- 1Key Laboratory of Microbial Diversity Research and Application of Hebei Province, College of Life Science/Institute of Life Science and Green Development, Hebei University, Baoding, China
- 2The James Hutton Institute, Invergowrie, Dundee, United Kingdom
- 3Key Laboratory of State Forestry Administration for Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, China
- 4Institute of Agricultural Resources and Regional Planning of CAAS, Beijing, China
- 5Institute of Agro-resources and Environment, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang, China
In order to investigated diversity and geographic distribitution of rhizobia associated with invasive Mimosa species, Mimosa nodules and soils around the plants were sampled from five provinces in southern China. In total, 361 isolates were obtained from Mimosa pudica and Mimosa diplotricha in 25 locations. A multi-locus sequence analysis (MLSA) including 16S rRNA, atpD, dnaK, glnA, gyrB, and recA identified the isolates into eight genospecies corresponding to Paraburkhleria mimosarum, Paraburkholderia phymatum, Paraburkholeria carbensis, Cupriavidus taiwanensis, Cupriavidus sp., Rhizobium altiplani, Rhizobium mesoamericanum, and Rhizobium etli. The majority of the isolates were Cupriavidus (62.6%), followed by Paraburkholderia (33.5%) and Rhizobium (2.9%). Cupriavidus strains were more predominant in nodules of M. diplotricha (76.2) than in M. pudica (59.9%), and the distribution of P. phymatum in those two plant species was reverse (3.4:18.2%). Four symbiotypes were defined among the isolates based upon the phylogeny of nodA-nifH genes, represented by P. mimosarum, P. phymatum–P. caribensis, Cupriavidus spp., and Rhizobium spp. The species affiliation and the symbiotype division among the isolates demonstrated the multiple origins of Mimosa rhizobia in China: most were similar to those found in the original centers of Mimosa plants, but Cupriavidus sp. might have a local origin. The unbalanced distribution of symbionts between the two Mimosa species might be related to the soil pH, organic matter and available nitrogen; Cupriavidus spp. generally dominated most of the soils colonized by Mimosa in this study, but it had a particular preference for neutral-alkaline soils with low fertility whereas. While Paraburkholderia spp. preferred more acidic and fertile soils. The Rhizobium spp. tended to prefer neutral–acidic soils with high fertility soils.
Introduction
Leguminous plants are important for their ability to fix-nitrogen in symbiosis with rhizobia, which makes them critical ecologically and economically. Ecologically, the symbiotic N-fixation not only supply the N nutrition to the host legume, but also enhance the soil N content by its root and shoot remnants (Wang et al., 2019). In addition, the wide distribution and specificity between the legume species and their microsymbionts make the same legume species form symbiosis with distinct rhizobial populations/species in the different geographic regions. Therefore, the growth of a legume plant in a certain region could enrich its corresponding rhizobia adapted to the local environment, e.g., the rhizobia are selected by both the host legumes and the soil conditions, mainly soil pH, nutrient (N, P, K and organic material) contents, and salinity (Wang et al., 2019). With the mentioned concern, characterization rhizobia associated with the same legume species grown in different regions will help to understand the evolution or diversification of rhizobia under the double selection from both the host plant and the soil condition, as well as help for screening the high effective rhizobial strains in agricultural sustainable development.
Mimosa species are able to form nitrogen fixing symbiotic associations with soil bacteria collectively termed “rhizobia” (Sprent, 2009; Sprent et al., 2017). Currently, rhizobia are found in two classes: Alpha-rhizobia including species in the well-known genus Rhizobium and other genera in the class Alphaproteobacteria, and Beta-rhizobia covering the symbiotic species in genera Paraburkholderia (splited from Burkholderia), Cupriavidus and Trinickia symbiotica in the class Betaproteobacteria (Gyaneshwar et al., 2011; Peix et al., 2015; Beukes et al., 2017; Sprent et al., 2017; Los Santos et al., 2018). Species in Mimosa genus and another large mimosoid genus Calliandra mainly nodulate with Beta-rhizobia, particularly Paraburkholderia and Trinickia, in its native range in South America, suggesting that the two partners co-evolved (Chen et al., 2005a; Bontemps et al., 2010; dos Reis et al., 2010; Los Santos et al., 2018; Silva et al., 2018). In addition, the three main invasive Mimosa species (M. diplotricha, C. Wright, M. pigra L., and M. pudica L., originated from the netotropics) in Asia, Australia and the Pacific region also preferred Beta-rhizobia for nodulation (Chen et al., 2001, 2003a,b, 2005b; Liu et al., 2007, 2011, 2012; Parker et al., 2007; Elliott et al., 2009; Andrus et al., 2012; Klonowska et al., 2012; Gehlot et al., 2013; Melkonian et al., 2014), and they are closely related to the microsymbionts of Mimosa and related genera in their original regions (Bournaud et al., 2013; da Silva et al., 2012; Mishra et al., 2012; Taulé et al., 2012; Platero et al., 2016; de Castro Pires et al., 2018). For exotic nodulating legumes, access to compatible rhizobial strains in new environments is a critical factor for their successful establishment, and hence, their ability to survive and spread will depend on the presence of compatible symbionts in the soil (Parker et al., 2007). Several studies have indicated that invasive legumes, such as Mimosa species and Dipogon lignosus, have been introduced into their invasive environments together with their symbionts (Liu et al., 2014). Although Alpha-rhizobia have occasionally been isolated from Mimosa species in South American, they either failed to nodulate their hosts of origin or did so ineffectively (Barrett and Parker, 2006; Elliott et al., 2009; Klonowska et al., 2012; Mishra et al., 2012). In addition, Alpha-rhizobia (Rhizobium or Ensifer species) appear to be the dominant symbionts of native Mimosa spp. in central Mexico, central Brazil and India, where the soils presented neutral –alkaline pH values (Wang et al., 1999; Gehlot et al., 2013; Baraúna et al., 2016; de Castro Pires et al., 2018). These discrepancies in symbiont preference between Mimosa species in different regions might be attributed to the soil characteristics, particularly pH, as the Beta-rhizobia, are highly tolerant to the low fertility acidic soils (dos Reis et al., 2010; de Castro Pires et al., 2018).
The herbaceous perennial legume Mimosa pudica was first introduced into Taiwan Province of China in 1645 as an ornamental plant (Wu et al., 2003) and it has been dispersed throughout the tropical and subtropical China. It is a plant serving as valuable bio-resource for various uses, such as green manure, fodder crops, honey source, as well as a medicine used in zoster therapy (Wang, 2014) and treating kidney disease. However, this naturalized plant is highly invasive causing considerable ecological damage e.g., by affecting the growth of grass lawns and as a common exotic weed in rice paddy fields (Guan et al., 2006). Therefore, M. pudica and M. diplotricha another invasive plant without history record, are considered to be serious pests widely dispersed in waste-grounds and city suburbs of China. It has been estimatied that the access to compatible rhizobial strains in new environments is a critical factor for the successful establishment of exotic nodulating legumes (Parker et al., 2007). For getting the compatible symbionts in the introduced region. The invasive legumes, such as Mimosa species (see above) and the invasive papilionoid legume Dipogon lignosus (L.) Verdc., may have been introduced into their invasive environments together with the symbionts from their original region (Liu et al., 2014). Or they may form the symbiosis with rhizobia adapted to the local environment and adopted the corresponding symbiotic genes through lateral gene transfer, like the cases of chickpea (Cicer arietinum L.) rhizobia in China (Zhang et al., 2017, 2018). Previously, Burkholderia spp. and Cupariavidus taiwanensis have been isolated from Mimosa species grown in China (Chen et al., 2001, 2005a; Liu et al., 2007, 2011, 2012). In which C. taiwanensis was recognized as native to Taiwan and the Burkholderia spp. were estimated as rhizobia introduced together with the host plants (Chen et al., 2005a). Furthermore, preference for Cupriavidus by M. pudica and Burkkholderia by M. pigra in Taiwan (Chen et al., 2005b), while Cupriavidus by M. diplotricha and Burkkholeria by M. pudica in Yunnan (Liu et al., 2012) demonstrated that both the host plants and the geographic regions affected the symbiosis combination between the legume and the rhizobia in the introduced regions. However, no soil conditions were considered in these previous studies about the Mimosa rhizobia in China.
In order to explore how the environmental factors and the host species influenced the composition and competitiveness of Mimosa symbionts, we performed this study to investigate the Mimosa symbionts in in different geographic regions for evaluating the competitiveness of different rhizobial species associated with invasive Mimosa spp. under varied soil traits (organic matter, N, P, K, and pH).
Materials and Methods
Isolates and Strains
Root nodules were sampled from M. pudica and two varieties of M. diplotricha var. inermis (Adelbert) Verdcourt. and var. diplotricha, growing in 25 locations of five Chinese provinces in the subtropical and tropic regions (Figure 1), as described previously (Liu et al., 2007). Root nodules were collected from three plant individuals at each site and were stored over silica gel in closed vials until their isolation in the laboratory (Vincent, 1970). Root nodule bacteria were isolated and purified from the nodules on yeast mannitol agar (YMA) using the standard procedure (Vincent, 1970). The nodule isolates obtained in this study were maintained in yeast mannitol broth (YMB) supplied with 20% (v/v) glycerol at −80°C.
