Abstract
Of all the ESKAPE pathogens, carbapenem-resistant and multidrug-resistant Acinetobacter baumannii is the leading cause of hospital-acquired and ventilator-associated pneumonia. A. baumannii infections are notoriously hard to eradicate due to its propensity to rapidly acquire multitude of resistance determinants and the virulence factor cornucopia elucidated by the bacterium that help it fend off a wide range of adverse conditions imposed upon by host and environment. One such weapon in the arsenal of A. baumannii is the outer membrane protein (OMP) compendium. OMPs in A. baumannii play distinctive roles in facilitating the bacterial acclimatization to antibiotic- and host-induced stresses, albeit following entirely different mechanisms. OMPs are major immunogenic proteins in bacteria conferring bacteria host-fitness advantages including immune evasion, stress tolerance, and resistance to antibiotics and antibacterials. In this review, we summarize the current knowledge of major A. baumannii OMPs and discuss their versatile role in antibiotic resistance and virulence. Specifically, we explore how OmpA, CarO, and OprD-like porins mediate antibiotic and amino acid shuttle and host virulence.
Introduction
The Gram-negative coccobacillus Acinetobacter baumannii is an aerobic opportunistic pathogen responsible for some of the most morbid hospital-acquired infections (Bouvet and Grimont, 1987; Peleg et al., 2008; Lin and Lan, 2014; Lee et al., 2017). The global threat from this pathogen comes from its high rate of resistance gene acquisition leading to rapid emergence of multidrug-resistant (MDR) clinical isolates (Abbott et al., 2013; Giammanco et al., 2017; Rodloff and Dowzicky, 2017). Increasing number of studies show frequent isolation of carbapenem and colistin-resistant strains of A. baumannii from clinical settings (Garnacho-Montero et al., 2005; Asadollahi et al., 2012; Zhao et al., 2015; Benmahmod et al., 2019; Pormohammad et al., 2020). Swift accumulation and dispersion of antibiotic resistance markers along with the ability to cause urinary tract infections, skin and soft tissue infections and wound infections (Sievert et al., 2013; Weiner et al., 2016; Giammanco et al., 2017) makes A. baumannii, a pathogen of great significance for both humans and animals (Sen and Joshi, 2016; van der Kolk et al., 2019). In light of this, the World Health Organization (WHO) categorized this organism as a priority-1 critical pathogen for which discovery of new treatment options is of utmost importance (World Health Organization [WHO], 2017). The potency of A. baumannii as a successful pathogen can be elaborated by the high number of deaths associated with its infection. A recent finding showed that bacteremia caused by multidrug-resistant (MDR) A. baumannii exhibited 56.2% mortality rate among infected patients (Zhou et al., 2019).
A plethora of virulence factors are elucidated by A. baumannii (Harding et al., 2018), some of which include, but are not limited to, porin proteins, efflux pumps, outer membrane vesicles, metal acquisition systems, secretion systems, phospholipases, and capsular polysaccharides (Lee et al., 2017; Sharma et al., 2019; Skariyachan et al., 2019). Recent advances in research have not only provided better knowledge of these determinants, but also shed light on how these can be used as potential drug targets (Bhattacharyya et al., 2017; Iyer et al., 2018). However, growth conditions like temperature, oxygen content, osmolarity, and media components regulate the expression of porins in A. baumannii (Hwa et al., 2010; Fernando and Kumar, 2012; Bazyleu and Kumar, 2014). Transcriptional and post-transcriptional regulatory networks (Kuo et al., 2017; Sharma et al., 2018) also determine the virulence and antibiotic resistance of A. baumannii.
Among the vast diversity of antibiotic resistance and virulence determinants and A. baumannii specific regulatory networks, one group of bacterial proteins, termed outer membrane proteins (OMPs) due to their localization, have been studied with utmost interest due to their distribution, functional relevance and stipulated role in both antibiotic resistance and virulence (Sato et al., 2017; Nie et al., 2020). OMPs in general are beta barrel-shaped monomeric or trimeric porins (Table 1) that allow diffusion of small molecules into and out of periplasmic space of Gram-negative bacteria (Nitzan et al., 1999; Nikaido, 2003; Slusky and Dunbrack, 2013). A. baumannii outer membrane holds scores of OMPs including OmpA, CarO, OprD- like OMPs, Omp 33-36 kDa, AbuO, TolB, DcaP, Oma87/BamA, NmRmpM, CadF, OprF, etc. (Borneleit and Kleber, 1991; Park et al., 2012; Srinivasan et al., 2015; Lee et al., 2017; Bhamidimarri et al., 2019; Rasooli et al., 2020). OMPs participate in a wide range of functions that assist the bacterium in enduring the harsh environmental conditions, in combating the threat posed by antimicrobial compounds (Limansky et al., 2002; del Mar Tomás et al., 2005; Dupont et al., 2005; Mussi et al., 2007; Choi et al., 2008a; Srinivasan et al., 2015; Wang et al., 2015), host (Choi et al., 2008b,c; Lee et al., 2008; Gaddy, 2010), and surprisingly, in degrading crude oil (Hanson et al., 1994). Immunization with A. baumannii OMPs ensued significant rise in protective immune parameters (McConnell et al., 2011; Alzubaidi and Alkozai, 2015; Bazmara et al., 2019) and antibodies against OMPs passively protected experimental animals (Goel and Kapil, 2001). Clinical studies frequently identify differential expression of OMPs in antibiotic resistant A. baumannii strains, establishing their role in conferring resistance (Cuenca et al., 2003; Yun et al., 2008; Vashist et al., 2010; Moganty et al., 2011; Mostachio et al., 2012). Here, we explore and summarize how antibiotic resistance and virulence in A. baumannii is mediated by different OMPs like OmpA, CarO and OprD.
TABLE 1
| Name of porin | Molecular weight | Structure | Proposed role in A. baumannii | References |
| OmpA | 28–36 kDa | Eight-stranded Beta barrel shaped | Cytotoxic protein. Mediates attachment to host cells via fibronectin. | Choi et al., 2005; Smani et al., 2012; Confer and Ayalew, 2013 |
| CarO | 25/29 kDa | Eight-stranded beta barrel shaped | Uptake of glycine and ornithine. Also implicated in carbapenem resistance. | Limansky et al., 2002; Siroy et al., 2005; Zahn et al., 2015; Zhu et al., 2019 |
| OprD/OccAB1 | 43 kDa | Eighteen-stranded beta-barrel shaped | Allows diffusion of basic amino acids and beta-lactam class of antibiotics into the cell. | Dupont et al., 2005; Zahn et al., 2016 |
| Omp33-36 | 33–36 kDa | Yet to be studied | Implicated in imipenem resistance. Induces apoptosis in host cells by activating caspases 3 and 9. | Clark, 1996; Rumbo et al., 2014 |
| AbuO | 50.2 kDa (Theoretical) | Three domains—four-stranded beta barrel, α-helical barrel and α- β mixed barrel | Homolog of E. coli TolC protein. Involved in pH and bile salt tolerance. | Srinivasan et al., 2015 |
| DcaP | 47–50 kDa | Sixteen-stranded beta-barrel shaped | An Omp with preference for anionic compounds. Involved in transport of phthalates into the cell. | Bhamidimarri et al., 2019 |
| OmpW | Yet to be studied | Eight-stranded beta-barrel shaped. (Theoretical) | Serves as a colistin binding site. Facilitates iron uptake into the cell. | Catel-Ferreira et al., 2016 |
Structure and function of major outer membrane proteins of A. baumannii.
OmpA
OmpA, a beta barrel-shaped monomeric protein (Park et al., 2011) is one of the most abundant OMPs (Gribun et al., 2003), which has been reported to impart drug resistance to A. baumannii by allowing slower diffusion of negatively charged beta-lactam antibiotics (Nitzan et al., 2002) and virulence (Sato and Nakae, 1991; Sato et al., 2017; Sánchez-Encinales et al., 2017) by its toxicity to host cells. Clinical isolates of A. baumannii overexpressing OmpA arbitrate higher morbidity and even mortality in patients (Sato et al., 2017; Sánchez-Encinales et al., 2017). The global repressor H-NS binds to the promoter region of OmpA gene and gene locus A1S_0316 and the two component system BfmSR function as a possible anti-repressor and repressor of OmpA in A. baumannii, respectively (Liou et al., 2014; Oh et al., 2020).
Role of OmpA in Antibiotic Resistance in A. baumannii
Being the most abundant porin in A. baumannii, the role of OmpA in antibiotic resistance was more prominent in disruption mutants of the gene, which showed increased susceptibility to nalidixic acid, chloramphenicol, aztreonam, imipenem, and meropenem. Besides diffusion, research indicates that OmpA possibly couples with efflux pumps and forces out antibacterial compounds from the periplasm (Smani et al., 2013; Fahmy et al., 2018; Tsai et al., 2020). OmpA also couples to A. baumannii peptidoglycan (PG) via its C-terminal region, where Asp271 and Arg286 bind to diaminopimelic acid of PG (Park et al., 2012). This binding may regulate outer membrane vesicle (OMV) production and the membrane stability in the bacteria (Moon et al., 2012). OMVs with OmpA in their membrane (Walzer et al., 2006; Jin et al., 2011; Yun et al., 2018) mediate antibiotic resistance by actively siphoning extracellular drugs (Agarwal et al., 2019). Recently, resistance to colistin, a last-resort antibiotic, was also attributed to the presence of OmpA in A. baumannii. An isogenic mutant of OmpA resulted in loss of cell wall integrity, thus making the bacterium 20-fold more sensitive to colistin (Kwon et al., 2019) and 5.3 fold more sensitive to trimethoprim (Kwon et al., 2017) than wild type A. baumannii. The distinctive role of OmpA in conferring antibiotic resistance thrusts researchers to discover novel antibacterials against the protein. In one study, a novel diazabicyclooctenone beta-lactamase inhibitor that inhibits major classes of carbapenemases and in turn potentiates sulbactam activity was shown to be OmpA-dependent (Iyer et al., 2018). OmpA blockers can function synergistically with last resort antibiotics like colistin in eradicating MDR strains of A. baumannii (Vila-Farrés et al., 2017; Parra-Millán et al., 2018). OmpA also interacts with antimicrobial peptides (AMPs) of diverse origin and confers resistance against them (Lin et al., 2015; Guo Y. et al., 2018). Minimum inhibitory concentrations of human AMP LL-37 and bovine AMP BMAP-28 increased upon binding to N-terminal region of OmpA. The multifaceted role of OmpA in A. baumannii membrane permeability and cell wall integrity indicates its potential as a candidate for novel antibacterial development via chemical genetic screens.
