- 1Department of Biomedical Science, Chosun University, Gwangju, South Korea
- 2Research Center for Proteineous Materials (RCPM), Chosun University, Gwangju, South Korea
Pine needles are used in several East Asian countries as food or traditional medicine. It contains functional components that exhibit a wide spectrum of pharmacological effects such as antioxidant, antimicrobial, anti-diabetic, and anti-inflammatory activities. We determined and characterized the novel antimicrobial peptides (AMPs) isolated from Pinus densiflora Sieb. et Zucc. The four active pine-needle (PN) peptides showed antimicrobial activity against foodborne bacteria with minimum inhibitory concentration (MIC) values within the range of 8–128 μg/ml. PN peptides showed no detectable hemolytic activity or cytotoxicity at the antimicrobial concentrations. The N-terminal amino acid sequence of the PN5 was identified using Edman degradation and Antimicrobial Peptide Database (APD) homology analysis showed that it was not identical to any other plant peptide. This suggests that PN5 can serve as an alternative therapeutic agent to be used in the food industry.
Introduction
Antimicrobial peptides (AMPs) play an important element in the innate immune system ranging from bacteria to plants, mammals, and insects (Hancock et al., 2016; Haney et al., 2019; Sathoff and Samac, 2019). Innate immunity is a defensive response in all multicellular organisms to combat pathogens. AMPs involved in these processes typically have a broad-spectrum activity against a wide range of Gram-positive and Gram-negative bacteria, fungi, and even certain viruses (Coates et al., 2018; Mookherjee et al., 2020). In the majority of cases, the mode of action of AMPs is related to the cytoplasmic membrane permeabilization. Plants are a source of bioactive compounds with various properties that are applicable in agriculture and medicine. Plants produced are short AMPs with a molecular mass less than 10 kDa; structurally, these are amphipathic and generally positively charged molecules at physiologically neutral pH values. They primarily play defensive roles such as acting as membrane-active antifungal, antibacterial, and antiviral agents (Tam et al., 2015; Dhama et al., 2018).
Many studies have been performed to evaluate pine needles (PN), and several compounds with antimicrobial activity have been detected (Ghaffari et al., 2019). The Korean red pine tree, Pinus densiflora, belongs to the family Pinaceae and is widely spread in East Asia (Korea, Japan, and China; Kim et al., 2020a). Various pine tree regions including needles, pollen, cones, and cortices are widely consumed as folk medicine, foods or dietary supplements for health promotion, antimicrobial, anti-inflammatory, and preservation effects (Hsu et al., 2006; Jeong et al., 2009). Pine bark protects collagen from the action of collagenase, while PNs exhibit anti-hypertensive effects and protect against oxidative DNA, protein, and lipid damage and oxidative stress-mediated apoptosis induced by hydroxyl radicals (Ku et al., 2007; Kim et al., 2010; Maimoona et al., 2011). Pine bark extracts are effective scavengers of free radicals and reactive oxygen species, can lower lipid levels in blood serum and may help to prevent disease and delay aging (Schafer et al., 2006; McGrath et al., 2015). PNs of form P. densiflora were used for folk medicine and for various disease prevention such as rheumatitis, hemorrhage, gastroenteric trouble, hypertension, and asthma (Kwak et al., 2006; Kim et al., 2020b). Recent scientific researches have shown that PNs form P. densiflora have antimicrobial, anti-viral, antioxidant, anti-mutagenic, anti-thrombosis, anti-asthmatic, and anti-inflammatory, and anti-cancer effects (Kwak et al., 2006; Park and Lee, 2011; Park et al., 2016; Ahn et al., 2018; Mostafa et al., 2018; Ha et al., 2020). However, the isolation and functional characterization of AMPs from PNs have been limited studies. Therefore, this study was conducted to identify and characterize AMPs from the needles of P. densiflora Sieb. et Zucc. We evaluated PN peptides to determine their antimicrobial ability against foodborne bacteria.
Materials and Methods
Pine Needle Collection and Extract Preparation
Fresh PNs of P. densiflora Sieb. et Zucc. were collected from Gok-Seong, Jeollanam-Do, Korea. The plant was initially determined based on its specific morphological observations, and the morphological and morphometric data were stored at the College of Natural Science and Public Health and Safety, Chosun University, Korea.
Pine needles of P. densiflora were rinsed with distilled water and then dried at 45°C for 5 h, and powdered using a mixer. The powder that passed through a 20-mesh sieve (850 μm) and was retained on a 40-mesh (450 μm) sieve, the mean particle size powder was stored in a sealed plastic bag at 25°C. PN powder (100 g) was added to 600 ml buffer (10 mM HEPES + 10 mM NaCl, pH 7.4) with continuous stirring at 25°C for 2 h. Thereafter, the extract was filtered and condensed under a vacuum in a rotary evaporator (N-1000VW, EYELA, Tokyo, Japan). The dried PNs extract was stored at 4°C until further analysis.
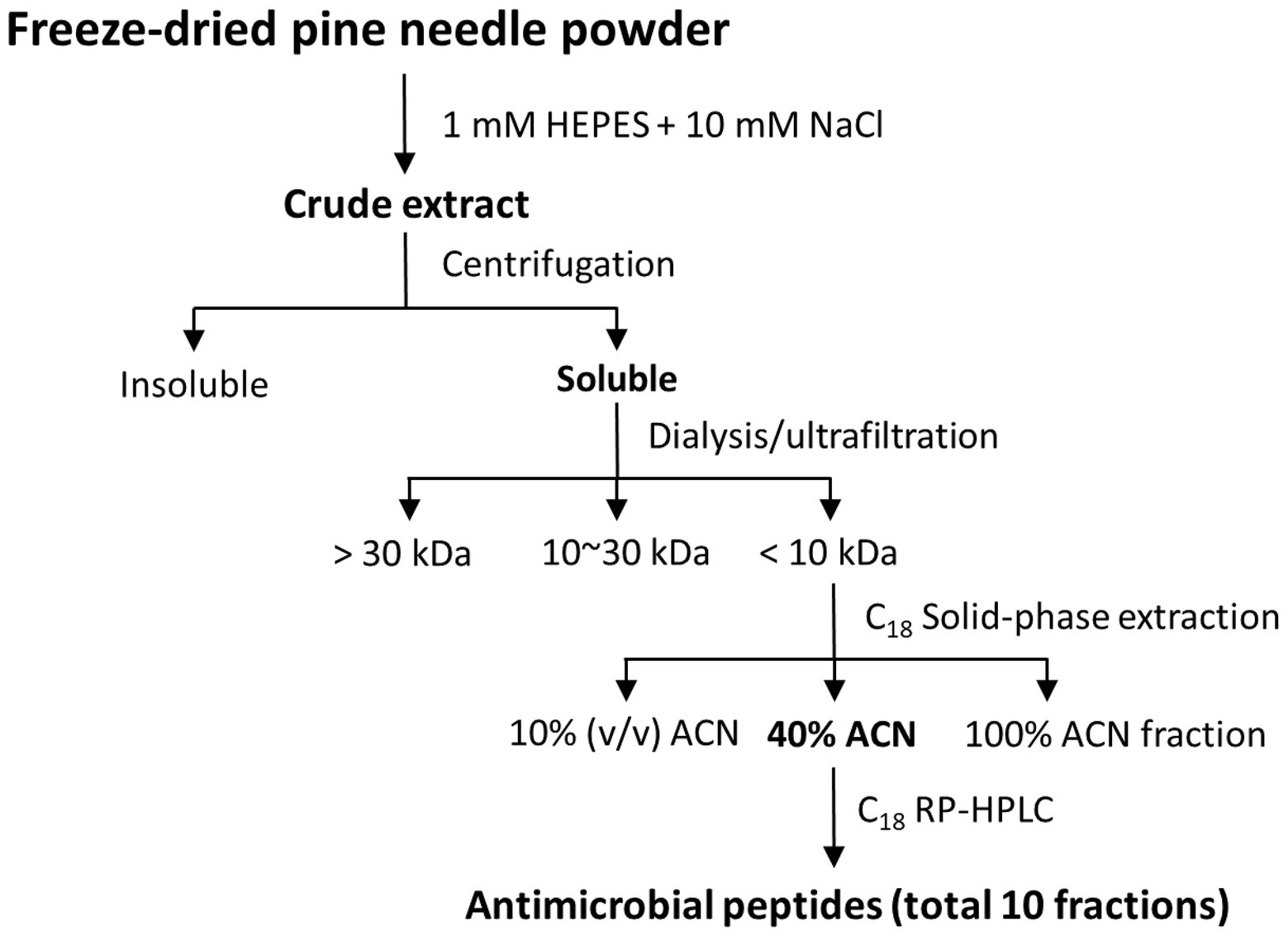
Purification of Antimicrobial Proteins
The dried extract sample was dissolved in distilled water and dialyzed using a molecular weight 1,000 membrane at 4°C overnight, and the dialyzed PN samples were purified using ultrafiltration (MW 30,000 and MW 1,000) and freeze-dried. The samples (MW < 10,000) were purified using an SPE 900 mg Lrg pore C18 column [Maxi-Clean™, Alltech Associates, Inc., Deerfield, IL, United States; 10% acetonitrile (ACN), 40% ACN, and 100% ACN]. The extract (40% ACN) was isolated using a reverse-phase C18 HPLC column [Jupiter 5u C18 300A, 250 mm (length) × 4.6 mm (inner diameter), 300 Å pore size, 5 μm particle size] on an HPLC system (Shimadzu Corporation, Kyoto, Japan) that had been equilibrated using 0.1% (v/v) trifluoroacetic acid (TFA, Merck, Kenilworth, NJ, United States) in water with 5% ACN. PN peptide fractions were eluted using a linear gradient of solvents A and B were 0.1% (v/v) TFA in water and 0.1% (v/v) TFA in acetonitrile, respectively. Elution was carried out using a linear gradient of 40% solvent B for 10 min, 40–65% for 25 min, and 65–95% for 45 min at a flow rate of 1 ml/min. The eluates were monitored by measuring the absorbance signal at 215 nm (Figure 1). Individual fractions were pooled and then were freeze-dried at −20°C. To confirm partly as a single peptide, freeze-dried fractions were successively re-subjected to a second C18 HPLC at the peak of the overlapping point. Each fraction was collected and subsequently assayed for antimicrobial activities. The purified PN peptides were confirmed as a single molecule using tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry.
