- 1Institute of Oceanic Research and Development, Tokai University, Shizuoka, Japan
- 2Department of Food and Nutrition, Japan Women’s University, Tokyo, Japan
- 3Research Institute of Green Science and Technology, Shizuoka University, Shizuoka, Japan
Bacteria must survive harsh environmental fluctuations at times and have evolved several strategies. “Collective” behaviors have been identified due to recent progress in single-cell analysis. Since most bacteria exist as single cells, bacterial populations are often considered clonal. However, accumulated evidence suggests this is not the case. Gene expression and protein expression are often not homogeneous, resulting in phenotypic heterogeneity. In extreme cases, this leads to bistability, the existence of two stable states. In many cases, expression of key master regulators is bimodal via positive feedback loops causing bimodal expression of the target genes. We observed bimodal expression of metabolic genes for alternative carbon sources. Expression profiles of the frlBONMD-yurJ operon driven by the frlB promoter (PfrlB), which encodes degradation enzymes and a transporter for amino sugars including fructoselysine, were investigated using transcriptional lacZ and gfp, and translational fluorescence reporter mCherry fusions. Disruption effects of genes encoding CodY, FrlR, RNaseY, and nucleoid-associated protein YlxR, four known regulatory factors for PfrlB, were examined for expression of each fusion construct. Expression of PfrlB-gfp and PfrlB-mCherry, which were located at amyE and its original locus, respectively, was bimodal; and disruption of ylxR resulted in the disappearance of the clear bimodal expression pattern in flow cytometric analyses. This suggested a role for YlxR on the bimodal expression of PfrlB. The data indicated that YlxR acted on the bimodal expression of PfrlB through both transcription and translation. YlxR regulates many genes, including those related to translation, supporting the above notion. Depletion of RNaseY abolished heterogenous expression of transcriptional PfrlB-gfp but not bimodal expression of translational PfrlB-mCherry, suggesting the role of RNaseY in regulation of the operon through mRNA stability control and regulatory mechanism for PfrlB-mCherry at the translational level. Based on these results, we discuss the meaning and possible cause of bimodal PfrlB expression.
Introduction
Bacterial “collective” behaviors of single cells have evolved to adapt to their harsh environments and have been identified as a result of recent progress in single-cell analysis (Veening et al., 2008; Bury-Moné and Sclavi, 2017). A growing number of examples show these behaviors (Kröger et al., 2011; Afroz et al., 2014; Kotte et al., 2014; Solopova et al., 2014; Norman et al., 2015; Leh et al., 2017; Kampf et al., 2018; Weiss et al., 2019). Heterogeneous expression of genes and proteins often leads to phenotypic heterogeneity. In extreme cases, this results in bistability, the existence of two stable states in a single population. For example, in Bacillus subtilis development of genetic competence for uptake of extracellular DNA, bimodal expression of the key master regulator ComK is observed, which leads to the differentiation of a fraction of cells among the cell population into the competent state (Maamar and Dubnau, 2005; Dubnau and Losick, 2006). When it comes to heterogeneous expression systems, bacteria sometime adopt a “bet-hedging strategy” where they differentiate into subpopulations in the same culture in order to facilitate adaptation to rapid environmental fluctuations (Veening et al., 2008; Norman et al., 2015). In this strategy, the cells to be adapted for the fluctuation with different phenotype have differentiated from sibling cells even before the environmental change. For example, Bacillus sporulation can be regarded as “bet-hedging” since the sporulating subpopulation prepares for more nutritionally harsh environments while the non-sporulating subpopulation retains the chance to re-initiate cell growth if more nutrients become available. In this case, highly heterogeneous phosphorylation of the master sporulation regulator Spo0A triggers the initiation of sporulation (Chastanet et al., 2010).
YlxR has characteristics specific to nucleoid-associated proteins (NAPs) and exhibits non-homogeneous expression (Browning et al., 2010; Ogura and Kanesaki, 2018). The heterogeneous expression of YlxR was revealed through microscopic observation of green fluorescent protein (GFP) expression by an YlxR–GFP fusion, although the biological consequence remains unclear (Ogura and Kanesaki, 2018). YlxR regulates transcription of more than 400 genes, including many metabolic genes (Ogura and Kanesaki, 2018). For example, in a B. subtilis ylxR-deletion mutant, expression of the frlBONMD-yurJ operon for amino sugar utilization and two arginine biosynthetic operons were enhanced (Deppe et al., 2011a, b). Furthermore, YlxR positively regulated the tsaEBD-containing operon through direct binding of YlxR to the operon promoter (Ogura et al., 2019; Figure 1). TsaEBD is an enzyme required for the synthesis of threonylcarbamoyl adenosine (t6A)-modified tRNA (Thiaville et al., 2015; Zhang et al., 2015). The t6A-modified tRNA is conserved in all domains of life, and its deficiency sometimes causes severe dysfunctions (Thiaville et al., 2016; Ogura et al., 2019). Expression of the ylxR-containing operon driven by the ylxS promoter (PylxS) itself requires cshA encoding a DEAD-box RNA helicase (Lehnik-Habrink et al., 2013; Ogura and Kanesaki, 2018). Proteomic analysis of B. subtilis revealed that CshA is lysine-acetylated (Kosono et al., 2015; Ogura and Asai, 2016). It has been reported that CshA associates with RNA polymerase (RNAP) and CshA-associated RNAP alters some of its own properties, such as its affinity to several sigma factors (Delumeau et al., 2011; Ogura and Asai, 2016). CshA acetylation is susceptible to pyruvate dehydrogenase (PDH) mutations in pdhABCD (Gao et al., 2002; Ogura and Asai, 2016). Disruption of the pdh genes reduces the intracellular acetyl-CoA pool and flux as a result of the loss of PDH activity, that is, the conversion of pyruvate to acetyl-CoA (Ogura et al., 2019). In B. subtilis, several lines of evidence suggest a relationship between low t6A and protein quality control, including PDH (Thiaville et al., 2015; Ogura et al., 2019). Therefore, t6A is required for a stable acetyl-CoA supply through control of PDH activity. Consequently, YlxR and CshA are concomitantly involved in the complex regulatory loop.
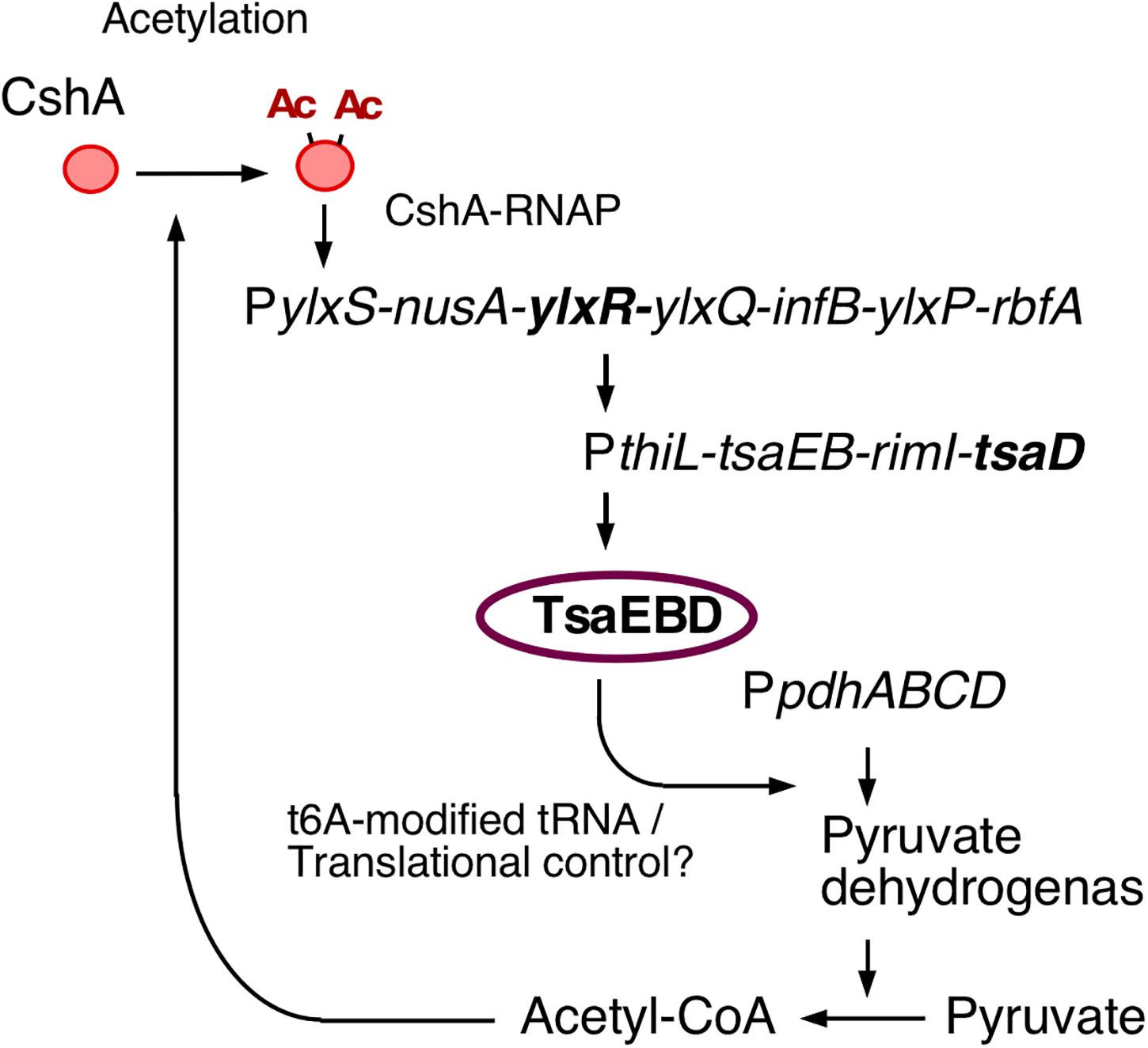
Figure 1. The indirect positive feedback loop of PylxS expression. The indicated pathways were previously identified: in vivo association of CshA with RNAP (Delumeau et al., 2011), CshA acetylation (Kosono et al., 2015; Ogura and Asai, 2016), and CshA-dependent PylxS regulation driving YlxR expression, which regulates transcriptional regulation of tsaEBD through the functional YlxR-binding to the promoter of tsaEBD, whose products are assembled and regulate pyruvate dehydrogenase translation (Ogura et al., 2019). Pyruvate dehydrogenase provides acetyl-CoA, which would be the acetyl moiety source for CshA acetylation. Arrows indicate transcription, translation, acetylation, enzymatic reaction, transcriptional activation, or metabolic reaction. Ac, acetyl moiety; RNAP, RNA polymerase.
