- 1Institute of Pathogenic Biology, Hengyang Medical College, University of South China, Hengyang, China
- 2Hunan Provincial Key Laboratory for Special Pathogens Prevention and Control, Pathogenic Biology Institute, University of South China, Hengyang, China
Chlamydia muridarum, an obligate intracellular pathogen, was used to establish a murine model of female upper genital tract infection by Chlamydia trachomatis. TC0668 in C. muridarum is a hypothetical chromosomal virulence protein that is involved in upper genital tract pathogenesis. The infection of mice with the C. muridarum TC0668-mutant (G216*) strain results in less pathological damage in the upper genital tract. In this study, an isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomics analysis was performed to identify differentially expressed proteins between TC0668 wild-type (TC0668wt) and TC0668 mutant (TC0668mut) strains at 6, 12, 18, and 24 h post-infection (p.i.). Of the 550 proteins differentially expressed at 18 h p.i., 222 and 328 were up-regulated and down-regulated, respectively, inTC0668mut-infected cells. The expression of seven up-regulated proteins (encoded by SRPRB, JAK1, PMM1, HLA-DQB1, THBS1, ITPR1, and BCAP31) and three down-regulated proteins (encoded by MAPKAPK2, TRAFD1, and IFI16) from the iTRAQ analysis were validated using quantitative real-time (qRT)-PCR. The qRT-PCR results were consistent with those of iTRAQ. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that the differentially expressed proteins primarily participated in inflammatory responses, fibrosis, metabolic processes, and complement coagulation cascades, and were mainly enriched in the phosphatidylinositol 3′-kinase (PI3K)/Akt, nuclear factor kappa-B (NF-κB), and other signaling pathways. Using western-blotting and immunofluorescence detection, significant differences in activation of the PI3K/Akt and NF-κB signaling pathways were observed between the TC0668wt- and TC0668mut-infected cells. Differentially expressed proteins linked with inflammation and fibrosis were used in a protein-protein interaction network analysis. The results suggest that TC0668 may play a pivotal role in C. muridarum-induced genital pathology by inducing inflammatory responses and fibrosis, which may involve the activation of the PI3K/Akt and NF-κB signaling pathways.
Introduction
Chlamydiae are Gram-negative obligate intracellular prokaryotic pathogens affecting humans and animals (Bachmann et al., 2014; Bommana and Polkinghorne, 2019). Female lower genital tract infection with Chlamydia trachomatis can spread to the upper genital tract, causing inflammatory pathologies such as hydrosalpinx that result in infertility (Land et al., 2009; Rodgers et al., 2011). However, the pathogenesis of C. trachomatis infection remains unclear. Although it is unknown whether Chlamydia muridarum causes disease in humans, it can be used to study the immunobiology of chlamydial infection and investigate the mechanisms underlying the pathogenesis in the urogenital tract (Morrison and Caldwell, 2002; Shah et al., 2005; Cheng et al., 2007; Chen et al., 2010).
Recent advancements in the manipulation of C. trachomatis plasmids and transformation of C. muridarum have revealed that the plasmid-encoded genes Pgp3 and Pgp5 are important virulence factors in C. muridarum for the induction of hydrosalpinx (Liu et al., 2014; Huang et al., 2015). Kari et al. showed that infection with plasmid-deficient C. trachomatis is highly attenuated in non-human primates, which induced an anti-chlamydial immune response (Kari et al., 2011). Then a transcriptional profiling of plasmid-bearing and plasmid-deficient C. trachomatis infected HeLa cells was conducted to explore the role of chlamydial plasmid in the host cell inflammatory response to infection and immune avoidance (Porcella et al., 2015). Additionally, TC0668, a chromosome-encoded hypothetical protein, is an important upper genital tract pathogenicity factor of C. muridarum, as TC0668 mutant strains (TC0668mut) exhibit attenuated virulence, involving reduced pathology in mice, compared with TC0668 wild-type strains (TC0668wt) (Chen et al., 2015; Conrad et al., 2016). Nevertheless, the molecular mechanisms of the TC0668 protein in the pathogenesis of C. muridarum need further investigation.
As TC0668 is a newly discovered virulence protein, with unknown homologous proteins and pathogenic mechanisms, an effective technical approach is needed for comprehensive screening and analysis of related molecules and signaling pathways that may be involved in the pathogenesis. In recent years, proteomics is a frontier method for investigating complex biological functions, which provide appropriate targets for researching on novel molecular biomarkers. In particular, isobaric tags for relative and absolute quantitation (iTRAQ) is an advanced high-throughput quantitative proteomics technique with high sensitivity, that has been rapidly developed and widely used to investigate the pathogenesis of many infectious agents (Jézéquel et al., 2019; Wu et al., 2019; Zhou et al., 2019). In this study, iTRAQ-based quantitative proteomic technology was used to screen and analyze the differentially expressed proteins in HeLa cells infected with C. muridarum TC0668wt and TC0668mut strains. Therefore, an iTRAQ-based proteomics analysis of TC0668 from C. muridarum may clarify its role in chlamydial pathogenesis.
We aimed to describe the proteomics profile of human epithelial HeLa cells infected with TC0668wt and TC0668mut strains, a pair of isogenic clones. The two clones differed in tc0668 genotypes. Briefly, TC0668mut strain carries the TC0668 G216* mutation, with the glycine (GGA) codon at position 216 changing to a stop codon (TGA) of TC0668 protein, and the rest of genome and plasmid of the two isogenic clones are identical (Chen et al., 2015; Conrad et al., 2016). Our results suggest that TC0668 participates in the induction of molecular responses and may be involved in the signaling pathways that underlie chlamydial pathogenesis.
Materials and Methods
Chlamydial Organisms and Cell Culture Conditions
The C. muridarum TC0668 wild-type strain (TC0668wt) and mutant strain (TC0668mut) are different only in the presence or absence of the chromosomal gene tc0668 and its function. G13.32.1 (retaining the wild-type tc0668 genotype of C. muridarum Nigg3 strain), and G13.11.1 (with a non-sense mutation in tc0668 resulting from a premature stop codon at the 216th codon position of TC0668 protein, defined as TC0668 G216*), a pair of isogenic clones, which had been subjected to plaque purification and deep whole-genome sequencing to ensure the genotype, were used as TC0668wt and TC0668mut strains in the experiments (Chen et al., 2015; Conrad et al., 2016). HeLa cells (human cervical carcinoma epithelial cells, CCL-2; American Type Culture Collection) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (DMEM-10) at 37°C in 5% CO2. Six-well plates containing HeLa cells were infected with TC0668wt or TC0668mut (multiplicity of infection [MOI] = 1), and the cell lysates were then collected at 6, 12, 18, and 24 h post-infection (p.i.).
Phase Detection of Chlamydia and TC0668 in Cells Infected With TC0668wt or TC0668mut
To assess the quantity and quality of organisms in parallel experiments, immunofluorescence assays (IFAs) and quantitative real-time PCR (qRT-PCR) were used. For the IFAs, HeLa cells grown on coverslips in 24-well tissue culture plates containing DMEM-10 with cycloheximide were infected with either C. muridarum TC0668wt or TC0668mut (MOI = 1). Briefly, cell samples were fixed with 4% paraformaldehyde for 30 min at 37°C, followed by permeabilizing with 0.1% Triton-100 dissolved in phosphate-buffered saline (PBS) for 15 min. After washing and blocking, the cells were subjected to antibody and chemical staining. A rabbit anti-chlamydial polyclonal antibody and a goat anti-rabbit IgG conjugated to the 488 dye (green, Jackson Immuno Research Laboratories) were used to visualize chlamydial inclusions. A mouse anti-TC0668 antibody and a goat anti-mouse IgG conjugated to the Cy3 dye (red; Jackson Immuno Research Laboratories) were used to visualize natural TC0668 proteins, which were only present in the cells infected with the TC0668wt strain (at 6, 12, 18, and 24 h p.i.). The nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). After incubation with primary and secondary antibodies for 1 h at 37°C and washing with PBS five times, observation and imaging were performed using an inverted fluorescence microscope.
qRT-PCR analysis was used to determine the copy number of target Chlamydia genes. Chlamydia 16S rRNA was used as the internal reference to control the changing in the number of pathogens during the life cycle. The tc0668 gene expression level was also assessed in cells, while pgp8, a steadily expressed Chlamydia plasmid gene in the growth and proliferation, were regarded as the control and compared with tc0668 (Ferreira et al., 2013). TRIZOL lysate (Invitrogen, Carlsbad, CA, USA) was added to extract total RNA in enzyme-free tubes, and then the same amount of templates was combined with the SYBR Green Premix with ROX Reference Dye (TIANGEN, Beijing, China). Assays were performed in using a LightCycle 96 Instrument (Roche, Basel, Switzerland). All samples were amplified as follows: 50°C for 5 min and 95°C for 10 min; followed by 40 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 20 s. After amplification, data were analyzed using the 2−ΔCt method. Primer sequences of amplified genes are shown in Table 1.
Protein Extraction and iTRAQ Labeling
Infected cells were harvested and then lysed with SDT buffer (4% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol [DTT], and 100 mM Tris-HCl). The lysate was sonicated, boiled for 15 min, and centrifuged at 14,000 × g for 40 min. The protein level in the supernatant was quantified with a Bicinchoninic Acid (BCA) Protein Assay Kit (Bio-Rad, CA, USA). iTRAQ assays were performed according to the manufacturer's instructions (Applied Biosystems; Ross et al., 2004).
