- 1Medical Technology Institute of Xuzhou Medical University, Xuzhou, China
- 2Beijing Institute of Radiation Medicine, Beijing, China
- 3Department of Laboratory Medicine, Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 4College of Life Sciences, Anhui Agricultural University, Hefei, China
Escherichia coli O157:H7 is regarded as one of the most harmful pathogenic microorganisms related to foodborne diseases. This paper proposes a rapid-detection biosensor for the sensitive and quantitative analysis of E. coli O157:H7 in biological samples by surface-enhanced Raman scattering (SERS)-based lateral flow immunoassay (LFIA). A novel gold-shell silica-core (SiO2/Au) nanosphere (NP) with monodispersity, good stability, and excellent SERS activity was utilized to prepare high-performance tags for the SERS-based LFIA system. The SiO2/Au SERS tags, which were modified with two layers of Raman reporter molecules and monoclonal antibodies, effectively bind with E. coli O157:H7 and form sandwich immune complexes on the test lines. E. coli O157:H7 was quantitatively detected easily by detecting the Raman intensity of the test lines. Under optimal conditions, the limit of detection (LOD) of the SiO2/Au-based SERS-LIFA strips for the target bacteria was 50 cells/mL in PBS solution, indicating these strips are 2,000 times more sensitive than colloidal Au-based LFIA strips. Moreover, the proposed assay demonstrated high applicability in E. coli O157:H7 detection in biological samples, including tap water, milk, human urine, lettuce extract and beef, with a low LOD of 100 cells/mL. Results indicate that the proposed SERS-based LFIA strip is applicable for the sensitive and quantitative determination of E. coli O157:H7.
Introduction
As one of the major foodborne pathogens that cause food poisoning and serious illnesses worldwide, Escherichia coli O157:H7 is a significant threat to public health (Suaifan et al., 2017; Hassan et al., 2019). E. coli O157:H7 can exist in sewage-tainted water, contaminative milk, and meat products. It can cause bloody diarrhea, urinary tract infection, and hemolytic uremic syndrome, and its infectious dose is as low as 10 cells (Song et al., 2016; Shin et al., 2018). Rapid and sensitive methods that could timely diagnose this pathogen are the key to reduce the spread of infection and guarantee food safety at the source. The traditional microbiological culture method is considered as the standard method for bacterial detection (Hu et al., 2016; Li et al., 2019). However, it is labor intensive and time consuming, usually needing more than 24 h of culture and analysis. Some modern methods, such as enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), bioluminescence signal, DNA sequencing-based test, and mass spectrometry (MS), have been recently developed and successfully applied in laboratory testing (Jia et al., 2015; Guo et al., 2016; Sauget et al., 2017; Liu et al., 2018). These methods are fairly rapid and accurate but have some disadvantages, such as tedious procedures, sample pretreating, and strict laboratory conditions (Law et al., 2014). Thus, a sensitive and convenient point-of-care testing (POCT) must be developed for E. coli O157:H7 detection.
Lateral flow immunoassay (LFIA), which possesses the advantages of short testing time, low cost, user-friendly format, and portability in diverse applications, has been considered one of the most promising POCT methods (Wang et al., 2018b, 2019a; Zhang et al., 2019; Mahmoudi et al., 2020). However, the inherent defects of conventional LFIA are poor quantitative ability and limited detection sensitivity, which both depend on colorimetric analysis. Increasing attention has been paid to surface-enhanced Raman Scattering (SERS)-based LFIA technology, which combines the high sensitivity and quantitative analysis of SERS technology and the rapidity and convenience of LIFA (Hwang et al., 2016; Lee et al., 2019; Wang et al., 2019b). SERS-based LFIA uses Raman reporter-labeled SERS nanomaterials instead of colloidal Au as the signal tags, thereby providing strong and stable SERS signals (Jia et al., 2018; Khlebtsov et al., 2018). SERS-based LFIA has been successfully applied in the sensitive and quantitative analysis of various targets, such as tumor biomarkers, heart disease biomarkers, toxins, DNA markers, and viruses (Maneeprakorn et al., 2016; Wang X. et al., 2017; Zhang et al., 2018a,b; Lu et al., 2020). However, some difficulties must be overcome in detecting E. coli O157:H7 via SERS-based LFIA. First, the pores of the NC membrane of LFIA strips are easily blocked by the size of E. coli (generally above 1 μm), resulting in strong non-specific signal or false-positive outcome on the test line. Second, food or clinical samples contain abundant matrix interferences, which may affect the flow of tags on the strips and decrease the sensitivity of the SERS-based LFIA system (Cho et al., 2015; Wang et al., 2020). Thus, high-performance SERS tags with excellent stability in complex samples and good dispersibility on the strip are needed to solve these problems.
In this study, a new type of dual Raman molecules loaded with Au-shell SiO2-core nanospheres (SiO2/Au NPs) was synthesized and introduced into the LFIA system as high-performance SERS labels for E. coli detection. The SiO2/Au tags possessed great dispersity and stability in complex samples and provided stable and intense SERS signal on the test zone of the LFIA system, which could meet the actual requirements for bacterial detection using SERS-based LFIA. By recording the SERS signal of the test line of the proposed SERS-based LFIA, E. coli O157:H7 could be quantitatively detected within 15 min. The sensitivity of the SiO2/Au-based SERS-LFIA strips for E. coli O157:H7 detection was 50 cells/mL in PBS solution, which was 2000 times higher than that of colloidal Au-based LFIA strips. Furthermore, the proposed SERS-based LFIA strips were successfully applied to tap water, milk, human urine, lettuce extract and beef samples. A low limit of detection (LOD) of 100 cells/mL was achieved because of the prominent stability of the synthesized SiO2/Au tags, indicating their good practicability in food security and clinical diagnosis.
Experimental
Chemicals and Materials
Chlorauric acid tetrahydrate (HAuCl4⋅4H2O), hydroxylamine hydrochloride, sodium borohydride, ammonia solution (28%, w/w) and trisodium citrate were purchased from Sinopharm Chemical Reagent Co. (Shanghai, China)1. Tetraethoxysylane (TEOS), polyethyleneimine (PEI), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), polyvinylpyrrolidone (PVP, 40 K), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), MES monohydrate, bovine serum albumin (BSA), phosphate buffer (PB, 0.1 M) and Tween-20 were purchased from Sigma-Aldrich (Shanghai, China)2. Fetal bovine serum (FBS) and phosphate buffer saline (PBS, 10 mM) were obtained from Thermo Fisher (Shanghai, China)3. Goat anti-mouse IgG antibody and mouse monoclonal antibody to E. coli O157:H7 were bought from ACTHTEAM (Changzhou, China)4. Sample pad, absorbent pad, conjugate pad, PVC bottom plate and 50 nm colloidal Au NPs were bought from Jieyi Biotechnology Co. (Shanghai, China)5. Nitrocellulose membrane were purchased from Sartorius (Shanghai, China)6. E. coli O157:H7 (ATCC 35150), enteroaggregative E. coli (EAEC, BNCC340663), enteroinvasive E. coli (EIEC, BNCC340975), enteropathogenic E. coli (EPEC, BNCC186732), enterotoxigenic E. coli (ETEC, BNCC186736), and E. coli BL21 (BNCC353806) was obtained from BeiNa Biotechnology Co., Ltd. (Beijing, China)7. The clinical isolates of Staphylococcus aureus (S. aureus), Salmonella enterobacter (S. enterobacter), Shigella flexneri (S. flexneri), Listeria monocytogenes (L. monocytogenes), Vibrio cholerae (V. cholerae), and Enterococcus faecalis (E. faecalis) were obtained from The Affiliated Hospital of Xuzhou Medical University.
Instruments
Transmission electron microscope (TEM) images were obtained with a Hitachi H-7650 TEM operated at 150 kV. The high-resolution transmission electron microscope (HRTEM) images were acquired via a Philips Tecnai G2 F20 microscope at an acceleration voltage of 200 kV. Scanning electron microscope (SEM) images were obtained from a JTOL JSM-7001F microscope operated at 10 kV. UV-Vis spectra were obtained with a Shimadzu 2600 spectrometer. Zeta potentials were measured by a Malvern Nano-ZS90 Zetasizer. Raman spectra was recorded by a B&W Tek portable Raman spectrometer (i-Raman Plus BWS465-785H). All the samples were excited with a 785 nm laser, which laser power and acquisition time at each spot was 10 mW and 10 s, respectively.
