- 1Shengli Clinical Medical College of Fujian Medical University, Fuzhou, China
- 2Clinical Microbiology Laboratory, Fujian Provincial Hospital, Fuzhou, China
- 3Department of Pulmonary and Critical Care Medicine, Fujian Provincial Hospital, Fuzhou, China
Background: The increase in carbapenem-resistant Klebsiella pneumoniae (CRKP), especially the emergence of tigecycline-resistant K. pneumoniae (KP), is a serious public health concern. However, the underlying mechanism of tigecycline resistance is unclear. In this study, we evaluated the role of the CusS-CusR two-component system (TCS), which is associated with copper/silver resistance, in tigecycline resistance in CRKP.
Methods: Following the in vitro evolution of tigecycline-resistant KP, the minimum inhibitory concentrations of tigecycline were determined using the micro-broth dilution method. RNA sequencing and data analysis were performed to identify differentially expressed genes. Quantitative PCR (qPCR) was performed to verify the genes of interest. Genes associated with tigecycline resistance, such as ramR, tex (T), and tet (A), were detected by PCR, and then mutants were confirmed by sequencing. Additionally, the efflux pump-associated genes soxS, oqxA, oqxB, acrE, and acrF were also analyzed by qPCR. CusR was deleted and complemented by the suicide vector pKO3-Km plasmid and pGEM-T-easy plasmid, respectively.
Results: Nine strains of KP were evaluated in our study. Strains A2 and A3 were evolved from A1, B2, and B3 were evolved from B1, and C2 and C3 were evolved from C1. The tigecycline minimum inhibitory concentration for A1, B1, and C1 was 0.5 μg/mL; that for A2, B2, and C3 was 16.0 μg/mL; and that for A3, B3, and C3 was 32.0 μg/mL. RNA-sequencing and qPCR confirmed that the differentially expressed genes cusE, cusS, cusR, cusC, cusF, cusB, and cusA showed higher expression in C2 and C3 than in C1. Genes related to the efflux pump AcrAB-TolC showed higher expression in B2 and B3 than in B1. No mutants of ramR, tex (T), or tet (A) were detected. SoxS, oqxA, oqxB, acrE, and acrF did not show increased expression in any group. After deletion and complementation of cusR among C3, the MIC of tigecycline decreased to 4 μg/mL, and then recovered to 32 μg/mL. The expression of cusFBCA, correspondingly decreased and increased significantly.
Conclusion: In addition to its primary function in resistance to copper/silver, the CusS-CusR two-component system is associated with CRKP resistance to tigecycline.
Introduction
In recent decades, multiple drug-resistant and carbapenem-resistant Enterobacteriaceae have increased in abundance, particularly Klebsiella pneumoniae (KP), Escherichia coli, Enterobacterial spp., Proteus spp., and Serratia mucosa, greatly limiting the efficacy of antibiotic treatment. Tigecycline is currently widely used in clinical practice as an anti-infective treatment and has become the most important antibiotic for treating carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Enterobacteriaceae. However, A. baumannii, E. coli, and K. pneumoniae have shown resistance to tigecycline. Additionally, various mechanisms have been reported to confer tigecycline resistance, with most studies focusing on resistance-nodulation cell division transporters, mainly among AcrAB-TolC efflux pumps, while other pumps and various control pathways are also reported to be associated with tigecycline resistance (Pournaras et al., 2016; Chiu et al., 2017; Ye et al., 2017; He et al., 2018).
Sheng et al. (2014) determined the tigecycline resistance mechanism in 26 strains of tigecycline-non-susceptible KP and found that the resistance of nine strains could not be explained by existing mechanisms. Recent studies revealed a co-regulatory effect between antibiotic resistance and metal resistance. Wang et al. (2017) reported that the CzcR-CzcS two-component system (TCS) in Pseudomonas aeruginosa participates in metal detoxification and antibiotic resistance, mainly by co-regulating cross-resistance between Zn (II) and carbapenems antibiotics. Perron et al. (2004) reported that in P. aeruginosa, the CzcR-CzcS TCS induced resistance to cadmium, zinc, and cobalt through over-expression of the efflux pump system, resulting in an increase in the MICs of carbapenems. In a study of metal resistance and its relationship with antibiotic resistance, Pal et al. (2017) found that metal resistance and antibiotic resistance genes were located at the same site in the cell, and a single resistance mechanism (such as the efflux pump) could result in resistance to either antibiotics or metal. Hölzel et al. (2012) studied the relationship between heavy metals and bacterial antibiotic resistance in pig manure, and revealed that the presence of copper and zinc was significantly correlated with the increased drug resistance rate of β-lactam antibiotics, while the presence of mercury was significantly correlated with the low drug resistance of E. coli to β-lactams, aminoglycosides, and other antibiotics. These studies suggest that bacterial resistance to metals is associated with antibiotic resistance.
