- 1Lehrstuhl für Technische Mikrobiologie, Technische Universität München, Freising, Germany
- 2Bayerisches Zentrum für Biomolekulare Massenspektrometrie (BayBioMS), Freising, Germany
The ability of certain Pseudomonas (P.) species to grow or persist in anoxic habitats by either denitrification, acetate fermentation, or arginine fermentation has been described in several studies as a special property. Previously, we had isolated strains belonging to the species P. lundensis, P. weihenstephanensis, and P. fragi from anoxic modified atmosphere packaged (MAP) minced beef and further proved their anaerobic growth in vitro on agar plates. This follow-up study investigated the anaerobic growth of two strains per respective species in situ on inoculated chicken breast filet under 100% N2 modified atmosphere. We were able to prove anaerobic growth of all six strains on chicken breast filet with cell division rates of 0.2–0.8/day. Furthermore, we characterized the anaerobic metabolic lifestyle of these Pseudomonas strains by comparative proteomics, upon their cultivation in meat simulation media, which were constantly gassed with either air or 100% N2 atmospheres. From these proteomic predictions, and respective complementation by physiological experiments, we conclude that the Pseudomonas strains P. fragi, P. weihenstephanensis, P. lundensis exhibit a similar anaerobic lifestyle and employ arginine fermentation via the arginine deiminase (ADI) pathway to grow anaerobically also on MAP meats. Furthermore, glucose fermentation to ethanol via the ED-pathway is predicted to enable long term survival but no true growth, while respiratory growth with nitrate as alternative electron acceptor or glucose fermentation to acetate could be excluded due to absence of essential genes. The citric acid cycle is partially bypassed by the glyoxylate shunt, functioning as the gluconeogenetic route without production of NADH2 under carbon limiting conditions as e.g., in packaged meats. Triggered by an altered redox balance, we also detected upregulation of enzymes involved in protein folding as well as disulfide bonds isomerization under anoxic conditions as a counteracting mechanism to reduce protein misfolding. Hence, this study reveals the mechanisms enabling anaerobic grow and persistence of common meat-spoiling Pseudomonas species, and further complements the hitherto limited knowledge of the anaerobic lifestyle of Pseudomonas species in general.
Introduction
The genus Pseudomonas currently comprises more than 240 valid published and correct named species (Parte, 2018; Parte et al., 2020), out of which several species have been identified as key players of food spoilage: P. fluorescens, P. putida, P. fragi, P. lundensis, and P. weihenstephanensis are spoiling meat (Ercolini et al., 2007; Hilgarth et al., 2019) but also cheese and raw milk (Wiedmann et al., 2000; Morales et al., 2005; Marchand et al., 2009; Martin et al., 2011; Scatamburlo et al., 2015; Neubeck et al., 2016; Meng et al., 2017). Their high spoilage potential is due to proteolytic and lipolytic enzymes, which are released out of the cell, initiating changes in the physicochemical and organoleptic properties of products (Alquati et al., 2002; Rajmohan et al., 2002; Marchand et al., 2009; Stoeckel et al., 2016).
Most Pseudomonas species are described as obligately aerobic microorganisms, which are unable to grow under conditions of complete oxygen exclusion (Garrity et al., 2005). Thus, anoxic food packaging technologies such as vacuum and modified atmosphere packaging (MAP) are powerful tools to limit spoilage by Pseudomonas species (Wang et al., 2018). Nevertheless, it is known that some Pseudomonas species such as P. aeruginosa and P. denitrificans are able to grow even under strictly anoxic conditions (O2 < 1 ppm) by either usage of alternative electron acceptors (mainly nitrate respiration) (Carlson and Ingraham, 1983), arginine fermentation (Vander Wauven et al., 1984) or pyruvate fermentation (Eschbach et al., 2004). Reports in the literature of anaerobic growth of meat-spoiling Pseudomonas spp. are scarce. A study by Wang et al. (2017) demonstrated anaerobic growth of several P. fragi strains in tryptone soy broth media. Other studies identified P. fragi strains in vacuum packed (VP) beef samples (Ercolini et al., 2011; Jääskeläinen et al., 2016), whereas De Filippis et al. (2019) proved those strains to be viable and metabolically active on VP beef, although those characteristics were strongly strain dependent (De Filippis et al., 2019).
