- 1Departamento de Microbiologia, Facultad de Ciencias Bioquimicas y Farmaceuticas, Instituto de Biologia Molecular y Celular de Rosario (IBR, CONICET), Universidad Nacional de Rosario, Rosario, Argentina
- 2Laboratory of Molecular Microbiology and Structural Biochemistry, CNRS UMR 5086, University of Lyon, Lyon, France
Several Acinetobacter strains are important nosocomial pathogens, with Acinetobacter baumannii as the species of greatest concern worldwide due to its multi-drug resistance and recent appearance of hyper-virulent strains in the clinical setting. Acinetobacter colonization of the environment and the host is associated with a multitude of factors which remain poorly characterized. Among them, the secretion systems (SS) encoded by Acinetobacter species confer adaptive advantages depending on the niche occupied. Different SS have been characterized in this group of microorganisms, including T6SS used by several Acinetobacter species to outcompete other bacteria and in some A. baumannii strains for Galleria mellonella colonization. Therefore, to better understand the distribution of the T6SS in this genus we carried out an in-depth comparative genomic analysis of the T6SS in 191 sequenced strains. To this end, we analyzed the gene content, sequence similarity, synteny and operon structure of each T6SS loci. The presence of a single conserved T6SS-main cluster (T6SS-1), with two different genetic organizations, was detected in the genomes of several ecologically diverse species. Furthermore, a second main cluster (T6SS-2) was detected in a subgroup of 3 species of environmental origin. Detailed analysis also showed an impressive genetic versatility in T6SS-associated islands, carrying VgrG, PAAR and putative toxin-encoding genes. This in silico study represents the first detailed intra-species comparative analysis of T6SS-associated genes in the Acinetobacter genus, that should contribute to the future experimental characterization of T6SS proteins and effectors.
Introduction
The Acinetobacter genus comprises a heterogeneous group of strictly aerobic Gram-negative bacterial organisms endowed with great metabolic versatility. This genus includes numerous non-pathogenic environmental species as well as some with significant pathogenic potential, notably Acinetobacter baumannii, frequently associated with disease in the context of hospital-acquired infections (McConnell et al., 2013). Although infections due to multi-drug resistant (MDR) Acinetobacter species are a serious health threat worldwide, the knowledge about the mechanisms that enable them to colonize the host and hospital environments is still scarce (Antunes et al., 2014; Harding et al., 2018).
Bacteria use several secretory mechanisms to export effector proteins into the environment or straight into target cells (Costa et al., 2015; Galán and Waksman, 2018). Among them, the multicomponent type VI secretion system (T6SS), which is structurally related to the cell-puncturing device of the T4 bacteriophage, was described in Gram-negative bacteria, including Acinetobacter (Weber et al., 2017; Elhosseiny and Attia, 2018). The T6SS dynamic machinery permits the injection of toxic effector proteins into prey cells in a contact-dependent manner. The concomitant expression of cognate immunity proteins prevents self-intoxication (Benz and Meinhart, 2014; Alcoforado et al., 2015). T6SS effectors with diverse enzymatic activities have been identified, including the membrane-, cell wall-, or nucleic acid-targeting antibacterial effectors and the heterogenous group of eukaryote-targeting effectors (Lien and Lai, 2017).
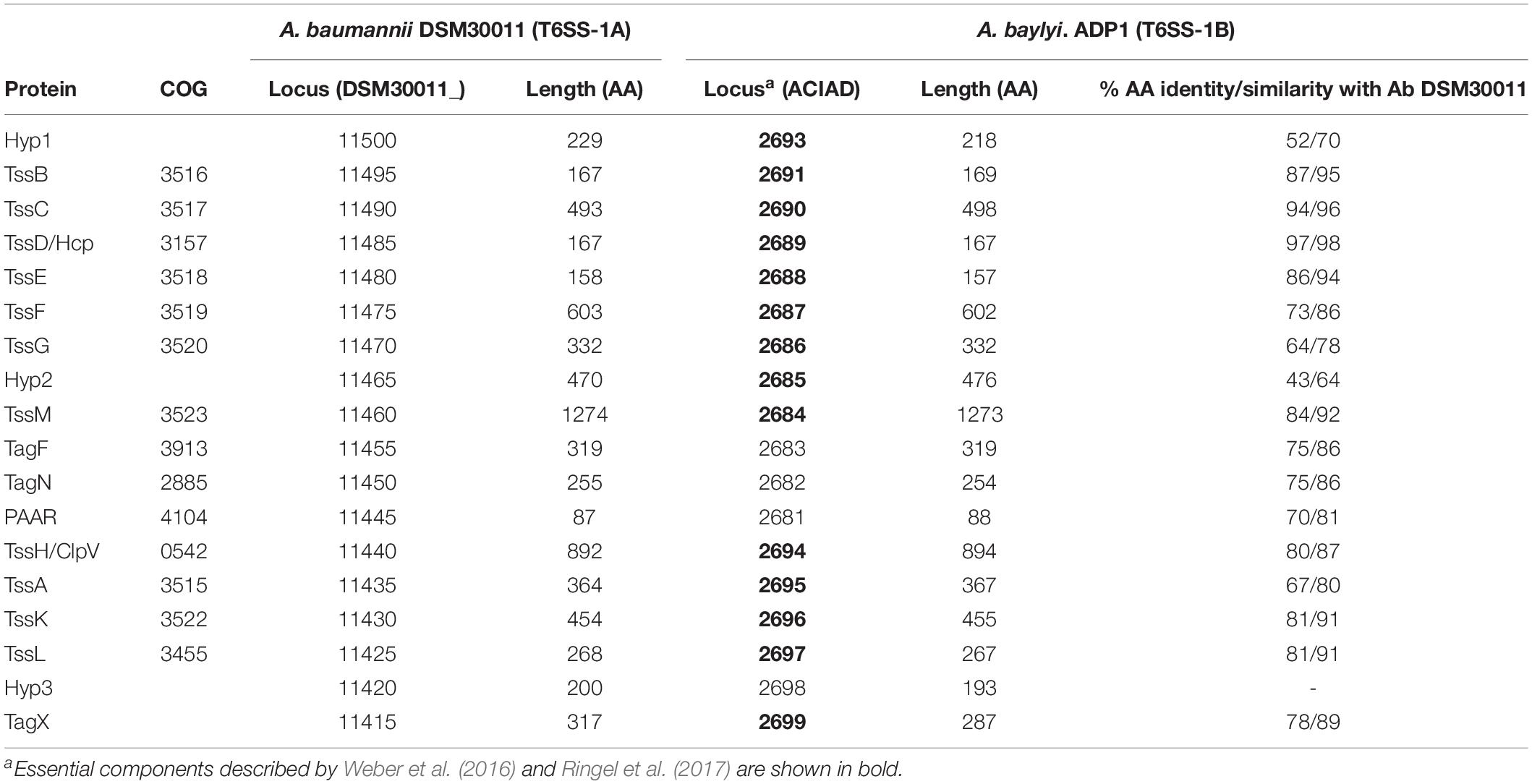
The T6SS apparatus is assembled from a set of core components proteins (Boyer et al., 2009), which comprise the minimal machinery necessary for its functionality. The genes coding for the core components (tss) are frequently organized within one genetic cluster. Most T6SS gene clusters contain additional genes (T6SS-associated genes; tag), the function of most remains unknown. Previous studies using a limited number of Acinetobacter species predicted the presence of a conserved T6SS gene cluster with two different genetic organizations (Weber et al., 2013). This locus, hereafter referred as T6SS main cluster (T6MC; Figure 1A and Table 1), is described to encompass 18 genes (Carruthers et al., 2013; Weber et al., 2013). Fourteen genes showed a significant homology at the protein level with T6SS components present in other bacteria, and were thus initially named following the nomenclature proposed by Shalom et al. (2007), namely tss and tag genes. The only gene that does not follow this nomenclature is that encoding a PAAR-domain protein, which has been accordingly dubbed just as PAAR gene (Weber et al., 2016). It was recently shown in A. baylyi ADP1 that one of the four genes with non-attributable function (ACIAD2699) coded for a peptidoglycan hydrolase (TagX), facilitating the passage of the T6SS apparatus through the cell wall (Weber et al., 2016). Despite the high levels of homology observed between most T6MCs, the genes encoding the VgrG and PAAR-repeat domain proteins (usually more than 1 of each per genome) are scattered throughout the genomes and conservation between Acinetobacter strains is low (Eijkelkamp et al., 2014). The regions flanking these genes are known as VgrG and PAAR islands (De Maayer et al., 2011) and represent evolutionary hot spots for genes that encode effector proteins. Genes encoding the cognate immunity proteins are usually present in these regions as well. These regions account for strain specificity regarding T6SS-related components (Unterweger et al., 2014).
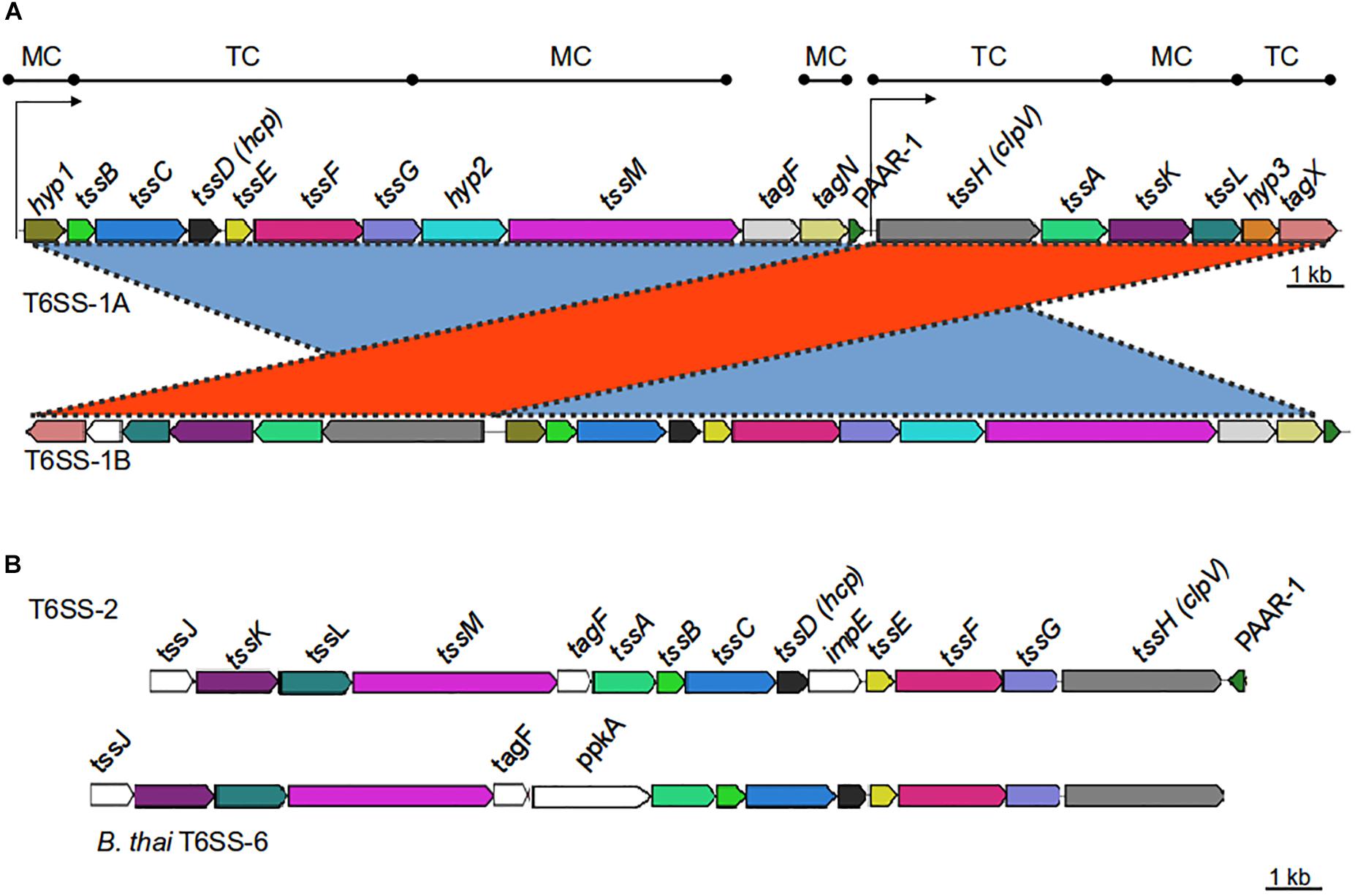
Figure 1. Genetic organization of T6SS-main loci in the Acinetobacter genus. (A) T6SS-1-main gene cluster (for details refer to Table 1). Gene products are classified according to their participation in the membrane complex (MC) or tail complex (TC) of the T6SS apparatus. Thin arrows represent putative promoter regions. (B) T6SS-2-main gene cluster (for details refer to Table 4). B. thailandiensis T6SS-6 main cluster is shown for comparison. Genes encoding proteins with amino acidic identity > 25% and query coverage > 50% were represented with the same color.
