- 1College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 2Institute of Comparative Medicine, Yangzhou University, Yangzhou, China
- 3Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
The emergence of tet(X) and carbapenemase genes in Enterobacterales pose significant challenges to the treatment of infectious diseases. Convergence of these two categories of genes in an individual pathogen would deteriorate the antimicrobial resistance (AMR) crisis furthermore. Here, tigecycline-resistant Enterobacterales strains were isolated and detected with carbapenemase genes, characterized by antimicrobial susceptibility testing, PCR, conjugation assay, whole genome sequencing, and bioinformatics analysis. Three tigecycline-resistant isolates consisting of one plasmid-mediated tet(X4)-bearing Escherichia fergusonii and two chromosomal tet(X6)-bearing Proteus cibarius were recovered from chicken feces. The tet(X4) was located on a conjugative IncX1 plasmid pHNCF11W-tetX4 encoding the identical structure as reported tet(X4)-bearing IncX1 plasmids in Escherichia coli. Among two P. cibarius strains, tet(X6) was located on two similar chromosomal MDR regions with genetic contexts IS26-aac(3)-IVa-aph(4)-Ia-ISEc59-tnpA-tet(X6)-orf-orf-ISCR2-virD2-floR-ISCR2-glmM-sul2 and IS26-aac(3)-IVa-aph(4)-Ia-ISEc59-tnpA-tet(X6)-orf-orf-ISCR2-glmM-sul2. Apart from tet(X6), P. cibarius HNCF44W harbored a novel transposon Tn6450b positive for blaNDM–1 on a conjugative plasmid. This study probed the genomic basis of three tet(X)-bearing, tigecycline-resistant strains, one of which coharbored blaNDM–1 and tet(X6), and identified P. cibarius as the important reservoir of tet(X6) variants. Emergence of P. cibarius encoding both blaNDM–1 and tet(X6) reveals a potential public health risk.
Introduction
Antimicrobial resistance (AMR) is a serious threat to public health globally. Carbapenem and tigecycline are regarded as vital antimicrobials reserved for clinical use due to their broad antibacterial spectrum. However, the increasing spread of carbapenemase-encoding genes has rendered carbapenem-resistant Enterobacteriaceae (CRE) infection a great threat (Du et al., 2018; Tacconelli et al., 2018). Plasmid-mediated resistance genes tet(X3) and tet(X4) conferring high-level resistance to tigecycline in Enterobacterales and Acinetobacter has been found to be ubiquitous in animals and food of animal origin (Bai et al., 2019; He et al., 2019; Sun et al., 2019; Li et al., 2020c). This undermines the efficacy of tigecycline as the last-resort drug in treating MDR bacterial infections, especially those caused by carbapenem-resistant, Gram-negative bacteria, such as CRE.
Initially, tet(X) genes were mainly found in Bacteroides species and bacteria derived from environmental microbiota (Forsberg et al., 2015; Tian et al., 2019). Alarmingly, novel tet(X) variants are being identified from the pathogens of animal, food, and human origins (Li et al., 2019; Wang et al., 2019; He D. et al., 2020; Liu et al., 2020; Peng et al., 2020), which implies that tet(X) is expanding from commensals to pathogens (Aminov, 2013). Thus, the convergence of mobile tigecycline-resistant tet(X) and carbapenem-resistant genes in Enterobacterales may be inevitable. Acinetobacter strains coharboring tet(X) and blaNDM–1 were identified from samples of waterfowl and dairy cows recently (Cui et al., 2020; He T. et al., 2020), highlighting the potential convergence risk of both critical resistance genes among bacteria. However, Enterobacterales carrying both tet(X) and blaNDM–1 is unknown, which constitutes a more severe concern. To cover this knowledge gap, we performed a tigecycline-resistant Enterobacterales screening pilot program and identified a Proteus cibarius coharboring both tet(X6) and blaNDM–1, implying that wide surveillance of Enterobacterales conferring resistance to carbapenems and tigecycline among bacteria should be performed worldwide.
