- 1Ocean College, Zhejiang University, Hangzhou, China
- 2College of Life Sciences, Zhejiang University, Hangzhou, China
- 3Cardiovascular Health Department, AstraZeneca Trading Co., Ltd., Wuxi, China
- 4Key Laboratory of Vector Biology and Pathogen Control of Zhejiang Province, College of Life Science, Huzhou University, Huzhou, China
Strain HM190, a moderate halophile, was isolated from rhizosphere soil of the mangrove Kandelia obovata in Fugong village, China. The 16S ribosomal RNA (rRNA) gene sequence and the results of phylogenetic analysis revealed that strain HM190 belonged to the genus Streptomyces and had the highest sequence similarity of 99.79% to Streptomyces heilongjiangensis NEAU-W2T. The complete genome of strain HM190 comprised 7,762,826 bp in a linear chromosome with 71.97% G + C content. According to antiSMASH analysis, a total of 30 biosynthetic gene clusters (BGCs) were predicted to be involved in secondary metabolism, 12 of which were responsible for the production of polyketide- and non-ribosomal peptide-derived secondary metabolites. Gene cluster 5 was responsible for macrolide biosynthesis in a strain-specific 126,331-bp genomic island belonging to the left-arm region. Combined genomics–metabolomics analysis led to the discovery of three 22-membered macrolides (compounds 1–3). Their structures were elucidated by using spectroscopic techniques including high-resolution electrospray ionization mass spectroscopy (HRESIMS) and nuclear magnetic resonance (NMR). The absolute configurations of compounds 1–3 were determined by the X-ray single crystal diffraction and NMR data analysis. All three compounds displayed moderate cytotoxic activities toward tumor cell lines HepG2, A549, and HCT116.
Introduction
Mangrove ecosystems cover about 60–75% of the world’s tropical and subtropical coastlines and grow in saline coastal sediment habitats at transition zones with ocean, fresh water, and land (Holguin et al., 2001). The tidal action in these areas causes large changes in temperature, oxygen, and salinity levels during the day, which leads to the formation of unique microbial communities (Kathiresan and Bingham, 2001). The phylogenetic diversity of mangrove microbial communities has been well established based on 16S ribosomal RNA (rRNA) analytical approaches, including PCR cloning (Li et al., 2011), denaturing gradient gel electrophoresis (Tian et al., 2008), and pyrosequencing (Santos et al., 2011). Recently, various mangrove-derived microorganisms have attracted increasing attention in the drug discovery field, especially those of the genus Streptomyces, as important sources of biologically active compounds with diverse structures (Xu, 2011; Thatoi et al., 2013; Blunt et al., 2015).
The genus Streptomyces belongs to the family Streptomycetaceae, the order Streptomycetales, and the class Actinobacteria, and was originally described in Waksman and Henrici (1943). At the time of writing, 854 Streptomyces species with validly published names had been proposed according to List of Prokaryotic names with Standing in Nomenclature1. Some of these Streptomyces strains have been isolated from a wide range of marine habitats, including mangroves (Yan et al., 2010), marine sediments (Zhao et al., 2009), seawater (Zhu et al., 2011), sponges (Huang et al., 2016), algae (Girão et al., 2019), and corals (Alfredo et al., 2017). Streptomyces constitutes an important source of new natural products because its strains contain a large number of biosynthetic gene clusters (BGCs) associated with the production of multiple secondary metabolites (Hwang et al., 2014; Liu et al., 2018). Since the first complete genome of Streptomyces coelicolor A3(2) was sequenced and reported in Bentley et al. (2002), increasing numbers of Streptomyces genomes have been sequenced and deposited in public databases in recent years (Studholme, 2016), which has led to an increase in genomics–metabolomics studies of this genus (Xu et al., 2019). However, most of these genomes were draft genomes. The number of complete genomes of Streptomyces has been limited because of their high G + C content, which results in shorter reads and much higher error rates during genome sequencing (Ioanna et al., 2012; Zhong et al., 2013).
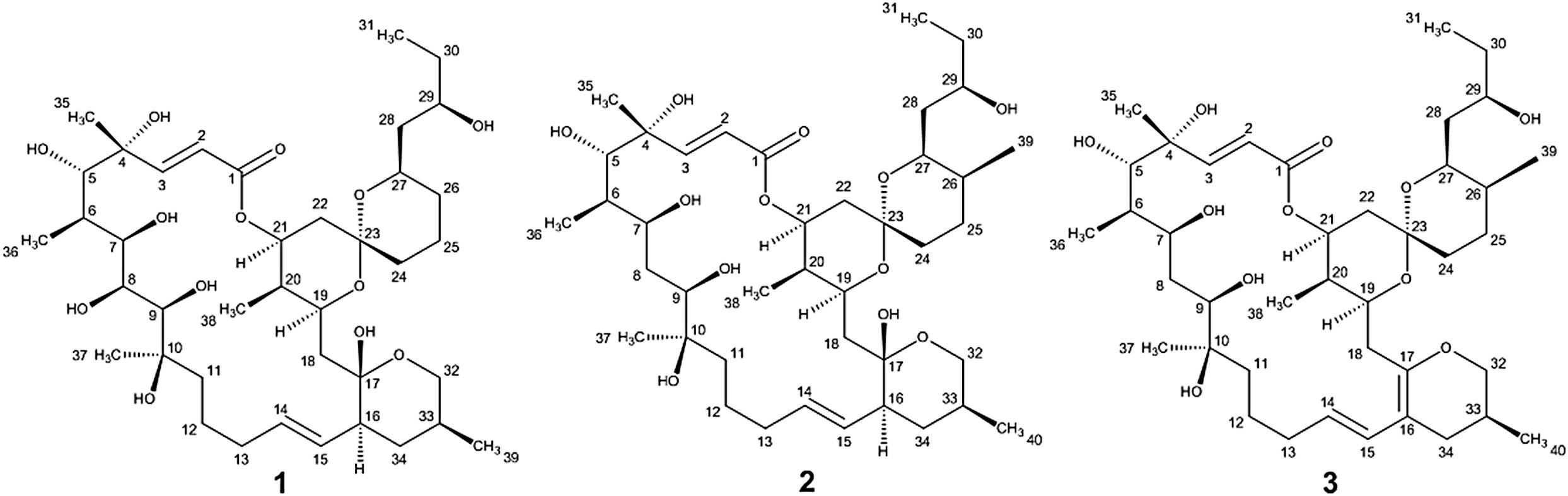
During our work focused on discovering microorganism diversity from mangrove sample, Strain HM190, as a novel species of the genus Streptomyces, was isolated from rhizosphere soil of the mangrove Kandelia obovata in Fugong village, Zhangzhou, China. The complete genome of the strain was sequenced, and 30 gene clusters were predicted to be involved in the biosynthesis of secondary metabolites using the antiSMASH 5.0 software (Blin et al., 2019). Genome analysis of the isolate indicated that gene cluster 5 was probably involved in the production of a compound structurally similar to the macrolide of apoptolidin, based on its high similarity to the type I polyketide synthase (PKS) from the apoptolidin BGC of Nocardiopsis sp. FU 40 (Du et al., 2011). As a result, three 22-membered macrolides (compounds 1–3, Figure 1) were isolated from its fermentation broth. The absolute configuration of compound 1 was confirmed by X-ray single-crystal diffraction. The absolute configurations of compounds 2 and 3 were established through detailed nuclear magnetic resonance (NMR) data analysis in comparison with compound 1 and other related compounds. Herein, we describe the isolation, structure elucidation, and cytotoxic activity of the three compounds.
Materials and Methods
General Experimental Procedures
Silica gel plates HSGF254 (Yantai Chemical Industry Research Institute, Yantai, China) were used for thin-layer chromatography (TLC). Column chromatography was performed with commercial silica gel (QingDao HaiYang Chemical Group Co., QingDao, China, 200-300 mesh). High-performance liquid chromatography (HPLC) separation was performed on semipreparative HPLC (Agilent 1100, Zorbax SB-C18, 5 μm, 250 × 9.4 mm inner diameter; 1.5 mL/min; 210 nm; Agilent, Palo Alto, CA, United States). NMR spectra were measured with a Bruker DRX-400 spectrometer (Bruker, Rheinstetten, Germany). The HRESIMS spectra were taken on a Q-TOF Micro LC-MS-MS mass spectrometer (Waters Co., Milford, MA, United States). Optical rotation was measured on a Perkin-Elmer 341 polarimeter (Perkin-Elmer, Suzhou, China). IR spectra were recorded on a Nicolet Magna FT-IR 750 spectrometer (Nicolet, Tokyo, Japan). UV spectra were recorded on a Varian CARY 300 BIO spectrophotometer (Varian, Cary, NC, United States).
Isolation and Identification of Strain HM190
Strain HM190 was isolated from a soil sample collected from rhizosphere soil of the mangrove K. obovata of Fugong village (117° 57′ N 24° 24′ E), in Zhangzhou, China, which was placed into clean plastic buckets and stored at 4°C until use. Serially diluted (10-fold dilutions each) samples were made and spread on the five selective isolation media by the traditional dilution-plating method: HV (humic acid-vitamin agar medium), M7 (glycerine-peptone agar medium), GS (Gauze’s modified medium no. 1), ISP2 (ISP medium no. 2), and MA (marine agar 2216 medium). All of the media were supplemented with 25 mg/L of nalidixic acid and 50 mg/L of nystatin. After 7 days of incubation at 30°C, a white-pigmented colony was picked from MA plates and named as HM190. After repeated plate streaking on the same medium, pure strains were obtained from individual colonies and preserved at −80°C as suspension with 25% (v/v) glycerol for further use (Ye et al., 2019).