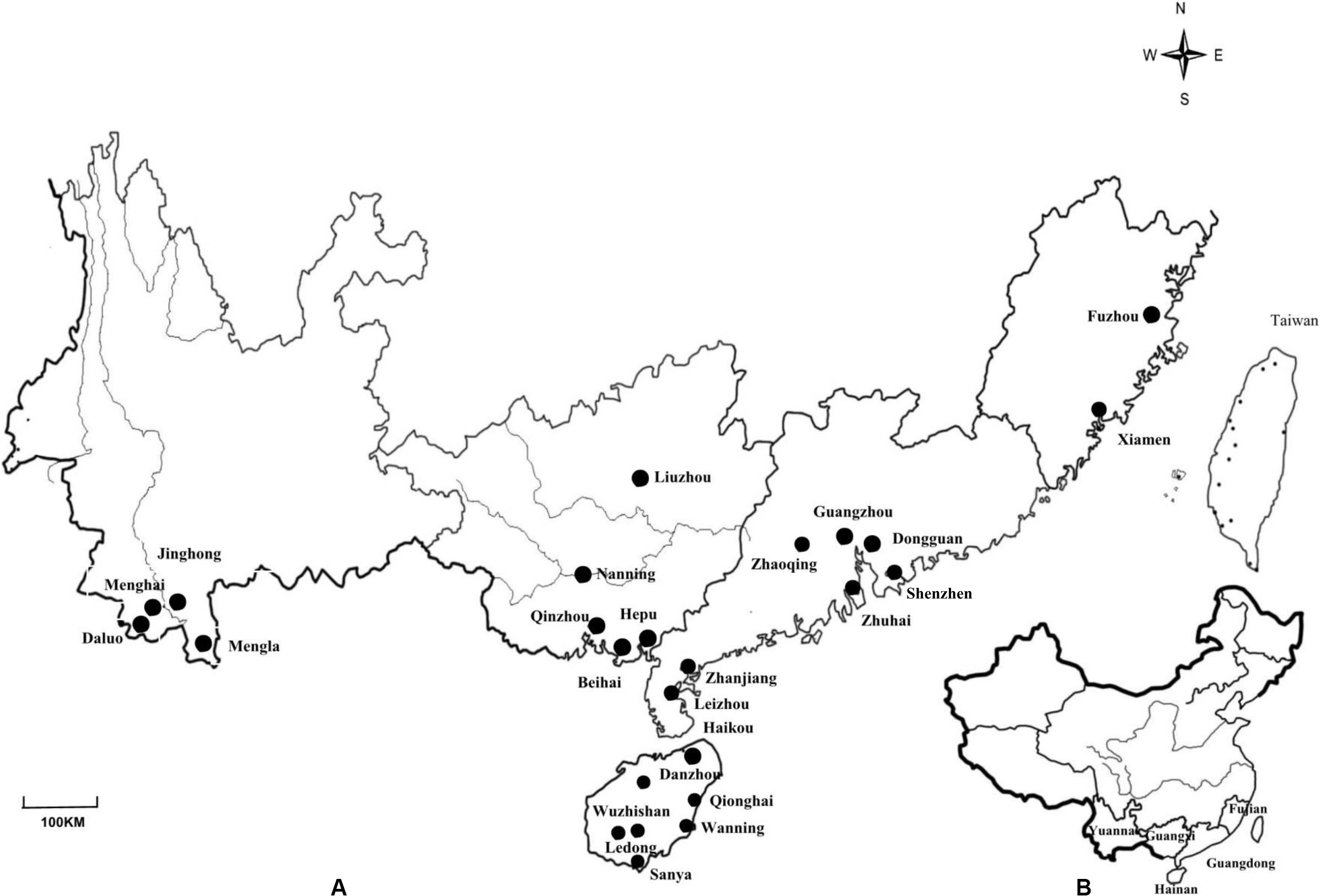
Figure 1. (A) Map of Yunnan, Guangxi, Guangdong, Fujian and Hainan Provinces in China showing the locations of the 25 sites where nodules from invasive Mimosa species were sampled. (B) Map showing the location of the sampled provinces in China.
Molecular Typing of Rhizobia
For grouping the isolates by genomic analysis, total DNA of each isolate and the reference strains Paraburkholderia mimosarum LMG23256T, Paraburkholderia phymatum LMG21445T, Paraburkholderia caribensis LMG18531T, Cupriavidus taiwanensis LMG19424T, Cupriavidus sp. SWF66294 (Liu et al., 2011) was extracted from 5 mL of culture in YMB (Vincent, 1970). The extracted genomic DNA was used as template DNA for BOX-AIR and the PCR-based RFLP (restriction fragment length polymorphism) of 16S rRNA gene (rDNA). The rDNA primers were fD1 (5′-AGAGTTTGATCCTGGCTCAGA-3′) and rD1 (5′-AAGGAGGTGATCCAGCC-3′) (Weisburg et al., 1991). The BOXAIR primer 5′-CTA CGG CAA GGC GAC GCT GAC G-3′ (Versalovic et al., 1991) was used for BOX-PCR. Both PCRs were carried out in a total volume of 25 μL of the reaction mixture with the PCR procedure of Nick et al. (1999) and the products were checked by electrophoresis in 1% (w/v) of a garose gel.
For analysis of RFLP, aliquot (5–10 μL, depending on the concentration) of PCR products was digested separately with the restriction endonucleases Hae III (GG∣CC), Rsa I (GT∣AC), Hif I (G∣ANTC), and Msp I (C∣CGG) (Laguerre et al., 1994) as specified by the manufacturer with an excess of enzyme (5 U per reaction). The restriction fragments were separated by horizontal electrophoresis in agarose (2%, w/v) gels (14 cm in length) at 80 V for 3 h and were visualized by staining with ethidium bromide. Strains or isolates with different RFLP patterns were designated into distinct rDNA types. The BOX-AIR products were separated by electrophoresis in 1.5% (w/v) agarose gels containing ethidium bromide and were photographed under UV light. The BOX profiles were distinguished by their different band patterns, e.g., the isolates sharing the same pattern were designed as the same BOX pattern.
Characterization of Whole Cell Protein by SDS-PAGE
Bacterial strains were grown until the end of the exponential phase at 28°C for 2 days on YMA. The cells were collected and washed twice in 10 mM Tris–HC1, pH 7.6, the pellet obtained by centrifugation (5000 × g, 10 min at 4°C) was weighed and the cells were resuspended in 10 mM Tris–HC1 to a concentration of 10 mg ml–1 using. Then, the same volume of 2 × treatment buffer (0.5 g of SDS, 3 ml of glycerol, 1 ml of 2-mercaptoethanol, 4 mg of bromophenol blue, 2 ml of 1 M Tris-hydrochloride, and distilled water to make a final volume of 10 ml at pH 6.8) was added. The samples were incubated at 100°C for 20 min and immediately stored at −20°C after cooled on ice. The SDS-polyacrylamide gel (200 mm × 200 mm × 1 mm) were used for electrophoresis according to Laemmli (1970). The samples were incubated at 100°C for 10 min before the sample loading. Twenty-five samples per gel were subjected to the discontinuous slab gel electrophoresis at 250 V in an SDS-Tris-glycine buffer system, as described by Laemmli (1970). The protein patterns were visualized by silver staining (Tan et al., 1997). The protein bands were scanned with a Densitometer Extra-Scanner and strains sharing the identical band patterns were designed into the same SDS-PAGE pattern.
Phylogenetic Analyses of Housekeeping Genes and Symbiotic Genes
The 16S rRNA amplified by PCR as described above was purified and sequenced directly (Weisburg et al., 1991) commercially in the Beijing Genomics Institute (BGI). The sequences acquired in this study were aligned with related sequences extracted from GenBank using Clustal W (Thompson et al., 1997). Maximum likelihood phylogenetic trees were constructed and were bootstrapped with 1000 pesudo-replicates using Mega 6.1 (Tamura et al., 2013).
Multilocus sequence analysis (MLSA) based on the five housekeeping genes atpD (encoding for the ATP synthase beta-chain), recA (recombinase A), dnaK (DnaK chaperone), gyrB (DNA gyrase, beta-subunit), and glnA (glutamine synthetase I) widely used to differentiate rhizobial species (Vinuesa et al., 2005; Martens et al., 2007, 2008) was also employed in the present study. The five genes were independently amplified using corresponding primer pairs reported in previous studies (Payne et al., 2005; Vinuesa et al., 2005; Andam and Parker, 2007; Martens et al., 2008), or designed in this study (Supplementary Table S1). The PCR products were checked by electrophoresis in 1% (w/v) agarose gel. After purified with the Solarbio DNA purification kit (Beijing Solarbio Science and Technology Co., Ltd.), the amplicons were sequenced directly using the same primers in BGI mentioned above. The separated sequences of recA and the combined sequences of atpD, glnA, gyrB, and dnaK, and their combined sequences were aligned using Clustal W with those from type strains of the defined bacterial species (obtained from the NCBI database). Distance calculation and construction of the gene phylograms were performed using the Maximum likelihood method and the bootstrapping algorithms with 1000 pseudo-replicates were carried out in MEGA 6.0 (Tamura et al., 2013). Phylogenies were also constructed using the concatenated sequences of 16S rRNA and the five housekeeping genes by Maximum likelihood method.
Fragments of the symbiosis genes nifH and nodA genes were amplified and sequenced using primers reported previously (Haukka et al., 1998; Laguerre et al., 2001; Liu et al., 2012) as well as with the new primers designed in this study (Supplementary Table S1). The visualization purification and sequencing of the nifH and nodA amplicons were performed same as that mentioned for the housekeeping genes. The sequences were deposited in the NCBI database and were used for alignment and construction of the phylogenies using the same methods described above for the 16S rRNA gene.
The obtained nucleic acid sequences were submitted in GENEBANK, and the accession numbers in this paper was MT337483 as list in Supplementary Table S2.
Nodulation Tests
A total of 98 representative strains were used in the nodulation tests that were selected according to their affiliations of genotypes based on the results of 16S rRNA sequencing, protein patterns in SDS-PAGE, and genomic fingerprinting by BOX-PCR. M. pudica seeds were scarified using concentrated sulfuric acid for 10 min, rinsed several times with sterile water, and then surface-sterilized in 3.2% (w/v) sodium hypochlorite followed by several rinses with sterile water. They were then placed on 0.8% water-agar at 4°C for 3 days, and after germinated at 28°C until the seedlings developed roots of 0.5–1 cm in length. Two seedlings then were transplanted into a sterile glass tubes (30 cm × 200 cm) with nitrogen-free plant nutrient solution (Vincent, 1970) in 0.8% agar. The seedlings were then inoculated separately with 0.1 mL liquid cultures of each test strain (about 108 cells mL–1). Five replicates were used and controls without inoculation were included. The plants were placed in a growth cabinet under conditions described previously (Zhang et al., 2012). The representative strains Paraburkholderia spp. SWF66044, SWF66029, and C. taiwanensis SWF66166, SWF66194, and SWF66322 from Liu et al. (2012) were also used for cross-inoculation tests with ten other leguminous species: Glycine max (Linn.) Merr., Pisum sativum L., Galega officinalis L., Phaseolus vulgaris Linn., Vigna unguiculata L. Walp, Lotus corniculatus L., Medicago sativa L., Trifolium repens L., Macroptilium atropurpureum (Moc. and Sessé ex DC.) Urb. and Leucaena leucocephala (Lam.) de Wit. Seed treatments and inoculation details were the same as described above. Plants were checked for nodule formation at 35 d after inoculation.
