- 1Systems Microbiology and Natural Products Laboratory, University of California, Davis, Davis, CA, United States
- 2Bayer, Pittsburg, PA, United States
- 3Microbiology Graduate Group, University of California, Davis, Davis, CA, United States
- 4Biochemistry, Molecular, Cellular, and Developmental Biology Graduate Group, University of California, Davis, Davis, CA, United States
- 5Department of Energy, Joint Genome Institute, Berkeley, CA, United States
- 6Greenlight Biosciences, Inc., Medford, MA, United States
- 7Department of Biology, University of Oregon, Eugene, OR, United States
Cyanobacteria are found in most illuminated environments and are key players in global carbon and nitrogen cycling. Although significant efforts have been made to advance our understanding of this important phylum, still little is known about how members of the cyanobacteria affect and respond to changes in complex biological systems. This lack of knowledge is in part due to our dependence on pure cultures when determining the metabolism and function of a microorganism. We took advantage of the Culture Collection of Microorganisms from Extreme Environments (CCMEE), a collection of more than 1,000 publicly available photosynthetic co-cultures maintained at the Pacific Northwest National Laboratory, and assessed via 16S rRNA amplicon sequencing if samples readily available from public culture collection could be used in the future to generate new insights into the role of microbial communities in global and local carbon and nitrogen cycling. Results from this work support the existing notion that culture depositories in general hold the potential to advance fundamental and applied research. Although it remains to be seen if co-cultures can be used at large scale to infer roles of individual organisms, samples that are publicly available from existing co-cultures depositories, such as the CCMEE, might be an economical starting point for such studies. Access to archived biological samples, without the need for costly field work, might in some circumstances be one of the few remaining ways to advance the field and to generate new insights into the biology of ecosystems that are not easily accessible. The current COVID-19 pandemic, which makes sampling expeditions almost impossible without putting the health of the participating scientists on the line, is a very timely example.
Introduction
Cyanobacteria are photosynthetic prokaryotes that are found in the majority of illuminated habitats and are known to be some of the most morphologically diverse prokaryotes on our planet (Whitton and Potts, 2000). The global cyanobacterial biomass is estimated to total ∼3 × 1014 g of carbon (Garcia-Pichel et al., 2003) and cyanobacteria may account for 20–30% of Earth’s primary photosynthetic productivity (Pisciotta et al., 2010). The efficient photosynthetic machinery of cyanobacteria has inspired growing interest in the utilization of cyanobacteria and cyanobacteria containing co-cultures in microbial fuel cells (Zhao et al., 2012; Gajda et al., 2015). In addition to having a global effect on the carbon cycle, cyanobacteria-mediated nitrogen fixation has been estimated to supply 20–50% of the nitrogen input in some marine environments (Karl et al., 1997). A detailed comprehension of cyanobacteria and their role in global carbon and nitrogen cycling is therefore indispensable for a multi-scalar and holistic understanding of these globally important nutrient cycles.
Besides their ecological relevance, cyanobacteria have potential applications in biotechnology: cyanobacteria facilitate the assimilation of carbon dioxide, a cheap and abundant substrate, to synthesize a variety of value-added compounds with industrial relevance (Al-Haj et al., 2016). Although monocultures have dominated in microbial biomanufacturing, controlled co-cultures have been recognized as valuable alternatives, due to their potential for reducing the risk of costly contaminations and in some cases enabling increasing product yield (Wang et al., 2015; Yen et al., 2015; Padmaperuma et al., 2018). Numerous cyanobacterial strains have been investigated for their potential to produce bioactive compounds, biofertilizer, biofuels, and bioplastics (Abed et al., 2009; Woo and Lee, 2017; Miao et al., 2018); and co-expression of non-cyanobacterial genes as well as co-cultivation of cyanobacteria with non-photosynthetic bacteria has resulted in self-sustained systems and improved desirable cyanobacterial phenotypes (de-Bashan et al., 2002; Subashchandrabose et al., 2011; Formighieri and Melis, 2016). Genes coding for enzymes capable of catalyzing reactions that result in unique products, such as modified trichamide, a cyclic peptide suggested to protect the bloom-forming Trichodesmium erythraeum against predation (Sudek et al., 2006); and prochlorosins, a family of lanthipeptides with diverse functions that are synthesized by various strains of Prochlorococcus and Synechococcus (Li et al., 2010; Cubillos-Ruiz et al., 2017), have been identified from cyanobacterial genomes (Zarzycki et al., 2013; Kleigrewe et al., 2016). It is very likely that de novo genome assembly from metagenomic data will facilitate the discovery of novel enzymes from cyanobacteria that are recalcitrant to current isolation and cultivation techniques. Although metagenome-derived genomes hold great potential to enhance our knowledge about genomic dark matter, improved techniques to isolate and enable axenic culturing of microorganisms that are currently considered as “unculturable,” as well as new genetic tools to study non-axenic cultures will be necessary in order to fully access the biotechnological potential of cyanobacteria.
Culture collections provide the possibility of preserving microbial isolates over extended periods of time without introducing significant genetic changes (McCluskey, 2017) and they provide easy access to these isolates and their associated metadata (Boundy-Mills et al., 2015). Although culture collections hold enormous potential for capturing and preserving microbial biodiversity, there are numerous challenges in maintaining these biological depositories. With recent advances in DNA sequencing technologies and the accessibility of 16S rRNA gene-based microbial community profiling, we are now well positioned to re-inventory, and standardize existing culture collections, which will be essential for preserving and cataloging the planet’s microbial biodiversity.
To explore the potential of culture collections, specifically those that maintain samples of microbial co-cultures, we reexamined the biodiversity of 26 historical phototrophic samples from the Culture Collection of Microorganisms from Extreme Environments (CCMEE). While some of the samples, selected for this project were studied previously (Supplementary Table 1) using cloned-based 16S rRNA profiling and morphological characterization (Camacho et al., 1996; Miller and Castenholz, 2000; Nadeau and Castenholz, 2000; Nadeau et al., 2001; Dillon et al., 2002; Dillon and Castenholz, 2003; Norris and Castenholz, 2006; Toplin et al., 2008), the diversity and the overall community assemblage of these co-cultures have not yet been characterized. We selected samples from environments with distinct and extreme physical properties from across the globe, suggesting each co-culture would yield a unique microbial consortium. Although reasonable to assume that these consortia have changed over time in composition and function (due to their cultivation), it is very likely that results obtained during this work will still provide insights into the microbial biodiversity of extreme habitats, some of which may no longer be accessible.
Materials and Methods
Sample Collection and Sample Description
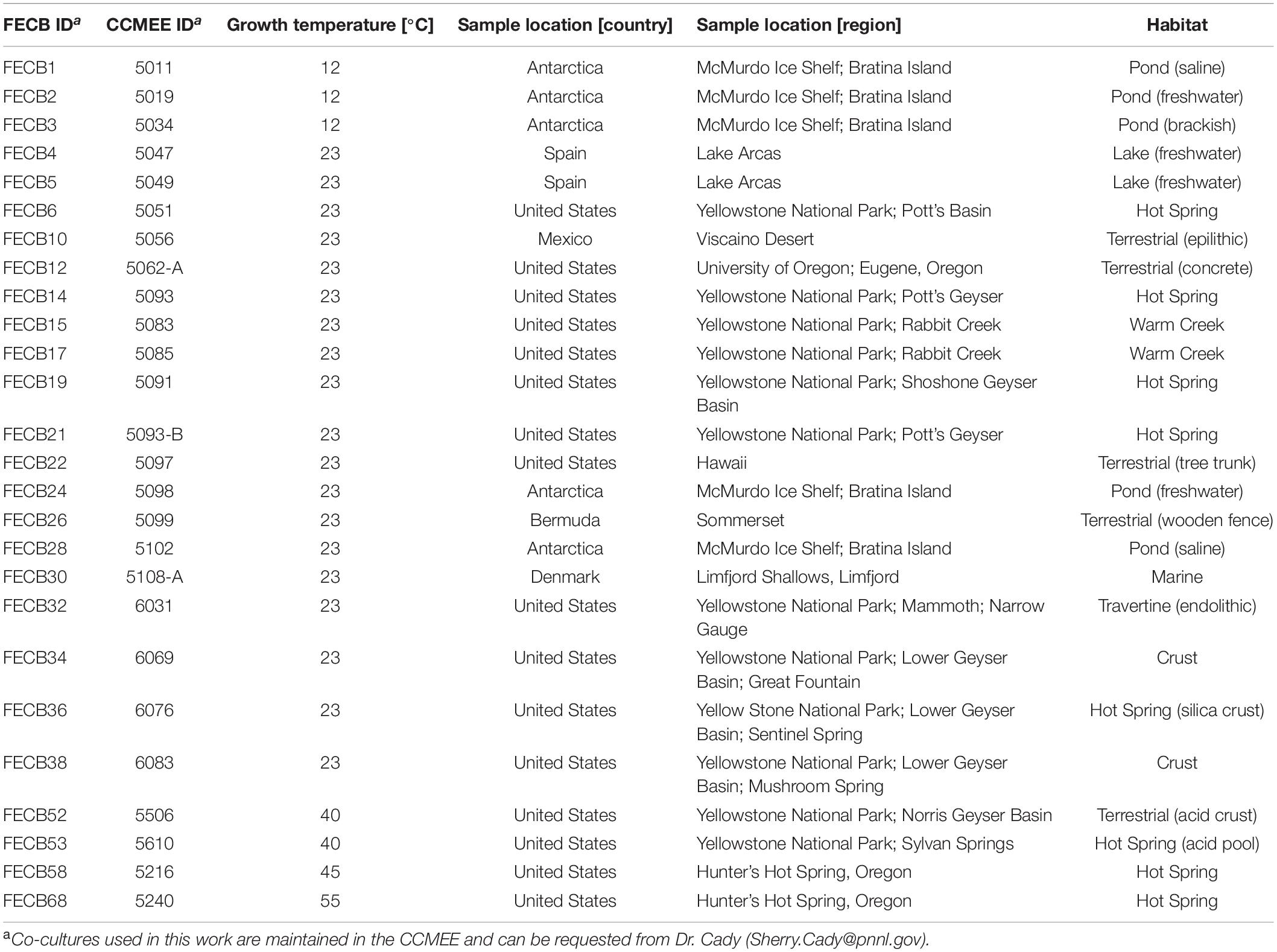
Co-cultures selected for this study are part of a larger culture collection and were collected from different locations (Table 1) between 1988 and 2002. Isolates were collected using sterile techniques, kept in the dark and stored on ice as soon as possible. Samples were transported to the laboratory where aliquots were prepared preservation at −80°C and cultivation. For this study, co-cultures were selected from the CCMEE to cover a variety of geographical locations (Supplementary Figure 1) as well as a range of different ecosystems (Table 1). Due to the lack of a consistent usage of terminology to describe the sampling sites, we categorized co-cultures according to the geographical location (e.g., Antarctica, Bermuda, Denmark, Mexico and Spain) and based on the general description of the ecosystems (i.e., creek, crust, freshwater, hot spring, marine, saline pond, terrestrial, travertine, and tree bark) from where the co-cultures were collected. In addition, we used the growth medium and temperature (i.e., 12, 23, 40, 45, and 55°C) at which available co-cultures have been maintained by the CCMEE curators to categorize the selected co-cultures.
