- Department of Agricultural Chemistry, National Taiwan University, Taipei, Taiwan
The type VI secretion system (T6SS) is a widespread bacterial nanoweapon used for delivery of toxic proteins into cell targets and contributes to virulence, anti-inflammatory processes, and interbacterial competition. In the model phytopathogenic bacterium Pseudomonas syringae pv. tomato (Pst) DC3000, two T6SS gene clusters, HSI-I and HSI-II, were identified, but their functions remain unclear. We previously reported that hcp2, located in HSI-II, is involved in competition with enterobacteria and yeast. Here, we demonstrated that interbacterial competition of Pst DC3000 against several Gram-negative plant-associated bacteria requires mainly HSI-II activity. By means of a systematic approach using in-frame deletion mutants for each gene in the HSI-II cluster, we identified genes indispensable for Hcp2 expression, Hcp2 secretion and interbacterial competition ability. Deletion of PSPTO_5413 only affected growth in interbacterial competition assays but not Hcp2 secretion, which suggests that PSPTO_5413 might be a putative effector. Moreover, PSPTO_5424, encoding a putative σ54-dependent transcriptional regulator, positively regulated the expression of all three operons in HSI-II. Our discovery that the HSI-II gene cluster gives Pst DC3000 the ability to compete with other plant-associated bacteria could help in understanding a possible mechanism of how phytopathogenic bacteria maintain their ecological niches.
Introduction
Natural habitats do not always provide suitable conditions for microorganisms to survive; therefore, bacteria have evolved various secretion systems to adapt to changing environments by translocating effector proteins or DNA into the surrounding milieu or target cells (Costa et al., 2015). The type VI secretion system (T6SS) has been found in more than 25% of sequenced Gram-negative bacteria, and some strains even harbor multiple copies of T6SS-associated genes, which suggests its conserved but specialized roles in bacterial survival (Bingle et al., 2008; Coulthurst, 2019). Genes coding for core components of T6SS are usually clustered together and are considered acquired via horizontal gene transfer (Sarris et al., 2010; Borgeaud et al., 2015; Salomon et al., 2015; Thomas et al., 2017). Functional and structural analyses indicated that T6SS could assemble into a tubular structure similar to a contractile phage tail, in which an Hcp inner tube capped by VgrG/PAAR spike proteins and wrapped by a VipA/B (also known as HsiB/C, or TssB/C) sheath is polymerized on a membrane-associated baseplate complex (Lossi et al., 2013). A conformational change in the baseplate complex stimulates sheath contraction, thus leading to propelling of an Hcp/VgrG/PAAR puncturing device through cell membranes for effector secretion (Basler et al., 2012; Shneider et al., 2013; Zoued et al., 2016). Upon firing, the contracted VipA/B sheath is disassembled by a specialized AAA + ATPase ClpV to recycle VipA/B subunits for assembly of a new T6SS apparatus. The phage tail-like structure is stabilized by a membrane core complex containing TssL, M, and J-like proteins, and this complex could also function as a channel for inner tube passage after sheath contraction (Brunet et al., 2015). To quickly and efficiently respond to the environmental stimuli, T6SS in a subset of bacterial species can be regulated by threonine phosphorylation and posttranslational regulation via serine/threonine kinase PpkA, phosphatase PphA, and a forkhead-associated (FHA) domain-containing protein, Fha (Mougous et al., 2007; Lin et al., 2014).
After the term T6SS was coined in 2006 (Mougous et al., 2006; Pukatzki et al., 2006), the requirement of T6SS for virulence against eukaryotic hosts was described in diverse pathogens (Coulthurst, 2019). However, the contribution of T6SS to interbacterial competitiveness toward various bacterial species via delivery of toxic proteins into target cells highlights its pivotal role in growth fitness in the natural environment containing diverse microbiota (Hood et al., 2010; Hachani et al., 2016; Bernal et al., 2017; Allsopp et al., 2019). T6SS is also involved in limiting colonization and intestinal inflammation to maintain a non-pathogenic, symbiotic relationship of a pathobiont in the gastrointestinal tract within the host (Chow and Mazmanian, 2010), secretion of unknown proteins that impair formation of effective nodules on pea and vetch in Rhizobium leguminosarum (Bladergroen et al., 2003), fostering competence-mediated horizontal gene transfer in Vibrio cholerae (Blokesch, 2015; Borgeaud et al., 2015) and anti-fungal activities (Haapalainen et al., 2012; Marchi et al., 2013; Trunk et al., 2018). In addition to the contact-dependent T6SS activities described above, some T6SSs could function to deliver effectors into the extracellular environments for uptake of particular metal ions such as Zn2+ (Wang et al., 2015; Si et al., 2017), Mn2+ (Si et al., 2017), Fe2+ (Lin et al., 2017), and Cu2+ (Han et al., 2019), which further expands the functions of this versatile and multipurpose nanoweapon.
In addition to the core components involved in apparatus assembly and activation, T6SS functions also rely on secreted proteins, termed T6SS effectors. Secretome, bioinformatics and genetic analyses revealed T6SS effectors such as anti-eukaryotic or anti-bacterial toxins and extracellular metallophores. According to the mode of delivery, T6SS effectors can be divided into cargo and specialized effectors: the former non-covalently associate with specific Hcp, VgrG or a PAAR-containing protein, and the latter contain an effector domain covalently fused to the C-terminal domain of Hcp, VgrG or a PAAR protein (Durand et al., 2014). Binding to the narrow lumen of the inner tube (40Å in diameter) formed by stacked hexameric Hcp rings, Hcp-mediated cargo effectors are usually small (<25 kDa) (Silverman et al., 2013). In certain scenarios, chaperones or adaptors (such as proteins containing DUF1795, DUF2169, or DUF4123 domain) are also required for effector recruitment and secretion by T6SS (Coulthurst, 2019). Genes coding for cargo effectors and cognate chaperones/adaptors are likely to be found in the vicinity of hcp, vgrG, or paar genes (Durand et al., 2014; Ma et al., 2018; Coulthurst, 2019).
Pseudomonas syringae pathovar tomato (Pst) DC3000, a causative agent of tomato speck disease, has become one of the well-known model organisms to study plant–microbe interactions. The pathogenicity and virulence of Pst DC3000 depend on a functional type III secretion system, which delivers effectors into host cells to block the plant defense system or interfere with normal metabolism in planta (Xin and He, 2013). In silico analysis of genomes from six pathovars of P. syringae revealed that Pst DC3000 as well as Pst T1, P. s. pv. tabaci (Psta) ATCC 11528, and P. s. pv. oryzae 1–6 contain two putative T6SS clusters, Hcp secretion island 1 (HSI-I) and HSI-II. The expression of an icmF (tssM) ortholog in HSI-II of Pst DC3000 has been demonstrated (Sarris et al., 2010; Barret et al., 2011). Secretome analysis revealed that Hcp2 encoded in the HSI-II cluster was secreted in an icmF-dependent manner and plays a role in competition ability against enterobacteria and yeast. However, Hcp2 is not involved in Pst DC3000 virulence on tomato and Arabidopsis (Haapalainen et al., 2012). Nevertheless, the biological role of T6SS in phytopathogenic Pst DC3000 remains largely unknown and needs further investigation.
Dissection of T6SS function by using systematical mutagenesis of each gene in the T6SS cluster has been successfully applied in Agrobacterium tumefaciens, Edwardsiella tarda, and V. cholerae (Zheng and Leung, 2007; Zheng et al., 2011; Lin et al., 2013). In this study, to gain a broad view of T6SS activity in Pst DC3000, we first investigated whether HSI-I and HSI-II are both functional in interbacterial competition ability. After redefining the content of the HSI-II cluster, we analyzed mutated genes in this gene cluster. Detection of the hallmark T6SS-secreted protein Hcp2 combined with interbacterial competition assay allowed us to confirm core components and identify a regulator and a putative effector encoded in the HSI-II gene cluster. Our data shed light on understanding the role of T6SS in Pst DC3000 and also provide tools for further dissecting T6SS functions in Pst DC3000.
Materials and Methods
Bacterial Strains, Plasmids and Culture Media
Bacterial strains and plasmids used in this study are in Supplementary Table S1. Pst DC3000 strains were cultured in King’s B medium (KBM) supplemented with appropriate antibiotics or in hrp-inducing minimal media (HMM) substituted with different carbon sources at 28°C. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C. The concentrations of antibiotics used were for gentamicin, 10 μg/mL; kanamycin, 50 μg/mL; rifampicin, 50 μg/mL; and tetracycline, 30 μg/mL.
Construction of Mutations in Pst DC3000
The upstream and downstream sequences of each gene to be deleted were PCR-amplified with the primers listed in Supplementary Table S2. Crossover PCR was then performed to obtain a DNA fragment containing the upstream and downstream sequences for each gene and cloned into the suicide vector pK18mobsac (Schafer et al., 1994). The successful clones were then used to select deletion mutants in a SacB-based procedure as described (Haapalainen et al., 2012). From annotation information provided by the Pseudomonas Genome Database1, ΔHSI-I and ΔHSI-II were constructed by deleting PSPTO_2542 to 2554 and PSPTO_5414 to 5427, respectively.
Construction of Plasmids for GUS Reporter Assay
Two and three promoters were predicted in HSI-I and HSI-II gene clusters, respectively (Supplementary Figure S1). The 500 bp fragments upstream of PSPTO_2542, PSPTO_2539, PSPTO_5427, PSPTO_5434, and PSPTO_5435 amplified with the primers listed in Supplementary Table S2 were cloned into pCHUB78, which contains a promoterless uidA downstream of a multiple cloning site, to obtain pHSI-Ip (PHSI-I:uidA), phcp1p (Phcp1:uidA), pHSI-II-ip (PHSI-II-i:uidA), pHSI-II-iip (PHSI-II-ii:uidA), and pHSI-II-hcp2p (Phcp2:uidA). The resulting plasmids were then introduced into Pst DC3000, which was then grown in KBM from OD600 = 0.1 to 0.8 before cells were collected for 4-methylumbelliferyl-β-D-glucuronide (4-MUG)-based β-glucuronidase (GUS) reporter assay (Gallagher, 1992). The relative GUS activity is represented as a ratio of GUS activity obtained from the tested strain and the corresponding control strain carrying an empty vector.
