- 1State Key Laboratory for Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Disease, National Institute for Communicable Disease Control and Prevention, China CDC, Beijing, China
- 2Wuhan Centers for Disease Prevention and Control, Wuhan, China
- 3State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 4College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 5Department of Laboratory Medicine, Beijing Hospital, National Center of Gerontology, Beijing, China
- 6School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 7Department of Medical, Microbiology, School of Medicine, Cardiff University, Cardiff, United Kingdom
Escherichia coli sequence type 131 (ST131) is well known for its multidrug resistance profile. Carbapenems have been considered the treatment of choice for E. coli ST131 infections, and resistance to carbapenems is emerging due to the acquisition of carbapenemase-encoding genes. In this study, 45 carbapenem-resistant E. coli strains were collected in a hospital. The resistance mechanisms, plasmid profiles, and genetic relatedness of these strains were determined. Phylogenetic relationships between these strains were assessed by molecular profiling and aligned with patient clinical details. The genetic context of blaKPC–2 was analyzed to trace the potential dissemination of blaKPC–2. The 45 carbapenem-resistant E. coli ST131 strains were closely related. Initially prevalent only in a single ward, ST131 subsequently spread to other ward, resulting in a respiratory infection outbreak of carbapenem-resistant E. coli ST131. Eight of the 30 patients died within 28 days of the first isolation of E. coli ST131. The blaKPC–2-positive plasmid profiles suggest that the carbapenem resistance was due to the acquisition by E. coli ST131 of transmissible plasmids pE0272_KPC and pE0171_KPC carrying blaKPC–2. Additionally, diverse multidrug resistance elements were transferred and rearranged between these plasmids mediated by IS26. Our research indicates that clinical attention should be paid to the importance of E. coli ST131 in respiratory infections and the spread of blaKPC-carrying E. coli ST131.
Introduction
Escherichia coli sequence type 131 (ST131) is one of the most widespread and successful extra-intestinal pathogenic E. coli (ExPEC) clones that can cause community- and hospital-acquired urinary tract infections (UTIs), abdominal and pelvic infections, and bacteremia with high morbidity and mortality (Nicolas-Chanoine et al., 2014). E. coli ST131 has developed quinolone resistance associated with mutations within the gyrA and parC genes, as well as resistance against extended-spectrum β-lactam antibiotics, mainly due to the acquisition of CTX-M-15 (Price et al., 2013; Petty et al., 2014; Ben Zakour et al., 2016). Thus, E. coli ST131 has raised public health concerns, due not only to its high pathogenicity potential, but also to its increasing antimicrobial resistance.
Currently, carbapenems have been considered the treatment of choice for E. coli ST131 infections. Although carbapenem resistance is rarely reported (Xu et al., 2015; Zhang et al., 2018), the rate at which bacteria are becoming resistant to carbapenems is slowly growing, due mainly to the acquisition of New Delhi metallo-β-lactamase (NDM) or oxacillinase (OXA)-48-like β-lactamases and Klebsiella pneumoniae carbapenemase (KPC) (Perry et al., 2011; Li et al., 2014; Peirano et al., 2014; Chen et al., 2016; Zhang et al., 2018). KPC is the most prevalent carbapenemase in K. pneumoniae, and blaKPC is typically found within the transposable elements of transferable plasmids in K. pneumoniae. Tn4401 is a Tn3-based transposon that carries the blaKPC gene (Naas et al., 2008). Tn1722 has also been found carrying blaKPC in K. pneumonia strains from China, which partially harbors the structure of Tn4401 (Shen et al., 2009). Thus far, there are only sporadic reports on the detection of blaKPC in E. coli ST131, and these few reports are unrelated to outbreaks (Peirano et al., 2014; Baraniak et al., 2015; Piazza et al., 2016).
Extraintestinal infections of E. coli occur mainly in the urinary tract. To date, there have been few reports of E. coli ST131 causing respiratory infections. A recent study reported the emergence of CTX-M-15-producing E. coli ST131, which caused hospital-acquired pneumonia in regions of Asia (Cha et al., 2017). The virulence factors of ExPEC are broad and associated with site-specific infections. Invasion of brain endothelium proteins, outer membrane protein A, the bacterial capsule fimH, and cytotoxic necrotizing factor 1 (cnf1) were associated with invasion of the blood-brain barrier (Croxen and Finlay, 2010). Salmochelin receptor iroN, adhesin factors sfa/foc and papGIII, and toxin factors hlyC and cnf1 were found to be significantly over-represented in pneumonia E. coli strains, compared with infectious strains found in other sites (La Combe et al., 2019).
Here, we report on a molecular epidemiological study of a nosocomial outbreak of respiratory infections caused predominantly by carbapenem-resistant E. coli ST131 in a Chinese hospital during 2012 and 2013. We demonstrate that the underlying resistance was due to the acquisition by E. coli ST131 of multiple transmissible plasmids carrying blaKPC–2.
Materials and Methods
Ethics Statement
The human specimens were acquired with consent from the patients. This study was reviewed and approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, China CDC, according to the medical research regulations of the Ministry of Health, China. This research was conducted within China.
Strain Collection
On July 1, 2012, one strain of E. coli was isolated from a sputum sample from a pneumonia patient presenting with expiratory dyspnea in the geriatrics department at a hospital in Beijing. This strain showed resistance to carbapenems. Since then, carbapenem-resistant E. coli strains were frequently isolated in four wards of the geriatric and neurology departments until December 28, 2013. A total of 45 strains were included in the study. Some of the E. coli strains were collected from the same patient on different days. The bacterial strains were cultured on Luria-Bertani (LB) (Oxoid, Wesel, Germany) plates at 37°C for 24 h.
MIC Determination and Carbapenem-Resistance Gene Screening
To determine the susceptibility of 45 strains to antibiotics, minimum inhibitory concentrations (MICs) were determined by broth microdilution, according to the protocol of the Clinical and Laboratory Standards Institute1.