A number of VgrG proteins have been characterized in A. baumannii (Weber et al., 2016; Fitzsimons et al., 2018). They all share a common domain structure (Figure 2A), with the typical N-terminal part constituted by domains commonly found in the bacteriophagic components gp44 and gp5, which altogether compose the VgrG domain (COG3501) defining the VgrG superfamily (cl27827). This domain is followed by a DUF2345 domain (pfam10106, superfamily cl27827) close to the C-terminus, responsible for the binding of the effector (Flaugnatti et al., 2016), and a C-terminal stretch of variable aminoacidic composition and length, which could even be totally absent. In Vibrio cholerae, the C-terminal portion of a number of these proteins, known as evolved VgrGs, displays an additional domain that functions itself as an effector (Pukatzki et al., 2007; Ma and Mekalanos, 2010).
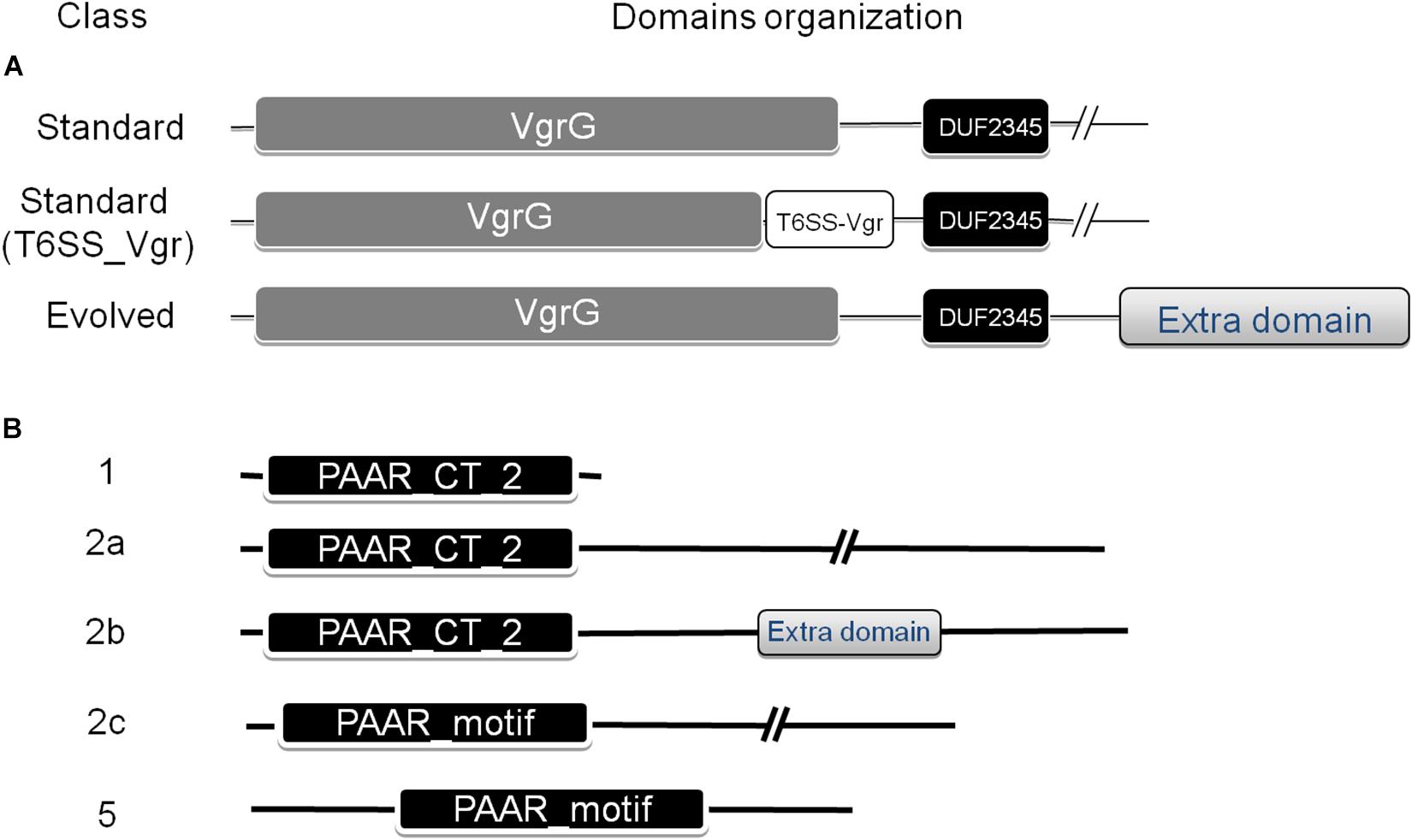
Figure 2. Domain architecture of VgrG (A) and PAAR repeat proteins (B) found in the Acinetobacter genus. The NCBI-CDD (Marchler-Bauer et al., 2015) was used for the domain search. The classification defined by Shneider et al. (2013), which categorized PAAR proteins into seven classes based on domain architecture, was used. Additional subclasses were defined for PAAR-2 proteins (see text for details).
Accessory proteins such as the PAAR-repeat domain protein are also part of most T6SS and have been shown to play a significant role in T6SS-mediated bacterial competition in A. baylyi (Shneider et al., 2013). The PAAR-repeat domain was found to dock onto the terminus of the pilus-like structure formed by VgrG and have been shown to interact with VgrGs and effector proteins, thus functioning as adaptors (Bondage et al., 2016).
The PAAR_like domain superfamily (cl21497; NCBI conserved domains database, CDD) includes proteins bearing PAAR motifs (pfam05488) or the DUF4150 domain (pfam13665). Tandemly repeated PAAR motifs constitute a PAAR domain (cd14671), which has been subclassified in 8 subgroups (cd14737–cd14744), according to the presence of additional N- and C-terminal domains with various predicted functions. Shneider et al. (2013) have classified PAAR-domain containing proteins in 7 classes according to their domain architecture. Following this scheme, Eijkelkamp et al. (2014) have previously detected the presence of PAAR proteins belonging to classes 1 (PAAR-1, N-terminal PAAR domain and no additional domains), 2 (PAAR-2, N-terminal PAAR domain and C-terminal extension of variable length) and 5 (PAAR-5, central PAAR domain) in a group of 8 A. baumannii strains (Figure 2B). As described for VgrG proteins, some PAAR proteins have evolved to contain virulence activity in their C-termini (“evolved PAAR”; Shneider et al., 2013).
T6SS involvement in eliminating competitors has been reported in a number of Acinetobacter species. A. nosocomialis M2 and A. baylyi ADP1 were found to utilize the T6SS for dueling against Escherichia coli (Carruthers et al., 2013; Shneider et al., 2013). In particular, A. baylyi ADP1 encodes 5 different effectors that provoke different degrees of E. coli cells lysis (Ringel et al., 2017), namely a putative metallopeptidase (Tpe1), a peptidoglycan-hydrolyzing amidase (Tae1), a phospholipase (Tle1), and two effectors (Tse1, and Tse2) representing new classes of effectors for which no enzymatic activity could be predicted or deduced from the lysis phenotype. Subsequent prey-DNA uptake has been detected, providing a competitive advantage to those strains expressing the T6SS (Ringel et al., 2017), and opening the possibility for the acquisition, among others, of antimicrobial resistance genes (Cooper et al., 2017). Moreover, A. baumannii strains DSM30011, ATCC17978 and AB307-0294 were able to outcompete other A. baumannii species as well as E. coli and P. aeruginosa, a bacterial pathogen relevant in mixed nosocomial infections (Weber et al., 2013; Repizo et al., 2015; Fitzsimons et al., 2018). Recently, toxins acting as peptidoglycan hydrolases (LysM), amidases (Tse4), nucleases (Rhs2 and Tse2), lipases (Tse1) and two with unknown function (Tse3 and Rhs1) have been characterized in A. baumannii (Weber et al., 2016; Fitzsimons et al., 2018), and many other effectors predicted using bioinformatics (Repizo et al., 2015; Fitzsimons et al., 2018), suggesting these microorganisms possess a significant arsenal of secreted toxins and immunity proteins.
In addition to an established role of the T6SS in bacterial competition, that could be advantageous in Acinetobacter colonization of specific environments such as the hospital setting, for some strains the T6SS seems to participate in host colonization, according to assays performed with the insect infection model Galleria mellonella (Gebhardt et al., 2015; Repizo et al., 2015). A recent report suggests that the presence of a functional T6SS contributes to infections in immune-compromised patients and those with implanted medical devices (Kim et al., 2017). In this work, the prevalence of the T6SS on 162 A. baumannii clinical isolates obtained from patients with bacteremia was also analyzed (Kim et al., 2017). The hcp gene was detected in 31.5% of the isolates and its presence showed a clear affiliation to particular STs. This observation is in agreement with genomic comparative analysis of phylogenetically- and epidemiologically-related A. baumannii MDR strains showing that the T6MC is only present in particular populations (Wright et al., 2014; Jones et al., 2015). Also, it suggests that this system is not critical in the conditions prevailing in the nosocomial environment. Indeed, the T6SS in A. baumannii is clearly tightly regulated, often repressed to promote conjugation and dissemination of MDR-carrying plasmids within a bacterial population (Di Venanzio et al., 2019).
In this study, we aimed at extending the present knowledge on T6SS core and accessory genes and toxins in the Acinetobacter genus. Therefore, we carried out an in-depth in silico comparative analysis of these clusters in different Acinetobacter species to better understand the composition and distribution of T6SS loci in this genus.
Materials and Methods
In silico Identification of the T6SS Loci
We conducted a comparative genomic analysis including Acinetobacter strains available the NCBI-GenBank database. Those strains with a complete genome sequence were included in this analysis. In order to have a full representation of the genus, for those Acinetobacter species with no complete genome we decided to work with draft genomes (Supplementary Table S1). We then extracted genomic and proteomic data corresponding to 191 strains and a local database was constructed. Of note, 110 genomes corresponded to A. baumannii strains and 81 to non-baumannii species, totalizing 42 accepted species and 9 strains so far not assigned to any known species. The proteins encoded within the T6MC from A. baumannii DSM30011 (Table 1) which carries a functional T6SS (Repizo et al., 2015) were used as query to perform BlastP-sequence similarity searches (Altschul et al., 1990) against the local database. With these data the corresponding T6SS loci were identified. Furthermore, each of the clusters was manually inspected to corroborate its genetic integrity, and those strains showing incomplete/non-functional clusters were accordingly informed (Supplementary Table S1). Visual representation of the alignments using nucleotide similarities (tblastx) of the T6SS loci were carried out with MultiGeneBlast (default parameters; Medema et al., 2013). Calculations of GC content1 and Codon adaptation index (CAI; coRdon) were performed in order to infer if studied regions were acquired by horizontal gene transfer. The CAI of each T6SS gene was normalized using the average CAI of all genes encoded in the corresponding genome. These values were compared with the CAIs obtained for those genes encoding ribosomal proteins in each case, generally accepted as highly adapted to the set of codons available in a particular genome. Statistical analysis (unpaired t-test) indicated for each of these genomes a significant difference between both data sets (p < 0.0001).
