- 1Institute of Oceanography, Minjiang University, Fuzhou, China
- 2Fujian Universities Key Laboratory for Plant-Microbe Interaction, College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
Marine fungi of the genus Penicillium are rich resources of secondary metabolites, showing a variety of biological activities. Our anti-bacterial screening revealed that the crude extract from a coral-derived fungus Penicillium steckii P2648 showed strong activity against some pathogenic bacteria. Genome sequencing and mining uncovered that there are 28 secondary metabolite gene clusters in P2648, potentially involved in the biosynthesis of antibacterial compounds. Chemical isolation and structural determination suggested citrinin is the dominant component of the crude extracts of P2648, and our further tests confirmed that citrinin showed excellent activities against various pathogenic bacteria. Moreover, the gene cluster containing a homolog of the polyketide synthase CitS was identified as the citrinin biosynthesis gene cluster through genetic analysis. Interestingly, three isoquinoline alkaloids were unexpectedly activated and isolated from the Δcits mutant and structural determination by using high-resolution electron spray ionization mass spectroscopy (HRESIMS), 1D, and 2D NMR. Further antibacterial assays displayed that compounds 1 and 2, but not compound 3, showed moderate activities against two antibiotic-resistant pathogenic bacteria with minimum inhibitory concentration (MIC) of 16–32 μg/ml. In conclusion, our results demonstrated that citrinin and isoquinoline alkaloids represent as the major antibacterial agents in the coral-associated fungus P. steckii P2648, and our genomic and chemical analyses present evidence in support of P. steckii P2648 as a potent natural products source for anti-bacterial drug discovery.
Introduction
Bacterial resistance to antibiotics, such as extended-spectrum β-lactamase (ESBL)-producing Escherichia coli, has become a global public health issue (Lee et al., 2006). The global spread of bacterial resistance significantly increased patient mortality and morbidity due to the shortage of suitable antibiotics (Peirano and Pitout, 2019). Given the richness and diversity in chemistry and biological activity, the fungal secondary metabolites are regarded as inexhaustible sources for new antibacterial drugs (Abouelhassan et al., 2019). Species in the genus Penicillium, which are world widely distributed and mostly saprophytic, have since been considered as valuable resources of life-saving medicine and agrochemicals (Nielsen et al., 2017). The genus Penicillium consists of more than 350 of species, however, only a very limited number of them have been exploited for bioactive secondary metabolites. Among them, Penicillium rubens (also known as Penicillium chrysogenum) is one of the most prominent species and famous for its outstanding secondary metabolite penicillin, a paramount antibiotic for the therapy of infectious disease since World War II (Penalva et al., 1998). In addition, some other Penicillium fungi had been developed to produce series of pharmaceutical compounds, including the cholesterol-lowering drugs statins from Penicillium citrinin (Manu and Rogozea, 2016), mycophenolic acid from Penicillium brevicompactum (Anand and Srivastava, 2020), anti-cancer auranthine from Penicillium aurantiogriseum (Kalinina et al., 2018), and anti-Inflammation (3R, 7R)-7-acetoxyl-9-oxo-de-o-methyllasiodiplodin produced by Penicillium sp. (Liu et al., 2019).
Isoquinoline alkaloids constitute the largest classes of secondary metabolites with more than 2,500 defined structures, mainly produced by higher plants (Sato et al., 2007). Since the early 19th century, their complex structures and significant biological activities have attracted considerable attentions from researchers worldwide. However, so far, only a limited number of isoquinoline compounds were isolated from filamentous fungi, such as fumisoquins from Aspergillus fumigatus (Baccile et al., 2016), spathullins A and B from Penicillium spathulatum Em19 (Nord et al., 2019), imizoquins from Aspergillus flavus (Khalid et al., 2018), chaetolines A and B from Chaetomium sp (Ancheeva et al., 2018), and benzoisoquinoline-9-one from Peyronellaea sp. FT431(Li et al., 2019), and most were identified with anti-bacterial activities.
In our study, the genome of a coral-derived fungus Penicillium steckii P2648 was revealed by high-throughput sequencing, and founded to encode 28 of secondary metabolite gene clusters. The mycotoxin citrinin was identified as the major secondary metabolite of P2648 and showed antibacterial activities. The coding gene of citrinin cluster (cits) was genetically disrupted from P2648 genome. Besides, the disruption of cits compromised citrinin biosynthesis but caused the unanticipated accumulations of two anti-bacterial isoquinoline alkaloid compounds (Compound 1 and 2). Their chemical structures were classified by combining use of 1D, 2D-NMR, and MS. Taken together, our study demonstrated that citrinin endows the coral-derived P. steckii P2648 with prominent anti-bacterial features, while blocking the biosynthesis of citrinin will generate another two novel anti-bacterial isoquinoline alkaloids. Moreover, genetic dereplication approach will shed novel light on the study of secondary metabolites in marine-derived fungi.
Materials and Methods
Fungal Identification
The fungal strain was isolated from a gorgonian which was collected from the South China Sea. For growth and conidiation, the fungal strain was cultured on the PDA agar (potato juice 20%, dextrose 2%, and agar 1%), YES (sucrose 15%, yeast extract 2%, and agar 1%), or GMM (sucrose 2%, NaNO3 3%, KH2PO4 1%, KCl 0.5%, MgSO4 0.1%, FeSO4 0.01%, and agar 1%) for 7 days at room temperature in the dark. For molecular identification of fungal strain, the genomic DNA of fungal strain was extracted from approximately 100 mg of mycelium, scraped from PDA plates, using a thermolysis method as previously reported (Zhang et al., 2010). The quality and quantity of DNA samples were measured with the NanoDrop Spectrophotometer (Thermo Scientific, Wilmington, Germany). The ITS fragment was amplified using the primer pair ITS1/ITS4 (Supplementary Table S2), and 18S rRNA gene was obtained from our sequencing data. A phylogenetic tree based on ITS and 18 s rDNA sequences was constructed by using the MAGA X with the neighbor-joining method (Kumar et al., 2018).
Genome Sequencing
Genome sequencing of P. steckii P2648 was performed using the second-generation Illumina seq 4000 platform at Majorbio, China. The sequencing libraries were constructed using Illumina Paired-End DNA sample Prep Kit. Reads were assembled from raw data by using SOAPdenovo v2.04 program with a range of Kmers (17–92). Gene predictions were performed by using Maker2 program. Gene model annotations were carried out through sequence alignment in Cluster of Orthologous Groups of proteins (COG) database. Prediction of putative secondary metabolite biosynthesis gene cluster was carried out using the online AntiSMASH software. To use single-copy genes to plot species tree, the protein-coding genes from P. steckii, Penicillium oxalicum, P. rubens, Penicillium griseofulvum, Penicillium brasilianum, Penicillium expansum, and Aspergillus nidulans were downloaded from NCBI NR database. The single-copy genes were obtained by using OrthoFinder software. The phylogeny tree was constructed inferred from the amino acid sequences of 5,884 single-copy genes by IQ-TREE based on maximum likelihood algorithm. The phylogenetic tree was displayed by using iTOL.
Gene Deletion
Gene deletion strategy in P2648 was slightly modified from that we previously established in P. oxalicum (Yao et al., 2015). Double-joint PCR was used to construct a gene deletion cassette, using the hph gene amplified from plasmid pSilent-1 as the selectable marker. All primers that were used to amplify the 5'- and 3'-flanks were listed in Supplementary Table S2 with the P2648 gDNA as the template. The entire gene deletion cassette was amplified with specific primers, using the 5'- and 3'-flanks for gene citS and hph mix as a template. The PCR products were transformed into the protoplasts of P2648 wild strain. For the preparation of protoplasts, approximately 109 cfu/ml of conidia was inoculated into 100 ml PDB Broth, shaken at 150 rpm for 12 h at 30°C. Mycelia were collected by using four-layers of cotton gauzes, transferred to a 50 ml tube, and resuspended in 20 ml lysis filter-sterilized solution that contained 12 mg/ml lysing enzymes (Sigma, St Louis, MO, United States), 50 mM KC1, 1 M NaCl, and 1 mM CaCl2. After shaken at 60 rpm, 30°C for 3–4 h, protoplasts were harvested by filtering through a two-layer of lens paper. The obtained protoplasts were washed twice with a solution of 0.5 M KC1, 50 mM CaC12, and 10 mmol Tris-HCl (pH 7.5). Fungal transformation and regeneration procedures were performed as described previously (Yao et al., 2015). Gene deletion mutants were preliminarily identified by PCR and then confirmed via RT-PCR. The primers used for verifying gene deletion mutants were listed in Supplementary Table S2.