To unveil the carbapenem-resistant mechanism, carbapenem resistance genes (blaOXA–48, blaKPC, blaVIM, blaIMP, and blaNDM) were screened, and the primers used for the PCR reactions are listed in Supplementary Table S1 (Poirel et al., 2011). Genomic DNA used as PCR template DNA was extracted from culture grown overnight on LB broth using the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China). Amplification was carried out with the following thermal cycling regimen: 3 min at 95°C, followed by 35 cycles of amplification consisting of 15 s at 95°C, 15 s at 52°C, and 60 s at 72°C, with 5 min at 72°C for the final extension. Amplicons were visualized after electrophoresis at 110 V for 30 min on a 0.7% agarose gel. All positive PCR products were sequenced using an ABI3730 sequencer (Applied Biosystems, Foster City, CA, United States), and their sequences were analyzed using the National Center for Biotechnology Information (NCBI) BLAST program.
XbaI-PFGE, S1-PFGE, and Southern Blot Hybridization
For strain typing, whole-cell E. coli DNA embedded in agarose gel plugs was digested with XbaI (Takara Bio Inc.) and was separated by Pulse-field gel electrophoresis (PFGE) using a CHEF-DR III apparatus (Bio-Rad, Hercules, CA, United States), as previously described (Beutin et al., 2005). Plasmids were analyzed and sized using the S1-PFGE nuclease method. Large fragments from the restriction enzyme digest were separated by PFGE using a CHEF-DR III apparatus (Bio-Rad) for 7 h at 6 V/cm and 14°C, with initial and final pulse times of 5 and 20 s, respectively. Southern hybridization was performed with the blaKPC–2 gene as a probe to ascertain the blaKPC–2-positive plasmid, using the electrochemiluminescence direct nucleic acid labeling and detection system per the manufacture’s operation protocol (GE Healthcare, Buckinghamshire, United Kingdom).
Genome Sequencing and Annotation
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, United States) for genome sequencing. DNA paired-end sequencing was performed on an Illumina HiSeq 2,000 (Illumina, San Diego, CA, United States) with a read length of 2 × 100 bp to obtain the draft genomes of 45 strains. The de novo genome was assembled from the Illumina data using Velvet ver. 1.2.10 (Zerbino and Birney, 2008). A Pacific Biosciences RSII DNA sequencing system (PacBio, Menlo Park, CA, United States) was used as the sequencing platform to sequence complete genomes of strains E0171, E0272, and E02162, which are representative of the main types of blaKPC–2-positive plasmids in the 45 strains, according to S1-PFGE southern blot profiles. De novo assembly of the Pacific Biosciences long reads was performed using the Hierarchical Genome Assembly Process (HGAP_Assembly.2) algorithm in the single molecule real-time (SMRT) Portal, with default parameters.
The assembled genome and plasmids were annotated using Prokka 1.12.21 (Seemann, 2014). The sequence type of the strains was predicted using BLAST with the multi-locus sequence typing (MLST) database2. The plasmid incompatibility groups (Inc.) were assigned using the PlasmidFinder Web tool (Carattoli et al., 2014). Antibiotic resistance genes were annotated using Resistance Gene Identifier (RGI) ver. 3.2.0 (Jia et al., 2017). The ISfinder database and Integron Database were used to annotate mobile elements (Siguier et al., 2006; Moura et al., 2009). The schematic map of linear comparison and circular diagram of sequenced plasmids were drawn using in-house Perl scripts and Inkscape version 0.923. The sequenced plasmids was compared using BLAST with nucleotide identity >90% and BLAST Ring Image Generator (BRIG) ver 0.95 (Alikhan et al., 2011). Sequence reads were mapped onto the backbone of plasmid pE0272_KPC, pE0171_KPC, and pE02162_KPC using Bowtie2 to determine the presence of plasmids in 45 strains (Langmead and Salzberg, 2012), and the mapped region on the plasmid was visualized using Artemis (Carver et al., 2012). The genes associated with virulence were determined by BLAST with a cutoff of nucleotide identity ≥95% and coverage ≥95% against the 38 ExPEC virulence genes (Dale and Woodford, 2015) (Supplementary Table S2). Fisher’s exact test was used to compare the distribution of virulence genes in strains isolated from sputum and urine samples. We considered p < 0.05 to be significant.
Phylogenetic Analysis
The kSNP alignment-free method was used to generate an independent phylogenetic tree based on single-nucleotide polymorphism (SNP) to interpret the phylogenetic relationships of the ST131 strains (Gardner et al., 2015). The draft genomes of 45 strains, together with 197 strains of E. coli ST131 downloaded from the NCBI database (Supplementary Table S3. The genomes from Sequence Read Archive database were assembled using Velvet ver. 1.2.10), were used as the input to kSNP version 3.0 with default parameters and a k-mer value of 19, as predicted by the kSNP-associated Kchooser script, to generate an independent phylogenetic tree based on SNPs (Gardner et al., 2015). The fimH, parC, and gyrA allelic profiles were obtained by BlastX according to the classification standard of Johnson et al. (2013).
A whole-genome, SNP-based phylogenetic analysis was performed to infer the phylogenetic relationships of the 45 strains. Strain FC10268 was selected as a reference genome; this strain was tightly clustered with the 45 strains in a phylogenetic tree of 242 ST131 E. coli strains. Bowtie2 was used to map the Illumina reads of the 45 strains to the genome of strain FC10268 and to generate BAM files for SNP calling using FreeBayes with a min-alternate-fraction of 0.2 (Garrison and Marth, 2012; Langmead and Salzberg, 2012). High-quality SNPs were filtered according to the following criteria: (1) quality scores >20; (2) >30 reads covering the SNP site; and (3) >70% of the reads support the SNP. SNPs from a manually curated plasmid contig using nucleotide BLAST were considered unreliable and were excluded from further analyses. MEGA6 software was used to infer the phylogenetic tree of 136 high-quality SNPs using the maximum parsimony (MP) method (Tamura et al., 2013). The MP tree was obtained using the Tree-Bisection-Regrafting algorithm with search level 1 (Nei and Kumar, 2000), in which the initial trees were obtained by random addition of sequences (ten replicates). The tree was drawn to scale, with branch lengths calculated using the average pathway method (Nei and Kumar, 2000), in which the dimensional units were the number of changes over the entire sequence. All trees were viewed using Interactive Tree of Life (iTOL) (Letunic and Bork, 2016).