Tricine-SDS-PAGE
Tricine-SDS-PAGE is an efficient method of separating low-molecular-mass peptides (16.5% polyacrylamide gel for peptide <10 kDa; Schagger and von Jagow, 1987). The purified PN peptides were resolved in a 16.5% tricine-SDS PAGE, followed by visualization with Coomassie Brilliant Blue G-250 staining.
Mass Spectrometry
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) was carried out using an Axima-CFR MALDI-TOF mass spectrometer (Kratos Analytical, Manchester, United Kingdom) as described by Pouvreau et al. (2001). The protein concentration of the purified PN peptide was determined by the Bradford assay using bovine serum albumin (BSA) as the calibration standard (Bradford, 1976).
PN Peptide Identification
The N-terminal amino acid (a.a.) sequences of the purified PN peptide was determined with an automated Edman degradation method using a pulse liquid automatic sequencer (Procise Model 491 HT protein sequencer; Applied Biosystems, Foster City, CA, United States) at the Korean Basic Science Institute (Seoul, Korea). The PN peptide sequences were compared to those in the Antimicrobial Peptide Database (APD), using the “APD3: Antimicrobial Peptide Calculator and Predictor” tool (Wang et al., 2016).
Antimicrobial Assay
For antimicrobial assays, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 15692, Staphylococcus aureus ATCC 25923, and Staphylococcus epidermidis ATCC 12228 were obtained from the American Type Culture Collection (Manassas, VA, United States). Salmonella typhimurium KCTC 1926 and Listeria monocytogenes KCTC 3710 were obtained from the Korean Collection for Type Cultures (Jeollabuk-do, Korea).
Minimum inhibitory concentrations (MICs) of purified PN peptides were assayed according to the Clinical and Laboratory Standards Institute recommendations (Wiegand et al., 2008). Briefly, bacterial suspension (5 × 105 CFU/ml), obtained by diluting an exponential growth phase culture, was then added into the 96-well plates containing 2-fold serial dilutions of each fraction, the plates were incubated for 18 h at 37°C. MICs of PN peptides were measured in optical density at 600 nm using Versamax™ ELISA Microplate Reader (Molecular Devices, Sunnyvale, CA, United States). PBS, culture media, and melittin were used as growth and growth inhibition controls. The MIC was defined as the lowest concentration of PN peptide that was able to inhibit microbial growth.
Hemolytic Activity
Hemolysis was determined using the mouse red blood cells (mRBCs). Fresh mRBCs were collected in PBS and washed three times with PBS (final mRBC concentration, 8% v/v). PN peptides were assessed at 200 μM final concentration and incubated for 60 min at 37°C. Hemolytic activity was monitored at an absorbance (A) of 414 nm and calculated as a positive control [0.1% Triton X-100 (Atriton)] and negative control [PBS (Ablank)]. The resulting values of cells that underwent hemolysis were calculated according to equation (Park et al., 2008):
Cytotoxicity
The cytotoxic effect of PN peptides was assessed against HaCaT (human keratinocytes) and RAW264.7 (mouse macrophages) cells cultured in 96-well plates at a density of 2 × 104 cells/well in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assays were used to evaluate cytotoxicity. PN peptides (0–200 μg/ml) were added with the cells for 24 h at 37°C. Subsequently, MTT (0.5 mg/ml) was added to each well and incubated for 4 h at 37°C After incubation, formazan crystals produced was dissolved in dimethyl sulfoxide, and the absorbance at 570 nm was measured and cytotoxicity was determined as a percentage of 100% cytotoxic control (0.1% Triton X-100; Park et al., 2008). Melittin was used as the control (reference) peptide.
Computational Analyses
3-D structural projections of PN5 were created online using the Mobyle@RPBS bioinformatics portal.1 The helical wheel projection was obtained using the online tool HeliQuest2 (Gautier et al., 2008).
Peptide Synthesis
PN5 peptides were synthesized using the fluorenylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis on a solid support of rink amide 4-methylbenzhydrylamine resin (Merck KGaA, Darmstadt, Germany) with Liberty microwave peptide synthesizer (CEM Co., Matthews, NC, United States; Kang et al., 2018). For coupling reactions, 0.1 M N-hydroxybenzotriazole and 0.45 M 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate in dimethylformamide (DMF) and 2 M N,N-diisopropylethylamine in N-methylpyrrolidone were used as coupling reaction solutions, and a 10-fold Fmoc-protected a.a. (Novabiochem, Läufelfingen, Switzerland) was added during all coupling reaction cycles. Fmoc from the Fmoc-protected synthetic peptide was cleaved with 20% (v/v) piperidine in DMF. After cleavage from resin, the crude PN peptides were purified using RP-HPLC on a Jupiter C18 column (250 mm × 21.2 mm, 15 μm, 300 Å) with a 0–60% ACN gradient in water containing 0.05% TFA. The purity of the peptides (>95%) was then determined by an analytical RP-HPLC using a Jupiter proteo C18 column (250 mm × 4.6 mm, 90 Å, 4 μm). The molecular weights of the synthetic peptides were detected by MALDI-TOF MS (MALDI II; Kratos Analytical, Inc., Chestnut Ridge, NY, United States).
Statistical Analysis
All experimental data represent mean ± standard deviation (SD). All the figures were obtained from several independent experiments and showed similar results. Differences among groups were analyzed using one-way ANOVA followed by Tukey’s multiple-comparison test.
