Abstract
Microbial communities associated to insect species are involved in essential biological functions such as host nutrition, reproduction and survivability. Main factors have been described as modulators of gut bacterial community, such as diet, habit, developmental stage and taxonomy of the host. The present work focuses on the complex changes that gut microbial communities go through when wild insects are introduced to artificial rearing conditions. Specifically, we analyzed the effect of the laboratory colonization on the richness and diversity of the gut bacteriome hosted by the fruit fly pest Anastrepha fraterculus sp. 1. Bacterial profiles were studied by amplicon sequencing of the 16S rRNA V3–V4 hypervariable region in gut samples of males and females, in teneral (1-day-old, unfed) and post-teneral (15-day-old, fed) flies. A total of 3,147,665 sequence reads were obtained and 32 bacterial operational taxonomic units (OTUs) were identified. Proteobacteria was the most abundant phylum (93.3% of the total reads) and, Wolbachia and Enterobacter were the most represented taxa at the genus level (29.9% and 27.7%, respectively, of the total read counts). Wild and laboratory flies showed highly significant differences in the relative abundances of bacteria. The analysis of the core bacteriome showed the presence of five OTUs in all samples grouped by origin, while nine and five OTUs were exclusively detected in laboratory and wild flies, respectively. Irrespective of fly origin or sex, a dominant presence of Wolbachia was observed in teneral flies, whereas Enterobacter was highly abundant in post-teneral individuals. We evidenced significant differences in bacterial richness and diversity among generations under laboratory colonization (F0, F1, F3 and F6) and compared to laboratory and wild flies, displaying also differential patterns between teneral and post-teneral flies. Laboratory and wild A. fraterculus sp. 1 harbor different gut bacterial communities. Laboratory colonization has an important effect on the microbiota, most likely associated to the combined effects of insect physiology and environmental conditions (e.g., diet and colony management).
Introduction
Insects can be considered as multiorganismal entities. Their microbiota accounts for up to 10% of the insect’s biomass and is involved in essential physiological roles modulating host fitness (Zilber-Rosenberg and Rosenberg, 2008; Feldhaar, 2011; Douglas, 2015; Morris, 2018; Carthey et al., 2019; Brown et al., 2020). Insect gut microbiome comprises obligate and facultative symbionts, opportunistic parasites, and mutualistic microbes (Bourtzis and Miller, 2003; Pontes and Dale, 2006), a complex structure that is mainly affected by host environment and taxonomy (Colman et al., 2012; Engel and Moran, 2013; Augustinos et al., 2019). Gut bacteria are particularly involved in metabolic pathways that enable the host to utilize nutrient-poor or unbalanced diets, providing essential amino acids (EAAs), vitamins, lipids and co-factors (Dillon and Dillon, 2004; Wu et al., 2006; Ankrah et al., 2017). Bacteria also participate in enzymatic functions, such as, hydrolysate of xylan, lipids, and esters, or fermentation of complex polysaccharides (Engel and Moran, 2013). In addition, gut bacteria are able to perform degradation of natural toxins such us phenolic compounds and complex terpenoids involved in plant defense (Hammer and Bowers, 2015; Berasategui et al., 2016; Jing et al., 2020) or chemical toxins facilitating host insecticide resistance (Vontas et al., 2011).
Tephritidae (Diptera) is a rather large family (ca. 5000 species, ref.) that includes about 70 species of fruit flies of economic importance. These species are considered pests because larvae develop inside a wide range of fruit species including many commercial ones (White and Elson-Harris, 1992). In most tephritid fruit flies examined so far, Enterobacteriaceae has been recognized as the dominant taxonomic group, also including a range of culturable and unculturable bacteria associated to the different life styles of the host species (Behar et al., 2008; Jurkevitch, 2011; Müller, 2013; Augustinos et al., 2015; Ventura et al., 2018). A greater richness of digestive symbiotic species is associated to polyphagous flies including microbes belonging to Proteobacteria and Firmicutes phyla (Morrow et al., 2015).
Previous studies on tephritids strongly suggested that symbionts associated to the digestive tract positively affect the host fitness. The majority of the studies have focused on the effect of adult and larval diet, host taxonomy and developmental stage (Augustinos et al., 2019; Noman et al., 2020). These studies showed that gut bacteria are able to improve parameters related with survivability and reproduction, such as male sexual performance, flight ability, female fecundity, foraging behavior and longevity under starvation (Ben-Yosef et al., 2008a,b; Ben-Ami et al., 2010; Gavriel et al., 2011; Augustinos et al., 2015; Kyritsis et al., 2017, 2019; Akami et al., 2019; reviewed by Noman et al., 2020). On the other hand, certain gut bacteria have been found to be pathogenic for the host (Behar et al., 2008).
Anastrepha fraterculus Wiedemann (Diptera: Tephritidae), known as the South American fruit fly, has a wide geographic distribution ranging from northern Mexico to central Argentina (Steck, 1999). This fruit fly pest is recognized as a complex of cryptic species composed by at least eight morphotypes (Selivon et al., 2005; Hernández-Ortiz et al., 2012, 2015; Devescovi et al., 2014). Particularly in Argentina, only one member of this complex is present, the A. fraterculus sp. 1 or Brazilian 1 morphotype (Selivon et al., 2005; Goday et al., 2006; Hernández-Ortiz et al., 2012). This morphotype is considered a serious threat to fruit production and trade in Argentina and several South American countries. Currently, only chemical control strategies or insect traps are implemented to manage the pest in integrated management programs, which prompted the development of specific and environmentally friendly methods such as the sterile insect technique (SIT). The SIT involves the mass rearing, sterilization and release of sterile insects to compete and mate with wild individuals resulting in non-viable offspring and a gradual reduction of the pest population (Metcalf and Luckmann, 1994; Hendrichs and Robinson, 2009). Significant progress has been made to set the basis of knowledge needed to implement the SIT against A. fraterculus. In particular, mass rearing protocols (Salles, 1999; Jaldo et al., 2001; Sobrinho et al., 2006; Vera et al., 2007; Hernández et al., 2010; Walder et al., 2014), irradiation procedures (Allinghi et al., 2007a,b) and behavioral and physiological studies to improve the competitiveness of sterile males (Segura et al., 2007, 2009, 2013; Abraham et al., 2011, 2013; Liendo et al., 2013; Vera et al., 2013; Bachmann et al., 2015, 2017, 2019) have been addressed. Nonetheless, previous studies seldom considered A. fraterculus as a multiorganismal entity and only Juárez et al. (2019) focused on the importance of gut bacteria in A. fraterculus sp. 1 physiology and behavior.
The implementation of a successful SIT strategy requires that insects adapt to mass rearing conditions without losing valuable traits that allow them to survive and mate in natural conditions (Ochieng-Odero, 1994; Cayol, 2000; Meats et al., 2009). During the introduction of a wild population to artificial rearing conditions (laboratory colonization), variations in the environment and diet could affect biological parameters of flies and the presence and abundance of symbionts that are key determinants of their sexual competitiveness and survival (Bartlett, 1984; Cayol, 2000). In natural habitats, the development of immature stages of fruit fly species takes place first inside the fruit and then, after pupation, buried into the ground. In laboratory conditions, the eggs are deposited by females in artificial oviposition units (OUs) then transferred to artificial larval diet and pupation substrate (Walder et al., 2014). Likewise, under laboratory rearing, adult flies are provided artificial food (including a protein supply and water offered ad libitum), which extensively differs from the available resources of the wild environment. Such artificial conditions (among others) could affect gut bacterial diversity and the vertical transmission of symbionts from parents to offspring, typically from mothers through the eggs when females are forced to lay eggs in artificial oviposition devices (Lauzon et al., 2009; Jaenike, 2012). Likewise, artificial rearing media (both at larval and adult stages) can potentially affect the diversity of bacteria in the gut, favoring the presence of specific taxonomic groups that are not common in nature.
In the present work, we hypothesized that the composition of gut bacterial taxa hosted by A. fraterculus sp. 1 is modified during the laboratory colonization and vary among diverse origins, maintaining a shared central core of bacteria. Our approach included the analysis of bacterial diversity and abundance regarding sex, feeding status and origin, studied in adult insects from a wild population, flies through the first six generations during the laboratory colonization process, and flies reared under laboratory conditions.
Materials and Methods
Laboratory Insects
Adult insects were obtained from the A. fraterculus colony kept at the Institute of Genetics “Ewald A. Favret” (IGEAF). This colony (hereafter LAB flies) was established in 2007 and maintained for ca. 60 generations. No wild material has been introduced to refresh its genetic background. This fruit fly strain derived from a colony kept at Estación Experimental Agroindustrial Obispo Colombres (Tucumán, Argentina) established in 1997 with wild pupae recovered from infested guavas (Psidium guajava, Myrtaceae) collected in Tafi Viejo (Tucumán, Argentina). Flies used in the present study were collected at the pupal stage. After emergence, adult insects were handled in the same way as described above and kept under the same experimental conditions.
Wild Insects Collected From Traps
Adult individuals of A. fraterculus sp. 1 were collected using McPhail traps lured with torula yeast. Traps were hanged from an ubajay tree (Hexachlamys edulis, Myrtaceae), located within INTA (Instituto Nacional de Tecnología Agropecuaria) experimental field (34°36′24.7″S 58°40′07.6″W), during the fruiting season (December) and were daily checked. Flies were collected with an aspirator and then taken to the laboratory, sorted by sex and their digestive tract immediately dissected (see below). The samples were named as WU (wild ubajay) (Supplementary Table S1).
Wild Insects Collected From Fruits
Feijoa (Acca sellowiana, syn. Feijoa sellowiana, Myrtaceae) fruits infested with A. fraterculus sp. 1 larvae were sampled in Hurlingham, Buenos Aires, Argentina (34°36′40.2″S 58°40′20.9″W). Fruits were transported to the laboratory and placed on perforated plastic trays which fit in larger trays with sand as substrate for pupation. Twice a week, pupae were recovered and transferred to 3-liter glass containers and kept under controlled environmental conditions (temperature [temp]: 25 ± 1°C; relative humidity [RH]: 70 ± 10%) until adult emergence.
Emerged adults were identified at the species level according to their morphology following Hernández-Ortiz et al. (2010) and Norrbom et al. (2012). Those flies that belong to A. fraterculus were placed in standard cages under controlled conditions (temp: 25°C ± 1°C; RH: 75 ± 5%; photoperiod: 14:10 [light:dark]) with water but no food. Fifteen adult individuals of each sex (males and females) were randomly sampled from the cage in two different feeding status: unfed individuals collected the first day after emergence (from now on called “teneral” [T] flies) and, 15-day-old (sexually mature and fed) individuals (from now on called “post-teneral” [PT] flies) (Figure 1). Post teneral flies were provided with water and artificial adult diet (a mix of hydrolyzed yeast:hydrolyzed corn:sugar in a 1:2:4 ratio, and vitamins [Dayamineral, Abbott]).
FIGURE 1
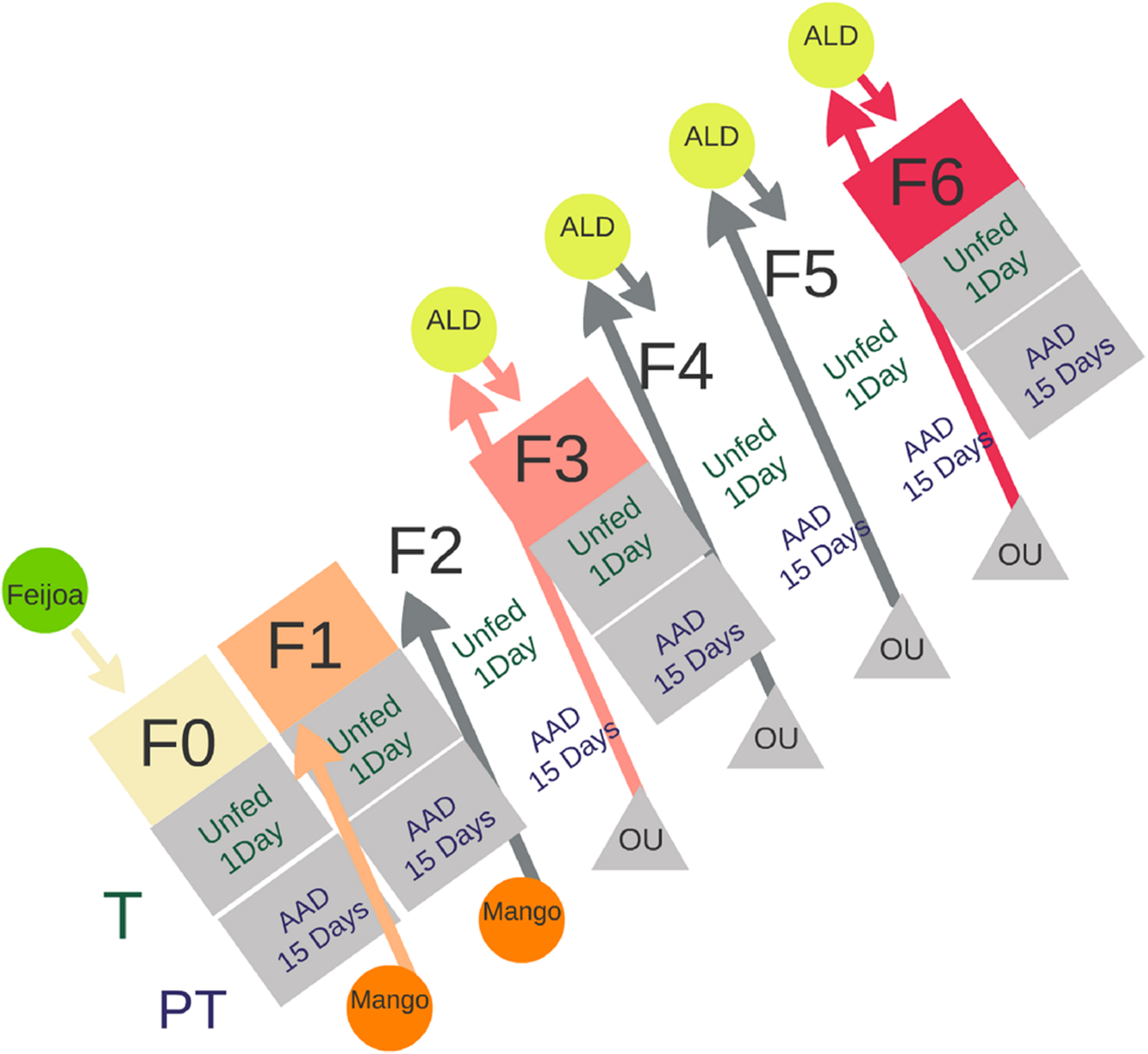
Schematic description of the protocol followed to establish an A. fraterculus sp. 1 laboratory colony using wild flies recovered from feijoa infested fruits (green circle). F0 to F6 indicate the generations of flies reared under laboratory conditions. T (teneral) and PT (post-teneral) individuals, represent feeding states of adult insects. Mango and artificial oviposition units (OUs) were used to collect eggs for the next generation. ALD, artificial larval diet; AAD, artificial adult diet.
