- State Key Laboratory of Biocontrol and Guangdong Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Effectors secreted by the type III protein secretion system (T3SS) of rhizobia are host-specific determinants of the nodule symbiosis. Here, we have characterized NopD, a putative type III effector of Bradyrhizobium sp. XS1150. NopD was found to possess a functional N-terminal secretion signal sequence that could replace that of the NopL effector secreted by Sinorhizobium sp. NGR234. Recombinant NopD and the C-terminal domain of NopD alone can process small ubiquitin-related modifier (SUMO) proteins and cleave SUMO-conjugated proteins. Activity was abolished in a NopD variant with a cysteine-to-alanine substitution in the catalytic core (NopD-C972A). NopD recognizes specific plant SUMO proteins (AtSUMO1 and AtSUMO2 of Arabidopsis thaliana; GmSUMO of Glycine max; PvSUMO of Phaseolus vulgaris). Subcellular localization analysis with A. thaliana protoplasts showed that NopD accumulates in nuclear bodies. NopD, but not NopD-C972A, induces cell death when expressed in Nicotiana tabacum. Likewise, inoculation tests with constructed mutant strains of XS1150 indicated that nodulation of Tephrosia vogelii is negatively affected by the protease activity of NopD. In conclusion, our findings show that NopD is a symbiosis-related protein that can process specific SUMO proteins and desumoylate SUMO-conjugated proteins.
Introduction
Various bacteria possess protein secretion systems, through which effectors are translocated into host cells. Type III (T3) effector proteins secreted via a needle-like type III secretion system (T3SS) are important virulence factors of pathogenic bacteria such as the plant pathogens Pseudomonas syringae and Xanthomonas campestris (Büttner, 2016; Deng et al., 2017). Functional T3SS have also been identified in various rhizobia, bacteria that establish a symbiotic relationship with legumes (Staehelin and Krishnan, 2015; Nelson and Sadowsky, 2015; López-Baena et al., 2016). Rhizobia, differentiated into bacteroids, reduce atmospheric nitrogen to ammonia in root nodules of host plants. Fixed nitrogen is delivered to the host plant in exchange of carbon assimilates and nutrients. Consequently, growth of legume crops does not depend on application of nitrogen fertilizer. Rhizobial infection and nodule initiation are controlled by various signals, including host flavonoids and rhizobial lipo-chitooligosaccharides, the so-called Nod factors (Perret et al., 2000; Oldroyd, 2013; Ferguson et al., 2019). Mutant analysis showed that several rhizobial T3 effectors of various strains also play a crucial role in establishment and maintenance of the symbiosis (Staehelin and Krishnan, 2015; Nelson and Sadowsky, 2015; López-Baena et al., 2016). However, besides secretion and translocation into host cells, only a few rhizobial T3 effectors have been biochemically characterized in detail. Examples of well-studied rhizobial effectors are the nodulation outer proteins NopE1/NopE2 (Wenzel et al., 2010; Schirrmeister et al., 2011), NopL (Bartsev et al., 2003; Bartsev et al., 2004; Zhang et al., 2011; Ge et al., 2016), NopM (Rodrigues et al., 2007; Kambara et al., 2009; Xin et al., 2012; Xu et al., 2018), NopP (Ausmees et al., 2004; Skorpil et al., 2005; Zhao et al., 2018; Sugawara et al., 2018), NopT (Dai et al., 2008; Dowen et al., 2009; Kambara et al., 2009; Fotiadis et al., 2012), and ErnA (Teulet et al., 2019).
Pattern recognition receptors of plants recognize structurally conserved microbial elicitors (PAMPs) to activate defense gene expression. In most cases, PAMP recognition results in PTI (Boller and Felix, 2009; Macho and Zipfel, 2014; Cao et al., 2017). T3 effectors translocated into plant cells often suppress PTI and some of them target pattern recognition receptors and downstream signaling components such as mitogen activated protein (MAP) kinases (Feng and Zhou, 2012). The T3 effector NopL of Sinorhizobium sp. (=Ensifer fredii) NGR234, for example, becomes multiply phosphorylated by MAP kinases and thereby inhibits MAP kinase signaling (Zhang et al., 2011; Ge et al., 2016). On the other hand, plants can recognize the presence or action of a specific T3 effector (avirulence protein) by a given intracellular disease resistance protein (nucleotide-binding/leucine-rich repeat receptor). This triggers a rapid and strong defense reaction that often culminates in programmed cell death, the so-called hypersensitive response. In this way, growth of invading pathogens is rapidly arrested and the T3 effector functions as an avirulence protein (ETI) (Cui et al., 2015). A strong hypersensitive response was also observed when the rhizobial effector protease NopT was expressed in the non-host plant tobacco (Nicotiana tabacum) (Dai et al., 2008; Fotiadis et al., 2012). Likewise, NopT and other rhizobial effectors (Staehelin and Krishnan, 2015) have a negative impact on nodule formation in certain host plants. In soybean (Glycine max), special forms of the disease resistance protein Rj2 are involved in blockage of nodule formation by specific Bradyrhizobium and Sinorhizobium strains in a T3SS-dependent manner (Yang et al., 2010; Sugawara et al., 2018). ETI-like defense responses were observed in a specific soybean cultivar (Rj4/Rj4 genotype) inoculated with B. elkanii USDA61 (Yasuda et al., 2016). Positional cloning revealed that the Rj4 gene encodes a thaumatin-like protein (Tang et al., 2016). Nodulation tests with rhizobia mutagenized with the Tn5 transposon indicated that Rj4-mediated nodulation blockage can be overcome by deletion of a putative T3 effector gene (BEL2_5 in USDA61, Faruque et al., 2015; MA20_12780 in B. japonicum Is-34, Tsurumaru et al., 2015).
Post-translational ubiquitination of proteins followed by degradation via the ubiquitin proteasome system regulates protein levels in eukaryotic cells. To suppress PTI, T3 effectors can interfere with the ubiquitin proteasome system. For example, T3 effectors of pathogenic bacteria can mimic the activity of ubiquitin ligases and therefore label PTI-related host proteins for proteasome-dependent degradation (Dudler, 2013; Banfield, 2015). Likewise, the E3 ubiquitin ligase NopM, a T3 effector of Sinorhizobium sp. NGR234, can dampen PAMP-induced generation of reactive oxygen species in Nicotiana benthamiana cells (Xin et al., 2012). Besides the ubiquitin system, effectors delivered to host cells may interfere with sumoylation, i.e. conjugation of a protein to a small ubiquitin-like modifier (SUMO) protein. Sumoylation in eukaryotic cells regulates various processes such as transcriptional regulation, intracellular localization, signal transduction, stress responses, cell cycle progression and protein stability. Sumoylation depends on a SUMO activating enzyme (E1), a SUMO conjugating enzyme (E2), and SUMO ligases (E3) that facilitate sumoylation. In addition, specific SUMO proteases such as Ulps are required for processing of SUMO to its major form (C-terminal di-glycine motif). SUMO-conjugated proteins can be deconjugated by SUMO proteases (desumoylases) and released SUMO can be recycled (Gareau and Lima, 2010).
Remarkably, bacterial effectors may possess SUMO protease activity. The T3 effector XopD of the plant pathogen X. campestris is a prototype of such a protease. XopD is a modular protein with a C-terminal SUMO protease domain that can process various plant SUMO isoforms (Hotson et al., 2003; Chosed et al., 2007; Kim et al., 2011). Moreover, XopD possesses deubiquitinase activity that depends on an unstructured ubiquitin-binding region, indicating a multi-functional enzyme (Pruneda et al., 2016). Proteolytic activity of XopD requires a catalytic triad (HDC residues) in the C-terminal SUMO protease (C48 cysteine peptidase) domain. In addition, DNA binding activity has been reported for XopD (Kim et al., 2008). Known plant target proteins of XopD proteins in Arabidopsis thaliana are transcription factors such as HFR1 (positive regulator of photomorphogenesis) (Tan et al., 2015) as well as DELLA proteins (negative regulators of gibberellin signaling) (Tan et al., 2014). In tomato (Solanum lycopersicum), XopD desumoylates the ethylene responsive transcription factor SIERF4 (Kim et al., 2013). Fluorescence-tagged XopD proteins expressed in plant cells are localized in nuclei and often accumulate in nuclear bodies that are referred to as nuclear foci in previous studies (Hotson et al., 2003). Nuclear bodies are distinct punctate structures in nuclei such as Cajal bodies and nuclear speckles (Morimoto and Boerkoel, 2013).
Several T3 effectors (or effector candidates) of rhizobia show certain sequence similarities with the C-terminal protease domain of XopD. The nodulation outer protein NopD (SFHH103_04358; CEO91485.1) of Sinorhizobium fredii HH103 was identified by mass spectrometry by comparing extracellular protein profiles from a T3SS-knockout mutant with the parent strain (Rodrigues et al., 2007). Mesorhizobium loti MAFF303099 secretes a related protein (mlr6316) in a T3SS-dependent manner. Mutant analysis and inoculation experiments with host plants suggested a possible symbiotic role of this protein in nodulation or nodulation competitiveness (Hubber et al., 2004; Sánchez et al., 2012). Moreover, the two recently identified bradyrhizobial proteins inducing Rj4-mediated nodulation blockage (BEL2_5, Faruque et al., 2015; MA20_12780, Tsurumaru et al., 2015) can be considered as NopD family proteins. On the molecular level, however, NopD proteins have not been studied yet.
