- 1Beijing Key Laboratory for Prevention and Control of Infectious Diseases in Livestock and Poultry, Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Department of Animal Science, University of Tennessee, Knoxville, TN, United States
- 3Key Laboratory of Animal Epidemiology of Ministry of Agriculture, College of Veterinary Medicine, China Agricultural University, Beijing, China
Avibacterium paragallinarum is the pathogen of infectious coryza, which is a highly contagious respiratory disease of chickens that brings a potentially serious threat to poultry husbandry. Iron is an important nutrient for bacteria and can be obtained from surroundings such as siderophores and hemophores. To date, the mechanisms of iron acquisition and heme utilization as well as detailed regulation in A. paragallinarum have been poorly understood. In this study, we investigated the transcriptomic profiles in detail and the changes of transcriptomes induced by iron restriction in A. paragallinarum using RNA-seq. Compared with the iron-sufficiency control group, many more differentially expressed genes (DEGs) and cellular functions as well as signaling pathways were verified in the iron-restriction group. Among these DEGs, the majority of genes showed decreased expression and some were found to be uniquely present in the iron-restriction group. With an in-depth study of bioinformatic analyses, we demonstrated the crucial roles of the Hut protein and DUF domain-containing proteins, which were preferentially activated in bacteria following iron restriction and contributed to the iron acquisition and heme utilization. Consequently, RT-qPCR results further verified the iron-related DEGs and were consistent with the RNA-seq data. In addition, several novel sRNAs were present in A. paragallinarum and had potential regulatory roles in iron homeostasis, especially in the regulation of Fic protein to ensure stable expression. This is the first report of the molecular mechanism of iron acquisition and heme utilization in A. paragallinarum from the perspective of transcriptomic profiles. The study will contribute to a better understanding of the transcriptomic response of A. paragallinarum to iron starvation and also provide novel insight into the development of new antigens for potential vaccines against infectious coryza by focusing on these iron-related genes.
Introduction
Infectious coryza, caused by Gram-negative bacterium Avibacterium paragallinarum, is a severe respiratory disease of chickens that brings enormous economic losses for the poultry industry worldwide (Blackall, 1999). A. paragallinarum is classified into three serovars—A, B, and C—according to the Page schemes. In South Africa, it is considered as one of the most serious diseases, with C3 being the predominant serovar (Bragg et al., 1996; Boucher et al., 2014). In China, serovars A, C, and B were identified for the first time in 1987, 1995, and 2003, respectively (Zhang et al., 2003; Sun et al., 2018; Xu et al., 2019). Several factors are responsible for the pathogenicity of A. paragallinarum. Among them, hemagglutinin (HA) has been regarded as the most key virulence factor because of its participation in tissue adhesion (Barnard et al., 2008). Vaccination remains the most effective preventive measure against A. paragallinarum. However, vaccination failures always occur with the emergence of new serovars or serovar variants (Blackall, 1999; Bragg, 2005). In addition, the advent of multidrug bacterial resistance is deemed one of the most alarming situations worldwide. Therefore, it is urgent for the development of novel broad-spectrum prophylactic agents against A. paragallinarum. To date, the molecular mechanisms of colonization, growth, and virulence of A. paragallinarum in chicken upper respiratory tract are still poorly understood.
Iron is an essential micronutrient for most bacteria, which plays essential roles in bacterial colonization, growth, and virulence. Unfortunately, the concentration of free iron in the host is not enough to support the growth of bacteria (del Rio et al., 2005). In order to achieve effective iron homeostasis, bacteria have evolved sophisticated iron-acquisition systems to scavenge iron from their surroundings (siderophores, hemophores, or host-molecule-binding proteins) to ensure adequate supplies (Parrow et al., 2013). The siderophore transport system, which is capable of acquiring free inorganic iron, consists of both siderophores and TonB-dependent transferrin receptors (TBDR) on the outer membrane of bacteria (Chu et al., 2010). In A. paragallinarum, Abascal et al. (2009) have identified three up-expressed outer membrane proteins acting as transferrin receptor and iron transport proteins in bacteria cultured in iron-restricted medium, which are regulated by the Fur protein (Abascal et al., 2009). They speculate that these proteins are associated with the colonization of bacteria in an iron-limited host environment. Besides transferrin and lactoferrin, enterobactin is also proved to be a crucial siderophore. Studies have confirmed that the growth ability of many bacteria has been enhanced in the presence of enterobactin, including not only enteric bacteria such as Escherichia coli and Campylobacter jejuni, but also respiratory bacteria such as Hemeophilus parainfluenzae and Bordetella pertussis (Williams et al., 1990; Thulasiraman et al., 1998; Anderson and Armstrong, 2004; Horiyama and Nishino, 2014). In terms of ferric enterobactin receptor, we have previously demonstrated that CfrA and CfrB contribute to iron acquisition in Campylobacter (Zeng et al., 2009; Xu et al., 2010). Until now, the mechanisms of iron acquisition by siderophores and the detailed regulation in A. paragallinarum are still on the preliminary stage and remain to be further explored.
Remarkably, the availability of free iron is strongly limited in vertebrate hosts, causing insufficient iron acquisition by siderophores, especially for pathogenic bacteria (Sekine et al., 2016). Thus, the heme utilization system has been well developed in bacteria to better obtain iron. Over the past decades, the utilization of heme is validated to be a common mechanism employed by pathogens. For example, heme is preferred as an iron source over ferrous iron for some Gram-positive bacteria such as Staphylococcus aureus and Streptococcus pyogenes (Rouault, 2004; Skaar et al., 2004; Zhu et al., 2008). For Gram-negative pathogens, proteins encoded within the heme transport operon have been studied such as Vibrio anguillarum (Sekine et al., 2016). To acquire heme from hemoproteins, bacteria have developed sophisticated heme acquisition machineries. Gram-negative pathogens can also utilize an outer TonB-dependent membrane receptor protein to transport heme into the periplasm, and then periplasmic heme-binding proteins transport heme to the cytoplasm (Contreras et al., 2014; Richard et al., 2019). In the cytoplasm, heme can be degraded by heme-degrading enzymes, which remove iron from the heme (Sekine et al., 2016). Nevertheless, the research on heme uptake and utilization by A. paragallinarum is rare.
Small regulatory RNAs (sRNAs) act as post-transcriptional regulators controlling bacterial adaptation to environmental changes (Koeppen et al., 2016; Gerrick et al., 2018). Their regulatory functions have become the center of attention in a variety of bacteria, such as E. coli, Salmonella, and Pseudomonas aeruginosa (Koeppen et al., 2016; Chareyre and Mandin, 2018; El Mouali et al., 2018). sRNAs play crucial roles in regulating outer membrane protein expression, iron ion balance, community induction, and bacterial pathogenicity. The regulatory mechanism involves binding to target bacterial mRNAs or directly interact with target proteins (Repoila and Darfeuille, 2009). For instance, sRNAs RyhB of enteric bacteria and PrrF of P. aeruginosa are highly upregulated during iron starvation, which are able to suppress mRNAs encoding iron-containing proteins (Masse et al., 2005; Reinhart et al., 2015). Recently, MrsI is identified to be important for iron-sparing response in Mycobacterium tuberculosis (Gerrick et al., 2018). To date, sRNA studies in A. paragallinarum have been limited, let alone the interaction between any sRNA and its targets.
The identification and quantification of differentially expressed genes (DEGs) can be conducted at a transcriptomic level by using RNA sequencing (RNA-seq) technology. This transcriptomic approach has identified 13 upregulated genes encoding for proteins exposed to iron-restriction stress at the bacterial surface of Hemeophilus (Glässerella) parasuis (Alvarez-Estrada et al., 2018). In addition, RNA-seq can also be used to characterize sRNAs, including their secondary structure and predicted mRNA targets. Koeppen et al. (2016) have characterized differentially packaged sRNAs in outer membrane vesicle secreted by P. aeruginosa. Nevertheless, a comprehensive analysis of the transcriptomic changes in A. paragallinarum to iron-restriction stress has yet to be performed.
To get a deeper understanding of the biological pathways and main transcripts involved in iron-restriction stress as well as exploring the specific molecular mechanism behind iron acquisition and heme utilization in A. paragallinarum, we used RNA-seq for gaining information about gene and sRNA abundance in A. paragallinarum under iron-restriction condition. To the best of our knowledge, this is the first report regarding the transcriptomic and bioinformatic characterizations of iron acquisition and heme utilization in A. paragallinarum in response to iron starvation.
Materials and Methods
Bacterial Strain and Growth Conditions
The strain 3005 of the A. paragallinarum serogroup C was used in this study, which was isolated from China in 2018. As outlined in Supplementary Figure 1, tryptic soya broth (TSB) and tryptic soya broth agar (TSA) that were added with 10% chicken serum and 0.0025% nicotinamide adenine dinucleotide (NAD) were used for the propagation and maintenance of the strain. Then, A. paragallinarum was grown under in vitro culture conditions with iron restriction (R) or without iron restriction (C, as a control). For the iron-restriction condition, A. paragallinarum cells were inoculated at 1:100 in 200 ml minimal essential medium alpha (MEMα) (Invitrogen) with 0.0025% (w/v) NAD (Palyada et al., 2004). For the control condition, an extra 40 μM FeSO4 was added in the cultures based on the related reference (Palyada et al., 2004). The cells were grown in a shaking incubator at 37°C until reaching an optical density (OD) of 0.6-1.0 at 600 nm (exponential phase of growth), and samples were harvested by centrifugation, followed immediately by being resuspended in RNA later (Ambion, Carlsbad, CA, United States). Samples were then stored at −80°C. Three replicates were made under each condition.
RNA Extraction and Sample Preparation
Total RNA for RNA sequencing was extracted using TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions, and the contaminating DNA was removed by treatment with DNase I (TaKaRa) and confirmed by negative PCR amplification results. Finally, RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies).
Library Preparation and Illumina Sequencing
Two micrograms of total RNA from each sample was used for RNA-seq transcriptome library preparation. After ribosomal RNA (rRNA) was depleted, mRNA was purified and fragmented. Subsequently, first- and second-strand cDNAs were synthesized and ligated with adapters. cDNA library preparation was performed sequentially with the TruSeq RNA Sample Prep Kit from Illumina (San Diego, CA, United States) following the manufacturer’s protocol. After quantified by TBS380 (Picogreen), paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq × 10 (2 × 150 bp read length). The processing of original images to sequences, base-calling, and quality value calculations were performed using the Illumina GA Pipeline (version 1.6), in which 150 bp paired-end reads were obtained. A Perl program was written to select clean reads by removing low-quality sequences, reads with more than 5% of N bases (unknown bases), and reads containing adaptor sequences.
Mapping and Alignment of Sequence Reads
After the low-quality reads were trimmed and the adapters were removed, the clean reads were mapped and aligned onto the whole reference genome (A. paragallinarum strain AVPG2015, GenBank accession nr NC_011852.1) using Bowtie 2.0.
