- Department of Biological Science, The University of Tulsa, Tulsa, OK, United States
Food poisoning due to the consumption of Staphylococcus aureus contaminated food is a major health problem worldwide. In this study, we sequenced the genomes of ten plasmid-bearing S. aureus strains isolated from retail beef, chicken, turkey, and pork. The chromosomes of the strains varied in size from 2,654,842 to 2,807,514 bp, and a total of 25 plasmids were identified ranging from 1.4 to 118 kb. Comparative genomic analysis revealed similarities between strains isolated from the same retail meat source, indicating an origin-specific genomic composition. Genes known to modulate attachment, invasion, and toxin production were identified in the 10 genomes. Strains from retail chicken resembled human clinical isolates with respect to virulence factors and genomic islands, and retail turkey and pork isolates shared similarity with S. aureus from livestock. Most chromosomes contained antimicrobial resistance, heavy metal resistance, and stress response genes, and several plasmids contained genes involved in antimicrobial resistance and virulence. In conclusion, the genomes of S. aureus strains isolated from retail meats showed an origin-specific composition and contained virulence and antimicrobial resistance genes similar to those present in human clinical isolates.
Introduction
The bacterium Staphylococcus aureus can incite life-threatening infections in both humans and animals (Tong et al., 2015). Although nosocomial infections are common, the acquisition of methicillin-resistant S. aureus (MRSA) from communities and livestock is also responsible for clinical cases and the spread of antimicrobial resistant (AMR) strains (Miller, 2010; Mediavilla et al., 2012). The virulence factors, exoenzymes, and toxins produced by S. aureus genomes are responsible for pathogenesis (Tam and Torres, 2019). Staphylococcal food poisoning (SFP) related to consumption of contaminated food is a major problem worldwide, and staphylococcal enterotoxins are responsible for clinical symptoms (Hennekinne et al., 2012; Lin et al., 2016). In the United States, S. aureus remains responsible for about 241,148 annual cases of domestically acquired foodborne illness (Hedberg, 2011), which is typified by nausea and vomiting.
Both humans and animals play an important role in food product contamination during preparation and storage (Lozano et al., 2016), and food handlers are regarded as a major factor in contamination (Castro et al., 2016). A high prevalence of S. aureus contamination in retail meat and dairy products has been reported (Kitai et al., 2005; Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015b; Ge et al., 2017), and the most common clonal complexes were CC5 (chicken) and CC398 (turkey) (Waters et al., 2011; Thapaliya et al., 2017). A high incidence of toxin genes in food isolates indicates the potential virulence of these S. aureus strains (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a; Song et al., 2015; Ge et al., 2017; Gaerste-Díaz et al., 2018). Previous studies from our laboratory showed up to 37% staphylococcal contamination in retail chicken liver and gizzards (Abdalrahman and Fakhr, 2015) and 57% in poultry products (Abdalrahman et al., 2015a). Approximately 80% of retail beef liver samples, 50% of beef cuts, and 43% of pork samples were contaminated with S. aureus (Abdalrahman et al., 2015b). A high prevalence of staphylococcal toxin genes (e.g., seg, sei, lukE-lukD, hla, and hld) was reported in these strains (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). Furthermore, spa typing of selected strains from retail chicken, turkey, chicken liver, and gizzards indicated a human origin (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a).
Multidrug resistant S. aureus strains, including MRSA, are prevalent in food products (Abdalrahman et al., 2015a; Ge et al., 2017; Sergelidis and Angelidis, 2017), especially in retail turkey meats (Abdalrahman et al., 2015a; Thapaliya et al., 2017). Livestock-associated MRSA can also cause clinical infections in humans (Larsen et al., 2017), and S. aureus strains in food products are often resistant to one or more antibiotics (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b; Jamali et al., 2015; Al-Ashmawy et al., 2016; Ge et al., 2017; Wu et al., 2018). S. aureus strains are generally screened for MRSA by PCR using mecA and mecC, and the recently characterized mecB may also play a role in methicillin resistance (Becker et al., 2018; Carretto et al., 2018). Recently, vancomycin-resistant S. aureus (VRSA) has become prevalent in retail meat products (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). VRSA, VISA (intermediate resistance to vancomycin), and phenotypic MRSA strains (resistant to oxacillin and cefoxitin) have also been reported in retail meat and food products (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). MRSA that lack mecA have been recovered from clinical specimens (Elhassan et al., 2015). In S. aureus, plasmids play important roles in the transfer and acquisition of antibiotic resistance genes (Haaber et al., 2017; Bukowski et al., 2019), which are often encoded by transposons, integrative conjugative elements (ICEs), staphylococcal chromosome cassettes (SCCs), and SaPI (Haaber et al., 2017). Furthermore, phage-related sequences in S. aureus may also encode genes for antimicrobial resistance (Baba et al., 2002, 2008; Xia and Wolz, 2014).
Whole genome sequencing (WGS) of S. aureus has focused primarily on clinical isolates (Baba et al., 2008; Shajari et al., 2017; Dweba et al., 2018), including MRSA (Dweba et al., 2018; Turner et al., 2019), and VRSA (Shajari et al., 2017). WGS has facilitated comparative analyses of S. aureus virulence factors, AMR, and pathogenicity islands (Baba et al., 2002, 2008; Nguyen et al., 2015; Bosi et al., 2016; Planet et al., 2017; You et al., 2018). A comparative study of 64 S. aureus clinical isolates revealed a conserved core genome of 1441 genes (Bosi et al., 2016). Furthermore, S. aureus pathogenicity islands (νSaα, νSaβ, and SaPI) encode exotoxin, lipoprotein, serine protease, and enterotoxin genes (Baba et al., 2002, 2008; Tam and Torres, 2019). Differences in the genomic composition of S. aureus pathogenicity islands and the type VII secretion system were also reported (Baba et al., 2002, 2008; Warne et al., 2016) and may contribute to differential pathogenicity.
Many studies describe the isolation and characterization of S. aureus strains from retail food products. However, few studies have used WGS to characterize strains from food products (Mossong et al., 2015; Ge et al., 2017; Sivaraman et al., 2017), and most of these focused on MRSA or strains related to food poisoning outbreaks (Mossong et al., 2015; Ge et al., 2017; Sivaraman et al., 2017). To our knowledge, WGS comparisons are lacking for S. aureus isolated from retail meats. We previously isolated multiple S. aureus strains from retail meat products and screened them for virulence and antimicrobial susceptibility (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). The aims of this study were to sequence the whole genome of a selected set of plasmid-bearing S. aureus strains previously isolated from various retail meats, and to perform comparative genomics analysis to explore their potential virulence, AMR, and origin.
Materials and Methods
Bacterial Cultures and DNA Isolation
The 10 S. aureus strains used in this study were previously isolated from retail beef (n = 3), chicken (n = 4), turkey (n = 2), and pork (n = 1); all strains contained plasmid DNA based on alkaline lysis and PFGE (Table 1; Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). S. aureus strains were selected to represent various retail meat sources (beef, beef liver, chicken, chicken liver, chicken gizzard, pork, and turkey), different plasmid sizes previously determined by PFGE, and various plasmid rep types (Neyaz, 2019; Neyaz et al., 2020). DNA isolation and sequencing were carried out as described previously with minor modifications (Marasini and Fakhr, 2016a,b,c, 2017a,b; Neyaz et al., 2019). Briefly, S. aureus cells were grown in tryptic soy agar (TSA) at 37°C for 16–24 h and then used for genomic and plasmid DNA isolation. The DNeasy Blood and Tissue kit (QIAGEN Inc., Valencia, CA, United States) was used for genomic DNA isolation. The Qubit dsDNA HS Assay kit (Life Technologies, Carlsbad, CA, United States) was used for DNA quantification with a Qubit 2.0 fluorimeter.
Library Preparation, Sequencing and Assembly
Sequencing libraries of genomic DNA were prepared and normalized with the Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA, United States) as recommended by the manufacturer. Sequencing of prepared libraries was conducted in the Illumina MiSeq platform using the Illumina MiSeq V2 Reagent kit and 2 × 250 cycles. Sequence assembly was conducted using the CLC Genomic Workbench v. 7.0 and the microbial genome finishing module. Identification, clustering and segregation of plasmid sequences from chromosomal sequences was performed with plasmidSPAdes (Antipov et al., 2016) and the PHASTER web server (Arndt et al., 2016). Several contigs were joined manually by consulting reference sequences. Assembled genomic and plasmid sequences were deposited in GenBank as listed in Table 1. Sequences were annotated using the NCBI Prokaryotic Genome Annotation pipeline. RAST1 (Overbeek et al., 2014) and PATRIC v. 3.5.392 (Wattam et al., 2014) tools were used to annotate whole genomic sequences of S. aureus for comparative genomic analysis.
Multilocus Sequence and spa Typing
Assembled FASTA files of all sequenced S. aureus strains were analyzed. Multilocus sequence typing (MLST) and single locus typing (spa typing) of genomic sequences were conducted using MLST 2.03 (Larsen et al., 2012) and spaTyper 1.04 (Bartels et al., 2014), respectively.
Comparative Genomic Analysis
Genomic comparisons among S. aureus strains were obtained by BLASTn analysis5. Nucleotide difference (ND) trees were created using NDtree-1.2 (Joensen et al., 2014; Kaas et al., 2014; Leekitcharoenphon et al., 2014) and FASTA files of the 10 S. aureus strains (this study) and the following reference strains from GenBank: S. aureus MRSA252 (accession no. BX571856.1), Mu50 (NC_002758.2), JH1 (CP000736.1), MW2 (NC_003923.1), COL (NC_002951.2), and N315 (NC_002745.2). GenBank files (.gbk files, RAST-annotated) of chromosomal and plasmid sequences were used for pangenome analysis in the GView Server6. Fifteen genomic sequences, including five from GenBank, were used for pangenome analysis. Similarly, pangenome analysis was performed for small plasmids (<5 kb), whereas the analysis on larger plasmids (>5 kb) was performed using RAST-annotated sequences in the GView Server. Values for percent identity cutoff (90), alignment length cutoff (100), and e-values (<1e–10) were used for BLASTn analysis in the GView Server. Genomic features of sequences were illustrated using PATRIC (3.5.39), the GView Server (display genomic feature tool), and CLC genomic workbench v. 12 (QIAGEN Bioinformatics). Genome tree report (based on genomic blast) of S. aureus strains (10453 genomes) from NCBI7 was obtained (retrieved on August 07, 2020) and phylogenetic trees from genome tree report were further analyzed in NCBI Genome workbench (version 3.4.1)8.
Identification of Virulence Factors and AMR Genes
Assembled FASTA files of whole genomic sequences derived from the CLC workbench were used for virulence gene prediction using VFanalyzer9 (Liu et al., 2019), and the results were tallied with RAST- and PATRIC-annotated genomic features. Curated S. aureus virulence gene sequences from VFDB10 (Liu et al., 2019) were manually blasted against whole genome sequences of all 10 S. aureus strains. RAST- and PATRIC-annotated virulence genes that were identified in the virulence, defense and diseases subsystems and stress response genes (stress response subsystem) were listed and compared.
Raw FASTA files or assembled contigs of whole genome sequences were used in RGI11, ResFinder 3.112 (Zankari et al., 2012), and CARD analysis (PATRIC annotation pipeline) for the prediction of AMR genes. Island viewer 413 (Bertelli et al., 2017) was used for the identification and visualization of genomic islands. Genomic arrangements in νSaα, νSaβ, and SaPI genomic islands and the Type VII secretion system were illustrated using genomic features from Seed Viewer (RAST)14. S. aureus NCTC8325 was used as a reference strain for studying the Type VII secretion system.
