- 1Department of Horticulture and Landscape Architecture, Northeast Agricultural University, Harbin, China
- 2Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Northeast Region), Ministry of Agriculture and Rural Affairs, Northeast Agricultural University, Harbin, China
Diazotrophs are important soil components that help replenish biologically available nitrogen (N) in the soil and contribute to minimizing the use of inorganic N fertilizers in agricultural ecosystems. However, there is little understanding of how diazotrophs respond to intercropping and soil physicochemical properties in cucumber continuous cropping systems. In this study, using the nifH gene as a marker, we have examined the impacts of seven intercropping plants on diazotrophic community diversity and composition compared to a cucumber continuous cropping system during two cropping seasons. The results showed that intercropping increased the abundance of the nifH gene, which was negatively correlated with available phosphorous in the fall. Diazotrophic diversity and richness were higher in the rape–cucumber system than in the monoculture. Multivariate regression tree analysis revealed that the diversity of the diazotrophic communties was shaped mainly by soil moisture and available phosphorous. Skermanella were the dominant genera in all of the samples, which increased significantly in the mustard–cucumber system in the fall. There was no effect of intercropping on the structure of the diazotrophic community in this case. Non-metric multidimensional scaling analysis showed that cropping season had a greater effect than intercropping on the community structure of the diazotrophs. Overall, our results suggest that intercropping altered the abundance and diversity rather than the structure of the diazotrophic community, which may potentially affect the N fixation ability of continuous cropping systems.
Introduction
The decline of biodiversity has negative impacts on ecosystem function which related to biomass production and nutrient cycling (Cardinale et al., 2012). Modern agricultural practices trend toward crop monocultures, which results in the simplification of the components of agricultural systems (Cook, 2006; Zhou et al., 2017). The continuous monocropping system, in which the same crop is repeatedly cropped in the same field year after year, is commonly adopted in the growing and harvesting of cucumber (Cucumis sativus L.) (Jin et al., 2020). However, monocropping is not sustainable in the long term, because it may eventually lead to reductions in the yield and quality of the crop, a phenomenon that has been described as “soil sickness” (Yu et al., 2000).
Large amounts of chemical fertilizers have been used to increase plant biomass and grain yield in monocropping systems (Li et al., 2016). Nitrogen (N) is a critical plant nutrient, and cucumber has a very high requirement for it. Long-term continuous cropping of cucumber has been found to increase soil available nitrogen and decrease soil enzyme activities (Zhou and Wu, 2015). Application of N fertilizer promotes cucumber biomass and improves cucumber yields, however, it also incurs a significant cost to the producer and could have undesirable environmental consequences. Environmental concerns around the application of N fertilizers include increased greenhouse gas emissions, soil acidification, and groundwater contamination (Ridley et al., 2004; Hayden et al., 2010; Reardon et al., 2014). Thus, reducing N fertilizer inputs and developing new cultural practices for the improvement of agroecosystem balance is very important (Yang L. et al., 2019).
Biological nitrogen fixation (BNF) is a major source of soil nitrogen, in which atmospheric nitrogen can be reduced to ammonia by diazotrophs using nitrogenase (Yin et al., 2018). The nifH gene, which encodes the reductase subunit of nitrogenase, has been widely used to study diazotroph communities in terrestrial ecosystems (Hu et al., 2018; Chen et al., 2019). Soil diazotrophic community diversity and composition are sensitive to many factors related to microbial biomass (Hayden et al., 2010), soil physical and chemical properties (Poly et al., 2001; Nelson and Mele, 2006; Hayden et al., 2010; Wakelin et al., 2010), and sampling season (Mergel et al., 2001; Pereira e Silva et al., 2011). Soil diazotrophs are also sensitive to agricultural practices such as plowing, cropping systems and fertilization (Hayden et al., 2010; Berthrong et al., 2014). Although numerous factors may affect diazotrophic communities, these specific factors do not have the same effects on community diversity and composition in different cropping systems (Poly et al., 2001).
Intercropping is the growing of more than one crop species in the same field during the growing season, and it is becoming more and more important to improve soil quality and relieve soil sickness (Zhou et al., 2011; Li and Wu, 2018). Compared with monocropping, intercropping was found to increase the diversity and populations of soil bacterial communities (Li et al., 2013; Yang et al., 2016; Yu et al., 2019). The abundance and diversity of nifH genes showed variations in continuous and rotational soybean cropping systems (Xiao et al., 2010). Intercropping can facilitate the mobilization and uptake of N, P, and K through interspecific rhizosphere interactions (Li et al., 2003a, b; Inal et al., 2007). Several studies have indicated that intercropping can reduce nitrate leaching and the release of nitrate pollution to groundwater (Li et al., 2005; Whitmore and Schroder, 2007). However, little is known about the effects of intercropping on the diazotrophic communities in the soils of continuous cropping systems.
Previously, several vegetable species belonging to Leguminosae (Thorsted et al., 2006), Brassicaceae (Lai et al., 2019), Gramineae (Xu et al., 2013, 2015), and Compositae (Hamida et al., 2016) have been shown to have overall beneficial effects on component crop production during intercropping. Therefore, in this study, we selected seven plants from those families and intercropped with cucumber to investigate their effects on the diazotrophic community in the soils of continuous cucumber cropping systems. The diversity and composition of the diazotrophic communities were determined by Illumine MiSeq sequencing, and the abundance of diazotrophs was determined by quantitative PCR analysis of the nifH gene. Our previous studies showed that both cucumber growth and yield were adversely affected by continuous monocultures, while intercropping was able to both improve cucumber yield and increase the diversity of the bacterial and fungal communities (Chang et al., 2017; Li and Wu, 2018). Thus, we hypothesize that: (1) intercropping might affect the diversity and composition of the diazotrophic community; and (2) the change in the diazotrophic community are related to soil physicochemical properties.
Materials and Methods
Experimental Design
The experiments were conducted in a greenhouse at the Horticultural Experimental Station of the Northeast Agricultural University, Harbin, China (45°41′ N, 126° 37′ E). The field soils were subjected to continuous cropping of cucumber for 3 years. The physicochemical properties of the soil before the experiments have been previously described by Li and Wu (2018).
Cucumber seedlings with three true leaves were transplanted into the field, with 12 cucumber seedlings per row. The experiments were carried out in a randomized block design, with three replicate block for each treatment. There were eight treatments in the experiment: alfalfa (A), trifolium (T), wheat (W), rye (Ry), chrysanthemum (C), rape (Ra), and mustard (M) were intercropped with cucumber as an experimental group, and cucumber monoculture (CM) as the control. First cropping of cucumber was grown in 2015. Spring and fall experiments were conducted from April to June and from July to October, respectively. Twelve cucumber were planted in one row as the test district. There were two protective rows on both sides. Ten days following the planting of the cucumber, the intercrop seeds were sown on the outside of the rows of cucumber at a distance of approximately 0.10 m. The distance between the two rows cucumber was 0.50 m (Figure 1).
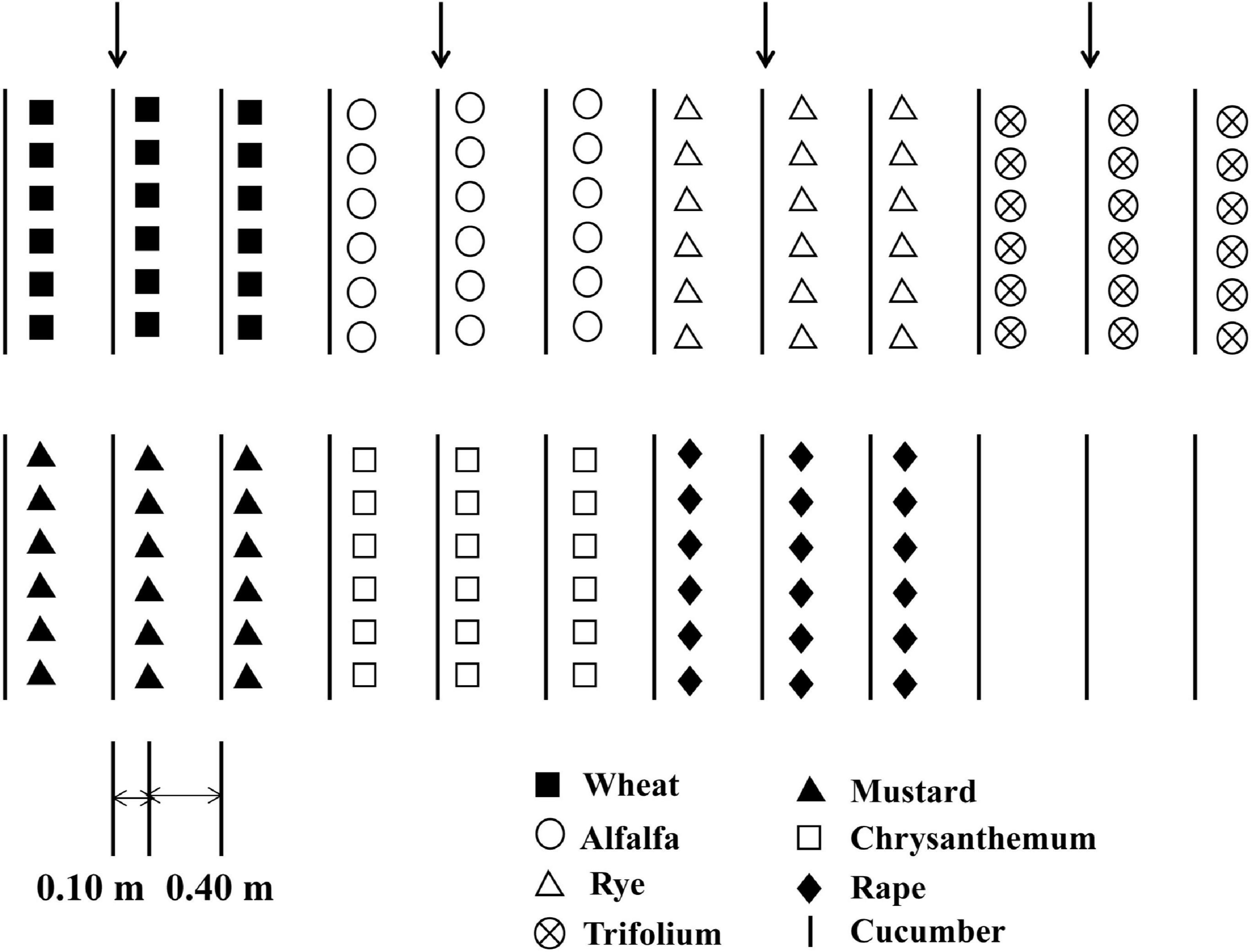
Figure 1. Diagram showing different intercropping systems and the cucumber monoculture system in the greenhouse experiment.