Fermentation, Extraction, Isolation, and Structure Determination
The fungus Δcits mutant was cultivated in the rice medium (80 g of rice, 3 g of natural sea salt, and 100 ml of H2O) in 1 L Erlenmeyer flask (20 flasks) at room temperature for 4 weeks. The fermented rice substrates were extracted three times with EtOAc (300 ml every time for each flask) and two times with MeOH (300 ml for each flask). The organic extracts were combined and concentrated under vacuum to afford a total extract (5.0 g), which was subjected to silica gel column chromatography (CC) using a step gradient elution with EtOAc-petroleum ether (0–100%) and then with MeOH-EtOAc (0–50%) to yield four fractions (Fr.1–Fr.4). Fr.3 was further subjected to silica gel CC, and eluted with CHCl2-MeOH (40:1, v/v) to obtain subfractions Fr.3-1–Fr.3-4, Fr.3-2 was further purified by semi-preparative high performance liquid chromatography (HPLC) eluted with MeOH-H2O (40:60, v/v) to give compounds 1 (14.0 mg) and 2 (19.0 mg). Fr.3-3 purified by Sephadex LH-20 CC with mixtures of CHCl2-MeOH (1:1, v/v) to yield compound 3 (12.0 mg).
Optical rotations were recorded on a JASCO P-1020 digital polarimeter. IR was acquired on a Nicolet-Nexus-470 spectrometer with of KBr Pellet Method. The 1D and 2D NMR spectra were recorded in MeOD on a JEOL JEM-ECP NMR spectrometer with tetramethyl silane (TMS) as the internal standard. All NMR assignments were based on the 1H-1H COSY, HSQC, and heteronuclear multiple bond correlation (HMBC) spectroscopic data. High-resolution electron spray ionization mass spectroscopy (HRESIMS) data were acquired on a Thermo MAT95XP high-resolution mass spectrometer (Thermo Fisher Scientific, United States). HPLC analysis and purification were performed on a Hitachi L-2000 HPLC system coupled with a Hitachi L-2455 photodiode array detector and using an analytical Kromasil column (250 mm × 4.6 mm, 5 mm) and a semi-prepared C18 column (250 mm × 10 mm, 5 mm), respectively. The mobile phase consisted of 0.1% formic acid in acetonitrile (A) and 0.1% formic acid in water (B) with a flow rate of 1 ml/min with 10 ml injection volume, and recorded at 254 nm.
Antibacterial Activities Tests
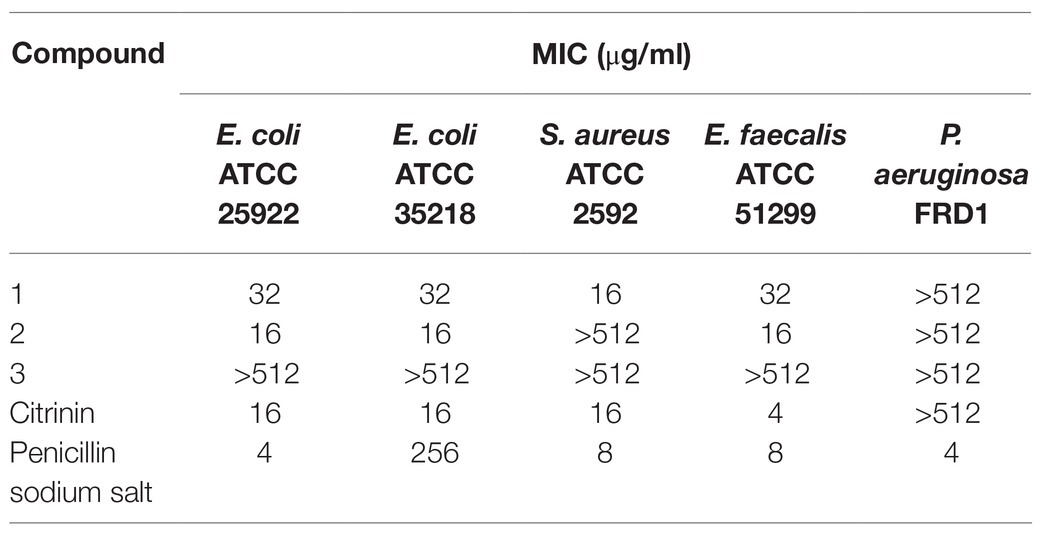
The tested pathogenic bacterial strains include three common zoonotic pathogens (E. coli 25922, Staphylococcus aureus ATCC 2592, and Pseudomonas aeruginosa FRD1), two antibiotic-resistant bacteria [ESBL-producing E. coli (ATCC 35218) and vancomycin-resistant Enterococcus faecalis ATCC 51299]. The minimum inhibitory concentration (MIC) was determined by using a broth microdilution method according to the standards and guidelines recommended by Clinical and Laboratory Standards Institute (2012). Penicillin G sodium salt was used as anti-bacterial drug positive control.
Results and Discussion
Genome Sequencing
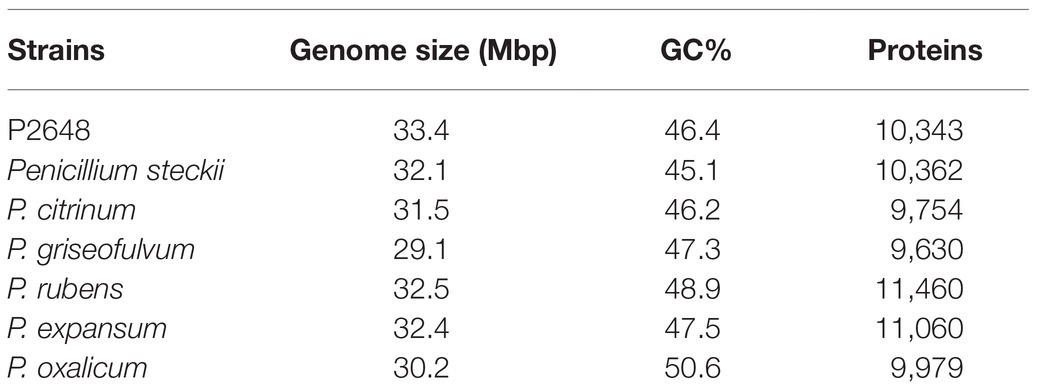
During our screening of the marine-derived fungi for producing anti-bacterial natural products, the crude extract of gorgonian-derived fungus P2648 showed strong inhibition on the growth of both Gram-negative E. coli and Gram-positive S. aureus (MIC 50–100 μg/ml). Based on morphological characteristics and phylogenetic analysis of 18S rRNA and ITS sequence, P2648 was classified into the genus Penicillium with the highest sequence similarity (99.8%) to P. steckii (Supplementary Figure S1). Until now, no genome of marine-derived P. steckii had been sequenced. To investigate comprehensively the potential of marine-derived fungus P2648 in production of secondary metabolites, genomic DNA was isolated from the mycelia of strain P2648 and sequenced by using the Illumina Hiseq 4000 sequencing platform. The obtained genome of P2648 was assembled into 961 scaffolds of about 33.4 megabases (Mb), which is larger than that of model P. steckii strain IBT 24891 (Table 1). And, compared to other fungi in Penicillium genus, P2648 has a relatively larger genome in size (Table 1). A total of 12,098 protein-encoding genes were predicted, which comparable to both soil-derived P. steckii and Penicillium fungi. Among them, 10,343 (85.49%) genes have functional annotations in the COG database (Supplementary Table S1). Referring to the functional classification of COG, and among them, 279 genes (2.7%) are involved into secondary metabolite biosynthesis, transport, or metabolism (Supplementary Table S1).
To better understand the evolutional relationship, phylogenetic analysis was performed by using whole-genome alignment between P2648 and its relatives. The result showed that six Penicillium species and P2648 formed a clade with a high bootstrap support, in which P2648 showed closest relationship with P. steckii (Figure 1A). Therefore, together multi-loci DNA sequence and whole-genome alignment phylogenetic analysis confirmed that P2648 was one of P. steckii species. Sequence-synteny analyses between P. steckii and P2648 revealed that most of scaffolds showed synteny, however, extensive genetic divergences existed in the two P. steckii strains that were from different geographical regions (Figure 1B).