Phylogenetic Analysis Among Virulence Genes
Protein sequences for clfA, fnbA, and spa genes were downloaded from the RAST server. Protein sequence alignments were conducted with MUSCLE (Edgar, 2004)15 and analyses for maximum likelihood were conducted in Mega X (Kumar et al., 2018) with a bootstrap value of 100.
Results
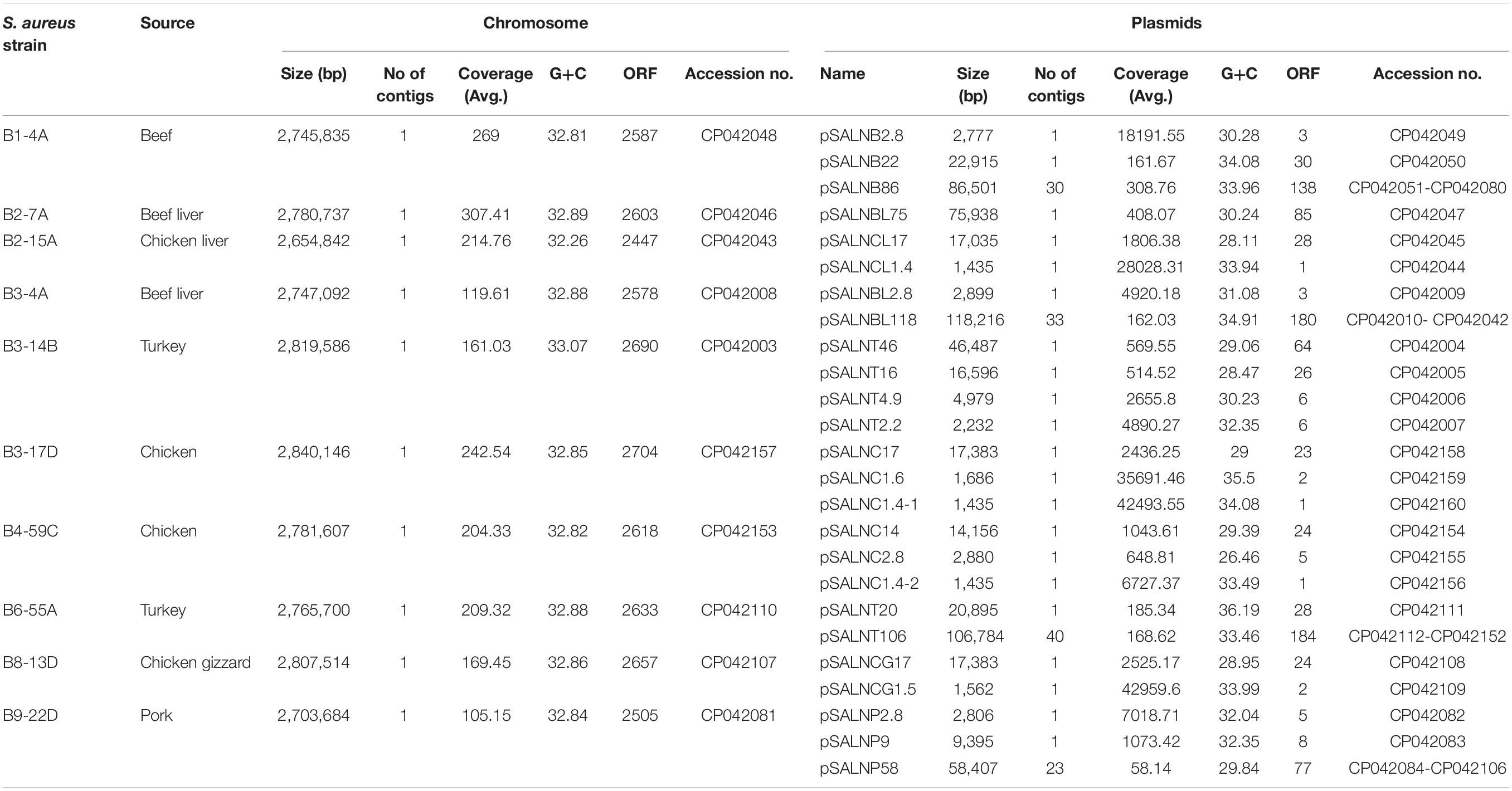
Features of Genomic and Plasmid DNA Sequences
Details for the ten sequenced S. aureus strains including size, contig number, and G+C ratio are shown in Table 1. Chromosomes ranged from 2,654,842 to 2,807,514 bp, and the average G+C ratio was 32.26–33.07. Twenty-five plasmids were detected and sequenced from the 10 S. aureus strains, and all harbored one or more plasmids (Table 1). Plasmids ranged from 1.4 to 118 kb with a mean G+C ratio of 26.46–36.19. The mean ORF length ranged from 2447 to 2704 bp. Between 1 and 6 ORFs were identified in small plasmids (<5 kb), and 6–184 ORFs were present in larger plasmids (>5 kb).
Comparative Genomics
Sequenced genomes from the same retail meat product showed greater similarity (%) to each other than sequences from different meats (Figure 1A). Sequence similarity values obtained from BLASTn analysis ranged from 97.93 to 99.99% (Figure 1A). Despite their variable origin (retail chicken, chicken liver, and chicken gizzard), all chicken strains shared similar MLST and spa types (ST5, t002). Similar results were found among beef (ST1159, t091), pork (ST9, t3446), and turkey isolates (ST398, t034). Nucleotide difference (ND) trees were created from genomic sequences, and NDtree v. 1.2 revealed clusters specific for meat sources (Figure 1B). Genome tree report (Supplementary Figure S1) showed that retail beef strains from our laboratory clustered together and most of the strains in neighboring clusters were clinical isolates from human host (Supplementary Figure S2 and Supplementary Table S1). Only two strain G08M and G11F in the neighboring clusters were from cattle (mastitis) which were isolated from milk. All retail chicken isolates from our study clustered with S. aureus strains mainly from broiler chicken (Supplementary Figure S3 and Supplementary Table S2). Strain B4-59C was found in the same cluster with S. aureus ED98 which has been studied in a previous report (Lowder et al., 2009). Blast analysis showed 99.98% similarity between genomic sequences of B4-59C and ED98 (Supplementary Figure S3). Retail pork isolate B9-22D was found to be clustered with mainly pork and cow isolates in Genome Tree report (Supplementary Figure S4 and Supplementary Table S3). Neighboring clusters included strains mainly from poultry and livestock. Likewise, most strains found in cluster with S. aureus strain B3-14B and B6-55A (retail turkey isolates) were from turkey source (Supplementary Figure S5 and Supplementary Table S4). Sequences were annotated and then analyzed for virulence, AMR and stress response genes (Figure 2B). Pangenome analysis showed differences between the S. aureus genomes from retail meats and genomes from reference clinical isolates (Figure 2A), most notably in hypothetical proteins, mobile elements and phage proteins (Supplementary Table S5).
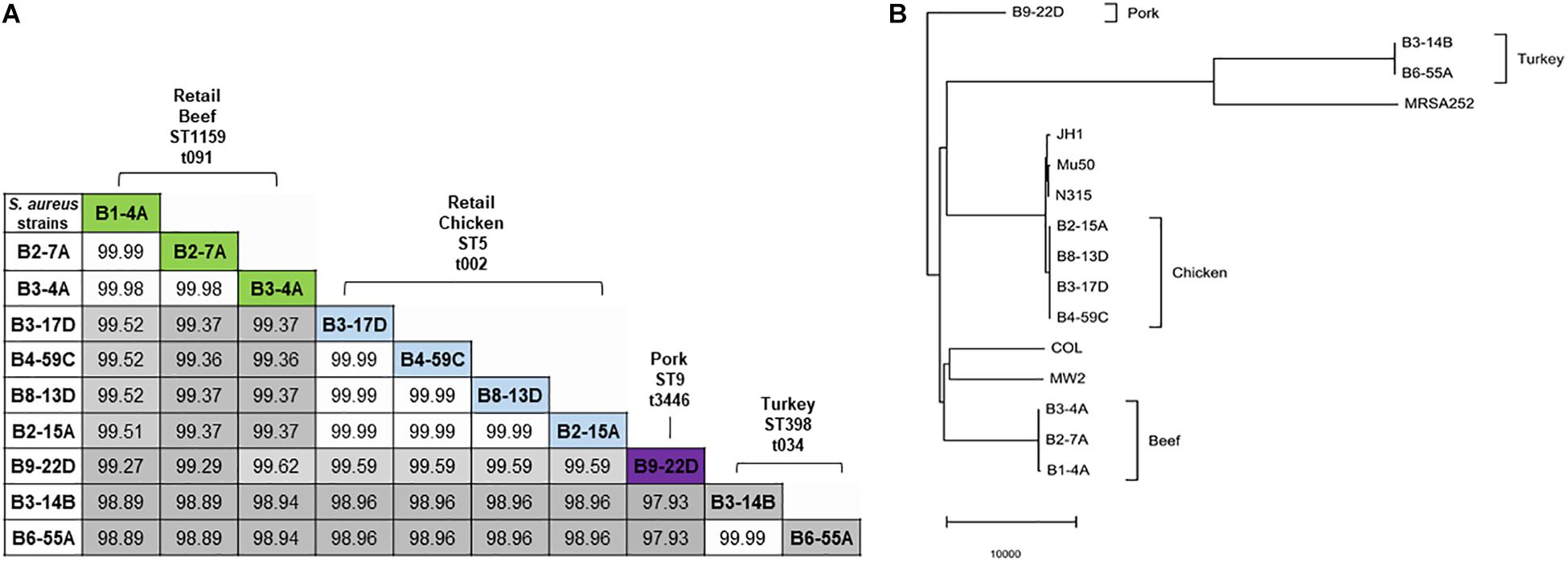
Figure 1. (A) Nucleotide sequence similarity in 10 S. aureus genomes. Sequence similarity values (%) are highlighted in gray (lowest value) to white (highest value). MLST types are indicated with prefix ST, and spa types with prefix t. (B) Nucleotide difference tree generated with S. aureus genomic sequences and NDtree v. 1.2.
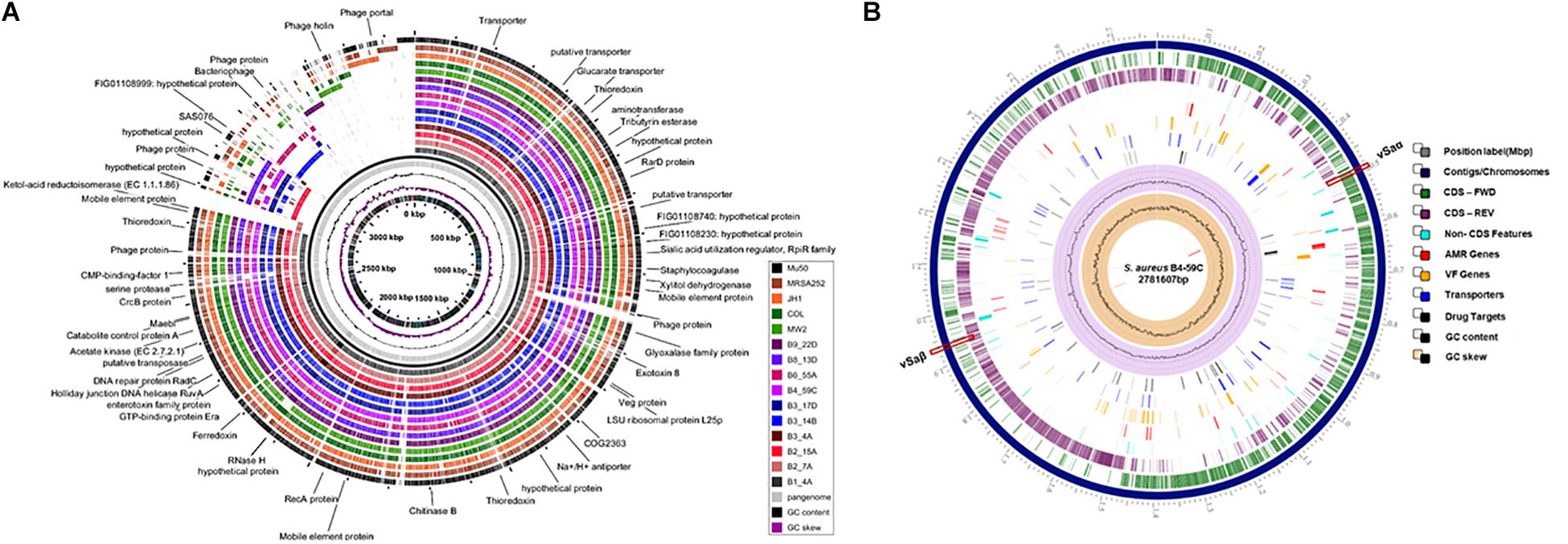
Figure 2. (A) Pangenome analysis of the 10 S. aureus strains described in this study and reference strains Mu50, MW2, JH1, MRSA252, and COL. Each circle depicts the sequence of a single strain. The pangenome (gray circle), GC content and GC skew are shown in the center. (B) Characteristics of the S. aureus B4-59C genome using PATRIC. AMR, antimicrobial resistance; VF, virulence factor.
