- 1MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI, United States
- 2Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, United States
- 3Environmental Genomics and Systems Biology and Molecular Biophysics and Integrated Bioimaging Divisions, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 4Department of Microbiology & Molecular Genetics, Michigan State University, East Lansing, MI, United States
Two proteins found in cyanobacteria contain a C-terminal domain with homology to the small subunit of rubisco (RbcS). These small subunit-like domains (SSLDs) are important features of CcmM, a protein involved in the biogenesis of carboxysomes found in all β-cyanobacteria, and a rubisco activase homolog [activase-like protein of cyanobacteria (ALC)] found in over a third of sequenced cyanobacterial genomes. Interaction with rubisco is crucial to the function of CcmM and is believed to be important to ALC as well. In both cases, the SSLD aggregates rubisco, and this nucleation event may be important in regulating rubisco assembly and activity. Recently, two independent studies supported the conclusion that the SSLD of CcmM binds equatorially to L8S8 holoenzymes of rubisco rather than by displacing an RbcS, as its structural homology would suggest. We use sequence analysis and homology modeling to examine whether the SSLD from the ALC could bind the large subunit of rubisco either via an equatorial interaction or in an RbcS site, if available. We suggest that the SSLD from the ALC of Fremyella diplosiphon could bind either in a vacant RbcS site or equatorially. Our homology modeling takes into account N-terminal residues not represented in available cryo-electron microscopy structures that potentially contribute to the interface between the large subunit of rubisco (RbcL) and RbcS. Here, we suggest the perspective that binding site variability as a means of regulation is plausible and that the dynamic interaction between the RbcL, RbcS, and SSLDs may be important for carboxysome assembly and function.
Introduction
Due to the evolution of a carbon concentrating mechanism (CCM), cyanobacteria are able to significantly contribute to global carbon fixation, despite the comparatively low atmospheric carbon dioxide (CO2) levels relative to their first appearance on Earth some 3.5 billion years ago (Schopf, 2012; Whitton and Potts, 2012). The CCM serves to significantly increase the flux of inorganic carbon into proteinaceous bacterial microcompartments called carboxysomes. Carboxysomes serve to encapsulate rubisco, and the shell acts as a semipermeable barrier to CO2 escape, allowing rubisco to function under high substrate levels (Dou et al., 2008). In the case of β-carboxysomes, which are present in cyanobacteria that express form 1B rubisco, synthesis occurs from the inside-out beginning with condensation of rubisco and carboxysomal protein CcmM into a liquid matrix (Cameron et al., 2013; Niederhuber et al., 2017; Wang et al., 2019).
The structure of CcmM is key to its nucleation of carboxysomal cargo. The C-terminus of CcmM contains three to five repeats of a domain that is homologous (around 60–70% similarity) to the small subunit of rubisco (RbcS)—denoted a small subunit-like domain (SSLD) (Price et al., 1993; Ludwig et al., 2000). The SSLD repeats domain-containing portion of CcmM can also be independently translated through an internal ribosome entry site. Both CcmM forms—full-length M58 and truncated M35—are necessary for normal carboxysome biogenesis (Long et al., 2010, 2011).
SSLDs were implicated in the interaction between rubisco and CcmM (Long et al., 2007; Cot et al., 2008) and were long hypothesized to bind in place of RbcS in rubisco complexes (Espie and Kimber, 2011; Long et al., 2011; Rae et al., 2013). However, recent structural work on SSLDs demonstrates equatorial binding of CcmM to L8S8 rubisco holoenzymes (Ryan et al., 2018; Wang et al., 2019). This binding appears to be driven largely by electrostatic interactions, and affinity for CcmM is not affected even when RbcS binding is partially compromised (Ryan et al., 2018).
SSLDs also appear as C-terminal domains in rubisco activase homologs [activase-like protein of cyanobacteria (ALC)] found in many cyanobacteria (Zarzycki et al., 2013). Recently, the ALC was shown to localize proximal to rubisco in the carboxysome and induce rubisco aggregation, much like M35 (Lechno-Yossef et al., 2019). Together, these findings provide strong evidence that the SSLD of ALC binds to rubisco and can induce coalescence of rubisco.