Interbacterial Competition Assay
To investigate the interbacterial competition ability contributed by HSI-I and HSI-II, a GFP-expressing plasmid was introduced into each competitor bacterial strain indicated in Figure 1. For checking the role of each gene in the HSI-II cluster, E. coli MG1655 and Psph 1448a were chosen as representative competitors. Cells from overnight cultures of Pst DC3000 wild type, its derivatives, and competitor bacterial strains were collected, washed and adjusted to 1 × 107 cfu/mL. Strains of Pst DC3000 were then mixed with each competitor in a 10:1 ratio, except for quantification of Psph 1448a surviving after co-incubation (a 1:1 ratio was used). Ten microliters of each mixture were spotted onto 0.22 μm nitrocellulose membranes placed on KBM plates and incubated for 2 days at 28°C. The GFP signals for each colony were recorded under a fluorescent stereomicroscope (Nikon Instruments Inc., Melville, NY, United States). The number of each bacterial strain in a mixed culture was counted by a method described previously (Hood et al., 2010).
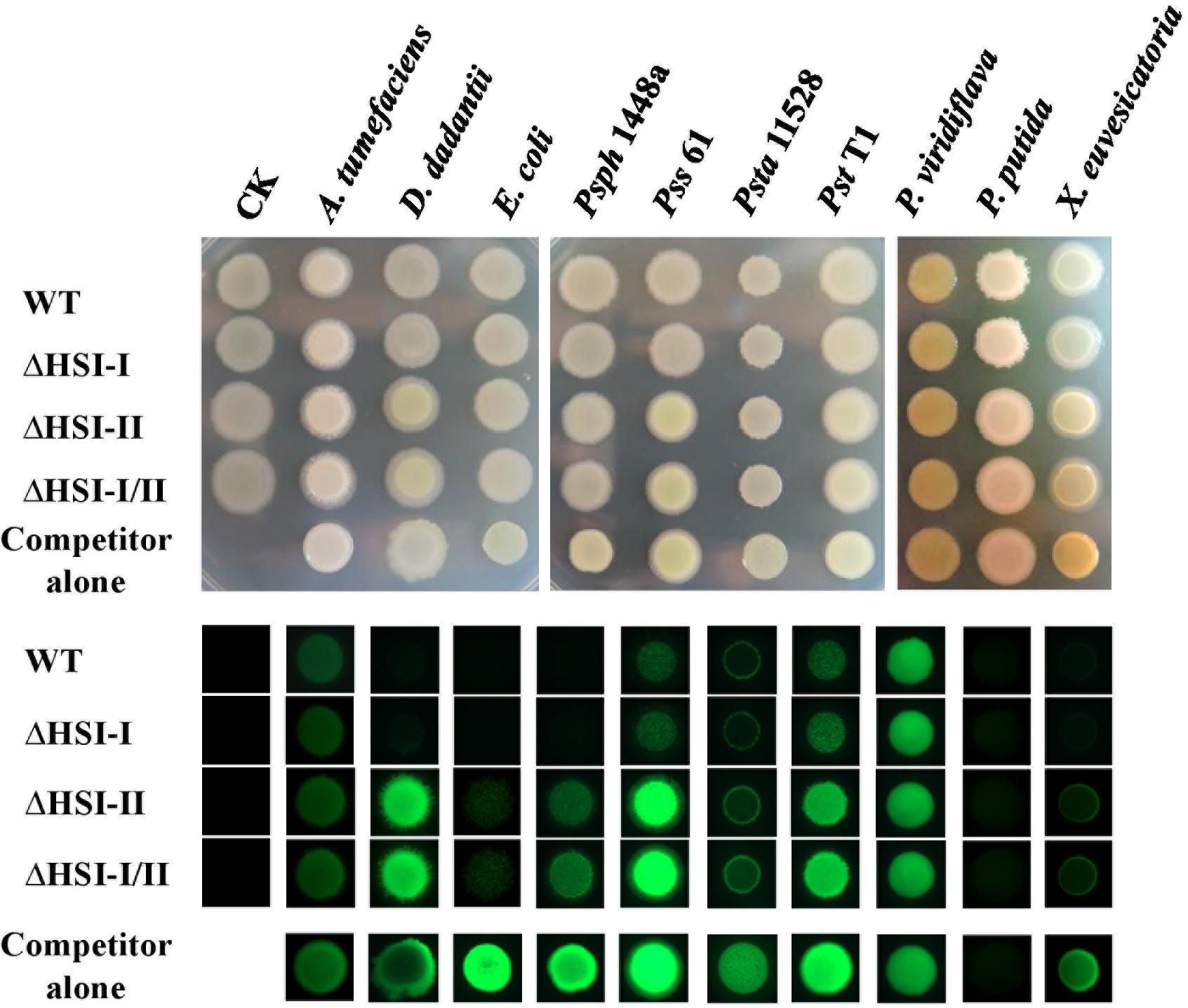
Figure 1. The HSI-II gene cluster is involved in the interbacterial competition ability of Pst DC3000. GFP-expressing competitor bacterial strains, including Agrobacterium tumefaciens C58, Dickeya dadantii CAS9, E. coli MG1655, P. savastanoi pv. phaseolicola (Psph) 1448a, P. syringae pv. syringae (Psy) 61, P. s. pv. tabaci (Psta) 11528, Pst T1, P. viridiflava, P. putida A7, and X. euvescatoria XvT147 were mixed with Pst DC3000 wild type (WT) or its derivative mutants in a 1:1 ratio. A 5 μL amount of the mixed bacterial solution was dropped onto KBM plates, then incubated at 28°C. Colony morphology was photographed 2 days after co-incubation (upper panel), and the GFP signals were detected under a fluorescent microscope (lower panel).
Live/dead Staining of Bacterial Cells
Interbacterial competition assay between Pst DC3000 and E. coli MG1655 was performed as described above. Two days after incubation at 28°C, survival rate of Pst DC3000 and E. coli MG1655 in a mixed (Pst: E. coli = 1:1) or pure culture was determined with a BacLight Bacterial Viability Kit (Invitrogen, Grand Island, NY, United States) as described previously (Hood et al., 2010); survival rate (%) was calculated as [(live cells/total cells) × 100].
Analysis of Secreted Proteins
Overnight cultures of different Pst DC3000 strains were subcultured into fresh KBM at OD600 = 0.1. After growing to OD600 = 0.8 at 28°C, culture media and bacterial cells were separated by centrifugation at 4,000 rpm, and then the supernatants were passed through 0.22 μm syringe filters. Secreted proteins were precipitated from culture media by using 10% trichloroacetic acid and washed with ice-cold acetone twice. The cell pellets and secreted proteins underwent Western blot analysis with anti-Hcp2 and anti-RpoA (RNA polymerase alpha subunit) antisera as primary antibodies and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG as the secondary antibody. Signals of the target proteins were detected by using Immobion Chemiluminescent HRP substrate (Millipore Corp., Bilerica, MA, United States) with a G: BOX system (Syngene, Frederick, MD, United States).
Results
Deletion of the HSI-II but Not HSI-I Gene Cluster Decreased Fluorescence Signals of Several Plant-Associated Gram-Negative Bacteria Harboring GFP Markers
Previous study showed that Pst DC3000 confers growth advantages over E. coli and yeast in a hcp2-dependent manner (Haapalainen et al., 2012). Which T6SS gene cluster (HSI-I and HSI-II) is responsible for this anti-bacterial and anti-yeast activities remains unclear. Moreover, whether T6SS is required for Pst DC3000 to survive in its natural niche full of other plant-associated bacteria and how HSI-I and HSI-II gene clusters are involved in T6SS apparatus assembly and effector delivery in Pst DC3000 remain unknown. To attempt to answer these questions, we first deleted HSI-I and HSI-II gene clusters singly and in combination to examine whether interbacterial competition activities of Pst DC3000 were altered. For a quick screen, a plasmid carrying constitutively expressed green fluorescence protein (gfp) was transformed into the selected plant-associated bacteria, including Agrobacterium tumefaciens C58, Dickeya dadantii CAS9, Pst T1, P. s. pv. syringae (Pss) 61, P. savastanoi pv. phaseolicola (Psph) 1448A, P. s. pv. tabaci (Psta) 11528, P. viridiflava, P. putida A7, and Xanthomonas euvescatoria XvT147 as well as E. coli MG1655. These competitor bacterial strains were then individually mixed with cells of the Pst DC3000 wild type and derived mutants at a 10:1 ratio. Two days after co-incubation on KBM at 28°C, changes in GFP intensity were observed under a fluorescent stereomicroscope. As shown in Figure 1, regardless of strain of Pst DC3000 co-incubated, the colonies of A. tumefaciens C58 or P. viridiflava looked similar under visible or UV light in terms of GFP fluorescence signal. However, the GFP signal intensity was decreased to various degrees when the bacterial competitors D. dadantii CAS9, E. coli MG1655, Psph 1448a, Pss 61, Psta 11528, Pst T1, and X. euvesicatoria XvT147 were co-incubated with Pst DC3000 wild type and ΔHSI-I versus incubation alone or co-incubation with ΔHSI-II and ΔHSI-I/II. Unexpectedly, GFP signal was not detected in P. putida A7; however, smaller colonies could be observed when P. putida A7 was co-incubated with the Pst DC3000 wild type and ΔHSI-I, which suggests their ability to inhibit growth or spreading of P. putida A7 (Figure 1). Therefore, deletion of HSI-II but not HSI-I could decrease the fluorescence signal of various plant-associated bacteria harboring GFP markers, which suggests that HSI-II is responsible for interbacterial competition activity of Pst DC3000 against E. coli and plant-associated bacteria. Further characterization focusing on HSI-II gene cluster should help understand how T6SS confers growth advantages over other Gram-negative bacteria in Pst DC3000.