VgrG, PAAR, and Toxin-Proteins Search
A. baumannii DSM30011 encodes 4 VgrG proteins of variable length. However, they all share the basic domain architecture, with a conserved N-terminal VgrG domain. Therefore, the VgrG domain of the protein encoded by DSM30011_13325 (amino acids 23–631) was used as query for a bioinformatic search of homologous proteins in Acinetobacter strains contained in our local database. Regarding PAAR-proteins, a homology search was performed using as queries the PAAR-1 and PAAR-2 encoded by A. baumannii DSM30011 (DSM30011_11445 and DSM30011_14800), and the PAAR-5 encoded by A. baylyi ADP1 (ACIAD0051). Besides, representative proteins found at the NCBI-CDD were used as query in a BlastP-homology search for DUF4150-domain containing proteins. Finally, proteins acting as putative toxins in the vgrG and PAAR gene islands were searched with BlastP (e < 0.0001), using as query T6SS-reported toxins identified in Acinetobacter spp. (Weber et al., 2016; Ringel et al., 2017; Fitzsimons et al., 2018) and in other bacteria (Lien and Lai, 2017; Ma et al., 2018), both targeting prokaryotic and eukaryotic cells.
Annotation of Proteins With Unknown Function
A number of bioinformatic analyses were conducted in order to infer the role of T6SS-main components and proteins encoded in the hcp and vgrG islands with unknown function. A search for conserved domains was done by means of the Batch Web-CD search tool (Marchler-Bauer et al., 2015). SignalP 4.0 (Petersen et al., 2011) and TMHMM Server v.2.02 were used to predict signal peptides and trans-membrane helices, respectively.
Analysis of Genetic Context of PAAR and vgrG Islands
In order to analyze the genetic context of the PAAR (not located within T6MCs) and vgrG genes, the accession numbers of the 5 downstream and upstream genes were parsed using an R script designed ad hoc and the feature table files of the strains under analysis. Then, a search for conserved domains carried by proteins encoded in the PAAR and vgrG islands was done using the Batch Web-CD search tool (Marchler-Bauer et al., 2015), and each of them was assigned to a superfamily. Then, the presence or absence of a particular domain superfamily was used as a binary score (present = 1, absent = 0) to perform a hierarchical clustering of PAAR or vgrG islands using “pvcluster” R package (Suzuki and Shimodaira, 2006), with binary distance and average agglomerative clustering. Only genetic islands encoding proteins with conserved superfamily domains found at least 3 times in our database were considered. Clustering reliability was assessed by bootstrapping with 1,000 repetitions. The resulting dendrogram was displayed using iTOL (Letunic and Bork, 2011). Those gene islands sharing a similar genetic context were grouped in PAAR or VgrG gene neighborhoods (PGNs and VGNs, respectively).
Results and Discussion
T6SS Activity in Acinetobacter Strains
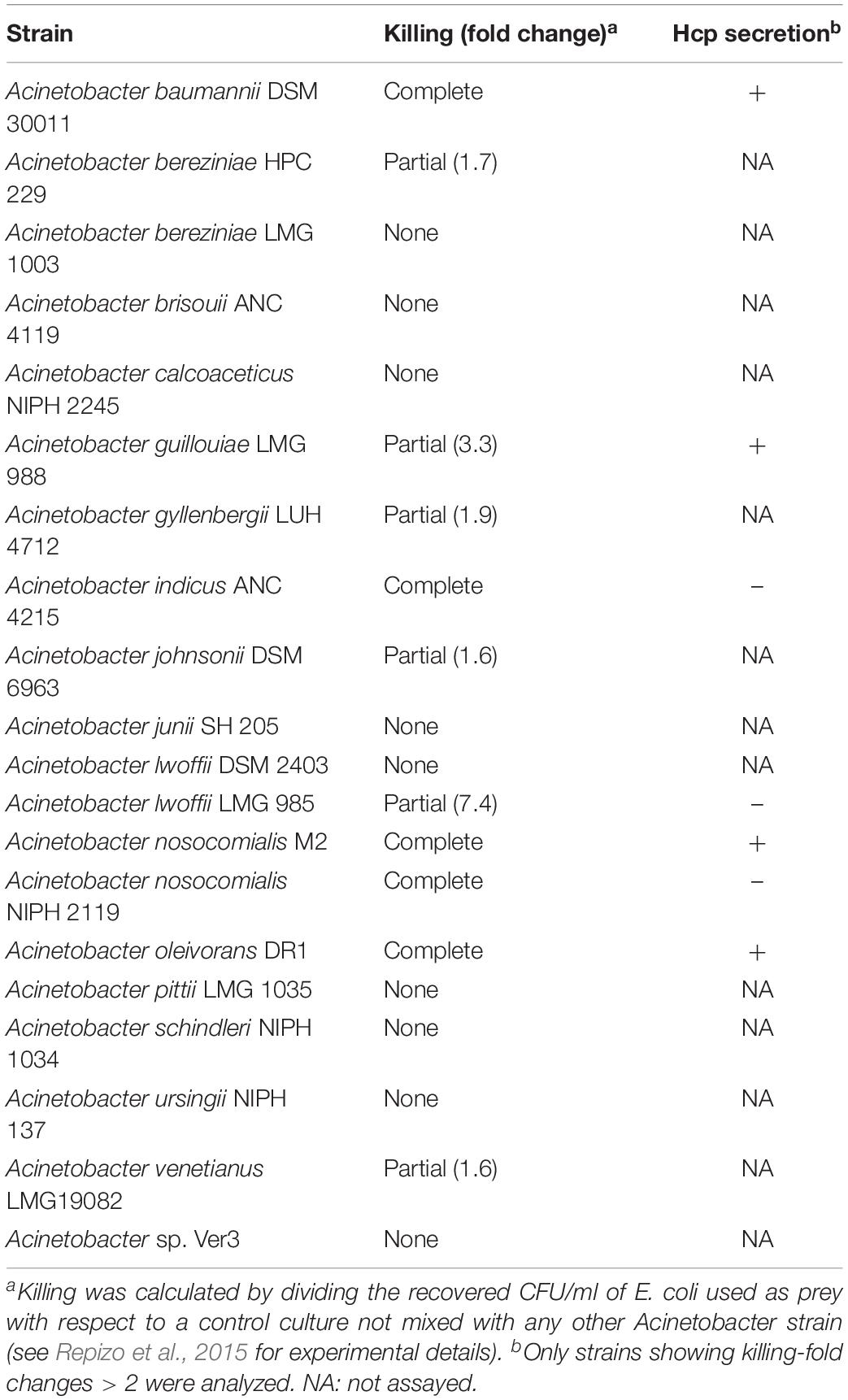
T6SS activity has been previously explored in a limited number of Acinetobacter spp. (see Introduction). In order to further investigate its function in a larger set of strains, a total number of 20 strains (Table 2) that were available in our laboratory collection were grown and tested. The ability of each Acinetobacter strain to eliminate E. coli was used as a proxy for an operative T6SS, as previously reported (Carruthers et al., 2013; Shneider et al., 2013; Weber et al., 2013; Repizo et al., 2015; Fitzsimons et al., 2018). Obtained results indicated that 11 out of 20 strains were able to outcompete E. coli (Table 2), of which A. guillouiae and A. oleivorans have never been reported as T6SS-positive strains. T6SS involvement in the observed killing was corroborated by Hcp-secretion assays (Supplementary Figure S1). In view of the observed variability in the killing capacity of different Acinetobacter spp. we decided to conduct a bioinformatic search that could shed light on the observed differences.
Identification and Prevalence of Orthologous T6SS Loci in the Acinetobacter Genus
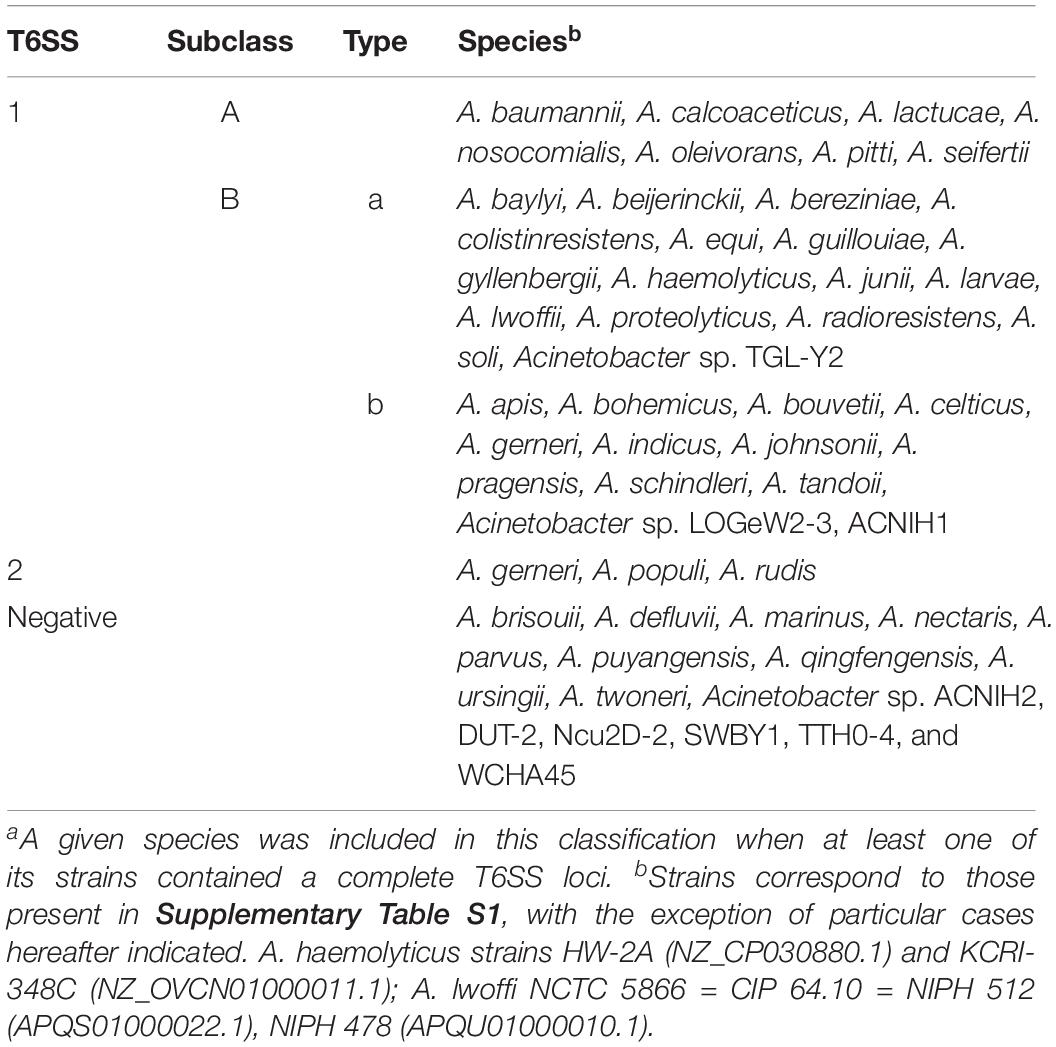
Bioinformatic analysis exerted on a local database including 191 Acinetobacter genomes (Supplementary Table S1) revealed the presence of genes encoding a T6SS in most Acinetobacter species (33 out of 42; Table 3). In some cases, such as A. lwoffi and A. haemolyticus, we were not able to detect a T6SS loci for those strains included in our local database. However, when performing a broader search against the complete NCBI-Protein database, some strains encoded a T6MC. Therefore, we decided to include these species in Table 3 as well.