Strong Cation Exchange (SCX) and Nano Liquid Chromatography Coupled With Tandem Mass Spectrometry (LC-MS/MS) Analysis
iTRAQ labeled peptides were fractionated using SCX chromatography with an AKTA Purifier system (GE Healthcare; Huang et al., 2019; Qin et al., 2019). The dried peptide mixture was reconstituted and acidified with 2 mM buffer A and loaded onto a PolySULFOETHYL 4.6 × 100 mm column. Peptides were eluted at a flow rate of 1 mL/min with a gradient of 0% buffer B for 25 min, 0–10% buffer B during 25–32 min, 10–20% buffer B during 32–42 min, 20–45% buffer B during 42–47 min, 45–100% buffer B during 47–52 min, 100% buffer B during 52–60 min, and buffer B was reset to 0% after 60 min. The elution was monitored by absorbance at 214 nm, and 30 fractions were collected every 1 min. Collected fractions were distributed into 15 fractions and desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 mL, Sigma), concentrated by vacuum centrifugation, and reconstituted in 40 μL of 0.1% (v/v) trifluoroacetic acid.
Each fraction was then injected for nanoLC-MS/MS analysis, which was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was operated in positive ion mode coupled to Easy nLC (Thermo Fisher Scientific) for 60 min. MS data were acquired using a data-dependent top 10 method. Automatic gain control (AGC) target was set to 1e6, and maximum inject time to 50 ms. Dynamic exclusion duration was 60.0 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and resolution for HCD spectra was set to 17,500 at m/z 200, and isolation width was 2 m/z. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Proteomic and Bioinformatics Analysis
Protein identification and iTRAQ were performed with the Mascot (version 2.2; Matrix Science, Massachusetts, USA) and Proteome Discoverer software (version 1.4; Thermo Scientific, Massachusetts, USA) against the UniProt Human Database (133549 sequences, downloaded on March 3rd, 2013). The parameters used included: mass tolerance = 20 ppm, MS/MS tolerance = 0.1 Da, enzyme = trypsin, missed cleavage = 2, oxidation (M), iTRAQ 8plex (Y) as the potential variable modifications, and carbamidomethyl (C), iTRAQ8plex (N-term), iTRAQ 8plex (K) as the fixed modifications. Decoy database search was used to calculate the false discovery rate (FDR) for peptide identification, using a screening criterion of FDR ≤ 0.01. Differential protein expression was defined as an iTRAQ ratio between the two groups of > 1.2 or < 0.83. All the differentially expressed proteins of the proteomics were analyzed via UniProt (http://www.uniprot.org/).
Venn diagram was constructed to analyze the common differentially expressed proteins between two strains at 6, 12, 18, and 24 h p.i. Gene Ontology (GO) analysis (version go_201608.obo; www.geneontology.org) was used to evaluate the biological significance of the differentially expressed proteins. GO analysis provide controlled vocabularies for the description of the biological process, molecular function, and cellular component of gene products by organism databases, and the differentially expressed proteins that are involved in similar processes, function and components were divided into corresponding clusters. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to investigate potential biological pathways using online software (KEGG Automatic Annotation Server [KAAS]). Information on protein–protein interactions (PPIs) of the studied proteins was retrieved using IntAct software (http://www.ebi.ac.uk/intact/main.xhtml) and Search Tool for the Retrieval of Interacting Genes/Proteins software (STRING; http://string-db.org/). The results were imported into Cytoscape5 software (version 3.2.1; RT-https://cytoscape.org/) to visualize and analyze the functional PPI networks.
qRT-PCR Validation of Proteomics Results
To validate the proteomics results, qRT-PCR analysis was performed to determine the mRNA expression levels of 10 differentially expressed proteins (with the corresponding gene names SRPRB, JAK1, PMM1, HLA-DQB1, THBS1, ITPR1, BCAP31, MAPKAPK2, TRAFD1, and IFI16) at random. Primer sequences of amplified genes are shown in Table 1. The general qRT-PCR steps were the same as described above. Total RNAs of TC0668wt and TC0668mut were collected at 18 h p.i. Chlamydia 16S rRNA was used to normalize the mRNA levels of the target genes. Samples were amplified as follows: 50°C for 5 min and 95°C for 10 min; followed by 45 cycles of 95°C for 5 s, 57°C for 10 s, and 72°C for 20 s.
Validation of KEGG Analysis
Western blotting and IFA were used for validation of the KEGG analysis. For western blotting, proteins from TC0668wt- and TC0668mut-infected cells at 6, 12, 18, and 24 h p.i were obtained as follows. First, the cells in each well (six-well plate) were added with 100 μL radioimmunoprecipitation assay (Solarbio, Beijing, China) lysis buffer, 1 μL of the protease inhibitor phenylmethylsulfonyl fluoride (Solarbio, Beijing, China) and phosphatase inhibitor cocktail (CWBIO, Beijing, China) for phospho-Akt (p-Akt). The cells lysate were removed to 2 mL Eppendorf tubes on ice and then centrifuged at 15,294 × g at 4°C for 10 min. The supernatants were analyzed using 12% SDS-polyacrylamide gel electrophoresis (PAGE), and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was blocked using 5% skim milk with Tris-buffered saline plus Tween 20 (TBST) buffer for 2 h, and then incubated with rabbit raised monoclonal PI3K, Akt, p-Akt, p53, and NF-κB (p65) antibodies (all from CST, Danvers, MA, USA) at 37°C for 1 h. The membrane was incubated with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (Sigma-Aldrich, St. Louis, Missouri, USA) at 37°C for 1 h. Visualization was conducted with an Enhanced Chemiluminescence (ECL) Kit (Santa Cruz Biotech, Santa Cruz, CA, USA). All proteins were normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control after the belts were scanned by gel quantification software (ImageJ 2.0.0-rc-54, Java 1.8.0-66, https://imagej.net/Downloads), and the relative intensity of target protein was calculated finally.
To further demonstrate the translocation of NF-κB (p65) into cell nuclei, IFAs were performed with TC0668wt- and TC0668mut-infected HeLa cells at 6, 12, 18, and 24 h p.i. The primary antibody was rabbit raised monoclonal p65 (CST) antibody, the secondary antibody was goat anti-rabbit IgG conjugated to the 488 dye (green; Jackson Immuno Research Laboratories), and DAPI dye core (blue) was used to label cell nuclei. Visualization and imaging (inverted fluorescence microscopy) were performed as mentioned above.
Statistical Analysis
Data on mRNA levels were analyzed using the 2−ΔCt method, and the data obtained by western blotting in three independent experiments were analyzed using one-way analysis of variance [ANOVA]. Experimental data were all statistically analyzed using SPSS software (version 13.0; SPSS, Chicago, IL, USA). Significant A (including t-tests and one-way ANOVA) was performed to analyze the experimental data of proteomics. A P-value < 0.05 was considered statistically significant.
Results
Equal Numbers of Inclusions but Not TC0668 Were Detected in TC0668wt- and TC0668mut-Infected Cells
HeLa cells infected with C. muridarum TC0668wt and TC0668mut strains were imaged at 6, 12, 18, and 24 h p.i. using an optical microscope (Figure 1). At 12, 18, and 24 h p.i, the inclusions (green) of both strains were visualized, but no obvious green fluorescence was observed in either group at 6 h p.i. No differences in inclusion formation was observed between two strains at 12, 18, and 24 h p.i, whereas TC0668 (red) was only observed on the membrane of chlamydial elementary bodies (EBs) or reticulate bodies (RBs) in the TC0668wt group and not in the TC0668mut group at any detected time points.
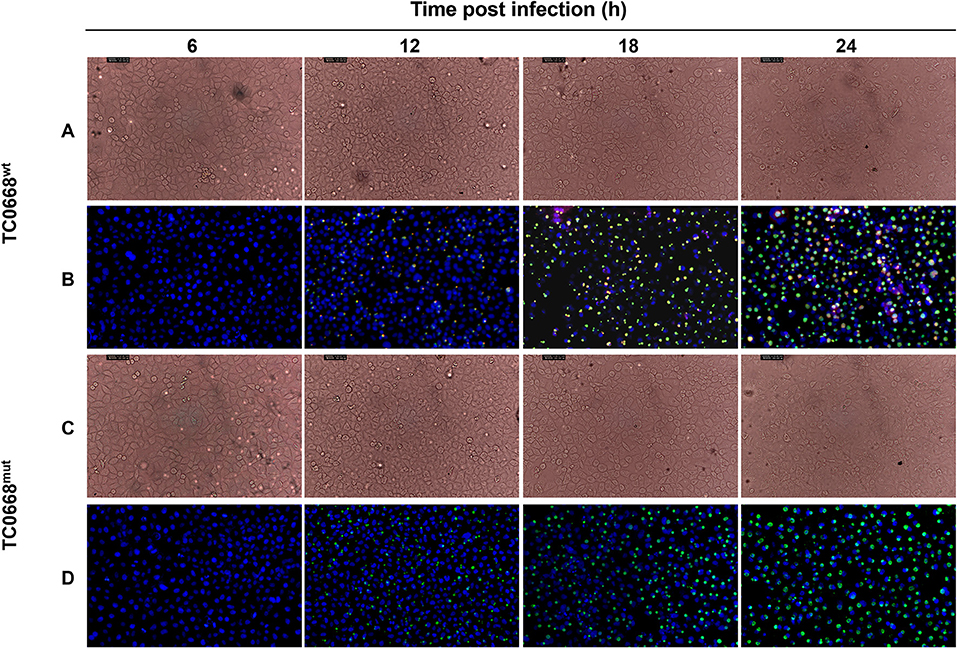
Figure 1. Indirect immunofluorescence assay (IFA) of HeLa cells infected with C. muridarum TC0668wt or TC0668mut strains. With 2.5 × 105 IFU/well inoculum (MOI = 1), Chlamydia-infected HeLa cells were photographed using optical microscopy at 6, 12, 18, and 24 h p.i. Chlamydial inclusion bodies (green) are visible in both TC0668wt- and TC0668mut-infected cells, whereas the TC0668 protein (red) is only visible in TC0668wt-infected HeLa cells. Magnification, ×200. TC0668wt-infected HeLa cells were photographed by using phase contrast microscopy (A) and IFA (B) at 6, 12, 18, and 24 h p.i. TC0668mut-infected HeLa cells were photographed by using phase contrast microscopy (C) and IFA (D) at 6, 12, 18, and 24 h p.i.
