- 1State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Argonaute proteins (Agos) from thermophilic archaea are involved in several important processes, such as host defense and DNA replication. The catalytic mechanism of Ago from different microbes with great diversity and genome editing potential is attracting increasing attention. Here, we describe an Argonaute from hyperthermophilic Ferroglobus placidus (FpAgo), with a typical DNA-guided DNA endonuclease activity but adopted with only a short guide 15–20 nt length rather than a broad guide selectivity for reported Agos. FpAgo performed the precise cleavage of phosphodiester bonds between 10 and 11 nt on the target strand (counting from the guide strand) guided strictly by 5′-phosphorylated DNA at temperatures ranging from 75 to 99°C. The cleavage activity was regulated by the divalent cations Mn2+, Mg2+, Co2+, and Ni2+. In addition, FpAgo possesses guide/target mismatch tolerance in the seed region but is sensitive to mismatches in the 3′-guide region. Notably, the EMSA assay revealed that the FpAgo-guide-target ternary complex exhibited a stronger binding affinity for short 15 and 16 nt guide DNAs than longer guides. Moreover, we performed structural modeling analyses that implied the unique PAZ domain of FpAgo for 3′-guide recognition and binding to affect guide length specificity. This study broadens our understanding of thermophilic Agos and paves the way for their use in DNA manipulation.
Introduction
Argonaute proteins (Agos) are widely present in all domains of life. Eukaryotic Argonaute proteins (eAgos) are the best studied and have been demonstrated to be involved in RNA interference using single-stranded RNA (ssRNA) guides (Ketting, 2011; Schirle and MacRae, 2012; Swarts et al., 2014b; Hegge et al., 2018). Recent studies have revealed that prokaryotic Argonaute proteins (pAgos) participate in host defense against invasive genetic elements and function in completing DNA replication (Swarts et al., 2015; Jolly et al., 2020; Kuzmenko et al., 2020). Unlike eAgos, pAgos exhibit high diversity in catalytic and biological functions, from the guide/target nuclei acid type, binding/cleavage activity, to pathways involved (Hegge et al., 2018). Due to its programmable endonuclease activity, researchers are interested in mining new pAgos and extending pools for genome-editing applications.
To date, only a few pAgos derived from thermophilic archaea and mesophilic bacteria have been well characterized (Swarts et al., 2014a, 2015; Kaya et al., 2016; Zander et al., 2017; Cao et al., 2019; Chong et al., 2019; Hegge et al., 2019; Kuzmenko et al., 2019; Kim et al., 2020; Olina et al., 2020; Liu et al., 2021b). Some Agos can act as programmable endonucleases to cleave targets between nucleotides 10 and 11 using a divalent cation to catalyze the hydrolysis of the phosphodiester bond that lies across nucleotides 10 and 11 of the base-pairing guide (Swarts et al., 2015; Jolly et al., 2020; Kuzmenko et al., 2020). This guide-directed specific cleavage is similar to the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein (Cas) system (Marraffini and Sontheimer, 2010; Jinek et al., 2012; Taly et al., 2013; Barrangou and Marraffini, 2014; Zhang, 2019). Compared to its analogous CRISPR-Cas nucleases, the precise recognition capacity of Ago is mediated by guide-target pairing without requiring the presence of any specific motifs of the target sequence, which makes Ago target any DNA fragment of interest. Furthermore, short DNA or RNA oligonucleotides, usually at 21 nt, utilized by Agos as guide molecules, are much easier and cheaper to synthesize than longer RNA guides required for Cas nucleases. Due to the unparalleled simplicity, programmability, and specific cleavage activity, Agos hold powerful potential for genetic manipulation. An artificial restriction enzyme platform based on Pyrococcus furiosus Argonaute (PfAgo) was developed to recognize and cleave DNA sequences at virtually any arbitrary site (Enghiad and Zhao, 2017). Furthermore, thermophilic pAgos have been recently used as programmable tools for specific target detection and rare single nucleotide variant enrichment in molecular diagnosis applications (He et al., 2019; Xun et al., 2019; Liu et al., 2021a; Song et al., 2020).
Due to the increasing importance of Agos in nucleic acid manipulation, we set out to identify novel Agos that possess desired biochemical properties with high specificity. In this study, an Ago from the hyperthermophilic archaeon Ferroglobus placidus (FpAgo) was biochemically characterized. The results demonstrated that FpAgo functions as a DNA-guided DNA endonuclease and shows a specific preference for guide utilization on the 3′-terminal. These new enzyme properties would provide new catalytic applications in the manipulation of DNA.
Materials and Methods
Sequence Analysis
To discover new potential pAgos candidates, BLAST, based on PfAgo amino acid sequences, was performed in the NCBI database, and the sequences were selected for phylogenetic analysis using MEGA 7.0.
FpAgo Expression and Purification
The His-tagged (N-terminal) FpAgo gene with optimized codons was synthesized and cloned into the pET-28a (+) plasmid to yield the expression plasmid pET-28a (+)-FpAgo by Sangon Biotech (Shanghai, China). The expression plasmid was transformed into E. coli BL21 (DE3) host strain. FpAgo was overexpressed and purified as previously described (Chong et al., 2019). The expression of recombinant FpAgo was induced by 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) at 20°C for 20 h, and the resulting protein was purified using an Ni-NTA affinity column. The purity of FpAgo was determined using SDS-PAGE. The protein concentration was determined using the BCA method. The purified FpAgo was stored in 15% (v/v) glycerol at −80°C until further use.
Enzymatic Activity Assay
For the standard activity assays, a total of 400 nM FpAgo was mixed with synthetic 2,000 nM guides, and 800 nM targets in reaction buffer (15 mM Tris/HCl pH 8, 250 mM NaCl) supplemented with 0.5 mM Mn2+ and incubated for 15 min at 95°C, except when otherwise indicated. The optimal temperature for FpAgo activity was assessed at temperatures ranging from 65 to 99°C. The effects of divalent metal ions on FpAgo activity were measured in the presence of Ca2+, Co2+, Cu2+, Mg2+, Mn2+, Zn2+, Ni2+, or EDTA. The effects of NaCl concentration on FpAgo activity were investigated using various NaCl concentrations (50–2000 mM). The preference for the 5′- end nucleotide of guide DNA (gDNA) was tested by providing DNA guides containing a 5′-dC, dT, dA, or dG. The influence of 5′- modifications of gDNA on FpAgo activity was assessed by different chemical modifications on the 5′-terminal (-P, -OH, -NH2, -SH, -Biotin, -Cy3, -Cy5, -BHQ1, -ROX, -FAM, -VIC). The optimal gDNA length adopted by FpAgo was measured in the presence of 11–21 nt gDNA. The minimum length of the target required for FpAgo catalytic activity was determined in the presence of 14–60 nt target DNA. A single mismatch was introduced in the gDNA from positions 2–15, respectively, to test the guide-target mismatch tolerance. All cleavage experiments were performed in triplicates. Reactions were stopped by rapid cooling to 4°C, followed by the addition of a loading buffer (Sangon Biotech, Shanghai, China) at a 1:1 ratio (v/v). The samples were then resolved on a 16% denaturing polyacrylamide gel. The gels were stained with GelRed (Biotium, United States). The nucleic acids were visualized using Tanon 3500BR (Shanghai, China) and quantitatively analyzed using Quantity One software (Bio-Rad, United States). Two guides (FW-gDNA and RV-gDNA) were designed to target 100 bp regions (29% GC content) of the pUC19 plasmid for the plasmid cleavage assay. FpAgo (400 nM), gDNA (2,000 nM), plasmid (1 μg), and 0.5 mM Mn2+ was mixed in reaction buffer. Reaction mixtures were incubated for 15 min at 75, 85, and 95°C. Samples were mixed with 5× loading buffer (Generay, Shanghai, China) before being resolved on 1% agarose gels. All nucleic acids used in this study are listed in Supplementary Tables 1, 2.
