- 1Institute of Biochemistry, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany
- 2Center for Structural Studies, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany
- 3Institute of Pharmaceutical Biology and Biotechnology, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany
- 4Institute for Molecular Medicine, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany
- 5Molecular Proteomics Laboratory, BMFZ, Heinrich-Heine-University-Düsseldorf, Düsseldorf, Germany
Lantibiotics are a growing class of antimicrobial peptides, which possess antimicrobial activity against mainly Gram-positive bacteria including the highly resistant strains such as methicillin-resistant Staphylococcus aureus or vancomycin-resistant enterococci. In the last decades numerous lantibiotics were discovered in natural habitats or designed with bioengineering tools. In this study, we present an insight in the antimicrobial potential of the natural occurring lantibiotic nisin H from Streptococcus hyointestinalis as well as the variant nisin H F1I. We determined the yield of the heterologously expressed peptide and quantified the cleavage efficiency employing the nisin protease NisP. Furthermore, we analyzed the effect on the modification via mass spectrometry analysis. With standardized growth inhibition assays we benchmarked the activity of pure nisin H and the variant nisin H F1I, and their influence on the activity of the nisin immunity proteins NisI and NisFEG from Lactococcus lactis and the nisin resistance proteins SaNSR and SaNsrFP from Streptococcus agalactiae COH1. We further checked the antibacterial activity against clinical isolates of Staphylococcus aureus, Enterococcus faecium and Enterococcus faecalis via microdilution method. In summary, nisin H and the nisin H F1I variant possessed better antimicrobial potency than the natural nisin A.
Introduction
Lantibiotics (lanthionine containing antibiotics) are a growing class of antimicrobial peptides (AMPs), which posses antimicrobial activity even against highly resistant strains such as methicillin-resistant Staphylococcus aureus (MRSA) or vancomycin-resistant enterococci (VRE) and some are already in pre-clinical trials (Mota-Meira et al., 2000; Jabes et al., 2011; Dawson and Scott, 2012; Crowther et al., 2013; Dischinger et al., 2014; Ongey et al., 2017; Brunati et al., 2018; Sandiford, 2019). Lantibiotics are peptides, containing 19–38 amino acids and are mainly produced by Gram-positive bacteria (Klaenhammer, 1993; Sahl and Bierbaum, 1998). In the last decades an increasing number of lantibiotic gene clusters were found by data-mining approaches using tools such as BAGEL4 (van Heel et al., 2018).
The best studied lantibiotic is nisin, which was first discovered in 1928 by Rogers and Whittier and belongs to the class I lantibiotics (Rogers, 1928; Rogers and Whittier, 1928; Arnison et al., 2013). It is used in the food industry since 1953 and obtained the status as generally recognized as safe (GRAS) in 1988 from the Food and Drug Administration (FDA) (Delves-Broughton et al., 1996). The class I lantibiotic nisin contains 34 amino acids and five (methyl)-lanthionine rings. These (methyl)-lanthionine rings require multiple posttranslational modifications (PTMs) which are introduced in the precursor peptide. First, the serine and threonine residues in the core peptide are dehydrated by a specific dehydratase NisB (lantibiotic class I LanB dehydratase) (Kaletta and Entian, 1989; Karakas Sen et al., 1999; Koponen et al., 2002; Ortega et al., 2015; Repka et al., 2017). The next step is a Michael-type condensation of dehydrated residues with the thiol group of a cysteine residue, thereby forming (methyl)-lanthionine rings, guided in a regio- and stereospecific manner by the cyclase NisC (class I lantibiotic cyclase) (Okeley et al., 2003; Li et al., 2006; Li and van der Donk, 2007; Repka et al., 2017). These characteristic (methyl)-lanthionine rings give lantibiotics high heat stability, resistance against proteolytic digestion and are responsible for the nanomolar antimicrobial activity (Gross and Morell, 1967; Rollema et al., 1995; Chan et al., 1996; Lu et al., 2010; Oppedijk et al., 2016).
The sequence of nisin A can be subdivided into three parts (see Figure 1). The N-terminal part with ring A, B, and C is responsible for binding to the cell wall precursor lipid II (Hsu et al., 2004). The hinge region is very flexible and allows reorientation of the C-terminal part to insert into the membrane (van Heusden et al., 2002; Hasper et al., 2004; Wiedemann et al., 2004; Medeiros-Silva et al., 2018), while changes in this region have a strong impact on the target antimicrobial activity (Zhou et al., 2015; Zaschke-Kriesche et al., 2019b). After penetrating the membrane, the C-terminal part with ring D and E forms a stable pore with a stoichiometry of eight nisin and four lipid II molecules, which subsequently leads to rapid cell death (Hasper et al., 2004; Wiedemann et al., 2004; Alkhatib et al., 2014a; Medeiros-Silva et al., 2018).
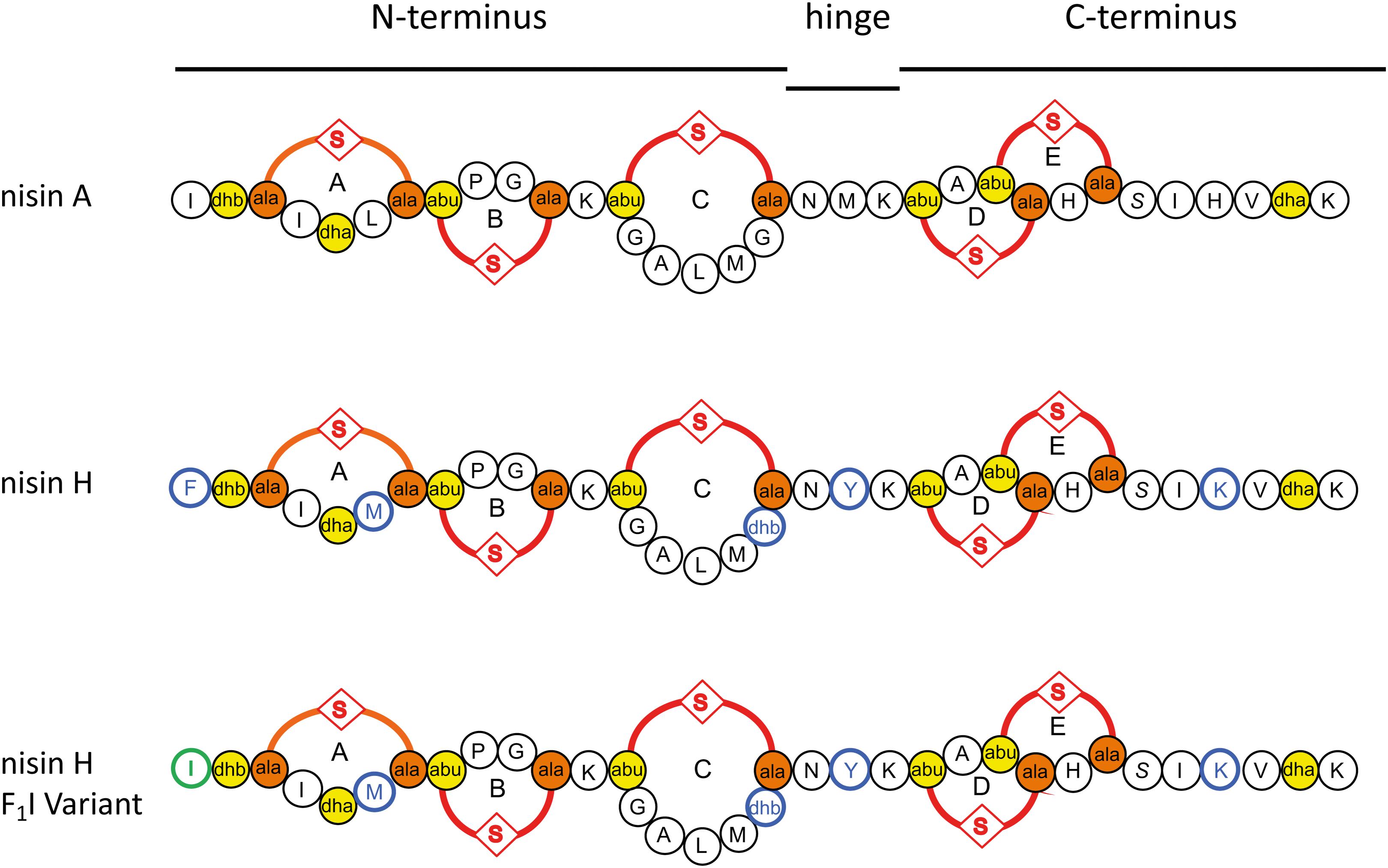
Figure 1. Schematic overview of the used lantibiotics nisin A, nisin H, and the nisin H F1I variant. Point mutations in nisin H and nisin H F1I in comparison to nisin A are highlighted in blue. Dehydrated amino acids and cysteine residues involved in (methyl)-lanthionine ring formation (ring A, B, C, D, and E) are labeled in yellow and orange. The mutation of the nisin H F1I variant is highlighted in green.
Unfortunately, some bacteria established resistance mechanism against lantibiotics. For instance lantibiotic producing strains express the immunity system LanI and LanFEG (Alkhatib et al., 2014a,b), which prevent a suicidal effect after the lantibiotic is secreted. In the case of nisin A from Lactococcus lactis these proteins are called NisI and NisFEG. But also non-lantibiotic producing strains showed resistance against lantibiotics like Streptococcus agalactiae COH1, which arises from the expression of the membrane-anchored peptidase SaNSR and the ABC transporter SaNsrFP (Khosa et al., 2013, 2016a,b; Reiners et al., 2017).
Several natural nisin variants have been discovered and up to now eight are known. First of all nisin A from L. lactis (Rogers and Whittier, 1928), nisin Z from L. lactis NIZO 221 86 (Mulders et al., 1991), nisin F from L. lactis F10 (de Kwaadsteniet et al., 2008), nisin Q from L. lactis 61-14 (Zendo et al., 2003), nisin O1 to O4 from Blautia obeum A2-162 (Hatziioanou et al., 2017), nisin U and U2 from Streptococcus uberis 42 and D536 (Wirawan et al., 2006), nisin P from Streptococcus gallolyticus subsp. pasteurianus (Zhang et al., 2012; Wu et al., 2014), nisin J from Staphylococcus capitis APC 2923 (O’Sullivan et al., 2020) and nisin H from Streptococcus hyointestinalis DPC 6484 (O’Connor et al., 2015).
