- 1Department for Integrative Risk Assessment, Division for Risk Assessment, Data and Statistics, AGES – Austrian Agency for Health and Food Safety, Vienna, Austria
- 2Université de Lorraine, CNRS, LCPME, Nancy, France
The dissemination of antimicrobial resistance (AMR) is one of the biggest challenges faced by mankind in the public health domains. It is currently favored by a lack of confinement between waste disposal and food production in the environmental compartment. To date, much effort has been devoted into the elucidation and control of cell-associated propagation of AMR. However, substantial knowledge gaps remain on the contribution of cell-free DNA to promote horizontal transfers of resistance genes in wastewater and downstream environments. Cell free DNA, which covers free extracellular DNA (exDNA) as well as DNA encapsulated in vesicles or bacteriophages, can persist after disinfection and promote gene transfer in the absence of physical and temporal contact between a donor and recipient bacteria. The increasing water scarcity associated to climatic change requires developing innovative wastewater reuse practices and, concomitantly, a robust evaluation of AMR occurrence by implementing treatment technologies able to exert a stringent control on AMR propagation in downstream environments exposed to treated or non-treated wastewater. This necessarily implies understanding the fate of ARGs on various forms of cell-free DNA, especially during treatment processes that are permissive to their formation. We propose that comprehensive approaches, investigating both the occurrence of ARGs and their compartmentalization in different forms of cellular or cell-free associated DNA should be established for each treatment technology. This should then allow selecting and tuning technologies for their capacity to limit the propagation of ARGs in any of their forms.
Introduction: Colliding Challenges in Health and Sustainable Development
With 10 millions expected deaths per year by 2050, the mortality due to antimicrobial resistance (AMR) will surpass all known human diseases, making the fight against AMR one of the biggest challenges faced by Mankind in order to preserve the effectiveness of our modern medicine (de Kraker et al., 2016; O’Neill, 2016; Nature Microbiology supplement, 20161). The emergence and dissemination of AMR is an unavoidable aspect of bacterial evolution following the consumption of antibiotics (Courvalin, 2005), but the excessive and inappropriate use of antimicrobials, together with the lack of innovation in discovering new antimicrobials, is driving modern medicine to the edge of a new dark age resembling the pre-antibiotic era (Levy and Marshall, 2004; Roberts and Simpson, 2008). This has led national and international organizations to take action in the public health and veterinary/farming domains for a better and confined usage of antimicrobials. But, are the proposed antibiotic stewardship and action plans sufficient for controlling the global increase of AMR? Surely not, as the spread of resistant bacteria (ARB) and their resistance genes (ARGs) over ecosystem boundaries appears now to be the dominant contributing factor for AMR dissemination (Collignon et al., 2018). Antimicrobial therapies exert selective processes allowing ARB to proliferate in microbiotas. Considering the continuum between human activities and the environment, AMR-determinants are also continuously released in anthropogenically-impacted environments where ARB can persist, accumulate, transfer their ARGs to indigenous microbes, and finally re-enter the food chain and be transferred to human and animal commensals and pathogens (Davies and Davies, 2010). Facing the global population growth, the environment is increasingly exposed to pollutants including ARB and ARGs. Concomitantly, the shortage in resources promotes an increasing demand for reuse and especially for recycled water. If water scarcity is another of the big challenges faced by humanity, the increasing need for wastewater reuse may also promote shortcuts in the AMR dissemination routes with bigger risks for human and animal health. Wastewater treatment plant (WWTP) effluents are at the interface between human activities and the downstream environment, therefore tackling water scarcity through water reuse will necessarily imply a better understanding of the AMR dissemination mechanisms to control them properly. Limiting the discharge of ARB in the downstream environment is obviously a critical point but this may not be enough to prevent the spread of ARGs toward and among environmentally adapted bacteria that may become reservoirs of resistances. Indeed, if controlling the discharge of ARB should provide a mean to limit the spread of ARGs involving cell-contact mechanism (e.g., conjugation), it just forgets to consider non-cell associated ARG transfer. As a matter of fact, non-cell associated ARG transfer is largely underestimated and we propose that cell-free sources of DNA are adequately considered for the risk assessment of ARG dissemination.
Cell-Free DNA as a Neglected Source of Args
The dissemination of ARB and ARGs in effluent-impacted environments is of concern when considering the safety aspects of reclaimed water (Hong et al., 2018). The efficiency of the processes implemented for treating effluents has been assessed by various techniques relying either on the culturability of ARB or on molecular approaches to estimate the abundance of ARGs. As a matter of fact, both approaches are not equivalent, because ARGs are not always associated with cultivable ARB (Figure 1). On the one hand, cell-associated ARGs can reside in bacteria that are non-cultivable (lack of proper culture medium, “viable but non-cultivable state,” or dead cells). On the other hand, there is also a non-negligible amount of ARGs carried on cell-free DNA (i.e., exDNA, bacteriophage-associated DNA, and membrane vesicle-associated DNA). This raises two interconnected questions with respect to wastewater reuse and associated wastewater treatment processes: (i) Is reclaimed wastewater adequately safe if a treatment process eliminates ARB below detection limits while ARGs are still present and quantifiable in the effluent? The correct answer will depend to some extent on the ability of ARB for regrowth, but undoubtedly also on the fate of ARGs encoded on cell-free DNA. Consequently, the second question arises: (ii) what is the persistence of ARGs associated to cell-free DNAs and their probability to be transferred and expressed in living bacteria? In the following sections, three main mechanisms leading to cell-free DNAs and their uptake by living bacteria, namely natural transformation of bacteria with free exDNA, transduction by bacteriophages and related particles, and DNA transfer by membrane vesicles (MVs), will be discussed in the context of the horizontal gene transfer (HGT) of ARGs. At present it is believed that, in the environment, HGT is mostly driven by conjugative elements, which explains why they have focused much attention so far (Sørensen et al., 2005). However, this lack of interest for alternative HGT pathways involving cell-free DNA probably reflects the technical difficulties to study such pathway rather than a real estimation of their implication. Actually, in spite of contributing to the global occurrence of ARGs in the environment, cell-free DNAs may well represent a significant source for ARG dissemination that suffers a lack of attention although it should be considered as a hazard in the context of the wastewater reuse.