Differential Gene Analysis of RNA-Seq
The raw data were processed for the gene data analysis using the normalized transcripts per million mapped reads (TPM) values using RSEM, and differential gene expression was obtained by DESeq2. Based on the negative binomial distribution model, heterogenetic analysis among samples was obtained by controlling the fold change (FC) and p value statistical methods. The screening conditions of this study were | log2FC| ≥ 1 and p < 0.05. An analysis of variance (ANOVA), using the Benjamini–Hochberg multiple testing correction, was performed to identify genes significantly differentially expressed (p < 0.05). Significantly differential expressed genes with fold change ≥ 2 were subjected to further Gene Ontology (GO) and pathway analysis.
Bioinformatics and Statistical Analyses
Differentially expressed genes were classified for the categories using annotations of GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with the DAVID online software1. The threshold of significantly enriched genes was determined according to the corrected p (Benjamini) (p ≤ 0.05). The default parameters (hypergeometric algorithm, Benjamini–Hochberg multiple testing correction) were performed to take overrepresentation analyses. GO terms (biological process, cellular component, and molecular function) and KEGG pathways terms with p-Values of ≤0.05 were considered to be significantly enriched with DEGs. The heatmaps of DEGs were achieved with the pheatmap packages from R.
sRNA Analysis
Small regulatory RNAs prediction was obtained by the current mainstream prokaryotic analysis software Rockhopper2, which was mainly based on the base sequencing coverage information. Besides, the RNAfold program in the Vienna RNA packages was used to predicate the secondary structure of sRNA based on the minimum free energy.
Data Deposition
The raw sequencing files from this study were deposited into the NCBI SRA repository and the SRA accession number was SRR13291129.
Validation of DEGs by Quantitative Real-Time PCR
Differentially expressed genes identified by the above-described method were validated using quantitative real-time PCR (qPCR). The gyrA was used as a reference control (Wen et al., 2016). The information on the primers is listed in Supplementary Table 1. The qPCR programs for VY92_RS06600, VY92_RS09125, VY92_RS09790, VY92_RS03730, and VY92_RS03735 were as follows: 1 cycle of 95°C for 3 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 40 s. The qPCR programs for VY92_RS01655, VY92_RS00335, VY92_RS05725, and gyrA were as follows: 1 cycle of 95°C for 3 min; 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 40 s. Gene expression levels were calculated as previously described (Huo et al., 2020). Data analysis was conducted by using two-way analysis of variance with GraphPad Prism (ver. 5.0). p < 0.05 represents statistical significance. The results were shown as the means ± standard deviations of three independent experiments.
Results
Quality Control of Sequencing Data and Mapping
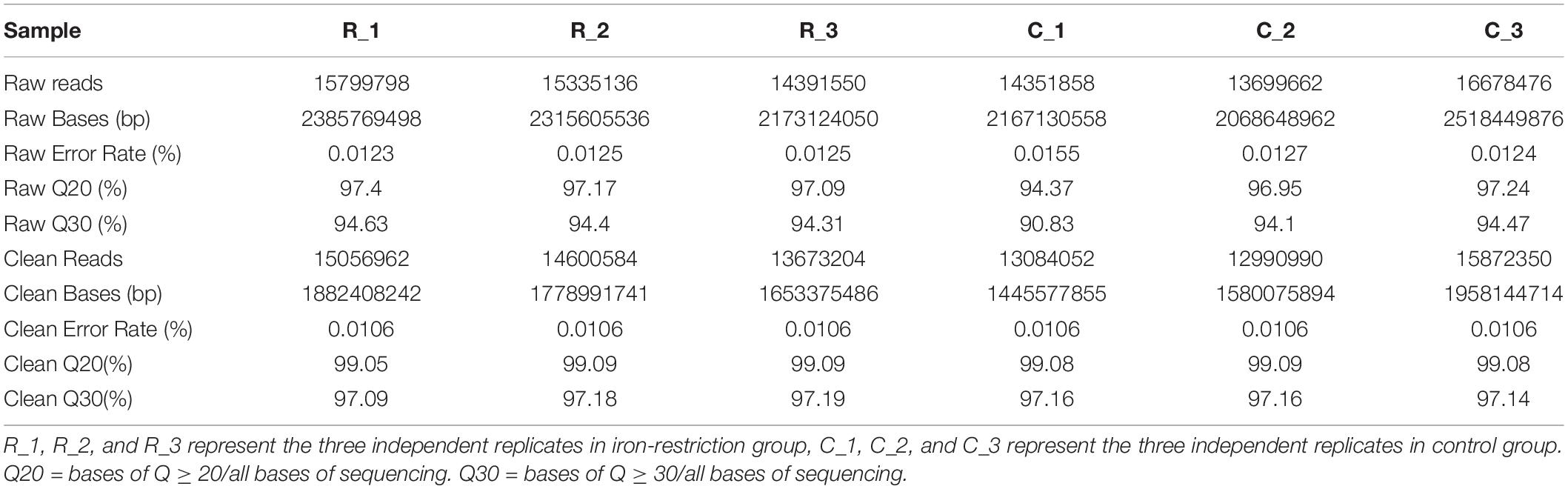
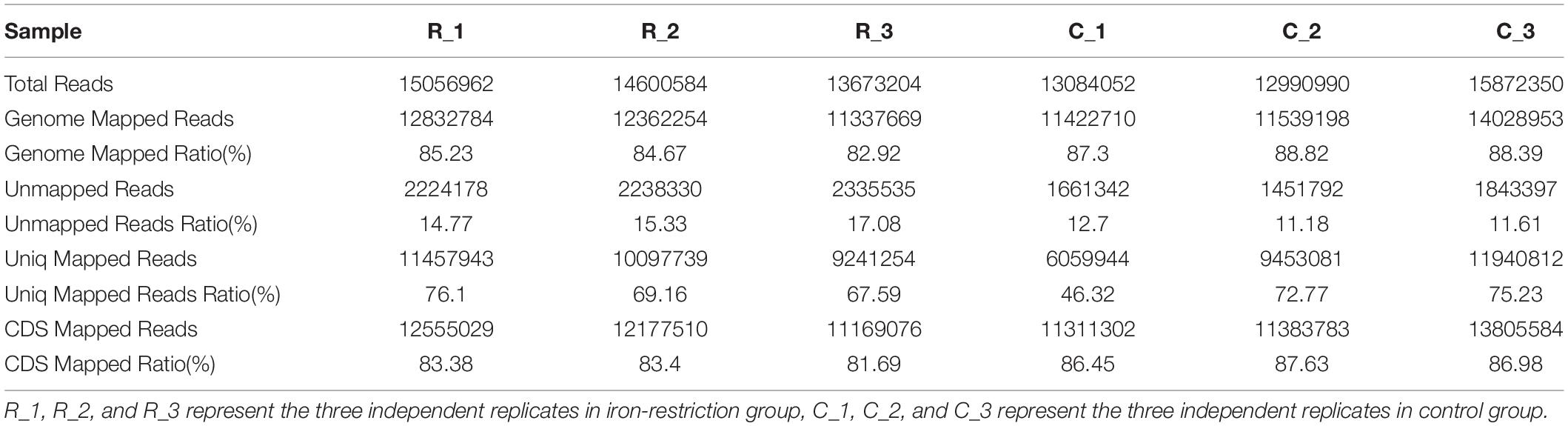
As outlined in Supplementary Figure 1, the cells were grown and harvested at 5 h for RNA sequencing based on the measurement of OD values via bacteria growth assay (Supplementary Table 2). Then, the quality control of sequencing data was taken. As shown in Table 1, 13.70 to 16.68 M raw reads were generated for each sample, which the RawQ20 and Raw30 ratios were all above 90%. High-quality reads were obtained and used for further analyses after quality assessment using the FastQC tool and data preprocessing. After the quality control, clean reads with numbers averaging between 13.00 and 15.87 M were obtained per sample, in which the CleanQ20 and CleanQ30 ratios reached more than 95%. Besides, we also assessed the nucleotide distribution of both raw and clean data and found that sequences were all roughly distributed at about 150 bp (Supplementary Figure 2).
In order to quantify the gene expression in samples, sequence mapping and alignment analysis of high-quality clean reads was taken at first by comparing with the reference genome (A. paragallinarum strain AVPG2015, GenBank accession nr NC_011852.1). As shown in Table 2, more than 80% of reads from all experimental groups could be aligned to the reference genome, and unique mapped reads ratio ranged from 46.32 to 76.1%. Besides, 81.69 to 87.63% of these clean reads were distributed in coding regions. The distribution and numbers of gene expression are shown in Figure 1A. In the density diagram of gene expression, high- and low-abundance genes could be shown and formed an obvious peak in each group. Furthermore, the TPM of genes was shown to be similar between these two groups, indicating the stability of the expression levels. Thus, the sequencing data conformed to the requirements of subsequently bioinformatics analysis.
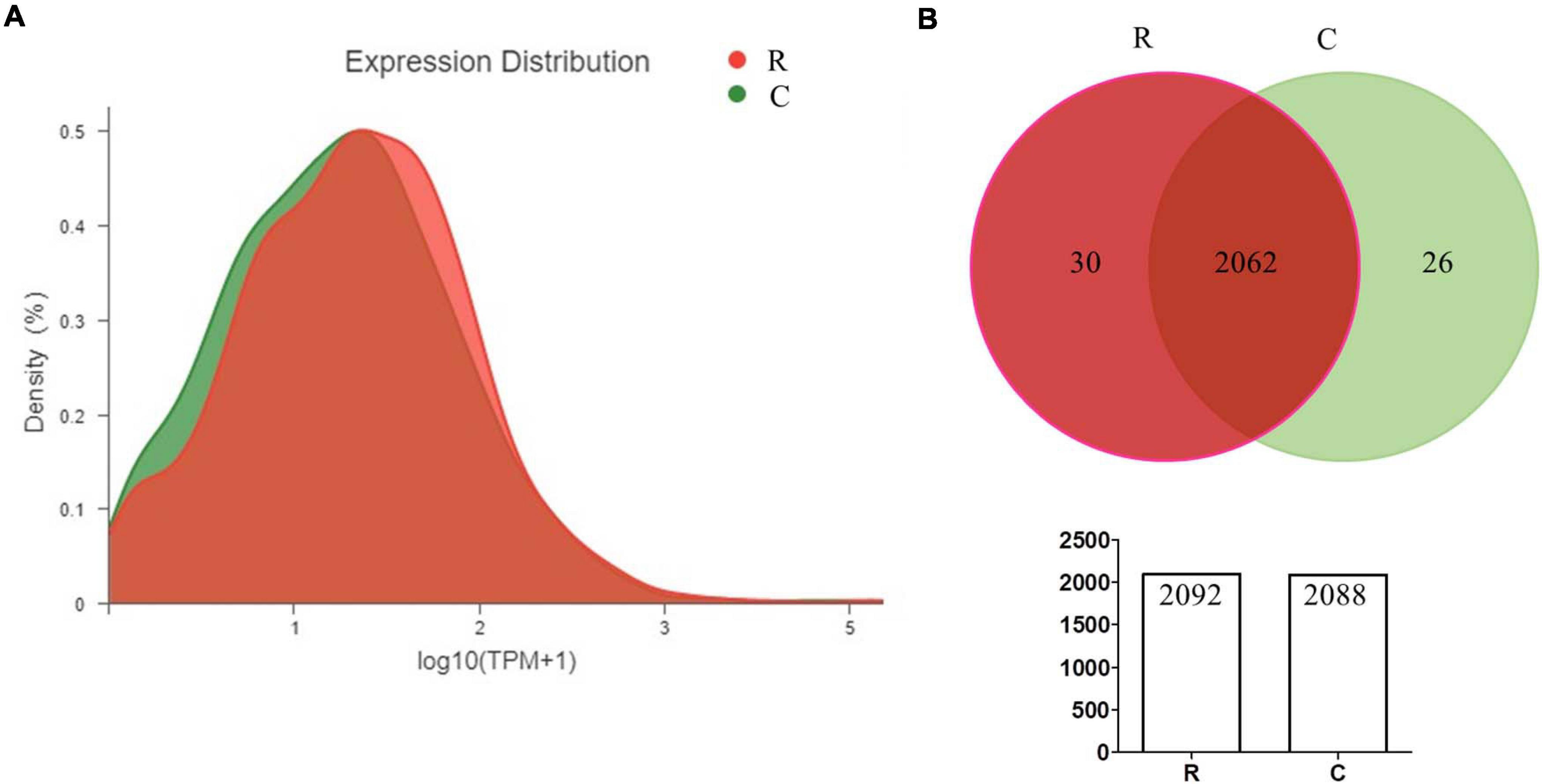
Figure 1. Quantitative statistics of gene expression (A,B). The density diagram and Venn diagram showing the distribution of gene expression and the overlap of genes in A. paragallinarum under conditions with iron restriction or without iron restriction, respectively.