Wild flies that emerged in the laboratory were considered the first generation of artificial rearing (F0). This population was used to reproduce insects and obtain the following generations (F1 to F6) (Figure 1). In order to offer an adequate oviposition substrate, females from F0 to F2 generations were provided ripe mango fruits (Mangifera indica, Anacardiaceae). Mangoes were washed with neutral detergent and tap water and then immersed in a 0.06% w/v sodium benzoate solution before exposure to females. Exposure lasted 24 h. Fruit were then placed in sealed boxes with a layer of vermiculite as pupation substrate. Pupae were recovered two times per week and used to establish the next generation. Further generations (F3 to F6) were obtained using artificial OUs according to Parreño et al. (2014) (Supplementary Table S1). OUs consisted of cylindrical plastic vials (2 cm in height, 2.5 cm in diameter) filled with water colored with red food dye (Fleibor, Tablada, Buenos Aires, Argentina) and covered with Parafilm M (Pechiney Plastic Packaging, Chicago, IL, United States). After 24–48 h, eggs were recovered from the OUs and transferred to containers (4 cm × 2.5 cm × 3 cm) filled with artificial larval diet (Percent composition [% w/v] of the following supplies: Torula type B (Bioserve Inc.) 6.0; sugar 6.0; wheat germ 6.0; cholesterol 0.05; agar–agar 3.0; methyl paraben 0.1; Be Na 0.1; water 70 to 80; and HCl 36% 0.5 ml to maintain the pH between 3.7 and 4.0 [Salles, 1999]). These containers were placed over a thin layer of vermiculite as pupation substrate. Pupae were recovered two times per week and used to establish the next generation.
Digestive Tract Dissection and DNA Isolation
Adult flies were washed twice in 70% ethanol and sterilized distilled water. The gut and the subesophageal bulb were identified and dissected with sterile dissecting forceps in PBS 1X under a stereoscopic microscope (Olympus SZ30, 40× zoom) according to Caetano et al. (2006). We performed three replicates for each evaluated condition with five guts per sample (see more details in Figure 1 and Supplementary Table S1). We considered three main factors: gender; origin (F0, LAB and WU) and feeding status (1-day-old and unfed individuals, and 15-day-old and fed flies); except in the case of WU for which feeding status was unknown.
Total DNA extractions of gut samples were performed following Juárez et al. (2019) based on the procedure described by Baruffi et al. (1995) with the following modifications related to sample size: (i) all volumes were reduced to half; (ii) final elution was reduced to 10 μL of TE buffer (Tris base 10 mM; EDTA 1 mM, pH 8.1). Briefly, five guts were ground and incubated (65°C) in a lysis buffer (NaCl 100 mM, Sucrose 200 mM, Tris-HCl pH 9.1 100 mM, EDTA 50 mM, SDS 0.5%) with proteinase K (final concentration of 100 μg/ml) (USB). The incubation was stopped by adding potassium acetate 8M and centrifuged. The recovered supernatant was treated with RNase (1 mg/ml) (USB). DNA was precipitated with 100% ethanol and centrifuged, the pellet was washed with 100% ethanol and 70% ethanol, and finally dried and resuspended in TE buffer.
Library Preparation and Illumina MiSeq Sequencing
The quantity and quality of the extracted DNAs were measured using NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, United States). A quantity of 50 ng of DNA per sample was used as template to generate amplicons corresponding to the V3–V4 hypervariable region of the bacterial 16S rRNA gene. A first round of PCR amplification was performed using KAPA HiFi HotStart PCR Kit (Kapa Biosystems) and MiSeq primers 341F and 805R (Klindworth et al., 2013). Negative controls were included in DNA extractions and PCRs were performed under the same conditions as the rest of the samples but without any genetic material. No amplicons were obtained from these negative controls. PCR products obtained were separated in a 1.2% w/v agarose gel electrophoresis to verify the size. The amplification products were visualized in Bio-Rad’s Gel DocTM XR + system. Positive PCR fragments were then purified from primers and primer dimers using a 20% PEG, 2.5 M NaCl solution, centrifuged at 14.000 × g for 20 min and the precipitate was washed twice with 125 μl of a 70% v/v ethanol solution and centrifuged at 14.000 × g for 10 min as previously described (Ntougias et al., 2016). The dried precipitates were suspended in 15 μl of sterile deionized water and the concentration was measured with a Quawell Q5000 micro-volume UV-Vis spectrophotometer, diluted up to 10 ng/μl and used as template in a second round of PCR. In this step, indexed adapters were added to the ends of the 16S rDNA amplicons, as well as the Illumina adaptors. The combinatorial use of index primers resulted in unique samples that were pooled and sequenced on one Illumina MiSeq run. The resulting amplicons were cleaned-up by AMPure XP beads (Agencourt, United Kingdom) and diluted to 2.66 ng/μl. Finally, they were pooled equimolarly and mixed into indexed library following the 16S-metagenomic library preparation guide 15044223-b (Illumina Inc., 2013). Massive amplicon sequencing was performed using an Illumina MiSeq sequencing platform by Macrogen.
Data Analysis
The pre-processing of reads was carried out using USEARCH v10. Paired Fastq files were assembled by using algorithms implemented in USEARCH v10 using -fastq_mergepairs command with -fastq_maxdiffs, -fastq_pctid, -fastq_minmergelen, and -fastq_maxmergelen options set at default values. All reads were trimmed and filtered by quality using -fastq_filter, with the -fastq_maxee option set at 1.0, and -fastx_uniques commands. Unique sequences were identified, and all samples were clustered at increasing similarities of 97% using UPARSE-OTU algorithm (Edgar, 2013). Using this algorithm, chimera filtering and operational taxonomic units (OTUs) clustering were carried out simultaneously. For the clustering, a minimum abundance (value = 2) was used discarding singletons. For the OTU Table trimming, we defined 0.001 as the minimum frequency for an OTU. The OTU frequency was calculated as follow: (number of count reads for an OTU/total number of count reads)∗100. For the OTU Table trimming, we defined 0.001 as the minimum frequency for an OTU. The taxonomy assignment was performed against a reference database (SILVA release 119; Quast et al., 2013). UNCROSS2 algorithm was run to detect and filter crosstalk (Edgar, 2018).
Diversity estimates including observed OTUs and Good’s coverage were calculated using final count data. Richness (Chao1), diversity (Simpson and Shannon), dominance (Berger–Parker) and evenness (Pielou) indices of alpha diversity, which reflect the diversity of individual samples were calculated based on “vegan” R package (Oksanen et al., 2019) and were plotted using the “ggplot2” R Package (Wickham, 2016).
Phylogenetic diversity (Faith index) was estimated using the Picante package in R (Kembel et al., 2010). Alpha diversity indices were compared by pairwise Kruskal–Wallis test in R (R Core Team, 2020). Core bacterial OTUs shared by LAB, WU, F0 and F1–F6 were identified by comparing OTUs from the different origins following Andongma et al. (2019).
Beta diversity was analyzed to evaluate the similarity of bacterial communities from different locations using Generalized UniFrac distance (Chen et al., 2012) and visualized via Non-metric Multidimensional scaling (NMDS) plot using the RHEA pipeline in R (Lagkouvardos et al., 2017). A permutational multivariate analysis of variance using distance matrices was calculated using “adonis” function from “vegan” R package to determine significance differences between the separated groups. Statistically significant differences between samples were identified with permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001), p-values of PERMANOVA test indicating the significance of group separations and the dissimilarity scale of the grid, d = 0.1 means that the distance between two grid lines represent approximately 10% dissimilarity between the samples. The Bonferroni–Hochberg method was used to correct for multiple PERMANOVA testing. The Non-parametric Wilcoxon rank sum test (Kruskal, 1957) was used to perform pair-wise comparisons of mean relative abundance of bacteria between gut samples.
Results
Overall Data Analysis
After an ultimate and strict trimming process, a total of 3,147,665 high quality reads were achieved from a total of 66 A. fraterculus sp. 1 gut samples. A set of 32 bacterial OTUs were identified, clustered at 97% sequence similarity (Table 1 and Supplementary Table S1). Members of three bacterial phyla were identified: Patescibacteria, Proteobacteria and Firmicutes. Proteobacteria was the most abundant taxonomic group (93.3% of reads), followed by Firmicutes (6.5%) and Patescibacteria (0.2%). Within Proteobacteria, two taxonomic classes dominated: Alpha- and Gammaproteobacteria. At the genus level, Wolbachia and Enterobacter were the most abundant taxa (29.9% and 27.7% of the obtained reads, respectively), followed by Providencia (8.3%), Aeromonas (7.1%), Citrobacter (5.9%) and Burkholderia–Caballeronia–Paraburkholderia (4.4%) (Table 1). Good’s coverage index was 98%, suggesting that the majority of bacterial phylotypes in the insect digestive tract were included in this study. In addition, three OTUs (15, 124 and 19) were placed in three distinct phylogenetic positions, showing lower than 97% similarity to known described species of the genera Raoultella sp., Klebsiella sp. and the phylum of Saccharibacteria, respectively. For OTU19 taxonomic assignment was not feasible below the phylum level.
TABLE 1
| OTU ID | Read counts | % read counts | Phylum | Class | Order | Family | Genus |
| OTU 1 | 942423 | 29.9 | Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | Wolbachia |
| OTU 2 | 860684 | 27.3 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter |
| OTU 3 | 141265 | 4.5 | Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | Aeromonas |
| OTU 4 | 259733 | 8.3 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Providencia |
| OTU 5 | 67142 | 2.1 | Firmicutes | Bacilli | Lactobacillales | Enterobacteriaceae | Enterococcus |
| OTU 6 | 72793 | 2.3 | Proteobacteria | Gammaproteobacteria | Betaproteobacteriales | Burkholderiaceae | Burkholderia–Caballeronia–Paraburkholderia |
| OTU 7 | 32959 | 1.1 | Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus |
| OTU 8 | 35181 | 1.1 | Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | Caulobacter |
| OTU 9 | 64879 | 2.1 | Proteobacteria | Gammaproteobacteria | Betaproteobacteriales | Burkholderiaceae | Burkholderia–Caballeronia–Paraburkholderia |
| OTU 10 | 81701 | 2.6 | Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | Aeromonas |
| OTU 11 | 41643 | 1.3 | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus |
| OTU 12 | 27726 | 0.9 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Lactococcus |
| OTU 13 | 29721 | 0.9 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Xanthobacteraceae | Bradyrhizobium |
| OTU 14 | 20969 | 0.7 | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas |
| OTU 15 | 65801 | 2.1 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Raoultella |
| OTU 16 | 10523 | 0.3 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Serratia |
| OTU 17 | 12157 | 0.4 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | Mesorhizobium |
| OTU 18 | 5227 | 0.2 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter |
| OTU 19 | 6261 | 0.2 | Saccharibacteria | Unknown | Unknown | Unknown | Unknown |
| OTU 20 | 19166 | 0.6 | Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus |
| OTU 21 | 5358 | 0.2 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | Mesorhizobium |
| OTU 22 | 4207 | 0.1 | Proteobacteria | Alphaproteobacteria | Acetobacterales | Acetobacteraceae | Commensalibacter |
| OTU 24 | 8207 | 0.3 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus |
| OTU 26 | 8951 | 0.3 | Proteobacteria | Gammaproteobacteria | Betaproteobacteriales | Neisseriaceae | Neisseria |
| OTU 31 | 4088 | 0.1 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter |
| OTU 35 | 6508 | 0.2 | Firmicutes | Bacilli | Lactobacillales | Enterobacteriaceae | Enterococcus |
| OTU 44 | 3323 | 0.1 | Proteobacteria | Gammaproteobacteria | Betaproteobacteriales | Neisseriaceae | Neisseria |
| OTU 52 | 5783 | 0.2 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter |
| OTU 65 | 184274 | 5.9 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Citrobacter |
| OTU 124 | 107352 | 3.4 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Klebsiella |
| OTU 136 | 4119 | 0.1 | Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Rheinheimera |
| OTU 168 | 7541 | 0.2 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter |
Representation and classification of OTUs identified in A. fraterculus sp. 1 gut bacteriome. The OTUs highlighted compose the A. fraterculus sp. 1 gut bacterial core proposed in the present work.
