- 1Institute of Entomology, Guizhou University, The Provincial Key Laboratory for Agricultural Pest Management of Mountainous Region, Guiyang, China
- 2Institute of Fungus Resources, Guizhou University, Guiyang, China
Background: Frankliniella occidentalis (Thysanoptera: Thripidae) is a highly rasping-sucking pest of numerous crops. The entomogenous fungi of Lecanicillium spp. are important pathogens of insect pests, and some species have been developed as commercial biopesticides. To explore Lecanicillium spp. resources in the development of more effective F. occidentalis controls, efficient barcode combinations for strain identification were screened from internal transcribed spacers (ITS), SSU, LSU, TEF, RPB1, and RPB2 genes.
Results: Six genes were used to reconstruct Lecanicillium genus phylogeny. The results showed that ITS, TEF, RPB1, and RPB2 could be used to identify the strains. All phylogenetic trees reconstructed by free combination of these four genes exhibited almost the same topology. Bioassay studies of a purified conidial suspension further confirmed the infection of second-instar nymphs and adult female F. occidentalis by seven Lecanicillium strains. L. attenuatum strain GZUIFR-lun1405 was the most virulent, killing approximately 91.67% F. occidentalis adults and 76.67% nymphs after a 7-day exposure. L. attenuatum strain GZUIFR-lun1405 and L. cauligalbarum strain GZUIFR-ZHJ01 were selected to compare the fungal effects on the number of eggs laid by F. occidentalis. The number of F. occidentalis nymphs significantly decreased when F. occidentalis adults were treated with L. cauligalbarum strain GZUIFR-ZHJ01.
Conclusions: The combination of ITS and RPB1 could be used for fast recognition of Lecanicillium spp. This is the first report of the pathogenicity of L. attenuatum, L. cauligalbarum, L araneogenum, and L. aphanocladii against F. occidentalis. Additionally, L. cauligalbarum strain GZUIFR-ZHJ01 caused high F. occidentalis mortality and inhibited the fecundity of the pest.
Introduction
With the development of molecular methods, it has become easier for biodiversity researchers to identify species by DNA barcoding than by morphological methods (Verkley et al., 2013; Vu et al., 2019). The success of DNA barcoding has facilitated the characterization of bacterial biodiversity in every nook and cranny on the planet (Mau et al., 2014; Afshinnekoo et al., 2015). Compared to bacteria, fungi are difficult to definitively identify because their genomes contain large amounts of non-coding and repetitive DNA (Mohanta and Bae, 2015). Internal transcribed spacer (ITS) sequences are useful for delineating many fungal species (Irinyi et al., 2015; Vu et al., 2016). Unfortunately, ITS sequences from more fungal species are required in GenBank (Hawksworth and Lücking, 2017), and some of the submitted sequences are of poor quality (Nilsson et al., 2006; Vu et al., 2016). Therefore, multilocus (TEF, RPB1, RPB2, LSU, and SSU) sequence data has been used to identify the phylogenetic relationship of some fungi, including as the Ophiocordycipitaceae (Sung et al., 2007; Kepler et al., 2017). However, it would be laborious if analysis of six genes was required to identify each newly collected strain. Some scholars suggest that ITS regions and at least one protein encoding gene with sufficient genetic variation, such as TEF or RPB2, should be sufficient for strain identification instead of using conserved 28S or 18S rDNA sequences (Jeewon and Hyde, 2016). Therefore, identifying the minimum ITS and gene combination required for accurate strain identification will save a lot of time.
Frankliniella occidentalis (Pergande) is a highly polyphagous herbivore that is an economically pernicious pest of many crops and results in especially serious losses to the vegetable and flower industries (van Rijn et al., 1995; Reitz et al., 2011). F. occidentalis feeds on the tender parts of crops and flowers or oviposits, which reduces crop productivity and quality (Shipp et al., 2000). The pest also facilitates the transmission of plant viruses, including tomato spotted-wilt virus and impatiens necrotic-spot virus (Whitfield et al., 2005; Ogada and Poehling, 2015). F. occidentalis was first described in California, United States (Morse and Hoddle, 2006). Facilitated by increasing international agriculture exchanges, F. occidentalis subsequently became a major worldwide pest after the first pesticide-resistant strain was discovered in the 1970s (Mouden et al., 2017; Reitz et al., 2020). F. occidentalis was first reported as an invasive pest in Beijing in 2003 (Zhang et al., 2003). It then gradually spread to many provinces in China, where it has been a dominant pest of floriculture and vegetable crops (Yang et al., 2015). At present, F. occidentalis can be found in more than 10 provinces throughout China (Zhao et al., 2017). Additionally, it has been predicted that F. occidentalis could spread to, and successfully overwinter in, more northern areas of China because of climate change (Wu et al., 2017).
The most common F. occidentalis control measures are chemical pesticides. However, their efficacy is often poor because F. occidentalis tends to reside within the flowers and buds, where it is difficult to reach with spray (Loughner et al., 2005; Demirozer et al., 2012; Mouden et al., 2017). Moreover, varying degrees of resistance are developing due to the overuse of pesticides (Zhao et al., 1995; Bielza et al., 2007; Wang et al., 2011; Gao et al., 2012). Therefore, biological control has received increasing amounts of attention. Representative entomopathogenic fungi for F. occidentalis, including Beauveria bassiana (Gao et al., 2012; Wu et al., 2015; Lee et al., 2017), Metarhizium anisopliae (Ansari et al., 2007; Skinner et al., 2012; Koupi et al., 2015), Lecanicillium lecanii (Schreiter et al., 1994; Vestergaard et al., 1995; Gouli et al., 2009), and Isaria fumosorosea (Ansari et al., 2008; Kivett et al., 2016), are of particular interest to researchers (Wu et al., 2018). Unfortunately, very little is known about the ability of various Lecanicillium species other than typical species of L. lacani to control F. occidentalis. Therefore, identifying new entomopathogenic fungi, and fast recognition of existing species capable of controlling F. occidentalis, are priorities for our research group because of the needs and demands of the majority of crop growers (Wu et al., 2018).
As insect pathogens, Lecanicillium spp. are effective biological control agents for numerous plant diseases, insect pests, and plant-parasitic nematodes (Goettel et al., 2008). To date, almost 15 Lecanicillium spp.-based commercial preparations have been, or are currently being, developed (Faria and Wraight, 2007). Kepler et al. (2017) concluded that Lecanicillium should be incorporated into Akanthomyces and formally transferred a number of Lecanicillium species, including L. attenuatum, L. muscarium, and L. sabanense. However, Lecanicillium were not very compatible with Akanthomyces. In addition, in later research, the name Lecanicillium was used again, for example L. testudineum and L. restrictum (Crous et al., 2018), L. subprimulinum (Huang et al., 2018), L. coprophilum (Su et al., 2019). To better use Lecanicillium to control F. occidentalis, we use the previous taxon to carry out our work.
Presently, more than 30 Lecanicillium species have been formally described and listed in the Fungorum Index1. Considering that the information provided by ITS limited, researchers have used different genes to identify new Lecanicillium species. This approach successfully identified two new Lecanicillium species (L. araneogenum and L. uredinophilum) using multilocus (TEF, RPB1, RPB2, LSU, and SSU) sequence data (Park et al., 2016; Chen et al., 2017). Two other new species (L. sabanense and L. cauligalbarum) were identified based on combined multilocus and ITS sequence phylogenetic analysis (Chiriví-Salomón et al., 2015; Zhou et al., 2018a). Therefore, screening for the most efficient gene combinations for fungal strains identification will have particular significance. These data will facilitate the exploitation of Lecanicillium spp. resources in a more time and budget efficient manner.
Materials and Methods
Entomopathogenic Fungi
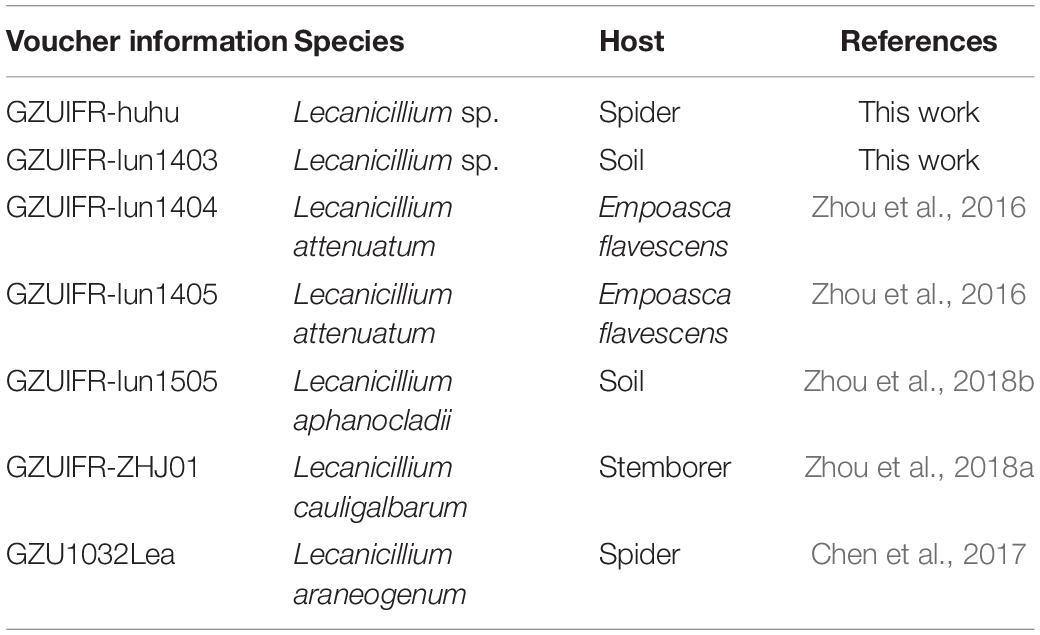
Samples were collected between 2014 and 2018 from forests or tea plantations in Guizhou Province, China. Specimens were preliminarily identified as Lecanicillium spp. by their morphology. Some specimens had been identified and some had not (Table 1). All strains were deposited in GZAC, Guizhou University, Guiyang. The details of the strains are listed in Table 1.