To facilitate future work, we assigned new unique FECB identifiers (FECB for Functional Encyclopedia of Cyanobacteria) to the selected co-cultures (Table 1). When requesting aliquots for future work from the CCMEE, these new FECB identifiers should be used.
FECB1 (CCMEE ID 5011) and FECB3 (CCMEE ID 5034) were collected from saline and brackish melt ponds and were dominated by phototrophic cyanobacteria previously classified as Oscillatoria sp. (Nadeau et al., 2001). FECB2 (CCMEE ID 5019) was collected from a freshwater pond and its composition was not investigated prior to our efforts. FECB4 (CCMEE ID 5047; AP1) and FECB5 (CCMEE ID 5049; AO21) were also isolated from freshwater and the dominant photosynthetic organisms within these samples were classified previously by 16S rRNA sequence analysis as relatives of Pseudanabaena limnetica and Oscillatoria cf. tenuis, respectively (Camacho et al., 1996). FECB6 (CCMEE ID 5051), FECB14 (CCMEE ID 5093; WT-97 Cal), FECB15 (CCMEE ID 5083), and FECB19 (CCMEE ID 5091; Y-97) were collected from diverse hot springs within Yellowstone National Park (YNP) (Table 1). FECB10 [CCMEE ID 5056; M88-VD (1)] was collected as epiliths (Dillon et al., 2002). FECB17 (CCMEE ID 5085; RC-97 Cal) and FECB36 (CCMEE ID 6076) were isolated from Rabbit Creek and a crust in the Sentinel Spring Meadows in YNP, respectively, and dominant phototrophs of these co-cultures were characterized previously as Calothrix spp. (Dillon and Castenholz, 2003). FECB22 (CCMEE ID 5097; HW-91) and FECB26 (CCMEE ID 5099; B77-scy, j) were collected from a tree trunk and a wooded fence, respectively. FECB24 (CCMEE ID 5098; AN-90) was collected from a shallow melt pond (∼10 m2) in the Victoria Valley, Antarctica, whereas FECB28 (CCMEE ID 5102) was collected from a saline melt pond on Bratina Island, Antarctica (Nadeau and Castenholz, 2000). FECB32 (CCMEE ID 6031), FECB34 (CCMEE ID 6069) and FECB38 (CCMEE ID 6083) were endoliths collected from subsurface (1–5 mm depths) travertine deposits in YNP (Norris and Castenholz, 2006). FECB53 (CCMEE ID 5610) was collected from Sylvan Springs in YNP. Temperature and pH at FECB53’s sampling site were determined to be 40°C and pH4, conditions which are considered to be too harsh to actively support growth of cyanobacteria, and Toplin et al. (2008) reported the thermo-acidophilic red algae Cyanidioschyzon as a highly abundant phototropic strain in this sample. FECB58 (CCMEE ID 5216; OH-9-45C) and FECB68 (CCMEE ID 5240; OH-2-55C) were collected from Hunter’s Hot Spring in Oregon and in 2000 Miller and Castenholz reported the isolation of several thermophilic clones belonging to the genus Synechococcus from these samples (Miller and Castenholz, 2000).
Growth of Co-cultures
To obtain sufficient biomass for subsequent DNA analysis, 100 μL of each co-culture were transferred to 25 mL of sterile BG11 media (Allen and Stanier, 1968). For FECB52 and FECB53 BG11 was substituted by Cyanidium medium (Castenholz, 1981). Co-cultures were subjected to a 12 h diurnal light/dark cycle while grown over 28 days at a temperature similar to the temperature that was measured at the location where the sample was collected. Growth temperature for each sample is indicated in Table 1.
DNA Extraction and 16S rRNA Gene Amplification
Total microbial DNA was extracted from 500 μL of each photosynthetic co-culture using the FastDNA SPIN Kit for Soil (MP Biomedical, Solon, OH, United States) according to the manufacturer’s instructions. Extracted DNA was quantified via fluorescence (Qubit; Thermo Scientific, United States) and the hypervariable V4 region of the 16S rRNA gene was amplified from extracted DNA using the primer set 515F/805R (515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 805R: 5′-GGACTACHVGGGTWTCTAAT-3′). The forward primer included an 11 bp barcode to allow multiplexing of samples during sequencing. The barcode sequence for each sample is listed in Supplementary Table 2. Subsequent PCR reactions were performed using the 5PRIME HotMasterMix amplification mix (QIAGEN, Beverly, MA, United States) with the following PCR conditions: initial denaturation for 90 s at 94°C, followed by 30 amplification cycles (45 s at 94°C, 60 s at 60°C, and 90 s at 72°C) followed by a final extension step of 72°C for 10 min. Amplification products were cooled to 4°C. Samples were sequenced at the Department of Energy’s Joint Genome Institute (JGI; 1) according to JGI’s standard operating procedure using Illumina’s MiSeq platform and v3 chemistry.
Sequence Data Analysis
Raw sequencing data were downloaded from the JGI’s Genome Portal2 were they are deposited and accessible under the project ID 1032475. Data were decompressed and de-interleaved using the 7-zip software3 and an in-house script, respectively. De-interleaved files were subsequently processed using MOTHUR version 1.38.1 (Schloss et al., 2009; Kozich et al., 2013). Paired-end reads were combined using the make.contigs command. Sequences with ambiguous base calls and sequences longer than 325 bp were removed using screen.seqs. Duplicate sequences were merged using unique.seqs, and the resulting unique sequences were aligned to the V4 region of the SILVA database (v123) (Quast et al., 2013). Chimeras were removed using UCHIME (Edgar et al., 2011) and quality filtered sequences were taxonomically classified at 80% confidence to the GreenGenes reference taxonomy (release gg_13_5_99) (McDonald et al., 2012). Non-prokaryotic sequences were removed and the dist.seqs command was used to calculate pairwise distances between the aligned sequences. The resulting pairwise distance matrix was used to cluster sequences into operational taxonomic units (OTUs) with a 97% sequence identity cut-off using UCLUST (Edgar, 2010). The most abundant sequence of each OTU was picked as the representative sequence. OTUs were taxonomically classified using the classify.otu command using the GreenGenes reference taxonomy (release gg_13_5_99). Shannon and Simpson estimators were calculated in MOTHUR (Schloss et al., 2009).
In order to visualize the overall compositional differences between the co-cultures, an uncorrected pairwise distance matrix was generated using the dist.seqs command in MOTHUR and a tree was generated using Clearcut (version 1.0.9) (Evans et al., 2006). A cladogram from the resulting tree file was constructed and visualized using iTOL (Letunic and Bork, 2016). Cluster designations were assigned at a branch length of 0.05, with branch length indicating the (number of differences/overall length of branches) between two samples. Samples whose branches split at a distance >0.05 were considered as part of the same cluster (Figure 1).
