- 1Joint Faculty of Veterinary Medicine, Laboratory of Veterinary Public Health, Yamaguchi University, Yamaguchi, Japan
- 2Joint Faculty of Veterinary Medicine, Laboratory of National BioResource Project Paramecium, Yamaguchi University, Yamaguchi, Japan
- 3Department of Research Infrastructure, National BioResource Project of Japan Agency for Medical Research and Development, Tokyo, Japan
Legionella pneumophila, an intracellular human pathogen, establishes intracellular relationships with several protist hosts, including Paramecium caudatum. L. pneumophila can escape the normal digestion process and establish intracellular relationships in Paramecium. In this study, we identify new Paramecium strains that significantly reduce the number of L. pneumophila during infection. As a result, stable intracellular relationships between L. pneumophila and these Paramecium strains were not observed. These digestion-type Paramecium also showed high efficiency for Escherichia coli elimination compared to other strains of Paramecium. These results suggest that the digestion-type strains identified have high non-specific digestion activity. Although we evaluated the maturation process of Legionella-containing vacuoles (LCVs) in the Paramecium strains using LysoTracker, there were no discriminative changes in these LCVs compared to other Paramecium strains. Detailed understanding of the mechanisms of high digestion efficiency in these strains could be applied to water purification technologies and L. pneumophila elimination from environmental water.
Introduction
Legionella pneumophila is the causative agent of Legionnaires’ disease, which causes serious pneumonia in humans. Cases of human infection occur by inhalation of bacterial-contaminated aerosols, which are produced in artificial aquatic environments (Fliermans et al., 1981; Tachibana et al., 2013). L. pneumophila exists in natural aquatic environments and often establishes relationships with various species of protists (Boamah et al., 2017; Best and Abu Kwaik, 2018). Free-living amebae and ciliates, such as Tetrahymena, are well-known natural hosts of L. pneumophila (Rowbotham, 1980; Fields et al., 1984). It has also been reported that L. pneumophila’s relationships with protist hosts contribute to its survival in certain environments (Winiecka-Krusnell and Linder, 1999). We have previously reported that the freshwater ciliates Paramecium spp. could be natural reservoirs of L. pneumophila (Watanabe et al., 2016), and have described some mechanisms of establishment of the intracellular relationship between L. pneumophila and Paramecium (Nishida et al., 2018; Watanabe et al., 2018). However this relationship remains largely unknown.
Generally, Paramecium shows high phagocytic uptake activity, and internalized bacteria are usually digested immediately after feeding. However, L. pneumophila is able to avoid digestion and establish intracellular relationships inside Paramecium food vacuoles. Previously, we have identified bacterial genes involved in the accomplishment of intracellular survival and analyzed the mechanisms of action of the respective products (Watanabe et al., 2016, 2018; Nishida et al., 2018). We have shown that these genes contribute to resistance to digestive enzyme (Watanabe et al., 2018), or to avoiding egestion by Paramecium hosts (Nishida et al., 2018). These previous studies suggest that L. pneumophila utilizes these genes and obtains the appropriate niches to survive by isolating themselves from an environment with many stress factors. In addition, the high mobility and cell division rates of Paramecium could be huge advantages for the spread of L. pneumophila in natural environments. However, it is not clear what, if any, benefits Paramecium gains by harboring L. pneumophila. Although there is no evidence of mutualistic symbiosis between Paramecium and L. pneumophila, it is not uncommon to find that endosymbiotic bacteria provide some benefit to their protist hosts. For example, some strains of Paramecium develop salinity tolerance and heat shock tolerance when maintaining Holospora spp. within their nuclei as an obligate symbiont (Smurov and Fokin, 1998; Hori and Fujishima, 2003; Fujishima et al., 2005; Hori et al., 2008). Meanwhile, Holospora spp. are not essential symbionts for the host’s survival. Furthermore, maintaining these symbionts causes negative effects such as inhibition of growth, lethal cell burst, and functional failure of a micronucleus in some situations (Görtz and Fujishima, 1983; Fujishima and Kodama, 2012). It has also been reported that the endosymbionts Neochlamydia and Protochlamydia provide protection against L. pneumophila infection to their amebal hosts (Ishida et al., 2014; Maita et al., 2018; König et al., 2019). These findings indicate that there is a complicated relationship between hosts and parasites or symbionts.
While the stable intracellular relationship between L. pneumophila and Paramecium for at least 48 h was clearly observed in our infection assay, another type of relationship was also observed (Watanabe et al., 2016). Depending on the combination of strains of L. pneumophila and Paramecium, a cytotoxic relationship was observed; in some combinations, intracellular L. pneumophila killed the host Paramecium, thereby stable intracellular relationships were not observed (Watanabe et al., 2016). We analyzed the cytotoxic strains of L. pneumophila and identified the gene, L. pneumophila endosymbiosis-modulating factor A (lefA), involved in cytotoxicity toward Paramecium (Watanabe et al., 2016). Using lefA, L. pneumophila is able to disrupt unsuitable Paramecium hosts and escape from the intracellular lifestyle and revert to the free-living lifestyle. It seems it is important for L. pneumophila to achieve the opportunity to encounter suitable hosts. These our previous studies suggest that the relationship between L. pneumophila and Paramecium is complicated and diverse.
In this study, we hypothesized that in some cases the intracellular relationship could not be established because of digestion of Legionella by Paramecium, and attempted to identify the Paramecium strains showing such features (Supplementary Figure S1). We searched combinations of L. pneumophila and Paramecium strains which show the high digestion efficiency by infection tests using 60 strains of Paramecium. Furthermore, we analyzed the mechanisms of the Paramecium digestion of L. pneumophila by comparison to other Paramecium strains.
Materials and Methods
Bacterial Strains and Culture Conditions
Legionella pneumophila Philadelphia-1 (Phi-1) and Ofk308 were maintained as frozen glycerol stocks and cultured at 37°C on either N-(2-acetamido)-2-aminoethanesulfonic acid-buffered charcoal yeast extract (BCYE) agar or in the same medium without agar and charcoal (AYE). Escherichia coli DH5α were cultured in either lysogeny broth (LB) or on 1.5% LB agar. If necessary, chloramphenicol (10 μg/mL) was included in the media. Green florescent protein (GFP) expression in L. pneumophila was induced by adding isopropyl-β-D-thiogalactopyranoside (1 mM) to AYE.
Paramecium Strains and Culture Conditions
All Paramecium strains used in this study were provided by the Symbiosis Laboratory, Yamaguchi University, with support from the National Bioresource Project (NBRP)1. Paramecium strains were cultured and maintained as previously described (Watanabe et al., 2016). In brief, the culture medium used for Paramecium was 2.5% (w/v) fresh lettuce juice in Dryl’s solution (Dryl, 1959), inoculated with a non-pathogenic strain of Klebsiella pneumoniae. The cultivation of Paramecium strains was performed at 25°C. Cells at the stationary phase of growth (24 h after the last feeding) were used for all experiments.
