- 1Department of Biomolecular Sciences, Biotechnology Section, University of Urbino “Carlo Bo”, Urbino, Italy
- 2Department of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
Unlike human isolates, environmental Escherichia coli isolates have not been thoroughly investigated for the diversity and transferability of antibiotic-resistant plasmids. In this study, antibiotic-resistant strains from marine sediment (n = 50) and clams (n = 53) were analyzed (i) for their plasmid content using a PCR-based plasmid replicon typing (PBRT) kit and (ii) for the transferability of plasmid-associated antibiotic resistance (AR) traits by mating experiments. Fifteen of the thirty replicons targeted by the PBRT kit were detected in the isolates; 8/15 were identified in both sediment and clam isolates, although at different frequencies. The most frequent replicons in sediment (74%) and in clam strains (66%) alike, were FIA, FIB, or FII, which are associated with the IncF group, followed by the I1α replicon, which was more frequent in clam (24.5%) than in sediment (10%) strains. More than 50% of the strains contained multiple replicons; although 15 were untypable, S1-PFGE analysis demonstrated that 14/15 carried no plasmids. All cryptic strains were successfully typed and were positive for IncF or IncI replicons. Antibiotic-resistant strains accounted for 63% of all isolates and were significantly (p < 0.05) more frequent in phylogroup A. Most (35%) multidrug-resistant (MDR) strains belonged to phylogroup A, too. Although 25/26 MDR strains were positive for IncF plasmids (the exception being a clam strain), the FII-FIB rep combination was predominant (63%) among the sediment isolates, whereas most clam isolates (40%) carried the FII replicon alone. In mating experiments, selected MDR strains carrying FIB, FII, and I1α replicons, used as the donors, transferred multiple ARs together with the IncF or IncI plasmids at high frequency. Since IncI plasmids are common in E. coli and Salmonella enterica isolates from poultry, our findings suggest an animal origin to the E. coli clam strains carrying IncI plasmids. They also suggest a role for IncI plasmids in the spread of ARs among environmental Enterobacteriaceae and, through the food chain, to human isolates. In conclusion, the PBRT kit proved to be a useful tool to identify plasmids carrying antibiotic-resistant genes and to shed light on the factors underpinning their diffusion.
Introduction
Antimicrobial resistance – a major and topical clinical issue – has also become an environmental concern owing to the growing spread and ubiquity of resistance genes and bacteria (Bengtsson-Palme et al., 2018). The human and animal microbiota appears to be the primary reactor where antibiotic resistance develops, due to exposure to the selective pressure of antibiotics administered for infection treatment or prophylaxis (Baquero et al., 2008; Rolain, 2013). Moreover, the gut may be a reservoir of resistance genes, where horizontal genetic transfer (HGT) among different microbial species may easily occur (Rolain, 2013). Human and animal antibiotic-resistant (AR) bacteria released into wastewater find their way to the soil and to water environments; in particular, water is a favorable habitat for interactions and gene exchanges among micro-organisms, the dissemination of resistance genes or bacteria, and the transmission of waterborne infectious disease (Finley et al., 2013; Jang et al., 2017). Enterobacteriaceae are found in a wide range of environments. The presence of Escherichia coli – a common bacterium of the human and animal intestinal microbiota – in natural water bodies has long been interpreted as indicating fecal contamination. More recently, E. coli has been demonstrated to be highly adaptable and to be able to survive and replicate outside the host, in water, soil, sediment and vegetables (Delaquis et al., 2007; Bergholz et al., 2011; Frank et al., 2011; Jang et al., 2017). Commensal as well as pathogenic E. coli strains resistant to several antibiotics have been recovered in the marine environment (Vignaroli et al., 2013, 2016; Charnock et al., 2014; Drali et al., 2018). Antibiotic resistance in E. coli species is of particular concern because of the growing prevalence of multidrug-resistant (MDR) strains involved in human and animal infections. Moreover, the incidence of AR commensal E. coli isolates in healthy humans is increasing worldwide (Broaders et al., 2013; Rolain, 2013), contributing to the emergence and spread of AR pathogens. AR clinical isolates of E. coli are also frequently related to those from animals, suggesting that both the food chain and food animals may be a source of MDR strains (Rolain, 2013; Lazarus et al., 2015; Berg et al., 2017).
Besides antibiotic exposure and HGT events, the acquisition of resistance genes by E. coli isolates is also affected by their genetic background. Some phylogenetic groups (i.e., A and D) are more prone to develop antibiotic resistances, and strains belonging to the same phylogroup and sequence type (ST) often share the same antibiotic resistance profile (Tenaillon et al., 2010). E. coli population structure can provide useful information on strain origin, since different phylogroups predominate in distinct ecological niches (Tenaillon et al., 2010). The acquisition of plasmids, which enhance resistance gene dissemination, is believed to play a key role in the growing prevalence of MDR E. coli strains (Mathers et al., 2015). In Enterobacteriaceae, some plasmids associated with specific resistance determinants are predominant in specific geographic areas; they are able to replicate in a wide host range (e.g., IncA/C and IncL/M plasmids) and their dissemination relies on antimicrobial selective pressure (Rozwandowicz et al., 2018). Other plasmids are only found in closely related hosts and are maintained by the bacterial cell because they encode virulence factors enhancing bacterial adaption and fitness (Carattoli, 2009).
This study was undertaken to determine: (i) the distribution and prevalence of major plasmid replicons in E. coli isolated from clams and marine sediment using the PCR-based replicon typing (PBRT) kit and (ii) the involvement of specific plasmids in the conjugal transfer of antibiotic resistance from environmental E. coli.
