- 1Anhui Provincial Key Laboratory of Microbial Pest Control, Anhui Agricultural University, Hefei, China
- 2School of Life Sciences, University of Science and Technology of China, Hefei, China
- 3School of Plant Protection, Anhui Agricultural University, Hefei, China
G proteins are critical modulators or transducers in various transmembrane signaling systems. They play key roles in numerous biological processes in fungi, including vegetative growth, development of infection-related structures, asexual conidiation, and virulence. However, functions of G proteins in entomopathogenic fungi remain unclear. Here, we characterized the roles of MrGPA1, a G-protein subunit Gαi, in conidiation, stress resistance, and virulence in Metarhizium robertsii. MrGPA1 was localized in the mitochondria. MrGpa1 deletion resulted in a significant reduction (47%) in the conidiation capacity, and reduced expression of several key conidiation-related genes, including fluG, flbD, brlA, wetA, phiA, and stuA. Further, MrGpa1 disruption resulted in decreased fungal sensitivity to UV irradiation and thermal stress, as determined based on conidial germination of ΔMrGpa1 and wild-type (WT) strains. Chemical stress analysis indicated that MrGpa1 contributes to fungal antioxidant capacity and cell wall integrity, but is not involved in tolerance to antifungal drug and osmotic stress. Importantly, insect bioassays involving (topical inoculation and injection) of Galleria mellonella larvae revealed decreased virulence of ΔMrGpa1 strain after cuticle infection. This was accompanied by decreased rates of appressorium formation and reduced expression of several cuticle penetration-related genes. Further assays showed that MrGpa1 regulated intracellular cyclic AMP (cAMP) levels, but feeding with cAMP could not recover the appressorium formation rate of ΔMrGpa1. These observations suggest that MrGpa1 contributes to the regulation of conidiation, UV irradiation, thermal stress response, antioxidant capacity, and cell wall integrity in M. robertsii. This gene is also involved in insect cuticle penetration during infection. These findings raise the possibility of designing powerful strategies for genetic improvement of M. robertsii conidiation capacity and virulence for killing pests.
Introduction
G protein with GTP-hydrolase activity is a type of signaling protein that binds to guanine nucleotides (Robishaw and Berlot, 2004). It participates in signal transduction pathways linking activated cell-surface receptors with intracellular effectors, including adenylate cyclase and phospholipase through a series of signaling cascades involved in the regulation of physiological and biochemical processes (Ortiz-Urquiza and Keyhani, 2015; Chakravorty and Assmann, 2018). In fungi, G protein is associated with sexual and asexual reproduction, virulence, and response to external signal stimuli (Ivey et al., 1996; Liu and Dean, 1997; Guo et al., 2016b).
The heterotrimeric G protein is composed of three subunits α, β, and γ, wherein α subunit binds to GDP, and β and γ subunits form a heterodimer (Birnbaumer, 2007; Lambert, 2008; McIntire, 2009). When the heterotrimeric G protein is stimulated by a G-protein-coupled receptor (GPCR) that senses external signals, GDP is exchanged for GTP, and Gα and Gβγ complexes dissociate (Wedegaertner, 2012). Then, Gα-GTP and Gβγ act on the respective downstream effectors (Barren and Artemyev, 2007). The cycle is reset by the hydrolysis of GTP to GDP, and Gα recombining with Gβγ and GPCR (Gilchrist et al., 1999; Slessareva and Dohlman, 2006).
In mammals, G-protein α (GPA) subunits are divided into four classes, Gαs, Gαi, Gαq, and Gα12, based on the amino acid sequence identity (Neer, 1995). Further, the Gαi family is composed of four subfamilies, Gαi, Gαo, Gαt, and Gαz. The functions of Gαi family proteins are diverse, and include regulation of adenylyl cyclase, K+ and Ca+ channels, and cGMP phosphodiesterase activities (Simon et al., 1991). The conserved functional motif of Gαi protein is characterized by possession of N-myristoylation and ADP-ribosylation (Buss et al., 1987).
The functions of the GPA subunit have been characterized in some fungi. In Saccharomyces cerevisiae, two kinds of Gα proteins have been identified (Gpa1 and Gpa2). Gpa1 is involved in pheromone regulation (Jahng et al., 1988), while Gpa2 regulates pseudohyphal development via cyclic AMP (cAMP)-dependent pathways and heat resistance (Kubler et al., 1997). However, these proteins have different functions in other filamentous fungi. For example, Fusarium oxysporum f. sp. cubense possesses three Gα proteins (Gα-fga1, -fga2, and -fga3), and the deletion of encoding genes leads to phenotypic defects in colony morphology, reduced conidiation, increased heat tolerance, reduced virulence, and decreased intracellular cAMP levels (Jain et al., 2002; Guo et al., 2016a, b). Further, three different Gα proteins control unique signal transduction pathways in Magnaporthe grisea, influencing fungal vegetative growth, conidiation, conidium attachment, appressorium formation, mating, and pathogenicity (Liu and Dean, 1997; Zhang et al., 2012). GNA-1 protein is also required for the extension of basal hypha, growth, conidiation, and formation of female reproductive structures in Neurospora crassa (Ivey et al., 1996; Yang and Borkovich, 1999). In entomopathogenic fungus, some upstream and downstream genes for Gpa, such as GPCRs gene BbGpcr3, and regulators of the G protein signaling (RGS) genes Bbrgs1 and Mrcag8, have been only characterized (Fang et al., 2007, 2008; Ying et al., 2013), but the function of Gpa in insect pathogenic fungi is poorly understood. Thus, it is necessary to characterize the Gα proteins, and figure out whether the Gα proteins are involving in vegetative growth, conidiation, stress resistance, and virulence in entomopathogenic fungi, such as Metarhizium robertsii.
Metarhizium robertsii, an important entomopathogenic fungus, has been developed as an environmentally friendly alternative to chemical insecticides (Frazzon et al., 2000; Lord, 2005; Wang and Wang, 2017). Unfortunately, commercialized broad application of M. robertsii formulations is limited by the low conidiation rate, failure of conidia germination under high-temperature and UV stress, slow killing speed, and inconsistent field performance (Faria and Wraight, 2007; Fang et al., 2012; Muniz-Paredes et al., 2017). Genetic improvements of this mycoinsecticide require extensive understanding of the molecular mechanisms and M. robertsii genes involved in stress tolerance and virulence (Zhang and Feng, 2018).
In the current study, we aimed to investigate the role of Gα proteins in M. robertsii. BLASTP search of against the assembled draft genome sequence of M. robertsii identified four putative Gα proteins. Among these Gα proteins, MrGPA1 (EFZ00892) shared the highest identity (96.32%) with GNA-1. We then characterized the biological function of MrGPA1 by constructing and analyzing MrGpa1 gene deletion mutant. We show that MrGpa1 influences conidiation, stress resistance, virulence, and intracellular cAMP levels in M. robertsii.
Materials and Methods
Fungal Strains and Culture
In the present study, M. robertsii strain ARSEF 23 was the wild-type (WT) strain. All M. robertsii strains were inoculated onto potato dextrose agar (PDA, 20% potato, 2% glucose, and 2% agar, w/v), and cultured at 25°C for 10 days. Conidial suspensions were obtained by vortex-mixing in 0.05% (v/v) Tween-80, and filtered through sterile non-woven fabric to remove mycelial debris.
Sequence Analysis
To construct the phylogenetic tree of GPA proteins and analyze the structural domains of guanine nucleotide-binding site, amino acid sequences of the GPA subunits were downloaded from the National Center for Biotechnology Information1, and phylogenetic analysis was performed using MEGAX software2.
Construction of MrGPA1-GFP Fusion Vector and Analysis of Subcellular Localization of MrGPA1
To monitor subcellular localization of MrGpa1, gfp and MrGpa1 gene fragments were amplified by polymerase chain reaction (PCR), using gfp-F/gfp-R, and gfpMrGpa1-F/gfpMrGpa1-R primers (Supplementary Table S1), high-fidelity Taq DNA polymerase (KOD Plus Neo, Toyobo, Osaka, Japan), and M. robertsii genomic DNA as a template. The amplification products were inserted into the EcoRI restriction site in pDHt-SK-bar vector (kindly provided by Dr. Chengshu Wang; the vector conferred resistance against glufosinate-ammonium) (Fang et al., 2006) containing a strong promoter and terminator to generate vector pDHt-MrGpa1-gfp for Agrobacterium tumefaciens transformation. The corresponding transformants resistant to glufosinate ammonium were obtained, and verified by PCR using the primers gfp-F and gfp-R (Supplementary Table S1).
The MrGPA1-GFP strain was cultured on sabouraud dextrose agar medium containing yeast extract (SDAY, 4% glucose, 1% peptone, 2% agar, and 1% yeast extract powder, w/v) at 25°C for 2 days. The hyphae were then washed off the plate with sterile water and mixed with 500 nM MitoTracker Red CMXRos (Invitrogen, Shanghai, China), a dye specific to mitochondria. Subcellular localization of MrGPA1 was evaluated using a laser scanning confocal microscopy (LSCM, Zeiss LSM880). Before using laser scanning confocal microscopy, Wolf PSORT software3 was used for prediction subcellular localization by analyzing protein sequence of MrGPA1.
