- 1Unit of Animal Infectious Diseases, State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2College of Biomedicine and Health, Huazhong Agricultural University, Wuhan, China
- 3Key Laboratory of Preventive Veterinary Medicine in Hubei Province, The Cooperative Innovation Center for Sustainable Pig Production, Wuhan, China
- 4Key Laboratory of Development of Veterinary Diagnostic Products, Ministry of Agriculture, Wuhan, China
Influenza virus infection can alter the composition of the gut microbiota, while its pathogenicity can, in turn, be highly influenced by the gut microbiota. However, the details underlying these associations remain to be determined. The H7N9 influenza virus is an emerging zoonotic pathogen which has caused the death of 616 humans and has incurred huge losses in the poultry industry. Here, we investigated the effects of infection with highly pathogenic H7N9 on gut microbiota and determined potential anti-influenza microbes. 16S rRNA sequencing results show that H7N9 infection alters the mouse gut microbiota by promoting the growth of Akkermansia, Ruminococcus 1, and Ruminococcaceae UCG-010, and reducing the abundance of Rikenellaceae RC9 gut group and Lachnoclostridium. Although the abundance of Akkermansia muciniphila is positively related to H7N9 infection, the oral administration of cultures, especially of pasteurized A. muciniphila, can significantly reduce weight loss and mortality caused by H7N9 infection in mice. Furthermore, oral administration of live or pasteurized A. muciniphila significantly reduces pulmonary viral titers and the levels IL-1β and IL-6 but enhances the levels of IFN-β, IFN-γ, and IL-10 in H7N9-infected mice, suggesting that the anti-influenza role of A. muciniphila is due to its anti-inflammatory and immunoregulatory properties. Taken together, we showed that the changes in the gut microbiota are associated with H7N9 infection and demonstrated the anti-influenza role of A. muciniphila, which enriches current knowledge about how specific gut bacterial strains protect against influenza infection and suggests a potential anti-influenza probiotic.
Introduction
Influenza is a global infectious disease caused by influenza viruses, which are single-stranded negative-sense RNA viruses belonging to the Orthomyxoviridae family (Steinhauer and Skehel, 2002). The influenza epidemic occurs every year, and intermittent pandemics have broken out over the past century, making it a serious threat to global public health (Cui et al., 2017). Moreover, influenza viruses have a high degree of variability and can continuously reassort to form new subtypes that can be highly pathogenic to humans and animals. For example, in March 2013, a novel avian-origin influenza A(H7N9) virus emerged in China, leading to the deaths of three patients (Gao et al., 2013). Since then, the H7N9 avian influenza virus has continued to circulate in China (Qi et al., 2018). As of May 2020, a total of 1,568 influenza A(H7N9) human cases and 616 deaths have been officially reported (Food Agriculture Organization of the United Nations, 2019; World Health Organization, 2020).
Numerous studies have confirmed the close relationship between the pathogenicity of influenza and intestinal microbiota. Influenza infection can alter the structure of the gut microbiota. For example, influenza A viruses have been shown to provoke the quantitative depletion of small intestine microbiota and exacerbate secondary Salmonella infection (Deriu et al., 2016; Yildiz et al., 2018). Changes in the gut microbiota can also visibly affect the outcome of influenza infection (Ichinohe et al., 2011). In recent years, some articles have confirmed that oral administration of Lactobacillus, Bifidobacterium, etc. can significantly improve influenza clinical symptoms by regulating the body's immune response (Iwabuchi et al., 2011; Waki et al., 2014; Kawahara et al., 2015). Although several studies have reported the close relationship between influenza and the gut microbiota, many details remain largely unknown, especially regarding the relationship between emerging influenza viruses, such as H7N9, and the gut microbiota.
In this study, to analyze the effects of H7N9 infection on the mouse gut microbiota, 16S rRNA sequencing of fecal microorganisms was performed. We found that the abundances of several gut microbes were significantly altered following H7N9 infection. Akkermansia muciniphila was highly enriched in H7N9 infected mice, suggesting that A. muciniphila is related to H7N9 pathogenicity. However, a growing body of research has indicated that A. muciniphila can exert a beneficial role in host defense against various diseases (Everard et al., 2013; Bian et al., 2019) and has great potential as a next-generation probiotic after Lactobacillus and Bifidobacterium (Feng et al., 2019). Therefore, we further evaluated the effects of A. muciniphila on the pathogenicity of the H7N9 influenza virus and analyzed the probable mechanisms underlying these effects.
Materials and Methods
Bacteria and Viruses
A. muciniphila ATCC BAA-835 and Lactobacillus gasseri ATCC 33323 were obtained from BeNa Culture Collection Co., Ltd. (Suzhou, China). A. muciniphila was streaked out on a BD Brain Heart Infusion (BHI) agar (Becton Dickinson, Heidelberg, Germany) supplemented with 0.25% mucin (Sigma-Aldrich, St. Louis, MO, USA), while L. gasseri was streaked out on MRS agar (Qingdao Hope Biotechnology Co., Ltd., Qingdao, China). Cultures were incubated under anaerobic conditions at 37°C for 48 h (Derrien et al., 2004). Bacterial colonies were washed with 1 mL PBS and resuspended, and 100 μL of the bacterial solution was plated onto the corresponding agar media. After 48 h of incubation at 37°C in an anaerobic jar, A. muciniphila from each plate swabbed and suspended in 1 mL BHI containing 15% glycerol and 0.4% mucin, and L. gasseri was likewise swabbed and suspended in 1 mL MRS broth with 15% glycerol. For either sample, the bacterial suspensions from each plate were mixed and then divided into equal parts and frozen at −80°C. To determine the colony forming units (CFU), one aliquot for each bacterial species was serially diluted and plated onto the corresponding agar media. Bacterial colonies were counted after 48 h (Greer et al., 2016).
Influenza virus A/chick/Guangxi/YL01/2017 (H7N9) virus was propagated in the allantoic fluid of 10-day-old chicken embryos and purified as previously described (Suzuki et al., 1996). Purified H7N9 virus was stored at −80°C until use. The HA cleavage motif of the virus met the criteria for a highly pathogenic avian influenza virus, showing high pathogenicity in mice.
Influenza virus A/Puerto Rico/8/1934 (H1N1; PR8) virus was propagated in the allantoic fluid of 10-day-old chicken embryos. The purified PR8 virus was stored at −80°C until use.
Animals
The infection experiments were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals Monitoring Committee of Hubei Province, China, and the protocol was approved by the Scientific Ethics Committee of Huazhong Agricultural University (Permit Number: HZAUMO-2019-018). All efforts were made to minimize the suffering of the animals. Eight-week-old female C57BL/6 mice were obtained from the Experimental Animal Center of China Three Gorges University and the total number of mice used in animal experiments is 298. All animal experiments were performed under animal biosafety level 3 (ABSL3) conditions.
Mouse Infection for the Collection of Fecal Samples
A total of 12 C57BL/6 mice were randomly divided into an NC group (negative control; mock infection using PBS) and an H7N9 infection group (n = 6 mice/group). After being anesthetized by intraperitoneal injection with a mix of ketamine/xylazine (100 and 5 mg/kg, respectively) in 100 μL of sterile PBS, each mouse was intranasally inoculated with 1 × 104 EID50 of H7N9 influenza virus or an equivalent volume of PBS. At 0 and 7 days post-infection, fecal samples were collected, and immediately stored at −80°C for total DNA extraction and sequencing.
Antibiotic Treatment
The mice were treated for 3 days with an antibiotic solution (ATB) via sterile drinking water, including ampicillin (1 g/L), vancomycin (0.25 g/L), neomycin sulfate (1 g/L), and streptomycin (5 g/L) (Routy et al., 2018). ATB treatment was discontinued 1 d prior to gavage with A. muciniphila.
Oral Administration of A. muciniphila and Mouse Infection
Thirty ATB-pretreated mice were randomly assigned to three groups (n = 10 mice/group): the PBS group, the A. muciniphila group, and the pasteurized A. muciniphila group. Each mouse received a daily oral administration of 200 μL PBS or an equivalent volume of PBS containing 1 × 108 CFU of live or pasteurized A. muciniphila. After 8 days, all mice were intranasally inoculated with H7N9 influenza virus (1 × 104 EID50 per mouse). The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection).
In another experiment, 63 ATB-pretreated mice were randomly assigned to three groups (n = 21 mice/group): the PBS group, the A. muciniphila group, and the pasteurized A. muciniphila group. The gavage and infection procedures were similarly performed. At 0, 3, and 5 d after infection, the lungs and blood of mice were collected (n = 5 for each time point in each group). At each time point, the lung viral titers and cytokine concentrations of the mice in each group were determined. Additional mice per group were sacrificed at 0 and 6 days after infection (n = 3 per time point), and their lungs were used for histological examination.