In this study we focused on the natural nisin H variant (Figure 1). We used a standardized workflow for the characterization of lantibiotics, previous described in Lagedroste et al. (2019) to determine the impact on the expression, modification and antimicrobial properties of this nisin variant. We tested further the antimicrobial activity against some pathogen strains from Staphylococcus aureus, Enterococcus faecium, and Enterococcus faecalis using the microdilution method. As a reference we used the wild-type version of nisin A expressed using the same experimental setup. Furthermore, we exchanged the phenylalanine at position one (F1) of nisin H to isoleucine, which is the natural amino acid of nisin A at this position (Figure 1). This position one was previously analyzed in nisin A and showed a major impact on different levels of the characterization (Lagedroste et al., 2019).
Materials and Methods
Microorganisms and Culture Conditions
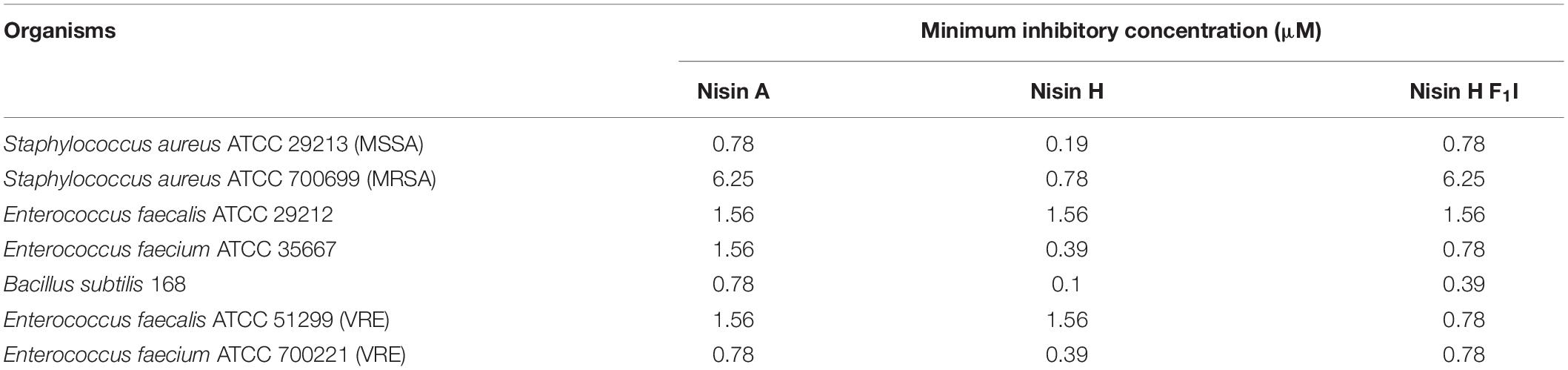
Cultures of L. lactis NZ9000 (Kuipers et al., 1997) containing the plasmids for immunity and resistance proteins were grown in M17 medium (Terzaghi and Sandine, 1975) at 30°C supplemented with 0.5% glucose [GM17 and the appropriate antibiotics descripted in Alkhatib et al. (2014a, b), Khosa et al. (2016b), Reiners et al. (2017), Lagedroste et al. (2019)]. For pre-nisin secretion, the L. lactis strain NZ9000 was grown in minimal medium (Jensen and Hammer, 1993) at 30°C supplemented with 0.5% glucose and the appropriate antibiotics. All bacteria used for minimum inhibitory concentration (MIC) determination of nisin variants [Bacillus subtilis 168; S. aureus: MSSA strain ATCC 29213, MRSA/VISA strain ATCC 700699; E. faecium: ATCC 35667, ATCC 700221 (vancomycin resistant); E. faecalis: ATCC 29212, ATCC 51299 (vancomycin resistant)] were cultivated in Mueller-Hinton broth (MHB) at 37°C and shaking at 150 rpm.
Cloning of Nisin H and the F1I Variant
Nisin H was created as described in Reiners et al. (2017). The substitution of the phenylalanine at position one (F1I) to an isoleucine was performed by site-directed mutagenesis. Here, we used the following primers (forward: 5′-GTGCATCACCACGCTTTACAAGTATTTCGC-3′; reverse: 5′-GCGAAATACTTGTAAAGCGTGGTGATGCAC-3′). After sequence analysis a competent L. lactis NZ9000 strain was transformed with the resulting plasmid via electroporation (Holo and Nes, 1989). The L. lactis NZ9000 strain already contain a vector (pil3-BTC) encoding for the modification and secretion proteins (Rink et al., 2005).
Expression, Purification and Activation of Pre-nisin Variants
The precursor of nisin H and its variant were expressed and purified as previously described (Abts et al., 2013; Alkhatib et al., 2014b; Lagedroste et al., 2017, 2019). Briefly, for pre-nisin secretion, the L. lactis strain NZ9000 was grown in minimal medium (Jensen and Hammer, 1993) supplemented with 0.5% glucose and 5 μg/ml of each erythromycin and chloramphenicol at 30°C. Cells were induced with 10 ng/ml nisin at an OD600 of 0.4 and further grown overnight at 30°C without shaking. After harvesting the cells, the 0.45 μm-filtered supernatant was loaded onto a HiTrap SP HP cation exchange chromatography column. After washing with 50 mM lactic acid, the buffer was changed to 50 mM Hepes pH 7 via gradient and the final elution was done with 50 mM Hepes pH 7, 500 mM NaCl buffer. Elution fractions were concentrated in a 3 kDa cutoff filter. With a soluble version of NisP, the activation of all variants was performed overnight at 8°C (Lagedroste et al., 2017). The yield and cleavage efficiency determination was done by RP-HPLC (Agilent Technologies 1260 Infinity II) with a LiChrospher WP 300 RP-18 end-capped column and an acetonitrile/water solvent system (Abts et al., 2013; Lagedroste et al., 2017, 2019).
MALDI-TOF Analysis: Dehydration and (Methyl)-Lanthionine Ring Analysis
With MALDI-TOF analysis we analyzed the modification state of nisin H and its variant. Dehydrations are directly visible in the spectra, due to the loss of mass (−18 Da). For the determination of the presence of (methyl)-lanthionine rings, we used the organic coupling agent CDAP (1-cyano-4 dimethylaminopyridinium tetrafluoroborate) that binds to free cysteine residues (Wu and Watson, 1998). The reaction of the coupling agent to free cysteine side chains would results in an increased mass in the spectra. The analysis was performed as previously descripted (Lagedroste et al., 2019). The samples were analyzed with MALDI-TOF (UltrafleXtreme, Bruker Daltonics, Bremen, Software: Compass 1.4) in positive mode.
Tandem Mass Spectrometric Analysis of Nisin H and Nisin H F1I
Nisin H and the nisin H F1I variant were purified using solid phase extraction (Oasis HLB, Waters) and finally resuspended in 0.1% trifluoroacetic acid. The samples were first subjected to liquid chromatography on a rapid separation liquid chromatography system (Ultimate 3000, Thermo Fisher Scientific) using an 1 h gradient and C18 columns as described (PMID 24646099) and further analyzed by a QExactive Plus mass spectrometer (Thermo Fisher Scientific) coupled via a nano-source electrospray interface. First, a precursor spectrum was acquired at a resolution of 140,000 (advanced gain control target 3E6, maximum ion time 50 ms, scan range 200–2000 m/z, profile data type). Subsequently, up to four 4–6-fold charged precursors were selected by the quadrupole (2 m/z isolation window), fragmented by higher-energy collisional dissociation (normalized collision energy 30) and recorded at a resolution of 35,000 (advanced gain control target 1E5, maximum ion time 50 ms, available scan range 200–2000 m/z, centroid data type).
Recorded spectra were analyzed by the MASCOT search engine (version 2.4.1, Matrix Science) and searches triggered by Proteome Discoverer (version 2.4.0.305, Thermo Fisher Scientific). A database was generated for the searches including 1000 randomly generated sequence entries each 34 amino acid long) and the sequences of nisin H and nisin H F1I. Methionine oxidation and dehydration of serine and threonine residues were considered as variable modifications and the precursor mass tolerance set to 10 ppm and the fragment mass tolerance set to 0.02 Da. For peptide validation, the Fixed Value PSM validator was used (1% false discovery rate) and the IMP-ptmRS node for site validation (PMID 22073976). No random sequences were found by the search.
Determination of the Antimicrobial Activity by Growth Inhibition Assay
The determination of the antimicrobial activity of the different nisin variants was tested using a growth inhibition assay. The used strains were described in Alkhatib et al. (2014a, b), Reiners et al. (2017), and Lagedroste et al. (2019).
Briefly, the L. lactis NZ9000 strains were grown in M17 medium (Terzaghi and Sandine, 1975) at 30°C supplemented with 0.5% glucose (GM17 and the appropriate antibiotics) overnight with 1 ng/ml nisin. In a 96-well plate, a serial dilution of the nisin variant was applied and incubated with the test strains at a final OD600 of 0.1 for 5 h at 30°C. Later on, the optical density was measured at 584 nm via 96-well plate reader BMG. The normalized optical density was plotted against the logarithm of the nisin concentration and the resulting inhibitory concentration (IC50), represents the value where 50% of the cells died in the presence of the different nisin variants. By dividing the IC50 values of strains expressing the immunity or resistance proteins from the IC50 value of the sensitive strain we calculated the fold of immunity/resistance, which is an indicator for the recognition of nisin H or its variant by the immunity or resistance proteins.
Minimum Inhibitory Concentration Determination of Nisin Variants
Nisin variants were tested for antibacterial capabilities against B. subtilis and different strains from S. aureus, E. faecium, and E. faecalis using the microdilution method, according to the recommendations of Clinical and Laboratory Standards Institute (2012). Briefly, fresh cultures prepared from overnight cultures were incubated until exponential phase (OD ∼ 0.6) and seeded at 5 × 104 CFU/well in 96-well round-bottom microplates, in a total volume of 100 μL containing twofold serially diluted test peptides. Moxifloxacin was used as a positive control. Plates were incubated statically and aerobically for 24 h at 37°C. MICs were determined macroscopically by identifying the least concentration of peptides that resulted in complete inhibition of bacterial visual growth.
