- 1Quorum Sensing Laboratory, Centre for Research in Infectious Diseases, School of Chemical and Biotechnology, SASTRA Deemed to be University, Thanjavur, India
- 2Faculty of Dentistry, The University of Hong Kong, Pok Fu Lam, Hong Kong
Vibrio cholerae, the etiological agent of cholera, employs quorum sensing (QS) pathways to control the expression of virulence factors, including the production of cholera toxin and biofilm formation. Acquired antibiotic resistance in V. cholerae draws attention to the development of novel therapeutics that counteract virulence, rather than the viability of the pathogen. In this context, we explored the anti-infective potential of rare marine Actinobacteria (RMA) from a mangrove ecosystem. Here, we report the effects of Micromonospora sp. RMA46 against V. cholerae in vitro. The RMA46 organic extract was non-bactericidal to V. cholerae cells and non-cytotoxic to macrophage RAW264.7 cell lines. RMA46 inhibited the formation of V. cholerae biofilms and downregulated the QS global switches LuxO and HapR, as well as other virulence genes including ct, tcp, and hapA. In silico molecular docking simulation of RMA46 ethyl acetate extract with LuxO and HapR revealed that 2-methoxy-4-vinylphenol and hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione could interact with the active sites of LuxO and HapR and potentially inhibit them. This study highlights Micromonospora sp. RMA46 as a potential source of anti-infectives against V. cholerae.
Introduction
Vibrio cholerae, a Gram-negative bacterium from the family Vibrionaceae, causes cholera, a self-limiting acute diarrheal disease (Zhu and Mekalanos, 2003). This contagious disease spreads primarily through the fecal–oral route and by consumption of contaminated food and water (Sack et al., 2004; Clemens et al., 2017). V. cholerae is classified into more than 200 serogroups on the basis of their lipopolysaccharide antigens, yet only two serogroups, O1 and O139, are associated with epidemic outbreaks of cholera (Wang et al., 2016). It is estimated that about 2.86 million people are affected by cholera in endemic countries, resulting in 95,000 deaths annually (Ali et al., 2015).
Antibiotic therapy is crucial for the treatment of cholera, as it abates the period of diarrhea, the amount of rehydration fluids, and the density of vibrios excreted in the stools (World Health Organization [WHO], 2019). Over the years, the rapid emergence of extensive drug-resistant (XDR) and multidrug-resistant (MDR) V. cholerae strains possess a great public health threat, owing to their association with treatment failure and potential dissemination of resistance among other pathogens through mobile genetic elements (MGEs) (Chowdhury et al., 2016; Clemens et al., 2017; Verma et al., 2019; Gladkikh et al., 2020). The above caveats compel the need for developing novel strategies to thwart V. cholerae infection. Numerous studies have focused on developing compounds that target other fitness tactics of bacteria instead of developing newer antibiotics (Hema et al., 2016, 2017).
V. cholerae demonstrates extraordinary fitness by coordinated group behavior in expressing virulence factors through quorum sensing (QS) pathways (Miller and Mekalanos, 1984; Hammer and Bassler, 2003). There are two major parallel QS systems in V. cholerae comprising cholera autoinducer-1 (CAI-1), autoinducer 2 (AI-2), and their corresponding cognate inner membrane receptors (CqsS and LuxP/Q) (Miller et al., 2002; Rutherford et al., 2011). These individual receptors converge the information to a global response regulator, LuxO, in response to the low and high concentrations of their respective signals. Under low signal strength (LSS), the receptors function as kinases and phosphorylate LuxO, which activates the transcription of quorum regulatory RNAs (Qrrl-4) and controls the translation of two master regulators, AphA and HapR. The qrr1-4 small RNAs upregulate the expression of AphA and concurrently repress the expression of HapR (Rutherford et al., 2011). In turn, AphA regulates the expression of biofilm activator (VpsT), toxin-coregulated pilus (TCP), and cholera toxin (CTX), commencing the process of adhesion and production of CTX in humans (Lenz et al., 2004; Yang et al., 2010; Rutherford et al., 2011). These processes are key to the colonization of this bacterium in the intestine and form microcolonies. By contrast, under high signal strength (HSS), the receptors function as phosphatases, leading to dephosphorylation and inactivation of LuxO. This results in the downregulation of virulence and biofilm formation by the expression of hemagglutinin/protease A (HapA). HapA expression facilitates the dissemination of bacteria from the intestinal cells. Thus, LSS promotes infection, whereas HSS promotes bacterial detachment from the intestinal cells, resulting in dissemination (Zhu et al., 2002; Hammer and Bassler, 2003).
Anti-virulence therapy using synthetic and natural molecules that interfere with QS pathways in pathogens has been explored extensively. These molecules function either as an inhibitor or an agonist of the factors involved in QS signaling, affecting their biosynthesis, detection, signal transduction, and even as enzymes that inactivated the QS signal molecules (Defoirdt, 2018). Such QS molecules that occur naturally have been sourced from various plants, microorganisms, and higher organisms from a wide range of terrestrial and marine ecosystems. Among microorganisms, the phylum Actinobacteria has been a prodigious source of anti-virulence agents, providing a range of chemicals that interfere with the QS system of various human pathogens (Sarveswari and Solomon, 2019). In this work, we isolated and screened 42 rare marine Actinobacteria (RMA) from mangrove soil against rugose V. cholerae HYR14 biofilm and virulence. The organic solvent of RMA displaying a significant anti-biofilm activity was also analyzed for its potential to inhibit virulence mechanism through gene expression studies. Finally, the potential interaction of the active principles identified in the organic extract of RMA with the V. cholerae global response regulators LuxO and HapR was analyzed through in silico docking studies.
Materials and Methods
Target Bacterial Strains and Culture Conditions
Vibrio cholerae biofilm-forming rugose strain, HYR14 obtained from NICED, India, was used throughout this study (Chowdhury et al., 2016). The strain was cultured in thiosulfate–citrate–bile salts–sucrose (TCBS) agar to confirm cell viability and propagated under standard growth conditions in Luria–Bertani (LB) broth at 37°C. An inoculum size of 0.2 (OD595) ≈ 105 cells were used for all the experiments (Hema et al., 2016).
Soil Sampling From Mangrove Ecosystem
Mangrove soil was sampled from Muthupet Mangrove Reserve, Tamil Nadu, India (Supplementary Table S1). Soil samples were collected using sterile containers from a site that was 20 m away from the shore and with 20 m depth. The samples were transported to the laboratory under cooling conditions and stored at 4°C in a dry condition prior to examination (Hong et al., 2009). The physio-chemical parameters such as salinity and temperature were measured using a handheld refractometer (Erma, Japan) during sample collection. The pH of the samples was determined using Eutech pH meter (Oakton, United States).
Pretreatment of Mangrove Soil and Selective Isolation of Rare Marine Actinobacteria
The soil was sieved through a large mesh to eliminate organic particles and was homogenized using a mortar and pestle. About 1 g of the homogenized samples was pretreated by 120°C for 60 min (dry heat) (Pisano et al., 1986) and by 1.5% of phenol for 30 min at 30°C (Pisano et al., 1986) and through moist heating with sterilized mangrove water at 50°C for 15 min (Takahashi, 2004). The pretreated soil samples were then serially diluted [1:10 (v/v) ratio] with sterile saline solution, and about 100 μl was spread plated on various isolation media. For selective isolation of RMA, International Streptomyces Project medium (ISP1 to ISP7) (Shirling and Gottlieb, 1966), actinomycetes isolation agar, starch casein agar, and humic acid–vitamin agar (Hayakawa and Nonomura, 1987) (HiMedia Laboratories, Mumbai, India) (Supplementary Table S2) supplemented with the filter-sterilized cycloheximide (40 μg/ml) and nalidixic acid (40 μg/ml) were used. The inoculated Petri plates were then incubated at 28°C for 4–6 weeks and regularly checked for the growth of actinomycetes-like colonies. Single discrete actinomycetes-like colonies grown on spread plated isolation media were re-subcultured on to ISP4 and actinomycetes isolation agar and further incubated at 28°C for the isolation of pure colonies (Shirling and Gottlieb, 1966). Collection of pure colonies of the RMA isolates was stored at 4°C on ISP4 agar slants and also as lyophilized cultures for short-term and long-term storages, respectively.
Cultural, Morphological, and Biochemical Characterization of Rare Marine Actinobacteria Isolates
The RMA isolates collected from various isolation media were subjected to morphological characterization and biochemical analysis (Pridham and Gottlieb, 1948; Shirling and Gottlieb, 1966). Light microscopy was used to observe the morphologies of Actinobacteria isolates including the arrangement of mycelium and formation of spores by Gram’s staining. The culture characteristics, including growth patterns on different media, pigmentation, colony color, presence, and color of aerial and substrate mycelium on various ISP media (ISP1 to ISP7), were determined.
Fermentation and Ethyl Acetate Extract Preparation
International Streptomyces Project medium no. 2 (ISP2) (0.5 g of peptone, 0.3 g of malt extract, 0.3 g of yeast extract, and 1.0 g of dextrose pH 6.2 ± 0.2) was used as fermentation medium for the exploration of the secondary metabolites produced by the 42 mangrove Actinobacteria isolates. The ISP2 medium was sterilized by autoclave at 121°C at 15 lb for 20 min. Initially, about four to five colonies from pure culture were inoculated into modified ISP2 media supplemented with 2.0 g/l of calcium carbonate in an Erlenmeyer conical flask and incubated at 150 rpm at 28°C for 7 days. After the incubation period, 5% (v/v) of well-grown culture from the calcium carbonate-supplemented ISP2 medium was inoculated into ISP2 media in an Erlenmeyer conical flask. The flask was incubated at 150 rpm at 28°C for 14 days. The uninoculated ISP2 medium served as a medium control, to ensure purity. After fermentation, the spent medium of the isolates was centrifuged at 5,000 rpm for 10 min, and the culture supernatant was filtered through a 0.2 μm nitrocellulose filter. The cell-free extracts were stored at -20°C. For the extraction of secondary metabolites by ethyl acetate, the cell-free filtrate was extracted twice with an equal volume of ethyl acetate, and the organic extract was condensed by evaporating ethyl acetate using a rotational evaporator (Heidolph Laborata, 4001) (Balasubramanian et al., 2017). The organic extract was resuspended in ethyl acetate and was used for further studies.
