- 1Department of Chemical and Process Engineering, University of Canterbury, Christchurch, New Zealand
- 2Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand
- 3Maurice Wilkins Center for Molecular Biodiscovery, Auckland, New Zealand
- 4Geomicrobiology Research Group, Department of Geothermal Sciences, GNS Science, Taupō, New Zealand
- 5School of Biological Sciences, University of Canterbury, Christchurch, New Zealand
Members of the genus Methylacidiphilum, a clade of metabolically flexible thermoacidophilic methanotrophs from the phylum Verrucomicrobia, can utilize a variety of substrates including methane, methanol, and hydrogen for growth. However, despite sequentially oxidizing methane to carbon dioxide via methanol and formate intermediates, growth on formate as the only source of reducing equivalents (i.e., NADH) has not yet been demonstrated. In many acidophiles, the inability to grow on organic acids has presumed that diffusion of the protonated form (e.g., formic acid) into the cell is accompanied by deprotonation prompting cytosolic acidification, which leads to the denaturation of vital proteins and the collapse of the proton motive force. In this work, we used a combination of biochemical, physiological, chemostat, and transcriptomic approaches to demonstrate that Methylacidiphilum sp. RTK17.1 can utilize formate as a substrate when cells are able to maintain pH homeostasis. Our findings show that Methylacidiphilum sp. RTK17.1 grows optimally with a circumneutral intracellular pH (pH 6.52 ± 0.04) across an extracellular range of pH 1.5–3.0. In batch experiments, formic acid addition resulted in no observable cell growth and cell death due to acidification of the cytosol. Nevertheless, stable growth on formic acid as the only source of energy was demonstrated in continuous chemostat cultures (D = 0.0052 h−1, td = 133 h). During growth on formic acid, biomass yields remained nearly identical to methanol-grown chemostat cultures when normalized per mole electron equivalent. Transcriptome analysis revealed the key genes associated with stress response: methane, methanol, and formate metabolism were differentially expressed in response to growth on formic acid. Collectively, these results show formic acid represents a utilizable source of energy/carbon to the acidophilic methanotrophs within geothermal environments. Findings expand the known metabolic flexibility of verrucomicrobial methanotrophs to include organic acids and provide insight into potential survival strategies used by these species during methane starvation.
Introduction
The aerobic methane-oxidizing bacteria (methanotrophs) are able to grow exclusively on methane (CH4) as their sole source of carbon and energy (Whittenbury et al., 1970). They provide vital ecosystem function by serving as the primary biological sink for methane (Chistoserdova, 2015; Knief, 2015) and are of biotechnological interest in the development of commercial gas-to-liquid (Kalyuzhnaya et al., 2015) and proteinaceous feedstock (Strong et al., 2016) bioprocesses. Methanotrophs oxidize methane to methanol via a particulate or soluble methane monooxygenase enzyme (pMMO/sMMO) and subsequently yield reducing equivalents (e.g., NADH) for cellular respiration and biosynthesis through the oxidation of methanol to carbon dioxide. The gammaproteobacterial (Type I) and alphaproteobacterial (Type II) methanotrophs assimilate the intermediate formaldehyde using the ribulose monophosphate (RuMp) or serine pathways (Hanson and Hanson, 1996), respectively. In contrast, methanotrophs within the phylum Verrucomicrobia (genera Methylacidiphilum and Methylacidimicrobium) oxidize methanol directly to formate (Keltjens et al., 2014) and grow autotrophically by fixing carbon dioxide (CO2) via the Calvin-Benson-Bassham cycle (Khadem et al., 2012c). For all known methanotrophs, the enzyme formate dehydrogenase (FDH) catalyzes the terminal step of methane oxidation, yielding NADH and CO2. There is considerable heterogeneity in the structure and composition of bacterial formate dehydrogenases with multiple copies of FDH-encoding genes commonly found in the genomes of methanotrophic and methylotrophic bacteria (Ferry, 1990; Yoch et al., 1990; Ward et al., 2004).
While the majority of studies have emphasized that methanotrophic bacteria have limited metabolic flexibility (Whittenbury et al., 1970; Smith and Hoare, 1977; Shishkina and Trotsenko, 1982; Wood et al., 2004; Kelly et al., 2005), several investigations have now overturned the paradigm of “obligate methanotrophy” (Dedysh and Dunfield, 2010). A few strains, notably Methylocella silvestris and other Methylocella species, utilize simple organic acids, alcohols, and short-chain alkane gases for growth (Dedysh et al., 2005; Crombie and Murrell, 2014). Aerobic H2 metabolism has also been reported in several methanotrophs (Chen and Yoch, 1987; Shah et al., 1995; Hanczar et al., 2002; Hakobyan et al., 2020), and hydrogenase-encoding genes are ubiquitously distributed in methanotroph genomes (Greening et al., 2016). In the acidophilic verrucomicrobial methanotrophs, chemolithoautotrophic growth on H2 has been reported (Mohammadi et al., 2016, 2019; Carere et al., 2017), with mixotrophic growth (H2 and CH4) proposed to provide a competitive advantage over obligate methanotrophy at the oxic/anoxic soil boundaries of acidic geothermally heated soils (Carere et al., 2017). A recent report that Methylacidiphilum fumariolicum SolV can utilize ethane and propane gases has further expanded the suit of substrates that can support the survival of these methanotrophs in situ (Picone et al., 2020).
Methanotrophs thrive at the interface of many oxic/anoxic habitats (e.g., peat bogs, forest soils, wetlands, rice paddies, geothermal, and volcanic environments; Dunfield et al., 2007; Singh et al., 2010; Knief, 2015) where the availability of oxidant (O2), energy, and carbon resources for growth fluctuate (Knief et al., 2003; Tavormina et al., 2010). In these habitats, organic polymers (i.e., lignocellulose) may anaerobically decompose into organic acids, alcohols, molecular hydrogen (H2), and carbon dioxide (CO2) and subsequently diffuse into oxic environments to facilitate aerobic growth. In general, low molecular weight organic acids display low concentrations and a short residence time in soils (Angeles et al., 2006); however, the accumulation of formic (0.65 mM), acetic (0.26 mM), and lactic acids (0.085 mM) in acidic wetland soil porewaters (Küsel et al., 2008) is illustrative that these compounds represent available substrates for microbial growth in situ.
Organic acids, such as formic acid, however, are potentially harmful to acidophilic species because they can function as uncouplers of the respiratory chain. In acidic conditions (pH < 3), diffusion of the protonated form (i.e., CHOOH; pKa 3.74) into the cell is followed by rapid proton dissociation and cytosolic acidification (i.e., CHOO− + H+) that ultimately can collapse the proton motive force governing respiratory-linked ATP synthesis (Alexander et al., 1987; Kishimoto et al., 1990; Ciaramella et al., 2005; Baker-Austin and Dopson, 2007; Lund et al., 2014). Accordingly, the genomes of many acidophilic species encode for enzymes to actively degrade organic acids. Thus, although cells may gain energy from their oxidation, the genomic constituents of organic acid degradation may primarily serve a role in pH homeostasis. Despite the ubiquity of FDH in sequenced acidophilic methanotroph genomes (Methylacidiphilum spp., Methylacidimicrobium spp.), no studies have demonstrated the metabolic capability to grow using formate or other organic acids as a sole energy source (Op den Camp et al., 2009; van Teeseling et al., 2014).
In this work, we investigated growth and intracellular pH homeostasis in response to formate/formic acid addition within the thermoacidophilic methanotroph Methylacidiphilum sp. RTK17.1. Demonstrating that Methylacidiphilum sp. RTK17.1 can grow organoautotrophically on formic acid expands the known spectrum of substrates used by verrucomicrobial methanotrophs to include organic acids and provides important new insights into the physiology and ecology of these acidophiles.
Materials and Methods
Cultivation of Methylacidiphilum sp. RTK17.1
Methylacidiphilum sp. RTK17.1 was previously isolated from geothermally heated soils sampled from Rotokawa, New Zealand (Carere et al., 2017), and shares 99% 16S rRNA gene sequence identity to Methylacidiphilum infernorum V4. All batch cultivations were performed at 50°C in a V4 mineral medium as described previously (Dunfield et al., 2007) but with the addition (0.2 μM) of rare earth elements lanthanum and cerium (Pol et al., 2014) unless otherwise specified. To determine the pH range for the growth of Methylacidiphilum sp. RTK17.1, 4 ml of pH-adjusted V4 media (range: pH 0.5–6.0; H2SO4) was made, added to (20 ml) Balch tubes (Bellco), and sealed with butyl rubber stoppers prior to sterilization. After autoclaving (121°C, 15 psi, 20 min), air headspaces were supplemented with methane (~10% v/v) and CO2 (~5% v/v). Tubes were inoculated with exponentially growing Methylacidiphilum sp. RTK17.1 cells to an initial OD600nm of 0.02. The maximum specific growth rates (μmax) were determined from the exponential phase of cultures spectrophotometrically (600 nm). The concentration of CH4 in the headspace was monitored with a PeakPerformer gas chromatograph (Peak Laboratories) outfitted with a HayeSepD column and equipped with a flame ionizing detector (FID) by injecting 1 ml samples via a gas-tight syringe (SGE Analytical Science, Melbourne).
