- 1College of Animal Science, South China Agricultural University, Guangzhou, China
- 2Lingnan Guangdong Laboratory of Modern Agriculture, Guangzhou, China
- 3Guangdong Engineering Research Center for Vector Vaccine of Animal Virus, Guangzhou, China
- 4South China Collaborative Innovation Center for Poultry Disease Control and Product Safety, Guangzhou, China
- 5USDA, Agriculture Research Service, Avian Disease and Oncology Laboratory, East Lansing, MI, United States
H9N2 avian influenza virus (AIV) infection in chickens is often accompanied by secondary bacterial infection, but the mechanism is unclear. The aim of the present study was to reveal that mechanism and explore non-antibiotic treatment. 16s rRNA sequencing and metabonomics were performed in the intestinal contents of chickens infected with H9N2 AIV or H9N2 AIV and fed with ageratum-liquid (AL) to reveal the metabolite that promote intestinal Escherichia coli (E. coli) proliferation caused by H9N2 AIV, as well as to determine the regulatory effect of AL. It was found that H9N2 AIV infection led E. coli to become the dominant gut microbe and promoted E. coli translocation from the intestinal tract to the visceral tissue through the damaged intestinal barrier. H9N2 AIV infection induces inflammation in the intestinal mucosa and promotes the secretion and release of nitrate from the host intestinal epithelium. In addition, nitrate promoted E. coli proliferation in the inflamed intestinal tract following H9N2 AIV infection. Furthermore, Chinese herbal medicine AL can restore intestinal homeostasis, inhibit the production of nitrate in the intestinal epithelium and effectively prevent the proliferation and translocation of E. coli in the intestines. This is the first report on the mechanism of E. coli secondary infection induced by H9N2 AIV, where herbal medicine AL was shown to have a good preventive effect on the secondary infection.
Introduction
Avian influenza virus (AIV) belongs to the influenza virus A genus of the Orthomyxoviridae family (Marc, 2016). The genetic material of AIV is composed of single-stranded negative-stranded RNA, which is divided into 8 independent fragments (Resa-Infante et al., 2011). AIV is divided into the H1-16 and N1-9 subtypes, according to the antigenic types of hemagglutinin (HA) and neuraminidase on the envelope (Lee et al., 2016). Due to the different pathogenicity of the different subtypes, AIV can have high and low pathogenicity. Highly pathogenic AIV mainly includes the H5 and H7 subtypes and low pathogenic AIV the H9N2 subtype (Gharaibeh, 2008; Pantin-Jackwood and Swayne, 2009).
H9N2 AIV is widespread in all parts of the world and was first isolated from turkeys in 1966 (Peacock et al., 2019). The biggest harm of H9N2 AIV infection in poultry is the induction of immunosuppression and destruction of their immune system. H9N2 AIV can be isolated from host tissues, including the trachea, lung, brain, spleen, pancreas, cloacal cavity and intestinal tract, and causes inflammation and enteric problems to the hosts (Li et al., 2018). H9N2 AIV is prone to secondary infection of bacteria and other pathogens, the most common of which is Escherichia coli (E. coli) infection, which is associated with high mortality in poultry (Barbour et al., 2009; Samy and Naguib, 2018; Nguyen et al., 2019). The H9N2 AIV subtype poses a significant public health threat, as it can replicate in permissive mammalian tissues without prior adaptation (Samir et al., 2019). Furthermore, several influenza viruses that infect humans all originate from variants of H9N2 AIV (Lam et al., 2013; Chen et al., 2014); hence, the potential harmful effects of H9N2 AIV on aquaculture and human health and safety cannot be ignored.
A large number of studies have shown that viral infections can destroy the balance of intestinal microorganisms and cause inflammation (Kotler et al., 1993; Groves et al., 2018; Kritas et al., 2020). The influenza virus can not only change the respiratory tract flora, but also affect the composition of intestinal microorganisms located far away from the respiratory tract; the reason for that is not clear (Qin et al., 2015). So far, only a few articles have reported that H9N2 AIV infection can affect poultry intestinal microorganisms (Yitbarek et al., 2018a,c). Previously, H9N2 AIV was reported to affect intestinal microbiota, barrier structure injury and inflammatory intestinal disease in the chicken ileum, but the mechanism of H9N2 AIV-induced E. coli proliferation remains unclear (Li et al., 2018). It was recently reported that the intestinal epithelium of the host produces certain active substances under the condition of inflammation to provide growth factors for conditional pathogenic bacteria in the intestinal tract (Haberman et al., 2014). The metabolic substances of the microbiota can affect the immune homeostasis, but also the sensitivity of the host to several immune-mediated diseases, and the regulatory substances of the host epithelium can provide a colonization environment for the microbiota (Rooks and Garrett, 2016). Gut microbiota uses their substrates to generate energy for cellular processes and for growth, the microbiota produces several metabolites that influence human health and metabolism (Ramakrishna, 2013). We therefore speculated that intestinal metabolites also play a key role in inducing the proliferation of E. coli in H9N2 AIV-infected chickens. Most importantly, to date, there has been no report on the relationship between intestinal metabolism and intestinal microbiota disorder caused by H9N2 AIV.
H9N2 infection can induce inflammation in chickens. Antibiotics are often used in the clinic to treat inflammation; however, although antibiotics can kill bacteria, they can also cause damage to the beneficial flora of the intestine, aggravating intestinal injury due to its broad spectrum (Li et al., 2018). A large number of studies have shown that Chinese herbal medicine can have a favorable effect on the regulation of the intestinal tract; an example of such medicine is ageratum-liquid (AL), which has been shown to exert a significant curative effect against gastrointestinal influenza (Lu et al., 2011; Liu et al., 2014). AL has been shown to regulate CD4+ and CD8+ cells in Peyer’s patch and suppress tumor necrosis factor (TNF)-α levels in enteric homogenates to improve the diarrhea caused by Salmonella typhimurium in mice (He et al., 2006; Ko et al., 2011; Chen et al., 2016). Our previous study showed that AL can effectively prevent bacterial translocation following H9N2 infection in mice (Lu et al., 2019). In our previous study, it was also demonstrated that H9N2 AIV infection could affect intestinal microbiota, barrier structure injury, and inflammatory intestinal disease in the chicken ileum (Li et al., 2018). We therefore aimed at exploring whether AL has a good therapeutic effect on intestinal flora disorder and bacterial infection induced by H9N2 AIV in chickens.
The purpose of this study was to explore the relationship between intestinal flora disorder and intestinal metabolism, in order to reveal the cause of secondary E. coli infection induced by H9N2 AIV infection in chickens. In addition, through the identification of the efficacy of AL in the treatment of intestinal flora disorder, inflammation and intestinal bacterial translocation induced by H9N2 AIV infection, the new data for the clinical treatment of secondary bacterial infection caused by H9N2 AIV could be provided.
Materials and Methods
Ethics Approval
The animal study protocol was approved by the South China Agricultural University Committee of Animal Experiments (approval ID, SYXK-2014-0136). The experiments were performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Animals and Viral Infection
One-day-old specific pathogen free (SPF) chickens and SPF chicken embryos were purchased from the SPF Experimental Animal Center of Guangdong Dahuanong Co., Ltd. The H9N2 SH01 strain was isolated and preserved in our laboratory. A total of 32 9-day-old healthy SPF chickens with a uniform body weight were selected and divided into four groups: The mock, H9N2 AIV infection (H9N2), AL feeding (AL), and H9N2 AIV infection with feeding AL (H9N2-AL) groups. H9N2 AIV infection was carried out by nasal drip and the infection dose of the H9N2 virus was 3-fold 106TCID50/0.1 mL, and AL was administered at the same time as the viral infection, at a dose of 2.5 mL/kg/day for 3 days, and then put into a negative pressure isolator for routine feeding. At the same time of viral infection, 0.1 mL phosphate buffer saline (PBS) was dripped into nasal cavity of chickens in mock group and AL group.
Detection of Virus Shedding
The cloacal swab was frozen and thawed repeatedly 3 times, and the virus RNA in the sample was extracted, according to the instructions of the AxyPrep humoral virus DNA/RNA small extraction kit (Axygen, CA, United States). A pair of identification primers were designed according to the HA gene of the H9N2 virus; the upstream sequence was 5′-CAAGATGGAAGTAGTATCACT-3′ and the downstream sequence was 5′-TTGCCAATTATATACAAATGT-3′. The extracted RNA was used as a template for the preparation of a one-step reverse transcription system and detected by RT-qPCR, as previous reported (Su et al., 2019).
Detection of Viral Load by RT-qPCR
The tissue samples (lung and ileum) of the same quality were collected, and the total RNA in the tissue was extracted by TRIzol (Invitrogen, Carlsbad, CA, United States) reagent method, according to the manufacturer’s instructions. The total RNA of the obtained tissue sample was reverse transcribed into cDNA, according to the instructions of the genome removal reverse transcription kit (TiANGEN, Beijing, China). Detection of the viral copy number in each tissue was assessed by quantitative RT-PCR (RT-qPCR). The primers used for constructing standard curves were as follows: 5′-ACTCGATGAGCATGACGCAA-3′ and 5’-GGTCCCGTTCCGAATTGTCT-3’. RT-qPCR quantification of the viral load of each organ was conducted using standard curves, as previously described (Zhang et al., 2015).
16s rRNA High Throughput Sequencing of Intestinal Microbiota and Bioinformatics Analysis
The DNA of the ileum contents was extracted using a genomic DNA extraction kit. The V3 and V4 regions of bacterial 16S rDNA were amplified by RT-qPCR. The primers used were as follows: 5′-CCTACGGRRBGCASCAGKVRVGAAT-3′ and 5′-GGACTACNVGGGTWTCTAATCC-3′; a connector with Index was simultaneously added to the end of the fragment, and then a sequencing library was constructed. After the library was qualified, 2 × 300 bp double-terminal sequencing was carried out according to the instructions of the Illumina MiSeq instrument. The forward and backward reads obtained by sequencing were spliced, the splicing sequences containing N were filtered out and the sequences of > 200 bp were retained. The retained sequences were qualitatively filtered and the chimeric sequences deleted; the sequence operational taxonomic unit (OTU) cluster analysis was then performed using VSEARCH (1.9.6; with the similarity set to 97%). The representative sequence of OTU was then annotated and analyzed by RDP classifier Bayesian algorithm, and the species composition of each sample was calculated. Based on the results of OTU, the subsequent diversity difference analysis was carried out. Mothur (v.1.30) software was used for A index analysis (Schloss et al., 2009). Principal component analysis (PCA), principal coordinates analysis (PCoA), non-metric multidimensional scaling (NMDS) and unweighted pair group method with arithmetic mean (UPGMA) analysis was performed to analyze the diversity among different groups. The dominant bacterial community difference between groups was detected using Linear discriminant analysis effect size (LEfSe). LEfSe analysis was performed to analyze the significant differences between groups.
Detection of Gene Expression in Ileal Tissue by RT-qPCR
The cDNA templates of ileal tissue samples were prepared, and RT-qPCR was used to detect gene expression. The PCR system used 20 μL volume according to the instructions of the kit (Roche Diagnostics, Montreal, QC, Canada), including 10 μL of 2 x SYBR Green qPCR Master Mix, 1 μL forward primer and 1 μL reversed primer, 1 μL RNA and 7 μL ddH2O; the primers of detecting the gene expression in ileal tissue were listed in Table 1. The cycling conditions were as follows: 30 cycles at 95°C for 10 min; 95°C for 10 s, 60°C for 30 s, 95°C for 15 s; 60°C for 1 min and 95°C for 15 s. At the end of the reaction, according to the CT value of the target gene and the internal reference gene, the relative expression of the target gene was calculated by the 2–ΔΔCT method (Zhang et al., 2019).
Red/ET Engineering-Related Strains and Plasmids
The E. coli Nissle1917 (EcN) strain, E. coli NeonGreen strain [GB05, ampicillin (amp) resistance modified by the NeonGreen fluorescence labeling gene], pSC101-BAD-gbaA-amp plasmid (pSC101 vector, amp resistance, red γβαA under PBAD promoter), pSC101-BAD-cre-tet plasmid [pSC101 vector, tetracycline (tet) resistance, Cre under PBAD promoter] and pR6K-lox71-cm-lox66 plasmid were all donated by Professor Zhang Youming (German College of Shandong University, Qingdao, Shandong).
Tissue Bacteria Isolation
The aseptic isolated liver, lung and mesentery were homogenized. Homogenate (100 μL) was mixed on a Luria broth (LB) plate, and the bacterial solution was coated and cultured in a 37°C incubator for 12 h. When the bacterial growth was observed on the plate, the colony growth was judged to be positive, while the non-colony growth was judged to be negative.