Correlation Between Soil Types and Distribution of Rhizobial Groups
In order to evaluate the influence of soil characters on the symbiosis between Mimosa spp. and different rhizobial types, soil, and root nodules were sampled intensively from 13 locations (59 sites) including Hepu, Beihai, and Nanning city in Guangxi (GXh,GXb, and GXn), Zhangjiang, Leizhou, Mazhang, and Foshan town in Guangdong (Gzj, Gl, Gm, and Gf), Jinhong and Mangshi town in Yunan (Yj and Ym) and Ledong, Wuzhishan, Wanning and Danzhou in Hainan (Hl, Hw, Hwn, and Hd), which were main districts for Mimosa speies and habitats for diverse rhizobia. For most locations, four or more sites with minimum distance of 5 km between them were samples, except the location Mangshi town in Yunnan where rhizobial strains were isolated from only one sampling site. Soils were sampled compositely from the root zone of nodule sampled plants (5–20 cm in depth). The soil samples were dried and milled until they could pass through an 80-mesh sieve. Soil alkali-hydrolysable N, available P (using Bray’s hydrochloric acid fluoride ammonium by extraction method), and available K (by ammonium acetate extraction plus flame photometry) were determined with the standard procedures (Du and Gao, 2006). Soil pH was measured using a pH meter (Mettler Toledo) by suspending 5 g soil in 5 mL of distilled water, and organic matter was measured using the potassium dichromate volumetric method (Du and Gao, 2006). Rhizobial isolation, and genus/rRNA type identification by PCR-based RFLP of 16S rRNA gene were performed same as mentioned above.
Based on the soil characters, the soil samples in the 59 sites were sorted into soil types by SPSS 13.0 (SPSS Inc., Chicago, IL, United States), in terms of their pH values and the nutritional characteristics, including organic matter (OM), alkali-hydrolysable N, available P, and available K.
The data was standardized, then using construct UPGMA dendrogram (Sneath and Sokal, 1973) for soil clustering. Principal component analysis (PCA) on a correlation matrix was used to evaluate the distribution of the different rhizobial rRNA types in the 59 sites to see if they correlated with the soil characteristics. Data analysis and graphs were performed using Past 3.0.
Results
Isolation and Genotyping of the Rhizobia
In total, 361 strains were isolated from the nodules of M. pudica and M. diplotricha sampled in the 25 locations in southern China (Figure 1 and Table 1). The majority of the isolates were obtained from M. pudica (83.7%), and minor from M. diplotricha (16.3%), which most likely reflects the relative abundance of these two plant species in the sampling sites. By 16S rRNA PCR-RFLP analysis, six rRNA types were revealed (Table 1), which were recognized as members of Paraburkholderia (three rRNA types with 55, 57, and 9 strains; 33.5%), Cupriavidus (two rRNA of types with 98 and 128 strains; 62.6%) and Rhizobium (a single rRNA type with 9 strains; 3.9%). Paraburkholderia genotypes I and II, Cupriavidus genotypes I and II, as well as Rhizobium genotype were isolated from both M. pudica and M. diplotricha; while. Paraburkholderia genotype III was only isolated from M. pudica.
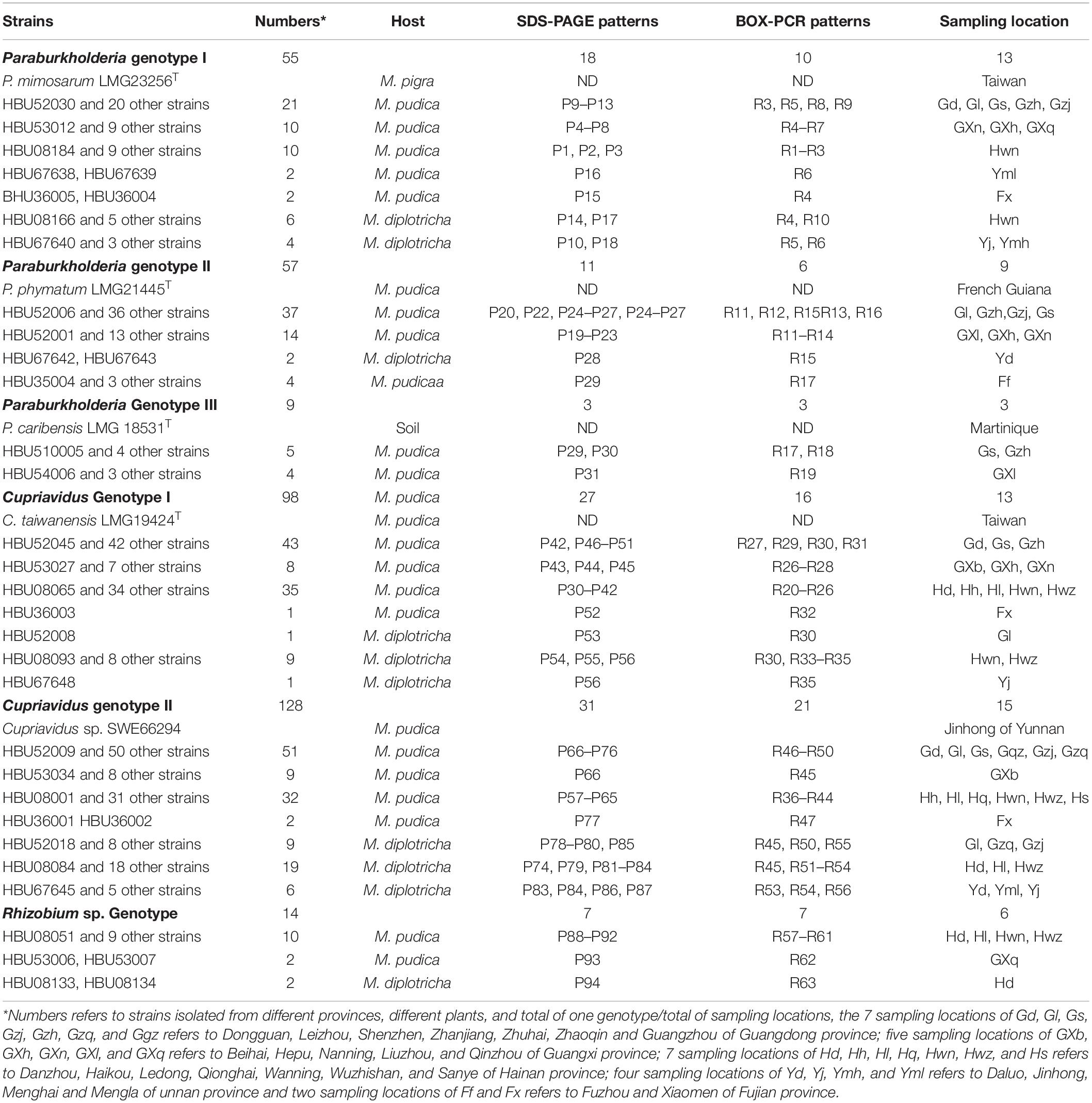
Table 1. Occurrence of rhizobial genotypes isolated from two invasive Mimosa species in southern China.
The multivariate statistical analysis (Supplementary Table S4) for conducting population distribution of the six genotypes associated with M. pudica and M. diplotricha in the four provinces (Guangdong, Guangxi, Hainan, Yunnan, and Yunnan data also from previous study in Liu et al., 2012) of China showed no significant difference (p = 0.094, >0.05), but it was significantly different (p = 0.039, <0.05) for distribution of the three genera Paraburkholderia, Cupriavidus, and Rhizobium in the four provinces, for example, the Rhizobium isolates were mainly from Hainan.
Fingerprinting of the Isolates for Estimation of Genetic Diversity
In analyses of genetic diversity, a total of 97 protein profiles and 51 BOX PCR profiles were distinguished among the 361 strains (Table 1), revealing a high level of diversity among them. Paraburkholderia rRNA type I contains 18 protein profiles and 10 BOX PCR profiles; Paraburkholderia rRNA type II contains 11 protein profiles and 6 BOX PCR profiles; and Paraburkholderia rRNA type III contains 3 protein profiles and 3 BOX PCR profiles; Cupriavidus rRNA type I contains 27 protein profiles and 16 BOX PCR profiles; and Cupriavidus rRNA type II contains 31 protein profiles and 9 BOX PCR profiles. Rhizobium rRNA type contains 7 protein profiles and 7 BOX PCR profiles. The greater number of patterns in the Cupriavidus populations suggested that they were more diverse than the Paraburkholderia populations.
Phylogenies by MLSA and Affiliation of the Isolates
Out of the total collection, 34 strains (Supplementary Table S2) were selected according to their different rRNA types, protein patterns, BOX profiles, host species and collected sites for full 16S rRNA and housekeeping gene sequencing. The relationships of the Mimosa isolates were high similar in 16S rRNA gene phylogeny (Supplementary Figure S2) and in the MLSA-based phylogeny deduced from the concatenated sequences of 16S rRNA and the five housekeeping genes (Figure 2B). Five groups were defined among the beta-rhizobial isolates at the species level (similarities ≥96.4%), which corresponded to (1) P. mimosarum (Paraburkholderia rRNA type I), (2) P. phymatum (Paraburkholderia rRNA type II), (3) P. caribensis (Paraburkholderia rRNA type III), (4) C. taiwanensis (Cupriavidus rRNA types I), and (5) Cupriavidus sp. SWF66294 (Cupriavidus rRNA types II). The Rhizobium isolates obtained in this study were grouped into three species corresponding to R. etli, R. mesoamericanum, and R. altiplani. The phylogenies of the individual housekeeping genes (atpD, recA, dnaK, gyrB, and glnA) (Supplementary Figures S1–S5) were generally consistent with that of 16S rRNA gene and the MLSA (Figure 2), except the isolates of R. etli that was a unique linage separated from all the defined species in atpD phylogenetic tree (Supplementary Figure S3).