It is worth noting the particular case of A. towneri DSM 14962 = CIP 107472, the only strain of this species which genome is available at the non-redundant protein database (NCBI). It encodes a complete T6SS-main cluster, but the tssH gene is interrupted by an insertion sequence (IS). No orphan copy of the tssH gene was detected, and thus A. towneri was thereby considered as a T6SS-deficient strain. Sequencing of other strains is needed to determine if this is the rule for this species. A number of Acinetobacter species (8 out of 42), namely A. brisouii, A. defluvii, A. marinus, A. nectaris, A. parvus, A. puyangensis, A. qingfengensis, and A. ursingii do not encode a T6MC (Supplementary Table S1). Lack of T6SS main genes was also observed for 6 out of 9 Acinetobacter spp. (ACNIH2, DUT-2, Ncu2D-2, SWBY1, TTH0-4, and WCHA45).
Regarding the prevalence of the T6MCs in A. baumannii, 27 out of 110 strains showed a complete deletion of the T6SS-main cluster, while 15 contained gene frameshifts, partial deletions or insertions resulting in an abnormal T6MC (Supplementary Table S1), very likely conducting to a non-functional apparatus. T6SS genes loss has been already documented in MDR A. baumannii clinical strains (Wright et al., 2014; Jones et al., 2015; Kim et al., 2017), suggesting that this system is not critical for survival in the nosocomial environment. Possible hypotheses favoring this genetic loss are higher chances of evasion from the host immune system and/or the lower requirement for interbacterial competition over the course of antibiotic therapy (Repizo, 2017). Furthermore, silencing of the T6SS was recently shown to be critical for horizontal gene transfer through conjugation, which is crucial for antimicrobial resistance spread (Di Venanzio et al., 2019).
Classification of the T6SS-Main Clusters in Acinetobacter Species
Bioinformatic analyses revealed the presence of two types of T6MC in Acinetobacter species, here designated as T6SS-1 and T6SS-2 (Figure 1 and Supplementary Figure S2, and Tables 1, 4). Different bacterial species encoding more than one T6MC have been previously reported (Boyer et al., 2009). These two loci differ in genetic composition but both encode the essential components of the T6SS apparatus. The TSS6-1 is the more ubiquitous T6MC in the Acinetobacter genus (30 out of 32 T6SS-proficient species; Table 3) and its gene composition is coincident with that reported for A. baumannii by Boyer et al. (2009). We established that the main characteristics that define the Acinetobacter T6SS-1 are: (1) No hcp genes are encoded outside the core cluster; (2) None of its genes code for evolved-Hcp proteins, as previously reported in other bacterial species (Blondel et al., 2009; Ma et al., 2017a); (3) no vgrG genes are encoded within the main cluster, in contrast to many other bacteria (Boyer et al., 2009).
A search for T6SS-1-core proteins in non-Acinetobacter species, using as query A. baumannii DSM30011 conserved components (TssB, TssC, TssK, TssL) indicated the presence of homologous proteins (identity > 45%) in members of several genera, namely Alkanindiges, Psychrobacter, Achromobacter, Chitinimonas, and Cupriavidus, and as previously noticed with those which are frequently associated to plants such as Ralstonia, Burkholderia, and Xanthomonas (Boyer et al., 2009; Spiewak et al., 2019). However, it is important to notice that the genetic architecture of T6SS-core clusters in these species is diverse. It has already been demonstrated that the T6SS mediates bacterial interactions with host plants, through the secretion of effectors which act in symbiosis, biofilm formation, virulence, and interbacterial competition (Ryu, 2015). Given the fact that several Acinetobacter species have been isolated from plants, including the enviromental isolate A. baumannii DSM30011 (Repizo et al., 2017), it is not surprising that they might have originally acquired T6SS-core genes from other plant associated bacteria and subsequently modeled these clusters to the actual genetic organization.
The T6SS loci encoded by most bacteria are organized in operons, suggesting a coregulated expression (Bernard et al., 2010). Therefore, we investigated the presence of putative transcriptional units in the A. baumannii T6SS-1 main cluster. According to bioinformatic predictions available at ProOpDb (Taboada et al., 2012), two transcriptional blocks might be present in these loci, the first encompassing hyp1-PAAR genes and the second including tssH-tagX genes (Figure 1A). The separation of this locus into two transcriptional units might explain the existence of two different genetic arrangements among T6SS-1 gene clusters (Figure 1A):
(i) T6SS-1A: In this case, all the T6MC genes are encoded in the same DNA strand. This genetic architecture is conserved in Acinetobacter species belonging or phylogenetically related to the calcoaceticus-baumannii complex (Table 3).
(ii) T6SS-1B: In this T6MC the tssH-tagZ gene block is divergently located to the hyp1-PAAR gene block (Figure 1A). This is the more ubiquitous organization (23 out of 32 species; Table 3) and for clarity, we further classified it in two types depending on the presence (T6SS-1Ba) or not (T6SS-1Bb) of the gene encoding a PAAR domain containing-protein within the locus. In line with this observation, the PAAR domain-protein encoded in the main cluster is not essential for T6SS functioning in A. baylyi (Weber et al., 2016). This is not surprising since more than one gene is usually present per Acinetobacter genome (see below).
We observed a strain clustering that was in line with the proposed sub-classification (Supplementary Figure S2 heat map), with some exceptions mostly corresponding to ACB complex species carrying T6SS-1A loci that did not group with the larger block of T6SS-1A ACB strains. This might be an indication of gene acquisition by horizontal gene transfer. Furthermore, ST affiliation of each A. baumannii strain was included in this same figure, showing a sub-clustering, which correlated with the ST classification (Pasteur Scheme).
The role played by three of the genes present in the Acinetobacter T6SS-1 encoding cluster is still unknown (ACIAD2693, ACIAD2685, and ACIAD2698), although deletion of any of the former two, coding for hypothetical proteins 1 (Hyp1) and 2 (Hyp2), affects the functionality of the system (Weber et al., 2016; Ringel et al., 2017). We therefore decided to investigate if we could infer putative functions based on bioinformatic predictions carried out with the DSM30011 homologous proteins. We observed that Hyp1 (DMS30011_11500) bears no putative transmembrane segments and was predicted to carry a signal peptide which might direct it to the periplasm (Supplementary Figure S3A). Interestingly, we were able to identify in DSM30011 Hyp1 several features of its signal peptide sequence, which are characteristic of lipoproteins. It possesses a C23 residue which is part of a lipobox motif (PROSITE pattern PS51257), a positively charged K5 residue and a stretch of hydrophobic and uncharged amino acids residues between position 7 and 22 (Zückert, 2014). These structural characteristics resemble those of TssJ, which has been proposed to anchor the membrane complex (MC) to the inner membrane (IM) through its interaction with TssM (Felisberto-Rodrigues et al., 2011). Moreover, TssJ bears a lipobox motif which is critical for the association of the mature protein with the outer membrane through acylation of the N-terminal cysteine residue (Aschtgen et al., 2008). However, no protein showing homology with TssJ could be identified (Weber et al., 2013). Therefore, we propose that Hyp1 encoded within the A. baumannii T6MC could fulfill a role similar to that played by TssJ.
In enteroaggregative E. coli and in several other bacteria, TagL associates with the TssJLM membrane complex (Aschtgen et al., 2010). TagL is embedded in the IM through 3 transmembrane regions. It also bears a central cytoplasmic loop and a C-terminal periplasmic domain with a functional peptidoglycan-binding motif (Aschtgen et al., 2010), which tethers the T6SS to the cell wall. The peptidoglycan binding motif may also be encoded in a separate subunit, TagN. We and others (Ringel et al., 2017) observed that this is the case in Acinetobacter species. A. baumannii DSM300011 TagN bears an OmpA_C-like peptidoglycan binding-domain (152–254; cd07185; Supplementary Figure S4A) and a putative exportation signal (cleavage site between positions 27 and 28: ALA-QP). The obvious question is which Acinetobacter protein fulfills the role played by the N-terminal portion of E. coli TagL. Topology predictions performed on Hyp2 (DSM30011_11465) indicates that this could be a good candidate to play this role, since it contains two putative transmembrane domains in the N-terminal followed by a long periplasmic region (Supplementary Figure S3B). Recent predictions consistent with this hypothesis have been made when analyzing the topology of a homologous protein (TagZ) encoded by Burkholderia, Paraburkholderia and related species (Spiewak et al., 2019), which share a T6SS-1 main cluster with a similar gene composition but with a different arrangement with respect to Acinetobacter spp.
Hyp3 (DSM30011_11420) contains a C-terminal domain (PRK03427) found in the E. coli cell division protein ZipA, which is tied to the membrane through a short N-terminal membrane-anchored domain. Correspondingly, Hyp3 bears a putative transmembrane domain in its N-terminal region (residues 21–43; Supplementary Figure S3C) with the rest of the protein protruding toward the periplasm. Despite the fact that this protein shows low conservation in the Acinetobacter genus, its predicted topology is conserved. Weber et al. (2016) have suggested that TagX interaction with other components of the T6SS apparatus may control its enzymatic activity, allowing for precise spatial regulation of PG degradation. It is therefore possible that Hyp3 fulfils this role but experimental work needs to be carried out to test this hypothesis.
VgrG Protein Diversity in Acinetobacter spp.
