- 1One Health Center for Zoonoses and Tropical Veterinary Medicine and Center for Conservation Medicine and Ecosystem Health, Department of Biomedical Sciences, Ross University School of Veterinary Medicine, Basseterre, Saint Kitts and Nevis
- 2School of Animal, Rural and Environmental Sciences, Nottingham Trent University, Nottingham, United Kingdom
- 3Kalba Bird of Prey Centre, Kalba, Sharjah, United Arab Emirates
- 4Breeding Centre for Endangered Arabian Wildlife, Sharjah, United Arab Emirates
Endangered species recovery plans often include captive breeding and reintroduction, but success remains rare. Critical for effective recovery is an assessment of captivity-induced changes in adaptive traits of reintroduction candidates. The gut microbiota is one such trait and is particularly important for scavengers exposed to carcass microbiomes. We investigated husbandry-associated differences in the gut microbiota of two Old World vulture species using 16S RNA gene amplicon sequencing. Increased abundance of Actinobacteria occurred when vultures were fed quail but not rat or chicken. Conversely, diet preparation (sanitization) had no effect, although bacterial diversity differed significantly between vulture species, likely reflective of evolved feeding ecologies. Whilst the relative lack of influence of a sanitized diet is encouraging, changes in bacterial abundance associated with the type of prey occurred, representing a dietary influence on host–microbiome condition warranting consideration in ex situ species recovery plans. Incorporation of microbiome research in endangered species management, therefore, provides an opportunity to refine conservation practice.
Introduction
For diverse reasons, many attempts to breed and subsequently reintroduce endangered species into their natural habitat from captivity have not been successful (Bowkett, 2009; Conde et al., 2013; Willoughby et al., 2015). One potential reason is the loss of adaptive traits (Araki et al., 2007; Willoughby et al., 2015), which are not only encoded by the host genetic architecture but also by the host-associated microbiome. The gut microbiome could be considered such an adaptive trait, representing a substantial community of microorganisms (and their collective genes) which play vital roles in host physiology (West et al., 2019) and potentially influences reintroduction success (Redford et al., 2012). In turn, the microbiome is under both genetic and environmental control, with diet acting as a pivotal determinant of gut microbial assembly (Spor et al., 2011). Over the past decade, knowledge of microbial symbionts in host health and disease has increased considerably. However, animal microbiome research has only recently been introduced as a perspective for modern conservation and species recovery practices (Redford et al., 2012; Chong et al., 2019; Trevelline et al., 2019; West et al., 2019).
Species recovery often necessitates movement of animals for translocation or captive breeding, but typically involves biosecurity protocols and anti-microbial prophylaxis (West et al., 2019), which are at odds with current appreciation for the symbiotic host–microbiome relationship. Hence, a paradigm shift is required to not only include microbial research as a fundamental component in species recovery programs, but to also consider co-extinction of host-associated microbes an undesirable outcome (Trevelline et al., 2019; West et al., 2019). In particular, the influence of husbandry factors on the gut microbiome of captive animals and consequently their health (and post-release survival) is poorly understood (Chong et al., 2019; Trevelline et al., 2019; West et al., 2019), notably in regard to specialized taxa.
Vultures are such specialists, well-known for their intimate interactions with pathogens. These obligate scavengers remove carcasses from the environment, and provide important ecosystem functions (Safford et al., 2019). Yet, vultures are now among the most threatened group of birds, suffering global population declines of >80% (Safford et al., 2019). Consequently, vultures have become the focus of intensive conservation efforts (Safford et al., 2019). Critical to vultures is their ability to safely consume carrion in varying stages of decomposition; an adaptation which is integrally linked to their gut microbiota (Roggenbuck et al., 2014). However, the gut microbiota of many vulture species remains largely uncharacterized with little known regarding the impact of consumption of sanitized food stuffs on the vulture microbiome in wild and captive settings.
Materials and Methods
The aim of the current study was to investigate the potential impact of diet preparation on the specialized, luminal-bacterial alliance of two species of Old World vultures, the Griffon (Gyps fulvus) and Egyptian vulture (Neophron percnopterus). This was achieved by characterization of the luminal-microbiome using high-throughput amplicon sequencing of DNA form fecal samples collected after provision of diets prepared under divergent conditions. A secondary objective was identified post hoc, whereby prey type provisioning associated with fecal sample characterization permitted the post hoc investigation of the impact of prey type on luminal microbiota.
Ethics Statement
This project was approved by the Nottingham Trent University’s School of Animal, Rural and Environmental Science Ethics Review Group (ARE76).
Study Population, Experimental Design of Diets, and Sample Collection
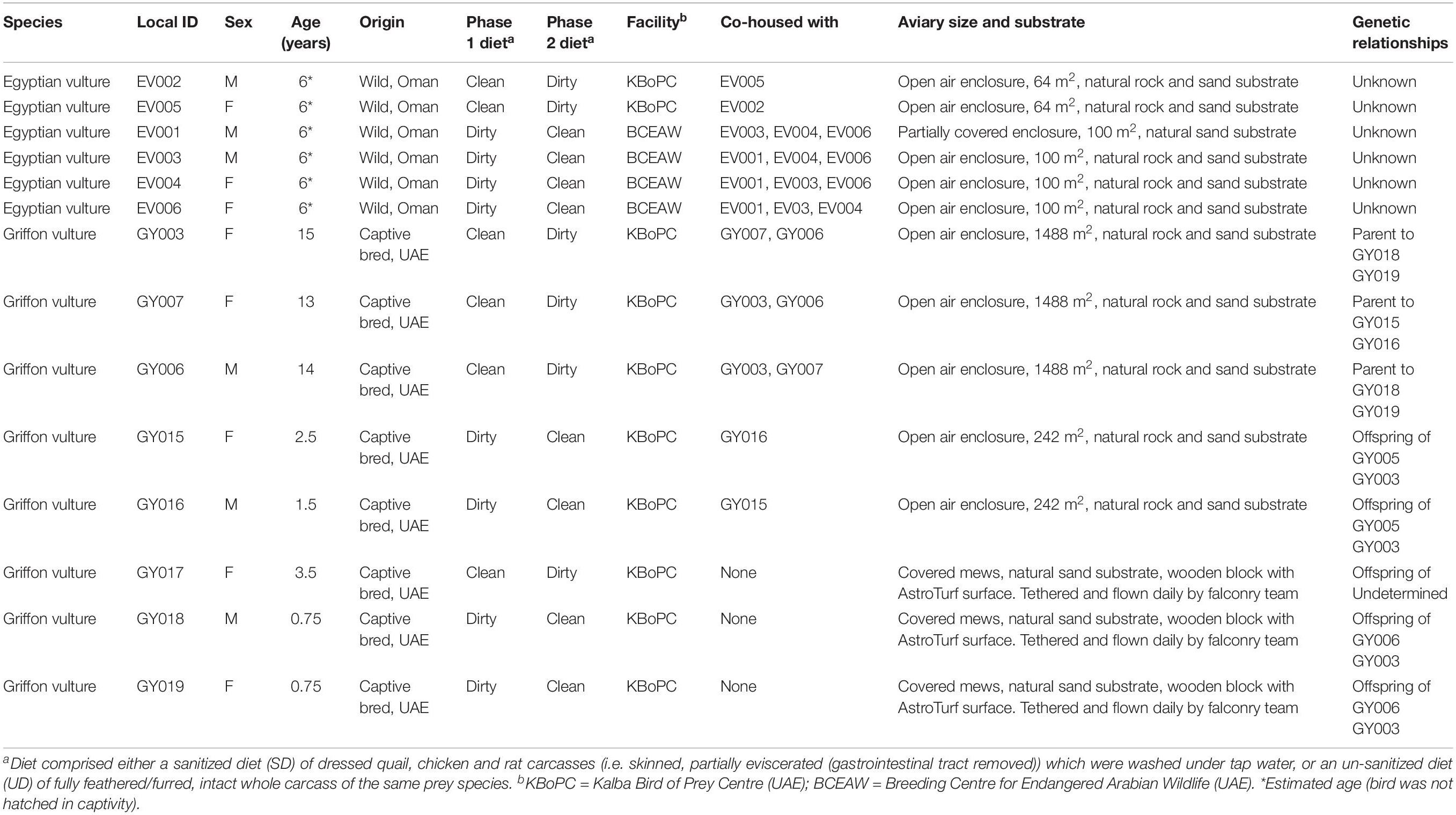
