- 1Department of Chemistry, University of Helsinki, Helsinki, Finland
- 2VTT Technical Research Centre of Finland, Espoo, Finland
The uptake of nickel [Ni(II)] by Paenibacillus sp., Methylobacterium sp., Paraburkholderia sp., and Pseudomonas sp. strains isolated from a boreal bog was studied using batch experiments. All strains removed Ni(II) from the solution and the uptake efficiency was affected by the nutrient source, incubation temperature, time, and pH. As highest Ni uptake (with a maximum Kd of 1890 L/kg DW) was recorded for the Pseudomonas sp. strains, these bacteria were used in the following protein expression (SDS-PAGE and MALDI-TOFF), transmission electron microscopy (TEM) and EDS experiments. In addition, Freundlich and Langmuir sorption isotherms were determined. In the Ni(II) treated cells, dense crystalline intra-cellular accumulations were observed in TEM examinations, which were identified as Ni accumulations using EDS. SDS-PAGE and MALDI-TOFF spectra of Ni(II) treated cells showed several changes in the protein profiles, which can indicate active accumulation of Ni in these bacteria. Concurrently, we observed Ni(II) uptake to follow Freundlich and Langmuir isotherms, suggesting straight cellular biosorption in addition to the intra-cellular accumulation. The role of cellular (cell membrane and cell wall) functional groups involved in Ni(II) binding were therefore studied using Fourier transformation infrared spectroscopy. These analyses supported the potential role of the alcoholic hydroxyl, carboxyl and amine groups in Ni(II) binding in these bacteria, therefore suggesting two different Ni(II) uptake mechanisms; (i) intra-cellular accumulation [possibly connected to detoxification of Ni(II)], and (ii) straight biosorption on cell membrane/wall functional groups.
Introduction
Nickel (Ni) occurs extensively in Earth’s crust and is found in various mineral forms (e.g., pentlantide) (Cempel and Nikel, 2006). Significant amounts of Ni are released into the environment as a result of human activities, including mining, energy production (combustion of coal, diesel, etc.) and industrial use of Ni (e.g., production of ferronickel) (Nriagu, 1990). Industrial processes generally increase Ni mobility and may result in potentially large releases to the environment through surface and groundwater flows. In addition, the potentially high mobility of the long-lived Ni radioactive isotopes, 59Ni and 63Ni, found in spent nuclear fuel, are of concern for the long-term safety of spent nuclear fuel bedrock disposal (Hjerpe et al., 2010).
In the environment, Ni normally occurs at the oxidation state +2, but oxidation states 0, +1, and +3 also exist (Cempel and Nikel, 2006). Ni2+ (aq) ion is the dominant form throughout the pH and Eh range of most natural waters (Supplementary Figure 1). Ni forms complexes with inorganic ligands, such as halides, sulfides, sulfates, hydroxides and carbonates, and with organic oxygen, nitrogen, and sulfur compounds (Nieminen et al., 2007). Common Ni salts, e.g., chloride, nitrate and sulfate, dissolve in water, whereas carbonate and oxalate complexes are less soluble, and hydroxide and sulfide complexes are practically insoluble in water (Nieminen et al., 2007). In the environment under neutral and alkaline conditions, the mobility of Ni is considered to be low and Ni is bound on sediment or soil particles by adsorption (i.e., surface complex formation/absorption to ion exchange sites), or removed from the liquid phase by precipitation or co-precipitation (Cempel and Nikel, 2006). Under acidic conditions and in the presence of dissolved organic matter, the mobility of Ni typically increases as Ni readily forms rather stable complexes with organic ligands (Nieminen et al., 2007).
In the environment, Ni accumulates in biota and occurs inherently in food chains, and its concentration in a particular food chain may increase as a result of regional pollution (Cempel and Nikel, 2006; Nieminen et al., 2007). In low quantities, Ni is essential for humans, animals, plants and microorganisms, but high concentrations of Ni have adverse effects on biota (IARC, 2012). Although Ni is not a cumulative poison, the higher concentrations may be toxic and even carcinogenic to humans and animals (IARC, 2012). For humans and other animals, the major source of Ni exposure is oral consumption, as Ni readily accumulates to various plants. There are several species of plants, such as members of the Brassicaceae family (Jaffré et al., 2013), known to accumulate Ni in particularly high concentrations. Ni is also an essential nutrient for microorganisms and is known to play an important role in the biology of some bacteria, fungi and algae. Ni acts as a co-factor and participates in a variety of cellular enzymatic reactions. Several microbial enzymes have Ni as a co-factor, including urease, [NiFe]-hydrogenase, carbon monoxide dehydrogenase, acetyl-coenzyme-A decarbonylase/synthase, superoxide dismutases, and glyoxylases (Mobley and Hausinger, 1989; Hausinger, 1994; Mulrooney and Hausinger, 2003; Sukdeo et al., 2004).
Many microbes have an ability to bind Ni and can have various Ni uptake mechanisms, including biosorption on cell surfaces (based on ion-exchange, complexation, and physical adsorption), active intracellular accumulation and/or extracellular precipitation (White et al., 1995; Ledin and Pedersen, 1996; López et al., 2000; Sar et al., 2001; Mulrooney and Hausinger, 2003). Microbial (e.g., bacterial) cell walls have several potentially metal-binding functional groups, such as carboxyl, hydroxyl, sulfate, phosphate, and amine groups (White et al., 1995; Ledin and Pedersen, 1996; López et al., 2000; Sar et al., 2001). In addition, especially under low metal loading conditions sulfhydryl groups (-SH) may also be important for trace metal adsorption (Yu et al., 2014). The efficiency of Ni biosorption is dependent on various cell external factors, such as solution pH, ion activity (including competing ions), organic material contents (including complexing agents), temperature and the content of cell metabolic products in the extracellular solution (which may cause metal precipitation) (Tsezos et al., 1996; Veglió and Beolchini, 1997; Veglió et al., 1997; López et al., 2000).
For example, Pseudomonas aeruginosa has been reported to bind Ni, and cell wall phosphoryl, carboxyl or carbonyl groups were suggested as the major groups participating in Ni and other cation uptake in this bacterium (Sar et al., 2001). Ni uptake has also been reported in Ps. fluorescens 4F39 (López et al., 2000). However, in this bacterium uptake was assumed through ion exchange and precipitation on cell surfaces (López et al., 2000). In addition, Ni biosorption has also been reported, e.g., by the bacteria Arthrobacter sp. (Veglió et al., 1997); Pseudomonas sp. BP 7/26 and BP 7/15, Alcaligenes eutrophus CH34 and ER 121, Ps. mendocina AS 302, Methylobacillus sp. MB 127 (Tsezos et al., 1995) and the fungus Rhizopus arrhizus (Fourest and Roux, 1992).
In addition to straight biosorption/precipitation on cell walls, many microbes are capable of accumulating Ni inside the cell, and so far, two distinct uptake mechanisms have been identified, i.e., ATP-binding cassette (ABC)-type transporter systems and Ni-specific permeases (membrane transport proteins) (Eitinger and Mandrand-Berthelot, 2000). The first and best described ABC-type transporter for Ni was reported in Escherichia coli (Navarro et al., 1993; Mulrooney and Hausinger, 2003; Cherrier et al., 2005; Schalk et al., 2011; Chivers, 2015). Other bacteria, such as Brucella suis (Jubier-Maurin et al., 2001), Actinobacillus pleuropneumoniae (Bossé et al., 2001), Helicobacter pylori (Hendricks and Mobley, 1997), Yersinia pseudotuberculosis, Y. pestis, and Y. enterocolitica (Sebbane et al., 2002), have also been found to contain corresponding Ni-specific ABC-type transporter systems (Mulrooney and Hausinger, 2003). Ni-specific permeases have been identified, e.g., in Ralstonia eutropha (Eberz et al., 1989; Eitinger and Friedrich, 1991; Wolfram et al., 1995), Bradyrhizobium japonicum (Fu et al., 1994), Bacillus sp. (Maeda et al., 1994), and the yeast Schizosaccharomyces pombe (Eitinger et al., 2000).
In Helicobacter pylori, the Ni accumulation pathway involves a TonB-dependent transporter (TBDT) FrpB4 and a permease NixA (Schauer et al., 2007; Schalk et al., 2011). TBDT FrbB4 is used for Ni uptake through the outer-membrane (Schauer et al., 2007; Schalk et al., 2011). In the periplasmic space, Ni may bind to a specific protein and passes the cytoplasmic membrane by the NixA permease or another metal transporter leading to increased urease activity (Schauer et al., 2007; Schalk et al., 2011). In another Helicobacter species, H. mustelae, two distinct Ni pathways have been proposed Stoof et al. (2010) and Schalk et al. (2011). In the first one, Ni accumulation involves a TBDT NikH and a permease NixA through the outer- and inner-membrane, respectively (Stoof et al., 2010; Schalk et al., 2011). For the second, only the inner membrane ABC transporter (FecDE-CeuE) has been identified and the mechanism involved in Ni uptake across the outer membrane is unknown (Stoof et al., 2010; Schalk et al., 2011). In addition, non-specific Ni accumulation in the cytoplasm has been reported (Snavely et al., 1989, 1991; Smith and Maguire, 1995). For example, Smith and Maguire (1995) identified a CorA Mg2+ transporter system (metal inorganic transport) in several Gram negative bacteria. It has been suggested that CorA also mediates uptake of Ni2+ and Co2+ in addition to Mg2+ uptake (Snavely et al., 1989, 1991).
Significant environmental Ni deposits are found in the boreal and arctic regions (Mudd, 2010), including the Talvivaara Ni-Zn-Cu-Co mine area deposit, located in the Sotkamo area, Finland. This deposit comprises one of the largest known Ni resources in Europe (Loukola-Ruskeeniemi and Lahtinen, 2013). Large quantities of mine process waters and solid waste materials (e.g., waste rock, leached ore), having high potential for adverse environmental impacts, especially in the surrounding water systems, are found in the mining areas (Pöyry Finland Oy, 2017). A particular problem is the increased surface area of the waste rock exposed to the circulating waters, resulting in the dissolution of minerals and the leaching of toxic heavy metals like Ni. In addition, the sulfide minerals often present on mine areas are sources for acid mine drainage (AMD), which may then further dissolve potentially-toxic metals from surrounding materials and cause contamination of surrounding water resources (Akcil and Koldas, 2006). The aim of the present work was to study the interaction of Ni with newly isolated, previously unidentified bacterial strains isolated from boreal environment and to estimate their potential in the biopurification of Ni especially in the northern environment. In the northern, boreal regions, ombrotrophic bogs represent unique ecological biotopes, with distinct microbial populations. However, so far there is only limited knowledge about the heavy metal metabolism of the microbes inhabiting these northern environments. The bacterial strains newly isolated in this research, are adapted to the northern climate and environment. Thus, these bacteria have high potential for site-specific biopurification of Ni especially in the heavy-metal contaminated mine areas found in the boreal region and in the present study this potential was assessed in enhanced and controlled environment.
Materials and Methods
Description of the Sampling Site
In Finland, spent nuclear fuel will be stored of in deep bedrock disposal repository located on Olkiluoto Island on the western coast of Finland (Posiva, 2012). As a result of post-glacial land uplift, new bogs will be form on the repository area during the next 6000 years (Haapanen et al., 2013). At the same time period, the first possible emissions from the repository may be release to the surface ecosystem based on the biosphere safety assessments (Posiva, 2012). The sampling site, Lastensuo bog, is located about 25 km west from the Olkiluoto Island. This area has been used as a reference site for the spent nuclear fuel disposal research, representing boreal ombrotrophic bog (e.g., Mäkilä and Grundström, 2008; Lusa et al., 2015a,b,c, 2016a,b). Lastensuo is a raised bog/hummock–hollow pine bog (Mäkilä and Grundström, 2008), with an area of 4.4 km2 and the average depth of 6.3 m (Mäkilä and Grundström, 2008). The catchment area is till forest and the edges of the mire have been ditched. The middle part of the mire is near-treeless. Vegetation in the mire includes, e.g., slender sedge and fewflower sedge (Mäkilä and Grundström, 2008). The two main peat types are sphagnum peat and carex peat. The average degree of decomposition (von Post’s scale) is 3.4, and the annual peat growth rate is 1.1 mm/y (Mäkilä and Grundström, 2008). The bottom of the mire is mainly clay and sand. Gyttja occurs in the middle part of mire on the top of clay subsoil (Mäkilä and Grundström, 2008). The bog layers can be divided in to moss (0 m), peat (0.5–5.0 m), gyttja (5.5–6.0 m), and clay (6.7–7.0 m) (Lusa et al., 2015b). The pH of the bog profile increases with bog layer depth and the pH values vary from 3.1 (surface moss) to 5.3 (clay) (Lusa, 2017). Correspondingly the highest total Ni concentrations [15 mg/kg DW (dry weight)] are found in the gyttja layer (Lusa, 2017). High Ni concentrations (7.7 mg/kg DW) are in addition found in the moss layer. In the peat layers, significantly lower, < 2.3 mg/kg DW, Ni concentrations are found. Major cation (Na, K, Mg, Ca) concentrations are between 113 and 5178 mg/kg DW, with highest concentrations measured for Ca and lowest for Na (Lusa, 2017). The average salt concertation calculated as a sum of Na, K, Mg, and Ca is approximately 3224 mg/kg DW. In the pore water lower concentrations between 0.47 and 3.91 mg/L are found (Lusa, 2017).
Sampling
The samples were collected from the middle part of Lastensuo bog in early summer, 2nd of June, 2015. The sample plot was located on the treeless part of the bog with two small pools near the sample area. The samples for the isolation of bacteria were taken from the surface Sphagnum moss and from the lowest part of the bog peat layer (4.5–5.0 m). Samples were collected into 50 mL sterile centrifuge tubes using sterile tools and aseptic working methods. Peat samples were retrieved using a Russian peat corer with a nest length of 50 cm and diameter of 15 cm. The sample collection tubes were sealed with Parafilm, transported to the laboratory in cooling boxes, stored frozen at −18°C and thawed immediately before use.
The temperature of each bog layer was recorded immediately after the core sample was taken to the surface in June 2015. The temperature of the surface layer was + 10.3°C. In the lower layers, the temperature was relatively constant, with an average of + 6.6°C. In May 2015 the average temperature at the west-coastal region of Finland (Pori region) was approximately + 9°C and in June + 12°C in 2015 (Finnish Meteorological Institute [FMI], 2015). Typical average temperature in this region in July is around + 16–20°C (Finnish Meteorological Institute [FMI], 2015).
Isolation of Bacteria
Six bacterial strains were isolated from the moss (surface) and peat (4.5–5.0 m) samples by mixing two grams of thawed, wet sample with 0.01 mol/L sterile phosphate buffer (pH 7), where after the samples were incubated aerobically for 24 h at room temperature in the dark. The resulting suspension was serially diluted in 10-fold steps to a dilution of up to 10–5. Aliquots of 100 μL from dilutions 10–3 and 10–5 were spread on Tryptone Soya Agar (TSA, Merckoplate) (15% Casein peptone, 5% Soya peptone, 5% NaCl; pH 7.3), R2A Agar (R2A, Merckoplate) (0.5% Proteose peptone, 0.5% Casein peptone, 0.5% Yeast extract, 0.5% Glucose, 0.5% Soluble starch, 0.3% Sodium pyruvate, 0.3% K2HPO4, 0.024% MgSO4; pH 7.2), Potato Dextrose Agar (PDA, Merckoplate) (4% Potato peptone, 20% Glucose; pH 5.6) and Plate Count Agar (PCA, Merckoplate) (5% Casein peptone, 2.5% Yeast Extract, 1% Glucose; pH 7.0). The plates were further incubated aerobically at 20°C for 1 week in the dark. After the incubation, pure cultures were prepared from isolated colonies by consecutive re-cultivation on corresponding agar plates.
Bacterial cells from each pure culture were Gram stained and examined using a light microscope (Nikon ECLIPSE E200) with 1000-fold magnification. Catalase activity was tested using 3% hydrogen peroxide. When bubbles were formed, the bacterium was determined positive for catalase. Oxidase activity was tested using 1% Kovács oxidase reagent. In addition, RapID ONE and RapID NF Plus (Remel) systems, and ERIC electronic code were used according to the manufacturer’s instructions for characterizing the utilization of substrates by the isolated bacterial strains.