To explore whether A. muciniphila plays a beneficial role in mice infected with PR8 influenza virus, 21 ATB-pretreated mice were randomly assigned to three groups (n = 7 mice/group): the PBS, A. muciniphila or pasteurized A. muciniphila group. The gavage procedures were similarly performed as described in Figure 4A. After 8 days, each mouse was intranasally inoculated with 1 × 103 TCID50 of the PR8 influenza virus. The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection).
In order to investigate the effect of LPS (Lipopolysaccharide isolated from Escherichia coli) on influenza infection, 21 ATB-pretreated mice were randomly assigned to three groups (n = 7 mice/group): PBS, LPS or pasteurized A. muciniphila group. Mice in the PBS group were orally administered 200 μL PBS and those in the pasteurized A. muciniphila group were administered 200 μL PBS containing 1 × 108 CFU of pasteurized A. muciniphila per day. In addition, the mice in the LPS group received daily oral administration of 200 μL PBS containing the same dose of LPS as in 1 × 108 CFU pasteurized A. muciniphila (determined by the limulus reagent) (Liu et al., 2011). After 8 days, each mouse was intranasally inoculated with 1 × 104 EID50 of the H7N9 influenza virus. The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection).
Oral Administration of L. gasseri and Mouse Infection
A total of 14 ATB-pretreated mice were randomly assigned to two groups (n = 7 mice/group): PBS and L. gasseri group. Each mouse received a daily oral administration of 200 μL PBS or an equivalent volume of PBS containing 1 × 108 CFU of L. gasseri. After 8 days, all mice were intranasally inoculated with 1 × 104 EID50 of the H7N9 influenza virus. The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection).
Mouse Serum Collection
Blood samples of mice at 0, 3, and 5 days after infection were placed at 37°C for 30 min, centrifuged at 4,000 rpm for 5 min, and the supernatant were collected. The serum samples were stored at −80°C until use.
RNA Extraction and Real-Time Polymerase Chain Reaction (qRT-PCR)
Whole lungs were homogenized in PBS (1 mL/lung) and then centrifuged at 8,000 rpm. The supernatants were collected and stored at −80°C until use. Total RNA of the supernatants were extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's instructions. Total RNA (4 μg) was reverse transcribed using AMV reverse transcriptase (Takara Bio, Shiga, Japan). qRT-PCR was performed using an ABI ViiA 7 instrument (Applied Biosystems, Foster City, CA, USA) and a SYBR Green PCR Kit (Roche, Basel, Switzerland). All primers used in qRT-PCR are listed in Supplementary Tables 1, 2. The GAPDH housekeeping gene was used as a reference to normalize the amount of mRNA in each sample.
Determining Lung Viral Titers
To determine viral titers, 10-day-old SPF embryonated chicken eggs were inoculated using diluted supernatants of lung homogenates [as mentioned in section RNA Extraction and Real-Time Polymerase Chain Reaction (qRT-PCR)]. After 72 h at 37°C, the allantoic fluid was harvested for the hemagglutination test. Virus titers were calculated using the method described by Reed and Muench (Cook et al., 2018).
Analyses of Cytokine Concentrations
The concentration of cytokines, including IFN-β, IFN-γ, IL-1β, TNF-α, IL-6, and IL-10, in the supernatants of lung homogenates and in the serum were detected using an enzyme-linked immunosorbent assay kit (NeoBioscience Technology Company, Dakewe Biotechnology Company, Shenzhen, China).
Lung Histopathology
The lungs were fixed in 4% paraformaldehyde and were embedded in paraffin. Slices were stained with hematoxylin and eosin (H&E) for histopathological analysis. The lung sections were assessed using an AxioVert 200 M (Zeiss, Oberkochen, Germany) optical microscope.
Measurement of Influenza-Specific Antibody Responses
Eight mice were divided into PBS gavage and pasteurized A. muciniphila gavage group (n = 4/group), and the gavage procedure was performed as described in Figure 4A, each mouse was intranasally inoculated with 5 × 102 EID50 of H7N9 influenza virus (non-lethal dose). Blood samples of all mice were collected at 14 and 21 days after infection. The influenza-specific antibodies in the serum was measured by ELISA as previously described (Akache et al., 2018; Stark et al., 2019). Briefly, 96-well ELISA plates were coated with UV-inactivated H7N9 influenza virus at 4°C overnight. The plates were blocked with 2% non-fat milk in PBS-Tween 20 (0.05%; PBS-T) for 1 h at 37°C. After washing the plates five times with PBS-T, 3.162-fold serially diluted samples in PBS-T were added in 100 μL volumes and incubated for 1 h at 37°C. The plates were washed five times with PBS-T, and HRP goat anti-mouse IgG antibody (PM Biotechnology Co., Ltd, Wuhan, China) (diluted 1:5,000, 100 μL/well) was added for 1 h at 37°C. After five washes with PBS-T, TMB substrate (KPL) was added and the reaction was stopped by the addition of 2 N H2SO4. Serum IgG titers were defined as the dilution that resulted in an optical density of 0.1 at 450 nm (OD450). This cutoff value represents the value of the mean OD450 plus twice the standard deviations (SD) of the normal mouse serum samples tested at 1:100 dilution.
Extraction of Genomic DNA and Sequencing of the 16S rRNA Gene
Genomic DNA was extracted from the fecal samples (as mentioned in section Mouse Infection for the Collection of Fecal Samples) using the CTAB/SDS method. The DNA concentration and purity were determined by electrophoresis on 1% agarose gels. The concentration of DNA was adjusted to 1 ng/μL by dilution with sterile water.
The 16S rRNA/18S rRNA and internal transcribed spacer (ITS) segments of distinct regions (16SV4/16SV3/16SV3V4/16SV4V5, 18SV4/18SV9, ITS1/ITS2, and ArcV4) were amplified using specific barcode primers (16SV4: 515F, 806R; 18SV4: 528F, 706R; 18SV9: 1380F, 1510R). All PCR reactions were carried out using Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA).
Loading buffer (containing SYBR Green) was mixed with an aliquot of PCR products to a final concentration of 1×, and electrophoresis was performed on 2% agarose gel for detection. Samples corresponding to a single bright band observed between 400 and 450 bp in electrophoresis were chosen for further purification and these PCR products were mixed in equidense ratios. Then, the mixture of PCR products was purified using a Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
Sequencing libraries were generated using a TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer's recommendations, and index codes were added. Library quality was assessed on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). Finally, the library was sequenced on an Illumina HiSeq 2500 platform, and 250-bp paired-end reads were generated (BioProject: PRJNA596326; Submission: SUB6749693).
Operational Taxonomic Unit (OUT) Cluster and Species Annotation
Sequence analyses were performed using the UPARSE software (UPARSE v.7.0.1001, http://drive5.com/uparse/). Sequences with ≥97% similarity were assigned to the same OTUs. The representative sequence for each OTU was screened for further annotation.
For each representative sequence, the Silva Database (version 132; https://www.arb-silva.de/), based on the Ribosomal Database Project (RDP) classifier algorithm, was used (version 2.2; http://rdp.cme.msu.edu/) to annotate taxonomic information.
In order to study the phylogenetic relationship between different OTUs, as well as the differences among the dominant species in different samples (groups), multiple sequence alignments were conducted using the MUSCLE software (version 3.8; http://www.drive5.com/muscle/).
OTU abundance information was normalized using a standard sequence number corresponding to the sample with the lowest number of sequences. Subsequent analyses of the alpha diversity and beta diversity were all performed based on these normalized output data.
Alpha and Beta Diversity
Alpha diversity was applied to analyze the complexity of species diversity in a sample using 6 indices: observed species, Chao1, Shannon, Simpson, ACE, and Good's coverage. All the indices in our samples were calculated using QIIME (version 1.7.0) and displayed with the R software (version 2.15.3).
Beta diversity analysis was used to evaluate the differences in species complexity across the samples. The beta diversity of both weighted and unweighted UniFracs were calculated using QIIME software (version 1.7.0).
The cluster analysis was preceded by principal component analysis (PCA), which was applied to reduce the dimension of the original variables using the FactoMineR package and ggplot2 package in the R software (version 2.15.3).
Principal coordinate analysis (PCoA) was performed to obtain the principal coordinates and visualize complex, multidimensional data. A distance matrix of weighted or unweighted UniFracs among the samples obtained before being transformed into a new set of orthogonal axes, through which the maximum variation factor is demonstrated by the first principal coordinate, and the second maximum variation factor by the second principal coordinate, and so on. PCoA analysis was determined by the WGCNA package, stat packages, and ggplot2 package in the R software (version 2.15.3).