Identification of L. pneumophila-Digesting Paramecium Strains
As a first screening assay, 60 Paramecium strains were infected with L. pneumophila Ofk308 in 1.5 mL tubes at an multiplicities of infection (MOI) of 10,000 according to a previously reported method (Watanabe et al., 2016) with slight modifications. Tubes were incubated at 25°C for 24 h. After incubation, samples were treated for 30 min at 50°C to purge the K. pneumoniae fed to the Paramecium. Ten microliters of each sample was spotted onto GVPC agar (BCYE agar supplemented with glycine; Wako, 3 mg/mL), containing vancomycin HCl (Wako, 1 μg/mL), polymyxin B (Wako, 80 IU/mL), and sulfate cycloheximide (Wako, 80 μg/mL). As a second screening assay, the Paramecium strains selected during the first screening assay were infected with L. pneumophila Ofk308 in the same manner as a first screening assay. Incubated samples were cultured on GVPC agar after serial dilution to determine the colony forming units (CFU). The reduction of bacterial loads were evaluated by classification into three classes: -; no reduction of bacterial load (50–100%), ±; low reduction of bacterial load (less than 50%), +; high reduction of bacterial load (less than 10%) compared to control (cultured Legionella in the medium without Paramecium).
Determination of the Size, Food Vacuole Formation, and Cell Division Rate of Paramecia
The sizes of Paramecium multimicronucleatum Y-2, YM-25, and Paramecium caudatum RB-1 were observed using a FluoView FV100 confocal laser scanning microscope (Olympus). To evaluate the capacity of food vacuole formation, 1,000 Paramecium cells were cultured with 4.55 × 107 particles of microspheres (Fluoresbrite® YG Carboxylate Microspheres 1.00 μm, Polysciences) for 30 min. Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Subsequently, samples were washed twice with PBS and applied to a glass slide which had 0.07 mm gap with a cover glass (Matsunami Glass). Food vacuoles in 20 Paramecium cells were counted under light microscopy (Olympus, BX51). Data were represented as averages based on triplicate samples from three identical experiments. To evaluate the cell division rate, the numbers of cells at the start of culture were aligned in 100 cells/mL and cultured without adding any bacteria. After incubation at 25°C for 24 h, to count the numbers of cells again, cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Subsequently, samples were washed twice with PBS and applied to a glass slide described above. With the number of cells at the start of culture defined as 100%, the cell numbers after 24 h are shown as relative values. Data were represented as averages based on triplicate samples from three identical experiments.
Evaluation of Digestion Ability of Intracellular Bacteria
The Paramecium strains Y-2, YM-25, and RB-1 were infected with L. pneumophila or E. coli at MOIs of 1,000, 10,000, and 100,000 in the same manner as in the screening assay described above. After incubation at 25°C for 24 h, CFU were determined by serial dilution on BCYE or LB agar containing chloramphenicol. Data were represented as averages based on triplicate samples from three identical experiments.
Determination of Mobility of Paramecia
The mobility (swimming velocity) of Y-2, YM-25, and RB-1 was measured according to a previously reported method (Fujishima et al., 2005) with slight modifications. Briefly, each live Paramecium strain was applied to a glass slide which had 0.07 mm gap with a cover glass (Matsunami Glass). This slide was placed on the stage of microscope and pictures of the cells under dark-field illumination were taken with an exposure time of 0.5 s. The lengths of the swimming loci of the cells were measured to calculate mean swimming velocities. The loci of 20 cells were measured in each. Data were represented as averages based on three identical experiments.
LysoTracker Analysis
Green florescent protein-expressing L. pneumophila were added to Y-2, YM-25, and RB-1 at an MOI of 10,000 and then incubated at 25°C for 30 min or 120 h. Samples were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Subsequently, samples were washed twice with PBS. Fluorescent images were obtained using a FluoView FV100 confocal laser scanning microscope. LysoTracker (Life Technologies) was used after fixation for 30 min at a concentration of 50 nM. Data were represented as averages based on triplicate samples from three identical experiments.
Statistical Analysis
Statistical analyzes were performed using Student’s t-test in Figures 2, 4, 5 or the Tukey–Kramer test in Figure 3. Data comprised the averages of three identical experiments. Standard deviations were represented as error bars.
Results
The Identification of Paramecium Strains Showing High Digestion Efficiency
First, we aimed to identify the strains of Paramecium that showed high digestion efficiency of L. pneumophila. Sixty strains of Paramecium were examined in the first screening infection assay (Figure 1), and 29 strains of Paramecium showed digestion of L. pneumophila Ofk308 to varying degrees. These 29 strains were selected for the second screening assay (Table 1). In the second screening infection assay, only two Paramecium strains, P. multimicronucleatum Y-2 and P. multimicronucleatum YM-25, caused remarkable reduction of the bacterial load (Table 1). Both Y-2 and YM-25 are larger in body size compared to other Paramecium strains, especially P. caudatum RB-1 (Figure 2A). RB-1 was employed as a control host strain in subsequent examinations, since RB-1 has been analyzed in our previous studies and its characteristics have been described in detail (Watanabe et al., 2016, 2018). Indeed, RB-1 showed no reduction of L. pneumophila in our screening infection assay (Table 1), corresponding to the results observed in our previous studies (Watanabe et al., 2016).
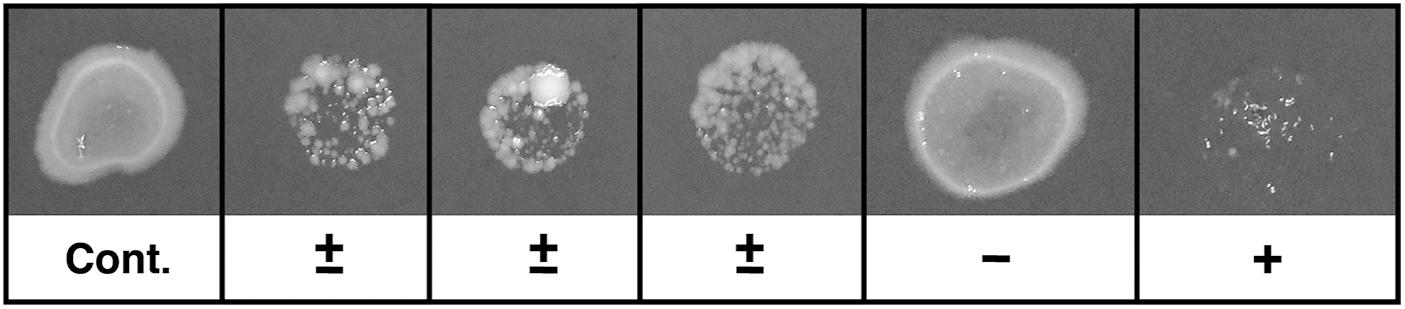
Figure 1. Typical example of screening test. Ten microliters of each sample were spotted onto GVPC agar. The agar plates were incubated at 37°C. The relative colony volumes were evaluated by comparison with the control (without Paramecium). Control and typical example of samples were evaluated as -; no reduction of bacterial load (50–100%), ±; low reduction of bacterial load (less than 50%), +; high reduction of bacterial load (less than 10%) compared to control.