SYTOX Green Nucleic Acids Binding Assay
The cells of NZ9000Cm were grown overnight in GM17 supplemented with 5 mg/ml chloramphenicol. The overnight culture was diluted to an OD600 of 0.1 in fresh media and the cultures were grown until the OD600 reaches 0.3. The SYTOX green dye was added at a final concentration of 2.5 mM according to the manual of the manufacturer (Invitrogen). After reaching a stable baseline (∼200 s) we added 100 nM of the nisin variants. The fluorescence signal was measured at an excitation wavelength of 504 nm and emission wavelength of 523 nm, respectively (using a fluorolog Horiba III). After a stable baseline was reached, the nisin variant was added and the fluorescence was monitored over an additional time period. The measurement was performed at 30°C.
Results
O’Connor et al. (2015) described a new natural nisin variant from S. hyointestinalis DPC 6484 and named it nisin H. In following, we compared nisin A and its natural variant nisin H, both heterologously produced in L. lactis, following the protocol of lantibiotic characterization (Lagedroste et al., 2019). We also included the nisin H F1I variant.
The characterization starts with the expression, secretion and purification of the lantibiotic and its comparison to nisin A. The heterologously expressed and secreted nisin A and the variants nisin H and nisin H F1I can be purified with high purity as observed on the Tricine-SDS-PAGE gel (Figure 2A). Nisin A was purified with a yield of 6.0 ± 0.3 mg/L of cell culture (Lagedroste et al., 2019), nisin H was expressed and purified with a yield of 5.3 ± 0.6 mg/L of cell culture, which is identical within experimental error. The nisin H F1I variant displayed a slightly reduced yield of 4.9 ± 0.2 mg/L of cell culture (Figure 2B and Table 1).
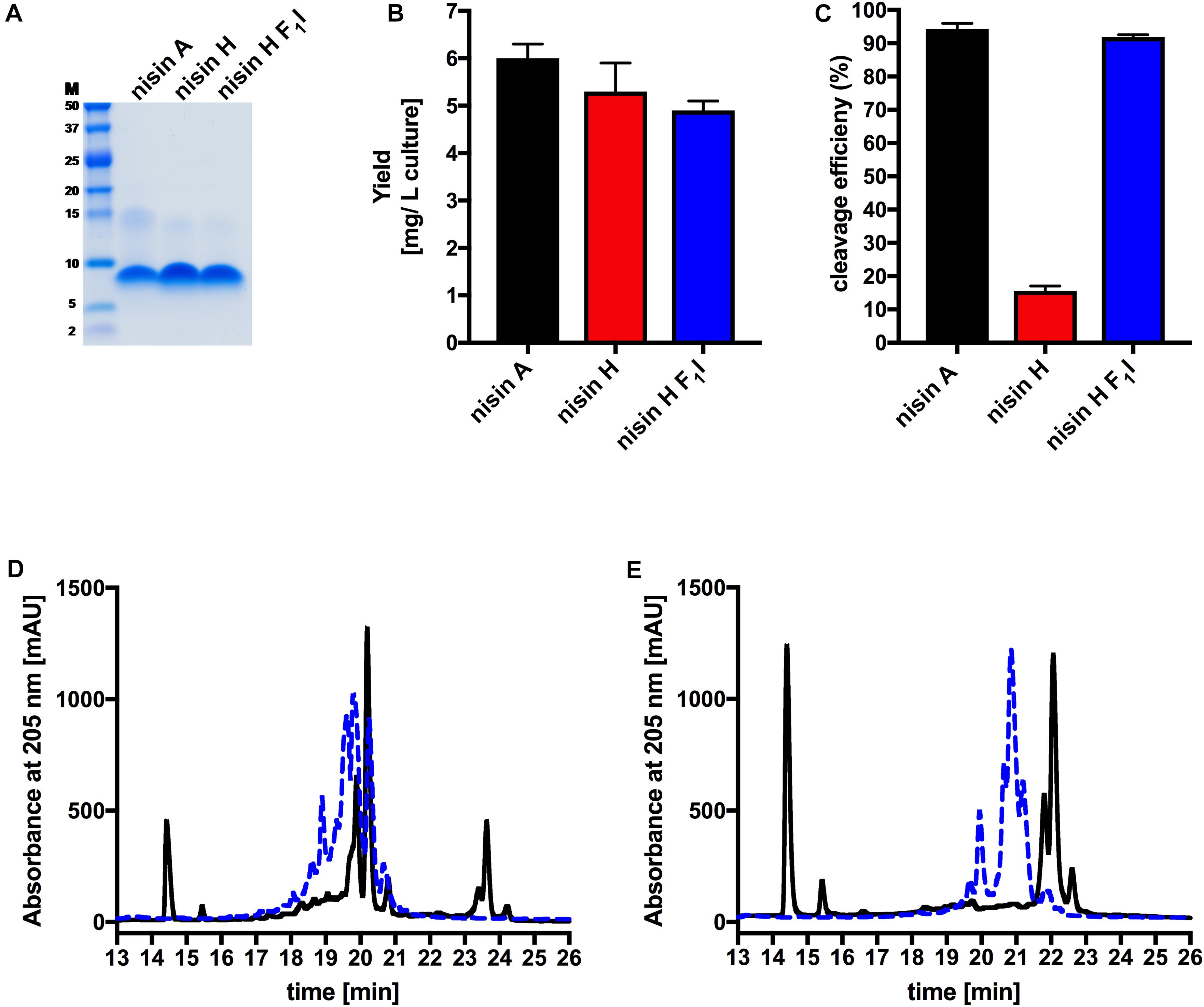
Figure 2. Determination of purity, yield and cleavage efficiency of the pre-nisin variants. (A) Purity of the purified nisin A and the variants nisin H and nisin H F1I (Marker: Precision Plus Protein Dual Xtra standards Bio-Rad). (B) Yields after purification of nisin A and their corresponding variants via cation-exchange chromatography. (C) Quantification of the cleavage efficiency of NisP. (D) Chromatogram of nisin H. (E) Chromatogram of the nisin H F1I variant. The pre-nisin variants before NisP cleavage were shown by blue dashed lines and after NisP cleavage by black lines. Error bars represent the standard deviation of at least three biological replicates.
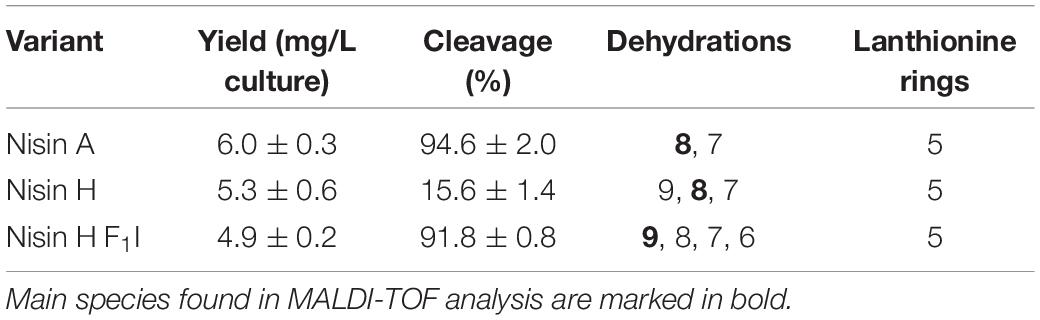
Table 1. Determination of the yield, cleavage efficiency, dehydrations, and (methyl)-lanthionine ring formation for nisin A, nisin H, and nisin H F1I.
An important step prior to the activity assay is the cleavage of the leader sequence form the pre-nisin variants, resulting in biologically active compounds. For the cleavage reaction, we used the peptidase NisP and monitored the cleavage efficiency via RP-HPLC (Figures 2C–E).
Intriguingly, the natural variant nisin H showed only a low cleavage efficiency of 15.6 ± 1.4%, compared to nisin A with 94.6 ± 2.0% (Figure 2C and Table 1). In comparison to nisin A, nisin H contains a phenylalanine at the first position (O’Connor et al., 2015), which apparently leads to a significant reduction in cleavage efficiency. The first residue of nisin A is an isoleucine, and as demonstrated before (Lagedroste et al., 2019), the introduction of aromatic residues at position one results in a clearly reduced cleavage efficiency. To counteract the lower cleavage efficiency of nisin H by NisP, we created a mutant of nisin H, in which the phenylalanine was substituted by isoleucine and termed it nisin H F1I. For this variant, nisin H F1I, the cleavage efficiency of the pre-lantibiotic was restored with an efficiency of 91.8 ± 0.8% (Figure 2C and Table 1), which corresponds to levels previously observed for nisin A. We monitored the cleavage via RP-HPLC, the pre-nisin elutes normally between 18 and 22 min (shown as blue dashed lines, Figures 2D,E). After cleavage by NisP, the leader peptide can be detected at 14.5 and 15.5 min in the chromatogram (shown as black lines, Figures 2D,E). For nisin H there was a high amount of uncleaved nisin H visible (eluting from 18 to 21 min) and only a small amount of cleaved product at 23–24 min (black lines, Figure 2D). For the nisin H F1I variant, high amounts of leader peptide and cleaved product could be detected in the chromatogram, indicating high cleavage efficiency (black lines, Figure 2E). This efficiency was similar as observed for nisin A and in line with previous results that the position one is important for the cleavage reaction (Lagedroste et al., 2019). Thus, we assume, that the four other mutations naturally occurring in nisin H (compared to nisin A) do not interfere with cleavage, however the isoleucine at position one does.