For phylogenetic studies, the isolate was grown in MB (marine broth 2216 medium) for 7 days at 30°C. The 16S rRNA gene sequence amplification was performed by using universal primers 27F [5′-AGAGTTTGATCCTGGCTCAG-3′] and 1492R [5′-ACGGCTACCTTGTTACGACTT-3′] (Anzai et al., 2000). Amplification reactions were prepared in a 25 μL final reaction volume containing 12.5 μL of PCR SuperMix, 11 μL of distilled water, 0.5 μL of each primer, and 0.5 μL of extracted DNA template. PCR was performed under the following conditions: 30 cycles of 94°C/5 min, 4°C/30 s, 55°C/30 s, 72°C/75 s and a final extension of 72°C/10 min. PCR products were ligated to vector pMD 19-T (TaKaRa) and cloned into Escherichia coli DH5a for sequencing, and the almost-complete sequence of the 16S rRNA gene sequence was obtained. The obtained sequence was assembled with DNASTAR SeqMan (LaserGene, Madison, WI, United States). Further, the 16S rRNA gene sequence was analyzed by submitting to the NCBI2 and the EzTaxon-e server3 (Altschul et al., 1997; Yoon et al., 2017). The multiple sequences were aligned with Clustal W (Thompson et al., 1994). Phylogenetic trees were constructed with the MEGA 5.0 software package (Molecular Evolutionary Genetics Analysis, version 5.0) (Tamura et al., 2011) by using neighbor-joining (Saitou and Nei, 1987), minimum-evolution (Rzhetsky and Nei, 1992), and maximum-likelihood (Felsenstein, 1981). Bootstrap analysis (1000 replicates) was used to evaluate the trees topology. Kimura two-parameter model was used for phylogeny construction and evolutionary distances analysis (Kimura, 1981). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain HM190 is MN897722.
Genome Sequencing and Annotation
For DNA isolation, strain HM190 was inoculated into MB medium and grown at 30°C with shaking (200 r/min) for 7 days. High-quality genomic DNA was extracted using the Bacteria Genomic DNA Extraction Kit (DongSheng Biotech) according to the manufacturer’s instructions. The genome was sequenced using the PacBio RS II platform and the Illumina HiSeq 4000 platform (Beijing Genomics Institute) (163-fold). Four SMRT cells Zero-Mode Wave guide arrays of sequencing were used by the PacBio platform to generate the sub-reads set. The PacBio sub-reads (length < 1 kb) were removed. To improve the accuracy of the genome sequence, the GATK4 and the SOAP (SOAP2, SOAPsnp, SOAPindel) tool packages were used to make single-base corrections. To confirm the presence of any plasmid, the filtered Illumina reads were mapped using the SOAP to the bacterial plasmid database5. The gene prediction was performed by the glimmer 36 with hidden Markov models. The open reading frames (ORFs) were annotated by the Rapid Annotation using Subsystem Technology (RAST) server online (Overbeek et al., 2014). The Clusters of Orthologous Groups (COG) database was used for general function annotation. For the identification of secondary metabolism gene clusters, the antiSMASH 5.0 program was used (Blin et al., 2019). Whole Genome Shotgun project of Streptomyces sp. HM190 has been deposited at DDBJ/ENA/GenBank under the accession CP047318.
Fermentation, Extraction, and Isolation of Strain HM190
Strain HM190 was inoculated into a 1000-mL flask containing 250 mL of the seed culture medium consisting of glucose (0.4%), yeast extract (0.4%), malt extract (1.0%), and CaCO3 (0.2%), with a pH of 7.2. After incubated at 30°C for 3 days on a rotary shaker operating at 250 r/min, the 5.0% seed culture broth was added to a 50 L fermenter (containing 30 L of fermentation medium) incubation for 7 days at 30°C. The fermentation medium component was the same as the isolation medium of MB (marine broth 2216 medium).
The final 30 L of fermentation broth was centrifuged to separate supernatant and bottomed mycelial cake. The mycelial cake was subsequently extracted with MeOH (3 L). The supernatant passed through a Diaion HP-20 resin column (Mitsubushi Chemical Co., Ltd., Tokyo, Japan) and eluted with 95% EtOH. The MeOH extract and the EtOH eluent were concentrated under reduced pressure at 55°C to yield a crude extract (30 g). The crude extract was chromatographed on a silica gel column (Qingdao Haiyang Chemical Group, Qingdao, China; 200-300 mesh) eluted with CHCl3/MeOH (100:0, 98:2, 95:5, 90:10, 80:20, 70:30, 60:40, and 50:50, v/v) to give six fractions (Fr.1–6) based on the TLC profiles. After the Fr.5 was concentrated under reduced pressure at 53°C and dried in vacuo, the material was subjected to a Sephadex LH-20 gel column (GE Healthcare, Glies, United Kingdom) and eluted with CHCl3/MeOH (1:1, v/v), to yield two fractions (Fr.5-1 and Fr.5-2) by using TLC detection. The Fr.5-1 was further chromatographed on a silica gel column and eluted with CHCl3/MeOH (100:0, 98:2, 95:5, 90:10, 85:15, 80:20, 75:25, and 70:30 v/v) to give four fractions (Fr.5-1-1–Fr.5-1-4). The Fr.5-1-1 was analyzed and purified by semi-preparative reversed-phase HPLC and eluted with CH3CN/H2O (60:40, v/v) to get compound 1 (tR 28.5 min, 15.2 mg). The Fr.5-1-3 was subjected to a Sephadex LH-20 column eluted with CHCl3/MeOH (1:1, v/v) to give three fractions (Fr.5-1-3a, Fr.5-1-3b, and Fr.5-1-3c). The Fr.5-1-3c was further separated by semi-preparative reversed-phase HPLC and eluted with CH3CN/H2O (48:52, v/v) to get compounds 2 (tR 37.0 min, 9.8 mg) and 3 (tR 45.0 min, 5.8 mg), respectively.
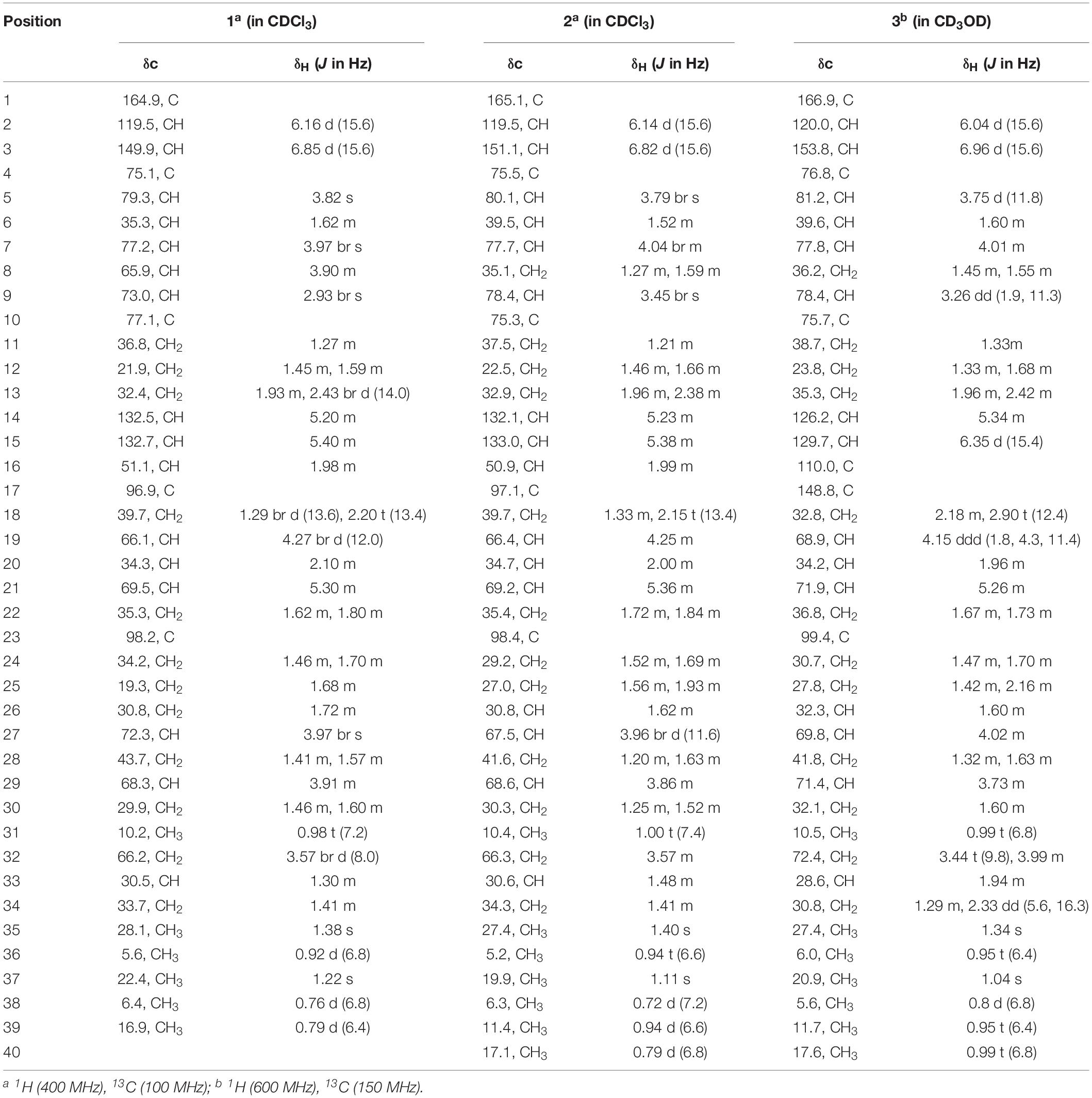
Compound 1: white powder; [α] = −4 (c 0.05, EtOH); IR (KBr) νmax 3441.9, 2931.3, 1701.1, 1647.0, 1419.2, 1392.8, and 1287.2 cm–1; UV (EtOH) λmax (log ε) 202 (3.93), 217 (3.82) nm; 1H (400 MHz) and 13C NMR (100 MHz) data shown in Table 1; positive HRESIMS m/z 765.4359 [M + Na]+, (calcd. for C39H66NaO13, 765.4396).
Compound 2: white powder; [α] = −6 (c 0.05, EtOH); IR (KBr) νmax 3440, 2900, 1700, 1640, 1450, 1380, 1260, 1180, 1080, and 980 cm–1; UV (MeOH) λmax 223 nm (ε 8,900); 1H (400 MHz) and 13C NMR (100 MHz) data shown in Table 1; positive HRESIMS m/z 763.4546 [M + Na]+, (calcd. for C39H66NaO13, 763.4603).