The frlBONMD-yurJ operon is driven by PfrlB, which encodes metabolic enzymes fructoselysine-6-P-glycosidase from frlB and fructosamine kinase from frlD, and the FrlMNO-YurJ transporter for amino sugars, including fructoselysine. In the current study, expression profiles of the frlBONMD-yurJ operon were investigated using four fusion constructs. Two were transcriptional lacZ and gfp fusions at the ectopic locus amyE. The third was a transcriptional gfp fusion located at its original chromosomal location. The fourth fusion construct was a translational mCherry fusion at its original locus. Disruption effects of “genes encoding” CodY, FrlR, and YlxR, three known transcription factors for PfrlB, were examined for expression of each fluorescent reporter fusion in flow cytometric analyses (Molle et al., 2003; Belitsky and Sonenshein, 2008; Deppe et al., 2011b). The two gfp fusions showed heterogeneous expression profiles. The expression of PfrlB-mCherry was bimodal, but disruption of ylxR resulted in the disappearance of its clear bimodal expression. RNaseY is known to likely degrade mRNA of the frlBONMD-yurJ operon (Lehnik-Habrink et al., 2011), and depletion of the RNaseY-encoding gene resulted in abolishment of the heterogeneous expression of PfrlB-gfp, but not bimodal one of PfrlB-mCherry. These findings suggested a translational level of PfrlB regulation and a role for YlxR on the bimodal expression of PfrlB. Finally, we discuss the implications and possible causes of the bimodal expression of PfrlB.
Materials and Methods
Strains, Media, and PCR
All Bacillus subtilis strains used in this study are shown in Table 1. One-step competence medium (MC) (Kunst et al., 1994), Schaeffer’s sporulation medium (SM) (Schaeffer et al., 1965), Luria–Bertani (LB Lennox) medium (Difco, MI, United States), and Antibiotic medium 3 (Difco) were used. Antibiotic concentrations were described previously (Ogura and Tanaka, 1996; Ogura et al., 1997). Synthetic oligonucleotides were commercially prepared by Tsukuba Oligo Service (Ibaraki, Japan) and are listed in Supplementary Table S1. For PCR-mediated construction of strains and plasmids, PrimeSTAR MAX DNA polymerase (Takara Co., Shiga, Japan) was used. For screening of recombinant DNA during plasmid construction, LA PCR DNA polymerase (Takara Co.) was used.
Plasmid Construction
The plasmids used in this study are listed in Table 1. For PCR, B. subtilis chromosomal DNA was used as template. To construct pIS284-frlB, pIS284-acsA, and pIS284-codV, PCR products were amplified using the oligonucleotides pairs pIS-frlB-F-E/pIS-frlB-R-B, pIS-acsA-F-E/pIS-acs-R-B, and pIS-codV-Eco/pIS-codV-Bam; digested with EcoRI/BamHI; and cloned into pIS248 treated with the same enzymes (Tsukahara and Ogura, 2008). To construct pUKM504-frlR, PCR products were amplified using the oligonucleotides pair pUKM-frlR-B/pUKM-frlR-H, digested with BamHI/HindIII, and cloned into a pUKM504 plasmid treated with the same enzymes (Ogura and Tanaka, 1996). To construct pfrlB-SD-gfp, oligonucleotide pairs pIS-frlB-F-E/PfrlB-(SD)-gfp-R and gfp(SD)-F/gfp-Xba-R were used for amplification of the genomic region and gfp from genomic DNA of strains 168 and OAM-N32, respectively (Ogura, 2016). Here, SD means Shine–Dalgarno sequence. After the combination of the both PCR products in the PCR using oligonucleotide pair pIS-frlB-F-E/gfp-Xba-R, the fragment digested with EcoRI/XbaI was cloned into pSG1194 without dsRed generated by digestion with the same restriction enzyme pair (Feucht and Lewis, 2001). To construct pfrlB-mCherry, PCR products were amplified using the oligonucleotide pair mChe-frlB-F-H/mChe-frlB-R-E, digested with HindIII and EcoRI, and cloned into pNG621 treated with the same enzymes (Doherty et al., 2010).
Strain Construction
To construct OAM914, OAM933, and OAM934, the plasmids pIS284-frlB, pIS284-acsA, and pIS284-codV were transformed to the wild-type (WT) strain 168; and among the resultant chloramphenicol resistant colonies, those with amylase non-producing phenotype were selected on the LB agar plate containing 1% starch. To construct OAM913, OAM922, and OAM927, the plasmids pUKM504-frlR, pfrlB-SD-gfp, and pfrlB-mCherry were transformed to 168. To construct strains carrying the amyE:PfrlB-gfp, amyE:PacsA-gfp, amyE:PcodV-gfp, and amyE:PilvB-gfp fusions, first, PCR products were amplified from strain OAM914 carrying amyE:PfrlB-lacZ, OAM933 carrying amyE:PacsA-lacZ, OAM934 carrying amyE:PcodV-lacZ, and FU676 carrying amyE:PilvB-lacZ, using the oligonucleotide pairs amyE-RR/PfrlB-(SD)-gfp-R, amyE-RR/PacsA-(SD)-gfp-R, amyE-RR/PcodV-(SD)-gfp-R, and amyE-RR/PilvB-(SD)-gfp-R, respectively. Second, PCR products were amplified from the strain OAM-N32 using the oligonucleotide pair gfp(SD)-F/amyE-FF (Ogura, 2016). Each fragment for the promoters and the amplified gfp-amyE cassette was combined in a final PCR using the oligonucleotide pair amyE-FF/amyE-RR. The final PCR products were transformed into B. subtilis 168, and the chromosomal structure of the transformant was verified by PCR analysis using appropriate primers.
β-Galactosidase Analysis
Growth conditions and β-galactosidase analysis procedures were previously described (Ogura and Asai, 2016; Ogura and Kanesaki, 2018).
Microscopic Observations
Cells were picked up from flesh colony on LB agar plate and inoculated to 1 ml of LB medium in L-tube. The tube was shaken for 14 h at 37°C. One hundred microliters of the culture was centrifuged, and 80 μl of the supernatant was removed. The cells were then resuspended in the remaining 20 μl. Portions (2 μl) of each sample were mounted on glass slides treated with 0.1% (wt/vol) poly-L-lysine (Sigma-Aldrich, MO, United States). Microscopy was performed with an Olympus BX51 phase contrast and fluorescence microscope with a 100 × PLAN-N objective (Olympus, Tokyo, Japan). Images were captured using a CoolSNAP HQ charge-coupled device camera (Nippon Roper, Tokyo, Japan) and Metavue 4.6r8 software (Universal Imaging, PA, United States).
Flow Cytometry Analysis
All the strains were streaked on LB agar plates supplemented with specific antibiotics and incubated overnight. The resulting single colony was picked up and grown overnight in 1 ml of LB medium in L-tube at 37°C. Cells were washed and resuspended in phosphate-buffered saline (PBS) and directly measured on BD LSRFortessa (Becton–Dickinson, CA, United States) with an argon laser (488 nm) and yellow green laser (561 nm). For each sample, the green fluorescent signal or mCherry signal of 30,000 cells was collected by a bandpass (BP) filter (530/30 nm, 610/20 nm). The fluorescent intensity was calculated in arbitrary units (AUs). All the captured data were further analyzed with FlowJo version 7.6.5 software (TreeStar, CA, United States).