Efficacy of Rare Marine Actinobacteria Cell-Free Extract Against Vibrio cholerae HYR14 Biofilm
The antibiofilm activity of various dilutions of the 42 RMA cell-free extracts (50, 25, 12.50, 6.25, and 3.13%) was investigated. One hundred microliters (final volume) of LB broth containing various concentrations of RMA cell-free extract was aliquoted into a 96-well microtiter plate. Ten microliters of an overnight-grown culture of V. cholerae HYR14 strain that has been diluted up to 1:100 (OD595 = 0.2) was inoculated into each well, and the plates were incubated at 37°C for 24 h. Controls were included for all the experiments. After the incubation period, the plates were subjected to crystal violet staining. The medium and the planktonic cells present in the 96-well microtiter plates were aspirated at the end of the incubation period. The non-adherent HYR14 cells present in the wells were discarded by washing the wells thrice with sterile 1× phosphate-buffered saline. The microtiter plates were then air-dried for 20 min at ambient temperature to fix the adherent biofilms. To the dried wells, 100 μl of 0.2% (w/v) crystal violet was added and incubated for 20 min at room temperature. After the incubation period, the excess stain was washed off with sterile water twice, and the plates were air-dried completely. The bound crystal violet was solubilized by pipetting 100 μl of 33% glacial acetic acid into each well (Kaur et al., 2016). The absorbance of the wells was measured at 595 nm using an iMarkTM microplate absorbance reader (Bio-Rad Laboratories, Inc., United States).
Nucleic Acid-Based Identification of Rare Marine Actinobacteria Strain Demonstrating Vibrio cholerae Biofilm Inhibitory Activity
The extraction of the genomic DNA of the RMA strain exhibiting a significant inhibitory activity against the V. cholerae HYR14 biofilm was performed using commercially available DNeasy Blood and Tissue kits (Qiagen, Germany) as per the manufacturer-recommended protocol (Ahmed et al., 2008). Until further use, the eluted DNA was stored at -20°C. Nested PCR targeting the 16SrRNA of the eubacterial genome was done using primers: forward primer (U1): 5′-TTGGAGAGTTTGATCCTGGCTC-3′ and reverse primer U4: 5′-GGACTACCAGGGTATCTA-3′ for the first round of nested PCR and forward primer (U2): 5′-TTGGAGAGTTTGATCCTGGCTC-3′) and reverse primer (U3): 5′-GCGGCTGGCACGTAGTTAG-3′ for the second round of nested PCR (Therese et al., 1998). The reaction conditions were set as described in Kharel (Sitaula) et al. (2018). The amplified product was then cyclo-sequenced, purified, and loaded to ABI3200 genetic analyzer (Applied Biosystems, United States) and sequenced as described previously (Aarthi et al., 2011). The obtained sequence was analyzed using BioEdit software (version 7.0.5.3) and was compared with 16S rRNA sequence database of National Center for Biotechnology Information (NCBI) using nucleotide BLAST (Altschul et al., 1990). The phylogenetic tree was constructed using the neighbor-joining method (Perrière and Gouy, 1996) and was validated using bootstrap analysis. The sequence was submitted to GenBank for accession number.
Quantification of Biofilm Formation and Eradication
To evaluate the effect of RMA46 organic extract on mature V. cholerae biofilm, 100 μl of fresh LB broth was aliquoted into a 96-well microtiter plate. This was inoculated with 10 μl of an overnight-grown culture of V. cholerae HYR14 strain that had been diluted up to 1:100 (OD595 = 0.2). The microtiter plate was incubated at 37°C for 24 h. After the incubation period, the non-adherent planktonic cells were aspirated gently without disturbing the biofilm. To the wells, 100 μl (final volume) of LB broth containing various concentrations of RMA46 organic extract (50–0.13 mg/ml) was aliquoted. The microtiter plate was incubated again at 37°C for 24 h. Following the incubation period, crystal violet assay was performed to evaluate the potential of RMA46 organic extract on eradication of V. cholerae biofilm. Untreated and vehicle controls were also included for all experiments (Balasubramanian et al., 2017).
To quantify the effect of RMA46 organic extract on the formation of V. cholerae biofilm, 100 μl (final volume) of LB broth containing various concentrations of RMA46 organic extract (50–0.13 mg/ml) was aliquoted into 96-well microtiter plate. Ten microliters of an overnight-grown culture of V. cholerae HYR14 strain that has been diluted up to 1:100 (OD595 = 0.2) was inoculated into each well, and the plates were incubated at 37°C for 24 h. After the incubation period, the plates were subjected to crystal violet staining. Untreated and vehicle controls were also included for all experiments.
For crystal violet staining, the medium and the planktonic cells present in the 96-well microtiter plates were aspirated at the end of the incubation period. The non-adherent HYR14 cells present in the wells were discarded by washing the wells thrice with sterile 1 × phosphate-buffered saline. The microtiter plates were then air-dried for 20 min at ambient temperature to fix the adherent biofilms. To the dried wells, 100 μl of 0.2% (w/v) crystal violet was added and incubated for 20 min at room temperature. After the incubation period, the excess stain was washed off with sterile water twice, and the plates were air-dried completely. The bound crystal violet was solubilized by pipetting 100 μl of 33% glacial acetic acid into each well (Kaur et al., 2016). The absorbance of the wells was measured at 595 nm using an iMarkTM microplate absorbance reader (Bio-Rad Laboratories, Inc., United States).
Confocal Laser Scanning Microscopic Analysis
V. cholerae HYR14 (OD595 = 0.1) and RMA46 ethyl acetate extract at a concentration of (9.3 mg/ml) were inoculated into LB medium in cell culture plate containing sterile borosilicate coverslips and were incubated statically at 37°C for 24 h. After 24 h, the suspension was aspirated and removed carefully. The biofilms established on the coverslips were rinsed with 0.9% NaCl solution. Stock solutions of SYTOTM 9 green fluorescent stain (Thermo Fisher Scientific) and propidium iodide red fluorescent dyes (BacLight, Invitrogen Ltd.) were prepared at 1:1 ratio and stored at −20°C until further use. Briefly, 200 μl of the dyes was added to the coverslips and incubated at room temperature for 15 min under dark conditions. After incubation, the dyes were aspirated, and the coverslips were carefully washed with 0.9% NaCl. The washed coverslips were air-dried at ambient temperature and fixed on a sterile glass slide. These slides were observed under a 40× objective using a confocal laser scanning microscope (Olympus FLUOVIEW, FV1000). The biomass, average thickness, and roughness coefficient of biofilms were analyzed using COMSTAT package in MATLAB R2017b (Heydorn et al., 2000).
Cell Viability Assay
The effect of the RMA46 ethyl acetate extract on host cell viability was assayed using murine macrophages RAW264.7, cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (w/v), and 1% L-glutamine using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. About 1 × 104 RAW264.7 cells/well were seeded in a 96-well microtiter plate and incubated at 37°C in a CO2 incubator for 24 h. After the incubation period, the cells were treated with various concentrations (0.13, 0.39, and 9.375 mg/ml) of Micromonospora sp. RMA46 ethyl acetate extract and incubated for another 24 h at 37°C in a CO2 incubator. After 24 h, 1 mg/ml of MTT was added, and the microtiter plate was incubated at 37°C for 3 h. Dimethyl sulfoxide (DMSO) was added to dissolve the formazan crystals, and absorbance was read at 570 nm using a microplate reader spectrophotometer (Synergy H1) (Kartik et al., 2020).
Extraction of RNA and Quantitative Analysis of Virulence Gene Expression in Vibrio cholerae HYR14
Total RNA was extracted from the RMA46 extract-treated and untreated V. cholerae HYR14 from stationary phase culture (24 h) using RNeasy® Protect Bacteria Mini Kit (Qiagen), according to the manufacturer’s guidelines. The purity and integrity of the isolated RNA were checked using NanoDrop (Thermo Fisher Scientific, United States). Preparation of cDNA was performed using the iScriptTM cDNA Synthesis kit with a concentration of 100 ng following the manufacturer’s guidelines. The following conditions were set for cDNA synthesis; 24°C for 5 min (annealing), 42°C for 30 min (extension), and 85°C for 5 min (inactivation). The effect of RMA46 ethyl acetate extract on the virulence gene expression in the HYR14 strain was analyzed by qRT-PCR using primers listed in Supplementary Table S4. The 16s rRNA was used as the reference gene. The relative gene expression was calculated using the 2–Δ Δ CT method (Vezzulli et al., 2015; Hema et al., 2017).
Gas Chromatography–Mass Spectrometry Analysis
Separation and identification of the active compounds present in the ethyl acetate extract of RMA46 were performed with a PerkinElmer Clarus 500 GC-MS system. Helium was employed as a carrier gas at a flow rate of 1 ml/min, and the injector temperature was set at 280°C. The program of the column temperature was held at 50°C for 1 min and increased at 10°C/min to 150°C (1 min hold), to 250°C for 1 min hold, and then at 15°C/min to 300°C (3 min hold). The mass spectrometer was operated at a mass range of 40–450 amu. One microliter of RMA46 extract dissolved in ethyl acetate was injected into the system. The separated compounds were identified by comparing their spectra with those in the National Institute of Standards and Technology (NIST) mass spectral library (Ravichandiran et al., 2012).
Computational Studies
Two-dimensional structures of the compounds identified in the Micromonospora sp. RMA46 ethyl acetate were drawn using MarvinSketch1. The ligands were prepared for molecular docking using LigPrep (Schrödinger, LLC, New York, NY, 2020). The primary structure of the ATP binding domain of LuxO (Q9KT84) was retrieved from Uniprot2. Protein BLAST (BLASTp) was performed for the query sequence against Protein Data Bank (PDB) to identify the structurally similar homolog. The QS signal integrator LuxO of Photobacterium angustum with the PDB accession number 5EP0 was chosen as the template. Homology modeling was performed using the protein structure prediction tool implemented in BioLuminate (Schrödinger, LLC, New York, NY, 2020). The modeled protein was subjected to a constrained energy minimization using GROMACS (Kutzner et al., 2019), and the quality of the modeled structure was further evaluated using the SAVES server3. The experimentally solved structure of HapR protein (2PBX) was downloaded and used for molecular docking simulations. Both the LuxO and HapR proteins were prepared using the protein preparation wizard (Sastry et al., 2013). A grid box was generated and was centered on the binding sites of the target proteins, and the docking simulation was performed using extra precision (XP) docking algorithm implemented in Glide (Schrödinger, LLC, New York, NY, 2020) (Friesner et al., 2006).
Statistical Analysis
GraphPad Prism software version 8.0.2 (GraphPad Software Inc., San Diego, CA, United States) was used for analyzing the data statistically. The significance was analyzed by Student t-test with p set at p ≤ 0.05. All the assays were performed in triplicates, and the results have been expressed as mean ± SD.
Results
Mangrove Soil Is Rich in Morphologically Distinct Rare Marine Actinobacteria
Non-Streptomyces like colonies that were selectively subcultured for the isolation of pure RMAs produced a wide range of colonies from round convex to dry, raised, irregularly edged colonies. These colonies also produced varied colony pigments from white, yellow, pink, orange, brownish-orange, gray, and black. Out of the 42 isolates, only one isolate (RMA44) produced diffusible pigment. Light microscopy revealed that a variety of structures including coccus, rod-coccus, coryneform, and short- and long-branched sometimes filamented hyphal formations. Many of these hyphal-producing strains also formed oval spores. The pattern of hyphae and spore formation also varied significantly among RMA isolates. A few isolates changed colony pigmentation from orange to black as the colonies got older (Supplementary Table S3). This showed that the mangrove soil supported a rich diversity of Actinobacteria.