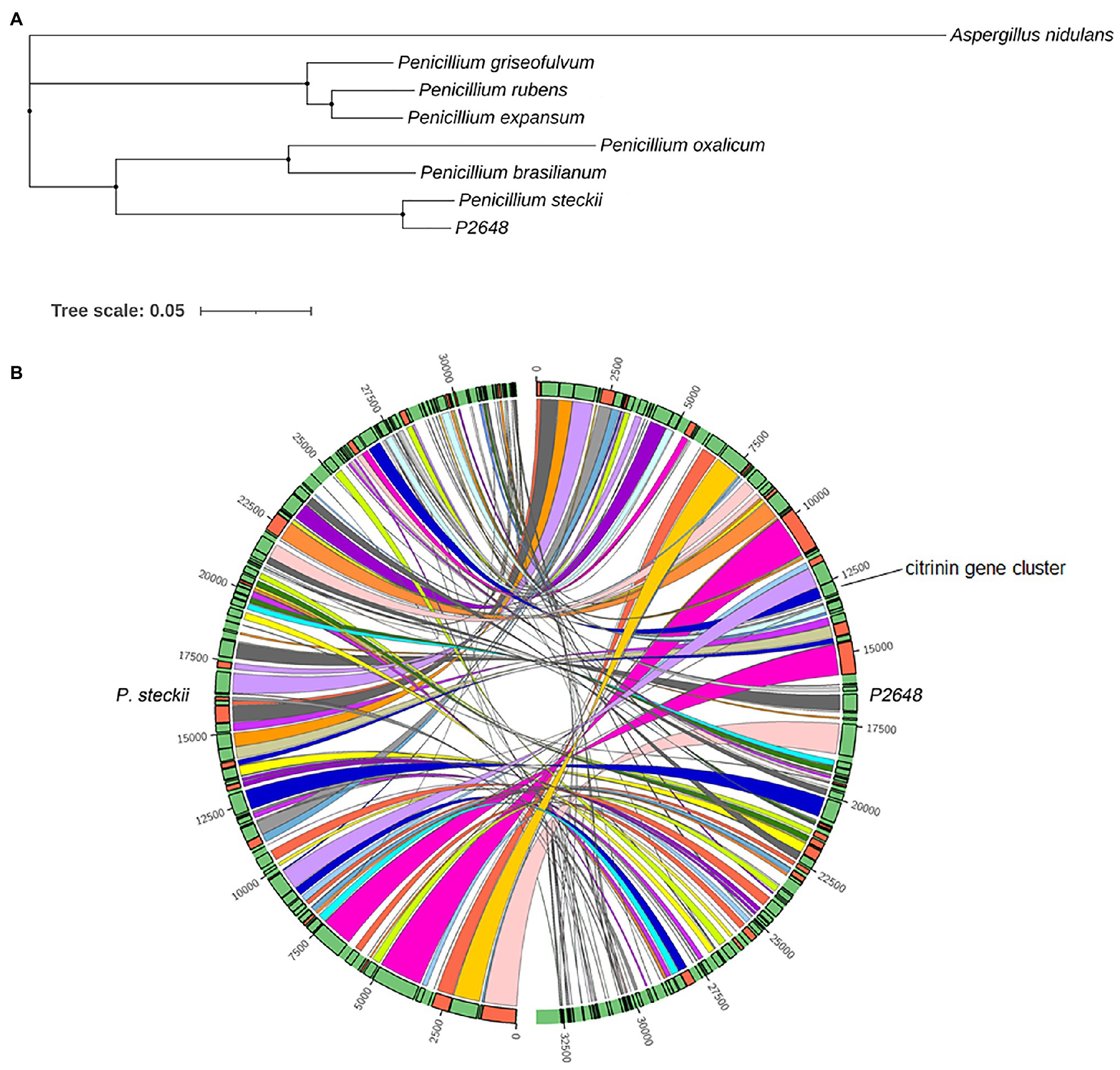
Figure 1. Phylogenetic and synteny analysis of P2648 with other fungal species. (A) The maximum likelihood phylogeny was constructed inferred from the amino acid sequences of 5,884 single-copy genes by IQ-TREE. The phylogenetic tree was displayed by using iTOL. The single-copy genes were obtained by OrthoFinder tool. Aspergillus nidulans from Aspergillus genus was used as the outgroup; (B) Sequence-synteny analyses between marine-derived P2648 and soil-species, and the Circo plots represent syntenic blocks between P2648 and Penicillium steckii.
Secondary Metabolite Gene Cluster
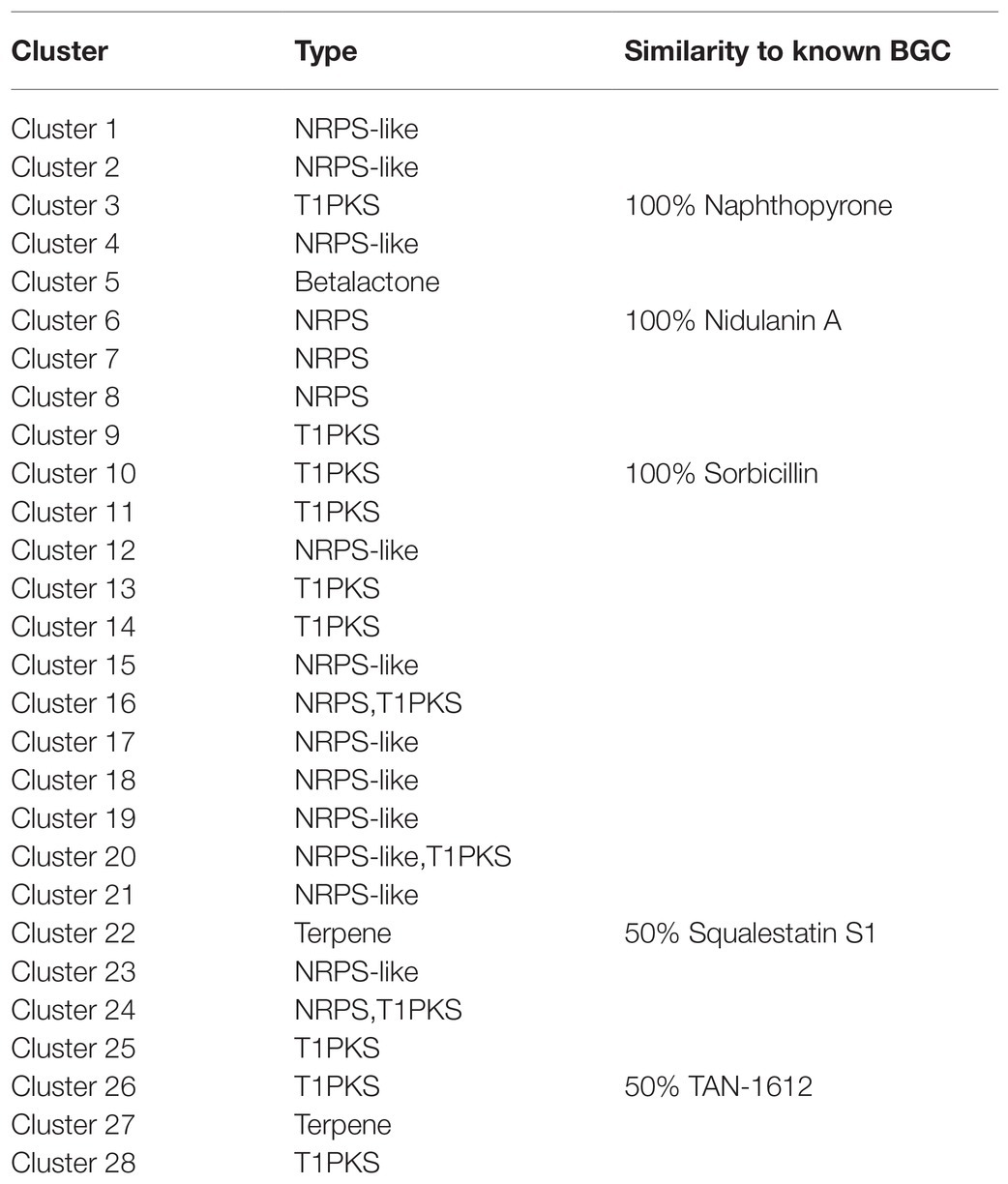
The secondary metabolites of filamentous fungi constitute a rich resource of bioactive natural products with antibiotic activities, such as penicillin from P. chrysogenum. Interestingly, genes encoding biosynthetic pathway for secondary metabolite are often located in a gene cluster on chromosome to form the secondary metabolite biosynthetic gene clusters (SMBGC; Keller, 2015). Mining P2684 genome for gene clusters involved in the biosynthesis of secondary metabolites was performed by using the online antiSMASH tool. A total of 28 (SMBGCs) were identified from the genome of P2648, including nine of polyketide (PKS), 10 of non-ribosomal peptide (NRPS)-like (NRPS-like), four of NRPS, two of terpene (TE), three of PKS-NRPS hybrid, and one betalactone (Table 2).
