- 1PhD Degree Program in Environmental Science, Environmental Science Research Center, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 2Department of Environmental Science, Faculty of Science and Technology, Chiang Mai Rajabhat University, Chiang Mai, Thailand
- 3Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 4Research Center in Bioresources for Agriculture, Industry and Medicine, Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
Thailand is an agricultural country. However, agricultural productivity relies on the heavy use of herbicides, especially paraquat. Paraquat accumulation is emerging as a problem in an ever-growing portion of agricultural land. Paraquat residues are toxic to plants, animals, and aquatic organisms in the environment. Biological remediation is a process that can mitigate agricultural chemical contaminants. One of the interesting bioremediators is bacteria. Not only do certain soil bacteria remediate paraquat, but some of them also possess plant growth-promoting properties, which provide advantages in field application. Thus, this study aimed to screen soil bacteria that could degrade paraquat and, at the same time, promote plant growth. Bacteria were isolated from paraquat-treated agricultural soil in Mueang Kaen Pattana municipality, Chiang Mai province, Thailand. On the basis of morphological and 16S rDNA sequence analyses, the selected bacterium was identified as Bacillus aryabhattai strain MoB09. It is capable of growing in nitrogen-free media. B. aryabhattai growth and paraquat degradation were found to be optimum at pH 7 and 30°C. This selected strain also possessed plant growth-promoting abilities, including indole production, siderophore production, phosphate solubilization, and 1-aminocyclopropane-1-carboxylic acid deaminase activity. Paraquat degradation was also evaluated in pot experiments of cowpea (Vigna unguiculata). It was found that this strain could remediate the paraquat residue in both sterilized and non-sterilized soils. The cowpea plants grown in paraquat-contaminated soil with B. aryabhattai showed longer root and shoot lengths than those grown in soil without bacterial inoculation. In addition, B. aryabhattai also promoted the growth of cowpea under induced drought stress. These results suggested that B. aryabhattai could be applied to mitigate paraquat residue in soil and also to promote plant productivity for the organic crop production.
Introduction
Vegetables have long been consumed worldwide, and the trend for vegetable consumption as clean and healthy food is currently increasing. However, vegetable production relies heavily on the use of pesticides for crop protection, increase in production yields, and product quality improvement as well as product appearance. Cowpea (Vigna unguiculata) is an annual herbaceous legume from the genus Vigna. Due to its tolerance for sandy soil and low rainfall, it is an important crop in the semiarid regions across Africa and Asia. In addition, cowpea has been reported to be highly sensitive to water and herbicide stress (Oyewole et al., 2017; Silva et al., 2020).
The global uses of herbicides, chemical compounds that are toxic to plants and aim for the elimination of unwanted weeds, have been increasing significantly over time (Gianessi, 2013). Paraquat (1,1-dimethyl-4,4-bipyridyl dichloride) is a herbicide that has been widely used in agriculture for more than 60 years, and but at the present, it has been banned or disallowed in many countries (Bromilow, 2004). It is a non-selective herbicide that diverges electron flow from the photosystem, resulting in the production of lethally reactive free radicals, superoxide radicals, which damage plant tissues (Reczek et al., 2017). Paraquat may be classified as immobile herbicides according to its strong adsorption on soil particles. However, inappropriate use of paraquat, for instance, repeated and excessive (beyond the recommended concentration) use, over the years may cause an accumulation of paraquat in the environment. Environmental contaminations of paraquat have been reported over the world, for example, Indonesia (Ardiwinata et al., 2019), Brazil (Veríssimo et al., 2018), and Thailand (Teerakun et al., 2020). Although the concentrations of paraquat contamination in some areas are still below the tolerance limit, long-term exposure to those paraquat residues might result in harmful effects on humans and mammals and the food chain (Huang et al., 2019).
Several processes have been reported for a removal of those residues, e.g., biodegradation, photodegradation, oxidation, flocculation, filtration, and adsorption. However, biological remediation, the so-called biodegradation, is a more economical and environment-friendly approach. Although naturally occurring soil microbes can alter those residual chemicals, this process generally takes time to achieve. Depending upon the texture and soil composition, natural microorganisms can only remediate less than 1% of residual paraquat in the soil (Roberts et al., 2002). Thus, it is necessary to enhance paraquat-degrading microorganisms for restoration of paraquat-contaminated sites. Many bacteria capable of degrading pesticides have been isolated and identified such as Bacillus, Pseudomonas, Flavobacterium, Arthrobacter, Diaphorobacter, Klebsiella, Ochrobactrum, Agrobacterium, and others (Ataikiru et al., 2020).
Microorganisms also perform an important role in supplying nutrients to plants and reducing the demand of chemical fertilizers (Cakmakci et al., 2006). It is commonly known that microorganisms are capable of plant growth regulator productions such as auxins, gibberellins, siderophores, phosphate-solubilizing substances (Khalid et al., 2009), and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity (Penrose and Glick, 2003). In addition, the use of bacteria as inoculants simultaneously increased phosphate uptake by the plant and increased crop yields. Many bacterial strains from the genera Pseudomonas, Bacillus, Rhizobium, Burkholderia, Achromobacter, Agrobacterium, Micrococcus, Enterobacter, Flavobacterium, and Erwinia have been known as the producer of the powerful phosphate solubilizers (Esitken et al., 2003). The mechanism for mineral phosphate solubilization is the production of organic acids and acid phosphatases. Indole-3-acetic acid (IAA) is one of the most physiologically active plant growth regulators produced from L-tryptophan metabolism by soil fungi and bacteria (Muhammad and Frankenberger, 1991). Siderophores are the important compounds for the survival and growth of bacteria in the soil and in aqueous environments. It has been reported that siderophore mediated iron transport found mostly in Gram-negative bacteria and also in Gram-positive bacteria such as Bacillus, Staphylococcus, and Streptomyces. However, siderophores and their substituted derivative structures varied from one species to another (Kannahi and Senbagam, 2014). Besides, it was found that Pantoea ananatis, Pseudomonas putida, Brevibacillus agri, Bacillus subtilis, and Bacillus megaterium were growth boosters for the plant (endophyte) as well as the area of rooting (rhizosphere). Some Bacillus species also possess both plant growth-regulating and herbicide-degrading properties. For example, Bacillus sp. ACD-9 could degrade acetochlor and show phosphate-solubilizing activity on maize seedlings (Li et al., 2020).
Other than pollutants remaining in soil, environmental factors could also generate various stresses, which affect plant growth and development. Environmental factors such as drought and salinity are of interest. Their effects on crop yield and quality for many plants have been reported (Salehi-Lisar and Bakhshayeshan-Agdam, 2016). Interestingly, some bacteria showed dual-plant-benefit properties, both growth promotion and stress protection, such as Bacillus fortis strain SSB21, which could promote plant growth and contains osmolyte production to improve salinity tolerance in Capsicum annum L. (Yasin et al., 2018). Bacillus methylotrophicus CSY-F1 produces plant growth-promoting substances and alleviates drought stress in cucumber (Cucumis sativus) grown in soil with high ferulic acid (Hou et al., 2018). Bacillus aryabhattai SRB02 could promote the growth and heat resistance of soybean by production of phytohormones (Park et al., 2017).
These described bacteria could play vital roles as bio-fertilizers, bio-stimulants, bio-protectants, and biodegradators (Andriani et al., 2017). However, to our knowledge, multi-plant growth-promoting, drought-protective, and paraquat-degrading abilities from individual bacteria have yet been observed. Thus, this study aims for the discovery of a reasonable potential strain and its optimal culture condition, which could be acclimatized to the target polluted sites and enhance the bioremediation process and increase crop production.
Materials and Methods
Chemicals Used
Paraquat dichloride hydrate standard was obtained from Sigma-Aldrich, Merck, Germany. Solvents for HPLC were purchased from SM Chemical Supplies Co., Ltd. All chemicals were of the highest purity that is commercially available.
Isolation and Identification of Paraquat-Degrading Bacteria
Soil specimens were collected from an agricultural area in Mueang Kaen Pattana municipality, Chiang Mai province, Thailand. This area is under heavy chemical use, including paraquat. One kilogram of the soil samples was collected from the first 10–15 cm depth, pooled, and sieved. Samples were air-dried and stored in sterile plastic bags at 4°C until use. The paraquat-degrading bacteria were screened for molybdenum-reducing ability and for paraquat utilization capacity.
Screening by Molybdenum-Reducing Method
Ten grams of the soil samples were firstly suspended in sterile tap water, and 0.1 ml of the suspended soil was transferred to low phosphate molybdate agar (LPMA) (pH 7.0) containing glucose (1%), (NH4)2SO4 (0.3%), MgSO4⋅7H2O (0.05%), NaCl (0.5%), yeast extract (0.05%), Na2MoO4⋅2H2O (0.242%) Na2HPO4 (0.05%), and agar (1.5%) (Yunus et al., 2009) and further incubated at room temperature for 48 h. The presence of blue colonies indicates the presence of molybdenum-reducing bacteria, which might have paraquat-degrading properties. The intense blue colonies were repeatedly streaked on low phosphate media (LPM) agar to obtain the axenic bacterial culture. Molybdenum-reducing bacterial strains were collected for further colony identification.
Screening by Enrichment Method
For the isolation of bacteria using paraquat as the sole carbon and energy source, mineral salt medium (MSM) (pH 7.0), consisting of 2 g of (NH4)2SO4 (0.2%), KH2PO4 (0.15%), Na2HPO4 (0.15%), MgSO4⋅7H2O (0.02%), CaCl2⋅2H2O (0.001%), and FeSO4⋅7H2O (0.0001%), was used (Singh and Walker, 2006). Paraquat was added in MSM for enrichment. Two grams of each soil sample was added to 20 ml of MSM containing 0.5 g/L paraquat and incubated in the dark at 30°C under a shaking condition (150 rpm), for 7 days. Five milliliters of these suspensions was then transferred to fresh MSM containing 1 g/L paraquat and further incubated for 7 days. The plates were incubated at 37°C for 24 h. Colonies were collected for further colony purification.
Screening for Paraquat Degradation of the Isolated Bacterial Strains
Paraquat degradation of the isolated bacterial strains was performed in a 250-ml tube containing 100 ml of nutrient broth (NB medium) (pH 7.0), consisting of peptone (0.5%), yeast extract (0.3%), and 0.5% NaCl. The tubes were then incubated for 7 days, under continuous stirring on a rotary shaker at 150 rpm. Bacterial growth was followed by taking a sample of 2 ml of cultures after every 24 h until 72 h of incubation. The paraquat concentration was evaluated spectrophotometrically based on the AOAC Official Method 969.09 (Tsai and Chen, 2013). One milliliter of each standard concertation and sample was mixed with 1 ml of sodium dithionite (0.04%) in 1 M NaOH; then the mixture was diluted to 10 ml in a volumetric flask. The absorbance at 600 nm was immediately measured by using a spectrophotometer.