Core Bacteriome Analysis
A total of five OTUs (Rheinheimera, Enterobacter [OTU2], Acinetobacter [OTU31], Enterococcus and Providencia) made up the core bacterial community (36.1% of read counts). We defined the core bacteriome of A. fraterculus sp. 1 as constituted by taxonomic units present in all the samples grouped by origin (LAB, WU, including samples involved in the laboratory colonization assay F0 and F1–F6) (Figure 2A and Table 1). The relative abundance of taxonomic groups from the core bacteriome varied in the examined samples ranging from 9.5% reads (40% samples) in average for LAB, 7.9% (50% of the samples) in average for WU, 6.1% (50% of the samples in average) in average for F0 and 6.7% (33.3% of the samples) in average for F1–F6 (Figure 2A and Supplementary Tables S2A,B). In addition, when LAB and WU were compared, six OTUs (Rheinheimera, Klebsiella [OTU124], Enterobacter [OTU2], Acinetobacter [OTU31], Enterococcus and Providencia) were shared. Furthermore, nine OTUs (Wolbachia, Bradyrhizobium, Sphingomonas, two OTUs of Mesorhizobium, Acinetobacter [OTU18], Staphylococcus, Burkholderia–Caballeronia–Paraburkholderia [OTU6] and Caulobacter) were found in LAB but not in WU samples and, five OTUs (Lactobacillus, Lactococcus, Raoultella [OTU15], Enterobacter [OTU168] and Citrobacter) were detected in WU samples but not in LAB flies (Figure 2B; see details in Supplementary Tables S2A,B). Additionally, when total values were compared, five OTUs (Enterobacter [OTU2], Wolbachia, Rheinheimera, Enterobacter [OTU168] and Citrobacter) were represented in at least 40% of the analyzed samples (Figure 2C). This overall analysis showed two out of five OTUs (Enterobacter [OTU2] and Wolbachia) with a high percentage of read counts and the other three OTUs (Rheinheimera, Enterobacter [OTU168] and Citrobacter) were represented by a low percentage of read counts (Figure 2C and Table 1).
FIGURE 2
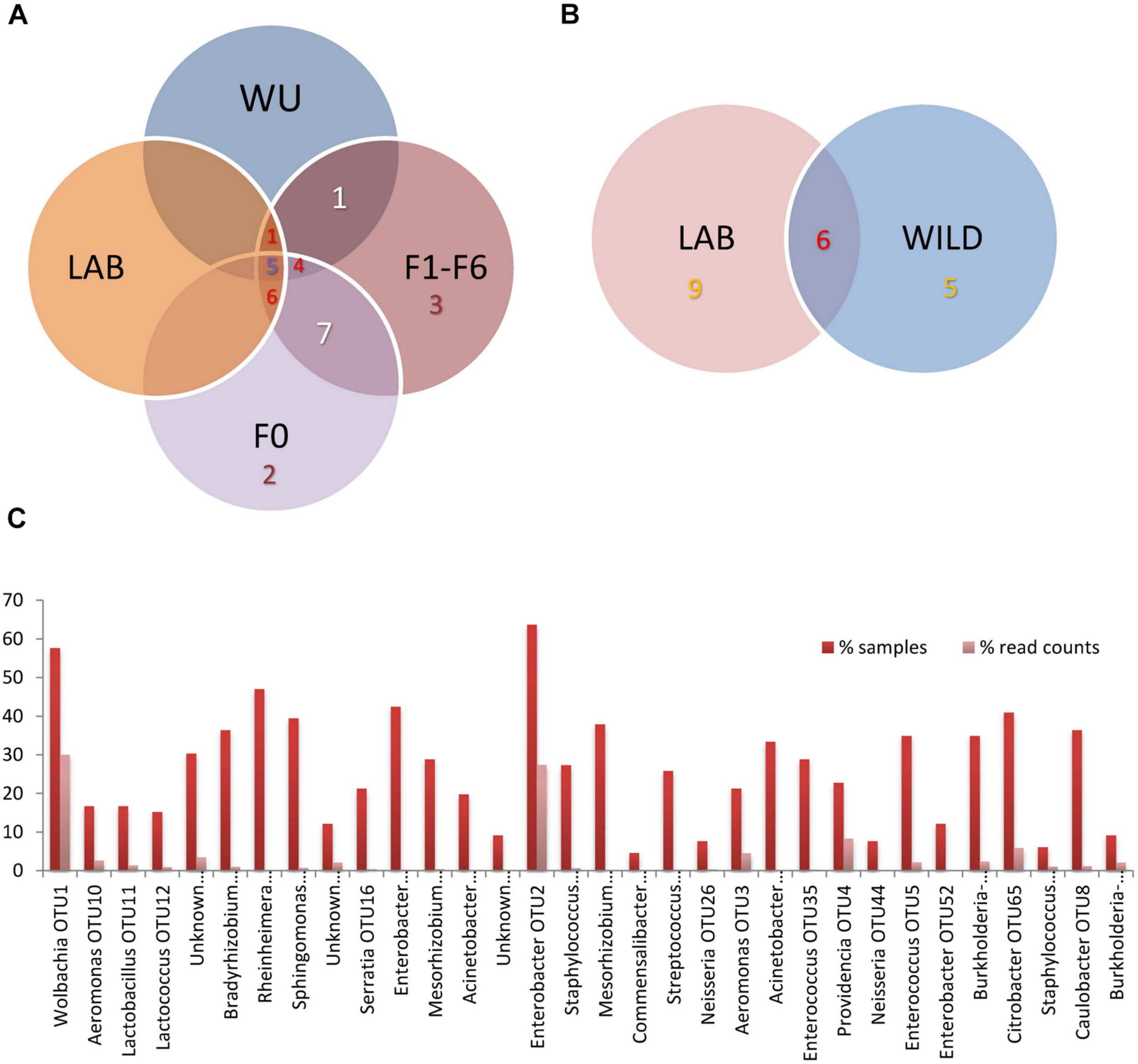
Distribution of bacterial OTUs among A. fraterculus sp. 1 gut samples. (A) samples pooled in four groups (LAB, WU, F0, and F1–F6) according to their origin. (B) Samples grouped in LAB and WILD (F0 + WU). LAB, individuals from the laboratory colony; WU, wild individuals from ubajay with unknown feeding status; F0, individuals collected from infested feijoa fruits; F1–F6, individuals from generations 1, 3 and 6 reared under laboratory conditions. (C) OTU representation: % of samples and reads per OTU considering all analyzed samples (N = 66) (See more details in Supplementary Tables S2A–D).
Gut Bacterial Community and Sex
Statistical comparisons of samples grouped by sex evidenced that there are not significant differences in the gut bacteriome composition between females and males of A. fraterculus (UniFrac distance; PERMANOVA p > 0.05; Supplementary Figure S1). Males and females showed the same distribution of bacterial OTUs and abundance, with Enterobacter and Wolbachia observed in a high relative abundance (Supplementary Figure S2), and similar values of Chao1, Shannon and Simpson indices (Supplementary Figures S3A,B,D). In addition, non-significant differences were observed between sexes for evenness (Pielou index), phylogenetic diversity (Faith index), and dominance (Berger–Parker index) (Supplementary Figures S3C,E,F). Therefore, sex was not considered for further analyses performed in this study.
Effect of the Feeding Status and Fly Origin on the Gut Bacteriome
Significant differences in gut bacterial composition were observed between individuals with different feeding status (teneral [T] and post-teneral [PT]) (UniFrac distance; PERMANOVA p < 0.05; Figure 3A). In addition, we observed a differential distribution of bacterial OTUs and abundance between T and PT samples. Wolbachia was detected at a high prevalence (60.0% of reads) in T samples, whereas, Enterobacter (42.0% of reads), Providencia (18.3% of reads) and Aeromonas (15.2% of reads) dominated the gut bacteriome of PT individuals (Supplementary Figure S4). Furthermore, differential values of Chao1 and Simpson indices were observed between T and PT samples, showing higher values in T flies (Supplementary Figure S5). Additionally, evenness (Pielou index), phylogenetic diversity (Faith index), and dominance (Berger–Parker index) evidenced the same trends of significant differences between T and PT flies (Supplementary Figures S5C,E,F). Conversely, Shannon index showed no significant differences between T and PT flies (Supplementary Figure S5B).
FIGURE 3
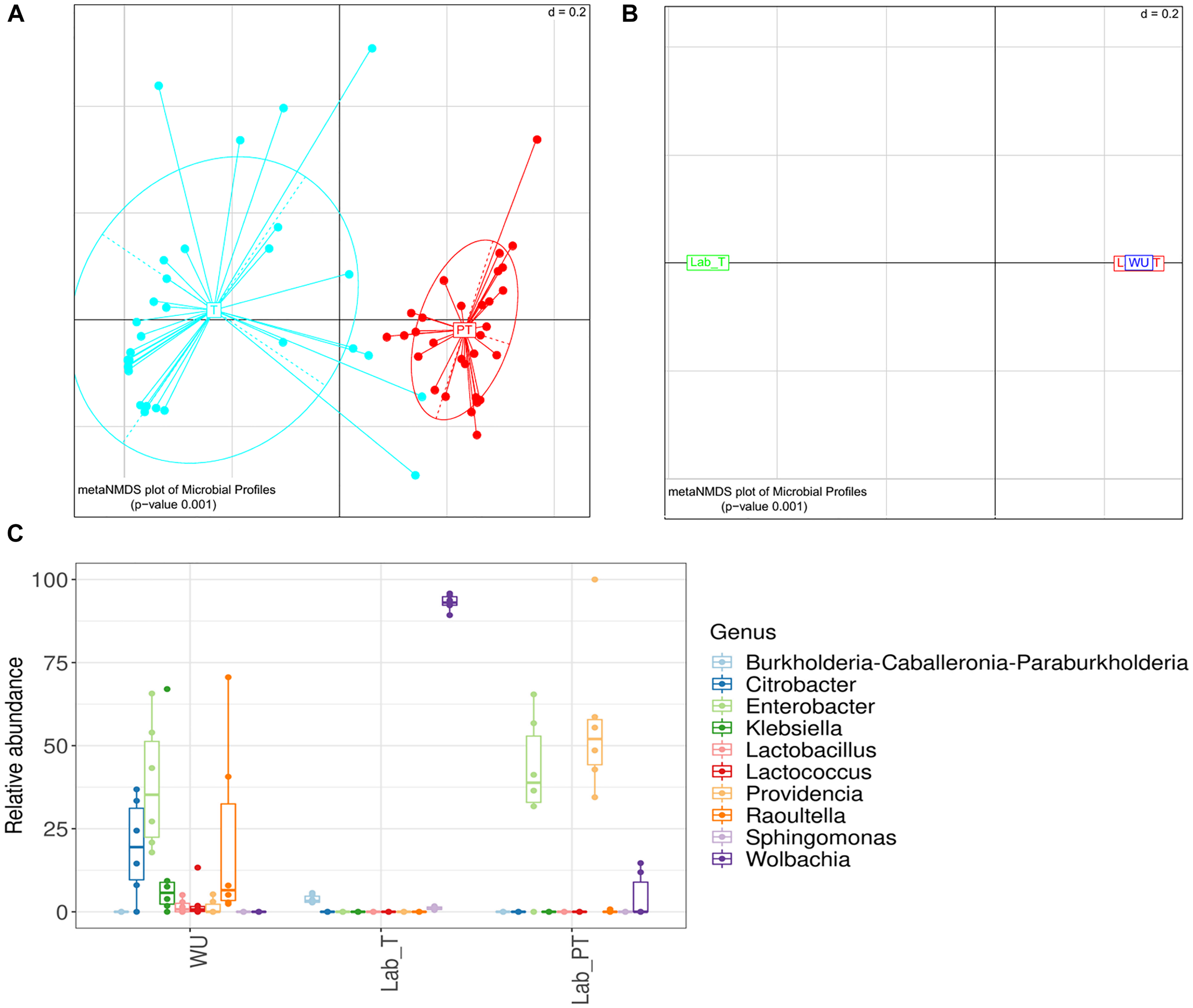
Effect of the feeding status and fruit fly origin on the gut bacteriome. (A,B) Meta-Non-metric Multidimensional scaling (meta NMDS) plot representing sample groups according to: (A) the feeding status (T, teneral; PT, post-teneral); (B) the origin (LAB, laboratory individuals with different feeding status [T, teneral; PT, post-teneral]; WU, wild individuals from ubajay with unknown feeding status). Significance p-value from PERMANOVA analysis; d = 0.2. (C) Relative abundance of bacterial genera associated to the digestive tract of flies from two different environmental origins (Lab, laboratory samples with different feeding status (T and PT); WU, wild samples from ubajay. Error bars correspond to standard error of the mean.
In relation to the fly origin and feeding status, significant differences were observed among gut samples from the laboratory (LAB_T and LAB_PT), and wild flies (WU) collected in ubajay trees with unknown feeding status (UniFrac distance; PERMANOVA p < 0.05; Figure 3B). The gut bacteriome of WU samples was dominated by Enterobacter OTU2 (37.86% of reads), Raoultella (21.75% of reads) and Citrobacter (19.50% of reads) (Supplementary Table S2B) whereas LAB_PT gut bacteria was mainly represented by Enterobacter OTU2 (39.02%) and Providencia (56.09%) (Supplementary Table S2C). LAB_T gut community was dominated by Wolbachia (93.12% of reads) (Figure 3C).
Bacterial richness and diversity of teneral (T) and post-teneral (PT) flies from LAB, and WU showed significant differences in paired comparisons. Chao1 index and phylogenetic diversity (Faith index) showed non-significant differences between Lab_T and WU flies (Figure 4A and Supplementary Figure S6D). However, WU showed significantly higher values of Shannon index than LAB_T (Figure 4B). The same statistical differences were also observed for the Simpson index and evenness (Pielou index) (Supplementary Figures S6A,B). Congruently, Lab_T showed significantly higher values of dominance (Berger–Parker index) than WU (Supplementary Figure S6C).
