- Department of Microbiology, University of Illinois at Urbana-Champaign, Urbana, IL, United States
In bacteria, DnaA is the most conserved DNA replication initiator protein. DnaA is a DNA binding protein that is part of the AAA+ ATPase family. In addition to initiating chromosome replication, DnaA can also function as a transcription factor either as an activator or repressor. The first gene identified to be regulated by DnaA at the transcriptional levels was dnaA. DnaA has been shown to regulate genes involved in a variety of cellular events including those that trigger sporulation, DNA repair, and cell cycle regulation. DnaA’s dual functions (replication initiator and transcription factor) is a potential mechanism for DnaA to temporally coordinate diverse cellular events with the onset of chromosome replication. This strategy of using chromosome replication initiator proteins as regulators of gene expression has also been observed in archaea and eukaryotes. In this mini review, we focus on our current understanding of DnaA’s transcriptional activity in various bacterial species.
Introduction
DnaA is a multifunctional protein that can serve as a master regulator in bacteria. DnaA is composed of four structural domains with some species-specific variations: (I) protein–protein interaction domain, (II) linker domain, (III) AAA+ ATPase domain, and (IV) DNA binding (helix–turn–helix) domain (Fujikawa et al., 2003; Duderstadt and Berger, 2013) (Figure 1A). The two functions of DnaA that are best understood are as an initiator of chromosome replication and as a transcription factor. DnaA and/or the onset of chromosome replication have been linked to cell size regulation and chromosome segregation; however, details about these links remain unclear (Lobner-Olesen et al., 1989; Hill et al., 2012; Mera et al., 2014). As a replication initiator, DnaA opens the origin of replication (ori) by binding at specific DNA sequences referred to as DnaA boxes (Fuller and Kornberg, 1983; Bramhill and Kornberg, 1988). This function has been extensively studied, and mechanistic questions continue to be articulated (recent reviews; Hansen and Atlung, 2018; Frandi and Collier, 2019; Leonard et al., 2019; Ozaki, 2019; Kohiyama, 2020). In this mini review, we focus on the transcriptional activity of DnaA and its role in modulating various cellular events (Figure 1B). DnaA has been shown to have a global transcriptional impact because it regulates the expression levels of other global regulators involved in cell cycle progression and developmental processes (Figure 2). The specific genes found in DnaA’s transcriptional regulon vary significantly depending on the bacterial species. To begin, we will discuss the ability of DnaA to autoregulate its levels as a way to provide context for the importance of its transcriptional activity.
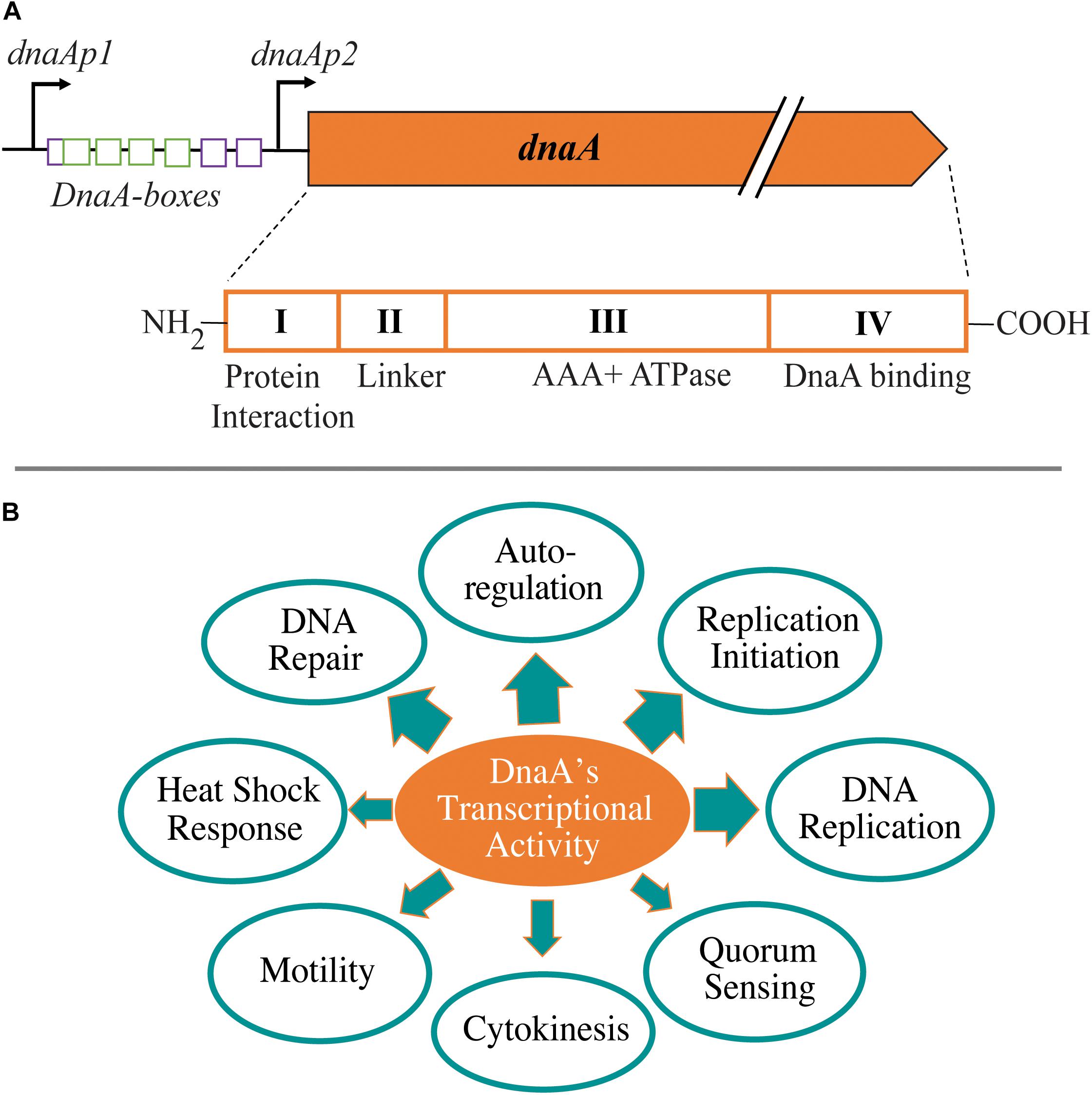
Figure 1. Cellular impact from transcriptional activity of DnaA. (A) Schematic of the promoter region of dnaA in Escherichia coli and conserved domains of the DnaA protein. Within the promoter region, purple boxes depict ATP-specific DnaA boxes and green boxes have no nucleotide-bound DnaA specificity. (B) Cellular processes from different bacterial species shown to be influenced by DnaA’s transcriptional activity. Thickness of arrow is an approximate representation of the amount of evidence that exists for that particular process.