Synthesis of Dual DTNB-Modified SiO2/Au NPs
The fabrication principle of dual DTNB-modified SiO2/Au NPs is shown in Scheme 1A. The SiO2/DTNB/Au/DTNB NPs were synthesized using our previously reported seed-mediated growth method with some modifications. First, SiO2 NPs were prepared via a modified Stöber method (Wang C. et al., 2017). Subsequently, 0.5 mL of SiO2 NPs (7 mg/mL) were added to 100 mL of deionized water and ultrasonically treated for 10 min. Then, 10 mL of PEI solution (0.5 mg/mL) was added into the mixture and ultrasonically treated for 40 min to form SiO2/PEI NPs. The obtained SiO2/PEI NPs were centrifuged and washed two times using deionized water to remove excess PEI. Afterward, 3–5 nm Au NPs were prepared in accordance with the method of Fang et al. (2010) and modified with DTNB (30 μM). The obtained SiO2/PEI NPs were added into 150 mL of 3–5 nm Au/DTNB NPs and ultrasonically treated for 30 min, forming SiO2/seed/DTNB microspheres. The obtained SiO2/seed/DTNB NPs were centrifuged and washed using deionized water to eliminate the superfluous 3–5 nm Au/DTNB NPs and kept in 5 mL of anhydrous ethyl alcohol.
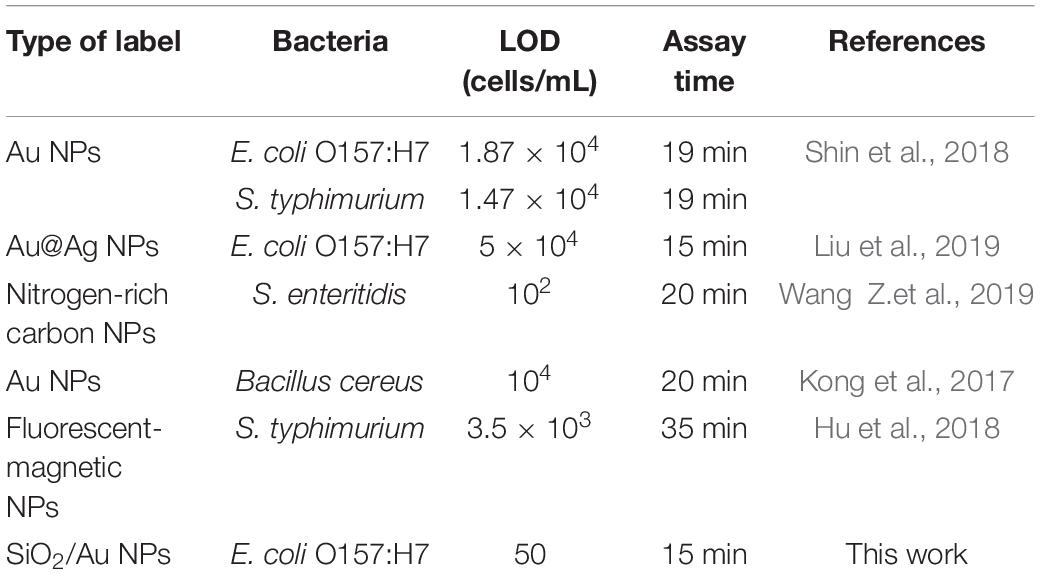
Scheme 1. Schematic of the preparation of dual DTNB-modified SiO2/Au NPs (A), SiO2/Au SERS tags (B), and SERS-based LFIA strip (C).
The SiO2/DTNB/Au NPs were synthesized with an ultrasonic cleaner (800 W power) within 5 min. In brief, 2.5 mL of the obtained SiO2/seed/DTNB NPs were added into 100 mL of aqueous solution containing hydroxylamine hydrochloride (0.5 mg/mL) and PVP (2 mg/mL). The mixture was ultrasonically treated for 5 min and 206 μL of HAuCl4 (1 wt.%) was added and ultrasonically treated for another 5 min. Then, the obtained SiO2/DTNB/Au NPs were centrifuged and washed two times using deionized water to remove the superfluous PVP and kept in 5 mL of anhydrous alcohol. Finally, the obtained SiO2/DTNB/Au NPs were ultrasonically treated with DTNB ethanol solution (50 μL, 10 mM) for 1 h to form dual DTNB-modified SiO2/Au NPs.
Preparation of Dual DTNB-Modified SiO2/Au SERS Tags
Dual DTNB-modified SiO2/Au NP SERS tags were prepared via EDC/sulfo-NHS, where the DTNB-COOH group is combined with the amide group of the antibody (Scheme 1B). In brief, 1 mL of the dual DTNB-modified SiO2/Au NPs were centrifuged and washed using deionized water to eliminate the superfluous DTNB and alcohol. Afterward, the pellet was resuspended in 1 mL of MES buffer (0.1 M, pH 5.5). Freshly prepared EDC (10 mM, 100 μL) and NHS (100 mM, 20 μL) were added into the mixture and ultrasonically treated for 15 min. Subsequently, the mixture was centrifuged and resuspended in 1 mL of 0.05% PBST solution (PBS containing 0.05% Tween-20, v/v). E. coli O157:H7 detection antibody (20 μg) was added into the mixture and incubated for 2 h. Then, 100 μL of 10% BSA (w/v) was added to block the unreacted carboxyl sites of SERS labels for an additional 1 h at 37°C. The functionalized SERS labels were pelleted by centrifugation and resuspended in 500 μL of a solution containing Tris–HCl (50 mM, pH 8.0), 0.5% Tween-20 (v/v), 1% BSA (w/v), 10% sucrose (w/v), and 0.1% PVP (w/v). The SERS tags were evenly pipetted onto the glass fiber paper and dried at 37°C to form conjugate pads.
Preparation of Dual DTNB-Modified SiO2/Au-Based SERS-LFIA Strips
The SERS-based LFIA strip was composed of four sections: sample pad, absorbent pad, conjugate pad with SERS tags, and NC membrane (test and control lines). E. coli O157:H7 capture antibody (0.4 mg/mL) and goat anti-mouse IgG antibody (1 mg/mL) were drawn onto the NC membrane pad separately and dried at 37°C for 1 h. Then, the four sections were assembled onto a PVC bottom plate in sequence. The integrated membrane was cut into 3 mm-wide strips with a paper cutter and stored in a dry container until use.
Preparation of Bacterial Sample
The standard strains were cultivated in 5 mL of Luria–Bertani broth at 37°C for 5 h. The E. coli O157:H7 concentration was verified via plate count method. In brief, the original bacterial solution was diluted 105, 106, and 107 times into 1 mL of sterile water, and 200 μL of which was added onto blood agar plates and cultured for 12 h, as shown in Supplementary Figure S1. The original E. coli O157:H7 concentration was 1.145 × 108 cells/mL.
E. coli O157:H7 Detection Using Dual DTNB-Modified SiO2/Au-Based SERS-LFIA Strips
The detection process of the LFIA strips is shown in Scheme 1C. E. coli O157:H7 was diluted in running buffer (PBS containing 20% FBS and 0.5% Tween-20, v/v), with concentrations ranging from 50 cells/mL to 107 cells/mL, as positive samples. The running buffer without E. coli O157:H7 was used as the blank control. Under the capillary effect, the sample solution (100 μL) moved toward the absorbent pad when dropped onto the sample pad. The test process was completed within 15 min. The SERS signals of the test lines were tested with a portable Raman spectrometer, with integration time and laser power of 10 s and 10 mW, respectively.
E. coli O157:H7 Detection in Biological Samples
The preparation of lettuce extract and beef samples was done as described by Li et al. (2020). E. coli O157:H7 was spiked into tap water, milk, human urine, the lettuce extract and beef samples, with concentrations ranging from 50 cells/mL to 107 cells/mL. The detection process was similar to that of E. coli in running buffer but needed an additional diluent (FBS containing 2.5% Tween-20, v/v). In brief, 80 μL of the sample was adequately mixed with 20 μL diluent. The mixture was then pipetted onto the sample pad of the SERS-based LIFA strips. Twelve min later, the test lines were tested with a portable Raman spectrometer.
