- 1Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
- 2Department of Food Science & Technology, Jinan University, Institute of Food Safety & Nutrition, Jinan University, Guangzhou, China
The accurate and rapid classification of Salmonella serovars is an essential focus for the identification of isolates involved in disease in humans and animals. The purpose of current research was to identify novel sensitive and reliable serovar-specific targets and to develop PCR method for Salmonella C2 serogroups (O:8 epitopes) in food samples to facilitate timely treatment. A total of 575 genomic sequences of 16 target serovars belonging to serogroup C2 and 150 genomic sequences of non-target serovars were analysed by pan-genome analysis. As a result, four and three specific genes were found for serovars Albany and Hadar, respectively. Primer sets for PCR targeting these serovar-specific genes were designed and evaluated based on their specificity; the results showed high specificity (100%). The sensitivity of the specific PCR was 2.8 × 101–103 CFU/mL and 2.3 × 103–104 CFU/mL for serovars Albany and Hadar, respectively, and the detection limits were 1.04 × 103–104 CFU/g and 1.16 × 104–105 CFU/g in artificially contaminated raw pork samples. Furthermore, the potential functions of these serovar-specific genes were analysed; all of the genes were functionally unknown, except for one specific serovar Albany gene known to be a encoded secreted protein and one specific gene for serovars Hadar and Albany that is a encoded membrane protein. Thus, these findings demonstrate that pan-genome analysis is a precious method for mining new high-quality serovar-targets for PCR assays or other molecular methods that are highly sensitive and can be used for rapid detection of Salmonella serovars.
Introduction
Salmonella is one of crucial foodborne pathogen that causes illness worldwide, including diarrhoea, gastroenteritis, typhoid, paratyphoid, septicaemia, and other clinical syndromes (Dekker and Frank, 2015). Pigs, poultry and their eggs, and cattle could be infected by Salmonella (Rodriguez et al., 2006; Xu et al., 2017), which can be disseminate to humans via ingestion of contaminated pork, chicken, eggs, beef, and milk (Kerouanton et al., 2013; Bonardi, 2017). Salmonellosis represents a serious occupational and public health hazard. Characteristic lipopolysaccharide, which is composed of lipid-A and a major O-antigen with side chains of repeating units of sugar residues, is a dominant cause for Salmonellosis (Sannigrahi et al., 2020). On the basis of the O antigen, Salmonella has been divided into 46 serogroups that markedly differ in their virulence (Davies et al., 2013); the isolates of D, B, C1, C2, and E serogroups take up a great majority of foodborne outbreaks (Graziani et al., 2017). Several studies have undertaken mechanism analysis and developed detection methods for Salmonella serovars Enteritidis (Wagner and Hensel, 2011), Typhimurium (Maurischat et al., 2015), and Derby (Castelijn et al., 2013), since they top the list of the most prevalent serotypes (Ni et al., 2018). However, little information about the other serovars is available, particularly for the infective serotypes that belong to the C2 serogroup. For example, S. Hadar is a host-non-specific serotype that causes infection in both humans and animals. It has been identified previously in hospital outbreaks and confirmed as the fourth most frequently isolated Salmonella serovar in Germany (Weidebotjes et al., 1998; Deshpande et al., 2015). Furthermore, instances of multidrug resistance in S. Albany have increased in recent years (Doublet et al., 2003). In order to reduce the prevalence of Salmonella, establishment rapid and feasible detection methods is essential for the identification of high-risk Salmonella serovars.
Traditional serotyping methods are based on slide and tube agglutination tests using O and H antigen-specific anti-sera (Herrera-León et al., 2007). However, these measures are costly, labour-intensive, time-consuming and insensitive, with certain isolates remaining partially typed or untyped, attributed to the loss of somatic and flagellar antigens. Nucleic acid amplification tests (NAATs), with the advantages of their high-speed, convenient operation, high-sensitivity and-specificity, have been extensively applied in different fields including the food industry, agriculture, and environmental sciences (Lee et al., 2019). Selection of appropriate pathogen genes is key to the sensitivity and specific detection capabilities of NAATs. Various genes have been used to detect Salmonella serogroups and serovars including SNSL254_A2005 for Salmonella C2 serogroups (Liu et al., 2011), STM4495 for S. Typhimurium (Liu et al., 2012), sdfI for S. Enteritidis (De Freitas et al., 2010), ISR2 for S. Infantis (Akiba et al., 2011), and Newp2 for S. Newport (Bugarel et al., 2017)—some of which may also be present in non-target strains.
With the recent advancements in high-throughput sequencing technology, an increasing number of whole genome sequences for Salmonella are available online. Through bioinformatics analysis, the resulting genomic information elucidates the diversity of Salmonella serovars. The pan-genome is the sum of a core genome which constitutes of genes present in all of the strains analysed and a dispensable genome that comprises genes present in some but not all of the strains analysed (Tettelin et al., 2008; Kim et al., 2020). We obtain molecular targets by selecting the co-existence genes in the target serovar of Salmonella and excluding the unspecific genes that are distributed among non-target serovars. Based on our prior research on the prevalence of Salmonella in China (Yang et al., 2015a, b, 2016, 2020a, b), more than 1,400 strains of Salmonella with 42 serovars have been isolated. We undertook the present study with the goal of identifying serovar-specific molecular targets for the most common serovars in serogroup C2 by pan-genome analysis of Salmonella genome sequences. Furthermore, practical PCR verification was conducted with a large number of bacterial strains and the specificity, sensitivity, and reliability of the PCR were also assessed. We evaluated the specificity targets of various Salmonella serovars by PCR and analysed the potential functions of these target genes. These data were analysed to obtain high-quality candidates for NAATs, as the use of this PCR-typing method permits for the identification of Salmonella serovars can be save a large abundance of time and money, and its accuracy is the same as the traditional serotyping methods.
Materials and Methods
Tested Strains
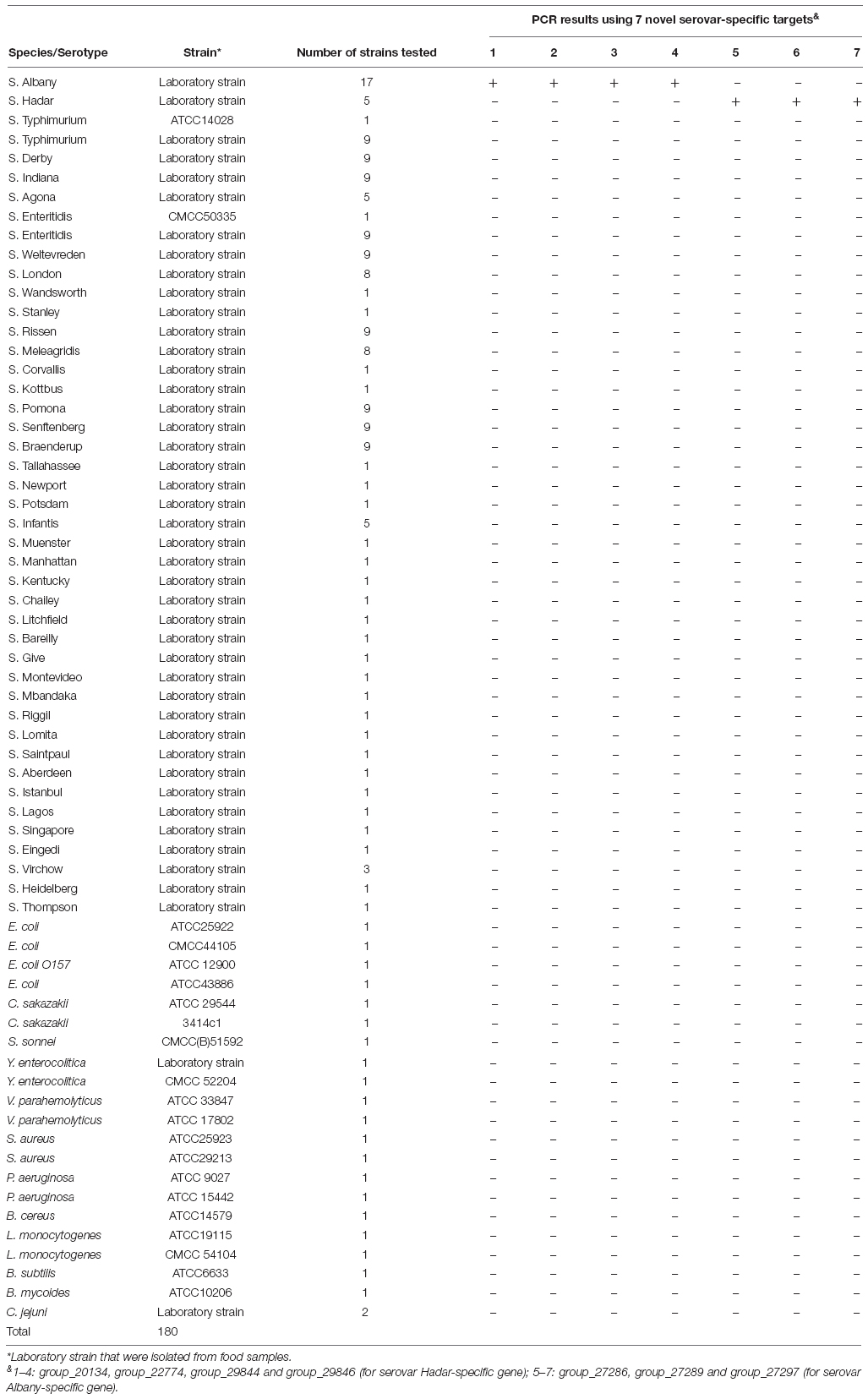
One hundred fifty-eight Salmonella strains which were serotyped by traditional methods, and twenty-two non-Salmonella strains were used for assessing the specificity in our study (Table 1). Sixteen and four strains were acquired from The American Type Culture Collection (ATCC, Manassas, VA, United States) and National Centre for Medical Culture Collection (CMCC, Beijing, China), and other strains were isolated from food sources in our laboratory from 2011 to 2014.