Results
YlxR-Mediated PfrlB Expression at the amyE Locus
The frlBONMD-yurJ operon is subject to the severe YlxR-dependent transcription repression according to previous YlxR-transcriptome analysis (Wiame et al., 2004; Deppe et al., 2011a, b; Ogura and Kanesaki, 2018). To confirm this repression, the PfrlB-lacZ transcriptional fusion at amyE was constructed, and the influence of ylxR disruption on PfrlB expression was measured. Expression of PfrlB-lacZ was clearly repressed by YlxR as its expression in the ylxR-disruptant strain increased four-fold compared with that in the WT strain (Figure 2). We note that sporulation media were used for the lacZ and previous transcriptome analysis experiments. With the use of PylxS-gfp, no YlxR-expressing (YlxR-ON) cells were observed during growth in SM (Ogura and Kanesaki, 2018); however, YlxR-ON cells are observed 30% of the cell population when grown in LB medium (Figure 3; supporting information in Ogura and Kanesaki, 2018). In addition, heterogeneous ylxR expression was subject to positive autoregulation of ylxR as disruption of ylxR resulted in no expression of PylxS-gfp (Figure 3). This autoregulation is reported to be indirect and mediated by CshA (Figure 1; Ogura and Kanesaki, 2018). This was confirmed by our observation that cshA disruption also abolished PylxS-gfp expression (Figure 3). This suggested that YlxR-regulated PfrlB expression may also be heterogeneous in LB medium. In the previous study of PfrlB, M9 medium supplemented with Amadori products (fructosamines) was used, and thus growth profile showed diauxie probably due to two carbon sources, glucose and Amadori products (Deppe et al., 2011b). However, we were not able to produce Amadori products efficiently (see section “Discussion”). Thus, to avoid diauxie and difficulties for preparing M9 with Amadori products, we used LB medium for further investigation using fluorescence reporter. To examine possible heterogeneous expression of frlB, we constructed a transcriptional PfrlB-gfp fusion at the amyE locus. As shown in Figure 4, the autonomous SD sequence of frlB failed to function due to the long distance between the SD sequence and the initiation codon of gfp. The strain bearing this fusion showed approximately 30% GFP-ON cells in the microscopic analysis with the remaining 70% being GFP-OFF cells (Figure 4). We note that the expression was observed during early stationary phase of growth in LB medium, i.e., overnight culture. To confirm this observation, flow cytometry analysis of the strain bearing PfrlB-gfp at amyE was performed. As shown in Figures 5A1,_D1, distinct bimodal expression patterns of GFP expression were seen in WT strains after 14 and 24 h of culturing. However, disruption of ylxR resulted in a 10% increase in the frequency of GFP-ON cells among the culture population (Figure 5A4). This was consistent with the YlxR repression of the lacZ fusion, because total fluorescence increased 2.5-fold. Recently, it has been reported that GFP expression driven by an IPTG-dependent promoter to some extent shows heterogeneous expression, which is due to “noise” in gene expression (Cao and Kuipers, 2018). To examine the expression of other promoters, we randomly selected three (PacsA, PilvB, and PcodV) and created transcriptional GFP fusions, and then we evaluated their expression by flow cytometry. All three of the promoter fusions showed homogeneous expression (Figure 5E). These results indicated that the observed bimodal expression of PfrlB-gfp was derived from regulated expression specific for PfrlB, not simply from gene and protein expression “noise.”
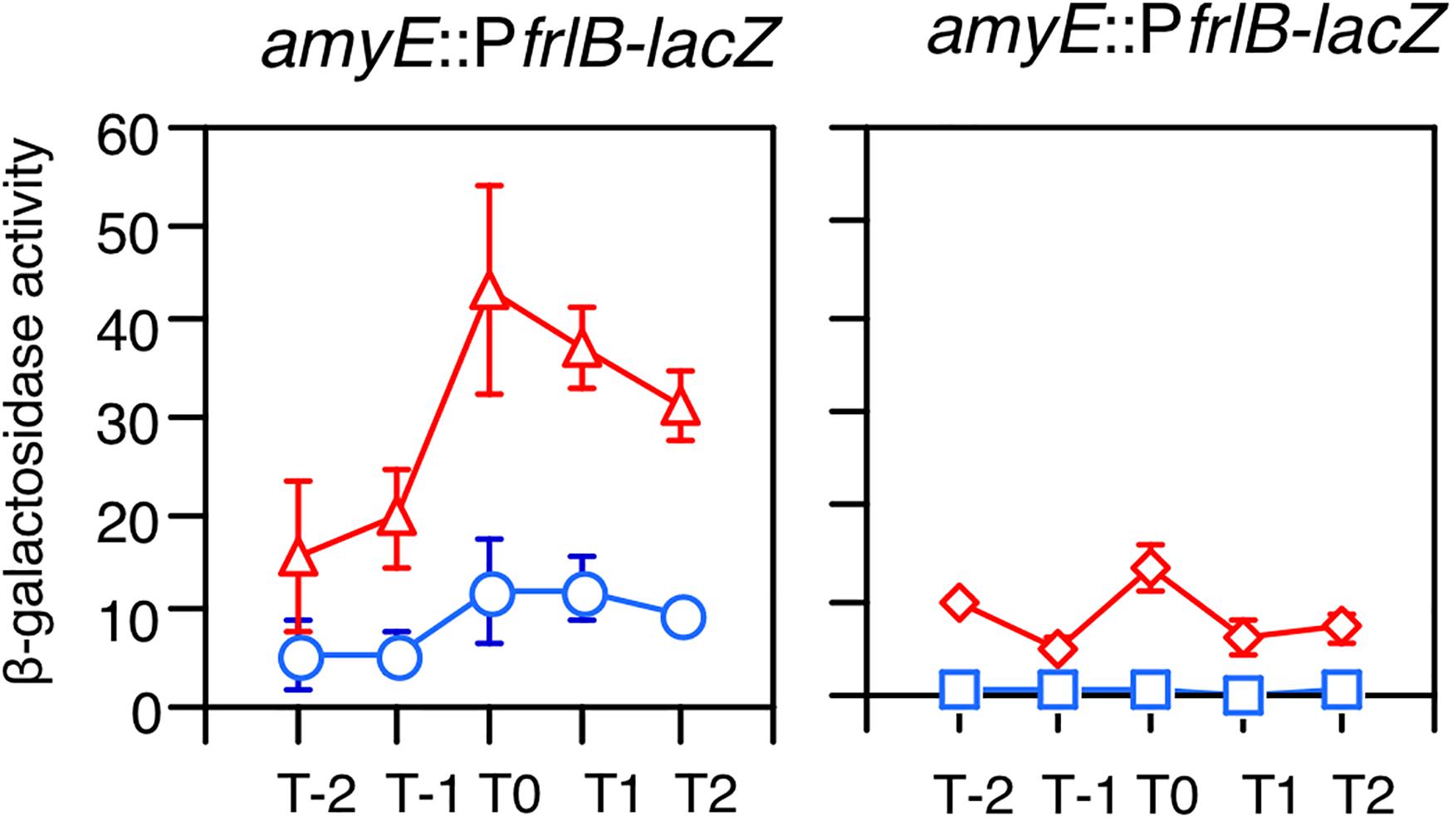
Figure 2. Expression of PfrlB-lacZ. Strains were grown in sporulation medium and sampled hourly. The x axis represents the growth time in hours relative to the end of vegetative growth (T0). Means from three independent experiments and the standard deviations are shown. Left graph: circles, wild type (OAM914); triangles, ylxR (OAM915). Right graph: diamonds, codY (OAM917); squares, frlR (OAM916).
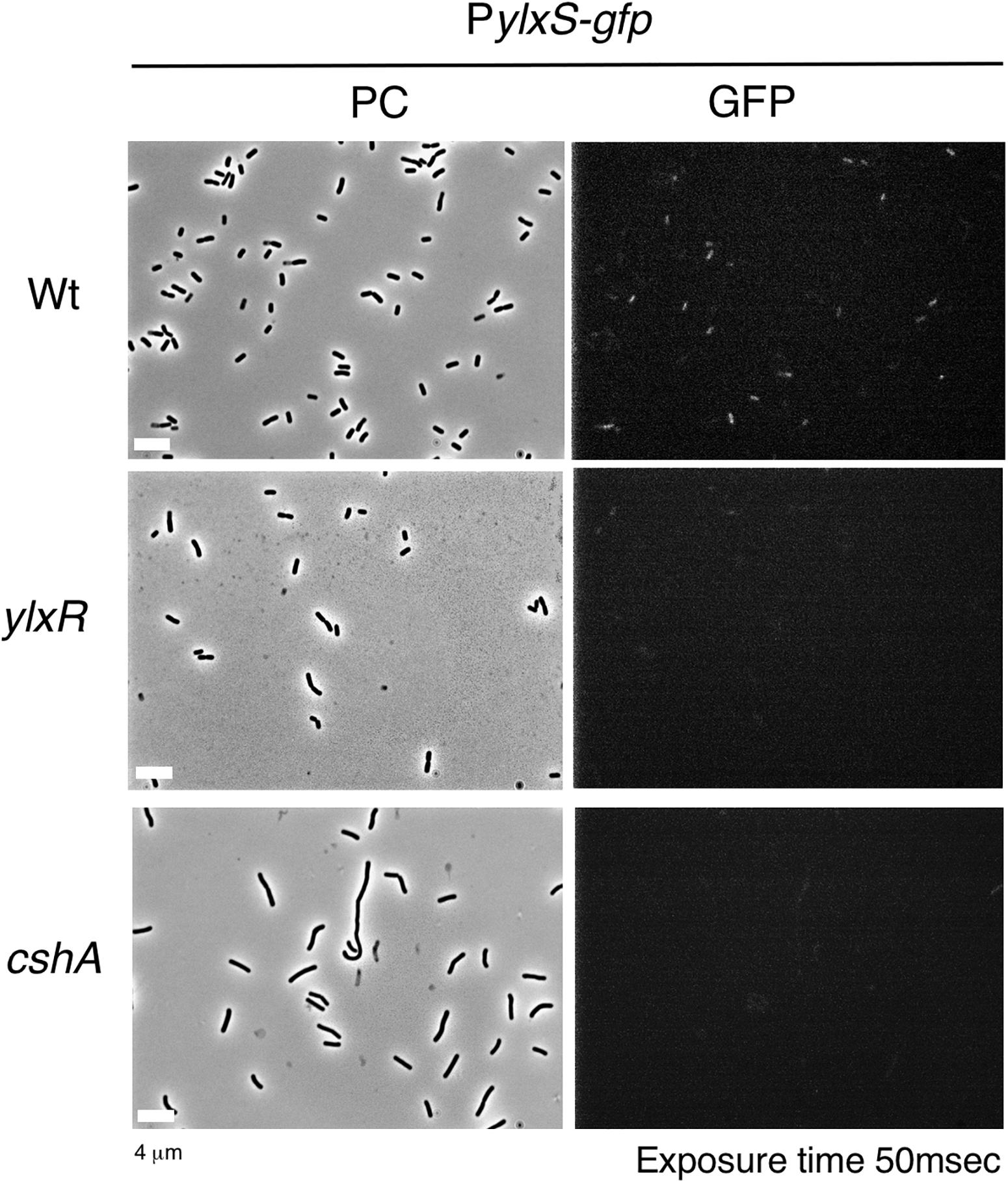
Figure 3. Expression of PylxS-gfp. Strains bearing amyE:PylxS-gfp, wild type (OAM818), cshA (OAM938), and ylxR (OAM939) were grown in LB medium in L-tubes. After 14 h, the cells were sampled and processed. Representative micrographs of the microscopic observation are shown. PC, phase contrast; GFP, green fluorescent protein; LB, Luria–Bertani. GFP fluorescence was visualized using a WIB filter set (Olympus). Image processing and data analysis were performed using Adobe Photoshop CS5.