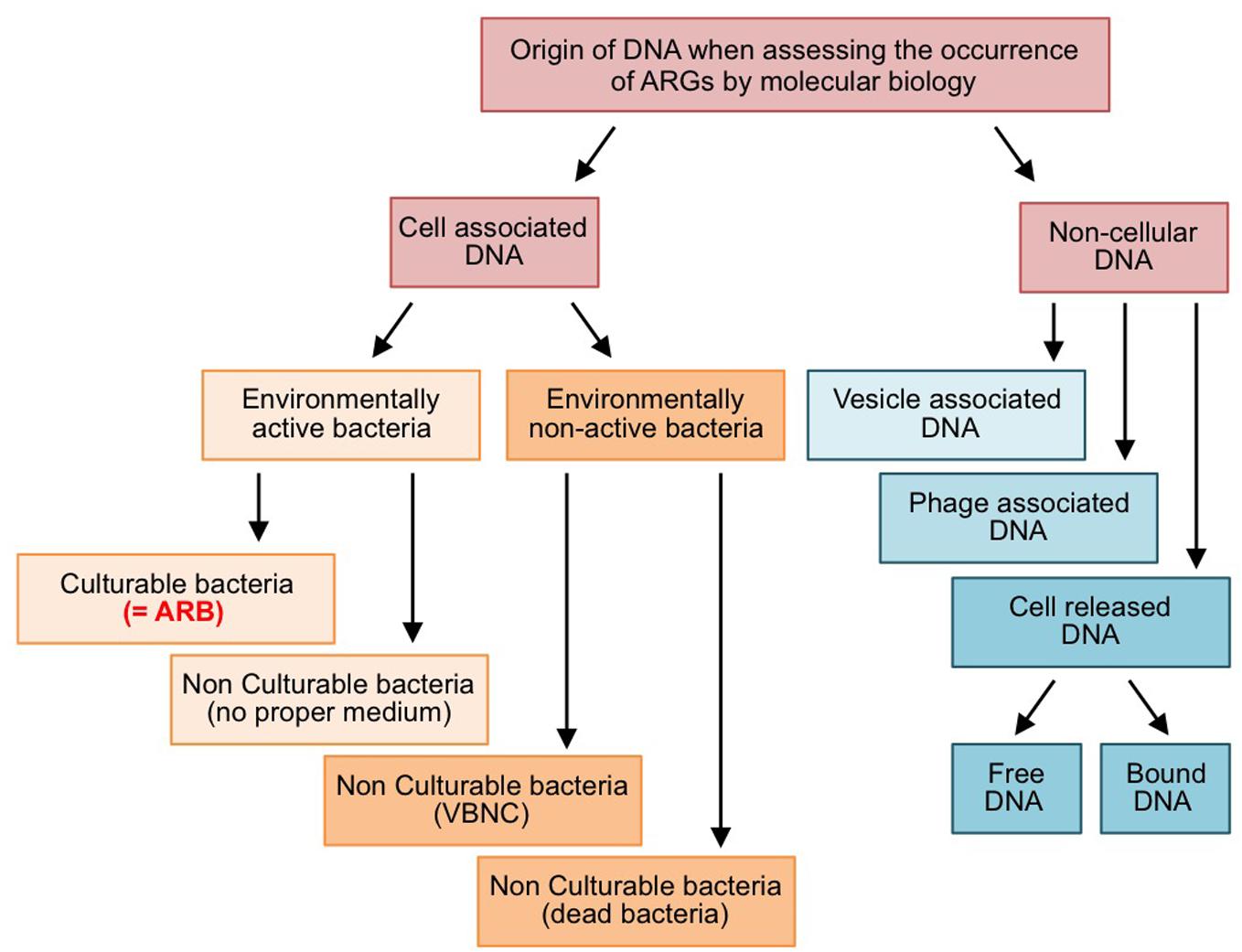
Figure 1. Forms and origins of ARGs quantified by molecular biology approaches. VBNC: viable but non-culturable bacteria.
Membrane Vesicle-Mediated Horizontal Gene Transfer
Lately, the formation of MVs by bacteria has received increasing attention, as they seem to be involved in the transport of a myriad of biomolecules between bacteria (Grüll et al., 2018; Toyofuku et al., 2019). Several types of MVs can be distinguished. In Gram-negative bacteria, budding of the outer membrane (OM) results in the formation of 10–300 nm wide OM vesicles (OMVs) trapping periplasmic material. Several non-exclusive mechanisms promoting local OM shape alterations have been proposed to explain OMV formation (Gaudin et al., 2013; Toyofuku et al., 2015; Roier et al., 2016). It starts with a loss of linkage between the OM and peptidoglycan, which then allows OM budding to happen. This is further promoted by various factors affecting the curvature of the membranes such as the accumulation of OM proteins, the turgor pressure, the OM rigidity due to the accumulation saturated fatty acids, the anionic charge repulsion between saccharide chains of lipopolysaccharides (LPS), the wrong localization of phospholipids in the outer layer of the OM (instead of LPS), and the insertion of amphiphilic molecules in the OM (e.g., quinolones). Sometimes, plasma membranes are dragged along during OM budding, thus leading to double layered outer-inner membrane vesicles (O-IMVs) with cytoplasmic content, including DNA (Pérez-Cruz et al., 2015). MVs are also produced by Gram-positive bacteria following localized cell wall damages with the concomitant action of the turgor pressure (Toyofuku et al., 2017), and were also observed in Archaea (Gaudin et al., 2013). The size and the relative amount of MVs tend to increase in antimicrobial-stressed environments which may favor OM destabilization and budding (Fulsundar et al., 2014; Devos et al., 2017).
Several important functions have been attributed to bacterial MVs (Toyofuku et al., 2015). MVs appear as a major component of biofilm matrices and they may be involved in bacterial cell protection by adsorbing antimicrobial agents, including bacteriophages (Schooling and Beveridge, 2006). MVs also seem to fulfill multiple roles depending on the kind of biological material they trap and transport: cell-cell communication (transport of signaling molecules), virulence (transport of toxins), electron transfer (transport of redox-active proteins), and finally HGT (transport of DNA) (Toyofuku et al., 2017; Grüll et al., 2018), for which the size distribution of MV-packaged DNA ranges from a few base pairs to several dozen kilobases depending on the species and its physiological state (Klieve et al., 2005; Biller et al., 2017; Bitto et al., 2017). Several studies reported MV-assisted transformation of chromosomal and plasmid DNA, between either Gram-negative bacteria, Gram-positive bacteria or Archaea, and even from bacteria to the nuclear fraction of eukaryotic cells (Bitto et al., 2017). In 2011, Rumbo et al. (2011) reported the MV-assisted transfer of the plasmid-borne blaOXA24 resistance gene from two clinical isolates of Acinetobacter baumannii into the susceptible strain ATCC 17978, therefore demonstrating that ARGs are technically transferable by MV. However, demonstration of MV-mediated HGT remains relatively sporadic and solely assayed in pure cultures, where both the environmental dimension and frequency of the phenomenon remain unexplored (Domingues and Nielsen, 2017).
The global occurrence of ARGs in environmental MVs is a subject that is largely unexplored, and even less is known regarding MVs in WWTP effluents or in effluent-impacted downstream environments. This probably comes from the technical challenges of separating MVs from bacteriophages as they share common properties for isolation (Biller et al., 2017), raising the question of the accuracy of what was once attributed to bacteriophages. However, it can be taken for granted that (i) MVs are produced by numerous bacteria, (ii) they can transport any kind of DNA and this should include ARGs, (iii) with a membrane fusion-based mechanisms for MV-assisted DNA transformation, no taxonomic limitation are expected regarding the DNA uptake process, (iv) maintaining the acquired DNA in bacterial cell is likely to meet the same limitations as for natural transformation (see below), and (v) the budding process is favored in antibiotic-stress environments. Putting these observations together, the implication of MVs in the dissemination of ARGs in WWTPs and downstream environments is likely but its incidence remains to be elucidated. With this respect, structures once known as “biological colloids” or “liposomes-like biogenic materials,” probably MVs, have already been described in wastewater effluents and sometime reported to affect the operation and the quality of treatment processes (Song et al., 2010; Barry et al., 2014; Toyofuku et al., 2015). Second, considering that WWTP combines both MV forming bacteria and stressing molecules (e.g., antibiotics), it is of prime importance to determine: (i) whether the induction of MV formation can occur during the wastewater treatment processes, (ii) where it occurs during the process, (iii) whether a given advanced treatment process could eliminate or retain MVs in the final effluents, and finally (iv) the fate of the MVs discharged from WWTPs in downstream (aquatic and terrestrial) environments.