Gene Deletion and Complementation
To disrupt MrGpa1 gene, the 5′- and 3′-flanking regions of MrGpa1 were obtained by using MrGpa1-5F/MrGpa1-5R and MrGpa1-3F/MrGpa1-3R primers, genomic DNA (using the Plant Genomic DNA Kit; Tiangen, Beijing, China) extracted as a PCR template and high-fidelity Taq DNA polymerase (KOD Plus Neo, Toyobo, Osaka, Japan). The amplification products were then inserted into the pDHt-SK-bar vector (containing glufosinate resistance gene) (Fang et al., 2006) digested with the SmaI, BamHI, and XbaI restriction enzymes to generate vector pDHt-MrGpa1-bar for A. tumefaciens transformation (Fang et al., 2006). ΔMrGpa1 strains were obtained by selection for glufosinate resistance, and two fragments containing the MrGpa1- upstream-bar-MrGpa1-downstream cassette subsequently verified by PCR and reverse-transcription (RT)-PCR using primer pairs MrGpa1-F/MrGpa1-R, upMrGpa1-F/upMrGpa1-R, dnMrGpa1-F/dnMrGpa1-R, gpd-F/gpd-R, and bar-F/bar-R (Supplementary Table S1).
For gene complementation, the entire MrGpa1 gene and the 1000-bp upstream sequence and 600-bp downstream sequence were inserted into vector pDHt-SK-ben (containing benomyl resistance gene) digested with the SpeI restriction enzyme for fungal transformation. The 3050-bp fragment was ectopically integrated into ΔMrGpa1 strain by the same method as that used for gene deletion. Complemented strains (cpΔMrGpa1) were obtained by selection for benomyl resistance, and verified by PCR using primer pairs MrGpa1-F/MrGpa1-R and ben-F/ben-R (Supplementary Table S1).
Phenotype Assays
For phenotype assays, these experiments were performed with three technical and biological replicates per strain (WT, ΔMrGpa1, and cpΔMrGpa1).
Fungal conidiation ability was evaluated as previously described (Meng et al., 2017). Briefly, 30 μl of conidial suspension (1 × 106 conidia/ml) was spread on PDA plate (35-mm diameter). After culturing at 25°C for 14 days, the conidia on each plate were collected into 30 mL of 0.05% Tween-80 by vortex-mixing, and conidial density was determined using a hemocytometer and converted to the number of conidia per square centimeter of colony.
To evaluate the fungal vegetative growth, 1 μl of WT, ΔMrGpa1, and cpΔMrGpa1 conidial suspensions (1 × 107 conidia/ml) was spotted on PDA and 1/4 SDAY (1/4 dilution of SDAY) media, and incubated in the dark at 25°C for 10 days. Colony diameters were then measured.
For conidial germination assay, 10 μl of conidial suspension (5 × 106 conidia/ml) were spread on PDA medium. The conidial germination was observed by microscope (Olympus BX 51, Tokyo, Japan) at 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24 h after incubated at 25°C. Conidia are considered to be germinated when the length of the germ tube reaches or longer than the length of the conidia (Wang et al., 2014). Three hundred conidia were counted at least per plate and the germination rates were calculated by comparing the number of germinated conidia with the 300 counted conidia, and the median germinate time (GT50) was calculated using the SPSS software.
For heat stress tolerance assays, 1 ml of conidial suspensions (5 × 106 conidia/ml) of WT, ΔMrGpa1, and cpΔMrGpa1 strains were placed in 1.5-ml Eppendorf tubes, and then incubated in a water bath at 42 or 28°C (as control) for 1 h. Then 10 μl of the suspension were spread on PDA medium, incubated at 25°C. Conidial germination was observed under a microscope (Olympus BX 51, Tokyo, Japan) after 16 and 24 h. Three hundred conidia were counted at least by per plate and the relative germination rates were calculated by comparing the number of germinated conidia with had not been heat stressed (Wang et al., 2019).
To determine fungal tolerance to ultraviolet B (UV-B) light, 10 μl of conidial suspensions (5 × 106 conidia/ml) of WT, ΔMrGpa1, and cpΔMrGpa1 was taken to PDA medium. The plates were then exposed to UV-B irradiation (312-nm wavelength at 100 μJ cm–2) using HL-2000 Hybrilinker (UVP, CA, United States) (Yao et al., 2010) or exposed to sunlight (as control). Relative UV-B tolerance was assessed and calculated by aforementioned methods to assess relative germination rate of tolerance to UV-B.
To examine the fungal tolerance to chemical stress, 1 μl conidial suspensions (1 × 107 conidia/ml) of WT, ΔMrGpa1, and cpΔMrGpa1 strains were spotted onto PDA medium containing carbendazim (2 μg/ml), NaCl (0.5 M), H2O2 (2 mM), or Congo red (2 μg/ml), and incubated in the dark for 10 days at 25°C. Colony diameter was then measured and the relative inhibition rate was calculated (Ying and Feng, 2011; Wang et al., 2017).
To assess the effects of MrGpa1 disruption on virulence, bioassays with Galleria mellonella larvae (RuiQing Bait, Shanghai, China) were performed as described previously (Zhou et al., 2018). The larvae were immersed in conidial suspension (1 × 106 conidia/ml) for 90 s or injected (into the hemocoel) with 10 μl of conidial suspensions (1 × 105 conidia/ml) and incubated at 25°C, each treatment was performed in triplicate, with 18 larvae in each group. The experiment was repeated three times. Larva mortality was evaluated every 24 h, and the median lethal time (LT50) was calculated using the SPSS software.
The appressorium formation assay was performed as described previously (Gao et al., 2013). Briefly, to test the appressorium formation on a hydrophobic surface, 1 ml of conidial suspension (1 × 106 conidia/ml) in MMGly (minimal medium amended with 1% glycerol) was spread on a sterile plastic Petri dishes (3.5-cm diameter), followed by 24 and 48 h incubation at 25°C. At least 300 conidia of each strain were evaluated microscopically, and the induction rates of appressorium formation were quantified by observing different microscopic fields (inverted microscope, Olympus IX 71, Tokyo, Japan).
Quantitative RT-PCR (RT-qPCR)
To analyze the expression of conidiation-related genes, 200 μl of conidial suspensions (1 × 107 conidia/ml) of WT, ΔMrGpa1, and cpΔMrGpa1 strains were plated on PDA medium, and cultured in the dark at 25°C for 2.5 days. The samples were collected and milled in liquid nitrogen to extract total RNA. To analyze the expression of virulence genes related to cuticle infection, G. mellonella larvae were dipped in conidial suspensions (5 × 107 conidia/ml) of WT, ΔMrGpa1, and cpΔMrGpa1 strains for 1.5 min, transferred to 25°C for 48 h, and then placed in liquid nitrogen for total RNA extraction. Total RNA was extracted by using Trizol reagent (Invitrogen, Foster City, CA, United States). cDNA was obtained by using the PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China), and used as a template for RT-qPCR. The gene expression analysis was performed by using the CFBR96TM Real-Time PCR System (Bio-Rad, Hercules, CA, United States) and SYBR® PremixEx TaqTM II (TaKaRa). Three biological repeats of each treatment were analyzed. The qPCR primers are listed in Supplementary Table S2. The expression of the gpd gene (MAA_07675, encoding glyceraldehyde 3-phosphate dehydrogenase) was used as an internal control (Fang and Bidochka, 2006). The relative gene expression was calculated by using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
cAMP Assay
To measure the intracellular cAMP levels of WT, ΔMrGpa1, and cpΔMrGpa1 strains, the method was performed previously (Liu et al., 2012). The cAMP levels were measured by high-performance liquid chromatography (HPLC) analysis as described previously (Liu et al., 2016).
To test whether exogenous cAMP could recover appressorium formation rate of ΔMrGpa1, the conidial suspensions (1 × 106 conidia/ml) were amended with the final concentration of 5 mM cAMP and left for appressorium formation on a hydrophobic surface as mentioned above.
Statistical Analysis
All data are presented using GraphPad Prism version 6.0. Data are expressed as the mean ± standard error (SE) of the mean, from three biological replicates. Statistical analysis was performed by one-way analysis of variance (ANOVA). For multiple comparisons, Tukey’s multiple comparison test was used to analyze statistical the significance. p < 0.05 was considered to be significant, and p < 0.01 was considered to be extremely significant.
Results
Sequence Characteristics of MrGPA1 From M. robertsii
We chose the sequences of GNA-1 proteins of the model fungus N. crassa as reference sequences to retrieve their M. robertsii orthologs. We thus identified four genes encoding GPA subunit [GenBank accession numbers EFZ00892.1, EFY98464.1, EFY99066.2, and EFZ00060.2, named MrGpa1, MrGpa2, MrGpa3, and MrGpa4 (i.e., the M. robertsii Gpa1, Gpa2, Gpa3, and Gpa4 genes), respectively] in the genome of M. robertsii ARSEF 23.