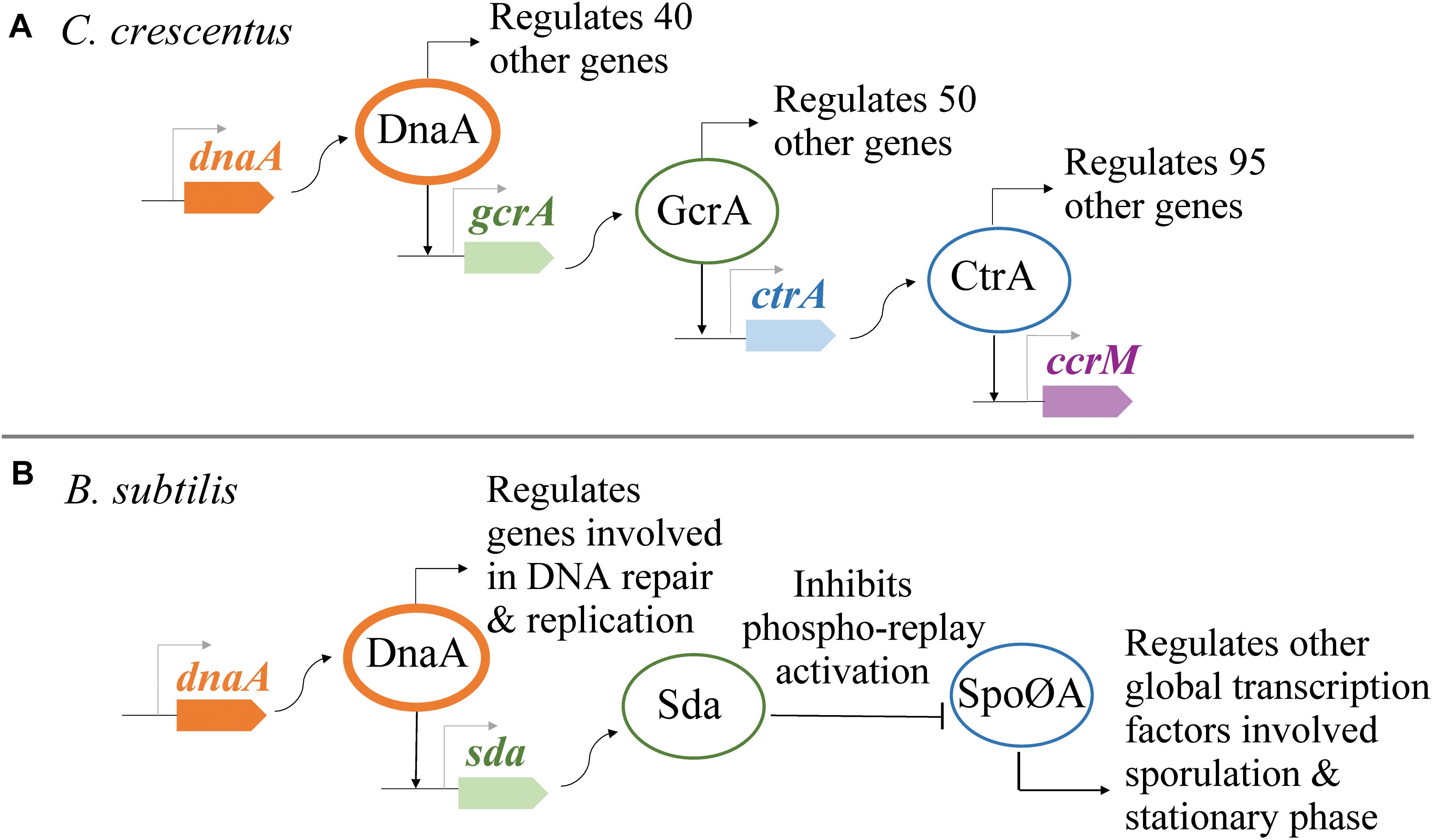
Figure 2. DnaA regulates the transcription of other global regulators. (A) In Caulobacter crescentus, DnaA is part of a genetic network responsible for regulating the forward progression of the cell cycle. (B) In Bacillus subtilis, DnaA indirectly inhibits the phosphorylation of Spo0A, a global regulator of sporulation and entrance to stationary phase.
Autoregulation of DnaA Expression
The ability of DnaA to autoregulate its transcription was discovered at a time when the direct role of DnaA in chromosome replication was not yet clear. Early characterizations of temperature-sensitive dnaA mutants of Escherichia coli revealed that the copy number of ori decreased, while DnaA levels increased simultaneously at nonpermissive temperatures, thus accurately predicting that DnaA had a positive involvement in the onset of chromosome replication and a negative involvement in regulating its own synthesis (Hanna and Carl, 1975; Hansen and Rasmussen, 1977). DnaA’s ability to repress its own transcription was later confirmed using in vitro and in vivo analyses. In vitro, DnaA was shown to directly bind dnaA’s promoter region using gel shift assays and DNase footprinting methods (Braun et al., 1985; Wang and Kaguni, 1987). In vivo, two strategies were used to show that DnaA represses its own transcription. First, increased cellular levels of DnaA (expressed from inducible promoters) were shown to decrease the activity of the dnaA promoter, and second, decreased DnaA levels able to bind the dnaA promoter (by adding dnaA binding sites on a plasmid that titrated DnaA levels away) were shown to increase the activity of the dnaA promoter (Atlung et al., 1985; Braun et al., 1985; Kucherer et al., 1986; Hansen et al., 1987).
In E. coli, the expression of dnaA is regulated from two promoters: dnaAp1 and dnaAp2 ((Hansen E. B. et al., 1982; Hansen F. G. et al., 1982) (Figure 1A). DnaA can repress its own expression from both promoters with dnaAp2 being a threefold stronger promoter than dnaAp1 (Atlung et al., 1984, 1985; Braun et al., 1985; Kucherer et al., 1986; Chiaramello and Zyskind, 1990). A 9-mer DnaA box with the consensus sequence TTATCCACA was first identified between dnaAp1 and dnaAp2 explaining the mechanism of autoregulation (Hansen F. G. et al., 1982). Intriguingly, the elimination of this DnaA box on dnaA’s promoter did not fully eliminate the autoregulation of dnaA expression, leading to the hypothesis of indirect mechanisms (Polaczek and Wright, 1990; Smith et al., 1997). The mystery was solved by the identification of DnaA-ATP-specific DnaA boxes (AGATCT) that were involved in full repression of dnaA expression (Speck et al., 1999). In addition to the autoregulation by DnaA, the transcription of E. coli dnaA is regulated at multiple levels and by various other proteins, including DNA methylation, second messenger ppGpp, growth rate, SeqA, Fis, IciA (ArgP), and QseB (Braun and Wright, 1986; Kucherer et al., 1986; Campbell and Kleckner, 1990; Chiaramello and Zyskind, 1990; Polaczek and Wright, 1990; Zyskind and Smith, 1992; Lu et al., 1994; Froelich et al., 1996; Lee et al., 1996; Hansen and Atlung, 2018; Riber and Lobner-Olesen, 2020; Wu et al., 2021).