Horizontal Gene Transfer by Natural Transformation
Natural transformation describes the physiologically regulated uptake and genomic integration of free exDNA by competent bacteria (Nielsen and Van Elsas, 2019). Approximately 130 bacterial species from almost all branches of the bacterial tree of life, including important human and animal pathogens and soil bacteria, have been characterized as being able to develop competence under naturally occurring environmental conditions (Johnston et al., 2014; Woegerbauer et al., 2015). The DNA uptake process either utilizes distinct species-specific uptake sequences or is sequence-independent (Nielsen and Van Elsas, 2019). Efficient transformation depends on sequence similarity between the incoming linear DNA and the recipient genome for homologous recombination (Lorenz and Wackernagel, 1994). The efficiency of recombination decreases in a log-linear relationship with increasing sequence divergence between DNA partner molecules (Ray et al., 2009), dropping below the detection limit if sequence identity is lower than ca. 75% (Townsend et al., 2003). Two additional recombination processes concerning bacterial transformation, homology facilitated illegitimate recombination (HFIR; de Vries and Wackernagel, 2002), and short patch double illegitimate recombination (SPDIR; Harms et al., 2016), with relaxed sequence identity requirements, have been identified but are supposed to occur at insignificant rates in natural bacterial populations (Nielsen and Van Elsas, 2019). However, for the formation of mosaic ARGs, HFIR is thought to play a decisive role (Woegerbauer et al., 2015). Natural transformation does not require physical contact between donor and recipient bacteria but allows large tempo-spatial separation between the genetic source of information and the recipient (Johnston et al., 2014).
Antibiotic resistance genes are prevalent in WWTPs (Reinthaler et al., 2003; da Costa et al., 2006; Auerbach et al., 2007; Novo and Manaia, 2010; Munir et al., 2011; Yuan et al., 2014) and in their effluents (Liu et al., 2018). Initially attributed to intracellular DNA, it was demonstrated that ARGs encoded on free exDNA persist throughout different WWTP compartments (1–16 ng/ml) and ARG-associated exDNA is present in substantial amounts even in purified discharged effluents (Zhang et al., 2018). Although WWTPs reduce ARG loads by up to 3 orders of magnitude (Auerbach et al., 2007; Zhang et al., 2009; Munir et al., 2011), 105–109 copies of ARGs per liter (total DNA) of effluent are generally discharged (Gao et al., 2012; Zhang et al., 2018), which can foster the spread of ARGs in receiving environments (Proia et al., 2016). Upon entry into the wastewater environment, a fraction of exDNA adsorbs to particulate material, rendering it inaccessible to nuclease (Breazeal et al., 2013). A rapid degradation of DNA from external sources (e.g., from decomposing plant leaves or from spikes with purified plasmid DNA) is observed in non-sterile soils (Romanowski et al., 1992; Pote et al., 2005). A similar fate of exDNA originating from WWTP effluents is expected if reclaimed water is used for irrigation of agricultural soils. However, a small fraction of the inoculated exDNA is usually adsorbed to sand particles, clay and other soil colloids (Nielsen et al., 2007). Some DNA is also tightly bound to humic acids (Crecchio and Stotzky, 1998). These interactions stabilize dsDNA fragments in soil ecosystems and render them inaccessible for DNases (Nielsen et al., 2007). ExDNA fragments have therefore been reported to be detectable for up to 2 years in agricultural soil environments (Gebhard and Smalla, 1999; Nielsen et al., 2007). Although bound exDNA remains to be accessible for uptake by competent bacteria, the observed long-term physical persistence of DNA fragments appears to be in contrast to a shorter-term availability of exDNA for transformation of competent bacteria as observed in soil microcosm experiments (Nielsen et al., 2007). Actively excreted exDNA is a core structural component of bacterial biofilms especially in activated sludge (Dominiak et al., 2011). High abundances of extracellular ARGs (1.7 × 103–4.2 × 108 copies per gram of dry weight) have been observed in sludge (Zhang et al., 2013). Exposure to sub-inhibitory concentrations of antibiotics leads to increased ARG concentrations and higher amounts of exDNA in the exposed ecosystem (Johnson et al., 2013; Lewenza, 2013; Doroshenko et al., 2014; Hathroubi et al., 2015; Schilcher et al., 2016), thus making exDNA an important source of genetic information fueling HGT by natural transformation (Wang et al., 2002; Merod and Wuertz, 2014).
While natural transformation in wastewater environments is believed to occur, informations on transformation frequencies, and conditions that may induce competence of bacteria in these habitats are scarce. Agricultural soils and associated crop plants are prominent downstream environments exposed to ARGs encoded on exDNA if irrigated with treated WWTP effluents. A limiting factor for efficient transformation is the availability of free exDNA (Nielsen and Van Elsas, 2019). However, the situation is unclear as fragments of exDNA show long-term persistence (several years) in soil and other natural environments (Overballe-Petersen et al., 2013), and despite adsorption to soil particles exDNA is capable to transform competent cells in these environments (Romanowski et al., 1993; Thomas and Nielsen, 2005; Morrissey et al., 2015). HGT by natural bacterial transformation in soils is assumed as an infrequent process (Nielsen et al., 1998; Pietramellara et al., 2006; Pietramellara et al., 2007). However, this may be an underestimation due to the substantial number of non-cultivable soil bacteria that may be able to develop competence (Pietramellara et al., 2009).
Environmental conditions in soil adverse to bacterial growth and proliferation are expected to be the rule rather than the exception. HGT via transformation, however, may occur in the absence of host cell proliferation. There is still a substantial lack of thorough understanding of the real, microhabitat-specific, impact of these processes in relation to the selective forces that operate in these ecological niches. Bioinformatics analyses of soil metagenomes provide strong evidence for the importance of HGT for bacterial biodiversity in natural soil habitats. It must be stressed that the impact and long-term effects of natural transformation – which is considered a rare event in soil environments – is not determined by the frequency of the event but by selection (Nielsen and Townsend, 2004; Townsend et al., 2012; Nielsen et al., 2014). Stochastic survival, dissemination and natural selection will determine the outcome of natural transformation (Nielsen and Van Elsas, 2019).
Bacteriophage-Assisted DNA Transfer
Transduction is an HGT mechanism involving bacterial viruses, i.e., bacteriophages (or phages) that represent the most abundant class of microorganisms on Earth (1030–1032 phages vs. 1029–1031 bacteria; Chibani-Chennoufi et al., 2004; Brabban et al., 2005). After their injection into a bacterial cell, phage genomes can either enter into an active replicative state, leading to the production of new phages (lytic state), or into a dormancy state (lysogenic state). The latter case is restricted to temperate bacteriophages, which integrate into the host genome as a prophage. Classically, there are two distinct mechanisms of transduction. The “generalized transduction,” mediated by either lytic or temperate (lysogenic) phages, allows transferring any part of a bacterial genome form one bacteria to another, by accidentally packaging bacterial DNA fragments instead of the phage genome. On the other hand, the “specialized transduction,” solely mediated by temperate phages, leads to the HGT of the few bacterial genes located in the vicinity of the prophage insertion sites in the bacterial genome. Here, the inaccurate excision of prophage genome leads to the formation of chimerical prophage-bacterial DNA molecules that are packaged in the newly formed virions. When virions contain bacterial DNA, they retain their “nucleic acid syringe” function, allowing the packaged bacterial DNA to be injected into a new recipient bacterium (Calendar, 2012). A successful gene transduction event requires the injected DNA, or part of it, to integrate into the bacterial host genome to be maintained and expressed. Linear bacterial DNA fragments can be integrated in the new host genome thanks to its homologous or illegitimate recombination machineries, whereas transduced mobile genetic elements (MGEs) such as transposons or plasmids can ensure their maintenance by transposition or self-replication, respectively (Berg et al., 1983; Jiang and Paul, 1998; Calendar, 2012). It is worth noting that gene transfer can also be mediated by phage-resembling particles encoded by bacterial chromosomes, called Gene Transfer Agents (GTAs; Grüll et al., 2018). Lately, a new phenomenon called “lateral transduction,” initially described in Staphylococcus aureus prophages, seems to be involved in gene transfers in a wide range of bacterial species (Chen et al., 2018; Chiang et al., 2019). Briefly, upon entry in lytic state, some prophages do not start their activation by an excision from the chromosome but undergo an in situ genome replication. Then, DNA packaging begins while the prophages are still integrated in the chromosome, and may proceeds beyond the prophage boundaries, using over hundreds kilobases of adjacent chromosomal DNA as a substrate for filling capsids. The gene transfer frequencies based on lateral transduction appeared to be higher than those corresponding to generalized or specialized transductions.