In this work, we have characterized NopD of Bradyrhizobium sp. XS1150. NopD can process specific plant SUMO proteins and desumoylate SUMO-conjugated proteins. Moreover, we provide evidence that NopD expressed in planta is targeted to nuclei where it accumulates in nuclear bodies. NopD activity induces ETI-like plant responses, namely cell death in tobacco and reduced nodule formation on roots of the legume Tephrosia vogelii.
Materials and Methods
Strains, Plasmids and Primers
Information on strains and plasmids used in this study is provided in Supplementary Table S1. Plasmids were constructed according to standard methods with restriction enzymes and PCR-based methods. Primers are listed in Supplementary Table S2.
Identification of a T3SS Gene Cluster and a nopD Gene in Bradyrhizobium sp. XS1150
Bradyrhizobium sp. XS1150 was isolated from a nodule of a peanut plant (Arachis hypogaea cv. Liaoning Silihong) at a suburban field close to Guangzhou, China (23.38920N, 113.39900E). Strain XS1150 is resistant to 10 μg/mL chloramphenicol and efficiently grows in various media (Supplementary Text 1). Genomic DNA of strain XS1150 was shotgun-sequenced by the company Ai Jian Genomics (Guangzhou, China) using the Illumina GAIIx system (Illumina). Genes on scaffolds were predicted by the Prodigal v2_60 software. Database comparisons were performed using the Basic Local Alignment Search Tool (BLAST) at the NCBI homepage1. The draft genome sequence of strain XS1150 has been deposited at DDBJ/ENA/GenBank (whole genome shotgun sequencing project NFUH00000000.1; Bioproject PRJNA385724). Using putative rhizobial T3SS genes and predicted T3 effectors (Staehelin and Krishnan, 2015) as query sequences, a T3SS gene cluster (in scaffold 201) and a nopD gene (in scaffold 90) were identified in XS1150. The coding sequence of nopD was PCR-cloned and confirmed by Sanger sequencing (accession number MF100854). Amino acid sequence alignment of the C-terminal part of NopD with related rhizobial proteins and the Xanthomonas effector XopD was performed with DNAstar. Lasergene.v7. A corresponding phylogentic tree was constructed with MEGA5 software using the neighbor-joining method and default setting. Bootstrap analysis was performed with 1000 replications (Tamura et al., 2011).
Functional Analysis of the NopD Secretion Signal Sequence
To analyze functionality of the N-terminal secretion signal of NopD, the N-terminal secretion signal sequence (residues 1–50) of the effector NopL produced by Sinorhizobium sp. NGR234 was replaced by the corresponding N-terminal sequence of NopD. A DNA fragment consisting of the nopL promoter from NGR234, the nopD sequence (encoding amino acid residues 1–50) fused to nopL (encoding amino acid residues 51–338) was cloned into the RK2-derived cloning vector pFAJ1703 (Dombrecht et al., 2001). The plasmid, named pFAJ-NopD:NopL, was then mobilized into NGRΩnopL (NGR234 derivative with an Ω interposon in the nopL gene; Marie et al., 2003) and NGRΩrhcN (Ω interposon in the rhcN gene and thus lacking a functional T3SS; Viprey et al., 1998). Bacterial cultures (180 rpm, 27°C) were treated with 1 μM apigenin and harvested 45 h later. Proteins from culture supernatants were precipitated with 10% (w/v) trichloroacetic acid and used for SDS-PAGE, Ponceau staining and Western blot analysis with a previously prepared antibody against NopL (Zhang et al., 2011). Details are described in Supplementary Text 1.
Recombinant Proteins Expressed in Escherichia coli
Escherichia coli BL21 (DE3) cells carrying a given plasmid were used for expression of recombinant proteins. Purification of proteins was carried out according to the manufacturer’s protocol for affinity chromatography of native proteins (for 6 × His-tagged proteins: Ni-NTA magnetic agarose beads from Qiagen, Germantown, MA, United States; for GST fusion proteins: glutathione agarose beads from Novagen, Madison, WI, United States). Purified proteins were subjected to SDS-PAGE, Western blot analysis or enzyme tests.
SDS-PAGE and Western Blot Analysis
Proteins were separated by SDS-PAGE on 12% polyacrylamide gels and stained with Coomassie Brilliant Blue G-250. For Western blot analysis, proteins were separated onto nitrocellulose membranes. Membranes were incubated with commercially available antibodies against protein tags, with an antibody recognizing NopL of Sinorhizobium sp. NGR234 (Zhang et al., 2011) or against an antibody recognizing a C-terminal part of NopD of strain Bradyrhizobium sp. XS1150. For preparation of the anti-NopD antibody, recombinant NopD (residues 640–1017) with an N-terminal 6 × His tag was expressed in E. coli BL21 (DE3) and the purified protein was used for immunization of a rabbit. After incubation with horseradish peroxidase-conjugated second antibodies, Western blots were developed with 3,3′-diaminobenzidine (Boster, Wuhan, China) or by electrochemiluminescence detection reagents (Amersham GE Healthcare, Little Chalfont, United Kingdom) according to the supplier’s protocols.
Peptidase and Isopeptidase Activity Assays
For the in vitro peptidase assay, purified substrates (GST fused to various SUMO-Gly-Gly-3HA) were incubated with purified 6 × His-tagged test proteins in elution buffer used for purification of 6 × His-tagged proteins (20 mM Tris–HCl, pH 7.9, containing 500 mM imidazole and 0.5 M NaCl) for 30 min at 30°C. Enzyme assays were performed with: (i) full-length NopD; (ii) NopD-C, the C-terminal domain of NopD (residues 640–1017 with an N-terminal methionine); and (iii) NopD-C972A, a NopD variant with a cysteine-to-alanine substitution in the catalytic core. Reaction mixtures were then analyzed on Western blots with an anti-GST antibody. Removal of the C-terminal 3HA tag resulted in a clear band shift.
For the isopeptidase assay, a commonly used in vitro desumoylation assay was performed with sumoylated RanGAP of Homo sapiens (Matunis et al., 1996). To prepare the substrates, different SUMO proteins (processed forms with terminal Thr-Gly-Gly residues) were conjugated to the acceptor RanGAP (with an N-terminal 6 × His tag and a C-terminal Myc tag) by using recombinant E1 and E2 proteins of A. thaliana. Purified 6 × His-tagged AtSAE1 (E1), AtSAE2 (E1) and AtUbc9 (E2) were prepared for this purpose. The sumoylation reaction was carried out in a total volume of 100 μl with 8 μg of RanGAP-Myc-6 × His, 8 μg of GST-SUMO(TGG), 1 μg of AtSAE1-His6, 1 μg of AtSAE2-His6 and 2 μg of AtUbc9-His6 in 50 mM Tris–HCl buffer (pH 7.8) containing 100 mM NaCl, 15% glycerol, 5 mM ATP, and 10 mM MgCl2 at 22°C for 6 or 8 h. No sumoylated RanGAP was formed when the SUMO protease XopD of X. campestris (also expressed as 6 × His tagged protein) was added to the reaction mixture. The reaction products containing RanGAP conjugated to different SUMO proteins were then incubated (30°C; 30 min) with 0.1 μg of 6 × His-tagged enzymes (NopD, NopD-C and NopD-C972A). Removal of SUMO from sumoylated RanGAP-Myc-His6 forms was analyzed on Western blots with an anti-Myc antibody.
Expression of NopD and Variants in Plant Cells
Agrobacterium-mediated transient gene expression in tobacco (N. tabacum cv. Xanthi) was used for expression of NopD and enzymatically inactive NopD-C972A (cysteine-to-alanine substitution in the catalytic core). In subcellular localization studies, NopD fused to YFP were expressed in A. thaliana protoplasts. In a similar way, NopD variants fused to YFP were analyzed, namely (i) enzymatically inactive NopD-C972A, (ii) NopD-N, the N-terminal domain of NopD (residues 1–390), (iii) NopD-NΔ2-53, a NopD-N variant lacking residues 2–53, (iv) NopD-NΔ2-60, a NopD-N variant lacking residues 2–60; (v) NopD-TR, the tandem repeat domain of NopD (residues 391–720 with an N-terminal methionine), and (vi) NopD-C, the C-terminal protease domain of NopD (residues 640–1017 with an N-terminal methionine). ARF4 (auxin response factor 4 of A. thaliana) fused to RFP served as nuclear marker. Details on protein expression in tobacco and A. thaliana are shown in Supplementary Text 1.
Construction of XS1150 Mutants
The mutant XS1150ΩrhcST (lacking a functional T3SS) was constructed by inserting an ΩSpe interposon into the T3SS apparatus gene rhcS of strain Bradyrhizobium sp. XS1150. Strain XS1150ΔnopD, a nopD-deficient mutant of XS1150, was constructed by replacing the nopD coding sequence with an ΩSpe interposon. Strain XS1150ΔnopD+nopD is a derivative of XS1150ΔnopD in which the nopD gene (including a 1-kb promoter region) was re-introduced. The mutant XS1150ΔnopD+nopD-C972A was constructed in a similar way to obtain a strain that produces an enzymatically inactive NopD variant (substitution of cysteine residue 927 by alanine). Details on the mutant construction procedure are provided in Supplementary Text 1 and Supplementary Figure S1).