Compound 3: white powder; [α] = −5 (c 0.05, EtOH); IR (KBr) νmax 3450, 2950, 1720, 1660, 1460, 1390, 1280, 1180, 1100, 980 cm–1; UV (MeOH) λmax 247 nm (ε 13,500); 1H (600 MHz) and 13C NMR (150 MHz) data shown in Table 1; positive HRESIMS m/z 745.4437 [M + Na]+, (calcd. for C39H66NaO13, 745.4497). The IR and UV data of compounds 2 and 3 from Akira H. et al.
X-Ray Crystallography
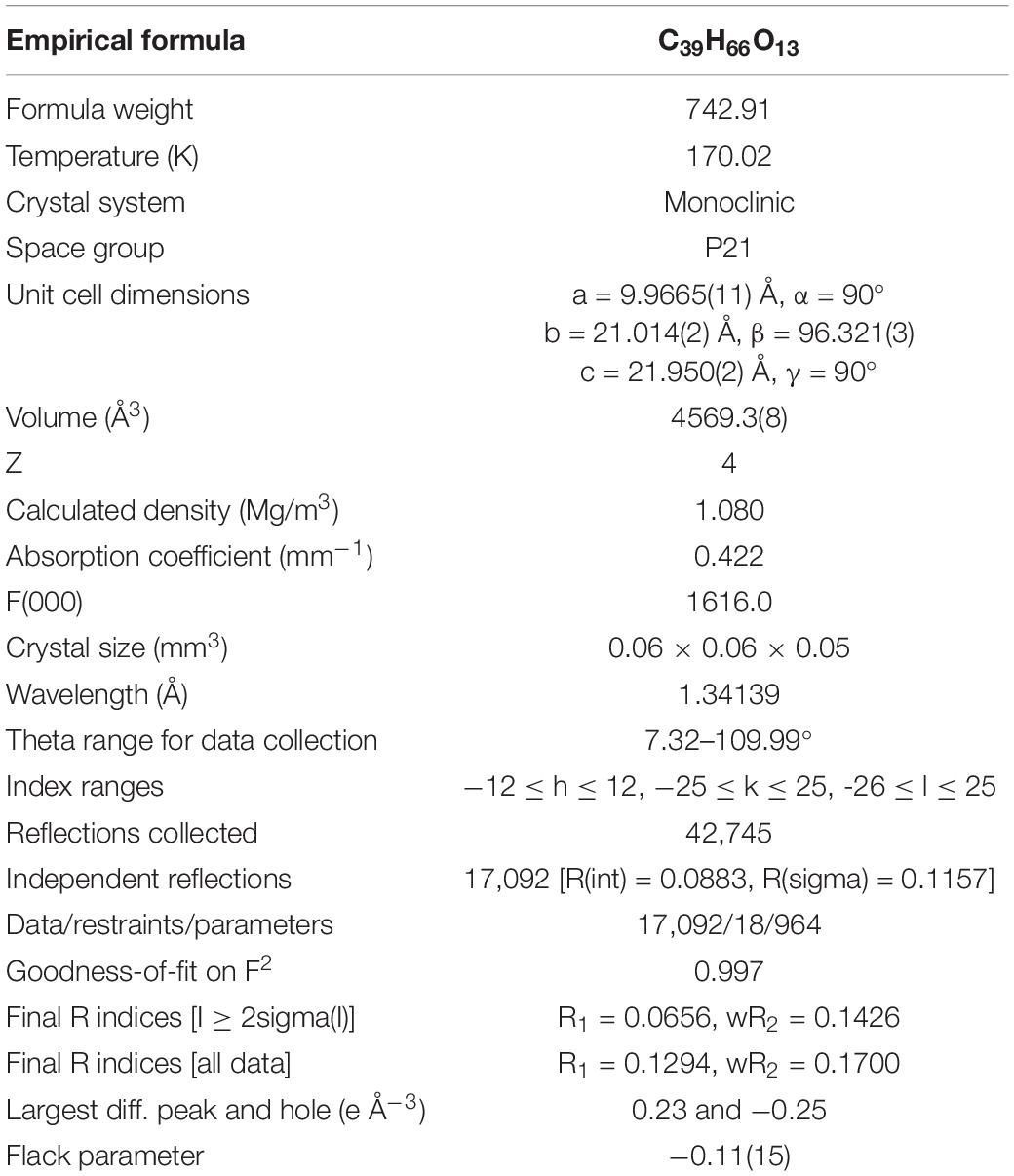
Small seed crystals of compound 1 were produced by controlling evaporation of solutions at room temperature over 2 weeks in CHCl3/MeOH (60:40, v/v). Re-crystallization at 4°C to induce the production of high-quality crystals for X-ray diffraction was conducted for 20 days. A suitable crystal was selected and mounted on a Bruker D8 Venture diffractometer. The crystal was kept at 170.02 K during data collection. The structure was solved with the ShelXT (Sheldrick, 2015) structure solution program using Intrinsic Phasing. Calculations were made with the ShelXL (Kratzert et al., 2015) refinement package using least squares minimization as implementation in Olex2 (Dolomanov et al., 2009). Crystallographic data (excluding structure factor) for compound 1 has been deposited with the Cambridge Crystallographic Data Center under the deposition number CCDC 1990958.
Biological Assay
The antimicrobial activities of compounds 1–3 against pathogenic bacteria Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus, and pathogenic fungus Candida albicans were detected with the minimum inhibitory concentrations (MICs) method recommended by the Clinical and Laboratory Standards Institute (Wang et al., 2019). Amphotericin B (an antifungal antibiotic) and gentamicin (an antibacterial antibiotic) were used as a positive control.
The cytotoxic activities of compounds 1–3 were investigated against the human colon tumor cell line HCT116, the human lung carcinoma cell line A549, and the hepatocellular carcinoma cell line HepG2 in vitro by the CCK8 (cell counting kit-8) colorimetric method. The cell lines were incubated in a 5% CO2 incubator at 37°C for 4 h under Dulbecco’s Modified Eagle’s Medium (DMEM) solution containing 10% calf serum. The adherent cells of the logarithmic growth stage were digested and seeded in a 96-well culture plate at a density of 1 × 104 cells per/well. Then the test samples and controls [Frame1] were added to the medium and cultivated for 48 h. Further the CCK8 (Dojindo, Kumamoto, Japan) reagent was added to the medium and cultured for 3 h. Cell viability was detected by the absorbance at 450 nm using a SpectraMax M5 microplate reader (Molecular Devices Inc., Sunnyvale, CA, United States) (Wang et al., 2009). Doxorubicin was used as a positive control. Cell solution (dimethylsulfoxide) was tested as a negative control. The inhibitory rate of cell proliferation was expressed as IC50 value.
Results and Discussion
16S rRNA Gene Sequence and Phylogenetic Analysis
PCR was used to determine the 16S rRNA gene sequence (1451 bp, NCBI GenBank accession number: MN897722) of strain HM190. The results of the analysis indicated that the isolate belonged to the genus Streptomyces and showed the highest sequence similarities to Streptomyces heilongjiangensis NEAU-W2T (99.79%) and Streptomyces neyagawaensis NRRL B-3092T (99.59%). In the phylogenetic analysis based on a neighbor-joining tree (Figure 2), strain HM190 fell within the cluster of the genus Streptomyces and formed a coherent clade with S. heilongjiangensis NEAU-W2T. The clade had firm bootstrap support and represented an independent lineage. Similar results were obtained by using the maximum-parsimony and maximum-likelihood trees (Supplementary Figures S26, S27).
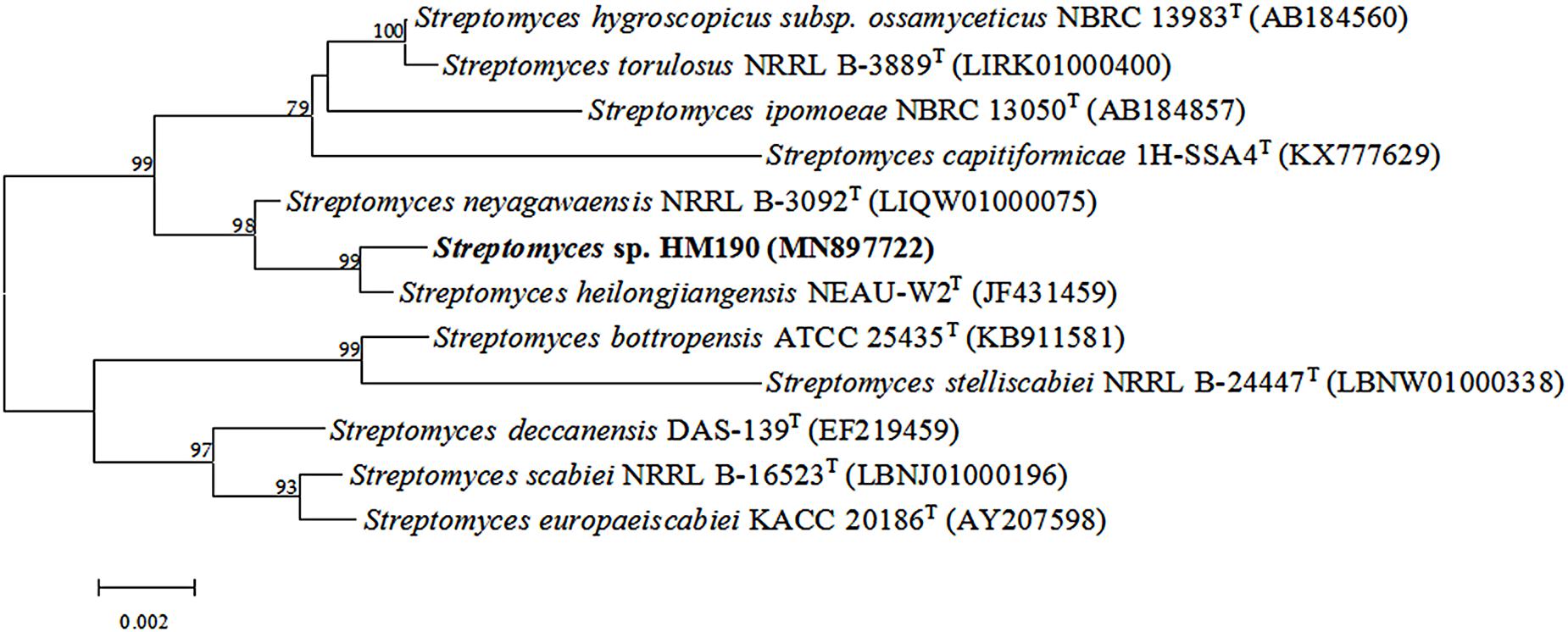
Figure 2. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationships among strain HM190 and other closely related species of the genus Streptomyces. Bootstrap values are expressed as a percentage of 1000 replicates and only those higher than 50% are given at the branch points. Bar, 0.002 substitutions per nucleotide position.
