- Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India
The nucleotide alarmone (p)ppGpp, signaling the stringent response, is known for more than 5 decades. The cellular turnover of the alarmone is regulated by RelA/SpoT homolog (RSH) superfamily of enzymes. There are long RSHs (RelA, SpoT, and Rel) and short RSHs [small alarmone synthetases (SAS) and small alarmone hydrolases (SAH)]. Long RSHs are multidomain proteins with (p)ppGpp synthesis, hydrolysis, and regulatory functions. Short RSHs are single-domain proteins with a single (p)ppGpp synthesis/hydrolysis function with few exceptions having two domains. Mycobacterial RelZ is a dual-domain SAS with RNase HII and the (p)ppGpp synthetase activity. SAS is known to impact multiple cellular functions independently and in accordance with the long RSH. Few SAS in bacteria including RelZ synthesize pGpp, the third small alarmone, along with the conventional (p)ppGpp. SAS can act as an RNA-binding protein for the negative allosteric inhibition of (p)ppGpp synthesis. Here, we initially recap the important features and molecular functions of different SAS that are previously characterized to understand the obligation for the “alarmone pool” produced by the long and short RSHs. Then, we focus on the RelZ, especially the combined functions of RNase HII and (p)ppGpp synthesis from a single polypeptide to connect with the recent findings of SAS as an RNA-binding protein. Finally, we conclude with the possibilities of using single-stranded RNA (ssRNA) as an additional therapeutic strategy to combat the persistent infections by inhibiting the redundant (p)ppGpp synthetases.
Introduction
In 1969, Cashel and Gallant first identified the nucleotide alarmone molecules, guanosine-5', 3'-pentaphosphate (pppGpp) and guanosine-5', 3' -tetraphosphate (ppGpp), from amino acid-starved Escherichia coli (Cashel and Gallant, 1969). Intracellular levels of (p)ppGpp are controlled by RelA/SpoT homolog (RSH) proteins as a response to various external and internal stresses encountered by the organisms (Chatterji and Ojha, 2001; Potrykus and Cashel, 2008; Srivatsan and Wang, 2008; Wu and Xie, 2009; Roghanian et al., 2019). This is a direct pathway of stringent response in which the (p)ppGpp signals the massive switch from energy-consuming to energy-conserving process (Potrykus and Cashel, 2008; Abranches et al., 2009; Kriel et al., 2012; Gaca et al., 2013; Hauryliuk et al., 2015; Liu et al., 2015; Steinchen and Bange, 2016). In Gram-negative organisms, beta and gamma subgroups of proteobacteria carry two such enzymes where the main role of RelA is (p)ppGpp synthesis and SpoT in hydrolysis. SpoT can also synthesize (p)ppGpp and is therefore bifunctional (Xiao et al., 1991; Gentry and Cashel, 1996). In Gram-positive organisms, there is a single bifunctional Rel enzyme which synthesizes and degrades (p)ppGpp (Mittenhuber, 2001; Jain et al., 2006; Srivatsan and Wang 2008; Takada et al., 2020).
Apart from these classical, long, multidomain RSHs, few small RSH homologs were identified in organisms ranging from bacteria to plants. They are mostly monodomain, monofunctional proteins either with short alarmone synthetase (SAS) or short alarmone hydrolase (SAH) activity (Sun et al., 2010; Atkinson et al., 2011; Jimmy et al., 2020). The discovery of SAS and SAH opened a new line of research, to understand the indirect pathways of stress response induced by cues such as cell wall antibiotics, acid, alkali, hydrogen peroxide, ethanol, etc. (Horsburgh and Moir, 1999; Cao et al., 2002; Mascher et al., 2003; Thackray and Moir, 2003; Weinrick et al., 2004; D’Elia et al., 2009; Kim et al., 2012; Geiger et al., 2014; Pando et al., 2017). We have identified a dual-domain SAS in Mycobacterium smegmatis with RNase HII and (p)ppGpp synthesis activity (Murdeshwar and Chatterji, 2012).