Based on antiSMASH prediction, we found that P. steckii P2648 has the potential to produce sorbocillin (Table 2 and Figure 2). Sorbocillin and its derivatives, sorbicillinoids are a class of biologically active and structurally diverse fungal polyketides (Salo et al., 2016). Cluster 10 is consisted of three genes, two polyketide synthase (gene04233 and gene04234), and one FAD-dependent monooxygenase (gene04235). The two-polyketide synthases 04233 and 04234 showed high sequence similarities with the highly reducing iterative PKS SorA and the non-reducing iterative PKS SorbB, respectively (Figure 2). In sorbocillin biosynthesis, these two megaenzymes were reported to assemble the polyketide skeleton. And, the FAD-dependent monooxygenase 04235 from Cluster 10 showed high sequence similarity with the post-modification enzyme SobC (Figure 2). Together, our in silico analysis suggested that P2648 could produce sorbocillin and its analogs.
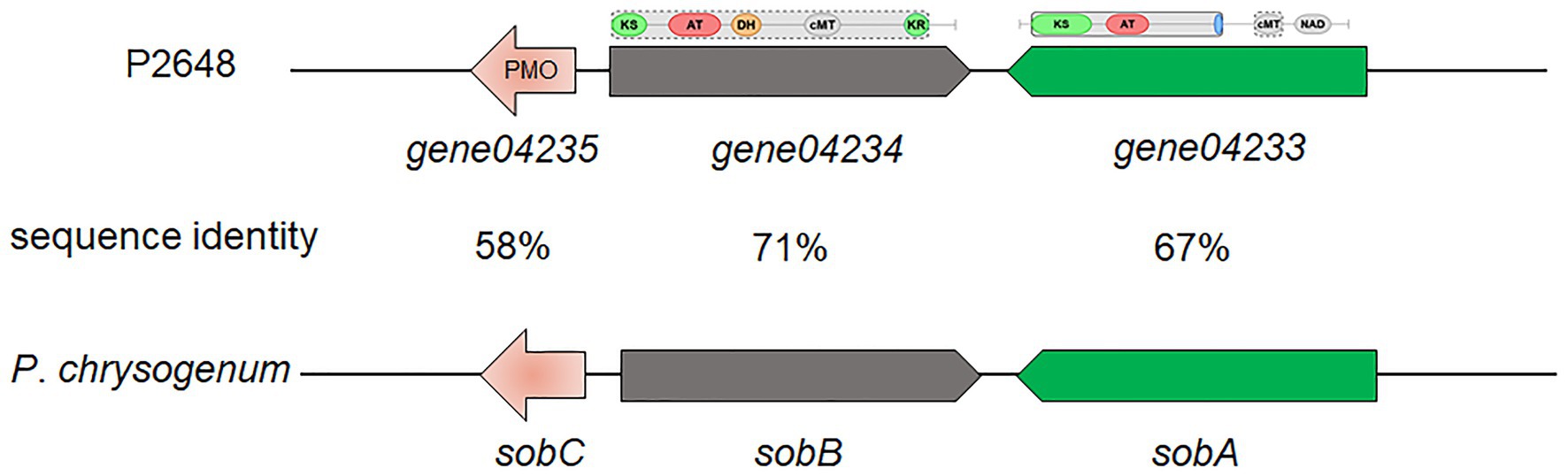
Figure 2. The putative sorbocillin biosynthetic gene cluster of P2648 and the comparison of this cluster with the sorbocillin cluster reported for P. chrysogenum. PMO, monooxygenase; KS, ketosynthase domain; AT, acyltransferase; DH, dehydratase domain; cMT, carbon methyltransferase; KR, ketoreductase domain; and NAD, male sterility protein.
The polyketide cluster encoded by Cluster 3 showed 100% sequence similarity to the naphthopyrone cluster from A. nidulans, which responsible for biosynthesis of the yellow conidial wall pigment (Mayorga and Timberlake, 1992). In addition, Cluster 6 was predicted to biosynthesize secondary metabolites with structurally similar to nidulanin A and Cluster 22 showed high similarity to the terpene cluster squalestatin S1. Cluster 26 is composed of six genes, encoding FAD-dependent monooxygenase (gene10770), oxidoreductase (gene10771), o-methyltransferase (gene10772), monooxygenase (gene10773), non-reducing polyketide synthase (gene10774), and a transcription factor (gene10775). Among them, four of genes showed high sequence similarities with their counterparts involved in biosynthesis of TAN-1612 (Li et al., 2011). Therefore, we hypothesized that Cluster 26 potentially synthesize similar polyketide compounds.
Intriguingly, secondary metabolite products encoded by most of these SMBGCs in P2648 genome could not be assigned. In particular, three PKS/NRPS hybrid SMBGCs (Cluster 16, Cluster 20, and Cluster 24) were identified in P2648 genome, their secondary metabolite products remain to be determined. Also, we predicted 10 NRPS-like BGC gene clusters in P2648, no any compound could be assigned by bioinformatic analysis. Together, we have performed a comprehensive analysis and provide valued information on gene clusters for secondary metabolite biosynthesis by using both antiSMASH and gene cluster homology comparisons.
Citrinin Is the Major Anti-bacterial Compound Produced by the Coral-Derived Fungus P. steckii P2648
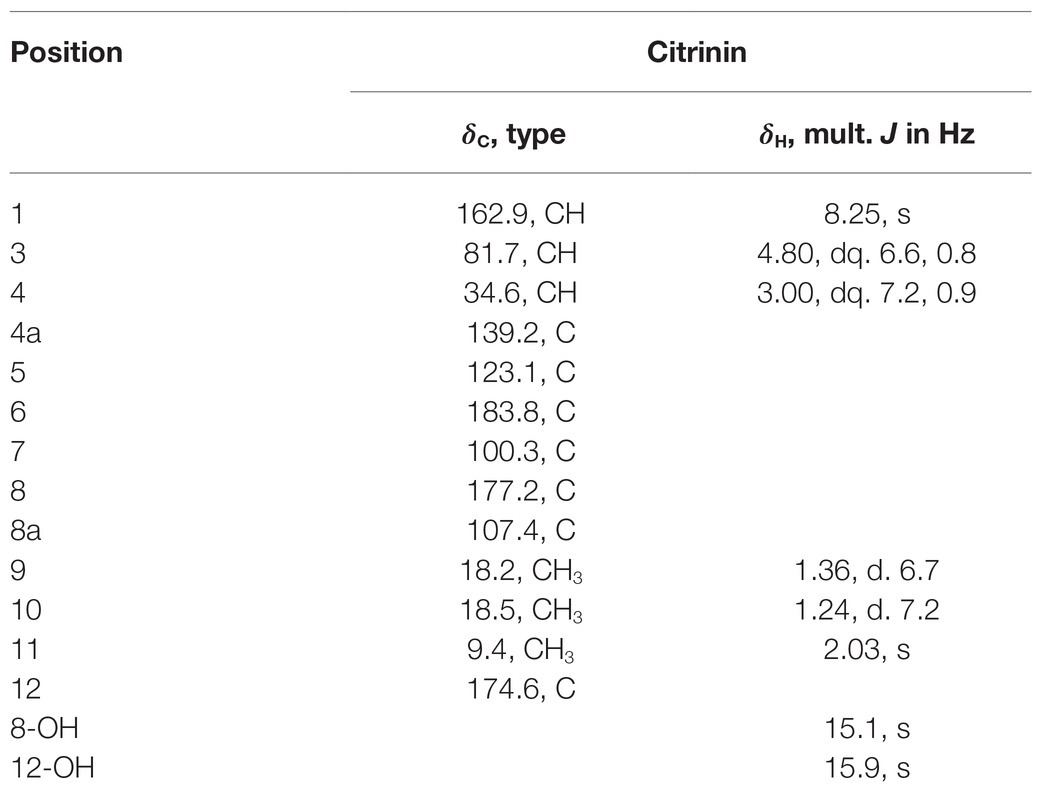
To chemically identify the antibacterial agents, scaled fermentation of P2648 was performed by using a solid rice medium for 10 days. The ethyl acetate extracts of the fermentation were fractionated by using repeated positive and negative silica chromatography. A golden-yellow needle crystal (14 g) was obtained, which accounting for the largest population of total organic extracts (about 67%). Its structure was then identified to be as citrinin by comparing its 1H NMR and 13C NMR (Table 3) with previously reported NMR data (Blanc et al., 1995).
