- 1Department of Biological Sciences, Louisiana State University, Baton Rouge, LA, United States
- 2Department of Infectious Diseases, University of Georgia College of Veterinary Medicine, Athens, GA, United States
- 3Center for Vaccines and Immunology, University of Georgia College of Veterinary Medicine, Athens, GA, United States
Colistin is a “last resort” antibiotic for treatment of infections caused by some multidrug resistant Gram-negative bacterial pathogens. Resistance to colistin varies between bacterial species. Some Gram-negative bacteria such as Burkholderia spp. are intrinsically resistant to very high levels of colistin with minimal inhibitory concentrations (MIC) often above 0.5 mg/ml. We have previously shown DedA family proteins YqjA and YghB are conserved membrane transporters required for alkaline tolerance and resistance to several classes of dyes and antibiotics in Escherichia coli. Here, we show that a DedA family protein in Burkholderia thailandensis (DbcA; DedA of Burkholderia required for colistin resistance) is a membrane transporter required for resistance to colistin. Mutation of dbcA results in >100-fold greater sensitivity to colistin. Colistin resistance is often conferred via covalent modification of lipopolysaccharide (LPS) lipid A. Mass spectrometry of lipid A of ΔdbcA showed a sharp reduction of aminoarabinose in lipid A compared to wild type. Complementation of colistin sensitivity of B. thailandensis ΔdbcA was observed by expression of dbcA, E. coli yghB or E. coli yqjA. Many proton-dependent transporters possess charged amino acids in transmembrane domains that take part in the transport mechanism and are essential for function. Site directed mutagenesis of conserved and predicted membrane embedded charged amino acids suggest that DbcA functions as a proton-dependent transporter. Direct measurement of membrane potential shows that B. thailandensis ΔdbcA is partially depolarized suggesting that loss of protonmotive force can lead to alterations in LPS structure and severe colistin sensitivity in this species.
Introduction
Colistin (polymyxin E) is a last resort antibiotic for treatment of infections caused by Gram-negative pathogenic bacteria (Paterson and Harris, 2016). Discovered in 1947 (Ainsworth et al., 1947), polymyxins were rarely used internally due to nephrotoxicity (Koch-Weser et al., 1970). However, their use has increased recently due to ineffectiveness of approved antibiotics against multidrug-resistant bacteria, including carbapenemase-producing Enterobacteriaceae such as Klebsiella pneumoniae (Queenan and Bush, 2007). However, many Gram-negative bacteria are intrinsically resistant to colistin and plasmid-acquired resistance has recently been reported (Liu et al., 2016; McGann et al., 2016; Rolain et al., 2016; Sun et al., 2018).
The genus Burkholderia is a group of highly adaptable, Gram-negative bacteria that includes a number of animal and plant pathogens (Lipuma, 2010; Waters, 2012). In terms of human infections, Burkholderia is extremely difficult to treat due to resistance to a number of commonly used antibiotics, arising from the presence of numerous multidrug resistance efflux pumps as well as restricted permeation (Podnecky et al., 2015; Krishnamoorthy et al., 2017). Burkholderia spp. exhibit extremely high intrinsic polymyxin resistance with minimal inhibitory concentrations (MIC) often exceeding 500 μg/ml (Loutet and Valvano, 2011), two orders of magnitude greater than that observed for most Gram-negative species. Burkholderia thailandensis is closely related to Burkholderia pseudomallei, the cause of melioidosis (Wiersinga et al., 2018), and both species are highly resistant to colistin and most other antibiotics (Krishnamoorthy et al., 2019). While B. thailandensis is considered a suitable surrogate for B. pseudomallei and only rarely causes infections in humans, it is infectious in a number of mammalian tissue culture, murine, insect and plant models and possesses virulence factors and drug resistance mechanisms that are found in its more virulent relatives (Gallagher et al., 2013). The existence of an ordered transposon library also makes B. thailandensis a valuable model organism to study Burkholderia virulence (Gallagher et al., 2013).
Cationic peptides produced by the immune system and antibiotics such as colistin interact electrostatically with negatively charged outer membrane of Gram negative bacteria. A common mechanism that Burkholderia spp. share with other Gram-negative species involves expression of a biosynthetic pathway that results in the modification of LPS lipid A with aminoarabinose (Ara4N). This amine-containing group neutralizes the negative charge of lipid A inhibiting colistin binding (Simpson and Trent, 2019). The remarkably high colistin resistance of Burkholderia spp. is likely due to a combination of a number of factors such as LPS modifications, as well as hopanoid synthesis (Malott et al., 2012), secreted metalloproteases (Loutet and Valvano, 2011) and synthesis of antioxidant putrescine (El-Halfawy and Valvano, 2014).
The DedA/Tvp38 membrane protein family (DedA family for short) is a highly conserved protein family that remains poorly characterized. There are currently 27,035 individual sequences in the protein database across 8547 species belonging to the “SNARE-associated PF09335” family of proteins (PFAM 31.0). We have characterized members of the DedA family in Escherichia coli and Borrelia burgdorferi (Thompkins et al., 2008; Liang et al., 2010; Sikdar and Doerrler, 2010; Boughner and Doerrler, 2012; Doerrler et al., 2013; Sikdar et al., 2013; Kumar and Doerrler, 2014, 2015; Kumar et al., 2016). The DedA family includes E. coli YqjA and YghB; putative proton dependent transporters that together are required for normal growth and cell division (Thompkins et al., 2008; Sikdar and Doerrler, 2010) and resistance to a number of antibiotics and biocides (Kumar and Doerrler, 2014) while YqjA is alone required for alkaline tolerance (Kumar and Doerrler, 2015). Both YqjA and YghB possess essential membrane embedded charged amino acids (Kumar and Doerrler, 2014; Kumar et al., 2016) that are present in proton-dependent transporters belonging to the major facilitator superfamily and other families (Noumi et al., 1997; Abramson et al., 2004; Adler and Bibi, 2004; Sigal et al., 2005; Fluman et al., 2012; Holdsworth and Law, 2012). While the reasons for this are unclear, DedA family proteins are required for polymyxin and/or antimicrobial peptide resistance of Salmonella enterica (Shi et al., 2004), Neisseria meningitidis (Tzeng et al., 2005), E. coli (Weatherspoon-Griffin et al., 2011), K. pneumoniae (Jana et al., 2017) and Enterobacter cloacae (Huang et al., 2019).
The energetic requirements of lipid A modification required for colistin resistance are not well characterized. Periplasmic modification of lipid A by ArnT requires cytoplasmic synthesis and inner membrane transport of undecaprenyl-P-Ara4N (Simpson and Trent, 2019). The transport of this lipid-linked intermediate in E. coli is carried out by membrane transporters designated ArnE and ArnF which are similar to small multidrug resistance protein EmrE (Yan et al., 2007). Dephosphorylation and (possibly) recycling of undecaprenyl pyrophosphate is carried out by the membrane protein UppP/BacA that displays low similarity to major facilitator superfamily transporter MdfA and other transporters (El Ghachi et al., 2018; Workman et al., 2018). We have previously demonstrated that proton motive force (PMF)-dependent resistance to EmrE and MdfA substrates such as methyl viologen and ethidium bromide is compromised in an E. coli dedA family mutant (3). Our hypothesis is that transport and/or recycling of undecaprenyl-P-Ara4N and undecaprenyl-P, respectively, is inefficient in DedA family mutants resulting in production of lipid A with lower amounts of Ara4N. In this work, we characterized a B. thailandensis DedA family mutant and observed altered lipid A with reduced amounts of Ara4N compared to the parent strain. In addition, we demonstrate the essentiality of membrane embedded charged amino acids in the DedA protein for colistin resistance. Further, we show that membrane potential is reduced in this mutant and membrane potential is itself required for colistin resistance. These results collectively suggest that the transport activity of DedA and tight control of PMF is required for lipid A modification with Ara4N and colistin resistance of B. thailandensis.
Materials and Methods
Culture Conditions
Burkholderia thailandensis E264 and transposon mutants disrupting the dbcA gene were acquired from the Manoil lab (University of Washington)1 (Gallagher et al., 2013). E. coli cultures were grown in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract and 1% NaCl). B. thailandensis was grown in LB or cation adjusted Mueller-Hinton broth 2 (MH2; typical final pH 7.3. Sigma-Aldrich). Antibiotics colistin, ampicillin (Amp) 100 μg/ml, kanamycin (Kan) 30 μg/ml (E. coli) or 100 μg/ml (B. thailandensis), tetracycline (Tet) 12.5 μg/ml, and trimethoprim (Tmp) 100 μg/ml were purchased from Sigma-Aldrich or VWR. Cultures were grown at 37°C unless otherwise indicated.
Deletion of B. thailandensis E264 dbcA
For targeted mutagenesis of Bth_I1321 (herein referred to as dbcA; NCBI GenBank: ABC36705.1), we used natural transformation of PCR fragments (Thongdee et al., 2008). All oligonucleotides used in PCR were purchased from Sigma-Aldrich and are listed in Supplementary Table S2. Using P1F and P1R, a 1064 bp upstream region of DbcA was PCR-amplified using Q5 DNA polymerase (New England BioLabs). Primers P2F and P2R were used to amplify 980 bp downstream of dbcA (Supplementary Table S2). DNA elements carrying the dhfrIIa gene encoding for trimethoprim resistance (TmpR) was PCR amplified from pUC18T-mini-Tn7T-Tmp plasmid using TmpF and TmpR primers. The plasmid, pUC18T-mini-Tn7T-Tmp was a gift from Dr. Colin Manoil (University of Washington). Primers P1R and TmpF have 40 bp overlapping ends and so do primers P2F and TmpR as shown in Supplementary Table S2 (underlined text). All three amplified fragments were ligated in one step using the Gibson Assembly kit (New England Biolabs). One microliter of the ligated product was used as a template for PCR amplification using P1F and P2R primers. The amplified product (2986 bp) was confirmed by 1% agarose gel electrophoresis and purified using QIAquick Gel Extraction Kit (Qiagen).