Small alarmone synthetases play an important role to maintain the basal level of (p)ppGpp, which in turn induces the virulence of the pathogenic bacteria. The “(p)ppGpp pool” produced by the long and short RSH enzymes (Ronneau and Hallez, 2019) and the consecutive guanosine triphosphate (GTP) depletion are the key factors determining the formation of bacterial persister cells (Fung et al., 2020). Therefore, understanding the SAS-mediated synthesis and regulation of (p)ppGpp is the need of the hour to modify the current antibacterial therapy.
Salient Features of Small Alarmone Synthetases
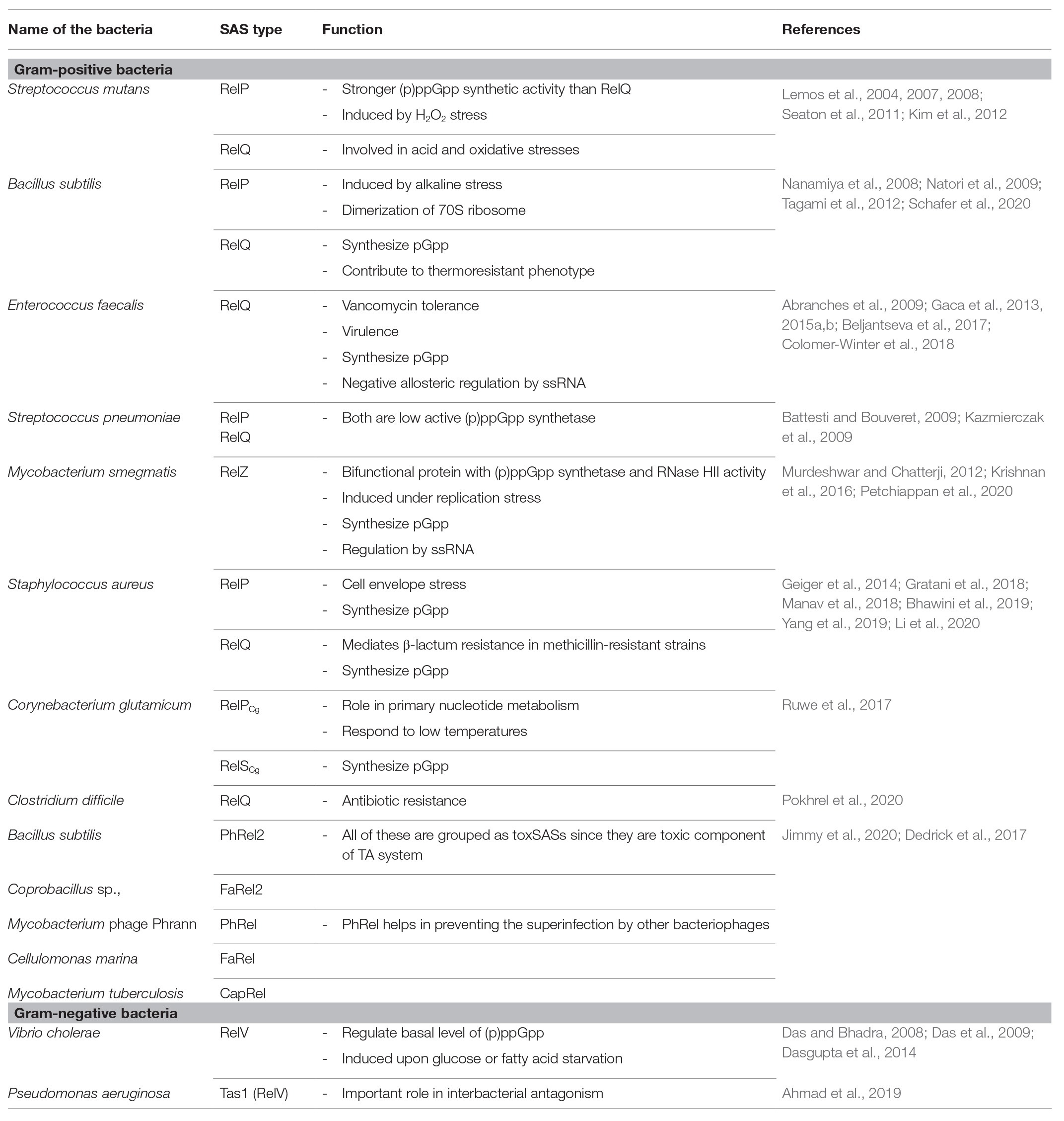
Small alarmone synthetases were identified in bacteria, such as Streptococcus mutans, Bacillus subtilis, Enterococcus faecalis, Streptococcus pneumoniae, Mycobacterium smegmatis, Staphylococcus aureus, Corynebacterium glutamicum, Clostridium difficile, Vibrio cholerae, and Pseudomonas aeruginosa. There are two highly homologous SAS proteins in bacteria and are named as RelP (SAS2, YwaC) and RelQ (SAS1, YjbM). Jimmy et al. (2020) reported the recent classification of SAS and identified 30 subfamilies. The functions of five of these subgroup enzymes were experimentally validated (Table 1) and found to be present in toxin–antitoxin (TA) system (Jimmy et al., 2020). The list of previously characterized bacterial SAS is given in Table 1. Their domain structures are given in Figure 1.
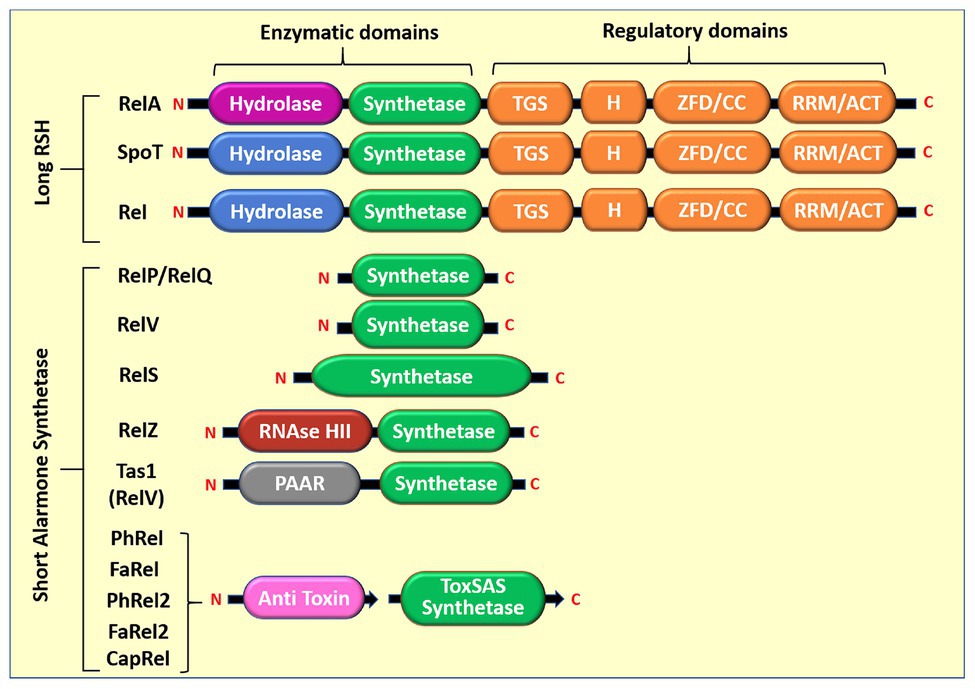
Figure 1. Domain structure of long RelA/SpoT homolog (RSH) and short alarmone synthetase (SAS). SAS proteins have only (p)ppGpp synthetic domain (~25–29 kDa), the hydrolysis and regulatory domains are absent. RelS is a 39.8-kDa protein with extended synthetase domain than other SAS. RelZ is a 64.5-kDa protein with RNase HII domain. The hydrolysis and regulatory domains are TGS, ThrRs, GTPase, and SpoT; H, helical domain; ZFD, zinc finger domain; CC, conserved cysteine; RRM, ribosome recognition motif; ACT, aspartokinase, chorismate mutase, and TyrA. Tas1 synthetase is a toxin effector domain, and proline-alanine-alanine-arginine (PAAR) is a toxin delivery domain; PhRel, FaRel, PhRel2, FaRel2, and CapRel are known as ToxSAS because of their presence in toxin–antitoxin (TA) module.
Molecular Functions of Small Alarmone Synthetases
Different SAS have different roles because they are induced by different signals (Figure 2). RSH is activated mostly under starvation and to the intracellular imbalances involving LPS biosynthesis and ADP metabolism (Roghanian et al., 2019), whereas SAS may respond to various types of environmental stimuli (Figure 2). Maintaining the basal level of (p)ppGpp is important for protection against different kinds of stresses, especially antibiotics stress. Most of the SAS proteins prefer guanosine diphosphate (GDP) to GTP as a substrate (Murdeshwar and Chatterji, 2012; Geiger et al., 2014; Gaca et al., 2015b). Rel and SAS are involved in the allosteric regulation of guanosine and GTP biosynthesis (Gaca et al., 2013; Bittner et al., 2014; Kriel et al., 2014).