Further bioinformatics analysis indicated that MrGpa1 (MAA-03488) is a single copy gene encoding GPA subunit (353-aa protein) in M. robertsii. A BLASTP search of MrGPA1 homologs in NCBI revealed that the protein shares 100% amino acid similarity with GPA from Metarhizium acridum (XP_007811324), Metarhizium anisopliae (KFG82129), Metarhizium brunneum (XP_014542423), and Metarhizium rileyi (OAA44756). Phylogenetic tree of GPA proteins from Metarhizium spp., and related fungal species was constructed with S. cerevisiae as an outgroup (Supplementary Figure S1A). All GPA proteins from the genus Metarhizium formed an independent branch (100% support value). These Metarhizium GPA proteins are closely related to GPA proteins from Pochonia chlamydosporia (99.72%), Moelleriella libera (99.43%), and Purpureocillium lilacinum (99.15%).
A conserved domain database4 search demonstrated that MrGPA1 contains a highly conserved guanine nucleotide-binding site (34-347-aa protein), which is the key identification domain of the GPA subunit (Supplementary Figure S1B). Furthermore, homologous alignment revealed the presence of N-myristoylation and ADP-ribosylation sites (two conserved positions in the GPAi subunit) in the conserved functional motif of MrGPA1. Hence, MrGPA1 is a member of the Gαi family.
MrGPA1 Is a Mitochondria Protein
To investigate the subcellular localization of MrGPA1, the Wolf PSORT software was first used. The analysis predicted that the protein is localized in the mitochondria. To verify this prediction, we generated MrGpa1-gfp strain (Supplementary Figures S2A,B). As shown in LSCM images in Figure 1, the mitochondria in hypha is stained with a mitochondrial dye (red), in a punctate patterns, while punctate green fluorescence was also observed in vegetative hyphae. Red and green fluorescence was detected and overlapped (Figure 1), suggesting that MrGPA1 is a mitochondria protein.
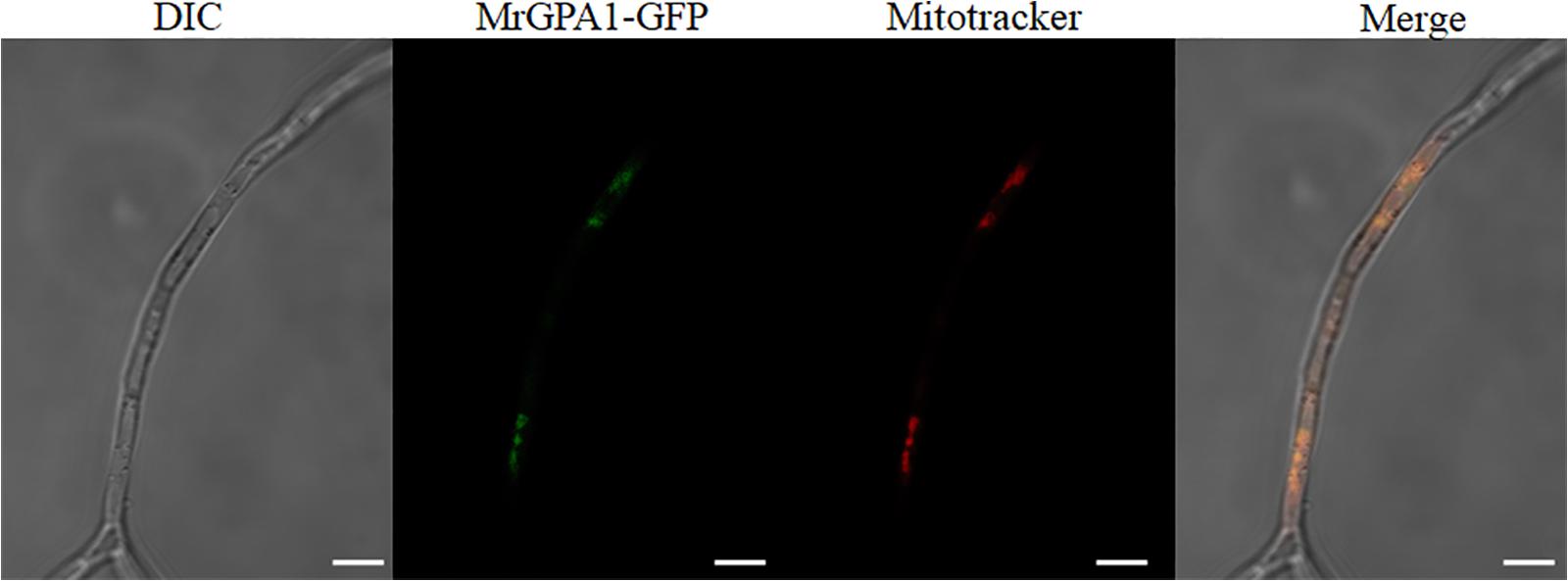
Figure 1. MrGPA1 is a mitochondria protein. LSCM images (scale bars: 10 μm) of the cellular location of MrGPA1, showing that MrGPA1 is located in the mitochondria, with the GFP fluorescence signal apparent in hyphae.
Construction of MrGpa1 Knockout and Complementation
The result indicated the presence of a 1, 222-bp fragment corresponding to the partial MrGpa1 gene sequence in the WT and cpΔMrGpa1 stains, but not in the ΔMrGpa1 strain. In addition, a partial 806-bp bar gene fragment was detected in ΔMrGpa1 and cpΔMrGpa1 strains, and a partial 785-bp ben gene fragment was detected in the cpΔMrGpa1 strain. Furthermore, PCR analysis indicated the presence of a fragment containing upstream sequence of MrGpa1 and a partial bar gene (2689 bp) and a fragment containing downstream sequence of MrGpa1 and a partial bar gene (1975 bp), and detected by using the primer sets up MrGpa1-F/upMrGpa1-R and dnMrGpa1-F/dnMrGpa1-R, respectively, in the ΔMrGpa1 strain. Finally, RT-PCR analysis verified the loss or regain of the MrGpa1 gene expression in ΔMrGpa1 and cpΔMrGpa1 strains, accordingly. These observations indicated a successful construction of the MrGpa1 knockout and complementation strains (Supplementary Figure S2).
MrGpa1 Contributes to Fungal Conidiation but Is Not Involved in Vegetative Growth
To examine the effect of MrGpa1 on the growth and development of M. robertsii, we evaluated mycelial growth and conidial yield of WT and mutant strains on PDA and 1/4 SDAY medium, respectively. The 14-day-old colonies of WT, ΔMrGpa1, and cpΔMrGpa1 strains formed 6.18 × 107, 3.28 × 107, and 6.55 × 107 conidia/cm–2, respectively (Figure 2A). The loss of MrGpa1 caused a significant, 47% reduction in conidiation, but little difference in the growth rate of WT, ΔMrGpa1, and cpΔMrGpa1 strains on PDA and 1/4 SDAY media was apparent (Figures 2A,B). We also examined the expression of genes involved in conidiation in M. robertsii by RT-qPCR. The expression of fluG, flbD, brlA, wetA, phiA, and stuA genes in the MrGpa1 strain was significantly reduced compared with that in the WT and cpΔMrGpa1 strains (Figure 2C). Collectively, these observations indicate that while MrGPA1 plays an important role in the conidiation of M. robertsii, it is not involved in vegetative growth.