Genome Sequence and AntiSMASH Analysis
The whole genome sequence of Streptomyces sp. HM190 was assembled using the PacBio RSII and Illumina HiSeq400 platforms. High-quality clean data of size 1266 Mb with a total genome size of 9.10 Mb were generated and assembled into a linear chromosome. The complete genome consisted of 776,2826 bp composing a linear chromosome with 71.97% G + C content. No plasmid was identified. A total of 6971 protein-coding genes were predicted. Of these, 4728 genes (67.8%) were annotated by querying the COG database. A map of the chromosome and the COG functional categories of strain HM190 are shown in Figure 3. For further accurate secondary metabolism BGC (SMBGC) mining analysis, 30 SMBGCs of strain HM190 were proposed using antiSMASH 5.0 (Blin et al., 2019), occupying 15.4% of the chromosome (Supplementary Table S2).
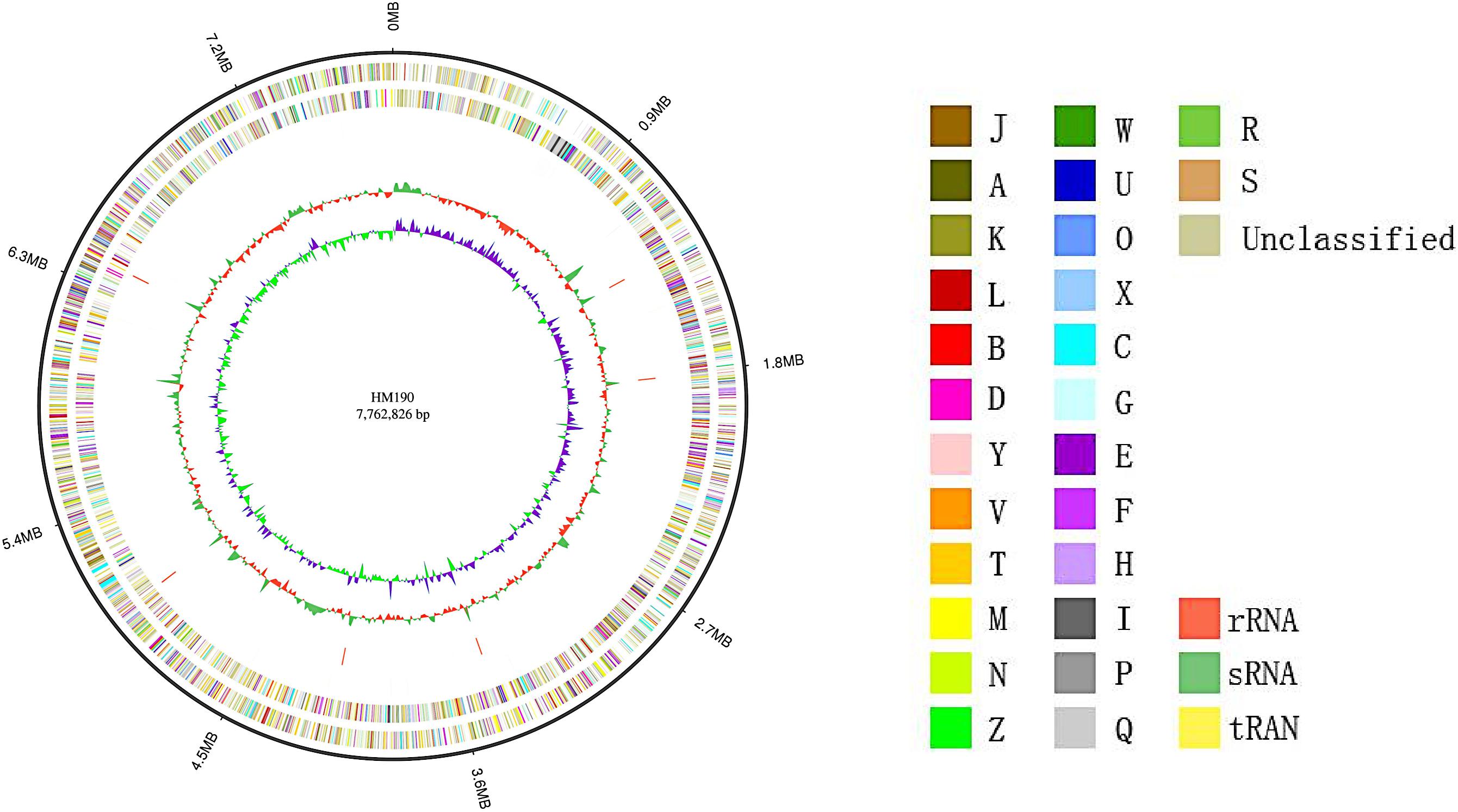
Figure 3. Schematic representation of the complete genome of Streptomyces sp. HM190. The outer scale is numbered in intervals of 0.9 Mb from the right to the left ends. From outer to inner: circle 1 (solid line) shows the genome size; in circles 2 and 3 (forward and reverse strands), the predicted protein-coding regions are colored according to clusters of orthologous groups classification (Supplementary Table S1); circles 4 and 5 (forward and reverse strands) show the distribution of rRNA operons (red), sRNA operons (green), and tRNA operons (yellow); circle 6 shows G + C content; and circle 7 indicates the GC skew.
Twelve of the 30 BGCs were predicted to be responsible for the production of PKS and non-ribosome peptide- (NRPS) derived secondary metabolites, including one type I PKS (cluster 5), one type II PKS (cluster 14), two type III PKS (clusters 2, 16), two NRPS (clusters 3, 21), and six hybrid BGCs (clusters 1, 4, 6, 10, 15, 23), which possessed genes encoding more than one type of biosynthetic enzyme. Cluster 5 encoded for a type I PKS that is probably responsible for the biosynthesis of a 20/21-membered macrolide of apoptolidin. A sequence similarity of 74.5% was obtained between cluster 5 and the Nocardiopsis sp. FU 40 apoptolidin gene cluster (NCBI accession number: JF819834) based on BLASTN analysis. According to Du et al. (2011), the apoptolidin gene cluster of Nocardiopsis sp. FU 40 produces 10 apoptolidin and isoapoptolidin compounds, and the biosynthetic pathway was confirmed. Cluster 14 was probably involved in the production of a hiroshidine-like compound, based on its high similarity to the type II PKS enzyme from the hiroshidine BGC from Streptomyces hiroshimensis (Moon et al., 2019). Gene clusters 2 and 16 encoded, respectively, for type III PKSs that resembled the alkylresorcinol and germicidin PKSs from Streptomyces spp. (Funabashi et al., 2008; Becerril et al., 2018). Cluster 3 encoded for an NRPS that probably synthesized scabichelin (Kodani et al., 2013). Cluster 21 was similar to the Salinispora tropica CNB-440 gene cluster associated with biosynthesis of the anticancer agent salinosporamide A (Fenical et al., 2009). Based on its high similarity to the recently discovered naphthyridinomycin BGC from Streptomyces lusitanus, cluster 1 was probably involved in the production of a naphthyridinomycin-like compound (Pu et al., 2013). Gene clusters 4 and 15 were predicted to be responsible for biosynthesis of gaudimycin- and glycosyl ester-type compounds, respectively (Kallio et al., 2008; Wang et al., 2015). Cluster 6 encoded for a type II PKS that probably synthesized granaticin (Ichinose et al., 1998). Cluster 10 encoded for a 2H-furan type PKS based on its high similarity to the Streptomyces sp. 88-682 gene cluster (Sun et al., 2009). Six gene clusters (8, 9, 11, 20, 22, 29) within the genome of strain HM190 were observed related to saccharide biosynthesis; cluster 11 also harbored genes encoding for melanin formation. Several other secondary metabolites were potentially produced by strain HM190, including two siderophore molecules encoded by clusters 12 and 27 (cluster 12 showed high similarity to desferrioxamine B biosynthetic genes). Bacteriocin or other unspecified ribosomally synthesized and post-translationally modified peptides were encoded by clusters 13 and 25 (cluster 13 showed high similarity to bottromycin A2 biosynthetic genes), cluster 7 (ectoine), cluster 17 (lanthipeptide or lasso peptide), cluster 18 (indole), cluster 19 (halogenide), cluster 28 [linear azol(in)e-containing peptide], and clusters 24, 26, and 30 (terpenes).
Structure Elucidation
Based on the COG categorization of the genome of strain HM190, there were a considerable number of genes related to secondary metabolite biosynthesis, transport, and catabolism (Figure 3). Together with the antiSMASH analysis, strain HM190 appeared to be an isolate with prolific potential for the production of various new and unique secondary metabolites. In order to assess this, the strain was grown for scale-up culture in 30 L of fermentation medium for 7 days at 30°C. Results of HPLC analysis of the crude extract are shown in Figure 4. Compounds 1–3 were purified and subjected to detailed structural analysis.