Role of OmpA in A. baumannii Adherence and Invasion of Host Cells
Besides their distinguishable role in antibiotic resistance, OMPs confer virulence to A. baumannii. The bacterium is capable of invading and persisting in host epithelial and immune cells (Figure 1). The primary requisite for invasion is to adhere to the host cells, which is mediated by many virulence factors expressed by A. baumannii viz., OmpA (Nie et al., 2020), BapA (Brossard and Campagnari, 2012), fimbrial like protrusions (Mortensen and Skaar, 2012). A. baumannii adherence to host cells can be both host cell and bacterial sequence-type specific. For instance, two types of adherence patterns have been elucidated in A. baumannii; dispersed adherence of bacteria to the host cells, and adherence of clusters of bacteria at localized areas of the host cells (Lee et al., 2006). Bacterial clusters can be a result of amyloidogenic BAP protein mediated biofilm formation. Interestingly, OmpA specifically mediates bacterial binding to healthy cells than cancerous cells (Choi et al., 2008c). A. baumannii cells devoid of OmpA were found to be less virulent to human airway epithelium due to decreased adherence to cells (Gaddy, 2010) and formed weaker biofilms (Gaddy et al., 2009; Lin et al., 2020).
FIGURE 1
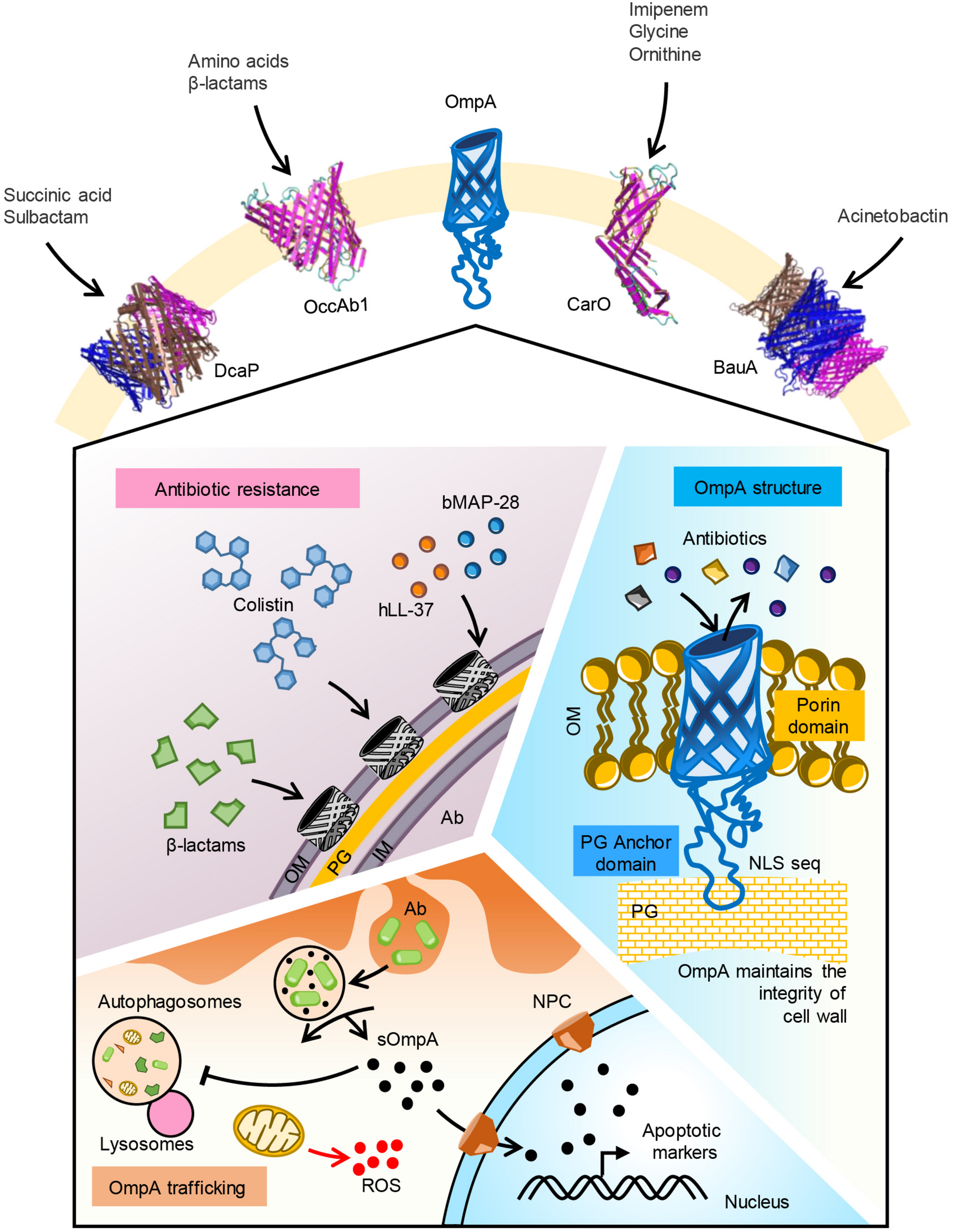
Function of major OMPs in A. baumannii. Five major OMPs with their substrate specificity is shown. OmpA porin mediates uptake of β-lactam and colistin antibiotics. Human antimicrobial peptide LL-37 and bovine AMP BMAP-28 may bind to the porin. OmpA pore size is optimal for uptake of iron siderophores like acenitobactin. A coiled loop in the C-terminal region of the protein binds to peptidoglycan maintaining the membrane integrity. Phagocytosed A. baumannii releases OmpA into the cytoplasm of host cells. Secreted OmpA inhibits lysosome fusion to autophagosomes, enters into mitochondria inducing ROS generation. Secreted OmpA actively localizes to nucleus via nuclear pore complex where it activates apoptotic markers like caspases. Ab, A. baumannii; OM, outer membrane; PG, peptidoglycan; IM, inner membrane; sOmpA, secreted OmpA; ROS, reactive oxygen species.
Following adherence to host cells, A. baumannii invades into the cell cytoplasm. The bacterial penetration into epithelial cells is microfilament- and microtubule-dependent following zipper-like mechanism (Choi et al., 2008c). Upon internalization, A. baumannii cells localize to membrane-bound vacuoles and finally traffic to the nucleus. Bacterial cells actively divide and finally kill the host cell to release into the blood stream. In this process, OmpA actively assists bacterial invasion, although by unknown mechanisms (Kim et al., 2016). Iron homeostasis is found to be a key factor regulating the survival of A. baumannii in the cytoplasm (Gaddy, 2010). Other adherence factors like BapA are not found to mediate invasion of A. baumannii (Brossard and Campagnari, 2012; De Gregorio et al., 2015) indicating that OmpA specific pathways regulate A. baumannii virulence. The functional dynamics of such a feature for a protein can be emphasized succinctly by looking into immune responses in the host.
Immune Response Configuration Against A. baumannii OmpA
In healthy individuals, A. baumannii cells in the blood stream and airways are actively phagocytosed by circulatory or tissue-resident immune cells like macrophages, neutrophils, dendritic cells (DCs), etc. (Harding et al., 2018) before leading to fulminant A. baumannii sepsis, although the latter is the case frequently encountered in immunocompromised patients. A. baumannii induces host cell cytotoxicity by targeting mitochondrial system and nuclear localization. In epithelial cells, OmpA induces the surface expression of Toll-like receptor 2 and the production of inducible nitric oxide synthase (Kim et al., 2008). Phagocytosed bacteria release several structural and cytoplasmic proteins that induce cytotoxicity directly (e.g., Type VI secretion system effector enzymes, and toxins) or indirectly by activating caspases (e.g., OmpA). In macrophages and epithelial cells, OmpA triggers autophagy, albeit incomplete, by preventing the fusion of autophagosomes with lysosomes, activating MAPK/JNK signaling pathway (Kim et al., 2008; An and Su, 2019) and enhancing the levels of phosphorylated JNK, p38, ERK and c-Jun (An et al., 2019). Early response in DCs treated with OmpA includes augmenting the expression of CD40, CD54, CD80, CD86 and MHC-I and II surface markers. The marker expression is accompanied by secretion of Th-1 promoting IL-12 (Lee et al., 2007). However, upon prolonged exposure, secreted OmpA in DCs targets mitochondria and induces production of reactive oxygen species (ROS) (Lee et al., 2008 and 2010). ROS stimulates early-onset apoptosis and delayed-onset necrosis in DCs, thus impairing T-cell response against A. baumannii. Besides induction of ROS, OmpA in conjunction with carbonic anhydrase IX stimulates DCs to generate potent anti-tumor immune response against renal cell carcinoma (Kim B.R. et al., 2011; Kim D.Y. et al., 2011; Kim et al., 2012).