For transformation of B. thailandensis E264 with the purified PCR product, a defined medium (DM) was made consisting of 0.25X M63 supplemented with 0.2% glucose, 0.4% glycerol, 1 mM MgSO4, 1 μg/ml thiamine, and six amino acids- leucine, isoleucine, valine, tryptophan, glutamic acid, and glutamine (40 μg/ml each) (19). E264 was grown overnight in LB. The culture was then diluted 1:100 (∼50 μl) in 5 ml of DM media and grown with shaking at 37°C until an optical density at 600 nm of ∼0.6 was reached. 1 ml of culture was centrifuged for 1 min and resuspended in 200 μl of DM. 50 μl aliquots of resuspended cells were mixed with either 0, 100, or 200 ng of the PCR product in a 5 μl volume. The mixtures were incubated without agitation for 30 min at room temperature, and then 1 ml of DM was added and incubated for more than 6 h with shaking at 37°C. The cells were washed twice with 1 ml DM, then resuspended in 250 μl of DM. Different dilutions were made and plated on LB supplemented with Tmp 100 μg/ml to select for recombinants and incubated at 37°C. Genomic DNA was extracted from TmpR clones using Easy-DNA kit (Invitrogen). Genomic DNA was used as a template and primers – Confirm FW and Confirm REV were used to confirm the recombinants. Primers- Seq FW and Seq REV were used to amplify the region, and it was cloned into pBBR1MCS-2 (Kovach et al., 1995) in XhoI and HindIII sites and plasmid specific primers M13FW and M13REV were used to sequence the entire XhoI/HindIII fragment. The ΔdbcA:TmpR replacement was confirmed by DNA Sequencing at the LSU Genomic facility. Removal of the trimethoprim-resistance cassette from TmpR clones was carried out using pFlpTet (Garcia et al., 2013) leaving an FRT scar (ΔdbcA:FRT). The plasmid pFlpTet was a gift from Dr. Erin C. Garcia (University of Kentucky College of Medicine) and was cured at 39°C prior to mutant selection. Some experiments used the strain ΔdbcA:TmpR while others utilized ΔdbcA:FRT and the strain used is indicated in the respective figure legends. The ΔdbcA:TmpS clone was sequenced and confirmed as described above. Genomic DNA was extracted from WT, TmpR, and TmpS clones using Easy-DNA kit (Invitrogen). Genomic DNA was used as a template and primers Seq FW and Seq REV were used to confirm the recombinants (Supplementary Figure S1).
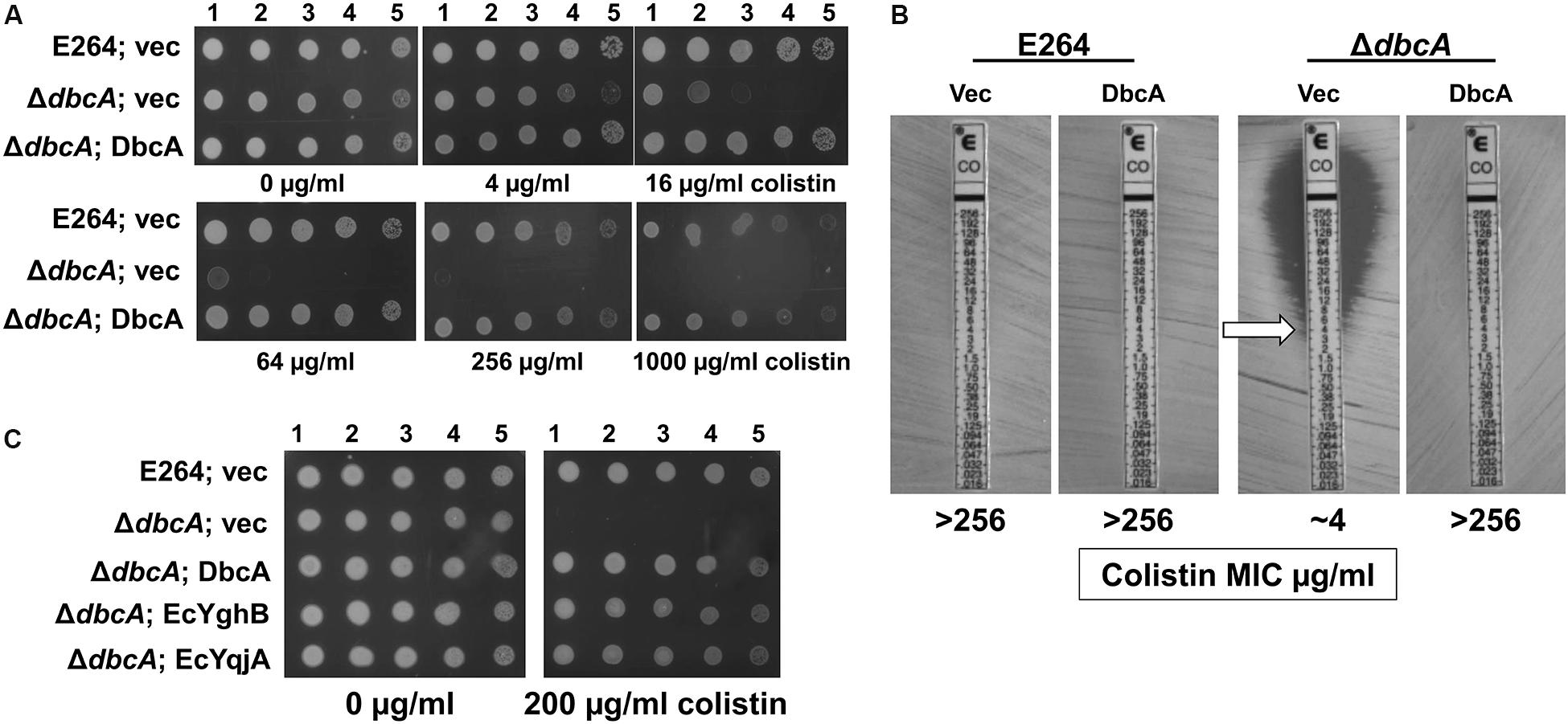
Figure 1. Sensitivity of Burkholderia thailandensis E264 ΔdbcA to colistin. (A) 1:10 dilutions of indicated log-phase grown strains were spotted and grown on MH2 growth medium containing 100 μg/ml kanamycin and either 0, 4, 16, 64, 256, or 1000 μg/ml of colistin. (B) MIC was determined for indicated strains using colistin E-test strips (Biomerieux). Arrow indicates approximate MIC. (C) ΔdbcA was transformed with control vector (vec) or pRP101 (dbcA), pRP102 (EcyqjA) or pRP103 (EcyghB) directing constitutive expression of the indicated genes and log phase cells were diluted and spotted onto plates containing 100 μg/ml kanamycin and either 0 or 200 μg/ml of colistin. Plates were incubated 24 h at 37°C.

Figure 2. Amino acid similarity between Escherichia coli and B. thailandensis DedA family proteins. Amino acid alignment of DbcA, EcYghB and EcYqjA using Clustal Omega (Sievers and Higgins, 2014). Functional acidic (E39, D51; YghB numbering) and basic (R130, R136; YghB numbering) amino acids are in bold font (Kumar and Doerrler, 2014; Kumar et al., 2016). Solid lines represent predicted transmembrane helices of DbcA and EcYghB using TMHMM (Krogh et al., 2001). Residues 156–175 (dashed line) are hydrophobic in nature but the software does not predict a true transmembrane helix. This region may instead be a “re-entrant” helix, similar to what was observed in the crystal structures of UppP/BacA (El Ghachi et al., 2018; Workman et al., 2018). Residues 218–220 of DbcA (dotted line) do not align with the beginning of the same helix of YghB. An asterisk indicates positions that have a single, fully conserved residue. A colon indicates conservation between strongly similar amino acids. A period indicates conservation between weakly similar amino acids.
Site-Directed Mutagenesis
For making point mutants in dbcA gene, we cloned the dbcA gene under the inducible rhamnose promoter in pSCrhaB2 vector (Cardona and Valvano, 2005) using primers FWdbcAhis and REVdbcAhis in such a way to add a hexahistidine tag at the C terminus (Iiyama et al., 2011). The plasmid pScrhaB2 was a generous gift of Dr. Josephine Chandler (University of Kansas, Dept. of Molecular Biosciences). Site-specific mutations were created according to a previously published protocol (Kumar and Doerrler, 2014). The primers with site specific mutations (Supplementary Table S2) were used to amplify the entire vector containing the dbcA gene. The PCR products were digested with DpnI, purified with a Qiagen kit and used to transform competent XL1-Blue cells. Transformants were screened by gene specific primers. Point mutations were confirmed by DNA sequencing at the LSU College of Science Genomic Facility. Membrane preparation and Western blotting using an anti-pentahis antibody (Qiagen) was performed as previously described (Kumar and Doerrler, 2014).
Transformation and Complementation Analysis
For transformation of E. coli, a common heat shock method was used (Froger and Hall, 2007). For B. thailandensis, washes and electroporation was carried out at room temperature, which improved transformation efficiency (Tu et al., 2016). B. thailandensis was grown in LB with shaking at 37°C until an optical density at 600 nm of ∼0.6 was reached. The cells were washed once with water and twice with 10% glycerol. The cells were resuspended with ∼300 μl of 10% glycerol to a concentration 2–3∗1010 cells/ml. A 50 μl aliquot was mixed with ∼500 ng of plasmid DNA and electroporated using a Bio-Rad MicroPulser using a 0.2 cm cuvette and 2.5 kV voltage setting. One ml of warm SOC media was added immediately after the pulse and incubated for 1.5 h with shaking at 37°C. The cells were centrifuged and resuspended in 100 μl of SOC, plated on LB plates with appropriate antibiotics, and incubated at 37°C for up to 48 h.
For complementation experiments, all genes were cloned under a constitutive lac promoter of an expression vector, pBBR1MCS-2 (Kovach et al., 1995) and strains selected in the presence of Kan 100 μg/ml (B. thailandensis) or 30 μg/ml (E. coli). The unmodified plasmid was used as vector control in all experiments. E. coli DedA genes yqjA (EcyqjA) and yghB (EcyghB) were cut from pBAD-yqjA and pBAD-yghB (Sikdar et al., 2013) with XhoI and HindIII and ligated into a similarly digested pBBR1MCS-2 vector using T4 DNA ligase resulting in pRP102 and pRP103, respectively. B. thailandensis DbcA was PCR amplified from genomic DNA of B. thailandensis using FWdbcA and REVdbcA primers and ligated into XhoI and HindIII sites of pBBR1MCS-2 resulting in pRP101. For construction of kanamycin-sensitive strain BC202KS, the kanamycin resistance cassette of BC204 (ΔyghB:KanR) (Thompkins et al., 2008) was removed using Flp recombinase expressed from plasmid pCP20 (Cherepanov and Wackernagel, 1995; Datsenko and Wanner, 2000; Boughner and Doerrler, 2012). Following curing of pCP20 at 42°C, the resulting strain was transduced to tetracycline resistance using a p1vir lysate prepared from strain BC203 (ΔyqjA:TetR) (Thompkins et al., 2008), resulting in kan-sensitive BC202KS (Supplementary Table S1).
Microscopy
Overnight cultures of E. coli were diluted 1:100 in fresh LB media with suitable antibiotics and additives, and grown to OD600 ∼ 0.6 at 30°C in a shaking incubator. Ten μL of cells were applied to a 1% agarose coated glass slide for imaging. A Leica DM6B-Z deconvolution microscope was used for all the differential interference contrast (DIC) micrographs. Observations were made by a 100X, 1.44-numerical-aperture oil immersion objective lens (HC PL APO). The images were captured through Hamamatsu C11440-22C, 16 bit camera and recorded using Leica Application Suite X (LAS X) software.