16S rRNA Gene Sequencing and Phylogenetic Analysis
For the identification of bacterial isolates by sequencing, one colony of bacterial biomass was collected directly from the growth plate of each isolate using a sterile loop. The bacterial mass was suspended in the lysis buffer SL1 and Enhancer solution SE of the NucleoSpin Soil DNA extraction kit (Macherey-Nagel), after which the DNA was extracted according to the manufacturer’s instructions. The DNA was subsequently sent for PCR amplification, fragment purification, and sequencing to Macrogen, Inc., Netherlands, where PCR amplification and sequencing was conducted in both directions using primers 27f, 1492r, 518f, and 800r according to Macrogen’s protocol. Sequences were imported into Geneious Pro v. 10.1.3 (Biomatters, Ltd., Auckland, New Zealand) and processed for phylogenetic analyses. The sequences were assembled and edited by hand. In order to identify the isolated bacteria, the obtained 16S rRNA gene sequences were compared to the sequences deposited in GenBank using the blast tool in Geneious Pro. The closest matching sequences as well as relevant reference sequences were aligned using the ClustalW (Thompson et al., 1994) alignment tool in Geneious Pro. A phylogenetic tree was calculated from the alignments using the Geneious Tree Builder tool Jukes and Cantor (1969) substitution model and the topology of the tree was tested by bootstrap analyses of 1000 random resampling. The sequences have been deposited in GenBank under accession numbers MK290398-MK290403.
Microbial Culture Conditions and Performance of Biosorption Experiments
Batch Experiments
Six isolated bacterial strains, Paenibacillus sp. IV-0-L, KV-0-YR and VV-0-L, Methylobacterium sp. P4-5-LR, Paraburkholderia sp. RP-0-BL and Pseudomonas sp. V4-5-SB, were selected for the Ni uptake experiments. Paenibacillus sp. IV-0-L, KV-0-YR and VV-0-L, and Paraburkholderia sp. RP-0-BL were isolated from surface moss layer and Methylobacterium sp. P4-5-LR, and Pseudomonas sp. V4-5-SB from 4.5 to 5.0 m peat layer. In addition, Pseudomonas sp. PS-0-L and T5-6-I (Lusa et al., 2016a, b), previously isolated from the surface moss (PS-0-L) and gyttja layer (T5-6-I) of the Lastensuo bog, were used. All bacteria were able to grow on PCA growth plates (PCA, Merckoplate, pH 7), which were selected for following stock cultures to standardize the experiments. Stocks were cultured aerobically on sterile PCA growth plates at 20°C in the dark and the colonies were transferred onto new plates weekly.
The uptake of Ni(II) by all isolated bacteria was studied using batch experiments in two different growth media. Medium A consisted of 1% Tryptone + 0.5% NaCl (pH 7) and Medium B of 1% Yeast extract + 0.5% NaCl (pH 7). These media and pH 7 were chosen for the uptake experiments as the effects of the two main components (Peptone∼Tryptone and Yeast extract) of the stock PCA (pH 7) medium was to be studied. This is because, the used bacterial strains were originally isolated on media, which all contained peptone. pH 7 was chosen, as it well corresponds to the average pH values of the natural ground waters and surface waters found in Finland, with pH between 4 and 9, highest values found in the deep ground waters (Niemi and Raateland, 2007; Posiva, 2012). The 0.5% NaCl concentration was chosen to represent to the average salt concentration of Lastensuo bog (see section “Description of the Sampling Site”). The cells were cultivated at + 4 and +20°C using incubation times of 7 and 14 days. In all uptake experiments, 14 Bq/mL of 63Ni(II) with a carrier concentration of 10–10 mol/L, corresponding to an initial Ni concentration of 6 × 10–10 mol/L was used. The average bacterial mass in the experiments was 0.56 g DW (dry weight)/L. Due to the weak growth of Methylobacterium sp. P4-5-LR a higher mass of on average 8.5 g DW/L was used. Before addition of the bacteria to the solutions, microbial suspension was prepared by adding cultivated biomass from the plates to sterile 0.01 mol/L phosphate buffer (pH 7). The mass concentration of the bacterial suspension was determined by absorbance at 600 nm.
All suspensions were incubated for 7–14 days at 4 or 20°C in the dark, after which the suspensions were centrifuged at 5000 × g for 15 min at room temperature. The supernatant was used for the liquid scintillation determination of 63Ni activity using a Tri-Carb (PerkinElmer) liquid scintillation counter. In addition, suspensions without added bacteria were prepared accordingly and measured to assure that no sorption of Ni on laboratory equipment, filters or nutrient broth solutions occurred.
The removal of Ni(II) from the solution by bacterial cells was calculated from the difference between initial and final Ni concentration in the solution and expressed as distribution coefficient (Kd, L/kg DW): Kd = [(Ci – Cf)/Cf] × (V/m), where Ci (Bq/L) and Cf (Bq/L) are the initial 63Ni activity and final activity concentration of the solution, V (L) is the solution volume, and m (kg DW) is the sample mass at t = 0.
Based on batch experiments, Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I were selected for further studies due to their high Ni(II) uptake capacity.
The Effect of pH on Ni(II) Uptake
The effect of pH on Ni(II) uptake by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I was investigated using 0.1 mol/L NaCl at + 20oC with 7 days incubation. The 0.1 mol/L (∼5 g/L) NaCl concentration was chosen as it closely corresponds to the average salt concentration found in the Lastensuo bog from where the bacterial strains used in this study were isolated from (see section “Description of the Sampling Site”). In addition, intracellular Ni accumulation was assumed low under these scarce nutrient conditions and uptake was therefore assumed to occur principally through biosorption on bacterial cell surfaces. The pH of the samples was adjusted between 1.2 and 11.7 using 1 mol/L NaOH or HCl. This pH range corresponds to the average pH (pH 4–9) found in the natural ground and surface waters in Finland (Niemi and Raateland, 2007; Posiva, 2012). Higher pH values were not used, as at higher pH, Ni starts to precipitate as hydroxides (Nieminen et al., 2007). After 7 days incubation, the biomass was separated by centrifugation at 5000 × g for 15 min at room temperature and the final 63Ni concentration of the broth was determined using a liquid scintillation counter as descried above for batch experiments and Ni bacterial uptake was expressed as Kd (L/kg DW). In addition, equilibrium pH of the samples was measured.
The Effect of Ni(II) on Bacterial Growth
The effect of Ni(II) on bacterial growth was tested using Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I with 10–6, 10–3, and 2 × 10–2 mol/L Ni(II) amendment. The growth of Ni(II) treated cells was compared to the cells grown without Ni(II) amendment. In these experiments, Medium A and a temperature of + 20°C was used. The cells were incubated in the dark on an orbital shaker (120 rpm) for 2 days and sampled regularly (see Figure 4) for absorbance measurement of the growth solution. The wavelength of 600 nm was used in the measurements.
Ni(II) Sorption Isotherms
Sorption isotherms of Pseudomonas sp. V4-5-SB and PS-0-L were determined in 0.1 mol/L NaCl solution buffered with 0.01 mol/L MOPS [3-(N-morpholino)propanesulfonicacid] or TRIS [tris(hydroxylmethyl)aminomethane] at pH 5.7 and 8.7, representing the slightly acidic surface waters and basic ground waters typically found in Finland (Niemi and Raateland, 2007; Posiva, 2012). The 0.1 mol/L (∼5 g/L) NaCl concentration was chosen to represent to the average salt concentration of Lastensuo bog (see section “Description of the Sampling Site”). The initial Ni concentration of the samples was adjusted to between 10–10 and 10–3 mol/L. Bacterial suspensions were prepared for pre-incubation by transferring bacterial colonies with a sterile loop from the growth plates into Medium A, after which the suspensions were incubated for 16 h on an orbital shaker (120 rpm) at 20°C, during which the bacterial cultures reached the plateau phase. After the incubation the absorbance of the bacterial cultures was recorded at 600 nm and 0.5 g DW/L of each bacterium was used in sorption isotherm experiments. The isotherm samples were incubated for 6 days, after which the cells were removed by centrifugation at 5000 × g for 15 min at the room temperature and the supernatant was used for the liquid scintillation measurement of the remaining 63Ni concentration and final pH of the solution. All experiments were done in triplicate. Standard samples were prepared similarly to the actual samples, except without bacterial addition. The standards were then further used to exclude the possible Ni sorption to the buffer molecules from the results. In addition, MOPS and TRIS were chosen, as Ni retention on these molecules has been previously reported non-existent or low (Perrin and Dempsey, 1974).
In order to optimize the design of sorption isotherms and to establish the most appropriate correlation, experimental isotherm data of Pseudomonas sp. V4-5-SB and PS-0-L were fitted to Langmuir and Freundlich isotherm models.
The linear equation for Langmuir adsorption isotherm (Langmuir, 1916) is expressed as:
where q is the equilibrium concentration of metal in biomass (mg/g DW), Ce is the equilibrium concentration of metal in solution (mg/L), b (L/g DW), and qm (mg/g DW) are Langmuir constants for forming single layer. A plot 1/q vs. 1/Ce should give a straight line, if the sorption process conforms to the Langmuir isotherm. Constants b and qm are evaluated from the slope and intercept.
The linear equation for Freundlich adsorption isotherm (Freundlich, 1906) is expressed as:
where q is the equilibrium concentration of metal in biomass (mg/g DW), Ce is the equilibrium concentration of metal in solution (mg/L), Kf is a constant related to adsorption capacity and n is a constant related to adsorption intensity. If the plot log q vs. log Ce yields a straight line it implies that the sorption process conforms to the Freundlich isotherm. Constants Kf and n are evaluated from the slopes and intercepts.
The Ni uptake capacity (q, mg/g DW) was calculated from the Ni concentration of 10–3 mol/L according to the following equation:
where V is the sample volume (L), Ci is the initial metal ion concentration (mg/L), Ce is the equilibrium metal ion concentration (mg/L) and m is the dry weight of the biomass (g).
TEM and EDS Analyses
The morphology of Pseudomonas sp. V4-5-SB and PS-0-L incubated with 9 × 10–4 mol/L (∼53 mg/L) stable Ni(II) in Medium A was examined using transmission electron microscopy (TEM; Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki). For these experiments, the maximum Ni concentration below lethal dose (<DL) was chosen based on the growth experiments (see section “The Effect of Ni(II) on Bacterial Growth”) to estimate the effects of Ni concentrations found in the highly contaminated mine environments. E.g., in the above mentioned Talvivaara mine area unprocessed ores and leaching heaps, extremely high Ni concentrations, around 1000 mg/kg are found (Tuovinen et al., 2018). In addition, samples without Ni amendment were prepared. The cells were incubated at + 20°C for 6 days, which after the cell mass was separated as described above for batch experiments. After centrifugation, 1 ml of 5% glutaraldehyde was added to the bacterial pellet and allowed to stand at room temperature for 2 h. Thereafter, the fixed cells were centrifuged (5000 × g, 15 min) and transferred to 0.01 mol/L phosphate buffer.
Cells were dehydrated through an ethanol series and embedded in Taab hard epon and polymerized at 60°C. Thin sections were cut using a Leica ultracut UCT ultramicrotome (Leica Mikrosysteme GmbH, Austria) and collected on single-slot copper grids. Section thickness of 60 nm was used for the morphological examinations. The sections, unstained and stained with uranyl acetate and lead citrate, were examined using Jeol JEM-1400 TEM under standard operating conditions.
Energy dispersive X-ray spectroscopy (EDS; Department of Chemistry, University of Helsinki) was used for the determination of elemental compositions of bacterial biomass incubated with Ni. EDS samples were cultured as described for the TEM samples above. After incubation, the cells were separated by centrifugation at 5000 × g for 15 min at room temperature followed by washing of the remaining bacterial pellet twice with 0.01 mol/L phosphate buffer. Thereafter, the pre-frozen bacterial pellet (at −18°C) was lyophilized using an Alpha 1-4 LSCbasic lyophilizer (Christ). The lyophilized bacterial mass was fixed to the sample stub/support with conductive carbon tape and both the sample and plug were coated with 10 nm carbon layer using Leica EM ACE200 coating system (Leica Microsystems). Qualitative elemental analysis of samples was performed using Oxford INCA 350 Energy dispersive X-ray microanalysis system at 20 kV under standard operating conditions. The detection limit of EDS is estimated to be about 0.1 wt%.
Fourier Transform Infrared Spectroscopy
Fourier transform infrared (FTIR) spectroscopy [Bruker ALPHA FTIR Spectrometer (Platinum ATR)] was used to study the functional groups present on the cell surfaces of Pseudomonas sp. strains (V4-5-SB, PS-0-L, and T5-6-I). In these experiments, lyophilized biomass was used and the samples were cultured and prepared as described above for EDS. The IR spectra were recorded from Ni(II) treated samples, as well as from samples without Ni(II) amendment.
Protein Expression in the Presence of Ni(II)
The stress responses generated by Ni(II) addition were examined to identify potentially important proteins involved in Ni(II) metabolism. SDS-PAGE and MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization – Time of flight) – mass spectrometry were used to study the proteins expressed in Pseudomonas sp. strains (V4-5-SB, PS-0-L, and T5-6-I) in the presence of Ni (9 × 10–4 mol/L) and compared to the cells grown without Ni(II) amendment. Samples were prepared similarly to the TEM samples described above, except of a shorter incubation time of 4 days. The soluble protein fractions from control untreated and Ni(II) treated cells were isolated using the BugBuster method according to the manufacturer’s instructions.
The isolated proteins were separated with SDS-PAGE using pre-cast ClearPage SDS 10% gels with constant voltage of 175 V for 45 min. SDS-PAGE samples were prepared following the manufacturer’s instructions for ClearPage gels, using 3 μL of 10 × DTT per 13 μL of sample volume. Equal amounts of protein (ca. 3 μg) were loaded onto each well. TEO-Tricine (ClearPAGE SDS Running Buffer) was used as running buffer. The proteins were visualized using the Blue BANDIT protein stain and the visible protein lines of gel were compared to the protein standard (The ClearPAGE Two-Color SDS Marker, CBS Scientific).
Additionally, MALDI-TOF was used for the mass determination of proteins isolated from Pseudomonas sp. PS-0-L by using the BugBuster method. For MALDI-TOFF the samples were sent to the Proteomics Unit, Viikki, University of Helsinki, where the protein solutions were purified and concentrated using the ZipTip –method and MALDI-TOF was run in sinapic acid/acetonitrile buffer.
Statistical Analyses
To study the statistical difference between the different growth conditions, i.e., the difference between nutrient broths A and B and the temperatures + 4 and +20°C the analysis of variance was performed using Microsoft Excel Data Analysis and one-way ANOVA at the p < 0.05 level. Analysis of variance was done for all studied bacteria separately for temperature and nutrient broth. In addition, the ANOVA analysis for the statistical differences in the uptake between different bacterial groups (Pseudomonas, Paenibacillus, Paraburkholderia/Methylobacterium) was done. Kurtosis and skewness of the data was tested to examine the normality of the data. As both skewness and kurtosis were found to differ from zero, log transformed data was used for the one-way ANOVA tests of the data.
Results
Characterization of the Microbial Isolates
Based on the 16S rRNA gene sequences, the isolated bacteria affiliated with genera Paenibacillus (KV-0-YR, IV-0-L, and VV-0-L), Methylobacterium (P4-5-LR), Paraburkholderia (RP-0-BL), and Pseudomonas (V4-5-SB) (Figure 1). The utilization of substrates by these strains was tested using the RapID ONE and RapID NF Plus systems (Remel) (Table 1). Five of the newly isolated bacteria were stained Gram negative (Paenibacillus sp. IV-0-L and VV-0-L; Methylobacterium sp. P4-5-LR; Paraburkholderia sp. RP-0-BL; and Pseudomonas sp. V4-5-SB) and one Gram positive (Paenibacillus sp. KV-0-YR) (Table 1). Two of the three Paenibacillus (IV-0-L and VV-0-L) isolates were found Gram negative, although Paenibacillus strains are usually stained Gram positive. However, also previously (Grady et al., 2016) Paenibacillus have been shown to have mixed Gram staining, and they may also stain Gram negative.