Results
Purification of AMPs From Pine Needles
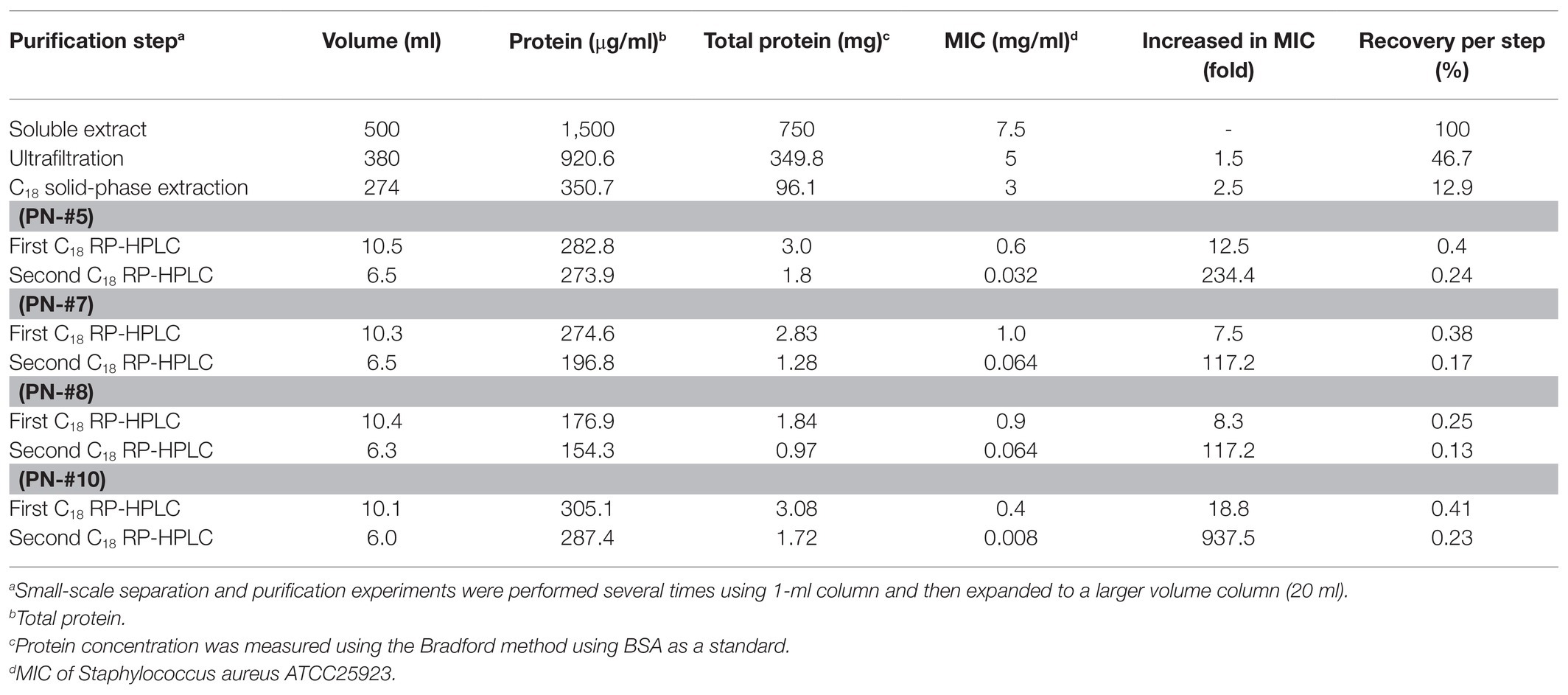
AMP fractions were purified from PNs in five steps: extraction, dialysis, ultrafiltration, purification using SPE 900 mg Lrg pore C18 column, and C18 reverse-phase HPLC (Figure 1). By varying the concentration of the sample separated using Tricine gel electrophoresis, small proteins with sizes of 1–3.5 kDa were obtained (Figure 2A). The antimicrobial activity of the dialyzed samples was tested against pathogenic bacteria (E. coli, S. aureus, P. aeruginosa, S. aureus, and S. epidermidis) and bacterial strains that cause food poisoning (S. typhimurium and L. monocytogenes). Results displayed that the dialyzed samples exhibited potent antimicrobial activity toward all tested bacteria at low concentrations (>5 mg/ml; Figure 2B; Supplementary Figure S3). The dialyzed samples were isolated using ultrafiltration (MW 30,000 and 10,000). According to the results shown in Supplementary Figure S1, all ultrafiltered fractions (>30, 10–30, and <10 kDa) inhibited E. coli and S. aureus. Among these fractions, the antimicrobial activity of the sample ≤10 kDa in size was the highest. In addition, this fraction was most effective in inhibiting S. aureus (2.5 mg/ml) compared to E. coli (5 mg/ml) and showed the same results as the dialyzed samples. After confirming the antimicrobial activity against L. monocytogenes and S. typhimurium of the ultrafiltered fractions ≤10 kDa, we found that these samples retained the same activity as the dialyzed samples (Supplementary Figure S3). As shown in Table 1, the MIC value increased considerably after the first purification step using ultrafiltration (1.5-fold compared to the soluble extract). The total protein content (512 mg) was lower than that of the initial soluble extract of PNs (1,250 mg).
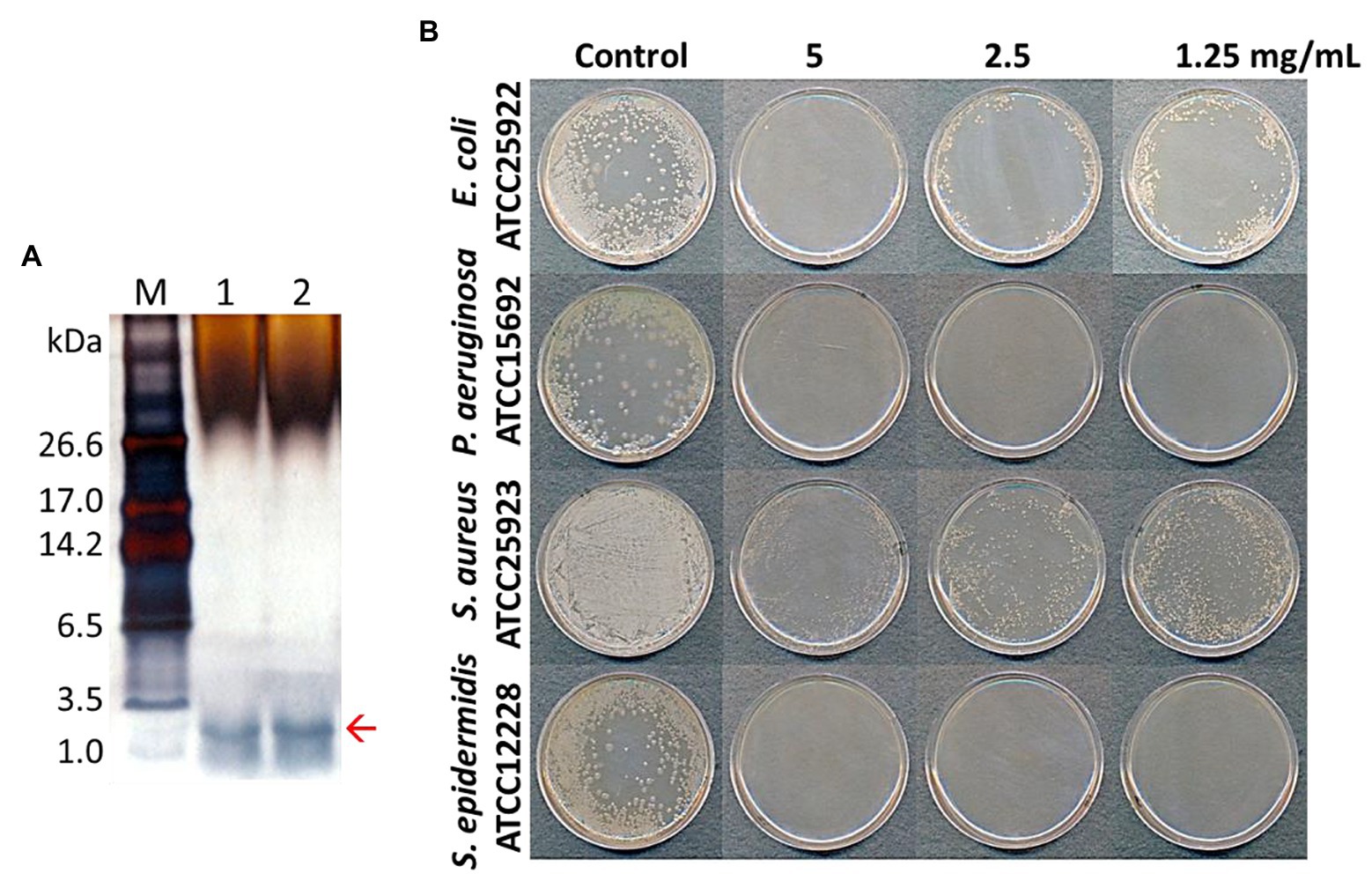
Figure 2. Tricine-SDS-PAGE analysis of total protein extracted from pine needles and evaluation of their antibacterial activity against bacterial strains [lane M; molecular weight size marker, Lanes 1 and 2: total extracted proteins from pine needles (5 and 10 μg, respectively)] (A); Antimicrobial activity of extracted protein fraction against Escherichia coli, Pseudomonas aeruginosa, S. aureus, and Staphylococcus epidermidis (B). The red arrow indicates the expected PN peptides.