Identification and Quantification of Differentially Expressed Genes
According to the quantification of gene expressions, we analyzed 2,566 transcripts, including 2,445 genes encoding proteins and 121 genes encoding sRNAs (Supplementary Data Sheet 1). By taking the Venn analysis, we found that there were 2,062 common genes in these two groups (Figure 1B). In addition, 30 genes were uniquely expressed in A. paragallinarum under iron-restriction condition and 26 genes were uniquely expressed in A. paragallinarum under the control condition, respectively.
Based on the criteria of | log2FC| ≥ 1 and p-adjust < 0.05, the numbers of genes that up- and downregulated their abundance were summarized. As shown in Figure 2 and Supplementary Data Sheet 1, 122 DEGs were identified significantly between the iron-restriction group and the control group. Compared with the iron-sufficiency control group, iron restriction could significantly downregulate 68 genes, including 65 genes encoding for proteins and 3 genes encoding for sRNAs, which include autotransporter outer membrane beta-barrel domain-containing protein and other proteins. Meanwhile, 54 genes were significantly upregulated in the iron-restriction condition than in the control, which consisted of 50 genes encoding for proteins and 4 genes encoding for sRNAs. Among these 54 differentially upregulated genes, 19 genes could directly evolve into sophisticated iron-acquisition systems, such as iron utilization receptors, ABC transporters, and TonB energy systems (Table 3). Additionally, several iron-regulated operons were identified for the gene clustering analysis, suggesting that a number of differentially up-expressed genes might be cotranscribed. There were several operons involved in ABC transporters: one that contained hemoglobin/transferrin/lactoferrin receptor (VY92_RS00325) and ABC transporter ATP-binding protein (VY92_RS00330 and VY92_RS00335), one that contained Fe(3+)-hydroxamate ABC transporter permease FhuB (VY92_RS05675) and iron-hydroxamate transporter ATP-binding subunit (VY92_RS05685), and another one that contained ABC transporter ATP-binding protein (VY92_RS05745) and ABC transporter permease (VY92_RS05750). An operon was shown to be involved in heme utilization systems that contained heme utilization protein HutZ (VY92_RS03730), heme utilization cytosolic carrier protein HutX (VY92_RS03735), and TonB-dependent receptor (VY92_RS03740). An operon was shown to be involved in iron transporter and iron permease that contained iron transporter (VY92_RS05765), iron permease (VY92_RS05770), and iron-sulfur cluster carrier protein ApbC (VY92_RS05775). There was also an operon that was involved in TonB energy systems, containing energy transducer TonB (VY92_RS11145) and TonB system transport proteins ExbD and ExbB (VY92_RS11150 and VY92_RS11155). Interestingly, 98 genes encoding for numerous domains of unknown function (DUF) domain-containing proteins were discovered in A. paragallinarum. Among these genes, there were 57 (58%) upregulated and 17 (17%) downregulated DUF genes in the iron-restriction group. Furthermore, significant differences of gene expressions were shown in three upregulated DUF genes, which encoded DUF454, DUF4198, and DUF2318 domain-containing proteins, respectively. There was also one DUF gene that encodes DUF1345 domain-containing protein that was significantly downregulated in the iron-restriction group than in the control group.
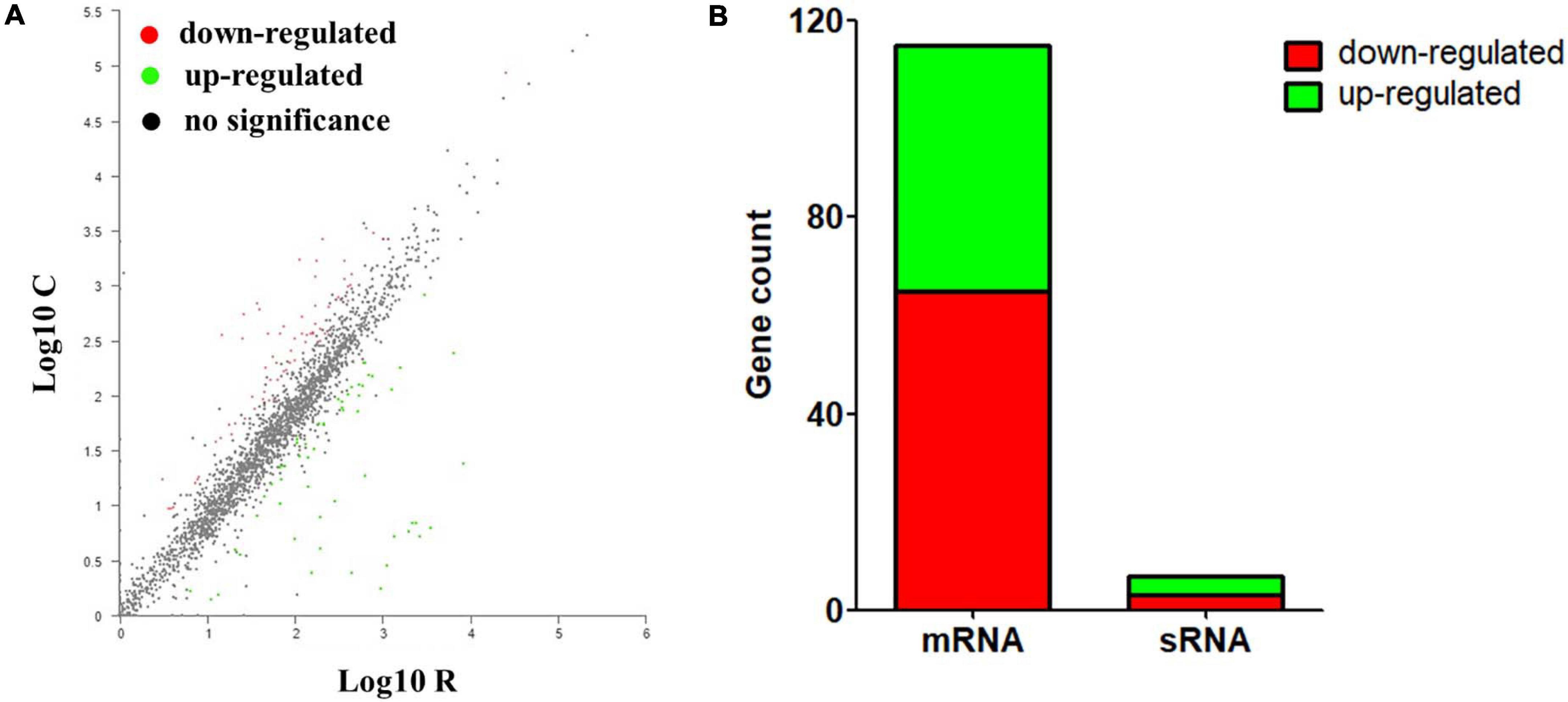
Figure 2. Identification and quantification comparison of differentially abundant genes in A. paragallinarum under iron-restriction condition compared with the control condition. (A) Scatter plots of gene abundance ratio in iron-restriction versus control conditions. (B) Bar chart showing upregulated differentially expressed genes (DEGs) (green) and downregulated DEGs (red) in A. paragallinarum under iron-restriction condition compared with the control condition (iron restriction vs control).

Table 3. Summary of 19 differentially up-regulated genes that could directly evolve into sophisticated iron-acquisition systems in Av. paragallinarum under iron-restriction condition compared to control condition.
In the case of 115 significantly DEGs encoding proteins, the hierarchical cluster analysis of these DEGs was carried out for understanding the relationships between genes across all 6 samples (Supplementary Figure 3). Above all, it suggests that iron restriction could result in significant changes of gene expressions in A. paragallinarum and seems to be likely to suppress expressions of the majority of genes in bacteria.
Functional Annotations and Bioinformatic Characterization of DEGs in A. paragallinarum Under Iron-Restriction Condition Using GO Enrichment Analysis
Based on the identification and quantification of DEGs, we further performed the functional annotations and bioinformatic characterization of DEGs in A. paragallinarum under iron restriction using GO enrichment analysis through the DAVID online analysis system. Firstly, we took the GO annotations analysis of these DEGs. As shown in Figure 3A and Supplementary Data Sheet 2, upregulated genes could be clustered into three categories, including six kinds of biological processes, five kinds of cellular components, and six kinds of molecular functions. Then, the GO enrichment analysis of these upregulated genes was carried out and the top 20 terms are summarized in Figure 3B and Supplementary Data Sheet 3. Twenty-two upregulated genes enriched in the 13 GO biological process terms, 22 upregulated genes enriched in the 5 GO cellular component terms, and 45 upregulated genes actively participated in the 27 GO molecular function terms were present in A. paragallinarum under the iron-restriction condition, respectively. Among them, three biological process terms that were involved in the response to macromolecule localization, protein localization, and establishment of protein localization were significantly enriched. Another three cellular component terms for protein binding were significantly enriched, which were mainly involved in the cell outer membrane, external encapsulating structure part, and membrane. Besides, biological process items for the transcription factor activity and sequence-specific DNA binding, nucleic acid binding transcription factor activity, and ATPase activity were significantly enriched.