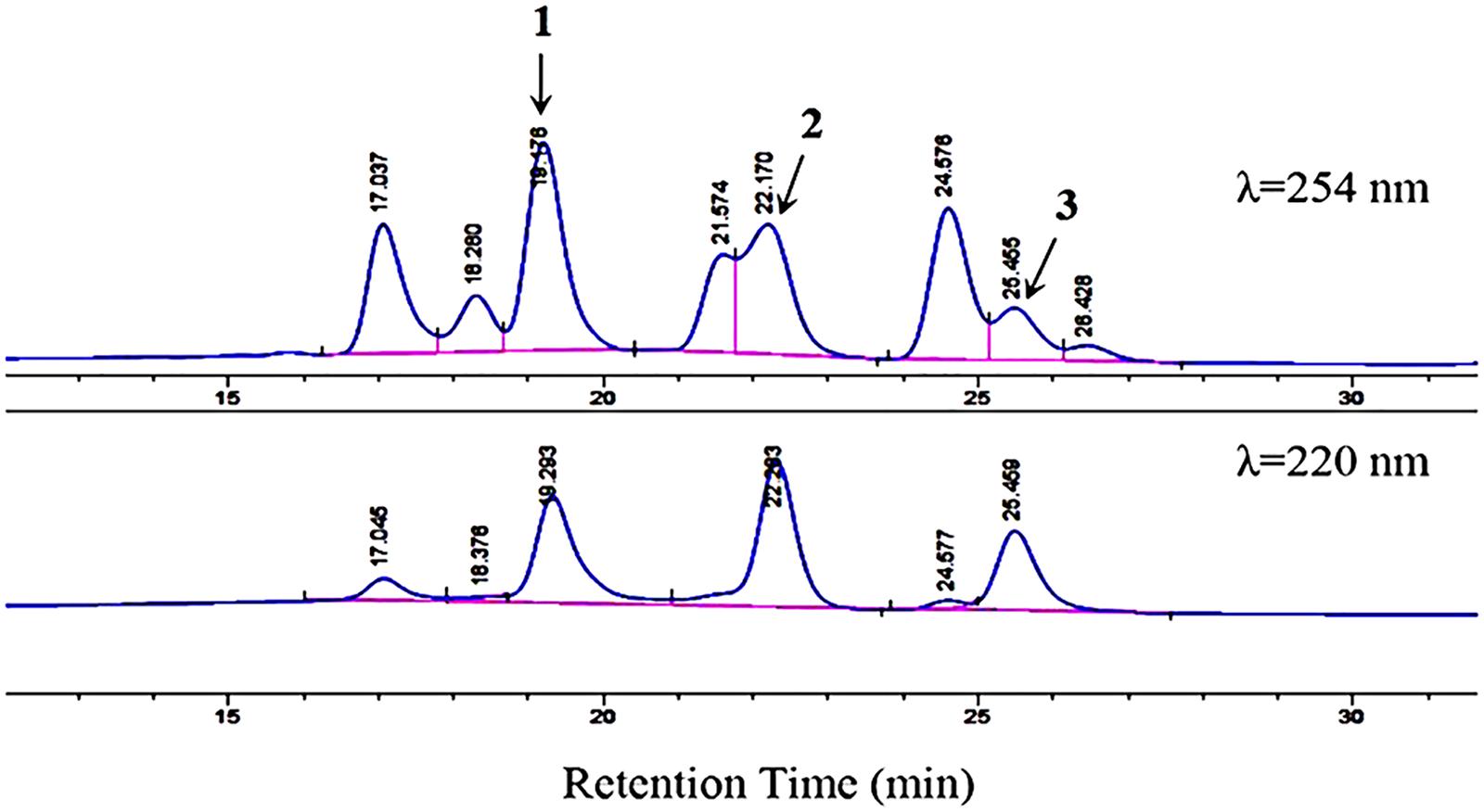
Figure 4. HPLC analysis of compounds 1–3 with a mobile phase of CH3CN/H2O (70:30, v/v). Flow rate, 1.5 mL/min. Detection wavelengths, 254 and 220 nm. The figure does not include the full chromatogram map of the crude extract but only an expansion of it.
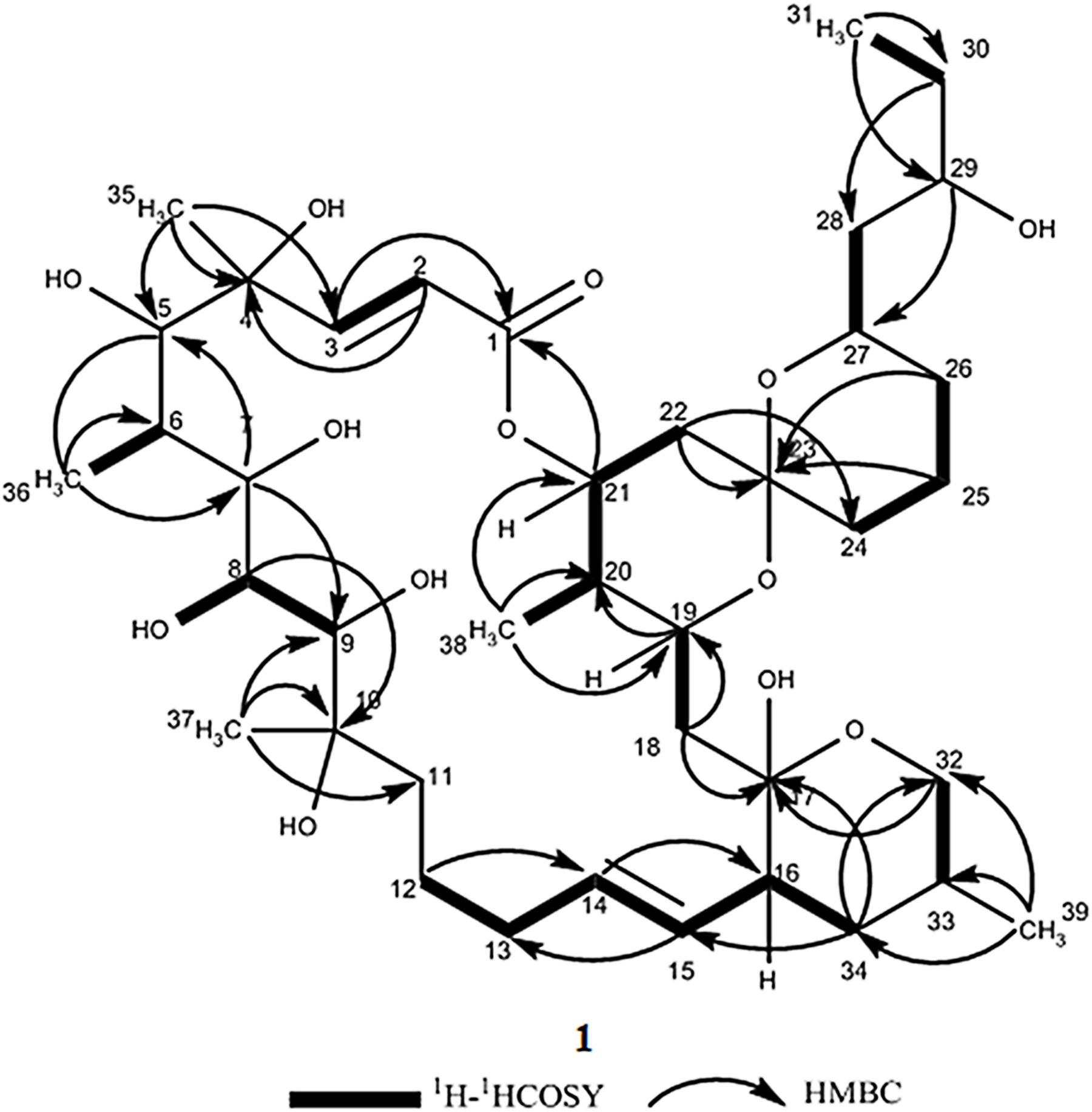
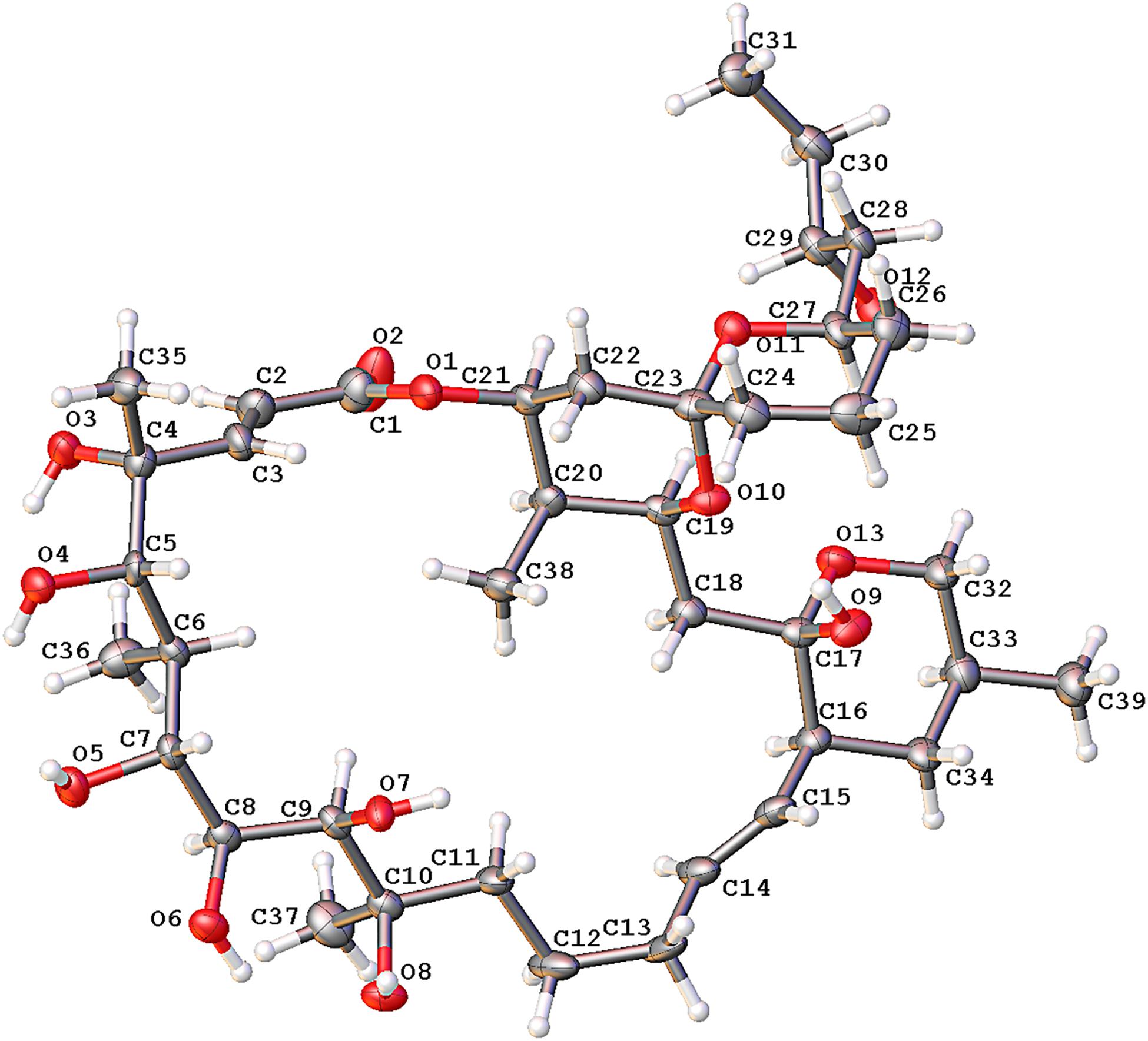
Compound 1 was obtained as a white powder with a molecular formula of C39H66O13 (seven degrees of unsaturation) based on high-resolution electrospray ionization mass spectroscopy (HRESIMS) data (m/z 765.4359 [M + Na]+, calcd. 765.4396). The 1H and 13C NMR (Table 1), distortionless enhancement by polarization transfer (DEPT) and heteronuclear single quantum coherence (HSQC) spectra for 1 indicated the presence of two double bonds (δC 149.9, 119.5, 132.5, and 132.7); five quaternary carbons (δC 164.9, 98.2, 96.9, 77.1, and 75.1), including one carbonyl carbon (δC 164.9); six methyl groups (δC 28.1, 22.4, 16.9, 10.2, 6.4, and 5.6); 12 methylene groups, one of which was oxygenated (δC 66.2); and 16 methine groups, eight of which were oxygenated. Detailed analysis of the 1H–1H COSY (homonuclear chemical shift correlation spectroscopy) and heteronuclear multiple bond correlation (HMBC) spectra revealed that 1 was a spiroketal 22-membered macrolide with a dihydropyran moiety (Figure 5). The spiroketal ring system was established based on the HMBC correlations of H-22 with C-23, of H-22 with C-24, of H-25 with C-23, and of H-26 with C-23, together with the 1H-1H COSY correlations of H-21 with H-22, H-24 with H-25, and H-25 with H-26. The HMBC correlations from H-32 to C-17, from H-34 to C-17 and C-32, from H-34 to C-15, and from H-39 to C-32, C-33, and C-34 revealed the presence of a dihydropyran moiety and the fusion of this dihydropyran moiety to the 22-membered macrolide system. This result was supported by the 1H-1H COSY correlations H-16/H-34 and H-32/H-33. From these unambiguous one- and two-dimensional correlation NMR spectral data, the structure of 1 was assigned (Figure 1) and determined to represent a new 22-membered macrolide. These data were similar to those of phthoramycin, which was previously isolated from Streptomyces sp. WK-1875 (Nakagawa et al., 1989). Compound 1 had six methyl groups, whereas phthoramycin contained seven methyl groups. Comparisons of the NMR data between 1 and phthoramycin indicated that 1 differed from phthoramycin by the absence of a methyl substituent at C-26 and the presence of a hydroxy group at C-8.
The molecular formula of compound 2 was established as C40H68O12 (seven degrees of unsaturation) by the HRESIMS data (m/z 763.4546 [M + Na]+, calcd. 763.4603). The molecular formula of compound 2 revealed that it had the same weight as the known kaimonolide A (Akira et al., 1989). Based on detailed NMR data analysis, the structure of compound 2 was found to show the same planar structure with kaimonolide A. Compound 3 was isolated as an analog of compound 2. The molecular formula of compound 3 was established as C40H66O11 (eight degrees of unsaturation) from the HRESIMS data (m/z 745.4437 [M + Na]+, calcd. 745.4497). The 1H and 13C NMR spectral data for 2 and 3 are shown in Table 1. Comparisons of the molecular formulas and NMR data of 2 and 3 indicated that 3 was a dehydrated product of 2, resulting from the formation of a double bond at C-16/C-17. It should be noted that during our studies, compound 2 would have gradually changed to 3 in CDCl3 solution (Akira et al., 1989). Compounds 2 and 3 also closely resembled phthoramycin and cytovaricin B, which have been reported in Streptomyces spp. (Sakurai et al., 1983; Nakagawa et al., 1989).
Absolute Configurations of Compounds 1–3