Each cucumber seedling was intercropped with (I) 30 wheat and rye seedlings, respectively, (II) 40 trifolium and alfalfa seedlings, respectively, (III) 3 rape and mustard seedlings, respectively, (IV) 5 chrysanthemum seedlings. The seedlings were irrigated with groundwater twice a week. NPK compound fertilizer (16:16:8) was applied with 300 kg ha–1 in each plot. The weeds were removed manually and no pesticide were applied. A more detailed description of the experiment is available in the references cited (Chang et al., 2017; Li and Wu, 2018).
Soil Sampling, Microbial Biomass, and Soil Enzyme Activities Analysis
A total of 48 soil samples were collected from 24 test districts on June 30 and October 6. The soil microbial biomass C and N were measured by using the chloroform fumigation-extraction method (Brookes et al., 1985; Vance et al., 1987). The soil urease activity was measured according to Zhou et al. (2011). The soil protease activity was measured as described by Ladd et al. (1976).
DNA Extraction and nifH Gene Copy Number Quantification
Soil DNA was extracted from 0.5 g soil using a powerful DNA Isolation Kit (Mo Bio Laboratories Inc., Carlsbad, CA, United States) according to the manufacturer’s instructions. DNA quality was assessed using 1% agarose gel electrophoresis. nifH gene copy numbers were determined using qPCR assays in an IQ5 real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, United States) with Po1F/Po1R (Poly et al., 2001). The PCR reaction of diazotroph including 10 μl of 2 × Real SYBR Mixture (Tiangen Beijing, China), 0.8 μl of 10 μM forward and reverse primers, 7.2 μl of sterilized MilliQ water, and 2 μl of DNA template. A negative control reactions contained 2 μl of H2O instead of DNA. Amplification was initiated by denaturation at 95°C for 3 min, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 45 s, extension at 72°C for 30 s, and a final elongation at 72°C for 10 min, and the plate was read at 83°C. The specificity of the amplicon was verified by melting curve analysis and agarose gel electrophoresis.
Illumina Miseq Sequencing and Bioinformatics Analysis
The nifH gene was amplified with the primer pairs nifH-F/nifH-R (Rösch et al., 2002). The reaction composition and thermal conditions of the PCR were conducted according to the method of Fan et al. (2019). The PCR products obtained were purified with an Agarose Gel DNA purification kit (TaKaRa Bio), and triplicate PCR amplifications for each sample were conducted and pooled as a PCR product and then sequenced on an Illumina Miseq platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw sequences were processed using the QIIME-1.9.1 pipeline (Edgar, 2013). The low-quality sequences were removed, as described previously by Fan et al. (2019), and the remaining sequences were further translated into amino acids using the FunGene pipeline. The translated protein and chimera sequences that did not match the nifH protein sequence were discarded. The remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 95% similarity using Uparse software (version 7.0.1090). The raw data were uploaded to the NCBI SRA database with the submission accession number SRP159857. The read numbers of all the samples were normalized to the same sequencing depth, and the Chao1, Observed species, and Shannon and Simpson indices were chosen to evaluate alpha diversity (Yang Y. et al., 2019).
Data Analysis
Mean comparison (microbial biomass C and N, soil enzyme activities, nifH gene abundance, number of OTUs, alpha-diversity) of the different treatments was performed based on the Tukey’s honest significance difference (HSD) test at the 0.05 probability level. The nifH gene abundance was analyzed using two-way analysis of variance (ANOVA) with season, cropping system, and their interaction as the fixed factors. These statistical analyses were conducted with SAS 9.2 software (SAS Institute Inc., Cary, NC, United States). Spearman correlation coefficients were calculated in order to test the relationships between the soil physicochemical properties, the nifH gene abundance, and the relative abundance of genera in SPSS software (Version 17.0). Non metric multidimensional scaling (NMDS) and redundancy analysis (RDA) were conducted to reveal the structure of the diazotroph community and the relationship between soil environmental factors and soil diazotroph community abundances. Analysis of similarity (Anosim) and non-parametric MANOVA (Adonis) were used to compare the differences in the microbial communities of the two cropping seasons using the Bray–Curtis distance and 999 permutations, and were carried out in R using the vegan package. A multivariate regression tree analysis was performed to identify the most important soil factors for diazotroph diversity using R-package “mvpart.”
Results
Microbial Biomass and Soil Enzyme Activities
Compared with the monoculture system, the wheat–cucumber system in the two growing seasons, and rye-cucumber and chrysanthemum–cucmber systems in the fall significantly increased microbial biomass C. The microbial biomass C of alfalfa–cucumber and chrysanthemum–cucmber systems was significantly lower than those of the monoculture in the spring. The chrysanthemum–cucmber and mustard–cucumber systems significantly increased the microbial biomass N in the two growing seasons compared with the monoculture. The microbial biomass N of alfalfa–cucumber, trifolium–cucumber, wheat-cucumber, and rape–cucumber systems was significantly lower than those of the monoculture in the fall. Compared with the monoculture system, rape–cucumber system significantly increased the urease activity in the fall and all intercropping systems significantly increased the protease activity in the two growing seasons (Supplementary Table 1).
Abundance of the nifH Gene and Its Correlation With the Soil Properties
Compared with the cucumber monoculture, the nifH gene abundance significantly increased under the intercropping systems in the two growing seasons except for the mustard–cucumber system in the fall. Moreover, the trifolium–cucumber system had the highest nifH gene abundance in the two growing seasons. Two-way analysis showed that season, cropping system and their interaction had a significant effect on nifH gene abundance (Figure 2). No significant correlation was observed between nifH gene abundance and soil properties measured in the spring. In the fall, a significant negative correlation was observed between nifH gene abundance and available phosphorous (AP). Regardless of the season, nifH gene abundance was positively correlated with pH and nitrate (Table 1).
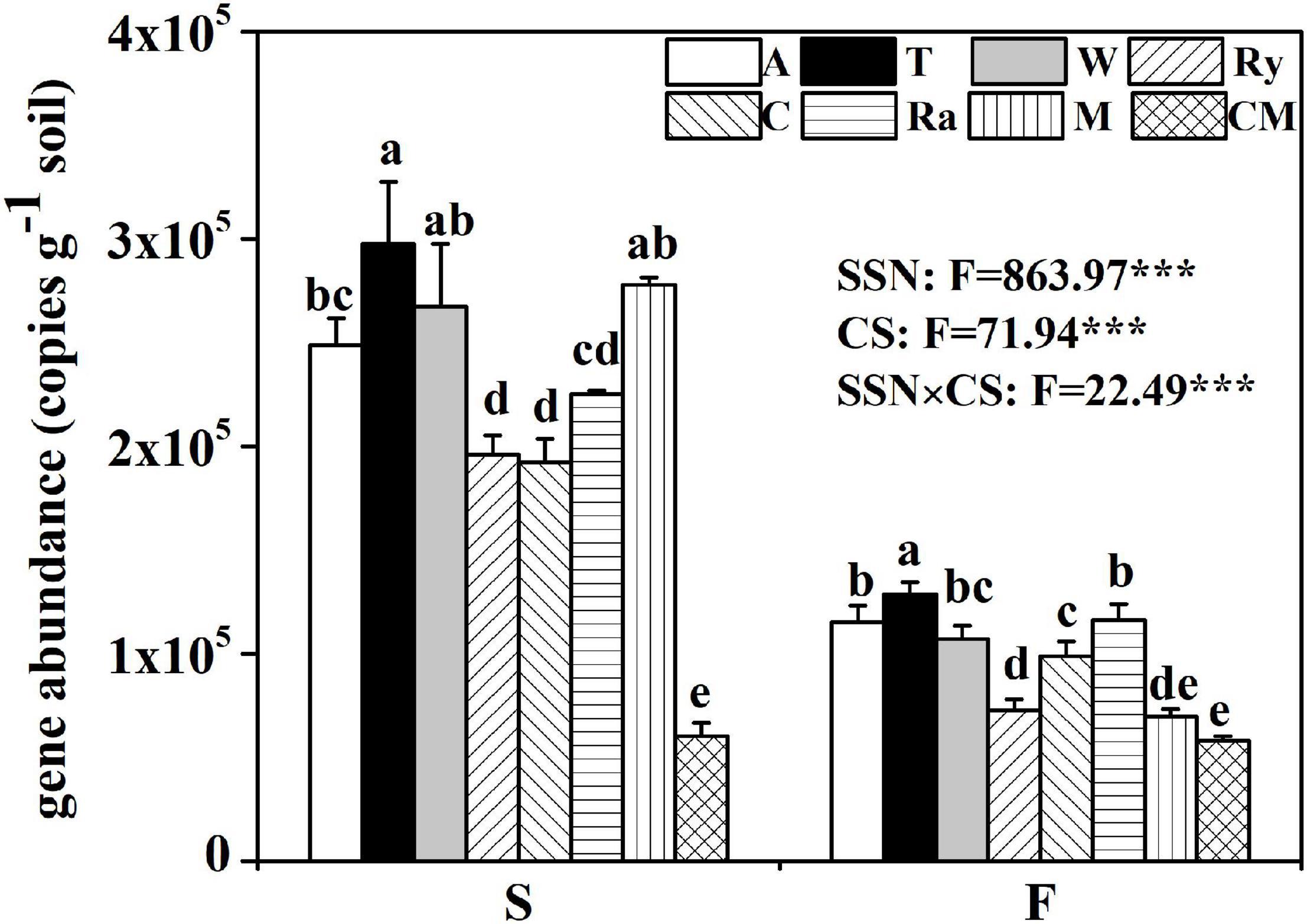
Figure 2. Abundance of the nifH gene among eight cropping systems in the spring (S) and fall (F) cropping seasons. Data with different letters in each column indicate significant differences among the samples (p < 0.05, one-way ANOVA). F-value from two way ANOVA analysis of effects of season and cropping system on nifH gene abundance. SSN, season; CS, cropping system.