Electrophoretic Mobility Shift Assay (EMSA)
FpAgo was incubated with ssDNA guides (400 nM FpAgo preloaded with 2,000 nM guides) in reaction buffer at 95°C for 15 min and then cooled to 25°C. After pre-incubation, 800 nM 45 nt FAM-labeled target DNA was added to a final 1:5:2 ratio (protein:guide:target) and incubated for 1 h at 37°C. Samples were then mixed with 10× loading buffer (50% [v/v] glycerol and 0.1% [w/v] bromophenol blue) and resolved by 8% native polyacrylamide gel electrophoresis (PAGE) with Tris-Borate-EDTA (TBE) buffer, at room temperature. Nucleic acids were visualized using a Tanon 3500BR microscope (Shanghai, China).
Site-Directed Mutagenesis
Site-directed mutagenesis of the DEDD catalytic tetrad of FpAgo to alanine was performed using a whole plasmid PCR method. The plasmid pET-28a (+)-FpAgo was used as the initial template for PCR amplification using PrimeSTAR Max DNA Polymerase (Takara, Dalian, China), and mutations were introduced using the corresponding primers listed in Supplementary Table 3. The PCR products were then treated with DpnI for an hour at 37°C, followed by direct transformation into competent E. coli BL21 (DE3) cells. The successful introduction of the desired mutants was confirmed by DNA sequencing (Genewiz, Inc., Suzhou, China).
Homology Modeling and Structural Analysis of PAZ Domain of FpAgo
The homology model of FpAgo was performed with RosettaCM (Song et al., 2013) using the binary structure of MjAgo with the 5′-P gDNA (PDB ID: 5G5T, identity 40.2%) as the template (Willkomm et al., 2017). A total of 1,000 models were built, and the best 10% of models by energy were identified and clustered. The PAZ domain alignments of FpAgo were analyzed using PyMol software.
Results
Sequence Analysis of FpAgo
To mine new functional resources in pAgos, we performed phylogenetic analysis by aligning the well-studied PfAgo amino acid sequences in the NCBI database. Phylogenetic analysis revealed that in addition to two well-characterized hyperthermophilic pAgos, MjAgo and MfAgo, FpAgo was most closely related to PfAgo (Figure 1A). Multiple sequence alignments showed that FpAgo has a conserved catalytic tetrad (DEDX) in the PIWI domain, indicating that FpAgo may be catalytically active (Figure 1B).
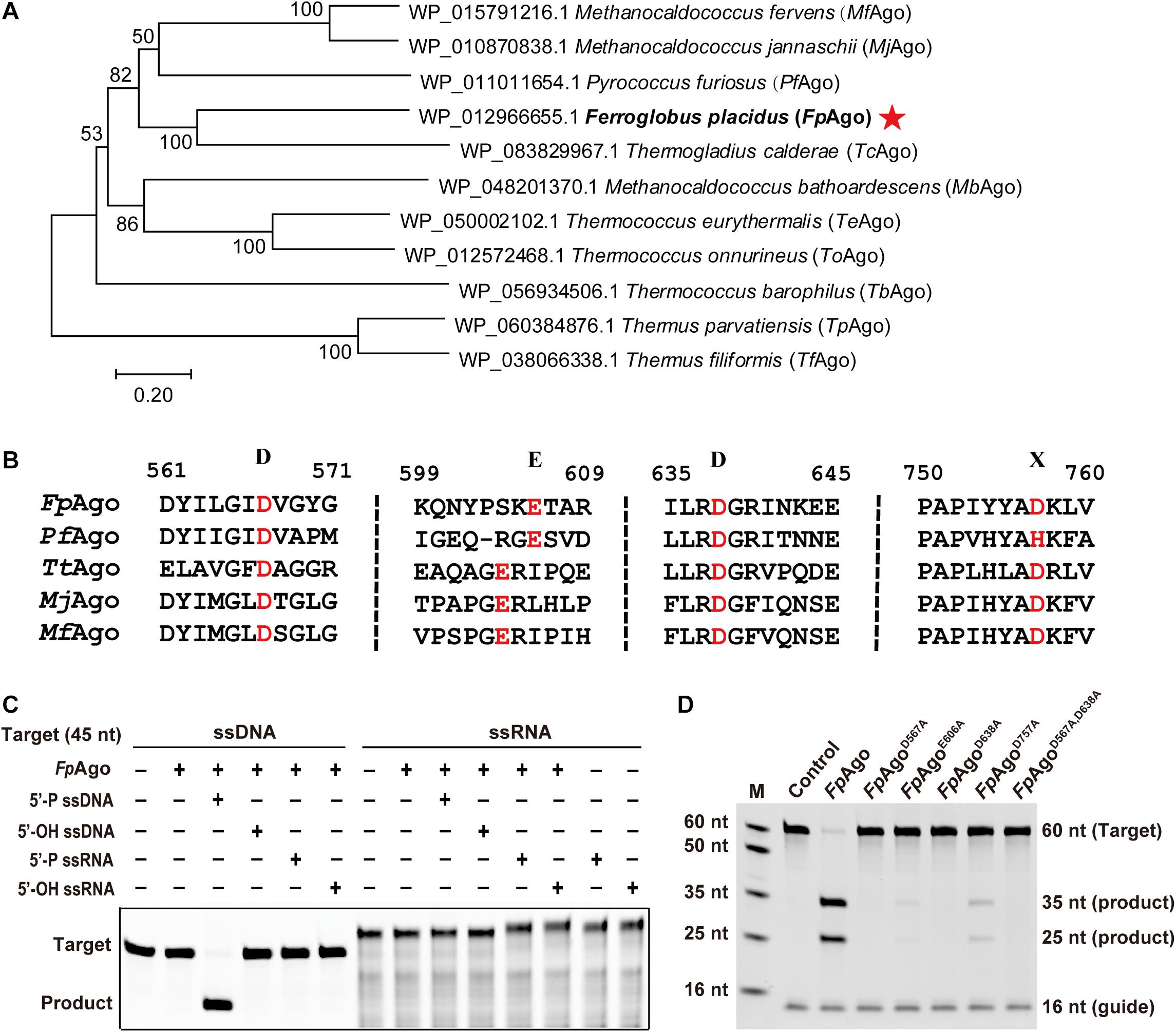
Figure 1. Phylogenetic analysis and cleavage activity assay of FpAgo. (A) Neighbor-joining phylogenetic tree analysis of FpAgo (marked by asterisks) based on amino acid sequences. The numbers at the nodes indicate the bootstrap values for neighbor-joining analysis of 1,000 resampled data sets. The scale bar represents the evolutionary distance between species. (B) Multiple sequence alignment of conserved amino acid residues (marked by red) of the DEDX tetrad localized in the PIWI domain of pAgos. (C) Guide and target preferences of FpAgo. Cleavage activity assays of FpAgo were performed with 45 nt FAM-labeled ssDNA or ssRNA target in reaction buffer containing 0.5 mM Mg2+ for 15 min at 95°C. (D) Cleavage activity of FpAgo and its DEDD mutants. M, ssDNA marker; nt, nucleotide. The “Control” sample contains no protein.
FpAgo Mediates DNA-Guided DNA Cleavage
The recombinant FpAgo and its mutants were heterologously expressed in E. coli BL21 (DE3), and the soluble fraction was purified and analyzed by SDS-PAGE. The purified protein showed a clear band, consistent with the predicted molecular weight of recombinant FpAgo (94 kDa) (Supplementary Figure 1). Next, we tested the purified recombinant FpAgo endonuclease activity using an in vitro cleavage assay. We used 16 nt RNA or DNA containing a 5′-P or 5′-OH group as guides to cleave complementary 45 nt FAM-labeled ssDNA or ssRNA as targets, respectively. The cleavage assays verified that FpAgo does not show DNA-guided cleavage of RNA targets and only uses 5′-P guides to cleave the DNA target (Figure 1C). To experimentally identify the residues involved in FpAgo activity, site-directed mutants were constructed. FpAgo mutants D567A and D638A completely abolished FpAgo activity, while E606A and D757A severely curtailed cleavage activity (Figure 1D), indicating that these four residues are the critical amino acid sites for FpAgo activity.