Genomic Sequences of Salmonella Species
Considering the number of scaffolds exceed 200, the genome sequence presumably has large gaps. Thus, the number of scaffold less than or equal to 200 for representative strains of the genome sequences are applied to analysis in this study. The 16 serovars belonging to serogroup C2 were represented by 575 isolates, with 1–248 isolates for each serovar. An additional 150 isolates from non-target serovars were also included. All 725 Salmonella genomes were downloaded from NCBI GenBank (NIH, Bethesda, MD, United States) in nucleotide FASTA format. A complete listing of the 725 genomes including GenBank identifier, serogroup, serovar, isolation source, geographic location, collection date, genome size, GC%, the number of scaffold etc. are listed in Supplementary Table 1.
Phylogenetic Analysis
The core-genome alignment of Salmonella was implemented using Harvest v1.1.2 software (Treangen et al., 2014) with the S. Albany ATCC 51960 genome as a reference. Conducting recombination and removing the putative recombined regions were implemented Genealogies Unbiased By recomBinations In Nucleotide Sequences (Gubbins, Croucher et al., 2015). Single nucleotide polymorphisms (SNPs) data were obtained from the recombination-free core-genome alignment by the script online at https://github.com/sanger-pathogens/snp-sites. The maximum-likelihood (ML) phylogenetic tree was established on the connected core SNPs by Random Axelerated Maximum Likelikhood (RAxML v8.2.10) in the GTRGAMMA model (1000 bootstrap) (Stamatakis, 2014). Results were visualised with Interaction Tree Of Life (iTOL, Letunic and Bork, 2016).
Pan-Genome Analysis and Identification of Serovar-Specific Genes
All analysed genome sequences were re-annotated using Prokka v1.11 (Seemann, 2014). With a BlastP (NCBI/NIH) identity cut-off of 70%, the Prokka output was utilised to establish the pan-genome by Roary v3.11.2 (Page et al., 2015). The absence/presence profile of all genes across all samples was transferred into a 0/1 matrix with a local script. Based on the 0/1 matrix, the core genes (presenting in over 99% Salmonella genomes) and serovar-specific genes were screened, presenting in over 95% target serovar strains (considered soft-core genes) and in less than 5% of other serovar strains (considered non-related) (Buchanan et al., 2017; Thépault et al., 2017). Functional classification of the core genes were assigned to the Gene Ontology (Go) terms (Ashburner et al., 2000). The specificity of serovar-specific genes were further confirmed by BLAST against the NCBI nucleotide sequence database.
Functional Analysis of Serovar-Specific Genes
The serovar-specific genes were annotated in public databases including the Non-redundant (NR), Kyoto Encyclopaedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups (COG), Virulence Factors Database (VFDB), Comprehensive Antibiotic Resistance Database (CARD), ResFinder, and Antibiotic Resistance Gene-ANNOTation (AGR-ANNOT). TMHMM Server, SignalP, and Phobius were also implemented to confirm whether these genes encode membrane and/or secretory proteins.
Evaluation of Specificity and Sensitivity for Novel Serovar-Specific Molecular Targets
The novel serovar-specific target genes were applied to design primer sets (Table 2) using Oligo 7 software and synthesised by Generay Biotech Co., Ltd. (Shanghai, China). The specificity of the primer sets were tested against the 180 strains listed in Table 1. Genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Shanghai, China) according to the manufacturer’s instructions. The purity and concentration of 50 μL genomic DNA samples were measured by Qubit® 3.0 Fluorometer (Life Invitrogen, United States) and stored at -20°C before being used as a template for PCR. Each 25 μL PCR amplification mixture consisted 12.5 μL of buffer (2×, Novoprotein Scientific Inc., Shanghai, China), 1 μL each of the forward and reverse primer (5 μM), 1 μL of template DNA, and sterile distilled water (filled to a final volume). The PCR conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30–60 s, and final extension at 72°C for 10 min. The PCR products were analyse via 1.5% agarose gel electrophoresis and visualised using a UV transilluminator (GE 138 Healthcare, WI, United States).
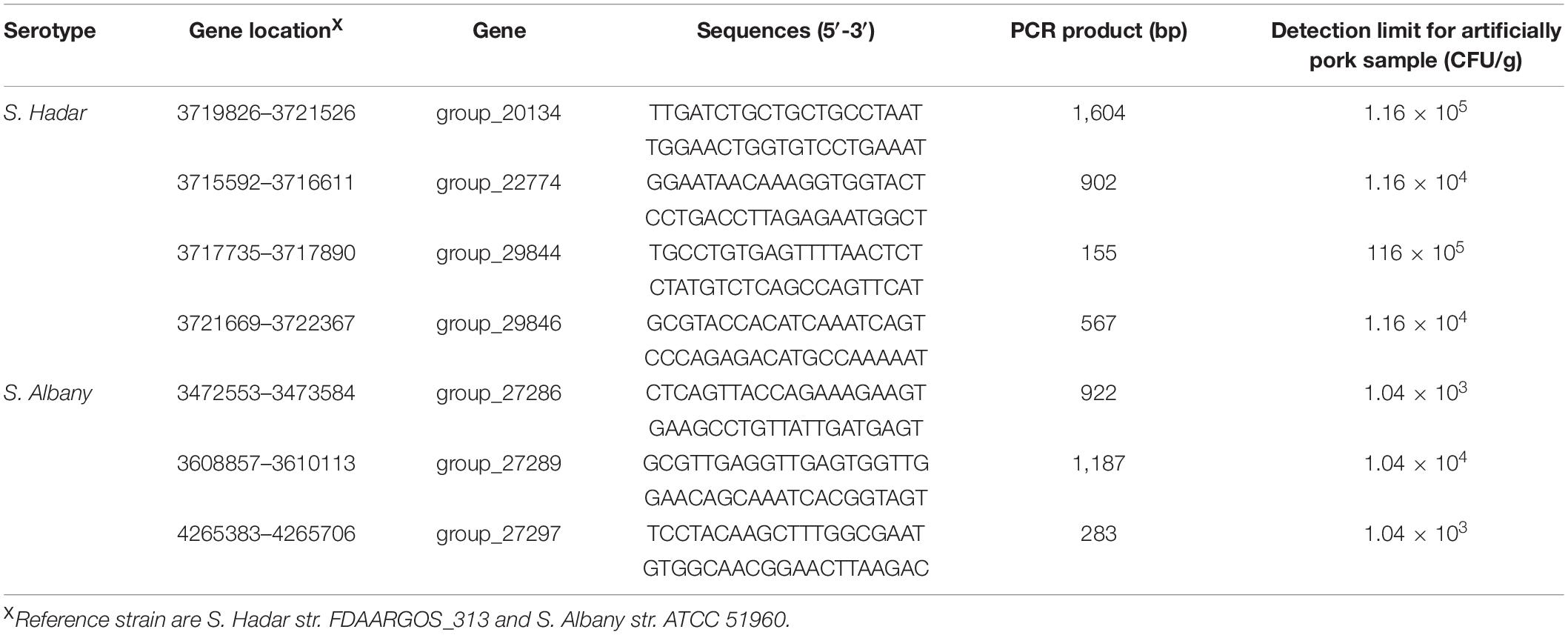
Table 2. Specific primer sets and sensitivity of PCR for artificially contaminated raw pork samples with Salmonella serovar Hadar and Albany.
