- Department of Microbiology and Immunology, University of Miami Miller School of Medicine, Miami, FL, United States
CexE is a 12 kDa protein that was originally reported to be present in just three strains of enterotoxigenic Escherichia coli (ETEC); a frequent cause of diarrheal illnesses worldwide. However, an examination of sequenced genomes has revealed that CexE is actually present in a majority of ETEC strains. In addition, homologs of CexE are present in enteroaggregative E. coli (EAEC), Yersinia enterocolitica, Providencia alcalifaciens, and Citrobacter rodentium. Although it has been hypothesized that CexE and its homologs are virulence factors, this has yet to be tested. Thus the primary aim of this study was to determine if these proteins contribute to pathogenicity. Our secondary aim was determine if they are secreted coat proteins. Here we report that all neonatal mice infected with a wild-type strain of C. rodentium perished. In contrast a cexE mutant was significantly attenuated with 45% neonate survival. In adult mice the wild-type strain reached significantly higher loads in the large intestines and were shed in greater numbers than cexE mutants. Secretion of the CexE homolog in EAEC is dependent upon an atypical Type I secretion system that accepts its client from the periplasm rather than the cytoplasm. Insertion mutants of cexC, the putative ATPase of the CexE secretion system, were attenuated in our murine model. In vitro we found that CexC is required for the secretion of CexE to the outer membranes of both ETEC and C. rodentium. Secretion is not constitutive because CexE accumulates in the periplasm when the two pathogens are cultured under noninducing conditions. Although secretion conditions differ between ETEC and C. rodentium, secreted CexE remains predominantly associated with the outer membranes of both species. In aggregate these findings demonstrate that CexE is a secreted coat protein and virulence factor that promotes colonization of host intestinal tissues by enteric pathogens.
Introduction
Approximately half a million infants and children perish from diarrheal diseases every year (World Health Organization, 2017). For citizens of low–income countries diarrheal diseases are projected to remain among the ten leading causes of death through 2030 (Mathers and Loncar, 2006). Of the many viral and bacterial pathogens that cause diarrhea enterotoxigenic Escherichia coli (ETEC) is one of the most frequent causes with at least 280 million people sickened annually; most in low-income nations (Qadri et al., 2005). ETEC causes diarrhea by elaborating enterotoxins that disrupt the normal flow of ions across the apical membranes of enterocytes. This disruption leads to a net loss of water into the lumen of the intestinal tract resulting in profuse watery diarrhea (Fleckenstein et al., 2010). In some instances ETEC induced diarrhea results in severe dehydration that can result in death if not adequately addressed.
Enterotoxins are the penultimate cause of diarrhea but ETEC must also colonize the gastrointestinal tract to cause disease. Colonization is itself dependent upon the expression of pili that function as adherence factors facilitating the direct attachment of ETEC to enterocytes (Evans et al., 1978; Kernéis et al., 1992). Although ETEC are heterogeneous and there are numerous antigenically distinct pilus types, many are positively regulated at the level of transcription by the ETEC virulence regulator Rns (CfaD) (Caron et al., 1989; Caron and Scott, 1990; Bodero et al., 2008; Bodero and Munson, 2016). In vitro DNase I footprinting studies have shown that each Rns activated pilin promoter contains a Rns DNA binding site immediately upstream of the promoter’s -35 hexamer (Munson and Scott, 1999; Bodero et al., 2008; Bodero and Munson, 2016). In most cases the promoter proximal binding site is usually accompanied by one or more additional Rns binding sites further upstream. However site directed mutagenesis studies have shown that occupancy of the promoter proximal site has the greatest contribution to promoter activation (Munson and Scott, 1999; Bodero and Munson, 2016).
An in silico analysis of the genome of ETEC strain H10407 -a strain that has been used in several human challenge studies- led to the identification of two Rns binding sites upstream of cexE (Dupont et al., 1971; Evans et al., 1978; Satterwhite et al., 1978; Pilonieta et al., 2007; Crossman et al., 2010). As with pilin promoters, one of the binding sites was located immediately upstream of the -35 hexamer of cexEp and the other further upstream. As expected from the positions of its binding sites, Rns was found to activate the expression of cexE. Although cexE does not encode a pilin nor is it required for the expression of pili, its regulation by the virulence regulator Rns implies that cexE encodes a virulence factor.
As in ETEC, the expression of CexE homologs in Citrobacter rodentium and enteroaggregative E. coli (EAEC) is positively regulated by virulence regulators with homology to Rns (Sheikh et al., 2002; Hart et al., 2008). In C. rodentium cexECr is under the control of RegA while AggR regulates the expression of dispersin, the homolog of CexE in EAEC (Sheikh et al., 2002; Nishi et al., 2003; Hart et al., 2008). Of the three only dispersin has been characterized beyond the level of expression. Dispersin is a secreted protein that associates with the external face of the outer membrane and its NMR structure suggests that it associates with the outer membrane via electrostatic interactions (Velarde et al., 2007). Dispersin has further been shown to decrease adherence to biotic and abiotic surfaces, and reduce EAEC aggregation (Sheikh et al., 2002). Collectively these effects result in greater dispersal of EAEC in vitro. Whether or not these effects contribute to pathogenesis is unknown because to date there have been no in vivo studies of these putative virulence factors. This is partly due to the fact that ETEC and EAEC do not naturally infect small laboratory animals. However, C. rodentium is natural murine pathogen and a well-developed model for enteric infections (Mundy et al., 2005). Therefore, we used murine C. rodentium challenge studies to interrogate the function of CexE in vivo.
Materials and Methods
Bacterial Growth Conditions
Bacteria were cultured aerobically at 37°C in CFA medium (Evans et al., 1977), Lysogeny Broth (LB; HIMEDIA), or Iscove’s Modified Dulbecco’s Medium (IMDM; Thermo Fisher Scientific). For hypoxic growth bacteria were cultured in sealed CyroELITETM cyrogenic vials (Wheaton) at 37°C in IMDM. Growth media were supplemented with 50 μg/ml kanamycin, 50 μg/ml hygromycin, or 100 μg/ml ampicillin as appropriate.
Bacterial Strains and Plasmids
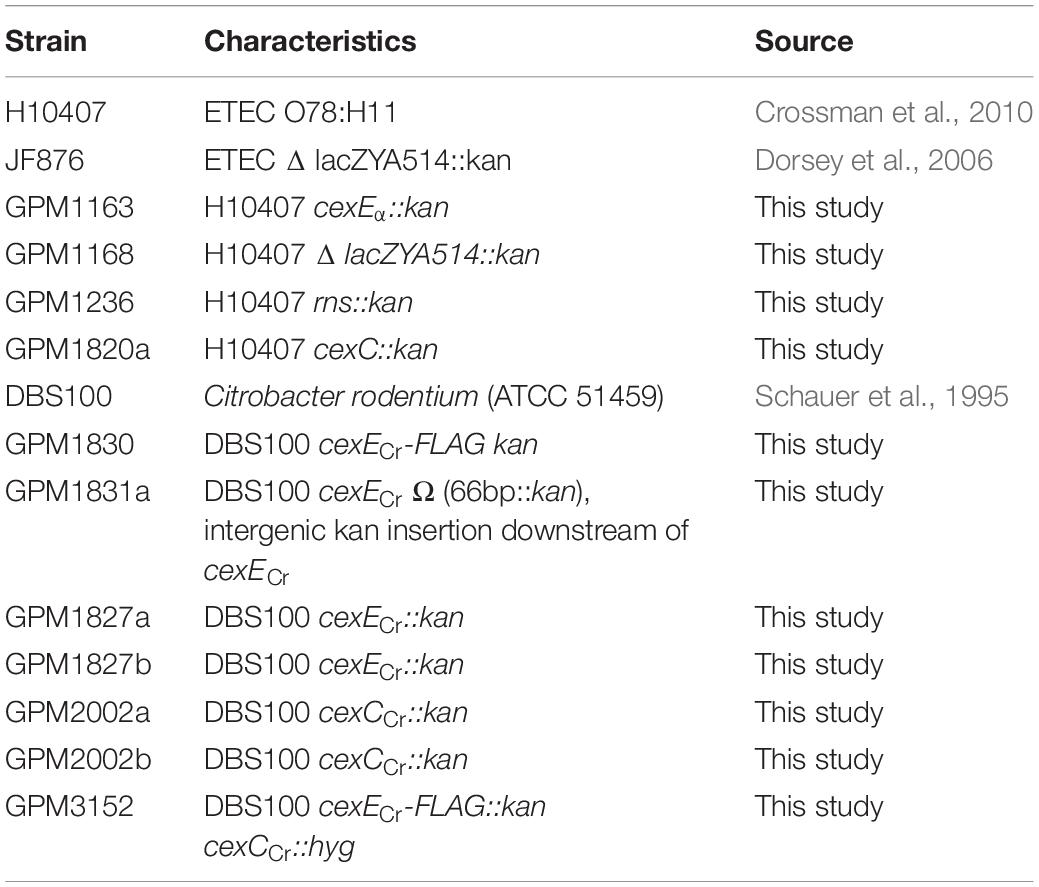