Plant Growth Promotion of the Isolated Bacterial Strains
The isolated bacteria were cultured for the measurement of plant growth-promoting substances such as IAA, siderophore, phosphate solubilization, and ACC deaminase activity.
IAA Production
Indole-3-acetic acid production of the isolated bacteria was determined spectrophotometrically as described previously (Lasudee et al., 2018). A 5-mm-diameter agar plug of bacteria-grown nutrient agar was transferred into 5 ml of nutrient broth containing 2 mg/ml of L-tryptophan (Gordon and Weber, 1951). Tubes were incubated at 28 ± 0.2°C with continuous shaking at 110 rpm for 5 days. The supernatant was collected by centrifugation at 11,000 rpm for 5 min. IAA concentration was quantified by mixing 1 ml of the supernatant with 2 ml of Salkowski’s reagent (Glickmann and Dessaux, 1995). The mixer was then incubated in the dark. A pink color indicated IAA production, which was measured for optical density at 530 nm using a spectrophotometer. The IAA concentration was approximated based on a calibration curve of pure IAA standard.
Siderophore Production
Siderophore production of all isolate bacteria was detected by chrome azurol S (CAS) assay (Schwyn and Neilands, 1987). A 5-mm-diameter agar plug of bacteria-grown nutrient agar was placed on CAS agar and incubated for 7 days at 28 ± 0.2°C in a dark room. An orange zone around the agar plugs indicated positive siderophore production.
Phosphate-Solubilizing Activity
Phosphate-solubilizing activity of all isolate bacteria was measured on Pikovskaya (PVK) agar (Pikovskaya, 1948), containing tricalcium phosphate (0.5%). A 5-mm-diameter agar plug of bacteria-grown nutrient agar was inoculated to PVK agar plates and incubated at 28 ± 0.2°C for 7 days. The visibility of a clear zone around the agar plug showed positive phosphate-solubilizing activity (Nautiyal, 1999).
ACC Deaminase Activity
1-aminocyclopropane-1-carboxylic acid deaminase activity was detected by using the method reported by Palaniyandi et al. (2014). The B. aryabhattai strain MoB09 was grown on minimal media (Dworkin and Foster, 1958) without a nitrogen source (negative control), minimal media supplemented with ammonium sulfate (positive control), and minimal media supplemented with ACC at a final concentration of 3 mmol/L. All agar plates were incubated at 30°C for 7 days. Growth on the additional ACC medium showed positive ACC deaminase activity.
Characterization and Identification of the Isolated Bacterial Strains
The selected bacterial isolates were identified by the 16S rRNA gene sequence analysis. Genomic DNA was extracted using a TIANamp Bacteria DNA Kit (Tiangen) according to the manufacturer’s instructions. The 16S rRNA gene from genomic DNA was amplified by polymerase chain reaction (PCR) using bacterial universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The PCR was carried out in an MJ Mini Personal Thermal Cycler (Bio-Rad) in 50 μl reactions containing 10 μl of 5 × Phusion Green HF buffer (Thermo Scientific), 1 μl of 10 mM DNTP (Bioline), 1.0 μl of 10 mM primer 27f, 1.0 μl of 10 mM primer 1492R, 1.5 μl of DMSO, 0.5 μl of 2 U/μl Phusion Hot Start II DNA polymerase (Thermo Scientific), and approximately 100–250 ng of template DNA. PCR was carried out at an initial denaturation step at 98°C for 30 s, followed by 30 cycles at 98°C for 7 s, 50°C for 20 s, and 72°C for 45 s and a final extension step at 72°C for 7 min. The amplicons were purified with the Universal DNA Purification Kit (Tiangen) and sequenced in both directions by using the same primers. The sequences were compared by aligning the result with the sequences in GenBank using the Basic Local Alignment Search Tool (BLAST) search program at the National Center for Biotech Information (NCBI), with the closely related Bacillus strains based on 16S rRNA gene sequences. Phylogenetic analysis was performed based on the neighbor-joining method with MEGA X software. The sequence identity matrix analysis was performed with BioLign software ver. 4.0.6.2 (Lau et al., 2020).
Optimization of Paraquat Degradation
To optimize paraquat degradation, some important abiotic factors were chosen according to Ouided and Abderrahmane (2013). The optimum pH level was tested at 5.0, 6.0, 7.0, 8.0, and 9.0, and the optimum temperature was observed at 30, 37, and 40°C. All treatments were performed in a 250-ml tube containing 100 ml of NB medium. Utilization of paraquat as a sole carbon source was performed in a 250-ml tube containing 100 ml of MSM, pH 7.0 and 30°C. The tubes were then incubated for 7 days, under continuous stirring on a rotary shaker at 150 rpm. Bacterial growth was followed by taking a sample of 2 ml of cultures after every 24 h until 72 h of incubation. The remaining paraquat was measured by HPLC according to the methods of Wahyu (2013). The HPLC system used for analyzing paraquat was Agilent HPLC 1260 Infinity II with diode array detection (DAD) (Agilent). Analytical column was a C18 column (4.6 × 150 mm) from Vertical. The mobile phase consisted of 5 g of NaCl in 600 ml distilled water, where the pH level was pre-adjusted to 3.0 with HCl, and then the solution was mixed with 400 ml of acetonitrile. Flow rate of the mobile phase was fixed at 1.0 ml/m. DAD wavelength was 257 nm. Twenty microliters each of paraquat standard and sample was injected into the chromatographic column by an autosampler system. Chromatograms were recorded, and the peak area was quantitatively measured with a computer software from the manufacturer.
Paraquat Degradation and Growth Promotion of Cowpea (V. unguiculata)
Paraquat degradation in pot experiments with cowpea (V. unguiculata) was conducted in order to examine the effects of bacterial inoculation on plant growth and paraquat degradation. Soil samples (1.5 kg) were spiked with paraquat to a concentration of 200 mg/kg. Cowpea (V. unguiculata) was used as a representative of dicotyledon plants. Cowpea (V. unguiculata) seeds (Chia Tai Brand, Chia Tai Company Limited, Thailand) were surface sterilized by being immersed sequentially in 2% (v/v) sodium hypochlorite for 1 min, 95% (v/v) ethanol for 1 min, and 70% (v/v) ethanol for 1 min and then washed with sterile distilled water for 1 min; these steps were repeated three times (Lasudee et al., 2018). Five decontaminated seeds were randomly selected to check for surface sterility on nutrient agar. Cell suspension was prepared from the selected strain grown in nutrient broth. Surface-sterilized seeds were mixed with 108 CFU/ml of the bacteria and incubated on a shaker at 120 rpm at 30°C for 16–18 h before being sown. Seedlings were watered once a day with sterile distilled water for 7 days at room temperature. Seven-day-old seedlings with two leaves were transferred to a pot.
The experiment was carried out in a pot (15-cm diameter × 15-cm height) containing 1.5 kg of soil and being kept in a closed room at room temperature for 6 weeks, with five replicates per treatment. Pots were arranged in a completely randomized arrangement in a room. Tap water was supplied once a day to full container capacity and kept at 1 cm above soil level. The following treatments were investigated: (1) sterilized soil with paraquat, (2) sterilized soil with paraquat and seeds mixed with bacterial cell suspension, (3) non-sterilized soil, (4) non-sterilized soil with paraquat, and (5) non-sterilized soil with paraquat and seeds mixed with bacterial cell suspension. At the end of the experiment (6 weeks after sowing), the following growth parameters were recorded: fresh weight (g), dry weight (g), root length (cm), total length (cm), leaf length (cm), and paraquat residue in soil, which was extracted and estimated by LC-MS (In house method, 1997).
Growth Promotion of Cowpea (V. unguiculata) in Drought Conditions
Growth promotion of cowpea (V. unguiculata) in drought conditions and under well-watered conditions of the selected strain was performed in a pot experiment using the same protocol as described above. The treatments for drought tolerance promotion included (1) drought-induced treatment, (2) drought-induced treatment with seeds mixed with bacterial cell suspension, (3) well-watered conditions, and (4) well-watered conditions with seeds mixed with bacterial cell suspension. Drought stress was induced by completely withholding water starting on 35 for 10 days. The water deficit treatment was imposed through withholding water from 35 to 45 days after emergence. At the end of the experiment (day 45 after sowing), the maximum water stress was reached after 10 days of treatment; the following growth parameters were recorded: fresh weight (g), dry weight (g), root length (cm), total length (cm), leaf length (cm), chlorophyll content (mg/g dry weight), and proline content (mmol/dry weight). Proline content was determined by the rapid colorimetric method (Bates et al., 1973). Chlorophyll content was determined spectrophotometrically based on the standard method of Arnon (1949).
Results
Screening and Identifying the Paraquat-Degrading Bacteria
According to the variety of bacteria in a soil, it is difficult to isolate desired bacteria from a single method. In this study, paraquat-degrading bacteria were isolated from two methods: molybdenum-reducing ability using LPMA and paraquat utilization capacity using MSM containing paraquat. It was found that 12 morphologically different isolates were selected from LPMA and assigned as MoB strains. The blue colonies on LPMA indicated molybdenum-reducing ability (Figure 1). For screening with MSM containing paraquat, 10 different morphological colonies were selected and assigned as MSM strains. Colony morphology, Gram staining, and light compound microphotographs of those isolates were shown in the Supplementary Material. The 22 selected isolates from both screening methods were then examined for their paraquat degradation.
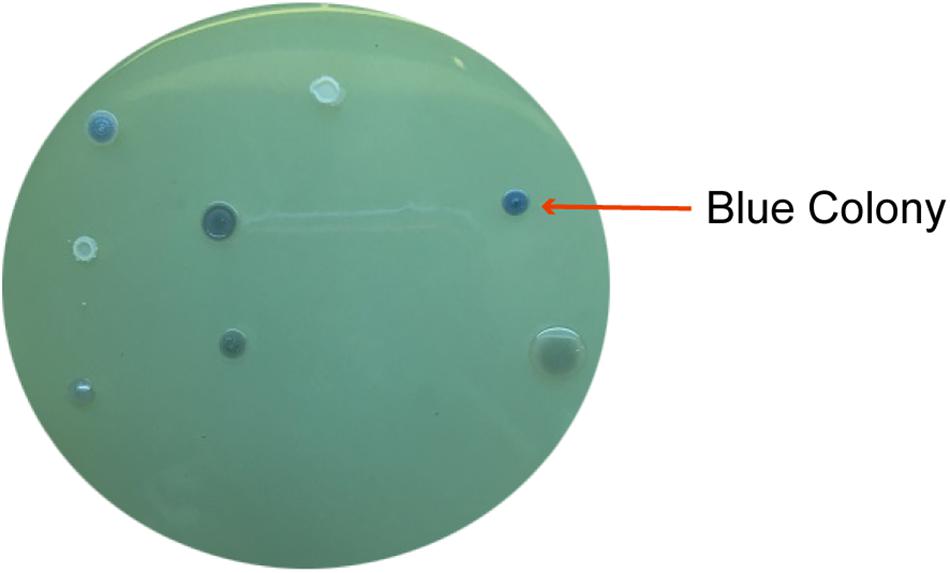
Figure 1. Several blue colonies on LPMA indicated the molybdenum-reducing ability of the isolated bacteria.