Mangrove-Derived Rare Marine Actinobacteria Extract Could Attenuate Vibrio cholerae HYR14 Biofilm
The cell-free culture supernatants of all the 42 RMA isolates were screened for an anti-biofilm activity. The isolates RMA2, RMA4, RMA15, RMA42, RMA44, RMA45, RMA46, and RMA49 exhibited a > 50% biofilm inhibitory activity (Figure 1A). However, the cell-free culture supernatant of RMA46 exhibited a consistent level of ≥ 30% inhibition of HYR14 biofilm without a bactericidal activity at all concentrations (v/v) chosen in the study (Figure 1B and Supplementary Figure S1). Therefore, the strain RMA46 was chosen for further investigations.
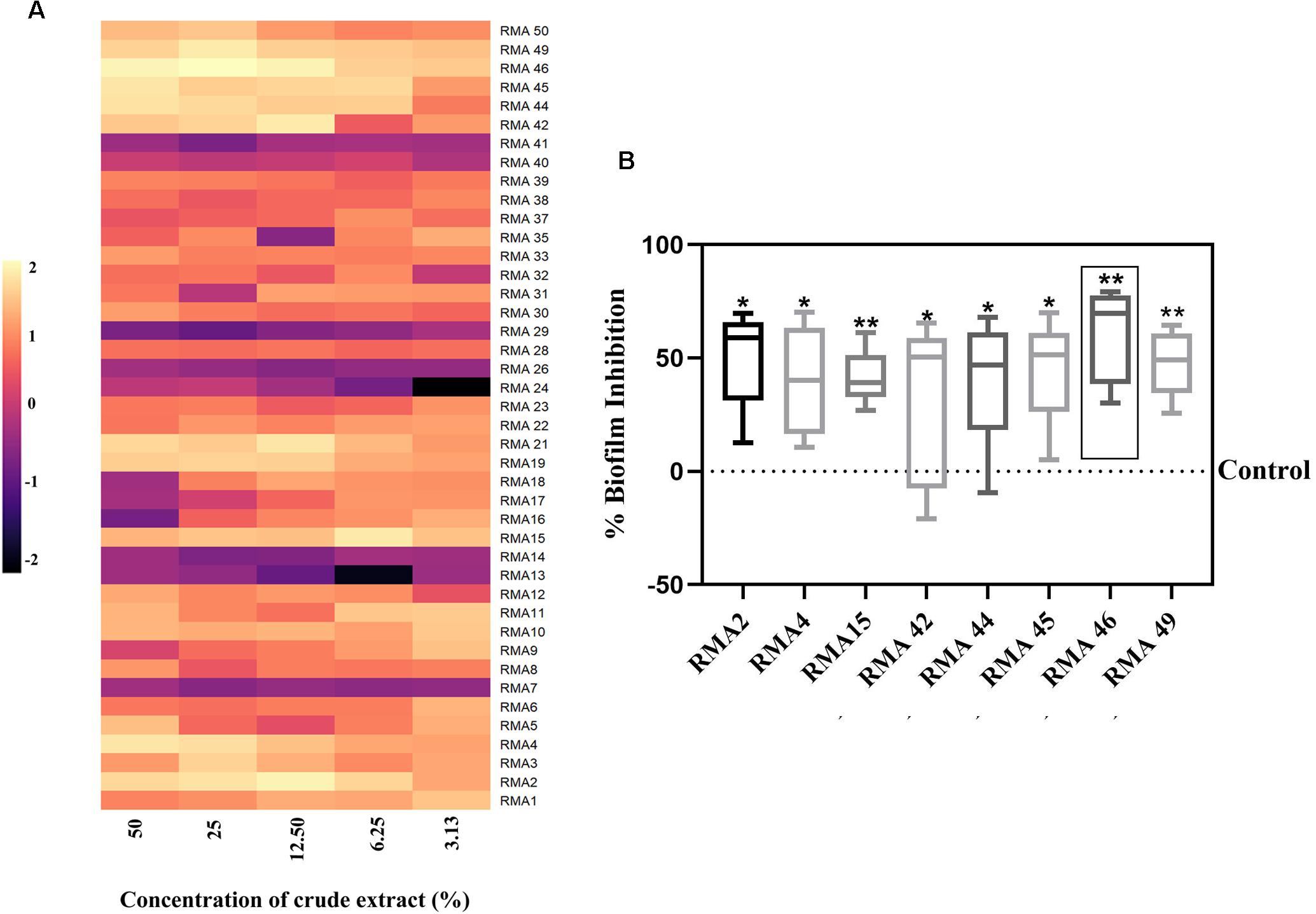
Figure 1. (A) Effect of various concentrations of cell-free extract of 42 rare marine Actinobacteria (RMA) [50, 25, 12.50, 6.25, and 3.13% (v/v)] on 24 h biofilm of Vibrio cholerae HYR14. (B) Comparison of various concentrations [50, 25, 12.50, 6.25, and 3.13% (v/v)] of cell-free extracts of selected RMA strains that demonstrated significant HYR14 biofilm inhibitory activity. RMA46 displays the maximum (p < 0.01) inhibition than do all the other strains. A one-sample t-test was used for significance analysis. *p < 0.05, and **p < 0.01.
Isolate RMA46 Belongs to the Genus Micromonospora
The complete 16S rRNA sequence of the RMA46 strains (724 bp) determined in this study was deposited in GenBank database (accession number MK788184.1). To study the evolutionary relationship, similarity search of 16S rRNA sequence of RMA46 was performed against 16S rRNA database in National Center for Biotechnology Information (NCBI). The closely related sequences of RMA46 were used to construct the phylogenetic tree. The result showed that the strain RMA46 is closely related to Micromonospora sp. 171111 (accession number EF538723) and was clustered together in the phylogenetic tree (Supplementary Figure S2).
Micromonospora sp. RMA46 Ethyl Acetate Extract Effectively Inhibits Biofilm Formation by HYR14 Than by Eradicating Preformed Biofilms
The RMA46 organic extract (50–0.13 mg/ml) was evaluated for its Vibrio cholerae HYR14 biofilm inhibitory/disruptive potential. The RMA46 ethyl acetate extract inhibited HYR14 biofilms ≥ 50% at low concentrations (0.39–0.13 mg/ml). At higher concentrations (0.59–50 mg/ml), the RMA46 ethyl acetate extract effectively inhibited > 70% of the HYR14 biofilm formation (Figure 2A). Overall, the data were consistent to show a dose-dependent increase in the inhibitory potential of RMA46 ethyl acetate extract against the V. cholerae HYR14 biofilm. On the contrary, the RMA46 ethyl acetate extract was unable to eradicate > 50% of preformed biofilm (Figure 2B). Because the biofilm inhibitory concentration (BIC) of RMA46 ethyl acetate extract ranges from 50 to 90%, three effective BICs were chosen – BIC50 (0.13 mg/ml), BIC60 (0.39 mg/ml), and BIC90 (9.38 mg/ml) – to validate anti-infective potential.
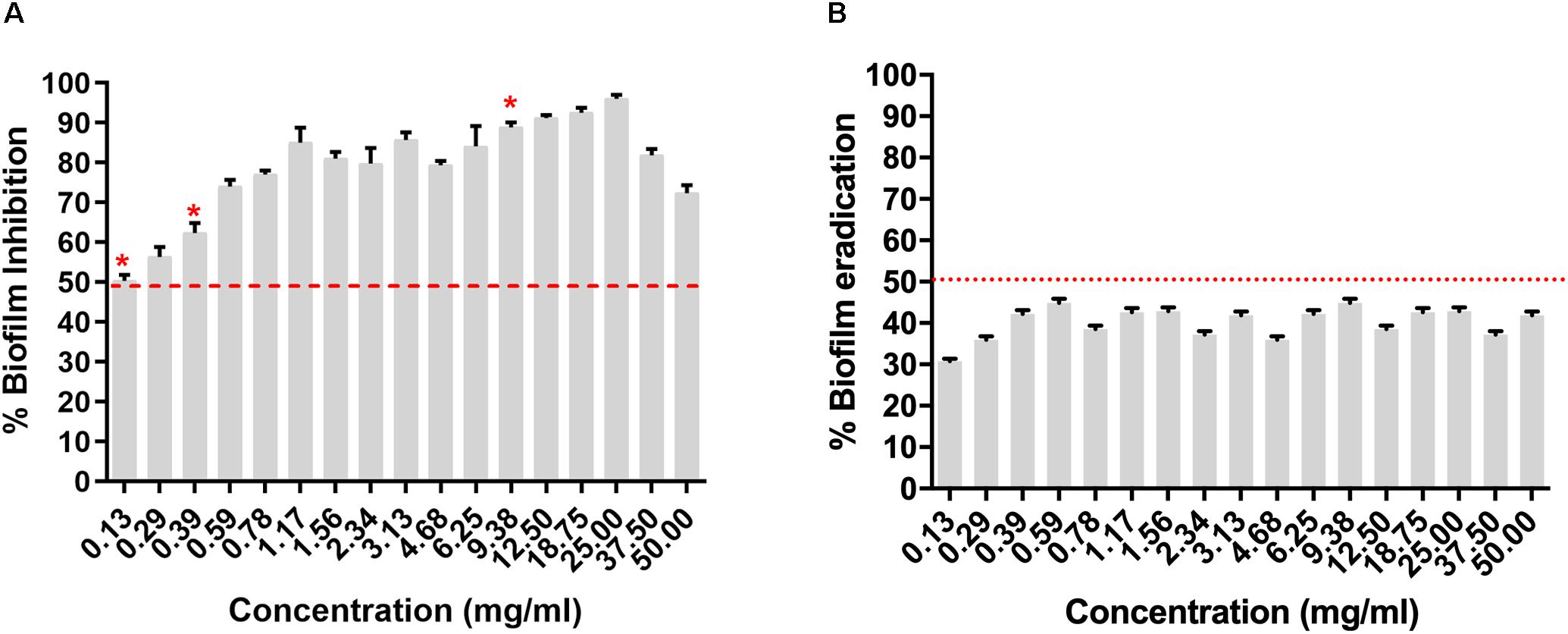
Figure 2. (A) Dose-dependent inhibition of Micromonospora sp. RMA46 organic extract on the formation of Vibrio cholerae HYR14 biofilm. (B) Dose-dependent effect of RMA46 organic extract on preformed biofilm V. cholerae HYR14 biofilm. * Effective concentrations chosen for further studies.