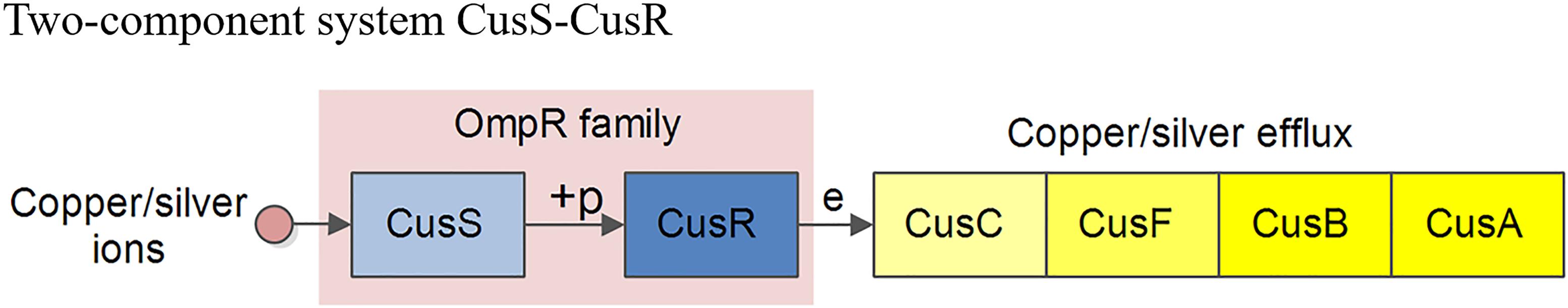
The CusS-CusR TCS is associated with bacteria resistant to the heavy metals copper/silver, in which copper/silver ions stimulate the phosphorylation of the outer membrane protein, CusS; the phosphorylated CusS phosphorylates the translation regulator, CusR. The phosphorylated CusR promotes higher expression of the genes encoding the efflux pump CusCFBA, which pumps the copper/silver out of the bacteria, thus leading to bacteria that are resistant to heavy metals. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of CusS-CusR TCS is shown in Figure 1.
However, only a few studies have focused on the tolerance of K. pneumoniae to heavy metals and its association with antibiotic resistance. Accordingly, in this study, we induced three forms of carbapenem-resistant Klebsiella pneumoniae (CRKP) resistance to tigecycline in vitro and identified the differentially expressed genes using transcriptome sequencing. In particular, we focused on the CusS-CusR TCS to determine its potential role in tigecycline resistance in nine CRKP strains that evolved from the three parental strains KPN222 (A), KPN114 (B), and KPN315 (C).
Materials and Methods
Antimicrobial Susceptibility Testing
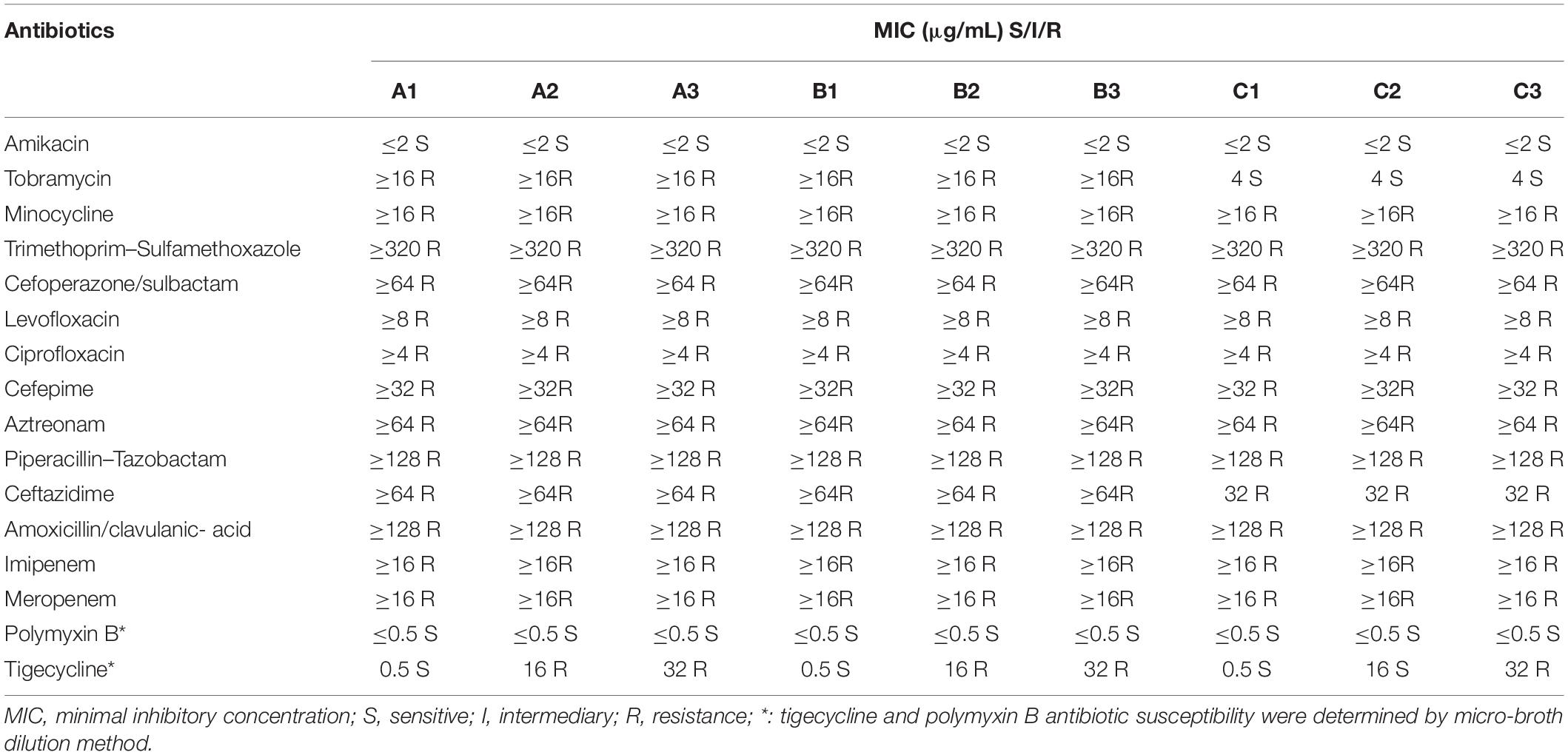
Antibiotic susceptibility testing was performed using the Vitek 2 system (BioMérieux, Marcy-l Étoile, France). The MICs of tigecycline and polymyxin B were determined using the micro-broth dilution method (BIO-KONT Co., Ltd., Wenzhou, China). The breakpoints of tigecycline and polymyxin B were interpreted according to the Clinical and Laboratory Standards Institute guidelines (Kuti et al., 2018). E. coli ATCC25922 and P. aeruginosa ATCC27853 were used as quality controls.
Laboratory Evolution of Tigecycline-Resistant Mutants
Independent single colonies of the CRKP strains KPN222 (A), KPN114 (B), and KPN315 (C) were grown overnight at 37°C. The cultures were then inoculated into Luria–Bertani (LB) agar with serially increasing concentrations of tigecycline, starting at a value equal to half of the MIC of the original strain and doubled when bacteria grew on the plates; the final concentration of tigecycline was 256 μg/mL. The resistant strains were transferred to a blood agar plate and continuously transferred 10 times, and then the MIC of tigecycline was determined to obtain the stable resistant strain. The tigecycline resistant strains were stored at −80°C until analysis.
RNA Extraction, Sequencing, and Data Analysis