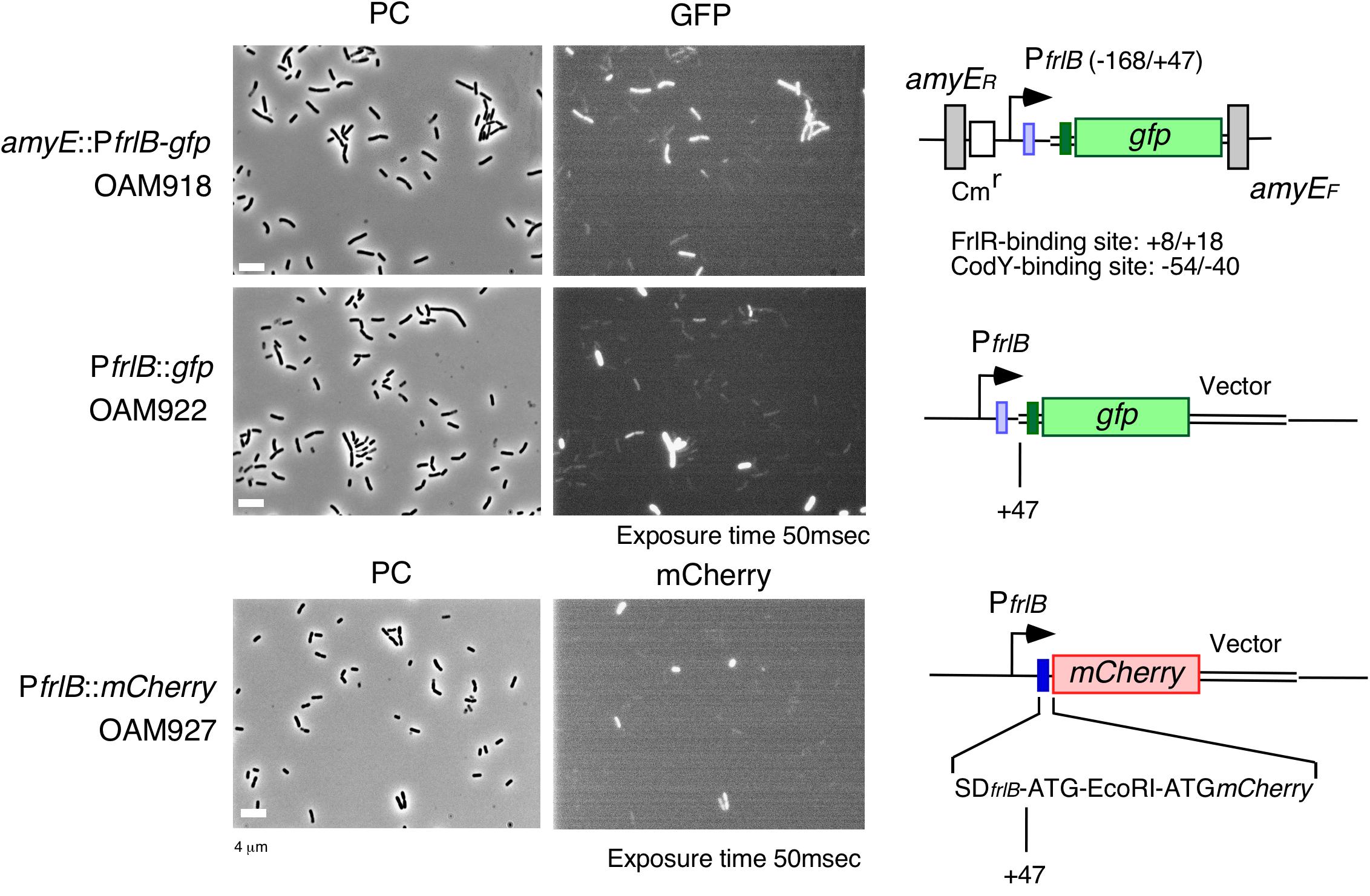
Figure 4. Expression of PfrlB-gfp fusions located at its own and ectopic chromosomal regions and PfrlB-mCherry. Structures of two gfp and mCherry fusions are schematically depicted alongside the corresponding micrographs. The large and small boxes on the line represent the open reading frames and Shine–Dalgarno (SD) sequences (blue for frlB and green for gfp). The pale box represents an SD sequence that failed to function. The bent arrow indicates the promoter. All the strains retain the intact frlB gene. The numbers indicate the nucleotide positions relative to the transcription start nucleotide. Strains were grown in LB medium in L-tubes. PC, phase contrast; GFP, green fluorescent protein; and mCherry, red fluorescent protein derived from Discosoma sp. GFP and mCherry fluorescence were visualized using WIB and WIG filter sets (Olympus, Tokyo, Japan), respectively. Image processing and data analysis were performed using Adobe Photoshop CS5. Representative images are shown.
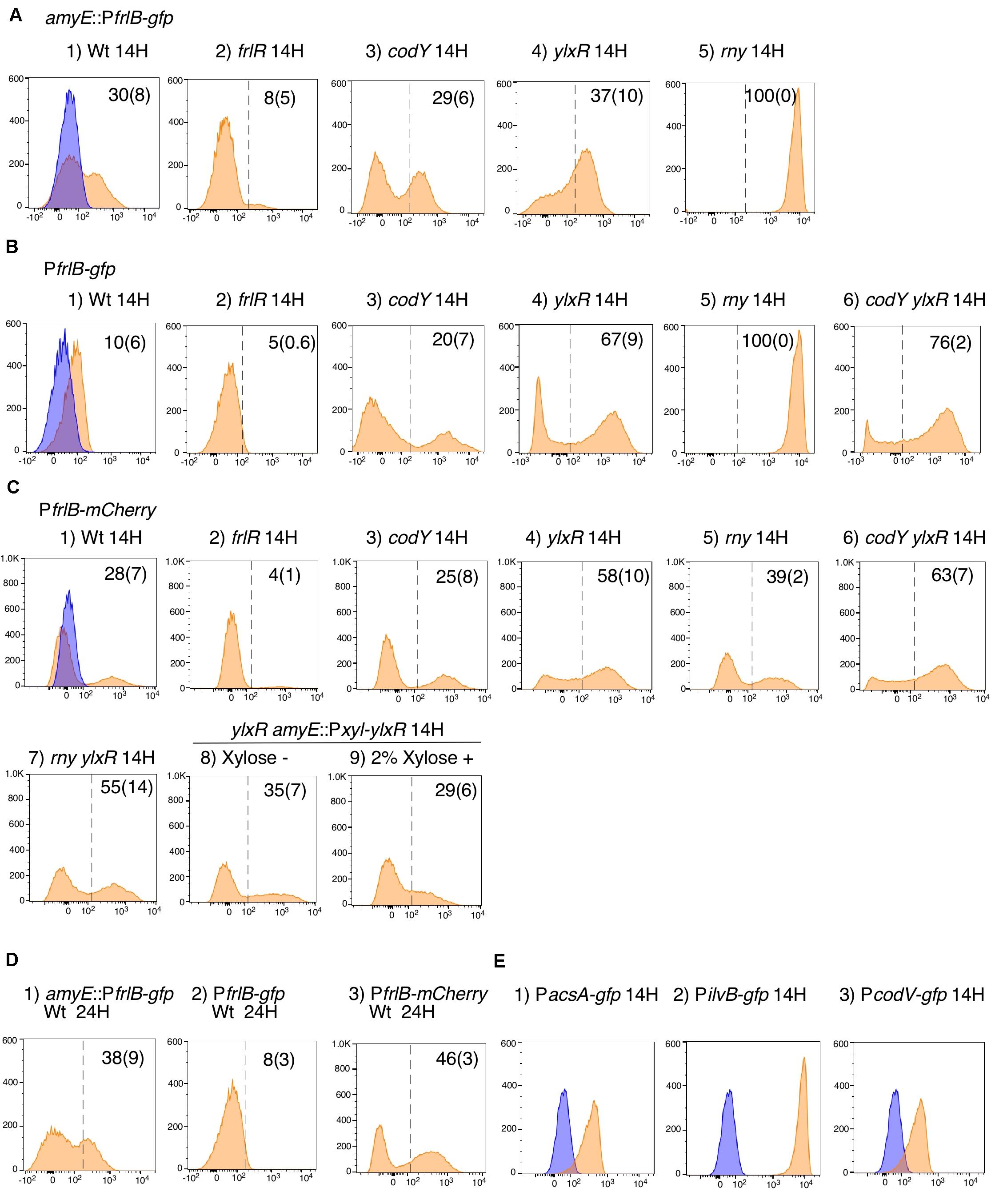
Figure 5. Flow cytometry analysis of three fluorescent PfrlB fusions. Strains as follows were grown in LB medium. (A) amyE:PfrlB-gfp. (1) OAM918; (2) OAM919; (3) OAM920; (4) OAM921; (5) OAM940. (B) PfrlB-gfp. (1) OAM922; (2) OAM923; (3) OAM924; (4) OAM925; (5) OAM941; (6) OAM926. (C) PfrlB-mCherry. (1) OAM927; (2) OAM928; (3) OAM929; (4) OAM930; (5) OAM942; (6) OAM931; (7) OAM943; (8 and 9) OAM944. (D) Results of longer cultivation time for three wild-type fusions. (E) (1) OAM935; (2) OAM937; (3) OAM936. X- and Y-axes indicate fluorescence intensity and cell numbers, respectively. The biexponential transformation was applied to display the flow cytometry data and X-axis is in “logicle” scale (Parks et al., 2006). In B4 and B6, to show cells with very weak fluorescence intensity, the X-axis is expanded to 10– 3 but not 10– 2. Mean percentages of fluorescence-positive cells from three independent experiments are shown with standard deviations in parentheses. The dotted lines indicate the ends of fluorescence-negative cell fractions (also as blue fractions) obtained from measurement using the control strain 168. Typical patterns are shown. “H” indicates incubation time in hours. LB, Luria–Bertani.