As recent findings indicate that SSLDs do not displace RbcS in L8S8 rubisco holoenzymes and instead bind equatorially, it is puzzling why there would be conservation of the RbcS-like secondary and tertiary structure elements that facilitate interactions with the large subunit of rubisco (RbcL). Some of the conserved residues from RbcS may fill repurposed roles in the SSLD-unique equatorial binding position, thus driving conservation of these features. Others, though, suggest that RbcS displacement may be possible. We decided to homology model the SSLD and RbcL and systematically compare the interfaces in order to evaluate the plausibility of equatorial versus RbcS substitution as a mode of binding (Ryan et al., 2018; Wang et al., 2019). We modeled the SSLD found in the ALC of Fremyella diplosiphon (FdALC SSLD) and analyzed the number and type of predicted interactions and free energy of solvation when the SSLD binds at the RbcS site (i.e., binds an empty site or displaces RbcS) or binds equatorially. We suggest that while equatorial binding was favored for CcmM in Synechococcus elongatus PCC 7942 (hereafter Syn7942), which lacks an ALC homolog, the FdALC SSLD had similar interface features in both positions. We propose that the FdALC could bind either equatorially or in place of RbcS and suggest that the current models of equatorial SSLD may be a part of a larger set of possibilities depending on specific proteins, for example, whether or not the cyanobacterium contains an ALC and perhaps is reflective of the recently uncovered diversity of cyanobacterial RbcL subunits (Lechno-Yossef et al., 2019).
Materials and Methods
Protein Homology Modeling
The structures of the Fremyella proteins were generated by homology modeling. For FdALC SSLD, the Swiss Model web server1 (Bertoni et al., 2017; Waterhouse et al., 2018) was used to generate a model for amino acid residues 317–424 based on Syn7942 CcmM SSLD1 in the reduced (PDB: 6HBB) form as well as Synechococcus sp. PCC 6301 (hereafter Syn6301) RbcS (PDB: 1RBL, Chain M). Additionally, a homology model of Fremyella L8S8 rubisco was made using Syn6301 rubisco (PDB: 1RBL) as a template. Alignment scores between two sequences were calculated using the LAlign webserver2 in order to evaluate candidate template structures and to compare primary structure conservation.
Multiple Sequence Alignment of Activase-Like Protein of Cyanobacteria Small Subunit-Like Domains
The multiple sequence alignment (MSA) of 141 ALCs from cyanobacteria described in Lechno-Yossef et al. (2019) was trimmed to remove the ATPase domain, then the remaining regions (linker and SSLD) were realigned with a low gap cost at the end of sequences in CLC Sequence Viewer. This allowed for alignment of the SSLD region despite significant variations in the sizes of linkers between species, which were then trimmed to match the SSLD region identified in FdALC (residues 317–424, corresponding to residues 1–107 of the SSLD). An MSA was also generated for RbcS for each of the 128 organisms that had both a full-length SSLD and an annotated RbcS sequence. MSA for RbcS and SSLD were visualized and compared using HMM logos (Schuster-Böckler et al., 2004) generated on Skylign3.
Analysis of Protein–Protein Interactions
Using the homology model for L8S8 Fremyella rubisco, the two FdALC SSLD models were aligned to RbcS1 in PyMol, and structures were generated containing each SSLD replacing RbcS1. Another structure aligned Syn7942 CcmM SSLD1 in complex with rubisco (PDB: 6HBC) to the Fremyella rubisco, and then FdALC SSLD was aligned to the CcmM SSLD resulting in a Fremyella rubisco model with FdALC SSLD in the M position (Figure 1A). The Syn7942 CcmM SSLD1 structure was also used to replace the RbcS1 position in the Syn6301 rubisco L8S8 structure. Local refinement of structures was performed using Rosetta 3.4. Structures were subjected to the docking prepack protocol followed by the generation of 1,000 decoys using the docking protocol in docking local refine mode with the SSLD as the mobile target (Gray et al., 2003; Wang et al., 2005; Chaudhury et al., 2011). Based on interface score, the top 200 structures were clustered by pairwise RMSD with a 1 Å cutoff using energy-based clustering in Rosetta 3.4 (Hosseinzadeh et al., 2017). In all cases, the structure with the lowest interface score belonged to the largest cluster and was selected for use in downstream analysis.