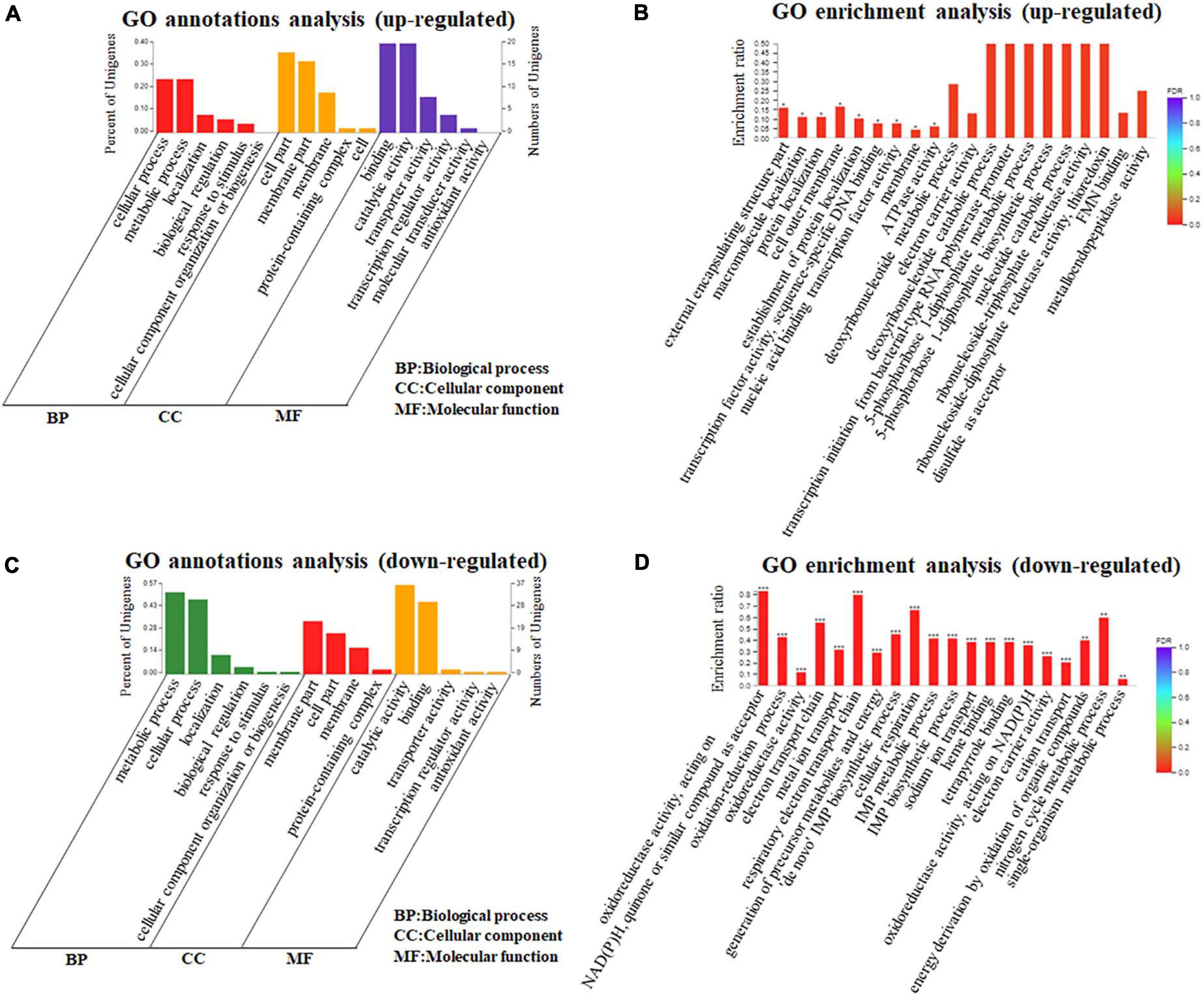
Figure 3. Gene Ontology (GO) annotations and enrichment analysis of DEGs in A. paragallinarum under iron-restriction condition compared with the control condition. (A,B) GO annotations and enrichment analysis of upregulated genes were analyzed, respectively. (C,D) GO annotations and enrichment analysis of downregulated genes were analyzed, respectively (*P < 0.05; **P < 0.01; ***P < 0.001).
In addition, the functional annotations and bioinformatic characterization of downregulated genes in A. paragallinarum under the iron-restriction condition were taken by GO enrichment analysis. As shown in Figure 3C and Supplementary Data Sheet 2, the downregulated genes could also be classified into three categories, which consisted of six kinds of biological processes, four kinds of cellular components, and five kinds of molecular functions. The GO enrichment analysis of these downregulated genes was also conducted and the top 20 terms are summarized in Figure 3D and Supplementary Data Sheet 4. The data showed that there were 391 downregulated genes enriched in the 75 GO biological process terms, 54 downregulated genes enriched in the 8 GO cellular component terms, and 185 downregulated genes actively participated in the 50 GO molecular function terms in A. paragallinarum under the iron-restriction condition, respectively. Additionally, 14 biological process items and 6 molecular function items were significantly enriched, which mainly participated in the oxidation–reduction process; electron transport chain; respiratory electron transport chain; generation of precursor metabolites and energy; “de novo” inosine-5’-monophosphate (IMP) biosynthetic process; cellular respiration; IMP metabolic process; IMP biosynthetic process; sodium ion transport; cation transport; energy derivation by oxidation of organic compounds; nitrogen cycle metabolic process; single-organism metabolic process; metal ion homeostasis; oxidoreductase activity; acting on NAD(P)H, quinone, or similar compound as acceptor; heme binding; tetrapyrrole binding; acting on NAD(P)H; and electron carrier activity. However, no cellular component item was significantly enriched for these downregulated genes. Taken together, the identification of a larger number of DEGs between the iron-restriction group and the control group using DEGs GO enrichment analysis suggests that iron restriction can result in changes of a variety of biological processes and cellular component terms as well as molecular functions that are related to DEGs in A. paragallinarum.
Functional Annotations and Bioinformatic Characterization of DEGs in A. paragallinarum Under Iron-Restriction Condition Using KEGG Pathway Enrichment Analysis
In order to further detect the related pathways in which these DEGs in A. paragallinarum under the iron-restriction condition were involved in, the KEGG pathway analysis was performed. As shown in Figure 4A, upregulated genes could be classified into four KEGG categories, including four kinds of metabolism, one kind of genetic information processing, two kinds of environmental information processing, and one kind of cellular process. Then, KEGG enrichment analysis of these upregulated genes was carried out. As shown in Figure 4B and Supplementary Data Sheet 5, 17 upregulated genes were enriched in the 11 KEGG pathway items such as ABC transporters, pyrimidine metabolism, purine metabolism, quorum sensing, nicotinate and nicotinamide metabolism, and so on. However, no KEGG pathway item was significantly enriched.
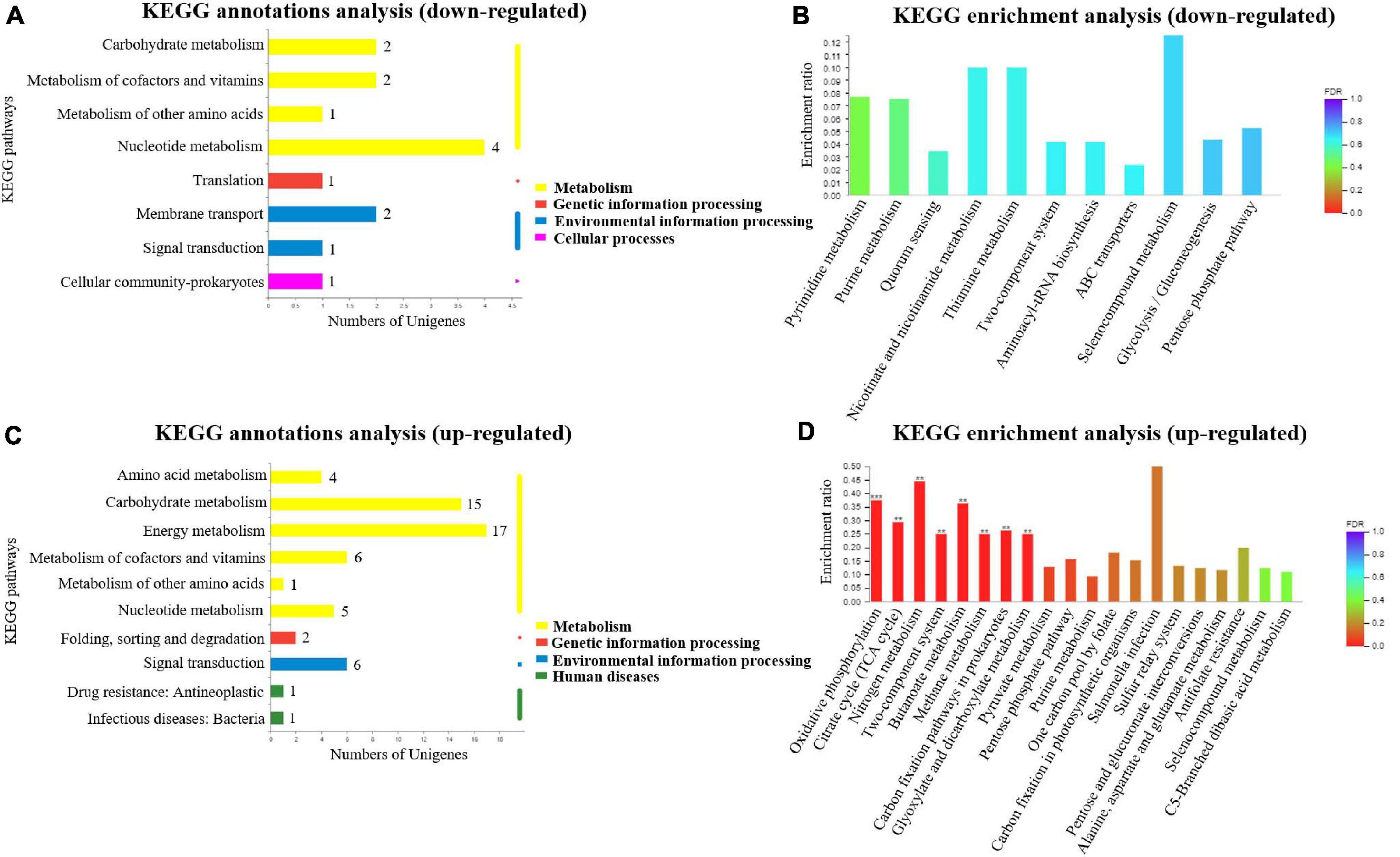
Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations and enrichment analysis of DEGs in A. paragallinarum under iron-restriction condition compared with the control condition. (A,B) KEGG annotations and enrichment analysis of upregulated genes were performed, respectively. (C,D) KEGG annotations and enrichment analysis of downregulated genes were performed, respectively (**P < 0.01; ***P < 0.001).
By taking KEGG pathway analysis of downregulated genes in A. paragallinarum under iron-restriction condition, we found that the downregulated genes could also be classified into four KEGG categories, including six kinds of metabolism, one kind of genetic information processing, one kind of environmental information processing, and two kinds of human diseases (Figure 4C). Furthermore, KEGG enrichment analysis showed that 75 downregulated genes were enriched in the 30 KEGG pathway items. Eight of them exhibited dramatic enrichment that were related to oxidative phosphorylation, citrate cycle (TCA cycle), nitrogen metabolism, two-component system, butanoate metabolism, methane metabolism, carbon fixation pathways in prokaryotes, and glyoxylate and dicarboxylate metabolism (Figure 4D and Supplementary Data Sheet 6). Above all, the results of KEGG enrichment analysis demonstrate that iron restriction can result in changes of a series of biological metabolic pathways and signal transduction pathways that are associated with DEGs in A. paragallinarum.