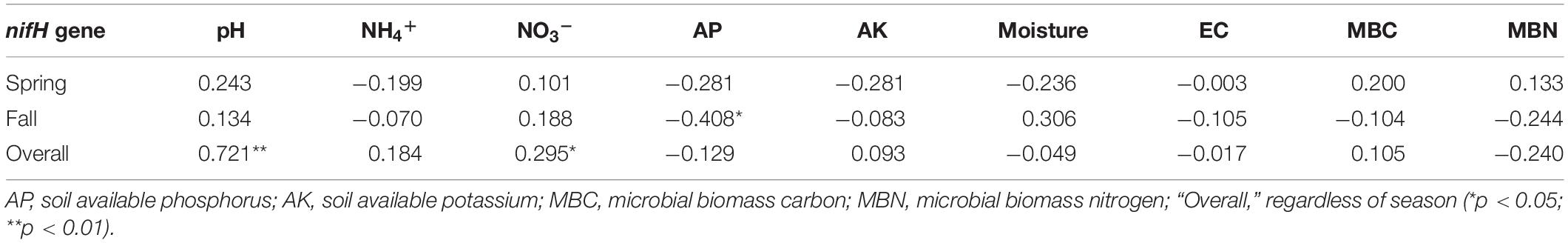
Table 1. Correlation coefficients for relationships between nifH gene abundance and soil physicochemical properties.
Diazotrophic Community Diversity and Composition
The alpha-diversity indices of the diazotrophic community are listed in Table 2. The number of OTUs in the rape–cucumber system was significantly higher than that in the monoculture in either the spring or the fall. Compared with the monoculture system, the mustard–cucumber system in the spring and the wheat–cucumber and chrysanthemum–cucumber systems in the fall had significantly increased the numbers of OTUs. The Shannon indices of the wheat–cucumber, rape–cucumber, and mustard–cucumber systems were significantly higher than those of the monoculture in both the spring and the fall. However, the Simpson index showed the opposite trend to the Shannon index. The Chao1 index was used to estimate the diazotrophic richness. The Chao1 index of the rape-cucumber system was significantly higher than those of the monoculture in the spring and the fall. The coverage of the diazotrophic community was more than 96%, showing that the sequencing depth in this report was enough to cover the soil diazotrophic community (Table 2). Two-way analysis showed that both the number of OTUs and the alpha-diversity of the diazotrophic community were affected mainly by the cropping system and the interaction between season and cropping system (Table 2). Multivariate regression tree analysis indicated that diazotrophic diversity and richness were mainly shaped by soil moisture in the spring and available phosphorus in the fall (Figure 3).
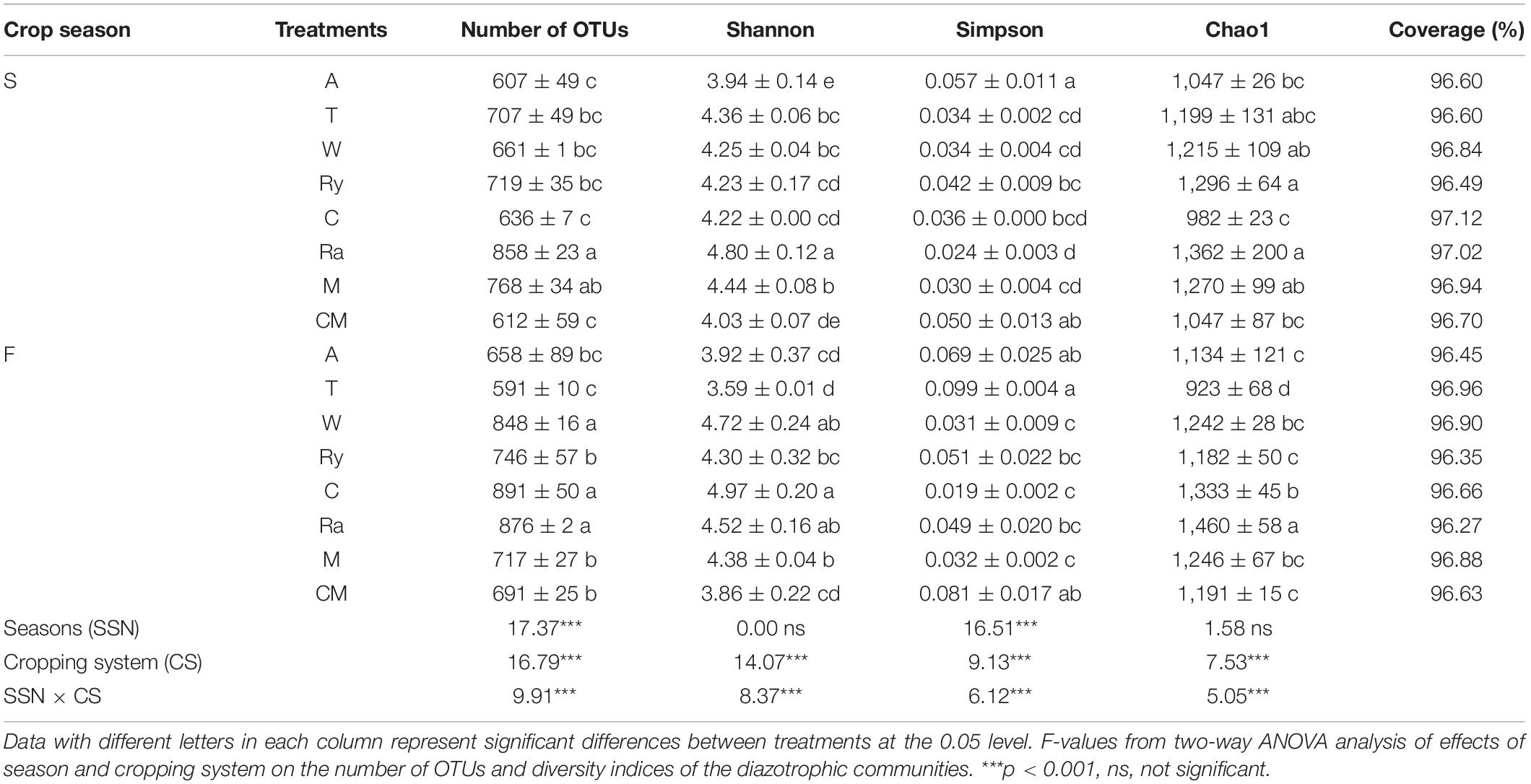
Table 2. Number of OTUs and diversity indices of the diazotrophic communities in the soils of cucumber crops for all treatments.
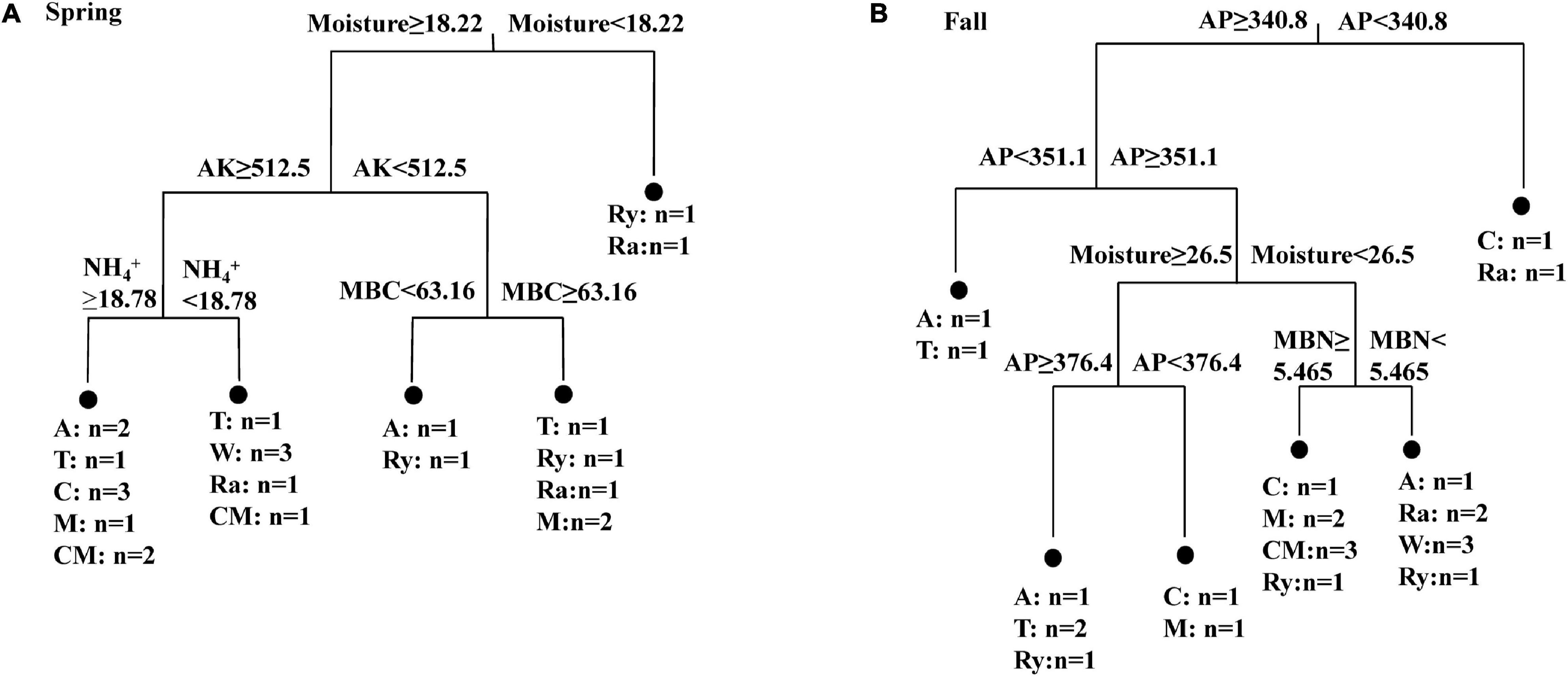
Figure 3. Multivariate regression tree analysis of alpha diversity (OTUs, Chao 1, Shannon and Simpson indices) of diazotrophs and soil physicochemical properties in the (A) Spring and (B) Fall cropping seasons. Treatments and the number of samples included in the analysis are shown at the bottom. AK, available potassium; AP, available phosphorus; MBC, microbial biomass carbon; MBN, microbial biomass nitrogen.
All of the samples were dominated to a great degree by the phylum Proteobacteria, with relative abundances ranging from 67.11 to 95.50%, followed by Verrucomicrobia (11.39–0.94%), and Cyanobacteria (4.17–0.24%). Compared with the monoculture, the relative abundances of these phyla showed no significant changes in response to the intercropping system except for the Cyanobacteria in the chrysanthemum–cucumber system in the spring (Figure 4A).