In this study, the small (≤10 kDa) sample was treated as follows. The active ultrafiltered fractions of the sample ≤10 kDa were applied to an SPE 900 mg Lrg pore C18 column. The fractions from solid-phase extraction were collected using the following stepwise gradient: 10, 40, and 100% ACN (w/w). The 40% ACN (w/w) fraction was found to have antimicrobial activity (Supplementary Figures S2, S3C). C18 solid-phase extraction resulted in a 1.6-fold increased MIC value. The total protein (34.8 mg) decreased and the recovery was 2.8% of that for the soluble extract (Table 1). The active 40%-ACN (w/w) fractions were subjected to C18 RP-HPLC twice. Ten main peaks with retention times of 12–25 min were detected, which were eluted over a 40–65% gradient (Figure 3A). These peak fractions were checked for antimicrobial activity and the active PN peptide peaks were collected, pooled (Figure 3B), and confirmed using Tricine-SDS-PAGE, which resulted in one peptides band with molecular weights of 1–3.5 kDa (Figure 3A). The first elution yield of total protein obtained after the RP-HPLC step was drastically reduced (0.25–0.41%), although the MIC value against S. aureus was higher than that of the soluble extract (7.5–18.8-fold). The second RP-HPLC step increased the MIC value of the antibacterial peptides (PN-#5, PN-#7, PN-#8, and PN-#10) 117.2–937.5-fold compared to the initial soluble extract, with a recovery of 0.13–0.24% compared to the soluble extract (Table 1).
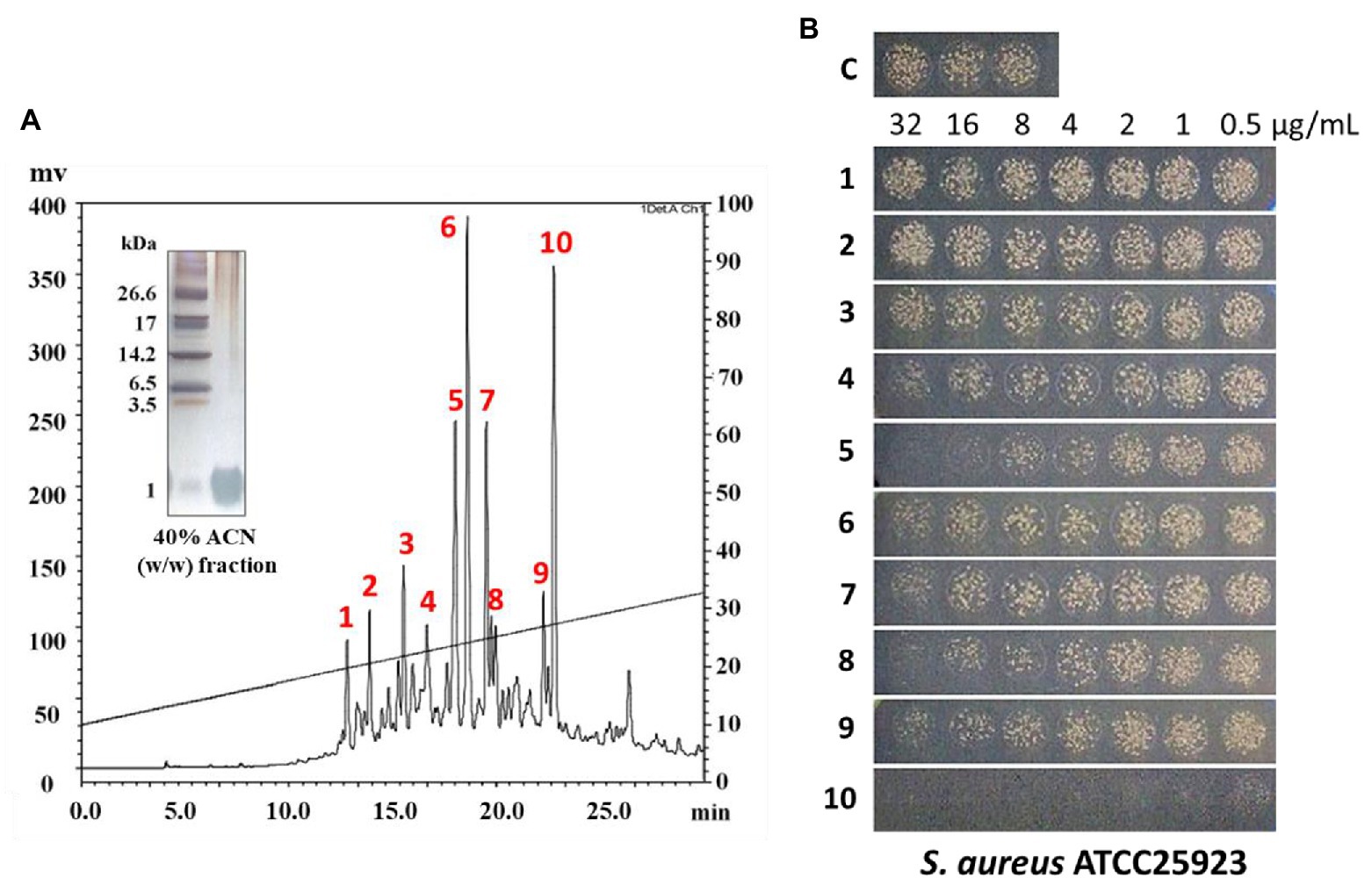
Figure 3. RP-HPLC analysis of peptides from 40% (w/w) ACN fractions of pine needles and estimation of their antimicrobial activity against Staphylococcus aureus. RP-HPLC chromatograms (absorbance was monitored at 215 nm) (A); Peak fractions from 40% (w/w) ACN fractions of pine needles exhibiting antimicrobial activity against Staphylococcus aureus (B).
The PN peptide fractions with potent antimicrobial activity (PN-#5, PN-#7, PN-#8, and PN-#10) were analyzed using RP-HPLC (Figure 4A) and MALDI TOF-MS (Figure 4B). MALDI TOF-MS analysis showed that PN-#5, PN-#7, PN-#8, and PN-#10 exhibited m/z peaks at 1346.0, 1567.4, 1715.4, and 1815.5, respectively (Figure 4B).