The next step was to determine the modification state of the heterologous produced nisin H and its F1I variant. As the natural variant nisin H contains ten possible residues in the core peptide that can be dehydrated, we were curious to determine if the modification machinery of nisin A was able to modify the peptide as efficiently (O’Connor et al., 2015). In the MALDI-TOF spectra, a dominant species of eightfold dehydrated residues was observed for nisin H, followed by a minor species containing nine- and sevenfold dehydrations. The possible 10-fold dehydrated species however was not observed (Figure 3A and Table 1). Furthermore no CDAP-coupling products were observed, which indicates that all cysteine residues are linked in (methyl)-lanthionine rings. We proved the functionality of the assay with unmodified pre-nisin A as demonstrated in Lagedroste et al. (2019). Thus, the modification enzymes were able to modify nisin H proving the promiscuity of the nisin modification machinery. Interestingly, the nisin H F1I variant showed a dominant ninefold dehydrated species in comparison the nisin H wild-type (Figure 3B and Table 1) and also showed no CDAP-coupling products, indicating that all cysteine residues are closed to (methyl)-lanthionine rings. The difference in the dehydration pattern for the nisin H F1I variant indicates a different accessibility of the serine and/or threonine residues in the core peptide for at least the dehydratase NisB. To validate which serine or threonine residues is dehydrated, we performed a tandem mass spectrometric analysis of nisin H and the F1I variant. Here we found that the Thr2 partially escape the dehydration in nisin H. Only in the small amount of the ninefold dehydrated species the Thr2 is dehydrated, in all other species we found a mix in the dehydration pattern, where Thr2 partially escape the dehydration. For example in the eightfold species we have a mix of dehydrated Thr2 or Ser33. In the nisin H F1I variant the Thr2 was in all species dehydrated, which suggests that, the phenylalanine at position one in nisin H is critical for the dehydratase NisB. Ser29 was never dehydrated in the found species.
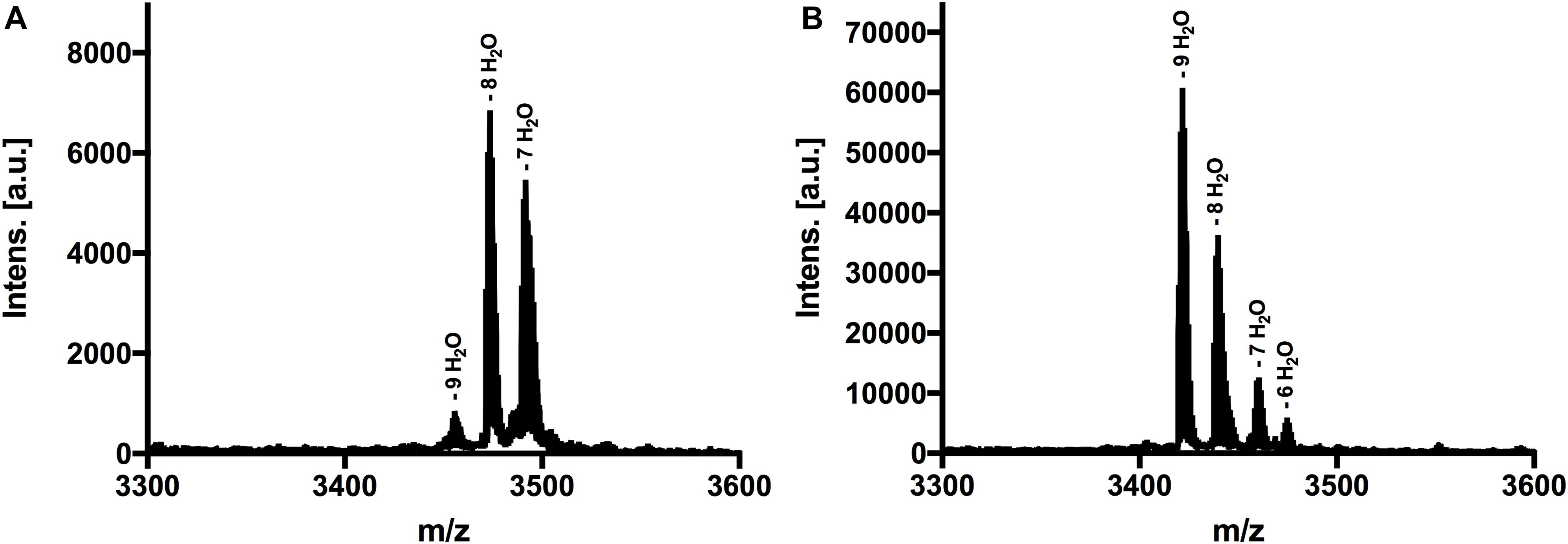
Figure 3. MALDI-TOF analysis to determine dehydrations and (methyl)-lanthionine ring formations. (A) MS spectra of nisin H, threated with CDAP. Dominant species was eightfold dehydrated. (B) MS spectra of nisin H F1I variant, threated with CDAP. Dominant species was ninefold dehydrated. Both variants showed no coupling products, indicating that all cysteine residues were involved in (methyl)-lanthionine rings.
Lantibiotics are very potent and possess an antimicrobial activity in the nanomolar range (Gross and Morell, 1967; Rollema et al., 1995; Chan et al., 1996; Lu et al., 2010; Oppedijk et al., 2016). To verify this potential for nisin H and the nisin H F1I variant we used a standardized growth inhibition assay and first screened against the nisin sensitive L. lactis strain NZ9000Cm. Here, Cm stands for chloramphenicol resistance, which arises from the empty plasmid, which was transformed. In comparison to nisin A (IC50 value: 4.8 ± 0.7 nM), the heterologous expressed variant nisin H possessed a comparable IC50 value (5.3 ± 1.0 nM) (Figure 4 and Table 2). Both values are in line with previously determined IC50 values for the strain NZ9000Cm (Reiners et al., 2017). The effect of the nisin H F1I variant was more pronounced. For the NZ9000Cm sensitive strain we calculated an IC50 value of 14.2 ± 0.2 nM, approximately threefold lower than the wild-type nisin H (Figure 4 and Table 2).
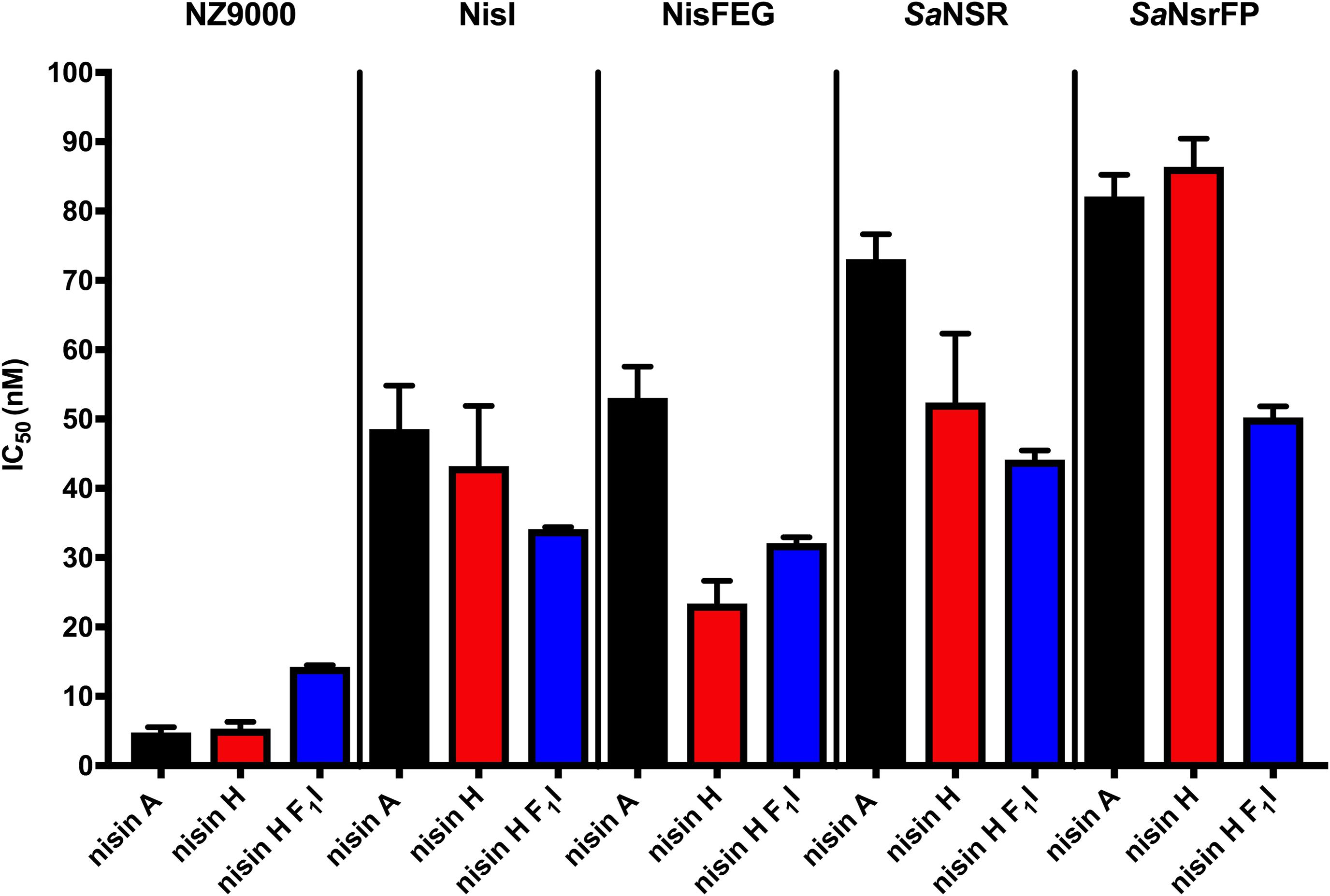
Figure 4. Growth inhibition assays in the presence of nisin A and the nisin H varants. The lantibiotic nisin A (black bars), the heterologous expressed nisin H (red bars) and the position 1 variant of nisin H F1I (blue bars) were used for growth inhibition (IC50) with the strains NZ9000Cm, NZ9000NisI, NZ9000NisFEG, NZ9000SaNSR, and NZ9000SaNsrFP. Values represent the average of at least five biological independent measurements and the errors report the standard deviation of the mean (SDM).
