- 1School of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
- 3Department of Food Science and Technology, Jinan University, Guangzhou, China
- 4College of Food Science, South China Agricultural University, Guangzhou, China
Antimicrobial resistance has become a major public health threat. Food-related Staphylococcus species have received much attention due to their multidrug resistance. The cfr gene associated with multidrug resistance has been consistently detected in food-derived Staphylococcus species. In this retrospective study, we examined the prevalence of cfr-positive Staphylococcus strains isolated from poultry meat in different geographical areas of China from 2011 to 2016. Two cfr-positive Staphylococcus delphini strains were identified from poultry meat in China. Comparative and whole-genome analyses were performed to characterize the genetic features and overall antimicrobial resistance genes in the two S. delphini isolates 245-1 and 2794-1. Whole-genome sequencing showed that they both harbored a novel 20,258-bp cfr-carrying Tn558 transposon derivative on their chromosomes. The Tn558 derivative harbors multiple antimicrobial resistance genes, including the transferable multiresistance gene cfr, chloramphenicol resistance gene fexA, aminoglycoside resistance genes aacA-aphD and aadD, and bleomycin resistance gene ble. Surprisingly, within the Tn558 derivative, an active unconventional circularizable structure containing various resistance genes and a copy of a direct repeat sequence was identified by two-step PCR. Furthermore, core genome phylogenetic analysis revealed that the cfr-positive S. delphini strains were most closely related to S. delphini 14S03313-1 isolated from Japan in 2017 and 14S03319-1 isolated from Switzerland in 2017. This study is the first report of S. delphini harboring a novel cfr-carrying Tn558 derivative isolated from retail food. This finding raises further concerns regarding the potential threat to food safety and public health safety. The occurrence and dissemination of similar cfr-carrying transposons from diverse Staphylococcus species need further surveillance.
Introduction
In recent years, resistance in bacteria has spread worldwide and presents a serious threat to human health. Linezolid is an oxazolidinone antibiotic and is considered as the last-resort antibiotic for the treatment of infections caused by multidrug-resistant (MDR) Gram-positive pathogens, including Staphylococcus species (Wilson et al., 2008). The antibiotic targets the P site in the peptidyl transferase center of the 23S ribosomal RNA of the 50S ribosomal subunit, acting on this target and blocking protein synthesis (Aoki et al., 2002). In fact, due to the synthetic nature of the drug, resistance to this antibiotic is rare. However, the cfr gene could mediate resistance to linezolid (Long et al., 2006). This gene encodes a methyltransferase that catalyzes the posttranscriptional methylation of adenosine at nucleotide position 2503 (Escherichia coli numbering) in 23S rRNA, which replaced the target of binding for linezolid (Corinna et al., 2005; Giessing et al., 2009; Anna et al., 2016). However, due to overlapping binding sites, cfr methylation also confers resistance to four other classes of antimicrobial agents and results in the PhLOPSA multiresistance phenotype, including resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A compounds (Long et al., 2006; Anna et al., 2016). Interestingly, cfr is often associated with erm, fexA, lsa(B), and tet(L), which can assist in co-selecting the cfr gene and in its spread (Shen et al., 2013; Mendes et al., 2014).
Generally, the cfr gene is often associated with mobile genetic elements (MGEs) (plasmids, integrative, and conjugative elements or transposons), which have great potential for dissemination (Shen et al., 2013). Tn558 is one of these bacterial transposons and was first identified on the plasmid pSCFS2 harboring the antimicrobial resistance gene (ARG) fexA from Staphylococcus lentus (Kehrenberg and Schwarz, 2005). Currently, this transposon is often harbored with cfr, and derivatives of Tn558 usually carry other acquired ARGs (Kehrenberg et al., 2007; Li et al., 2018). Therefore, this transposon plays an important role as vectors in the spread of transposon-borne ARGs.
Members of the genus Staphylococcus are widespread in nature and play vital roles in disease causation in humans and animals (McGavin and Heinrichs, 2012; Vrbovská et al., 2020). Among these species, Staphylococcus delphini is a pathogen that causes animal and human infections (Magleby et al., 2019; Ruiz-Ripa et al., 2019). It belongs to the Staphylococcus intermedius group and was first described in purulent skin lesions of dolphins (Varaldo et al., 1988). S. delphini is further separated into two subgroups, groups A and B, based on the phylogenetic analysis of the sodA, hsp60, and nuc genes and DNA–DNA hybridization (Sasaki et al., 2007). Although this staphylococcal species is poorly documented due to misidentification with S. intermedius, it has been isolated from humans and a wide range of diseased animals, including domestic pigeons, camels, horses, magpies, cinereous vultures, and mustelids, which serve as the natural hosts of S. delphini group A (Devriese et al., 2005; Sasaki et al., 2007; Sledge et al., 2010; Guardabassi et al., 2012; Sudagidan and Aydin, 2012; Stull et al., 2014; Magleby et al., 2019; Ruiz-Ripa et al., 2019).
In this retrospective study, we examined the prevalence of cfr-positive Staphylococcus isolates in poultry meat from 2011 to 2016. We determined the complete genome sequence of cfr-positive S. delphini and described their phenotypic and genotypic profiles. This is the first report of a Tn558 derivative-embedded cfr in S. delphini isolated from retail food.
Materials and Methods
Bacterial Isolation
From July 2011 to June 2016, we collected 4,300 retail food samples from supermarkets, fairs, and farmer markets, covering most of the provincial capitals of China (Supplementary Figure 1), and isolated 1,581 Staphylococcus strains, including Staphylococcus aureus, Staphylococcus argenteus, S. delphini, Staphylococcus epidermidis, and other staphylococci from 1,063 positive samples from all the sampling sites (Wu et al., 2018a,b). During the retrospective study of cfr-positive Staphylococcus species among these isolates, the cfr-positive strains 245-1 and 2794-1 were isolated from frozen duck wings in Guangzhou 2013 and frozen duck legs in Kunming 2014, respectively. The isolates were further identified as S. delphini by the MALDI-TOF/MS system (Bruker, Bremen, Germany) (Decristophoris et al., 2011).