Furthermore, we have previously investigated the diversity development of Pseudomonas spp. during minced beef spoilage and isolated certain anaerobic P. fragi, P. lundensis and P. weihenstephanensis strains (Hilgarth et al., 2019). Although these species had been previously described as strictly aerobic at that time, that study could demonstrate anaerobic growth for isolates of the three species in vitro on agar plates. However, the potential of those meat-spoiling Pseudomonas species to either grow or only persist under anoxic conditions has not yet been investigated in detail and the underlying mechanism remained unknown in the previous work. Consequently, this study addresses this question by performing a full-proteomic study in meat simulation media, providing insights into the aerobic and anaerobic metabolism of the isolated Pseudomonas strains from Hilgarth et al. (2019). Furthermore, we checked the in situ growth of all strains on anoxically packaged chicken meat (100% N2).
Materials and Methods
Strain Selection
Two strains of each meat-spoiling species P. lundensis (TMW2. 1732/TMW2.2076), P. weihenstephanensis (TMW2.2077/TMW2. 1728), and P. fragi (TMW2.2081/TMW2.2082) were chosen for this study. All strains were isolated in previous studies from minced meat (Hilgarth et al., 2019) or beef steak (Hilgarth et al., 2018a) initially packaged under modified atmosphere (>70% O2, >20% CO2). Isolation of anaerobically growing Pseudomonas strains has been performed at late storage time of the meat products when oxygen had already been depleted within the packages. P. aeruginosa DSM 1117 was used as a positive control for anaerobic nitrate respiration and substrate utilization tests.
DNA Extraction and Sequencing
DNA extraction was performed using the E.Z.N.A. bacterial DNA Kit (Omega Bio-Tek, Norcross, GA) following the manufacturer’s instructions. Isolated DNA was sent for sequencing to the microbiome core facility of the Institute of Food & Health (TUM Freising, Germany) for Illumina MiSeq shotgun sequencing.
The NCBI accession numbers for all strains are as followed: P. lundensis TMW2.1732 (JACJCQ000000000), P. lundensis TMW2.2076 (JAAEBS000000000), P. weihenstephanensis TM W2.2077 (JAAEBW000000000), P. weihenstephanensis TMW2. 1728 (JAAEBV000000000), P. fragi TMW2.2081 (JAAEBQ 000000000), and P. fragi TMW2.2082 (JAAEBR000000000).
Test of Anaerobic Growth in situ on Chicken Breast
Chicken breast filet, packaged under a modified atmosphere containing 70% O2 and 30% CO2 were obtained from a local discounter with a use-by date assigned to >3 days. Packages were opened and meat was cut into equally thick, square pieces (36 cm2) under sterile conditions. For meat inoculation, cells were pre-cultured oxically, overnight in brain heart infusion (BHI) media (Roth, Karlsruhe, Germany) at 25°C. Before harvesting, cultures were adapted to chilled conditions by shaking at 4°C for 4 h. Afterward, cells were washed twice with quarter-strength ringer’s solution (Merck, Darmstadt, Germany). The prepared meat pieces were inoculated on both sides with 100 μl of OD600 = 10 of one of the strains. Cells were distributed homogeneously on the meat surface using a sterile spatula. Inoculation was performed at high cell counts (approx. log10/cm2 = 6) to outcompete the autochthonous microbiome of purchased chicken meat, which was log10/cm2 = 2.0–4.9, depending on the package. Inoculated meat pieces were packaged under an oxygen free atmosphere (100% N2) within polypropylene (PP) trays (ES-Plastic, Hutthurm, Germany) coated with an oxygen impermeable barrier film ethylen-vinylalcohol (EVOH) (oxygen transmission rate 0.5 cc/m2/24 h) using the packaging machine Rotarius VG (VarioVAC, Zarrentin, Germany). CO2 in the MAP (as mostly used in industrial packages) was omitted to avoid any additional effects of this gas and strictly focus on the anoxic conditions effect. To ensure complete anoxic conditions, an oxygen scavenger AnaeroGen 2.5 liter (Thermo Fisher Scientific, Waltham, MA) was added to each package, containing three meat pieces (3 replicates). Packages were stored at 4°C until sampling.