Validation of DEGs in A. paragallinarum Under Iron-Restriction Condition Through qPCR Analysis
Based on the transcriptomic results, some crucial genes involved in the iron homeostasis showed significant difference between the iron-restriction group and the control group. In order to further validate the results, qPCR was carried out to detect the expressions of up- and downregulated genes in A. paragallinarum under iron-restriction condition. Here, three upregulated genes (VY92_RS03730, VY92_RS03735, and VY92_RS00335) and four downregulated genes (VY92_RS06600, VY92_RS09125, VY92_RS09790, and VY92_RS01655) were selected as representative DEGs for qPCR. Among them, VY92_RS03730, VY92_RS03735, and VY92_RS00335 were annotated as the heme utilization protein HutZ, heme utilization cystosolic carrier protein HutX, and ABC transporter ATP-binding protein, respectively. VY92_RS06600, VY92_RS09125, VY92_RS09790, and VY92_RS01655 were annotated as the H-type ferritin, cytochrome c nitrite reductase pentaheme subunit, succinate dehydrogenase/fumarate reductase iron–sulfur subunit, and autotransporter outer membrane beta-barrel domain-containing protein, respectively. As shown in Figure 5A, the iron-restriction group showed significantly increased mRNA levels of all three upregulated genes than the control group. In addition, the expressions of four downregulated genes were lower in the iron-restriction group than in the control group, and a significant difference could be seen in VY92_RS06600 and VY92_RS09125 (Figure 5B). Taken together, the results are in accordance with the above RNA-seq and suggest the distinct gene expressions of these iron-related DEGs in A. paragallinarum under iron restriction.
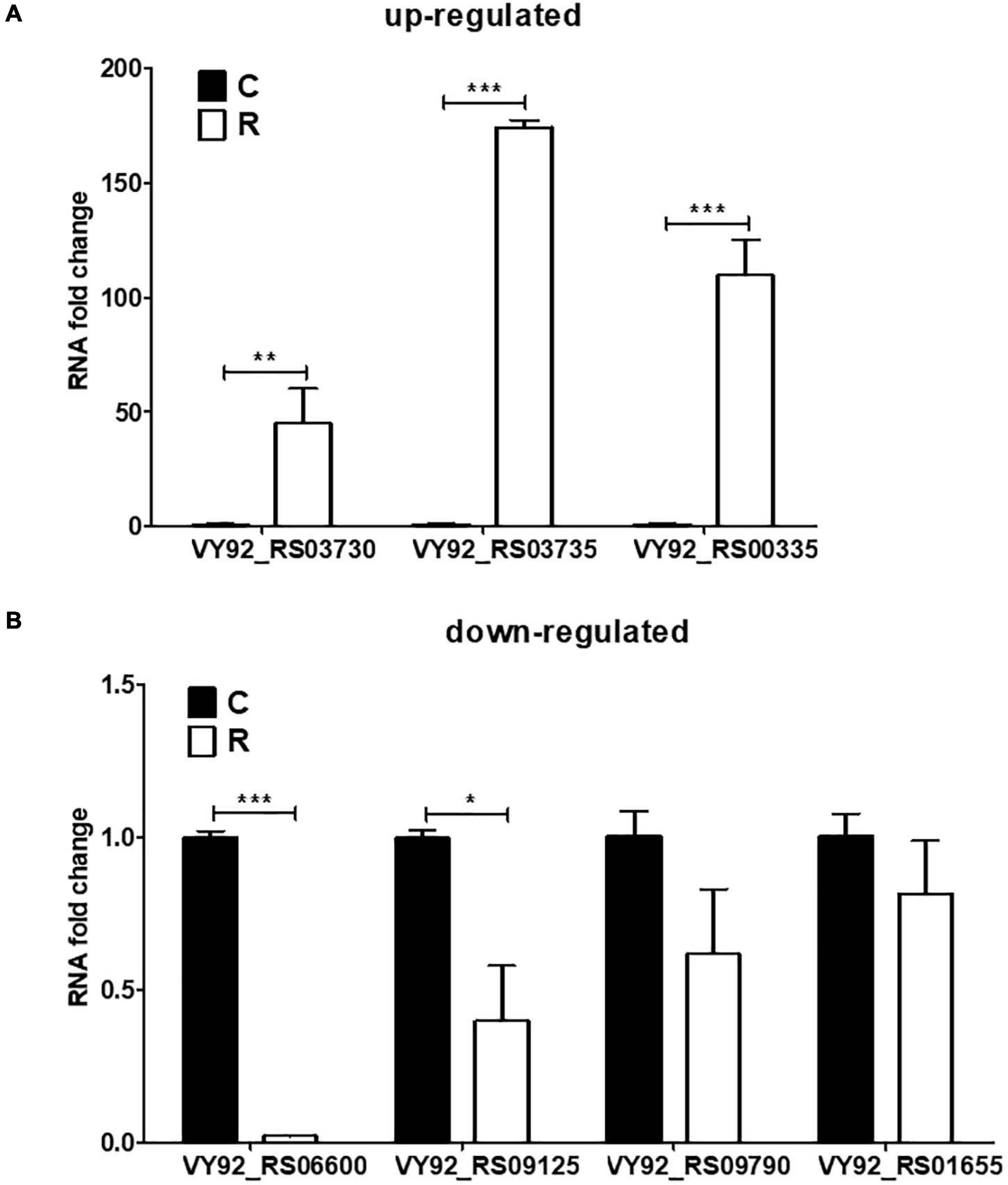
Figure 5. Validation of DEGs in A. paragallinarum under iron-restriction condition compared with the control condition by qPCR. (A,B) The mRNA levels of three upregulated genes and four downregulated genes in A. paragallinarum under iron-restriction condition compared with the control condition were determined by real-time PCR (N = 3), respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
Identification and Bioinformatic Characterization of sRNAs in A. paragallinarum Under Iron-Restriction Condition
In previous results of the identification and quantification of genes in this study, 121 genes encoding sRNAs were verified. We further performed bioinformatic characterization of these sRNAs in A. paragallinarum under the iron-restriction condition. Firstly, we summarized the length of these sRNAs. As shown in Figure 6 and Supplementary Data Sheet 7, the length of each sRNA was smaller than 500 nt, and most of them had the length of 101 to 150 nt. Subsequently, we determined if these sRNAs were likely to form stable secondary structures. Here, we selected four upregulated sRNAs (sRNA0022, sRNA0080, sRNA0084, and sRNA0103) and three downregulated sRNAs (sRNA0004, sRNA0036, and sRNA0099) in A. paragallinarum under the iron-restriction condition. As shown in Figure 7, these sRNAs could form desirable stem-loop secondary structures.
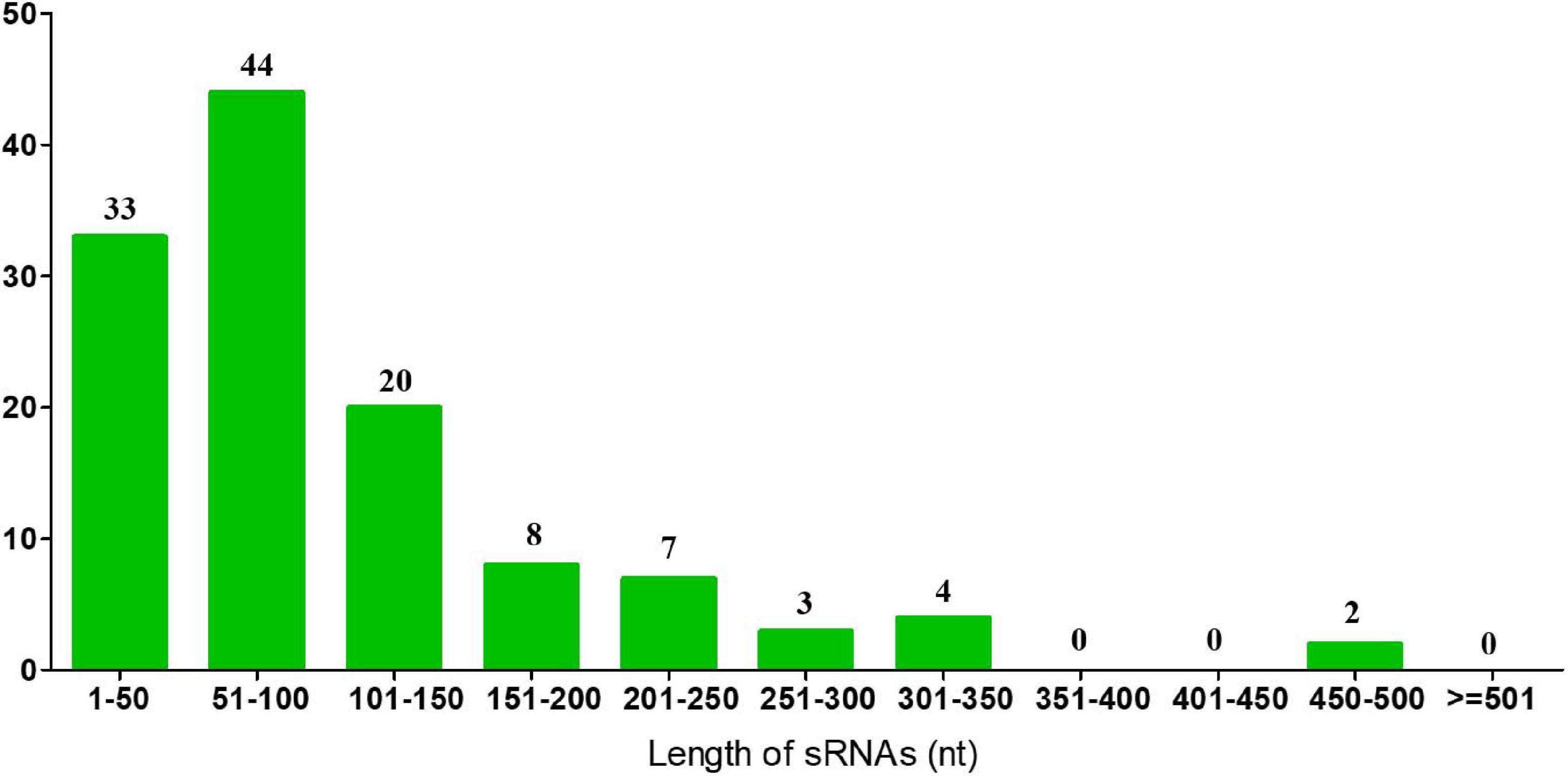
Figure 6. Identification of sRNAs in A. paragallinarum. The length of all sRNAs in A. paragallinarum was bioinformatically determined.