Identification of the Citrinin Biosynthetic Gene Cluster
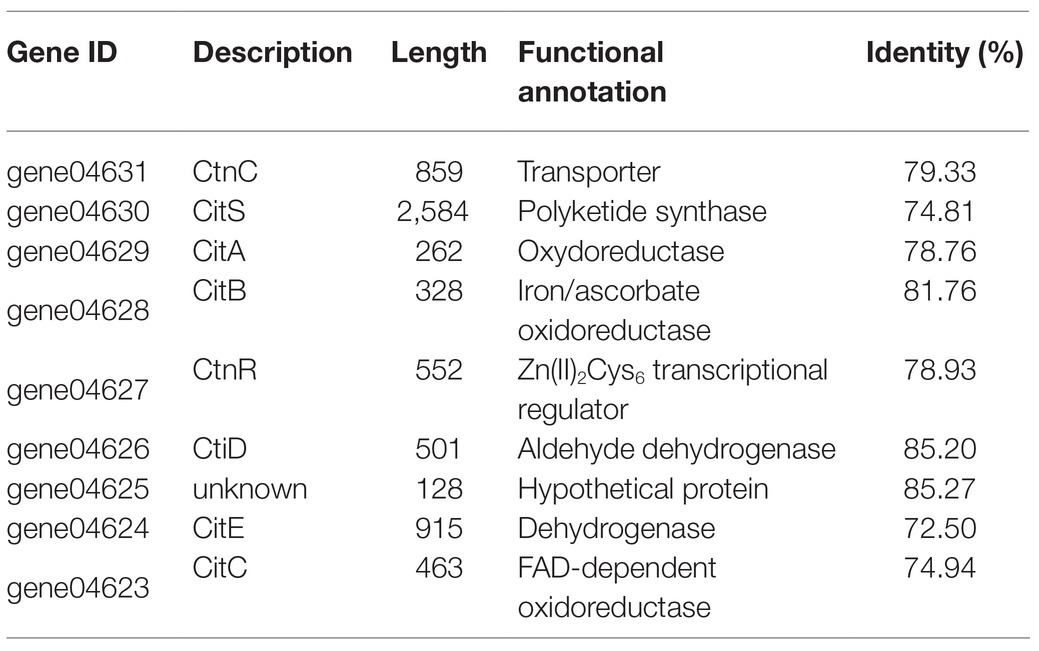
No gene cluster for citrinin biosynthesis could be ascertained by antiSMASH analysis (Table 2). Therefore, to identify the gene cluster for citrinin biosynthesis, a manual BLAST search by using CitS coding gene from Monascus ruber as a query against the P. steckii P2648 genome was performed, and results showed that a putative citrinin polyketide cluster containing nine genes were identified (Table 4). And, the entire citrinin gene cluster showed synteny between P2648 and P. steckii. The gene04630 (the homolog of cits) encodes the polyketide synthase, synthesizing triply methylated pentaketide skeleton by using methylmalonyl-SCoA as the building block (He and Cox, 2016). The methyl at 12-site of methylated pentaketide is progressively oxidized to carboxylic acid by non-heme iron oxidase CitB, FAD-dependent oxidoreductase CitC, and followed by CitD. Then, the 3-carbonyl is reduced to ether by the dehydrogenase CitE. CtnR and CtnC are the pathway-specific transcription factor and transporter, respectively. To confirm the function of the putative polyketide cluster in the citrinin biosynthesis, the open reading frame (ORF) region of PKS gene cits was deleted from P. steckii wild-type strain P2648 (WT) genome by replacing with hygromycin phosphate transferase (hph) gene. A total of 21 hygromycin-resistance transformations were obtained by three cycles of genetic transformation, and two deletion mutants were further verified through RT-PCR with gene-specific primers (Figure 3A). Absence of cits gene had no detectable defects on both vegetative growth and conidiation of P2648 on various media (Figure 3B). As anticipated, the Δcits mutant produced a significant difference in secondary metabolite profiles when compared with WT (Figure 4). Citrinin dominates the fermentation extracts of WT, but no citrinin can be detected in Δcits, suggesting that CitS is essential for citrinin biosynthesis. Besides, intriguingly, as shown in Figure 4, several peaks with retention time from 26 to 28 min were emerged and enhanced in Δcits when compared with WT.
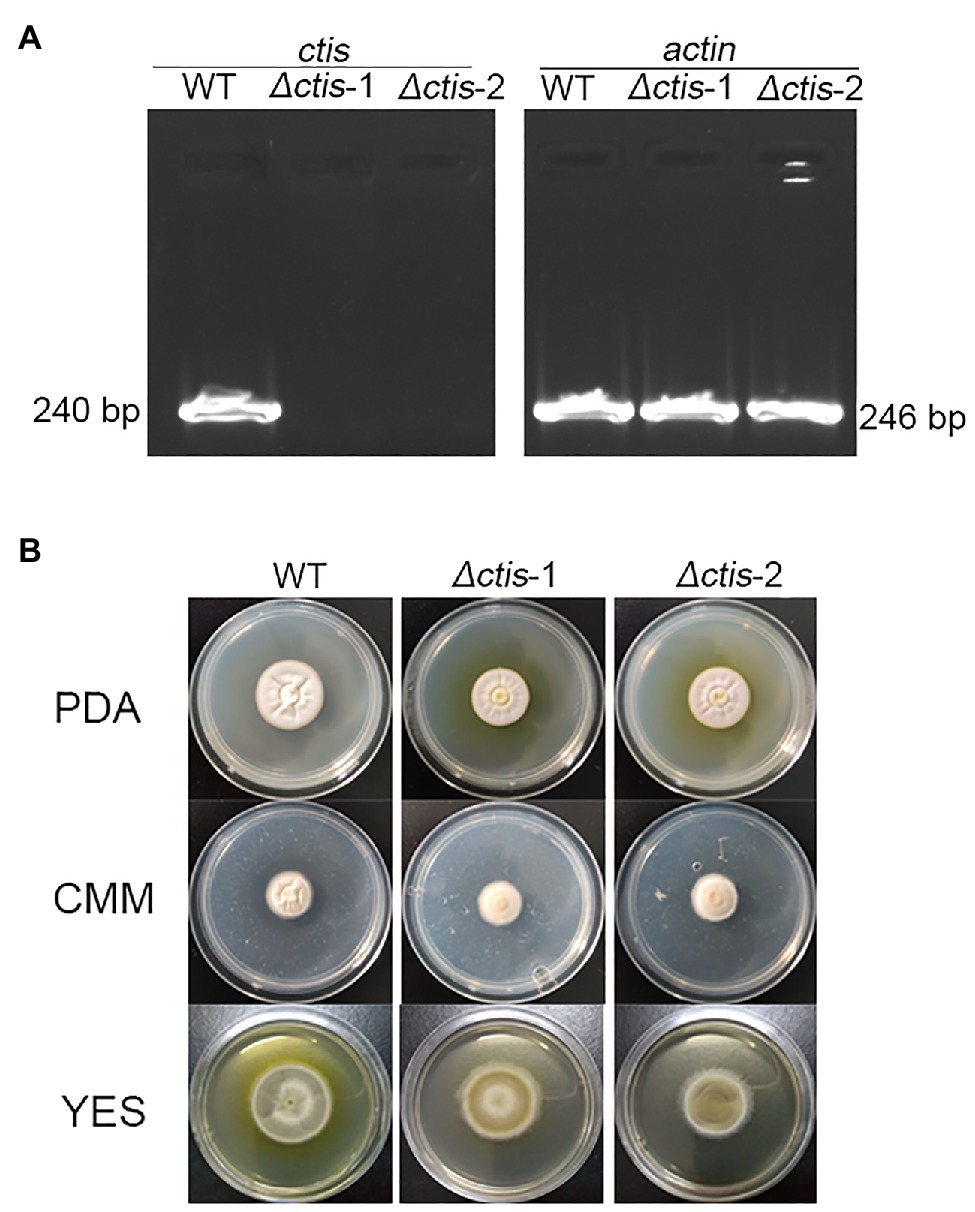
Figure 3. Deletion of citS gene in P2648. (A) RT-PCR verification of gene deletion, the transcripts of actin gene were used as the control; (B) Conidia of Δcits and wild-type (WT) were inoculated on the PDA, CMM, and YES plates, culturing at 30°C for 7 days.