Materials and Methods
Bacterial Isolates
A total of 16 chicken fecal samples were collected from four chicken farms in Henan province, China, in 2019. Tigecycline-resistant Enterobacterales were selected on MacConkey agar plates containing tigecycline (4 mg/L). The species of purified tigecycline-resistant isolates was identified by 16S rRNA gene sequencing and further confirmed by analyzing WGS data. The tet(X) and carbapenem-resistant genes were determined by PCR with reported primers (Dallenne et al., 2010; He et al., 2019).
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing (AST) of tet(X)-positive tigecycline-resistant strains against 13 antibiotics was performed based on the broth microdilution method with Escherichia coli ATCC 25922 as the quality control and interpreted according to CLSI guidelines (CLSI, 2018). The tigecycline non-susceptibility in Enterobacterales was interpreted with an MIC of ≥4 mg/L (Marchaim et al., 2014).
Conjugation Assay and S1-PFGE
A conjugation assay was conducted using rifampicin-resistant E. coli C600 (Rifr) as the recipient to investigate the transferability of tet(X) and blaNDM–1. The donor and recipient strains were mixed at a ratio of 1:4 on a filter, followed by culturing on LB agar plates at 30 and 37°C, respectively, overnight and screening on MacConkey plates containing tigecycline (4 mg/L) or meropenem (2 mg/L) with rifampicin (300 mg/L). Transconjugants were confirmed with PCR. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) was utilized to characterize the plasmid profiles.
Whole Genome Sequencing and Bioinformatics Analysis
Genomid DNA (gDNA) was purified with the TIANamp Genomic DNA Kit and quantified by Qubit 4 Fluorometer. Short-read Illumina sequencing and long-read Nanopore sequencing were combined to generate complete genome sequences (Li et al., 2018). The de novo assembled sequences were annotated by RAST1. Antibiotic resistance genes (ARGs) and plasmid replicon types were determined using the ResFinder and PlasmidFinder2. BRIG and Easyfig were used to compare the plasmid and MDR structures (Alikhan et al., 2011; Sullivan et al., 2011).
Functional Confirmation of tet(X6) and Phylogenetic Analysis
To confirm the resistance function of the identified tet(X6) variant, TA-cloning and AST were performed. Briefly, the new variant, together with its natural promoter sequence, was amplified by PCR using primers HNCF44W-F/AGCGAACAAGAATATGACTTTACT and HNCF44W-R/CGCCTTTCTGTTTTATAGATTCAT, cloned into a pMD19-T vector and transformed chemically into E. coli DH5α. The resistance phenotype of tet(X6) was tested by measuring the MICs of different tetracycline antibiotics. Phylogenetic analysis of Tet(X) amino acid sequences were conducted in MEGA X using the neighbor-joining method (Kumar et al., 2018).
Nucleotide Sequence Accession Numbers
The complete sequences obtained in this study have been deposited in the GenBank database under BioProject number PRJNA625637. The novel blaNDM–1-bearing Tn6450b was also deposited in the NCBI database (MT701524).
Results and Discussion
Identification of Tigecycline-Resistant Enterobacterales
Three tigecycline-resistant, tet(X)-positive Enterobacterales strains were isolated from 16 samples (18.8%), which consisted of Escherichia fergusonii HNCF11W carrying tet(X4), P. cibarius HNCF43W, and HNCF44W positive for a tet(X6) variant. Notably, HNCF44W also carried the blaNDM–1 gene. Antimicrobial susceptibility testing revealed that the three tigecycline-resistant strains were resistant to multiple antibiotics, including kanamycin, florfenicol, amoxicillin, and tetracyclines, showing high-level resistance to tigecycline (Supplementary Table S1). In addition, the NDM-producing strain HNCF44W was also resistant to meropenem (>16 mg/L). S1-PFGE indicated that two plasmids of ca. 130 and 55 kb were observed in tet(X4)-bearing E. fergusonii HNCF11W, and one plasmid of ca. 140 kb existed in tet(X6)-bearing P. cibarius HNCF44W, but no plasmid existed in P. cibarius HNCF43W (Supplementary Figure S1). To uncover the genetic locations of tet(X) and blaNDM–1, complete genome sequences of the three strains were finished successfully.