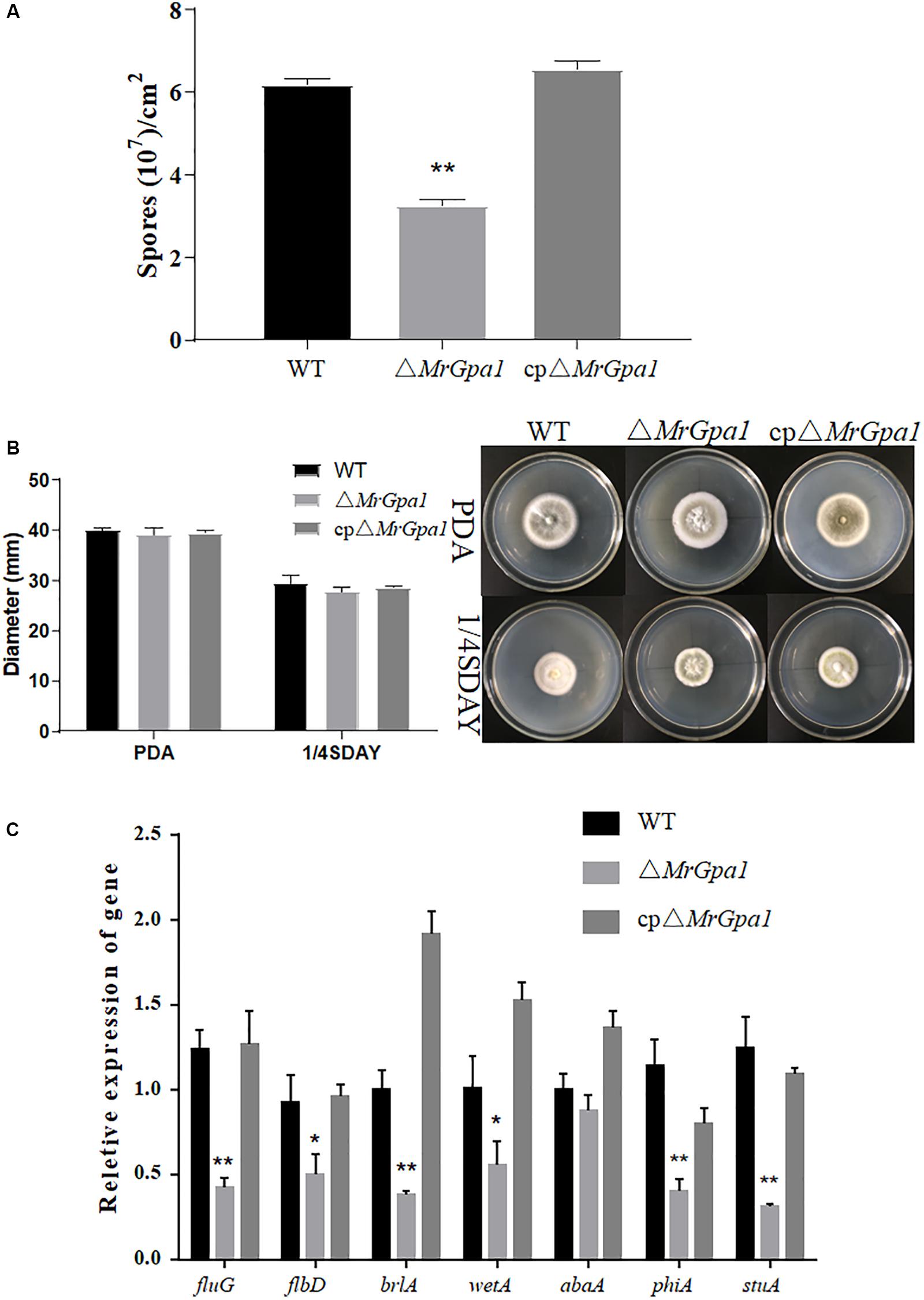
Figure 2. MrGpa1 contributes to fungal conidiation but is not involved in vegetative growth. (A) Conidial yields of three fungal strains after 14-days, growth on PDA at 25°C. (B) Colony diameters and phenotype of three fungal strains on PDA and 1/4SDAY media after 10-days, growth at 25°C. (C) RT-qPCR analysis of the relative expression of conidiation-related genes in 2.5-day-old PDA cultures of three fungal strains. *p < 0.05, **p < 0.01.
MrGpa1 Is Important for Heat and UV Stresses Tolerance, and Is Involved in Antioxidant Capacity and Cell Wall Integrity of M. robertsii
The GT50 values for WT, ΔMrGpa1, and cpΔMrGpa1 were 11.99 h, 6.16 h (p < 0.01, compared with WT strain), and 12.16 h, respectively (Figure 3A). Conidial germination speed of ΔMrGpa1 strain on PDA medium was significantly faster than that of WT and cpΔMrGpa1 strains.
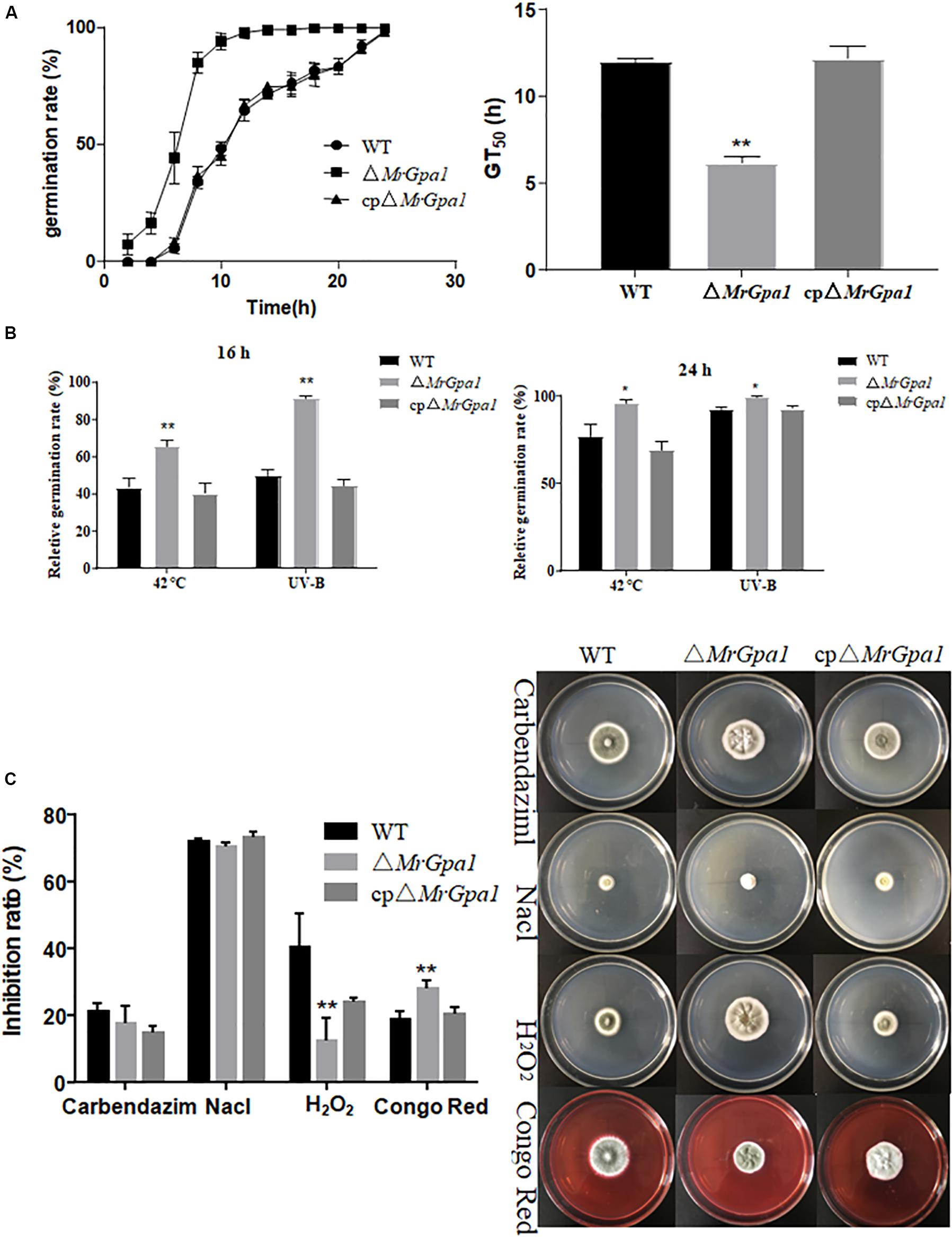
Figure 3. MrGpa1 is required for fungal stress response. (A) Germination rate and GT50 values for three fungal strains after growth on PDA. (B) Relative germination rate of three fungal strains after exposure to heat stress and UV-B treatment. (C) Inhibition ratio and colony phenotype values for three fungal strains, on PDA medium containing Carbendazim (2 μg/ml), NaCl (0.5 M), H2O2 (10 mM), or Congo red (2 mg/ml). *p < 0.05, **p < 0.01.
To investigate the effects of MrGpa1 deletion on UV irradiation and thermal stress, the relative germination rate of conidia exposed to these stresses was determined 16 or 24 h after stress exposure. We found that the sensitivity of ΔMrGpa1 strain to 42°C heat stress was reduced. For example, compared with the WT, ΔMrGpa1 germination rates at 16 and 24 h increased by 51% (p < 0.01) and 24% (p < 0.05), respectively (Figure 3B). Similar results were obtained for conidial tolerance of UV irradiation; compared with the WT, ΔMrGpa1 germination rates at 16 h increased by 83% (p < 0.01), but only by 8% (p < 0.05) at 24 h (Figure 3B). Hence, it appears that MrGPA1 plays an important role in conidial tolerance of both UV irradiation and thermal stress.
To evaluate the role of MrGpa1 in fungal growth under different chemical stress conditions, we investigated the mycelial growth of the WT and mutant strains on PDA containing carbendazim, NaCl, H2O2, or Congo red. The antioxidant capacity and cell wall integrity of ΔMrGpa1 strain were significantly different than those of the WT and cpΔMrGpa1 strains. For instance, compared with the WT, the relative inhibition of ΔMrGpa1 growth was decreased by 68.6% (p < 0.01) on PDA containing H2O2, while the sensitivity to Congo red was increased by 47.4% (p < 0.01). However, the relative inhibition of ΔMrGpa1 growth in the presence of carbendazim and NaCl was not markedly different from that of the control strains (Figure 3C). These observations indicate that MrGpa1 contributes to fungal antioxidant capacity and cell wall integrity, but is not involved in antifungal ability and osmotic stress.