Four Egyptian vultures (Neophron percnopterus) and 7 Griffon vultures (Gyps fulvus) housed at the Kalba Bird of Prey Centre (KBoPC) along with 4 Egyptian vultures housed at the Breeding Centre for Arabian Wildlife (BCEAW), both located in the United Arab Emirates (UAE), were used in this study (Table 1). To represent typical captive dietary provision (Gaengler and Clum, 2015), two dietary conditions were implemented in a semi-randomized cross-over study design. Birds were fed either a sanitized diet (SD) comprising an overall weekly mixture of dressed quail, chicken and rat carcasses [i.e., skinned, partially eviscerated (gastrointestinal tract removed)] which were washed under tap water, or an un-sanitized diet (UD) of fully feathered/furred, intact whole carcass of the same prey species. Daily rations comprised only single prey species, and the species consumed each day were recorded for the duration of the study. No intervention in terms of the choice of prey species offered per day was performed in order to best replicate normal husbandry conditions for captive vultures. Diets (sanitized or un-sanitized; see Supporting Information for further details) were fed for a period of 4 weeks with fecal sampling in the following (fifth) week. A 2-week washout period was then implemented, during which time the birds were fed a mixture of prey items prepared as per standard husbandry practices at each facility. This mixed diet included both dressed carcasses and intact prey items of the same species as fed during the study period. After the washout period, birds were fed the alternative diet for 4 weeks before fecal sample collection in the fifth week (with daily prey species consumed recorded as previously described).
Fresh fecal samples (approximately 2 g/bird) were collected by scraping or syringe suction from the surface (see Supporting Information). We collected multiple samples per bird during the sampling week on an opportunistic basis, i.e., when a bird was seen to defecate (therefore confirming ownership and freshness) and the fecal matter was accessible (i.e., having been voided onto a surface amenable for sampling) the sample was collected. All voidings meeting this sampling criteria were collected during the week of sampling. Samples were transferred into sterilized containers and then stored at −20°C for an average of 60 (maximum 114) days prior to transport to the laboratory (ABC Labs, Dubai, United Arab Emirates).
DNA Extraction, Amplification, and Sequencing
Total bacterial community DNA extraction from each distinct fecal sample followed the conventional phenol–chloroform protocol (Pitcher et al., 1989). DNA size and integrity were assessed on 1% agarose electrophoresis gels. DNA extracts were then subject to Illumina MiSeq sequencing targeting the V4-16S rRNA gene region. The variable regions were amplified using a modified version (Apprill et al., 2015; Walters et al., 2015; Parada et al., 2016) of the original 515F-806R primer pair (Caporaso et al., 2011, 2012) and pooled libraries were constructed following the protocol as described by Kozich et al. (2013). Libraries were sequenced using 250 bp paired-end sequencing chemistry on an Illumina MiSeq platform as described previously (Kozich et al., 2013).
16S rRNA Sequence Read Processing
Pre-processing of sequencing data was done using scripts from the Microbiome Helper 16S Workflow (Comeau et al., 2017) and included stitching paired-end reads with PEAR (v0.9.10) (Zhang et al., 2014), quality assessment with FastQC (v0.11.5) (Andrews, 2010) and filtering based on read length and quality. The quality threshold score was set at 37 over at least 90% of the bases and reads shorter than 250 bp were removed. Following read filtering, potentially chimeric reads were screened out using VSEARCH (v1.11.1) (Rognes et al., 2016), which implements the UCHIME algorithm (Edgar et al., 2011). In this study, the filtered reads were classified into different operational taxonomic units (OTUs) following two approaches. First, we used an open-reference algorithm (Rideout et al., 2014) which clusters reads against a reference sequence collection (≥97% sequence similarity) and subsequently clusters sequences that do not match the sequence database de novo. The OTU table generated by this approach was used for all diversity and taxonomic analyses. The reference sequence collection used was the v.13_8 of the GreenGenes 16S rRNA gene database (DeSantis et al., 2006). OTUs having <0.1% of the total number of reads were filtered out and the OTU tables were rarefied to a minimal number of reads (11 150 seq).
Statistical Analysis
Bacterial Composition According to Vulture Species and Diet Preparation
To assess sampling depth coverage and species heterogeneity in each sample, alpha diversity metrics were employed on rarefied OTU tables using observed species (i.e., total OTUs per sample) and Shannon’s diversity indexes. Beta-diversity was assessed by calculating unweighted and weighted UniFrac and Bray–Curtis distances (Lozupone et al., 2011), which were tested for significant differences between sample categories using non-parametric ANOSIM tests with 999 permutations on non-rarefied data. Relative abundances of OTUs at different taxonomic levels were assessed using non-parametric Kruskal–Wallis test with false discovery rate (FDR) correction for multiple testing. Our threshold for significance was P < 0.05. Analysis was done using scripts from QIIME (Caporaso et al., 2010), STAMP (Parks et al., 2014), and RStudio (RStudio Team, 2015). Differences in taxonomic relative abundance for each phylum between dietary conditions (UD vs. SD) and different prey types were tested using generalized linear models, with dietary conditions, prey type and vulture species as fixed effects, and individuals from different facilities as nested random effects. Likelihood tests were used for comparisons of the models to one another and to a null model that included only the nested random factor. Similarly, we tested for an effect of vulture species on alpha diversity measures (observed number of OTUs and Shannon diversity index) in the fecal samples by comparing a linear mixed-effects model that included vulture species, dietary condition and prey type to one that included only dietary condition and prey type. These analyses were carried out in the “lmer package” in R.