Unweighted pair group method with arithmetic mean (UPGMA) clustering was performed as a type of hierarchical clustering method to interpret the distance matrix using average linkage and was conducted using QIIME software (version 1.7.0).
Statistical Analysis
Survival rates after H7N9 infection were estimated by the Kaplan–Meier analysis, and their homogeneity was evaluated by a log-rank test. Statistical significance was tested by a two-way ANOVA or a Wilcoxon–Mann–Whitney test. A P-value less than 0.05 (P < 0.05) in these analyses indicated significant difference.
Results
H7N9 Influenza Virus Infection Significantly Alters the Composition of Gut Microbiota in Mice
To determine whether H7N9 infection alters the composition of the gut microbiota in mice, fecal microbes from mock-infected mice (NC group) and H7N9 infected mice (H7N9 group) were analyzed by 16S rRNA sequencing. To study the species composition of each sample, the effective tags of all samples were clustered with OTUs (operational taxonomic units) with a 97% similarity, and species annotation was performed on the representative sequences of the OTUs. As shown by the rarefaction curves, the sequencing depth was great enough to represent all the bacterial diversity (Figure 1A). The richness and uniformity of the species in the samples are reflected in the rank abundance curve (Figure 1B). Based on an ANOSIM test, we found that the microbial composition of the feces 7 days after H7N9 infection showed noticeable changes not only compared to feces before infection (R = 0.546, P = 0.005) but also compared to the feces of the NC group (R = 0.541, P = 0.005; Figures 1C,D). Meanwhile, the PCoA plots and the UPGMA analysis also demonstrated that the fecal microbiomes of mice in the H7N9 infection group at 7 days post-infection were different from those of the other groups (Figures 1E,F).
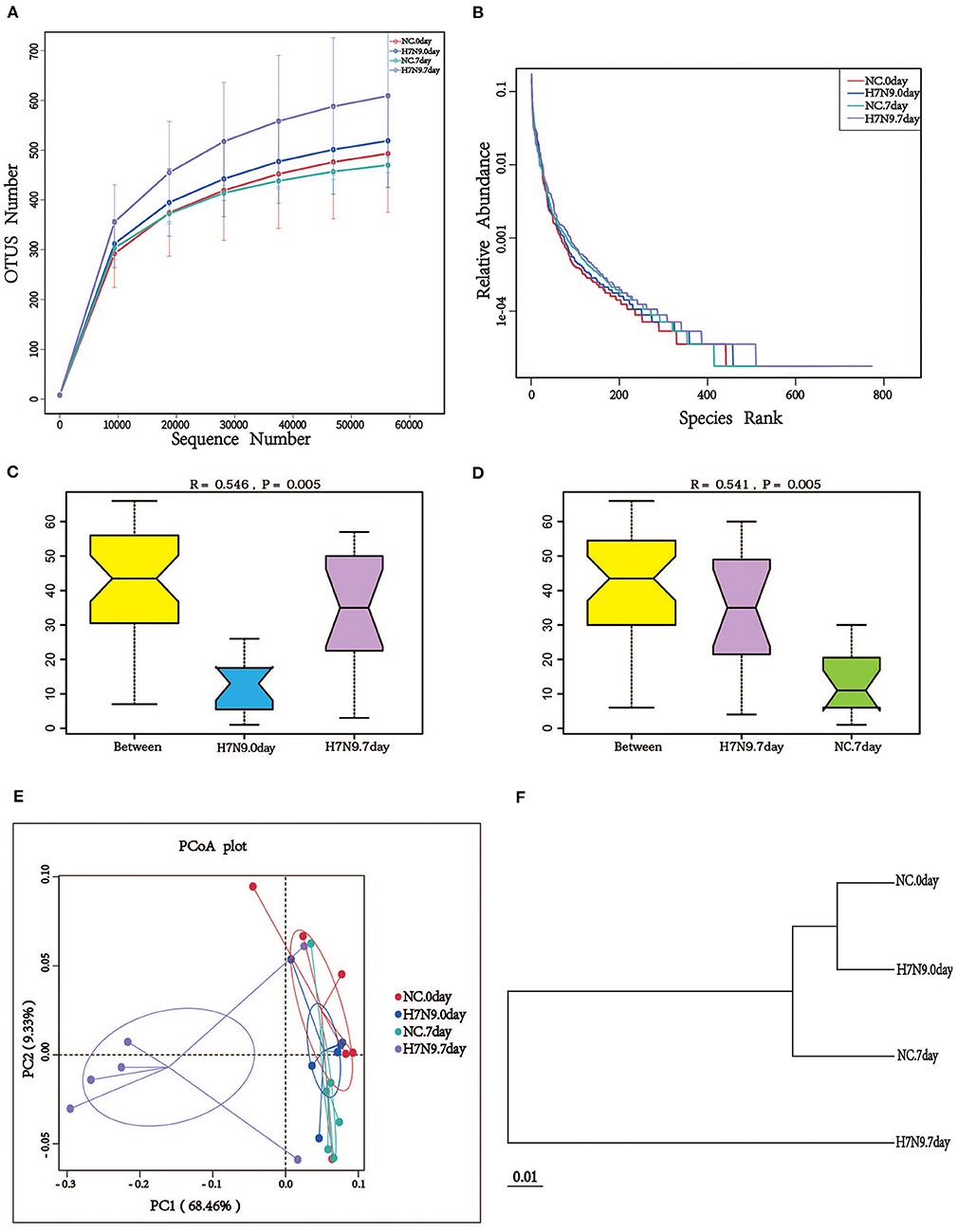
Figure 1. H7N9 influenza virus infection significantly alters the composition of mouse gut microbiota. (A) The rarefaction curves; the abscissa is the number of sequencing samples randomly extracted from a sample, and the ordinate is the number of OTUs that can be constructed based on the sequencing counts. (B) The ranks of abundance curves; the abscissa is the serial number sorted by OTU abundance, and the ordinate is the relative abundance of the corresponding OTUs. The ANOSIM analysis between (C) the H7N9.0 group and the H7N9.7 group, and (D) the NC.7 group and the H7N9.7 group; the ordinate is the rank of the distance between the samples and the abscissa, which is the result from the two groups, and the other two are the results in the respective groups. (E) The PCoA plot based on the weighted UniFrac distances of the 16S rRNA genes. (F) UPGMA tree based on the weighted UniFrac distance matrix.
A. muciniphila and L. gasseri Are Modulated by H7N9 Infection
Alterations in the characteristics of gut microbes associated with H7N9 infection were analyzed at the phylum and genus levels. As shown in Figure 2A, at the phylum level, H7N9 infection led to greater abundance of Proteobacteria and Verrucomicrobia and lower abundance of Bacteroidetes. In addition, the differences between bacterial taxonomic composition at the genus level are shown in the relative abundance map and heat map (Figures 2B,C). The results indicate that the abundances of Akkermansia, Ruminococcus 1, Ruminococcaceae UCG-010, [Eubacterium] ruminantium group, Stenotrophomonas, etc. increased more significantly in the H7N9 infection group than in the other groups. By contrast, the abundances of the Rikenellaceae RC9 gut group, Lachnoclostridium, Tyzzerella, etc., decreased in the H7N9 infection group.