PCR Detection
The presence of the resistance gene cfr was identified by PCR and Sanger sequencing (Kehrenberg et al., 2009). The presence of the two direct repeats (DRs) and circular intermediate translocatable units (TUs) was detected by PCR and inverse PCR (the primers and conditions are shown in Table 2). To minimize the detection of artificial products, a high-fidelity polymerase (PrimeSTAR GXL DNA Polymerase, Takara, Dalian, China) and an 8-min elongation step were used (Tansirichaiya et al., 2016). The amplicons obtained by PCR and inverse PCR experiments were subjected to Sanger sequencing.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) were determined using a standard broth dilution method according to the CLSI guidelines with S. aureus ATCC 29213 as a quality control strain (Weinstein and Clinical and Laboratory Standards Institute, 2018). The MICs for all of the following antimicrobials were determined: FFC, florfenicol; CHL, chloramphenicol; CLI, clindamycin; TIA, tiamulin; LZD, linezolid; K, kanamycin; ERY, erythromycin; FOX, cefoxitin; VAN, vancomycin; RIP, rifampicin; and DAP, daptomycin. The MIC breakpoints of each antibiotic, except florfenicol, were used as recommended by the current CLSI guidance (Weinstein and Clinical and Laboratory Standards Institute, 2018). For florfenicol, the results were interpreted according to the Veterinary CLSI (VET01-A5).
Whole-Genome Sequence and Analysis
Genomic DNA for whole-genome sequencing was extracted from the cfr-positive strains using a genomic extraction kit (Magen Biotech, Guangzhou, China) according to the manufacturer’s instructions. Whole-genome sequencing of the cfr-positive strains was performed using the Illumina HiSeq Xten platform (800-bp paired-end reads with 100-fold average coverage) and a PacBio Sequel II sequencing instrument (100-fold average read depth). The chromosome sequences were assembled into one scaffold using the software SMRT Portal, version 3.2.0. The genomic DNA annotation was performed in Prokka NCBI-BLASTP/BLASTX (Torsten, 2014). The single-nucleotide polymorphisms (SNPs) between strains 245-1 and 2794-1 were identified with Snippy software1.
The acquired antibiotic resistance genes were identified by ResFinder 3.02 and were further verified through a BLAST search against the Comprehensive Antibiotic Resistance Database (Ea et al., 2012). The genetic environment of the cfr gene was analyzed using BLAST3, followed by visualization of the comparative cfr multiresistance region (MRR) with Easyfig, v2.2.2 (Sullivan et al., 2011).
Phylogenetic Analysis
All publicly available draft genome sequences of S. delphini strains were acquired (22 strains with at least 50 × read coverage), and core SNP alignments were produced via Snippy using the S. delphini 8086 complete genome sequence (ASM30811v1) as a reference (see text footnote 1). The maximum-likelihood (ML) phylogenetic tree was constructed with RAxML-NG based on the ML optimality criterion (Kozlov et al., 2019). The locations of recombined regions on each branch were detected, and this tree was reconstructed by ClonalFrameML (Didelot and Wilson, 2015). FigTree, v1.4.3, was used to finalize the tree visualization (Morariu et al., 2008).