Results and Discussion
Characterization of Dual DTNB-Modified SiO2/Au SERS Tags
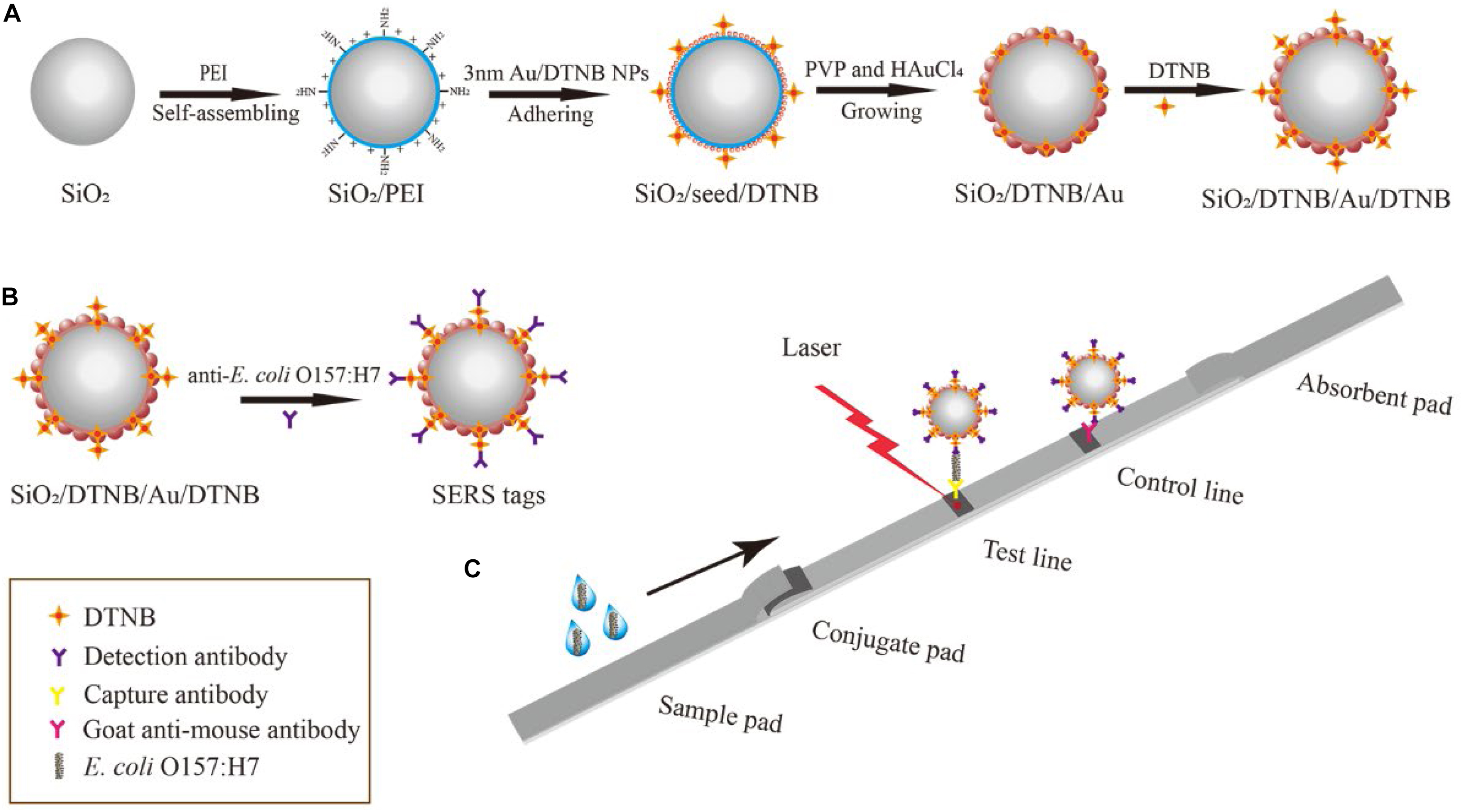
The products synthesized at different stages were analyzed via TEM and SEM to characterize their structure and morphology. The monodispersed SiO2 NPs (Figure 1A) prepared using the modified Stöber method had a uniform diameter size of approximately 150 nm. After the SiO2 NPs were coated with PEI layer, the 3–5 nm Au/DTNB NPs densely adhered on the surface of the SiO2/PEI NPs (Figure 1B). These small Au NPs acted as nucleation sites for Au shell synthesis. Figure 1C displays the TEM image of the obtained SiO2/DTNB/Au NPs, whose size increased from 150 nm to approximately 180 nm. The Au shell was approximately 15 nm thick. Figures 1B,C illustrate the strong difference between the SiO2/seed NPs and the SiO2/Au NPs. Figures 1D,F show the HRTEM and SEM images of the SiO2/DTNB/Au NPs, respectively, displaying the rough surface of the SiO2/DTNB/Au NPs. The dense Au/DTNB NPs obviously increased in size and formed a rough Au shell on the surface of the SiO2 particles, thereby ensuring high SERS performance. Figure 1E illustrates the HRTEM image of a part of Au shell. The distance of the crystal planes was 0.235 nm, corresponding to the Au(111) plane (Switzer et al., 2016; Wang et al., 2016).
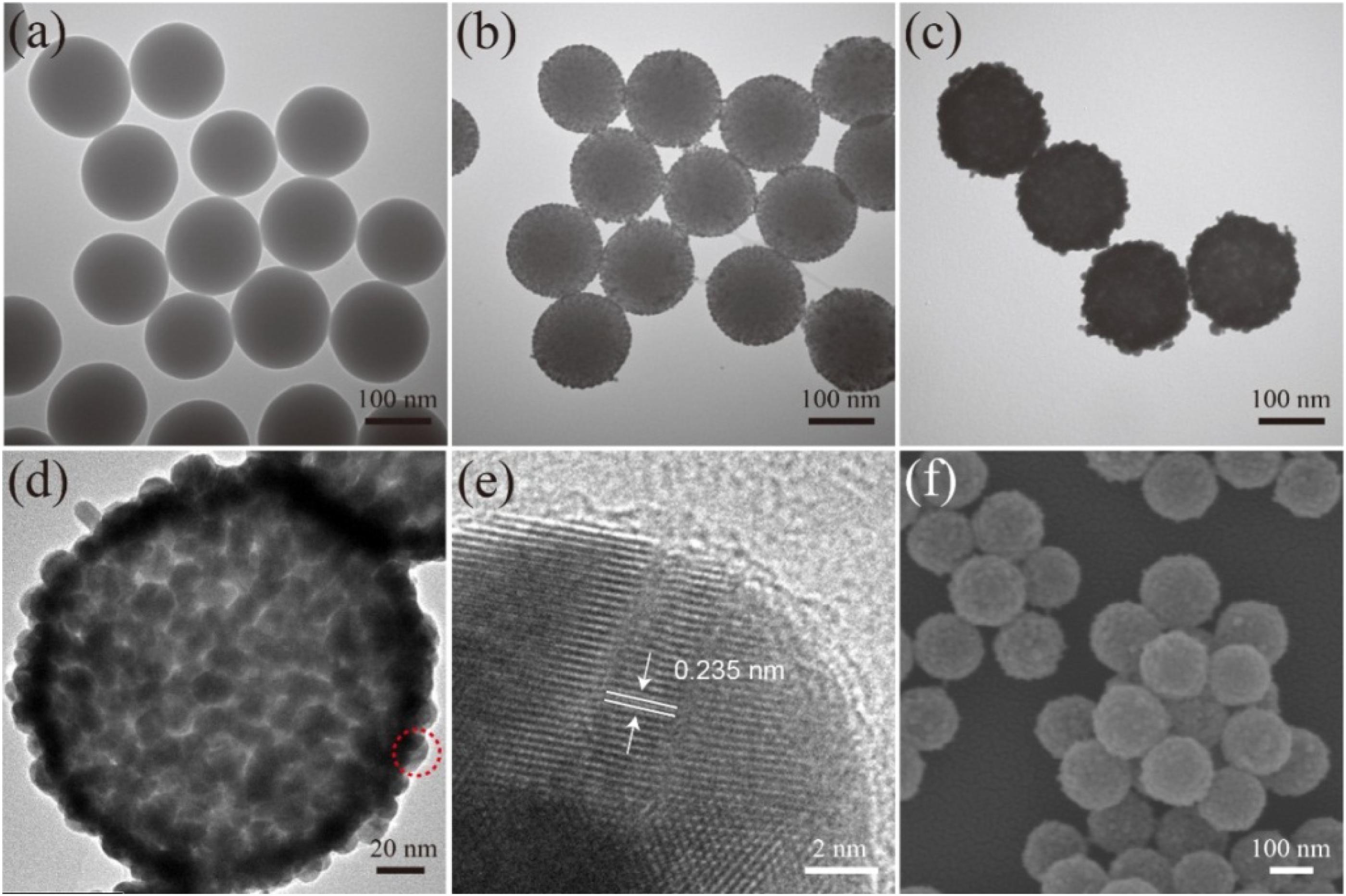
Figure 1. Characterization of synthesis SiO2/Au NPs. TEM images of SiO2 (a), SiO2/PEI-seed (b), and SiO2/Au (c). HRTEM images (d,e) and SEM image (f) of SiO2/Au NPs.
Figure 2A shows the element mapping of the SiO2/Au NPs. Si (green) and O (blue) were surrounded by dense Au (red), indicating that the SiO2 microspheres were successfully coated with an Au shell. The synthesized products at different stages were measured in deionized water with a UV-vis spectrometer, as shown in Figure 2B. The SiO2 NPs and SiO2/PEI NPs did not show distinct absorption peaks (curves a and b), whereas the SiO2/PEI-seed had a distinct absorption peak at approximately 564 nm (curve c). This finding suggested that several small Au/DTNB seeds were successfully absorbed on the surface of SiO2 NPs. After an Au shell was formed, the absorption peak in the UV-vis spectra shifted to 642 nm, and its intensity increased simultaneously (curve d). As shown in Figure 2C, the synthesized products at different stages were tested in deionized water with a zeta potential analyzer. The zeta potential of the SiO2 NPs increased from −47.8 to 53.0 mV when the PEI was modified on the silicon spheres. This change confirmed that the surface of the SiO2 NPs successfully absorbed a layer of positive-charged PEI. The zeta potential became negative again when small Au/DTNB NPs were attached to the SiO2/PEI particles (−11.5 mV). The negative potential increased further when an Au shell was complete formed (−22.4 mV).