Besides E. coli, autoregulation by DnaA directly binding at its promoter region has been confirmed in Bacillus subtilis (Ogura et al., 2001), Vibrio harveyi (Berenstein et al., 2002), Streptomyces lividans (Zakrzewska-Czerwinska et al., 1994; Jakimowicz et al., 2000), and in slow-growing mycobacteria (Salazar et al., 2003). In Pseudomonas putida, 12 DnaA boxes were identified on its dnaA promoter region (Fujita et al., 1989). However, when the levels of DnaA from P. putida were increased (by inducible expression from plasmid), repression of dnaA expression was not observed in P. putida. Interestingly, the overexpression of dnaA (only the open reading frame) from E. coli under the same broad host range plasmid construct in P. putida does result in repression of dnaA’s transcription (Ingmer and Atlung, 1992). The function, if any, of the 12 DnaA boxes in P. putida dnaA promoter remains to be determined. Likewise, in Caulobacter crescentus, the role that DnaA plays in its transcriptional regulation remains unclear. Transcription of dnaA in C. crescentus has been proposed to be regulated by methylation status of the promoter region and by a cis-acting element upstream of the -35 promoter region (Collier et al., 2007; Cheng and Keiler, 2009; Felletti et al., 2019; Frandi and Collier, 2019).
Nucleotide Switch Regulates the Transcriptional Activity of DnaA
Both activities of DnaA (transcription factor and replication initiator) can be modulated using an ATP-dependent molecular switch. In E. coli, DnaA can be found inside the cell tightly bound to ATP (Kd 30 nM) or bound to ADP (Kd 100 nM) (Sekimizu et al., 1987). The protein Hda (homologous to DnaA) and chromosomal loci datA promote hydrolysis of DnaA–ATP to DnaA–ADP in E. coli (Kato and Katayama, 2001; Kasho and Katayama, 2013). The cellular ratio of DnaA–ATP to DnaA–ADP changes as the cell progresses over the cell cycle. Although DnaA–ATP and DnaA–ADP bind ori, only DnaA–ATP can open ori and initiate replication in E. coli (Sekimizu et al., 1987; Mizushima et al., 1996). As a transcription factor, the nucleotide switch has been shown to turn DnaA into a stronger repressor or stronger activator depending on the bound nucleotide. The ability of DnaA’s transcriptional activity to be regulated with this switch was first identified in the autoregulation of dnaA’s transcription in E. coli (Speck et al., 1999). DnaA–ATP is able to fully repress the expression of dnaA, whereas DnaA–ADP can only repress up to 40%. By distinguishing the nucleotide bound to DnaA, four new DnaA boxes were identified in the promoter region of dnaA with specificity for DnaA–ATP and potential cooperative binding (Speck et al., 1999).
The second example of this switch involves the synthesis of DNA substrates in E. coli. Ribonucleotide reductase (RNR) encoded by nrdAB catalyzes the reduction of ribonucleotides to deoxyribonucleotides (Thelander and Reichard, 1979). In E. coli, DnaA regulates the expression of nrdAB by binding the promoter region that contains three DnaA boxes: two boxes do not have specificity for the nucleotide-bound DnaA, and one box is specific for DnaA–ATP (Tuggle and Fuchs, 1986; Augustin et al., 1994; Jacobson and Fuchs, 1998; Olliver et al., 2010). DnaA can activate or repress the expression of nrdAB based on the nucleotide bound to DnaA and based on the levels of DnaA. For instance, cells expressing DnaA variants that are deficient for ATP binding or hyperactive for ATPase activity display an increased expression of nrdAB (Gon et al., 2006; Babu et al., 2017). Thus, high levels of DnaA-ATP repress the transcription of nrdAB presumably by precluding RNA polymerase from binding the promoter region (Olliver et al., 2010). Conversely, DnaA–ADP or low levels of DnaA–ATP bound at the high-affinity DnaA boxes in nrdAB promoter region can activate nrdAB transcription by stabilizing the RNA polymerase–DNA complex (Augustin et al., 1994; Olliver et al., 2010). Although the expression of RNR coincides with the onset of chromosome replication (Sun and Fuchs, 1992), DnaA seems to only regulate the expression levels and not necessarily the timing of nrdAB expression (Sun et al., 1994; Olliver et al., 2010). Interestingly, the expression of dnaA and nrdAB in E. coli share two other regulators besides DnaA: Fis and IciA (Augustin et al., 1994; Froelich et al., 1996; Lee et al., 1996; Han et al., 1998). In B. subtilis and C. crescentus, DnaA has also been proposed to act as a transcriptional regulator of genes encoding RNR (Goranov et al., 2005; Hottes et al., 2005), although no detailed analyses has been performed to determine if DnaA’s nucleotide switch is also involved.
In C. crescentus, DnaA-ADP (not DnaA-ATP) has been proposed to activate the transcription of a set of three essential genes encoding proteins involved in cell cycle regulation: FtsZ (tubulin-like protein essential for cell division), MipZ (inhibitor of FtsZ polymerization), and GcrA (global transcription factor) (Fernandez-Fernandez et al., 2011). The expression of the hyperactive replication initiator variant DnaAR357A resulted in C. crescentus cells over-initiating replication (Fernandez-Fernandez et al., 2011; Wargachuk and Marczynski, 2015). The corresponding variant in E. coli (DnaAR334A) was shown to bind ATP but was unable to hydrolyze ATP (Nishida et al., 2002). In C. crescentus, DnaAR357A variant was shown to lose its ability to activate the transcription of ftsZ, mipZ, and gcrA (Hottes et al., 2005; Fernandez-Fernandez et al., 2011). In B. subtilis, most chromosomal regions that bind DnaA displayed higher specificity for DnaA-ATP over DnaA-ADP in vitro (Smith and Grossman, 2015), suggesting a potential wide usage of DnaA’s nucleotide switch to modulate the transcriptional activity of DnaA. However, the mechanism(s) that regulate differences in DNA binding specificity between DnaA–ATP and DnaA–ADP remain unclear.
DnaA’s Global Transcriptional Regulation
Identifying the complete transcriptional regulon of DnaA is complex due to the essential function of DnaA as a replication initiator and the role that replication initiation plays in the progression of the cell cycle. Work on C. crescentus and B. subtilis utilized innovative approaches to separate DnaA’s transcriptional activity from its function as a replication initiator (Hottes et al., 2005; Washington et al., 2017). One major finding from these whole-cell transcriptional analyses is that the transcriptional activity of DnaA can have a global effect on the cell. Aside from DnaA regulating the expression of various important genes, DnaA has also been found to be part of genetic networks where DnaA (directly or indirectly) regulates the expression of other transcription factors that are themselves global regulators (Figure 2). A global analysis of DnaA’s transcriptional activity remains to be determined for E. coli.
