C., Trent, M. S., et al. (2014). Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5, e1313–e1314. doi: 10.1128/mBio.01313-1314
van Ulsen, P., Rahman, S. U., Jong, W. S. P., Daleke-Schermerhorn, M. H., and Luirink, J. (2014). Type V secretion: from biogenesis to biotechnology. Biochim. Biophys. Acta 1843, 1592–1611. doi: 10.1016/j.bbamcr.2013.11.006
Willett, J. L. E., Ruhe, Z. C., Goulding, C. W., Low, D. A., and Hayes, C. S. (2015). Contact-dependent growth inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J. Mol. Biol. 427, 3754–3765. doi: 10.1016/j.jmb.2015.09.010
Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G., and Court, D. L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983. doi: 10.1073/pnas.100127597
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M., and Aravind, L. (2012). Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 7:18. doi: 10.1186/1745-6150-7-18
Keywords: type V secretion system, contact-dependent growth inhibition, Acinectobacter baumannii, biofilm, cell adhesion ability
Citation: Roussin M, Rabarioelina S, Cluzeau L, Cayron J, Lesterlin C, Salcedo SP and Bigot S (2019) Identification of a Contact-Dependent Growth Inhibition (CDI) System That Reduces Biofilm Formation and Host Cell Adhesion of Acinetobacter baumannii DSM30011 Strain. Front. Microbiol. 10:2450. doi: 10.3389/fmicb.2019.02450
Received: 25 July 2019; Accepted: 11 October 2019;
Published: 30 October 2019.
Edited by:
Ignacio Arechaga, University of Cantabria, SpainReviewed by:
Alfonso Soler-Bistue, CONICET Institute of Biotechnological Research (IIB-INTECH), ArgentinaMarco Maria D’Andrea, University of Rome Tor Vergata, Italy
Copyright © 2019 Roussin, Rabarioelina, Cluzeau, Cayron, Lesterlin, Salcedo and Bigot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzana P. Salcedo, c3V6YW5hLnNhbGNlZG9AaWJjcC5mcg==; Sarah Bigot, c2FyYWguYmlnb3RAaWJjcC5mcg==
†These authors share last authorship
 Morgane Roussin
Morgane Roussin Sedera Rabarioelina1
Sedera Rabarioelina1 Suzana P. Salcedo
Suzana P. Salcedo Sarah Bigot
Sarah Bigot