- 1Key Laboratory of Molecular Microbiology and Technology, Ministry of Education, TEDA Institute of Biological Sciences and Biotechnology, Nankai University, Tianjin, China
- 2The Third Central Hospital of Tianjin, Tianjin Key Laboratory of Extracorporeal Life Support for Critical Diseases, Tianjin Institute of Hepatobiliary Disease, Tianjin, China
- 3Department of Vascular Surgery, Tianjin Hospital, Tianjin, China
- 4Tianjin Children’s Hospital, Third Central Clinical College of Tianjin Medical University, Tianjin, China
Enterobacter cloacae is a well-characterized opportunistic pathogen that is closely associated with various nosocomial infections. The O-antigen, which is one of the most variable constituents on the cell surface, has been used widely and traditionally for serological classification of many gram-negative bacteria. E. cloacae is divided into 30 serotypes, based on its O-antigen diversity. In this study, by using genomic and comparative-genomic approaches, we analyzed the O-antigen gene clusters of 26 E. cloacae serotypes in depth. We also identified the sero-specific gene for each serotype and developed a multiplex polymerase chain reaction (PCR) method. The sensitivity of the assay was 0.1 ng for genomic DNA and 103 colony forming units for pure cultures. The assay reliability was evaluated by double-blinded testing with 81 clinical strains. Furthermore, we established a valid, genome-based tool for in silico serotyping of E. cloacae. By screening 431 E. cloacae genomes deposited in GenBank, 304 were classified into current antigenic scheme, and 112 were allocated into 55 putative novel serotypes. Our results represent the first genetic basis of the O-antigen diversity and variation of E. cloacae, providing a rationale for studying the O-antigen associated evolution and pathogenesis of this bacterium. In addition, we extended the current serotyping system for E. cloacae, which is important for detection and epidemiological surveillance purposes for this important pathogen.
Introduction
Enterobacter cloacae, which is ubiquitous in soil, water, and sewage, is a well-known human opportunistic pathogen that is frequently responsible for nosocomial infections contributing to bacteremia, endocarditis, septic arthritis, osteomyelitis, skin/soft tissue infections, and lower respiratory tract, urinary tract, and intra-abdominal infections (Fata et al., 1996; Mezzatesta et al., 2012; Davin-Regli and Pagès, 2015). E. cloacae has been implicated repeatedly as a nosocomial pathogen in neonatal units, and several outbreaks of E. cloacae infections have been reported (Marra et al., 2011; Qureshi et al., 2011; Pestourie et al., 2014). In recent decades, E. cloacae has emerged as the third most frequent and lethal Enterobacteriaceae species involved in bloodstream infections (Wang et al., 2017). Moreover, with the extensive use of broad-spectrum antibiotics over extended periods of time, the increasing prevalence of multidrug-resistant isolates in different populations has become a growing concern (Mezzatesta et al., 2012; Annavajhala et al., 2019).
Lipopolysaccharide (LPS), which is a hallmark structural entity, is essential for membrane stability and cell survival and is a key virulence determinant for many gram-negative bacterial species. LPS molecules are typically composed of three segments: lipid A anchoring LPS to the outer membrane; a core oligosaccharide, that is a non-repeating oligosaccharide commonly consisting of monosaccharides as such as heptose and keto-deoxyoctulosonate; and O-antigen (O-polysaccharide), which is a polymer of repeating oligosaccharide (O-units), each ranging from two to seven residues from a broad range sugars and their derivatives (Valvano, 2003; Merino et al., 2016). In many cases, the O-antigen contributes the most to cell-surface diversity in gram-negative bacteria, thus offering a selective advantage in its specific niche (Reeves, 1992; Wang et al., 2010), and is also an key virulence factor associated with bacterial pathogenesis (March et al., 2013; Sarkar et al., 2014; Caboni et al., 2015). In particular, considerable variation of the O-antigen composition provides a basis for serotyping schemes with many gram-negative bacteria, which has been recognized one of the most important cell constituents in typing strains, and a basic tool utilized in outbreak investigations and epidemiological survey (Kenyon et al., 2017; DebRoy et al., 2018; Guo et al., 2018; Qian et al., 2018).
The major genes for O-antigen synthesis are generally clustered at a chromosomal locus that maps between two housekeeping genes, namely O-antigen gene cluster (O-AGC). These genes are commonly classified into three main classes: nucleotide sugar precursor synthesis genes for sugars that are specific to the particular polysaccharide; glycosyltransferase genes that are associated with the O-unit assemblies and are specific for donor and acceptor sugars, and generate a specific linkage between them; and O-unit processing genes for O-unit translocation and polymerization. Furthermore, three different pathways are known for their involvement in O-antigen synthesis, which are generally named after the proteins involved: the Wzx/Wzy-dependent pathway, the ATP-binding cassette (ABC) transporter (Wzm/Wzt)-dependent pathway, and the synthase-dependent pathway (Keenleyside and Whitefield, 1996; Samuel and Reeves, 2003; Liu et al., 2008, 2014). All O-antigen biosynthesis pathways are initiated by the transfer of a sugar phosphate from an NDP-sugar to the carrier lipid, undecaprenyl phosphate (Und-P), forming Und-PP-sugar (Reeves and Wang, 2002). In the Wzx/Wzy pathway, sugars are transferred one by one from the respective sugar nucleotides to Und-PP-sugar by glycosyltransferases to form O-unit, then, the Und-PP-linked O-units are flipped by the flippase protein, Wzx, across the inner membrane to the periplasm, where the O-unit is polymerized by the polymerase protein, Wzy, to generate the polymer (Reeves and Wang, 2002). In the ABC transporter pathway, the O-antigen is synthesized directly on the Und-PP-sugar, and the translocation of the Und-PP linked O-antigen is carried out by an ABC transporter. The ABC transporter is typically composed of two transmembrane domains (Wzm) and two nucleotide binding domains (Wzt), with the former forming the translocation channel and the latter driving the transport cycle by hydrolyzing ATP (Cuthbertson et al., 2007; Whitfield and Trent, 2014). Following translocation and polymerization, the resultant O-antigen is then attached to the lipid A-core by the ligase, WaaL, to generate mature LPS molecules (Han et al., 2011; Ruan et al., 2012), and the LPS will be transported to the outer membrane by the Lpt pathway (Silhavy et al., 2010).
In Sakazaki and Namioka (1960) firstly reported on the serology of 170 E. cloacae strains, and 53 O-antigens and 56 H-antigens of E. cloacae were distinguished in agglutination tests. In 1983, an antigenic scheme comprising 28 heat-stable O-antigen types, which is still currently accepted, was developed by Gaston et al. (1983), followed by the subsequent addition of another two serotypes1.
The O-AGC of E. cloacae has been reported to be located between two housekeeping genes, galF and gnd, and shows perfect correlations with each O-antigen structure in several isolates (Filatov et al., 2014; Perepelov et al., 2014, 2015, 2016, 2017; Han et al., 2017). These previous studies shed clear genetic and evolutionary information regarding O-AGC of E. cloacae. However, the isolates used in those studies were not reference strains and their serotypes were not indicated. Here, we present a detailed analysis of the O-AGCs of 26 E. cloacae reference strains with available O-serotypes. Moreover, a sero-specific multiplex polymerase chain reaction (PCR) assay was developed, and its specificity and sensitivity were evaluated. We also screened the serotype distribution of 431 isolates with available genomes deposited in GenBank, using the sero-specific genes characterized in this study, and 55 putative novel gene clusters were characterized by us, extending dramatically the current antigenic scheme of E. cloacae. Our current work provides a valuable framework for further assessing the evolution of E. cloacae, and the developed molecular-serotyping assay gives a potential for molecular diagnostics and epidemiological surveillance of this important pathogen.
Materials and Methods
Bacterial Strains and Genomic-DNA Extraction
Details for all bacterial strains used in this study are summarized in Table 1. These strains included 26 reference strains with known serotypes and 81 clinical isolates. Sixteen other strains from eight species within the Enterobacteriaceae family were used to assess the specificity of our multiplex PCR. All strains were grown overnight in Luria-Bertani medium at 37°C with shaking, and genomic DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol.