Nodulation Tests
Tephrosia vogelii was used to characterize the symbiotic phenotypes of the constructed mutants (XS1150ΔnopD, XS1150ΔnopD+nopD, XS1150ΔnopD+nopD-C972A and XS1150ΩrhcST) as compared to the parent strain Bradyrhizobium sp. XS1150. Information on performed nodulation tests can be found in Supplementary Text S1. Statistical analysis was performed by Kruskal–Wallis tests considering each plastic jar unit (1 plant) as a replicate.
Accession Numbers
Sequences used for DNA constructs of this study have the following accession numbers in sequence databases: Draft genome of Bradyrhizobium sp. (B. guangdongense) XS1150: NFUH00000000 (BioProject PRJNA385724); NopD of Bradyrhizobium sp. XS1150: MF100854; NopL of Sinorhizobium sp. NGR234: NC_000914; AtSUMO1 of A. thaliana: AEE85259; AtSUMO2 of A. thaliana: NM_124898; AtSUMO3 of A. thaliana: NM_124899; AtSUMO5 of A. thaliana: NM_128836; HuSUMO1 of H. sapiens: AK311840; HuSUMO2 of H. sapiens: AK311837; HuSUMO4 of H. sapiens: AB205057; PvSUMO of Phaseolus vulgaris: XM_007146455; GmSUMO of G. max: NM_001248279; Smt3 of Saccharomyces cerevisiae: CP020194; AtSAE1 of A. thaliana: BT000094; AtUbc9 of A. thaliana: NM_001202641; AtSAE2 of A. thaliana: BT003377; RanGAP of H. sapiens: NM_001317930.1; ARF4 of A. thaliana: NP_200853.
Results
Identification of nopD in the Genome of Bradyrhizobium sp. XS1150
Bradyrhizobium sp. XS1150 was isolated from a peanut (A. hypogaea) nodule at a suburban field close to Guangzhou, China. Re-inoculation tests resulted in efficient nodule formation that promoted growth of peanuts. Strain XS1150 induced also nodules on roots of T. vogelii (Supplementary Figure S2). Whole-genome shotgun sequencing revealed that XS1150 is a Bradyrhizobium strain (tentatively named Bradyrhizobium guangdongense). The sequences (670 contigs; totally 7624764 nucleotides) were submitted to the DDBJ/ENA/GenBank database (accession number NFUH00000000). Sequence homology searches indicated that the XS1150 genome possesses a T3SS gene cluster that contains the transcriptional regulator gene ttsI and the putative effector genes nopL, nopE1, and nopP. Moreover, a sequence homologous to the effector gene nopAR of B. japonicum USDA122 (=bll1840 in strain USDA110; Tsukui et al., 2013) was found in the T3SS gene cluster of XS1150 (Supplementary Figure S3).
Using the C-terminal protease domain sequence of nopD from Sinorhizobium fredii HH103 (accession number CEO91485.1) as query sequence, an additional putative effector gene of XS1150 was identified outside the T3SS gene cluster of XS1150. The NopD protein of strain XS1150 possesses a calculated molecular weight of 111.45 kDa. It consists of an N-terminal domain (residues 1–390), a tandem repeat (TR) domain with 7 repeats (residues 391–720; the first 6 repeats contain 49 residues, the last one contains 36 residues) and a C-terminal protease domain (residues 721–1017) (Figure 1A). The C-terminal protease domain of NopD shows sequence similarities to NopD of strain HH103 and to various other putative rhizobial effectors (such as BEL2_5 of USDA61, MA20_12780 of Is-34, mlr6316 of MAFF303099, bll8244 of USDA110, blr1693 of USDA110, blr1705 of USDA110). The C-terminal protease domains of these rhizobial proteins could be aligned to the Xanthomonas effector XopD, a SUMO protease of the C48 cysteine peptidase family (Supplementary Figure S4). The alignment allowed prediction of conserved residues (catalyctic triad) required for SUMO protease activity. Based on the obtained alignment, a corresponding phylogenetic tree was constructed (Figure 1B).
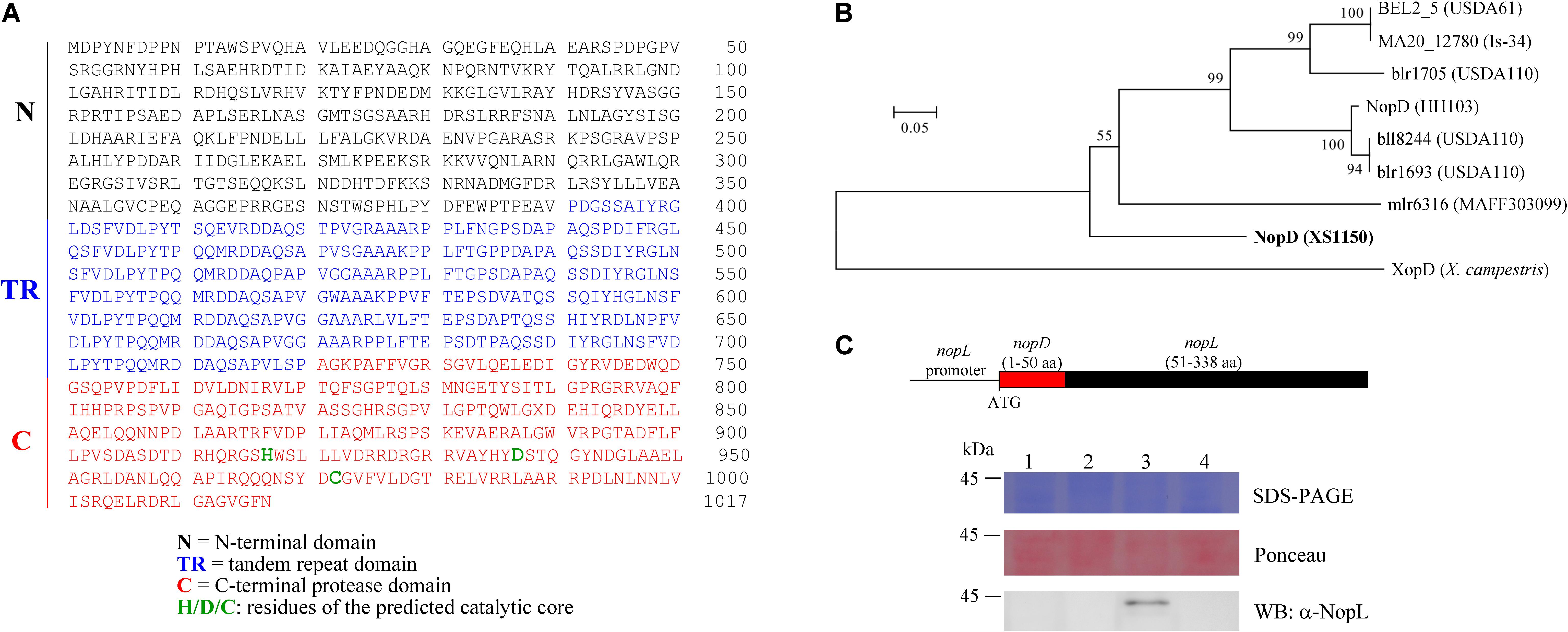
Figure 1. NopD of Bradyrhizobium sp. XS1150. (A) Amino acid sequence of NopD, a modular protein that consists of an N-terminal domain (N), a tandem repeat domain (TR) and a C-terminal protease domain (C) with histidine, aspartic acid and cysteine residues (predicted catalytic triad). (B) Phylogenetic analysis of NopD family proteins based on a conserved C-terminal region. The residues 835–982 of NopD were aligned with a selection of related rhizobial proteins (B. elkanii USDA61, B. japonicum Is-34, B. japonicum USDA110, M. loti MAFF303099 and Sinorhizobium fredii HH103) and with XopD of X. campestris pv. campestris 8004. The alignment and accession numbers of the proteins are shown in Supplementary Figure S4. The tree was constructed with MEGA5 software. Bootstrap values are indicated next to branches. The scale bar represents 0.05 substitutions per site. (C) T3SS-dependent secretion of a chimeric NopD-NopL protein. The DNA construct of plasmid pFAJ-NopD:NopL is shown on the top of the panel. The plasmid was mobilized into the NGRΩnopL and NGRΩrhcN mutants of Sinorhizobium sp. NGR234. Equal amounts of secreted proteins from culture supernatants were analyzed by SDS-PAGE and Ponceau staining (loading control of Western blot). NopD-NopL was immunodetected by an antibody against NopL. Lane 1, NGRΩnopL; lane 2, NGRΩrhcN; lane 3: NGRΩnopL carrying pFAJ-NopD:NopL; lane 4, NGRΩrhcN carrying pFAJ-NopD:NopL.
NopD Possesses a Functional Secretion Signal Sequence
Bioinformatic analysis with EffectiveDB (Eichinger et al., 2016) predicted that NopD possesses an N-terminal secretion signal sequence required for T3SS-dependent secretion. To confirm this prediction, we prepared a plasmid (named pFAJ-NopD:NopL) to express a chimeric NopD-NopL protein in the mutants NGRΩnopL and NGRΩrhcN of Sinorhizobium sp. NGR234. NGRΩnopL is a knockout mutant deficient in synthesis of the NopL effector and NGRΩrhcN lacks a functional T3SS. Figure 1C shows a schematic view of the expressed construct. Western blot analysis with an anti-NopL antibody indicated presence of the NopD-NopL protein in the culture supernatant of NGRΩnopL carrying pFAJ-NopD:NopL. However, no corresponding Western blot signals were observed for protein preparations from the culture supernatant of NGRΩrhcN carrying pFAJ-NopD:NopL (Figure 1C). These findings indicate that NopD possesses an N-terminal secretion signal sequence that is recognized by the T3SS of strain NGRΩnopL.