All these observations in the host indicate that OmpA might be toxic in nature, unlike other outer membrane proteins. The toxicity of A. baumannii OmpA can be attributed to its unique structural features and vast diversity of alleles (Viale and Evans, 2020) with conserved domains. OmpA protein is comprised of two domains; an N-terminal 8-stranded β-barrel domain and a C-terminal periplasmic peptidoglycan binding domain. OmpA possesses a basic amino-acid rich signal termed nuclear localization signal (NLS) ‘KTKEGRAMNRR’ (Choi et al., 2007) on its C-terminal domain. Karyopherin β family proteins on nuclear pore complex recognize the NLS via its lysine (K) residues and shuttle OmpA to nucleus from cytoplasm. It can thus be speculated that OmpA devoid of NLS can be a non-toxic variant. Studies to reduce the host cell toxicity of recombinant A. baumannii revealed the importance of both N and C-terminal regions and the importance of lysine residues in NLS sequence. A synthetic OmpA with mutations at K320 and K322 to Alanine, replacing “NADEEFWN” sequence with “YKYDFDGVNRGTRGTSEEGTL” and deleting N-terminal signal sequence and “VVQPGQEAAAPAAAQ” at C-terminal resulted in least toxic but highly immunogenic OmpA (Jahangiri et al., 2017). In addition to NLS, the N-terminal region of OmpA is bioinformatically predicted to be more immunogenic (Darbandian and Sefid, 2016). The epitopes “QIQDSEHSGKMVAKRQ” at position 100–115 and “HTSFDKLPEGGRAT” at position 125–138 are delineated best by Ellipro software. A peptide at N-terminal region located at 24–50 position, “VTVTPLLLGYTFQDSQHNNGGKDGNLT” alone is immunogenic and elicited similar levels serum antibodies like OmpA (Mehdinejadiani et al., 2019). It is clear that OmpA is toxic to host when secreted and when intact, it provides antibiotic resistance to the bacterium. Whether these observations can be implied to the clinical manifestations of A. baumannii is yet to be elucidated.
CarO
Carbapenem susceptibility porin or CarO was first reported in imipenem (IMP) sensitive A. baumannii isolates that acquired resistance upon the loss of a 29 kDa protein (Limansky et al., 2002). CarO is an 8-stranded beta barrel-shaped outer membrane channel protein that does not have a continuous channel (Mussi et al., 2005 and 2011; Siroy et al., 2005; Zahn et al., 2015) but mediates influx of beta lactams (selectively imipenem) into A. baumannii (Mussi et al., 2005). However, contradicting these observations, liposome model system embedded with CarO revealed its ability to transport small amino acids such as glycine and ornithine, but not carbapenem antibiotics (Zahn et al., 2016). Despite this lonesome tangential observation, the excessive evidence from diverse research groups denotes the role of CarO in antibiotic resistance.
CarO is classified into two sub-groups; CarOa and CarOb; of which CarOb exhibits a two-fold greater specificity for IMP (Catel-Ferreira et al., 2011). However, there has been a recent call to rethink the CarO classification system based on phylogenetic analysis (Catel-Ferreira et al., 2011; Novovic et al., 2015). So far, at least six polymorphic variants of CarO have been reported to co-exist in A. baumannii populations with varied specificities to imipenem, highlighting the importance of the protein. The alterations in CarO gene are posited to be a result of rapid adaptation of A. baumannii to diverse habitats and hosts. Besides gene alterations, conformational changes in primary structure, intra-genic insertion sequences (Lee et al., 2011), posttranscriptional (Kuo et al., 2017) and transcriptional (Fonseca et al., 2013; Cardoso et al., 2016) regulation dramatically affect CarO function (summarized in Supplementary Table 1). The recent identification that CarO is significantly up-regulated in an Hfq deletion mutant strain of A. baumannii indicate that it is kept under post-transcriptional control by the bacterium to regulate its expression in response to the changing environment (Kuo et al., 2017). Finally, the occasional isolation of antibiotic resistant strains with a loss of CarO gene signifies the diversity of resistance mechanisms in A. baumannii (Li et al., 2015). In contrast to these studies linking carbapenem resistance to the loss of CarO, there are a few reports of the presence of CarO porin on the OM of carbapenem resistant clinical isolates of A. baumannii. However, this can possibly be explained by the “porin-localized toxin inactivation” model, where carbapenemases like Oxa-23 interact with the periplasmic region of OMPs like CarO or OmpA to act as an efficient selective filter to inactivate incoming antibacterial compounds (Li et al., 2015; Wu et al., 2016; Royer et al., 2018).
The clinical relevance of CarO has also been ascertained by many hospital epidemiological studies. These revealed that there is a prevalence of CarO deficiency amongst carbapenem resistant isolates expressing BlaOXA (Pajand et al., 2013; Abbasi et al., 2020) and TEM-1 (Nan et al., 2018) genes among the hospital isolates of A. baumannii. Various carbapenem resistant clinical isolates demonstrated a disruption in the CarO gene by insertion sequences like ISAba1, ISAba125, ISAba825, ISAba10, ISAba15, and ISAba36 (Mussi et al., 2005; Lee et al., 2011; Ravasi et al., 2011; Kim and Ko, 2015; Khorsi et al., 2018; Mirshekar et al., 2018). When exposed to a high concentration of monovalent cations, A. baumannii release a variety of OMPs including CarO into the surrounding media and becomes more tolerant to IMP stress. This finding implicated that MICs of antibiotics determined in vitro may not help eradicate A. baumannii infection from within the host system, especially in the case of urinary tract infections where there is the presence of a high concentration of monovalent cations like NaCl and KCl (Hood et al., 2010).
The immunological role of CarO protein in A. baumannii is studied inadequately. Sato et al. (2017) showed that clinical A. baumannii isolates expressing higher CarO mRNA levels negatively regulated TNF-α, IL-6 and IL-8 in lung epithelial cells. Recently, CarO has been linked to A. baumannii adhesion and virulence in host cells via inhibition of NF-kβ signaling (Zhang et al., 2019). However, the significance of this observation is debatable as the strain used in the study is ATCC 19606 where expression of CarO is significantly lesser than that of clinical strains (Sato et al., 2017).
OprD
OprD was first identified during outer membrane investigations of carbapenem resistant A. baumannii isolates (Dupont et al., 2005). It is an orthologous protein to a porin involved in the basic amino acid and imipenem transport in Pseudomonas aeruginosa (Hancock and Brinkman, 2002). Crystallographic studies of a conserved P. aeruginosa OprD revealed a monomeric 18-stranded β-barrel structure characterized by a very narrow pore constriction (Biswas et al., 2007). The amino acid conservation at structural domains between A. baumannii and P. aeruginosa OprD porins indicate its putative function in A. baumannii. However, sequence and homology analysis of A. baumannii OprD showed that it belongs to P. aeruginosa OprQ, a protein involved in resisting low-iron or magnesium and low oxygen stresses (Catel-Ferreira et al., 2012). Recombinant A. baumannii OprD did not conduct antibiotics but partially bound to Fe2+ and Mg2+ cations. An isogenic deletion mutant of A. baumannii OprD did not affect MICs of β-lactams (Smani and Pachón, 2013), but in A. baylyi spp., a significant reduction in MIC of imipenem, ertapenem and meropenem is observed (Morán-Barrio et al., 2017). Despite these two heralding reports on lack of relationship between OprD and antibiotic resistance in A. baumannii, single nucleotide polymorphisms and insertional elements in OprD have been frequently identified in MDR A. baumannii signifying its role in resistance. For instance, Yang et al., 2015; Liu et al., 2016 and Lai et al., 2018 showed SNP clusters in OprD in MDR and tigecycline-resistant A. baumannii, respectively. Downregulation of OprD was observed in MDR (Asai et al., 2014) and pan drug-resistant (Cuenca et al., 2011) A. baumannii clinical strains. Insertion of mobile element ISAba1 upstream to the gene was also associated with increased carbapenem MICs of A. baumannii sequence type 107 strains (Costa et al., 2019). OprD was renamed to OccAB1 by Zahn et al. (2016), while solving its crystal structure. In their work, Zahn et al., resolved structures of four carboxylate channels OccAB1, 2, 3, and 4 and showed that OccAB1 has the largest channel size with corresponding high rates of small-molecule shuttle, including amino acids, sugars, and antibiotics, contrary to previous observations. The particularly large pore size of OccAB1 facilitates the objective translocation of both positive and negative substrates at low energy cost (Benkerrou and Ceccarelli, 2018). On the other hand, OccAB2, OccAB3, and OccAB4 mediate hydroxycinnamate (Smith et al., 2003), vanillate (Segura et al., 1999), and benzoate (Clark et al., 2002) transport, respectively. Being induced by limitation of metal ions, it can be presumed that OccAB1 may play a significant role in combating host-induced nutritional immunity and stress survival.