Figure 2 shows that all the selected isolates could degrade paraquat in a range of approximately 10–45%. The highest paraquat-degrading ability was found in isolate MoB09, which was from the screening by the molybdenum-reducing method, followed by MoB01 and MSM02. A previous study reported that molybdenum-reducing microorganisms showed multiple detoxification capacities; for example, Klebsiella oxytoca strain SAW-5 could detoxify Mo and degrade glyphosate (Sabullah et al., 2016). Molybdenum-reducing Bacillus sp. strain Zeid 14 and Burkholderia sp. strain NENI-11 could utilize acrylamide as a source of electron donor for reduction and were able to grow on media containing acrylamide, acetamide, and acetonitrile (Mansur et al., 2016; Mohd et al., 2016). Enterobacter sp. strain Saw-1 and Bacillus sp. strain Neni-12 could consume the pesticide coumaphos as an alternative carbon source for growth (Sabullah et al., 2016; Rusnam and Gusmanizar, 2019). However, a previous study found that although some molybdenum-reducing bacteria can successfully utilize pesticides as a carbon source, those pesticides cannot be used as an electron donor for molybdenum reduction (Sabullah et al., 2016). Thus, screening by an enrichment method was also involved in this study. Our result, similar to those of previous studies, revealed that the enrichment method could aid successfully in the screening of xenobiotics-degrading isolates, for example, carbofuran- and paraquat-degrading bacteria (Ataikiru et al., 2020), glyphosate-degrading bacteria (Moneke et al., 2010), chlorpyrifos-degrading bacteria (Parmar et al., 2018), and organophosphorus insecticide diazinon-degrading bacteria (Cycoń et al., 2009). The enrichment method could assist in isolating pesticide-degrading strains and events from a non-pesticide impacted area (Ataikiru et al., 2020). Consequently, screening from various methods may enhance success of the desired potential strains.
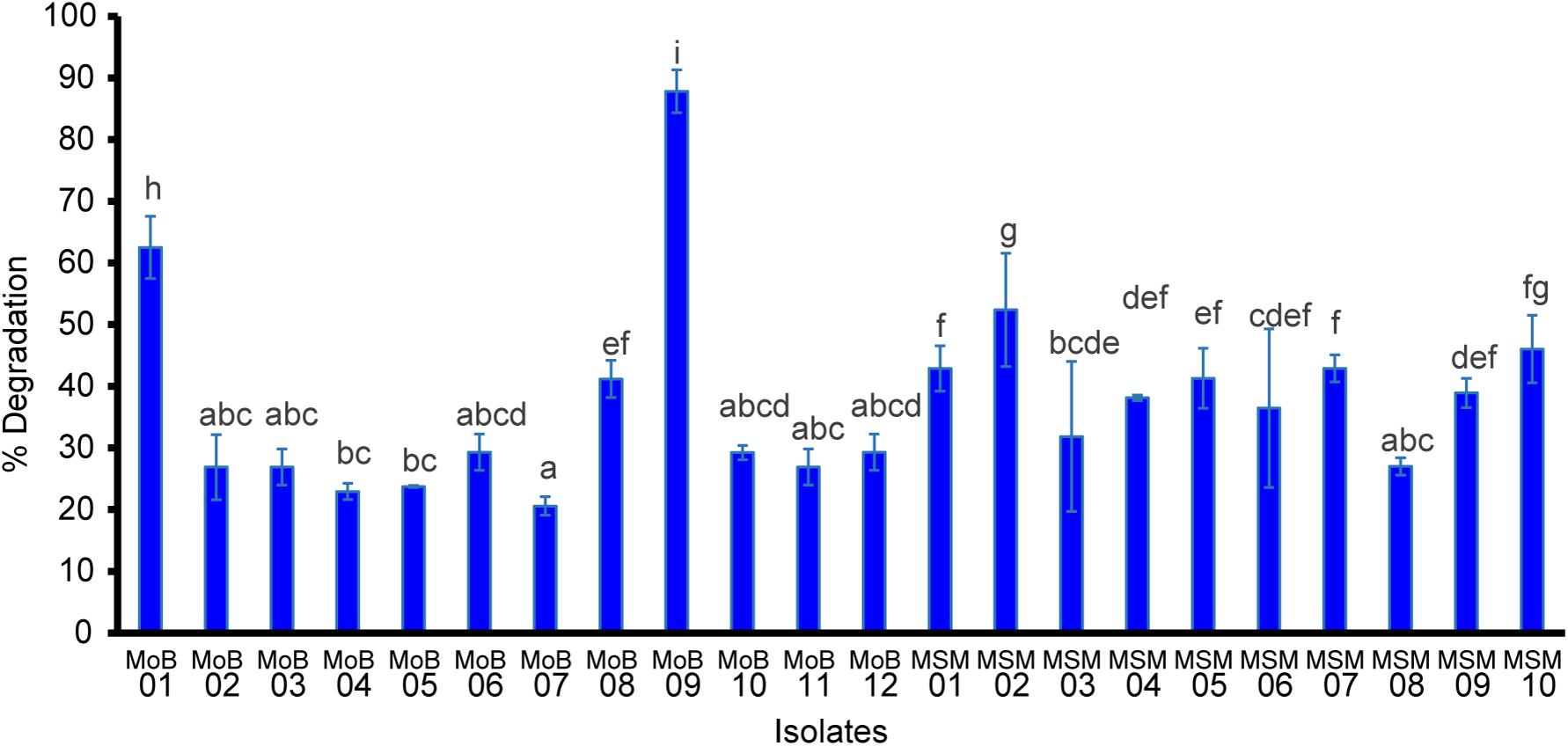
Figure 2. Paraquat degradation of the selected isolates. Data were presented as mean ± SD of three replicas. Letters indicate a significant difference (p < 0.05) using ANOVA with post hoc analysis.
In Vitro Plant Growth-Promoting Potential
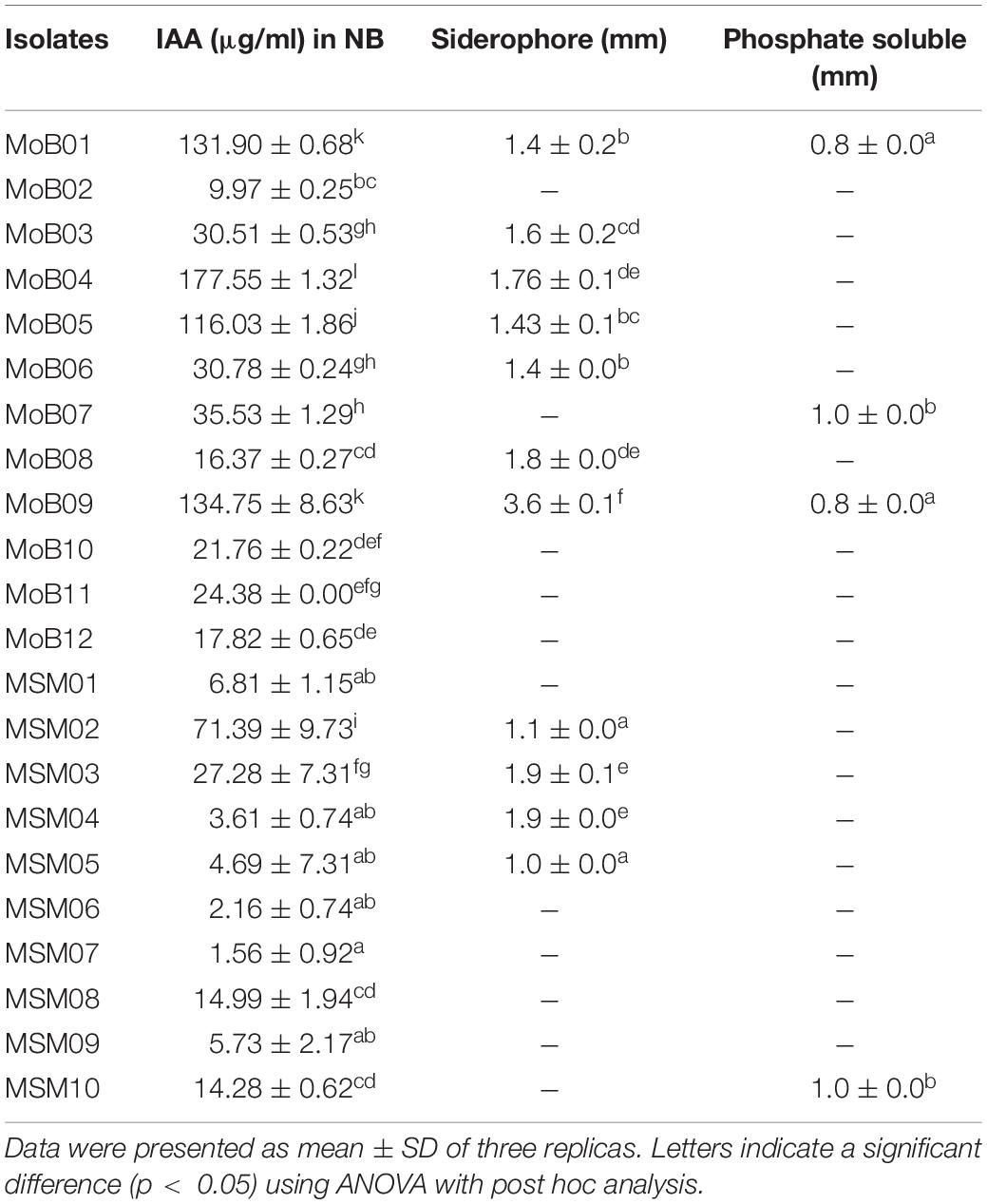
The B. aryabhattai strain MoB09 showed growth-promoting substance production comparable to that of the “Bacillus” group (Table 1). Previous studies reported that bacteria in the “Bacillus” group can produce plant growth-promoting substances. For instance, B. aryabhattai SRB02 produced 1.5 ng/100 ml of cytokinin, 2 ng/100 ml of abscisic acid, 2.25 ng/100 ml of gibberellins, and 5.7 μg/ml of IAA and induced growth promotion of soybeans (Park et al., 2017). B. megaterium STB1 produced IAA and cytokinins, which can promote the growth of tomatoes (Francisco et al., 2020). Bacillus amyloliquefaciens UCMB5113 produced 2.3 ± 0.1 μg/mL of IAA, which can promote Arabidopsis thaliana growth by auxin, gibberellin, cytokinin, and brassinosteroid production (Shashidar et al., 2017). B. amyloliquefaciens FZB45 produced 2.3 ± 0.1 μg/ml of IAA (Camilo and Joseph, 2010), while B. amyloliquefaciens HYD-B17, Bacillus licheniformis HYTAPB18, Bacillus thuringiensis HYDGRFB19, B. subtilis RMPB44 produced 7.2–32.5 g/mg of IAA. B. aryabhattai strain MoB09 also tested positive for gibberellin, cytokinin, P solubilization, and siderophore production. MoB09 tested positive for ACC deaminase activity (Supplementary Data). This strain can be grown on an ACC supplied medium, indicating that it can utilize ACC as a nitrogen source (Penrose and Glick, 2003). The ACC deaminase activity of strain MoB09 was added in the Supplementary Figure.