To identify the transcriptional regulon of DnaA in C. crescentus, Hottes et al. (2005) took advantage of the ability to synchronize the Caulobacter cell cycle in a strain with dnaA’s expression regulated from an inducible promoter. DnaA-dependent and DnaA-independent changes were identified by comparing transcriptional profiles of cells grown expressing dnaA with cells whose dnaA expression was delayed. This study focused on DnaA-dependent changes of genes whose expression increased during the transition from G1 to S phase. Of the 40 genes identified to be DnaA-dependent, 13 genes included DnaA boxes on their promoter region, and three genes (gcrA, ftsZ, and podJ) were shown in vitro to have promoter regions with affinity for purified His6DnaA. Notably, one of the DnaA-dependent genes identified was the gene encoding for GcrA, the global regulator of Caulobacter cell cycle (Holtzendorff et al., 2004) (Figure 2). The role of DnaA in the progression of the cell cycle has been predicted to be widely conserved among alpha proteobacteria (Panis et al., 2015). In the two alpha proteobacteria model systems C. crescentus (Hottes et al., 2005; Collier et al., 2006) and the plant symbiont Sinorhizobium meliloti (De Nisco et al., 2014), DnaA has been shown to be a key component of a closed genetic network that drives the progression through G1–S–G2.
In B. subtilis, the transcriptional regulon of DnaA has been characterized using various in vitro and in vivo high-throughput analyses (Goranov et al., 2005; Ishikawa et al., 2007; Cho et al., 2008; Breier and Grossman, 2009; Hoover et al., 2010; Smith and Grossman, 2015). To isolate the transcriptional activity of DnaA from DNA replication, Washington et al. (2017) eliminated DnaA’s essential role in replication by using a strain that initiates chromosome replication from the plasmids oriN by the plasmids replication initiator RepN. This B. subtilis strain has its native ori and the operon flanking it (dnaA–dnaN) knocked out. The expressions of dnaA and dnaN were engineered at different loci regulated by two different inducible promoters. Using this system, 91% of 339 total number of genes that displayed DnaA-dependent transcriptional regulation were shown to be indirectly regulated by DnaA via Sda. DnaA regulates the transcription of sda by directly binding at sda’s promoter region (Burkholder et al., 2001; Ishikawa et al., 2007; Breier and Grossman, 2009). Sda is an inhibitor of the phosphorelay that ultimately activates Spo0A, a global regulator of sporulation and stationary phase gene expression (Figure 2B) (Burkholder et al., 2001; Rowland et al., 2004; Whitten et al., 2007). Consistent with previous analyses, Washington et al. identified eight sets of genes that are directly regulated by DnaA: dnaA-dnaN, sda, yqeG-M, ywlC, ywcI-sacT, vpr, yyzF-yydABCD, and trmEF-rsmG-noc (Burkholder et al., 2001; Ogura et al., 2001; Ishikawa et al., 2007; Washington et al., 2017).
Transcriptional Activity of DnaA and its Role in DNA Replication
Replication Initiation
Aside from opening the double-stranded ori region, DnaA has been proposed to regulate the initiation of replication in three other ways that are dependent on its transcriptional activity. First, DnaA regulates the levels of the replication initiator by modulating its own expression in various bacterial species (as discussed in previous sections). Second, in C. crescentus, DnaA regulates the levels of the active form of the replication initiator (DnaA–ATP) by regulating the expression of hdaA (Collier and Shapiro, 2009). HdaA represses DnaA’s activity as a replication initiator by promoting the hydrolysis of DnaA–ATP to DnaA–ADP (Collier and Shapiro, 2009). Third, DnaA’s transcriptional activity can promote the replication of λ-derived plasmids via a potential direct interaction between DnaA and the β-subunit of RNA polymerase (RpoB) (Szalewska-Palasz et al., 1998b). In E. coli, DnaA’s binding at ori-λ is necessary for transcriptional activation of ori-λ and also for efficient expression of λ replication initiator proteins (Szalewska-Palasz et al., 1998a).
DNA Replication
DnaA as a transcription factor regulates the levels of substrates for DNA synthesis and of components of the replisome (molecular machinery required for chromosome replication). In E. coli, DnaA has been shown to repress the expression of the gua operon involved in purine biosynthesis (Tesfa-Selase and Drabble, 1992). The role of DnaA in regulating expression levels of nrdAB (involved in the last step of deoxynucleotide biosynthesis) has been shown in E. coli and proposed in C. crescentus (Tuggle and Fuchs, 1986; Augustin et al., 1994; Jacobson and Fuchs, 1998; Hottes et al., 2005). In E. coli and B. subtilis, DnaA regulates the transcription of dnaN (DNA polymerase III, β-subunit) commonly found in the same operon downstream of dnaA (Hansen F. G. et al., 1982; Ogura et al., 2001; Berenstein et al., 2002; Washington et al., 2017). In C. crescentus, predicted DnaA boxes are found upstream of dnaQ (DNA polymerase III, β-subunit) and dnaB (DNA helicase), both of which were identified as part of the DnaA transcriptional regulon by delaying dnaA transcription (Hottes et al., 2005).
DNA Repair
DNA repair plays an important role in DNA replication due to the relatively frequent replication-fork arrests that occur in bacteria even when growing under normal conditions (Cox et al., 2000). DnaA has been proposed to serve as a regulator for maintaining the integrity of the genome in response to DNA damage (Wurihan et al., 2018). In stationary phase, DnaA activates the transcription of polA (Quinones et al., 1997), the DNA polymerase in E. coli involved in DNA replication and DNA repair (Sharma and Smith, 1987; Savic et al., 1990; Quinones et al., 1997). DnaA along with LexA (global regulator of the SOS response) were shown to co-regulate the transcription of two key genes in the SOS regulon (Wurihan et al., 2018): uvrB (component of Nucleotide Excision Repair NER system) and recN [involved in repair of double-stranded breaks (Uranga et al., 2017)]. The expression of uvrB had previously been predicted to be regulated by DnaA based on DnaA boxes found upstream of uvrB (van den Berg et al., 1985; Arikan et al., 1986). In B. subtilis, the transcriptional activity of DnaA has been linked to the RecA-independent response to DNA stress (Goranov et al., 2005; Washington et al., 2017).
DnaA Beyond Chromosome Replication
Cytokinesis
Maintaining the integrity of the chromosome after each cell division requires exquisite coordination between chromosome replication and cytokinesis. In E. coli, the expression levels of the gene encoding FtsZ oscillate over the cell cycle reaching the highest levels at the same time when chromosome replication initiates (Garrido et al., 1993). The cell cycle-dependent expression of ftsZ and the identification of three DnaA boxes found upstream of ftsZ (within ftsQA) initially suggested DnaA’s involvement in ftsZ expression; however, the three DnaA boxes were later shown not to play a role in ftsZ transcription (Masters et al., 1989; Garrido et al., 1993; Smith et al., 1996). A more recent potential connection between DnaA’s transcriptional activity and cytokinesis involves MioC in E. coli in that DnaA regulates the transcription of mioC that is located next to ori (Lother et al., 1985; Rokeach and Zyskind, 1986; Nozaki et al., 1988). The protein MioC has been proposed to promote cell division independent of ori’s replication and segregation (Lies et al., 2015). However, mioC mutants display only a moderate cell division defect. In C. crescentus, DnaA was shown to bind ftsZ’s promoter region in vitro and its in vivo expression to be dependent on DnaA’s nucleotide switch (Hottes et al., 2005; Fernandez-Fernandez et al., 2011). In B. subtilis, DnaA was shown to bind the promoter region of ftsL (encodes membrane-associated Z-ring protein) in vivo using chromatin immunoprecipitation assays (Goranov et al., 2005). So far, the transcriptional activity of DnaA has not been shown to have a major impact with the timing of cytokinesis in any bacterial species. Our current understanding of the mechanism(s) that coordinate the timing of replication initiation with cytokinesis remains limited.