Susceptibility to Colistin and Other Antibiotics
For testing the susceptibility on solid medium, overnight cultures were freshly diluted 1:100 in LB or MH2 media with appropriate antibiotics and additives, and grown to OD600 ∼0.6 at 37°C in a shaking incubator. Five microliters of serially log10-diluted cells were spotted onto MH2 or LB agar plates containing antibiotics. Growth was analyzed after incubation for 24 h at 37°C. The MIC was measured with colistin E-test strips (Biomerieux). Overnight cultures were diluted 1:100 into fresh MH2 broth and grown to OD600 0.6. A 1:10 dilution was spread on MH2 plates to create a lawn of cells and the strip was applied to the plates and evaluated after 24 h at 37°C. All experiments were repeated at least three times.
Mass Spectrometry
For isolation of lipid A, cultures were grown at 37°C to an OD600 of ∼1.0. Lipid A chemical extraction was carried out after mild acidic hydrolysis of LPS as previously described (Zhou et al., 1999; Herrera et al., 2014). For visualization of lipid A by mass spectrometry, lipids were analyzed using MALDI-TOF (ABI 4700 Proteomic Analyzer) in the negative-ion linear mode as previously described (Zhou et al., 2010; Henderson et al., 2013). Briefly, lipid A samples were dissolved in a mixture of chloroform-methanol (4:1, vol/vol), and 1 μl of sample was mixed with 1 μl of matrix solution. The matrix consisted of 5-chloro-2-mercaptobenzothiazole (CMBT) (20 mg/mL) resuspended in chloroform-methanol-water (4:4:1, vol/vol/vol) mixed with saturated ammonium citrate (20:1, vol/vol). One μl of sample-matrix mixture was loaded on to MALDI target plate for final analysis.
Determination of Membrane Potential
Assessment of membrane potential (Δψ) using JC-1 dye was as described (Sikdar et al., 2013). Briefly, All strains were treated with 4 μM JC-1 dye in permeabilization buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 10 mM glucose), incubated in the dark at 30°C without shaking for 30 min and fluorescence measurements carried out using a JASCO FP-6300 spectrofluorometer. Strains treated with CCCP for 30 min served as a control for the loss of Δψ.
Results
Colistin Sensitivity of the B. thailandensisΔdbcA
By screening several sequence-defined B. thailandensis transposon mutants obtained from the Manoil lab (Gallagher et al., 2013) in genes encoding DedA family members we discovered that several with confirmed insertions in the gene Bth_I1321 (referred to herein as dbcA; DedA of Burkholderia required for colistin resistance) were highly sensitive to colistin and polymyxin B. To confirm this observation in a gene deletion strain, we created a B. thailandensis strain where the dbcA locus was replaced with a trimethoprim resistance (TmpR) gene (dhfrIIa). Gene replacement and removal of the TmpR cassette in strain ΔdbcA was confirmed by PCR and DNA sequencing (Supplementary Figure S1). While wild type strain E264 and complemented ΔdbcA grew in the presence of high concentrations of colistin (>1000 μg/ml), ΔdbcA carrying the control vector failed to grow at colistin concentrations greater than about 4 μg/ml (Figure 1). This was shown by spotting dilutions of log-phase cells (Figure 1A) and by using colistin E-test gradient strips (Figure 1B). Expression of E. coli yghB (EcyghB) or EcyqjA could restore growth of ΔdbcA in the presence of high concentrations of colistin (Figure 1C). DbcA displays moderate amino acid identity to EcYqjA (∼34%) and EcYghB (∼30%) (Figure 2).
Mass Spectrometry Analysis Reveals Reduced Levels of Ara4N-Modified Lipid A in ΔdbcA Mutant
Since lipid A is the initial interaction site of polymyxins, and lipid A modifications contribute to colistin resistance in numerous Gram-negative species, lipid A structure was analyzed by mass spectrometry. In the analysis of wild type B. thailandensis E264 harboring control and complementing vector as well as the ΔdbcA strain harboring complementing vector, we observed three major peaks (m/z ∼ 1670.01, 1801.49, and 1932.06) which correspond to pentaacylated lipid A, pentaacylated lipid A singly modified with Ara4N and pentaacylated lipid A doubly modified with Ara4N (Figures 3A,B,D). Lipid A species containing aminoarabinose are labeled with red font in Figure 3. While singly modified and unmodified lipid A are major species in these strains, there is a significant amount of doubly modified species as well. Strikingly, in ΔdbcA harboring control vector, doubly modified 2-Ara4N lipid A is undetectable and singly and unmodified lipid A are minor and major species, respectively (Figure 3C). There is also a detectable peak at m/z = 1444 which corresponds to unmodified tetraacylated lipid A. The depletion of Ara4N-modified lipid A may contribute to the observed colistin sensitivity in strain ΔdbcA. Predicted structures of lipid A species are shown in Figure 3E. Complete absence of Ara4N modified lipid A is not expected in B. thailandensis due to the demonstrated essentiality of the glycosyltransferase ArnT in Burkholderia cenocepacia (Ortega et al., 2007; Hamad et al., 2012). ArnT is also likely essential in B. thailandensis due to the absence of such a transposon mutant in the ordered library (Gallagher et al., 2013) and comparative Tn-Seq analysis (Gislason et al., 2017). Modification of lipid A with phosphoethanolamine was not detected, consistent with the absence of an EptA homolog in the B. thailandensis E264 genome (Kim et al., 2005). This observation in combination with the marked sensitivity to colistin of ΔdbcA is noteworthy and prompted us to analyze B. thailandensis DbcA protein and corresponding ΔdbcA strain in more detail.
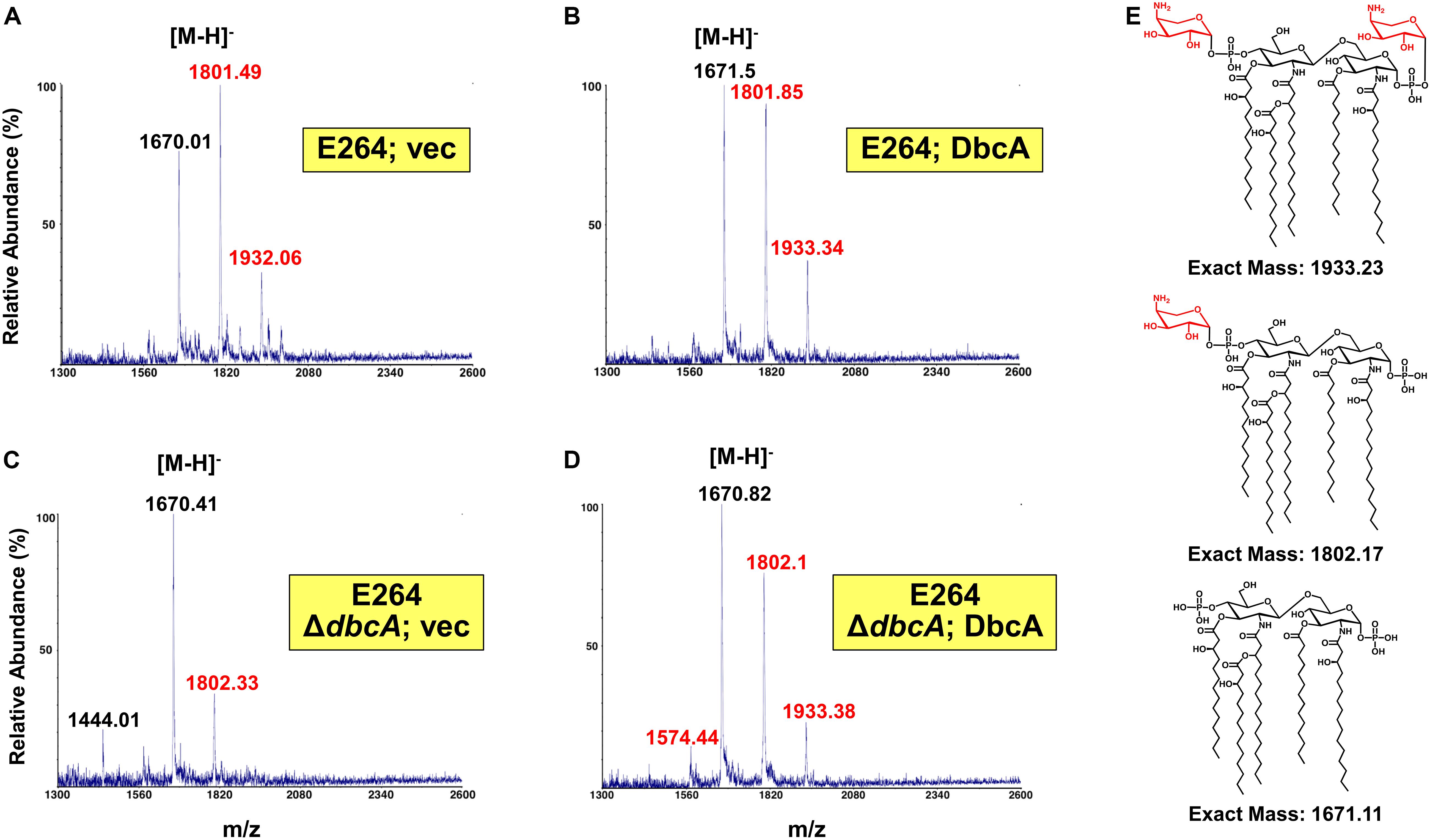
Figure 3. Mass spectrometry of lipid A isolated from B. thailandensis strains. (A–D) Lipid A extracted from the indicated strains was analyzed using a MALDI-TOF mass spectrometer (ABI 4700 Proteomic Analyzer) in the negative-ion linear mode. Species modified with Ara4N are labeled in red font. (E) Predicted structures of each observed species. Minor species of unmodified tetraacylated lipid A (m/z = 1444.01) and singly Ara4N-modified tetraacylated lipid A (m/z = 1574.44) were also detected.