Nucleotide Sequence Accession Numbers
The complete genomic sequences of 245-1 and 2794-1 have been deposited in GenBank: 245-1 (GenBank ID: CP063368) and 2794-1 (GenBank ID: CP063367).
Results
Phenotypic Characteristics of cfr-Positive S. delphini
In this study, 245-1 and 2794-1 displayed the same MDR profiles. Antimicrobial susceptibility testing showed that these strains were resistant to chloramphenicol, florfenicol, tiamulin, clindamycin, and linezolid, exhibiting a high level of resistance to florfenicol (MIC = 256 μg/ml), chloramphenicol (MIC > 128 μg/ml), and tiamulin (MIC > 128 μg/ml). Moreover, the isolates were susceptible to vancomycin, daptomycin, and rifampicin (Table 1).
Basic Genomic Information for cfr-Positive S. delphini
To understand the molecular characteristics and resistomes of the two strains of S. delphini, they were submitted for whole-genome sequencing. Basic information related to the complete genome sequence of cfr-positive S. delphini is shown in Figure 1. The chromosomes of 245-1 and 2794-1 consisted of 2,708,646 bp with 2,486 predicted ORFs along with 102 RNAs and 2,707,963 bp with 2,486 predicted ORFs along with 102 RNAs, respectively. The genome analysis of the complete chromosomal DNA revealed that there were 166 variants between the chromosomes of 245-1 and 2794-1, and there were multiple ARGs located on their chromosomes, including fexA (conferring resistance to chloramphenicol), aacA-aphD and aadD (resistance to aminoglycosides), ble (resistance to bleomycin), and the multiresistance gene cfr (resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A).
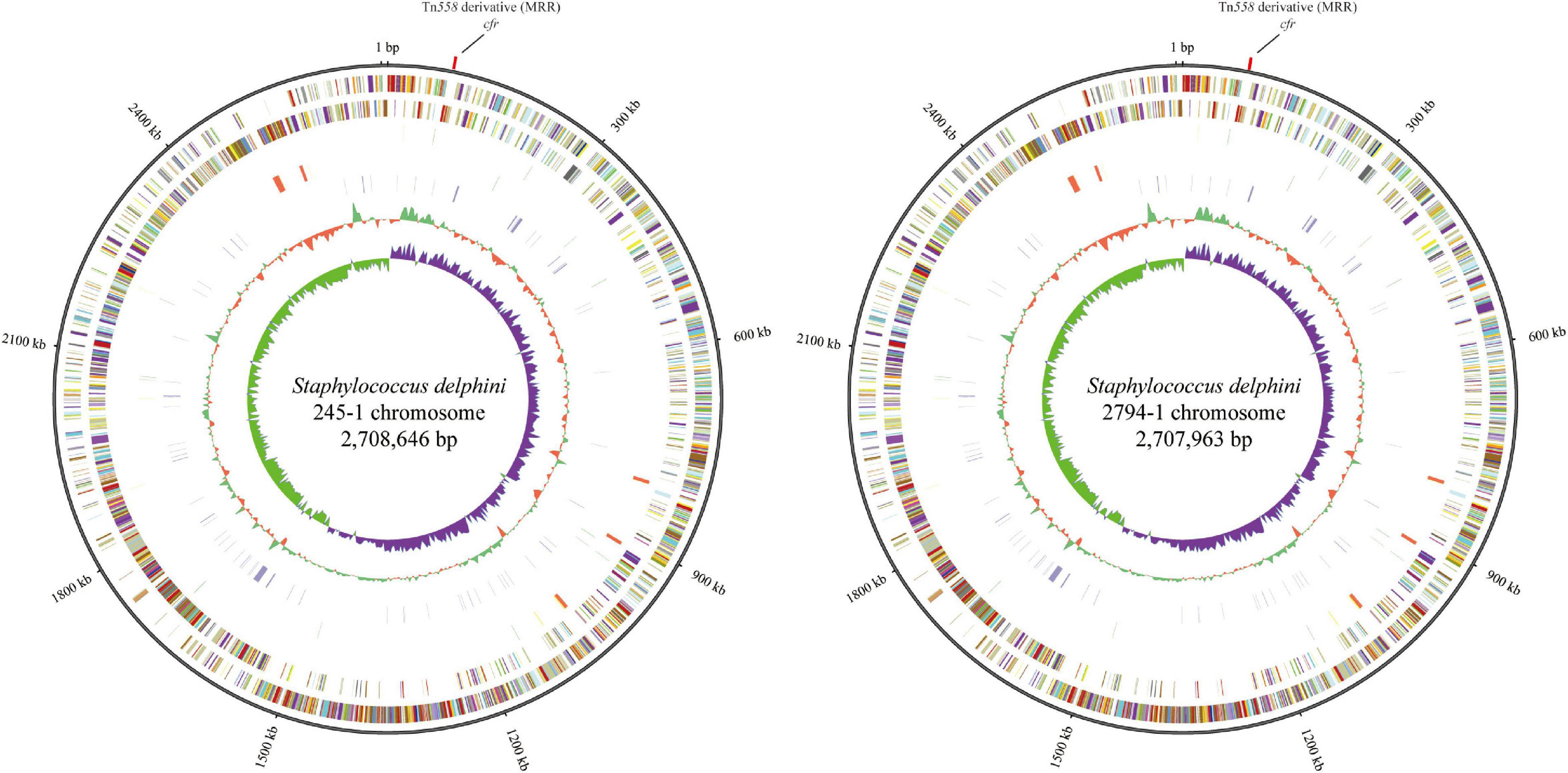
Figure 1. Circular representation of the cfr-positive Staphylococcus delphini 245-1 and 2794-1 genomes. From the outer to the inner circles in the chromosome circular map: slot 1 (ARGs) and slots 2–9 (slot 2, genome size; slot 3, forward strand gene, colored according to the cluster of orthologous groups classification; slot 4, reverse strand gene, colored according to the cluster of orthologous groups classification; slot 5, forward strand ncRNA; slot 6, reverse strand ncRNA; slot 7, repeat; slot 8, GC content; and slot 9, GC skew).
Core Genome Phylogenetic Analysis of cfr-Positive S. delphini
To further investigate the potential sources of cfr-positive S. delphini 245-1 and 2794-1, we performed a core genome phylogenetic analysis of all publicly available draft genome sequences of S. delphini strains. The phylogenetic analysis shows that 245-1 and 2794-1 are most closely related to S. delphini 14S03313-1 (GCA_002374125.1) isolated from Japan in 2017 and 14S03319-1 (GCA_002369675.1) isolated from Switzerland in 2017 (Figure 2). This phylogenetic analysis did not reveal the origin of 245-1 and 2794-1, indicating that the scarcity of genomic sequences may be the constraint, and further genomic sequencing is needed to identify the source of the cfr-positive strains.
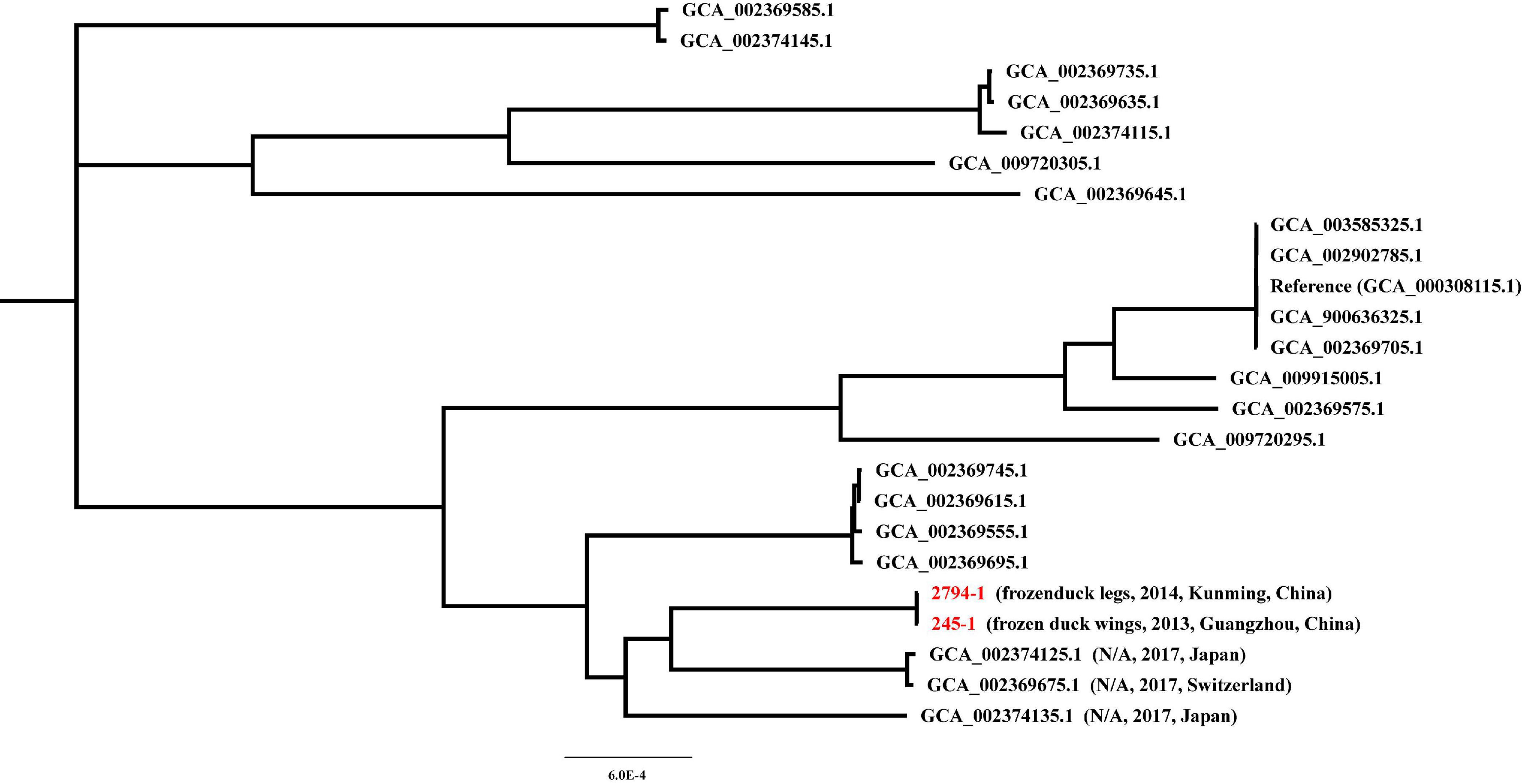
Figure 2. Maximum-likelihood (ML) core genome phylogeny of cfr-positive Staphylococcus delphini 245-1 and 2794-1 based on the ML method.
