Abstract
Deoxynivalenol (DON) is one of the most devastating and notorious contaminants in food and animal feed worldwide. A novel DON-degrading strain, Nocardioides sp. ZHH-013, which exhibited complete mineralization of DON, was isolated from soil samples. The intermediate products of DON generated by this strain were identified by high-performance liquid chromatography and ultra-performance liquid chromatography tandem mass spectrometry analyses. It was shown that, on an experimental level, 3-keto-DON was a necessary intermediate product during the conversion from DON to 3-epi-DON. Furthermore, the ZHH-013 strain could also utilize 3-epi-DON. This DON degradation pathway is a safety concern for food and feed. The mechanism of DON and 3-epi-DON elimination will be further studied, so that new enzymes for DON degradation can be identified.
Introduction
Deoxynivalenol (vomitoxin, DON) is one of the most important toxic secondary metabolites of mold. Wheat and corn are the main sources of DON in food and animal feed because they are highly important raw food materials that are susceptible to Fusarium infection (Xu and Nicholson, 2009). Issues with planting, processing, and warehouse management can cause DON pollution and reduce production yields (Bai and Shaner, 2004). DON has been found to be enriched in water from contaminated cereals during wet processing, in rainwater in the field, and in polluted water from animal farms (Wettstein and Bucheli, 2010; Gracia-Lor et al., 2020; Eagles et al., 2021).
In addition, DON derivatives are important sources of DON for animals and humans. Wheat cultivars and toxigenic Fusarium strains can convert DON into masked mycotoxins, such as 3-acetyl DON (3-ADON), 15-acetyl DON (15-ADON), DON-3-glucoside, DON-15-glucoside, DON-sulfate, and DON-glutathione (Miller and Arnison, 1986; Hassan et al., 2016; Jurisic et al., 2019). These masked mycotoxins can then be broken down into DON by gut microbes. Overexposure to DON can lead to intense vomiting in humans and animals, as well as damage to the immune and reproductive systems and disruption of developmental processes (Gunther et al., 2017; Lucke et al., 2017; Springler et al., 2017; Juan-García et al., 2019).
Among the solutions to DON pollution, disease-resistant plants and fungal biocontrol agents are more promising than antifungal agricultural chemicals. Lower disease incidence greatly reduces DON pollution during planting and raw material storage. In addition, it is necessary to develop efficient and environmentally friendly detoxification technologies for processing, waste treatment and ecological restoration.
Heat (Bretz et al., 2006) and ozone (Li et al., 2019) are widely studied physical and chemical DON elimination methods that are considered safe and environmentally friendly and achieve efficient degradation; however, the resulting degradation products are unstable and difficult to analyze because of their complexity, and these methods may reduce the nutritional quality of raw food materials (Santos Alexandre et al., 2018). Therefore, non-thermal methods, such as intense pulsed light (Chen et al., 2019), plasma-activated water (Chen et al., 2019), carbon nitride nested tubes (Bai et al., 2019), and the upconversion nanoparticle @TiO2 (Zhou et al., 2020), are of interest because they are environmentally friendly, efficient, and/or economical. These methods are good for processing foods with smooth surfaces or free DON in solvent, and may be useful in the sorting and pretreatment of raw food materials; however, their use to influence animal digestion processes, with the aim of eliminating unreleased or masked DON in food, is less feasible.
Compared with the methods mentioned above, DON biodegradation technologies are gentle, effective, and easy to scale up. Among them, microorganisms or their metabolic detoxification systems that exhibit DON degradation abilities are receiving the most attention.
Massive screening techniques using diverse methods have been conducted to identify DON-degrading microbes, several of which have high DON conversion ability. Over the last three decades, DON-degrading strains have been identified among genera including Eubacterium (Fuchs et al., 2002), Eggerthella (Gao et al., 2018), Desulfitobacterium (He et al., 2020), Devosia (He et al., 2016; Wang et al., 2017, 2019), Sphingomonas (Ito et al., 2013; He et al., 2017), Nocardioides (Ikunaga et al., 2011), Marmoricola (Ito et al., 2012), Bacillus (Li et al., 2011), Pelagibacterium (Zhang et al., 2020), and Aspergillus (He et al., 2008; Jaqueline and Eliana, 2010; Jaqueline et al., 2011; Yang et al., 2017).
Extensive epidemiological investigations have shown that many animals are insensitive to DON; however, monogastric animals, especially swine, are sensitive to DON. Rumen and gut microbes are considered to be effective at DON detoxification. Nevertheless, only a few DON-degrading strains have been isolated from rumens or chicken intestines, and they could only transform DON to de-epoxy DON (de-DON or DOM-1) under anaerobic conditions. Eubacterium sp. BBSH 797 was the first anaerobic DON-degrading strain isolated from the rumen fluid of a cannulated cow (He et al., 1992; Fuchs et al., 2002). When swine ate feed containing this strain, the negative effect of DON on pig intestines was controlled (Grenier et al., 2013). Eggerthella sp. DII-9, isolated from chicken intestines, can similarly transform DON (Gao et al., 2018). In the past, it was thought that the conversion of DON to DOM-1 could only be completed under anaerobic conditions; however, recent studies have shown that Desulfitobacterium sp. PGC-3-9 can transform DON under both anaerobic and aerobic conditions (He et al., 2020). The genus Devosia has been widely studied, and several species in this genus can detoxify DON. The DON transformation mechanism of Devosia sp. 17-2-E-8 is relatively well understood and involves two enzymes from the DON epimerization (Dep) pathway. DON is first converted to 3-keto-DON by DepA (Carere et al., 2018a), and then 3-keto-DON is transformed to 3-epi-DON by DepB (Carere et al., 2018b). Sphingomonas sp. KMS1 and S3-4 can utilize DON as a sole carbon source; the former strain could transform DON to 16-hydroxy-DON using a special cytochrome P450 system (Ito et al., 2013), while the latter strain could only convert DON to 3-keto-DON (3-oxo-DON) and 3-epi-DON (He et al., 2017). Recently, two novel strains, from the LZ-N1 consortium, namely, Pseudomonas sp. Y1 and Lysobacter sp. S1, were shown to exhibit sustained ability to transform DON into the metabolite 3-epi-DON, with no degradation products detected after 72 h (Zhai et al., 2019).
In this study, a new DON-degrading strain, designated Nocardioides sp. ZHH-013, was isolated from soil associated with a high incidence of root rot. We examined the DON-degrading activity of the species and assessed the detected intermediate products, 3-keto-DON and 3-epi-DON.
Materials and Methods
Soil Samples, Chemicals, and Media
Soil samples (n = 1360) were collected from wheat, corn, or forest fields at Jiangsu, Anhui, Hebei, Beijing, Liaoning, Heilongjiang, Nei Mongol, Yunnan, Gansu, and Xinjiang in China, to isolate DON-degrading microbes.
Deoxynivalenol (32943-5MG, Sigma-Aldrich, United States; 16.87 mM in water) was stored at −20°C.
Mineral salt medium (MM) (Ikunaga et al., 2011) was used for enrichment and DON degradation assays. R2A (Difco, United States), Luria-Bertani (LB) broth (Oxoid, United Kingdom), and tryptic soy broth (TSB; Oxoid) were used to culture the DON-degrading strains isolated from soil samples.
Screening, Enrichment, and Isolation of DON-Degrading Strains
Each soil sample (0.05–0.10 g) was suspended in 0.7 ml of MM with 168.74 μM DON and incubated at 30°C and 220 rpm for 21 days. Thereafter, 20-μl culture aliquots were added to 0.5 ml of MM containing 168.74 μM DON, followed by 21 days of incubation under the same conditions. This process was repeated at least three times. Samples were taken at each step, mixed with an equal volume of sterile 50% glycerol, and stored at 80°C. The concentration of DON in samples was then assessed by high-performance liquid chromatography (HPLC), as described below.
After a series of dilutions, final enriched cultures were spread onto R2A, LB, or TSB agar plates and incubated at 30°C for 14 days. Various colonies were selected and incubated in MM containing 168.74 μM DON at 30°C for 14 days. Thereafter, concentrations of DON were detected by HPLC. Pure cultures were mixed with an equal volume of sterile 50% glycerol and stored at −80°C. All screening and incubation processes were carried out under aerobic conditions.