Effects of Temperature, Metal Ion, and NaCl on FpAgo Cleavage Activity
To investigate the biochemical properties of FpAgo, we designed a 16 nt 5′-P gDNA to cleave a 60 nt ssDNA target (Figure 2A). We observed that the cleavage site at the target was located precisely between positions 10 and 11 from the 5′-end of the guide DNA, identical to all previously reported pAgo proteins. Considering that divalent metal ions are an essential co-factor for Agos activity, we investigated which divalent metal ions could support FpAgo cleavage activity. We found that FpAgo can widely utilize Mn2+, Mg2+, Co2+, and Ni2+ as cations, with the preference of Mn2+ > Mg2+ > Co2+ > Ni2+ (Figure 2C and Supplementary Figure 2). Here, we also observed that a high Mn2+ concentration (>2 mM) could inhibit the cleavage activity of FpAgo, while FpAgo decreased the activity to Mg2+ strength above 7 mM (Supplementary Figure 2). FpAgo is sensitive to NaCl, with 250 mM as the optimal concentration, whereas a higher NaCl concentration reduced activity (Figure 2D), indicating that NaCl plays an important role in maintaining the catalytic activity and stability of FpAgo.
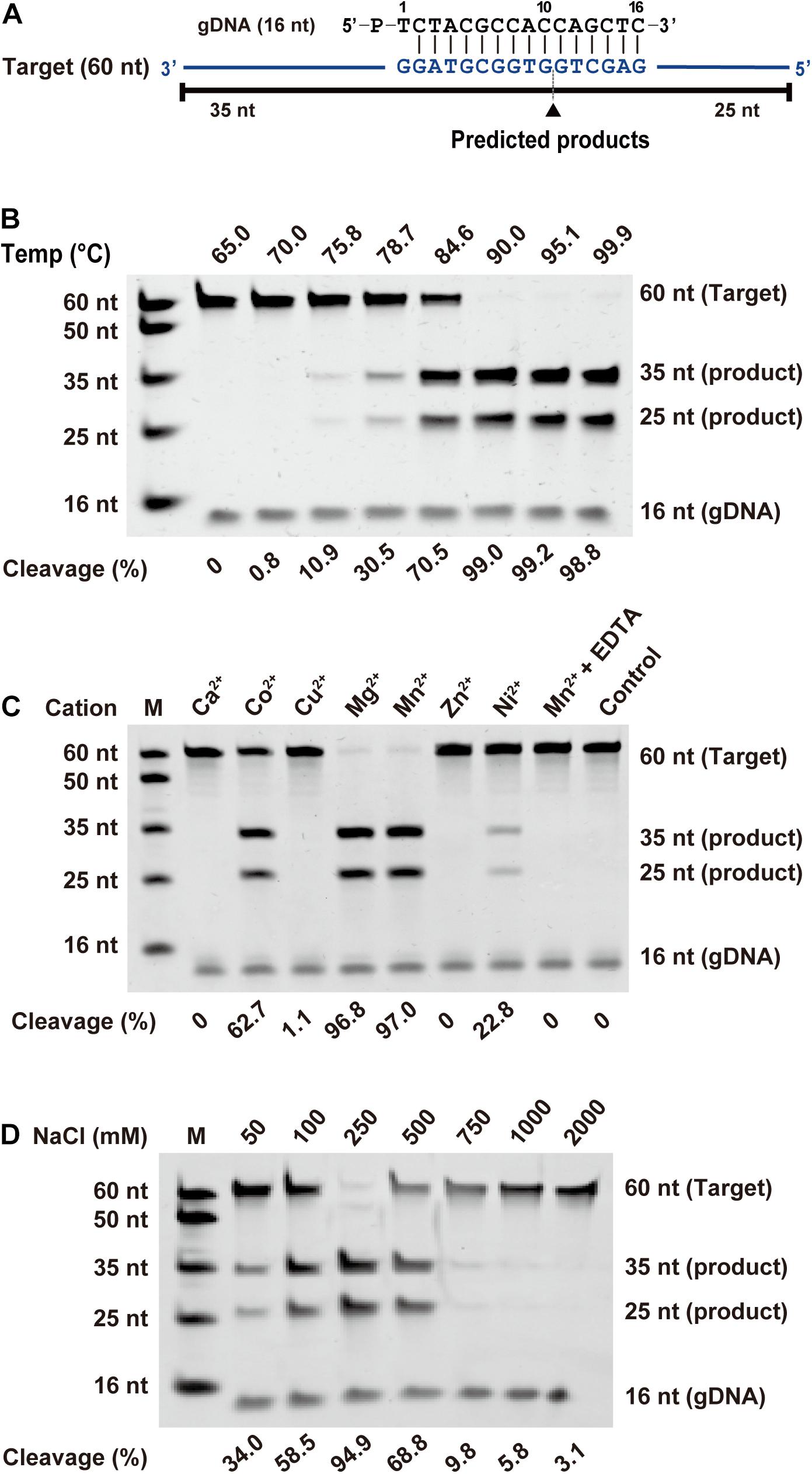
Figure 2. Effects of temperature, metal ion, and NaCl on FpAgo cleavage activity. (A) Synthetic ssDNA guide (black) and target (blue). Predicted cleavage positions are indicated with a black triangle, black lines indicate the predicted 25 and 35 nt cleavage products from 60 nt target. (B) The optimal temperature of FpAgo. (C) The divalent metal ions preferences of FpAgo. Substrate cleavage was performed in the presence of different divalent metal ions and EDTA with a final concentration of 0.5 mM. (D) Effect of NaCl concentrations on the enzyme activity of FpAgo. Cleavage experiments were carried out at the 1:5:2 molar ratio (400 nM FpAgo: 2000 nM guide: 800 nM target) in reaction buffer containing 0.5 mM Mn2+ for 15 min at 95°C.
Effects of 5′-End Nucleotide and Modification of gDNA on Cleavage Activity
As the preference for the 5′-end nucleotide of guides occurs in certain eAgos and pAgos (Lau et al., 2001; Frank et al., 2012; Willkomm et al., 2017), we tested whether FpAgo prefers a specific 5′-end nucleotide on the gDNA. According to our time course cleavage assays, FpAgo exhibited the highest initial reaction rate for 5′-dG at the 5′-end of the gDNA. Despite this preference, identical FpAgo cleavage activities were observed after incubation for 15 min with gDNAs containing a 5′-end dC, dT, dA, or dG (Figure 3A). Unlike MjAgo, which prefers a 5′-purine base of the gDNA (Willkomm et al., 2017), FpAgo exhibits a preference specific for 5′-dG but not 5′-dA.
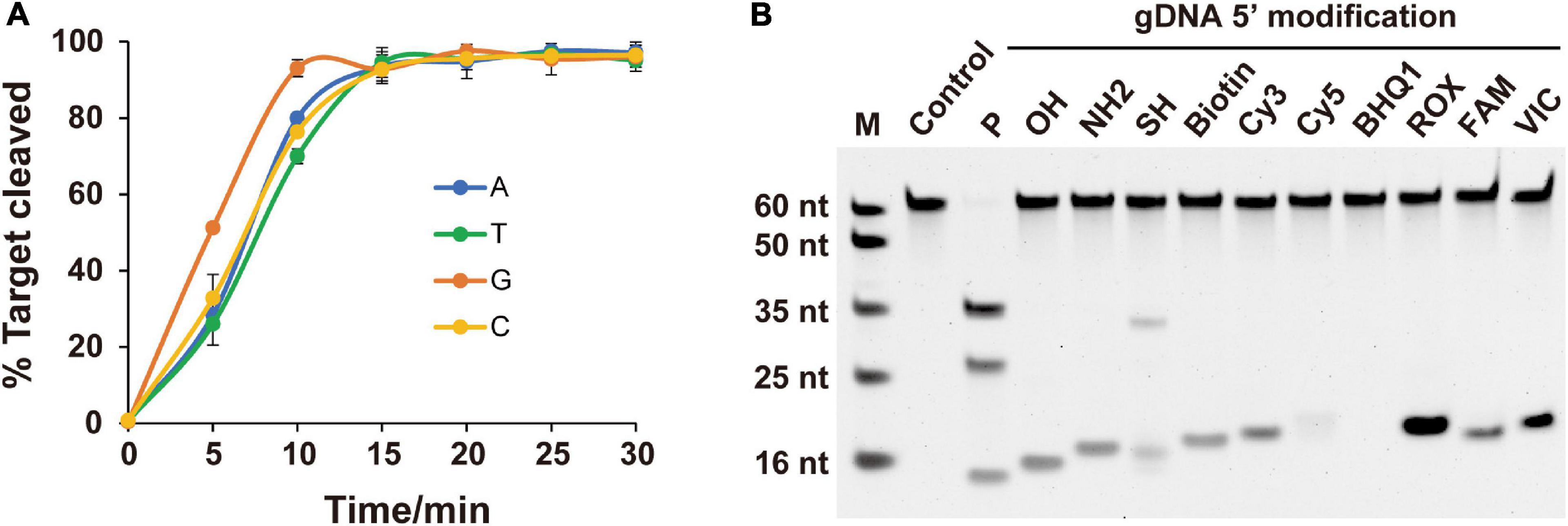
Figure 3. Effects of 5′-end nucleotides and modifications of gDNA on cleavage activity. (A) Cleavage kinetics of ssDNA target using 5′-P guided FpAgo. Error bars represent the standard deviation of three independent experiments. (B) Effects of 5′-end modification of gDNA on FpAgo cleavage activity.