Almost complete 16S rRNA gene sequences of isolate MoB01 (1,400 bp) and isolate MoB09 (1,414 bp) were obtained and submitted to GenBank1 under accession numbers MT919336 and MT919337, respectively. Phylogenetic analysis based on the neighbor-joining method displayed that both MoB01 and MoB09 were clustered in members of the genus Bacillus (Figure 3). The sequence identity matrix analysis from BioLign software showed the highest similarity (0.999) of isolate MoB09 to B. aryabhattai strain pgB (MK519199), followed by (0.998) Bacillus zanthoxyli strain 1910ICU241 (MT225770) and (0.997) B. megaterium strain JYW3 (MN161199), respectively. Thus, MoB09 was identified as B. aryabhattai strain MoB09. Isolate MoB01 was closely related to Bacillus cereus strain ATCC14579 (AF290547), which was reported as an opportunistic pathogen causing food poisoning such as diarrheal or emetic syndromes (Ivanova et al., 2003). Therefore, only isolate MoB09 was selected for further experiments.
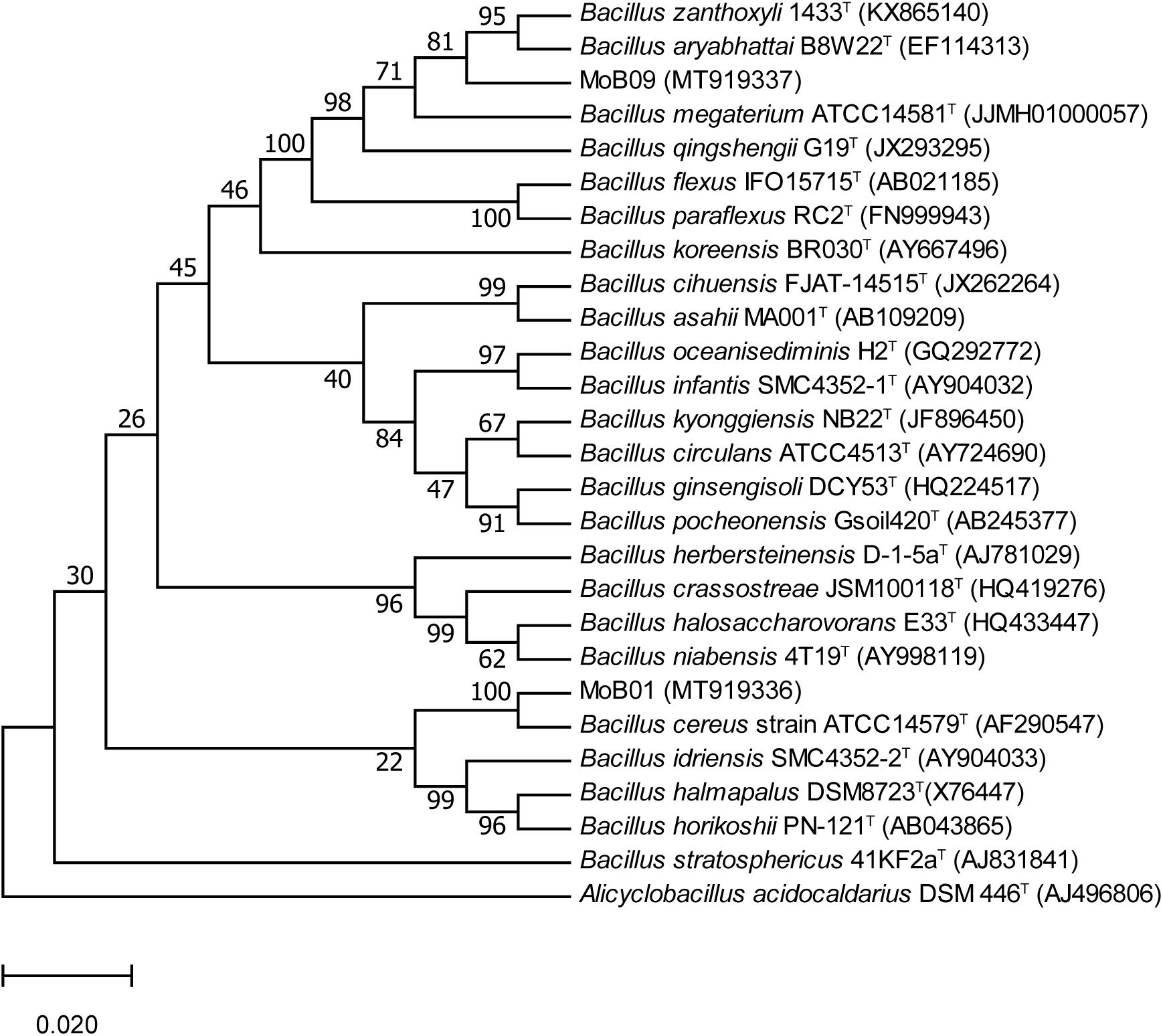
Figure 3. Phylogenetic tree of strains MoB01 and MoB09 with closely related Bacillus-type species based on 16S rRNA gene sequences, constructed using the neighbor-joining method. Bootstrap values (expressed as percentages of 1,000 replications) of above 50%.
Bacillus aryabhattai strain MoB09 performed comparable growth-promoting substance production to those of the “Bacillus” group. Previous studies reported that bacteria in the “Bacillus” group can produce plant growth-promoting substances such as B. aryabhattai SRB02, which produced 1.5 ng/100 ml of cytokinin, 2 ng/100 ml of abscisic acid, 2.25 ng/100 ml of gibberellins, and 5.7 μg/ml of IAA and promote soybean growth (Park et al., 2017). B. megaterium STB1 produced IAA and cytokinins and could promote growth of tomato (Francisco et al., 2020). B. amyloliquefaciens UCMB5113 produced 2.3 ± 0.1 μg/ml of IAA. It promoted A. thaliana growth by auxin, gibberellin, cytokinin, and brassinosteroid production (Shashidar et al., 2017). B. amyloliquefaciens FZB45 produced 2.3 ± 0.1 μg/ml of IAA (Camilo and Joseph, 2010), and B. amyloliquefaciens HYD-B17, B. licheniformis HYTAPB18, B. thuringiensis HYDGRFB19, and B. subtilis RMPB44 produced 7.2–32.5 g/mg protein of IAA. They were also positive for gibberellin, cytokinin, P solubilization, and siderophore production. In contrast, B. aryabhattai strain MoB09 produced 134.75 μg/ml of IAA and tested positive for siderophore production, phosphate solubilization, and ACC deaminase activity.
Biodegradation of Paraquat in Liquid Medium
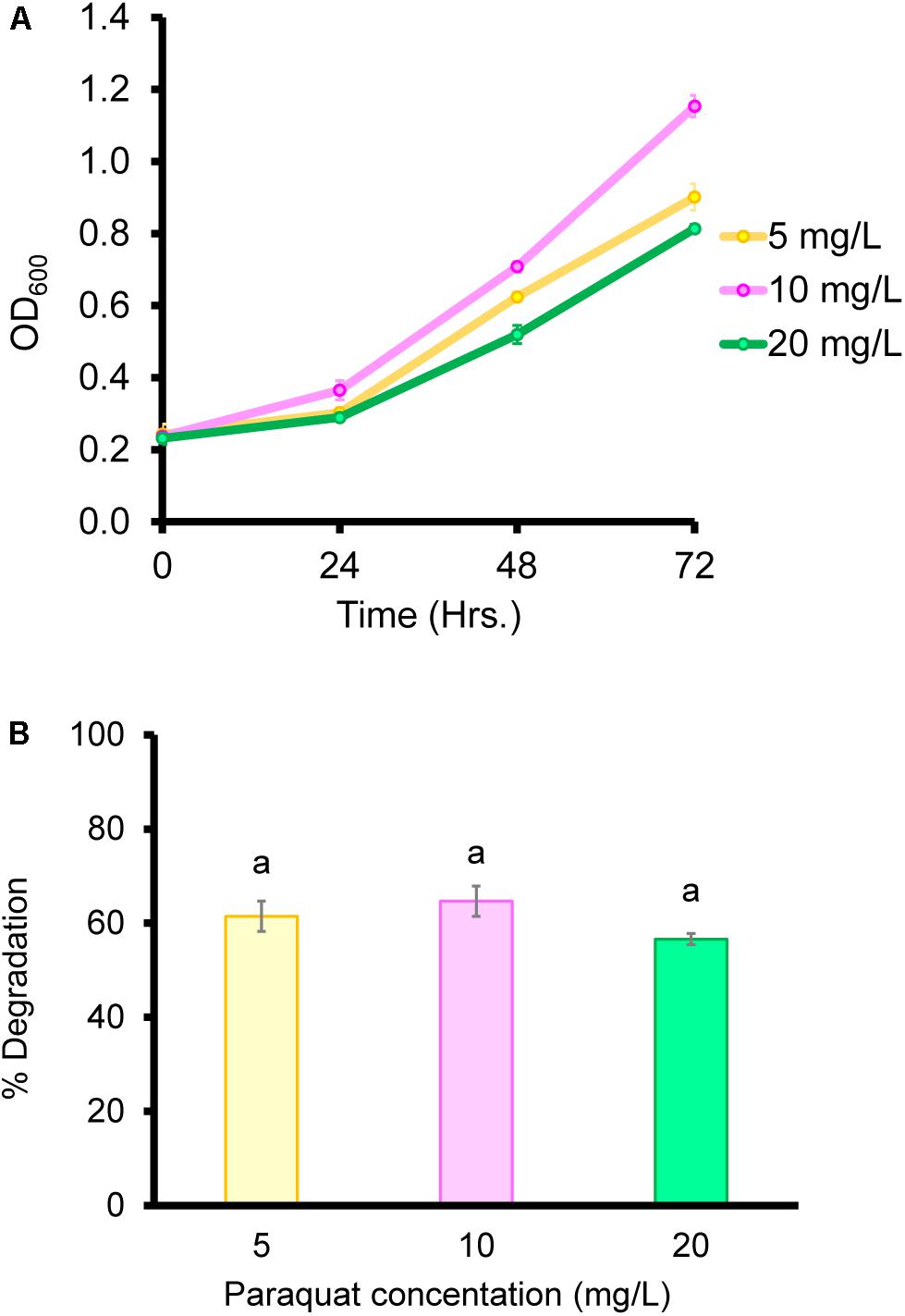
Biodegradation under controlled conditions is influenced by multiple factors such as temperature, pH, and pesticide concentration (Gangireddygari et al., 2017). In our study, the B. aryabhattai strain MoB09 showed maximum paraquat degradation at pH 7.0, 30°C and a paraquat concentration of less than 10 mg/L after 72 h of incubation (Figure 4). In addition, the B. aryabhattai strain MoB09 can utilize paraquat as a carbon source under controlled conditions (Figure 5).