Functionality of B. thailandensis DbcA in Escherichia coli
The alignment of DbcA with EcYqjA and EcYghB (Figure 2) includes conservation of important membrane embedded acidic (YqjA E39, D51) and basic (YqjA R130, R136) amino acids required for function (Kumar and Doerrler, 2014, 2015; Kumar et al., 2016). To begin to dissect the function of DbcA, we asked whether it could functionally complement altered cell division, temperature sensitivity, and biocide sensitivity of the E. coli mutant BC202 (ΔyqjA, ΔyghB) (Thompkins et al., 2008; Sikdar and Doerrler, 2010; Kumar and Doerrler, 2014). We cloned dbcA into pBBR1MCS-2 and the resulting plasmid pRP101 was introduced into parent strain W3110 and mutant BC202KS (a kanamycin-sensitive strain to allow for selection of Burkholderia shuttle vectors). pRP102 (EcyqjA) and pRP103 (EcyghB) served as positive controls in these experiments as they can complement these phenotypes of BC202 (Sikdar et al., 2013; Kumar and Doerrler, 2015). As shown in Figure 4, expression of DbcA in BC202KS can restore growth at 42°C (Figure 4A), resistance to biocides (Figure 4B) and normal cell division (Figure 4C) to the same degree as EcyqjA and EcyghB. We conclude that DbcA functions similarly to DedA family proteins YqjA and YghB in E. coli.
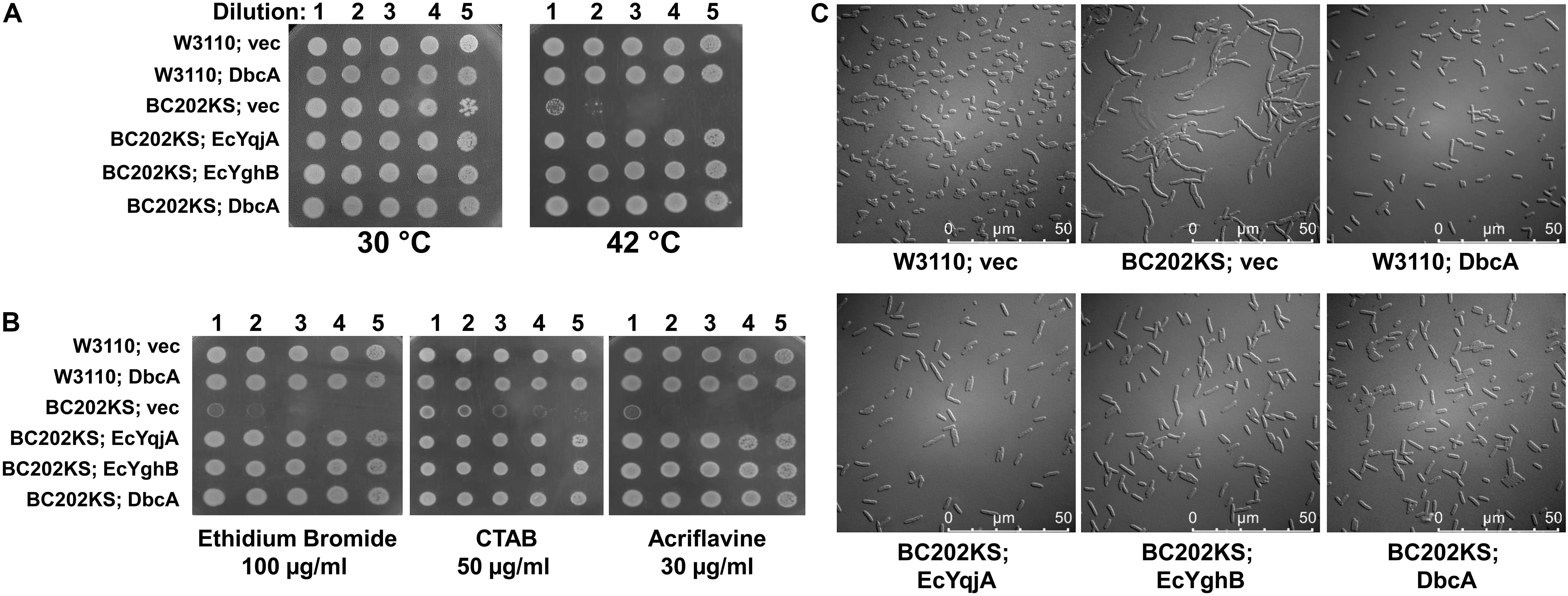
Figure 4. Complementation of E. coli BC202 with dbcA. (A) Restoration of growth of BC202KS at 42°C by plasmid expression of dbcA, EcyqjA or EcyghB. (B) Restoration of BC202KS resistance to ethidium bromide, CTAB (cetyltrimethyl ammonium bromide), and acriflavine by plasmid expression of dbcA, EcyqjA or EcyghB. (C) Restoration of normal cell division of BC202KS at 30°C by plasmid expression of dbcA, EcyqjA or EcyghB. Abbreviations: Vec, vector control. All strains were grown in LB medium.
Site-Directed Mutagenesis of DbcA Conserved, Charged Amino Acids
As stated above, DbcA has moderate amino acid identity to the E. coli DedA family proteins YqjA and YghB. This conservation includes charged amino acids E67, D79, R161, and R167 that are in comparable positions as YghB/YqjA E39, D51, R130 and R136, respectively (Figure 2), which are required for function (Kumar and Doerrler, 2014; Kumar et al., 2016). Acidic amino acids have been shown to be required for function of numerous secondary transporters including MdfA, NhaA, MdtM, and LacY (Gerchman et al., 1993; Noumi et al., 1997; Abramson et al., 2004; Adler and Bibi, 2004; Sigal et al., 2005; Fluman et al., 2012; Holdsworth and Law, 2012). Furthermore, the significance of membrane embedded basic amino acids such as arginine are well documented in the literature and play a role in a number of processes including regulation of redox potential (Cutler et al., 1989; Winn et al., 2002), voltage detection across a lipid bilayer (Jiang et al., 2003; Long et al., 2005; Tao et al., 2010), and proton transport (Cain and Simoni, 1989; Hellmer et al., 2003; Sigal et al., 2005). We changed each of these amino acids of DbcA to alanine to determine the effect on the ability of the expressed protein to restore colistin resistance to the mutant ΔdbcA. All strains grew well in the absence of colistin (Figure 5A). Complementation of growth in the presence of colistin was observed when wild type dbcA along with dbcA-E67A and dbcA-R161A was expressed. Less growth was seen in the presence of colistin when control vector, dbcA-D79A or dbcA-R167A were expressed. Therefore, we conclude that D79 and R167 likely play major roles in the transport mechanism of DbcA while E67 and R161 may play more minor roles. All proteins were found expressed in the membrane fraction of cells (Figure 5B) suggesting that mutant protein misfolding was not a major issue under our conditions. These results suggest that B. thailandensis DbcA functions as a proton-dependent membrane transporter.
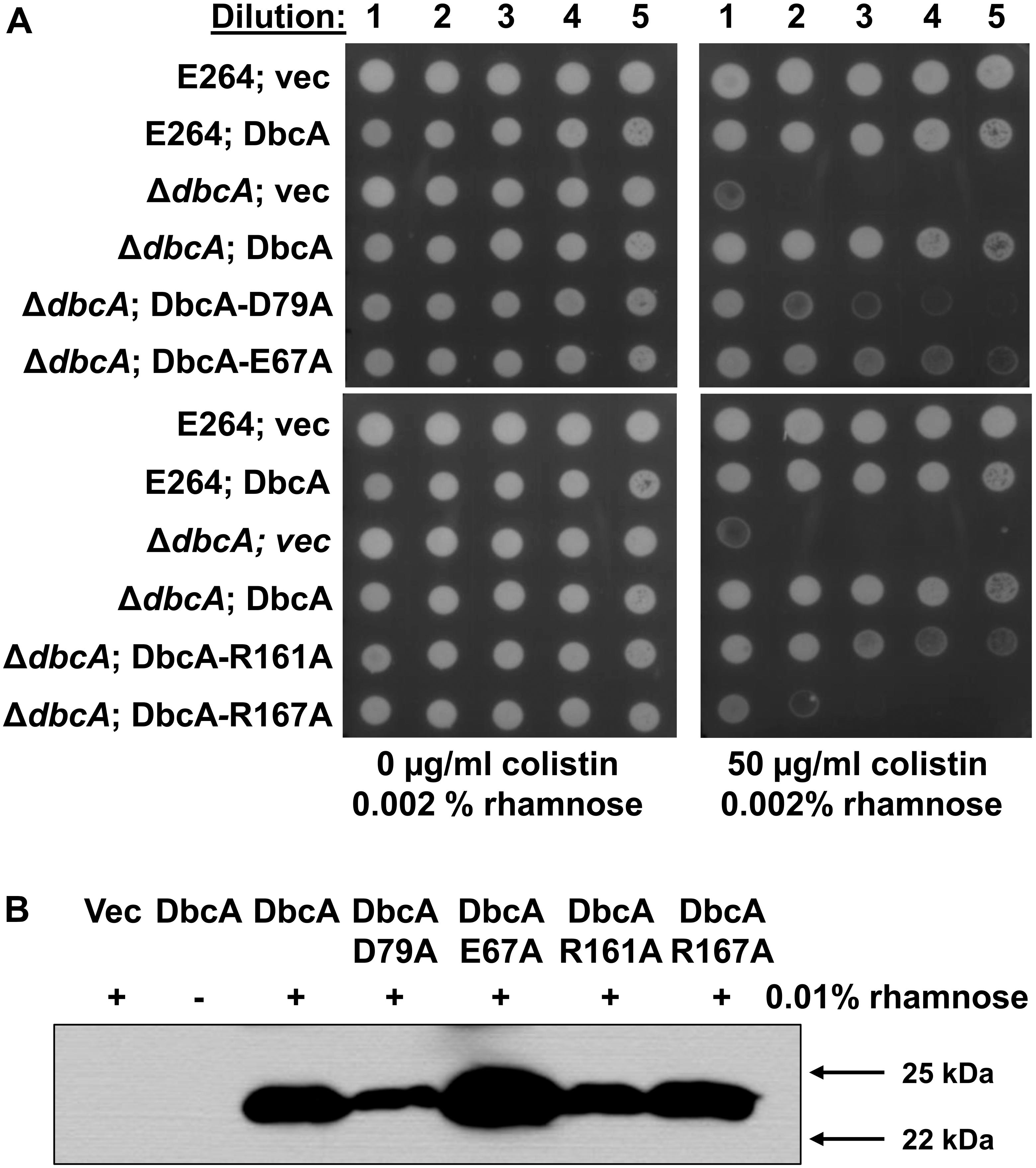
Figure 5. Site directed mutagenesis of B. thailandensis DbcA conserved, charged amino acids. (A) Dilutions of mid-log phase grown cells of the indicated strains were spotted on MH2 agar plates containing 0.002% rhamnose, 100 μg/ml Tmp and either 0 or 50 μg/ml colistin. This concentration of rhamnose was used to avoid toxicity caused by overexpression of dbcA in the wild type strain but was not sufficient for detection using Western blotting (see below). DbcA with mutation D79A or R167A were unable to restore growth in the presence of colistin to the extent of wild type DbcA. (B) Expression of dbcA and point mutants in membrane fractions of ΔdbcA as determined by Western blotting with anti-hexahistidine antibody. All experiments in this figure were conducted with strain ΔdbcA:FRT to allow for use of Tmp selection of vectors. Abbreviations: Vec, vector control. Sixty micrograms of membrane protein was loaded per lane and strains were grown in the presence of Tmp and 0.01% rhamnose.