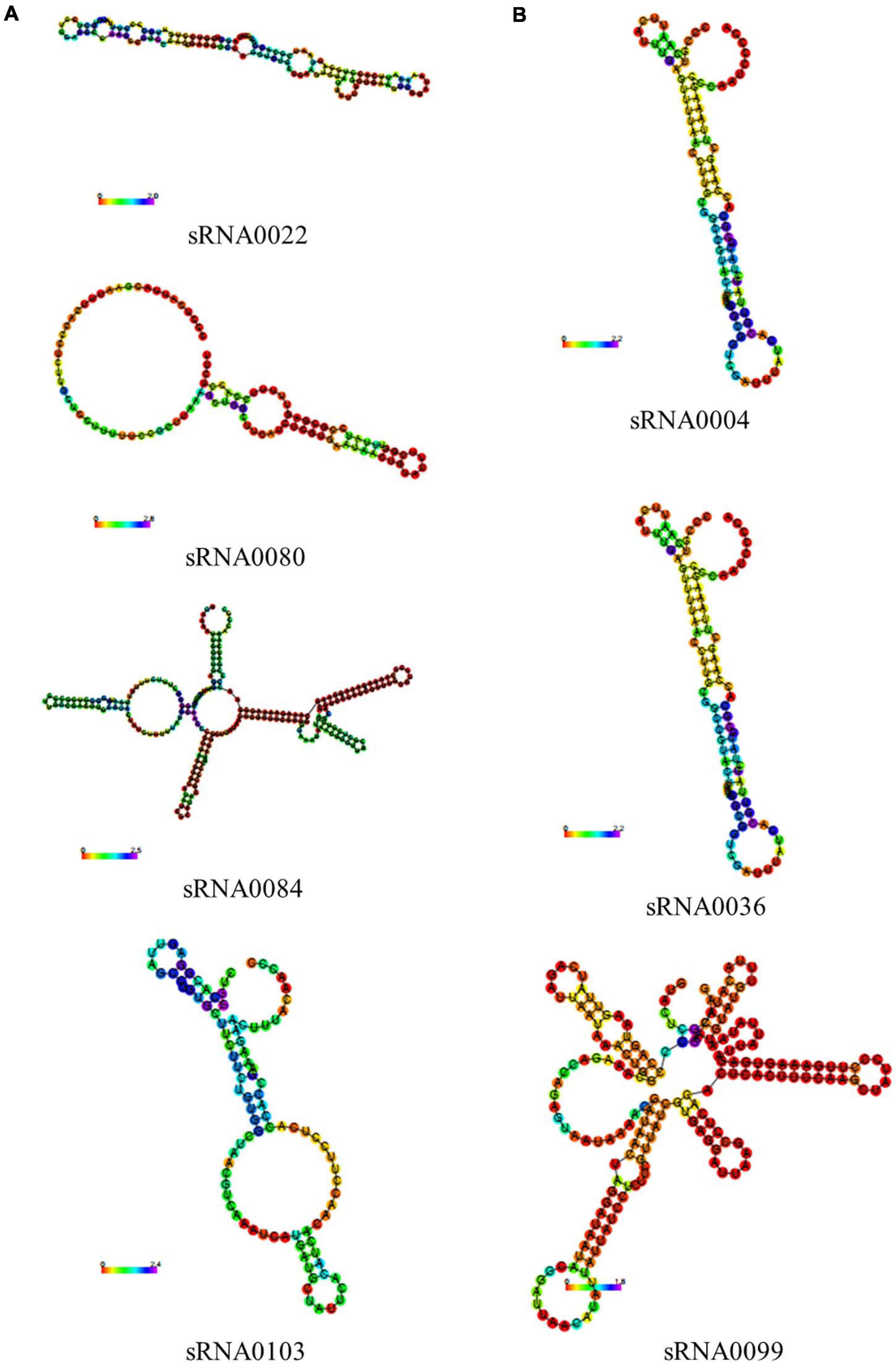
Figure 7. Prediction of secondary structures of several crucial sRNAs in A. paragallinarum under the iron-restriction condition. (A,B) The secondary structures of four upregulated genes and three downregulated sRNAs in A. paragallinarum under the iron-restriction condition compared with the control condition were predicted, respectively.
It is widely known that sRNA executes its regulatory function by pairing and binding to its target genes. We also identified potential interactions between these sRNAs and target mRNAs. As shown in Supplementary Data Sheet 8, a single sRNA could target various mRNAs, and the same target mRNA may also be regulated by different sRNAs. Among the four upregulated sRNAs, we found that they could target the genes mainly encoding the proteins such as DUF3277 domain-containing protein, Fic family protein, DUF1043 domain-containing protein, DUF1828 domain-containing protein, ABC transporter permease, ABC transporter ATP-binding protein, spermidine/putrescine ABC transporter permease, heme-binding protein, DNA helicase RecQ, DNA-binding protein, DNA repair protein RecO, cAMP-activated global transcriptional regulator CRP, transcriptional regulator MraZ, succinate dehydrogenase/fumarate reductase iron–sulfur subunit, autotransporter domain-containing protein, MurR/RpiR family transcriptional regulator, met regulon transcriptional regulator MetJ, and so on. In terms of three downregulated sRNAs, sRNA0004 and sRNA0036 tended to target the genes which coded for the proteins such as DUF1367 domain-containing protein, Fic family protein, two-component system sensor histidine kinase QseC, and so on. Interestingly, we could see that genes encoding the same protein such as DUF1367 domain-containing protein could be targeted by both upregulated sRNAs and downregulated sRNAs. Unfortunately, no target gene was detected for sRNA0099 in this study. Considering that the gene encoding for the Fic protein showed no significance of transcriptomic changes of expression between the iron-restriction group and the control group as well as being targeted by the both up- and downregulated sRNAs, we also used qPCR to further detect the mRNA levels of the related gene. As shown in Supplementary Figure 4, the results were also consistent with the above RNA-seq and showed no difference of mRNA levels of VY92_RS05725 encoding for the Fic protein between the iron-restriction group and the control group, which suggested the stable expressions of Fic protein in A. paragallinarum under iron restriction.
Furthermore, the potential interaction between sRNAs and qPCR-validated DEGs was examined. Among those eight DEGs, VY92_RS09790 and VY92_RS00335 were shown to be targeted by various sRNAs, especially VY92_RS09790 (Table 4). VY92_RS09790, one of the crucial downregulated genes in A. paragallinarum under the iron-restriction condition, could be targeted by 13 sRNAs, including upregulated sRNA0022 and sRNA0103. Notably, upregulated VY92_RS00335 could also be targeted by sRNA0022, indicating the potential regulatory role of sRNA0022 in A. paragallinarum under the iron-restriction condition. Taken together, these results validate that sRNAs have a potential role in the regulation of iron homeostasis in A. paragallinarum.
Discussion
Recently, transcriptomic and bioinformatic analyses, as a new frontier developed in life science, have been extensively used to study dynamic alterations in global transcriptomic profiles and explore the transcriptome response of bacteria to stimuli including nutritional stress (Ying et al., 2015). Therefore, we took RNA-seq to better understand gene expression profiles in A. paragallinarum under the iron-restriction condition. Here, we focused on the large quantities of bacterial genes regulated by iron restriction as well as possible functional activities. These analyses will be helpful for the in-depth realization of the molecular mechanisms involved in response to nutritional iron availability in A. paragallinarum to control cellular iron homeostasis when faced with iron starvation.
Bacteria are generally adaptable to changing circumstances such as various nutrient deficiencies or stresses. This adaption results in many physiological changes such as expression changes of global genes and induction of specific genes (Siegele and Kolter, 1992). Here, we also evaluated and compared the DEGs in A. paragallinarum under conditions with iron restriction and iron sufficiency as well as the related cellular functions and signaling pathways. Between these two conditions, we have revealed a number of distinct characteristics of global gene expression in bacteria. In comparison with iron sufficiency, iron restriction could lead to both upregulation and downregulation of genes, and it seems that the majority of genes showed decreased expression. Additionally, some genes were found to be uniquely present in the iron-restriction group. As previously reported, perhaps reduction of global gene expression and induction of unique genes are also helpful for A. paragallinarum to enter stationary phase by ceasing growth and retaining survivability for a long time (Miksch and Dobrowolski, 1995). Notably, several upregulated genes are demonstrated to be able to facilitate A. paragallinarum to effectively obtain iron for their growth (Abascal et al., 2009). In our study, when bacteria are in an iron-restriction environment, these upregulated gene products that we found can also be helpful for the release of iron from iron-binding proteins and compounds, promoting the acquisition and use of more iron sources by bacteria. The outer membrane receptors are generally induced by iron restriction and thought to enhance the rate of ferric-siderophore uptake, allowing bacteria to more effectively scavenge ferric-siderophores from their surroundings. Additionally, transport of ferric-siderophores through outer membrane receptors requires energy, and this energy is provided by the TonB energy system (Postle, 1993; Larsen et al., 1994; Higgs et al., 1998). Therefore, the increased expression of some proteins that are related to the TonB energy system in bacteria under the iron-restricted condition can facilitate the entry of ferric-siderophores across the outer membrane into the periplasm. TonB ExbB-ExbD proteins are found in many Gram-negative bacteria, mostly known to be found in the E. coli system (Braun et al., 1998; Palyada et al., 2004). In our study, the TonB system transport proteins ExbD and ExbB were found to be either iron stimulated and cotranscribed, and it is likely that the formation of the protein complex TonB ExbB–ExbD also existed. Due to that iron periplasm-to-cytosol transport requires ABC transporters on the cytosolic membrane, the transporter ATP-binding proteins are also upregulated to better contribute to the entry of iron into the cytoplasm as well as iron utilization for bacteria. Certainly, there are many complex regulatory mechanisms that were related to the use of iron, and we believe that our study will provide a good start for the iron regulation system in A. paragallinarum.
Furthermore, numerous cellular functions and signaling pathways were triggered or suppressed in A. paragallinarum under iron restriction in order to ensure iron homeostasis. Interestingly, several novel sRNAs were also elucidated in our study, while their expressions appeared to be different in A. paragallinarum under the iron-restriction condition compared with the iron-sufficiency condition. It suggests that these differences may reflect a survival strategy of A. paragallinarum in ensuring a rapid and accurate response to environmental iron changes. As far as we know, this is the first study of reporting comprehensive transcriptomic information in A. paragallinarum following iron restriction, with a bonus scene of the bioinformatic identification of new important sRNAs. In recent years, the iron-regulated proteins, especially the ferric siderophore outer membrane receptors, have been investigated as potential vaccine candidates (Alteri et al., 2009; Brumbaugh et al., 2013; Wang et al., 2019). Meaningfully, our study will also provide novel insight into the development of new antigens for potential vaccines against infectious coryza. The upregulated genes in A. paragallinarum under iron-restriction condition could be used for the preparation of gene-deleted bacterial strain and, thus, could be candidates for live attenuated vaccine and the genes themselves can also be regarded as candidates for subunit vaccines to control A. paragallinarum in poultry farms, just like other bacteria that have been previously reported (Zeng et al., 2009).