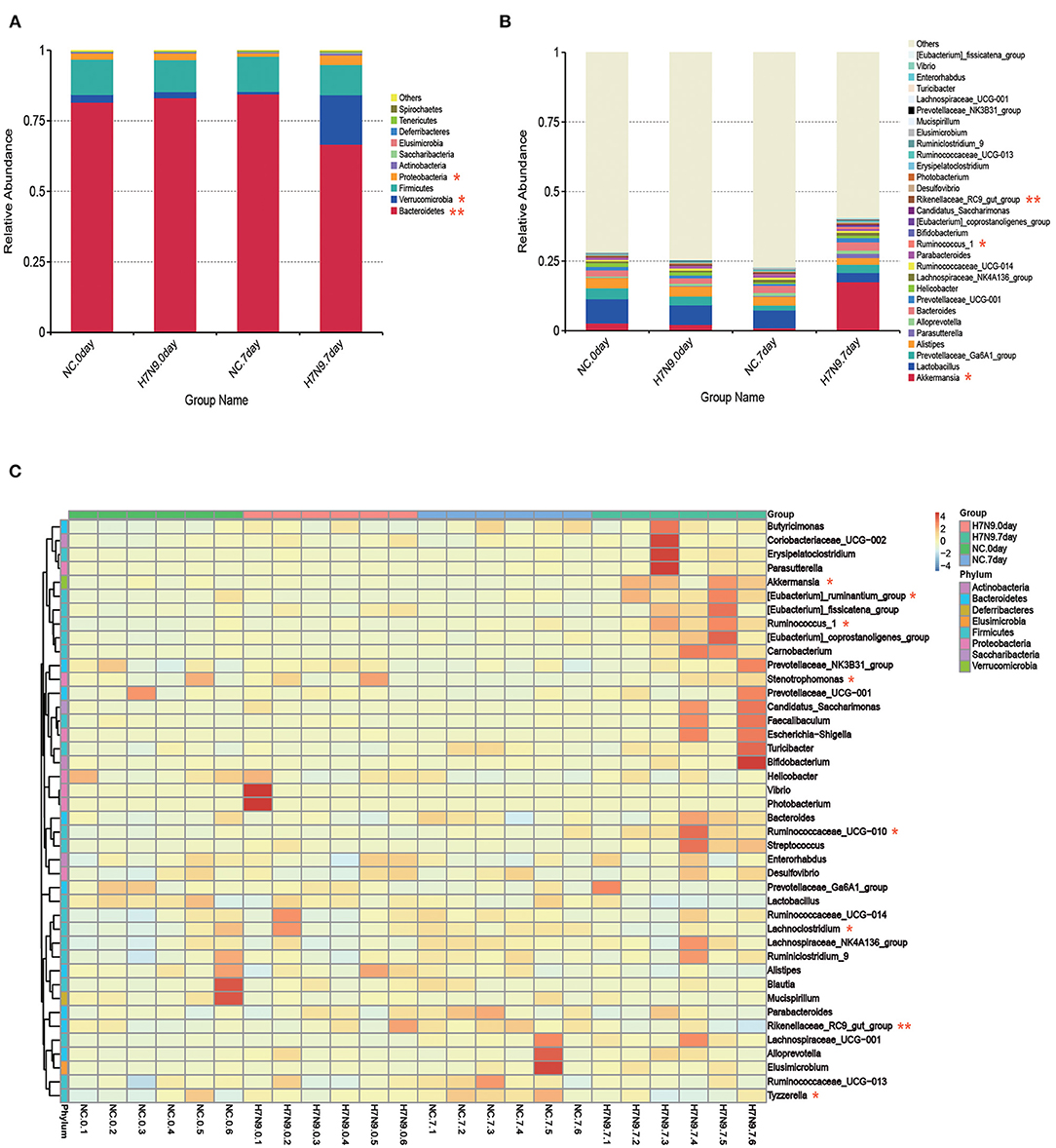
Figure 2. Fecal microbiota abundances at the phylum and genus levels for H7N9-infected mice and healthy controls. The relative abundance of the top (A) 10 phyla and (B) top 30 genera in each group. (C) Hierarchically clustered heatmap analysis at the genera level of each sample; the relative values for bacterial genera are indicated by color intensity. Statistical significance was tested by a Wilcoxon–Mann–Whitney test. *P < 0.05 and **P < 0.01.
To further identify the specific microbes associated with H7N9 infection, the fecal microbes were analyzed using the linear discriminant analysis (LDA) effect size (LEfSe) method between the uninfected and the H7N9-infected mice. The greatest differences (LDA >4) in taxa between the H7N9 infection group and the uninfected control group are displayed in Figure 3. The results show that the H7N9 infection group are enriched in A. muciniphila and have less L. gasseri, suggesting that these two bacteria are modulated by H7N9 infection.
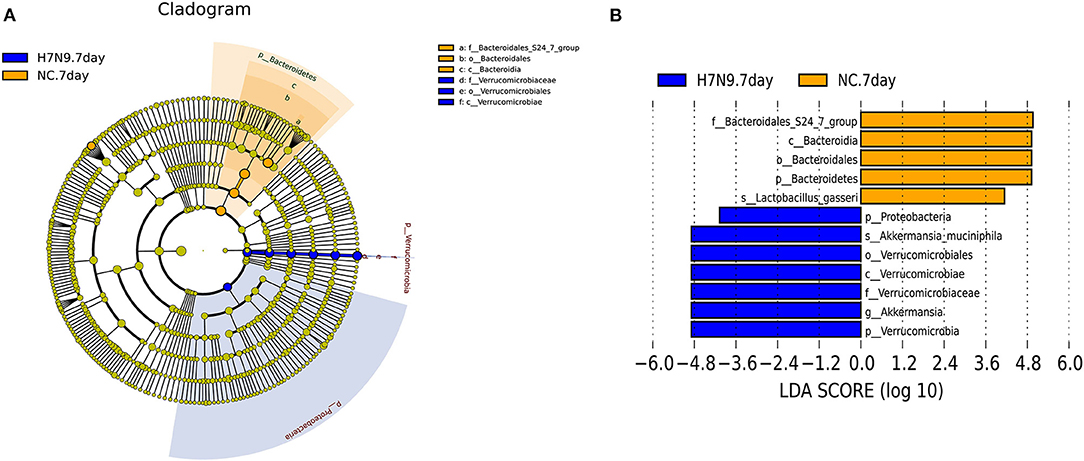
Figure 3. The taxa with different abundances between the H7N9 infected mice and healthy controls as identified by LEfSe. (A) A taxonomic cladogram obtained from an LEfSe analysis of sequences (relative abundance ≥0.5%). Biomarker taxa are highlighted by colored circles and shaded areas (H7N9-infected samples are shown in red, and uninfected samples are shown in green). Each circle's diameter reflects the abundance of that taxa in the community. (B) The taxa with different abundances in the H7N9-infected samples (red bars) and the uninfected samples (green bars). The length of the histogram represents the influence size of the different species (LDA score >4.0).
Oral Administration of A. muciniphila Improved Clinical Symptoms of H7N9 Infection
To evaluate the effects of A. muciniphila on the pathogenicity of the H7N9 influenza virus, ATB-treated female C57BL/6 mice were administered A. muciniphila by oral gavage and then intranasally infected with the H7N9 influenza virus (Figure 4A). The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection). The results show that oral administration of A. muciniphila and pasteurized A. muciniphila can increased the survival rate of mice (P = 0.1623, P = 0.0104) and alleviated mouse weight loss (P = 0.0352; Figures 4B,C). However, pasteurized A. muciniphila displayed better protective effects against H7N9 infection than viable A. muciniphila (Figure 4B). In order to eliminate the influence of LPS, the mice in the LPS group received daily oral administration the same dose of LPS as in 1 × 108 CFU pasteurized A. muciniphila before infection. As shown in Supplementary Figure 1, the survival and weight loss of infected mice gavage with LPS are almost the same as those in the PBS group. In addition, we also studied the role of L. gasseri in influenza infection. The resulted show that gavage with L. gasseri neither improved the survival rate nor reduced weight loss of the infected mice (Supplementary Figure 2). These discoveries support that the effects observed in A. muciniphila are actually specific to the genus and provide further evidence for the potential of A. muciniphila as an anti-influenza probiotic.