To define the correct absolute configuration of compound 1, a single crystal of compound 1 was prepared for X-ray single-crystal diffraction. Crystals of compound 1 (C39H66O13, CCDC 1990958) were almost colorless and monoclinic with typical dimensions 0.06 × 0.06 × 0.05 mm3. Compound 1 crystallized in space group P21 with unit cell parameters a = 9.9665(11), b = 21.014(2), c = 21.950(2) Å, and Z = 4 molecules per unit cell. The refined structure of compound 1 had final R index [I ≥ 2sigma(I)] values of R1 = 0.0656, wR2 = 0.1426, and final R index [all data] values of R1 = 0.1294, wR2 = 0.1700. Detailed crystal data and structure refinements are shown in Table 2. The quality of the crystal was high enough for absolute configuration analysis to be carried out (Palmer and Potter, 2008). The absolute structure Flack parameter was −0.11(15), which was equal to 0.0 with standard deviation of 15 (Flack, 1983), confirming that the structure of 1 had been assigned the correct absolute configuration. Figure 6 shows the ORTEP diagram of compound 1 according to the X-ray data.
Compound 2 was dehydrated at C16/C-17 to produce compound 3. Thus, we determined the absolute configurations of compounds 2 and 3 simultaneously. Based on the similarity of the NMR spectra of compounds 1–3, including chemical shifts and peak multiplicities (Table 1), the multiple stereocenters of 2 and 3 had the same absolute configuration as in compound 1, although within differences at C-26 and C-27, because of the addition of a methyl substituent at C-26. According to Bilyk et al. in 2019, the absolute configurations of 22- to 26-membered macrolides compounds were confirmed by the data of NMR spectra, single-crystal X-ray crystallography, and elegant total synthesis of representative. Detailed NMR data comparison revealed a striking structural similarity of compounds 2, 3 to kaimonolide A (Akira et al., 1989; Bilyk et al., 2019) and cytovaricin B (Sakurai et al., 1983; Yamashita et al., 1997). Finally, the absolute configurations of compounds 2 and 3 were established as shown in Figure 1.
Biosynthetic Pathway Analysis
In strain HM190, the predicted BGC of macrolide compounds 1–3 contained 15 individual ORFs, including eight type I PKS genes, three cytochrome P450 genes, one crotonyl-CoA reductase (CCR) gene, one transcriptional regulator gene, and two hypothetical protein genes (Du et al., 2011; Bilyk et al., 2019). PKS genes S1–S8 were proposed to be responsible for the biosynthesis of the polyketide core of compounds 1–3. A suggested module and domain organization and a proposed model for the PKS template assembly of the macrolide backbone are shown in Figure 7. PKS gene S2 encoded the putative protein possessing the probable initiating module. The next 13 extension modules were proposed to be encoded by genes S3–S8 and were ordered according to the predicted linear arrangement of the required domains. Finally, the terminating module protein was identified to be encoded by gene S8 by its terminal punctuation with a thioesterase domain. One additional PKS gene S1 containing a complete module sequence “KS-AT-KR-ER-DH-ACP” and CCR apparently encoded a free-standing isobutylmalonyl-ACP that assembled to the macrolide backbone of module 7 in gene S5. After the core skeletons were constructed, the structural diversity of both macrocyclization and spiroketalization was mainly due to the tailoring reactions of cytochrome P450 genes to complete the hydroxylation reactions.
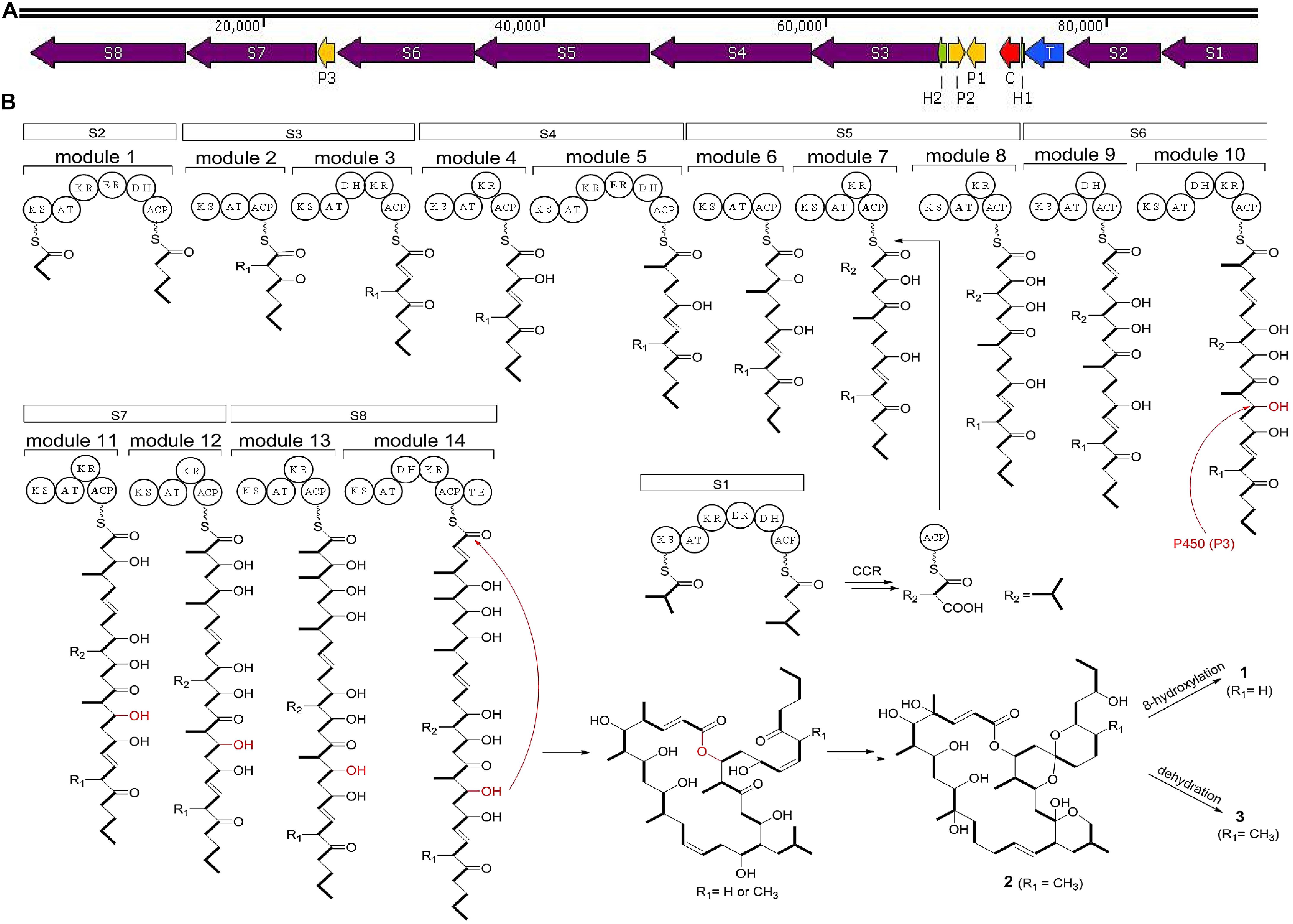
Figure 7. (A) Biosynthetic gene clusters of compounds 1–3 from strain HM190. S1–S8 include the PKS genes; P1–P3 account for the cytochrome P450 genes; C is a crotonyl-CoA reductase gene; T is a transcriptional regulator gene; H1 and H2 are hypothetical protein genes. (B) The proposed biosynthetic pathway for compounds 1–3. ACP, acyl carrier protein; KS, ketosynthase; AT, acyltransferase; KR, ketoreductase; ER, enoylreductase; DH, dehydratase; TE, thioesterase.