Altered Membrane Potential in ΔdbcA Mutant
According to chemiosmotic theory (Mitchell, 1961), the membrane PMF is equal to the sum of the charge difference across the membrane (ΔΨ) and the pH difference across the membrane (ΔpH). To examine the PMF in more detail, we measured the ΔΨ component of the PMF using dye JC-1. JC-1 is a membrane permeable dye that exhibits green fluorescence (530 nm) as a monomer but forms aggregates at the membrane in the presence of membrane potential, shifting its emission from green to red (595 nm). Therefore, relative membrane potential can be expressed as the ratio of red to green fluorescence (Jovanovic et al., 2006; Engl et al., 2011). We previously reported that E. coli strain BC202 (ΔyqjA, ΔyghB) displays compromised ΔΨ using this dye (Sikdar et al., 2013). B. thailandensisΔdbcA along with wild type harboring either control or complementing vector were grown to mid-log phase and treated with JC-1 dye. Cells treated with the proton ionophore CCCP were included as a control. All wild type and complemented mutant strains exhibited a consistent 595/530 ratio (Figure 6A). However, B. thailandensisΔdbcA displayed a lower ratio suggesting partial dissipation of the ΔΨ component of the PMF. This value could alter PMF-dependent processes required for colistin resistance. Non-complemented B. thailandensisΔdbcA was also compromised for motility (Figure 6B), and hypersensitive to CCCP (Figure 6C); consistent with partial depolarization of the membrane. This result suggests that loss of PMF can be associated with colistin sensitivity in B. thailandensis.
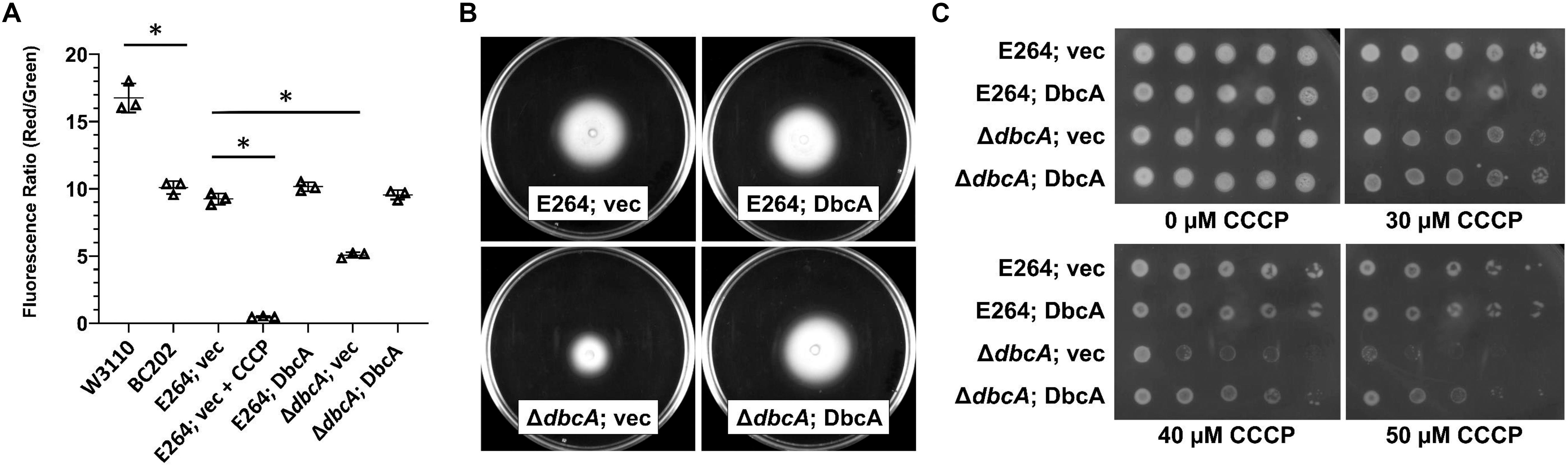
Figure 6. Measurement of membrane potential and CCCP sensitivity. (A) Assessment of membrane potential (Δψ) of B. thailandensis strains using JC-1 dye represented as the red (595 nm)/green (530 nm) ratio. BC202 was previously shown to be depolarized compared to W3110 (Sikdar et al., 2013) and is included here as a control. E264 (vec) treated with 25 μM CCCP for 30 min served as a control for the loss of Δψ. The graph was drawn using GraphPad Prism 8.1.1 software. Each bar represents the average and standard deviation of three biological replicates. Each experiment was repeated three times. Growth media included 0.0001% rhamnose. (B) Impaired motility in ΔdbcA:FRT strain. For the motility test assay, strains were grown overnight in MH2 and 100 μg/ml Tmp. Five microliters of the overnight culture were inoculated on MH2 media containing 100 μg/ml Tmp and solidified with 0.4% agar and incubated at 37°C for 48 h. No rhamnose was included in the growth media. (C) Sensitivity of ΔdbcA:FRT to CCCP. Dilutions of strains were spotted on MH2 plates containing 100 μg/ml Tmp, 0.001% rhamnose and the indicated concentration of CCCP and incubated overnight at 37°C. The experiments in panel A-C were conducted with strain ΔdbcA:FRT to allow use of Tmp selection (see Supplementary Table S1). Bars represent mean ± SD of three independent determinations and statistical significance was calculated by unpaired Student’s t-test using GraphPad Prism 8.1.1. ∗p < 0.001.
PMF Depletion Results in Colistin Sensitivity
In order to test if PMF depletion can directly cause sensitivity to colistin, we grew both parent strain B. thailandensis E264 and mutant ΔdbcA in the presence of CCCP and measured MIC of colistin. We used CCCP concentrations that we determined could reduce the ΔΨ but still allow for survival of the bacteria (Figure 6). As depicted in Figure 7, growth in the presence of CCCP results in significant sensitivity to colistin. The MIC of E264 reduced from unmeasurable to approximately 32 μg/ml or 2 μg/ml in the presence of 20 or 25 μM CCCP, respectively. The colistin MIC of ΔdbcA was reduced from 6 μg/ml to approximately 2 μg/ml or 0.75 μg/ml in the presence of 15 or 20 μM CCCP, respectively. This result is consistent with previous findings showing exposure to CCCP sensitizes other species of Gram-negative bacteria to colistin (Park and Ko, 2015; Ni et al., 2016; Osei Sekyere and Amoako, 2017; Baron and Rolain, 2018). These results strongly support our hypothesis of a requirement of PMF for efficient modification of lipid A with Ara4N and colistin resistance in B. thailandensis and likely other Gram-negative species (Figure 8). DedA family protein DbcA is required for PMF maintenance in this model.
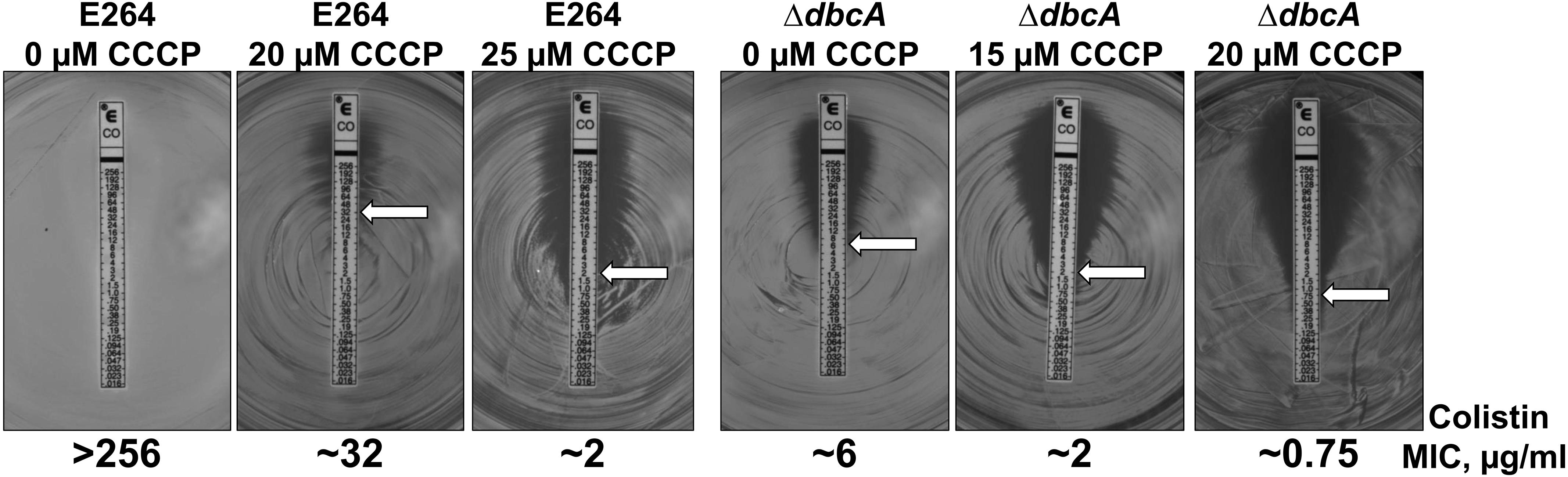
Figure 7. PMF dissipation with CCCP results in B. thailandensis colistin sensitivity. Colistin MIC was determined for indicated strains using E-test strips (Biomerieux). MH2 plates contained 0, 15, 20 or 25 μM CCCP as indicated and 100 μg/ml Tmp. White arrows indicate approximate MIC.