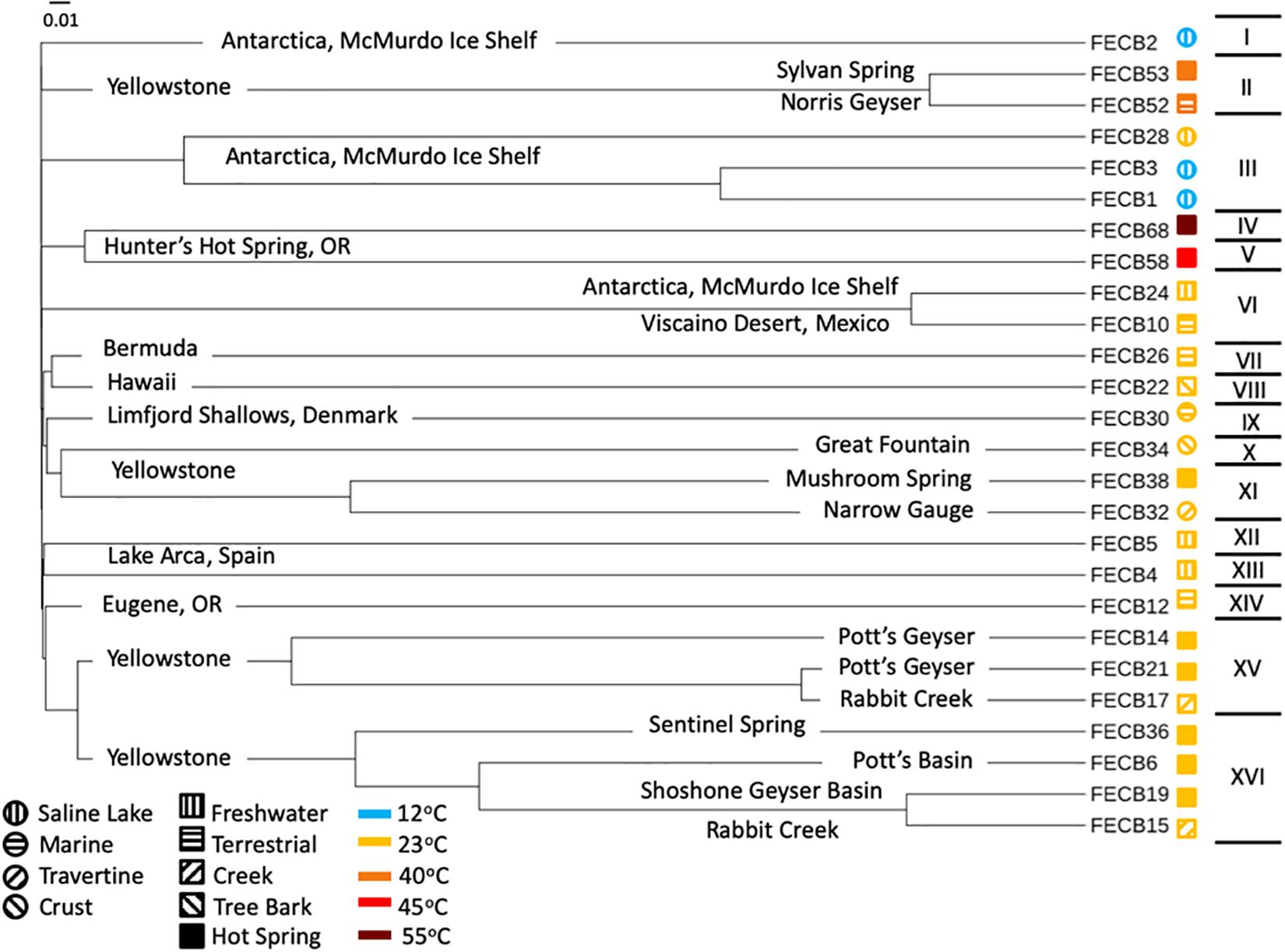
Figure 1. Cladogram of 16S rRNA based community composition of co-cultures under investigation. FECB identifier (sample ID) is provided for each co-culture. Sample location is indicated on the corresponding branch. Roman numerals on the right indicate the clusters identified at a branch cutoff of 0.05. Symbols (i.e., circles and squares) next to sample ID indicate habitat type and color indicates the temperatures at which sample was historically maintained in the CCMEE. Branch length indicates (number of differences/overall length of branches) between two samples.
Availability of Data and Material
Co-cultures subject to this study are publicly available through the CCMEE upon request by contacting Sherry Cady4 using the corresponding FECB ID or CCMEE ID (Table 1). Co-cultures can also be obtained from the Hess Lab at UC Davis. Sequences generated during this project have been deposited and are publicly available at NCBI’s SRA under the BioProject ID PRJNA401502. All other data is included in this published article and its Supplementary Information files.
The CCMEE is now maintained at the Northwest National Laboratory by Dr. Cady. The CCMEE is comprised of >1,200 co-cultures, including the co-cultures that were studied in the work presented here, and has been established to provide a valuable resource to the scientific community. Cultures that are part of the CCMEE can be requested from Sherry Cady1.
Results and Discussion
Prokaryotic Diversity and Eukaryotic Population of Co-cultures
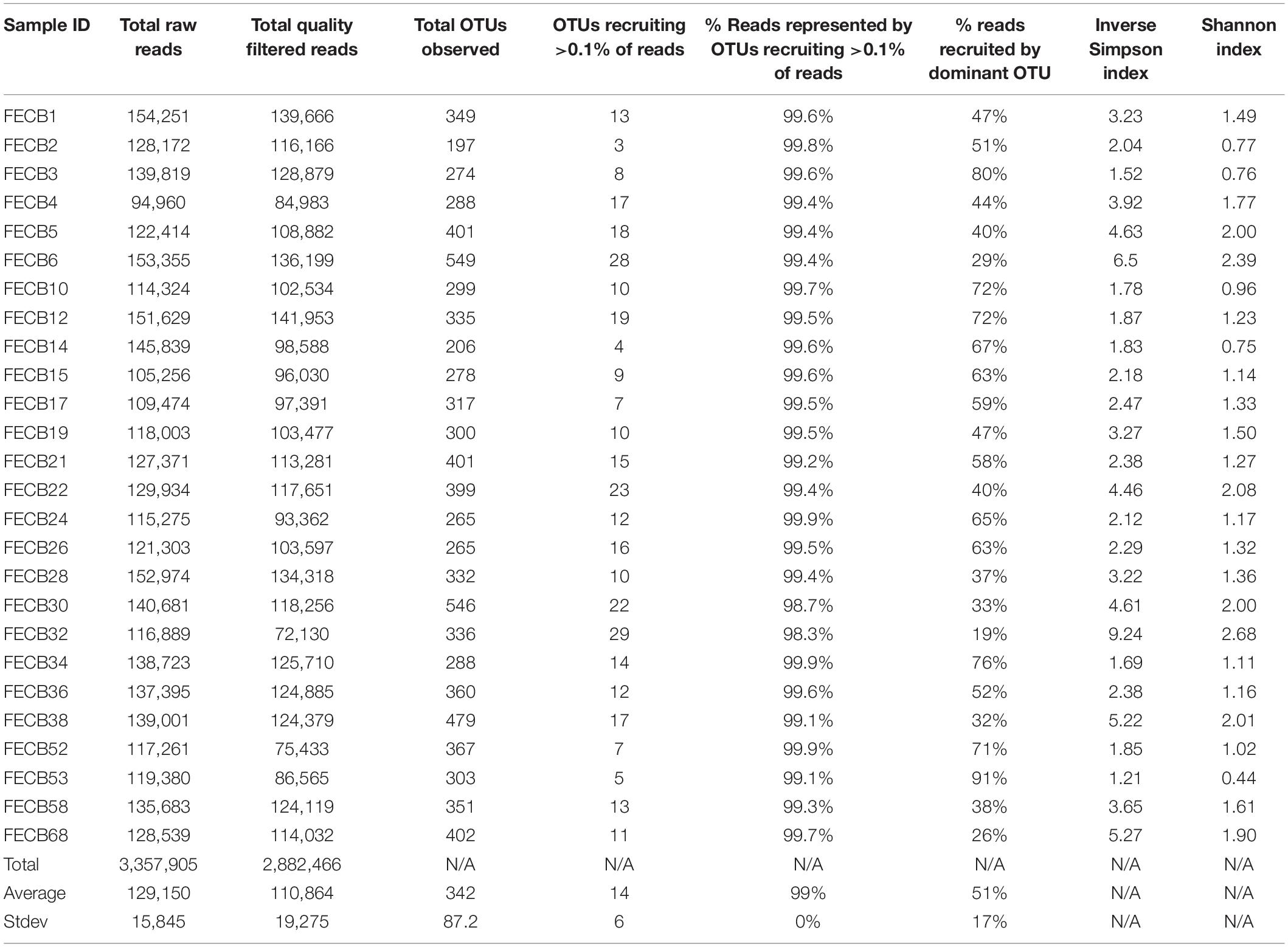
A total of 3,357,905 raw reads [mean (SD) = 129,150 (±15,845) reads per sample] were generated from the V4 region of the 16S rRNA gene (Table 2). Quality filtering removed ∼3.8% (±0.57%) of the raw reads from each sample due to insufficient quality. The remaining reads were assigned to a total of 5,785 distinct Operational Taxonomic Units (OTUs) based on 97% sequence identity (Supplementary Table 3).