Although HGT mediated by infectious phages and GTAs have been well studied in vitro, their occurrence in natural environments remains to be fully elucidated (Balcázar, 2018; Grüll et al., 2018). Metagenomic or qPCR-based studies investigating the content of phages/GTAs purified from natural environments impacted by human activities, animal/Human feces, or WWTP sludge and effluents have shown that both particles often carried ARGs (Muniesa et al., 2013; Calero-Cáceres et al., 2014; Ross and Topp, 2015; Colombo et al., 2016; Lekunberri et al., 2017; Brown-Jaque et al., 2018; Larrañaga et al., 2018; Wang et al., 2018). A recent work based on database mining has shown that phage genomes rarely encode ARGs by themselves, therefore suggesting that most phages/GTAs associated ARGs are on transduced bacterial DNA fragments (Enault et al., 2017). Although important for maintaining the transduced DNA in the new host, to date, the association of such transduced ARGs with MGEs has only be described recently (Yang et al., 2018).
Many environmental stressors, anthropogenic or not, have been demonstrated to induce phage proliferation within environmental microbial communities. These stressors are often of chemical nature: genotoxic compounds (e.g., antibiotics), metallic trace elements (e.g., Cu2+, Cd2+), polycyclic aromatic hydrocarbons (e.g., phenanthrene, pyrene), and polychlorinated biphenyls or pesticides (Danovaro et al., 2003; Jończyk et al., 2011; Lee et al., 2006; Allen et al., 2010); but can also be of physico-chemical nature such as temperature increase, UV radiation, and Ca2+ depletion or inorganic phosphate enrichment (Danovaro et al., 2003; Yue et al., 2012; Colomer-Lluch et al., 2014). These stressors are commonly encountered in WWTPs, as pollutants or as disinfection processes, and in the environments in which treated/untreated wastewater are discharged. Therefore, studying the ARG content of phages/GTAs released from wastewater treatment facilities is of particular interest, to identify specific pollutants and processes favoring the formation of transducing particles, and evaluating the role of phages/GTAs in the dissemination of ARGs in the downstream environments (Calero-Cáceres et al., 2014).
Considering the environmental abundance of phages/GTAs and their in vitro ability to mobilize ARGs, they are assumed to play a significant role in the environmental dissemination of ARGs/MGEs as suggest by a few studies, especially in environments impacted by human activities (Kenzaka et al., 2007; Ross and Topp, 2015; Shousha et al., 2015). Unfortunately, the effective transfer of a specific ARG by transduction has never been formally shown in environmental settings (Ross and Topp, 2015). Consequently, the real chances of success of the transduction of ARGs, i.e., their stable integration into the genome of a recipient bacterium after being injected into the cell, have never been explored and the global propensity of the phenomenon to occur in the environment remains to be assessed quantitatively. The phage host range is believed to be one of the limiting parameters for transduction to occur. However, recent works in this field tend to show that phages exhibiting broad host ranges, sometime spanning multiple genera, are more common than anticipated (de Jonge et al., 2019). At this stage, it is worth distinguishing between the “penetrative-host-range,” which refers to the bacteria into which phages can deliver DNA (or RNA) without necessarily engendering a progeny, and the narrower “productive-host-range,” i.e., the bacteria in which infections effectively lead to the production and release of virions (Hyman and Abedon, 2010). Consequently, transducing phages can deliver DNA in larger host-ranges than their usually reported “productive-host-range.” Altogether, the global transducing properties of natural phage communities remain to be determined but, as recently suggested, broad-host-range transducing phages are likely frequent in the environment (Kenzaka et al., 2007; de Jonge et al., 2019).
Are Wastewater Treatment Effluents Hot Spots for Hgt Promoted by Cell-Free DNA?
Nowadays, it is well understood that HGT plays a significant role in the dissemination of ARGs in bacterial communities. However, to date much effort has primarily focused on elucidating HGT events promoted by MGEs, especially conjugative elements, which rely on direct cell-to-cell contacts (Sørensen et al., 2005). Actually, if conjugation is perceived as the key mechanism of HGT in soil (van Elsas et al., 2003; Heuer and Smalla, 2012, Zhang et al., 2014), there is currently no information available on the relative importance of transformation compared to conjugation and transduction (Nielsen and Van Elsas, 2019). Yet, HGT can also occur by alternative mechanisms comprising transduction, transformation and membrane vesicle-assisted transformation, for which the transfer of DNA can take place distantly from the donor host in time and space. This is particularly worrying in the context of wastewater reuse because conventional estimation of effluent quality based on the reduction of ARB will be of no help to estimate the occurrence of ARGs in free exDNA (solvated or associated to particles), and/or phage/vesicle fractions. The abundance of ARGs in all of these fractions tends to increase in anthropogenically stressed environments. This is also the case for WWTPs, which receive both ARB and antibiotics that have been reported to increase vesicle formation or phage production. At present, there is an obvious lack of empirical evidence assessing the type and extent of cell-free DNAs in the environment and their persistence in various WWTP processes. This results from the fact that most emphasis has been given to HGT by “conventional” MGEs and little case has been given to HGT by cell-free DNA. Considering the non-negligible probability of ARG dissemination by means of cell-free DNAs, there is an urgent need to assess both the persistence of such cell-free DNAs and the efficiency of the cognate HGT mechanisms along wastewater reuse processes, to limit the dissemination of ARGs and preserve the downstream environments (i.e., food chain and water cycles) from being exposed. Such assessment will necessarily require implementing an integrated approach to determine the distribution of ARGs in each biological compartment (free DNA, cell-, phage-, or vesicle-associated DNA), as exemplified in Figure 2. The critical point of such approach lies on the separation of each biological compartment before quantifying their ARG abundances using standard molecular methods. Knowing the occurrence of relevant ARGs in each biological compartment and estimating independently their respective HGT frequencies, will improve ARG risk assessment in the reuse context and, in the meantime, allow adapting processes and practices to target ARGs in specific biological compartments. To date, we miss a comprehensive and integrated approach comparing the relative contribution of conjugation, transformation, and phage-/vesicle-assisted transduction in the HGT of relevant ARGs, so as to identify the most effective mechanisms. Such approach necessarily implies microcosm-/reactor-based experiments implementing sensitive molecular strategies for the detection of so-called “rare events” in complex environmental matrices, as proposed by Bellanger et al. (2014) for conjugation, for instance. Next to the efficiency of the different transfer mechanisms, the persistence of the different biological entities (DNA, phages, and vesicles) is a key parameter that needs to be further evaluated in the environmental matrices considered. With this respect, it is worth mentioning that a recent work considering raw wastewater shown that ARGs in phages tend to be more resistant than those in bacteria to temperature and pH stresses, as well as UV irradiation or chlorination disinfection processes (Calero-Cáceres and Muniesa, 2016). This longer persistence of ARGs in phages has also been demonstrated in the downstream environment (Calero-Cáceres et al., 2017). In wastewater, Zhang et al. (2018) reported persistence of exDNA and observed a substantial increase of the abundance ratio of exDNA to internal DNA along the whole WWTP treatment process indicative for an amplification of ARGs encoded on free exDNA during wastewater treatment. Disinfection measures appear to increase the content of extracellular ARGs in WWTPs and corresponding effluents (Zheng et al., 2017; Liu et al., 2018; Yuan et al., 2019). A rapid degradation of DNA from external sources (e.g., from decomposing plant leaves or from spikes with purified plasmid DNA) is observed in non-sterile soils (Romanowski et al., 1992; Pote et al., 2005). A similar fate of exDNA originating from WWTP effluents is expected if reclaimed water is used for irrigation of agricultural soils. However, a small fraction of the inoculated exDNA is usually adsorbed to sand particles, clay and other soil colloids (Nielsen et al., 2007). Some DNA is also tightly bound to humic acids (Crecchio and Stotzky, 1998). These interactions stabilize dsDNA fragments in soil ecosystems and render them inaccessible for DNases (Nielsen et al., 2007). ExDNA fragments have therefore been reported to be detectable for up to 2 years in agricultural soil environments (Gebhard and Smalla, 1999; Nielsen et al., 2007). To the best of our knowledge, the persistence of MV in environmental matrices remains to be elucidated. Knowing the persistence of phages, vesicles and naked DNA, as well as their propensity to promote gene transfer will certainly be a step forward for assessing risk associated to ARG propagation, even if relatively rare. However, it should be kept in mind that ARG propagation is an intricate combination of DNA transfer, selection and clonal expansion of fit organisms. This means that a rare transfer event may become significant when considering the full scale of the receiving environment or the volumes of wastewater processed in WWTPs. Under such circumstances, even extremely rare transfer events may be rapidly amplified and disseminated in exposed bacterial populations. In summary, we propose to give more attention to cell-free transfer of ARGs that has been underestimated so far. Phages, vesicles and naked DNA share in common to possibly promote ARG transfer between bacteria that are distant in space and time. This rises the question of the efficiency of disinfection processes to control ARG propagation, since they solely inactivate ARB, while (i) alternative biological supports of ARGs may pass between cracks, (ii) some cell stresses may promote the release of DNA, phage and vesicles. Putting back these observations into the water reuse context, special care should be given to the identification of the ARG biological supports released in WWTP effluents so as to adapt the treatment processes consequently.