HSI-II Gene Cluster Is Expressed Under Both Nutrient-Rich and Poor Conditions
The type III secretion system of Pst DC3000 was induced only when cells were grown under a condition mimicking apoplastic growth in planta (i.e., in HMM at 18°C). To investigate how T6SS is regulated in Pst DC3000, each putative promoter in HSI-I and HSI-II clusters was transcriptionally fused to the uidA gene for a GUS reporter assay to monitor the promoter activity under different carbon sources. Consistent with a previous study (Sarris et al., 2010), the two promoters in HSI-I (Figures 2A,B) showed no activity when Pst DC3000 was grown in King’s medium B or HMM supplemented with all carbon sources tested, but three promoters in the HSI-II cluster (Figures 2C–E) were expressed under all tested conditions. The promoter region of the tssB/C-containing operon was expressed approximately fourfold higher than the other two operons (Figure 2D). Although the HSI-II gene cluster appears to be expressed constitutively in both minimal and nutrient-rich media, its expression was modulated by distinct carbon sources with fructose, arabinose and mannitol, in particular, conferring higher expression.
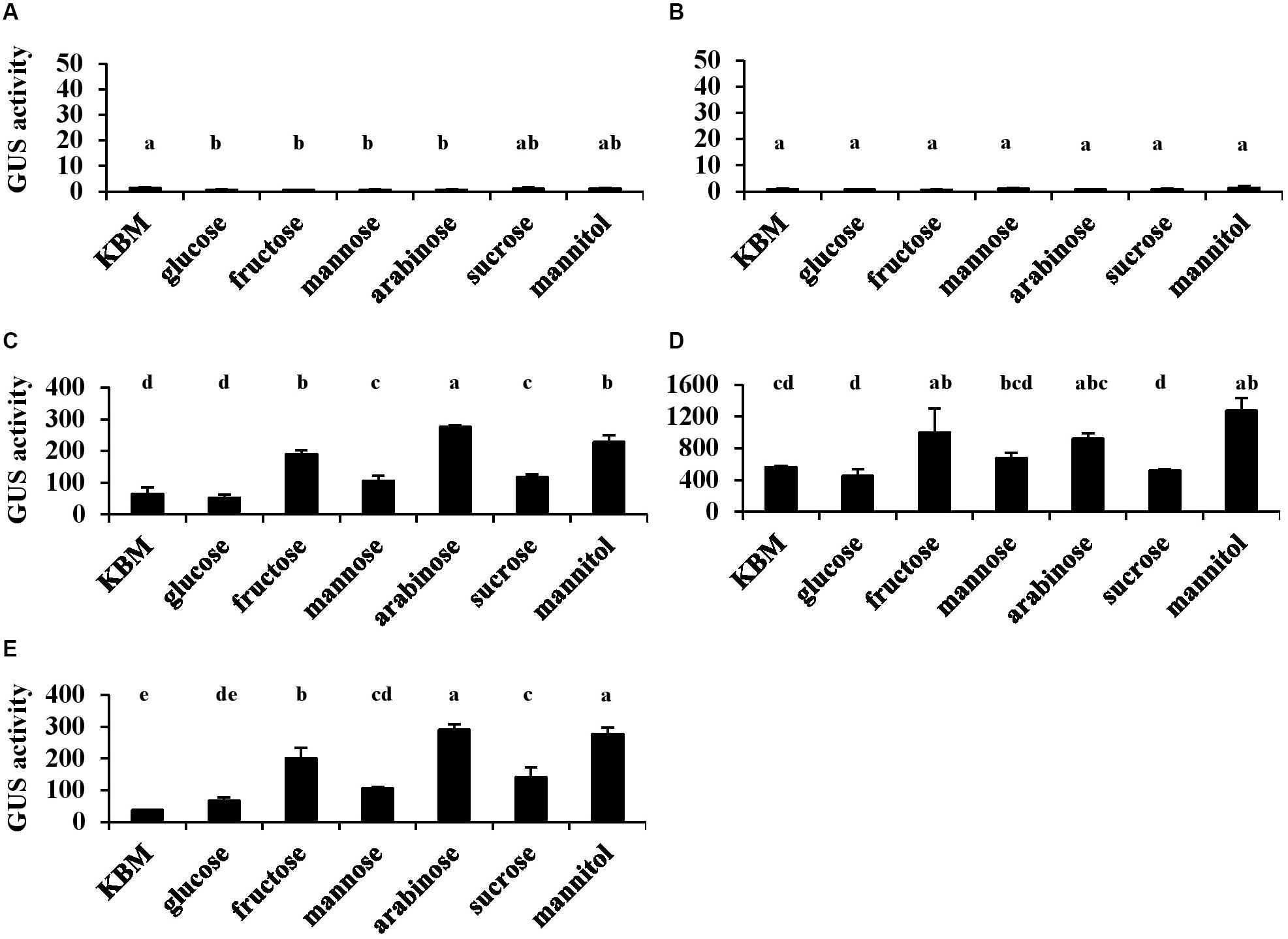
Figure 2. The activities of five putative promoters located in the HSI-I and HSI-II gene clusters of Pst DC3000. Each Pst DC3000 strain carrying uidA controlled by the 500 bp upstream sequence of PSPTO_2542 (A; PHSI-I), PSPTO_2539 (B; Phcp1 in HSI-I), PSPTO_5427 (C; PHSI-II-i), PSPTO_5434 (D; PHSI-II-ii), or PSPTO_5435 (E; Phcp2 in HSI-II) was cultured in King’s B medium (KBM) or hrp-minimal salt supplemented with different carbohydrates as indicated. Fold change of the GUS activity was calculated as described in section “Materials and Methods.” Data are mean ± SD (n = 3). Data analysis involved one-way ANOVA and Tukey’s HSD test. Different letters indicate significant difference between groups (p < 0.05). This experiment was performed three times with similar results.
HSI-II Gene Cluster Contains Three Transcriptional Units
In the Pseudomonas Genome Database, the HSI-II gene cluster was annotated as a region containing 24 genes, from PSPTO_5415 to PSPTO_5438, divided into six transcriptional units: PSTPO_5415-5427, PSPTO_5428, PSPTO_5645-5646, PSPTO_5430-5434, PSPTO_5435, and PSPTO_5436-5438. Information for each gene is provided in Table 1. In addition to using the tss/tag nomenclature system, gene names were also based on H2-T6SS of Pseudomonas aeruginosa PAO1, to which the HSI-II gene cluster is homologous. The putative transposase genes (PSPTO_5411, 5412, 5428, and 5440) were in or flanked this region, which infers possible horizontal gene transfer events (Figure 3A), and thus PSPTO_5413, PSPTO_5414, and PSPTO_5439 may also be part of this cluster. Moreover, the predicted functions of these genes based on use of HHPred and Phyre2 programs also signified their relevance to the T6SS secretion or interbacterial competition function: PSPTO_5413 encodes a putative protein with EF-hand, colicin and peptidase domains; PSPTO_5414 is a putative lipoprotein; and PSPTO_5439 was predicted as an Rhs domain-containing toxin (also summarized in Table 1). To verify the composition of the HSI-II gene cluster, the extent of the transcriptional units in this region was elucidated by using a series of primers to amplify the regions between two predicted transcriptional units (Figure 3A and Supplementary Table S3). When cells were grown in both rich and minimal media, we detected all transcripts except those containing intergenic regions between PSPTO_5412 and PSPTO_5413, and PSPTO_5427, and PSPTO_5646 could not be detected (Figure 3Ba,e). These data indicate that PSPTO_5413, PSPTO_5414, and PSPTO_5439 are indeed part of the HSI-II cluster: PSPTO_5413 and 5414 could be expressed with PSPTO_5415-5427 as an operon, PSPTO_5428 and PSPTO_5645-5646 belong to PSPTO_5430-5434 operon, and PSPTO_5435 and PSPTO_5439 are members of PSPTO_5436-5438 operon. The three major transcriptional units were hereafter named tssM2 (PSPTO_5427 to PSPTO_5413), tssB2/C2 (PSPTO_5434 to PSPTO_5428), and hcp2 (PSPTO_5435 to PSPTO_5439) operons.
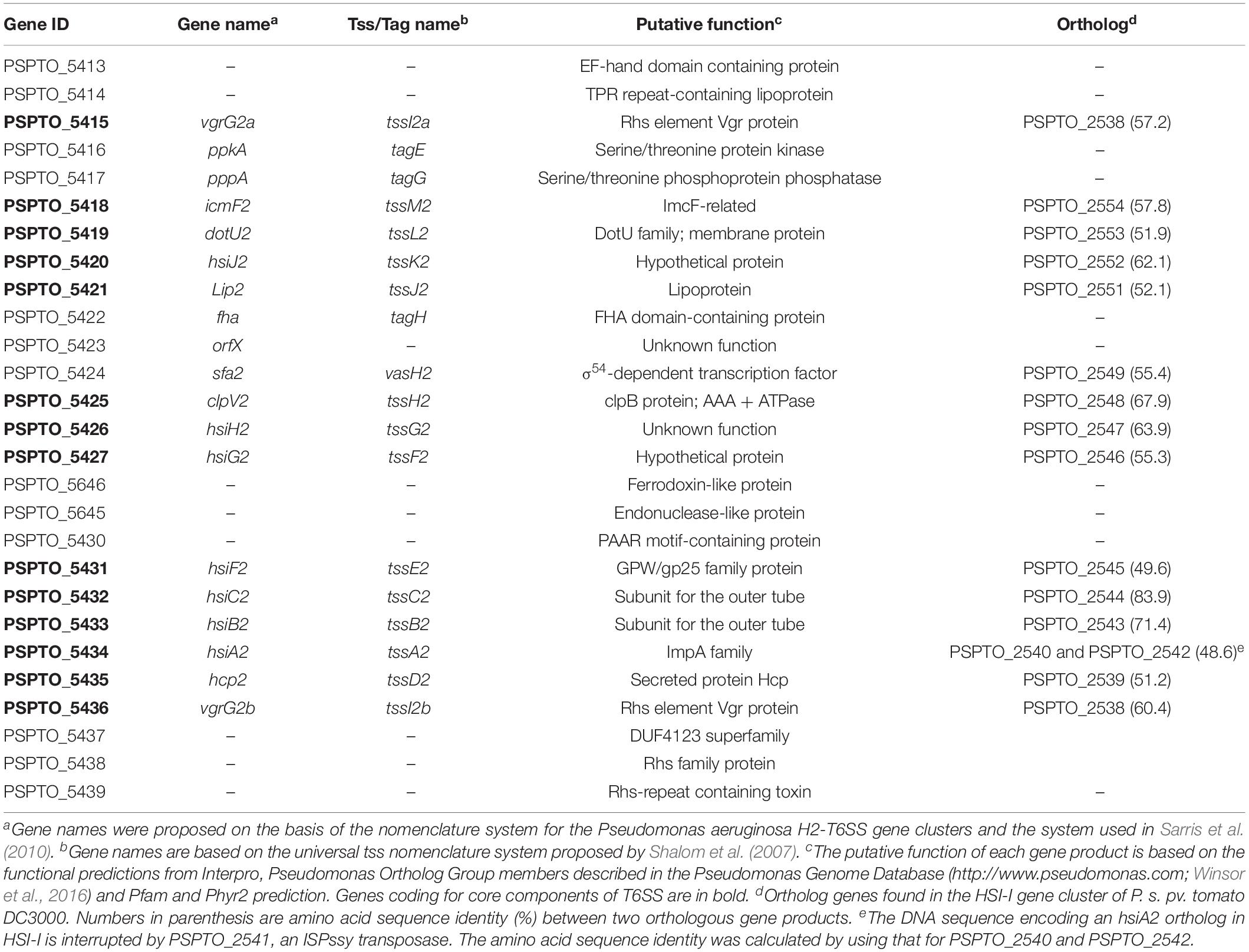
Table 1. Putative function of proteins encoded in the HSI-II gene cluster and their orthologs in the HSI-I gene cluster of Pseudomonas syringae pv. tomato DC3000.