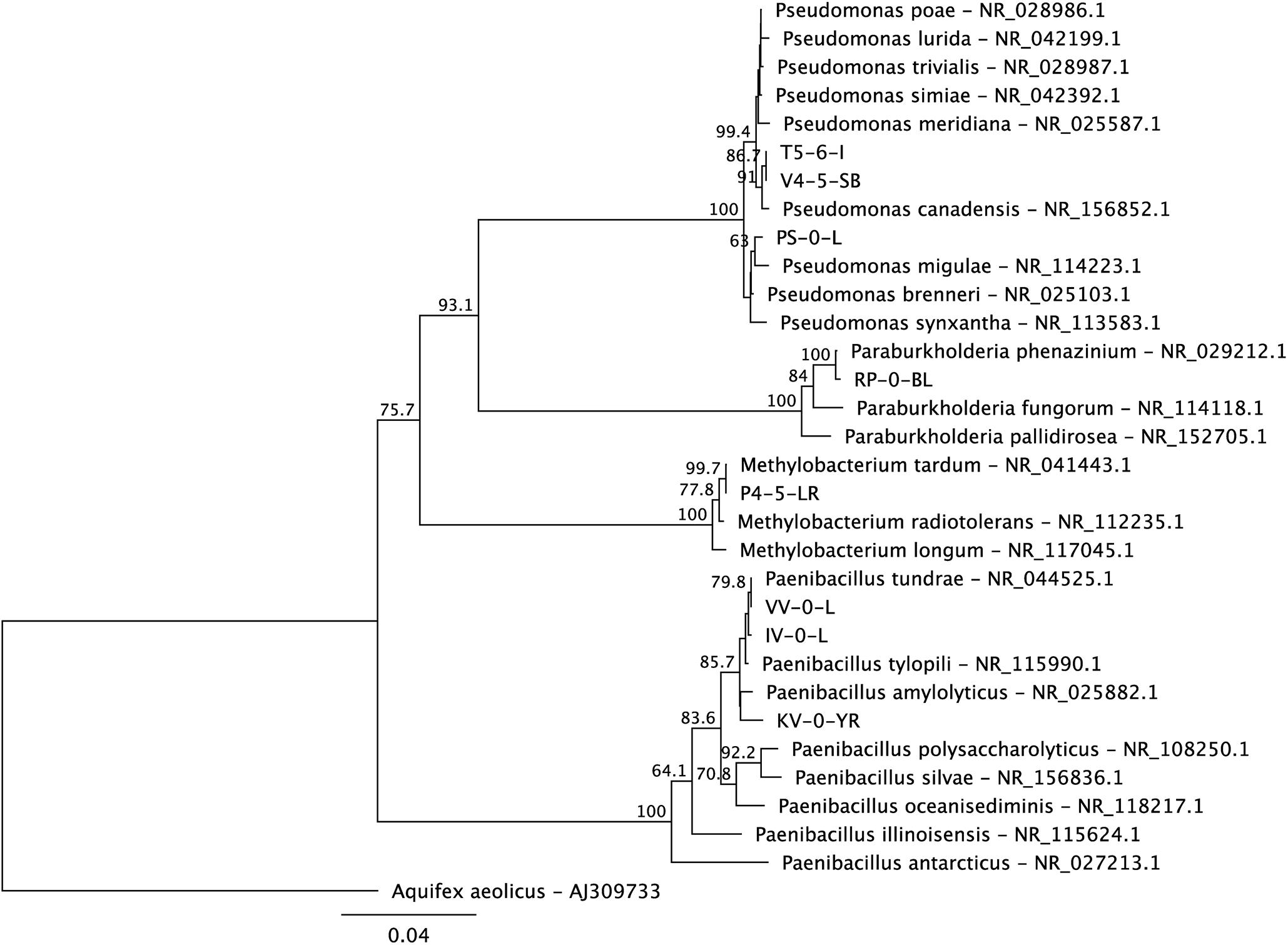
Figure 1. A maximum likelihood phylogenetic tree of the 16S rRNA genes of the new Lastensuo isolates in respect to the closest relative reference strains. The topology of the tree was tested by bootstrap analyses of 1000 random resamplings and nodes with > 50% bootstrap support are indicated. The scale bar represents number of nucleotide substitutions per site.
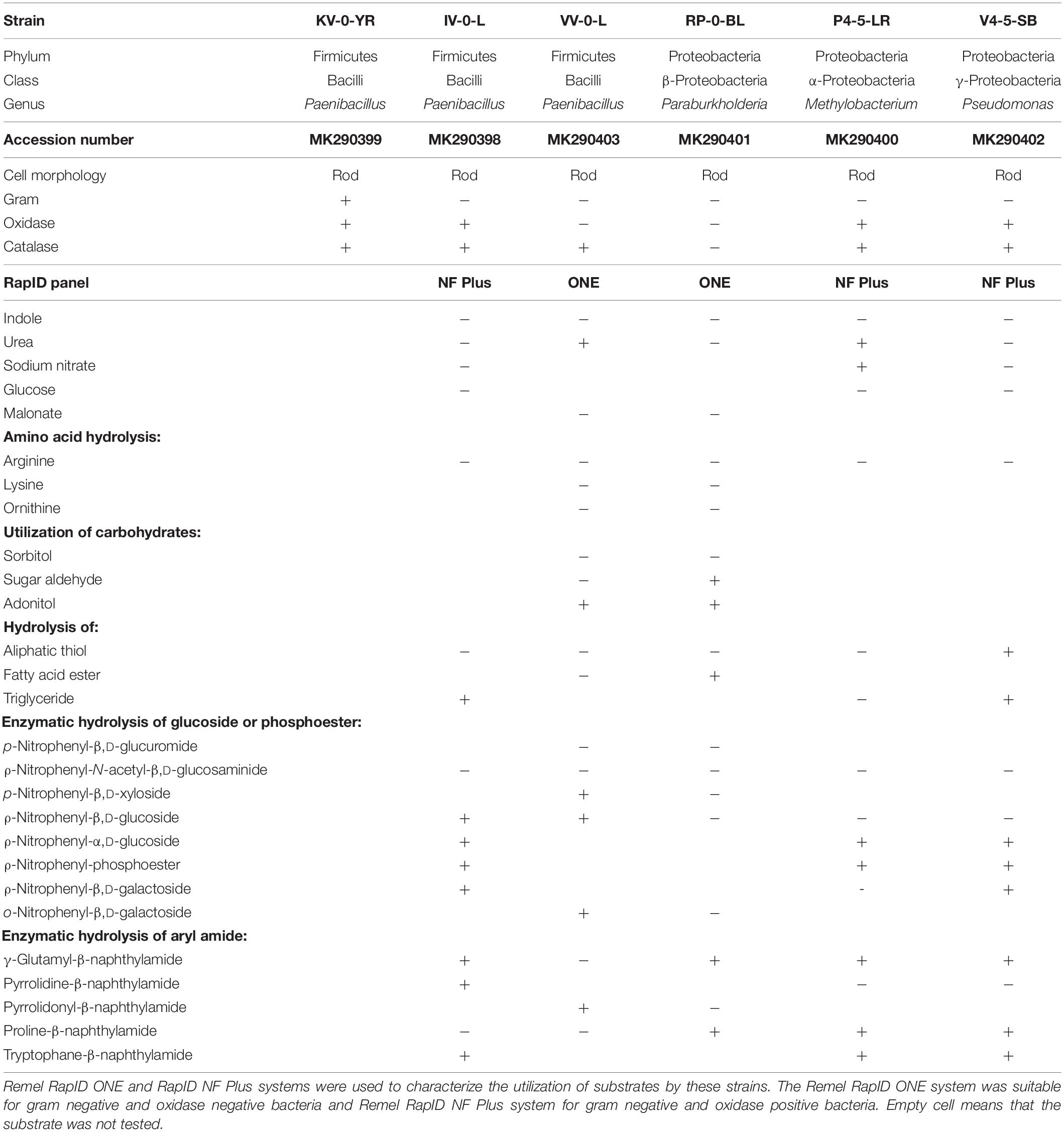
Table 1. Biochemical characteristic of Paenibacillus sp. KV-0-YR, Paenibacillus sp. IV-0-L, Paenibacillus sp. VV-0-L, Paraburkholderia sp. RP-0-BL, Methylobacteria sp. P4-5-LR, and Pseudomonas sp. V4-5-SB.
Of the Paenibacillus sp. strains, VV-0-L was oxidase negative and strains KV-0-YR and IV-0-L were oxidase positive. Due to the Gram negative staining of Paenibacillus sp. strains IV-0-L and VV-0-L, these were further characterized with RapID panels designed for Gram negative bacteria. The substrate utilization patterns of these bacteria were similar, with the exception that Paenibacillus sp. IV-0-L could hydrolyze γ-glutamyl-β-naphthylamide and Paenibacillus sp. VV-0-L could not. In turn, Paenibacillus VV-0-L was able to utilize nitrate, while Paenibacillus sp. IV-0-L was not.
Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I had partially similar substrate utilization patterns. However, only Pseudomonas sp. V4-5-SB showed positive oxidase activity, while Pseudomonas sp. PS-0-L and T5-6-I (Lusa et al., 2016a) were oxidase negative. None of the tested Pseudomonas sp. strains (V4-5-SB, PS-0-L, and T5-6-I) were able to utilize glucose. In addition, Pseudomonas sp. V4-5-SB was unable to utilize nitrate while Pseudomonas sp. PS-0-L and T5-6-I used nitrate (Lusa et al., 2016a). All Pseudomonas sp. strains hydrolyzed triglyceride, but the hydrolysis of aliphatic thiol was variable. Overall, Pseudomonas sp. PS-0-L appears to be more similar to Pseudomonas sp. T5-6-I, and Pseudomonas sp. V4-5-SB differed from these two strains in the utilization of substrates. The results of the 16S rRNA gene sequences, Gram staining, catalase and oxidase activity tests and RapID systems for Pseudomonas sp. PS-0-L and T5-6-I have been previously reported by Lusa et al. (2016a).
Uptake of Ni(II) by Paenibacillus sp., Methylobacterium sp., Paraburkholderia sp., and Pseudomonas sp. Strains
The effect of temperature, incubation time, nutrient conditions, and pH on Ni(II) removal by the isolated bacteria was studied using batch experiments. Since there is only limited knowledge about the metabolism of the bacteria inhabiting the northern bogs and because the ability of these bacteria to interact with Ni is not known, batch experiments were conducted in an enhanced and controlled environment. All studied bacterial strains removed Ni(II) from the solution and the uptake efficiency was affected by nutrient source, incubation time and temperature (Figure 2). The highest Ni uptake was shown by Pseudomonas sp. PS-0-L in Medium A at + 20°C after 7 days incubation. Compared to Pseudomonas sp. V4-5-SB (max Kd 640 L/kg DW) and T5-6-I (max Kd 420 L/kg DW), Ni uptake was three- to five-fold higher in Pseudomonas sp. PS-0-L. In PS-0-L, a maximum Kd value of 1890 L/kg DW was recorded and this strain showed higher Ni removal compared to the other two Pseudomonas sp. strains irrespective of the used growth media, temperature or incubation time. In all Pseudomonas sp. strains, the average Ni uptake was observed to increase with increasing incubation temperature from + 4 to + 20°C. In addition, longer incubation time (14 days, compared to 7 days) decreased Ni(II) removal in Medium A for all studied strains. Only Pseudomonas sp. T5-6-I removed Ni(II) more efficiently after 14 days incubation, compared to the 7 days incubation at + 20°C. Contrary to Medium A, in Medium B incubation time and temperature had no clear effect on Ni(II) uptake by any of the studied Pseudomonas sp. strains. In Medium B, the highest Ni(II) uptake was shown by Pseudomonas sp. PS-0-L (max Kd 529 L/kg DW) and T5-6-I (max Kd 92 L/kg DW) at + 20°C using 14 days incubation time. For Pseudomonas sp. V4-5-SB the maximum Kd value of 248 L/kg DW was observed after 7 days incubation.
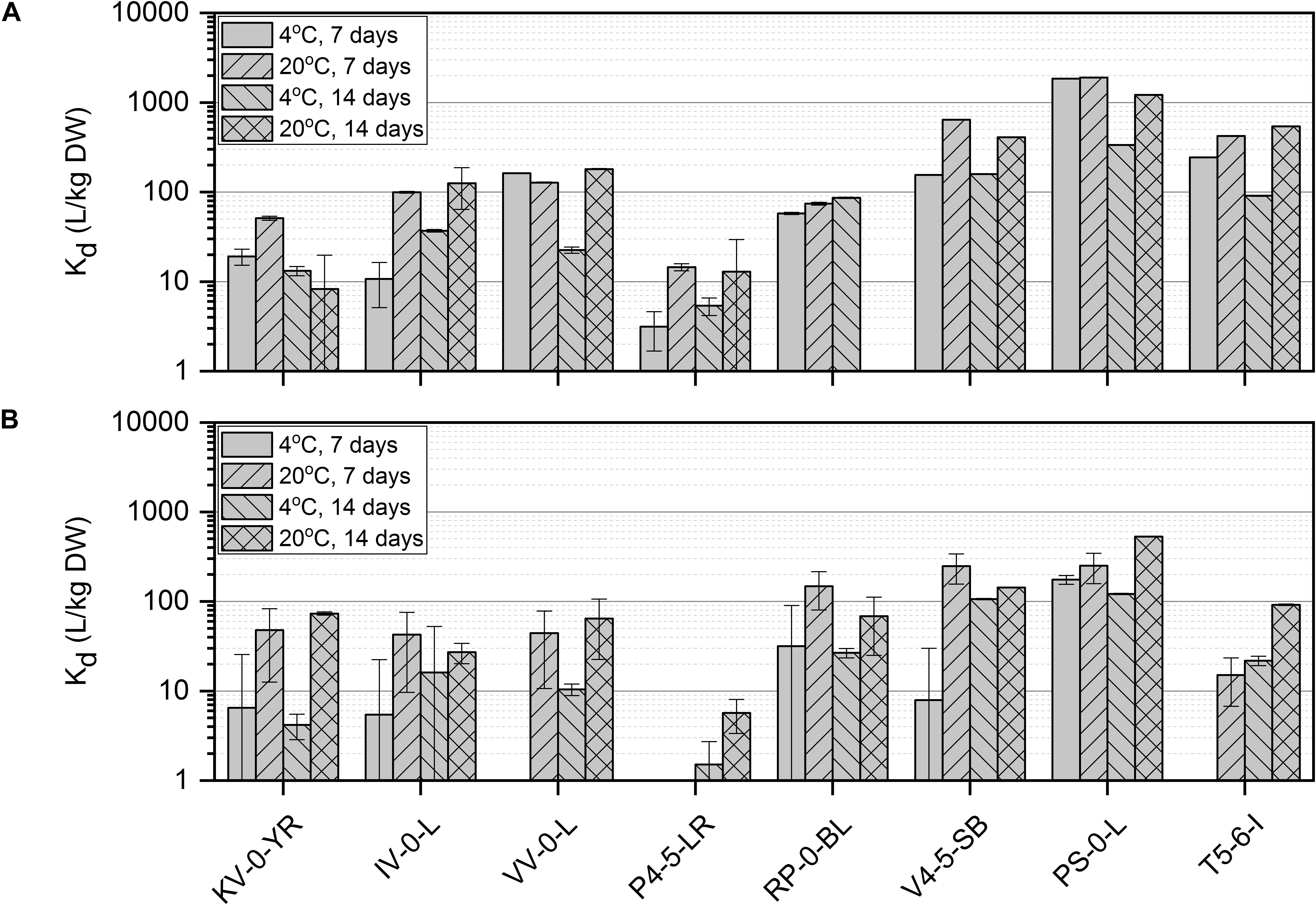
Figure 2. Ni(II) uptake (initial Ni(II) concentration 6 × 10–10 mol/L) by isolated Paenibacillus sp. KV-0-YR, IV-0-L, and VV-0-L; Methylobacterium sp. P4-5-LR; Paraburkholderia sp. RP-0-BL; and Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I in Media A (A) and B (B) at +4 and +20°C with 7 and 14 days incubation time. Values represent the geometric means of triplicate trials and uncertainty bars are the standard error of the geometric mean.
The highest uptake by the studied Paenibacillus sp. strains (IV-0-L and VV-0-L) was observed in Medium A, with maximum Kd values of 125 L/kg DW (+ 20°C, 14 days) and 180 L/kg DW (+ 20°C, 14 days) for IV-0-L and VV-0-L, respectively. For Paenibacillus sp. KV-0-YR, the highest uptake (max Kd 73 L/kg DW at + 20°C, 14 days) was observed in Medium B.
For Paraburkholderia sp. RP-0-BL the maximum Kd of 148 L/kg DW was observed at + 20°C after 7 days incubation in Medium B. For Methylobacterium sp. P4-5-LR only low Ni(II) uptake was observed, with a maximum uptake of 15 L/kg DW at + 20°C after 7 days incubation in Medium A. Compared to the other studied bacterial strains, Methylobacterium sp. P4-5-LR showed significantly lower Ni(II) retention regardless of the growth medium, temperature, or incubation time used than any of the other strains. In Medium B, Ni(II) uptake by Methylobacterium sp. P4-5-LR was non-existent, with recorded Kd values between 2 and 6 L/kg DW.