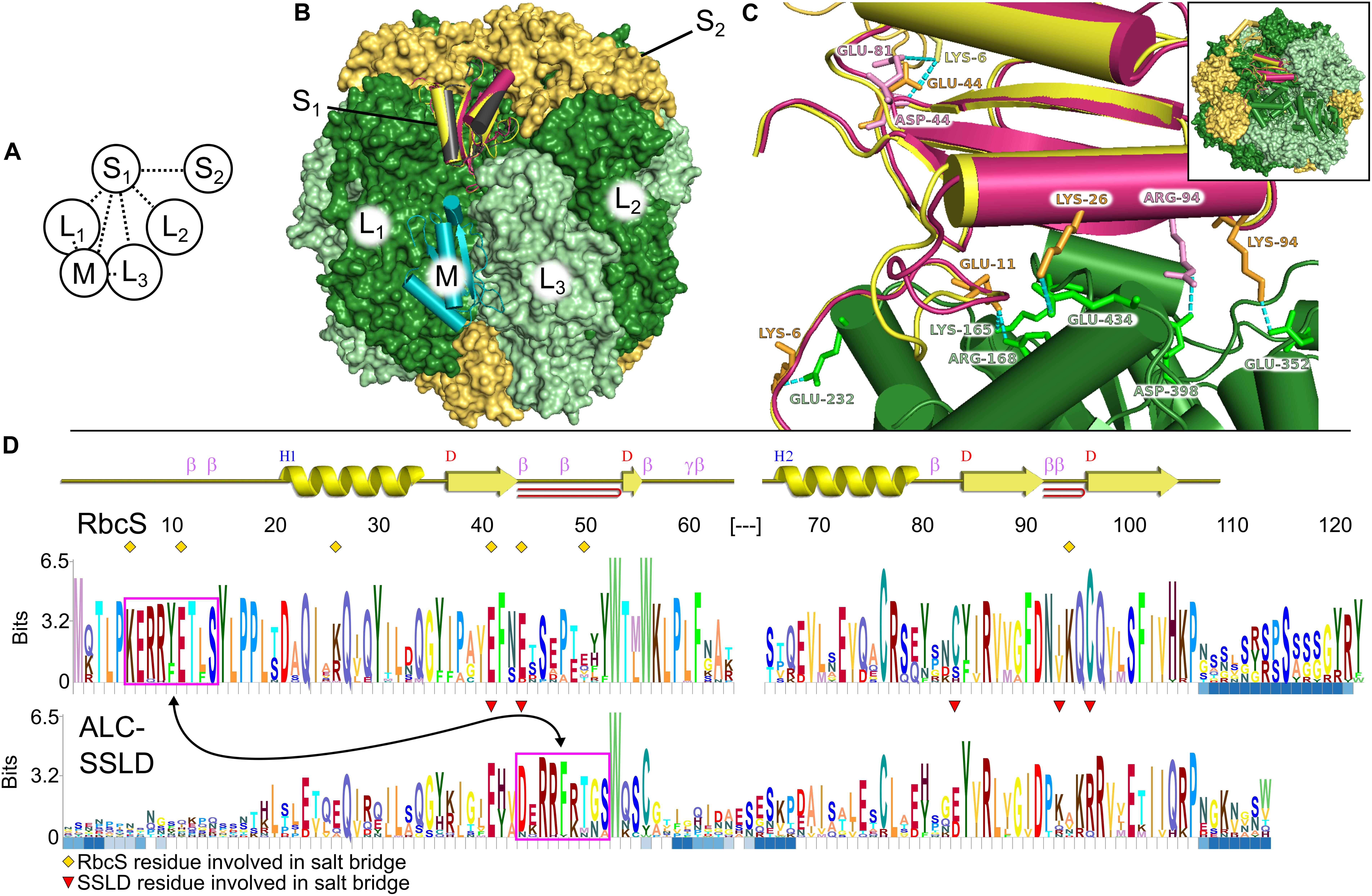
Figure 1. Structural comparison of small subunit of rubisco (RbcS) and small subunit-like domain (SSLD). (A) Schematic diagram of the interactions that a single RbcS (S1) or SSLD (M) has in a rubisco holoenzyme. Numbering based on van Lun et al. (2011). (B) Alignment of the SSLD found in activase-like proteins of cyanobacteria (ALCs) (black is based on an SSLD model, burgundy on an RbcS model) to the RbcS (yellow) or SSLD (teal) binding sites of an L8S8 rubisco holoenzyme. (C) Interaction between the large subunit of rubisco (RbcL1) (green) and RbcS (yellow) or SSLD (burgundy) in the context of the RubisCO holoenzyme. Labeled residues highlight residues predicted to interact in a salt bridge interaction (Supplementary Table S1), with label color based on the subunit it is from. Numbering for the SSLD is based on the trimmed region beginning at residue 317 in the full-length FdALC. Dashed cyan lines show the predicted interacting atoms. Inset indicates a zoomed-out view of the holoenzyme. (D) HMM-Logo highlighting the areas of sequence conservation between RbcS and the SSLD of ALC across cyanobacteria containing both. A schematic for the secondary structure of the homology model for RbcS from Fremyella is presented above the two logos. Blue squares below each residue depict regions with gaps in significant portions of the multiple sequence alignment (MSA). Magenta boxes, connected by an arrow, indicate a motif found in both MSAs. Yellow diamonds indicate the RbcS residues involved in salt bridge interactions in the Fremyella homology model, while red triangles indicate ALC-SSLD residues involved in salt bridge interactions in the S1 position in the homology model (Supplementary Table S1).