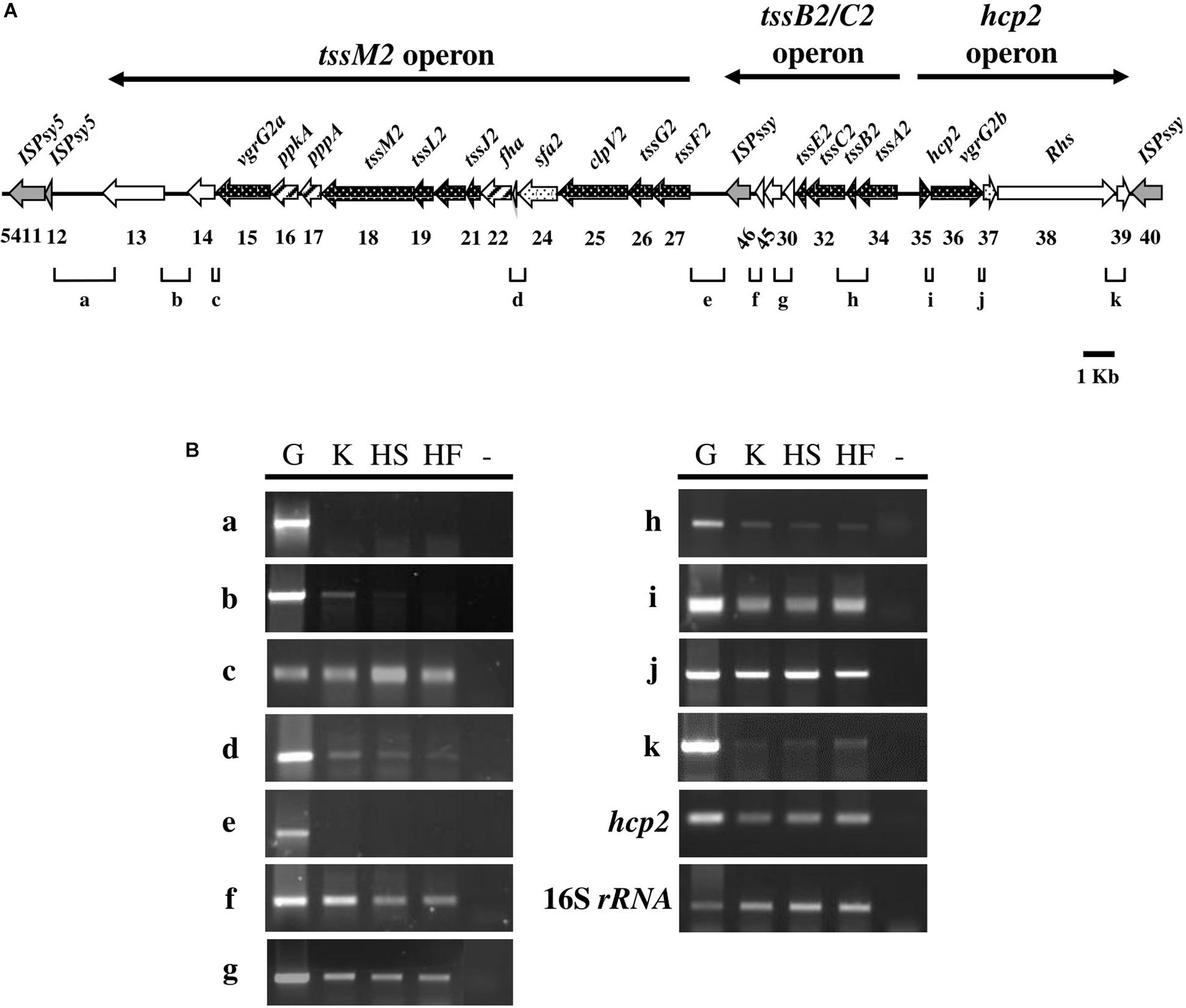
Figure 3. Transcript mapping of the HSI-II gene cluster in Pst DC3000. (A) Schematic diagram exhibiting 27 genes (PSPTO_5413 to PSPTO_5439) of the HSI-II gene cluster and the intergenic regions “a∼k” analyzed in (B). Black arrows indicate open reading frames (ORFs) orthologous to the core T6SS components (names are given above and gene ID numbers are shown below) and genes coding for putative transposases are shaded gray. Dotted arrows represent ORFs whose orthologs can be found in the HSI-I gene cluster. Arrows with diagonal lines represent ORFs orthologous to the regulatory T6SS components. The arrows above the diagram represent the extent of the three transcripts/operons, namely tssM2, tssB2/C2, and hcp2 operons. (B) Semi-quantitative RT-PCR analysis of transcriptional units in the HSI-II gene cluster. Three culture media were used to investigate the expression of each intergenic region. K, KBM; HS, Hrp minimal salt (HMS) supplemented with sucrose; HF, HMS supplemented with fructose. The expression of hcp2 was a positive control and 16S rRNA was an internal control. “G” and “-” indicate genomic DNA and a negative control, respectively. This experiment was repeated using three individual RNA samples with similar results.
Deletion of Most Genes in HSI-II Gene Cluster Changed Hcp2 Secretion
In addition to the genes coding for the core components of T6SS, the putative attributes of other non-conserved genes in the HSI-II gene cluster for T6SS functions are listed in Table 1. To determine whether those genes are critical for T6SS functions, we constructed in-frame deletion mutants for each gene in the HSI-II cluster and obtained single mutants for 26 genes, except PSPTO_5646. Because Hcp2 is the only marker protein of T6SS secretion function detected so far in Pst DC3000 (Haapalainen et al., 2012), Hcp2 secretion was analyzed to assess the effect of these mutants on T6SS secretion. Cellular Hcp2 proteins accumulated in most of the mutants, except no or few Hcp signals were detected in cell pellets of Δ5435 (Δhcp2), Δ5424 with deletion of sfa2 encoding a σ54-dependent transcriptional regulator, and Δ5427 with deletion of tssF2 (Figures 4A,B, “Cell Pellet”). Regardless of normal Hcp2 levels inside the cells, deletion of genes coding for core components affected the ability of Pst DC3000 to secrete Hcp2. Although both PSPTO_5415 and PSPTO_5436 encode VgrG homologs, only deletion of PSPTO_5436 resulted in loss of Pst DC3000 ability to secrete Hcp2. Deletion of PSPTO_5416 and 5422, encoding the regulatory components PpkA and Fha, respectively, also impaired Hcp2 secretion. However, repression of Hcp2 secretion by PSPTO_5417, coding for a PppA ortholog, was not observed in Pst DC3000 (Mougous et al., 2007; Fritsch et al., 2013; Lin et al., 2014). In addition to the core structural genes (tssA2-M2) and positive regulatory genes (ppkA and fha), Δ5423, representing deletion of an accessory gene orfX with unknown function, also impaired Hcp2 secretion (Figures 4A,B, “Secreted protein”).