Diversity in A. baumannii OMP Architecture, Expression, and Function
Besides the above three OMPs, a variety of proteins are identified in the outer membrane of A. baumannii with varied expressions and distinctive roles. One of the most abundant OMPs in A. baumannii is Omp33. Crystal structure of the protein revealed its function as a putative gated channel contributing to low permeability of the outer membrane (Abellón-Ruiz et al., 2018). Intracellular A. baumannii in the host cell expresses DcaP OMP in abundance. Crystallographic studies on DcaP revealed a trimeric porin structure with affinity to dicarboxylic acids and sulbactam (Bhamidimarri et al., 2019). The most abundant OMP under osmotic stress in A. baumannii is Omp38 (Jyothisri et al., 1999). Intracellular A. baumannii secretes Omp38, which localizes to the mitochondria stimulating the release of proapoptotic molecules such as cytochrome c and apoptosis-inducing factor (Choi et al., 2005). Oxidative stress in A. baumannii induces expression of AbuO, a homolog of Escherichia coli TolC OMP involved in resistance to amikacin, carbenicillin, ceftriaxone, meropenem, streptomycin, and tigecycline, and hospital-based disinfectants like benzalkonium chloride and chlorhexidine (Srinivasan et al., 2015). Under iron-limiting conditions, a 76-kDa iron-repressible OMP termed fhuE was overexpressed in A. baumannii to facilitate uptake of xenosiderophores desferricoprogen, rhodotorulic acid and desferrioxamine B (Funahashi et al., 2012). Besides fhuE, two other siderophore (acinetobactin) uptake proteins, BfnH and BauA are also elucidated in the outer membrane of A. baumannii (Sefid and Rasooli, 2012; Aghajani et al., 2019). The translation initiation factor EF-Tu, typically a cytoplasmic protein, is also found to localize in the outer membrane in A. baumannii (Dallo et al., 2012). Membrane associated EF-Tu binds to DsbA protein in the periplasm and assists in disulfide bonding during protein folding (Premkumar et al., 2014). Externally, EF-Tu binds to fibronectin thus mediating host cell adhesion (Dallo et al., 2012; Harvey et al., 2019). Decreased expression of a 33–36 kDa OMP (Clark, 1996; del Mar Tomás et al., 2005) and a 29 kDa (Jeong et al., 2009) is associated with imipenem resistance among A. baumannii. Serodiagnostic studies revealed an antigenic 34.4-kDa OMP specific to sera from A. baumannii infected patients (Islam et al., 2011). Upregulation of this protein along with downregulation of CarO and OprD was found to mediate imipenem resistance (Luo et al., 2011). The protein along with OmpA and TonB-dependant copper receptor was identified as fibronectin binding protein during infection (Smani et al., 2012). In silico exploration into the genome of A. baumannii revealed a nuclease (NucAb), BamA (Oma87), FilF, and TolB in the outer membrane as potential vaccine targets. Immunization with these recombinant proteins protected mice from lethal challenge with A. baumannii (Singh et al., 2014, 2016 and Singh et al., 2017; Garg et al., 2016; Song et al., 2018; Rasooli et al., 2020). In another effort toward developing a subunit vaccine against A. baumannii infections, a fusion protein of OmpK and Omp22 was synthesized which provided significantly greater protection against A. baumannii challenge in mice than those immunized with either of the two proteins individually (Huang et al., 2016; Guo et al., 2017; Guo S.J. et al., 2018).
Conclusion
One of the critical gaps in combating A. baumannii is deciphering its overall membrane permeability. Significant progress has been made during the last decade in our understanding of how A. baumannii OMPs mediate antibiotic resistance and virulence in the host cells. Remarkable breakthroughs have also been made in understanding the regulatory mechanisms behind OMP expression and the mechanisms of antibiotic uptake. However, these efforts fall short in many aspects. The knowledge about in vitro or in vivo OMP assembly and folding dynamics in lipid bilayers is scarce. The crystal structure of most studied A. baumannii OMP, OmpA is still elusive, although NLS domain structure has been resolved. Due to its complex structure and hydrophobic loops in its structure, expression and purification of recombinant OmpA presents various hurdles. The solution of crystal structure is decisive in molecular dynamic studies tracking the antibiotic entry and exit through OMPs. Many questions still remain unanswered. How does the beta barrel assembly complex in A. baumannii function? What are the different chaperones mediating OMP folding in A. baumannii? Does the expression of OMPs in A. baumannii depend on transcription factors alone or is it small RNA mediated? Although in small number, concerted efforts are directed toward solving these problems. Crystal structures of CarO1, CarO2, OccAB1 through 4, DcaP, PiuA, Omp33 BauA have been resolved. Understanding the magnitude of posttranscriptional regulation in A. baumannii OMP synthesis is a necessary goal, as this aspect has been overlooked till date. In the next few years, it is likely that A. baumannii OMP compendium will be resolved with novel insights into its structure, diversity, biogenesis, and expression, furthering our efforts in confronting antibiotic resistance and virulence in A. baumannii.
Statements
Author contributions
SU, AS, and RP conceptualized the manuscript and contributed the ideas on the texts. SU and AS wrote the first draft of the manuscript. SU and RP edited the subsequent versions. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.589234/full#supplementary-material
References
1
AbbasiE.GoudarziH.HashemiA.ChiraniA. S.ArdebiliA.GoudarziM.et al (2020). Decreased carO gene expression and OXA-type carbapenemases among extensively drug-resistant Acinetobacter baumannii strains isolated from burn patients in Tehran, Iran.Acta Microbiol. Immunol Hung.10.1556/030.2020.01138[Epub ahead of print].
2
AbbottI.CerqueiraG. M.BhuiyanS.PelegA. Y. (2013). Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies.Expert Rev. Anti. Infect. Ther.11395–409. 10.1586/eri.13.21
3
Abellón-RuizJ.ZahnM.BasléA.van den BergB. (2018). Crystal structure of the Acinetobacter baumannii outer membrane protein Omp33.Acta Crystallogr D Struct. Biol. Struct. Biol.74(Pt 9), 852–860. 10.1107/S205979831800904X
4
AgarwalB.KarthikeyanR.GayathriP.RameshBabuB.AhmedG.JagannadhamM. V. (2019). Studies on the mechanism of multidrug resistance of Acinetobacter baumannii by proteomic analysis of the outer membrane vesicles of the bacterium.J. Proteins Proteo101–15. 10.1007/s42485-018-0001-4
5
AghajaniZ.RasooliI.Mousavi GargariS. L. (2019). Exploitation of two siderophore receptors, BauA and BfnH, for protection against Acinetobacter baumannii infection.APMIS127753–763. 10.1111/apm.12992
6
AlzubaidiA. N.AlkozaiZ. M. (2015). Immunogenic properties of outer membrane protein of Acinetobacter baumannii that loaded on chitosan nanoparticles.Am. J. Bio Med.359–74.
7
AnZ.HuangX.ZhengC.DingW. (2019). Acinetobacter baumannii outer membrane protein A induces HeLa cell autophagy via MAPK/JNK signaling pathway.Int. J. Med. Microbiol.30997–107. 10.1016/j.ijmm.2018.12.004
8
AnZ.SuJ. (2019). Acinetobacter baumannii outer membrane protein 34 elicits NLRP3 inflammasome activation via mitochondria-derived reactive oxygen species in RAW264. 7 macrophages.Microbes Infect.21143–153. 10.1016/j.micinf.2018.10.005
9
AsadollahiP.AkbariM.SoroushS.TaherikalaniM.AsadollahiK.SayehmiriK.et al (2012). Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients.Burns381198–1203. 10.1016/j.burns.2012.04.008
10
AsaiS.UmezawaK.IwashitaH.OhshimaT.OhashiM.SasakiM.et al (2014). An outbreak of blaOXA-51-like- and blaOXA-66-positive Acinetobacter baumannii ST208 in the emergency intensive care unit.J. Med. Microbiol.63(Pt 11), 1517–1523. 10.1099/jmm.0.077503-0
11
BazmaraH.RasooliI.JahangiriA.SefidF.AstanehS. D. A.PayandehZ. (2019). Antigenic properties of iron regulated proteins in Acinetobacter baumannii: an in silico approach.Int. J. Pept. Res. Ther.25205–213. 10.1007/s10989-017-9665-6
12
BazyleuA.KumarA. (2014). Incubation temperature, osmolarity, and salicylate affect the expression of resistance-nodulation-division efflux pumps and outer membrane porins in Acinetobacter baumannii ATCC19606T.FEMS Microbiol. Lett.357136–143. 10.1111/1574-6968.12530
13
BenkerrouD.CeccarelliM. (2018). Free energy calculations and molecular properties of substrate translocation through OccAB porins.Phys. Chem. Chem. Phys.208533–8546. 10.1039/c7cp08299a
14
BenmahmodA. B.SaidH. S.IbrahimR. H. (2019). Prevalence and Mechanisms of Carbapenem Resistance among Acinetobacter baumannii clinical isolates in Egypt.Microb. Drug Resist.25480–488. 10.1089/mdr.2018.0141
15
BhamidimarriS. P.ZahnM.PrajapatiJ. D.SchlebergerC.SöderholmS.HooverJ.et al (2019). A multidisciplinary approach toward identification of antibiotic scaffolds for Acinetobacter baumannii.Structure27268.e6–280.e6. 10.1016/j.str.2018.10.021
16
BhattacharyyaT.SharmaA.AkhterJ.PathaniaR. (2017). The small molecule IITR08027 restores the antibacterial activity of fluoroquinolones against multidrug-resistant Acinetobacter baumannii by efflux inhibition.Int. J. Antimicrob. Agents50219–226. 10.1016/j.ijantimicag.2017.03.005
17
BiswasS.MohammadM. M.PatelD. R.MovileanuL.van den BergB. (2007). Structural insight into OprD substrate specificity.Nat. Struct. Mol. Biol.141108–1109. 10.1038/nsmb1304
18
BorneleitP.KleberH. P. (1991). “The outer membrane of acinetobacter: structure-function relationships,” in The Biology of Acinetobacter, Federation of European Microbiological Societies Symposium Series, Vol. 57, edsTowner K. J.Bergogne-BérézinE.FewsonC. A. (Boston, MA: Springer). 10.1007/978-1-4899-3553-3_18
19
BouvetP. J.GrimontP. A. (1987). Identification and biotyping of clinical isolates of Acinetobacter.Ann. Inst. Pasteur Microbiol.138569–578. 10.1016/0769-2609(87)90042-1
20
BrossardK. A.CampagnariA. A. (2012). The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells.Infect. Immun.80228–233. 10.1128/IAI.05913-11
21
CardosoJ. P.CayôR.GirardelloR.GalesA. C. (2016). Diversity of mechanisms conferring resistance to β-lactams among OXA-23-producing Acinetobacter baumannii clones.Diagn. Microbiol. Infect. Dis.8590–97. 10.1016/j.diagmicrobio.2016.01.018
22
Catel-FerreiraM.CoadouG.MolleV.MugnierP.NordmannP.SiroyA.et al (2011). Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii.J. Antimicrob. Chemother.662053–2056. 10.1093/jac/dkr267
23
Catel-FerreiraM.NehméR.MolleV.ArandaJ.BouffartiguesE.ChevalierS.et al (2012). Deciphering the function of the outer membrane protein OprD homologue of Acinetobacter baumannii.Antimicrob. Agents Chemother.563826–3832. 10.1128/AAC.06022-11
24
Catel-FerreiraM.MartiS.GuillonL.JaraL.CoadouG.MolleV.et al (2016). The outer membrane porin OmpW of Acinetobacter baumannii is involved in iron uptake and colistin binding.FEBS Lett.590224–231. 10.1002/1873-3468.12050
25
ChoiC. H.HyunS. H.KimJ.LeeY. C.SeolS. Y.ChoD. T.et al (2008a). Nuclear translocation and DNAse I-like enzymatic activity of Acinetobacter baumannii outer membrane protein A.FEMS Microbiol. Lett.28862–67. 10.1111/j.1574-6968.2008.01323.x
26
ChoiC. H.HyunS. H.LeeJ. Y.LeeJ. S.LeeY. S.KimS. A.et al (2008b). Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity.Cell Microbiol.10309–319. 10.1111/j.1462-5822.2007.01041.x
27
ChoiC. H.LeeJ. S.LeeY. C.ParkT. I.LeeJ. C. (2008c). Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells.BMC Microbiol.8:216. 10.1186/1471-2180-8-216
28
ChoiC. H.LeeE. Y.LeeY. C.ParkT. I.KimH. J.HyunS. H.et al (2005). Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells.Cell Microbiol.71127–1138. 10.1111/j.1462-5822.2005.00538.x
29
ChoiC. H.LeeJ. S.LeeJ. C. (2007). “A versatile role of outer membrane protein a in pathogenesis of Acinetobacter baumannii,” in Proceedings of the Microbiological Society of Korea Conference, (South Korea: The Microbiological Society of Korea), 107–108.