NopD Is a SUMO Protease
A truncated NopD protein (residues 640–1017) was expressed in E. coli in order to produce a polyclonal antibody against NopD. The protein with a 6 × His-tag was purified by nickel affinity purification and then used for immunization of a rabbit. Full-length His-tagged NopD, albeit to a lesser extent, could also be expressed in E. coli. After purification by nickel affinity chromatography, a band corresponding to the expected molecular weight was detected with the prepared antibody. In addition, faster migrating bands (presumably degraded NopD forms) were observed (Supplementary Figure S5).
As the C-terminal protease domain of NopD proteins is related to the Xanthomonas T3 effector XopD, we expected that NopD possesses SUMO protease activity. We expressed full-length NopD, NopD-C972A (substitution of the predicted catalytic core cysteine residue to alanine) and NopD-C (C-terminal protease domain) in E. coli as 6 × His-tagged proteins. SUMO proteins with a GST tag at the N- terminus and three HA tags at the C-terminus (directly following the C-terminal Gly-Gly residues) were also expressed in E. coli. Such GST-SUMO1-3HA fusion proteins were prepared for various SUMOs from A. thaliana (AtSUMO1, AtSUMO2, AtSUMO3 and AtSUMO5), soybean (GmSUMO), common bean (PvSUMO), human (HuSUMO1, HuSUMO2 and HuSUMO4) and yeast (S. cerevisiae; Smt3). The native proteins, purified by affinity chromatography, were then used for hydrolytic tests. As shown in Figure 2, full-length NopD and NopD-C, but not NopD-C972A, had the capacity to release the three HA tag from GST-AtSUMO1-3HA (A. thaliana), GST-AtSUMO2-3HA (A. thaliana), GST-GmSUMO-3HA (soybean) and GST-PvSUMO-3HA (common bean). However, the two other Arabidopsis SUMO isoforms (GST-AtSUMO3-3HA and GST-AtSUMO5-3HA) were not cleaved in this assay. Likewise, SUMO isoforms from human (GST-HuSUMO1-3HA, GST-HuSUMO2-3HA, GST-HuSUMO4-3HA) as well as GST-Smt3-3HA from yeast were no substrates for NopD or NopD-C. Hence, NopD and NopD-C could process the C-terminal end of specific plant SUMO proteins.
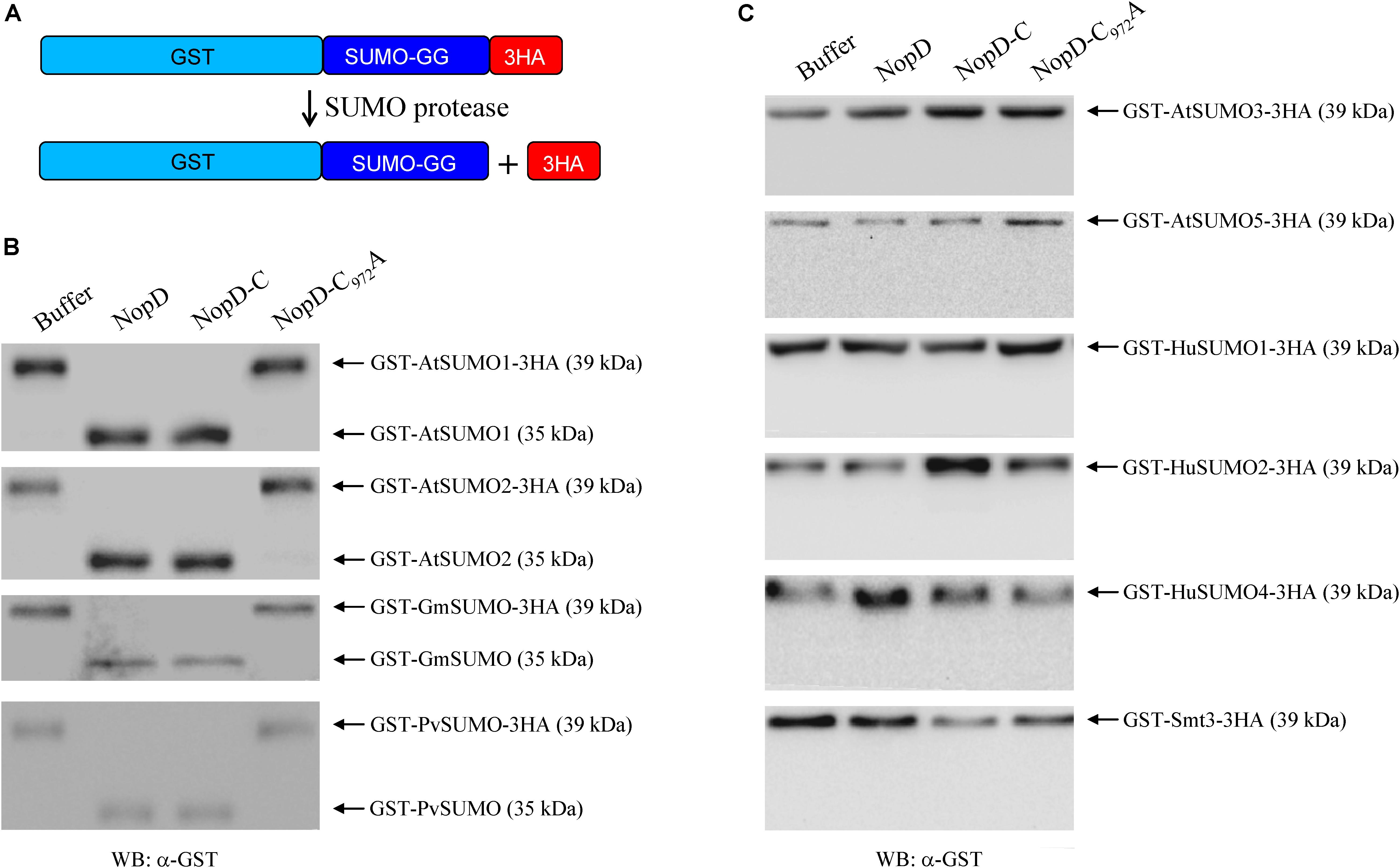
Figure 2. Peptidase activity of 6 × His-tagged NopD and NopD-C. (A) Schematic view of the test system that is based on release of the C-terminal 3HA tag from a given GST-SUMO protein by NopD and NopD-C (C-terminal protease domain of NopD). Reaction mixtures were incubated at 30°C for 1 h. 6 × His-tagged NopD-C972A (enzymatically inactive variant) and buffer (no enzyme) were used as negative controls. (B) Analysis of reaction mixtures on Western blots probed with an anti-GST antibody. Incubation of GST-AtSUMO1-3HA, GST-AtSUMO2-3HA, GST-GmSUMO1-3HA and GST-PvSUMO1-3HA with 6 × His-tagged NopD or NopD-C resulted in band shifts, indicating release of the C-terminal 3HA tag. Incubation with NopD-C972A or buffer alone did not result in a band shift. (C) Results for reactions with GST-SUMO-3HA proteins that were not cleaved by NopD or variants (no obvious band shift).
SUMO proteases not only process SUMO proteins but can also remove SUMO from SUMO conjugated acceptor proteins. To investigate whether NopD has such isopeptidase activity, we cloned Arabidopsis genes of the sumoylation cascade, namely AtSAE1, AtSAE2 and AtUbc9. These genes were subsequently expressed in E. coli as 6 × His-tagged proteins and purified. Similarly, we prepared the acceptor protein RanGAP of H. sapiens (with an N-terminal 6 × His tag and a C-terminal Myc tag) and AtSUMO1 in its processed form (TGG), fused to an N-terminal GST tag. The recombinant proteins were used to obtain sumoylated RanGAP. Formation of an AtSUMO1-RanGAP conjugate was not observed when the known SUMO protease XopD was added to the reaction (Figure 3A). Other SUMO-RanGAP conjugates were prepared in a similar way (Supplementary Figure S6). The proteins were then used for isopeptidase activity tests with NopD and variants. Reactions with NopD and NopD-C resulted in desumoylation of AtSUMO1-RanGAP, AtSUMO2-RanGAP, GmSUMO-RanGAP or PvSUMO-RanGAP. In contrast, NopD-C972A did not show enzyme activity (Figure 3B). All other conjugates (AtSUMO3-RanGAP, AtSUMO5-RanGAP, HuSUMO1-RanGAP, HuSUMO2-RanGAP, HuSUMO4-RanGAP and Smt3-RanGAP) remained intact when incubated with NopD or NopD-C (Figure 3C). Hence, isopeptidase activities of NopD and NopD-C were similar to those obtained with GST-SUMO-3HA proteins.