Plasmid Conjugal Transfer Experiment
To verify the mobility of the blaKPC–2-carrying plasmid, plasmid conjugal transfer experiments were carried out using rifampicin-resistant E. coli EC600 as the recipient, and strains E0272 and E0171 as the donors. Overnight cultures of the donor strain (600 μL) and recipient strain (400 μL) were mixed and collected by centrifugation (2,500 rcf, 10 min). The mixture was incubated on LB agar plates for 24 h at 37°C. LB agar plates containing rifampicin (100 μg/mL) plus meropenem (2 μg/mL) were used for the selection of carbapenemase-positive E. coli transconjugants. To determine the presence of the plasmid in transconjugants, primers were designed based on the three backbone genes of each plasmid (Supplementary Table S4) and amplified by PCR as described above. The PCR products were detected using agarose gel electrophoresis.
Data Availability
The Illumina sequence reads from all strains were deposited in the Sequence Read Archive database under accession numbers PRJNA514844. The complete nucleotide sequences of pE0171_KPC, pE0272_2, pE0272_KPC, and pE02162_KPC were submitted to GenBank under accession numbers MK370988, MK370989, MK370990, and MK370991, respectively.
Results
Nosocomial Outbreak of Carbapenem-Resistant E. coli Infections in a Geriatric Department
From July 2012 to December 2013, a total of 45 carbapenem-resistant E. coli strains were collected from 30 patients. The 45 strains were all isolated from infections, 34 of which (75.6%) were from sputum or nasopharyngeal samples, eight (17.8%) were from urine samples, two (4.4%) were isolated from blood cultures, and one (2.2%) isolate was from an ascitic fluid sample of a patient presenting with sepsis (Figure 1 and Supplementary Table S5). Of these 30 patients, 27 had been administered biapenem at 0.6 g per day for 3–30 days. Eight patients died within 28 days of the first isolation of carbapenem-resistant E. coli (Supplementary Table S5). All 45 strains exhibited medium to high MICs of carbapenem (Supplementary Table S5). All (100%) were resistant to ertapenem, 35 (77.8%) were resistant to imipenem, and 25 (51.1%) were resistant to meropenem (Supplementary Table S5). Furthermore, these strains exhibited resistance to most other classes of antibiotics, such as non-carbapenem β-lactams, quinolones, aminoglycosides, and tetracyclines, but were sensitive to chloramphenicol (Supplementary Table S5).
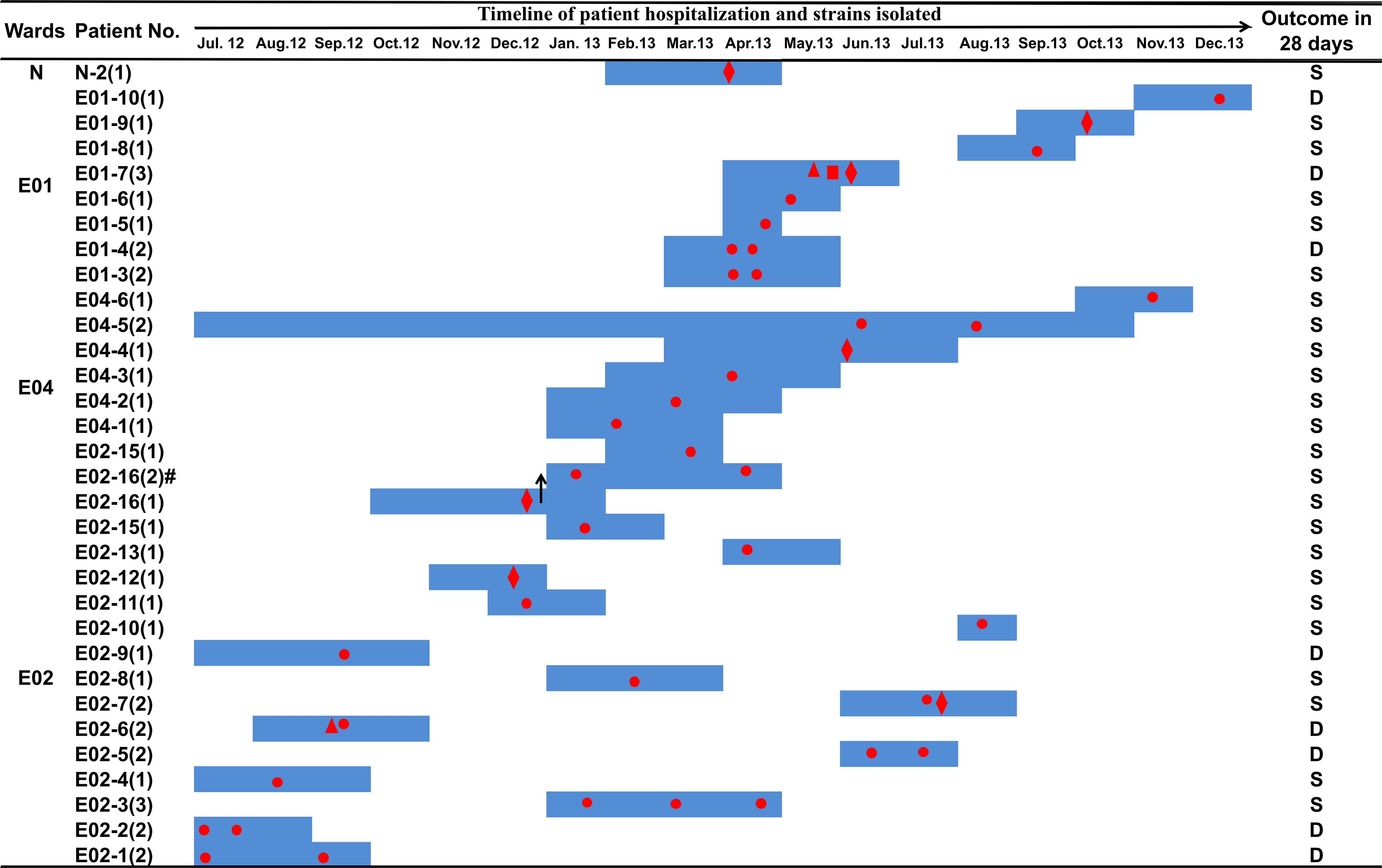
Figure 1. Clinical features of patients from whom ST131 E. coli carrying blaKPC–2 was isolated. Dark blue boxes indicate the period of hospitalization. Red dots indicate the time of blaKPC–2-positive E. coli isolation. The shapes of the red dots indicate the type of samples: circle, sputum or nasopharyngeal swab sample; diamond, urine; triangle, blood; square, drainage. The black arrows indicate a change of ward: patient number E02-16 was transferred from ward E02 to ward E04 on December 27, 2012. Abbreviations: D, death; S, survival.