MrGpa1 Plays an Important Role in Insect Cuticle Penetration via Appressorium Formation
We next used G. mellonella bioassays to assess the consequences of MrGpa1 deletion on fungal virulence. In topical infection bioassays, the mean lethal times to death (LT50) in insects infected with ΔMrGpa1, WT, and cpΔMrGpa1 strains were 7.2 ± 0.45, 11.8 ± 0.54, and 8.3 ± 0.71 days, respectively, with a significant (p < 0.05) attenuation of virulence in G. mellonella (Figures 4A,B). The treatment with ΔMrGpa1 also resulted in an increased survival rate of the larvae compared with the WT and cpΔMrGpa1 strains treatment. By contrast, in the injection bioassays, we did not observe any differences in LT50 values between larvae infected with ΔMrGpa1 (3.69 ± 0.15 days), and WT (3.53 ± 0.14 days) or cpΔMrGpa1 strains (3.57 ± 0.12 days) (Figures 4C,D).
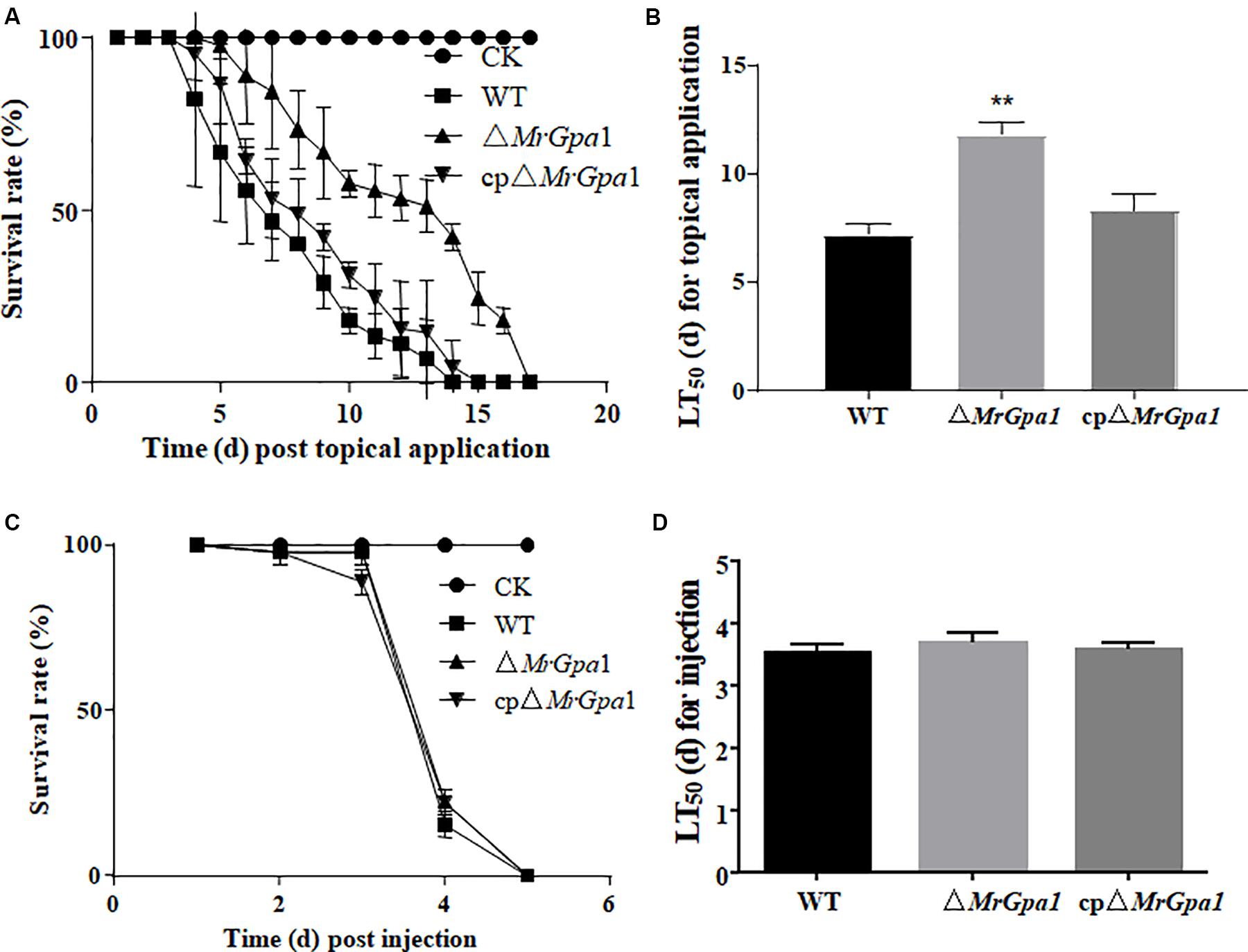
Figure 4. MrGpa1 deletion impacts fungal virulence. (A) Survival of G.mellonella after topical application of conidial suspensions from three fungal strains. The control insects were treated with sterile water. (B) LT50 (days) of three fungal strains after topical inoculation of larvae. (C) Survival of G. mellonella after injection of conidial suspension of three fungal strains. The control insects were treated with sterile water. (D) LT50 (days) of three fungal strains after injection. **p < 0.01.
We then determined the expression of insect virulence-related genes during cuticle penetration by RT-qPCR. Indeed, the expression of several genes involved in the adhesion (mad1, 45% expression in ΔMrGpa1 strain compared with the WT strain), appressorium formation (mpl1, 72%, and gpa, 52%), and cuticle penetration (pr1A, 93%, and pr1C, 96%) was significantly decreased in the ΔMrGpa1 strain compared with their expression in the WT strains (Figure 5A).
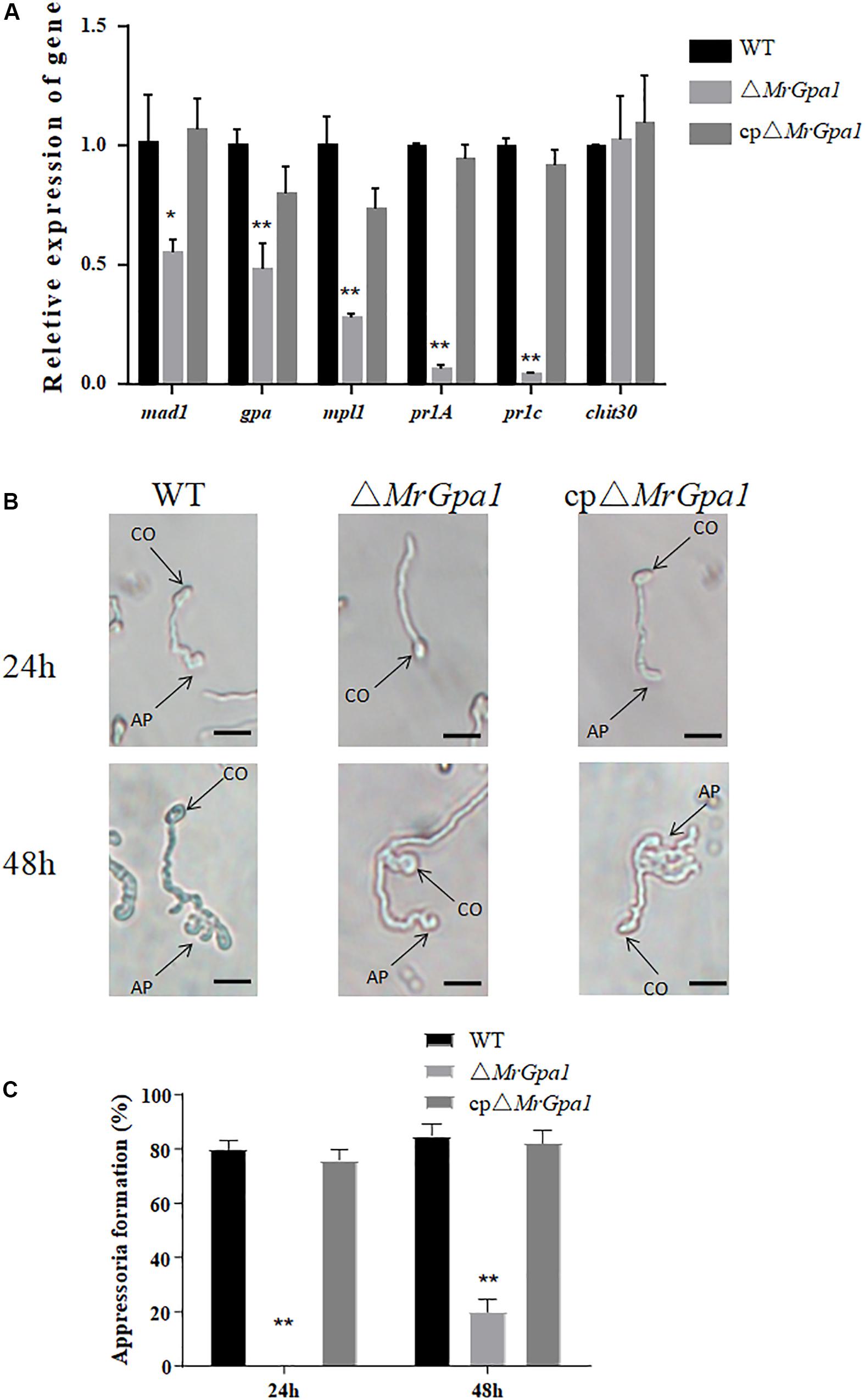
Figure 5. MrGpa1 deletion affects appressorium formation. (A) RT-qPCR analysis for the relative expression of virulence-associated genes of three fungal strains in vivo (24 h after inoculation of G. mellonella larvae). (B) Microscopic analysis and percentage appressorium formation by fungal strains induced on a plastic hydrophobic surface. Scale: 10 μm. (C) Percentage of appressorium formation in vitro. *p < 0.05, **p < 0.01. CO, conidium, AP, appressorium.