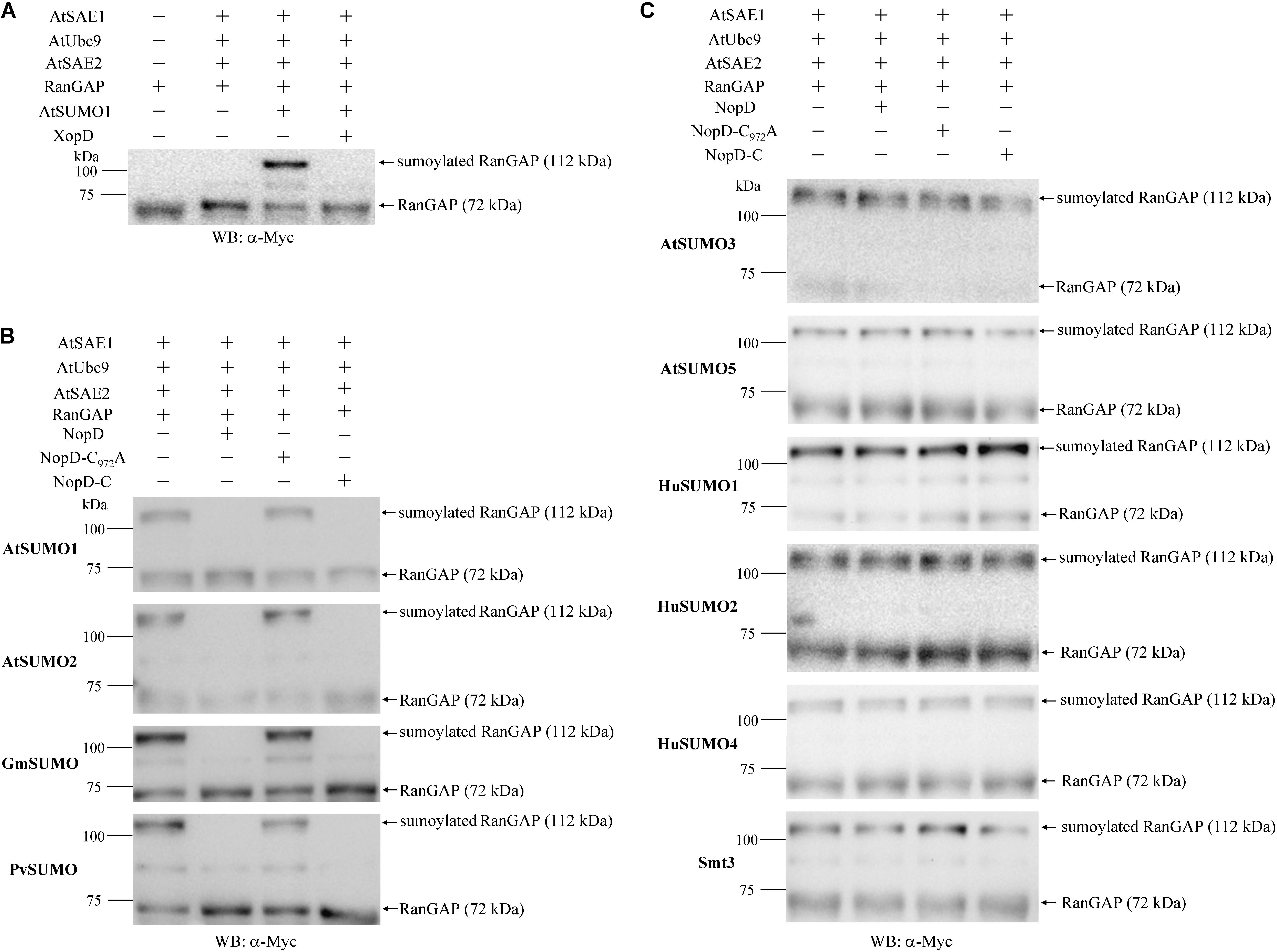
Figure 3. Isopeptidase activity of 6 × His-tagged NopD and NopD-C. SUMO-RanGAP conjugates were prepared to analyze the desumoylation activity of NopD and NopD-C. (A) Preparation of an AtSUMO1-RanGAP conjugate in an in vitro sumoylation system with indicated 6 × His-tagged Arabidopsis proteins and human RanGAP (with 6 × His and Myc tags). The obtained AtSUMO1-RanGAP conjugate was not formed in the presence of 6 × His-tagged XopD. Western blot analysis of sumoylated RanGAP and RanGAP was performed with an anti-Myc antibody. Similar SUMO-RanGAP conjugates were obtained for other SUMO proteins (see Supplementary Figure S6). (B) NopD and NopD-C show SUMO isopeptidase activity for indicated SUMO-RanGAP conjugates. Incubation with 6 × His tagged NopD and NopD-C resulted in desumoylation of the conjugate, whereas NopD-C972A (enzymatically inactive) or buffer (no enzyme) showed no effects. Reaction mixtures were analyzed on Western blots probed with an anti-Myc antibody. (C) Western blot results of reactions with indicated SUMO-RanGAP conjugates that were not desumoylated by NopD or NopD-C under the same test conditions.
NopD but Not NopD-C972A Induces Cell Death in Tobacco
To study effects of NopD in living plant cells, we transiently expressed NopD in tobacco cells. Agrobacterium tumefaciens carrying binary vectors containing the CaMV 35S promoter and a given nopD sequence were infiltrated into leaves of tobacco plants. Remarkably, a rapid cell death response was induced by expression of NopD. The strength of the hypersensitive response was comparable to that induced by the effector NopT of Sinorhizobium sp. NGR234 (Dai et al., 2008). However, NopD-C972A expression in tobacco did not cause cell death (Figure 4A). These data indicate that the observed hypersensitive response depended on the protease activity of NopD.
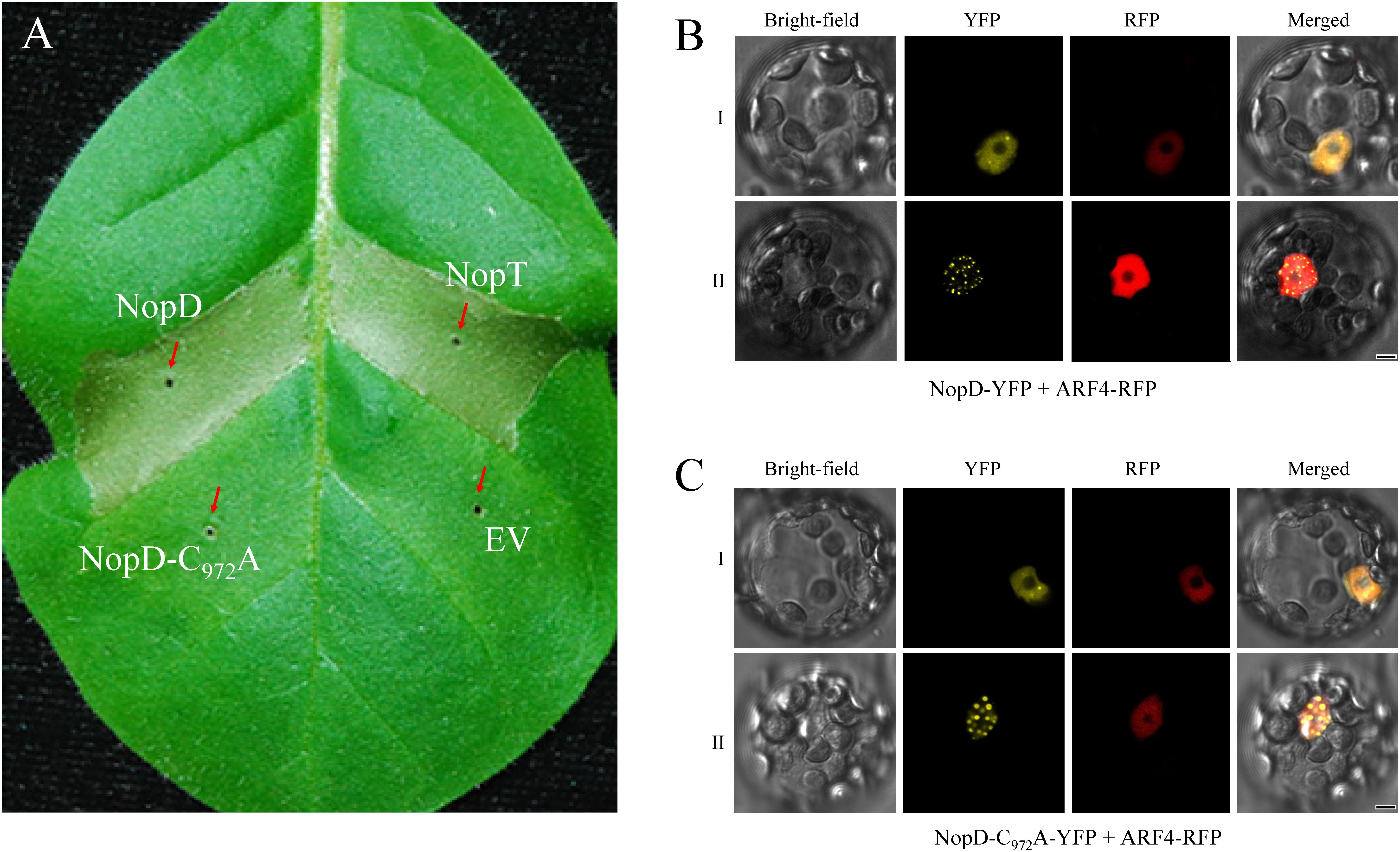
Figure 4. Expression of NopD and NopD-C972A in plant cells. (A) NopD induces cell death in tobacco. Expression of NopD and NopD-C972A in different sections of a tobacco leaf was performed by infiltration of A. tumefaciens carrying appropriate plasmids (pCAMBIA1302 derivatives). NopT (a T3 effector of Sinorhizobium sp. NGR234) was used for comparison. The photograph was taken 48 h after bacterial infiltration. Cell death (necrotic tissue) was observed for NopD and NopT, whereas expression of NopD-C972A showed no visible effects. (B,C) Subcellular localization of YFP-tagged NopD and NopD-C972A in Arabidopsis protoplasts. The nuclear marker ARF4 fused to RFP was co-expressed. Cells were analyzed with a confocal microscope for red fluorescence (RFP) emission, yellow fluorescence (YFP) emission and under bright-field illumination (18 h after transformation). YFP fluorescence was evenly distributed throughout the nucleus (type I cells) or preferentially in nuclear bodies (type II cells).