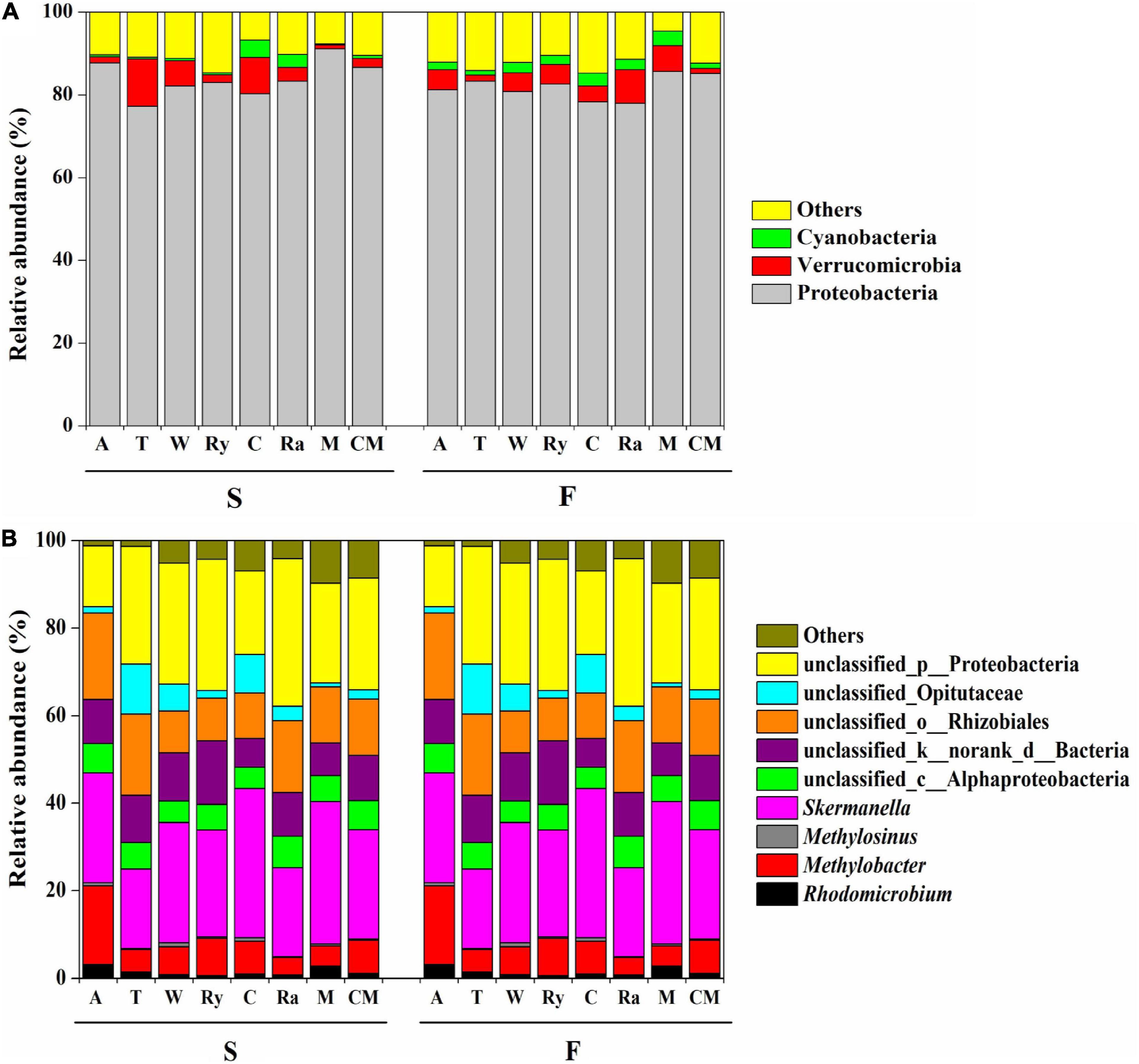
Figure 4. Relative abundances (%) of (A) diazotrophic phyla and (B) genera among eight cropping systems in the spring (S) and fall (F) cropping seasons. “Others” includes phyla and genera below 0.5% relative abundance.
Figure 4B shows the relative abundances of the four main soil diazotrophic genera in the two growing seasons. We found that different intercropping regimes resulted in different changes in the diazotrophic communities at the genus level in the two growing seasons. For example, the rye–cucumber system significantly increased the relative abundance of Rhodomicrobium in the fall, and the alfalfa–cucumber system significantly increased the relative abundances of Methylobacter and Methylosinus in spring compared with the monoculture. The mustard–cucumber system significantly increased the relative abundance of Skermanella in the fall compared with the monoculture (Supplementary Figure 1).
The results of the Spearman correlation analysis showed that several properties had significant relationships with the relative abundance of some diazotrophic genera in individual seasons (Figures 5A,B). In the spring, the relative abundance of Methylobacter and Rhodomicrobium were significantly positively correlated with AP and AK, respectively; the relative abundance of Methylosinus and Skermanella were significantly negatively correlated with moisture and nitrate, respectively (Figure 5A). In the fall, the relative abundances of Methylobacter and Methylosinus were significantly negatively correlated with moisture (Figure 5B).
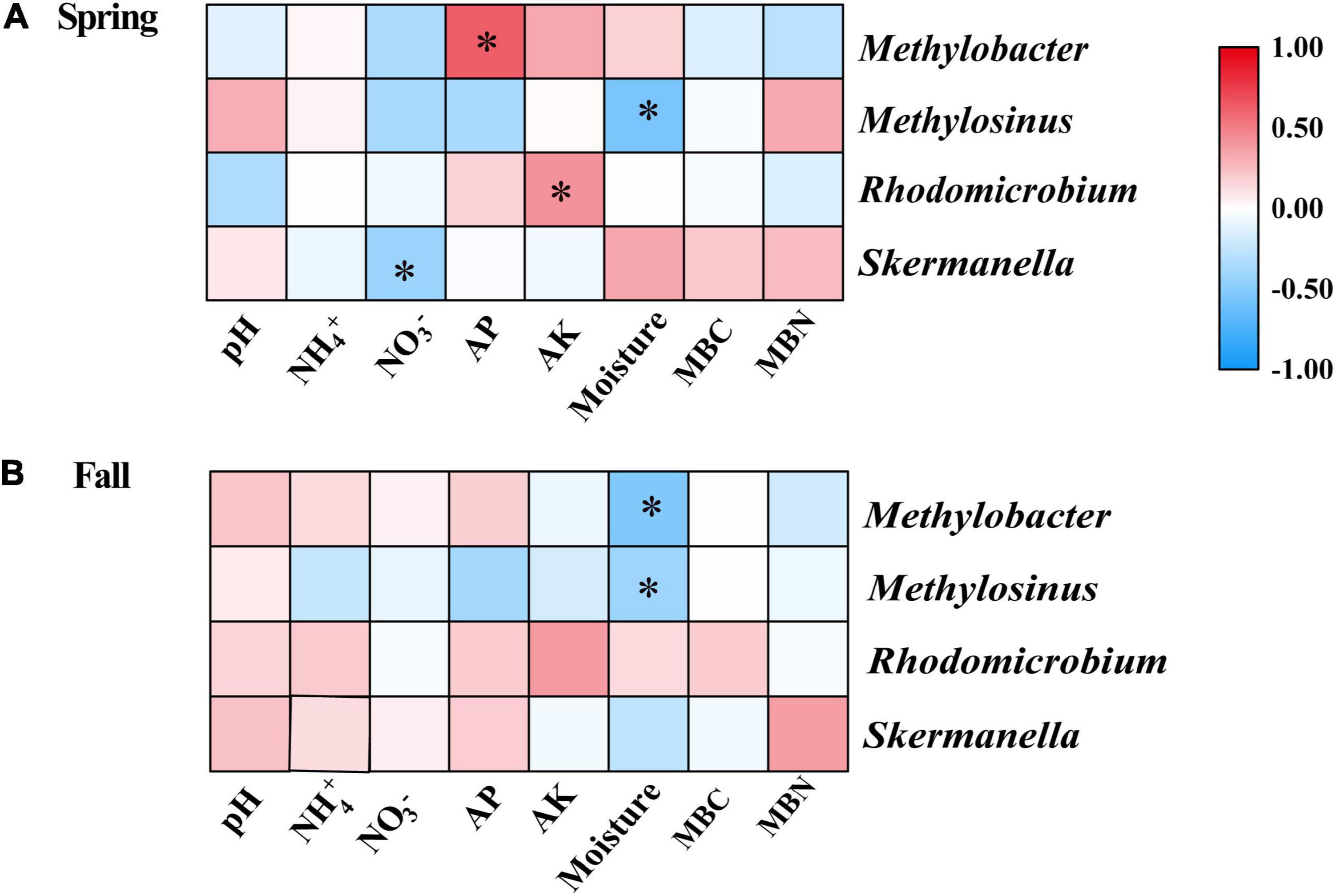
Figure 5. Spearman correlation analysis of the relative abundances of dominant diazotrophic genera and soil physicochemical variables in the (A) Spring and (B) Fall cropping seasons. *p < 0.05. AK, available potassium; AP, available phosphorus; MBC, microbial biomass carbon; MBN, microbial biomass nitrogen.
Structure of the Diazotrophic Community and Its Correlation With Soil Properties
Non-metric multidimensional scaling (NMDS) analysis based on the abundance of OTUs revealed differences in the structure of the diazotrophic community (Figures 6A,B). The NMDS plot based on the Euclidean distance dissimilarity showed that the three replicates per treatment were not situated close together, and the soil diazotrophic communities of all samples were distinctly different in the two growing seasons (Table 3). Moreover, there was no obvious distinction between the intercropped systems and the monoculture in the fall (Figure 6B), but the rape–cucumber system was separated from the monoculture in the spring (Figure 6A). Redundancy analysis (RDA) (Figure 7) and the Monte Carlo permutation test (Table 4) were carried out in order to examine the relationship between environmental factors and the soil diazotrophic community composition. Among the environmental factors, AK was significantly correlated with the soil diazotrophic community composition in the two growing seasons (Table 4). Moisture and AP were significantly correlated with the composition of the soil diazotrophic community in the spring and pH, NO3–, and MBC were significantly correlated with the soil diazotrophic community composition in the fall. However, NH4+ and MBN were not significantly correlated with soil diazotrophic community composition (Table 4).