Genome Characterization of Tigecycline-Resistant E. fergusonii
The tet(X4)-bearing E. fergusonii harbored one chromosome (4,584,794 bp, ST10543); one p0111-like plasmid pHNCF11W-130 kb (130,643 bp) harboring aadA1, floR, tet(A), dfrA14, blaTEM–209, blaOXA–10, cmlA1, arr-3, and qnrS1; and one typical IncX1 plasmid pHNCF11W-tetX4 (57,105 bp) encoding tet(X4) in the genetic structure of IS26-abh-tet(X4)-ISCR2-virD2-floR-△ISCR2 coupled with aadA2, lnu(F), tet(A), and blaSHV–12. A BLASTn search of pHNCF11W-tetX4 against the nr database retrieved other tet(X4)-bearing plasmids p1916D18-1 (59,353 bp), p1916D6-2 (59,351 bp), and pYY76-1-2 (57,104 bp) of swine and cattle origins, sharing nearly identical sequences (Supplementary Figure S2). Both pHNCF11W-tetX4 and pHNCF11W-130 kb were transferred into the recipient strain, demonstrating that IncX1-type tet(X4)-bearing plasmids were mobilizable and spread in Enterobacterales of animal origin (Chen et al., 2019). Although tet(X4) was initially found in plasmids of IncFIB and IncQ1 types (He et al., 2019; Sun et al., 2019), IncX1 plasmids are common in various Enterobacterales and associated with various resistance genes (Dobiasova and Dolejska, 2016; Li et al., 2020b), which potentially increases the transmission of tet(X4) between different Enterobacterales.
Characterization of Proteus cibarius Coharboring tet(X6) and blaNDM–1
HNCF43W harbored a 3,965,977 bp chromosome. HNCF44W contained a 3,929,176 bp chromosome and one blaNDM–1-bearing plasmid pHNCF44W_NDM-1 of 139,490 bp in size. A conjugation assay failed to recover transconjugants positive for tet(X6). The tet(X6) gene was located on large MDR regions (IS26-aac(3)-IVa-aph(4)-Ia-ISEc59-tnpA-tet(X6)-orf-orf-ISCR2-virD2-floR-ISCR2-glmM-sul2 and IS26-aac(3)-IVa-aph(4)-Ia-ISEc59-tnpA-tet(X6)-orf-orf-ISCR2-glmM-sul2, respectively) embedded in chromosomes of both P. cibarius strains (Figure 1A), and the MDR regions may derive from recombination of tet(X6)-bearing elements and reported plasmids p19110F47-2 (CP046044) and pJXP9 (MK673549) (Supplementary Figure S3). The two tet(X6)-bearing MDR regions differed by virD2-floR-ISCR2, which was a potential mobile segment (Doublet et al., 2005). ISCR2 is the major mobile element associated with tet(X3), tet(X4), and tet(X5) (Bai et al., 2019; He et al., 2019; Sun et al., 2019; Li et al., 2020a). Recently, three reports demonstrated tet(X6) could be embedded in ICEs or other MDR regions, all of which harbor ISCR2 (He D. et al., 2020; Liu et al., 2020; Peng et al., 2020), suggesting it may facilitate the transfer of tet(X6) to other plasmids or chromosomes.