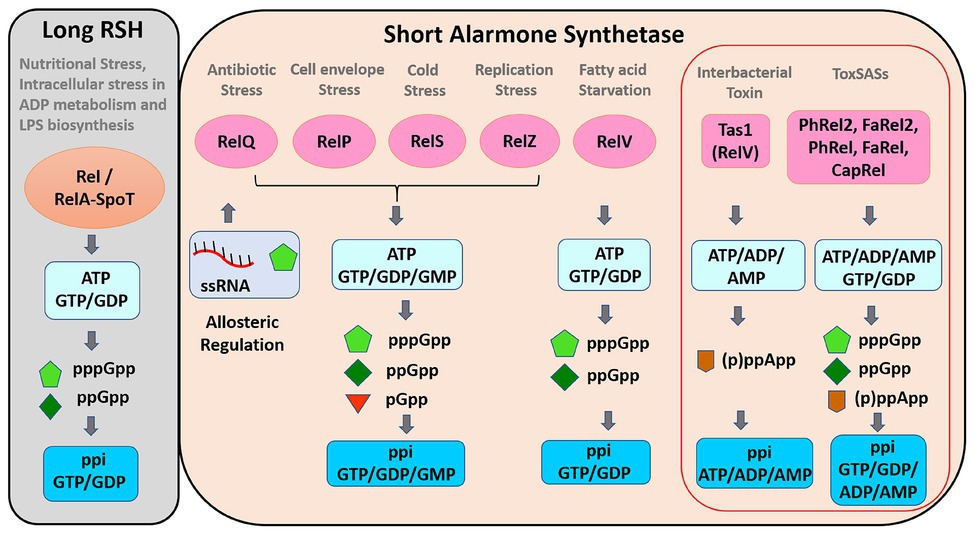
Figure 2. Functions of long RSH and short alarmone synthetase. Long RSH, Rel/RelA-SpoT, synthesize pppGpp and ppGpp using GTP/GDP as substrates during nutritional stress. SAS protein expression is induced by various stress signals. SAS Rels synthesize pGpp in addition to pppGpp and ppGpp using GTP/GDP/GMP as substrates. pppGpp and ssRNA bind to RelQ and mediate the allosteric regulation. The pppGpp synthesis activity of RelZ is also inhibited by RNA and pppGpp. Tas1 synthesize (p)ppApp using ATP/ADP/AMP as substrates. ToxSASs (not Tas1) synthesize both (p)ppGpp and (p)ppApp. ToxSASs are represented within red outlined box.
RelP and RelQ
RelP and RelQ share nearly 50% sequence similarity at the amino acid level. relP/relQ genes are upregulated due to various stress cues, such as cell envelope (Cao et al., 2002; D’Elia et al., 2009; Geiger et al., 2014), alkali (Nanamiya et al., 2008), ethanol (Pando et al., 2017), high salt, acidic, heat, and hydrogen peroxide (Thackray and Moir, 2003; Weinrick et al., 2004; Kim et al., 2012; Zweers et al., 2012). The first SAS proteins (RelP and RelQ) were identified in S. mutans (Lemos et al., 2004, 2007). During oxidative and acidic stress, RelP helped to slow the growth of the bacteria (Kim et al., 2012). Rel inactivation did not yield a lethal phenotype of S. mutans, and the basal level of (p)ppGpp was not increased through RelP and RelQ-dependent (p)ppGpp synthesis (Lemos et al., 2007). This could be due to the existence of an alternative mechanism for (p)ppGpp degradation in Streptococci (Lemos et al., 2007). In B. subtilis, RelP and RelQ have growth phase-dependent regulation. relQ is mainly transcribed in mid-exponential phase and it slows down its expression in the late-exponential phase; in addition, the relP is highly induced at this phase (Nanamiya et al., 2008). The (p)ppGpp synthesis of B. subtilis RelP is induced by alkaline stress (Nanamiya et al., 2008). In E. faecalis, only RelQ synthesizes (p)ppGpp apart from Rel (Abranches et al., 2009). The ∆relAQ strain showed significant sensitivity to vancomycin, ampicillin, and norfloxacin (Abranches et al., 2009; Gaca et al., 2013). In E. faecalis, (p)ppGpp-mediated antibiotic resistance happens at a concentration below the required value to mount stringent response. Staphylococcus aureus contains RelP and RelQ homologs. The expression of these two SAS is induced upon cell wall stress with vancomycin and ampicillin. The presence of three (p)ppGpp synthetases plays a significant role in the development of methicillin-resistant S. aureus (MRSA). Like the RelP of S. mutans, the RelP of S. aureus is also a more potent (p)ppGpp synthetase (Geiger et al., 2014). Clostridium difficile has a RelQ that is induced by antibiotic stress. There is a 2-fold upregulation of relQ after exposure to ampicillin and clindamycin, which explains the role of RelQ in antibiotic survival (Pokhrel et al., 2020).
Crystal structures of RelP and RelQ from B. subtilis and S. aureus revealed the homotetramer structures with highly similar monomers and homologs of (p)ppGpp synthetase domains. RelQ activity is inhibited by ssRNA (Beljantseva et al., 2017) and positively regulated by pppGpp (not ppGpp), whereas RelP is not allosterically regulated by (p)ppGpp. This is because of the difference in the conformation of the substrate binding site of these proteins. The RelQ, homotetramer of B. subtilis, has a distinct cleft in its center for the binding of two allosteric (p)ppGpp molecules (Steinchen et al., 2015, 2018; Steinchen and Bange, 2016). RelP has been shown to influence the formation of ribosome dimers to inactivate the translation of metabolic pathway (Tagami et al., 2012).
RelS
Corynebacterium glutamicum has two SAS proteins (Figure 2), represented as RelPCg and RelSCg (actRel subgroup). The SAS protein encoded by cg2324 is named as RelS and shares sequence similarity with the (p)ppGpp synthetase domain of RelQ. (p)ppGpp synthesis activity is not found for RelPCg. The maximum activity of the RelSCg is obtained at a temperature below optimum; therefore, it is assumed that (p)ppGpp is synthesized in response to low temperatures (Ruwe et al., 2017).
ToxSASs
Many SAS subfamilies were identified in conserved bicistronic operon of TA system from actinobacteria, firmicutes, and proteobacteria. Five of these SAS were demonstrated to be the toxic component of the TA system and hence named as toxSASs (Jimmy et al., 2020). They are B. subtilis PhRel2, Coprobacillus sp., FaRel2, Mycobacterium phage Phrann PhRel, Cellulomonas marina FaRel, and Mycobacterium tuberculosis CapRel (Figures 1, 2). The toxicity of the toxSASs was neutralized by the six adjacent antitoxin proteins, among which five are specific to corresponding toxSASs and C. marina FaRel2 can neutralize all the five toxSASs. The specific function of the toxSASs is not identified, except of PhRel (also known as Gp29), which plays a role in preventing the superinfection by other bacteriophages (Dedrick et al., 2017).