30
ClarkR. B. (1996). Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein.J. Antimicrob. Chemother.38245–251. 10.1093/jac/38.2.245
31
ClarkT. J.MomanyC.NeidleE. L. (2002). The benPK operon, proposed to play a role in transport, is part of a regulon for benzoate catabolism in Acinetobacter sp. strain ADP1.Microbiology148(Pt 4), 1213–1223. 10.1099/00221287-148-4-1213
32
ConferA. W.AyalewS. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity.Vet. Microbiol.163207–222. 10.1016/j.vetmic.2012.08.019
33
CostaR. F.CayôR.MatosA. P.GirardelloR.MartinsW.Carrara-MarroniF. E.et al (2019). Temporal evolution of Acinetobacter baumannii ST107 clone: conversion of blaOXA-143 into blaOXA-231 coupled with mobilization of ISAba1 upstream occAB1.Res. Microbiol.17053–59. 10.1016/j.resmic.2018.07.001
34
CuencaF. F.PascualA.MartínezL. M.ConejoM. C.PereaE. J. (2003). Evaluation of SDS-polyacrylamide gel systems for the study of outer membrane protein profiles of clinical strains of Acinetobacter baumannii.J. Basic Microbiol.43194–201. 10.1002/jobm.200390022
35
CuencaF. F.SmaniY.Gómez-SánchezM. C.Docobo-PérezF.Caballero-MoyanoF. J.Domínguez-HerreraJ.et al (2011). Attenuated virulence of a slow-growing pandrug-resistant Acinetobacter baumannii is associated with decreased expression of genes encoding the porins CarO and OprD-like.Int. J. Antimicrob. Agents38548–549. 10.1016/j.ijantimicag.2011.08.002
36
DalloS. F.ZhangB.DennoJ.HongS.TsaiA.HaskinsW.et al (2012). Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin.ScientificWorldJournal2012:128705. 10.1100/2012/128705
37
DarbandianP.SefidF. (2016). Evaluation of OMP A (Outer Membrane Protein A) linear and conformational epitopes in Acinetobacter baumannii.Int. J. Adv. Biotechnol. Res.7:3.
38
De GregorioE.Del FrancoM.MartinucciM.RoscettoE.ZarrilliR.Di NoceraP. P. (2015). Biofilm-associated proteins: news from Acinetobacter.BMC Genomics16:933. 10.1186/s12864-015-2136-6
39
del Mar TomásM.BeceiroA.PérezA.VelascoD.MoureR.VillanuevaR.et al (2005). Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii.Antimicrob. Agents Chemother.495172–5175. 10.1128/AAC.49.12.5172-5175.2005
40
DupontM.PagèsJ. M.LafitteD.SiroyA.BolletC. (2005). Identification of an OprD homologue in Acinetobacter baumannii.J. Proteome Res.42386–2390. 10.1021/pr050143q
41
FahmyI. L.AminM. H.MohamedF. A.HashemA. G. (2018). Colonization of multi-drug resistant (MDR) Acinetobacter baumannii isolated from tertiary hospitals in Egypt and the possible role of the outer membrane protein (OmpA.).Int. Res. J. Pharm.9103–110.
42
FernandoD.KumarA. (2012). Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606.J. Antimicrob. Chemother.67569–572. 10.1093/jac/dkr519
43
FonsecaE. L.ScheideggerE.FreitasF. S.CiprianoR.VicenteA. C. (2013). Carbapenem-resistant Acinetobacter baumannii from Brazil: role of CarO alleles expression and blaOXA-23 gene.BMC Microbiol.13:245. 10.1186/1471-2180-13-245
44
FunahashiT.TanabeT.MiharaK.MiyamotoK.TsujiboH.YamamotoS. (2012). Identification and characterization of an outer membrane receptor gene in Acinetobacter baumannii required for utilization of desferricoprogen, rhodotorulic acid, and desferrioxamine B as xenosiderophores.Biol. Pharm. Bull.35753–760. 10.1248/bpb.35.753
45
GaddyJ. A. (2010). Acinetobacter baumannii Virulence Attributes: The Roles of Outer Membrane Protein A, Acinetobactin-mediated Iron Acquisition Functions, and Blue Light Sensing Protein A.Doctoral dissertation, Miami University, Oxford.
46
GaddyJ. A.TomarasA. P.ActisL. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells.Infect. Immun.773150–3160. 10.1128/IAI.00096-09
47
GargN.SinghR.ShuklaG.CapalashN.SharmaP. (2016). Immunoprotective potential of in silico predicted Acinetobacter baumannii outer membrane nuclease. NucAb.Int. J. Med. Microbiol.3061–9. 10.1016/j.ijmm.2015.10.005
48
Garnacho-MonteroJ.Ortiz-LeybaC.Fernández-HinojosaE.Aldabó-PallásT.CayuelaA.Marquez-VácaroJ. A.et al (2005). Acinetobacter baumannii ventilator-associated pneumonia: epidemiological and clinical findings.Intensive Care Med.31649–655. 10.1007/s00134-005-2598-0
49
GiammancoA.CalàC.FascianaT.DowzickyM. J. (2017). Global assessment of the activity of tigecycline against multidrug-resistant gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial.mSphere2:e00310-16. 10.1128/mSphere.00310-16
50
GoelV. K.KapilA. (2001). Monoclonal antibodies against the iron regulated outer membrane proteins of Acinetobacter baumannii are bactericidal.BMC Microbiol.1:16. 10.1186/1471-2180-1-16
51
GribunA.NitzanY.PechatnikovI.HershkovitsG.KatcoffD. J. (2003). Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii.Curr. Microbiol.47434–443. 10.1007/s00284-003-4050-4
52
GuoS. J.RenS.XieY. E. (2017). Recombinant expression and purification of the OmpK outer membrane protein of Acinetobacter baumannii.J. Pathog. Biol.12837–839.