Post hoc Analysis According to Prey Type (Regardless of Diet Condition)
Effect of prey type appeared as an important variable during analysis described in 2.5.1. As such, records of prey consumed each day were subsequently matched to instances where a fecal sample had been produced and collected on the following day. This time lag was considered appropriate on the basis of a known ∼21 h mean digesta retention time determined in a separate study with this population of Griffon vultures (Daneel et al., 2019). Griffon vultures had fecal samples matched to a total of 18 quail-feeding days, and 12 rat-feeding days. Egyptian vultures had fecal samples matched to a total of 2 quail-feeding days, 12 chicken-feeding days, 5 rat-feeding days, and 3 fasting days. The effect of prey type was tested by modeling phylum abundance measures against prey type consumed the day prior to sample collection, regardless of vulture species or preparation condition of the diets. These analyses were carried out in the “lmer package” in R.
Results
We collected 52 fecal samples from the 15 birds in our cross-over study design; each bird was sampled at least once per dietary condition (range 1–5 samples per condition), with an average of 4 samples per bird being collected.V4-16S rRNA gene sequencing and subsequent quality filtering generated 5,293,884 high-quality sequences, with an average of 101,805 reads per sample (minimum 11,150; maximum 867,136 reads per sample). Using a threshold of 97% identity, sequences clustered into 533 OTUs with an average of 236 ± 62 OTUs retrieved in Griffon vulture samples and 180 ± 77 OTUs in Egyptian vulture samples.
Bacterial Composition According to Vulture Species and Diet Preparation
No significant impact of diet preparation (i.e., sanitization) was detected (P = 0.1454) for either vulture species. Nonetheless, patterns of change were detectable at the taxonomic family level in our birds whereby a general trend toward reduced abundance under sanitized dietary conditions was observed (Supplementary Figure 1).
Vulture species significantly affected fecal bacterial richness (P < 0.05) and Shannon diversity index was significantly different between vulture species (Figure 1; P < 0.01), but no overall effect of vulture species (P = 0.546) nor diet (P = 0.1454) or prey type (P = 0.2707) were observed in the full mixed-effects model. The gut bacterial community composition in both Griffon and Egyptian vultures was characterized by the dominance of genera within the phyla Firmicutes (58.4%) and Proteobacteria (36.6%) (Figure 2A). Within Firmicutes, sequences were classified into seven families with an abundance of >1% of total reads (Figure 2B). Clostridia dominated the bacterial community, represented by Clostridiaceae (17%) and Peptostreptococcaceae (16%). Fusobacteria (2.4%), Actinobacteria (1%) and Cyanobacteria (0.1%) were minor contributors to the vulture’s gut bacterial composition and Bacteroidetes represented 1.5% of the microbiome in the studied Griffon and Egyptian vultures.
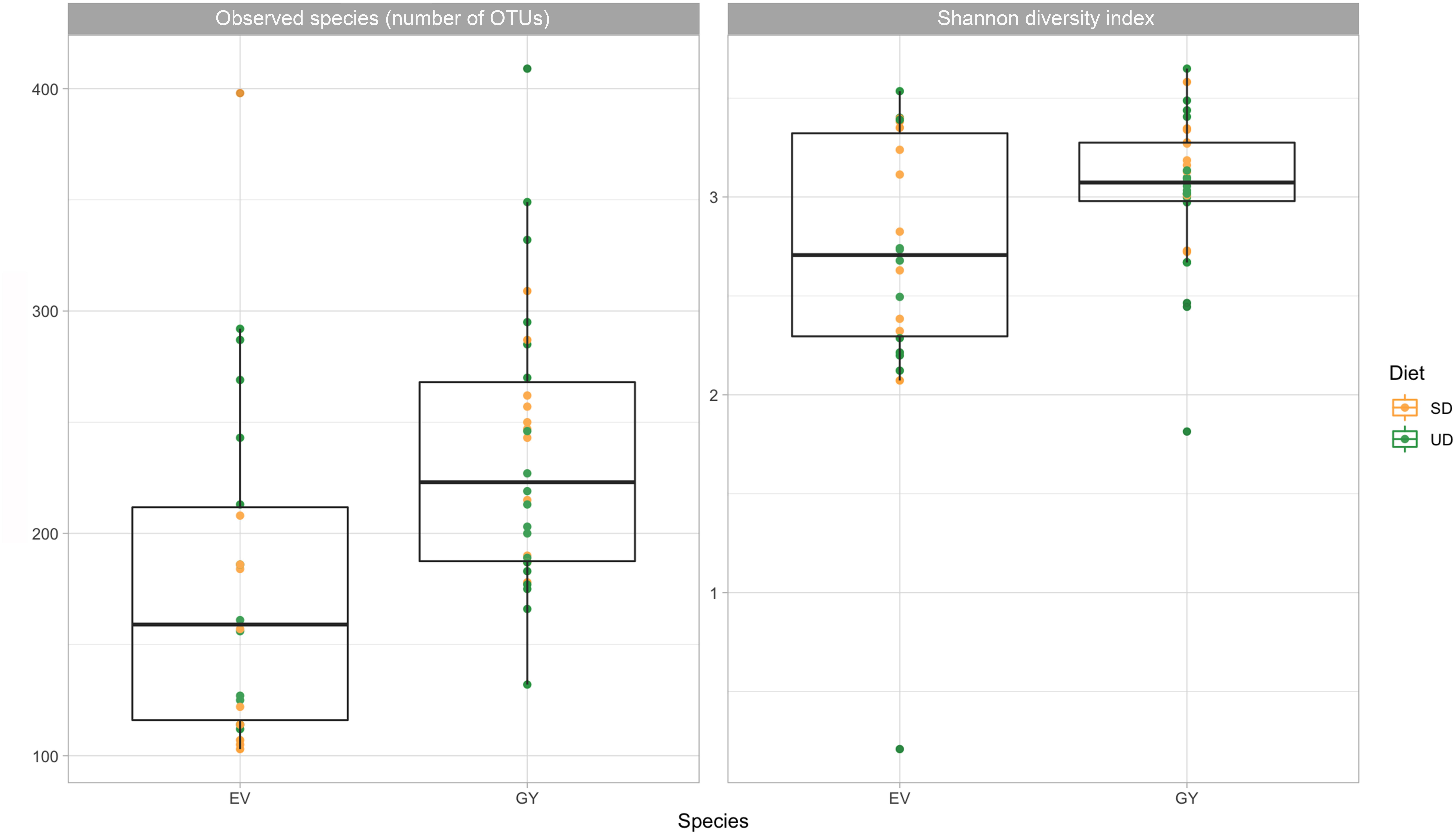
Figure 1. Variation in gut bacterial diversity between Egyptian and Griffon vultures. Alpha diversity based on rarefied data, measured by observed species and Shannon diversity Index, plotted for 52 fecal samples of two Old World vulture species (EV = Egyptian vulture, six individuals, n = 22 samples; GY = Griffon vulture, seven individuals, n = 30 samples). Statistical testing showed significant difference in observed species (Wilcoxon, P < 0.05) and Shannon diversity (Wilcoxon, P < 0.05) between both vulture species. Vultures were fed either a sanitized diet (SD) consisting of skinned, de-gutted and washed rats, chicken and quail, or un-sanitized diet (UD) consisting of intact whole rats, chicken and quail. No significant difference were observed between diets.