RelV
RelV (relA-like (p)ppGpp synthetase domain coding gene in vibrios) shared poor homology with RelP and RelQ, because the bacteria itself are phylogenetically different from firmicutes, but there is a high conservation of amino acid residues in the synthetase domains of RelV, RelP, and RelQ. In V. cholerae, RelV can produce (p)ppGpp upon glucose or fatty acid starvation (Das and Bhadra, 2008; Das et al., 2009; Dasgupta et al., 2014). Another RelV subfamily homolog Tas1 was identified in P. aeruginosa. Tas1 RSH domain is encoded within a large conserved T6SS cluster (type 6 secretion system) and fused to a toxin delivery domain (Figure 2), which exhibits its toxic effect on another competitor cell, thus playing an important function in interbacterial antagonism (Ahmad et al., 2019).
RelZ (MS_RHII-RSD)
In M. smegmatis, MSMEG_5849 codes for a bifunctional protein MS_RHII-RSD (renamed as RelZ), which has a C-terminal RSD domain similar to the other SAS but is different from them due to the presence of N-terminal RNase HII domain in the same polypeptide chain (Figure 3). RelZ efficiently hydrolyze RNA–DNA hybrids (Murdeshwar and Chatterji, 2012) and R-loops (Krishnan et al., 2016). R-loops have a major role in replication–transcription conflicts and lead to stalled arrays of RNA polymerase to block the replication fork movement, thereby promoting replication stress (Drolet, 2006; Poveda et al., 2010; Stirling et al., 2012). This stress can be efficiently managed by two mechanisms: R-loop removal by RNase HII (Aguilera and García-Muse, 2012) and destabilization of stalled RNA polymerase by (p)ppGpp synthesis (Cashel et al., 1996; Ross et al., 2013). RelZ possesses both these important activities (RNase HII and (p)ppGpp synthetase) in a single polypeptide. Our previous study (Krishnan et al., 2016) showed that under UV stress, RelZ removes the accumulated R-loops in RNase H-deficient E. coli, and relZ expression is upregulated in M. smegmatis to remove the R-loops generated due to UV stress. Based on these results, we proposed a model to explain the function of RelZ. Upon UV stress, the levels of RelZ increase within the cell. Any R-loops formed are removed by the RNase HII and (p)ppGpp helps to destabilize the stalled RNA polymerase via an unknown mechanism to rescue the cells from replication stress (Krishnan et al., 2016). In addition, RelZ mediates antibiotic tolerance in M. smegmatis but does not impact biofilm formation significantly (Petchiappan et al., 2020).
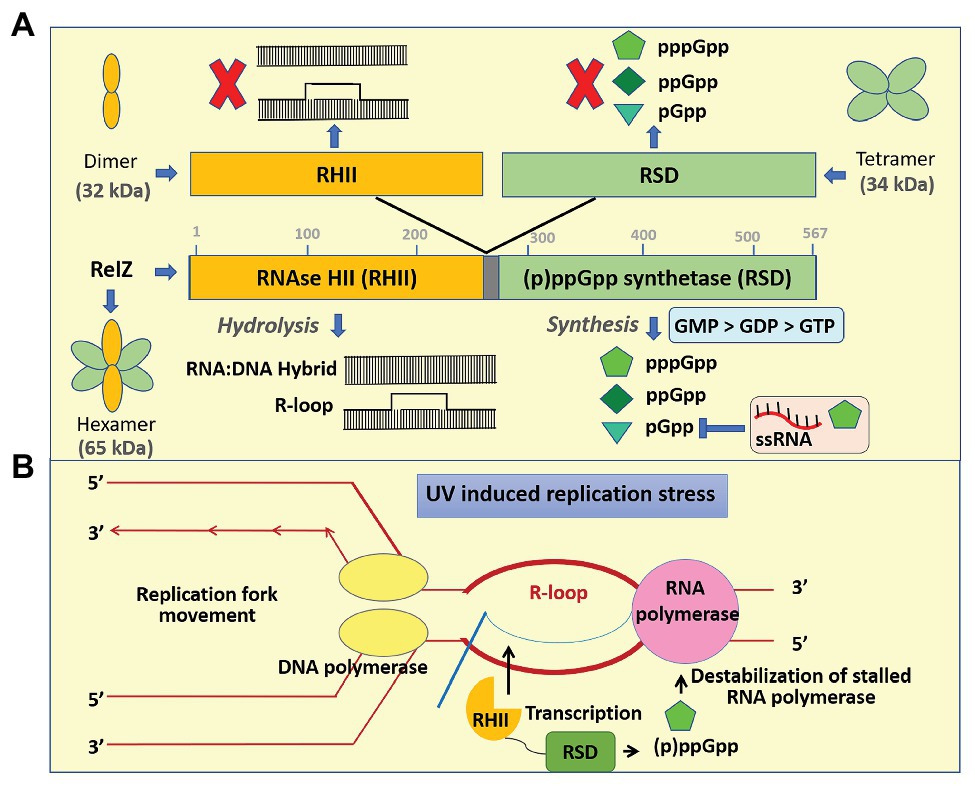
Figure 3. Mechanism of regulation and functions of RelZ. (A) RelZ contains an RNAse HII (RHII) domain in tandem with the (p)ppGpp synthetase domain (RSD, RelA SpoT nucleotidyl transferase domain). Full-length protein in cis has both RNAse HII activity and (p)ppGpp synthetase activity whereas neither the purified domain variants in isolation nor on trans complementation can function independently. RelZ can hydrolyze RNA:DNA hybrid as well as R-loop. It can synthesize pGpp, ppGpp, and pppGpp having the substrate preferences as GMP > GDP > GTP. pGpp synthesis is negatively regulated by ssRNA and high concentrations of pppGpp. From our earlier studies on active site mutational analysis, gel filtration chromatography followed by native PAGE revealed that the N-terminal RHII domain is monomeric and C-terminal RSD domain is tetrameric upon isolation. The full-length active protein is, however, hexamer in solution. We also found that RelZ and all the mutant variants of the full-length RelZ remain as hexameric form in solution. (B) In our previous study, we demonstrated that RelZ can hydrolyze R-loops in Escherichia coli exposed to UV stress. RelZ gene expression was upregulated under UV stress, and this gene-deleted strain showed increased R-loop accumulation as compared to the wild type. Based on these results, we proposed a model for the physiological function of RelZ. UV stress leads to increased R-loop formation and replication–transcription conflicts. Under UV stress, RelZ expression is upregulated than the conventional RNase HI and HII. Its RHII activity removes the R-loops and the stalled RNA polymerase is destabilized indirectly by (p)ppGpp. Thus, RelZ plays an important role during R-loop-induced replication stress response in Mycobacterium smegmatis. (B) is adapted and redrawn from Krishnan et al. (2016).