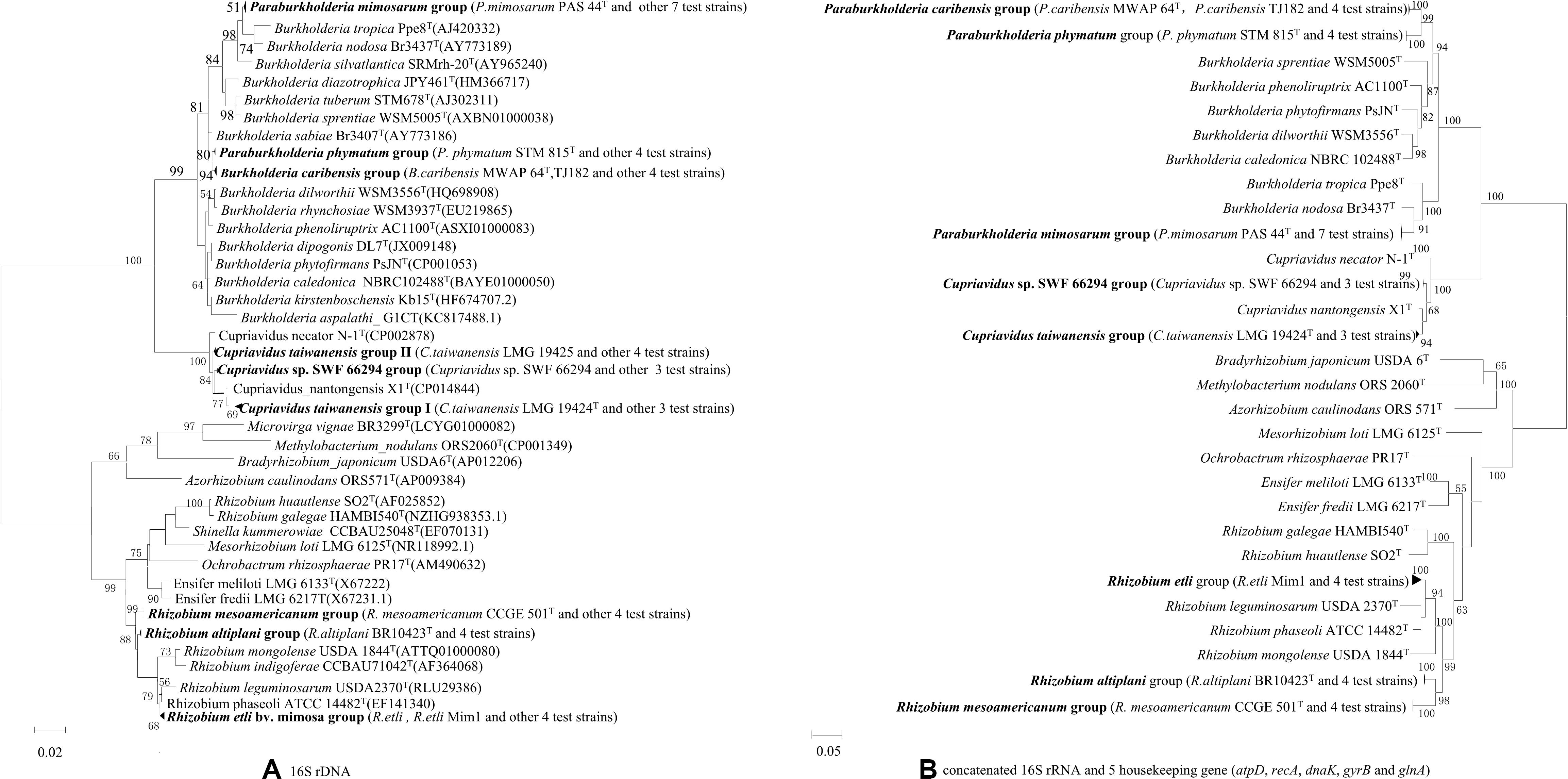
Figure 2. Phylogenies of 16S rRNA gene and the concatenated 16S rRNA and five housekeeping genes (atpD, recA, dnaK, gyrB, and glnA) in rhizobial strains isolated from nodules on invasive Mimosa spp. in southern China. The Maximum likelihood phylogenies are based on (A) 16S rRNA gene sequences (1320 bp); (B) concatenated 16S rRNA and five housekeeping gene sequences (3706 bp).
Sequencing and Phylogenetic Analysis of Symbiosis Genes
This analysis were performed for the 34 representative strains mentioned above. Four nodA and four nifH lineages were defined among them (Figure 3 and Supplementary Figure S6). The phylogenies of the nodA and nifH genes of the isolates were the same and they were incongruent in several cases with that of the housekeeping genes, e.g., both the Alpha-and Beta-rhizobia formed two clades and they were intermingled. P. caribensis strains and P. phymatum strains shared similar symbiosis genes, while P. mimosarum presented another lineage (Figure 3 and Supplementary Figure S6). The strains in both Cupriavidus genotypes I and II, including C. taiwanensis LMG19424T, formed the third lineage in the symbiosis gene phylogeny, which was inserted between the two lineages of the Paraburkholderia species. All the three Rhizobium species identified in the present study shared the same symbiosis gene and formed the forth lineage represented by that of R. mesoamericanum STM3625 isolated from M. pudica in Mexico.

Figure 3. Phylogenetic tree based on nodA gene sequences (230 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 5% substitutions per site.
Nodulation Test
All 98 representative strains selected from different groups according to their patterns in 16S rRNA PCR-RFLP, SDS-PAGE protein and BOX-PCR profiles (Table 1 and Liu et al., 2012) formed nodules on M. pudica. Cross-inoculation studies showed that only the invasive mimosoid legume, L. leucocephala could nodulate with all five test strains (Paraburkholderia spp. SWF66044, SWF66029, C. taiwanensis SWF66166, SWF66194, and SWF66322), which is in keeping with its neotropical origin, close relatedness to Mimosa, and its known promiscuity.
Correlation Between Sampling Sites and the Distribution of Mimosa Symbionts
Soil characteristics in the 59 sample sites at 13 locations are presented in Supplementary Material (Supplementary Table S3). Briefly, among these sites, the soils pH ranged from 5.2 to 7.78; soil organic matter ranged from 3.51 to 43.82 mg kg–1; alkali-hydrolysable N ranged from 1.49 to 82.16 mg kg–1; available P ranged from 0.94 to 88.98 mg kg–1; and available K ranged from 4.34 to 321.84 mg kg–1 (Supplementary Table S3).
The locations and the soil parameters within each sampling site were analyzed via cluster analysis by SPSS and PCA, which revealed main four groups with distinct soil patterns, an unusual soil site as another type for Cupriavidus spp. surviving (because it is distant from other soil point) was not considered (Figure 4). We obtained 200 rhizobial strains from the 59 sites and they were identified into four groups as Cupriavidus spp., P. mimosarum, P. phymatum, and Rhizobium spp., and each group grown soil nutrition and pH range displayed as Table 2. In PCA, only the existence (not abundance) of the different group in each site was considered, and they were in relation to their spatial distribution (Figure 4 and Table 2). The soil PCA resulted in three components with eigenvalues greater than one which explained 75.3% of the total variance (first component: 47.7%, second component: 27.6%, third component: 14.2%). However, the third component did not provide any further information above the first two components, and hence was excluded from the interpretation. The first component revealed the positive correlation of OM and available N, and a contrasting correlation of these with pH (loading factors = 0.58, 0.60, and 0.37, respectively) with soil types I, II, III, and IV; the second component is characterized by a positive correlation of available P and available K (loading factors = 0.55, 0.73). On the PCA scatter plot, the four soil patterns are almost completely separated.
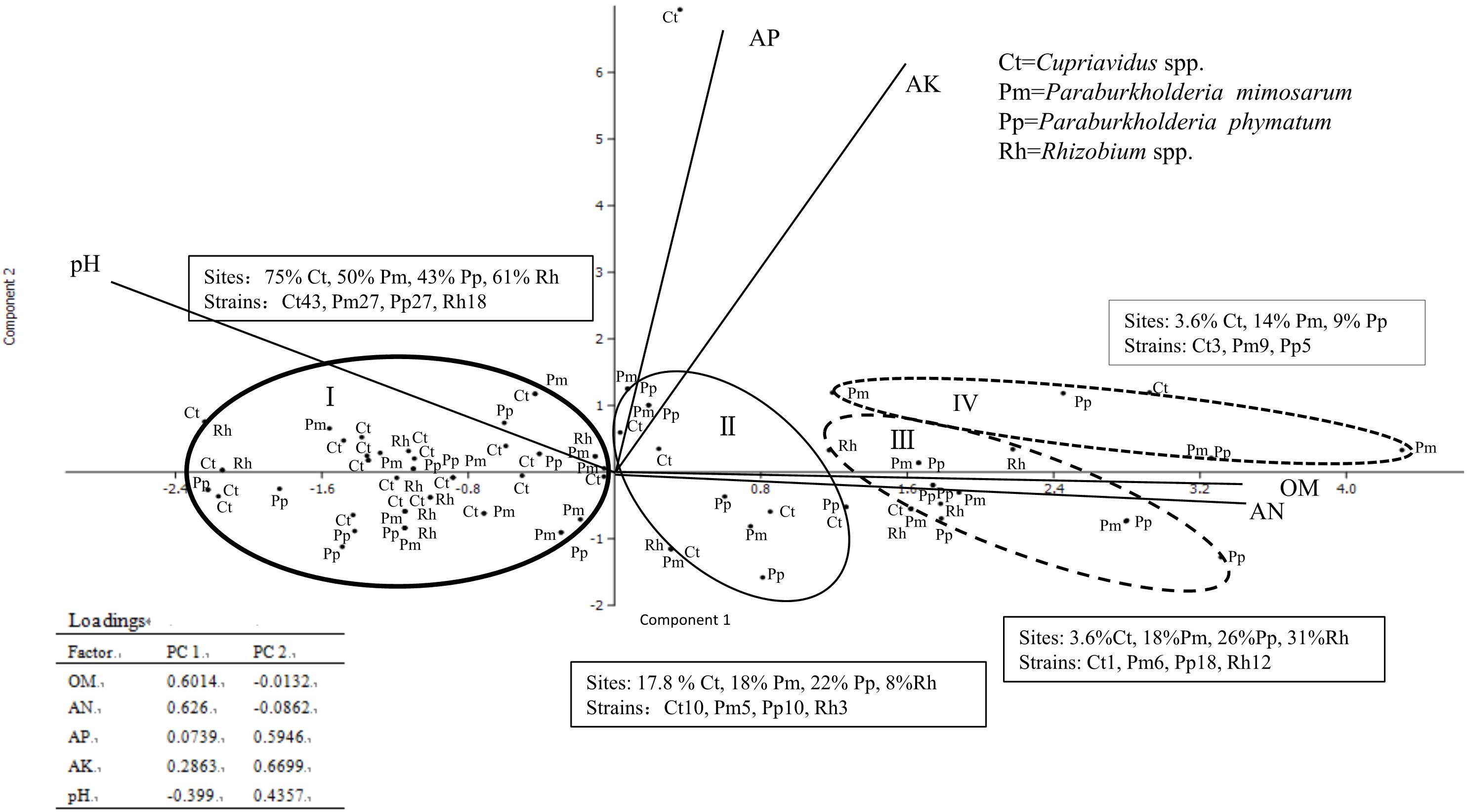
Figure 4. Comparison of the distribution of four communities of rhizobial types associated with invasive Mimosa species in southern China by principal component analysis of total N, available K, available P, precipitation and pH. The principal component one revealed correlations of available K with available P and pH. The principal component two is characterized by precipitation and total N and organic matter. Ellipses represent 90% confidence limits. The word in text box means each occupied sites percent and species constitute in different soil types.