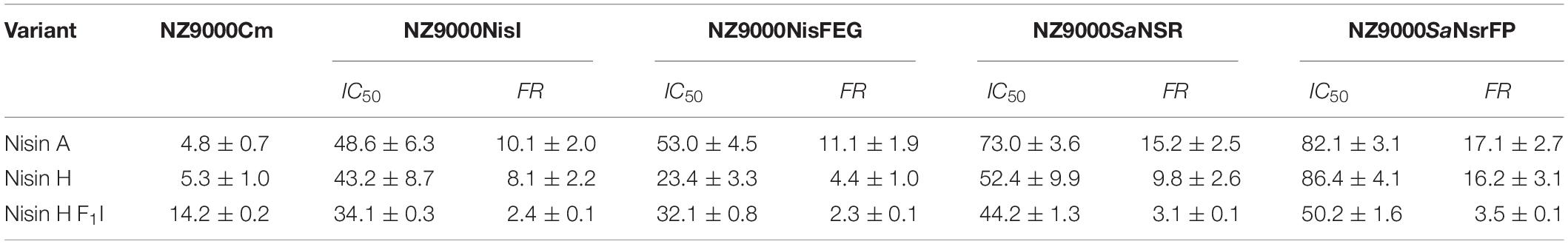
Table 2. IC50 values (nM) for nisin A, nisin H, and nisin H F1I with the corresponding fold of resistance (FR) against the strains NZ9000Cm, NZ9000NisI, NZ9000NisFEG, NZ9000SaNSR, and NZ9000SaNsrFP.
To test the effect of the nisin variants on the immunity proteins NisI (Alkhatib et al., 2014a) and NisFEG (Alkhatib et al., 2014b), as well as the resistance proteins SaNSR (Khosa et al., 2016a) and SaNsrFP (Reiners et al., 2017), we expressed each of them in a plasmid-based system in a L. lactis NZ9000 strain. We termed these strains NZ9000NisI, NZ9000NisFEG, NZ9000SaNSR and NZ9000SaNsrFP, respectively.
Nisin A displayed an IC50 value of 48.6 ± 6.3 nM against strain NZ9000NisI and 53.0 ± 4.5 nM against strain NZ9000NisFEG. For the resistance strains NZ9000SaNSR and NZ9000SaNsrFP nisin A displayed IC50 values of 73.0 ± 3.6 and 82.1 ± 3.1 nM, respectively (Figure 4 and Table 2). By comparing theses values, we calculated the fold of immunity/resistance to 10.1 ± 2.0 for NZ9000NisI, 11.1 ± 1.9 for NZ9000NisFEG, 15.2 ± 2.5 for NZ9000SaNSR and 17.1 ± 2.7 for NZ9000SaNsrFP (Table 2). After the first screen against the sensitive strain NZ9000Cm, nisin H and its variant were used to screen against the strains expressing the immunity or resistance proteins.
Nisin H revealed an IC50 value of 43.2 ± 8.7 nM against the NZ9000NisI strain, similar to nisin A, and a fold of immunity of 8.1 ± 2.2. Against the NZ9000NisFEG strain we determined an IC50 value of 23.4 ± 3.3 nM for nisin H which displayed a fold of resistance of 4.4 ± 1.0. Against the NZ9000SaNSR strain we obtained an IC50 value of 52.4 ± 9.9 nM, resulting in a fold of resistance of 9.8 ± 2.6. Nisin H showed an IC50 value of 86.4 ± 4.0 nM for the NZ9000SaNsrFP strain, resulting in a fold of resistance of 16.2 ± 3.1 (Figure 4 and Table 2). NZ9000SaNsrFP showed the highest fold of resistance for nisin H [in-line with a previous report (Reiners et al., 2017)]. Intriguingly, strain NZ9000NisFEG showed a reduced immunity and consequently we propose that nisin H is not recognized as efficiently as nisin A. Even NZ9000SaNSR had a reduced resistance. That could be due to the exchange of His31 against lysine in the C-terminal part of nisin H (Figure 1).
Surprisingly, all strains displayed a reducing resistance/immunity effect for the nisin H F1I variant in comparison to nisin A and also, with exception of NZ9000FEG for nisin H. Against the NZ9000NisI strain we determined an IC50 value of 34.1 ± 0.3 nM for the nisin H F1I variant, with a fold of resistance 2.4 ± 0.1, which is nearly threefold lower than for nisin A. We obtained an IC50 value of 32.1 ± 0.8 nM against the ABC transporter NZ9000NisFEG, with a fold of resistance 2.3 ± 0.1 (Figure 4 and Table 2). Nisin H F1I showed for the resistance strain NZ9000SaNSR and NZ9000SaNsrFP an IC50 value of 44.2 ± 1.3 and 50.2 ± 1.6 nM, respectively. The calculated folds of resistance were 3.1 ± 0.1 and 3.5 ± 0.1, respectively (Figure 4 and Table 2) and both are fivefold less than the observed fold of resistances for nisin A.
Since a similar activity for nisin H and nisin A was observed it became obvious that both exhibit the same mode of action. In the case of nisin A this combines growth inhibition with pore formation in the membrane with subsequent cell death. To test this we performed a SYTOX assay previously used for nisin A (Reiners et al., 2017). Here the SYTOX dye was added to L. lactis cells and displayed an increased fluorescence signal upon binding of DNA which is released from the cell upon pore formation (Roth et al., 1997). We use 100 nM of nisin A, nisin H and nisin H F1I variant respectively and observed an almost instant fluorescence increase similar to the signal increase observed for nisin A (Figure 5). This shows that nisin H as well as its F1I variant form pores in the membrane of L. lactis cell.
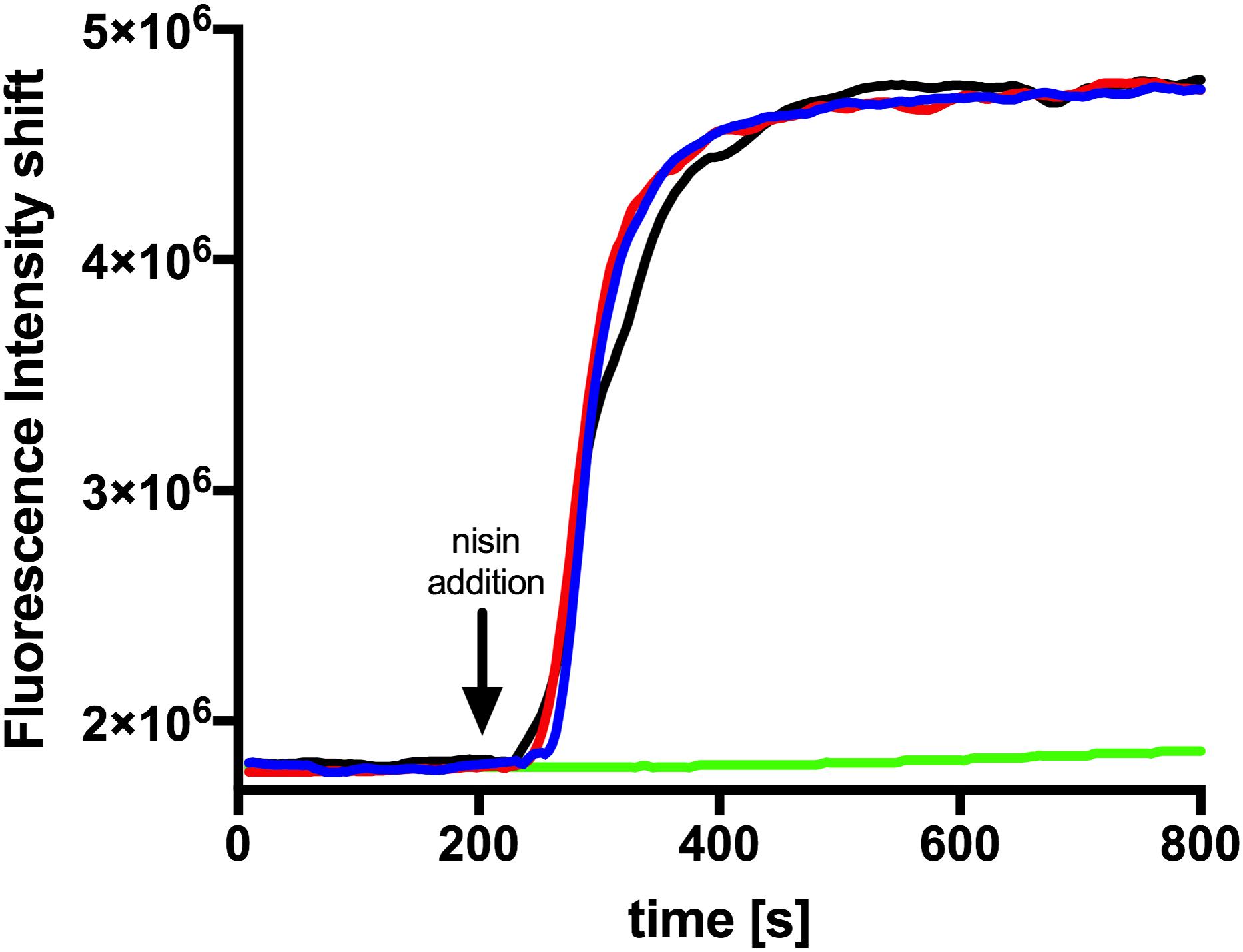
Figure 5. Nisin mediated pore formation, visualized with the SYTOX green assay. The NZ9000Cm strain incubated with the SYTOX dye. After a stable baseline (∼200 s), one of the nisin variants (100 nM) was added (indicated with an arrow). The fluorescence signal was measured using a fluorolog (Horiba III). The rapid increase in fluorescence indicates pore formation. The black line represents the addition of nisin A, the red line nisin H, and the blue line nisin H F1I. As a control we added buffer shown as green line.
The nisin variants were further tested for antibacterial capabilities against B. subtilis and different pathogenic strains from S. aureus, E. faecium, and E. faecalis using the microdilution method, according to the recommendations of CLSI (2012). Here we found that nisin H and the F1I variant performed almost identically or in most cases even better than the natural nisin A. Especially against the MSSA and MRSA strains, nisin H had significant lower MIC values of 0.19 and 0.78 μM, in comparison to 0.78 and 6.25 μM for nisin A, respectively (Table 3). Also, against E. faecium ATCC 35667, B. subtilis 168 as well as E. faecium ATCC 700221 (VRE), nisin H showed more potency with about two to eightfold lower MIC values than nisin A [0.39, 0.1, and 0.39 μM compared to 1.56, 0.78, and 0.78 μM for nisin A, respectively (Table 3)]. While the nisin H F1I variant and nisin A had similar MIC values against both MSSA and MRSA strains, the nisin H F1I variant only performed better than nisin A or nisin H against E. faecalis ATCC 51299 (VRE) with a MIC value of 0.78 μM compared to 1.56 μM for nisin A and nisin H. Against E. faecium ATCC 35667 and B. subtilis 168 nisin H F1I was less efficient than nisin H, but still better than nisin A (Table 3).