For most reported Agos, cleavage activity is strictly dependent on the presence of a 5′-P on the guide strand, although few Agos can use both 5′-P and 5′-OH guides for target cleavage (Cao et al., 2019; Chong et al., 2019; Kuzmenko et al., 2019). To determine the dependence of the 5′-end nucleotide modification on FpAgo cleavage activity, we tested 11 different chemical modifications on the 5′-terminal of the guide DNA. The results demonstrated that 5′-P was required for FpAgo cleavage activity (Figure 3B). When FpAgo was loaded with 5′-SH gDNA, an uncanonical product band was observed on the gel. However, this brighter band likely resulted from the 5′-SH gDNA oxidation due to its instability under the tested conditions (Supplementary Figure 3).
Effects of Target and gDNA Length on Cleavage Activity
Previous structure-based mechanism studies on TtAgo have demonstrated that for sufficient guide-target pairing, at least 15 bp DNA target formation, is required for TtAgo switching from cleavage-incompatible to cleavage-compatible complexes (Sheng et al., 2014). In this study, the minimum guide length was studied. Strikingly, in contrast to previously reported eAgos and pAgos that function over a wide range of gDNA lengths between and 15–30 nt long, FpAgo efficiently cleaves targets only with short gDNA of 15 and 16 nt length (Figure 4A). The optimal gDNA length was specific to 15–16 nt, and a more extended guide significantly decreased the cleavage efficiency. EMSA showed that the FpAgo-guide-target ternary complex exhibited a stronger binding affinity for short 15 nt and 16 nt gDNAs (Figure 4B). In addition, we tested the target length to influence the cleavage. As expected, FpAgo cut target DNAs with a minimal size of 16 nt. No cleavage activity loss was observed when the target was extended to 60 nt (Figure 4C).
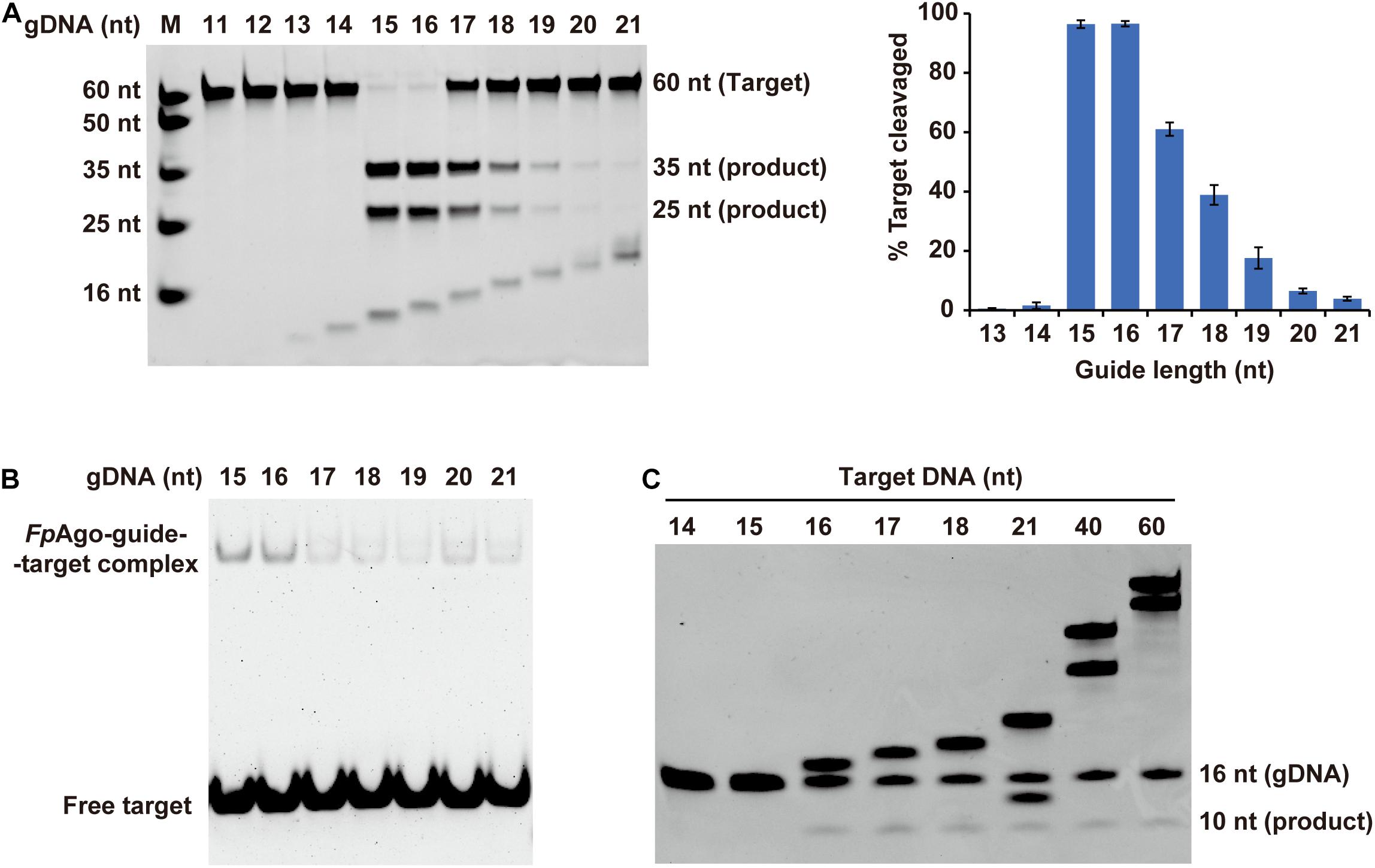
Figure 4. Effect of target and gDNA length on FpAgo cleavage activity. (A) The gDNAs of 11–21 nt in lengths were tested for cleavage efficiency in vitro. Histogram data were calculated from gels quantitatively analyzed by Quantity One software. Error bars represent the SD of three independent experiments. (B) The affinities of FpAgo form binary complexes with different length of 5′-P guide DNAs. (C) Minimum size of target used by FpAgo. ssDNA targets of 14–18, 21, 40, and 60 nt in lengths were tested for cleavage efficiency in vitro. All cleavage experiments were carried out at the 1:5:2 molar ratio (400 nM FpAgo: 2000 nM guide: 800 nM target) in reaction buffer containing 0.5 mM Mn2+ for 15 min at 95°C.
Effects of Mismatches on the Cleavage Activity of FpAgo
Agos use small ssRNA or ssDNA guides to identify ssRNA or ssDNA targets through Watson-Crick base-pairing interactions, and mismatches between gDNA and target sequences may have significant effects on target recognition and cleavage (Wang et al., 2008; Schirle and MacRae, 2012; De et al., 2013). Previous studies have demonstrated that a single mismatch in the seed region of TtAgo (positions 2–8) showed impaired cleavage activity (Wang et al., 2008). To investigate the effects of nucleotide mismatch in the guide-target duplex on FpAgo activity, we introduced a single nucleotide mismatch at positions 2–15 in the 16 nt guide strand for cleavage of a KRAS G12D ssDNA target. The KRAS WT sequence was used as a control, paired completely with the above guides (Figures 5A,B). We observed that FpAgo-mediated target cleavage was sensitive to single-base mismatches at positions 11–15 of gDNA. In contrast, mismatches in the seed region had little or no effect on the efficiency of target cleavage by FpAgo. Notably, we observed that certain gDNAs, such as gM3, gM12, gM3, and gM15, resulted in lower cleavage efficiency even for the KRAS WT target. Surprisingly, a single mismatch at position 7 led to higher cleavage activity than that without mismatch in the guide-target duplex (Figure 5C).