FIGURE 4
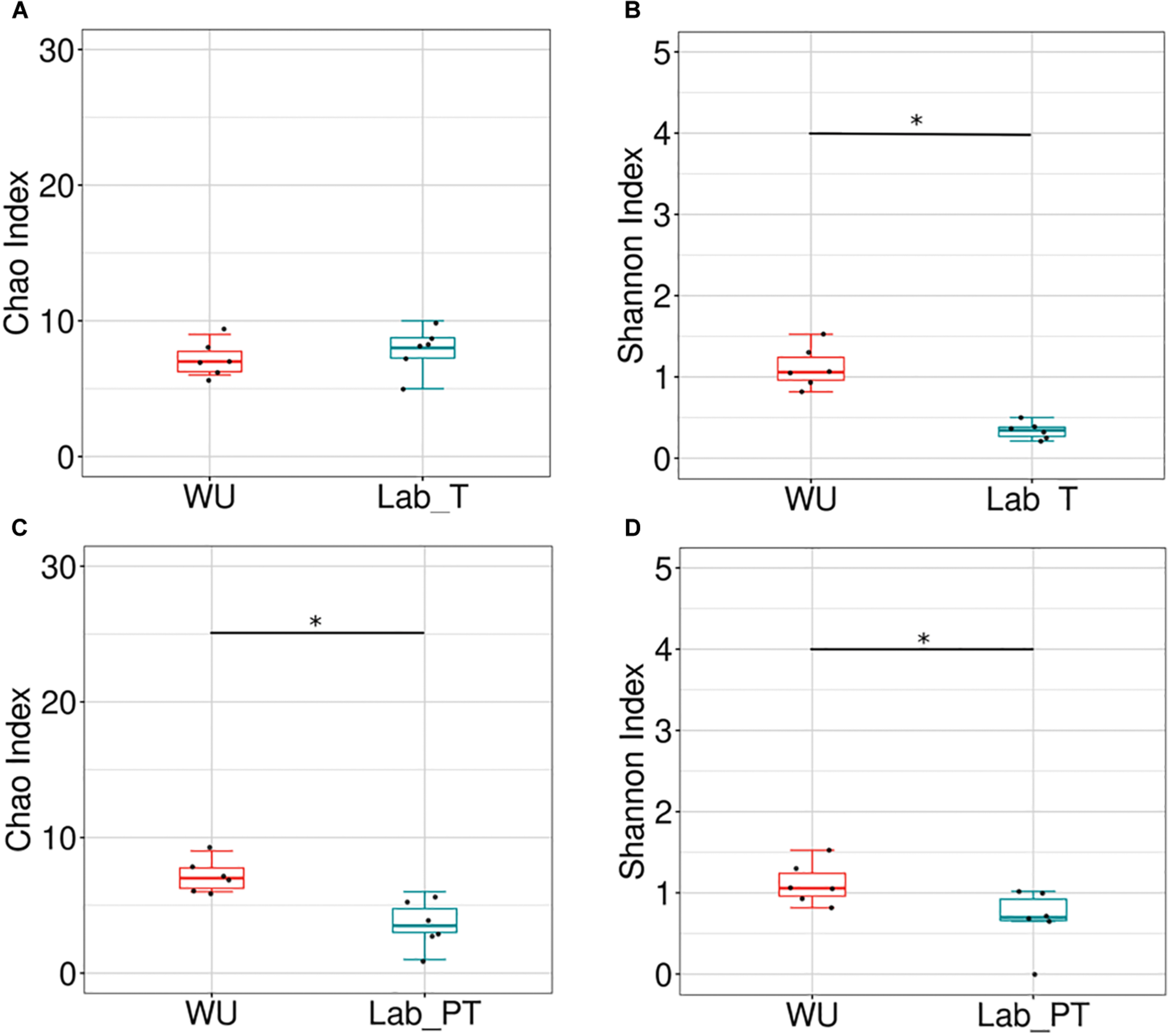
Effect of the fruit fly origin and feeding status on the gut bacterial community. Box plots representing bacterial richness and diversity among origins. Chao index (A,C) and Shannon index (B,D) in teneral (T) and post-teneral (PT) individuals. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony. Dots indicate observed values and box plots depict means and standard deviation of the data. Bars with asterisks above boxes indicate significant p-values (paired comparisons, Kruskal–Wallis test).
In Lab_PT vs. WU comparisons, WU samples evidenced significantly higher values of Chao1, Shannon indices and phylogenetic diversity (Faith index) (Figures 4C,D and Supplementary Figure S6H). Conversely, additional analyses of diversity (Simpson index), evenness (Pielou index), and dominance (Berger–Parker index) showed non-significant differences between compared samples (Supplementary Figures S6E–G).
Changes in Gut Bacteriome During Laboratory Colonization
Significant differences in the gut bacterial profile were observed across generations under artificial rearing conditions (F0–F6) both in teneral (T) and post-teneral (PT) flies (UniFrac distance; PERMANOVA p < 0.05; Figures 5A,B). Considering T flies from F0–F6, we observed that bacterial profiles differed between generations, and in all cases, were different from LAB_T and WU flies (PERMANOVA; p < 0.05; Table 2 and Figure 5A). For PT flies, F0–F6 generations differed in paired comparisons between them and among LAB_PT, and WU. Interestingly, bacterial profiles of teneral and post-teneral F6 were significantly different from teneral and post-teneral LAB samples, respectively (Table 2 and Figures 5A,B).
FIGURE 5
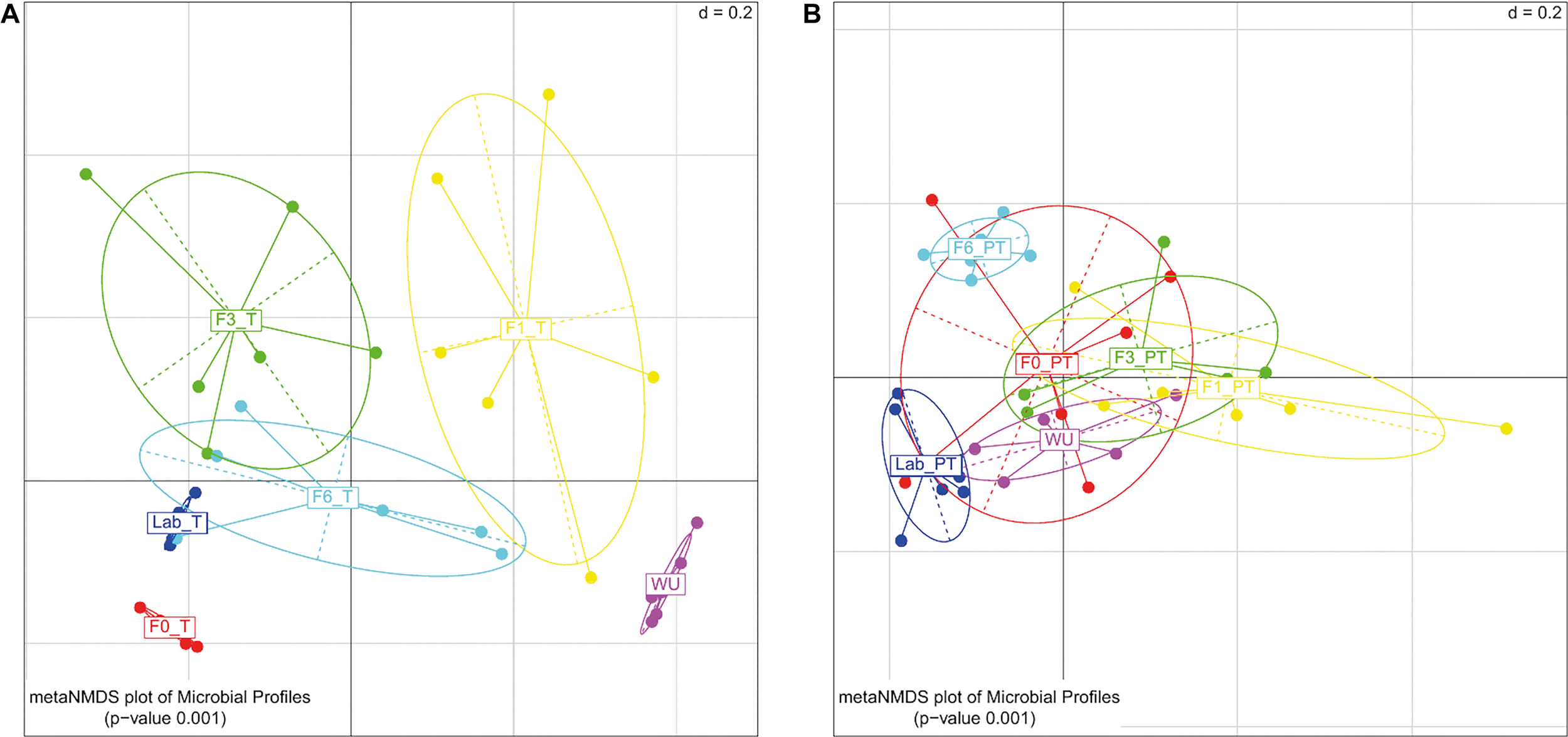
Gut-associated bacterial communities of A. fraterculus sp. 1 during laboratory colonization. Meta-Non-metric Multidimensional scaling (metaNMDS) plot of bacterial profile representing samples grouped by generation under artificial rearing (F0, F1, F3, and F6) and feeding status. (A) Teneral (T) samples. (B) Post teneral (PT) samples. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony. Significance p-value from PERMANOVA analysis (d = 0.2).
TABLE 2
| Comparison | p-Value | Corr. p-value |
| F0_T-F1_T | 0.005 | 0.006 |
| F0_T-F3_T | 0.001 | 0.003 |
| F0_T-F6_T | 0.003 | 0.0045 |
| F0_T-LAB_T | 0.003 | 0.005 |
| F0_T-WU_unk | 0.003 | 0.0045 |
| F1_T-F3_T | 0.005 | 0.0057 |
| F1_T-F6_T | 0.009 | 0.010 |
| F1_T-LAB_T | 0.002 | 0.006 |
| F1_T-WU_unk | 0.005 | 0.00625 |
| F3_T-F6_T | 0.002 | 0.021 |
| F3_T-LAB_T | 0.004 | 0.005 |
| F3_T-WU_unk | 0.004 | 0.006 |
| F6_T-LAB_T | 0.01 | 0.011 |
| F6_T-WU_unk | 0.004 | 0.0066 |
| LAB_T-WU_unk | 0.001 | 0.006 |
| F0_PT-F1_PT | 0.017 | 0.024 |
| F0_PT-F3_PT | 0.021 | 0.024 |
| F0_PT-F6_PT | 0.017 | 0.0225 |
| F0_PT-LAB_PT | 0.004 | 0.008 |
| F0_PT-WU_unk | 0.019 | 0.023 |
| F1_PT-F3_PT | 0.053 | 0.053 |
| F1_PT-F6_PT | 0.005 | 0.0125 |
| F1_PT-LAB_PT | 0.001 | 0.0085 |
| F1_PT-WU_unk | 0.007 | 0.011 |
| F3_PT-F6_PT | 0.002 | 0.0064 |
| F3_PT-LAB_PT | 0.004 | 0.0085 |
| F3_PT-WU_unk | 0.004 | 0.0428 |
| F6_PT-LAB_PT | 0.002 | 0.01 |
| F6_PT-WU_unk | 0.003 | 0.0056 |
| LAB_PT-WU_unk | 0.004 | 0.012 |
| F0_T-F1_T | 0.005 | 0.006 |
| F0_T-F3_T | 0.001 | 0.003 |
| F0_T-F6_T | 0.003 | 0.0045 |
| F0_T-LAB_T | 0.003 | 0.005 |
PERMANOVA analysis – pair-wise comparisons.
The gut bacteriome associated with T flies was dominated by Wolbachia sp. (>50% of the total reads, found in all samples), with the exception of F1_T, in which, members of Burkholderia–Caballeronia–Paraburkholderia (OTU9) were the most dominant taxon (32.2% of reads) (Figures 6A–C and Supplementary Table S2C). Gammaproteobacteria was the most representative class in PT samples (>75% of reads, found in all samples) (Figures 6D,E). The same taxonomic classes with a differential relative abundance were observed in F0–F6_PT flies compared to F0–F6_T flies (Figures 6B,E). At the genus level, Wolbachia was detected in a low relative abundance in PT flies (<15%, detected in 50% of the samples) (Supplementary Tables S2C,D). Significant differences were observed when the Wolbachia relative abundance was compared between T and PT flies in each generation (Wilcoxon Rank Sum Test; p < 0.05), except for F1_T vs F1_PT (Supplementary Table S4). In addition, Wolbachia was not detected in WU samples (Figure 6F and Supplementary Tables S2A,B).