The effect of pH on Ni(II) uptake by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I was in addition investigated at the pH-range of 1.2–11.7. In all studied Pseudomonas sp. strains a strong effect of pH on Ni(II) adsorption was observed (Figure 3) and Ni uptake increased with the increasing pH. The maximum Kd values were 502, 807, and 461 L/kg DW for V4-5-SB, PS-0-L, and T5-6-I, respectively. The highest Ni uptake was observed at pH 6–9 depending on strain. As mentioned above, this pH range well corresponds to the average pH values of the natural ground and surface waters found in Finland (Niemi and Raateland, 2007; Posiva, 2012).
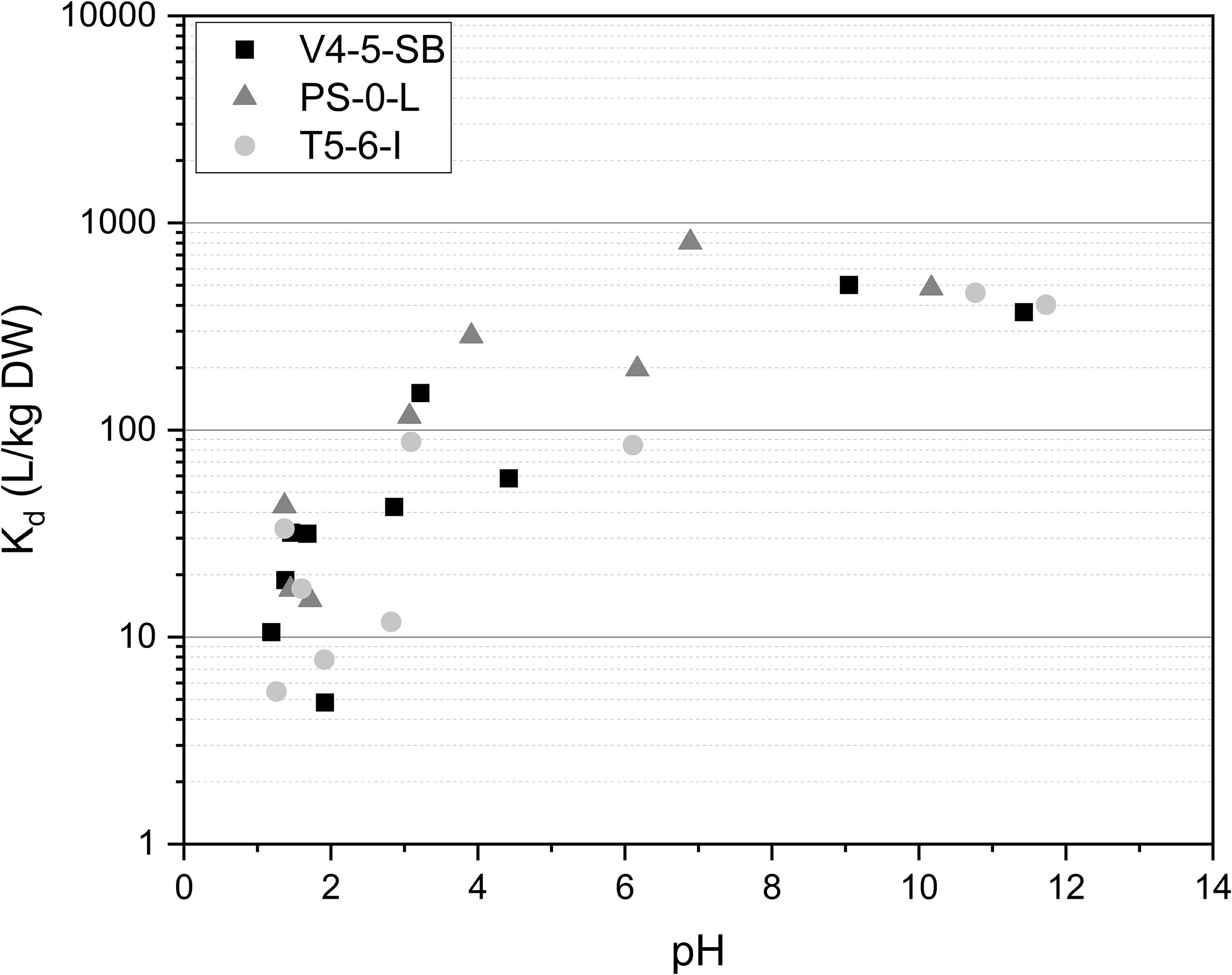
Figure 3. The effect of pH on Ni uptake by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I in 0.1 mol/L NaCl at pH-range 1.2–11.7. Based on pH-Eh diagram (Supplementary Figure 1) Ni is found as hydroxide above pH 10.
Bacterial Growth Under Ni(II) Stress
The effect of 10–6, 10–3, and 2 × 10–2 mol/L concentration of Ni(II) on bacterial growth was tested with Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I using a 2-day growth period and an incubation temperature of + 20°C. In these experiments, no significant differences between the growth of cells with 10–6 and 10–3 mol/L Ni(II) and cells without Ni(II) were observed (Figure 4). In both treatments, the exponential increase in cell growth was observed after a lag phase between 60 and 720 min (1–12 h). However, no bacterial growth was observed at the highest Ni(II) concentration (2 × 10–2 mol/L) with tested Pseudomonas sp. V4-5-SB, PS-0-L and T5-6-I, indicating a potential toxicity for the bacteria.
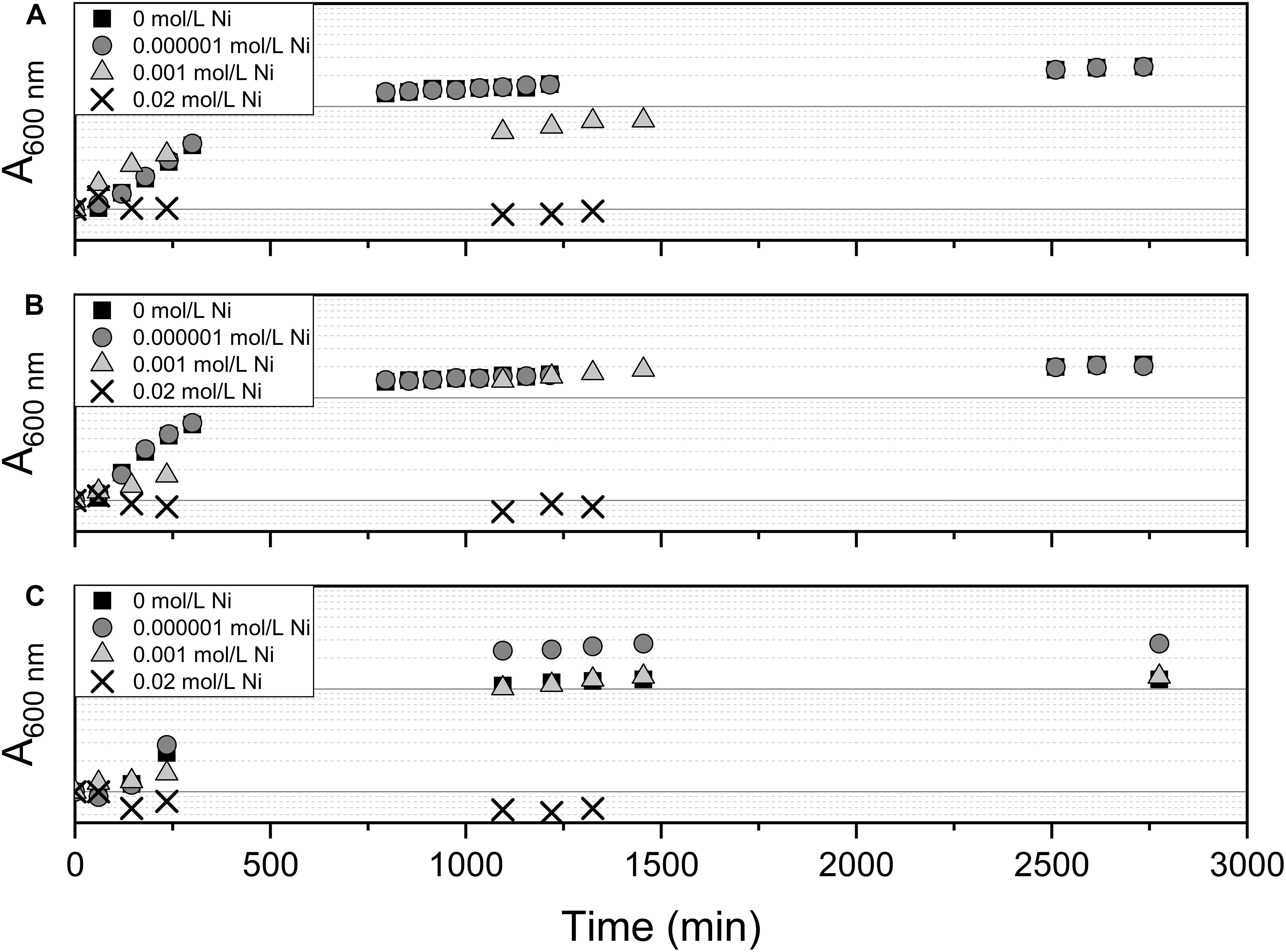
Figure 4. The effect of Ni on bacterial growth by Pseudomonas sp. V4-5-SB (A), PS-0-L (B), and T5-6-I (C) during the 2-day growth period in Medium A. Tested Ni concentrations were 0, 0.000001, 0.001, and 0.02 mol/L.
Morphology and Elemental Composition of the Cells
The morphology of Pseudomonas sp. V4-5-SB and PS-0-L Ni(II) treated cells was studied using TEM. This was done for more detailed information on Ni localization either on the bacterial cell walls or inside the cells. In the TEM images dense crystalline formations were observed inside the bacterial cells after incubation with 9 × 10–4 mol/L Ni(II) solution (Figures 5C,D) in both studied bacteria. In the cells grown without Ni(II) amendment, corresponding crystalline formations were not observed (Figures 5A,B). In the corresponding EDS spectra (Figures 5E,F), the Kα and Lα X-ray peaks of Ni are seen at 7.5 and −0.85 keV, respectively. However, the intensities of Ni X-ray peaks remained relatively low in the measured spectra. In addition to Ni, other elements detected by EDS were carbon, oxygen, phosphorus, potassium, sulfur, chlorine, sodium, magnesium, and calcium. Two major elements, potassium and phosphorus, are probably from the pre-treatment step of the samples and from the phosphate buffer used. Other elements are naturally present on the bacterial surfaces.
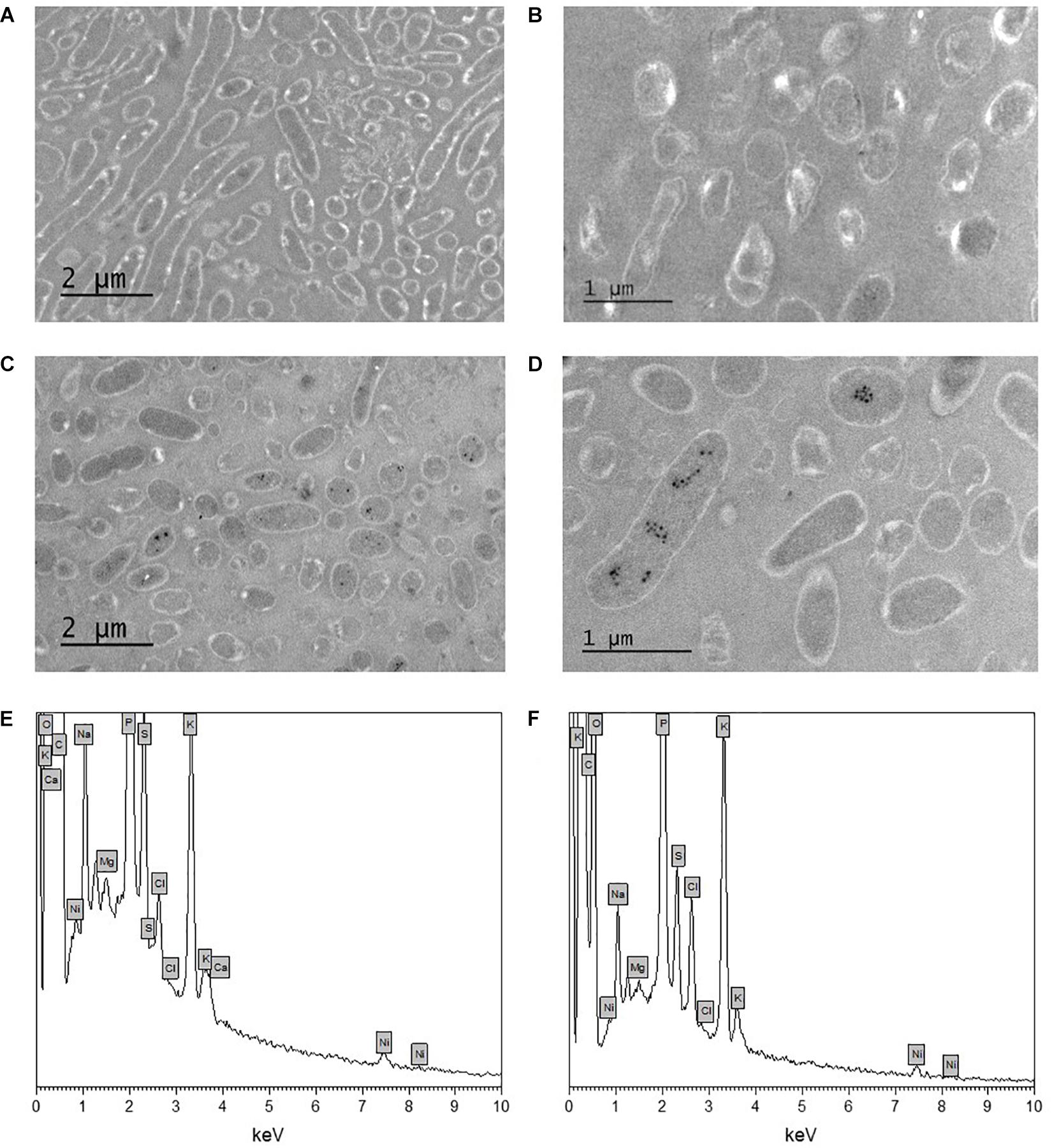
Figure 5. Transmission electron microscopy (TEM) images of unstained Pseudomonas sp. V4-5-SB (A,C) and PS-0-L (B,D) cells incubated without nickel (A,B) and with nickel (9 × 10–4 mol/L) (C,D), and EDS spectra of Pseudomonas sp. V4-5-SB (E) and PS-0-L (F) representing the presence of nickel in bacterial biomass.
The Effect of Ni(II) on Protein Expression
The stress responses of Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I to the presence of Ni(II) were investigated using SDS-PAGE (V4-5-SB, PS-0-L, and T5-6-I) and MALDI-TOFF (PS-0-L). In the SDS-PAGE experiments, no major differences were observed in the expressed protein profiles by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I induced by Ni(II) amendment, compared to the samples incubated without Ni(II) (Figure 6A). However, the intensities (corresponding to the concentrations) of the expressed proteins were clearly lower at the approximate regions of ∼60–71 and ∼40–45 kDa in Pseudomonas sp. V4-5-SB induced by the presence of Ni(II). A slight decrease in the range of ∼32 kDa in the line intensities was also observed in Pseudomonas sp. T5-6-I incubated with Ni(II), compared to the cells without Ni(II) (Figure 6A). As the resolution of the SDS-PAGE is relatively low, when using crude extracts with several proteins present and especially on the low kDa proteins, MALDI-TOFF was used for better resolution of proteins with low molecular mass. In the MALDI-TOFF spectra (Figures 6B,C), several additional protein peaks were observed at the approximate region of 13–40 kDa in PS-0-L incubated with Ni(II) (Figure 6C), compared to the sample without Ni(II) (Figure 6B).