NeonGreen-Tagged Bacteria Isolation and Quantification
A total of 36 9-day-old healthy SPF chickens with a uniform body weight were purchased and divided into three groups: The mock, H9N2 AIV infection (H9N2) and H9N2 AIV infection with feeding AL (H9N2-AL) groups. At the age of 9 days, the infection dose of the H9N2 virus was 3-fold 106 TCID50/0.1 mL, and AL was fed to the chickens at the same time as the viral infection at a dose of 2.5 mL/kg/day, which lasted 3 days. On day 3 after viral infection, all chickens were fed NeonGreen bacteria at a dose of 300 mL 1 × 109 CFU/mL per chicken, which was routinely raised in a negative pressure isolator.
Samples were taken at 12, 24, 36 and 48 h after the administration of NeonGreen bacteria. The mixture containing ileum, liver, lung and mesentery inner wall and lumen tissues was aseptically collected and homogenized in a sterile homogenate tube filled with 1 mL saline. Homogenate mixture (100 μL) was applied to the LB&Amp plate, and the bacterial solution was coated in a 37°C incubator for 12 h. When the bacterial growth on the plate was observed, the colony growth was judged to be positive, while the non-colony growth was judged to be negative.
RT-qPCR was performed for the count of NeonGreen E. coli bacteria. The total volume of DNA extracted was derived from enteric contents, mesentery, lung and liver. The DNA was adjusted to the same concentrations. DNA standards were prepared from E. coli strains carrying plasmids with E. coli fragment inserts, which was isolated from a poultry farm (GenBank No. MG602206). The abundance of the gene was evaluated by multiplying the number of copies per well by the total volume of DNA per well. The total reaction volume of 20 μL contained 1.0 μL DNA, 10.0 μL SYBR Green qPCR Mix (Roche Diagnostics, Shanghai, China), 0.5 μL of each primer and 8 μL H2O. Samples were amplified by the following conditions: Initial denaturation at 94°C for 2 min, 40 cycles of heat denaturation at 94°C for 10 s, primer annealing at 60°C for 40 s. Fluorescence signal acquisition was set at 60°C. The primers for E. coli amplification were as follows: 5′-GTTAATACCTTTGCTCATTGA-3′ and 5′-ACCAGGGTATCTTAATCCTGTT-3′.
Metabonomic Analysis of Intestinal Contents
The collected ileum contents were sent to Shanghai Matt painting Biotechnology Co., Ltd., for non-targeted metabonomics. Gas chromatography time-of-flight mass spectrometry based on silanization derivation was used as the analytical chromatographic mass. A metabolite database, JIALIB, was established through the identification of high-purity chemical standards of metabolites. The statistical analysis of the data was performed based on conventional multivariate statistical methods, such as PCA, Partial Least Square Discriminant Analysis (PLS-DA) and Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA). The metabolic pathway enrichment analysis used a specific hypergeometric algorithm to enrich the information of metabolites, weigh their contribution and identify key metabolic networks.
Construction of EcN Amp Resistant Strain (EcN:Amp)
In order to screen and count the strains in the later stage, the resistant high-copy plasmid was transformed into the target strain to carry the corresponding resistance function. In the present study, the pMD-19T (Control Insert) plasmid was electrotransformed to make EcN carry Amp resistance. The specific transfer steps of EcN were conducted as previously described (Bian et al., 2012).
Construction of EcN Mutants (ΔmoaA and ΔAZG)
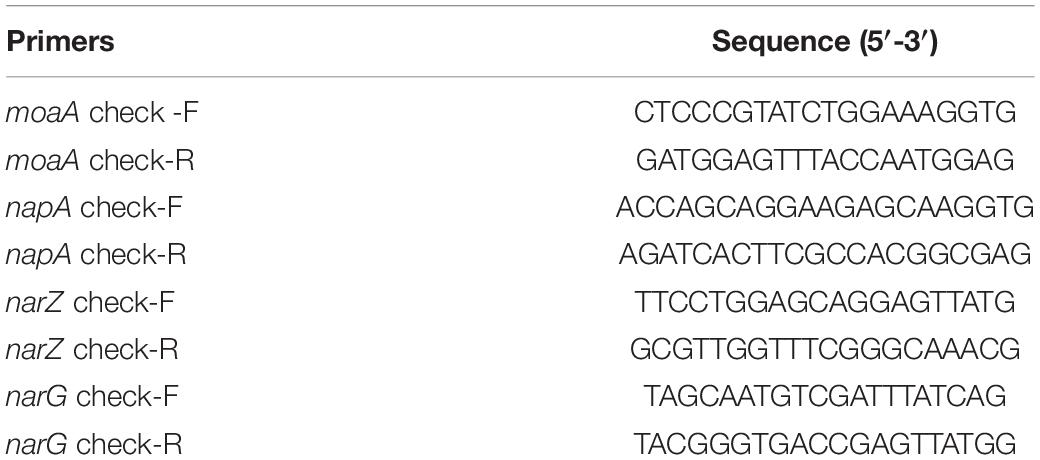
In order to construct nitrate metabolism deletion strains, molybdenum cofactor genes moaA and nitrate reductase genes napA, narZ and narG in the nitrate metabolism were deleted (Winter et al., 2013), the target genes were replaced by lox71-cm-lox66 fragments by RedE/T recombination engineering (Wang et al., 2016), the resistant fragments between lox sites were then deleted by a site-specific recombination system, and the target genes were repeatedly knocked out using this strategy to obtain gene deletion strains. In this experiment, ΔmoaA mutant only replaced the moaA gene with the cm resistance gene, while the ΔAZG mutant in turn knocked out the napA and narZ genes, and finally replaced the narG gene with the chloramphenicol resistance gene. The specific steps of constructing mutants were conducted as previously described (Autieri et al., 2007; Liu et al., 2016). The primer sequences for target gene replacement were designed based on the lox site of the pR6K-lox71-cm-lox66 plasmid sequence and the target knockout gene sequences were shown in Table 2. According to the position of the homologous arm of the linear substrate, identification primers were designed at both ends of the homologous arm of the target knockout gene in EcN to identify the sequence changes before and after substitution and knockout by RT-qPCR. The primer sequences of the identification of recombinants by RT-qPCR were shown in Table 3.
In vitro Competitive Testing of Mutant Strains
EcN:Amp and EcN strain ΔmoaA were simultaneously added to the cell culture flask containing 10 mL LB liquid medium at 1:1, and their final concentration was 1 × 104 CFU/mL. At the same time, KNO3 (100 μM) was added to the culture system in the treatment group, repeated 3 times in each treatment group, sealed in an anaerobic incubator and transferred to a 37°C incubator for culture. After 24 h, the number of EcN:Amp and EcN ΔmoaA strains in each culture bottle was counted. The viable bacterial count of 3 mutant strains (EcN:Amp, EcN ΔmoaA and EcN ΔAZG) was determined, the bacterial liquid was diluted 10 times, the sample homogenate with suitable 3-5 continuous dilutions was selected, and the 1 mL was poured into the plate containing antibiotics ampicillin and chloramphenicol; 2 plates were made for each dilution, and the dilution was compared at the same time. Following incubation at 37°C for 18–24 h, the plate colony count was determined, and the plate record of colony numbers between 15–150 CFU was selected. Finally, the competitive index (CI) of EcN:Amp and EcN strain ΔmoaA with or without nitrate treatment was calculated. The competitive culture of EcN:Amp and EcN ΔAZG was carried out according to the above steps, and the CI of EcN:Amp and EcN strain ΔAZG was calculated with or without nitrate treatment.
Verification of Nitrate Metabolic Function in vivo
A total of 16 9-day-old healthy SPF chickens with uniform body weight were divided into four groups: The mock, H9N2 AIV infection, feeding S-methyl-isothiourea (SMT) after H9N2 AIV infection, and feeding AL after H9N2 AIV infection groups. At the age of 9 days, the infection dose of the H9N2 virus was 3-fold 106 TCID50/0.1 mL, and the mixed bacterial solution of EcN:Amp and EcN ΔmoaA was administered at the same time as the viral infection at a dose of 100 μL/per chicken, 1 × 1010 CFU/mL; SMT and AL were fed to them on day 1 after viral infection. The dose of SMT was 10 mg/per/d and that of AL 2.5 mL/kg, lasting for 3 days, and put into the negative pressure isolator for routine feeding. The above animal experiments were repeated with EcN:Amp and EcN strain ΔAZG.
On day 5 after viral infection, the ileum tissue near the ileocecal junction was cut and sampled. Following rinsing with phosphate-buffered saline (PBS), the intestinal tissue was lysed using cell and tissue lysate reagent, and the nitrate content in the intestinal tissue was detected, according to the instructions of the total nitric oxide detection kit. In addition, the contents at the junction of the blind were collected in the aseptic environment and, following weighing, each sample was diluted with PBS, according to a fixed mass-volume ratio. The sample was then used as the original solution. Next, the number of bacteria was identified and the competition index was calculated.
Results
Determination of Tissue Viral Load in SPF Chickens Following H9N2 AIV Infection and AL Treatment
Cloacal swabs were collected on day 3 after viral infection for the RT-PCR detection of the HA gene of H9N2 AIV. Results showed that the cloacal swabs collected from SPF chickens infected with the virus can be amplified into HA fragments of H9N2 AIV, indicating that SPF chickens were infected successfully in this experiment (Supplementary Figure 1). Lung and ileum tissues were collected on days 5 and 12 after H9N2 AIV infection, and the viral load was determined by RT-qPCR. The results showed that the virus could be detected in the lungs and ileum at both time points, indicating that the influenza virus could proliferate effectively in SPF chickens and intestinal epithelial cells (P < 0.01, Figure 1). The feeding of AL 5 days after viral infection in SPF chickens could significantly reduce the viral load in the lung and ileum epithelium (P < 0.01 and P < 0.05, Figure 1), indicating that AL might exert its antiviral effect by affecting intestinal function.
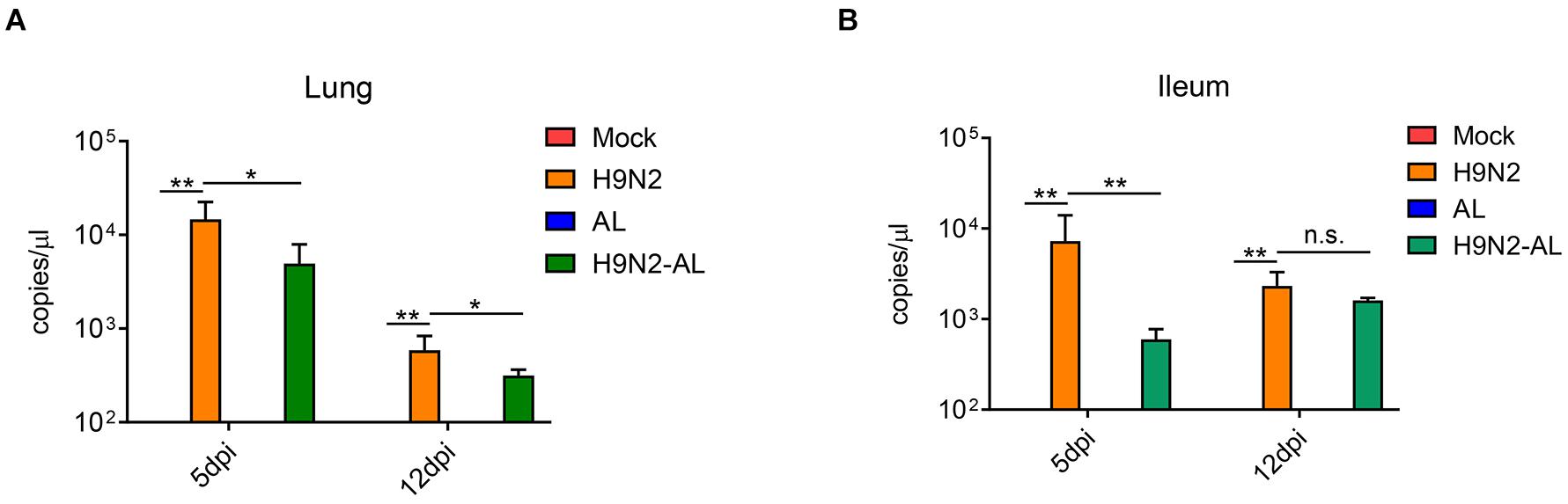
Figure 1. Determination of viral load in lung and ileum tissue 5 and 12 days after H9N2 AIV infection in SPF chickens by RT-qPCR. (A) Viral load in the lung 5 and 12 days after H9N2 AIV infection in chickens. (B) Viral load in the ileum 5 and 12 days after H9N2 AIV infection in chickens. Data are presented as the mean ± standard deviation (n = 3). The differences between groups were analyzed using ANOVA. *P < 0.05, **P < 0.01 and n.s. represents not significant; dpi, day post infection; AL, ageratum-liquid.