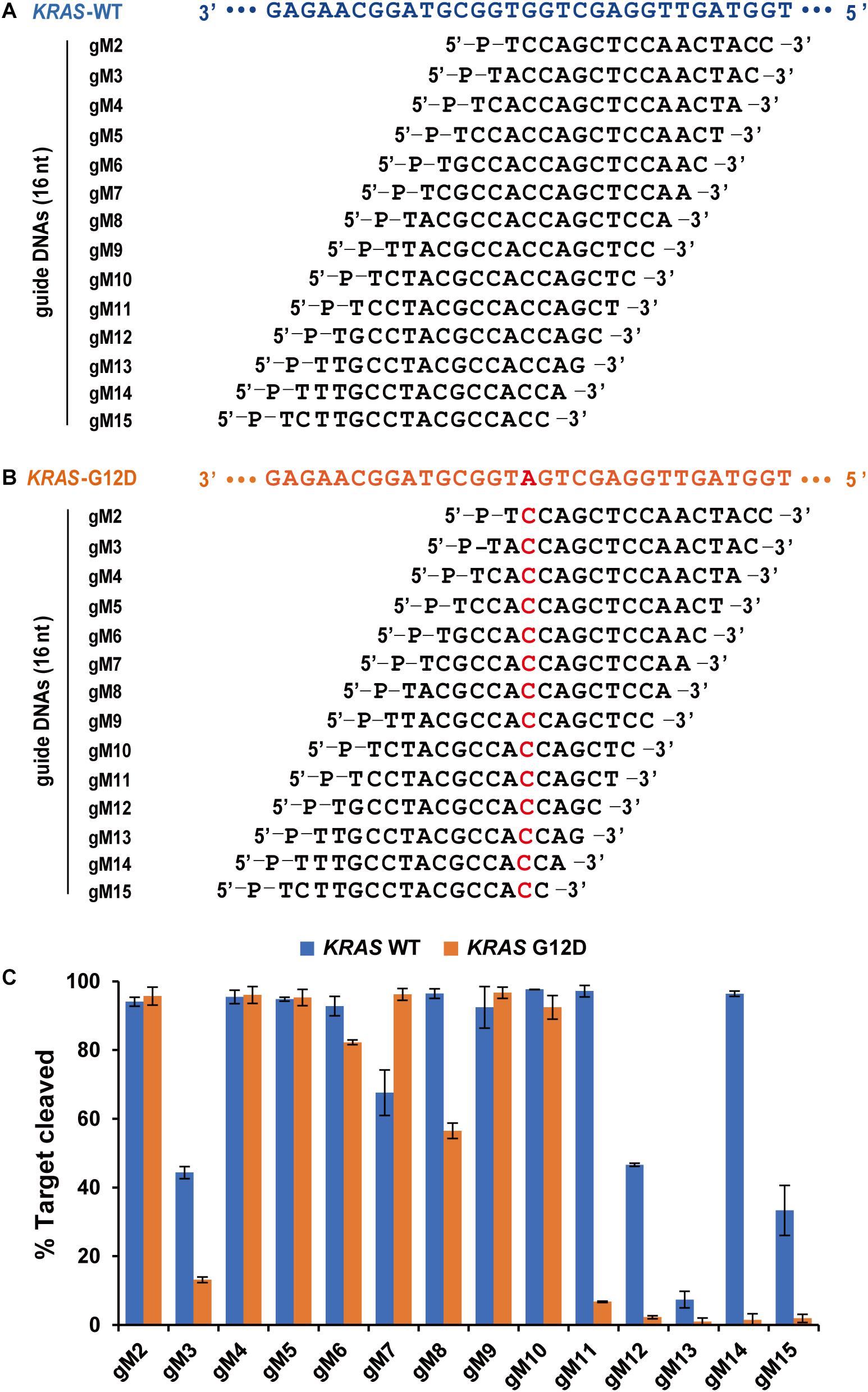
Figure 5. Effects of mismatches on the cleavage activity of FpAgo. Sequences of designed guides corresponding to KRAS WT target (A) and mutant target (B). All guides carry a base “T” as the first base based on the previous structural studies that 5′-end base of the guide strand does not participate in target pairing. (C) Cleavage efficiencies of WT KRAS DNA and KRAS G12D DNA mediated by guides. All cleavage experiments were carried out at the 1:5:2 molar ratio (400 nM FpAgo: 2,000 nM guide: 800 nM target) in reaction buffer containing 0.5 mM Mg2+ for 15 min at 95°C.
Homology Modeling and Structural Analysis of PAZ Domain of FpAgo
Based on the above results on specific guide binding properties, we suggest that the unique structural conformation may guide the selection of guide length and mismatch tolerance of FpAgo, especially for its interaction with the 3′-terminal of gDNA. Although we have tried to obtain the crystal structure, it is challenging to collect informatics data. Next, we performed homology modeling analysis to explain the structural details. Despite low primary sequence similarities, the overall fold of Agos is conserved in structure and consists of four canonical domains. Previous structural studies on Agos established that the 3′-end of the guide strand is anchored within the PAZ domain (Wang et al., 2008; Schirle and MacRae, 2012; De et al., 2013). Although FpAgo has high sequence similarity with PfAgo, FpAgo has an extra loop compared to PfAgo (Figure 6A). We further compared the PAZ domain between FpAgo and the MjAgo binary complex (MjAgo carries a guide DNA strand). Interestingly, the stick-out loop in FpAgo was vertically set in the middle of the guide 3′-end and PAZ pocket and likely narrowed the channel to bind the 3′-end of the guide (Figure 6B). We also performed sequence alignment analysis of FpAgo PAZ domains with other Agos adopting long guides for cleavage activity. The results showed significant residue divergence in FpAgo (Supplementary Figure 4), indicating that the FpAgo PAZ domain has an additional conformation compared with other Agos. Thus, the unique extra loop employed by FpAgo may explain the preference for FpAgo to use shorter guides because longer guides are easily interfered with by the loop. This loop may also contribute to the sensitivity to mismatch at the 3′-end of the gDNA.