Figure 2. (A) Element mapping of SiO2/Au NPs (i–iv). (B) UV-vis spectra of SiO2 (curve a), SiO2/PEI (curve b), SiO2/PEI-seed (curve c), and SiO2/Au (curve d). (C) Zeta potential of SiO2, SiO2/PEI, SiO2/PEI-seed, and SiO2/Au. (D) Raman intensity of SiO2/seed/DTNB/Au NPs with different concentrations of HAuCl4: 0.01, 0.02, 0.03, 0.04, 0.05, and 0.06 mM. (E) Raman spectra of SiO2/PEI-seed/DTNB (curve a), SiO2/Au/DTNB (curve b), and SiO2/DTNB/Au/DTNB (curve c).
The method for forming Au shell is basically seed-mediated growth. The thickness of the Au shell could be easily controlled by adjusting the concentration of HAuCl4 when the amount of SiO2/PEI-seed NPs remained constant. Supplementary Figure S2 exhibits the representative TEM images of SiO2/seed/DTNB/Au NPs, which were fabricated with different concentrations of HAuCl4 when other conditions were kept invariable. The 3–5 nm Au/DTNB NPs attached to the SiO2 spheres gradually increased in size and finally connected with one another to completely form an Au shell with different thicknesses when the concentrations of HAuCl4 increased from 0.01 to 0.06 mM. The Raman intensity of the SiO2/seed/DTNB/Au NPs was tested to investigate the influence of Au shell thickness on SERS performance. Figure 2D exhibits the SERS activity of the SiO2/seed/DTNB/Au NPs with different Au shell thicknesses. Compared with the poor SERS performance of SiO2/seed NPs, the SERS enhancement ability of the SiO2/seed/DTNB/Au NPs gradually increased along with the thickening of the Au shell. However, the SERS ability of SiO2/seed/DTNB/Au NPs decreased when the Au shell overgrew, decreasing the hot spots and nanogaps on the surface of the SiO2/Au NPs. Therefore, the SiO2/Au NPs prepared with 0.05 mM HAuCl4 was chosen as the ideal material for SERS application because its Au shell was almost complete and the SERS performance was optimal. Notably, we selected DTNB as the Raman reporter molecule to build SiO2/Au SERS tags, because DTNB can be easily modified on the Au shell through Au–S covalent bonding and provide a large Raman cross section (Song et al., 2018; Zhang C. et al., 2018; Pang et al., 2019).
Figure 2E shows the SERS activity of the SiO2/PEI-seed, SiO2/Au, and SiO2/DTNB/Au NPs. The SERS activity of the SiO2/PEI-seed NPs was fairly weak but increased when an Au shell was completely formed. The SERS activity further increased as DTNB was modified inside the Au shell (Rong et al., 2018). This phenomenon could be due to two reasons. First, a mass of hot spots existed in the rough Au shell. Second, increased amounts of DTNB molecules were embedded inside the Au shell. However, once the amount of DTNB molecules on the surface of the SiO2/PEI-seeds was in excess, an Au shell could not be completely fabricated outside the SiO2 NPs, as shown in Supplementary Figure S3A. Compared with common colloidal Au NPs, the SiO2/PEI-seed/DTNB/Au NPs could work in high ion solution and be centrifuged repeatedly without agglutinating, indicating their good stability (Supplementary Figure S3B). Supplementary Figure S4 illustrates the Raman intensities of the dual DTBN-modified SiO2/Au NPs detected 30 times in five separate batches. The difference in SERS intensities was 2.76%, indicating good repeatability.
Optimization of SERS-Based LFIA Strips
Dual DTNB-modified SiO2/Au NPs are very suitable as high-performance SERS probe for LFIA-based bacterial detection due to their excellent SERS performance, superior dispersity, and good stability and repeatability. In this study, E. coli O157:H7 was used to evaluate the performance of the SiO2/Au-based SERS-LFIA strips. As shown in Scheme 1B, an E. coli O157:H7 detection antibody was conjugated with functionalized carboxyl groups of SiO2/Au NPs via carbodiimide chemistry (Wang et al., 2015). The antibody concentration was saturated to ensure effectiveness and stability. The unreacted carboxyl sites of the SiO2/Au NPs were blocked by saturated BSA to decrease the non-specific adsorption of immune-SiO2/Au NPs on the NC membrane. As shown in Supplementary Figure S5A, the zeta potential of the SiO2/Au NPs decreased from −21.5 to −25.5 mV after antibody modification. Supplementary Figure S5B displays the TEM image of E. coli O157:H7 combined with the antibody-labeled SiO2/Au NPs. These results proved the successful conjugation of the anti-E. coli O157:H7 antibody with the SiO2/Au NPs.
Herein, the SiO2/Au NPs acted as a multifunctional material for visual and SERS detection of target bacteria on the LFIA strips. The usage conditions (such as type of NC membrane, running buffer ingredient, and antibody concentration on the test line) of the LFIA strips must be optimized to achieve their best effect on bacterial detection. Two large-aperture NC membranes (CN 95 and CN 140, with pore sizes of 15 and 8 μm, respectively) were tested because the size of the SiO2/Au-bacteria complexes are generally larger than 1 μm. Supplementary Figure S6 shows that the these complexes could pass through the wide channels of the CN 95 membrane, whereas many SiO2/Au agglomerates were blocked at the junction of the CN 140 membrane and conjunction pad. The accuracy and repeatability of the proposed LFIA strip was seriously influenced by this blocking phenomenon. Therefore, the CN 95 membrane was chosen as the ideal NC membrane of the SiO2/Au-based SERS-LFIA strip. The next optimizing experiment was the running buffer, which could substantially affect the immunoreaction of the test lines and the flow rate of the SiO2/Au NPs. PBS and 0.5% PBST (PBS containing 0.5% Tween-20, v/v) were considered because our previous work indicated that PBS solution has no influence on the activity of antibody-labeled nanotags (Qie et al., 2019; Wang et al., 2019c). The moderate concentration of Tween-20 ensured the smooth surface of the SiO2/Au-bacterial complexes, enabling easy delivery along the strips. However, the non-specific binding at the test line increased when the strips detected the running buffer without bacteria, as shown in Supplementary Figure S7A. FBS was added into the running buffer to decrease the non-specific combination on the test line. The 0.5% PBST buffer containing 20% FBS (v/v) suppressed the non-specific binding more effectively than that containing 10% FBS (v/v), which did not affect the specific binding of the test line. Therefore, 0.5% PBST containing 20% FBS (v/v) was chosen as the running buffer. The sensitivity of the LFIA strips was also affected by the antibody concentration of the test lines. Supplementary Figure S7B indicates that the test line with 0.4 mg/mL capture antibody had the highest signal-to-noise ratio of the SERS signal. In this study, all the optimal conditions were utilized to achieve superior performance of the SiO2/Au-based SERS-LFIA strips for pathogen detection.
Quantitative Analysis of E. coli O157:H7 by SiO2/Au-Based SERS-LFIA Strip
Under optimal conditions (such as NC membrane, running buffer and antibody concentration of test line), E. coli O157:H7 sample concentrations ranging from 50 cells/mL to 107 cells/mL were analyzed using the SiO2/Au-based SERS-LFIA strip. Figure 3A displays the photo of the SiO2/Au-based SERS-LFIA strip testing for E. coli O157:H7 detection under different concentrations. When the bacterial concentration was high, the dark color of the SiO2/Au NPs immobilizing on the test lines could be clearly observed. This color gradually faded away as the E. coli concentration decreased. The visual sensitivity of the dark test lines was 105 cells/mL, and the color was barely visible when the E. coli concentration was lower than 105 cells/mL. Thus, quantitative detection of E. coli O157:H7 was difficult to conduct with high sensitivity due to the colorimetric change in the test line. However, highly sensitive analysis of E. coli O157:H7 was feasible when the SERS signals of these test lines were measured and analyzed using the proposed SiO2/Au-based SERS-LIFA strips. Figure 3B illustrates the Raman spectra of the test line with different E. coli O157:H7 concentrations ranging from 50 cells/mL to 107 cells/mL in running buffer. The SERS signal intensity declined with the decrease in E. coli O157:H7 concentrations. The main peak at 1,331 cm–1 was still distinct when the E. coli O157:H7 concentrations decreased to 50 cells/mL compared with the blank control. Under SERS detection mode, the Raman intensity of the test lines was measured to quantitatively detect E. coli O157:H7. As displayed in Figure 3C, the calibration curves of the SiO2/Au-based SERS-LFIA strips were constructed using the Raman intensity from their test lines and the E. coli O157:H7 concentrations. Error bars showed the standard deviation (SD) of five detections. The LOD of E. coli O157:H7 analyzed using the LFIA strips was 50 cells/mL based on the IUPAC formula (LOD = yblank + 3 × SDblank, where yblank and SDblank are the average Raman intensity and the SD of the blank control, respectively) (Hu et al., 2017; Putnin et al., 2019). The SiO2/Au-based SERS-LFIA strips were characterized by SEM to prove that the SERS signal came from the SERS tags fixed on the test line. Figure 3D illustrates the SEM image of the test line of the negative sample. The color and SERS signal remained unchanged, and SERS tags could not be observed on the test line. Figure 3E displays the SEM image of the test line of the positive sample. The color of the test line became dark, and the Raman intensity was enhanced as the SERS tags were gradually immobilized on the test area. These results directly proved that SiO2/Au-bacterial complexes were immobilized on the test zone by forming sandwich-like immunocomplexes.