Samples for CFU determination were taken after 0, 3, 5, and 7 days. Packages were opened within a Bactron anaerobic chamber (Sheldon, Cornelius, United States) containing <5% H2 and >95% N2 atmosphere. Each meat piece was transferred into a sterile 50 ml falcon tube containing quarter-strength ringer’s solution and homogenized by vertexing for 2 min. Afterward, the cell suspension was diluted serial times and each dilution was plated on BHI plates. Cultivation of plates was performed anoxically at 25°C for 48 h. Significant growth on meat was checked by a t-test between log10 (CFU/cm2) values obtained at day 0 with CFU values of the other days (day 0–3, day 0–5, and day 0–7; significance level p < 0.05). The recovery/identity of inoculated strains on meat was proven by a random amplified polymorphic DNA (RAPD) PCR (Hilgarth et al., 2018b), performed with 12 colonies of each replicate after 7 days.
Experimental Design of the Proteomic Study
In order to exclude interfering background microbiota and high numbers of detected meat proteins, all strains were cultivated in vitro in sterile glass bottles containing 400 ml of a meat simulation (MS) media as previously employed and described by Kolbeck et al. (2019). In detail, MS media consists of 12.5 g/l meat extract, 0.05 mM Tween80, 0.5% glycerol and 2 μg/ml hemin chloride adjusted to a pH of 5.8 with 100% lactic acid. MS medium was inoculated with an optical density of 0.1 at 590 nm with freshly prepared and washed precultures as described above. Bacteria were cultivated for 48 h, 120 rpm at 25°C ± 2°C. During cultivation, MS media were constantly aerated with either 100% N2 or air. Growth of the strains was monitored over whole cultivation period and samples for proteomic were taken at logarithmic growth phase.
Proteomic Sample Preparation, Measurement, and Data Analysis
Samples for the proteomic study were prepared as described previously (Kolbeck et al., 2020). Briefly, cells were lysed, protein concentrations were determined; 100 μg proteins were reduced, carbamidomethylated and digested with trypsin. The resulting peptide mixture was desalted and finally resuspended to a concentration of 0.1 μg∗μl–1. LC-MS/MS data acquisition was performed on an Ultimate 3000 RSLCnano system coupled to a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Waltham, MA). All operating parameters were set equal to Kolbeck et al. (2020). Identification and quantification of proteins was done by the software MaxQuant (version 1.6.3.4) (Tyanova et al., 2016a) using its built-in search engine Andromeda (Cox et al., 2011). All build in options and operating parameters of MaxQuant were set equal as described by Kolbeck et al. (2020).
Proteomic Data Interpretation
The open-source software Perseus (Tyanova et al., 2016b) was used for statistical data analysis. Therefore, proteins were filtered for high-confidence (false-discovery rate cutoff < 1% based on peptide-spectrum match (PSM) level and protein level, removal of reverse, contaminant and “only-identified by site” proteins) and grouped into biological replicates. Label free quantification (LFQ) values were log2 transformed and a Welch t-test was performed revealing statistic significantly differential regulated proteins between the two cultivation conditions air and N2 (p < 0.01 and log2 > 2). All obtained proteins were further confirmed by manual sequence searches using the basic local alignment search tool (BLAST) provided by NCBI (Altschul et al., 1990). Different annotation programs (NCBI, TIGR, SEED, KEGG) as well as manual curation was applied to group the identified proteins into functional categories, revealing whole metabolic pathways being differentially regulated between oxic and anoxic cultivation of our Pseudomonas strains.
API 20 NE Test
An API NE 20 test (BioMérieux, Nürtingen, Germany) was performed following the manufacturer’s instructions. Results from the NO3, TRP, Glu were read out after 24 h, while other results were read out after 48 h of incubation of the stripe.
In vitro Test for Anaerobic Arginine and Glucose Fermentation
Arginine and glucose metabolism were checked performing a plate assay. Therefore, 20 mM L-arginine (Sigma-Aldrich GmbH, Darmstadt, Germany) or 20 mM glucose monohydrate (Merck, Darmstadt, Germany) was supplemented to 5 g∗l–1 NaCl (Roth, Karlsruhe, Germany), 2 g∗l–1 KH2PO4 (Merck, Darmstadt, Germany), 1 g∗l–1 peptone from soy (Roth, Karlsruhe, Germany) and 12 mg∗l–1 phenol red (Thermo Fisher Scientific, Waltham, MA). The latter was added as a pH indicator, changing from yellow to red-purple in case of alkalization, as expected from arginine degradation. All chemicals were added after autoclaving by sterile filtering with 0.02-mm pore size. The final pH was adjusted to 6.0. Medium without arginine or glucose monohydrate was used as control.