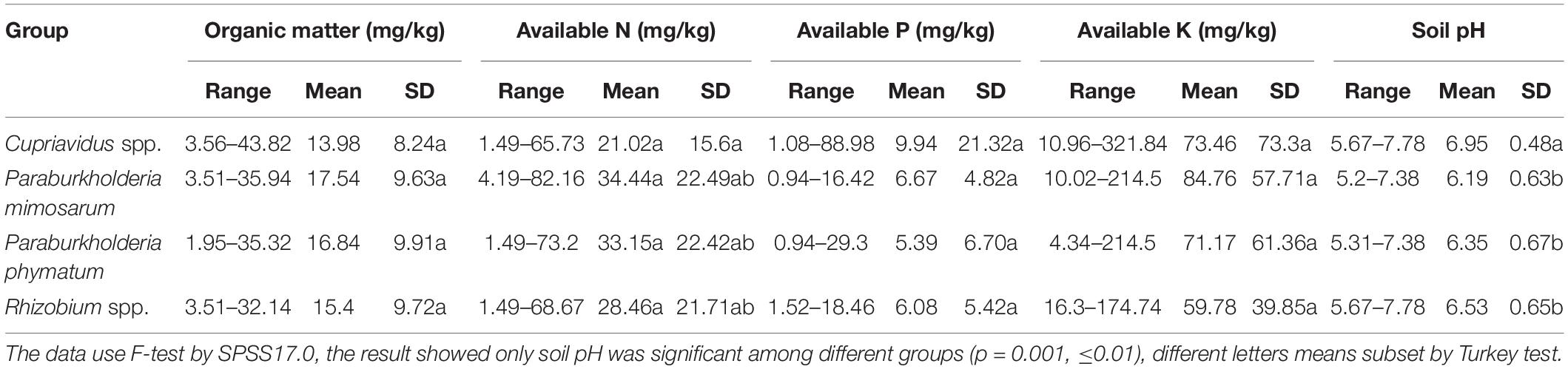
Table 2. Characteristics of the habitats of four rhizobial groups from invasive Mimosa species in southern China.
In total, the 59 sampling sites were plotted onto the soil-site PCA (Figure 4), and the rhizobial species that were isolated from each site and isolates numbers are indicated in Supplementary Table S3. Four soil types corresponded to the localization site of the different rhizobial types associated with Mimosa in southern China. Soil category I (32 sites, 115 isolates) was the major soil type characterized by low fertility, relatively high available P, and neutral-alkaline pH and occupied by the majority of the Mimosa symbionts obtained in this study. In this type of soil, the rhizobial community contained 37.4% Cupriavidus, 23.5% P. mimosarum, 23.5% P. phymatum and 15.5% Rhizobium spp. strains which covered 75% of the Cupriavidus isolates, 50% of the P. mimosarum, 43% of the P. phymatum and 62% of the Rhizobium spp. strains. Soil category II (10 sites, 28 isoates) harbored 35.7% Cupriavidus, 17.9% P. mimosarum, 35.7% P. phymatum, and 10.7% Rhizobium spp. strains; it characterized by intermediate fertility and neutral pH. Soil category III (11strains, 37 isoates) tended to acid-neutral pH, with lower fertility and lower available P and K; it contained 2.7% Cupriavidus (1 strain), 16.2% P. mimosarum, 48.6% P. phymatum, and 32.5% Rhizobium spp. strains. Soil category IV (5 sites, 17 isoates) was quite acidic with high fertility, but with low available P; it harbored the rhizobial community with 17.6% Cupriavidus, 52.9% P. mimosarum and 29.5% P. phymatum strains.
Within all the 59 sites, Cupriavidus strains habitat in 29 soil sites (49.2% of total sites), but occupied 75%, 17.8%, 3.6, and 3.6% of the sites in soil categories I through IV, respectively. P. mimosarum strains habitat in 22 soil sites (37.3% of total sites), and scattered on 50, 18, 18, and 14% of the sites in categories I through IV, respectively, P. phymatum strains survive in 23 soil sites (about 39% of total sites), dispersed 43, 22, 26, and 14% of the sites in the soil categories I through IV, respectively, Rhizobium spp. strains habitat in 13 soil sites (22% of total sites) belonging to the soil categories I through III, appearing in 61, 3, and 12% of the sites, respectively.
Discussion
Mimosa-Nodulating Rhizobial Community in Southern China
Based upon the MLSA resules (Figure 2), the six rRNA types of Mimosa rhizobia defined by PCR-based RFLP of 16S rRNA gene (Table 1), could be indentified as 8 species: Paraburkholderia rRNA type I as P. mimosarum, Paraburkholderia rRNA type II as P. phymatum, Paraburkholderia rRNA type III as P. caribensis, Cupriavidus rRNA types I as C. taiwanensis, Cupriavidus rRNA types II as Cupriavidus sp., Rhizobium sp. genotype as R. etli, R. mesoamericanum, and R. altiplani. These identifications might imply that the PCR-RFLP of 16S rRNA is an efficient method to identify the current beta-rhizobial species, but it is unable to differentiate the alpha-rhizobial species, as evidenced in many previous studies (for example, Huo et al., 2019; Li et al., 2019).
Although four of the six beta-rhizobial genotypes/species defined among the symbionts of Mimosa diplotricha and M. pudica in southern China (Table 1) were commonly associated with Mimosa species in both their native and invasive regions. The Cupriavidus sp. represented by strains SWF66294 covering 128 isolates was unique in China. The presence of identical nodA and nifH in Cupriavidus sp. and in C. taiwanensis might be evidence that lateral transfer of symbiosis gene between these two species has happened, as reported in the Lotus-nodulation Mesorhizobium species (Bamba et al., 2019). The situation in the three Rhizobium species was similar. In addition, the intermingling of the Alpha-and Beta-rhizobial clades in the phylogenies of symbiosis genes (Figure 3 and Supplementary Figure S6) also demonstrating the lateral transfer of symbiosis gene between these two rhizobial categories, as estimated previously (see review of Andrews et al., 2018). Therefore, new symbionts of Mimosa has been evolved in China under the double selection from host plant (for symbiotic gene background) and the soil conditions (for survival in the local sites). Previously Gehlot et al. (2013) estimated that invasive Mimosa spp. do not interact with the symbionts of native legumes; however, the Mimosa species could get novel symbiosis adapted to their invaded region by lateral transfer of the symbiosis genes from its known rhizobia to the native relatives. Cupriavidus sp. may constitute a new species intermediate between C. taiwanensis and C. nantongensis (Sun et al., 2016), but more analyses are required to clarify its species affiliation, such as DNA-DNA hybridization (DDH) and/or average nucleotide identity (ANI) with the closest type strains.
Cupriavidus taiwanensis is a very common symbiont of invasive Mimosa species in South East Asia (Chen et al., 2001, 2003b, 2005a; Elliott et al., 2009; Andrus et al., 2012; Liu et al., 2012, 2011; Gehlot et al., 2013), and it could be the dominant symbiont in some locations for M. diplotricha and M. pudica, but not for M. pigra (Chen et al., 2001, 2003b, 2005b; Klonowska et al., 2012). So, different Mimosa species may have distinct preferences for their symbionts (Chen et al., 2005a, b; Elliott et al., 2009). Although Cupriavidus is less commonly isolated in the native regions of Mimosa, C. taiwanensis has been isolated from M. pudica (Barrett and Parker, 2006; Mishra et al., 2012) and M. asperata (Andam et al., 2007) in Americas. Moreover, Cupriavidus strains closely related to C. necator and C. pinatubonensis were the dominant symbionts of native Mimosa in Uruguay (Platero et al., 2016). All these results imply that C. taiwanensis might originated in the Americans and was introduced to China together with the Mimosa plants.