Genetic Environment of cfr Located on a Novel Tn558 Transposon Derivative
Genomic mining revealed that the cfr gene, along with four other ARGs, namely, fexA, aacA-aphD, aadD, and ble, was located on a 20,258-bp (62,847–83,104 nt on the chromosomes of 245-1 and 2794-1 in Figure 1) MRR on the chromosomes. Further BLAST analysis showed that the ARGs aacA-aphD, aadD, ble, and cfr were flanked by two DRs oriented in the same direction within the MRR and that the two DRs both belonged to Tn558 (Figure 3). The presence of the two DRs was further identified by PCR assays followed by sequencing of the amplicons (primers shown in Table 2). Both DRs were 1,326 bp in size, except for 18-bp exchanges in DRB compared to DRA. DRA contained partial fexA (430 bp) and orf138 sequences, while DRB comprised partial orf1 (430 bp) and orf2 sequences. Further analysis revealed that the single-nucleotide exchange TAG (orf138) → TAC (orf2) caused the termination codon to mutate to a Tyr codon, resulting in an extension of the open reading frame that transformed orf138 to orf2.
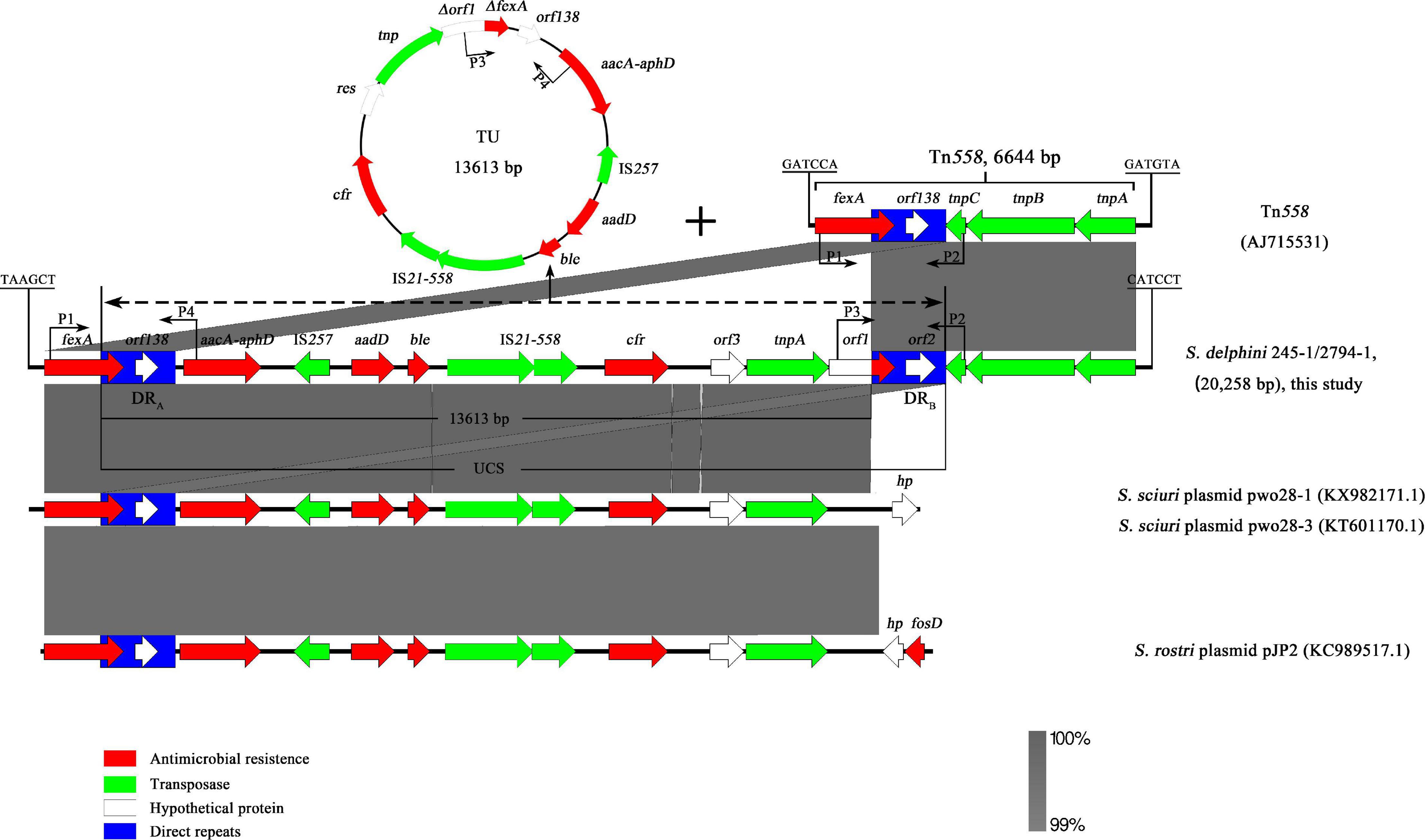
Figure 3. Schematic presentation of the environment of the cfr gene on chromosomes 245-1 and 2794-1 and the formation of translocatable units (TUs) mediated by direct repeats (DRs). The TUs derived from the region between DRA and DRB and the remaining structures after the excision of unconventional circularizable structures on chromosomes. Areas shaded in gray indicate homologous regions of ≥99% nucleotide sequence identity. Arrows indicate the orientations of the open reading frames, and the colors are based on their predicted gene functions. Frames with blue represent DRs. “Delta” represents a truncated gene. The figure is drawn to scale.