53
GuoS. J.RenS.XieY. E. (2018). Evaluation of the protective efficacy of a fused OmpK/Omp22 protein vaccine Candidate against Acinetobacter baumannii Infection in Mice.Biomed. Environ. Sci.31155–158. 10.3967/bes2018.019
54
GuoY.XunM.HanJ. (2018). A bovine myeloid antimicrobial peptide (BMAP-28) and its analogs kill pan-drug-resistant Acinetobacter baumannii by interacting with outer membrane protein A (OmpA).Medicine97:e12832. 10.1097/MD.0000000000012832
55
HancockR. E.BrinkmanF. S. (2002). Function of Pseudomonas porins in uptake and efflux.Annu. Rev. Microbiol.5617–38. 10.1146/annurev.micro.56.012302.160310
56
HansonK. G.KaleV. C.DesaiA. J. (1994). The possible involvement of cell surface and outer membrane proteins of Acinetobacter sp. A3 in crude oil degradation.FEMS Microbiol. Lett.122275–279. 10.1111/j.1574-6968.1994.tb07180.x
57
HardingC. M.HennonS. W.FeldmanM. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence.Nat. Rev. Microbiol.1691–102. 10.1038/nrmicro.2017.148
58
HarveyK. L.JarockiV. M.CharlesI. G.DjordjevicS. P. (2019). The diverse functional roles of elongation factor Tu (EF-Tu) in microbial pathogenesis.Front. Microbiol.10:2351. 10.3389/fmicb.2019.02351
59
HoodM. I.JacobsA. C.SayoodK.DunmanP. M.SkaarE. P. (2010). Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations.Antimicrob. Agents Chemother.541029–1041. 10.1128/AAC.00963-09
60
HuangW.YaoY.WangS.XiaY.YangX.LongQ.et al (2016). Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii.Sci. Rep.6:20724. 10.1038/srep20724
61
HwaW. E.SubramaniamG.MansorM. B.YanO. S.Gracie, AnbazhaganD.et al (2010). Iron regulated outer membrane proteins (IROMPs) as potential targets against carbapenem-resistant Acinetobacter spp. isolated from a Medical Centre in Malaysia.Indian J. Med. Res.131578–583.
62
IslamA. H.SinghK. K.IsmailA. (2011). Demonstration of an outer membrane protein that is antigenically specific for Acinetobacter baumannii.Diagn. Microbiol. Infec.t Dis.6938–44. 10.1016/j.diagmicrobio.2010.09.008
63
IyerR.MoussaS. H.Durand-RévilleT. F.TommasiR.MillerA. (2018). Acinetobacter baumannii OmpA is a selective antibiotic permeant porin.ACS Infect. Dis.4373–381. 10.1021/acsinfecdis.7b00168
64
JahangiriA.RasooliI.OwliaP.FooladiA. A.SalimianJ. (2017). In silico design of an immunogen against Acinetobacter baumannii based on a novel model for native structure of outer membrane protein A.Microb. Pathog.105201–210. 10.1016/j.micpath.2017.02.028
65
JeongH. W.CheongH. J.KimW. J.KimM. J.SongK. J.SongJ. W.et al (2009). Loss of the 29-kilodalton outer membrane protein in the presence of OXA-51-like enzymes in Acinetobacter baumannii is associated with decreased imipenem susceptibility.Microb. Drug Resist.15151–158. 10.1089/mdr.2009.0828
66
JinJ. S.KwonS. O.MoonD. C.GurungM.LeeJ. H.KimS. I.et al (2011). Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles.PLoS One6:e17027. 10.1371/journal.pone.0017027
67
JyothisriK.DeepakV.RajeswariM. R. (1999). Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii.FEBS lett.44357–60. 10.1016/s0014-5793(98)01679-2
68
KhorsiK.MessaiY.AmmariH.HamidiM.BakourR. (2018). ISAba36 inserted into the outer membrane protein gene carO and associated with the carbapenemase gene blaOXA-24-like in Acinetobacter baumannii.J. Glob. Antimicrob. Resist.15107–108. 10.1016/j.jgar.2018.08.020
69
KimB. R.YangE. K.KimD. Y.KimS. H.MoonD. C.LeeJ. H.et al (2012). Generation of anti-tumour immune response using dendritic cells pulsed with carbonic anhydrase IX-Acinetobacter baumannii outer membrane protein A fusion proteins against renal cell carcinoma.Clin. Exp. Immunol.16773–83. 10.1111/j.1365-2249.2011.04489.x
70
KimB. R.YangE. K.KimS. H.MoonD. C.KimH. J.LeeJ. C.et al (2011). Immunostimulatory activity of dendritic cells pulsed with carbonic anhydrase IX and Acinetobacter baumannii outer membrane protein A for renal cell carcinoma.J. Microbiol.49115–120. 10.1007/s12275-011-1037-x
71
KimD. Y.KimB.KimS. H.LeeJ.KimJ. C.ParkC. H.et al (2011). 242 Acinetobacter baumannii outer membrane protein a enhance immune responses to renal cell carcinoma vaccines targeting carbonic anhydrase IX.J. Urol.185:e98. 10.1016/j.juro.2011.02.311
72
KimD. H.KoK. S. (2015). ISAba15 inserted into outer membrane protein gene carO in Acinetobacter baumannii.J. Bacteriol. Virol.4551–53. 10.4167/jbv.2015.45.1.51
73
KimS. A.YooS. M.HyunS. H.ChoiC. H.YangS. Y.KimH. J.et al (2008). Global gene expression patterns and induction of innate immune response in human laryngeal epithelial cells in response to Acinetobacter baumannii outer membrane protein A.FEMS Immunol. Med. Microbiol.5445–52. 10.1111/j.1574-695X.2008.00446.x
74
KimS. W.OhM. H.JunS. H.JeonH.KimS. I.KimK.et al (2016). Outer membrane Protein A plays a role in pathogenesis of Acinetobacter nosocomialis.Virulence7413–426. 10.1080/21505594.2016.1140298
75
KuoH. Y.ChaoH. H.LiaoP. C.HsuL.ChangK. C.TungC. H.et al (2017). Functional Characterization of Acinetobacter baumannii Lacking the RNA Chaperone Hfq.Front. Microbiol.8:2068. 10.3389/fmicb.2017.02068
76
KwonH. I.KimS.OhM. H.NaS. H.KimY. J.JeonY. H.et al (2017). Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain.J. Antimicrob. Chemother.723012–3015. 10.1093/jac/dkx257
77
KwonH. I.KimS.OhM. H.ShinM.LeeJ. C. (2019). Distinct role of outer membrane protein A in the intrinsic resistance of Acinetobacter baumannii and Acinetobacter nosocomialis.Infect. Genet. Evol.6733–37. 10.1016/j.meegid.2018.10.022
78
LaiC. C.ChenC. C.LuY. C.ChuangY. C.TangH. J. (2018). In vitro activity of cefoperazone and cefoperazone-sulbactam against carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa.Infect. Drug. Resist.1225–29. 10.2147/IDR.S181201
79
LeeC. R.LeeJ. H.ParkM.ParkK. S.BaeI. K.KimY. B.et al (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options.Front. Cell Infect. Microbiol.7:55. 10.3389/fcimb.2017.00055
80
LeeJ. C.KoertenH.van den BroekP.BeekhuizenH.WolterbeekR.van den BarselaarM.et al (2006). Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells.Res. Microbiol.157360–366. 10.1016/j.resmic.2005.09.011
81
LeeJ. S.ChoiC. H.KimJ. W.LeeJ. C. (2010). Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting.J. Microbiol.48387–392. 10.1007/s12275-010-0155-1
82
LeeJ. S.KimJ. W.ChoiC. H.LeeW. K.ChungH. Y.LeeJ. C. (2008). Anti-tumor activity of Acinetobacter baumannii outer membrane protein A on dendritic cell-based immunotherapy against murine melanoma.J. Microbiol.46221–227. 10.1007/s12275-008-0052-z
83
LeeJ. S.LeeJ. C.LeeC. M.JungI. D.JeongY. I.SeongE. Y.et al (2007). Outer membrane protein A of Acinetobacter baumannii induces differentiation of CD4+ T cells toward a Th1 polarizing phenotype through the activation of dendritic cells.Biochem. Pharmacol.7486–97. 10.1016/j.bcp.2007.02.012
84
LeeY.KimC. K.LeeH.JeongS. H.YongD.LeeK. (2011). A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the bla(OXA-23) gene and disrupting the outer membrane protein gene CarO in Acinetobacter baumannii.Antimicrob. Agents Chemother.55361–363. 10.1128/AAC.01672-09
85
LiQ.LiZ.QuY.LiH.XingJ.HuD. (2015). A study of outer membrane protein and molecular epidemiology of carbapenem-resistant Acinetobacter baumannii.Zhonghua Wei Zhong Bing Ji Jiu Yi Xue27611–615. 10.3760/cma.j.issn.2095-4352.2015.07.014
86
LimanskyA. S.MussiM. A.VialeA. M. (2002). Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance.J. Clin. Microbiol.404776–4778. 10.1128/jcm.40.12.4776-4778.2002
87
LinM. F.LanC. Y. (2014). Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside.World J. Clin. Cases2787–814. 10.12998/wjcc.v2.i12.787
88
LinM. F.LinY. Y.LanC. Y. (2020). Characterization of biofilm production in different strains of Acinetobacter baumannii and the effects of chemical compounds on biofilm formation.PeerJ8:e9020. 10.7717/peerj.9020
89
LinM. F.TsaiP. W.ChenJ. Y.LinY. Y.LanC. Y. (2015). OmpA binding mediates the effect of antimicrobial peptide LL-37 on Acinetobacter baumannii.PLoS One10:e0141107. 10.1371/journal.pone.0141107
90
LiouM. L.SooP. C.LingS. R.KuoH. Y.TangC. Y.ChangK. C. (2014). The sensor kinase BfmS mediates virulence in Acinetobacter baumannii.Microbiol. Immunol. Infec.47275–281. 10.1016/j.jmii.2012.12.004