Insects
F. occidentalis were collected from vegetable plants in the Guiyang area of Guizhou Province, China, and were used to establish a laboratory colony. The individuals were reared and maintained on kidney bean pods in the laboratory of the Institute of Entomology, Guizhou University. The thrips were reared in an artificial-climate chest, with a photoperiod of 14 h light, 10 h dark at 25 ± 1°C and relative humidity of 70 ± 5%.
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted as previously described (Zhou et al., 2018a). The ITS, SSU, LSU, TEF, RPB1, and RPB2 regions were amplified using previously described primers (Zhou et al., 2018a).
Comparison of Combination Identification
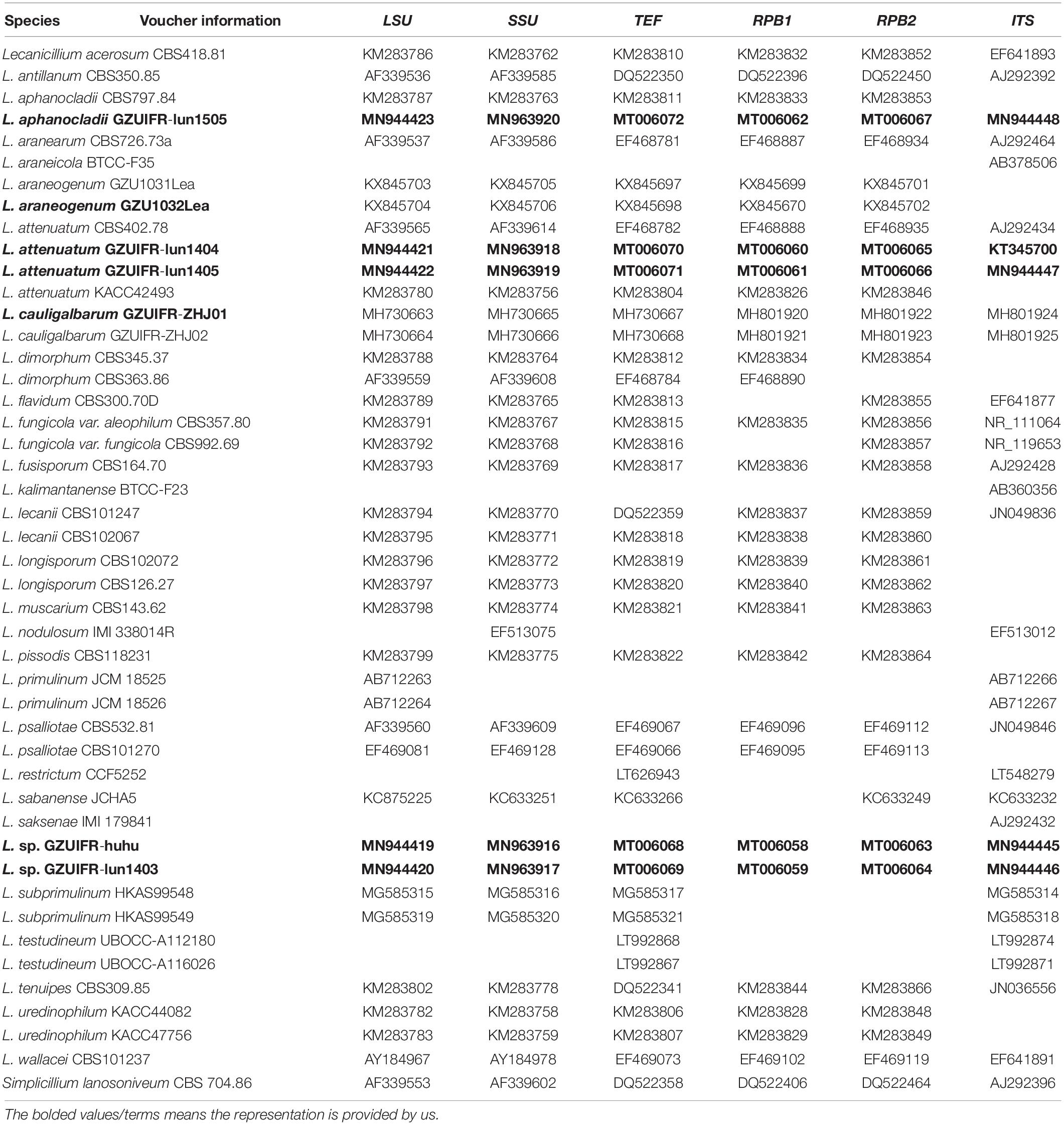
DNA sequences used in this study were edited using the LASERGENE software (version 6.0, DNASTAR). Sequences downloaded from GenBank2 comprised 29 Lecanicillium taxa and one sequence from Simplicillium lanosoniveum CBS 704.86, which was used as the outgroup (Huang et al., 2018). A total of 46 sequence submissions were selected (Table 2). Multiple sequence alignments for TEF, RPB1, and RPB2 were performed in MAFFT with the default settings (Katoh et al., 2009). Multiple sequence alignments for ITS, LSU, and SSU were conducted using the MUSCLE algorithm (Edgar, 2004) from MEGA 6 (Tamura et al., 2013). A multiple alignment of the combined partial sequences was assembled with MEGA 6 (Tamura et al., 2013) and SEQUENCEMATRIX 1.7.8 (Vaidya et al., 2011). The command “hompart” in PAUP∗ 4.0b10 was used for assessment of concordance amongst the genes and the ITS region (Swofford, 2001).
Bayesian inference (BI) was performed using MRBAYES 3.2 (Ronquist et al., 2012) and maximum likelihood (ML) analysis was performed using RAxML (Alexandros, 2014). The ML and BI analyses were performed for each of the six genes (ITS, LSU, SSU, TEF, RPB1, and RPB2). Portfolio analysis was performed for ML and BI analyses on the basis of single gene analysis. Nucleotide substitution models were determined by MrModeltest 2.3 (Nylander et al., 2004). For BI, 10 000 000 generations were performed, with one tree selected every 500th generation. For ML, the GTRGAMMA model was used, and a bootstrap analysis with 1,000 replicates was performed to assess statistical support for the tree topology. Phylogenetic trees were viewed with TREEGRAPH3.
Bioassay
The virulence of Lecanicillium spp. strains against second-instar nymphs and adult females was tested. Second, the biometrics of the adults and the number of nymphs hatched under following treatment with each strain treatment were compared. Strains that significantly reduced the number of nymphs underwent preliminary exploration of thrip fecundity to assess whether the strain directly affected fertility and hatchability.
Preparation of Spore Suspension
The strains were routinely grown on PDA. Plates were incubated at 25°C for 14-day and aerial conidia were harvested by flooding the plate with 0.025% Tween-80. The conidial suspension was vortexed for 1 min and filtered through double layers of lens paper. The final spore concentrations were resuspended to 5 × 108 conidia ml–1 as determined by counting using a hemocytometer (Shanghai Qiujing Biochemical Reagent Instrument Co., Ltd.).
Inoculation of Adult Females
Females were inoculated using the leaf dipping method with some modifications (bean pods were used instead of leaves) (Zhang et al., 2008). The bean pods were cut longitudinally into long strips of equal size. They were placed in the prepared spore suspension for 15 s, then removed and allowed to dry naturally. The pods were fixed with a paper clip placed in the fingertip tube. Twenty female adults (2–3-day old) F. occidentalis were transferred with suction trap into the fingertip tube which was then sealed with cotton wool. The experiment was repeated four times, and 0.025% Tween-80 solution was used as a control. The pods were placed in a light incubator (Jiangnan Instrument, model: RXZ-380A) with a temperature of 25 ± 1 C, 70% relative humidity, and a 12 h light/dark photoperiod. The thrip death was observed, and the number of deaths recorded daily for 7 days. Thrips were considered dead if they were unable to stand upright and/or move forward when probed by a hair pencil tip. At the end of the experiment, dead adults were removed and placed in a petri dish with wet filter paper at the bottom for wet culture. Those that showed white mycelia grow around the thrips and typical conidiogenous structure observed by microscope were considered effectively infected.