Motility, Quorum Sensing, and Heat Shock Response
The transcriptional activity of DnaA has been linked to other developmental processes. The first observation connecting DnaA to motility was in a temperature-sensitive dnaA mutant of E. coli. When grown at its permissive temperature, this mutant strain expressed significantly lower levels of flagellin that resulted in loss of motility (Mizushima et al., 1994). DnaA’s regulation was later shown to not directly affect the expression of flagellin but rather indirectly via the expression of the gene (flhD) encoding the flagellar transcriptional regulator FlhD (Mizushima et al., 1997). Notably, the quorum sensing regulators QseB and QseC in E. coli have been shown to regulate the expression of dnaA and flhD (Sperandio et al., 2002; Wu et al., 2021). In the nitrogen-fixing symbiont S. meliloti, the transcriptional activity of DnaA has been proposed to coordinate growth phase with quorum sensing. DnaA was shown to activate the transcription of nurR (encodes a LuxR-like solo regulator) by binding the promoter region during exponential growth and high nutrient availability (McIntosh et al., 2019). NurR activates the transcription of sinR, which encodes the major regulator of N-acyl-homoserine lactones (AHL) production (Calatrava-Morales et al., 2018). The transcriptional activity of DnaA has also been connected to E. coli heat shock response. DnaA was shown to bind the promoter region and repress the transcription of the gene encoding the sigma factor RpoH (Wang and Kaguni, 1989).
Future Directions for Replication Initiators
This review has focused on DnaA as the key chromosome replication initiator in bacteria. Replication initiators are different in the other domains of life such as archaea and eukaryotes. Interestingly, the replication initiators in archaea and in eukaryotes have also been shown to serve as regulators of gene expression. In eukaryotes, chromosome replication is initiated by the origin recognition complex (ORC) composed of six subunits (Bell and Dutta, 2002). In human cells, replication initiator proteins have been shown to regulate the transcription of genes involved in cell division (Hossain and Stillman, 2016). Replication initiation in archaea resembles the molecular machinery of ORC in eukaryotes (Makarova and Koonin, 2013). In the archaeon Sulfolobus islandicus, replication initiator proteins were shown to bind the promoter regions and regulate the expression of genes involved in DNA damage response (Sun et al., 2018). The ability of replication initiators to regulate gene expression in eukaryotes and archaea are a relatively recent discovery that continues to accumulate supporting data (Popova et al., 2018; Hu et al., 2020).
In this mini review, several aspects of the transcriptional activity of DnaA were discussed including autoregulation, its mechanism of regulation, and its role in chromosome replication and other key cellular events. Many exciting questions about the transcriptional activity of DnaA remain to be answered. For instance, DnaA has been shown to act as an activator and a repressor of transcription (Messer and Weigel, 1997). Is there a common mechanism that differentiates and regulates these two opposing functions of DnaA? Furthermore, bipartite oris flanking the dnaA gene (as in B. subtilis and Helicobacter pylori; Krause et al., 1997; Donczew et al., 2012) suggest that DnaA can act as a replication initiator and transcription factor when bound at the same chromosomal locus upstream of dnaA. How are these two different functions of DnaA coordinated and differentiated over the cell cycle? Most of the work done on DnaA’s nucleotide switch has been performed in E. coli. How widespread is DnaA’s nucleotide switch used to regulate DnaA’s transcriptional activity in other bacterial species? To conclude, the transcriptional activity of DnaA has only been characterized in a few bacterial species. Within these few, the specifics of DnaA’s transcriptional regulon vary significantly. Thus, the characterization of replication initiators’ roles in coordinating various cellular events will continue to provide exciting results for years to come.
Author Contributions
All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Eric Morgan for critically reading this manuscript and our funding NIH R01GM133833.
References
Arikan, E., Kulkarni, M. S., Thomas, D. C., and Sancar, A. (1986). Sequences of the E. coli uvrB gene and protein. Nucleic Acids Res. 14, 2637–2650.
Atlung, T., Clausen, E. S., and Hansen, F. G. (1985). Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200, 442–450. doi: 10.1007/bf00425729
Atlung, T., Clausen, E., and Hansen, F. G. (1984). Autorepression of the dnaA gene of Escherichia coli. Adv. Exp. Med. Biol. 179, 199–207. doi: 10.1007/978-1-4684-8730-5_20
Augustin, L. B., Jacobson, B. A., and Fuchs, J. A. (1994). Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J. Bacteriol. 176, 378–387. doi: 10.1128/jb.176.2.378-387.1994
Babu, V. M. P., Itsko, M., Baxter, J. C., Schaaper, R. M., and Sutton, M. D. (2017). Insufficient levels of the nrdAB-encoded ribonucleotide reductase underlie the severe growth defect of the Deltahda E. coli strain. Mol. Microbiol. 104, 377–399. doi: 10.1111/mmi.13632
Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374.