91
LiuL.CuiY.ZhengB.JiangS.YuW.ShenP.et al (2016). Analysis of tigecycline resistance development in clinical Acinetobacter baumannii isolates through a combined genomic and transcriptomic approach.Sci. Rep.6:26930. 10.1038/srep26930
92
LuoL.JiangX.WuQ.WeiL.LiJ.YingC. (2011). Efflux pump overexpression in conjunction with alternation of outer membrane protein may induce Acinetobacter baumannii resistant to imipenem.Chemotherapy5777–84. 10.1159/000323620
93
McConnellM. J.Domínguez-HerreraJ.SmaniY.López-RojasR.Docobo-PérezF.PachónJ. (2011). Vaccination with outer membrane complexes elicits rapid protective immunity to multidrug-resistant Acinetobacter baumannii.Infect. Immun.79518–526. 10.1128/IAI.00741-10
94
MehdinejadianiK.BandehpourM.HashemiA.RanjbarM. M.TaheriS.JalaliS. A.et al (2019). In Silico design and evaluation of Acinetobacter baumannii outer membrane protein a antigenic peptides as vaccine candidate in immunized mice.Iran J. Allergy Asthma Immunol.18655–663. 10.18502/ijaai.v18i6.2178
95
MirshekarM.ShahcheraghiF.AziziO.SolgiH.BadmastiF. (2018). Diversity of Class 1 integrons, and disruption of carO and dacD by insertion sequences among Acinetobacter baumannii Isolates in Tehran. Iran.Microb. Drug. Resist.24359–366. 10.1089/mdr.2017.0152
96
MogantyR. R.VashisttJ.TiwariV.KapilA. (2011). Differential expression of Outer membrane proteins in early stages of meropenem-resistance in Acinetobacter baumannii.J. Integrat. OMICS1281–287. 10.5584/jiomics.v1i2.67
97
MoonD. C.ChoiC. H.LeeJ. H.ChoiC. W.KimH. Y.ParkJ. S.et al (2012). Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles.J. Microbiol.50155–160. 10.1007/s12275-012-1589-4
98
Morán-BarrioJ.CameranesiM. M.RellingV.LimanskyA. S.BrambillaL.VialeA. M. (2017). The Acinetobacter outer membrane contains multiple specific channels for carbapenem β-lactams as revealed by kinetic characterization analyses of imipenem permeation into Acinetobacter baylyi Cells.Antimicrob. Agents Chemother.61:e01737-16. 10.1128/AAC.01737-16
99
MortensenB. L.SkaarE. P. (2012). Host-microbe interactions that shape the pathogenesis of Acinetobacter baumannii infection.Cell Microbiol.141336–1344. 10.1111/j.1462-5822.2012.01817.x
100
MostachioA. K.LevinA. S.RizekC.RossiF.ZerbiniJ.CostaS. F. (2012). High prevalence of OXA-143 and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. isolates in Brazil.Int. J. Antimicrob. Agents39396–401. 10.1016/j.ijantimicag.2012.01.021
101
MussiM. A.LimanskyA. S.RellingV.RavasiP.ArakakiA.ActisL. A.et al (2011). Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel.J. Bacterial.1934736–4748. 10.1128/JB.01533-10
102
MussiM. A.LimanskyA. S.VialeA. M. (2005). Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins.Antimicrob. Agents Chemother.491432–1440. 10.1128/AAC.49.4.1432-1440.2005
103
MussiM. A.RellingV. M.LimanskyA. S.VialeA. M. (2007). CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake.FEBS Lett.5815573–5578. 10.1016/j.febslet.2007.10.063
104
NanL. I.ZhangH. J.WangY.MengQ. I.GuoW. X.WangZ.et al (2018). Study on the drug resistance of clinical strain of Acinetobacter baumannii mediated by TEM-1 beta-lactamases and porin CarO.Tianjin Med. J.46246–250.
105
NieD.HuY.ChenZ.LiM.HouZ.LuoX.et al (2020). Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection.J. Biomed. Sci.27:26. 10.1186/s12929-020-0617-7
106
NikaidoH. (2003). Molecular basis of bacterial outer membrane permeability revisited.Microbiol. Mol. Biol. Rev.67593–656. 10.1128/mmbr.67.4.593-656.2003
107
NitzanY.DeutschE. B.PechatnikovI. (2002). Diffusion of beta-lactam antibiotics through oligomeric or monomeric porin channels of some gram-negative bacteria.Curr. Microbiol.45446–455. 10.1007/s00284-002-3778-6
108
NitzanY.PechatnikovI.Bar-ElD.WexlerH. (1999). Isolation and characterization of heat-modifiable proteins from the outer membrane of Porphyromonas asaccharolytica and Acinetobacter baumannii.Anaerobe543–50. 10.1006/anae.1998.0181
109
NovovicK.MihajlovicS.VasiljevicZ.FilipicB.BegovicJ.JovcicB. (2015). Carbapenem-resistant Acinetobacter baumannii from Serbia: revision of CarO classification.PLoS One10:e0122793. 10.1371/journal.pone.0122793
110
OhK. W.KimK.IslamM. M.JungH. W.LimD.LeeJ. C.et al (2020). Transcriptional Regulation of the Outer Membrane Protein A in Acinetobacter baumannii.Microorganisms8:706. 10.3390/microorganisms8050706
111
PajandO.RezaeeM. A.NahaeiM. R.MahdianR.AghazadehM.SoroushM. H.et al (2013). Study of the carbapenem resistance mechanisms in clinical isolates of Acinetobacter baumannii: comparison of burn and non-burn strains.Burns391414–1419. 10.1016/j.burns.2013.03.024
112
ParkJ. S.LeeW. C.ChoiS.YeoK. J.SongJ. H.HanY. H.et al (2011). Overexpression, purification, crystallization and preliminary X-ray crystallographic analysis of the periplasmic domain of outer membrane protein A from Acinetobacter baumannii.Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun.67(Pt 12), 1531–1533. 10.1107/S1744309111038401
113
ParkY. K.JungS. I.ParkK. H.KimS. H.KoK. S. (2012). Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea.Int. J. Antimicrob. Agents3981–85. 10.1016/j.ijantimicag.2011.08.006
114
Parra-MillánR.Vila-FarrésX.Ayerbe-AlgabaR.VareseM.Sánchez-EncinalesV.BayóN.et al (2018). Synergistic activity of an OmpA inhibitor and colistin against colistin-resistant Acinetobacter baumannii: mechanistic analysis and in vivo efficacy.J. Antimicrob. Chemother.733405–3412. 10.1093/jac/dky343
115
PelegA. Y.SeifertH.PatersonD. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen.Clin. Microbiol. Rev.21538–582. 10.1128/CMR.00058-07
116
PormohammadA.MehdinejadianiK.GholizadehP.NasiriM. J.MohtavinejadN.DadashiM.et al (2020). Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis.Microb. Pathog.139:103887. 10.1016/j.micpath.2019.103887
117
PremkumarL.KurthF.DuprezW.GrøftehaugeM. K.KingG. J.HaliliM. A.et al (2014). Structure of the Acinetobacter baumannii dithiol oxidase DsbA bound to elongation factor EF-Tu reveals a novel protein interaction site.J. Biol. Chem.28919869–19880. 10.1074/jbc.M114.571737
118
RasooliI.AbdolhamidiR.JahangiriA.Darvish Alipour AstanehS. (2020). Outer membrane protein, Oma87 Prevents Acinetobacter baumannii Infection.Int. J. Pept. Res. Ther.1–8. 10.1007/s10989-020-10056-0[Epub ahead of print].
119
RavasiP.LimanskyA. S.RodriguezR. E.VialeA. M.MussiM. A. (2011). ISAba825, a functional insertion sequence modulating genomic plasticity and bla(OXA-58) expression in Acinetobacter baumannii.Antimicrob. Agents Chemother.55917–920. 10.1128/AAC.00491-10
120
RodloffA. C.DowzickyM. J. (2017). Antimicrobial susceptibility among european gram-negative and gram-positive isolates collected as part of the tigecycline evaluation and surveillance Trial (2004-2014).Chemotherapy621–11. 10.1159/000445022
121
RoyerS.de CamposP. A.AraújoB. F.FerreiraM. L.GonçalvesI. R.BatistãoD.et al (2018). Molecular characterization and clonal dynamics of nosocomial blaOXA-23 producing XDR Acinetobacter baumannii.PLoS One13:e0198643. 10.1371/journal.pone.0198643
122
RumboC.TomásM.MoreiraE. F.SoaresN. C.CarvajalM.SantillanaE.et al (2014). The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells.Infect. Immun.824666–4680. 10.1128/IAI.02034-14
123
Sánchez-EncinalesV.Álvarez-MarínR.Pachón-IbáñezM. E.Fernández-CuencaF.PascualA.Garnacho-MonteroJ.et al (2017). Overproduction of outer membrane protein A by Acinetobacter baumannii as a risk factor for nosocomial pneumonia. Bacteremia, and Mortality Rate Increase.J. Infect. Dis.215966–974. 10.1093/infdis/jix010
124
SatoK.NakaeT. (1991). Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance.J. Antimicrob. Chemother.2835–45. 10.1093/jac/28.1.35
125
SatoY.UnnoY.KawakamiS.UbagaiT.OnoY. (2017). Virulence characteristics of Acinetobacter baumannii clinical isolates vary with the expression levels of omps.J. Med. Microbiol.66203–212. 10.1099/jmm.0.000394
126
SefidF.RasooliI. (2012). “Cleft analysis Of BauA outer membrane protein in Acinetobacter Baumannii,” in Bioinformatics conference, (Tehran: Shahed University).
127
SeguraA.BünzP. V.D’ArgenioD. A.OrnstonL. N. (1999). Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter.J. Bacteriol.1813494–3504.
