- 1Department of Biological Sciences, Florida International University, Miami, FL, United States
- 2Biomolecular Sciences Institute, Florida International University, Miami, FL, United States
Wolbachia are maternally transmitted bacterial endosymbionts, carried by approximately half of all insect species. Wolbachia prevalence in nature stems from manipulation of host reproduction to favor the success of infected females. The best known reproductive modification induced by Wolbachia is referred to as sperm-egg Cytoplasmic Incompatibility (CI). In CI, the sperm of Wolbachia-infected males cause embryonic lethality, attributed to paternal chromatin segregation defects during early mitotic divisions. Remarkably, the embryos of Wolbachia-infected females “rescue” CI lethality, yielding egg hatch rates equivalent to uninfected female crosses. Several models have been discussed as the basis for Rescue, and functional evidence indicates a major contribution by Wolbachia CI factors. A role for host contributions to Rescue remains largely untested. In this study, we used a chemical feeding approach to test for CI suppression capabilities by Drosophila simulans. We found that uninfected females exhibited significantly higher CI egg hatch rates in response to seven chemical treatments that affect DNA integrity, cell cycle control, and protein turnover. Three of these treatments suppressed CI induced by endogenous wRi Wolbachia, as well as an ectopic wMel Wolbachia infection. The results implicate DNA integrity as a focal aspect of CI suppression for different Wolbachia strains. The framework presented here, applied to diverse CI models, will further enrich our understanding of host reproductive manipulation by insect endosymbionts.
Introduction
Endosymbiosis is a specialized form of interaction, with one organism dwelling inside the cells and tissues of another (Archibald, 2015). The bacterium Wolbachia pipientis is one of the most widespread endosymbionts, carried by half or more of all insect species (Weinert et al., 2015; Sazama et al., 2017). Wolbachia are gram negative bacteria that belong to the alpha-protobacterial class Rickettsiales. Wolbachia are maternally transmitted, with the efficacy of transmission dependent on the bacteria being loaded into eggs (Breeuwer and Werren, 1990; Zchori-Fein et al., 1998; Hadfield and Axton, 1999; Stouthamer et al., 1999; Rasgon and Scott, 2003; Veneti et al., 2004; Ferree et al., 2005; Serbus et al., 2008; Fast et al., 2011). Wolbachia commonly modify host reproduction to favor the success of infected females. This is accomplished by induction of parthenogenesis, male killing, feminization and sperm-egg cytoplasmic incompatibility (Yen and Barr, 1971; Rousset et al., 1992; Stouthamer et al., 1993; Jiggins et al., 1998; Hurst et al., 1999; Werren et al., 2008). Though Wolbachia interactions with their host appear generally commensal, this extent of host manipulation classifies Wolbachia bacteria as reproductive parasites.
Cytoplasmic incompatibility (CI) is the most widely known of all Wolbachia-induced reproductive manipulations (Hoffmann and Turelli, 1997; Werren, 1997; Stouthamer et al., 1999). CI is characterized by embryonic lethality in crosses between uninfected females and Wolbachia-infected males (Hertig, 1936; Laven, 1959; Yen and Barr, 1971, 1973; Noda, 1984; Hsiao and Hsiao, 1985; Wade and Stevens, 1985; Hoffmann et al., 1986; Binnington and Hoffmann, 1989). By contrast, Wolbachia-infected females are compatible with both uninfected and Wolbachia-infected males, with viable progeny produced by both types of crosses. The ability of embryos from Wolbachia-infected females to survive the Wolbachia-modified sperm is known as “Rescue.” Compatibility is conferred by specific pairings of sperm modification (mod) and rescue capacity (resc) associated with different Wolbachia strains (Turelli, 1994; Werren, 1997; McGraw and O’Neill, 1999; Charlat et al., 2001; Poinsot et al., 2003; Duron et al., 2006; Zabalou et al., 2008; Nor et al., 2013). With infected females favored by elimination of incompatible embryos, the CI/Rescue paradigm effectively drives host population replacement in natural populations as well as in applied, vector management scenarios (Turelli and Hoffmann, 1991, 1995; Riegler et al., 2005; Hoffmann et al., 2011; Kriesner et al., 2013; Schuler et al., 2013; Schmidt et al., 2017; Turelli et al., 2018).
The cellular basis of CI has been a point of interest for many years. Cytological experiments indicate that mitotic defects are a consensus feature of CI across Wolbachia-host systems. Specifically, paternal chromatin remains at the metaphase plate while maternal chromatin segregates to opposite poles in anaphase, resulting in chromosome bridging and aneuploidy (O’Neill and Karr, 1990; Reed and Werren, 1995; Callaini and Riparbelli, 1996; Lassy and Karr, 1996; Callaini et al., 1997; Tram and Sullivan, 2002; Landmann et al., 2009; Bonneau et al., 2018b). Studies from Nasonia and Drosophila simulans have suggested these mitotic defects are produced from a timing mismatch between male and female pronuclei at the first mitotic division, which must be reconciled in order to enable Rescue (Callaini et al., 1997; Tram and Sullivan, 2002; Landmann et al., 2009). A separate line of work implicated Wolbachia-induced oxidative damage to spermatocyte DNA as a contributor to CI lethality (Brennan et al., 2012) with the implication that DNA damage prevention and/or repair in the embryo is important in conferring Rescue. This model is consistent with a body of literature on Wolbachia and induction of oxidative stress (Brennan et al., 2008; Xi et al., 2008; Hughes et al., 2011; Andrews et al., 2012; Pan et al., 2012; Bian et al., 2013; Zug and Hammerstein, 2015).
The most recent model is that Wolbachia CI factors (Cifs) are responsible for both CI and Rescue (Beckmann et al., 2017; LePage et al., 2017; Shropshire et al., 2018). Bioinformatic predictions and transgenic studies using yeast and Drosophila melanogaster models, have indicated that CifB proteins have deubiquitlase activity (CidB), nuclease activity (CinB), or both (CndB) (Beckmann and Fallon, 2013; Beckmann et al., 2017, 2019a,b; Lindsey et al., 2018; Chen et al., 2019). CifB is required to induce CI (LePage et al., 2017; Bonneau et al., 2018b, 2019; Chen et al., 2019; Meany et al., 2019), possibly by affecting sperm chromatin remodeling during spermatogenesis (Beckmann et al., 2019b). By contrast, CifA can Rescue classical CI phenotypes induced by Wolbachia (Shropshire et al., 2018), as well as CI associated with dual expression of CifA and CifB in transgenic males (Chen et al., 2019; Shropshire and Bordenstein, 2019). Proteomic evidence also indicates that CidA binding modifies CidB targeting (Beckmann et al., 2019b).
One caveat of the CifA findings to date is that D. melanogaster exhibits transient CI, mainly in association with newly eclosed males (Reynolds and Hoffmann, 2002; Yamada et al., 2007; LePage et al., 2017), particularly for first-emerging males of a population (Yamada et al., 2007). Depending upon the host strain used, initial hatch rates of 5–50% increase to 50–80% by day 3, and become normal by day 5 (Reynolds and Hoffmann, 2002; Yamada et al., 2007; LePage et al., 2017). This CI decline appears attributable to the host background, as wRi Wolbachia transinfected into D. melanogaster also elicit a mild CI response, allowing on the order of 70% egg hatch (Boyle et al., 1993). By contrast, progeny produced by wRi Wolbachia CI range from 0 to 6% of normal for newly eclosed males, 15–30% in 7–9 day old males, and 40–50% in 14-day old males, depending upon the experiment (Hoffmann et al., 1986; Turelli and Hoffmann, 1995). Thus, it is unclear if D. melanogaster CI represents mild induction of the CI defect, a background environment that is already highly permissive/enabling of Rescue, or both. As transgenic studies of Cif function in CI and Rescue have only just begun, it is not known to what extent Cif-related mechanisms in D. melanogaster represent that of other hosts exhibiting severe, Wolbachia-induced CI defects.
A long-standing question has been to what extent host factors contribute to the mechanism of Rescue. If Cif proteins act exclusively in terms of a toxin-antitoxin system, with CifA suppressing CifB function via direct binding (Beckmann et al., 2019a), no host involvement is required for Rescue. Consistent with the toxin–antitoxin model, direct binding has been demonstrated for multiple cognate pairs of CifA and CifB proteins (Beckmann et al., 2017, 2019b; Chen et al., 2019). Complementary studies did not identify contributions by additional Wolbachia-generated factors (Bonneau et al., 2018a, 2019; Perlmutter and Bordenstein, 2020), though host genetic background differences are reported to alter CI severity (Boyle et al., 1993; Poinsot et al., 1998). It remains possible that host mechanisms, to some extent, run in support of, in parallel to, or independently of bacterial effectors in the context of Rescue. Cardinium endosymbionts carried by Encarsia wasps have been credited with inducing their own forms of CI and Rescue (Hunter et al., 2003; Penz et al., 2012; Mann et al., 2017), in the apparent absence of Cif proteins altogether (Lindsey et al., 2018; Doremus and Hunter, 2020). Two additional non-Wolbachia CI systems were recently identified (Doremus and Hunter, 2020), in Lariophagus wasps (König et al., 2019) as well as in Brontispa beetles (Takano et al., 2017). While it is possible that each endosymbiont has a self-contained mechanism for CI and Rescue, another possibility is that these functionally convergent phenotypes are due to endosymbiont effects on conserved, cellular processes of the host. As such, the extent of host involvement in the process of Rescue merits examination.
Since Rescue involves preventing and/or repairing CI defects, this study examined whether D. simulans females have the capacity to alter developmental outcomes of CI embryos. If this is possible, then modifying the function of the relevant host pathways should confer CI suppression, evident as increased hatch rates for uninfected embryos that are otherwise subject to CI lethality. To test this, uninfected D. simulans females were exposed to chemicals that alter candidate cellular processes, previously implicated in CI and Rescue. CI induced by the endogenous wRi Wolbachia strain, as well as a transinfected wMel Wolbachia strain, were investigated. Egg hatch data were evaluated in light of existing CI/Rescue models, as described below.
Materials and Methods
Fly Stocks and Rearing Conditions
The wRi Wolbachia strain, endogenous to D. simulans, used in this study was originally described by Hoffman, Turrelli, and Simmons (Hoffmann et al., 1986). The uninfected D. simulans strain (w–) is of the same genetic background, as it was this original line cured of Wolbachia with tetracycline. The wMel trans-infected line was created with Wolbachia from D. melanogaster (Poinsot et al., 1998), backcrossed into the cured fly stock for six generations to standardize the D. simulans genetic background. We previously confirmed the identity of wRi and wMel in D. simulans, and verified that the wMel transinfection matches the standard wMel strain carried by most D. melanogaster stocks (Christensen et al., 2016).
All the flies used in this study were maintained at 25°C on a 12 h light/dark cycle using an Invictus Drosophila incubator (Genessee Scientific, United States). Flies were raised in standard 6oz square bottom polyethylene bottles containing 25–30 ml of fly food (described later). Each stock bottle was seeded by approximately 80–100 flies, a mixture of both male and female, and incubated for 3–6 days. After this period, flies were either transferred to a new bottle or discarded. Flies used to seed all bottles were discarded by 12–15 days of age. To collect virgin flies, stock bottles were completely cleared, and rechecked by eye to verify the absence of flies. Newly eclosed flies were collected 5–8 h later using standard CO2 gas pads. Males and females were separated and temporarily stored in narrow polypropylene vials until loading into treatment vials/plates, as described below. To avoid damage from prolonged exposure to CO2, fly sorting was limited to a 20–25 min time frame.
Microbial 16S rRNA Gene Sequencing
To determine the infection status in D. simulans flies, microbial 16S rRNA gene sequencing was performed. Ovaries were dissected from uninfected, wRi-infected and wMel-infected D. simulans females, followed by sequencing of region V1–V3 of the 16S rRNA gene, carried out on an Illumina MiSeq as previously described (Christensen et al., 2019). Each sample represents ovarian content from 20 flies. The raw sequencing data are available at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA663645.
Food Preparation
The stock food used in this study was made as per a standard Bloomington Stock Center recipe, described earlier1 (Camacho et al., 2017). For chemical feeding assays, concentrated stock solutions of each chemical were prepared using an appropriate solvent, dependent on necessary final concentration. To make chemical food for independent experiments, the appropriate amount of stock solution was mixed with melted food and mixed thoroughly by stirring. This food was transferred to either vials or plate wells immediately, before cooling and solidifying. The same amount of solvent was mixed with standard food to create a parallel “control food” condition in each experiment. Vial-based trials contained 3–5 ml of food per vial, while in the 24-well plate format, each well contained 800 μL of food.