FIGURE 6
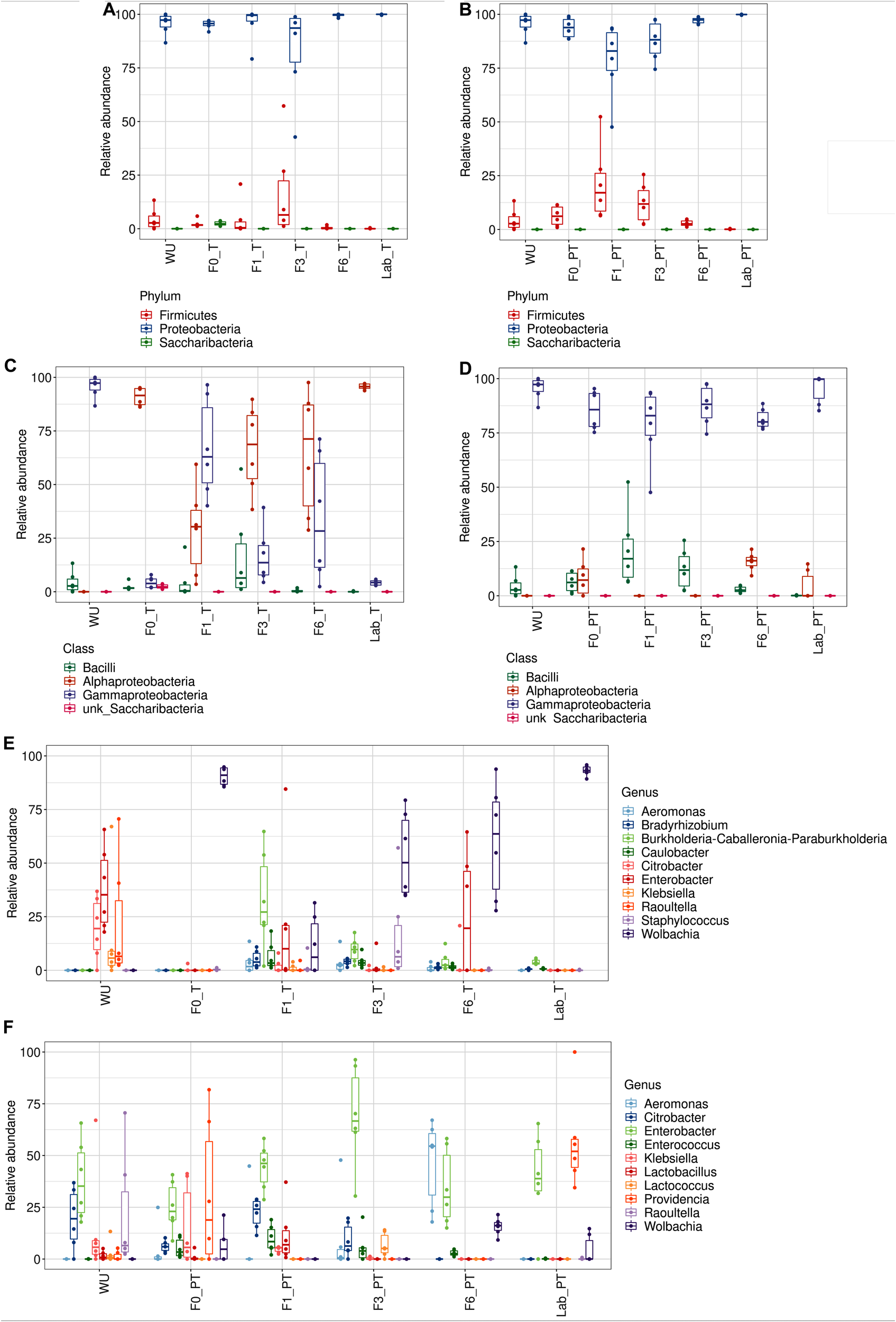
Gut-associated bacterial communities during of A. fraterculus sp. 1 laboratory colonization and feeding status. Relative abundance of the ten main OTUs identified. Taxonomic identification at the Phylum, Class and Genus levels for teneral (T) (A–C, respectively) and post-teneral (PT) (D–F, respectively) flies from F0, F1, F3, F6 generations under laboratory conditions. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony.
Klebsiella sp. and Providencia were noticeably abundant in F0_PT (>20%). In addition, we detected Citrobacter highly represented in WU, which was also found in F1_T and F6_T flies and in PT samples from F0, F1, and F3.
Teneral (T) and post-teneral (PT) flies presented different patterns of Chao1, Shannon and Simpson indices across the laboratory colonization process. For T flies, F1 and F3 showed the highest Chao1, Shannon and Simpson values (Figures 7A,B, Supplementary Table S3, and Supplementary Figure S7A) and F3 resulted significantly different from F0 and LAB (for the three indices, Figures 7A,B and Supplementary Figure S7A). In addition, LAB_T showed significant differences in Chao1 index compared to F0, F1 and F3. LAB_T and WU showed the lowest values for this index. In the case of Shannon and Simpson indices analysis, differentiation between paired comparisons of LAB_T and F1–F6_T flies and LAB_T-WU was detected. LAB_T and F0_T flies displayed the lowest Shannon index values but non-significant differences were detected between them (Figure 7B and Supplementary Figure S7A). With regard to PT flies, significant differences were observed between LAB_PT and WU, and each paired comparisons of LAB_PT with F0, F1, and F6 PT for Chao1 index. Differential values were also observed in F0–F1, F0–F3, F1–F3, and F3–F6 comparisons for this parameter. In sum, T flies showed a tendency to a reduction of Chao1, Shannon, Simpson, Faith and Pielou indices, as generations under artificial rearing increased (F1–F6), congruently with an increase of dominance estimated by Berger–Parker index (Figures 7A,B and Supplementary Figures S7B–D). Laboratory flies showed the lowest values for Shannon, Simpson, Chao1, phylogenetic diversity (Faith) and evenness (Pielou) estimators together with the highest values recorded for Berger–Parker index. This tendency was not detected in PT flies (Figures 7C,D and Supplementary Figure S7).
FIGURE 7
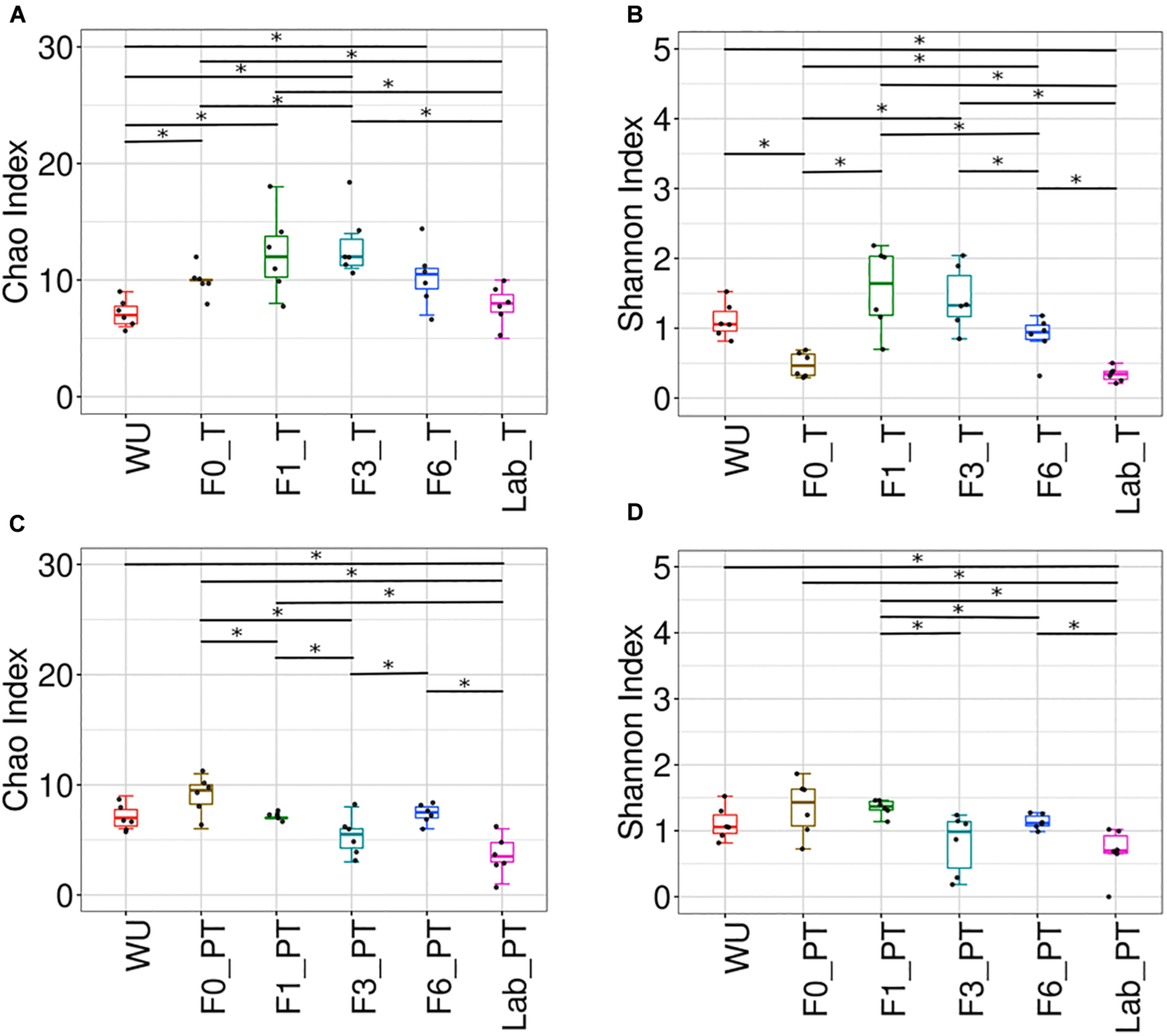
Bacterial community of the A. fraterculus sp. 1 digestive tract and laboratory colonization process. Bacterial richness and diversity associated to the digestive tract of F0–F6 individuals and laboratory flies. Chao index (A,C) and Shannon index (B,D) in teneral (T) and post-teneral (PT) individuals. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony. See Figure 4 for dots and box plots description. Bars with asterisks above boxes indicate significant p-values (paired comparisons, Kruskal–Wallis test).
Discussion
In the present work, we analyzed the bacterial community associated with the digestive tract of A. fraterculus sp. 1 adults using 16S rRNA amplicon sequencing. We were able to identify 32 OTUs in this gut bacteriome. From these classified taxonomic units, 29 OTUs were described at the genus level, one OTU (OTU19) remains unclassified under phylum level, identified as unknown Saccharibacteria and two other OTUs (15 and 124) were placed in two distinct taxonomic positions, showing lower similarity than 97% with Raoultella sp. and Klebsiella sp., respectively.
Overall analyses suggested that the gut bacterial profile was dominated by Proteobacteria, particularly by Alpha and Gammaproteobacteria; followed by Firmicutes and Saccharibacteria. Within the Gammaproteobacteria, the Enterobacteriaceae family was dominant in the gut of A. fraterculus, in agreement with previous reports in Tephritidae fruit flies (Behar et al., 2008; Jurkevitch, 2011; Müller, 2013; Morrow et al., 2015; Ventura et al., 2018; Augustinos et al., 2019; Koskinioti et al., 2019). The most abundant genera, were Wolbachia (Alphaproteobacteria) and Enterobacter (Gammaproteobacteria), followed by other members of the Gammaproteobacteria such as Providencia, Aeromonas, Citrobacter, Burkholderia–Caballeronia–Paraburkholderia, Klebsiella and Raoultella. Tephritid literature reports these genera of Proteobacteria with specific functions: Wolbachia with cytoplasmic incompatibility and male killing (Boller and Bush, 1974; Riegler and Stauffer, 2002; Zabalou et al., 2004, 2009; Apostolaki et al., 2011; Conte et al., 2019; Devescovi et al., 2019; Mateos et al., 2020), Enterobacter, Citrobacter, Burkholderia, Klebsiella and Raoultella with nitrogen metabolism (Murphy et al., 1988; Behar et al., 2005; Raza et al., 2020), and Providencia with pathogenic effects (Msaad Guerfali et al., 2018; Ksentini et al., 2019). In addition, Acinetobacter and Rheinheimera (Gammaproteobacteria) were detected in low abundance (0,1% of total reads) but in a high percentage of samples (33.33 and 46.97% of total samples, respectively) and wide distribution (Lab, WU, F0, F1–F6 flies). Despite Acinetobacter having been reported in several studies on fruit flies, its role is so far unclear (Kounatidis et al., 2009; Deutscher et al., 2019).
The gut bacteriome of Anastrepha genus was previously addressed using different tools. In A. ludens, A. obliqua, A. serpentina, and Anastrepha striata, Escherichia was one of the dominant genera (Ventura et al., 2018). Nonetheless, Escherichia was not identified in our samples, nor in A. grandis, A. ludens, and two morphotypes of A. fraterculus studied by Augustinos et al. (2019). For A. fraterculus, Proteobacteria dominate the gut bacterial community according to Augustinos et al. (2019) and our results. However, Müller (2013) found that the gut bacteriome of a Brazilian wild population of A. fraterculus was dominated by Actinobacteria. This author targeted a different region of the 16S rRNA gene, which could partially explain the differences; however, the main explanation is probably associated to environmental variation (Augustinos et al., 2019).
Despite some differences in the three studies on A. fraterculus, a rather clear pattern seems to emerge with Enterobacteriaceae being consistently the most abundant family associated to A. fraterculus, and particularly Enterobacter as the genus with the highest abundance. Recent studies have revealed the importance of Enterobacter spp. associated to Tephritidae (Kyritsis et al., 2017, 2019; Noman et al., 2020). As an example of essential contribution of this symbiont to fruit fly physiology, multiple traits have been addressed, including: nitrogen fixation and pectinolytic activity (Behar et al., 2005; Aharon et al., 2012) as well as, provision of essential and non-essential amino acids and vitamins (Azis et al., 2019) and its role on host behavior and fitness (Hamden et al., 2013; Augustinos et al., 2015; Kyritsis et al., 2017, 2019; Raza et al., 2020). These beneficial effects might explain why adding Enterobacter spp. to the larval diet in C. capitata improved pupal and adult productivity, as well as a faster development, particularly of males (Hamden et al., 2013; Augustinos et al., 2015). Further studies on the physiological role of Enterobacter spp. in A. fraterculus sp. 1 will bring valuable information to be applied to mass rearing protocols and environmentally safe control strategies.
Within Firmicutes, Enterococcus was the most abundant genus in the A. fraterculus gut bacterial community. A recent publication revealed that diet enriched with Enterococcus reduced the duration of the larval stage, increased pupal weight, and increased longevity in Bactrocera dorsalis (Khaeso et al., 2017). Finally, within the Saccharibacteria phylum, we detected an unclassified taxon that was exclusively found in F0_T flies. Similar results were observed by Koskinioti et al. (2019) who detected an unknown family of the order Saccharimonadales in the gut symbiotic communities of Bactrocera oleae wild samples.
Our results evidenced the presence of five common bacterial OTUs (Enterobacter [OTU2], Providencia, Enterococcus, Rheinheimera and Acinetobacter [OTU31]) described as the core bacteriome associated to the gut of A. fraterculus sp. 1 adult individuals. Digestive core bacteria and their contribution to the host biology have been addressed in other Tephritidae species including C. capitata, B. minax, and Zeugodacus cucurbitae (Wang et al., 2014; Yong et al., 2017; Andongma et al., 2019; reviewed by Deutscher et al., 2019). Particularly, our study showed an initial evaluation of the taxonomic composition of A. fraterculus sp. 1 core bacteria, with a restricted sampling considering different origins. In addition, we detected non-shared bacteria between laboratory and wild populations (WU, F0) and a population under adaptation (F1–F6), which could bring useful information to perform a further characterization and selection of bacterial isolates with potential benefits to improve mass rearing protocols and fitness of adults in the field in assistance to the SIT development against A. fraterculus sp. 1.