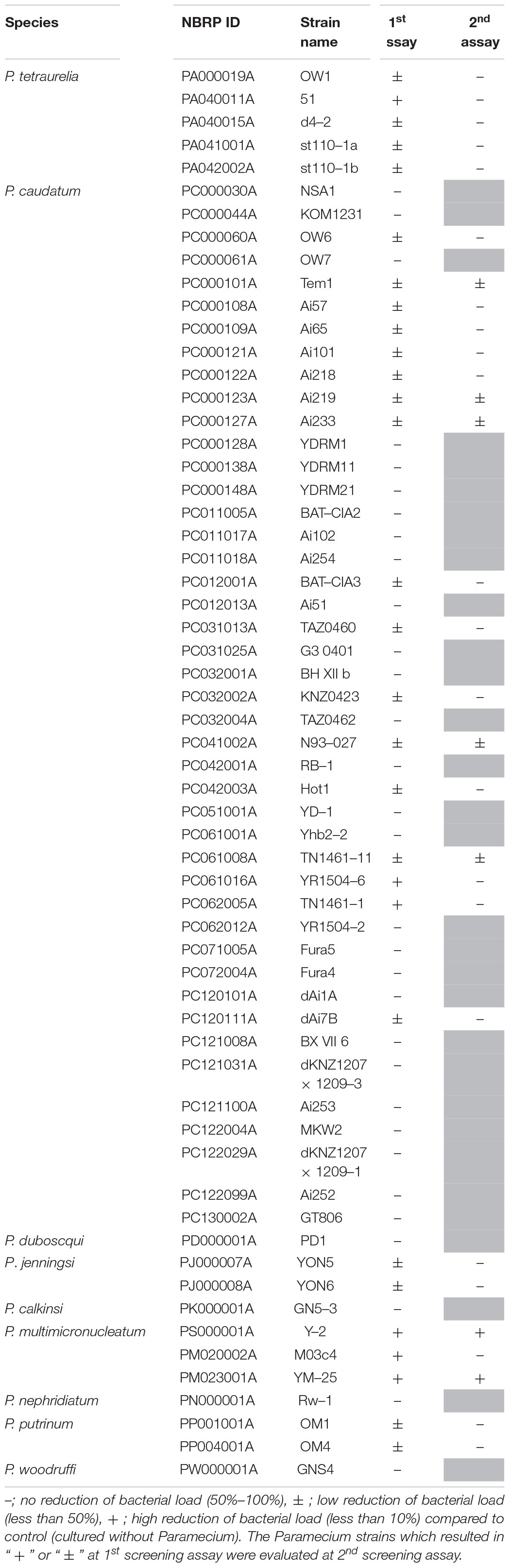
Table 1. The results of screening assay to identify Paramecium strains that show digestion for L. pneumophila Ofk308.
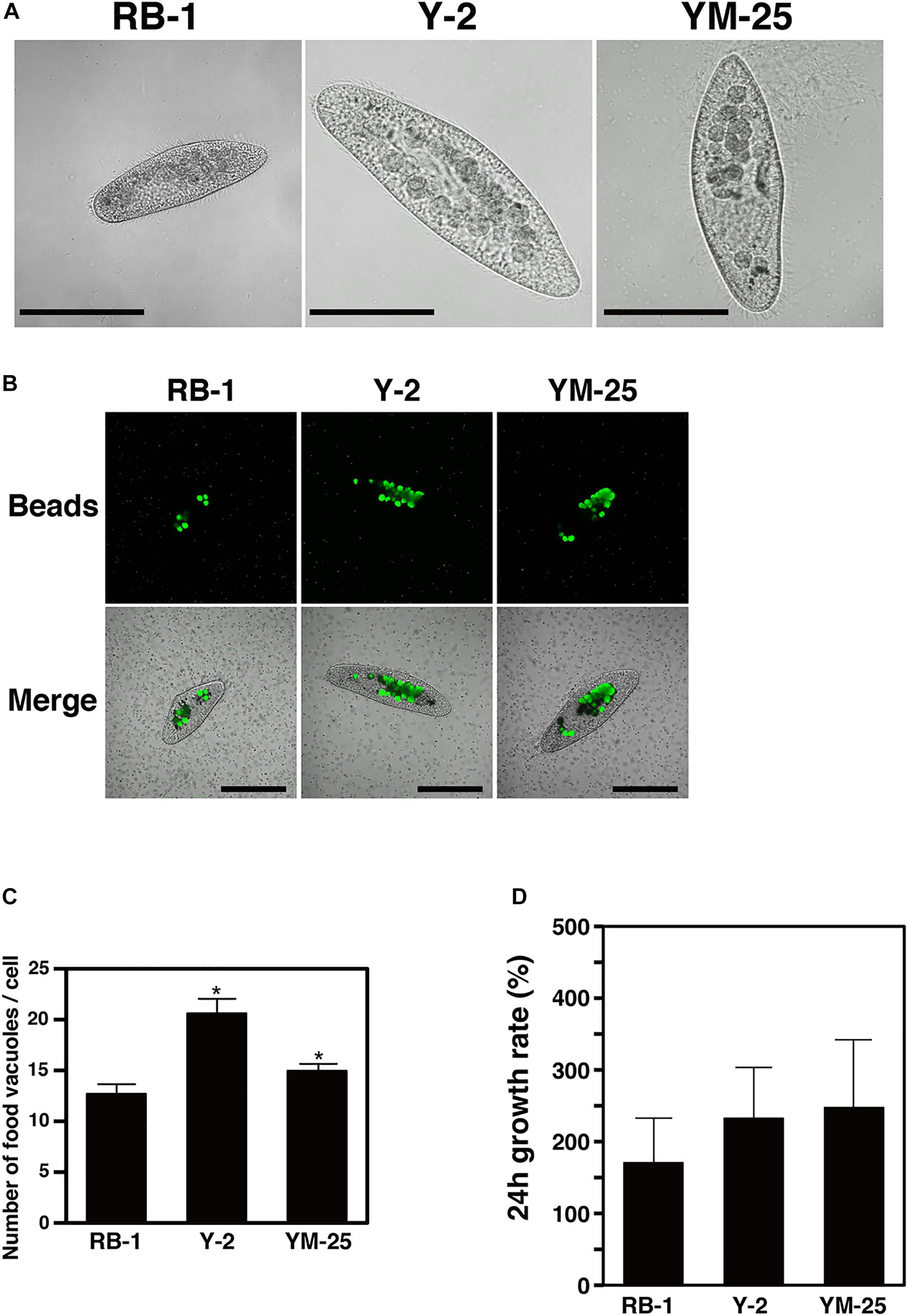
Figure 2. The size, food vacuole formation, and cell division rate of Y-2 and YM-25 strains. (A) Microscopic appearance of each strain. Scale bars represent 100 μm. (B,C) The average number of food vacuoles formed in each Paramecium strain. Paramecium were incubated with microbeads for 30 min and the number of food vacuoles in 20 Paramecium cells were counted. Data are the averages based on triplicate samples from three identical experiments and error bars represent standard deviations. Significant differences compared with RB-1 are indicated by asterisks (*P < 0.05). Scale bars represent 100 μm. (D) The cell division rate of each Paramecium strain. With the number of cells at the start of culture defined as 100%, the cell numbers after 24 h are shown as relative values. Data are the averages based on triplicate samples from three identical experiments and error bars represent standard deviations.