Subcellular Localization of NopD and Variants in Plant Nuclei
To investigate the subcellular localization of NopD in plant cells, NopD variants fused to YFP were expressed in Arabidopsis protoplasts. The constructs were expressed from the CaMV 35S promoter. Analysis of transformed protoplasts by confocal microscopy revealed that fluorescence of NopD-YFP appeared in nuclei although no classic nuclear localization signal was found in NopD (Figure 4B). The ARF4 protein of A. thaliana fused to RFP was used as nuclear marker. We noticed that the distribution of NopD-YFP in the nucleus was of two types: (i) fluorescence distributed evenly throughout the nucleus and (ii) fluorescence predominantly localized to nuclear bodies as reported previously for the effector Xanthomonas effector XopD (Hotson et al., 2003). Over time, the strength of fluorescence signals increased in the nuclear bodies, suggesting that NopD-YFP was first homogeneously localized in the nucleus and then re-localized to the nuclear bodies. The enzymatically inactive variant NopD-C972A fused to YFP showed a similar nuclear localization pattern (Figure 4C). In contrast, NopD-N (residues 1–390) fused to YFP did not accumulate in nuclear bodies but was localized in the nucleus (with strong fluorescence signals in the nucleolus), suggesting the presence of a cryptic nuclear localization signal in the N-terminal domain of NopD. Removal of N-terminal residues from NopD-N (T3SS secretion signal sequence) had no impact, i.e. localization of YFP-tagged NopD-N lacking residues 2–53 (NopD-NΔ2-53) or 2–60 (NopD-NΔ2-60) was not different from NopD-N. YFP-tagged NopD-TR (tandem repeat domain of NopD; residues 391–720) and NopD-C (C-terminal SUMO protease domain; residues 640–1017) were evenly distributed in the cell like YFP alone (Supplementary Figure S7).
NopD Negatively Affects Nodulation of the Host Plant T. vogelii
To explore symbiotic effects of NopD during symbiosis, a nopD deletion mutant of Bradyrhizobium sp. XS1150, named XS1150ΔnopD, was constructed (Supplementary Figure S1). Nodulation tests with various legumes revealed that T. vogelii is a host plant that differently responds to XS1150 and XS1150ΔnopD. The mutant induced significantly more nodules and the nodule biomass per plant was also increased. These findings suggest that NopD functions as asymbiotic effector that negatively affects the symbiosis between strain XS1150 and T. vogelii. Compared to the parent strain XS1150, less nodules and a lower nodule biomass per plant were observed when T. vogelii plants were inoculated with the T3SS knockout mutant XS1150ΩrhcST. These findings suggest that uncharacterized effectors of XS1150 show symbiosis-promoting effects (Figure 5).
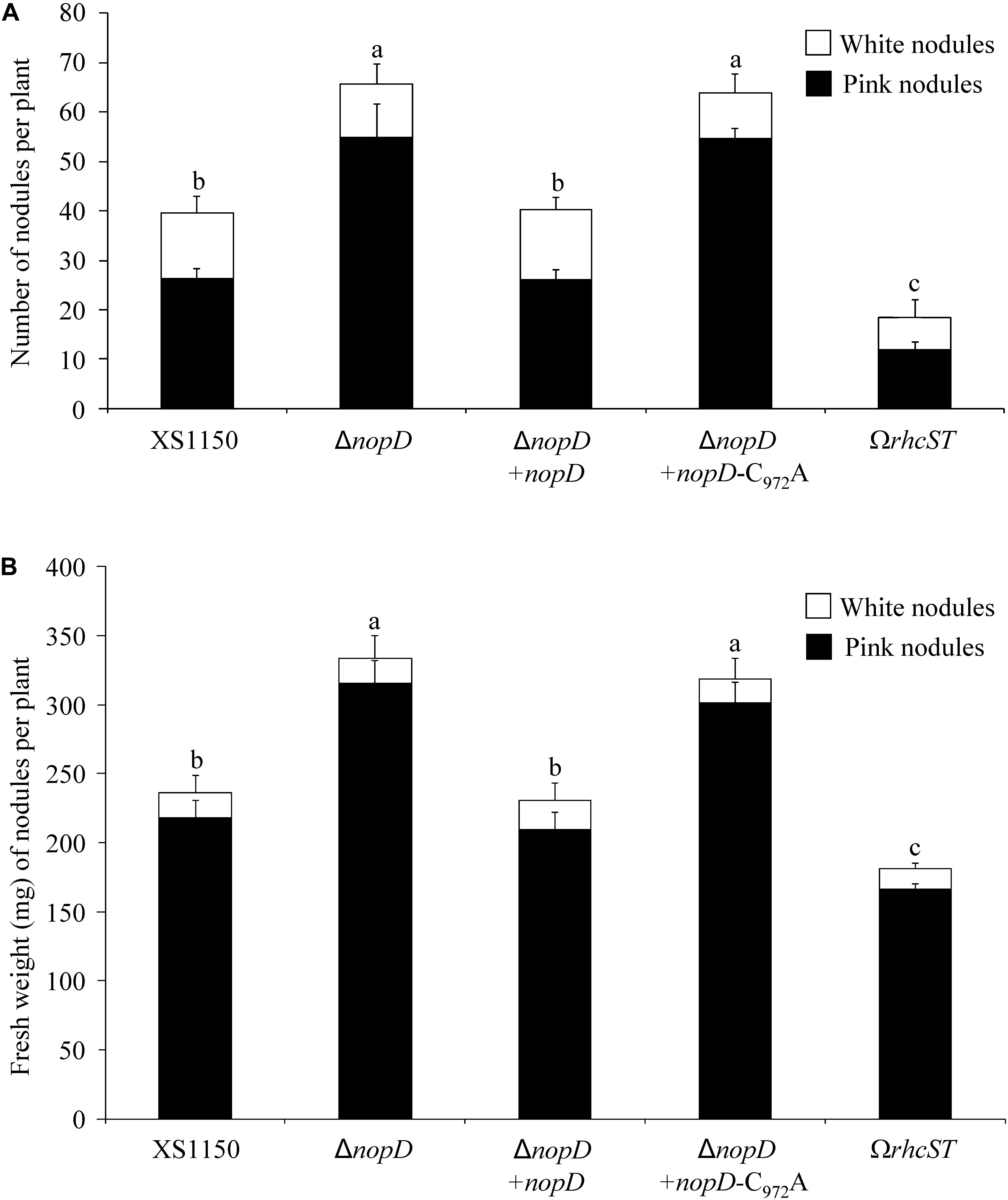
Figure 5. Symbiotic phenotype of Bradyrhizobium sp. XS1150 and constructed mutants on the host plant T. vogelii. Data shown are the results of a representative nodulation test. Plants were inoculated with indicated strains and harvested 36 days later. Data indicate means ± SE (8 jars; n = 8). Different letters indicate significant differences (Kruskal–Wallis tests, P < 0.02). (A) Nodule number (number of pink nodules and white nodules per plant). (B) Nodule biomass (fresh weight of pink and white nodules per plant). Abbreviations: XS1150, Bradyrhizobium sp. XS1150 (wild-type); ΔnopD, XS1150ΔnopD (nopD knockout mutant); ΔnopD+nopD, XS1150ΔnopD+nopD (rescued nopD knockout mutant); ΔnopD+nopD-C972A, XS1150ΔnopD+nopD-C972A (nopD knockout mutant expressing NopD-C972A); ΩrhcST, XS1150ΩrhcST (mutant lacking a functional T3SS).
Re-introduction of nopD into the XS1150ΔnopD mutant resulted in a nopD expressing strain (XS1150ΔnopD+nopD). Moreover, we introduced a modified nopD sequence (C972A substitution) into XS1150ΔnopD to create a mutant that produces enzymatically inactive NopD-C972A and named the strain XS1150ΔnopD+nopD-C972A (Supplementary Figure S1). As expected, nodulation of XS1150ΔnopD+nopD on T. vogelii roots resulted in nodulation parameters comparable to strain XS1150, indicating that the wild-type phenotype was restored. In contrast, the symbiotic phenotype of strain XS1150ΔnopD+nopD-C972A was not different from XS1150ΔnopD (Figure 5). Hence, the cysteine residue 972 required for SUMO protease activity was indispensable for the NopD effect in the interaction with T. vogelii.
Discussion
In this study, we have characterized a putative T3 effector of Bradyrhizobium sp. XS1150. NopD and the C-terminal domain alone (NopD-C) show SUMO processing activity and SUMO deconjugation activity. The cysteine residue 972 of NopD was found to be essential for enzyme activity in these tests. We propose to use the protein name NopD for all rhizobial T3 effectors with an enzymatically active SUMO protease domain (C48 or Ulp1 peptidase family) even if other domains in these effectors are different or absent.