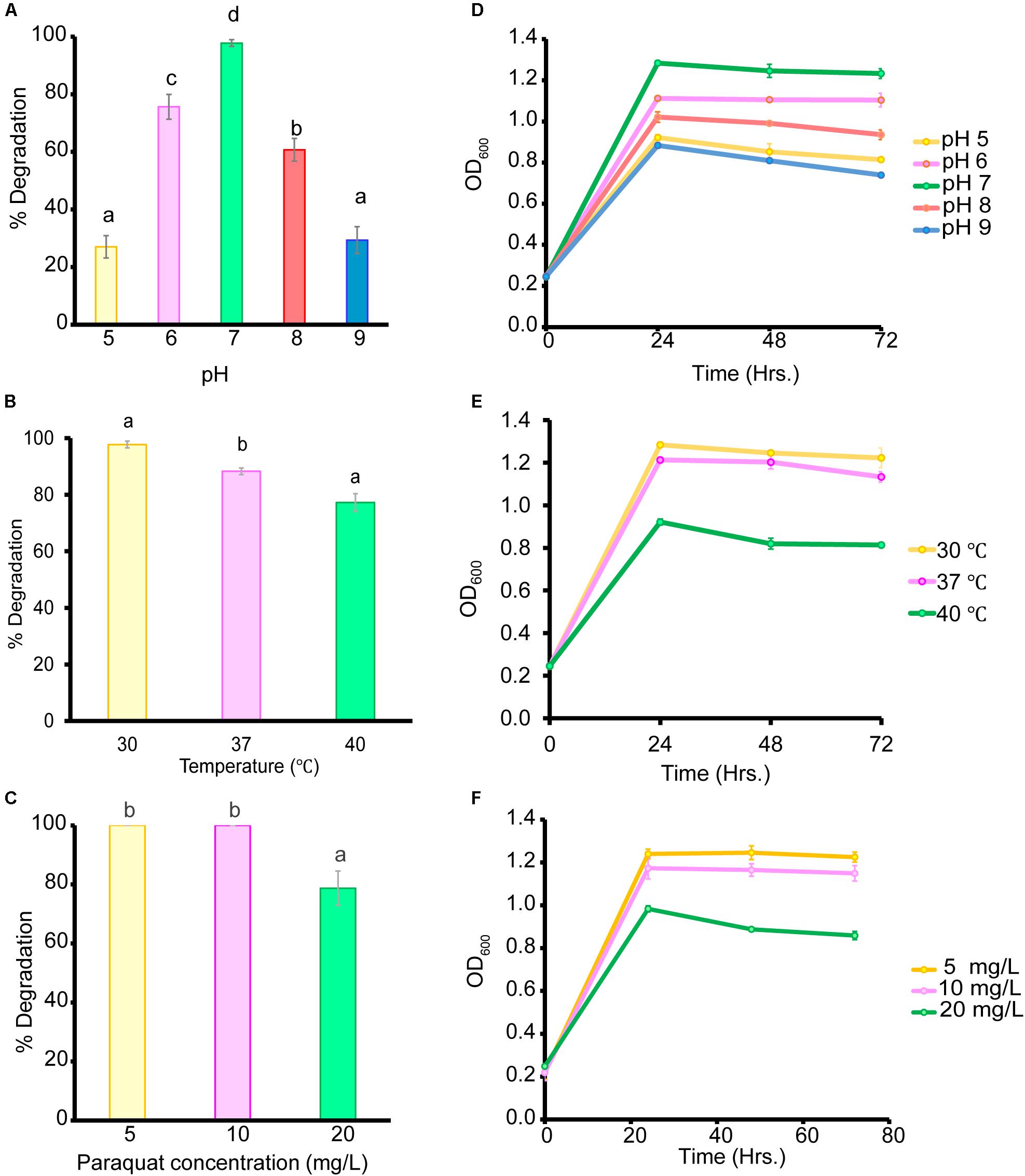
Figure 4. Effect of initial pH on paraquat degradation (A) and growth of bacteria (D), cultivation temperature on paraquat degradation (B), and growth of bacteria (E) and initial paraquat concentration on paraquat degradation (C) and growth of bacteria (F).
Interestingly, some Bacillus spp. have been reported for their paraquat degradation. For instance, B. cereus was able to utilize 10 mg/L of paraquat as the sole carbon and nitrogen sources in synthetic media (Tu and Bollen, 1968). The Bacillus altitudinis strain XT12 can degrade 40.58% of 5 μg/ml methyl viologen (Binghua et al., 2015). The genetic level of paraquat resistance was evaluated in B. subtilis. The study revealed that in the case of the sigM gene, encoding for extra cytoplasmic function factors plays an important role in bacterial resistance toward paraquat oxidative stress (Cao et al., 2005).
The genus Bacillus is well known for its xenobiotic degradation and plant growth promotion abilities. For example, B. megaterium strains CCT 7729 and CCT 7730 were found to degrade 49.7 and 62.5% of mesotrione in 14 h (Dobrzanski et al., 2018), B. thuringiensis strain SG4 was found to degrade 85.0% of cypermethrin in 15 days (Pankaj et al., 2020) and 90% of chlorpyrifos 3,5,6-trichloro-2-pyridinol in 8 days (Samina et al., 2009). Our study supports the reports from those previous studies. In the controlled condition, B. aryabhattai strain MoB09 showed potential for paraquat degradation. This isolate could degrade almost 100% of 10 mg/ml of paraquat in 72 h. A previous study revealed that bacteria can degrade paraquat in two ways. First, they use paraquat as the sole nitrogen or carbon source, and second, they transform paraquat into low-toxicity or non-toxic products through co-metabolism (Huang et al., 2019).
Even though the B. aryabhattai strain MoB09 can grow in minimal media by using paraquat as its sole carbon source, growth and paraquat degradation were lower than that in NB. Since this strain was isolated by the molybdenum-reducing method, the paraquat degradation of this strain may have occurred via metabolic transformation, which would require extra electron donors such as glucose (Wu et al., 2013). The addition of glucose can enhance degradation of paraquat in Achromobacter xylosoxidans and Streptomyces sp. (Hashemi et al., 2015). The impact of paraquat concentration on the growth of organisms was also previously reported. They found that the biomass of the microbes was negatively correlated with paraquat concentration (Andy et al., 2015), which coincides with the results in this study. Paraquat showed toxic effects; the highest OD600 was obtained from the lowest paraquat concentration of 5 mg/L, and the lowest OD600 from the highest paraquat concentration of 20 mg/L. A higher paraquat concentration of 100 mg/L was also evaluated, but the B. aryabhattai strain MoB09 could not survive (data not shown). Although the metabolic product from paraquat degradation in this study was not evaluated, the metabolic pathways of paraquat in microorganisms have been suggested elsewhere (Huang et al., 2019). However, the paraquat metabolic pathways of B. aryabhattai strain MoB09 may be different from those found in that study, which is interesting and should be investigated in future works.
Application of paraquat-degrading bacteria with plant growth-promoting abilities is like killing two birds with one stone. The benefits are not only the resolution of the environmental concerns but also the enhancement of crop productivity, which is critical for non-agrochemical production. Nevertheless, the real challenge for the bioremediation process is the application of the isolated strain in situ under uncontrolled conditions. Accordingly, paraquat degradation and plant growth promotion of B. aryabhattai strain MoB09 were performed in cowpea under a pot experiment.
Paraquat Degradation and Growth Promotion of Cowpea (V. unguiculata) Under Paraquat Stress
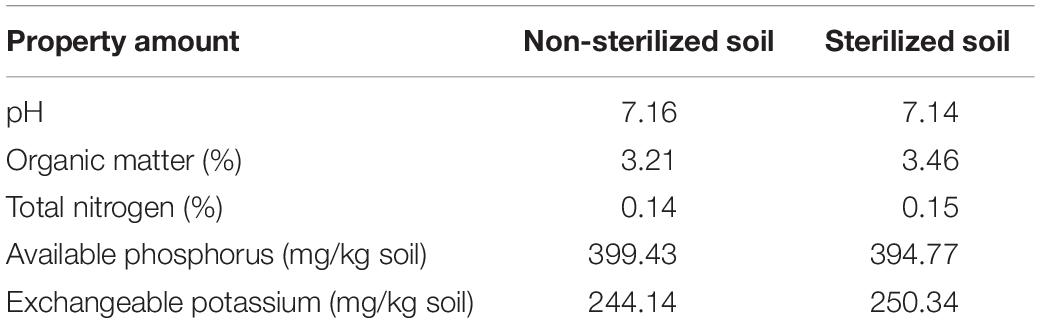
Cowpea (V. unguiculata) is sensitive to herbicide (Silva et al., 2020) and could serve as a model to perform the ability of bacterial isolates to promote plant growth under paraquat stress in pot experiments. The experiments were conducted in non-sterile and sterile soils. The properties of the soils used in this study are shown in Table 2. Cowpea seedlings inoculated with B. aryabhattai strain MoB09 significantly increased in stem length, root length, stem weight, and root weight in both sterilized and non-sterilized soils with paraquat (Figures 6, 7). In sterilized soil, B. aryabhattai strain MoB09 was the sole bacterium that promoted the growth of cowpea and remediated paraquat. In contrast, in non-sterilized soil with paraquat and B. aryabhattai strain MoB09, the growth parameters of cowpea, including fresh weight, dry weight, root length, total length, and leaf length, were higher than those in non-sterilized soil with paraquat only and non-sterilized soil alone (control). This result indicated that B. aryabhattai strain MoB09 may provide synergistic activities with other native microorganisms in the soil. However, the microbial community in the soil should be monitored in future works, in order to understand the population dynamic and survival of the inoculated bacteria.