Virulence Factors
Virulence and toxin genes were predicted and identified using VFanalyzer and RAST annotation, and the resulting patterns reflected the meat origin of the isolates (Figures 3A,B and Supplementary Table S6). Among genes related to adherence, differences were seen for collagen adhesion (cna), sdrD, and sdrE. The sdrD and sdrE genes were harbored by both beef and chicken isolates, whereas cna was only present in turkey isolates. Genes encoding clfA and eap/map were present in all RAST-annotated sequences but were not observed with VFanalyzer. Comparative analysis of protein sequences derived from cflA, fnbA, and spa genes revealed differences that were inferred from maximum likelihood analysis (Figures 4A–C). spa gene has been annotated as pseudogene in NCBI Prokaryotic Genome Annotation Pipeline (PGAP) for all retail chicken isolates. Retail chicken isolates grouped with clinical reference strains; however, beef, pork and turkey isolates were genetically distinct from most clinical strains (Figures 4A–C). Comparative analysis of the genomic sequences on the individual gene level supports the results obtained from the nucleotide difference tree created using whole genome sequences. Other adherence-related genes, including atl, ebh, clfB, ebp, efb, fnbB, icaA-D, icaR, and sdrC, were present in all sequenced strains. Among the exoenzymes, serine protease genes (splA, splB, splC, splD, and splF) were present in beef and chicken isolates; however, splE was only identified in beef isolates. Genes for cysteine protease, staphylocoagulase, and thermonuclease were present in all strains, but staphylokinase was absent.

Figure 3. (A) Virulence genes in S. aureus strains are indicated by the following colors: red, present using VFanalyzer; brown, present in RAST-annotated genomes; white, absent; and pink, truncated. (B) RAST-annotated genes identified in subsystems of virulence, defense, and disease. Strain names in (A,B) are highlighted based on source as follows: green, retail beef; blue, retail chicken; purple, pork; and gray, turkey.
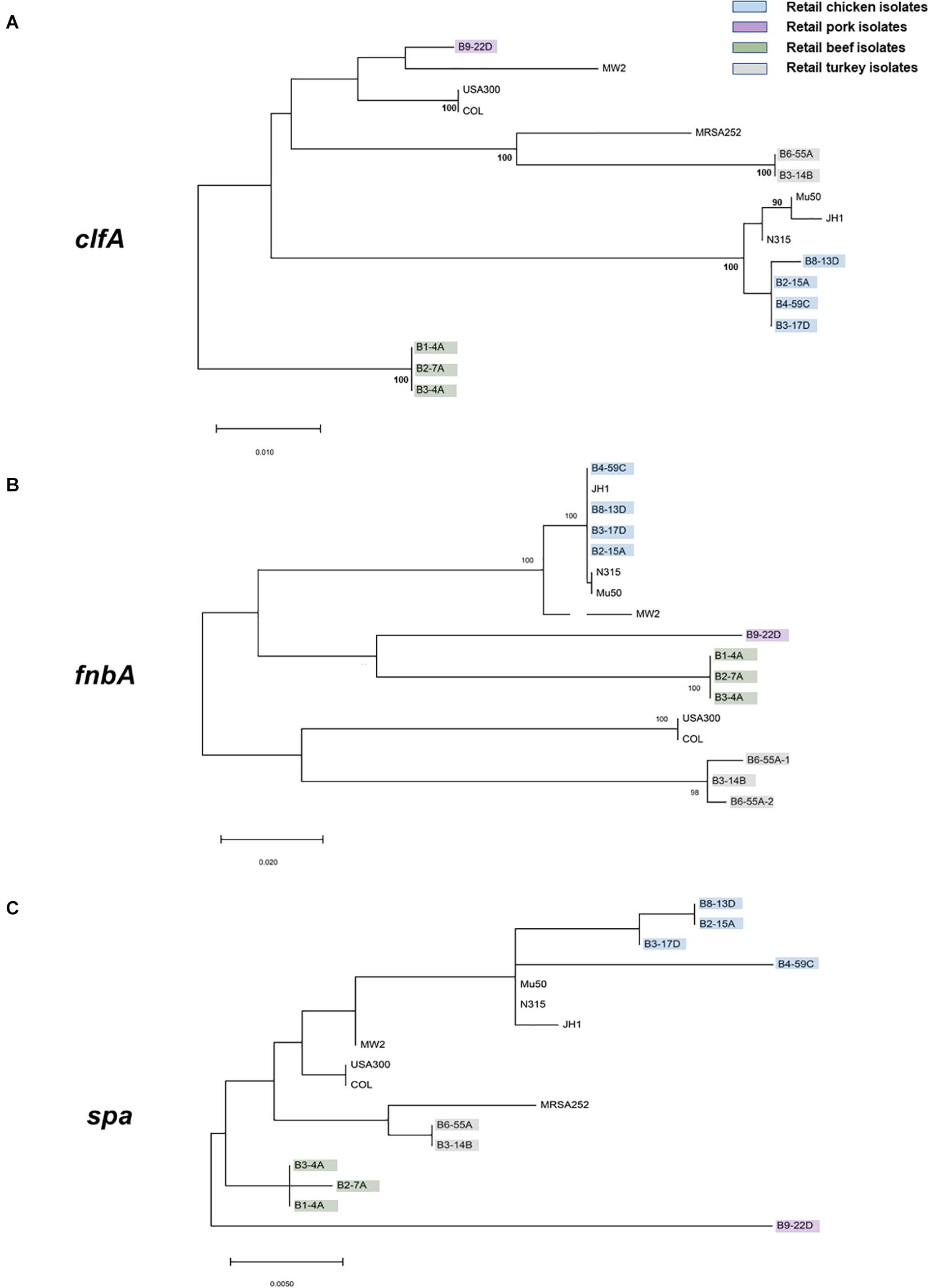
Figure 4. Maximum likelihood trees (bootstrap values of 100) derived from the predicted protein sequences of (A) clfA, (B) fnbA, and (C) spa genes. S. aureus strains characterized in this study are highlighted with the following colors: green, retail beef; blue, retail chicken; purple, pork; and gray, turkey. All remaining isolates are clinical reference strains.
Genes encoding sphingomyelinase (Supplementary Table S6) and hemolysin (hly/hla, hld, and hlgA-C) were identified in all strains, and a truncated hlb was present in chicken isolates (RAST annotation). Exotoxin genes (set6, set7 set10, set11, set13, and set14) were found in all strains, and chicken and pork isolates harbored one or more enterotoxin genes such as seg, sei, Ψent1/2 selm, seln, slo, and selq. Leukotoxin genes lukD and lukE were present in beef and chicken isolates, but absent in pork and turkey isolates. lukF-PV and lukS-PV were identified in all 10 RAST-annotated sequences; however, BLASTp annotated these as LukG and LukH leukotoxins in retail beef, chicken, and pork isolates. Isolates from retail turkey contained genes for β-pore-forming cytolysin and leucocidin S.
All 10 genomes contained genes related to the Type VII secretion system (e.g., ssaA, esxA, esaA, esaB, and essA-C). Beef strains also contained esaC and esxB and higher numbers of repeats than reference strain S. aureus NCTC8325 (Figure 5A). Although chicken and pork isolates shared similarity with reference strain NCTC8325, they contained proteins with unidentified functions instead of esaE, esxD, or essD (Figure 5A). Retail turkey isolates lacked esxC, esxB and the SAV0291 homolog, and exhibited differences in the repeat regions.
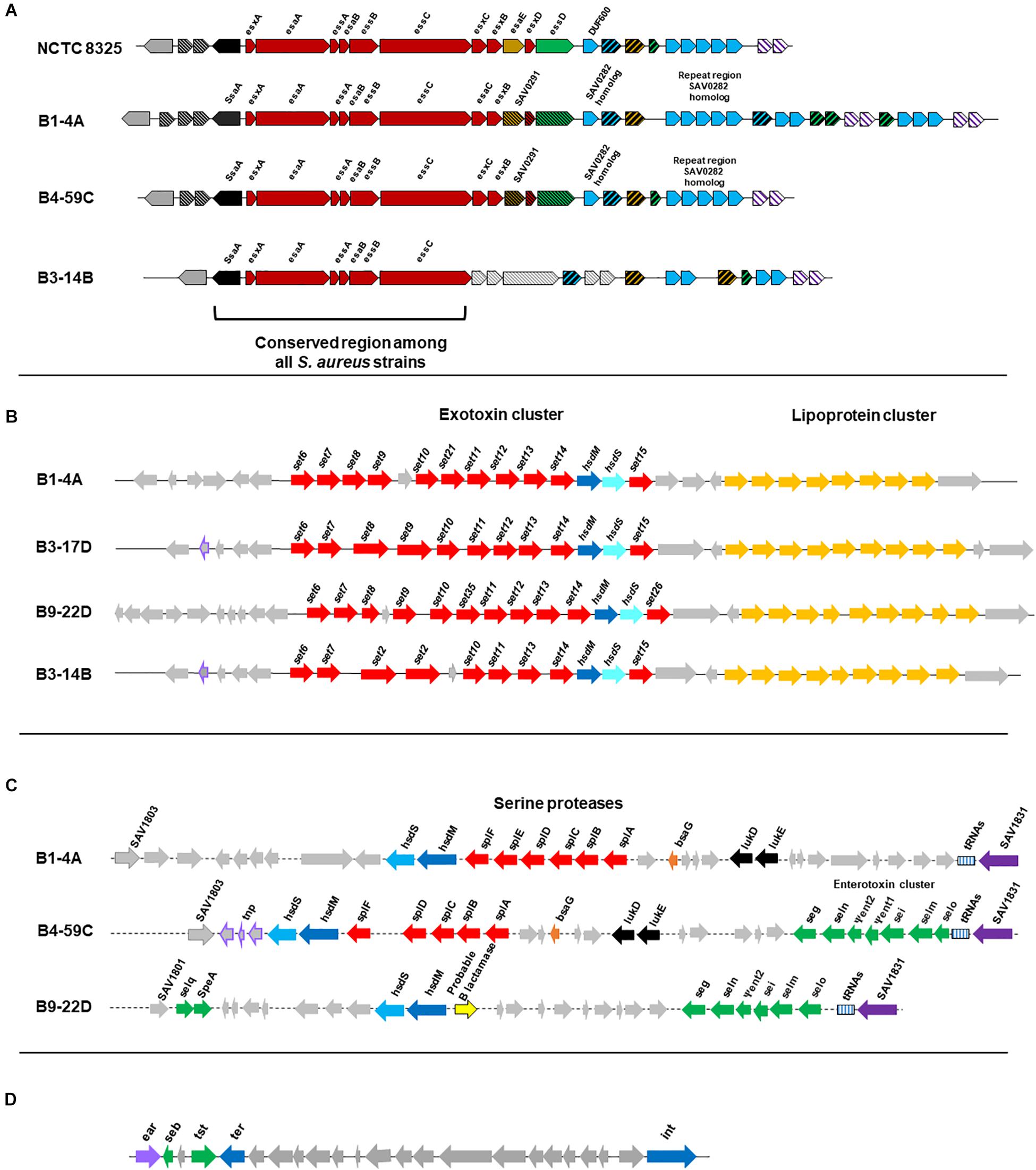
Figure 5. (A) Organization of gene clusters encoding the Type VII secretion system in S. aureus strains according to RAST annotation. The functional map for reference strain S. aureus NCTC8325 was adapted from a previous study (Warne et al., 2016). Cross-hatched and gray arrows indicate proteins with unknown functions. S. aureus strain B1-4A is a beef isolate that shares similarity with beef strains B2-7A and B3-4A, and the chicken isolate B4-59C is similar to the other three chicken strains (B3-17D, B8-13D, and B2-15A) and the pork isolate B9-22D. The map for the turkey isolate B3-14B is shown and shares similarity with B6-55A, which is also from turkey. (B) Organization of the νSaα genomic island and exotoxin cluster (set genes) in S. aureus strains. Genes were predicted from VFanalyzer and drawn to scale based on the genomic arrangement in RAST seed viewer. Arrows represent the following: red, coding sequence (CDS) for exotoxins; yellow, CDS for tandem lipoproteins; purple boundary, transposase; and gray, hypothetical proteins with unknown functions. S. aureus strains include representative isolates from beef (B1-4A), chicken (B3-17D), pork (B9-22D), and turkey (B3-14B). (C) νSaβ genomic island with serine proteases and enterotoxin gene cluster (seg, sei, Ψent1/2, selm, seln, selo, selq) in S. aureus strains. Genes were predicted from VFanalyzer and drawn to scale using the arrangement observed with the RAST seed viewer. Arrows represent the following: red, CDS for serine proteases; green, CDS for enterotoxins; purple, putative transposase; and gray, hypothetical proteins. S. aureus strains include representative isolates from beef (B1-4A), chicken (B4-59C), and pork (B9-22D). (D) S. aureus pathogenicity island (SaPI) carrying the gene for toxic shock syndrome toxin-1 and seb in S. aureus strain B4-59C. Genes were predicted from VFanalyzer and drawn to scale using the genomic arrangement observed with RAST seed viewer.