To determine the mechanism of the virulence defect of ΔMrGpa1 strain, we then assayed appressorium formation on a hydrophobic surface. We observed that the loss of MrGpa1 impaired appressorium differentiation, compared with the control strains. Specifically, 24 h after induction, ΔMrGpa1 strain did not form appressoria, while the appressorium formation rate of the WT strain was 80% (Figure 5B). Further, 48 h after induction, the appressorium formation rate of ΔMrGpa1 was only approximately 20% and were significantly reduced (by 76.5%) compared with WT strains (Figure 5C). Therefore, MrGpa1 plays an important role in cuticle penetration by impacting appressorium formation.
MrGpa1 Regulates Intracellular cAMP Levels, but Further Feeding With cAMP Cannot Recover the Appressorium Formation Rate of ΔMrGpa1
To determine whether MrGpa1 regulates cAMP levels in M. robertsii, the intracellular cAMP levels were measured in the hyphal stage. We found that the intracellular cAMP levels of ΔMrGpa1 were significant decreased by 52.8% (p < 0.01), compared with WT strains (Figure 6A). The result indicated that MrGpa1 is a positive regulator in intracellular cAMP levels of M. robertsii. However, the appressorium formation rate of ΔMrGpa1 cannot recover with the addition of 5 mM exogenous cAMP (Figure 6B).
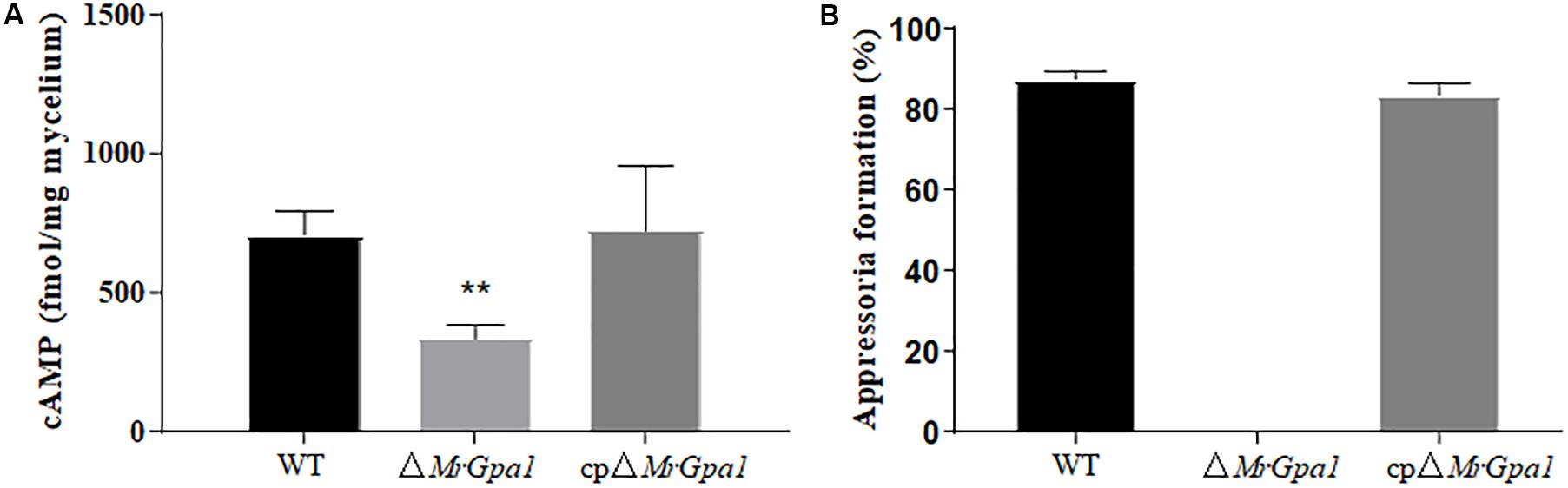
Figure 6. (A) Quantification of intracellular cAMP in the hyphal stage. (B) After 24 h incubation, the percentage of appressorium formation by adding exogenous 5 mM cAMP. **p < 0.01.
Discussion
G-proteins are key components of various signal transduction pathways and play show important biological roles in the control of cell proliferation, behavior, and development in higher organisms (Ivey et al., 1996; Regenfelder et al., 1997; Harashima and Heitman, 2005). In the current study, we described the identification of MrGpa1 gene encoding the Gαi subunit in M. robertsii, and showed that this Gαi protein plays an important role in conidiation, stress resistance, and virulence in the host fungus.
In filamentous fungi, genes encoding three GPA subunits were reported in N. crassa, F. oxysporum f. sp. cubense, Aspergillus nidulans, and M. grisea (Ivey et al., 1996; Hicks et al., 1997; Liu and Dean, 1997; Guo et al., 2016b). However, while genes of four G protein α subunits were identified in M. robertsii, two of the open reading frames (ORFs) are highly similar (MrGpa2 and MrGpa4). Further, the existence of Gαi has been demonstrated in filamentous fungi including the M. robertsii in this study, but not in the yeasts S. cerevisiae or Schizosaccharomyces pombe (Kubler et al., 1997; Kallal and Fishel, 2000).
Previously, to be activated by GPCR that sense external signals in plasma membrane (PM), G protein was considered to locate at the PM (Marrari et al., 2007). However, recent studies revealed G-protein localization beyond the PM, e.g., in the mitochondrion, endoplasmic reticulum, and Golgi apparatus (Michaelson et al., 2002; Nair et al., 2017). Wolf PSORT prediction in the current study indicated a possible location of MrGPA1 in the mitochondrion. Indeed, by using a fluorescent protein fusion, we showed here that MrGPA1 is located in the mitochondria, which is consistent with location of Gαi in HEK293T cells (Lyssand and Bajjalieh, 2007). It have been reported that the roles of mitochondria in fungi is involved in the aging, conidiation, tolerance to adverse stresses and pathogenesis (Lorin et al., 2006; Kretschmer et al., 2012; Kitagaki and Takagi, 2014; Khan et al., 2015). For example, previous report in Magnaporthe oryzae shows that mitochondrial fission protein MoFis1 mediates conidiation and is required for full virulence (Khan et al., 2015). Moreover, Kretschmer et al. (2012) found that defects in mitochondrial influence virulence from Ustilago maydis. Therefore, the biological roles of MrGPA1 localized to mitochondria are further investigated in the present study.
According to multiple studies, GPA subunits play a role in the conidial yield in some fungi. For example, the conidiation of magB deletion mutant of M. grisea and Δfga1 mutant of F. oxysporum f. sp. cubense is impaired (Liu and Dean, 1997; Guo et al., 2016b). A similar phenomenon was also observed in the current study. Because a conserved conidiation regulatory pathway in M. robertsii contains Mr-BrlA, Mr-AbaA, and Mr-WetA (Zeng et al., 2017), these key genes were downregulated in the MrGpa1 deletion mutant too. All these indicated that the MrGpa1 gene is involved in the conidiation of M. robertsii by regulating the expression of conidiation-related genes.
We also observed that ΔMrGpa1 strain is more tolerant to UV irradiation and thermal stress than the WT and cpΔMrGpa1 strains. This observation was consistent with findings for F. oxysporum f. sp. cubense and N. crassa (Yang and Borkovich, 1999; Guo et al., 2016b). According to the two cited studies, intracellular cAMP levels are reduced in the Gαi deletion mutants, suggesting that the cAMP pathway might be involved in the response to thermal stress and tolerance of UV irradiation in some fungi.