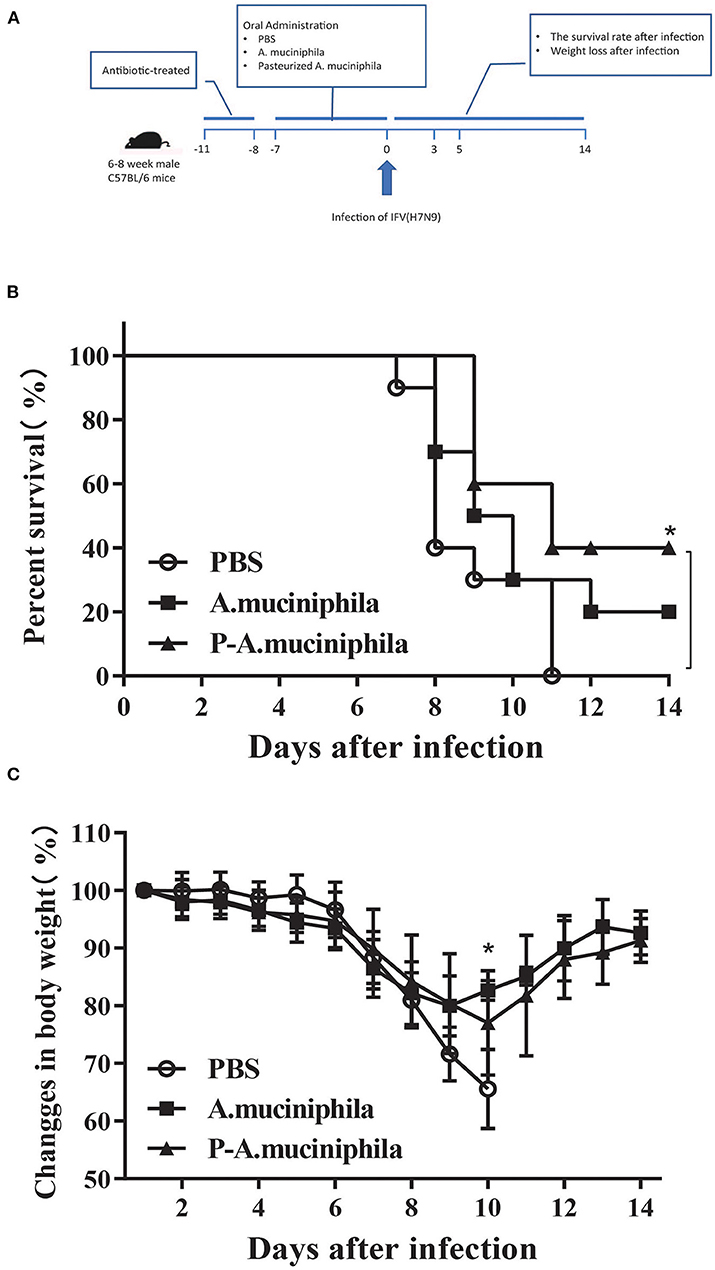
Figure 4. Oral administration of A. muciniphila improved the symptoms of H7N9 infection in mice. (A) Experimental procedure: 30 mice were pretreated with ATB and then randomly assigned to three groups (n = 10 mice/group). The three groups received daily oral administration of PBS, viable A. muciniphila, or pasteurized A. muciniphila (P-A. muciniphila). The gavage dosage was 200 μL PBS, or equivalent volumes of PBS containing 1 × 108 CFU of live or pasteurized A. muciniphila. After 8 days, each mouse was intranasally inoculated with 1 × 104 EID50 of the H7N9 influenza virus. The survival and body weights of the mice were monitored daily for 15 days (0–14 days post-infection). (B) Survival rate and (C) weight loss of the H7N9-infected mice. Data are shown as the mean ± SD (n = 10 for each group). *P < 0.05, mean the comparison between the PBS group and P-A. muciniphila group. Weight changes: two-way ANOVA; survival assay: log-rank test (Mantel–Cox). All experiments were performed at least twice under similar conditions and yielded similar results.
Oral Administration of Pasteurized A. muciniphila Suppressed the Proliferation of H7N9 Virus in vivo and Improved Lung Pathology
Next, we investigated whether oral administration of A. muciniphila can influence the replication of the H7N9 virus in vivo. Firstly, qRT-PCR was performed to analyze the mRNA levels of the viral nucleoprotein (NP) in mouse lungs. The results show that oral administration of A. muciniphila, either live or pasteurized, significantly decreased NP mRNA expression (P = 0.0138 and P = 0.0022) in the lungs at day 5 post-infection (Figure 5A). Secondly, virus titers in the lung were detected by inoculating 10-day-old SPF embryonated chicken eggs. Similarly, at day 5 post-infection, the lung viral titers (P = 0.0350 and P = 0.0045) in the mice treated with live or pasteurized A. muciniphila were significantly lower than those in the mice treated with only PBS (Figure 5B). Further, to determine whether oral administration of A. muciniphila can improve the lung pathology caused by H7N9 infection, lung histopathological analysis was performed by hematoxylin and eosin staining. Compared to the A. muciniphila group and the pasteurized A. muciniphila group, the lungs of the PBS group showed more severe inflammatory cell infiltration, alveolar atrophy, and fibrosis (Figure 5C).
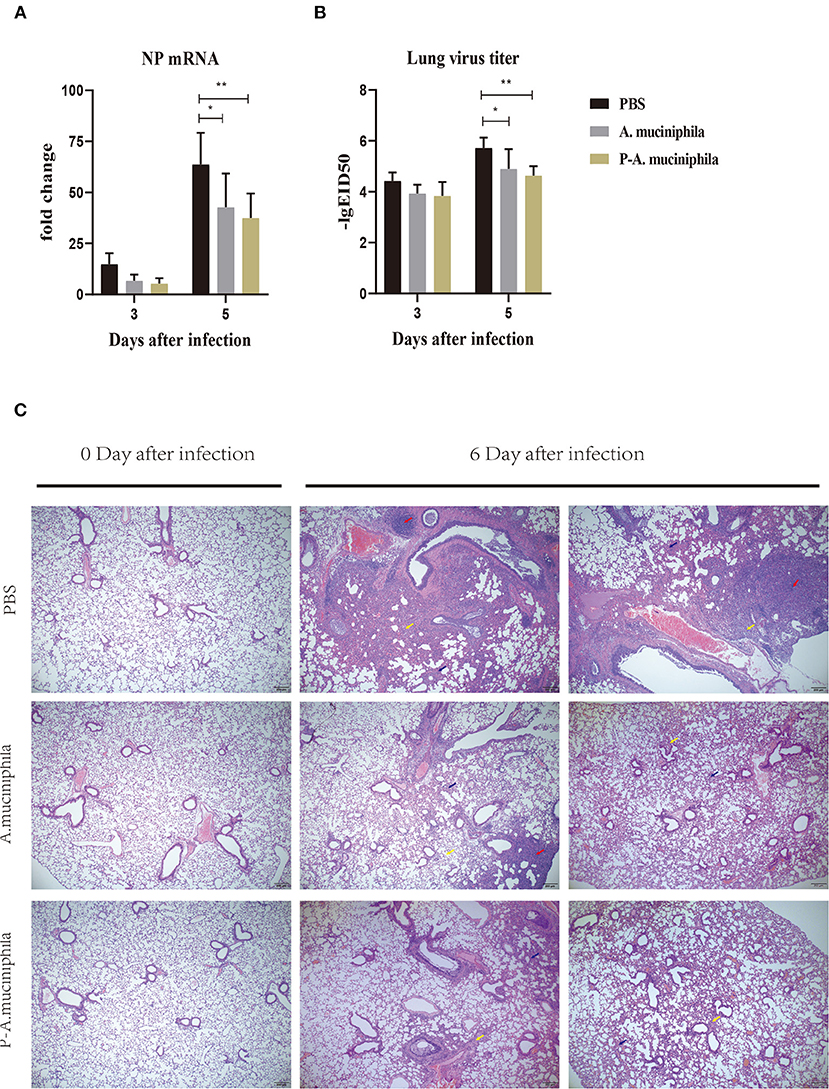
Figure 5. Oral administration of A. muciniphila suppressed the multiplication of H7N9 virus in vivo and improved lung pathology. The experimental procedure is described in Figure 4. H7N9 infected mice (n = 5) were sacrificed at 0, 3, and 5 days after infection. Their lungs were then homogenized in PBS (1 mL/lung). (A) NP mRNA levels were measured by qRT-PCR. (B) Virus titers were measured using 10-day-old SPF embryonated chicken eggs. Data are shown as the means ± SD (n = 5, for each time point per group). *P < 0.05 by two-way ANOVA. (C) Histological examination (H&E staining) of the lungs. Lungs of the PBS group, the A. muciniphila group and the pasteurized A. muciniphila group (n = 3, for each time point per group, at 0 and 6 days after infection) were removed and fixed in 4% paraformaldehyde. Paraffin-embedded sections (5-mm thick) were stained with H&E. Representative images are shown. All experiments were performed at least twice under similar conditions and yielded similar results. Blue arrows, alveolar atrophy; red arrows, fibrosis; yellow arrows, inflammatory cell infiltration. **P < 0.01.
Oral Administration of Pasteurized A. muciniphila Significantly Improved Immune Responses in H7N9-Infected Lungs
To further analyze the mechanism underlying the protective role of A. muciniphila against H7N9 infection, we assessed the cytokine responses induced by oral administration of A. muciniphila. The concentrations of IFN-β, IFN-γ, IL-1β, IL-10, and TNF-α in the lungs were measured by ELISA. As shown in Figure 6, compared with PBS group, the levels of pulmonary IFN-β, IFN-γ, and IL-10 significantly increased at 3 or 5 days post-infection in the mice administered with live (P = 0.0425, P = 0.0111, and P = 0.0400) or pasteurized A. muciniphila (P = 0.0076, P = 0.0241, and P = 0.0126). In contrast, IL-1β (P = 0.0324, P = 0.0126) and IL-6 (P = 0.0340, P = 0.0077) in lung was significantly lower at 5 days after infection. Especially, our data indicated that there are lower lung IL-6 and higher lung IFN-β in mice administered with pasteurized A. muciniphila. We also found that the levels of IL-6 significantly decreased (P = 0.0076, P = 0.0043) in the blood of mice administered with pasteurized A. muciniphila at 5 days post-infection. Further, compared with the other two groups, the level of TNF-α (P = 0.0001, P = 0.0131) in the blood of mice administered with pasteurized A. muciniphila was significantly down-regulated. Although serum IL-1β levels did not change, the serum levels of other cytokines (IFN-β, IFN-γ, and IL-10) significantly increased in the A. muciniphila group (P = 0.0449, P = 0.0362, and P = 0.0369) and the pasteurized A. muciniphila group (P = 0.0191, P = 0.0132, and P = 0.0224), which is roughly consistent with the changes in lung cytokine levels (Supplementary Figure 3). In addition, to investigate the effect of the bacterial administrations on humoral antibody development, the titers of influenza-specific IgG in mouse serum were detected at 14 and 21 d post-infection. We found that there was no difference in the levels of influenza-specific IgG between PBS group and pasteurized A. muciniphila group (Supplementary Figure 4). These results indicate that oral administration of A. muciniphila can improve the innate immune response to H7N9 infection, suggesting that the anti-influenza role of A. muciniphila depends on its anti-inflammatory and immunoregulatory capacity.