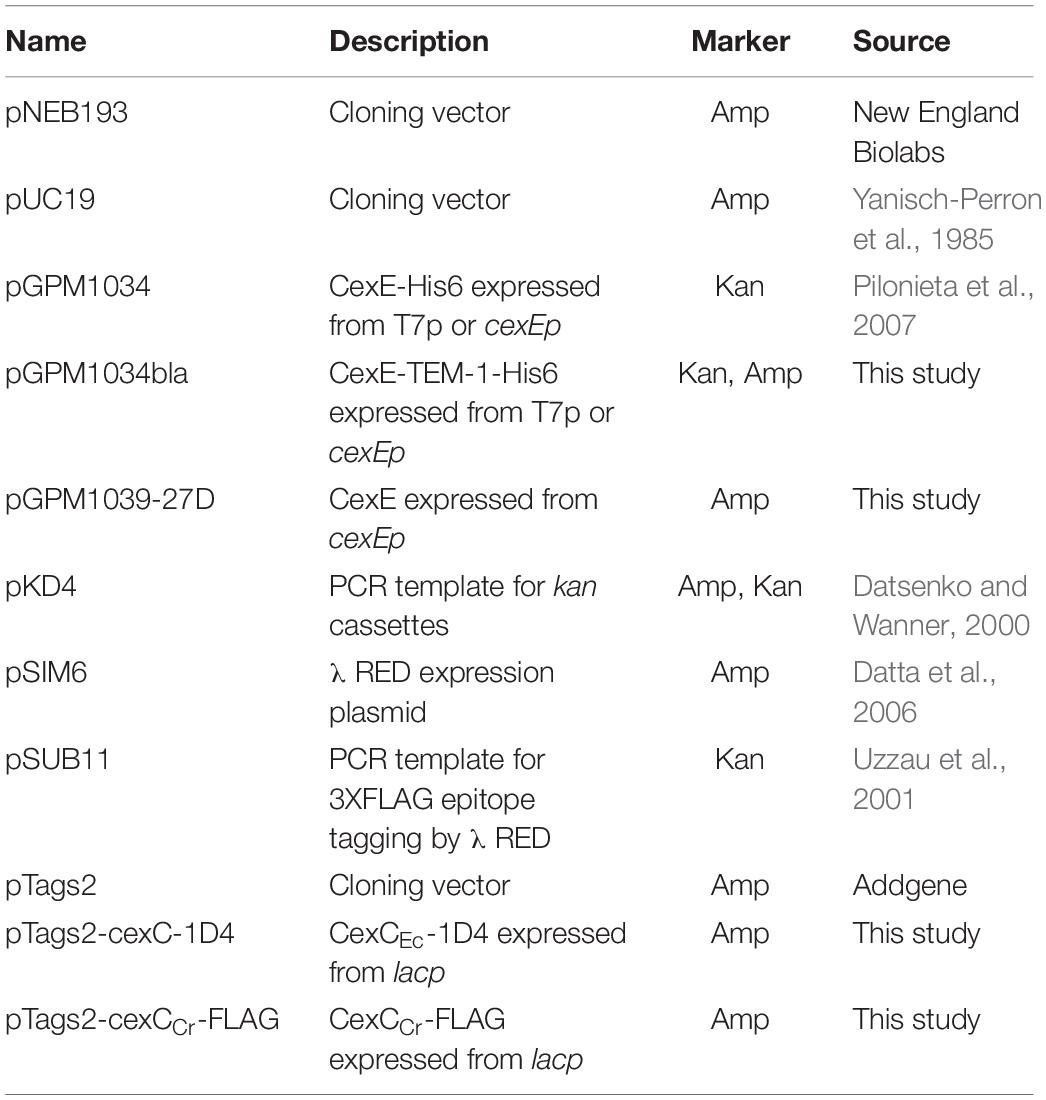
Strains, plasmids, and primer sequences are listed in Tables 1–3 respectively. Plasmid pGPM1034bla expresses a CexE-TEM-1 fusion protein from cexEp or T7p. It was constructed by amplification of the β-lactamase gene from pUC19 with oligonucleotide primer pair 1126/1127. The PCR product was digested with XhoI then ligated into the same site of pGPM1034. Plasmid pGPM1039-27D expresses CexE from cexEp. It was constructed by amplification of the cexE gene from 394/478. The PCR product was digested with BamHI and XbaI then ligated into the previously digested pNEB193 vector. HiFi DNA assembly was used to construct plasmid pTags2-cexC-1D4 and pTags2-cexCCr-FLAG which expresses CexC-1D4 (∼24 kDa) and CexCCr-FLAG (∼27 kDa), respectively, from lacp. For cexC the vector backbone was amplified from pTags2 with primers 1533/1534. CexC was amplified from H10407 with primers 1531/1532. For cexCCr the vector backbone was amplified from pTags2 with primers 1415/1416. CexCCr was amplified from DBS100 with primers 2142/2143. The PCR products were assembled with NEB HiFi. λRED mediated recombineering was used for the construction of the following strains as previously described (Datsenko and Wanner, 2000; Datta et al., 2006). Kanamycin resistance cassettes targeting cfaD and cexEα were amplified from pKD4 with primer pairs 608/625 and 622/623, respectively. A cassette for the disruption of the lac operon was amplified from JF876 with primers 108/518. Electroporation of the cassettes into H10407 / pSIM6 resulted in recombinants GPM1236, GPM1163, and GPM1168, respectively. Cassettes for epitope tagging, disruption, or insertion downstream of cexECr were amplified from pSUB11 with primer pairs 1178/1179, 1177/1179, and 1193/1179, respectively. Electroporation of the cassettes into DBS100/pSIM6 resulted in recombinants GPM1830, GPM1827, and GPM1831a, respectively. Cassettes for disrupting cexCCr were amplified from pSUB11 with primer pair 1673/74 and pTags2 with primer pair 2136/37. Electroporation into DBS100 / pSIM6 resulted in recombinant GPM2002 and GPM3152, respectively. Recombinants were cured of λRED expression plasmids by passage at 42°C. Insertions were verified by PCR with primers flanking the insertion sites.
Quantitative Bacterial Adherence Assays
HCT-8 cells, a human ileocecal epithelial cell line, was purchased from the American Type Culture Collection (ATCC CCL-244). HCT-8 cells were grown in IMDM supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. Approximately 24 h before infection HCT-8 cells were seeded in 24-well tissue culture plates at ca. 105 cells per well. Bacteria were cultured in CFA medium to an OD600 of 0.7–0.8. Bacteria were collected from 500 μl of culture by centrifugation then resuspended in 1 ml IMDM, 10% FBS. The tissue culture wells were aspirated, washed once with PBS, then 200 μl IMDM, 10% FBS was added to each well followed by 50 μl of the bacterial suspension for an MOI of ca. 50. After a 1 h attachment period culture medium was aspirated and wells were washed four times with PBS to remove nonadherent bacteria. Adherent bacteria were collected in PBS, 0.1% (vol/vol) Triton X-100 and enumerated by serial dilution and plating. To control for differences between inocula adherent CFUs were normalized to inocula.
Trichloroacetic Acid (TCA) Precipitation of Culture Supernatants
H10407/pNEB193 was cultured in either CFA or IMDM to stationary phase. Bacteria were removed from the culture medium by centrifugation and successive filtration through 0.45 and 0.20 micron syringe tip filters. TCA was added to the clarified supernatants at a final concentration of 10% (vol/vol). Proteins were precipitated overnight at 4°C then pelleted for 8 min. at 21,000 × g. Protein pellets were washed twice with 500 μl ice-cold acetone to remove residual TCA, air-dried for 15 min. at room temperature, then resuspended in 60 μl of 1x SDS-PAGE loading buffer with 100 mM β-mercaptoethanol.
Proteinase K Digests
Bacteria were collected from 400 μl of stationary phase cultures by centrifugation at 21,000 g for 3 min. and resuspended in 75 μl of either PBS or PBS, 10 mM EDTA, 1% (vol/vol) Triton X-100. Suspensions were equilibrated at 37°C for 1 h prior to addition of proteinase K to a final concentration of 200 μg/ml. Some samples received 11 mM phenylmethylsulfonyl fluoride (PMSF) prior to enzyme addition. All samples were incubated for 1 h at 37°C with continuous gentle rocking. Enzyme activity was terminated by addition of PMSF followed by 25 μl of 4x SDS-PAGE loading buffer with β-mercaptoethanol.
Immunoblots
Whole-cell lysates were subject to SDS-PAGE, transferred to PVDF membranes, and blocked in TBS-Blotto (25 mM TrisCl pH 7.6, 150 mM NaCl, 5% (wt/vol) powdered nonfat milk. Antibodies against CexEα were produced by immunization of rabbits (Proteintech Group, Inc.) with purified CexEα -His6 and used at a dilution of 1:5000 in TBS-Blotto with 0.05% vol./vol. Tween20 (Pilonieta et al., 2007). Anti-DnaK (ab69617) and anti-β-lactamase (ab12251) antibodies were purchased from AbCam and used at dilutions of 1:10,000. Anti-FLAG (F1804) was purchased from Sigma-Aldrich and used at a dilution of 1:10,000 HRP conjugated goat anti-rabbit (sc-2030) and goat anti-mouse (115-036-062) antibodies were purchased from Santa Cruz Biotechnology and Jackson ImmunoResearch Laboratories, respectively. Secondary antibodies were used at a dilutions of 1:10,000. Chemiluminescence was detected with an Odyssey FC Imaging System (LI-COR Biosciences). Densitometry analysis was performed utilizing ImageJ software.
In vivo Studies
All mice were bred and housed under barrier conditions in the Division of Veterinary Resources of the University of Miami Miller School of Medicine. Mice were regularly screened for specific common pathogens. C57Bl/6 mice were inoculated orogastrically with C. rodentium strains in PBS. Adults were inoculated with a 22-gauge, round-tipped feeding needle. Infant mice were used at 15 days of age and inoculated with PE-10 tubing (polyethylene tubing with an outside diameter of 0.61 mm) attached to a 30-gauge needle. Administered CFUs were determined by serial dilution and plating of inocula. Infant mice were weaned at 3½ weeks of age. On select days fecal pellets were collected from adult mice and homogenized in sterile ddH2O, diluted, and plated on MacConkey agar with kanamycin. Organs of adult mice were homogenized in PBS using a OMNI International Tissue Homogenizer (Kennesaw, GA, United States) for 2 min at medium speed. Organs from infant mice were homogenized in HBSS using a Seward Biomaster 80 Stomacher (Brinkman, Westbury, NY, United States) for 4 min. at high speed. Homogenates were diluted and plated on MacConkey agar plates with kanamycin. CFUs were normalized to the weight of fecal pellets and organs.