Inoculation of the Second-Instar Nnymphs
The leaf dipping method was adapted by cutting the leaves of common bean into small round blocks of about 1 cm2. The leaves of the common bean were soaked in the prepared spore suspension for 15 s and removed and dried naturally. Thirty second-instar nymphs were placed into a 25-mL conjoined plastic dip box (4.3 cm × 3.1 cm × 3 cm, with mesh glued to the holes at the top). The experiment was repeated four times, and 0.025% Tween-80 solution was used as the control. Nymphs were placed into a light incubator with 25 ± 1°C, relative humidity of 70%, and a 12 h light/dark cycle. Thrip death was observed, and the number of deaths was recorded daily for 7 days. At the end of the experiment, dead thrips were removed and placed in a petri dish with wet filter paper at the bottom for wet culture. Those that showed spore structure after incubation were considered effectively infected.
Effects on F. occidentalis Fertility and Hatchability
Eggs of thrips are laid inside the bean pod tissue and are not easily visualized. Therefore, the number of eggs could not be determined. The difference in hatchability was determined by comparing the number of nymphs that appeared after the same number of thrips had laid their eggs over the same period of time. There were two experiments, the first involved treating the bean pods within which thrips had deposited eggs, and the other involved treating bean pods without eggs. The difference resulting from treatment after eggs were laid represented the difference in hatchability, while the difference resulting from the treatment before eggs were laid represented the difference caused by both fertility and hatchability.
In the first experiment, 20 female adult (2–3-day old) F. occidentalis were transferred by suction trap into the fingertip tube and sealed with cotton wool. After 24 h, the pods were placed into the spore suspension for 15 s and transferred into a new fingertip tube to identify the effects on the hatchability of eggs in pods. In the second experiment, pods were placed into the spore suspension for 15 s and transferred into a new fingertip tube. Twenty F. occidentalis adults female (2–3-day old) were transferred by suction trap into the fingertip tube, which was immediately sealed with cotton wool. Each experiment was repeated nine times, and 0.025% Tween-80 solution was used as the control. The pods were placed in a light incubator with a temperature of 25 ± 1°C, relative humidity of 70%, and 12 h light/dark cycle. The number of nymphs that emerged after 7-d was recorded.
Statistical Analyses
F. occidentalis mortality was analyzed by one-way ANOVA using SPSS software (version 18.0; SPSS, Chicago, IL, United States). Tukey’s honestly significant difference test (P < 0.05) was used to analyze the survival rate of the adults and nymphs and the number of nymphs that hatched. Inverse sine square root transformation was used for the percentage data before the ANOVA analysis. All data are presented as mean ± SE. The corrected percent mortality was calculated by the Abbott’s formula (Abbott, 1925).
Results
Sequencing and Phylogenetic Analysis