The numbers of food vacuoles that formed intracellularly within 30 min in strains Y-2 and YM-25 were higher than the numbers formed in RB-1 (Y-2, 20.6 ± 1.56/cell, P = 0.0038; YM-25, 15.0 ± 0.62/cell, P = 0.028, versus RB-1, 12.7 ± 0.90/cell) (Figures 2B,C). A different degree of the numbers of food vacuoles was also observed in between Y-2 and YM-25. Although we expected the cell division rate to be linked to the digestion efficiency, when we determined the 24-hour division rates, there were no significant differences (P > 0.05) between the cell division rates of Y-2, YM-25, and RB-1 (Y-2, 232.56 ± 70.9%; YM-25, 247.56 ± 94.4%; RB-1, 170.67 ± 62.1%) (Figure 2D). The mobility of strains was also determined and no significant differences were observed (Supplementary Figure S2).
Detailed Evaluation of Digestion Ability of Intracellular Bacteria
Next, we analyzed the number of intracellular bacteria in detail. Y-2, YM-25, and RB-1 were infected with L. pneumophila Ofk308 at various MOIs, and the CFU of L. pneumophila were counted 24 h after infection. At all MOIs, Y-2 and YM-25 showed larger bacterial reduction by digestion than RB-1. At an MOI of 100,000, Y-2 showed the largest digestion efficiency of the three strains (Figure 3A). We checked the digestion efficiency of other L. pneumophila strains to consider whether these results were confined to L. pneumophila Ofk308. Y-2, YM-25, and RB-1 were infected with L. pneumophila Phi-1 at various MOIs, and the Phi-1 CFU were counted 24 h after infection. Although the total number of Phi-1 CFU was different from that of Ofk308 in all Paramecium strains, there was no remarkable difference between the two strains of L. pneumophila (Figure 3B). We also performed the infection assay with E. coli, which is usually digested immediately by Paramecium. As a result, E. coli was also digested by Y-2 and YM-25 at a higher rate compared to the RB-1 strain (Figure 3C). These results indicate that Y-2 and YM-25 have a significantly higher digestive activity than RB-1, which is not specific to intracellular L. pneumophila.
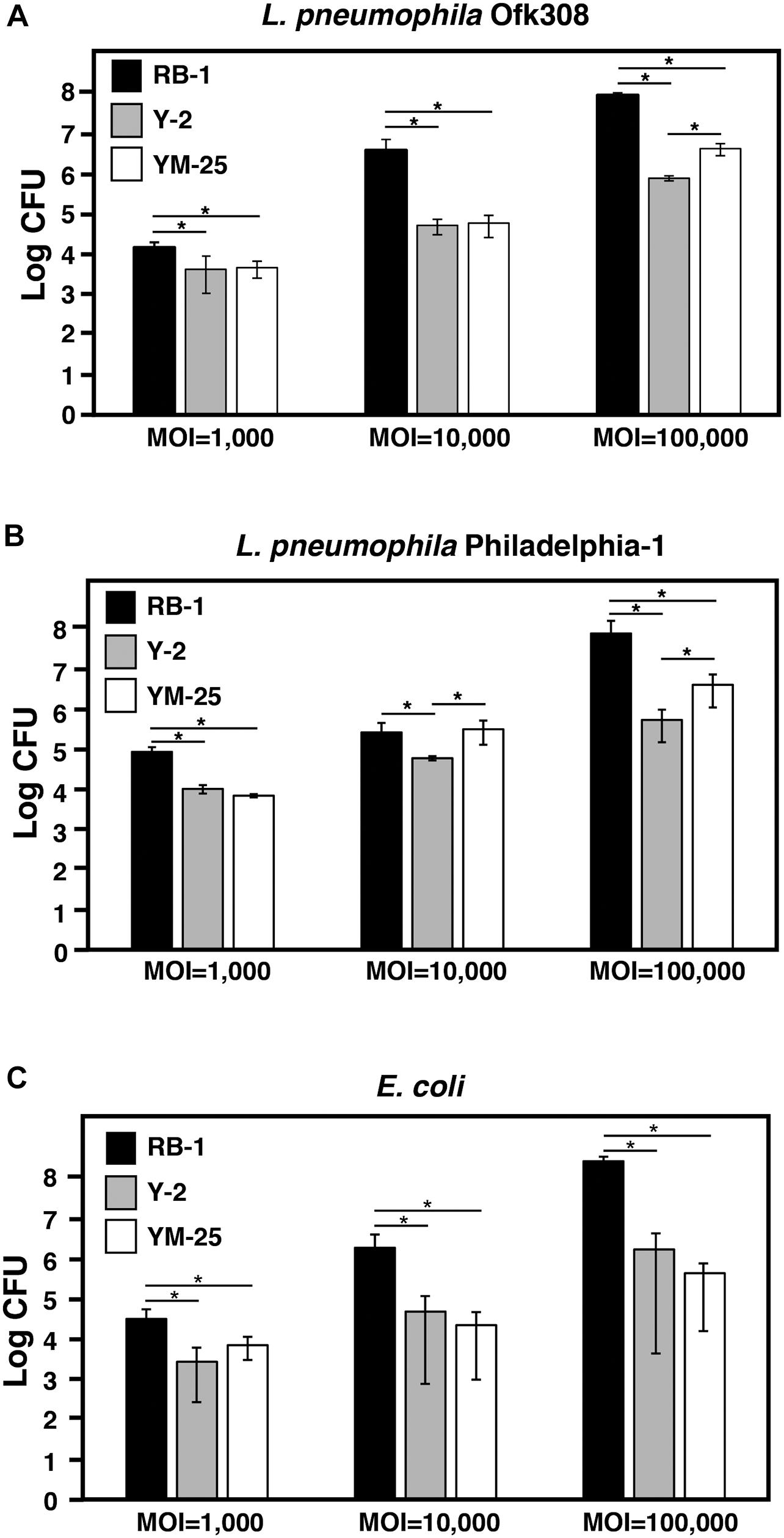
Figure 3. Evaluation of digestion of L. pneumophila or E. coli by RB-1, Y-2, and YM-25. RB-1, Y-2, and YM-25 were infected with L. pneumophila Ofk308 (A), Phi-1 (B), or E. coli (C) in 1.5 mL tubes at an MOI of 1,000, 10,000, or 100,000. Tubes were incubated at 25°C for 24 h. Samples were cultured on BCYE agar (A,B) or LB agar (C) after serial dilution to determine the CFU. Data are the averages based on triplicate samples from three identical experiments and error bars represent standard deviations. Significant differences are indicated by asterisks (*P < 0.05).