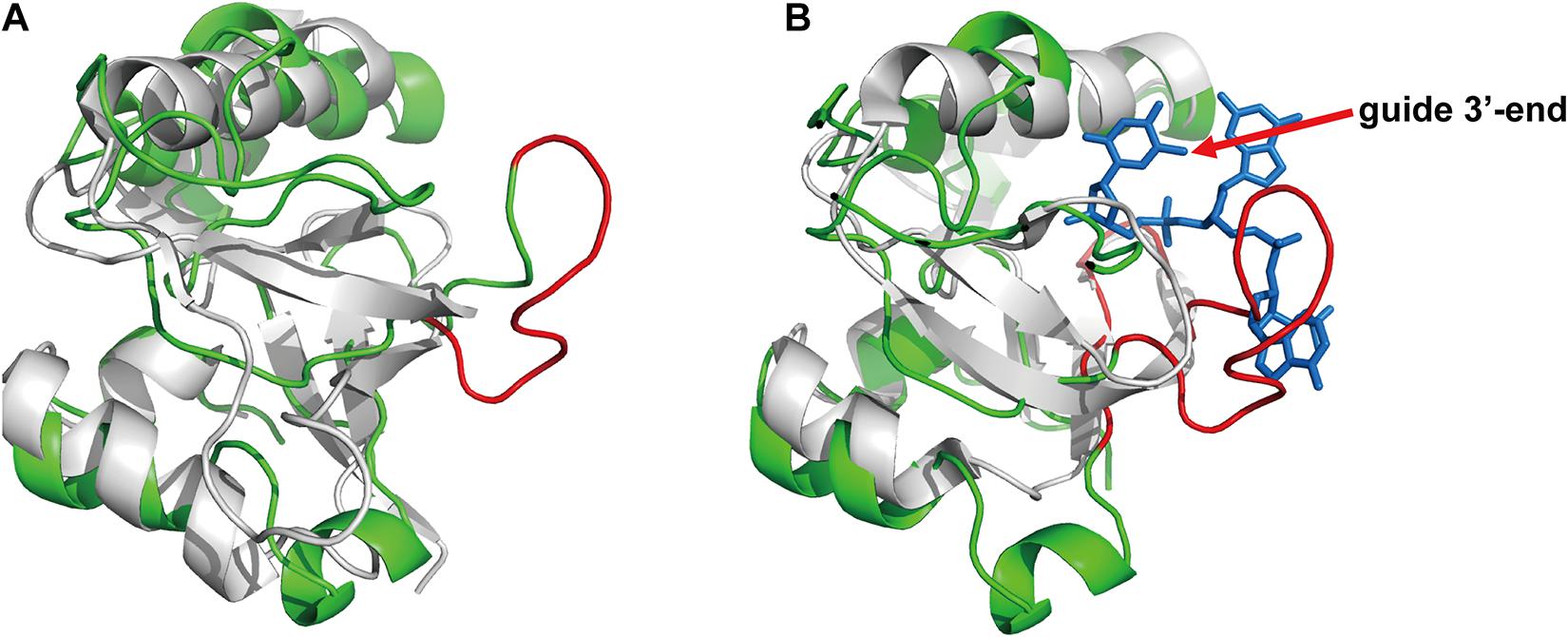
Figure 6. The PAZ domain comparison of the FpAgo with PfAgo and MjAgo via homology modeling. (A) The comparison of the FpAgo (green) with apo PfAgo (gray). (B) The comparison of the FpAgo (green) with MjAgo binary complex (gray) carrying a guide (blue). Only the last three nucleotides locating in the 3′-end of the guide strand were shown. A special loop employed by FpAgo is highlighted in red.
FpAgo Mediates Cleavage of Double-Stranded Plasmids
To test whether FpAgo cleaves plasmid DNA, FpAgo was incubated with its pUC19 plasmid in the absence or presence of guides. Like PfAgo, FpAgo could nick plasmids in the absence of gDNA at 75 and 85°C, whereas the guide-independent plasmid nicking activity could be suppressed at 95°C (Figure 7). Guide-independent processing activity has also been observed for CbAgo and KmAgo, and is suppressed at elevated temperatures (Kuzmenko et al., 2019; Liu et al., 2021b). When FpAgo was loaded with a single gDNA, FpAgo nicked or broke the plasmid DNA, generating open circular or linearized plasmids. The accumulation of linearized plasmids in the presence of RV-gDNAs but not FW-gDNAs may be because their efficiency is different, and the plasmid is unstable at elevated temperatures. When supplied with one pair of gDNAs, the efficiency of plasmid linearization was increased, in particular, by increasing the temperature of the reaction (Figure 7), which likely facilitated plasmid DNA melting and enhanced FpAgo activity at elevated temperatures. These results demonstrate that FpAgo could efficiently mediate DNA-guided cleavage of dsDNA plasmids if both strands of the plasmid DNA are targeted.
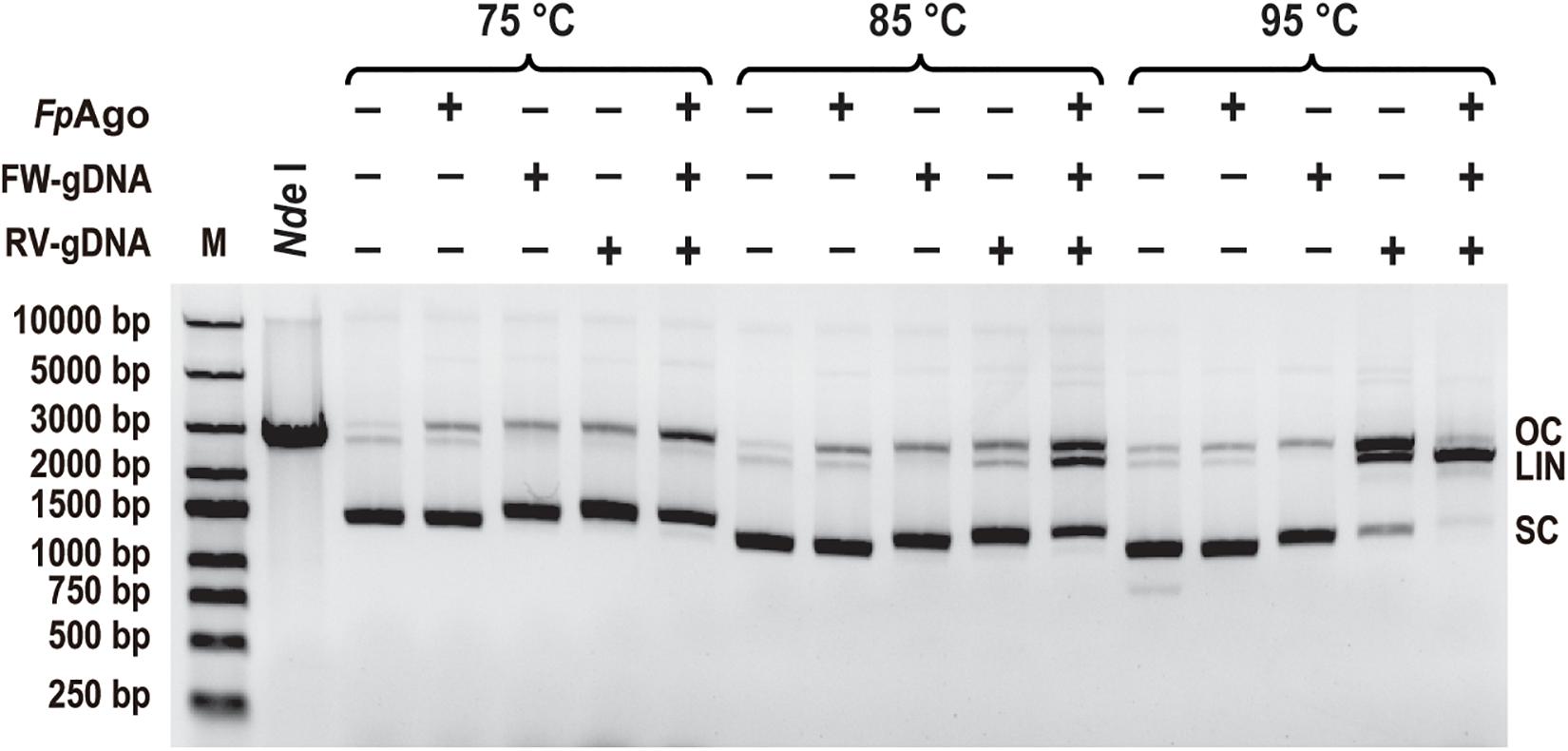
Figure 7. Plasmid cleavage by FpAgo. FpAgo-mediated plasmid cleavage after incubation for 15 min at different temperatures. NdeI lane: The extracted plasmid was digested with NdeI for 2 h, followed by 1% agarose gel electrophoresis. FW/RV-gDNA, forward and reverse guide DNA corresponded to a target site in plasmid pUC19; SC, supercoiled plasmid; LIN, linearized plasmid; OC, open circular plasmid.