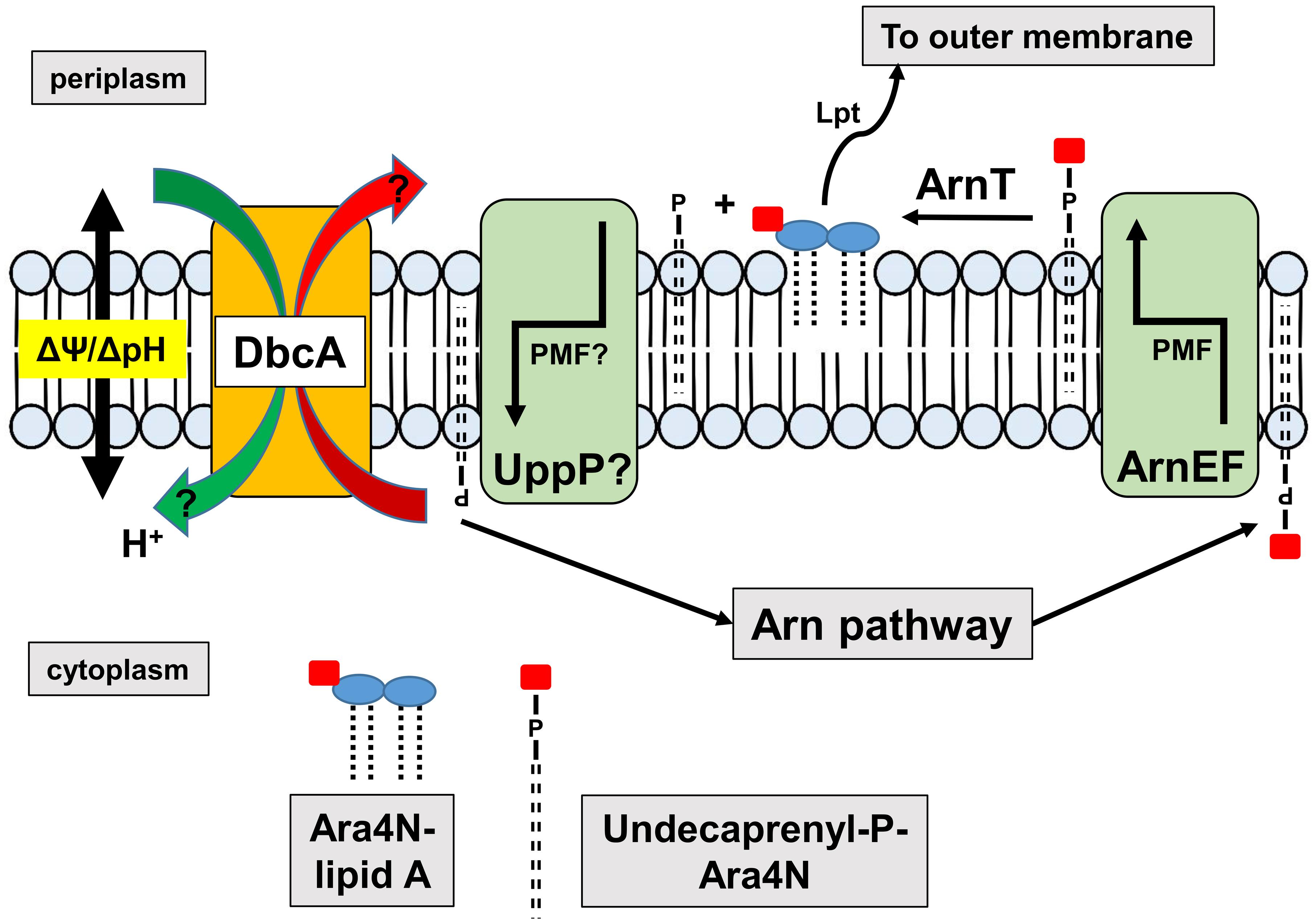
Figure 8. Requirement of DedA family and PMF for efficient synthesis of Ara4N-lipid A. The Arn operon produces undecaprenyl-P-Ara4N that is found initially in the cytoplasmic leaflet of the inner membrane. The EmrE-like transporters ArnEF catalyze the transbilayer movement of undecaprenyl-P-Ara4N to the periplasmic surface of the inner membrane (Yan et al., 2007) where it is a substrate of the glycosyltransferase ArnT (Petrou et al., 2016; Tavares-Carreon et al., 2016). ArnT modifies lipid A with Ara4N on the periplasmic surface of the inner membrane, followed by transport to the outer membrane by the lipopolysaccharide transport pathway (Lpt) (Li et al., 2019; Owens et al., 2019) contributing to colistin resistance. Undecaprenyl-P produced is recycled to the cytoplasm possibly by UppP/BacA, the undecaprenyl pyrophosphate phosphatase, which bears similarity to MdfA and other transporters (El Ghachi et al., 2018; Workman et al., 2018). Both MdfA (Fluman et al., 2012) and EmrE (Robinson et al., 2017) are proton-dependent transporters suggesting that the PMF may be required for both the transbilayer movement of undecaprenyl-P-Ara4N and the recycling of undecaprenyl-P. Perturbation of the PMF in DedA family mutants may interfere with either or both of these steps. We have shown that DedA family proteins are likely proton-dependent transporters and efflux of MdfA and EmrE substrates is compromised in the E. coli DedA family mutant BC202, resulting in hypersensitivity to these compounds (Kumar and Doerrler, 2014, 2015).
Discussion
Multidrug resistant bacterial infections pose an enormous public safety risk and are a challenge to modern medicine, made worse by a lack of new antimicrobial drugs (O’Neill, 2014; World Health Organization [WHO]∗, 2014). The emergence of carbapenem-resistant Enterobacteriaceae is of particular concern and has forced the reintroduction of colistin therapy as a last resort treatment. Colistin is an antimicrobial peptide belonging to the polymyxin family that can cause numerous side effects including nephrotoxicity (Koch-Weser et al., 1970). Moreover, some epidemic clones of K. pneumoniae have acquired colistin resistance by LPS modification via cationic substitution. Other Enterobacteriaceae such as Proteus mirabilis and Serratia marcescens are naturally resistant to polymyxins due to the constitutive expression of the arnBCADTEF operon and/or the eptB gene (Poirel et al., 2017). The role of the DedA family in providing intrinsic resistance to polymyxins in Burkholderia spp. has not been investigated prior to this study. Here we demonstrate that the loss by mutation of DedA homolog DbcA lowers the colistin MIC of B. thailandensis to 5–10 μg/ml or at least 100-fold, indicating that this protein plays a key role in determining the resistance phenotype in this species. Although dedA family genes have been identified in screens for colistin sensitivity in several species (Shi et al., 2004; Tzeng et al., 2005; Weatherspoon-Griffin et al., 2011; Jana et al., 2017; Huang et al., 2019), this work represents the first characterization of the role of the conserved DedA protein family in resistance to colistin and the first such study in any Burkholderia species.
Resistance to colistin in Gram-negative bacteria is usually conferred by activation of pathways that lead to the covalent modification of cell surface LPS, causing loss of electrostatic binding by the cationic antibiotic (Raetz et al., 2007; Olaitan et al., 2014). In K. pneumoniae and other species, these pathways are controlled by the PhoPQ and PmrAB two-component signaling pathways that lead to modification of lipid A with Ara4N and phosphoethanolamine (Olaitan et al., 2014; Poirel et al., 2017). While the energy requirements of the Ara4N pathway have not been characterized extensively, certain steps likely require the PMF (Figure 8). For example, it is known that transbilayer movement of the undecaprenyl-P-Ara4N from the cytoplasm to the periplasm require the EmrE-like transporters ArnEF (Yan et al., 2007) which are predicted to utilize the PMF. In addition, the undecaprenyl pyrophosphate phosphatase UppP/BacA displays amino acid similarity to the drug efflux pump MdfA and other secondary transporters and therefore may catalyze the transbilayer movement of undecaprenyl-P back to the cytoplasm in a PMF-dependent manner (El Ghachi et al., 2018; Workman et al., 2018). Therefore, tight control of PMF may be required for efficient modification of lipid A and resistance to colistin.
Extreme polymyxin resistance is a hallmark of Burkholderia species (Loutet and Valvano, 2011). While wild type enteric bacteria display resistance to low μg/ml concentrations of polymyxins (MIC 0.2–2 μg/ml) and resistant variants reach MIC up to 128 μg/ml2, Burkholderia species are often resistant to mg/ml concentrations and beyond (Burtnick and Woods, 1999; Thwaite et al., 2009; Loutet et al., 2011). In most resistant species, lipid A modification with aminoarabinose plays a major role, but it is possible that Burkholderia spp. employ other mechanisms including secreted proteases, utilization of efflux pumps, synthesis of putrescine, and hopanoid biosynthesis (Burtnick and Woods, 1999; Loutet and Valvano, 2011; Malott et al., 2012; El-Halfawy and Valvano, 2014). It is possible that the ΔdbcA mutation affects more than one resistance mechanism. The glycosyltransferase ArnT catalyzes the periplasmic modification of lipid A with Ara4N (Raetz et al., 2007; Petrou et al., 2016; Tavares-Carreon et al., 2016). While Ara4N synthesis is non-essential in most species and regulated by environmental cues (Raetz et al., 2007), ArnT is essential in Burkholderia spp. (Ortega et al., 2007; Hamad et al., 2012; Gislason et al., 2017) and appears to be required for LPS export (Hamad et al., 2012). For this reason, it is not possible to assess the colistin resistance of B. thailandensis in the complete absence of Ara4N modification without utilizing second site suppressors. The existence of a B. thailandensis mutant that inefficiently modifies lipid A with Ara4N will prove to be a valuable tool to study the role of the Ara4N pathway in LPS biogenesis in Burkholderia spp.
The DedA family of membrane proteins is widely distributed in nature, found in all kingdoms. Until recently, most of what is known about the family came from studies in bacteria due to pleiotropic phenotypes associated with null mutations. These phenotypes include defects in growth, cell division and sensitivity to alkaline pH, antibiotics and membrane penetrating dyes (Thompkins et al., 2008; Liang et al., 2010; Sikdar and Doerrler, 2010; Sikdar et al., 2013; Kumar and Doerrler, 2014, 2015). The correction of these phenotypes by growth in slightly acidic pH and the presence of membrane embedded charged amino acids suggest that members of the DedA family are proton-dependent transporters required for PMF maintenance (Sikdar et al., 2013; Kumar and Doerrler, 2014, 2015; Kumar et al., 2016). While there is no published structure of any DedA family protein, they do have an evolutionary relationship and, indeed may share structural similarity to proteins of the LeuT family of transporters (Khafizov et al., 2010; Keller et al., 2014). One of the earliest reports on a eukaryotic DedA family found the protein, called Tvp38, associated with tSNARE in Tlg-2 containing Golgi compartments in yeast (Inadome et al., 2007). Recent studies using CRISPR screening has demonstrated that a human DedA protein, known as TMEM41B, plays a role in autophagosome formation (Moretti et al., 2018; Morita et al., 2018; Shoemaker et al., 2019). While the eukaryotic proteins of the DedA family containing the so-called VTT domain (for VMP, TMEM41, Tvp38) (Morita et al., 2018) are distantly related to their bacterial counterparts, and a functional relationship has not been established, the presence of an absolutely conserved glycine residue suggests an evolutionary relationship (Tabara et al., 2019).