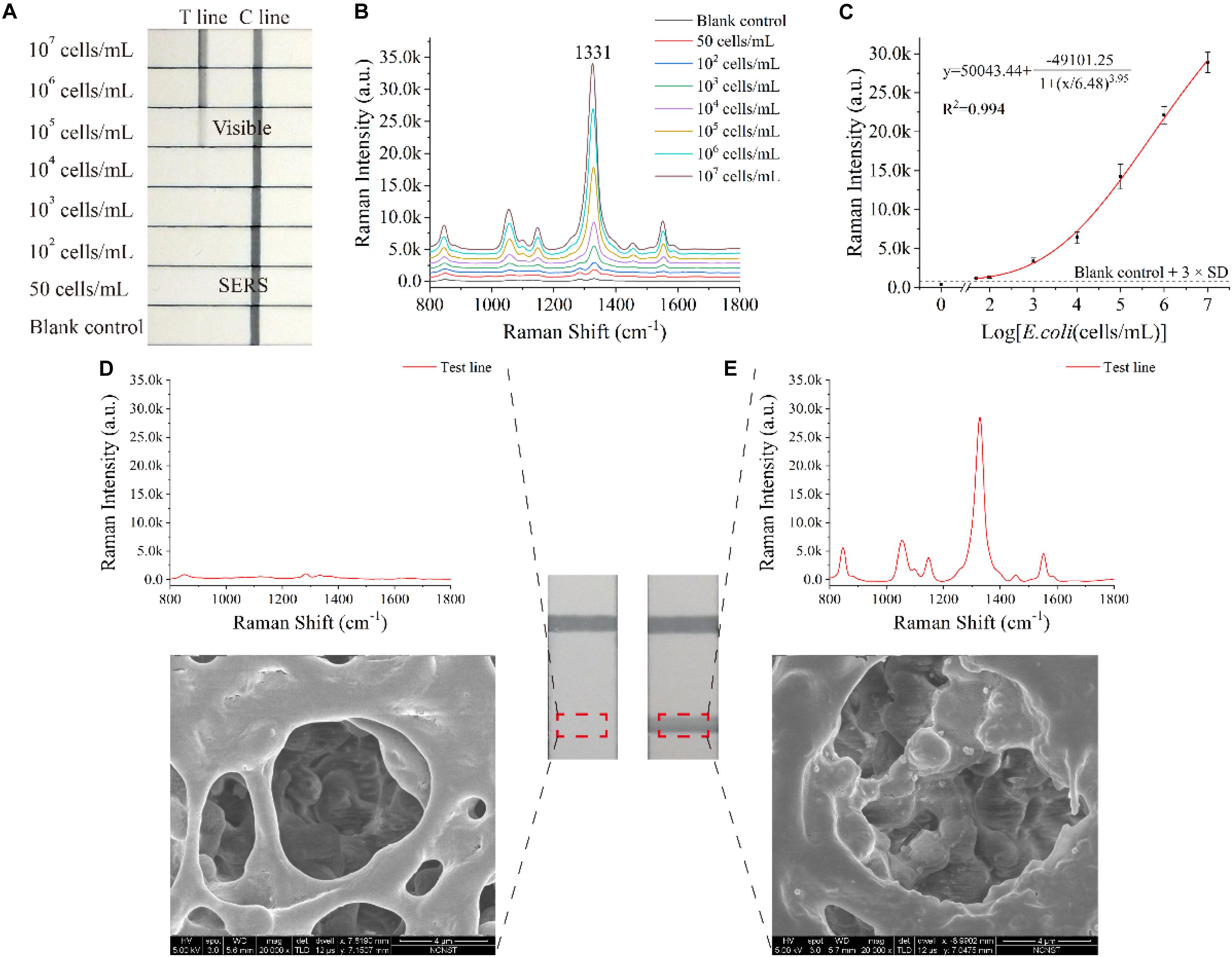
Figure 3. Analytical performance of the dual DTNB-modified SiO2/Au-based SERS-LFIA strips. Photographs (A), SERS spectra of the test lines (B), and corresponding calibration curve (C) of the LIFA strips for E. coli O157:H7 detection. SERS signals, SEM images, and photographs of the test lines of the blank control (D) and the positive control (E).
An ordinary SERS-based LFIA strip was prepared by utilizing the same paired E. coli O157:H7 antibody and replacing the SiO2/Au NPs with common colloidal Au NPs (50 nm) to prove the superior performance of the proposed SiO2/Au-based SERS-LFIA strips. The preparation of colloidal Au SERS tags-based LFIA strips and the detection method of E. coli O157:H7 are described in Supplementary Material. As shown in Supplementary Figure S8, the LOD of the colloidal Au SERS tags-based LFIA strip was 105 cells/mL on vision, while that of E. coli O157:H7 under SERS detection mode was 104 cells/mL. Thus, the SERS sensitivity of SiO2/Au-based SERS-LFIA strip was 2,000 and 200 times higher than the visual and SERS sensitivity of colloidal Au SERS tags-based LFIA strip, respectively. Moreover, the SiO2/Au based SERS-LFIA strip enabled fast bacterial detection within 15 min, which included a 12 min chromatographic process and 3 min SERS analysis time.
Other indexes, such as specificity, reproducibility, and stability, were also considered to further assess the performance of the SiO2/Au-based SERS-LFIA strips. Several enteropathogenic bacteria, such as S. aureus, S. enteritidis, S. flexneri, L. monocytogenes, V. cholera, and E. faecalis, were used as interferents to test the specificity of the proposed assay. All the bacterial samples were detected by the developed strip and prepared at a concentration of approximately 107 cells/mL. Figure 4A shows that the Raman intensity of all these non-target bacteria was close to the intensity of the blank control, but the signal of E. coli O157:H7 group was obvious on the test line. Moreover, other non-O157:H7 serotype of E. coli such as E. coli BL21, EAEC, EIEC, EPEC, and ETEC were detected to further verify the specificity of the SiO2/Au-based SERS-LFIA strips, as shown in Figure 4B. Results indicate excellent specificity of the SiO2/Au-based SERS-LFIA strips for target bacterial detection. The reproducibility of these strips was studied by testing tap-water samples, which were spiked with different E. coli O157:H7 concentrations. Five independent experiments were performed to estimate 107 and 104 cells/mL of E. coli O157:H7 samples. Supplementary Figure S9 shows that the relative SD values of the SERS intensity were 5.03% and 6.92%, respectively, indicating high reliability and reproducibility of the prepared SERS-LFIA strips.