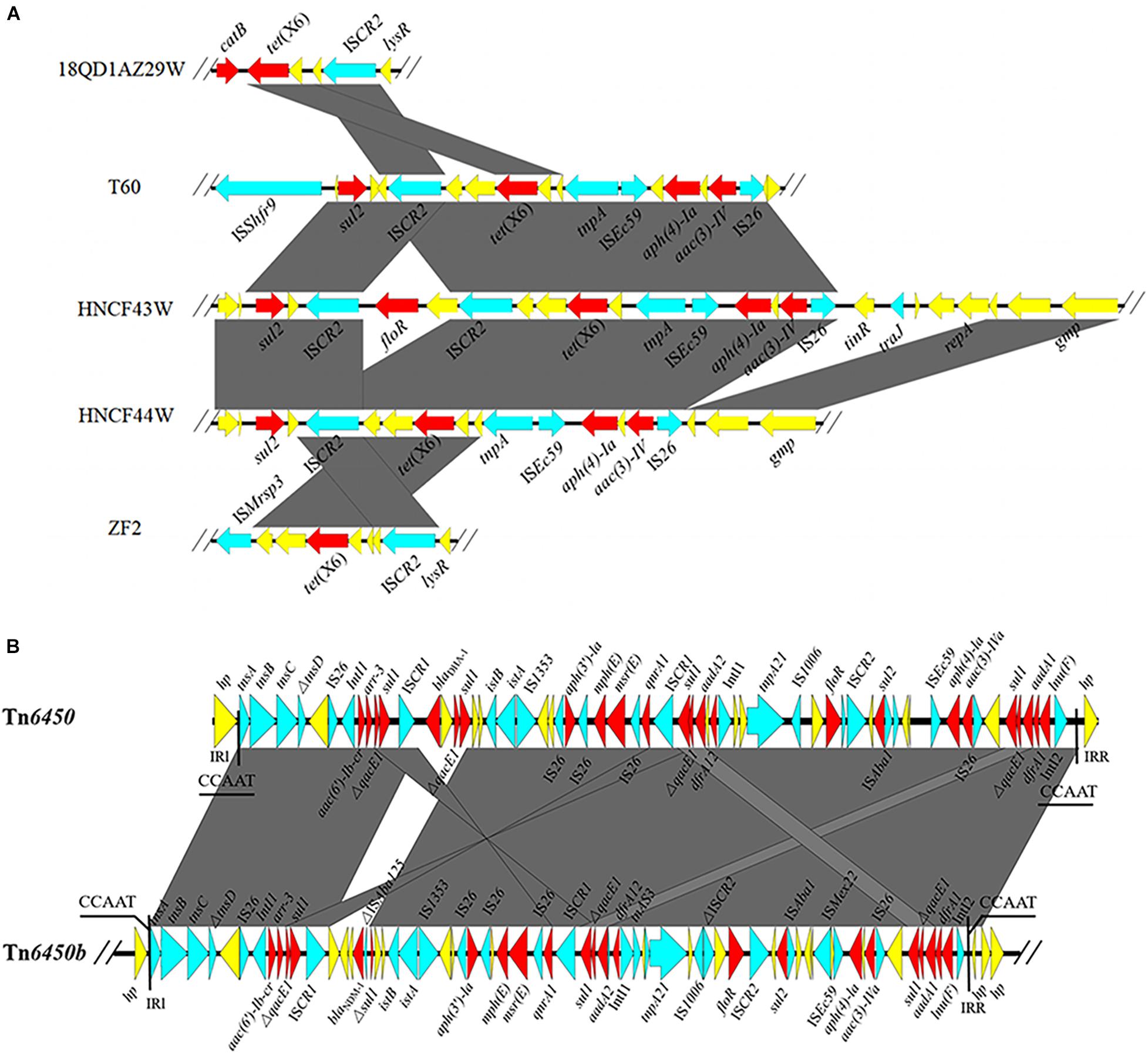
Figure 1. (A) Genetic environments of tet(X6) from different strains. (B) Linear alignment of blaNDM–1-bearing Tn6450b in pHNCF44W_NDM-1 and reported Tn6450. The red arrows denote the resistance genes; the blue arrows represent the mobile elements; the yellow arrows signify other CDSs. The shadow parallelograms stand for homologous genetic regions.