Sequencing and Bioinformatics Analysis
Whole-genome sequencing (WGS) of 26 E. cloacae reference strains was performed with Solexa paired-end sequencing technology. In general, the genomic DNA were sheared, polished, and prepared using the Illumina Sample Preparation Kit. Genomic libraries were constructed containing 500 bp paired-end inserts, and sequencing was then performed via Solexa sequencing technology (Illumina, Inc.) for ∼100-fold coverage. The reads obtained were assembled using the de novo genome-assembly program, Velvet, to generate a multi-contig draft genome. Gaps within the O-AGCs were closed by directed PCR, and the products were sequenced using BigDye terminator chemistry on ABI 3730 capillary sequencers.
The Artemis program (Rutherford et al., 2000) was used for annotation and the lockMaker program (Henikoff et al., 1995) was used to identify conserved motifs. The BLAST and PSI-BLAST programs (Altschul et al., 1997) were used to search available databases, including GenBank2 and the Pfam protein motif databases3. The TMHMM v2.0 analysis program4 was used to identify potential transmembrane domains within protein sequences. O-AGC sequences, between galF and gnd genes of each strain, were retrieved from the genomes for further analysis.
Development of a Multiplex PCR Assay
All sero-specific primers were designed based on the wzy gene sequences determined in this study, except for serotype O23, for which the wzt gene was targeted (Table 2). The specificity of each individual primer pair was confirmed using the BLAST program and was subsequently validated by a single PCR amplification, using the strains listed in Table 1. Each PCR was performed in a 25 μl reaction mixture containing 50 ng genomic DNA, 1 × Goldstar PCR buffer, 0.04 mM deoxynucleoside triphosphates, 0.1 μM each primer, and 1 unit Goldstar DNA polymerase. The PCR program used was as follows: denaturation at 95° C for 10 min; 30 cycles of denaturation at 95° C for 30 s, annealing at 55° C for 30 s, and extension at 72° C for 1 min; followed by a final extension at 72° C for 5 min.
The multiplex PCR assay consisted of three groups (Table 2). Group 1 comprised serotypes O1, O3, O4, O5, O6, O7, O8, and O9/10/11; group 2 comprised serotypes O12, O13, O14, O15 O16, O17, O18, and O19; and group 3 comprised serotypes O20, O21, O22, O23, O24, O26, O27, and O30. The PCR mixtures and the PCR program used were the same as those for the singleplex PCR amplifications.
Construction of an in silico Serotyping Program
A Python script was constructed for E. cloacae serotyping using genomic data (Supplementary Data S1). Generally, a database was first generated based on the sero-specific genes characterized and tested in this study, i.e., the wzy genes for 25 of the 26 serotypes and the wzt gene for serotype O23. Next, genomic assemblies were employed to a BLASTn search against the database with an identity cutoff of >99%. The script outputs contained the best-matching genes via BLASTn analysis, as well as the identity level between sero-specific gene(s) and homologous genes(s) in the query genome, which enabled determination of the exact serotype.
Nucleotide Sequence Accession Number
The DNA sequences of the O-AGCs from all 26 E. cloacae reference strains were deposited in GenBank database under accession numbers MK595714 to MK595739.
Results
Analysis of the E. cloacae O-AGCs
Twenty-six O-AGCs of E. cloacae reference strains collected from National Collection of Type Cultures, United Kingdom (NCTC) were obtained via genome sequencing. All O-AGCs are located between two housekeeping genes, galF and gnd, and range from 4,473 to 16,323 bp, with all genes being transcribed from galF and gnd (except the fdtC gene of O27) and five to 16 open reading frames (ORFs) being encoded. Generally, the main three classes of genes within the O-AGC were annotated in each serotype. In addition, several pyruvyl transferases, acetyl transferases, and hypothetical protein encoding genes were also assigned for individual strains. Figure 1 shows a schematic representation of all 26 O-AGCs, and the characteristics of all ORFs within each O-AGC are summarized in Supplementary Table S1.
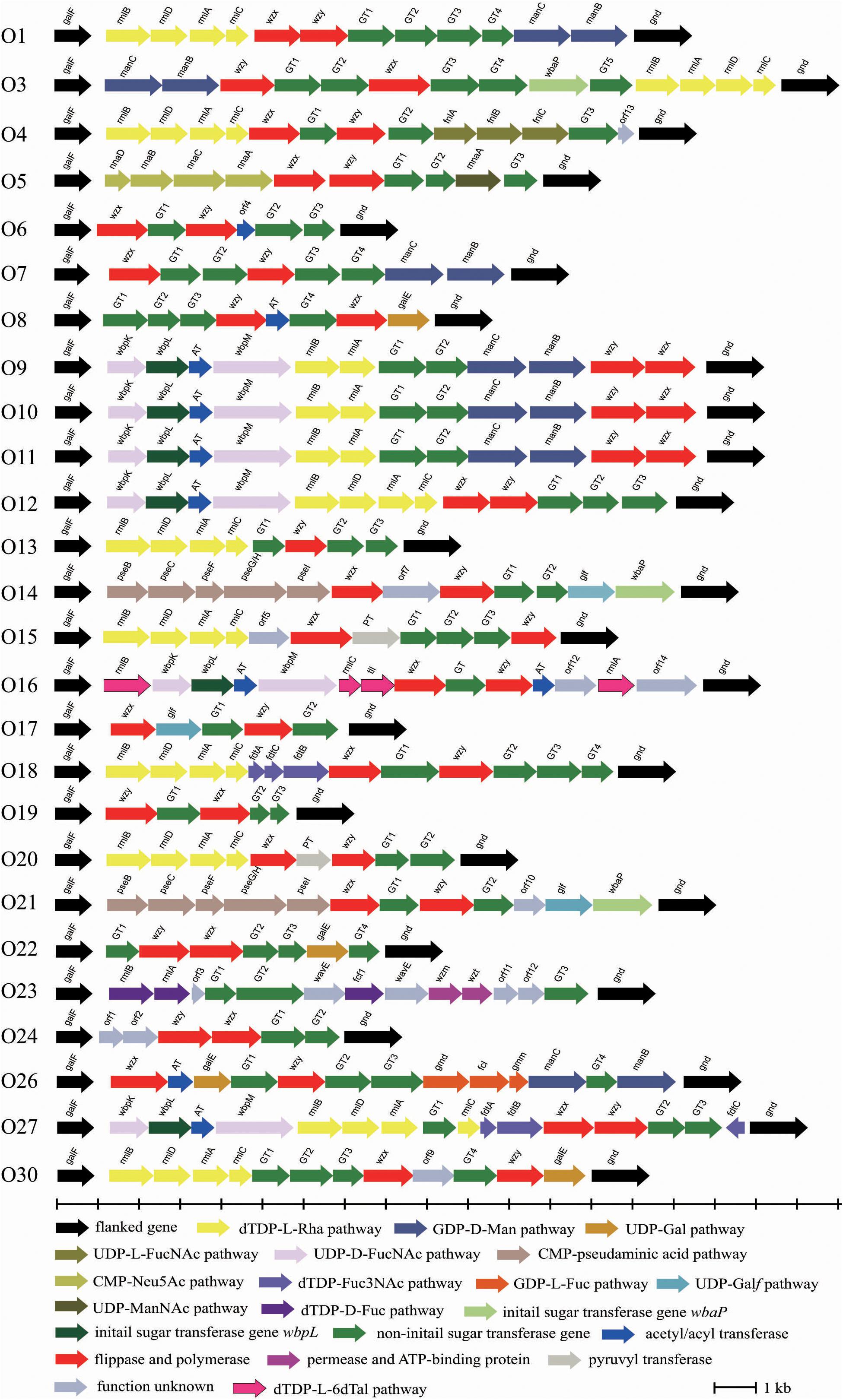
Figure 1. Schematic diagram of the O-AGCs identified from the 26 E. cloacae reference strains. Genes are represented by arrows and colored according to the gene key at the bottom with gene names indicated above each arrow.
Some normal sugars, including D-GlcNAc, D-Glc, D-GlcA, and D-GalA, are also found in other structures in the Enterobacteriaceae family and the biosynthesis genes are normally found at various loci outside the O-AGCs. Here, the biosynthesis pathway of 13 rare occurring sugars was proposed based on the occurrence of their corresponding nucleotide sugar precursor synthesis genes (Figure 2).