α Diversity Index Statistics and β Diversity Analysis of Gut Microbiota Following H9N2 AIV Infection and AL Treatment in SPF Chickens
α diversity reflects the species richness and diversity of a single sample (Mancini et al., 2018). The Chao and Ace index indicate species richness (Xiao Joe et al., 2019). As shown in Supplementary Figures 2A,B, there was no significant change in community species richness on the fifth day after H9N2 AIV infection (P > 0.05), indicating that H9N2 AIV infection did not cause the change of intestinal community species richness. The species richness of the AL group increased significantly compared with the mock group (P < 0.05), and that of the H9N2-AL group increased significantly compared to the H9N2 AIV infection group, indicating that AL could increase the intestinal community species richness (Supplementary Figures 2A,B). Shannon and Simpson index measure species diversity; the higher the Shannon index value, the higher the species diversity, while the opposite is true for the Simpson index (Kim et al., 2017). As shown in Supplementary Figures 2C,D, the species diversity of the intestinal flora decreased significantly on day 5 in the H9N2 AIV infection group, while the species diversity of the intestinal flora in the AL and H9N2-AL groups increased significantly compared to the mock and H9N2 infection groups (P < 0.01), indicating that H9N2 AIV infection can destroy intestinal flora diversity, while AL can restore it. OTU analysis-based dilution and grade abundance curves in different groups were shown in Supplementary Figures 2E,F. As compared with the mock and H9N2 AIV infected groups, the sample curve of the group fed with AL and the group fed with AL following infection was broader on the horizontal axis, indicating that the species composition of the AL and H9N2-AL groups was relatively rich and the species composition was more specific compared with mock and H9N2 AIV infected groups.
β diversity analysis, which included PCA, PCoA, NMDS and UPGMA analysis, showed that the samples of the same treatment group were concentrated in one area, while the sample distances of different groups were concentrated in different areas, with marked differences between the mock and H9N2 AIV infection groups at the 5 day time point, which indicated that the similarity of samples between the mock and H9N2 AIV infection groups was low. By contrast, the sample distance between the single feeding AL and H9N2 AIV infection with feeding AL groups were close to each other, indicating a high species similarity between the two groups (Supplementary Figures 2G–I). According to the distance, the trend of the four groups of samples was divided into 3 parts that represent the effects of different treatments on the similarity of intestinal flora composition in chickens, indicating that H9N2 AIV infection caused changes in intestinal diversity and AL could also improve intestinal flora diversity (Supplementary Figure 2J).
Taxonomic Analysis of Gut Microbiota Following H9N2 AIV Infection and AL Treatment in SPF Chickens
According to the sequencing results, sequences with a similarity of > 97% were defined as an OTU, with each OTU corresponding to a representative sequence. As shown in Figure 2A, the number of OTUs of each sample in each group was almost the same, with a total of 139 OTUs in 12 samples. A Venn map of the four groups showed that there were 5, 3, 1 and 3 unique OTUs in the mock, H9N2 AIV infection, AL feeding and H9N2 AIV infection with feeding AL groups, respectively, at 5 days post infection, indicating that different treatments can cause changes in the intestinal flora and produce corresponding unique flora (Figure 2B).
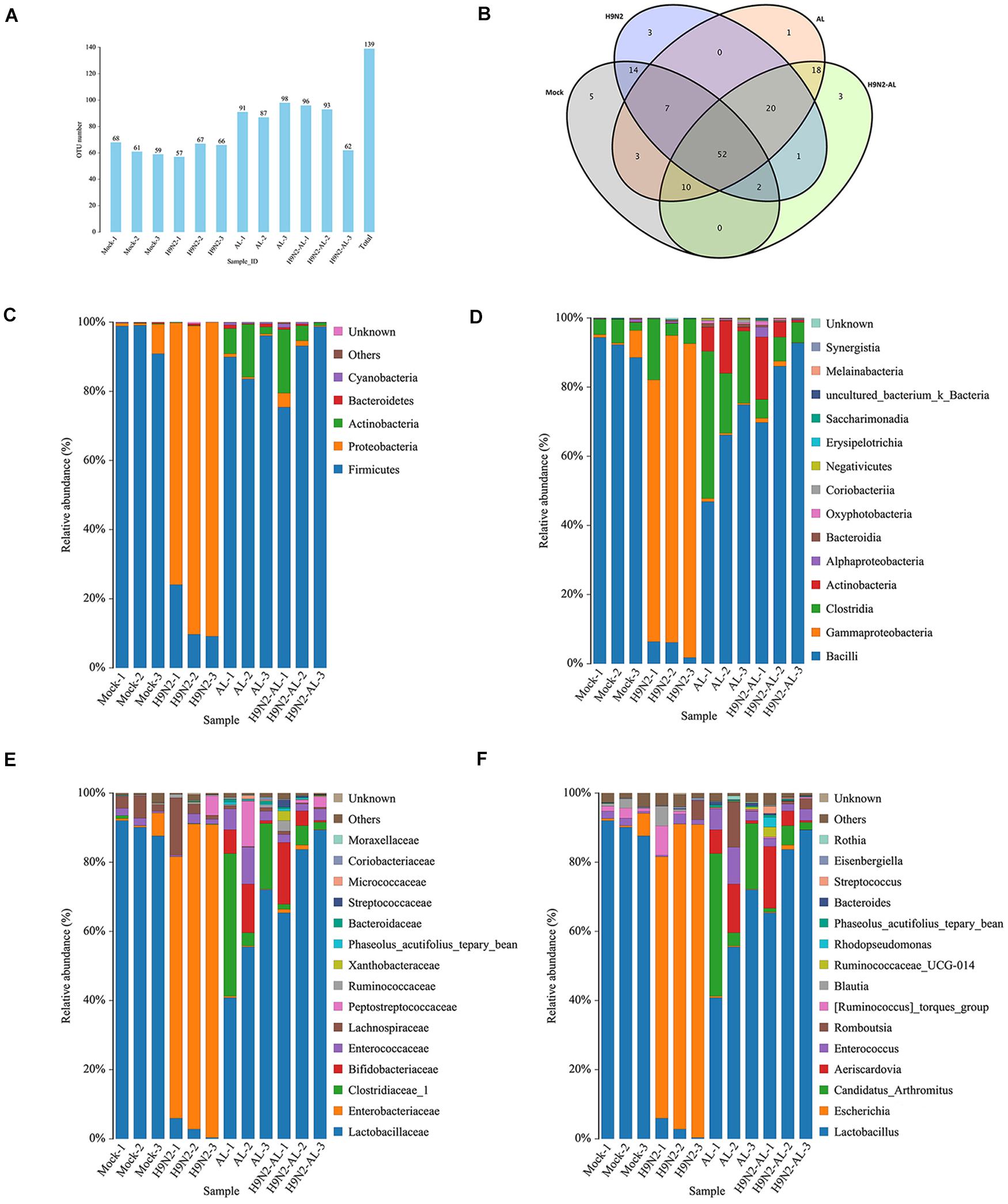
Figure 2. Taxonomic analysis of gut microbiota 5 days after H9N2 AIV infection and AL treatment in SPF chickens. (A) Distribution map of OTU number in samples from different treatment groups. (B) Venn diagrams of OTU in different treatment groups. Relative abundance of intestinal microflora at the (C) phylum, (D) class, (E) family and (F) genus levels in different treatment groups. AL, ageratum-liquid; OTU, operational taxonomic unit (OTU).
By comparing the OTU sequences with the reference database, the composition of the flora at each classification level was obtained. As shown in Figure 2C, at the phylum level, the dominant flora in the mock group was Firmicutes, with an average relative abundance of 96%. Secondly, the relative abundance of Proteobacteria reached 3%. Following infection with H9N2 AIV, the relative abundance of Proteobacteria became the dominant flora, with an average relative abundance of 85%, while that of Firmicutes decreased to 14%, indicating that H9N2 AIV could cause intestinal flora disorder and lead to the proliferation of Proteobacteria. In the AL group, the average relative abundance of the phylum Firmicutes accounted for 89%, followed by Actinobacteria, with an average relative abundance of 8%. In the H9N2-AL group, the phylum Firmicutes returned to the dominant flora, with an average relative abundance of 89%, followed by Actinobacteria with an average of 8%, and Proteobacteria with an average of 2%. The above results suggested that H9N2 AIV can induce intestinal flora disorder and cause a high proliferation of Proteobacteria, while AL can improve the intestinal microecological environment, inhibit the proliferation of Proteobacteria and promote the proliferation of Firmicutes and Actinobacteria into the dominant flora.
In the mock group, at the class, family and genus levels, the first dominant flora was Bacilli, Lactobacillaceae and Lactobacillus, respectively. The second dominant flora was Gammaproteobacteria, Enterobacteriaceae and Escherichia. The average relative abundance of Lactobacillus and Escherichia in the mock group was 90 and 2.5%, respectively (Figures 2D–F). On day 5 following H9N2AIV infection, the relative abundance of Lactobacillus decreased to 3%, while that of Escherichia increased to 85% (Figure 2F). In the AL group, at the genus level, the main flora was Lactobacillus, Candidatus-Arthromitus, Aeriscardovia, Enterococcus and Romboutsia, with their relative abundance at 56, 21, 7, 6 and 4%, respectively (Figure 2F). In the H9N2-AL group, the main flora was Lactobacillus, Aeriscardovia, Candidatus-Arthromitus and Enterococcus, with a relative abundance of 79, 7.5, 3 and 2.5% at the genus level, respectively (Figure 2F).
In combination, H9N2 AIV infection can destroy intestinal homeostasis, causing an increase in intestinal opportunistic pathogens, like Escherichia and inhibiting the growth of beneficial bacteria, like Lactobacillus. AL can improve intestinal homeostasis, restore the micro-ecological environment destroyed by H9N2 AIV, and lead to Lactobacillus becoming the dominant flora again.
LefSe Analysis of Gut Microbiota Following H9N2 AIV Infection and AL Treatment in SPF Chickens
In order to further screen the biomarker in the changes of intestinal flora caused by different treatments, LefSe analysis was performed. With a linear discriminant analysis (LDA) score of > 4.0 as the threshold, the species were screened from the phylum to the genus level. As shown in Figure 3A, the biomarkers used for the mock group were Bacilli, Lactobacillaceae, Lactobacillus and Lactobacillales. The biomarkers used for the H9N2 AIV infection group on day 5 were Escherichia, Enterobacteriaceae, Enterobacteriales, Lachnospiraceae, Ruminococcus-gauvreauii and Blautia. The biomarkers used for the AL group were Clostridiaceae, Candidatus-Arthromitus, Actinobacteria, Phaseolus acutifolius (tepary beean), Veillonellaceae, Phaseolus acutifolius (tepary bean), Megamonas and Faecalibacterium. The biomarkers used for the H9N2-AL group were Bifidobacteriaceae, Bifidobacteriales, Aeriscardovia and Enterobacter on day 5 post infection. According to the biological classification of bacteria and LDA score, the biomarker for each treatment group was further screened. Lactobacillus, Escherichia, Candidatus-Arthromitus and Aeriscardovia were the biomarkers used for the mock, H9, AL and H9N2-AL groups, respectively.
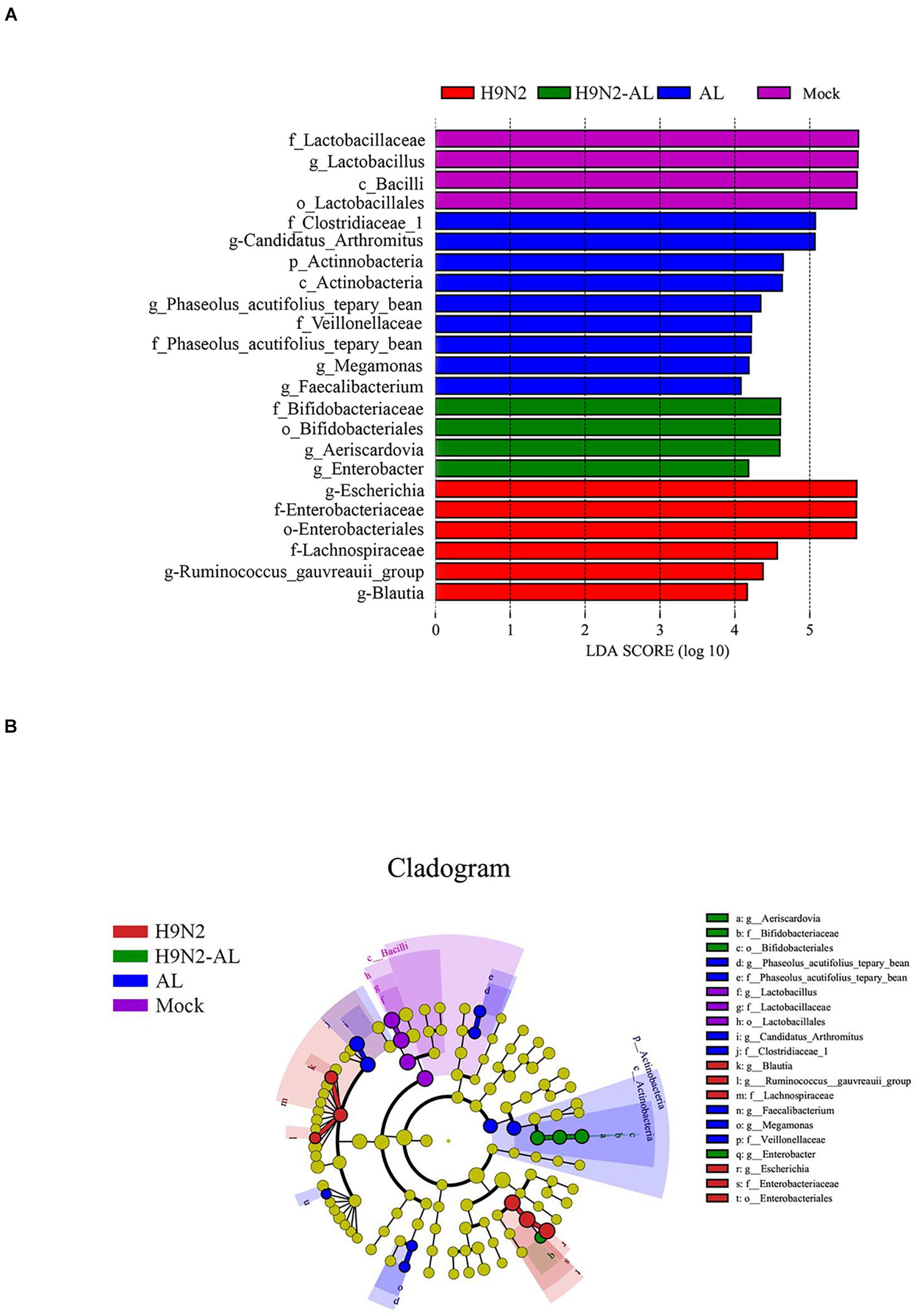
Figure 3. LefSe analysis of gut microbiota at 5 days after H9N2 AIV infection and AL treatment in SPF chickens. (A) Histogram of LDA value distribution in different treatment groups. (B) LEfSe analysis of evolutionary branching diagram in different treatment groups. AL, ageratum-liquid; LDA, linear discriminant analysis; LefSe, LDA effect size.