According to previous reports, the repressors CodY and FrlR bind to the promoter region of frlB (Molle et al., 2003; Belitsky and Sonenshein, 2008; Deppe et al., 2011b). The PfrlB-lacZ and PfrlB-gfp at amyE constructs that we generated for the current study contained both binding sites. Moreover, disruption of either codY or frlR has been reported to abate the repression of fusion expression at amyE (Belitsky and Sonenshein, 2008; Deppe et al., 2011b). To confirm these observations, we disrupted frlR and codY in the strains expressing PfrlB-lacZ and PfrlB-gfp at amyE. Both β-galactosidase (beta-Gal) and flow cytometric analyses showed that the disruption of frlR almost completely abolished the expression of both PfrlB-lacZ and PfrlB-gfp (Figures 2, 5A2). This suggested that under the conditions we used, FrlR acted on PfrlB as an activator, not repressor. This was contrary to the previous report by Deppe et al. (2011b). Both the beta-Gal and flow cytometric analyses showed that disruption of codY had no detectable influence on the lacZ and gfp expression (Figures 2, 5A3), again contrary to a previous report, where minimal medium with ammonium or that supplemented with 16 amino acids was used (Belitsky and Sonenshein, 2008). We note that in synthetic medium containing casamino acid (MC medium), basal expression of PfrlB-lacZ in the WT strain was approximately 10 Miller units, and a slight increase (approximately 2.5-fold) was observed in the codY disruptant (data not shown). However, this enhanced rate of the fusion expression was 500-fold lower than the value reported by Belitsky and Sonenshein (2008). We will argue this difference (see section “Discussion”).
YlxR- and CodY-Mediated PfrlB-gfp Expression at the Original Chromosomal Locus
In a previous report, expression of PfrlB (yurP)-lacZ in its original chromosomal context is 10-fold enhanced by codY disruption when cultured in minimal glucose-glutamine medium containing a mixture of 16 additional amino acids (Molle et al., 2003). In the transcriptome (ChIP-to-chip and DNA microarray) analyses using the cells grown in SM, i.e., detection of intact mRNA, 20-fold enhancement of frlB mRNA amount was observed by codY disruption (Molle et al., 2003). Therefore, we constructed a transcriptional PfrlB-gfp fusion at its original locus. Heterogeneous expression of the fusion was still observed by microscopic analysis (Figure 4). In the middle row in Figure 4, about 10% of GFP-ON cells were observed. The fusion expression was then analyzed by flow cytometry. The heterogeneous expression of PfrlB-gfp was observed, but not in a bimodal fashion (Figures 5B1,_D2). The observation of 10% GFP-ON cell fraction was consistent with that in microscopy. As expected, disruption of frlR in this strain resulted in almost complete abolishment of PfrlB-gfp expression (Figure 5B2). In the case of codY disruption, the frequency of the GFP-ON cells was increased two-fold (Figure 5B3). It should be noted that average intensity of fluorescence among the GFP-ON cells increased 10-fold in the codY disruptant. These findings were consistent with a previous report of approximately a 10-fold enhancement of PfrlB-lacZ fusion expression (Molle et al., 2003). Disruption of ylxR in this strain resulted in a seven-fold larger subpopulation of GFP-ON cells among the population, and the bimodal fashion of expression was restored (Figure 5B4). Our earlier results suggested the possibility that CodY and YlxR may function cooperatively (Ogura and Kanesaki, 2018). To examine this possibility, we constructed a codY/ylxR double mutant and measured the frequency of GFP-ON cells among the population. A non-synergistic additive increase was observed, suggesting CodY and YlxR are not in the same regulatory line. Moreover, the frequency of GFP-ON cells was not 100% in the double mutant, suggesting that an unknown factor may function in this double mutant (Figure 5B6, see below, section on RNaseY).
YlxR-Mediated PfrlB-mCherry Translational Fusion Expression at the Original Locus
As detailed in the Supplementary Table S1 in Ogura and Kanesaki (2018), YlxR regulates the expression of more than 400 genes, including translation-related genes such as tsaD (encodes a component of the enzyme required for t6A modification of tRNA), rrnE-16S (encodes an rRNA), and rpsNB (encodes ribosomal protein S14). Therefore, it is possible that YlxR acts not only on the transcription of PfrlB but also on its translation. To explore this possibility, we constructed a translational fusion of frlB with the fluorescence protein gene mCherry at its original chromosomal position. In the construct, the mCherry protein was added with three amino acids to the N-terminus and expressed with the frlB SD sequence as required for its translational initiation (Figure 4). Microscopic analysis revealed heterogeneous expression of PfrlB-mCherry, as expected (Figure 4). In flow cytometric analysis, a subpopulation of PfrlB-mediated mCherry-ON cells was represented by a distinct peak, indicating bimodal expression of mCherry (Figures 5C1,_D3). We note that a higher rate of mCherry-ON cells was detected by flow cytometry than that by microscopic analysis. Rapid fading of mCherry with an N-terminal adduct (a half-life of approximately 3 seconds according to the analysis of continuous photographing of fluorescence images; data not shown) resulted in more efficient detection of fluorescence by flow cytometry. The distinctly different expression profiles between transcriptional PfrlB-gfp and translational PfrlB-mCherry fusions, both of which are located at its original locus, suggested regulation other than transcriptional regulation, that is, at the post-transcriptional level including translation. Disruption of ylxR in the strain bearing the mCherry fusion caused severe decline of the sharp bimodal expression, suggesting that frlB-mediated bimodal expression of mCherry required functioning YlxR (Figure 5C4). To confirm this, a complementation test of ylxR disruption by xylose-inducible ylxR was performed. Without xylose, frequency and expression profile of mCherry-ON cells were similar to those in the WT, suggesting complementation of ylxR disruption probably due to leaky expression of Pxyl-ylxR (Figure 5C8). Further induction of ylxR with xylose showed slightly decreased frequency of mCherry-ON cells and reduced total fluorescence of the mCherry fusion to 30% of that in the absence of xylose, leading to change of the expression profile (Figure 5C9). A cell population with fluorescence intensity more than 103 almost disappeared by addition of xylose (Figure 5C8 vs. 5C9). These experiments showed that the severe decline of the sharp bimodal expression is indeed caused by the ylxR disruption, but not polar effect. It should be noted that in the ylxR disruptant, still significant mCherry-OFF cells were observed, which suggested the uncovered regulatory mechanism in the frlB expression. Disruption of frlR also almost completely abolished the mCherry expression, as expected (Figure 5C2), but disruption of codY had no noticeable effect on mCherry expression, suggesting a negligible if any increase in transcription of mCherry in the codY disruptant (Figure 5B vs. 5C). Disruption of codY in association with the disruption of ylxR enhanced frequency of mCherry-ON cells slightly, compared with that seen for the disruption of ylxR alone (Figure 5C6). This suggested that the enhancement of transcription due to the disruption of codY, as observed in the case of PfrlB-gfp located at its original locus, did not result in the higher frequency of mCherry-ON cells among the population than that in the single ylxR disruptant.
frlB Expression in the rny Depletion Mutant
Expression of PfrlB is reported to be negatively regulated by RNaseY, which is encoded by rny and has endonuclease activity for many mRNAs (Lehnik-Habrink et al., 2011). That cleavage triggers mRNA degradation. Although the entire mRNA structure between frlB itself and PfrlB-gfp transcriptional fusions is different, those mRNAs share the common short upstream untranslated region in structure (Figure 4). Therefore, expression of PfrlB-gfp fusion at either amyE or the original locus may be increased in the rny-depleted strain. Consistent with this expectation, we observed 100% GFP-ON cells from the population among the two strains bearing both PfrlB-gfp and rny-depletion (Figures 5A5,_B5). These findings indicated that RNaseY acted through the degradation of PfrlB-driven mRNA and was perhaps responsible for the bimodal expression profiles of these fusions. Consequently, the depletion of rny may not have affected frlB expression at the level of translation. To examine this, the rny depletion was introduced into a strain bearing PfrlB-mCherry, and the expression was evaluated in flow cytometric analyses. As expected, the depletion of rny only slightly affected the bimodal expression of PfrlB-mCherry, i.e., 10% enhancement of frequency of mCherry-ON cells (Figure 5C5). This indicated that the depletion of rny affected the expression of frlB transcriptional fusions profoundly, but not significantly its translational control. Moreover, the frequency of mCherry-ON cells in the rny depleted strain with the ylxR disruption was similar to that in the ylxR single disruptant (Figure 5C7). This result is also consistent with the observation that RNaseY did not affect translational frlB fusion expression.