All E. coli strains carried the blaKPC–2 gene, while other carbapenem resistance genes were not detected (Supplementary Table S5), and all strains shared highly similar XbaI-PFGE patterns (Supplementary Figure S1A), suggesting that they may belong to the same clone. S1-PFGE southern blot hybridization demonstrated that blaKPC–2 was located on plasmids of various sizes, ranging from 130 to 270 kb (Supplementary Figures S1B,C). Most strains carried a single blaKPC–2-positive plasmid, although strain E02-11 contained two blaKPC–2-positive plasmids. According to the sizes of the blaKPC–2-positive plasmids, there were two main plasmid types in 45 strains: a 147-kb plasmid, which was the predominant blaKPC–2-positive plasmid (56%, 25/45), and a 130-kb plasmid (13%, 6/45). Strain E02162 carried the largest blaKPC–2-positive plasmid.
Genomic Characterization of the Outbreak Strains
We sequenced the 45 strains on an Illumina HiSeq 2000; details of the Illumina reads are provided in Supplementary Table S6. In silico MLST analysis showed that all 45 strains belonged to group ST131. We generated a phylogenetic tree based on SNPs using a draft genome of the 45 strains along with 197 E. coli ST131 strains from the NCBI database. The 242 total ST131 strains were divided into clades A–C (Supplementary Figure S2). The 45 strains collected from the patients clustered to the C clade, characterized by a fimH30 allele and the fluoroquinolone resistance alleles gyrA1AB and parC1aAB (Ben Zakour et al., 2016). We confirmed that our strains also harbored the fimH30 allele and gyrA1AB, and parC1aAb. Clade C was further divided into two subclades: C1, characterized by different blaCTX–M alleles, and C2, characterized by the blaCTX–M–15 gene (Ben Zakour et al., 2016). The 45 strains were clustered in the C1 subclade, and four strains possessed blaCTX–M–3 (Supplementary Figure S2). Furthermore, our strains were tightly clustered with the human clinical strain FC10268 from Beijing, which we used as a reference genome (Supplementary Figure S2).
We further analyzed the phylogenetic relationships of the 45 carbapenem-resistant E. coli ST131 strains by comparison with the FC10268 reference genome. The strains were clustered into two clusters, separated by six SNPs (Figure 2). The six SNPs were all found in coding regions, including one synonymous and five non-synonymous SNPs (Supplementary Table S7). Cluster I contained eight strains isolated from ward E02, two strains from ward E01, and one from ward E04. All of the strains from ward E02 in Cluster I were isolated before September 15, 2012, indicating that these strains spread within ward E02 only during the early period and were subsequently transmitted to ward E04 and E01.
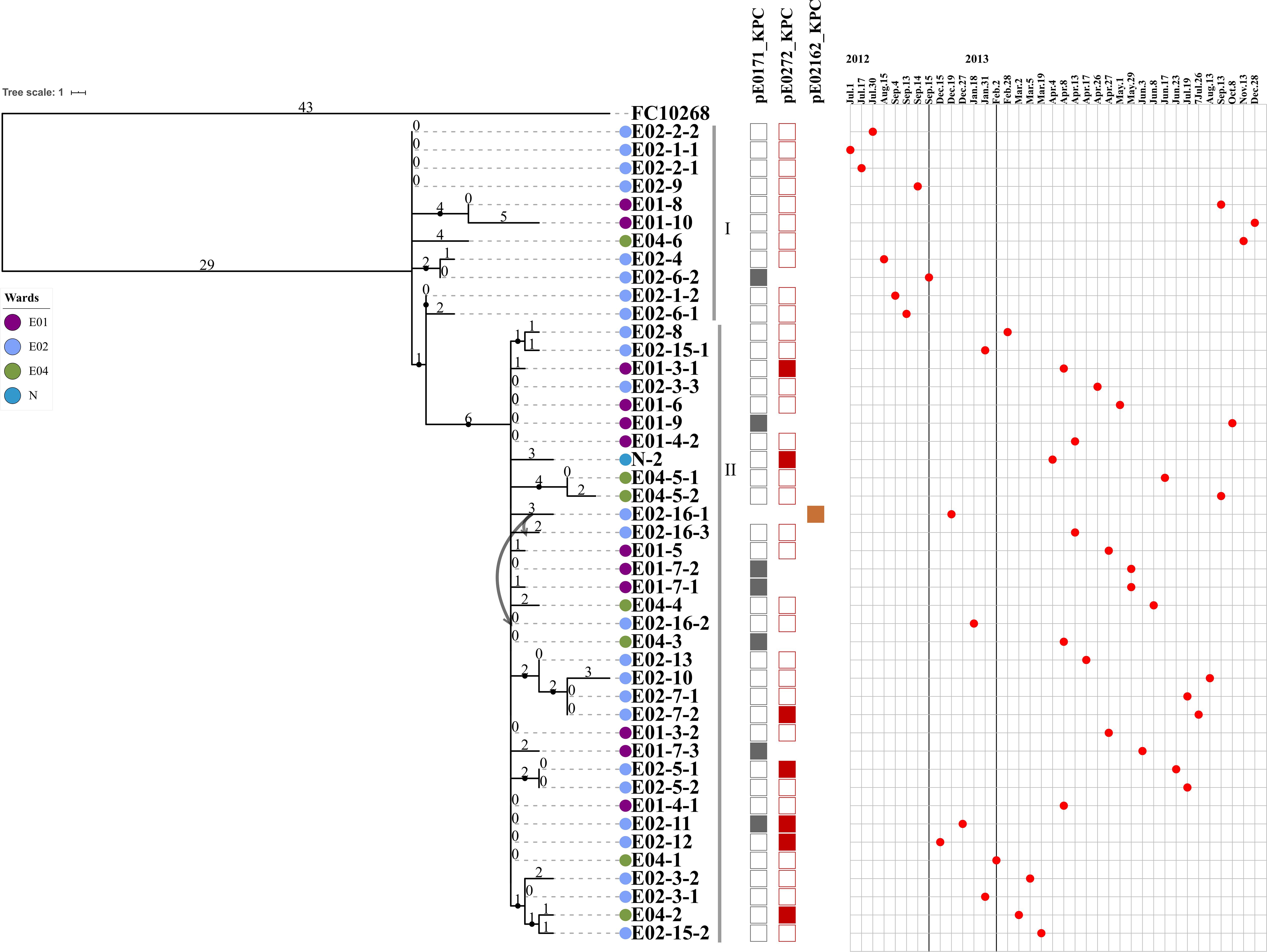
Figure 2. The maximum parsimony phylogenetic tree based on SNP and information about 45 ST131 E. coli strains and the reference genome FC10268. Tree number four out of the ten most parsimonious trees (length = 136) is shown. The consistency index, retention index, and composite index were each 1.000000 for all sites and parsimony-informative sites. Only bootstrap (1,000 replicates) values greater than 60% are shown. The strain IDs are labeled using three strings of characters, separated by a hyphen. The first string of characters represents the ward of the patient from whom the strain was isolated, E01, E02, E04, and N1. The second string of characters represents the patient number. The third string of characters represents the strain isolated from that patient. Lavender indicates the E01 ward, light blue indicates the E02 ward, green indicates the E04 ward, and dark blue indicates the N1 ward. The curved lines indicate that patients E02-16 were transferred from the E02 ward to the E04 ward. The SNP number is marked on the middle of branch. The strains of Cluster I and Cluster II are marked with gray vertical lines. Strains carrying the pE0171_KPC plasmid backbone, the pE0272_KPC plasmid backbone, or the pE02162_KPC plasmid backbone are indicated by gray, red, or brown squares, respectively. The blaKPC–2-positive plasmid in the strain is displayed above the column of corresponding color-filled squares. The strains are arranged in the chronological order in which the samples from which they were isolated were collected.