These structures, as well as the Fremyella rubisco model and PDB: 6HBC, were analyzed using the Profunc web server4 (Laskowski, 2017). Interactions involving the RbcS and SSLD were compared for each structure. Further analyses were performed using the Pisa web server5 (Krissinel and Henrick, 2007) to calculate the solvation free energy gain (ΔG) upon formation of the interfaces for each structure.
Binding at the RbcS1 Position
Each RbcS in an L8S8 assembly forms four unique protein–protein interfaces, three with the surrounding RbcL subunits and a fourth with a proximal RbcS (Figures 1A,B). The number of interfaces and predicted residue-level interactions were comparable to results from molecular dynamic simulations using Chlamydomonas reinhardtii rubisco (van Lun et al., 2011) and crystal structures (Knight et al., 1990). RbcS interactions with its nearest RbcL (RbcL1; Figure 1A) are substantial, burying ∼1,600 Å2 of surface area, with five predicted salt bridges, 13 hydrogen bonds and a free energy of solvation of −5.2 kcal⋅mol–1 (Table 1). The remaining interfaces bury less area and contain fewer bond interactions but favorably contribute to the overall L8S8 rubisco (Table 1, Column 5–FdRbcS).
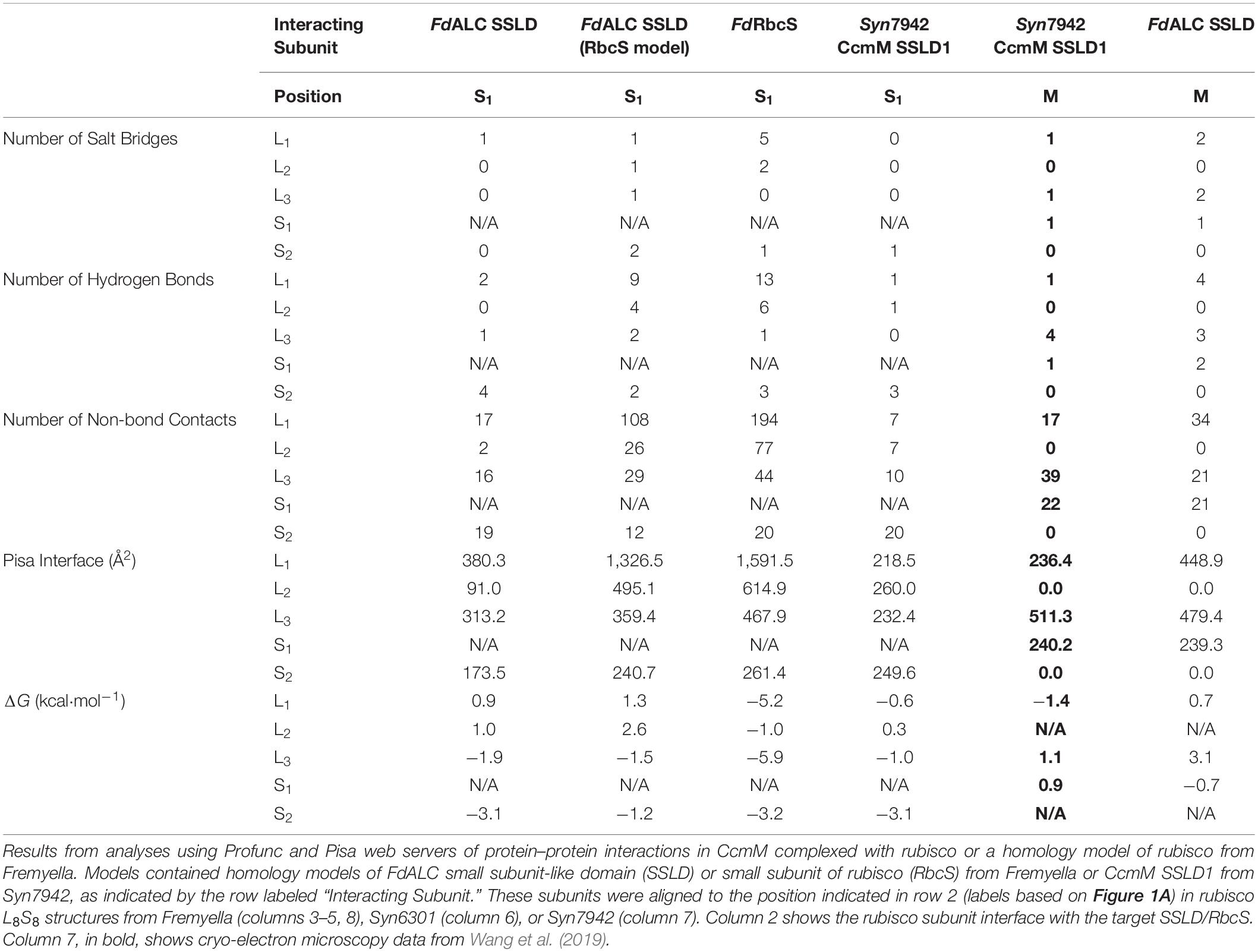
Table 1. Predicted small subunit interactions of rubisco from Fremyella diplosiphon and Synechococcus elongatus PCC 7942.