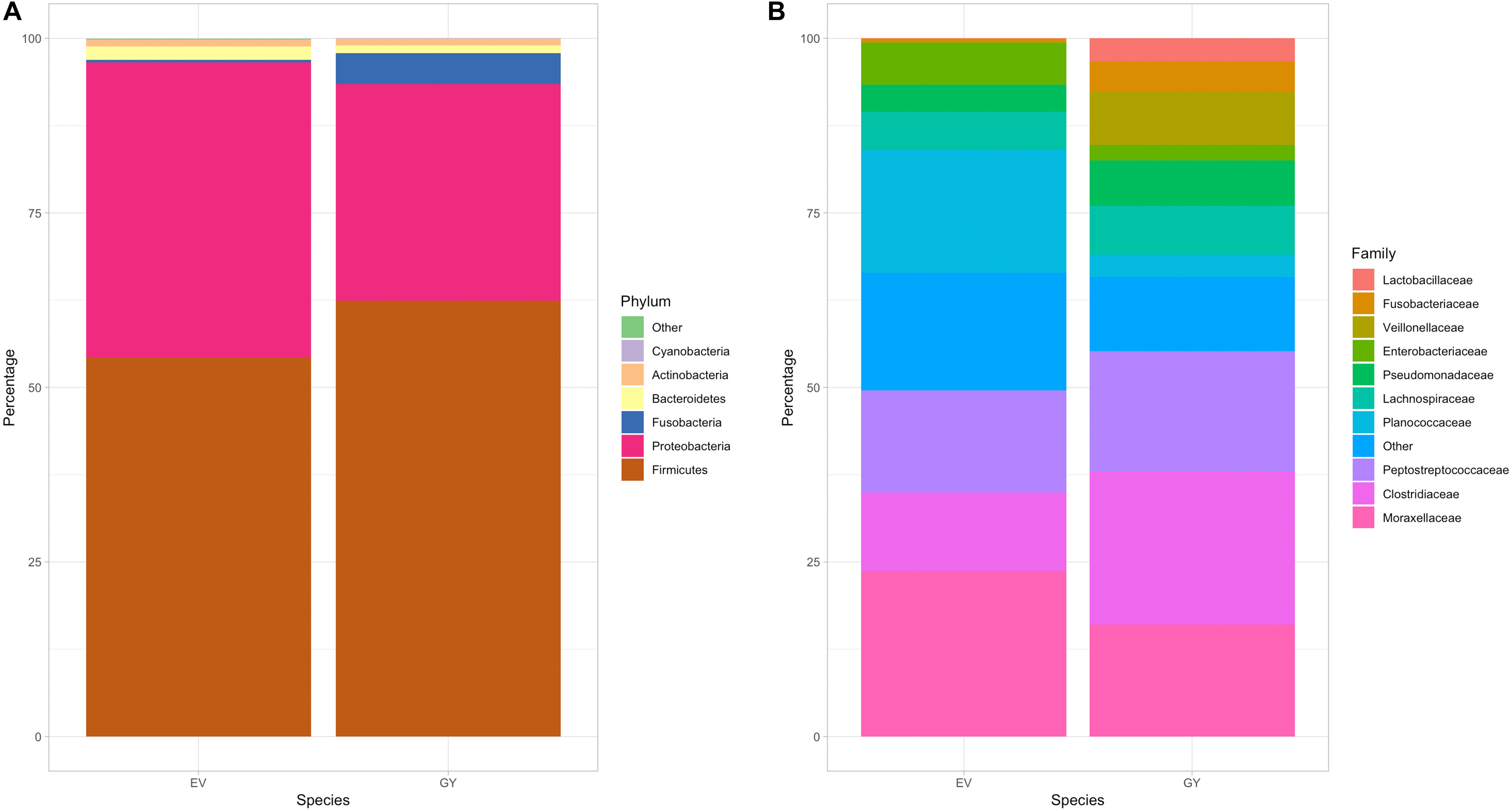
Figure 2. Gut bacterial composition of Egyptian and Griffon vultures. Taxonomic bacterial profile of 52 fecal samples from Egyptian (EV; six individuals, n = 22 samples) and Griffon vultures (GY; seven individuals, n = 30 samples) at phylum (A; left) and family (B; right) level. Of 75 families classified, only 14 with an abundance >1% of total reads are displayed.
Structural differences in bacterial community composition between species were also observed (Figure 3 and Supplementary Figures 2, 3). These differences were apparent at phylum level with a significantly higher relative abundance of Firmicutes (Welch’s t-test, q = 0.018) in Griffon vultures and of Proteobacteria (Welch’s t-test, q = 0.025) in Egyptian vultures (Supplementary Figure 4). Additionally, although not statistically significant, Fusobacteria were observed in a higher abundance and Bacteroidetes in lower abundance in Griffon vultures. No other metadata included in the mixed-effects models (age, location, aviary) had a significant impact on the gut bacterial diversity.