The isolates were grown in high-osmolality LB broth at 37°C until the early exponential phase, and then RNA was extracted using the RNApure Bacteria kit (CWBio Co., Beijing, China) according to the manufacturer’s protocol. The concentration of RNA was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). RNA sequencing was performed using a HiSeq 2000 sequencer (Illumina, San Diego, CA, United States) and de novo assembly was carried out using Trinity software1. BLASTn (Altschul et al., 1990) was used to analyze Unigene NT with a Blastx (Altschul et al., 1990) or Diamond (Buchfink et al., 2015) search on Unigene for NR; KOG, KEGG, and SwissProt were used to analyze Blast2GO (Conesa et al., 2005) and the NR annotation results for Gene Ontology (GO) annotation. Bowtie 2 (Langmead and Salzberg, 2012) was used to compare the clean reads with the corresponding sequences in Unigene, and RSEM (Li and Dewey, 2011) was then used to calculate the gene expression levels of each sample. The PossionDis method was used for differential gene detection.
Quantitative PCR and Standard PCR
The expression levels of the CusS-CusR TCS were determined using quantitative qPCR. The primers used for the CusS-CusR TCS and other genes associated with tigecycline resistance are shown in Table 1. The primers used for the AcrAB-TolC efflux pump-associated genes soxS, acrB, rarA, ramA, ramR, marA, tolC, and acrA were obtained from the literature (He et al., 2015). The primers associated with the SdiA-AcrEF and RarA-OqxAB efflux pump are also shown in Table 2. Total bacterial RNA was extracted using the RNA Pure Bacteria kit (CWBIO Co., Beijing, China). cDNA was then synthesized using a Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). Finally, quantitative PCR (qPCR) was performed with the UltraSYBR Mixture kit (CWBIO Co., Beijing, China) using a Cobas z 480 analyzer (LightCycler 480, Roche, Basel, Switzerland) with initial incubation at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. The melting curve fluorescence was evaluated at five times per degree Celsius from 60 to 95°C. Each reaction was carried out in triplicate. For all samples, a housekeeping gene (16S rRNA-F: 5′-GTGCCAGCMGCCGCGGTAA-3′, 16S rRNA-R: 5′-GGCGTGGACTTCCAGGGTATCT-3′) (Chen et al., 2007) was used to normalize the expression of genes. The threshold cycle (CT) numbers were confirmed using the detection system software, and the data were analyzed using the 2ΔΔCt method. Gene expression levels were determined and compared with those of wild-type strains (A1, B1, and C1, expression = 1).