Microscopic Analysis of Cells Bearing PfrlB-mCherry Translational Fusion and amyE:PylxS-gfp
Flow cytometric analysis suggested that YlxR mediated negative control of PfrlB-mCherry. Moreover, as YlxR expression is heterogeneous (Ogura and Kanesaki, 2018), PfrlB-mCherry-expressing cells may be differentiated from YlxR-OFF cells. To examine this possibility, we constructed the strain with PylxS-gfp at amyE and PfrlB-mCherry. Unfortunately, PylxS-gfp fluorescence was very weak, resulting in flow cytometric analysis detecting only a few percent of GFP-ON cells among the population of cells (0.5–5% in three independent observations), which was lower than that observed in the microscopic analysis. This may be due to detection of auto-fluorescence derived from the intracellular molecules like NADPH and aromatic amino acids from cells with no GFP-expressing strain, i.e., the control strain 168, in flow cytometry. Discrimination of weak but significant fluorescence signals from auto-fluorescence is difficult in flow cytometry. Contrary to this, in microscopic analysis, we did not observe auto-fluorescence derived from the strain without the gfp gene under the conditions we used (data not shown). Therefore, we had to explore the possibility using the microscopic analysis. As shown in Figure 6, most mCherry-ON cells lacked PylxS-gfp expression; however, a small fraction of mCherry-ON cells did exhibit associated PylxS-gfp expression. Among the mCherry-ON cells, distribution of PylxS-gfp expression was low compared with that among the mCherry-OFF cells with the difference being statistically meaningful (see legend to Figure 6). These results suggested that the mCherry-ON cells were often differentiated from cells that did not expressed YlxR. This was consistent with the results obtained from the flow cytometric analysis.
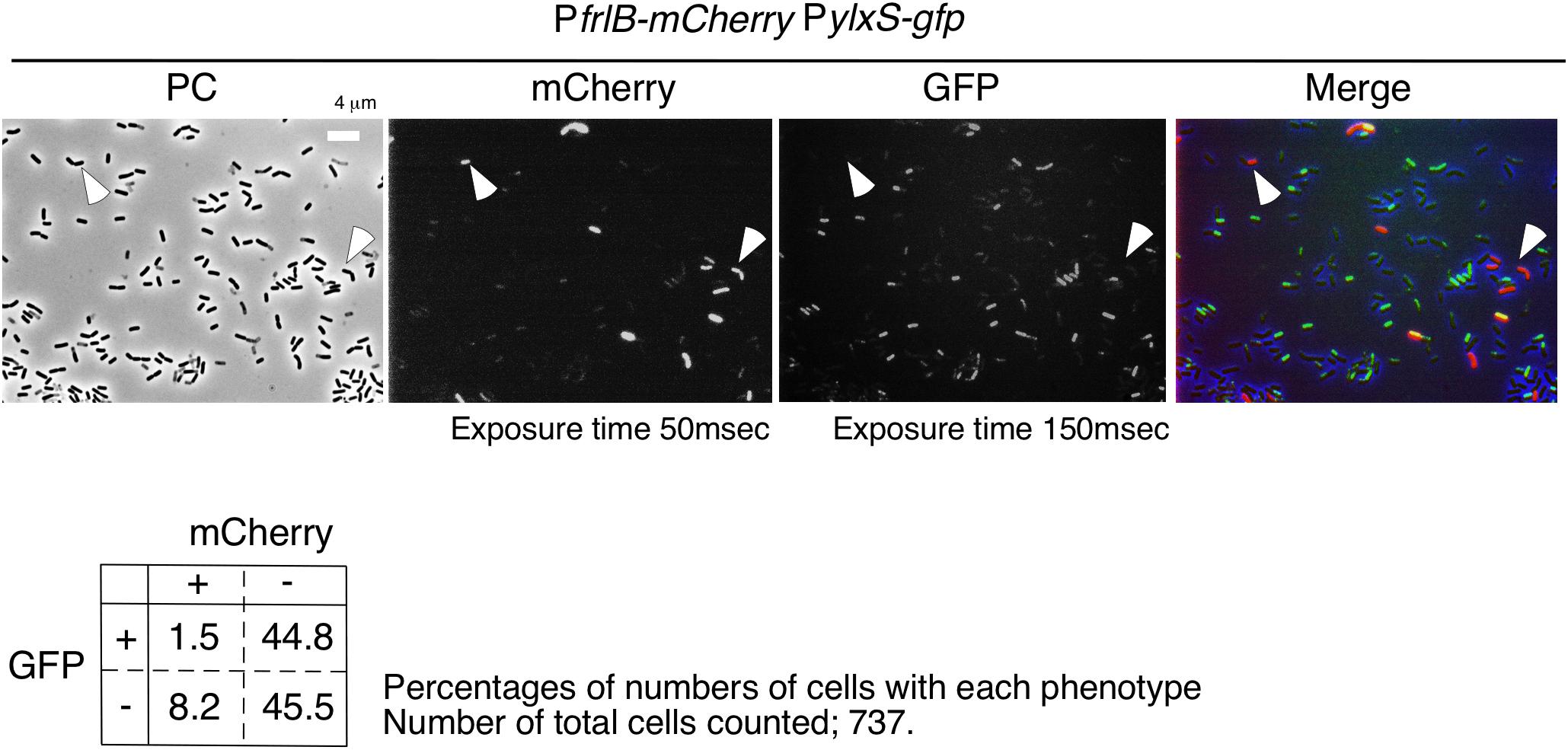
Figure 6. Expression of gfp and mCherry in cells bearing PylxS-gfp and PfrlB-mCherry. OAM932 was grown in LB medium in L-tubes. After 14 h of incubation, cells were sampled and processed. Representative micrographs from the microscopic observation are shown. PC, phase contrast; GFP, green fluorescent protein; mCherry, red fluorescent protein; LB, Luria–Bertani. GFP and mCherry fluorescence were visualized using WIB and WIG filter sets (Olympus), respectively. Arrowheads indicate mCherry-ON/GFP-OFF cells. Image processing and data analysis were performed using Adobe Photoshop CS5. The merged micrographs are in shown with pseudocolor, red, mCherry; green, GFP. Results of the quantitative analysis are shown below the photos. Chi-square test of independence was performed to compare the frequencies or proportions among variables in four types of cells with respect to mCherry and GFP. Chi-square value, p-value, and degrees of freedom (df) value were 30.19, 3.93 × 10– 8, and 1, respectively. This indicated that the chi-square statistic was at a significant level.
Discussion
In this study, we observed bimodal expression of PfrlB-mCherry. Gene products of the frlB operon are used for the utilization of amino sugars. However, to the best of our knowledge, there are no reports on whether sporulation or LB media contain amino sugars. In synthetic MC medium, which does not contain amino sugars, frlB expression did not change compared with that in sporulation or LB media (Ogura, unpublished results). These results indicated that some cells differentiate into PfrlB-expressing cells, even though there is no availability of amino sugars. This means that the Bacillus cells adopt a bet-hedging strategy with respect to nutritional fluctuation. At the transcription level, the observed bimodal or heterogeneous expression of PfrlB located at ectopic or original locus was caused by mRNA degradation triggered by RNaseY; however, this regulation was restricted for the transcriptional fusion expression through mRNA stability control. Based on our current study, we suggest that YlxR bimodal expression may result in the bimodal expression of the frlB operon through regulation at both the transcription and post-transcription levels including translation. Similar to YlxR in Bacillus subtilis, Escherichia coli nucleoid-associated proteins Ler and H-NS cause bimodal expression from the enterocyte effacement (LEE) pathogenicity loci (Leh et al., 2017). The exact mechanism underlying ylxR bimodal expression is not known. Generally, positive feedback regulation is responsible for the bimodal expression of a gene (Veening et al., 2008; Norman et al., 2015; Bury-Moné and Sclavi, 2017). Our previous reports showed that YlxR is subject to an indirect positive feedback loop (Ogura and Asai, 2016; Ogura and Kanesaki, 2018; Ogura et al., 2019). This loop may be responsible for the bimodal expression of YlxR (Figures 1, 7).