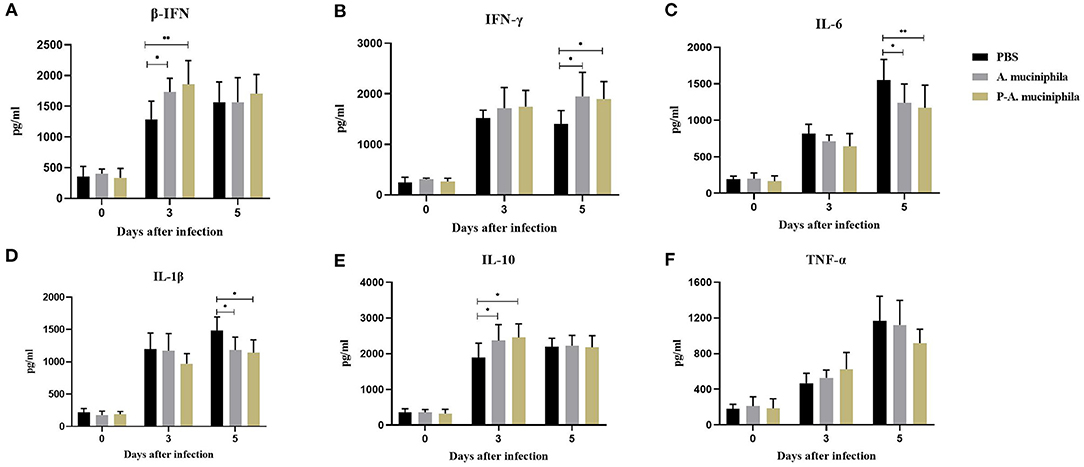
Figure 6. Oral administration of A. muciniphila significantly improved the immune responses in H7N9-infected mouse lungs. The concentrations of (A) IFN-β, (B) IFN-γ, (C) IL-6, (D) IL-1β, (E) IL-10, and (F) TNF-α in the lung were measured by ELISA. The data are shown as the means ± SD (n = 5, for each time point per group). *P < 0.05 and **P < 0.05 via two-way ANOVA. All experiments were performed at least twice under similar conditions and yielded similar results.
Discussion
Numerous studies have shown that influenza infections can result in changes in the composition of intestinal microbes, for example, mice infected with H1N1 influenza virus will cause a significant blooming of Proteobacteria (Deriu et al., 2016) and Bacteroidetes, while Firmicutes will be significantly reduced (Groves et al., 2018). A previous study has shown that Elusimicrobia and Proteobacteria in the intestines of H7N9 influenza virus-infected mice became more abundant while Bacteroidetes became less abundant (Zhang et al., 2020). A recent study has also revealed that H7N9 influenza virus-infected patients had fewer Bacteroidetes and higher levels of Proteobacteria (Qin et al., 2015). In general, influenza A virus infection in mice usually leads to the consumption of intestinal anaerobic bacteria, such as Lactobacillus and Segmented Filamentous Bacteria (SFB) (Wang et al., 2014), and followed by secondary infections of Enterobacteriaceae (Escherichia coli or Salmonella) (Deriu et al., 2016). One of the reasons for the change may be caloric restriction caused by the decrease of food intake after infection (Bartley et al., 2017). In addition, interferon production in the lungs have also been reported to alter the composition of intestinal microbes (Wang et al., 2014; Deriu et al., 2016). Similar to previous studies (Fuglsang et al., 2018a,b), we found that the abundance of A. muciniphila significantly increased after influenza infection. A recent literature has shown that influenza infection enhanced intestinal Muc2 levels (Deriu et al., 2016), which may be related to the upregulation of A. muciniphila, due to muc2 is an essential ingredient to maintain the growth of A. muciniphila. However, the role of A. muciniphila in influenza virus infection has not yet been reported.
Akkermansia muciniphila has been widely recognized as a special next-generation beneficial microorganism. A. muciniphila is the first member and the only current representative of Verrucomicrobia, which is widely distributed in human and animal intestines (Derrien et al., 2010). A. muciniphila colonizes the intestines early in life and reaches adult levels within 1 year (Collado et al., 2007), accounting for ~1–4% of the total number of microorganisms (Derrien et al., 2008). Numerous studies have confirmed that A. muciniphila treatment can significantly improve inflammatory bowel disease (Bian et al., 2019) and metabolic diseases (Everard et al., 2013; Shin et al., 2014; Katiraei et al., 2019). It also has beneficial effects on other special diseases, such as atherosclerosis (Li et al., 2016) and liver inflammation (Wu et al., 2017). Our previous study showed that A. muciniphila was up-regulated more significantly in the mice that died of H7N9 influenza virus infection, suggesting that A. muciniphila could exert an adverse effect (Zhang et al., 2020). However, in this study, we confirmed that A. muciniphila does not aggravate but instead reduce the severity of H7N9 infection. It is indeed very interesting and surprising that the endogenous up-regulation of A. muciniphila after fatal influenza infection hardly shows a protective effect, thus, more research needs to be done to understand the reason of this phenomenon.
In addition, we discovered that pasteurization of A. muciniphila enhanced its capacity to reduce the severity of influenza infection. Coincidentally, this interesting phenomenon has also been found in several other diseases (Plovier et al., 2017; Depommier et al., 2019). To gain further insights into the reasons for this difference, we compared the virus replication, lung pathology and cytokine production among the PBS group, the A. muciniphila group and the pasteurized A. muciniphila group. We found that oral administration of A. muciniphila, either live or pasteurized, significantly suppress the proliferation of influenza virus in vivo, improve lung pathology, and affect the production of immune factors. Notably, the change of certain cytokines in pasteurized A. muciniphila group (such as IL-6, TNF-α, and IFN-β) is more significant (Figures 4, 6 and Supplementary Figure 3). These suggest that the beneficial effects of A. muciniphila could be due to some of its components but not to its proliferation in the gut. Considering that A. muciniphila is Gram-negative bacteria, and LPS is an important component of the cell wall with stimulation of the physiological function of toll-like receptors. Thus, we investigated the effect of LPS on influenza infection. The results indicate that oral administration with an equivalent amount of LPS did not have an anti-influenza effect (Supplementary Figure 1). In addition, a previous study has shown that A. muciniphila exacerbates S. typhimurium-induced intestinal inflammation by disturbing the host mucus homeostasis (Ganesh et al., 2013). Therefore, we speculate that the reason pasteurized A. muciniphila is better than live bacteria is that abundant live A. muciniphila weakens the beneficial effects of the specific components by disturbing host mucus homeostasis. Certainly, these inferences still need to be explored further.