Pseudomonas strains were precultured in BHI as described above, washed twice in sterile quarter-strength ringer’s solution and adjusted to an OD600 = 0.5. 20 μl of the prepared precultures were dripped on plates containing arginine and no arginine or glucose and no glucose. The experiment was performed twice, as plates were cultivated once under oxic and once under anoxic conditions at 25°C. Anoxic plates were packaged into sterile PP-EVOH trays with 100% N2 containing an oxygen scavenger as described above. After 4 days, color change was validated taking standardized pictures using a colonyDoc-It imaging station (VWR, Darmstadt, Germany). P. aeruginosa DSM 1117 was used as a positive control.
In vitro Test for Anaerobic Nitrate Respiration
Dissimilatory nitrate reduction was checked by a physiological assay, monitoring growth of our Pseudomonas strains in presence or absence of NO32–. Therefore, a minimal medium was prepared containing 4.75 g∗l–1 K2HPO4 (Merck, Darmstadt, Germany), 4.55 g∗l–1 KH2PO4, 2 g∗l–1 yeast extract (Roth, Karlsruhe, Germany) and 20 mM glucose monohydrate. Medium was eighter supplemented with 20 mM NaNO3 or no NaNO3 and pH was adjusted to 6.69. After autoclaving, media was aliquoted into microplates and kept in an anaerobic chamber for 48 h to become anoxic prior to inoculation. Plates were inoculated with an OD600 = 0.1 of a freshly prepared preculture as described above. Growth was measured using a Fluostar Omega microplate reader (BMG LABTECH GmbH, Ortenberg, Germany) after 3 days of incubation within the anaerobic chamber at 25°C. P. aeruginosa DSM 1117 was used as a positive control for nitrate respiration. Significant growth differences were defined based on a two-side open t-test (p < 0.01) between samples containing NaNO3 and no NaNO3.
Results
In situ Growth on Anoxically Packaged Chicken Breast Filet