Two software programs (MRBAYES 3.2 and RAxML) were used to identify seven strains (Table 1) based on ITS, SSU, LSU, TEF, RPB1, and RPB2 alone. Among these, ITS, TEF, RPB1, and RPB2 were validated and useful for identifying the strains (phylogenetic incongruence with statistical measures of confidence and Supplementary Figures 1–4). The phylogenetic relationship analysis revealed that LSU and SSU could not be used for identifying the strains alone because of the small interspecific differences. The ITS + TEF + RPB1 + RPB2 partial sequence-based phylogenetic tree was formed using almost all Lecanicillium species (L. evansii was not in NCBI) and one Simplicillium species (S. lanosoniveum) as outgroup (Figure 1). The sequence dataset consisted of 2,735 bases, including inserted gaps (ITS: 523 bp; TEF: 772 bp; RPB1: 555 bp; and RPB2: 885 bp). L. attenuatum GZUIFR-lun1404 and L. attenuatum GZUIFR-lun1405 formed one branch in a polytomy together with a clade containing L. attenuatum (BI posterior probabilities 0.99, ML bootstrap 98%). Lecanicillium sp. GZUIFR- lun1403 formed one branch in a polytomy together with a clade containing L. lecanii (BI posterior probabilities 0.98, ML bootstrap 97%). L. araneogenum GZU1032Lea and L. araneogenum, L. aphanocladii GZUIFR-lun1505 and L. aphanocladii, and L. cauligalbarum GZUIFR-ZHJ01 and L. cauligalbarum, formed independent branches (BI posterior probabilities 1, ML bootstrap 100%). Lecanicillium sp. GZUIFR-huhu formed one branch in a polytomy together with a clade containing L. tenuipes (BI posterior probabilities 1, ML bootstrap 100%) (Figure 1). The other phylogenetic trees exhibited almost the same topology (Supplementary Figures 5–9 and Figure 2). No significant differences in topology were observed between BI and ML phylogenies.
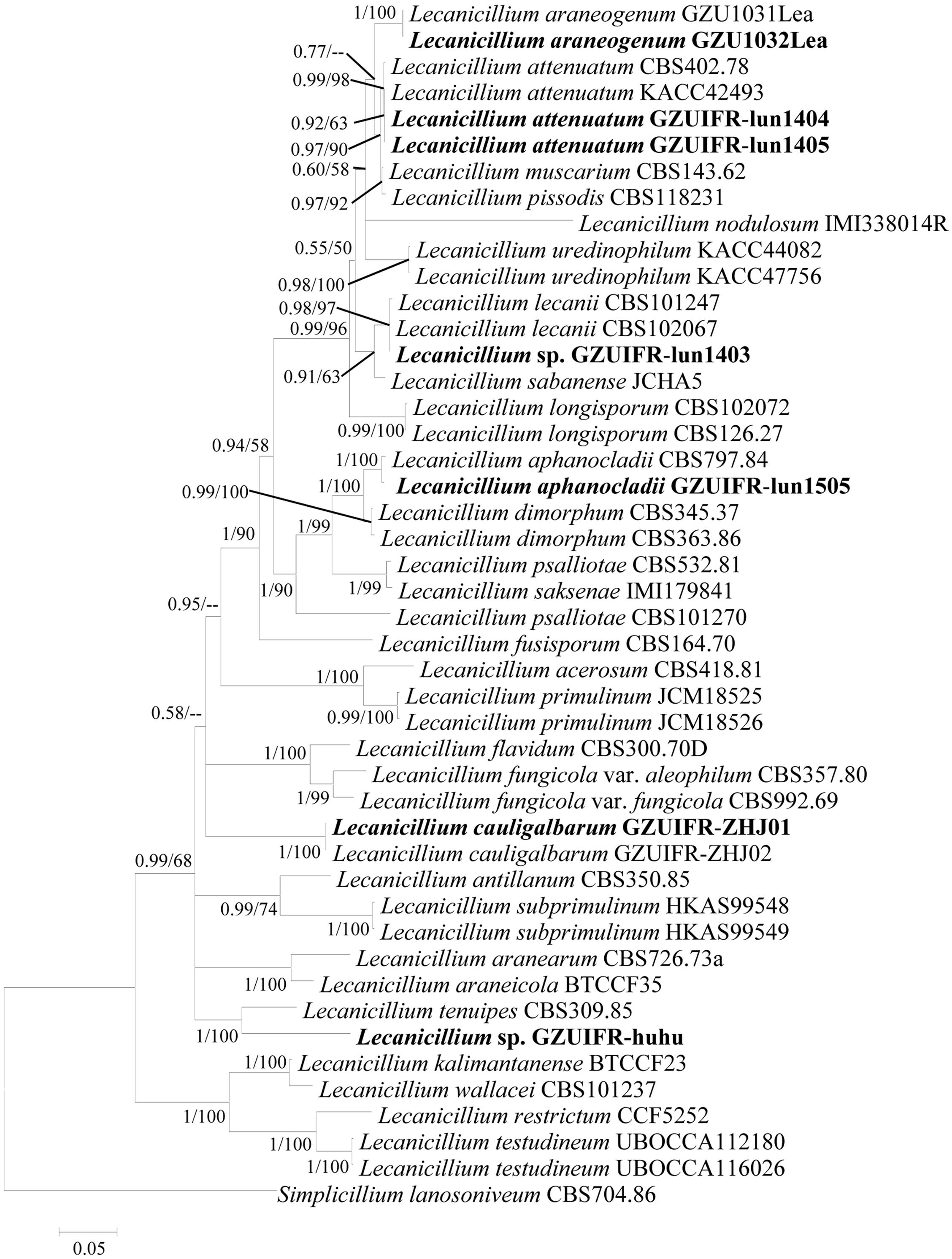
Figure 1. Phylogenetic analysis of isolated strains and related species based on partial ITS + TEF + RPB1 + RPB2 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
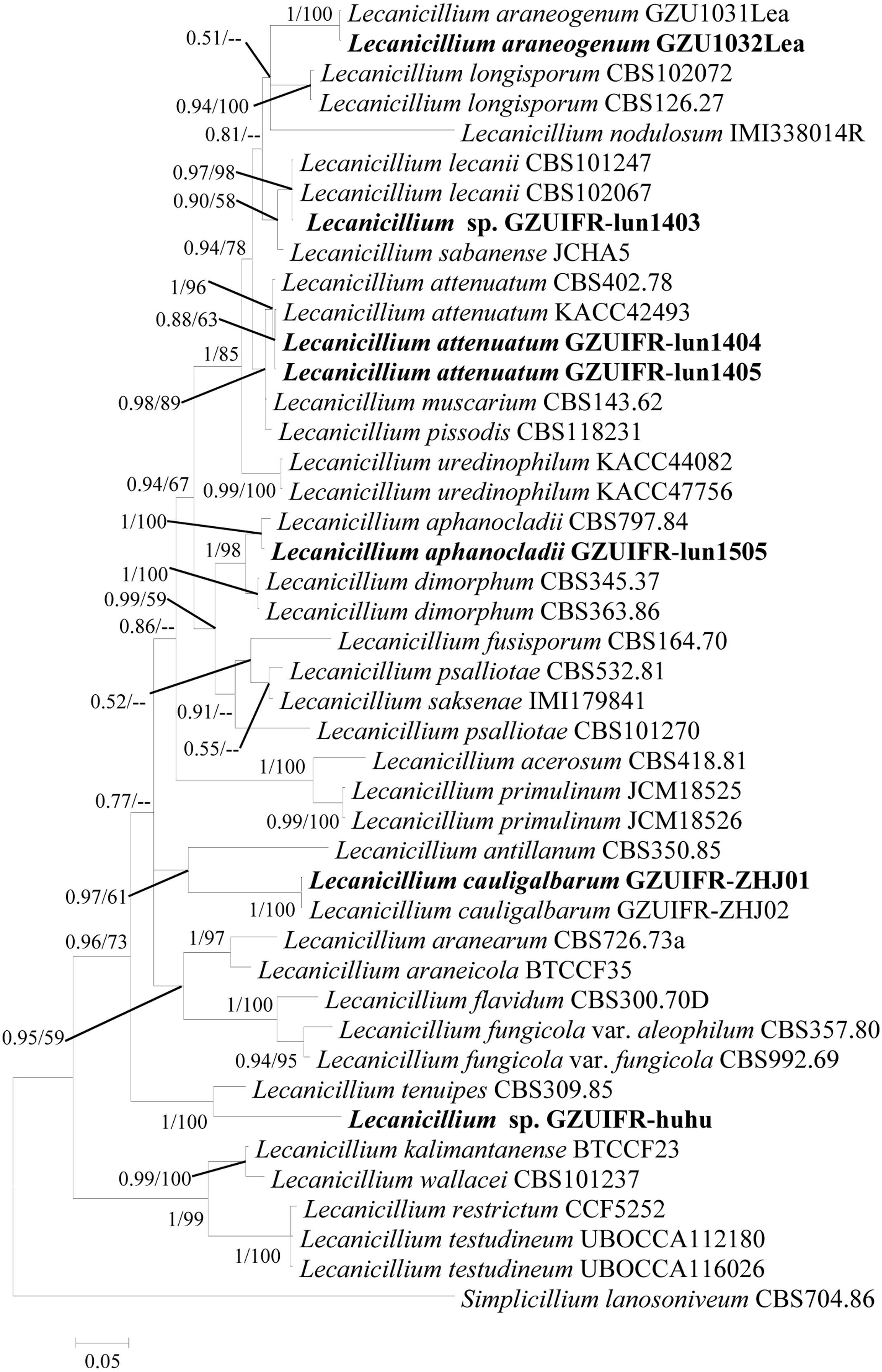
Figure 2. Phylogenetic analysis of isolated strains and related species derived from partial ITS + RPB1 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Virulence Comparison of the Seven Fungal Strains Infecting F. occidentalis
Lecanicillium sp. GZUIFR-huhu was non-virulent to F. occidentalis. The other six strains were pathogenic to F. occidentalis, but their virulence varied (Figure 3 and Supplementary Table 1). L. attenuatum strains GZUIFR-lun1404 and GZUIFR-lun1405 were the most virulent of the six strains from the five species. Approximately 8.33% of adult thrips survived after a 7-day exposure to these two L. attenuatum strains. Five strains showing effectiveness against F. occidentalis adult females were also pathogenic to second-instar nymphs. L. attenuatum strain GZUIFR-lun1405 was the most virulent, with approximately 23.33% of nymphs surviving after a 7-day exposure (Figure 4 and Supplementary Table 2).
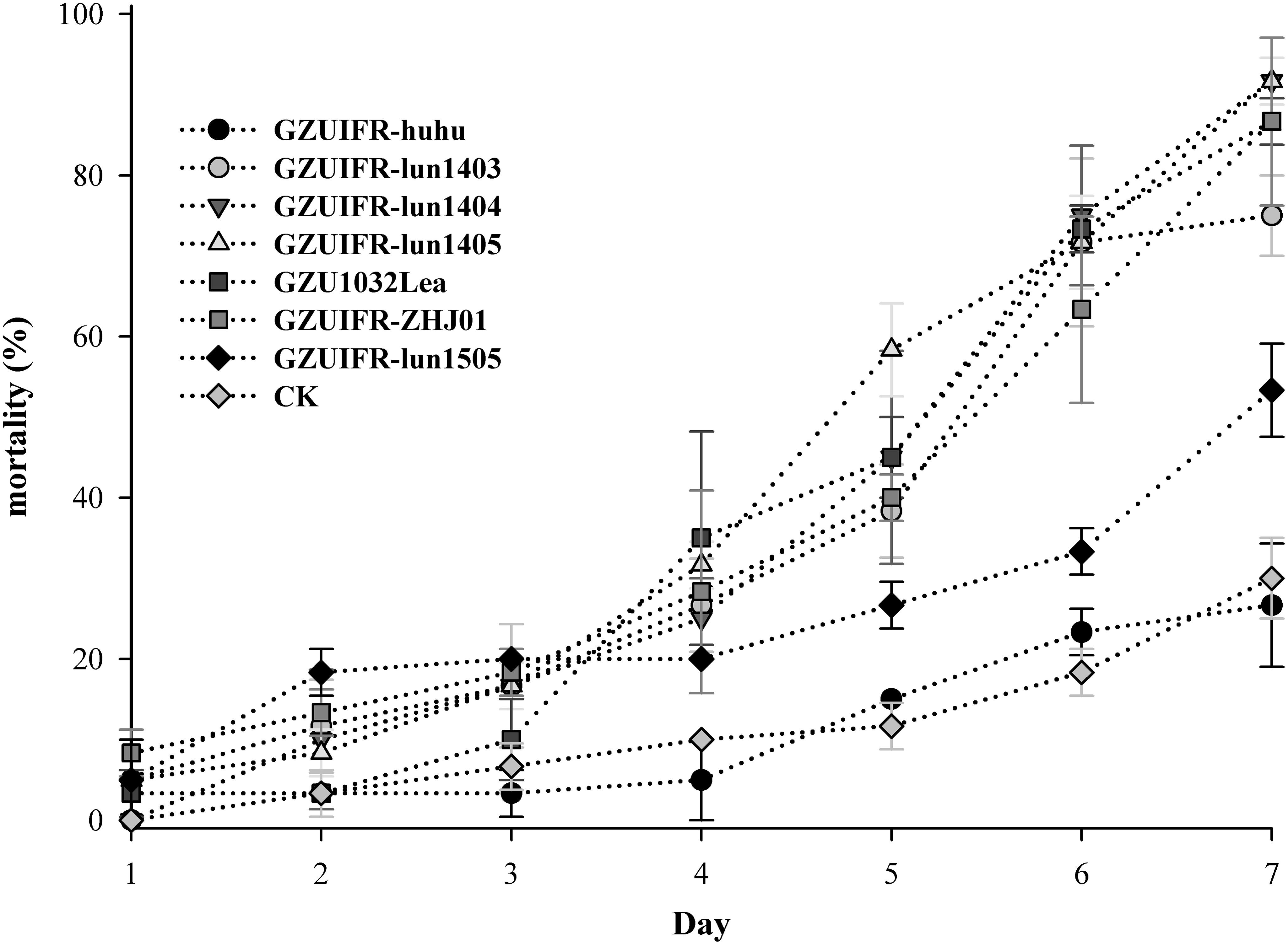
Figure 3. Mortality (%) of F. occidentalis adults after inoculation with different fungal species (mean ± SE).