When RbcS is replaced with the SSLD-modeled FdALC SSLD, most salt bridges are lost (Table 1, Column 3–FdALC SSLD; Supplementary Table S1), as are many hydrogen bonds. This is particularly true at the L1-S1 interface, where the absence of the crucial N-terminal loop (residues 3–17) of RbcS in the SSLD accounts for three of the four missing salt bridges at this interface, as well as the significant reduction of major buried surface area (Knight et al., 1990; Ryan et al., 2018). SSLDs have two features that may play a role in this interaction. First, SSLDs have a poorly conserved flexible linker at their N-terminus that could be involved in non-specific interactions. Additionally, a portion of the N-terminal loop in RbcS involved in L1-S1 interactions is positionally displaced in the primary structures of SSLDs (Figure 1D, magenta box) (Ludwig et al., 2000). Notably, the structural position of this region corresponds to a helix in the SSLD structures but a loop in RbcS (Ryan et al., 2018; Wang et al., 2019). This “displaced motif” region is conserved and resembles the important lost motif of the N-terminus of RbcS but was not noted in Ryan et al. (2018) nor Wang et al. (2019) possibly because without significant backbone rearrangement, this motif is unlikely to be positioned to bind in the same way and its conservation could be attributed to its role in binding at the SSLD equatorial interface. Overall, our modeling with the truncated SynCcmM SSLD template is consistent with the experimental observations that the SSLD structure has a significant loss of favorable binding interactions at the RbcS interface.
When the FdALC SSLD is modeled using RbcS as a template, part of the linker at the N-terminus of the SSLD (residues 1–17) is included in the model. Analysis of the FdALC SSLD in complex with rubisco suggested the potential for conservation of significantly more interactions [Table 1, Column 4–FdALC SSLD (RbcS model)]. Compared to the native RbcS, each interface buries slightly less area (∼80% of that observed for RbcS) and is predicted to contain fewer hydrogen bonds and non-bonding contacts. Many salt bridges are potentially maintained or are similar for a total of five compared to the eight found in RbcL–RbcS (Supplementary Table S1). For example, the SSLD model loses two salt bridges that contribute to the three structural checkpoints of the L1-S interface described in van Lun et al. (2011) (Figure 1C); this FdALC SSLD model is predicted to form a novel salt bridge with L3, and K6 forms an additional salt bridge with S2 instead of L1. Although the SSLD of ALCs shows many regions of relatively low conservation, the regions that are important for RbcS interactions are generally well conserved even in the SSLD (Figure 1D), with the notable uncertainty of the displaced N-terminal motif (magenta box) and linker. This suggests that when the flexible linker domain is also taken into account, FdALC SSLD could occupy an empty RbcS site.
Binding at the Equatorial (M) Position
As reported in Wang et al. (2019), SynCcmM SSLD1 forms favorable interactions with L8S8 rubisco in an equatorial position (we refer to this as position M). It forms a salt bridge with each rubisco subunit it contacts (L1, L3, and S1) and forms some hydrogen bonds (Table 1, Column 7–Syn7942 CcmM SSLD1). The ΔG at these three interfaces are less favorable overall than those calculated for the four interfaces of FdALC SSLD (Table 1, Column 3) and SynCcmM SSLD1 in the S1 position (Table 1, Column 6–Syn7942 CcmM SSLD1). However, considering the presence of three salt bridges and that all RbcS positions were occupied in Ryan et al. (2018), these data are consistent with the model that SSLDs would bind equatorially rather than displace an engaged RbcS subunit.