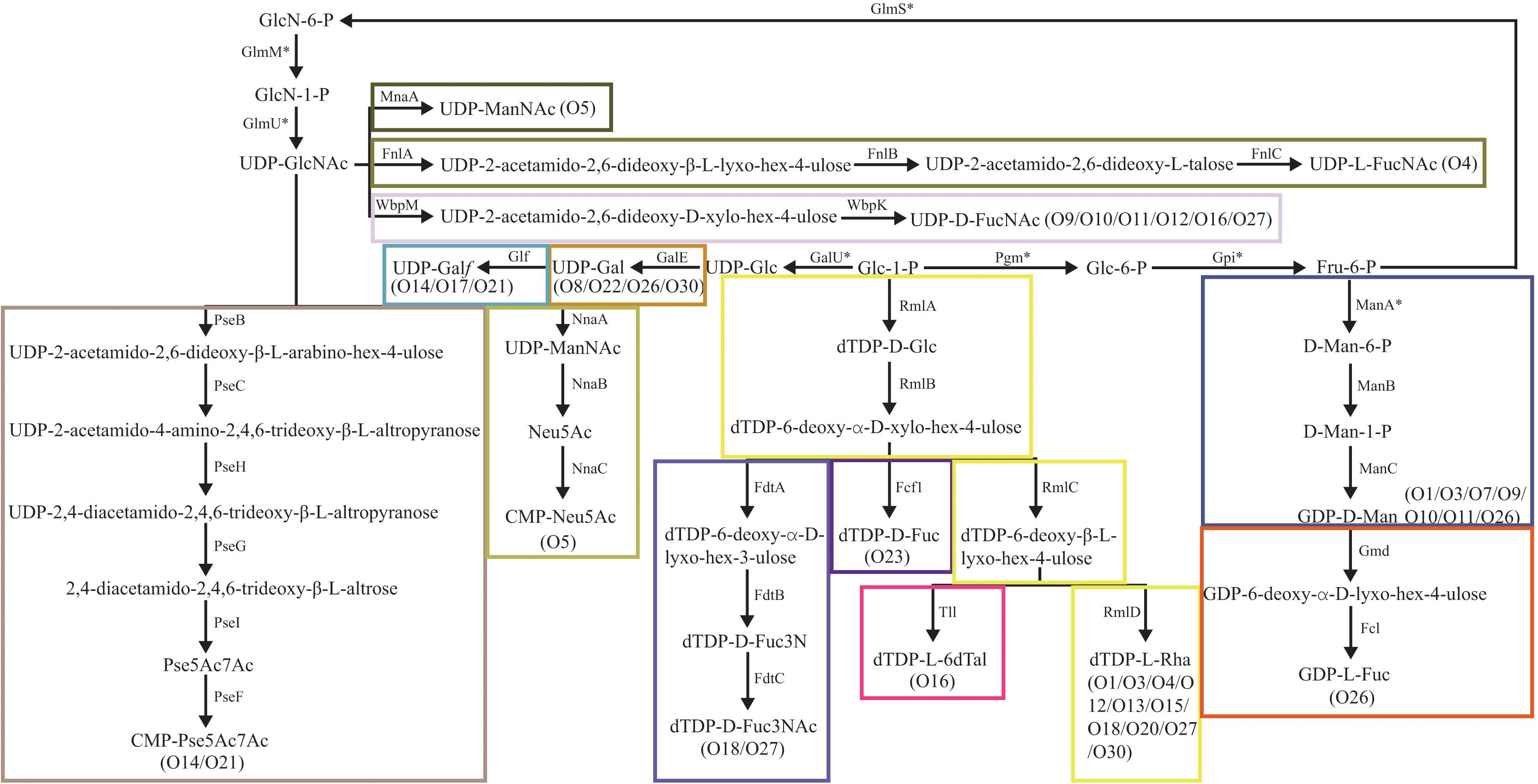
Figure 2. Biosynthesis pathways for the putative sugars in the E. cloacae O-antigens. MnaA, UDP-N-acetylglucosamine-2-epimerase (Campbell et al., 2000); FnlA, 4,6-dehydratase, 3- and 5-epimerase; FnlB, reductase; FnlC, C-2 epimerase (Kneidinger et al., 2003); GalE, UDP-glucose-4-epimerase (Samuel and Reeves, 2003); Glf, UDP-galactopyranose mutase (Nassau et al., 1996); GalU, UTP-glucose-1-phosphate uridylyltransferase (Bonofiglio et al., 2005); GlmU, UDP-N-acetyl-glucosamine pyrophosphorylase (Mengin-Lecreulx and van Heijenoort, 1993); GlmM, phosphoglucosamine mutase (Mengin-Lecreulx and van Heijenoort, 1996); GlmS, L-Glutamine:D-fructose-6-phosphate amidotransferase (Badet et al., 1987); Pgm, phosphoglucomutase (Lu and Kleckner, 1994); Gpi, Glucose-6-phosphate isomerase (Winkler, 1970); NnaA, UDP-N-acetylglucosamine-2-epimerase; NnaB, N-acetylneuraminic acid synthetase; NnaC, CMP-N-acetylneuraminic acid synthetase (Annunziato et al., 1995); RmlA, glucose-1-phosphate thymidylyltransferase (Zuccotti et al., 2001); RmlB, dTDP-D-glucose 4,6-dehydratase (Allard et al., 2001); RmlC, dTDP-4-keto-6-deoxy-D-glucose 3,5-epimerase (Giraud et al., 1999a); RmlD, dTDP-6-deoxy-L-mannose-dehydrogenase (Giraud et al., 1999b); ManA, phosphomannose isomerase; ManB, phosphomannomutase; ManC, mannose-1-phosphate guanylyltransferase (Samuel and Reeves, 2003); Gmd, GDP-mannose-4,6-dehydratase (Somoza et al., 2000; Kneidinger et al., 2001); Fcl, GDP-L-fucose synthetase (Rosano et al., 2000); FdtA, dTDP-6-deoxy-hex-4-ulose isomerase; FdtB, dTDP-6-deoxy-D-xylo-hex-3-ulose aminase; FdtC, dTDP-D-Fuc3N acetylase (Pfoestl et al., 2003); WbpM, UDP-D-GlcNAc 4,6-dehydratase; WbpK, 4-reductase (King et al., 2009); PseB, C6 dehydratase/C5 epimerase; PseC, aminotransferase; PseH, N-acetyltransferase; PseG, nucleotidase; PseI, condensase; PseF, cytidylyltransferase (Schoenhofen et al., 2006); Fcf1, dTDP-6-deoxy-D-xylo-hex-4-ulopyranose reductase (Wang et al., 2008); Tll, dTDP-6-deoxy-L-lyxo-4-hexulose reductase (Nakano et al., 2000).
The O-antigen synthesis pathway is initiated by transfer of a sugar phosphate from an NDP-sugar to Und-P. In most E. coli and Shigella strains, and in a high proportion of Salmonella strains, WecA, encoded by wecA gene of the enterobacterial common antigen gene cluster, mediates this step by transferring UDP-GlcNAc to Und-P (Alexander and Valvano, 1994; Rick et al., 1994; Al-Dabbagh et al., 2016). Among the 26 serotypes, 17 (65%) possess no initial transferase (IT) gene, which probably means that O-antigen synthesis is mediated by WecA in those serotypes. Indeed, we discovered a homolog of the wecA gene by screening each genome for all of them (data not shown). In serotypes O9 to O12, O16, and O27, the homologs of wbpL gene were found, whose product has been identified as an IT and transferred UDP-D-FucNAc to Und-P to initiate O-antigen synthesis in Pesudomonas aeruginosa (Rocchetta et al., 1998). We also assigned wbpM and wbpK genes in these serotypes, whose products were characterized collectively in terms of UDP-D-FucNAc formation in P. aeruginosa (King et al., 2009). Thus, we deduced that the initial sugar of the O-antigens of O9 to O12, O16, and O27 is very likely D-FucNAc. Another IT gene, wbaP, was assigned to O3, O14, and O21. WbaP was previously identified as an IT that transfer Gal-1-P to Und-P to initiate O-antigen synthesis (Reeves et al., 2013). We therefore propose that the initial sugar of the O-antigens of these three serotypes should be D-Gal.
Three different pathways have been reported for O-antigen synthesis. For E. cloacae, 96% (25 of 26 serotypes) O-AGCs contain wzx/wzy genes, meaning very likely that most E. cloacae strains utilize the Wzx/Wzy-dependent pathway for O-antigen translocation and polymerization. The only exception is O23, which possess wzm/wzt genes instead of wzx/wzy genes, suggesting that the O23-antigen is synthesized via the ABC transporter (Wzm/Wzt)-dependent pathway. An anomaly here is that only wzy gene is annotated in O13, and we propose that wzx gene of O13 must be located elsewhere in the chromosome. This atypical feature has been reported in other strains, such as Klebsiella K11 and K34 (Pan et al., 2015), and Salmonella serotypes A, B, and D1 (Wang et al., 2002).