In the M. robertsii, MrGpa1 deletion resulted in a marked attenuation of virulence in the G. mellonella model, with an increased LT50 value (by 63.9%) compared with that of the WT strain. Further analysis indicated that the reduced virulence is associated with an impaired appressorium differentiation in the mutant. This was similar to the effect of magB deletion in M. grisea, namely, blocked appressorium formation in a deletion strain (Liu and Dean, 1997). The virulence of plant pathogenic fungi is regulated by multiple pathways, such as the mitogen activated protein kinase (MAPK) cascades and the cAMP-PKA pathway (Dean, 1997). Further, the Gαi family contributes to pathogenicity in many plant pathogenic fungi. For example, the pathogenicity of the deletion strains M. grisea ΔmagB, Botryils cinerea Δbcg1, and F. oxysporum f. sp. cubense Δfga1 is markedly reduced (Liu and Dean, 1997; Gronover et al., 2001; Guo et al., 2016b). Moreover, MAPK cascades and cAMP-PKA pathway are also involved in virulence of entomopathogenic fungus (Fang et al., 2009; Jin et al., 2012; Chen et al., 2016). In this study, the decrease of the intracellular cAMP levels in ΔMrGpa1 strains demonstrates that MrGpa1 involved in cAMP-PKA pathway. However, feeding with cAMP cannot recover the appressorium formation rate of ΔMrGpa1. In contrast to magB deletion in M. grisea, the appressorium differentiation is dependent on cAMP (Liu and Dean, 1997). We speculated that MrGpa1 located in mitochondria and magB are different G proteins, and MrGpa1 is involved in not only cAMP-PKA pathway, but also other regulatory pathways affecting on appressorium differentiation, such as MAKP cascades or RGS pathway.
Conclusion
MrGPA1 is located in the mitochondria of M. robertsii cell and is a member of Gαi family. MrGPA1 controls unique signal transduction pathways, and thus plays important role in conidiation, stress resistance, virulence, and intracellular cAMP levels in that fungus. These findings raise the possibility of designing powerful strategies for genetic improvement of M. robertsii conidiation capacity and virulence for killing pests.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
BH and ZW conceived and designed the study. YT and HW wrote the manuscript, conducted the experiments, and analyzed the data. ZL did a part of the experiments. BH edited the manuscript and supervised the project. All authors read and approved the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2017YFD0200400) and the National Natural Science Foundation of China (Grant Nos. 31972332 and 31772226).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Chengshu Wang for providing fungus material and gene deletion vectors. This manuscript has been released as a pre-print at Tong et al. (2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01251/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.megasoftware.net/
- ^ https://wolfpsort.hgc.jp
- ^ http://www.ebi.ac.uk/interpro/
References
Barren, B., and Artemyev, N. O. (2007). Mechanisms of dominant negative G-protein alpha subunits. J. Neurosci. Res. 85, 3505–3514. doi: 10.1002/jnr.21414
Birnbaumer, L. (2007). Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 alpha subunits plus betagamma dimers. Biochim. Biophys. Acta 1768, 772–793. doi: 10.1016/j.bbamem.2006.12.002
Buss, J. E., Mumby, S. M., Casey, P. J., Gilman, A. G., and Sefton, B. M. (1987). Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proc. Natl. Acad. Sci. U.S.A. 84, 7493–7497. doi: 10.1073/pnas.84.21.7493
Chakravorty, D., and Assmann, S. M. (2018). G protein subunit phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochem. J. 475, 3331–3357. doi: 10.1042/BCJ20160819
Chen, X., Xu, C., Qian, Y., Liu, R., Zhang, Q., Zeng, G., et al. (2016). MAPK cascade-mediated regulation of pathogenicity, conidiation and tolerance to abiotic stresses in the entomopathogenic fungus Metarhizium robertsii. Environ. Microbiol. 18, 1048–1062. doi: 10.1111/1462-2920.13198
Dean, R. A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. doi: 10.1146/annurev.phyto.35.1.211
Fang, W., Azimzadeh, P., and St Leger, R. J. (2012). Strain improvement of fungal insecticides for controlling insect pests and vector-borne diseases. Curr. Opin. Microbiol. 15, 232–238. doi: 10.1016/j.mib.2011.12.012
Fang, W., and Bidochka, M. J. (2006). Expression of genes involved in germination, conidiogenesis and pathogenesis in Metarhizium anisopliae using quantitative real-time RT-PCR. Mycol. Res. 110(Pt 10), 1165–1171. doi: 10.1016/j.mycres.2006.04.014
Fang, W., Pava-ripoll, M., Wang, S., and St Leger, R. (2009). Protein kinase a regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fungal Genet. Biol. 46, 277–285. doi: 10.1016/j.fgb.2008.12.001
Fang, W., Pei, Y., and Bidochka, M. J. (2006). Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 52, 623–626. doi: 10.1139/w06-014
Fang, W., Pei, Y., and Bidochka, M. J. (2007). A regulator of a G protein signalling (RGS) gene, cag8, from the insect-pathogenic fungus Metarhizium anisopliae is involved in conidiation, virulence and hydrophobin synthesis. Microbiology 153(Pt 4), 1017–1025. doi: 10.1099/mic.0.2006/002105-0
Fang, W., Scully, L. R., Zhang, L., Pei, Y., and Bidochka, M. J. (2008). Implication of a regulator of G protein signalling (BbRGS1) in conidiation and conidial thermotolerance of the insect pathogenic fungus Beauveria bassiana. FEMS Microbiol. Lett. 279, 146–156. doi: 10.1111/j.1574-6968.2007.00978.x
Faria, M. R. D., and Wraight, S. P. (2007). Mycoinsecticides and Mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43, 237–256. doi: 10.1016/j.biocontrol.2007.08.001
Frazzon, A. P., da Silva Vaz Junior, I., Masuda, A., Schrank, A., and Vainstein, M. H. (2000). In vitro assessment of Metarhizium anisopliae isolates to control the cattle tick Boophilus microplus. Vet. Parasitol. 94, 117–125. doi: 10.1016/s0304-4017(00)00368-x
Gao, Q., Shang, Y., Huang, W., and Wang, C. (2013). Glycerol-3-phosphate acyltransferase contributes to triacylglycerol biosynthesis, lipid droplet formation, and host invasion in Metarhizium robertsii. Appl. Environ. Microbiol. 79, 7646–7653. doi: 10.1128/AEM.02905-13
Gilchrist, A., Bunemann, M., Li, A., Hosey, M. M., and Hamm, H. E. (1999). A dominant-negative strategy for studying roles of G proteins in vivo. J. Biol. Chem. 274, 6610–6616. doi: 10.1074/jbc.274.10.6610
Gronover, C. S., Kasulke, D., Tudzynski, P., and Tudzynski, B. (2001). The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact 14, 1293–1302. doi: 10.1094/mpmi.2001.14.11.1293
Guo, L., Yang, L., Liang, C., Wang, J., Liu, L., and Huang, J. (2016a). The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 93, 29–38. doi: 10.1016/j.pmpp.2015.12.003
Guo, L., Yang, Y., Yang, L., Wang, F., Wang, G., and Huang, J. (2016b). Functional analysis of the G-protein α subunits FGA1 and FGA3 in the banana pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 94, 75–82. doi: 10.1016/j.pmpp.2016.04.003
Harashima, T., and Heitman, J. (2005). Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 4557–4571. doi: 10.1091/mbc.e05-05-0403
Hicks, J. K., Yu, J. H., Keller, N. P., and Adams, T. H. (1997). Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16, 4916–4923. doi: 10.1093/emboj/16.16.4916
Ivey, F. D., Hodge, P. N., Turner, G. E., and Borkovich, K. A. (1996). The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7, 1283–1297. doi: 10.1091/mbc.7.8.1283
Jahng, K. Y., Ferguson, J., and Reed, S. I. (1988). Mutations in a gene encoding the alpha subunit of a Saccharomyces cerevisiae G protein indicate a role in mating pheromone signaling. Mol. Cell. Biol. 8, 2484–2493. doi: 10.1128/mcb.8.6.2484
Jain, S., Akiyama, K., Mae, K., Ohguchi, T., and Takata, R. (2002). Targeted disruption of a G protein alpha subunit gene results in reduced pathogenicity in Fusarium oxysporum. Curr. Genet. 41, 407–413. doi: 10.1007/s00294-002-0322-y
Jin, K., Ming, Y., and Xia, Y. X. (2012). MaHog1, a Hog1-type mitogen-activated protein kinase gene, contributes to stress tolerance and virulence of the entomopathogenic fungus Metarhizium acridum. Microbiology 158(Pt 12), 2987–2996. doi: 10.1099/mic.0.059469-0