The mutations of tex (T) and tex (A), which are associated with tetracycline resistance, were detected using the standard PCR. The DreamTaq PCR Master Mix kit (Thermo Fisher Scientific) was used and the reaction system contained 25.0 μL of 2 × Dream Taq PCR Master Mix, 0.4 μL of forward and reverse primers (10 mol/L), and 4.0 μL of DNA template supplemented with 50.0 μL ddH2O. The reaction conditions were as follows: 95°C for 1 min; followed by 30 cycles of 95°C at 30 s, 50°C for 30 s, and 72°C for 1 min; with a final extension at 72°C for 8 min. All qPCR and standard PCR products were confirmed using agarose gel electrophoresis and sequencing.
Deletion and Complementation of the Gene cusR
Based on the homologous recombination technology, the deletion strain of the △cusR gene was constructed using the suicide vector pKO3-Km plasmid, and the gene fragment containing the activation region and the transcriptional termination region of the coding region of the C-△cusR gene was amplified. The gene fragment was cloned into the pGEM-T-easy plasmid and transferred to the deletion strain to construct the complemented strain of the C-△cusR gene. The primers are shown in Table 4.
Results
Antibiotic Susceptibility of the Strains
All strains were resistant to tobramycin, minocycline, trimethoprim–sulfamethoxazole, cefoperazone/sulbactam, levofloxacin, ciprofloxacin, cefepime, aztreonam, piperacillin–tazobactam, ceftazidime, amoxicillin/clavulanic acid, imipenem, and meropenem. The strains were only sensitive to amikacin and polymyxin B with MICs of 2 and 0.5 μg/mL, respectively. The tigecycline MIC of A1, B1, and C1 was 0.5 μg/mL; that of A2, B2, and C2 was 16 μg/mL; and that of A3, B3, and C3 was 32 μg/mL. Antibiotic susceptibility of the primary strains and induced strains is shown in Table 2.
RNA Sequencing
The differentially expressed genes identified by RNA-sequencing are shown in Table 3 and Supplementary Material. Unigene 28_All was 4997 bp long and the NCBI BLAST revealed that it contained cusE, cusS, cusR, cusC, and cusF. Unigene274_All was 1090 bp long and contained cusA. Unigene1733_All was 1111 bp long and contained cusB. These three genes showed higher expression in C2 and C3 than in C1. Unigene1142_All was 2137 bp long, annotated to the transcriptional regulatory gene ramA in the genes encoding the AcrAB-TolC efflux pump, and highly expressed in B2 and B3, with B2/B1 log2 fold-change = 3.31 and B3/B1 log2 fold-change = 3.61.