For sensitivity testing, 10-fold serial dilutions (107–100 CFU/mL) of S. Hadar strain SA39(5) and S. Albany strain SA48(32) were subjected to DNA extraction. The PCR amplification were implemented as mentioned above except 2 μL of each genome DNA was used as a template.
Artificial Contamination of Raw Pork Samples
The S. Hadar strain SA39(5) and S. Albany strain SA48(32) were enriched in LB broth at 37°C overnight, and the 10-fold serial dilutions of different concentrations of cultures were prepared. Twenty-five grams of pork determined to be negative for Salmonella by standard culture methods was homogenised in 225 mL of sterile saline to obtain the matrix. Subsequently, 9 mL homogenate was spiked with 1 mL of various concentrations of a specific Salmonella serovar to final levels of approximately 108–101 CFU/g samples. DNA was extracted from 1 mL of the mixture and analysed by PCR under the same conditions. Non-inoculated pork meat was used as the negative control and all assays were performed independently, in triplicate.
Results
Phylogenetic Analysis of Salmonella
The 725 selected Salmonella isolates differed in the 1,803 core-genome SNPs. The ML phylogenetic tree was established based on the connected core SNPs. Isolates were distributed across serovars and all isolates belonged to serovar Albany and Hadar are clustered respectively. Notably, 80.34% (94/117) and 57.66% (143/248) strains belonged to serovar Kentucky and Newport are clustered, respectively (Figure 1). Isolates that fell within different serovars were highly diverse, indicating that an evolutionary difference might exist between these serovars.
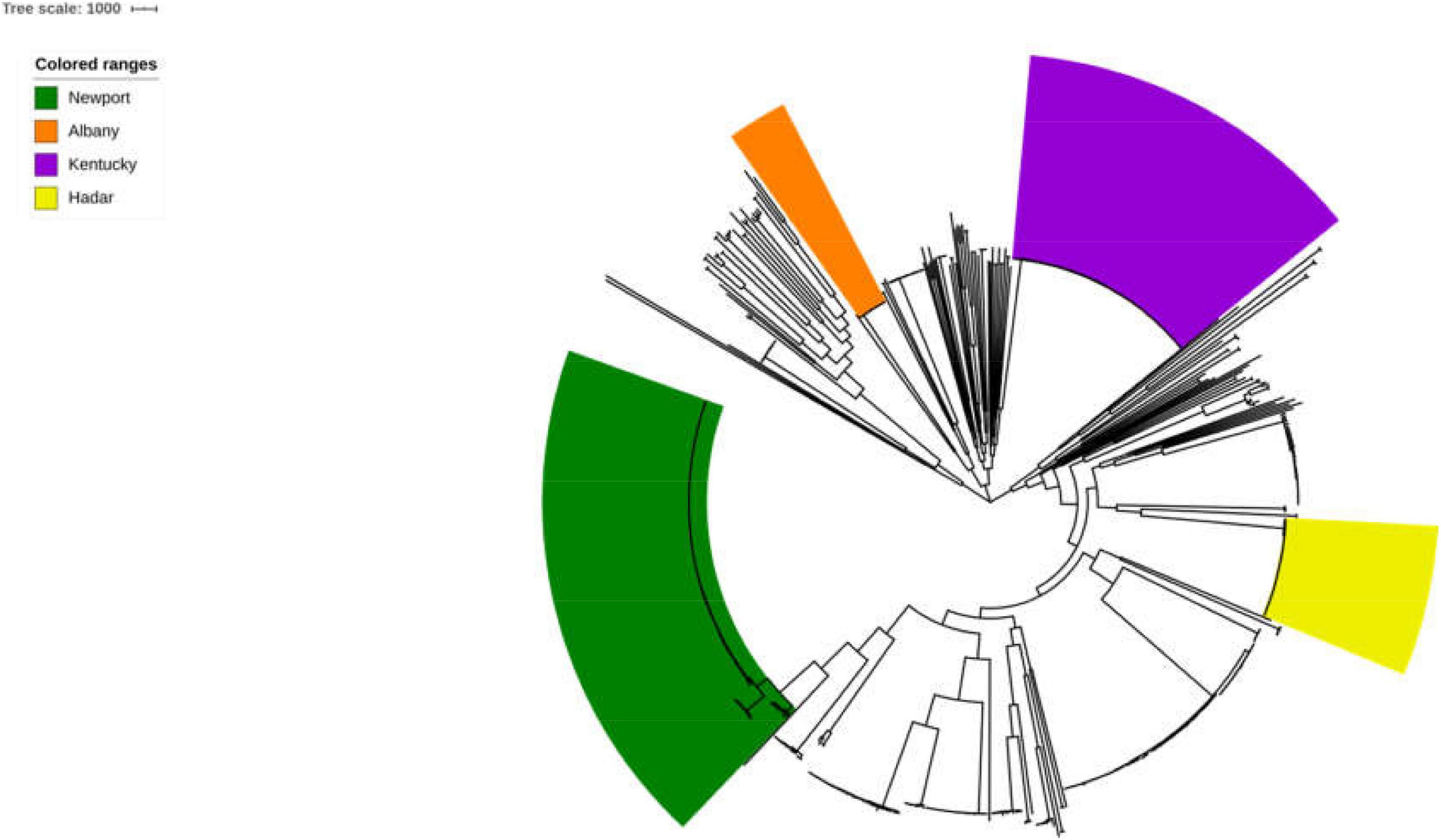
Figure 1. Phylogenetic analysis of Salmonella. The ML phylogenetic tree was established based on the connected core 1,803 core-genome SNPs from725 selected Salmonella strains. Strains were distributed across serovars and 100% (17/17), 100% (41/41), 80.34% (94/117), and 57.66% (143/248) strains belonged to serovar Albany, Hadar, Kentucky, and Newport are clustered, respectively.