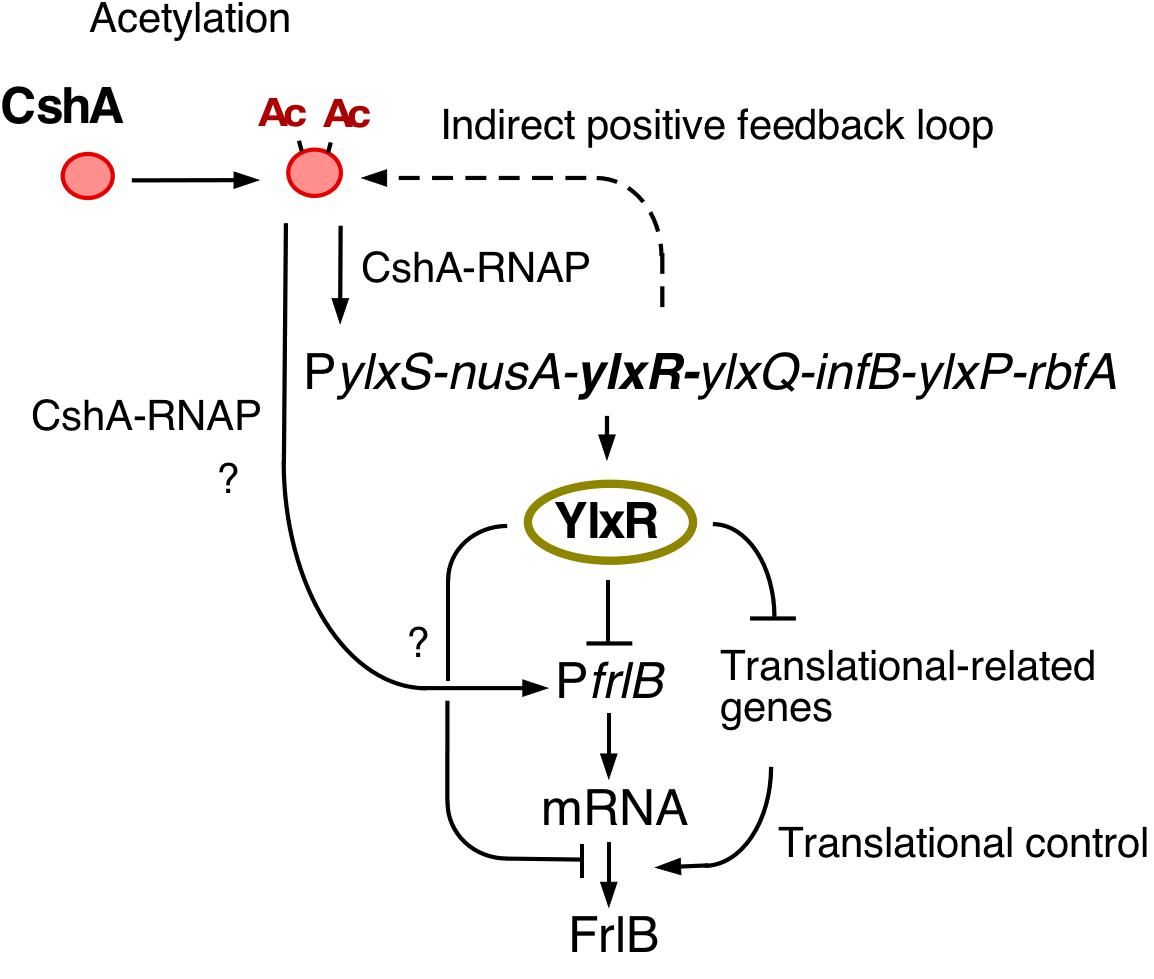
Figure 7. Hypothetical schematic for the regulation of frlB expression. Arrows and T-bars indicate positive and negative actions, respectively. Ac, acetyl moiety; RNAP, RNA polymerase.
Disruption of codY enhanced the expression of the transcriptional PfrlB-gfp fusion but did not affect the expression of the translational fusion. Considering the changes in the expression profiles between both fusions also suggested the regulation of frlB was at the post-transcriptional level including translation. Two possible routes of YlxR-dependent translational regulation of frlB were feasible (Figure 7). First, YlxR itself may function in frlB translation. It is possible that YlxR binds to frlB mRNA and thereby affects its translation as the crystal stereo-structure of YlxR suggests RNA-binding by YlxR (Osipiuk et al., 2001). Second, the YlxR-regulon contains several translation regulatory factors, including rrnE-16S and rpsNB, whose expression is directly or indirectly repressed by YlxR (Ogura and Kanesaki, 2018). Thus, disruption of ylxR enhances the expression of these genes. This may lead to the activation of frlB translation, which would then result in the disruption of the distinct bimodal expression profile of frlB.
The expression of ylxR is dependent on cshA, and therefore, disruption cshA would be expected to increase frlB expression as the cshA disruption would lead to decreased expression of YlxR, the negative regulator of frlB (Figures 1, 7). However, according to a previous transcriptome analysis of the cshA disruptant, the frlB operon is severely suppressed (Lehnik-Habrink et al., 2013). We also observed a significant decrease in the expression of frlB in the preliminary RNA-seq analysis using the cshA disruptant with approximately a 70% reduction compared with that in the WT strain (Ogura and Kanesaki, unpublished results). In addition, flow cytometry analysis using PfrlB-gfp at amyE revealed no GFP fluorescence expression in the cshA mutant (Ogura, unpublished results). Consequently, we speculated that cshA may positively regulate the expression of frlB, independent of ylxR (Figure 7). This should be clarified in future analyses.
A previous study presented a model where FrlR was a repressor and suggested that the inducer fructosamine-6-phosphate may inhibit the repressor activity of FrlR, leading to expression of the frlB operon (Deppe et al., 2011b). Our experimental data are inconsistent with that model. First, FrlR acted on the expression of PfrlB as an activator, not a repressor. FrlR belongs to the GntR bacterial transcription family, and that family includes several transcriptional activators (Blancato et al., 2008; Wiethaus et al., 2008; Edayathumangalam et al., 2013; Brambilla and Sclavi, 2015), which reinforces our conclusion. Second, in the previous study one of Amadori products, fructose-arginine was synthesized and used to show that it was an inducer of the frlB operon through acting on FrlR (Deppe et al., 2011b). The protocol for the synthesis of fructose-arginine in the previous paper is ambiguous, and we therefore used a modified procedure. Nuclear magnetic resonance spectroscopy analysis of the reaction products indicated small but significant amounts of reaction products (Shindo, unpublished results). However, we added the reaction products to culture media of strains with both types of the PfrlB-gfp fusions, which resulted in no influence on gfp expression (Ogura, unpublished results). Based on these data, we concluded that FrlR functions as an activator and that fructose-arginine is not an inducer of PfrlB expression at least in LB medium.
We observed that CodY only functioned when the target frlB promoter was located in its original chromosomal position as disruption of codY did not affect frlB expression from the amyE locus. However, in an earlier report, PfrlB-lacZ expression at amyE increased in the codY disruptant (Belitsky and Sonenshein, 2008). The fusion used in the earlier report bears the longer promoter region of frlB (-202/ + 90) than that used in this study (-168/ + 47). The difference may have led to the discrepancy in the results. Under the conditions we used, however, codY disruption influenced frlB expression at its original position, but not its ectopic amyE position. Thus, the function of CodY might be dependent on the chromosomal position of the target gene frlB. This is not unprecedented as several other examples have been reported (Bryant et al., 2014; Brambilla and Sclavi, 2015).
There are few known cases of bimodal expression of metabolic genes. In E. coli, when cells are transferred from glucose-containing media to media with a different carbon-source medium, such as malate, which is used for gluconeogenesis, most of the cells remain dormant in a persister state; however, a subpopulation that is prepared to use malate and the gluconeogenesis pathway appears among the cell population (Kotte et al., 2014). In Pseudomonas putida, when glycerol is the sole carbon source, bistable expression of the glycerol utilization operon occurs due to the repressor of that operon (Nikel et al., 2015). These are examples of metabolic bet-hedging strategies. In addition, when cells were grown using some carbon-source, such as D-xylose for E. coli and myo-inositol for Salmonella, expression of the operon promoters for their utilization is bimodal (Kröger et al., 2011; Afroz et al., 2014). In Lactococcus lactis, carbon diauxie (glucose to cellobiose) results in the bimodal expression from the promoter of the genes encoding the sugar phospho-transfer system (PTS) for cellobiose/lactose (Solopova et al., 2014). Interestingly this bimodal expression during diauxie disappears with the disruption of CcpA, which is a master regulator of carbon catabolites in gram-positive bacteria and represses that PTS promoter (Fujita, 2009). Therefore, bimodal expression of frlB in B. subtilis deserves further study, since the substrate of FrlB is not the most favorable carbon source but is sometimes provided. For example, this is the case in rhizospheres where it is actually preferred by Gram-positive bacteria of the family Bacillaceae.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. further inquiries can be directed to the corresponding author.
Author Contributions
MO performed experiments and wrote the manuscript. KS performed experiments. YK performed statistical analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by JSPS KAKENHI Grant Number 18K05415 and the Research Program of the Institute of Oceanic Research and Development.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge Support Center for Medical Research and Education, Tokai University, for excellent technical support of flow cytometry analyses by Y. Okada and Y. Iida. The authors also thank the technical aid of Chiharu Takagi (Japan Women’s University).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02024/full#supplementary-material
References
Afroz, T., Biliouris, K., Kaznessis, Y., and Beisel, C. L. (2014). Bacterial sugar utilization gives rise to distinct single-cell behaviors. Mol. Microbiol. 93, 1093–1103.
Belitsky, B. R., and Sonenshein, A. L. (2008). Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190, 1224–1236. doi: 10.1128/jb.01780-07
Blancato, V. S., Repizo, G. D., Suárez, C. A., and Magni, C. (2008). Transcriptional regulation of the citrate gene cluster of Enterococcus faecalis involves the GntR family transcriptional activator CitO. J. Bacteriol. 190, 7419–7430. doi: 10.1128/jb.01704-07
Brambilla, E., and Sclavi, B. (2015). Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 5, 605–614. doi: 10.1534/g3.114.016139
Browning, D. F., Grainger, D. C., and Busby, S. J. (2010). Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13, 773–780. doi: 10.1016/j.mib.2010.09.013
Bryant, J. A., Sellars, L. E., Busby, S. J. W., and Lee, J. (2014). Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 42, 11383–11392. doi: 10.1093/nar/gku828
Bury-Moné, S., and Sclavi, B. (2017). Stochasticity of gene expression as a motor of epigenetics in bacteria: from individual to collective behaviors. Res. Microbiol. 168, 503–514. doi: 10.1016/j.resmic.2017.03.009
Cao, H., and Kuipers, O. P. (2018). Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in Bacillus subtilis. Microbial. Cell Fact 17:134.