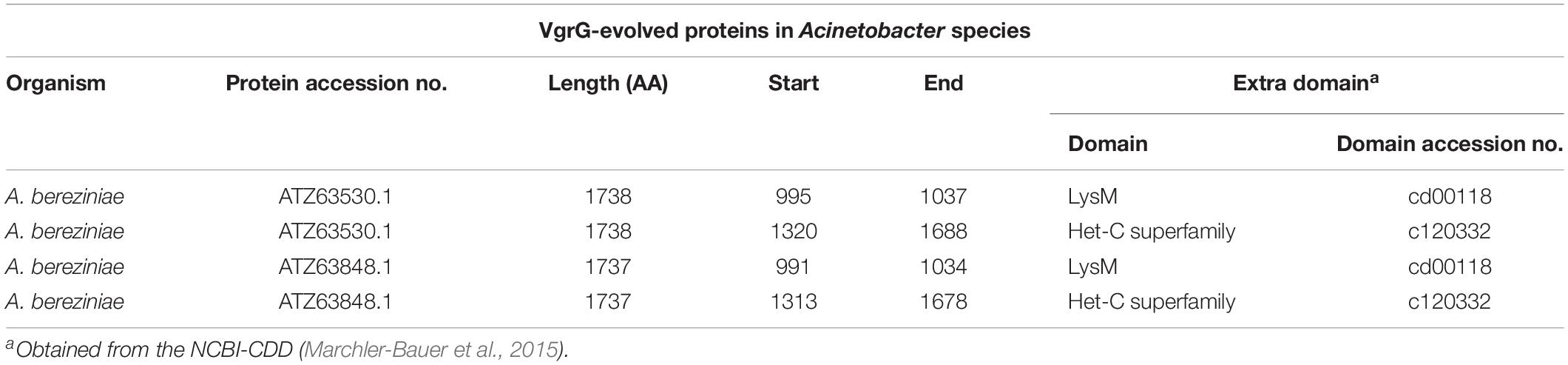
Varying numbers of genes located outside the Acinetobacter T6MC encode putative VgrG proteins (Weber et al., 2013). These T6SS-associated regions additionally encode a variable number of accessory and hypothetical proteins that act as effectors and immunity proteins (see Toxins section). VgrG proteins encoded in Acinetobacter genomes under study were investigated (see section Materials and Methods). As result, 534 homologous proteins were detected (average vgrG genes per genome = 2.79; Supplementary Table S2) throughout the Acinetobacter genus. In correlation with the lack of T6SS structural components, 7 out of 8 T6SS- Acinetobacter species (with A. ursingii as the exception; Table 3) do not encode vgrG genes. Lack of vgrG genes was also observed for the aforementioned T6SS-deficient Acinetobacter sp. Ncu2D-2, TTH0-4 and WCHA45 (Supplementary Table S1). Interestingly, species such as A. celticus, A. equi, A. pragensis, and A. radioresistens encode only 1 vgrG gene, whereas A. bereziniae carries 13 (Supplementary Table S2). A. baumannii T6SS-proficient strains usually contain between 2 and 4 copies. Interestingly, some Acinetobacter strains, show a split vgrG locus encoding separate N-terminal and C-terminal VgrG domains. This split among VgrG domains resembled the case of the bacteriophage spike proteins, and supported the proposed common evolutionary origin of these two membrane-penetrating devices (Pukatzki et al., 2007; Zoued et al., 2014). In addition, 3 of the identified VgrG sequences in our database contain putative effector domains in their C-termini (Supplementary Tables S2, S3). These evolved-VgrGs, which were identified in A. bereziniae and A. proteolyticus, encode both LysM and Het-C domains (Table 5). While the former (cd00118) is responsible for protein binding to peptidoglycan (Buist et al., 2008), the latter (pfam07217, cl20332 superfamily) is found in proteins regulating self/non-self-recognition in filamentous fungi (Wu et al., 1998). Antifungal effectors dependent on the T6SS have been recently described in Serratia marcescens (Trunk et al., 2018), therefore it is possible that those Acinetobacter VgrG proteins bearing a Het-C domain fulfill a similar function. In sum, these predictions suggest that some of these VgrGs may have additional functions, adding further levels of complexity to the Acinetobacter T6SS toxins repertoire.
The Newly Identified T6SS-2 Is Present in Three Environmental Acinetobacter Species
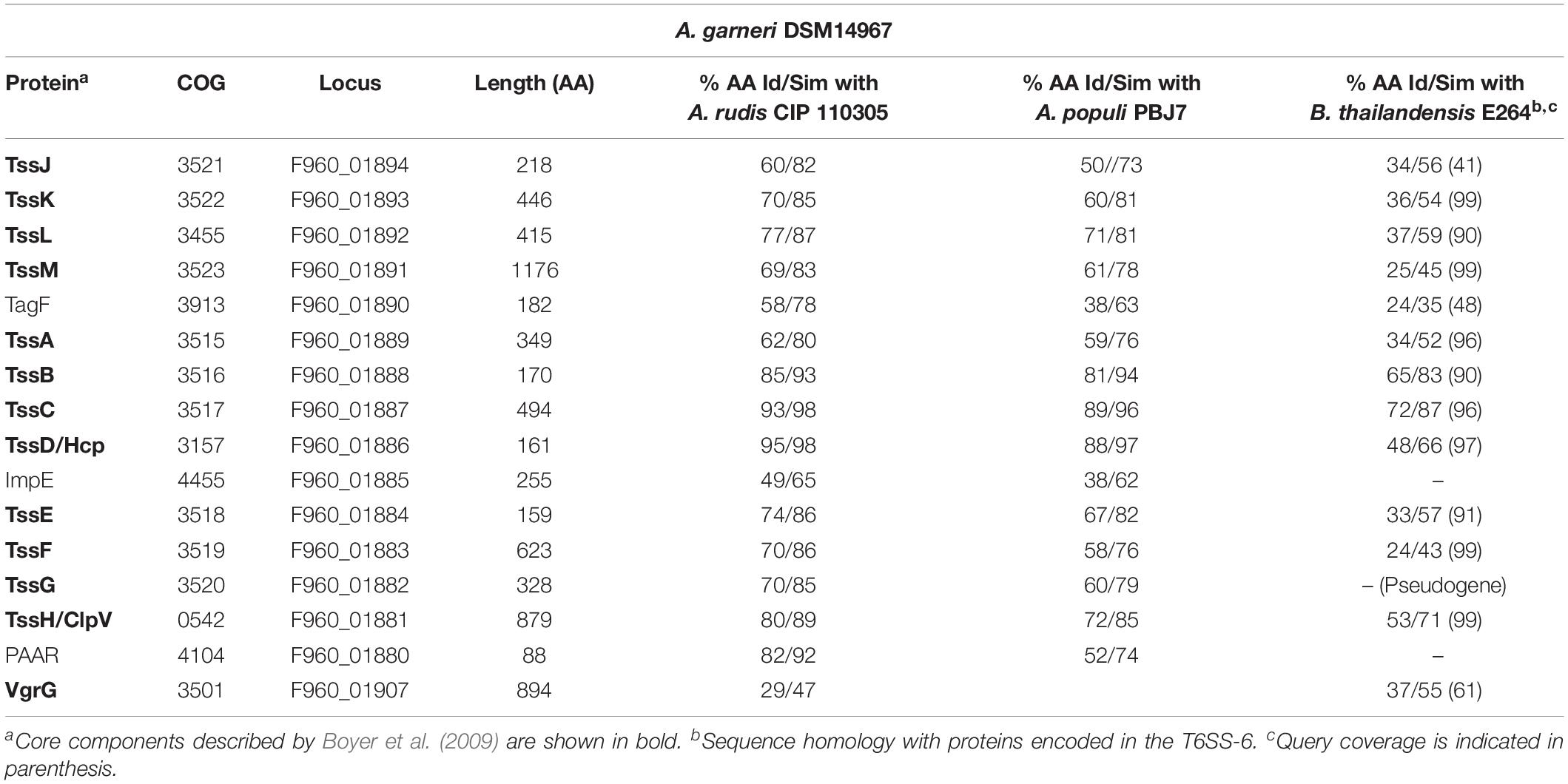
As previously mentioned, a T6MC with a different gene composition (T6SS-2) was also detected. We identified the T6SS-2 locus in A. gerneri DSM 14967 (also encoding a T6SS-1Bb cluster), isolated from activated sludge (Carr et al., 2003); in A. rudis CIP 110305 isolated from raw milk, and in A. populi PBJ7, isolated from a bark canker of Populus euramericana (Table 4 and Supplementary Table S1). It is composed of 15 genes, including the 13 T6SS-core genes described by Boyer et al. (2009); Figure 1B and Table 4). It shares 13 orthologous genes with the T6SS-1 main cluster, with tssJ and impE as genes specific of the T6SS-2 cluster. Key components (TssB, TssC, TssK, TssL) are highly conserved (aminoacidic identity > 60%) among the three aforementioned Acinetobacter species (Table 4). Remarkably, the PAAR gene is encoded in the complementary strand in A. gerneri and A. rudis, whereas in A. populi it is located in the same strand as the rest of the cluster. This observation is in line with the documented variability of PAAR genes location in the Acinetobacter T6SS-1 core clusters.
T6SS-2 organization resembles that of the gene cluster encoding the T6SS-6 described in Burkholderia species (Figure 1B and Table 4; Schwarz et al., 2014) with genes encoding VgrG proteins, putative toxins and cognate immunity proteins all located in the proximity of the main cluster in these three Acinetobacter strains. Still, important differences in genetic composition of the main cluster are observed between them such as the presence of the impE gene in the Acinetobacter T6SS-2, which is part of the T6MC in Rhizobium leguminosarum (Bladergroen et al., 2003), and the absence of the ppkA gene, encoding a kinase involved in post-translational regulation of the system in P. aeruginosa (Basler et al., 2013). Interestingly, the A. gerneri T6SS-2 specific components (meaning those for which no homologous proteins are encoded in the T6SS-1 cluster also encoded in its genome), namely TssJ, TssL and ImpE, show the highest identity percentages (between 35 and 50%) with proteins present in members of the Moraxella and Neisseria genera, which carry an unreported T6SS cluster with a different genetic structure. Analysis of G + C% of the T6SS-2 region in each of the three above mentioned Acinetobacter strains shows a significant deviation from the genomic average (35.8% vs. 37.9% for A. gerneri DSM14967; 33.3% vs. 39.2% for A. rudis CIP110305; and 32.6% vs. 40.2% for A. populi PBJ7). Moreover, average codon adaptation index calculations for each of these T6SS-2 clusters indicate a significant deviation (p < 0.0001) from the results obtained with the corresponding ribosomal proteins of each organism. Overall, these predictions imply a recent acquisition of this cluster by horizontal gene transfer from an unknown bacterial donor, thereby suggesting that the T6SS-1 and T6SS-2 clusters are evolutionarily distinct.
Remarkably, Acinetobacter species carrying the T6SS-2-core cluster encode several VgrG proteins with an additional domain (T6SS_Vgr; Supplementary Table S3) not frequently detected in VgrG present in other members of this genus (Figure 2A). The T6SS_Vgr domain (pfam13296) overlaps with the C-terminal part of the VgrG domain and is usually located before the start of the DUF2345 domain. Of note, A. populi, which is the only Acinetobacter species under study carrying uniquely a T6SS-2 main cluster, solely encodes VgrG proteins (total number = 3) with this domain architecture in its genome (Supplementary Table S2). On the other hand, A. gerneri (T6SS-1 and -2 clusters; total number = 9) and A. rudis (incomplete T6SS-1 and complete T6SS-2 clusters; total number = 8) inspected strains encode VgrG proteins falling into both domains architectures (Figure 2A and Supplementary Tables S2, S3). Furthermore, we detected that these types of VgrGs are encoded by genes which are usually in close proximity to the T6SS-2 main cluster. It is tempting to speculate that VgrGs bearing the T6SS_Vgr domain are somehow related to the apparatus encoded by T6SS-2-main cluster. An additional evidence supporting this hypothesis is that the most similar VgrG proteins outside the Acinetobacter genus are encoded by members of the Moraxella genus (40% average identity), in agreement with what has been observed for proteins encoded in the T6SS-2 main cluster (see above). This suggests that the complete system (T6MC plus VgrG proteins) has been acquired from species of the Moraxella genus.
Overall, these observations suggest that the Acinetobacter T6SS-2 cluster has a unique genetic organization and composition so far not described in other bacteria. Future work needs to be directed toward understanding the role that this T6SS plays in the biology of these Acinetobacter spp.
Three Major Classes of PAAR Domain Containing-Proteins Are Found in Acinetobacter spp.