Active site mutational studies of RelZ revealed that inactivation of one domain does not affect the activity of the other domain. However, the purified subdomains are nonfunctional when separated and expressed independently (Figure 3). This kind of domain interdependence was extensively characterized, and the results showed that the full-length RelZ is essential for its function and it is a hexamer (Krishnan et al., 2016). The synthetic subdomain of RelZ is a tetramer in solution like the other solved structures of RelP and RelQ (Steinchen et al., 2015, 2018, 2020; Steinchen and Bange, 2016). Petchiappan et al. (2020) showed that RelZ prefers guanosine monophosphate (GMP) as a substrate and synthesizes pGpp. To understand the difference between pGpp and ppGp, the reaction mixture was treated with NaOH that hydrolyzes only pGpp. From thin layer chromatography, it was shown that Rel hydrolyzes pGpp to GMP and pyrophosphate as evidenced by the comigration of the radiolabeled product with the purified pyrophosphate whereas RelZ showed weak hydrolysis. We found that ssRNA inhibits RelZ-mediated pGpp synthesis, but R-loop did not show any effect (Petchiappan et al., 2020). The pGpp synthesis activity of the RelZ is inhibited by pppGpp whereas ppGpp and pGpp did not have significant effect. Therefore, we infer that the cellular pppGpp levels determine the RelZ-mediated synthesis, whereas ssRNA and pppGpp carefully regulate it. The altered cell surface properties of ΔrelZ strain indicated that RelZ plays a role in cell wall metabolism (Petchiappan et al., 2020).
Mycobacterium tuberculosis has a SAS encoding (p)ppGpp synthetase, Rv1366. But it has no RNase H domain and it is incapable of synthesizing (p)ppGpp (Nanamiya et al., 2008; Weiss and Stallings, 2013; Bag et al., 2014). Few RHII-RSD dual-domain orthologs were identified from Mycobacteria; Mycobacterium vanbaalenii (YP_995923.1), Mycobacterium tusciae (ZP_09680741.1), and Mycobacterium gilvum (YP_001132882.1). However, RelZ is the only dual-domain mycobacterial SAS characterized so far. RelZ type of SAS with RNase H and (p)ppGpp synthetase domains are found only in the environmental species and they are absent in the pathogenic species of mycobacteria.
SAS Synthesize pGpp
Recently, SAS proteins but not Rel are found to use GMP as a substrate and synthesize pGpp, a third alarmone which makes the alarmone group representation from (p)ppGpp to (pp)pGpp (Gaca et al., 2015b). pGpp can function like (p)ppGpp as well and may have different functions which is not regulated by (p)ppGpp (Gaca et al., 2015a). The pGpp can be hydrolyzed by Rel, like the hydrolysis of (p)ppGpp (Gaca et al., 2015b; Yang et al., 2019). In B. subtilis, RelP and RelQ are shown to synthesize ppGp or pGpp. (Tagami et al., 2012). RelQ from E. faecalis is an efficient producer of pGpp (Gaca et al., 2015a). RelQ and RelP of S. mutans showed much weaker pGpp synthesis activity upon comparison with RelQEf. RelP and RelQ of S. aureus and RelSCg of C. glutamicum synthesize pGpp along with (p)ppGpp. ppGpp/pGpp effectively reduce the intracellular levels of GTP and these guanine nucleotides are synthesized only when RelA is inactive in the cells (Ruwe et al., 2017). The synthesis of pGpp will become relevant only when the GMP levels in the cells are increased like GTP level. Such kind of GMP accumulation has been reported in B. subtilis (Liu et al., 2015). It was also speculated that pGpp may be involved in stretching the stress response after the depletion of GTP and GDP in the cell (Gaca et al., 2015b; Ruwe et al., 2017). However, pGpp regulates the purine synthesis but does not involve in ribosome biogenesis (Tagami et al., 2012; Yang et al., 2020).
SAS Synthesize (p)ppApp
Recent studies by Ahmad et al. (2019) and Jimmy et al. (2020) revealed that SAS not only synthesize ppGpp but also synthesize ppApp. In P. aeruginosa, a secreted toxic effector of T6SS was identified as Tas1. Though the crystal structure of Tas1 is similar to the other (p)ppGpp synthetases, it does not synthesize (p)ppGpp but produces (p)ppApp (Ahmad et al., 2019). Another SAS that produces (p)ppApp was identified in C. marina FaRel. The toxicity of this toxSAS is mediated by ppGpp and ppApp followed by the depletion of intracellular GTP and ATP pools (Jimmy et al., 2020).
SAS Bind to ssRNA
Hauryliuk and Atkinson (2017) reviewed the RNA-binding properties of SAS. Beljantseva et al. (2017) discovered that RelQEf activity is inhibited when it binds to ssRNA. RNA binds to RelQ in a sequence-specific manner with GGNGG, a putative Shine–Dalgarno-like consensus sequence. pppGpp strongly counteracts the inhibition by RNA and destabilizes the RNA:RelQ complex. In this way, RelQ has both enzyme activity and RNA-binding property. In a RelQ:RNA complex, (p)ppGpp synthesis and pppGpp binding are mutually incompatible. Hence, there is a possibility that the RelQ:RNA complex acts a regulatory switch between inactive and active forms of the enzyme. ssRNA and pppGpp compete with each other to bind into the central cleft of the homotetramer, but this property is not conserved in RelP of S. aureus, because pppGpp is not an allosteric regulator of RelP. The central cleft in the RelP tetramer could be an allosteric site bound by other small molecules (Manav et al., 2018; Steinchen et al., 2018).
The RNA-binding property of RelQ can be compared with that of RelZ since the ssRNA inhibits the activity of RelZ (Petchiappan et al., 2020). Since RelZ is involved in R-loop-mediated replication stress (Krishnan et al., 2016), (p)ppGpp synthesis can occur by sensing the R-loops. Once the RNase H cleaves the R-loop into dsDNA and ssRNA (Dutta et al., 2011), the replication stress is relieved and hence the (p)ppGpp synthesis stops. This could be the reason for ssRNA showing inhibitory effect on RelZ-mediated alarmone synthesis. Structural analysis of RelZ is in progress to understand the RelZ:ssRNA complex.