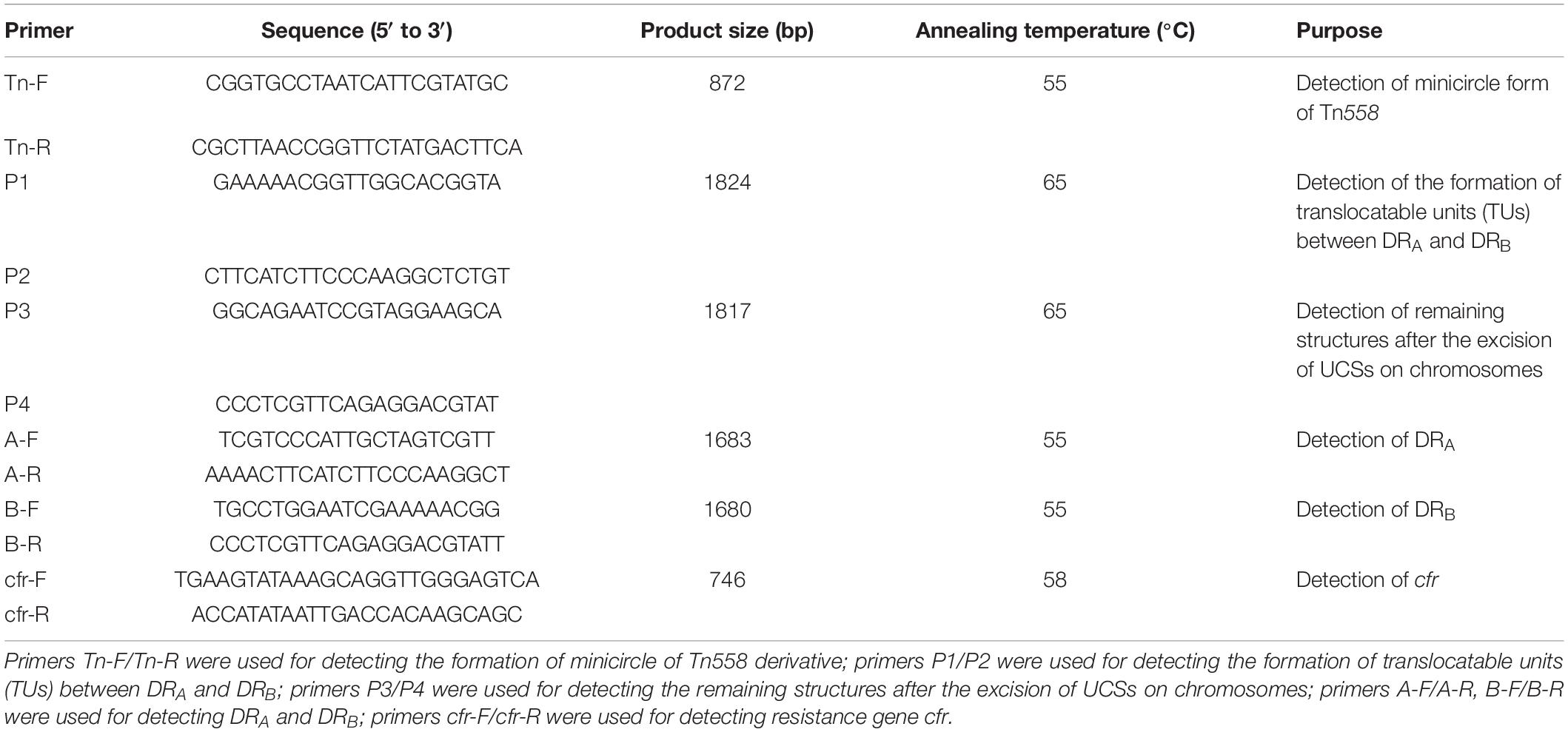
Table 2. Primers used for detecting antibiotics resistance genes, the circular forms and the structures not included in the corresponding region of the unconventional circularizable structures.
To further determine whether these unknown DRs in 245-1 and 2794-1 could mediate the formation of circular intermediate TUs, inverse PCR (P3, P4) was performed, followed by sequencing of the amplicons. Two identical PCR products (1,824 bp) were acquired from 245-1 and 2794-1, including a copy of DRB, orf138, and part of fexA, as determined by sequencing (Figure 3). The TUs (13,613 bp) resulted from the recombination between DRA and DRB, including multi-ARGs and one copy of DRA. The PCRs (P1, P2) containing one copy of DRB detected the remaining structures after the excision of unconventional circularizable structures (UCSs) on chromosomes, and the results were consistent with the inverse PCR results (Figure 3). Importantly, the remaining structures were Tn558. These results confirmed the excision and cyclization of the structure (Figure 3). Further BLAST analysis revealed that the left ΔfexA-UCS exhibited 99.88% nucleotide identity to the corresponding region of the plasmids pWo28-1 (KX982171.1) and pWo28-3 (KY601170.1) from Staphylococcus sciuri and the plasmid pJP2 (KC989517.1) from Staphylococcus rostri lacking DRB (Figure 3).
The sequence alignment analysis showed that the cfr MRR consisted of a Tn558 homologous region (6,644 bp) and a 13,613-bp region (Figure 3). This arrangement is a novel derivative of the Tn558 transposon. Compared to the fexA, orf138, tnpC, tnpB, and tnpA genes in Tn558 (Kehrenberg and Schwarz, 2005), a closer inspection of the Tn558 derivative showed that several nucleotide exchanges were identified in fexA (14 bp), tnpB (20 bp), and tnpA (14 bp), except for orf138. To further explain the genetic environment of the cfr MRR in this study, the plasmids pWo28-1 (KX982171.1) and pWo28-3 (KY601170.1) from S. sciuri and plasmid pJP2 (KC989517.1) from S. rostri are also shown in Figure 3. Analysis of the regions flanking the Tn558 derivative insertion in the chromosome identified a reading frame encoding a putative protein of 114 aa (62,844–62,846 and 83,105-83,443 nt on chromosomes 245-1 and 2794-1) that shared 98.54% nucleotide identity with a 148-aa DNA repair protein from Macrococcus canis (CP021059.1) (Gobeli et al., 2017). Additionally, a minicircle of Tn558, an indication of Tn558 having transposition activity, was identified via PCR (primers shown in Table 2) and sequencing of the derivative.