In order to more intuitively judge the changes in the microflora caused by different treatments, an evolutionary branching tree was made, and the circles radiated from inside to outside, representing the taxonomic level from the phylum to the genus, respectively. As shown in Figure 3B, the species with statistical differences at > 3 consecutive classification levels were Lactobacillus, Escherichia and Aeriscardovia. In combination, H9N2 AIV infection caused intestinal flora disorder, and the possible bacterial biomarkers for the mock and infection groups were Lactobacillus and Escherichia, respectively. After feeding the infection group with AL, the biomarker was Aeriscardovia.
Effects of H9N2 AIV Infection and AL Treatment on Tight Junction of Ileum Epithelium of SPF Chickens
In order to explore the barrier function of H9N2 AIV on intestinal mucosal epithelial cells, the mRNA expression of scaffold protein ZO-1, and transmembrane proteins Claudin 3 and Occludin was measured. As shown in Figure 4, the mRNA expression level of genes ZO-1 and Occludin was significantly decreased (5 dpi, P < 0.01), while that of Claudin 3 did not change significantly (P > 0.05), indicating that H9N2 AIV infection can cause the destruction of tight junction proteins in intestinal epithelial cells. There was no significant difference in the expression of intestinal epithelial tight junction proteins between the mock and AL groups (P > 0.05), while AL treatment in the infection group could upregulate the ZO-1 and Occludin, as compared with the infection group (P < 0.01 and P < 0.05), indicating that AL can protect the structure and function of the intestinal epithelial mucosa to a certain extent.
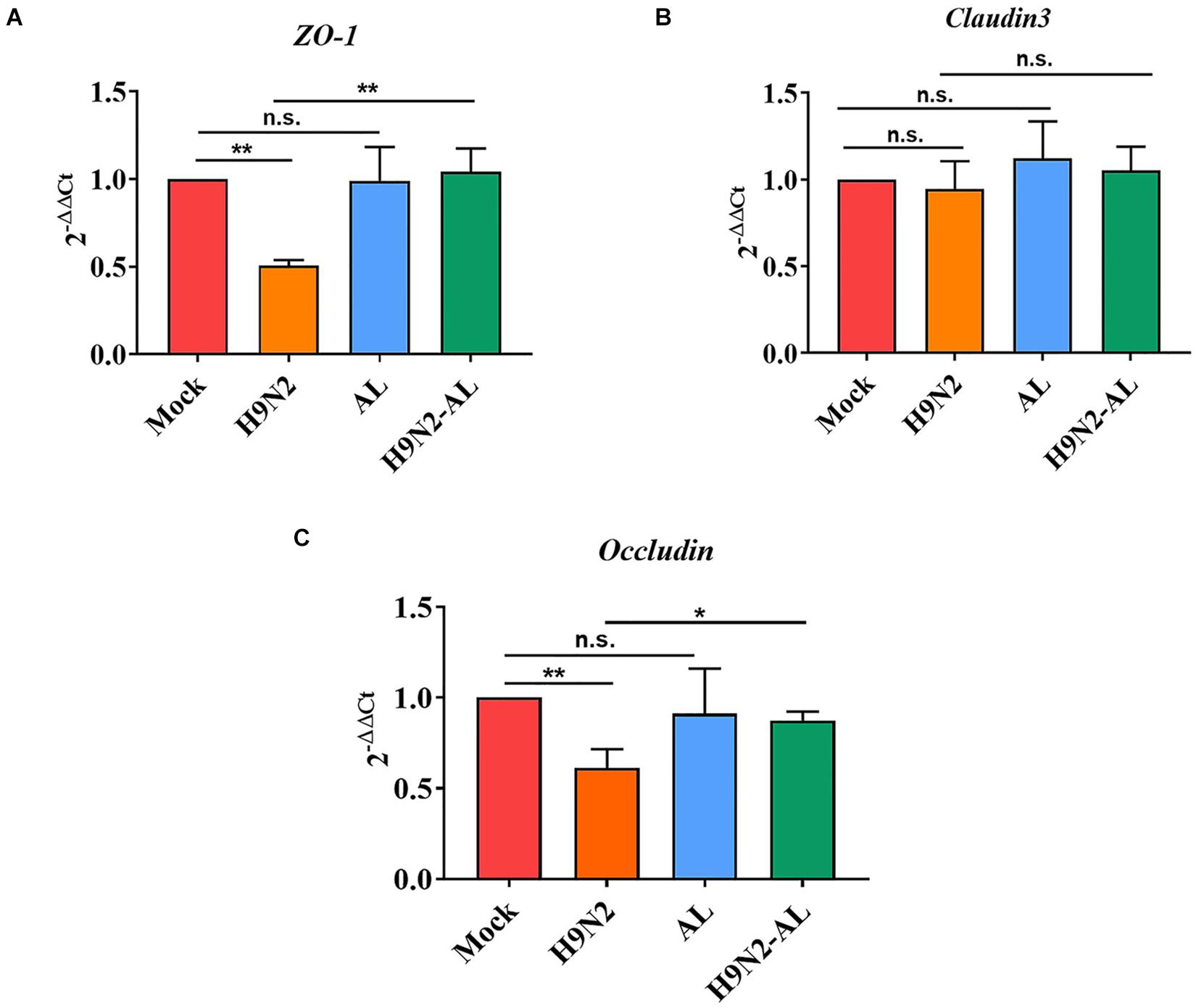
Figure 4. Effect of H9N2 AIV infection and AL treatment on mRNA expression of tight junction protein in the ileum epithelium of SPF chicken 5 days post-infection. Detection of mRNA expression of tight junction proteins (A) ZO-1, (B) Claudin 3 and (C) Occludin in ileum epithelial cells by RT-qPCR. Data are presented as the mean ± standard deviation (n = 3). The differences between groups were analyzed using ANOVA. *P < 0.05 and **P < 0.01. n.s., not significant. AL, ageratum-liquid.
Effects of H9N2 AIV Infection and AL Treatment on Intestinal Bacterial Translocation in SPF Chickens
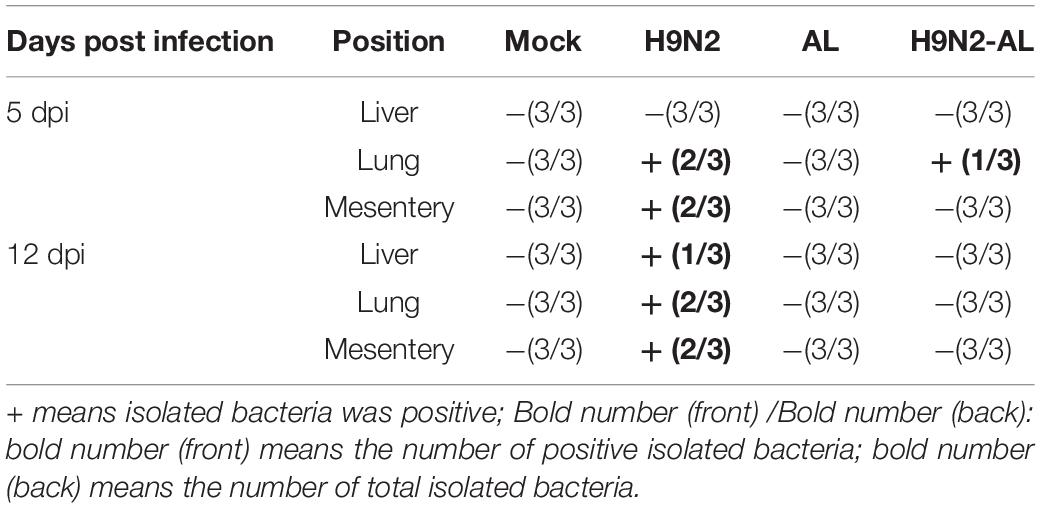
H9N2 AIV infection is often accompanied by secondary bacterial infection in visceral organs. The present results showed that bacteria were isolated from the lungs and mesentery at 5 dpi and were positive in all tissues at 12 dpi, while in the H9N2-AL group, bacteria were only isolated from the lungs at 5 dpi (Table 4). These findings suggested that H9N2 AIV infection can cause bacterial secondary infection in tissues and organs and that AL can prevent infection.
Following H9N2 AIV infection, bacteria can be isolated in the mesentery, lung and liver, indicating that there is a phenomenon of bacterial translocation (Table 4). Our previous results also showed that H9N2 AIV infection can cause the destruction of intestinal barrier function (Figure 4). It was therefore speculated that H9N2 AIV infection causes intestinal homeostasis disorder and intestinal opportunistic bacterial translocation, leading to bacterial secondary infection. To further test this speculation, the fluorescence-labeled bacterial strain NeonGreen was used to trace bacterial translocation. Results showed that NeonGreen-tagged bacteria were isolated from the intestinal cavity, indicating that the flora entered the intestine. In the H9N2 AIV infection group, NeonGreen-tagged bacteria was positive in the mesentery and lung at 24-48 h, and in the liver at 36-48 h, indicating that the H9N2 AIV infection promoted bacterial translocation to the mesentery and other tissues through the intestinal wall. Bacteria were isolated only in the lungs at 36 h in the H9N2-AL group, indicating that AL could effectively inhibit bacterial translocation through the intestinal wall to mesentery, lung and liver (Supplementary Table 1). Furthermore, the NeonGreen-tagged E. coli bacteria count in different treatment groups was shown in Supplementary Figure 3. Results showed that the count of NeonGreen-tagged E. coli from intestinal cavity in H9N2-NeonGreen group was significantly higher than that in NeonGreen group and H9N2-AL-NeonGreen group at 12 h, 24 h, 36 h and 48 h. The count of NeonGreen-tagged E. coli from mesentery in H9N2-NeonGreen group was gradually increasing from 24 h to 48 h, however, no NeonGreen-tagged E. coli was detected in NeonGreen and H9N2-AL-NeonGreen groups. Besides, the count of NeonGreen-tagged E. coli was gradually increasing from 24 h to 48 h in lung, however, no NeonGreen-tagged E. coli was detected in NeonGreen and H9N2-AL- NeonGreen groups at 12 h, 24 h and 48h, although the NeonGreen-tagged E. coli was detected in H9N2-AL-NeonGreen group at 36 h, while the count was significantly lower than that in the H9N2-NeonGreen group. Similarly, the count of NeonGreen-tagged E. coli in liver was gradually higher from 36 h to 48 h in H9N2-NeonGreen group, but no NeonGreen-tagged E. coli was detected in NeonGreen group and H9N2-AL- NeonGreen group from 12 h to 48 h, illustrating that H9N2 AIV infection causes NeonGreen-tagged E. coli translocation, leading to bacterial secondary infection, while AL could effectively inhibit E. coli translocation through the intestinal wall to mesentery, lung and liver.
In combination, H9N2 AIV infection promotes bacterial translocation from the intestinal cavity to the mesentery, lung and liver and causes secondary bacterial infection. AL can effectively prevent bacterial translocation induced by H9N2 AIV.