RMA46 Ethyl Acetate Extract Affects the Thickness and Biomass of Vibrio cholerae Biofilm
Investigation on the effect of RMA46 ethyl acetate extract on HYR14 biofilm formation on coverslips was performed using confocal laser microscopy and COMSTAT analysis. Treatment with BIC90 (9.38 mg/ml) revealed a detached biofilm matrix, whereas the untreated control sample showed highly condensed and compact biofilms (Figure 3A). COMSTAT analysis revealed that the untreated V. cholerae formed thicker biofilms (107.7 μm) with higher biomass (45.4 μm3/μm2) and a roughness coefficient of 0.27. Yet the RMA46 ethyl acetate treated HYR14 biofilms was 23.2 μm and 4.2 μm3/μm2 in thickness and biomass, respectively. The roughness coefficient of the RMA46 treated HYR14 biofilm is 1.30. These results signify that RMA46 ethyl acetate leads to an irregular distribution and reduced biomass of HYR14 than does the untreated HYR14 biofilm (Figure 3B).
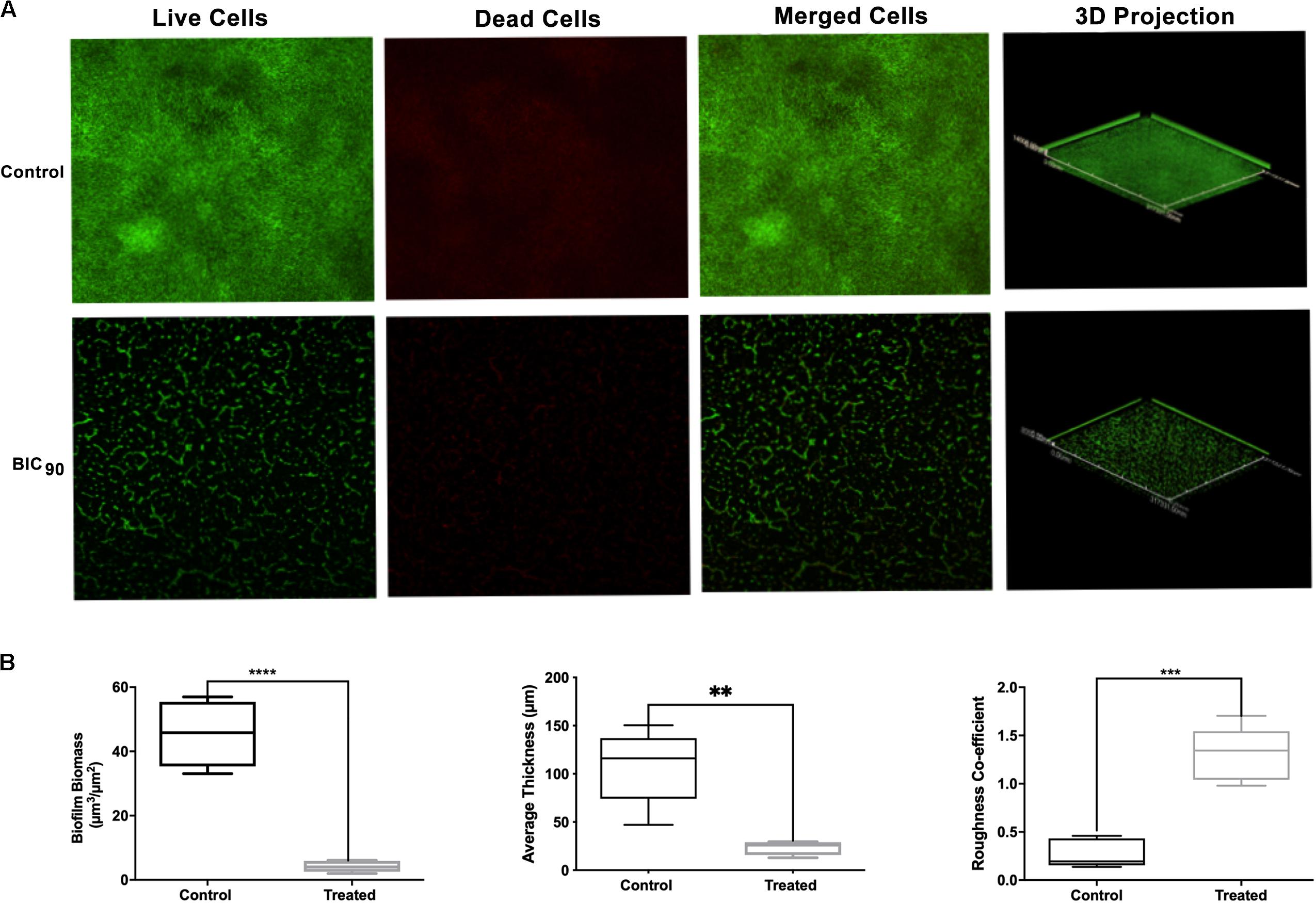
Figure 3. (A) Confocal laser scanning microscopy of HYR14 biofilm in the presence of the RMA46 extract, visualized by red and green fluorescent vital dyes. (B) COMSTAT analysis on the biomass, average thickness, and roughness coefficient of HYR14 biofilm treated with RMA46 organic extract. Student’s t-test was used for significance analyses. *p < 0.05 was considered significant. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
RMA46 Ethyl Acetate Extract Does Not Affect Mammalian Cell Viability
The effect of ethyl acetate extract of RMA46 on the viability of host cells was assessed against macrophage RAW264.7 cell lines. Three concentrations, representing BIC50, BIC60, and BIC90 (0.13, 0.39, and 9.37 mg/ml, respectively), were chosen to investigate the extract’s lethality for 24 h. In all treatments compared with the control (untreated macrophage RAW264.7 cells), the viability of the cells was not affected, which indicated the non-toxic nature of the extract without affecting the cell viability (Figure 4).
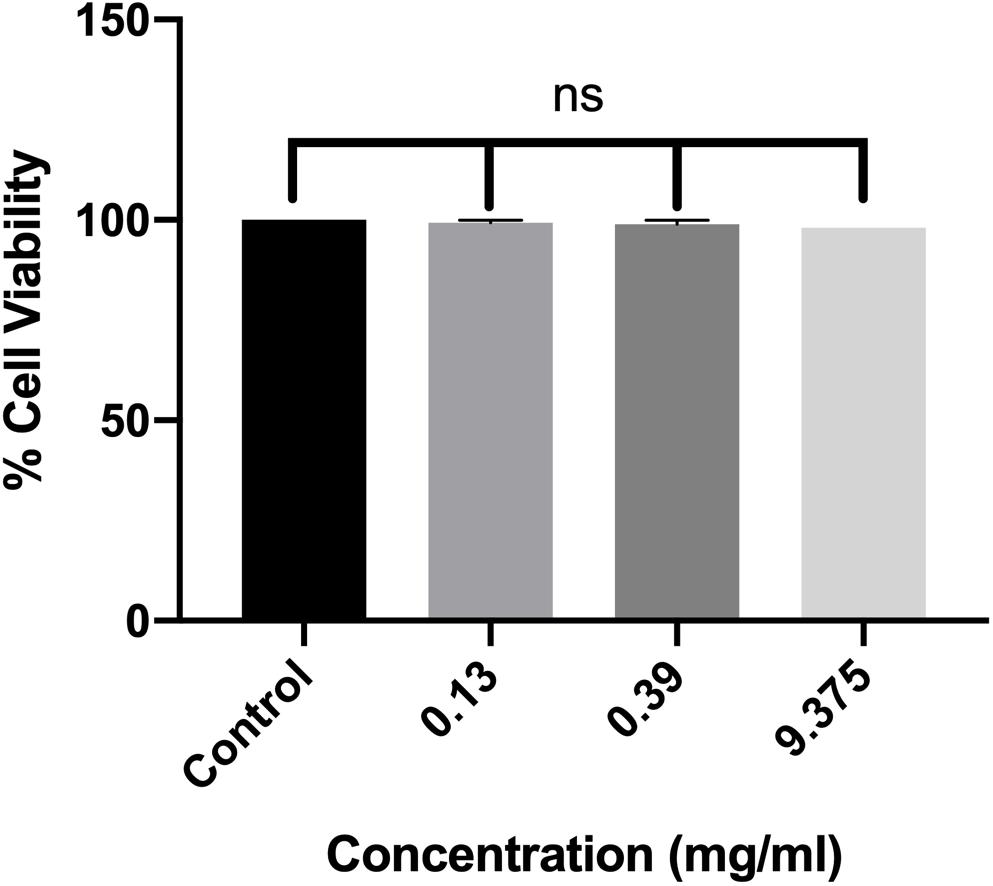
Figure 4. Dose-dependent effect of RMA46 ethyl acetate extract on the cell viability. Experiments were performed in triplicates, and difference in mean values was compared with that of the untreated experiment control. One-way ANOVA was used for multiple comparison analyses. ns denotes not significant.
RMA46 Extract Interferes With HYR14 Molecular Machinery
The real-time analysis of the expression pattern of V. cholerae HYR14 global switches (LuxO and HapR) and its downstream virulence genes – ct, tcp, small regulatory RNAs (qrr2 and qrr4), aphA, and hapA – were quantified differentially under RMA46 organic (ethyl acetate) extract induced/uninduced state. At LSS, the global switch, LuxO is switched on to regulate the transcription of many other vital genes including the tcp and ct to encode toxin coregulated pilus and CTX, respectively. The small regulatory rRNA termed as qrr1-4 along with Hfq (RNA chaperone) binds with mRNA transcripts of hapR. HapR repression leads to the formation of biofilm. Similarly, at HSS, the de-repression of HapR leads to the suppression of biofilm formation and upregulation of protease production, causing dissemination of V. cholerae. In this regard, the expression analysis at BIC50 showed a 2–3 log2 fold reduction in the expression of LuxO and HapR and its downstream virulence genes hapA, tcp, and ct. Similarly, the treated (BIC60) cells showed a > 2 log2 fold downregulation in the expression of the global regulators (LuxO and HapR) and virulence genes (tcp and ct). Overall, the data were significant to show that RMA46 extract interferes with the HYR14 molecular machinery in a dose-dependent manner to downregulate to > 2–5 log2 fold, which confirmed that the entire virulence mechanism of V. cholerae has been switched off (Figures 5A,B).