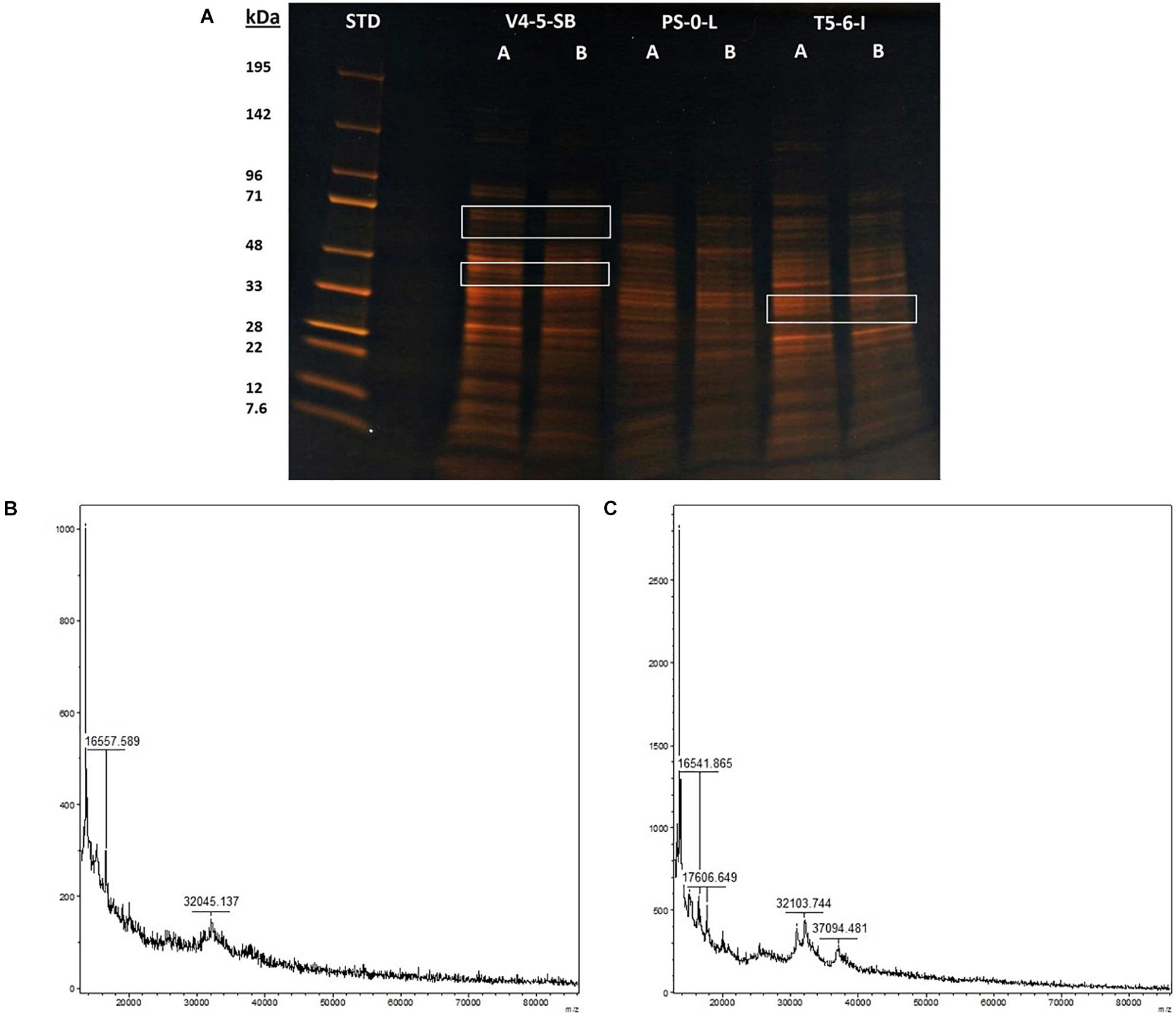
Figure 6. Protein profiles resolved on a 10% acryl amide SDS-PAGE gel, lanes marked with (A) and (B) indicate bacterial cells incubated without Ni (A) and with Ni (B). Equal amounts of protein (ca. 3 μg) were loaded onto each well. MALDI-TOF spectra of PS-0-L (B) control and (C) Ni treated cells.
Ni(II) Sorption Isotherms
Due to the high Ni(II) uptake observed in the batch Kd experiments, Pseudomonas sp. V4-5-SB and PS-0-L were used in the following sorption isotherm experiments. Both Langmuir and Freundlich isotherm models were tested and the Ni uptake by the tested bacterial strains were found to follow both equations (Figure 7). The Langmuir model gave a good fit for the Ni(II) uptake for both strains at pH 5.7 and 8.7 with the R2 values of 0.997 and 0.986, and 0.987 and 0.989, for V4-5-SB and PS-0-L at pH 5.7 and 8.7, respectively. At pH 5.7 and 8.7, the qm values of Pseudomonas sp. V4-5-SB were 0.00494 and 0.00496 mg/g DW, and the b values were 468 and 242 L/g DW (Table 2), respectively. For Pseudomonas sp. PS-0-L, the qm values were 0.0265 and 0.00756 mg/g DW, and the b values were 46.3 and 73.6 L/g DW at pH 5.7 and 8.7, respectively.
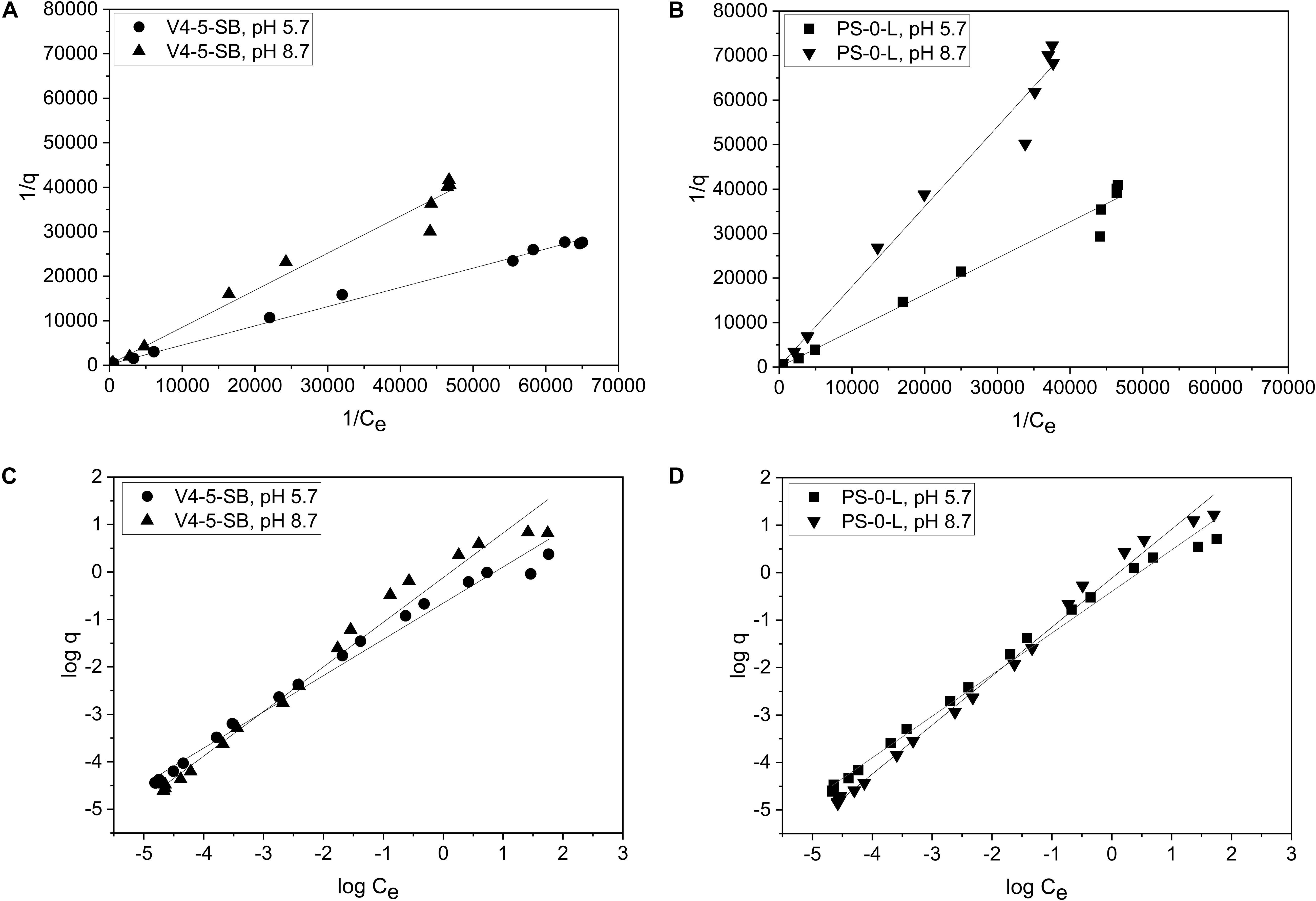
Figure 7. The biosorption isotherms of Pseudomonas sp. V4-5-SB (A,C) and PS-0-L (B,D) at pH 5.7 and 8.7. Values represent the geometric means of triplicate trials and the uncertainty bars are the standard error of the geometric mean. Graphs (A,B) present the plot 1/q vs. 1/Ce and a straight line implies that the sorption process conforms to the Langmuir isotherm. Graphs (C,D) present the plot log q vs. log Ce and a straight line implies that the sorption process conforms to the Freundlich isotherm.
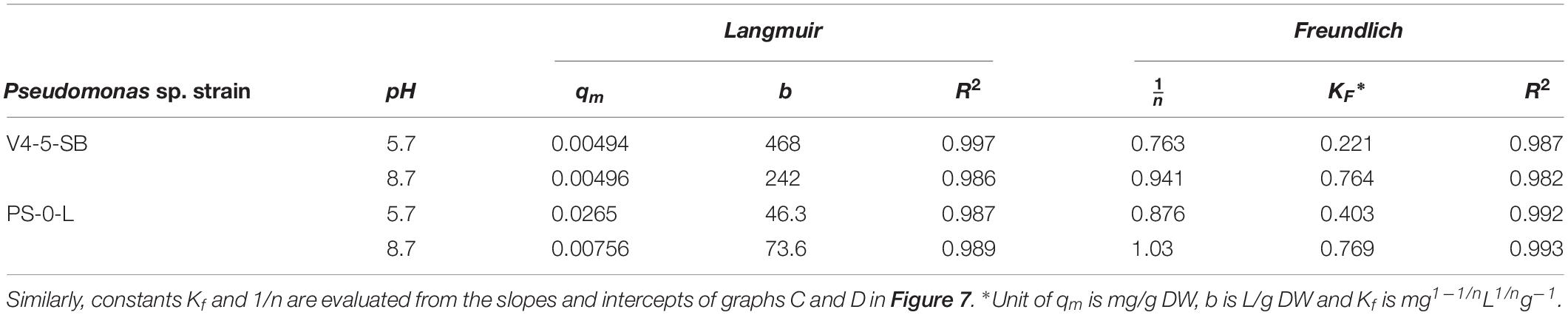
Table 2. Constants qm and b evaluated from the intercepts and slopes of graphs A and B in Figure 7.
The Freundlich isotherm model showed also a good fit for the Ni(II) uptake by Pseudomonas sp. V4-5-SB and PS-0-L at pH 5.7 and 8.7 with the R2 values of 0.987 and 0.982; and 0.992 and 0.993, for V4-5-SB (at pH 5.7 and 8.7) and PS-0-L, respectively. At pH 5.7, the Freundlich adsorption constant (Kf value) of Pseudomonas sp. V4-5-SB was 0.221 mg L1/n g–1 and the 1/n value 0.763 (Table 2). Correspondingly, at pH 8.7, the Kf value was 0.764 mg L1/n g–1 and the 1/n value was 0.941. For Pseudomonas sp. PS-0-L, Kf values were 0.403 mg L1/n g–1 and 0.769 mg L1/n g–1; and the 1/n values were 0.876 and 1.03 at pH 5.7 and 8.7, respectively.
At low initial Ni concentrations, Freundlich isotherms behaved ideally but after 10–4 mol/L a slight deflection in the Freundlich isotherms at both pH 5.7 and 8.7 was observed (Figures 7A,B). In addition, the equilibrium uptake capacity (q) of Pseudomonas sp. V4-5-SB and PS-0-L increased with increasing solution pH and initial Ni concentration. The highest q values were observed as the initial Ni concentration of 10–3 mol/L was used. The corresponding q values for Pseudomonas sp. V4-5-SB and PS-0-L were 2.4 and 5.2 mg/g DW at pH 5.7, and 6.6 and 16.8 mg/g DW at pH 8.7, respectively.
FTIR Analysis
As Ni(II) uptake conformed both Freundlich and Langmuir isotherms, FTIR analysis was performed to obtain more information on the functional groups present on the bacterial cell surfaces and possibly participating Ni(II) uptake in the Pseudomonas sp. strains V4-5-SB, PS-0-L, and T5-6-I. In these experiments, no significant differences were observed in the functional groups of different Pseudomonas sp. strains, and alcoholic and carboxylic hydroxyl, amine, alkane and phosphate groups were observed in all bacteria (Figure 8 and Table 3). In addition, nickel was not found to affect the presence of functional groups on the cell surface. All bacteria showed absorption peaks around 3300 cm–1, which indicate O-H and N-H stretching and shows the presence of alcoholic hydroxyl groups and amine groups in these bacteria. In addition, the bands at 1500 and 1200 cm–1 can be assigned to the N-H stretch and C-N stretch caused by amine groups. In addition, in all cells, a band at about 1000–1100 cm–1 was observed, representing a P = O stretch caused by a phosphate group. The overall spectral analysis supports the potential role of alcoholic hydroxyl, carboxylic carbonyl, and amine groups in Ni(II) binding in the Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I strains.
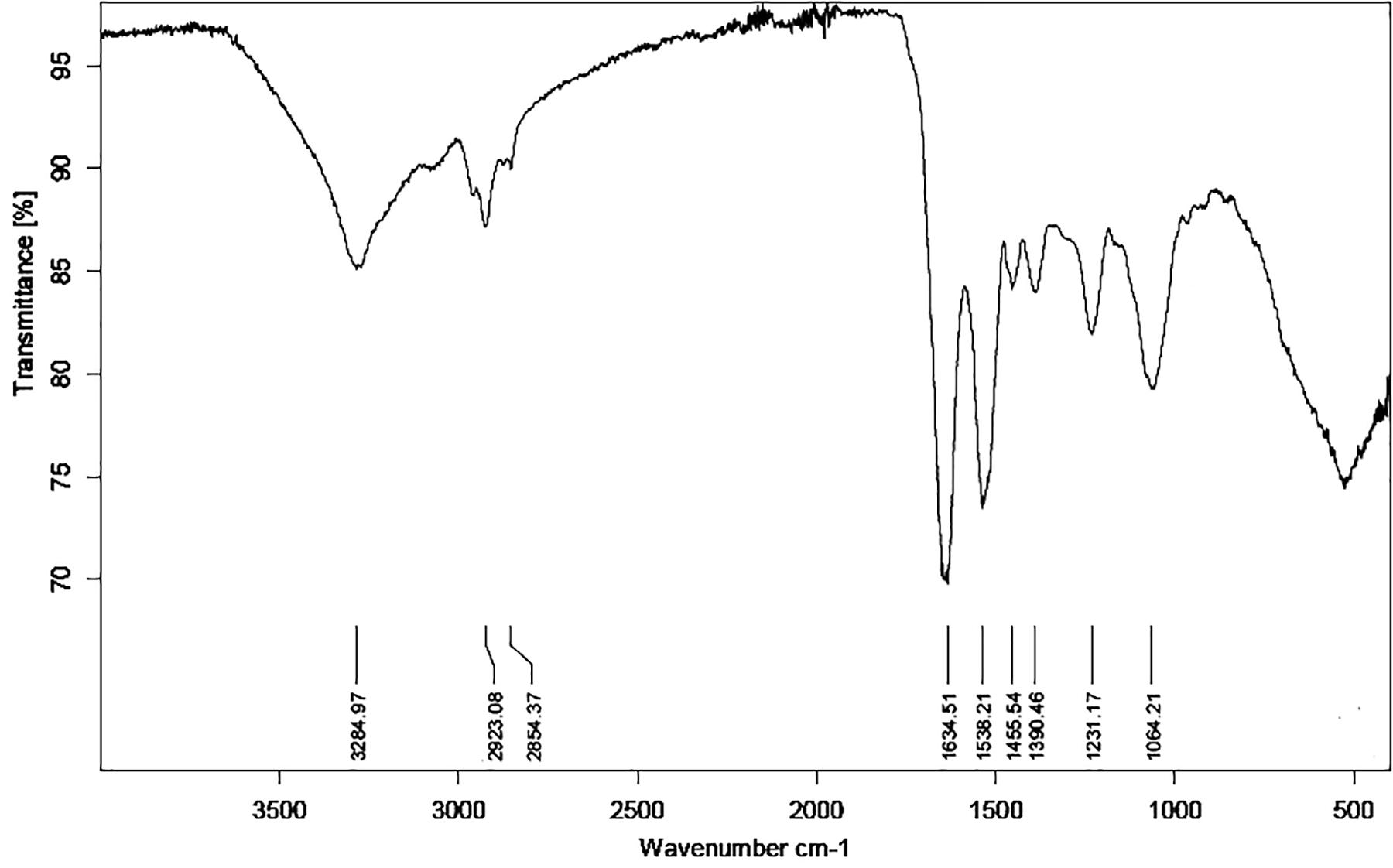
Figure 8. Fourier transformation infrared spectroscopy (FTIR) spectrum of Pseudomonas sp. PS-0-L. The spectra of Pseudomonas sp. V4-5-SB and T5-6-I were similar to that of PS-0-L and the presence of nickel did not change the situation (spectra are not shown).