Histopathology Scoring
The distal 1 cm of the colons of infected mice were collected and fixed in 10% neutral buffered formalin. The fixed distal colons were embedded in paraffin, sectioned into three equal parts, and stained with hematoxylin and eosin. Total surface area of the sections were blindly scored twice using light microscopy by evaluation of submucosal edema, PMN infiltration, and epithelial necrosis. Briefly, pathology was scored as WNL (0, <1% of surface area affected), mild (1, <5% surface area affected), moderate (2, 5–10% surface area affected), severe (3, 10–40% surface area affected), and extensive (4, >40% surface area affected). Samples scored differently amongst rankings with a difference of 1 were averaged. No samples differed by more than one rank between the duplicate scorings. This method was adapted from (Yu et al., 2017).
Growth Curves
To test if the kanamycin insertion into the cexE or cexC genes disrupts bacteria we constructed growth curves of WT bacteria and compared them to their respective mutants. Due to the variation in bacterial size biomass indicators such as optical density can be misleading (Swinnen et al., 2004). Because of this we used viable count measurements via serial dilution and plating to accurately determine population lag times. Fifty microliter baffled flasks of IMDM were inoculated with bacteria and grown over at 37°C with agitation at 200 rpm. The bacteria were then diluted to an OD600 of 0.05 in new baffled flasks with fresh IMDM. Approximately every 30 min samples were taken and enumerated by serial dilution and plating. Growth curves were obtained by plotting CFU/ml against time. Slopes of the bacterial growth curve were analyzed by nonlinear regression to generate a semilog line.
Statistical Analysis
One-way ANOVA with Tukey’s multiple comparisons test, Student’s t-test, Mann-Whitney U-test, nonlinear regression, and Mantel-Cox log rank test. (GraphPad Prism Version 8.1.2 was utilized for statistical analysis).
Ethics Statement
All animal experiments were approved by and performed in accordance with the University of Miami Institutional Animal Care and Use Committee guidelines.
Results
CexE Is Broadly Distributed Across ETEC Strains
CexE was initially identified as a member of the Rns regulon by an in silico search of the genome of ETEC strain H10407 for Rns binding sites (Pilonieta et al., 2007). In the intervening years over 200 ETEC genomes have been sequenced and deposited in NCBI’s databases; most as whole genome shotgun (WGS) sequences. To gauge the distribution of CexE across ETEC strains we ran iterative BLASTN searches to identify cexE homologs. After each search the most distal member was used as the query for a subsequent search until no additional members were found. BLASTP searches were not effective because automated annotation pipelines apparently have a high failure rate with regards to the identification of cexE alleles. The reasons for this are unclear as these genes are open reading frames of ca. 360 bp; most with recognizable Shine-Dalgarno sequences upstream of ATG start codons.
Of the 250 strains that were identified as ETEC –either explicitly in the record definition lines or by the presence of genes encoding the heat-labile (LT) and/or heat-stable (ST) enterotoxins– 76% were found to harbor cexE alleles. The prevalence of cexE alleles is similar to the genes encoding LT and ST which are present in 64 and 78% of ETEC strains, respectively. The 190 alleles of cexE encode eleven distinct polypeptides (Figure 1). Hereafter these will be distinguished from each other by Greek letter subscripts starting with CexEα that was first identified by Pilonieta et al. (2007). Protein sequences and multi-sequence alignment are presented in Supplementary Figures S1, S2. Each polypeptide is likely transported to the periplasm via the general secretory pathway because each contains a predicted signal peptide at its amino terminus (Pugsley, 1993; Bendtsen et al., 2004). For CexEα, this prediction has been confirmed experimentally (Pilonieta et al., 2007).
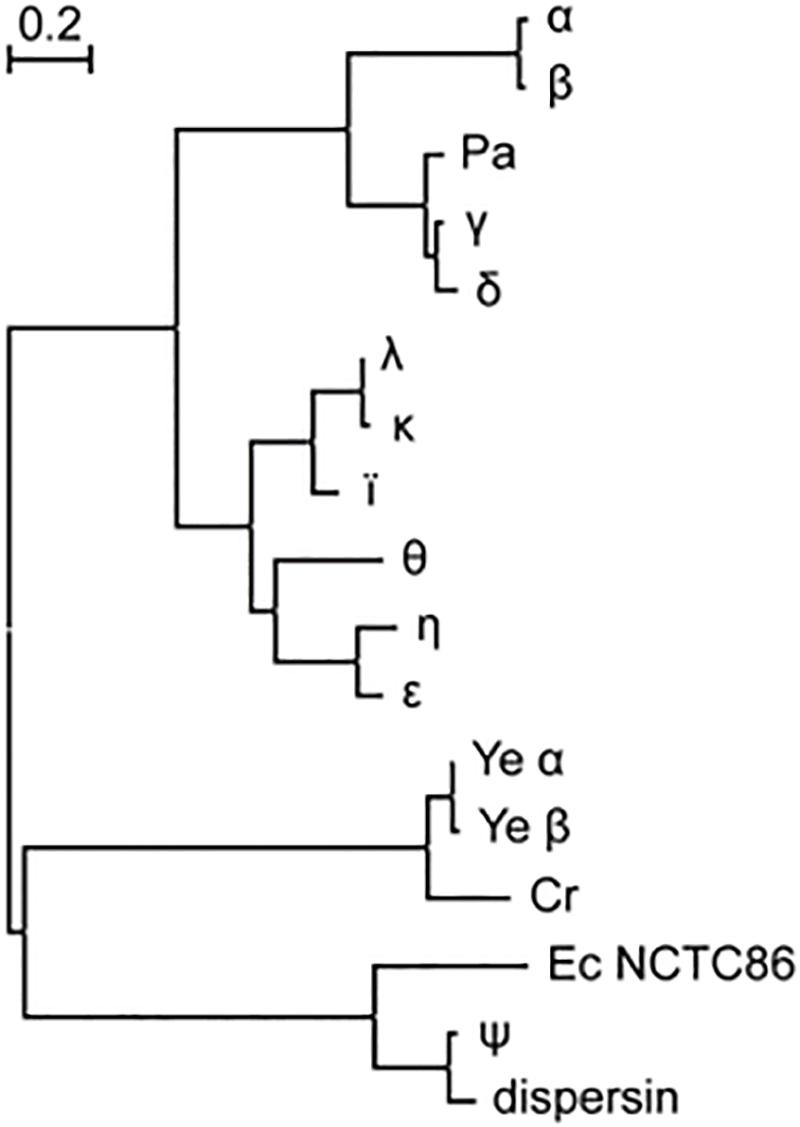
Figure 1. CexE homologs are prevalent amongst ETEC and a subset of other enteric pathogens. Proteins from ETEC strains are designated with a single Greek letter. dispersin is from EAEC strain 042. Ec NCTC86 is the strain of E. coli that was originally isolated by Theodor Escherich in 1885. The phylogram was constructed using the maximum likelihood method on a multiple sequence alignment generated by the MUSCLE algorithm; both of which were run within the MEGA7 software package (22). Cr, C. rodentium; Pa, P. alcalifaciens; Ye, Y. enterocolitica.
With the lone exception of CexEψ all ETEC variants of CexE cluster within the same clade. Within the CexEψ clade are two homologs that were found in 60% of the 162 Yersinia enterocolitica strains in NCBI’s WGS databases. In contrast to the situation in ETEC there is little variation between Y. enterocolitica strains; 93% of the polypeptides are identical to Yeα found in a strain of Y. enterocolitica (ERL04757) that was isolated from human blood (Reuter et al., 2014). Other species of Yersinia apparently lack CexE homologs. A CexE homolog is also present in the two complete genomes of Citrobacter rodentium as well as in one isolate of Providencia alcalifaciens. The former is a murine enteric pathogen while the latter has been described as an emerging enteric pathogen of humans (Schauer and Falkow, 1993; Murata et al., 2001; Yoh et al., 2005). Notably the P. alcalifaciens homolog resides within a clade of ETEC proteins while the Y. enterocolitica and C. rodentium homologs are more closely related to dispersin of enteroaggregative E. coli (EAEC) (Figure 1). Also within the dispersin clade is CexEψ from ETEC as well as a polypeptide from E. coli strain NCTC86. The latter was isolated by Theodor Escherich in 1885 as he sought to identify the cause of neonatal dysentery (Méric et al., 2016; Khetrapal et al., 2017).
CexEα Is a Conditional Coat Protein in ETEC
Like CexEα, dispersin of EAEC has an amino terminal signal peptide and enters the periplasm through the general secretory pathway (Pilonieta et al., 2007). Subsequently dispersin crosses the outer membrane but remains associated with the cell as a coat protein bound by electrostatic interactions (Sheikh et al., 2002; Velarde et al., 2007). To determine if CexEα and CexECr are also coat proteins proteinase K was used as a membrane impermeable probe of their subcellular distribution. For these experiments we also chose culture media that resulted in moderate to robust expression of the target proteins. When ETEC was cultured in IMDM CexEα associated with intact and permeabilized cells was digested by the protease with equivalent efficiency (Figure 2A, bottom). This indicates that the majority of CexEα is deposited on the outer membrane and that there is little to no intracellular pool. In addition the amount of cell-free protein was negligible as it was below detection even after TCA precipitation of IMDM supernatants (Figure 3A). However, the translocation of CexEα across the outer membrane is conditional because it remained intracellular when ETEC was cultured in CFA medium (Figure 2A, top).