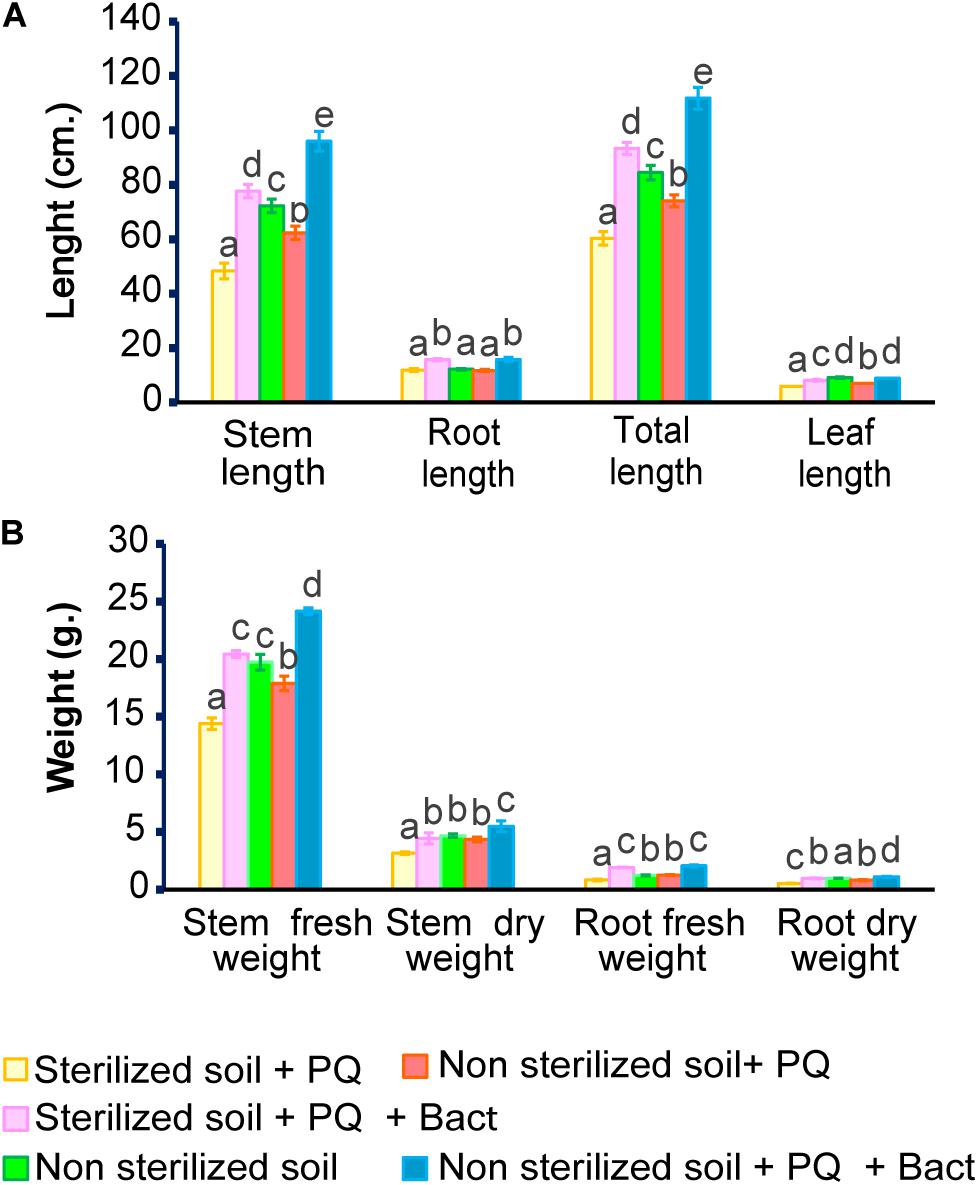
Figure 6. Growth parameters of cowpea (V. unguiculata) in soil with/without paraquat. (A) Stem, root, leaf and total length. (B) Fresh weight, dry weight of stem and root.

Figure 7. Growth of cowpea (V. unguiculata) (6 weeks age) in soil with/without paraquat in the pot experiment.
The residues of paraquat in the soil ranged from 5 to 200 mg/kg. Therefore, the maximum value (200 mg/kg) was applied as the initial paraquat concentration in this study (FAO/WHO, 1971). Paraquat residue in soil was determined from the treatment of non-sterilized soil with paraquat and non-sterilized soil with paraquat and B. aryabhattai strain MoB09. It was found that 0.05 mg/kg soil of paraquat remains in non-sterilized soil with paraquat and B. aryabhattai strain MoB09, which was lower than that in non-sterilized soil without B. aryabhattai strain MoB09 (0.18 mg/kg soil).
Growth Promotion of Cowpea (V. unguiculata) Under Drought Conditions in Soil by B. aryabhattai
Apart from residual pollutants remaining in the soil, environmental factors also generate various stresses, which affect plant growth and development. Environmental factors such as drought and salinity are of interest, as they affect the crop yield and quality of many plants, including cowpea (V. unguiculata L. Walp.). Cowpea is a suitable model for testing drought resistance due to its high sensitivity to water stress during flowering and pod-filling stages (Oyewole et al., 2017). Our study found that the completely withholding watering significantly suppresses various growth parameters, such as shoot length, root length, biomass weight, and chlorophyll content and enhances proline production (Figures 8, 9).
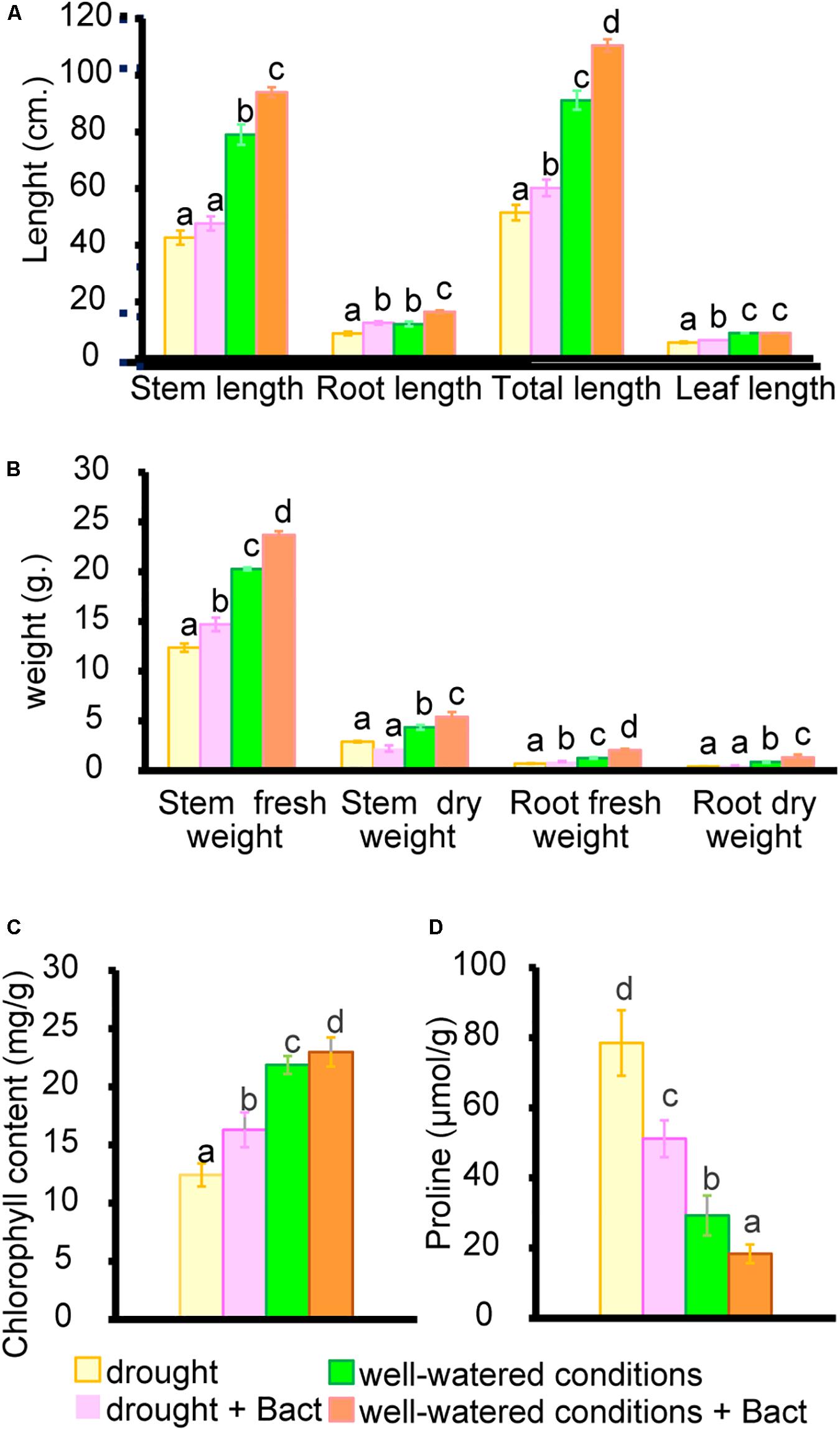
Figure 8. Growth parameters and proline content of cowpea (V. unguiculata) in well-watered and drought conditions with/without B. aryabhattai MoB09. (A) Stem root leaf and total length. (B) Fresh weight, dry weight of stem and root. (C) Chlorophyll contect and (D) Proline contents.
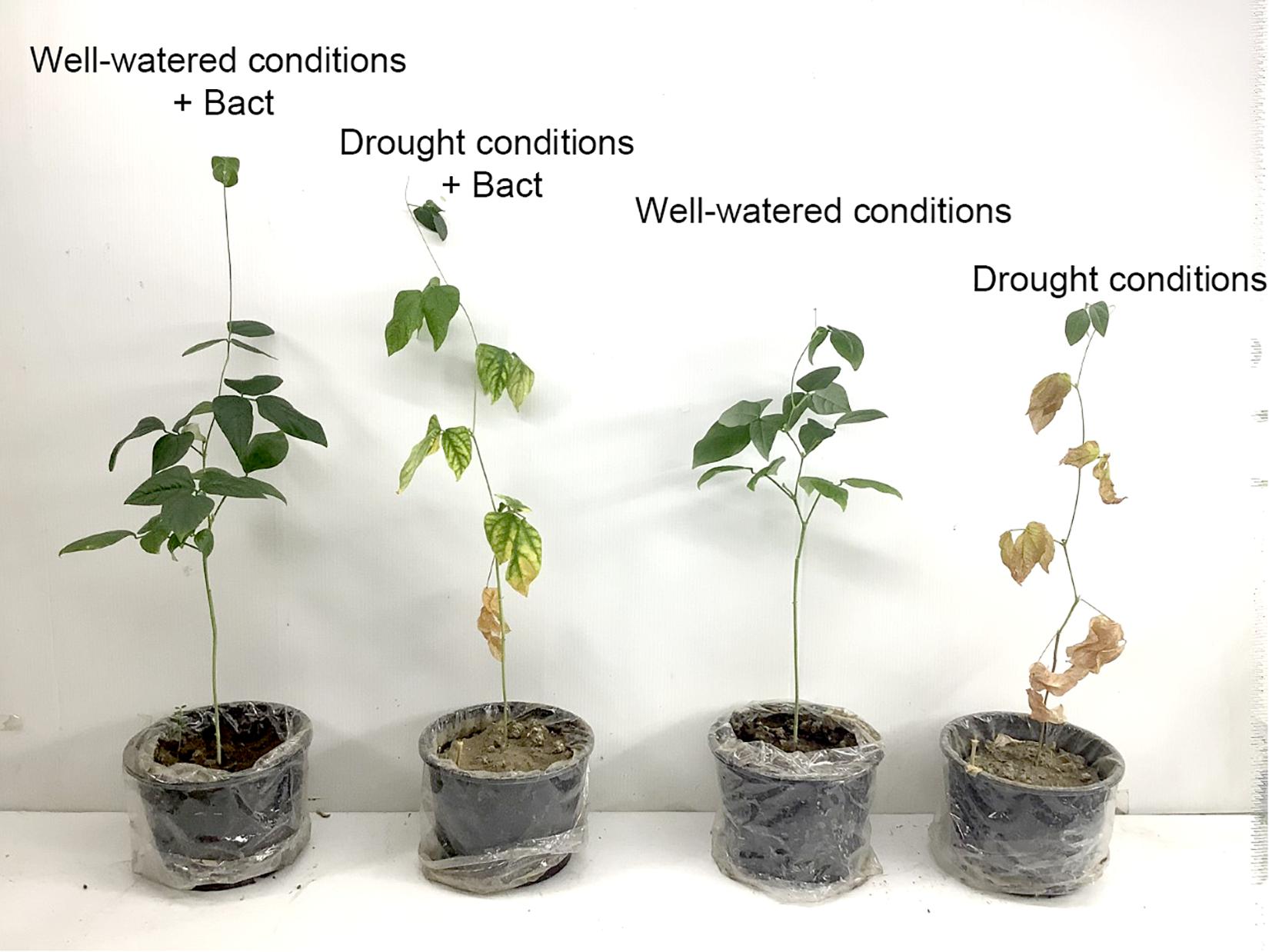
Figure 9. Growth of cowpea (V. unguiculata) in well-watered and drought conditions with/without B. aryabhattai MoB09 in day 45 after sowing.