Identification of Serovar-Specific Genes in Salmonella
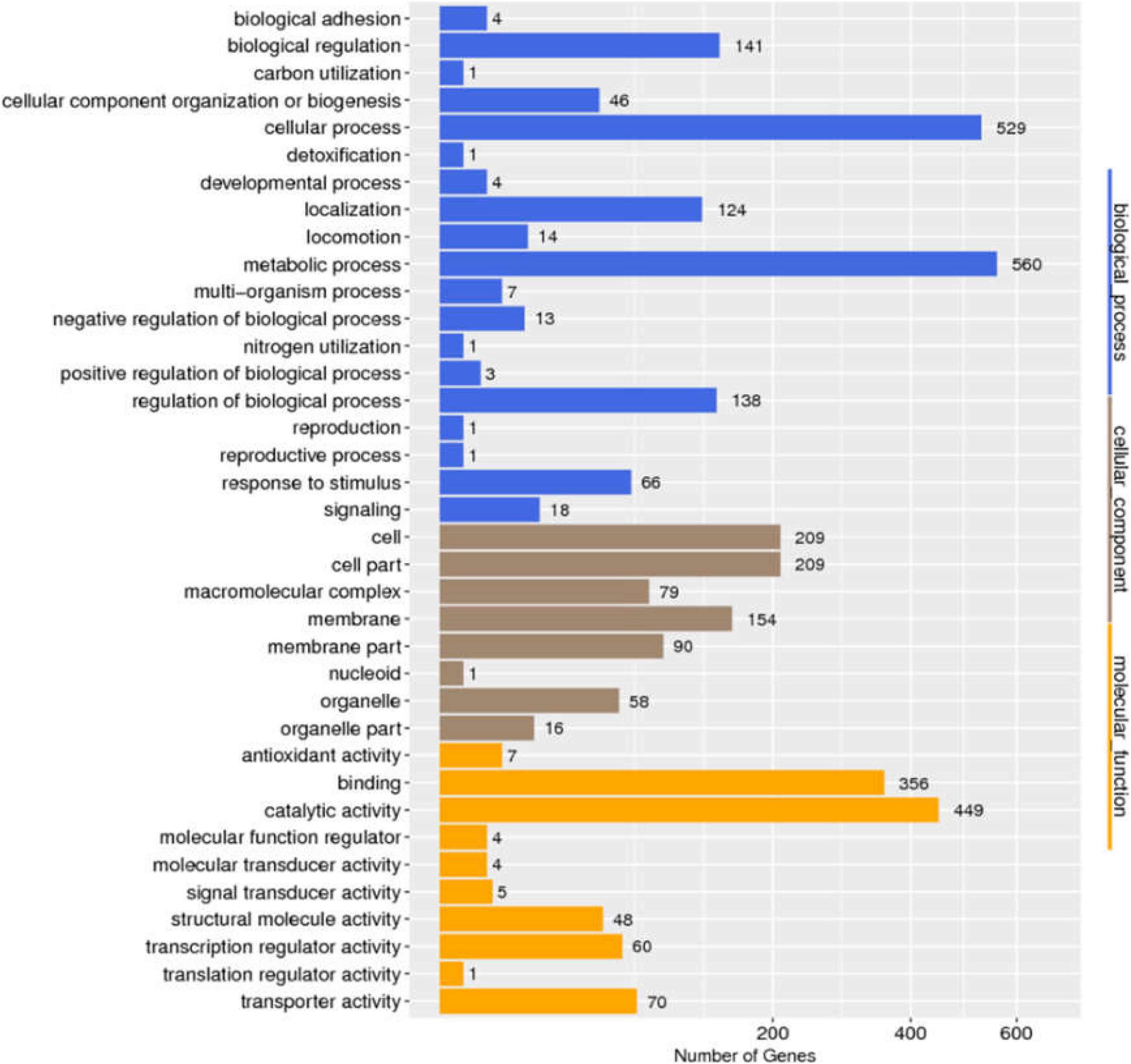
We determined the size and distribution of the Salmonella pan-genome across the 725 genome sequences and the analysis revealed a set of 1,318 conserved genes that were universally present within ≥99% Salmonella genomes. Functional profiles of conserved genes were determined in Figure 2. Furthermore, a group of 1,190 genes were present in 95% ≤ strains < 99%, which was called the soft-core genes of Salmonella. Besides, 2,904 and 30,032 genes were found in 15% ≤ strains < 95% and 0% ≤ strains < 15% of the isolates of Salmonella. Consistent with previous report (Chanda et al., 2020), the Salmonella showed an open pan-genome structure, and that the size of pan-genome continue to expand with addition sequenced genomes, but its conserved genes will remain stable (Supplementary Figure S1).
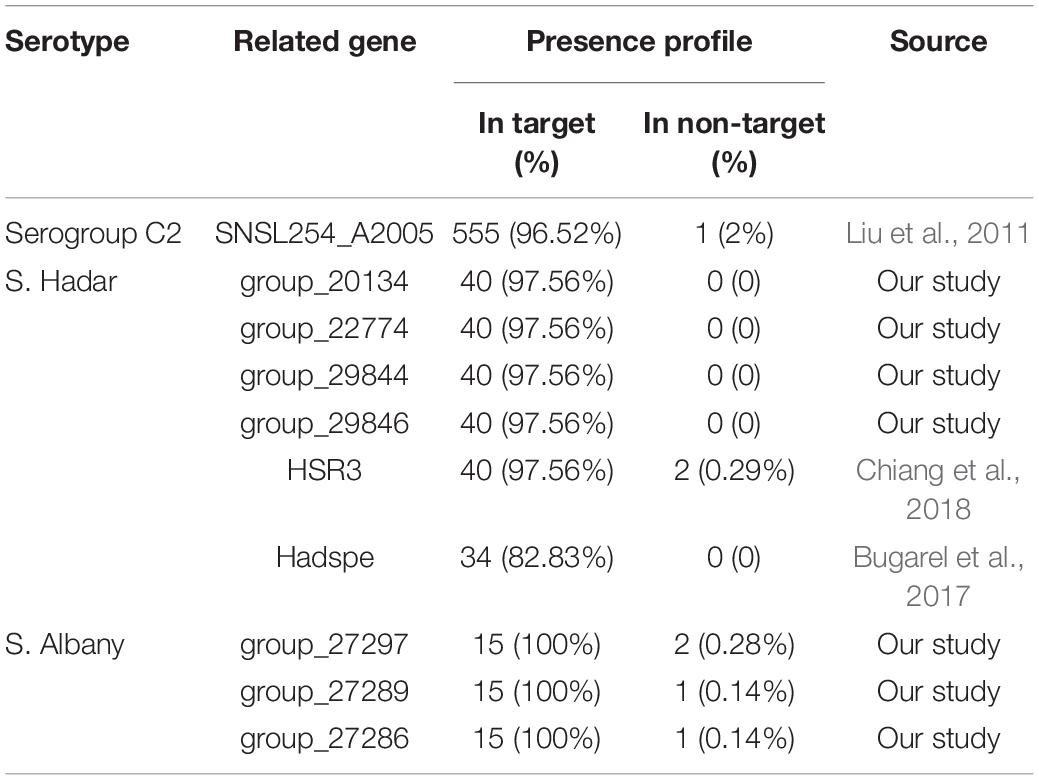
Based on the gene presence/absence profile among the 725 Salmonella genomes, 10 fragments were found to be specific to serogroups C2 and serovar Hadar and Albany. For other serovars of Salmonella serogroup C2, there is no novel single targets because the percentage of genomic sequences of strains for the genes presence in target-serovar were less than 85% and in non-target serovars were more than15%. Notably, four (group_20134, group_22774, group_29844, and group_29846) and three (group_27286, group_27289, and group_27297) specific gene markers were found in serovar Hadar and Albany. However, SNSL254_A2005 is a previously reported marker in serogroups C2 and HSR3 and Hadspe are previously reported markers in Hadar. The percentage of genomic sequences of strains for the present in target and non-target serovars are shown in Table 3.
Characterisation of Serovar-Specific Genes
One serovar albany-specific gene (group_27289) is linked with the function of defence mechanisms (Supplementary Table S2). However, all other serovar-specific genes were annotated uncharacterised functions. We went on to predict whether these genes participate in secretion. According to the prediction pipeline described in previous studies (refs), potentially secreted extracellular proteins and cell membrane proteins were identified from serovar-specific genes (Supplementary Table S2). The group_29846 gene (for serovar Hadar-specific gene) and group_27297 gene (for serovar Albany-specific gene) are encoded membrane proteins. Group _27286 (for serovar albany-specific gene) is encoded as a secreted protein. In addition, the seven specific genes were not related to the virulence-associated genes or the antibiotic resistance-associated genes.