FdALC SSLD also shows potential for interaction with the equatorial position, although the ΔG values calculated from these preliminary models are relatively less favorable. When bound at M, it is predicted to form a greater number of salt bridges and more hydrogen bonds compared to the SynCcmM SSLD1 (Table 1, Column 8–FdALC SSLD).
Discussion
Here, we present a prediction that the SSLDs found in cyanobacteria may be able to substitute for RbcS in binding RbcL; we propose that this could occur in addition to recently demonstrated equatorial binding (Ryan et al., 2018; Wang et al., 2019). Our analysis considers the SSLDs found in the absolutely conserved carboxysomal protein CcmM and the SSLDs found in the ALC, which is present in a subset of ecophysiologically diverse cyanobacteria. In the case of the FdALC SSLD, we found that its predicted binding with RbcL when substituting for the RbcS may be more favorable than that of the SSLD of CcmM of Syn7942, an organism that lacks the ALC. Although we find that the FdALC SSLD also could engage at the equatorial site, it may be a less favorable interaction than that observed for the SSLD of CcmM (Ryan et al., 2018; Wang et al., 2019). We suggest that the SSLD of FdALC has the potential for both equatorial binding and binding in the RbcS position. For Syn7942, the model organism used by Wang et al. (2019) and more closely related to Thermosynechococcus elongatus (Ryan et al., 2018), which also lacks an ALC, the predicted interface appears to point more favorably toward the equatorial binding found in vitro. Thus, it is possible that both the type of SSLD and the organism (i.e., class of RbcL) could influence whether the SSLD binds exclusively equatorially, especially for interactions with L8S8 rubisco holoenzymes. Additionally, in no case did we find that the SSLD bound better than the native RbcS, supporting the view that SSLDs cannot displace RbcS, though they might bind if sites are available.
In quantifications of protein abundance in cyanobacteria, a shortfall of RbcS relative to RbcL has been reported (Long et al., 2007, 2011; Sun et al., 2019). This finding suggests that isoforms other than the L8S8 rubisco holoenzyme may be present in vivo, although further substantiating analyses and investigations across multiple species as well as conditions are needed. Experiments in Syn7942 report five to six RbcS for eight RbcL, suggesting that vacant RbcS binding sites could be available in vivo (Long et al., 2011; Sun et al., 2019). Additionally, we suggest it may be possible that the RbcL:RbcS ratio could be dynamically regulated with impacts on both enzyme activity and the binding position of RbcS. Moreover, rubisco undergoes numerous posttranslational modifications that regulate its activity and subunit interactions (reviewed in Grabsztunowicz et al., 2017). In plants, phosphorylation influences rubisco kinetics (Lohrig et al., 2009) and modulate the interactions between RbcL, RbcS, and rubisco activase (Guitton and Mache, 1987; Aggarwal et al., 1993). Phosphorylation reversibly alters surface electrostatics potentially affecting the extent of equatorial SSLD interations. Notably, both the large and small subunits of cyanobacterial rubisco are targets of phosphorylation (Mikkat et al., 2014; Spät et al., 2015; Angeleri et al., 2016). These factors suggest mechanisms by which RbcS and SSLDs could be targets of dynamic regulation.
Throughout the evolution of diverse organisms containing Form I rubisco, a RbcL–RbcS fusion has never been observed. Given the importance of rubisco for survival, this is a significant clue to the potential for dynamic regulation of the RbcS:RbcL stoichiometry in cyanobacteria, potentially by the SSLDs. Though recent observations suggest that SSLDs bind primarily equatorially, we propose the possibility of a dynamic relationship between multiple binding locations. Such dynamics could play a large role in the nucleation of carboxysomes, which is fascinating given the observed impact that SSLD-containing proteins have on carboxysome morphology (Long et al., 2011; Rohnke et al., 2018). These features would seem to depend heavily on the availability of RbcS binding locations, the flexibility of the protein structures, redox state, posttranslational modifications, the species, the composition of the linker, the type of SSLD, and potentially even the subtype of RbcL (Lechno-Yossef et al., 2019), factors to consider in future investigations on the interaction(s) between rubisco and SSLDs.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
BR designed and conducted the research, analyzed and interpreted the data, and wrote the manuscript. CK and BM designed the research, analyzed and interpreted the data, and wrote the manuscript.