Discussion
We focused in this study on the natural variant nisin H and the nisin H F1I mutant. Nisin H was discovered from the gut-derived strain S. hyointestinalis DPC6484 in 2015 by O’Connor et al. (2015). Here, we showed the heterologous expression of nisin H and the F1I variant with the NICE-system in L. lactis (Eichenbaum et al., 1998; Mierau and Kleerebezem, 2005; Rink et al., 2005; Zhou et al., 2006; Lagedroste et al., 2019) and extended the characterization in terms of cleavage efficiency by the protease NisP and the antimicrobial activity against the immunity proteins NisFEG (Alkhatib et al., 2014b) and NisI (Alkhatib et al., 2014a), as well as the resistance proteins SaNSR (Khosa et al., 2013; Khosa et al., 2016a,b) and SaNsrFP (Khosa et al., 2016a; Reiners et al., 2017). We further tested for antibacterial capabilities against B. subtilis and different pathogenic strains from S. aureus, E. faecium, and E. faecalis.
Both lantibiotics, nisin H and the F1I variant were purified in high amounts and purity with 5.3 ± 0.6 mg/L of cell culture for nisin H and 4.9 ± 0.2 mg/L of cell culture for nisin H F1I variant, respectively (Figure 2B and Table 1). In comparison, the homologous expression of nisin H in S. hyointestinalis and nisin A in L. lactis NZ9700 results in a very low amount of 0.15 mg/L of cell culture and 0.50 mg/L of cell culture (O’Connor et al., 2015), respectively, which demonstrates the enormous potential of the NICE-system, for lantibiotic and even non-lantibiotic expression (Eichenbaum et al., 1998; Mierau and Kleerebezem, 2005; Rink et al., 2005; Zhou et al., 2006; Lagedroste et al., 2019).
An important step in the maturation of a lantibiotic is the cleavage of the leader peptide to become biological active. The cleavage efficiency of the natural substrate nisin A was determined with 94.6 ± 2.0%. For nisin H the cleaving efficiency was drastically reduced to 15.6 ± 1.4%. The first residue of nisin A is an isoleucine, while the corresponding residue in nisin H is phenylalanine and as demonstrated before (Lagedroste et al., 2019), aromatic residues prevent efficient cleavage likely by interfering with the S1′ binding pocket of NisP. With the nisin H F1I variant, the cleavage efficiency was restored to 91.8 ± 0.8% (Figures 1, 2C and Table 1). This indicated that the other point mutations naturally occurring in nisin H did not affect cleavage by NisP. With respect to other natural nisin variants, NisP cleavage could be a critical step. For example nisin O1 to O4 from B. obeum A2-162 (Hatziioanou et al., 2017) has a tyrosine or a threonine, respectively, at position one, which should also result in a low NisP cleavage efficiency. Natural variants such as nisin U (Wirawan et al., 2006), nisin J (O’Sullivan et al., 2019, 2020), nisin Q (Zendo et al., 2003), nisin Z (Mulders et al., 1991) and nisin F (de Kwaadsteniet et al., 2008) have an isoleucine and nisin U2 (Wirawan et al., 2006) and nisin P (Zhang et al., 2012; Wu et al., 2014) a valine at position one, which should result in high NisP cleavage efficiency.
Furthermore, we made a sequence alignment with Clustal Omega (Madeira et al., 2019) for the NshP (the natural protease for the nisin H cleavage) from S. hyointestinalis and NisP from L. lactis to see if there is any difference in the active site, which could be the reason for the reduced cleavage efficiency (Figure 6A and Supplementary Figure 1). Here the three important residues His306, Asp259, and Ser512 which build up the catalytic triad in NisP are conserved in NshP. We also calculated a homology model of NshP based on the known NisP structure (PDB code 4MZD) using Phyre2 (Kelley et al., 2015; Figure 6B and Supplementary Figure 2). Here, no significant differences are found within the overall fold as well as the active site which would explain the difference in the cleavage site. This is intriguing since the recognition site within the leader peptide differs between nisin A (sequence is ASPR) and nisin H (sequence is ASTR) (see Supplementary Figure 3). This suggests that the proteases NisP and NshP recognize their substrate by small difference in their active site.
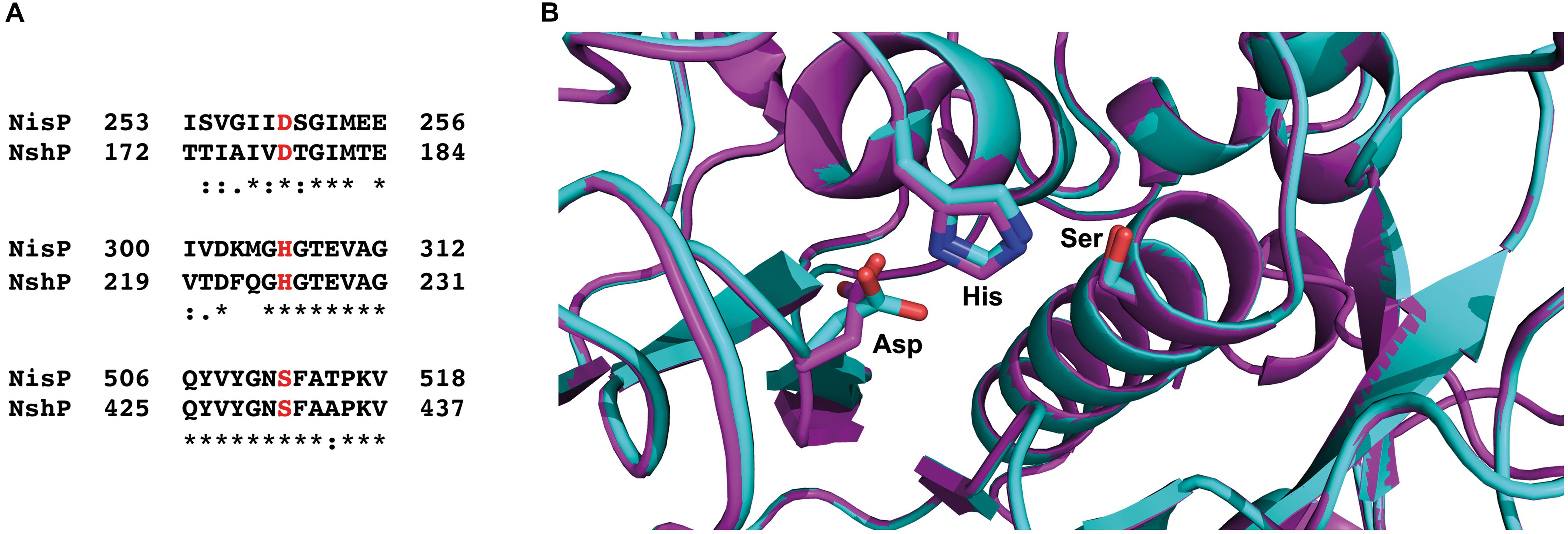
Figure 6. Sequence alignment of NisP from Lactococcus lactis and NshP from Streptococcus hyointestinalis. (A) The important residues from the active site (Asp259, His306, and Ser512 for NisP and Asp178, His225, and Ser431 for NshP) are marked in red, shown together with the close neighborhood residues. The sequence alignment was done with Clustal Omega (Madeira et al., 2019). (B) Zoom-in into the active sites of NshP (magenta) and NisP (cyan). The important residues for the catalytic activity (Asp259, His306, and Ser512 for NisP and Asp178, His225, and Ser431 for NshP) are shown in stick representation. Figure was generated using Pymol (2015). For a full alignment and a corresponding homology model, please see Supplementary Material.
Nevertheless, for an efficient cleavage, (methyl)-lanthionine rings have to be present. This even holds true in light of the presence of all (methyl)-lanthionine rings, which is generally assumed as the prerequisite for fast and efficient conversion of the pre-nisin to modified nisin (Plat et al., 2011; Lagedroste et al., 2017). To check the amount of dehydrations, necessary for (methyl)-lanthionine ring formation we applied MALDI-TOF analysis. The loss of water molecules within the peptide is directly visible in the reduced molecular weight, but not the (methyl)-lanthionine ring formation. Here we used 1-cyano-4 dimethylaminopyridinium tetrafluoroborate (CDAP) (Wu and Watson, 1998), which binds to free cysteine residues, indicating that these cysteines are not involved in a (methyl)-lanthionine ring. For both lantibiotics, nisin H and the nisin H F1I variant no CDAP coupling products were found, indicating that no (methyl)-lanthionine ring is lacking (Figure 3 and Table 1). Nisin H has 10 possible dehydration sites and is predicated to be ninefold dehydrated when expressed homologous (O’Connor et al., 2015). A minor species of nine dehydrations was found, but the dominant species was eightfold dehydrated. The dehydration pattern of the F1I variant is changed in comparison to nisin H. Here we determined a dominant ninefold species (Figure 3 and Table 1). This provides a hint, that position two of wild-type nisin H might not have been previously dehydrated, due to steric hindrance of the phenylalanine. To validate which serine or threonine residues is dehydrated, we perform a tandem mass spectrometric analysis of nisin H and the F1I variant. Here we found that the Thr2 partially escape the dehydration in nisin H. In the nisin H F1I variant the Thr2 was in all species dehydrated, which gives a hint that, the phenylalanine at position one in nisin H is critical for the dehydratase NisB. This is in line with previous data from the I1F variant of nisin A, where the dominant species was sevenfold dehydrated and not eightfold as wild-type nisin A (Lagedroste et al., 2019). It has also been reported for the natural nisin Z (Mulders et al., 1991), that the I1W mutation showed a partial inhibition of dehydration of the Thr2 (Breukink et al., 1998), which could also be the case for nisin H with the aromatic phenylalanine at position one, resulting in eight dehydrations. A dehydration of position Ser29 normally goes in line with the lack of ring E (Lubelski et al., 2009), which drastically reduces the antimicrobial activity of nisin A against the sensitive NZ9000Cm strain (Alkhatib et al., 2014b; Khosa et al., 2016a; Reiners et al., 2017). Since the activity was high for nisin H and the F1I variant, we expected that Ser29 was not dehydrated and tandem mass spectrometric analysis supported this.