qRT-PCR was used to detect target RNAs of C. muridarum TC0668wt and TC0668mut strains at 6, 12, 18, and 24 h p.i (Figure 2). Pgp8, an essential plasmid gene, is stably expressed during the chlamydial development cycle, and it was used as a control to evaluate tc0668 expression at the transcriptional level. Both tc0668 (tc0668/16S rRNA) and pgp8 (pgp8/16S rRNA) were gradually upregulated from 0 to 18 h p.i and stabilized at 18 h p.i. Figure 2 illustrates that the pgp8 gene copy numbers in the two groups were approximately equivalent. However, tc0668 was not detected in the TC0668mut group, while it was detected in the TC0668wt group (one-way ANOVA, P < 0.05).
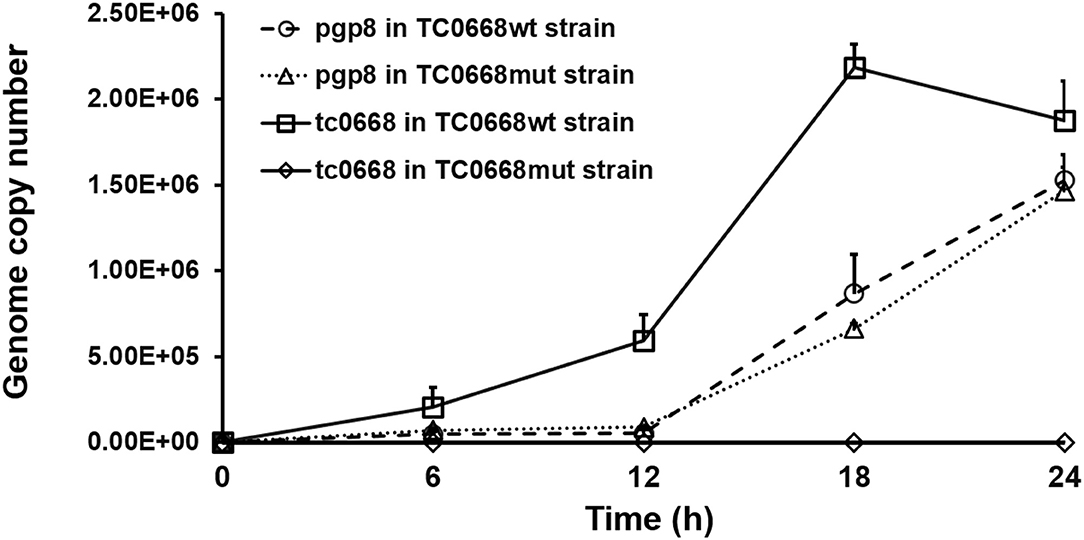
Figure 2. qRT-PCR analysis of tc0668 gene copy number in C. muridarum TC0668wt- and TC0668mut-infected HeLa cells. With 1 × 106 IFU/well inoculum (MOI = 1), the copy number of the C. muridarum gene tc0668 was determined using qRT-PCR, and the C. muridarum plasmid gene pgp8 was used as the control. 16S rRNA was used to normalize tc0668 and pgp8 signals. Three biological replicates of each time point were performed, and points represent mean and standard errors. Copy number differences of tc0668 between TC0668wt- and TC0668mut-infected cells were statistically significant (one-way ANOVA, P < 0.05).
Comparative Proteomics Analysis Using iTRAQ
To identify the differentially expressed proteins between TC0668wt- and TC0668mut-infected cells, protein profiles of these two strains were compared at 6, 12, 18, 24 h p.i. As presented in Figure 3A, iTRAQ analysis of the TC0668wt- and TC0668mut-infected cell proteome showed 76,704 queries in the database, and resulted in 5,534 identified proteins, 33,662 peptides using Mascot software. According to the protein quantification criteria (cut-off values of 1.2-fold for up-regulation and 0.83-fold for down-regulation), compared to TC0668wt-infected cells, 265, 270, 222, and 197 up-regulated proteins and 306, 245, 328, and 214 down-regulated proteins (totaling 571, 515, 550, and 411 modulated proteins) were identified in the TC0668mut-infected cells at 6, 12, 18, and 24 h p.i., respectively. Figures 3B,C show the trends over time of differentially expressed proteins in TC0668wt and TC0668mut-infected samples, respectively.
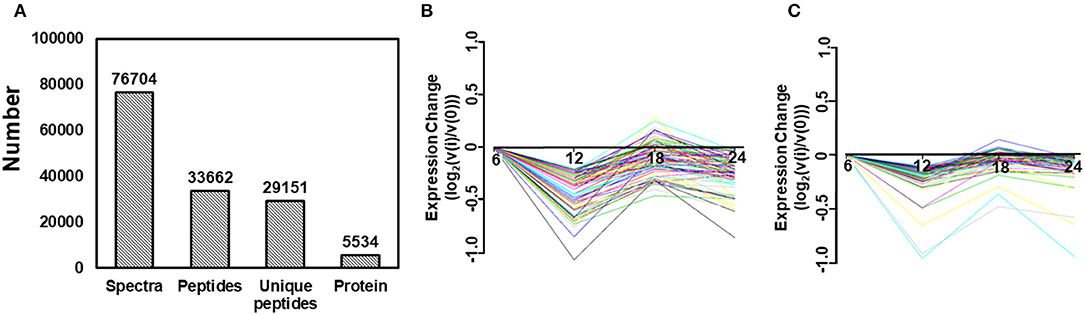
Figure 3. Quantitative proteomic analysis of HeLa cells infected with C. muridarum TC0668wt or TC0668mut strains. (A) Basic statistics of proteome results from iTRAQ. Spectra, secondary mass spectra after quality control; Unique peptides, identified peptides that belong to only a group of proteins; and protein, identified proteins using Mascot 2.3.02 software. (B) Trends of differentially expressed proteins in TC0668wt-infected cells at 6, 12, 18, and 24 h p.i. (C) Trends of differentially expressed proteins in TC0668mut-infected cells at 6, 12, 18, and 24 h p.i.
A four-way Venn diagram was constructed to analyze the numbers of differentially expressed proteins between TC0668wt- and TC0668mut-infected cells (P < 0.05) that were unique or common to the different time points (Figure 4). Only 226, 243, 201, and 169 differentially expressed proteins were unique at 6, 12, 18, and 24 h p.i., respectively; 378 were shared by any two of the four time points. One hundred and twenty-four were shared by any three of the four time points; and 20 were common to all four time points.
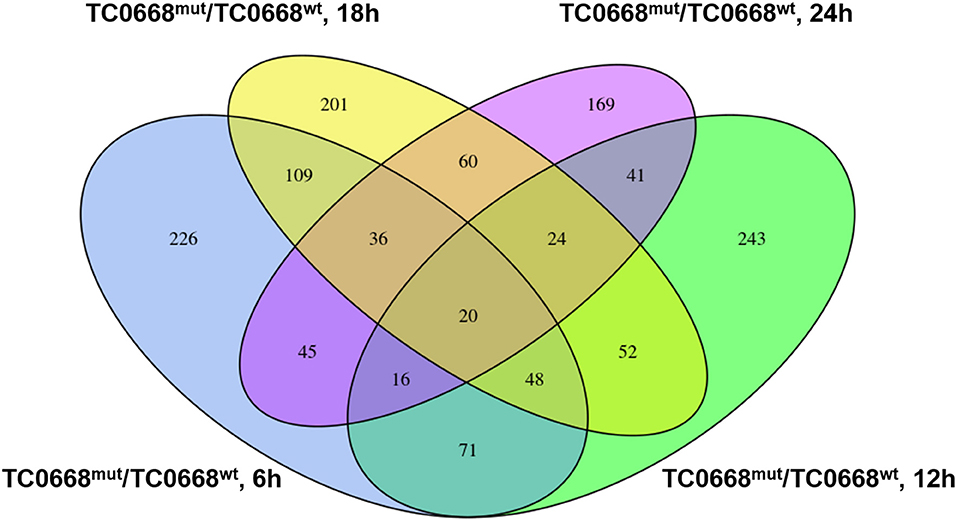
Figure 4. Four-way Venn diagram of the total number of proteins significantly differentially expressed (P < 0.05) between HeLa cells infected with C. muridarum TC0668wt or TC0668mut strains at 6, 12, 18, and 24 h p.i. Numbers of shared or unique proteins are indicated at the intersections of the circles in the Venn diagram.