The first strain of Cluster II, E02-12, was isolated on December 15, 2012, after which the Cluster II strains became dominant in ward E02, suggesting that the Cluster II strains acquired genetic variation that adapted them to external stresses. Historically, patient E02-16, from whom the blaKPC–2-positive strain E02-16-1 was isolated, was transferred from ward E02 to E04 on December 27, 2012. A second blaKPC–2-positive strain E02-16-2 was isolated from patient E02-16 on January 18, 2013, while the patient was resident in ward E04. Then, more blaKPC–2-positive strains were isolated from patients in ward E04. We speculate that the transfer of patient E02-16 led to the spread of Cluster II strains in ward E04. Subsequently, the Cluster II strains were found in four wards, suggesting concomitant transmission across different ward.
The genes associated with virulence were determined by BLAST against the 38 ExPEC virulence genes and nine virulence factors detected in the 45 strains (Supplementary Table S2). The nine genes encode the adhesin siderophore receptor, the E. coli common pilus, the type 1 fimbriae, the periplasmic iron binding protein, the yersiniabactin receptor, an outer membrane receptor, β-glucoronidase, a pathogenicity island marker, and D-serine deaminase (Supplementary Tables S2, S8). The distribution of these nine genes in strains from sputum and urine samples did not show significant differences (p > 0.05; Supplementary Table S8).
Genetic Context of Carbapenem Resistance by Plasmid Sequencing
We selected three strains E0171, E0272, and E02162, which were representative for the main types of blaKPC–2-positive plasmids in the 45 strains, according to the S1-PFGE southern blot profiles, to be sequenced using the Pacific Biosciences RSII DNA sequencing system. Complete genomes were obtained. Strain E02-7-2 contained two plasmids (pE0272_KPC and pE0272_2), strain E01-7-1 contained one plasmid (pE0171_KPC), and strain E02-16-2 contained one plasmid (pE02162_KPC).
Plasmid pE0272_KPC, carrying blaKPC–2, is a 146,156-bp plasmid containing 191 predicted open reading frames (ORFs) and was assigned to IncQ1 (Supplementary Figure S3). It contains a 104-Kb backbone that shares 90% identity with 68 plasmid sequences in the NCBI database with a query coverage of greater than 50% (Supplementary Table S9). Of these 68 plasmids, 59 were from K. pneumoniae, three from E. coli, and seven from other species (Supplementary Table S9). Hence, the pE0272_KPC backbone is mainly distributed in and possibly derived from K. pneumoniae. The multidrug resistance (MDR) region of plasmid pE0272_KPC was composed of mobile elements flanked by two IS26 sequences and contained a diversity of antibiotic resistance genes. The blaKPC–2 was located on an IS26-based composite transposon in pE0272_KPC, which was generated from a Tn1722-based unit transposon flanked by the insertion of two directly repeated IS26 elements, resulting in truncation of resolvase of Tn3 and Tn1722.
The second plasmid in strain E02-7-2 is pE0272_2, a 113,709-bp plasmid with a G+C content of 54%, and is assigned to multi-replicons, including IncFIA and IncFIB. The backbone of pE0272_2 shared a 62.5-Kb backbone with 70 plasmids from the NCBI database (Supplementary Table S9). Of these 70 plasmids, 60 were found in E. coli, five in K. pneumoniae, and the rest in other species (Supplementary Table S9). Therefore, pE0272_2 is likely to be of E. coli origin, in contrast to pE0272_KPC. Plasmid pE0272_2 does not contain blaKPC–2.