Phylogenetic Analysis of Nocardioides sp. ZHH-013
The partial 16S rRNA gene sequence of the ZHH-013 strain was amplified by PCR using primers 27f and 1492r (Zhang et al., 2017). The resulting amplicon was an approximately 1381-bp 16S rDNA sequence (GenBank Accession No. MW493343). Similar sequences were identified using BLAST (Johnson et al., 2008) and 16S-based ID apps in the EzBioCloud server (Yoon et al., 2017). Neighbor-joining phylogenetic trees were constructed using MEGA X software (Kumar et al., 2018). Alignment was conducted using the ClustalW program (Higgins et al., 1994). Bootstrap analysis was performed with 1,000 replications.
DON Degradation Assay
To obtain an initial culture, a single ZHH-013 colony was inoculated in 700 μl of MMD and grown under aerobic conditions at 30°C for 8–12 days.
To assess DON degradation by a high-density culture of the ZHH-013 strain, 3 ml of TSB medium was mixed with the initial culture (1%) and grown at 30°C, with agitation at 220 rpm for 7 days. Next, culture densities were adjusted to an OD600 of 0.4–0.6, inoculated in 100 ml of TSB, and incubated at 30°C and 220 rpm for 5 days.
Next, 50 ml of fermentation broth was transferred to a 50-ml centrifuge tube and centrifuged at 14,400 × g for 3 min at 4°C. The supernatant was collected and filtered through a 0.22-μm filter (Millipore, Cork, Ireland) to generate a cell-free supernatant. Additionally, the cell pellets were washed three times with sterilized water and resuspended in 5 ml of MM. Next, DON was added to the samples at the final concentrations of 0.169, 1.687, 3.375, and 33.747 mM. All samples were incubated at 30°C with 220 rpm for 10 and 24 h.
In addition to the fermentation broth supernatant and the cell pellets, we also assessed lysed cells. Cells were lysed using a sonic disrupter, and lysates centrifuged at 20,700 × g (4°C, 20 min) to obtain supernatants. Aliquots were left untreated or filtered using 0.22-μm filters. DON (168.74 μM) was added to the samples and incubated at 30°C and 220 rpm for 10 and 24 h.
All samples were stored at −80°C before analysis by HPLC or ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS), as described below.
Preparation and Purification of 3-keto-DON and 3-epi-DON
To prepare 3-keto-DON as the standard control (Carere et al., 2018a), the synthesis gene of depA from Devosia sp. 17-2-E-8 was purchased from GENEWIZ (Su Zhou, China). The gene was cloned into pET-28a (+) and then transformed into Escherichia coli BL21(DE3). After ensuring inducible expression and purification by Ni-nitrilotriacetic acid chromatography, DepA was mixed with 0.1 mM KH2PO4, 0.1 mM NaOH, 1 mM Ca2+, and 100 μM pyrroloquinoline quinone, and then incubated at 30°C for 20 h. Thereafter, a triple volume of methanol was added and samples were left at −20°C for 20 min. After centrifugation (4°C, 20 min), the supernatant was collected. The solvent was removed using vacuum freeze-drying equipment (Martin Christ, Osterode am Harz, Germany), and 3-keto-DON was dissolved in 100 μl of methanol. The 3-keto-DON standard was stored at −80°C before HPLC or UPLC-MS/MS analysis.
Preparation of 3-keto-DON produced by ZHH-013 strain referred to the analysis method as described above. Preparation of 3-epi-DON produced by ZHH-013 strain refers to the method described by Ikunaga et al. (2011) with modification. Wet Nocardioides sp. ZHH-013 cells were resuspended in 1 ml of MM (OD600 = 1.0) containing 10 mg of DON, and incubated for 41 h at 30°C. After centrifugation at 20,700 × g (4°C, 20 min), the supernatant was collected and filtered using a 0.22-μm filter. The solvent was drawn off completely using vacuum freeze-drying equipment (Martin Christ) and the sample was resuspended in 100 μl of methanol. Analytical HPLC (described in the next section) was used to prepare 3-epi-DON and 3-keto-DON. All elution fractions of 3-epi-DON and 3-keto-DON were mixed, respectively. After sample enrichment by vacuum freeze-drying (Martin Christ) and resuspension in 100 μl of methanol, all degradation products were stored at −80°C before HPLC or UPLC-MS/MS analysis.
HPLC Analysis of DON, 3-epi-DON, and 3-keto-DON
Equal volumes of samples were mixed well with methanol stored at −20°C. Before HPLC analysis, samples were filtered using 0.22-μm filters.
The HPLC system (Shimadzu, Kyoto, Japan) consisted of an LC-20AT pump and an SPD-20A UV/VIS detector. A reverse phase column (Agilent TC-C18, 4.6 mm × 250 mm, 80 Å, 5 μm) was used. To detect DON and its derivatives, the mobile phase comprised methanol and water (15:85, v/v) at a flow rate of 1.0 ml/min. To detect 3-keto-DON, the mobile phase comprised methanol and water (1:2, v/v). UV/VIS detection was performed at a wavelength of 220 nm. The column was heated to 40°C.
UPLC-MS/MS Analysis of DON, 3-epi-DON, and 3-keto-DON
UPLC-MS/MS analysis was performed using a Nexera UHPLC LC-30A (Shimadzu) UPLC system coupled with TripleTOF5600 (AB Sciex, United States). For C18 separation, mobile phase A was acetonitrile, and mobile phase B was 0.5% formic acid in water. The column was an HSS T3 column (150 mm × 3 mm, 1.8 μm; Waters) operated at 40°C. The flow rate was 300 μl/min and the injection volume was 1 μl. Gradient conditions were as follows: 0–10 min, A: 0 to 50%, B: 100 to 50%; 10–13 min, A: 50 to 95%, B: 50 to 5%; 13–14 min, A: 95 to 0%, B: 5 to 100%; 14–15 min, A: 0 to 0%, B: 100 to 100%.
The mass spectrometer was operated in positive ion mode with ion spray voltage floating, 5500 V; ion source gas 1 50 psi; ion source gas 2 50 psi; and curtain gas at 25 psi and source temperature, 500°C. Sample analysis was performed by information-dependent acquisition, with a 200-ms time-of-flight (TOF)-MS scan from 100 to 1,500 Da, followed by an MS/MS scan in high-sensitivity mode from 50 to 1,500 Da of the top 20 precursor ions from the TOF-MS scan.
Results and Discussion
Screening, Identification, and Characterization of Nocardioides sp. ZHH-013
Microbes are the main decomposers in nature and soil is the richest source of strains with degradation activity. Over the last three decades, especially the past 3 years, several strains that can efficiently transform DON have been reported. Nocardioides sp. WSN05-2, isolated from wheat field soil, was the first Nocardioides strain demonstrated to degrade DON as a carbon source (Ikunaga et al., 2011). It completely converted DON after 10 days of incubation and no signal of a typical trichothecene skeleton was detected by nuclear magnetic resonance (NMR) analysis (Ikunaga et al., 2011); however, no more public information has been released about the DON degradation function of this genus, despite several researchers continuing to screen for DON degrading microorganisms.
Enrichment culture of 1,360 soil samples was conducted; after three rounds of screenings, 24 samples with DON-degrading activity were isolated. Among them, 13 samples exhibited DON degradation characteristics similar to those reported for Devosia. The remaining 11 samples could completely transform DON. After 10 sub-inoculations, Culture Sample No. 13 showed stable and efficient DON-degrading activity. Microbial diversity analysis of Sample No. 13 showed that Nocardioides (45.20%) and Paracoccus (27.25%) were the main components at the genus level (unpublished data).