The blaNDM–1 gene in HNCF44W was located on an MDR plasmid pHNCF44W_NDM-1 carrying resistance genes aac(6′)-Ib, arr-3, qacE1, sul1, sul2, aph(3′)-la, mph(E), msr(E), qnrA1, aadA1, aadA2, dfrA1, dfrA12, floR, aac(3)-IVa, and lnu(F). No plasmid replicon was identified, but pHNCF44W_NDM-1 was very similar to plasmid plas1.1.2 (CP047114) from Proteus mirabilis of swine origin; it differed mainly in the blaNDM–1-bearing region (Figure 2). The blaNDM–1 gene and other resistance genes were found in a 65-kb MDR region, showing high similarity to transposon Tn6450 but with blaDHA–1 replaced by blaNDM–1 (Chen et al., 2018), designated as Tn6450b here (Figure 1B). Other similar plasmids or transposons were found in the nr database, including p16Pre36-NDM from Providencia rettgeri, pHFK418-NDM from P. mirabilis, pPrY2001 from P. rettgeri, p16Pre36-NDM from P. rettgeri, p06-1619-1 from P. rettgeri, and Tn6450 from P. mirabilis. The plasmid pHNCF44W_NDM-1 carrying blaNDM–1 was successfully transferred into E. coli EC600 at 30°C. Notably, ISCR2-lysR-floR-virD2-△ISCR2 structure in pHNCF44W_NDM-1 was also found in a tet(X6)-bearing chromosomal segment, rendering this a Tn6450b-bearing plasmid with the potential ability to capture tet(X6) under certain circumstances, which warrants further surveillance.
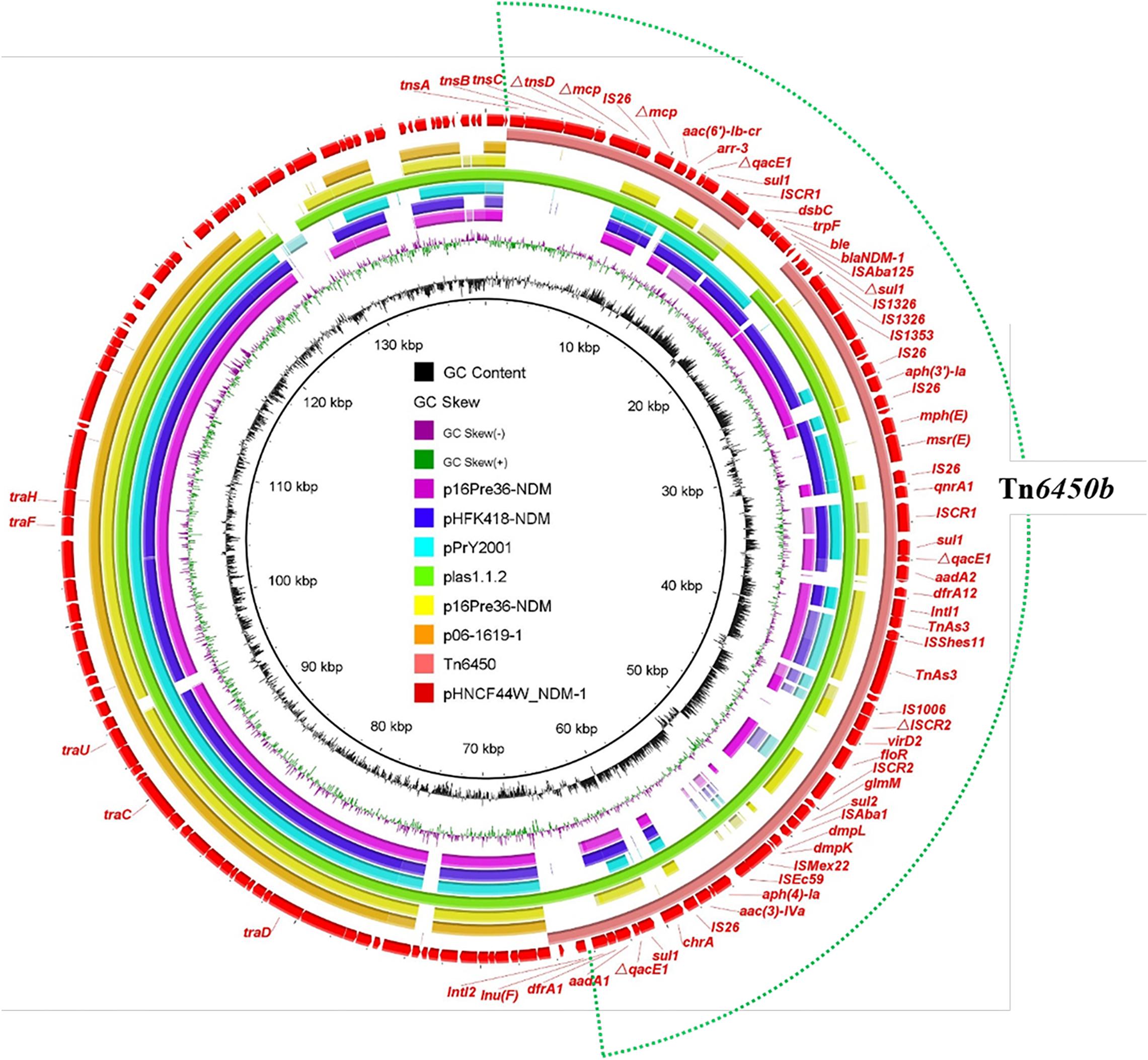
Figure 2. Alignment of the plasmid pHNCF44W_NDM-1 identified in this study with other similar plasmids. The outermost red circle denotes the reference plasmid pHNCF44W_NDM-1. The location of Tn6450b is highlighted in the dotted circle.