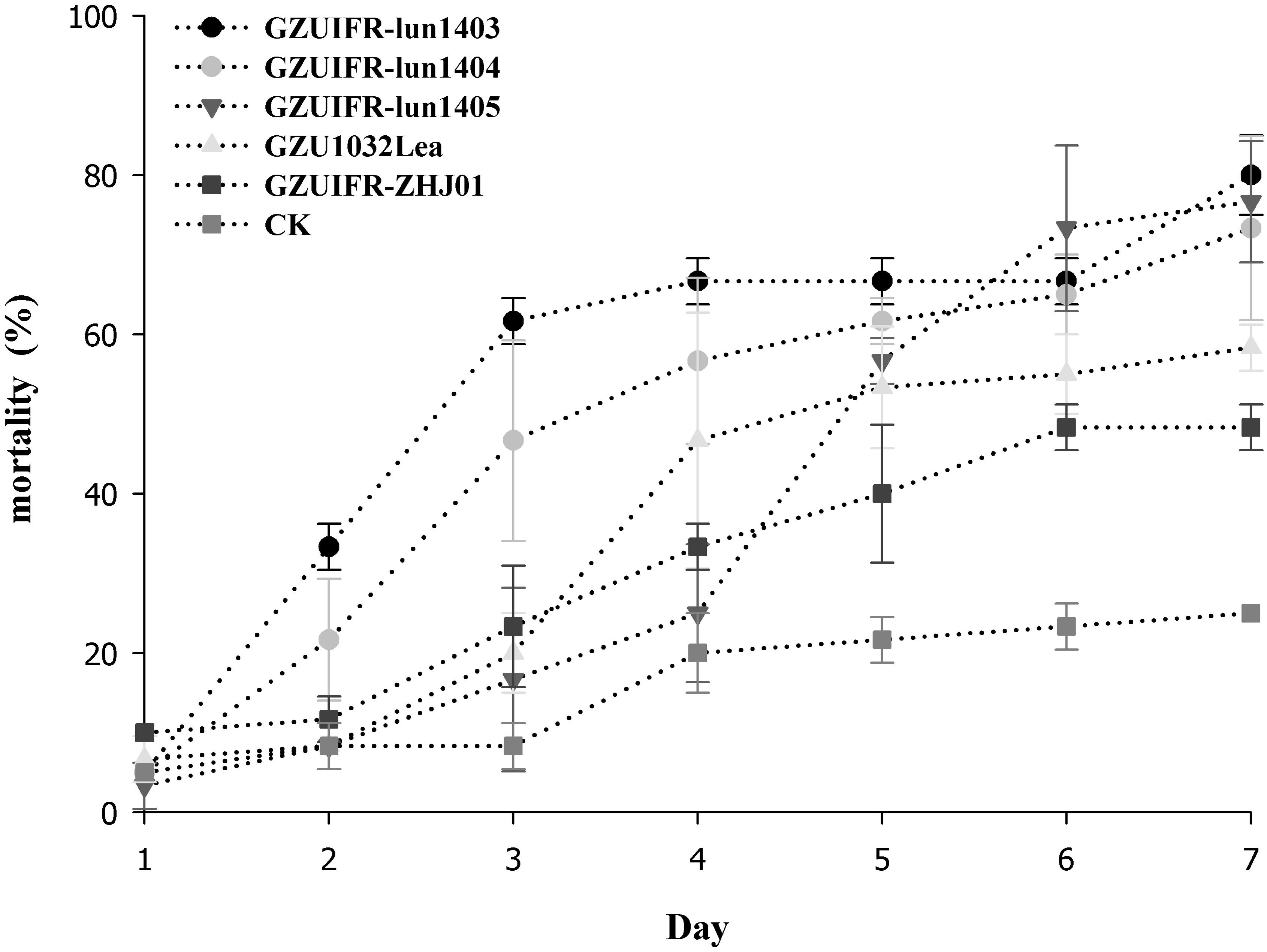
Figure 4. Mortality (%) of F. occidentalis second instar nymphs after inoculation with different fungal species (mean ± SE).
Differences were found in the number of nymphs hatched by thrips after exposure to different strains. There were no significant differences among Lecanicillium sp. GZUIFR-huhu, L. attenuatum strain GZUIFR-lun1405, and control groups. The group treated with L. cauligalbarum strain GZUIFR-ZHJ01 had the fewest survivors, and their number of survivors was significantly fewer than those in other treatments and in the control group (F7,23 = 28.798; p < 0.0001) (Figure 5). L. attenuatum strain GZUIFR- lun1405 and L. cauligalbarum strain GZUIFR-ZHJ01 were selected for future analysis on the thrips fecundity. There was no significant difference among the number of nymphs hatched from experimental and control groups (F2,26 = 7.925; p = 0.0023) (Figure 6). However, the number of F. occidentalis nymphs significantly decreased when adult F. occidentalis were treated with L. cauligalbarum strain GZUIFR-ZHJ01 (F2,26 = 54.117; p < 0.0001) (Figure 6). These results show that the L. cauligalbarum strain GZUIFR-ZHJ01 affected fertility of the thrips.
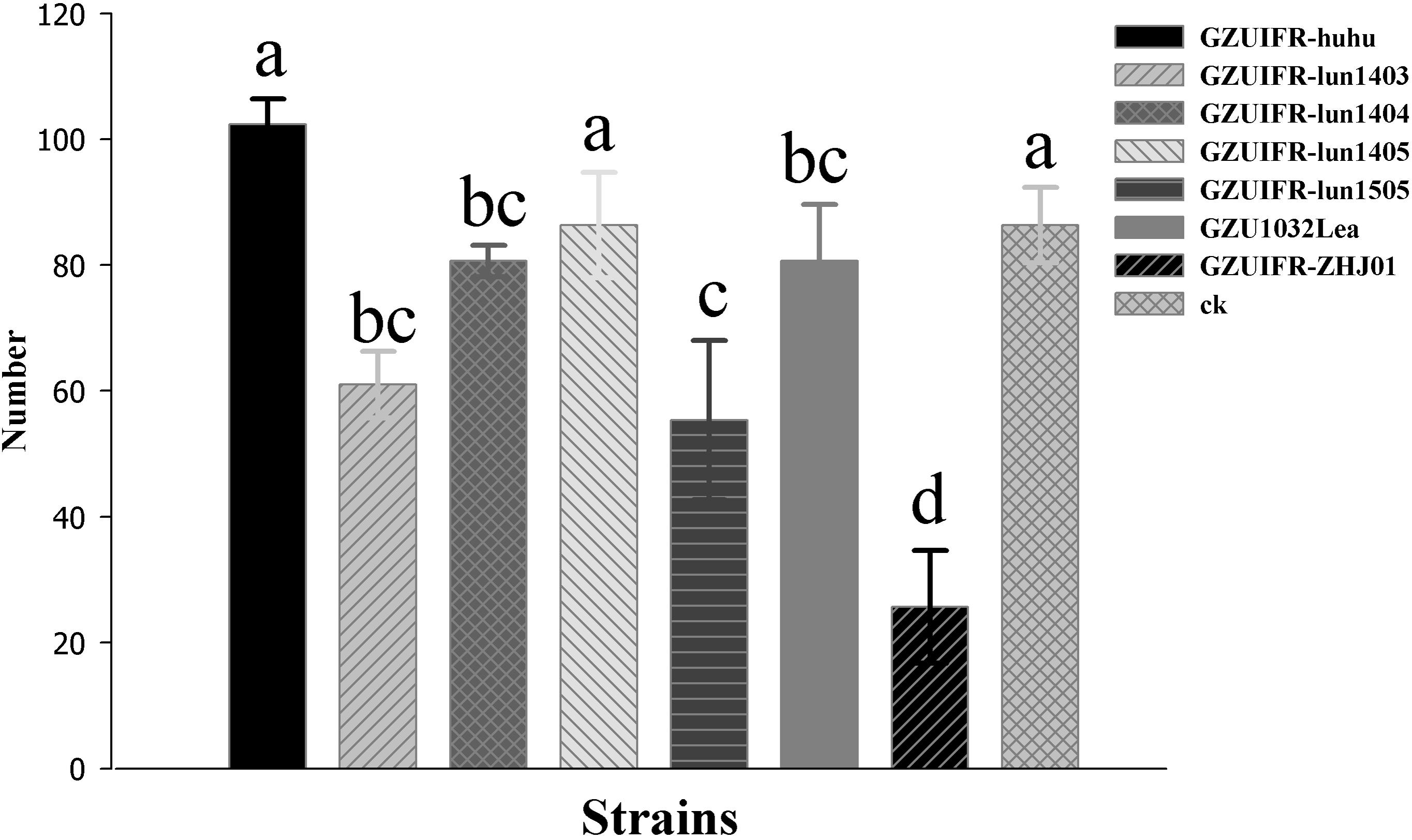
Figure 5. The number of nymphs produced by F. occidentalis adult females after the bioassay experiment. Bars represented by the same letter are not significantly different by Tukey–Kramer grouping for least squares means (a = 0.05) (F7,23 = 28.80; p = 0.0001).