Discussion
Naturally, S. delphini is widely susceptible to clinically relevant classes of antibiotics. In a previous study from Denmark, among 55 S. delphini isolates recovered from mink, only some isolates were resistant to tetracycline (51%), penicillin (47%), and erythromycin (20%), whereas all the isolates tested susceptible to a vast majority of the antimicrobials assayed, including cefoxitin (Nikolaisen et al., 2017). In 2019, Magleby et al. also reported the first human case of S. delphini infection and found that the isolate exhibited low MIC values for all the antimicrobials assayed, including oxacillin (Magleby et al., 2019). Remarkably, the multiresistance gene cfr was shown to encode Cfr, an RNA methyltransferase that affects the binding of at least five chemically unrelated antimicrobial classes, namely, phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics, ultimately leading to a multidrug resistance phenotype (Long et al., 2006). Thus, the emergence and the global spread of the multiresistance gene cfr reduce the efficacy of a number of antibiotics in the control of Gram-positive bacteria. In this study, we identified the cfr gene in two food-related S. delphini strains. To the best of our knowledge, this study is the first report of the cfr gene existing in S. delphini. Furthermore, the cfr gene was located in an MRR with a number of antibiotic genes (fexA, aacA-aphD, aadD, and ble). The coexistence of cfr and other ARGs limits the choice of antibiotic therapy and may lead to the co-selection of these genes even without direct selection pressure, thereby increasing the retention and dissemination of these ARGs in Staphylococcus.
In this study, MRRs, including cfr and other ARGs, were confirmed as novel derivatives of the Tn558 transposon. Tn558 is a 6.6-kb bacterial transposon. It was first identified on the plasmid pSCFS2 harboring ARG fexA from S. lentus, and then numerous derivatives harboring numerous ARGs were found (Kehrenberg and Schwarz, 2005; Kehrenberg et al., 2007; Li et al., 2018). With a few exceptions, cfr is often harbored in the Tn558 transposon as coexisting with other ARGs, such as fexA, mecA, erm(A/B/C), tet(K/L/M), and drf(K/G) in the plasmids pSCFS3, pSCFS6, and pSCFS7 in previous studies (Witte and Cuny, 2011), but in this study, the derivative of the Tn558 transposon harbored cfr, fexA, aacA-aphD, aadD, and ble on the chromosomes. In addition, the additional DRs within the Tn558 derivative further confirm the particularity of this transposon. As previously reported for Tn558 derivatives, there are no inverted repeats at the ends and no duplication of the target sequence at the integration site of the Tn558 derivative. The typical 6-bp core sequences 5′-GATGTA-3′ at the left-end junction and 5′-GATCCA-3′ at the right-end junction were replaced by 5′-CATCCT-3′ and 5′-TAAGCT-3′ in the novel derivative. The disappearance of target duplication and the alteration of the typical core sequences may have occurred during the transposition process (Diaz-Aroca et al., 1987; Murphy, 1990). Moreover, the reading frame, including the insertion site of the Tn558 derivative, is similar to the protein containing the Tn558 site, and the excision of TUs in this Tn558 derivative could lead to the formation of Tn558, indicating that the DRA and DRB in this study may be involved in the evolution of Tn558 and that this derivative may be the ancestor of Tn558 (Kehrenberg and Schwarz, 2005). Although multiple conjugation assays failed, the presence of a circular Tn558 structure is indicative of the functional activity, suggesting that this novel Tn558 derivative is a transposable element and may mediate the transfer of the cfr gene in the process of transposition (Kehrenberg and Schwarz, 2005).
Generally, the cfr gene often coexists with other ARGs on transposons or plasmids and is often in close proximity to insertion sequences (ISs), such as IS21-558, IS256, or ISEnfa4, which play a crucial role in the mobility of cfr (Witte and Cuny, 2011; Wendlandt et al., 2015). These mobile structures have been detected among several Gram-positive bacteria, such as staphylococci, Enterococcus faecalis, Macrococcus caseolyticus, Jeotgalicoccus pinnipedialis, Bacillus spp., and Streptococcus suis, as well as in Gram-negative bacteria, such as E. coli and Proteus vulgaris (Shen et al., 2013). However, mobile structures can form UCSs (Palmieri et al., 2013). UCSs lack recombinase genes and can be excised in circular form due to the extensive DRs flanking the DNA segment undergoing excision (Locke et al., 2012; Palmieri et al., 2012, 2013). Thus, they are very important for the horizontal transmission of ARGs. In this study, the ARGs aacA-aphD, aadD, ble, and cfr, bracketed by DRs, formed a novel genetically mobile structure. The particular genetic structures identified by the analysis were referred to as UCSs. Two-step PCR results indicated that this structure can be looped out and excised from the chromosome, leading to the formation of Tn558 (Figure 3), which suggests that the DR is active and involved in the mobility of the Tn558-carried cfr gene in this study. Further BLAST analysis revealed that the left ΔfexA-UCS exhibited 99.88% nucleotide identity to the corresponding region of the plasmids pWo28-1 (KX982171.1) and pWo28-3 (KY601170.1) from S. sciuri and plasmid pJP2 (KC989517.1) from S. rostri lacking DRB (Figure 3). Therefore, the DRA and DRB in this study, similar to ISs, might facilitate the dissemination and accumulation of ARGs in Tn558 (Palmieri et al., 2013; Harmer et al., 2014). Of course, the functions of these two unknown DRs still need to be further studied and explored in the future.
Unconventional circularizable structures are widely distributed in Gram-negative and Gram-positive bacteria and play an important role in the dissemination of ARGs (Palmieri et al., 2013; Chanchaithong et al., 2019). The DRs in UCSs are usually long and are more than 100 times longer than the att sites functioning in traditional MGEs (Frost et al., 2005). The DRs may contain genes, such as erm(B), mef, (macrolide efflux), and ofr138 in this study, but they are not involved in transposition (Locke et al., 2012; Hao et al., 2019). The exact mechanism of mobilization has not been determined, although hypotheses have been proposed (Azpiroz et al., 2011). This transfer mechanism may be similar to that of IS26 via site-specific recombination, including a multistep process that requires the formation of a TU, precise excision of the TU, and integration targeting the preexisting DR (Harmer et al., 2014; Harmer and Hall, 2015). The endogenous instability of UCSs endows the encompassed niche adaptation determinants with the ability to be transferred. Moreover, they are often carried by MGEs, which prompts the updating of MGEs (such as the derivative of Tn558) and further accelerates the spread of UCSs. Furthermore, the presence of DRs on this novel cfr-carrying Tn558 derivative may accelerate the spread and persistence of ARGs among staphylococci and exacerbate the threat of superbugs, such as methicillin-resistant S. aureus. The proliferation of the transferable ARG cfr kidnapped by transposons or other MGEs has impaired the efficiency of oxazolidiones in clinical settings and threatens public health (Li et al., 2018).