Functional Analysis of tet(X6) and Its Evolution
Recently, several reports have characterized different tet(X6) variants from diverse bacteria (He D. et al., 2020; Liu et al., 2020; Peng et al., 2020; Xu et al., 2020). The tet(X6) variant carried by HNCF44W and HNCF43W is the same as that reported in P. cibarius from swine (Liu et al., 2020). The expression of tet(X6) in the cloning vector pMD19-T-tet(X6) in E. coli DH5α conferred resistance to tetracycline (64 mg/L) and tigecycline (16 mg/L) with 32-fold increments compared to that of DH5α-pMD19-T, indicating that this tet(X6) variant can confer tigecycline resistance (Supplementary Table S2). The 1137 bp tet(X6) variant encoded 378 amino acids and showed 84.7, 66.3, 84.9, 79.9, 87.8, 93.7, and 98.7% amino acid sequence identity to Tet(X), Tet(X1), Tet(X2), Tet(X3), Tet(X4), Tet(X5), and Tet(X6), respectively (Figure 3) with 5, 8, and 16 amino acid substitutions to Tet(X6) (MN507533, T60), Tet(X6) (CP025402, pMS8345A), and Tet(X6) (NZ_CP047050, 18QD1AZ29W).

Figure 3. The phylogenetic tree of Tet(X) variants. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using MEGA X and are in the units of the number of amino acid substitutions per site.
Phylogenetic analysis of Tet(X) indicated that of all reported Tet(X6) variants clustered in a clade, most related to Tet(X5) (Figure 3). Although most tet(X6) variants were found in Proteus and Acinetobacter spp., analysis of the NCBI database shows that E. coli positive for tet(X6) existed as early as 2003 in Denmark (JWJM01000174), indicating the transmission of tet(X6) occurred previously. Furthermore, chromosomal tet(X6)-bearing ICEs and MDRs are found in Proteus, Acinetobacter, and Myroides phaeus (He D. et al., 2020; Liu et al., 2020; Peng et al., 2020). Plasmid-mediated tet(X6) in Proteus, Acinetobacter, and E. coli also emerged according to published reports (Cui et al., 2020; Li et al., 2020c; Xu et al., 2020), implying tet(X6) is at the stage of being expanded via different mobile elements. This highlights that tet(X6) variants would be the next possible prominent tet(X) allele, approaching the widespread situation of tet(X4) in E. coli (Bai et al., 2019; He et al., 2019; Sun et al., 2019). Nomenclature of tet(X) by different allele numbers is not recommended because many of the alleles are 85–100% aa identical3, but an identical tet(X) naming system may not benefit research communications and AMR surveillance, especially as different tet(X) alleles possess different resistance phenotypes, genetic contexts, hosts, and evolutionary routes as more research reveals. For example, tet(X4) was found to be more common in E. coli than tet(X6) owing to the fact that tet(X4) has merged into conjugative plasmids adapted in E. coli. Also, the plasmids coharboring both blaNDM–1 and tet(X) in Acinetobacter spp. (Cui et al., 2020) could be divided into tet(X3)- or tet(X6)-bearing plasmids, which could make it difficult to decipher the accurate different genetic contexts if only tet(X) represented the two alleles simultaneously. To better investigate the molecular epidemiology, genomic evolution, and functional analysis and avoid confusion in tet(X) designations, a clear nomenclature scheme acceptable to researchers worldwide should be proposed as soon as possible. Because mcr genes exist in certain bacterial species as housekeeping genes and certain alleles leap over species boundary limits to Enterobacterales via mobile elements like Tn6330 (Li et al., 2016; Shen et al., 2020), tet(X) genes may behave similarly because they exist as core genes in certain bacteria and transfer to Enterobacterales by mobile elements like ISCR2. Thus, the mcr nomenclature scheme may be a good example when it comes to tet(X) designation (Partridge et al., 2018). More research should be performed to put forward a widely accepted tet(X) allele numbering system to facilitate research communications.