The NopD-NopL fusion protein was secreted by the T3SS of strain NGRΩnopL whereas no Western blot signal was observed for the T3SS-deficient mutant NGRΩrhcN. These findings indicate that the N-terminal sequence of NopD is a functional secretion signal sequence as predicted by EffectiveDB (Eichinger et al., 2016). T3SS-dependent NopD secretion by XS1150 and translocation into legume cells remains to be experimentally confirmed. Support for translocation into host cells is provided by our findings that NopD can target plant SUMO proteins and that nodulation of T. vogelii was negatively affected by the catalytic cysteine residue 972 in NopD. Remarkably, NopD and NopD-C (C-terminal protease domain) both showed a rigid substrate preference for specific plant SUMO proteins, i.e. they can process only specific GST-SUMO-3HA proteins (AtSUMO1, AtSUMO2, GmSUMO and PvSUMO). A similar preference for the same plant SUMO protein was observed when NopD was used in a SUMO deconjugation assay with sumoylated RanGAP. All SUMO proteins processed by NopD contain a C-terminal recognition motif previously identified for XopD (A35-R29-M7L6H5Q4T3G2G1; numbers following the amino acid residues indicate positions relative to the cleavage site; see Supplementary Figure S8; Chosed et al., 2007). Other SUMO proteins lacking this motif (particularly M7L6H5 residues) were not processed by NopD or XopD. Hence, NopD and XopD appear to possess a similar substrate preference for SUMOs and thus may target plant proteins that are sumoylated in a similar way. However, the N-terminal parts of NopD and XopD proteins are rather different (residues 1–720 of NopD show only 17% amino acid sequence identity with XopD of X. campestris pv. campestris strain 8004).
Although various rhizobial T3 effectors have been identified (Staehelin and Krishnan, 2015), subcellular localization analysis in plant cells has been only performed for few effectors of Sinorhizobium sp. NGR234 (Dowen et al., 2009; Ge et al., 2016; Xu et al., 2018) and Bradyrhizobium sp. ORS3257 (Teulet et al., 2019). Fluorescence-tagged NopD was found to be targeted to the plant nucleus (Figure 4) although the protein apparently lacks a classic nuclear localization signal. We suggest that NopD possesses a cryptic nuclear localization signal in its N-terminus as YFP-tagged NopD-N (residues 1–390) also showed nuclear localization. NopD and NopD-C972A (Figure 4), but not other NopD variants (Supplementary Figure S7), accumulated in nuclear bodies. Hence, subnuclear localization depended on full-length NopD whereas protease activity was not required for accumulation of NopD in nuclear bodies. The Xanthomonas T3 effector XopD expressed in plant cells may also accumulate in nuclear bodies (Hotson et al., 2003). Co-expression of XopD proteins with a given target protein (SlERF4 or HFR1) resulted in co-localization of both proteins in nuclear bodies (Kim et al., 2013; Tan et al., 2015).
Like T3 effectors from pathogens, rhizobial effectors are expected to suppress plant defense reactions, thereby promoting rhizobial infection, nodule formation and survival of bacteroids in nodules (Staehelin and Krishnan, 2015; Nelson and Sadowsky, 2015; Cao et al., 2017). On the other hand, rhizobial effectors can have negative effects on symbiosis with certain legumes. The role of NopD in the interaction between strain XS1150 and T. vogelii suggests that the protein is an asymbiotic effector similar to ETI-inducing avirulence proteins in plant-pathogen interactions. Likewise, the hypersensitive reaction of tobacco cells elicited by NopD expression can be considered as an ETI response. The proteolytically inactive NopD-C972A variant did not elicit cell death, however. This finding suggests indirect effector recognition through desumoylation of a NopD substrate that perhaps functions as sensor for disease resistance protein-mediated ETI (Cui et al., 2015). Nodulation tests with T. vogelii showed that the symbiotic effector activity of NopD also depended on cysteine residue 972. This finding suggests that abnormal desumoylation events caused by proteolytically active NopD were not favorable for nodulation of this plant. The NopD-dependent nodulation phenotype of T. vogelii is reminiscent of the incompatible interaction between Rj4/Rj4 soybeans and B. elkanii USDA61 and B. japonicum Is-34. The C-terminal regions of the BEL2_5 (USDA61) and MA20_12780 (Is-34) proteins are related to NopD (Figure 1B) and thus are predicted to possess SUMO protease activity. Mutant strains lacking these proteins gained the ability to induce nodules on Rj4/Rj4 soybeans, suggesting that NopD family proteins possess asymbiotic effector activity on soybean genotypes expressing Rj4 (Faruque et al., 2015; Tsurumaru et al., 2015; Yasuda et al., 2016). Rj4 encodes a specific thaumatin-like protein that only differs in few amino acids from homologs (Tang et al., 2016).
Taken together, we have identified and characterized NopD of Bradyrhizobium sp. XS1150. NopD is a modular protein that consists of at least three different units: (i) an N-terminal domain which appears to be required for T3SS-dependent secretion and that also contains information for NopD targeting into plant nuclei; (ii) a middle tandem repeat domain, and (iii) a C-terminal protease domain which targets specific plant SUMO proteins. The protease activity of NopD was required for cell death induction in tobacco and negatively affected nodule formation of T. vogelii. Future work will be required to identify SUMO-conjugated targets of NopD in T. vogelii or other host legumes.
Data Availability Statement
The nucleotide sequences generated for this study can be found in the GenBank database (Bioproject PRJNA385724 and accession MF100854). Non-cropped images of SDS-PAGE gels and Western blots are shown in Supplementary Material.
Author Contributions
Q-WX, Z-PX, and CS conceived and designed the experiments. Q-WX, JB, JC, Q-YH, YW, YL, and ZZ performed the experiments. Q-WX, JB, JC, Q-YH, YW, YL, ZZ, Z-PX, and CS analyzed the data. Q-WX, CW, Z-PX, and CS wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant 31470197), by the Department of Science and Technology of Guangdong Province, China (grants 2013B020302002 and 2013B051000043), by the Science Foundation of the State Key Laboratory of Biocontrol (grants SKLBC322017A09 and SKLBC322018A10), and by the Guangdong Key Laboratory of Plant Resources (2017B030314023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Min Chen, Di Zhang, and Zhen Chen (Sun Yat-sen University) are acknowledged for initial help in identification and characterization of NopD. We thank Xuan-Qiang Liang (Guangdong Academy of Agricultural Sciences, China) for peanut plants. Fang Wang, Jian Li, Nan Yao, and Da Luo (Sun Yat-sen University, China), Guo-Liang Wang (Hunan Agricultural University, China), William J. Broughton (University of Geneva, Switzerland) and Jan Michiels (Katholieke Universiteit Leuven, Belgium) kindly provided plasmids or strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00386/full#supplementary-material
Abbreviations
CaMV, cauliflower mosaic virus; E1, SUMO activating enzyme; E2, SUMO conjugating enzyme; E3, SUMO ligase; ETI, effector-triggered immunity; GST, glutathione S-transferase; PAMP, pathogen-associated molecular pattern; PTI, PAMP-triggered immunity; RFP, red fluorescence protein; SUMO, small ubiquitin-related modifier; T3, type III (effector); T3SS, type III protein secretion system; Ulp, ubiquitin-like protein-specific protease; YFP, yellow fluorescence protein.