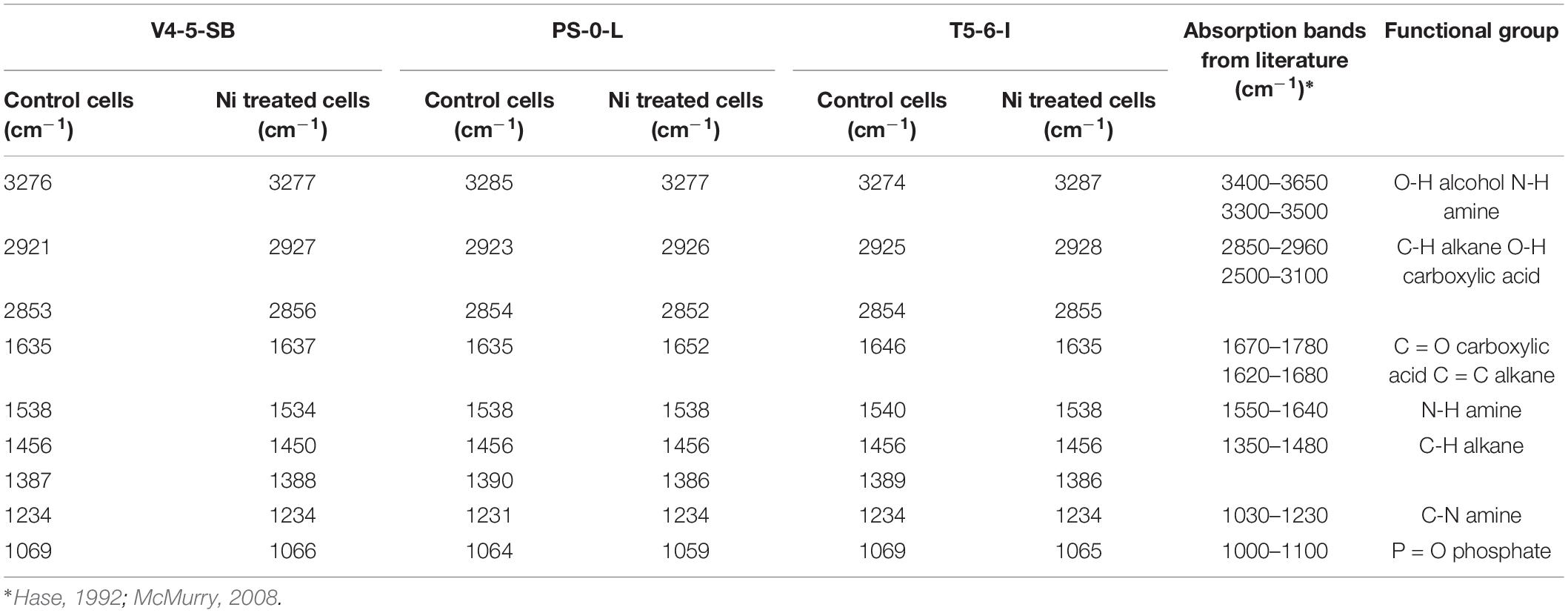
Table 3. Major absorption peaks observed in the FTIR spectra of Pseudomonas sp. V4-5-SB, PS-0-L and T5-6-I (incubated without and with Ni addition) and possible functional groups involved in Ni binding on cell surface.
Statistical Analyses
Based on the analysis of variance (ANOVA), the different growth conditions (i.e., nutrient broth and temperature) affected the Ni uptake. The differences in the Kd values in Media A and B, and at temperatures + 4 and + 20°C were found statistically significant at the p < 0.05 level (Table 4). In addition, the difference in the Ni uptake between different bacterial groups [i.e., Pseudomonas, Paenibacillus, Paraburkholderia/Methylobacterium (other bacteria)] were statistically significant at 0.05 level (Table 4).
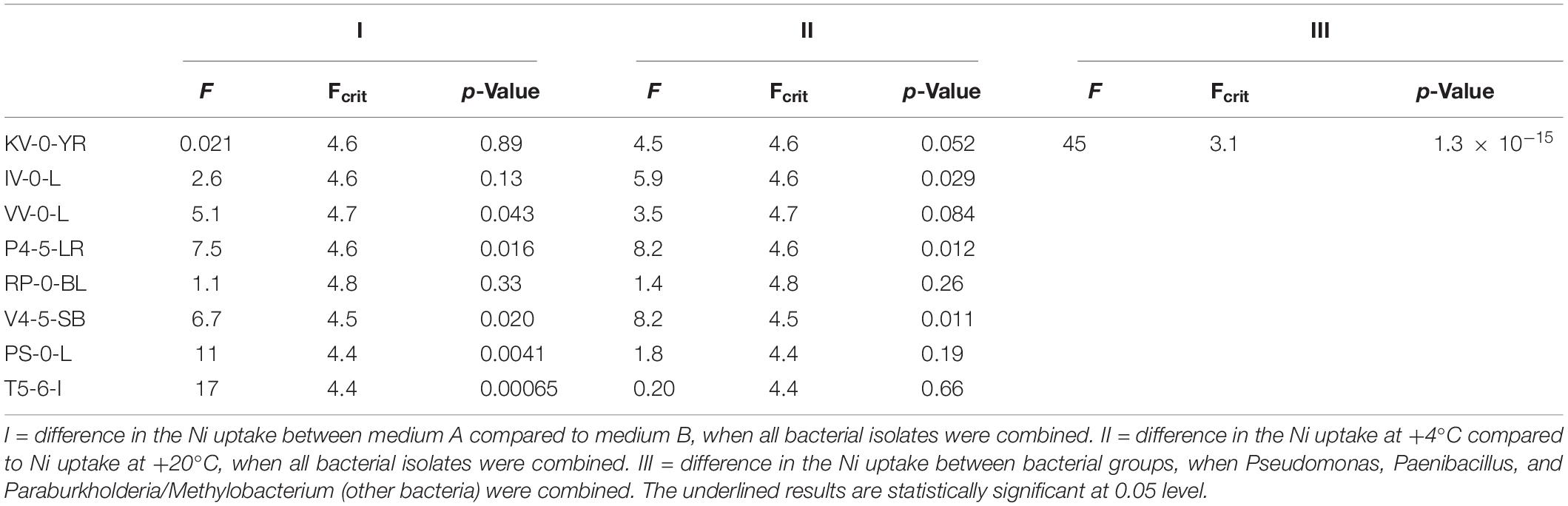
Table 4. Statistically significant differences in the Ni uptake between the different bacterial strains analyzed by ANOVA.
Discussion
Ni mining and other industrial, as well as domestic use can cause increased, potentially toxic emissions of Ni to the environment. In addition, nuclear energy production results in long-lived, possibly highly mobile radioactive isotopes of Ni, 59Ni and 63Ni, potentially harmful for the biosphere. Significant environmental Ni deposits are found, e.g., in North America and Russia (Mudd, 2010), in the boreal and arctic regions with significant portions of the world’s mires, bogs, and other peatlands (Joosten and Clarke, 2002). In the boreal region, ombrotrophic bogs represent ecological biotopes, with distinct microbial populations, but so far, only limited knowledge is available about the metabolism of the microorganisms inhabiting these areas. Many environmental bacteria can bind Ni and other heavy metals and two types of uptake processes, biosorption and bioaccumulation, have been presented (e.g., Wang and Chen, 2009). In biosorption, the functional groups of the bacterial cell wall, such as hydroxyl, carboxyl and phosphate groups, participate in metal binding either by adsorption, ion-exchange, precipitation, or complexation (White et al., 1995; Ledin and Pedersen, 1996; López et al., 2000; Sar et al., 2001). In contrast to biosorption, in bioaccumulation, metal is transported through the bacterial cell wall and membrane and the transport is typically linked to the cell metabolism and is consequently a substantially slower process than direct biosorption on to cell walls. In addition to the characteristics of a specific bacterial strain, the efficiency of metal uptake is dependent on various external factors, such as pH, competing ions, organic material, cell metabolism and temperature, because the adsorption reaction can change as a function of changing conditions (Tsezos et al., 1996; Veglió and Beolchini, 1997; Veglió et al., 1997; López et al., 2000; Jiang et al., 2004).
In the present study, new bacterial strains were isolated from the acidic, nutrient-poor, boreal Lastensuo bog. The isolated strains belonged to the genera Paenibacillus (KV-0-YR, IV-0-L, and VV-0-L), Methylobacterium (P4-5-LR), Paraburkholderia (RP-0-BL), and Pseudomonas (V4-5-SB). Previously, we characterized the bacterial community of this bog by 454 pyrosequencing and found 40 different bacterial phyla in the peat profiles from the surface to the 6 m depth (Tsitko et al., 2014). Acidobacteria and α- and γ-Proteobacteria dominated the surface community. In addition, Chloroflexi, Bacteroidetes, Verrucomicrobia, Planctomycetes, and Spirochaeta formed the majority of the bacterial phyla present in this bog. These bacteria were, however, not captured in pure cultures in our study.
In the current work, the uptake of Ni by Paenibacillus (KV-0-YR, IV-0-L, and VV-0-L), Methylobacterium (P4-5-LR), Paraburkholderia (RP-0-BL), and Pseudomonas (V4-5-SB) was studied using batch experiments under favorable growth conditions for the isolated bacteria. Ni uptake varied substantially between different bacterial genera and even between different bacterial strains of the same genera. In Medium A, Ni uptake was observed to increase in the order Methylobacterium < Paenibacillus ≤ Paraburkholderia < Pseudomonas, with the average Kd values of 8.4676, 80, 81, and 676 L/kg DW in this medium, respectively. Pseudomonas had the highest average Ni uptake (average Kd 222 L/kg DW) also in Medium B. Paraburkholderia and Paenibacillus showed intermediate uptake of Ni(II) in Medium B, with Paraburkholderia showing generally higher Ni retention, with an average Kd of 64 L/kg DW, than Paenibacillus (average Kd 35 L/kg DW) in this growth medium. In Medium B, Ni removal by Methylobacterium was only marginal (average Kd 3.2 L/kg DW). The bacteria used in this study, had substantial variation in their substrate utilization patterns and metabolism, which presumably also affects their ability to remove Ni(II) under variable nutritional conditions. However, the ability of a strain to use a specific substrate, was not the most significant Ni(II) uptake controlling factor in the tested bacterial strains. Pseudomonas sp. PS-0-L and T5-6-I had very similar substrate utilization patterns, with only minor exceptions. Pseudomonas sp. V4-5-SB differed from these two strains in its ability to utilize different substrates, e.g., Pseudomonas sp. V4-5-SB had positive oxidase activity and was unable to utilize nitrate, while Pseudomonas sp. PS-0-L and T5-6-I were oxidase negative and capable of nitrate utilization. Nevertheless, the average Ni(II) uptake was at the same level for Pseudomonas sp. V4-5-SB and Pseudomonas sp. T5-6-I, while the average Kd values of Pseudomonas sp. PS-0-L differed substantially from the values of V4-5-SB and T5-6-I. Pseudomonas sp. PS-0-L had the highest Ni(II) uptake with an average Kd of 970 L/kg DW. In the other two Pseudomonas sp. strains a significantly lower average uptake, 240 L/kg DW, of Ni(II) was observed.
Because the highest Ni uptake was observed in Pseudomonas sp. V4-5-SB, PS-0-L and T5-6-I, these bacteria were selected for more detailed uptake studies. Solution pH has an important role in the metal uptake of bacteria, affecting both the metal speciation and solution chemistry, as well as surface chemistry of bacterial cell membranes and walls (Vijayaraghavan and Yun, 2008). As the pH increases, deprotonation of functional groups, such as carboxyl and phosphate groups, lead to the net negative charge of bacterial surfaces and additionally increase the electrochemical attraction and cation adsorption on bacterial cell membrane or cell wall (Jiang et al., 2004; Vijayaraghavan and Yun, 2008). In the present study, the effect of pH on Ni(II) removal by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I was examined at the pH range of 1.2–11.7. This pH range includes the natural pH range of most boreal soils and other environmental habitats (pH 6–9). On the other hand, at pH over 9, precipitation of metal species takes place, removing free metal cations from the solution and from the reach of the bacterial cells (Nieminen et al., 2007). In our research, the increase in the solution pH increased Ni(II) uptake by Pseudomonas sp. V4-5-SB, PS-0-L and T5-6-I, and the highest Ni uptake was observed in solution pH ∼6–9. As energy-independent interaction mechanisms, such as direct biosorption on cell wall functional groups, would be affected by pH, the observed increase in Ni(II) uptake with the increasing pH may indicate cationic Ni(II) biosorption on to the bacterial cell wall. Previously, the isoelectric points (IEP) of several Gram positive and Gram negative bacteria have been determined and the reported IEPs have varied from 1.75 to 4.15 (Harden and Harris, 1952). For Pseudomonas cyanogenes (strain 795), Ps. convexa, Ps. aeruginosa and Ps. pyocyaneus, IEP values between 2.2 and 3.3 have been reported (Winslow and Upton, 1926; Harden and Harris, 1952; Yee and Fein, 2001). These values are within the observed increase in the Ni uptake at pH range 2–4 in the current work, which indicates negative surface charge of Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I at the pH above 4 and the cationic Ni(II) biosorption on to the bacterial cell wall. However, the observed Ni(II) uptake of Pseudomonas sp. V4-5-SB and PS-0-L were lower in 0.1 mol/L NaCl at pH ∼7 (Kd 502 and 807 L/kg DW) compared to the results in Medium A at + 20°C at pH 7 after 7 days incubation (Kd 640and 1890 L/kg DW). This may indicate, e.g., intra-cellular bioaccumulation of Ni or secretion of Ni-complexing metabolites by Pseudomonas sp. V4-5-SB and PS-0-L in addition to Ni(II) biosorption under these conditions.
The Ni(II) cell wall biosorption was further examined using adsorption isotherms. Adsorption isotherms are commonly used to describe the relation between the equilibrium concentration of an adsorbate [Ni(II)] and an adsorbent (bacterial cell) (Ho et al., 2002; Sparks, 2003). The use of isotherms requires their proper understanding and critical interpretation. Recently, linear regression analysis has generally been applied to determine the most suitable adsorption model because it analyzes the adsorption system and verifies the theoretical assumptions of isotherm model (Ayawei et al., 2017). However, the use of empirical models, e.g., Langmuir and Freundlich models, is limited and the previous studies cannot be directly applied to the systems obtained under conditions different or more complex than those studied. Generally, the shape of isotherms has been used to evaluate the capacity and affinity of sorption surface (Sparks, 2003). Isotherm shapes have also been used to estimate the number of different sorption sites of a particular surface taking part in the sorption process. We used both the Langmuir and Freundlich model equations to describe the absorption system in the two Pseudomonas sp. strains V4-5-SB and PS-0-L. Typically, the Langmuir model can be applied for sorption on monolayer surfaces (Wang and Chen, 2009). In this model, it is assumed that the metal ions are chemically adsorbed at a fixed number of well-defined sites; there is no interaction between the ions; all adsorption sites are energetically equivalent (homogeneous surface) and each site can accommodate one ion. However, the Langmuir model does not take into account that the surface is rarely homogeneous; the adsorbed ions are not necessarily inert, and the adsorption mechanism is clearly not the same for the first ions to adsorb to a surface as for the last one (Vijayaraghavan et al., 2006; Ayawei et al., 2017). Langmuir constants, qm and b, represent sorption capacity and bonding energy of absorption (affinity), respectively (Wang and Chen, 2009; Oves et al., 2013). In general, low b values are reflected in the steep slope of a sorption isotherm, indicating a high affinity. Thus, for a good sorbent typically a high qm value and a steep isotherm slope (i.e., low b) is observed (Wang and Chen, 2009). The experimental data of Pseudomonas sp. V4-5-SB and PS-0-L followed the Langmuir adsorption isotherm. The b value for Pseudomonas sp. V4-5-SB and PS-0-L at pH 5.7 were 468 and 46.3 L/g DW and the qm values were 0.00494 and 0.0265 mg/g DW, respectively. At higher pH (8.7), the corresponding b values were 242 and 73.6 L/g DW and the qm values were 0.00496 and 0.00756 mg/g DW. The Freundlich model can be applied for sorption on heterogeneous surfaces or surfaces supporting sites of varied affinities (multilayer sorption) (Volesky, 1990; Ho et al., 2002; Wang and Chen, 2009). This isotherm gives an expression which defines the surface heterogeneity and the exponential distribution of active sites (Ayawei et al., 2017). It is assumed that the stronger binding sites are occupied first and that the binding strength decreases with the increasing degree of binding site occupation (Vijayaraghavan et al., 2006). Biosorption equilibrium constant, Kf, from the Freundlich sorption model represents sorption ability and n indicates biosorption intensity (Wang and Chen, 2009; Oves et al., 2013). However, Kf parameters cannot be compared among samples whenever the 1/n parameters differ (Chen et al., 1999). It is also not possible to compare Kf values from literature if they were derived from different units of q and Ce (Chen et al., 1999). Feundlich isotherm is not sufficient at very high concentrations of adsorbate because in reality the mass-ratio of adsorbate/adsorbent has an asymptotic maximum as concentration increases without limitations (Ayawei et al., 2017). Experimental sorption data of Pseudomonas sp. V4-5-SB and PS-0-L also followed, in addition to the Langmuir model, the Freundlich adsorption isotherm. The Kf value for Pseudomonas sp. V4-5-SB and PS-0-L at pH 5.7 were 0.221 and 0.403 mg L1/n g–1 and the 1/n values were 0.763 and 0.876, respectively. At higher pH (8.7), the corresponding Kf values were 0.764 and 0.769 mg L1/n g–1 and the 1/n values were 0.941 and 1.03. Both models, Freundlich and Langmuir, confirmed the higher sorption capacity and more intensive Ni(II) biosorption for Pseudomonas sp. PS-0-L compared to Pseudomonas sp. V4-5-SB.