An enriched culture of Sample No. 13 was plated on various media (LB, TSA, R2A, or MM) using the dilution method. Consequently, the ZHH-013 strain was isolated and a 1,381-bp sequence of its 16S rDNA was used for phylogenetic analysis. Sequences with the highest similarities to the ZHH-013 strain were as follows: Nocardioides vastitatis 21Sc5-5T (97.68%), Nocardioides kongjuensis A2-4T (97.39%) Nocardioides nitrophenolicus NSP 41T (97.32%), Nocardioides caeni MN8T (97.25%), Nocardioides albidus THG-S11.7T (97.25%), Nocardioides flava THG-DN5.4T (97.18%), and Nocardioides pelophilus THG-T63T (97.18%). A phylogenetic tree, constructed using the neighbor-joining method, showed that the ZHH-013 strain belongs to the genus Nocardioides (Figure 1). The 16S rRNA similarity was lower than the recommended classification threshold of 98.65% (Kim et al., 2014); hence, we inferred that the ZHH-013 strain may be a new Nocardioides species. The sequence similarity between ZHH-013 and other DON-degrading Nocardioides strains (Sato et al., 2012) was less than or equal to 92.27%.
FIGURE 1
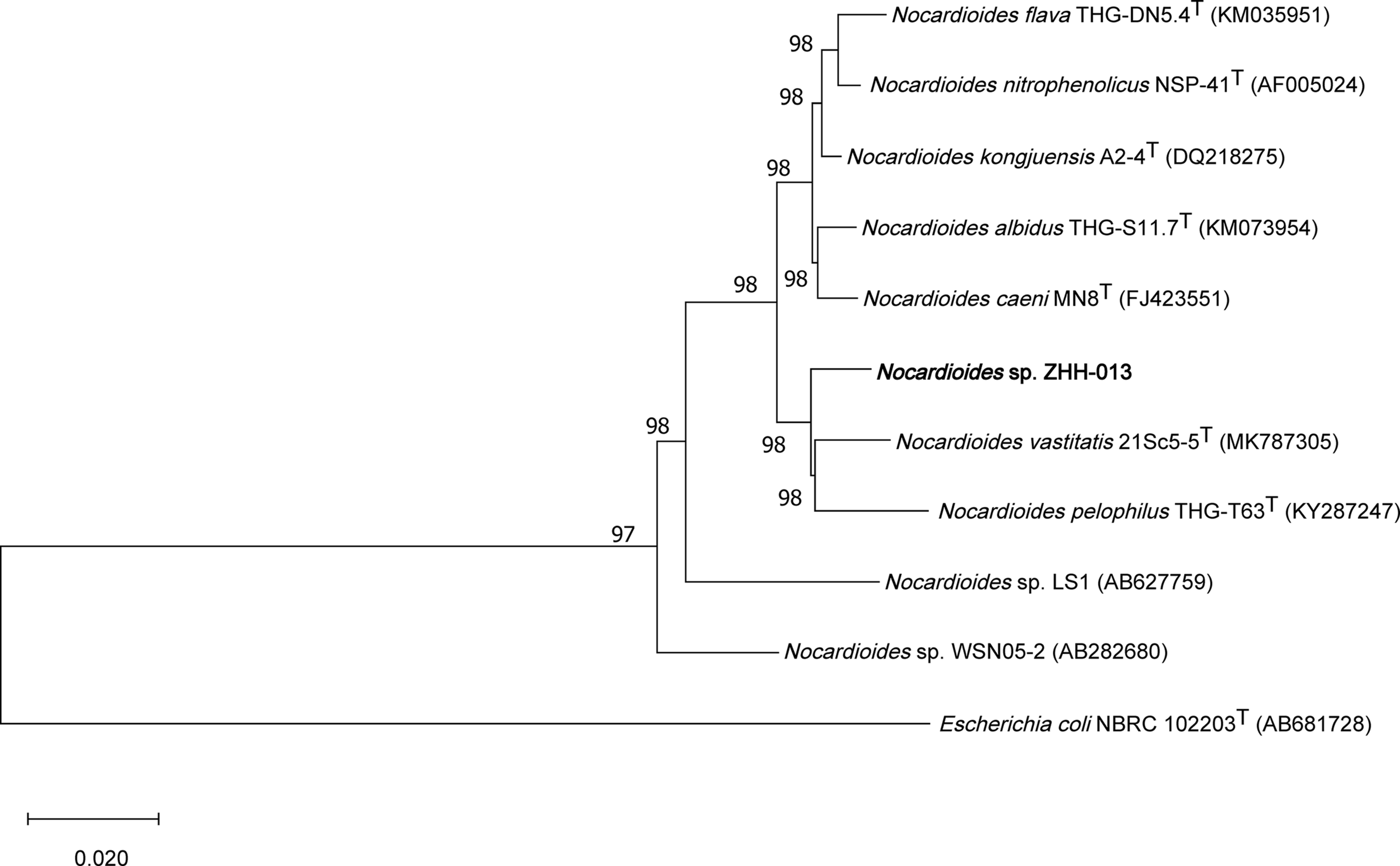
Phylogenetic tree, including Nocardioides sp. ZHH-013, based on neighbor-joining analysis of 16S rDNA sequences.
The ZHH-013 strain is a Gram-positive, aerobic, rod-shaped, non-spore-forming bacterium that is 0.28–0.29 × 0.86–0.87 μm in size (Figure 2). Colonies were white and translucent on R2A plates, and yellowish white on LB or TSA plates. The ZHH-013 strain could use DON as a sole carbon source, whereas related species (N. vastitatis 21Sc5-5T, N. kongjuensis A2-4T, N. nitrophenolicus NSP 41T, N. caeni MN8T, and N. albidus THG-S11.7T) could not.
FIGURE 2
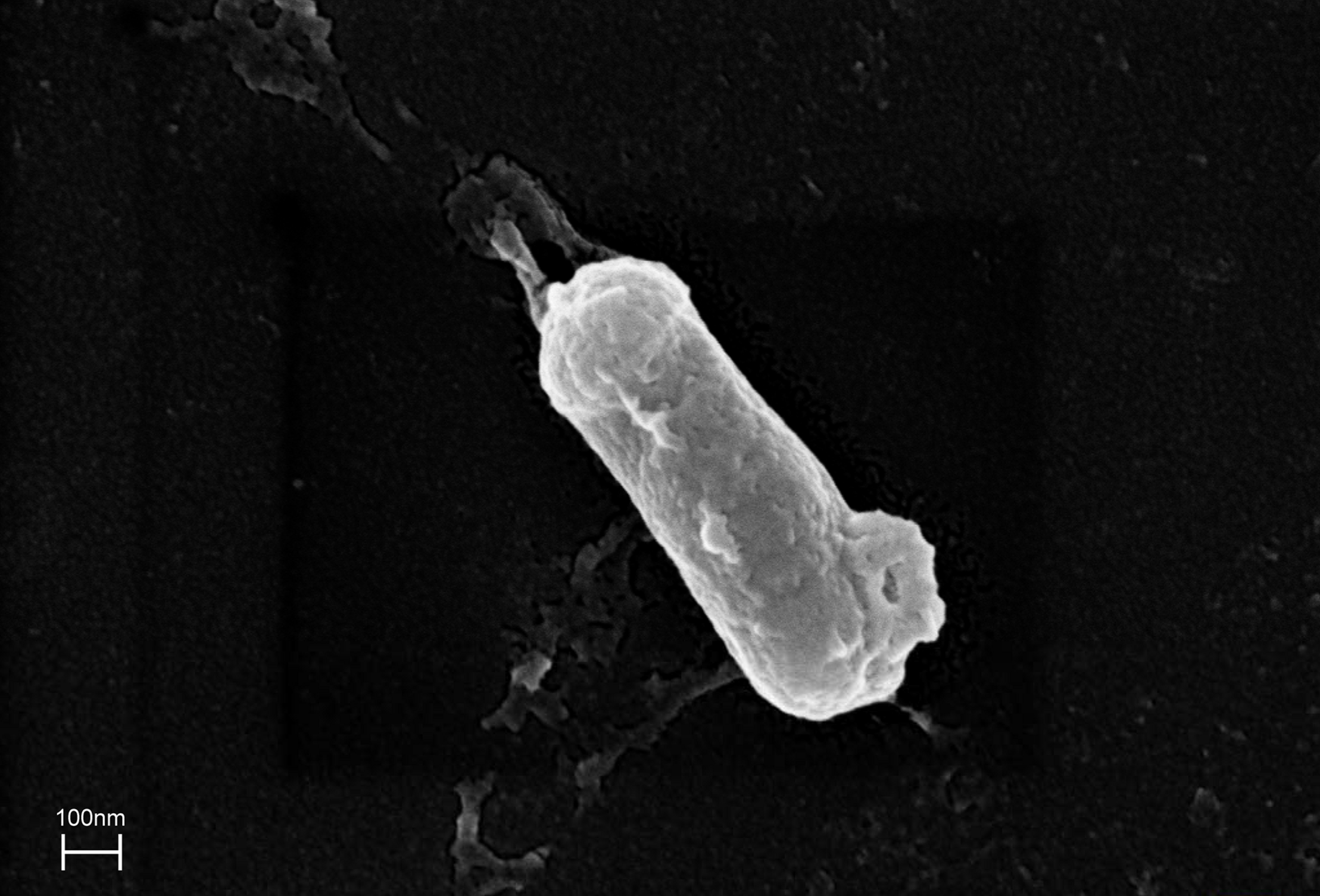
Morphological characteristics of Nocardioides sp. ZHH-013 cell (×50,000).