We have shown that a DedA family protein is required for intrinsic colistin resistance in B. thailandensis. Colistin sensitivity may be due to the requirement of the DedA family for proper PMF maintenance. We show that B. thailandensis ΔdbcA has lower membrane potential compared to the wild type. The colistin hypersensitivity seen with exposure to CCCP (Figure 7) also suggests that membrane potential is critical for colistin resistance in B. thailandensis. Membrane depolarization caused by CCCP has been shown to sensitize numerous Gram-negative species to colistin (Park and Ko, 2015; Ni et al., 2016; Osei Sekyere and Amoako, 2017; Baron and Rolain, 2018). Metabolically less active Pseudomonas aeruginosa biofilm cells were also more readily killed by colistin compared to metabolically active biofilm cells (Pamp et al., 2008). It is possible that the membrane depolarization of ΔdbcA has a negative impact on PMF dependent transporters that contribute directly or indirectly to colistin resistance. Efflux pump activity has recently been shown to be linked to lipid A modification with Ara4N and polymyxin resistance in B. thailandensis (Krishnamoorthy et al., 2019). Collectively, these results underscore the importance of maintenance of PMF in resistance to polymyxins as well as provide fresh insight into the roles of the widely distributed DedA family of membrane proteins.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
WD, PP, and SK: conception or design of the study. PP, CB, CS, MD, CH, MT, and WD: the acquisition, analysis, or interpretation of the data. PP, MT, and WD: writing of the manuscript.
Funding
This work was supported by the LSU Foundation Biotransport Research Support Fund (WD), and the NIH grants AI138576, AI076322, and AI064184 (MT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Megan Justice, Dr. Gregg Pettis (LSU Department of Biological Sciences), Dr. Ann Grove (LSU Department of Biological Sciences), and Dr. Nathan Gilbert (LSU Department of Biological Sciences) for their valuable suggestions and discussions. We would also like to thank Dr. Marcia Newcomer (LSU Department of Biological Sciences) and Dr. David Burk (Pennington Biomedical Research Center) for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02532/full#supplementary-material
Footnotes
References
Abramson, J., Iwata, S., and Kaback, H. R. (2004). Lactose permease as a paradigm for membrane transport proteins (Review). Mol. Membr. Biol. 21, 227–236. doi: 10.1080/09687680410001716862
Adler, J., and Bibi, E. (2004). Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J. Biol. Chem. 279, 8957–8965. doi: 10.1074/jbc.M313422200
Ainsworth, G. C., Brown, A. M., and Brownlee, G. (1947). Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature 159:263. doi: 10.1038/160263a0
Baron, S. A., and Rolain, J. M. (2018). Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and Gram-negative bacteria. J. Antimicrob. Chemother. 73, 1862–1871. doi: 10.1093/jac/dky134
Boughner, L. A., and Doerrler, W. T. (2012). Multiple deletions reveal the essentiality of the DedA membrane protein family in Escherichia coli. Microbiology 158(Pt 5), 1162–1171. doi: 10.1099/mic.0.056325-0
Burtnick, M. N., and Woods, D. E. (1999). Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 43, 2648–2656. doi: 10.1128/aac.43.11.2648
Cain, B. D., and Simoni, R. D. (1989). Proton translocation by the F1F0ATPase of Escherichia coli. Mutagenic Analysis of the a subunit. J. Biol. Chem. 264, 3292–3300.
Cardona, S. T., and Valvano, M. A. (2005). An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54, 219–228. doi: 10.1016/j.plasmid.2005.03.004
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli - Tc(R) and Km(R) cassettes with the option of FLP-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-a
Cutler, R. L., Davies, A. M., Creighton, S., Warshel, A., Moore, G. R., Smith, M., et al. (1989). Role of arginine-38 in regulation of the cytochrome c oxidation-reduction equilibrium. Biochemistry 28, 3188–3197. doi: 10.1021/bi00434a012
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Doerrler, W. T., Sikdar, R., Kumar, S., and Boughner, L. A. (2013). New functions for the ancient DedA membrane protein family. J. Bacteriol. 195, 3–11. doi: 10.1128/JB.01006-12
El Ghachi, M., Howe, N., Huang, C. Y., Olieric, V., Warshamanage, R., Touze, T., et al. (2018). Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis. Nat. Commun. 9:1078. doi: 10.1038/s41467-018-03477-5
El-Halfawy, O. M., and Valvano, M. A. (2014). Putrescine reduces antibiotic-induced oxidative stress as a mechanism of modulation of antibiotic resistance in Burkholderia cenocepacia. Antimicrob. Agents Chemother. 58, 4162–4171. doi: 10.1128/AAC.02649-14
Engl, C., Beek, A. T., Bekker, M., de Mattos, J. T., Jovanovic, G., and Buck, M. (2011). Dissipation of proton motive force is not sufficient to induce the phage shock protein response in Escherichia coli. Curr. Microbiol. 62, 1374–1385. doi: 10.1007/s00284-011-9869-5
Fluman, N., Ryan, C. M., Whitelegge, J. P., and Bibi, E. (2012). Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol. Cell 47, 777–787. doi: 10.1016/j.molcel.2012.06.018
Froger, A., and Hall, J. E. (2007). Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 6:253. doi: 10.3791/253
Gallagher, L. A., Ramage, E., Patrapuvich, R., Weiss, E., Brittnacher, M., and Manoil, C. (2013). Sequence-defined transposon mutant library of Burkholderia thailandensis. MBio 4:e00604-13. doi: 10.1128/mBio.00604-13
Garcia, E. C., Anderson, M. S., Hagar, J. A., and Cotter, P. A. (2013). Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol. Microbiol. 89, 1213–1225. doi: 10.1111/mmi.12339
Gerchman, Y., Olami, Y., Rimon, A., Taglicht, D., Schuldiner, S., and Padan, E. (1993). Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 1212–1216. doi: 10.1073/pnas.90.4.1212
Gislason, A. S., Turner, K., Domaratzki, M., and Cardona, S. T. (2017). Comparative analysis of the Burkholderia cenocepacia K56-2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia. Microb. Genom. 3:e000140. doi: 10.1099/mgen.0.000140
Hamad, M. A., Di Lorenzo, F., Molinaro, A., and Valvano, M. A. (2012). Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia. Mol. Microbiol. 85, 962–974. doi: 10.1111/j.1365-2958.2012.08154.x
Hellmer, J., Teubner, A., and Zeilinger, C. (2003). Conserved arginine and aspartate residues are critical for function of MjNhaP1, a Na+/H+ antiporter of M. jannaschii. FEBS Lett. 542, 32–36. doi: 10.1016/s0014-5793(03)00332-6
Henderson, J. C., O’Brien, J. P., Brodbelt, J. S., and Trent, M. S. (2013). Isolation and chemical characterization of lipid a from gram-negative bacteria. J. Vis. Exp. 79:e50623. doi: 10.3791/50623
Herrera, C. M., Crofts, A. A., Henderson, J. C., Pingali, S. C., Davies, B. W., and Trent, M. S. (2014). The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio 5:e2283-14. doi: 10.1128/mBio.02283-14
Holdsworth, S. R., and Law, C. J. (2012). Functional and biochemical characterisation of the Escherichia coli major facilitator superfamily multidrug transporter MdtM. Biochimie 94, 1334–1346. doi: 10.1016/j.biochi.2012.03.001
Huang, L., Feng, Y., and Zong, Z. (2019). Heterogeneous resistance to colistin in Enterobacter cloacae complex due to a new small transmembrane protein. J. Antimicrob. Chemother. 74, 2551–2558. doi: 10.1093/jac/dkz236
Iiyama, K., Man Lee, J., Kusakabe, T., Yasunaga-Aoki, C., and Shimizu, S. (2011). Improvement in pHERD vectors to express recombinant proteins tagged with hexahistidine at either the NH2 or COOH- terminal in Pseudomonas aeruginosa. J. Insect Biotechnol. Sericol. 80, 57–61. doi: 10.11416/jibs.80.2-057
Inadome, H., Noda, Y., Kamimura, Y., Adachi, H., and Yoda, K. (2007). Tvp38, Tvp23, Tvp18 and Tvp15: novel membrane proteins in the Tlg2-containing Golgi/endosome compartments of Saccharomyces cerevisiae. Exp. Cell Res. 313, 688–697. doi: 10.1016/j.yexcr.2006.11.008
Jana, B., Cain, A. K., Doerrler, W. T., Boinett, C. J., Fookes, M. C., Parkhill, J., et al. (2017). The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci. Rep. 7:42483. doi: 10.1038/srep42483
Jiang, Y., Ruta, V., Chen, J., Lee, A., and MacKinnon, R. (2003). The principle of gating charge movement in a voltage-dependent K+ channel. Nature 423, 42–48. doi: 10.1038/nature01581
Jovanovic, G., Lloyd, L. J., Stumpf, M. P., Mayhew, A. J., and Buck, M. (2006). Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281, 21147–21161. doi: 10.1074/jbc.M602323200
Keller, R., Ziegler, C., and Schneider, D. (2014). When two turn into one: evolution of membrane transporters from half modules. Biol. Chem. 395, 1379–1388. doi: 10.1515/hsz-2014-0224
Khafizov, K., Staritzbichler, R., Stamm, M., and Forrest, L. R. (2010). A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry 49, 10702–10713. doi: 10.1021/bi101256x
Kim, H. S., Schell, M. A., Yu, Y., Ulrich, R. L., Sarria, S. H., Nierman, W. C., et al. (2005). Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genom. 6:174. doi: 10.1186/1471-2164-6-174
Koch-Weser, J., Sidel, V. W., Federman, E. B., Kanarek, P., Finer, D. C., and Eaton, A. E. (1970). Adverse effects of sodium colistimethate. manifestations and specific reaction rates during 317 courses of therapy. Ann. Intern. Med. 72, 857–868.