Analysis of Legionella-Containing Phagosomes
To understand whether Y-2 and YM-25 show differences in their properties of food vacuole maturation during bacterial digestion, we evaluated the acidification and forms of Legionella-containing vacuoles (LCVs) using LysoTracker and GFP-expressing L. pneumophila. In our previous studies, it was confirmed that the acidification of LCVs in RB-1 is not inhibited during infection of L. pneumophila Phi-1 which established a stable intracellular relationship (Watanabe et al., 2016). The strain shows resistance against acidification of LCVs and survives within the LCVs. In this study, the LCVs of Y-2 and YM-25 also showed acidification during Phi-1 infection as with that of RB-1 (Figures 4A,B). Furthermore, we have previously reported that the cytotoxic-type L. pneumophila strain, Ofk308, partially inhibits LCV acidification and enlarges LCVs in RB-1. This modification of LCVs by Ofk308 is likely to correlate with cytotoxicity toward Paramecium hosts since the mutant strain of Ofk308 which shows no cytotoxicity toward Paramecium hosts failed to modify the LCVs. In the Y-2 and YM-25 strains, Ofk308 did not produce modifications of LCVs, such as inhibition of acidification and enlargement, during infection (Figures 5A,B).
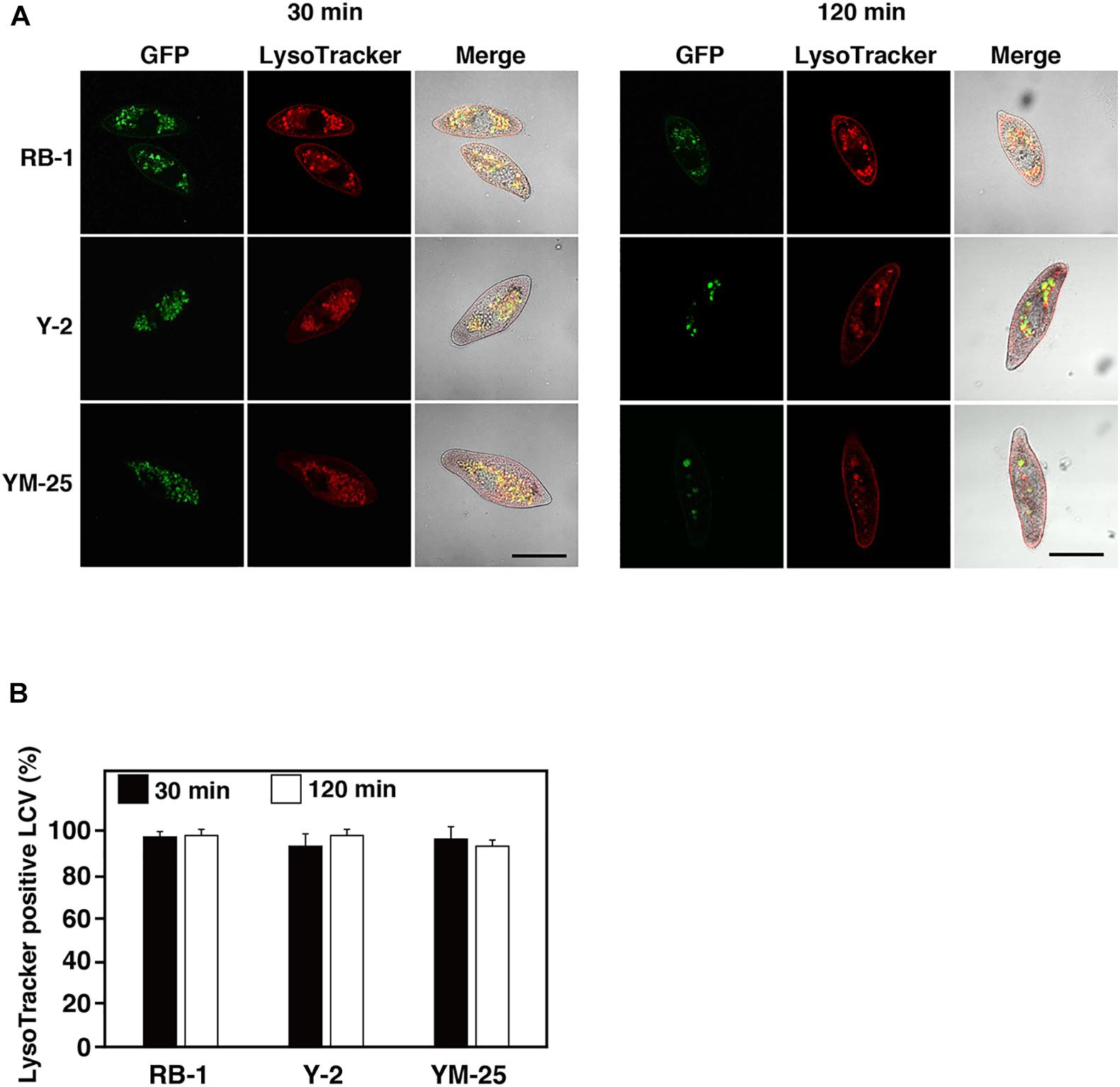
Figure 4. Maturation of Legionella-containing vacuoles (LCVs) in RB-1, Y-2, and YM-25 infected with L. pneumophila Phi-1. (A) LCV maturation 30 or 120 min after infection was evaluated using LysoTracker. L. pneumophila Phi-1 was added to RB-1, Y-2, and YM-25 at an MOI of 10,000. Scale bars represent 100 μm. (B) Relative LysoTracker-positive LCV percentages are shown, where the total of all LCVs is 100%. Data are the averages based on triplicate samples from three identical experiments and error bars represent standard deviations.