Chemostat cultivation was performed to investigate the growth of Methylacidiphilum sp. RTK17.1 on methanol and formic acid. For these experiments, cells were cultivated in a bioreactor with a working volume of 2 L (LABFORS 3; max volume 3.6 L) at pH 2.5, 45°C, and with continuous agitation at 450 rpm. A gas headspace composition of 2% O2 and 2% CO2 (v/v; balance N2) was supplied continuously at 150 ml min−1 via mass flow control throughout the duration of these experiments. Initially, cells were grown on methanol (12.4 mM) in a fed-batch mode, such that exhaustion of methanol (as determined spectrophotometrically via cessation of cell growth) prompted repeated dosing of additional methanol back to a concentration of 12.4 mM. Once cell density reached OD600nm ~0.5, the bioreactor was switched to chemostat operation for continuous growth experiments. During chemostat growth experiments, V4 mineral medium was supplemented with either 12.4 mM methanol or 12.4 mM formic acid and supplied at a constant flow rate of 10.4 ml h−1 (D = 0.0052 h−1, td = 133 h). After achieving a steady state, cultures were monitored over a period of several days in order to determine biomass productivity data. During this time, biomass samples of Methylacidiphilum sp. RTK17.1 were harvested directly from the bioreactor for transcriptome sequencing and high-performance liquid chromatography (HPLC) analysis. Steady state methanol and formic acid concentrations (from feed medium and bioreactor samples) were quantified using a Thermo UltiMate 3000 HPLC (Dionex) equipped with an Aminex HPX-87H column (300 × 7.8 mm) and refractive index and UV vis (210 nm) detectors. A 5 mM H2SO4 mobile phase was provided at a constant flowrate of 0.5 ml min−1. Resultant chromatogram profiles were integrated using tools embedded within the Chromeleon 7.2 SR5 software. Effluent biomass (500 ml) was collected over this period for cell dry weight determinations. For this analysis, cell pellets were collected by centrifugation (10,000 rpm, 10 min, 25°C; Eppendorf Centrifuge 5810 R) and then dried to constant mass using a Labconco FreeZone 2.5 freeze dryer.
Bioenergetic Determinations
Cell suspensions of Methylacidiphilum sp. RTK17.1 were prepared in V4 medium (pH 2.5) from methane-grown cultures to a final OD600nm 0.8–1.0 as previously described (Carere et al., 2017). Previously, the energization of cell suspensions via the catabolism of intracellular glycogen reservoirs was confirmed by O2 consumption assays using a Digital model 10 Oxygen electrode (Rank Brother ltd., United Kingdom) in the presence of cyanide m-chlorophenyl hydrazone (CCCP), the glycolytic inhibitor iodoacetamide (IAA), and the respiratory chain inhibitor potassium cyanide (KCN; Carere et al., 2017).
Cytosolic pH, membrane potential (Δψ), and intracellular volume determinations were performed as described previously (Cook et al., 1996). Briefly, the intracellular volume (3.45 ± 0.59 μl mg protein−1) was estimated from the difference between the partitioning of 3H2O and [7-14C]benzoic acid. The Δψ was calculated from the uptake of the lipophilic cation [14C]tetraphenylphosphonium (TPP+) according to the Nernst relationship. Non-specific isotope binding was estimated from cells, which had been treated with valinomycin (10 μM) and/or nigericin (10 μM) for 25 min. The ∆pH was determined from the distribution of [carboxyl-14C]salicylic acid (56 mCi mmol−1) using the Henderson-Hasselbalch equation (Riebeling et al., 1975), and Z∆pH was calculated as 62 mV × ∆pH. Following incubation for 20 min at 50°C, Methylacidiphilum sp. RTK17.1 cell suspensions were centrifuged through a silicon oil mixture (equal parts Dexter Hysol 550 and 560; Hysol Co., Oleam, NY) in microcentrifuge tubes (13,000 × g, 5 min), and 20-μl samples of supernatants were removed. The tubes and contents were then frozen (−20°C), and the bottoms (containing cell pellets) were subsequently removed with dog nail clippers. Supernatants and cell pellets were dissolved in scintillation fluid (Optiphase Hisafe 2; Scitech Biolab) and counted.
Next, the influence of formic acid dosing on intracellular pH homeostasis was determined. To perform these experiments, formic acid was added to non-growing, but energized, cultures of Methylacidiphilum sp. RTK17.1 (methane-grown) at concentrations ranging from 0.01 to 1,000 mM. Cells were pelleted by centrifugation, suspended in V4 medium (OD600nm ~1.0) supplemented with formic acid (pH adjusted to 2.5 with 1 M H2SO4), and incubated for 20 min at 50°C prior to intracellular pH determinations (as described above).
Formate Dehydrogenase Assays
Biomass (1 l) collected from Methylacidiphilum sp. RTK17.1 cells grown in batch on methane (Carere et al., 2017) was used to determine the activity of formate dehydrogenase at different pH values. Briefly, 1 l of cells (OD600nm 0.810) was pelleted by centrifugation at 4,500 rpm for 20 min at 4°C and stored at −80°C until required for further processing. The resultant cell pellet was then suspended in a lysis buffer (10 ml) containing 50 mM Tris-HCl (pH 8.0), 10 μM methyl viologen, and 25 mM β-mercaptoethanol and then disrupted by sonication at 15 μF (10 × 30 s) on ice. Cell lysis (>99%) was confirmed microscopically, after which the crude lysate was centrifuged at 4,000 rpm for 30 min (4°C) to collect cell debris. The resultant cell-free extracts (2 ml) were then anaerobically transferred into N2-flushed stoppered glass assay tubes containing 2 ml of lysis buffer supplemented with 10 mM formate, gassed/degassed three times with N2 (3 min/3 min), and preincubated at 50°C for 10 min. An assay solution was prepared containing 100 mM select phosphate buffer (pH 5.5–8.0), 2 mM methyl viologen, 20 mM formate, and 10 mM β-mercaptoethanol and then gassed/degassed three times with N2 (3 min/3 min). Enzymatic reactions were then initiated by the addition of 100 μl of preincubated cell-free extracts into assay solution-containing tubes, and the time-dependent reduction of methyl viologen was recorded spectrophotometrically (600 nm). All assays were performed in triplicate, and the activity of formate dehydrogenase was presented as a percentage of the rate observed at pH 7.
Transcriptome Sequencing
Samples for transcriptome sequencing were harvested (10 ml) from exponential (methanol) and steady-state (methanol, formic acid) phases of chemostat experiments. Methylacidiphilum sp. RTK17.1 cells were pelleted by centrifugation at 5,000 × g (15 min, 4°C), suspended in 300 μl of RNAlater Stabilization solution (ThermoFisher Scientific) and then stored at −20°C until required for further analysis (as per the manufacturer’s recommended protocols). Total RNA was extracted using the Mo Bio PowerBiofilm RNA Isolation kit following the manufacturers recommended protocol, eluted into 100 μl of ddH2O, and quantified using the Qubit HS assay kit (Thermo Fisher Scientific). Following this, 2 μg of RNA was transferred into RNAstable preservation medium (Sigma Aldrich) and dehydrated at room temperature before transport for transcriptome sequencing (Custom Science; China). Upon receipt of samples, RNA integrity was confirmed using an Agilent 2100 Bioanalyser.
Following total RNA extraction, ribosomal RNAs were removed using the Ribo-Zero rRNA removal kit (bacteria), and the quality of the remaining RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent). Library construction was performed using the TruSeq Stranded Total RNA Sample Prep (microbe) Kit (Illumina), and sequencing was performed using an Illumina HiSeq2500 platform. From this, an average of 10,452,057 raw untrimmed reads were obtained for each of the conditions sampled (methanol exponential, methanol steady state, and formic acid steady state). These reads were then analyzed using the Artificial Intelligence RNA-Seq pipeline (Sequentia Biotech, Barcelona, Spain; Vara et al., 2019), which were reduced to an average of 9,154,571 reads following quality filtering and trimming. Retained paired-end reads (100 bp) were then mapped to the genome of Methylacidiphilum infernorum strain V4 (GCA_000019665.1; Hou et al., 2008).
Differential gene expression profiles and accompanying statistical analysis was performed to investigate transcriptional regulation using edgeR (Robinson et al., 2009). Synonymous conditions (methanol exponential, methanol steady state) were grouped as replicates for differential gene expression analysis and compared with the formic acid growth condition. Where indicated, comparisons to methane-grown Methylacidiphilum sp. RTK17.1 cultures refer to transcriptomes described previously (accession numbers GSM3872525-GSM3872529; Carere et al., 2019). Using these additional transcriptomes, samples were partitioned into five groups [formic acid (n = 1), methanol (n = 2), CH4 with O2 limitation (n = 2), CH4 with O2 limitation and N2-fixing (n = 2), and CH4 with excess O2 (n = 2)] to estimate the biological coefficient of variation within the dataset. Using tools within edgeR, multi-dimensional scaling and sample distance matrix analysis then was performed to verify that formic acid, methanol, and methane growth conditions were sufficiently different from one another. Expression values are provided as either fragments per kilobase million (FPKM; Supplementary Table S1) values (Mortazavi et al., 2008) or Z-normalized FPKM (Hart et al., 2013). Raw and processed transcriptome sequence files (accession number GSE145277) are available in the Gene Expression Omnibus (GEO) under accession numbers GSM4314091–GSM4314093.
Results and Discussion
Methylacidiphilum sp. RTK17.1 Maintains a Circumneutral Cytosolic pH That Is Sensitive to Organic Acid-Induced pH Stress
Methylacidiphilum sp. RTK17.1 was observed to grow methanotrophically between pH 1.0 and 6.0 (pHopt: ~2.5; μmax: 0.015 h−1; Figure 1A) in batch culture experiments. These results approximate reports of growth in the other known verrucomicrobial thermoacidophilic methanotrophs: Methylacidiphilum infernorum V4 (Dunfield et al., 2007), Methylacidiphilum kamchatkensis Kam1 (Islam et al., 2008), Methylacidiphilum fumariolicum SolV (Pol et al., 2007). Although growth down to pH 0.8 has been reported in Methylacidiphilum fumariolicum SolV, growth at values of pH < 1.0 was not observed in cultures of Methylacidiphilum sp. RTK17.1 and appears to be a trait more common with the mesophilic strains of the Methylacidiphilales (van Teeseling et al., 2014).