Hut Protein Is Required for Iron Acquisition and Heme Utilization in A. paragallinarum
It is well known that heme utilization systems have essential roles in bacterial iron acquisition and pathogenicity (Shi et al., 2019). A heme uptake system generally consists of the TonB-dependent outer membrane receptor protein, periplasmic binding protein, and inner membrane-associated ABC transporter (Rio et al., 2005). Despite extensive studies on the mechanism of heme transport from outside to the cytoplasm, proteins that participated in heme utilization remain unknown. Nowadays, Hut (heme utilization) protein has been identified in Vibrio cholerae according to genome sequence and bioinformatics-based predictions (Uchida et al., 2019). Uchida et al. have recently purified and characterized HutZ as a heme-degrading enzyme in V. cholerae, which releases iron into the cytoplasm (Uchida et al., 2012). As a unique heme storage protein, HutZ is critical for heme utilization and can cleave heme to biliverdin via verdoheme (Uchida et al., 2019). Besides, it is reported that HutZ is also required for biofilm formation and contributes to the pathogenicity of Edwardsiella piscicida (Shi et al., 2019). HutX, a cytoplasmic heme transport protein for HutZ, facilitates the transfer of heme to HutZ via a specific protein–protein interaction (Sekine et al., 2016). In A. paragallinarum, the function of the Hut protein family has not yet been elucidated. In this study, HutZ and HutX proteins were also identified in A. paragallinarum. RNA-seq analysis and qPCR successfully confirmed the preferential expressions of these proteins in A. paragallinarum under iron-restriction condition, indicating that heme uptake may be an important route for iron acquisition in A. paragallinarum. Herein, our results provide a clue that the heme transport system can play an essential role in iron acquisition as well as HutZ and HutX proteins are considered to be necessary for heme utilization in A. paragallinarum. Just like other bacteria mentioned above, this HutZ in A. paragallinarum can also be considered as a heme-degrading enzyme that releases iron into the cytoplasm and HutX is regarded as a cytoplasmic heme transport protein for HutZ, which facilitates the iron acquisition and heme utilization in A. paragallinarum. These heme acquisition-associated genes might be potential candidate genes for the development of subunit vaccines to control A. paragallinarum. Nevertheless, the regulatory mechanism of heme utilization by HutZ and HutX is worthy of further investigation.
Dominant Role of DUF Domain-Containing Proteins in A. paragallinarum Under Iron Restriction
Even when sequencing data is being increased, there are also a lot of genes or proteins that are uncharacterized and their domain functions are unknown (Nabi et al., 2020). These proteins are regarded as proteins having DUFs (Finn et al., 2016; El-Gebali et al., 2019). DUFs exist in a variety of species, such as around 1,500 in eukaryotes and 2,704 in bacteria (Goodacre et al., 2013). Evolutionary conservation suggests that many of the DUFs have vital functions. For example, DUF59, one of the most common DUFs, occurs in both bacteria and eukaryotes. Though the precise biochemical function of DUF59 remains unclear, previous studies have provided insights into its physiological roles. For instance, proteins containing DUF59 participate in many processes, which include maturation of iron–sulfur (FeS) proteins and intracellular iron homeostasis in S. aureus (Mashruwala et al., 2016). Recently, DUF143 has been considered to be essential in E. coli when cells are starved and its deletion in bacteria shows no obvious phenotype (Baba et al., 2006; Hauser et al., 2012). Until now, few studies characterize the DUF domain-containing proteins in A. paragallinarum. In this study, most of the genes encoding for numerous DUF domain-containing proteins were identified in A. paragallinarum. Among these proteins, majority of them were upregulated and only a few were downregulated in bacteria under the condition of iron restriction. Remarkably, DUF454, DUF4198, and DUF2318 domain-containing proteins were significantly promoted, while DUF1345 domain-containing protein was significantly suppressed in the iron-restriction group than in the control group. According to the sRNA analysis, we speculate that the expression pattern of several DUF domain-containing proteins could be regulated by sRNAs. This is the first research to characterize DUF domain-containing proteins in A. paragallinarum, implying that various DUF domain-containing proteins may have positive roles in transcriptional regulation of physiological processes to cope with environmental iron restriction. Further experimentation will be needed to scrutinize the molecular and functional aspects of these crucial DUF domain-containing proteins in detail.
Stable Expression of Fic Protein Might Be Attributed to the Regulatory of sRNAs in A. paragallinarum Under Iron Restriction
Fic (filamentation induced by cAMP) protein is extensively present in bacteria, with a single member found in animals (Lu et al., 2016). It plays essential roles in drug tolerance, bacterial pathogenicity, and stress response. Previous studies have shown that Fic domain containing protein is able to post-translationally modify proteins (Mukherjee et al., 2011; Feng et al., 2012; Castro-Roa et al., 2013; Harms et al., 2016, 2017). In addition, Fic protein in non-pathogenic bacteria has been implicated in the regulation of bacterial cell division and programmed death (Worby et al., 2009). For instance, a mutation of the fic gene in E. coli can result in filamentation (Utsumi et al., 1982, 1983). Although Fic protein has attracted considerable interest for its functions in bacterial infections and stress responses, their functions and related molecular mechanisms in iron homeostasis in A. paragallinarum had been unknown until recently. Here, we found that this protein was present in A. paragallinarum. However, there was no significance of gene expression of this protein between the iron-restriction group and the control group. Thus, it seems that iron- restriction may not result in the abnormal expression of Fic protein in A. paragallinarum. Identification and bioinformatic characterization of sRNAs further confirmed that Fic protein could be regulated not only by upregulated sRNA0022 but also by downregulated sRNA0004 and sRNA0036 in bacteria under the iron-restriction condition compared with that under the control condition. Consequently, we speculate that Fic protein is still stably expressed in A. paragallinarum under iron restriction, which might be largely attributed to the existence of these regulatory sRNAs. Herein, our results demonstrate for the first time about the effects of iron restriction on the expression of Fic protein as well as the tight relationship between this protein and regulatory sRNAs in A. paragallinarum.
Conclusion
We investigate the more profound transcriptomic profiles for the first time and illustrate changes of transcriptomes in A. paragallinarum in response to starvation using RNA-seq. Compared with iron sufficiency, iron restriction results in many transcriptomic changes including changes of global gene expression and induction of specific genes. The bioinformatic analyses demonstrate that the Hut protein and DUF domain-containing proteins are preferentially activated in A. paragallinarum following iron restriction and have a positive role in iron acquisition and heme utilization in response to iron starvation. In addition, several novel sRNAs are identified in A. paragallinarum and have potential regulatory roles in iron homeostasis, which is likely to facilitate the stable expression of Fic protein. Researches on these genes will enrich our knowledge about the mechanisms of growth and pathogenesis of A. paragallinarum, which can be regarded as potential targets for the development of new prophylactic and therapeutic measures against A. paragallinarum.
Data Availability Statement
The data presented in the study are deposited in the (NCBI SRA) repository, accession number (SRR13291129).
Author Contributions
CH, FX, FL, DL, ZH, and HS carried out the experiments, analyzed the data, and wrote the manuscript. CH, JL, and HS designed the study and supervised the project. CH, XZ, FX, GL, YH, JL, and HS assisted in the data analysis and discussion. CH and HS drew the figures. All authors read and approved the final manuscript.
Funding
This research was supported by the Beijing Municipal Natural Science Foundation of China (Grant No. 6182010) and the National Key Research and Development Program of China (Grant No. 2016YFD0500804).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the staff of the Beijing Key Laboratory for Prevention and Control of Infectious Diseases in Livestock and Poultry, Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Science, China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.610196/full#supplementary-material
Footnotes
References
Abascal, E. N., Guerra, A. C., Vazquez, A. S., Tenorio, V. R., Cruz, C. V., Zenteno, E., et al. (2009). Identification of iron-acquisition proteins of Avibacterium paragallinarum. Avian Pathol. 38, 209–213. doi: 10.1080/03079450902912143
Alteri, C. J., Hagan, E. C., Sivick, K. E., Smith, S. N., and Mobley, H. L. T. (2009). Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathogens 5:e1000586. doi: 10.1371/journal.ppat.1000586
Alvarez-Estrada, A., Gutierrez-Martin, C. B., Rodriguez-Ferri, E. F., and Martinez-Martinez, S. (2018). Transcriptomics of Haemophilus (Glasserella) parasuis serovar 5 subjected to culture conditions partially mimetic to natural infection for the search of new vaccine antigens. BMC Vet. Res. 14:326. doi: 10.1186/s12917-018-1647-1641
Anderson, M. T., and Armstrong, S. K. (2004). The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186, 7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Barnard, T. G., Van Heerden, E., Bragg, R. R., and Albertyn, J. (2008). Haemophilus paragallinarum haemagglutinin: role in adhesion, serotyping and pathogenicity. Onderstepoort J. Vet. Res. 75, 11–16.
Blackall, P. J. (1999). Infectious coryza: overview of the disease and new diagnostic options. Clin. Microbiol. Rev. 12, 627–632. doi: 10.1128/cmr.12.4.627
Boucher, C. E., Theron, C. W., Jansen, A. C., and Bragg, R. R. (2014). Transcriptional profiling of chicken immunity-related genes during infection with Avibacterium paragallinarum. Vet. Immunol. Immunopathol. 158, 135–142. doi: 10.1016/j.vetimm.2013.12.004
Bragg, R. R. (2005). Effects of differences in virulence of different serovars of Haemophilus paragallinarum on perceived vaccine efficacy. Onderstepoort J. Vet. Res. 72, 1–6. doi: 10.4102/ojvr.v72i1.218
Bragg, R. R., Coetzee, L., and Verschoor, J. A. (1996). Changes in the incidences of the different serovars of Haemophilus paragallinarum in South Africa: a possible explanation for vaccination failures. Onderstepoort J. Vet. Res. 63, 217–226.
Braun, V., Hantke, K., and Koster, W. (1998). Bacterial iron transport: mechanisms, genetics, and regulation. Met. Ions Biol. Syst. 35, 67–145.