Arresting the (p)ppGpp synthetase activity using (p)ppGpp analogues is emerging as a clinically important method in eradicating persistent infections (de la Fuente-Núñez et al., 2014; Andresen et al., 2016; Petchiappan and Chatterji, 2017; Syal et al., 2017; Dutta et al., 2019). Similarly, the ssRNA-binding property of the SAS can be explored to regulate the SAS-mediated (p)ppGpp synthesis. Mutant huntingtin protein that causes Huntington’s disease was selectively and effectively inhibited by ss siRNA approach (Yu et al., 2012). According to Lima et al. (2012), the identification of potent ssRNA would provide an easy route to therapeutics than dsRNA. ssRNA do not require special formulations for tissue penetration (Bennett and Swayze, 2010), whereas the ds siRNAs need to undergo complex and expensive lipid formulations (Tao et al., 2011). Nucleic acids not only recognize specific target sequences by complementary base pairing but they can interact with proteins and this property is currently being explored in therapeutics (Roberts et al., 2020).
Conclusion
The co-evolution of SAS along with Rel, redundant (p)ppGpp synthetases, and multiple types of closely related alarmones in bacteria is intriguing. (p)ppGpp is a key factor for biofilm formation, antibiotic tolerance, virulence, and persistence in many pathogenic bacteria. Therefore, inhibition of (p)ppGpp synthesis will inhibit the long-term survival of the pathogen. Therefore, finding an inhibitor to prevent (p)ppGpp synthesis is of high therapeutic interest. In addition to that, ssRNA with specific binding sequence could be a supplementary therapeutic element to inhibit the SAS-dependent (p)ppGpp synthesis because SAS is an RNA-binding protein. The discovery of SAS has not only augmented the prospects of stringent response but also adds value to the upcoming field of RNA therapies.
Author Contributions
DC and SK conceptualized and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was partially supported by JC Bose fellowship, Department of Science and Technology, Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SK thanks the Department of Biotechnology for the fellowship.
References
Abranches, J., Martinez, A. R., Kajfasz, J. K., Chávez, V., Garsin, D. A., and Lemos, J. A. (2009). The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256. doi: 10.1128/JB.01726-08
Aguilera, A., and García-Muse, T. (2012). R loops: from transcription by products to threats to genome stability. Mol. Cell 46, 115–124. doi: 10.1016/j.molcel.2012.04.009
Ahmad, S., Wang, B., Walker, M. D., Tan, H. R., Sogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9
Andresen, L., Varik, V., Tozawa, Y., Jimmy, S., Lindberg, S., Tenson, T., et al. (2016). Auxotrophy-based high throughput screening assay for the identification of Bacillus subtilis stringent response inhibitors. Sci. Rep. 6:35824. doi: 10.1038/srep35824
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Bag, S., Das, B., Dasgupta, S., and Bhadra, R. K. (2014). Mutational analysis of the (p)ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. Arch. Microbiol. 196, 575–588. doi: 10.1007/s00203-014-0996-9
Battesti, A., and Bouveret, E. (2009). Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191, 616–624. doi: 10.1128/JB.01195-08
Beljantseva, J., Kudrin, P., Andresen, L., Shingler, V., Atkinson, G. C., Tenson, T., et al. (2017). Negative allosteric regulation of Enterococcus faecalis small Alarmone Synthetase RelQ by single stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 114, 3726–3731. doi: 10.1073/pnas.1617868114
Bennett, C. F., and Swayze, E. E. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293. doi: 10.1146/annurev.pharmtox.010909.105654
Bhawini, A., Pandey, P., Dubey, A. P., Zehra, A., Gopal, N., and Mishra, M. N. (2019). RelQ mediates the expression of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Front. Microbiol. 10:339. doi: 10.3389/fmicb.2019.00339
Bittner, A. N., Kriel, A., and Wang, J. D. (2014). Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J. Bacteriol. 196, 2067–2076. doi: 10.1128/JB.01471-14
Cao, M., Kobel, P. A., Morshedi, M. M., Wu, M. F., Paddon, C., and Helmann, J. D. (2002). Defining the Bacillus subtilis sigma (W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316, 443–457. doi: 10.1006/jmbi.2001.5372
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Cashel, M., Gentry, D. M., Hernandez, V. J., and Vinella, D. (1996). “The stringent response” in Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. ed. F. C. Neidhardt (Washington, DC: ASM Press), 1458–1496.
Chatterji, D., and Ojha, A. K. (2001). Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4, 160–165. doi: 10.1016/s1369-5274(00)00182-x
Colomer-Winter, C., Gaca, O. A., Chuang-Smith, O., Lemos, A. J., and Frank, K. L. (2018). Basal levels of (p)ppGpp differentially affect the pathogenesis of infective endocarditis in Enterococcus faecalis. Microbiology 164, 1254–1265. doi: 10.1099/mic.0.000703
D’Elia, M. A., Millar, K. E., Bhavsar, A. P., Tomljenovic, A. M., Hutter, B., Schaab, C., et al. (2009). Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem. Biol. 16, 548–556. doi: 10.1016/j.chembiol.2009.04.009
Das, B., and Bhadra, R. K. (2008). Molecular characterization of Vibrio cholerae DrelA DspoT double mutants. Arch. Microbiol. 189, 227–238. doi: 10.1007/s00203-007-0312-z
Das, B., Pal, R. R., Bag, S., and Bhadra, R. K. (2009). Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol. Microbiol. 72, 380–398. doi: 10.1111/j.1365-2958.2009.06653.x
Dasgupta, S., Basu, P., Pal, R. R., Bag, S., and Bhadra, R. K. (2014). Genetic and mutational characterization of the small alarmone synthetase gene relV of Vibrio cholerae. Microbiology 160, 1855–1866. doi: 10.1099/mic.0.079319-0