Chastanet, A., Vitkup, D., Yuan, G.-C., Norman, T. M., Liu, J. S., and Losick, R. (2010). Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 8486–8491. doi: 10.1073/pnas.1002499107
Delumeau, O., Lecointe, F., Muntel, J., Guillot, A., Guédon, E., Monnet, V., et al. (2011). The dynamic protein partnership of RNA polymerase in Bacillus subtilis. Proteomics 11, 2992–3001. doi: 10.1002/pmic.201000790
Deppe, V. M., Bongaerts, J., O’Connell, T., Maurer, K. H., and Meinhardt, F. (2011a). Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions. Appl. Microbiol. Biotech. 90, 399–406. doi: 10.1007/s00253-010-3083-4
Deppe, V. M., Klatte, S., Bongaerts, J., Maurer, K. H., O’Connell, T., and Meinhardt, F. (2011b). Genetic control of amadori product degradation in Bacillus subtilis via regulation of frlBONMD expression by FrlR. Appl. Environ. Microbiol. 77, 2839–2846. doi: 10.1128/aem.02515-10
Doherty, G. P., Fogg, M. J., Wilkinson, A. J., and Lewis, P. J. (2010). Small subunits of RNA polymerase: localization, levels and implications for core enzyme composition. Microbiology 156, 3532–3543. doi: 10.1099/mic.0.041566-0
Dubnau, D., and Losick, R. (2006). Bistability in bacteria. Mol. Microbiol. 61, 564–572. doi: 10.1111/j.1365-2958.2006.05249.x
Edayathumangalam, R., Wu, R., Garcia, R., Wang, Y., Wang, W., Kreinbring, C. A., et al. (2013). Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT. Proc. Natl. Acad. Sci. U.S.A. 110, 17820–17825. doi: 10.1073/pnas.1315887110
Feucht, A., and Lewis, P. J. (2001). Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 264, 289–297. doi: 10.1016/s0378-1119(01)00338-9
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotech. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Gao, H., Jiang, X., Pogliano, K., and Aronson, A. I. (2002). The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184, 2780–2788. doi: 10.1128/jb.184.10.2780-2788.2002
Hayashi, K., Kensuke, T., Kobayashi, K., Ogasawara, N., and Ogura, M. (2006). Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol. Microbiol. 59, 1714–1729. doi: 10.1111/j.1365-2958.2006.05059.x
Kampf, J., Gerwig, J., Kruse, K., Cleverley, R., Dormeyer, M., Grünberger, A., et al. (2018). Selective pressure for biofilm formation in Bacillus subtilis: differential effect of mutations in the master regulator SinR on bistability. mBio 9:e1464-18.
Kosono, S., Tamura, M., Suzuki, S., Kawamura, Y., Yoshida, A., Nishiyama, M., et al. (2015). Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10:e0131169. doi: 10.1371/journal.pone.0131169
Kotte, O., Volkmer, B. V., Radzikowski, J. L., and Heinemann, M. (2014). Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol. Syst. Biol. 10:736. doi: 10.15252/msb.20135022
Kröger, C., Srikumar, S., Ellwart, J., and Fuchs, T. M. (2011). Bistability in myo-inositol utilization by Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 1427–1435. doi: 10.1128/jb.00043-10
Kunst, F., Msadek, T., and Rapoport, G. (1994). “Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis,” in Regulation of Bacterial Differentiation, eds P. J. Piggot and C. P. Moran Jr. P. Youngman (Washington, DC: ASM Press), 1–20.
Leh, H., Khodr, A., Bouger, M. C., Sclavi, B., Rimsky, S., and Bury-Moné, S. (2017). Bacterial-chromatin structural proteins regulate the bimodal expression of the locus of enterocyte effacement (LEE) pathogenicity island in enteropathogenic Escherichia coli. mBio 8:e0773-17.
Lehnik-Habrink, M., Rempeters, L., Kovács, ÁT., Wrede, C., Baierlein, C., Krebber, H., et al. (2013). DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J. Bacteriol. 195, 534–544. doi: 10.1128/jb.01475-12
Lehnik-Habrink, M., Schaffer, M., Mäder, U., Diethmaier, C., Herzberg, C., and Stülke, J. (2011). RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol. Microbiol. 81, 1459–1473. doi: 10.1111/j.1365-2958.2011.07777.x
Maamar, H., and Dubnau, D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56, 615–624. doi: 10.1111/j.1365-2958.2005.04592.x
Molle, V., Nakaura, Y., Shivers, R. P., Yamaguchi, H., Losick, R., Fujita, Y., et al. (2003). Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185, 1911–1922. doi: 10.1128/jb.185.6.1911-1922.2003
Nikel, P. I., Romeo-Campero, F. J., Zeidman, J. A., Goni-Monero, A., and de Lorenzo, V. (2015). The glycerol-dependent metabolic persistence of Pseudomonas putida KT2440 reflects the regulatory logic of the GlpR repressor. mBio 6:e00340-15.
Norman, T. M., Lord, N. D., Paulsson, J., and Rosick, R. (2015). Stochastic switching of cell fate in microbes. Annu. Rev. Microbiol. 69, 381–403. doi: 10.1146/annurev-micro-091213-112852
Ogura, M. (2016). Post-transcriptionally generated cell heterogeneity regulates biofilm formation in Bacillus subtilis. Genes Cells 21, 335–349. doi: 10.1111/gtc.12343
Ogura, M., and Asai, K. (2016). Glucose induces ECF sigma factor genes, sigX and sigM, independent of cognate anti-sigma factors through acetylation of CshA in Bacillus subtilis. Front. Microbiol. 7:918. doi: 10.3389/fmicb.2016.01918
Ogura, M., and Kanesaki, Y. (2018). Newly identified nucleoide-associated-like protein YlxR regulates metabolic gene expression in Bacillus subtilis. mSphere 3:e0501-18.
Ogura, M., Ohshiro, Y., Hirao, S., and Tanaka, T. (1997). A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179, 6244–6253. doi: 10.1128/jb.179.20.6244-6253.1997
Ogura, M., Sato, T., and Abe, K. (2019). YlxR, which is involved in glucose-responsive metabolic changes, regulates expression of for protein quality control of pyruvate dehydrogenase. Front. Microbiol. 10:923. doi: 10.3389/fmicb.2016.00923
Ogura, M., and Tanaka, T. (1996). Transcription of Bacillus subtilis degR is D-dependent and suppressed by multicopy proB through D. J. Bacteriol. 178, 216–222. doi: 10.1128/jb.178.1.216-222.1996
Osipiuk, J., Górnicki, P., Maj, L., Dementieva, I., Laskowski, R., and Joachimiak, A. (2001). Streptococcus pneumonia YlxR at 1.35 A shows a putative new fold. Acta Crystallogr. D Biol. Crystallogr. 57, 1747–1751. doi: 10.1107/s0907444901014019
Parks, D. R., Roederer, M., and Moore, W. A. (2006). A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytomet. A 69, 541–551. doi: 10.1002/cyto.a.20258
Schaeffer, P., Millet, J., and Aubert, J. (1965). Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 54, 704–711.
Solopova, A., van Gestel, J., Weissing, F. J., Bachmann, H., Teusink, B., Kok, J., et al. (2014). Bet-hedging during bacterial diauxic shift. Proc. Natl. Acad. Sci. U.S.A. 111, 7427–7432. doi: 10.1073/pnas.1320063111
Thiaville, P. C., El Yacoubi, B., Köhrer, C., Thiaville, J. J., Deutsch, C., Iwata-Reuyl, D., et al. (2015). Essentiality of threonylcarbamoyladenosine (t(6)A), a universal tRNA modification, in bacteria. Mol. Microbiol. 98, 1199–1221. doi: 10.1111/mmi.13209
Thiaville, P. C., Legendre, R., Rojas-Benítez, D., Baudin-Baillieu, A., Hatin, I., Chalancon, G., et al. (2016). Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 3, 29–45. doi: 10.15698/mic2016.01.473
Tojo, S., Satomura, T., Morisaki, K., Yoshida, K., Hirooka, K., and Fujita, Y. (2004). Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA. J. Bacteriol. 186, 7971–7979. doi: 10.1128/jb.186.23.7971-7979.2004
Tsukahara, K., and Ogura, M. (2008). Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 8:8. doi: 10.1186/1471-2180-8-8
Veening, J. W., Smits, W. K., and Kuipers, O. P. (2008). Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210. doi: 10.1146/annurev.micro.62.081307.163002
Weiss, C. A., Hoberg, J. A., Liu, K., Tu, B. P., and Winkler, W. C. (2019). Single-cell microscopy reveals that levels of cyclic di-GMP vary among Bacillus subtilis subpopulations. J. Bacteriol. 201:e00247-19.
Wiame, E., Duquenne, A., Delpierre, G., and Van Schaftingen, E. (2004). Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis. FEBS Lett. 577, 469–472. doi: 10.1016/j.febslet.2004.10.049
Wiethaus, J., Schubert, B., Pfänder, Y., Narberhaus, F., and Masepohl, B. (2008). The GntR-like regulator TauR activates expression of taurine utilization genes in Rhodobacter capsulatus. J. Bacteriol. 190, 487–493. doi: 10.1128/jb.01510-07
Keywords: amino sugar utilization, bimodal expression, autoregulation of ylxR, Bacillus subtilis, bet-hedging strategy
Citation: Ogura M, Shindo K and Kanesaki Y (2020) Bacillus subtilis Nucleoid-Associated Protein YlxR Is Involved in Bimodal Expression of the Fructoselysine Utilization Operon (frlBONMD-yurJ) Promoter. Front. Microbiol. 11:2024. doi: 10.3389/fmicb.2020.02024
Received: 07 July 2020; Accepted: 30 July 2020;
Published: 21 August 2020.
Edited by:
Peter Graumann, Philipps University of Marburg, GermanyReviewed by:
Kazutake Hirooka, Fukuyama University, JapanFabian M. Commichau, Brandenburg University of Technology Cottbus-Senftenberg, Germany
Copyright © 2020 Ogura, Shindo and Kanesaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mitsuo Ogura, b2d1cmFtQHNjYy51LXRva2FpLmFjLmpw
 Mitsuo Ogura
Mitsuo Ogura Kazutoshi Shindo2
Kazutoshi Shindo2 Yu Kanesaki
Yu Kanesaki