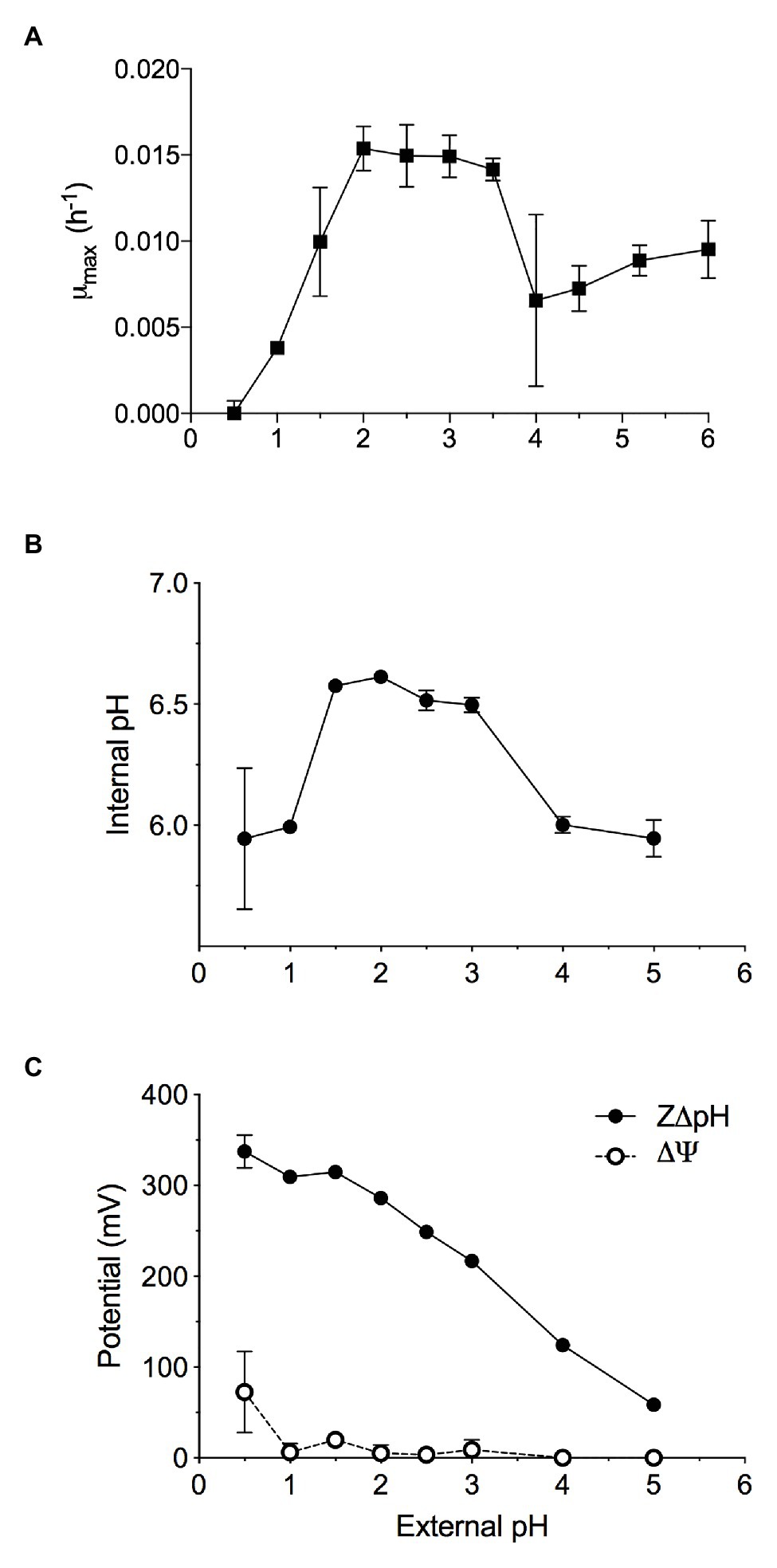
Figure 1. The maximal growth rate of methane-grown Methylacidiphilum sp. RTK17.1 in batch culture is highly dependent on maintaining pH homeostasis. Optimal growth was observed where extracellular 1.5 < pH < 4.0, resulting in the maintenance of an internal pH 6.55 ± 0.05. (A) Maximum specific growth rates observed in batch cultures of Methylacidiphilum sp. RTK17.1 (10% CH4 v/v in air) at 50°C in V4 medium at different pHs (range pH 0.5–6.0). (B) The corresponding internal pH and; (C) membrane potential (Δψ) and (Z∆pH) as a function of external pH are shown. Error bars represent the standard deviation of triplicate measurements.
Similar to neutralophilic bacteria, acidophiles must maintain circumneutral intracellular pH in order to remain viable (Baker-Austin and Dopson, 2007). However, very little is understood about how acidophilic species manage the considerable pH gradients that exist between the extracellular environment and the cytosol. We sought to investigate pH homeostasis in Methylacidiphilum sp. RTK17.1 in response to external acid stress. When non-growing, but energized, Methylacidiphilum sp. RTK17.1 cells at pH 2.5 display an intracellular pH of 6.52 ± 0.04 (Figure 1B). Likewise, similar intracellular pH values (6.55 ± 0.05) were obtained at extracellular values between pH 1.5 and 3.0. These circumneutral values approximate the known cytoplasmic pH for other acidophilic bacteria, including Acidithiobacillus ferrooxidans (pH 6.5), Acidithiobacillus thiooxidans (pH ~7), and Acidiphilium acidophilum (pH 6.0; Baker-Austin and Dopson, 2007) but differ markedly from the archaeon Picrophilus oshimae, which maintains an intracellular value of pH 4.6 when extracellular pH is <4.0 (van de Vossenberg et al., 1998). When Methylacidiphilum sp. RTK17.1 was incubated at pH values analogous to a decreased observed rate of growth (1.0 > pH > 3.0), cytosolic acidification (pH 5.97 ± 0.13) was observed. A similar degree of cytosolic acidification was observed in cell suspensions following treatment with nigericin/valinomycin (10 μM each; Figure 2A). The lack of complete pH gradient dissipation is consistent with previous reports of uncoupler treatment and has been attributed to both the limited cation membrane permeability and high cytoplasmic buffering capacity characteristic of acidophiles (Goulbourne et al., 1986; Baker-Austin and Dopson, 2007).
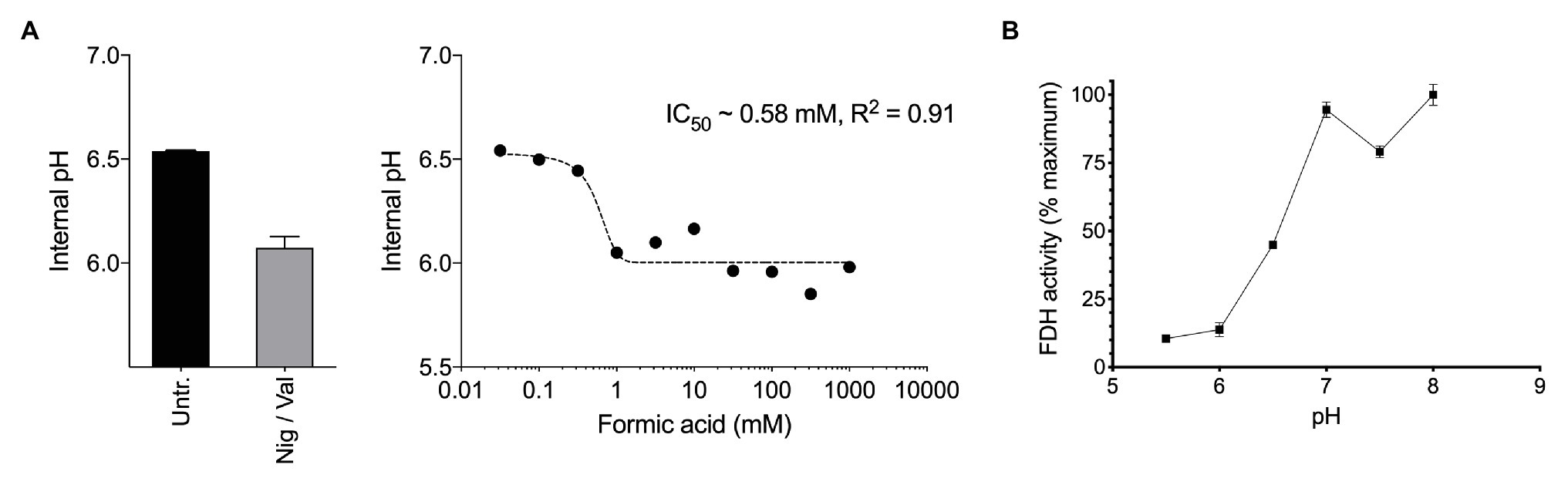
Figure 2. (A) Exposure of methane-grown batch cultures of Methylacidiphilum sp. RTK17.1 to increasing concentrations of formic acid (pka = 4.3) leads to cytosol acidification. A nonlinear fit was used to estimate the half maximal inhibitory concentration of formic acid (IC50 = 0.58 mM, R2 = 0.91). The internal pH of untreated (Untr.) cell suspensions and samples treated with 10 μM each nigericin and valinomycin (Nig/Val) are shown. (B) Formate dehydrogenase (FDH) activity, as determined from crude cell extracts of methane-grown Methylacidiphilum sp. RTK17.1 cultures is maximal at the circumneutral pH values characteristic of cytosolic pH optima. Results are presented as a percentage of the rate at pH 7. Error bars represent the standard deviation of triplicate measurements.
The chemical gradient of protons (∆pH) across the cell membrane is a major contributor to the proton motive force (PMF; PMF = Δψ − Z∆pH) driving ATP production via the electron transport chain. This large ∆pH is actively maintained in acidophiles and is further supported by a “reversed” membrane potential (Δψ) that is cytosol (inside)-positive relative to the extracellular (outside) environment. This is opposite to the cytosol-negative Δψ of neutralophiles. Indeed, at extracellular pH values between pH 2 and 3, Methylacidiphilum sp. RTK17.1 maintains a minimal membrane potential (Δψ = 5.86 mV, Figure 1C) that is “reversed.” These data are comparable to values reported for other known acidophiles, including A. ferrooxidans and Alicyclobacillus acidocaldarius (Krulwich et al., 2011), and Thiobacillus acidophilus (Zychlinsky and Matin, 1983). The inside positive Δψ contributes to inhibiting the entry of extracellular protons and detracts from the large proton motive force formed by this ΔpH.