Quantitative PCR
The relative expression levels (fold-change) of genes related to the AcrAB-TolC efflux pump (soxS, acrB, rarA, ramA, ramR, marA, tolC, and acrA) in A2/A1 and A3/A1, B2/B1 and B3/B1, and C2/C1 and C3/C1 are shown in Figure 2. The expression level of AcrAB-TolC efflux pump-related factors was the highest in group B. The relative expression levels (fold-change) of genes related to the CusS-CusR TCS (cusF, cusB, cusC, cusA, cusS, cusR, and cusE) in A2/A1 and A3/A1, B2/B1 and B3/B1, and C2/C1 and C3/C1 are shown in Figure 3. CusS-CusR TCS showed the highest expression in group C.
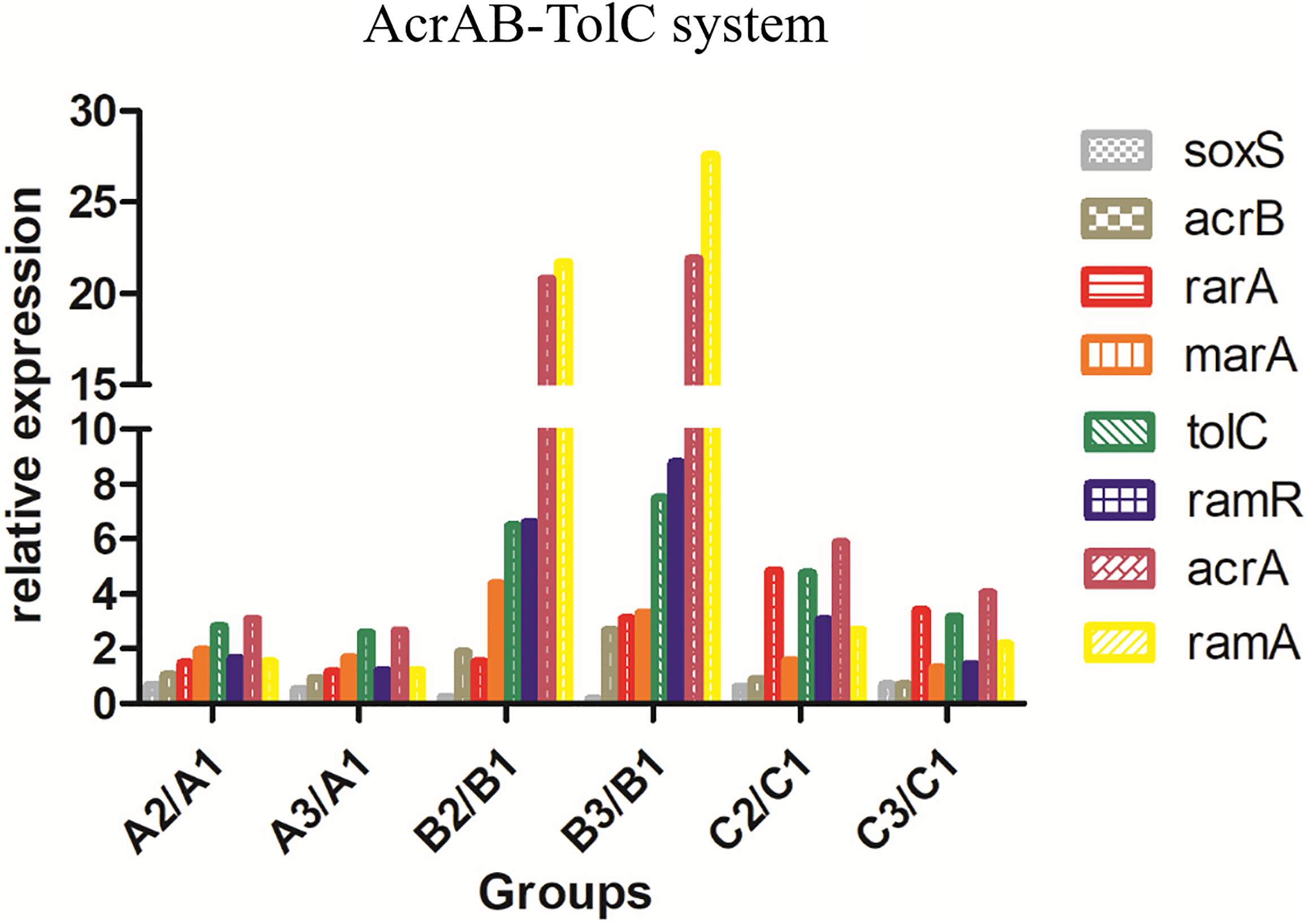
Figure 2. Relative expression of AcrAB-TolC in groups A, B, and C. Efflux pump AcrAB-TolC-associated genes: soxS, acrB, rarA, ramA, ramR, marA, tolC, and acrA. Group B showed higher expression than groups A and C.
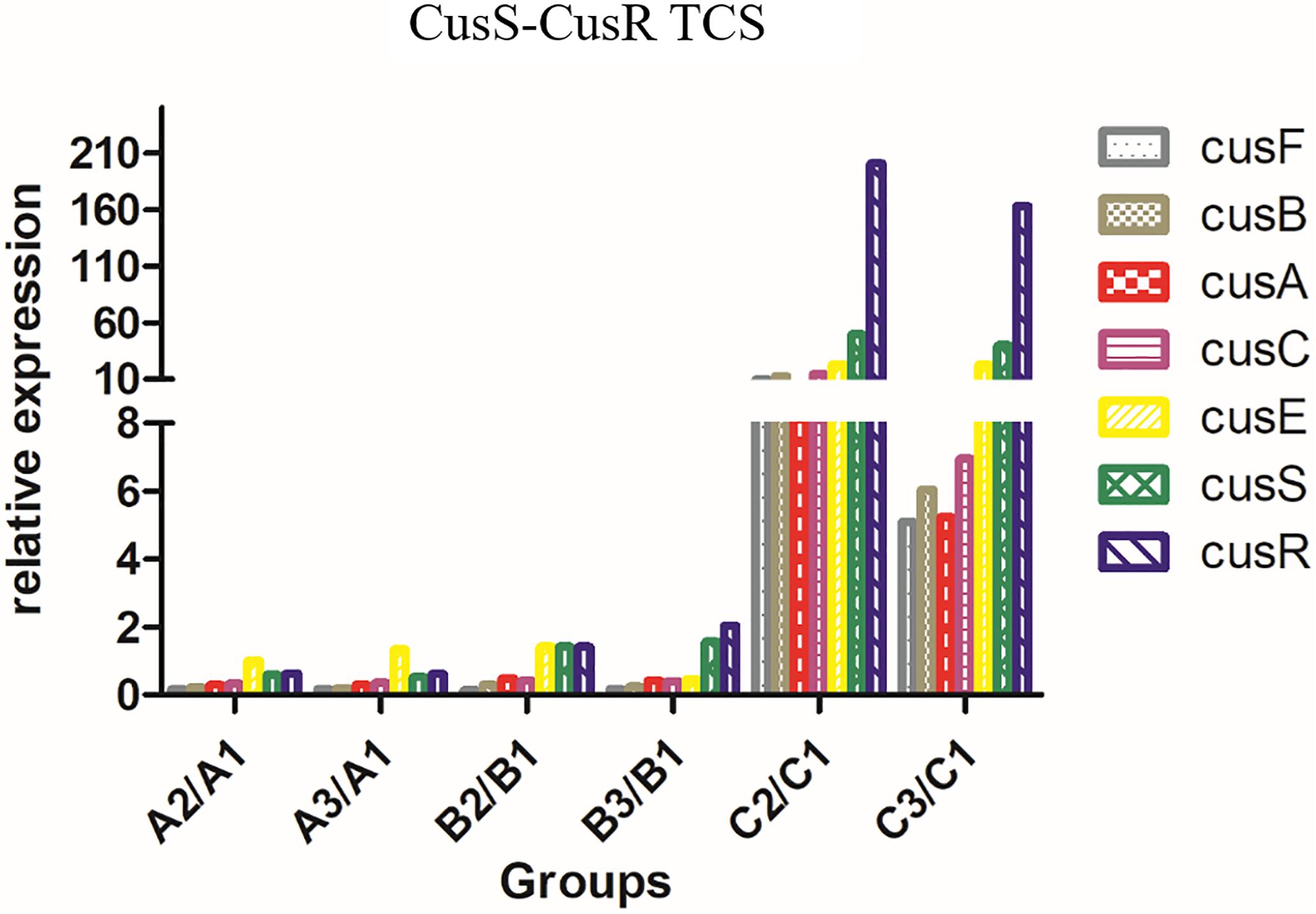
Figure 3. Relative expression of CusS-CusR TCS in groups A, B, and C. CusS-CusR TCS associated genes: cusF, cusB, cusC, cusA, cusS, cusR, and cusE. Group C showed higher expression than groups A and B.
Tigecycline Resistance-Associated Genes
We detected the AcrEF-TolC efflux pump and OqxAB-TolC efflux pump in A2, A3, B2, B3, C2, and C3. In contrast, the acrE, acrF, oqxA, and oqxB genes did not show higher expression than A1, B1, and C1. The ramR, tex (T), and tet (A) genes contained no mutations.
Effect of Deletion and Complementation
Successfully built a non-trace cusS gene deletion strain △C3 and the complemented strain C-△C3, and then verified by RT-PCR of the cusR gene in ΔC3 showing no expression. The tigecycline MIC of △C3 was decreased to 4 μg/mL and that of C-△C3 was recovered to 32–4 μg/mL, and the expression of cusFBCA correspondingly decreased and increased significantly.
Discussion
In the present study, we successfully screened three groups of tigecycline-resistant strains by RNA-sequencing, followed by annotation with biological information. We found that the AcrAB-TolC efflux pump system was highly expressed in group B, indicating that this system plays an essential role in the development of CRKP resistance to tigecycline. The AcrAB-TolC efflux pump is involved in resistance-nodulation cell division and has been reported to be resistant to several types of antibiotics (Zhong et al., 2014; Castanheira et al., 2016; Yuhan et al., 2016). However, Nielsen et al. (2014) reported that the IS5 insertion