Materials and Methods
Bacterial Strains
A total of 103 E. coli isolates (53 from clams and 50 from marine sediments) were used in the study. The 53 clam strains were selected from a collection of 141 strains isolated from Venus clams collected in Italy in the middle Adriatic Sea, which had previously been characterized for antibiotic resistance phenotype and phylogroup (Vignaroli et al., 2016). The 50 sediment strains were isolated from samples collected at three coastal sites (SE, PN, and API) at a depth of 4–12 m (from latitude 43°45.300′N, longitude 13°12.630′E, to latitude 43°39.0′N, longitude 13°22.0′E) near the areas where the clams had been harvested.
To detach bacteria from sediment, aliquots (5 g) of each sample were suspended in 20 mL sterile seawater, vortexed and sonicated (3 times, 1 min per cycle) as described previously (Luna et al., 2010; Vignaroli et al., 2013). The resuspensions were filtered by the membrane filter technique and E. coli strains were isolated in the selective medium mFC agar (BBL, Becton Dickinson & Co., Sparks, MD, United States) (Vignaroli et al., 2013).
Strain Identification and Typing
Presumptive E. coli colonies from sediment samples were identified by the molecular approach based on PCR amplification of the species-specific uidA gene (McDaniels et al., 1996). The PCR methods developed by Clermont et al. (2011, 2013) allowed each E. coli isolate to be assigned to a phylogenetic group or cryptic clade.
Antimicrobial Susceptibility and PCR Detection of Class 1 Integrons and Antibiotic Resistance Genes
Strain susceptibility to ampicillin (10 μg), cefotaxime (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), tetracycline (30 μg), chloramphenicol (30 μg), nalidixic acid (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg) and streptomycin (10 μg) was assessed by the disk diffusion method according to CLSI recommendation (Clinical and Laboratory Standards Institute (CLSI), 2017). Strains resistant to β-lactams were analyzed for extended spectrum β-lactamase (ESBL) production by screening and confirmatory tests (Clinical and Laboratory Standards Institute (CLSI), 2017). In brief, a standard disk diffusion test was performed in which the β-lactam antibiotic disks of both cefotaxime and ceftazidime, alone and in combination with clavulanate were used. A ≥5-mm increase in a zone diameter for either antimicrobial agent tested in combination with clavulanate vs. the zone diameter of the agent when tested alone indicated the ESBL production by the strain (Clinical and Laboratory Standards Institute (CLSI), 2017).
E. coli ATCC 25922 was the reference strain in all antimicrobial susceptibility assays. Resistant strains were screened by PCR for the following determinants: blaTEM, blaSHV, and blaCTX–M (ESBL-encoding genes) for β-lactam resistance; tet(A) for tetracycline resistance; dfrA1 for trimethoprim/sulfamethoxazole resistance; and strA, strB, aadA, and ant(3″) for aminoglycoside resistance. Primers and PCR conditions were as reported previously (Vignaroli et al., 2012, 2013).
MDR strains were analyzed by PCR for the intI1 integrase gene and the variable region of the class 1 integron using primers and PCR conditions described previously (Vignaroli et al., 2012). The resistance genes linked to class 1 integron were characterized by sequencing (GATC Biotech Cologne, Germany) the cassette amplicons and by nucleotide sequence analysis using the Basic Local Alignment Search Tool (BLAST)1.
Plasmid Typing
The PBRT 2.0 kit (Diatheva, Fano, Italy) which has been used for plasmid identification and to type the major resistance plasmids found in Enterobacteriaceae (Carloni et al., 2017), was applied in our study. The amplicons recognized by the PBRT kit were analyzed by capillary electrophoresis with an AATI Fragment Analyzer (Agilent, Santa Clara, CA, United States).
Where necessary, amplicons were purified using MinElute PCR purification kit (Qiagen, Hilden, Germany) and directly sequenced by the Sanger method using BigDye Terminator v. 1.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Vilnius, Lithuania) on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, United States). The consensus sequences obtained were submitted to the pMLST database for allele variant identification2.
The stx1, stx2, and eae genes of shiga toxin-producing E. coli (STEC) strains were identified using STEC FLUO detection kit (Diatheva, Fano, Italy) according to the manufacturer’s instructions; the strains that were positive for stx and/or eae genes underwent O-serogroup identification (O157, O111, O26, O103, and O145) using the STEC Serotypes FLUO kit (Diatheva).
S1-PFGE
S1-nuclease pulsed field gel electrophoresis (S1-PFGE) was performed to determine plasmid size and compare the plasmid profile of strains. Total DNA embedded in 0.8% agarose plugs was incubated for 30 min at 25°C with 100 U Aspergillus oryzae S1 nuclease (Takara Bio Inc., Shiga, Japan). The plugs were loaded on a 1% agarose gel using 0.5X TBE running buffer. Electrophoresis was performed in a CHEF-mapper system (Bio-Rad Laboratories, Inc., CA, United States) with the pulse time increasing from 1 to 25 s for 17 h at 14°C and 200 V (6 V/cm). The Low Range PFG Marker (0.1–200 kb) and Lambda Ladder PFG Marker (50–1,000 kb) from New England Biolabs (Ipswich, MA, United States) were used as molecular size markers.
Conjugation Experiments
Conjugal transfer of tetracycline and β-lactam resistance was performed by filter mating using the protocol previously described (Vignaroli et al., 2011). The E. coli 1816 (a mutant of E. coli C600, lactose-non-fermenting, resistant to nalidixic acid and rifampicin) was used as the recipient strain. Transconjugants were selected on Brain Heart Infusion agar (BHIA) (Oxoid, Basingstoke, United Kingdom) supplemented with tetracycline (20 μg/mL), rifampicin (50 μg/mL), and nalidixic acid (50 μg/mL). Transfer frequency was expressed as number of transconjugants per recipient. Transconjugants were first confirmed by three passages on MacConkey agar (Oxoid) containing all three antibiotics at the concentrations used for selection. Plasmid acquisition was assessed by comparing the S1-PFGE profiles of transconjugant and donor and confirmed using the PBRT kit.