Effects of H9N2 AIV Infection and AL Treatment on Intestinal Metabolism of SPF Chickens
A multivariate model was used to construct a multivariate control chart to evaluate the validity of each sample data. Results showed that the biological variability of metabolic group samples was small and could be used for follow-up data analysis (Supplementary Figure 4A). By comparing the data of metabolic substances obtained by sequencing with the database of standard metabolic substances, 154 metabolic substances, 45 metabolic ratios and 211 unknown substances were obtained (Supplementary Figure 4B). The 154 substances annotated were chemically classified as follows: Amino acids (31%), carbohydrates (20%), organic acids (14%), nucleotides (9%), fatty acids (7%), lipids (6%), alkylamines (4%), alcohols (3%) and others (6%) (Supplementary Figure 4C). The 3D-PCA and 3D-PLS-DA showed that the sample clustering trends between the mock and H9N2 infection groups, or between H9N2 infection and H9N2-AL groups were different (Supplementary Figures 4D,E). In combination, both H9N2 AIV and AL can cause changes in the intestinal metabolic spectrum. OPLS-DA and VIP index volcano maps were constructed to analyze the changes in the metabolic spectrum between the mock and H9N2 AIV infection groups, and between the H9N2 AIV infection and H9N2-AL groups. Visualization of overall metabolite profile differences between the mock and H9N2 AIV infection groups or between the H9N2 and H9N2-AL groups were illustrated in Supplementary Figures 4F–I. Total 11 differential metabolites were screened out in the H9N2 AIV infection group, as compared with the mock group (Table 5). Total 5 differential metabolites were screened out in the H9N2-AL group, as compared with the H9N2 group (Supplementary Table 2).
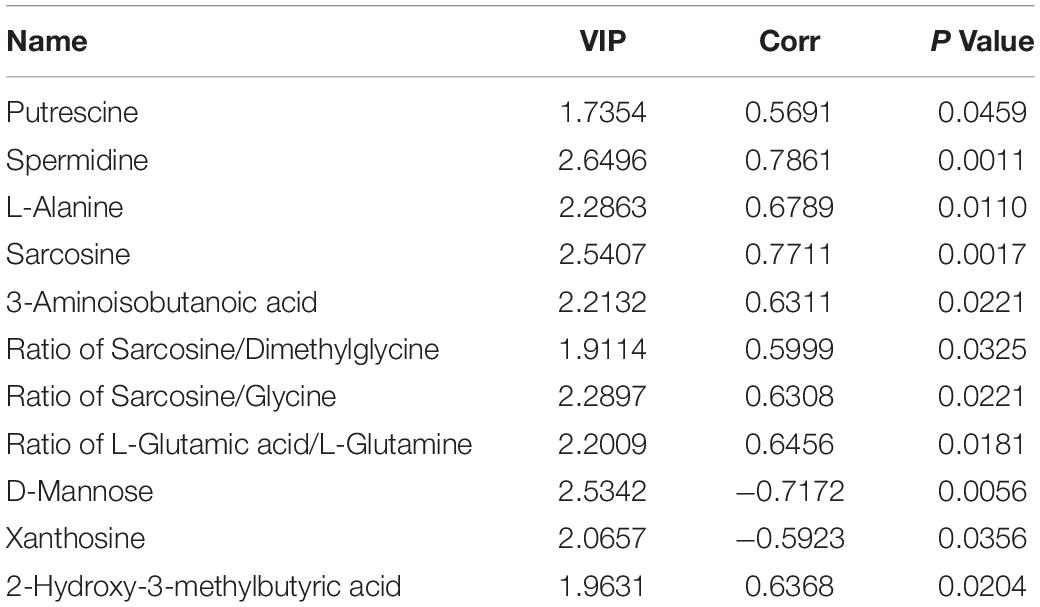
Table 5. The differential metabolites were screened out in the H9N2 AIV infection group, as compared with the mock group (log2FC > 1.5, P-Value < 0.05, VIP > 1).
The heatmap provides an overview of the global metabolic profiles among all samples, revealing the relative variations of each individual metabolite across all study samples in different treatment groups (Figure 5A). Overview of pathway analysis between H9N2 AIV infection group and the mock group, or between H9N2-AL group and H9N2 AIV infection group were shown in Figures 5B,C. In order to further determine the direct relationship between intestinal bacteria and metabolic changes, bacterial biomarker Lactobacillus from the mock group and Escherichia from the H9N2 AIV infection group were selected and screened according to the absolute value of a correlation coefficient of > 0.5. A total of 6 metabolites were found to be associated with Lactobacillus and Escherichia, in which the L-Glutamic acid/L-Glutamine ratio was positively correlated with Escherichia (Figure 5D).
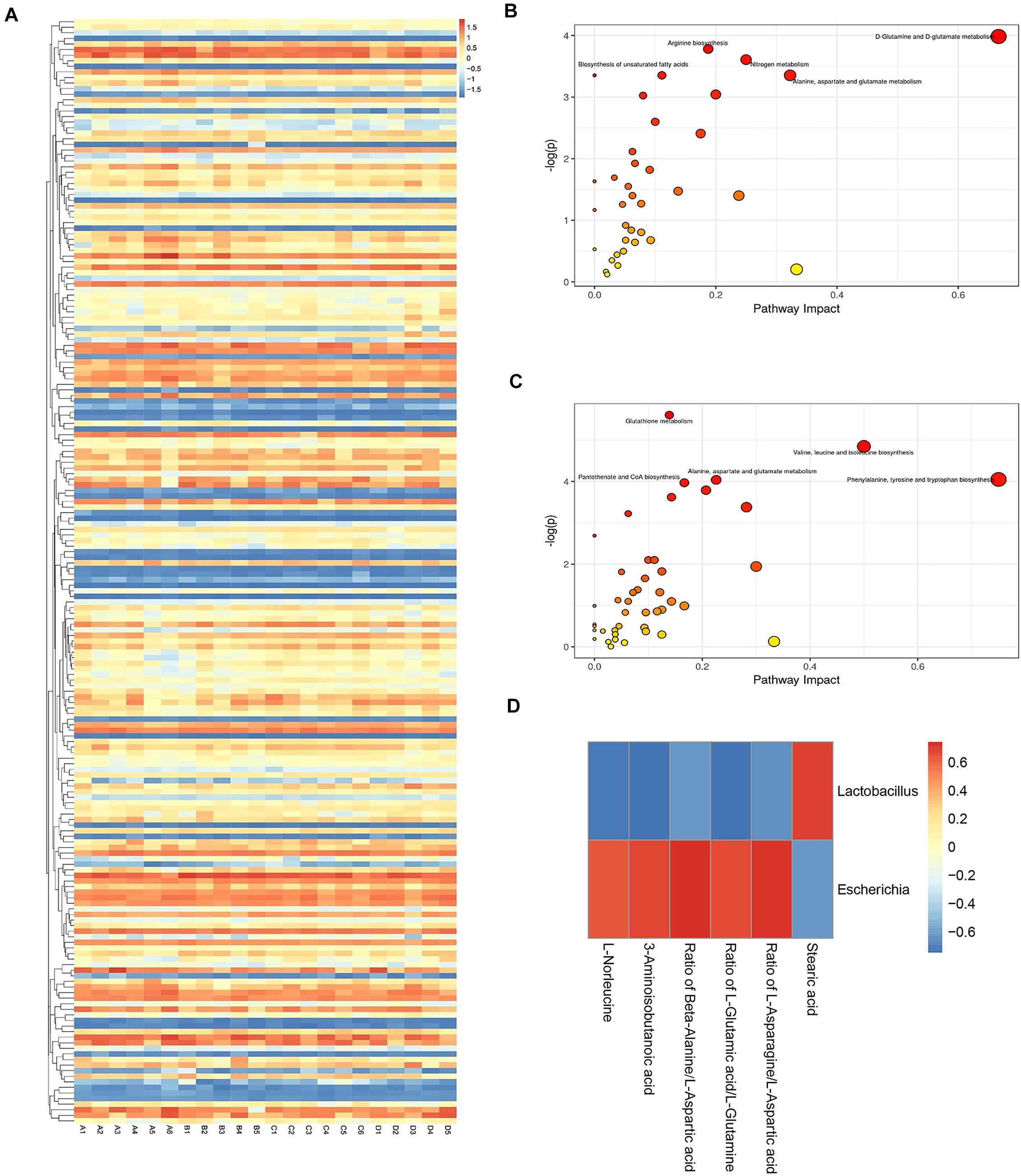
Figure 5. Metabolite profile visualization, metabolic pathway enrichment analysis and analysis of the correlation between intestinal flora and metabolic changes. (A) Heatmap depicting the overall metabolic variations among different treatment groups. (B) Differential metabolic pathway between the mock and H9N2 AIV infection groups. (C) Differential metabolic pathway between H9N2 AIV infection and H9N2-AL groups. (D) Corrplot of association analysis of microflora and metabolites. The number of sequencing samples in each group was ≥ 5. (A), mock group; (B), H9N2 AIV infection group; (C), AL feeding group; (D), after H9N2 infection group with feeding AL group.
Effects of H9N2 AIV Infection and AL Treatment on Inflammatory Gene Expression of TNF-α, IFN-γ and iNOS in the Intestinal Mucosa of SPF Chickens
It was reported that iNOS is the sole source of nitric oxide during inflammation and TNF-α and IFN-γ are important inflammatory cytokines, and IFN-γ can induce the production of iNOS (Winter et al., 2013). The expression of TNF-α, IFN-γ and iNOS were therefore determined by RT-qPCR. The results showed that the gene expression of TNF-α and IFN-γ was significantly increased following H9N2 AIV infection (P < 0.01, Figures 6A,B). The gene expression of iNOS was also significantly increased following H9N2 AIV infection (P < 0.01; Figure 6C), while that of TNF-α, IFN-γ and iNOS was significantly downregulated in the H9N2 AIV infection group with AL feeding, as compared with the infection group (P < 0.01 and P < 0.05, Figures 6A–C). The results illustrated that H9N2 AIV infection increased the expression of inflammatory cytokines in intestinal epithelial cells, and that AL could inhibit the inflammation induced by the H9N2 AIV infection.
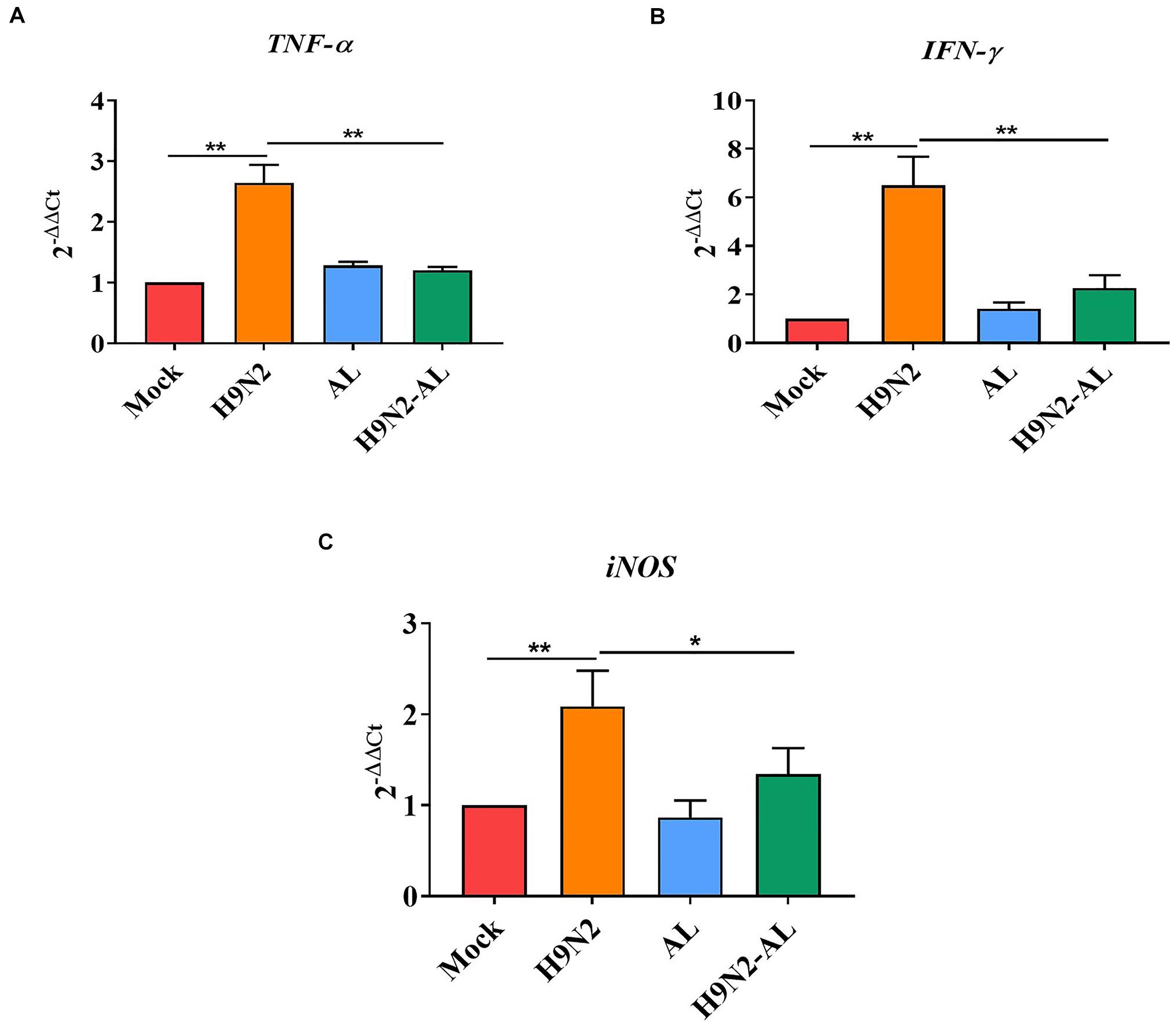
Figure 6. mRNA expression of pro-inflammatory genes in the intestinal mucosal epithelial cells of the ileum in different treatment groups. mRNA expression of (A) TNF-α, (B) IFN-γ and (C) iNOS in the intestinal mucosal epithelial cells of the ileum in different treatment groups 5 days post-H9N2 AIV infection. Data are presented as the mean ± standard deviation (n = 3). The differences between groups were analyzed using ANOVA. *P < 0.05 and **P < 0.01. AL, ageratum-liquid.