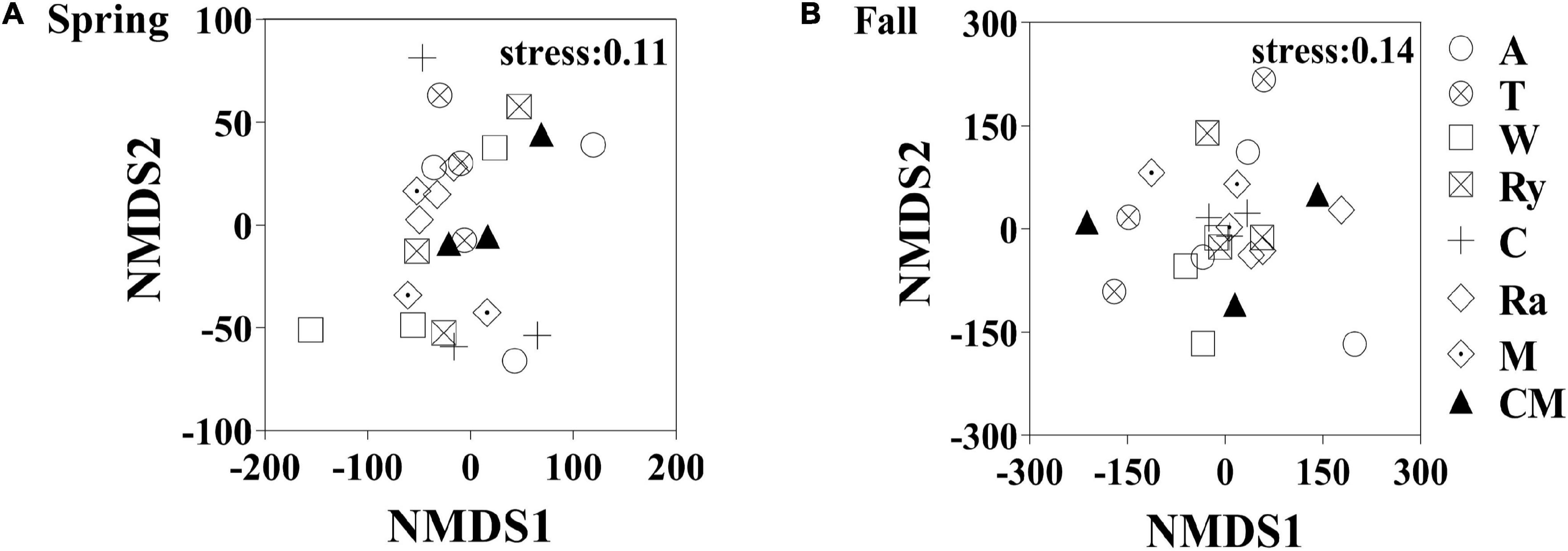
Figure 6. Non-metric multidimensional scaling (NMDS) analysis of the diazotrophic communities in the (A) spring (S) and (B) Fall (F) cropping seasons.

Table 3. Dissimilarity comparison of the soil diazotrophic community structure between the spring and fall cropping seasons.
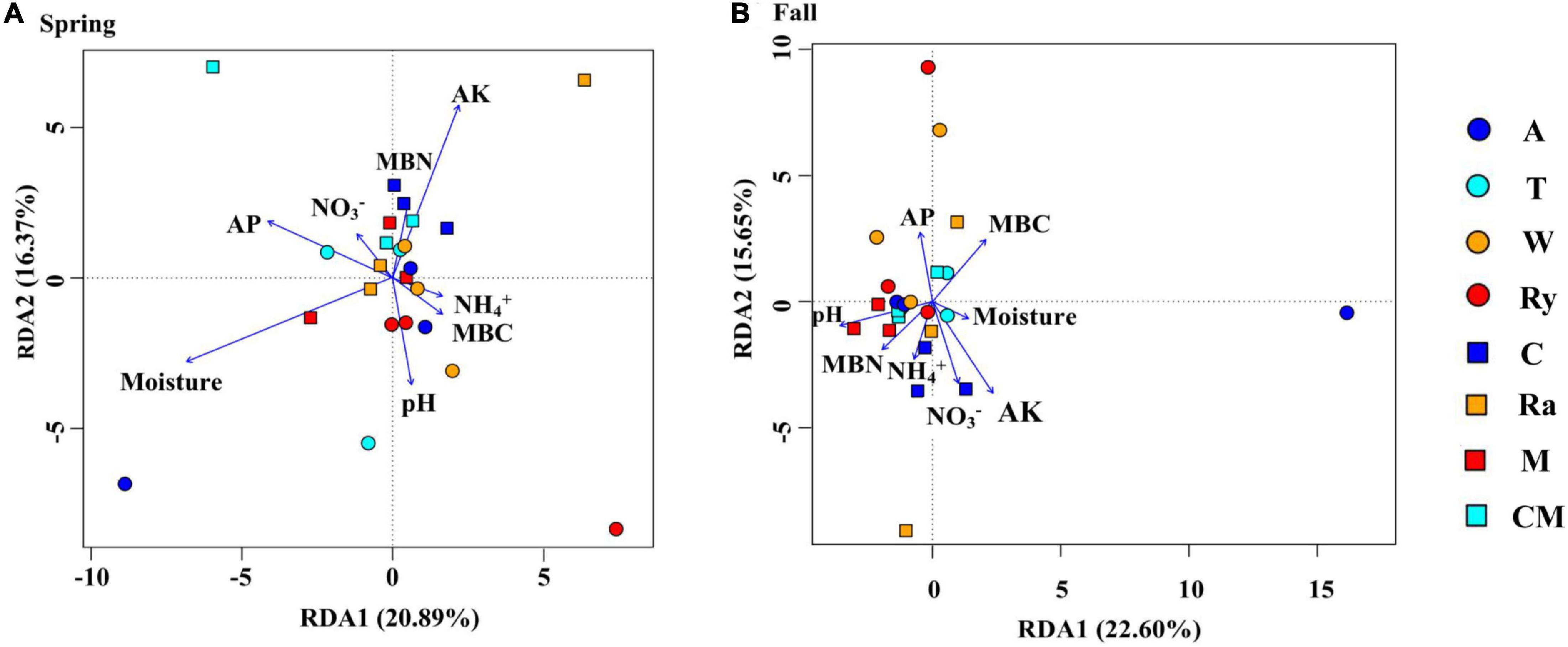
Figure 7. Redundancy analysis (RDA) showing correlations between soil diazotrophic communities and soil environmental factors during the (A) Spring and (B) Fall cropping seasons in the control (CM), alfalfa (A), trifolium (T), wheat (W), chrysanthemum (C), rye (Ry), mustard (M), and rape (Ra) intercropping systems.
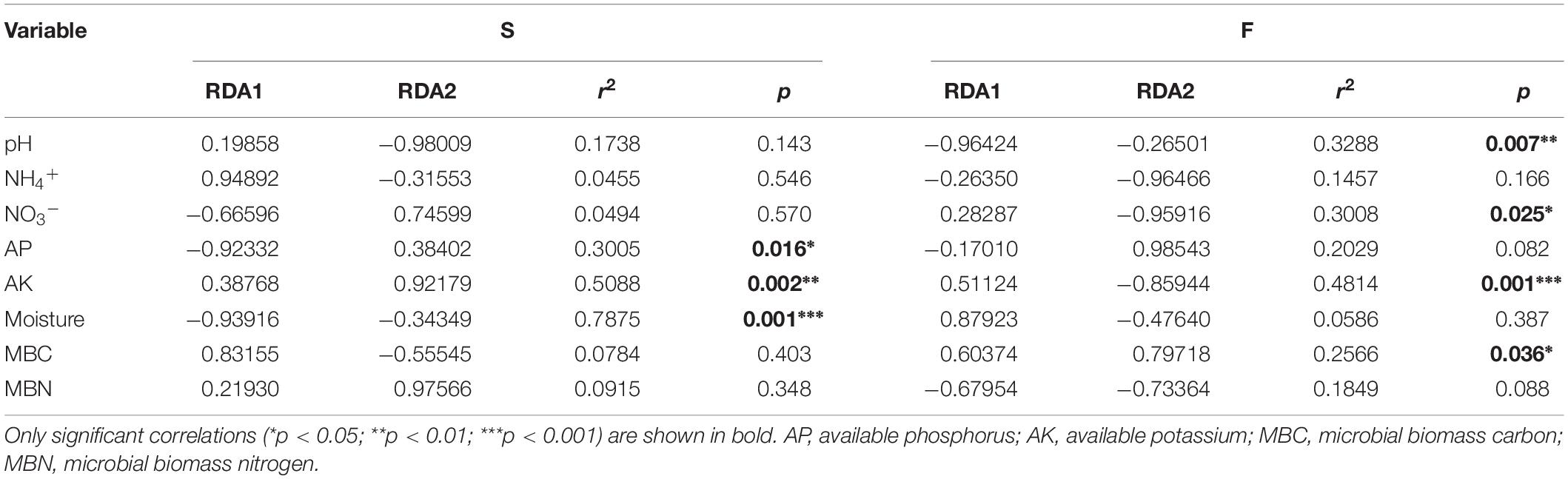
Table 4. Monte Carlo permutation test of environmental factors and soil diazotrophic community composition.
Discussion
Effect of Intercropping on Soil Diazotrophic Abundance
Intercropping is a sustainable farming model that have an effect on crop growth (Hauggaard-Nielsen and Jensen, 2005), microbial communities (Li et al., 2016), and microclimatic conditions (Cong et al., 2015). BNF is a major source of soil nitrogen, in which atmospheric nitrogen can be reduced to ammonia by diazotrophs using nitrogenase (Yin et al., 2018). In this study, we found that intercropping can lead to an increase in nifH gene abundance compared with monoculture systems (Figure 2). Previous study also found that intercropping can promote the population of bacteria associated with nitrogen fixing (Chen et al., 2018). Enhanced diazotrophic abundance in the present study may be caused by interspecific facilitations in the intercropping systems. It has been reported that plant species could alter the soil bacterial communities (Rooney and Clipson, 2008; Junier et al., 2009). Our observation that the trifolium–cucumber system had the highest nifH gene abundance in the two growing seasons (Figure 2). Normally, nitrogen fixation rate is correlated with nifH gene abundance (Reed et al., 2010), and it is possible that the higher nifH gene abundance, due to the presence of symbiotic N-fixers in cucumber intercropped with leguminous crops (Yang Y. et al., 2019), may improve the nitrogen fixation in such treated soil. However, Rocca et al. (2015) found that the higher abundance of the gene did not always lead to higher metabolic rates in soils. Therefore, the BNF rate of intercropping systems needs to be determined in future studies.