The DON degradation rate was directly proportional to the amount of ZHH-013 strain. After 48 h of incubation, the DON degradation rate was 80% at an OD600 of 0.4 and only 40% after 10-fold dilution. After 10 h of co-incubation with DON, DON was not degraded by ZHH-013 fermentation broth supernatant or filtered or moist heat sterilized (121°C, 20 min) by supernatant of ZHH-013 cell lysate (Figure 3). Thus, the complete degradation capacity of DON may rely on live cells.
FIGURE 3
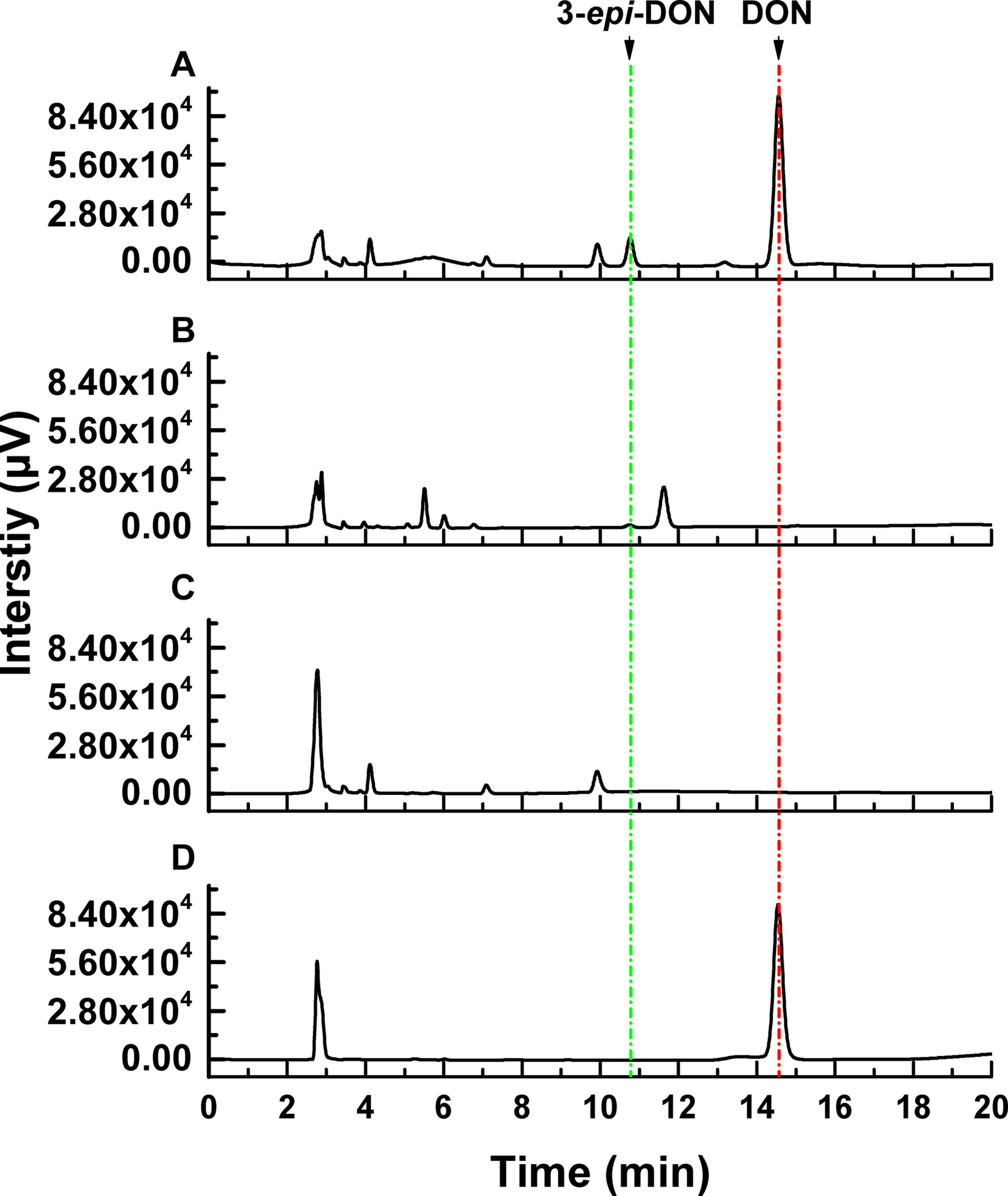
High-performance liquid chromatography analysis of DON degradation products after incubation of DON with (A) filtered intracellular supernatant treated, (B) intracellular supernatant, and (C) untreated Nocardioides sp. ZHH-013 cells. (D) The 168.74 μM DON standard was used as control.
Characterization of DON Biodegradation by Nocardioides sp. ZHH-013
Researchers have confirmed DON detoxification mechanisms in several organisms, including animals and plants, and most of these processes rely on microorganisms (Fuchs et al., 2002; Ito et al., 2012; Gao et al., 2018). The five main degradation pathways detected in microorganisms are as follows: (1) DON to DOM-1; (2) DON to 3-ADON or 15-ADON; (3) DON to 3-keto DON; (4) DON to 16-DON; and (5) two-step transformation: DON to 3-keto-DON and 3-keto-DON to 3-epi-DON. Some other strains, such as Aspergillus niger As-D.1 (Yang et al., 2017), exhibit different mechanisms; however, the degradation products have not been clearly explained based on detailed data. Moreover, the ZHH-013 strain exhibited a new degradation mechanism that differed from the five mechanisms mentioned above.
Deoxynivalenol degradation products generated by ZHH-013 were detected by HPLC and UPLC-MS/MS. 3-epi-DON appeared as an early DON degradation product (Figures 4, 5). Based on MS and MS/MS data indicating a compound with a retention time (RT) of 3.09 min (Supplementary Figure 2A), the predominant ion at m/z 297.13 was consistent with a molecular ion of [DON + H]+ (Supplementary Figures 2B,C), which was similar to that of DON (Supplementary Figures 1B,C). 3-epi-DON has previously been identified as a DON degradation product, based on MS and NMR data (Ikunaga et al., 2011; He et al., 2015b). After continued incubation, the peak for 3-epi-DON surprisingly disappeared (Figure 5B and Supplementary Figure 5) and we concluded that ZHH-013 could also utilize 3-epi-DON. These results resemble the findings reported for the strain, WSN05-2 (Ikunaga et al., 2011), which can utilize 3-epi-DON as a sole carbon source.