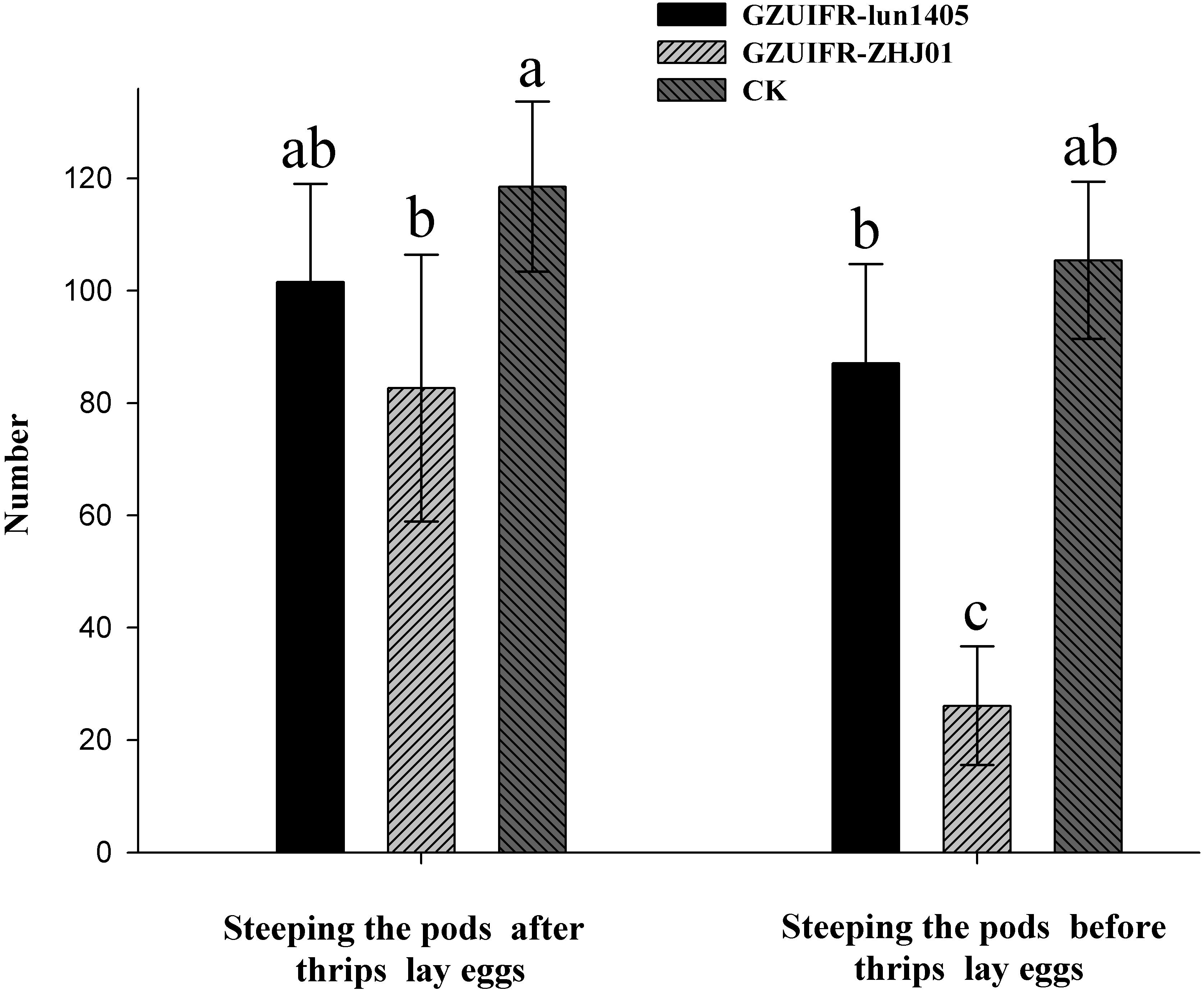
Figure 6. The numbers of F. occidentalis nymphs produced following different treatments. Bars represented by the same letter are not significantly different by Tukey–Kramer grouping for least squares means (a = 0.05) (F5,52 = 30.13; p = 0.0001).
Discussion
DNA barcoding is proposed as an effective way to delineate and distinguish closely related species (Vu et al., 2019). A major advantage of DNA barcoding is that it usually does not require a trained technician and an experienced professional taxonomist. As part of Ophiocrdcipitaceae, Lecanicillium species are identified by ITS and multilocus (TEF, RPB1, RPB2, LSU, and SSU) sequence data. In general, combining barcodes shows clear benefits for species discrimination and provides higher discriminatory power than a single locus (Vu et al., 2016). However, we found no substantial benefit of using multilocus barcodes. In the present study, LSU and SSU could not successfully identify all species of Lecanicillium alone. Only TEF, RPB1, and RPB2 were found to be useful for identifying all species alone. Our phylogenetic analyses reveal that an RPB1- based single locus tree showed better topology than did those based on TEF and RPB2. However, any one of these genes combined with ITS could also clearly distinguish the Lecanicillium genus. We believe a combination of ITS and RPB1 is optimal for the efficient recognition of Lecanicillium spp. based on the richness of each sequence. Moreover, ITS and RPB1 data already cover almost all species in the genus.
The two phylogenetic trees created in this study (Figures 1, 2) were formed with almost all Lecanicillium species. In previous reports, species aggregated with their corresponding sequences and maintained high approval ratings (Table 1). Lecanicillium sp. strain GZUIFR-lun1403 was identified as L. lecanii based on phylogenetic results. Although the Lecanicillium sp. strain GZUIFR-huhu was close to L. tenuipes in the phylogenetic tree, the frequency of base substitutions indicated a clear distinction from L. tenuipes (Figure 1). Therefore, we considered Lecanicillium sp. strain GZUIFR-huhu could be one new species. In all phylogenetic trees two L. psalliotae strains were not together in one branch. This indicated that the L. psalliotae strains require additional confirmation of the current taxon name by CBS.
Currently, the microbial agents used for F. occidentalis control are mainly Beauveria bassiana sensu lato (s.l.), Metarhizium anisopliae s.l., Isaria fumosorosea, and Lecanicillium lecanii strains (Wu et al., 2018). We found that L. attenuatum, L. cauligalbarum, L. araneogenum, and L. aphanocladii were pathogenic to F. occidentalis, but their virulence varied. This is the first report detailing the pathogenicity of these strains to F. occidentalis. The different host origin of our Lecanicillium strains do not appear to be directly associated with F. occidentalis virulence. Considering that L. lecanii has pathogenicity to many other thrips, we believe that the virulence of these strains to other thrips deserves further examination (Wu et al., 2018). In addition to direct pathogenicity, sublethal effects of the examined fungi on thrips, including fecundity, longevity, and feeding ability, may be the most important aspects of exposure to microbial control agents (Wu et al., 2018). Zhang et al. (2015) reported that a strain of B. bassiana, GZGY-1-3, significantly reduced the reproductive success of F. occidentalis and the subsequent fitness of their progeny. Additionally, one M. anisopliae strain significantly reduced the fecundity, egg fertility, and longevity of Megalurothrips sjostedti adults (Ekesi and Maniania, 2000). In this article, L. cauligalbarum strain GZUIFR-ZHJ01 did not produce the highest mortality rate in adults or nymphs, but significantly reduced the number of eggs laid by F. occidentalis. This effect could be an important factor to measure in the development of future F. occidentalis control projects. It is possible that the fungus competes with the host for nutrients and, consequentially, interferes with egg fertility at the physiological level. This sublethal effect could partially offset the relatively slow death rate of fungal infection compared to conventional insecticides and may play an important role in the control of thrips (Wu et al., 2018). The additional sublethal effects of the L. cauligalbarum strain GZUIFR-ZHJ01 on F. occidentalis need to be individually verified.
Conclusion
Based on the performance of single barcodes and their combinations, we recommend combination of ITS and RPB1 as the best barcodes for Lecanicillium identification. These two sequences individually do not currently cover all Lecanicillium species, so a combination of ITS and RPB1 should be used for fast recognition of Lecanicillium spp. We have provided the first demonstration of the virulence of several Lecanicillium spp. against F. occidentalis, such as L. attenuatum, L. cauligalbarum, L araneogenum, and L. aphanocladii. In addition, the L. cauligalbarum strain GZUIFR-ZHJ01 caused both high F. occidentalis mortality and inhibited the fecundity of the pest.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Frankliniella occidentalis samples were maintained at our greenhouse at Institute of Entomology, Guizhou University, China – no permission was required.
Author Contributions
YMZ performed the experiments, analyzed data, and wrote the article. XZ provided strains of fungus and revised the manuscript. JRZ designed the experiments and revised the manuscript. TJ analyzed the data. JQX performed the experiments. All the authors reviewed the manuscript.