Development of a Multiplex PCR Assay
Compared with the nucleotide sugar precursor synthesis genes and glycosyltransferase genes, the O-antigen processing genes (wzx/wzy and wzm/wzt) are much more highly serotype-determinative (Li and Reeves, 2000; Ballmer et al., 2007). We constructed neighbor-joining phylogenetic trees for wzx and wzy, which showed high diversity levels among the different serotypes, except for O9/O10/O11, of which the O-AGCs shared 100% identity (Supplementary Figure S1). Therefore, wzy was selected as the target gene in terms of primer design for 25 of the 26 serotypes. Because wzy is lacking in the O-AGC of O23, wzt was selected instead. The 24 primer pairs were divided into three groups to generate target DNAs (Table 2).
The multiplex PCR method was tested against each of the 26 O-standard E. cloacae reference strains and 16 strains of other species within the Enterobacteriaceae family (Table 1). In the presence of each target strain, only the corresponding sero-specific primer pair worked, and only one band of the expected size was generated (Figure 3). The amplicons ranged in size from 211 to 1,137 bp in length (Table 2). The representative E. cloacae strains belonging to other serotypes or other bacterial strains did not generate PCR products of the correct size. The results showed that all 24 primer pairs were specific and compatible in the multiplex PCR runs.
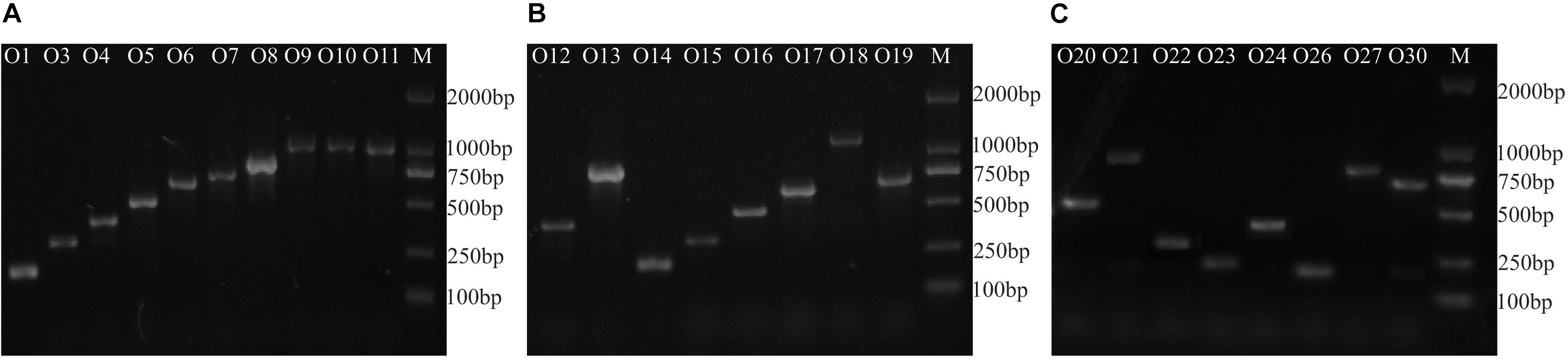
Figure 3. Agarose gel electrophoresis of representative Multiplex PCR products from 26 E. cloacae reference strains. M, DL2000 DNA molecular standard. (A) products of group 1, (B) products of group 2, and (C) products of group 3.
Furthermore, O3 from group 1, O17 from group 2, and O26 from group 3 were selected to determine the detection limit of our multiplex PCR assay. To determine the detection limit, serial 10-fold dilutions (10 ng to 0.1 pg) of genomic DNA from each strain were tested, which showed that the sensitivity of our assay was 0.1 ng for genomic DNA. To determine the sensitivity for pure cultures, the three serotypes were serially diluted 10-fold from 108 down to 100 colony forming units (CFUs) and used as templates for multiplex PCR. Our test demonstrated that positive signals could be generated for templates containing 103 CFUs of pure culture.
A double-blinded test using 81 E. cloacae strains with unknown serotypes was performed to evaluate our multiplex PCR system. Among them, 73 were typeable, including 11 assigned to O1, 24 assigned to O3, 12 assigned to O9/10/11, and 10 assigned to O13, with the other serotypes each representing <10%. This result was confirmed to be correct by ABI3730 sequencing. The distribution of serotypes is generally consistent with Gaston’s study by using agglutination test against 300 clinical isolates (Gaston et al., 1983), with exception of O8, which accounted for >13% in his investigation.
In silico Serotyping of Genomic Data for E. cloacae Strains
To evaluate our molecular serotyping scheme, we downloaded 431 E. cloacae genomes from GenBank and screened them using all sero-specific genes identified in our study. Among them, 304 could be assigned to certain serotypes, with O3 representing the predominant group (38%), followed by O8 (15%) and O13 (10%), and other serotypes assigned each <7%. The result of our in silico analysis is also in line with the allocation of serotypes studied by Gaston et al. (1983).
Among the remaining 127 genomes, the O-AGC was either not found or was too fragmented for 15 genomes, thus, these genomes were excluded from further analysis. The genetic region between galF and gnd in 112 strains was then extracted and analyzed, and 55 novel putative O-AGCs (temp 1–55) were obtained (Supplementary Table S2 and Supplementary Figure S2).
Discussion
At present, the O-antigen structure has only been elucidated for one reference strain (NCTC 11579, serotype O10), by Wilkinson’s group (Moule et al., 1989). The O-AGC of O10 in this study showed a perfect correlation with the structure (Figure 4A). In general, ManB and ManC, combined with ManA, are responsible for the formation of GDP-D-Man, the nucleotide sugar precursor of D-Man, whereas manA is always located outside of the O-AGC (Samuel and Reeves, 2003). WbpM and WbpK are responsible for the formation of UDP-D-FucNAc, the nucleotide sugar precursor of D-FucNAc (King et al., 2009). The products of two glycosyl transferase genes are proposed for the synthesis of two D-Man-(α1→2)-D-Man linkages and one D-Man-(β1→3)-D-FucNAc linkage, however, the exact functions of each could not be inferred. The presence of wzx and wzy genes probably means that the O-antigen of O10 is synthesized by the Wzx/Wzy-dependent pathway. We noticed that there is a D-Glc side branch attached to the β D-Man residue of the backbone of O10 antigen. This is commonly mediated by the Gtr process, which is involved by three enzymes, GtrA, GtrB, and GtrC, with all genes (gtrA/B/C) always being clustered in prophage genomes. GtrA and GtrB are highly conserved among different serotypes, and GtrC is unique to each serotype and is therefore the sero-specific glucosyl transferase (Allison and Verma, 2000; Wang et al., 2007). By screening the genome sequence of O10, we observed gtrA and gtrB homologs, but gtrC could not be annotated due to its low identity shared with the analogs. However, we consider that the gene just downstream of gtrB is most likely a gtrC gene unique to E. cloacae O10, as the protein encoded by it possesses 12 potential transmembrane domains, as predicted using TMHMM v2.0, being consistent with the topology of GtrC of Shigella flexneri (Korres and Verma, 2004). We also characterized the gtr gene set in O9 and O11, respectively. Pairwise comparison showed that the % identity level of GtrA among the three serotypes is 86–100, and GtrB 93–96, however, the % identity level of GtrC, the serotype determinant, ranges only from 30 to 34. In addition, O9, O10, and O11 possess almost identical O-AGCs with > 99% overall identity (Figure 4A), suggesting that the O-AGCs of these serotypes may be recently transferred from one isolate to the others and that their O-antigens very likely contain identical backbones. Although O9/O10/O11 appeared to represent an antigenic group, the distinct numbers for them were still be retained as high-titer-specific sera could be prepared by absorption (Gaston et al., 1983). Therefore, the minor antigenic difference among these serotypes must be accounted for by the variations in the side branches or modifications encoded by genes located elsewhere in the chromosome. On the other hand, the possibility could not be entirely excluded that each of the three serotypes possesses unique O-antigen structure, since a few gene-product pairs share 99% identity level which may influence the activity of them due to non-synonymous mutations, as the case in E. coli O9/O9a (Kido and Kobayashi, 2000).