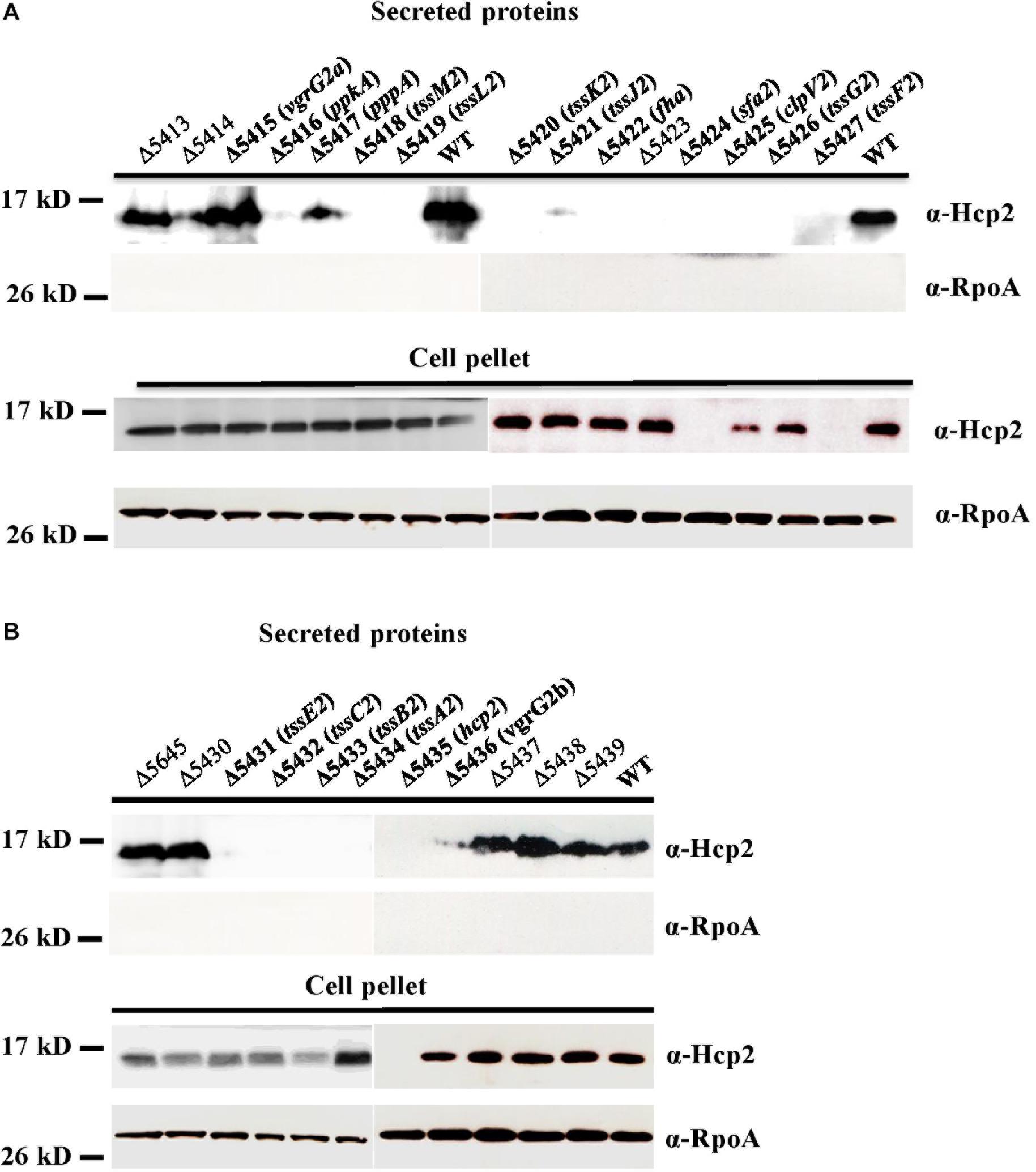
Figure 4. Secretion assay differentiates the role of each gene in the HSI-II gene cluster. Pst DC3000 wild type and mutants with deletion of each gene in the tssM2 operon (A) and in the tssB2/C2 and hcp2 operons (B) were grown in KBM at 28°C from OD600 0.1 to 0.8. Western blot analysis was used to detect Hcp2 in the culture media (secreted proteins) and cell pellets with anti-Hcp2 antiserum. RpoA was an internal control for cytoplasmic proteins (equal loading) and a negative control for secreted proteins.
Deletion of Most Genes in the HSI-II Gene Cluster Reduced the Interbacterial Competition Ability of Pst DC3000
Deletion of most genes in the tssM2 operon (PSPTO_5414 to PSPTO_5427) reduced the ability of Pst DC3000 to compete against several plant-associated Gram-negative bacteria (Figure 1). This phenomenon was also observed when the hcp2 operon was deleted, so the contribution of the tssM2 and hcp2 operons to interbacterial competition was similar on the basis of bacterial counts after co-incubation (Supplementary Figure S2). Among the Gram-bacteria tested in Figure 1, Psph 1448a and E. coli MG1655 were selected as representatives for competitors with and without functional T6SS, respectively, and used to determine the interbacterial competition ability of each single mutant of the HSI-II gene cluster. Pst DC3000 growth did not differ when mutants were co-incubated with Psph 1448a or E. coli MG1655 or incubated alone (Supplementary Figure S3). However, all mutants found impaired in Hcp secretion lost or reduced their ability to cause growth inhibition of the competitors, whereas the mutants with slightly reduced or wild-type levels of Hcp2 secretion (Δ5414, Δ5417, Δ5645, Δ5430, Δ5437, Δ5438, and Δ5439) retained the same interbacterial competition activity (Figure 5). Of note, deletion of PSPTO_5415 reduced its ability to antagonize Psph 1448a but retained the wild-type antibacterial activity to E. coli MG1655 (Figures 5A,B). Hence, mutants with malfunction in Hcp2 secretion did not always alter their ability to inhibit growth of E. coli MG1655 or Psph 1448a. Furthermore, PSPTO_5413, which is not required for Hcp2 secretion, was critical for the competition ability of Pst DC3000, which suggests that PSPTO_5413 may be a T6SS effector targeting competitors for growth fitness.
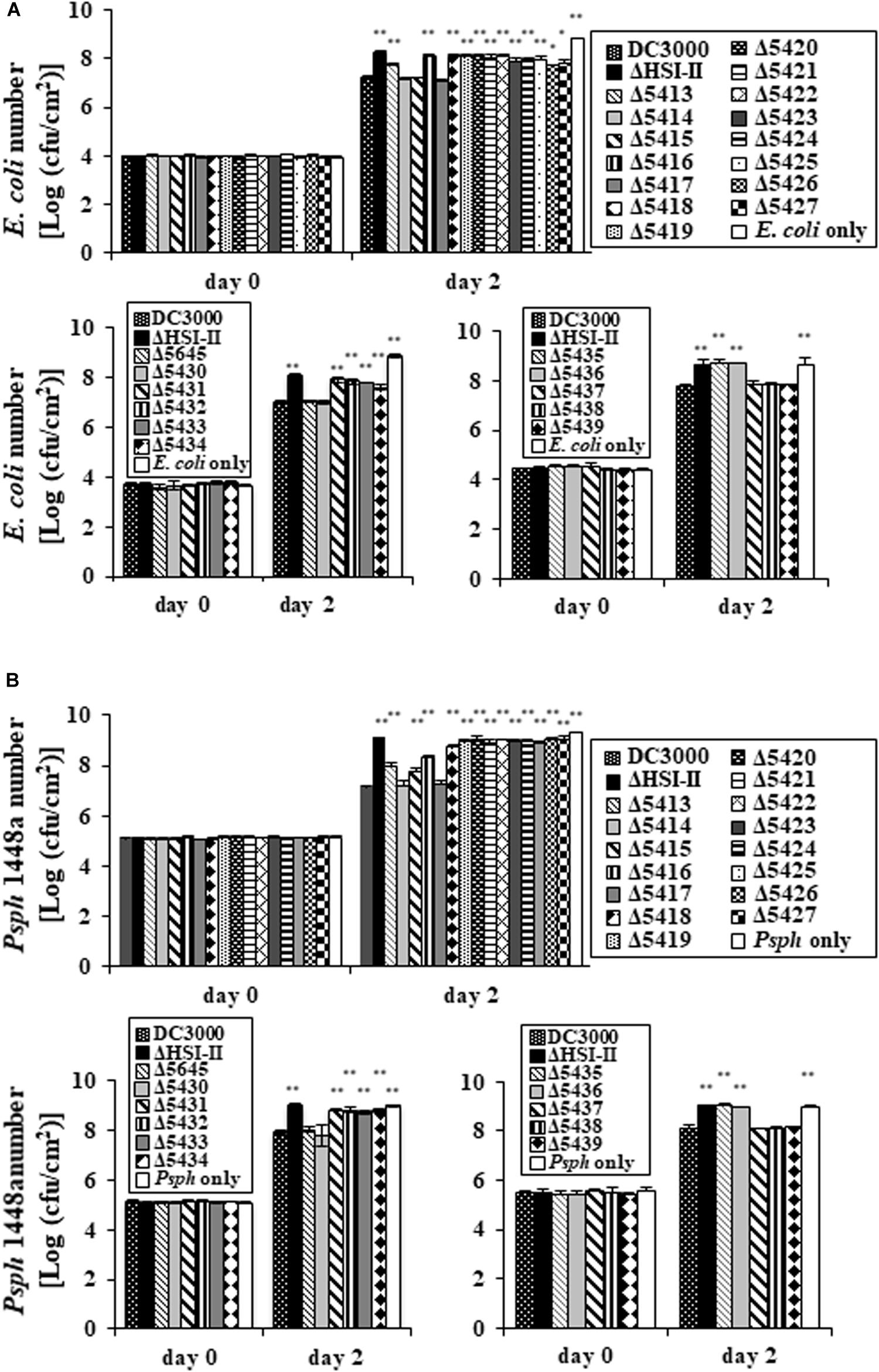
Figure 5. Deletion of each gene in the HSI-II gene cluster affects the interbacterial competition activity of Pst DC3000. After co-incubation with GFP-expressing E. coli MG1655 or Psph 1448a for 2 days, the competition ability of each mutant was evaluated by calculating the bacterial number of E. coli (A) and Psph 1448a (B). “E. coli only” and “Psph only” indicate bacterial numbers without co-incubation with any Pst DC3000 strain. Data analysis involved Student’s t-test. Data are mean ± SD (n = 3) *P < 0.05; **P < 0.01 compared with the wild type. This experiment was conducted three times with similar results.
HSI-II–Reduced Competitor Growth Is a Cell Contact-Dependent and Bacteriostatic Effect
T6SS-mediated interbacterial competition is likely mediated in a cell contact-dependent manner as experimentally demonstrated in Aeromonas hydrophila, Burkholderia thailandensis, P. aeruginosa, and V. cholerae (Russell et al., 2014). To determine whether the observed interbacterial competition activity of Pst DC3000 requires cell–cell contact, with a competition assay, we placed a 0.22 μm nitrocellulose membrane between E. coli MG1655 and Pst DC3000. When E. coli and Pst DC3000 were mixed together without a membrane, HSI-II-dependent inhibition of E. coli MG1655 growth was detected (Figure 6A, “Contact”). In contrast, this HSI-II-dependent reduction of E. coli cell count was not detectable when a membrane was placed between E. coli MG1655 and Pst DC3000 (Figure 6A, “No contact”). The same results were obtained when different competing bacteria were tested (data not shown). Also, E. coli growth did not decrease much, which suggests that the HSI-II-mediated reduction of competitor growth may be a weak bacteriocidal or a bacteriostatic effect. To answer this question, we calculated the survival rates of both Pst DC3000 and E. coli MG1655 during co-inoculation by using a BacLight Bacterial Viability Kit. Survival rates of Pst DC3000 (Figure 6B) and E. coli MG1655 (Figure 6C) did not differ when the two bacteria were co-incubated with each other or incubated alone. Thus, cell-to-cell contact was required for HSI-II–mediated toxicity of Pst DC3000, and this interbacterial competition activity was a bacteriostatic effect.