While the mechanisms used by acidophiles to maintain pH homeostasis permit respiration and growth in acidic environments (Baker-Austin and Dopson, 2007), the ∆pH has a detrimental effect of increasing susceptibility of these bacteria to organic acid-induced pH stress. This is because the conjugate acid-base pairing of organic compounds (e.g., formate/formic acid) strongly favors the protonated form in acidic environments. When organic acids diffuse from an acidic extracellular environment across the cell membrane and into the cytosol (circumneutral pH), the conjugate-base anion dissociates, releasing protons (Baker-Austin and Dopson, 2007; Krulwich et al., 2011). At elevated organic acid concentrations, these liberated protons can act as respiratory chain uncouplers by collapsing the ∆pH. For the acidophilic methanotrophs occupying oxic/anoxic interfaces in acidic geothermal environments, this scenario poses a problem, as these species are likely to encounter organic acids. It is unknown how acidophilic methanotrophs cope with organic acid-induced pH stress, but given that formate oxidation is the terminal step common to all aerobic methanotrophs, they should be poised to gain energy from environmental fluxes of formic acid.
Therefore, we next investigated how formic acid stress affected cytosolic pH and formate dehydrogenase (FDH) activity in Methylacidiphilum sp. RTK17.1. In a dose-dependent manner, we observed that the addition of formic acid to non-growing cultures of Methylacidiphilum sp. RTK17.1 resulted in cytosolic acidification (Figure 2A), which resembled external pH stress and uncoupler-treatment experiments. Intracellular pH decreased from pH 6.52 to 6.05 with the addition of 1 mM formic acid. We interpret the absence of further cytosolic acidification with increasing formic acid concentrations (up to 1 M) as a consequence of cytoplasmic buffering that has previously been reported (Zychlinsky and Matin, 1983; Goulbourne et al., 1986; Baker-Austin and Dopson, 2007).
Formate dehydrogenase activities from crude cell extracts using the artificial electron acceptor methy-viologen were maximal between pH 7.0 and 8.0; consistent with the internal pH optima (Figure 2B). FDH activities, however, decreased to 13.8% of the maximum value at pH < 6.0. This shows that the FDH of Methylacidiphilum sp. RTK17.1 is less active at the pH values mirroring cytosolic acidification and therefore could be vulnerable to formic acid-induced pH stress. Although very little is known about the pH dependence of FDH in acidophiles, similar pH-dependent activities have been reported in several neutralophiles (Schauer and Ferry, 1982; Axley et al., 1990), anaerobes (Liu and Mortenson, 1984), and fungi (Altaş et al., 2017). In addition, the presence of several acid-labile SH groups have been attributed to the circumneutral pH optima (pH 6.5–7.5) displayed by the NAD+-dependent FDH of Methylosinus trichosporium OB3b (Trotsenko and Murrell, 2008). Collectively, these data indicate that a formic acid-induced pH stress, of sufficient magnitude, can “overwhelm” the catabolic machinery of Methylacidiphilum sp. RTK17.1, thus precipitating a nonrecoverable collapse of the proton motive force. The absence of growth on formic acid, in repeated Methylacidiphilum sp. RTK17.1 batch experiments, is further evidence of organic acid-induced cytosolic acidification and is consistent with reports that formic acid does not support growth in M. infernorum V4 (Dunfield et al., 2007), and inhibits growth in Methylacidiphilum fumariolicum SolV at acidic pH values (Pol et al., 2007).
Stable Growth of Methylacidiphilum sp. RTK17.1 on Formic Acid Is Possible in Continuous Culture
The lack of detectable growth on formate/formic acid by Methylacidiphilum sp. RTK17.1, although rationalized as a failure of pH homeostatic machinery, poses a metabolic quandary for this taxon. Formate clearly serves as an intermediate in the oxidation of methane to CO2 via an FDH-catalyzed reaction (Dunfield et al., 2007; Khadem et al., 2012c), yet conversely appears not to support growth as a sole substrate. Indeed, the oxidation of formate as a sole energy source should theoretically be capable of supporting CO2 fixation and growth in Methylacidiphilum spp. as it does with some neutralophilic methanotrophs (Kemp and Quayle, 1967; Bowman, 2014). To test this hypothesis, we attempted to grow Methylacidiphilum sp. RTK17.1 on formic acid in a steady-state continuous culture, the principle being that chemostat growth would minimize induced pH stress under formic acid-limiting growth conditions. To do this, we initially grew Methylacidiphilum sp. RTK17.1 on methanol in a chemostat before switching to formic acid in the nutrient feed. In contrast to comparable batch experiments at 12.4 mM formic acid, our findings show that Methylacidiphilum sp. RTK17.1 is capable of sustained (chemostat) growth using formic acid as the sole metabolizable source of energy (Figure 3). As evidenced by the absence of their detection in chemostat effluent streams (<0.15 mM), methanol and formic acid were the growth-limiting nutrients in their respective steady-state growth conditions (Figure 3). Biomass yields (as determined by cell dry weight; gCDW) were 7.83 (±0.41) gCDW mol−1 during growth on methanol and decreased by 63% to 2.86 (±0.27) gCDW mol−1 during growth on formic acid (Table 1). We did not assess the extent of intracellular glycogen produced as this has been previously reported in Methylacidiphilum spp. RTK17.1 (Carere et al., 2019) and SolV (Khadem et al., 2012b). It is noteworthy to mention that during both methanol‐ and formic acid-dependent chemostat growth, CO2 was continuously supplied in excess. While we cannot confirm that the oxidation of formic acid was able to supply both the necessary reducing equivalents (i.e., NADH) and inorganic carbon (CO2) required for cell growth, a previous report of methane-dependent growth in Methylacidiphilum fumariolicum SolV has shown that the oxidation of methane alone is able to sustain growth as long as evolved CO2 in the headspace reaches a minimum threshold concentration of 0.3% (v/v; Khadem et al., 2011). Biomass yields (g mol−1) on methanol and formic acid were ~115 and ~42%, respectively, of the values previously reported during growth of Methylacidiphilum sp. RTK17.1 on methane (Carere et al., 2017, 2019). However, on a per mole electron equivalent basis (YCDW/e), biomass yields were greater for both methanol‐ and formic acid-grown cultures than for methane grown cells (Table 1). This likely reflects the additional energetic input necessary to catalyze the oxidation of methane to methanol via methane monooxygenase (CH4 + O2 + [NAD(P)H + H+]/QH2 → CH3OH + NAD(P)+/Q + H2O; Carere et al., 2017).
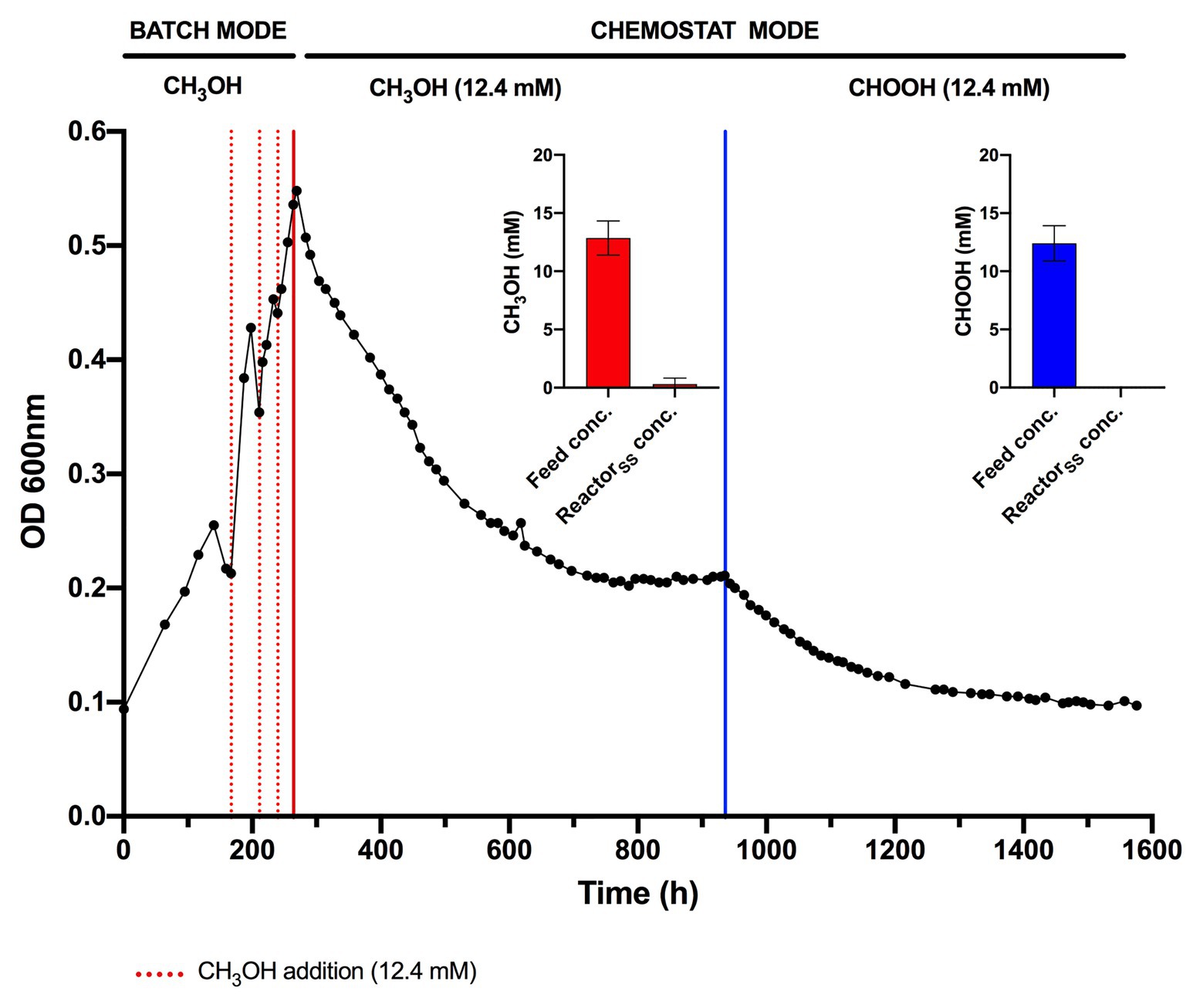
Figure 3. Chemostat cultivation of Methylacidiphilum sp. RTK17.1 on methanol (CH3OH) and formic acid (CHOOH). For all conditions, cells were grown on V4 medium at 45°C, pH 2.5 with an agitation of 450 rpm and continuous gas supply (2% O2, 2% CO2, balance N2 (v/v); 150 ml min−1). Dashed red lines denote the supplemental addition of methanol (12.4 mM) during batch growth. Solid red and blue lines indicate the transition to chemostat mode (D = 0.0052 h−1) on methanol and formic acid, respectively (supplied at 12.4 mM for both). Inset panels denote the influent feed concentration (feed conc.) and steady-state reactor concentration (ReactorSS conc.) of methanol (red) and formic acid (blue) detected by high-performance liquid chromatography (HPLC) during chemostat operation. Error bars represent the standard deviation of triplicate measurements.