Footnotes
References
Ausmees, N., Kobayashi, H., Deakin, W. J., Marie, C., Krishnan, H. B., Broughton, W. J., et al. (2004). Characterization of NopP, a type III secreted effector of Rhizobium sp. strain NGR234. J. Bacteriol. 186, 4774–4780. doi: 10.1128/jb.186.14.4774-4780.2004
Banfield, M. J. (2015). Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell. Microbiol. 17, 18–25. doi: 10.1111/cmi.12385
Bartsev, A. V., Boukli, N. M., Deakin, W. J., Staehelin, C., and Broughton, W. J. (2003). Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 554, 271–274. doi: 10.1016/s0014-5793(03)01145-1
Bartsev, A. V., Deakin, W. J., Boukli, N. M., McAlvin, C. B., Stacey, G., Malnoë, P., et al. (2004). NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134, 871–879. doi: 10.1104/pp.103.031740
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Büttner, D. (2016). Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. doi: 10.1093/femsre/fuw026
Cao, Y., Halane, M. K., Gassmann, W., and Stacey, G. (2017). The role of plant innate immunity in the legume-rhizobium symbiosis. Annu. Rev. Plant Biol. 68, 535–561. doi: 10.1146/annurev-arplant-042916-041030
Chosed, R., Tomchick, D. R., Brautigam, C. A., Mukherjee, S., Negi, V. S., Machius, M., et al. (2007). Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J. Biol. Chem. 282, 6773–6782. doi: 10.1074/jbc.m608730200
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dai, W. J., Zeng, Y., Xie, Z. P., and Staehelin, C. (2008). Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of Rhizobium sp. strain NGR234. J. Bacteriol. 190, 5101–5110. doi: 10.1128/JB.00306-08
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Dombrecht, B., Vanderleyden, J., and Michiels, J. (2001). Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant Microbe Interact. 14, 426–430. doi: 10.1094/mpmi.2001.14.3.426
Dowen, R. H., Engel, J. L., Shao, F., Ecker, J. R., and Dixon, J. E. (2009). A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J. Biol. Chem. 284, 15867–15879. doi: 10.1074/jbc.M900519200
Dudler, R. (2013). Manipulation of host proteasomes as a virulence mechanism of plant pathogens. Annu. Rev. Phytopathol. 51, 521–542. doi: 10.1146/annurev-phyto-082712-102312
Eichinger, V., Nussbaumer, T., Platzer, A., Jehl, M. A., Arnold, R., and Rattei, T. (2016). EffectiveDB: updates and novel features for a better annotation of bacterial secreted proteins and type III, IV, VI secretion systems. Nucleic Acids Res. 44, D669–D674. doi: 10.1093/nar/gkv1269
Faruque, O. M., Miwa, H., Yasuda, M., Fujii, Y., Kaneko, T., Sato, S., et al. (2015). Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 81, 6710–6717. doi: 10.1128/AEM.01942-15
Feng, F., and Zhou, J. M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15, 469–476. doi: 10.1016/j.pbi.2012.03.004
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M. B., Su, H., Jones, C. H., et al. (2019). Legume nodulation: the host controls the party. Plant Cell Environ. 42, 41–51. doi: 10.1111/pce.13348
Fotiadis, C. T., Dimou, M., Georgakopoulos, D. G., Katinakis, P., and Tampakaki, A. P. (2012). Functional characterization of NopT1 and NopT2, two type III effectors of Bradyrhizobium japonicum. FEMS Microbiol. Lett. 327, 66–77. doi: 10.1111/j.1574-6968.2011.02466.x
Gareau, J. R., and Lima, C. D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. doi: 10.1038/nrm3011
Ge, Y. Y., Xiang, Q. W., Wagner, C., Zhang, D., Xie, Z. P., and Staehelin, C. (2016). The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 67, 2483–2494. doi: 10.1093/jxb/erw065
Hotson, A., Chosed, R., Shu, H., Orth, K., and Mudgett, M. B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50, 377–389. doi: 10.1046/j.1365-2958.2003.03730.x
Hubber, A., Vergunst, A. C., Sullivan, J. T., Hooykaas, P. J., and Ronson, C. W. (2004). Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54, 561–574. doi: 10.1111/j.1365-2958.2004.04292.x
Kambara, K., Ardissone, S., Kobayashi, H., Saad, M. M., Schumpp, O., Broughton, W. J., et al. (2009). Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 71, 92–106. doi: 10.1111/j.1365-2958.2008.06507.x
Kim, J. G., Stork, W., and Mudgett, M. B. (2013). Xanthomonas type III effector XopD desumoylates tomato transcription factor SIERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13, 143–154. doi: 10.1016/j.chom.2013.01.006
Kim, J. G., Taylor, K. W., Hotson, A., Keegan, M., Schmelz, E. A., and Mudgett, M. B. (2008). XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20, 1915–1929. doi: 10.1105/tpc.108.058529
Kim, J. G., Taylor, K. W., and Mudgett, M. B. (2011). Comparative analysis of the XopD type III secretion (T3S) effector family in plant pathogenic bacteria. Mol. Plant Pathol. 12, 715–730. doi: 10.1111/j.1364-3703.2011.00706.x
López-Baena, F. J., Ruiz-Sainz, J. E., Rodríguez-Carvaja, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:755. doi: 10.3390/ijms17050755
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Marie, C., Deakin, W. J., Viprey, V., Kopciñska, J., Golinowski, W., Krishnan, H. B., et al. (2003). Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 16, 743–751. doi: 10.1094/mpmi.2003.16.9.743
Matunis, M. J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. doi: 10.1083/jcb.135.6.1457
Morimoto, M., and Boerkoel, C. F. (2013). The role of nuclear bodies in gene expression and disease. Biology (Basel) 2, 976–1033. doi: 10.3390/biology2030976
Nelson, M. S., and Sadowsky, M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6:491. doi: 10.3389/fpls.2015.00491
Oldroyd, G. E. (2013). Speak, friend, and enter: signaling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Perret, X., Staehelin, C., and Broughton, W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64, 180–201. doi: 10.1128/mmbr.64.1.180-201.2000
Pruneda, J. N., Durkin, C. H., Geurink, P. P., Ovaa, H., Santhanam, B., Holden, D. W., et al. (2016). The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 63, 261–276. doi: 10.1016/j.molcel.2016.06.015
Rodrigues, J. A., López-Baena, F. J., Ollero, F. J., Vinardell, J. M., del Rosario Espuny, M., Bellogín, R. A., et al. (2007). NopM and NopD are rhizobial nodulation outer proteins: identification using LC-MALDI and LC-ESI with a monolithic capillary column. J. Proteome Res. 6, 1029–1037. doi: 10.1021/pr060519f
Sánchez, C., Mercante, V., Babuin, M. F., and Lepek, V. C. (2012). Dual effect of Mesorhizobium loti T3SS functionality on the symbiotic process. FEMS Microbiol. Lett. 330, 148–156. doi: 10.1111/j.1574-6968.2012.02545.x
Schirrmeister, J., Friedrich, L., Wenzel, M., Hoppe, M., Wolf, C., Göttfert, M., et al. (2011). Characterization of the self-cleaving effector protein NopE1 of Bradyrhizobium japonicum. J. Bacteriol. 193, 3733–3739. doi: 10.1128/JB.00437-11
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Staehelin, C., and Krishnan, H. B. (2015). Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochem. J. 470, 263–274. doi: 10.1042/BJ20150518
Sugawara, M., Takahashi, S., Umehara, Y., Iwano, H., Tsurumaru, H., Odake, H., et al. (2018). Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity. Nat. Commun. 9:3139. doi: 10.1038/s41467-018-05663-x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, C. M., Li, M. Y., Yang, P. Y., Chang, S. H., Ho, Y. P., Lin, H., et al. (2015). Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopDXcc8004. PLoS One 10:e0117067. doi: 10.1371/journal.pone.0117067
Tan, L., Rong, W., Luo, H., Chen, Y., and He, C. (2014). The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 204, 595–608. doi: 10.1111/nph.12918
Tang, F., Yang, S., Liu, J., and Zhu, H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 170, 26–32. doi: 10.1104/pp.15.01661
Teulet, A., Busset, N., Fardoux, J., Gully, D., Chaintreuil, C., Cartieaux, F., et al. (2019). The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U.S.A. 116, 21758–21768. doi: 10.1073/pnas.1904456116
Tsukui, T., Eda, S., Kaneko, T., Sato, S., Okazaki, S., Kakizaki-Chiba, K., et al. (2013). The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 79, 1048–1051. doi: 10.1128/AEM.03297-12
Tsurumaru, H., Hashimoto, S., Okizaki, K., Kanesaki, Y., Yoshikawa, H., and Yamakawa, T. (2015). A putative type III secretion system effector encoded by the MA20_12780 gene in Bradyrhizobium japonicum Is-34 causes incompatibility with Rj4 genotype soybeans. Appl. Environ. Microbiol. 81, 5812–5819. doi: 10.1128/AEM.00823-15
Viprey, V., Greco, A. D., Golinowski, W., Broughton, W. J., and Perret, X. (1998). Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28, 1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x
Wenzel, M., Friedrich, L., Göttfert, M., and Zehner, S. (2010). The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol. Plant Microbe Interact. 23, 124–129. doi: 10.1094/MPMI-23-1-0124
Xin, D. W., Liao, S., Xie, Z. P., Hann, D. R., Steinle, L., Boller, T., et al. (2012). Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 8:e1002707. doi: 10.1371/journal.ppat.1002707
Xu, C. C., Zhang, D., Hann, D. R., Xie, Z. P., and Staehelin, C. (2018). Biochemical properties and in planta effects of NopM, a rhizobial E3 ubiquitin ligase. J. Biol. Chem. 293, 15304–15315. doi: 10.1074/jbc.RA118.004444
Yang, S., Tang, F., Gao, M., Krishnan, H. B., and Zhu, H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 18735–18740. doi: 10.1073/pnas.1011957107
Yasuda, M., Miwa, H., Masuda, S., Takebayashi, Y., Sakakibara, H., and Okazaki, S. (2016). Effector-triggered immunity determines host genotype-specific incompatibility in legume-rhizobium symbiosis. Plant Cell Physiol. 57, 1791–1800. doi: 10.1093/pcp/pcw104
Zhang, L., Chen, X. J., Lu, H. B., Xie, Z. P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Keywords: effector, legume, nitrogen fixation, nodulation, protease, small ubiquitin-related modifier, symbiosis, type III protein secretion system
Citation: Xiang Q-W, Bai J, Cai J, Huang Q-Y, Wang Y, Liang Y, Zhong Z, Wagner C, Xie Z-P and Staehelin C (2020) NopD of Bradyrhizobium sp. XS1150 Possesses SUMO Protease Activity. Front. Microbiol. 11:386. doi: 10.3389/fmicb.2020.00386
Received: 21 November 2019; Accepted: 21 February 2020;
Published: 20 March 2020.
Edited by:
Benjamin Gourion, UMR2594 Laboratoire Interactions Plantes-Microorganismes (LIPM), FranceReviewed by:
Jung-Gun Kim, Stanford University, United StatesFrancisco Javier López-Baena, University of Seville, Spain
Shin Okazaki, Tokyo University of Agriculture and Technology, Japan
Copyright © 2020 Xiang, Bai, Cai, Huang, Wang, Liang, Zhong, Wagner, Xie and Staehelin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Ping Xie, eGllenBpbmdAbWFpbC5zeXN1LmVkdS5jbg==; Christian Staehelin, Y3N0QG1haWwuc3lzdS5lZHUuY24=
 Qi-Wang Xiang
Qi-Wang Xiang Juan Bai
Juan Bai Jie Cai
Jie Cai Yan Wang
Yan Wang Zhi Zhong
Zhi Zhong Christian Wagner
Christian Wagner Christian Staehelin
Christian Staehelin