Plasmid pE02162_KPC in strain E02-16-2 is a 275,546-bp plasmid containing 347 predicted ORFs (Supplementary Figure S4) and is assigned to multi-replicons, including IncFIA, IncFIB, and IncQ1. Plasmid pE02162_KPC is a composite of plasmids pE0272_KPC and pE0272_2 (>98% query coverage, >99% identity) and has two sets of genes involved in replication, plasmid partition, and conjugative transfer. The MDR regions of pE02162_KPC share 99% identity with pE0272_KPC and pE0272_2, both of which are composed of IS26-mediated transposons, although they differ in arrangements and the copy number of mobile elements (Figure 3). Each MDR region of pE02162_KPC is joined to the backbones of pE0272_KPC and pE0272_2 in a head-to-tail fashion (Figure 3). The relationship between the plasmid backbone and the MDR regions indicate that pE0272_KPC and pE0272_2 might be integrated into strain E02-16-2 as pE02162_KPC in the MDR regions, mediated by IS26 (Figure 3 and Supplementary Figure S5). The blaKPC–2 in pE02162_KPC was located on the same IS26-based composite transposon as pE0272_KPC, with three copies.
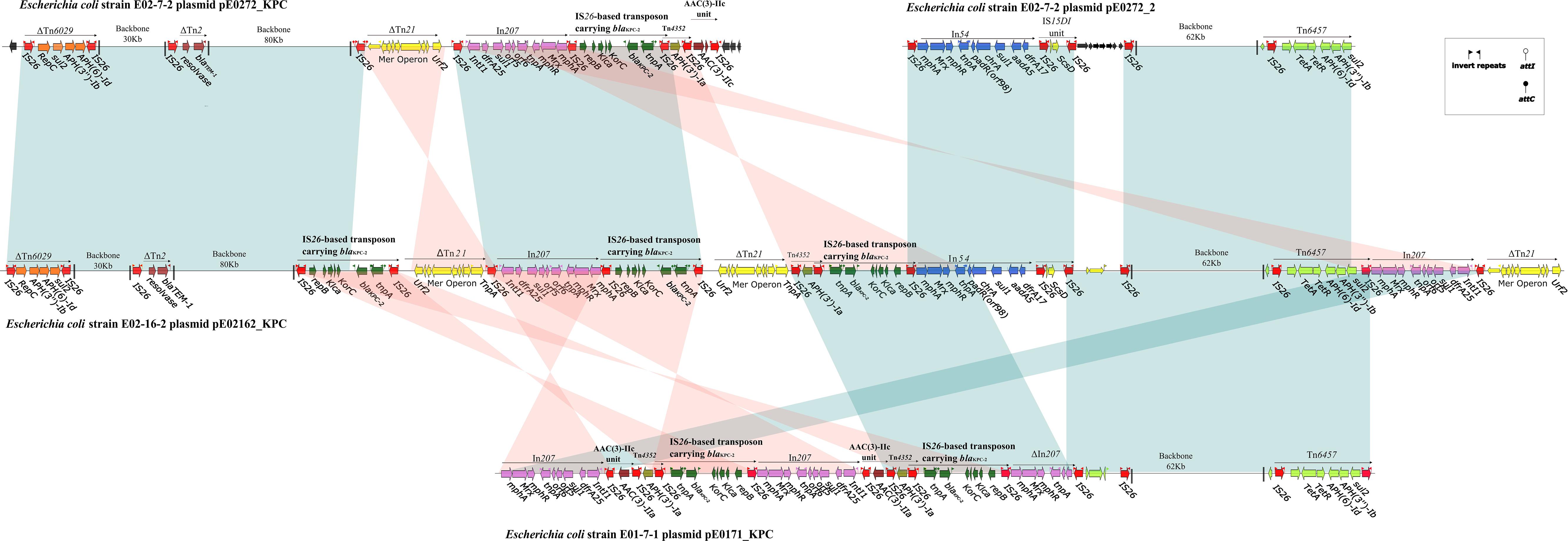
Figure 3. Linear comparison of sequenced plasmids. Genes are denoted by arrows and are colored based on the gene function classification. Shaded regions denote regions of homology (>99% nucleotide similarity). The backbone sequence is denoted by a shortened black line, not to scale.
Strain E01-7-1 contained pE0171_KPC, a 133,709-bp plasmid assigned to multi-replicons, including IncFIA and IncFIB (Supplementary Figure S6). The pE0171_KPC backbone shared 99% sequence identity with pE0272_2 but shared no identity with pE0272_KPC (Figure 3 and Supplementary Figure S5). Therefore, pE0171_KPC is also likely to be of E. coli origin. The MDR region of pE0171_KPC has identical IS26-mediated transposons as pE0272_KPC and pE02162_KPC, indicating that pE0171_KPC acquired antibiotic resistance genes, including blaKPC–2, via mediation by IS26. The MDR region of pE0171_KPC might have originated from the resolution of the plasmid cointegrate or the transmission of IS26-mediated transposons.
Illumina reads mapping combined with hybridization results showed that five strains contained the backbone of pE0171_KPC and 37 strains contained the backbone of pE0272_KPC and pE0171_KPC in the remaining 42 strains (Figure 2). Since most of the strains contained the pE0272_KPC sequence, and since the plasmid is mainly distributed in K. pneumoniae, it is possible that E. coli acquired this plasmid from K. pneumoniae. Seven strains with pE0171_KPC as blaKPC–2-positive plasmid were isolated after September 15, 2012 (Figure 2), suggesting that plasmid pE0272_2 acquired blaKPC–2 from pE0272_KPC, producing plasmid pE0171_KPC, and then disseminated. We further demonstrated that two blaKPC–2-carrying plasmids, pE0272_KPC and pE0171_KPC, were both successfully transferred to E. coli EC600. The transconjugants exhibited significantly reduced carbapenem susceptibility (Supplementary Table S10).
Discussion
In this study, we examined an outbreak of infections caused by a carbapenem-resistant E. coli ST131 clone that persisted for 18 months in a hospital in Beijing. E. coli ST131 caused predominantly (76%) respiratory infections; the remainder of infections (24%) were UTIs or bacteremia. ST131 is a well-known clone causing UTIs but has been less frequently reported in causing respiratory and other infections. Our study is the first report of a nosocomial outbreak of ST131 carrying blaKPC–2. E. coli, especially ST131, has been shown to spread between and within hospitals and communities (Nicolas-Chanoine et al., 2014). Furthermore, ST131 can be transmitted via direct host-to-host contact and even via domestic pets (Johnson et al., 2009; Hilty et al., 2012). Our study demonstrates multiple challenges in controlling MDR ST131 infections in hospitals and communities. In 2013, we communicated our preliminary results to the hospital, which in turn adopted stricter infection control measures. The transmission of ST131 containing blaKPC–2 was controlled, and no E. coli carrying blaKPC–2 was detected in sputum samples during the following year.