In the present study, P. mimosarum, P. phymatum, and P. caribensis comprised 15.2, 15.8, amd 2.5% of the total isolates from 14, 11, and 3 sample sites, respecitively, P. mimosarum and P. phymatum have wide distribution in both the invaded and the original regions of the Mimosa species and P. caribensis (Bontemps et al., 2010; dos Reis et al., 2010; Chen et al., 2005a, b, 2006; Elliott et al., 2007, 2009; Liu et al., 2011, 2012; Mishra et al., 2012; Gehlot et al., 2013; Lardi et al., 2017). In addtion, its greater abundance in Yunnan Province (Liu et al., 2011, 2012) than in neighbor provinces examined in the present study demonstrated that P. mimosarum strains were more adapted to the soil in Yunnan. P. caribensis was originally described for non-symbiotic strains isolated from soil (Achouak et al., 1999) and symbionts of Mimosa belonging to this species were isolated lately in China (Chen et al., 2003b; Liu et al., 2012). So, again, its possible that novel symbiont of Mimosa may have evolved in China after this plant was introduced.
Rhizobium was isolated as minor group from the Mimosa species in this study (Table 1), and most of them (12 of the 14 strains) were from Hainan Province, with few isolates from Yunnan (Liu et al., 2012) and Guangxi (Table 1). Based upon the MLSA results (Figure 2) these strains were idertified as R. mesoamericanum, R. etli, and R. altiplani, while all of them were identified as sv. mimosa according to the nodA and nifG phylogenies (Figure 3 and Supplementary Figure S6). Previously, R. etli sv. mimosae has isolated from invasive Mimosa species in low frequency (Chen et al., 2001, 2003b, 2005a; Elliott et al., 2009; Klonowska et al., 2012; Mishra et al., 2012; Melkonian et al., 2014), while both R. etli and R. mesoamericanum were found to be predominate in Mimosa symbionts in Mexico, the second largest center of Mimosa diversity (Wang et al., 1999; Bontemps et al., 2016). The exception is R. altiplani, which is relatively common in central Brazil (Baraúna et al., 2016; de Castro Pires et al., 2018), but has never previously been isolated from Mimosa in its invasive range regions. So, the Mimosa-nodulation Rhizobium species in China might be also introduced together with their hosts, but they are not so adapted to the conditions in the invasive regions.
In addtition to the species definition, great genetic diversity represented by the 94 protein patterns and 63 BOX-PCR patterns (Table 1) was revealed in this study among the Alpha- and Beta-rhizobial symbionts isolated from only two Mimosa species. This might be related to the vast area of the sampling locations, which covered diverse soil types and forced the diversification of rhizobia for their survival.
In summary, five beta-rhizobial species, especially Cupriavidus genotype II, with great genetic diversity as the dominant microsymbionts and three Rhizobium species as minor microsymbionts for M. pudica and M. diplotricha plants were detected in a vast sampling area in China. Most of them have been found in both the invasive regions and the centers of origin of these plants, but Cupriavidus genotype II might be a novel symbiont for Mimosa species evolved in China.
Soil Variables and the Distribution of Rhizobial Genotypes Associated With Mimosa Species
The worldwide distribution of various symbionts isolated from invasive Mimosa species may be the result of selection by soil characteristics and other ecological factors (Elliott et al., 2009; Melkonian et al., 2014; de Castro Pires et al., 2018). In the present study, the community composition in the five provinces were unbalanced: P. mimosarum was more abundant in Yunnan; P. phymatum was more in Guangdong and Guangxi; Rhizobium stains were mostly from Hainan; and the minor group P. caribensis was only isolated from M. pudica plants grown in Guangdong and Guangxi. These geographic distributions implied an interaction among the plants, the rhizobial species, and the environment factors, as described for rhizobia associated with soybean (Zhang et al., 2011).
In the SPSS analysis and the principal component analysi (Figure 4), the correlation of Cupriavidus spp. and Rhizobium spp. with the infertile alkaline soil type (category I) and neutral soil (category II), and the accommodating of Paraburkholderia spp. in more acidic soils (category III and IV) were consistent with the previous reports that have shown that C. taiwanensis is abundant as a Mimosa symbiont in neutral to basic pH soils (Table 2), often with relatively high fertility (Elliott et al., 2009; Klonowska et al., 2012; Mishra et al., 2012; Gehlot et al., 2013). In contrast to Cupriavidus, Paraburkholderia strains can tolerate acidic soils (Stopnisek et al., 2014), and indeed become dominant symbionts with the compatible legumes; it was similar for rhizobia nodulating with mimosoid and papilionoid legumes (Garau et al., 2009; dos Reis et al., 2010; Howieson et al., 2013; Liu et al., 2014; Lemaire et al., 2015, 2016a,b). The present study has clearly illustrated that pH appears to be the most important environmental factor in helping to explain the distribution of Mimosa symbionts in southern China (Table 2, p = 0.001, ≤0.01). For example, although higher soil fertility and N concentration improved the competitive nodulation of Paraburkholderia over Cupriavidus and Rhizobium on Mimosa spp. in reduced N concentrations/low fertility growth media (Elliott et al., 2009), this was not the case in the present study wherein the dominant symbionts in the alkaline-neutral soils (soil types I and II) were overwhelmingly Cupriavidus regardless of their low fertility. The opposite was the case for the acidic soils wherein the Paraburkholderia strains dominated even though the soils were relatively fertile.
Taken together, Cupriavidus spp. are the most common and competitive rhizobial type in southern China due to its ability to grow in the widest range of pH, soil nutrition/fertility, and soil moisture levels (from drought to flooding), and maybe also to its tolerance to heavy metals (Klonowska et al., 2012). C. taiwanensis was recognized as strong stress resistant bacteria able to survive and grow at phenol concentrations up to 900 mg/L (Chen et al., 2004). It is also particularly dominant in islands like Taiwan (Chen et al., 2003b), New Caledonia (Klonowska et al., 2012), and Hainan (Table 1). Certainly it is possible that the similar climate and soils in these islands have created habitats ideal for the two invasive Mimosa species (M. pudica, M. diplotricha), where the soils are also rich in P and K, and would favor C. taiwanensis rather than (for instance) Rhizobium.
On the other hand, the dominance of Cupriavidus as a Mimosa symbionts appears to be a phenomenon that occurs mainly in invasive ecosystems. For example, in some natural ecosystems, such as central Mexico (Bontemps et al., 2016), the Indian Thar Desert (Gehlot et al., 2013), and even in parts of central Brazil (Baraúna et al., 2016; de Castro Pires et al., 2018), where soils are neutral to alkaline, Alpha-rhizobia can be the dominant symbionts of the native/endemic Mimosa species. Indeed, Cupriavidus species are normally minor group or absent in their centers of origin (Bontemps et al., 2010; dos Reis et al., 2010), de Castro Pires et al., 2018). The exceptions come from Texas where a widespread species M. asperata was nodulated only with C. taiwanensis-like bacteria (Andam et al., 2007), and from Uruguay in which native/endemic Mimosa species grown in slightly acidic soils of a heavy metal mining area were exclusively nodulated with C. necator- and C. pinatubonensis-like bacteria with nod genes divergent from C. taiwanensis (Platero et al., 2016). Interestingly, the Uruguayan native/endemic Mimosa appeared to be incapable of nodulating effectively with Paraburkholderia strains suggesting that they had co-evolved with their Cupriavidus symbionts in a manner similar to that described for species in central Brazil and central Mexico with Paraburkholderia and Alpha-rhizobia symbionts, respectively (Bontemps et al., 2010, 2016).
In summary, Cupriavidus strains are highly adaptable and competitive symbionts of the two Mimosa species in an invasive context, particularly when soils are neutral-alkaline, but they even retain a high degree of competitiveness in soils less optimal for it (i.e., slightly acidic and with low levels of OM and N). The disparity between the dominance of Cupariavidus in an invasive environment, and its sporadic occurrence in their native regions is still not clearly explained, but it could be related to the fact that most Mimosa species are not capable of nodulating effectively with C. taiwanensis (Elliott et al., 2007; dos Reis et al., 2010). Interestingly, its preferred hosts, M. pudica and M. diplotricha, although common in lowland areas of the neotropics (Barrett and Parker, 2005, 2006; Mishra et al., 2012), are mainly restricted to disturbed sites (Bontemps et al., 2010; Baraúna et al., 2016). Therefore, it is possible that the same soil factors which encouraging the invasiveness of its Mimosa plants (fertile ground subject to anthropogenic disturbance) also created a niche which favoring C. taiwanensis over its usual competitors for nodulation in its native region (i.e., Paraburkholderia species).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the Supplementary Material, Supplementary Table S2.
Author Contributions
XYL conceived and designed the study. SHY, BJY, HYW, and FW collected the nodules from Mimosa and isolated the rhizobia strains. WDC and ZKL performed the soil nutrients detection, HJL conduct the PCA analysis, XYL and EKJ wrote the manuscript and edited the manuscript. All authors read and approved the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (grant nos. 30970005, 31370051, and 31360003), China Agricultural Research System-Green Manure (CARS-22), Key Bioengineering Discipline of Hebei Province (no. 1050-5030023) and the Public Service Sectors (agriculture) Research and Special Funds (grant no. 201303061), the Key Discipline Project for Biotechnology of Hebei Province, and Hebei University Construction Project for Comprehensive Strength Promotion of Midwest colleges and universities, Hebei Grass Industry Innovation team of Modern Agricultural Industry Technology System (HBCT2018050204).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Clare Gough (Laboratoire des Interactions Plantes-Microorganismes (LIPM), Institut National de la Recherche Agronomique, Castanet-Tolosan, France) for reviewing the manuscript. At end, we also thank En Tao Wang (ENCB-IPN, Mexico) a lot for helping to revise this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.563389/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree based on recA gene sequences (382 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using the Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 5% substitutions per site.
Supplementary Figure 2 | Phylogenetic tree based on gyrB gene sequences (432 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using the Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 10% substitutions per site.
Supplementary Figure 3 | Phylogenetic tree based on atpD gene sequences (434 bp) showing the groups ofthe rhizobial strains isolated from Mimosa spp. The tree was constructed by using the Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 5% substitutions per site.
Supplementary Figure 4 | Phylogenetic tree base on dnaK gene sequences (260 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using the Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 10% substitutions per site.
Supplementary Figure 5 | Phylogenetic tree based on glnA gene sequences (874 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 20% substitutions per site.