Changes in gut bacterial composition during laboratory colonization were strongly associated to the feeding status and generation. After six generations of laboratory rearing, the bacterial community is still changing and seems to maintain the differentiation from LAB flies. In our experiment, females from F0, F1, and F2 did not lay eggs in artificial OUs, and thus were offered mangoes. Artificial units were used from F3 onward. Our results are in line with previous studies conducted in other fruit fly species (Deutscher et al., 2019) and are strongly associated to the feeding behavior. Accordingly, the lack of differences in gut bacterial composition between females and males obtained in our work and in other related articles (Wang et al., 2014; Morrow et al., 2015; Augustinos et al., 2019) supported a substantial role of nutrition on the digestive microbiota composition, however, the bacterial diversity associated to different origins remained to be an important point of differentiation. These findings were also supported by studies performed in B. oleae (Koskinioti et al., 2019; revised by Deutscher et al., 2019).
We identified two OTUs (OTU22 assigned to Commensalibacter and OTU19, classified as an unknown member of the Saccharibacteria phylum) exclusively present in teneral wild flies from feijoa host fruit. These OTUs were no longer detected in F1 onward. Similarly, Citrobacter (OTU65) and Klebsiella (OTU124) were found in high abundance in wild adult flies (WU) and wild flies from feijoa (F0), but then slowly decreased as laboratory colonization progressed (F1 to F6); not being detected at all in LAB fruit fly guts. In other fruit fly species, Citrobacter [as all Enterobacteriaceae (Octavia and Lan, 2014)] has been shown to be capable of reducing nitrate to nitrite (Borenshtein and Schauer, 2006). Furthermore, the analysis of tryptic soy broth culture filtrates of Citrobacter freundii isolated from A. ludens showed that it contained greater amounts of nitrogenated compounds (Robacker and Bartelt, 1997). Because in nature nitrogen is a rather scarce resource (Mattson, 1980) and most tephritids need protein to achieve sexual maturation, gut bacteria may play a key role by making nitrogen available to their hosts. However, under laboratory conditions flies had unlimited access to a highly rich peptide source (hydrolyzed yeast) and the presence of some of these bacterial groups may no longer be essential. This might indicate that these OTUs are relevant for the host fitness in nature but are probably not needed under laboratory conditions. Alternatively, vertical transmission of these OTUs could be compromised under artificial rearing (Sacchetti et al., 2014; Augustinos et al., 2019; Deutscher et al., 2019). Under this scenario, some gut bacteria might have disadvantageous conditions under the laboratory, and this situation could have negatively impacted on their diversity. However, more work is needed to address potential negative selection on specific bacterial groups as a consequence of drastic environmental changes, such as those suffered by wild flies when are brought to the laboratory and breed under captivity. Understanding the physiological role of these OTUs might shed light on important attributes of wild flies that are lost because of domestication.
Teneral flies of the F0 generation had the lowest values of diversity estimated through different indices among F0–F6 generations; whereas F1 and F3 showed the highest values for these parameters. This may be related with stochastic drift processes emerging after disturbs and associated to environmental factors that may have compromised the dominance of Wolbachia at least in F1, right after the introduction of the host to novel conditions (Staubach et al., 2013). After F3, and in F6 and LAB teneral flies, Wolbachia dominated the gut bacteria community. On the other hand, in post-teneral flies (15-day-old, fed individuals) the community is dominated by Enterobacter. This might indicate that the community is stabilized in post-teneral flies, and this status would be less exposed to environmental stressors. In agreement with our results, a recent study performed in A. obliqua (whole body) showed Wolbachia sp. more abundant in larvae and Enterobacter sp. in adults (Gallo-Franco and Toro-Perea, 2020).
Changes in the composition of digestive bacteria during the domestication process could affect the physiology and behavior of the host eventually leading to limitations in the context of the SIT, which requires a productive mass rearing but also sexually competitive. Previous studies in C. capitata highlighted the contribution of gut bacterial symbionts associated to the digestive tract to male sexual performance, flight ability and longevity under starvation and enhancement of the SIT (Niyazi et al., 2004; Ben-Yosef et al., 2008a; Ben-Ami et al., 2010; Gavriel et al., 2011; Augustinos et al., 2015; Kyritsis et al., 2017). Understanding the replacement of gut bacteria associated with domestication is an initial step to determine which bacterial symbionts are the most affected. This characterization would allow to design domestication protocols that either maintain key players in the gut of domesticated insects or restore them as part of the rearing process (i.e., use specific bacteria as dietary probiotics). Because the insect gut microbiome includes not only the gut bacteriome, but also virus, protozoa, fungi, yeasts that might interact not only with the host but also among them (Gurung et al., 2019), future studies should aim at disentangling these complex interactions and enrich our understanding of the physiology and behavior of this important fruit pest of South America.
Conclusion
Our work revealed that the origin and feeding status could shape the gut bacterial community of A. fraterculus adults. We observed a dynamic interaction between A. fraterculus and its microbiota. Upon emergence, the gut is dominated by Wolbachia but as flies feed and age, other genera such as Enterobacter, Providencia, and Citrobacter become more abundant, and the whole community more diverse, reaching a seemingly stable composition. We evidenced gradual changes during the first steps of the laboratory colonization process keeping, however, a degree of differentiation between flies under adaptation and a well-established laboratory strain. The taxonomic identification of the gut bacterial community of A. fraterculus sp. 1 from Argentina and the analysis of key factors modeling the structure and composition of the gut bacteriome provide valuable and novel information. Understanding the dynamic interaction between a tephritid host and digestive bacterial symbionts will enable to improve environmentally safe control strategies against fruit fly pest species.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. 16S rRNA gene sequences reported in this study have been deposited in the NCBI under BioProject number PRJNA624812.
Author contributions
SL, LP, GT, KB, and DS conceived and designed the study. JS, LP, FM, RR, EA, and PS conducted the experiments. JS, DS, SL, and GT analyzed the results. JS, DS, SL, KB, JC, and GT drafted the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by funds from the International Atomic Energy Agency through the Research Contracts N°17041 to DS and N°17074 to GT; the Agencia Nacional de Promoción Científica y Tecnológica (Argentina) through the project Foncyt-PICT 2013-0054 to DS; and the National Institute of Agricultural Technology (INTA) through the projects AEBIO-242411 (module pests) and PNBIO 11031044 to SL. JS was supported by Erasmus Mundus International Study Programme, European Union (EU) for a training visit to GT laboratory (University of Patras, Greece) (Sep-Dec 2018).
Acknowledgments
This study benefitted from discussions at International Atomic Energy Agency funded meetings for the Coordinated Research Projects “Use of symbiotic bacteria to reduce mass-rearing costs and increase mating success in selected fruit pests in support of SIT application,” “Comparing rearing efficiency and competitiveness of sterile male strains produced by genetic, transgenic or symbiont-based technologies” and “Colony management of insects for sterile insect technique application.”
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.570960/full#supplementary-material
Supplementary Figure 1A. fraterculus sp. 1 digestive bacterial community and sex. Meta-Non-metric Multidimensional scaling (metaNMDS) plot of bacterial profile representing samples grouped by sex: male in blue; female in red color. PERMANOVA analysis p-value (d = 0.2).
Supplementary Figure 2Relative abundance of OTUs (at genus level) of bacterial community associated to the digestive tract of flies grouped by sex.
Supplementary Figure 3Bacterial diversity found in A. fraterculus sp. 1 digestive tract of adult individuals grouped by sex. (A) Chao index. (B) Shannon index. (C) Faith index. (D) Simpson index. (E) Pielou index and (F) Berger index. See Figure 4 for dots and box plots description.
Supplementary Figure 4Relative abundance of OTUs (at genus level) of bacterial community associated to the digestive tract of flies grouped by feeding status: T (teneral); PT (post-teneral).
Supplementary Figure 5Bacterial diversity found in A. fraterculus sp. 1 digestive tract of adult individuals grouped by feeding status: T (teneral); PT (post-teneral). (A) Chao index. (B) Shannon index. (C) Faith index. (D) Simpson index. (E) Pielou index and (F) Berger index. See Figure 4 for dots and box plots description. Bars with asterisks above boxes indicate significant p-values (paired comparisons, Kruskal–Wallis test).
Supplementary Figure 6Bacterial diversity among origins - additional parameters. Simpson index (A,E); Pielou index (B,F); Berger-Parker index (C,G); and Faith index (D,H) in teneral (T) and post-teneral (PT) individuals. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony. Dots indicate observed values and box plots depict means and standard deviation of the data. Bars with asterisks above boxes indicate significant p-values (paired comparisons, Kruskal–Wallis test).
Supplementary Figure 7Bacterial diversity during laboratory colonization - additional parameters. Simpson index (A,E); Pielou index (B,F); Berger–Parker index (C,G); and Faith index (D,H) in teneral (T) and post-teneral (PT) individuals. WU, wild individuals from ubajay with unknown feeding status; Lab, individuals from the laboratory colony. See Figure 4 for dots and box plots description. Bars with asterisks above boxes indicate significant p-values (paired comparisons, Kruskal–Wallis test).
Supplementary Table 1Identification of gut samples and description of rearing conditions of A. fraterculus sp. 1 laboratory colonies and wild flies used in this work. F0–F6 indicates samples from generations under laboratory colonization. Lab, laboratory samples; WU, wild samples from ubajay; T, teneral feeding status; PT, post-teneral feeding status; M, male; F, female; Unk, unknown, without information on age, feeding status, and diet (at larval and adult stages).
Supplementary Table 2OTU description and representation in A. fraterculus sp. 1 gut bacteriome. (A) OTU representation – Percentage of samples. (B) OTU representation – Percentage of read counts. (C) OTU representation – Percentage of read counts considering feeding status. (D) OTU representation in % of samples considering feeding status. OTUs belonging to the bacterial core proposed are highlighted.
Supplementary Table 3Bacterial diversity and richness (Shannon and Chao indices).
Supplementary Table 4Relative abundance of Wolbachia (OTU 1) pair-wise comparisons (teneral [T] vs post-teneral [PT]) using Wilcoxon Rank Sum Test. Group compared: generation F0–F6 and laboratory flies (Lab). Mean values of each compared group were considered to the statistical analysis.
References
1
Abraham S. Goane L. Cladera J. Vera M. T. (2011). Effects of male nutrition on sperm storage and remating behavior in wild and laboratory Anastrepha fraterculus (Diptera: Tephritidae) females.J. Insect Physiol.571501–1509. 10.1016/j.jinsphys.2011.08.004
2
Abraham S. Liendo M. C. Devescovi F. Peralta P. A. Yusef V. Ruiz J. et al (2013). Remating behavior in Anastrepha fraterculus (Diptera: Tephritidae) females is affected by male juvenile hormone analog treatment but not by male sterilization.Bull. Entomol. Res.103310–317. 10.1017/S0007485312000727
3
Aharon Y. Pasternak Z. Ben-Yosef M. Behar A. Lauzon C. Yuval B. et al (2012). Phylogenetic, metabolic, and taxonomic diversities shape Mediterranean fruit fly microbiotas during ontogeny.Appl. Environ. Microbiol.79303–313. 10.1128/aem.02761-12
4
Akami M. Andongma A. A. Zhengzhong C. Nan J. Khaeso K. Jurkevitch E. et al (2019). Intestinal bacteria modulate the foraging behavior of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae).PLoS One14:e210109. 10.1371/journal.pone.0210109
5
Allinghi A. Calcagno G. Petit-Marty N. Gómez Cendra P. Segura D. Vera T. et al (2007a). Compatibility and competitiveness of a laboratory strain of Anastrepha Fraterculus (Diptera: Tephritidae) after irradiation treatment.Florida Entomol.9027–32. 10.1653/0015-4040(2007)90[27:cacoal]2.0.co;2
6
Allinghi A. Gramajo C. Willink E. Vilardi J. C. (2007b). Induction of Sterility in Anastrepha Fraterculus (Diptera: Tephritidae) by gamma radiation.Florida Entomol.9096–102. 10.1653/0015-4040(2007)90[96:iosiaf]2.0.co;2
7
Anderson M. J. (2001). Permutation tests for univariate or multivariate analysis of variance and regression.Can. J. Fish. Aquat. Sci.58626–639. 10.1139/cjfas-58-3-626
8
Andongma A. A. Wan L. Dong Y.-C. Wang Y.-L. He J. Niu C.-Y. (2019). Assessment of the Bacteria community structure across life stages of the Chinese Citrus Fly, Bactrocera minax (Diptera: Tephritidae).BMC Microbiol.19:1–9. 10.1186/s13049-019-0683-6
9
Ankrah N. Y. D. Luan J. Douglasa A. E. (2017). Cooperative metabolism in a threepartner insect-bacterial symbiosis revealed by metabolic modeling.J. Bacteriol.1991–15. 10.1128/JB.00872-16
10
Apostolaki A. Livadaras I. Saridaki A. Chrysargyris A. Savakis C. Bourtzis K. (2011). Transinfection of the olive fruit fly Bactrocera oleae with Wolbachia: towards a symbiont-based population control strategy.J. Appl. Entomol.135546–553. 10.1111/j.1439-0418.2011.01614.x
11
Augustinos A. A. Kyritsis G. A. Papadopoulos N. T. Abd-Alla A. M. M. Cáceres C. Bourtzis K. (2015). Exploitation of the medfly gut microbiota for the enhancement of sterile insect technique: use of Enterobacter sp. in larval diet-based probiotic applications.PLoS One10:e136459. 10.1371/journal.pone.0136459
12
Augustinos A. A. Tsiamis G. Cáceres C. Abd-Alla A. M. M. Bourtzis K. (2019). Taxonomy, diet, and developmental stage contribute to the structuring of gut-associated bacterial communities in tephritid pest species.Front. Microbiol.10:2004. 10.3389/fmicb.2019.02004
13
Azis K. Zerva I. Melidis P. Caceres C. Bourtzis K. Ntougias S. et al (2019). Biochemical and nutritional characterization of the medfly gut symbiont Enterobacter sp. AA26 for its use as probiotics in sterile insect technique applications.BMC Biotechnol.19:90. 10.1186/s12896-019-0584-9
14
Bachmann G. E. Devescovi F. Nussenbaum A. L. Cladera J. L. Fernández P. C. Vera M. T. et al (2017). Male sexual enhancement after methoprene treatment in Anastrepha fraterculus (Diptera: Tephritidae): a sustained response that does not fade away after sexual maturation.J. Insect Physiol.1017–14. 10.1016/j.jinsphys.2017.06.009
15
Bachmann G. E. Devescovi F. Nussenbaum A. L. Milla F. H. Shelly T. E. Cladera J. L. et al (2019). Mate choice confers direct benefits to females of Anastrepha fraterculus (Diptera: Tephritidae).PLoS One14:e214698. 10.1371/journal.pone.0214698
16
Bachmann G. E. Segura D. F. Devescovi F. Juárez M. L. Ruiz M. J. Vera M. T. et al (2015). Male sexual behavior and pheromone emission is enhanced by exposure to guava fruit volatiles in Anastrepha fraterculus.PLoS One10:e124250. 10.1371/journal.pone.0124250
17
Bartlett A. C. (1984). “Genetic changes during insect domestication,” in Advances and Challenges In Insect Rearing, edsKingE. G.LepplaN. C. (Atlanta, GA: Southern Region, U.S. Dept. of Agriculture), 2–8.