Nisin H showed nearly the same activity as nisin A against the sensitive NZ9000Cm strain but the nisin H F1I variant is roughly threefold less active. For the immunity protein NisI, it was revealed that nisin H has an identical activity like nisin A within experimental error. However, the nisin H F1I variant exhibited a lower IC50 value of 34.1 ± 0.3 nM and due to the weaker wild-type activity more than a threefold lower fold of resistance (2.4 ± 0.1 compared to 8.1 ± 2.2) (Figure 4 and Table 2). NisI recognizes the N-terminus of nisin (Wiedemann et al., 2001) and the lower IC50 could be due to the fact that Thr2 is dehydrated in the nisin H F1I variant in contrast to nisin H. An additional change is the leucine at position 6 against methionine in nisin H and the nisin H F1I variant, which could be responsible for the better recognition by NisI.
The immunity protein NisFEG, in comparison to nisin A, showed a strong reduction in immunity in the presence of nisin H and the nisin H F1I variant. NisFEG recognizes the C-terminus of nisin (Alkhatib et al., 2014b), which indicates that the point mutations of nisin H affect NisFEG. So, we suppose that nisin H and the nisin H F1I variant are not recognized and subsequently transported out of the membrane like nisin A.
The resistance protein SaNSR also recognizes the C-terminus of nisin (Khosa et al., 2016a), and cleaves nisin between the positions 28 and 29. Other studies showed that mutations in this area of the nisin molecule, e.g., S29P or C28P strongly reduce the efficiency of SaNSR (Field et al., 2019; Zaschke-Kriesche et al., 2019a). We assume that the exchange of His31 against lysine in nisin H and the nisin H F1I variant (Figure 1) has the same effect on SaNSR thereby lowering the resistance efficiency to an IC50 value of 52.4 ± 2.6 nM for nisin H and 44.2 ± 1.3 nM for the nisin H F1I variant, respectively (Figure 4 and Table 2).
For the resistance protein SaNsrFP, we observed an activity for nisin H identical to nisin A. SaNsrFP recognizes the N-terminus of nisin (Reiners et al., 2017), which is affected due to the different dehydration pattern in wild-type nisin H in comparison to the nisin H F1I variant. The nisin H F1I variant showed a lower IC50 value of 50.2 ± 1.6 nM, compared to 86.4 ± 4.1 nM for nisin H. This effect is even more pronounced when comparing the fold of resistances of 16.2 ± 3.1 for nisin H to 3.5 ± 0.1 for the nisin H F1I variant, respectively (Figure 4 and Table 2).
This study demonstrated again that only a complete characterization of a lantibiotic reveals the full antimicrobial potential. Based on the IC50 value of the sensitive NZ9000Cm strain the F1I variant might be classified as weakly antimicrobial active, but with respect to the immunity and resistance proteins, it becomes more interesting, due to its high activity even against the immunity proteins NisI and NisFEG from L. lactis and the nisin resistance proteins SaNSR and SaNsrFP from S. agalactiae COH1. Against the tested pathogenic bacteria, we found that nisin H and the nisin H F1I variant performed almost identically or in the most cases even better than the natural nisin A. Nisin H displayed high antimicrobial potential against both methicillin-resistant and –susceptible the S. aureus strains, both vancomycin-resistant and -susceptible, E. faecium strains, as well as B. subtilis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LS and SS conceived and directed this study. JR and ML conducted the expression, purification, MS-analysis, and the growth inhibition experiments. JG performed the SYTOX experiments. GP and KS performed the tandem mass spectrometric analysis. EA and RK performed the MIC experiments. JR, ML, SS, and LS wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemein- schaft (DFG, grant Schm1279/13-1 to LS). The Center for Structural studies is funded by the DFG (Grant number 417919780 to SS). Project number 270650915 (Research Training Group GRK2158, TP2a to RK and TP4a to SS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Institute of Biochemistry for fruitful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.573614/full#supplementary-material
References
Abts, A., Montalban-Lopez, M., Kuipers, O. P., Smits, S. H., and Schmitt, L. (2013). NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52, 5387–5395. doi: 10.1021/bi4008116
Alkhatib, Z., Lagedroste, M., Fey, I., Kleinschrodt, D., Abts, A., and Smits, S. H. (2014a). Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS One 9:e102246. doi: 10.1371/journal.pone.0102246
Alkhatib, Z., Lagedroste, M., Zaschke, J., Wagner, M., Abts, A., Fey, I., et al. (2014b). The C-terminus of nisin is important for the ABC transporter NisFEG to confer immunity in Lactococcus lactis. Microbiologyopen 3, 752–763. doi: 10.1002/mbo3.205
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160.
Breukink, E., Van Kraaij, C., Van Dalen, A., Demel, R. A., Siezen, R. J., De Kruijff, B., et al. (1998). The orientation of nisin in membranes. Biochemistry 37, 8153–8162. doi: 10.1021/bi972797l
Brunati, C., Thomsen, T. T., Gaspari, E., Maffioli, S., Sosio, M., Jabes, D., et al. (2018). Expanding the potential of NAI-107 for treating serious ESKAPE pathogens: synergistic combinations against Gram-negatives and bactericidal activity against non-dividing cells. J. Antimicrob. Chemother. 73, 414–424. doi: 10.1093/jac/dkx395
Chan, W. C., Leyland, M., Clark, J., Dodd, H. M., Lian, L. Y., Gasson, M. J., et al. (1996). Structure-activity relationships in the peptide antibiotic nisin: antibacterial activity of fragments of nisin. FEBS Lett. 390, 129–132. doi: 10.1016/0014-5793(96)00638-2
Clinical and Laboratory Standards Institute (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th Edn. Wayne PA: Clinical and Laboratory Standards Institute.
Crowther, G. S., Baines, S. D., Todhunter, S. L., Freeman, J., Chilton, C. H., and Wilcox, M. H. (2013). Evaluation of NVB302 versus vancomycin activity in an in vitro human gut model of Clostridium difficile infection. J. Antimicrob. Chemother. 68, 168–176. doi: 10.1093/jac/dks359
Dawson, M. J., and Scott, R. W. (2012). New horizons for host defense peptides and lantibiotics. Curr. Opin. Pharmacol. 12, 545–550. doi: 10.1016/j.coph.2012.06.006
de Kwaadsteniet, M., Ten Doeschate, K., and Dicks, L. M. (2008). Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 74, 547–549. doi: 10.1128/aem.01862-07
Delves-Broughton, J., Blackburn, P., Evans, R. J., and Hugenholtz, J. (1996). Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69, 193–202. doi: 10.1007/bf00399424
Dischinger, J., Basi Chipalu, S., and Bierbaum, G. (2014). Lantibiotics: promising candidates for future applications in health care. Int. J. Med. Microbiol. 304, 51–62. doi: 10.1016/j.ijmm.2013.09.003
Eichenbaum, Z., Federle, M. J., Marra, D., De Vos, W. M., Kuipers, O. P., Kleerebezem, M., et al. (1998). Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64, 2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998
Field, D., Blake, T., Mathur, H., Pm, O. C., Cotter, P. D., Paul Ross, R., et al. (2019). Bioengineering nisin to overcome the nisin resistance protein. Mol. Microbiol. 111, 717–731.
Gross, E., and Morell, J. L. (1967). The presence of dehydroalanine in the antibiotic nisin and its relationship to activity. J. Am. Chem. Soc. 89, 2791–2792. doi: 10.1021/ja00987a084
Hasper, H. E., De Kruijff, B., and Breukink, E. (2004). Assembly and stability of nisin-lipid II pores. Biochemistry 43, 11567–11575. doi: 10.1021/bi049476b
Hatziioanou, D., Gherghisan-Filip, C., Saalbach, G., Horn, N., Wegmann, U., Duncan, S. H., et al. (2017). Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology 163, 1292–1305. doi: 10.1099/mic.0.000515
Holo, H., and Nes, I. F. (1989). High-Frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989
Hsu, S. T. D., Breukink, E., Tischenko, E., Lutters, M. A. G., De Kruijff, B., Kaptein, R., et al. (2004). The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967. doi: 10.1038/nsmb830
Jabes, D., Brunati, C., Candiani, G., Riva, S., Romano, G., and Donadio, S. (2011). Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob. Agents Chemother. 55, 1671–1676. doi: 10.1128/aac.01288-10
Jensen, P. R., and Hammer, K. (1993). Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59, 4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993
Kaletta, C., and Entian, K. D. (1989). Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J. Bacteriol. 171, 1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989
Karakas Sen, A., Narbad, A., Horn, N., Dodd, H. M., Parr, A. J., Colquhoun, I., et al. (1999). Post-translational modification of nisin. The involvement of NisB in the dehydration process. Eur. J. Biochem. 261, 524–532. doi: 10.1046/j.1432-1327.1999.00303.x
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Khosa, S., Alkhatib, Z., and Smits, S. H. (2013). NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 394, 1543–1549. doi: 10.1515/hsz-2013-0167
Khosa, S., Frieg, B., Mulnaes, D., Kleinschrodt, D., Hoeppner, A., Gohlke, H., et al. (2016a). Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6:18679.
Khosa, S., Lagedroste, M., and Smits, S. H. (2016b). Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NSR. Front. Microbiol. 7:504. doi: 10.3389/fmicb.2016.00504
Klaenhammer, T. R. (1993). Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12, 39–85. doi: 10.1016/0168-6445(93)90057-g
Koponen, O., Tolonen, M., Qiao, M., Wahlstrom, G., Helin, J., and Saris, P. E. J. (2002). NisB is required for the dehydration and NisC for the lanthionine formation in the post-translational modification of nisin. Microbiology 148, 3561–3568. doi: 10.1099/00221287-148-11-3561
Kuipers, O. P., De Ruyter, P. G., Kleerebezem, M., and De Vos, W. M. (1997). Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15, 135–140. doi: 10.1016/s0167-7799(97)01029-9
Lagedroste, M., Reiners, J., Smits, S. H. J., and Schmitt, L. (2019). Systematic characterization of position one variants within the lantibiotic nisin. Sci. Rep. 9:935.