element increased the MIC of tigecycline by four-fold and was not dependent on the functional AcrAB-TolC efflux pump, suggesting that the AcrAB-TolC efflux pump is not the only mechanism causing tigecycline resistance. Hladicz et al. (2017) reported that RamR is a repressor that prevents the overexpression of ramA; when ramR is mutated, ramA is overexpressed, leading to tigecycline resistance; this mechanism was confirmed by Li et al. (2017). However, in the current study, no mutation in ramR was detected.
Zhang et al. (2018) reported that impairment of AcrAB-TolC function upregulates acrEF quinolone resistance in E. coli. Thus, we also detected acrE and acrF, but no increase in expression was observed. Juan et al. (2016) evaluated 43 strains each of tigecycline-non-susceptible KP and tigecycline-susceptible KP, and found that the overexpression of OqxAB efflux pumps was a major cause of tigecycline-non-susceptible KP resistance to tigecycline. However, the evolved strains in this study did not show an increase in their expression. Furthermore, the genes tex(T) and tet(A), which are associated with tigecycline resistance, also did not contain mutations, indicating that these genes were not involved in tigecycline resistance in groups A, B, and C.
Andrade (Andrade et al., 2018) showed that multidrug-resistant CTX-M-(15, 9, and 2) – and KPC-2-producing Enterobacter hormaechei and Enterobacter asburiae isolates possessed a set of acquired heavy metal tolerance genes including a chromosomal sil operon (for acquired silver resistance). Yuan et al. (2019) also indicated that mercury/silver resistance genes were associated with antibiotic resistance. Elkrewi et al. (2017) reported that cryptic Ag + resistance pertaining to the sil operon is prevalent and readily activated in particular genera (Enterobacter and Klebsiella). Conroy et al. (2010) reported that CusCFBA had a narrow substrate spectrum transporting Cu (I) and Ag (I); in this research, the MIC of tigecycline and the expression of cusCFBA presented significant changes after deletion and complementation of cusR. The cus system was clearly elaborated by Pal et al. (2017), however, further research is necessary to verify whether it is involved in cross-resistance or co-resistance to metal and resistance to antibiotics. In conclusion, we considered that in group C, the CusS-CusS TCS was activated and resistance was induced in C2/C3 strains to tigecycline.