To estimate the microbial diversity within each sample, rarefaction analyses were performed (Supplementary Figure 2) and diversity indices were calculated (Table 2). The Inverse Simpson index (Simpson, 1949; Morris et al., 2014) of the samples ranged between 1.21 and 9.24 with the lowest and highest indices calculated for FECB53 and FECB32, respectively (Table 2), illustrating that co-cultures investigated during this project represented co-cultures from a wide range of diversity. Not surprisingly, the diversity in the co-cultures under investigation appeared to be negatively correlated with the proportion of reads recruited by the dominant OTU of each sample (Pearson r = −0.8806; p < 0.01). Although samples ranked slightly differently based on their diversity, when diversity was calculated using the Shannon index (Kim et al., 2017), the overall trend remained the same (Table 2).
The presence of eukaryotic microorganisms in each co-culture was estimated using mitochondrial reads resulting from the 16S rRNA sequencing (Supplementary Table 4). FECB52 had the greatest percentage of mitochondrial DNA at 0.29% of total reads. FECB2, FECB4, FECB6, FECB14, FECB17, FECB26, FECB30, and FECB68 all contained no mitochondrial reads. Average mitochondrial reads as a percentage of all reads averaged 0.02% across all 26 samples.
Ubiquity of Cyanobacteria and Proteobacteria Within Photosynthetic Co-cultures
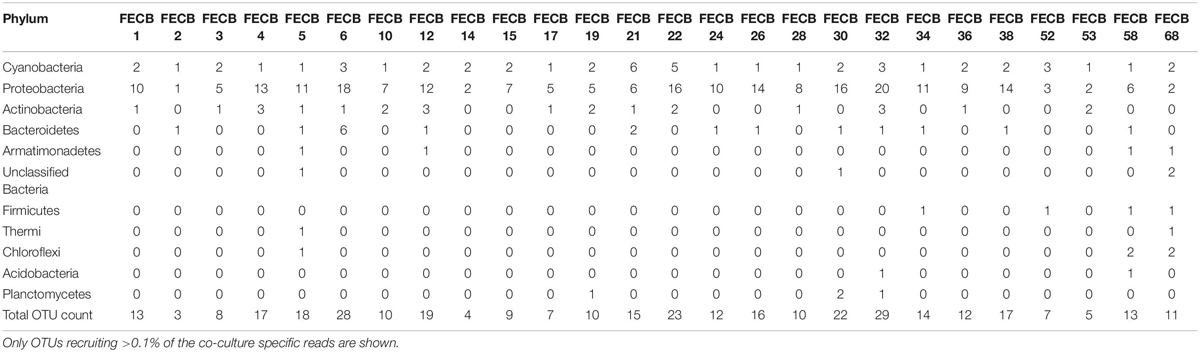
While the microbial communities of the co-cultures under investigation varied greatly, cyanobacteria and proteobacteria co-occurred in all 26 of the community assemblages. Community composition analysis revealed that each of the co-cultures contained at least one OTU [mean (SD) = 2 (±1.23)] that recruited (>0.1% of the co-culture specific reads and that was classified as Cyanobacteria (Table 3). The only other phylum present in each of the individual 26 co-cultures and represented by at least one OTU recruiting (>0.1% of the reads was the Proteobacteria phylum (Table 3). In contrast, only three samples, namely FECB5, FECB30, and FECB68, contained OTUs that recruited >0.1% of the sample specific reads and that could not be classified at the phylum level or at a higher taxonomic resolution (Table 3). It is possible that the relatively high abundance of non-classified phyla might contribute to the separation of these samples into distinct clusters (i.e., cluster XII, IX, and IV; Figure 1). In addition to their ubiquity, Cyanobacteria and Proteobacteria also recruited the majority of the reads in all but four (i.e., FECB2, FECB12, FECB58, and FECB68) of the samples under investigation (Figure 2 and Supplementary Table 5). In FECB2 and FECB12 the majority of the reads were recruited by OTUs classified as members of the phylum Bacteroidetes (recruiting 50.6 and 72% of the reads, respectively), whereas within FECB58 and FECB68, Armatimonadetes (38.3%) and Chloroflexi (25.9%) were identified as the most abundant phyla (Figure 2 and Supplementary Table 5). The fact that these samples were dominated by phyla other than the Cyanobacteria or Proteobacteria may also help to explain why these samples form distinct clusters (cluster I, XIV and V, IV, respectively; Figure 1).
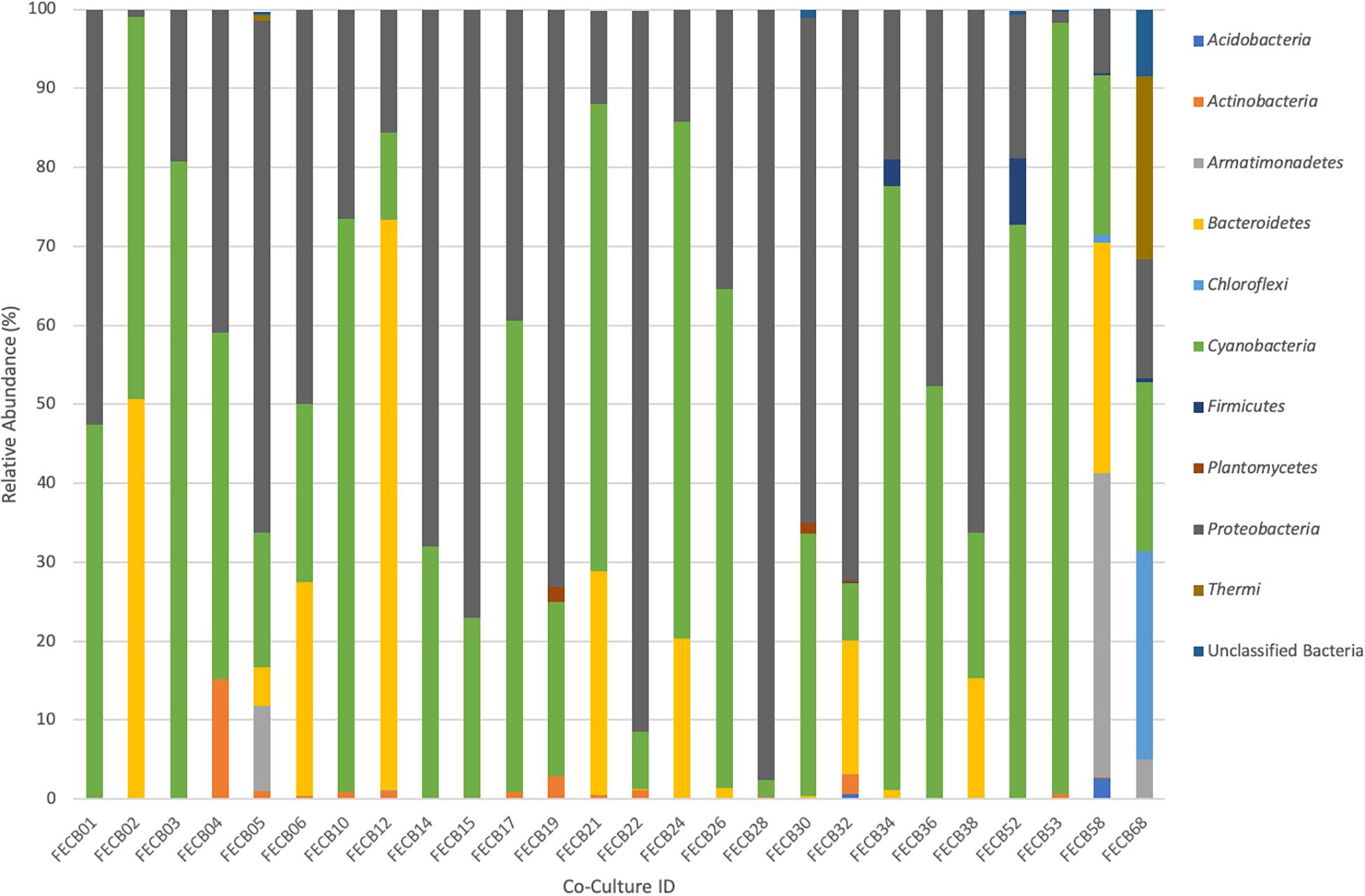
Figure 2. Relative abundance of phyla associated with phototrophic co-cultures. 16S rRNA based community profile. Only phyla recruiting >0.1% of the reads in at least one of the co-cultures are shown.