Dose Response Curve Preparation
To determine the appropriate feeding concentration for each chemical, a range of 6–7 doses was empirically tested for each compound. The range of concentrations used was based on information available in existing literature. Each chemical was diluted to the appropriate concentration in beakers containing 10 mL standard food with Brilliant Blue G food added (Acros Organics). All content was mixed thoroughly for 30 s, then divided equally into two treatment vials and cooled under the fume hood for ∼2 h to prevent condensation from collecting along the sides of the vials. In each case, one treatment vial was immediately used, and the second vial was plugged with rayon, wrapped in foil, and stored in a sealed container at 4°C for use 6 days later.
To carry out dose-response testing, six uninfected male and six uninfected female D. simulans flies were incubated in the first set of treatment vials. On the 6th day, the flies were transferred to the second corresponding treatment vials for an additional 6 days. Adult mortality, egg lay, egg hatch and larval development were qualitatively scored for treatment vials and corresponding controls across the 12-day period. Treatment vials equivalent to the control were scored as “+.” Conditions exhibiting loss of 40% or more relative to the control, of eggs, larvae and/or pupae, were separately noted. Defects occurring during later days (Zchori-Fein et al., 1998; Rasgon and Scott, 2003; Veneti et al., 2004; Ferree et al., 2005; Serbus et al., 2008; Fast et al., 2011) of treatment were scored as “some,” and consistent developmental defects across the 12-day span were scored as “–.” The highest concentration of chemical with no adverse effect on flies as per the above criteria was selected for subsequent feeding assays. Dose response curves for dual drug treatment combinations were carried out similarly. Two independent biological replicates were performed for all dose-response experiments.
CI Suppression Tests
In the Vial-Based Format
Virgin D. simulans flies were incubated for 3 days in vials containing 3–5 ml of food. Females were split between treatment food and control food conditions, whereas infected males were exposed to standard food only. In all cases, flies were grouped, with 15–20 flies per vial. On day 3, male and female flies were transferred to fresh vials of standard food for an 8-h mating period. Depending upon the experiment, this was done as single pair matings or as mass matings of 30–40 flies, using equal numbers of males and females. Afterward, male flies were discarded and female flies were returned their original treatment vials. At day 4, individual females were split up into separate vials that sustained their existing treatment conditions, with the addition of blue food coloring to the food to improve egg visibility.
In the Plate Assay Format
A detailed description is provided in Additional File S1. Briefly, Corning 24-well plates (Cat# 3738) were set up with 800 μL of fly food per well. 10 D. simulans virgin females were added to each well and incubated for 3 days. Uninfected females were added to eight standard food wells for use as the CI control, and to eight treatment wells to test for chemical suppression of CI. Infected females were added to eight standard food wells for use as a Rescue control. Wolbachia-infected males were incubated separately on standard food for 3 days, at a density of 45 flies or fewer per vial. At day 3, the females were transferred to a new plate carrying standard food and mated to 10 infected males for 8 h. Afterward, males were removed, and female flies returned to their respective wells in the original treatment plate. At day 4, females were transferred to a fresh plate that contained the same treatment condition per well, along with blue food coloring.
For Both Assay Formats
Female flies were discarded at day 5, followed by scoring of egg hatch at day 6. Two or more independent biological replicates were performed for each plate assay experiment. For rigor and consistency across the plate assays, only plates that showed a 12% or lower hatch rate for the CI control were scored.
Statistical Analysis
Chi Square tests of goodness of fit were performed manually as per standard procedures (McDonald, 2014). A Bonferonni correction was applied, so that alpha values were scaled to the number of data categories analyzed (McDonald, 2014). Z′ values were calculated as previously (Zhang et al., 1999; Serbus et al., 2012). As previously, the IBM SPSS v.23 analysis package was used for all other statistical test (Field, 2013; Christensen et al., 2019). The data were analyzed for normality using the Shapiro–Wilk test and for homogeneity of variance by Levene’s test (Shapiro and Wilk, 1965; Lim and Loh, 1996; Mohd Razali and Bee, 2011). For the data with normal distribution, mean differences were evaluated using a t-test if variance was homogeneous, and Welch’s t-test if it was not (McDonald, 2014; Rietveld and van Hout, 2015). If the data did not fit the normal distribution, the Mann–Whitney U-test was performed for data showing homogeneity of variance, and an independent t-test was performed with bootstrapping as an approximation when variance was uneven (LaFlair et al., 2015; Rietveld and van Hout, 2015). To assess the power of different sample sizes, we also used a bootstrap procedure in MATLABTM (Mathworks, Natick, MA, United States) that randomly sub-samples from the data to determine the sample size required to meet specified p-values (Christensen et al., 2019).
Results
Verification of D. simulans Endosymbiont Identity by 16S rRNA Analysis
The D. simulans flies used in this study carry the Wolbachia strain wRi as a natural infection (Hoffmann et al., 1986), or wMel as a transinfected strain (Poinsot et al., 1998). Female flies from these strains have been shown by DNA staining to exhibit nucleoids in their germline cells that are consistent with Wolbachia infection (Serbus and Sullivan, 2007; Christensen et al., 2016). These lines have also been confirmed as PCR-positive for the Wolbachia surface protein (Wsp) gene, and the Wolbachia strain identities have been confirmed by sequencing (Christensen et al., 2016). Use of these detection methods, while consistent with expected Wolbachia identities, does not rule out the possible presence of other bacterial endosymbionts.
To independently confirm the identity of the germline bacteria carried by Wolbachia-infected flies, 16S rRNA microbiome analyses were carried out as previously described (Christensen et al., 2019). Ovary tissue samples were analyzed from both uninfected and Wolbachia-infected D. simulans lines. The data indicate Wolbachia spp. as the predominant taxon carried by both wRi- and wMel-infected tissues, with 94.5–98.3% of the reads representing the Wolbachia genus (Figure 1) (Supplementary Table S1) (Additional File S2). The other non-Wolbachia taxa detected in the Wolbachia-infected ovary samples paralleled that of the uninfected control (Figure 1). As this was a non-sterile assay, these signatures likely reflect the microbiome of the cuticle, the body cavity and residual contamination of dissection equipment (Christensen et al., 2019). Notably, the Wolbachia-infected samples show no evidence of other Drosophila-resident symbionts such as Spiroplasma. Thus, Wolbachia endosymbionts represent the vast majority, if not all, of the microbiome carried by maternal germline cells that generate Rescue-capable eggs.
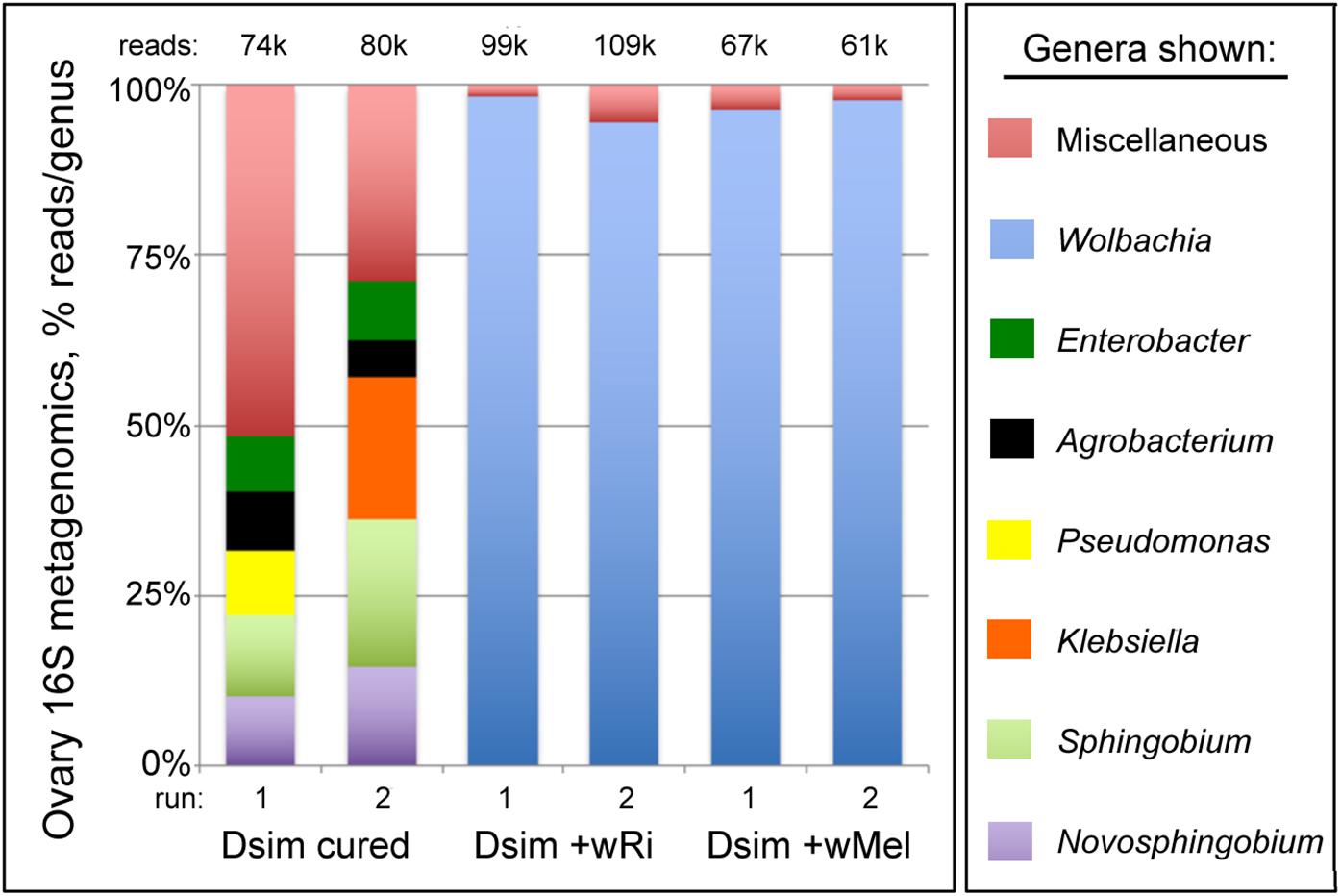
Figure 1. 16S rRNA microbiome profiles associated with Drosophila simulans ovary tissues. Uninfected and Wolbachia-infected tissues are shown. Top five most abundant genera that equal or exceed 1% abundance per sample are shown. For further details, see Supplementary Table S1 and Additional File S2.
Confirmation of Baseline CI and Rescue Phenotypes From D. simulans
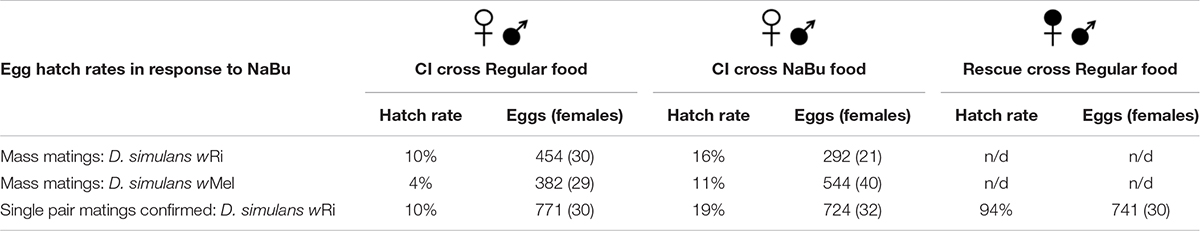
Previous studies have demonstrated that natural (wRi) as well as transinfected (wMel) Wolbachia strains induce robust CI and Rescue effects in D. simulans (Hoffmann et al., 1986; Binnington and Hoffmann, 1989; Poinsot et al., 1998). To determine the strength of CI and Rescue in current laboratory settings, group mating assays were performed and egg hatch outcomes were scored for individual females. Crosses of infected females to infected males, referred to as the Rescue, yielded a 92% hatch rate. This was not significantly different from the uninfected Control cross (p > 0.05; adjusted α = 0.0083) (Table 1). By contrast, the CI egg hatch rates ranged from 4 to 10% for wMel and wRi. This represents a significant reduction in hatch rates, as compared to both Control and Rescue crosses (p < 0.001; adjusted α = 0.0083) (Table 1). This outcome is consistent with the expectation of strong CI and Rescue phenotypes associated with D. simulans.
Wolbachia infection has been reported to alter pheromone release and perception by other species of Drosophila, leading to altered mating patterns (Ringo et al., 2011; Schneider et al., 2019). Thus, it is formally possible that low egg hatch in incompatible crosses, normally attributed to wRi-induced CI lethality, instead reflects a failure to mate. To distinguish between these possibilities, single pair matings were set up, with egg hatch scored only for vials in which mating was visually confirmed. In these experiments, hatch rates remained low for CI crosses (10%) as compared to Control crosses (90%) (Table 1) (p < 0.001; adjusted α = 0.0125). The results of these mating-confirmed crosses closely parallels that shown above for group matings. This demonstrates that the low hatch rates currently associated with wRi-induced CI are due to the reduced viability of CI eggs laid by uninfected D. simulans females.