FIGURE 4
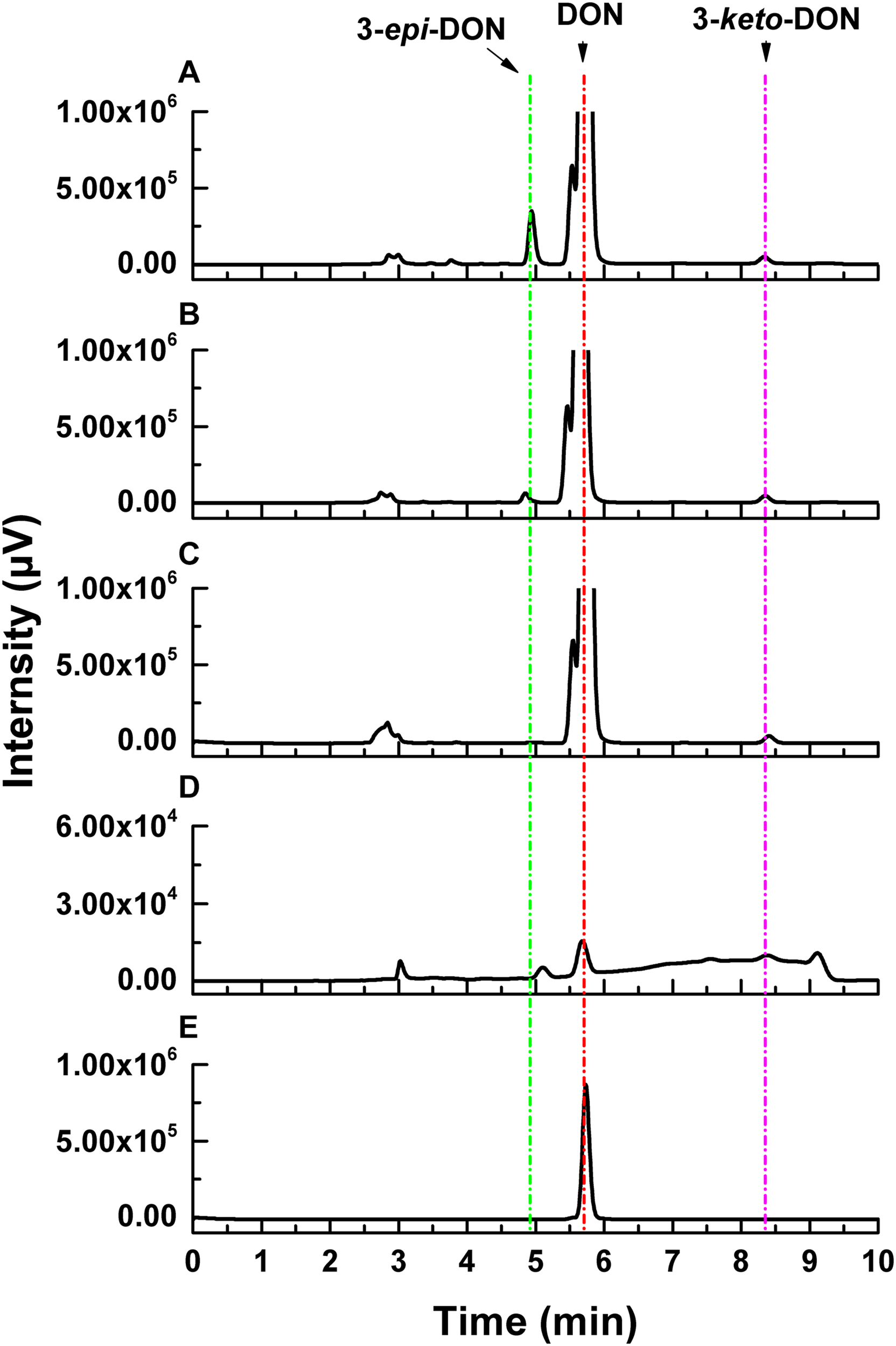
High-performance liquid chromatography analysis of DON degradation by Nocardioides sp. ZHH-013 after (A) 8 h, (B) 4 h, (C) 0.5 h, and of (D) 3-keto-DON produced by DepA. (E) The 3.37 mM DON standard.
FIGURE 5
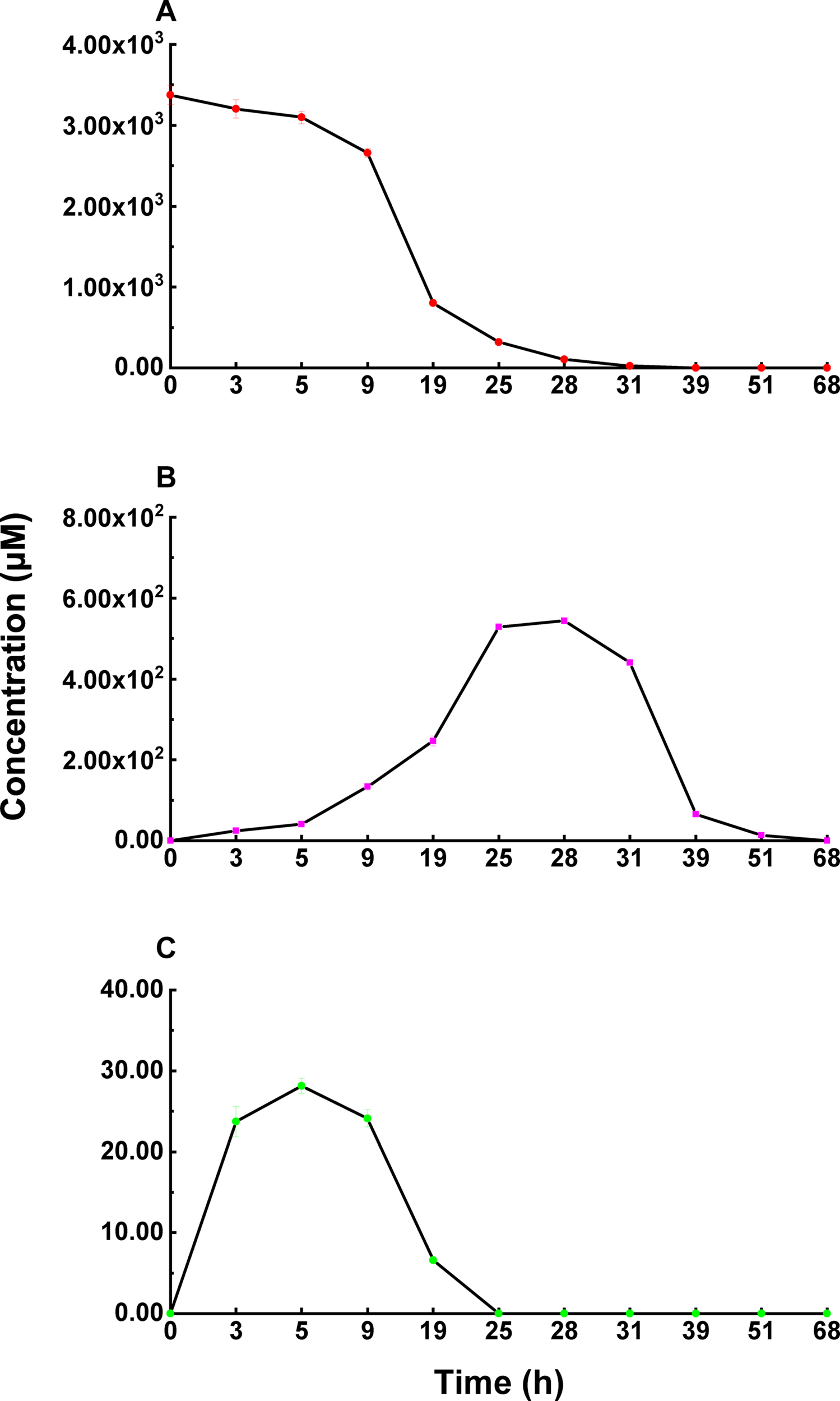
The concentration changes of 3-epi-DON (B) and 3-keto-DON (C) in the process of DON (A) degradation by Nocardioides sp. ZHH-013.