18
Baruffi L. Damiani G. Guglielmino C. R. Bandii C. Malacrida A. R. Gasperi G. (1995). Polymorphism within and between populations of Ceratitis: Comparison between RAPD and multilocus enzyme electrophoresis data.Heredity74425–437. 10.1038/hdy.1995.60
19
Behar A. Yuval B. Jurkevitch E. (2005). Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata.Mol. Ecol.142637–2643. 10.1111/j.1365-294X.2005.02615.x
20
Behar A. Yuval B. Jurkevitch E. (2008). Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity.J. Insect Physiol.541377–1383. 10.1016/j.jinsphys.2008.07.011
21
Ben-Ami E. Yuval B. Jurkevitch E. (2010). Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance.ISME J.428–37. 10.1038/ismej.2009.82
22
Ben-Yosef M. Behar A. Jurkevitch E. Yuval B. (2008a). Bacteria-diet interactions affect longevity in the medfly - Ceratitis capitata.J. Appl. Entomol.132690–694. 10.1111/j.1439-0418.2008.01330.x
23
Ben-Yosef M. Jurkevitch E. Yuval B. (2008b). Effect of bacteria on nutritional status and reproductive success of the Mediterranean fruit fly Ceratitis capitata.Physiol. Entomol.33145–154. 10.1111/j.1365-3032.2008.00617.x
24
Berasategui A. Shukla S. Salem H. Kaltenpoth M. (2016). Potential applications of insect symbionts in biotechnology.Appl. Microbiol. Biotechnol.1001567–1577. 10.1007/s00253-015-7186-9
25
Boller E. F. Bush G. L. (1974). Evidence for genetic variation in populations of the european cherry fruit fly, Rhagoletis cerasi (Diptera: Tephritidae) based on physiological parameters and hybridization experiments.Entomol. Exp. Appl.3279–293. 10.1111/j.1570-7458.1974.tb00345.x
26
Borenshtein D. Schauer D. B. (2006). The Genus Citrobacter in The Prokaryotes.New York, NY: Springer. 90–98. 10.1007/0-387-30746-x_5
27
Bourtzis K. Miller T. A. (eds) (2003). Insect Symbiosis. Bocaraton, FL: CRC Press LLC.
28
Brown J. J. Mihaljevic J. R. Des Marteaux L. Hrček J. (2020). Metacommunity theory for transmission of heritable symbionts within insect communities.Ecol. Evol.101703–1721. 10.1002/ece3.5754
29
Caetano F. H. Solferini V. N. De Britto F. B. Lins D. S. Aluani T. Brito V. G. et al (2006). Ultra morphology of the digestive system of Anastrepha fraterculus and Ceratitis capitata (Diptera Tephritidae).Braz. J. Morphol. Sci.23455–462.
30
Carthey A. J. R. Blumstein D. T. Gallagher R. V. Tetu S. G. Gillings M. R. (2019). Conserving the holobiont.Funct. Ecol.2019764–776. 10.1111/1365-2435.13504
31
Cayol J. P. (2000). “Changes in sexual behavior and life history traits of tephritid species caused by mass-rearing processes,” in Fruit Flies (Tephritidae): Phylogeny and Evolution of Behavior, edsAlujaM.NorrbomA. (Boca Raton, FL: CRC Press), 843–860. 10.1201/9781420074468.ch31
32
Chen J. Bittinger K. Charlson E. S. Hoffmann C. Lewis J. Wu G. D. et al (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances.Bioinformatics282106–2113. 10.1093/bioinformatics/bts342
33
Colman D. R. Toolson E. C. Takacs-Vesbach C. D. (2012). Do diet and taxonomy influence insect gut bacterial communities?Mol. Ecol.215124–5137. 10.1111/j.1365-294X.2012.05752.x
34
Conte C. A. Segura D. F. Milla F. H. Augustinos A. Cladera J. L. Bourtzis K. et al (2019). Wolbachia infection in Argentinean populations of Anastrepha fraterculus sp1: Preliminary evidence of sex ratio distortion by one of two strains.BMC Microbiol.19(Suppl. 1):289. 10.1186/s12866-019-1652-y
35
Core Team R. (2020). R: A Language and Environment For Statistical Computing.Vienna: R Foundation for Statistical Computing.
36
Deutscher A. T. Chapman T. A. Shuttleworth L. A. Riegler M. Reynolds O. L. (2019). Tephritid-microbial interactions to enhance fruit fly performance in sterile insect technique programs.BMC Microbiol.19(Suppl. 1):287. 10.1186/s12866-019-1650-0
37
Devescovi F. Abraham S. Roriz A. K. P. Nolazco N. Casta-neda R. Tadeo E. et al (2014). Ongoing speciation within the Anastrepha fraterculus cryptic species complex: the case of the andean morphotype.Entomol. Exp. Appl.152238–247. 10.1111/eea.12219
38
Devescovi F. Conte C. A. Augustinos A. Martinez E. I. C. Segura D. F. Cáceres C. et al (2019). Symbionts do not affect the mating incompatibility between the Brazilian-1 and Peruvian morphotypes of the Anastrepha fraterculus cryptic species complex.Sci. Rep.91–12. 10.1038/s41598-019-54704-y
39
Dillon R. J. Dillon V. M. (2004). The gut bacteria of insects: nonpathogenic Interactions.Annu. Rev. Entomol.4971–92. 10.1146/annurev.ento.49.061802.123416
40
Douglas A. E. (2015). Multiorganismal Insects: diversity and function of resident microorganisms.Annu. Rev. Entomol.6017–34. 10.1146/annurev-ento-010814-020822
41
Edgar R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads.Nat. Methods10996–998. 10.1038/nmeth.2604
42
Edgar R. C. (2018). UNCROSS2: identification of cross-talk in 16S rRNA OTU tables.bioRxiv[Preprint].10.1101/400762
43
Engel P. Moran N. A. (2013). The gut microbiota of insects - diversity in structure and function.FEMS Microbiol. Rev.37699–735. 10.1111/1574-6976.12025
44
Feldhaar H. (2011). Bacterial symbionts as mediators of ecologically important traits of insect hosts.Ecol. Entomol.36533–543. 10.1111/j.1365-2311.2011.01318.x
45
Gallo-Franco J. J. Toro-Perea N. (2020). Variations in the bacterial communities in Anastrepha obliqua (Diptera: Tephritidae) according to the insect life stage and host plant.Curr. Microbiol.771283–1291. 10.1007/s00284-020-01939-y
46
Gavriel S. Jurkevitch E. Gazit Y. Yuval B. (2011). Bacterially enriched diet improves sexual performance of sterile male Mediterranean fruit flies.J. Appl. Entomol.135564–573. 10.1111/j.1439-0418.2010.01605.x
47
Goday C. Selivon D. Perondini A. L. P. Greciano P. G. Ruiz M. F. (2006). Cytological characterization of sex chromosomes and ribosomal DNA location in Anastrepha species (Diptera, Tephritidae).Cytogenet. Genome Res.11470–76. 10.1159/000091931
48
Gurung K. Wertheim B. Falcao Salles J. (2019). The microbiome of pest insects: it is not just bacteria.Entomol. Exp. Appl.167156–170. 10.1111/eea.12768
49
Hamden H. M’saad Guerfali M. Fadhl S. Saidi M. Chevrier C. (2013). Fitness improvement of mass-reared sterile males of Ceratitis capitata (Vienna 8 strain) (Diptera: Tephritidae) after gut enrichment with probiotics.J. Econ. Entomol.106641–647. 10.1603/ec12362
50
Hammer T. J. Bowers M. D. (2015). Gut microbes may facilitate insect herbivory of chemically defended plants.Oecologia1791–14. 10.1007/s00442-015-3327-1
51
Hendrichs J. Robinson A. (2009). “Sterile Insect Technique,” in Encyclopedia of Insects, 2nd Edn, edsReshV. H.CardeR. T. (Amsterdam: Elsevier Inc), 953–957. 10.1016/B978-0-12-374144-8.00252-6
52
Hernández E. Escobar A. Bravo B. Montoya P. (2010). Chilled packing systems for fruit flies (Diptera: Tephritidae) in the sterile insect technique.Neotrop. Entomol.39601–607. 10.1590/S1519-566X2010000400021
53
Hernández-Ortiz V. Bartolucci A. F. Morales-Valles P. Frías D. Selivon D. (2012). Cryptic Species of the Anastrepha fraterculus Complex (Diptera: Tephritidae): a multivariate approach for the recognition of south american morphotypes.Ann. Entomol. Soc. Am.105305–318. 10.1603/an11123
54
Hernández-Ortiz V. Canal N. A. Tigrero Salas J. O. Ruíz-Hurtado F. M. Dzul-Cauich J. F. (2015). Taxonomy and phenotypic relationships of the Anastrepha fraterculus complex in the mesoamerican and pacific neotropical dominions (Diptera, tephritidae).Zookeys201595–124. 10.3897/zookeys.540.6027
55
Hernández-Ortiz V. Guillén J. López L. (2010). “Taxonomía e Identificación de Moscas de la Fruta de Importancia Económica en América,” in Moscas de la Fruta: Fundamentos y Procedimientos para su Manejo, edsMontoyaP.ToledoJ.HernándezE. (Mexico: S y G editores), 49–80.
56
Illumina Inc. (2013). 16S Metagenomic Sequencing Library Preparation - Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. In 16S Metagenomic Sequencing Library Preparation Manual.San Diego, CA: Illumina. Inc, 1–28.
57
Jaenike J. (2012). Population genetics of beneficial heritable symbionts.Trends Ecol. Evol.27226–232. 10.1016/j.tree.2011.10.005
58
Jaldo H. E. Gramajo M. C. Willink E. (2001). Mass Rearing of Anastrepha fraterculus (Diptera: Tephritidae): A Preliminary Strategy.Florida Entomol.84:716. 10.2307/3496407
59
Jing T. Z. Qi F. H. Wang Z. Y. (2020). Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision?Microbiome81–20. 10.1186/s40168-020-00823-y
60
Juárez M. L. Pimper L. E. Bachmann G. E. Conte C. A. Ruiz M. J. Goane L. et al (2019). Gut bacterial diversity and physiological traits of Anastrepha fraterculus Brazilian-1 morphotype males are affected by antibiotic treatment.BMC Microbiol.19:283. 10.1186/s12866-019-1645-x
61
Jurkevitch E. (2011). Riding the Trojan horse: Combating pest insects with their own symbionts.Microb. Biotechnol.4620–627. 10.1111/j.1751-7915.2011.00249.x
62
Kembel S. W. Cowan P. D. Helmus M. R. Cornwell W. K. Morlon H. Ackerly D. D. et al (2010). Picante: R tools for integrating phylogenies and ecology.Bioinformatics261463–1464. 10.1093/bioinformatics/btq166
63
Khaeso K. Andongma A. A. Akami M. Souliyanonh B. Zhu J. Krutmuang P. et al (2017). Assessing the effects of gut bacteria manipulation on the development of the oriental fruit fly, Bactrocera dorsalis (Diptera; Tephritidae).Symbiosis7497–105. 10.1007/s13199-017-0493-4
64
Klindworth A. Pruesse E. Schweer T. Peplies J. Quast C. Horn M. et al (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies.Nucl. Acids Res.411–11. 10.1093/nar/gks808
65
Koskinioti P. Ras E. Augustinos A. A. Tsiamis G. Beukeboom L. W. Cáceres C. et al (2019). The effects of geographic origin and antibiotic treatment on the gut symbiotic communities of Bactrocera oleae populations.Entomol. Exp. Appl.167197–208. 10.1111/eea.12764
66
Kounatidis I. Crotti E. Sapountzis P. Sacchi L. Rizzi A. Chouaia B. et al (2009). Acetobacter tropicalis is a major symbiont of the olive fruit fly (Bactrocera oleae).Appl. Environ. Microbiol.753281–3288. 10.1128/aem.02933-08
67
Kruskal W. H. (1957). Historical notes on the wilcoxon unpaired two-sample test.J. Am. Statist. Assoc.52356–360. 10.1080/01621459.1957.10501395
68
Ksentini I. Gharsallah H. Sahnoun M. Schuster C. Hamli Amri S. Gargouri R. et al (2019). Providencia entomophila sp. nov., a new bacterial species associated with major olive pests in Tunisia. PLoS One14:e0223943. 10.1371/journal.pone.0223943
69
Kyritsis G. A. Augustinos A. A. Cáceres C. Bourtzis K. (2017). Medfly gut microbiota and enhancement of the sterile insect technique: Similarities and differences of Klebsiella oxytoca and Enterobacter sp. AA26 probiotics during the larval and adult stages of the VIENNA 8D53+ genetic sexing strain.Front. Microbiol.8:2064. 10.3389/fmicb.2017.02064
70
Kyritsis G. A. Augustinos A. A. Ntougias S. Papadopoulos N. T. Bourtzis K. Cáceres C. (2019). Enterobacter sp. AA26 gut symbiont as a protein source for Mediterranean fruit fly mass-rearing and sterile insect technique applications.BMC Microbiol.19(Suppl. 1):288. 10.1186/s12866-019-1651-z
71
Lagkouvardos I. Fischer S. Kumar N. Clavel T. (2017). Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons.PeerJ5:e2836. 10.7717/peerj.2836
72
Lauzon C. R. McCombs S. D. Potter S. E. Peabody N. C. (2009). Establishment and Vertical Passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumoniae through All Life Stages of the Mediterranean Fruit Fly (Diptera: Tephritidae).Ann. Entomol. Soc. Am.10285–95. 10.1603/008.102.0109
73
Liendo M. C. Devescovi F. Bachmann G. E. Utgs M. E. Abraham S. Vera M. T. et al (2013). Precocious sexual signalling and mating in Anastrepha fraterculus (Diptera: Tephritidae) sterile males achieved through juvenile hormone treatment and protein supplements.Bull. Entomol. Res.1031–13. 10.1017/S0007485312000442
74
Mateos M. Martinez H. Lanzavecchia S. B. Conte C. Morán-Aceves B. M. Toledo J. et al (2020). Wolbachia pipientis associated with tephritid fruit fly pests: from basic research to applications.Front. Microbiol.11:1080. 10.3389/fmicb.2020.01080
75
Mattson W. J. (1980). Herbivory in relation to plant nitrogen content.Annu. Rev. Ecol. Syst.11119–161. 10.1146/annurev.es.11.110180.001003
76
Meats A. Streamer K. Gilchrist A. S. (2009). Bacteria as food had no effect on fecundity during domestication of the fruit fly, Bactrocera tryoni.J. Appl. Entomol.133633–639. 10.1111/j.1439-0418.2009.01420.x
77
Metcalf R. L. Luckmann W. H. (1994). Introduction to Insect Pest Management.Hoboken, NJ: John Wiley and Sons.