The E. coli ST131 subclone H30 (fimH-based putative clonal lineages) is a major UTI pathogen that has been associated with multiple drug-resistance profiles (Tchesnokova et al., 2019). The outbreak we report here was also due to subclone H30, and our strains also presented with an MDR phenotype, highlighting the difficulty of treating respiratory tract infections, clinical failure due to inappropriate antibiotic treatment, and subsequent persistence of the clone within the hospital.
The E. coli ST131 strains described here were initially prevalent only in ward E02. Six SNPs separated the two clusters. Cluster II replaced Cluster I and became dominant in ward E02. Using whole-genome sequencing, we infer that the transfer of the patient carrying E. coli with blaKPC–2 triggered the spread of the ST131 clone, causing an outbreak in multiple wards.
To our knowledge, this is the first time the spread of blaKPC–2 has been fully described by genetically characterizing plasmid sequences from a carbapenem-resistant E. coli outbreak. There were three types of blaKPC–2-carrying plasmids found in E. coli ST131 in this outbreak: K. pneumoniae-derived plasmid pE0272_KPC, E. coli-derived plasmid pE0171_KPC, and a composite plasmid pE02162_KPC. Based on the distribution of pE0272_KPC-like plasmids, pE0272_KPC and, by association, the blaKPC–2 gene were presumably derived from K. pneumoniae. The E. coli plasmid pE0171_KPC backbone sequence has been frequently found in pathogenic E. coli (Ho et al., 2015; Li et al., 2015). The pE0171_KPC plasmid could acquire blaKPC–2 from a K. pneumoniae-derived plasmid containing the same MDR region but different backbone as pE0272_KPC. Furthermore, these plasmids have been shown to be self-transmissible, with an intact transfer region, suggesting that these plasmids maintain the ability to transfer between different strains carrying blaKPC–2.
The third blaKPC–2-containing plasmid pE02162_KPC is likely to be cointegrated by E. coli-derived and K. pneumoniae-derived plasmids during the hospital outbreak, as only one of the strains carried the plasmid. Plasmid cointegrate formation is a type of plasmid evolution involved in the development of antimicrobial resistance, virulence, and other traits (Wang et al., 2013). Plasmid p17-15-vir previously reported was a super hybrid plasmid with multiple replicons and carrying antimicrobial resistance genes, virulence determinants and conjugational systems, resulting in enhanced ability of transfer and risk (Li et al., 2020). IS26 appears to play a vital role in formation and resolution of plasmid cointegrate (He et al., 2015). Our results indicated that the pE02162_KPC plasmid is the product of IS26-mediated cointegration of pE0272_2 and pE0272_KPC. The cointegrate could subsequently resolve, with carbapenem resistance gene assigned to pE0272_2, resulting in pE0171_KPC. An E. coli-derived plasmid would be more suitable for the spread of blaKPC–2 in E. coli.
In conclusion, E. coli ST131 with blaKPC–2 was a multi-drug resistant epidemic clone during the nosocomial infection outbreak that can cause hospital-acquired pneumonia infections. Acquisition of multiple transmissible blaKPC–2-carrying plasmids resulted in a carbapenem-resistant ST131 clone that could rapidly disseminate in hospitals. Our results indicated that close monitoring of blaKPC–2-positive ST131 in hospitals is necessary for the control and proper treatment of ST131 infections.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository, repositories, and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JLi and JF conceived the study and designed experimental procedures. LG, MY, XC, XZ, JC, XB, and YZ performed the experiments. NT, WZ, and RL analyzed the data. DC, HZ, and HX contributed reagents and materials. JF, KS, NT, JLi, RL, TW, JLu, and JX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81861138053, 31870134, 31761133004, and 81861138004) and the Ministry of Science and Technology (2018ZX10733-402).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Richard Bonnet from Department of Bacteriology, University of Auvergne and Hong-Yu Ou from Microbial Bioinformatics Group, State Key Laboratory of Microbial Metabolism, and Shanghai Jiao Tong University for guidance on the analysis of strain evolutionary relationships.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02068/full#supplementary-material
Footnotes
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Baraniak, A., Izdebski, R., Fiett, J., Herda, M., Derde, L. P., Bonten, M. J., et al. (2015). KPC-like carbapenemase-producing Enterobacteriaceae colonizing patients in europe and israel. Antimicrob. Agents Chemother. 60, 1912–1917. doi: 10.1128/AAC.02756-2715
Ben Zakour, N. L., Alsheikh-Hussain, A. S., Ashcroft, M. M., Khanh Nhu, N. T., Roberts, L. W., Stanton-Cook, M., et al. (2016). Sequential Acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. MBio 7, e347–e316. doi: 10.1128/mBio.00347-316
Beutin, L., Kaulfuss, S., Herold, S., Oswald, E., and Schmidt, H. (2005). Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43, 1552–1563. doi: 10.1128/JCM.43.4.1552-1563.2005
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Cha, M. K., Kang, C. I., Kim, S. H., Thamlikitkul, V., So, T. M., Ha, Y. E., et al. (2017). Emergence and dissemination of ST131 Escherichia coli isolates among patients with hospital-acquired pneumonia in Asian countries. Microb. Drug. Resist. 23, 79–82. doi: 10.1089/mdr.2016.0009
Chen, D., Gong, L., Walsh, T. R., Lan, R., Wang, T., Zhang, J., et al. (2016). Infection by and dissemination of NDM-5-producing Escherichia coli in China. J. Antimicrob. Chemother. 71, 563–565. doi: 10.1093/jac/dkv352
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Dale, A. P., and Woodford, N. (2015). Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J. Infect. 71, 615–626. doi: 10.1016/j.jinf.2015.09.009
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: sNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Garrison, E., and Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv [Preprint]. https://arxiv.org/abs/1207.3907.