Construction of EcN Mutant and in vitro Competitive Tests of Mutant Strains
Nitrogen is an essential element for living organisms, and nitrate is one of the most important forms of nitrogen (Cabello et al., 2004). Metabolic pathway enrichment analysis showed that the nitrogen metabolism pathway was positively correlated with H9N2 AIV infection (Figure 7A), further analysis showed that L-Glutamic acid was significantly upregulated while L-Glutamine was significantly downregulated in the nitrogen metabolism pathway. Since that we have revealed that the L-Glutamic acid/L-Glutamine ratio was positively correlated with Escherichia (Figure 5D), we speculated that the nitrogen metabolism pathway may play an important role in the proliferation of Escherichia. Since a previous study showed that nitrate promotes the growth of E. coli in the inflamed gut (Winter et al., 2013), and it was confirmed in the present study that the H9N2 AIV infection could induce inflammation (Figure 6), moreover, nitrate is a key regulator in the nitrogen metabolism pathway which was positively correlated with H9N2 AIV infection (Figure 7A), it was speculated that the proliferation of E. coli caused by H9N2 AIV infection may be associated with nitrate.
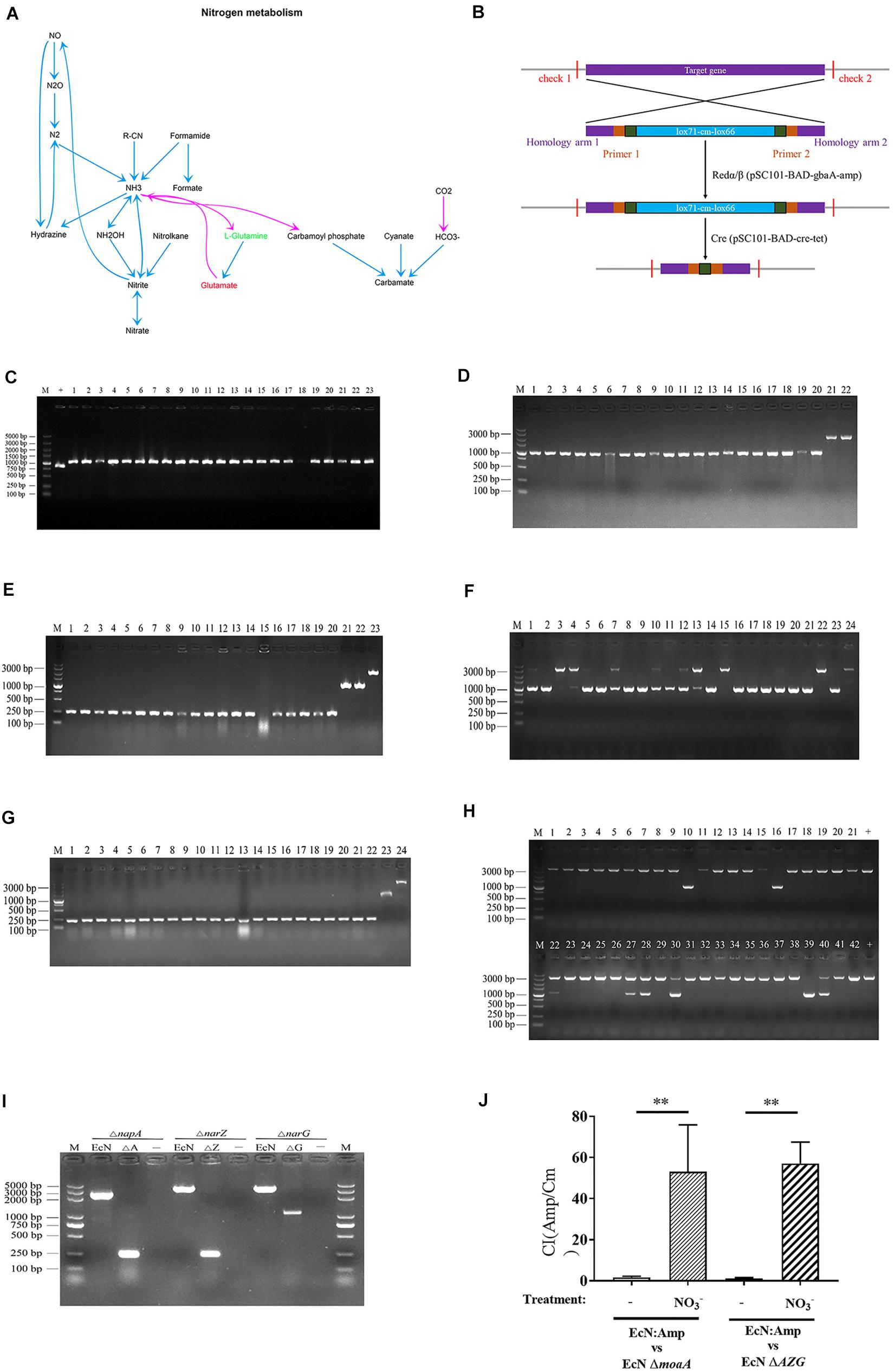
Figure 7. Construction of EcN mutant and in vitro competitive testing of mutant strains. (A) Metabolic pathway enrichment analysis of nitrogen metabolism in H9N2 AIV infection group, as compared with the mock group. (B) Schematic diagram of the process of knockout bacterial genes using Red/ET recombinant engineering technology and a site-specific system. (C) Identification of E. coli moA mutant (ΔmoaA) by RT-qPCR. M represents the 5000 DNA marker, + the positive control of wild type E. coli (EcN WT; size, 840 bp), and 1-23 the recombinants of EcN moaA (lox71-cm-lox66; size, 1057 bp). (D) Identification of recombinant replacement of EcN napA gene. M represents the 3000 DNA marker, 1-20 the recombinants of EcN napA (lox71-cm-lox66; size, 1071 bp), and 21-22 the positive control of EcN WT (size, 2354 bp). (E) Identification of resistance gene deletion of EcN napA mutant (ΔnapA) by RT-qPCR. M represents the 3000 DNA marker, 1-20 the knockout of resistance gene deletion (ΔnapA; 243 bp), 21-22 the recombinants of EcN napA (lox71-cm-lox66; 1071 bp) and 23 the positive control of EcN WT (2,354 bp). (F) Recombinant replacement identification of EcN ΔnapA narZ. M represents the 3000 DNA marker, 1-23 the recombinants of EcN ΔnapA narZ (lox71-cm-lox66; 1071 bp), 24 the positive control of EcN WT (3,608 bp). (G) Identification of resistance gene deletion of EcN ΔnapA ΔnarZ by RT-qPCR. M represents the 3000 DNA marker, 1-22 the knockout of resistance gene deletion (ΔnapA ΔnarZ; 244 bp), 23 the recombinants of EcN ΔnapA ΔnarZ (lox71-cm-lox66; 1071 bp), 24 the positive control of EcN WT (3608 bp). (H) Recombinant replacement identification of EcN ΔnapA ΔnarZΔnarG. M represents the 3000 DNA marker, 1-21 and 22-42 the recombinants of EcN ΔnapA ΔnarZΔnaG (lox71-cm-lox66), 10, 16 and 39 the successfully constructed EcN ΔnapA ΔnarZ narG (1082 bp), + the positive control of EcN WT (3622 bp). (I) Identification of resistance gene deletion of EcN ΔnapA ΔnarZ ΔnarG by RT-qPCR. The size of the positive control of EcN WT for EcN ΔnapA, EcN ΔnapA narZ and EcN ΔnapA ΔnarZΔnarG was 2,354, 3,608 and 3,622 bp. The size of EcN ΔnapA, EcN ΔnapA narZ and EcN ΔnapA ΔnarZΔnarG was 243, 244 and 1,082 bp. M represents the 5000 DNA marker and - the negative control. (J) Mutant Competitiveness Index. Data are presented as the mean ± standard deviation (n = 3). The differences between groups were analyzed using ANOVA. **P < 0.01. EcN, E. coli Nissle1917; E. coli, Escherichia coli.
It has been reported that MOA and AZG are the key genes for the proliferation of E. coli using nitrate (Winter et al., 2013). Next, EcN ΔmoaA and ΔAZG mutants were further constructed according to the basic process for constructing a mutant strain (Figures 7B–I). The CI of EcN:Amp with EcN ΔmoaA and EcN ΔAZG strains was further determined. The results showed that, under anaerobic conditions, the growth competitiveness index of the EcN:Amp and function-deficient strains tended to be 1, indicating that the growth rates of the 2 strains were similar in the same culture system (Figure 7J). After the addition of NO3– to the culture system, the CI between the EcN:Amp and EcN ΔmoaA strains, or between the EcN:Amp and EcN ΔAZG strains significantly increased (Figure 7J); this indicated that the growth rate of EcN:Amp was significantly faster than that of the two gene-deficient bacteria, illustrating that EcN:Amp could markedly proliferate with NO3–.
Nitrate Promotes the Proliferation of E. coli in vivo
In order to explore whether the H9N2 AIV infection can induce the production of nitrate in intestinal epithelial cells and promote the proliferation of E. coli, an iNOS inhibitor, SMT, was used to evaluate the content of nitrate in the intestinal epithelium. The results showed that the nitrate content in the intestinal epithelium increased significantly following H9N2 AIV infection (P < 0.01), while SMT could significantly reduce the H9N2 AIV-induced nitrate content (P < 0.01). AL could also significantly reduce the production of H9N2 AIV-induced nitrate (P < 0.01, Figure 8A), indicating that H9N2 AIV infection could induce the production of nitrate in intestinal epithelial cells and that AL exerted the effects of an iNOS inhibitor.
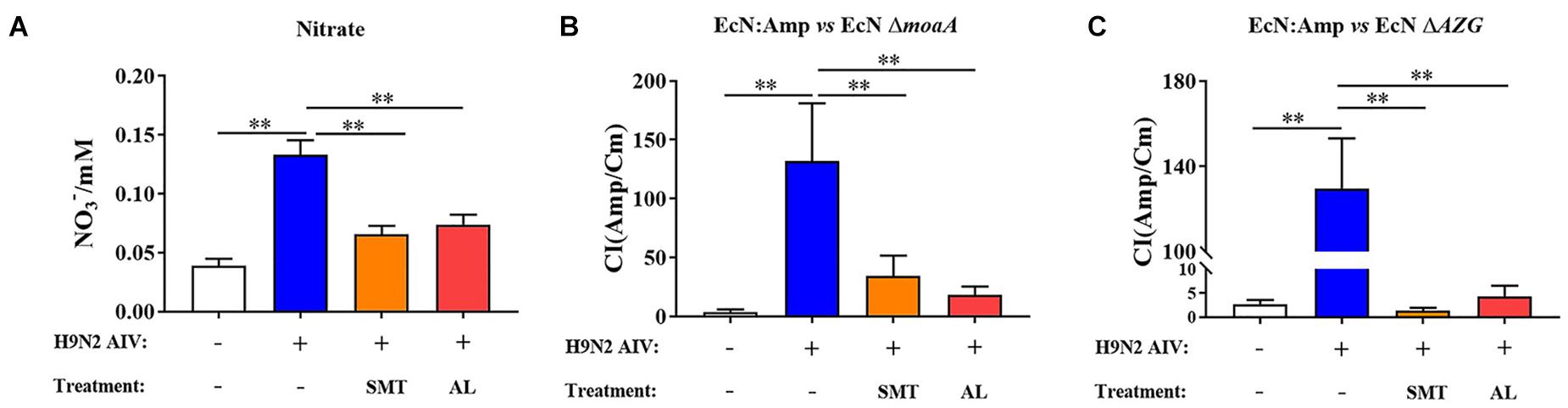
Figure 8. Verification of nitrate metabolism in vivo. (A) Nitrate content of intestinal epithelium. SMT is an iNOS inhibitor added to evaluate the nitrate content produced by intestinal epithelium. (B) CI of EcN:Amp and EcN moaA. (C) CI between EcN:Amp and EcN AZG. Data are presented as the mean ± standard deviation (n = 3). The differences between groups were analyzed using ANOVA. **P < 0.01. SMT, S-methyl-isothiourea; iNOS, nitric oxide synthase; CI, competitive index; EcN, E. coli Nissle1917.