Hybridization Assays
The plasmid location of the tet(A) gene was investigated in the donors, the transconjugants and the recipient E. coli 1816 by hybridization assays after S1-PFGE and Southern blotting. DNA was blotted onto positively charged nylon membranes (Bio-Rad Laboratories) and hybridized with a biotin-labeled tet(A) probe using North2SouthTM Chemiluminescent Hybridization and Detection Kit (Thermo Fisher Scientific, Rockford, IL, United States), according to the manufacturer instructions.
Statistical Analysis
Differences in the prevalence of the replicon types were analyzed by the χ2 test. The significance of the association between a replicon type and resistance to a specific antibiotic or belonging to a specific phylogroup was analyzed by Fisher’s test. A P-value < 0.05 was considered significant.
Results
A total number of 103 E. coli isolates, obtained from clam and sediment samples collected at roughly the same three sites (API, SE, and PN) were analyzed in this study. The 53 E. coli clam strains belonged to different phylogroups; most (n = 31) were from clams harvested next to the API site, 10 came from site SE and 12 from site PN. Most isolates belonged to phylogroup A (n = 21), B1 (n = 7) or D (n = 6) whereas six were cryptic (Vignaroli et al., 2016; Table 1).
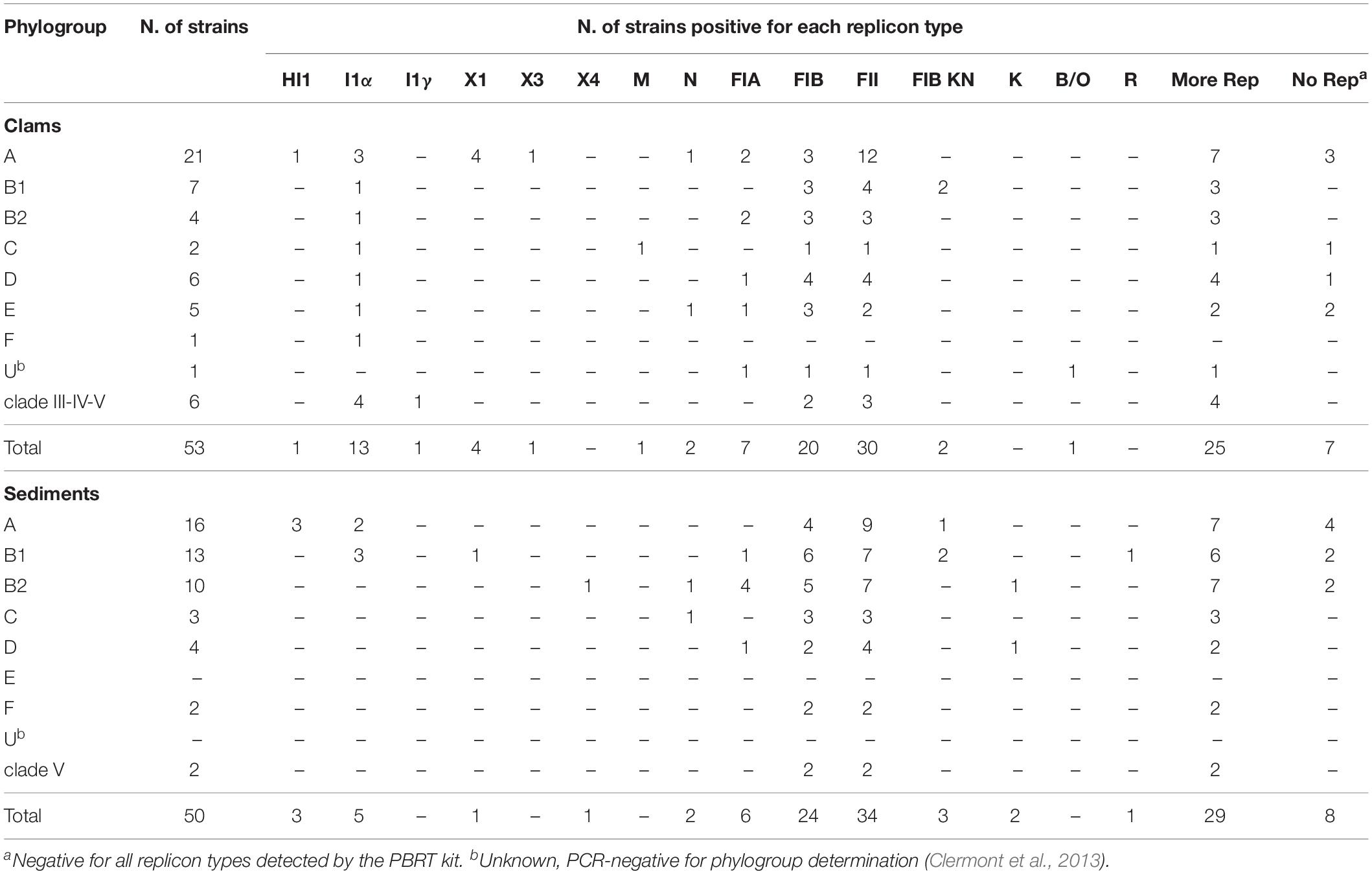
Table 1. Prevalence of the replicons detected by the PBRT kit in E. coli isolates from clams and sediments.
Analysis of the 50 isolates from sediment samples grown on mFC agar and identified as E. coli showed that most of them (n = 41) were from site PN and that they belonged to all phylogroups except E and F. There were only two cryptic strains, both of clade V, from site PN (Table 1).