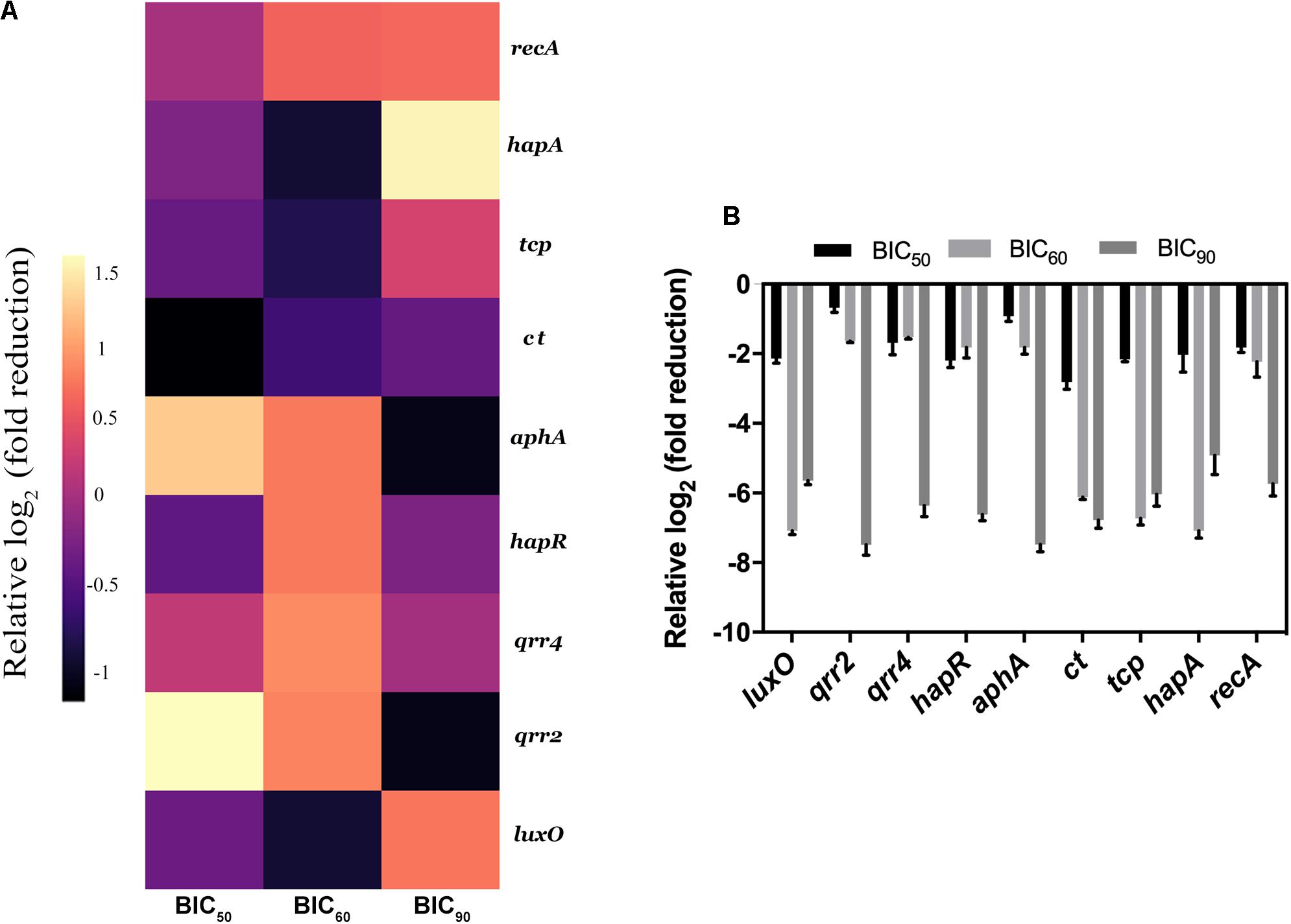
Figure 5. Expression analysis of Vibrio cholerae virulence genes by quantitative PCR. (A) Heat map of real-time qRT-PCR to show the effect of RMA46 organic extract on virulence gene expression in V. cholerae. (B) Comparison between the various inhibitory concentration of RMA46 extract on HYR14 virulence-associated genes.
Metabolic Profiling of RMA46 and Their Molecular Interaction With Active Sites of LuxO and HapR
With the use of gas chromatography–mass spectrometry (GC–MS), a total of 43 secondary metabolites were identified in the RMA46 extract belonging to different categories of secondary metabolites including alkanes (4.6%), alkenes (23.2%), carboxylic acids (25.5%), esters (6.9%), dairy ketone (4.6%), phenols (2.6%), and terpene (2.3%). The extract also contained aromatic monocyclic and heterocyclic compounds (20.9%) and other hydrocarbons (4.6%). Interestingly, the RMA46 extract also contained an oxaspiro compound (2.3%) (Table 1 and Supplementary Figure S3).
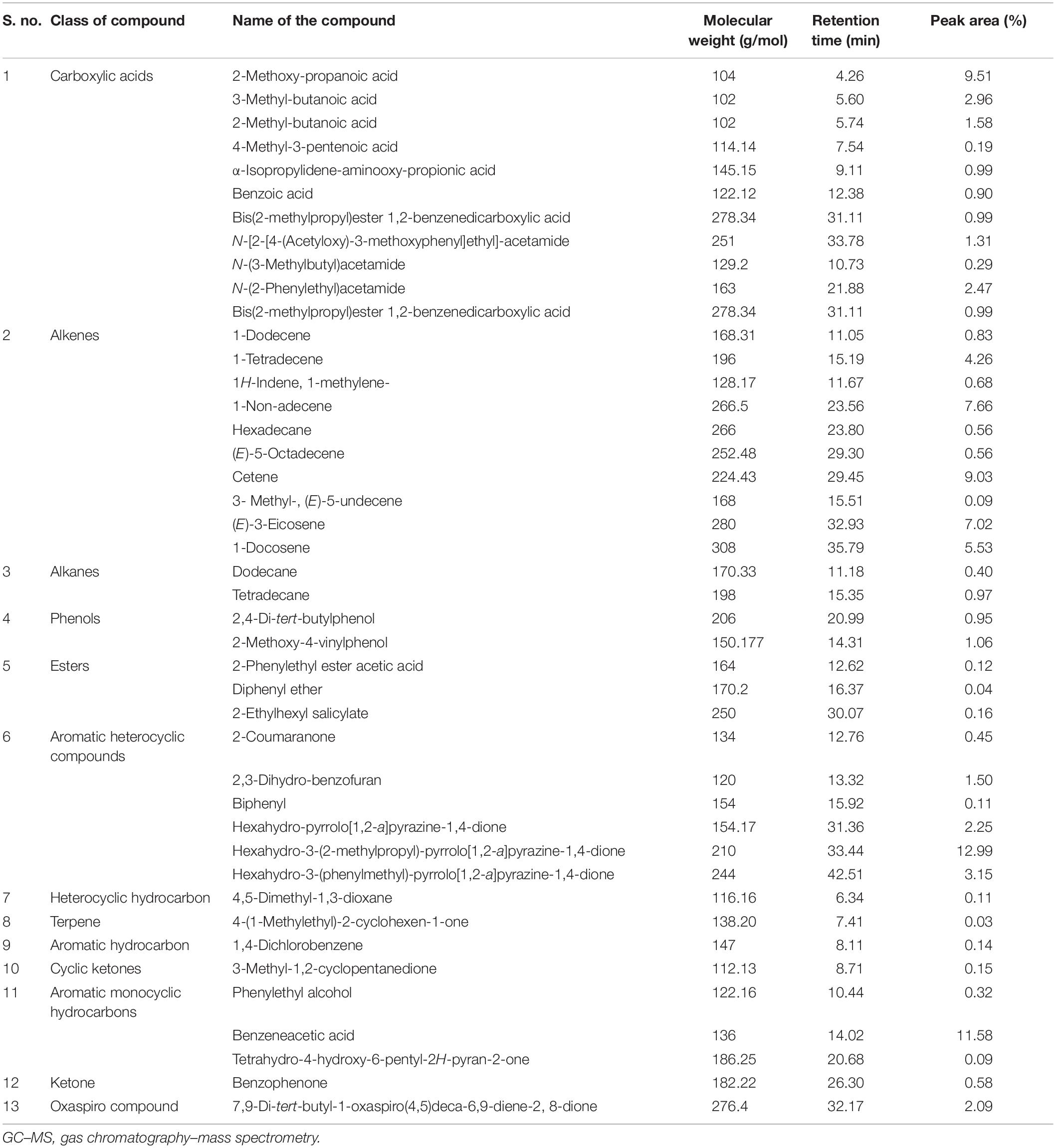
Table 1. List of compounds identified in Micromonospora sp. RMA46 ethyl acetate extract using GC–MS.
QS global regulators: LuxO and HapR drive the virulence mechanism including biofilm and CTX production in V. cholerae in response to low and high environmental signals. Because an experimentally solved structure of LuxO of V. cholerae is not available, we used a 1.6-Å resolution X-ray structure of the LuxO receiver-catalytic domain of Photobacterium angustum, which has 69% of sequence identity as a template for modeling V. cholerae LuxO (Figure 6). Any gaps observed during core modeling were subjected to loop modeling using Schrödinger PRIME (Jacobson et al., 2004). Then, the modeled protein was subjected to a constrain-based energy minimization using GROMACS. The Ramachandran plot of the modeled protein showed that PHI and PSI angles of the modeled proteins are within the allowed ranges, and the modeled protein was acceptable and hence used for further docking studies. For HapR, an existing X-ray structure of 2.2-Å resolution with putative ligand-binding domains was used. The small molecules identified in the RMA46 ethyl acetate extract identified using GC–MS was docked with the active sites of LuxO and HapR (Figure 7). Compound 2-methoxy-4-vinylphenol demonstrated an increased Glide score (-4.130) and Glide energy (-20.408 kcal/mol) with LuxO protein than did other molecules (Figure 8). The interaction pattern was observed as the ligand forms hydrogen bonding with the most crucial amino acid residues (GLN363 and TYR139) of the receiver-catalytic domain of ATP binding domain of LuxO. Similarly, the ligand, hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione, was found to have the highest affinity with the HapR protein with a Glide score of -4.543 and Glide energy of -30.465 kcal/mol and formed hydrogen bond interaction with SER165 amino acid.