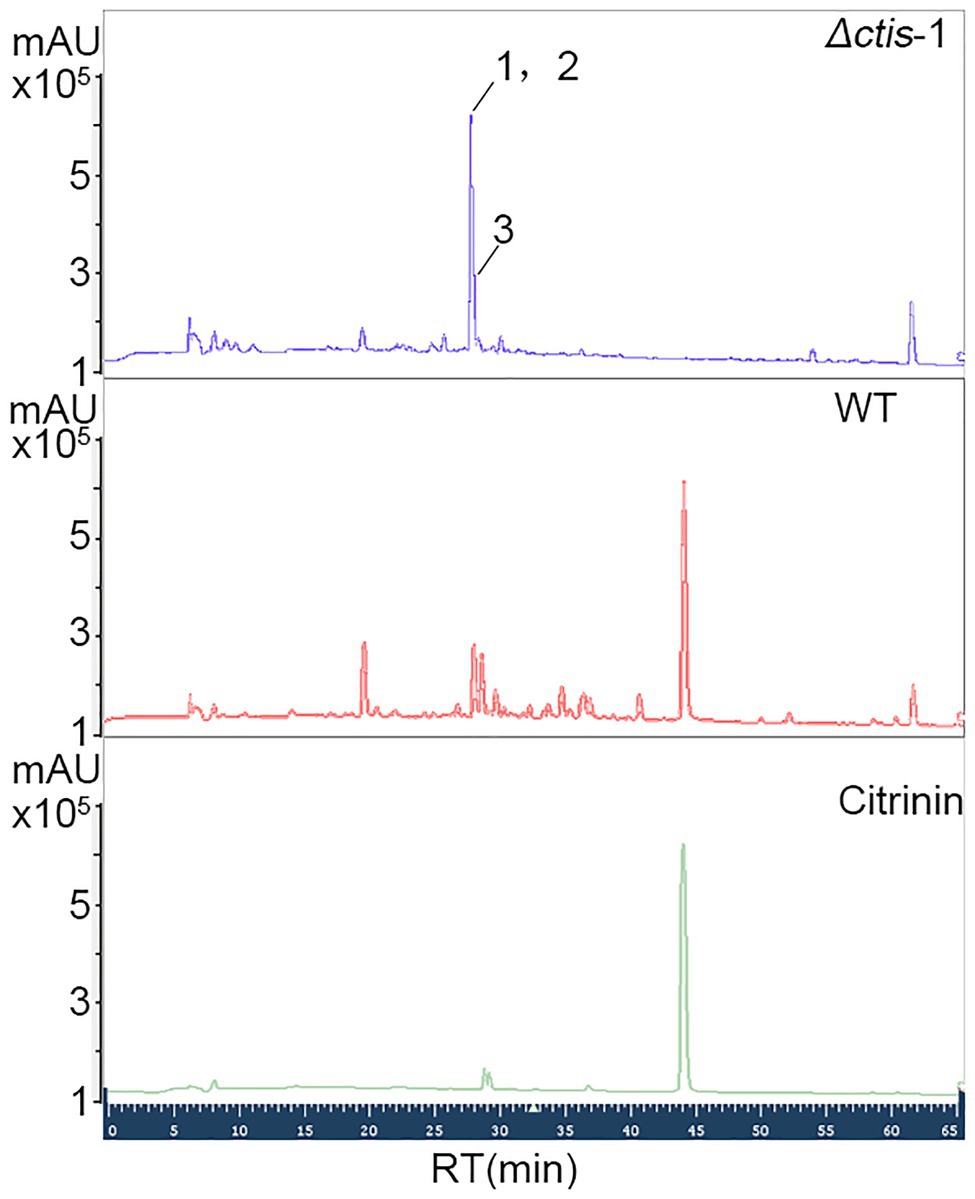
Figure 4. High performance liquid chromatography (HPLC) analysis of secondary metabolite profiles of both Δcits mutant and WT strains. Conidia of Δcits and WT were inoculated in the rice medium and cultured at room temperate for 21 days, the secondary metabolites were extracted by using EtOAc, and then analyzed by HPLC.
Three Isoquinoline Alkaloids Were Unexpectedly Accumulated in the Δcits Mutant
To further mine the induced metabolite potentials by obstructing citrinin synthesis, the secondary metabolites of Δcits were harvested by scaled fermentation. The organic extracts were fractionated by repeated silica gel chromatography, Sephadex LH-20, and semi-preparative HPLC, and finally obtained compounds 1–3 (Figure 5).
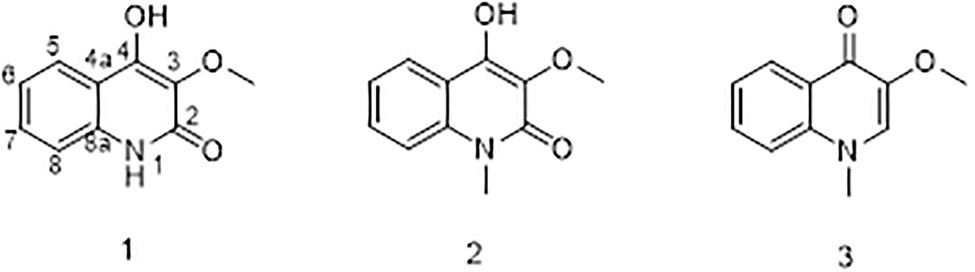
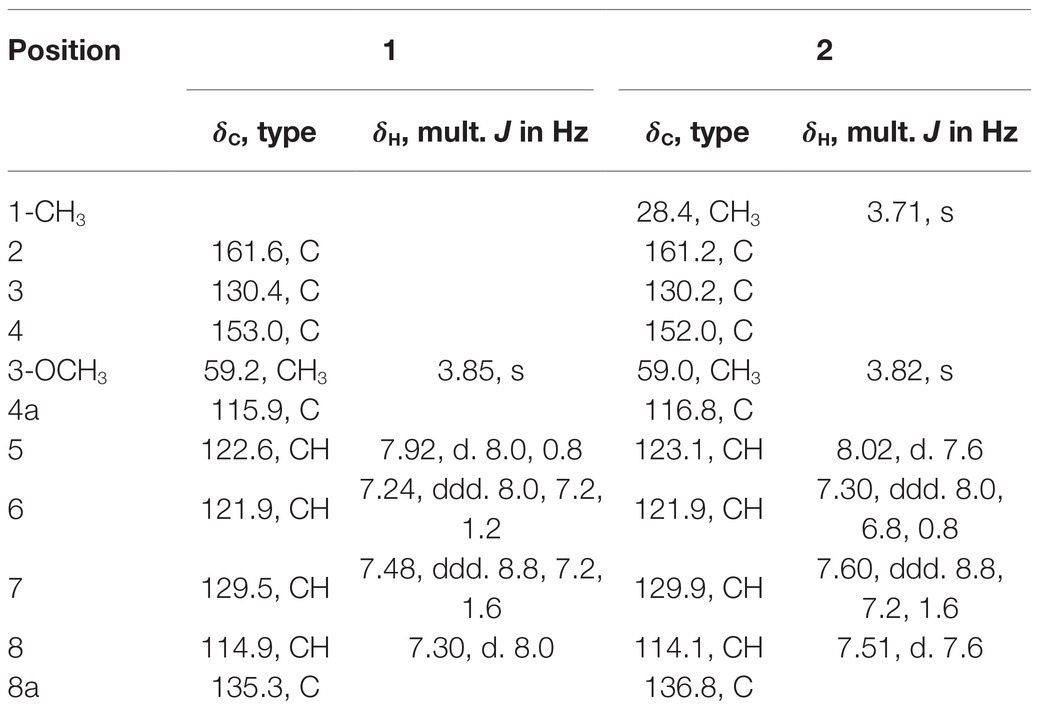
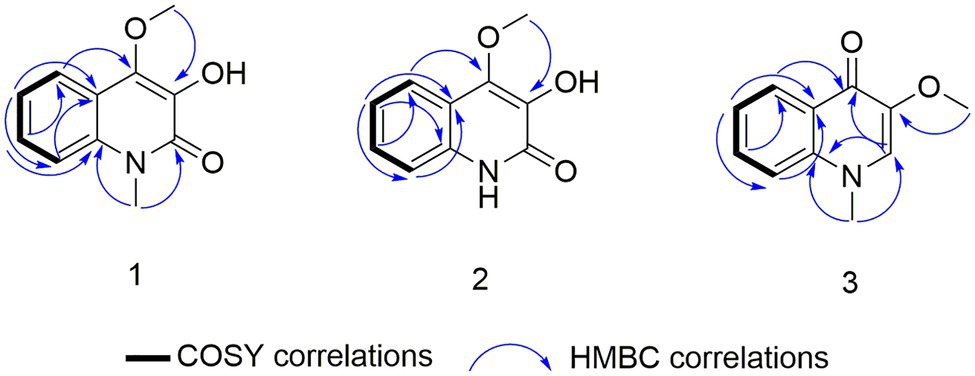
Compound 1 was obtained as a white powder. Its molecular formula was determined to be C10H9NO3 on the basis of the HRESIMS (m/z 192.0653 [M + H]+; calcd for C10H10NO3, 192.0655) and the 13C NMR data. The 1H NMR data (Table 5) displayed four aromatic proton signals at δH 7.92 (1H, d, J = 8.0, 0.8 Hz, H-5),7.48 (1H, ddd, J = 7.2, 8.8, 1.2 Hz, H-7), 7.30 (1H, d, J = 8.0 Hz, H-8), and 7.24 (1H, ddd, J = 7.2, 8.0, 1.2 Hz, H-6); and an oxygenated singlet methyl proton resonance at δH 3.85. Additionally, the 13C and HSQC spectra showed 10 carbon signals, attributable to one ester group carbonyl resonating at δC 161.6, eight aromatic or olefinic carbon signals at δC 153.0, 135.3, 130.4, 129.5, 122.6, 121.9, 115.9, and 114.9, and one oxygenated methyl signal at δC 59.2. The aforementioned NMR data indicated the presence of one 1,2 disubstituted aromatic ring and one olefinic bond and these groups accounted for six out of the seven degrees of unsaturation, requiring one additional ring in 1. Analysis of the COSY spectrum showed correlations of H-5/H-6, H-6/H-7, and H-7/H-8 (Figure 6). Based on the HMBC cross-peaks from H-5 to C-4/C-7/C-8a (Figure 6), from H-6 to C-4a/C-5/C-8a/C-8, from H-7 to C-5/C-8/C-8a, as well as from H-8 to C-4/C-4a/C-6 were used to connect the aromatic ring with the olefinic bond. The HMBC correlations from oxygenated methyl proton δH 3.85 to C-3 (δC 130.4) demonstrated the groups of ▬OCH3 was anchored at C-3. Herein, compound 1 was elucidated as an isoquinoline alkaloid, 4-hydroxy-3-methoxy-2(1H)-quinolinone.
Compound 2 was also obtained as a white powder with the molecular formula C11H11NO3 from HRESIMS (m/z 206.0808 [M + H]+; calcd for C11H12NO3, 206.0812), suggesting that 2 has one CH2 mass unit more than 1. A careful comparison of the 1H and 13C NMR spectra of 2 (Table 6) with those of 1 showed a close structural relationship, with the only difference being the oxygenated methyl proton signal at δH 3.71 and the oxygenated methyl signal at δC 28.4. According to the HMBC cross-peaks from ▬NCH3 (δH 3.71) to C-2 and C-8a, the oxygenated methyl group was attached to the amide group. Therefore, compound 2 was characterized as 4-hydroxy-3-methoxy-1-methyl-2(1H)-quinolinone.