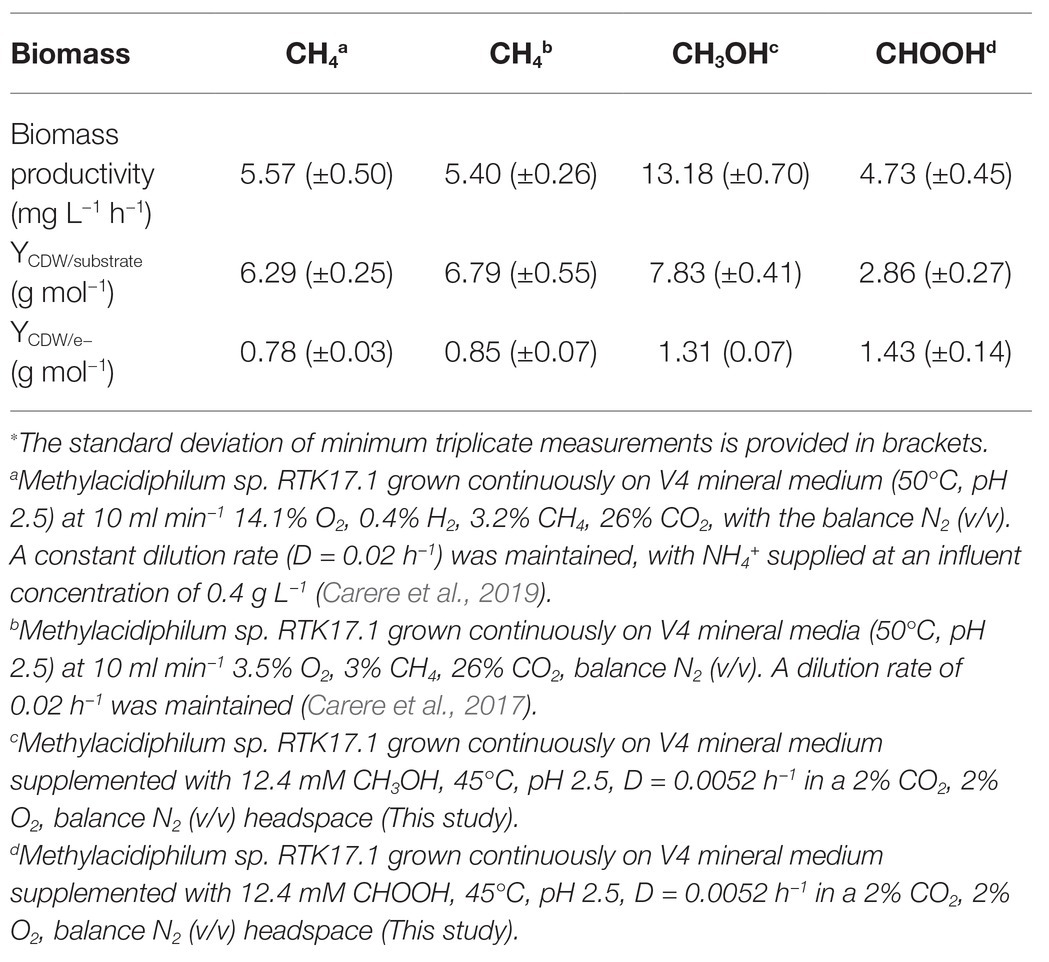
Table 1. Steady state biomass productivity of Methylacidiphilum sp. RTK17.1 during growth on methane (CH4), methanol (CH3OH), and formic acid (CHOOH)∗.
Transcriptome Analysis Reveals Key Changes in the Expression of Genes Relating to Energy/Carbon Metabolism and pH Homeostasis
Transcriptome analysis was performed on steady-state Methylacidiphilum sp. RTK17.1 cultures to determine whether genes associated with pH homeostasis, energy metabolism, and/or carbon metabolism were regulated in response to growth on formic acid (Figure 4). To account for the lack of replicates in the formic acid growth condition, complementary expression analyses were performed against methane-grown cultures described previously (Carere et al., 2019). In response to the transition from methanol to formic acid, 188 genes were identified as significantly upregulated, and 193 genes were significantly downregulated [Log2FC ≥ |2|, p < 0.001; false discovery rate (FDR) < 0.05].
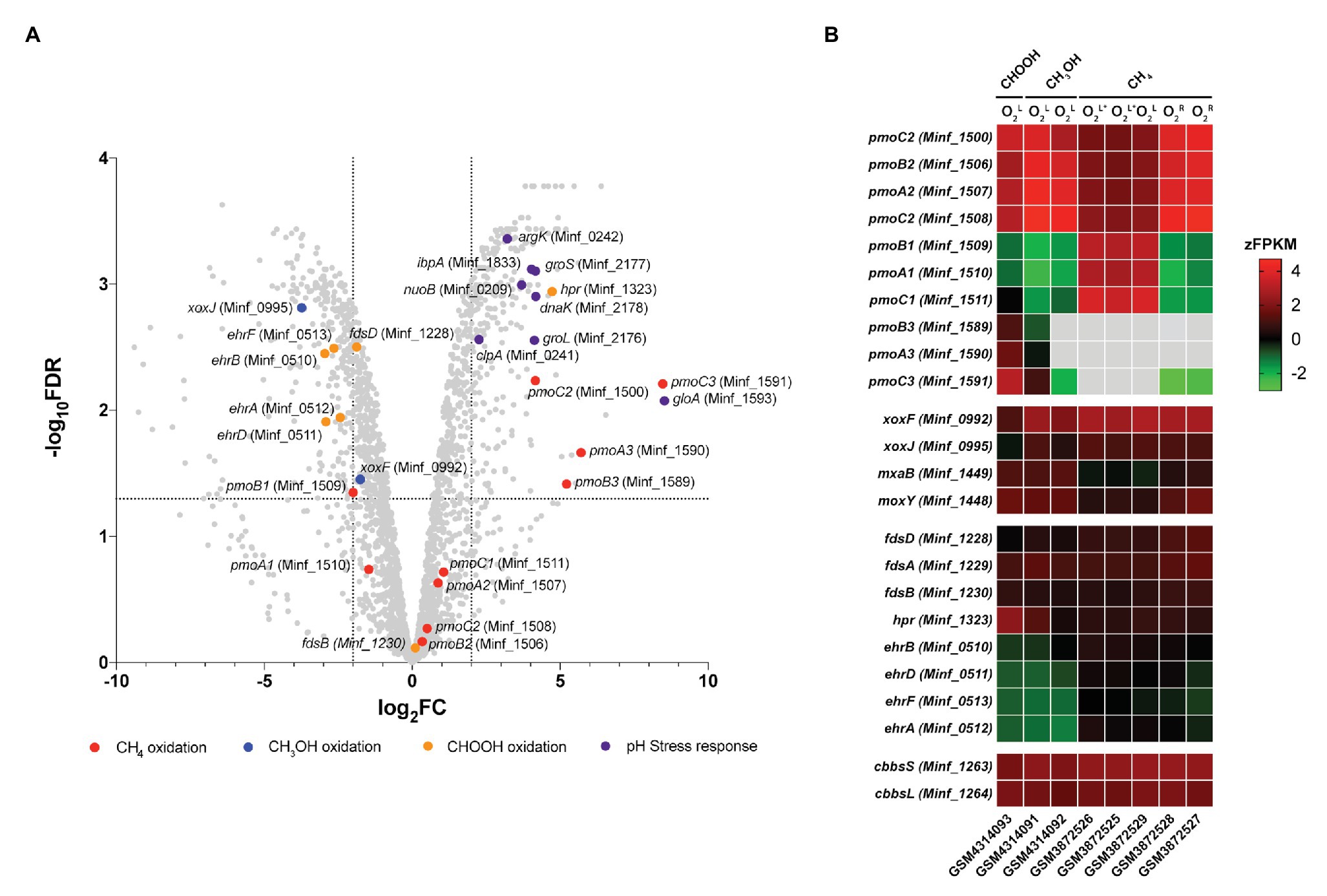
Figure 4. Differential gene expression profiles of chemostat-grown cultures of Methylacidiphilum sp. RTK17.1 grown on methanol (CH3OH) and formic acid (CHOOH). (A) Volcano plot showing differential gene expression changes following the transition from growth on methanol to growth on formic acid. Fold-change values (log2FC) and false discovery rates (FDR) are calculated using methylotrophic growth as the reference condition. Each gene is represented by a gray dot and genes of interest are highlighted as per the legend. Dashed horizontal and vertical lines signify FDR = 0.001 and log2FC = ∣2∣, respectively. (B) Heat map of transcript abundance for key genes encoding the structural subunits of enzymes participating in methane oxidation (pmoBAC; particulate methane monooxygenase), methanol oxidation (xoxFJ, mxaB, and moxY), formate oxidation (fdsDAB, hpr, and ehrBDAF), and carbon dioxide fixation (cbbsSL). Expression data is displayed as the z-normalization of fragment counts per kilobase million transcripts (zFPKM; Hart et al., 2013) for steady-state formic acid (CHOOH, GSM4314093), methanol (CH3OH, GSM4314092, GSM4314091), and methane (CH4, GSM3872525-GSM3872529) grown cultures under oxygen limited (O2L), oxygen, and ammonia limited (O2L∗) and oxygen replete (O2R) condition. Gray boxes denote expression values where zFPKM < −3.