The 20 differentially expressed proteins that were common to all four time points were analyzed using UniProt database. Two inflammation-related proteins were up-regulated (encoded by INPP5D) or down-regulated (encoded by MAPKAPK2) in TC0668mut-infected cells. Four proteins related to signaling pathways were up-regulated (encoded by INPP5D and MYO10) or down-regulated (encoded by ITGA6 and MAPKPKA2). Three proteins with binding functions were up-regulated (encoded by GNL3L) or down-regulated (encoded by CYB5R4 and CWC15) in TC0668mut-infected cells (Supplementary Materials).
qRT-PCR Validation of Differentially Expressed Proteins
To validate the iTRAQ-based proteomics results, qRT-PCR analysis was performed on 10 selected proteins that were differentially expressed between TC0668mut- and TC0668wt-infected cells. mRNA expression levels of seven up-regulated proteins (encoded by SRPRB, JAK1, PMM1, HLA-DQB1, THBS1, ITPR1, and BCAP31) and three down-regulated proteins (encoded by MAPKAPK2, TRAFD1, and IFI16) were validated using qRT-PCR. As shown in Figure 5, in TC0668mut-infected cells, the mRNA expression levels of the seven up-regulated proteins were significantly increased, whereas the mRNA expression levels of the three down-regulated proteins were significantly reduced (P < 0.05). Thus, the qRT-PCR results (regarding transcript levels) are consistent with those of the proteomics analysis.
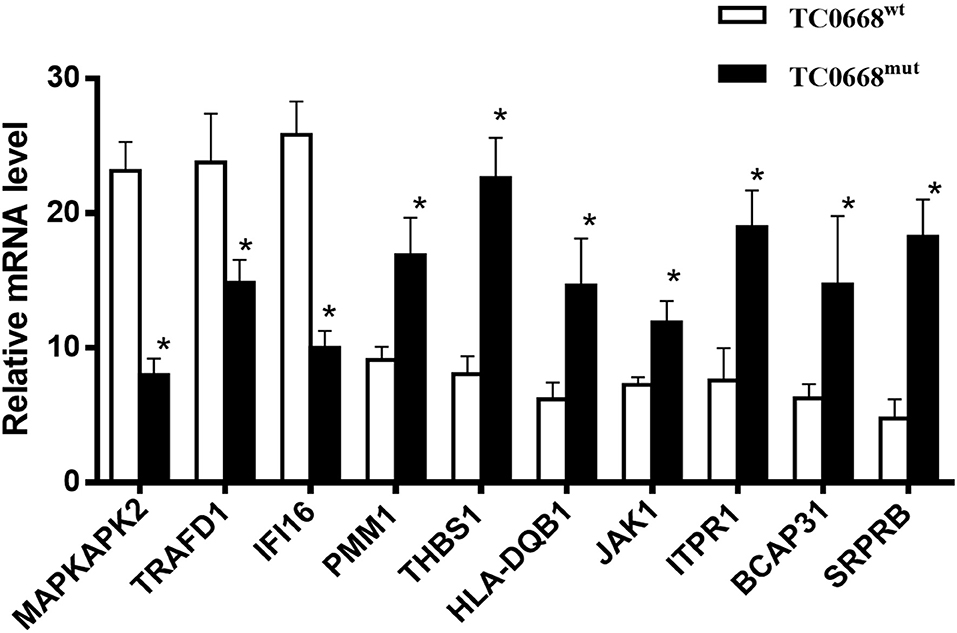
Figure 5. Validation of profiling data with qRT-PCR. mRNA expression levels of seven up-regulated (encoded by SRPRB, JAK1, PMM1, HLA-DQB1, BCAP31, ITPR1, and THBS1) and three down-regulated (encoded by MAPKAPK2, TRAFD1, and IFI16) proteins in C. muridarum TC0668mut-infected HeLa cells were determined using qRT-PCR at 18 h p.i, and compared with those of C. muridarum TC0668wt-infected group. mRNA levels from three replicates for each group are expressed as mean and the standard errors. *represents that all copy number differences between TC0668wt and TC0668mut were statistically significant (t test, P < 0.05).
GO and KEGG Analyses of Differentially Expressed Proteins Between TC0668wt- and TC0668mut-Infected Cells
GO annotation of the 550 differentially expressed proteins at 18 h p.i. was analyzed using Blast2GO software. The analysis revealed the diversity of biological processes related to the differentially expressed proteins (Figure 6A), including cellular process, metabolic process, immune system process, and biological regulation. It also showed that these differentially expressed proteins are involved in a large number of molecular functions (Figure 6B), such as binding, catalytic activity, transporter activity, and molecular function regulation, as well as being associated with multiple cellular components (Figure 6C), including the cell, cell part, organelle, and membrane.
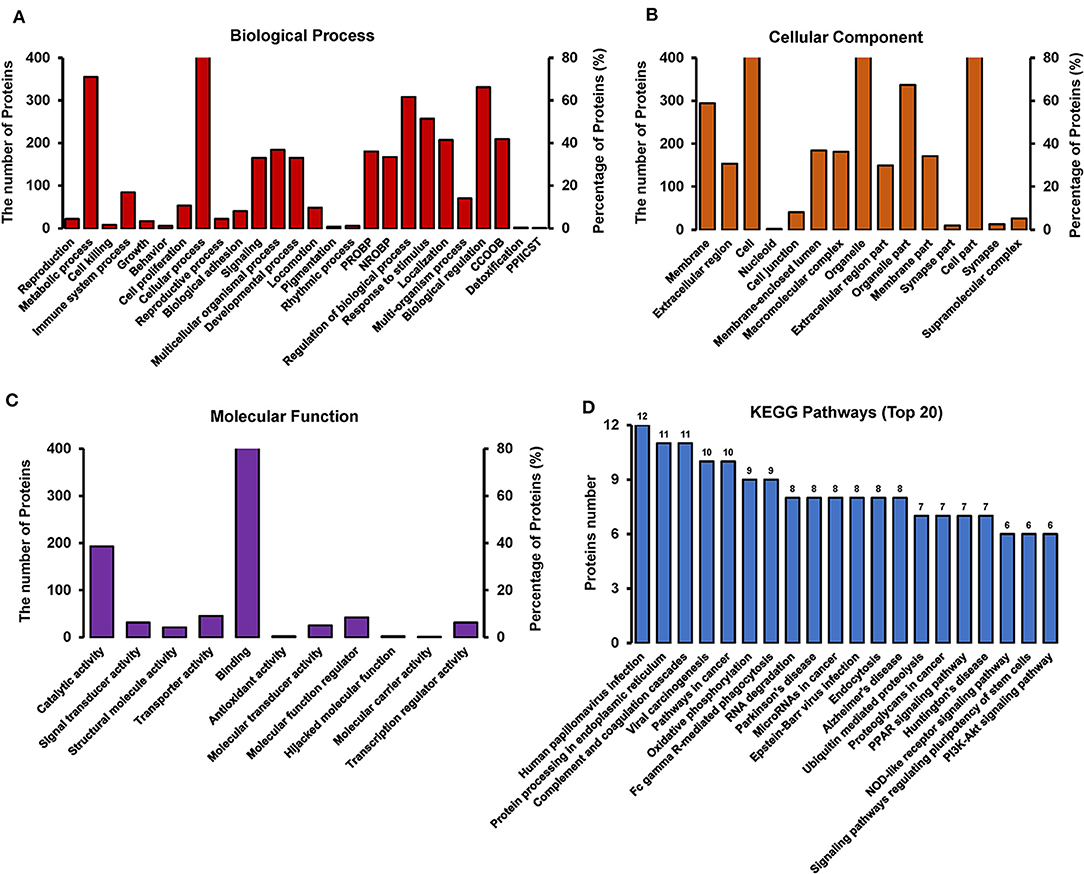
Figure 6. GO and KEGG pathway analysis of differentially expressed proteins between TC0668wt and TC0668mut strains at 18 h p.i (P < 0.05). (A) Biological process. The items “PROBP,” “NROBP,” “CCOOB,” and “PPIICST” are the abbreviations of “positive regulation of biological process,” “negative regulation of biological process,” “cellular component organization or biogenesis,” and “presynaptic procell involved in chemical synaptic transmission,” respectively. (B) Cellular component. The item “cell” including all related cell components besides the parallel cell part, membrane, nucleoid and so on. (C) Molecular function. (D) KEGG pathway analysis.
KAAS software was used to identify the potential biological pathways. The 550 differentially expressed proteins were allocated to 240 KEGG pathways. The pathways are mainly involved in oxidative phosphorylation, endocytosis, complement coagulation cascades, and the PPAR, PI3K-Akt, NOD-like receptor, and other signaling pathways (Figure 6D). Collectively, the findings provide relevant information to guide future research on the molecular basis of the role of TC0668 in C. muridarum pathogenesis.
Western Blotting and IFA Validation of Proteomics Analysis
Based on the proteomics results, differentially expressed proteins were significantly enriched in several signaling pathways. We further verified the activation of the PI3K/Akt and NF-κB signaling pathways, which may be involved in the pathogenesis of chlamydial infections, based on the expression of PI3K, p-Akt, p53 (a tumor suppressor that can be downregulated by activation of PI3K/Akt pathway), and NF-κB (p65). As shown in Figure 7A, cells infected with C. muridarum TC0668wt strains for 6, 12, 18, and 24 h displayed a significantly higher level of PI3K compared to TC0668mut-infected cells at all four time points (P < 0.05). The relative expression level of p-Akt (p-Akt/Akt) in the TC0668wt-infected cells was significantly higher than that in the TC0668mut-infected cells (P < 0.05) (Figure 7B). In contract, the level of p53 was significantly lower in the TC0668wt-infected cells compared to the TC0668mut-infected cells during chlamydial infection (P < 0.05) (Figure 7C).