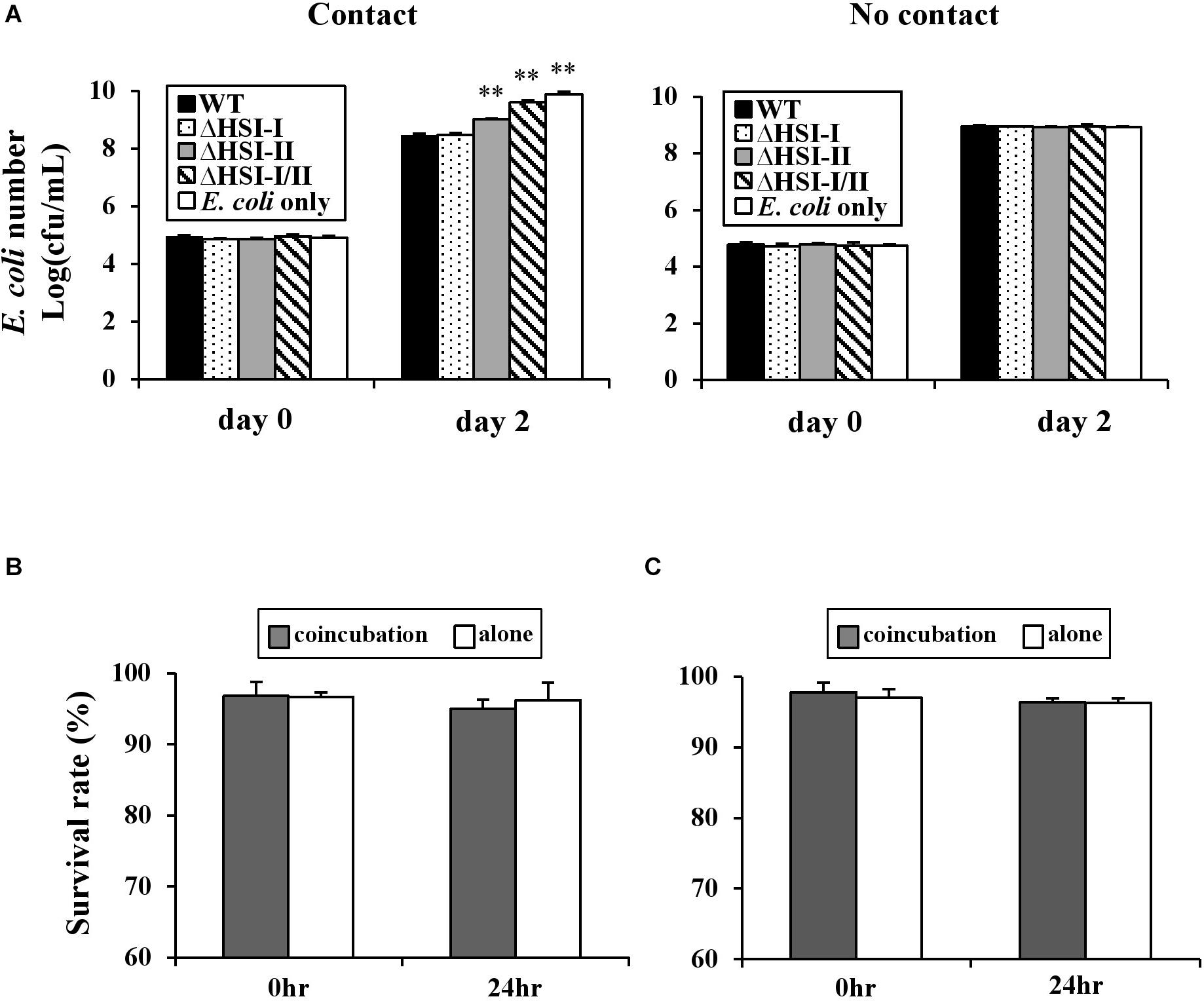
Figure 6. HSI-II-mediated antibacterial toxicity depends on cell contact and is a bacteriostatic effect. (A) The GFP-labeled Pst DC3000 and its derivative mutants were co-incubated with (contact) or separated from the test strain E. coli MG1655 by a 0.22 μm filter membrane (no contact). Two days after incubation at 28°C, E. coli numbers in each treatment were measured. “E. coli only” indicates bacterial number without co-incubation with any Pst DC3000 strain. Survival rate of Pst DC3000 (B) and E. coli K-12 MG1655 (C) in a mixed (Pst: E. coli = 10:1) or pure culture calculated with a BacLight Bacterial Viability Kit. Data are mean ± SD (n = 3) and were analyzed with Student’s t-test. ∗∗P < 0.01 compared with the wild type. This experiment was conducted three times with similar results.
PSPTO_5424 Encodes a Transcriptional Regulator Controlling the Expression of the HSI-II Gene Cluster
PSPTO_5424 was annotated as a Sfa ortholog, a putative σ54-dependent transcriptional regulator, and could control T6SS gene expression in several strains of V. cholorae and P. aeruginosa PAO1 (Pukatzki et al., 2006; Sana et al., 2013). Data in Figure 4 implied that Sfa2 mediates the expression of Hcp2; however, whether the other two operons in the HSI-II gene cluster are also regulated by Sfa2 remain uncharacterized. Thus, we used GUS reporter assay to check the promoter activities of tssM2 and tssB2/C2 operons in the presence or absence of Sfa2. Similar to the hcp2 operon, the promoter activities of these two operons were decreased in Δsfa2 as compared with the wild type, which indicates that Sfa2 not only affects the expression of hcp2 but also participates in regulating the other two operons in the HSI-II gene cluster (Figure 7).
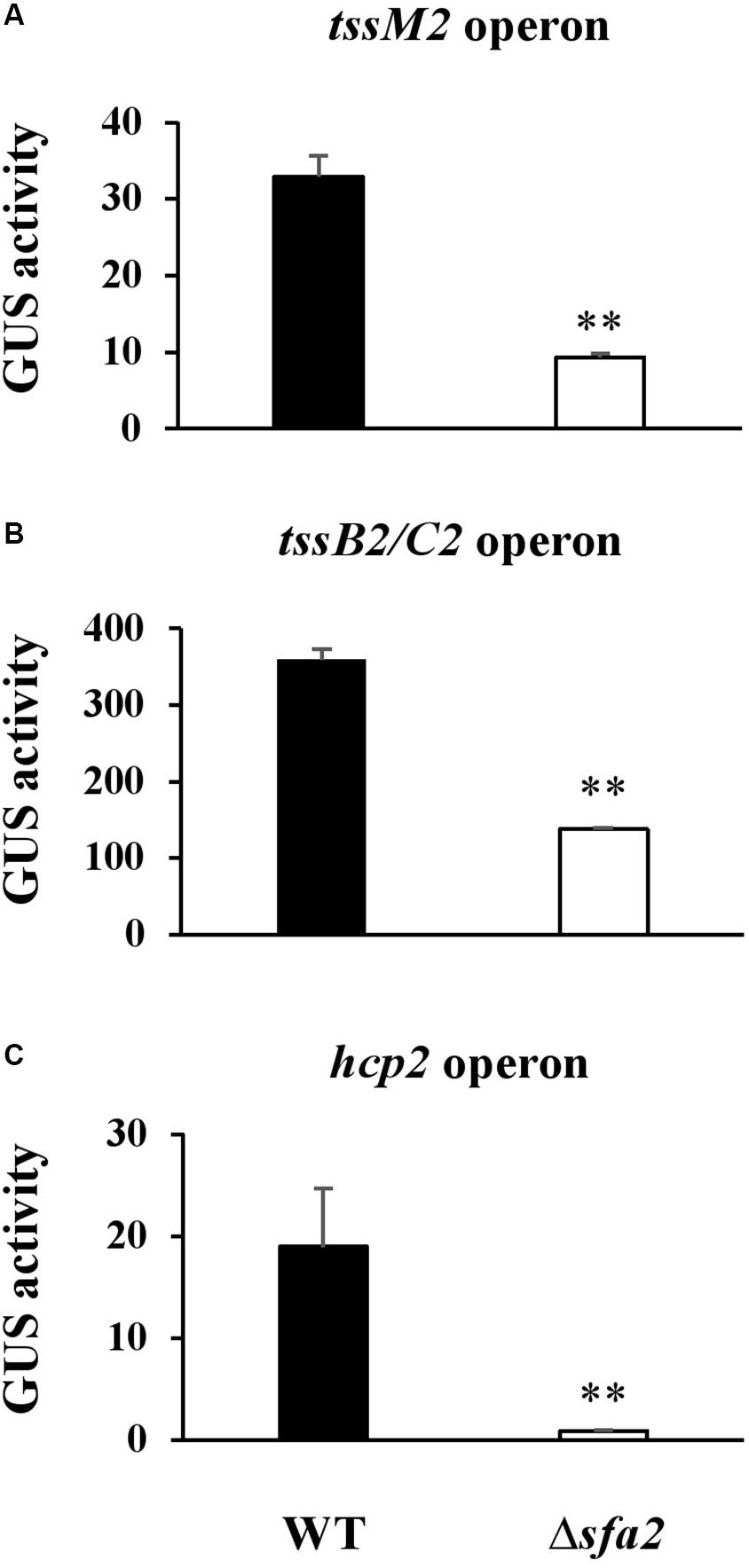
Figure 7. Sfa2 positively regulates the expression of three operons in the HSI-II gene cluster. Pst DC3000 or Δ5424 (Sfa2) carrying uidA controlled by the 500 bp upstream sequence of (A) tssM2, (B) tssB2/C2 or (C) hcp2 operon were cultured in KBM from OD600 = 0.1 to 0.8 before bacterial cells were collected for determining GUS activity. Fold change of the GUS activity was calculated as described in section “Materials and Methods.” Data are mean ± SD (n = 3) ∗∗P < 0.01 compared with the wild type. This experiment was conducted three times with similar results.