Genomic Islands
The genomic island νSaα, containing exotoxin set genes and a tandem lipoprotein cluster, was present in all genomes; however, differences were observed in set gene arrangement (Figure 5B). The νSaβ island, which encodes the enterotoxin gene cluster (egc), was present in chicken and pork isolates (Figure 5C). Beef isolates lacked an enterotoxin gene cluster but possess a serine protease cluster. Interestingly, the toxic shock syndrome toxin gene (tsst) was encoded by the SaPI genomic island in strains B4-59C (Figure 5D) and B3-17D.
AMR and Stress Response Genes
Various AMR genes were identified in the sequenced genomes (Figure 6A). The tetracycline resistance (TetR) gene tetM was present in the chromosome of turkey isolates, and tetK, tetL, and tetT were plasmid-encoded. Beef and chicken isolates harbored similar AMR genes (Figure 6A). All strains encoded bacitracin stress response genes bceA, bceB, bceR, and bceS (Figure 3B). Genes related to multidrug resistance, fluoroquinolone resistance (parC parE, gyrA gyrB) and multidrug reistance efflux pumps (acrB, MATE family MDR pump) were present in all strains (Figure 3B). The fosfomycin-resistance gene fosB was present in chicken and pork isolates but absent in beef and turkey isolates (Figure 3B). All strains harbored genes related to cobalt/zinc/cadmium resistance (CzcD, CzrB, and TR) and the stress response subsystem (Table 2). Genes encoding nitric oxide reductase (EC1.7.99.7) and a quinol-dependent gene were only present in turkey isolates.
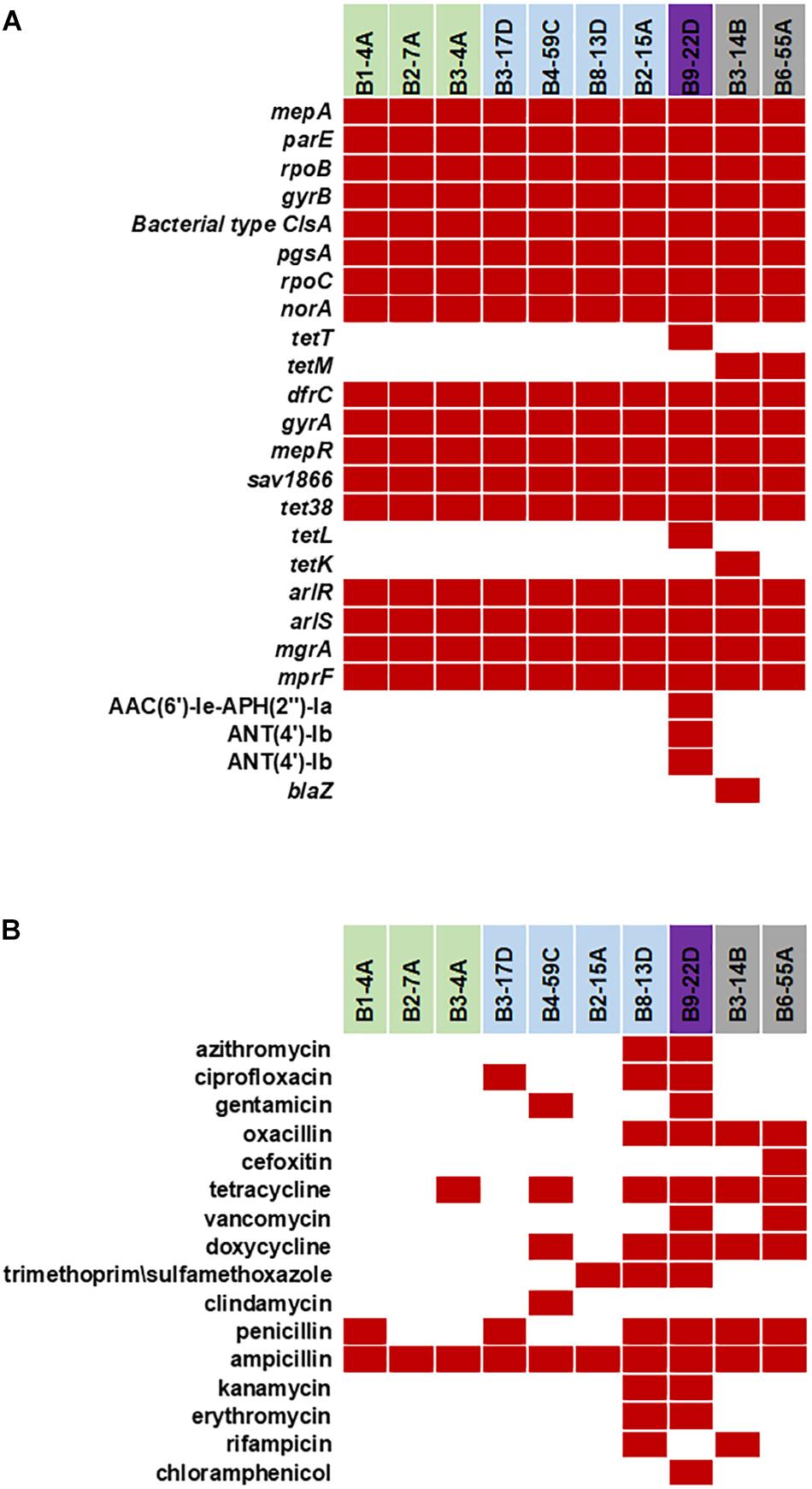
Figure 6. (A) Antimicrobial resistance genes in the 10 sequenced S. aureus strains. Red solid blocks indicate that a gene was present. (B) Antimicrobial resistance pattern in the 10 foodborne S. aureus strains. AMR screening was reported previously (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b) (Red block – resistant, white block – sensitive).
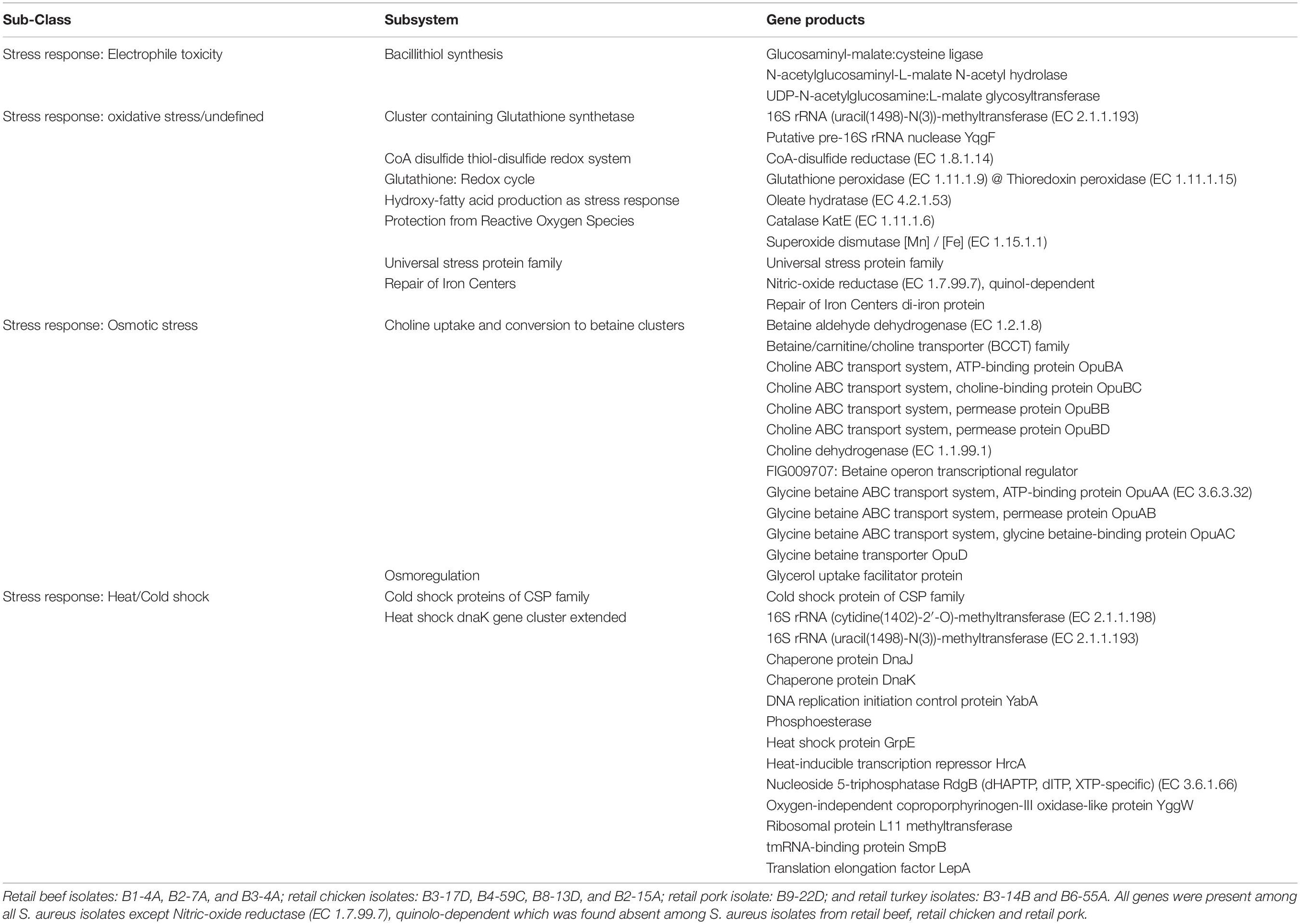
Table 2. Common stress response genes found in genomes of S. aureus strains (RAST subsystem – stress response).