Demonstrating Use of Small Molecule Inhibitors to Suppress CI Phenotypes
CI embryos have previously been shown to exhibit defective incorporation of maternal histones into paternal DNA (Landmann et al., 2009). We reasoned that chromatin-modifying compounds may also be able to confer CI suppression in a manner analogous to natural Rescue. It is known that acetylation of histones lowers their affinity for DNA and loosens chromatin structure, whereas removal of acetyl groups by histone de-acetylase (HDAC) enzymes reverses this effect (Pasyukova and Vaiserman, 2017). Since HDAC inhibitor compounds are commonly used in animal models and clinical settings (Drummond et al., 2005; Ganai et al., 2016; Tandon et al., 2016; Pasyukova and Vaiserman, 2017; Salcedo Magguilli, 2017), an array of well-established compounds is available for testing. Thus, we established a chemical feeding protocol to test a role for chromatin remodeling in CI suppression.
One of the most well-known HDAC inhibitors is the non-toxic, short chain fatty acid butyric acid, or sodium butyrate (NaBu) (Cousens et al., 1979; Davie, 2003). Dose-response assays were performed to determine the maximum tolerable NaBu dosage for adult D. simulans. Doses within a 200-fold range were tested, and the highest dose which did not substantially alter developmental phenotypes was identified (Supplementary Table S2). This dose was used for all subsequent tests for NaBu impact on CI hatch rates. It is not known what stage(s) of oogenesis are important for conferring Rescue ability upon embryos. Thus, uninfected females were kept on drug food throughout the duration of the experiment except during matings with CI males. Males were incubated and mated on control food only to prevent ingestion of the compound (Figure 2).
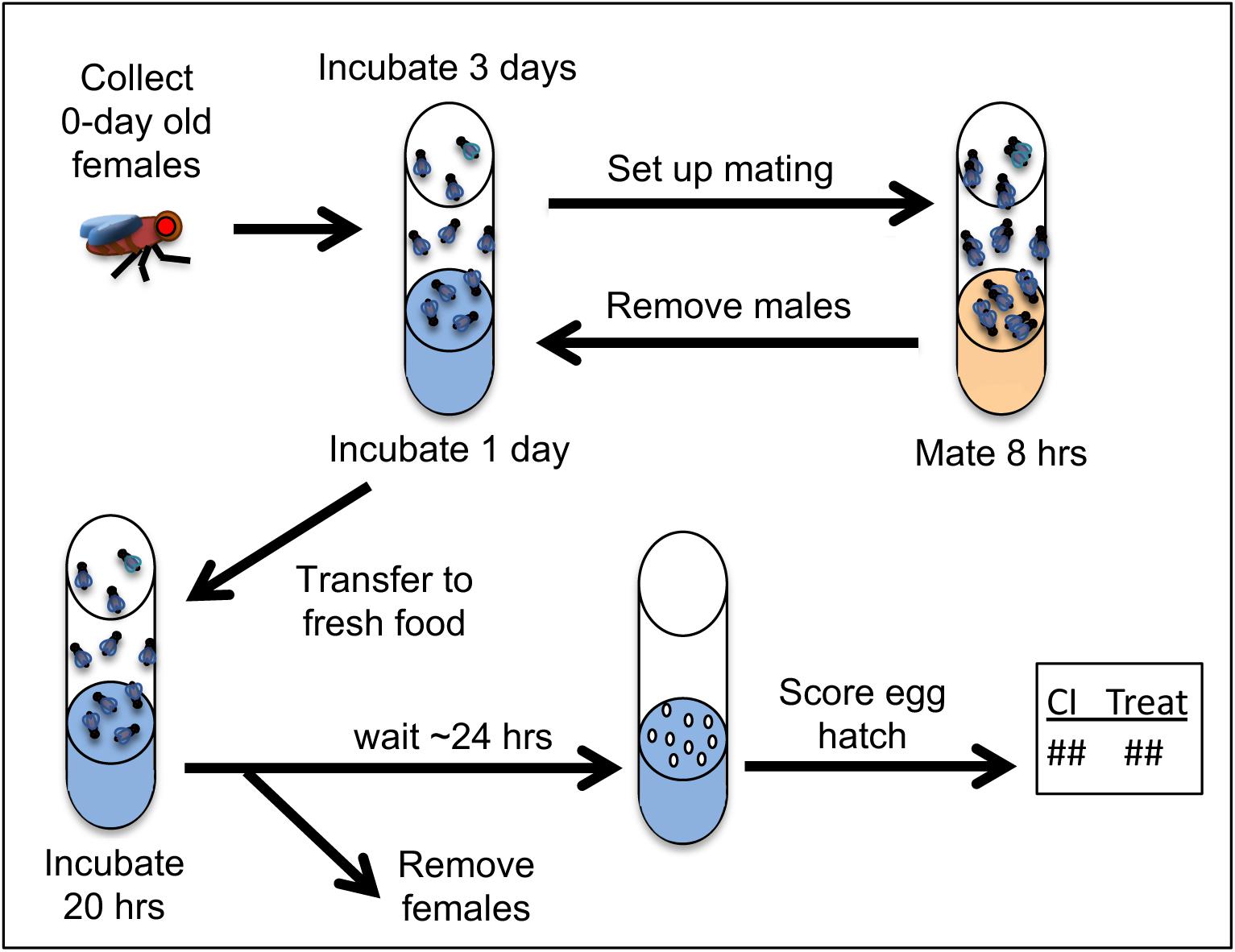
Figure 2. Procedures used for CI- and Rescue-related crosses. Drug feedings were carried out on blue food where specified in the protocol timeline. For details on food preparation, please see section “Materials and Methods” and Additional File S1.
Group mating experiments were performed to determine the impact of NaBu on CI hatch rates associated with wRi and wMel Wolbachia. For wRi, the CI hatch rate was 1.6-fold higher for the NaBu treated vials when compared to control food (p = 0.0122; adjusted α = 0.0125) (Table 2). For wMel, the CI hatch rate on NaBu food was more than twice that of control food conditions (p < 0.001 adjusted α = 0.0125). To further verify whether the increased egg hatch rates with NaBu feeding were due to unanticipated changes in the mating behavior, single pair matings were also performed. The data confirmed a 94% hatch rate for Rescue crosses, as compared to a 10% hatch rate for CI (p < 0.001; adjusted α = 0.0083). For the NaBu treatment condition, CI hatch rates were nearly double that of the CI control (p < 0.001; adjusted α = 0.0083) (Table 2). Taken together, these data indicate that the HDAC inhibitor NaBu induces CI suppression in uninfected females.
Scaling Up Screening of Small Molecule Inhibitors to Test for CI Suppression
The observation that CI can be partially suppressed by NaBu raises potential questions of whether targeting other host factors could confer CI suppression as well. Assessing this possibility requires screening of more compounds, including sufficient replicates to distinguish CI suppression effects. To this end, we developed a plate-based feeding assay for analyzing small molecule effects on CI hatch rates, based upon existing adult screening methods (Markstein et al., 2014) that were previously optimized for use in Wolbachia assays (Christensen et al., 2019). In the context of 24-well plates, a maximum of 8 wells can be analyzed per condition for CI, CI+treatment and Rescue. Ensuring that results would be relevant to those of a naturally robust CI system, wRi-infected males were used to perform all plate assay matings.
To determine whether chemical feeding in a plate assay format is an effective means of identifying CI suppression, the HDAC inhibitor NaBu was retested across five biologically independent plate replicates. All plate replicates indicated consistently higher egg hatch for the CI+NaBu condition (20–30%) than was seen in the CI control condition (11–14%) (p-value range: <0.001 to 0.003) (Figure 3A) (Supplementary Tables S3, S4). It is also possible to consider the data from the perspective of plate-based cell screens, where the quality of such assays is typically described in terms of its Z′ factor. Positive Z′ values, ranging between 0 and 1, only result when the average values for the controls are separated by more than three times the standard deviation for each treatment (Zhang et al., 1999; Birmingham et al., 2009; Serbus et al., 2012). According to this analysis, data from the five NaBu plate replicates returned Z′ values ranging from 0.76 to 0.89 (Supplementary Table S5). The CI+NaBu condition also occupied the intermediate “hit” range, consistently distinguishable from the CI control (Figure 3B). This demonstrates that the plate-based feeding assay reproducibly identifies CI-suppressing treatments.
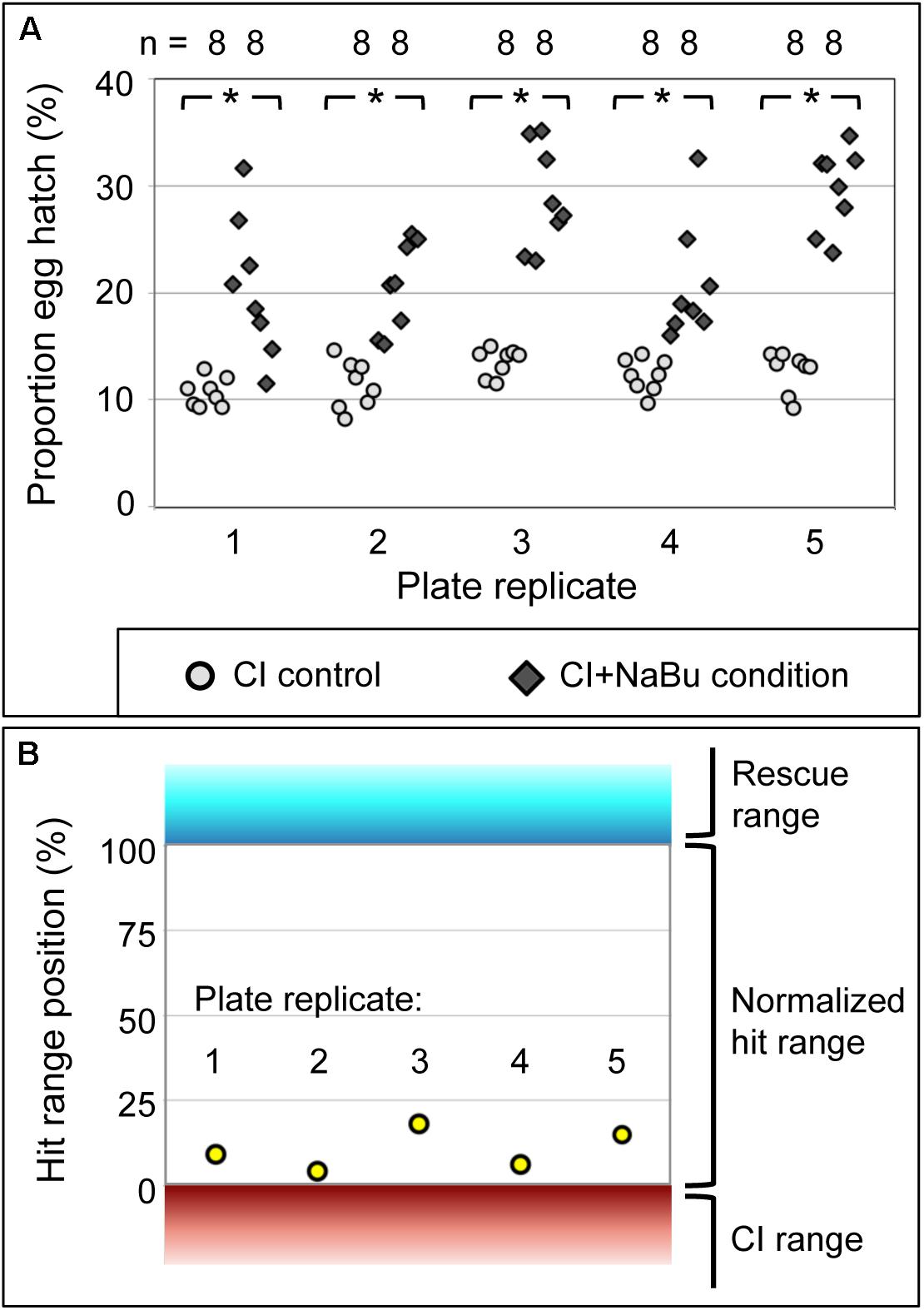
Figure 3. The impact of NaBu treatment on CI egg hatch. (A) Hatch rate data from assay plates using wRi-infected D. simulans flies. Each symbol represents data from a single well. *p < 0.005. (B) NaBu impact on CI, in terms of conventional Z′ analysis. Range boundaries of the CI control (red) and Rescue control (cyan) are indicated. The normalized “hit range” between controls is shown in white. Yellow dots: average hatch rate for the CI+NaBu condition per screening plate, normalized to the range between CI and Rescue controls. For further details, see Supplementary Tables S4, S5.