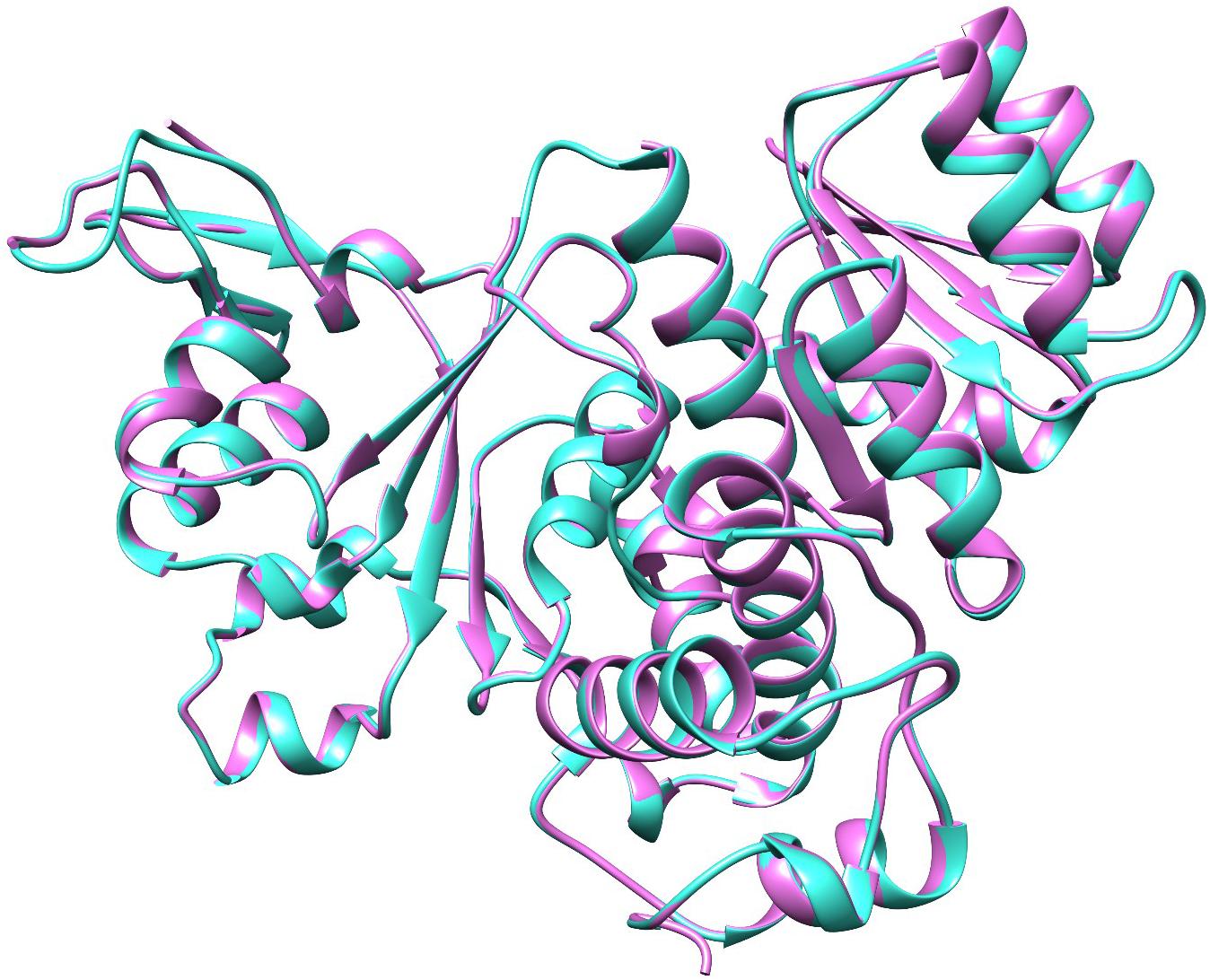
Figure 6. Molecular superimposition of LuxO of Photobacterium angustum (template) and V. cholerae. The LuxO of P. angustum is represented in purple color and the LuxO of V. cholerae in turquoise color.
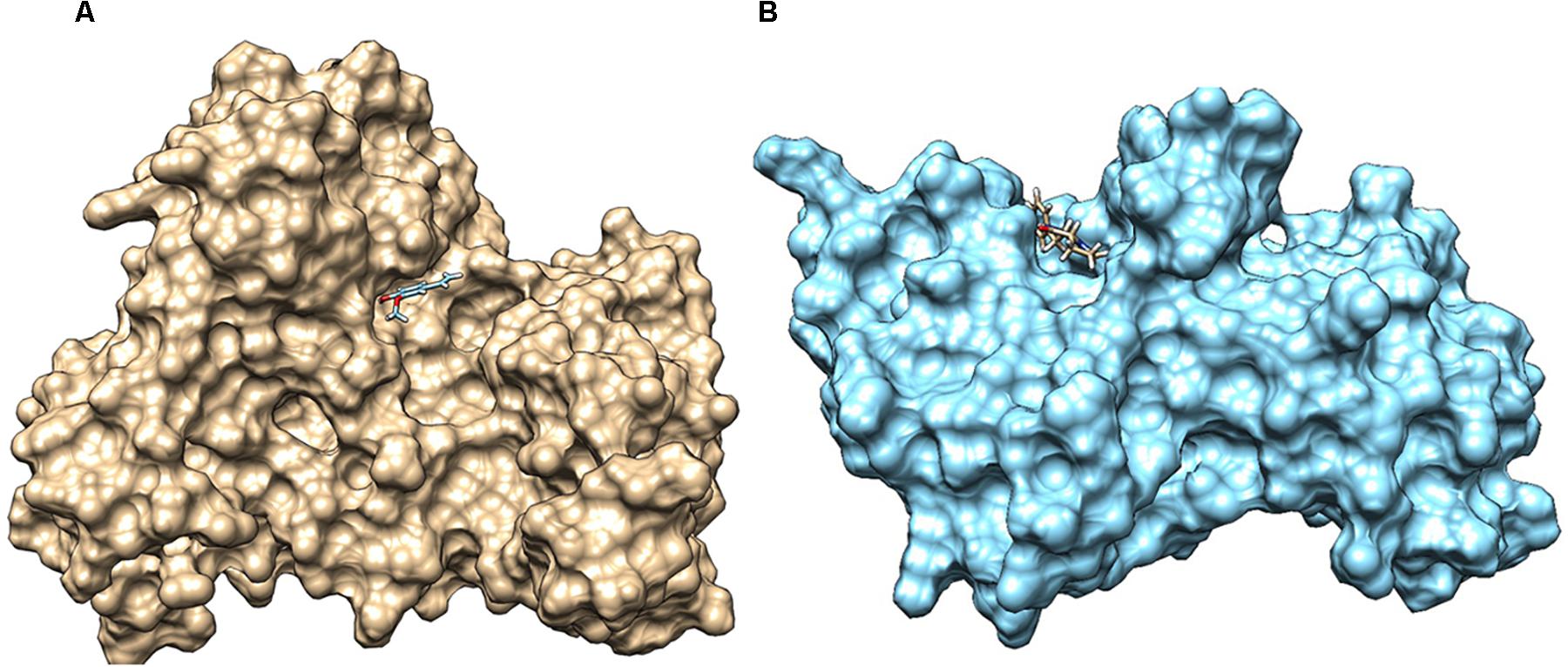
Figure 7. (A) Docked complex of 2-methoxy-4-vinylphenol with LuxO of Vibrio cholerae, which forms hydrogen bond interaction with TYR129 and GLN363. (B) Interaction of hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione with HapR of V. cholerae, forming hydrogen bond with SER165. The V. cholerae response regulators are represented as surface (LuxO, gold color; and HapR, blue color), and the small molecules are represented as sticks.
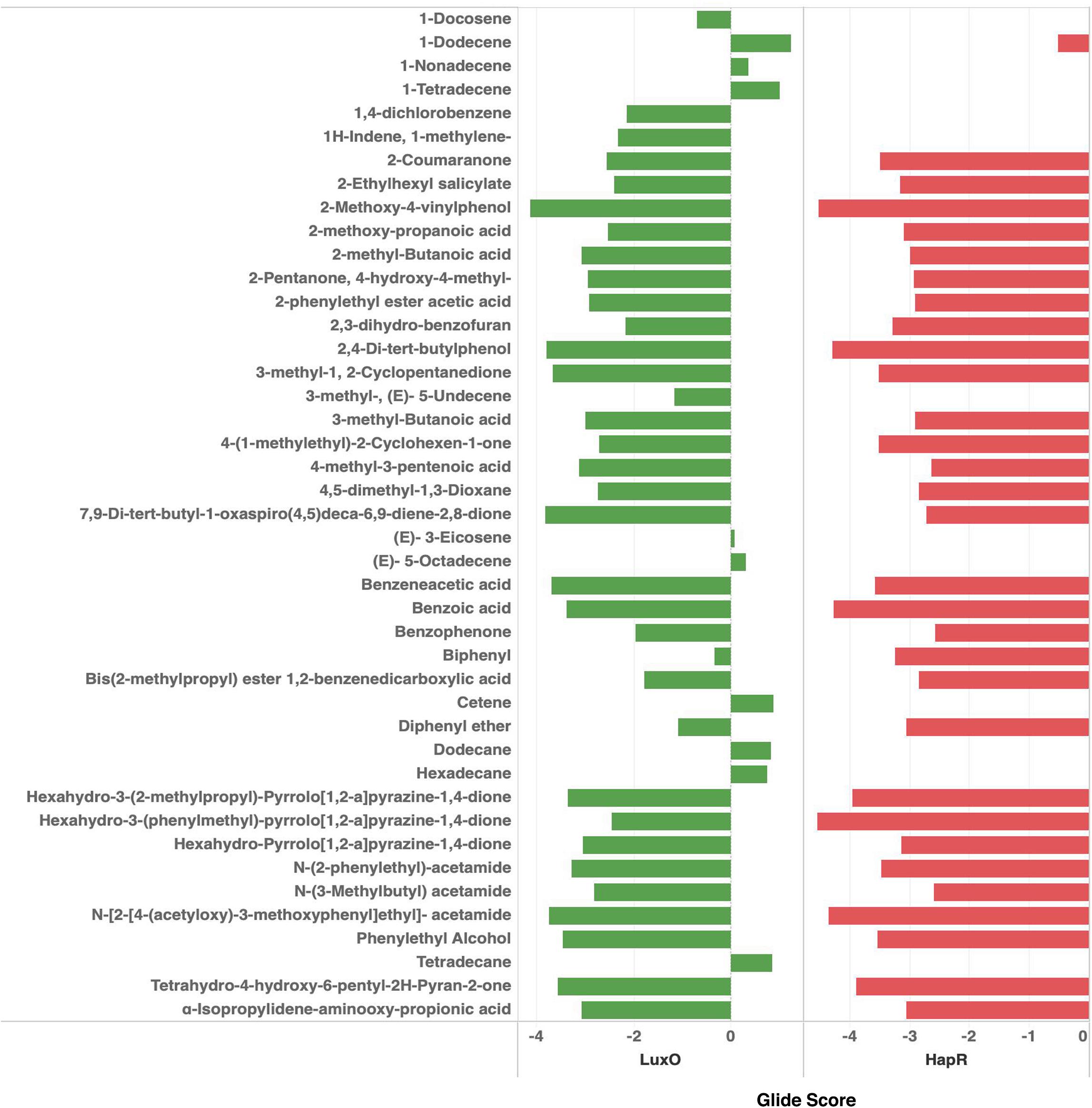
Figure 8. Docking scores of the bioactive compounds identified in the ethyl acetate extract of RMA46 with LuxO and HapR of Vibrio cholerae.