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio 6:e00762-15. doi: 10.1128/mBio.00762-15
Hilty, M., Betsch, B. Y., Bogli-Stuber, K., Heiniger, N., Stadler, M., Kuffer, M., et al. (2012). Transmission dynamics of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin. Infect Dis. 55, 967–975. doi: 10.1093/cid/cis581
Ho, W. S., Yap, K. P., Yeo, C. C., Rajasekaram, G., and Thong, K. L. (2015). The Complete sequence and comparative analysis of a multidrug-resistance and virulence multireplicon IncFII Plasmid pEC302/04 from an extraintestinal pathogenic Escherichia coli EC302/04 indicate extensive diversity of IncFII plasmids. Front. Microbiol. 6:1547. doi: 10.3389/fmicb.2015.01547
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Johnson, J. R., Miller, S., Johnston, B., Clabots, C., and Debroy, C. (2009). Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and Urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47, 3721–3725. doi: 10.1128/JCM.01581-1589
Johnson, J. R., Tchesnokova, V., Johnston, B., Clabots, C., Roberts, P. L., Billig, M., et al. (2013). Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect Dis. 207, 919–928. doi: 10.1093/infdis/jis933
La Combe, B., Clermont, O., Messika, J., Eveillard, M., Kouatchet, A., Lasocki, S., et al. (2019). Pneumonia-specific Escherichia coli with distinct phylogenetic and virulence profiles. France, 2012-2014. Emerg. Infect Dis. 25, 710–718. doi: 10.3201/eid2504.180944
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, J., Lan, R., Xiong, Y., Ye, C., Yuan, M., Liu, X., et al. (2014). Sequential isolation in a patient of Raoultella planticola and Escherichia coli bearing a novel ISCR1 element carrying blaNDM-1. PLoS One 9:e89893. doi: 10.1371/journal.pone.0089893
Li, J. J., Spychala, C. N., Hu, F., Sheng, J. F., and Doi, Y. (2015). Complete nucleotide sequences of bla(CTX-M)-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob. Agents Chemother. 59, 3002–3007. doi: 10.1128/AAC.04772-4714
Li, R., Cheng, J., Dong, H., Li, L., Liu, W., Zhang, C., et al. (2020). Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 55:105952. doi: 10.1016/j.ijantimicag.2020.105952
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Naas, T., Cuzon, G., Villegas, M. V., Lartigue, M. F., Quinn, J. P., and Nordmann, P. (2008). Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52, 1257–1263. doi: 10.1128/AAC.01451-1457
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford: Oxford University Press.
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-113
Peirano, G., Bradford, P. A., Kazmierczak, K. M., Badal, R. E., Hackel, M., Hoban, D. J., et al. (2014). Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg. Infect. Dis. 20, 1928–1931. doi: 10.3201/eid2011.141388
Perry, J. D., Naqvi, S. H., Mirza, I. A., Alizai, S. A., Hussain, A., Ghirardi, S., et al. (2011). Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66, 2288–2294. doi: 10.1093/jac/dkr299
Petty, N. K., Ben Zakour, N. L., Stanton-Cook, M., Skippington, E., Totsika, M., Forde, B. M., et al. (2014). Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U.S.A. 111, 5694–5699. doi: 10.1073/pnas.1322678111
Piazza, A., Caltagirone, M., Bitar, I., Nucleo, E., Spalla, M., Fogato, E., et al. (2016). Emergence of Escherichia coli sequence type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a long-term care and rehabilitation facility (LTCRF) in Northern Italy. Adv. Exp. Med. Biol. 901, 77–89. doi: 10.1007/5584_2015_5017
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Price, L. B., Johnson, J. R., Aziz, M., Clabots, C., Johnston, B., Tchesnokova, V., et al. (2013). The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 4, e377–e313. doi: 10.1128/mBio.00377-313
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-269
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tchesnokova, V., Riddell, K., Scholes, D., Johnson, J. R., and Sokurenko, E. V. (2019). The uropathogenic Escherichia coli subclone sequence type 131-H30 is responsible for most antibiotic prescription errors at an urgent care clinic. Clin. Infect. Dis. 68, 781–787. doi: 10.1093/cid/ciy523
Wang, P., Zhang, C., Zhu, Y., Deng, Y., Guo, S., Peng, D., et al. (2013). The resolution and regeneration of a cointegrate plasmid reveals a model for plasmid evolution mediated by conjugation and oriT site-specific recombination. Environ. Microbiol. 15, 3305–3318. doi: 10.1111/1462-2920.12177
Xu, Y., Gu, B., Huang, M., Liu, H., Xu, T., Xia, W., et al. (2015). Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. J Thorac Dis 7, 376–385. doi: 10.3978/j.issn.2072-1439.2014.12.33
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Keywords: blaKPC–2, Escherichia coli ST131, outbreak, respiratory infection, transmissible plasmids
Citation: Gong L, Tang N, Chen D, Sun K, Lan R, Zhang W, Zhou H, Yuan M, Chen X, Zhao X, Che J, Bai X, Zhang Y, Xu H, Walsh TR, Lu J, Xu J, Li J and Feng J (2020) A Nosocomial Respiratory Infection Outbreak of Carbapenem-Resistant Escherichia coli ST131 With Multiple Transmissible blaKPC–2 Carrying Plasmids. Front. Microbiol. 11:2068. doi: 10.3389/fmicb.2020.02068
Received: 21 May 2020; Accepted: 06 August 2020;
Published: 11 September 2020.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Katharina Schaufler, University of Greifswald, GermanyRuichao Li, Yangzhou University, China
Copyright © 2020 Gong, Tang, Chen, Sun, Lan, Zhang, Zhou, Yuan, Chen, Zhao, Che, Bai, Zhang, Xu, Walsh, Lu, Xu, Li and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Li, bGlqdWFuQGljZGMuY24=; Jie Feng, ZmVuZ2pAaW0uYWMuY24=
†These authors have contributed equally to this work
 Lin Gong
Lin Gong Na Tang
Na Tang Dongke Chen5†
Dongke Chen5† Ruiting Lan
Ruiting Lan Wen Zhang
Wen Zhang Min Yuan
Min Yuan Jie Che
Jie Che Timothy R. Walsh
Timothy R. Walsh Jianguo Xu
Jianguo Xu Juan Li
Juan Li Jie Feng
Jie Feng