Funding
This study was supported by the National Science Foundation of China (Project number: 31860507) and Guizhou international science and technology cooperation base [Project number: G(2016)5802].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Charlesworth Group (http://charlesworth.com.cn/) for editing a draft of this manuscript. This manuscript has been released as a pre-print at research square (Zhou et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.561381/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of the isolated strains and related species derived from partial ITS sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 2 | Phylogenetic analysis of the isolated strains and related species derived from partial TEF sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 3 | Phylogenetic analysis of the isolated strains and related species derived from partial RPB1 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 4 | Phylogenetic analysis of the isolated strains and related species derived from partial RPB2 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 5 | Phylogenetic analysis of the isolated strains and related species deriving from partial ITS + TEF sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 6 | Phylogenetic analysis of the isolated strains and related species derived from partial ITS + RPB2 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 7 | Phylogenetic analysis of the isolated strains and related species derived from partial ITS + TEF + RPB1 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 8 | Phylogenetic analysis of the isolated strains and related species derived from partial ITS + TEF + RPB2 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Figure 9 | Phylogenetic analysis of the isolated strains and related species derived from partial ITS + RPB1 + RPB2 sequences. Statistical support values (≥0.5/50%) are shown at the nodes for Bayesian inference posterior probabilities/maximum-likelihood bootstrap support.
Supplementary Table 1 | Comparison of the influences of test strains on the mortality rate (%) of F. occidentalis adults.
Supplementary Table 2 | Comparison of the influences of test strains on the mortality rate (%) of F. occidentalis second instar nymphs.
Abbreviations
Spp., species pluralis; GZAC, the Institute of Fungal Resources of Guizhou University; rDNA, ribosomal DNA; LSU, the large subunits of the rDNA; SSU, the small subunits of the rDNA; TEF, the transcription elongation factor-1 α; ITS, ITS1–5.8S rDNA–ITS2 region, the first and the internal transcribed spacers; RPB1, DNA-directed RNA polymerase II subunit rpb1; RPB2, DNA-directed RNA polymerase II subunit rpb2.
Footnotes
- ^ http://www.indexfungorum.org
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://treegraph.bioinfweb.info/
References
Abbott, W. S. (1925). A method of computing the effectiveness of and insecticide. J. Econom. Entomol. 18, 265–267.
Afshinnekoo, E., Meydan, C., Chowdhury, S., Jaroudi, D., Boyer, C., Bernstein, N., et al. (2015). Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst. 1, 72–87. doi: 10.1016/j.cels.2015.01.001
Alexandros, S. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Ansari, M. A., Brownbridge, M., Shah, F. A., and Butt, T. M. (2008). Efficacy of entomopathogenic fungi against soil-dwelling life stages of western flower thrips, Frankliniella occidentalis, in plant-growing media. Entomol. Exp. Appl. 127, 80–87. doi: 10.1111/j.1570-7458.2008.00674.x
Ansari, M. A., Shah, F. A., Whittaker, M., Prasad, M., and Butt, T. M. (2007). Control of western flower thrips (Frankliniella occidentalis) pupae with Metarhizium anisopliae in peat and peat alternative growing media. Biol. Control 40, 293–297. doi: 10.1016/j.biocontrol.2006.12.007
Bielza, P., Quinto, V., Contreras, J., Torné, M., Martín, A., and Espinosa, P. J. (2007). Resistance to spinosad in the western flower thrips. Frankliniella occidentalis (Pergande), in greenhouses of south-eastern Spain. Pest Manag. Sci. 63, 682–687. doi: 10.1002/ps.1388
Chen, W. H., Han, Y. F., Liang, Z. Q., and Jin, D. C. (2017). Lecanicillium araneogenum sp. nov., a new araneogenous fungus. Phytotaxa 305, 29–34. doi: 10.11646/phytotaxa.305.1.4
Chiriví-Salomón, J. S., Danies, G., Restrepo, S., and Sanjuan, T. (2015). Lecanicillium sabanense sp. nov. (Cordycipitaceae) a new fungal entomopathogen of coccids. Phytotaxa 234, 63–74. doi: 10.11646/phytotaxa.234.1.4
Crous, P. W., Wingfield, M. J., Burgess, T. I., Hardy, G. E. S. J., Gené, J., Guarro, J., et al. (2018). Fungal Planet description sheets: 716-784. Persoonia 40, 240–393. doi: 10.3767/persoonia.2018.40.10
Demirozer, O., Tyler-Julian, K., Funderburk, J., Leppla, N., and Reitz, S. (2012). Frankliniella occidentalis (Pergande) integrated pest management programs for fruiting vegetables in Florida. Pest Manag. Sci. 68, 1537–1545. doi: 10.1002/ps.3389
Edgar, R. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ekesi, S., and Maniania, N. K. (2000). Susceptibility of Megalurothrips sjostedti developmental stages to Metarhizium anisopliae and the effect of infection on feeding, adult fecundity, egg fertility and longevity. Entomol. Exp. Appl. 94, 229–236.
Faria, M. R., and Wraight, S. P. (2007). Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43, 237–256. doi: 10.1016/j.biocontrol.2007.08.001
Gao, Y. L., Lei, Z. R., and Reitz, S. R. (2012). Western flower thrips resistance to insecticides: detection, mechanisms, and management strategies. Pest Manag. Sci. 68, 1111–1121. doi: 10.1002/ps.3305
Gao, Y. L., Reitz, S. R., Wang, J., Xu, X. N., and Lei, Z. R. (2012). Potential of a strain of the entomopathogenic fungus Beauveria bassiana (Hypocreales: Cordycipitaceae) as a biological control agent against western flower thrips, Frankliniella occidentalis (Thysanoptera: Thripidae). Biocontrol. Sci. Technol. 22, 491–495. doi: 10.1080/09583157.2012.662478
Goettel, M. S., Koike, M., Kim, J. J., Aiuchi, D., Shinya, R., and Brodeur, J. (2008). Potential of Lecanicillium spp. for management of insects, nematodes and plant diseases. J. Invertebr. Pathol. 98, 256–261. doi: 10.1016/j.jip.2008.01.009
Gouli, V. V., Gouli, S. Y., Skinner, M., and Shternshis, M. V. (2009). Effect of the entomopathogenic fungi on mortality and injury level of western flower thrips. Frankliniella occidentalis. Archiv. Phytopathol. Plant Protect. 42, 118–123. doi: 10.1080/03235400600951829
Hawksworth, D. L., and Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5:28752818. doi: 10.1128/microbiolspec.FUNK-0052-2016
Huang, S. K., Maharachchikumbura, S. S. N., Jeewon, R., Bhat, D. J., Phookamsak, R., Hyde, K. D., et al. (2018). Lecanicillium subprimulinum (Cordycipitaceae, Hypocreales), a novel species from Baoshan, Yunnan. Phytotaxa 348, 99–108. doi: 10.11646/phytotaxa.348.2.4
Irinyi, L., Serena, C., Garcia-Hermoso, D., Arabatzis, M., Desnos-Ollivier, M., Vu, D., et al. (2015). International society of human and animal mycology (ISHAM)-ITS reference DNA barcoding database-the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 53, 313–337. doi: 10.1093/mmy/myv008
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi-recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677.
Katoh, K., Asimenos, G., and Toh, H. (2009). Multiple alignment of DNA sequences with MAFFT, Methods Mol. Biol. 537, 39–64.
Kepler, R. M., Luangsa-ard, J. J., Hywel-Jones, N. L., Quandt, C. A., Sung, G. H., Rehner, S. A., et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus. 8, 335–353. doi: 10.5598/imafungus.2017.08.02.08
Kivett, J. M., Cloyd, R. A., and Bello, N. M. (2016). Evaluation of entomopathogenic fungi against the western flower thrips (Thysanoptera: Thripidae) under laboratory conditions. J. Entomol. Sci. 51, 274–291. doi: 10.18474/jes16-07.1
Koupi, N., Ghazavi, M., Kamali, K., and Baniameri, V. (2015). Virulence of Iranian isolates of Metarhizium anisopliae on western flower thrips. Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae). J. Crop Protect. 5, 131–138. doi: 10.18869/modares.jcp.5.1.131
Lee, S. J., Kim, S., Kim, J. C., Lee, M. R., Hossain, M. S., Shin, T. S., et al. (2017). Entomopathogenic Beauveria bassiana granules to control soil-dwelling stage of western flower thrips. Frankliniella occidentalis (Thysanoptera: Thripidae). BioControl 62, 639–648. doi: 10.1007/s10526-017-9818-8
Loughner, R. L., Warnock, D. F., and Cloyd, R. A. (2005). Resistance of greenhouse, laboratory, and native populations of western flower thrips to spinosad. Hortscience 40, 146–149.