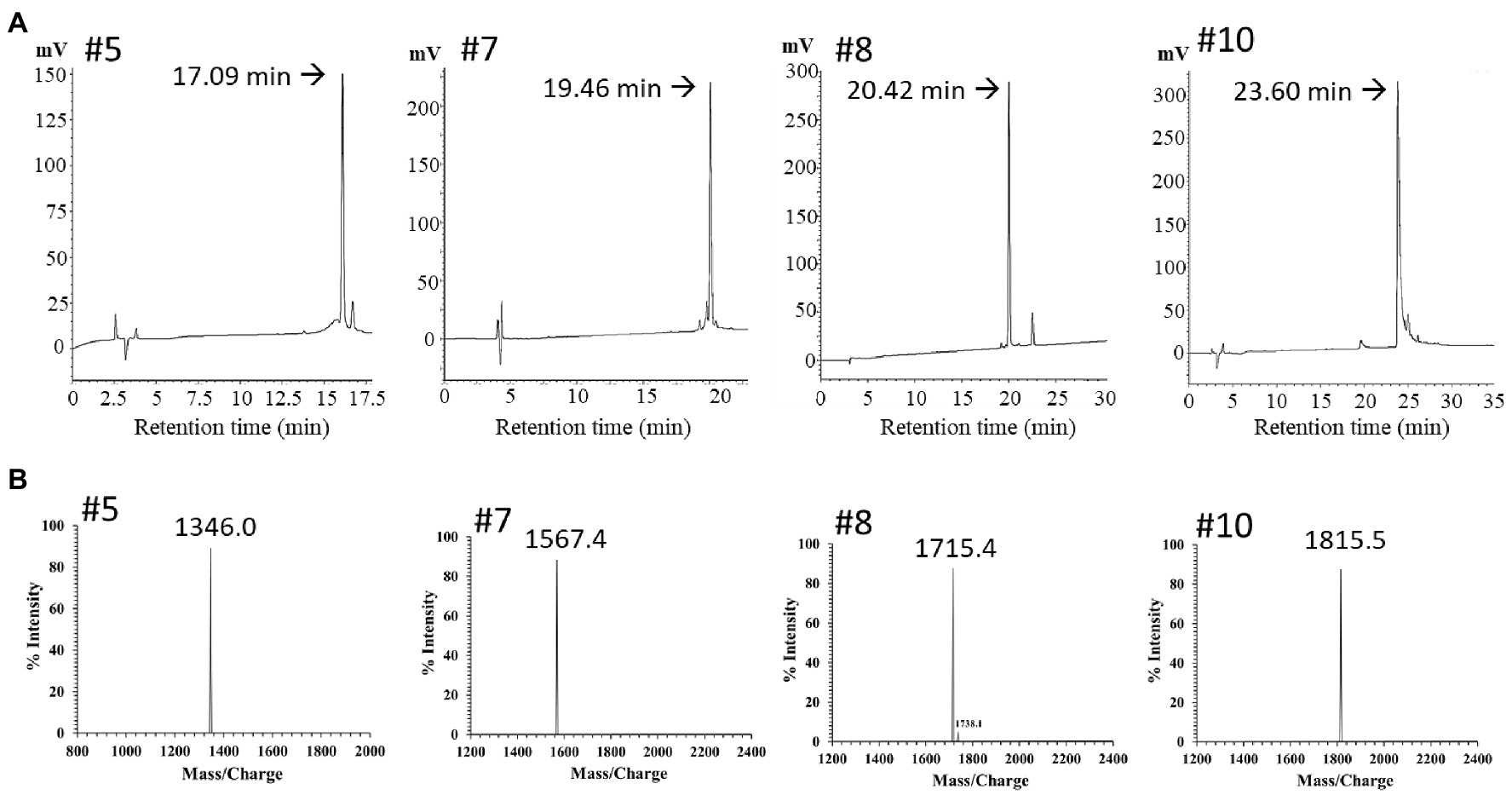
Figure 4. RP-HPLC and mass spectrum of purified peak peptides (PN-#5, PN-#7, PN-#8, and PN-#10). Chromatogram of AMPs in the 40% ACN fraction obtained using HPLC (A); Mass spectra of purified peak peptides using MALDI-TOF/MS (B).
Antimicrobial Assay of PN Peptides
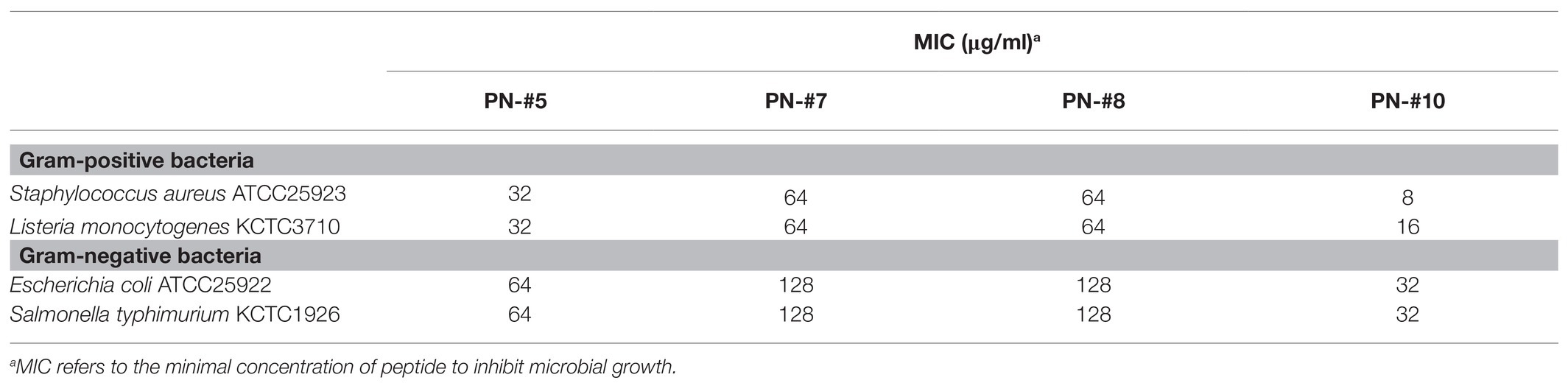
The bactericidal activities of PN peptides (PN-#1–10) were tested against S. aureus and E. coli (Figure 3B). None of the peptide fractions, except PN-#10 (data not shown), showed antimicrobial activity against E. coli up to a concentration of 32 μg/ml. In contrast, PN-#5, PN-#7, PN-#8, and PN-#10 showed particularly potent activities against S. aureus (Figure 3B). PN-#1, PN-#2, and PN-#3 exhibited no activity against S. aureus and E. coli. We determined the MIC values of PN-#5, PN-#7, PN-#8, and PN-#10 for S. aureus, E. coli, L. monocytogenes, and S. typhimurium (Table 2). The MIC values of the PN peptides ranged from 8 to 128 μg/ml. Of all PN peptides from PNs, PN-#10 showed the best activity against all tested bacteria, with MIC values of 8, 16, 32, and 32 μg/ml for S. aureus, L. monocytogenes, E. coli, and S. typhimurium, respectively. As a result, all PN peptides were more susceptible to Gram-positive bacteria (S. aureus and L. monocytogenes) than Gram-negative bacteria (Table 2).
Hemolytic Activity and Cytotoxicity of PN Peptides
The toxicity of the AMPs against eukaryotic cells was evaluated based on their lytic ability at tested concentrations (0–200 μg/ml; Figure 5A). The percentage of hemolysis was determined by measuring the amount of mouse hemoglobin released after incubation with PN-#5, PN-#7, PN-#8, PN-#10, or melittin (the reference AMP). The PN peptides did not cause hemolysis even at a concentration of 200 μg/ml. In contrast, melittin induced hemolysis even at the lowest concentration (3 μg/ml).
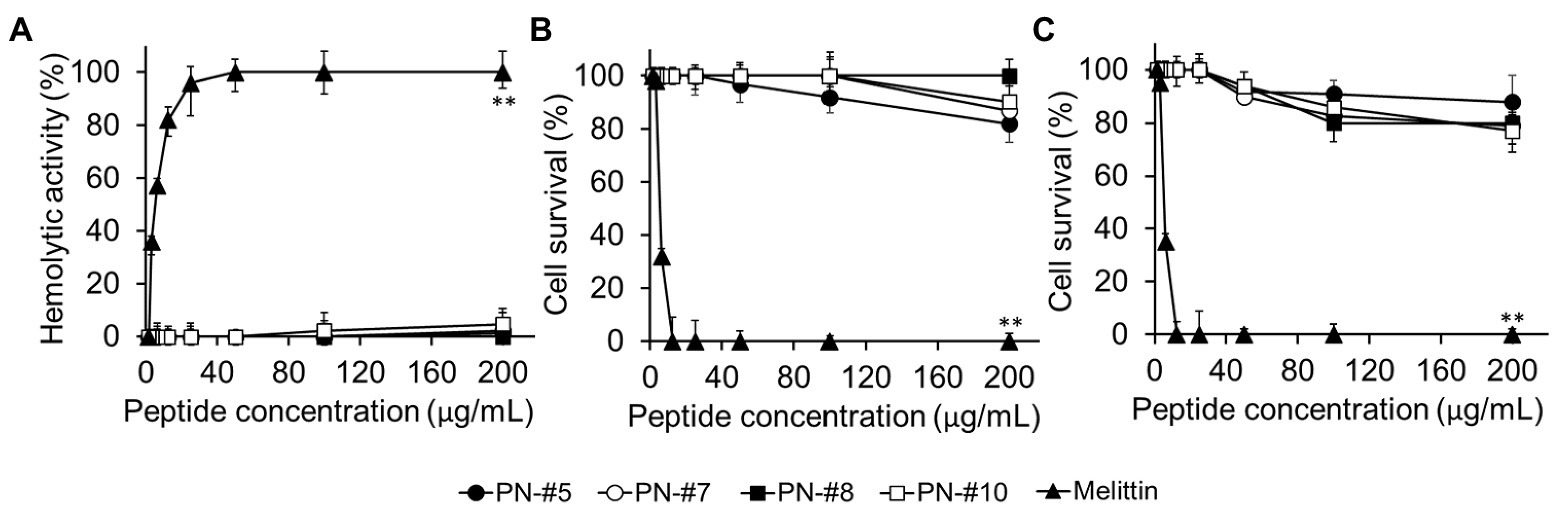
Figure 5. Hemolysis in mRBCs and cytotoxicity against RAW264.7 murine macrophages and HaCaT human keratinocyte cells. Hemolytic activity against mRBCs (n = 3 per condition) (A). Cytotoxic activity against RAW264.7 (n = 4 per condition) (B); and HaCaT (n = 3 per condition) (C) cells. Statistical analysis was performed by one-way ANOVA. **p < 0.01 vs. melittin.