Plasmids in S. aureus Strains
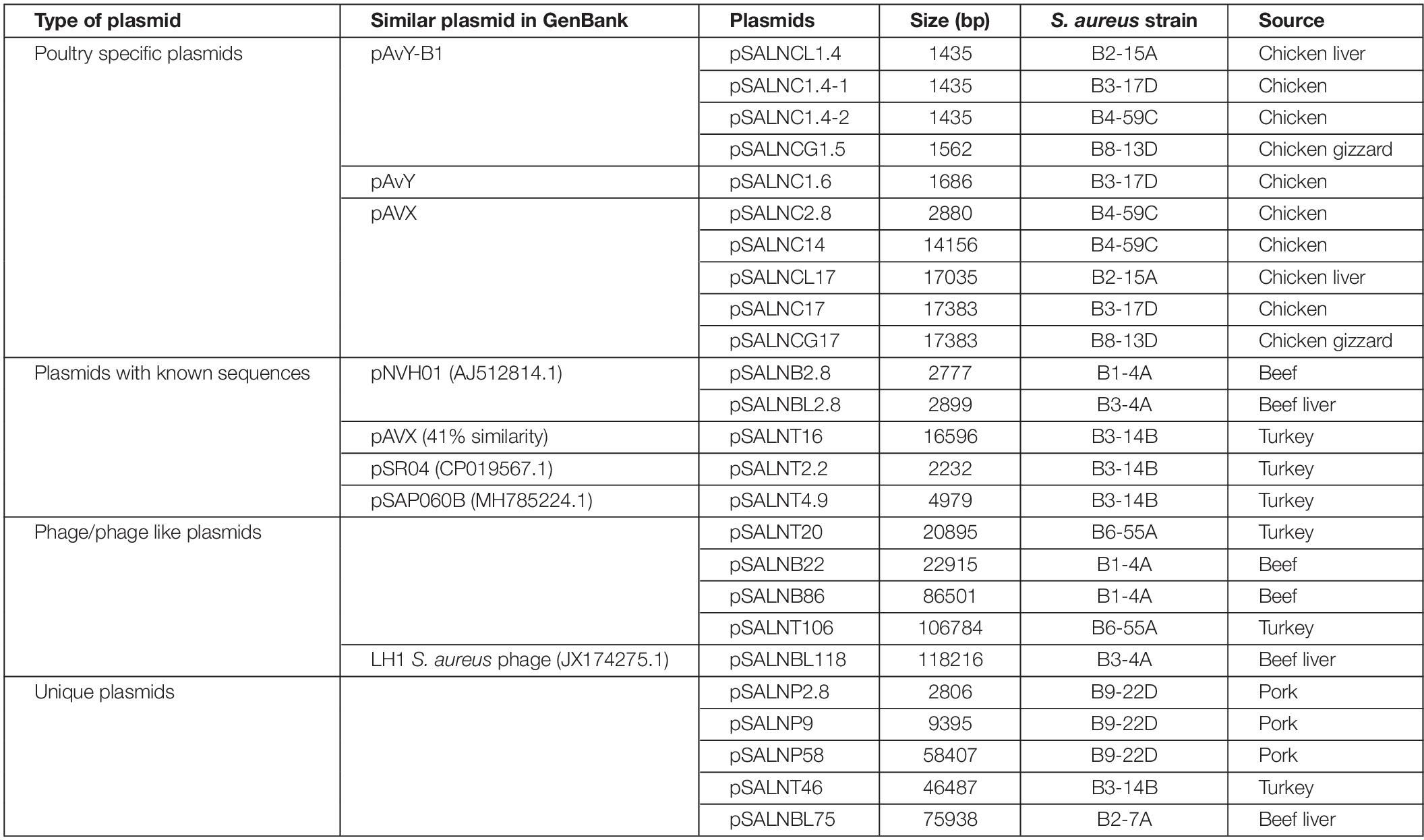
The plasmids sequenced in this study were previously characterized by S1 nuclease PFGE (Marasini and Fakhr, 2014; Cornell et al., 2018). A total of 25 plasmids were sequenced including 10 small (<5 kb) plasmids (Tables 1, 3). Five small plasmids were <2 kb (1435–1686 bp) and harbored genes annotated as hypothetical proteins (Supplementary Table S7). These plasmids were identified as poultry-specific pAvY-B1 (Bukowski et al., 2019) (pSALNCL1.4, pSALNC1.4-1, pSALNC1.4-2, and pSALNCG1.5) and pAvY (Bukowski et al., 2019) plasmids (pSALNC1.6), which were reported previously in a recent report (Bukowski et al., 2019; Table 3). The remaining five small plasmids harbored replication and hypothetical proteins (Supplementary Figures S6A,B and Supplementary Table S7). pSALNB2.8 and pSALNBL2.8 were related to pNVH01 (AJ512814.1) from horses (Bjorland et al., 2003), pSALNT2.2 was similar to pSR04 (CP019567.1) from humans, and pSALNC2.8 was identified as a pAVX (Bukowski et al., 2019) plasmid (Table 3). pSALNBL2.8 and pSALNB2.8 harbored the small multidrug resistance (SMR) family protein (Supplementary Figure S6A and Supplementary Table S7), and pSALNC2.8 contained a staphopain A precursor gene (Supplementary Figure S6B). Interestingly, pSALNP2.8, and pSALNP9 did not show strong identity to other plasmid sequences deposited in GenBank.
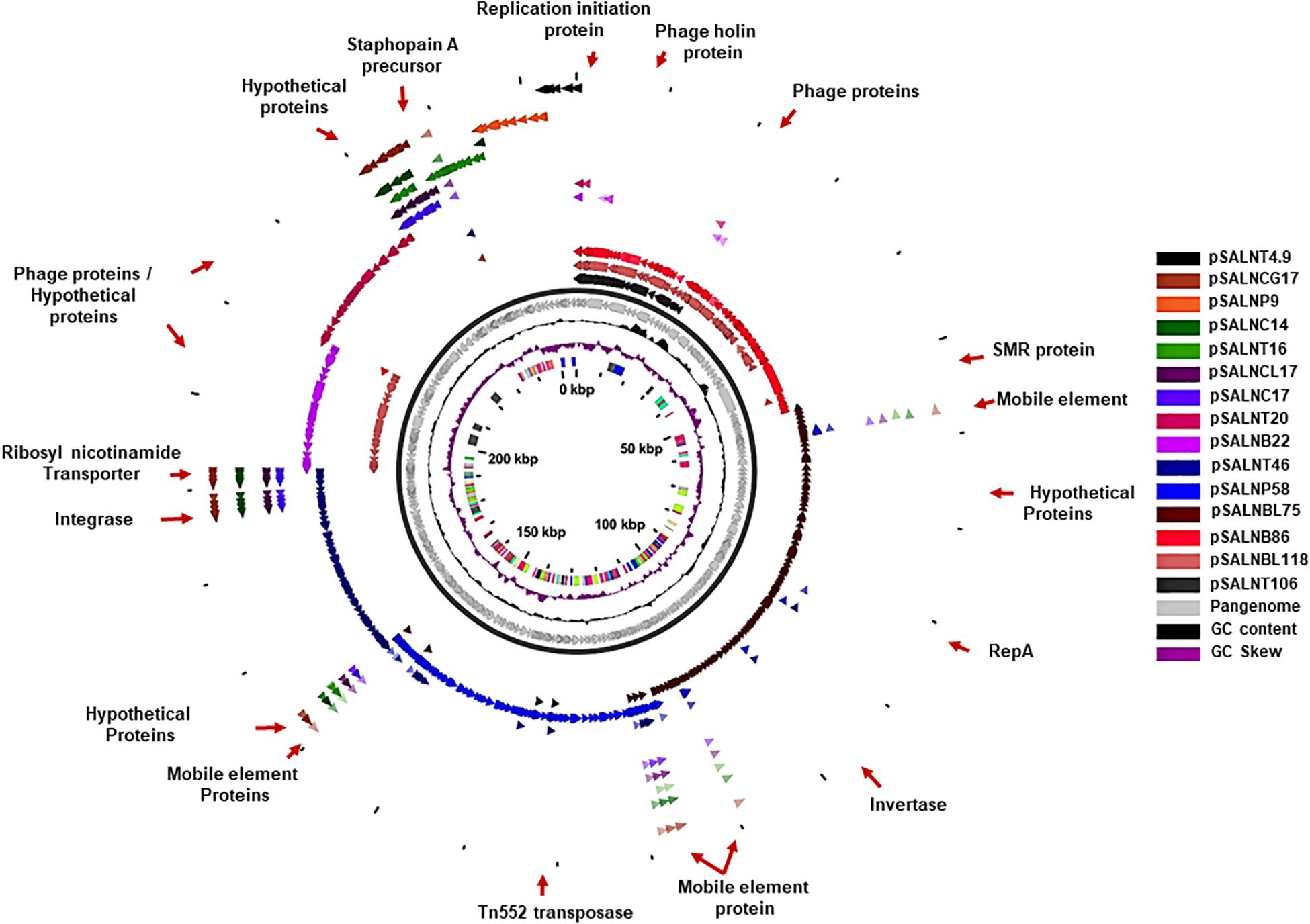
The larger plasmids (>5 kb) shared genes for mobile elements, hypothetical proteins, and integrase (Figures 7, 8 and Supplementary Table S8). TetR genes were identified in pSALNP9 [tetL and tetT] and pSALNT4.9 (tetK) (Supplementary Figures S7A,B). Moreover, pSALNT4.9 was related to pSAP060B (MH785224.1) with query cover 78% and 96.69% similarity; both plasmids contain TetR genes. Among the five plasmids ranging from 14,156 to 17,383 kb, the pAVX type plasmids (pSALNCL17, pSALNC17, pSALNC14, and pSALNCG17) were similar in genomic composition (Supplementary Table S8); however, pSALNT16 from turkey shared only 41% similarity with pAVX plasmids in GenBank. Three plasmids from retail chicken isolates, pSALNCL17, pSALNCG17 and pSALNC17, encoded staphopain A precursor gene (Supplementary Figures S8A,B). pSALNP58 shared 72% similarity with SAP068A (GQ900421.1) from hospital-acquired S. aureus strains. All remaining >5 kb plasmids encoded phage-related sequences; an example is pSALNBL118, which is 98% similar to the LH1 S. aureus phage (JX174275.1) sequence (Table 3).