Discussion
FpAgo Can Cleave Both ssDNA and dsDNA Target Directed by gDNA
Prokaryotic Argonaute proteins has been shown to have more functional and mechanistic diversity than eAgos (Swarts et al., 2014b; Hegge et al., 2018). Previous attempts have been made to discover pAgos with genome-editing capabilities, and some pAgos from mesophilic bacteria have been described. The cleavage of double-stranded DNA (dsDNA) mediated by mesophilic Agos is likely dependent on the local sequence context because the Agos lack DNA helicase activity (Hegge et al., 2018, 2019; Cao et al., 2019; Kuzmenko et al., 2019; Liu et al., 2021b). Therefore, increasing substrate accessibility when using mesophilic pAgos in biotechnology is regarded as a key challenge. The dsDNA cleavage by hyperthermophilic Agos is considered as two independent processes of ssDNA nicks since dsDNA denatures at elevated temperatures (Swarts et al., 2015). Here, we describe a novel FpAgo from the hyperthermophilic F. placidus. FpAgo functions as a DNA-guided DNA endonuclease that requires divalent metal ions such as Mn2+, Mg2+, Co2+, or Ni2+ for its activity. Although FpAgo prefers Mn2+ over Mg2+ for cleavage, similar to most Agos, FpAgo displays a higher tolerance for Mg2+ over Mn2+. Moreover, FpAgo efficiently cleaves double-stranded plasmids at elevated temperatures, demonstrating its potential application in dsDNA manipulation. To best of knowledge, specific DNA helicase can enhance thermophilic TtAgos activity by strand invasion and unwinding of dsDNA (Hunt et al., 2018). And no putative DNA helicase in proximity (within 5 kb of FpAgo) was found in the F. placidus genome via bioinformatic analysis. We proposed the optimal growing temperature for F. placidus, which would be sufficient for the unwinding dsDNA without the requirement of helicase.
FpAgo Exhibits a Preference for a 5′-dG Base at the 5′ End of the gDNA
A previous analysis of the nucleotide composition of ssDNA sequences co-purified with TtAgo revealed that TtAgo has a strong bias for a 5′-end deoxycytidine. However, in vitro cleavage assays showed that all guides containing a 5′-dC, dT, dA, or dG resulted in similar target cleavage activities (Swarts et al., 2014a). The 5′-end base of the guide strand and 1st base of the target splayed out in separate binding pockets, respectively, was observed in the crystal structure of TtAgo (Sheng et al., 2014). The identical target cleavage activities could be explained by the fact that the 5′-end base of the guide strand does not participate in target pairing. Interestingly, MjAgo-mediated cleavage efficiency is significantly higher using 5′-purine guides than 5′-pyrimidines guides (Willkomm et al., 2017). For FpAgo, we found that a guide with a 5′-dG supports higher cleavage efficiency than the other three types of guides. Earlier findings imply that the higher affinity of 5′-dG ternary FpAgo-guide complexes resulted in the increase of cleavage efficiency by increasing the number of ternary FpAgo-guide-target complexes compared to other guides (Deerberg et al., 2013; Willkomm et al., 2016, 2017). A moderate AT-bias near the 5′-end and downstream of the target cleavage site was observed for mesophilic CbAgo when expressed and purified from E. coli cells, indicating that their biogenesis might depend on the melting of these DNA regions in the context of dsDNA (Kuzmenko et al., 2020). The 5′-dC preference of thermophilic TtAgo in vivo was thought to be the result of specific guide processing or preferential 5′ nucleoside selection by TtAgo (Swarts et al., 2014a). Taken together, the 5′-dG has a preference for hyperthermophilic FpAgo, suggesting that the efficiency of gDNA processing does not strongly depend on the GC content, especially for thermophilic pAgos.
FpAgo Tolerates Mismatches in the Seed Region but Is Sensitive to Mismatches in the 3′-Terminal of gDNA
It is generally acknowledged that even a single mismatch in the seed region reduces cleavage activity, whereas mismatches in the 3′-guide strands are usually tolerated without significant loss of cleavage activity (Dahlgren et al., 2008; Wang et al., 2008; Parker et al., 2009; Salomon et al., 2015). In this study, we observed that mismatches in the seed region did not significantly affect FpAgo cleavage activity, and conversely, even stimulated target DNA cleavage (for gM7). Coincidentally, a recent study on zebrafish Argonaute2 demonstrated that a mismatch involving guide-RNA position 6 enhances slicing activity, whereas, without the mismatch, cleavage is essentially abolished (Chen et al., 2017). Together, these findings suggest that some seed mismatches have opposite effects on cleavage activity. In contrast, for mismatches in the 3′-guide strand, target cleavage was either almost abolished or strongly reduced by FpAgo, and similar effects were recently reported for TtAgo, CbAgo, and LrAgo (Kuzmenko et al., 2019; Song et al., 2020). Surprisingly, FpAgo was less active with a perfectly complementary target mediated by gDNAs of gM3, gM12, gM3, and gM15. These results suggest that the cleavage activity of FpAgo depends on the sequences of the guide and target.
gDNA Recognition by the Distinctive FpAgo PAZ Domain
Both eAgos and long pAgos generally consist of four conserved structure domain architectures, namely, MID (middle) domain, PIWI (P-body-induced wimpy testes) domain, PAZ (PIWI-Argonaute-Zwille) domain, and N domain. The MID and PAZ domains usually form binding pockets that anchor the 5′- and 3′-ends of the guide strand, respectively. The PIWI domain contains a catalytic motif that mediates target strand cleavage (Schirle and MacRae, 2012; Hegge et al., 2018). Ago-mediated cleavage of targets by guide strands up to 36 nt in length could be rationalized by the guide to adopting an alternative trajectory to allow 3′-end insertion into the PAZ binding pocket. In addition, the open nucleic acid-binding channel, between the PAZ domain and PIWI domain, which encompasses the 3′-end of the guide strand, is accessible to the outside and permissive to looping-out longer 3′-end (Wang et al., 2008). FpAgo adopts a shorter guide strand for cleavage activity, indicating a distinct catalytic pattern. Previous structural studies based on TtAgo showed that the retained or released state of the 3′-end of the guide strand within the PAZ binding pocket transforms the TtAgo ternary complex from a cleavage-incompatible conformation to a cleavage-compatible conformation (Sheng et al., 2014). Our structural analysis indicates that the severely reduced activity mediated by long guide strands may be explained by the fact that FpAgo employs a distinct PAZ domain that restricts the 3′-ends of the long guide strand inserted into the PAZ binding pocket, which may result in the instability of guide-target duplex hybridization. Our EMSA assays also showed that the FpAgo-guide-target ternary complex exhibited a weaker binding affinity for long guide strands. Indeed, structural work on FpAgo and its binary and ternary complexes is needed to elucidate the molecular basis for the short guide strand in the future.
Conclusion
This study identified a novel hyperthermophilic FpAgo that cleaves DNA targets in a 5′-P gDNA-dependent sequence-specific manner. Starting from a minimal gDNA length of 15 nt, FpAgo only accepted gDNA lengths of less than 21 nt. FpAgo prefers the 5′-dG of the guide strand. The single mismatch near the 3′-terminal of gDNA rather than the seed region curtails the FpAgo cleavage activity. The structure modeling analysis implicates a unique loop of the PAZ domain that may be responsible for the specificity of FpAgo for guide binding. Furthermore, FpAgo efficiently mediates DNA-guided DNA cleavage of double-stranded plasmids at elevated temperatures. These findings broaden our understanding of thermophilic Agos and pave the way for their use in DNA manipulations in the future.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: National Center for Biotechnology Information (NCBI) Protein, https://www.ncbi.nlm.nih.gov/protein/, WP_012966655.1.
Author Contributions
XG performed the research, data analysis, and preparation of the manuscript. YF and QL provided guidance, designed the experiments, and revised the manuscript. YS assisted with cleavage assays and data analysis. LC and FH performed structural modeling analysis. All authors participated in the project proposal and approved the final version of the manuscript.
Funding
This work was funded by grants from the Natural Science Foundation of China (31770078), the Ministry of Science and Technology (2020YFA0907700) and the National Key Research & Development Program of China (2018YFA0900403).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.654345/full#supplementary-material
References
Barrangou, R., and Marraffini, L. A. (2014). CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244.
Cao, Y., Sun, W., Wang, J., Sheng, G., Xiang, G., Zhang, T., et al. (2019). Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 degrees C. Cell Discov. 5:38.
Chen, G. R., Sive, H., and Bartel, D. P. (2017). A seed mismatch enhances Argonaute2-catalyzed cleavage and partially rescues severely impaired cleavage found in fish. Mol. Cell 68, 1095–1107.e5.
Chong, Y., Liu, Q., Huang, F., Song, D., and Feng, Y. (2019). Characterization of a recombinant thermotolerant Argonaute protein as an endonuclease by broad guide utilization. Bioresour. Bioprocess. 6:21.