de la Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2014). Broad-Spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathol. 10:e1004152. doi: 10.1371/journal.ppat.1004152
Dedrick, R. M., Jacobs-Sera, D., Bustamante, C. A., Garlena, R. A., Mavrich, T. N., Pope, W. H., et al. (2017). Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2:16251. doi: 10.1038/nmicrobiol.2016.251
Drolet, M. (2006). Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation, and DNA topology. Mol. Microbiol. 59, 723–730. doi: 10.1111/j.1365-2958.2005.05006.x
Dutta, N. K., Klinkenberg, L. G., Vazquez, M. J., Segura-Carro, D., Colmenarejo, G., Ramon, F., et al. (2019). Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 5:eaav2104. doi: 10.1126/sciadv.aav2104
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E., and Nudler, E. (2011). Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543. doi: 10.1016/j.cell.2011.07.034
Fung, D. K., Barra, J. T., Schroeder, J. W., Ying, D., and Wang, J. D. (2020). A shared alarmone-GTP switch underlies triggered and spontaneous persistence. bioRxiv [Preprint] doi: 10.1101/2020.03.22.002139
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015a). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/JB.02577-14
Gaca, A. O., Kajfasz, J. K., Miller, J. H., Liu, K., Wang, J. D., Abranches, J., et al. (2013). Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio 4:e00646–13. doi: 10.1128/mBio.00646-13
Gaca, A. O., Kudrin, P., Colomer-Winter, C., Beljantseva, J., Liu, K., Anderson, B., et al. (2015b). From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small Alarmone Synthetase of Enterococcus faecalis. J. Bacteriol. 197, 2908–2919. doi: 10.1128/JB.00324-15
Geiger, T., Kastle, B., Gratani, F. L., Goerke, C., and Wolz, C. (2014). Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 196, 894–902. doi: 10.1128/JB.01201-13
Gentry, R. D., and Cashel, M. (1996). Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19, 1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x
Gratani, F. L., Horvatek, P., Geiger, T., Borisova, M., Mayer, C., Grin, I., et al. (2018). Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 14:e1007514. doi: 10.1371/journal.pgen.1007514
Hauryliuk, V., and Atkinson, G. C. (2017). Small Alarmone Synthetases as novel bacterial RNA-binding proteins. RNA Biol. 14, 1695–1699. doi: 10.1080/15476286.2017.1367889
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Horsburgh, M. J., and Moir, A. (1999). sM, an ECF RNA polymerase sigma factor of Bacillus subtilis, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32, 41–50. doi: 10.1046/j.1365-2958.1999.01323.x
Jain, V., Saleem-Batcha, R., China, A., and Chatterji, D. (2006). Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 15, 1449–1464. doi: 10.1110/ps.062117006
Jimmy, S., Saha, C. K., Kurata, T., Stavropoulos, C., Oliveira, S., Koh, A., et al. (2020). A widespread toxin-antitoxin system exploiting growth control via alarmone signalling. Proc. Natl. Acad. Sci. U. S. A. 117, 10500–10510. doi: 10.1073/pnas.1916617117
Kazmierczak, K. M., Wayne, K. J., Rechtsteiner, A., and Winkler, M. E. (2009). Roles of rel in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72, 590–611. doi: 10.1111/j.1365-2958.2009.06669.x
Kim, J. N., Ahn, S.-J., Seaton, K., Garrett, S., and Burne, A. R. (2012). Transcriptional organization and physiological contributions of the relQ operon of Streptococcus mutans. J. Bacteriol. 194, 1968–1978. doi: 10.1128/JB.00037-12
Kriel, A., Bittner, A. N., Kim, S. H., Liu, K., Tehranchi, A. K., Zou, W. Y., et al. (2012). Direct regulation of GTP homeostasis by (p) ppGpp: a critical component of viability and stress resistance. Mol. Cell 48, 231–241. doi: 10.1016/j.molcel.2012.08.009
Kriel, A., Brinsmade, S. R., Tse, J. L., Tehranchi, A. K., Bittner, A. N., Sonenshein, A. L., et al. (2014). GTP Dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid Auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J. Bacteriol. 196, 189–201. doi: 10.1128/JB.00918-13
Krishnan, S., Petchiappan, A., Singh, A., Bhatt, A., and Chatterji, D. (2016). R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol. Microbiol. 102, 168–182. doi: 10.1111/mmi.13453
Lemos, J. A., Brown, T. A., and Burne, R. A. (2004). Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72, 1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004
Lemos, J. A., Lin, V. K., Nascimento, M. M., Abranches, J., and Burne, R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x
Lemos, J. A., Nascimento, M. M., Lin, V. K., Abranches, J., and Burne, R. A. (2008). Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190, 5291–5299. doi: 10.1128/JB.00288-08
Li, L., Bayer, A. S., Cheung, A., Lu, L., Abdelhady, W., Donegan, N. P., et al. (2020). The stringent response contributes to persistent methicillin-resistant Staphylococcus aureus endovascular infection through the purine biosynthetic pathway. J. Infect. Dis. 222, 1188–1198. doi: 10.1093/infdis/jiaa202
Lima, W. F., Prakash, T. P., Murray, H. M., Kinberger, G. A., Li, W., Chappell, A. E., et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150, 883–894. doi: 10.1016/j.cell.2012.08.014
Liu, K., Bittner, A. N., and Wang, J. D. (2015). Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. doi: 10.1016/j.mib.2015.01.012
Manav, M. C., Beljantseva, J., Bojer, M. S., Tenson, T., Ingmer, H., Hauryliuk, V., et al. (2018). Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP. J. Biol. Chem. 293, 3254–3264. doi: 10.1074/jbc.RA117.001374
Mascher, T., Margulis, N. G., Wang, T., Ye, R. W., and Helmann, J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol.Microbiol. 50, 1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x
Mittenhuber, G. J. (2001). Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3, 585–600.