Funding
This work was supported by the United States Department of Energy (Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, grant no. DE-FG02-91ER20021 to BM and CK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Sigal Lechno-Yossef for critical discussion of the data and interpretation of results and Dr. Kaillathe “Pappan” Padmanabhan of the Macromolecular Computing Facility in the Michigan State University’s Department of Biochemistry and Molecular Biology for assistance with the Rosetta installation and the use of Linux clusters.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00187/full#supplementary-material
Footnotes
- ^ https://swissmodel.expasy.org
- ^ https://embnet.vital-it.ch/software/LALIGN_form.html
- ^ http://skylign.org/
- ^ https://www.ebi.ac.uk/thornton-srv/databases/profunc/
- ^ https://www.ebi.ac.uk/msd-srv/prot_int/
References
Aggarwal, K. K., Saluja, D., and Sachar, R. C. (1993). Phosphorylation of rubisco in Cicer rietinum: non-phosphoprotein nature of rubisco in Nicotiana tabacun. Phytochemistry 34, 329–335. doi: 10.1016/0031-9422(93)80004-C
Angeleri, M., Muth-Pawlak, D., Aro, E. M., and Battchikova, N. (2016). Study of O-phosphorylation sites in proteins involved in photosynthesis-related processes in Synechocystis sp. strain PCC 6803: application of the SRM approach. J. Proteome Res. 15, 4638–4652. doi: 10.1021/acs.jproteome.6b00732
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., and Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7:10480. doi: 10.1038/s41598-017-09654-8
Cameron, J. C., Wilson, S. C., Bernstein, S. L., and Kerfeld, C. A. (2013). Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155, 1131–1140. doi: 10.1016/j.cell.2013.10.044
Chaudhury, S., Berrondo, M., Weitzner, B. D., Muthu, P., Bergman, H., and Gray, J. J. (2011). Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One 6:e0022477. doi: 10.1371/journal.pone.0022477
Cot, S. S.-W., So, A. K.-C., and Espie, G. S. (2008). A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 190, 936–945. doi: 10.1128/JB.01283-07
Dou, Z., Heinhorst, S., Williams, E. B., Murin, C. D., Shively, J. M., and Cannon, G. C. (2008). CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283, 10377–10384. doi: 10.1074/jbc.M709285200
Espie, G. S., and Kimber, M. S. (2011). Carboxysomes: cyanobacterial rubisco comes in small packages. Photosynth. Res. 109, 7–20. doi: 10.1007/s11120-011-9656-y
Grabsztunowicz, M., Koskela, M. M., and Mulo, P. (2017). Post-translational modifications in regulation of chloroplast function: recent advances. Front. Plant Sci. 8:240. doi: 10.3389/fpls.2017.00240
Gray, J. J., Moughon, S., Wang, C., Schueler-Furman, O., Kuhlman, B., Rohl, C. A., et al. (2003). Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299. doi: 10.1016/S0022-2836(03)00670-3
Guitton, C., and Mache, R. (1987). Phosphorylation in vitro of the large subunit of the ribulose-1,5-bisphosphate carboxylase and of the glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 166, 249–254. doi: 10.1111/j.1432-1033.1987.tb13509.x
Hosseinzadeh, P., Bhardwaj, G., Mulligan, V. K., Shortridge, M. D., Craven, T. W., Pardo-Avila, F., et al. (2017). Comprehensive computational design of ordered peptide macrocycles. Science 358, 1461–1466. doi: 10.1126/science.aap7577
Knight, S., Andersson, I., and Brändén, C.-I. (1990). Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2⋅4 Å resolution: subunit interactions and active site. J. Mol. Biol. 215, 113–160. doi: 10.1016/S0022-2836(05)80100-7
Krissinel, E., and Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. doi: 10.1016/j.jmb.2007.05.022
Laskowski, R. A. (2017). The ProFunc function prediction server. Methods Mol. Biol. 1611, 75–95. doi: 10.1007/978-1-4939-7015-5_7
Lechno-Yossef, S., Rohnke, B. A., Belza, A. C. O., Melnicki, M. R., Montgomery, B. L., and Kerfeld, C. A. (2019). Cyanobacterial carboxysomes contain an unique rubisco-activase-like protein. New Phytol. 225, 793–806. doi: 10.1111/nph.16195
Lohrig, K., Müller, B., Davydova, J., Leister, D., and Wolters, D. A. (2009). Phosphorylation site mapping of soluble proteins: bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta 229, 1123–1134. doi: 10.1007/s00425-009-0901-y
Long, B. M., Badger, M. R., Whitney, S. M., and Price, G. D. (2007). Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282, 29323–29335. doi: 10.1074/jbc.M703896200