Mau, R. L., Liu, C. M., Aziz, M., Schwartz, E., Dijkstra, P., Marks, J. C., et al. (2014). Linking soil bacterial biodiversity and soil carbon stability. ISME J. 9, 1477–1480. doi: 10.1038/ismej.2014.205
Mohanta, T. K., and Bae, H. (2015). The diversity of fungal genome. Biol. Proc. Online 17:8. doi: 10.1186/s12575-015-0020-z
Morse, J. G., and Hoddle, M. S. (2006). Invasion biology of thrips. Annu. Rev. Entomol. 51, 67–89. doi: 10.1146/annurev.ento.51.110104.151044
Mouden, S., Sarmiento, K. F., Klinkhamer, P. G. L., and Leiss, K. A. (2017). Integrated pest management in western flower thrips: past, present and future. Pest Manag. Sci. 73, 813–822. doi: 10.1002/ps.4531
Nilsson, R. H., Ryberg, M., Kristiansson, E., Abarenkov, K., Larsson, K., and Koljalg, U. (2006). Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS One 1:e59. doi: 10.1371/journal.pone.0000059
Nylander, J. A. A., Ronquist, F., Huelsenbeck, J. P., Nieves-Aldrey, J., and Buckley, T. (2004). Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67. doi: 10.1080/10635150490264699
Ogada, P. A., and Poehling, H. M. (2015). Sex-specific influences of Frankliniella occidentalis (Western Flower Thrips) in the transmission of Tomato Spotted Wilt Virus (Tospovirus). J. Plant Dis. Prot. 122, 264–274.
Park, M. J., Hong, S. B., and Shin, H. D. (2016). Lecanicillium uredinophilum sp. nov. associated with rust fungi from Korea. Mycotaxon 130, 997–1005. doi: 10.5248/130.997
Reitz, S. R., Gao, Y. L., and Lei, Z. R. (2011). Thrips: pests of concern to China and the United States. Agric. Sci. China 10, 867–892. doi: 10.1016/s1671-2927(11)60344-1
Reitz, S. R., Gao, Y. L., Kirk, W. D. J., Hoddle, M. S., Leiss, I. A., and Funderburk, J. E. (2020). Invasion biology, ecology, and management of western flower thrips. Annu. Rev. Entomol. 65, 17–37. doi: 10.1146/annurev-ento-011019-024947
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schreiter, G., Butt, T., Beckett, A., Vestergaard, S., and Moritz, G. (1994). Invasion and development of Verticillium lecanii in the western flower thrips. Frankliniella occidentalis. Mycol. Res. 98, 1025–1034.
Shipp, J. L., Wang, K., and Binns, M. R. (2000). Economic injury levels for western flower thrips (Thysanoptera: Thripidae) on greenhouse cucumber. J. Econom. Entomol. 93, 1732–1740. doi: 10.1603/0022-0493-93.6.1732
Skinner, M., Gouli, S., Frank, C. E., Parker, B. L., and Kim, J. S. (2012). Management of Frankliniella occidentalis (Thysanoptera: Thripidae) with granular formulations of entomopathogenic fungi. Biol. Control. 63, 246–252. doi: 10.1016/j.biocontrol.2012.08.004
Su, L., Zhu, H., Guo, Y., Du, X., Guo, J., Zhang, L., et al. (2019). Lecanicillium coprophilum (Cordycipitaceae. Hypocreales), a new species of fungus from the feces of Marmota monax in China. Phytotaxa 387, 55–62.
Sung, G. H., Hywel-Jones, N. L., Sung, J. M., Luangsa-ard, J. J., Shrestha, B., and Spatafora, J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.57.01
Swofford, D. L. (2001). Paup∗: Phylogenetic Analysis using Parsimony (and other methods) 4.0. b10. Massachusetts: Sunderland.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mole. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vaidya, G., Lohman, D. J., and Rudolf, M. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180.
van Rijn, P. C. J., Mollema, C., and Steenhuis-Broers, G. M. (1995). Comparative life history studies of Frankliniella occidentalis and Thrips tabaci (Thysanoptera: Thripidae) on cucumber. Bull. Entomol. Res. 85, 285–297.
Verkley, G. J. M., Quaedvlieg, W., Shin, H. D., and Crous, P. W. (2013). A new approach to species delimitation in Septoria. Stud. Mycol. 75, 213–305. doi: 10.3114/sim0018
Vestergaard, S., Gillespie, A. T., Butt, T. M., Schreiter, G., and Eilenberg, J. (1995). Pathogenicity of the hyphomycete fungi Verticillium lecanii and Metarhizium anisopliae to the western flower thrips. Frankliniella occidentalis. Biocontr. Sci. Technol. 5, 185–192. doi: 10.1080/09583159550039909
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Vu, D., Groenewald, M., Szöke, S., Cardinali, G., Eberhardt, U., Stielow, B., et al. (2016). DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Wang, Z. H., Hou, W. J., Hao, C. Y., Wu, Q. J., Xu, B. Y., and Zhang, Y. J. (2011). Monitoring the insecticide resistance of the field populations of western flower thrips. Frankliniella occidentalis in Beijing area. Chin. J. Appl. Entomol. 48, 542–547.
Whitfield, A. E., Ullman, D. E., and German, T. L. (2005). Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43, 459–489. doi: 10.1146/annurev.phyto.43.040204.140017
Wu, S. Y., Gao, Y. L., Xu, X. N., Goettel, M. S., and Lei, Z. R. (2015). Compatibility of Beauveria bassiana with Neoseiulus barkeri for control of Frankliniella occidentalis. J. Integr. Agric. 14, 98–105. doi: 10.1016/s2095-3119(13)60731-5
Wu, S. Y., Tang, L. D., Fang, F., Li, D. G., Yuan, X., Lei, Z. R., et al. (2018). Screening, efficacy and mechanisms of microbial control agents against sucking pest insects asthrips. Adv. Insect Physiol. 55, 201–219. doi: 10.1016/bs.aiip.2018.07.005
Wu, S., Tang, L., Zhang, X., Xing, Z., Lei, Z., and Gao, Y. (2017). A decade of a thrips invasion in China: lessons learned. Ecotoxicology 27, 1032–1038. doi: 10.1007/s10646-017-1864-6
Yang, X. M., Lou, H., Sun, J. T., Zhu, Y. M., Xue, X. F., and Hong, X. Y. (2015). Temporal genetic dynamics of an invasive species, Frankliniella occidentalis (Pergande), in an early phase of establishment. Sci. Rep. 5:11877. doi: 10.1038/srep11877
Zhang, S., Kono, S., Murai, T., and Miyata, T. (2008). Mechanisms of resistance to spinosad in the western flower thrip. Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae). Insect Sci. 15, 125–132.
Zhang, T., Reitz, S. R., Wang, H., and Lei, Z. (2015). Sublethal effects of Beauveria bassiana (Ascomycota: Hypocreales) on life table parameters of Frankliniella occidentalis (Thysanoptera: Thripidae). J. Econom. Entomol. 108, 975–985. doi: 10.1093/jee/tov091
Zhang, Y. J., Wu, Q. J., Xu, B. Y., and Zhu, G. R. (2003). The occurrence and damage of Frankliniella occidentalis (Thysanoptera: Thripidae): a dangerous alien invasive pest in Beijing. Plant Protect. 29, 58–59.
Zhao, G. Y., Liu, W., Brown, J. M., and Knowles, H. O. (1995). Insecticide resistance in field and laboratory strains of western flower thrips (Thysanoptera: Thripidae). J. Econom. Entomol. 88, 1164–1170.
Zhao, X., Reitz, S. R., Yuan, H., Lei, Z., Paini, D. R., and Gao, Y. (2017). Pesticide-mediated interspecific competition between local and invasive thrips pests. Sci. Rep. 7:40512. doi: 10.1038/srep40512
Zhou, Y. M., Zhi, J. R., Ye, M., Zhang, Z. Y., Yue, W. B., and Zou, X. (2018a). Lecanicillium cauligalbarum sp. nov. (Cordycipitaceae. Hypocreales), a novel fungus isolated from a stemborer in the Yao Ren National Forest Mountain Park, Guizhou. MycoKeys 43, 59–74. doi: 10.3897/mycokeys.43.30203
Zhou, Y. M., Zhi, J. R., Zhang, X., Zou, X., and Yang, K. (2018b). Identification and pathogenicity test of a new soil-derived fungus strain of Lecanicillium to Ephestia elutella (Lepidoptera: Pyralidae). Acta Entomol. Sin. 61, 505–510.
Zhou, Y. M., Zou, X., Qu, J. J., Zhou, J. X., and Li, J. (2016). Identification of a parasitic Lecanicillium of tea lesser leafhopper and optimization of sporulation conditions. Microbiol. Chin. 43, 935–941.
Keywords: entomopathogenic fungi, fast recognition, Frankliniella occidentalis, Lecanicillium, pathogenicity
Citation: Zhou YM, Zou X, Zhi JR, Xie JQ and Jiang T (2020) Fast Recognition of Lecanicillium spp., and Its Virulence Against Frankliniella occidentalis. Front. Microbiol. 11:561381. doi: 10.3389/fmicb.2020.561381
Received: 12 May 2020; Accepted: 01 October 2020;
Published: 22 October 2020.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Jing-Zu Sun, Chinese Academy of Sciences, ChinaJavad Karimi, Ferdowsi University of Mashhad, Iran
Yulin Gao, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2020 Zhou, Zou, Zhi, Xie and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junrui Zhi, YWdyLmpyemhpQGd6dS5lZG4uY24=
 Yeming Zhou
Yeming Zhou Xiao Zou
Xiao Zou Junrui Zhi1*
Junrui Zhi1*