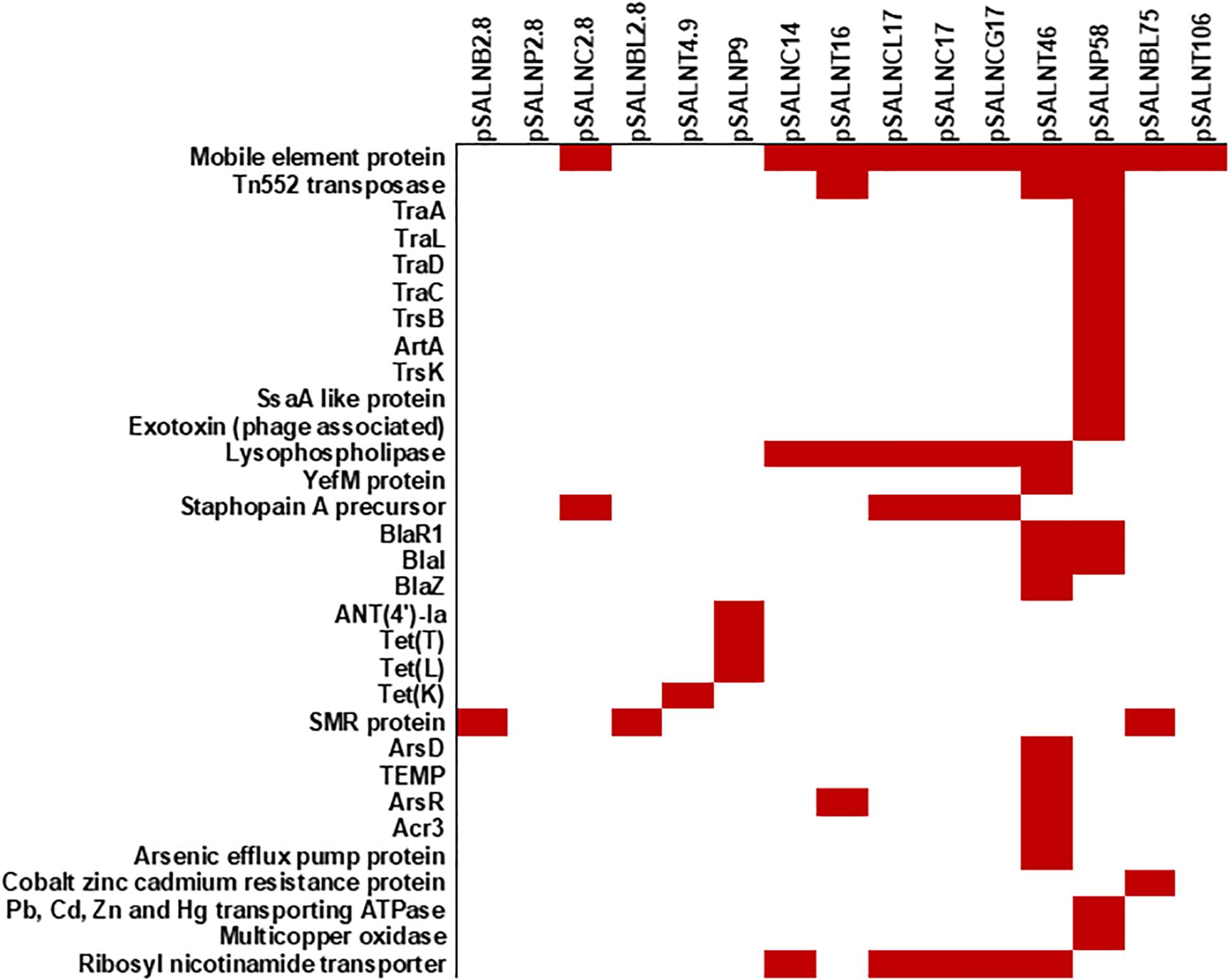
Figure 8. Plasmid-encoded genes potentially responsible for transfer, virulence, antimicrobial resistance, and heavy metal resistance in S. aureus strains. Solid red rectangles indicate that the gene was present. All gene products were annotated in RAST. Small (<5 kb) plasmids with hypothetical proteins (pSALNCL1.4, pSALNC1.4-1, pSALNC1.4-2, pSALNCG1.5, pSALNC1.6, and pSALNT2.2) and phage-like plasmids harboring only phage proteins (pSALNT20, pSALNB22, pSALNB86, and pSALNBL118) were excluded.
pSALNT46 encoded β-lactamase (BlaZ family) and arsenate resistance genes (Supplementary Figure S9A). Similarly, blaI, blaR, and Tn552 transposase genes were encoded by pSALNT46 and pSALNP58. Plasmid pSALNP58 harbored heavy metal transporting ATPase, aminoglycoside N(6′)-acetyltransferase, and traC/D/L and trsB. The small multi-drug resistance (SMR) family protein gene was encoded by pSALNBL75 (Supplementary Figure S9B). The pSALNT20 sequence was comprised primarily of unique phage genes (Supplementary Figure S10B), and other phage-related genes were encoded by pSALNB22, pSALNB86, pSALNT106, and pSALNBL118 (Supplementary Figure S10 and Supplementary Table S8).
Discussion
The S. aureus strains used in this study were previously isolated from retail meat products and characterized for enterotoxin genes and antimicrobial susceptibility (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). Although the number of S. aureus strains used here was relatively small (n = 10), an origin-specific genomic composition for the strains was clearly observed. Genomic comparisons (Figure 1A) and clustering of S. aureus strains (Figure 1B) indicate that strains from the same meat origin (e.g., beef, chicken, turkey or pork) shared a similar genomic composition. Genome report tree showed the clustering of genomes of retail chicken isolate from our study with poultry isolates, retail pork isolate with pork isolates, and retail turkey isolates with turkey isolates from GenBank (Supplementary Figure S1–S5 and Supplementary Tables S2–S5). This support that these isolates originated from respective animal sources either due to systemic infection, colonization or during slaughtering and processing. Interestingly, retail beef isolates didn’t cluster with any other strains, but neighboring cluster mainly includes strains from human sources. Most of the whole genome sequenced S. aureus sequences in GenBank are from human sources. Hence, it is not surprising to see most of the strains from retail meat sources clustering with human strains. Meanwhile, previous reports from our laboratory had also showed that S. aureus strains mainly originated from retail beef (retail beef liver) were similar to S. aureus strains from human sources (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b).
NDtree constructed using our strains and few reference clinical strains has shown the proximity of retail chicken isolates with JH1, Mu50, and N315 clinical strains (Figure 1B). Retail chicken isolates and these clinical strains are also found in neighboring clusters in Genome report tree (Supplementary Figure S1). The arrangement of the νSaα genomic island in the four retail chicken isolates was similar to the Type I νSaα genomic island in clinical strain S. aureus N315 (Baba et al., 2002, 2008). Retail chicken isolates B3-17D and B4-59C harbored the toxic shock syndrome toxin gene (tsst), which was 100% similar to tsst in clinical strain N315. Interestingly, prior studies suggested a human origin for S. aureus isolated from retail chicken (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a). However, the origin of poultry ST5 clade has been suggested to be due to the recent human to poultry host jump with adaptation in poultry hosts (Lowder et al., 2009). The Genome tree report showed the clustering of WGS retail chicken isolates from our laboratory with genomes of S. aureus strains from chicken in GenBank (Supplementary Figure S3 and Supplementary Table S2). Strain B4-59C shared higher genomic similarity with strain ED98 (Supplementary Figure S3). Many functional genes, found in human clinical strains which play role in human host pathogenesis, had been documented to have mutation to create pseudogenes among poultry associated S. aureus strains (Lowder et al., 2009). Acquisition of novel mobile genetic element had also played major role for genetic diversification among poultry specific ST5 S. aureus strains. Meanwhile, enhanced resistance toward the chicken heterophils was found by poultry specific S. aureus ED98 (Lowder et al., 2009). Many genes including spa and asp1 were found to be pseudogenes in annotation from PGAP for all our retail chicken isolates similar like S. aureus ED98. Presence of poultry specific plasmids (pAvY-B1, pAvY, pAVX) (Table 3) in retail chicken isolates from our study which supports their poultry associated origin. Although the presence of similar virulence genes is predicted in genomes of retail chicken isolates from this study with reference clinical strains, their virulence on chicken or human host have not been assayed which remains a limitation of this study and warrants for future investigation.
The pork isolate was assigned to ST9, a MLST group prevalent in pigs (Neela et al., 2009), which supports a livestock origin for pork isolate B9-22D. Many S. aureus strains found in cluster with B9-22D were isolated from pork skin and pig source (Supplementary Figure S4 and Supplementary Table S3). The turkey isolates were assigned to ST398, that also includes a dominant type of livestock-associated (LA) MSRA (Smith and Pearson, 2011) in CC398. Differences in virulence and AMR genes were previously observed in LA-MRSA (CC398) and community-associated (CA)-MRSA strains in a mouse model (Randad et al., 2019). Although ST398 MRSA had been well documented for its involvement in clinical cases in livestock and human, methicillin susceptible ST398 (MSSA) has also been reported as emerging public health threat (Valentin-Domelier et al., 2011). ST398 strains causing human infection are found not only as livestock associated but also found in livestock free environment (Diene et al., 2017). Previous studies had shown major factor influencing for pathogenesis in human hosts is the presence of prophages in S. aureus genome carrying virulence related genes (Diene et al., 2017; Kashif et al., 2019). Although, two retail turkey isolates used in this study (B3-14B and B6-55A) lack mec genes, phenotypic resistance toward oxacillin and cefoxitin was observed for these strains (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). In this study, we observed that virulence factors in LA-CC398 S. aureus strains IHW398-1 and IHW398-2 (Randad et al., 2019) were similar to B3-14B and B6-55A. Presence of tetM gene, Tn916 and lack of φ3 prophage indicate livestock origin of these strains. In genome report tree, both retail turkey isolates cluster with S. aureus strains (MRSA as well as MSSA) from turkey source (Supplementary Figure S5). Interestingly, strain B3-14B from our study is found highly similar to SA33924 strain that had been described as human adapted ST398 (blood stream infection isolate) in a previous report (Diene et al., 2017).
The S. aureus strains sequenced herein encoded virulence-related genes associated with adherence, enzyme secretion, immune system evasion, toxins, and leucocidins. Lipoproteins are known to function in immune system stimulation and invasion in human cells (Nguyen et al., 2015), which were also harbored by all our strains. Furthermore, the type VII secretion system genes esxA, esaA, esaB, and essA-C (Warne et al., 2016) were present in all strains. Meanwhile, these strains also lack many virulence related genes and phage proteins found in studied reference clinical strains (Supplementary Table S6). Presence of virulence related genes and AMR genes might play role in pathogenesis and adaptation in animals and human hosts. However, the virulence potential of these strains has not been assayed in this study which limits our assertion for their pathogenic nature in animals or human hosts. Enterotoxin genes were identified in retail chicken and pork strains (Figure 5C), which suggests a potential role in enterotoxin-mediated food poisoning.
The S. aureus strains in this study were resistant to antimicrobial compounds (Figure 6B; Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). Despite having mecA, mecB or mecC in their genome, all sequenced strains were resistant to ampicillin. Several strains (B8-13D, B9-22D, B3-14B, and B6-55A) found to be phenotypic MRSA and were resistant to oxacillin (Abdalrahman and Fakhr, 2015; Abdalrahman et al., 2015a,b). Antimicrobial resistance genes detected in plasmids sequenced in our study (Figures 6A,B) correlate with the phenotypic antibiotic resistance patterns seen in the S. aureus strains harboring them (Figure 6B). Ampicillin resistance might be partially mediated by pSALNT46 and pSALNP58, which contain Tn552-encoded blaI/blaR genes. The pork isolate B9-22D was kanamycin resistant, and the aminoglycoside 4′-nucleotidyltransferase gene borne on pSALNP9 might impart Km resistance. Strains B3-4A, B4-59C, B8-13D, B9-22D, B3-14B, and B6-55A were TetR and encode tet(38) in their genome (Figures 3B, 6A). The gene tetM is commonly encoded by Tn916 and Tn5801 in S. aureus from humans or by Tn916 in animals (De Vries et al., 2009). The two turkey isolates harbor tetM in Tn916; furthermore, pSALNT4.9 and pSALNP9 plasmids in B9-22D and B3-14B were shown to encode tetK, tetL, and tetT. Plasmids pSALNB2.8 and pSALNBL75 from retail beef isolates harbored genes encoding SMR proteins; this is significant because SMR family proteins confer resistance to antispetics and quaternary ammonium products (Bay and Turner, 2009; Costa et al., 2013). The presence of genes on plasmids for arsenic efflux pump protein, cobalt zinc cadmium resistance protein, Pb, Cd, Zn, and Hg transporting ATPase, multicopper oxidase, SMR proteins might play possible roles in AMR and heavy metal resistance mechanisms of their hosts. It also emphasizes the role of plasmids in dissimination of resistance genes among S. aureus strains. The unique genomic composition of pSALNP2.8 and pSALNP9 plasmids when compared to all available plasmid sequences in GenBank warrants further studies in regards to plasmids from retail pork isolates. Phage or phage-like sequences are prevalent in S. aureus strains and are important for horizontal gene transfer (Goerke et al., 2009). Plasmids pSALNB22, pSALNT20, pSALNB86, pSALNBL75, pSALNT46, and pSAlNT106 contained phage-related proteins, and these plasmids might be important in the transmission of virulence factors.