Specificity and Sensitivity Assessment of Serovar-Specific Genes by PCR
A total of 158 Salmonella strains and 22 non-Salmonella strains (Table 1) were applied to assess the specificity of the primer sets designed based on serovar-specific fragments by PCR (Table 2). The proper detectable amplicon were observed from Salmonella serovar Hadar and Albany strains, but no amplification was obtained with DNA from all non-target strains. Thus, four Hadar-specific and three Albany-specific primer sets were accurately detected of Salmonella serovar Hadar and Albany (Table 1).
The analytical sensitivity test showed that the detection limit of the primers designed based on the group_27297 and group_27286 primers was approximately 28 CFU/mL (Figure 4), whereas that of the group_29844 primer based on Salmonella serovar Hadar was approximately 2.3 × 104 CFU/mL (Figure 2). Others showed a detection limit of 2.8–2.3 × 103 CFU/mL (Figures 3, 4).
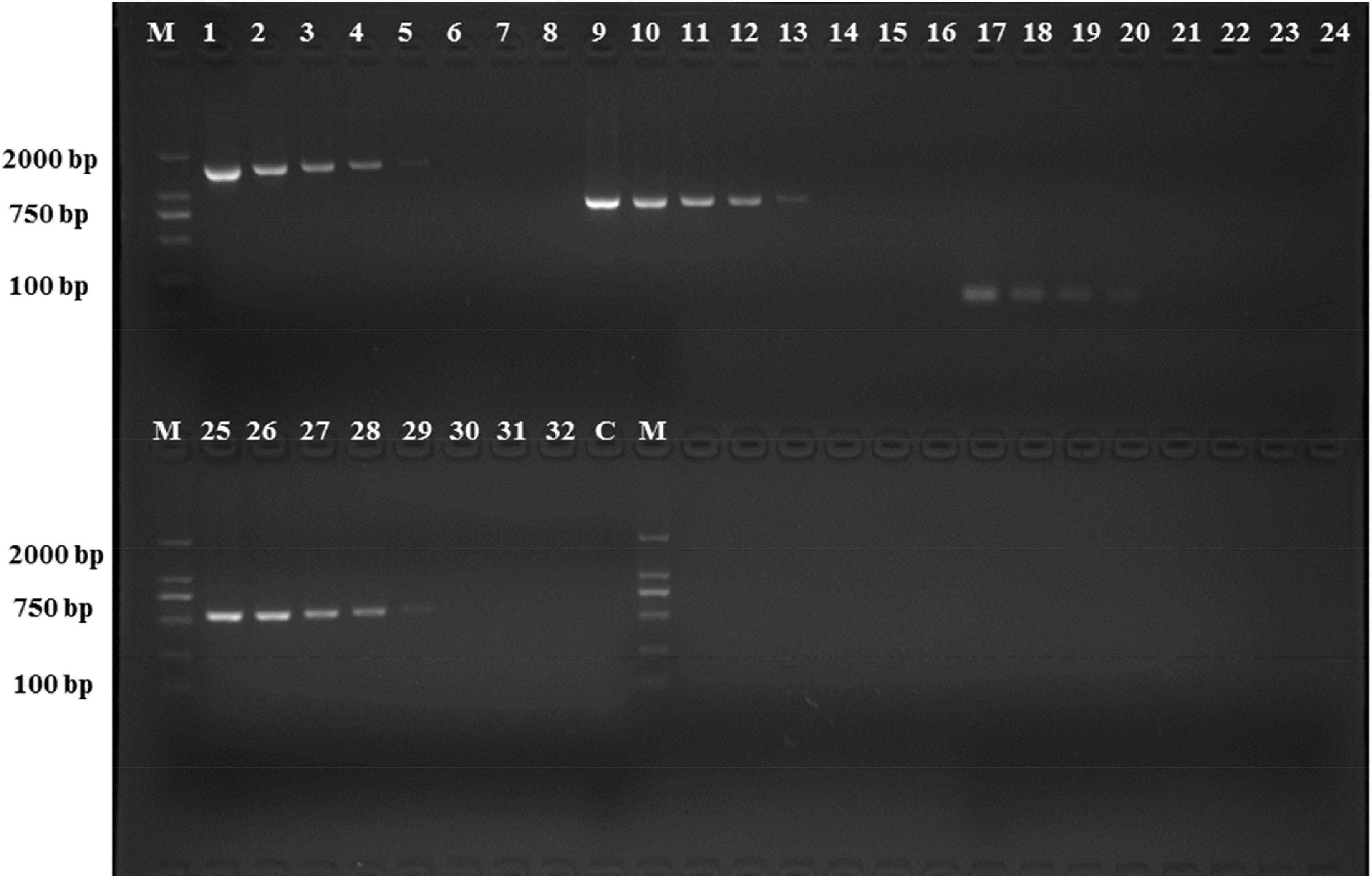
Figure 3. PCR detection sensitivity using dilutions of a pure culture of S. Hadar strain SA39(5). Lane M: DSTM 2000 marker; Lane C: negative control (double-distilled H2O); lane 1–8, 9–16, 17–24, and 25–32 primer sets based ongroup_20134, group_22774, group_29844, and group_29846 genes, respectively (concentrations ranging from 2.3 × 107 to 100 CFU/mL).
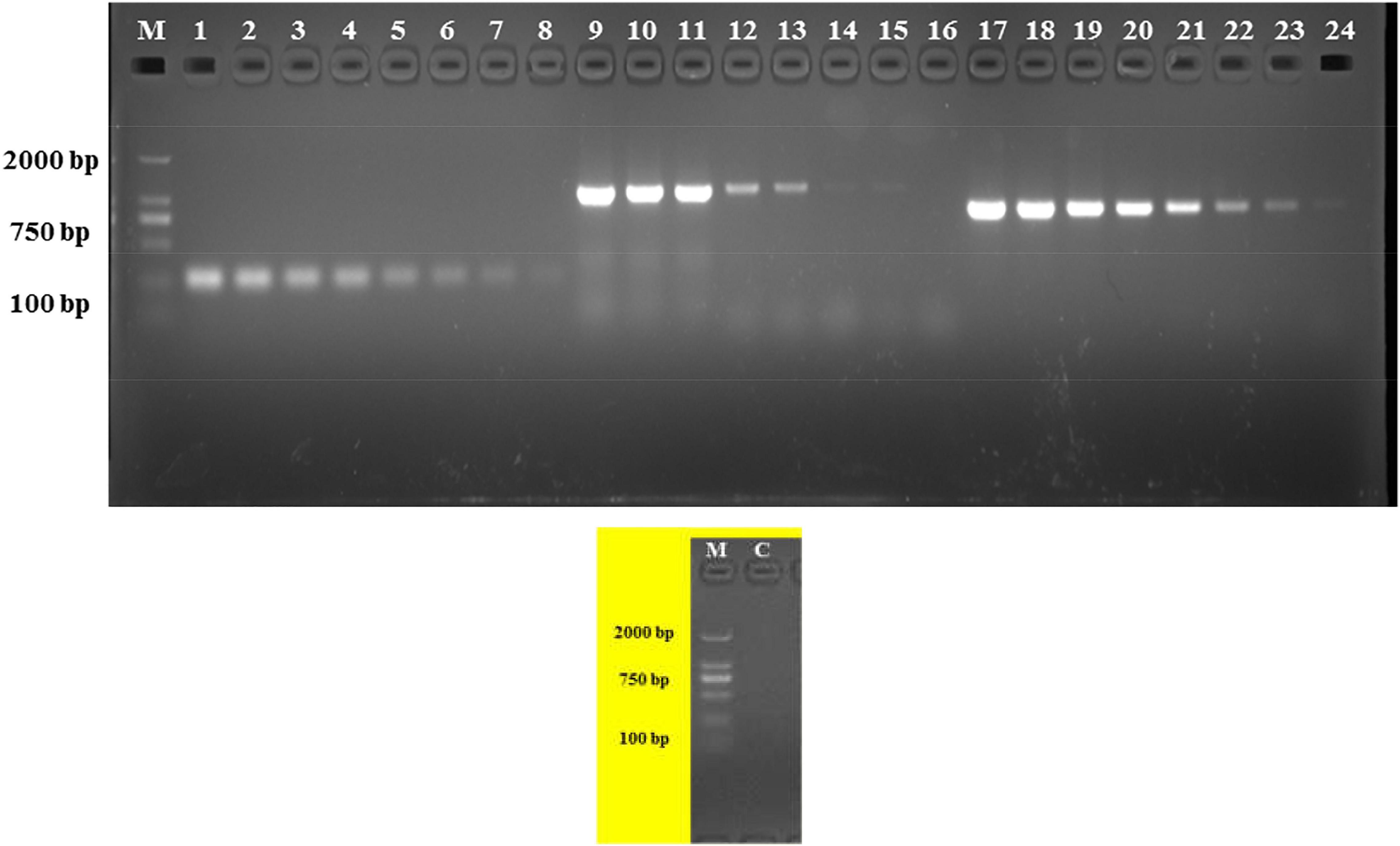
Figure 4. PCR detection sensitivity using dilutions of a pure culture of S. Albany strain SA48(32). Lane M: DSTM 2000 marker; Lane C: negative control (double-distilled H2O); lane 1–8, 9–16, and 17–24 primer sets based on group_27297, group_27289 and group_27286 genes, respectively (concentrations ranging from 2.8 × 107 to 100 CFU/mL).