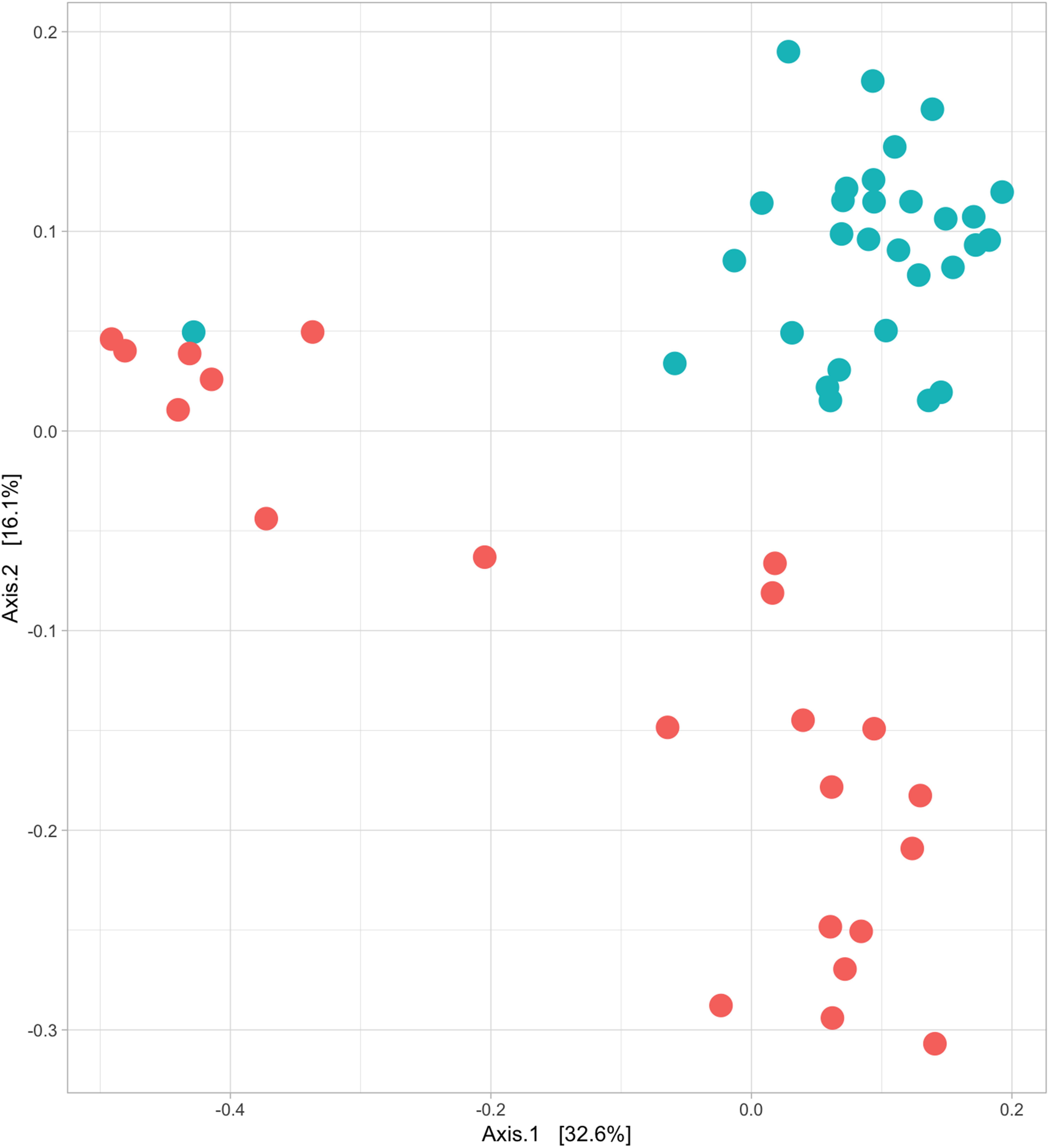
Figure 3. Egyptian and Griffon vultures exhibit different bacterial communities. Beta diversity; principal coordinate analysis visualizing the clustering of bacterial communities of 52 fecal samples from Egyptian (six individuals, n = 22 samples; red) and Griffon vultures (seven individuals, n = 30 samples; blue) based on unweighted UniFrac dissimilarity matrix. Vulture species exhibited minor overlap (ANOSIM; R = 0.545, P = 0.001).
Post hoc Analysis According to Prey Type (Regardless of Diet Condition)
Griffon vultures exhibited a higher relative abundance of Actinobacteria (represented by 53 OTUs) when fed quail (P = 0.02; n = 18 samples) compared to when fed rats (n = 12 samples) (Figure 4). No equivalent effect of prey type was detectable for Egyptian vultures. The increase of Actinobacteria could be attributed to an increase in abundance of seven OTUs assigned to Coriobacteriaceae (Genus Rhodococcus, ∼21% of sequences assigned to Actinobacteria) and one OTU assigned to Nocardiaceae (∼ 24% of sequences assigned to Actinobacteria).
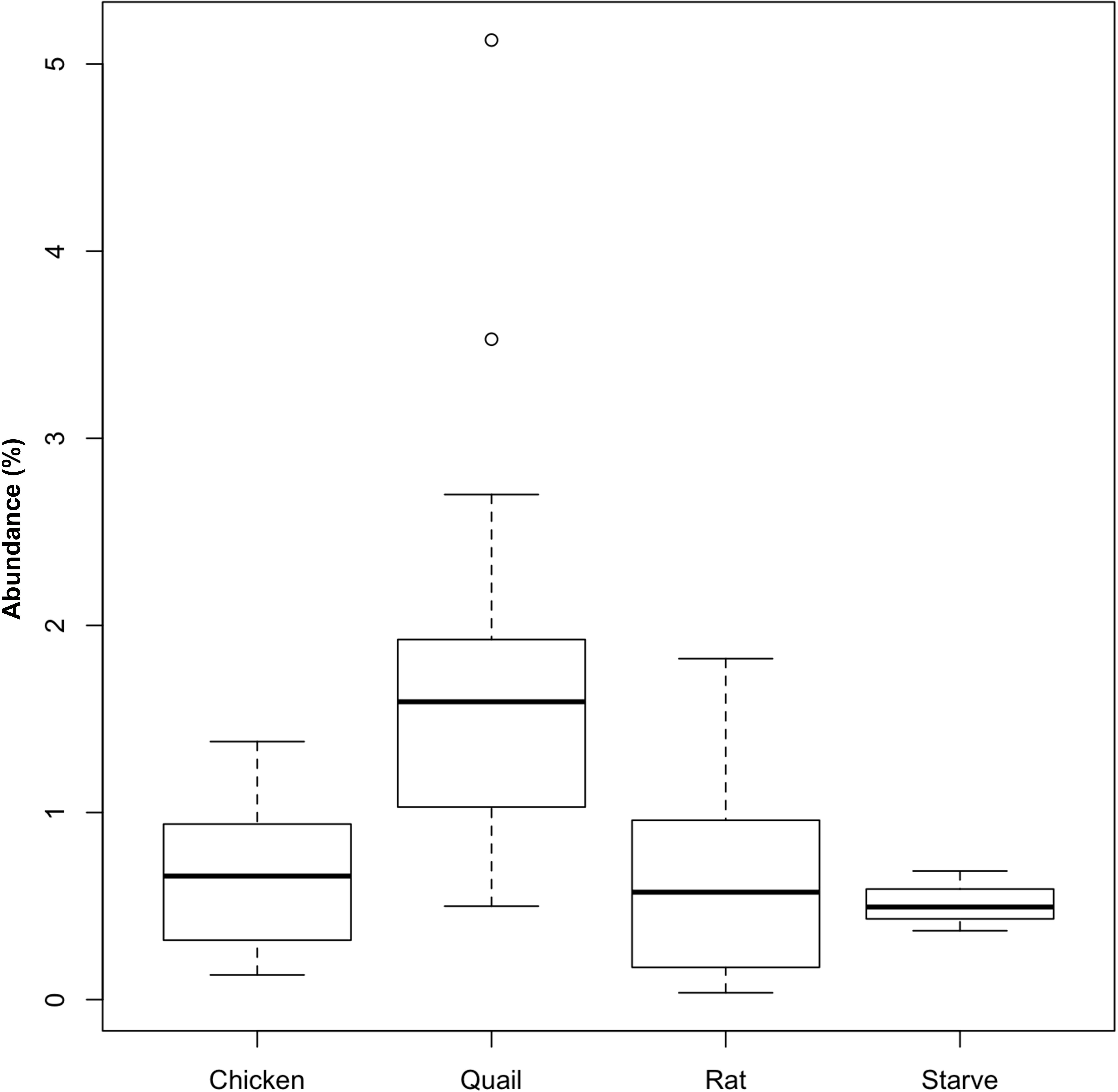
Figure 4. Relative abundance of Actinobacteria in the fecal bacterial community of vultures varied according to prey type. Boxplots showing the relative abundance of Actinobacteria in fecal samples from Griffon vultures (seven individuals, n = 30 samples) fed either rat (n = 12 samples) or quail (n = 18 samples), and Egyptian vultures (six individuals, n = 22 samples) fed either quail (n = 2 samples), rat (n = 5 samples), or chicken (n = 12 samples), or following a ‘fasted’ day (n = 3 samples). For quail and rat prey types, fecal Actinobacteria abundance data from both vulture species were combined, but differences between prey type were only statistically significant for Griffon vultures (P = 0.02). No statistical differences were detected between the four prey types fed to Egyptian vultures.