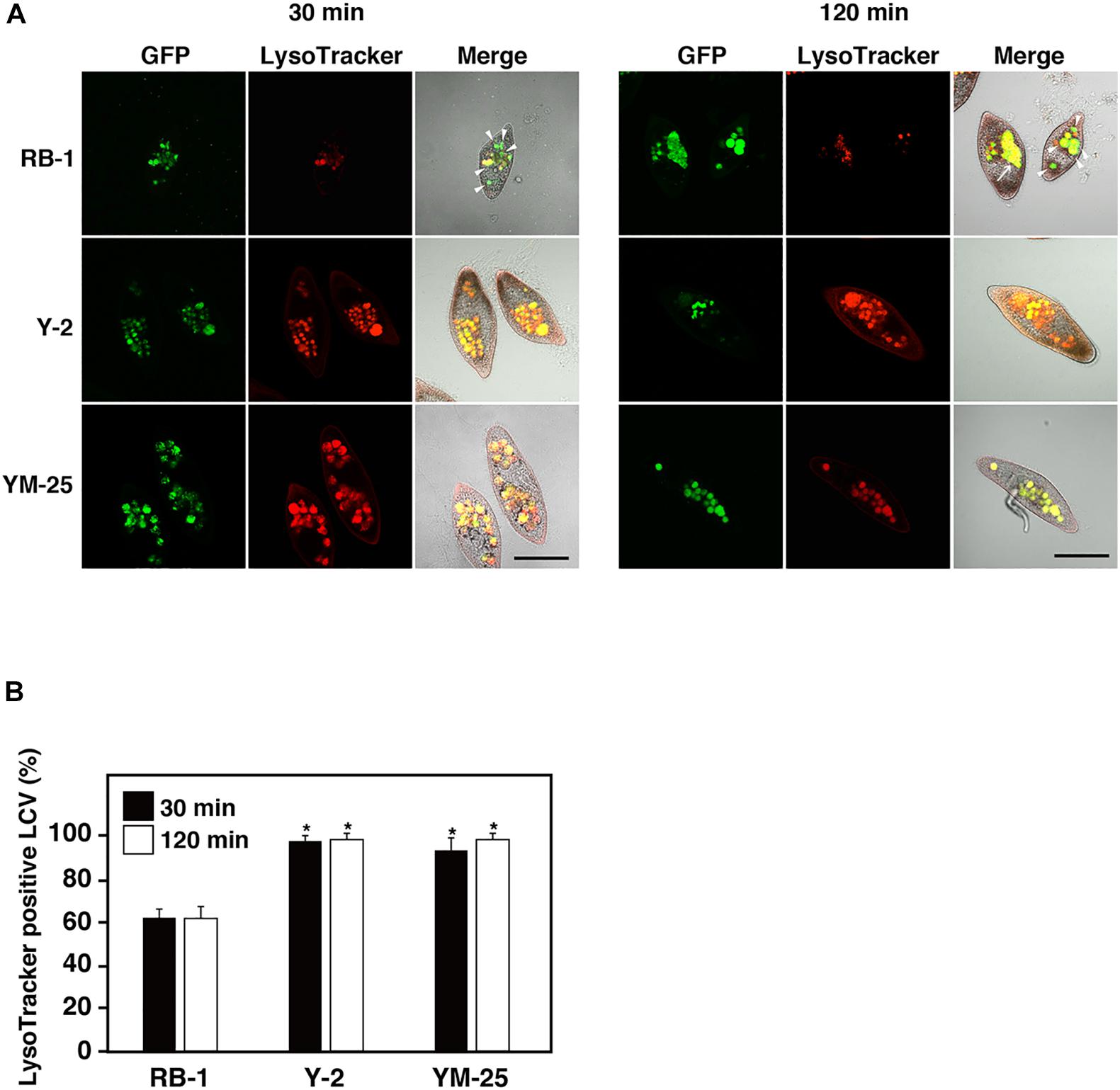
Figure 5. Maturation of LCVs in RB-1, Y-2, and YM-25 infected with L. pneumophila Ofk308. (A) LCV maturation 30 or 120 min after infection was evaluated using LysoTracker. L. pneumophila Ofk308 was added to RB-1, Y-2, and YM-25 at an MOI of 10,000. The arrowheads indicate LysoTracker-negative LCVs. Arrows indicate enlarged LCVs. Scale bars represent 100 μm. (B) Relative LysoTracker-positive LCVs percentages are shown, where the total of all LCVs is 100%. Data are the averages based on triplicate samples from three identical experiments and error bars represent standard deviations. Significant differences compared with RB-1 are indicated by asterisks (*P < 0.05).
Discussion
The relationship between L. pneumophila and protist hosts could be an important factor in human infection. Many types of protist hosts, including Paramecium, which are thought to be reservoirs of L. pneumophila (Boamah et al., 2017), may exist ubiquitously in the environment (Foissner, 2007; Oliverio et al., 2020). In addition to L. pneumophila, relationships between protist hosts and intracellular bacteria, such as Holospora in Paramecium and Mycobacterium or Neochlamydia in Acanthamoeba, have been reported (Amann et al., 1991; Fokin et al., 1996; Cirillo et al., 1997; Horn et al., 2000; Goy et al., 2007; Fujishima and Kodama, 2012). The ecology and mechanisms of these intracellular relationships remain largely unclear. Exhaustive research using numerous protist strains is necessary to reveal these perspectives. To this end, we employed many Paramecium strains obtained from the NBRP to analyze the intracellular relationships between Paramecium and L. pneumophila.
In this study, we performed screening infection tests using 60 strains of 10 species of Paramecium host. As a result, we identified two strains of Paramecium which fail to establish the intracellular relationship with L. pneumophila due to their potent digestion: P. multimicronucleatum Y-2 and P. multimicronucleatum YM-25. Since another P. multimicronucleatum strain, M03c4, did not show this digestion of L. pneumophila, it appears that there is no tendency of digestion within this species and that the ability of digestion may not depend on species. Their responses to intracellular L. pneumophila are likely to be largely attributable to each strain’s characteristics. These results were also observed in our previous study (Watanabe et al., 2016). The syngen and mating type of Y-2 has not yet been classified, thus the relationship or affinity between Y-2 and YM-25 is unknown. Y-2 and YM-25 display a seemingly larger body size compared to other Paramecium strains (Figure 2A) and, probably due to this feature, they are able to establish and maintain a large number of food vacuoles (Figures 2B,C). It is assumed that the first step of the digestion process is the uptake of extracellular components, including bacteria, and that active formation of food vacuoles becomes an advantage for the effective digestion of bacteria. However, considering that there was a difference in the level of formation of food vacuoles between in Y-2 and in YM-25, this aspect may contribute to the effective digestion partially. Be that as it may, since we have been able to obtain and examine only three strains of P. multimicronucleatum in this study, it is impossible to conclude this issue here. In the further study, a comparative analysis of each strain’s characteristics using more P. multimicronucleatum strains would reveal it.
During the initial stage of screening, we expected that the high digestion efficiency of Y-2 and YM-25 was specific to L. pneumophila; however, they showed same level of digestion for L. pneumophila Ofk308 and Phi-1 strains in a relatively short amount of time (Figures 3A,B). These results indicate that Y-2 and YM-25 are able to reduce intracellular bacteria due to inhibition of some mechanisms which are involved in establishment of the intracellular relationship or due to complicated systems which control the intracellular growth of L. pneumophila. In mammalian hosts, Legionella-replication permissive cells are also restricted. While human cell lines such as THP-1 and HeLa have been used for Legionella research (Daisy et al., 1981; Takemura et al., 2000), in mice, their capability as a host of Legionella depends on their strain (Yamamoto et al., 1992; Losick et al., 2009). Macrophages isolated from naip5 mutant A/J mice are recognized as a convenient Legionella host model (Yoshida et al., 1991; Wright et al., 2003). In these infection models, both the host and bacteria factors that are involved in intracellular growth have been identified and studied in detail (Yoshida et al., 1991; Lightfield et al., 2008; Vinzing et al., 2008). If such critical factors of Paramecium are identified in further analysis, they could explain the mechanisms involved in the high digestion efficiency for L. pneumophila in Y-2 and YM-25. Comparative genomic analysis of Y-2, YM-25, and RB-1 would also be helpful to reveal the differences in intracellular relationship with L. pneumophila.