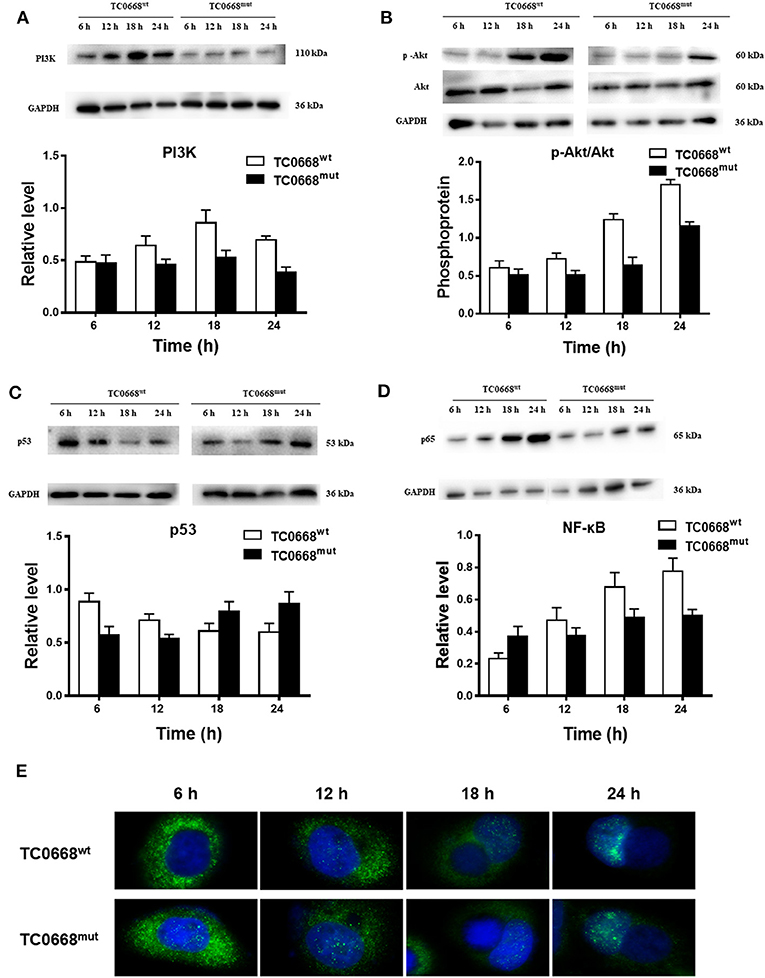
Figure 7. Activation of NF-κB and PI3K/Akt signal pathways as determined using western blotting and IFA. With 1 × 106 IFUs/well inoculum (six-well plate) or 2.5 × 105 IFUs/well inoculum (24-well plate), expression of PI3K, p-Akt, p53, and NF-κB (p65) representing activation of PI3K/Akt and NF-κB signal pathways were determined using western blotting or IFA, respectively. Gel quantification software was used to calculate the relative intensity of the corresponding signals. The relative value (target protein/GAPDH) differences of PI3K, p53, and NF-κB (p65) molecules and the relative expression level of p-Akt (p-Akt/Akt) between TC0668wt- and TC0668mut-infected cells were statistically significant (one-way ANOVA, P < 0.05). (A) Expression of PI3K molecules in TC0668mut- and TC0668wt-infected cells were determined using western blotting at 6, 12, 18, and 24 h post-infection. (B) Expression of p-Akt and total Akt in TC0668mut- and TC0668wt-infected cells were determined using western blotting at 6, 12, 18, and 24 h post-infection. (C) Expression of p53 molecules in the TC0668mut- and TC0668wt-infected cells at 6, 12, 18, and 24 h post-infection. (D) Expression of p65 molecules in the TC0668mut- and TC0668wt-infected cells at 6, 12, 18, and 24 h post-infection. (E) NF-κB molecules in the cytoplasm and nuclei of TC0668mut- and TC0668wt-infected HeLa cells at 6, 12, 18, and 24 h post-infection. DAPI dye core (blue), NF-κB fluorescence secondary antibody is 488 dye (green).
As depicted in Figure 7D, NF-κB (p65) expression in TC0668wt-infected cells was significantly higher at all four time points than that in TC0668mut-infected cells (P < 0.05). It was noticeable that NF-κB (p65) expression was significantly increased in TC0668wt-infected cells at 18 h p.i. To further evaluate the role of the NF-κB signaling pathway in the TC0668 virulence mechanism, the colocalization of NF-κB molecules and the nuclei of cells infected with either the TC0668wt or TC0668mut strains at 6, 12, 18, and 24 h p.i were visualized using IFAs (Figure 7E). NF-κB molecules (green) in TC0668wt-infected cells were localized outside the nuclei (blue) at 6 h p.i, and became increasingly localized to the nuclei as the infection progressed. Finally, NF-κB completely overlapped with the cell nuclei at 24 h p.i. NF-κB molecules also increased in the nuclei of TC0668mut-infected cells, but some green fluorescence was still maintained outside the nuclei and it only partially overlapped with the cell nuclei at 24 h p.i.
Inflammation- and Fibrosis-Related Proteins Differentially Expressed Between TC0668wt- and TC0668mut-Infected HeLa Cells
Tables 2, 3 show inflammation- and fibrosis-related proteins with differential expression (P < 0.05) between TC0668wt- and TC0668mut-infected cells at 18 h p.i. A total of 36 pro-inflammatory proteins and 27 fibrosis-related proteins presenting a clear fold-change (TC0668mut/TC0668wt) of ≥1.2 or ≤ 0.83 are shown with functional annotation, and they may play roles in chlamydial pathogenesis.
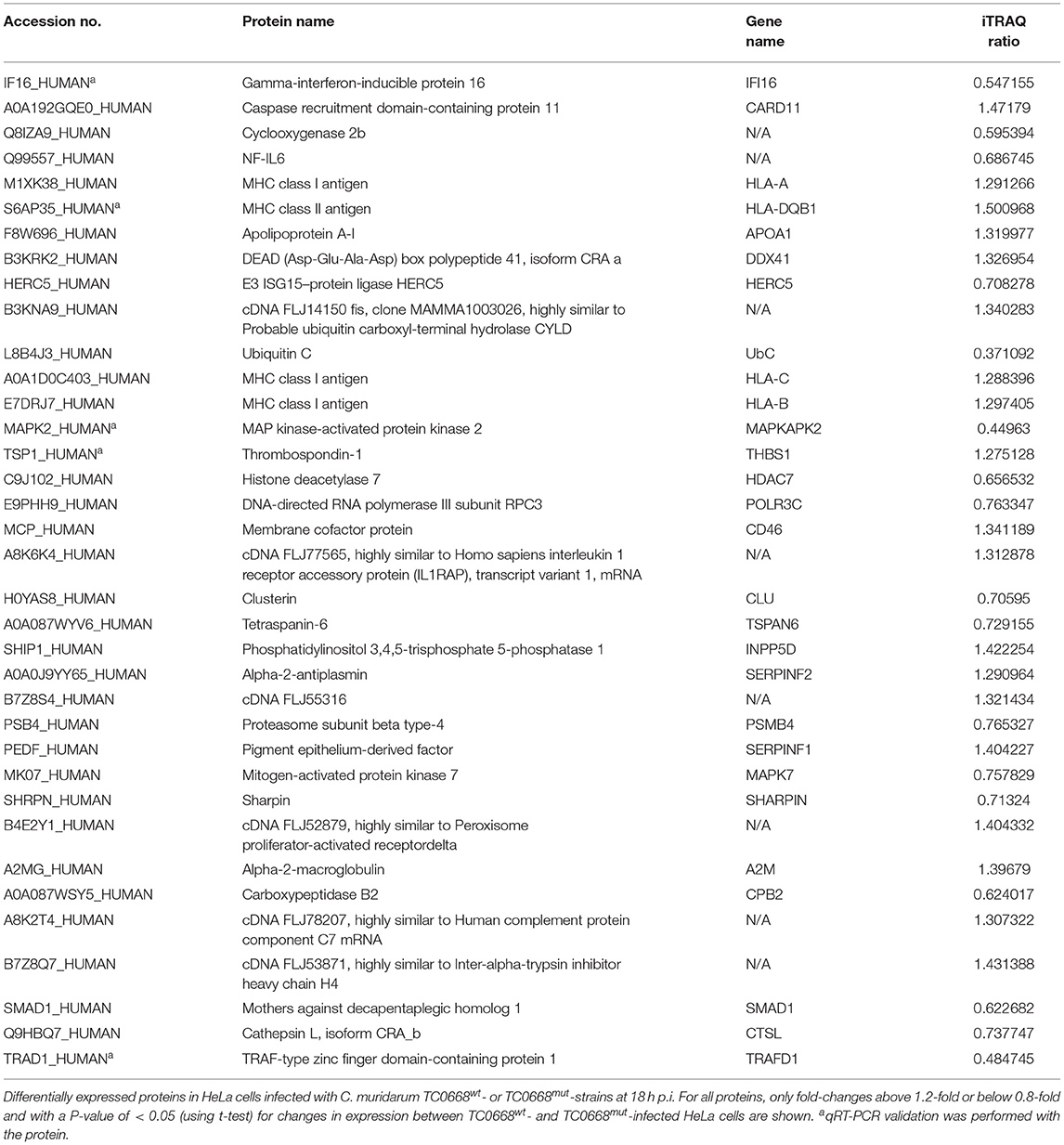
Table 2. Differentially expressed proteins related with inflammation in TC0668mut- vs. TC0668wt-infected HeLa cells at 18 h p.i.