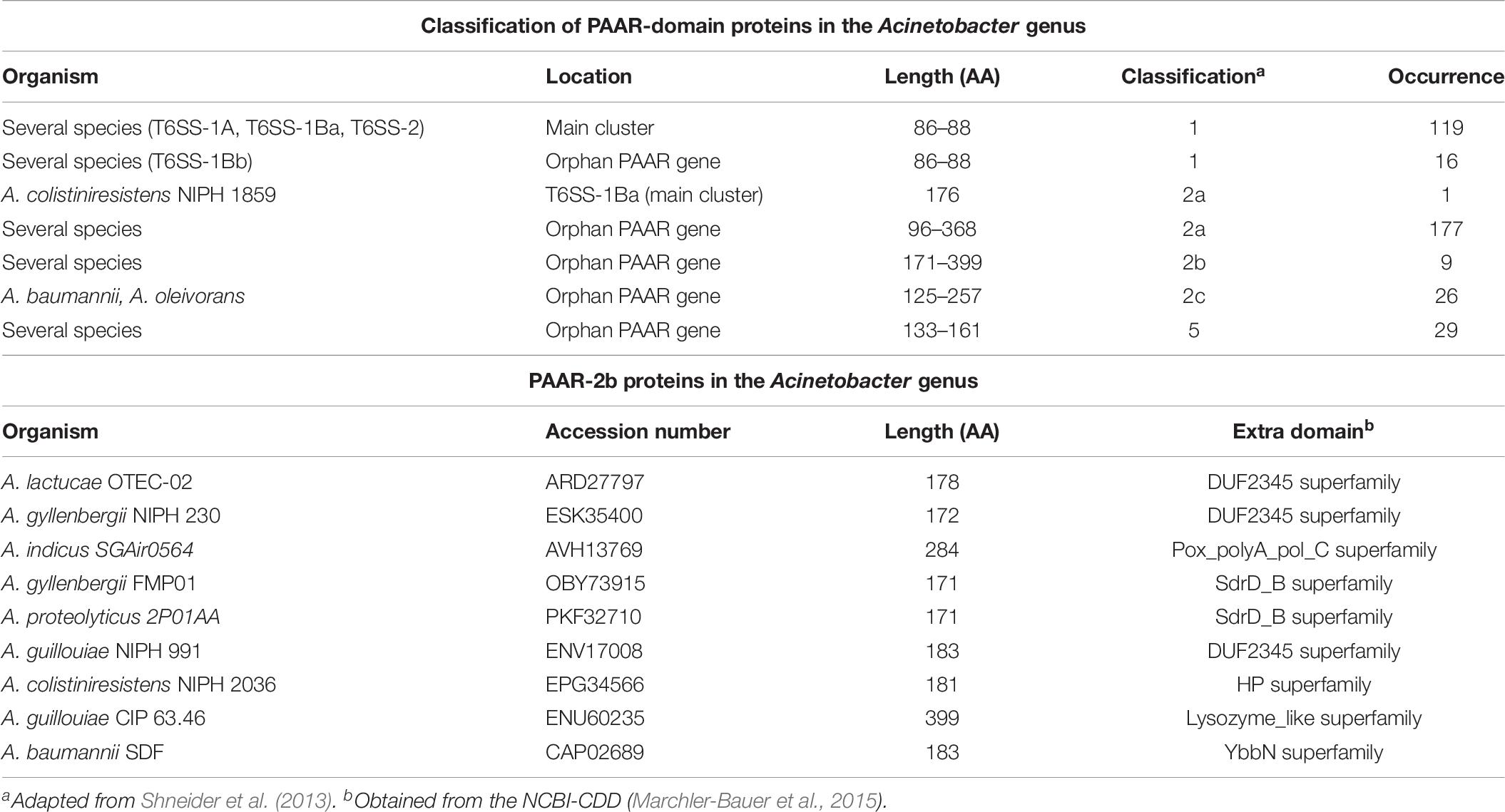
A bioinformatic search for PAAR proteins in members of the Acinetobacter genus was conducted (see section Materials and Methods). As result, 377 homologous proteins were detected (average PAAR genes per genome = 1.97; Supplementary Table S4) throughout the Acinetobacter genus. An interesting case is that of A. baumannii SDF which carries 10 PAAR genes (Eijkelkamp et al., 2014), namely 1 PAAR-1, 7 PAAR-2, and 2 PAAR-5. Remarkably, one of the PAAR-2 proteins (CAP02972) is encoded in a plasmid (p2ABSDF, CU468232) as part of a locus carrying two vgrG genes and several putative toxins, such as a potential amidase (CAP02973). Plasmid-borne T6SS genes have been described in other bacteria such as Pantoea ananatis (Shyntum et al., 2014), but so far not in Acinetobacter species. On the other hand, 7 out of 8 T6SS– Acinetobacter species (with A. ursingii as the exception; Table 3) do not encode any PAAR gene, in correlation with the lack of the structural components of the T6SS. Lack of PAAR genes was also observed for all the aforementioned T6SS– Acinetobacter members still not assigned to any species (Supplementary Table S1).
By means of this analysis we also determined that the majority of Acinetobacter PAAR proteins (Figure 2B and Table 6) belong to Shneider’s classes 1 (PAAR_CT_2 domain, cd14744 of NCBI-CDD), 2 (PAAR_CT_2 domain or PAAR_motif), and 5 (PAAR_motif; Supplementary Table S5). Interestingly, the PAAR proteins encoded within the T6SS-main clusters all belong to class 1, with the exception of A. colistinresistens NIPH 1859 which encodes a PAAR-2 protein (ENX34299) of 176 amino acids (Table 5 and Supplementary Table S4) and no other PAAR-1 elsewhere in its genome. Through this analysis we also observed that all Acinetobacter species bearing a T6SS-1Bb cluster (lacking a PAAR gene in the main cluster), encode an orphan PAAR-1 elsewhere in the genome. With the exception of A. indicus SGAir0564 (3 PAAR genes in total) and A. gerneri DSM 14967 = CIP 107464 (6 PAAR genes in total; also encoding a T6SS-2 main cluster) this is the only PAAR gene copy they carry in their genomes. This suggests that the PAAR-1 proteins are the T6SS-apparatus dedicated proteins in Acinetobacter species.
We also noticed that PAAR-2 proteins could be sub-classified into 3 classes (2a to 2c) according to their domain organization (Figure 2B), with those belonging to class 2b bearing additional domains (evolved-PAAR proteins; Table 6). Some of the latter carry a DUF2345 domain (pfam10106), usually part of VgrG proteins, suggesting it could fulfill an adapter role. Others bear domains probably acting as toxins, such as those belonging to the Lysozyme-like superfamily (cl00222). However, only 9 PAAR-2b proteins were detected and according to predictions in most cases the additional domain is not complete (Supplementary Table S5). Another aspect that is worth-commenting is that PAAR-2c proteins were mostly encoded by A. baumannii strains (25 out of 26; Supplementary Table S4).
With respect to PAAR-5 proteins, they are the less represented with only 29 members among Acinetobacter strains under study (Table 6). This is coincident with previous analysis performed on 1,353 bacterial PAAR proteins (24 occurrences, Shneider et al., 2013). Another aspect that is interesting to mention, is that PAAR-5 genes are usually located in tandem with a PAAR-2 gene (Supplementary Table S4) as has been described for A. baylyi ADP1 (CAG67033 and CAG67034; Shneider et al., 2013) and that these regions usually encode toxin genes (see below).
The genetic variability within each PAAR class was lastly investigated, based on the identity percentage of PAAR proteins under study with respect to A. baumannii DSM30011 PAAR-1 (Supplementary Table S4). We observed a significant level of conservation among PAAR-1 proteins encoded within the T6MC-1A and 1Ba (%Id 64–100), which was also evidenced from the heatmap (Supplementary Figure S2). These levels of conservation were also detected for 12 out of 15 PAAR-1 proteins present in the genomes of strains carrying the T6MC-1Bb (%Id 66–82; with respect to Acinetobacter sp. ACNIH1 PAAR-1). High conservation levels were also observed for PAAR-2c proteins (%Id 83–87), mainly encoded in A. baumannii strains. In turn, other groups showed an important variability, such as PAAR-2a (Id% 30–88) and PAAR-5 (Id% 40–100; with respect to A. baylyi ADP1 PAAR-5), and as expected PAAR-2b (%Id 31–85), since they carry an extra domain. It is tempting to speculate that those PAAR proteins fulfilling a structural role tend to be more conserved than those probably acting as toxin-chaperones (see below).
Two Undescribed PAAR-Rhs Proteins Probably Acting as Toxins
Since none of the PAAR proteins identified through this analysis bore a DUF4150 domain, also found in proteins belonging to the PAAR_like domain superfamily, a search for proteins carrying this domain was conducted. By this means, two proteins present in A. gerneri DSM 14967 = CIP 107464 (ENV33198) and A. equi (ALH95481) with significant homology were detected. These proteins additionally carried RhsA domains (cl27255; Supplementary Figure S4B) and thus could be categorized as PAAR-3 proteins according to a previous classification (Shneider et al., 2013). A predicted toxin domain (pfam15633) of the ADP-ribosyltransferase superfamily was detected in ENV33198. PAAR-3 proteins carrying this toxin domain (categorized as “protein-modifying”) have been identified in 16 bacterial species (Ma et al., 2017b). The genes encoding these PAAR-3 proteins, are preceded by genes coding for a protein carrying a DUF2169 domain and a “short” VgrG (<750 aa, lacking a DUF2345 domain). We found that these clusters are also present in bacteria belonging to diverse genera such as Bordetella, Burkholderia, Variovorax, Alcalinovorax, and Pseudomonas. Furthermore, a DUF2169 domain-carrying protein fulfill an adapter role specific for the Tde2 toxin (AAK89757; carrying a DUF4150 and a Ntox15 DNase domain) in Agrobacterium tumefaciens (Bondage et al., 2016). Interestingly, the gene encoding for the DUF2169-domain–containing protein is always located between the vgrG2 and tde2 genes. Conservation in gene cluster organization across diverse Proteobacterial lineages has also been reported. All these data are on favor of the hypothesis that the Tde effector is stabilized and carried by its cognate adaptor/chaperone, which loads the effector onto the C terminus of VgrG for secretion across bacterial membranes (Bondage et al., 2016). It is thus tempting to speculate that the DUF2169 domain-carrying protein present in Acinetobacter strains is involved in the loading of PAAR-3 partners.
T6SS-Associated Toxin Repertoire in Acinetobacter spp.