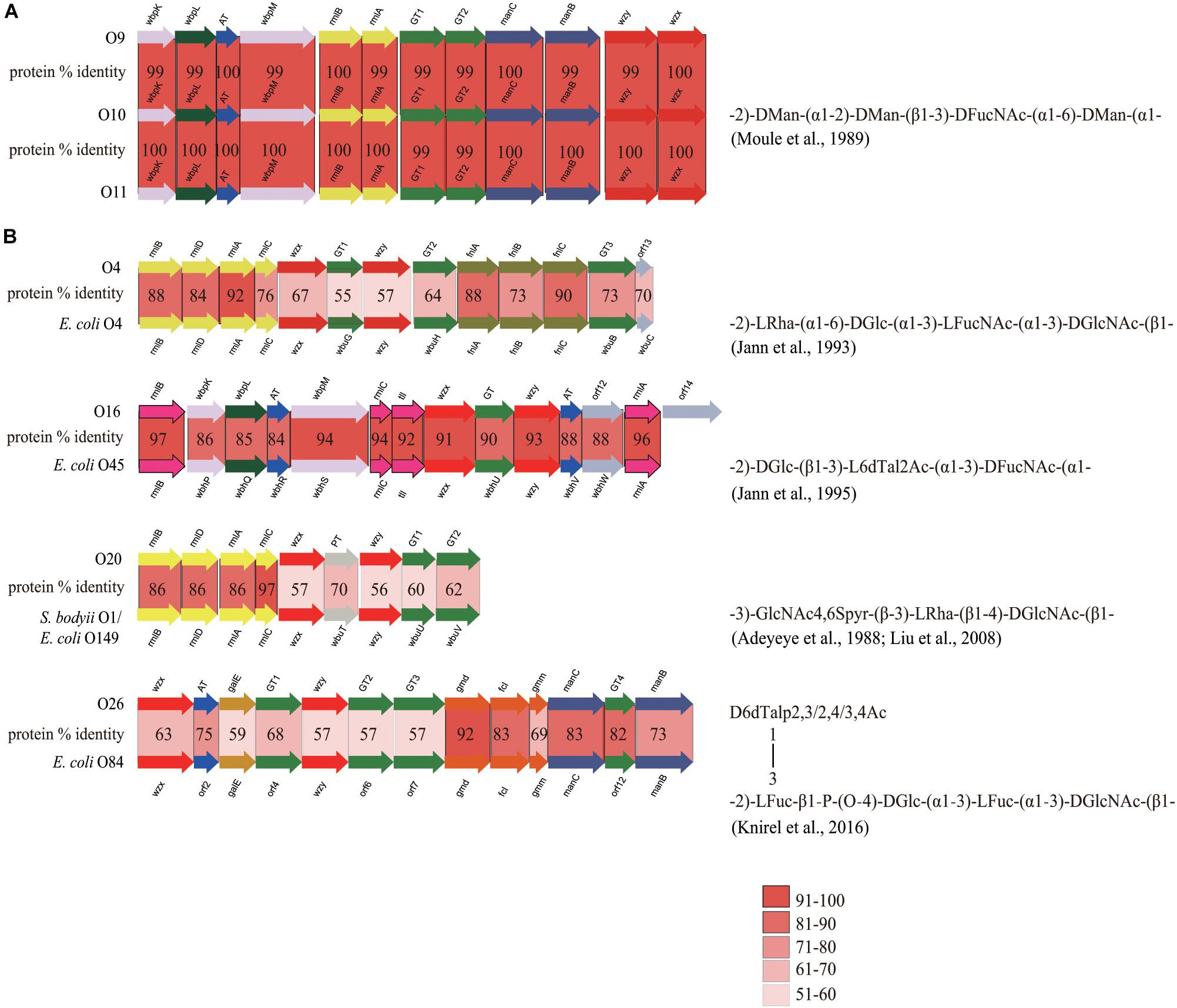
Figure 4. Comparison of closely related O-AGCs within E. cloacae strains and between E. cloacae and E. coli strains. (A) Comparison of O-AGCs of E. cloacae O9/O10/O11, with the O-antigen structure of O10 being shown. (B) Comparison of O-AGCs of E. cloacae O4 and E. coli O4, O16 and E. coli O45, O20 and E. coli O149/S. bodyii type 1, and O26 and E. coli O84, with the O-antigen structures of the latter in each group being shown.
In addition, we also observed a few strains whose O-AGCs are closely related to those of some serotypes of E. coli: they are O4 and E. coli O4 (Jann et al., 1993), O16 and E. coli O45 (Jann et al., 1995), O20 and E. coli O149/S. bodyii type 1 (Adeyeye et al., 1988; Liu et al., 2008), and O26 and E. coli O84 (Knirel et al., 2016) (Figure 4B). Overall, strains in each group share almost identical gene order, and significant protein identity level (55–97%). In these pairs of O-AGCs in E. cloacae and E. coli excluding the O16/E. coli O45 group, the average level of protein identity encoded by nucleotide sugar precursor synthesis genes, glycosyltransferase genes, and O-unit processing genes, is 82, 64, and 59%, respectively. We assume that each pair has evolved from a gene cluster located in a common ancestor, but that the three classes of genes underwent different selective pressures after divergence, as is the case in some Salmonella O-antigens (Liu et al., 2014). For the O16/E. coli O45 group, the overall identity of each pair of gene products is similar (84–97%), and we suppose that the O-AGCs of them probably also originated from a common ancestor recently and still underwent rapidly-evolving events. Elucidation of the O-antigen structures of more E. cloacae serotypes will undoubtedly enhance our understanding of the evolution of O-AGCs of this bacterium, as well as the genetic relatedness of intra- and inter-species.
Efforts have been made to develop a method for timely clinical diagnosis, epidemiological surveillance, and outbreak detection for E. cloacae, including biotyping, phage typing, and ribotyping (Gaston, 1987; Garaizar et al., 1991; Weischer et al., 1993). However, these assays are not highly discriminatory and reproducible methods for E. cloacae typing. Subsequently, a multilocus sequence-typing scheme was presented (Miyoshi-Akiyama et al., 2013) and employed to characterize E. cloacae isolates (Viau et al., 2017; Wang et al., 2017; Miao et al., 2019). More recently, WGS-based methods were presented, showing enhanced discriminative power and shedding new insights into the phylogeny and resistance mechanisms of E. cloacae (Chavda et al., 2016; Beyrouthy et al., 2018); however, the WGS data analysis has not been fully standardized.
Conventional serotyping by using agglutination test is always delicate, laborious, time-consuming, and expensive. For decades, several molecular assays targeting sero-specific genes and showing fast, reliable, and cost-effective detection were developed for bacterial serotyping (Azzari et al., 2010; Lin et al., 2011; Bai et al., 2015). Compared to normal PCR and Taqman probe-based real-time PCR methods that amplify individual or only at most four target gene(s), the multiplex PCR assay could simultaneously detect multiple targets in a single reaction, and generate the same accuracy while saving time and effort. Because of these advantages, multiplex PCR has been applied widely for the detection of many bacterial strains (van der Veer et al., 2018; Kwack et al., 2020; Collins et al., 2020). In the past few years, several WGS-based in silico serotyping approaches have been presented and showed better resolution compared to conventional methods, and have been utilized for epidemiological investigation and tracing (Thrane et al., 2016; Ibrahim and Morin, 2018; Wu et al., 2019). However, all of those studies are based on a key prerequisite that is the full and deep understanding of the O-antigen and the genetic basis for its diversity/variation. To date, the O-antigens and O-AGCs (or the capsular antigen and its genetic determinant) of several pathogenic species, especially in the Enterobacteriaceae family, including Escherichia coli (Iguchi et al., 2015), Shigella (Liu et al., 2008), Salmonella (Liu et al., 2014), Klebsiella (Pan et al., 2015), and Yersinia pseudotuberculosis (Kenyon et al., 2017) have been characterized in depth.
Although more O-antigen structures need to be elucidated to support our study, the work here, for the first time, presented the genetic basis regarding the O-antigen diversity and variation of E. cloacae, which also may partially help in understanding the evolution of this important pathogen. It should be noted, however, that using a conventional agglutination test or our multiplex PCR assay targeting only the present serotype groups, 10–23% isolates could not be assigned to certain serotypes, meaning that other novel serotypes are still evolving and remain to be discovered. Indeed, a large number of putative novel serotypes were characterized by screening the E. cloacae genomes deposited in GenBank. The antigenic scheme for E. cloacae has not been updated since the 1990s; therefore, our current findings have expanded the existing serotyping system for E. cloacae, which is significant for detection and epidemiological surveillance purposes for this important pathogen.