Detection of Sereotype Salmonella in Artificially Contaminated Raw Pork Samples
Artificially contaminated raw pork samples were used to evaluate the sensitivity, specificity, and reliability of the primer sets designed based on serovar-specific genes. As shown in Table 2, the novel target-specific Albany serovar achieved a limit of detection as low as 1.04 × 103–104 CFU/g via PCR and target-specific Hadar serovar achieved a limit of detection as low as 1.16 × 104–105 CFU/g via PCR.
Discussion
At present, the Kauffmann-White scheme of serotyping is still the most commonly used method for the identification of Salmonella. This conventional method is challenging in its complicated control of serum quality and lengthy, while the role of NAATs such as PCR is clear in their rapid identification of foodborne diseases. The usefulness of the DNA amplification method is predicated on the selection of the appropriate target sequence and specificity of the primer sets. A few serovar-specific fragments for Salmonella are available except for those related to genes of the O and H antigens such as STM4495 for S. Typhimurium (Liu et al., 2012), sdfI for S. Enteritidis (De Freitas et al., 2010), ISR2 for S. Infantis (Akiba et al., 2011), and Newp2 for S. Newport (Bugarel et al., 2017), some of which may also be present in non-target strains due to the low specificity of targets—easily causing false-positive and false-negative results. Thus, additional novel specific molecular targets for Salmonella serovars must be identified and applied.
The comparative genomic method provides an available and efficient approach to obtain specific molecular targets for different pathogens. Unlike the complicated process of suppression subtractive hybridisation (requires restriction endonuclease, ligase, vector plasmid, and hybridisation) used for identifying specific targets to detect Salmonella serovar of Enteritidis and Pullorum (Agron et al., 2001; Li et al., 2009), the comparative genomic method only requires the collection of genomic sequences, classification target and non-target strains, and identification specific targets only present in target strains but absent in non-target strains by computers. Pan-genome analysis has been used to identify specific markers in bacteria, such as serotype, virulence, and antibiotic resistance, because bacterial phenotypes are usually associated with specific genes acquired through horizontal gene transfer (Buchanan et al., 2017; Laing et al., 2017). These genes are most frequently accessory genes in a bacterial species and are only present in bacterial strains that show corresponding phenotypes (McInerney et al., 2017). In order to obtain targets with high reliability and feasibility, we herein employed the pan-genome analysis approach to identify serovar-specific molecular targets for the detection of serogroup C2.
In a previous study, the SNSL254_A2005 gene (hypothetical protein) was evaluated as a PCR target for Salmonella serogroup C2 detection and the results demonstrated that among 4 Salmonella C2 isolates of 2 serovars and 103 non-target isolates, the SNSL254_A2005-based PCR detection showed 100% inclusivity and 100% exclusivity (Liu et al., 2011). In our study, the percentage of genomic sequences of strains for the SNSL254_A2005 gene presence in serogroup C2 and in other serogroups was 96.52% (555/575) and 2% (1/150), respectively, based on the gene presence/absence profile by pan-genome analysis. Thus, according to our results and previous studies, it can be concluded that the SNSL254_A2005 gene is an effective specificity target for Salmonella serogroup C2 detection.
Currently, O and H antigen genes are ordinary targets for the detection of Salmonella serovars based on PCR (Yang et al., 2012; Maurischat et al., 2015). The major drawbacks of this approach are the utilisation of several targets for the detection of one serotype. In this study, we obtained seven novel single targets to detect Salmonella serovar Hadar and Albany. The HSR3 and Hadspe (hypothetical protein) genes were evaluated as PCR targets for Salmonella serovar Hadar detection and the results showed high specificity (100%) of this gene (Bugarel et al., 2017; Chiang et al., 2018). Compared to previous reports, the percentage of genomic sequences of strains for the four novel genes (group_20134, group_22774, group_29844, and group_29846) and HSR3 specific for serovar Hadar presence in target strains was higher (97.56%, 40/41) than in the Hadspe (82.93, 34/41), whereas in non-target serovars, the HSR3 genes were observed in serovars Paratyphi B and Tees (0.29%, 2/684). To the best of our knowledge, no primer set has been reported for the specific detection of serovar Albany. In the current study, the group_27286, group_27289, and group_27297 genes were identified to be specific to serovar Albany. The percentage of genomic sequences of strains for the gene presence in serovar Albany was 100% (15/15) and in other serovars was 0.14 (1/710), 0.14 (1/710), and 0.28 (2/710). These three genes were present in one strain for serovar Newport (0.4%, 1/248) and one strain for serovar Kentucky (0.85, 1/117) was also observed in the group_27297 gene. In addition, Newp and Newspe genes were evaluated as PCR targets for Salmonella serovar Newport detection in a previous report and the results showed almost all of tested strains belonged to serovar Newport, Hadar, Bovismorbificans, Kottbus, Blockley, Manhattan, Litchfield, Glostrup, and several strains belonged to serovar Virchow and Muenchen were amplified by Newp gene; in order to distinguish serovar Newport and Hadar, the Newspe marker was designed, which showed 93.3% (28/30) strains for Newport and only S. Blockey were amplified Newspe gene (Chiang et al., 2018). This result was also observed in our study. The percentage of genomic sequences of strains for the Newp gene in target and non-target strains was 87.5% (217/248) and 23.69% (113/477), respectively, and 87.8% (36/41) strains for S. Hadar also had this gene, but when the Newspe gene was added, although S. Hadar did not show a cross-reaction, there were still some serovars harbouring of the Newspe gene, in which the percentage of genomic sequences of strains for the Newp gene in target and non-target strains was 78.3% (188/248) and 23.69% (169/477), respectively. Based on our research, there is no ideal specific gene for S. Newport. The results suggest that some molecular targets previously considered specific may inevitably result in elimination with the expansion of genome databases. We also analysed the potential functions of serovar-specific genes. Consistent with previous reports (Kim et al., 2006; Liu et al., 2012; Zhang et al., 2018), all of the selected specific genes for the serovar are encoded hypothetical proteins or putative proteins. Therefore, future studies on the functions of the selected seven genes are necessary for a better understanding of Salmonella serovar Hadar and Albany.