Firmicutes Dominate Photosynthetic Co-cultures From Hot Springs
Firmicutes abundances calculated for co-cultures from hot spring samples were higher compared to those calculated for co-cultures from other environments. OTUs assigned to the Firmicutes phylum were detected above the applied cut-off level of 0.1% in only five of the twenty-six co-cultures under investigation (Table 3). Interestingly, these samples (i.e., FECB32, FECB34, FECB52, FECB58, and FECB68) are co-cultures collected from hot springs or from deposits within hot springs, with FECB52, FECB58, and FECB68 being maintained in culture at temperatures >40°C. OTU000073 (classified as Alicyclobacillus tolerans), OTU00082 (classified as members of the genus Paenibacillus), OTU000154 (classified as Geobacillus vulcani), and OTU000158 (classified as a member of the Bacillaceae family) recruited 5.9, 3.4, 0.5, and 0.4% of the reads generated from FECB52, FECB34, FECB68, and FECB58, respectively (Supplementary Table 3). Alicyclobacillus tolerans and Geobacillus vulcani have been described previously as aerobic spore-forming thermophiles and have been isolated from lead–zinc ores (Karavaiko et al., 2005) and hot springs (Nazina et al., 2004) located in Russia, respectively. Members of the genus Paenibacillus have been isolated from a wide variety of environments and some Paenibacillus species have been found to promote crop growth directly via biological nitrogen fixation, phosphate solubilization, production of the phytohormone indole-3-acetic acid; and they have been identified as a potential source of novel antimicrobial agents (Grady et al., 2016).
Photosynthetic Co-cultures From Antarctica and YNP to Study Adaptation to Increased Radiation, Low Temperatures and Oligotrophic Growth Conditions
Microbial adaptation to extreme environments and the molecular framework that enable microorganisms to survive and thrive in the presence of increased rates of radiation, low temperatures and in the absence of nutrients has fascinated the scientific community for decades and remains poorly understood. In an attempt to provide a better basis of the taxonomic make-up of co-cultures that were collected from ecosystems that are characterized by these extremes, we included co-cultures from Antarctica and YNP in this study (Table 1). OTU-based comparison of Antarctica and YNP co-cultures revealed between 197 (FECB2) and 549 (FECB6) distinct OTUs [mean (SD) 342 (87.2) OTUs] based on 97% sequence similarity (Table 2). The number of OTUs that recruited >0.1% of all reads ranged from 3 to 29 OTUs, with FECB2 and FECB32 having the lowest and highest OTU count, respectively (Table 2). FECB2 was dominated by an OTU classified as Hymenobacter, which recruited all Bacteroidetes-specific reads generated from this sample (Tables 3, 4). The genus Hymenobacter contains several pigmented bacteria that have been isolated from Antarctica and have been reported to possess increased resistance to radiation (Oh et al., 2016; Marizcurrena et al., 2017), which might explain their increased abundance in FECB2, a co-culture isolated from an environment known to possess increased levels of UV radiation. Taking this into consideration, FECB2 and its individual community members could be a potential target for future studies to enhance our understanding of processes that infer resistance to radiation and DNA damage. The second most abundant OTU in FECB2, recruiting 48% of the generated samples, was classified as Phormidium sp. (Supplementary Table 3), a cyanobacterial genus that has been reported to dominate aquatic microbial mats from Antarctica (Jungblut et al., 2005; Strunecky et al., 2012). Representative isolates from this genus have been proposed previously as cost-effective options for industrial carotenoid production (Shukla and Kashyap, 2003), suggesting that FECB2 may hold the potential for this process.
FECB32 is a mixed culture isolated from an ancient travertine at Mammoth in YNP. Our analysis indicated that FECB32 contained 29 OTUs that each accounted for >0.1% of the reads generated (Table 2). Fifteen of these OTUs recruited >1% of all reads and four OTUs collectively accounted for ∼60% of the reads generated from this co-culture (Supplementary Table 5). These four OTUs were classified as Sphingopyxis alaskensis, Chelativorans sp., and as members of the Chitinophagaceae and Comamonadaceae families, recruiting ∼19, 13, 17, and 11% of the reads, respectively (Supplementary Tables 3,4). S. alaskensis is a Gram-negative bacterium found in relatively high abundance in oligotrophic regions of the ocean (Vancanneyt et al., 2001; Cavicchioli et al., 2003) and it has been studied in great detail as a model system for marine bacteria, specifically to understand microbial adaptation to cold or oligotrophic environments (Lauro et al., 2009; Ting et al., 2010). The Chitinophagaceae family contains a wide phylogenetic diversity with many of its members being mesophilic. However, Chitinophagaceae have been reported to grow optimally at temperatures of 55°C and higher (Anders et al., 2014; Hanada et al., 2014).
Photosynthetic Co-cultures Containing the Deep-Branching Candidate Phylum Melainabacteria
Extreme environments similar to those on early Earth are often proposed to hold critical information about the historical progression of life on our planet and a niche that encompasses those physical stresses is the endolithic environment of rocks (Norris and Castenholz, 2006). Phylogenetic analysis of the heterotrophic population associated with FECB32, which was isolated from travertine deposited by hot springs in YNP, found that sequences from MLE-12 (OTU000109) recruited ∼2% of the sample specific sequences (Supplementary Table 3). This rendered MLE-12, previously assigned to the deep-branching candidate phylum Melainabacteria (Di Rienzi et al., 2013), as the eleventh most abundant organism in this photosynthetic co-culture. It has been proposed previously that Melainabacteria, which is commonly found in aquatic habitats, separated from the cyanobacteria before the latter acquired photosynthetic capabilities (Di Rienzi et al., 2013). Hence FECB32 might be a particularly valuable co-culture to generate new insights into the evolution of and relationship between the phylogenetically closely related Cyanobacteria and Melainabacteria.
Interestingly, OTU000109 was also detected in FECB36 and FECB38 (Supplementary Table 3), although at significantly lower abundance (<0.001%). FECB36 and FECB38 were similar to FECB32 in that they were isolated from sites in YNP. Interestingly, FECB32 and FECB38 cluster together (cluster IX) suggesting similar overall microbial community profiles, but separately from FECB36 (Figure 1). The only additional samples that contained OTUs classified as Melainabacteria, recruiting >0.1% of the generated reads, were FECB58 and FECB68 with ∼0.9 and ∼0.2% of their reads to this deeply branched phylum, respectively (Supplementary Table 5). It seems noteworthy that FECB58 and FECB68 were also isolated from hot springs and clustered closely together based on their overall microbiome composition (Clusters V and IV, respectively; Figure 1).
The McMurdo Dry Valley Lake System, a Physically Highly Stable Lacustrine System
The McMurdo Dry Valley (MDV) is one of the most extreme deserts on Earth, and although the importance of microbial communities for biogeochemical cycles of this region is widely accepted, the microbial ecology of the MDV remains poorly understood (Chan et al., 2013). FECB3, originating from a brackish pond on Bratina Island, was dominated by OTU000003, which recruited 80.3% of all reads (Supplementary Table 5). OTU000003 was classified as the cyanobacterium Phormidium pseudopriestleyi, previously reported to dominate microbial mats of the anoxic zone of Lake Fryxell, Antarctica (Jungblut et al., 2015). The second and third most abundant OTUs in FECB3 were OTU000015 and OTU000061, respectively (Supplementary Table 5). Both OTU000015 and OTU000061 were classified as Rhodobacteriaceae and recruited 9.2 and 8.2% of the reads generated for FECB3. Whereas a taxonomic classification of OTU000015 was not possible beyond the family level, OTU000061 was classified as Paracoccus marcusii, a Gram-negative organism that displays a bright orange color due to the synthesis of carotenoids such as astaxanthin (Harker et al., 1998).
Photosynthetic Co-cultures From Hunter’s Hot Spring, Oregon
FECB58 and FECB68 were both isolated from Hunters Hot Spring in Oregon, United States and they shared similar microbial community members. Despite their similar community profile, abundances of the dominant OTUs associated with these two hot spring co-cultures were remarkably different. FECB58 was dominated by three OTUs (OTU000014, OTU000024, and OTU000033). OTU000014 was classified as OS-L, an uncultured representative of the phylum Armatimonadetes, OTU000024 which was classified as belonging to the Bacteroidetes phylum, and OTU000033 which was classified as Thermosynechococcus. These OTUs contributed 38, 29, and 20% of the reads generated from FECB58, respectively. Whereas OTU000014 recruited ∼4.9% of all reads generated from FECB68, representing the sixth most abundant OTU in the FECB68 community, OTU000024 and OTU000033 were only present at an abundance <0.0001% in FECB68 (Supplementary Table 3).