Data Availability Statement
The datasets generated for this study can be found in the GenBank database under accession numbers MK595714 to MK595739.
Author Contributions
XG and TH conceived the project and prepared the manuscript. YL and XW prepared the strain samples, preformed genome sequencing and bioinformatic analyses. JH conducted the in silico serotyping program and analyses. CX developed the multiplex PCR assay and performed the double-blinded test. All authors read and approved the final manuscript.
Funding
This work was supported by Tianjin Municipal Natural Science Foundation Grant (17JCYBJC24300 and 15JCQNJC11500), research grants from key projects of health industry of Tianjin Health Department (12KG108 and 2011KR05), and National Key Programs for Infectious Diseases of China (2017ZX10303405-001 and 2017ZX10104002-001-006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00727/full#supplementary-material
Footnotes
- ^ https://www.phe-culturecollections.org.uk/products/bacteria/detail.jsp?refId=NCTC+11932&collection=nctc and https://www.phe-culturecollections.org.uk/products/bacteria/detail.jsp?refId=NCTC+11933&collection=nctc
- ^ www.ncbi.nlm.nih.gov/genbank
- ^ pfam.sanger.ac.uk
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0/
References
Adeyeye, A., Jansson, P. E., Lindberg, B., Abaas, S., and Svenson, S. B. (1988). Structural studies of the Escherichia coli O-149 O-antigen polysaccharide. Carbohydr. Res. 176, 231–236. doi: 10.1016/0008-6215(88)80134-4
Al-Dabbagh, B., Olatunji, S., Crouvoisier, M., El Ghachi, M., Blanot, D., Mengin-Lecreulx, D., et al. (2016). Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie 127, 249–257. doi: 10.1016/j.biochi.2016.06.005
Alexander, D. C., and Valvano, M. A. (1994). Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176, 7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994
Allard, S. T., Giraud, M. F., Whitfield, C., Graninger, M., Messner, P., and Naismith, J. H. (2001). The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J. Mol. Biol. 307, 283–295. doi: 10.1006/jmbi.2000.4470
Allison, G. E., and Verma, N. K. (2000). Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8, 17–23.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3398–3402. doi: 10.1093/nar/25.17.3389
Annavajhala, M. K., Gomez-Simmonds, A., and Uhlemann, A. C. (2019). Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 10:44. doi: 10.3389/fmicb.2019.00044
Annunziato, P. W., Wright, L. F., Vann, W. F., and Silver, R. P. (1995). Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coli K1. J. Bacteriol. 177, 312–319. doi: 10.1128/jb.177.2.312-319.1995
Azzari, C., Moriondo, M., Indolfi, G., Cortimiglia, M., Canessa, C., Becciolini, L., et al. (2010). Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 5:e9282. doi: 10.1371/journal.pone.0009282
Badet, B., Vermoote, P., Haumont, P. Y., Lederer, F., and LeGoffic, F. (1987). Glucosamine synthetase from Escherichia coli: purification, properties, and glutamine-utilizing site location. Biochemistry 26, 1940–1948. doi: 10.1021/bi00381a023
Bai, X., Liu, Z., Ji, S., Gottschalk, M., Zheng, H., and Xu, J. (2015). Simultaneous detection of 33 Streptococcus suis serotypes using the luminex xTAGR assayTM. J. Microbiol. Methods. 117, 95–99. doi: 10.1016/j.mimet.2015.07.018
Ballmer, K., Korczak, B. M., Kuhnert, P., Slickers, P., Ehricht, R., and Hächler, H. (2007). Fast DNA serotyping of Escherichia coli by use of an oligonucleotide microarray. J. Clin. Microbiol. 45, 370–379. doi: 10.1128/JCM.01361-06
Beyrouthy, R., Barets, M., Marion, E., Dananché, C., Dauwalder, O., Robin, F., et al. (2018). Novel enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg. Infect. Dis. 24, 1505–1515. doi: 10.3201/eid2408.180151
Bonofiglio, L., Garcia, E., and Mollerach, M. (2005). Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr. Microbiol. 51, 217–221. doi: 10.1007/s00284-005-4466-0
Caboni, M., Pédron, T., Rossi, O., Goulding, D., Pickard, D., Citiulo, F., et al. (2015). An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei. PLoS Pathog. 11:e1004749. doi: 10.1371/journal.ppat.1004749
Campbell, R. E., Mosimann, S. C., Tanner, M. E., and Strynadka, N. C. (2000). The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry 39, 14993–15001. doi: 10.1021/bi001627x
Chavda, K. D., Chen, L., Fouts, D. E., Sutton, G., Brinkac, L., Jenkins, S. G., et al. (2016∗). Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. MBio 7:e2093-16. doi: 10.1128/mBio.02093-16
Collins, M. E., Popowitch, E. B., and Miller, M. B. (2020). Evaluation of a novel multiplex PCR panel compared to quantitative bacterial culture for the diagnosis of lower respiratory tract infections. J. Clin. Microbiol. doi: 10.1128/JCM.02013-19
CrossRef Full Text Epub ahead of print.
Cuthbertson, L., Kimber, M. S., and Whitfield, C. (2007). Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534. doi: 10.1073/pnas.0705709104
Davin-Regli, A., and Pagès, J. M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392. doi: 10.3389/fmicb.2015.00392
DebRoy, C., Fratamico, P. M., and Roberts, E. (2018). Molecular serogrouping of Escherichria coli. Anim. Health Res. Rev. 22, 1–16. doi: 10.1017/S1466252317000093
Fata, F., Chittivelu, S., Tessler, S., and Kupper, Y. (1996). Gas gangrene of the arm due to Enterobacter cloacae in a neutropenic patient. South. Med. J. 89, 1095–1096. doi: 10.1097/00007611-199611000-00014
Filatov, A. V., Wang, M., Wang, W., Perepelov, A. V., Shashkov, A. S., Wang, L., et al. (2014). Structure and genetics of the O-antigen of Enterobacter cloacae C6285 containing di-N-acetyllegionaminic acid. Carbohydr. Res. 392, 21–24. doi: 10.1016/j.carres.2014.01.012
Garaizar, J., Kaufmann, M. E., and Pitt, T. L. (1991). Comparison of ribotyping with conventional methods for the type identification of Enterobacter cloacae. J. Clin. Microbiol. 29, 1303–1307.
Gaston, M. A. (1987). Evaluation of a bacteriophage-typing scheme for Enterobacter cloacae. J. Med. Microbiol. 24, 291–295. doi: 10.1099/00222615-24-4-291
Gaston, M. A., Bucher, C., and Pitt, T. L. (1983). O serotyping scheme for Enterobacter cloacae. J. Clin. Microbiol. 18, 1079–1083.