Kallal, L., and Fishel, R. (2000). The GTP hydrolysis defect of the Saccharomyces cerevisiae mutant G-protein Gpa1(G50V). Yeast 16, 387–400. doi: 10.1002/(sici)1097-0061(20000330)16:5<387::aid-yea525<3.0.co;2-u
Khan, I. A., Ning, G., Liu, X., Feng, X., Lin, F., and Lu, J. (2015). Mitochondrial fission protein MoFis1 mediates conidiation and is required for full virulence of the rice blast fungus Magnaporthe oryzae. Microbiol. Res. 178, 51–58. doi: 10.1016/j.micres.2015.06.002
Kitagaki, H., and Takagi, H. (2014). Mitochondrial metabolism and stress response of yeast: applications in fermentation technologies. J. Biosci. Bioeng. 117, 383–393. doi: 10.1016/j.jbiosc.2013.09.011
Kretschmer, M., Klose, J., and Kronstad, J. W. (2012). Defects in mitochondrial and peroxisomal beta-oxidation influence virulence in the maize pathogen Ustilago maydis. Eukaryot. Cell 11, 1055–1066. doi: 10.1128/ec.00129-12
Kubler, E., Mosch, H. U., Rupp, S., and Lisanti, M. P. (1997). Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272, 20321–20323. doi: 10.1074/jbc.272.33.20321
Lambert, N. A. (2008). Dissociation of heterotrimeric g proteins in cells. Sci. Signal. 1:re5. doi: 10.1126/scisignal.125re5
Liu, S., and Dean, R. A. (1997). G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant Microbe Interact 10, 1075–1086. doi: 10.1094/mpmi.1997.10.9.1075
Liu, S., Peng, G., and Xia, Y. (2012). The adenylate cyclase gene MaAC is required for virulence and multi-stress tolerance of Metarhizium acridum. BMC Microbiol 12:163. doi: 10.1186/1471-2180-12-163
Liu, X., Qian, B., Gao, C., Huang, S., Cai, Y., Zhang, H., et al. (2016). The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol. Plant Microbe Interact 29, 496–507. doi: 10.1094/MPMI-11-15-0259-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lord, J. C. (2005). From metchnikoff to monsanto and beyond: the path of microbial control. J. Invertebr. Pathol. 89, 19–29. doi: 10.1016/j.jip.2005.04.006
Lorin, S., Dufour, E., and Sainsard-Chanet, A. (2006). Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim. Biophys. Acta 1757, 604–610. doi: 10.1016/j.bbabio.2006.03.005
Lyssand, J. S., and Bajjalieh, S. M. (2007). The heterotrimeric G protein subunit G alpha i is present on mitochondria. FEBS Lett. 581, 5765–5768. doi: 10.1016/j.febslet.2007.11.044
Marrari, Y., Crouthamel, M., Irannejad, R., and Wedegaertner, P. B. (2007). Assembly and trafficking of heterotrimeric G proteins. Biochemistry 46, 7665–7677. doi: 10.1021/bi700338m
McIntire, W. E. (2009). Structural determinants involved in the formation and activation of G protein betagamma dimers. Neurosignals 17, 82–99. doi: 10.1159/000186692
Meng, H., Wang, Z., Wang, Y., Zhu, H., and Huang, B. (2017). Dicer and argonaute genes involved in RNA interference in the entomopathogenic fungus Metarhizium robertsii. Appl. Environ. Microbiol. 83:e03230-16. doi: 10.1128/AEM.03230-16
Michaelson, D., Ahearn, I., Bergo, M., Young, S., and Philips, M. (2002). Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol. Biol. Cell 13, 3294–3302. doi: 10.1091/mbc.e02-02-0095
Muniz-Paredes, F., Miranda-Hernandez, F., and Loera, O. (2017). Production of conidia by entomopathogenic fungi: from inoculants to final quality tests. World J. Microbiol. Biotechnol. 33:57. doi: 10.1007/s11274-017-2229-2
Nair, R. R., Kiran, A., and Saini, D. K. (2017). G protein signaling, journeys beyond the plasma membrane. J. Indian Inst. Sci. 97, 95–108. doi: 10.1007/s41745-016-0012-2
Neer, E. J. (1995). Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80, 249–257. doi: 10.1016/0092-8674(95)90407-7
Ortiz-Urquiza, A., and Keyhani, N. O. (2015). Stress response signaling and virulence: insights from entomopathogenic fungi. Curr. Genet. 61, 239–249. doi: 10.1007/s00294-014-0439-9
Regenfelder, E., Spellig, T., Hartmann, A., Lauenstein, S., Bolker, M., and Kahmann, R. (1997). G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16, 1934–1942. doi: 10.1093/emboj/16.8.1934
Robishaw, J. D., and Berlot, C. H. (2004). Translating G protein subunit diversity into functional specificity. Curr. Opin. Cell Biol. 16, 206–209. doi: 10.1016/j.ceb.2004.02.007
Simon, M. I., Strathmann, M. P., and Gautam, N. (1991). Diversity of G proteins in signal transduction. Science 252, 802–808. doi: 10.1126/science.1902986
Slessareva, J. E., and Dohlman, H. G. (2006). G protein signaling in yeast: new components, new connections, new compartments. Science 314, 1412–1413. doi: 10.1126/science.1134041
Tong, Y., Wu, H., Liu, Z., Wang, Z., and Huang, B. (2020). G-protein subunit Gαi in mitochondria, MrGPA1, affects conidiation, stress resistance, and virulence of entomopathogenic fungus Metarhizium robertsii. Preprint (Version 1). Available online at: https://doi.org/10.21203/rs.2.23319/v1 accessed(February 12, 2020).
Wang, C., and Wang, S. (2017). Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 62, 73–90. doi: 10.1146/annurev-ento-031616-035509
Wang, Y., Wang, T., Qiao, L., Zhu, J., Fan, J., Zhang, T., et al. (2017). DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 101, 4215–4226. doi: 10.1007/s00253-017-8197-5
Wang, Z., Jiang, Y., Li, Y., Feng, J., and Huang, B. (2019). MrArk1, an actin-regulating kinase gene, is required for endocytosis and involved in sustaining conidiation capacity and virulence in Metarhizium robertsii. Appl. Microbiol. Biotechnol. 103, 4859–4868. doi: 10.1007/s00253-019-09836-6
Wang, Z. X., Zhou, X. Z., Meng, H. M., Liu, Y. J., Zhou, Q., and Huang, B. (2014). Comparative transcriptomic analysis of the heat stress response in the filamentous fungus Metarhizium anisopliae using RNA-Seq. Appl. Microbiol. Biotechnol. 98, 5589–5597. doi: 10.1007/s00253-014-5763-y
Wedegaertner, P. B. (2012). G protein trafficking. Subcell. Biochem. 63, 193–223. doi: 10.1007/978-94-007-4765-4_11
Yang, Q., and Borkovich, K. A. (1999). Mutational activation of a Galphai causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151, 107–117.
Yao, S.-L., Ying, S.-H., Feng, M.-G., and Hatting, J. L. (2010). In vitro and in vivo responses of fungal biocontrol agents to gradient doses of UV-B and UV-A irradiation. BioControl 55, 413–422. doi: 10.1007/s10526-009-9265-2
Ying, S. H., and Feng, M. G. (2011). A conidial protein (CP15) of Beauveria bassiana contributes to the conidial tolerance of the entomopathogenic fungus to thermal and oxidative stresses. Appl. Microbiol. Biotechnol. 90, 1711–1720. doi: 10.1007/s00253-011-3205-7
Ying, S. H., Feng, M. G., and Keyhani, N. O. (2013). A carbon responsive G-protein coupled receptor modulates broad developmental and genetic networks in the entomopathogenic fungus, Beauveria bassiana. Environ. Microbiol. 15, 2902–2921. doi: 10.1111/1462-2920.12169
Zeng, G., Chen, X., Zhang, X., Zhang, Q., Xu, C., Mi, W., et al. (2017). Genome-wide identification of pathogenicity, conidiation and colony sectorization genes in Metarhizium robertsii. Environ. Microbiol. 19, 3896–3908. doi: 10.1111/1462-2920.13777
Zhang, H., Wang, M., Wang, W., Li, D., Huang, Q., Wang, Y., et al. (2012). Silencing of G proteins uncovers diversified plant responses when challenged by three elicitors in Nicotiana benthamiana. Plant Cell Environ. 35, 72–85. doi: 10.1111/j.1365-3040.2011.02417.x
Zhang, L. B., and Feng, M. G. (2018). Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl. Microbiol. Biotechnol. 102, 4995–5004. doi: 10.1007/s00253-018-9033-2
Keywords: Metarhizium robertsii, Gαi protein, appressorium, conidiation, virulence
Citation: Tong Y, Wu H, Liu Z, Wang Z and Huang B (2020) G-Protein Subunit Gαi in Mitochondria, MrGPA1, Affects Conidiation, Stress Resistance, and Virulence of Entomopathogenic Fungus Metarhizium robertsii. Front. Microbiol. 11:1251. doi: 10.3389/fmicb.2020.01251
Received: 08 March 2020; Accepted: 15 May 2020;
Published: 16 June 2020.
Edited by:
Yuxian Xia, Chongqing University, ChinaReviewed by:
Nancy Keller, University of Wisconsin-Madison, United StatesMengyao Niu, University of Wisconsin-Madison, United States, in collaboration with reviewer NK
Jinkui Yang, Yunnan University, China
Zhibing Luo, Southwest University, China
Copyright © 2020 Tong, Wu, Liu, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Huang, Ymh1YW5nQGFoYXUuZWR1LmNu
†These authors have contributed equally to this work
 Youmin Tong1†
Youmin Tong1† Bo Huang
Bo Huang