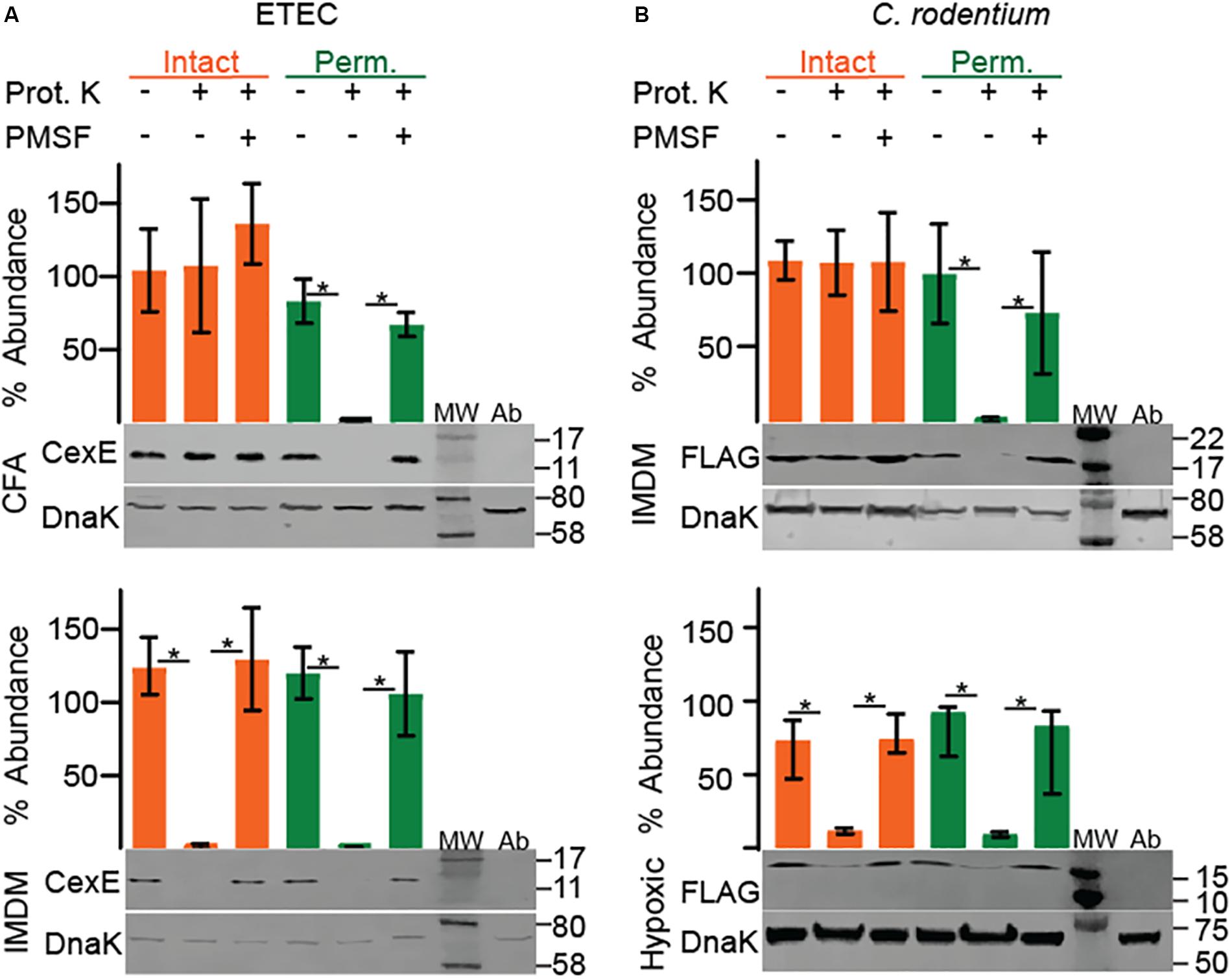
Figure 2. Culture medium and oxygen levels influence the subcellular distribution of CexE. Intact cells that present CexE on their outer membranes render it sensitive to proteinase K digestion as determined by Western blots and quantification. Cells that require permeabilization for digestion of CexE have failed to transport it across the outer membrane. (A) Permeabilization was required for the digestion of CexEα when ETEC H10407 was grown in CFA but not IMDM. (B) FLAG tagged CexECr was retained within the envelope of C. rodentium when GPM1830 was cultured aerobically in IMDM but not when cultured in IMDM under hypoxic conditions. Representative blots are shown. Quantification of CexE digestion relative to DnaK. For quantification n = 3, *P < 0.05 by Student’s t-test. Prot. K, proteinase K; MW, molecular weight ladder; Ab, antibody specificity controls. Antibody specificity control lanes were loaded with whole cell lysates of ETEC strain GPM1163 (cexEα::kan) or C. rodentium strain DBS100 which lacks the FLAG epitope.
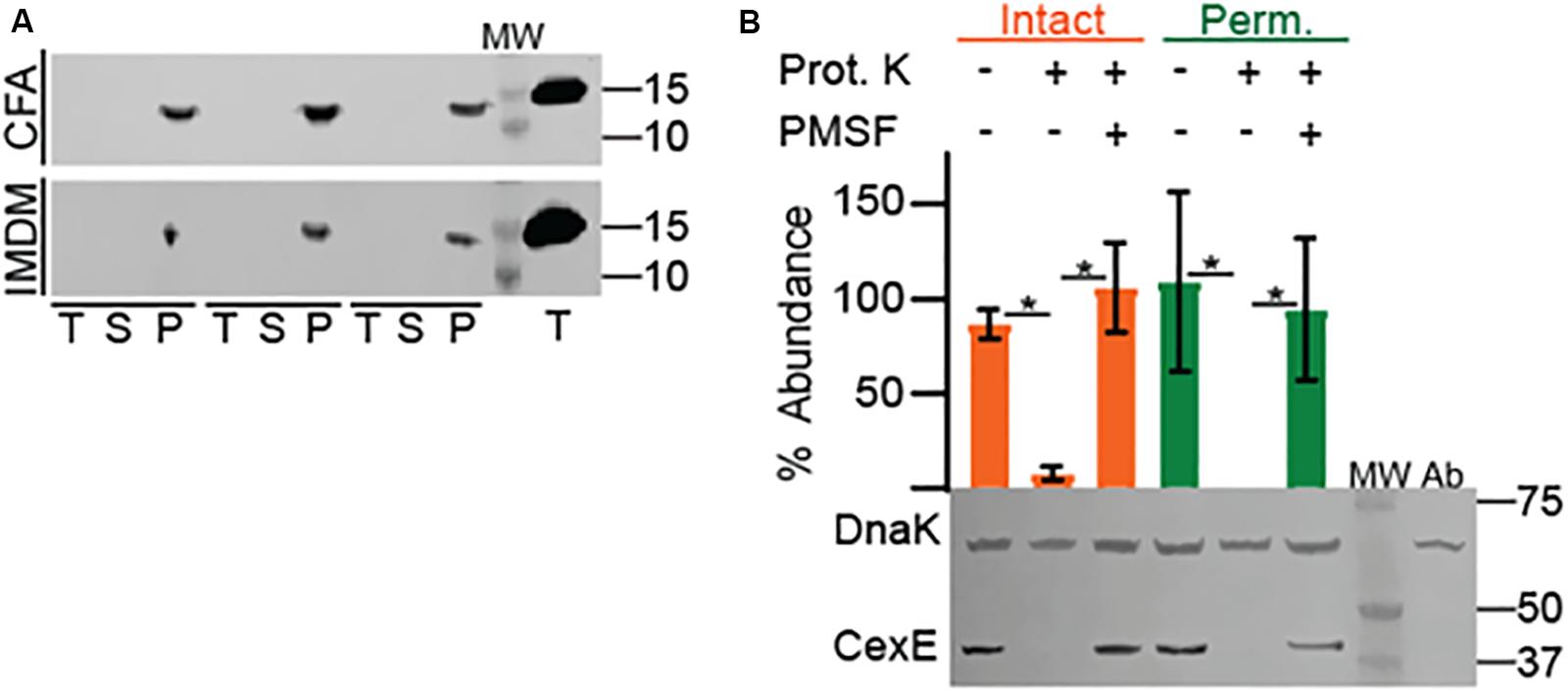
Figure 3. Association of native CexEα and a 43 kDa CexEα -TEM-1 fusion protein with the outer membrane of ETEC. (A) H10407 was cultured in either CFA or IMDM. Cell pellets (P), culture supernatants (S), and TCA precipitates (T) probed for CexEα. The far-right lane contains the TCA precipitate of 10ng of purified CexEα -H6. (B) Translocation of CexEα -TEM-1 (β-lactamase) to the surface of ETEC cultured in IMDM. Representative Western blot of whole cell lysates of ETEC strain GPM1163 (cexEα ::kan)/pGPM1034bla probed with anti-CexE and DnaK antibodies. Quantification of CexEα -TEM-1 digestion relative to DnaK. For quantification n = 3, *P < 0.0001 by Student’s t-test. Prot. K, proteinase K; MW, molecular weight ladder; Ab, antibody specificity control with whole cell lysates of GPM1163 lacking pGPM1034bla.
In contrast, CexECr-FLAG was not translocated across the outer membrane when C. rodentium was cultured in IMDM or LB (Figure 2B, top and Supplementary Figure S3). It is unlikely that the <3 kDa FLAG epitope interfered with the translocation of CexECr because fusion of TEM-1 –a 30 kDa protein– to the carboxy terminus of CexEα did not inhibit its translocation (Figure 3B). We also did not observe secretion of CexECr-FLAG when C. rodentium was cultured in CFA with 45 mM NaHCO3, or LB with 45 mM NaHCO3 in the presence or absence of 4 mM bile salts. Culturing on LB agar plates also failed to induce secretion. Likewise CexECr-FLAG was not secreted in M9 minimal media supplemented with 0.4% glucose or glycerol, DMEM, or RPMI with GlutaMAXTM. Varying the temperature from 30° to 42°C also had no effect. We also unsuccessfully attempted to induce CexECr secretion using unconventional methods such as culturing C. rodentium in homogenates of C57Bl/6 intestinal tissue and/or feces to mimic in vivo conditions. However when C. rodentium was cultured under hypoxic conditions CexECr associated with intact cells was efficiently digested (Figure 2B bottom). Thus, reduced oxygen levels –as would be expected in lumen of the large intestine– induces the secretion of CexECr to the outer membrane. These results also demonstrate that the secretion of CexE is not linked to its expression. Rather secretion is regulated and responsive to environmental cues and/or growth conditions. Although secretion conditions differ between ETEC and C. rodentium, secreted CexE remains predominantly associated with the outer membranes of both species.
CexEα and CexECr Do Not Significantly Modulate Adherence
Since CexEα is a coat protein we sought to determine if it affects adherence. Prior to doing so we determined that the growth rates of ETEC and C. rodentium cexE mutants were equivalent to the WT parent strain of each (Supplementary Figures S4A–C). In adherence assays we observed that a cexEα mutant was more adherent to HCT-8 cells than WT (Figure 4A). However, the difference was insignificant and in stark contrast to the effects of an insertion in rns that abolishes expression of both CexEα and the adhesive CFA/I pilus. These results suggest that CexEα does not significantly modulate the adherence of ETEC to HCT-8 cells even when the pathogen is cultured in IMDM; a condition in which CexEα coats the outer membrane. The difference between WT C. rodentium and a cexECr mutant was also insignificant (Figure 4B).