Dahlgren, C., Zhang, H. Y., Du, Q., Grahn, M., Norstedt, G., Wahlestedt, C., et al. (2008). Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 36:e53. doi: 10.1093/nar/gkn190
De, N., Young, L., Lau, P. W., Meisner, N. C., Morrissey, D. V., and Macrae, I. J. (2013). Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell 50, 344–355. doi: 10.1016/j.molcel.2013.04.001
Deerberg, A., Willkomm, S., and Restle, T. (2013). Minimal mechanistic model of siRNA-dependent target RNA slicing by recombinant human Argonaute 2 protein. Proc. Natl. Acad. Sci. U. S. A. 110, 17850–17855. doi: 10.1073/pnas.1217838110
Enghiad, B., and Zhao, H. (2017). Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 6, 752–757. doi: 10.1021/acssynbio.6b00324
Frank, F., Hauver, J., Sonenberg, N., and Nagar, B. (2012). Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 31, 3588–3595. doi: 10.1038/emboj.2012.204
He, R., Wang, L., Wang, F., Li, W., Liu, Y., Li, A., et al. (2019). Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. (Camb.) 55, 13219–13222. doi: 10.1039/c9cc07339f
Hegge, J. W., Swarts, D. C., Chandradoss, S. D., Cui, T. J., Kneppers, J., Jinek, M., et al. (2019). DNA-guided DNA cleavage at moderate temperatures by clostridium butyricum Argonaute. Nucleic Acids Res. 47, 5809–5821. doi: 10.1093/nar/gkz306
Hegge, J. W., Swarts, D. C., and Van Der Oost, J. (2018). Prokaryotic Argonaute proteins: novel genome-editing tools? Nat. Rev. Microbiol. 16, 5–11. doi: 10.1038/nrmicro.2017.73
Hunt, E. A., Evans, T. C., and Tanner, N. A. (2018). Single-stranded binding proteins and helicase enhance the activity of prokaryotic argonautes in vitro. PLoS One 13:e0203073. doi: 10.1371/journal.pone.0203073
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jolly, S. M., Gainetdinov, I., Jouravleva, K., Zhang, H., Strittmatter, L., Bailey, S. M., et al. (2020). Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell 182, 1545–1559.e18.
Kaya, E., Doxzen, K. W., Knoll, K. R., Wilson, R. C., Strutt, S. C., Kranzusch, P. J., et al. (2016). A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U. S. A. 113, 4057–4062. doi: 10.1073/pnas.1524385113
Ketting, R. F. (2011). The many faces of RNAi. Dev. Cell 20, 148–161. doi: 10.1016/j.devcel.2011.01.012
Kim, S. Y., Jung, Y., and Lim, D. (2020). Argonaute system of Kordia jejudonensis is a heterodimeric nucleic acid-guided nuclease. Biochem. Biophys. Res. Commun. 525, 755–758. doi: 10.1016/j.bbrc.2020.02.145
Kuzmenko, A., Oguienko, A., Esyunina, D., Yudin, D., Petrova, M., Kudinova, A., et al. (2020). DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637. doi: 10.1038/s41586-020-2605-1
Kuzmenko, A., Yudin, D., Ryazansky, S., Kulbachinskiy, A., and Aravin, A. A. (2019). Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 47, 5822–5836. doi: 10.1093/nar/gkz379
Lau, N. C., Lim, L. P., Weinstein, E. G., and Bartel, D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. doi: 10.1126/science.1065062
Liu, Q., Guo, X., Xun, G., Li, Z., Chong, Y., Yang, L., et al. (2021a). Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. gkab274. doi: 10.1093/nar/gkab274
Liu, Y., Li, W., Jiang, X., Wang, Y., Zhang, Z., Liu, Q., et al. (2021b). A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 49, 1597–1608. doi: 10.1093/nar/gkaa1278
Marraffini, L. A., and Sontheimer, E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190. doi: 10.1038/nrg2749
Olina, A., Kuzmenko, A., Ninova, M., Aravin, A. A., Kulbachinskiy, A., and Esyunina, D. (2020). Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 17, 677–688. doi: 10.1080/15476286.2020.1724716
Parker, J. S., Parizotto, E. A., Wang, M., Roe, S. M., and Barford, D. (2009). Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell 33, 204–214. doi: 10.1016/j.molcel.2008.12.012
Salomon, W. E., Jolly, S. M., Moore, M. J., Zamore, P. D., and Serebrov, V. (2015). Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95. doi: 10.1016/j.cell.2015.06.029
Schirle, N. T., and MacRae, I. J. (2012). “Structure and mechanism of Argonaute proteins,” in Eukaryotic RNases and their Partners in RNA Degradation and Biogenesis, Part B, eds F. Guo and F. Tamanoi (Cambridge, MA: Academic Press), 83–100. doi: 10.1016/b978-0-12-404741-9.00004-0
Sheng, G., Zhao, H., Wang, J., Rao, Y., Tian, W., Swarts, D. C., et al. (2014). Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U. S. A. 111, 652–657. doi: 10.1073/pnas.1321032111
Song, J., Hegge, J. W., Mauk, M. G., Chen, J., Till, J. E., Bhagwat, N., et al. (2020). Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus Argonaute with applications in cancer diagnostics. Nucleic Acids Res. 48:e19. doi: 10.1093/nar/gkz1165
Song, Y., Dimaio, F., Wang, R. Y., Kim, D., Miles, C., Brunette, T., et al. (2013). High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. doi: 10.1016/j.str.2013.08.005
Swarts, D. C., Hegge, J. W., Hinojo, I., Shiimori, M., Ellis, M. A., Dumrongkulraksa, J., et al. (2015). Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 43, 5120–5129. doi: 10.1093/nar/gkv415
Swarts, D. C., Jore, M. M., Westra, E. R., Zhu, Y., Janssen, J. H., Snijders, A. P., et al. (2014a). DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261. doi: 10.1038/nature12971
Swarts, D. C., Makarova, K., Wang, Y., Nakanishi, K., Ketting, R. F., Koonin, E. V., et al. (2014b). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753. doi: 10.1038/nsmb.2879
Taly, V., Pekin, D., Benhaim, L., Kotsopoulos, S. K., Le Corre, D., Li, X., et al. (2013). Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 59, 1722–1731. doi: 10.1373/clinchem.2013.206359
Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl, T., and Patel, D. J. (2008). Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926. doi: 10.1038/nature07666
Willkomm, S., Oellig, C. A., Zander, A., Restle, T., Keegan, R., Grohmann, D., et al. (2017). Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2:17035.
Willkomm, S., Zander, A., Grohmann, D., and Restle, T. (2016). Mechanistic insights into archaeal and human Argonaute substrate binding and cleavage properties. PLoS One 11:e0164695. doi: 10.1371/journal.pone.0164695
Xun, G., Liu, Q., Chong, Y., Li, Z., Guo, X., Li, Y., et al. (2019). The stepwise endonuclease activity of a thermophilic Argonaute protein. bioRxiv:821280. doi: 10.1101/821280
Zander, A., Willkomm, S., Ofer, S., Van Wolferen, M., Egert, L., Buchmeier, S., et al. (2017). Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2:17034.
Keywords: Argonaute, thermophilic archaea, Ferroglobus placidus, endonuclease, structural analysis
Citation: Guo X, Sun Y, Chen L, Huang F, Liu Q and Feng Y (2021) A Hyperthermophilic Argonaute From Ferroglobus placidus With Specificity on Guide Binding Pattern. Front. Microbiol. 12:654345. doi: 10.3389/fmicb.2021.654345
Received: 16 January 2021; Accepted: 23 April 2021;
Published: 09 June 2021.
Edited by:
Yoshizumi Ishino, Kyushu University, JapanReviewed by:
Likui Zhang, Yangzhou University, ChinaShuang-yong Xu, New England Biolabs, United States
Copyright © 2021 Guo, Sun, Chen, Huang, Liu and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Liu, bGl1cWlhbjEwMThAc2p0dS5lZHUuY24=; Yan Feng, eWZlbmcyMDA5QHNqdHUuZWR1LmNu
 Xiang Guo
Xiang Guo Yingying Sun
Yingying Sun Liuqing Chen
Liuqing Chen Fei Huang
Fei Huang Qian Liu
Qian Liu Yan Feng
Yan Feng