Discussion
Our study represents the first ever empirical investigation of the hypothesis that captive dietary conditions could influence gut microbiota of an obligate scavenger (Blanco, 2014; Roggenbuck et al., 2014), with findings in support of a modifying role for prey type, but not diet preparation. In contrast to previously suggested links between feeding ground sanitization status and raptor gut microbiota (Gangoso et al., 2009; Blanco, 2014), no significant impact of diet preparation (sanitization) was detected. Rather, it appears that increased sanitization in zoos (Crissey et al., 2001), compared to free-ranging habitats, is unlikely to compromise vulture gut bacterial diversity. Nonetheless, the trend toward reduced bacterial abundance under sanitized dietary conditions aligns with the inoculation theory and warrants investigation utilizing larger, longitudinal studies.
Considering the bacterial composition observed, Bacteroidetes, typically a major phylum in many species including birds (Ley et al., 2008; Waite and Taylor, 2014), was only a minor contributor of the microbiome in our Griffon and Egyptian vultures. This is in accordance with the low proportions (<1%) of this phylum in three other Old World (Meng et al., 2017) and a New World vulture species (Rodrigues De Carvalho et al., 2003; Roggenbuck et al., 2014). Members of the Bacteroidetes are known to thrive on the plethora of complex polysaccharides that constitute “dietary fiber” (Thomas et al., 2011) and are correspondingly represented in lower proportions in species with higher dietary protein intake (Becker et al., 2014). Hence, this likely reflects vultures’ carnivorous nature and may explain their divergence from other (non-carnivorous) avian gut microbiomes. Inter-specific differences in bacterial composition detected in our study and others (Roggenbuck et al., 2014; Waite and Taylor, 2014; Meng et al., 2017) emphasize the need for caution in extrapolation of data between different vulture species, supporting recent calls to increase fundamental knowledge of animal microbiomes on a species-specific basis (Trevelline et al., 2019; West et al., 2019), including in conservation biology (Redford et al., 2012).
Diet specialization, along with phylogeny, is considered integral in shaping microbial diversity in a healthy vertebrate’s gut (Ley et al., 2008; Waite and Taylor, 2014). In the wild, Griffon vultures access the carcass directly during group feeding bouts to obtain protein- and fat-rich tissues, whereas the smaller Egyptian vultures rely on scraps of tissue picked up from the area surrounding the carcass (Kruuk, 1967; Hertel, 1994). Egyptian vultures also include insects in their diet, pick at bare bones, and have unusual coprophagic tendencies (Kruuk, 1967; Negro et al., 2002). This likely contributes toward a noteworthy fiber intake of plant (e.g., prey digestive tracts, feces) and animal (e.g., skin, bone, chitin, connective tissue) origin. This different feeding ecology could explain the lower proportions of (fat-adapted) Firmicutes and the relatively higher (fiber-adapted) Bacteroidetes detected in Egyptian vultures. A greater abundance of Enterococcaceae (associated with increased fiber intake and decreased Lactobacillaceae) [associated with decreased protein intake (Clarke et al., 2012)] in the Egyptian vulture could also reflect an evolved adaptation to these differences in feeding ecology. Likewise, fibrous prey components from the un-sanitized diets (e.g., skin, digestive tracts) may facilitate population growth of organisms associated with carbohydrate substrates such as Bacteroidaceae (Thomas et al., 2011) (observed here with a numerically higher abundance). Comparisons between free-ranging and captive birds using equivalent sampling and analyses techniques to avoid bias have not yet been conducted for Griffon and Egyptian vultures. Our findings serve as a valuable starting point for future comparative studies.
Unlike previous findings (Waite and Taylor, 2015), age, location, and aviary had no significant impact on the gut bacterial diversity. Importantly, data from co-housed birds did not cluster together and no clustering was apparent on the basis of housing location, despite multiple environmental differences (e.g., substrates, vegetation, aviary size, husbandry protocols, and neighboring species). Although similar to observations in New World vultures (Roggenbuck et al., 2014) and other avian species (Ley et al., 2008), this effect had to date been untested in Old World vultures. This demonstrates the resilience of vulture microbiota to captivity-related environmental and husbandry factors, whereby the vulture’s microbiome was most reflective of their carnivorous lifestyle.
As captive birds represent potential source populations for wild population recovery efforts, this resilience is of particular significance. However, our finding of a significant impact of one particular prey type (quail) requires further consideration as it represents a potentially important husbandry-associated influence on vulture microbiome. Quail may have acted as an inoculation source of Actinobacteria for Griffon vultures. This prey type has been shown to have a notably high abundance of Actinobacteria (Su et al., 2014) in contrast to the microbiome of rats (Li et al., 2017) and chickens (Oakley et al., 2014) that only includes Actinobacteria as a minor contributor. The lack of equivalent effect in Egyptian vultures may relate to our study design, which was not established to test this hypothesis and therefore our finding in Griffon vultures was not based on an experimental design established for the purpose of testing this. The relatively balanced split between fecal samples associated with quail and only one other prey species (rat) was fortunate, but the low number of days when the birds were fed other prey types may have impacted our ability to detect their influence. In contrast, Egyptian vultures were only fed quail on two occasions that could be temporally associated with samples used in analysis. Chicken was, however, associated with 12 samples but no influence of this prey type on fecal microbiome was detectable. Consideration is also required of the duration of prey type exposure. Our post hoc analysis of fecal samples evaluated according to the prey type consumed on the day prior to fecal voiding assumes that this ∼24 h period was sufficient to elicit an acute bacterial response. Although not commonly reported, there is evidence to demonstrate a rapid response to diet changes and that such acute bacterial changes are detectable within 24 h of feeding (Wu et al., 2011), thereby supporting our analytical approach.