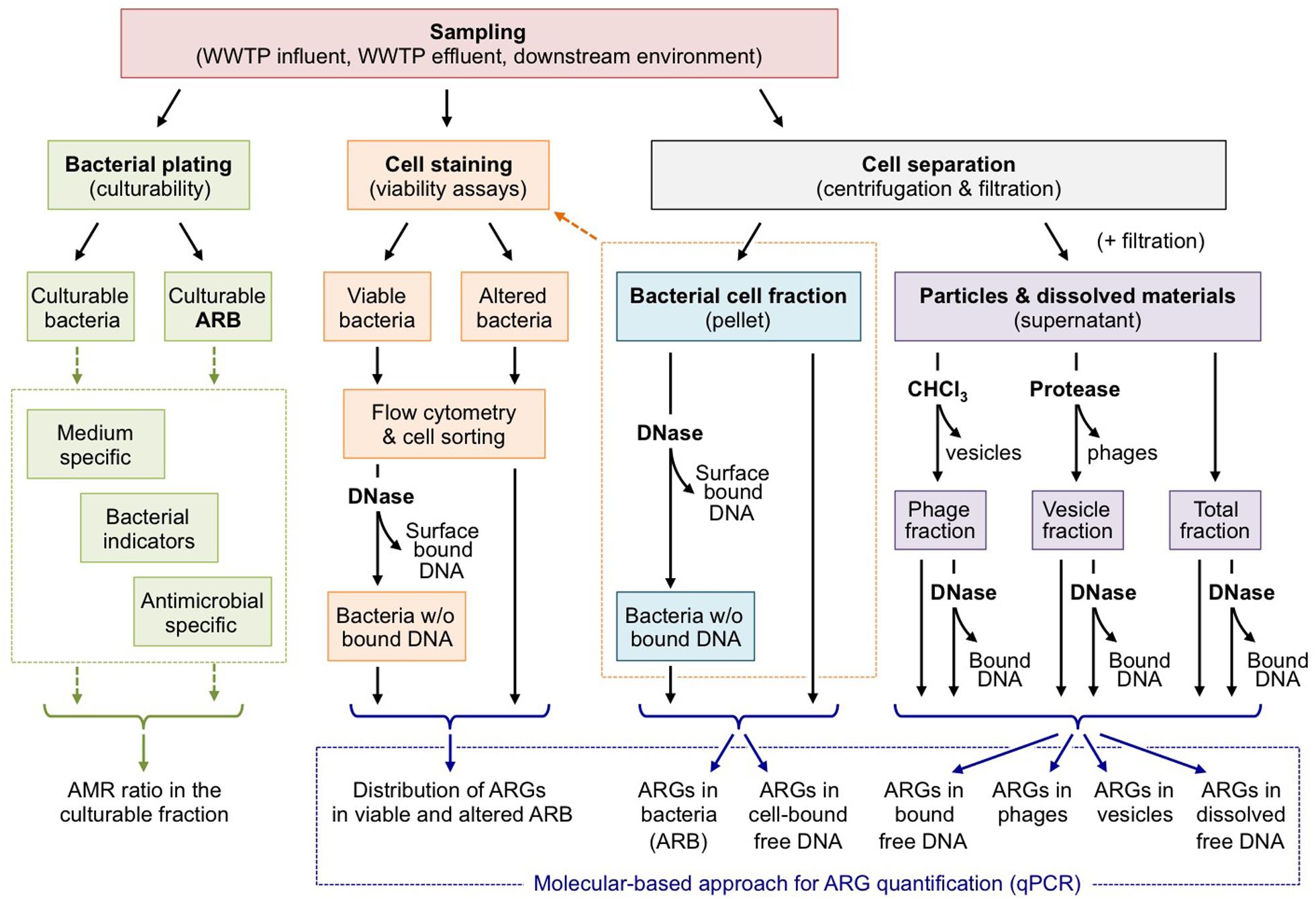
Figure 2. Assessing the distribution of ARGs in the different compartments of wastewater effluents. As stated by Biller et al. (2017) or Soler et al. (2015) MVs and phages remain difficult to separate. This figure should be understood as a working hypothesis.
Author Contributions
MW, XB, and CM equally contributed to this manuscript under the supervision of CM.
Funding
CM and XB wish to thank the French Agency for Food, Environmental and Occupational Health & Safety (Project “AquaResist,” PNREST Anses, 2018/1/052, and Project “PHARG,” PNREST Anses, 2017-3 ABR 12), and the Horizon 2020 EU Funding Research and Innovation program (Project “DSWAP,” Prima call, grant # 1822) for research funding. MW acknowledges the funding by the Austrian Ministry of Health and the Austrian Ministry of Agriculture for the projects MARGINS-I (GZ: BMASGK-74602/0005-IX/B/15/2019 and DaFNE Nr. 101446) and MARGINS-II (GZ: BMASGK-74602/0006-IX/B/15/2019 and DaFNE Nr. 101447).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript is based upon work carried out in the framework of the COST Action ES1403: New and emerging challenges and opportunities in wastewater reuse (NEREUS), supported by COST (European Cooperation in Science and Technology).
Footnotes
- ^ Resistance ascends the political summit. Nat. Microbiol. 1:16223; Davies, S. C. (2011). Annual Report of the Chief Medical Officer, Vol. 2, Infections and the Rise of Antimicrobial Resistance (Department of Health, 2013) (http://go.nature.com/2ecdeKA); Report to the President on Combating Antibiotic Resistance (President’s Council of Advisors on Science and Technology, 2014) (https://www.cdc.gov/drugresistance/pdf/report-to-the-president-on-combating-antibiotic-resistance.pdf); Global Action Plan on Antimicrobial Resistance (WHO, 2015) (http://go.nature.com/2bbijap); A Scientific Roadmap for Antibiotic Discovery (The Pew Charitable Trusts, 2016) (http://go.nature.com/2eofNsf); Tackling Drug-Resistant Infections Globally: Final Report and Recommendations (Review on Antimicrobial Resistance, 2016) (http://go.nature.com/2e3bkMJ); Draft Political Declaration of the High-level Meeting of the General Assembly on Antimicrobial Resistance (UN, 2016) (http://go.nature.com/2e3bMdF); CARA: The conscience of antimicrobial resistance accountability The Center For Disease Dynamics, Economics & Policy (2016) (http://go.nature.com/2dGFhQv).