Conclusion
To the best of our knowledge, this study is the first report of S. delphini harboring a novel cfr-carrying Tn558 derivative. The constant occurrence of the cfr gene in new staphylococcal host species underlines its strong transmissibility and wide distribution. This finding raises further concerns regarding the potential threat to food safety and public health safety. The occurrence and the dissemination of similar cfr-carrying transposons from diverse Staphylococcus species need further surveillance.
Data Availability Statement
The complete genomic sequences of 245-1 and 2794-1 have been deposited in GenBank: 245-1 (GenBank ID: CP063368) and 2794-1 (GenBank ID: CP063367).
Author Contributions
QW, JZ, SW, and TL conceived and designed the experiments. FZ, JH, and JD performed the experiments. FZ, SW, and RY analyzed the data. YD, LX, MC, and JW contributed reagents, materials, and analysis tools. FZ, SW, and JW contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We would like to acknowledge the financial support provided by the National Key Research and Development Program of China (2019YFC1606300), the National Natural Science Foundation of China (No. 31801657), the Key Research and Development Program of Guangdong Province (2018B020205001), and GDAS’ Special Project of Science and Technology Development (2020GDASYL-20200401002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JS declared a shared affiliation with one of the authors JW to the handling editor at the time of review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.598990/full#supplementary-material
Supplementary Figure 1 | Sample collection locations for this study in China.
Footnotes
- ^ https://github.com/tseemann/snippy
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Anna, S., Lazaris, A., Kinnevey, P., Brennan, O. M., Brennan, G. I, O’Connell, B., et al. (2016). First report of cfr-carrying plasmids in the pandemic sequence type 22 methicillin-resistant Staphylococcus aureus Staphylococcal cassette chromosome mec type IV clone. Antimicrob. Agents Chemother. 60, 3007–3015. doi: 10.1128/aac.02949-15
Aoki, H., Ke, L. Z., Poppe, S. M., Poel, T. J., Weaver, E. A., Gadwood, R. C., et al. (2002). Oxazolidinone antibiotics target the P site on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 46, 1080–1085. doi: 10.1128/aac.46.4.1080-1085.2002
Azpiroz, M. F., Bascuas, T., and Lavina, M. (2011). Microcin H47 system: an Escherichia coli small genomic island with novel features. PLoS One 6:e0026179. doi: 10.1371/journal.pone.0026179
Chanchaithong, P., Perreten, V., and Schwendener, S. (2019). Macrococcus canis contains recombinogenic methicillin resistance elements and the mecB plasmid found in Staphylococcus aureus. J. Antimicrob. Chemother. 74, 2531–2536. doi: 10.1093/jac/dkz260
Corinna, K., Schwarz, S., Jacobsen, L., Hansen, L. H., and Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x
Decristophoris, P., Fasola, A., Benagli, C., Tonolla, M., and Petrini, O. (2011). Identification of Staphylococcus intermedius Group by MALDI-TOF MS. Syst. Appl. Microbiol. 34, 45–51. doi: 10.1016/j.syapm.2010.11.004
Devriese, L. A., Vancanneyt, M., Baele, M., Vaneechoutte, M., De Graef, E., Snauwaert, C., et al. (2005). Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 55(Pt 4), 1569–1573. doi: 10.1099/ijs.0.63413-63410
Diaz-Aroca, E., Mendiola, M. V., Zabala, J. C., and de la Cruz, F. (1987). Transposition of IS91 does not generate a target duplication. J. Bacteriol. Bacteriol. 169, 442–443.
Didelot, X., and Wilson, D. J. (2015). ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11:e1004041. doi: 10.1371/journal.pcbi.1004041
Ea, Z., Henrik, H., Salvatore, C., Martin, V., Simon, R., Ole, L., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644.
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Giessing, A. M., Jensen, S. S., Rasmussen, A., Hansen, L. H., Gondela, A., Long, K., et al. (2009). Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15, 327–336. doi: 10.1261/rna.1371409
Gobeli, S., Cotting, K., Gomez Sanz, E., Collaud, A., Thomann, A., Brodard, I., et al. (2017). Macrococcus canis sp. nov., a skin bacterium associated with infections in dogs. Int. J. Syst. Evol. Microbiol. 67, 621–626. doi: 10.1099/ijsem.0.001673
Guardabassi, L., Schmidt, K. R., Petersen, T. S., Espinosa-Gongora, C., Moodley, A., Agerso, Y., et al. (2012). Mustelidae are natural hosts of Staphylococcus delphini group A. Vet. Microbiol. 159, 351–353. doi: 10.1016/j.vetmic.2012.04.004
Hao, W. B., Shan, X. X., Li, D. X., Schwarz, S., Zhang, S. M., Li, X. S., et al. (2019). Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J. Antimicrob. Chemother. 74, 1771–1775. doi: 10.1093/jac/dkz109
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-1815
Harmer, C. J., Moran, R. A., Hall, R. M., and Bush, K. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-1814
Kehrenberg, C., Aarestrup, F. M., and Schwarz, S. (2007). IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51, 483–487. doi: 10.1128/AAC.01340-06-1346
Kehrenberg, C., Cuny, C., Strommenger, B., Schwarz, S., and Witte, W. (2009). Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53, 779–781. doi: 10.1128/aac.01376-08-1378
Kehrenberg, C., and Schwarz, S. (2005). Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Chemother. 49, 813–815. doi: 10.1128/aac.49.2.813-815.2005
Kozlov, A., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Li, S.-M., Zhou, Y.-F., Li, L., Fang, L.-X., Duan, J.-H., Liu, F.-R., et al. (2018). Characterization of the multi-drug resistance gene cfr in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front. Microbiol. 9:2925. doi: 10.3389/fmicb.2018.02925
Locke, J. B., Rahawi, S., Lamarre, J., Mankin, A. S., and Shaw, K. J. (2012). Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 56, 332–340. doi: 10.1128/AAC.05420-5411
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06-136
Magleby, R., Bemis, D. A., Kim, D., Carroll, K. C., Castanheira, M., Kania, S. A., et al. (2019). First reported human isolation of Staphylococcus delphini. Diagn. Microbiol. Infect. Dis. Microbiol. Infect. Dis 94, 274–276. doi: 10.1016/j.diagmicrobio.2019.01.014
McGavin, M. J., and Heinrichs, D. E. (2012). The staphylococci and staphylococcal pathogenesis. Front. Cell. Infect. Microbiol. 2:66. doi: 10.3389/fcimb.2012.00066
Mendes, R. E., Deshpande, L. M., and Jones, R. N. (2014). Linezolid update: Stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updates 17, 1–12.