Furthermore, the Freundlich adsorption isotherms of Pseudomonas sp. V4-5-SB and PS-0-L showed a slight deflection at both pH 5.7 and 8.7. At low initial Ni concentrations, isotherms were observed to behave ideally. Such behavior can be explained by the high affinity of the adsorbent for the adsorptive at lower Ni(II) concentrations and with decreasing affinity as the concentration increases. Additionally, the deflection of isotherms may result from the saturation of the exchange sites or from the retention of Ni(II) on multiple sorption sites. Generally, Freundlich sorption isotherms tend to fit experimental data better at low concentrations, while Langmuir isotherms fit the data better at higher concentrations (Richter et al., 1989). This was also observed in our experimental data. However, adsorption isotherms are descriptions of experimental data and more specific conclusions require support of molecular investigations, e.g., the use of spectroscopic techniques.
Ni biosorption over a range of metal ion concentrations with Pseudomonas sp. V4-5-SB and PS-0-L at pH 5.7 and 8.7 showed that the adsorption of Ni(II) was dependent on the initial Ni(II) concentration. The values of equilibrium uptake capacities (q) by Pseudomonas sp. V4-5-SB and PS-0-L increased as the pH of the solution and the amount of Ni(II) increased. The highest q values were achieved with the initial Ni(II) concentration of 10–3 mol/L. The q values by Pseudomonas sp. V4-5-SB and PS-0-L were 2.4 and 5.2 mg/g DW at pH 5.7, and 6.6 and 16.8 mg/g DW at pH 8.7, respectively. Previously, Ni(II) uptake has been studied with several bacteria, such as E. coli, Pseudomonas sp., Bacillus sp., Arthrobacteria sp., Sterptomyces sp., Alcaligenes eutrophus, under varying experimental conditions. The Ni biosorption capacities were reported to be between 0.8 and 55 mg/g DW at initial Ni concentrations of 10–3–10–2 mol/L (Mattuschka and Straube, 1993; Tsezos et al., 1995; Veglió et al., 1997; López et al., 2000; Öztürk et al., 2004; Selatnia et al., 2004; Ansari and Malik, 2007; Öztürk, 2007). In addition, a very high value of 100 mg/g DW by Ps. fluorescens 4F39 was reported, with initial Ni concentration of 10–3 mol/L at pH 9 (López et al., 2000). In our study, the Ni(II) uptake capacities fell in the middle of the typical uptake capacity range, being fivefold lower than the maximum capacity reported by López et al. (2000) for Ps. fluorescens 4F39. In our research, the highest Ni(II) uptake was shown by Pseudomonas sp. PS-0-L with the equilibrium Ni(II) biosorption capacity of approximately 17 mg/g DW at pH 8.7 with the initial Ni(II) concentration of 10–3 mol/L. However, all strains removed Ni(II) to a certain extent, implying that the capacity for Ni(II) uptake may be wide spread in soil microbial communities.
The role of cellular functional groups in the Ni(II) binding by Pseudomonas sp. V4-5-SB, PS-0-L, and T5-6-I was studied using FTIR spectroscopy. The functional groups found on Pseudomonas sp. cells were similar regardless of strain and all strains had alcoholic hydroxyl, amine, carboxylic carbonyl and hydroxyl, as well as phosphate and alkane groups. Of these groups, alcoholic hydroxyl, carboxylic hydroxyl and amine groups may participate Ni(II)binding, but as amine groups typically are protonated under physiological pH, the alcoholic and carboxylic hydroxyl groups most probably would act as the main Ni(II) binding groups in these bacterial cells, with lower sorption capacity found for the amino groups. This assumption is supported by the observations obtained using sorption isotherm approach, in which Ni biosorption and retention on multiple sorption sites was indicated. Previously, Choudhary and Sar (2009) have reported carboxyl, amide, and phosphate groups of Pseudomonas sp. J007 for being the dominant functional groups involved in bacteria-metal (Ni, Co, Cu, Cd) interactions. In addition, the presence of carboxyl and amine groups on the surfaces of Ps. aeruginosa (Kang et al., 2007) and carboxyl and phosphate groups on Pseudomonas sp. MTCC 3087 (Kazy et al., 2006) have been reported. Further, Yu et al. (2014) have suggested that sulfhydryl groups (-SH) may also control the binding of metals onto bacterial walls under low metal-loading conditions.
We studied the morphology of Pseudomonas sp. V4-5-SB and PS-0-L in Ni(II) treated cells using TEM. As stated above, López et al. (2000) have reported accumulation of dense Ni structures on cell walls of Ps. fluorescens 4F39 at pH 9. Additionally, Ps. aeruginosa has been shown to bind Ni onto the cell envelope region (Sar et al., 2001). On the basis of these previous observation on Ps. fluorescens and Ps. aeruginosa and on our results using FTIR, we assumed that Ni(II) attached to the bacterial cell membrane or cell wall. However, TEM images showed dense intra-cellular Ni accumulations in the cells of Pseudomonas sp. V4-5-SB and PS-0-L treated with Ni(II), but no significant Ni deposits were observed on or in the close vicinity of the cell membrane/cell wall of these bacteria. EDS analysis confirmed the presence of the Ni in these bacteria although at low concentrations. To our knowledge, clear Ni structures/crystals inside the bacterial cells or in cytoplasm have not been reported previously. However, copper (Cu2+) and cadmium (Cd2+) uptake have been detected in and around the cell periphery of Pseudomonas sp. J007, with a small fraction of metal in the cytoplasm (initial metal conc. 5 × 10–4 mol/L) (Choudhary and Sar, 2009). In addition, fractionation experiments showed low nickel (Ni2+) distribution in the cytoplasm (Choudhary and Sar, 2009). Divalent heavy metal cations are structurally very similar (radius, charge) and are expected to behave in a similar way, supporting our finding of Ni(II) accumulations inside bacterial cells. It is also known that several bacteria have uptake systems for Ni and other heavy metals (Nies, 1999; Eitinger and Mandrand-Berthelot, 2000). For example, Smith and Maguire (1995) identified CorA Mg2+ transporter system (metal inorganic transport) in several Gram negative bacteria. In addition to magnesium, it has been suggested that CorA also mediates uptake of Ni2+ and Co2+ (Snavely et al., 1989, 1991). Certain ABC-type transporters and transporters of the HoxN family have been identified to possibly convey different divalent cations (Nies, 1999). The Ni crystals observed in the TEM images of our study, may indicate active uptake of Ni(II). This was also supported by the several changes observed in the protein expression profiles of these bacteria. However, the relatively high Ni concentrations used in our study, may have activated the detoxification mechanism in the bacterial cells. This mechanism could be further utilized for bio-purification applications in contaminated sites.
We used the incubation temperatures of +4 and +20°C in our experiments and + 20°C was chosen to represent the average daytime surface temperatures found in southwestern Finland in June (Finnish Meteorological Institute [FMI], 2015). The lower temperature was chosen to represent the temperature of the bottom layers of the bog. All bacterial strains used in this study removed Ni(II) from the solution more efficiently at + 20°C. The average uptake at this temperature was 89% higher than the average uptake observed at + 4°C. Similarly, at this temperature, the bacterial growth rate was higher compared to that at the lower temperature of + 4°C, which is likely to affect the efficiency of Ni(II) removal by biosorption and/or bioaccumulation as the cell densities at the time = 0 were taken into account in the calculations. Moreover, the presence of Ni(II) (10–6–10–3 mol/L) was not found to affect bacterial growth, indicating that the possible toxic effect of Ni(II) is either surpassed by the bacterial metabolism (e.g., precipitation of Ni inside the cells through detoxification reactions) or the used Ni(II) concentrations were below the toxic levels. Growing bacteria may accumulate Ni inside the cell as part of their metabolism or release metabolites into the solution that may affect Ni solubility. In addition, the increasing amount of biomass provides more surface area and functional groups for Ni(II) biosorption to potentially occur on cell surfaces.
In addition to the effect of temperature on cell growth rate, incubation time affects the growth phase of bacteria. After the exponential growth phase, the biomass typically reaches a plateau phase in which growth rate and death rate are equal. After a certain period of time, the amount of bacterial mass begins to decrease and cell death prevails (death phase). In the current research, Ni(II) uptake varied with different bacterial strains depending on the incubation time. As we increased the incubation time from 7 to 14 days, we observed 43% decrease in Ni(II) uptake by the studied bacteria, which may result from increased cell death and cell lysis in the cultures. This supports the active Ni(II) uptake hypothesis for the bacteria used in the present study, as Ni is potentially released from the cells after cell lysis, resulting in decreased overall uptake. If direct biosorption on cell walls would occur, decrease after cell lysis would not be expected as biosorption occurs between the absorbent and cell wall functional groups regardless of whether the cells are alive or dead. Previously, we studied interaction of Ni(II) with other bacterial isolates (Pseudomonas, Paraburkholderia, Rhodococcus, Paenibacillus) from same Lastensuo bog (Lusa et al., 2016a) and observed that Ni(II) uptake to increase as the incubation time was increased from 1–3 to 7–14 days, which suggested that the 7-day incubation time is optimal, especially in the case of Pseudomonas sp. PS-0-L and T5-6-I, which is in line with the results obtained also in the current study.
Our results obtained from the batch uptake studies under different nutrient conditions, temperature and pH as well as the spectroscopic data using EDS and finally the data obtained using electron microscopy (TEM) all supported Ni accumulation inside the bacterial cells used in this study. In addition, several changes in the protein profiles in the bacteria treated with Ni(II) were observed in the MALDI-TOFF spectra, which can indicate active accumulation of Ni in these bacteria. However, at the same time the uptake of Ni(II) was observed to follow the Langmuir and Freundlich isotherm models, which are typically used to describe the direct sorption (or in our case biosorption). The sorption isotherms indicated Ni(II) retention on multiple sorption sites. In addition, FTIR analysis showed the presence of the alcoholic hydroxyl, carboxylic carbonyl and hydroxyl, amine, alkane, and phosphate groups on bacterial surfaces. Based on these results we propose a distinct uptake mechanisms of Ni(II) in the Pseudomonas sp. strains used in this work; (i) intra-cellular accumulation (e.g., through detoxification mechanism) and (ii) direct biosorption on cell membrane or wall functional groups. Even though the proportions of Ni(II) removed from the solution by bioaccumulation and biosorption could not be fully concluded in our study, the fact that Ni was detected inside the bacterial cells, and that the sorption isotherm model could (at least partly) describe the uptake process implies in two different uptake mechanisms present in these bacteria.
Conclusion
All studied bacteria were able to remove Ni(II) from the solution and the uptake was affected by incubation time, temperature, pH as well as nutrient solution used. Temperature, incubation time, and nutrient source also affected the overall bacterial growth rate, affecting presumably also Ni(II) removal. Even though the isolated bacteria belonged to the minority of the whole bacterial population of the bog, the fact that they all were able to remove Ni(II) from solution indicates that this is a common feature for bacteria found in this environment and that it is most likely that soil bacteria are capable to influence the biogeochemical behavior of Ni(II) in the northern, boreal environment also under in situ conditions. Based on our 63Ni batch uptake experiments as well as the results obtained from EDS, TEM, FTIR, and MALDI-TOFF studies, we suggest that in the studied bacterial strains (especially the Pseudomonas sp. strains), Ni(II) is taken up inside the cells using an active mechanisms and in addition, part of the Ni(II) is retained on the cell membrane/cell wall functional groups through biosorption.
Data Availability Statement
The datasets generated for this study can be found in NCBI GenBank, accession numbers MK290398–MK290403.
Author Contributions
JK and ML participated in the design of the study. JK isolated new bacterial strains, characterized them and extracted DNA, performed all biosorption experiments, transmission electron microscopy (TEM), and Fourier transform infrared (FTIR) analyses, carried out the statistical analyses, and prepared the morphological samples. ML and MB performed a phylogenetic analysis and wrote this part of the manuscript and supervised the project. MK characterized the morphological samples with Energy dispersive X-ray spectroscopy (EDS). JK wrote the manuscript with support from ML and MB. All authors contributed to the interpretation of the results, provided critical feedback and contributed to the final manuscript.
Funding
This work was supported by the KAUTE Foundation (Grant Number 201700417) and Emil Aaltonen Foundation (Grant Number 180103O).
Conflict of Interest
MB is employed by the company VTT Technical Research Centre of Finland, Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mervi Lindman from Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki for technical assistance in TEM imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02677/full#supplementary-material
References
Akcil, A., and Koldas, S. (2006). Acid mine drainage (AMD): causes, treatment and case studies. J. Clean. Environ. 14, 1139–1145. doi: 10.1016/j.jclepro.2004.09.006
Ansari, M., and Malik, A. (2007). Biosorption of nickel and cadmium by metal resistant bacterial isolates from agricultural soil irrigated with industrial wastewater. Bioresour. Technol. 98, 3149–3153. doi: 10.1016/j.biortech.2006.10.008
Ayawei, N., Ebelegi, A. N., and Wankasi, D. (2017). Modelling and interpretation of adsorption isotherms. J. Chem. 2017:11. doi: 10.1155/2017/3039817
Bossé, J. T., Gilmour, H. D., and MacInnes, J. I. (2001). Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183, 1242–1247. doi: 10.1128/JB.183.4.1242-1247.2001
Cempel, M., and Nikel, G. (2006). Nickel: a review of its sources and environmental toxicology. Polish J. Environ. Stud. 15, 375–382.
Chen, Z., Xing, B., and McGill, W. B. (1999). A unified sorption variable for environmental applications of the freundlich equation. J. Environ. Qual. 28, 1422–1428. doi: 10.2134/jeq1999.00472425002800050005x
Cherrier, M. V., Martin, L., Cavazza, C., Jasquamet, L., Lemaire, D., Gaillard, J., et al. (2005). Crystallographic and spectroscopic evidence for high affinity binding of FeEDTA(H2O)– to the periplasmic nickel transporter NikA. J. Am. Chem. Soc. 127, 10075–10082. doi: 10.1021/ja0518530
Chivers, P. T. (2015). Nickel recognition by bacterial importer proteins. Metallomics 7, 590–595. doi: 10.1074/jbc.M002232200
Choudhary, S., and Sar, P. (2009). Characterization of a metal resistant Pseudomonas sp. isolated from uranium mine for its potential in heavy metal (Ni2+, Co2+, Cu2+, and Cd2+) sequestration. Bioresour. Technol. 100, 2482–2492. doi: 10.1016/j.biortech.2008.12.015
Eberz, G., Eitinger, T., and Friedrich, B. (1989). Genetic determinants of a nickel-species transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J. Bacterol. 171, 1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989
Eitinger, T., Degen, O., Böhnke, U., and Müller, M. (2000). Nic1p, a relative of bacterial transition metal permeases in Schizosaccharomyces pombe, provides nickel ion for urease biosynthesis. J. Biol. Chem. 275, 18029–18033. doi: 10.1074/jbc.M001978200
Eitinger, T., and Friedrich, B. (1991). Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J. Biolo. Chem. 266, 3222–3227.