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., and Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Krishnamoorthy, G., Leus, I. V., Weeks, J. W., Wolloscheck, D., Rybenkov, V. V., and Zgurskaya, H. I. (2017). Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into gram-negative bacteria. MBio 8:e1172-17. doi: 10.1128/mBio.01172-17
Krishnamoorthy, G., Weeks, J. W., Zhang, Z., Chandler, C. E., Xue, H., Schweizer, H. P., et al. (2019). Efflux pumps of burkholderia thailandensis control the permeability barrier of the outer membrane. Antimicrob. Agents Chemother. 63:e00956-19. doi: 10.1128/AAC.00956-19
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Bradley, C. L., Mukashyaka, P., and Doerrler, W. T. (2016). Identification of essential arginine residues of Escherichia coli DedA/Tvp38 family membrane proteins YqjA and YghB. FEMS Microbiol. Lett. 363:fnw133. doi: 10.1093/femsle/fnw133
Kumar, S., and Doerrler, W. T. (2014). Members of the conserved DedA family are likely membrane transporters and are required for drug resistance in Escherichia coli. Antimicrob. Agents Chemother. 58, 923–930. doi: 10.1128/AAC.02238-13
Kumar, S., and Doerrler, W. T. (2015). Escherichia coli YqjA, a member of the conserved DedA/Tvp38 membrane protein family, is a putative osmosensing transporter required for growth at alkaline pH. J. Bacteriol. 197, 2292–2300. doi: 10.1128/JB.00175-15
Li, Y., Orlando, B. J., and Liao, M. (2019). Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490. doi: 10.1038/s41586-019-1025-6
Liang, F. T., Xu, Q., Sikdar, R., Xiao, Y., Cox, J. S., and Doerrler, W. T. (2010). BB0250 of Borrelia burgdorferi is a conserved and essential inner membrane protein required for cell division. J. Bacteriol. 192, 6105–6115. doi: 10.1128/JB.00571-10
Lipuma, J. J. (2010). The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23, 299–323. doi: 10.1128/CMR.00068-09
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Long, S. B., Campbell, E. B., and Mackinnon, R. (2005). Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908. doi: 10.1126/science.1116270
Loutet, S. A., Mussen, L. E., Flannagan, R. S., and Valvano, M. A. (2011). A two-tier model of polymyxin B resistance in Burkholderia cenocepacia. Environ. Microbiol. Rep. 3, 278–285. doi: 10.1111/j.1758-2229.2010.00222.x
Loutet, S. A., and Valvano, M. A. (2011). Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front. Cell Infect. Microbiol. 1:6. doi: 10.3389/fcimb.2011.00006
Malott, R. J., Steen-Kinnaird, B. R., Lee, T. D., and Speert, D. P. (2012). Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans. Antimicrob. Agents Chemother. 56, 464–471. doi: 10.1128/AAC.00602-11
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Mitchell, P. (1961). Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148. doi: 10.1038/191144a0
Moretti, F., Bergman, P., Dodgson, S., Marcellin, D., Claerr, I., Goodwin, J. M., et al. (2018). TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 19:e45889. doi: 10.15252/embr.201845889
Morita, K., Hama, Y., Izume, T., Tamura, N., Ueno, T., Yamashita, Y., et al. (2018). Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217, 3817–3828. doi: 10.1083/jcb.201804132
Ni, W., Li, Y., Guan, J., Zhao, J., Cui, J., Wang, R., et al. (2016). Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant gram-negative bacteria. Antimicrob. Agents Chemother. 60, 3215–3218. doi: 10.1128/AAC.00248-16
Noumi, T., Inoue, H., Sakurai, T., Tsuchiya, T., and Kanazawa, H. (1997). Identification and characterization of functional residues in a Na+/H+ antiporter (NhaA) from Escherichia coli by random mutagenesis. J. Biochem. 121, 661–670. doi: 10.1093/oxfordjournals.jbchem.a021637
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
O’Neill, J. (2014). The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Franklin Lakes, NJ: Becton Dickinson.
World Health Organization [WHO]∗ (2014). Antimicrobial Resistance : Global Report on Surveillance 2014. Geneva: World Health Organization.
Ortega, X. P., Cardona, S. T., Brown, A. R., Loutet, S. A., Flannagan, R. S., Campopiano, D. J., et al. (2007). A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 189, 3639–3644. doi: 10.1128/JB.00153-07
Osei Sekyere, J., and Amoako, D. G. (2017). Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front. Microbiol. 8:228. doi: 10.3389/fmicb.2017.00228
Owens, T. W., Taylor, R. J., Pahil, K. S., Bertani, B. R., Ruiz, N., Kruse, A. C., et al. (2019). Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553. doi: 10.1038/s41586-019-1039-0
Pamp, S. J., Gjermansen, M., Johansen, H. K., and Tolker-Nielsen, T. (2008). Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240. doi: 10.1111/j.1365-2958.2008.06152.x
Park, Y. K., and Ko, K. S. (2015). Effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on killing Acinetobacter baumannii by colistin. J. Microbiol. 53, 53–59. doi: 10.1007/s12275-015-4498-5
Paterson, D. L., and Harris, P. N. (2016). Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis. 16, 132–133. doi: 10.1016/S1473-3099(15)00463-6
Petrou, V. I., Herrera, C. M., Schultz, K. M., Clarke, O. B., Vendome, J., Tomasek, D., et al. (2016). Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid a glycosylation. Science 351, 608–612. doi: 10.1126/science.aad1172
Podnecky, N. L., Rhodes, K. A., and Schweizer, H. P. (2015). Efflux pump-mediated drug resistance in Burkholderia. Front. Microbiol. 6:305. doi: 10.3389/fmicb.2015.00305
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07
Raetz, C. R., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid a modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Robinson, A. E., Thomas, N. E., Morrison, E. A., Balthazor, B. M., and Henzler-Wildman, K. A. (2017). New free-exchange model of EmrE transport. Proc. Natl. Acad. Sci. U.S.A. 114, E10083–E10091. doi: 10.1073/pnas.1708671114
Rolain, J. M., Kempf, M., Leangapichart, T., Chabou, S., Olaitan, A. O., Le Page, S., et al. (2016). Plasmid-mediated mcr-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and laos. Antimicrob. Agents Chemother. 60, 6994–6995. doi: 10.1128/AAC.00960-16
Shi, Y., Cromie, M. J., Hsu, F. F., Turk, J., and Groisman, E. A. (2004). PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53, 229–241. doi: 10.1111/j.1365-2958.2004.04107.x
Shoemaker, C. J., Huang, T. Q., Weir, N. R., Polyakov, N., Schultz, S. W., and Denic, V. (2019). CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 17:e2007044. doi: 10.1371/journal.pbio.2007044
Sievers, F., and Higgins, D. G. (2014). Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116. doi: 10.1007/978-1-62703-646-7-6
Sigal, N., Vardy, E., Molshanski-Mor, S., Eitan, A., Pilpel, Y., Schuldiner, S., et al. (2005). 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry 44, 14870–14880. doi: 10.1021/bi051574p
Sikdar, R., and Doerrler, W. T. (2010). Inefficient Tat-dependent export of periplasmic amidases in an Escherichia coli strain with mutations in two DedA family genes. J. Bacteriol. 192, 807–818. doi: 10.1128/JB.00716-09
Sikdar, R., Simmons, A. R., and Doerrler, W. T. (2013). Multiple envelope stress response pathways are activated in an Escherichia coli strain with mutations in two members of the DedA membrane protein family. J. Bacteriol. 195, 12–24. doi: 10.1128/JB.00762-12
Simpson, B. W., and Trent, M. S. (2019). Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416. doi: 10.1038/s41579-019-0201-x
Sun, J., Zhang, H., Liu, Y. H., and Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Tabara, L. C., Vincent, O., and Escalante, R. (2019). Evidence for an evolutionary relationship between Vmp1 and bacterial DedA proteins. Int. J. Dev. Biol. 63, 67–71. doi: 10.1387/ijdb.180312re
Tao, X., Lee, A., Limapichat, W., Dougherty, D. A., and MacKinnon, R. (2010). A gating charge transfer center in voltage sensors. Science 328, 67–73. doi: 10.1126/science.1185954
Tavares-Carreon, F., Fathy Mohamed, Y., Andrade, A., and Valvano, M. A. (2016). ArnT proteins that catalyze the glycosylation of lipopolysaccharide share common features with bacterial N-oligosaccharyltransferases. Glycobiology 26, 286–300. doi: 10.1093/glycob/cwv095
Thompkins, K., Chattopadhyay, B., Xiao, Y., Henk, M. C., and Doerrler, W. T. (2008). Temperature sensitivity and cell division defects in an Escherichia coli strain with mutations in yghB and yqjA, encoding related and conserved inner membrane proteins. J. Bacteriol. 190, 4489–4500. doi: 10.1128/JB.00414-08
Thongdee, M., Gallagher, L. A., Schell, M., Dharakul, T., Songsivilai, S., and Manoil, C. (2008). Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Appl. Environ. Microbiol. 74, 2985–2989. doi: 10.1128/AEM.00030-08
Thwaite, J. E., Humphrey, S., Fox, M. A., Savage, V. L., Laws, T. R., Ulaeto, D. O., et al. (2009). The cationic peptide magainin II is antimicrobial for Burkholderia cepacia-complex strains. J. Med. Microbiol. 58(Pt 7), 923–929. doi: 10.1099/jmm.0.008128-0
Tu, Q., Yin, J., Fu, J., Herrmann, J., Li, Y., Yin, Y., et al. (2016). Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency. Sci. Rep. 6, 24648. doi: 10.1038/srep24648
Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M., et al. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187, 5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005
Waters, V. (2012). New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr. Pharm. Des. 18, 696–725. doi: 10.2174/138161212799315939
Weatherspoon-Griffin, N., Zhao, G., Kong, W., Kong, Y., Morigen, Andrews-Polymenis, H., et al. (2011). The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J. Biol. Chem. 286, 5529–5539. doi: 10.1074/jbc.M110.200352
Wiersinga, W. J., Virk, H. S., Torres, A. G., Currie, B. J., Peacock, S. J., Dance, D. A. B., et al. (2018). Melioidosis. Nat. Rev. Dis. Primers 4, 17107. doi: 10.1038/nrdp.2017.107
Winn, P. J., Ludemann, S. K., Gauges, R., Lounnas, V., and Wade, R. C. (2002). Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. U.S.A. 99, 5361–5366. doi: 10.1073/pnas.082522999
Workman, S. D., Worrall, L. J., and Strynadka, N. C. J. (2018). Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling. Nat. Commun. 9, 1159. doi: 10.1038/s41467-018-03547-3548
Yan, A., Guan, Z., and Raetz, C. R. (2007). An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem. 282, 36077–36089. doi: 10.1074/jbc.M706172200
Zhou, P., Altman, E., Perry, M. B., and Li, J. (2010). Study of matrix additives for sensitive analysis of lipid a by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 76, 3437–3443. doi: 10.1128/AEM.03082-09
Zhou, Z., Lin, S., Cotter, R. J., and Raetz, C. R. (1999). Lipid a modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274, 18503–18514. doi: 10.1074/jbc.274.26.18503
Keywords: colistin, antibiotic resistance, lipopolysaccharide, membrane protein, proton motive force
Citation: Panta PR, Kumar S, Stafford CF, Billiot CE, Douglass MV, Herrera CM, Trent MS and Doerrler WT (2019) A DedA Family Membrane Protein Is Required for Burkholderia thailandensis Colistin Resistance. Front. Microbiol. 10:2532. doi: 10.3389/fmicb.2019.02532
Received: 21 August 2019; Accepted: 21 October 2019;
Published: 05 November 2019.
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilReviewed by:
Herbert P. Schweizer, University of Florida, United StatesJohn Dallas Boyce, Monash University, Australia
Copyright © 2019 Panta, Kumar, Stafford, Billiot, Douglass, Herrera, Trent and Doerrler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William T. Doerrler, d2RvZXJyQGxzdS5lZHU=
†Present address: Sujeet Kumar, Department of Microbiology, The Ohio State University, Columbus, OH, United States
 Pradip R. Panta1
Pradip R. Panta1 Sujeet Kumar
Sujeet Kumar William T. Doerrler
William T. Doerrler