Our result agrees with reports from a previous study that drought stress affected all physiological parameters in plants, especially a significant decrease in chlorophyll a, chlorophyll b, and total chlorophyll contents (Mafakheri et al., 2010). Proline accumulation has been reported as a mitigation mechanism for drought stress in cowpea (Zegaoui et al., 2017). Enhancement in proline biosynthesis helps in balancing osmotic potential and maintaining cell turgor pressure (Chun et al., 2018).
Bacillus species such as B. amyloliquefaciens HYD-B17, B. licheniformis HYTAPB18, B. thuringiensis HYDGRFB19, and B. subtilis RMPB44, have been reported for their plant growth-promoting and positive effects on drought tolerance in maize (Vardharajula et al., 2011). Bacillus spp. have been applied in agriculture for a long time. Formulated and sporulated cultures of B. thuringiensis (Bt) are widely used for pest control (Crickmore, 2006). One of the advantages for the application of Bacillus spp. is their endospore-forming ability, which provides long-term stability and survival comparable with those of agrochemicals and other bacteria (Borriss, 2011).
Conclusion
A great number of microorganisms have been reported for their paraquat degradation. However, there are hardly any bacteria capable of breaking down paraquat in soil, promoting plant growth, and enhancing drought tolerance altogether. Our results suggest that the B. aryabhattai MoB09 bacterium, which is screened from soil under heavy use of chemicals, can be very beneficial as a bioremediation agent in paraquat-contaminated sites and as a biofertilizer. It could promote the growth of cowpea (V. unguiculata) in soil under induced conditions of drought stress. The mechanisms of plant growth promotion and mitigation of drought stress as well as their plant growth-promoting hormone production will be evaluated in a future work. In addition, the application of this bacterium in situ will be conducted in a future study to remediate paraquat contamination in field trials.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
PI: conceptualization, methodology, formal analysis, investigation, and writing—original draft. PP: conceptualization, funding acquisition, writing—review and editing, and resources. JP: writing—review and editing. WP-A: writing—review and editing and resources. CP: supervision, conceptualization, project administration, funding acquisition, resources, and writing—review and editing.
Funding
This work was supported by the National Research Council of Thailand (NRCT) through the budget of the fiscal year of 2018 of Chiang Mai Rajabhat University and financially supported by the Graduate School of Chiang Mai University and the Research Center in Bioresources for Agriculture, Industry and Medicine, Chiang Mai University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Mr. Jamrern Nuichaiwong for providing the sampling sites. We would like to thank Mr. Phichit Phromsenjai, Department of Chemistry, Faculty of Science and Technology, Chiang Mai Rajabhat University, for their support in agrochemicals analysis and Ms. Wichaya Singtorn and Mr. Krisana Lasudee for their help in the laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.604662/full#supplementary-material
Footnotes
References
Andriani, L. T., Aini, L. Q., and Hadiastono, T. (2017). Glyphosate biodegradation by plant growth promoting bacteria and their affect to paddy germination in glyphosate contaminated soil. J. Degrade Min. Land Manage. 5, 995–1000. doi: 10.15243/jdmlm.2017.051.995
Andy, I. E., Edu, G. S., Bassey, I. U., Markson, A. A., Umana, E. I., and Udo, S. E. (2015). Biodegradation of paraquat. J. Biopestic. Envi. 1, 80–85.
Ardiwinata, A. N., Harsanti, E. S., Kurnia, A., Sulaeman, E., Fauriah, R., and Paputri, D. M. W. (2019). Contamination of paraquat residues in soil and water from several provinces in Indonesia. AIP Conference Proc. 2120:040024. doi: 10.1063/1.5115662
Arnon, D. T. (1949). Copper enzymes in isolated chloroplast polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Ataikiru, T. L., Okerentugba, P. O., and Okpokwasili, G. C. (2020). Identification of carbofuran and paraquat degrading microorganisms from South Asian. J. Res. Microbiol 7, 40–52. doi: 10.9734/sajrm/2020/v7i130164
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water stress studies. Plant Soil. 39, 205–207. doi: 10.1007/BF00018060
Binghua, L., Yang, L., Luo, Y., Pu, X., and Li, J. (2015). Screening. Identification and Degradation Characteristics of the Strain XT12 of Degrading Methyl Viologen. Genom. Appl. Biol. 4, 29.
Borriss, R. (2011). “Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents in agriculture,” in Bacteria in Agrobiology: Plant Growth Responses, ed. D. Maheshwari (Berlin: Springer), 41–76. doi: 10.1007/978-3-642-20332-9_3
Bromilow, R. H. (2004). Paraquat and sustainable agriculture. Pest Manag. Sci. 60, 340–349. doi: 10.1002/ps.823
Cakmakci, R., Donmez, F., Aydin, A., and Sahin, F. (2006). Growth promotion of plants by plant growth-promoting rhizobacteria under greenhouse and two different field soil condition. Soil Biol. Biochem. 38, 1482–1487. doi: 10.1016/j.soilbio.2005.09.019
Camilo, A. R., and Joseph, W. K. (2010). Plant growth promotion by Bacillus amyloliquefaciens FZB45 depends on inoculum rate and P-related. Biol Fertil Soils. 46, 835–844. doi: 10.1007/s00374-010-0488-2
Cao, M., Charles, M. M., and John, D. H. (2005). Bacillus subtilis paraquat resistance is directed by σM, an extracytoplasmic function sigma factor, and is conferred by YqjL and BcrC. J.Bacteriol. 187, 2948–2956. doi: 10.1128/JB.187.9.2948-2956.2005
Chun, S. C., Paramasivan, M., and Chandrasekaran, M. (2018). Proline accumulation influenced by osmotic stress in arbuscular mycorrhizal symbiotic plants. Front. Microbiol. 9:2525. doi: 10.3389/fmicb.2018.02525
Crickmore, N. (2006). Beyond the spore–past and future developments of Bacillus thuringiensis as a biopesticide. J. Appl. Microbiol 101, 616–619. doi: 10.1111/j.13652672.2006.02936.x
Cycoń, M., Wójcik, M., and Piotrowska-Seget, Z. (2009). Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere 76, 494–501. doi: 10.1016/j.chemosphere.2009.03.023
Dobrzanski, T., Gravina, F., Steckling, B., Olchanheski, L. R., Sprenger, R. F., and Espírito Santo, B. C. (2018). Bacillus megaterium strains derived from water and soil exhibit differential responses to the herbicide mesotrione. PLoS One 13:e0196166. doi: 10.1371/journal.pone.0196166
Dworkin, M., and Foster, J. W. (1958). Experiments with some microorganism which utilize ethane and hydrogen. J. Bacteriol. 175, 592–601. doi: 10.1128/JB.75.5.592-603.1958
Esitken, A., Karlıdag, H., Ercisli, S., Turan, M., and Sahin, F. (2003). The effect of spraying a growth promoting bacterium on the yield, growth and nutrient element composition of leaves of apricot (PrunusarmeniacaL. cv. Hacihaliloglu). Aust. J. Agric. Res. 5, 377–380. doi: 10.1071/AR02098
FAO/WHO (1971). Diquat. In: 1970 Evolution of some pesticide residues in food, Rome, Food and Agriculture Organization of the United Nations. Document Numberswho/Food Add/71.42AGP:1970/M/12/1., Rome, FAO.
Francisco, X. N., Anabel, G. H., Bernard, R. G., and Márcio, J. R. (2020). Plant growth-promoting activities and genomic analysis of the stress-resistant Bacillus megaterium STB1, a bacterium of agricultural and biotechnological interest. Appl. Biotechnol. Rep. 25, e00406. doi: 10.1016/j.btre.2019.e00406
Gangireddygari, V. S. R., Kalva, P. K., Ntushelo, K., Bangeppagari, M., Tchatchou, A. D., and Bontha, R. R. (2017). Influence of environmental factors on biodegradation of quinalphos by Bacillus thuringiensis. Environ. Sci. Eur. 29, 1–10. doi: 10.1186/s12302-017-0109-x
Gianessi, L. P. (2013). The increasing importance of herbicides in worldwide crop production. Pest Manag. Sci. 69, 1099–1105. doi: 10.1002/ps/3598
Glickmann, E., and Dessaux, Y. (1995). A critical examination of the specificity of the Salkowski reagent forindolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. doi: 10.1128/AEM.61.2.793-796.1995
Gordon, A., and Weber, P. (1951). Colorimetric estimation of indole acetic acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Hashemi, S. S., Aliasgharzad, N., Khakvar, R., and Oustan, S. (2015). Isolation and identification of paraquat degrading bacteria from Tabriz soil. J. Soil Biol. 3, 117–127.