Conclusion
To conclude, we characterized three tet(X)-bearing Enterobacterales strains of chicken origin and found that one P. cibarius coharbored both blaNDM–1 and tet(X6). To the best of our knowledge, this is the first report of convergence of tet(X6) and blaNDM–1 genes in Enterobacterales, highlighting the importance of continuous surveillance of tigecycline- and carbapenem-resistant Enterobacterales of animal origin, which may act as the potential reservoir of clinical Enterobacterales resistant to tigecycline and carbapenems.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
RL and ZW: conceptualization, writing – review and editing, and supervision. YaL and RL: methodology. YaL and QW: investigation. QW, KP, and YuL: data curation and visualization. YaL: writing – original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20180900), the National Natural Science Foundation of China (31872523), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01940/full#supplementary-material
Footnotes
- ^ https://rast.nmpdr.org/
- ^ http://www.genomicepidemiology.org/
- ^ http://faculty.washington.edu/marilynr/
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Aminov, R. I. (2013). Evolution in action: dissemination of tet(X) into pathogenic microbiota. Front. Microbiol. 4:192. doi: 10.3389/fmicb.2013.00192
Bai, L., Du, P., Du, Y., Sun, H., Zhang, P., Wan, Y., et al. (2019). Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Euro Surveill. 24:1900340.
Chen, C., Wu, X. T., He, Q., Chen, L., Cui, C. Y., Zhang, Y., et al. (2019). Complete sequence of a tet(X4)-harboring IncX1 plasmid, pYY76-1-2, in Escherichia coli from a cattle sample in China. Antimicrob. Agents Chemother. 63:e01528-19.
Chen, Y. P., Lei, C. W., Kong, L. H., Zeng, J. X., Zhang, X. Z., Liu, B. H., et al. (2018). Tn6450, a novel multidrug resistance transposon characterized in a Proteus mirabilis isolate from chicken in China. Antimicrob. Agents Chemother. 62:e02192-17.
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100. Wayne, PA: CLSI.
Cui, C. Y., Chen, C., Liu, B. T., He, Q., Wu, X. T., Sun, R. Y., et al. (2020). Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob. Agents Chemother. 64:e02502-19.
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dobiasova, H., and Dolejska, M. (2016). Prevalence and diversity of IncX plasmids carrying fluoroquinolone and beta-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 71, 2118–2124. doi: 10.1093/jac/dkw144
Doublet, B., Schwarz, S., Kehrenberg, C., and Cloeckaert, A. (2005). Florfenicol resistance gene floR is part of a novel transposon. Antimicrob. Agents Chemother. 49, 2106–2108. doi: 10.1128/aac.49.5.2106-2108.2005
Du, X., He, F., Shi, Q., Zhao, F., Xu, J., Fu, Y., et al. (2018). The rapid emergence of tigecycline resistance in blaKPC-2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front. Microbiol. 9:648. doi: 10.3389/fmicb.2018.00648
Forsberg, K. J., Patel, S., Wencewicz, T. A., and Dantas, G. (2015). The tetracycline destructases: a novel family of tetracycline-inactivating enzymes. Chem. Biol. 22, 888–897. doi: 10.1016/j.chembiol.2015.05.017
He, D., Wang, L., Zhao, S., Liu, L., Liu, J., Hu, G., et al. (2020). A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J. Antimicrob. Chemother. 75, 1159–1164. doi: 10.1093/jac/dkaa012
He, T., Li, R., Wei, R., Liu, D., Bai, L., Zhang, L., et al. (2020). Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM-1 of dairy cow origin. J. Antimicrob. Chemother. 2020:dkaa182.