All tested Pseudomonas strains exhibited significant but limited anaerobic growth on chicken breast after >3 days of cultivation at 4°C (Figure 1). P. lundensis TMW2.1732 and P. weihenstephanensis TMW2.1728 exhibited significant growth after 3 days, P. lundensis TMW2.2076 and both P. fragi strains after 5 days and P. weihenstephanensis TMW2.2077 after 7 days. The cell division rates were low for all Pseudomonas strains (0.4/d) except for P. lundensis TMW2.1732, which exhibited a cell division rate of 0.8/d.
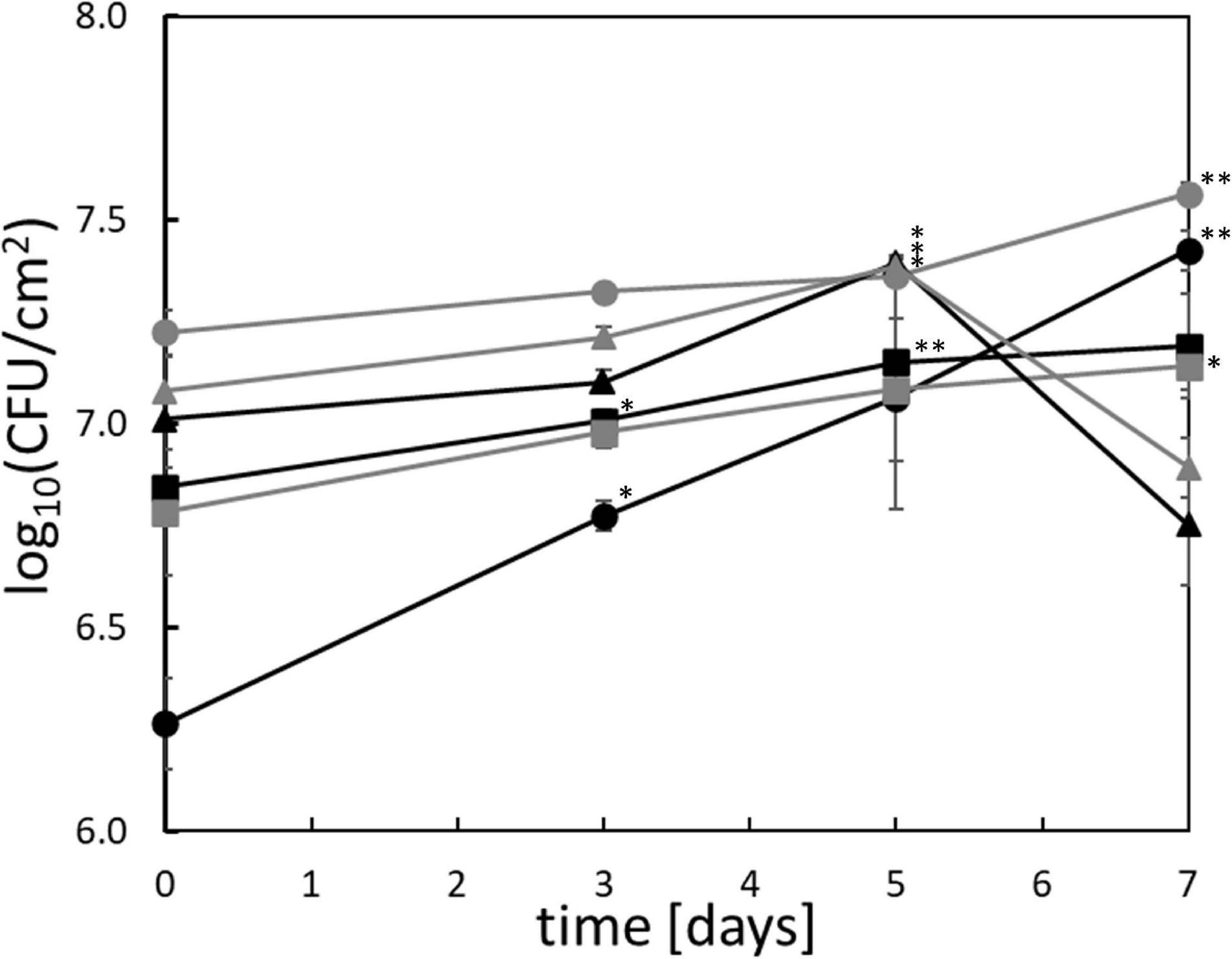
Figure 1. Anaerobic growth of Pseudomonas in situ on chicken meat. The six strains ( ) P. lundensis TMW2.1732, (
) P. lundensis TMW2.1732, ( ) P. lundensis TMW2.2076, (
) P. lundensis TMW2.2076, ( ) P. weihenstephanensis TMW2.2077, (
) P. weihenstephanensis TMW2.2077, ( ) P. weihenstephanensis TMW2.1728, (
) P. weihenstephanensis TMW2.1728, ( ) P. fragi TMW2.2081, and (
) P. fragi TMW2.2081, and ( ) P. fragi TMW2.2082 were checked for their anaerobic growth on chicken breast filet over 7 days at 4°C. ∗significant (p = 0.05–0.01) or ∗∗highly significant (p ≤ 0.01) log10(CFU/cm2) compared to the log10(CFU/cm2) on day 0. SE = standard error based one three independent replicates.
) P. fragi TMW2.2082 were checked for their anaerobic growth on chicken breast filet over 7 days at 4°C. ∗significant (p = 0.05–0.01) or ∗∗highly significant (p ≤ 0.01) log10(CFU/cm2) compared to the log10(CFU/cm2) on day 0. SE = standard error based one three independent replicates.
Growth and Comparative Proteomic Analysis of Pseudomonas spp. in vitro in MS Media Under Oxic and Anoxic Conditions
Growth in MS-medium was determined by monitoring the optical density (OD = 600 nm) (Supplementary Figure 1). Similar to growth on meat, the anoxically growth in MS media was also limited for all strains, reaching a maximal OD600 of 0.22. Most strains, except for P. fragi TMW2.2081, began to enter stationary phase after 48 h of anoxic cultivation. Furthermore, an increase in pH values of the MS-media of approx. 0.65 was detected within 48 h under anoxic cultivation for all strains analyzed (Supplementary Figure 2).