Severe influenza infection (for example, infection with highly pathogenic avian influenza virus H5N1, H7N9, etc.) can cause acute respiratory infection and lead to a “cytokine storm,” which induces significant immunopathology and severe disease outcomes (Liu et al., 2016; Guo and Thomas, 2017). The cytokines associated with the “cytokine storm” mainly include IL-1β, TNF-α, IL-6, etc., and their levels can reflect the severity of highly pathogenic avian influenza virus infection (Peiris et al., 2009). Therefore, suppressing these cytokines should improve the outcomes of highly pathogenic avian influenza virus infection. A. muciniphila has been reported to exert beneficial effects mainly through improving host metabolic functions, participating in host immune regulation, and suppressing inflammatory responses (Shang et al., 2017; Zhang et al., 2019). In this study, our data showed that oral administration of A. muciniphila decrease the levels of IL-6 and IL-1β in the lungs and the levels of IL-6 and TNF-α in the blood during the course of H7N9 infection, suggesting that suppressing these cytokines is one of the mechanisms by which A. muciniphila protects the host against influenza virus infection. In addition, IFN-β, a type I interferon, plays an important role for protecting the host against influenza viruses (Liedmann et al., 2014; Downey et al., 2017). IFN-γ, mainly produced by activated T cells (including Th0, Th1 cells, and almost all CD8+ T cells), NK cells, and professional antigen-presenting cells (APCs), is an important lymphokine involved in immune regulation (Schroder et al., 2004). IFN-γ has been reported to play an important role in influenza A virus-specific CD8+ T cell homeostasis and its transport to the site of virus-induced pathology (Turner et al., 2007). Here, we found that the expression of IFN-β and IFN-γ were further enhanced in the A. muciniphila group and the pasteurized A. muciniphila group. This may be another important anti-influenza mechanism of A. muciniphila. Considering that A. muciniphila can induce intestinal adaptive immune responses, including immunoglobulin G1 (IgG1) and antigen-specific T cell responses (Ansaldo et al., 2019), we investigated whether the oral administration of pasteurized A. muciniphila can promote the humoral immune responses. The titers of influenza-specific IgG in the mouse serum were detected at 14 and 21 days post-infection. However, we found that there was no difference in the levels of influenza-specific IgG between the PBS and the pasteurized A. muciniphila groups (Supplementary Figure 4). Thus, we speculate that the anti-influenza role of A. muciniphila mainly depends on its anti-inflammatory and immunoregulatory capacities.
In addition to its protective role against the H7N9 influenza virus infection, our research shows that oral administration of pasteurized A. muciniphila also reduced the mortality and alleviated the weight loss in mice infected with the PR8 influenza virus (Supplementary Figure 5), suggesting that A. muciniphila could have a broad-spectrum anti-influenza effect. A previous study has shown that A. muciniphila can significantly improve metabolic disease without visible side effects in human trials (Depommier et al., 2019), indicating that A. muciniphila has a good safety profile. In addition, pasteurization does not eliminate but instead enhances the benefits of A. muciniphila, indicating that the use of A. muciniphila has great advantages in the manufacturing and storage. Therefore, we believe that A. muciniphila has great potential to be developed into an anti-influenza microecological preparation. For example, it can be used as a preventive agent before infection or can be used continuously after infection to reduce inflammatory storms during the influenza season. However, our study only demonstrates the anti-influenza effects of A. muciniphila in mice. Whether A. muciniphila plays a beneficial role in other animals or in humans infected with influenza requires further studies.
Furthermore, our data show that the gut L. gasseri profile is also altered by H7N9 infection. However, unlike A. muciniphila, the abundance of L. gasseri was significantly reduced by H7N9 infection in mice. Many studies have confirmed that several species of Lactobacillus, including L. delbrueckii ssp. bulgaricus, L. rhamnosus, L. plantarum, L. casei, etc. (Bae et al., 2018; Kumova et al., 2019; Takahashi et al., 2019), contribute to host defenses against influenza virus infection by boosting host immune responses, suggesting that L. gasseri may also play a beneficial role during influenza infection. In order to verify this hypothesis, we also investigated the role of L. gasseri in influenza-infected mice, following the procedures used for investigating the effects of A. muciniphila. However, the data show that L. gasseri did not protect mice against influenza virus infection (Supplementary Figure 2).
In summary, our study describes alterations to the gut microbiota associated with highly pathogenic H7N9 influenza virus infection. More importantly, we captured a positive correlation between intestinal A. muciniphila and influenza virus infection, and further demonstrated that A. muciniphila can improves host defense against the influenza virus. Furthermore, our data suggest that the anti-influenza role of A. muciniphila is most likely due to its anti-inflammatory and immunoregulatory properties. The current study not only enriches our understanding of the interactions between influenza virus infection and the gut microbiota, but also identified a novel anti-influenza microbe, thereby laying the foundation for the development of anti-influenza probiotics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA596326; https://www.ncbi.nlm.nih.gov/, SUB6749693.
Ethics Statement
The animal study was reviewed and approved by the Scientific Ethics Committee of Huazhong Agricultural University (Permit Number: HZAUMO-2019-018).
Author Contributions
XH, QZ, and MJ designed the research, analyzed the data, and wrote the manuscript with the help of all authors. The experiments were performed mainly by XH, and some experiments were performed with the assistance of YZ, YY, WG, XS, and LY. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the National Key Research and Development Program (2017YFD0501001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.586476/full#supplementary-material
References
Akache, B., Stark, F. C., Jia, Y., Deschatelets, L., Dudani, R., Harrison, B. A., et al. (2018). Sulfated archaeol glycolipids: comparison with other immunological adjuvants in mice. PLoS ONE 13:e0208067. doi: 10.1371/journal.pone.0208067
Ansaldo, E., Slayden, L. C., Ching, K. L., Koch, M. A., Wolf, N. K., Plichta, D. R., et al. (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. doi: 10.1126/science.aaw7479
Bae, J. Y., Kim, J. I., Park, S., Yoo, K., Kim, I. H., Joo, W., et al. (2018). Effects of Lactobacillus plantarum and Leuconostoc mesenteroides probiotics on human seasonal and avian influenza viruses. J. Microbiol. Biotechnol. 28, 893–901. doi: 10.4014/jmb.1804.04001
Bartley, J. M., Zhou, X., Kuchel, G. A., Weinstock, G. M., and Haynes, L. (2017). Impact of age, caloric restriction, and influenza infection on mouse gut microbiome: an exploratory study of the role of age-related microbiome changes on influenza responses. Front. Immunol. 8:1164. doi: 10.3389/fimmu.2017.01164
Bian, X., Wu, W., Yang, L., Lv, L., Wang, Q., Li, Y., et al. (2019). Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front. Microbiol. 10:2259. doi: 10.3389/fmicb.2019.02259
Collado, M. C., Derrien, M., Isolauri, E., de Vos, W. M., and Salminen, S. (2007). Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767-70. doi: 10.1128/AEM.01477-07
Cook, J. C., Wu, H., Aleo, M. D., and Adkins, K. (2018). Principles of precision medicine and its application in toxicology. J. Toxicol. Sci. 43, 565-77. doi: 10.2131/jts.43.565
Cui, L., Fang, J., Ooi, E. E., and Lee, Y. H. (2017). Serial metabolome changes in a prospective cohort of subjects with influenza viral infection and comparison with dengue fever. J. Proteome Res. 16, 2614–2622. doi: 10.1021/acs.jproteome.7b00173
Depommier, C., Everard, A., Druart, C., Plovier, H., Van Hul, M., Vieira-Silva, S., et al. (2019). Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103. doi: 10.1038/s41591-019-0495-2
Deriu, E., Boxx, G. M., He, X., Pan, C., Benavidez, S. D., Cen, L., et al. (2016). Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type i interferons. PLoS Pathog. 12:e1005572. doi: 10.1371/journal.ppat.1005572
Derrien, M., Collado, M. C., Ben-Amor, K., Salminen, S., and W. M. de Vos. (2008). The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74, 1646–1648. doi: 10.1128/AEM.01226-07
Derrien, M., van Passel, M. W., van de Bovenkamp, J. H., Schipper, R. G., de Vos, W. M., and Dekker, J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 1, 254–268. doi: 10.4161/gmic.1.4.12778
Derrien, M., Vaughan, E. E., Plugge, C. M., and de Vos, W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476. doi: 10.1099/ijs.0.02873-0
Downey, J., Pernet, E., Coulombe, F., Allard, B., Meunier, I., Jaworska, J., et al. (2017). RIPK3 interacts with MAVS to regulate type I IFN-mediated immunity to influenza A virus infection. PLoS Pathog. 13:e1006326. doi: 10.1371/journal.ppat.1006326
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Feng, S., Liu, Y., Huang, Y., Zhao, J., Zhang, H., Zhai, Q., et al. (2019). Influence of oral administration of Akkermansia muciniphila on the tissue distribution and gut microbiota composition of acute and chronic cadmium exposure mice. FEMS Microbiol. Lett. 366:fnz160. doi: 10.1093/femsle/fnz160
Food and Agriculture Organization of the United Nations (2019). Food and Agriculture Organization of the United Nations. H7N9 Situation Update. www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html (accessed December 4, 2019).