FECB68 was dominated by 6 OTUs (i.e., OTU000028, OTU000030, OTU000036, OTU000049, OTU000065, and OTU000014) recruiting ∼25.7, 23.1, 20.4, 14.3, 7.6, and 4.9% of the reads, respectively. OTU000028 was classified as belonging to the genus Chloroflexus, whereas OTU000030 and OTU000036 were classified as representative of the genus Meiothermus and Gloeobacter, respectively. Chloroflexus is an anoxygenic phototrophic bacterium that grows at temperatures up to 70°C (Castenholz, 2015) and forms yellow-orange-greenish mats in association with cyanobacteria (Hanada, 2014). Members of the cyanobacterial genus Gloeobacter lack thylakoids, and have been proposed to host the earliest ancestors, or a missing link, in the cyanobacteria lineage (Saw et al., 2013). Thus, FECB68 offers a unique opportunity to investigate interspecies interaction between a member of these basal cyanobacteria and the thermophilic phototroph Chloroflexus, represented by OTU000028 in this co-culture. As outlined in a recent review by Castenholz (2015), Hunter’s Hot Spring located in Oregon is one of the most studied hot springs in the world and a large repertoire of work has been conducted on this habitat over the last 40 years. However, most of this work was performed prior to the advent of recent molecular and -omics techniques.
Photosynthetic Co-cultures From Lignocellulosic Surfaces With Potential to Fix Nitrogen and Degrade Aromatic Compounds
FECB22 and FECB26 are mesophilic co-cultures collected from similar habitats (i.e., from tree bark and a wooden fence) from two locations (i.e., Hawaii and Bermuda) approximately 9,000 kilometers apart from each other (Supplementary Figure 1 and Table 1). Diversity index calculation placed these two samples in the mid-range of the diversity spectrum of the 26 co-cultures analyzed for this study. The inverse Simpson and Shannon index was calculated at 4.46 and 2.08 for FECB22 and 2.29 and 1.32 for FECB26, respectively (Table 2). Within FECB22, 23 OTUs were identified as individually recruiting more than 0.1% of the generated reads. In contrast, FECB26 contained only 16 OTUs that recruited more than 0.1% of the reads each (Supplementary Table 5). FECB22, scraped from tree bark in Hawaii, was dominated by 11 OTUs, each recruiting (>1% of the reads. The most abundant OTU (OTU000017) was classified as a member of the Mycoplana, a genus that contains bacteria capable of aromatic compound degradation (Urakami et al., 1990), and it recruited 40.2% of the reads. OTU000042 (classified as Rhizobium leguminosarum), OTU000045 (classified as Acetobacteraceae), and OTU000072 (classified as Cyanobacteria), were the next most abundant OTUs, recruiting 17.1, 16.3, and 5.5% of the reads generated from FECB22, respectively. Rhizobium leguminosarum is a well-studied α-proteobacterium capable of N2-fixation and “rhizobia” have been suggested repeatedly to facilitate more sustainable agricultural practices through their symbiosis with legumes, reducing the need for nitrogen fertilizer (Marek-Kozaczuk et al., 2017). It remains to be seen if OTU000042 provides N2 to the other organisms in this co-culture or if it consumes all of the fixed N2 itself. Acetobacteraceae are α-proteobacteria often associated with low pH environments and are known for their ability to efficiently synthesize biological cellulose (Rozenberga et al., 2016; Semjonovs et al., 2017). Furthermore, Acetobacteraceae have been reported before as some of the dominant players in photosynthetic consortia during soil formation (Mapelli et al., 2011). It would be interesting to explore the agricultural and chemical potential of a minimalistic co-culture composed of the four OTUs (i.e., OTU000017, OTU000042, OTU000045, and OTU000072) that dominated FECB22, as they may combine the ability to degrade aromatic compounds and synthesize cellulose while removing nitrogen from the atmosphere. FECB26, on the other hand, was dominated by OTU000010, which recruited 63.2% of the reads generated and it was identified as an unclassified member of the Nostocales; a phylogenetic group known for their functional and morphological diversity. Members of the Sphingomonadaceae (i.e., OTU000041 and OTU000062), phototropic α-proteobacteria often found in high abundance in environments previously thought to support mostly the growth of cyanobacteria (Tahon and Willems, 2017), contributed to a total of 25.6% of the generated reads. Most interestingly, OTU000017 was also detected within FECB26 recruiting ∼1.6% of the reads. It is possible that OTU000017 facilitates a metabolic reaction in which aromatic compounds typically associated with the decomposition of woody material under aerobic conditions are utilized.
Conclusion
Culture collections can provide easy access to biological samples without the need for extensive resources by the requesting individual, subsequently facilitating new studies and ultimately advancing our understanding of phylogenetic and functional biodiversity. While these collections present increased access to typically hard to acquire samples, there is lost diversity due to cultivation bias, but it remains to be understood exactly how prevalent and consistent the loss of diversity is sample to sample. Although care is taken to mimic the native environmental conditions of each sample in the cultivation process, there are real world factors that either cannot be mimicked in a lab setting or are unknown to researchers. More work is needed to assess this cultivation bias and to develop techniques to minimize the effects. Although some of the diversity of the original microbial community might have been lost due to a cultivation bias, the 16S rRNA based community fingerprints of the 26 photosynthetic co-cultures described here provide a first in-depth glimpse into the taxonomic and functional diversity of communities from extreme environments that were considered for a long time as too harsh to support the growth of complex microbial communities. The extreme conditions that are associated with the habitats from which these co-cultures were collected offer the unique opportunity to study the molecular mechanisms that support the growth of these extremophilic co-cultures and their role in global carbon and nitrogen cycling. Co-cultures from the CCMEE, and data presented here, also provide a first opening to enhance our understanding of the origin of oxygenic photosynthesis and aerobic respiration in Cyanobacteria, an area that is currently still poorly understood (Soo et al., 2017). Furthermore, an in-depth understanding of these co-cultures holds the potential to discover novel microbial proteins that might render current agricultural, industrial and medical processes more economical and sustainable, for example by promoting or inhibiting plant and microbial growth.
The heterogeneity of the physical parameters reported for the sites where the samples presented in this work were collected, highlights a major challenge (i.e., standardization of protocols) associated with environmental samples and their corresponding metadata (i.e., data describing conditions at each sampling site), specifically when collected during independent sampling efforts. Fortunately, with recent advances in data technologies, the task of data acquisition and dissemination has become less of a challenge. In order to make the best use of these technologies defining a set of minimal information parameters to be recorded during the collection of an environmental sample is of great importance. Similar efforts have been successfully implemented by the Genomic Standards Consortium (GSC) for microbial genomes and metagenomes in the form of the “minimum information about a genome sequence” (MIGS) (Field et al., 2008) and are enforced when describing a novel microbial species (Kampfer et al., 2003).
The identification of Minimum Information about a Co-Culture Sample (MICCS) would be a significant step in standardizing sample acquisition and maintenance, increasing the value of current and future microbial samples collected from the environment. Developing MICCS and applying them to co-cultures currently available from existing culture depositories is beyond the scope of the work presented here, but we hope that the results presented here will contribute to the initiation of this process and stimulate broad involvement and support from the scientific community and various funding agencies.
In summary, we encourage the scientific community to take advantage of the CCMEE and the data we generated during this pilot study. Both data and samples from which these data were generated are publicly available from the CCMEE for further in-depth analyzes and investigations. Future work which might provide a more detailed picture of the microbe-microbe interactions in these co-cultures and their role in the global carbon and nitrogen cycle.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA401502.
Author Contributions
CS, CB, RC, DC, MH, and ST wrote the manuscript. RC and MH designed the experiment. EH and MH performed the experiment. MB, TG, MH-S, EH, MH, NS, and ST generated the data. MB, CS, CB, MC, DC, JG, TG, MH-S, EH, MH, and NS analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the College of Agricultural and Environmental Science and the Microbiology and Biochemistry, Molecular, Cellular, and Developmental Biology Graduate Group at University of California Davis (Davis, CA, United States) and the United States Department of Energy (DOE) Joint Genome Institute (JGI) in Walnut Creek, CA, United States. Work conducted by the JGI, a DOE User Facility, is supported by DOE’s Office of Science under Contract No. DE-AC02-05CH11231.