Diazotrophic community abundance is sensitive to the soil physicochemical characteristics (Hayden et al., 2010). In our study, a significant negative correlation was observed between nifH gene abundance and AP in the fall (Table 1). Phosphorus is one of the crucial factors controlling nitrogen fixation due to the high demand for adenosine triphosphate in BNF (Hill, 1988; Vitousek et al., 2002; Reed et al., 2011). A previous study has shown that soil nitrogenase activity was positively correlated with soil total phosphorus (Han et al., 2019). Thus, we suggest that the lower nifH gene abundance of rye–cucumber, mustard–cucumber, and monoculture systems in the fall may be attributed to the higher AP, while the nitrogenase activity in these treatments may higher than in others. Regardless of season, pH was positively significantly correlated with the nifH gene abundance (Table 1). Soil pH is a key factor affecting nifH gene abundance, and a high pH value favors BNF (Nelson and Mele, 2006). Thus, we deduced that the higher abundance of the nifH gene observed in the spring than in the fall was ascribed to the higher soil pH in the spring. A similar result was reported by Pereira e Silva et al. (2013), who found that pH was positively correlated with nifH gene abundance in soils with pH values ranging from 4.3 to 7.7. The change of pH may influence the bioavailability of carbon or nitrogen sources in soil which may be one possible reason why pH value affect the growth of diazotrophs (Lauber et al., 2009). N availability may have a negative impact on diazotroph abundance (Reardon et al., 2014; Zhalnina et al., 2015). In our study, however, the NH4+ availability was not different between the intercropping and monoculture except for the rye-cucumber system in the spring (Li and Wu, 2018). The increase in soil protease activities in all intercropping systems were not accompanied by increased in soil NH4+ availability, probably due to the plant uptake or N losses. Moreover, our study found that nifH gene abundance was positively correlated with NO3– regardless of the season (Table 1). This might be due to the fact that soils with high total nitrogen may be helpful for sustaining better plant growth and thus supply more plant carbon substrates to nourish diazotrophs, as the latter are highly dependent on the availability of carbon (Han et al., 2019).
It has been shown that temporal variation can affect the diazotrophic abundance (Pereira e Silva et al., 2011, 2013). Consistently, we observed significant seasonal changes of the nifH gene abundance in the intercropping systems (Figure 2). Temperature may induce the variation of energy influx in different seasons due to its effect on soil bacterial communities (Pettersson and Bååth, 2003; Pereira e Silva et al., 2013). BNF is an energy consuming process (Pereira e Silva et al., 2013), it may trend to increase with increasing temperature and energy input, supporting the higher nifH gene abundance in the spring. All these results indicate that the abundance of diazotroph is not affected by any single factor, but by several correlated factors.
Effect of Intercropping on Diazotrophic Communities
Intercropping could increase plant diversity, different plant species usually secrete different kinds and amounts of root exudates, which can exert specific effects on soil microbial community diversity and composition (Grayston et al., 1998; Arafat et al., 2017; Chen et al., 2018; Li and Wu, 2018; Rahman et al., 2020). We observed that the alpha-diversities of diazotrophic communities in the rape–cucumber and mustard–cucumber systems were significantly higher than that of the monoculture (Table 2). Rape and mustard both belong to the Brassicaceae, and they could release glucosinolates along with their degradation products through root exudation, which can directly impact soil microbes (Choesin and Boerner, 1991; Schreiner and Koide, 1993; Hanschen et al., 2015; Jin et al., 2019). A previous study has shown that glucosinolates alter bacterial communities through an increased relative abundance of bacteria able to use these compounds as an additional C-source (Hanschen et al., 2015). Thus, our results imply that specific compounds in the Brassicaceae may affect diazotrophic community diversity. Soil microbial diversity is closely related to nutrient cycling (Giuditta et al., 2019). In our study, we found that the activities of N cycle related enzymes (urease and protease) in rape–cucumber system were highest in the fall (Supplementary Table 1), while the increase in activities of urease and protease did not cause an increase in available NH4+ (Li and Wu, 2018). We suggest this might be due to the high plant uptake, which needs to be investigated in further studies.
Multivariate regression tree analysis showed that diazotrophic diversity and richness were mainly shaped by soil moisture in the spring (Figure 3A). This result was in line with a previous finding reported by Che et al. (2018). Water content can affect soil microbial communities directly or indirectly by affecting the availability and quality of organic carbon (Che et al., 2018). Soil P nutrient is one of the important factors in regulating the change of diazotrophic diversity and richness (Yang L. et al., 2019). Similar result was found in our present study where the diazotrophic diversity in the fall was mainly changed by soil AP content (Figure 3B). Taken together, our results suggest that both the soil moisture and AP are important factors in regulating the changes of the diazotrophic diversity and richness.
The nifH community was dominated by Proteobacteria in terms of relative abundance, followed by Verrucomicrobia and Cyanobacteria, whereas the presence of other phyla were found to be negligible in our study (Figure 4A). This finding is in line with those of a previous study, namely, that Proteobacteria were one of the most commonly diazotrophs found in terrestrial ecosystems (Zehr et al., 2003). In addition, a study by Quesada et al. (1997) showed that Cyanobacteria play key roles in BNF in rice-field soils. However, the abundance of soil bacterial at the phylum level does not always reveal the functional features of sites, and, consequently, further analysis we focus on the genus level.
In this study, we discovered that Skermanella were the most abundant diazotrophic genera in all of the soil samples tested (Figure 4B). Skermanella belong to the Proteobacteria phylum, which are obligate chemo-organotrophs and facultative anaerobes, unable to fix nitrogen (Zhu, 2014). We found that the relative abundances of Skermanella in the mustard–cucumber system were slightly increased in spring and significantly increased in the fall compared with the monoculture (Supplementary Figure S1). This may have been due to the lower level of soil NO3– in the mustard–cucumber system, since the relative abundance of Skermanella was negatively correlated with NO3– concentration (Figure 5A). Moreover, a previous study has shown that the presence of Skermanella was correlated with the level of the phenolic-related compound gamma-aminobutyric acid GABA from Arabidopsis root exudates (Badri et al., 2013). Arabidopsis and mustard both belong to the Brassicaceae, and this component may also be present in the root exudates of mustard, which affects the abundance of Skermanella.
Methylobacter and Methylosinus belong to Type I and Type II methanotrophs, respectively (Zhang et al., 2014). Both methanotrophs possess nitrogen fixing genes, and both are able to fix N2 under laboratory experimental conditions (Auman et al., 2001; Boulygina et al., 2002; Dedysh et al., 2004). The significant correlations between soil moisture and the abundance of Methylobacter in the fall (Figure 5B) and of Methylosinus in both growing seasons (Figures 5A,B) implied that these two diazotrophic genera were very sensitive to soil moisture. The alfalfa–cucumber system significantly increased the relative abundances of Methylobacter and Methylosinus in spring compared with the monoculture (Supplementary Figure 1). Specific flavonoids in the root exudates of leguminous crops, which act as signaling molecules to attract N-fixing bacteria, might be the reason for the diazotrophic abundance changes in alfalfa–cucumber system (Broughton et al., 2003). Rhodomicrobium as photosynthetic bacteria, have been shown to be capable of fixing N (Lindstrom et al., 1950). The significant correlations between the relative abundance of Rhodomicrobium and soil AK (Figure 5A) imply that this diazotrophic genus is sensitive to the soil concentration of potassium nutrients.
The Bradyrhizobium genus is ubiquitous in soil, and includes symbiotic N-fixing bacterial species and free-living soil diazotrophs. The relative abundance of Bradyrhizobium was found to be lower than 0.5% in our study (data not shown), and this lower relative abundance may be attributed to the higher pH of the soil in our study. Previous studies have reported that indigenous Bradyrhizobium abundance decreased with an increase in soil pH, especially when the soil pH was greater than 6.0 (Tang and Robson, 1993; Rossum et al., 1994; Zhalnina et al., 2013). Yet, when soil nitrogen was not limiting to the plant, this symbiosis did not occur (Omrane and Chiurazzi, 2009). Compared with the result of the study by Lin et al. (2018), the soil nitrogen content found in our study was higher. This may have been the reason why the genera measured in our study were non-symbiotic diazotrophs. Further research is required to evaluate their N-fixing capacity.
NMDS and RDA showed that the structure of the soil diazotrophic community did not change significantly between intercropped and monocultured soils (Figures 6, 7). However, the diazotrophic community structure changed significantly in the two growing seasons of spring and fall (Table 3). The pH of the soil did not differ significantly among the treatments, but increased in the fall compared with the spring. This suggests that pH was the predominant factor driving seasonal changes in the diazotroph community. Moreover, previous studies have shown that temperature affected the structure of soil diazotrophic communities (Wang et al., 2015; Lin et al., 2018). In northeastern China, significant differences in temperature exist between the two growing seasons. Therefore, we also speculate that the variation in diazotrophic community structure in the two growing seasons is related to temperature. Poly et al.’s (2001) study showed that the composition of N-fixers is influenced by the soil physicochemical characteristics. In our study, however, the soil pH, AK, and moisture, which primarily related to the diazotrophic communities were identical between the intercropped and monocultured systems. Thus, the finding that the structure of the soil diazotrophic community was not significantly affected by intercropping may have been a result of the lack of an effect of intercropping on the soil physicochemical characteristics in continuous cropping soil. Further experiments need to be carried out over the longer term in order to confirm these results.
Conclusion
In summary, the results of our study have shown that intercropping increases nifH gene abundance. The rape–cucumber system significantly increased diazotrophic diversity and richness. However, intercropping had no effect on diazotrophic community structure. Moreover, we found that changes in the soil diazotrophic community were related to environmental factors, indicating that the effect of intercropping on the soil diazotrophic community was indirectly affected by the physicochemical properties of the soil. Changes in abundance and diversity rather than the structure of the diazotrophic community in intercropping systems might potentially affect the N-fixing ability of continuous cropping systems, and this hypothesis needs to be explored further.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP159857.
Author Contributions
FW contributed to design this experiment. HG and SL performed the experiment. HG analyzed the data and wrote the manuscript. All authors have read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.630302/full#supplementary-material
Supplementary Figure 1 | Relative abundance (%) of the domiant genera among eight cropping systems in spring (S) and fall (F) cropping seasons.
Supplementary Table 1 | Soil physicochemical properties and soil enzymes among eight cropping systems in spring (S) and fall (F) cropping seasons.