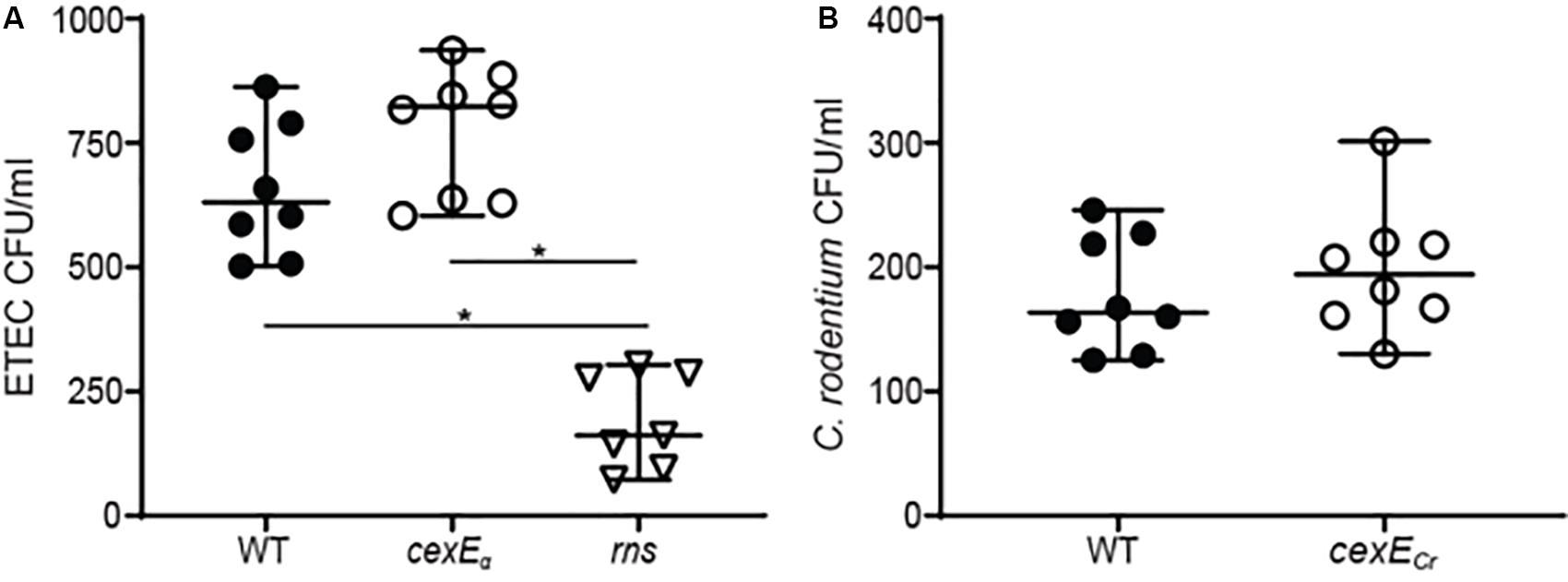
Figure 4. CexEα and CexECr do not significantly modulate adherence to mammalian cells. Adherent bacteria were enumerated after a 1 h infection of HCT-8 cells in IMDM 10% fetal bovine serum. (A) The difference between GPM1168 (WT ETEC) and a GPM1163 (cexEα ::kan) was not statistically significant. In contrast, GPM1236 (rns::kan) was significantly impaired in its ability to adhere to HCT-8 cells. (B) The difference between GPM1831a (WT C. rodentium) and GPM1827a (cexECr::kan) was not statistically significant. Graphs display median and 95% CI. Symbols represent samples collected from separate wells. *P < 0.0001 by Tukey’s multiple comparisons test.
CexC Is Necessary for the Secretion of CexE
In EAEC the secretion of dispersin is dependent on the Aat secretion system encoded by aatPABCD (Nishi et al., 2003) and homologs of each gene are present in both ETEC and C. rodentium. Although aatPABCD are contiguous in the prototypical EAEC strain 042 (Nishi et al., 2003), in both ETEC and C. rodentium cexPABC and cexD are always separated by at least several KBs and in some strains they occur on separate virulence plasmids. To test if CexE secretion is dependent on the putative Cex secretion system we chose to disrupt cexC since insertions in the terminal gene of the cexPABC operon would not have polar effects. As expected secretion of CexE was abolished in cexC mutants of both ETEC and C. rodentium because CexE was not digested by proteinase K unless their outer membranes were chemically permeabilized (Figures 5A,B). Complementation of the cexC mutants restored translocation of CexE across the outer membranes of both pathogens but vector controls did not (Figures 5C–F). Thus, CexC –the putative ATPase of the Cex secretion system– is required for the secretion of CexE in both ETEC and C. rodetium; as is the case for AatC and dispersin of EAEC (Nishi et al., 2003).
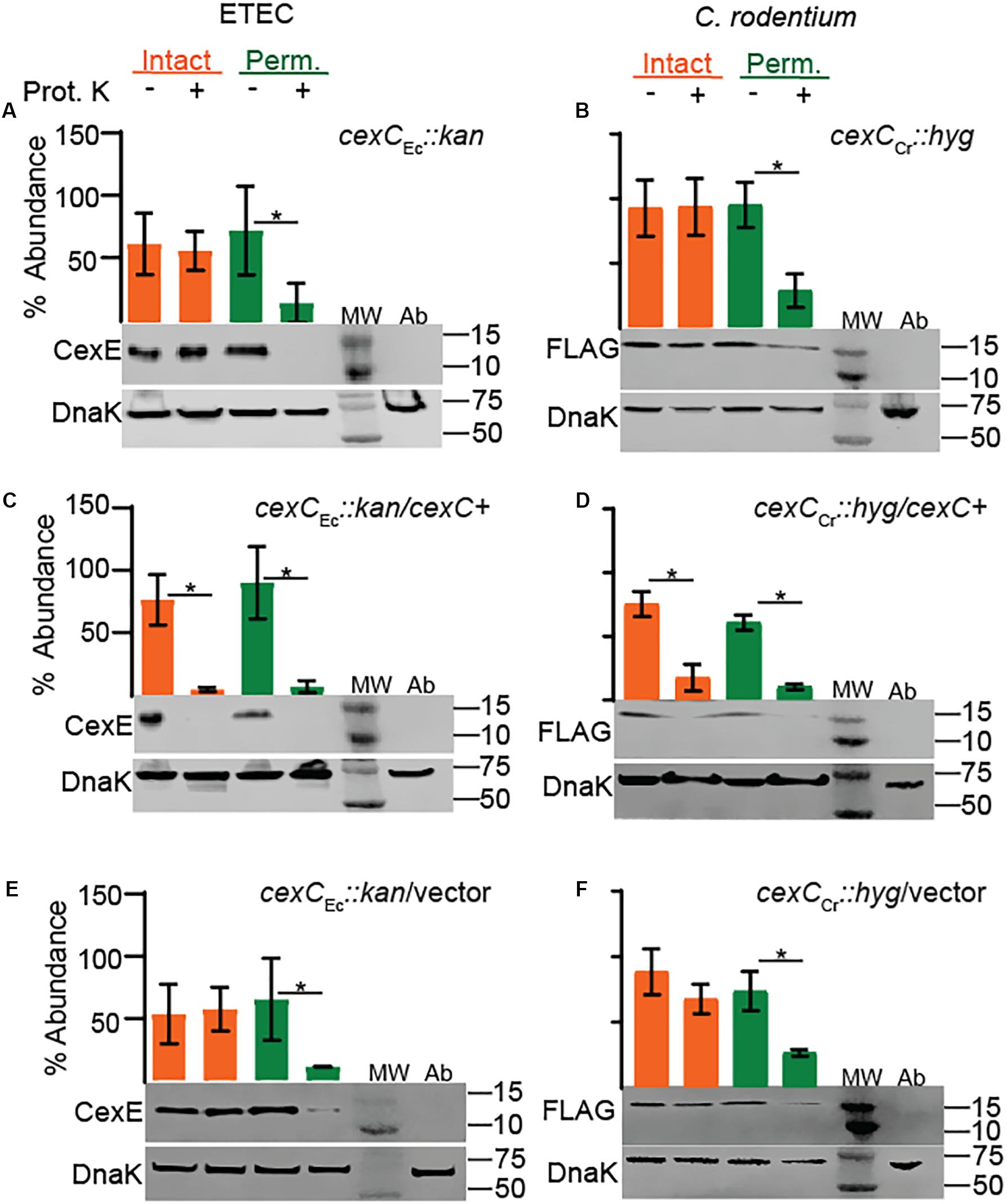
Figure 5. CexC is required for the secretion of CexE in both ETEC and C. rodentium. CexE secretion is abolished in cexC mutants of (A) ETEC strain GPM1820a and (B) C. rodentium strain GPM3152. Complementation with CexC expression plasmids (C) pCexC-1D4 or (D) pCexC-FLAG restored secretion of CexE to the outer membrane of both pathogens. CexCCr-FLAG is not visible because it is approximately 27 kDa. (E,F) As expected the vector control pTags2 failed to restore secretion. Quantification of CexEα digestion relative to DnaK. For quantification n = 3–4, *P < 0.05 by Student’s t-test. Prot. K, proteinase K; MW, molecular weight ladder; Ab, antibody specific controls. Antibody specificity control lanes were loaded with whole cells lysates of ETEC strain GPM1163 (cexEα ::kan) or C. rodentium strain DBS100 which lacks the FLAG epitope.