The cytotoxicity of the peptides against RAW264.7 murine macrophages and HaCaT keratinocytes is shown in Figures 5B,C, respectively. The PN peptides at a concentration of 200 μg/ml caused less than 20% cytotoxicity in the RAW264.7 and HaCaT cell lines. These results suggested that the PN peptides are not toxic in the MIC range. In contrast, melittin, the reference AMP, caused 100% lysis at a concentration of 12.5 μg/ml (Figures 5B,C).
PN5 Identification
To identify the peptides and their a.a. sequence, PN-#5 was analyzed N-terminal a.a. sequence analysis with an Edman degradation. The a.a. sequence of the purified PN5 peptide was determined to be FKFLARTGKFL. BLASTp database search of the PN5 peptide was carried out to identify regions of similarity between NCBI sequence databases. The results revealed that the a.a. sequence FKFLARTGKFL has only two a.a. differences from FKYLQRTGKFL of transposase (NCBI seq. Id WP_065256059) from Moraxella lacunata.
For potential AMP prediction and their similarities to database-defined AMPs, the “APD3: Antimicrobial Peptide Calculator and Predictor” tool of the APD was used to identify PN peptides (Wang et al., 2016). The PN5 sequence showed 50% similarity with Temporin-HB2 from the Hubei gold-striped pond frog Pelophylax hubeiensis found in China (APD ID: AP02838, FLPFLAGLFGKIF), and Temporin-1Ce from the bronze frog Rana clamitans (APD ID: AP00108, FLPFLATLLSKVL). However, the sequence of PN5 did not identify any other plant protein or peptide, revealing that PN5 is a novel plant AMP. Figure 4B (#5) displays the singly charged potassium adducts [M+K]+ of PN5 with an m/z value of 1,347, equivalent to 1,328 Da, was determined using MALDI-TOF/MS. These results and the theoretical molecular weight of PN5 agree well with the determined a.a. sequence.
PN5’s Secondary Structure and in silico Analysis
PN5 has an α-helical amphipathic structure, as suggested by the helical wheel projection in Figure 6A. In addition, the PN5 peptide carried a net charge (3) and was hydrophobic (H, 0.465). The ExPASy tool (SIB Bioinformatics Resource Portal) and APD3 (Antimicrobial Peptide Calculator and Predictor) were used to obtain the physicochemical properties of PN5 such as theoretical isoelectric point (pI, 11.17), hydrophobic moment (μH, 0.471), protein-binding potential (Boman index, 0.64 kcal/mol), and grand average of hydropathicity (GRAVY, 0.400). The PN5 instability index was estimated to be −20.01, indicating a stable peptide under both in vitro and in vivo conditions. These results with in silico prediction were consistent with the results obtained using PyMOL, which predicted that PN5 assumes an α-helical conformation (Figure 6B).
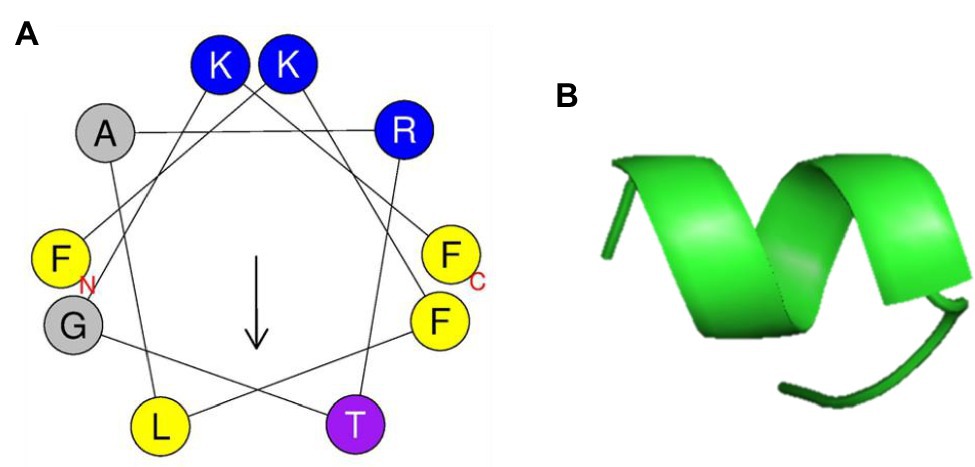
Figure 6. Helical wheel projection and three-dimensional structure projection of PN5 peptide. Helical wheel projection of PN5 was generated using HeliQuest (http://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) (A). Simulations of the three-dimensional structures of peptides generated using PyMOL (B).
PN5 Peptide Synthesis, Antimicrobial Activity, and Toxicity
In general, amidated peptides have a greater tendency to generate an α-helical structure than the non-amidated peptides. Chemically synthesized peptides with C-terminal amidation showed strong inhibitory activity against bacteria and low hemolytic activity (Strandberg et al., 2007; Shahmiri et al., 2015). Thus, we synthesized PN5 peptide (FKFLARTGKFL) and PN5 with an amidated C-terminus (FKFLARTGKFL-NH2), based on the N-terminal sequence results.
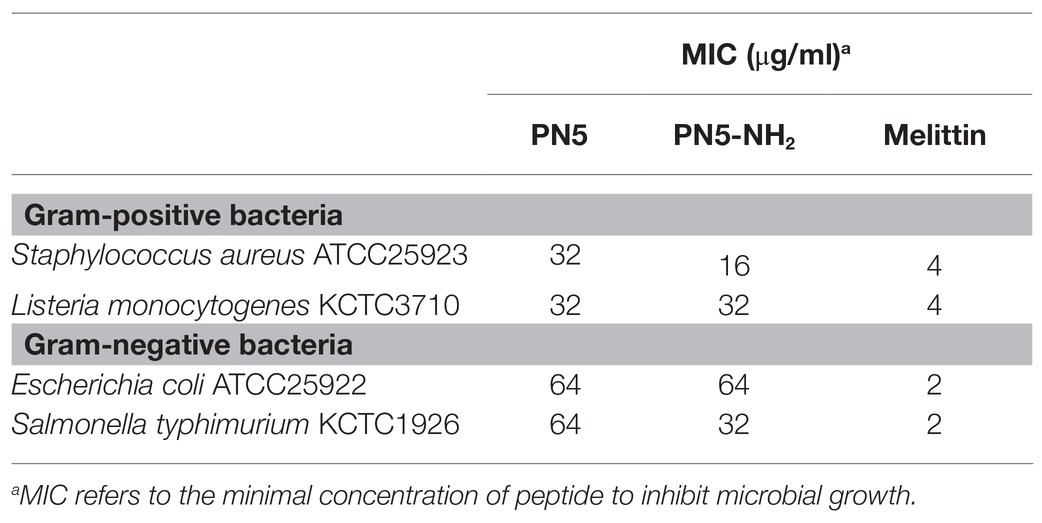
The antimicrobial activities of the synthetic PN5 and PN5-NH2 peptide were evaluated against two Gram-negative (E. coli and S. typhimurium) and two Gram-positive (S. aureus and L. monocytogenes) bacteria. The synthetic PN5 peptide showed the same antimicrobial activity as that did the purified PN5 peptide (PN-#5; Table 3). PN5-NH2 peptide showed better antimicrobial activity than PN-#5, against S. aureus, L. monocytogenes, E. coli, and S. typhimurium with MICs of 16, 32, 64, and 32 μg/ml, respectively (Table 3).