The primers for the PCR assays were designed according to the sequences of the seven specific genes and specificity was verified using 158 isolates with 42 Salmonella serovars in China and other genera. All primer sets showed perfect specificity in the PCR assays (Table 1). We determined the detection limit of the PCR assays using seven primer sets and different concentrations of pure Salmonella cultures. The detection limit for primer sets to serovar Albany and Hadar were 2.8 × 101–3 CFU/mL and 2.3 × 103–4 CFU/mL, respectively (Figure 4); these results are in agreement with those reported by Kong et al. (2013), Chen et al. (2015), and Chin et al. (2017). The sensitivity, specificity, and reliability of the PCR were further confirmed in S. Albany and Hadar in artificially contaminated raw pork samples. The detection limit was 1.04 × 103–4 CFU/g and 1.16 × 104–5 CFU/g in raw pork samples, which was in the same range as that of other targets by PCR (Beaubrun et al., 2012). Therefore, it can be applied for rapid convenient, specific and sensitive detection of Salmonella serovar Albany and Hadar by PCR using seven serovar-specific genes in food.
Conclusion
In conclusion, pan-genome analysis is an effective method to identify molecular targets specific to Salmonella serovars and the identified genes can be used as targets for molecular typing and identification of Salmonella serovars. The accuracy of the PCR results showed that it was a suitable method to explore the differences among serogroups. Future studies are needed to identify other single specific targets for the identification of other Salmonella serovars through pan-genome analysis for use as a new target for improved high-throughput detection methods crucial for the effective treatment and prevention of transmission from animal food to humans.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
QY contributed to design the experiments, analysed the data, and wrote the manuscript. QW, YD, and MC contributed to revise the manuscript. YS, FL, XX, CW, and BZ acquired the data. RP, SZ, JZ, XY, and LX played an important role in interpreting the results. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFC1600403), the Local Innovative and Research Teams Project of Guangdong PEARL River Talents Program (2017BT01S174), the Natural Science Foundation of Guangdong Province, China (2018A030313442), and the GDAS’ Project of Science and Technology Development (2018GDASCX-0401).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.605984/full#supplementary-material
Supplementary Figure 1 | Distribution of pan-genome.
Supplementary Table 1 | 725 genomic sequences of Salmonella serovar were divided into serogroup C2 and others databanks.
Supplementary Table 2 | Serovar-specific genes for Salmonella serogroups C2.
References
Agron, P. G., Walker, R. L., Kinde, H., Sawyer, S. J., Hayes, D. C., Wollard, J., et al. (2001). Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. App. Environ. Microb. 67, 4984–4991. doi: 10.1128/aem.67.11.4984-4991.2001
Akiba, M., Kusumoto, M., and Iwata, T. (2011). Rapid identification of Salmonella enterica serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by multiplex PCR. J. Microbiol. Methods 85, 9–15. doi: 10.1016/j.mimet.2011.02.002
Ashburner, M., Ball, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., et al. (2000). Gene ontology: tool for the untification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29.
Beaubrun, J. J. G., Cheng, C. M., Chen, K. S., Ewing, L., Wang, H., Agpaoa, M. C., et al. (2012). The evaluation of a PCR-based method for identification of Salmonella enterica serotypes from environmental samples and various food matrices. Food Microbiol. 31, 199–209. doi: 10.1016/j.fm.2012.03.016
Bonardi, S. (2017). Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol. Infect. 145, 1513–1526. doi: 10.1017/s095026881700036x
Buchanan, C. J., Webb, A. L., Mutschall, S. K., Kruczkiewicz, P., Barker, D. O. R., Hetman, B. M., et al. (2017). A genome-wide association study to identify diagnostic markers for human pathogenic Campylobacter jejuni strains. Front. Microbiol. 8:1224. doi: 10.3389/fmicb.2017.01224
Bugarel, M., Tudor, A., Loneragan, G. H., and Nightingale, K. K. (2017). Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J. Microbiol. Methods 134, 14–20. doi: 10.1016/j.mimet.2016.12.011
Castelijn, G. A. A., Parabirsing, J. A., Zwietering, M. H., Moezelaar, R., and Abee, T. (2013). Surface behaviour of S. Typhimurium, S. Derby, S. Brandenburg and S. Infantis. Vet. Microbiol. 161, 305–314. doi: 10.1016/j.vetmic.2012.07.047
Chanda, Y., Alam, M. A., and Singh, S. (2020). Pan-genomic analysis of the species Salmonella enterica: identification of core essential and putative essential genes. Gene Rep. 20:100669. doi: 10.1016/j.genrep.2020.100669
Chen, Q. M., Tao, T. T., Bie, X. M., Lu, Y. J., Lu, F. X., Zhai, L. G., et al. (2015). Mining for sensitive and reliable species-specific primers for PCR for detection of Cronobacter sakazakii by a bioinformatics approach. J. Dairy Sci. 98, 5091–5101. doi: 10.3168/jds.2015-9304
Chiang, Y. C., Wang, H. H., Ramireddy, L., Chen, H. Y., Shih, C. M., Lin, C. K., et al. (2018). Designing a biochip following multiplex polymerase chain reaction for the detection of Salmonella serovars Typhimurium, Enteritidis, Infantis, Hadar, and Virchow in poultry products. J. Food Drug Anal. 26, 58–66. doi: 10.1016/j.jfda.2016.11.019
Chin, W. H., Sun, Y., Høgberg, J., Quyen, T. L., Engelsmann, P., Wolff, A., et al. (2017). Direct PCR-A rapid method for multiplexed detection of different serotypes of Salmonella in enriched pork meat samples. Mol. Cell. Probes 7, 24–32. doi: 10.1016/j.mcp.2016.11.004
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Davies, M. R., Broadbent, S. E., Harris, S. R., Thomson, N. R., and Der Woude, M. W. V. (2013). Horizontally acquired glycosyltransferase operons drive Salmonellae lipopolysaccharide diversity. PLoS Genet. 9:e1003568. doi: 10.1371/journal.pgen.1003568
De Freitas, C. G., Santana, ÂP., Silva, P. H. C. D., Goncalves, V. S. P., Barros, M. D. A. F., Torres, F. A. G., et al. (2010). PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 139, 15–22. doi: 10.1016/j.ijfoodmicro.2010.02.007
Dekker, J. P., and Frank, K. M. (2015). Salmonella, Shigella, and yersinia. Clin. Lab. Med. 35, 225–246.
Deshpande, A., Curran, E. T., Jamdar, S., Inkster, T., and Jones, B. (2015). Historical outbreak of Salmonella hadar. J. Hosp. Infect. 91, 171–175. doi: 10.1016/j.jhin.2015.05.014
Doublet, B., Lailler, R., Meunier, D., Brisabois, A., Boyd, D., Mulvey, M. R., et al. (2003). Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9, 585–591.
Graziani, C., Losasso, C., Luzzi, I., Ricci, A., Scavia, G., and Pasquali, P. (2017). “Chapter 5 – Salmonella,” in Foodborne Diseases (Third Edition), eds C. E. R. Dodd, T. Aldsworth, R. A. Stein, D. O. Cliver, and H. P. Riemann (Cambridge, MA: Academic Press), 133–169.