CexECr Is a Secreted Virulence Factor in vivo
To determine whether or not CexE is a virulence factor we used C. rodentium for in vivo studies because it is a natural murine pathogen; unlike EAEC and ETEC. We orogastrically inoculated C57BL/6 mice with C. rodentium strains GPM1831a –which carries a kanamycin cassette in an intergenic region downstream of cexE and was considered our WT strain– or GPM1827a (cexECr::kan). We inoculated a third group of mice with GPM2002a (cexCCr::kan) to evaluate if CexECr is secreted in vivo. As gauged by fecal shedding on days three and six post-inoculation, the three strains of bacteria colonized the mice equally well (Figure 6A). However the amount of shed WT bacteria continued to increase while that of the mutants plateaued, resulting in significant differences between WT and mutant strains on day nine (Figure 6A). Not surprisingly, day nine fecal shedding was mirrored by the intestinal loads of WT and mutant strains on day ten; particularly in the large intestine where C. rodentium preferentially colonizes (Figures 6B–E; Silberger et al., 2017). The results were reproduced in another infection experiment, with an independent cexECr mutant (GPM1827b) (Supplementary Figures S5A–C). We also observed and scored the pathology of the distal large intestine and found that that WT bacteria were associated with more intestinal pathology than either mutant (Figures 6F,G and Supplementary Figure S5D).
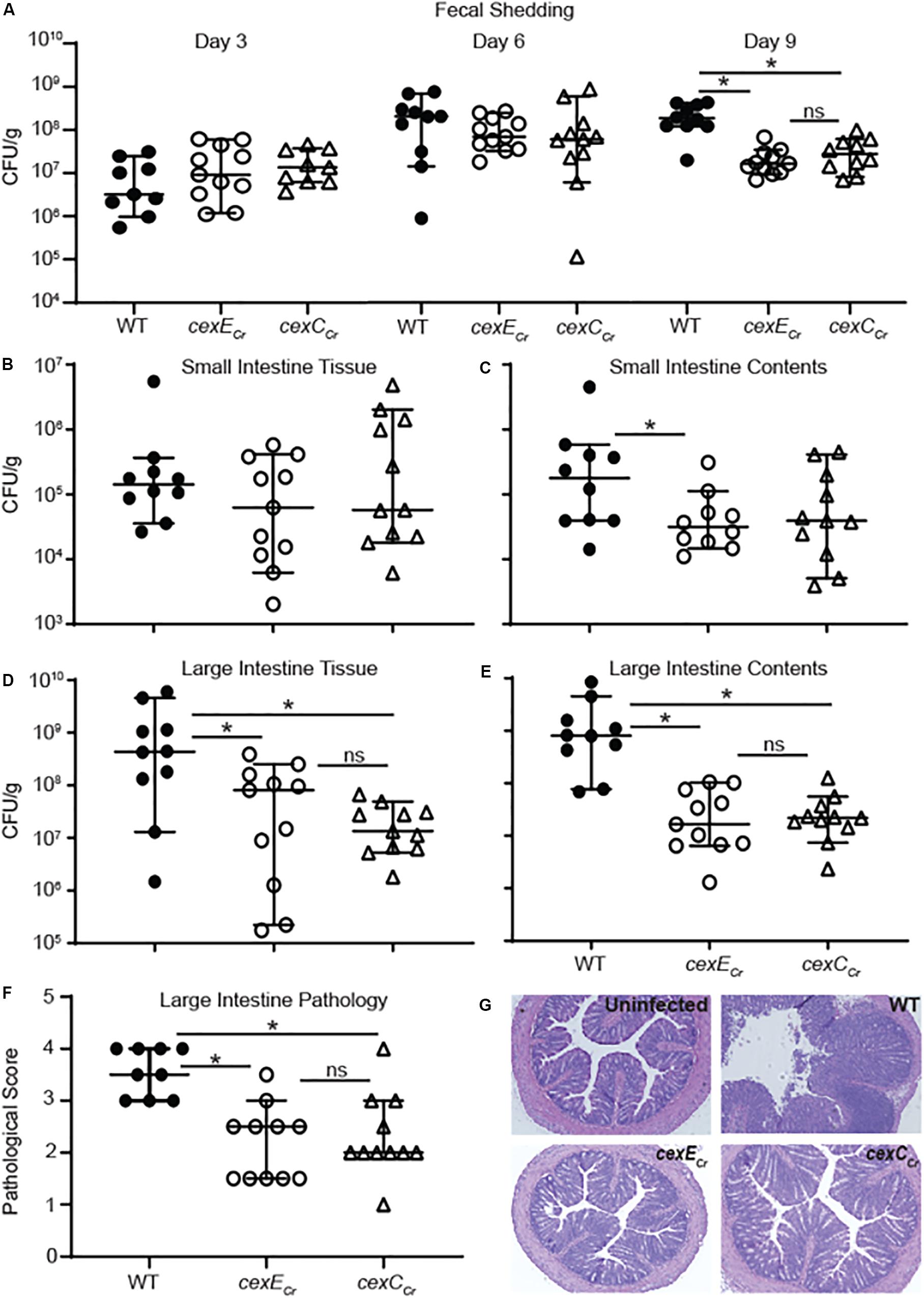
Figure 6. Intestinal and fecal loads of mice infected with WT C. rodentium and cex mutants. C57BL/6 mice were orogastrically inoculated with 108 CFU of either GPM1831a (WT C. rodentium), GPM1827a (cexECr::kan), or GPM2002a (cexCCr::kan). (A) Fecal pellets were collected on the days indicated post-inoculation. (B–E) Intestinal tissue and contents were harvested 10 days post-inoculation. CFUs were normalized to sample mass. (F,G) Histological scoring and representative images of H&E-stained colon sections at day 10. Medians and 95% CI are shown. n = 9–10 mice per group, *P < 0.05 by Mann-Whitney U-test.
Across all measurements the differences between the cexECr and cexCCr mutants were statistically insignificant (Figure 6). This suggest that CexCCr is required for secretion of CexECr in vivo and is consistent with our in vitro results (Figure 5). A separate experiment with an independent cexCCr mutant (GPM2002b) replicated the day 9 fecal shedding results accompanied by a trend of fewer mutant bacteria in the large intestine and less pathology than the WT strain (Supplementary Figure S6). Over the course of these infections mice were also monitored for weight loss. Consistent with previous C. rodentium murine studies, we observed only mild and transient weight loss (Bhinder et al., 2013). We further disaggregated the data based on the sex of the animals and found no trends to suggest a gender-bias in infections (data not shown). Additionally these results were not restricted to C57BL/6 mice as similar results were observed when 129X1/SvJ mice were used (Supplementary Figure S7).
Our results demonstrating that CexECr contributes the pathogenicity of C. rodentium were further confirmed in an infant mouse model (Figure 7). For technical reasons the small intestines were not analyzed but the loads of both the WT and mutant strain in the large intestine increased over the course of the experiment (Figure 7A). However, the loads of WT strain significantly exceeded those of the cexECr mutant on each of the three sampling days (Figure 7A). Although immunocompetent adult mice are able to clear C. rodentium infections, all infant mice infected with the WT strain perished (Figure 7B; Diaz et al., 2017). In contrast a significant number of the mice infected with the cexECr strain survived for the duration of the experiment. In aggregate these results demonstrate that CexECr is a secreted virulence factor of C. rodentium that results in higher intestinal burdens and increased virulence in both adult and infant mice.
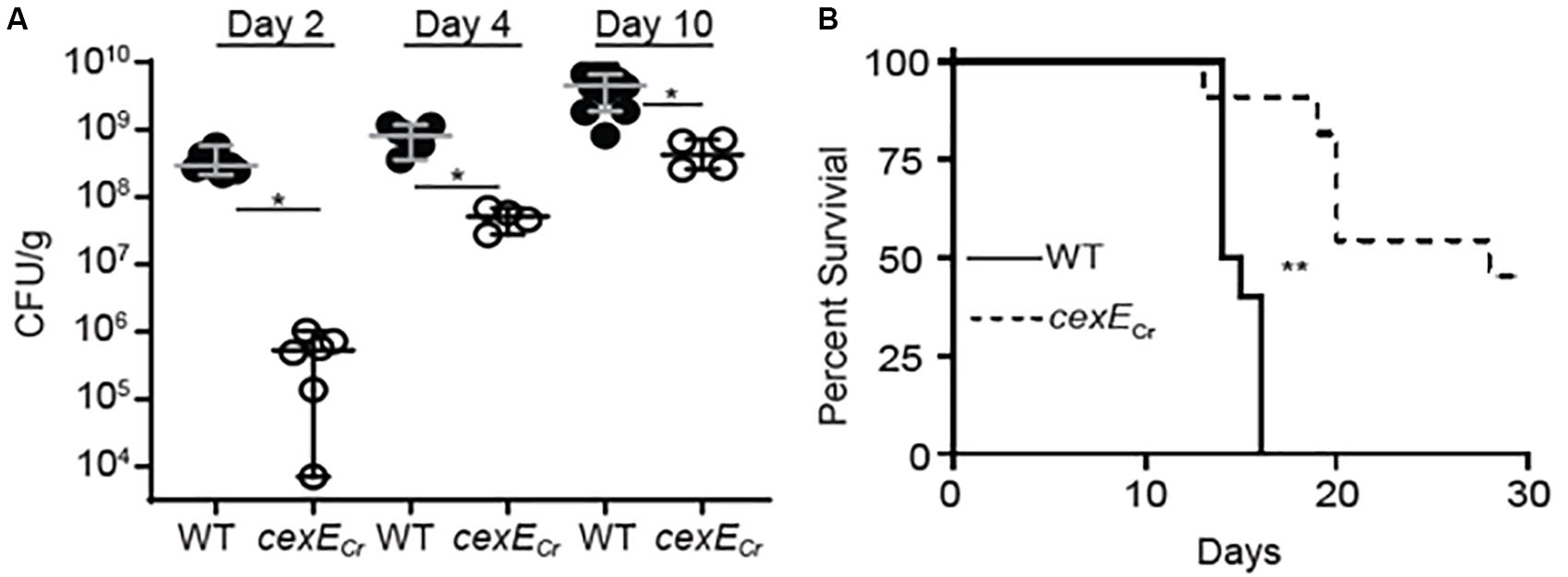
Figure 7. C. rodentium cexECr mutants are attenuated in neonatal mice. Fifteen-day old C57Bl/6 mice were infected with 107 CFU of GPM1831a (WT C. rodentium) or a GPM1827a (cexECr::kan) in groups of 12 and 6, respectively. (A) C. rodentium colon titers, normalized to tissue mass, were determined on the days indicated post-inoculation. Median and 95% CI are shown. n = 4–12 mice per group, *P < 0.05 by Mann-Whitney U-test. (B) In a separate experiment neonate survival was monitored out to 30 days post-inoculation. n = 10–11 animals per group, **P = 0.0005 by log rank (Mantel-Cox) test.