Fuglsang, E., Krych, L., Nielsen, D. S., Frøkiær, H., and Reading, P. C. (2018b). Influenza A virus infection alters gut microbiota composition in juvenile mice. J. Inf. Dis. Trav. Med. 2:000118. doi: 10.23880/JIDTM-16000118
Fuglsang, E., Pizzolla, A., Krych, L., Nielsen, D. S., Brooks, A. G., Frokiaer, H., and Reading, P. C. (2018a). Changes in gut microbiota prior to influenza A virus infection do not affect immune responses in pups or juvenile mice. Front. Cell Infect. Microbiol. 8:319. doi: 10.3389/fcimb.2018.00319
Ganesh, B. P., Klopfleisch, R., Loh, G., and Blaut, M. (2013). Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 8:e74963. doi: 10.1371/journal.pone.0074963
Gao, R., Cao, B., Hu, Y., Feng, Z., Wang, D., Hu, W., et al. (2013). Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897. doi: 10.1056/NEJMoa1304459
Greer, R. L., Dong, X., Moraes, A. C., Zielke, R. A., Fernandes, G. R., Peremyslova, E., et al. (2016). Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat Commun. 7:13329. doi: 10.1038/ncomms13329
Groves, H. T., Cuthbertson, L., James, P., Moffatt, M. F., Cox, M. J., and Tregoning, J. S. (2018). Respiratory disease following viral lung infection alters the murine gut microbiota. Front. Immunol. 9:182. doi: 10.3389/fimmu.2018.00182
Guo, X. J., and Thomas, P. G. (2017). New fronts emerge in the influenza cytokine storm. Semin. Immunopathol. 39, 541–550. doi: 10.1007/s00281-017-0636-y
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Iwabuchi, N., Xiao, J. Z., Yaeshima, T., and Iwatsuki, K. (2011). Oral administration of Bifidobacterium longum ameliorates influenza virus infection in mice. Biol. Pharm. Bull. 34, 1352–1355. doi: 10.1248/bpb.34.1352
Katiraei, S., de Vries, M. R., Costain, A. H., Thiem, K., Hoving, L. R., van Diepen, J. A., et al. (2019). Akkermansia muciniphila exerts lipid-lowering and immunomodulatory effects without affecting neointima formation in hyperlipidemic APOE*3-Leiden.CETP Mice. Mol. Nutr. Food Res. 64:e1900732. doi: 10.1002/mnfr.201900732
Kawahara, T., Takahashi, T., Oishi, K., Tanaka, H., Masuda, M., Takahashi, S., et al. (2015). Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol. Immunol. 59, 1–12. doi: 10.1111/1348-0421.12210
Kumova, O. K., Fike, A. J., Thayer, J. L., Nguyen, L. T., Mell, J. C., Pascasio, J., et al. (2019). Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLoS Pathog. 15:e1008072. doi: 10.1371/journal.ppat.1008072
Li, J., Lin, S., Vanhoutte, P. M., Woo, C. W., and Xu, A. (2016). Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe−/− mice. Circulation 133, 2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645
Liedmann, S., Hrincius, E. R., Guy, C., Anhlan, D., Dierkes, R., Carter, R., et al. (2014). Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat. Commun. 5:5645. doi: 10.1038/ncomms6645
Liu, N., Liu, J. T., Ji, Y. Y., and Lu, P. P. (2011). Rosiglitazone regulates c-reactive protein-induced inflammatory responses via glucocorticoid receptor-mediated inhibition of p38 mitogen-activated protein kinase-toll-like receptor 4 signal pathway in vascular smooth muscle cells. J. Cardiovasc. Pharmacol. 57, 348–356. doi: 10.1097/FJC.0b013e31820a0e67
Liu, Q., Zhou, Y. H., and Yang, Z. Q. (2016). The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol. Immunol. 13, 3–10. doi: 10.1038/cmi.2015.74
Peiris, J. S., Cheung, C. Y., Leung, C. Y., and Nicholls, J. M. (2009). Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 30, 574–584. doi: 10.1016/j.it.2009.09.004
Plovier, H., Everard, A., Druart, C., Depommier, C., Van Hul, M., Geurts, L., et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236
Qi, W., Jia, W., Liu, D., Li, J., Bi, Y., Xie, S., et al. (2018). Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J. Virol. 92:e00921-17. doi: 10.1128/JVI.00921-17
Qin, N., Zheng, B., Yao, J., Guo, L., Zuo, J., Wu, L., et al. (2015). Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 5:14771. doi: 10.1038/srep14771
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillere, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706
Schroder, K., Hertzog, P. J., Ravasi, T., and Hume, D. A. (2004). Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189. doi: 10.1189/jlb.0603252
Shang, Q., Sun, W., Shan, X., Jiang, H., Cai, C., Hao, J., et al. (2017). Carrageenan-induced colitis is associated with decreased population of anti-inflammatory bacterium, Akkermansia muciniphila, in the gut microbiota of C57BL/6J mice. Toxicol. Lett. 279, 87–95. doi: 10.1016/j.toxlet.2017.07.904
Shin, N. R., Lee, J. C., Lee, H. Y., Kim, M. S., Whon, T. W., Lee, M. S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735. doi: 10.1136/gutjnl-2012-303839
Stark, F. C., Akache, B., Ponce, A., Dudani, R., Deschatelets, L., Jia, Y., et al. (2019). Archaeal glycolipid adjuvanted vaccines induce strong influenza-specific immune responses through direct immunization in young and aged mice or through passive maternal immunization. Vaccine 37, 7108–7116. doi: 10.1016/j.vaccine.2019.07.010
Steinhauer, D. A., and Skehel, J. J. (2002). Genetics of influenza viruses. Annu. Rev. Genet. 36, 305–332. doi: 10.1146/annurev.genet.36.052402.152757
Suzuki, T., Sometani, A., Yamazaki, Y., Horiike, G., Mizutani, Y., Masuda, H., et al. (1996). Sulphatide binds to human and animal influenza A viruses, and inhibits the viral infection, Biochem. J. 318 (Pt 2), 389–393. doi: 10.1042/bj3180389
Takahashi, E., Sawabuchi, T., Kimoto, T., Sakai, S., and Kido, H. (2019). Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1 feeding enhances humoral immune responses, which are suppressed by the antiviral neuraminidase inhibitor oseltamivir in influenza A virus-infected mice. J. Dairy Sci. 102, 9559–9569. doi: 10.3168/jds.2019-16268
Turner, S. J., Olivas, E., Gutierrez, A., Diaz, G., and Doherty, P. C. (2007). Disregulated influenza A virus-specific CD8+ T cell homeostasis in the absence of IFN-gamma signaling. J. Immunol. 178, 7616–7622. doi: 10.4049/jimmunol.178.12.7616
Waki, N., Yajima, N., Suganuma, H., Buddle, B. M., Luo, D., Heiser, A., et al. (2014). Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett. Appl. Microbiol. 58, 87–93. doi: 10.1111/lam.12160
Wang, J., Li, F., Wei, H., Lian, Z. X., Sun, R., and Tian, Z. (2014). Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 211, 2397–2410. doi: 10.1084/jem.20140625
World Health Organization (2020). Influenza at the Human-Animal Interface Summary and Assessment. https://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_08_05_2020.pdf?ua=1 (accessed May 8, 2020).
Wu, W., Lv, L., Shi, D., Ye, J., Fang, D., Guo, F., et al. (2017). Protective effect of Akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front. Microbiol. 8:1804. doi: 10.3389/fmicb.2017.01804
Yildiz, S., Mazel-Sanchez, B., Kandasamy, M., Manicassamy, B., and Schmolke, M. (2018). Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 6:9. doi: 10.1186/s40168-017-0386-z
Zhang, Q., Hu, J., Feng, J. W., Hu, X. T., Wang, T., Gong, W. X., et al. (2020). Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol. 21:99. doi: 10.1186/s13059-020-02007-1
Keywords: gut microbiota, Akkermansia muciniphila, influenza virus, H7N9, anti-influenza role
Citation: Hu X, Zhao Y, Yang Y, Gong W, Sun X, Yang L, Zhang Q and Jin M (2021) Akkermansia muciniphila Improves Host Defense Against Influenza Virus Infection. Front. Microbiol. 11:586476. doi: 10.3389/fmicb.2020.586476
Received: 23 July 2020; Accepted: 21 December 2020;
Published: 02 February 2021.
Edited by:
Sunil Kumar Lal, Monash University Malaysia, MalaysiaReviewed by:
Gabriel Vinderola, Facultad de Ingeniería Química, Universidad Nacional del Litoral (FIQ-UNL), ArgentinaShashank Tripathi, Indian Institute of Science (IISc), India
Caio Tavares Fagundes, Federal University of Minas Gerais, Brazil
Copyright © 2021 Hu, Zhao, Yang, Gong, Sun, Yang, Zhang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Zhang, MDV5aXNhbkAxNjMuY29t; Meilin Jin, am1sODMyOEAxMjYuY29t
 Xiaotong Hu
Xiaotong Hu Ya Zhao
Ya Zhao Yong Yang1
Yong Yang1 Xiaomei Sun
Xiaomei Sun Li Yang
Li Yang Qiang Zhang
Qiang Zhang Meilin Jin
Meilin Jin