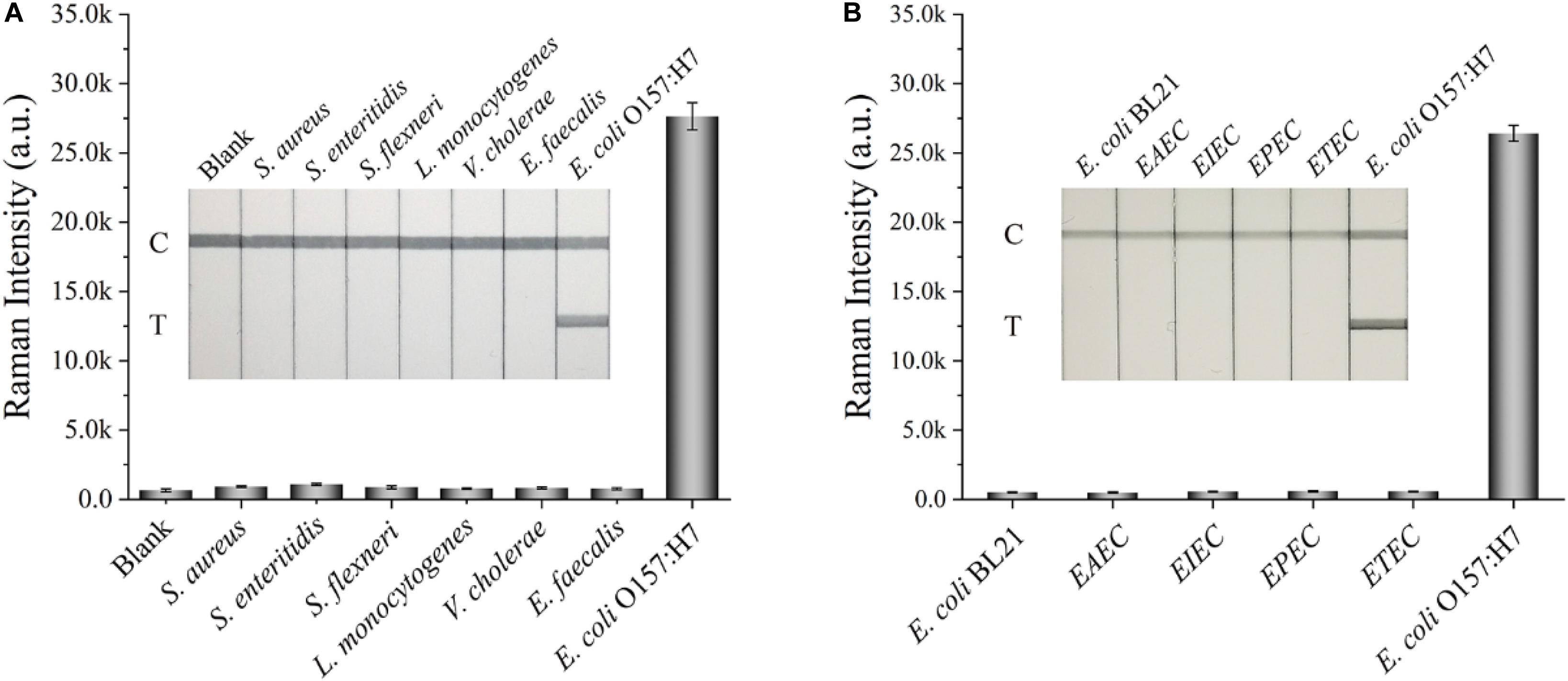
Figure 4. Specificity of the dual DTNB-modified SiO2/Au-based SERS-LFIA strip. Images of the strips and corresponding Raman intensities of the test lines for several enteropathogenic bacteria (A) and other non-O157 serotype E. coli (B). The concentration of these bacteria was approximately 107 cells/mL.
Detection of E. coli O157:H7 in Tap Water and Biological Samples
The proposed assay detected E. coli O157:H7 in tap water and biological samples, including milk and human urine, to evaluate the stability and practicability of the proposed LFIA strips. E. coli O157:H7 was spiked into these samples with different concentrations ranging from 50 cells/mL to 107 cells/mL. In the detection process, 80 μL of the sample solution was mixed with 20 μL of the diluent (FBS containing 2.5% Tween-20, v/v) and pipetted onto the sample pad of the SiO2/Au-based SERS-LFIA strip. Using the proposed LFIA system, E. coli O157:H7 was accurately detected in the biological samples. As shown in Figure 5, the Raman signals of these positive samples were distinguished but slightly weaker than those in the running buffer. Moreover, no Raman signals could be achieved in the negative groups. The LODs of E. coli O157:H7 in tap water (Figure 5A), milk (Figure 5B), and human urine (Figure 5C) were as low as 100 cells/mL. Such low values met the clinic test requirement (Ho et al., 2019). To further verify the practicality of the SiO2/Au-based SERS-LFIA strips, E. coli O157:H7 was detected in other food samples, such as lettuce extract and beef. As shown in Supplementary Figure S10, the sensitivity of the SiO2/Au-based SERS-LFIA strip was not influenced by the food samples. It was the high stability of the SiO2/Au particles that ensured the superior anti-interference ability of this method for biological samples.
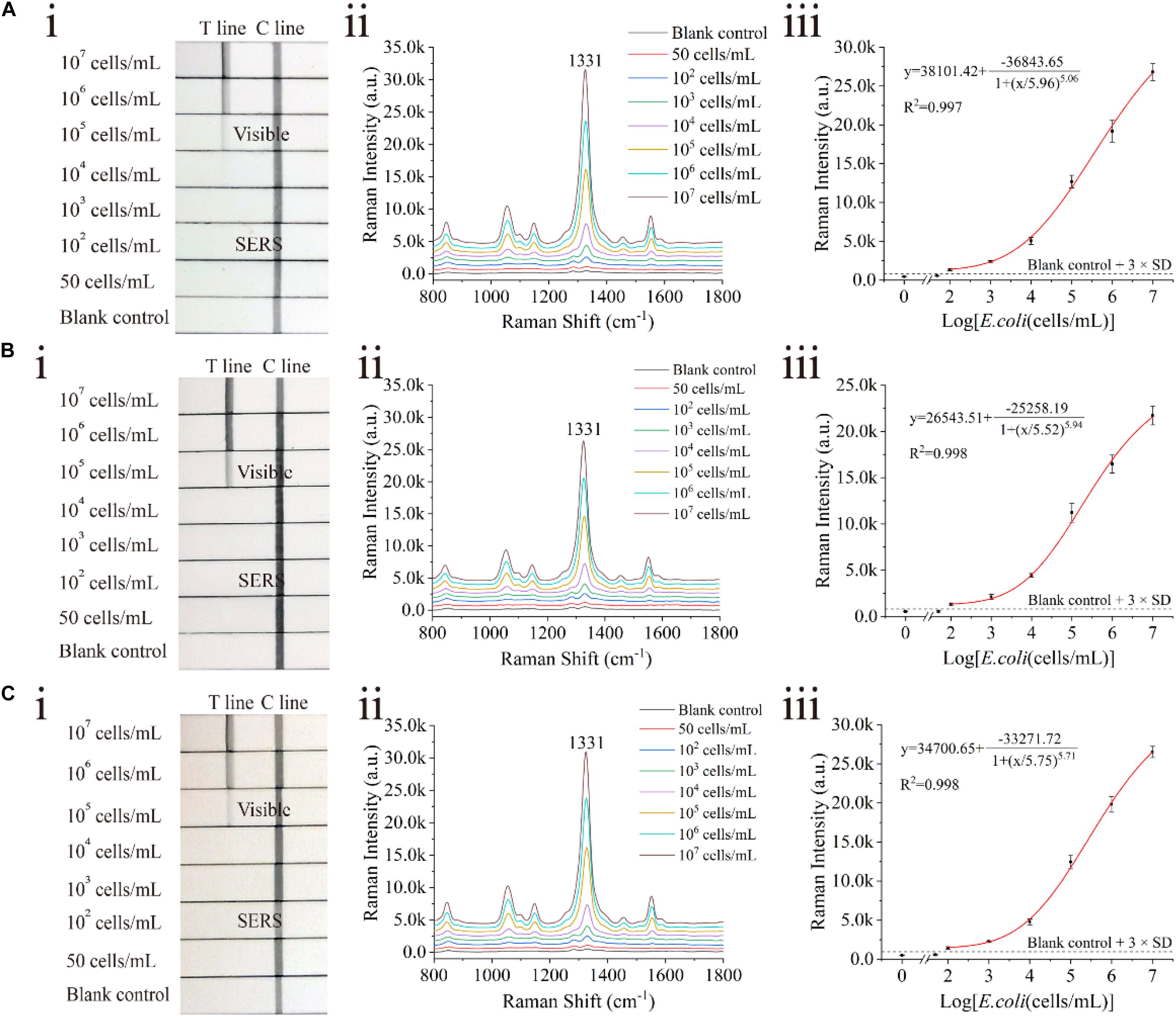
Figure 5. Analytical performance of dual DTNB-modified SiO2/Au-based SERS-LFIA strip in biological samples. Photographs (i), Raman intensities of the test lines (ii), and corresponding calibration curves (iii) of the LFIA strips for E. coli O157:H7 detection in tap water (A), milk (B), and human urine (C).
Table 1 shows the comparison between results from the current study and those from other reported LFIAs for bacterial detection. The SiO2/Au based SERS-LFIA strip considerably improved the detection sensitivity of the target bacteria in comparison with other previously reported LFIA system (102–105 cells/mL), especially the commercial colloidal Au strip (105–107 cfu/mL) (Gast et al., 2003; Kong et al., 2017; Hu et al., 2018; Shin et al., 2018; Wang et al., 2018a; Huang et al., 2019; Zhang et al., 2020). In addition, the SiO2/Au-LFIA strip could be successfully used for quantitative detection of E. coli O157:H7 in biological samples, making the POCT promising for sensitive testing of pathogenic bacteria. A notable detail that the great reproducibility and specificity of this strip were too dependent on the specific E. coli antibody utilized on the SiO2/Au labels and test lines. In theory, sensitive detection could also be conducted on other pathogenic bacteria via the SiO2/Au-LFIA strip by using their corresponding specific antibodies.