The in vitro functional verified strains EcN:Amp and EcN ΔmoaA, and EcN:Amp and EcN ΔAZG were fed to SPF chickens at 1:1, and the CI of the 2 pairs of strains was determined to evaluate the intestinal epithelial environment. As shown in Figure 8B, following H9N2 AIV infection, the CI of EcN:Amp and EcN ΔmoaA increased significantly, indicating that the intestinal environment can produce growth factors that can lead to EcN:Amp growth, but cannot meet the EcN ΔmoaA growth requirements. Both SMT and AL reduced the CI, indicating that this growth factor was nitrate. Further experiments showed that, following H9N2 AIV infection, the CI of EcN:Amp and EcN ΔAZG significantly increased, since the EcN ΔAZG strain was a nitrate deficient strain, which, again, indicated that H9N2 AIV infection could induce the production of nitrate in the intestinal epithelium and provide an electron acceptor for the growth of E. coli. Following H9N2 AIV infection, both SMT and AL could significantly reduce the CI between strains (Figure 8C), indicating that AL exerted an effect associated with iNOS inhibitors, such as SMT.
In combination, the results showed that H9N2 AIV infection can promote the production of nitrate in intestinal epithelial cells, nitrate can provide electron acceptors for the growth of E. coli in the intestine, and AL can inhibit the production of nitrate in the intestinal tract.
Discussion
The host and intestinal microorganisms are symbiotes, and pathogen infection affects changes in intestinal microorganisms. In recent years, the association between influenza viruses and intestinal microorganisms has attracted considerable attention. For example, studies have shown that the number of Proteobacteria increases in the feces of mice infected with the influenza virus (mainly Enterobacteriaceae) (Deriu et al., 2016). Subsequent studies have shown that influenza virus infection can change the composition of intestinal microorganisms in mice and cause intestinal microbial imbalance (Yildiz et al., 2018). In 2015, it was reported that H7N9 viral infection caused changes in the composition of human intestinal microorganisms, in antibiotic and non-antibiotic treated patients, the abundance of E. coli ranked first and third, respectively (Qin et al., 2015; Groves et al., 2018). However, there are few reports on intestinal microbial disorders caused by H9N2 infection in poultry. It was found that H9N2 AIV can disrupt the composition of intestinal microorganisms, and viral infection can induce intestinal microbiota to regulate the immune response mediated by type I interferon, and then play an antiviral role (Yitbarek et al., 2018a,b,c). In the present study, it was found that intestinal E. coli rapidly proliferated and the intestinal mucosal barrier was damaged following H9N2 AIV infection in SPF chickens, which was consistent with the previous research results of our group (Li et al., 2018).
The destruction of the intestinal barrier increases intestinal permeability, and intestinal bacteria are easy to translocate through the barrier to visceral organs (Ma et al., 2018). In addition, certain studies have shown that HIV infection causes systemic microbial disorder and increased microbial translocation in the plasma (Xu et al., 2018). Therefore, it was speculated that the increased E. coli in the intestinal tract caused by H9N2 AIV may translate to other organs. As expected, our results showed that bacteria could indeed be isolated from the liver and other visceral tissues following H9N2 AIV infection. In order to further confirm whether the microbiota in the viscera were translocated from the intestinal tract, NeonGreen-tagged bacteria were administrated into the gastrointestinal tract and used to trace the translocation of E. coli from the intestinal tract to the mesentery, lung and liver, in order to illustrate whether the bacterial secondary infection in SPF chickens infected with H9N2 was due to intestinal flora disorder and then result to the E. coli translocate to the visceral organs. Furthermore, the NeonGreen-tagged E. coli bacteria count in different treatment groups was detected by RT-qPCR to further illustrate that H9N2 AIV infection causes NeonGreen-tagged E. coli translocation, leading to bacterial secondary infection. This explains the causal relationship among H9N2 virus, intestinal microbial imbalance and bacterial secondary infection.
H9N2 AIV infection damages the intestinal mucosal barrier in chickens, resulting in the increase of E. coli and its translocation to other organs. Therefore, in-depth understanding of the causes of E. coli proliferation is key to exploring the mechanism of bacterial secondary infection. E. coli is a facultative anaerobic bacteria that belongs to the Proteobacteria of the Escherichia genus. E. coli is a conditional pathogen in the intestinal tract and has a symbiotic relationship with the body. Under normal circumstances, E. coli is harmless to the body; however, when a disease occurs in the host, E. coli proliferates to become harmful bacteria. Proteobacteria, as a feature of intestinal flora imbalance, is often associated with some inflammatory diseases, such as ulcerative colitis (Carvalho et al., 2012; Maharshak et al., 2013). This phenomenon suggests that the proliferation of facultative anaerobes in the intestinal tract may be driven by oxygen or other respiratory electron receptors, thus putting forward the “oxygen hypothesis” (Rigottier-Gois, 2013). In the colitis animal model group, the decrease of intestinal short-chain fatty acids caused intestinal epithelial cells to grow on glycolysis and lactic acid fermentation (Kelly et al., 2015; Byndloss et al., 2017). As a result, intestinal epithelial cells released lactate into the intestinal cavity and the epithelial cells did not consume oxygen, causing oxygen molecules to be released into the intestinal cavity and the intestinal tract to become a micro-oxygen environment and promote the proliferation of E. coli (Takaishi et al., 2008; Hughes et al., 2017). Symbiotic Enterobacteriaceae can use formate as an electron donor and oxygen as an electron acceptor for respiratory metabolism (Hughes et al., 2017). In addition, Enterobacteriaceae can use the nitrate produced by the host as an electron acceptor. Under healthy conditions, E. coli is a symbiotic bacteria in the intestine. When inflammation occurs, the abundance of E. coli in intestinal tract significantly increased and use nitrate for metabolic competitive growth, which is an independent metabolic pathway (Winter et al., 2013). Therefore, there is an important association among the imbalance of intestinal flora, the proliferation of Enterobacteriaceae and the metabolite.
The present study was the first to perform intestinal metabonomics to study the changes of metabolites in the intestinal lumen following H9N2 AIV infection. It was found that the L-Glutamic acid/L-Glutamine ratio was significantly positive correlated with H9N2 infection group, as compared with the mock group. Further correlation analysis showed that E. coli was correlated with the L-Glutamic acid/L-Glutamine ratio. Combined with metabonomic pathway enrichment analysis, it was found that the metabolic pathways associated with L-Glutamic acid and L-Glutamine was nitrogen metabolism. It has been reported that IFN-γ can induce the expression of the NOS2 gene in the intestinal epithelium and can encode inducible iNOS, while iNOS can catalyze the formation of nitric oxide from arginine in the intestinal tract (Salzman et al., 1996). According to the metabonomics results, it was inferred that the imbalance of intestinal bacteria caused by the H9N2 AIV infection may have been caused by the anaerobic metabolism and proliferation of Enterobacteriaceae using nitrate as an electron acceptor.
In order to test that hypothesis, interferon and other genes, as well as nitrate content, were first determined in the intestinal epithelium. Our results showed that H9N2 AIV could upregulate the mRNA level of TNF-α, IFN-γ and iNOS, and the nitrate content in the intestinal epithelium was also significantly increased, indicating that E. coli may proliferate through nitrate metabolism in the inflamed intestinal tract. In order to determine the association of nitrate metabolism and proliferation of E. coli in chickens, nitrate-deficient mutant E. coli was constructed by genetic engineering recombination technology, and the metabolic environment in the intestinal epithelium was judged by the growth of functional deficient strains. The present study showed that H9N2 AIV infection can increase the growth competition index between normal nitrate metabolic and mutant strains, indicating that E. coli in the intestine proliferates markedly using nitrate produced by the intestinal epithelium as a respiratory electron. Therefore, this study elucidated the mechanism of secondary E. coli infection in chickens caused by H9N2 AIV infection that H9N2 AIV infection induces inflammation in the intestinal mucosa and further promotes the secretion and release of nitrate from the host intestinal epithelium, E. coli in the intestine proliferates massively with nitrate as the electron acceptor and the E. coli in the intestine translocate to visceral tissue through the damaged intestinal barrier, resulting in secondary bacterial infection. The intestinal microorganism plays an important role in innate immune response in poultry, and therefore the regulatory relationship among E. coli, nitrate and innate immune needs to be further studied.
In the breeding industry, the highly prevalent abuse of chemical drugs and antibiotics poses a severe threat to humans. The emergence of super resistant strains, livestock and poultry products, drug residues, and environmental pollution is becoming an increasingly serious problem. Traditional Chinese herbal medicines have been widely used as sources of novel antiviral drugs. For that reason, the present study used the Chinese medicine AL to regulate secondary bacterial infection caused by H9N2 AIV. Our results showed that AL could regulate the changes in intestinal bacteria, restore the dominant position of Lactobacillus, and inhibit the bacterial disorder caused by H9N2 AIV, and particularly the abnormal proliferation of E. coli. It has been reported that Agastache rugosa inhibits the expression of iNOS in osteocytes activated by TNF-α and IL-1β (Oh et al., 2005). Our results of this study indicated that AL can alleviate intestinal damage, reduce inflammation and iNOS expression, and then reduce the production of nitrate in intestinal epithelial cells. In general, AL can regulate the expression of inflammatory genes in the intestinal epithelium of the host, and then regulate the composition of intestinal microorganisms and inhibit the abnormal proliferation of E. coli by inhibiting metabolite nitrate. Furthermore, AL can repair intestinal mucosal damage and provide a strong protection for E. coli translocation.
Conclusion
In conclusion, the present study firstly revealed the mechanism of H9N2 AIV-induced secondary E. coli infection. In briefly, H9N2 AIV infection caused intestinal microbial disorder and made Escherichia become the dominant flora, promoted bacterial translocation from intestinal tract to visceral tissue through the damaged intestinal barrier, induced inflammation in the intestinal mucosa and promoted the secretion and release of nitrate from the host intestinal epithelium. Importantly, the nitrate was the reason for the growth of E. coli in the inflammatory intestinal tract following H9N2 AIV infection. In addition, AL can restore the homeostasis of the intestinal flora, improve the injury degree of intestinal epithelial mucosa, inhibit inflammatory genes and the production of nitrate in the intestinal epithelium, effectively preventing the proliferation and translocation of E. coli in the intestine. This study provided effective theoretical guidance for the prevention and control of H9N2 AIV-induced secondary bacterial infection.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA625215.
Ethics Statement
The animal study was reviewed and approved by the South China Agricultural University Committee of Animal Experiments (approval ID, SYXK-2014-0136). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XZ, HL, and WL conceived study, designed the experiments, and analyzed the data. QZ, CW, ZX, and XC performed the experiments and analyzed the data. XZ wrote the manuscript. HZ and QX supervised the experiments and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chief Expert Project of Agricultural Industry Technology System in Guangdong Province (grant no. 2019KJ128), Creation of Triple Chimeric Vaccine (rIBV-ND-H9) based on attenuated Avian Infectious bronchitis virus D90 (grant no. 2017KZDM008), Special Project of National Modern Agricultural Industrial Technology System (grant no. CARS-41), Guangdong Provincial Promotion Project on Preservation and Utilization of Local Breed of Livestock and Poultry, the China Postdoctoral Science Foundation (grant no. 2019M652922), and the Natural Science Foundation of Guangdong Province (grant no. 2019A1515012006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer WR declared a shared affiliation, with no collaboration, with the authors to the handling editor at the time of review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.555739/full#supplementary-material
References
Autieri, S. M., Lins, J. J., Leatham, M. P., Laux, D. C., Conway, T., and Cohen, P. S. (2007). L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 DeltafucAO and E. coli Nissle 1917 DeltafucAO mutants in the mouse intestine and in M9 minimal medium. Infect. Immun. 75, 5465–5475. doi: 10.1128/IAI.00822-07
Barbour, E. K., Mastori, F. A., Abdel Nour, A. M., Shaib, H. A., Jaber, L. S., Yaghi, R. H., et al. (2009). Standardisation of a new model of H9N2/Escherichia coli challenge in broilers in the Lebanon. Vet. Ital. 45, 317–322.