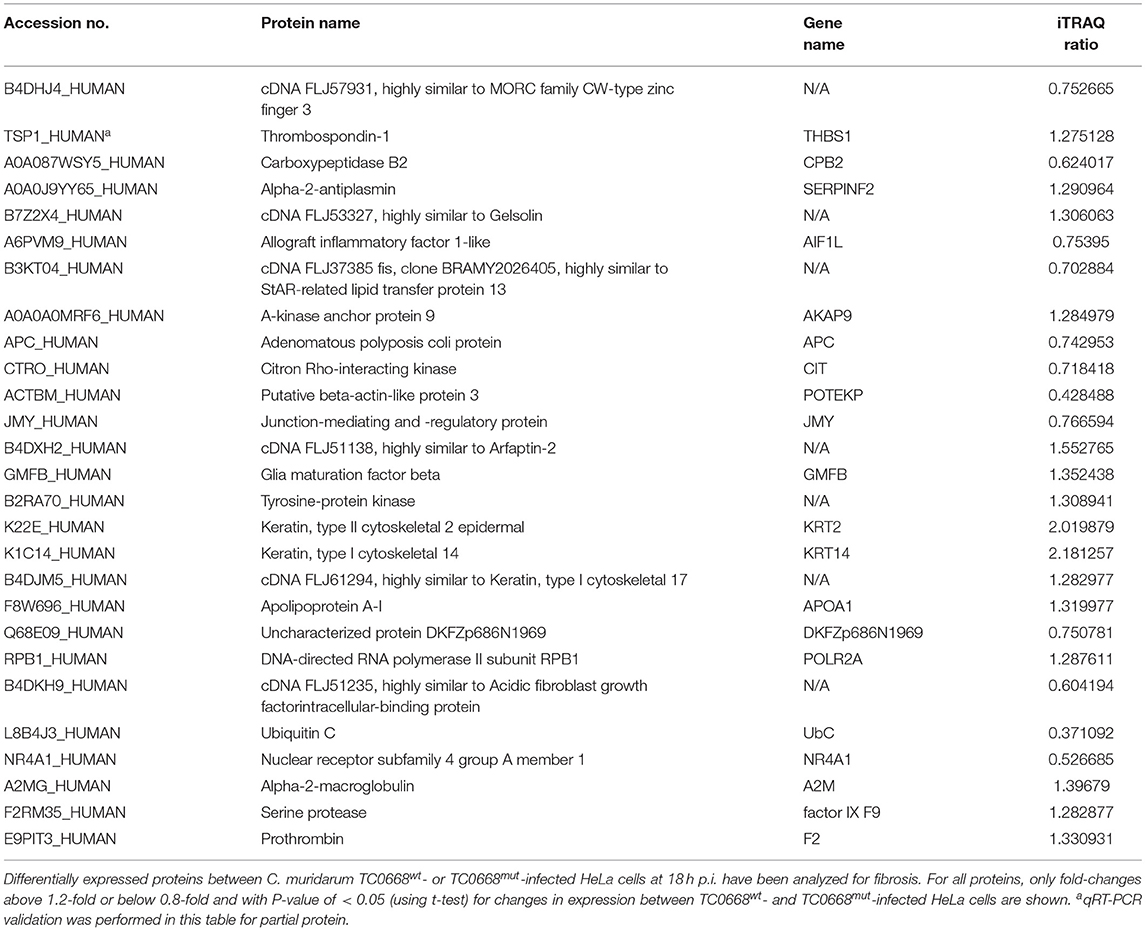
Table 3. Differentially expressed proteins related with fibrosis in TC0668mut- vs. TC0668wt-infected HeLa cells at 18 h p.i.
As shown in Table 2, proteins that regulate type I interferon production (encoded by IFI16, DDX41, HERC5, UbC, HLA-A, HLA-DQB1, and POLR3C) and the CARD11 gene (encoding caspase recruitment domain-containing protein 11), which positively regulates NF-κB activity, were all more highly expressed in TC0668wt-infected cells. Nine proteins involved in interleukin (IL) production (encoded by IFI16, HLA-C, HLA-B, THBS1, MAPKAPK2, INPP5D, HDAC7, CD46, and APOA1) and four proteins that affect TNF production (encoded by IFI16, MAPKAPK2, CD46, and CLU) were also differentially expressed. Furthermore, multiple other differentially expressed proteins (encoded by TSPAN6, PSMB4, SERPINF1, MAPK7, SHARPIN, A2M, CPB2, SMAD1, CTSL, TRAFD1, and SERPINF2) were found to be related to inflammatory responses.
The differentially expressed fibrosis-related proteins in Table 3 include three proteins that are involved in the regulation of fibrinolysis (encoded by THBS1, CPB2, and SERPINF2), 11 proteins that participate in supramolecular fiber organization (encoded by AIF1L, AKAP9, APC, CIT, POTEKP, JMY, GMFB, KRT2, KRT14, APOA1, and DKFZp686N1969), three proteins that are associated with fibrin clot formation (A2M, factor IX F9, and F2), and three proteins that play important roles in the response to fibroblast growth factor (encoded by POLR2A, UbC, and NR4A1).
PPI assays were used to investigate the interaction between proteins, it will deepen the understanding of protein structure and function, especially that we are interested in. Therefore, the differentially expressed proteins (P < 0.05) related to inflammation and fibrosis, which might play important roles in Chlamydia-induced upper genital tract damage were employed in a PPIs network analysis using the STRING database. As shown in Figure 8, ubiquitin C (UbC) and alpha-2-macroglobulin (A2M), the most connected differentially expressed proteins, interacted directly with multiple proteins such as NF-IL6, cDNA FLJ14150, and cDNA FLJ53871. Proteins with high connectivity may be key points affecting the entire system of metabolism or signaling transduction pathways during chlamydial infection. Thus, a better understanding of the molecular activity of TC0668 can be gained by combining the results of PPI network analysis and GO annotations, which could facilitate further research on the molecular mechanisms underlying C. muridarum infections.
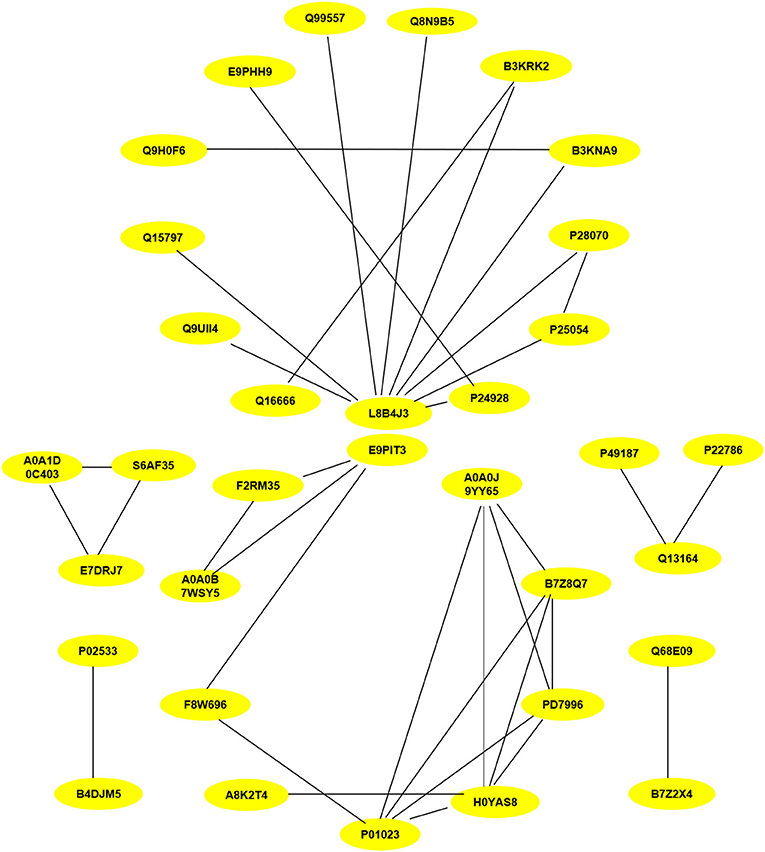
Figure 8. Protein-protein interaction network of statistically differentially expressed proteins (P < 0.05) associated with inflammation and fibrosis at 18 h p.i. The proteins interconnectivity of two categories (inflammation and fibrosis) are shown. The yellow circles marked with symbols combining numbers and letters, represent the protein ID of differentially expressed proteins screened by comparison of TC0668mut-infected and TC0668wt-infected HeLa cells. The corresponding information of proteins can be analyzed via UniProt (https://www.uniprot.org/), each protein ID corresponds to a protein. For example, the protein ID “P01023” corresponds to the protein “pha-2-macroglobulin”.
Discussion
Studying protein function has become more relevant in the post-genomic era because the main embodiment of function in organisms is proteins. Additionally, the ability of naturally occurring plasmid-free chlamydial strains to establish infection drives the enthusiasm to investigate the role of chromosomal proteins in chlamydial pathogenesis (Kari et al., 2011; Sigar et al., 2014). Previous studies demonstrated that TC0668mut strains are highly attenuated and cause less upper genital tract diseases in murine models compared to TC0668wt strains (Chen et al., 2015; Conrad et al., 2016; Shao et al., 2017). The pathogenesis differences regarding in vivo infections between TC0668mut and TC0668wt strains suggest that TC0668 contributes to a robust infection and the induction of pathological inflammation (Chen et al., 2015; Conrad et al., 2016). However, the role of TC0668 in inducing inflammatory responses remains unclear. To better understand the role of TC0668 in the pathogenesis of C. muridarum, we investigated the biological functions of differentially expressed proteins associated with TC0668 expression using an iTRAQ-based quantitative proteomics analysis of HeLa cells infected with TC0668wt or TC0668mut strains.