Karlovsky speculated that Nocardioides sp. WSN05-2 may provide a high ratio of oxidation and reduction rates, leading to low accumulation of the intermediate product (3-keto-DON) (Karlovsky, 2011); however, no experimental evidence was published. Using the ZHH-013 strain, we observed very little accumulation of DON degradation product, 3-keto-DON, unlike reports of experiments using DepA obtained from Devosia sp. 17-2-E-8 (Figures 4, 5C); however, after further analysis, a new peak (RT = 4.06 min) was identified as 3-keto-DON. MS and MS/MS data from that compound revealed a predominant ion at m/z 293.10, consistent with the molecular ion [DON–2–H]– (Supplementary Figures 4B,C), similar to that of 3-keto-DON prepared from DON using DepA (Supplementary Figures 3B,C). The difference of 2 atomic mass units between the ions at m/z 293.10 [DON–2H–H]– and m/z 297.13 [DON + H]+ indicates that two protons were absent. 3-keto-DON disappeared when DON was depleted, and no 3-keto-DON accumulation was found when 3-epi-DON was degraded. Therefore, we presumed that DON must have been converted to 3-keto-DON before then being converted to 3-epi-DON (Figure 6) and that the efficient conversion ability of Nocardioides sp. ZHH-013 may have led to low accumulation of 3-keto-DON.
FIGURE 6
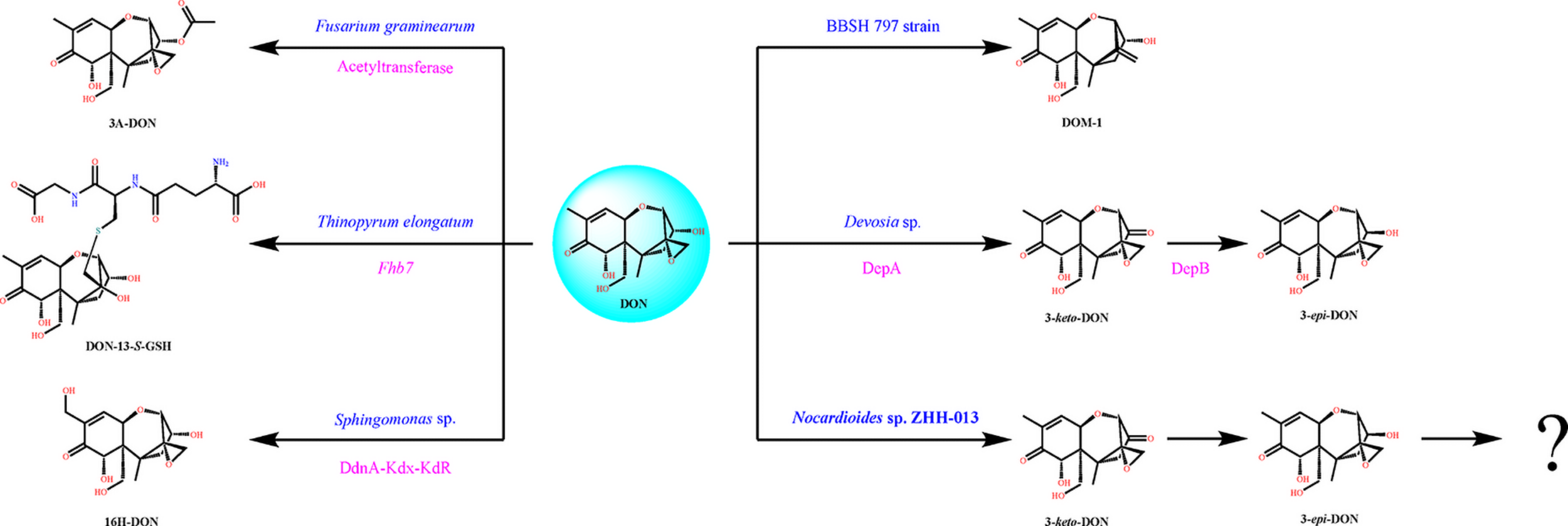
Typical DON degradation pathways of different microorganisms including Nocardioides sp. ZHH-013. The blue characters denote microorganism name, and the rose red characters denote identified gene or enzyme.
The toxicity of DON degradation products is a core question for strain or enzyme applications. The toxicity of DON has been fully studied, while in vitro and vivo toxicological data on its derivatives remain very scarce. The 12,13-epoxide ring, hydroxyl group at C-15, hydroxyl group at C-3, and double bond between C-9 and C-10 are crucial DON toxic groups (McCormick et al., 2011). DON inhibits nucleic acid synthesis by binding to peptidyl transferase in the 60S ribosomal subunit, inducing ribotoxic stress, and activating MAPKinases (Maresca, 2013; Garreau de Loubresse et al., 2014). Unlike DON, 3-epi-DON does not activate MAPKinases (Pierron et al., 2016). The toxicity of DON derivatives created by microorganisms has been tested using BrdU bioassays (Widestrand et al., 1999; Sundstøl et al., 2004). The toxicity of 3-epi-DON is substantially lower than that of DON and its derivatives, while excessive exposure to 3-epi-DON still caused organ lesions in mice (He et al., 2015a). Moreover, exposure to 3-epi-DON caused slight changes in intestinal explants (Pierron et al., 2016); however, 3-epi-DON showed almost no hepatotoxicity or immunotoxicity in in vivo pig experiments (Bracarense et al., 2020). Therefore, 3-epi-DON may represent a low-toxicity, safe DON degradation product in mice and pigs. The toxicity of 3-epi-DON is potentially due to the 12,13-epoxide ring and the double bond between C-9 and C-10 (Wei and McLaughlin, 1974). Therefore, elimination of 3-epi-DON may be necessary. Nocardioides sp. ZHH-013 metabolizes 3-epi-DON, and we speculate that it may induce the oxygen heterocyclic form of 3-epi-DON to undergo a ring-opening reaction in one or more steps, and that the toxicity of DON can thereby be eliminated.
Conclusion
Biodegradation is an effective strategy to reduce toxin exposure risks and economic losses. There are five important issues that have been hindering the development of industrial bacteria and enzyme preparations: (1) the lack of strains or enzyme sources, (2) the safety of the transformed products, (3) the lack of compound toxicology data, (4) the safety of the strains, and (5) the industrial application of the strains and enzymes.
In this paper, we report a novel DON-degrading strain, Nocardioides sp. ZHH-013, isolated in China. ZHH-013 can degrade DON to 3-keto-DON and 3-epi-DON, achieving DON mineralization. The DON degradation mechanism of this strain may inspire the development of attenuated agents or bio-detoxifiers. Determining the mechanisms underlying the efficient generation and degradation of 3-epi-DON may lead to the discovery of new enzymes. Furthermore, there is potential to improve enzyme activity, expression level, and adaptability via enzyme engineering technology, which would help to realize the application of DON-degrading enzymes to eliminate DON pollution in food/feed fields or to restore DON-polluted bodies of water on farms.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW493343.
Author contributions
HoZ and HeZ performed the experiments. FZ and YB designed and performed the synergism experiments and analyzed the data. XQ, XW, and XX designed the research and collected soil samples. HoZ, FZ, and HL revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Young Scientist Start-up funding of Beijing Forestry University (No. BLX201910), the China Postdoctoral Science Foundation (No. M670178), the National Key Research and Development Program of China (2016YFD0501409-02), and the China Agriculture Research System (CARS-41).
Acknowledgments
We would like to thank Chenghang Sun, from the Chinese Academy of Medical Sciences & Peking Union Medical College, China, and Qiliang Lai, Third Institute of Oceanography, Ministry of Natural Resources, China, for their assistance in microbe identification. We would like to thank Mengting Liu for experimental assistance. We would also like to thank Yang Wang, from the Academy of National Food and Strategic Reserves Administration, China, for reagent preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.658421/full#supplementary-material
Abbreviations
- 15-ADON
15-acetyl DON
- 3-epi-DON
3-epi-deoxynivalenol
- 3-keto-DON
3-keto-deoxynivalenol
- Dep
DON epimerization
- DOM-1
de-epoxy DON
- DON
deoxynivalenol
- HPLC
high-performance liquid chromatography
- LB
Luria-Bertani
- MM
mineral salt medium
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NMR
nuclear magnetic resonance
- RT
retention time
- TSB
tryptic soy broth
- UPLC
ultra-performance liquid chromatography.