All 103 strains were subjected to plasmid replicon typing; the results are reported in Table 1. Overall, 15 of the 30 replicons detected by PBRT kit were found in the strains; 8/15 were shared by clam and sediment strains, although at different frequencies. FIA, FIB, and FII, carried by IncF family plasmids, were the most frequent types, followed by replicon I1α, which is associated with IncI plasmids. IncF plasmids which were detected in 74% of sediment and 66% of clam strains carried single or multiple IncF replicons. In particular, the rep combination FII-FIB was significantly (p = 0.03) more frequent in sediment (54%) as well as in clam (28.6%) strains. More than 50% of strains contained multiple replicons, whereas 15 (8 from sediments and 7 from clams) were untypable by the PBRT kit. S1-PFGE analysis showed that 14/15 untypable strains did not contain plasmids. Interestingly, replicon I1α was more frequent among clam (24.5%) than among sediment strains (10%), but the difference was not significant. All cryptic clade strains (8) were typable by the kit and were positive only for IncF and/or IncI family replicons; 50% of these strains were positive only for FIB and FII replicons, whereas the remaining 50% carried I1α in addition to I1γ (clade III, E. coli ISZ 201), or FII (clade V, E. coli ISZ 272). In three E. coli clam strains (ISZ 45, ISZ 211, and ISZ 275), the PCR products of the FIA replicon were smaller (from ∼411 to ∼440 bp) than those of the control strain (462 bp). Sequencing and comparison with the pMLST database disclosed that all were FIA replicons carrying short deletions; in particular, E. coli ISZ 45 and E. coli ISZ 211 bore the FIA6 allele, whereas E. coli ISZ 275 carried the FIA5 allele; compared with the FIA2 allele of the control strain E. coli ISZ 35, the FIA6 and FIA5 alleles showed a deletion of 55 and 22 bp, respectively.
The STEC FLUO kit for the stx1, stx2, and eae virulence genes, to detect shiga toxin-producing E. coli showed that only E. coli PN37 from sediment was positive for gene eae, but that it did not belong to any of the serogroups detected by the kit.
The results of antimicrobial susceptibility testing showed that resistant isolates were significantly (p < 0.05) more numerous in phylogroup A (75.6%) than in the other phylogroups (56%). The prevalence of clam and sediment strains resistant to the different antimicrobials is reported in Figure 1. Among the 53 clam strains, resistance to tetracycline was the most frequent (62%; n = 33), followed by resistance to ampicillin (43%; n = 23) and streptomycin (26%; n = 14) (Figure 1), in line with a previous report (Vignaroli et al., 2016). Resistance to tetracycline was significantly more frequent (p = 0.0015) among clam than among sediment strains (Figure 1). Multidrug resistance was detected in 15 strains (28%), most of which belonged to phylogroup A (47%), B2 (20%) or E (13%) and was not significantly associated with any phylogroup. The percentage of resistant strains positive to the relevant resistance genes is shown in Figure 2. Of the 23 ampicillin-resistant strains, 21 (91%) were ESBL producers and 19 of them (83%) carried a gene (blaTEM) encoding a TEM-type β-lactamase, whereas 24 out of 33 (73%) tetracycline-resistant strains were positive for tet(A). In the streptomycin-resistant strains (n = 14) the genes strA and strB were recovered at a frequency of 86% (n = 12) and 57% (n = 8), respectively, whereas the ant(3″) gene was found in 21% (n = 3) of clam strains and aadA (50%; n = 7) was significantly (p = 0.03) more common in clam than in sediment strains (Figure 2).
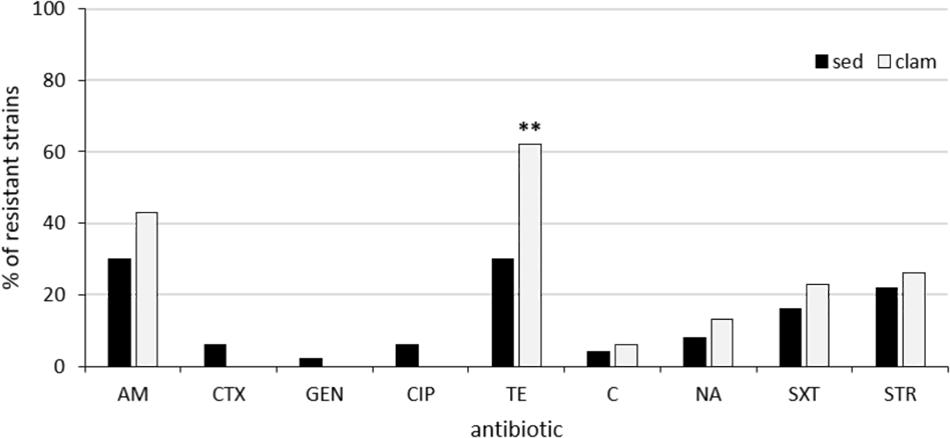
Figure 1. Prevalence of resistant strains among clam and sediment isolates. AM, ampicillin; CTX, cefotaxime; GEN, gentamicin; CIP, ciprofloxacin; TE, tetracycline; C, chloramphenicol; NA, nalidixic acid; SXT, trimethoprim/sulfamethoxazole; STR, streptomycin. **Highly statistically significant (p = 0.0015).