Berenstein, D., Olesen, K., Speck, C., and Skovgaard, O. (2002). Genetic organization of the Vibrio harveyi DnaA gene region and analysis of the function of the V. harveyi DnaA protein in Escherichia coli. J. Bacteriol. 184, 2533–2538. doi: 10.1128/jb.184.9.2533-2538.2002
Bramhill, D., and Kornberg, A. (1988). Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52, 743–755. doi: 10.1016/0092-8674(88)90412-6
Braun, R. E., and Wright, A. (1986). DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol. Gen. Genet. 202, 246–250. doi: 10.1007/bf00331644
Braun, R. E., O’day, K., and Wright, A. (1985). Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40, 159–169. doi: 10.1016/0092-8674(85)90319-8
Breier, A. M., and Grossman, A. D. (2009). Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J. Bacteriol. 191, 486–493. doi: 10.1128/jb.01294-08
Burkholder, W. F., Kurtser, I., and Grossman, A. D. (2001). Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104, 269–279. doi: 10.1016/s0092-8674(01)00211-2
Calatrava-Morales, N., Mcintosh, M., and Soto, M. J. (2018). Regulation mediated by N-Acyl homoserine lactone quorum sensing signals in the rhizobium-legume symbiosis. Genes (Basel) 9:263. doi: 10.3390/genes9050263
Campbell, J. L., and Kleckner, N. (1990). E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979. doi: 10.1016/0092-8674(90)90271-f
Cheng, L., and Keiler, K. C. (2009). Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. J. Bacteriol. 191, 4268–4275. doi: 10.1128/jb.00362-09
Chiaramello, A. E., and Zyskind, J. W. (1990). Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J. Bacteriol. 172, 2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990
Cho, E., Ogasawara, N., and Ishikawa, S. (2008). The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet. Syst. 83, 111–125. doi: 10.1266/ggs.83.111
Collier, J., and Shapiro, L. (2009). Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in caulobacter. J. Bacteriol. 191, 5706–5716. doi: 10.1128/jb.00525-09
Collier, J., Mcadams, H. H., and Shapiro, L. (2007). A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 17111–17116. doi: 10.1073/pnas.0708112104
Collier, J., Murray, S. R., and Shapiro, L. (2006). DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25, 346–356. doi: 10.1038/sj.emboj.7600927
Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J., and Marians, K. J. (2000). The importance of repairing stalled replication forks. Nature 404, 37–41. doi: 10.1038/35003501
De Nisco, N. J., Abo, R. P., Wu, C. M., Penterman, J., and Walker, G. C. (2014). Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 111, 3217–3224. doi: 10.1073/pnas.1400421111
Donczew, R., Weigel, C., Lurz, R., Zakrzewska-Czerwinska, J., and Zawilak-Pawlik, A. (2012). Helicobacter pylori oriC–the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 40, 9647–9660. doi: 10.1093/nar/gks742
Duderstadt, K. E., and Berger, J. M. (2013). A structural framework for replication origin opening by AAA+ initiation factors. Curr. Opin. Struct. Biol. 23, 144–153. doi: 10.1016/j.sbi.2012.11.012
Felletti, M., Omnus, D. J., and Jonas, K. (2019). Regulation of the replication initiator DnaA in Caulobacter crescentus. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 697–705. doi: 10.1016/j.bbagrm.2018.01.004
Fernandez-Fernandez, C., Gonzalez, D., and Collier, J. (2011). Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus. PLoS One 6:e26028. doi: 10.1371/journal.pone.0026028
Frandi, A., and Collier, J. (2019). Multilayered control of chromosome replication in Caulobacter crescentus. Biochem. Soc. Trans. 47, 187–196. doi: 10.1042/bst20180460
Froelich, J. M., Phuong, T. K., and Zyskind, J. W. (1996). Fis binding in the dnaA operon promoter region. J. Bacteriol. 178, 6006–6012. doi: 10.1128/jb.178.20.6006-6012.1996
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Fujita, M. Q., Yoshikawa, H., and Ogasawara, N. (1989). Structure of the dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol. Gen. Genet. 215, 381–387. doi: 10.1007/bf00427033
Fuller, R. S., and Kornberg, A. (1983). Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. U.S.A. 80, 5817–5821. doi: 10.1073/pnas.80.19.5817
Garrido, T., Sanchez, M., Palacios, P., Aldea, M., and Vicente, M. (1993). Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 12, 3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x
Gon, S., Camara, J. E., Klungsoyr, H. K., Crooke, E., Skarstad, K., and Beckwith, J. (2006). A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25, 1137–1147. doi: 10.1038/sj.emboj.7600990
Goranov, A. I., Katz, L., Breier, A. M., Burge, C. B., and Grossman, A. D. (2005). A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U.S.A. 102, 12932–12937. doi: 10.1073/pnas.0506174102
Han, J. S., Kwon, H. S., Yim, J. B., and Hwang, D. S. (1998). Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in E. coli. Mol. Gen. Genet. 259, 610–614. doi: 10.1007/s004380050854
Hanna, M. H., and Carl, P. L. (1975). Reinitiation of deoxyribonucleic acid synthesis by deoxyribonucleic acid initiation mutants of Escherichia coli: role of ribonucleic acid synthesis, protein synthesis, and cell division. J. Bacteriol. 121, 219–226. doi: 10.1128/jb.121.1.219-226.1975
Hansen, E. B., Hansen, F. G., and Von Meyenburg, K. (1982). The nucleotide sequence of the dnaA gene and the first part of the dnaN gene of Escherichia coli K-12. Nucleic Acids Res. 10, 7373–7385.
Hansen, F. G., and Atlung, T. (2018). The DnaA tale. Front. Microbiol. 9:319. doi: 10.3389/fmicb.2018.00319NODOI
Hansen, F. G., and Rasmussen, K. V. (1977). Regulation of the dnaA product in Escherichia coli. Mol. Gen. Genet. 155, 219–225. doi: 10.1007/bf00393163
Hansen, F. G., Hansen, E. B., and Atlung, T. (1982). The nucleotide sequence of the dnaA gene promoter and of the adjacent rpmH gene, coding for the ribosomal protein L34, of Escherichia coli. EMBO J. 1, 1043–1048. doi: 10.1002/j.1460-2075.1982.tb01294.x
Hansen, F. G., Koefoed, S., Sorensen, L., and Atlung, T. (1987). Titration of DnaA protein by oriC DnaA-boxes increases dnaA gene expression in Escherichia coli. EMBO J 6, 255–258. doi: 10.1002/j.1460-2075.1987.tb04747.x
Hill, N. S., Kadoya, R., Chattoraj, D. K., and Levin, P. A. (2012). Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 8:e1002549. doi: 10.1371/journal.pgen.1002549
Holtzendorff, J., Hung, D., Brende, P., Reisenauer, A., Viollier, P. H., Mcadams, H. H., et al. (2004). Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304, 983–987. doi: 10.1126/science.1095191
Hoover, S. E., Xu, W., Xiao, W., and Burkholder, W. F. (2010). Changes in DnaA-dependent gene expression contribute to the transcriptional and developmental response of Bacillus subtilis to manganese limitation in Luria-Bertani medium. J. Bacteriol. 192, 3915–3924. doi: 10.1128/jb.00210-10
Hossain, M., and Stillman, B. (2016). Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of cyclin E gene transcription. Elife 5:e12785.
Hottes, A. K., Shapiro, L., and Mcadams, H. H. (2005). DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58, 1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x
Hu, Y., Tareen, A., Sheu, Y. J., Ireland, W. T., Speck, C., Li, H., et al. (2020). Evolution of DNA replication origin specification and gene silencing mechanisms. Nat. Commun. 11:5175.
Ingmer, H., and Atlung, T. (1992). Expression and regulation of a dnaA homologue isolated from Pseudomonas putida. Mol. Gen. Genet. 232, 431–439. doi: 10.1007/bf00266248
Ishikawa, S., Ogura, Y., Yoshimura, M., Okumura, H., Cho, E., Kawai, Y., et al. (2007). Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 14, 155–168. doi: 10.1093/dnares/dsm017
Jacobson, B. A., and Fuchs, J. A. (1998). Multiple cis-acting sites positively regulate Escherichia coli nrd expression. Mol. Microbiol. 28, 1315–1322. doi: 10.1046/j.1365-2958.1998.00897.x
Jakimowicz, D., Majka, J., Lis, B., Konopa, G., Wegrzyn, G., Messer, W., et al. (2000). Structure and regulation of the dnaA promoter region in three Streptomyces species. Mol. Gen. Genet. 262, 1093–1102. doi: 10.1007/pl00008652
Kasho, K., and Katayama, T. (2013). DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941. doi: 10.1073/pnas.1212070110
Kato, J., and Katayama, T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. doi: 10.1093/emboj/20.15.4253
Kohiyama, M. (2020). Research on DnaA in the early days. Res. Microbiol. 171, 287–289. doi: 10.1016/j.resmic.2020.11.004
Krause, M., Ruckert, B., Lurz, R., and Messer, W. (1997). Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274, 365–380. doi: 10.1006/jmbi.1997.1404
Kucherer, C., Lother, H., Kolling, R., Schauzu, M. A., and Messer, W. (1986). Regulation of transcription of the chromosomal dnaA gene of Escherichia coli. Mol. Gen. Genet. 205, 115–121.