After analyzing the synthetic PN5 activity, to confirm the increased toxicity of PN5-NH2, hemolysis and cytotoxicity were confirmed under the same conditions as those used for the purified PN-#5 peptide (Supplementary Figure S4). The synthetic PN5 peptide showed only approximately 5% hemolysis activity at a high concentration of 200 μg/ml, which is similar to that of the PN-#5 peptide (Supplementary Figure S4A). The PN5 peptide caused only 19% cytotoxicity up to a concentration of 200μg/ml in the HaCaT cell line (Supplementary Figure S4B). These results show that the synthetic PN5 peptide is not toxic in the MIC range, which is consistent with the toxicity of the PN-#5 peptide. Thus, the PN5 peptide appears to be a good candidate as an AMP.
Discussion
Recent studies on PN extract have been reported as food and health supplements, as well as food preservatives (Mahajan et al., 2016; Wu et al., 2017). Contaminated food by the pathogenic microorganisms is a major common form of food degradation, and is often responsible for the occurrence of food-borne illnesses such as infection, food poisoning, toxic shock syndrome, and sepsis (Bintsis, 2017; Chen and Alali, 2018; Riley, 2020). Increasing the use of chemical preservatives can effectively prevent the survival and proliferation of most food-borne bacteria; however, drawbacks related with the safety of chemical preservatives are on the rise (Halla et al., 2018; Silva et al., 2018). Recently, as the demand for foods with health benefits has increased, the amount of chemical preservatives used in foods has decreased; thus, there is a need for natural preservatives. As bacterial resistance to antibiotics is increasing, many studies have been performed to develop effective and nontoxic antimicrobial substances that do not induce antimicrobial resistance. Many studies are being conducted to develop new natural antibacterial substances in various plant extracts for safe food preservation and to investigate whether they have antibacterial activity in various microorganisms including food-borne bacteria (Hintz et al., 2015; Mostafa et al., 2018). Recent studies reported new AMPs (peptides consisting of 10–14 a.a. residues) isolated from the Mediterranean medical plant Charybdis, with similar antimicrobial activity (25–45 μ/ml in S. aureus and P. aeruginosa) as that of PN peptides (Cunsolo et al., 2020). In this study, the PN peptide showed potent antimicrobial activity against some representative food poisoning pathogens, particularly S. aureus (Figure 3B; Table 2). PN peptides had better antimicrobial activity against Gram-positive bacteria than Gram-negative bacteria, and showed the highest ability to kill S. aureus. This may be related to differences in bacterial cell structure between Gram-positive and Gram-negative bacteria. The cell wall of Gram-negative bacteria is thinner but more complex than that of Gram-positive bacteria. It has been suggested that differences in antimicrobial activity toward some antimicrobial agents between Gram-positive and Gram-negative bacteria are due to the low permeability of Gram-negative bacteria to antibacterial compounds through the outer cell wall. The outer cell wall with low permeability of Gram-negative bacteria can interfere with the accumulation in the cytoplasmic membrane, thereby reducing the killing efficiency of the antimicrobial compound (Zgurskaya et al., 2015; Breijyeh et al., 2020). Besides transporting small molecules such as nutrients, the membranes of Gram-negative bacteria are resistant to chemical entry (Wu et al., 2016; Tiz et al., 2018). Therefore, developing alternative substances is essential for eradicating these bacteria using natural AMPs.
PN peptides isolated from PNs from P. densiflora Sieb. et Zucc. exhibited strong antimicrobial activity against foodborne bacteria. Moreover, the AMPs PN-#5, PN-#7, PN-#8, and PN-#10 exhibited no cytotoxicity at the antimicrobial concentrations. Given these results, PN5 might become an effective drug candidate for developing antimicrobial agents in the food and pharmaceutical industries. In addition, PN5 peptide can be used as a natural food additive. Future studies can be directed at a.a. substitution on the primary structure, to obtain more active AMPs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JL: study design, experiment, and data analysis. HKK: study design, experiment, data analysis, and writing – original draft. HC: methodology, visualization, and resources. YP: investigation, resources, and writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (No. 2019R1A2B5B03070330 and NRF-2017M3A9E4077206) and Institute for Information & Communications Technology Promotion (IITP) grant funded by the Korean Government (MSIT; No. 2017-0-01714, Development of Antimicrobial Peptide using Deep Learning).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.662462/full#supplementary-material
Footnotes
References
Ahn, C., Jang, Y. J., Kim, J. W., Park, M. J., Yoo, Y. M., and Jeung, E. B. (2018). Anti-asthmatic effects of volatile organic compounds from Chamaecyparis obtusa, Pinus densiflora, Pinus koraiensis, and Larix kaempferi wood panels. J. Physiol. Pharmacol. 69, 933–941. doi: 10.26402/jpp.2018.6.07
Bintsis, T. (2017). Foodborne pathogens. AIMS Microbiol. 3, 529–563. doi: 10.3934/microbiol.2017.3.529
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Breijyeh, Z., Jubeh, B., and Karaman, R. (2020). Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 25:1340. doi: 10.3390/molecules25061340
Chen, L., and Alali, W. (2018). Editorial: recent discoveries in human serious foodborne pathogenic bacteria: resurgence, pathogenesis, and control strategies. Front. Microbiol. 9:2412. doi: 10.3389/fmicb.2018.02412
Coates, M., Blanchard, S., and Macleod, A. S. (2018). Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PLoS Pathog. 14:e1007353. doi: 10.1371/journal.ppat.1007353
Cunsolo, V., Schicchi, R., Chiaramonte, M., Inguglia, L., Arizza, V., Cusimano, M. G., et al. (2020). Identification of new antimicrobial peptides from the Mediterranean medicinal plant Charybdis pancration (Steinh.) Speta. Antibiotic 9:747. doi: 10.3390/antibiotics9110747
Dhama, K., Karthik, K., Khandia, R., Munjal, A., Tiwari, R., Rana, R., et al. (2018). Medicinal and therapeutic potential of herbs and plant metabolites/extracts countering viral pathogens-current knowledge and future prospects. Curr. Drug Metab. 19, 236–263. doi: 10.2174/1389200219666180129145252
Gautier, R., Douguet, D., Antonny, B., and Drin, G. (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24, 2101–2102. doi: 10.1093/bioinformatics/btn392
Ghaffari, T., Kafil, H. S., Asnaashari, S., Farajnia, S., Delazar, A., Baek, S. C., et al. (2019). Chemical composition and antimicrobial activity of essential oils from the aerial parts of Pinus eldarica grown in Northwestern Iran. Molecules 24:3203. doi: 10.3390/molecules24173203
Ha, T. K. Q., Lee, B. W., Nguyen, N. H., Cho, H. M., Venkatesan, T., Doan, T. P., et al. (2020). Antiviral activities of compounds isolated from Pinus densiflora (pine tree) against the influenza A virus. Biomol. Ther. 10:711. doi: 10.3390/biom10050711
Halla, N., Fernandes, I. P., Heleno, S. A., Costa, P., Boucherit-Otmani, Z., Boucherit, K., et al. (2018). Cosmetics preservation: a review on present strategies. Molecules 23:1571. doi: 10.3390/molecules23071571
Hancock, R. E., Haney, E. F., and Gill, E. E. (2016). The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334. doi: 10.1038/nri.2016.29
Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2019). Reassessing the host defense peptide landscape. Front. Chem. 7:43. doi: 10.3389/fchem.2019.00043
Hintz, T., Matthews, K. K., and Di, R. (2015). The use of plant antimicrobial compounds for food preservation. Biomed. Res. Int. 2015:246264. doi: 10.1155/2015/246264
Hsu, B., Coupar, I. M., and Ng, K. (2006). Antioxidant activity of hot water extract from the fruit of the Doum palm. Hyphaene thebaica. Food Chem. 98, 317–328. doi: 10.1016/j.foodchem.2005.05.077
Jeong, J. B., Seo, E. W., and Jeong, H. J. (2009). Effect of extracts from pine needle against oxidative DNA damage and apoptosis induced by hydroxyl radical via antioxidant activity. Food Chem. Toxicol. 47, 2135–2141. doi: 10.1016/j.fct.2009.05.034
Kang, H. K., Seo, C. H., Luchian, T., and Park, Y. (2018). Pse-T2, an antimicrobial peptide with high-level, broad-spectrum antimicrobial potency and skin biocompatibility against multidrug-resistant Pseudomonas aeruginosa infection. Antimicrob. Agents Chemother. 62, e01493–e01418. doi: 10.1128/AAC.01493-18
Kim, K. J., Hwang, E. S., Kim, M. J., Park, J. H., and Kim, D. O. (2020a). Antihypertensive effects of polyphenolic extract from Korean red pine (Pinus densiflora Sieb. et Zucc.) bark in spontaneously hypertensive rats. Antioxidants 9:333. doi: 10.3390/antiox9040333
Kim, N. Y., Jang, M. K., Lee, D. G., Yu, K. H., Jang, H., Kim, M., et al. (2010). Comparison of methods for proanthocyanidin extraction from pine (Pinus densiflora) needles and biological activities of the extracts. Nutr. Res. Pract. 4, 16–22. doi: 10.4162/nrp.2010.4.1.16
Kim, W., Park, C., Park, J., Cheong, H., and Kim, S. J. (2020b). Pine needle hexane extract promote cell cycle arrest and premature senescence via p27KIP1 upregulation gastric cancer cells. Food Sci. Biotechnol. 29, 845–853. doi: 10.1007/s10068-019-00730-5
Ku, C. S., Sathishkumar, M., and Mun, S. P. (2007). Binding affinity of proanthocyanidin from waste Pinus radiata bark onto proline-rich bovine achilles tendon collagen type I. Chemosphere 67, 1618–1627. doi: 10.1016/j.chemosphere.2006.11.037
Kwak, C. S., Moon, S. C., and Lee, M. S. (2006). Antioxidant, antimutagenic, and antitumor effects of pine needles (Pinus densiflora). Nutr. Cancer 56, 162–171. doi: 10.1207/s15327914nc5602_7
Mahajan, D., Bhat, Z. F., and Kumar, S. (2016). Pine needles (Cedrus deodara (Roxb.) loud.) extract as a novel preservative in cheese. Food Packag. Shelf Life 7, 20–25. doi: 10.1016/j.fpsl.2016.01.001
Maimoona, A., Naeem, I., Saddiqe, Z., and Jameel, K. (2011). A review on biological, nutraceutical and clinical aspects of French maritime pine bark extract. J. Ethnopharmacol. 133, 261–277. doi: 10.1016/j.jep.2010.10.041
Mcgrath, K. C., Li, X. H., Mcrobb, L. S., Heather, A. K., and Gangoda, S. V. S. (2015). Inhibitory effect of a French maritime pine bark extract-based nutritional supplement on TNF-α-induced inflammation and oxidative stress in human coronary artery endothelial cells. Evid. Based Complement. Alternat. Med. 2015:260530. doi: 10.1155/2015/260530
Mookherjee, N., Anderson, M. A., Haagsman, H. P., and Davidson, D. J. (2020). Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug. Discov. 19, 311–332. doi: 10.1038/s41573-019-0058-8
Mostafa, A. A., Al-Askar, A. A., Almaary, K. S., Dawoud, T. M., Sholkamy, E. N., and Bakri, M. M. (2018). Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi J. Biol. Sci. 25, 361–366. doi: 10.1016/j.sjbs.2017.02.004
Park, S. C., Kim, M. H., Hossain, M. A., Shin, S. Y., Kim, Y., Stella, L., et al. (2008). Amphipathic alpha-helical peptide, HP (2-20), and its analogues derived from helicobacter pylori: pore formation mechanism in various lipid compositions. Biochim. Biophys. Acta 1778, 229–241. doi: 10.1016/j.bbamem.2007.09.020
Park, J. S., and Lee, G. H. (2011). Volatile compounds and antimicrobial and antioxidant activities of the essential oils of the needles of Pinus densiflora and Pinus thunbergii. J. Sci. Food Agric. 91, 703–709. doi: 10.1002/jsfa.4239
Park, J., Lee, B., Choi, H., Kim, W., Kim, H. J., and Cheong, H. (2016). Antithrombosis activity of protocatechuic and shikimic acids from functional plant Pinus densiflora Sieb. Et Zucc needles. J. Nat. Med. 70, 492–501. doi: 10.1007/s11418-015-0956-y
Pouvreau, L., Gruppen, H., Piersma, S. R., Van Den Broek, L. A., Van Koningsveld, G. A., and Voragen, A. G. (2001). Relative abundance and inhibitory distribution of protease inhibitors in potato juice from cv. Elkana. J. Agric. Food Chem. 49, 2864–2874. doi: 10.1021/jf010126v
Riley, L. W. (2020). Extraintestinal foodborne pathogens. Annu. Rev. Food Sci. Technol. 11, 275–294. doi: 10.1146/annurev-food-032519-051618
Sathoff, A. E., and Samac, D. A. (2019). Antibacterial activity of plant defensins. Mol. Plant-Microbe Interact. 32, 507–514. doi: 10.1094/MPMI-08-18-0229-CR
Schafer, A., Chovanova, Z., Muchova, J., Sumegova, K., Liptakova, A., Durackova, Z., et al. (2006). Inhibition of COX-1 and COX-2 activity by plasma of human volunteers after ingestion of French maritime pine bark extract (Pycnogenol). Biomed. Pharmacother. 60, 5–9. doi: 10.1016/j.biopha.2005.08.006
Schagger, H., and Von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. doi: 10.1016/0003-2697(87)90587-2
Shahmiri, M., Enciso, M., and Mechler, A. (2015). Controls and constraints of the membrane-disrupting action of Aurein 1.2. Sci. Rep. 5:16378. doi: 10.1038/srep16378(2015)
Silva, C. C. G., Silva, S. P. M., and Ribeiro, S. C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 9:594. doi: 10.3389/fmicb.2018.00594
Strandberg, E., Tiltak, D., Ieronimo, M., Kanithasen, N., Wadhwani, P., and Ulrich, A. S. (2007). Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl. Chem. 79, 717–728. doi: 10.1351/pac200779040717
Tam, J. P., Wang, S., Wong, K. H., and Tan, W. L. (2015). Antimicrobial peptides from plants. Pharmacy 8, 711–757. doi: 10.3390/ph8040711
Tiz, D. B., Kikelj, D., and Zidar, N. (2018). Overcoming problems of poor drug penetration into bacteria: challenges and strategies for medicinal chemists. Expert Opin. Drug Discov. 13, 497–507. doi: 10.1080/17460441.2018.1455660
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wu, Y. P., Bai, J. R., Zhong, K., Huang, Y. N., and Gao, H. (2017). A dual antibacterial mechanism involved in membrane disruption and DNA binding of 2R,3R-dihydromyricetin from pine needles of Cedrus deodara against Staphylococcus aureus. Food Chem. 218, 463–470. doi: 10.1016/j.foodchem.2016.07.090
Wu, Y., Bai, J., Zhong, K., Huang, Y., Qi, H., Jiang, Y., et al. (2016). Antibacterial activity and membrane-disruptive mechanism of 3-p-trans-coumaroyl-2-hydroxyquinic acid, a novel phenolic compound from pine needles of Cedrus deodara, against Staphylococcus aureus. Molecules 21:1084. doi: 10.3390/molecules21081084
Keywords: Pinus densiflora, pine needles, antimicrobial peptide, ultrafiltration, foodborne bacteria
Citation: Lee J, Kang HK, Cheong H and Park Y (2021) A Novel Antimicrobial Peptides From Pine Needles of Pinus densiflora Sieb. et Zucc. Against Foodborne Bacteria. Front. Microbiol. 12:662462. doi: 10.3389/fmicb.2021.662462
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilReviewed by:
Eugene A. Rogozhin, Institute of Bioorganic Chemistry, Russian Academy of Sciences (RAS), RussiaAlba Pérez-Cataluña, Institute of Agrochemistry and Food Technology (IATA), Spain
Copyright © 2021 Lee, Kang, Cheong and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoonkyung Park, eV9rX3BhcmtAY2hvc3VuLmFjLmty
†These authors have contributed equally to this work and share first authorship
 Junho Lee1†
Junho Lee1† Yoonkyung Park
Yoonkyung Park