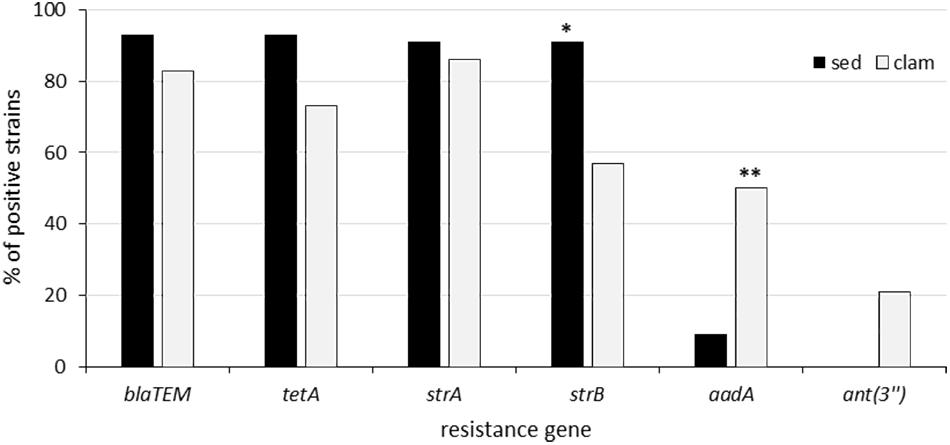
Figure 2. Prevalence of resistance genes among resistant clam and sediment strains. **Statistically significant (p = 0.03). *Not quite statistically significant (p = 0.09).
Altogether, 30% (n = 15) of sediment strains were resistant to tetracycline, 30% (n = 15) to ampicillin and 22% (n = 11) to streptomycin, in line with the results from the clam strains, which come from the same areas (Figure 1). The association between the occurrence of antimicrobial resistance and specific Inc family plasmids was then investigated. The only detected association was between ampicillin resistance and the IncF family in the clam isolates (p = 0.007), as shown in Figure 3.
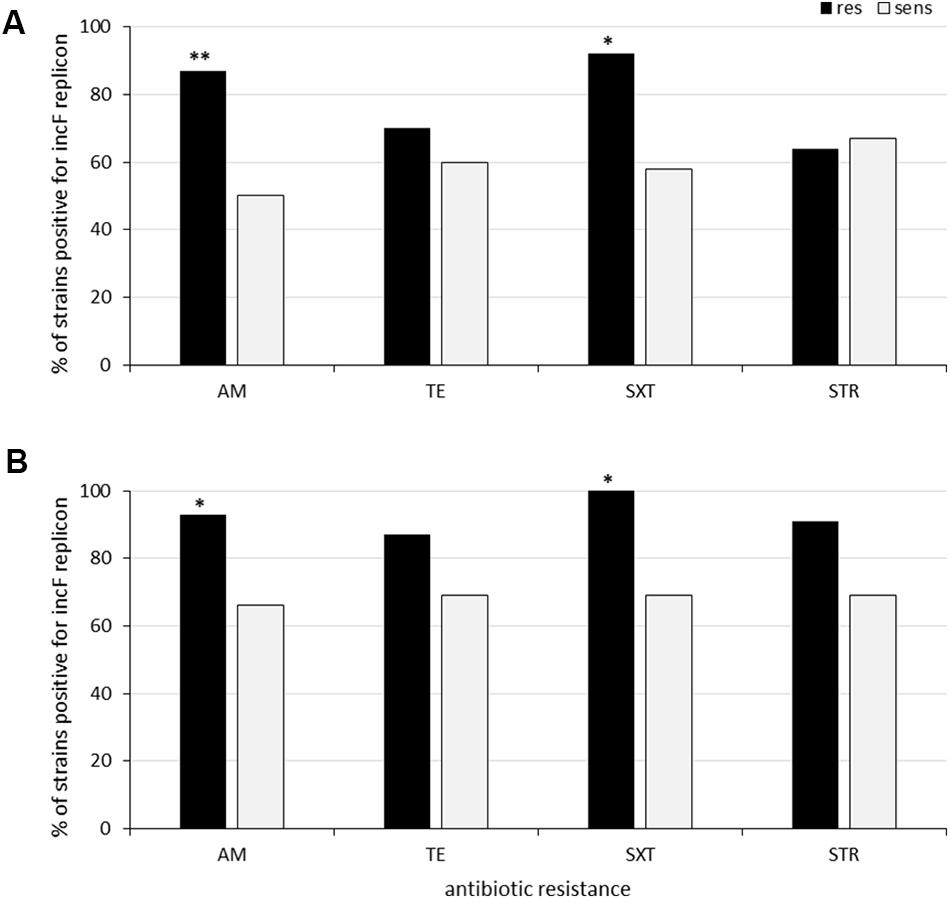
Figure 3. Association between antibiotic resistance and IncF family replicons among clam (A) and sediment isolates (B). **Statistically significant (p = 0.007). *Not quite statistically significant (p = 0.07–0.09).
Multidrug resistance (to three or more antibiotic classes) was also detected in 11 sediment strains (22%); most of them (82%) belonged to phylogroup B1 (36%), B2 (28%) or A (18%); however, as in the clam strains, multidrug resistance was associated with none of the phylogroups. All but one of the ampicillin-resistant strains (93%; n = 14) were positive to the ESBL production test and carried blaTEM gene. Two strains from the PN site were also positive for a gene encoding a SHV-type β-lactamase (blaSHV) and an ESBL-encoding gene (blaCTX–M), respectively. ESBL producers were recovered from all three sites, as displayed by the phenotypic screening. Moreover, 93% (n = 14) of tetracycline-resistant strains carried the tet(A) gene and all streptomycin-resistant isolates were positive for strA and strB at a high frequency (91%; n = 10), like the streptomycin-resistant clam strains (Figure 2). The ant(3″) gene was never detected, whereas a single strain (E. coli PN56) carried both aadA and dfrA1. All MDR sediment strains (n = 11) carried multiple IncF replicons, with the FII-FIB combination being predominant (64%). All MDR clam strains but one (n = 14) were also IncF-positive. Most of them (n = 6) carried the FII replicon alone, whereas three strains carried FII and FIB.