To determine the quantity of screening plates required for reproducible identification of a chemical suppressor of CI, the NaBu plate data were statistically analyzed. Data were collated and compared for every crosswise pairing of five independent screening plates, using data from 16 wells per condition in each case. Sub-sampling among the five plate replicates indicated that data combinations from any two plate replicates identified a significant difference between CI and CI+NaBu conditions (p < 0.001, n = 10 plate data combinations) (Supplementary Table S4). To further determine how many wells are required for significance, data were sub-sampled from within each of the paired plate datasets (Christensen et al., 2019). Data from eight or more wells per condition were sufficient to identify significant differences between CI and CI+NaBu conditions, when setting an alpha value at 0.05 (Figure 4A). Sampling from 11 or more wells per condition yielded a significant difference with alpha set at 0.01 (n = 10 plate data combinations) (Figure 4B). These data indicate that screening two chemical assay plates is sufficient to detect chemical suppressors of CI. In addition to corroborating NaBu-induced CI suppression, these plate assay data also created the foundation for testing additional candidate compounds.
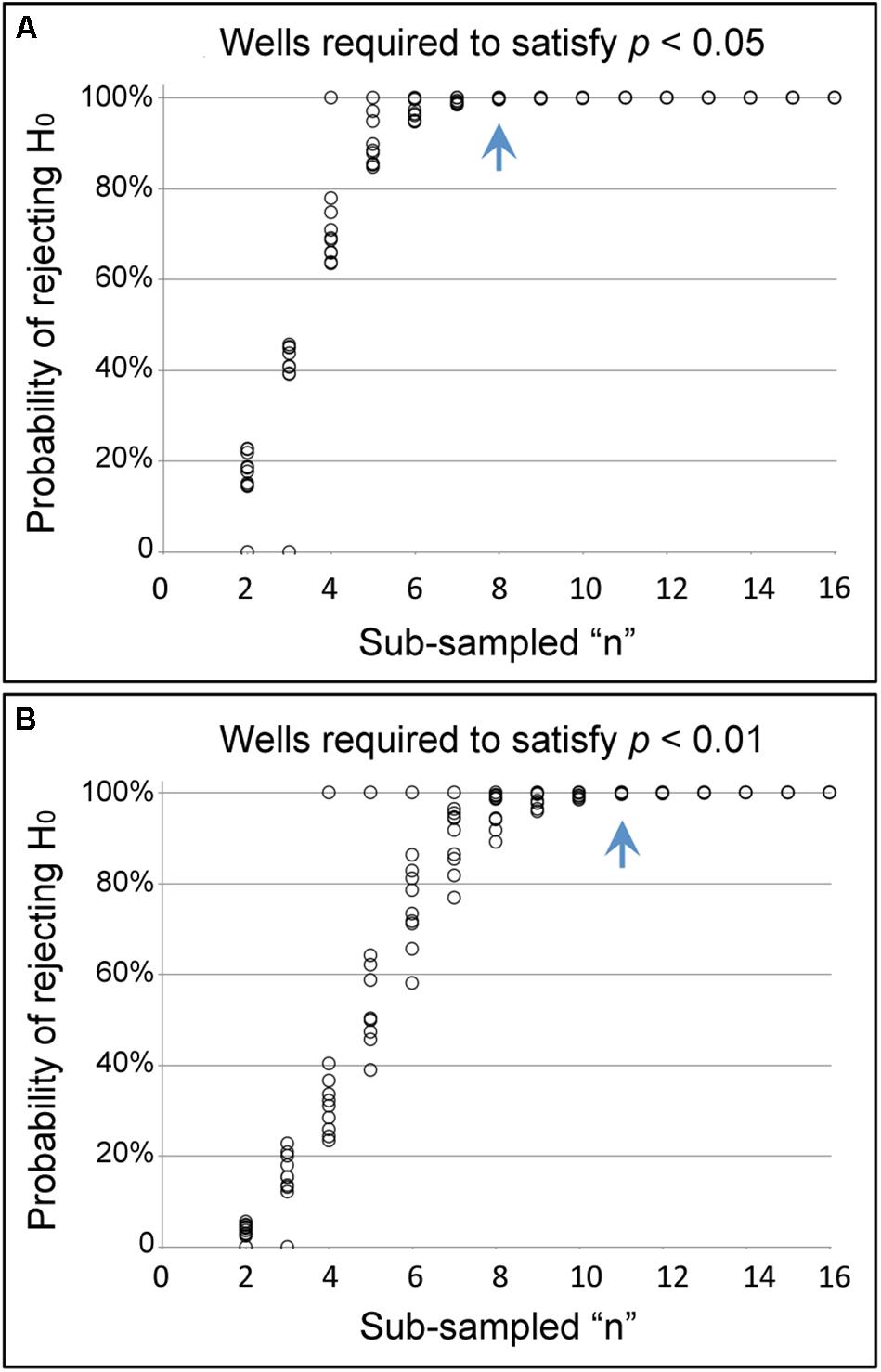
Figure 4. Identifying sufficient sample size in the pate assay format. The graphs show the likelihood of seeing a significant difference in hatch rate between CI and CI+NaBu conditions, as defined by (A) the conventional α-value of 0.05, as well as (B) the more stringent α-value of 0.01. The blue arrow indicates the number of wells at which the probability of rejecting the null hypothesis has reached 99.5% or higher for all sub-sampled datasets analyzed.
Testing for CI Suppression by Short-Chain Fatty Acids and Protein Acetylation Modifier
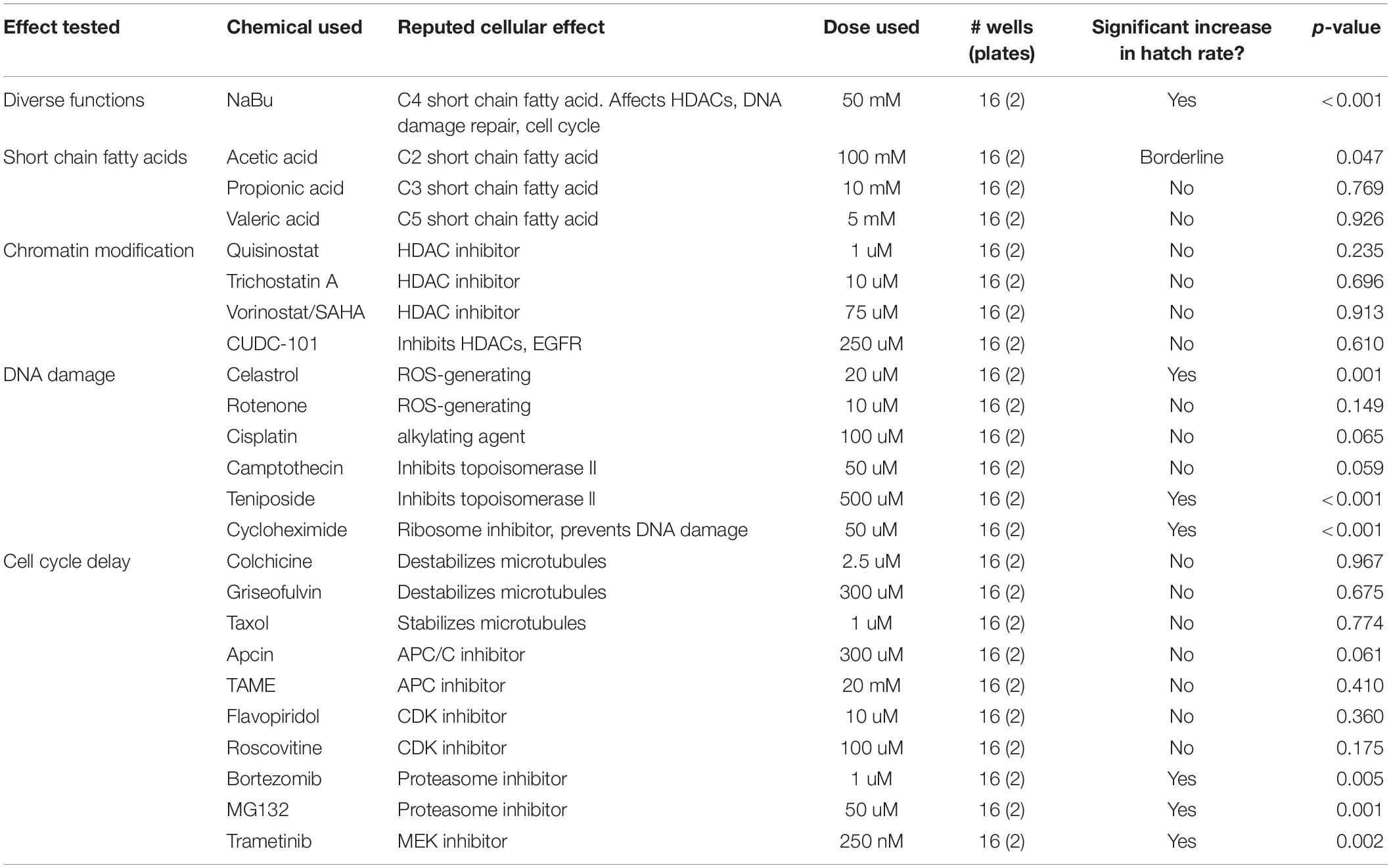
To further pursue the functional role of NaBu in CI suppression, the basic structure of this compound was considered. Since NaBu is a short-chain fatty acid, this opens the question of whether short-chain fatty acids generally exert CI-suppressing effects. To investigate this possibility, flies were fed with three other forms of short-chain fatty acids, specifically acetic, propionic and valeric acid, to test for suppression of wRi-induced CI in D. simulans. The doses used for each compound, as well as all others described below, were empirically determined in vial format (Supplementary Table S6), then tested for impact on CI hatch rates in the context of the plate-based assay. Results from these experiments indicated that acetic acid conferred borderline CI suppression abilities upon uninfected embryos (p = 0.047). Propionic acid and valeric acid had no significant effect on CI hatch rates (Table 3) (Supplementary Tables S7, S8). Thus, CI suppression is not a generalized effect associated with dietary short-chain fatty acids.
NaBu is best known for its impact on chromatin structure and has been credited with suppressing HDACs 1–5 and 7–9, representing the entirety of class I and IIa HDAC enzymes (Ganai et al., 2016; Tandon et al., 2016). To determine whether maternal HDAC function affects wRi-induced CI, an array of HDAC inhibitors was dose-optimized (Supplementary Table S6) and tested in the plate-based assay. The selected inhibitors included the class I and II HDAC inhibitors quisinostat (Arts et al., 2009) and trichostatin A, as well as the pan HDAC inhibitor vorinostat (SAHA) (Ganai et al., 2016; Tandon et al., 2016). The inhibitor CUDC-101, which targets class I and II HDAC as well as growth factor receptors, was also tested (Lai et al., 2010). Remarkably, none of these HDAC inhibitors exerted significant impact on CI hatch rates (Table 3) (Supplementary Tables S7, S8). To confirm that the fly stocks and assay parameters were still performing as expected, the effect of NaBu was retested in the plate-based format. The data confirmed that CI suppression by NaBu was still significant, with a p-value below 0.001 (Table 3) (Supplementary Figure S1 and Supplementary Tables S7–S9). Overall, these results do not support HDAC inhibition and associated chromatin remodeling as a generalized mechanism for CI suppression.
Testing Modifiers of DNA Damage for Maternal CI Suppression Effects
NaBu has also been shown to promote DNA repair, in part by indirectly increasing acetylation of histone H4 (Smerdon et al., 1982; Williamson et al., 2012; Mao and Wyrick, 2016). To test whether maternal DNA repair processes affect wRi-induced CI, an array of inhibitors was pursued. To activate the DNA repair response, agents that induce oxidative DNA damage were selected, specifically celastrol and rotenone (Sanders and Greenamyre, 2013; Xu et al., 2013; Moreira et al., 2019). The alkylating agent cisplatin, which also generates reactive oxygen, was included as well (Basu and Krishnamurthy, 2010; Podratz et al., 2011; Rezaee et al., 2013). Topoisomerase inhibitors were also used, including camptothecin, which prevents re-sealing of single stranded nicks by topoisomerase I, and teniposide, which prevents removal of topoisomerase II from DNA and induces degradation of the enzyme (Rowe et al., 1986; Hartmann and Lipp, 2006; Nitiss, 2009). As the ribosome inhibitor cycloheximide has been reported to prevent formation of single- and double-stranded DNA breaks (Yoshioka et al., 1987; Lorico et al., 1988), this drug was also tested.
Using optimized doses (Supplementary Table S6), the plate-based feeding assay indicated CI suppression for half of the treatment conditions used. A significant increase in hatch rate was observed for uninfected females exposed to celastrol, teniposide and cycloheximide, all associated with p-values of 0.001 or less (Table 3) (Supplementary Figure S1 and Supplementary Tables S7–S9). These data open a possible role for maternal processes that prevent and repair DNA damage in conferring CI suppression upon uninfected embryos.