We recognize some limitations to our study. The strains in group A did not show any changes in AcrAB-TolC efflux pump-related and CusS-CusR TCS-related gene expression. Additionally, genes associated with tigecycline resistance such as acrE, acrF, oqxA, and oqxB did not show increased expression, and tex (T) and tet (A) did not contain any mutations. Thus, other mechanisms may have caused A2 and A3 resistance to tigecycline, which requires additional analysis.
Conclusion
Despite these limitations, we found that the increased expression of the CusS-CusR TCS, which is associated with Cu and Ag resistance, mediated CRKP resistance to tigecycline, which may become a novel target of antibiotics. Thus, in cases where the common mechanisms are not identified as mediators of tigecycline resistance, the presence of high CusS-CusR TCS expression may be considered.
Data Availability Statement
The datasets generated for this study can be found in NCBI SRA https://www.ncbi.nlm.nih.gov/sra/PRJNA596084.
Author Contributions
YC, DC, and HL designed the experiments, analyzed the data, and wrote the manuscript. YZ, LX, YQ, HH, and DZ performed the experiments and analyzed the data. XL, XY, NX, XH, and FC analyzed the data.
Funding
This work was supported by the Scientific Innovation Project of Fujian Provincial Health and Family Planning Commission (Grant No. 2017-CX-3), the National Major Science and Technology Project for the Control and Prevention of Major Infectious Diseases of China (Grant No. 2017ZX10103004), the Fujian Medical Science and Technology Innovation Project (Grant No. 2016Y9005), the Fujian Provincial Natural Science Foundation Project (Grant No. 2019J01178), and the Startup Fund for scientific research, Fujian Medical University (Grant No. 2016QH116).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Bei Li from Basic Medical College of Hubei University of Medicine for providing the pKO3-Km plasmid and pGEM-T-easy plasmid.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03159/full#supplementary-material
DATA SHEET S1 | Transcriptome sequencing results of groups B and C.
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrade, L. N., Siqueira, T. E. S., Martinez, R., and Darini, A. L. C. (2018). Multidrug-Resistant CTX-M-(15, 9, 2) and KPC-2-Producing Enterobacter hormaechei and Enterobacter asburiae Isolates Possessed a Set of Acquired Heavy Metal Tolerance Genes Including a Chromosomal sil Operon (for Acquired Silver Resistance). Front. Microbiol. 9:539. doi: 10.3389/fmicb.2018.00539
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Castanheira, M., Deshpande, L. M., Mills, J. C., Jones, R. N., Soave, R., Jenkins, S. G., et al. (2016). Klebsiella pneumoniae Isolate from a New York City Hospital Belonging to Sequence Type 258 and Carrying blaKPC-2 and blaVIM-4. Antimicrob. Agents Chemother. 60, 1924–1927. doi: 10.1128/AAC.01844-15
Chen, S., Cui, S., McDermott, P. F., Zhao, S., White, D. G., Paulsen, I., et al. (2007). Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51, 535–542. doi: 10.1128/AAC.00600-06
Chiu, S. K., Huang, L. Y., Chen, H., Tsai, Y. K., Liou, C. H., Lin, J. C., et al. (2017). Roles of ramR and tet (A) Mutations in Conferring Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 61, e391–17. doi: 10.1128/AAC.00391-17
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Conroy, O., Kim, E. H., McEvoy, M. M., and Rensing, C. (2010). Differing ability to transport nonmetal substrates by two RND-type metal exporters. FEMS Microbiol. Lett. 308, 115–122. doi: 10.1111/j.1574-6968.2010.02006.x
Elkrewi, E., Randall, C. P., Ooi, N., Cottell, J. L., and O’Neill, A. J. (2017). Cryptic silver resistance is prevalent and readily activated in certain Gram-negative pathogens. J. Antimicrob. Chemother. 72, 3043–3046. doi: 10.1093/jac/dkx258
He, F., Fu, Y., Chen, Q., Ruan, Z., Hua, X., Zhou, H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. doi: 10.1371/journal.pone.0119064
He, F., Shi, Q., Fu, Y., Xu, J., Yu, Y., and Du, X. (2018). Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect. Genet. Evol. 66, 188–191. doi: 10.1016/j.meegid.2018.09.025
Hladicz, A., Kittinger, C., and Zarfel, G. (2017). Tigecycline Resistant Klebsiella pneumoniae Isolated from Austrian River Water. Int. J. Env. Res. Public Health 14:E1169. doi: 10.3390/ijerph14101169