Supplementary Figure 6 | Phylogenetic tree based on nifH gene sequences (253 bp) showing the groups of the rhizobial strains isolated from Mimosa spp. The tree was constructed by using the Maximum likelihood method. Bootstrap values are indicated (>50) in the main nodes in a bootstrap analysis of 1,000 replicates, 5% substitutions per site.
References
Achouak, W., Christen, R., Barakat, M., Martel, M.-H., and Heulin, T. (1999). Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int. J. Syst. Bacteriol. 49, 787–794. doi: 10.1099/00207713-49-2-787
Andam, C. P., Mondo, S. J., and Parker, M. A. (2007). Monophyly of nodA and nifH genes across Texan and Costa Rican populations of Cupriavidus nodule symbionts. Appl. Environ. Microbiol. 73, 4686–4690. doi: 10.1128/AEM.00160-07
Andam, C. P., and Parker, M. A. (2007). Novel alphaproteobacterial root nodule symbiont associated with Lupinus texensis. Appl. Environ. Microbiol. 73, 5687–5691. doi: 10.1128/AEM.01413-07
Andrews, M., De Meyer, S., James, E., Stępkowski, T., Hodge, S., Simon, M., et al. (2018). Horizontal Transfer of Symbiosis Genes within and Between Rhizobial Genera: occurrence and Importance. Genes 9:321. doi: 10.3390/genes9070321
Andrus, A. D., Andam, C., and Parker, M. A. (2012). American origin of Cupriavidus bacteria associated with invasive Mimosa legumes in the Philippines. FEMS Microbiol. Ecol. 80, 747–750. doi: 10.1111/j.1574-6941.2012.01342.x
Bamba, M., Aoki, S., Kajita, T., Setoguchi, H., Watano, Y., Sato, S., et al. (2019). Exploring genetic diversity and signatures of horizontal gene transfer in nodule bacteria associated with lotus japonicus in natural environments. Mol. Plant Microbe Interact. 32, 1110–1120. doi: 10.1094/MPMI-02-19-0039-R
Baraúna, A. C., Rouws, L. F. M., Simoes-Araujo, J. L., dos Reis Junior, F. B., Iannetta, P. P. M., Maluk, M., et al. (2016). Rhizobium altiplani sp. nov., isolated from effective nodules on Mimosa pudica growing in untypically alkaline soil in central Brazil. Int. J. Syst. Evol. Microbiol. 66, 4118–4124. doi: 10.1099/ijsem.0.001322
Barrett, C. F., and Parker, M. A. (2005). Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 28, 57–65. doi: 10.1016/j.syapm.2004.09.002
Barrett, C. F., and Parker, M. A. (2006). Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Society 72, 1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006
Beukes, C. W., Palmer, M., Manyaka, P., Chan, W. Y., Avontuur, J. R., van Zyl, E., et al. (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 8, 1–11. doi: 10.3389/fmicb.2017.01154
Bontemps, C., Elliott, G. N., Simon, M. F., Dos Reis Júnior, F. B., Gross, E., Lawton, R. C., et al. (2010). Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 19, 44–52. doi: 10.1111/j.1365-294X.2009.04458.x
Bontemps, C., Rogel, M. A., Wiechmann, A., Mussabekova, A., Moody, S., Simon, M. F., et al. (2016). Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol. 209, 319–333. doi: 10.1111/nph.13573
Bournaud, C., de Faria, S. M., dos Santos, J. M. F., Tisseyre, P., Silva, M., Chaintreuil, C., et al. (2013). Burkholderia Species Are the Most Common and Preferred Nodulating Symbionts of the Piptadenia Group (Tribe Mimoseae). PLoS One 8:e63478. doi: 10.1371/journal.pone.0063478
Chen, W. M., Chang, J. S., Wu, C. H., and Chang, S. C. (2004). Characterization of phenol and trichloroethene degradation by the rhizobium Ralstonia taiwanensis. Res. Microbiol. 155, 672–680. doi: 10.1016/j.resmic.2004.05.0045
Chen, W. M., de Faria, S. M., Straliotto, R., Pitard, R. M., Simões-Araùjo, J. L., Chou, J., et al. (2005a). Proof that Burkholderia Strains Form Effective Symbioses with Legumes: a Study of Novel Mimosa-Nodulating Strains from South America. Appl. Environ. Microbiol. 71, 7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005
Chen, W.-M., James, E. K., Chou, J.-H., Sheu, S.-Y., Yang, S.-Z., and Sprent, J. I. (2005b). Beta-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. 168, 661–675. doi: 10.1111/j.1469-8137.2005.01533.x
Chen, W. M., James, E. K., Coenye, T., Chou, J. H., Barrios, E., de Faria, S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56, 1847–1851. doi: 10.1099/ijs.0.64325-0
Chen, W. M., James, E. K., Prescott, A. R., Kierans, M., and Sprent, J. I. (2003a). Nodulation of Mimosa spp. by the beta-proteobacterium Ralstonia taiwanensis. Mol. Plant Microbe Interact. 16, 1051–1061. doi: 10.1094/MPMI.2003.16.12.1051
Chen, W. M., Laevens, S., Lee, T. M., Coenye, T., De Vos, P., Mergeay, M., et al. (2001). Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51, 1729–1735. doi: 10.1099/00207713-51-5-1729
Chen, W.-M., Moulin, L., Bontemps, C., Vandamme, P., Béna, G., and Boivin-Masson, C. (2003b). Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 185, 7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003
da Silva, K., Florentino, L. A., da Silva, K. B., de Brandt, E., Vandamme, P., and de Souza Moreira, F. M. (2012). Cupriavidus necator isolates are able to fix nitrogen in symbiosis with different legume species. Syst. Appl. Microbiol. 35, 175–182. doi: 10.1016/j.syapm.2011.10.005
de Castro Pires, R., dos Reis Junior, F. B., Zilli, J. E., Fischer, D., Hofmann, A., James, E. K., et al. (2018). Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant Soil 423, 411–428. doi: 10.1007/s11104-017-3521-5
dos Reis, F. B., Simon, M. F., Gross, E., Boddey, R. M., Elliott, G. N., Neto, N. E., et al. (2010). Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 186, 934–946. doi: 10.1111/j.1469-8137.2010.03267.x
Du S., and Gao, X. (2006). Technical Specification of Soil Analysis. Beijing: China Agriculture Press.
Elliott, G. N., Chen, W. M., Chou, J. H., Wang, H. C., Sheu, S. Y., Perin, L., et al. (2007). Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173, 168–180. doi: 10.1111/j.1469-8137.2006.01894.x
Elliott, G. N., Chou, J. H., Chen, W. M., Bloemberg, G. V., Bontemps, C., Martínez-Romero, E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11, 762–778. doi: 10.1111/j.1462-2920.2008.01799.x
Garau, G., Yates, R. J., Deiana, P., and Howieson, J. G. (2009). Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol. Biochem. 41, 125–134. doi: 10.1016/j.soilbio.2008.10.011
Gehlot, H. S., Tak, N., Kaushik, M., Mitra, S., Chen, W. M., Poweleit, N., et al. (2013). An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann. Bot. 112, 179–196. doi: 10.1093/aob/mct112
Guan, Z. B., Deng, W. H., Huang, Z. L., Huang, N. Y., Ai, L., and Li, C. (2006). A preliminary investigation on the alien invasive plants in Xishuangbanna. Trop. Agric. Sci. Technol. 29, 35–38.