Lee, Y. S., Kim, H., and Hwang, D. S. (1996). Transcriptional activation of the dnaA gene encoding the initiator for oriC replication by IciA protein, an inhibitor of in vitro oriC replication in Escherichia coli. Mol. Microbiol. 19, 389–396. doi: 10.1046/j.1365-2958.1996.485902.x
Leonard, A. C., Rao, P., Kadam, R. P., and Grimwade, J. E. (2019). Changing perspectives on the role of DnaA-ATP in orisome function and timing regulation. Front. Microbiol. 10:2009. doi: 10.3389/fmicb.2019.02009NODOI
Lies, M., Visser, B. J., Joshi, M. C., Magnan, D., and Bates, D. (2015). MioC and GidA proteins promote cell division in E. coli. Front. Microbiol. 6:516. doi: 10.3389/fmicb.2015.00516
Lobner-Olesen, A., Skarstad, K., Hansen, F. G., Von Meyenburg, K., and Boye, E. (1989). The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57, 881–889. doi: 10.1016/0092-8674(89)90802-7
Lother, H., Kolling, R., Kucherer, C., and Schauzu, M. (1985). dnaA protein-regulated transcription: effects on the in vitro replication of Escherichia coli minichromosomes. EMBO J. 4, 555–560. doi: 10.1002/j.1460-2075.1985.tb03664.x
Lu, M., Campbell, J. L., Boye, E., and Kleckner, N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. doi: 10.1016/0092-8674(94)90156-2
Makarova, K. S., and Koonin, E. V. (2013). Archaeology of eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol. 5:a012963.
Masters, M., Paterson, T., Popplewell, A. G., Owen-Hughes, T., Pringle, J. H., and Begg, K. J. (1989). The effect of DnaA protein levels and the rate of initiation at oriC on transcription originating in the ftsQ and ftsA genes: in vivo experiments. Mol. Gen. Genet. 216, 475–483. doi: 10.1007/bf00334393
McIntosh, M., Serrania, J., and Lacanna, E. (2019). A novel LuxR-type solo of Sinorhizobium meliloti, NurR, is regulated by the chromosome replication coordinator, DnaA and activates quorum sensing. Mol. Microbiol. 112, 678–698. doi: 10.1111/mmi.14312
Mera, P. E., Kalogeraki, V. S., and Shapiro, L. (2014). Replication initiator DnaA binds at the Caulobacter centromere and enables chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 111, 16100–16105. doi: 10.1073/pnas.1418989111
Messer, W., and Weigel, C. (1997). DnaA initiator–also a transcription factor. Mol. Microbiol. 24, 1–6. doi: 10.1046/j.1365-2958.1997.3171678.x
Mizushima, T., Koyanagi, R., Katayama, T., Miki, T., and Sekimizu, K. (1997). Decrease in expression of the master operon of flagellin synthesis in a dnaA46 mutant of Escherichia coli. Biol. Pharm. Bull. 20, 327–331. doi: 10.1248/bpb.20.327
Mizushima, T., Sasaki, S., Ohishi, H., Kobayashi, M., Katayama, T., Miki, T., et al. (1996). Molecular design of inhibitors of in vitro oriC DNA replication based on the potential to block the ATP binding of DnaA protein. J. Biol. Chem. 271, 25178–25183. doi: 10.1074/jbc.271.41.25178
Mizushima, T., Tomura, A., Shinpuku, T., Miki, T., and Sekimizu, K. (1994). Loss of flagellation in dnaA mutants of Escherichia coli. J. Bacteriol. 176, 5544–5546. doi: 10.1128/jb.176.17.5544-5546.1994
Nishida, S., Fujimitsu, K., Sekimizu, K., Ohmura, T., Ueda, T., and Katayama, T. (2002). A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277, 14986–14995. doi: 10.1074/jbc.m108303200
Nozaki, N., Okazaki, T., and Ogawa, T. (1988). In vitro transcription of the origin region of replication of the Escherichia coli chromosome. J. Biol. Chem. 263, 14176–14183. doi: 10.1016/s0021-9258(18)68202-x
Ogura, Y., Imai, Y., Ogasawara, N., and Moriya, S. (2001). Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183, 3833–3841. doi: 10.1128/jb.183.13.3833-3841.2001
Olliver, A., Saggioro, C., Herrick, J., and Sclavi, B. (2010). DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 76, 1555–1571. doi: 10.1111/j.1365-2958.2010.07185.x
Ozaki, S. (2019). Regulation of replication initiation: lessons from Caulobacter crescentus. Genes Genet. Syst. 94, 183–196. doi: 10.1266/ggs.19-00011
Panis, G., Murray, S. R., and Viollier, P. H. (2015). Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol. Rev. 39, 120–133. doi: 10.1093/femsre/fuu002
Polaczek, P., and Wright, A. (1990). Regulation of expression of the dnaA gene in Escherichia coli: role of the two promoters and the DnaA box. New Biol. 2, 574–582.
Popova, V. V., Brechalov, A. V., Georgieva, S. G., and Kopytova, D. V. (2018). Nonreplicative functions of the origin recognition complex. Nucleus 9, 460–473. doi: 10.1080/19491034.2018.1516484
Quinones, A., Wandt, G., Kleinstauber, S., and Messer, W. (1997). DnaA protein stimulates polA gene expression in Escherichia coli. Mol. Microbiol. 23, 1193–1202. doi: 10.1046/j.1365-2958.1997.2961658.x
Riber, L., and Lobner-Olesen, A. (2020). Inhibition of Escherichia coli chromosome replication by rifampicin treatment or during the stringent response is overcome by de novo DnaA protein synthesis. Mol. Microbiol. 114, 906–919. doi: 10.1111/mmi.14531
Rokeach, L. A., and Zyskind, J. W. (1986). RNA terminating within the E. coli origin of replication: stringent regulation and control by DnaA protein. Cell 46, 763–771. doi: 10.1016/0092-8674(86)90352-1
Rowland, S. L., Burkholder, W. F., Cunningham, K. A., Maciejewski, M. W., Grossman, A. D., and King, G. F. (2004). Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13, 689–701. doi: 10.1016/s1097-2765(04)00084-x
Salazar, L., Guerrero, E., Casart, Y., Turcios, L., and Bartoli, F. (2003). Transcription analysis of the dnaA gene and oriC region of the chromosome of Mycobacterium smegmatis and Mycobacterium bovis BCG, and its regulation by the DnaA protein. Microbiology (Reading) 149, 773–784. doi: 10.1099/mic.0.25832-0
Savic, D. J., Jankovic, M., and Kostic, T. (1990). Cellular role of DNA polymerase I. J. Basic Microbiol. 30, 769–784.