Hölzel, C. S., Müller, C., Harms, K. S., Mikolajewski, S., Schäfer, S., Schwaiger, K., et al. (2012). Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 113, 21–27. doi: 10.1016/j.envres.2012.01.002
Juan, C. H., Huang, Y. W., Lin, Y. T., Yang, T. C., and Wang, F. D. (2016). Risk Factors, Outcomes, and Mechanisms of Tigecycline-Nonsusceptible Klebsiella pneumoniae Bacteremia. Antimicrob. Agents Chemother. 60, 7357–7363. doi: 10.1128/AAC.01503-16
Kuti, J. L., Wang, Q., Chen, H., Li, H., Wang, H., and Nicolau, D. P. (2018). Defining the potency of amikacin against Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii derived from Chinese hospitals using CLSI and inhalation-based breakpoints. Infect. Drug Resist. 11, 783–790. doi: 10.2147/IDR.S161636
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, R., Han, Y., Zhou, Y., Du, Z., Wu, H., Wang, J., et al. (2017). Tigecycline Susceptibility and Molecular Resistance Mechanisms among Clinical Klebsiella pneumoniae Strains Isolated during Non-Tigecycline Treatment. Microb. Drug Resist. 23, 139–146. doi: 10.1089/mdr.2015.0258
Nielsen, L. E., Snesrud, E. C., Onmus-Leone, F., Kwak, Y. I., Avilés, R., Steele, E. D., et al. (2014). IS5 element integration, a novel mechanism for rapid in vivo emergence of tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58, 6151–6156. doi: 10.1128/AAC.03053-14
Pal, C., Asiani, K., Arya, S., Rensing, C., Stekel, D. J., Larsson, D. G. J., et al. (2017). Metal Resistance and Its Association with Antibiotic Resistance. Adv. Microb. Physiol. 70, 261–313. doi: 10.1016/bs.ampbs.2017.02.001
Perron, K., Caille, O., Rossier, C., Van Delden, C., Dumas, J. L., and Köhler, T. (2004). CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279, 8761–8768. doi: 10.1074/jbc.M312080200
Pournaras, S., Koumaki, V., Spanakis, N., Gennimata, V., and Tsakris, A. (2016). Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int. J. Antimicrob. Agents 48, 11–18. doi: 10.1016/j.ijantimicag.2016.04.017
Sheng, Z. K., Hu, F., Wang, W., Guo, Q., Chen, Z., Xu, X., et al. (2014). Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 58, 6982–6985. doi: 10.1128/AAC.03808-14
Wang, D., Chen, W., Huang, S., He, Y., Liu, X., Hu, Q., et al. (2017). Structural basis of Zn (II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa. PLoS Pathog. 13:e1006533. doi: 10.1371/journal.ppat.1006533
Ye, M., Ding, B., Qian, H., Xu, Q., Jiang, J., Huang, J., et al. (2017). In vivo development of tigecycline resistance in Klebsiella pneumoniae owing to deletion of the ramR ribosomal binding site. Int. J. Antimicrob. Agents 50, 523–528. doi: 10.1016/j.ijantimicag.2017.04.024
Yuan, L., Li, Z. H., Zhang, M. Q., Shao, W., Fan, Y. Y., and Sheng, G. P. (2019). Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci. Total Environ. 657, 1014–1022. doi: 10.1016/j.scitotenv.2018.12.088
Yuhan, Y., Ziyun, Y., Yongbo, Z., Fuqiang, L., and Qinghua, Z. (2016). Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of Acinetobacter baumannii and Klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 49, 165–171. doi: 10.1590/0037-8682-0411-2015
Zhang, C. Z., Chang, M. X., Yang, L., Liu, Y. Y., Chen, P. X., and Jiang, H. X. (2018). Upregulation of AcrEF in Quinolone Resistance Development in Escherichia coli When AcrAB-TolC Function Is Impaired. Microb. Drug Resist. 24, 18–23. doi: 10.1089/mdr.2016.0207
Keywords: Klebsiella pneumoniae, carbapenem resistance, tigecycline resistance, CusS-CusR two-component system, RNA sequencing
Citation: Chen D, Zhao Y, Qiu Y, Xiao L, He H, Zheng D, Li X, Yu X, Xu N, Hu X, Chen F, Li H and Chen Y (2020) CusS-CusR Two-Component System Mediates Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae. Front. Microbiol. 10:3159. doi: 10.3389/fmicb.2019.03159
Received: 19 August 2019; Accepted: 30 December 2019;
Published: 28 January 2020.
Edited by:
Henrietta Venter, University of South Australia, AustraliaReviewed by:
Christopher Rensing, Fujian Agriculture and Forestry University, ChinaJozsef Soki, University of Szeged, Hungary
Copyright © 2020 Chen, Zhao, Qiu, Xiao, He, Zheng, Li, Yu, Xu, Hu, Chen, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongru Li, bXV6aTEzMTEyMkAxNjMuY29t; Yusheng Chen, Y3lza3R6QDE2My5jb20=; Y2hlbnl1c2hlbmc4MjAxQDE2My5jb20=
†These authors share first authorship
 Dongjie Chen
Dongjie Chen Yunan Zhao1†
Yunan Zhao1†