Testing the Impact of Cell-Cycle Timing on Maternal CI Suppression
Exposure to NaBu has been shown to slow cell cycle timing (D’Anna et al., 1980; Lallemand et al., 1996). To test the effect of maternal cell cycle timing on wRi-induced CI, uninfected D. simulans were exposed to an array of complementary inhibitors. In attempt to slow the progression of mitosis by altering microtubule dynamics, the microtubule destabilizers colchicine and griseofulvin, as well as the microtubule stabilizer taxol were tested (Singh et al., 2008; Stanton et al., 2011). To slow anaphase onset and exit from mitosis, inhibitors of the anaphase promoting complex, apcin and TAME, were used (Zeng et al., 2010; Sackton et al., 2014). To inhibit the progress of mitosis and the cell cycle overall, the Cyclin dependent kinase inhibitors flavopiridol and roscovitine were used (Gray et al., 1999; Cicenas et al., 2015; Bailon-Moscoso et al., 2017), as well as the proteasome inhibitors bortezomib and MG132 (Goldberg, 2012; Rastogi and Mishra, 2012). To stall general re-entry into the cell cycle, the MAPKK (MEK) inhibitor trametinib was also used (Zeiser, 2014; Kurata et al., 2016).
After identifying appropriate doses (Supplementary Table S6), chemical manipulators of cell cycle timing were tested for CI suppression. The plate assay data indicated significantly increased CI hatch rates for bortezomib, MG132, and trametinib-fed females compared to control (p-value range: 0.001–0.005) (Table 3) (Supplementary Figure S1 and Supplementary Tables S7–S9). As cell cycle delays are a recognized consequence of DNA damage, resulting from checkpoint activation that allows damage repair (Chao et al., 2017), these data are consistent with a possible role for altered embryonic cell cycle timing in suppression of CI.
Re-testing CI-Suppressing Compounds Against Transinfected D. simulans
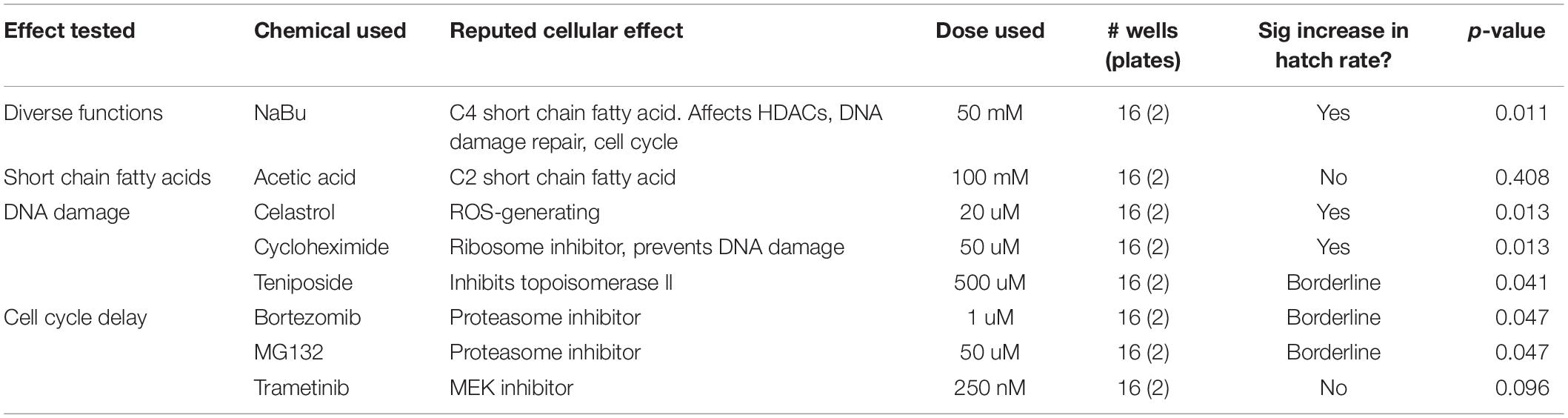
If the compounds that suppress wRi-induced CI act upon a network of conserved, Rescue-related maternal interactions, the effects would be expected to be applicable to other host-strain combinations. To this end, the chemicals identified above as hits were analyzed for suppression of wMel-induced CI as well, using flies from a transinfected stock population (Poinsot et al., 1998). Specifically, NaBu, celastrol, cycloheximide, teniposide, bortezomib, MG132, trametinib, and the initially borderline hit acetic acid (Table 3) were retested in the plate assay format. The same dosing and procedures were used as above, with the only difference being that wMel-infected males were used for CI induction.
The results indicated that NaBu, celastrol, and cycloheximide significantly elevated CI hatch rates for wMel-induced CI (p-value range: 0.011–0.013) (Table 4) (Supplementary Figure S2 and Supplementary Tables S10–S12). By contrast, teniposide, bortezomib, and MG132 treatments exhibited borderline CI suppression effects (p-value range 0.041–0.047). Trametinib and acetic acid did not induce any significant effects (Table 4) (Supplementary Tables S10, S11). This outcome distinguishes cellular responses associated with certain DNA damage and/or cell cycle timing regulators as general contributors to maternal suppression of CI in D. simulans.
Testing Combined Pathway Effects for Suppression of CI
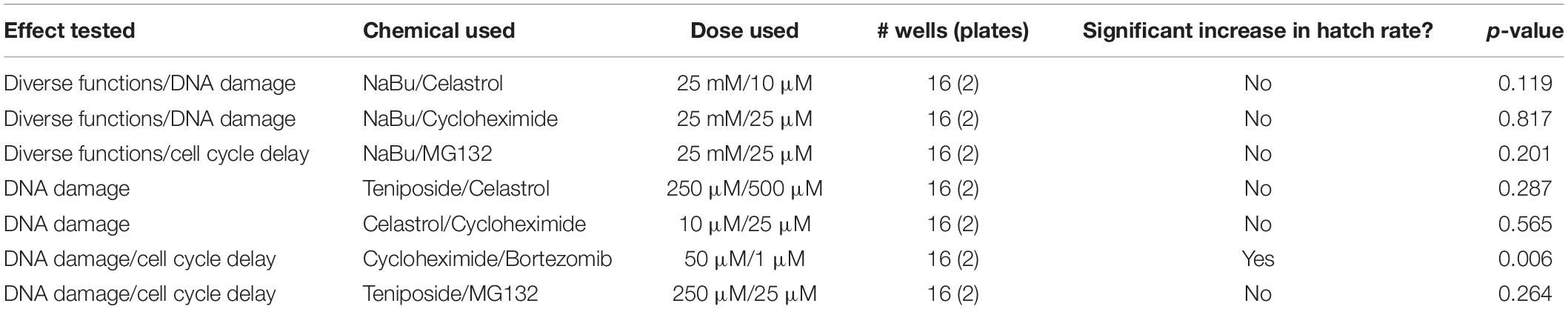
To further test the extent to which suppression of CI under these treatments is due to a shared network of pathway functions, a dual chemical treatment strategy was pursued. We focused on hits that elicited the most robust CI suppression across both systems tested, namely: NaBu, celastrol, and cycloheximide. Additional treatment combinations included compounds that exerted more modest effects, namely: teniposide, bortezomib, and MG132. As for the single drug trials, dose response curves were carried out for all pairwise chemical treatments on uninfected D. simulans flies (Supplementary Table S13). Nearly all treatment combinations required reduced drug dosing, as compared to singly administered treatments (Supplementary Table S6). The cycloheximide/bortezomib combination was the only case in which original treatment doses for both compounds could be tolerated additively, without adverse effects on fecundity, egg hatch or larval development (Supplementary Table S13).
After uninfected D. simulans females were treated with dual drug combinations, their egg hatch rates were compared against females raised on control food in the plate assay format. To ensure that the results would be representative of natural CI, wRi-infected males were used to induce CI in this series of experiments. The results indicated that the cycloheximide/bortezomib combination significantly increased the CI hatch rate as compared to the CI control (p = 0.006) (Table 5) (Supplementary Tables S14, S15). No other paired chemical treatments induced CI suppression (Table 5). It is possible that loss of CI suppression effects is attributable to reduced combinatorial doses of otherwise effective compounds. Regardless, the data indicate CI suppression by compounds that are traditionally associated with manipulation of protein synthesis and protein turnover.
Discussion
This study was designed to inform host capacity for modifying CI outcomes in the context of non-model organisms. A strong case is currently being made for involvement of Wolbachia Cif proteins in induction of CI and Rescue, based on analysis of D. melanogaster and yeast models (Beckmann et al., 2017, 2019b; LePage et al., 2017; Shropshire et al., 2018; Chen et al., 2019). This study, using natural and transinfected D. simulans, add to the complex biological underpinnings of embryonic lethality by indicating that maternal contributions to Rescue are also possible. Demonstrating CI suppression through chemical feeding of uninfected females, without invoking any effect or contribution by Wolbachia-supplied antitoxins, opens the possibility of a role for host contributions to Rescue. Fundamental to these questions is whether a core set of maternal mechanisms in insects can act to suppress CI across systems. Fortunately, the framework presented here enables broad investigation of diverse CI-Rescue systems in the future, including that of non-Wolbachia endosymbionts like Cardinium (Hunter et al., 2003; Penz et al., 2012; Mann et al., 2017; Takano et al., 2017; König et al., 2019; Doremus and Hunter, 2020).
Despite the reproducibility and statistical significance of CI-suppression effects by multiple drugs and drug combinations, the limitations of the current work are reflected by CI hatch rates which did not exceed three times that of CI-control hatch rates. The disparity between the 90%+ egg hatch frequencies of Wolbachia-induced Rescue and that of chemical CI suppression can be due to a variety of factors or experimental limitations. One important aspect could be the absence of Wolbachia-supplied Cif proteins. It is possible that maternal mechanisms act as a supplement to, or in coordination with Cif functions to a substantial extent. It also remains possible that CI-suppressing treatments mimic Cif effects, even in the absence of a usual role for host factors in Rescue. There are no known chemical treatments that will specifically mimic predicted Cif proteins at this time (Beckmann et al., 2017; Lindsey et al., 2018). Our attempt to alter broader functional networks of maternal proteins by use of multiple inhibitors was met with limited success, possibly due to dosage considerations, limited by systemic tolerances for dual treatments.
When interpreting the CI suppression data yielded by the current chemical screen, the technical limitations inherent to the method itself are important to consider. Whole body feedings lack the time and tissue-specific nuance afforded to Wolbachia in vivo. While dosing within the food was standardized for this study, it is not possible to control for local dosing to tissues/cells of the recipient organism. This is due to differences in ingestion, absorption, efflux, metabolism, and/or excretion rates, which are expected to vary in association with each cell type and each drug. A feeding assay also creates the possibility for side effects due to host microbiome impacts. Initial attempts to run this screen using standardized micro-injections were curtailed by observations that injected D. simulans flies stop laying eggs. Working with flies under anexic or gnotobiotic conditions presents its own set of complications (Koyle et al., 2016). For these reasons, seeing a response from this chemical screen is informative, whereas the lack of a response is not. Distinct from most drug screens, whose interpretations are limited by typically detrimental health effects on the test subject, the output of this screen leads to increased survival of otherwise ill-fated embryos.
This study was designed to address contributions of host chromatin remodeling, DNA damage repair and cell cycle timing impacts on CI suppression in vivo. Though little support was evident for chromatin remodeling in CI suppression, impacts on DNA integrity and cell cycle timing conferred significant increases in CI egg hatch. As such, our results at present do not readily distinguish between existing models of CI and Rescue, but instead opens consideration of networked models that may better reflect the cell biology of CI. DNA damage and cell cycle timing are well-known to be intrinsically connected, since DNA damage triggers a checkpoint mechanism that arrests the cell cycle (Alberts et al., 2015; Chao et al., 2017). Variation upon functions of the ubiquitin-proteasome system may further link these processes. DNA damage repair is facilitated by ubiquitination of histones and DNA repair pathway proteins, followed by their deubiquitination upon completion of the repair (Cohn and D’Andrea, 2008; Stadler and Richly, 2017; Uckelmann and Sixma, 2017). Ubiquitination and degradation of cyclins and other regulators is also fundamentally required for cell cycle progression (Bassermann et al., 2014; Alberts et al., 2015). The deubiquitylase and nuclease functions of Cif proteins converge upon these same processes (Beckmann and Fallon, 2013; Beckmann et al., 2017, 2019a,b; Lindsey et al., 2018; Chen et al., 2019), and expression studies in CI-inducing Cardinium have implicated the ubiquitin-proteasome system and DNA repair as well (Mann et al., 2017). The CI-suppressing compounds identified in this study reflect this continuum of function.
One question raised by this work is what distinguished the compounds that affected egg hatch for both wRi and wMel-induced CI, from those that affected wRi-induced CI only (Figure 5). As wRi and wMel encode different types of Cif proteins, Cin versus Cid, respectively (Beckmann and Fallon, 2013), it would be reasonable for CI associated with different strains to respond differently to the same CI-suppressing compounds. In general, this study found that “hit” compounds that suppressed wRi-induced CI exerted overall weaker effects on wMel-induced CI. It is not clear whether differences in Cif proteins are responsible, as other circumstantial explanations could also cause this outcome. Future tests of CI-suppressing compounds across more host-strain combinations will be needed to make a stronger correlative statement addressing this point.