Conflict of Interest
EH was employed by the company Bayer. DC was employed by the company Greenlight Biosciences.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would thank Drs. Jorge Rodrigues and John Meeks from UC Davis for providing valuable comments and suggestions on how to improve this manuscript. We would like to dedicate this publication to RC who passed away during the completion of this work after a long and satisfying journey in the world of Cyanobacteria. He was, and will remain, a great inspiration to many of us. This manuscript has been released as a pre-print at http://www.biorxiv.org (Shaw et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.572131/full#supplementary-material
Footnotes
- ^ http://www.jgi.doe.gov
- ^ http://genome.jgi.doe.gov/
- ^ http://www.7-zip.org
- ^ http://sherry.cady@pnnl.gov
References
Abed, R. M., Dobretsov, S., and Sudesh, K. (2009). Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 106, 1–12. doi: 10.1111/j.1365-2672.2008.03918.x
Al-Haj, L., Lui, Y. T., Abed, R. M., Gomaa, M. A., and Purton, S. (2016). Cyanobacteria as chassis for industrial biotechnology: progress and prospects. Life (Basel) 6:42. doi: 10.3390/life6040042
Allen, M. M., and Stanier, R. Y. (1968). Selective isolation of blue-green algae from water and soil. J. Gen. Microbiol. 51, 203–209. doi: 10.1099/00221287-51-2-203
Anders, H., Dunfield, P. F., Lagutin, K., Houghton, K. M., Power, J. F., Mackenzie, A. D., et al. (2014). Thermoflavifilum aggregans gen. nov., sp. nov., a thermophilic and slightly halophilic filamentous bacterium from the phylum Bacteroidetes. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1264–1270. doi: 10.1099/ijs.0.057463-0
Boundy-Mills, K., Hess, M., Bennett, A. R., Ryan, M., Kang, S., Nobles, D., et al. (2015). The United States culture collection network (USCCN): enhancing microbial genomics research through living microbe culture collections. Appl. Environ. Microbiol. 81, 5671–5674. doi: 10.1128/AEM.01176-15
Camacho, A., Garcia-Pichel, F., Vicente, E., and Castenholz, R. W. (1996). Adaptation to sulfide and to the underwater light field in three cyanobacterial isolates from Lake Arcas (Spain). FEMS Microbiol. Ecol. 21, 293–301. doi: 10.1111/j.1574-6941.1996.tb00126.x
Castenholz, R. W. (1981). “Isolation and cultivation of thermophilic cyanobacteria,” in Isolation and Identification of Bacteria, eds M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (Berlin: Springer), 236–246. doi: 10.1007/978-3-662-13187-9_11
Castenholz, R. W. (2015). Portrait of a geothermal spring, hunter’s hot springs, oregon. Life (Basel) 5, 332–347. doi: 10.3390/life5010332
Cavicchioli, R., Ostrowski, M., Fegatella, F., Goodchild, A., and Guixa-Boixereu, N. (2003). Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis). Microb. Ecol. 45, 203–217. doi: 10.1007/s00248-002-3008-6
Chan, Y., Van Nostrand, J. D., Zhou, J., Pointing, S. B., and Farrell, R. L. (2013). Functional ecology of an Antarctic Dry Valley. Proc. Natl. Acad. Sci. U.S.A. 110, 8990–8995. doi: 10.1073/pnas.1300643110
Cubillos-Ruiz, A., Berta-Thompson, J. W., Becker, J. W., van der Donk, W. A., and Chisholm, S. W. (2017). Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, E5424–E5433. doi: 10.1073/pnas.1700990114
de-Bashan, L. E., Bashan, Y., Moreno, M., Lebsky, V. K., and Bustillos, J. J. (2002). Increased pigment and lipid content, lipid variety, and cell and population size of the microalgae Chlorella spp. when co-immobilized in alginate beads with the microalgae-growth-promoting bacterium Azospirillum brasilense. Can. J. Microbiol. 48, 514–521. doi: 10.1139/w02-051
Di Rienzi, S. C., Sharon, I., Wrighton, K. C., Koren, O., Hug, L. A., Thomas, B. C., et al. (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2:e01102. doi: 10.7554/eLife.01102
Dillon, J. G., and Castenholz, R. W. (2003). The synthesis of the UV-screening pigment, scytonemin, and photosynthetic performance in isolates from closely related natural populations of cyanobacteria (Calothrix sp.). Environ. Microbiol. 5, 484–491. doi: 10.1046/j.1462-2920.2003.00436.x
Dillon, J. G., Tatsumi, C. M., Tandingan, P. G., and Castenholz, R. W. (2002). Effect of environmental factors on the synthesis of scytonemin, a UV-screening pigment, in a cyanobacterium (Chroococcidiopsis sp.). Arch. Microbiol. 177, 322–331. doi: 10.1007/s00203-001-0395-x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Evans, J., Sheneman, L., and Foster, J. (2006). Relaxed neighbor joining: a fast distance-based phylogenetic tree construction method. J. Mol. Evol. 62, 785–792. doi: 10.1007/s00239-005-0176-2
Field, D., Garrity, G., Gray, T., Morrison, N., Selengut, J., Sterk, P., et al. (2008). The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 26, 541–547. doi: 10.1038/nbt1360
Formighieri, C., and Melis, A. (2016). Sustainable heterologous production of terpene hydrocarbons in cyanobacteria. Photosynthesis Res. 130, 123–135. doi: 10.1007/s11120-016-0233-2
Gajda, I., Greenman, J., Melhuish, C., and Ieropoulos, I. (2015). Self-sustainable electricity production from algae grown in a microbial fuel cell system. Biomass Bioenergy 82, 87–93. doi: 10.1016/j.biombioe.2015.05.017
Garcia-Pichel, F., Belnap, J., Neuer, S., and Schanz, F. (2003). Estimates of global cyanobacterial biomass and its distribution. Algological Stud. 109, 213–227. doi: 10.1127/1864-1318/2003/0109-0213
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15:203. doi: 10.1186/s12934-016-0603-7
Hanada, S. (2014). “The phylum chloroflexi, the family Chloroflexaceae, and the related phototrophic families Oscillochloridaceae and Roseiflexaceae,” in Other Major Lineages of Bacteria and the Archaea, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 515–532. doi: 10.1007/978-3-642-38954-2_165
Hanada, S., Tamaki, H., Nakamura, K., and Kamagata, Y. (2014). Crenotalea thermophila gen. nov., sp. nov., a member of the family Chitinophagaceae isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1359–1364. doi: 10.1099/ijs.0.058594-0
Harker, M., Hirschberg, J., and Oren, A. (1998). Paracoccus marcusii sp. nov., an orange gram-negative coccus. Int. J. Syst. Bacteriol. 48(Pt 2), 543–548. doi: 10.1099/00207713-48-2-543
Jungblut, A. D., Hawes, I., Mackey, T. J., Krusor, M., Doran, P. T., Sumner, D. Y., et al. (2015). Microbial mat communities along an oxygen gradient in a perennially ice-covered Antarctic lake. Appl. Environ. Microbiol. 82, 620–630. doi: 10.1128/AEM.02699-15
Jungblut, A.-D., Hawes, I., Mountfort, D., Hitzfeld, B., Dietrich, D. R., Burns, B. P., et al. (2005). Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 7, 519–529. doi: 10.1111/j.1462-2920.2005.00717.x
Kampfer, P., Buczolits, S., Albrecht, A., Busse, H. J., and Stackebrandt, E. (2003). Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int. J. Syst. Evol. Microbiol. 53(Pt 3), 893–896. doi: 10.1099/ijs.0.02710-0
Karavaiko, G. I., Bogdanova, T. I., Tourova, T. P., Kondrat’eva, T. F., Tsaplina, I. A., Egorova, M. A., et al. (2005). Reclassification of ‘Sulfobacillus thermosulfidooxidans subsp. thermotolerans’ strain K1 as Alicyclobacillus tolerans sp. nov. and Sulfobacillus disulfidooxidans Dufresne et al. 1996 as Alicyclobacillus disulfidooxidans comb. nov., and emended description of the genus Alicyclobacillus. Int. J. Syst. Evol. Microbiol. 55(Pt 2), 941–947. doi: 10.1099/ijs.0.63300-0
Karl, D., Letelier, R., Tupas, L., Dore, J., Christian, J., and Hebel, D. (1997). The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388, 533–538. doi: 10.1038/41474
Kim, B. R., Shin, J., Guevarra, R., Lee, J. H., Kim, D. W., Seol, K. H., et al. (2017). Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Kleigrewe, K., Gerwick, L., Sherman, D. H., and Gerwick, W. H. (2016). Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat. Prod. Rep. 33, 348–364. doi: 10.1039/c5np00097a
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lauro, F. M., McDougald, D., Thomas, T., Williams, T. J., Egan, S., Rice, S., et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533. doi: 10.1073/pnas.0903507106
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, B., Sher, D., Kelly, L., Shi, Y., Huang, K., Knerr, P. J., et al. (2010). Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 10430–10435. doi: 10.1073/pnas.0913677107
Mapelli, F., Marasco, R., Rizzi, A., Baldi, F., Ventura, S., Daffonchio, D., et al. (2011). Bacterial communities involved in soil formation and plant establishment triggered by pyrite bioweathering on arctic moraines. Microb. Ecol. 61, 438–447. doi: 10.1007/s00248-010-9758-7