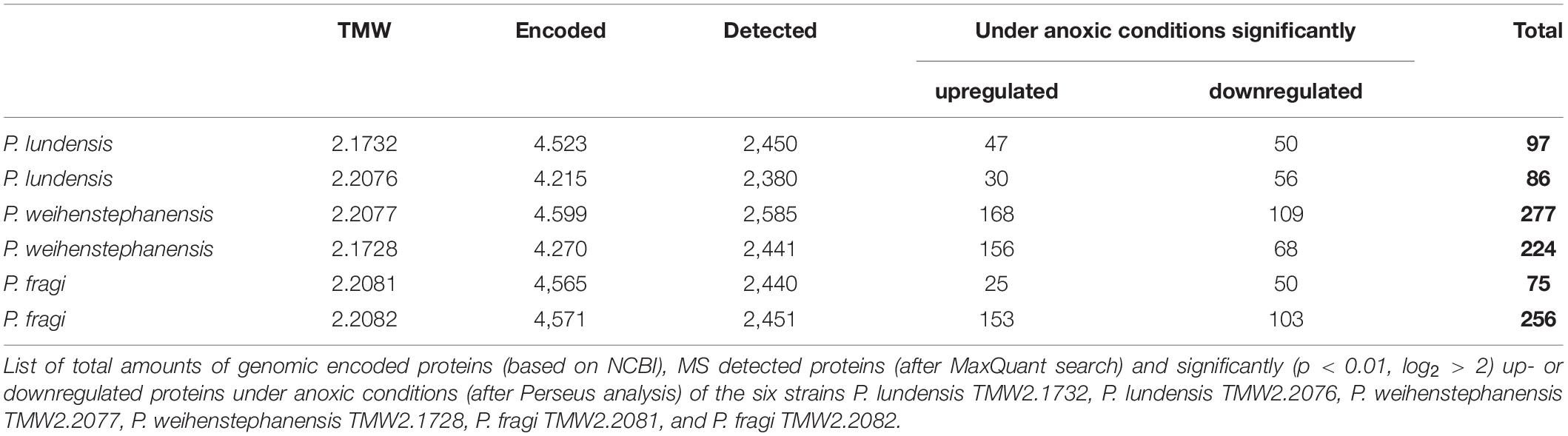
During exponential growth phase, samples for proteomic analysis were taken. Approximately 53–57% of the genomically predicted proteins could be detected by the mass spectrometer for each of the strains (Table 1). Both P. weihenstephanensis strains and P. fragi TMW2.2082 exhibited more than twice as much significantly regulated proteins compared to P. fragi TMW2.2081 and both P. lundensis strains. P. fragi TMW2.2081 was the strain with the lowest number of regulated proteins under the conditions tested. Overall, a high degree of regulation under the different atmospheres was detected for all Pseudomonas strains. A detailed list of differentially expressed single enzymes for each strain is provided in Supplementary Table 1.
Strains of all three Pseudomonas species exhibited similar differential proteomic expression patterns within the relevant major metabolic pathways indicating genus specific metabolic reaction. A detailed list of those metabolic pathways and their direction of regulation is provided in Table 2 for all strains. Important metabolic pathways regarding their energy metabolism were genomically predicted (Supplementary Table 2) and illustrated in separate figures, which further highlight the genus specific metabolic regulations according to proteomic data.
Figure 2A shows the genomically predicted respiratory chain of our Pseudomonas strains, which was similar to the previously described respiratory chain of P. aeruginosa by Liang et al. (2020). All cytochromes described for P. aeruginosa, except cytochrome caa3, could be identified in the genomes of our species. However, neither terminal oxidases cytochrome bd (CIO) and cytochrome bo3 (Cyo) with a low oxygen affinity nor terminal oxidases Cbb3-1 and Cbb3-2 with a high oxygen affinity were differentially expressed according to our proteomic study. Furthermore, enzymes for denitrification (NarGHIJ, NirS, NorCB, NosZ) or dissimilatory nitrate reduction (NarG, NIR) could not be found in the genome of our strains, except for P. fragi TMW2.2082, which encodes an assimilatory nitrate reductase, a small subunit of the nitrite reductase (NirD) as well as the enzyme NirE. However, we have not detected those proteins within the proteomic approach. Furthermore, our proteomic study revealed several enzymes for the usage of alternative electron donors to be strongly upregulated under anoxic conditions. The cytochrome bc1 complex and the protein cupredoxin were also upregulated under anoxic conditions, while cytochrome c and some other enzymes for usage of alternative electron donors were downregulated.