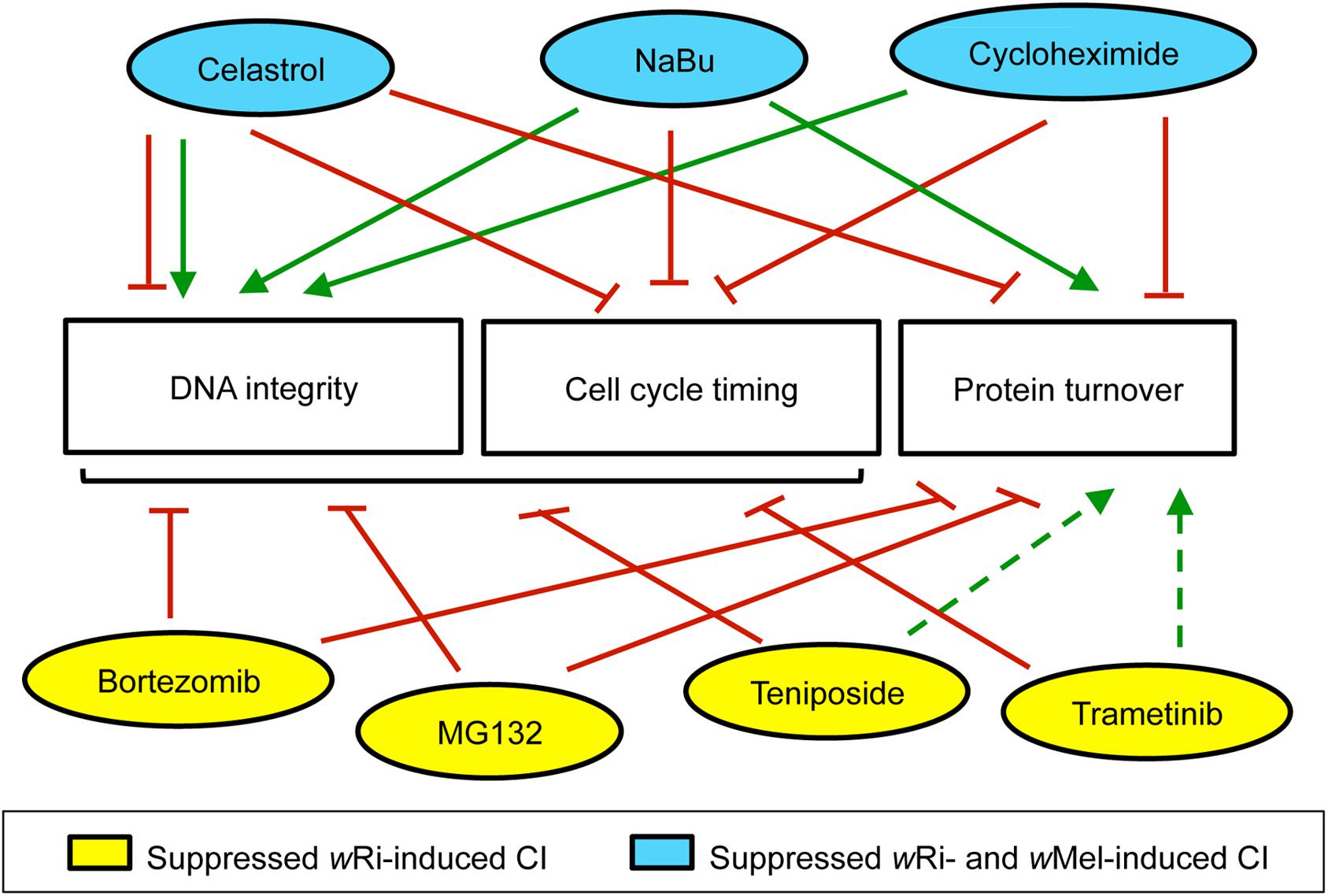
Figure 5. Summary of maternal impacts that significantly increased D. simulans CI hatch rates. This information is based upon available literature, summarized in Additional File S3. Green arrows: positive impact. Red lines: negative effect. Dotted lines: interpretation based upon partial datasets. Bracket: includes multiple categories.
Another perspective for interpretation regarding the cellular impacts of the compounds themselves. Is there anything distinct about the activity profile of compounds that suppress both wRi and wMel-induced CI? The use of well-known compounds in this study provides access to vast literature for interpreting reproducible CI suppression effects. Past studies of mammalian systems and cell lines provide a patchwork of information that informs effects on DNA integrity, cell cycle timing and protein turnover. According to existing literature, the compounds identified here as CI-suppressing agents act as consistent suppressors of cell cycle timing, but exert variable impacts on protein turnover, depending upon the compound (Additional File S3). Although neither of those functional profiles aligns with the strain-specific differences observed in CI egg hatch outcomes thus far (Figure 5).
Clearer associations are evident between existing literature and CI suppression outcomes in this study from the perspective of DNA integrity. The four compounds that suppress wRi-induced CI are known inducers of DNA damage, whereas the three compounds that suppress both wRi- and wMel-induced CI, reportedly support DNA integrity (Figure 5) (Additional File S3). Though celastrol can exert detrimental impacts on DNA (Han et al., 2018; Wang et al., 2020), it has also been shown to suppress radiation-induced damage (Xu et al., 2013; Moreira et al., 2019). Cycloheximide treatments prevent formation of single- and double-strand DNA breaks (Yoshioka et al., 1987; Lorico et al., 1988). NaBu protects DNA integrity by up-regulating antioxidant pathways and by facilitating DNA repair (Smerdon et al., 1982; Mao and Wyrick, 2016; El-Shorbagy, 2017). This suggests that DNA integrity is a dynamic, focal aspect of CI suppression with respect to different Wolbachia strains in D. simulans. Future analyses of CI and Rescue, from the perspective of both host and microbe promise to be informative in elucidating the molecular basis of this ecologically relevant mechanism.
Data Availability Statement
The datasets GENERATED for this study can be found in NCBI SRA https://www.ncbi.nlm.nih.gov/bioproject/PRJNA663645.
Author Contributions
AJMZM, AAS, SS, SC, and LS supervised the experiments. AJMZM, AAS, JGA, SS, AF, JH, and SC conducted the experiments. AJMZM, AAS, JH, SC, and LS wrote the manuscript. All authors designed the experiments and reviewed the manuscript.
Funding
This work was supported the FIU startup fund, the FIU Biology graduate program, the FIU College of Arts, Sciences and Education, the FIU McNair program (P217A170301), and NSF-IOS-SDS (1656811).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Serbus lab, past and present, William Sullivan, Michael Turelli, the FIU Department of Biological Sciences, and the FIU Biomolecular Sciences Institute for helpful discussions and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.576844/full#supplementary-material
Supplementary Figure 1 | Sub-sampling analyses, in tests for chemical suppression of wRi-induced CI. The likelihood of satisfying set p-values is shown for “hit” compounds identified by chemical screening. Compound treatments are color coded as indicated at the bottom of the figure. For NaBu, the data reached 100% satisfaction of the cutoff p < 0.05 at 12 wells, and p < 0.01 at 15 wells. For Teniposide, the data reached 100% satisfaction of the cutoff p < 0.05 at 11 wells, and p < 0.01 at 14 wells. All other conditions showed a 95%+ likelihood of satisfying p < 0.05, and an 80%+ likelihood of satisfying p < 0.01, at n = 16 wells.
Supplementary Figure 2 | Sub-sampling analyses, in tests for chemical suppression of wMel-induced CI. The likelihood of satisfying set p-values is shown for “hit” compounds identified by chemical screening. Compound treatments are color coded as indicated at the bottom of the figure. The data showed a 72–82% likelihood of satisfying p < 0.05, and a 47–72% likelihood of satisfying p < 0.01, at n = 16 wells.
Footnotes
References
Alberts, B., Johnson, A., Lewis, J., Morgan, D., Raff, M., Roberts, K., et al. (2015). Molecular Biology of the Cell. New York, NY: Garland Science.
Andrews, E. S., Crain, P. R., Fu, Y., Howe, D. K., and Dobson, S. L. (2012). Reactive oxygen species production and Brugia pahangi survivorship in Aedes polynesiensis with artificial Wolbachia infection types. PLoS Pathog. 8:e1003075. doi: 10.1371/journal.ppat.1003075
Arts, J., King, P., Mariën, A., Floren, W., Beliën, A., Janssen, L., et al. (2009). JNJ-26481585, a novel “second-generation” oral histone deacetylase inhibitor, shows broad-spectrum preclinical antitumoral activity. Clin. Cancer Res. 15, 6841–6851. doi: 10.1158/1078-0432.ccr-09-0547
Bailon-Moscoso, N., Cevallos-Solorzano, G., Romero-Benavides, J. C., and Orellana, M. I. (2017). Natural compounds as modulators of cell cycle arrest: application for anticancer chemotherapies. Curr. Genomics. 18, 106–131. doi: 10.2174/1389202917666160808125645
Bassermann, F., Eichner, R., and Pagano, M. (2014). The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta 1843, 150–162. doi: 10.1016/j.bbamcr.2013.02.028
Basu, A., and Krishnamurthy, S. (2010). Cellular responses to cisplatin-induced DNA damage. J. Nucleic Acids 2010:201367.
Beckmann, J. F., Bonneau, M., Chen, H., Hochstrasser, M., Poinsot, D., Merçot, H., et al. (2019a). The toxin-antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet. 35, 175–185. doi: 10.1016/j.tig.2018.12.004
Beckmann, J. F., Sharma, G. D., Mendez, L., Chen, H., and Hochstrasser, M. (2019b). The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. eLife 8:e50026.
Beckmann, J. F., and Fallon, A. M. (2013). Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 43, 867–878. doi: 10.1016/j.ibmb.2013.07.002
Beckmann, J. F., Ronau, J. A., and Hochstrasser, M. (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2:17007.
Bian, G., Joshi, D., Dong, Y., Lu, P., Zhou, G., Pan, X., et al. (2013). Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 340, 748–751. doi: 10.1126/science.1236192
Binnington, K. C., and Hoffmann, A. A. (1989). Wolbachia-like organisms and cytoplasmic incompatibility in Drosophila simulans. J. Invert Pathol. 54, 344–352. doi: 10.1016/0022-2011(89)90118-3
Birmingham, A., Selfors, L. M., Forster, T., Wrobel, D., Kennedy, C. J., Shanks, E., et al. (2009). Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6, 569–575. doi: 10.1038/nmeth.1351
Bonneau, M., Atyame, C., Beji, M., Justy, F., Cohen-Gonsaud, M., Sicard, M., et al. (2018a). Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat. Commun. 9:319.
Bonneau, M., Landmann, F., Labbé, P., Justy, F., Weill, M., and Sicard, M. (2018b). The cellular phenotype of cytoplasmic incompatibility in Culex pipiens in the light of cidB diversity. PLoS Pathog. 14:e1007364. doi: 10.1371/journal.ppat.1007364
Bonneau, M., Caputo, B., Ligier, A., Caparros, R., Unal, S., Perriat-Sanguinet, M., et al. (2019). Variation in Wolbachia cidB gene, but not cidA, is associated with cytoplasmic incompatibility mod phenotype diversity in Culex pipiens. Mol. Ecol. 28, 4725–4736. doi: 10.1111/mec.15252
Boyle, L., O’Neill, S. L., Robertson, H. M., and Karr, T. L. (1993). Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260, 1796–1799. doi: 10.1126/science.8511587
Breeuwer, J. A., and Werren, J. H. (1990). Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346, 558–560. doi: 10.1038/346558a0
Brennan, L. J., Haukedal, J. A., Earle, J. C., Keddie, B., and Harris, H. L. (2012). Disruption of redox homeostasis leads to oxidative DNA damage in spermatocytes of Wolbachia-infected Drosophila simulans. Insect Mol. Biol. 21, 510–520. doi: 10.1111/j.1365-2583.2012.01155.x
Brennan, L. J., Keddie, B. A., Braig, H. R., and Harris, H. L. (2008). The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS One 3:e2083. doi: 10.1371/journal.pone.0002083
Callaini, G., Dallai, R., and Riparbelli, M. G. (1997). Wolbachia-induced delay of paternal chromatin condensation does not prevent maternal chromosomes from entering anaphase in incompatible crosses of Drosophila simulans. J. Cell Sci. 110(Pt 2), 271–280.
Callaini, G., and Riparbelli, M. G. (1996). Fertilization in Drosophila melanogaster: centrosome inheritance and organization of the first mitotic spindle. Dev. Biol. 176, 199–208. doi: 10.1006/dbio.1996.0127
Camacho, M., Oliva, M., and Serbus, L. R. (2017). Dietary saccharides and sweet tastants have differential effects on colonization of. Biol. Open 6, 1074–1083. doi: 10.1242/bio.023895
Chao, H. X., Poovey, C. E., Privette, A. A., Grant, G. D., Chao, H. Y., Cook, J. G., et al. (2017). Orchestration of DNA damage checkpoint dynamics across the human cell cycle. Cell Syst. 5, 445.e5–459.e5.