Giraud, M. F., Gordon, F. M., Whitfield, C., Messner, P., McMahon, S. A., and Naismith, J. H. (1999a). Purification, crystallization and preliminary structural studies of dTDP-6-deoxy-D-xylo-4- hexulose 3,5-epimerase (RmlC), the third enzyme of the dTDP-L-rhamnose synthesis pathway, from Salmonella enterica serovar typhimurium. Acta Crystallogr. D. Biol. Crystallogr. 55, 706–708. doi: 10.1107/s0907444998015042
Giraud, M. F., McMiken, H. J., Leonard, G. A., Messner, P., Whitfield, C., and Naismith, J. H. (1999b). Overexpression, purification, crystallization and preliminary structural study of dTDP-6-deoxy-L-lyxo-4-hexulose reductase (RmlD), the fourth enzyme of the dTDP-L-rhamnose synthesis pathway, from Salmonella enterica serovar Typhimurium. Acta Crystallogr. D. Biol. Crystallogr. 55, 2043–2046. doi: 10.1107/s0907444999012251
Guo, X., Wang, M., Wang, L., Wang, Y., Chen, T., Wu, P., et al. (2018). Establishment of a molecular serotyping scheme and a multiplexed luminex-based array for Enterobacter aerogenes. Front. Microbiol. 9:501. doi: 10.3389/fmicb.2018.00501
Han, R., Perepelov, A. V., Wang, Y., Filatov, A. V., Wang, M., Shashkov, A. S., et al. (2017). Structural and genetic characterization of the O-antigen of Enterobacter cloacae C5529 related to the O-antigen of E. cloacae G3054. Carbohydr. Res. 44, 49–52. doi: 10.1016/j.carres.2017.02.006
Han, W., Wu, B., Li, L., Zhao, G., Woodward, R., Pettit, N., et al. (2011). Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J. Biol. Chem. 287, 5357–5365. doi: 10.1074/jbc.M111.308486
Henikoff, S., Henikoff, J. G., Alford, W. J., and Pietrokovski, S. (1995). Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 163, GC17–GC26. doi: 10.1016/0378-1119(95)00486-P
Ibrahim, G. M., and Morin, P. M. (2018). Salmonella serotyping using whole genome sequencing. Front. Microbiol. 9:2993. doi: 10.3389/fmicb.2018.02993
Iguchi, A., Iyoda, S., Kikuchi, T., Ogura, Y., Katsura, K., Ohnishi, M., et al. (2015). A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 22, 101–107. doi: 10.1093/dnares/dsu043
Jann, B., Shashkov, A., Torgov, V., Kochanowski, H., Seltmann, G., and Jann, K. (1995). NMR investigation of the 6-deoxy-L-talose-containing O45, O45-related (O45rel), and O66 polysaccharides of Escherichia coli. Carbohydr. Res. 278, 155–165. doi: 10.1016/0008-6215(95)00243-6
Jann, B., Shashkov, A. S., Kochanowski, H., and Jann, K. (1993). Structural comparison of the O4-specific polysaccharides from E. coli O4:K6 and E. coli O4:K52. Carbohydr. Res. 248, 241–251. doi: 10.1016/0008-6215(93)84131-o
Keenleyside, W. J., and Whitefield, C. (1996). A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J. Biol. Chem. 271, 28581–28592. doi: 10.1074/jbc.271.45.28581
Kenyon, J. J., Cunneen, M. M., and Reeves, P. R. (2017). Genetics and evolution of Yersinia pseudotuberculosis O-specific polysaccharide: a novel pattern of O-antigen diversity. FEMS Microbiol. Rev. 41, 200–217. doi: 10.1093/femsre/fux002
Kido, N., and Kobayashi, H. (2000). A single amino acid substitution in a mannosyltransferase, WbdA, converts the Escherichia coli O9 polysaccharide into O9a: generation of a new O-serotype group. J. Bacteriol. 182, 2567–2573. doi: 10.1128/jb.182.9.2567-2573.2000
King, J. D., Kocíncová, D., Westman, E. L., and Lam, J. S. (2009). Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312. doi: 10.1177/1753425909106436
Kneidinger, B., Graninger, M., Adam, G., Puchberger, M., Kosma, P., Zayni, S., et al. (2001). Identification of two GDP-6-deoxy-D-lyxo-4-hexulose reductases synthesizing GDP-Drhamnose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 276, 5577–5583. doi: 10.1074/jbc.M010027200
Kneidinger, B., O’Riordan, K., Li, J., Brisson, J., Lee, J., and Lam, J. (2003). Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-D-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. Implications for the UDP-N-acetyl-L-fucosamine biosynthetic pathway. J. Biol. Chem. 278, 3615–3627. doi: 10.1074/jbc.M203867200
Knirel, Y. A., Qian, C., Senchenkova, S. N., Guo, X., Shashkov, A. S., Chizhov, A. O., et al. (2016). Structure of the β-Lfucopyranosyl phosphate-containing O-specific polysaccharide of Escherichia coli O84. Int. J. Biol. Macromol. 88, 578–585. doi: 10.1016/j.ijbiomac.2016.04.025
Korres, H., and Verma, N. K. (2004). Topological analysis of glucosyltransferase GtrV of Shilella flexneri by a dual reporter system and identification of a unique reentrant loop. J. Biol. Chem. 279, 22469–22476. doi: 10.1074/jbc.M401316200
Kwack, W. G., Lim, Y. J., Kwon, K. H., Chung, J. W., and Oh, J. Y. (2020). Outcomes and clinical relevance of stool multiplex bacterial polymerase chain reaction in patients with acute diarrhea: single center experience. Korean. J. Intern. Med. 35, 300–309. doi: 10.3904/kjim.2017.189
Li, Q., and Reeves, P. R. (2000). Genetic variation of dTDP-L-rhamnose pathway genes in Salmonella enterica. Microbiology 136, 2291–2307. doi: 10.1099/00221287-146-9-2291
Lin, A., Nguyen, L., Lee, T., Clotilde, L. M., Kase, J. A., Son, I., et al. (2011). Rapid O serogroup identification of the ten most clinically relevant STECs by Luminex microbead-based suspension array. J. Microbiol. Methods 87, 105–110. doi: 10.1016/j.mimet.2011.07.019
Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V., Senchenkova, S. N., Reeves, P., et al. (2014). Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 38, 56–89. doi: 10.1111/1574-6976.12034
Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V., Senchenkova, S. N., Wang, Q., et al. (2008). Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32, 627–653. doi: 10.1111/j.1574-6976.2008.00114.x
Lu, M., and Kleckner, N. (1994). Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 176, 5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994
March, C., Cano, V., Moranta, D., Llobet, E., Pérez-Gutiérrez, C., Tomás, J. M., et al. (2013). Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One 8:e56847. doi: 10.1371/journal.pone.0056847
Marra, A. R., Camargo, L. F., Pignatari, A. C., Sukiennik, T., Behar, P. R., Medeiros, E. A., et al. (2011). Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J. Clin. Microbiol. 49, 1866–1871. doi: 10.1128/JCM.00376-11
Mengin-Lecreulx, D., and van Heijenoort, J. (1993). Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 175, 6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993
Mengin-Lecreulx, D., and van Heijenoort, J. (1996). Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 271, 32–39. doi: 10.1074/jbc.271.1.32
Merino, S., Gonzalez, V., and Tomás, J. M. (2016). The first sugar of the repeat units is essential for the Wzy polymerase activity and elongation of the O-antigen lipopolysaccharide. Future Microbiol. 11, 903–918. doi: 10.2217/fmb-2015-0028
Mezzatesta, M. L., Gona, F., and Stefani, S. (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7, 887–902. doi: 10.2217/fmb.12.61
Miao, M., Wen, H., Xu, P., Niu, S., Lv, J., Xie, X., et al. (2019). Genetic diversity of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in eastern China. Front. Microbiol. 9:3341. doi: 10.3389/fmicb.2018.03341
Miyoshi-Akiyama, T., Hayakawa, K., Ohmagari, N., Shimojima, M., and Kirikae, T. (2013). Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358
Moule, A. L., Kuhl, P. M., Galbraith, L., and Wilkinson, S. G. (1989). Structure of the O-specific polysaccharide from Enterobacter cloacae strain N.C.T.C. 11579 (serogroup O10). Carbohydr. Res. 186, 287–293. doi: 10.1016/0008-6215(89)84042-x
Nakano, Y., Suzuki, N., Yoshida, Y., Nezu, T., Yamashita, Y., and Koga, T. (2000). Thymidine diphosphate-6-deoxy-L-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-L-talose from Actinobacillus actinomycetemcomitans. J. Biol. Chem. 275, 6806–6812. doi: 10.1074/jbc.275.10.6806
Nassau, P. M., Martin, S. L., Brown, R. E., Weston, A., Monsey, D., McNeil, M. R., et al. (1996). Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 178, 1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996
Pan, Y. J., Lin, T. L., Chen, C. T., Chen, Y. Y., Hsieh, P. F., Hsu, C. R., et al. (2015). Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5, 15573. doi: 10.1038/srep15573
Perepelov, A. V., Filatov, A. V., Wang, M., Shashkov, A. S., Wang, L., and Knirel, Y. A. (2016). Structure and gene cluster of the O-antigen of Enterobacter cloacae G3421. Carbohydr. Res. 427, 55–59. doi: 10.1016/j.carres.2016.03.008
Perepelov, A. V., Wang, M., Filatov, A. V., Guo, X., Shashkov, A. S., Wang, L., et al. (2014). Structural and genetic studies of the O-antigen of Enterobacter cloacae G2277. Carbohydr. Res. 387, 10–13. doi: 10.1016/j.carres.2014.01.001
Perepelov, A. V., Wang, M., Filatov, A. V., Guo, X., Shashkov, A. S., Wang, L., et al. (2015). Structure and genetics of the O-antigen of Enterobacter cloacae G3054 containing di-N-acetylpseudaminic acid. Carbohydr. Res. 407, 59–62. doi: 10.1016/j.carres.2015.01.015
Perepelov, A. V., Xu, G., Filatov, A. V., Zhang, X., Shashkov, A. S., Wang, M., et al. (2017). Structure and gene cluster of the O-antigen of Enterobacter cloacae C4115. Carbohydr. Res. 448, 110–114. doi: 10.1016/j.carres.2017.05.021
Pestourie, N., Garnier, F., Barraud, O., Bedu, A., Ploy, M. C., and Mounier, M. (2014). Outbreak of AmpC β-lactamase-hyper-producing Enterobacter cloacae in a neonatal intensive care unit in a French teaching hospital. Am. J. Infect. Control 42, 456–458. doi: 10.1016/j.ajic.2013.11.005
Pfoestl, A., Hofinger, A., Kosma, P., and Messner, P. (2003). Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-a-D-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 278, 26410–26417. doi: 10.1074/jbc.M300858200
Qian, C., Du, Y., Li, H., Wu, P., Wang, L., Wei, Y., et al. (2018). Development of rapid and simple experimental and in silico serotyping systems for Citrobacter. Future Microbiol. 13, 1511–1522. doi: 10.2217/fmb-2018-0187
Qureshi, Z. A., Paterson, D. L., Pakstis, D. L., Adams-Haduch, J. M., Sandkovsky, G., Sordillo, E., et al. (2011). Risk factors and outcome of extended-spectrum beta-lactamase-producing Enterobacter cloacae bloodstream infections. Int. J. Antimicrob. Agents. 37, 26–32. doi: 10.1016/j.ijantimicag.2010.09.009
Reeves, P. R. (1992). Variation in O-antigens, niche-specific selection and bacterial populations. FEMS Microbiol. Lett. 100, 509–516. doi: 10.1111/j.1574-6968.1992.tb14085.x
Reeves, P. R., Cunneen, M. M., Liu, B., and Wang, L. (2013). Genetics and evolution of the Salmonella galactose-initiated set of o antigens. PLoS One 8:e69306. doi: 10.1371/journal.pone.0069306
Reeves, P. R., and Wang, L. (2002). Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264, 109–135.