Hou, X., Wu, F., Wang, X. J., Sun, Z. T., Zhang, Y., Yang, M. T., et al. (2018). Bacillus methylotrophicus CSY-F1 alleviates drought stress in cucumber (Cucumis sativus) grown in soil with high ferulic acid levels. Plant Soil 431, 89–105. doi: 10.1007/s11104-018-3748-9
Huang, Y., Zhan, H., Bhatt, P., and Chen, S. (2019). Paraquat degradation from contaminated environments: current achievements and perspectives. Front. Microbiol. 10:1754. doi: 10.3389/fmicb.2019.01754
Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., et al. (2003). Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91. doi: 10.1038/nature01582
Kannahi, M., and Senbagam, N. (2014). Studies on siderophore production by microbial isolates obtained from rhizosphere soil and its antibacterial activity. J. chem. Pharm. 6, 1142–1145.
Khalid, A., Arshad, M., Shaharoona, B., and Mahmood, T. (2009). “Plant growth promoting rhizobacteria and sustainable agriculture,” in Microbial Strategies for Crop Improvement, eds M. S. Khan, A. Zaidi, and J. Musarrat 133–160.
Lasudee, K., Tokuyama, S., Lumyong, S., and Pathom-aree, W. (2018). Actinobacteria associated with arbuscular mycorrhizal funneliformis mosseae Spores, taxonomic characterization and their beneficial traits to plants: evidence Obtained from mung bean (Vigna radiata) and thai jasmine rice (Oryza sativa). Front. Microbiol. 9:1247. doi: 10.3389/fmicb.2018.01247
Lau, T. T. V., Tan, J. A. M. A., Puthucheary, S. D., Puah, S. M., and Chua, K. H. (2020). Genetic relatedness and novel sequence types of clinical Aeromonas dhakensis from Malaysia. Braz. J. Microbiol. 51, 909–918. doi: 10.1007/s42770-020-00239-8
Li, H., Wang, Y., Fu, J., Hu, S., and Qu, J. (2020). Degradation of acetochlor and beneficial effect of phosphate-solubilizing Bacillus sp. ACD-9 on maize seedlings. Biotech 10, 67. doi: 10.1007/s13205-020-2056-2
Mafakheri, A., Siosemardeh, A. F., Bahramnejad, B., Struik, P. C., and Sohrabi, Y. (2010). Effect of drought stress on yield, proline and chlorophyll contents in three chickpea cultivars.aust. J. Crop. Sci. 4:580.
Mansur, R., Gusmanizar, N., Dahalan, F. A., Masdor, N. A., Ahmad, S. A., Shukor, M. S., et al. (2016). Isolation and characterization of a molybdenum-reducing and amide-degrading burkholderia sp. Strain neni-11 in soils from west sumatera, Indonesia. IIOAB J. 7, 28–40.
Mohd, A. A. S., Abu, Z. I. M., Ahmad, S. A., Halmi, M. I. E., Abdullah, S. R. S., Masdor, N. A., et al. (2016). A Molybdenum-reducing Bacillus sp. strain zeid 14 in Soils from sudan that could grow on amides and acetonitrile. Malaysian J. Soil Sci. 20, 111–134.
Moneke, A. N., Okpala, G. N., and Anyanwu, C. U. (2010). Biodegradation of glyphosate herbicide in vitro using bacterial isolates from four rice fields. Afr. J. Biotechnol. 9, 4067–4074.
Muhammad, A. W. T., and Frankenberger Jr. (1991). The Rhizosphere and Plant Growth. BSAR. 14, 327–334.
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate- solubilizing microorganism. FEMS Microbiol. Lett. 170, 165–170. doi: 10.1111/j.1574-6968.1999.tb13383.x
Ouided, B., and Abderrahmane, B. (2013). Isolation and characterization of glyphosate- degradingbacteria from different soils of Algeria. Afr. J. Microbiol. Res. 7, 5587–5595. doi: 10.5897/ajmr2013.6080
Oyewole, B. O., Olawuyi, O. J., Odebode, A. C., and Abiala, M. A. (2017). Influence of arbuscular mycorrhiza fungi (AMF) on drought tolerance and charcoal rot disease of cowpea. Biotechnol. Rep. 14, 8–15. doi: 10.1016/j.btre.2017.02.004
Palaniyandi, S. A., Damodharan, K., Yang, S. H., and Suh, J. W. (2014). Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of ‘Micro Tom’ tomato plants. J. Appl. Microbiol. 117, 766–773. doi: 10.1111/jam.12563
Pankaj, B., Yaohua, H., Wenping, Z., Anita, S., and Shaohua, C. (2020). Enhanced Cypermethrin Degradation Kinetics and Metabolic Pathway in Bacillus thuringiensis Strain SG4. Microorganisms. 8:223. doi: 10.3390/microorganisms8020223
Park, Y. G., Mun, B. G., Kang, S. M., Hussain, A., Shahzad, R., Seo, C. W., et al. (2017). Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS One 12:e0173203. doi: 10.1371/journal.pone.0173203
Parmar, K. J., Tomar, R. S., Parakhia, M. V., Malviya, B. J., Rathod, V. M., Thakkar, J. R., et al. (2018). Isolation and bio-analytical characterization of chloropyrifos degrading bacteria.J. Cell Tissue Res. 14:4641. doi: 10.21474/IJAR01/4234
Penrose, D. M., and Glick, B. R. (2003). Methods for isolating and characterizing ACC deaminase- containing plant growth-promoting rhizobacteria. Physiologia Plantarum 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 17, 362–370.
Reczek, C. R., Birsoy, K., Kong, H., Martínez-Reyes, I., Wang, T., Gao, P., et al. (2017). A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol. 13, 1274–1279. doi: 10.1038/nchembio.2499
Roberts, T. R., Dyson, J. S., and Lane, M. C. G. (2002). Deactivation of the biological activity of paraquat in the soil environment: a review of long-term environmental fate. J. Agric. Food Chem. 50, 3623–3631. doi: 10.1021/jf011323x
Rusnam and Gusmanizar, N. (2019). Isolation and Characterization of a molybdenum-reducing and coumaphos-degrading Bacillus sp. strain Neni-12 in soils from West Sumatera. Indonesia.JEMAT 7, 20–25. doi: 10.21315/tlsr2017.28.1.5
Sabullah, M. K., Rahman, M. F., Ahmad, S. A., Sulaiman, M. R., Shukor, M. S., Shamaan, N. A., et al. (2016). Isolation and characterization of a molybdenum-reducing and glyphosate - degrading Klebsiella oxytoca strain saw 5 in soils from sarawak. J. Agric. Sci. 38, 1–13. doi: 10.17503/agrivita.v38i1.654
Salehi-Lisar, S. Y., and Bakhshayeshan-Agdam, H. (2016). “Drought stress in plants: causes, consequences, and tolerance. Drought Stress Tolerance Plants. Cham, Springer.NODOI doi: 10.1007/978-3-319-28899-4_1
Samina, A., Fauzia, L., Qaiser, M. K., Zafar, M. K., and Samina, I. (2009). Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J. Hazard. Mater. 168, 400–405. doi: 10.1016/j.jhazmat.2009.02.059
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shashidar, A., Tarkowská, D., Rolčík, J., Novák, O., Palmero, D. V., Bejai, S., et al. (2017). Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using arabidopsis thaliana as host plant. Planta 245, 15–30. doi: 10.1007/s00425-016-2580-9
Silva, J. N., Costa, E. M., Pereira, L. S., Gonçalves, E. C. Z., Zuchi, J., and Jakelaitis, A. (2020). Cowpea yield and quality after application of desiccating herbicides. Res. J. Seed Sci. 42, 1545–2317. doi: 10.1590/2317-1545v42228204
Singh, B. K., and Walker, A. (2006). Microbial degradation of organophosphorus compounds. FEMS microb. reviews. 30, 428–471. doi: 10.1111/j.1574-6976.2006.00018.x
Teerakun, M., Reungsang, A., Chaowarat, M., and Saraphirom, P. (2020). Optimization of paraquat degradation with microbial consortium from contaminated soil using statistic method. Int. J. Geomech. 18, 73–79.
Tsai, W. T., and Chen, H. R. (2013). Adsorption kinetics of herbicide paraquat in aqueous solution onto a low-cost adsorbent, swine-manure-derived biochar. Int. J. Environ. Sci. Technol. 10, 1349–1356. doi: 10.1007/s13762-012-0174-z
Tu, C. M., and Bollen, W. B. (1968). Interaction between paraquat and microbes in soils. Weed. Res 8, 38–45. doi: 10.1111/j.1365-3180.1968.tb01399.x
Vardharajula, S., Zulfikar Ali, S., Grover, M., Reddy, G., and Bandi, V. (2011). Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 6, 1–14. doi: 10.1080/17429145.2010.535178
Veríssimo, G., Moreira, J. C., and Meyer, A. (2018). Paraquat contamination in surface waters of a rural stream in the mountain region in the state of Rio De Janeiro southeastern Brazil. J. Environ. Toxicol. Stud. 2, 1. doi: 10.16966/2576-6430.111
Wahyu, W. (2013). Development of method for residue analysis of three herbicides in the soil by high performance liquid chromatography (HPLC). J. Environ. Chem. Ecotoxicol. 5, 220–226.
Wu, Y., DiMaggio, P. A., Perlman, D. H., Zakian, V. A., and Garcia, B. A. (2013). Novel phosphorylation sites in the S. cerevisiae Cdc13 protein reveal new targets for telomere length regulation. J. Proteome Res. 12, 316–327. doi: 10.1021/pr300408v
Yasin, N. A., Akram, W., Khan, W. U., Ahmad, S. R., Ahmad, A., and Ali, A. (2018). Halotolerant plant-growth promoting rhizobacteria modulate gene expression and osmolyte production to improve salinity tolerance and growth in Capsicum annum L. Environ. Sci. Pollut. Res. 25, 23236–23250. doi: 10.1007/s11356-018-2381-8
Yunus, S. M., Hamim, H. M., Anas, O. M., Aripin, S. N., and Arif, S. M. (2009). Mo (VI) reduction to molybdenum blue by Serratia marcescens strain Dr. Y9. Pol. J. Microbiol. 58, 141–147.
Keywords: paraquat, plant growth-promoting, bioremediation, soil, pot experiment, contaminated agricultural soil
Citation: Inthama P, Pumas P, Pekkoh J, Pathom-aree W and Pumas C (2021) Plant Growth and Drought Tolerance-Promoting Bacterium for Bioremediation of Paraquat Pesticide Residues in Agriculture Soils. Front. Microbiol. 12:604662. doi: 10.3389/fmicb.2021.604662
Received: 10 September 2020; Accepted: 15 February 2021;
Published: 18 March 2021.
Edited by:
Guining Lu, South China University of Technology, ChinaReviewed by:
Mariusz Cycoń, Medical University of Silesia, PolandAbhishek Walia, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, India
Shaohua Chen, South China Agricultural University, China
Copyright © 2021 Inthama, Pumas, Pekkoh, Pathom-aree and Pumas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chayakorn Pumas, Y2hheWFrb3JuLnB1bWFzQGdtYWlsLmNvbQ==
 Phatcharida Inthama1
Phatcharida Inthama1 Pamon Pumas
Pamon Pumas Chayakorn Pumas
Chayakorn Pumas