Marek-Kozaczuk, M., Wdowiak-Wrobel, S., Kalita, M., Chernetskyy, M., Derylo, K., Tchorzewski, M., et al. (2017). Host-dependent symbiotic efficiency of Rhizobium leguminosarum bv. trifolii strains isolated from nodules of Trifolium rubens. Antonie Van Leeuwenhoek 110, 1729–1744. doi: 10.1007/s10482-017-0922-7
Marizcurrena, J. J., Morel, M. A., Brana, V., Morales, D., Martinez-Lopez, W., and Castro-Sowinski, S. (2017). Searching for novel photolyases in UVC-resistant Antarctic bacteria. Extremophiles 21, 409–418. doi: 10.1007/s00792-016-0914-y
McCluskey, K. (2017). A review of living collections with special emphasis on sustainability and its impact on research across multiple disciplines. Biopreserv. Biobank 15, 20–30. doi: 10.1089/bio.2016.0066
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Miao, R., Xie, H., and Lindblad, P. (2018). Enhancement of photosynthetic isobutanol production in engineered cells of Synechocystis PCC 6803. Biotechnol. Biofuels 11:267. doi: 10.1186/s13068-018-1268-8
Miller, S. R., and Castenholz, R. W. (2000). Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66, 4222–4229. doi: 10.1128/aem.66.10.4222-4229.2000
Morris, E. K., Caruso, T., Buscot, F., Fischer, M., Hancock, C., Maier, T. S., et al. (2014). Choosing and using diversity indices: insights for ecological applications from the German biodiversity exploratories. Ecol. Evol. 4, 3514–3524. doi: 10.1002/ece3.1155
Nadeau, T.-L., and Castenholz, R. W. (2000). Characterization of psychrophilic oscillatorians (cyanobacteria) from antarctic meltwater ponds. J. Phycol. 36, 914–923. doi: 10.1046/j.1529-8817.2000.99201.x
Nadeau, T.-L., Milbrandt, E. C., and Castenholz, R. W. (2001). Evolutionary relationships of cultivated antarctic oscillatorians (cyanobacteria). J. Phycol. 37, 650–654. doi: 10.1046/j.1529-8817.2001.037004650.x
Nazina, T. N., Lebedeva, E. V., Poltaraus, A. B., Tourova, T. P., Grigoryan, A. A., Sokolova, D., et al. (2004). Geobacillus gargensis sp. nov., a novel thermophile from a hot spring, and the reclassification of Bacillus vulcani as Geobacillus vulcani comb. nov. Int. J. Syst. Evol. Microbiol. 54(Pt 6), 2019–2024. doi: 10.1099/ijs.0.02932-0
Norris, T. B., and Castenholz, R. W. (2006). Endolithic photosynthetic communities within ancient and recent travertine deposits in Yellowstone National Park. FEMS Microbiol. Ecol. 57, 470–483. doi: 10.1111/j.1574-6941.2006.00134.x
Oh, T. J., Han, S. R., Ahn, D. H., Park, H., and Kim, A. Y. (2016). Complete genome sequence of Hymenobacter sp. strain PAMC26554, an ionizing radiation-resistant bacterium isolated from an Antarctic lichen. J. Biotechnol. 227, 19–20. doi: 10.1016/j.jbiotec.2016.04.011
Padmaperuma, G., Kapoore, R. V., Gilmour, D. J., and Vaidyanathan, S. (2018). Microbial consortia: a critical look at microalgae co-cultures for enhanced biomanufacturing. Crit. Rev. Biotechnol. 38, 690–703. doi: 10.1080/07388551.2017.1390728
Pisciotta, J. M., Zou, Y., and Baskakov, I. V. (2010). Light-dependent electrogenic activity of cyanobacteria. PLoS One 5:e10821. doi: 10.1371/journal.pone.0010821
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rozenberga, L., Skute, M., Belkova, L., Sable, I., Vikele, L., Semjonovs, P., et al. (2016). Characterisation of films and nanopaper obtained from cellulose synthesised by acetic acid bacteria. Carbohydr. Polym. 144, 33–40. doi: 10.1016/j.carbpol.2016.02.025
Saw, J. H., Schatz, M., Brown, M. V., Kunkel, D. D., Foster, J. S., Shick, H., et al. (2013). Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kilauea Caldera, Hawai’i. PLoS One 8:e76376. doi: 10.1371/journal.pone.0076376
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Semjonovs, P., Ruklisha, M., Paegle, L., Saka, M., Treimane, R., Skute, M., et al. (2017). Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Appl. Microbiol. Biotechnol. 101, 1003–1012. doi: 10.1007/s00253-016-7761-8
Shaw, C., Brooke, C. G., Connolly, M. P., Garcia, J. A., Harmon-Smith, M., Shapiro, N., et al. (2020). Phototrophic co-cultures from extreme environments: community structure and potential value for fundamental and applied research. bioRxiv [Preprint]. doi: 10.1101/427211
Shukla, S. P., and Kashyap, A. K. (2003). An assessment of biopotential of three cyanobacterial isolates from Antarctic for carotenoid production. Indian J. Biochem. Biophys. 40, 362–366.
Soo, R. M., Hemp, J., Parks, D. H., Fischer, W. W., and Hugenholtz, P. (2017). On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 355, 1436–1440. doi: 10.1126/science.aal3794
Strunecky, O., Elster, J., and Komarek, J. (2012). Molecular clock evidence for survival of Antarctic cyanobacteria (Oscillatoriales, Phormidium autumnale) from Paleozoic times. FEMS Microbiol. Ecol. 82, 482–490. doi: 10.1111/j.1574-6941.2012.01426.x
Subashchandrabose, S. R., Ramakrishnan, B., Megharaj, M., Venkateswarlu, K., and Naidu, R. (2011). Consortia of cyanobacteria/microalgae and bacteria: biotechnological potential. Biotechnol. Adv. 29, 896–907. doi: 10.1016/j.biotechadv.2011.07.009
Sudek, S., Haygood, M. G., Youssef, D. T., and Schmidt, E. W. (2006). Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 72, 4382–4387. doi: 10.1128/AEM.00380-06
Tahon, G., and Willems, A. (2017). Isolation and characterization of aerobic anoxygenic phototrophs from exposed soils from the Sor Rondane Mountains, East Antarctica. Syst. Appl. Microbiol. 40, 357–369. doi: 10.1016/j.syapm.2017.05.007
Ting, L., Williams, T. J., Cowley, M. J., Lauro, F. M., Guilhaus, M., Raftery, M. J., et al. (2010). Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 12, 2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x
Toplin, J. A., Norris, T. B., Lehr, C. R., McDermott, T. R., and Castenholz, R. W. (2008). Biogeographic and phylogenetic diversity of thermoacidophilic cyanidiales in Yellowstone National Park, Japan, and New Zealand. Appl. Environ. Microbiol. 74, 2822–2833. doi: 10.1128/AEM.02741-07
Urakami, T., Oyanagi, H., Araki, H., Suzuki, K.-I., and Komagata, K. (1990). Recharacterization and emended description of the genus Mycoplana and description of two new species, Mycoplana ramosa and Mycoplana segnis. Int. J. Syst. Evol. Microbiol. 40, 434–442. doi: 10.1099/00207713-40-4-434
Vancanneyt, M., Schut, F., Snauwaert, C., Goris, J., Swings, J., and Gottschal, J. C. (2001). Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. Evol. Microbiol. 51(Pt 1), 73–79. doi: 10.1099/00207713-51-1-73
Wang, R., Xue, S., Zhang, D., Zhang, Q., Wen, S., Kong, D., et al. (2015). Construction and characteristics of artificial consortia of Scenedesmus obliquus-bacteria for S. obliquus growth and lipid production. Algal Res. 12, 436–445. doi: 10.1016/j.algal.2015.10.002
Whitton, B. A., and Potts, M. (2000). The Ecology of Cyanobacteria: their Diversity in time and Space. Alphen aan den Rijn: Kluwer Academic.
Woo, H. M., and Lee, H. J. (2017). Toward solar biodiesel production from CO2 using engineered cyanobacteria. FEMS Microbiol. Lett. 364:fnx066. doi: 10.1093/femsle/fnx066
Yen, H.-W., Chen, P.-W., and Chen, L.-J. (2015). The synergistic effects for the co-cultivation of oleaginous yeast-Rhodotorula glutinis and microalgae-Scenedesmus obliquus on the biomass and total lipids accumulation. Bioresource Technol. 184, 148–152. doi: 10.1016/j.biortech.2014.09.113
Zarzycki, J., Axen, S. D., Kinney, J. N., and Kerfeld, C. A. (2013). Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64, 787–798. doi: 10.1093/jxb/ers294
Keywords: biodiversity, biotechnology, culture collection, cyanobacteria, extreme environments, fundamental research, microbial ecology and diversity
Citation: Shaw C, Brooke C, Hawley E, Connolly MP, Garcia JA, Harmon-Smith M, Shapiro N, Barton M, Tringe SG, Glavina del Rio T, Culley DE, Castenholz R and Hess M (2020) Phototrophic Co-cultures From Extreme Environments: Community Structure and Potential Value for Fundamental and Applied Research. Front. Microbiol. 11:572131. doi: 10.3389/fmicb.2020.572131
Received: 12 June 2020; Accepted: 13 October 2020;
Published: 06 November 2020.
Edited by:
Daniel Ducat, Michigan State University, United StatesReviewed by:
Ralf Steuer, Humboldt University of Berlin, GermanyAlison Gail Smith, University of Cambridge, United Kingdom
Copyright © 2020 Shaw, Brooke, Hawley, Connolly, Garcia, Harmon-Smith, Shapiro, Barton, Tringe, Glavina del Rio, Culley, Castenholz and Hess. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias Hess, bWhlc3NAdWNkYXZpcy5lZHU=
 Claire Shaw
Claire Shaw Charles Brooke
Charles Brooke Erik Hawley2
Erik Hawley2 Morgan P. Connolly
Morgan P. Connolly Javier A. Garcia
Javier A. Garcia Susannah G. Tringe
Susannah G. Tringe David E. Culley
David E. Culley Matthias Hess
Matthias Hess