Although 80% (n = 21) of MDR strains from both clam and sediment were positive for the integrase gene intI1, amplification of the variable region of the class 1 integron was obtained only from 20 and 9% of the clam and sediment strains, respectively. Sequence analysis of gene cassettes demonstrated three different arrangements: dfrA1-aadA1 and dfrA17-aadA5 in clam strains and dfrA12-aadA2 in sediment strains.
The 14 E. coli isolates from clams (n = 9) and sediments (n = 5) showing positivity for the IncI1α group were subjected to S1-PFGE. Analysis of the number and size of their plasmids (Table 2) indicated that all isolates harbored 1–3 plasmids, ranging in size from 75 to 145 kb. To assess the involvement of the IncIα group in the transfer of antibiotic resistance, six tetracycline-resistant isolates carrying the tet(A) gene were used as donors in conjugation experiments. E. coli ISZ 220 from clams and E. coli PN30 from sediments transferred tetracycline resistance to E. coli 1816 at the highest frequency (respectively, 4.8 × 10–3 and 1.2 × 10–7). Five transconjugants obtained from the two mating pairs were analyzed for their plasmid profile and resistance gene acquisition. In both mating assays the transconjugants acquired all the resistance determinants and the intI1 gene, but not all the plasmids (Table 2 and Supplementary Figure S1). In fact, S1-PFGE showed that E. coli ISZ 220 transferred two (∼110 and 55 kb) of its three plasmids; PBRT confirmed that the transconjugants were positive for replicons I1α and M, but not for replicon F. In contrast, E. coli PN30 transferred only the IncF plasmid, although it also carried an IncI1 plasmid, as shown by S1-PFGE and by PCR amplification of the replicons. The hybridization assays confirmed the location of the tet(A) gene on the larger plasmids of the two donors (on the 110 kb plasmid in E. coli ISZ 220 and on the 145 kb plasmid in E. coli PN30) and of the relevant transconjugants (Supplementary Figure S1).

Table 2. E. coli isolates from clams and sediments showing positivity for the IncI1α group: phylogroups, resistance genes and plasmid sizes.
Discussion
The rapid evolution and global diffusion of MDR Enterobacteriaceae (mostly E. coli and Klebsiella pneumoniae) is raising widespread concern. Specific successful bacterial clones such as E. coli ST131 and K. pneumoniae ST258 are major causes of hospital- and community-acquired infections (Mathers et al., 2015). For epidemiological purposes, epidemic clones are usually identified by their genetic background (e.g., ST determination), whereas plasmid DNA is ignored despite its important role in strain resistance traits. The association between high-risk clones and specific resistant plasmids involved in resistance gene dissemination has been demonstrated in clinical settings (Mathers et al., 2015; Roer et al., 2018). In contrast, environmental E. coli isolates have not been thoroughly investigated for the diversity and transferability of antibiotic resistance plasmids, despite the fact that assessment of their plasmid profile could help identify dangerous strains and their origin. Moreover, recurrent detection of some plasmid types could indicate their role in strain survival in specific habitats as well as in the spread of antibiotic resistance traits. In this study, 103 AR strains from clam and marine sediment samples, collected along the mid-Adriatic coast, were analyzed for their plasmid content and for the transferability of plasmid-associated resistance traits. The PBRT kit employed in the study proved useful to identify the most common plasmids. Even though 14.5% of strains were negative for all the replicons targeted by the kit, most (93%) were not typable because they did not contain plasmid DNA. IncF replicons were the most frequent in our isolates, in line with reports that IncF plasmids are the most common plasmids in Enterobacteriaceae, especially in E. coli species (Carattoli, 2009). In Europe IncF plasmids are described predominantly in human and animal isolates, not in environmental strains (Rozwandowicz et al., 2018). Consequently, their high prevalence in our clam and sediment isolates may reflect their human or animal origin.
IncF plasmids are typically multireplicon, often encoding FII together with FIA and/or FIB (Villa et al., 2010). The plasmids containing FII and FIA replicon types have been described in the epidemic E. coli strain ST131 and in K. pneumoniae ST258 clones (Mathers et al., 2015). In our E. coli strains, most IncF-positive isolates, particularly among sediment isolates, contained the FII-FIB combination or FII alone. In a recent study (Lambrecht et al., 2018), the FII-FIB combination was found to be predominant in commensal MDR E. coli from farm animals, mainly broilers. IncF plasmids are often associated with IncI plasmids, which are also common in E. coli and Salmonella enterica from poultry (Rozwandowicz et al., 2018). Moreover, IncF and IncI plasmids have been reported in association with MDR E. coli strains, mainly ESBL producers, from food animals (Xie et al., 2016). The prevalence of both IncF and IncI plasmids in our isolates, particularly from clams, strengthen the hypothesis that contamination of our clam harvesting areas came from animal sources.