The genomes of Methylacidiphilum spp. are known to encode up to three paralogous pMMO operons (pmoCAB; in addition to a fourth orphaned pmoC) that are phylogenetically divergent from one another by up to 50% amino acid identity (Op den Camp et al., 2009). This includes three pmoCAB operons in Methylacidiphilum spp. Kam1, SolV, V4, Phi, Fur, FdI, and RTK17.1, and only two complete copies of pmoCAB in strains Rib and Ice. Only one complete pmoCAB cluster has been identified in strain Yel, along with two truncated pmoAB and pmoCA clusters (Erikstad et al., 2019). Multiple pMMOs, with differing apparent Km values, are also characteristic of the type II methanotrophs (Baani and Liesack, 2008), and it has been proposed that this multiplicity provides survival advantages in niches where substrate concentrations may be variable. Likewise, in Methylacidiphilum spp. Kam1, SolV and V4, it has been shown that each of the pmoCAB operons are highly conserved and under intense purifying selection, further suggesting that they have evolved to fulfill distinct physiological roles (Kruse et al., 2019). Accordingly, the differential regulation of the pmoCAB1 and pmoCAB2 operons in Methylacidiphilum sp. RTK17.1 (Carere et al., 2019) and M. fumariolicum SolV (Khadem et al., 2012a) has previously been demonstrated in response to oxygen availability. Recently, expression of the pmoCAB3 operon was reported during the growth of M. fumariolicum SolV on methanol/ethane and methanol/propane (Picone et al., 2020). This led authors to suggest that pmoCAB3 expression was linked to the absence of methane or the presence of methanol and to note that expression levels increased when propane was supplied.
In this study, growth of Methylacidiphilum sp. RTK17.1 on methanol was accompanied by high levels of pmoCAB2 (Minf_1506–1508) expression. Considering that methylotrophic growth was oxygen limited, this result is consistent with our previous work (Carere et al., 2019). Congruently, the closely related, and proximally located, pmoCAB1 operon (homologous to M. infernorum V4 loci Minf_1509–1511) that is highly expressed during oxygen-replete growth on methane was only weakly transcribed. As observed in M. fumariolicum SolV (Picone et al., 2020), methylotrophic growth of Methylacidiphilum sp. RTK17.1 was accompanied by very weak expression of the pmoCAB3 operon (average FPKM = 331; Supplementary Table S1). Surprising, however, was the observation that formic acid-dependent growth strongly stimulated expression of the pmoCAB3 operon. When growing on formic acid, subunits of the particulate methane monooxygenase operon (pmoCAB3), corresponding to M. infernorum V4 loci Minf_1589–1591, were highly expressed (average FPKM = 18,422) and displayed the greatest degree of transcriptional upregulation (average: 6.5 Log2FC, p < 0.009) observed (Figure 4B; Supplementary Table S1). Although the physiological role of the pmoCAB3 operon remains unknown, these collective data suggest that it may encode a high-affinity methane monooxygenase that exhibits some promiscuity toward other alkanes. Given the challenges associated with the heterologous expression of pMMO (Chan et al., 2011), future biochemical and biophysical studies aiming to characterize this divergent pmoCAB3 operon may benefit from using a similar chemostat cultivation-dependent approach to enrich for its expression.
Transcripts for each of the genes required for the complete oxidation of CH4, carbon fixation, and glycogen synthesis were detected (Supplementary Table S1), thereby indicating that Methylacidiphilum sp. RTK17.1 remains primed for methanotrophic growth. Nevertheless, in response to formic acid-dependent growth, genes encoding the lanthanide-dependent methanol dehydrogenase, xoxF (Minf_0992; −1.72 Log2FC, p = 0.014) and xoxJ (Minf_0995; −3.69 Log2FC, p < 0.001), were down regulated. Transcriptional regulation of these XoxF-type methanol dehydrogenase genes has previously been reported in response to lanthanide availability in the methanotroph Methylorubrum extorquens AM1 (Good et al., 2019). Conversely, the NAD+-dependent formate dehydrogenase encoded by Minf_1323 (hpr) was upregulated (4.72 Log2FC, p < 0.001) and highly transcribed (FPKM: 7408) in response to growth on formic acid. The other putative formate dehydrogenases, including a hetero-multimeric Mo-containing formate dehydrogenase (Minf_1228–1231; fdsBAD) and a formate hydrogen lyase (ehrBDAF) were only weakly transcribed under all growth conditions (Supplementary Table S1). The presence of multiple formate dehydrogenases within methanotroph genomes is not unusual (Chistoserdova et al., 2004, 2007; Crowther et al., 2008); however little is known about the physiological role these enzymes may play in acidophilic methanotrophs. Differential expression of FDH genes in response to molybdenum or tungsten availability, in the facultative methylotroph Methylobacterium extorquens AM1, suggests that the presence of multiple FDHs may provide a means for increased ecological fitness (Chistoserdova et al., 2004).
Transcriptome analysis also revealed that several genes commonly associated with bacterial stress response were differentially expressed during growth on formic acid (Figure 4A). Methylacidiphilum sp. RTK17.1 upregulated the expression of molecular chaperone heat-shock proteins groES (Minf_2177; 4.16 Log2FC, p < 0.001), groEL (Minf_2176; 4.12 Log2FC, p < 0.001), dnaK (Minf_2178; 4.17 Log2FC, p < 0.001), and ipbA (Minf_1833; 4.03 Log2FC, p < 0.001) during growth on formic acid (Figure 4B). These proteins encode key factors that prevent the misfolding and aggregation of ribosome-bound polypeptides and have shown upregulation in response to acid (Len et al., 2004), oxidative and heat stress (Kitagawa et al., 2002). Likewise, homologs to the stress response protein-arginine kinase (mcsB; Minf_0242) and clpA unfoldase (Minf_0241) of Bacillus subtilis (Elsholz et al., 2011) were upregulated 3.21‐ and 2.26-fold, respectively, (p < 0.001). Interestingly, the most highly upregulated gene in response to growth on formic acid (8.52 Log2FC, p = 0.002) putatively encodes a methylmalonyl-CoA epimerase (Minf_1593, gloA). This enzyme is involved in the metabolism of propionate, branched-chain amino acids, odd-chain fatty acids, and the reversible conversion of (S)-methylmalonyl-CoA to succinyl-CoA. Regulation of gloA has previously been implicated in acid stress response in Propionibacterium acidipropionici (Guan et al., 2014); however, its role in Methylacidiphilum sp. RTK17.1 is not yet clear. One possibility is that the gloA transcription is linked to the proximate pmoCAB3 operon (Minf_1589-1591) that is strongly upregulated. Finally, although the use of transporters to catalyze active proton or cation transport is a common strategy for bacterial pH homeostasis (Slonczewski et al., 2009), transcriptional regulation is not always observed. In support of this, constitutive expression of genes encoding a K+ translocating ATPase (Minf_0033-0035; kpdCBA) was observed under all growth conditions. As observed in other acidophiles (Baker-Austin and Dopson, 2007), it seems likely Methylacidiphilum sp. RTK17.1 actively imports K+ (electrogenic uptake) as a strategy to generate its reversed membrane potential, thereby minimizing the inward flux of H+ and facilitating pH homeostasis.
Our findings show that the proton motive force (PMF) for this species is primarily generated by a pH gradient across the cellular membrane. In batch experiments, the addition of formic acid resulted in no observable cell growth and, correspondingly, acidification of the cytosol. Nevertheless, stable growth on formic acid as the sole source of metabolizable energy was demonstrated in continuous cultures following the transition from methanol-dependent growth. Under these conditions, biomass yields on formic acid were nearly equivalent on a per mole electron basis to methanol-grown cells. The transition to growth on formic acid, however, coincided with transcriptional upregulation of several genes associated with an acid-stress response. These results therefore highlight the advantages of using chemostats to complement batch-culture experiments for the physiological characterization of microbial species. This has been well demonstrated with Methylacidiphilum spp. where otherwise cryptic phenotypes, such as lithoautotrophic and mixotrophic growth on H2 (Mohammadi et al., 2016; Carere et al., 2017, 2019), and ammonia oxidation (Mohammadi et al., 2017) were identified via genome analysis but were not demonstrable in batch-culture experiments (Hou et al., 2008; Op den Camp et al., 2009; Khadem et al., 2012c). It is likely that further cryptic phenotypes can be discovered using chemostat-based setups, not only in acidophilic methanotrophic strains but also for other microbial strains (Tamburini and Mastromei, 2000). Finally, these results also show that low concentrations of formic acid represent a utilizable source of energy/carbon to the acidophilic methanotrophs that are commonly found within geothermal environments and adds to previous research (Mohammadi et al., 2016; Carere et al., 2017, 2019) showing that metabolic flexibility in aerobic methane-oxidizing bacteria (methanotrophs) likely enhances cell growth and survival in environments where methane resources are variable or limiting.