Herrera-León, S., Ramiro, R., Arroyo, M., Díez, R., Usera, M. A., and Echeita, M. A. (2007). Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res. Microbiol. 158, 122–127. doi: 10.1016/j.resmic.2006.09.009
Kerouanton, A., Rose, V., Weill, F. X., Granier, S. A., and Denis, M. (2013). Genetic diversity and antimicrobial resistance profiles of Salmonella enterica serotype derby isolated from pigs, pork, and humans in France. Foodborne Pathog. Dis. 10, 977–984. doi: 10.1089/fpd.2013.1537
Kim, H. J., Park, S. H., Lee, T., Nahm, B., Chung, Y. H., Seo, K. H., et al. (2006). Identification of Salmonella enterica serovar Typhimurium using specific PCR primers obtained by comparative genomics in Salmonella serovars. J. Food Prot. 69, 1653–1661. doi: 10.4315/0362-028x-69.7.1653
Kim, Y., Gu, C., Kim, H. U., and Lee, S. Y. (2020). Current status of pan-genome analysis for pathogenic bacteria. Curr. Opin. Biotech. 63, 54–62. doi: 10.1016/j.copbio.2019.12.001
Kong, X. H., Lu, Z. X., Zhai, L. G., Yao, S. L., Zhangm, C., Lv, F. X., et al. (2013). Mining and evaluation of new specific molecular targets for the PCR detection of Salmonella spp. genome. World J. Microb. Biot. 29, 2219–2226. doi: 10.1007/s11274-013-1387-0
Laing, C. R., Whiteside, M. D., and Gannon, V. P. J. (2017). Pan-genome analyses of the species Salmonella enterica, and identification of genomic markers predictive for species, subspecies, and serovar. Front. Microbiol. 8:1345. doi: 10.3389/fmicb.2017.01345
Lee, S. H., Park, S. M., Kim, B. N., Kwon, O. S., Rho, W. Y., and Jun, B. H. (2019). Emerging ultrafast nucleic acid amplification technologies for next-generation molecular diagnostics. Biosens. Bioelectron. 141:111448. doi: 10.1016/j.bios.2019.111448
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, 242–245.
Li, Q. C., Xu, Y. H., and Jiao, X. A. (2009). Identification of Salmonella pullorum genomic sequences using suppression subtractive hybridization. J. Microbiol. Biotechnol. 19, 898–903. doi: 10.4014/jmb.0812.694
Liu, B., Zhang, L. D., Zhu, X. N., Shi, C. L., Chen, J., Liu, W. B., et al. (2011). PCR identification of Salmonella serogroups based on specific targets obtained by comparative genomics. Int. J. Food Microbiol. 144, 511–518. doi: 10.1016/j.ijfoodmicro.2010.11.010
Liu, B., Zhou, X., Zhang, L., Liu, W., Dan, X., and Shi, C. (2012). Development of a novel multiplex PCR assay for the identification of Salmonella enterica Typhimurium and Enteritidis. Food Control 27, 87–93. doi: 10.1016/j.foodcont.2012.01.062
Maurischat, S., Baumann, B., Martin, A., and Malorny, B. (2015). Rapid detection and specific differentiation of Salmonella enterica subsp. enterica Enteritidis, Typhimurium and its monophasic variant 4,[5],12:i:- by real-time multiplex PCR. Int. J. Food Microbiol. 193, 8–14. doi: 10.1016/j.ijfoodmicro.2014.10.004
McInerney, J. O., McNally, A., and Connell, M. J. O. (2017). Why prokaryotes have pangenomes. Nat. Microbiol. 2:17040.
Ni, P. E., Xu, Q., Yin, Y., Liu, D., Zhang, J., Wu, Q., et al. (2018). Prevalence and characterization of Salmonella serovars isolated from farm products in Shanghai. Food Control 85, 269–275. doi: 10.1016/j.foodcont.2017.10.009
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Rodriguez, A., Pangloli, P., Richards, H. A., Mount, J. R., and Draughon, F. A. (2006). Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69, 2576–2580. doi: 10.4315/0362-028x-69.11.2576
Sannigrahi, S., Arumugasamy, S. K., and Mathiyarasu, J. K. S. (2020). Magnetosome-anti-Salmonella antibody complex based biosensor for the detection of Salmonella typhimurium. Mater. Sci. Eng. 114:111071. doi: 10.1016/j.msec.2020.111071
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tettelin, H., Riley, D. R., Cattuto, C., and Medini, D. (2008). Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477. doi: 10.1016/j.mib.2008.09.006
Thépault, A., Méric, G., Rivoal, K., Pascoe, B., Mageiros, L., Touzain, F., et al. (2017). Genome-wide identification of hostsegregating epidemiological markers for source attribution in Campylobacter jejuni. App. Environ. Microbiol. 83:e3085–16.
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524.
Wagner, C., and Hensel, M. (2011). Adhesive mechanisms of Salmonella enterica. Adv. Exp. Med. Biol. 715, 17–34. doi: 10.1007/978-94-007-0940-9_2
Weidebotjes, M., Kobe, B., Lange, C. C., and Schwarz, S. (1998). Molecular typing of Salmonella enterica subsp. enterica serovar Hadar: evaluation and application of different typing methods. Vet. Microbiol. 61, 215–227. doi: 10.1016/s0378-1135(98)00181-3
Xu, C., Ren, X., Feng, Z., Fu, Y., Hong, Y., Shen, Z., et al. (2017). Phenotypic characteristics and genetic diversity of Salmonella enterica serotype derby isolated from human patients and foods of animal origin. Foodborne Pathog. Dis. 14, 593–599. doi: 10.1089/fpd.2017.2278
Yang, X. J., Huang, J. H., Wu, Q. P., Zhang, J. M., Liu, S. R., Guo, W. P., et al. (2016). Prevalence, antimicrobial resistance and genetic diversity of Salmonella isolated from retail ready-to-eat foods in China. Food Control 60, 50–56. doi: 10.1016/j.foodcont.2015.07.019
Yang, X. J., Huang, J. H., Zhang, Y. X., Liu, S. R., Chen, L., Xiao, C., et al. (2020a). Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Yang, X. J., Li, H. G., Wu, Q. P., Zhang, J. M., and Chen, L. (2015b). Comparison of direct culture, immunomagnetic separation/culture, and multiplex PCR methods for detection of Salmonella in food. Food Sci. Technol. Res. 21, 671–675. doi: 10.3136/fstr.21.671
Yang, X. J., Wu, Q. P., Huang, J. H., Wu, S., Zhang, J. M., Xiao, C., et al. (2020b). Prevalence and characterization of Salmonella isolated from raw vegetablesin China. Food Control 109:10691.
Yang, X. J., Wu, Q. P., Zhang, J. M., Huang, J. H., Chen, L., Liu, S. R., et al. (2015a). Prevalence, enumeration, and characterization of Salmonella isolated from aquatic food products from retail markets in China. Food Control 57, 308–313. doi: 10.1016/j.foodcont.2015.03.046
Yang, Y. J., Wan, C. X., Xu, H. Y., Lai, W. H., Xiong, Y. H., Xu, F., et al. (2012). Development of a multiplexed PCR assay combined with propidium monoazide treatment for rapid and accurate detection and identification of three viable Salmonella enterica serovars. Food Control 28, 456–462. doi: 10.1016/j.foodcont.2012.05.061
Keywords: Salmonella, C2 serogroups, serovar-specific molecular targets, PCR, pan-genome analysis
Citation: Ye Q, Shang Y, Chen M, Pang R, Li F, Xiang X, Wang C, Zhou B, Zhang S, Zhang J, Yang X, Xue L, Ding Y and Wu Q (2021) Identification of Novel Sensitive and Reliable Serovar-Specific Targets for PCR Detection of Salmonella Serovars Hadar and Albany by Pan-Genome Analysis. Front. Microbiol. 12:605984. doi: 10.3389/fmicb.2021.605984
Received: 14 September 2020; Accepted: 22 February 2021;
Published: 16 March 2021.
Edited by:
Sheng Chen, City University of Hong Kong, Hong KongReviewed by:
Shizhong Geng, Yangzhou University, ChinaChunlei Shi, Shanghai Jiao Tong University, China
Copyright © 2021 Ye, Shang, Chen, Pang, Li, Xiang, Wang, Zhou, Zhang, Zhang, Yang, Xue, Ding and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Ding, ZGluZ3l1QGpudS5lZHUuY24=; Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors share first authorship
 Qinghua Ye1†
Qinghua Ye1† Moutong Chen
Moutong Chen Rui Pang
Rui Pang Fan Li
Fan Li Jumei Zhang
Jumei Zhang Liang Xue
Liang Xue Yu Ding
Yu Ding Qingping Wu
Qingping Wu