References
1
Bai G. Shaner G. (2004). Management and resistance in wheat and barley to Fusarium head blight.Annu. Rev. Phytopathol.42135–161. 10.1146/annurev.phyto.42.040803.140340
2
Bai X. Li H. Zhang Z. Zhang X. Wang C. Xu J. et al (2019). Carbon nitride nested tubes with graphene as a dual electron mediator in Z-scheme photocatalytic deoxynivalenol degradation.Catal. Sci. Technol.91680–1690. 10.1039/C9CY00209J
3
Bracarense A. P. F. L. Pierron A. Pinton P. Gerez J. R. Schatzmayr G. Moll W. D. et al (2020). Reduced toxicity of 3-epi-deoxynivalenol and de-epoxy-deoxynivalenol through deoxynivalenol bacterial biotransformation: in vivo analysis in piglets.Food Chem. Toxicol.140:111241. 10.1016/j.fct.2020.111241
4
Bretz M. Beyer M. Cramer B. Knecht A. Humpf H. U. (2006). Thermal degradation of the Fusarium mycotoxin deoxynivalenol.J. Agric. Food Chem.546445–6451. 10.1021/jf061008g
5
Carere J. Hassan Y. I. Lepp D. Zhou T. (2018a). The enzymatic detoxification of the mycotoxin deoxynivalenol: identification of DepA from the DON epimerization pathway.Microb. Biotechnol.111106–1111. 10.1111/1751-7915.12874
6
Carere J. Hassan Y. I. Lepp D. Zhou T. (2018b). The identification of DepB: an enzyme responsible for the final detoxification step in the deoxynivalenol epimerization pathway in Devosia mutans 17-2-E-8.Front. Microbiol.9:1573. 10.3389/fmicb.2018.01573
7
Chen D. Chen P. Cheng Y. Peng P. Liu J. Ma Y. et al (2019). Deoxynivalenol decontamination in raw and germinating barley treated by plasma-activated water and intense pulsed light.Food Bioproc. Tech.12246–254. 10.1007/s11947-018-2206-2
8
Eagles E. J. Benstead R. MacDonald S. Handy R. D. Hutchinson T. H. (2021). Environmental risks to freshwater organisms from the mycotoxins deoxynivalenol and zearalenone using species sensitivity distributions.Chemosphere267:129279. 10.1016/j.chemosphere.2020.129279
9
Fuchs E. Binder E. M. Heidler D. Krska R. (2002). Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797.Food Addit. Contam.19379–386. 10.1080/02652030110091154
10
Gao X. Mu P. Wen J. Sun Y. Chen Q. Deng Y. (2018). Detoxification of trichothecene mycotoxins by a novel bacterium, Eggerthella sp. DII-9.Food Chem. Toxicol.112310–319. 10.1016/j.fct.2017.12.066
11
Garreau de Loubresse N. Prokhorova I. Holtkamp W. Rodnina M. V. Yusupova G. Yusupov M. (2014). Structural basis for the inhibition of the eukaryotic ribosome.Nature513517–522. 10.1038/nature13737
12
Gracia-Lor E. Zuccato E. Hernández F. Castiglioni S. (2020). Wastewater-based epidemiology for tracking human exposure to mycotoxins.J. Hazard. Mater.382:121108. 10.1016/j.jhazmat.2019.121108
13
Grenier B. Bracarense A. P. F. L. Schwartz H. E. Lucioli J. Cossalter A. Moll W. et al (2013). Biotransformation approaches to alleviate the effects induced by Fusarium mycotoxins in swine.J. Agric. Food Chem.616711–6719. 10.1021/jf400213q
14
Gunther A. Siegrid D. B. Mathias D. Van I. F. An M. Siska C. (2017). Feed contamination with Fusarium mycotoxins induces a corticosterone stress response in broiler chickens.Poult. Sci.9614–17. 10.3382/ps/pew280
15
Hassan Y. Zhu H. Zhu Y. Zhou T. (2016). Beyond ribosomal binding: the increased polarity and aberrant molecular interactions of 3-epi-deoxynivalenol.Toxins8:261. 10.3390/toxins8090261
16
He C. Fan Y. Liu G. Zhang H. (2008). Isolation and identification of a strain of Aspergillus tubingensis with deoxynivalenol biotransformation capability.Int. J. Mol. Sci.92366–2375. 10.3390/ijms9122366
17
He J. W. Bondy G. S. Zhou T. Caldwell D. Boland G. J. Scott P. M. (2015a). Toxicology of 3-epi-deoxynivalenol, a deoxynivalenol-transformation product by Devosia mutans 17-2-E-8.Food Chem. Toxicol.84250–259. 10.1016/j.fct.2015.09.003
18
He J. W. Yang R. Zhou T. Boland G. J. Scott P. M. Bondy G. S. (2015b). An epimer of deoxynivalenol: purification and structure identification of 3-epi-deoxynivalenol.Food Addit. Contam.321523–1530. 10.1080/19440049.2015.1072771
19
He J. W. Hassan Y. I. Perilla N. Li X. Z. Boland G. J. Zhou T. (2016). Bacterial epimerization as a route for deoxynivalenol detoxification: the influence of growth and environmental conditions.Front. Microbiol.7:572. 10.3389/fmicb.2016.00572
20
He P. Young L. G. Forsberg C. (1992). Microbial transformation of deoxynivalenol (vomitoxin).Appl. Environ. Microbiol.583857–3863. 10.1128/AEM.58.12.3857-3863.1992
21
He W. J. Shi M. M. Yang P. Huang T. Yuan Q. S. Yi S. Y. et al (2020). Novel soil bacterium strain Desulfitobacterium sp. PGC-3-9 detoxifies trichothecene mycotoxins in wheat via de-epoxidation under aerobic and anaerobic conditions.Toxins12:363. 10.3390/toxins12060363
22
He W. J. Zhang L. Yi S. Y. Tang X. L. Yuan Q. S. Guo M. W. et al (2017). An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain.Sci. Rep.7:9549. 10.1038/s41598-017-08799-w
23
Higgins D. Thompson J. Gibson T. Thompson J. D. Higgins D. G. Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.Nucleic Acids Res.224673–4680. 10.1093/nar/22.22.4673
24
Ikunaga Y. Sato I. Grond S. Numaziri N. Yoshida S. Yamaya H. et al (2011). Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol.Appl. Microbiol. Biotechnol.89419–427. 10.1007/s00253-010-2857-z
25
Ito M. Sato I. Ishizaka M. Yoshida S. Koitabashi M. Yoshida S. et al (2013). Bacterial cytochrome P450 system catabolizing the Fusarium toxin deoxynivalenol.Appl. Environ. Microbiol.791619–1628. 10.1128/aem.03227-12
26
Ito M. Sato I. Koitabashi M. Yoshida S. Imai M. Tsushima S. (2012). A novel actinomycete derived from wheat heads degrades deoxynivalenol in the grain of wheat and barley affected by Fusarium head blight.Appl. Microbiol. Biotechnol.961059–1070. 10.1007/s00253-012-3922-6
27
Jaqueline G. B. Eliana B. F. (2010). Kinetics of deoxynivalenol degradation by Aspergillus oryzae and Rhizopus oryzae in submerged fermentation.J. Brazil. Chem. Soc.21710–714. 10.1590/S0103-50532010000400018
28
Jaqueline G. B. Larine K. Eliana B. F. (2011). Deoxynivalenol (DON) degradation and peroxidase enzyme activity in submerged fermentation.Ciência e Tecnologia de Alimentos31198–203. 10.1590/S0101-20612011000100030
29
Johnson M. Zaretskaya I. Raytselis Y. Merezhuk Y. McGinnis S. Madden T. L. (2008). NCBI BLAST: a better web interface.Nucleic Acids Res.36W5–W9. 10.1093/nar/gkn201
30
Juan-García A. Juan C. Tolosa J. Ruiz M. (2019). Effects of deoxynivalenol, 3-acetyl-deoxynivalenol and 15-acetyl-deoxynivalenol on parameters associated with oxidative stress in HepG2 cells.Mycotoxin Res.35197–205. 10.1007/s12550-019-00344-0
31
Jurisic N. Schwartz-Zimmermann H. E. Kunz-Vekiru E. Reisinger N. Klein S. Caldwell D. et al (2019). Deoxynivalenol-3-sulphate is the major metabolite of dietary deoxynivalenol in eggs of laying hens.World Mycotoxin J.12245–255. 10.3920/WMJ2018.2429
32
Karlovsky P. (2011). Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives.Appl. Microbiol. Biotechnol.91491–504. 10.1007/s00253-011-3401-5
33
Kim M. Oh H.-S. Park S.-C. Chun J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes.Int. J. Syst. Evol. Microbiol.64346–351. 10.1099/ijs.0.059774-0
34
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms.Mol. Biol. Evol.351547–1549.