Brumbaugh, A. R., Smith, S. N., and Mobley, H. L. T. (2013). Immunization with the Yersiniabactin receptor. FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect. Immun. 81, 3309–3316. doi: 10.1128/Iai.00470-413
Castro-Roa, D., Garcia-Pino, A., De Gieter, S., van Nuland, N. A. J., Loris, R., and Zenkin, N. (2013). The fic protein doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817. doi: 10.1038/nchembio.1364
Chareyre, S., and Mandin, P. (2018). Bacterial iron homeostasis regulation by sRNAs. Microbiol. Spectr. 6, 1–15. doi: 10.1128/microbiolspec.RWR-0010-2017
Chu, B. C., Garcia-Herrero, A., Johanson, T. H., Krewulak, K. D., Lau, C. K., Peacock, R. S., et al. (2010). Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23, 601–611. doi: 10.1007/s10534-010-9361-x
Contreras, H., Chim, N., Credali, A., and Goulding, C. W. (2014). Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol. 19, 34–41. doi: 10.1016/j.cbpa.2013.12.014
del Rio, M. L., Gutierrez-Martin, C. B., Rodriguez-Barbosa, J. I., Navas, J., and Rodriguez-Ferri, E. F. (2005). Identification and characterization of the TonB region and its role in transferrin-mediated iron acquisition in Haemophilus parasuis. FEMS Immunol. Med. Microbiol. 45, 75–86. doi: 10.1016/j.femsim.2005.02.008
El Mouali, Y., Gaviria-Cantin, T., Sanchez-Romero, M. A., Gibert, M., Westermann, A. J., Vogel, J., et al. (2018). CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 14:e1007401. doi: 10.1371/journal.pgen.1007401
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Feng, F., Yang, F., Rong, W., Wu, X., Zhang, J., Chen, S., et al. (2012). A Xanthomonas uridine 5’-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118. doi: 10.1038/nature10962
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Gerrick, E. R., Barbier, T., Chase, M. R., Xu, R., Francois, J., Lin, V. H., et al. (2018). Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proc. Natl. Acad. Sci. U S A. 115, 6464–6469. doi: 10.1073/pnas.1718003115
Goodacre, N. F., Gerloff, D. L., and Uetz, P. (2013). Protein domains of unknown function are essential in bacteria. mBio 5:e00744-13. doi: 10.1128/mBio.00744-713
Harms, A., Liesch, M., Korner, J., Quebatte, M., Engel, P., and Dehio, C. (2017). A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella. PLoS Genet. 13:e1007077. doi: 10.1371/journal.pgen.1007077
Harms, A., Stanger, F. V., and Dehio, C. (2016). Biological diversity and molecular plasticity of FIC domain proteins. Annu. Rev. Microbiol. 70, 341–360. doi: 10.1146/annurev-micro-102215-195245
Hauser, R., Pech, M., Kijek, J., Yamamoto, H., Titz, B., Naeve, F., et al. (2012). RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 8:e1002815. doi: 10.1371/journal.pgen.1002815
Higgs, P. I., Myers, P. S., and Postle, K. (1998). Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180, 6031–6038. doi: 10.1128/JB.180.22.6031-6038.1998
Horiyama, T., and Nishino, K. (2014). AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. doi: 10.1371/journal.pone.0108642
Huo, C., Xiao, J., Xiao, K., Zou, S., Wang, M., Qi, P., et al. (2020). Pre-Treatment with zirconia nanoparticles reduces inflammation induced by the pathogenic H5N1 influenza virus. Int. J. Nanomed. 15, 661–674. doi: 10.2147/IJN.S221667
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 12:e1005672. doi: 10.1371/journal.ppat.1005672
Larsen, R. A., Thomas, M. G., Wood, G. E., and Postle, K. (1994). Partial suppression of an Escherichia coli TonB transmembrane domain mutation (delta V17) by a missense mutation in ExbB. Mol. Microbiol. 13, 627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x
Lu, C., Nakayasu, E. S., Zhang, L. Q., and Luo, Z. Q. (2016). Identification of Fic-1 as an enzyme that inhibits bacterial DNA replication by AMPylating GyrB, promoting filament formation. Sci. Signal. 9:ra11. doi: 10.1126/scisignal.aad0446
Mashruwala, A. A., Bhatt, S., Poudel, S., Boyd, E. S., and Boyd, J. M. (2016). The DUF59 containing protein SufT is involved in the maturation of iron-sulfur (FeS) proteins during conditions of high FeS cofactor demand in Staphylococcus aureus. PLoS Genet. 12:e1006233. doi: 10.1371/journal.pgen.1006233
Masse, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Miksch, G., and Dobrowolski, P. (1995). Growth phase-dependent induction of stationary-phase promoters of Escherichia coli in different gram-negative bacteria. J. Bacteriol. 177, 5374–5378. doi: 10.1128/jb.177.18.5374-5378.1995
Mukherjee, S., Liu, X., Arasaki, K., McDonough, J., Galan, J. E., and Roy, C. R. (2011). Modulation of rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106. doi: 10.1038/nature10335
Nabi, R. B. S., Tayade, R., Imran, Q. M., Hussain, A., Shahid, M., and Yun, B. W. (2020). Functional insight of nitric-oxide induced DUF genes in Arabidopsis thaliana. Front. Plant Sci. 11:1041. doi: 10.3389/fpls.2020.01041
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729. doi: 10.1128/Jb.186.14.4714-4729.2004
Parrow, N. L., Fleming, R. E., and Minnick, M. F. (2013). Sequestration and scavenging of iron in infection. Infect. Immun. 81, 3503–3514. doi: 10.1128/IAI.00602-613
Postle, K. (1993). TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25, 591–601. doi: 10.1007/BF00770246
Reinhart, A. A., Powell, D. A., Nguyen, A. T., O’Neill, M., Djapgne, L., Wilks, A., et al. (2015). The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect. Immun. 83, 863–875. doi: 10.1128/IAI.02707-2714
Repoila, F., and Darfeuille, F. (2009). Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell 101, 117–131. doi: 10.1042/BC20070137
Richard, K. L., Kelley, B. R., and Johnson, J. G. (2019). Heme uptake and utilization by gram-negative bacterial pathogens. Front. Cell Infect. Microbiol. 9:81. doi: 10.3389/fcimb.2019.00081
Rio, S. J., Osorio, C. R., and Lemos, M. L. (2005). Heme uptake genes in human and fish isolates of Photobacterium damselae: existence of hutA pseudogenes. Arch. Microbiol. 183, 347–358. doi: 10.1007/s00203-005-0779-774
Rouault, T. A. (2004). Microbiology. pathogenic bacteria prefer heme. Science 305, 1577–1578. doi: 10.1126/science.1102975
Sekine, Y., Tanzawa, T., Tanaka, Y., Ishimori, K., and Uchida, T. (2016). Cytoplasmic heme-binding protein (HutX) from Vibrio cholerae is an intracellular heme transport protein for the heme-degrading enzyme, hutZ. Biochemistry 55, 884–893. doi: 10.1021/acs.biochem.5b01273
Shi, Y. J., Fang, Q. J., Huang, H. Q., Gong, C. G., and Hu, Y. H. (2019). HutZ is required for biofilm formation and contributes to the pathogenicity of Edwardsiella piscicida. Vet. Res. 50:76. doi: 10.1186/s13567-019-0693-694
Siegele, D. A., and Kolter, R. (1992). Life after log. J. Bacteriol. 174, 345–348. doi: 10.1128/jb.174.2.345-348.1992
Skaar, E. P., Humayun, M., Bae, T., DeBord, K. L., and Schneewind, O. (2004). Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628. doi: 10.1126/science.1099930
Sun, H., Xie, S., Li, X., Xu, F., Li, Y., Boucher, C. E., et al. (2018). Selection of Avibacterium paragallinarum page serovar B strains for an infectious coryza vaccine. Vet. Immunol. Immunopathol. 199, 77–80. doi: 10.1016/j.vetimm.2018.04.001
Thulasiraman, P., Newton, S. M., Xu, J., Raymond, K. N., Mai, C., Hall, A., et al. (1998). Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J. Bacteriol. 180, 6689–6696. doi: 10.1128/JB.180.24.6689-6696.1998
Uchida, T., Dojun, N., Ota, K., Sekine, Y., Nakamura, Y., Umetsu, S., et al. (2019). Role of conserved arginine in the heme distal site of HutZ from Vibrio cholerae in the heme degradation reaction. Arch. Biochem. Biophys. 677:108165. doi: 10.1016/j.abb.2019.108165
Uchida, T., Sekine, Y., Matsui, T., Ikeda-Saito, M., and Ishimori, K. (2012). A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem. Commun. (Camb) 48, 6741–6743. doi: 10.1039/c2cc31147j
Utsumi, R., Kawamukai, M., Obata, K., Morita, J., Himeno, M., and Komano, T. (1983). Identification of a membrane protein induced concurrently with cell filamentation by cyclic AMP in an Escherichia coli K-12 fic mutant. J. Bacteriol. 155, 398–401. doi: 10.1128/JB.155.1.398-401.1983
Utsumi, R., Nakamoto, Y., Kawamukai, M., Himeno, M., and Komano, T. (1982). Involvement of cyclic AMP and its receptor protein in filamentation of an Escherichia coli fic mutant. J. Bacteriol. 151, 807–812. doi: 10.1128/JB.151.2.807-812.1982
Wang, Y. Q., Wang, X. Y., Ali, F., Li, Z. Q., Fu, Y. Y., Yang, X. J., et al. (2019). Comparative extracellular proteomics of Aeromonas hydrophila reveals iron-regulated secreted proteins as potential vaccine candidates. Front. Immunol. 10:256. doi: 10.3389/fimmu.2019.00256
Wen, S., Chen, X., Xu, F., and Sun, H. (2016). Validation of reference genes for real-time quantitative PCR (qPCR) analysis of Avibacterium paragallinarum. PLoS One 11:e0167736. doi: 10.1371/journal.pone.0167736
Williams, P., Morton, D. J., Towner, K. J., Stevenson, P., and Griffiths, E. (1990). Utilization of enterobactin and other exogenous iron sources by Haemophilus influenzae, H. parainfluenzae and H. paraphrophilus. J. Gen. Microbiol. 136, 2343–2350. doi: 10.1099/00221287-136-12-2343
Worby, C. A., Mattoo, S., Kruger, R. P., Corbeil, L. B., Koller, A., Mendez, J. C., et al. (2009). The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103. doi: 10.1016/j.molcel.2009.03.008
Xu, F., Zeng, X., Haigh, R. D., Ketley, J. M., and Lin, J. (2010). Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J. Bacteriol. 192, 4425–4435. doi: 10.1128/JB.00478-410
Xu, Y., Cheng, J., Huang, X., Xu, M., Feng, J., Liu, C., et al. (2019). Characterization of emergent Avibacterium paragallinarum strains and the protection conferred by infectious coryza vaccines against them in China. Poult. Sci. 98, 6463–6471. doi: 10.3382/ps/pez531
Ying, B. W., Matsumoto, Y., Kitahara, K., Suzuki, S., Ono, N., Furusawa, C., et al. (2015). Bacterial transcriptome reorganization in thermal adaptive evolution. BMC Genom. 16:802. doi: 10.1186/s12864-015-1999-x
Zeng, X., Xu, F., and Lin, J. (2009). Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect. Immun. 77, 5437–5448. doi: 10.1128/IAI.00666-669
Zhang, P. J., Miao, M., Sun, H., Gong, Y., and Blackall, P. J. (2003). Infectious coryza due to Haemophilus paragallinarum serovar B in China. Aust. Vet. J. 81, 96–97. doi: 10.1111/j.1751-0813.2003.tb11445.x
Keywords: Avibacterium paragallinarum, iron acquisition, heme utilization, transcriptomes, differentially expressed genes
Citation: Huo C, Zeng X, Xu F, Li F, Li D, Li G, Hu Z, Hu Y, Lin J and Sun H (2021) The Transcriptomic and Bioinformatic Characterizations of Iron Acquisition and Heme Utilization in Avibacterium paragallinarum in Response to Iron-Starvation. Front. Microbiol. 12:610196. doi: 10.3389/fmicb.2021.610196
Received: 25 September 2020; Accepted: 02 February 2021;
Published: 04 March 2021.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Robert Bragg, University of the Free State, South AfricaSatoshi Wakai, Japan Agency for Marine-Earth Science and Technology, Japan
Copyright © 2021 Huo, Zeng, Xu, Li, Li, Li, Hu, Hu, Lin and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiling Sun, c3VuaHVpbGluZzAxQDE2My5jb20=
 Caiyun Huo
Caiyun Huo Ximin Zeng
Ximin Zeng Fuzhou Xu
Fuzhou Xu Fangbing Li1
Fangbing Li1 Yanxin Hu
Yanxin Hu Jun Lin
Jun Lin Huiling Sun
Huiling Sun