Conclusion
In conclusion, a new type of dual DTNB molecule-modified SiO2/Au NP was developed using an efficient PEI-mediated seed growth method. PEI was utilized to form an interlayer with positive charge into the nanostructure, which effectively absorbed a mass of 3–5 nm Au/DTNB NPs as seeds for inner DTNB loading and Au shell reduction. The obtained SiO2/Au nanocomposites possessed excellent stability and monodispersity and superior SERS properties. They were applied in the LFIA strips as high-performance SERS tags for bacterial detection in high sensitivity. The feasibility of the SiO2/Au-based SERS-LFIA system was verified by detecting E. coli O157:H7. The SiO2/Au-based SERS-LFIA strips were proven to be an effective tool for the specific detection of E. coli in biological samples and the SERS quantitative analysis of target bacteria on the test line of the strip under optimal experimental conditions. The LOD of E. coli was 50 cells/mL, indicating that these strips were 200 and 2,000 times more sensitive than the SERS and visual LOD of the colloidal Au SERS tags-based LFIA strips, respectively. Moreover, the high stability of the SiO2/Au particles ensured the good practicability of our proposed strip for biological samples. To our knowledge, this study was the first to utilize SiO2/Au-based SERS-LFIA strip for bacterial detection. Considering other advantages, such as shortened detection time, easy operation, great reproducibility, and low cost (<$1 per test), the proposed assay has a good potential for rapid and sensitive detection of pathogenic bacteria in biological samples.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
BG, SW, and CW designed and managed the project. LS, LX, RX, and ZZ performed all the experiments. LS and LX did the analysis of SERS immunoassay results. BG, CW, and LS wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the Major Infectious Diseases such as AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (2018ZX10712-001), the National Natural Science Foundation of China (Grant Nos. 81830101, 81471994, and 81871734), the Natural Science Foundation of Anhui Province (Grant No. 1908085QB85), Jiangsu Privincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), and Advanced Health Talent of Six-One Project of Jiangsu Province (LGY2016042).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Beijing Zhongkebaice Technology Service Co, for helping to conduct TEM and SEM analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.596005/full#supplementary-material
Footnotes
- ^ https://www.reagent.com.cn
- ^ https://www.sigmaaldrich.com
- ^ https://www.thermofisher.com
- ^ http://www.xinxinbio.com
- ^ http://www.joey-bio.com
- ^ https://www.sartorius.com.cn
- ^ http://www.bnbio.com
References
Cho, I. H., Bhandari, P., Patel, P., and Irudayaraj, J. (2015). Membrane filter-assisted surface enhanced Raman spectroscopy for the rapid detection of E. coli O157:H7 in ground beef. Biosens. Bioelectron. 64, 171–176. doi: 10.1016/j.bios.2014.08.063
Fang, Y., Guo, S., Zhu, C., Zhai, Y., and Wang, E. (2010). Self-assembly of cationic polyelectrolyte-functionalized graphene nanosheets and gold nanoparticles: a two-dimensional Heterostructure for hydrogen peroxide sensing. Langmuir 26, 11277–11282. doi: 10.1021/la100575g
Gast, R. K., Holt, P. S., Nasir, M. S., Jolley, M. E., and Stone, H. D. (2003). Detection of Salmonella enteritidis in incubated pools of egg contents by fluorescence polarization and lateral flow immunodiffusion. Poult. Sci. 82, 687–690. doi: 10.1093/ps/82.4.687
Guo, Q., Han, J. J., Shan, S., Liu, D. F., Wu, S. S., Xiong, Y. H., et al. (2016). DNA-based hybridization chain reaction and biotin-streptavidin signal amplification for sensitive detection of Escherichia coli O157:H7 through ELISA. Biosens. Bioelectron. 86, 990–995. doi: 10.1016/j.bios.2016.07.049
Hassan, A. H. A., Bergua, J. F., Morales-Narvaez, E., and Mekoci, A. (2019). Validity of a single antibody-based lateral flow immunoassay depending on graphene oxide for highly sensitive determination of E. coli O157:H7 in minced beef and river water. Food Chem. 297:124965. doi: 10.1016/j.foodchem.2019.124965
Ho, H. J., Tan, M. X., Chen, M. I., Tan, T. Y., Koo, S. H., Koong, A. Y. L., et al. (2019). Interaction between antibiotic resistance, resistance genes, and treatment response for urinary tract infections in primary care. J. Clin. Microbiol. 57:e0143-19. doi: 10.1128/JCM.00143-19
Hu, J., Jiang, Y. Z., Tang, M., Wu, L. L., Xie, H. Y., Zhang, Z. L., et al. (2018). Colorimetric-Fluorescent-magnetic nanosphere-based multimodal assay platform for Salmonella detection. Anal. Chem. 91, 1178–1184. doi: 10.1021/acs.analchem.8b05154
Hu, J., Jiang, Y. Z., Wu, L. L., Wu, Z., Bi, Y., Wong, G., et al. (2017). Dual-signal readout nanospheres for rapid point-of-care detection of ebola virus glycoprotein. Anal. Chem. 89, 13105–13111. doi: 10.1021/acs.analchem.7b02222
Hu, R.-R., Yin, Z.-Z., Zeng, Y.-B., Zhang, J., Liu, H.-Q., Shao, Y., et al. (2016). A novel biosensor for Escherichia coli O157:H7 based on fluorescein-releasable biolabels. Biosens. Bioelectron. 78, 31–36. doi: 10.1016/j.bios.2015.11.018
Huang, Z., Peng, J., Han, J., Zhang, G., Huang, Y., Duan, M., et al. (2019). A novel method based on fluorescent magnetic nanobeads for rapid detection of Escherichia coli O157:H7. Food Chem. 276, 333–341. doi: 10.1016/j.foodchem.2018.09.164
Hwang, J., Lee, S., and Choo, J. (2016). Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of Staphylococcal enterotoxin B. Nanoscale 8:C5nr07243c. doi: 10.1039/c5nr07243c
Jia, K., Adam, P. M., Marks, R. S., and Ionescu, R. E. (2015). Fixed Escherichia coli bacterial templates enable the production of sensitive SERS-based gold nanostructures. Sens. Actuat. B Chem. 211, 213–219. doi: 10.1016/j.snb.2015.01.049
Jia, X., Wang, C., Rong, Z., Li, J., Wang, K., Qie, Z., et al. (2018). Dual dye-loaded Au@Ag coupled to a lateral flow immunoassay for the accurate and sensitive detection of Mycoplasma pneumoniae infection. RSC Adv. 8, 21243–21251. doi: 10.1039/c8ra03323d
Khlebtsov, B. N., Bratashov, D. N., Byzova, N. A., Dzantiev, B. B., and Khlebtsov, N. G. (2018). SERS-based lateral flow immunoassay of troponin I by using gap-enhanced Raman tags. Nano Res. 12, 413–420. doi: 10.1007/s12274-018-2232-4
Kong, M., Shin, J. H., Heu, S., Park, J. K., and Ryu, S. (2017). Lateral flow assay-based bacterial detection using engineered cell wall binding domains of a phage endolysin. Biosens. Bioelectron. 96, 173–177. doi: 10.1016/j.bios.2017.05.010
Law, J. W., Ab Mutalib, N. S., Chan, K. G., and Lee, L. H. (2014). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Lee, S. H., Hwang, J., Kim, K., Jeon, J., Lee, S., Ko, J., et al. (2019). Quantitative serodiagnosis of scrub typhus using surface-enhanced raman scattering-based lateral flow assay platforms. Anal. Chem. 91, 12275–12282. doi: 10.1021/acs.analchem.9b02363
Li, J., Wang, C., Shi, L., Shao, L., Fu, P., Wang, K., et al. (2019). Rapid identification and antibiotic susceptibility test of pathogens in blood based on magnetic separation and surface-enhanced Raman scattering. Mikrochim. Acta 186:475. doi: 10.1007/s00604-019-3571-x
Li, Y., Lu, C., Zhou, S., Fauconnier, M.-L., Gao, F., Fan, B., et al. (2020). Sensitive and simultaneous detection of different pathogens by surface-enhanced Raman scattering based on aptamer and Raman reporter co-mediated gold tags. Sens. Actuat. B Chem. 317:182. doi: 10.1016/j.snb.2020.128182
Liu, H. B., Chen, C. Y., Zhang, C. N., Du, X. J., Li, P., and Wang, S. (2019). Functionalized Au(MBA) @Ag nanoparticles as an optical and SERS dual probe in a lateral flow strip for the quantitative detection of Escherichia coli O157:H7. J. Food Sci. 84, 2916–2924. doi: 10.1111/1750-3841.14766
Liu, Y., Singh, P., and Mustapha, A. (2018). High-resolution melt curve PCR assay for specific detection of E. coli O157:H7 in beef. Food Control 86, 275–282. doi: 10.1016/j.foodcont.2017.11.025