Rick, P. D., Hubbard, G. L., and Barr, K. (1994). Role of the rfe gene in the synthesis of the O8 Antigen in Escherichia coli K-12. J. Bacteriol. 176, 2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994
Rocchetta, H. L., Burrows, L. L., Pacan, J. C., and Lam, J. S. (1998). Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28, 1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x
Rosano, C., Bisso, A., Izzo, G., Tonetti, M., Sturla, L., De Flora, A., et al. (2000). Probing the catalytic mechanism of GDP-4-keto-6-deoxy-D-mannose Epimerase/Reductase by kinetic and crystallographic characterization of site-specific mutants. J. Mol. Biol. 303, 77–91. doi: 10.1006/jmbi.2000.4106
Ruan, X., Loyola, D. E., Marolda, C. L., Perez-Donoso, J. M., and Valvano, M. A. (2012). The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology 22, 288–299. doi: 10.1093/glycob/cwr150
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualisation and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Sakazaki, R., and Namioka, S. (1960). Serological studies on the Cloaca (Aerobacter) gruop of enteric bacteria. Jpn. J. Med. Sci. Biol. 13, 1–12.
Samuel, G., and Reeves, P. R. (2003). Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338, 2503–2519. doi: 10.1016/j.carres.2003.07.009
Sarkar, S., Ulett, G. C., Totsika, M., Phan, M. D., and Schembri, M. A. (2014). Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli. PLoS One 9:e94786. doi: 10.1371/journal.pone.0094786
Schoenhofen, I. C., McNally, D. J., Brisson, J. R., and Logan, S. M. (2006). Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16, 8C–14C. doi: 10.1093/glycob/cwl010
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Somoza, J. R., Menon, S., Schmidt, H., Joseph-McCarthy, D., Dessen, A., Stahl, M. L., et al. (2000). Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme’s catalytic mechanism and regulation by GDP-fucose. Structure 8, 123–135. doi: 10.1016/s0969-2126(00)00088-5
Thrane, S. W., Taylor, V. L., Lund, O., Lam, J. S., and Jelsbak, L. (2016). Application of WGS data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 54, 1782–1788. doi: 10.1128/JCM.00349-16
Valvano, M. A. (2003). Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452–s471. doi: 10.2741/1079
van der Veer, C., van Houdt, R., van Dam, A., de Vries, H., and Bruisten, S. (2018). Accuracy of a commercial multiplex PCR for the diagnosis of bacterial vaginosis. J. Med. Microbiol. 67, 1265–1270. doi: 10.1099/jmm.0.000792
Viau, R. A., Kiedrowski, L. M., Kreiswirth, B. N., Adams, M., Perez, F., Marchaim, D., et al. (2017). A comparison of molecular typing methods applied to Enterobacter cloacae complex: Sequencing. Rep-PCR, and MLST. Pathog. Immun. 2, 23–33. doi: 10.20411/pai.v2i1.99
Wang, L., Andrianopoulos, K., Liu, D., Popoff, M. Y., and Reeves, P. R. (2002). Extensive variation in the O-antigen gene cluster within one Salmonella enterica serogroup reveals an unexpected complex history. J. Bacteriol. 184, 1669–1677. doi: 10.1128/JB.184.6.1669-1677.2002
Wang, L., Wang, Q., and Reeves, P. R. (2010). The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 53, 123–152. doi: 10.1007/978-90-481-9078-2_6
Wang, Q., Ding, P., Perepelov, A. V., Xu, Y., Wang, Y., Knirel, Y. A., et al. (2008). Characterization of the dTDP-D-fucofuranose biosynthetic pathway in Escherichia coli O52. Mol. Microbiol. 70, 1358–1367. doi: 10.1111/j.1365-2958.2008.06449.x
Wang, S., Xiao, S. Z., Gu, F. F., Tang, J., Guo, X. K., Ni, Y. X., et al. (2017). Antimicrobial susceptibility and molecular epidemiology of clinical Enterobacter cloacae bloodstream isolates in Shanghai. China. PLoS One 12:e0189713. doi: 10.1371/journal.pone.0189713
Wang, W., Perepelov, A. V., Feng, L., Shevelev, S. D., Wang, Q., Senchenkova, S. N., et al. (2007). A group of Escherichia coli and Salmonella enterica O antigens sharing a common backbone structure. Microbiology 153, 2159–2167. doi: 10.1099/mic.0.2007/004192-0
Weischer, M., Kolmos, H. J., Kaufmann, M. E., and Rosdahl, V. T. (1993). Biotyping, phage typing, and O-serotyping of clinical isolates of Enterobacter cloacae. APMIS 101, 838–844. doi: 10.1111/j.1699-0463.1993.tb00189.x
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Winkler, H. H. (1970). Compartmentation in the induction of the hexose-6-phosphate transport system of Escherichia coli. J. Bacteriol. 101, 470–475.
Wu, Y., Lau, H. K., Lee, T., Lau, D. K., and Payne, J. (2019). In silico serotyping based on whole-genome sequencing improves accuracy of Shigella identification. Appl. Environ. Microbiol. 85:e00165-19. doi: 10.1128/AEM.00165-19
Zuccotti, S., Zanardi, D., Rosano, C., Sturla, L., Tonetti, M., and Bolognesi, M. (2001). Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidyltransferase. J. Mol. Biol. 313, 831–843. doi: 10.1006/jmbi.2001.5073
Keywords: Enterobacter cloacae, serotype, O-antigen, gene cluster, multiplex PCR
Citation: Li Y, Huang J, Wang X, Xu C, Han T and Guo X (2020) Genetic Characterization of the O-Antigen and Development of a Molecular Serotyping Scheme for Enterobacter cloacae. Front. Microbiol. 11:727. doi: 10.3389/fmicb.2020.00727
Received: 21 January 2020; Accepted: 27 March 2020;
Published: 28 April 2020.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Xin Deng, City University of Hong Kong, Hong KongGuang Zhao, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2020 Li, Huang, Wang, Xu, Han and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Han, aGFudGFvbWRAMTI2LmNvbQ==; Xi Guo, Z3VveGlAbmFua2FpLmVkdS5jbg==
†These authors have contributed equally to this work
 Yayue Li2†
Yayue Li2† Xi Guo
Xi Guo