Lagedroste, M., Smits, S. H. J., and Schmitt, L. (2017). Substrate specificity of the secreted nisin leader peptidase NisP. Biochemistry 56, 4005–4014. doi: 10.1021/acs.biochem.7b00524
Li, B., and van der Donk, W. A. (2007). Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin. J. Biol. Chem. 282, 21169–21175. doi: 10.1074/jbc.m701802200
Li, B., Yu, J. P., Brunzelle, J. S., Moll, G. N., Van Der Donk, W. A., and Nair, S. K. (2006). Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311, 1464–1467. doi: 10.1126/science.1121422
Lu, Y., Jiang, L., Chen, M., Huan, L., and Zhong, J. (2010). [Improving heat and pH stability of nisin by site-directed mutagenesis]. Wei Sheng Wu Xue Bao 50, 1481–1487.
Lubelski, J., Khusainov, R., and Kuipers, O. P. (2009). Directionality and coordination of dehydration and ring formation during biosynthesis of the lantibiotic nisin. J. Biol. Chem. 284, 25962–25972. doi: 10.1074/jbc.m109.026690
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641.
Medeiros-Silva, J., Jekhmane, S., Paioni, A. L., Gawarecka, K., Baldus, M., Swiezewska, E., et al. (2018). High-resolution NMR studies of antibiotics in cellular membranes. Nat. Commun. 9:3963.
Mierau, I., and Kleerebezem, M. (2005). 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717. doi: 10.1007/s00253-005-0107-6
Mota-Meira, M., Lapointe, G., Lacroix, C., and Lavoie, M. C. (2000). MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 44, 24–29. doi: 10.1128/aac.44.1.24-29.2000
Mulders, J. W., Boerrigter, I. J., Rollema, H. S., Siezen, R. J., and De Vos, W. M. (1991). Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201, 581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x
O’Sullivan, J. N., O’Connor, P. M., Rea, M. C., O’Sullivan, O., Walsh, C. J., Healy, B., et al. (2020). Nisin J, a novel natural nisin variant, is produced by Staphylococcus capitis Sourced from the human skin microbiota. J. Bacteriol 202. doi: 10.1128/JB.00639-19
O’Connor, P. M., O’Shea, E. F., Guinane, C. M., O’Sullivan, O., Cotter, P. D., Ross, R. P., et al. (2015). Nisin H is a new nisin variant produced by the gut-derived strain Streptococcus hyointestinalis DPC6484. Appl. Environ. Microbiol. 81, 3953–3960. doi: 10.1128/aem.00212-15
Okeley, N. M., Paul, M., Stasser, J. P., Blackburn, N., and Van Der Donk, W. A. (2003). SpaC and NisC, the cyclases involved in subtilin and nisin biosynthesis, are zinc proteins. Biochemistry 42, 13613–13624. doi: 10.1021/bi0354942
Ongey, E. L., Yassi, H., Pflugmacher, S., and Neubauer, P. (2017). Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol. Lett. 39, 473–482. doi: 10.1007/s10529-016-2279-9
Oppedijk, S. F., Martin, N. I., and Breukink, E. (2016). Hit ‘em where it hurts: the growing and structurally diverse family of peptides that target lipid-II. Biochim. Biophys. Acta 1858, 947–957. doi: 10.1016/j.bbamem.2015.10.024
Ortega, M. A., Hao, Y., Zhang, Q., Walker, M. C., Van Der Donk, W. A., and Nair, S. K. (2015). Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512. doi: 10.1038/nature13888
O’Sullivan, J. N., Rea, M. C., O’Connor, P. M., Hill, C., and Ross, R. P. (2019). Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 95:fiy241.
Plat, A., Kluskens, L. D., Kuipers, A., Rink, R., and Moll, G. N. (2011). Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 77, 604–611. doi: 10.1128/aem.01503-10
Reiners, J., Lagedroste, M., Ehlen, K., Leusch, S., Zaschke-Kriesche, J., and Smits, S. H. J. (2017). The N-terminal region of nisin is important for the BceAB-Type ABC Transporter NsrFP from Streptococcus agalactiae COH1. Front. Microbiol. 8:1643. doi: 10.3389/fmicb.2017.01643
Repka, L. M., Chekan, J. R., Nair, S. K., and Van Der Donk, W. A. (2017). Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 117, 5457–5520. doi: 10.1021/acs.chemrev.6b00591
Rink, R., Kuipers, A., De Boef, E., Leenhouts, K. J., Driessen, A. J., Moll, G. N., et al. (2005). Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44, 8873–8882. doi: 10.1021/bi050081h
Rogers, L. A. (1928). The inhibiting effect of streptococcus lactis on Lactobacillus Bulgaricus. J. Bacteriol. 16, 321–325. doi: 10.1128/jb.16.5.321-325.1928
Rogers, L. A., and Whittier, E. O. (1928). Limiting factors in the lactic fermentation. J. Bacteriol. 16, 211–229. doi: 10.1128/jb.16.4.211-229.1928
Rollema, H. S., Kuipers, O. P., Both, P., De Vos, W. M., and Siezen, R. J. (1995). Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 61, 2873–2878. doi: 10.1128/aem.61.8.2873-2878.1995
Roth, B. L., Poot, M., Yue, S. T., and Millard, P. J. (1997). Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63, 2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997
Sahl, H. G., and Bierbaum, G. (1998). Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52, 41–79. doi: 10.1146/annurev.micro.52.1.41
Sandiford, S. K. (2019). Current developments in lantibiotic discovery for treating Clostridium difficile infection. Expert Opin. Drug. Discov. 14, 71–79. doi: 10.1080/17460441.2019.1549032
Terzaghi, B. E., and Sandine, W. E. (1975). Improved medium for lactic Streptococci and Their Bacteriophages. Appl. Microbiol. 29, 807–813. doi: 10.1128/aem.29.6.807-813.1975
van Heel, A. J., De Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281.
van Heusden, H. E., De Kruijff, B., and Breukink, E. (2002). Lipid II induces a transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 41, 12171–12178. doi: 10.1021/bi026090x
Wiedemann, I., Benz, R., and Sahl, H. G. (2004). Lipid II-mediated pore formation by the peptide antibiotic nisin: a black lipid membrane study. J. Bacteriol. 186, 3259–3261. doi: 10.1128/jb.186.10.3259-3261.2004
Wiedemann, I., Breukink, E., Van Kraaij, C., Kuipers, O. P., Bierbaum, G., De Kruijff, B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. doi: 10.1074/jbc.m006770200
Wirawan, R. E., Klesse, N. A., Jack, R. W., and Tagg, J. R. (2006). Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72, 1148–1156. doi: 10.1128/aem.72.2.1148-1156.2006
Wu, J., and Watson, J. T. (1998). Optimization of the cleavage reaction for cyanylated cysteinyl proteins for efficient and simplified mass mapping. Anal. Biochem. 258, 268–276. doi: 10.1006/abio.1998.2596
Wu, Z., Wang, W., Tang, M., Shao, J., Dai, C., Zhang, W., et al. (2014). Comparative genomic analysis shows that Streptococcus suis meningitis isolate SC070731 contains a unique 105K genomic island. Gene 535, 156–164. doi: 10.1016/j.gene.2013.11.044
Zaschke-Kriesche, J., Behrmann, L. V., Reiners, J., Lagedroste, M., Groner, Y., Kalscheuer, R., et al. (2019a). Bypassing lantibiotic resistance by an effective nisin derivative. Bioorg. Med. Chem. 27, 3454–3462. doi: 10.1016/j.bmc.2019.06.031
Zaschke-Kriesche, J., Reiners, J., Lagedroste, M., and Smits, S. H. J. (2019b). Influence of nisin hinge-region variants on lantibiotic immunity and resistance proteins. Bioorg. Med. Chem. 27, 3947–3953. doi: 10.1016/j.bmc.2019.07.014
Zendo, T., Fukao, M., Ueda, K., Higuchi, T., Nakayama, J., and Sonomoto, K. (2003). Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 67, 1616–1619. doi: 10.1271/bbb.67.1616
Zhang, Q., Yu, Y., Velasquez, J. E., and Van Der Donk, W. A. (2012). Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 18361–18366. doi: 10.1073/pnas.1210393109
Zhou, L., Van Heel, A. J., and Kuipers, O. P. (2015). The length of a lantibiotic hinge region has profound influence on antimicrobial activity and host specificity. Front. Microbiol 6:11. doi: 10.3389/fmicb.2015.00011
Keywords: lantibiotics, nisin, nisin H, MS analysis, antimicrobial activity
Citation: Reiners J, Lagedroste M, Gottstein J, Adeniyi ET, Kalscheuer R, Poschmann G, Stühler K, Smits SHJ and Schmitt L (2020) Insights in the Antimicrobial Potential of the Natural Nisin Variant Nisin H. Front. Microbiol. 11:573614. doi: 10.3389/fmicb.2020.573614
Received: 17 June 2020; Accepted: 25 September 2020;
Published: 20 October 2020.
Edited by:
Des Field, University College Cork, IrelandReviewed by:
Oscar P. Kuipers, University of Groningen, NetherlandsStefano Donadio, Naicons Srl, Italy
Copyright © 2020 Reiners, Lagedroste, Gottstein, Adeniyi, Kalscheuer, Poschmann, Stühler, Smits and Schmitt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lutz Schmitt, bHV0ei5zY2htaXR0QGhodS5kZQ==; Sander H. J. Smits, c2FuZGVyLnNtaXRzQGhodS5kZQ==
†These authors have contributed equally to this work
 Jens Reiners
Jens Reiners Marcel Lagedroste
Marcel Lagedroste Julia Gottstein1
Julia Gottstein1 Sander H. J. Smits
Sander H. J. Smits Lutz Schmitt
Lutz Schmitt