Discussion
The emergence of antimicrobial resistance among Vibrio cholerae is a burden on public health especially in developing countries (Das et al., 2019). An increase in the incidence of multidrug resistance V. cholerae could hurt cholera-controlling measures within a community (Mushayabasa and Bhunu, 2012). To subdue the emergence of antibiotic resistance, natural compounds that exert anti-virulence properties have been recognized as an alternative strategy (Narendrakumar et al., 2019a). Marine-derived Actinobacteria are a rich source of secondary metabolites that provide unique carbon scaffolds for the synthesis of advanced therapeutic agents for pathogens (Hassan and Shaikh, 2017). More than 3,000 compounds have been reported from marine bacteria most prevalently from the phylum Actinobacteria, which are known to inhibit drug-resistant bacterial pathogens (Rahman et al., 2010). The density of saprophytic Actinobacteria is high in mangrove soil owing to the detritus ecosystem found throughout the year (Saravanakumar et al., 2016). In this study, Actinobacteria strains were isolated from mangrove soil by various physical and chemical pretreatments by using a wide range of selective isolation media. It was observed that pretreatment of soil did not significantly affect the isolation of Actinobacteria strains. However, the application of starch casein agar and ISP no. 4 medium was most effective in the isolation of the Actinobacteria. The phylum Actinobacteria is one of the largest among the prokaryotes and encompasses many genera that are morphologically and phenotypically diverse.
The preliminary identification using light microscopy and colony morphology showed that the isolates were morphologically diverse, indicating the isolation of various genus. The isolated RMAs were screened using HYR14, a rugose V. cholerae O1 Ogawa biofilm-forming strain isolated during an outbreak in India. The strain is resistant to ampicillin, furazolidone, nalidixic acid, streptomycin, ciprofloxacin, erythromycin, and norfloxacin (Chowdhury et al., 2016). Because the rugose phenomenon is stable even after passage into humans and their potential to cause epidemics, the multidrug-resistant HYR14 strain was chosen for the screening the RMAs that can inhibit biofilm formation by V. cholerae for the current study (Morris et al., 1996; Chowdhury et al., 2016).
Although marine Actinobacteria account for 46% of the total Actinobacteria density exerting virulence inhibitory potential (Sarveswari and Solomon, 2019), the exploration for anti-virulence agents against V. cholerae had been very limited. Previously, culture supernatant of arctic derived rare Actinobacteria, Nocardiopsis sp. A731, had been reported to exhibit a biofilm inhibitory activity against V. cholerae (Augustine et al., 2012). In our study, anti-biofilm phenotype-based screening of the RMA against HYR14 strain revealed that about 19% of the mangrove-derived isolates could inhibit the formation of V. cholerae biofilm. The study reinforces that the mangrove-derived Actinobacteria are a prospective source for anti-infectives against V. cholerae. Moreover, the HYR14 cell-free extract inhibits the formation of V. cholerae biofilms at a concentration (v/v) lesser than that of the Nocardiopsis sp. A731 reported by Augustine et al. (2012). In the same study, 200 μg/ml of Streptomyces sp. A745 diethyl ether fraction has been reported to suppress 60% of V. cholerae biofilm. Yet in our study, about 50% inhibition was observed at 130 μg/ml of RMA46 ethyl acetate extract. Although eight RMA isolates showed a significant activity against HYR14 biofilms, only the RMA46 strain that showed > 50% biofilm inhibition at least concentration was further subjected to nucleic acid-based identification. The sequence of the mangrove-derived Micromonospora sp. RMA46 was found to be closely related to the 16S rRNA sequence of another mangrove isolate Micromonospora sp. 171111. As both the strains were isolated from the mangrove region, the sequence similarity could reflect an environment selection among Micromonospora strains (Hong et al., 2009). The RMA46 ethyl acetate extract clearly eliminates the formation of HYR14 biofilm than its eradication. Previously, a non-significant impact on eliminating the preformed V. cholerae biofilms by natural products was attributed to their potential to affect the initial stages of biofilm formation including motility (Narendrakumar et al., 2019b). Hence, further studies with the V. cholerae HYR14 are needed to understand the mechanism of inhibition of biofilm formation.
The signal (autoinducer) strength influences the QS global switches (LuxO and HapR) to enhance the survivability and persistence of the intestinal pathogen, V. cholerae (Joelsson et al., 2007). At LSS, LuxO is positively regulated, and its synergistic interaction with the global response regulator, TsrA, enhances the production of Vibrio polysaccharide (VPS) TCP and CT to establish biofilm and virulence (Zheng et al., 2010).
When the signal strength is elevated (HSS), HapR is depressed, resulting in the upregulation of hemagglutinin/protease (HapA), facilitating the dispersal of V. cholerae cells within the biofilm matrix (Silva et al., 2003; Vance et al., 2003; Benitez and Silva, 2016). Also, HSS is directly proportional to the increased population size of Vibrio in an environment, which might lead to nutrient depletion. Interestingly, under nutrient-limited condition, the expression of HapA protease increases owing to the elevated intracellular cAMP pool. The cAMP binds to its allosteric receptor and exerts a positive feedback loop to activate CRP, RpoS, and HapR (Benitez and Silva, 2016). Our transcriptomic analysis revealed that the expression of major virulence genes and the response regulators of V. cholerae HYR14 is highly attenuated without affecting their growth in the presence of RMA46 ethyl acetate extract. This overall attenuation of the molecular machinery activated at LSS and HSS might be a combinatorial effect of two or more potential virulence attenuating bioactive molecules present in the RMA46 ethyl acetate extract against V. cholerae.
Genetically marine Actinobacteria are equipped to produce about 20 secondary metabolites (Lam, 2006). In this study, 43 secondary metabolites from various classes of chemical compounds have been identified. The RMA46 extract encompasses many chemicals from the class of carboxylic acid, and interestingly our group had previously reported the QS inhibitory effect of synthetically derived pyrazine-2-carboxylic acid on V. cholerae (Hema et al., 2017). The derivative of various other compounds reported in RMA46 has been reported to have other biological activities including antibacterial and anti-virulence activities against various other pathogens. This eventually exemplifies the potential of Micromonospora sp. RMA46 as a source for chemical structural templates for the development of anti-virulence agents. Docking of the 43 chemical compounds leads to the identification of 2-methoxy-4-vinylphenol and hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione to have the highest interacting capability with LuxO and HapR, respectively. The data corroborate our qRT-PCR data, which were also significant to show a complete blockage of virulence so as to prevent the colonization and dissemination of bacteria to persist infection. A derivative of 2-methoxy-4-vinylphenol has already been reported to have an inhibitory activity against LasIR/RhIIR circuit of Pseudomonas aeruginosa (Shah et al., 2019). Interestingly, hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione, also known as cyclo D-phenyalanyl-L-prolyl [Cyclo(Phe-Pro)], has been reported to attenuate the expression of genes associated with virulence in V. cholerae. Cyclo(Phe-Pro) inhibits the production of CTX and toxin coregulated pili in O1 El Tor strain by negatively affecting the transcription of genes tcpPH in V. cholerae (Bina and Bina, 2010). It was later shown that Cyclo(Phe-Pro) enhanced the ToxR-dependent transcription of leuO, which in turn repressed the downregulation of the ToxR regulon, altering the production of CT and TCP (Bina et al., 2013). However, the outcomes derived from this research support the bioprospecting of mangrove-derived RMA as sources of potential anti-infectives.
Conclusion
The pathogenesis of Vibrio cholerae is controlled via the QS-mediated communication process. The present study provides an insight in to the bioprospecting of mangrove-derived RMA to block QS-mediated response in V. cholerae. Micromonospora is one of the most productive genera among the phylum Actinobacteria in terms of the production of bioactive compounds. Yet the study of anti-virulence compounds with this genus is very limited. The Indian Ocean encompasses an astonishing source of marine microbial wealth, providing an opportunity to extensively exploit the RMA for the development of anti-virulence agents. The tapping of effective compounds from RMAs is achievable with the rapid development of modern technology in the field of sampling, isolation, and identification strategies that open up novel mechanisms to arrest the development of rapid dissemination of antimicrobial resistance and pathogenesis of human and animal pathogens. To conclude, the identification of the anti-virulence property of Micromonospora sp. is another example of Actinobacteria as a treasure trove for pharmaceutically important compounds.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HS conducted all the experiments described in this study. SK supported the acquisition of images. KS designed the computational work. AS and PN designed the study and supervised all the experiments. HS and SK drafted the manuscript and it was reviewed by SK, PN, and AS.