An inoculating or modifying role for prey type has previously been shown in other birds of prey, including kites (Blanco, 2014), falcons and owls (Bangert et al., 1988) and New World vultures (Roggenbuck et al., 2014), whereby microorganisms identified in the hindgut of these raptors were considered to originate directly from the diet consumed. It is not possible to ascertain whether our findings represent an adaptation or inoculation effect of the luminal microbiome by prey type in our study. However, either mechanism is a particularly intriguing possibility in scavengers, given that these species are generally considered to have evolved efficient strategies to protect themselves against such inoculation. Concurrently, research in mice and humans has demonstrated an association between increased abundance of Actinobacteria and obesity and the consumption of high-fat diets (Clarke et al., 2012) such that the macronutrient content of prey offered in captivity is likely an important factor to consider. The implications of our findings in Griffon vulture remain to be elucidated but nonetheless represents an important anthropogenic influence, whereby free-ranging vultures (of any species) would not typically include large proportions of quail in their diet. Moreover, the increased abundance of Nocardiaceae should be interpreted with caution as these ubiquitous environmental bacteria are more likely to be transient passengers in the gastro-intestinal tract of vultures upon quail intake. However, they have been shown to act as opportunistic pathogens (including the genus Rhodococcus) in immunocompromised hosts (Barka et al., 2016). Elucidation of the functional importance of Actinobacteria may be facilitated once the microbiome of free-ranging individuals is characterized.
Whereas the implications of increased Actinobacteria abundance are as yet unknown, bacterial alignment with species-specific feeding strategies is still tangible here. These inter-specific differences should be considered when evaluating host–microbiota interactions, especially for animals intended for release to the wild. The notable lack of large ungulate carcass feeding for captive vultures (Gaengler and Clum, 2015) is at odds with their evolved dietary specialization, and reliance on smaller whole prey species may introduce important, but as yet unquantified, differences in bacterial communities. Whilst it is possible that a captive-to-wild bacterial composition transition may occur following release, e.g., most recently evidenced in Tasmanian devil’s (Sarcophilus harrisii) (Chong et al., 2019), this represents another acclimatization process, amongst a suite of other physiological and behavioral adaptations, incurred by released individuals. Since pre-release conditioning and training is already considered vital to post-release success, it would appear prudent that reintroduction programs include monitoring for (and mitigation against) captivity-induced microbiome alterations prior to release, alongside optimization of other health parameters, rather than leaving microbial adaptation to occur post-release. Given the importance of the microbiome to host health, the value of integrating microbiome knowledge into ex situ breeding program management is hereby emphasized.
Combined, these findings highlight the importance of species- and husbandry-specific drivers in shaping the gut bacterial community and cautions against inter-specific extrapolations. Captive breeding programs aimed at propagating vultures for release can be encouraged by the relative lack of influence that a more sanitized diet had on vulture gut microbiota; hygiene procedures implemented to protect human health do not appear to compromise vulture bacterial composition. The nutritional and behavioral implications of feeding such a sanitized diet were beyond the scope of this study but are nonetheless vital considerations when formulating captive vulture diets. The importance of incorporating microbial research in conservation practice is evident; most notably an understanding of species- and environment-specific effects should be considered fundamental to advancing knowledge necessary for implementing best practice in species recovery.
Data Availability Statement
The 16S rRNA data sets generated in this study are made available and deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA621094 with BioSample accession numbers SAMN14501507 to SAMN14501558.
Ethics Statement
The animal study was reviewed and approved by Nottingham Trent University’s School of Animal, Rural and Environmental Science Ethics Review Group (ARE76).
Author Contributions
KW-T, SH, and GW-T conceived and designed the study with assistance from JB. KW-T and GW-T conducted the study, collected and prepared samples for laboratory analyses. AB completed all data analyses and interpretation, with input from SH and KW-T. KW-T and AB prepared and wrote the manuscript with input from all authors.
Funding
The research was funded with Quality Research funding of the School of Animal, Rural and Environmental Sciences (Nottingham Trent University, United Kingdom) and financial support from the Environment and Protected Areas Authority (EPAA, Sharjah, United Arab Emirates). We would also like to thank the One Health Center for Zoonoses and Tropical Veterinary Medicine of Ross University School of Veterinary Medicine for their financial support for open access publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank staff at Kalba Bird of Prey Centre and the Breeding Centre for Endangered Arabian Wildlife for their assistance during sample collection; in particular Mrs. Tanya McCormack, Mr. Qaisar Awan, Mr. Rahis Khan, and Mr. Indika Thilaksiri are acknowledged for their roles in this process. Dr. Alan McNally is gratefully acknowledged for completing the sequencing work at Nottingham Trent University’s School of Science and Technology. The authors thank His Highness Sheikh Dr. Sultan bin Mohammad al-Qasimi, Ruler of Sharjah and UAE Supreme Council Member, and Her Excellency Hana Saif Al Suwaidi, Chairperson of the Environment and Protected Areas Authority of Sharjah, for their support. The authors are grateful to colleagues at the School of Animal, Rural and Environmental Sciences (Nottingham Trent University) for their constructive feedback and assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01025/full#supplementary-material
References
Andrews, S. (2010). FastQC: A Quality-Control Tool for High-Throughput Sequence Data. Cambridge, United Kingdom: Babraham Institute.
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137.
Araki, H., Cooper, B., and Blouin, M. S. (2007). Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 318, 100–103. doi: 10.1126/science.1145621
Bangert, R. L., Ward, A. C. S., Stauber, E. H., Cho, B. R., and Widders, P. R. (1988). A survey of the aerobic bacteria in the feces of captive raptors. Avian Dis. 32, 53–62.
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/mmbr.00044-16
Becker, A., Hesta, M., Hollants, J., Janssens, G. P. J., and Huys, G. (2014). Phylogenetic analysis of faecal microbiota from captive cheetahs reveals underrepresentation of Bacteroidetes and Bifidobacteriaceae. BMC Microbiol. 14:43. doi: 10.1186/1471-2180-14-43
Blanco, G. (2014). Influence of diet on the gastrointestinal flora of wintering red kites. Eur. J. Wildl. Res. 60, 695–698. doi: 10.1007/s10344-014-0820-5
Bowkett, A. E. (2009). Recent captive-breeding proposals and the return of the ark concept to global species conservation. Conserv. Biol. 23, 773–776. doi: 10.1111/j.1523-1739.2008.01157.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chong, R., Grueber, C. E., Fox, S., Wise, P., Barrs, V. R., Hogg, C. J., et al. (2019). Looking like the locals - gut microbiome changes post-release in an endangered species. Anim. Microbiome 1, 1–10. doi: 10.1186/s42523-019-0012-4
Clarke, S. F., Murphy, E. F., Nilaweera, K., Ross, P. R., Shanahan, F., O’Toole, P. W., et al. (2012). The gut microbiota and its relationship to diet and obesity. Gut Microbes 3, 186–202. doi: 10.4161/gmic.20168
Comeau, A. M., Douglas, G. M., and Langille, M. G. I. (2017). Microbiome Helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127-16. doi: 10.1128/mSystems.00127-16
Conde, D. A., Colchero, F., Gusset, M., Pearce-Kelly, P., Byers, O., Flesness, N., et al. (2013). Zoos through the Lens of the IUCN red list: a global metapopulation approach to support conservation breeding programs. PLoS One 8:e80311. doi: 10.1371/journal.pone.0080311
Crissey, S. D., Slifka, K. A., Shumway, P. J., and Spencer, S. (2001). Handling Frozen/Thawed Meat and Prey Items Fed to Captive Exotic Animals A Manual of Standard Operating Procedures. Beltsville, MD: United States Department of Agriculture, National Agricultural Library.