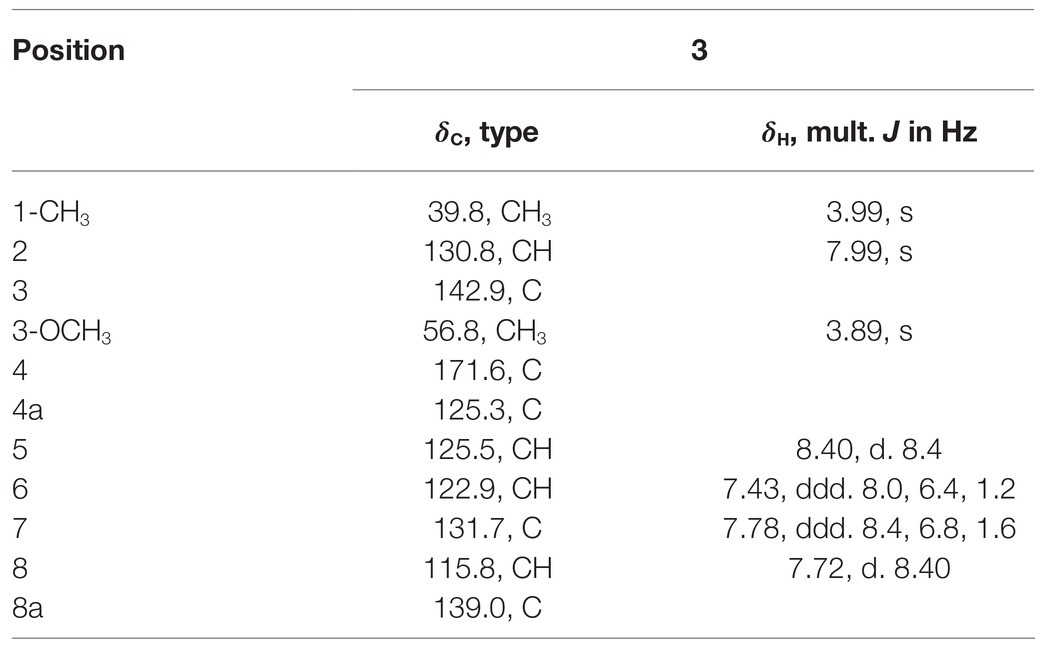
Compound 3 with the molecular formula C11H11NO2 as determined by HRESIMS (m/z 190.0858 [M + H]+; calcd for C11H12NO2, 190.0863), was isolated as the white powder. The 1H NMR spectrum (Table 6) was very similar to that of 2, with the notable difference being the addition of a singlet proton resonance at δH 7.99 in 3. Comparison of the 13C NMR spectra of 3 with those of 2 displayed an unsubstituted olefinic carbon signal at δC 130.8. Combining the HMBC correlations from ▬NCH3 (δH 3.71) to C-2 and C-8a and from ▬OCH3 (δH 3.89) to C-3, the structure of compound 3 was assigned as 3-methoxy-1-methyl-4(1H)-quinolone.
The unexpected accumulation of three isoquinoline alkaloids after the deletion of citS might cause by, at least, the redistribution of biosynthetic precursors. More specifically, in WT strain, citrinin is the dominant secondary metabolite, which obstructs the biosynthesis of other compounds by consuming most of the universal precursor substances, such as acetyl-CoA and malonyl-CoA. Elimination of the citrinin biosynthesis might increase the reserve pools of these precursors, which would be redirected into the biosynthesis pathway of isoquinoline alkaloids. Additionally, the deletion of citS also resulted in a low SM background, which consequently contributed to the discovery of those isoquinoline alkaloids with formerly low synthetic levels.
Biological Activity Assay
The antibacterial activities of three isoquinoline alkaloids and citrinin against a variety of pathogenic bacteria were assessed using the broth microdilution method to determine the MICs (Table 7).
Citrinin displayed excellent biological activities against all tested pathogenic bacteria with MIC of from 4 to 16 μg/ml, except for P. aeruginosa FRD1. Compound 1 exhibited an MIC of 16–32 μg/ml against all tested pathogens expect for P. aeruginosa FRD1, and compound 2 also showed an MIC of 16 μg/ml against three pathogenic bacteria, including two E. coli and one E. faecalis. The most noteworthy result is that both compounds 1 and 2 showed obvious activities against two drug-resistant microbial bacterial pathogens E. coli ATCC 35218 and E. faecalis ATCC 51299, implying a different action mechanism from common antibiotics. Compound 3 did not show detectable biological activities against any of tested pathogenic bacteria. Compared with compounds 1 and 2, hydroxy at position 4 was oxidized to a carbonyl in compound 3, suggesting that the 4-hydroxy is essential for the anti-bacterial activities of isoquinoline alkaloids.