Morariu, V., Srinivasan, B., Raykar, V., Duraiswami, R., and Davis, L. (2008). “Automatic online tuning for fast Gaussian summation,” in Proceedings of the Twenty-Second Annual Conference on Neural Information Processing Systems, Vancouver.
Murphy, E. (1990). “Properties of the site-specific transposable element Tn554,” in Molecular Biology of the Staphylococci, ed. R. P. Novick (Hoboken, NJ: Wiley).
Nikolaisen, N. K., Lassen, D. C. K. V., Chriél, M., Larsen, G., Jensen, V. F. K. R., and Pedersen, K. (2017). Antimicrobial resistance among pathogenic bacteria from mink (Neovison vison) in Denmark. Acta Vet. Scand. 59:60.
Palmieri, C., Mingoia, M., Massidda, O., Giovanetti, E., and Varaldo, P. E. (2012). Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm(B)- and aphA3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob. Agents Chemother. 56, 5994–5997. doi: 10.1128/aac.01487-1412
Palmieri, C., Mingoia, M., and Varaldo, P. E. (2013). Unconventional circularizable bacterial genetic structures carrying antibiotic resistance determinants. Antimicrob. Agents Chemother. 57, 2440–2441.
Ruiz-Ripa, L., Gomez, P., Alonso, C. A., Camacho, M. C., de la Puente, J., Fernandez-Fernandez, R., et al. (2019). Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in magpies and cinereous vultures in Spain. Microb. Ecol. Ecol. 78, 409–415. doi: 10.1007/s00248-019-01328-1324
Sasaki, T., Kikuchi, K., Tanaka, Y., Takahashi, N., Kamata, S., and Hiramatsu, K. (2007). Reclassification of phenotypically identified Staphylococcus intermedius Strains. J. Clin. Microbiol. 45, 2770–2778. doi: 10.1128/JCM.00360-07-367
Shen, J., Wang, Y., and Schwarz, S. (2013). Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092
Sledge, D. G., Danieu, P. K., Bolin, C. A., Bolin, S. R., Lim, A., Anderson, B. C., et al. (2010). Outbreak of neonatal diarrhea in farmed mink Kits (Mustella vison) associated with enterotoxigenic Staphylococcus delphini. Vet. Pathol. 47, 751–757. doi: 10.1177/0300985810364514
Stull, J. W., Slavic, D., Rousseau, J., and Weese, J. S. (2014). Staphylococcus delphini and methicillin-resistant S. pseudintermedius in Horses, Canada. Emerg. Infect. Dis. 20, 485–487. doi: 10.3201/eid2003.130139
Sudagidan, M., and Aydin, A. (2012). Virulence properties of Staphylococcus delphini strains isolated from domestic pigeons. Medycyna Weterynaryjna-Vet. Med. Sci. Pract. 68, 231–236.
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010.
Tansirichaiya, S., Mullany, P., and Roberts, A. (2016). PCR-based detection of composite transposons and translocatable units from oral metagenomic DNA. FEMS Microbiol. Lett. 363:fnw195. doi: 10.1093/femsle/fnw195
Varaldo, P. E., Kilpper-Bälz, R., Biavasco, F., Satta, G., and Schleifer, K. H. (1988). Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int. J. Syst. Evol. Microbiol. 38, 436–439. doi: 10.1099/00207713-38-4-436
Vrbovská, V., Sedláček, I., and Zeman, M. (2020). Characterization of Staphylococcus intermedius group isolates associated with animals from antarctica and emended description of Staphylococcus delphini. Microorganisms 8:204. doi: 10.3390/microorganisms8020204
Weinstein, M. P., and Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing. Geneva: Clinical and Laboratory Standards Institute.
Wendlandt, S., Shen, J., Kadlec, K., Wang, Y., Li, B., Zhang, W.-J., et al. (2015). Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol. 23, 44–54. doi: 10.1016/j.tim.2014.10.002
Wilson, D. N., Schluenzen, F., Harms, J. M., Starosta, A. L., Connell, S. R., and Fucini, P. (2008). The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U.S.A. 105, 13339–13344. doi: 10.1073/pnas.0804276105
Witte, W., and Cuny, C. (2011). Emergence and spread of cfr-mediated multiresistance in staphylococci: an interdisciplinary challenge. Future Microbiol. 6, 925–931. doi: 10.2217/fmb.11.69
Wu, S., Huang, J., Wu, Q., Zhang, F., Zhang, J., Lei, T., et al. (2018a). Prevalence and characterization of Staphylococcus aureus isolated from retail vegetables in China. Front. Microbiol. 9:1263. doi: 10.3389/fmicb.2018.01263
Keywords: Tn558, cfr, Staphylococcus delphini, unconventional circularizable structure, multidrug resistance
Citation: Zhang F, Wu S, Huang J, Yang R, Zhang J, Lei T, Dai J, Ding Y, Xue L, Wang J, Chen M and Wu Q (2021) Presence and Characterization of a Novel cfr-Carrying Tn558 Transposon Derivative in Staphylococcus delphini Isolated From Retail Food. Front. Microbiol. 11:598990. doi: 10.3389/fmicb.2020.598990
Received: 26 August 2020; Accepted: 07 December 2020;
Published: 15 January 2021.
Edited by:
Zhangqi Shen, China Agricultural University, ChinaReviewed by:
Jian Sun, South China Agricultural University, ChinaXiang-Dang Du, Henan Agricultural University, China
Rong Zhang, Zhejiang University, China
Copyright © 2021 Zhang, Wu, Huang, Yang, Zhang, Lei, Dai, Ding, Xue, Wang, Chen and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t; Moutong Chen, Y210b29uQGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Feng Zhang1,2†
Feng Zhang1,2† Shi Wu
Shi Wu Jumei Zhang
Jumei Zhang Yu Ding
Yu Ding Liang Xue
Liang Xue Juan Wang
Juan Wang Moutong Chen
Moutong Chen Qingping Wu
Qingping Wu