Gyaneshwar, P., Hirsch, A. M., Moulin, L., Chen, W.-M., Elliott, G. N., Bontemps, C., et al. (2011). Legume-Nodulating Betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microbe Interact. 24, 1276–1288. doi: 10.1094/MPMI-06-11-0172
Haukka, K., Lindström, K., and Young, J. (1998). Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 64, 419–426. doi: 10.1128/AEM.64.2.419-426.1998
Howieson, J. G., De Meyer, S. E., Vivas-Marfisi, A., Ratnayake, S., Ardley, J. K., and Yates, R. J. (2013). Novel Burkholderia bacteria isolated from Lebeckia ambigua - A perennial suffrutescent legume of the fynbos. Soil Biol. Biochem. 60, 55–64. doi: 10.1016/j.soilbio.2013.01.009
Huo, Y., Tong, W., Wang, J., Wang, F., Bai, W., Wang, E., et al. (2019). Rhizobium chutanense sp. nov., isolated from root nodules of phaseolus vulgaris in China. Int. J. Syst. Evol. Microbiol. 69, 2049–2056. doi: 10.1099/ijsem.0.003430
Klonowska, A., Chaintreuil, C., Tisseyre, P., Miché, L., Melkonian, R., Ducousso, M., et al. (2012). Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol. Ecol. 81, 618–635. doi: 10.1111/j.1574-6941.2012.01393.x
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Laguerre, G., Allard, M.-R., Revoy, F., and Amarger, N. (1994). Rapid Identification of Rhizobia by Restriction Fragment Length Polymorphism Analysis of PCR-Amplified 16S rRNA Genes. Appl. Environ. Microbiol. 60, 56–63. doi: 10.1128/AEM.60.1.56-63.1994
Laguerre, G., Nour, S. M., Macheret, V., Sanjuan, J., Drouin, P., and Amarger, N. (2001). Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147, 981–993. doi: 10.1099/00221287-147-4-981
Lardi, M., de Campos, S. B., Purtschert, G., Eberl, L., and Pessi, G. (2017). Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front. Microbiol. 8:1527. doi: 10.3389/fmicb.2017.01527
Lemaire, B., Chimphango, S. B. M., Stirton, C., Rafudeen, S., Honnay, O., Smets, E., et al. (2016a). Biogeographical patterns of legume-nodulating Burkholderia spp.: from African fynbos to continental scales. Appl. Environ. Microbiol. 82, 5099–5115. doi: 10.1128/AEM.00591-16
Lemaire, B., Dlodlo, O., Chimphango, S., Stirton, C., Schrire, B., Boatwright, J. S., et al. (2015). Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 91, 1–17. doi: 10.1093/femsec/fiu024
Lemaire, B., Van Cauwenberghe, J., Verstraete, B., Chimphango, S., Stirton, C., Honnay, O., et al. (2016b). Characterization of the papilionoid-Burkholderia interaction in the Fynbos biome: the diversity and distribution of beta-rhizobia nodulating Podalyria calyptrata (Fabaceae, Podalyrieae). Syst. Appl. Microbiol. 39, 41–48. doi: 10.1016/j.syapm.2015.09.006
Li, Y. H., Wang, R., Sui, X. H., Wang, E. T., Zhang, X. X., Tian, C. F., et al. (2019). Bradyrhizobium nanningense sp. nov., Bradyrhizobium guangzhouense sp. nov. and Bradyrhizobium zhanjiangense sp. nov., isolated from effective nodules of peanut in Southeast China. Syst. Appl. Microbiol. 42:126002. doi: 10.1016/j.syapm.2019.126002
Liu, W. Y. Y., Ridgway, H. J., James, T. K., James, E. K., Chen, W. M., Sprent, J. I., et al. (2014). Burkholderia sp. Induces Functional Nodules on the South African Invasive Legume Dipogon lignosus (Phaseoleae) in New Zealand Soils. Microb. Ecol. 68, 542–555. doi: 10.1007/s00248-014-0427-0
Liu, X. Y., Wei, S., Wang, F., James, E. K., Guo, X., Zagar, C., et al. (2012). Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol. Ecol. 80, 417–426. doi: 10.1111/j.1574-6941.2012.01310.x
Liu, X. Y., Wang, E. T., Li, Y., and Chen, W. X. (2007). Diverse bacteria isolated from root nodules of Trifolium, Crotalaria and Mimosa grown in the subtropical regions of China. Arch. Microbiol. 188, 1–14. doi: 10.1007/s00203-007-0209-x
Liu, X. Y., Wu, W., Wang, E. T., Zhang, B., Macdermott, J., and Chen, W. X. (2011). Phylogenetic relationships and diversity of β-rhizobia associated with Mimosa species grown in Sishuangbanna, China. Int. J. Syst. Evol. Microbiol. 61, 334–342. doi: 10.1099/ijs.0.020560-0
Los Santos, P., Palmer, M., Beukes, C., Steenkamp, E. T., Briscoe, L., Id, N. K., et al. (2018). Whole Genome Analyses Suggests that Burkholderia sensu lato Contains Two Additional Novel Genera Implications for the Evolution of Diazotrophy and Nodulation in the Burkholderiaceae. Gene 9, 1–23. doi: 10.3390/genes9080389
Martens, M., Dawyndt, P., Coopman, R., Gillis, M., De Vos, P., and Willems, A. (2008). Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 58, 200–214. doi: 10.1099/ijs.0.65392-0
Martens, M., Delaere, M., Coopman, R., De Vos, P., Gillis, M., and Willems, A. (2007). Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 57, 489–503. doi: 10.1099/ijs.0.64344-0
Melkonian, R., Moulin, L., Béna, G., Tisseyre, P., Chaintreuil, C., Heulin, K., et al. (2014). The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ. Microbiol. 16, 2099–2111. doi: 10.1111/1462-2920.12286
Mishra, R. P. N., Tisseyre, P., Melkonian, R., Chaintreuil, C., Miché, L., Klonowska, A., et al. (2012). Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 79, 487–503. doi: 10.1111/j.1574-6941.2011.01235.x
Nick, G., de Lajudie, P., Eardly, B. D., Suomalainen, S., Paulin, L., Zhang, X. P., et al. (1999). Sinorhizobium arboris sp nov and Sinorhizobium kostiense sp nov., isolated from leguminous trees in Sudan and Kenya. Int. J. Syst. Bacteriol. 49, 1359–1368. doi: 10.1099/00207713-49-4-1359
Parker, M. A., Wurtz, A. K., and Paynter, Q. (2007). Nodule symbiosis of invasive Mimosa pigra in Australia and in ancestral habitats: a comparative analysis. Biol. Invasions 9, 127–138. doi: 10.1007/s10530-006-0009-2
Payne, G. W., Vandamme, P., Morgan, S. H., LiPuma, J. J., Coenye, T., and Weightman, A. J. (2005). Development of a recA Gene-Based Identification Approach for the Entire Burkholderia Genus. Appl. Environ. Microbiol. 21, 3917–3927. doi: 10.1128/AEM.71.7.3917-3927.2005
Peix, A., Ramírez-bahena, M. H., Velázquez, E., Eulogio, J., Peix, A., Ram, M. H., et al. (2015). Critical reviews in plant sciences bacterial associations with legumes bacterial associations with legumes. Crit. Rev. Plant Sci. 34, 17–42. doi: 10.1080/07352689.2014.897899
Platero, R., James, E. K., Rios, C., Iriarte, A., Sandes, L., Zabaleta, M., et al. (2016). Novel Cupriavidus strains isolated from root nodules of native Uruguayan Mimosa species. Appl. Environ. Microbiol. 82, 3150–3164. doi: 10.1128/AEM.04142-15
Silva, V. C., Alves, P. A. C., Rhem, M. F. K., dos Santos, J. M. F., James, E. K., and Gross, E. (2018). Brazilian species of Calliandra Benth. (tribe Ingeae) are nodulated by diverse strains of Paraburkholderia. Syst. Appl. Microbiol. 41, 241–250. doi: 10.1016/j.syapm.2017.12.003
Sneath P. H. A., and Sokal, R. R. (1973). Numerial Taxonomy. The Principles and Practice of Classification. San Francisco, CA: W. H. Freeman.
Sprent, J. (2009). Legume Nodulation: A Global Perspective, 1st Edn. Hoboken, NJ: Wiley. doi: 10.1002/9781444316384
Sprent, J. I., Ardley, J., and James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56. doi: 10.1111/nph.14474
Stopnisek, N., Bodenhausen, N., Frey, B., Fierer, N., Eberl, L., and Weisskopf, L. (2014). Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ. Microbiol. 16, 1503–1512. doi: 10.1111/1462-2920.12211
Sun, L. N., Wang, D. S., Yang, E. D., Fang, L. C., Chen, Y. F., Tang, X. Y., et al. (2016). Cupriavidus nantongensis sp. nov., a novel chlorpyrifos-degrading bacterium isolated from sludge. Int. J. Syst. Evol. Microbiol. 66, 2335–2341. doi: 10.1099/ijsem.0.001034
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, Z.-Y., Xu, X.-D., Wang, E.-T., Gao, J.-L., Martinez-Romer, E., and Chen, W.-X. (1997). Phylogenetic and Genetic Relationships of Mesorhizobium tianshanense and Related Rhizobia. Int. J. Syst. Bacteriol. 874–879. doi: 10.1099/00207713-47-3-874
Taulé, C., Zabaleta, M., Mareque, C., Platero, R., Sanjurjo, L., Sicardi, M., et al. (2012). New betaproteobacterial Rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl. Environ. Microbiol. 78, 1692–1700. doi: 10.1128/AEM.06215-11
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Versalovic, J., Koeuth, T., and Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Vincent, J, M. (1970). A Manual for the Practical Study of the Root-Nodule Bacteria. Oxford: Blackwell Scientific.
Vinuesa, P., Silva, C., Lorite, M. J., Izaguirre-Mayoral, M. L., Bedmar, E. J., and Martínez-Romero, E. (2005). Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst. Appl. Microbiol. 28, 702–716. doi: 10.1016/j.syapm.2005.05.007
Wang, E. T., Chen, W. F., Tian, C. F., Young, J. P. W., and Chen, W. X. (2019). Ecology and Evolution of Rhizobia: Principles and Applications. Singapore: Springer Verlag. doi: 10.1007/978-981-32-9555-1
Wang, E. T., Rogel, M. A., Santos, A. G., Martinez-romero, J., and Cevallos, M. A. (1999a). Rhizobium etli bv. mimosae, a novel biovar isolated from Mimosa affinis. Int. J. Syst. Bacteriol. 49, 1479–1491. doi: 10.1099/00207713-49-4-1479
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/JB.173.2.697-703.1991
Wu, S., Chaw, S., and Rejmánek, M. (2003). Naturalized Fabaceae (Leguminosae) in Taiwan: the first approximation. Bot. Bull. Acad. Sin. 44, 59–66.
Zhang, J. J., Guo, C., Chen, W., DeLajudie, P., Zhang, Z., Shang, Y., et al. (2018). Mesorhizobium wenxiniae sp. nov., isolated from chickpea (Cicer arietinum L.) in China. Int. J. Syst. Bacteriol. 68, 1930–1936. doi: 10.1099/ijsem.0.002770
Zhang, J. J., Lou, K., Jin, X., Mao, P. H., Wang, E. T., Tian, C. F., et al. (2012). Distinctive Mesorhizobium populations associated with Cicer arietinum L. in alkaline soils of Xinjiang, China. Plant Soil 353, 123–134. doi: 10.1007/s11104-011-1014-5
Zhang, J. J., Yang, X., Guo, C., de Lajudie, P., Singh, R. P., Wang, E., et al. (2017). Mesorhizobium muleiense and Mesorhizobium gsp. nov. are symbionts of Cicer arietinum L. in alkaline soils of Gansu, Northwest China. Plant Soil 410, 103–112. doi: 10.1007/s11104-016-2987-x
Keywords: ecological distribution, Mimosa, phylogenetic diversity, rhizobia, soil conditions
Citation: Liu XY, You SH, Liu HJ, Yuan BJ, Wang HY, James EK, Wang F, Cao WD and Liu ZK (2020) Diversity and Geographic Distribution of Microsymbionts Associated With Invasive Mimosa Species in Southern China. Front. Microbiol. 11:563389. doi: 10.3389/fmicb.2020.563389
Received: 18 May 2020; Accepted: 11 September 2020;
Published: 28 October 2020.
Edited by:
Luisa I. Falcon, National Autonomous University of Mexico, MexicoReviewed by:
Angelica Cibrian-Jaramillo, National Polytechnic Institute of Mexico (CINVESTAV), MexicoMarcelo Simon, Brazilian Agricultural Research Corporation (EMBRAPA), Brazil
Copyright © 2020 Liu, You, Liu, Yuan, Wang, James, Wang, Cao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Liu, bGl1eGlhb3l1bmx5QDEyNi5jb20=
 Xiaoyun Liu
Xiaoyun Liu Shenghao You1
Shenghao You1 Euan K. James
Euan K. James