Similar levels of digestion efficiency were also shown to E. coli in Y-2 and YM-25 (Figure 3C). These results suggest that both Y-2 and YM-25 have a high, non-specific digestion ability for a wide variety of bacteria. Although the strain of E. coli used in this study is not an intracellular bacterium and is digested promptly by Paramecium, the total bacterial number of E. coli was similar to that of L. pneumophila, especially for the Phi-1 strain. These results may be due to L. pneumophila’s failure to grow in culture medium for Paramecium at 25°C (Watanabe et al., 2018), while E. coli grows well in these conditions. The extracellular growth of E. coli may therefore mask the difference between L. pneumophila and E. coli.
We have previous confirmed that the acidification of LCVs occurs normally during L. pneumophila Phi-1 infection in Paramecium (Watanabe et al., 2016). The maturation of food vacuoles can be observed as LysoTracker-positive LCVs. It seems that L. pneumophila showing the stable intracellular relationship in Paramecium can withstand acidification and resist digestion, and then establish intracellular niches inside Paramecium (Watanabe et al., 2016, 2018). The process of establishing intracellular relationships in Paramecium is different from that in mammalian macrophages (Vogel and Isberg, 1999; Hubber and Roy, 2010). In addition, L. pneumophila inhibits acidification of LCVs and induces enlarged LCVs characteristically, and finally, results in the appearance of cytotoxic effects for Paramecium hosts (Watanabe et al., 2016). Analysis of the process of LCV maturation would help to reveal the relationships between L. pneumophila and Paramecium. In the present study, YM-25 and Y-2 did not show inhibition of LCV acidification or enlargement of LCVs (Figure 5A). Their LCV maturation processes were similar to those of stable intracellular relationship-type combinations such as P. caudatum RB-1 infected with L. pneumophila Phi-1. Our LysoTracker assay revealed no discriminative LCVs. YM-25 and Y-2 may have other mechanisms that can explain their high digestion efficiency for bacteria.
Conclusion
We have revealed the peculiar Paramecium strains which do not establish intracellular relationships with L. pneumophila. Our results imply that not all Paramaecium hosts are able to accommodate and establish a stable intracellular relationship with L. pneumophila. The mechanisms and factors which dictate the success of the stable intracellular relationship between Paramecium and Legionella should be studied in detail in further studies. However, taking in another perspective, these Paramecium hosts can eliminate harmful or pathogenic bacteria, including Legionella, from environmental water. It may be possible that these Paramecium can be utilized in industrial water clarification applications and play a part in a novel system to control waterborne diseases. We are looking into the applications that may be developed from the results shown in this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
KW and MW conceived and designed the experiments. KW, YH, MS, and MW performed the experiments. KW, TS, and MW analyzed the data. MT and MF contributed reagents, materials, and analysis tools. KW and MW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported partly by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (C) under Grant Number 19K06383 and by the Kurita Water and Environment Foundation under Grant Number 18A024 to KW.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.596731/full#supplementary-material
Supplementary Figure 1 | The assumed relationships between host Paramecium and L. pneumophila. We have previously identified a stable intracellular relationship; Legionella is able to avoid the digestion and establish the intracellular relationship inside Paramecium food vacuoles. We have also identified the case that the intracellular relationship is unstable; intracellular Legionella kills host Paramecium and fails to establish the intracellular relationship. We hypothesized that there is another case that the intracellular relationship is not established because of digestion of Legionella by Paramecium hosts.
Supplementary Figure 2 | The swimming velocities of RB-1, Y-2, and YM-25. (A) Photomicrographs of swimming loci of RB-1, Y-2, and YM-25. Scale bar represents 500 μm. (B) The swimming velocities of RB-1, Y-2, and YM-25. The loci of 20 cells were measured in each experiment. Data are averages based on three identical experiments and error bars represent standard deviations.
Footnotes
References
Amann, R., Springer, N., Ludwig, W., Gortz, H. D., and Schleifer, K. H. (1991). Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351, 161–164. doi: 10.1038/351161a0
Best, A., and Abu Kwaik, Y. (2018). Evolution of the arsenal of Legionella pneumophila effectors to modulate protist hosts. mBio 9:e01313-18. doi: 10.1128/mBio.01313-18
Boamah, D. K., Zhou, G., Ensminger, A. W., and O’Connor, T. J. (2017). From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front. Cell. Infect. Microbiol. 7:477. doi: 10.3389/fcimb.2017.00477
Cirillo, J. D., Falkow, S., Tompkins, L. S., and Bermudez, L. E. (1997). Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65, 3759–3767. doi: 10.1128/IAI.65.9.3759-3767.1997
Daisy, J. A., Benson, C. E., McKitrick, J., and Friedman, H. M. (1981). Intracellular replication of Legionella pneumophila. J. Infect. Dis. 143, 460–464. doi: 10.1093/infdis/143.3.460
Dryl, S. (1959). Antigenic transformation in Paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J. Protozool. 6:25.
Fields, B. S., Shotts, E. B. Jr., Feeley, J. C., Gorman, G. W., and Martin, W. T. (1984). Proliferation of Legionella pneumophila as an intracellular parasite of the ciliated protozoan Tetrahymena pyriformis. Appl. Environ. Microbiol. 47, 467–471.
Fliermans, C. B., Cherry, W. B., Orrison, L. H., Smith, S. J., Tison, D. L., and Pope, D. H. (1981). Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41, 9–16.
Foissner, W. (2007). Protist diversity and distribution: some basic considerations. Biodivers. Conserv. 17, 235–242. doi: 10.1007/s10531-007-9248-5
Fokin, S. I., Brigge, T., Brenner, J., and Görtz, H. D. (1996). Holospora species infected the nuclei of Paramecium appear to belong into two groups of bacteria. Eur. J. Protistol. 32, 19–24. doi: 10.1016/S0932-4739(96)80072-1
Fujishima, M., Kawai, M., and Yamamoto, R. (2005). Paramecium caudatum acquires heat-shock resistance in ciliary movement by infection with the endonuclear symbiotic bacterium Holospora obtusa. FEMS. Microbiol. Lett. 243, 101–105. doi: 10.1016/j.femsle.2004.11.053
Fujishima, M., and Kodama, Y. (2012). Endosymbionts in paramecium. Eur. J. Protistol. 48, 124–137. doi: 10.1016/j.ejop.2011.10.002
Görtz, H. D., and Fujishima, M. (1983). Conjugation and meiosis of Paramecium caudatum infected with the micronucleus-specific bacterium Holospora elegans. Eur. J. Cell Biol. 32, 86–91.