Biological Activities
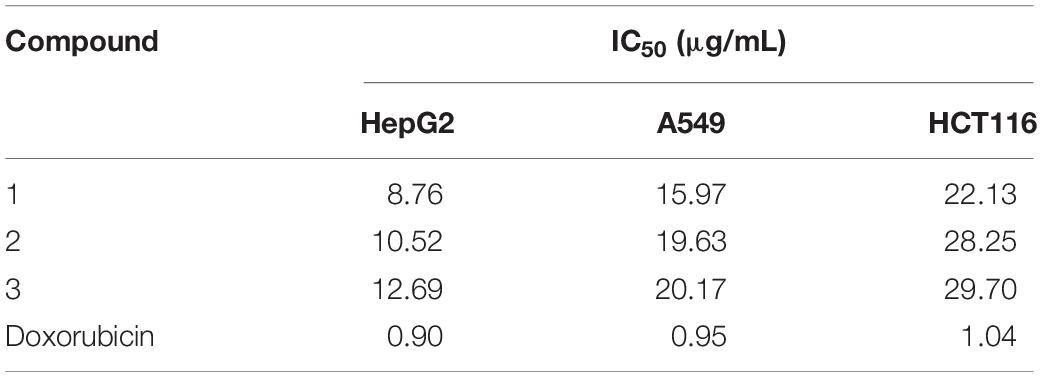
The cytotoxic activities of compounds 1–3 against HepG2, A549, and HCT116 human tumor cell lines are shown in Table 3. All the compounds suppressed the proliferation of the tested cells but exhibited different levels of activity. Compound 1 showed stronger cytotoxic activities than compounds 2 and 3, with a half maximal inhibitory concentration value (IC50) of 8.76 μg/mL against hepatocellular carcinoma cell line HepG2, with IC50 of 15.97 μg/mL against lung carcinoma cell line A549, and with IC50 of 22.13 μg/mL against colon tumor cell line HCT116.
The antibacterial and antifungal activities of the three compounds were detected against three human pathogens: Gram-positive bacterium methicillin-resistant S. aureus, Gram-negative bacterium K. pneumoniae, and fungus C. albicans. The MICs of compounds 1–3 were found to be >10 mg/mL, indicating that the three compounds had no biological activities against the tested pathogens.
Conclusion
During a study of microorganism diversity in mangrove samples, a white-pigmented colony was picked and named HM190. Detailed 16S rRNA gene sequencing and phylogenetic analysis identified the strain as Streptomyces. The whole-genome sequence of strain HM190 was obtained using the PacBio RSII and the Illumina HiSeq 4000 platforms by long-read single-molecule real-time sequencing technology. The assembled genome comprised a linear chromosome as a single contig of 7,762,826 bp with 71.97% G + C content. Based on the results of antiSMASH analysis, a total of 30 gene clusters were predicted to be involved in the biosynthesis of secondary metabolites; 12 of the 30 BGCs were responsible for the production of PKS- and NRPS-derived secondary metabolites. Several other secondary metabolites were also predicted by their BGCs. Gene cluster 5 was predicted to be involved in the production of an apoptolidin-like macrolide based on its high similarity of 74.5% to the type I PKS enzyme from the macrolide BGC of Nocardiopsis sp. FU 40 (Du et al., 2011). Furthermore, a chemical investigation was carried out on strain HM190, which led to the isolation of three 22-membered macrolides (compounds 1–3). The structures of the three compounds were determined based on the HRESIMS and NMR data. The absolute configurations of compounds 1–3 were established by the X-ray single-crystal diffraction and NMR data analysis. All three compounds showed moderate cytotoxic activities against hepatocellular carcinoma cell line HepG2, human lung carcinoma cell line A549, and human colon tumor cell line HCT116. A biosynthetic pathway for the three compounds was proposed on basis of the PKS gene cluster analysis.
Data Availability Statement
The datasets generated for this study can be found in the GenBank/EMBL/DDBJ and accession number for the 16S rRNA gene sequence of strain HM190 is MN897722. Whole Genome Shotgun project of Streptomyces sp. HM190 has been deposited at DDBJ/ENA/GenBank under the accession CP047318.
Ethics Statement
All studies of this article were processed with the standard of biosecurity and institutional safety procedures.
Author Contributions
YY conducted and performed the experiments and prepared the manuscript. JW and MW designed the experiments. XM and SW determined the compounds structures and the biosynthetic pathway analysis. NA performed the biological activities experiments. ZZ and RZ undertook the genome analysis. CY and YN performed the microbiology experiments. JZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Science and Technology Basic Resources Investigation Program of China (2017FY100300), the Sea Mountain Environmental Management Plan and APEI Construction Project Research (DY135-E2-2-05), and the Key Research and Development Program of Zhejiang Province (Grant No. 2020C02028).
Conflict of Interest
JZ was employed by AstraZeneca Trading Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Yu Liu (Life Science Institute of Zhejiang University) for performing NMR spectrometry.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01464/full#supplementary-material
The following are available online: the HRESIMS and NMR spectra (Supplementary Figures S1–S25) of compounds 1–3, the maximum-parsimony and maximum-likelihood phylogenetic trees (Supplementary Figures S26, S27) of strain HM190, the cluster of orthologous groups (COG) classification (Supplementary Table S1) and the antiSMASH-predicted BGCs (Supplementary Table S2) of strain HM190, and the crystal data (Supplementary Tables S3–S5) of compound 1.