35
Li M. Guan E. Bian K. (2019). Structure elucidation and toxicity analysis of the degradation products of deoxynivalenol by gaseous ozone.Toxins11:474. 10.3390/toxins11080474
36
Li X. Z. Zhu C. de Lange C. F. M. Zhou T. He J. Yu H. et al (2011). Efficacy of detoxification of deoxynivalenol-contaminated corn by Bacillus sp. LS100 in reducing the adverse effects of the mycotoxin on swine growth performance.Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess.28894–901. 10.1080/19440049.2011.576402
37
Lucke A. Doupovec B. Paulsen P. Zebeli Q. Böhm J. (2017). Effects of low to moderate levels of deoxynivalenol on feed and water intake, weight gain, and slaughtering traits of broiler chickens.Mycotoxin Res.33261–271. 10.1007/s12550-017-0284-z
38
Maresca M. (2013). From the gut to the brain: journey and pathophysiological effects of the food-associated trichothecene mycotoxin deoxynivalenol.Toxins5784–820. 10.3390/toxins5040784
39
McCormick S. P. Stanley A. M. Stover N. A. Alexander N. J. (2011). Trichothecenes: from simple to complex mycotoxins.Toxins3802–814. 10.3390/toxins3070802
40
Miller J. D. Arnison P. G. (1986). Degradation of deoxynivalenol by suspension cultures of the Fusarium head blight resistant wheat cultivar Frontana.Can. J. Plant Pathol.8147–150. 10.1080/07060668609501818
41
Pierron A. Mimoun S. Murate L. S. Loiseau N. Lippi Y. Bracarense A. P. F. L. et al (2016). Microbial biotransformation of DON: molecular basis for reduced toxicity.Sci. Rep.6:29105. 10.1038/srep29105
42
Santos Alexandre A. P. Vela-Paredes R. S. Santos A. S. Costa N. S. Canniatti-Brazaca S. G. Calori-Domingues M. A. et al (2018). Ozone treatment to reduce deoxynivalenol (DON) and zearalenone (ZEN) contamination in wheat bran and its impact on nutritional quality.Food Addit. Contam.351189–1199. 10.1080/19440049.2018.1432899
43
Sato I. Ito M. Ishizaka M. Ikunaga Y. Sato Y. Yoshida S. et al (2012). Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms.FEMS Microbiol. Lett.327110–117. 10.1111/j.1574-6968.2011.02461.x
44
Springler A. Hessenberger S. Reisinger N. Kern C. Nagl V. Schatzmayr G. et al (2017). Deoxynivalenol and its metabolite deepoxy-deoxynivalenol: multi-parameter analysis for the evaluation of cytotoxicity and cellular effects.Mycotoxin Res.3325–37. 10.1007/s12550-016-0260-z
45
Sundstøl E. G. Pettersson H. Lundh T. (2004). Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites.Food Chem. Toxicol.42619–624. 10.1016/j.fct.2003.11.006
46
Wang G. Wang Y. Ji F. Xu L. Yu M. Shi J. et al (2019). Biodegradation of deoxynivalenol and its derivatives by Devosia insulae A16.Food Chem.276436–442. 10.1016/j.foodchem.2018.10.011
47
Wang Y. Zhang H. H. Zhao C. Han Y. T. Liu Y. C. Zhang X. L. (2017). Isolation and characterization of a novel deoxynivalenol-transforming strain Paradevosia shaoguanensis DDB001 from wheat field soil.Lett. Appl. Microbiol.65414–422. 10.1111/lam.12790
48
Wei C. McLaughlin C. S. (1974). Structure-function relationship in the 12,13-epoxytrichothecenes novel inhibitors of protein synthesis.Biochem. Biophys. Res. Commun.57838–844. 10.1016/0006-291X(74)90622-6
49
Wettstein F. E. Bucheli T. D. (2010). Poor elimination rates in waste water treatment plants lead to continuous emission of deoxynivalenol into the aquatic environment.Water Res.444137–4142. 10.1016/j.watres.2010.05.038
50
Widestrand J. Lundh T. Pettersson H. Lindberg J. E. (1999). Cytotoxicity of four trichothecenes evaluated by three colorimetric bioassays.Mycopathologia147149–155.
51
Xu X. Nicholson P. (2009). Community ecology of fungal pathogens causing wheat head blight.Annu. Rev. Phytopathol.4783–103. 10.1146/annurev-phyto-080508-081737
52
Yang S. Wu Y. Yang J. Yan R. Bao Y. Wang K. et al (2017). Isolation and identification of an extracellular enzyme from Aspergillus Niger with deoxynivalenol biotransformation capability.Emir. J. Food Agric.29:742. 10.9755/ejfa.2017.v29.i10.1295
53
Yoon S. Ha S. Kwon S. Lim J. Kim Y. Seo H. et al (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies.Int. J. Syst. Evol. Microbiol.671613–1617. 10.1099/ijsem.0.001755
54
Zhai Y. Zhong L. Gao H. Lu Z. Bie X. Zhao H. et al (2019). Detoxification of deoxynivalenol by a mixed culture of soil bacteria with 3-epi-deoxynivalenol as the main intermediate.Front. Microbiol.10:30. 10.3389/fmicb.2019.02172
55
Zhang H. H. Wang Y. Zhao C. Wang J. Zhang X. L. (2017). Biodegradation of ochratoxin A by Alcaligenes faecalis isolated from soil.J. Appl. Microbiol.123661–668. 10.1111/jam.13537
56
Zhang J. Qin X. Guo Y. Zhang Q. Ma Q. Ji C. et al (2020). Enzymatic degradation of deoxynivalenol by a novel bacterium, Pelagibacterium halotolerans ANSP101.Food Chem. Toxicol.140:111276. 10.1016/j.fct.2020.111276
57
Zhou Y. Wu S. Wang F. Li Q. He C. Duan N. et al (2020). Assessing the toxicity in vitro of degradation products from deoxynivalenol photocatalytic degradation by using upconversion nanoparticles@TiO2 composite.Chemosphere238:124648. 10.1016/j.chemosphere.2019.124648
Summary
Keywords
deoxynivalenol, 3-epi-DON, 3-keto-DON, Nocardioides , biodegradation
Citation
Zhang H, Zhang H, Qin X, Wang X, Wang Y, Bin Y, Xie X, Zheng F and Luo H (2021) Biodegradation of Deoxynivalenol by Nocardioides sp. ZHH-013: 3-keto-Deoxynivalenol and 3-epi-Deoxynivalenol as Intermediate Products. Front. Microbiol. 12:658421. doi: 10.3389/fmicb.2021.658421
Received
25 January 2021
Accepted
18 June 2021
Published
19 July 2021
Volume
12 - 2021
Edited by
Hendrik Schäfer, University of Warwick, United Kingdom
Reviewed by
Barbara Weber, Biomin Holding GmbH, Austria; Roberta Fulthorpe, University of Toronto Scarborough, Canada
Updates
Copyright
© 2021 Zhang, Zhang, Qin, Wang, Wang, Bin, Xie, Zheng and Luo.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Zheng, zhengfei0718@sina.comHuiying Luo, luohuiying@caas.cn
†These authors have contributed equally to this work
This article was submitted to Microbiotechnology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.