Bian, X., Huang, F., Stewart, F. A., Xia, L., Zhang, Y., and Muller, R. (2012). Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. Chembiochem 13, 1946–1952. doi: 10.1002/cbic.201200310
Byndloss, M. X., Olsan, E. E., Rivera-Chavez, F., Tiffany, C. R., Cevallos, S. A., Lokken, K. L., et al. (2017). Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575. doi: 10.1126/science.aam9949
Cabello, P., Roldan, M. D., and Moreno-Vivian, C. (2004). Nitrate reduction and the nitrogen cycle in archaea. Microbiol. Sgm 150, 3527–3546. doi: 10.1099/mic.0.27303-0
Carvalho, F. A., Koren, O., Goodrich, J. K., Johansson, M. E., Nalbantoglu, I., Aitken, J. D., et al. (2012). Transient inability to manage Proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12, 139–152. doi: 10.1016/j.chom.2012.07.004
Chen, F., Wen, Q., Jiang, J., Li, H. L., Tan, Y. F., Li, Y. H., et al. (2016). Could the gut microbiota reconcile the oral bioavailability conundrum of traditional herbs? J. Ethnopharmacol. 179, 253–264. doi: 10.1016/j.jep.2015.12.031
Chen, H., Yuan, H., Gao, R., Zhang, J., Wang, D., Xiong, Y., et al. (2014). Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383, 714–721. doi: 10.1016/S0140-6736(14)60111-2
Deriu, E., Boxx, G. M., He, X. S., Pan, C., Benavidez, S. D., Cen, L. J., et al. (2016). Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog. 12:e1005572. doi: 10.1371/journal.ppat.1005572
Gharaibeh, S. (2008). Pathogenicity of an avian influenza virus serotype H9N2 in chickens. Avian Dis. 52, 106–110. doi: 10.1637/8108-090907-Reg
Groves, H. T., Cuthbertson, L., James, P., Moffatt, M. F., Cox, M. J., and Tregoning, J. S. (2018). Respiratory disease following viral lung infection alters the murine gut microbiota. Front. Immunol. 9:182. doi: 10.3389/fimmu.2018.00182
Haberman, Y., Tickle, T. L., Dexheimer, P. J., Kim, M. O., Tang, D., Karns, R., et al. (2014). Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633. doi: 10.1172/JCI75436
He, Y. H., Zhao, H. Y., Liu, Z. L., Lu, C., Luo, X. J., Lin, S. Q., et al. (2006). Effects of huoxiangzhengqi liquid on enteric mucosal immune responses in mice with Bacillus dysenteriae and Salmonella typhimurium induced diarrhea. World J. Gastroenterol. 12, 7346–7349. doi: 10.3748/wjg.v12.i45.7346
Hughes, E. R., Winter, M. G., Duerkop, B. A., Spiga, L., de Carvalho, T. F., Zhu, W. H., et al. (2017). Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21, 208–219. doi: 10.1016/j.chom.2017.01.005
Kelly, C. J., Zheng, L., Campbell, E. L., Saeedi, B., Scholz, C. C., Bayless, A. J., et al. (2015). Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671. doi: 10.1016/j.chom.2015.03.005
Kim, B. R., Shin, J., Guevarra, R., Lee, J. H., Kim, D. W., Seol, K. H., et al. (2017). Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Ko, S. J., Ryu, B., Kim, J., Hong, B. G., Yeo, I., Lee, B. J., et al. (2011). Effect of herbal extract granules combined with probiotic mixture on irritable bowel syndrome with diarrhea: study protocol for a randomized controlled trial. Trials 12:219. doi: 10.1186/1745-6215-12-219
Kotler, D. P., Reka, S., and Clayton, F. (1993). Intestinal mucosal inflammation associated with human immunodeficiency virus infection. Dig. Dis. Sci. 38, 1119–1127. doi: 10.1007/BF01295730
Kritas, S. K., Ronconi, G., Caraffa, A., Gallenga, C. E., Ross, R., and Conti, P. (2020). Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. J. Biol. Regul. Homeost. Agents 34, 9–14. doi: 10.23812/20-Editorial-Kritas
Lam, T. T., Wang, J., Shen, Y., Zhou, B., Duan, L., Cheung, C. L., et al. (2013). The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502, 241–244. doi: 10.1038/nature12515
Lee, D. H., Fusaro, A., Song, C. S., Suarez, D. L., and Swayne, D. E. (2016). Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology 488, 225–231. doi: 10.1016/j.virol.2015.11.023
Li, H. X., Liu, X. L., Chen, F. Y., Zuo, K. J., Wu, C., Yan, Y. M., et al. (2018). Avian influenza virus subtype H9N2 affects intestinal microbiota, barrier structure injury, and inflammatory intestinal disease in the chicken ileum. Viruses Basel 10:270. doi: 10.3390/v10050270
Liu, Q., Shen, Q., Bian, X., Chen, H., Fu, J., Wang, H., et al. (2016). Simple and rapid direct cloning and heterologous expression of natural product biosynthetic gene cluster in Bacillus subtilis via Red/ET recombineering. Sci. Rep. 6:34623. doi: 10.1038/srep34623
Liu, Y., Liu, W., Peng, Q. X., Peng, J. L., Yu, L. Z., and Hu, J. L. (2014). Protective effect of huoxiang zhengqi oral liquid on intestinal mucosal mechanical barrier of rats with postinfectious irritable bowel syndrome induced by acetic Acid. Evid. Based Complement. Alternat. Med. 2014:218383. doi: 10.1155/2014/218383
Lu, H., Zhang, L., Xiao, J., Wu, C., Zhang, H., Chen, Y., et al. (2019). Effect of feeding Chinese herb medicine ageratum-liquid on intestinal bacterial translocations induced by H9N2 AIV in mice. Virol. J. 16:24. doi: 10.1186/s12985-019-1131-y
Lu, Y., Li, D., and Tang, F. (2011). [Mechanism of huoxiang zhengqi extract for regulating the intestinal motility in rat model of diarrhea-predominant irritable bowel syndrome]. Zhongguo Zhong Xi Yi Jie He Za Zhi 31, 941–945.
Ma, H. D., Zhao, Z. B., Ma, W. T., Liu, Q. Z., Gao, C. Y., Li, L., et al. (2018). Gut microbiota translocation promotes autoimmune cholangitis. J. Autoimmun. 95, 47–57. doi: 10.1016/j.jaut.2018.09.010
Maharshak, N., Packey, C. D., Ellermann, M., Manick, S., Siddle, J. P., Huh, E. Y., et al. (2013). Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 4, 316–324. doi: 10.4161/gmic.25486
Mancini, M. V., Damiani, C., Accoti, A., Tallarita, M., Nunzi, E., Cappelli, A., et al. (2018). Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 18:126. doi: 10.1186/s12866-018-1266-9
Marc, D. (2016). Stop-codon variations in non-structural protein NS1 of avian influenza viruses. Virulence 7, 498–501. doi: 10.1080/21505594.2016.1175802
Nguyen, G. T., Rauw, F., Steensels, M., Ingrao, F., Bonfante, F., Davidson, I., et al. (2019). Study of the underlying mechanisms and consequences of pathogenicity differences between two in vitro selected G1-H9N2 clones originating from a single isolate. Vet. Res. 50:18. doi: 10.1186/s13567-019-0635-1
Oh, H. M., Kang, Y. J., Kim, S. H., Lee, Y. S., Park, M. K., Heo, J. M., et al. (2005). Agastache rugosa leaf extract inhibits the iNOS expression in ROS 17/2.8 cells activated with TNF-alpha and IL-1beta. Arch. Pharm. Res. 28, 305–310. doi: 10.1007/BF02977797
Pantin-Jackwood, M. J., and Swayne, D. E. (2009). Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev. Sci. Tech. 28, 113–136. doi: 10.20506/rst.28.1.1869
Peacock, T. H. P., James, J., Sealy, J. E., and Iqbal, M. (2019). A global perspective on H9N2 avian influenza virus. Viruses 11:620. doi: 10.3390/v11070620
Qin, N., Zheng, B., Yao, J., Guo, L., Zuo, J., Wu, L., et al. (2015). Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 5:14771. doi: 10.1038/srep14771
Ramakrishna, B. S. (2013). Role of the gut microbiota in human nutrition and metabolism.. J Gastroenterol. Hepatol. 28(Suppl. 4), 9–17. doi: 10.1111/jgh.12294
Resa-Infante, P., Jorba, N., Coloma, R., and Ortin, J. (2011). The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8, 207–215. doi: 10.4161/rna.8.2.14513
Rigottier-Gois, L. (2013). Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 7, 1256–1261. doi: 10.1038/ismej.2013.80
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Salzman, A., Denenberg, A. G., Ueta, I., O’Connor, M., Linn, S. C., and Szabo, C. (1996). Induction and activity of nitric oxide synthase in cultured human intestinal epithelial monolayers. Am. J. Physiol. 270(4 Pt 1), G565–G573. doi: 10.1152/ajpgi.1996.270.4.G565
Samir, A., Adel, A., Arafa, A., Sultan, H., and Hussein Ahmed, H. A. (2019). Molecular pathogenic and host range determinants of reassortant Egyptian low pathogenic avian influenza H9N2 viruses from backyard chicken. Int. J. Vet. Sci. Med. 7, 10–19. doi: 10.1080/23144599.2019.1637046
Samy, A., and Naguib, M. M. (2018). Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet. Sci. 5:23. doi: 10.3390/vetsci5010023
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Su, K., Ye, S., Li, Q., Xie, W., Yu, H., Qi, L., et al. (2019). Influenza A(H7N9) virus emerged and resulted in human infections in Chongqing, southwestern China since 2017. Int. J. Infect. Dis. 81, 244–250. doi: 10.1016/j.ijid.2019.02.011
Takaishi, H., Matsuki, T., Nakazawa, A., Takada, T., Kado, S., Asahara, T., et al. (2008). Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med. Microbiol. 298, 463–472. doi: 10.1016/j.ijmm.2007.07.016
Wang, H., Li, Z., Jia, R., Hou, Y., Yin, J., Bian, X., et al. (2016). RecET direct cloning and Redalphabeta recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 11, 1175–1190. doi: 10.1038/nprot.2016.054
Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. doi: 10.1126/science.1232467
Xiao Joe, J. T., Chiou, P. P., Kuo, C. Y., Jia Lin, J. H., Wu, J. L., and Lu, M. W. (2019). The microbiota profile and transcriptome analysis of immune response during metamorphosis stages in orange spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 90, 141–149. doi: 10.1016/j.fsi.2019.03.063
Xu, W., Luo, Z., Alekseyenko, A. V., Martin, L., Wan, Z., Ling, B., et al. (2018). Distinct systemic microbiome and microbial translocation are associated with plasma level of anti-CD4 autoantibody in HIV infection. Sci. Rep. 8:12863. doi: 10.1038/s41598-018-31116-y
Yildiz, S., Mazel-Sanchez, B., Kandasamy, M., Manicassamy, B., and Schmolke, M. (2018). Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 6:9. doi: 10.1186/s40168-017-0386-z
Yitbarek, A., Alkie, T., Taha-Abdelaziz, K., Astill, J., Rodriguez-Lecompte, J. C., Parkinson, J., et al. (2018a). Gut microbiota modulates type I interferon and antibody-mediated immune responses in chickens infected with influenza virus subtype H9N2. Benef. Microbes 9, 417–427. doi: 10.3920/BM2017.0088
Yitbarek, A., Taha-Abdelaziz, K., Hodgins, D. C., Read, L., Nagy, E., Weese, J. S., et al. (2018b). Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci. Rep. 8:13189. doi: 10.1038/s41598-018-31613-0
Yitbarek, A., Weese, J. S., Alkie, T. N., Parkinson, J., and Sharif, S. (2018c). Influenza A virus subtype H9N2 infection disrupts the composition of intestinal microbiota of chickens. FEMS Microbiol. Ecol. 94:fix165. doi: 10.1093/femsec/fix165
Zhang, X., Wu, B., Liu, Y., Chen, W., Dai, Z., Bi, Y., et al. (2015). Assessing the efficacy of an inactivated chicken anemia virus vaccine. Vaccine 33, 1916–1922. doi: 10.1016/j.vaccine.2015.02.066
Keywords: H9N2 AIV, gut microbiota, E. coli, secondary infection, metabolite, nitrate, ageratum-liquid
Citation: Zhang X, Zhao Q, Wu C, Xie Z, Ci X, Li H, Lin W, Zhang H and Xie Q (2020) Nitrate Is Crucial for the Proliferation of Gut Escherichia coli Caused by H9N2 AIV Infection and Effective Regulation by Chinese Herbal Medicine Ageratum-Liquid. Front. Microbiol. 11:555739. doi: 10.3389/fmicb.2020.555739
Received: 29 April 2020; Accepted: 12 October 2020;
Published: 30 October 2020.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Wenkai Ren, South China Agricultural University, ChinaDaxin Peng, Yangzhou University, China
Jue Liu, Beijing Academy of Agricultural and Forestry Sciences, China
Copyright © 2020 Zhang, Zhao, Wu, Xie, Ci, Li, Lin, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingmei Xie, cW14QHNjYXUuZWR1LmNu
 Xinheng Zhang
Xinheng Zhang Qiqi Zhao1,3,4
Qiqi Zhao1,3,4 Huanmin Zhang
Huanmin Zhang Qingmei Xie
Qingmei Xie