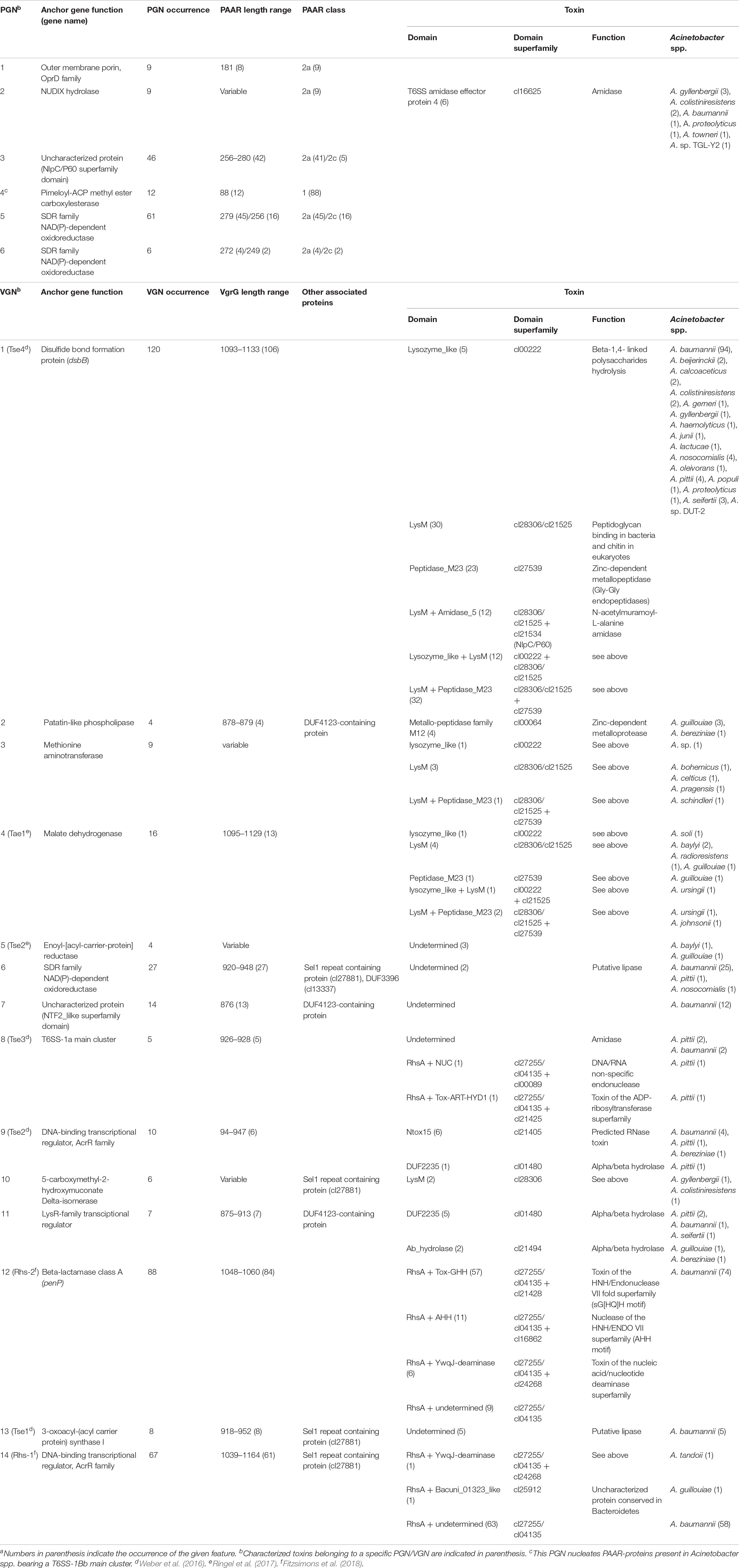
In order to search for putative toxin encoding genes in the proximity of vgrG and PAAR identified genes, a homology search using as query T6SS-reported toxins was performed (see section Materials and Methods and Supplementary Table S6). Gene neighborhoods of PAAR and vgrG genes were further investigated in order to analyze the existence of specific genomic spots where PAAR and vgrG islands are located, the Acinetobacter species encoding specific islands, the presence of different classes of PAAR or VgrG proteins linked to these neighborhoods and finally, the toxins associated to each of them. With all this information, a clustering approach was followed in order to distinguish either PAAR or VgrG proteins sharing common genetic environments (Supplementary Figures S5, S6). According to this analysis 6 PAAR- and 14 VgrG-gene neighborhoods (PGN and VGN, respectively) were defined (Supplementary Table S7), which are summarized in Table 7. We observed that a gene encoding for a SDR family NAD(P)-dependent oxidoreductase is part of several of these gene neighborhoods (PGN5, PGN6, and VGN6), suggesting this genome region might be a conserved spot for PAAR and vgrG islands. Moreover, 12 out of 15 PAAR-1 genes present in the genomes encoding a T6SS1-Bb-core cluster grouped in PGN4, whereas the remaining 3 (ENV70927, SNQ29620, and ENV33195) were located within mixed PAAR-vgrG islands not associated to any particular PGN/VGN (Supplementary Figure S5). The conservation of PGN4 reinforces the idea that the PAAR-1 proteins encoded in these clusters are devoted to the T6SS.
In correlation with toxin genes, we initially observed that vgrG islands were particularly enriched in genes encoding different classes of previously reported toxins whereas in PAAR islands mostly Tpe1 and Tae4 toxins were detected (Supplementary Table S6). In A. baylyi ADP1 the tpe1 gene is located downstream a PAAR-2 encoding gene (Ringel et al., 2017). In agreement with this data, our analysis detected only 1 tpe1 orthologous gene next to a vgrG gene whereas 20 were in close proximity to a PAAR gene (Supplementary Table S6). Furthermore, we observed that in 11 out of these 20 loci, there is a PAAR-5 gene upstream the PAAR-2 gene (Supplementary Table S6). This information suggests that these toxins are usually genetically linked to PAAR genes in Acinetobacter spp. Interestingly, A. colistineresistens encodes a Tpe1 toxin (ENX34300) in the T6MC next to an unusual PAAR-2a gene. While Tpe1 proteins are not particularly associated to any PGN, putative Tae4 proteins are encoded in the PGN2 (Supplementary Figure S5). We noticed that 6 out of the 9 clusters included in PGN2 encoded Tae4-amidase effectors accompanied by their immunity proteins (Tai4; Table 7). Of note, the Tae4-Tai4 pair (CAP02973-CAP02974) is encoded in A. baumannii SDF p2ABSDF. Furthermore, 5 PAAR genes (ATZ62304, ESK36633, OBY73307, ATZ64949, AMW78666; Supplementary Figure S5) were found to be accompanied by a gene encoding a protein with a VRR_Nuclease domain (cl22959) and followed by 1 to 4 genes bearing a DUF3396 domain (cl13337). These PAAR proteins all belong to class 2a and are between 176 and 179 aa long (Supplementary Table S4). It would be interesting to determine if they are responsible for the secretion of the putative nuclease effector and if the DUF3396-proteins are involved in this task as well.
When analyzing the different VGNs, we observed that VGN1 (the most prevalent), 3, 4, and 10 encoded peptidoglycan hydrolases, M12 and M23 domain peptidases, LysM-domain containing proteins and numerous proteins of unknown function. This is in agreement with a previous report including 23 Acinetobacter genomes (Fitzsimons et al., 2018). We also detected the presence of proteins with significant homology with toxins previously reported in Acinetobacter spp. (Weber et al., 2016; Ringel et al., 2017; Fitzsimons et al., 2018), associated to specific VGNs (Table 7). Several DUF4123 proteins were found in VGN2, 7 and 11, accompanied by a “short VgrG” (875–883 aa) and associated to Tle1 toxins (DUF2235) or proteins carrying a M12 metallo-peptidase family domain (pfam13688). The presence of DUF4123 proteins in VGNs has already been pinpointed by Fitzsimons et al. (2018). Ringel et al. (2017) identified in A. baylyi the Tap-1 chaperone (carrying a DUF4123 domain) necessary for the putative Tse2 toxin delivery. Therefore, it is possible that these proteins might play a role as chaperones for different toxin partners.
In this analysis we were also able to identify a vast repertoire of Rhs-domain containing proteins, which were found to be present in vgrG islands of 13 out of the 33 Acinetobacter T6SS-proficient species under study (Supplementary Table S6). Moreover, they are clearly conserved in A. baumannii strains (90 out of 110). Rhs-encoding genes were particularly grouped in the VGN8, 12, and 14 (Table 7), of which 174 showed homology at the protein level with Rhs1 and Rhs2 identified in A. baumannii AB307-0294 (Supplementary Table S6; Fitzsimons et al., 2018). We observed that while a group of 20 of these proteins belonging to the VGN14 showed an identity > 94% with the Rhs1 toxin of unknown function, 11 proteins (VGN12) were identical to the Rhs2 effector which carries an AHH nuclease domain (cl16862). This prompted us to analyze the variety of effector domains carried by the remaining Rhs-domain containing proteins (Supplementary Table S8). Remarkably, 59 candidates contained a Tox-GHH superfamily nuclease domain (cl21428) and 8 a YwqJ-deaminase superfamily domain (cl24268). Importantly, with the exception of the aforementioned PAAR-3 protein (ALH95481), none of these Rhs proteins bears a PAAR domain or a chaperone gene nearby, and thus their delivery mechanism remains unknown.
Diverse functions have been attributed to Rhs proteins in other bacteria such as motility, cellular toxicity, virulence in mice and insecticidal activity (Lien and Lai, 2017; Yang et al., 2018), and contact-dependent killing of other bacteria species in A. baumannii (Fitzsimons et al., 2018). Rhs proteins are capable of replacing the C-terminal end with a non-homologous alternative. By this means, they can switch between different toxin domains and thereby they have been included within the bacterial polymorphic toxin systems (Jamet and Nassif, 2015). Our findings indicate that the rhs genes associated with the Acinetobacter vgrG islands encode different C-terminal domains which might play different roles as toxins. To date none of these proteins have been implicated in A. baumannii virulence.
Conclusion
The T6SS fulfills critical roles in bacterial competition, bacterium–host interaction and other functions associated with bacterial physiology. We therefore conducted a genomic comparative analyses of 191 Acinetobacter strains which let us identify two putative gene clusters responsible for the synthesis of a functional T6SS. The T6SS-1 was widespread in the genome of most species, while the T6SS-2 was restricted to three environmental species. In this respect, it will be interesting to determine whether the T6SS-2 is active and the roles it plays. Future work will certainly focus on the identification and characterization of the secreted components of this system.
The finding that the T6SS-1 gene cluster was present in both pathogenic and non-pathogenic species raises the hypothesis that the T6SS may have evolved to play different roles in the Acinetobacter lifestyle. In this respect, the variable regions associated with PAAR and vgrG genes could account for specialization of the T6SS based on the needs of each species. A plethora of toxin-encoding genes was detected in these genomic regions, with those encoding Rhs-domain containing proteins clearly overrepresented. Although no proteins showing significant homology to previously characterized effectors targeting eukaryotic cells (Lien and Lai, 2017) were detected, these regions also encode an impressive number of proteins of unknown function. Hence, we cannot rule out the existence of novel effector functions still to be discovered.
The overall observations described above not only highlight both the high diversity and potential plasticity of the Acinetobacter T6SS genes at the species level, but also support the notion that this system could have evolved differential and even strain-specific roles related to the interaction with other cells in particular environments. We hope the results obtained in this work can provide a foundation for future characterization of the Acinetobacter T6SSs and its effectors.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov.
Author Contributions
GR and SS conceived and designed the work. JS conducted the experimental work. GR and ME conducted the bioinformatic analysis. GR, ME, and SS analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a FINOVI Young Researcher Grant awarded to SS and by grants from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT PICT-2017-3536) to GR. SS is a staff scientist of INSERM. GR and ME are staff members of CONICET.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02519/full#supplementary-material
FIGURE S1 | Hcp secretion assay. The presence of Hcp (indicative of a functional T6SS) in concentrated culture supernatants of the indicated Acinetobacter strains grown up to stationary phase in L-Broth was determined by 18% SDS-PAGE and Coomasie Blue staining (see Repizo et al., 2015 for experimental details). MM, molecular markers.
FIGURE S2 | Clustering of T6MC-proteins in 191 Acinetobacter strains. A. baumannii DSM30011 was used as reference. Strain subclustering according to T6MC classification is indicated. For A. baumannii strains, ST-classification according to the Pasteur scheme is also shown.
FIGURE S3 | (A–C) Putative transmembrane domains present in Acinetobacter T6SS proteins with unknown function (Hyp1-3). TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM) was used for predictions.
FIGURE S4 | (A) TagN domain structure. The NCBI-CDD was used for the domain search (Marchler-Bauer et al., 2015). (B) Domain architecture of PAAR-3 proteins found in Acinetobacter spp. The NCBI-CDD database was used for the domain search (Marchler-Bauer et al., 2015).
FIGURE S5 | Clustering of PAAR islands based on gene context. Conserved superfamily domains of proteins encoded within PAAR islands were used to define similar genetic contexts by clustering. Those gene islands sharing a similar genetic context were grouped in PAAR gene neighborhoods (PGN1-6, see Table 7 for detail). PAAR islands encoding putative toxins are indicated. Accession numbers corresponding to PAAR proteins encoded by A. baylyi ADP1 are boxed.
FIGURE S6 | Clustering of VgrG islands based on gene context. Conserved superfamily domains of proteins encoded within VgrG islands were used to define similar genetic contexts by clustering. Those gene islands sharing a similar genetic context were grouped in VgrG gene neighborhoods (VGN1-14, see Table 7 for detail). PAAR islands encoding putative toxins are indicated. Accession numbers corresponding to VgrG proteins encoded by A. baylyi ADP1 are boxed.
TABLE S1 | Strain under study.
TABLE S2 | BlastP-homology search for VgrG domain-containing proteins in Acinetobacter spp.