Conclusion
Very little is known about the pH homeostatic mechanisms used by acidophiles to accommodate the organic acids present within acidic environments. In this study, we have shown that formic acid represents a utilizable source of energy/carbon to the thermoacidophilic methanotroph, Methylacidiphilum sp. RTK17 at pH 2.5. During sustained growth, in response to the influx of formic acid across the cell membrane, Methylacidiphilum sp. RTK17.1 alters expression of key genes relating to energy/carbon metabolism and bacterial stress response. Findings reported in this study expand the known metabolic flexibility of verrucomicrobial methanotrophs to include organic acids while also highlighting the potential advantages of chemostat-culture experiments to characterize the physiology of acidophilic species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CC and MS conceived the study. CC, KHa, MS, and GC contributed to the experimental design. CC, LC, and KHo conducted the bioreactor and wet lab experiments. CC and KW performed the transcriptome analysis. CC, KHa, and GC performed the bioenergetic analysis. CC, KW, and MS wrote the manuscript with input from KHo, KHa, and GC. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by The Royal Society of New Zealand (Marsden grants to CC and KHa). CC and MS were also supported by an MBIE Smart Ideas grant (C05X1710). Work performed by KHo was supported by the MBIE Strategic Science Investment Funds granted to GNS Science.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Ngāti Tahu–Ngāti Whaoa is acknowledged as the iwi having mana whenua (customary rights) over the Rotokawa geothermal field, Methylacidiphilum sp. RTK17.1, and associated microorganisms. We thank Ngāti Tahu–Ngāti Whaoa Runanga Trust and Tauhara North No. 2 Trust for their support of our research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.651744/full#supplementary-material
References
Alexander, B., Leach, S., and Ingledew, W. J. (1987). The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the Acidophile Thiobacillus Ferrooxidans. Microbiology 133, 1171–1179. doi: 10.1099/00221287-133-5-1171
Altaş, N., Aslan, A. S., Karataş, E., Chronopoulou, E., Labrou, N. E., and Binay, B. (2017). Heterologous production of extreme alkaline thermostable NAD+-dependent formate dehydrogenase with wide-range pH activity from Myceliophthora thermophila. Process Biochem. 61, 110–118. doi: 10.1016/j.procbio.2017.06.017
Angeles, O. R., Johnson, S. E., and Buresh, R. J. (2006). Soil solution sampling for organic acids in rice paddy soils. Soil Sci. Soc. Am. J. 70, 48–56. doi: 10.2136/sssaj2005.0070
Axley, M. J., Grahame, D. A., and Stadtman, T. C. (1990). Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J. Biol. Chem. 265, 18213–18218. doi: 10.1016/S0021-9258(17)44740-5
Baani, M., and Liesack, W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. PNAS 105, 10203–10208. doi: 10.1073/pnas.0702643105
Baker-Austin, C., and Dopson, M. (2007). Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15, 165–171. doi: 10.1016/j.tim.2007.02.005
Bowman, J. P. (2014). “The family methylococcaceae” in The Prokaryotes: Gammaproteobacteria. eds. E. Rosenberg, E. F. Delong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 411–440.
Carere, C. R., Hards, K., Houghton, K. M., Power, J. F., Mcdonald, B., Collet, C., et al. (2017). Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J. 11, 2599–2610. doi: 10.1038/ismej.2017.112
Carere, C. R., Mcdonald, B., Peach, H. A., Greening, C., Gapes, D. J., Collet, C., et al. (2019). Hydrogen oxidation influences glycogen accumulation in a verrucomicrobial methanotroph. Front. Microbiol. 10:1873. doi: 10.3389/fmicb.2019.01873
Chan, S. I., Nguyen, H. H. T., Chen, K. H. C., and Yu, S. S. F. (2011). Overexpression and purification of the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Methods Enzymol. 495, 177–193. doi: 10.1016/B978-0-12-386905-0.00012-7
Chen, Y. P., and Yoch, D. C. (1987). Regulation of two nickel-requiring (inducible and constitutive) hydrogenases and their coupling to nitrogenase in Methylosinus trichosporium OB3b. J. Bacteriol. 169, 4778–4783. doi: 10.1128/JB.169.10.4778-4783.1987
Chistoserdova, L. (2015). Methylotrophs in natural habitats: current insights through metagenomics. Appl. Microbiol. Biotechnol. 99, 5763–5779. doi: 10.1007/s00253-015-6713-z
Chistoserdova, L., Crowther, G. J., Vorholt, J. A., Skovran, E., Portais, J.-C., and Lidstrom, M. E. (2007). Identification of a fourth Formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role offFormate oxidation in methylotrophy. J. Bacteriol. 189, 9076–9081. doi: 10.1128/JB.01229-07
Chistoserdova, L., Laukel, M., Portais, J.-C., Vorholt, J. A., and Lidstrom, M. E. (2004). Multiple Formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 186, 22–28. doi: 10.1128/JB.186.1.22-28.2004
Ciaramella, M., Napoli, A., and Rossi, M. (2005). Another extreme genome: how to live at pH 0. Trends Microbiol. 13, 49–51. doi: 10.1016/j.tim.2004.12.001
Cook, G. M., Russell, J. B., Reichert, A., and Wiegel, J. (1996). The intracellular pH of Clostridium paradoxum, an anaerobic, alkaliphilic, and thermophilic bacterium. Appl. Environ. Microbiol. 62, 4576–4579. doi: 10.1128/AEM.62.12.4576-4579.1996
Crombie, A. T., and Murrell, J. C. (2014). Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature 510, 148–151. doi: 10.1038/nature13192
Crowther, G. J., Kosály, G., and Lidstrom, M. E. (2008). Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 190, 5057–5062. doi: 10.1128/JB.00228-08
Dedysh, S. N., and Dunfield, P. F. (2010). “Facultative methane oxidizers” in Handbook of hydrocarbon and lipid microbiology. ed. K. N. Timmis (Cham: Springer), 1967–1976.
Dedysh, S. N., Knief, C., and Dunfield, P. F. (2005). Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670. doi: 10.1128/JB.187.13.4665-4670.2005
Dunfield, P. F., Yuryev, A., Senin, P., Smirnova, A. V., Stott, M. B., and Hou, S. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum verrucomicrobia. Nature 450, 879–882. doi: 10.1038/nature06411
Elsholz, A. K. W., Hempel, K., Michalik, S., Gronau, K., Becher, D., Hecker, M., et al. (2011). Activity control of the ClpC adaptor McsB in Bacillus subtilis. J. Bacteriol. 193, 3887–3893. doi: 10.1128/JB.00079-11
Erikstad, H.-A., Ceballos, R. M., Smestad, N. B., and Birkeland, N.-K. (2019). Global biogeographic distribution patterns of thermoacidophilic verrucomicrobia methanotrophs suggest allopatric evolution. Front. Microbiol. 10:1129. doi: 10.3389/fmicb.2019.01129
Ferry, J. G. (1990). Formate dehydrogenase. FEMS Microbiol. Rev. 7, 377–382. doi: 10.1111/j.1574-6968.1990.tb04940.x
Good, N. M., Moore, R. S., Suriano, C. J., and Martinez-Gomez, N. C. (2019). Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci. Rep. 9:4248. doi: 10.1038/s41598-019-41043-1
Goulbourne, E. Jr., Matin, M., Zychlinsky, E., and Matin, A. (1986). Mechanism of delta pH maintenance in active and inactive cells of an obligately acidophilic bacterium. J. Bacteriol. 166, 59–65. doi: 10.1128/JB.166.1.59-65.1986
Greening, C., Biswas, A., Carere, C. R., Jackson, C. J., Taylor, M. C., and Stott, M. B. (2016). Genome and metagenome surveys of hydrogenase diversity indicate H2 is a widely-utilised energy source for microbial growth and survival. ISME J. 10, 761–777. doi: 10.1038/ismej.2015.153
Guan, N., Shin, H.-D., Chen, R. R., Li, J., Liu, L., Du, G., et al. (2014). Understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the level of proteomics. Sci. Rep. 4:6951. doi: 10.1038/srep06951
Hakobyan, A., Zhu, J., Glatter, T., Paczia, N., and Liesack, W. (2020). Hydrogen utilization by Methylocystis sp. strain SC2 expands the known metabolic versatility of type IIa methanotrophs. Metab. Eng. 61, 181–196. doi: 10.1016/j.ymben.2020.05.003
Hanczar, T., Csaki, R., Bodrossy, L., Murrell, C. J., and Kovacs, K. L. (2002). Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch. Microbiol. 177, 167–172. doi: 10.1007/s00203-001-0372-4
Hanson, R. S., and Hanson, T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471. doi: 10.1128/MR.60.2.439-471.1996
Hart, T., Komori, H. K., Lamere, S., Podshivalova, K., and Salomon, D. R. (2013). Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14:778. doi: 10.1186/1471-2164-14-778
Hou, S., Makarova, K. S., Saw, J. H. W., Senin, P., Ly, B. V., and Zhou, Z. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum verrucomicrobia. Biol. Direct 3:26. doi: 10.1186/1745-6150-3-26
Islam, T., Jensen, S., Reigstad, L. J., Larsen, O., and Birkeland, N. K. (2008). Methane oxidation at 55 C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. PNAS 105, 300–304. doi: 10.1073/pnas.0704162105
Kalyuzhnaya, M. G., Puri, A. W., and Lidstrom, M. E. (2015). Metabolic engineering in methanotrophic bacteria. Metab. Eng. 29, 142–152. doi: 10.1016/j.ymben.2015.03.010
Kelly, D. P., Anthony, C., and Murrell, J. C. (2005). Insights into the obligate methanotroph Methylococcus capsulatus. Trends Microbiol. 13, 195–198. doi: 10.1016/j.tim.2005.03.003