Goy, G., Thomas, V., Rimann, K., Jaton, K., Prod’hom, G., and Greub, G. (2007). The neff strain of Acanthamoeba castellanii, a tool for testing the virulence of Mycobacterium kansasii. Res. Microbiol. 158, 393–397. doi: 10.1016/j.resmic.2007.01.003
Hori, M., Fujii, K., and Fujishima, M. (2008). Micronucleus-specific bacterium Holospora elegans irreversibly enhances stress gene expression of the host Paramecium caudatum. J. Eukaryot. Microbiol. 55, 515–521. doi: 10.1111/j.1550-7408.2008.00352.x
Hori, M., and Fujishima, M. (2003). The endosymbiotic bacterium Holospora obtusa enhances heat-shock gene expression of the host Paramecium caudatum. J. Eukaryot. Microbiol. 50, 293–298. doi: 10.1111/j.1550-7408.2003.tb00137.x
Horn, M., Wagner, M., Muller, K. D., Schmid, E. N., Fritsche, T. R., Schleifer, K. H., et al. (2000). Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146(Pt 5), 1231–1239. doi: 10.1099/00221287-146-5-1231
Hubber, A., and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283. doi: 10.1146/annurev-cellbio-100109-104034
Ishida, K., Sekizuka, T., Hayashida, K., Matsuo, J., Takeuchi, F., Kuroda, M., et al. (2014). Amoebal endosymbiont Neochlamydia genome sequence illuminates the bacterial role in the defense of the host amoebae against Legionella pneumophila. PLoS One 9:e95166. doi: 10.1371/journal.pone.0095166
König, L., Wentrup, C., Schulz, F., Wascher, F., Escola, S., Swanson, M. S., et al. (2019). Symbiont-mediated defense against Legionella pneumophila in Amoebae. mBio 10:e00333-19. doi: 10.1128/mBio.00333-19
Lightfield, K. L., Persson, J., Brubaker, S. W., Witte, C. E., von Moltke, J., Dunipace, E. A., et al. (2008). Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178. doi: 10.1038/ni.1646
Losick, V. P., Stephan, K., Smirnova, I. I., Isberg, R. R., and Poltorak, A. (2009). A hemidominant Naip5 allele in mouse strain MOLF/Ei-derived macrophages restricts Legionella pneumophila intracellular growth. Infect. Immun. 77, 196–204. doi: 10.1128/iai.01011-08
Maita, C., Matsushita, M., Miyoshi, M., Okubo, T., Nakamura, S., Matsuo, J., et al. (2018). Amoebal endosymbiont Neochlamydia protects host amoebae against Legionella pneumophila infection by preventing Legionella entry. Microb. Infect. 20, 236–244. doi: 10.1016/j.micinf.2017.12.012
Nishida, T., Hara, N., Watanabe, K., Shimizu, T., Fujishima, M., and Watarai, M. (2018). Crucial role of Legionella pneumophila TolC in the inhibition of cellular trafficking in the protistan host Paramecium tetraurelia. Front. Microbiol. 9:800. doi: 10.3389/fmicb.2018.00800
Oliverio, A. M., Geisen, S., Delgado-Baquerizo, M., Maestre, F. T., Turner, B. L., and Fierer, N. (2020). The global-scale distributions of soil protists and their contributions to belowground systems. Sci. Adv. 6:eaax8787. doi: 10.1126/sciadv.aax8787
Rowbotham, T. J. (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183.
Smurov, A. O., and Fokin, S. I. (1998). Resistance of Paramecium caudatum infected with endonuclear bacteria Holospora against salinity impact. Proc. Zool. Ins. RAS 276, 175–178.
Tachibana, M., Nakamoto, M., Kimura, Y., Shimizu, T., and Watarai, M. (2013). Characterization of Legionella pneumophila isolated from environmental water and ashiyu foot spa. Biomed. Res. Int. 2013:514395. doi: 10.1155/2013/514395
Takemura, H., Yamamoto, H., Kunishima, H., Ikejima, H., Hara, T., Kanemitsu, K., et al. (2000). Evaluation of a human monocytic cell line THP-1 model for assay of the intracellular activities of antimicrobial agents against Legionella pneumophila. J. Antimicrob. Chemother. 46, 589–594. doi: 10.1093/jac/46.4.589
Vinzing, M., Eitel, J., Lippmann, J., Hocke, A. C., Zahlten, J., Slevogt, H., et al. (2008). NAIP and Ipaf control Legionella pneumophila replication in human cells. J. Immunol. 180, 6808–6815. doi: 10.4049/jimmunol.180.10.6808
Vogel, J. P., and Isberg, R. R. (1999). Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2, 30–34. doi: 10.1016/s1369-5274(99)80005-8
Watanabe, K., Nakao, R., Fujishima, M., Tachibana, M., Shimizu, T., and Watarai, M. (2016). Ciliate Paramecium is a natural reservoir of Legionella pneumophila. Sci. Rep. 6:24322. doi: 10.1038/srep24322
Watanabe, K., Suzuki, H., Nishida, T., Mishima, M., Tachibana, M., Fujishima, M., et al. (2018). Identification of novel Legionella genes required for endosymbiosis in Paramecium based on comparative genome analysis with Holospora spp. FEMS. Microbiol. Ecol. 94:fiy162. doi: 10.1093/femsec/fiy162
Winiecka-Krusnell, J., and Linder, E. (1999). Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scand. J. Infect. Dis. 31, 383–385. doi: 10.1080/00365549950163833
Wright, E. K., Goodart, S. A., Growney, J. D., Hadinoto, V., Endrizzi, M. G., Long, E. M., et al. (2003). Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr. Biol. 13, 27–36.
Yamamoto, Y., Klein, T. W., and Friedman, H. (1992). Genetic control of macrophage susceptibility to infection by Legionella pneumophila. FEMS Microbiol. Immunol. 4, 137–145. doi: 10.1111/j.1574-6968.1992.tb04980.x
Keywords: Legionella pneumophila, Paramecium, intracellular relationships, digestion, Legionella-containing vacuoles
Citation: Watanabe K, Higuchi Y, Shimmura M, Tachibana M, Fujishima M, Shimizu T and Watarai M (2020) Peculiar Paramecium Hosts Fail to Establish a Stable Intracellular Relationship With Legionella pneumophila. Front. Microbiol. 11:596731. doi: 10.3389/fmicb.2020.596731
Received: 20 August 2020; Accepted: 06 October 2020;
Published: 23 October 2020.
Edited by:
Amparo Latorre, University of Valencia, SpainCopyright © 2020 Watanabe, Higuchi, Shimmura, Tachibana, Fujishima, Shimizu and Watarai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahisa Watarai, d2F0YXJhaUB5YW1hZ3VjaGktdS5hYy5qcA==
 Kenta Watanabe
Kenta Watanabe Yusei Higuchi1
Yusei Higuchi1 Masahiro Fujishima
Masahiro Fujishima Takashi Shimizu
Takashi Shimizu Masahisa Watarai
Masahisa Watarai