References
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Auerbach, E. A., Seyfried, E. E., and McMahon, K. D. (2007). Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 41, 1143–1151. doi: 10.1016/j.watres.2006.11.045
Balcázar, J. L. (2018). How do bacteriophages promote antibiotic resistance in the environment? Clin. Microbiol. Infect. 24, 447–449. doi: 10.1016/j.cmi.2017.10.010
Barry, M. C., Hristovski, K., and Westerhoff, P. (2014). Membrane fouling by vesicles and prevention through ozonation. Environ. Sci. Technol. 48, 7349–7356. doi: 10.1021/es500435e
Bellanger, X., Guilloteau, H., Bonot, S., and Merlin, C. (2014). Demonstrating plasmid-based horizontal gene transfer in complex environmental matrices: a practical approach for a critical review. Sci. Total. Environ. 493, 872–882. doi: 10.1016/j.scitotenv.2014.06.070
Berg, C. M., Grullón, C. A., Wang, A., Whalen, W. A., and Berg, D. E. (1983). Transductional instability of Tn5-induced mutations: generalized and specialized transduction of Tn5 by bacteriophage P1. Genetics 105, 259–263.
Biller, S. J., McDaniel, L. D., Breitbart, M., Rogers, E., Paul, J. H., and Chisholm, S. W. (2017). Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates. ISME J. 11, 394–404. doi: 10.1038/ismej.2016.134
Bitto, N. J., Chapman, R., Pidot, S., Costin, A., Lo, C., Choi, J., et al. (2017). Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 7:7072.
Brabban, A. D., Hite, E., and Callaway, T. R. (2005). Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog. Dis. 2, 287–303. doi: 10.1089/fpd.2005.2.287
Breazeal, M. V., Novak, J. T., Vikesland, P. J., and Pruden, A. (2013). Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 47, 130–140. doi: 10.1016/j.watres.2012.09.044
Brown-Jaque, M., Calero-Cáceres, W., Espinal, P., Rodríguez-Navarro, J., Miró, E., González-López, J. J., et al. (2018). Antibiotic resistance genes in phage particles isolated from human faeces and induced from clinical bacterial isolates. Int. J. Antimicrob. Agents. 51, 434–442. doi: 10.1016/j.ijantimicag.2017.11.014
Calendar, R., (ed.) (2012). The Bacteriophages (Vol. 1). Springer Science & Business Media. New York, NY: Plenum Press.
Calero-Cáceres, W., Méndez, J., Martín-Díaz, J., and Muniesa, M. (2017). The occurrence of antibiotic resistance genes in a mediterranean river and their persistence in the riverbed sediment. Environ. Pollut. 223, 384–394. doi: 10.1016/j.envpol.2017.01.035
Calero-Cáceres, W., and Muniesa, M. (2016). Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 95, 11–18. doi: 10.1016/j.watres.2016.03.006
Calero-Caìceres, W., Melgarejo, A., Colomer-Lluch, M., Stoll, C., Lucena, F., Jofre, J., et al. (2014). Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ. Sci. Technol. 48, 7602–7611. doi: 10.1021/es501851s
Chen, J., Quiles-Puchalt, N., Chiang, Y. N., Bacigalupe, R., Fillol-Salom, A., Chee, M. S. J., et al. (2018). Genome hypermobility by lateral transduction. Science. 362, 207–212. doi: 10.1126/science.aat5867
Chiang, Y. N., Penadés, J. R., and Chen, J. (2019). Genetic transduction by phages and chromosomal islands: the new and noncanonical. PLoS Pathog. 15:e1007878. doi: 10.1371/journal.ppat.1007878
Chibani-Chennoufi, S., Bruttin, A., Dillmann, M.-L., and Brüssow, H. (2004). Phage-host interaction: an ecological perspective. J. Bacteriol. 186, 3677–3686. doi: 10.1128/jb.186.12.3677-3686.2004
Collignon, P., Beggs, J. J., Walsh, T. R., Gandra, S., and Laxminarayan, R. (2018). Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health 2, e398–e405. doi: 10.1016/s2542-5196(18)30186-4
Colombo, S., Arioli, S., Guglielmetti, S., Lunelli, F., and Mora, D. (2016). Virome-associated antibiotic-resistance genes in an experimental aquaculture facility. FEMS Microbiol. Ecol. 92:fiw003. doi: 10.1093/femsec/fiw003
Colomer-Lluch, M., Jofre, J., and Muniesa, M. (2014). Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J. Antimicrob. Chemother. 69, 1265–1274. doi: 10.1093/jac/dkt528
Courvalin, P. (2005). Antimicrobial drug resistance: prediction is very difficult, especially about the future. Emerg. Infect. Dis. 11, 1503–1506. doi: 10.3201/eid1110.051014
Crecchio, C., and Stotzky, G. (1998). Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 30, 1061–1067. doi: 10.1016/s0038-0717(97)00248-4
da Costa, P. M., Vaz-Pires, P., and Bernardo, F. (2006). Antimicrobial resistance in Enterococcus spp. isolated in inflow, effluent and sludge from municipal sewage water treatment plants. Water Res. 40, 1735–1740. doi: 10.1016/j.watres.2006.02.025
Danovaro, R., Armeni, M., Corinaldesi, C., and Mei, M. L. (2003). Viruses and marine pollution. Mar. Pollut. Bull. 46, 301–304. doi: 10.1016/s0025-326x(02)00461-7
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Molec. Biol. Rev. 74, 417–433.
de Jonge, P. A., Nobrega, F. L., Brouns, S. J., and Dutilh, B. E. (2019). Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 27, 51–63.
de Kraker, M. E., Stewardson, A. J., and Harbarth, S. (2016). Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 13:e1002184. doi: 10.1371/journal.pmed.1002184
de Vries, J., and Wackernagel, W. (2002). Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl. Acad. Sci. U.S.A. 99, 2094–2099. doi: 10.1073/pnas.042263399
Devos, S., Van Putte, W., Vitse, J., Van Driessche, G., Stremersch, S., Van Den Broek, W., et al. (2017). Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ. Microbiol. 19, 3930–3937. doi: 10.1111/1462-2920.13793
Domingues, S., and Nielsen, K. M. (2017). Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 38, 16–21. doi: 10.1016/j.mib.2017.03.012
Dominiak, D. M., Nielsen, J. L., and Nielsen, P. H. (2011). Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13, 710–721. doi: 10.1111/j.1462-2920.2010.02375.x
Doroshenko, N., Tseng, B. S., Howlin, R. P., Deacon, J., Wharton, J. A., Thurner, P. J., et al. (2014). Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob. Agents Chemother. 58, 7273–7282. doi: 10.1128/aac.03132-14
Enault, F., Briet, A., Bouteille, L., Roux, S., Sullivan, M. B., and Petit, M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11:237. doi: 10.1038/ismej.2016.90
Fulsundar, S., Harms, K., Flaten, G. E., Johnsen, P. J., Chopade, B. A., and Nielsen, K. M. (2014). Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 80, 3469–3483. doi: 10.1128/aem.04248-13
Gao, P., Munir, M., and Xagoraraki, I. (2012). Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 42, 173–183. doi: 10.1016/j.scitotenv.2012.01.061
Gaudin, M., Gauliard, E., Schouten, S., Houel-Renault, L., Lenormand, P., Marguet, E., et al. (2013). Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Repep. 5, 109–116. doi: 10.1111/j.1758-2229.2012.00348.x
Gebhard, F., and Smalla, K. (1999). Monitoring field releases of genetically modified sugar beets for persistence of transgenic plant DNA and horizontal gene transfer. FEMS Microbiol. Ecol. 28, 261–272. doi: 10.1111/j.1574-6941.1999.tb00581.x
Grüll, M. P., Mulligan, M. E., and Lang, A. S. (2018). Small extracellular particles with big potential for horizontal gene transfer: membrane vesicles and gene transfer agents. FEMS Microbiol. Lett. 365:fny192.