Keltjens, J. T., Pol, A., Reimann, J., and Op den Camp, H. J. M. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98, 6163–6183. doi: 10.1007/s00253-014-5766-8
Kemp, M., and Quayle, J. (1967). Microbial growth on C1 compounds. Uptake of [14C]formaldehyde and [14C]formate by methane-grown Pseudomonas methanica and determination of the hexose labelling pattern after brief incubation with [14C]methanol. Biochem. J. 102, 94–102. doi: 10.1042/bj1020094
Khadem, A. F., Pol, A., Wieczorek, A. S., Jetten, M. S. M., and Op den Camp, H. J. M. (2012a). Metabolic regulation of “Ca. Methylacidiphilum Fumariolicum” SolV cells grown under different nitrogen and oxygen limitations. Front. Microbiol. 3:266. doi: 10.3389/fmicb.2012.00266
Khadem, A. F., Pol, A., Wieczorek, A., Mohammadi, S. S., Francoijs, K. J., and Stunnenberg, H. G. (2011). Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193, 4438–4446. doi: 10.1128/JB.00407-11
Khadem, A. F., van Teeseling, M. C. F., van Niftrik, L., Jetten, M. S. M., Op den Camp, H. J. M., and Pol, A. (2012b). Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph Ca. Methylacidiphilum fumariolicum SolV. Front. Microbiol. 3:345. doi: 10.3389/fmicb.2012.00345
Khadem, A. F., Wieczorek, A. S., Pol, A., Vuilleumier, S., Harhangi, H. R., and Dunfield, P. F. (2012c). Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J. Bacteriol. 194, 3729–3730. doi: 10.1128/JB.00501-12
Kishimoto, N., Inagaki, K., Sugio, T., and Tano, T. (1990). Growth inhibition of Acidiphilium species by organic acids contained in yeast extract. J. Ferment. Bioeng. 70, 7–10. doi: 10.1016/0922-338X(90)90021-N
Kitagawa, M., Miyakawa, M., Matsumura, Y., and Tsuchido, T. (2002). Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur. J. Biochem. 269, 2907–2917. doi: 10.1046/j.1432-1033.2002.02958.x
Knief, C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 6:1346. doi: 10.3389/fmicb.2015.01346
Knief, C., Lipski, A., and Dunfield, P. F. (2003). Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69, 6703–6714. doi: 10.1128/AEM.69.11.6703-6714.2003
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kruse, T., Ratnadevi, C. M., Erikstad, H.-A., and Birkeland, N.-K. (2019). Complete genome sequence analysis of the thermoacidophilic verrucomicrobial methanotroph “Candidatus Methylacidiphilum kamchatkense” strain Kam1 and comparison with its closest relatives. BMC Genomics 20:642. doi: 10.1186/s12864-019-5995-4
Küsel, K., Blöthe, M., Schulz, D., Reiche, M., and Drake, H. L. (2008). Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5, 1537–1549. doi: 10.5194/bg-5-1537-2008
Len, A., Harty, D., and Jacques, N. (2004). Stress-response proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150, 1339–1351. doi: 10.1099/mic.0.27008-0
Liu, C. L., and Mortenson, L. E. (1984). Formate dehydrogenase of Clostridium pasteurianum. J. Bacteriol. 159, 375–380. doi: 10.1128/JB.159.1.375-380.1984
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38, 1091–1125. doi: 10.1111/1574-6976.12076
Mohammadi, S., Pol, A., van Alen, T. A., Jetten, M. S. M., and Op den Camp, H. J. M. (2016). Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11, 945–958. doi: 10.1038/ismej.2016.171
Mohammadi, S. S., Pol, A., van Alen, T., Jetten, M. S. M., and Op den Camp, H. J. M. (2017). Ammonia oxidation and nitrite reduction in the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. Front. Microbiol. 8:1901. doi: 10.3389/fmicb.2017.01901
Mohammadi, S. S., Schmitz, R. A., Pol, A., Berben, T., Jetten, M. S. M., and Op den Camp, H. J. M. (2019). The acidophilic methanotroph Methylacidimicrobium tartarophylax 4AC grows as autotroph on H2 under microoxic conditions. Front. Microbiol. 10:2352. doi: 10.3389/fmicb.2019.02352
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., and Schouten, S. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Picone, N., Mohammadi, S. S., Waajen, A. C., van Alen, T. A., Jetten, M. S. M., Pol, A., et al. (2020). More than a methanotroph: a broader substrate spectrum for Methylacidiphilum fumariolicum SolV. Front. Microbiol. 11:604485. doi: 10.3389/fmicb.2020.604485
Pol, A., Barends, T. R. M., Dietl, A., Khadem, A. F., Eygensteyn, J., and Jetten, M. S. M. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. Rep. 16, 255–264. doi: 10.1111/1462-2920.12249
Pol, A., Heijmans, K., Harhangi, H. R., Tedesco, D., Jetten, M. S. M., and Op den Camp, H. J. M. (2007). Methanotrophy below pH1 by a new verrucomicrobia species. Nature 450, 874–878. doi: 10.1038/nature06222
Riebeling, V., Jungermann, K., and Thauer, R. K. (1975). The internal-alkaline pH gradient, sensitive to uncoupler and ATPase inhibitor, in growing Clostridium pasteurianum. Eur. J. Biochem. 55, 445–453. doi: 10.1111/j.1432-1033.1975.tb02181.x
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schauer, N. L., and Ferry, J. G. (1982). Properties of formate dehydrogenase in Methanobacterium formicicum. J. Bacteriol. 150, 1–7. doi: 10.1128/JB.150.1.1-7.1982
Shah, N. N., Hanna, M. L., Jackson, K. J., and Taylor, R. T. (1995). Batch cultivation of Methylosinus trichosporium OB3B: IV. Production of hydrogen-driven soluble or particulate methane monooxygenase activity. Biotechnol. Bioeng. 45, 229–238. doi: 10.1002/bit.260450307
Shishkina, V. N., and Trotsenko, Y. A. (1982). Multiple enzymic lesions in obligate methanotrophic bacteria. FEMS Microbiol. Lett. 13, 237–242. doi: 10.1111/j.1574-6968.1982.tb08264.x
Singh, B. K., Bardgett, R. D., Smith, P., and Reay, D. S. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. doi: 10.1038/nrmicro2439
Slonczewski, J. L., Fujisawa, M., Dopson, M., and Krulwich, T. A. (2009). Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 55, 1–317. doi: 10.1016/S0065-2911(09)05501-5
Smith, A. J., and Hoare, D. S. (1977). Specialist phototrophs, lithotrophs, and methylotrophs: a unity among a diversity of procaryotes? Bacteriol. Rev. 41, 419–448. doi: 10.1128/BR.41.2.419-448.1977
Strong, P. J., Kalyuzhnaya, M., Silverman, J., and Clarke, W. P. (2016). A methanotroph-based biorefinery: potential scenarios for generating multiple products from a single fermentation. Bioresour. Technol. 215, 314–323. doi: 10.1016/j.biortech.2016.04.099
Tamburini, E., and Mastromei, G. (2000). Do bacterial cryptic genes really exist? Res. Microbiol. 151, 179–182. doi: 10.1016/S0923-2508(00)00137-6
Tavormina, P. L., Ussler, W., Joye, S. B., Harrison, B. K., and Orphan, V. J. (2010). Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J. 4, 700–710. doi: 10.1038/ismej.2009.155
Trotsenko, Y. A., and Murrell, J. C. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229. doi: 10.1016/S0065-2164(07)00005-6
van de Vossenberg, J. L. C. M., Driessen, A. J. M., Zillig, W., and Konings, W. N. (1998). Bioenergetics and cytoplasmic membrane stability of the extremely acidophilic, thermophilic archaeon Picrophilus oshimae. Extremophiles 2, 67–74. doi: 10.1007/s007920050044
van Teeseling, M. C. F., Pol, A., Harhangi, H. R., van Der Zwart, S., Jetten, M. S. M., Op den Camp, H. J. M., et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. Nov. Appl. Environ. Microbiol. 80, 6782–6791. doi: 10.1128/AEM.01838-14
Vara, C., Paytuví-Gallart, A., Cuartero, Y., Le Dily, F., Garcia, F., Salvà-Castro, J., et al. (2019). Three-dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep. 28, 352–367.e359. doi: 10.1016/j.celrep.2019.06.037
Ward, N., Larsen, Ø., Sakwa, J., Bruseth, L., Khouri, H., Durkin, A. S., et al. (2004). Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. doi: 10.1371/journal.pbio.0020303
Whittenbury, R., Phillips, K. C., and Wilkinson, J. F. (1970). Enrichment, isolation and some properties of methane-utilizing bacteria. Microbiology 61, 205–218. doi: 10.1099/00221287-61-2-205
Wood, A. P., Aurikko, J. P., and Kelly, D. P. (2004). A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Rev. 28, 335–352. doi: 10.1016/j.femsre.2003.12.001
Yoch, D. C., Chen, Y. P., and Hardin, M. G. (1990). Formate dehydrogenase from the methane oxidizer Methylosinus trichosporium OB3b. J. Bacteriol. 172, 4456–4463. doi: 10.1128/JB.172.8.4456-4463.1990
Keywords: methanotroph, acidophile, pH homeostasis, Methylacidiphilum, formate, formic acid
Citation: Carere CR, Hards K, Wigley K, Carman L, Houghton KM, Cook GM and Stott MB (2021) Growth on Formic Acid Is Dependent on Intracellular pH Homeostasis for the Thermoacidophilic Methanotroph Methylacidiphilum sp. RTK17.1. Front. Microbiol. 12:651744. doi: 10.3389/fmicb.2021.651744
Edited by:
Marina G. Kalyuzhanaya, San Diego State University, United StatesReviewed by:
Svetlana N. Dedysh, Russian Academy of Sciences (RAS), RussiaHuub J. M. Op den Camp, Radboud University Nijmegen, Netherlands
Copyright © 2021 Carere, Hards, Wigley, Carman, Houghton, Cook and Stott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlo R. Carere, Y2FybG8uY2FyZXJlQGNhbnRlcmJ1cnkuYWMubno=
 Carlo R. Carere
Carlo R. Carere Kiel Hards
Kiel Hards Kathryn Wigley1
Kathryn Wigley1 Karen M. Houghton
Karen M. Houghton Matthew B. Stott
Matthew B. Stott