Plasmids harbored by S. aureus are known to carry various genes responsible for its survival and adaptation. Lineage specific distribution of plasmids has been suggested in regards to the presence of resistance and virulence determinants (McCarthy and Lindsay, 2012). Plasmids in S. aureus have been categorized according to rep gene families (McCarthy and Lindsay, 2012) as well as the presence of functional genes (Bukowski et al., 2019). Poultry specific plasmids (Group I) (Bukowski et al., 2019) like pAvY-B1 (pSALNCL1.4, pSALNC1.4-1, pSALNC1.4-2, and pSALNCG1.5) and pAvY plasmids (pSALNC1.6) are cryptic plasmids. Likewise, another small plasmid, pSALNT2.2, from a retail turkey isolate bears only a gene coding for a replication protein. Beneficiary roles of these cryptic plasmids for the S. aureus host strains are yet to be identified. One study has hypothesized that cryptic plasmids serve as flexible vectors to acquire mobile genomic elements which leads to the origination of plasmids with known functional genes (Attéré et al., 2017). Staphopain A (Laarman et al., 2012) and lysophospholipase (Daugherty and Low, 1993) are well documented virulence factors which are found prevalent in poultry specific pAvX plasmids sequenced in our study. pSALNC2.8 shares 100% genomic similarity with pSALNC17 and pSALNCG17 and other pAvX plasmids available in GenBank despite the discrepencies on their sizes. Although, some plasmids (pSALNB2.8, pSALNP2.8, pSALNC2.8, and pSALNBL2.8) have similar size (∼2.8 kb), origin specific differences could be seen among these plasmids (Supplementary Table S3).
In summary, staphylococcal food poisoning (SFP) remains a major health problem, and outbreaks have been documented around the globe (Argudín et al., 2010; Kadariya et al., 2014; Johler et al., 2015; Mossong et al., 2015; Vitale et al., 2015; Stewart, 2017; Thapaliya et al., 2017). Contamination of retail food products with S. aureus and the presence of enterotoxins in foodborne strains has increased the potential for SFP outbreaks (Crago et al., 2012; Abdalrahman et al., 2015a,b; Jamali et al., 2015; Mossong et al., 2015; Al-Ashmawy et al., 2016; Ge et al., 2017). The 10 S. aureus strains isolated from retail meat products in this study contain various virulence factors and AMR genes. Sequencing more strains from various retail meat products will be ideal to represent the population of S. aureus circulating in retail meat. As concluding remarks, genomic analysis supported that all sequenced S. aureus strains from retail meat products in this study originated from respective animal sources. Source specific differences in virulence factors and genomic composition was found among the strains in comparative genomic analysis. As S. aureus strains with potential viurlence factors and stress response genes are retained in retail meat products even after processng and storage conditions, these strains might play role in possible food poisioning outbreaks and other clinical cases. Hence, enhancement in food safety measures for reduction of foodborne pathogens including S. aureus contamination is required. The pathogenicity of these strains studied in this study has not been tested but remains possible and warrants further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MF: research design. AK and LN: experimental procedures, sequence assembly, and comparative genomics analysis. AK, LN, and MF: manuscript preparation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge financial support from the Research Office of The University of Tulsa (Tulsa, OK, United States) for granting LN a student research grant. The authors thank Dr. Daya Marasini for his technical assistance with the MiSeq run. LN is grateful to the Saudi Government for granting her a Ph.D. fellowship to educational expenses in the United States.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.574923/full#supplementary-material
Footnotes
- ^ http://rast.theseed.org/FIG/rast.cgi
- ^ https://www.patricbrc.org
- ^ https://cge.cbs.dtu.dk/services/MLST/
- ^ https://cge.cbs.dtu.dk/services/spatyper/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://server.gview.ca/
- ^ https://www.ncbi.nlm.nih.gov/genome/tree/154?
- ^ https://www.ncbi.nlm.nih.gov/tools/gbench/
- ^ http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer
- ^ http://www.mgc.ac.cn/VFs/
- ^ https://card.mcmaster.ca/analyze/rgi
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ http://www.pathogenomics.sfu.ca/islandviewer/
- ^ http://rast.theseed.org/FIG/rast.cgi
- ^ https://www.ebi.ac.uk/Tools/msa/muscle/
References
Abdalrahman, L., and Fakhr, M. (2015). Incidence, antimicrobial susceptibility, and toxin genes possession screening of Staphylococcus aureus in retail chicken livers and gizzards. Foods 4, 115–129. doi: 10.3390/foods4020115
Abdalrahman, L. S., Stanley, A., Wells, H., and Fakhr, M. K. (2015a). Isolation, virulence, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin sensitive Staphylococcus aureus (MSSA) strains from Oklahoma retail poultry meats. Int. J. Environ. Res. Public Health 12, 6148–6161. doi: 10.3390/ijerph120606148
Abdalrahman, L. S., Wells, H., and Fakhr, M. (2015b). Staphylococcus aureus is more prevalent in retail beef livers than in pork and other beef cuts. Pathogens 4, 182–198. doi: 10.3390/pathogens4020182
Al-Ashmawy, M. A., Sallam, K. I., Abd-Elghany, S. M., Elhadidy, M., and Tamura, T. (2016). Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathog. Dis. 13, 156–162. doi: 10.1089/fpd.2015.2038
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Argudín, M. Á, Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Attéré, S. A., Vincent, A. T., Paccaud, M., Frenette, M., and Charette, S. J. (2017). The role for the small cryptic plasmids as moldable vectors for genetic innovation in Aeromonas salmonicida subsp. salmonicida. Front. Genet. 8:211. doi: 10.3389/fgene.2017.00211
Baba, T., Bae, T., Schneewind, O., Takeuchi, F., and Hiramatsu, K. (2008). Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190, 300–310. doi: 10.1128/JB.01000-07
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K. I., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Bartels, M. D., Petersen, A., Worning, P., Nielsen, J. B., Larner-Svensson, H., Johansen, H. K., et al. (2014). Comparing whole-genome sequencing with sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 52, 4305–4308. doi: 10.1128/JCM.01979-14
Bay, D. C., and Turner, R. J. (2009). Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 9:140. doi: 10.1186/1471-2148-9-140
Becker, K., van Alen, S., Idelevich, E. A., Schleimer, N., Seggewiß, J., Mellmann, A., et al. (2018). Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 24, 242–248. doi: 10.3201/eid2402.171074
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Bjorland, J., Steinum, T., Sunde, M., Waage, S., and Heir, E. (2003). Novel plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob. Agents Chemother. 47, 3046–3052. doi: 10.1128/AAC.47.10.3046-3052.2003
Bosi, E., Monk, J. M., Aziz, R. K., Fondi, M., Nizet, V., and Palsson, B. Ø (2016). Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 113, E3801–E3809. doi: 10.1073/pnas.1523199113
Bukowski, M., Piwowarczyk, R., Madry, A., Zagorski-Przybylo, R., Hydzik, M., and Wladyka, B. (2019). Prevalence of antibiotic and heavy metal resistance determinants and virulence-related genetic elements in plasmids of Staphylococcus aureus. Front. Microbiol. 10:805. doi: 10.3389/fmicb.2019.00805
Carretto, E., Visiello, R., and Nardini, P. (2018). “Methicillin resistance in Staphylococcus aureus,” in Pet-to-Man Travelling Staphylococci: A World in Progress, ed. V. Savini (New York, NY: Macmillan Publishers), 225–235. doi: 10.1016/B978-0-12-813547-1.00017-0
Castro, A., Santos, C., Meireles, H., Silva, J., and Teixeira, P. (2016). Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. J. Infect. Public Health 9, 153–160. doi: 10.1016/j.jiph.2015.08.001
Cornell, C. R., Marasini, D., and Fakhr, M. K. (2018). Molecular characterization of plasmids harbored by actinomycetes isolated from the great salt plains of Oklahoma using PFGE and next generation whole genome sequencing. Front. Microbiol. 9:2282. doi: 10.3389/fmicb.2018.02282
Costa, S. S., Viveiros, M., Amaral, L., and Couto, I. (2013). Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol. J. 7, 59–71. doi: 10.2174/1874285801307010059
Crago, B., Ferrato, C., Drews, S. J., Svenson, L. W., Tyrrell, G., and Louie, M. (2012). Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in food samples associated with foodborne illness in Alberta, Canada from 2007 to 2010. Food Microbiol. 32, 202–205. doi: 10.1016/j.fm.2012.04.012
Daugherty, S., and Low, M. G. (1993). Cloning, expression, and mutagenesis of phosphatidylinositol-specific phospholipase C from Staphylococcus aureus: a potential staphylococcal virulence factor. Infect. Immun. 61, 5078–5089.