Sekimizu, K., Bramhill, D., and Kornberg, A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. doi: 10.1016/0092-8674(87)90221-2
Sharma, R. C., and Smith, K. C. (1987). Role of DNA polymerase I in postreplication repair: a reexamination with Escherichia coli delta polA. J. Bacteriol. 169, 4559–4564. doi: 10.1128/jb.169.10.4559-4564.1987
Smith, J. L., and Grossman, A. D. (2015). In vitro whole genome DNA binding analysis of the bacterial replication initiator and transcription factor DnaA. PLoS Genet. 11:e1005258. doi: 10.1371/journal.pgen.1005258
Smith, R. W., Mcateer, S., and Masters, M. (1996). The coupling between ftsZ transcription and initiation of DNA replication is not mediated by the DnaA-boxes upstream of ftsZ or by DnaA. Mol. Microbiol. 21, 361–372. doi: 10.1046/j.1365-2958.1996.6431365.x
Smith, R. W., Mcateer, S., and Masters, M. (1997). Autoregulation of the Escherichia coli replication initiator protein, DnaA, is indirect. Mol. Microbiol. 23, 1303–1315. doi: 10.1046/j.1365-2958.1997.3121675.x
Speck, C., Weigel, C., and Messer, W. (1999). ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 18, 6169–6176. doi: 10.1093/emboj/18.21.6169
Sperandio, V., Torres, A. G., and Kaper, J. B. (2002). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43, 809–821. doi: 10.1046/j.1365-2958.2002.02803.x
Sun, L., and Fuchs, J. A. (1992). Escherichia coli ribonucleotide reductase expression is cell cycle regulated. Mol. Biol. Cell 3, 1095–1105.
Sun, L., Jacobson, B. A., Dien, B. S., Srienc, F., and Fuchs, J. A. (1994). Cell cycle regulation of the Escherichia coli nrd operon: requirement for a cis-acting upstream AT-rich sequence. J. Bacteriol. 176, 2415–2426. doi: 10.1128/jb.176.8.2415-2426.1994
Sun, M., Feng, X., Liu, Z., Han, W., Liang, Y. X., and She, Q. (2018). An Orc1/Cdc6 ortholog functions as a key regulator in the DNA damage response in Archaea. Nucleic Acids Res. 46, 6697–6711. doi: 10.1093/nar/gky487
Szalewska-Palasz, A., Lemieszek, E., Pankiewicz, A., Wegrzyn, A., Helinski, D. R., and Wegrzyn, G. (1998a). Escherichia coli dnaA gene function and bacteriophage lambda replication. FEMS Microbiol. Lett. 167, 27–32. doi: 10.1016/s0378-1097(98)00363-2
Szalewska-Palasz, A., Wegrzyn, A., Blaszczak, A., Taylor, K., and Wegrzyn, G. (1998b). DnaA-stimulated transcriptional activation of orilambda: Escherichia coli RNA polymerase beta subunit as a transcriptional activator contact site. Proc. Natl. Acad. Sci. U.S.A. 95, 4241–4246. doi: 10.1073/pnas.95.8.4241
Tesfa-Selase, F., and Drabble, W. T. (1992). Regulation of the gua operon of Escherichia coli by the DnaA protein. Mol. Gen. Genet. 231, 256–264. doi: 10.1007/bf00279799
Thelander, L., and Reichard, P. (1979). Reduction of ribonucleotides. Annu. Rev. Biochem. 48, 133–158. doi: 10.1146/annurev.bi.48.070179.001025
Tuggle, C. K., and Fuchs, J. A. (1986). Regulation of the operon encoding ribonucleotide reductase in Escherichia coli: evidence for both positive and negative control. EMBO J. 5, 1077–1085. doi: 10.1002/j.1460-2075.1986.tb04325.x
Uranga, L. A., Reyes, E. D., Patidar, P. L., Redman, L. N., and Lusetti, S. L. (2017). The cohesin-like RecN protein stimulates RecA-mediated recombinational repair of DNA double-strand breaks. Nat. Commun. 8:15282.
van den Berg, E. A., Geerse, R. H., Memelink, J., Bovenberg, R. A., Magnee, F. A., and Van De Putte, P. (1985). Analysis of regulatory sequences upstream of the E. coli uvrB gene; involvement of the DnaA protein. Nucleic Acids Res. 13, 1829–1840.
Wang, Q. P., and Kaguni, J. M. (1987). Transcriptional repression of the dnaA gene of Escherichia coli by dnaA protein. Mol. Gen. Genet. 209, 518–525. doi: 10.1007/bf00331158
Wang, Q. P., and Kaguni, J. M. (1989). dnaA protein regulates transcriptions of the rpoH gene of Escherichia coli. J. Biol. Chem. 264, 7338–7344. doi: 10.1016/s0021-9258(18)83238-0
Wargachuk, R., and Marczynski, G. T. (2015). The Caulobacter crescentus homolog of DnaA (HdaA) also regulates the proteolysis of the replication initiator protein DnaA. J. Bacteriol. 197, 3521–3532. doi: 10.1128/jb.00460-15
Washington, T. A., Smith, J. L., and Grossman, A. D. (2017). Genetic networks controlled by the bacterial replication initiator and transcription factor DnaA in Bacillus subtilis. Mol. Microbiol. 106, 109–128. doi: 10.1111/mmi.13755
Whitten, A. E., Jacques, D. A., Hammouda, B., Hanley, T., King, G. F., Guss, J. M., et al. (2007). The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 368, 407–420. doi: 10.1016/j.jmb.2007.01.064
Wu, D., Baigalmaa, L., Yao, Y., Li, G., Su, M., Fan, L., et al. (2021). The Escherichia coli QseB/QseC signaling is required for correct timing of replication initiation and cell motility. Gene 773:145374. doi: 10.1016/j.gene.2020.145374
Wurihan, Gezi, Brambilla, E., Wang, S., Sun, H., Fan, L., et al. (2018). DnaA and LexA proteins regulate transcription of the uvrB gene in Escherichia coli: the role of DnaA in the control of the SOS regulon. Front. Microbiol. 9:1212. doi: 10.3389/fmicb.2018.01212NODOI
Zakrzewska-Czerwinska, J., Nardmann, J., and Schrempf, H. (1994). Inducible transcription of the dnaA gene from Streptomyces lividans 66. Mol. Gen. Genet. 242, 440–447. doi: 10.1007/bf00281794
Keywords: DnaA, replication initiation, transcription factor, cell cycle, sporulation, DNA repair, autoregulation
Citation: Menikpurage IP, Woo K and Mera PE (2021) Transcriptional Activity of the Bacterial Replication Initiator DnaA. Front. Microbiol. 12:662317. doi: 10.3389/fmicb.2021.662317
Received: 01 February 2021; Accepted: 19 April 2021;
Published: 01 June 2021.
Edited by:
Jianping Xie, Southwest University, ChinaReviewed by:
Justine Collier, University of Lausanne, SwitzerlandRodrigo Reyes, McGill University, Canada
Julie Anne Maupin-Furlow, University of Florida, United States
Copyright © 2021 Menikpurage, Woo and Mera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola E. Mera, cG1lcmFAaWxsaW5vaXMuZWR1
 Inoka P. Menikpurage
Inoka P. Menikpurage Kristin Woo
Kristin Woo Paola E. Mera
Paola E. Mera