Conclusion
In the study, we reported the draft genome of a coral-derived fungus P2648. Whole-genome phylogenetic analysis demonstrated that P2648 is a novel strain of P. steckii species. The genome data and our bioinformatic analysis firstly revealed that P. steckii contains a large number of secondary metabolite cluster without assigned compounds. Further, our chemical investigation indicated that citrinin was identified as the dominant antibacterial agent of P2648, and a CitS-containing SMBGC was proved to be involved in citrinin biosynthesis in P2648. Surprisingly, three isoquinoline alkaloids were accumulated after blocking the biosynthesis of citrinin, and two of isoquinoline alkaloids showed moderate activities against two antibiotic-resistant pathogenic bacteria. In conclusion, our genomic and chemical analyses present evidence in support of P. steckii P2648 as a potent natural products source for anti-bacterial drug discovery.
Data Availability Statement
The whole genome sequence of P. steckii P2648 has been deposited at NCBI database under the Bioproject number PRJNA671565.
Author Contributions
GY, ZW, and JC conceived and designed the research work. GY, HZ, XC, DL, and ZY performed the experiments and analyzed the data. GY wrote the manuscript. HZ, XC, ZW, and JC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Minjiang University (JAT190622, MYK19011, and MJY19019), China Ocean Mineral Resources R&D Association (DY135-B2-13), Project from Department of Fujian Science and Technology (2018N2001), and Program for Innovative Research Team in Science and Technology in Fujian Province University.
Conflict of Interest
The reviewer LL declared a shared affiliation, with no collaboration, with three of the authors DL, ZY, and ZW to the handling editor at the time of the review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Chang-Yun Wang from Ocean University of China for providing gorgonian materials. We also thank a lot for Yao-Yao Zheng for his excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.600991/full#supplementary-material
Supplementary Figure S1 | Phylogenetic tree analysis of P2648 and related filamentous fungi. Phylogenetic tree of P2648 and all related strains were reconstructed based on a 18S RNA gene alignment using the MEGA X software package with the neighbor-joining method. Bootstrap analysis (1,000 replications) was used to provide confidence estimates for phylogenetic tree topologies, and the percentage values are indicated at nodes.
Supplementary Table S1 | Protein-encoding gene annotations.
Supplementary Table S2 | Primers used in this study.
References
Abouelhassan, Y., Garrison, A. T., Yang, H., Chavez-Riveros, A., Burch, G. M., and Huigens, R. W. 3rd. (2019). Recent progress in natural-product-inspired programs aimed to address antibiotic resistance and tolerance. J. Med. Chem. 62, 7618–7642. doi: 10.1021/acs.jmedchem.9b00370
Anand, S., and Srivastava, P. (2020). Optimization strategies for purification of mycophenolic acid produced by Penicillium brevicompactum. Appl. Biochem. Biotechnol. 191, 867–880. doi: 10.1007/s12010-019-03204-w
Ancheeva, E., Mandi, A., Kiraly, S. B., Kurtan, T., Hartmann, R., Akone, S. H., et al. (2018). Chaetolines A and B, pyrano[3,2-f]isoquinoline alkaloids from cultivation of Chaetomium sp. in the presence of autoclaved Pseudomonas aeruginosa. J. Nat. Prod. 81, 2392–2398. doi: 10.1021/acs.jnatprod.8b00373
Baccile, J. A., Spraker, J. E., Le, H. H., Brandenburger, E., Gomez, C., Bok, J. W., et al. (2016). Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat. Chem. Biol. 12, 419–424. doi: 10.1038/nchembio.2061
Blanc, P. J., Laussac, J. P., Le Bars, J., Le Bars, P., Loret, M. O., Pareilleux, A., et al. (1995). Characterization of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 27, 201–213. doi: 10.1016/0168-1605(94)00167-5
Clinical and Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute (CLSI). Twenty-Second Informational Supplement. M100-S22.
He, Y., and Cox, R. J. (2016). The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 7, 2119–2127. doi: 10.1039/C5SC04027B
Kalinina, S. A., Kalinin, D. V., Hovelmann, Y., Daniliuc, C. G., Muck-Lichtenfeld, C., Cramer, B., et al. (2018). Auranthine, a benzodiazepinone from Penicillium aurantiogriseum: refined structure, absolute configuration, and cytotoxicity. J. Nat. Prod. 81, 2177–2186. doi: 10.1021/acs.jnatprod.8b00187
Keller, N. P. (2015). Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 11, 671–677. doi: 10.1038/nchembio.1897
Khalid, S., Baccile, J. A., Spraker, J. E., Tannous, J., Imran, M., Schroeder, F. C., et al. (2018). NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem. Biol. 13, 171–179. doi: 10.1021/acschembio.7b00731
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, S. Y., Kotapati, S., Kuti, J. L., Nightingale, C. H., and Nicolau, D. P. (2006). Impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species on clinical outcomes and hospital costs: a matched cohort study. Infect. Control Hosp. Epidemiol. 27, 1226–1232. doi: 10.1086/507962
Li, Y., Chooi, Y. H., Sheng, Y., Valentine, J. S., and Tang, Y. (2011). Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 133, 15773–15785. doi: 10.1021/ja206906d
Li, C., Sarotti, A. M., Wu, X., Yang, B., Turkson, J., Chen, Y., et al. (2019). An unusual benzoisoquinoline-9-one derivative and other related compounds with antiproliferative activity from hawaiian endophytic fungus Peyronellaea sp. FT431. Molecules 24:196. doi: 10.3390/molecules24010196
Liu, Y., Li, D., Jiang, Q., Zhang, Q., Liu, P., Wang, L., et al. (2019). (3R, 7R)-7-acetoxyl-9-oxo-de-O-methyllasiodiplodin, a secondary metabolite of Penicillium sp., inhibits LPS-mediated inflammation in RAW 264.7 macrophages through blocking ERK/MAPKs and NF-kappaB signaling pathways. Inflammation 42, 1463–1473. doi: 10.1007/s10753-019-01009-x
Manu, P., and Rogozea, L. (2016). The discovery of statins. Am. J. Ther. 23, e980–e981. doi: 10.1097/MJT.0000000000000467
Mayorga, M. E., and Timberlake, W. E. (1992). The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235, 205–212. doi: 10.1007/BF00279362
Nielsen, J. C., Grijseels, S., Prigent, S., Ji, B., Dainat, J., Nielsen, K. F., et al. (2017). Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2:17044. doi: 10.1038/nmicrobiol.2017.44
Nord, C., Levenfors, J. J., Bjerketorp, J., Sahlberg, C., Guss, B., Oberg, B., et al. (2019). Antibacterial isoquinoline alkaloids from the fungus Penicillium spathulatum Em19. Molecules 24:4616. doi: 10.3390/molecules24244616
Peirano, G., and Pitout, J. D. D. (2019). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Penalva, M. A., Rowlands, R. T., and Turner, G. (1998). The optimization of penicillin biosynthesis in fungi. Trends Biotechnol. 16, 483–489. doi: 10.1016/S0167-7799(98)01229-3
Salo, O., Guzman-Chavez, F., Ries, M. I., Lankhorst, P. P., Bovenberg, R. A. L., Vreeken, R. J., et al. (2016). Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl. Environ. Microbiol. 82, 3971–3978. doi: 10.1128/AEM.00350-16
Sato, F., Inui, T., and Takemura, T. (2007). Metabolic engineering in isoquinoline alkaloid biosynthesis. Curr. Pharm. Biotechnol. 8, 211–218. doi: 10.2174/138920107781387438
Yao, G., Li, Z., Gao, L., Wu, R., Kan, Q., Liu, G., et al. (2015). Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum. Biotechnol. Biofuels 8:71. doi: 10.1186/s13068-015-0253-8
Keywords: isoquinoline alkaloids, anti-bacterial activities, coral-derived fungus, genome sequencing, Penicillium steckii
Citation: Yao G, Chen X, Zheng H, Liao D, Yu Z, Wang Z and Chen J (2021) Genomic and Chemical Investigation of Bioactive Secondary Metabolites From a Marine-Derived Fungus Penicillium steckii P2648. Front. Microbiol. 12:600991. doi: 10.3389/fmicb.2021.600991
Edited by:
Jinwei Zhang, University of Exeter, United KingdomReviewed by:
Lijuan Liao, Fujian Agriculture and Forestry University, ChinaPeng Zhang, Chinese Academy of Agricultural Sciences (CAAS), China
Lu Liu, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Yao, Chen, Zheng, Liao, Yu, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangshan Yao, MjYxNkBtanUuZWR1LmNu; Jianming Chen, Y2hlbmptQHRpby5vcmcuY24=
 Guangshan Yao
Guangshan Yao Xiaofeng Chen1
Xiaofeng Chen1 Huawei Zheng
Huawei Zheng Zhi Yu
Zhi Yu Zonghua Wang
Zonghua Wang