Harms, K., Lunnan, A., Hulter, N., Mourier, T., Vinner, L., Andam, C. P., et al. (2016). Substitutions of short heterologous DNA segments of intragenomic or extragenomic origins produce clustered genomic polymorphisms. Proc. Natl. Acad. Sci. U.S.A. 113, 15066–15071. doi: 10.1073/pnas.1615819114
Hathroubi, S., Fontaine-Gosselin, S. E., Tremblay, Y. D., Labrie, J., and Jacques, M. (2015). Sub-inhibitory concentrations of penicillin G induce biofilm formation by field isolates of Actinobacillus pleuropneumoniae. Vet. Microbiol. 179, 277–286. doi: 10.1016/j.vetmic.2015.06.011
Heuer, H., and Smalla, K. (2012). Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36, 1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x
Hong, P.-Y., Julian, T. R., Pype, M. L., Jiang, S. C., Nelson, K. L., Graham, D., et al. (2018). Reusing treated wastewater: consideration of the safety aspects associated with antibiotic-resistant bacteria and antibiotic resistance genes. Water 10:244. doi: 10.3390/w10030244
Hyman, P., and Abedon, S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/s0065-2164(10)70007-1
Jiang, S. C., and Paul, J. H. (1998). Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64, 2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998
Johnson, L., Horsman, S. R., Charron-Mazenod, L., Turnbull, A. L., Mulcahy, H., Surette, M. G., et al. (2013). Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 13:115. doi: 10.1186/1471-2180-13-115
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Jończyk, E., Kłak, M., Miêdzybrodzki, R., and Górski, A. (2011). The influence of external factors on bacteriophages. Folia Microbiol. 56, 191–200. doi: 10.1007/s12223-011-0039-8
Kenzaka, T., Tani, K., Sakotani, A., Yamaguchi, N., and Nasu, M. (2007). High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level. Appl. Environ. Microbiol. 73, 3291–3299. doi: 10.1128/aem.02890-06
Klieve, A. V., Yokoyama, M. T., Forster, R. J., Ouwerkerk, D., Bain, P. A., and Mawhinney, E. L. (2005). Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 71, 4248–4253. doi: 10.1128/aem.71.8.4248-4253.2005
Larrañaga, O., Brown-Jaque, M., Quirós, P., Gómez-Gómez, C., Blanch, A. R., Rodríguez-Rubio, L., et al. (2018). Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. International 115, 133–141. doi: 10.1016/j.envint.2018.03.019
Lee, L. H., Lui, D., Platner, P. J., Hsu, S. F., Chu, T. C., Gaynor, J. J., et al. (2006). Induction of temperate cyanophage AS-1 by heavy metal–copper. BMC Microbiol. 6:17. doi: 10.1186/1471-2180-6-17
Lekunberri, I., Subirats, J., Borrego, C. M., and Balcázar, J. L. (2017). Exploring the contribution of bacteriophages to antibiotic resistance. Environm. Pollut. 220, 981–984. doi: 10.1016/j.envpol.2016.11.059
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10(12 Suppl.), S122–S129.
Lewenza, S. (2013). Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front. Microbiol. 4:21. doi: 10.3389/fmicb.2013.00021
Liu, S.-S., Qu, H.-M., Yang, D., Hu, H., Liu, W.-L., Qiu, Z.-G., et al. (2018). Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 136, 131–136. doi: 10.1016/j.watres.2018.02.036
Lorenz, M. G., and Wackernagel, W. (1994). Bacterial gene transfer by natural transformation in the environment. Microbiol. Mol Biol. Rev. 58, 5563–5602.
Merod, R. T., and Wuertz, S. (2014). Extracellular polymeric substance architecture influences natural genetic transformation of Acinetobacter baylyi in biofilms. Appl. Environ. Microbiol. 80, 7752–7757. doi: 10.1128/aem.01984-14
Morrissey, E. M., McHugh, T. A., Preteska, L., Hayer, M., Dijkstra, P., Hungate, B. A., et al. (2015). Dynamics of extracellular DNA decomposition and bacterial community composition in soil. Soil Biol. Biochem. 86, 42–49. doi: 10.1016/j.soilbio.2015.03.020
Muniesa, M., Colomer-Lluch, M., and Jofre, J. (2013). Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 8, 739–751. doi: 10.2217/fmb.13.32
Munir, M., Wong, K., and Xagoraraki, I. (2011). Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 45, 681–693. doi: 10.1016/j.watres.2010.08.033
Nielsen, K. M., Bohn, T., and Townsend, J. P. (2014). Detecting rare gene transfer events in bacterial populations. Front. Microbiol. 4:415. doi: 10.3389/fmicb.2013.00415
Nielsen, K. M., Bones, A. M., Smalla, K., and van Elsas, J. D. (1998). Horizontal gene transfer from transgenic plants to terrestrial bacteria - a rare event? FEMS Microbiol. Rev. 22, 79–103. doi: 10.1016/s0168-6445(98)00009-6
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37–53. doi: 10.1051/ebr:2007031
Nielsen, K. M., and Townsend, J. P. (2004). Monitoring and modeling horizontal gene transfer. Nat. Biotechnol. 22, 1110–1114. doi: 10.1038/nbt1006
Nielsen, K. M., and Van Elsas, J. D. (2019). “Horizontal gene transfer and microevolution in soil,” in Modern Soil Microbiology, 3rd Edn, eds J. D. Van Elsas, J. T. Trevors, A. S. Rosado, and P. Nannipieri (Boca Raton: CRC Press), 105–123. doi: 10.1201/9780429059186-7
Novo, A., and Manaia, C. M. (2010). Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 87, 1157–1166. doi: 10.1007/s00253-010-2583-6
O’Neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online at: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed November 13, 2019).
Overballe-Petersen, S., Harms, K., Orlando, L. A., Mayar, J. V., Rasmussen, S., Dahl, T. W., et al. (2013). Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci U.S.A. 110, 19860–19865. doi: 10.1073/pnas.1315278110
Pérez-Cruz, C., Delgado, L., López-Iglesias, C., and Mercade, E. (2015). Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS On 10:e0116896. doi: 10.1371/journal.pone.0116896
Pietramellara, G., Ascher, J., Borgogni, F., Ceccherini, M., Guerri, G., and Nannipieri, P. (2009). Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Ferti. Soils 45, 219–235. doi: 10.1007/s00374-008-0345-8
Pietramellara, G., Ascher, J., Ceccherini, M. T., Nannipieri, P., and Wenderoth, D. (2007). Adsorption of pure and dirty bacterial DNA on clay minerals and their transformation frequency. Biol. Fertil. Soils 43, 731–739. doi: 10.1007/s00374-006-0156-8
Pietramellara, G., Ceccherini, M. T., Ascher, J., and Nannipieri, P. (2006). Persistence of transgenic and not transgenic extracellular DNA in soil and bacterial transformation. Rivist. Biol. Biol. Forum 99, 37–68.
Pote, J., Rosse, P., Rosselli, W., Van, V. T., and Wildi, W. (2005). Kinetics of mass and DNA decomposition in tomato leaves. Chemosphere 61, 677–684. doi: 10.1016/j.chemosphere.2005.03.030
Proia, L., von Schiller, D., Sanchez-Melsio, A., Sabater, S., Borrego, C. M., Rodriguez-Mozaz, S., et al. (2016). Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ. Pollut. 210, 121–128. doi: 10.1016/j.envpol.2015.11.035
Ray, J. L., Harms, K., Wikmark, O. G., Starikova, I., Johnsen, P. J., and Nielsen, K. M. (2009). Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-fold over the genome. Genetics 182, 1165–1181. doi: 10.1534/genetics.109.103127
Reinthaler, F. F., Posch, J., Feierl, G., Wust, G., Haas, D., Ruckenbauer, G., et al. (2003). Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37, 1685–1690. doi: 10.1016/s0043-1354(02)00569-9
Roberts, L., and Simpson, S. (2008). Deadly defiance, in science, special issue “drug resistance. Science 321, 355–369.