Murdeshwar, M. S., and Chatterji, D. (2012). MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 194, 4003–4014. doi: 10.1128/JB.00258-12
Nanamiya, H., Kasai, K., Nozawa, A., Yun, C. S., Narisawa, T., Murakami, K., et al. (2008). Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304. doi: 10.1111/j.1365-2958.2007.06018.x
Natori, Y., Tagami, K., Murakami, K., Yoshida, S., Tanigawa, O., Moh, Y., et al. (2009). Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient of (p)ppGpp synthetase genes relA, yjbM, and ywaC. J. Bacteriol. 191, 4555–4561. doi: 10.1128/JB.00263-09
Pando, J. M., Pfeltz, R. F., Cuaron, J. A., Nagarajan, V., Mishra, M. N., Torres, N. J., et al. (2017). Ethanol-induced stress response of Staphylococcus aureus. Can. J. Microbiol. 63, 745–757. doi: 10.1139/cjm-2017-0221
Petchiappan, A., and Chatterji, D. (2017). Antibiotic resistance: current perspectives. ACS Omega 2, 7400–7409. doi: 10.1021/acsomega.7b01368
Petchiappan, A., Naik, S. Y., and Chatterji, D. (2020). RelZ-mediated stress response in Mycobacterium smegmatis: pGpp synthesis and its regulation. J. Bacteriol. 202:e00444–19. doi: 10.1128/JB.00444-19
Pokhrel, A., Poudel, A., Kory, B., Castro, K. B., Celestine, M. J., Oludiran, A., et al. (2020). The (p)ppGpp synthetase RSH mediates stationary phase onset and antibiotic stress survival in Clostridioides difficile. J. Bacteriol. 202:e00377–20. doi: 10.1128/JB.00377-20
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Poveda, A. M., Le Clech, M., and Pasero, P. (2010). Transcription and replication: breaking the rules of the road causes genomic instability. Transcription 1, 99–102. doi: 10.4161/trns.1.2.12665
Roberts, T. C., Langer, R., and Wood, M. J. A. (2020). Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov., 9, 673–694. doi: 10.1038/s41573-020-0075-7
Roghanian, M., Semsey, S., Løbner-Olesen, A., and Jalalvand, F. (2019). (p)ppGpp-mediated stress response induced by defects in outer membrane biogenesis and ATP production promotes survival in Escherichia coli. Sci. Rep. 9:2934. doi: 10.1038/s41598-019-39371-3
Ronneau, S., and Hallez, R. (2019). Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 43, 389–400. doi: 10.1093/femsre/fuz009
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T., and Gourse, R. L. (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429. doi: 10.1016/j.molcel.2013.03.021
Ruwe, M., Kalinowski, J., and Persicke, M. (2017). Identification and functional characterization of small Alarmone Synthetases in Corynebacterium glutamicum. Front. Microbiol. 8:1601. doi: 10.3389/fmicb.2017.01601
Schafer, H., Beckert, B., Frese, C. K., Shields, R. C., Kim, J. N., Ahn, S.-J., et al. (2020). Peptides encoded in the Streptococcus mutans RcrRPQ operon are essential for thermotolerance. Microbiology 166, 306–317. doi: 10.1099/mic.0.000887
Seaton, K., Ahn, S. J., Sagstetter, A. M., and Burne, R. A. (2011). A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacterial. 193, 862–874. doi: 10.1128/JB.01257-10
Srivatsan, A., and Wang, J. D. (2008). Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105. doi: 10.1016/j.mib.2008.02.001
Steinchen, W., and Bange, G. (2016). The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544. doi: 10.1111/mmi.13412
Steinchen, W., Nuss, A. M., Beckstette, M., Hantke, I., Driller, K., Sudzinova, P., et al. (2020). The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16:e1008275. doi: 10.1371/journal.pgen.1008275
Steinchen, W., Vogt, M. S., Altegoer, F., Giammarinaro, P. I., Horvatek, P., Wolz, C., et al. (2018). Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ. Sci. Rep. 8:2195. doi: 10.1038/s41598-018-206344
Steinchen, W., Schuhmacher, J. S., Altegoer, F., Fage, C. D., Srinivasan, V., Linne, U., et al. (2015). Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl. Acad. Sci. U. S. A. 112, 13348–13353. doi: 10.1073/pnas.1505271112
Stirling, P. C., Chan, Y. A., Minaker, S. W., Aristizabal, M. J., Barrett, I., Sipahimalani, P., et al. (2012). R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26, 163–175. doi: 10.1101/gad.179721.111
Sun, D., Lee, G., Lee, J. H., Kim, H. Y., Rhee, H. W., Park, S. Y., et al. (2010). A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17, 1188–1194. doi: 10.1038/nsmb.1906
Syal, K., Flentie, K., Bhardwaj, N., Maiti, K., Jayaraman, N., Stallings, C. L., et al. (2017). Synthetic (p) ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob. Agents Chemother. 61:e00443–17. doi: 10.1128/AAC.00443-17
Tagami, K., Nanamiya, H., Kazo, Y., Maehashi, M., Suzuki, S., Namba, E., et al. (2012). Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp0 mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. MicrobiologyOpen 1, 115–134. doi: 10.1002/mbo3.16
Takada, H., Roghanian, M., Murina, V., Dzhygyr, I., Murayama, R., Akanuma, G., et al. (2020). The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of ‘Long’ RelA-SpoT homolog enzymes by ribosomal complexes. Front. Microbiol. 11:277. doi: 10.3389/fmicb.2020.00277
Tao, W., Mao, X., Davide, J. P., Ng, B., Cai, M., Burke, P. A., et al. (2011). Mechanistically probing lipid-siRNA nanoparticle associated toxicities identifies Jak inhibitors effective in mitigating multifaceted toxic responses. Mol. Ther. 19, 567–575. doi: 10.1038/mt.2010.282
Thackray, P. D., and Moir, A. (2003). SigM, an extra cytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185, 3491–3498. doi: 10.1128/JB.185.12.3491-3498.2003
Weinrick, B., Dunman, P. M., McAleese, F., Murphy, E., Projan, S. J., Fang, Y., et al. (2004). Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186, 8407–8423. doi: 10.1128/JB.186.24.8407-8423.2004
Weiss, L. A., and Stallings, C. L. (2013). Essential roles for Mycobacterium Rel beyond the production of (p)ppGpp. J. Bacteriol. 95, 5629–5638. doi: 10.1128/JB.00759-13
Wu, J., and Xie, J. (2009). Magic spot: (p)ppGpp. J. Cell. Physiol. 220, 297–302. doi: 10.1002/jcp.21797
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990.
Yang, J., Anderson, B. W., Turdiev, A., Turdiev, H., Stevenson, D. M., Amador-Noguez, D., et al. (2020). Systemic characterization of pppGpp, ppGpp and pGpp targets in Bacillus reveals NahA converts (p)ppGpp to pGpp to regulate alarmone composition and signalling. bioRxiv [Preprint] doi: 10.1101/2020.03.23.003749
Yang, N., Xie, S., Tang, N.-Y., Choi, M. Y., Wang, Y., and Wat, R. M. (2019). The Ps and Qs of alarmone synthesis in Staphylococcus aureus. PLoS One 14:e0213630. doi: 10.1371/journal.pone.0213630
Yu, D., Pendergraff, H., Liu, J., Kordasiewicz, H. B., Cleveland, D. W., Swayze, E. E., et al. (2012). Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 150, 895–908. doi: 10.1016/j.cell.2012.08.002
Keywords: short alarmone, (p)ppGpp, pGpp, stress response, R-loop, replication stress, ssRNA, RNase HII
Citation: Krishnan S and Chatterji D (2020) Pleiotropic Effects of Bacterial Small Alarmone Synthetases: Underscoring the Dual-Domain Small Alarmone Synthetases in Mycobacterium smegmatis. Front. Microbiol. 11:594024. doi: 10.3389/fmicb.2020.594024
Edited by:
Gert Bange, University of Marburg, GermanyReviewed by:
Mohammad Roghanian, Umeå University, SwedenChristiane Wolz, University of Tübingen, Germany
Copyright © 2020 Krishnan and Chatterji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipankar Chatterji, ZGlwYW5rYXJAaWlzYy5hYy5pbg==
 Sushma Krishnan
Sushma Krishnan Dipankar Chatterji
Dipankar Chatterji