Footnotes
- ^ http://www.bacterio.net/streptomyces.html
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://eztaxon-e.ezbiocloud.net
- ^ https://www.broadinstitute.org/gatk/
- ^ http://www.ebi.ac.uk/genomes/plasmid.html
- ^ http://ccb.jhu.edu/software/glimmer/index.shtml
References
Akira, H., Hideki, O., Toshitaka, K., Mitsuru, N., Hiroshi, H., and Akira, I. (1989). Structure of kaimonolide A, a novel macrolide plant growth inhibitor from a Streptomyces strain. Agr. Biol. Chem. 53, 2831–2833. doi: 10.1271/bbb1961.53.2831
Alfredo, B., Sarmiento, V. A., Miguel, O., Pérez, V. I., Martín, J., Nuria, D. P., et al. (2017). Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 15:144. doi: 10.3390/md15050144
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anzai, Y., Kim, H., Park, J. Y., Wakabayashi, H., and Oyaizu, H. (2000). Phylogenetic affiliation of the Pseudomonads based on 16s rRNA sequence. Int. J. Syst. Evol. Microbiol. 50, 1563–1589. doi: 10.1099/00207713-50-4-1563
Becerril, A., Alvarez, S., Brana, A. F., Rico, S., Diaz, M., Santamaria, R. I., et al. (2018). Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS One 13:e0198145. doi: 10.1371/journal.pone.0198145
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bilyk, O., Samborskyy, M., and Leadlay, P. F. (2019). The biosynthetic pathway to ossamycin, a macrocyclic polyketide bearing a spiroacetal moiety. PLoS One 14:e0215958. doi: 10.1371/journal.pone.0215958
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, 81–87. doi: 10.1093/nar/gkz310
Blunt, J. W., Copp, B. R., Keyzers, R. A., Munro, M. H. G., and Michèle, R. P. (2015). Marine natural products. Nat. Prod. Rep. 32, 116–211. doi: 10.1039/C4NP00144C
Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., and Puschmann, H. (2009). Olex2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42, 339–341. doi: 10.1107/S0021889808042726
Du, Y., Derewacz, D. K., Deguire, S. M., Teske, J., and Bachmann, B. O. (2011). Biosynthesis of the apoptolidins in Nocardiopsis sp, Fu 40. Tetrahedron 67, 6568–6575. doi: 10.1016/j.tet.2011.05.106
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Fenical, W., Jensen, P. R., Palladino, M. A., Lam, K. S., Lloyd, G. K., and Potts, B. C. (2009). Discovery and development of the anticancer agent salinosporamide A (npi-0052). Bioorgan. Med. Chem. 17, 2175–2180. doi: 10.1016/j.bmc.2008.10.075
Flack, H. D. (1983). On enantiomorph-polarity estimation. Acta Crystallogr. 39, 876–881. doi: 10.1107/S0108767383001762
Funabashi, M., Funa, N., and Horinouchi, S. (2008). Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283, 13983–13991. doi: 10.1074/jbc.M710461200
Girão, M., Ribeiro, I., Ribeiro, T., Azevedo, I. C., Pereira, F., Urbatzka, R., et al. (2019). Actinobacteria isolated from Laminaria ochroleuca: a source of new bioactive compounds. Front. Microbiol. 10:683. doi: 10.3389/fmicb.2019.00683
Holguin, G., Vazquez, P., and Bashan, Y. (2001). The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol. Fert. Soils 33, 265–278. doi: 10.1007/s003740000319
Huang, X., Zhou, S., Huang, D., Chen, J., and Zhu, W. (2016). Streptomyces spongiicola sp. nov. an actinomycete derived from marine sponge. Int. J. Syst. Evol. Microbiol. 66, 738–743. doi: 10.1099/ijsem.0.000782
Hwang, K. S., Kim, H. U., Charusanti, P., Palsson, B. Ø, and Lee, S. Y. (2014). Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268. doi: 10.1016/j.biotechadv.2013.10.008
Ichinose, K., Bedford, D. J., Tornus, D., Bechthold, A., and Hopwood, D. A. (1998). The granaticin biosynthetic gene cluster of Streptomyces violaceoruber TU22: sequence analysis and expression in a heterologous host. Cell Chem. Biol. 5, 647–659. doi: 10.1016/S1074-5521(98)90292-7
Ioanna, P., Konstantinos, L., Jakob, J., Chen, I. A., Tatyana, S., Bahador, N., et al. (2012). The genomes online database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 40, 571–579. doi: 10.1093/nar/gkr1100
Kallio, P., Liu, Z., Mäntsälä, P., Niemi, J., and Metsä-Ketelä, M. (2008). Sequential action of two flavoenzymes, PGAe and PGAm, in angucycline biosynthesis: chemoenzymatic synthesis of gaudimycin C. Chem. Biol. 15, 157–166. doi: 10.1016/j.chembiol.2007.12.011
Kathiresan, K., and Bingham, B. L. (2001). Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 40, 81–251. doi: 10.1016/S0065-2881(01)40003-4
Kimura, M. (1981). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kodani, S., Bicz, J., Song, L., Deeth, R. J., Ohnishi-Kameyama, M., Yoshida, M., et al. (2013). Structure and biosynthesis of scabichelin, a novel tris-hydroxamate siderophore produced by the plant pathogen Streptomyces scabies 87.22. Org. Biomol. Chem. 11, 4686–4694. doi: 10.1039/C3OB40536B
Kratzert, D., Holstein, J. J., and Krossing, I. (2015). DSR: enhanced modelling and refinement of disordered structures with Shelxl. J. Appl. Crystallogr. 48, 933–938. doi: 10.1107/S1600576715005580
Li, M., Hong, Y. G., Cao, H. L., and Gu, J. D. (2011). Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the pearl river delta reflected by 16s rRNA and hydrazine oxidoreductase (hzo) encoding gene analyses. Ecotoxicology 20, 1780–1790. doi: 10.1007/s10646-011-0711-4
Liu, R., Deng, Z., and Liu, T. (2018). Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab. Eng. 50, 74–84. doi: 10.1016/j.ymben.2018.05.015
Moon, K., Xu, F., Zhang, C., and Seyedsayamdost, M. R. (2019). Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol. 14, 767–774. doi: 10.1021/acschembio.9b00049
Nakagawa, A., Miura, S., Imai, H., Imamura, N., and Omura, S. (1989). Structure and biosynthesis of a new antifungal antibiotic, phthoramycin. J. Antibiot. 42, 1324–1327. doi: 10.7164/antibiotics.42.1324
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, 206–214.
Palmer, R. A., and Potter, B. S. (2008). X-ray structures and absolute configurations of the antibiotics oligomycins A, B and C: inhibitors of ATP synthase. J. Chem. Crystallogr. 38, 243–253. doi: 10.1007/s10870-008-9317-y
Pu, J. Y., Peng, C., Tang, M. C., Zhang, Y., Guo, J. P., Song, L. Q., et al. (2013). Naphthyridinomycin biosynthesis revealing the use of leader peptide to guide nonribosomal peptide assembly. Org. Lett. 15, 3674–3677. doi: 10.1021/ol401549y
Rzhetsky, A., and Nei, M. (1992). Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J. Mol. Evol. 35, 367–375. doi: 10.1007/BF00161174
Saitou, N., and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. J. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sakurai, T., Kihara, T., and Isono, K. (1983). Structure of cytovaricin–acetonitrile (1:1), C47H80O16.C2H3N. Acta Crystallogr. 39, 295–297. doi: 10.1107/S010827018300445X
Santos, H. F. D., Cury, J. C., Flávia, L. D. C., Santos, A. L. D., Tiedje, J., Elsas, J. D. V., et al. (2011). Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6:e16943. doi: 10.1371/journal.pone.0016943
Sheldrick, G. M. (2015). Shelxt integrated space group and crystal structure determination. Acta Crystallogr. 71, 3–8. doi: 10.1107/S2053273314026370
Studholme, D. J. (2016). Genome update. Let the consumer beware: Streptomyces genome sequence quality. Microb. Biotechnol. 9, 3–7. doi: 10.1111/1751-7915.12344
Sun, Y., Hahn, F., Demydchuk, Y., Chettle, J., Tosin, M., Osada, H., et al. (2009). In vitro reconstruction of tetronate RK-682 biosynthesis. Nat. Chem. Biol. 6, 99–101. doi: 10.1038/nchembio.285
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. J. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thatoi, H., Behera, B. C., Mishra, R. R., and Dutta, S. K. (2013). Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: a review. Ann. Microbiol. 63, 1–19. doi: 10.1007/s13213-012-0442-7
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tian, Y., Liu, H. J., Zheng, T. L., Kwon, K. K., Kim, S. J., and Yan, C. L. (2008). PAHs contamination and bacterial communities in mangrove surface sediments of the jiulong river estuary. China. Mar. Pollut. Bull. 57, 707–715. doi: 10.1016/j.marpolbul.2008.03.011
Waksman, S. A., and Henrici, A. T. (1943). The nomenclature and classification of the actinomycetes. J. Bacteriol. 46, 337–341. doi: 10.1002/path.1700560116
Wang, J. D., Zhang, H., Ying, L. P., Wang, C. X., Jiang, N., Zhou, Y., et al. (2009). Five new epothilone metabolites from Sorangium cellulosum strain So0157-2. J. Antibiot. 62, 483–487. doi: 10.1038/ja.2009.55
Wang, R. J., Zhang, S. Y., Ye, Y. H., Yu, Z., Qi, H., Zhang, H., et al. (2019). Three new isoflavonoid glycosides from the mangrove-derived actinomycete Micromonospora aurantiaca 110B. Mar. Drugs 17:294. doi: 10.3390/md17050294
Wang, X., Reynolds, A. R., Elshahawi, S. I., Shaaban, K. A., and Thorson, J. S. (2015). Terfestatins B and C, new p-terphenyl glycosides produced by Streptomyces sp. RM-5–8. Org. Lett. 17, 2796–2799. doi: 10.1021/acs.orglett.5b01203
Xu, J. (2011). Biomolecules produced by mangrove-associated microbes. Curr. Med. Chem. 18, 5224–5266. doi: 10.2174/092986711798184307
Xu, L., Ye, K. X., Dai, W. H., Sun, C., Xu, L. H., and Han, B. N. (2019). Comparative genomic insights into secondary metabolism biosynthetic gene cluster distributions of marine Streptomyces. Mar. Drugs 17:498. doi: 10.3390/md17090498
Yamashita, N., Shin-Ya, K., Kitamura, M., Wakao, H., Furihata, K., Furihata, K., et al. (1997). Cytovaricin B, a new inhibitor of JAK-STAT signal transduction produced by Streptomyces torulosus. J. Antibiot. 50, 440–442. doi: 10.7164/antibiotics.50.440
Yan, L. L., Han, N. N., Zhang, Y. Q., Yu, L. Y., Chen, J., Wei, Y. Z., et al. (2010). Antimycin A18 produced by an endophytic Streptomyces albidoflavus isolated from a mangrove plant. J. Antibiot. 63, 259–261. doi: 10.1038/ja.2010.21
Ye, Y., Chen, C., Ren, Y., Wang, R., Zhang, C., Han, S., et al. (2019). Pseudomonas mangrovi sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 69, 377–383. doi: 10.1099/ijsem.0.003141
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., and Chun, J. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhao, X. Q., Li, W. J., Jiao, W. C., Li, Y., Yuan, W. J., Zhang, Y. Q., et al. (2009). Streptomyces xinghaiensis sp. nov. isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 59, 2870–2874. doi: 10.1099/ijs.0.009878-0
Zhong, X., Tian, Y., Niu, G., and Tan, H. (2013). Assembly and features of secondary metabolite biosynthetic gene clusters in Streptomyces ansochromogenes. Sci. China Life Sci. 56, 609–618. doi: 10.1007/s11427-013-4506-0
Keywords: Streptomyces sp. HM190, biosynthetic gene clusters, 22-membered macrolides, structure elucidation, cytotoxic activities
Citation: Ye Y, Anwar N, Mao X, Wu S, Yan C, Zhao Z, Zhang R, Nie Y, Zhang J, Wang J and Wu M (2020) Discovery of Three 22-Membered Macrolides by Deciphering the Streamlined Genome of Mangrove-Derived Streptomyces sp. HM190. Front. Microbiol. 11:1464. doi: 10.3389/fmicb.2020.01464
Received: 31 March 2020; Accepted: 04 June 2020;
Published: 26 June 2020.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Fernando Reyes, Fundación MEDINA, SpainLijiang Song, University of Warwick, United Kingdom
Andreas Bechthold, University of Freiburg, Germany
Copyright © 2020 Ye, Anwar, Mao, Wu, Yan, Zhao, Zhang, Nie, Zhang, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jidong Wang, ZXhjaXRld0AxNjMuY29t; Min Wu, d3VtaW5Aemp1LmVkdS5jbg==
 Yanghui Ye
Yanghui Ye Nusratgul Anwar2
Nusratgul Anwar2 Xuming Mao
Xuming Mao Shihua Wu
Shihua Wu Cen Yan
Cen Yan Min Wu
Min Wu