Discussion
As an archetypical phytopathogenic bacterium, Pst DC3000 has been studied extensively in terms of its biology, ecology and evolution. Genomic analyses of T6SS loci in all Pseudomonas spp. sequenced to date and from many divergent bacterial strains revealed several unique characteristics of T6SS in Pst DC3000: (1) it is one of the two strains (i.e., Pst DC3000 and P. putida KT2440) with two copies of a similar T6SS cluster (cluster 1.1 in Barret et al., 2011); (2) the HSI-II gene cluster contains a complete set of T6SS core and regulatory components, whereas HSI-I lacks genes encoding Fha, PphA, and PpkA; (3) the gene coding for a core component, HsiA/TssA, is interrupted by a transposase in the HSI-I gene cluster, probably leading to loss-of-function of this locus; (4) unlike the other P. syringae strains, Pst DC3000 does not possess the cluster 4B T6SS locus (Sarris et al., 2010; Barret et al., 2011). In this study, we demonstrated that the HSI-II cluster of Pst DC3000 encodes a functional T6SS conferring secretion and interbacterial competition activities. In contrast, no HSI-I activity could be detected in all assays performed, which again shows that HSI-I is inoperative. Although the HSI-II gene cluster is phylogenetically related to H2-T6SS of P. aeruginosa PAO1 (Bingle et al., 2008; Boyer et al., 2009), it does not contribute to virulence (Haapalainen et al., 2012), only interbacterial competition.
Unlike the type III secretion system of Pst DC3000, which is activated only in planta or under conditions mimicking apoplastic fluid (Huynh et al., 1989; Rico and Preston, 2008), all three operons in the HSI-II gene cluster were expressed constitutively in both rich and minimal media, although different carbon sources could affect their expression level. T6SS-mediated antibacterial toxicity may allow pathogens to secure their niches by competing with other bacteria in the environment or during infection, and different expression patterns of T6SS may reflect strategies of how to use the T6SS apparatus. For instance, V. cholerae uses T6SS as an offensive weapon, so constitutive expression of T6SS is usually prevalent in the environmental isolates, which are frequently exposed to the competitors or predators (Unterweger et al., 2012). However, T6SS serves as a defensive tool in P. aeruginosa, which detects the threat from other T6SS+ bacteria, and thus T6SS is stimulated when T6SS dueling occurs (Basler et al., 2013). Nevertheless, attempts to evaluate the growth fitness of the Pst DC3000 wild type and ΔHSI-II mutant using field soils containing abundant microorganisms was not successful because very low rates of both Pst DC3000 and its T6SS mutant were recovered from field soils after incubation for only 1 day (data not shown). In fact, records have linked higher survival of Pst to tomato seeds and host debris but not field soils (McCarter et al., 1983). With the weak performance of the HSI-II-mediated bacteriostatic effect (demonstrated by less reduction of competitor numbers in Figure 5), the continuous presence of other microorganisms during its life cycle likely puts much selective pressure on Pst DC3000, leading to its evolution into an entity constitutively expressing T6SS to maintain its population in the environment before host plants are present. The antibacterial activity is so weak that HSI-II–mediated growth advantage could only be detected when the circumstance is not as complicated as field soils or the bacterial concentration is higher, namely in a competition assay, seeds, or host debris.
T6SS effectors responsible for antibacterial activity have been identified in many bacteria, and many are encoded within the T6SS loci, which implies that their secretion strongly depends on the associated T6SS apparatus (Miyata et al., 2011; Russell et al., 2012; Ma et al., 2014; Whitney et al., 2014; Yang et al., 2015). Bioinformatics analysis predicted PSPTO_5413, PSPTO_5438, and PSPTO_5439 as VgrG-associated effectors (Hachani et al., 2014; Whitney et al., 2014; Liang et al., 2015), whereas PSPTO_5645/5646 could be effector/immunity pairs linked to the PAAR protein (PSPTO_5430) (Table 1). The antibacterial toxins are usually produced concurrently with their cognate immunity proteins to avoid self-intoxication (Russell et al., 2012). Thus, genes coding for immunity proteins are essential for protection against T6SS-dependent attack of neighboring sister cells, and mutations in immunity genes could be lethal in T6SS+ strains (Dong et al., 2013). That we could not obtain mutants of PSPTO_5646 also suggests that it could be the cognate immunity protein of PSPTO_5645. However, deletion of PSPTO_5645 has no effect on interbacterial competition ability, which suggests that PSPTO_5645 may not function as a potent antibacterial toxin or the chosen competitors (E. coli and Psph) are not the targets of PSPTO_5645.
Among four putative effector genes identified in this study, only deletion of PSPTO_5413 caused reduced interbacterial competition activity against E. coli and Psph. The evidence that PSPTO_5413 plays no role in mediating Hcp2 secretion is consistent with its accessory role not related to T6SS assembly. However, constitutively expressing PSPTO_5413 in E. coli MG1655 or Psph1448a did not reveal growth inhibition (data not shown). The lack of toxicity in the target cells may suggest that additional factors(s) from Pst DC3000 may be required for toxicity of PSPTO_5413, or subcellular location of ectopically expressed PSPTO_5413 may not be the site of its mode of action. Analysis of the amino acid sequence of PSPTO_5413 revealed the presence of an EF-hand motif and a domain highly similar to the catalytic domain of the antimicrobial peptidase lysostaphin. EF-hand motifs can be found in calcium-binding proteins, acting in Ca2+ homeostasis or calcium signaling (Yanez et al., 2012). Lysostaphin is a glycylglycine endopeptidase identified in Staphylococcus simulans, which can cleave the peptide bond between the third and fourth glycine residues of the pentagycine cross-link in the cell wall of S. aureus (Sabala et al., 2014). Despite limited information on the roles of EF-hand calcium-binding in prokaryotes (Michiels et al., 2002; Dominguez et al., 2015), the structural and biochemical characteristics of the lysin LysGH15 from Staphylococcal phage GH15 shed light on the possible mechanism underlying the toxic effect of PSPTO_5413 (Gu et al., 2014). PSPTO_5413 is likely a cell wall-targeting effector, whose activity is modulated dependent on calcium concentration via the function of the EF-hand motif. Because of all these characteristics and the presence of PSPTO_5413 orthologs in only a few Pseudomonas spp., this gene is unique and worthy of further dissection to clarify how PSPTO_5413 is involved in the interbacterial competition activity of Pst DC3000 and whether PSPTO_5414, coding for a lipoprotein, might be its cognate immunity protein.
In searching for putative functions of each gene in the HSI-II gene cluster, we identified proteins containing DUF4123 (PSPTO_5437) and recombination hotspot (Rhs) domains (PSPTO_5438 and 5439) (Table 1). DUF4123, together with DUF1759 and DUF2169, has been characterized as a domain associated with the chaperone/adaptor proteins of the T6SS cargo effectors and is required for effector delivery as well as the bacterial killing activity (Unterweger et al., 2015; Bondage et al., 2016; Cherrak et al., 2019). This conserved domain has been used to identify some novel T6SS effectors (Liang et al., 2015; Zepeda-Rivera et al., 2018). However, E. coli MG1655 or Psph 1448a co-incubated with Δ5437, Δ5438, and Δ5439 did not reduce bacterial growth, which suggests that these three genes do not participate in a growth advantage of Pst DC3000 over E. coli MG1655 or Psph 1448a (Figure 5). Besides PSPTO_5437, three DUF4123-containing proteins in Pst DC3000 are PSPTO_2537, PSPTO_3483, and PSPTO_3850. PSPTO_2537 is located in the HSI-I gene cluster, whose expression cannot be detected thus far. PSPTO_3483 and PSPTO_3850 are both located in an orphan VgrG island. Of note, genes coding for putative toxins are also present downstream of PSPTO_3483 and PSPTO_3850. Thus, future work will focus on understanding whether these proteins are involved in growth fitness of Pst DC3000 and can be uploaded onto cognate VgrG with the help of a DUF4123-containing chaperone encoded in the same island for successful delivery by T6SS.
Several reports have shown that the expression of genes coding for T6SS apparatus can be modulated by the σ54-dependent transcriptional regulators encoded within T6SS loci. In P. aeruginosa, Sfa2 negatively regulates H2-T6SS expression, but H3-T6SS expression is Sfa3-independent (Sana et al., 2013). In V. cholerae and A. hydrophila, the σ54-dependent transcriptional regulator VasH positively regulates the expression of hcp genes (Pukatzki et al., 2006; Suarez et al., 2008). Although the HSI-II gene cluster is phylogenetically related to H2-T6SS of P. aeruginosa, the regulatory activity of Sfa2 (PSPTO_5424) in Pst DC3000 is opposite to that of Sfa2 in P. aeruginosa. Discovery of effector proteins is important for understanding the mechanism underlying T6SS functions. Use of a vasH mutant expressing VasH under the control of an arabinose-inducible promoter revealed an effector VasX encoded outside the T6SS locus (Miyata et al., 2011). Because Pst DC3000 and V. cholerae both positively control the expression of T6SS by a σ54-dependent transcriptional regulator, the same strategy may be applicable for discovery of additional T6SS effectors in Pst DC3000.
Using a systematic mutagenesis approach, we identified genes located in the HSI-II cluster that are required for T6SS activities, including accumulation or secretion of Hcp2, and interbacterial competition (summarized in Supplementary Table S4). Although more work is required to elucidate the effector functions and the mechanisms underlying HSI-II–mediated interbacterial competition, this study identified the regulatory elements and genes responsible for expression, assembly, and function of T6SS-mediated interbacterial of Pst DC3000. From this and previous studies, we now know that instead of providing virulence activity, HSI-II-T6SS is likely used as a defensive weapon to protect Pst DC3000 against bacteria sharing the same ecological niches to ensure sufficient inoculum for successful infection when encountering appropriate host plants. At present, secretome and transcriptome analyses are undergoing, which should help us identify the effectors and regulatory components of HSI-II-T6SS for better understanding the mechanistic and biological functions of HSI-II in Pst DC3000.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
C-FC and N-CL conceived and designed the experiments. C-FC, C-YL, Y-YL, Y-HS, K-YC, and N-CL performed the experiments and analyzed the data. C-FC and N-CL wrote the manuscript.