Funding
The authors would like to thank the support of DST-SERB (EMR/2016/002296) and SASTRA Deemed to be University, Thanjavur, India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Thandavarayan Ramamurthy, National Chair, Translational Health Science and Technology Institute, for the kind gift of V. cholerae strain HYR14.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01393/full#supplementary-material
Footnotes
References
Aarthi, P., Harini, R., Sowmiya, M., Malathi, J., Therese, K. L., and Madhavan, H. N. (2011). Identification of bacteria in culture negative and polymerase chain reaction (PCR) positive intraocular specimen from patients with infectious endopthalmitis. J. Microbiol. Methods 85, 47–52. doi: 10.1016/j.mimet.2011.01.010
Ahmed, W., Huygens, F., Goonetilleke, A., and Gardner, T. (2008). Real-time PCR detection of pathogenic microorganisms in roof-harvested rainwater in Southeast Queensland, Australia. Appl. Environ. Microbiol. 74, 5490–5496. doi: 10.1128/AEM.00331-08
Ali, M., Nelson, A. R., Lopez, A. L., and Sack, D. A. (2015). Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9:e0003832. doi: 10.1371/journal.pntd.0003832
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Augustine, N., Kerkar, S., and Thomas, S. (2012). Arctic actinomycetes as potential inhibitors of Vibrio cholerae biofilm. Curr. Microbiol. 64, 338–342. doi: 10.1007/s00284-011-0073-4
Balasubramanian, S., Othman, E. M., Kampik, D., Stopper, H., Hentschel, U., Ziebuhr, W., et al. (2017). Marine sponge-derived Streptomyces sp. SBT343 extract inhibits staphylococcal biofilm formation. Front. Microbiol. 8:236. doi: 10.3389/fmicb.2017.00236
Benitez, J. A., and Silva, A. J. (2016). Vibrio cholerae hemagglutinin(HA)/protease: an extracellular metalloprotease with multiple pathogenic activities. Toxicon 115, 55–62. doi: 10.1016/j.toxicon.2016.03.003
Bina, R. X., Taylor, D. L., Vikram, A., Ante, V. M., and Bina, J. E. (2013). Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 4:e00366-13. doi: 10.1128/mBio.00366-13
Bina, X. R., and Bina, J. E. (2010). The cyclic dipeptide cyclo(phe-pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae. J. Bacteriol. 192, 3829–3832. doi: 10.1128/JB.00191-10
Chowdhury, G., Bhadra, R. K., Bag, S., Pazhani, G. P., Das, B., Basu, P., et al. (2016). Rugose atypical Vibrio cholerae O1 El Tor responsible for 2009 cholera outbreak in India. J. Med. Microbiol. 65, 1130–1136. doi: 10.1099/jmm.0.000344
Clemens, J. D., Nair, G. B., Ahmed, T., Qadri, F., and Holmgren, J. (2017). Cholera. Lancet 390, 1539–1549. doi: 10.1016/S0140-6736(17)30559-7
Das, B., Verma, J., Kumar, P., Ghosh, A., and Ramamurthy, T. (2019). Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine 38, A83–A92. doi: 10.1016/j.vaccine.2019.06.031
Defoirdt, T. (2018). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005
Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., et al. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196. doi: 10.1021/jm051256o
Gladkikh, A. S., Feranchuk, S. I., Ponomareva, A. S., Bochalgin, N. O., and Mironova, L. V. (2020). Antibiotic resistance in Vibrio cholerae El Tor strains isolated during cholera complications in Siberia and the Far East of Russia. Infect. Genet. Evol. 78:104096. doi: 10.1016/j.meegid.2019.104096
Hammer, B. K., and Bassler, B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–114. doi: 10.1046/j.1365-2958.2003.03688.x
Hassan, S. S., and Shaikh, A. L. (2017). Marine actinobacteria as a drug treasure house. Biomed. Pharmacother. 87, 46–57. doi: 10.1016/j.biopha.2016.12.086
Hayakawa, M., and Nonomura, H. (1987). Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509. doi: 10.1016/0385-6380(87)90108-7
Hema, M., Princy, S. A., Sridharan, V., Vinoth, P., Balamurugan, P., and Sumana, M. N. (2016). pyrizine-2-carboxylic acid and antibiotics against. RSC Adv. 6, 45938–45946. doi: 10.1039/C6RA04705J
Hema, M., Vasudevan, S., Balamurugan, P., and Adline Princy, S. (2017). Modulating the global response regulator, LuxO of V. cholerae quorum sensing system using a pyrazine dicarboxylic acid derivative (PDCApy): an antivirulence approach. Front. Cell. Infect. Microbiol. 7:441. doi: 10.3389/fcimb.2017.00441
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. doi: 10.1099/00221287-146-10-2395
Hong, K., Gao, A. H., Xie, Q. Y., Gao, H., Zhuang, L., Lin, H. P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. doi: 10.3390/md7010024
Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J. F., Honig, B., Shaw, D. E., et al. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351–367. doi: 10.1002/prot.10613
Joelsson, A., Kan, B., and Zhu, J. (2007). Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73, 3742–3746. doi: 10.1128/AEM.02804-06
Kartik, K. S., Muthusamy, S., Kadhirvel, S., and Mani, K. P. (2020). In-silico and in-vitro analysis of endocan interaction with statins. Int. J. Biol. Macromol. 146, 1087–1099. doi: 10.1016/j.ijbiomac.2019.09.235
Kaur, G., Balamurugan, P., Uma Maheswari, C., Anitha, A., and Princy, S. (2016). Combinatorial effects of aromatic 1, 3-disubstituted ureas and fluoride on in vitro inhibition of Streptococcus mutans biofilm formation. Front. Microbiol. 7:861. doi: 10.3389/fmicb.2016.00861
Kharel (Sitaula), R., Janani, M. K., Madhavan, H. N., and Biswas, J. (2018). Outcome of polymerase chain reaction (PCR) analysis in 100 suspected cases of infectious uveitis. J. Ophthalm. Inflamm. Infect. 8, 1–8. doi: 10.1186/s12348-017-0144-1
Kutzner, C., Páll, S., Fechner, M., Esztermann, A., de Groot, B. L., and Grubmüller, H. (2019). More bang for your buck: improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 40, 2418–2431. doi: 10.1002/jcc.26011
Lam, K. S. (2006). Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9, 245–251. doi: 10.1016/j.mib.2006.03.004
Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni, R. V., Wingreen, N. S., and Bassler, B. L. (2004). The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82. doi: 10.1016/j.cell.2004.06.009
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K., and Bassler, B. L. (2002). Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. doi: 10.1016/S0092-8674(02)00829-2
Miller, V. L., and Mekalanos, J. J. (1984). Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. U.S.A. 81, 3471–3475. doi: 10.1073/pnas.81.11.3471
Morris, J. G., Sztein, M. B., Rice, E. W., Nataro, J. P., Losonsky, G. A., Panigrahi, P., et al. (1996). Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174, 1364–1368. doi: 10.1093/infdis/174.6.1364
Mushayabasa, S., and Bhunu, C. P. (2012). Assessing the impact of increasing antimicrobial resistance of Vibrio cholerae on the future trends of cholera epidemic. ISRN Biomathematics 2012, 1–10. doi: 10.5402/2012/127492
Narendrakumar, L., Gupta, S., Sen Johnson, J. B., Ramamurthy, T., and Thomas, S. (2019a). Molecular adaptations and antibiotic resistance in Vibrio cholerae: a communal challenge. Microb. Drug Resist. 25, 1012–1022. doi: 10.1089/mdr.2018.0354
Narendrakumar, L., Theresa, M., Krishnankutty Chandrika, S., and Thomas, S. (2019b). Tryptanthrin, a potential biofilm inhibitor against toxigenic Vibrio cholerae, modulating the global quorum sensing regulator, LuxO. Biofouling 35, 1093–1103. doi: 10.1080/08927014.2019.1696315
Perrière, G., and Gouy, M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369. doi: 10.1016/0300-9084(96)84768-7
Pisano, M. A., Sommer, M. J., and Lopez, M. M. (1986). Application of pretreatments for the isolation of bioactive actinomycetes from marine sediments. Appl. Microbiol. Biotechnol. 25, 285–288. doi: 10.1007/BF00253664
Pridham, T. G., and Gottlieb, D. (1948). The utilization of carbon compounds by some actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–114. doi: 10.1128/jb.56.1.107-114.1948
Rahman, H., Austin, B., Mitchell, W. J., Morris, P. C., Jamieson, D. J., Adams, D. R., et al. (2010). Novel anti-infective compounds from marine bacteria. Mar. Drugs 8, 498–518. doi: 10.3390/md8030498
Ravichandiran, V., Shanmugam, K., Anupama, K., Thomas, S., and Princy, A. (2012). Structure-based virtual screening for plant-derived SdiA-selective ligands as potential antivirulent agents against uropathogenic Escherichia coli. Eur. J. Med. Chem. 48, 200–205. doi: 10.1016/j.ejmech.2011.12.015
Rutherford, S. T., Van Kessel, J. C., Shao, Y., and Bassler, B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011
Sack, D. A., Sack, R. B., Nair, G. B., and Siddique, A. (2004). Cholera. Lancet 363, 223–233. doi: 10.1016/S0140-6736(03)15328-7
Saravanakumar, K., Anburaj, R., Gomathi, V., and Kathiresan, K. (2016). Ecology of soil microbes in a tropical mangrove forest of south east coast of India. Biocatal. Agric. Biotechnol. 8, 73–85. doi: 10.1016/j.bcab.2016.08.010
Sarveswari, H. B., and Solomon, A. P. (2019). Profile of the intervention potential of the phylum actinobacteria toward quorum sensing and other microbial virulence strategies. Front. Microbiol. 10:73. doi: 10.3389/fmicb.2019.02073
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., and Sherman, W. (2013). Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234. doi: 10.1007/s10822-013-9644-8
Shah, M. D., Kharkar, P. S., Sahu, N. U., Peerzada, Z., and Desai, K. B. (2019). Potassium 2-methoxy-4-vinylphenolate: a novel hit exhibiting quorum-sensing inhibition in: Pseudomonas aeruginosa via LasIR/RhlIR circuitry. RSC Adv. 9, 40228–40239. doi: 10.1039/c9ra06612h
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Silva, A. J., Pham, K., and Benitez, J. A. (2003). Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149, 1883–1891. doi: 10.1099/mic.0.26086-0
Takahashi, Y. (2004). Exploitation of new microbial resources for bioactive compounds and discovery of new actinomycetes. Actinomycetologica 18, 54–61. doi: 10.3209/saj.18_54
Therese, K. L., Anand, A. R., and Madhavan, H. N. (1998). Polymerase chain reaction in the diagnosis of bacterial endophthalmitis. Br. J. Opthalmol. 82, 1078–1082. doi: 10.1136/bjo.82.9.1078
Vance, R. E., Zhu, J., and Mekalanos, J. J. (2003). A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71, 2571–2576. doi: 10.1128/IAI.71.5.2571-2576.2003
Verma, J., Bag, S., Saha, B., Kumar, P., Ghosh, T. S., Dayal, M., et al. (2019). Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 116, 6226–6231. doi: 10.1073/pnas.1900141116
Vezzulli, L., Stauder, M., Grande, C., Pezzati, E., Verheye, H. M., Owens, N. J. P., et al. (2015). GbpA as a novel qPCR target for the species-specificdetection of Vibrio cholerae O1, O139, Non-O1/Non-O139 in environmental, stool, and historical continuous plankton recorder samples. PLoS One 10:e0123983. doi: 10.1371/journal.pone.0123983
Wang, R., Yu, D., Yue, J., and Kan, B. (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 6, 1–8. doi: 10.1038/srep22733
World Health Organization [WHO] (2019). Cholera. Available online at: https://www.who.int/news-room/fact-sheets/detail/cholera (accessed January 29, 2020).
Yang, M., Frey, E. M., Liu, Z., Bishar, R., and Zhu, J. (2010). The virulence transcriptional activator aphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect. Immun. 78, 697–703. doi: 10.1128/IAI.00429-09
Zheng, J., Shin, O. S., Cameron, D. E., and Mekalanos, J. J. (2010). Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 21128–21133. doi: 10.1073/pnas.1014998107
Zhu, J., and Mekalanos, J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 647–656. doi: 10.1016/S1534-5807(03)00295-8
Keywords: anti-infectives, anti-virulence, anti-biofilm, rare marine Actinobacteria, Micromonospora species, marine compounds, Vibrio cholerae
Citation: Sarveswari HB, Kalimuthu S, Shanmugam K, Neelakantan P and Solomon AP (2020) Exploration of Anti-infectives From Mangrove-Derived Micromonospora sp. RMA46 to Combat Vibrio cholerae Pathogenesis. Front. Microbiol. 11:1393. doi: 10.3389/fmicb.2020.01393
Received: 02 April 2020; Accepted: 29 May 2020;
Published: 10 July 2020.
Edited by:
Jinwei Zhang, University of Exeter, United KingdomReviewed by:
Srinivasan Ramanathan, Fujian Agriculture and Forestry University, ChinaShunmugiah Karutha Pandian, Alagappa University, India
Copyright © 2020 Sarveswari, Kalimuthu, Shanmugam, Neelakantan and Solomon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasanna Neelakantan, cHJhc2FubmFAaGt1Lmhr; Adline Princy Solomon, YWRsaW5lcHJpbnp5QGJpb3RlY2guc2FzdHJhLmVkdQ==; YWRsaW5lcHJpbmN5QGdtYWlsLmNvbQ==
 Hema Bhagavathi Sarveswari
Hema Bhagavathi Sarveswari Shanthini Kalimuthu
Shanthini Kalimuthu Karthi Shanmugam
Karthi Shanmugam Prasanna Neelakantan
Prasanna Neelakantan Adline Princy Solomon
Adline Princy Solomon