78
Morris J. J. (2018). What is the hologenome concept of evolution? [version 1; peer review: 2 approved].F1000 Res.71–9. 10.12688/F1000RESEARCH.14385.1
79
Morrow J. L. Frommer M. Shearman D. C. A. Riegler M. (2015). The microbiome of field-caught and laboratory-adapted australian tephritid fruit fly species with different host plant use and specialisation.Microb. Ecol.70498–508. 10.1007/s00248-015-0571-1
80
Msaad Guerfali M. M. Djobbi W. Charaabi K. Hamden H. Fadhl S. Marzouki W. et al (2018). Evaluation of Providencia rettgeri pathogenicity against laboratory Mediterranean fruit fly strain (Ceratitis capitata).PLoS One13:e0196343. 10.1371/journal.pone.0196343
81
Müller F. A. (2013). Microbiota Intestinal de Larvas e Adultos de Anastrepha fraterculus (Wiedemann, 1830) (Diptera?: Tephritidae): Diversidade e efeito do Alimento. São Paulo: Universidade de São Paulo.
82
Murphy K. M. Mac Rae I. C. Teakle D. S. (1988). Nitrogenase activity in the queensland fruit fly, Dacus tryoni.Austr. J. Biol. Sci.41447–452. 10.1071/BI9880447
83
Niyazi N. Lauzon C. R. Shelly T. E. (2004). Effect of probiotic adult diets on fitness components of sterile male mediterranean fruit flies (Diptera: Tephritidae) under laboratory and field cage conditions.J. Econ. Entomol.971570–1580. 10.1603/0022-0493-97.5.1570
84
Noman M. S. Liu L. Bai Z. Li Z. (2020). Tephritidae bacterial symbionts: potentials for pest management.Bull. Entomol. Res.1101–14. 10.1017/S0007485319000403
85
Norrbom A. L. Korytkowski C. A. Zucchi R. A. Uramoto K. Venable G. L. McCormick J. et al (2012). Anastrepha and Toxotrypana: descriptions, illustrations, and interactive keys. Version: 9th April 2019. Available online at: http://delta-intkey.com/anatox/intro.htm(accessed February 24, 2020).
86
Ntougias S. Polkowska Z. Nikolaki S. Dionyssopoulou E. Stathopoulou P. Doudoumis V. et al (2016). Bacterial community structures in freshwater polar environments of svalbard.Microb. Environ.31401–409. 10.1264/jsme2.ME16074
87
Ochieng-Odero J. P. R. (1994). Does adaptation occur in insect rearing systems, or is it a case of selection, acclimatization and domestication?Insect Sci. Appl.151–7. 10.1017/s1742758400016696
88
Octavia S. Lan R. (2014). “The Family Enterobacteriaceae,” in The Prokaryotes, edsRosenbergE.DeLongE. F.LoryS.StackebrandtE.ThompsonF. (Berlin: Springer), 225–286. 10.1007/978-3-642-38922-1_167
89
Oksanen J. F. Blanchet G. Friendly M. Kindt R. Legendre P. McGlinn D. et al (2019). vegan: Community Ecology Package. R package version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan(accessed September 2, 2020).
90
Parreño M. A. Scannapieco A. C. Remis M. I. Juri M. Vera M. T. Segura D. F. et al (2014). Dynamics of genetic variability in Anastrepha fraterculus (Diptera: Tephritidae) during adaptation to laboratory rearing conditions.BMC Genet.15(Suppl. 2):S14.
91
Pontes M. H. Dale C. (2006). Culture and manipulation of insect facultative symbionts.Trends Microbiol.14406–412. 10.1016/j.tim.2006.07.004
92
Quast C. Pruesse E. Yilmaz P. Gerken J. Schweer T. Yarza P. et al (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools.Nucleic Acids Research41590–596. 10.1093/nar/gks1219
93
Raza M. F. Yao Z. Bai S. Cai Z. Zhang H. (2020). Tephritidae fruit fly gut microbiome diversity, function and potential for applications.Bull. Entomol. Res.110423–347. 10.1017/S0007485319000853
94
Riegler M. Stauffer C. (2002). Wolbachia infections and superinfections in cytoplasmically incompatible populations of the European cherry fruit fly Rhagoletis cerasi (Diptera, Tephritidae).Mol. Ecol.112425–2434. 10.1046/j.1365-294X.2002.01614.x
95
Robacker D. C. Bartelt R. J. (1997). Chemicals attractive to Mexican fruit fly from Klebsiella pneumoniae and Citrobacter freundii cultures sampled by solid-phase microextraction.J. Chem. Ecol.232897–2915. 10.1023/A:1022579414233
96
Sacchetti P. Ghiardi B. Granchietti A. Stefanini F. M. Belcari A. (2014). Development of probiotic diets for the olive fly: Evaluation of their effects on fly longevity and fecundity.Ann. Appl. Biol.164138–150. 10.1111/aab.12088
97
Salles L. A. (1999). Rearing of Anastrepha fraterculus (Wiedemann). Vienna: International Atomic Energy Agency (IAEA).
98
Segura D. Petit-Marty N. Sciurano R. Vera T. Calcagno G. Allinghi A. et al (2007). Lekking Behavior of Anastrepha fraterculus (Diptera: Tephritidae).Florida Entomol.90154–162.
99
Segura D. F. Cáceres C. Vera M. T. Wornoayporn V. Islam A. Teal P. E. A. et al (2009). Enhancing mating performance after juvenile hormone treatment in Anastrepha fraterculus: a differential response in males and females acts as a physiological sexing system.Entomologia Experimentalis et Applicata13175–84. 10.1111/j.1570-7458.2009.00830.x
100
Segura D. F. Utgés M. E. Liendo M. C. Rodríguez M. F. Devescovi F. Vera M. T. et al (2013). Methoprene treatment reduces the pre-copulatory period in Anastrepha fraterculus (Diptera: Tephritidae) sterile males.J. Appl. Entomol.137(Suppl.1), 19–29. 10.1111/j.1439-0418.2010.01534.x
101
Selivon D. Perondini A. L. P. Morgante J. S. (2005). A genetic-morphological characterization of two cryptic species of the Anastrepha fraterculus complex.Ann. Entomol. Soc. Am.98367–381. 10.1603/0013-8746(2005)098[0367:agcotc]2.0.co;2
102
Sobrinho R. B. Cáceres C. Islam A. Wornoayporn V. Enkerlin W. (2006). Improving mass rearing technology for south american fruit fly (Diptera: Tephritidae).Rev. Caatinga19310–316.
103
Staubach F. Baines J. F. Künzel S. Bik E. M. Petrov D. A. (2013). Host Species and environmental effects on bacterial communities associated with drosophila in the laboratory and in the natural environment.PLoS One8:e70749. 10.1371/journal.pone.0070749
104
Steck G. J. (1999). Taxonomic status of Anastrepha fraterculus.IAEA2285–299.
105
Ventura C. Briones-Roblero C. I. Hernández E. Rivera-Orduña F. N. Zúñiga G. (2018). Comparative analysis of the gut bacterial community of four Anastrepha fruit flies (Diptera: Tephritidae) based on pyrosequencing.Curr. Microbiol.75966–976. 10.1007/s00284-018-1473-5
106
Vera M. T. Ruiz M. J. Oviedo A. Abraham S. Mendoza M. Segura D. F. et al (2013). Fruit compounds affect male sexual success in the South American fruit fly, Anastrepha fraterculus (Diptera: Tephritidae).J. Appl. Entomol.137(Suppl.1), 2–10. 10.1111/j.1439-0418.2010.01516.x
107
Vera T. Abraham S. Oviedo A. Willink E. (2007). Demographic and Quality Control Parameters of Anastrepha fraterculus (Diptera: Tephritidae) Maintained Under Artificial Rearing.Florida Entomol.9053–57. 10.1653/0015-4040(2007)90[53:daqcpo]2.0.co;2
108
Vontas J. Hernández-Crespo P. Margaritopoulos J. T. Ortego F. Feng H. T. Mathiopoulos K. D. et al (2011). Insecticide resistance in Tephritid flies.Pesticide Biochem. Physiol.100199–205. 10.1016/j.pestbp.2011.04.004
109
Walder J. M. M. Morelli R. Costa K. Z. Faggioni K. M. Sanches P. A. Paranhos B. A. J. et al (2014). Large scale artificial rearing of Anastrepha sp. 1 aff. fraterculus (Diptera: Tephritidae) in Brazil.Sci. Agric.71281–286. 10.1590/0103-9016-2013-233
110
Wang A. Yao Z. Zheng W. Zhang H. (2014). Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing.PLoS One9:e106988. 10.1371/journal.pone.0106988
111
White I. M. Elson-Harris M. M. (1992). Fruit Flies Of Economic Significance: Their Identification And Bionomics.Wallingford: CAB International Wallingford UK.
112
Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis.New York, NY: Springer-Verlag,
113
Wu D. Daugherty S. C. Van Aken S. E. Pai G. H. Watkins K. L. Khouri H. et al (2006). Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters.PLoS Biol.4:1079–1092. 10.1371/journal.pbio.0040188
114
Yong H. S. Song S. L. Chua K. O. Lim P. E. (2017). Microbiota associated with Bactrocera carambolae and B. dorsalis (Insecta: Tephritidae) revealed by next-generation sequencing of 16S rRNA gene.Meta Gene11189–196. 10.1016/j.mgene.2016.10.009
115
Zabalou S. Apostolaki A. Livadaras I. Franz G. Robinson A. S. Savakis C. et al (2009). Incompatible insect technique: incompatible males from a Ceratitis capitata genetic sexing strain.Entomol. Exp. Appl.132232–240. 10.1111/j.1570-7458.2009.00886.x
116
Zabalou S. Riegler M. Theodorakopoulou M. Stauffer C. Savakis C. Bourtzis K. (2004). Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control.Proc. Natl. Acad. Sci. U.S.A.10115042–15045. 10.1073/pnas.0403853101
117
Zilber-Rosenberg I. Rosenberg E. (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution.FEMS Microbiol. Rev.32723–735. 10.1111/j.1574-6976.2008.00123.x
Summary
Keywords
NGS, taxonomic identification, core bacteriome, bacterial diversity, bacterial richness, fruit fly, SIT
Citation
Salgueiro J, Pimper LE, Segura DF, Milla FH, Russo RM, Asimakis E, Stathopoulou P, Bourtzis K, Cladera JL, Tsiamis G and Lanzavecchia SB (2020) Gut Bacteriome Analysis of Anastrepha fraterculus sp. 1 During the Early Steps of Laboratory Colonization. Front. Microbiol. 11:570960. doi: 10.3389/fmicb.2020.570960
Received
09 June 2020
Accepted
25 September 2020
Published
20 October 2020
Volume
11 - 2020
Edited by
Alfonso Benítez-Páez, Principe Felipe Research Center (CIPF), Spain
Reviewed by
Janisete Gomes Silva, Universidade Estadual de Santa Cruz, Brazil; Nelson Augusto Canal, Universidad del Tolima, Colombia
Updates
Copyright
© 2020 Salgueiro, Pimper, Segura, Milla, Russo, Asimakis, Stathopoulou, Bourtzis, Cladera, Tsiamis and Lanzavecchia.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia B. Lanzavecchia, lanzavecchia.silvia@inta.gob.ar
†These authors have contributed equally to this work
This article was submitted to Microbial Symbioses, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.