Resistance to multiple antimicrobial classes is common in E. coli (ECDC, 2019) and the prevalence of resistance to β-lactams, tetracycline and aminoglycosides in our clam and sediment strains is in line with earlier reports (Vignaroli et al., 2016; Pormohammad et al., 2019). Notably, antibiotic resistances are frequently associated with conjugative IncF or IncI plasmids (Rozwandowicz et al., 2018). Accordingly, in our study these Inc groups were detected at higher frequency in resistant than in susceptible strains; moreover, sediment strains exhibited a significant association between β-lactam resistance and the presence of IncF plasmids. This Inc group was also associated, besides the IncI group, with the conjugative transfer of tetracycline resistance from E. coli donors of both origin (clams and sediment). These plasmids were probably involved in multidrug resistance, since mating experiments resulted in the transfer of multiple resistance genes. Most MDR strains (∼80%) were positive for the class I integron and the co-transfer of the intI1 gene and of the resistance genes suggest the plasmid location of the integron. In particular, donor E. coli ISZ 220 (from clams) carried an integron with the dfrA1-aadA1 cassette that was probably linked to the ∼110 kb IncI plasmid transferred in mating assays. Plasmids containing this cassette array have been described more frequently in MDR Salmonella and MDR E. coli isolates from meat and food animals than in human isolates (van Essen-Zandbergen et al., 2009; Sunde et al., 2015). In contrast, the dfrA17-aadA5 cassette, which was found in a single clam strain, has typically been reported in isolates of human origin (Povilonis et al., 2010; Musumeci et al., 2012; Sunde et al., 2015). This finding may also be ascribed to contamination of the sampling areas with fecal bacteria mostly of animal origin. Therefore, the presence of a class I integron on conjugative plasmids contributes both to the emergence of MDR strains and to the dissemination of antibiotic resistance.
Conclusion
In conclusion, to the best of our knowledge this is one of the few studies focused on the prevalence of specific Inc group plasmids in E. coli isolates from secondary habitats (like clams and sediments). The PBRT kit, which has been developed for human isolates of Enterobacteriaceae, proved a useful tool to type the plasmids conferring antibiotic resistance on environmental E. coli isolates, to predict their origin and to formulate hypotheses on the contamination source. Moreover, these data could help correlate a plasmid type to strain adaptation and survival strategies outside the host, and provide further information on the spread of antibiotic-resistant plasmid families among Enterobacteriaceae in different settings.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
CV, BC, FA, and FB designed the experiments, analyzed the results, and drafted the manuscript. MM contributed to result interpretations. SS, EC, GM, and NC performed the sampling and the experiments.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by internal funding of Polytechnic University of Marche (Ricerca Scientifica di Ateneo 2019: “Trasferimento in vitro di geni di antibiotico-resistenza da ceppi di E. coli isolati da sedimento marino”).
Acknowledgments
We acknowledge Dr. Francesca Leoni of Istituto Zooprofilattico Sperimentale dell’Umbria e delle Marche at Laboratorio Nazionale di Riferimento (LNR) per il Controllo delle Contaminazioni Batteriche dei Molluschi Bivalvi Vivi (Ancona, Italy) for providing the E. coli strains isolated from clam samples. Part of the results of this study have been presented at the XXXIII SIMGBM Congress, June 2019, Firenze, Italy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01101/full#supplementary-material
Footnotes
References
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42, 68–80. doi: 10.1093/femsre/fux053
Berg, E. S., Wester, A. L., Ahrenfeldt, J., Mo, S. S., Slettemeås, J. S., Steinbakk, M., et al. (2017). Norwegian patients and retail chicken meat share cephalosporin-resistant Escherichia coli and IncK/blaCMY-2 resistance plasmids. Clin. Microbiol. Infect. 23, 407.e9–407.e15. doi: 10.1016/j.cmi.2016.12.035
Bergholz, P. W., Noar, J. D., and Buckley, D. H. (2011). Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl. Environ. Microbiol. 77, 211–219. doi: 10.1128/AEM.01880-10
Broaders, E., Gahan, C. G. M., and Marchesi, J. R. (2013). Mobile genetic elements of the human gastrointestinal tract: potential for spread of antibiotic resistance genes. Gut. Microbes. 4, 271–280. doi: 10.4161/gmic.24627
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carloni, E., Andreoni, F., Omiccioli, E., Villa, L., Magnani, M., and Carattoli, A. (2017). Comparative analysis of the standard PCR-Based Replicon Typing (PBRT) with the commercial PBRT-KIT. Plasmid 90, 10–14. doi: 10.1016/j.plasmid.2017.01.005
Charnock, C., Nordlie, A. L., and Hjeltnes, B. (2014). Toxin production and antibiotic resistances in Escherichia coli isolated from bathing areas along the coastline of the Oslo fjord. Curr. Microbiol. 69, 317–328. doi: 10.1007/s00284-014-0587-7
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Gordon, D. M., Brisse, S., Walk, S. T., and Denamur, E. (2011). Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ. Microbiol. 13, 2468–2477. doi: 10.1111/j.1462-2920.2011.02519.x
Clinical and Laboratory Standards Institute (CLSI) (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement. Wayne, PA: Technical report from the Clinical and Laboratory Standards Institute.
Delaquis, P., Bach, S., and Dinu, L. D. (2007). Behavior of Escherichia coli O157:H7 in leafy vegetables. J. Food Prot. 70, 1966–1974.