Discussion
We have found that a majority of ETEC strains harbor cexE alleles. Moreover, cexE homologs have been acquired by another pathovar of E. coli –EAEC– as well as three other species of enteric pathogens; P. alcalifaciens, Y. enterocolitica, and C. rodentium. For consistency with prior literature cexE homologs are referred to as dispersin when present in EAEC (Sheikh et al., 2002). These observations suggest a selective advantage for cexE and dispersin’s acquisition and maintenance. Previous studies with dispersin provide much of what is known about this family of proteins but have been limited to in vitro studies by the lack of a suitable animal model. In vitro dispersin coats the outer membrane of EAEC and has been proposed to promote dispersal by counteracting autoagglutination mediated by AAF fimbriae (Velarde et al., 2007). We have also found that CexE is a coat protein in both ETEC and C. rodentium. However the adherence of cexE mutants to mammalian HCT-8 cells was not significantly different than WT ETEC. The biological relevance of this result is uncertain and requires studies with additional cell lines, or ideally with intestinal tissues. We also did not observe significant differences in agglutination (data not shown) but note that dispersin’s pronounced effects were dependent on it is artificial overexpression from the strong T5 promoter (Sheikh et al., 2002).
All CexE variants and dispersin have predicted amino terminal signal peptides that would result in their transport to the periplasm via the general secretory pathway (Bendtsen et al., 2004). Such signal peptides are cleaved upon entry into the periplasm and cleavage of CexEα’s signal peptide has been experimentally verified (Pilonieta et al., 2007). This is apparently followed by a second translocation event that transports CexE to the external face of the outer membrane that is dependent upon by environmental cues and or growth conditions. For dispersin of EAEC this second translocation event requires five genes, aatPABCD (Nishi et al., 2003). Homologs of all five genes are also present in ETEC strains harboring cexE as well as C. rodentium, Y. enterocolitica, and P. alcalifaciens (unpublished observation). CexA/AatA likely form a conduit through the outer membrane because they are homologs of TolC; a protein that has been shown to form β-barrel pores in the outer membrane of E. coli and other gram-negative bacteria (Figure 8) (Koronakis et al., 2000, 2004). Type I secretion systems also utilize TolC or its homologs to transport client proteins (Wandersman and Delepelaire, 1990). However, such systems accept their clients from the cytoplasm and transport them in a single translocation event (Green and Mecsas, 2015). In contrast the secretion of both CexE and dispersin apparently involves two steps; one across the inner membrane through the SecYEG complex and a second across the outer membrane via their cognate secretion systems. CexB/AatB are predicted to be periplasmic proteins with amino-terminal signal peptides; although, some models suggest that their amino-termini are instead transmembrane helices (Tsirigos et al., 2015). CexP/AatP and CexD/AatD are predicted to be inner membrane proteins with significant extensions in the periplasm (Tsirigos et al., 2015). CexC/AatC are predicted to be cytoplasmic proteins and may energize translocation across the outer membrane because they contain the signatures of ATP binding cassettes (Nishi et al., 2003).
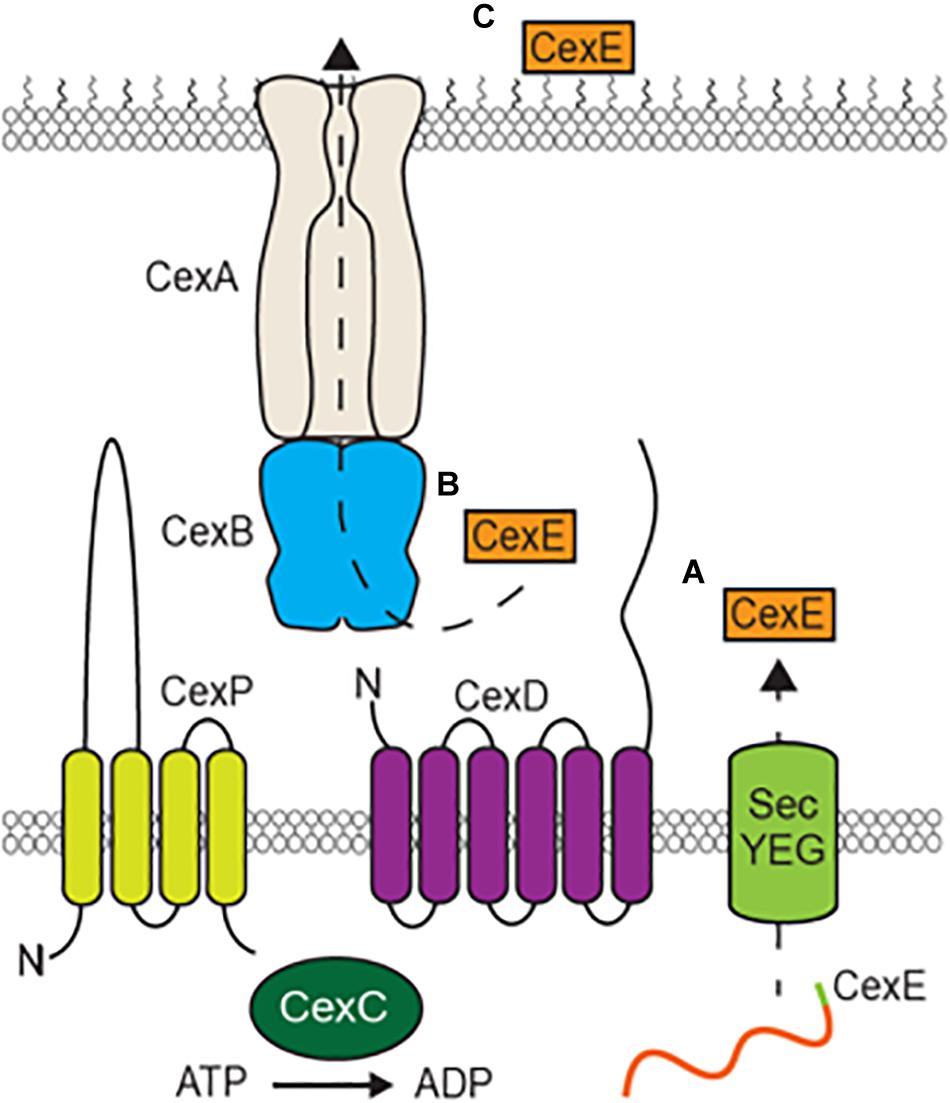
Figure 8. Model of CexE secretion by an atypical Type I secretion system. (A) The CexE polypeptide is transported from the cytoplasm to the periplasm by the SecYEG complex. (B) After cleavage of its signal peptide the CexPABCD secretion complex then transports CexE across the outer membrane (C) which it subsequently binds to.
As was previously observed with dispersin (Nishi et al., 2003), we found that translocation of CexE across the outer membrane was abolished in cexC mutants of both ETEC and C. rodentium. Cex-dependent secretion also takes place in vivo because we observed that cexECr and cexCCr mutants produced similar attenuated phenotypes. We also found that the secretion of CexE is not directly linked to its expression. Rather CexE accumulates in the periplasm unless the appropriate secretion conditions are met. The conditional secretion of CexE is also species specific since we observed that hypoxia triggers the secretion of CexE in C. rodentium but not ETEC. In either case it remains to be determined if conditional secretion is the result of differential regulation of the cex secretion and cexE genes or an actual sensing/regulatory mechanism within the secretory system.
After translocation across the outer membrane both CexE and dispersin remain associated with the bacterial envelope as coat proteins. Dispersin has been proposed to contribute to the pathogenicity of EAEC by facilitating the dispersal of the pathogen throughout the intestinal tract (Velarde et al., 2007). Although the dispersal model is an extrapolation of in vitro observations, it may explain the significantly higher loads of WT C. rodentium than the cexECr and cexCCr mutants that we observed in the large intestines of mice. The greater fecal shedding of the WT strain compared to the mutants is likely a consequence of these higher loads. Mechanistically it has been proposed that dispersin counteracts fimbriae mediated autoagglutination (Velarde et al., 2007). However, we did not observe similar effects with our ETEC cexE mutants and alternative mechanisms have not yet been excluded. In particular CexEα associates with ETEC outer membrane vesicles that have also been shown to facilitate the delivery of LT enterotoxin to the cytosol of mammalian cells (Horstman and Kuehn, 2000; Roy et al., 2011). Since CexEα colocalizes with LT in OMVs and OMVs deliver LT to host cells it is plausible that CexE also reaches host cells. Whether or not this contributes to pathogenesis remains to be determined. Nevertheless, our in vivo studies clearly demonstrate that CexE promotes colonization and/or survival of an enteric pathogen within the intestinal tract.
Data Availability Statement
The datasets generated for this study are available at NIH.FigShare.Com, https://doi.org/10.35092/yhjc.c.5015396.
Ethics Statement
The animal study was reviewed and approved by the University of Miami Institutional Animal Care and Use Committee.
Author Contributions
GM constructed phylogeny trees, analyzed sequence data, and obtained funding for the studies. ZR conducted in vitro adherence assays. ZR and KT conducted secretion assays and subsequent analyses. ZR, KT, LM, RM, and GM conducted the experiments with adult mice. BA did the experiments with neonatal mice. ZR, KT, and GM constructed the strains and plasmids. ZR, KT, LM, and GM designed the experiments and analyzed the data. ZR and GM wrote the manuscript with editorial assistance from KT, LM, and BA. All authors contributed to the article and approved the submitted version.