Daneel, A., Whitehouse-Tedd, K., Whitehouse-Tedd, G., Dierenfeld, E. S., and Janssens, G. P. J. (2019). “Differences in digestive traits between two Old World vultures,” in Proceedings of the 10th European Zoo Nutrition Conference, 17th - 20th January, (Marwell Zoo, UK: European Association of Zoos and Aquariums).
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Gaengler, H., and Clum, N. (2015). Investigating the impact of large carcass feeding on the behavior of captive Andean condors (Vultur gryphus) and its perception by Zoo Visitors. Zoo Biol. 34, 118–129. doi: 10.1002/zoo.21202
Gangoso, L., Grande, J. M., Lemus, J. A., Blanco, G., Grande, J., and Donázar, J. A. (2009). Susceptibility to infection and immune response in insular and continental populations of Egyptian vulture: implications for conservation. PLoS One 4:e0006333. doi: 10.1371/journal.pone.0006333
Hertel, F. (1994). Diversity in body size and feeding morphology within past and present vulture assemblages. Ecology 75, 1074–1084.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Li, D., Chen, H., Mao, B., Yang, Q., Zhao, J., Gu, Z., et al. (2017). Microbial biogeography and core microbiota of the rat digestive tract. Sci. Rep. 7, 1–16. doi: 10.1038/srep45840
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Meng, X., Lu, S., Yang, J., Jin, D., Wang, X., Bai, X., et al. (2017). Metataxonomics reveal vultures as a reservoir for Clostridium perfringens. Emerg. Microbes Infect. 6:e9. doi: 10.1038/emi.2016.137
Negro, J. J., Grande, J. M., Tella, J. L., Garrido, J., Hornero, D., Donazar, J. A., et al. (2002). An unusual source of essential carotenoids. Nature 416, 807–808.
Oakley, B. B., Lillehoj, H. S., Kogut, M. H., Kim, W. K., Maurer, J. J., Pedroso, A., et al. (2014). The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 360, 100–112. doi: 10.1111/1574-6968.12608
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Pitcher, D. G., Saunders, N. A., and Owen, R. J. (1989). Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8, 151–156.
Redford, K. H., Segre, J. A., Salafsky, N., Del Rio, C. M., and Mcaloose, D. (2012). Conservation and the microbiome. Conserv. Biol. 26, 195–197. doi: 10.1111/j.1523-1739.2012.01829.x
Rideout, J. R., He, Y., Navas-Molina, J. A., Walters, W. A., Ursell, L. K., Gibbons, S. M., et al. (2014). Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2“:e545. doi: 10.7717/peerj.545
Rodrigues De Carvalho, L., Macêdo Farias, L., Nicoli, J. R., Fernandes Silva, M. C., Santos Meira Corsino, A. T., De Lima, L. A. A., et al. (2003). Dominant culturable bacterial microbiota in the digestive tract of the American Black Vulture (Coragyps atratus Bechstein 1793) and search for antagonistic substances. Braz. J. Microbiol. 34, 218–224. doi: 10.1590/S1517-83822003000300007
Roggenbuck, M., Bærholm Schnell, I., Blom, N., Bælum, J., Bertelsen, M. F., Pontén, T. S., et al. (2014). The microbiome of New World vultures. Nat. Commun. 5:5498. doi: 10.1038/ncomms6498
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Safford, R., Andevski, J., Botha, A., Bowden, C. G. R., Crockford, N., Garbett, R., et al. (2019). Vulture conservation: the case for urgent action. Bird Conserv. Int. 29, 1–9. doi: 10.1017/S0959270919000042
Spor, A., Koren, O., and Ley, R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290. doi: 10.1038/nrmicro2540
Su, H., McKelvey, J., Rollins, D., Zhang, M., Brightsmith, D. J., Derr, J., et al. (2014). Cultivable bacterial microbiota of Northern Bobwhite (Colinus virginianus): a new reservoir of antimicrobial resistance? PLoS One 9:e99826. doi: 10.1371/journal.pone.0099826
Thomas, F., Hehemann, J. H., Rebuffet, E., Czjzek, M., and Michel, G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2:93. doi: 10.3389/fmicb.2011.00093
Trevelline, B. K., Fontaine, S. S., Hartup, B. K., and Kohl, K. D. (2019). Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B Biol. Sci. 286:20182448. doi: 10.1098/rspb.2018.2448
Waite, D. W., and Taylor, M. W. (2014). Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front. Microbiol. 5:223. doi: 10.3389/fmicb.2014.00223
Waite, D. W., and Taylor, M. W. (2015). Exploring the avian gut microbiota: current trends and future directions. Front. Microbiol. 6:673. doi: 10.3389/fmicb.2015.00673
Walters, W. A., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A. E., et al. (2015). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15
West, A. G., Waite, D. W., Deines, P., Bourne, D. G., Digby, A., McKenzie, V. J., et al. (2019). The microbiome in threatened species conservation. Biol. Conserv. 229, 85–98. doi: 10.1016/j.biocon.2018.11.016
Willoughby, J. R., Fernandez, N. B., Lamb, M. C., Ivy, J. A., Lacy, R. C., and Dewoody, J. A. (2015). The impacts of inbreeding, drift and selection on genetic diversity in captive breeding populations. Mol. Ecol. 24, 98–110. doi: 10.1111/mec.13020
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Keywords: gut microbiome, ex situ conservation, feeding ecology, husbandry, old world vultures, species recovery, prey diet
Citation: Becker AAMJ, Harrison SWR, Whitehouse-Tedd G, Budd JA and Whitehouse-Tedd KM (2020) Integrating Gut Bacterial Diversity and Captive Husbandry to Optimize Vulture Conservation. Front. Microbiol. 11:1025. doi: 10.3389/fmicb.2020.01025
Received: 15 November 2019; Accepted: 27 April 2020;
Published: 25 May 2020.
Edited by:
Lifeng Zhu, Nanjing Normal University, ChinaReviewed by:
Daniel P. R. Herlemann, Estonian University of Life Sciences, EstoniaMorten Schiøtt, Technical University of Denmark, Denmark
Copyright © 2020 Becker, Harrison, Whitehouse-Tedd, Budd and Whitehouse-Tedd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine M. Whitehouse-Tedd, S2F0aGVyaW5lLndoaXRlaG91c2V0ZWRkQG50dS5hYy51aw==
†These authors have contributed equally to this work
 Anne A. M. J. Becker
Anne A. M. J. Becker Stephen W. R. Harrison
Stephen W. R. Harrison Gerard Whitehouse-Tedd3
Gerard Whitehouse-Tedd3 Jane A. Budd
Jane A. Budd Katherine M. Whitehouse-Tedd
Katherine M. Whitehouse-Tedd