Lu, L., Yu, J., Liu, X., Yang, X., Zhou, Z., Jin, Q., et al. (2020). Rapid, quantitative and ultra-sensitive detection of cancer biomarker by a SERRS-based lateral flow immunoassay using bovine serum albumin coated Au nanorods. RSC Adv. 10, 271–281. doi: 10.1039/c9ra09471g
Mahmoudi, T., de la Guardia, M., and Baradaran, B. (2020). Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. TrAC Trends Analyt. Chem. 125:115842. doi: 10.1016/j.trac.2020.115842
Maneeprakorn, W., Bamrungsap, S., Apiwat, C., and Wiriyachaiporn, N. (2016). Surface-enhanced Raman scattering based lateral flow immunochromatographic assay for sensitive influenza detection. RSC Adv. 6, 112079–112085. doi: 10.1039/c6ra24418a
Pang, Y., Wang, C., Lu, L., Wang, C., Sun, Z., and Xiao, R. (2019). Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer. Biosens. Bioelectron. 130, 204–213. doi: 10.1016/j.bios.2019.01.039
Putnin, T., Ngamaroonchote, A., Wiriyakun, N., Ounnunkad, K., and Laocharoensuk, R. (2019). Dually functional polyethylenimine-coated gold nanoparticles: a versatile material for electrode modification and highly sensitive simultaneous determination of four tumor markers. Microchim. Acta 186:305. doi: 10.1007/s00604-019-3370-4
Qie, Z., Yan, W., Gao, Z., Meng, W., Xiao, R., and Wang, S. (2019). Ovalbumin antibody-based fluorometric immunochromatographic lateral flow assay using CdSe/ZnS quantum dot beads as label for determination of T-2 toxin. Mikrochim. Acta 186:816. doi: 10.1007/s00604-019-3964-x
Rong, Z., Xiao, R., Xing, S., Xiong, G., Yu, Z., Wang, L., et al. (2018). SERS-based lateral flow assay for quantitative detection of C-reactive protein as an early bio-indicator of a radiation-induced inflammatory response in nonhuman primates. Analyst 143, 2115–2121. doi: 10.1039/c8an00160j
Sauget, M., Valot, B., Bertrand, X., and Hocquet, D. (2017). Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 25, 447–455. doi: 10.1016/j.tim.2016.12.006
Shin, J. H., Hong, J., Go, H., Park, J., Kong, M., Ryu, S., et al. (2018). Multiplexed detection of foodborne pathogens from contaminated lettuces using a handheld multistep lateral flow assay device. J. Agric. Food Chem. 66, 290–297. doi: 10.1021/acs.jafc.7b03582
Song, C., Li, J., Liu, J., and Liu, Q. (2016). Simple sensitive rapid detection of Escherichia coli O157:H7 in food samples by label-free immunofluorescence strip sensor. Talanta 15, 42–47. doi: 10.1016/j.talanta.2016.04.054
Song, D., Yang, R., Fang, S., Liu, Y., Long, F., and Zhu, A. (2018). SERS based aptasensor for ochratoxin A by combining Fe3O4@Au magnetic nanoparticles and Au-DTNB@Ag nanoprobes with multiple signal enhancement. Mikrochim. Acta 185:491. doi: 10.1007/s00604-018-3020-2
Suaifan, G., Alhogail, S., and Zourob, M. (2017). Paper-based magnetic nanoparticle-peptide probe for rapid and quantitative colorimetric detection of Escherichia coli O157:H7. Biosens. Bioelectron. 92, 702–708. doi: 10.1016/j.bios.2016.10.023
Switzer, J. A., Hill, J. C., Mahenderkar, N. K., and Liu, Y. C. (2016). Nanometer-thick gold on silicon as a proxy for single-crystal gold for the electrodeposition of epitaxial cuprous oxide thin films. ACS Appl. Mater. Interf. 8, 15828–15837. doi: 10.1021/acsami.6b04552
Wang, C., Li, M., Li, Q., Zhang, K., Wang, C., Xiao, R., et al. (2017). Polyethyleneimine-mediated seed growth approach for synthesis of silver-shell silica-core nanocomposites and their application as a versatile SERS platform. RSC Adv. 7, 13138–13148. doi: 10.1039/c6ra28629a
Wang, X., Choi, N., Cheng, Z., Ko, J., Chen, L., and Choo, J. (2017). Simultaneous detection of dual nucleic acids using a SERS-Based lateral flow assay biosensor. Anal. Chem. 89, 1163–1169. doi: 10.1021/acs.analchem.6b03536
Wang, C., Shen, W., Rong, Z., Liu, X., Gu, B., Xiao, R., et al. (2019a). Layer-by-layer assembly of magnetic-core dual quantum dot-shell nanocomposites for fluorescence lateral flow detection of bacteria. Nanoscale. 57:e0143-19. doi: 10.1039/c9nr08509b
Wang, C., Wang, C., Wang, X., Wang, K., Zhu, Y., Rong, Z., et al. (2019b). Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interf. 11, 19495–19505. doi: 10.1021/acsami.9b03920
Wang, C., Xiao, R., Wang, S., Yang, X., Bai, Z., Li, X., et al. (2019c). Magnetic quantum dot based lateral flow assay biosensor for multiplex and sensitive detection of protein toxins in food samples. Biosens. Bioelectron. 146:111754. doi: 10.1016/j.bios.2019.111754
Wang, Z., Yao, X., Wang, R., Ji, Y., Yue, T., Sun, J., et al. (2019a). Label-free strip sensor based on surface positively charged nitrogen-rich carbon nanoparticles for rapid detection of Salmonella enteritidis. Biosens. Bioelectron. 132, 360–367. doi: 10.1016/j.bios.2019.02.061
Wang, C., Shen, W., Rong, Z., Liu, X., Gu, B., Xiao, R., et al. (2020). Layer-by-layer assembly of magnetic-core dual quantum dot-shell nanocomposites for fluorescence lateral flow detection of bacteria. Nanoscale 12, 795–807. doi: 10.1039/C9NR08509B
Wang, C., Wang, J., Li, M., Qu, X., Zhang, K., Rong, Z., et al. (2016). A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles. Analyst 141, 6226–6238. doi: 10.1039/c6an01105e
Wang, C., Xu, J., Wang, J., Rong, Z., Li, P., Xiao, R., et al. (2015). Polyethylenimine-interlayered silver-shell magnetic-core microspheres as multifunctional SERS substrates. J. Mater. Chem. C 3, 8684–8693. doi: 10.1039/c5tc01839k
Wang, R., Kim, K., Choi, N., Wang, X., Lee, J., Jeon, J. H., et al. (2018a). Highly sensitive detection of high-risk bacterial pathogens using SERS-based lateral flow assay strips. Sens. Actuat. B Chem. 270, 72–79. doi: 10.1016/j.snb.2018.04.162
Wang, R., Zhang, W., Wang, P., and Su, X. (2018b). A paper-based competitive lateral flow immunoassay for multi beta-agonist residues by using a single monoclonal antibody labelled with red fluorescent nanoparticles. Mikrochim. Acta 185:191. doi: 10.1007/s00604-018-2730-9
Zhang, B., Yang, X., Liu, X., Li, J., Wang, C., and Wang, S. (2020). Polyethyleneimine-interlayered silica-core quantum dot-shell nanocomposites for sensitive detection of Salmonella typhimurium via a lateral flow immunoassay. RSC Adv. 10, 2483–2489. doi: 10.1039/c9ra09252h
Zhang, C., Wang, C., Xiao, R., Tang, L., Huang, J., Wu, D., et al. (2018). Sensitive and specific detection of clinical bacteria via vancomycin-modified Fe3O4@Au nanoparticles and aptamer-functionalized SERS tags. J. Mater. Chem. B 6, 3751–3761. doi: 10.1039/c8tb00504d
Zhang, D., Huang, L., Liu, B., Ni, H., Sun, L., Su, E., et al. (2018a). Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 106, 204–211. doi: 10.1016/j.bios.2018.01.062
Zhang, D., Huang, L., Liu, B., Su, E., Chen, H.-Y., Gu, Z., et al. (2018b). Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuat. B Chem. 277, 502–509. doi: 10.1016/j.snb.2018.09.044
Keywords: SERS, lateral flow immunoassay, E. coli O157:H7, SiO2/Au, quantitative detection
Citation: Shi L, Xu L, Xiao R, Zhou Z, Wang C, Wang S and Gu B (2020) Rapid, Quantitative, High-Sensitive Detection of Escherichia coli O157:H7 by Gold-Shell Silica-Core Nanospheres-Based Surface-Enhanced Raman Scattering Lateral Flow Immunoassay. Front. Microbiol. 11:596005. doi: 10.3389/fmicb.2020.596005
Received: 19 August 2020; Accepted: 15 October 2020;
Published: 06 November 2020.
Edited by:
Dario De Medici, National Institute of Health (ISS), ItalyReviewed by:
Rolf Dieter Joerger, University of Delaware, United StatesJodi Woan-Fei Law, Guangdong University of Technology, China
Copyright © 2020 Shi, Xu, Xiao, Zhou, Wang, Wang and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongwen Wang, d2FuZ2Nob25nd2VuMTk4N0AxMjYuY29t; Shengqi Wang, c3F3YW5nQGJtaS5hYy5jbg==; Bing Gu, Z2IyMDAzMTEyOUAxNjMuY29t
†These authors have contributed equally to this work
 Luoluo Shi1,2,3†
Luoluo Shi1,2,3† Rui Xiao
Rui Xiao Chongwen Wang
Chongwen Wang Bing Gu
Bing Gu