Charlat, S., Calmet, C., and Merçot, H. (2001). On the mod resc model and the evolution of Wolbachia compatibility types. Genetics 159, 1415–1422.
Chen, H., Ronau, J. A., Beckmann, J. F., and Hochstrasser, M. (2019). A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl. Acad. Sci. U.S.A. 116, 22314–22321. doi: 10.1073/pnas.1914571116
Christensen, S., Camacho, M., Sharmin, Z., Momtaz, A. J. M. Z., Perez, L., Navarro, G., et al. (2019). Quantitative methods for assessing local and bodywide contributions to Wolbachia titer in maternal germline cells of Drosophila. BMC Microbiol. 19:206. doi: 10.1186/s12866-019-1579-3
Christensen, S., Pérez Dulzaides, R., Hedrick, V. E., Momtaz, A. J., Nakayasu, E. S., Paul, L. N., et al. (2016). Wolbachia endosymbionts modify drosophila ovary protein levels in a context-dependent manner. Appl. Environ. Microbiol. 82, 5354–5363. doi: 10.1128/aem.01255-16
Cicenas, J., Kalyan, K., Sorokinas, A., Stankunas, E., Levy, J., Meskinyte, I., et al. (2015). Roscovitine in cancer and other diseases. Ann. Transl. Med. 3:135.
Cohn, M. A., and D’Andrea, A. D. (2008). Chromatin recruitment of DNA repair proteins: lessons from the fanconi anemia and double-strand break repair pathways. Mol. Cell 32, 306–312. doi: 10.1016/j.molcel.2008.10.009
Cousens, L. S., Gallwitz, D., and Alberts, B. M. (1979). Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 254, 1716–1723.
D’Anna, J. A., Tobey, R. A., and Gurley, L. R. (1980). Concentration-dependent effects of sodium butyrate in Chinese hamster cells: cell-cycle progression, inner-histone acetylation, histone H1 dephosphorylation, and induction of an H1-like protein. Biochemistry 19, 2656–2671. doi: 10.1021/bi00553a019
Davie, J. R. (2003). Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133(7 Suppl.), 2485S–2493S.
Doremus, M. R., and Hunter, M. S. (2020). “The saboteur’s tools: common mechanistic themes across manipulative symbioses,” in Advances in Insect Physiology, eds K. M. Oliver and J. A. Russell (London: Academic Press.), 317–353.
Drummond, D. C., Noble, C. O., Kirpotin, D. B., Guo, Z., Scott, G. K., and Benz, C. C. (2005). Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45, 495–528.
Duron, O., Bernard, C., Unal, S., Berthomieu, A., Berticat, C., and Weill, M. (2006). Tracking factors modulating cytoplasmic incompatibilities in the mosquito Culex pipiens. Mol. Ecol. 15, 3061–3071. doi: 10.1111/j.1365-294x.2006.02996.x
El-Shorbagy, H. M. (2017). Potential anti-genotoxic effect of sodium butyrate to modulate induction of DNA damage by tamoxifen citrate in rat bone marrow cells. Cytotechnology 69, 89–102. doi: 10.1007/s10616-016-0039-2
Fast, E. M., Toomey, M. E., Panaram, K., Desjardins, D., and Kolaczyk, and Frydman, H. M. (2011). Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334, 990–992. doi: 10.1126/science.1209609
Ferree, P. M., Frydman, H. M., Li, J. M., Cao, J., Wieschaus, E., and Sullivan, W. (2005). Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1:e14. doi: 10.1371/journal.ppat.0010014
Field, A. (2013). Discovering Statistics Using IBM SPSS Statistics, 4th Edn. Newbury Park, CA: Sage.
Ganai, S. A., Ramadoss, M., and Mahadevan, V. (2016). Histone Deacetylase (HDAC) Inhibitors - emerging roles in neuronal memory, learning, synaptic plasticity and neural regeneration. Curr. Neuropharmacol. 14, 55–71. doi: 10.2174/1570159x13666151021111609
Goldberg, A. L. (2012). Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588. doi: 10.1083/jcb.201210077
Gray, N., Détivaud, L., Doerig, C., and Meijer, L. (1999). ATP-site directed inhibitors of cyclin-dependent kinases. Curr. Med. Chem. 6, 859–875.
Hadfield, S. J., and Axton, J. M. (1999). Germ cells colonized by endosymbiotic bacteria. Nature 402:482. doi: 10.1038/45002
Han, X. B., Tan, Y., Fang, Y. Q., and Li, F. (2018). Protective effects of celastrol against γ irradiation-induced oxidative stress in human umbilical vein endothelial cells. Exp. Ther. Med. 16, 685–694.
Hartmann, J. T., and Lipp, H. P. (2006). Camptothecin and podophyllotoxin derivatives: inhibitors of topoisomerase I and II - mechanisms of action, pharmacokinetics and toxicity profile. Drug Saf. 29, 209–230. doi: 10.2165/00002018-200629030-00005
Hertig, M. (1936). The rickettsia, Wolbachia pipientis (gen. et sp. n.) and associated inclusions of the mosquito, Culex pipiens. Parasitology 28, 453–486. doi: 10.1017/s0031182000022666
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457. doi: 10.1038/nature10356
Hoffmann, A. A., and Turelli, M. (1997). “Cytoplasmic incompatibility in insects,” in Influential Passengers: Microorganisms and Invertebrate Reproduction, eds S. O’Neill, A. A. Hoffmann, and J. H. Werren (Oxford: Oxford University Press), 42–80.
Hoffmann, A. A., Turelli, M., and Simmons, G. M. (1986). Unidirectional incompatibility between populations of Drosophila Simulans. Evolution 40, 692–701. doi: 10.2307/2408456
Hsiao, C., and Hsiao, T. H. (1985). Rickettsia as the cause of cytoplasmic incompatibility in the alfalfa weevil, Hypera postica. J. Invert Pathol. 45, 244–246. doi: 10.1016/0022-2011(85)90016-3
Hughes, G. L., Ren, X., Ramirez, J. L., Sakamoto, J. M., Bailey, J. A., Jedlicka, A. E., et al. (2011). Wolbachia infections in Anopheles gambiae cells: transcriptomic characterization of a novel host-symbiont interaction. PLoS Pathog. 7:e1001296. doi: 10.1371/journal.ppat.1001296
Hunter, M. S., Perlman, S. J., and Kelly, S. E. (2003). A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc. Biol. Sci. 270, 2185–2190. doi: 10.1098/rspb.2003.2475
Hurst, G. D. D., Jiggins, F. M., Schulenberg, J. H. G., Bertrand, D., West, S. A., and Goriacheva, I. I. (1999). Male-killing Wolbachia in two species of insect. Proc. R. Soc. Lond. B 266, 735–740.
Jiggins, F. M., Hurst, G. D. D., and Majerus, M. E. N. (1998). Sex ratio distortion in Acraea encedon (Lepidoptera: Nymphalidae) is caused by a male-killing bacterium. Heredity 81, 87–91. doi: 10.1046/j.1365-2540.1998.00357.x
König, K., Zundel, P., Krimmer, E., König, C., Pollmann, M., Gottlieb, Y., et al. (2019). Reproductive isolation due to prezygotic isolation and postzygotic cytoplasmic incompatibility in parasitoid wasps. Ecol. Evol. 9, 10694–10706. doi: 10.1002/ece3.5588
Koyle, M. L., Veloz, M., Judd, A. M., Wong, A. C., Newell, P. D., Douglas, A. E., et al. (2016). Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J. Vis. Exp. 113:54219.
Kriesner, P., Hoffmann, A. A., Lee, S. F., Turelli, M., and Weeks, A. R. (2013). Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9:e1003607. doi: 10.1371/journal.ppat.1003607
Kurata, K., Onoda, N., Noda, S., Kashiwagi, S., Asano, Y., Hirakawa, K., et al. (2016). Growth arrest by activated BRAF and MEK inhibition in human anaplastic thyroid cancer cells. Int. J. Oncol. 49, 2303–2308. doi: 10.3892/ijo.2016.3723
LaFlair, G. T., Egbert, J., and Plonsky, L. (2015). “A practical guide to bootstrapping descriptive statistics, correlations, T tests, and ANOVAs,” in Advancing Quantitative Methods in Second Language Research, 1st Edn, ed. L. Plonsky (New York, NY: Routledge), 378.
Lai, C. J., Bao, R., Tao, X., Wang, J., Atoyan, R., Qu, H., et al. (2010). CUDC-101, a multitargeted inhibitor of histone deacetylase, epidermal growth factor receptor, and human epidermal growth factor receptor 2, exerts potent anticancer activity. Cancer Res. 70, 3647–3656. doi: 10.1158/0008-5472.can-09-3360
Lallemand, F., Courilleau, D., Sabbah, M., Redeuilh, G., and Mester, J. (1996). Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem. Biophys. Res. Commun. 229, 163–169. doi: 10.1006/bbrc.1996.1774
Landmann, F., Orsi, G. A., Loppin, B., and Sullivan, W. (2009). Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 5:e1000343. doi: 10.1371/journal.ppat.1000343
Lassy, C. W., and Karr, T. L. (1996). Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mech. Dev. 57, 47–58. doi: 10.1016/0925-4773(96)00527-8
Laven, H. (1959). Speciation by cytoplasmic isolation in the Culex pipiens-complex. Cold Spring Harb. Symp. Quant. Biol. 24, 166–173. doi: 10.1101/sqb.1959.024.01.017
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Lim, T. S., and Loh, W. Y. (1996). A comparison of tests and equality of variances. Comput. Stat. Data Anal. 22, 287–301. doi: 10.1016/0167-9473(95)00054-2
Lindsey, A. R. I., Rice, D. W., Bordenstein, S. R., Brooks, A. W., and Newton, I. L. G. (2018). Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol. Evol. 10, 434–451. doi: 10.1093/gbe/evy012
Lorico, A., Toffoli, G., Boiocchi, M., Erba, E., Broggini, M., Rappa, G., et al. (1988). Accumulation of DNA strand breaks in cells exposed to methotrexate or N10-propargyl-5,8-dideazafolic acid. Cancer Res. 48, 2036–2041.
Mann, E., Stouthamer, C. M., Kelly, S. E., Dzieciol, M., Hunter, M. S., and Schmitz-Esser, S. (2017). Transcriptome sequencing reveals novel candidate genes for cardinium hertigii-caused cytoplasmic incompatibility and host-cell interaction. mSystems 2:e00141-17.
Mao, P., and Wyrick, J. J. (2016). Emerging roles for histone modifications in DNA excision repair. FEMS Yeast Res. 16:fow090. doi: 10.1093/femsyr/fow090
Markstein, M., Dettorre, S., Cho, J., Neumüller, R. A., Craig-Müller, S., and Perrimon, N. (2014). Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. U.S.A. 111, 4530–4535. doi: 10.1073/pnas.1401160111
McDonald, J. H. (2014). Handbook of Biological Statistics, 3rd Edn. Baltimore: Sparky House Publishing.
McGraw, E. A., and O’Neill, S. L. (1999). Evolution of Wolbachia pipientis transmission dynamics in insects. Trends Microbiol. 7, 297–302. doi: 10.1016/s0966-842x(99)01531-0
Meany, M. K., Conner, W. R., Richter, S. V., Bailey, J. A., Turelli, M., and Cooper, B. S. (2019). Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution 73, 1278–1295. doi: 10.1111/evo.13745
Mohd Razali, N., and Bee, W. Y. (2011). Power comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling tests. J. Stat. Model. Anal. 2, 21–33.
Moreira, H., Szyjka, A., Paliszkiewicz, K., and Barg, E. (2019). Prooxidative activity of celastrol induces apoptosis, DNA damage, and cell cycle arrest in drug-resistant human colon cancer cells. Oxid. Med. Cell Longev. 2019:6793957.
Nitiss, J. L. (2009). Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350. doi: 10.1038/nrc2607
Noda, H. (1984). Cytoplasmic incompatibility in a rice planthopper. J. Hered. 75, 345–348. doi: 10.1093/oxfordjournals.jhered.a109953
Nor, I., Engelstädter, J., Duron, O., Reuter, M., Sagot, M. F., and Charlat, S. (2013). On the genetic architecture of cytoplasmic incompatibility: inference from phenotypic data. Am. Nat. 182, E15–E24.
O’Neill, S. L., and Karr, T. L. (1990). Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348, 178–180. doi: 10.1038/348178a0
Pan, X., Zhou, G., Wu, J., Bian, G., Lu, P., Raikhel, A. S., et al. (2012). Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 109, E23–E31.