Long, B. M., Rae, B. D., Badger, M. R., and Price, G. D. (2011). Over-expression of the [beta]-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth. Res. 109, 33–45. doi: 10.1007/s11120-011-9659-8
Long, B. M., Tucker, L., Badger, M. R., and Price, G. D. (2010). Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 153, 285–293. doi: 10.1104/pp.110.154948
Ludwig, M., Sultemeyer, D., and Price, G. D. (2000). Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp PCC7002 (Cyanophyceae) and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 36, 1109–1119. doi: 10.1046/j.1529-8817.2000.00028.x
Mikkat, S., Fulda, S., and Hagemann, M. (2014). A 2D gel electrophoresis-based snapshot of the phosphoproteome in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 160, 296–306. doi: 10.1099/mic.0.074443-0
Niederhuber, M. J., Lambert, T. J., Yapp, C., Silver, P. A., and Polka, J. K. (2017). Superresolution microscopy of the β-carboxysome reveals a homogeneous matrix. Mol. Biol. Cell. 28, 2734–2745. doi: 10.1091/mbc.E17-01-0069
Price, G. D., Howitt, S. M., Harrison, K., and Badger, M. R. (1993). Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol. 175, 2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993
Rae, B. D., Long, B. M., Badger, M. R., and Price, G. D. (2013). Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some Proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379. doi: 10.1128/MMBR.00061-12
Rohnke, B. A., Singh, S. P., Pattanaik, B., and Montgomery, B. L. (2018). RcaE-dependent regulation of carboxysome structural proteins has a central role in environmental determination of carboxysome morphology and abundance in Fremyella diplosiphon. MSphere 3:e617. doi: 10.1128/mSphere.00617-17
Ryan, P., Forrester, T. J. B., Wroblewski, C., Kenney, T. M. G., Kitova, E. N., Klassen, J. S., et al. (2018). The small RbcS-like domains of the β-carboxysome structural protein, CcmM, bind rubisco at a site distinct from that binding the RbcS subunit. J. Biol. Chem. 294, 2593–2603. doi: 10.1074/jbc.RA118.006330
Schopf, J. W. (2012). “The fossil record of cyanobacteria,” in Ecology of Cyanobacteria II: Their Diversity in Space and Time, ed. B. A. Whitton (New York, NY: Springer), 15–36. doi: 10.1007/978-94-007-3855-3_2
Schuster-Böckler, B., Schultz, J., and Rahmann, S. (2004). HMM Logos for visualization of protein families. BMC Bioinformatics 5:7. doi: 10.1186/1471-2105-5-7
Spät, P., Maček, B., and Forchhammer, K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6:248. doi: 10.3389/fmicb.2015.00248
Sun, Y., Wollman, A. J. M., Huang, F., Leake, M. C., and Liu, L.-N. (2019). Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 31, 1648–1664. doi: 10.1105/tpc.18.00787
van Lun, M., van der Spoel, D., and Andersson, I. (2011). Subunit interface dynamics in hexadecameric rubisco. J. Mol. Biol. 411, 1083–1098. doi: 10.1016/j.jmb.2011.06.052
Wang, C., Schueler-Furman, O., and Baker, D. (2005). Improved side-chain modeling for protein–protein docking. Protein Sci. 14, 1328–1339. doi: 10.1110/ps.041222905
Wang, H., Yan, X., Aigner, H., Bracher, A., Nguyen, N. D., Hee, W. Y., et al. (2019). Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566, 131–135. doi: 10.1038/s41586-019-0880-5
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gkg520
Whitton, B. A., and Potts, M. (2012). “Introduction to the cyanobacteria,” in Ecology of Cyanobacteria II: Their Diversity in Space and Time, ed. B. A. Whitton (New York, NY: Springer), 1–13. doi: 10.1007/978-94-007-3855-3_1
Keywords: photosynthesis, cyanobacteria, carboxysomes, rubisco, CcmM, carbon dioxide fixation
Citation: Rohnke BA, Kerfeld CA and Montgomery BL (2020) Binding Options for the Small Subunit-Like Domain of Cyanobacteria to Rubisco. Front. Microbiol. 11:187. doi: 10.3389/fmicb.2020.00187
Received: 08 September 2019; Accepted: 27 January 2020;
Published: 28 February 2020.
Edited by:
Wendy Schluchter, The University of New Orleans, United StatesReviewed by:
G. Dean Price, Australian National University, AustraliaSabine Heinhorst, The University of Southern Mississippi, United States
Copyright © 2020 Rohnke, Kerfeld and Montgomery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheryl A. Kerfeld, Y2tlcmZlbGRAbGJsLmdvdg==; Beronda L. Montgomery, bW9udGcxMzNAbXN1LmVkdQ==
 Brandon A. Rohnke
Brandon A. Rohnke Cheryl A. Kerfeld
Cheryl A. Kerfeld Beronda L. Montgomery
Beronda L. Montgomery