References
Arafat, Y., Wei, X. Y., Jiang, Y. H., Chen, T., Saqib, H. S. A., Lin, S., et al. (2017). Spatial distribution patterns of root-associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 18:1727. doi: 10.3390/ijms18081727
Auman, A. J., Speake, C. C., and Lidstrom, M. E. (2001). nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 67, 4009–4016. doi: 10.1128/aem.67.9.4009-4016.2001
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., and Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512. doi: 10.1074/jbc.m112.433300
Berthrong, S. T., Yeager, C. M., Gallegos-Graves, L., Steven, B., Eichorst, S. A., Jackson, R. B., et al. (2014). Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl. Environ. Microb. 80, 3103–3112. doi: 10.1128/aem.04034-13
Boulygina, E. S., Kuznetsov, B. B., Marusina, A. I., Tourova, T. P., Kravchenko, I. K., Bykova, S. A., et al. (2002). A study of nucleotide sequences of nifH genes of some methanotrophic bacteria. Microbiology 71, 425–432.
Brookes, P. C., Landman, A., Pruden, G., and Jenkinson, D. S. (1985). Chloroform fumigation chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837e842.
Broughton, W. J., Zhang, F., Perret, X., and Staehelin, C. (2003). Signals exchanged between legumes and Rhizobium: agricultural uses and perspectives. Plant Soil 252, 129–137. doi: 10.1023/a:1024179717780
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., et al. (2012). Biodiversity loss and it impact on humanity. Nature 486, 59–67.
Chang, C. L., Fu, X. P., Zhou, X. G., and Wu, F. Z. (2017). Effects of seven different companion plants on cucumber productivity, soil chemical characteristics and Pseudomonas community. J. Integr. Agr. 16, 2206–2214. doi: 10.1016/s2095-3119(17)61698-8
Che, R. X., Deng, Y. C., Wang, F., Wang, W. J., Xu, Z. H., Hao, Y. B., et al. (2018). Autotrophic and symbiotic diazotrophs dominate nitrogen-fixing communities in Tibetan grassland soils. Sci. Total Environ. 639, 997–1006. doi: 10.1016/j.scitotenv.2018.05.238
Chen, J., Arafat, Y., Wu, L. K., Xiao, Z. G., Li, Q. S., Khan, M. A., et al. (2018). Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl. Soil Ecol. 124, 327–334. doi: 10.1016/j.apsoil.2017.11.010
Chen, J., Wang, P., Wang, C., Wang, X., Miao, L., Liu, S., et al. (2019). Dam construction alters function and community composition of diazotrophs in riparian soils across an environmental gradient. Soil Biol. Biochem. 132, 14–23. doi: 10.1016/j.soilbio.2019.01.020
Choesin, D. N., and Boerner, R. E. J. (1991). Allyl isothiocyanate release and the allelopathic potential of Brassica napus (Brassicaceae). Plant Physiol. Biochem. 78, 1083–1090. doi: 10.1002/j.1537-2197.1991.tb14516.x
Cong, W. F., Hoffland, E., Li, L., Six, J., Sun, J. H., Bao, X. G., et al. (2015). Intercropping enhances soil carbon and nitrogen. Glob. Change Biol. 21, 1715–1726. doi: 10.1111/gcb.12738
Cook, R. J. (2006). Toward cropping systems that enhance productivity and sustainability. Proc. Natl. Acad. Sci. U.S.A. 103, 18389–18394. doi: 10.1073/pnas.0605946103
Dedysh, S. N., Ricke, P., and Liesack, W. (2004). NifH and NifD phylogenies: an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150, 1301–1313. doi: 10.1099/mic.0.26585-0
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicons from microbial amplicon reads. Nat. Method 10, 996–998. doi: 10.1038/nmeth.2604
Fan, K., Delgado-Baquerizo, M., Guo, X. S., Wang, D. Z., Wu, Y. Y., Zhu, M., et al. (2019). Suppressed N fixation and diazotrophs after four decades of fertilization. Microbiome 7:143.
Giuditta, E., Marzaioli, R., Esposito, A., Ascoli, D., Stinca, A., Mazzoleni, S., et al. (2019). Soil microbial diversity, biomass, and activity in two pine plantations of southern italy treated with prescribed burning. Forests 11:19. doi: 10.3390/f11010019
Grayston, S. J., Wang, S. Q., Campbell, C. D., and Edwards, A. C. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378. doi: 10.1016/s0038-0717(97)00124-7
Hamida, A. O., Hoda, A. A., Mohamed, M. M., Sonaa, H. E., and Salwa, S. G. (2016). Control of meloidogyne incognita on potato plant and its associated weeds with some bioactive agents and chemicals under field conditions. Egypt. J. Biol. Pest Co. 26, 503–508.
Han, L. L., Wang, Q., Shen, J. P., Di, H. J., Wang, J. T., Wei, W. X., et al. (2019). Multiple factors drive the abundance and diversity of diazotrophic community in typical farmland soils of China. FEMS Microbiol. Ecol. 95:8.
Hanschen, F. S., Yim, B., Winkelmann, T., Smalla, K., and Schreiner, M. (2015). Degradation of biofumigant isothiocyanates and allyl glucosinolate in soil and their effects on the microbial community composition. PLoS One 10:e0132931. doi: 10.1371/journal.pone.0132931
Hauggaard-Nielsen, H., and Jensen, E. S. (2005). Facilitative root interactions in intercrops. Plant Soil 274, 237–250. doi: 10.1007/1-4020-4099-7_13
Hayden, H. L., Drake, J., Imhof, M., Oxley, A., Norng, S., and Mele, P. M. (2010). The abundance of nitrogen cycle genes amoA and nifH depends on land-uses and soil types in South-Eastern Australia. Soil Biol. Biochem. 42, 1774–1783. doi: 10.1016/j.soilbio.2010.06.015
Hill, S. (1988). How is nitrogenase regulated by oxygen? FEMS Microbiol Lett. 4, 111–129. doi: 10.1111/j.1574-6968.1988.tb02738.x
Hu, X. J., Liu, J. J., Zhu, P., Wei, D., Jin, J., Liu, X. B., et al. (2018). Long-term manure addition reduces diversity and changes community structure of diazotrophs in a neutral black soil of northeast China. J. Soil Sediment. 18, 2053–2062. doi: 10.1007/s11368-018-1975-6
Inal, A., Gunes, A., Zhang, F., and Cakmak, I. (2007). Peanut/maize intercropping induced changes in rhizosphere and nutrient concentrations in shoots. Plant Physiol. Bioch. 45, 350–356. doi: 10.1016/j.plaphy.2007.03.016
Jin, X., Wu, F. Z., and Zhou, X. G. (2020). Different toxic effects of ferulic and p-hydroxybenzoic acids on cucumber seedling growth were related to their different influences on rhizosphere microbial composition. Biol. Fert. Soils 56, 125–136. doi: 10.1007/s00374-019-01408-0
Jin, X., Zhang, J. H., Shi, Y. J., Wu, F. Z., and Zhou, X. G. (2019). Green manures of Indian mustard and wild rocket enhance cucumber resistance to Fusarium wilt through modulating rhizosphere bacterial community composition. Plant Soil 441, 283–300. doi: 10.1007/s11104-019-04118-6
Junier, P., Junier, T., Witzel, K. P., and Carú, M. (2009). Composition of diazotrophic bacterial assemblages in bean-planted soil compared to unplanted soil. Eur. J. Soil. Biol. 45, 153–162. doi: 10.1016/j.ejsobi.2008.10.002
Ladd, J. N., Brisbane, P. G., and Butler, J. H. A. (1976). Studies on soil fumigation. 3. Effects on enzyme-activities, bacterial numbers and extractable ninhydrin reactive compounds. Soil Biol. Biochem. 8, 255–260.
Lai, R. Q., Hu, H. Q., Wu, X. T., and Bai, J. J. (2019). Intercropping oilseed rape as a potential relay crop for enhancing the biological control of green peach aphids and aphid−transmitted virus diseases. Entomol. Exp. Appl. 167, 1–8. doi: 10.1002/9780470750988.ch1
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 75, 5111–5120. doi: 10.1128/aem.00335-09
Li, L., Tang, C. X., Renge, Z., and Zhang, F. S. (2003a). Chickpea facilitates phosphorus uptake by intercropped wheat from an organic phosphorus source. Plant Soil 248, 297–303. doi: 10.1007/978-94-010-0243-1_25
Li, L., Zhang, F., Li, X., Christie, P., Sun, J., Yang, S., et al. (2003b). Interspecific facilitation of nutrient uptake by intercropped maize and faba bean. Nutr. Cycl. Agroecosys. 65, 61–71.