Funding
This work was financially supported by the Ministry of Science and Technology (Grant Nos. 101-2313-B-002-058 and 108-2321-B-002-023) and National Taiwan University (109L891304), Taiwan, R.O.C.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Wen-Ling Deng for her kindness to share several phytopathogenic pathogens isolated in Taiwan with us. We also like to thank Dr. Erh-Min Lai for giving us critical comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01118/full#supplementary-material
Footnotes
References
Allsopp, L. P., Bernal, P., Nolan, L. M., and Filloux, A. (2019). Causalities of war: the connection between type VI secretion system and microbiota. Cell. Microbiol. 22:e13153. doi: 10.1111/cmi.13153
Barret, M., Egan, F., Fargier, E., Morrissey, J. P., and O’gara, F. (2011). Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology 157, 1726–1739. doi: 10.1099/mic.0.048645-0
Basler, M., Ho, B. T., and Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Blokesch, M. (2015). Competence-induced type VI secretion might foster intestinal colonization by Vibrio cholerae: intestinal interbacterial killing by competence-induced V. cholerae. Bioessays 37, 1163–1168. doi: 10.1002/bies.201500101
Bondage, D. D., Lin, J. S., Ma, L. S., Kuo, C. H., and Lai, E. M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940. doi: 10.1073/pnas.1600428113
Borgeaud, S., Metzger, L. C., Scrignari, T., and Blokesch, M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67. doi: 10.1126/science.1260064
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Brunet, Y. R., Zoued, A., Boyer, F., Douzi, B., and Cascales, E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11:e1005545. doi: 10.1371/journal.pgen.1005545
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L., and Cascales, E. (2019). Structure and activity of the type VI secretion system. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.PSIB-0031-2019
Chow, J., and Mazmanian, S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276. doi: 10.1016/j.chom.2010.03.004
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Coulthurst, S. (2019). The type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. doi: 10.1099/mic.0.000789
Dominguez, D. C., Guragain, M., and Patrauchan, M. (2015). Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 57, 151–165. doi: 10.1016/j.ceca.2014.12.006
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Fritsch, M. J., Trunk, K., Diniz, J. A., Guo, M., Trost, M., and Coulthurst, S. J. (2013). Proteomic identification of novel secreted antibacterial toxins of the Serratia marcescens type VI secretion system. Mol. Cell. Proteomics 12, 2735–2749. doi: 10.1074/mcp.M113.030502
Gallagher, S. R. (1992). GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. San Diego, CA: Academic Press.
Gu, J., Feng, Y., Feng, X., Sun, C., Lei, L., Ding, W., et al. (2014). Structural and biochemical characterization reveals LysGH15 as an unprecedented “EF-hand-like” calcium-binding phage lysin. PLoS Pathog. 10:e1004109. doi: 10.1371/journal.ppat.1004109
Haapalainen, M., Mosorin, H., Dorati, F., Wu, R. F., Roine, E., Taira, S., et al. (2012). Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194, 4810–4822. doi: 10.1128/JB.00611-12
Hachani, A., Allsopp, L. P., Oduko, Y., and Filloux, A. (2014). The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289, 17872–17884. doi: 10.1074/jbc.M114.563429
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Han, Y., Wang, T., Chen, G., Pu, Q., Liu, Q., Zhang, Y., et al. (2019). A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 15:e1008198. doi: 10.1371/journal.ppat.1008198
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Huynh, T. V., Dahlbeck, D., and Staskawicz, B. J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245, 1374–1377. doi: 10.1126/science.2781284
Liang, X., Moore, R., Wilton, M., Wong, M. J., Lam, L., and Dong, T. G. (2015). Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. U.S.A. 112, 9106–9111. doi: 10.1073/pnas.1505317112
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Lin, J. S., Ma, L. S., and Lai, E. M. (2013). Systematic dissection of the Agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One 8:e67647. doi: 10.1371/journal.pone.0067647
Lin, J. S., Wu, H. H., Hsu, P. H., Ma, L. S., Pang, Y. Y., Tsai, M. D., et al. (2014). Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens. PLoS Pathog. 10:e1003991. doi: 10.1371/journal.ppat.1003991
Lossi, N. S., Manoli, E., Forster, A., Dajani, R., Pape, T., Freemont, P., et al. (2013). The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 288, 7536–7548. doi: 10.1074/jbc.M112.439273
Ma, J., Sun, M., Pan, Z., Lu, C., and Yao, H. (2018). Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochim. Biophys. Acta Gen. Subj. 1862, 1635–1643. doi: 10.1016/j.bbagen.2018.04.010
Ma, L. S., Hachani, A., Lin, J. S., Filloux, A., and Lai, E. M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104. doi: 10.1016/j.chom.2014.06.002
Marchi, M., Boutin, M., Gazengel, K., Rispe, C., Gauthier, J. P., Guillerm-Erckelboudt, A. Y., et al. (2013). Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots. Environ. Microbiol. Rep. 5, 393–403. doi: 10.1111/1758-2229.12048
McCarter, S. M., Jones, J. B., Gitaitis, R. D., and Smitley, D. R. (1983). Survival of Pseudomonas syringae pv. tomato in association with tomato seed, soil, host tissue, and epiphytic weed hosts in Georgia. Phytopathology 73, 1393–1398.
Michiels, J., Xi, C., Verhaert, J., and Vanderleyden, J. (2002). The functions of Ca2+ in bacteria: a role for EF-hand proteins? Trends Microbiol. 10, 87–93. doi: 10.1016/s0966-842x(01)02284-3
Miyata, S. T., Kitaoka, M., Brooks, T. M., Mcauley, S. B., and Pukatzki, S. (2011). Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949. doi: 10.1128/IAI.01266-10
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Mougous, J. D., Gifford, C. A., Ramsdell, T. L., and Mekalanos, J. J. (2007). Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9, 797–803. doi: 10.1038/ncb1605
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Rico, A., and Preston, G. M. (2008). Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant Microbe Interact. 21, 269–282. doi: 10.1094/MPMI-21-2-0269
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Russell, A. B., Singh, P., Brittnacher, M., Bui, N. K., Hood, R. D., Carl, M. A., et al. (2012). A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549. doi: 10.1016/j.chom.2012.04.007
Sabala, I., Jagielska, E., Bardelang, P. T., Czapinska, H., Dahms, S. O., Sharpe, J. A., et al. (2014). Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans. FEBS J. 281, 4112–4122. doi: 10.1111/febs.12929
Salomon, D., Klimko, J. A., Trudgian, D. C., Kinch, L. N., Grishin, N. V., Mirzaei, H., et al. (2015). Type VI secretion system toxins horizontally shared between marine bacteria. PLoS Pathog. 11:e1005128. doi: 10.1371/journal.ppat.1005128
Sana, T. G., Soscia, C., Tonglet, C. M., Garvis, S., and Bleves, S. (2013). Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS One 8:e76030. doi: 10.1371/journal.pone.0076030
Sarris, P. F., Skandalis, N., Kokkinidis, M., and Panopoulos, N. J. (2010). In silico analysis reveals multiple putative type VI secretion systems and effector proteins in Pseudomonas syringae pathovars. Mol. Plant Pathol. 11, 795–804. doi: 10.1111/j.1364-3703.2010.00644.x
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Shalom, G., Shaw, J. G., and Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699. doi: 10.1099/mic.0.2007/006585-0
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Si, M., Wang, Y., Zhang, B., Zhao, C., Kang, Y., Bai, H., et al. (2017). The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. doi: 10.1016/j.celrep.2017.06.081
Silverman, J. M., Agnello, D. M., Zheng, H., Andrews, B. T., Li, M., Catalano, C. E., et al. (2013). Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell. 51, 584–593. doi: 10.1016/j.molcel.2013.07.025
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Thomas, J., Watve, S. S., Ratcliff, W. C., and Hammer, B. K. (2017). Horizontal gene transfer of functional type VI killing genes by natural transformation. mBio 8:e00654-17. doi: 10.1128/mBio.00654-17
Trunk, K., Peltier, J., Liu, Y. C., Dill, B. D., Walker, L., Gow, N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Unterweger, D., Kitaoka, M., Miyata, S. T., Bachmann, V., Brooks, T. M., Moloney, J., et al. (2012). Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One 7:e48320. doi: 10.1371/journal.pone.0048320
Unterweger, D., Kostiuk, B., Otjengerdes, R., Wilton, A., Diaz-Satizabal, L., and Pukatzki, S. (2015). Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 34, 2198–2210. doi: 10.15252/embj.201591163
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host Immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Whitney, J. C., Beck, C. M., Goo, Y. A., Russell, A. B., Harding, B. N., De Leon, J. A., et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542. doi: 10.1111/mmi.12571
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. doi: 10.1093/nar/gkv1227
Xin, X. F., and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. doi: 10.1146/annurev-phyto-082712-102321
Yanez, M., Gil-Longo, J., and Campos-Toimil, M. (2012). Calcium binding proteins. Adv. Exp. Med. Biol. 740, 461–482. doi: 10.1007/978-94-007-2888-2_19
Yang, W., Wang, L., Zhang, L., Qu, J., Wang, Q., and Zhang, Y. (2015). An invasive and low virulent Edwardsiella tarda esrB mutant promising as live attenuated vaccine in aquaculture. Appl. Microbiol. Biotechnol. 99, 1765–1777. doi: 10.1007/s00253-014-6214-5
Zepeda-Rivera, M. A., Saak, C. C., and Gibbs, K. A. (2018). A proposed chaperone of the bacterial type VI secretion system functions to constrain a self-identity protein. J. Bacteriol. 200:e00688-17. doi: 10.1128/JB.00688-17
Zheng, J., Ho, B., and Mekalanos, J. J. (2011). Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876. doi: 10.1371/journal.pone.0023876
Zheng, J., and Leung, K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x
Keywords: Pseudomonas syringe, tomato speck disease, type VI secretion system, interbacterial competition, Hcp2
Citation: Chien C-F, Liu C-Y, Lu Y-Y, Sung Y-H, Chen K-Y and Lin N-C (2020) HSI-II Gene Cluster of Pseudomonas syringae pv. tomato DC3000 Encodes a Functional Type VI Secretion System Required for Interbacterial Competition. Front. Microbiol. 11:1118. doi: 10.3389/fmicb.2020.01118
Received: 07 February 2020; Accepted: 05 May 2020;
Published: 03 June 2020.
Edited by:
Patrick Eichenberger, New York University, United StatesReviewed by:
Paul Stodghill, Robert W. Holley Center for Agriculture and Health, (USDA-ARS), United StatesMelanie J. Filiatrault, Agricultural Research Service (USDA), United States
Copyright © 2020 Chien, Liu, Lu, Sung, Chen and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nai-Chun Lin, bmxpbkBudHUuZWR1LnR3
 Ching-Fang Chien
Ching-Fang Chien Nai-Chun Lin
Nai-Chun Lin