Pasyukova, E. G., and Vaiserman, A. M. (2017). HDAC inhibitors: a new promising drug class in anti-aging research. Mech. Ageing Dev. 166, 6–15. doi: 10.1016/j.mad.2017.08.008
Penz, T., Schmitz-Esser, S., Kelly, S. E., Cass, B. N., Müller, A., Woyke, T., et al. (2012). Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8:e1003012. doi: 10.1371/journal.pgen.1003012
Perlmutter, J. I., and Bordenstein, S. R. (2020). Microorganisms in the reproductive tissues of arthropods. Nat. Rev. Microbiol. 18, 97–111. doi: 10.1038/s41579-019-0309-z
Podratz, J. L., Staff, N. P., Froemel, D., Wallner, A., Wabnig, F., Bieber, A. J., et al. (2011). Drosophila melanogaster: a new model to study cisplatin-induced neurotoxicity. Neurobiol. Dis. 43, 330–337. doi: 10.1016/j.nbd.2011.03.022
Poinsot, D., Bourtzis, K., Markakis, G., Savakis, C., and Merçot, H. (1998). Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150, 227–237.
Poinsot, D., Charlat, S., and Merçot, H. (2003). On the mechanism of Wolbachia-induced cytoplasmic incompatibility: confronting the models with the facts. Bioessays 25, 259–265. doi: 10.1002/bies.10234
Rasgon, J. L., and Scott, T. W. (2003). Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics 165, 2029–2038.
Rastogi, N., and Mishra, D. P. (2012). Therapeutic targeting of cancer cell cycle using proteasome inhibitors. Cell Div. 7:26. doi: 10.1186/1747-1028-7-26
Reed, K. M., and Werren, J. H. (1995). Induction of paternal genome loss by the paternal-sex-ratio chromosome and cytoplasmic incompatibility bacteria (Wolbachia): a comparative study of early embryonic events. Mol. Reprod. Dev. 40, 408–418. doi: 10.1002/mrd.1080400404
Reynolds, K. T., and Hoffmann, A. A. (2002). Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80, 79–87. doi: 10.1017/s0016672302005827
Rezaee, M., Sanche, L., and Hunting, D. J. (2013). Cisplatin enhances the formation of DNA single- and double-strand breaks by hydrated electrons and hydroxyl radicals. Radiat. Res. 179, 323–331. doi: 10.1667/rr3185.1
Riegler, M., Sidhu, M., Miller, W. J., and O’Neill, S. L. (2005). Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15, 1428–1433. doi: 10.1016/j.cub.2005.06.069
Rietveld, T., and van Hout, R. (2015). The t test and beyond: recommendations for testing the central tendencies of two independent samples in research on speech, language and hearing pathology. J. Commun. Disord. 58, 158–168. doi: 10.1016/j.jcomdis.2015.08.002
Ringo, J., Sharon, G., and Segal, D. (2011). Bacteria-induced sexual isolation in Drosophila. Fly 5, 310–315. doi: 10.4161/fly.5.4.15835
Rousset, F., Bouchon, D., Pintureau, B., Juchault, P., and Solignac, M. (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. Biol. Sci. 250, 91–98. doi: 10.1098/rspb.1992.0135
Rowe, T. C., Wang, J. C., and Liu, L. F. (1986). In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol. Cell Biol. 6, 985–992. doi: 10.1128/mcb.6.4.985
Sackton, K. L., Dimova, N., Zeng, X., Tian, W., Zhang, M., Sackton, T. B., et al. (2014). Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649. doi: 10.1038/nature13660
Salcedo Magguilli, M. (2017). Clinical development of histone deacetylase inhibitors. Medicographia 36, 300–310.
Sanders, L. H., and Greenamyre, J. T. (2013). Oxidative damage to macromolecules in human Parkinson’s disease and the rotenone model. Free Radic. Biol. Med. 62, 111–120. doi: 10.1016/j.freeradbiomed.2013.01.003
Sazama, E. J., Bosch, M. J., Shouldis, C. S., Ouelette, S. P., and Wesner, J. S. (2017). Incidence of Wolbachia in aquatic insects. Ecol. Evol. 7, 1165–1169.
Schmidt, T. L., Barton, N. H., Rašić, G., Turley, A. P., Montgomery, B. L., Iturbe-Ormaetxe, I., et al. (2017). Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 15:e2001894. doi: 10.1371/journal.pbio.2001894
Schneider, D. I., Ehrman, L., Engl, T., Kaltenpoth, M., Hua-Van, A., Le Rouzic, A., et al. (2019). Symbiont-driven male mating success in the neotropical Drosophila paulistorum Superspecies. Behav. Genet. 49, 83–98. doi: 10.1007/s10519-018-9937-8
Schuler, H., Bertheau, C., Egan, S. P., Feder, J. L., Riegler, M., Schlick-Steiner, B. C., et al. (2013). Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol. Ecol. 22, 4101–4111.
Serbus, L. R., Casper-Lindley, C., Landmann, F., and Sullivan, W. (2008). The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42, 683–707. doi: 10.1146/annurev.genet.41.110306.130354
Serbus, L. R., Landmann, F., Bray, W. M., White, P. M., Ruybal, J., Lokey, R. S., et al. (2012). A cell-based screen reveals that the albendazole metabolite, albendazole sulfone, targets Wolbachia. PLoS Pathog. 8:e1002922. doi: 10.1371/journal.ppat.1002922
Serbus, L. R., and Sullivan, W. (2007). A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 3:e190. doi: 10.1371/journal.ppat.0030190
Shapiro, S. S., and Wilk, M. B. (1965). An analysis of variance test for normality (complete samples). Biometrika 52, 591–611. doi: 10.2307/2333709
Shropshire, J. D., and Bordenstein, S. R. (2019). Two-By-One model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15:e1008221. doi: 10.1371/journal.pgen.1008221
Shropshire, J. D., On, J., Layton, E. M., Zhou, H., and Bordenstein, S. R. (2018). One prophage WO gene rescues cytoplasmic incompatibility in. Proc. Natl. Acad. Sci. U.S.A. 115, 4987–4991. doi: 10.1073/pnas.1800650115
Singh, P., Rathinasamy, K., Mohan, R., and Panda, D. (2008). Microtubule assembly dynamics: an attractive target for anticancer drugs. IUBMB Life 60, 368–375. doi: 10.1002/iub.42
Smerdon, M. J., Lan, S. Y., Calza, R. E., and Reeves, R. (1982). Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 257, 13441–13447.
Stadler, J., and Richly, H. (2017). Regulation of DNA repair mechanisms: how the chromatin environment regulates the DNA damage response. Int. J. Mol. Sci. 18:1715. doi: 10.3390/ijms18081715
Stanton, R. A., Gernert, K. M., Nettles, J. H., and Aneja, R. (2011). Drugs that target dynamic microtubules: a new molecular perspective. Med. Res. Rev. 31, 443–481. doi: 10.1002/med.20242
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Stouthamer, R., Breeuwert, J. A., Luck, R. F., and Werren, J. H. (1993). Molecular identification of microorganisms associated with parthenogenesis. Nature 361, 66–68. doi: 10.1038/361066a0
Takano, S. I., Tuda, M., Takasu, K., Furuya, N., Imamura, Y., Kim, S., et al. (2017). Unique clade of alphaproteobacterial endosymbionts induces complete cytoplasmic incompatibility in the coconut beetle. Proc. Natl. Acad. Sci. U.S.A. 114, 6110–6115. doi: 10.1073/pnas.1618094114
Tandon, N., Ramakrishnan, V., and Kumar, S. K. (2016). Clinical use and applications of histone deacetylase inhibitors in multiple myeloma. Clin. Pharmacol. 8, 35–44. doi: 10.2147/cpaa.s94021
Tram, U., and Sullivan, W. (2002). Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296, 1124–1126. doi: 10.1126/science.1070536
Turelli, M. (1994). Evolution of incompatibility-inducing microbes and their hosts. Evolution 48, 1500–1513. doi: 10.1111/j.1558-5646.1994.tb02192.x
Turelli, M., Cooper, B. S., Richardson, K. M., Ginsberg, P. S., Peckenpaugh, B., Antelope, C. X., et al. (2018). Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr Biol. 28, 963.e8–971.e8.
Turelli, M., and Hoffmann, A. A. (1991). Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353, 440–442. doi: 10.1038/353440a0
Turelli, M., and Hoffmann, A. A. (1995). Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140, 1319–1338.
Uckelmann, M., and Sixma, T. K. (2017). Histone ubiquitination in the DNA damage response. DNA Rep. 56, 92–101. doi: 10.1016/j.dnarep.2017.06.011
Veneti, Z., Clark, M. E., Karr, T. L., Savakis, C., and Bourtzis, K. (2004). Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl. Environ. Microbiol. 70, 5366–5372. doi: 10.1128/aem.70.9.5366-5372.2004
Wade, M. J., and Stevens, L. (1985). Microorganism mediated reproductive isolation in flour beetles (genus Tribolium). Science 227, 527–528. doi: 10.1126/science.3966160
Wang, H., Ahn, K. S., Alharbi, S. A., Shair, O. H., Arfuso, F., Sethi, G., et al. (2020). Celastrol alleviates gamma irradiation-induced damage by modulating diverse inflammatory mediators. Int. J. Mol. Sci. 21:1084. doi: 10.3390/ijms21031084
Weinert, L. A., Araujo-Jnr, E. V., Ahmed, M. Z., and Welch, J. J. (2015). The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. Biol. Sci. 282:20150249. doi: 10.1098/rspb.2015.0249
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Williamson, E. A., Wray, J. W., Bansal, P., and Hromas, R. (2012). Overview for the histone codes for DNA repair. Prog. Mol. Biol. Transl. Sci. 110, 207–227. doi: 10.1016/b978-0-12-387665-2.00008-0
Xi, Z., Gavotte, L., Xie, Y., and Dobson, S. L. (2008). Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics 9:1. doi: 10.1186/1471-2164-9-1
Xu, Z., Wu, G., Wei, X., Chen, X., Wang, Y., and Chen, L. (2013). Celastrol induced DNA damage, cell cycle arrest, and apoptosis in human rheumatoid fibroblast-like synovial cells. Am. J. Chin. Med. 41, 615–628. doi: 10.1142/s0192415x13500432
Yamada, R., Floate, K. D., Riegler, M., and O’Neill, S. L. (2007). Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics 177, 801–808. doi: 10.1534/genetics.106.068486
Yen, J. H., and Barr, A. R. (1971). New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232, 657–658. doi: 10.1038/232657a0
Yen, J. H., and Barr, A. R. (1973). The etiological agent of cytoplasmic incompatibility in Culex pipiens. J. Invertebr. Pathol. 22, 242–250. doi: 10.1016/0022-2011(73)90141-9
Yoshioka, A., Tanaka, S., Hiraoka, O., Koyama, Y., Hirota, Y., Ayusawa, D., et al. (1987). Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J. Biol. Chem. 262, 8235–8241.
Zabalou, S., Apostolaki, A., Pattas, S., Veneti, Z., Paraskevopoulos, C., Livadaras, I., et al. (2008). Multiple rescue factors within a Wolbachia strain. Genetics 178, 2145–2160. doi: 10.1534/genetics.107.086488
Zchori-Fein, E., Roush, R. T., and Rosen, D. (1998). Distribution of parthenogenesis-inducing symbionts in ovaries and eggs of Aphytis (Hymentoptera: Aphelinidae). Curr. Microbiol. 36, 1–8. doi: 10.1007/s002849900270
Zeng, X., Sigoillot, F., Gaur, S., Choi, S., Pfaff, K. L., Oh, D. C., et al. (2010). Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18, 382–395. doi: 10.1016/j.ccr.2010.08.010
Zhang, J. H., Chung, T. D., and Oldenburg, K. R. (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73. doi: 10.1177/108705719900400206
Keywords: Wolbachia, cytoplasmic incompatibility, CI, rescue, Drosophila
Citation: Momtaz AJMZ, Ahumada Sabagh AD, Gonzalez Amortegui JG, Salazar SA, Finessi A, Hernandez J, Christensen S and Serbus LR (2020) A Role for Maternal Factors in Suppressing Cytoplasmic Incompatibility. Front. Microbiol. 11:576844. doi: 10.3389/fmicb.2020.576844
Received: 27 June 2020; Accepted: 20 October 2020;
Published: 09 November 2020.
Edited by:
Mariana Mateos, Texas A&M University, United StatesReviewed by:
John Frederick Beckmann, Auburn University, United StatesMonica Rosenblueth, National Autonomous University of Mexico, Mexico
Copyright © 2020 Momtaz, Ahumada Sabagh, Gonzalez Amortegui, Salazar, Finessi, Hernandez, Christensen and Serbus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura R. Serbus, THNlcmJ1c0BmaXUuZWR1
 AJM Zehadee Momtaz1,2
AJM Zehadee Momtaz1,2 Abraham D. Ahumada Sabagh
Abraham D. Ahumada Sabagh Julian G. Gonzalez Amortegui
Julian G. Gonzalez Amortegui Laura R. Serbus
Laura R. Serbus