Li, Q. S., Wu, L. K., Chen, J., Khan, M. A., Luo, X. M., and Lin, W. X. (2016). Biochemical and microbial properties of rhizospheres under maize/peanut intercropping. J. Integr. Agr. 15, 101–110. doi: 10.1016/s2095-3119(15)61089-9
Li, S., and Wu, F. Z. (2018). Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 9:1521. doi: 10.3389/fmicb.2018.01521
Li, W. X., Li, L., Sun, J. H., Guo, T. W., Zhang, F. S., Bao, X. G., et al. (2005). Effects of intercropping and nitrogen application on nitrate present in the profile of an Orthic Anthrosol in Northwest China. Agr. Ecosyst. Environ. 105, 483–491. doi: 10.1016/j.agee.2004.07.008
Li, X. P., Mu, Y. H., Cheng, Y. B., Liu, X. G., and Nian, H. (2013). Effects of intercropping sugarcane and soybean on growth, rhizosphere soil microbes, nitrogen and phosphorus availability. Acta Physiol. Plant 35, 1113–1119. doi: 10.1007/s11738-012-1148-y
Lin, Y. X., Ye, G. P., Liu, D. Y., Ledgard, S., Luo, J. F., Fan, J. B., et al. (2018). Long-term application of lime or pig manure rather than plant residues suppressed diazotroph abundance and diversity and altered community structure in an acidic Ultisol. Soil Biol. Biochem. 123, 218–228. doi: 10.1016/j.soilbio.2018.05.018
Lindstrom, E. S., Tove, S. R., and Wilson, P. W. (1950). Nitrogen fixation by the green and purple sulfur bacteria. Science 112, 197–198. doi: 10.1126/science.112.2903.197
Mergel, A., Kloos, K., and Bothe, H. (2001). Seasonal fluctuations in the population of denitrifying and N2-fixing bacteria in an acid soil of a Norway spruce forest. Plant Soil 203, 145–160. doi: 10.1007/bf02220705
Nelson, D. R., and Mele, P. M. (2006). The impact of crop residue amendments and lime on microbial community structure and nitrogen-fixing bacteria in the wheat rhizosphere. Aust. J. Soil Res. 44, 319–329. doi: 10.1071/sr06022
Omrane, S., and Chiurazzi, M. (2009). A variety of regulatory mechanisms are involved in the nitrogen-dependent modulation of the nodule organo-genesis program in legume roots. Plant Signal Behav. 4, 1066–1068. doi: 10.4161/psb.4.11.9735
Pereira e Silva, M. C., Schloter-Hai, B., Schloter, M., van Elsas, J. D., and Salles, J. F. (2013). Temporal dynamics of abundance and composition of nitrogen-fixing communities across agricultural soils. PLoS One 8:e74500. doi: 10.1371/journal.pone.0074500
Pereira e Silva, M. C., Semenov, A. V., van Elsas, J. D., and Salles, J. F. (2011). Seasonal variations in the diversity and abundance of diazotrophic communities across soils. FEMS Microbiol. Ecol. 77, 57–68. doi: 10.1111/j.1574-6941.2011.01081.x
Pettersson, M., and Bååth, E. (2003). Temperature-dependent changes in the soil bacterial community in limed and unlimed soil. FEMS Microbiol. Ecol. 45, 13–21. doi: 10.1016/s0168-6496(03)00106-5
Poly, F., Ranjard, L., Nazaret, S., Gourbière, F., and Monrozier, L. J. (2001). Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67, 2255–2262. doi: 10.1128/aem.67.5.2255-2262.2001
Quesada, A., Leganés, F., and Fernández-Valiente, E. (1997). Environmental factors controlling N2-fixation in mediterranean ricefields. Microb. Ecol. 34, 39–48. doi: 10.1007/s002489900032
Rahman, M. K. U., Tan, S. C., Ma, C. L., Wu, F. Z., and Zhou, X. G. (2020). Exogenously applied ferulic acid and p-coumaric acid differentially affect cucumberrhizosphere Trichoderma spp. community structure and abundance. Plant Soil Environ. 66, 461–467. doi: 10.17221/681/2019-pse
Reardon, C. L., Gollany, H. T., and Wuest, S. B. (2014). Diazotroph community structure and abundance in wheat-fallow and wheat-pea crop rotations. Soil Biol. Biochem. 69, 406–412. doi: 10.1016/j.soilbio.2013.10.038
Reed, S. C., Cleveland, C. C., and Townsend, A. R. (2011). Functional ecology of free living nitrogen fixation: a contemporary perspective. Annu. Rev. Ecol. Evol. 42, 489–512. doi: 10.1146/annurev-ecolsys-102710-145034
Reed, S. C., Townsend, A. R., Cleveland, C. C., and Nemergut, D. R. (2010). Microbial community shifts influence patterns in tropical forest nitrogen fixation. Oecologia 164, 521–531. doi: 10.1007/s00442-010-1649-6
Ridley, A. M., Mele, P. M., and Beverly, C. R. (2004). Legume-based farming in Southern Australia: developing sustainable systems to meet environmental challenges. Soil Biol. Biochem. 36, 1213–1221. doi: 10.1016/j.soilbio.2004.04.007
Rocca, J. D., Hall, E. K., Lennon, J. T., Evans, S. E., Waldrop, M. P., Cotner, J. B., et al. (2015). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J. 9, 1693–1699. doi: 10.1038/ismej.2014.252
Rooney, D. C., and Clipson, N. (2008). Impact of sheep urine deposition and plant species on ammonia-oxidizing bacteria in upland grassland soil. Can. J. Microbiol. 54, 791–796. doi: 10.1139/w08-065
Rösch, C., Mergel, A., and Bothe, H. (2002). Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68, 3818–3829. doi: 10.1128/aem.68.8.3818-3829.2002
Rossum, D., Muyotcha, A., Hoop, B. M., Verseveld, H. W., Stouthamer, A. H., and Boogerd, F. C. (1994). Soil acidity in relation to groundnut-bradyrhizobium symbiotic performance. Plant Soil 163, 165–175. doi: 10.1007/bf00007965
Schreiner, R. P., and Koide, R. T. (1993). Mustards, mustard oils and mycorrhizas. Plant Physiol. 123, 107–113. doi: 10.1111/j.1469-8137.1993.tb04536.x
Tang, C., and Robson, A. D. (1993). pH above 6.0 reduces nodulation in Lupinus species. Plant Soil 152, 269–276. doi: 10.1007/bf00029097
Thorsted, M. D., Olesen, J. E., and Weiner, J. (2006). Mechanical control of clover improves nitrogen supply and growth of wheat in winter wheat/white clover intercropping. Eur. J. Agron. 24, 149–155. doi: 10.1016/j.eja.2005.07.004
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19:703e707.
Vitousek, P. M., Cassman, K., Cleveland, C., Crews, T., Field, C. B., Grimm, N. B., et al. (2002). Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry 57, 1–45.
Wakelin, S. A., Gupta, V. V. S. R., and Forrester, S. T. (2010). Regional and local factors affecting diversity, abundance and activity of free-living, N2-fixing bacteria in Australian agricultural soils. Pedobiologia 53, 391–399. doi: 10.1016/j.pedobi.2010.08.001
Wang, L., Yu, Z., Yang, J., and Zhou, J. (2015). Diazotrophic bacterial community variability in a subtropical deep reservoir is correlated with seasonal changes in nitrogen. Environ. Sci. Pollut. R. 22, 19695–19705. doi: 10.1007/s11356-015-5144-9
Whitmore, A. P., and Schroder, J. J. (2007). Intercropping reduces nitrate leaching from under field crops without loss of yield: a modelling study. Eur. J. Agron. 27, 81–88. doi: 10.1016/j.eja.2007.02.004
Xiao, C. H., Tang, H., Pu, L. J., Sun, D. M., Ma, J. Z., Yu, M., et al. (2010). Diversity of nitrogenase (nifH) genes pool in soybean field soil after continuous and rotational cropping. J. Basic Microb. 50, 373–379. doi: 10.1002/jobm.200900317
Xu, W. H., Wang, Z. G., and Wu, F. Z. (2015). The effect of D123 wheat as a companion crop on soil enzyme activities, microbial biomass and microbial communities in the rhizosphere of watermelon. Front. Microbiol. 6:899. doi: 10.3389/fmicb.2015.00899
Xu, W. H., Wu, F. Z., Chang, C. L., Liu, S. W., and Zhang, Y. (2013). Effects of wheat as companion cropping on growth, soil enzymes and disease resistance of watermelon. Allelopathy J. 32, 267–278.
Yang, L., Bai, J. S., Zeng, N. H., Zhou, X., Liao, Y. L., Lu, Y. H., et al. (2019). Diazotroph abundance and community structure are reshaped by straw return and mineral fertilizer in rice-rice-green manure rotation. Appl. Soil Ecol. 136, 11–20. doi: 10.1016/j.apsoil.2018.12.015
Yang, Y., Feng, X. M., Hu, Y. G., and Zeng, Z. H. (2019). The diazotrophic community in oat rhizosphere: effects of legume intercropping and crop growth stage. Front. Agric. Sci. Eng. 6:162–171. doi: 10.15302/J-FASE-2018212
Yang, Z., Yang, W., Li, S., Hao, J., Su, Z., Sun, M., et al. (2016). Variation of bacterial community diversity in rhizosphere soil of sole-cropped versus intercropped heat field after harvest. PLoS One 11:e0150618. doi: 10.1371/journal.pone.0150618
Yin, Y., Gu, J., Wang, X., Zhang, K., Hu, T., Ma, J., et al. (2018). Impact of copper on the diazotroph abundance and community composition during swine manure composting. Bioresour. Technol. 255, 257–265. doi: 10.1016/j.biortech.2018.01.120
Yu, H. J., Chen, S. C., Zhang, X. X., Zhou, X. G., and Wu, F. Z. (2019). Rhizosphere bacterial community in watermelon-wheat intercropping was more stable than in watermelon monoculture system under Fusarium oxysporum f. sp. niveum invasion. Plant Soil 445, 369–381. doi: 10.1007/s11104-019-04321-5
Yu, J. Q., Shou, S. Y., Qian, Y. R., Zhu, Z. Z., and Hu, W. H. (2000). Autotoxic potential of cucurbit crops. Plant Soil 223, 147–151.
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zhalnina, K., de Quadros, P. D., Gano, K. A., Davis-Richardson, A., Fagen, J. R., Brown, C. T., et al. (2013). Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front. Microbiol. 4:104. doi: 10.3389/fmicb.2013.00104
Zhalnina, K., Dias, R., de Quadros, P. D., Davis-Richardson, A., Camargo, F. A. O., Clark, I. M., et al. (2015). Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 69, 395–406. doi: 10.1007/s00248-014-0530-2
Zhang, X., Kong, J. Y., Xia, F. F., Su, Y., and He, R. (2014). Effects of ammonium on the activity and community of methanotrophs in landfill biocover soils. Syst. Appl. Microbiol. 37, 296–304. doi: 10.1016/j.syapm.2014.03.003
Zhou, X. G., Liu, J., and Wu, F. Z. (2017). Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 415, 507–520. doi: 10.1007/s11104-017-3181-5
Zhou, X. G., and Wu, F. Z. (2015). Changes in soil chemical characters and enzyme activities during continuous monocropping of cucumber (Cucumis sativus). Pak. J. Bot. 47, 691–697.
Zhou, X. G., Yu, G. B., and Wu, F. Z. (2011). Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Eur. J. Soil Biol. 47, 279–287. doi: 10.1016/j.ejsobi.2011.07.001
Keywords: cucumber, intercropping, diazotrophic community, nifH, continuous
Citation: Gao H, Li S and Wu F (2021) Impact of Intercropping on the Diazotrophic Community in the Soils of Continuous Cucumber Cropping Systems. Front. Microbiol. 12:630302. doi: 10.3389/fmicb.2021.630302
Received: 17 November 2020; Accepted: 08 March 2021;
Published: 31 March 2021.
Edited by:
Milko Alberto Jorquera, University of La Frontera, ChileReviewed by:
Mauricio Schoebitz, University of Concepcion, ChileXingwang Liu, China Agricultural University, China
Xuehao Chen, Yangzhou University, China
Copyright © 2021 Gao, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengzhi Wu, Znp3dTIwMDZAYWxpeXVuLmNvbQ==
 Huan Gao
Huan Gao Sen Li1,2
Sen Li1,2 Fengzhi Wu
Fengzhi Wu