HeLa cells have been widely used in chlamydial studies, which include mouse model genital pathogen C. muridarum, respiratory pathogen Chlamydia psittaci or Chlamydia pneumonia (Bulir et al., 2015; Koch-Edelmann et al., 2017; Gallegos et al., 2018). Most importantly, we have conducted some in vitro experiments both in the murine oviduct cell C57epi.1 and human cervical cell line HeLa, and found the results are consistent with each other (unpublished data). It's similar to previous study that both murine cells BM12.4 and human epithelial cells HeLa 229 infected with different chlamydial strains (C. trachomatis serovars A to H, L1 to L3, C. muridarum, C. pneumonia, and C. caviae) exhibited the same protein tyrosine phosphorylation patterns (Virok et al., 2005). It's practicable to apply human HeLa cells in our research.
Molecular databases have long been a powerful tool for the study of molecular evolution and the prediction of protein function. The iTRAQ results suggest that TC0668 influences many important proteins that are molecular switches involved in cell signal transduction and biological activity. Ten differentially expressed proteins (encoded by MAPKAPK2, TRAFD1, IFI16, SRPRB, JAK1, PMM1, HLA-DQB1, THBS1, ITPR1, and BCAP31) were selected for qRT-PCR analysis to validate the proteomic results using qRT-PCR, and the qRT-PCR results were consistent with the proteomics results. Hence, iTRAQ-based techniques can be key in proteomics analysis, potentially providing useful information to help to reveal regulatory mechanisms.
Protein function prediction and identification are useful to clarify mechanisms underlying changes due to specific physiological or pathological conditions. GO and KEGG analyses are often performed to investigate the potential biological roles of differentially expressed proteins (Zou et al., 2018; Cai et al., 2019). With regard to GO, the differentially expressed proteins were mainly involved in biological processes including cellular process, metabolic process, immune system process, and biological regulation, suggesting that the pathogenesis of TC0668wt strains might be regulated by complex signaling pathways. KEGG analysis showed that these differentially expressed proteins are significantly enriched in disease-related signaling pathways such as the PI3K/Akt and NF-κB signaling pathways. PI3K is a lipid kinase that phosphorylates inositol phospholipids, thereby controlling membrane lipid composition and regulating a range of processes within the cell, including vesicle transport and signal transduction (Fruman et al., 2017). Western blotting involving key molecules of the PI3K/Akt pathway showed that PI3K and p-Akt expression was significantly higher in TC0668wt-infected cells than TC0668mut-infected cells at all time points tested. As one of the effector molecules of the PI3K signaling pathway, p53 can repress genes involved in cell growth stimulation and regulate normal cell activities by mediating signal transduction. In contrast to PI3K and p-Akt expression, p53 expression was significantly lower in the TC0668wt-infected cells compared to the TC0668mut-infected cells. Previous research has shown that Chlamydia infection of HeLa cells promotes p53 degradation (Bensaad et al., 2006), which is consistent with the results of our study. Additionally, stable p53 expression in HeLa cells interferes with the growth of Chlamydia by regulating the energy metabolism of the host cells, thereby inhibiting the infection (Siegl et al., 2014). We therefore hypothesized that the TC0668wt strain may facilitate the degradation of p53 by activating the PI3K/Akt signaling pathway, thereby relieving the inhibition of chlamydial growth, promoting infection, and further enhancing chlamydial pathogenicity. Further studies involving the TC0668mut and TC0668wt strains are needed to address the effects of the PI3K/Akt-p53 axis on chlamydial growth and metabolism.
The NF-κB pathway is a ubiquitous and classical pathway responsible for mediating DNA transcription, innate and adaptive immunity, inflammation, and other cellular activities (Bakkar and Guttridge, 2010). In general, it exists freely in the cytoplasm, inhibited by IκB proteins. With a variety of stimuli, such as cellular stress or bacterial infection, activated NF-κB translocates to the nucleus and induces target gene expression, exerting transcriptional regulation. In our western blotting analysis of the NF-κB pathway, NF-κB (p65) expression increased over time as the infection progressed in both TC0668wt- and TC0668mut-infected cells, but NF-κB (p65) levels in TC0668wt-infected cells were significantly higher than those in TC0668mut-infected cells at all time points (P < 0.05). IFAs were used to further detect the translocated NF-κB in nuclei at different time points in the two groups of cells. It was found that, for the TC0668wt strain at 24 h, NF-κB fluorescence signals completely overlapped with the cell nuclei, whereas some NF-κB fluorescence signals were still located outside the nuclei at 24 h for the TC0668mut strain. Therefore, we hypothesized that TC0668 is involved in regulating the activation of the NF-κB transcription factor, thereby mediating the inflammatory response to C. muridarum.
Inflammation is considered one of the most common consequence of persistent chlamydial infections (Cheong et al., 2018; Hou et al., 2018; Jia et al., 2019). To investigate the potential roles of TC0668 in inducing inflammatory- and fibrosis-related pathogenesis, a PPI network analysis of differentially expressed proteins at 18 h p.i was performed. It is worth noting that the expression levels of various proteins related to the activities of chemokine, IL (such as IL-6 and IL-1), tumor necrosis factor and associated cytokines, and type-1 interferon were significantly different between TC0668wt- and TC0668mut-infected cells. Ubiquitin C (UbC), a highly connected protein, has been shown to be involved in several diseases and to mediate biological processes such as inflammation, programmed cell death, and proliferation (Chen et al., 2008). In addition, alpha-2-macroglobulin (A2M), another highly connected protein, plays a pivotal role in eukaryote innate immune responses by binding to and modulating biological molecules (James, 1980). Thus, these two highly connected proteiin may play key roles in triggering, regulating, and modifying cell signaling transduction related to TC0668-mediated biological processes during chlamydial infection.
At present, the prediction of protein function is a challenging problem in the study of PPI networks. However, our findings suggest that TC0668 induces proteins to trigger, modify, and regulate signaling pathways that lead to pathogenesis, and the findings therefore provide new insights into the pathogenicity and potential molecular signaling pathways of C. muridarum. The differential effects on PI3K and NF-κB signaling pathways may underlie the attenuated virulence exhibited by the TC0668mut strain compared to the TC0668wt strain to induce hydrosalpinx.
Conclusions
As a chlamydial protein, TC0668 participates in the induction of inflammation, fibrosis, metabolic processes, and other cellular activities by regulating various molecular responses and signaling pathways, playing important roles in the pathogenesis of C. muridarum. This study provides useful information on the role of TC0668 in C. muridarum pathogenicity.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YW: data preparation and interpretation, and writing the manuscript. EA: modifying the manuscript. NL: cell experimentation. XL: western blotting experiments. WX: extracting the proteins. AM: participation in language editing. ZL: intellectual contribution throughout the study. ZZ: intellectual contribution throughout the study and interpretation of data. All authors reviewed, read, and approved the final manuscript.
Funding
This work is supported by the grant from the National Natural Science Foundation of China (31570179), the Foundation of Hunan Provincial Key Laboratory for Special Pathogens Prevention and Control (No. 2014-5), the National student's innovation and entrepreneurship training program (201810555018). The Hunan Province Cooperative Innovation Center for Molecular Target New Drug Study (2015-351).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Institute of Pathogenic Biology, Medical College, University of South China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02553/full#supplementary-material
Supplementary Table 1. The differentially expressed proteins between TC0668wt- and TC0668mut-infected cells in Venn diagram.