TABLE S3 | NCBI web-Conserved domains search tool results for Acinetobacter VgrG proteins.
TABLE S4 | BlastP-homology search for PAAR proteins in the Acinetobacter genus.
TABLE S5 | NCBI web-Conserved domains search tool results for Acinetobacter PAAR proteins.
TABLE S6 | BlastP-homology search for toxins in PAAR and vgrG genetic islands in Acinetobacter spp.
TABLE S7 | List of PAAR and vgrG proteins grouped into the gene neighborhoods defined in this study.
TABLE S8 | NCBI web-Conserved domains search tool results for Acinetobacter Rhs proteins.
Footnotes
References
Alcoforado, D. J., Liu, Y. C., and Coulthurst, S. J. (2015). Molecular weaponry: diverse effectors delivered by the type VI secretion system. Cell. Microbiol. 17, 1742–1751. doi: 10.1111/cmi.12532
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antunes, L. C. S., Visca, P., and Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049-632X.12125
Aschtgen, M. S., Bernard, C. S., De Bentzmann, S., Lloubès, R., and Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531. doi: 10.1128/JB.00945-08
Aschtgen, M. S., Thomas, M. S., and Cascales, E. (2010). Anchoring the type VI secretion system to the peptidoglycan TssL, TagL, TagP, what else? Virulence 1, 535–540. doi: 10.4161/viru.1.6.13732
Basler, M., Ho, B. T., and Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Benz, J., and Meinhart, A. (2014). Antibacterial effector/immunity systems: It’s just the tip of the iceberg. Curr. Opin. Microbiol. 17, 1–10. doi: 10.1016/j.mib.2013.11.002
Bernard, C. S., Brunet, Y. R., Gueguen, E., and Cascales, E. (2010). Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192, 3850–3860. doi: 10.1128/JB.00370-10
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64. doi: 10.1094/mpmi.2003.16.1.53
Blondel, C. J., Jiménez, J. C., Contreras, I., and Santiviago, C. A. (2009). Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. doi: 10.1186/1471-2164-10-354
Bondage, D. D., Lin, J.-S., Ma, L.-S., Kuo, C.-H., and Lai, E.-M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940. doi: 10.1073/pnas.1600428113
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Carr, E. L., Kämpfer, P., Patel, B. K. C., Gürtler, V., and Seviour, R. J. (2003). Seven novel species of Acinetobacter isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53(Pt 4), 953–963. doi: 10.1099/ijs.0.02486-0
Carruthers, M. D., Nicholson, P. A., Tracy, E. N., and Munson, R. S. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388
Cooper, R. M., Tsimring, L., and Hasty, J. (2017). Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. eLife 6:e25950. doi: 10.7554/elife.25950
Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
De Maayer, P., Venter, S. N., Kamber, T., Duffy, B., Coutinho, T. A., and Smits, T. H. M. (2011). Comparative genomics of the type VI secretion systems of pantoea and erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 12:576. doi: 10.1186/1471-2164-12-576
Di Venanzio, G., Moon, K. H., Weber, B. S., Lopez, J., Ly, P. M., Potter, R. F., et al. (2019). Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc. Natl. Acad. Sci. U.S.A. 116, 1378–1383. doi: 10.1073/pnas.1812557116
Eijkelkamp, B. A., Stroeher, U. H., Hassan, K. A., Paulsen, I. T., and Brown, M. H. (2014). Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020
Elhosseiny, N. M., and Attia, A. S. (2018). Acinetobacter: an emerging pathogen with a versatile secretome review-article. Emerg. Microbes Infect. 7:33. doi: 10.1038/s41426-018-0030-4
Felisberto-Rodrigues, C., Durand, E., Aschtgen, M. S., Blangy, S., Ortiz-Lombardia, M., Douzi, B., et al. (2011). Towards a structural comprehension of bacterial type vi secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7:e1002386. doi: 10.1371/journal.ppat.1002386
Fitzsimons, T. C., Lewis, J. M., Wright, A., Kleifeld, O., Schittenhelm, R. B., Powell, D., et al. (2018). Identification of novel Acinetobacter baumannii type VI secretion system antibacterial effector and immunity pairs. Infect. Immun. 86:e297-18. doi: 10.1128/IAI.00297-18
Flaugnatti, N., Le, T. T. H., Canaan, S., Aschtgen, M. S., Nguyen, V. S., Blangy, S., et al. (2016). A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 99, 1099–1118. doi: 10.1111/mmi.13292
Galán, J. E., and Waksman, G. (2018). Protein-injection machines in Bacteria. Cell 172, 1306–1318. doi: 10.1016/j.cell.2018.01.034
Gebhardt, M. J., Gallagher, L. A., Jacobson, R. K., Usacheva, E. A., Peterson, L. R., Zurawski, D. V., et al. (2015). Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in acinetobacter baumannii. mBio 6, 1–12. doi: 10.1128/mBio.01660-15
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Jamet, A., and Nassif, X. (2015). New players in the toxin field: polymorphic toxin systems in bacteria. mBio 27, 897–905. doi: 10.1128/mbio.00285-15
Jones, C. L., Clancy, M., Honnold, C., Singh, S., Snesrud, E., Onmus-Leone, F., et al. (2015). fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin. Infect. Dis. 61, 145–154. doi: 10.1093/cid/civ225
Kim, J., Lee, J. Y., Lee, H., Choi, J. Y., Kim, D. H., Wi, Y. M., et al. (2017). Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 8, 1378–1389. doi: 10.1080/21505594.2017.1323164
Letunic, I., and Bork, P. (2011). Interactive tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Lien, Y.-W., and Lai, E.-M. (2017). Type VI secretion effectors: methodologies and biology. Front. Cell. Infect. Microbiol. 7:254. doi: 10.3389/fcimb.2017.00254
Ma, A. T., and Mekalanos, J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370. doi: 10.1073/pnas.0915156107
Ma, J., Pan, Z., Huang, J., Sun, M., Lu, C., and Yao, H. (2017a). The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence 8, 1189–1202. doi: 10.1080/21505594.2017.1279374
Ma, J., Sun, M., Dong, W., Pan, Z., Lu, C., and Yao, H. (2017b). PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 19, 345–360. doi: 10.1111/1462-2920.13621
Ma, J., Sun, M., Pan, Z., Lu, C., and Yao, H. (2018). Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochim. Biophys. Acta Gen. Subj. 1862, 1635–1643. doi: 10.1016/j.bbagen.2018.04.010
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
McConnell, M. J., Actis, L., and Pachón, J. (2013). Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37, 130–155. doi: 10.1111/j.1574-6976.2012.00344.x
Medema, M. H., Takano, E., and Breitling, R. (2013). Detecting sequence homology at the gene cluster level with multigeneblast. Mol. Biol. Evol. 30, 1218–1223. doi: 10.1093/molbev/mst025
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pukatzki, S., Ma, A. T., Sturtevant, D., Revel, A. T., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Repizo, G. D. (2017). Prevalence of Acinetobacter baumannii strains expressing the type 6 secretion system in patients with bacteremia. Virulence 8, 1099–1101. doi: 10.1080/21505594.2017.1346768
Repizo, G. D., Gagné, S., Foucault-Grunenwald, M. L., Borges, V., Charpentier, X., Limansky, A. S., et al. (2015). Differential role of the T6SS in acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265
Repizo, G. D., Viale, A. M., Borges, V., Cameranesi, M. M., Taib, N., Espariz, M., et al. (2017). The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol. Evol. 9, 2292–2307. doi: 10.1093/gbe/evx162
Ringel, P. D., Hu, D., and Basler, M. (2017). The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 21, 3927–3940. doi: 10.1016/j.celrep.2017.12.020
Ryu, C. M. (2015). Against friend and foe: type 6 effectors in plant-associated bacteria. J. Microbiol. 53, 201–208. doi: 10.1007/s12275-015-5055-y
Schwarz, S., Singh, P., Robertson, J. D., LeRoux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Shalom, G., Shaw, J. G., and Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699. doi: 10.1099/mic.0.2007/006585-0
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Shyntum, D. Y., Venter, S. N., Moleleki, L. N., Toth, I., and Coutinho, T. A. (2014). Comparative genomics of type VI secretion systems in strains of Pantoea ananatis from different environments. BMC Genomics 15:163. doi: 10.1186/1471-2164-15-163
Spiewak, H. L., Eberl, L., Shastri, S., Zhang, L., Schwager, S., Thomas, M. S., et al. (2019). Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. Microbiologyopen 9:e774. doi: 10.1002/mbo3.774
Suzuki, R., and Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Taboada, B., Ciria, R., Martinez-Guerrero, C. E., and Merino, E. (2012). ProOpDB: prokaryotic operon database. Nucleic Acids Res. 40, D627–D631. doi: 10.1093/nar/gkr1020
Trunk, K., Peltier, J., Liu, Y. C., Dill, B. D., Walker, L., Gow, N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Unterweger, D., Miyata, S. T., Bachmann, V., Brooks, T. M., Mullins, T., Kostiuk, B., et al. (2014). The vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 20, 130–137. doi: 10.1038/ncomms4549
Weber, B. S., Hennon, S. W., Wright, M. S., Scott, N. E., de Berardinis, V., Foster, L. J., et al. (2016). Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio. 7:e01253-16. doi: 10.1128/mBio.01253-16
Weber, B. S., Kinsella, R. L., Harding, C. M., and Feldman, M. F. (2017). The secrets of acinetobacter secretion. Trends Microbiol. 25, 532–545. doi: 10.1016/j.tim.2017.01.005
Weber, B. S., Miyata, S. T., Iwashkiw, J. A., Mortensen, B. L., Skaar, E. P., Pukatzki, S., et al. (2013). Genomic and functional analysis of the Type VI secretion system in acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142
Wright, M. S., Haft, D. H., Harkins, D. M., Perez, F., Hujer, K. M., Bajaksouzian, S., et al. (2014). New insights into dissemination and variation of the health care- associated pathogen Acinetobacter baumannii from genomic analysis. mBio. 5:e00963-13. doi: 10.1128/mBio.00963-13
Wu, J., Saupe, S. J., and Glass, N. L. (1998). Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 95, 12398–12403. doi: 10.1073/pnas.95.21.12398
Yang, X., Long, M., and Shen, X. (2018). Effector–immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities. Molecules 23:1009. doi: 10.3390/molecules23051009
Zoued, A., Brunet, Y. R., Durand, E., Aschtgen, M. S., Logger, L., Douzi, B., et al. (2014). Architecture and assembly of the Type VI secretion system. Biochim. Biophys. Acta Mol. Cell Res. 1843, 1664–1673. doi: 10.1016/j.bbamcr.2014.03.018
Keywords: comparative genomics, toxins, PAAR proteins, VgrG, type 6 secretion system, Acinetobacter
Citation: Repizo GD, Espariz M, Seravalle JL and Salcedo SP (2019) Bioinformatic Analysis of the Type VI Secretion System and Its Potential Toxins in the Acinetobacter Genus. Front. Microbiol. 10:2519. doi: 10.3389/fmicb.2019.02519
Received: 28 February 2019; Accepted: 18 October 2019;
Published: 01 November 2019.
Edited by:
Maria Alejandra Mussi, National Council for Scientific and Technical Research (CONICET), ArgentinaReviewed by:
Peng Luo, South China Sea Institute of Oceanology (CAS), ChinaGerman Matias Traglia, University of the Republic, Uruguay
Copyright © 2019 Repizo, Espariz, Seravalle and Salcedo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guillermo D. Repizo, cmVwaXpvQGlici1jb25pY2V0Lmdvdi5hcg==
 Guillermo D. Repizo
Guillermo D. Repizo Martín Espariz
Martín Espariz Joana L. Seravalle
Joana L. Seravalle Suzana P. Salcedo
Suzana P. Salcedo