128
SenB.JoshiS. G. (2016). Studies on Acinetobacter baumannii involving multiple mechanisms of carbapenem resistance.J. Appl. Microbiol.120619–629. 10.1111/jam.13037
129
SharmaA.DubeyV.SharmaR.DevnathK.GuptaV. K.AkhterJ.et al (2018). The unusual glycine-rich C terminus of the Acinetobacter baumannii RNA chaperone Hfq plays an important role in bacterial physiology.J. Biol. Chem.29313377–13388. 10.1074/jbc.RA118.002921
130
SharmaM.SapkotaJ.JhaB.MishraB.BhattC. P. (2019). Biofilm formation and extended-spectrum beta-lactamase producer among Acinetobacter species isolated in a tertiary care hospital: a descriptive cross-sectional study.JNMA J. Nepal. Med. Assoc.57424–428.
131
SievertD. M.RicksP.EdwardsJ. R.SchneiderA.PatelJ.SrinivasanA.et al (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the centers for disease control and prevention, 2009-2010.Infect. Control Hosp. Epidemiol.341–14. 10.1086/668770
132
SinghR.CapalashN.SharmaP. (2017). Immunoprotective potential of BamA, the outer membrane protein assembly factor, against MDR Acinetobacter baumannii.Sci. Rep.7:12411. 10.1038/s41598-017-12789-3
133
SinghR.GargN.CapalashN.KumarR.KumarM.SharmaP. (2014). In silico analysis of Acinetobacter baumannii outer membrane protein BamA as a potential immunogen.Int. J. Pure Appl. Sci. Technol.2132–39.
134
SinghR.GargN.ShuklaG.CapalashN.SharmaP. (2016). Immunoprotective efficacy of Acinetobacter baumannii outer membrane protein.FilF, Predicted In silico as a potential vaccine candidate.Front. Microbiol.7:158. 10.3389/fmicb.2016.00158
135
SiroyA.MolleV.Lemaître-GuillierC.VallenetD.Pestel-CaronM.CozzoneA. J.et al (2005). Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii.Antimicrob. Agents Chemother.494876–4883. 10.1128/AAC.49.12.4876-4883.2005
136
SkariyachanS.TaskeenN.GantaM.Venkata KrishnaB. (2019). Recent perspectives on the virulent factors and treatment options for multidrug-resistant Acinetobacter baumannii.Crit. Rev. Microbiol.45315–333. 10.1080/1040841X.2019.1600472
137
SluskyJ. S.DunbrackR. L.Jr. (2013). Charge asymmetry in the proteins of the outer membrane.Bioinformatics292122–2128. 10.1093/bioinformatics/btt355
138
SmaniY.Dominguez-HerreraJ.PachónJ. (2013). Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii.J. Infect. Dis.2081561–1570. 10.1093/infdis/jit386
139
SmaniY.McConnellM. J.PachónJ. (2012). Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells.PLoS One7:e33073. 10.1371/journal.pone.0033073
140
SmaniY.PachónJ. (2013). Loss of the OprD homologue protein in Acinetobacter baumannii: impact on carbapenem susceptibility.Antimicrob. Agents Chemother.57:677. 10.1128/AAC.01277-12
141
SmithM. A.WeaverV. B.YoungD. M.OrnstonL. N. (2003). Genes for chlorogenate and hydroxycinnamate catabolism (hca) are linked to functionally related genes in the dca-pca-qui-pob-hca chromosomal cluster of Acinetobacter sp. strain ADP1.Appl. Environ. Microbiol.69524–532. 10.1128/aem.69.1.524-532.2003
142
SongX.ZhangH.ZhangD.XieW.ZhaoG. (2018). Bioinformatics analysis and epitope screening of a potential vaccine antigen TolB from Acinetobacter baumannii outer membrane protein.Infect. Genet. Evol.6273–79. 10.1016/j.meegid.2018.04.019
143
SrinivasanV. B.VaidyanathanV.RajamohanG. (2015). AbuO, a TolC-like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance.Antimicrob. Agents Chemother.591236–1245. 10.1128/AAC.03626-14
144
TohidiniaM.MoshtaghiounS. M.SefidF.FalahatiA. (2019). Functional exposed amino acids of CarO analysis as a potential vaccine candidate in Acinetobacter Baumannii.Int. J. Peptide Res. Thera.26, 1185–1197. 10.1007/s10989-019-09923-2
145
TsaiY. K.LiouC. H.LinJ. C.FungC. P.ChangF. Y.SiuL. K. (2020). Effects of different resistance mechanisms on antimicrobial resistance in Acinetobacter baumannii: a strategic system for screening and activity testing of new antibiotics.Int. J. Antimicrob. Agents55:105918. 10.1016/j.ijantimicag.2020.105918
146
van der KolkJ. H.EndimianiA.GraubnerC.GerberV.PerretenV. (2019). Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii.J. Glob. Antimicrob. Resist.1659–71. 10.1016/j.jgar.2018.08.011
147
VashistJ.TiwariV.KapilA.RajeswariM. R. (2010). Quantitative profiling and identification of outer membrane proteins of beta-lactam resistant strain of Acinetobacter baumannii.Proteome Res.91121–1128. 10.1021/pr9011188
148
VialeA. M.EvansB. A. (2020). Microevolution in the major outer membrane protein OmpA of Acinetobacter baumannii.Microb. Genom.6:e000381. 10.1099/mgen.0.000381
149
Vila-FarrésX.Parra-MillánR.Sánchez-EncinalesV.VareseM.Ayerbe-AlgabaR.BayóN.et al (2017). Combating virulence of Gram-negative bacilli by OmpA inhibition.Sci. Rep.714683. 10.1038/s41598-017-14972-y
150
WalzerG.RosenbergE.RonE. Z. (2006). The Acinetobacter outer membrane protein A (OmpA) is a secreted emulsifier.Environ. Microbiol.81026–1032. 10.1111/j.1462-2920.2006.00994.x
151
WangT.YuanQ.LingB. (2015). Analysis of the expression of the outer membrane protein in carbapenem-resistant Acinetobacter baumannii.Int. J. Lab. Med.20152295–2296.
152
WeinerL. M.WebbA. K.LimbagoB.DudeckM. A.PatelJ.KallenA. J.et al (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011-2014.Infect. Control Hosp. Epidemiol.371288–1301. 10.1017/ice.2016.174
153
World Health Organization [WHO] (2017). Global Priority list of Antibiotic-Resistant Bacteria to Guide Researach, Discovery and Development of New Antibiotics.Geneva: WHO.
154
WuX.ChavezJ. D.SchweppeD. K.ZhengC.WeisbrodC. R.EngJ. K.et al (2016). In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075.Nat. Commun.7:13414. 10.1038/ncomms13414
155
YangH.HuangL.BarnieP. A.SuZ.MiZ.ChenJ.et al (2015). Characterization and distribution of drug resistance associated β-lactamase, membrane porin and efflux pump genes in MDR A. baumannii isolated from Zhenjiang. China.Int. J. Clin. Exp. Med.815393–15402.
156
YunS. H.ChoiC. W.ParkS. H.LeeJ. C.LeemS. H.ChoiJ. S.et al (2008). Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition.J. Microbiol.46720–727. 10.1007/s12275-008-0202-3
157
YunS. H.ParkE. C.LeeS. Y.LeeH.ChoiC. W.YiY. S.et al (2018). Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug-resistant clinical strain, Acinetobacter baumannii DU202.Clin. Proteomics15:28. 10.1186/s12014-018-9204-2
158
ZahnM.BhamidimarriS. P.BasléA.WinterhalterM.van den BergB. (2016). Structural insights into outer membrane permeability of Acinetobacter baumannii.Structure24221–231. 10.1016/j.str.2015.12.009
159
ZahnM.D’AgostinoT.ErenE.BasléA.CeccarelliM.van den BergB. (2015). Small-molecule transport by CarO, an abundant eight-stranded β-Barrel outer membrane protein from Acinetobacter baumannii.J. Mol. Biol.4272329–2339. 10.1016/j.jmb.2015.03.016
160
ZhangL.LiangW.S-GXuMeiJ.DiY. Y.LanH. H.et al (2019). CarO promotes adhesion and colonization of Acinetobacter baumannii through inhibiting NF-êB pathways.Int. J. Clin. Exp. Med.122518–2524.
161
ZhaoS. Y.JiangD. Y.XuP. C.ZhangY. K.ShiH. F.CaoH. L.et al (2015). An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China.Ann. Clin. Microbiol. Antimicrob.14:7. 10.1186/s12941-015-0066-4
162
ZhouH.YaoY.ZhuB.RenD.YangQ.FuY.et al (2019). Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: a retrospective study from a Chinese hospital.Medicine98:e14937. 10.1097/MD.0000000000014937
163
ZhuL. J.ChenX. Y.HouP. F. (2019). Mutation of CarO participates in drug resistance in imipenem-resistant Acinetobacter baumannii.J. Clin. Lab. Anal.33:e22976. 10.1002/jcla.22976
Summary
Keywords
OmpA, CARO, OprD, antibiotic resistance, virulence
Citation
Uppalapati SR, Sett A and Pathania R (2020) The Outer Membrane Proteins OmpA, CarO, and OprD of Acinetobacter baumannii Confer a Two-Pronged Defense in Facilitating Its Success as a Potent Human Pathogen. Front. Microbiol. 11:589234. doi: 10.3389/fmicb.2020.589234
Received
31 July 2020
Accepted
11 September 2020
Published
06 October 2020
Volume
11 - 2020
Edited by
Benjamin Andrew Evans, University of East Anglia, United Kingdom
Reviewed by
Brock Aaron Arivett, Middle Tennessee State University, United States; Shakiba Darvish Alipour Astaneh, Semnan University, Iran
Updates
Copyright
© 2020 Uppalapati, Sett and Pathania.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjana Pathania, ranjana.pathania@bt.iitr.ac.in
This article was submitted to Antimicrobials, Resistance and Chemotherapy, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.