Roier, S., Zingl, F. G., Cakar, F., and Schild, S. (2016). Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microb. Cell. 3, 257–259. doi: 10.15698/mic2016.06.508
Romanowski, G., Lorenz, M. G., Sayler, G., and Wackernagel, W. (1992). Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl. Environ. Microbiol. 58, 3012–3019. doi: 10.1128/aem.58.9.3012-3019.1992
Romanowski, G., Lorenz, M. G., and Wackernagel, W. (1993). Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl. Environ. Microbiol. 59, 3438–3446. doi: 10.1128/aem.59.10.3438-3446.1993
Ross, J., and Topp, E. (2015). Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 81, 7905–7913. doi: 10.1128/aem.02363-15
Rumbo, C., Fernández-Moreira, E., Merino, M., Poza, M., Mendez, J. A., Soares, N. C., et al. (2011). Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 3084–3090. doi: 10.1128/aac.00929-10
Schilcher, K., Andreoni, F., Dengler Haunreiter, V., Seidl, K., Hasse, B., and Zinkernagel, A. S. (2016). Modulation of Staphylococcus aureus biofilm matrix by subinhibitory concentrations of clindamycin. Antimicrob. Agents Chemother. 60, 5957–5967. doi: 10.1128/aac.00463-16
Schooling, S. R., and Beveridge, T. J. (2006). Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957. doi: 10.1128/jb.00257-06
Shousha, A., Awaiwanont, N., Sofka, D., Smulders, F. J., Paulsen, P., Szostak, M. P., et al. (2015). Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl. Environ. Microbiol. 81:4600. doi: 10.1128/aem.00872-15
Soler, N., Krupovic, M., Marguet, E., and Forterre, P. (2015). Membrane vesicles in natural environments: a major challenge in viral ecology. ISME J. 9:993.
Song, G., Wang, J., Chiu, C. A., and Westerhoff, P. (2010). Biogenic nanoscale colloids in wastewater effluents. Environ. Sci. Technol. 44, 8216–8222. doi: 10.1021/es101208b
Sørensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol 3, 700–710. doi: 10.1038/nrmicro1232
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Townsend, J. P., Bohn, T., and Nielsen, K. M. (2012). Assessing the probability of detection of horizontal gene transfer events in bacterial populations. Front. Microbiol. 3:27. doi: 10.3389/fmicb.2012.00027
Townsend, J. P., Nielsen, K. M., Fisher, D. S., and Hartl, D. L. (2003). Horizontal acquisition of divergent chromosomal DNA in bacteria: effects of mutator phenotypes. Genetics 164, 13–21.
Toyofuku, M., Cárcamo-Oyarce, G., Yamamoto, T., Eisenstein, F., Hsiao, C. C., Kurosawa, M., et al. (2017). Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 8:481.
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Toyofuku, M., Tashiro, Y., Hasegawa, Y., Kurosawa, M., and Nomura, N. (2015). Bacterial membrane vesicles, an overlooked environmental colloid: Biology, environmental perspectives and applications. Adv. Colloid Interf. Sci. 226(Pt A), 65–77. doi: 10.1016/j.cis.2015.08.013
van Elsas, J. D., Turner, S., and Bailey, M. J. (2003). Horizontal gene transfer in the phytosphere. New Phytol. 157, 525–537. doi: 10.1046/j.1469-8137.2003.00697.x
Wang, B. Y., Chi, B., and Kuramitsu, H. K. (2002). Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 17, 108–112. doi: 10.1046/j.0902-0055.2001.00001.x
Wang, M., Liu, P., Zhou, Q., Tao, W., Sun, Y., and Zeng, Z. (2018). Estimating the contribution of bacteriophage to the dissemination of antibiotic resistance genes in pig feces. Environ. Pollut. 238, 291–298. doi: 10.1016/j.envpol.2018.03.024
Woegerbauer, M., Kuffner, M., Kopacka, I., Domingues, S., Steinwider, J., Nielsen, K. M., et al. (2015). Impact of Mosaic Genes on the Risk Assessment of GMOs. Austrian: Federal Ministry of Health. Available online at: https://www.verbrauchergesundheit.gv.at/gentechnik/gruen/mosaic_genes_bericht_201511192.pdf?72a1u5 (accessed March 31, 2020).
Yang, Y., Shi, W., Lu, S. Y., Liu, J., Liang, H., Yang, Y., et al. (2018). Prevalence of antibiotic resistance genes in bacteriophage DNA fraction from Funan River water in Sichuan. China. Sci. Total Environ. 626, 835–841. doi: 10.1016/j.scitotenv.2018.01.148
Yuan, Q. B., Guo, M. T., and Yang, J. (2014). Monitoring and assessing the impact of wastewater treatment on release of both antibiotic-resistant bacteria and their typical genes in a Chinese municipal wastewater treatment plant. Environ. Sci. Process Impacts 16, 1930–1937. doi: 10.1039/c4em00208c
Yuan, Q. B., Huang, Y. M., Wu, W. B., Zuo, P., Hu, N., Zhou, Y. Z., et al. (2019). Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 131:104986. doi: 10.1016/j.envint.2019.104986
Yue, W. F., Du, M., and Zhu, M. J. (2012). High temperature in combination with UV irradiation enhances horizontal transfer of stx2 gene from E. coli O157: H7 to non-pathogenic E. coli. PLoS One 7:e31308. doi: 10.1371/journal.pone.0031308
Zhang, M., Pereira e Silva, M., Chaib De Mares, M., and van Elsas, J. D. (2014). The mycosphere constitutes an arena for horizontal gene transfer with strong evolutionary implications for bacterial-fungal interactions. FEMS Microbiol. Ecol. 89, 516–526. doi: 10.1111/1574-6941.12350
Zhang, T., Zhang, M., Zhang, X., and Fang, H. H. (2009). Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environ. Sci. Technol. 43, 3455–3460. doi: 10.1021/es803309m
Zhang, Y., Li, A., Dai, T., Li, F., Xie, H., Chen, L., et al. (2018). Cell-free DNA: a neglected source for antibiotic resistance genes spreading from WWTPs. Environ. Sci. Technol. 52, 248–257. doi: 10.1021/acs.est.7b04283
Zhang, Y., Snow, D. D., Parker, D., Zhou, Z., and Li, X. (2013). Intracellular and extracellular antimicrobial resistance genes in the sludge of livestock waste management structures. Environ. Sci. Technol. 47, 10206–10213. doi: 10.1021/es401964s
Keywords: wastewater reuse, antibiotic resistance, horizontal gene transfer, free DNA, transduced DNA, membrane vesicles associated DNA
Citation: Woegerbauer M, Bellanger X and Merlin C (2020) Cell-Free DNA: An Underestimated Source of Antibiotic Resistance Gene Dissemination at the Interface Between Human Activities and Downstream Environments in the Context of Wastewater Reuse. Front. Microbiol. 11:671. doi: 10.3389/fmicb.2020.00671
Received: 16 December 2019; Accepted: 24 March 2020;
Published: 21 April 2020.
Edited by:
Ed Topp, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Maite Muniesa, University of Barcelona, SpainJinxin Liu, University of California, Davis, United States
Eddie Cytryn, Agricultural Research Organization (ARO), Israel
Copyright © 2020 Woegerbauer, Bellanger and Merlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christophe Merlin, Y2hyaXN0b3BoZS5tZXJsaW5AdW5pdi1sb3JyYWluZS5mcg==
 Markus Woegerbauer
Markus Woegerbauer Xavier Bellanger
Xavier Bellanger Christophe Merlin
Christophe Merlin