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, R., Liu, Z., Peng, K., Liu, Y., Xiao, X., and Wang, Z. (2019). Cooccurrence of Two tet(X) variants in an empedobacter brevis strain of shrimp origin. Antimicrob. Agents Chemother. 63:e01636-19.
Li, R., Lu, X., Liu, Z., Liu, Y., Xiao, X., and Wang, Z. (2020a). Rapid detection and characterization of tet(X4)-positive Escherichia coli strains with nanopore sequencing. J. Antimicrob. Chemother. 75, 1068–1070. doi: 10.1093/jac/dkz528
Li, R., Lu, X., Peng, K., Liu, Z., Li, Y., Liu, Y., et al. (2020b). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet (X)-bearing plasmidome among bacteria. mSystems 5:e00134-20.
Li, R., Peng, K., Li, Y., Liu, Y., and Wang, Z. (2020c). Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci. Total Environ. 733:139306. doi: 10.1016/j.scitotenv.2020.139306
Li, R., Xie, M., Dong, N., Lin, D., Yang, X., Wong, M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7, 1–9.
Li, R., Xie, M., Lv, J., Wai-Chi Chan, E., and Chen, S. (2016). Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 72, 696–699.
Liu, D., Zhai, W., Song, H., Fu, Y., Schwarz, S., He, T., et al. (2020). Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J. Antimicrob. Chemother. 75, 1428–1431.
Marchaim, D., Pogue, J. M., Tzuman, O., Hayakawa, K., Lephart, P. R., Salimnia, H., et al. (2014). Major variation in MICs of tigecycline in gram-negative Bacilli as a function of testing method. J. Clin. Microbiol. 52, 1617–1621. doi: 10.1128/jcm.00001-14
Partridge, S. R., Di Pilato, V., Doi, Y., Feldgarden, M., Haft, D. H., Klimke, W., et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. doi: 10.1093/jac/dky262
Peng, K., Li, R., He, T., Liu, Y., and Wang, Z. (2020). Characterization of a porcine Proteus cibarius strain co-harbouring tet(X6) and cfr. J. Antimicrob. Chemother. 75, 1652–1654. doi: 10.1093/jac/dkaa047
Shen, Y., Zhang, R., Schwarz, S., Wu, C., Shen, J., Walsh, T. R., et al. (2020). Farm animals and aquaculture: significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 22, 2469–2484. doi: 10.1111/1462-2920.14961
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327.
Tian, Z., Liu, R., Zhang, H., Yang, M., and Zhang, Y. (2019). Developmental dynamics of antibiotic resistome in aerobic biofilm microbiota treating wastewater under stepwise increasing tigecycline concentrations. Environ. Int. 131:105008. doi: 10.1016/j.envint.2019.105008
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64:e01326-19.
Keywords: tigecycline resistance, carbapenem resistance, coexistence, plasmid, chromosome
Citation: Li Y, Wang Q, Peng K, Liu Y, Li R and Wang Z (2020) Emergence of Carbapenem- and Tigecycline-Resistant Proteus cibarius of Animal Origin. Front. Microbiol. 11:1940. doi: 10.3389/fmicb.2020.01940
Received: 09 June 2020; Accepted: 23 July 2020;
Published: 14 August 2020.
Edited by:
Zhi Ruan, Zhejiang University, ChinaReviewed by:
Séamus Fanning, University College Dublin, IrelandFang He, Zhejiang Provincial People’s Hospital, China
Congming Wu, China Agricultural University, China
Copyright © 2020 Li, Wang, Peng, Liu, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruichao Li, cmNobDg4QHl6dS5lZHUuY24=; Zhiqiang Wang, enF3YW5nQHl6dS5lZHUuY24=
 Yan Li
Yan Li Qian Wang1
Qian Wang1 Yuan Liu
Yuan Liu Ruichao Li
Ruichao Li Zhiqiang Wang
Zhiqiang Wang