De Vries, L. E., Christensen, H., Skov, R. L., Aarestrup, F. M., and Agersø, Y. (2009). Diversity of the tetracycline resistance gene tet (M) and identification of Tn916- and Tn5801-like (Tn 6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 64, 490–500. doi: 10.1093/jac/dkp214
Diene, S. M., Corvaglia, A. R., François, P., van der Mee-Marquet, N., Amirault, P., Lehiani, O., et al. (2017). Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics 18:133. doi: 10.1186/s12864-017-3516-x
Dweba, C. C., Zishiri, O. T., and El Zowalaty, M. E. (2018). Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect. Drug Resist. 11, 2497–2509. doi: 10.2147/IDR.S175967
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elhassan, M. M., Ozbak, H. A., Hemeg, H. A., Elmekki, M. A., and Ahmed, L. M. (2015). Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed. Res. Int. 2015:895860. doi: 10.1155/2015/895860
Gaerste-Díaz, Y. C., Lozano-Zarain, P., Torres, C., Castro González, N. P., Rocha-Gracia, R., and del, C. (2018). Genotyping of antimicrobial resistance and virulence in Staphylococcus isolated from food of animal origin in Mexico. Indian J. Microbiol. 58, 525–528. doi: 10.1007/s12088-018-0745-x
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010–2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191, 3462–3468. doi: 10.1128/JB.01804-08
Haaber, J., Penadés, J. R., and Ingmer, H. (2017). Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 25, 893–905. doi: 10.1016/j.tim.2017.05.011
Hedberg, C. W. (2011). Foodborne illness acquired in the United States. Emerg. Infect. Dis. 17, 1338–1340. doi: 10.3201/eid1707.110019
Hennekinne, J. A., De Buyser, M. L., and Dragacci, S. (2012). Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36, 815–836. doi: 10.1111/j.1574-6976.2011.00311.x
Jamali, H., Paydar, M., Radmehr, B., Ismail, S., and Dadrasnia, A. (2015). Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control 54, 383–388. doi: 10.1016/j.foodcont.2015.02.013
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johler, S., Weder, D., Bridy, C., Huguenin, M.-C., Robert, L., Hummerjohann, J., et al. (2015). Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. J. Dairy Sci. 98, 2944–2948. doi: 10.3168/jds.2014-9123
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:104984. doi: 10.1371/journal.pone.0104984
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. Biomed Res. Int. 2014:827965. doi: 10.1155/2014/827965
Kashif, A., McClure, J. A., Lakhundi, S., Pham, M., Chen, S., Conly, J. M., et al. (2019). Staphylococcus aureus ST398 virulence is associated with factors carried on prophage ?Sa3. Front. Microbiol. 10:2219. doi: 10.3389/fmicb.2019.02219
Kitai, S., Shimizu, A., Kawano, J., Sato, E., Nakano, C., Kitagawa, H., et al. (2005). Prevalence and characterization of Staphylococcus aureus and enterotoxigenic Staphylococcus aureus in retail raw chicken meat throughout Japan. J. Vet. Med. Sci. 67, 269–274. doi: 10.1292/jvms.67.269
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Laarman, A. J., Mijnheer, G., Mootz, J. M., Van Rooijen, W. J. M., Ruyken, M., Malone, C. L., et al. (2012). Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J. 31, 3607–3619. doi: 10.1038/emboj.2012.212
Larsen, J., Petersen, A., Larsen, A. R., Sieber, R. N., Stegger, M., Koch, A., et al. (2017). Emergence of livestock-associated methicillin-resistant Staphylococcus aureus bloodstream infections in Denmark. Clin. Infect. Dis. 65, 1072–1076. doi: 10.1093/cid/cix504
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leekitcharoenphon, P., Nielsen, E. M., Kaas, R. S., Lund, O., and Aarestrup, F. M. (2014). Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:87991. doi: 10.1371/journal.pone.0087991
Lin, J., Lin, D., Xu, P., Zhang, T., Ou, Q., Bai, C., et al. (2016). Non-hospital environment contamination with Staphylococcus aureus and methicillin-resistant Staphylococcus aureus: proportion meta-analysis and features of antibiotic resistance and molecular genetics. Environ. Res. 150, 528–540. doi: 10.1016/j.envres.2016.06.040
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Lowder, B. V., Guinane, C. M., Zakour, N. L. B., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Lozano, C., Gharsa, H., Ben Slama, K., Zarazaga, M., and Torres, C. (2016). Staphylococcus aureus in animals and food: methicillin resistance, prevalence and population structure. A review in the African continent. Microorganisms 4:12. doi: 10.3390/microorganisms4010012
Marasini, D., and Fakhr, M. K. (2014). Exploring PFGE for detecting large plasmids in Campylobacter jejuni and Campylobacter coli isolated from various retail meats. Pathogens 3, 834–844. doi: 10.3390/pathogens3040833
Marasini, D., and Fakhr, M. K. (2016a). Complete genome sequences of Campylobacter jejuni strains OD267 and WP2202 isolated from retail chicken livers and gizzards reveal the presence of novel 116-kilobase and 119-kilobase megaplasmids with type VI secretion systems. Genome Announc. 4:e01060-16. doi: 10.1128/genomeA.01060-16
Marasini, D., and Fakhr, M. K. (2016b). Complete genome sequences of the plasmid-bearing Campylobacter coli strains HC2-48, CF2-75, and CO2-160 isolated from retail beef liver. Genome Announc. 4:e01004-16. doi: 10.1128/genomeA.01004-16
Marasini, D., and Fakhr, M. K. (2016c). Whole-genome sequencing of a Campylobacter jejuni strain isolated from retail chicken meat reveals the presence of a megaplasmid with Mu-like prophage and multidrug resistance genes. Genome Announc. 4:e00460-16. doi: 10.1128/genomeA.00460-16
Marasini, D., and Fakhr, M. K. (2017a). Complete genome sequences of Campylobacter jejuni strains isolated from retail chicken and chicken gizzards. Genome Announc. 5:e01351-17. doi: 10.1128/genomea.01351-17
Marasini, D., and Fakhr, M. K. (2017b). Complete genome sequences of plasmid-bearing Campylobacter coli and Campylobacter jejuni strains isolated from retail chicken liver. Genome Announc. 5:e01350-17. doi: 10.1128/genomeA.01350-17
McCarthy, A. J., and Lindsay, J. A. (2012). The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 12:104. doi: 10.1186/1471-2180-12-104
Mediavilla, J. R., Chen, L., Mathema, B., and Kreiswirth, B. N. (2012). Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 15, 588–595. doi: 10.1016/j.mib.2012.08.003
Miller, L. G. (2010). Community-associated methicillin resistant Staphylococcus aureus. Issues Infect. Dis. 6, 1–20. doi: 10.1159/000298753
Mossong, J., Decruyenaere, F., Moris, G., Ragimbeau, C., Olinger, C. M., Johler, S., et al. (2015). Investigation of a staphylococcal food poisoning outbreak combining case-control, traditional typing and whole genome sequencing methods, Luxembourg, June 2014. Eurosurveillance 20:30059. doi: 10.2807/1560-7917.ES.2015.20.45.30059
Neela, V., Zafrul, A. M., Mariana, N. S., Van Belkum, A., Liew, Y. K., and Rad, E. G. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140.
Neyaz, L. (2019). Comparative Genomics of Plasmid-Bearing Staphylococcus aureus Strains Isolated from Various Retail Meats. Ph.D. dissertation, The University of Tulsa, Tulsa, Ok.
Neyaz, L., Karki, A. B., and Fakhr, M. K. (2019). The whole-genome sequence of plasmid-bearing Staphylococcus argenteus strain B3-25B from retail beef liver encodes the Type VII secretion system and several virulence factors. Microbiol. Resour. Announc. 8:e00962-19. doi: 10.1128/MRA.00962-19
Neyaz, L., Rajagopal, N., Wells, H., and Fakhr, M. K. (2020). Molecular characterization of Staphylococcus aureus plasmids associated with strains isolated from various retail meats. Front. Microbiol. 11:223. doi: 10.3389/fmicb.2020.00223
Nguyen, M. T., Kraft, B., Yu, W., Demicrioglu, D. D., Hertlein, T., Burian, M., et al. (2015). The νSaα specific lipoprotein like cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human cells. PLoS Pathog. 11:e1004984. doi: 10.1371/journal.ppat.1004984
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Planet, P. J., Narechania, A., Chen, L., Mathema, B., Boundy, S., Archer, G., et al. (2017). Architecture of a Species: phylogenomics of Staphylococcus aureus. Trends Microbiol. 25, 153–166. doi: 10.1016/j.tim.2016.09.009
Randad, P. R., Dillen, C. A., Ortines, R. V., Mohr, D., Aziz, M., Price, L. B., et al. (2019). Comparison of livestock-associated and community-associated Staphylococcus aureus pathogenicity in a mouse model of skin and soft tissue infection. Sci. Rep. 9:6774. doi: 10.1038/s41598-019-42919-y
Sergelidis, D., and Angelidis, A. S. (2017). Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen. Lett. Appl. Microbiol. 64, 409–418. doi: 10.1111/lam.12735
Shajari, G., Khorshidi, A., and Moosavi, G. (2017). Vancomycin resistance in Staphylococcus aureus strains. Arch. Razi Inst. 90, 107–110.
Sivaraman, G. K., Vanik, D., Visnuvinayagam, S., Prasad, M. M., and Ravishankar, C. N. (2017). Draft genome sequence of a methicillin resistant Staphylococcus aureus isolate (sequence type 1) from seafood. Genome Announc. 5:e01193-17. doi: 10.1128/genomeA.00776-17
Smith, T. C., and Pearson, N. (2011). The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11, 327–339. doi: 10.1089/vbz.2010.0072
Song, M., Bai, Y., Xu, J., Carter, M. Q., Shi, C., and Shi, X. (2015). Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 195, 1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020
Stewart, G. C. (2017). Staphylococcal Food Poisoning, Foodborne Diseases, 3rd Edn. (Amsterdam: Elsevier Inc), 367–380. doi: 10.1016/B978-0-12-385007-2.00018-8
Tam, K., and Torres, V. J. (2019). Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 7:10.1128/microbiolspec.GPP3-0039-2018. doi: 10.1128/microbiolspec.gpp3-0039-2018
Thapaliya, D., Forshey, B. M., Kadariya, J., Quick, M. K., Farina, S. O., Brien, A., et al. (2017). Prevalence and molecular characterization of Staphylococcus aureus in commercially available meat over a one-year period in Iowa, USA. Food Microbiol. 65, 122–129. doi: 10.1016/j.fm.2017.01.015
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Turner, N. A., Sharma-Kuinkel, B. K., Maskarinec, S. A., Eichenberger, E. M., Shah, P. P., Carugati, M., et al. (2019). Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218. doi: 10.1038/s41579-018-0147-4
Valentin-Domelier, A. S., Girard, M., Bertrand, X., Violette, J., François, P., Donnio, P. Y., et al. (2011). Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: an emerging human-adapted subclone? PLoS One 6:e28369. doi: 10.1371/journal.pone.0028369
Vitale, M., Scatassa, M. L., Cardamone, C., Oliveri, G., Piraino, C., Alduina, R., et al. (2015). Staphylococcal food poisoning case and molecular analysis of toxin genes in Staphylococcus aureus strains isolated from food in sicily, Italy. Foodborne Pathog. Dis. 12, 21–23. doi: 10.1089/fpd.2014.1760
Warne, B., Harkins, C. P., Harris, S. R., Vatsiou, A., Stanley-Wall, N., Parkhill, J., et al. (2016). The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics 17:222. doi: 10.1186/s12864-016-2426-7
Waters, A. E., Contente-Cuomo, T., Buchhagen, J., Liu, C. M., Watson, L., Pearce, K., et al. (2011). Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 52, 1227–1230. doi: 10.1093/cid/cir181
Wattam, A. R., Abraham, D., Dalay, O., Disz, T. L., Driscoll, T., Gabbard, J. L., et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591. doi: 10.1093/nar/gkt1099
Wu, S., Huang, J., Wu, Q., Zhang, J., Zhang, F., Yang, X., et al. (2018). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9:2767. doi: 10.3389/fmicb.2018.02767
Xia, G., and Wolz, C. (2014). Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 21, 593–601. doi: 10.1016/j.meegid.2013.04.022
You, Y., Song, L., Nonyane, B. A. S., Price, L. B., and Silbergeld, E. K. (2018). Genomic differences between nasal Staphylococcus aureus from HOG slaughterhouse workers and their communities. PLoS One 13:e0193820. doi: 10.1371/journal.pone.0193820
Keywords: S. aureus, plasmids, retail meat, comparative genomics, whole genome sequencing
Citation: Karki AB, Neyaz L and Fakhr MK (2020) Comparative Genomics of Plasmid-Bearing Staphylococcus aureus Strains Isolated From Various Retail Meats. Front. Microbiol. 11:574923. doi: 10.3389/fmicb.2020.574923
Received: 22 June 2020; Accepted: 06 October 2020;
Published: 23 October 2020.
Edited by:
Sophia Johler, University of Zurich, SwitzerlandReviewed by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyPatrick Rik Butaye, Ross University School of Veterinary Medicine, Saint Kitts and Nevis
Copyright © 2020 Karki, Neyaz and Fakhr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed K. Fakhr, bW9oYW1lZC1mYWtockB1dHVsc2EuZWR1
†These authors have contributed equally to this work
 Anand B. Karki
Anand B. Karki Leena Neyaz†
Leena Neyaz† Mohamed K. Fakhr
Mohamed K. Fakhr