Drali, R., Berrazeg, M., Zidouni, L. L., Hamitouche, F., Abbas, A. A., Deriet, A., et al. (2018). Emergence of mcr-1 plasmid-mediated colistin-resistant Escherichia coli isolates from seawater. Sci. Total Environ. 642, 90–94. doi: 10.1016/j.scitotenv.2018.05.387
ECDC (2019). European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe 2018. Stockholm: ECDC, doi: 10.2900/22212
Finley, R. L., Collignon, P., Larsson, D. G., McEwen, S. A., Li, X. Z., Gaze, W. H., et al. (2013). The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710. doi: 10.1093/cid/cit355
Frank, C., Werber, D., Cramer, J. P., Askar, M., Faber, M., der Heiden, M., et al. (2011). Epidemic profile of shiga-toxin–producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365, 1771–1780. doi: 10.1056/NEJMoa1106483
Jang, J., Hur, H. G., Sadowsky, M. J., Byappanahalli, M. N., Yan, T., and Ishii, S. (2017). Environmental Escherichia coli: ecology and public health implications - a review. J. Appl. Microbiol. 123, 570–581. doi: 10.1111/jam.13468
Lambrecht, E., Van Meervenne, E., Boon, N., Van de Wiele, T., Wattiau, P., Hermanet, L., et al. (2018). Characterization of cefotaxime- and ciprofloxacin-resistant commensal Escherichia coli originating from belgian farm animals indicates high antibiotic resistance transfer rates. Microb. Drug Resist. 24, 707–717. doi: 10.1089/mdr.2017.0226
Lazarus, B., Paterson, D. L., Mollinger, J. L., and Rogers, B. A. (2015). Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 60, 439–452. doi: 10.1093/cid/ciu785
Luna, G. M., Vignaroli, C., Rinaldi, C., Pusceddu, A., Nicoletti, L., Gabellini, M., et al. (2010). Extra-intestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 76, 5659–5668. doi: 10.1128/AEM.03138-09
Mathers, A. J., Peirano, G., and Pitout, J. D. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
McDaniels, A. E., Rice, E. W., Reyes, A. L., Johnson, C. H., Haugland, R. A., and Stelma, G. N. Jr. (1996). Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and beta-D-glucuronidase. Appl. Environ. Microbiol. 62, 3350–3354.
Musumeci, R., Rausa, M., Giovannoni, R., Cialdella, A., Bramati, S., Sibra, B., et al. (2012). Prevalence of plasmid-mediated quinolone resistance genes in uropathogenic Escherichia coli isolated in a teaching hospital of northern Italy. Microb. Drug Resist. 18, 33–41. doi: 10.1089/mdr.2010.0146
Pormohammad, A., Nasiri, M. J., and Azimi, T. (2019). Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect. Drug Resist. 12, 1181–1197. doi: 10.2147/IDR.S201324
Povilonis, J., Šeputienë, V., Ružauskas, M., Šiugždinienë, R., Virgailis, M., Pavilonis, A., et al. (2010). Transferable class 1 and 2 integrons in Escherichia coli and Salmonella enterica isolates of human and animal origin in Lithuania. Foodborne Pathog. Dis. 7, 1185–1192. doi: 10.1089/fpd.2010.0536
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., and Røder, B. L. (2018). Escherichia coli Sequence Type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18
Rolain, J. M. (2013). Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 4:173. doi: 10.3389/fmicb.2013.00173
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Sunde, M., Simonsen, G. S., Slettemeås, J. S., Böckerman, I., and Norström, M. (2015). Integron, plasmid and host strain characteristics of Escherichia coli from humans and food included in the Norwegian antimicrobial resistance monitoring programs. PLoS One 10:e0128797. doi: 10.1371/journal.pone.0128797
Tenaillon, O., Skurnik, D., Picard, B., and Denamur, E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8, 207–217. doi: 10.1038/nrmicro2298
van Essen-Zandbergen, A., Smith, H., Veldman, K., and Mevius, D. (2009). In vivo transfer of an incFIB plasmid harbouring a class 1 integron with gene cassettes dfrA1-aadA1. Vet. Microbiol. 137, 402–407. doi: 10.1016/j.vetmic.2009.02.004
Vignaroli, C., Di Sante, L., Leoni, F., Chierichetti, S., Ottaviani, D., Citterio, B., et al. (2016). Multidrug-resistant and epidemic clones of Escherichia coli from natural beds of Venus clam. Food Microbiol. 59, 1–6. doi: 10.1016/j.fm.2016.05.003
Vignaroli, C., Luna, G. M., Pasquaroli, S., Di Cesare, A., Petruzzella, R., Paroncini, P., et al. (2013). Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ. Sci. Technol. 47, 13772–13780. doi: 10.1021/es4019139
Vignaroli, C., Luna, G. M., Rinaldi, C., Di Cesare, A., Danovaro, R., and Biavasco, F. (2012). New sequence types and multidrug resistance among pathogenic Escherichia coli isolates from coastal marine sediments. Appl. Environ. Microbiol. 78, 3916–3922. doi: 10.1128/AEM.07820-11
Vignaroli, C., Zandri, G., Aquilanti, L., Pasquaroli, S., and Biavasco, F. (2011). Multidrug-resistant enterococci in animal meat and faeces and co-transfer of resistance from an Enterococcus durans to a human Enterococcus faecium. Curr. Microbiol. 62, 1438–1447. doi: 10.1007/s00284-011-9880-x
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Keywords: PBRT kit, plasmid, Inc group, replicon, antimicrobial resistance, Escherichia coli
Citation: Citterio B, Andreoni F, Simoni S, Carloni E, Magnani M, Mangiaterra G, Cedraro N, Biavasco F and Vignaroli C (2020) Plasmid Replicon Typing of Antibiotic-Resistant Escherichia coli From Clams and Marine Sediments. Front. Microbiol. 11:1101. doi: 10.3389/fmicb.2020.01101
Received: 30 January 2020; Accepted: 04 May 2020;
Published: 27 May 2020.
Edited by:
Margherita Sosio, Naicons Srl, ItalyReviewed by:
Michael Benedik, Texas A&M University, United StatesJason Patrick Folster, Centers for Disease Control and Prevention (CDC), United States
Copyright © 2020 Citterio, Andreoni, Simoni, Carloni, Magnani, Mangiaterra, Cedraro, Biavasco and Vignaroli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Citterio, YmFyYmFyYS5jaXR0ZXJpb0B1bml1cmIuaXQ=
†These authors have contributed equally to this work
 Barbara Citterio
Barbara Citterio Francesca Andreoni
Francesca Andreoni Serena Simoni
Serena Simoni Elisa Carloni1
Elisa Carloni1 Mauro Magnani
Mauro Magnani Gianmarco Mangiaterra
Gianmarco Mangiaterra Carla Vignaroli
Carla Vignaroli