Eitinger, T., and Mandrand-Berthelot, M.-A. (2000). Nickel transport systems in microorganisms. Arch. Microbiol. 173, 1–9. doi: 10.1007/s002030050001
Finnish Meteorological Institute [FMI] (2015). Kuukausitilastot. Available at: http://ilmatieteenlaitos.fi/kuukausitilastot (accessed January 2, 2019)
Fourest, E., and Roux, J.-C. (1992). Heavy metal biosorption by fungal mycelial by-products: mechanisms and influence of pH. Appl. Microbiol. Biotechnol. 37, 399–403. doi: 10.1007/BF00211001
Fu, C., Javedan, S., Moshiri, F., and Maier, R. J. (1994). Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc. Natl. Acad. Sci. U.S.A. 91, 5099–5103. doi: 10.1073/pnas.91.11.5099
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z.-C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15:203. doi: 10.1186/s12934-016-0603-7
Haapanen, R., Aro, L., Helin, J., Ikonen, A. T. K., and Lahdenperä, A. M. (2013). Studies on Reference Mires: 1. Lastensuo and Pesänsuo in 2010–2011. Working Report 2012–102. Posiva Oy. Eurajoki. Finland. Available at: http://www.posiva.fi (accessed January 12, 2019).
Harden, V., and Harris, J. O. (1952). The isoelectric point of bacterial cells. J. Bacteriol. 65, 198–202.
Hausinger, R. P. (1994). Nickel enzymes in microbes. Sci. Tot. Environ. 148, 157–166. doi: 10.1016/0048-9697(94)90392-1
Hendricks, J. K., and Mobley, H. L. T. (1997). Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J. Bacteriol. 179, 5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997
Hjerpe, T., Ikonen, A., and Broed, R. (2010). Biosphere Assessment Report 2009. Posiva Report 2010-03. Posiva Oy. Eurajoki. Finland. Available at: http://www.posiva.fi (accessed January 12, 2019).
Ho, Y. S., Porter, J. F., and McKay, G. (2002). Equilibrium isotherm studies for the sorption of divalent metal ions onto peat: copper, nickel and lead single component systems. Water Air Soil Poll. 141, 1–33. doi: 10.1023/A:1021304828010
IARC (2012). IARC Monographs Monographs on the Evaluation of Carcinogenic Risks to Humans: Nickel and Nickel Compounds. Lyon: IARC.
Jaffré, T., Pillon, Y., Thomine, S., and Merlot, S. (2013). The metal hyperaccumulators from New Caledonia can broaden our understanding of nickel accumulation in plants. Front. Plant Sci. 4:279. doi: 10.3389/fpls.2013.00279
Jiang, W., Saxena, A., Song, B., Ward, B. B., Beveridge, T. J., and Myneni, S. C. B. (2004). Elucidation of functional groups on Gram-positive and Gram-negative bacterial surfaces using infrared spectroscopy. Langmuir 20, 11433–11442. doi: 10.1021/la049043
Joosten, H., and Clarke, D. (2002). Wise use of mires and peatlands. Int. Mire Conserv. Group Int. Peat Soc. 304, 32–33.
Jubier-Maurin, V., Rodrigue, A., Ouahrani-Bettache, S., Layssac, M., Mandrand-Bethelot, M.-A., Köhler, S., et al. (2001). Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183, 426–434. doi: 10.1128/JB.183.2.426-434.2001
Jukes, T. H., and Cantor, C. R. (1969). Evolution of Protein Molecules. New York, NY: Academic Press.
Kang, S.-Y., Lee, J.-U., and Kim, K.-W. (2007). Biosorption of Cr(III) and Cr(VI) onto the cell surface of Pseudomonas aeruginosa. Biochem. Eng. J. 36, 54–58. doi: 10.1016/j.bej.2006.06.005
Kazy, S., Das, S., and Sar, P. (2006). Lanthanum biosorption by a Pseudomonas sp.: equilibrium studies and chemical characterization. J. Ind. Microbiol. Biotechnol. 33, 773–783. doi: 10.1007/s10295-006-0108-1
Langmuir, I. (1916). The adsorption of gases on plane surface of glass, mica and platinum. J. Am. Chem. Soc. 40, 1361–1368.
Ledin, M., and Pedersen, K. (1996). The environmental impact of mine wastes - Roles of microorganisms and their significance in treatment of mine wastes. Earth Sci. Rev. 41, 67–108. doi: 10.1016/0012-8252(96)00016-5
López, A., Lázaro, N., Priego, J. M., and Marqués, A. M. (2000). Effect of pH on the biosorption of nickel and other heavy metals by Pseudomonas fluorescens 4F39. J. Indust. Microbiol. Biotechnol. 24, 146–151. doi: 10.1038/sj.jim.2900793
Loukola-Ruskeeniemi, K., and Lahtinen, H. (2013). Multiphase evolution in the black shale-hosted Ni-Cu-Zn-Co deposit at Talvivaara, Finland. Ore Geol. Rev. 52, 85–99. doi: 10.1016/j.oregeorev.2012.10.006
Lusa, M. (2017). Sorption Experiments of Ag, Cl, Cs, I, Mo, Ni, Pb, Se, Sn, Sr and Tc – Lastensuo Bog Samples and Poosjärvi and Kivijärvi Sediments Samples. Working Report 2017-35. Eurajoki: Posiva. doi: 10.1016/j.oregeorev.2012.10.006
Lusa, M., Bomberg, M., Ahola, H., Knuutinen, J., and Lehto, J. (2015a). Sorption of radioiodide in an acidic, nutrient-poor boreal bog: insights into the microbial activity. J. Environ. Radioact. 143, 110–122. doi: 10.1016/j.jenvrad.2015.02.022
Lusa, M., Bomberg, M., Aromaa, H., Knuutinen, J., and Lehto, J. (2015b). The Microbial impact on the sorption behaviour of selenite in an acidic, nutrient-poor boreal bog. J. Environ. Radioact. 147, 85–96. doi: 10.1016/j.jenvrad.2015.05.014
Lusa, M., Bomberg, M., Virtanen, S., Lempinen, J., Aromaa, H., Knuutinen, J., et al. (2015c). Factors affecting the sorption of cesium in a nutrient-poor boreal bog. J. Environ. Radioact. 147, 22–32. doi: 10.1016/j.jenvrad.2015.05.005
Veglió, F., Beolchini, F., and Gasbarro, A. (1997). Biosorption of toxic metals: an equilibrium study using free cells of Arthrobacter sp. Process. Biochem. 32, 99–105. doi: 10.1016/S0032-9592(96)00047-7
Lusa, M., Lehto, J., Aromaa, H., Knuutinen, J., and Bomberg, M. (2016a). Uptake of radioiodide by Paenibacillus sp., Pseudomonas sp., Burkholderia sp. and Rhodococcus sp. isolated from a boreal, nutrient-poor bog. J. Environ. Sci. 44, 26–37. doi: 10.1016/j.jes.2015.08.026
Lusa, M., Lehto, J., and Bomberg, M. (2016b). The uptake of Ni2+ and Ag+ by bacterial strains isolated from a boreal nutrient-poor bog. AIMS Microbiol. 2, 120–137. doi: 10.3934/microbial.2016.2.120
Maeda, M., Hidaka, M., Nakamura, A., Masaki, H., and Uozumi, T. (1994). Cloning, sequencing, and expression of thermophilic Bacillus strain TB-90 urease gene complex in Escherichia coli. J. Bacteriol. 176, 432–442. doi: 10.1128/jb.176.2.432-442.1994
Mäkilä, M., and Grundström, A. (2008). Turpeen Ikä ja Kerrostumisnopeus Lounais-Suomen Soilla. Posiva WR 2008–12. Eurajoki: Posiva.
Mattuschka, B., and Straube, G. (1993). Biosorption of metals by a waste biomass. J. Chem. Tech. Biotechnol. 58, 57–63. doi: 10.1002/jctb.280580108
McMurry, J. (2008). Organic Chemistry, Chapter 12, Structure Determination: Mass Spectrometry and Infrared Spectroscopy, 7th Edn. Boston, MA: Thomson Learning.
Mobley, H., and Hausinger, R. (1989). Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108.
Mudd, G. M. (2010). Global trends and environmental issues in nickel mining: sulfides versus laterites. Ore Geol. Rev. 38, 9–26. doi: 10.1016/j.oregeorev.2010.05.003
Mulrooney, S. B., and Hausinger, R. P. (2003). Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27, 239–261. doi: 10.1016/S0168-6445(03)00042-1
Navarro, C., Wu, L.-F., and Mandrand-Berthelot, M.-A. (1993). The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Molec. Microbiol. 9, 1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x
Niemi, J., and Raateland, A. (2007). River water quality in the finnish eurowaternet. Boreal Environ. Res. 12, 571–584.
Nieminen, T. M., Ukonmaanaho, L., Rausch, N., and Shotyk, W. (2007). Chapter 1: biogeochemistry of nickel and its release into the environment. Metal Ions Life Sci. 2, 1–30.
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Nriagu, J. (1990). Global metal pollution: poisoning the biosphere. Environment 7, 7–33. doi: 10.1080/00139157.1990.9929037
Oves, M., Khan, M. S., and Zaidi, A. (2013). Biosorption of heavy metals by Bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi J. Biol. Sci. 20, 121–129. doi: 10.1016/j.sjbs.2012.11.006
Öztürk, A. (2007). Removal of nickel from aqueous solution by the bacterium Bacillus thuringiensis. J. Hazard. Mater. 147, 518–523. doi: 10.1016/j.jhazmat.2007.01.047
Öztürk, A., Artan, T., and Ayar, A. (2004). Biosorption of nickel(II) and copper(II) ions from aqueous solution by Streptomyces coelicolor A3(2). Colloid Surf B Biointerf. 34, 105–111. doi: 10.1016/j.colsurfb.2003.11.008
Posiva, (2012). Safety Case for the Disposal of Spent Nuclear Fuel at Olkiluoto - Description of the Disposal System 2012 (Report POSIVA 2012-05. Eurajoki: Posiva.
Pöyry Finland Oy, (2017). Terrafame Oy. Sulkemissuunnitelma YVA-selostus 24.8.2017. Available at: www.ymparisto.fi/yva ? YVA-hankkeet, in Finnish (accessed July 10, 2019).
Richter, E., Schutz, W., and Myers, A. L. (1989). Effect of adsorption equation on prediction of multicomponent adsorption equilibria by the ideal adsorbed solution theory. Chem. Engine. Sci. 44, 1609–1616. doi: 10.1016/0009-2509(89)80003-X
Sar, P., Kazy, S. K., and Singh, S. P. (2001). Intracellular nickel accumulation by Pseudomonas aeruginosa and its chemical nature. Lett. App. Microbiol. 32, 257–261. doi: 10.1046/j.1472-765X.2001.00878.x
Schalk, I. J., Hannauer, M., and Braud, A. (2011). New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 13, 2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x
Schauer, K., Gouget, B., Carriere, M., Labigne, A., and de Reuse, H. (2007). Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63, 1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x
Sebbane, F., Mandrand-Bethelot, M.-A., and Simonet, M. (2002). Genes encoding specific nickel transport systems flank the chromosomal urease locus of pathogenic Yersiniae. J. Bacteriol. 184, 5706–5713. doi: 10.1128/JB.184.20.5706-5713.2002
Selatnia, A., Madani, A., Bakhti, M. Z., Kertous, L., Mansouri, Y., and Yous, R. (2004). Biosorption of Ni2+ from aqueous solution by a NaOH-treated bacterial dead Streptomyces rimosus biomass. Miner. Eng. 17, 903–911. doi: 10.1016/j.mineng.2004.04.002
Smith, R., and Maguire, M. (1995). Distribution of the CorA Mg2+ transport system in gram-negative bacteria. J. Bacterial. 177, 1638–1640. doi: 10.1128/jb.177.6.1638-1640.1995
Snavely, M., Florer, J., Miller, C., and Maguire, M. (1989). Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J. Bacteriol. 171, 4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989
Snavely, M., Gravina, S., Cheung, T., and Miller, C. (1991). Magnesium Transport in Salmonella typhimurium. regulation of mgtA and mgtB expression. J. Biolog. Chem. 266, 824–829.
Sparks, D. L. (2003). Environmental Soil Chemistry, Chapter 5 Sorption Phenomena on Soils, 2nd Edn. San Diego: Elsevier Science.
Stoof, J., Kuipers, E. J., Klaver, G., and van Vliet, A. H. (2010). An ABC transporter and a TonB ortholog contribute to Helicobacter mustelae nickel and cobalt acquisition. Infect. Immun. 78, 4261–4267. doi: 10.1128/IAI.00365-10
Sukdeo, N., Clugston, S. L., Daub, E., and Honek, J. F. (2004). Distinct classes of glyoxalase I: metal specificity of the yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitides enzymes. Biochem. J. 384, 111–117. doi: 10.1042/BJ20041006
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tsezos, M., Remoudaki, E., and Angelatou, V. (1995). A systematic study on equilibrium and kinetics of biosorptive accumulation. the case of Ag and Ni. intern. Biodeterior. Biodegrad. 1995, 126–153. doi: 10.1016/0964-8305(95)00049-B
Tsezos, M., Remoudaki, E., and Angelatou, V. (1996). A study of the effects of competing ions on the biosorption of metals. Intern. Biodeterior. Biodegrad. 1996, 19–26.
Tsitko, I., Lusa, M., Lehto, J., Parviainen, L., Ikonen, A., Lahdenperä, A.-M., et al. (2014). The variation of microbial communities in a depth profile of an acidic, nutrient-poor boreal bog in southwestern Finland. Open J. Ecol. 4, 832–859. doi: 10.4236/oje.2014.413071
Tuovinen, H., Pelkonen, M., Lempinen, J., Pohjolainen, E., Read, D., Solatie, D., et al. (2018). Behaviour of metals during bioheap leaching at the talvivaara mine, Finland. Geosciences 8:66. doi: 10.3390/geosciences8020066
Veglió, F., and Beolchini, F. (1997). Removal of metals by biosorption: a review. Hydrometallurgy 44, 301–316. doi: 10.1016/S0304-386X(96)00059-X
Vijayaraghavan, K., Padmesh, T. V. N., Palanivelu, K., and Velan, M. (2006). Biosorption of nickel(II) ions onto Sargassum wightii: application of two-parameter and three-parameter isotherm models. J. Hazard. Mater. B 133, 304–308. doi: 10.1016/j.jhazmat.2005.10.016
Vijayaraghavan, K., and Yun, Y. S. (2008). Bacterial biosorbents and biosorption. Biotech. Adv. 26, 266–291. doi: 10.1016/j.biotechadv.2008.02.002
Volesky, B. (1990). “Removal and recovery of heavy metals by biosorption,” in Biosorption of Heavy Metals, ed. B. Volesky, (Florida: CRC press).
Wang, J., and Chen, C. (2009). Biosorbents for heavy metals removal and their future. Biotech. Adv. 27, 195–225. doi: 10.1016/j.biotechadv.2008.11.002
White, C. E., Wilkinson, S. C., and Gadd, G. M. (1995). The role of microorganisms in biosorption of toxic metals and radionuclides. Intern. Biodeterior. Biodegrad. 1995, 17–40. doi: 10.1016/j.biotechadv.2018.07.002
Winslow, C., and Upton, M. (1926). Electrophoretic migration of various types of vegetative cells. J. Bact. 11, 367–392.
Wolfram, L., Friedrich, B., and Eitinger, T. (1995). The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J. Bacteriol. 177, 1840–1843. doi: 10.1128/jb.177.7.1840-1843.1995
Yee, N., and Fein, J. (2001). Cd adsorption onto bacterial surface: a universal adsorption edge? Geochim. Cosmochim. Acta 65, 2037–2042. doi: 10.1016/S0016-7037(01)00587-7
Keywords: nickel uptake, adsorption, sphagnum peat, heavy metal contamination, biopurification
Citation: Knuutinen J, Bomberg M, Kemell M and Lusa M (2019) Ni(II) Interactions in Boreal Paenibacillus sp., Methylobacterium sp., Paraburkholderia sp., and Pseudomonas sp. Strains Isolated From an Acidic, Ombrotrophic Bog. Front. Microbiol. 10:2677. doi: 10.3389/fmicb.2019.02677
Received: 18 February 2019; Accepted: 04 November 2019;
Published: 26 November 2019.
Edited by:
Shuhei Ono, Massachusetts Institute of Technology, United StatesReviewed by:
Amy Michele Grunden, North Carolina State University, United StatesLeslie James Robbins, Yale University, United States
Copyright © 2019 Knuutinen, Bomberg, Kemell and Lusa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenna Knuutinen, amVubmEua251dXRpbmVuQGhlbHNpbmtpLmZp
 Jenna Knuutinen
Jenna Knuutinen Malin Bomberg
Malin Bomberg Marianna Kemell
Marianna Kemell Merja Lusa
Merja Lusa