References
Bachmann, N. L., Polkinghorne, A., and Timms, P. (2014). Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22, 464–472. doi: 10.1016/j.tim.2014.04.013
Bakkar, N., and Guttridge, D. C. (2010). NF-kappaB signaling: a tale of two pathways in skeletal myogenesis. Physiol. Rev. 90, 495–511. doi: 10.1152/physrev.00040.2009
Bensaad, K., Tsuruta, A., Selak, M. A., Vidal, M. N., Nakano, K., Bartrons, R., et al. (2006). TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120. doi: 10.1016/j.cell.2006.05.036
Bommana, S., and Polkinghorne, A. (2019). Mini review: antimicrobial control of chlamydial infections in animals: current practices and issues. Front. Microbiol. 10:113. doi: 10.3389/fmicb.2019.00113
Bulir, D. C., Waltho, D. A., Stone, C. B., Liang, S., Chiang, C. K., Mwawasi, K. A., et al. (2015). Chlamydia Outer Protein (Cop) B from Chlamydia pneumoniae possesses characteristic features of a type III secretion (T3S) translocator protein. BMC Microbiol. 15:163. doi: 10.1186/s12866-015-0498-1
Cai, Q., Wang, G., Li, Z., Zhang, L., Fu, Y., Yang, X., et al. (2019). SWATH based quantitative proteomics analysis reveals Hfq2 play an important role on pleiotropic physiological functions in Aeromonas hydrophila. J. Proteomics 195, 1–10. doi: 10.1016/j.jprot.2018.12.030
Chen, C., Zhou, Z., Conrad, T., Yang, Z., Dai, J., Li, Z., et al. (2015). In vitro passage selects for Chlamydia muridarum with enhanced infectivity in cultured cells but attenuated pathogenicity in mouse upper genital tract. Infect. Immun. 83, 1881–1892. doi: 10.1128/IAI.03158-14
Chen, L., Lei, L., Chang, X., Li, Z., Lu, C., Zhang, X., et al. (2010). Mice deficient in MyD88 develop a Th2-dominant response and severe pathology in the upper genital tract following Chlamydia muridarum infection. J. Immunol. 184, 2602–2610. doi: 10.4049/jimmunol.0901593
Chen, S. M., Zhang, H. X., Li, Y. G., Wang, D. M., Zhang, G. H., and Tan, C. J. (2008). Expression of ubiquitin in peripheral inflammatory cells from patients with coronary artery disease. J. Int. Med. Res. 36, 1227–1234. doi: 10.1177/147323000803600609
Cheng, W., Shivshankar, P., Li, Z., Chen, L., Yeh, I. T., and Zhong, G. (2007). Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect. Immun. 76, 515–522. doi: 10.1128/iai.01064-07
Cheong, H. C., Lee, C. Y. Q., Cheok, Y. Y., Shankar, E. M., Sabet, N. S., Tan, G. M. Y., et al. (2018). CPAF, HSP60 and MOMP antigens elicit pro-inflammatory cytokines production in the peripheral blood mononuclear cells from genital Chlamydia trachomatis-infected patients. Immunobiology 224, 34–41. doi: 10.1016/j.imbio.2018.10.010
Conrad, T. A., Gong, S., Yang, Z., Matulich, P., Keck, J., Beltrami, N., et al. (2016). The chromosome-encoded hypothetical protein TC0668 is an upper genital tract pathogenicity factor of Chlamydia muridarum. Infect. Immun. 84, 467–479. doi: 10.1128/IAI.01171-15
Ferreira, R., Borges, V., Nunes, A., Borrego, M. J., and Gomes, J. P. (2013). Assessment of the load and transcriptional dynamics of Chlamydia trachomatis plasmid according to strains' tissue tropism. Microbiol. Res. 168, 333–339. doi: 10.1016/j.micres.2013.02.001
Fruman, D. A., Chiu, H., Hopkins, B. D., Bagrodia, S., Cantley, L. C., and Abraham, R. T. (2017). The PI3K pathway in human disease. Cell 170, 605–635. doi: 10.1016/j.cell.2017.07.029
Gallegos, K. M., Taylor, C. R., Rabulinski, D. J., Del Toro, R., Girgis, D. E., Jourha, D., et al. (2018). A synthetic, small, sulfated agent is a promising inhibitor of Chlamydia spp. infection in vivo. Front. Microbiol. 9:3269. doi: 10.3389/fmicb.2018.03269
Hou, S., Yue, L., Xu, R., Zhu, C., Shan, S., Wang, H., et al. (2018). Chlamydia muridarum plasmid induces mouse oviduct pathology by promoting chlamydial survival and ascending infection and triggering host inflammation. Eur. J. Dermatol. 28, 628–636. doi: 10.1684/ejd.2018.3399
Huang, C., Yang, X., Zeng, B., Zeng, L., Gong, X., Zhou, C., et al. (2019). Proteomic analysis of olfactory bulb suggests CACNA1E as a promoter of CREB signaling in microbiota-induced depression. J. Proteomics 194, 132–147. doi: 10.1016/j.jprot.2018.11.023
Huang, Y., Zhang, Q., Yang, Z., Conrad, T., Liu, Y., and Zhong, G. (2015). Plasmid-encoded Pgp5 is a significant contributor to Chlamydia muridarum induction of hydrosalpinx. PLoS ONE 10:e0124840. doi: 10.1371/journal.pone.0124840
James, K. (1980). Alpha2 macroglobulin and its possible importance in immune systems. Trends Biochem. Sci. 5, 43–47. doi: 10.1016/S0968-0004(80)80094-6
Jézéquel, P., Guette, C., Lasla, H., Gouraud, W., Boissard, A., Guerin-Charbonnel, C., et al. (2019). iTRAQ-based quantitative proteomic analysis strengthens transcriptomic subtyping of triple-negative breast cancer tumors. Proteomics e1800484. doi: 10.1002/pmic.201800484. [Epub ahead of print].
Jia, L., Sun, F., Wang, J., Gong, D., and Yang, L. (2019). Chlamydia trachomatis ct143 stimulates secretion of proinflammatory cytokines via activating the p38/MAPK signal pathway in THP-1 cells. Mol. Immunol. 105, 233–239. doi: 10.1016/j.molimm.2018.12.007
Kari, L., Whitmire, W. M., Olivares-Zavaleta, N., Goheen, M. M., Taylor, L. D., Carlson, J. H., et al. (2011). A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J. Exp. Med. 208, 2217–2223. doi: 10.1084/jem.20111266
Koch-Edelmann, S., Banhart, S., Saied, E. M., Rose, L., Aeberhard, L., Laue, M., et al. (2017). The cellular ceramide transport protein CERT promotes Chlamydia psittaci infection and controls bacterial sphingolipid uptake. Cell Microbiol 19:12752. doi: 10.1111/cmi.12752
Land, J., Van Bergen, J., Morré, S., and Postma, M. (2009). Epidemiology of Chlamydia trachomatis infection in women and the cost-effectiveness of screening. Hum. Reprod. Update 16, 189–204. doi: 10.1093/humupd/dmp035
Liu, Y., Huang, Y., Yang, Z., Sun, Y., Gong, S., Hou, S., et al. (2014). Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect. Immun. 82, 5327–5335. doi: 10.1128/IAI.02576-14
Morrison, R. P., and Caldwell, H. D. (2002). Immunity to murine chlamydial genital infection. Infect. Immun. 70, 2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002
Porcella, S. F., Carlson, J. H., Sturdevant, D. E., Sturdevant, G. L., Kanakabandi, K., Virtaneva, K., et al. (2015). Transcriptional profiling of human epithelial cells infected with plasmid-bearing and plasmid-deficient Chlamydia trachomatis. Infect. Immun. 83, 534–543. doi: 10.1128/IAI.02764-14
Qin, Y., Jiang, X., Li, W., Li, J., Tian, T., Zang, G., et al. (2019). Chronic mild stress leads to aberrant glucose energy metabolism in depressed Macaca fascicularis models. Psychoneuroendocrinology 107, 59–69. doi: 10.1016/j.psyneuen.2019.05.007
Rodgers, A. K., Budrys, N. M., Gong, S., Wang, J., Holden, A., Schenken, R. S., et al. (2011). Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertil. Steril. 96, 715–721. doi: 10.1016/j.fertnstert.2011.06.021
Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., et al. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169. doi: 10.1074/mcp.M400129-MCP200.
Shah, A. A., Schripsema, J. H., Imtiaz, M. T., Sigar, I. M., Kasimos, J., Matos, P. G., et al. (2005). Histopathologic changes related to fibrotic oviduct occlusion after genital tract infection of mice with Chlamydia muridarum. Sex. Transm. Dis. 32, 49–56. doi: 10.1097/01.olq.0000148299.14513.11
Shao, L., Zhang, T., Liu, Q., Wang, J., and Zhong, G. (2017). Chlamydia muridarum with mutations in chromosomal genes tc0237 and/or tc0668 are deficient in colonizing mouse gastrointestinal tract. Infect. Immun. 85:e00321-17. doi: 10.1128/IAI.00321-17
Siegl, C., Prusty, B. K., Karunakaran, K., Wischhusen, J., and Rudel, T. (2014). Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep. 9, 918–929. doi: 10.1016/j.celrep.2014.10.004
Sigar, I. M., Schripsema, J. H., Wang, Y., Clarke, I. N., Cutcliffe, L. T., Seth-Smith, H. M., et al. (2014). Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog. Dis. 70, 61–69. doi: 10.1111/2049-632X.12086
Virok, D. P., Nelson, D. E., Whitmire, W. M., Crane, D. D., Goheen, M. M., and Caldwell, H. D. (2005). Chlamydial infection induces pathobiotype-specific protein tyrosine phosphorylation in epithelial cells. Infect. Immun. 73, 1939–1946. doi: 10.1128/IAI.73.4.1939-1946.2005.
Wu, H., Zhang, S., Huo, C., Zou, S., Lian, Z., and Hu, Y. (2019). iTRAQ-based proteomic and bioinformatic characterization of human mast cells upon infection by the influenza A virus strains H1N1 and H5N1. FEBS Lett. 593, 2612–2627. doi: 10.1002/1873-3468.13523
Zhou, X., Shi, F., Zhou, L., Zhou, Y., Liu, Z., Ji, R., et al. (2019). iTRAQ-based proteomic analysis of fertile and sterile flower buds from a genetic male sterile line 'AB01' in Chinese cabbage (Brassica campestris L. ssp. pekinensis). J. Proteomics 204:103395. doi: 10.1016/j.jprot.2019.103395
Keywords: Chlamydia muridarum (C. muridarum), isobaric tags for relative and absolute quantitation (iTRAQ), quantitative proteomics, TC0668, infection
Citation: Wang Y, Arthur EW, Liu N, Li X, Xiang W, Maxwell A, Li Z and Zhou Z (2019) iTRAQ-Based Quantitative Proteomics Analysis of HeLa Cells Infected With Chlamydia muridarum TC0668 Mutant and Wild-Type Strains. Front. Microbiol. 10:2553. doi: 10.3389/fmicb.2019.02553
Received: 20 August 2019; Accepted: 22 October 2019;
Published: 07 November 2019.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Guangming Zhong, The University of Texas Health Science Center at San Antonio, United StatesYimou Wu, University of South China, China
Chunfu Yang, National Institute of Allergy and Infectious Diseases (NIAID), United States
Copyright © 2019 Wang, Arthur, Liu, Li, Xiang, Maxwell, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhou Zhou, c3VzaWV6aG91OTk1MDNAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yingzi Wang
Yingzi Wang Emmanuel Wirekoh Arthur1,2†
Emmanuel Wirekoh Arthur1,2†