Funding
Research reported in this publication was supported by the NIAID of the National Institutes of Health under award number (AI128164 and AI110810). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01374/full#supplementary-material
References
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bhinder, G., Sham, H. P., Chan, J. M., Morampudi, V., Jacobson, K., and Vallance, B. A. (2013). The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. J. Vis. Exp. 19:e50222. doi: 10.3791/50222
Bodero, M. D., Harden, E. A., and Munson, G. P. (2008). Transcriptional regulation of subclass 5b fimbriae. BMC Microbiol. 8:180. doi: 10.1186/1471-2180-8-180
Bodero, M. D. R., and Munson, G. P. (2016). The virulence regulator rns activates the expression of CS14 Pili. Genes 7:120. doi: 10.3390/genes7120120
Caron, J., Coffield, L. M., and Scott, J. R. (1989). A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86, 963–967. doi: 10.1073/pnas.86.3.963
Caron, J., and Scott, J. R. (1990). A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 58, 874–878. doi: 10.1128/iai.58.4.874-878.1990
Crossman, L. C., Chaudhuri, R. R., Beatson, S. A., Wells, T. J., Desvaux, M., Cunningham, A. F., et al. (2010). A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192, 5822–5831. doi: 10.1128/JB.00710-10
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Datta, S., Costantino, N., and Court, D. L. (2006). A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115. doi: 10.1016/j.gene.2006.04.018
Diaz, L. A., Altman, N. H., Khan, W., Serhan, C. N., and Adkins, B. (2017). Specialized proresolving mediators rescue infant mice from lethal citrobacter rodentium infection and promote immunity against reinfection. Infect. Immun. 85:e0464-17. doi: 10.1128/IAI.00464-17
Dorsey, F. C., Fischer, J. F., and Fleckenstein, J. M. (2006). Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell. Microbiol. 8, 1516–1527. doi: 10.1111/j.1462-5822.2006.00736.x
Dupont, H. L., Formal, S. B., Hornick, R. B., Snyder, M. J., Libonati, J. P., Sheahan, D. G., et al. (1971). Pathogenesis of Escherichia coli Diarrhea. N. Engl. J. Med. 285, 1–9. doi: 10.1056/NEJM197107012850101
Evans, D. G., Evans, D. J., and Tjoa, W. (1977). Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 18, 330–337. doi: 10.1128/iai.18.2.330-337.1977
Evans, D. G., Satterwhite, T. K., Evans, D. J., DuPont, H. L., and DuPont, H. L. (1978). Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect. Immun. 19, 883–888. doi: 10.1128/iai.19.3.883-888.1978
Fleckenstein, J. M., Hardwidge, P. R., Munson, G. P., Rasko, D. A., Sommerfelt, H., and Steinsland, H. (2010). Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microb. Infect. 12, 89–98. doi: 10.1016/j.micinf.2009.10.002
Green, E. R., and Mecsas, J. (2015). Bacterial secretion systems – an overview. Am. Soc. Microbiol. 4, 1–32. doi: 10.1128/microbiolspec.VMBF-0012-2015
Hart, E., Yang, J., Tauschek, M., Kelly, M., Wakefield, M. J., Frankel, G., et al. (2008). RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 76, 5247–5256. doi: 10.1128/IAI.00770-08
Horstman, A. L., and Kuehn, M. J. (2000). Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275, 12489–12496. doi: 10.1074/JBC.275.17.12489
Kernéis, S., Chauvière, G., Darfeuille-Michaud, A., Aubel, D., Coconnier, M. H., Joly, B., et al. (1992). Expression of receptors for enterotoxigenic Escherichia coli during enterocytic differentiation of human polarized intestinal epithelial cells in culture. Infect. Immun. 60, 2572–2580. doi: 10.1128/iai.60.7.2572-2580.1992
Khetrapal, V., Mehershahi, K. S., and Chen, S. L. (2017). Complete genome sequence of the original Escherichia coli Isolate, strain NCTC86. Genome Announc. 5:e00243-17. doi: 10.1128/genomeA.00243-17
Koronakis, V., Eswaran, J., and Hughes, C. (2004). Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73, 467–489. doi: 10.1146/annurev.biochem.73.011303.074104
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. doi: 10.1038/35016007
Mathers, C. D., and Loncar, D. (2006). Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3:e442. doi: 10.1371/journal.pmed.0030442
Méric, G., Hitchings, M. D., Pascoe, B., and Sheppard, S. K. (2016). From Escherich to the Escherichia coli genome. Lancet. Infect. Dis. 16, 634–636. doi: 10.1016/S1473-3099(16)30066-4
Mundy, R., MacDonald, T. T., Dougan, G., Frankel, G., and Wiles, S. (2005). Citrobacter rodentium of mice and man. Cell. Microbiol. 7, 1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x
Munson, G. P., and Scott, J. R. (1999). Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181, 2110–2117. doi: 10.1128/jb.181.7.2110-2117.1999
Murata, T., Iida, T., Shiomi, Y., Tagomori, K., Akeda, Y., Yanagihara, I., et al. (2001). A large outbreak of foodborne infection attributed to Providencia alcalifaciens. J. Infect. Dis. 184, 1050–1055. doi: 10.1086/323458
Nishi, J., Sheikh, J., Mizuguchi, K., Luisi, B., Burland, V., Boutin, A., et al. (2003). The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 278, 45680–45689. doi: 10.1074/jbc.M306413200
Pilonieta, M. C., Bodero, M. D., and Munson, G. P. (2007). CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 189, 5060–5067. doi: 10.1128/JB.00131-07
Pugsley, A. P. (1993). The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57, 50–108. doi: 10.1128/mmbr.57.1.50-108.1993
Qadri, F., Svennerholm, A.-M., Faruque, A. S. G., and Sack, R. B. (2005). Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18, 465–483. doi: 10.1128/cmr.18.3.465-483.2005
Reuter, S., Connor, T. R., Barquist, L., Walker, D., Feltwell, T., Harris, S. R., et al. (2014). Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. U.S.A. 111, 6768–6773. doi: 10.1073/pnas.1317161111
Roy, K., Hamilton, D. J., Munson, G. P., and Fleckenstein, J. M. (2011). Outer membrane vesicles induce immune responses to virulence proteins and protect against colonization by enterotoxigenic Escherichia coli. Clin. Vaccine Immunol. 18, 1803–1808. doi: 10.1128/CVI.05217-11
Satterwhite, T. K., Evans, D. G., Dupont, H. L., and Evans, D. J. (1978). Role of Escherichia Coli colonisation factor antigen in acute diarrhśa. Lancet 2, 181–184. doi: 10.1016/S0140-6736(78)91921-9
Schauer, D. B., and Falkow, S. (1993). Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61, 2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993
Schauer, D. B., Zabel, B. A., Pedraza, I. F., O’Hara, C. M., Steigerwalt, A. G., and Brenner, D. J. (1995). Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J. Clin. Microbiol. 33, 2064–2068. doi: 10.1128/jcm.33.8.2064-2068.1995
Sheikh, J., Czeczulin, J. R., Harrington, S., Hicks, S., Henderson, I. R., Le Bouguénec, C., et al. (2002). A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Invest. 110, 1329–1337. doi: 10.1172/JCI16172
Silberger, D. J., Zindl, C. L., and Weaver, C. T. (2017). Citrobacter rodentium: a model enteropathogen for understanding the interplay of innate and adaptive components of type 3 immunity. Mucosal Immunol. 10, 1108–1117. doi: 10.1038/mi.2017.47
Swinnen, I. A. M., Bernaerts, K., Dens, E. J. J., Geeraerd, A. H., and Van Impe, J. F. (2004). Predictive modelling of the microbial lag phase: a review. Int. J. Food Microbiol. 94, 137–159. doi: 10.1016/j.ijfoodmicro.2004.01.006
Tsirigos, K. D., Peters, C., Shu, N., Käll, L., and Elofsson, A. (2015). The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407. doi: 10.1093/nar/gkv485
Uzzau, S., Figueroa-Bossi, N., Rubino, S., and Bossi, L. (2001). Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 98, 15264–15269. doi: 10.1073/pnas.261348198
Velarde, J. J., Varney, K. M., Inman, K. G., Farfan, M., Dudley, E., Fletcher, J., et al. (2007). Solution structure of the novel dispersin protein of enteroaggregative Escherichia coli. Mol. Microbiol. 66, 1123–1135. doi: 10.1111/j.1365-2958.2007.05985.x
Wandersman, C., and Delepelaire, P. (1990). TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. U.S.A. 87, 4776–4780. doi: 10.1073/pnas.87.12.4776
World Health Organization (2017). Diarrhoeal Disease. Available online at: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease (accessed March 20, 2020).
Yanisch-Perron, C., Vieira, J., and Messing, J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119. doi: 10.1016/0378-1119(85)90120-9
Yoh, M., Matsuyama, J., Ohnishi, M., Takagi, K., Miyagi, H., Mori, K., et al. (2005). Importance of Providencia species as a major cause of travellers’ diarrhoea. J. Med. Microbiol. 54, 1077–1082. doi: 10.1099/jmm.0.45846-0
Keywords: ETEC, C. rodentium, pathogenesis, virulence factors, CexE
Citation: Rivas ZP, Talbot KM, Merselis LC, McCormack RM, Adkins B and Munson GP (2020) CexE Is a Coat Protein and Virulence Factor of Diarrheagenic Pathogens. Front. Microbiol. 11:1374. doi: 10.3389/fmicb.2020.01374
Received: 21 January 2020; Accepted: 28 May 2020;
Published: 30 June 2020.
Edited by:
Leonard Peruski, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
James Paul Nataro, University of Virginia, United StatesFernando Navarro-Garcia, Centro de Investigación y Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), Mexico
James Michael Fleckenstein, Washington University in St. Louis, United States
Copyright © 2020 Rivas, Talbot, Merselis, McCormack, Adkins and Munson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George P. Munson, Z211bnNvbkBtaWFtaS5lZHU=
 Zachary P. Rivas
Zachary P. Rivas Kacey M. Talbot
Kacey M. Talbot Leidy C. Merselis
Leidy C. Merselis Ryan M. McCormack
Ryan M. McCormack Becky Adkins
Becky Adkins George P. Munson
George P. Munson