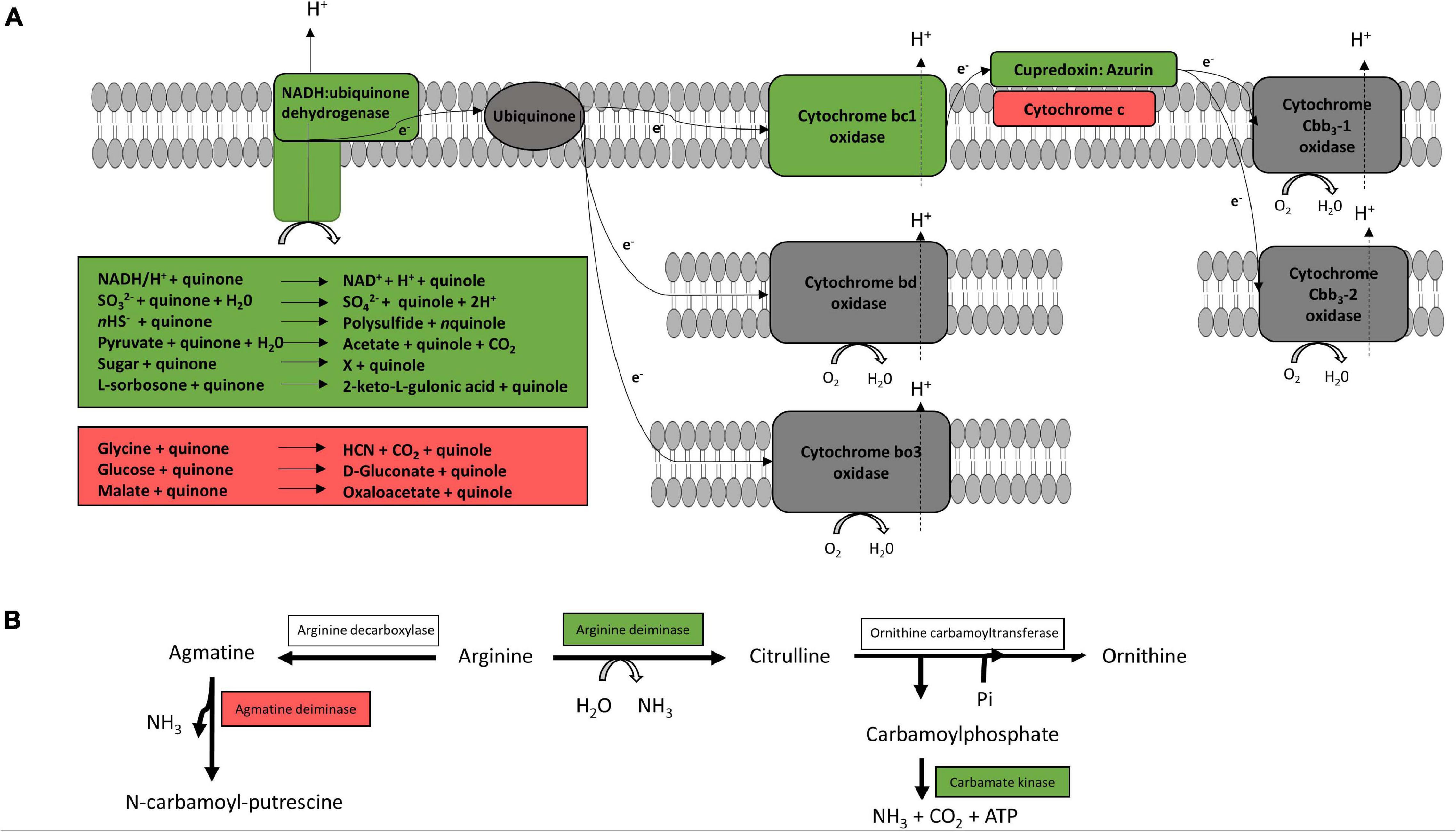
Figure 2. Predicted respiratory chain and arginine metabolism. The respiratory chain setup (A) and arginine metabolism (B) of the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082 were predicted based on our genomic study and differentially expressed enzymes were marked. Enzymes marked in green were upregulated and enzymes marked in red downregulated under anoxic conditions for at least one strain.
Figure 2B illustrates the genomically predicted arginine deiminase (ADI) pathway and enzymes differentially regulated according to our proteomic data. We identified ADI as well as the carbamate kinase to be upregulated under anoxic conditions while the agmatine deiminase was downregulated under anoxic conditions.
The Entner-Doudoroff pathway and pyruvate metabolism are illustrated in Figure 3. Almost all enzymes responsible for gluconate and 2-ketogluconate uptake and degradation and the lactate dehydrogenase were downregulated under anoxic conditions. However, enzymes for glucose uptake and degradation to pyruvate were constitutively expressed under both conditions, while enzymes for the conversion of pyruvate to ethanol were upregulated under anoxic conditions.
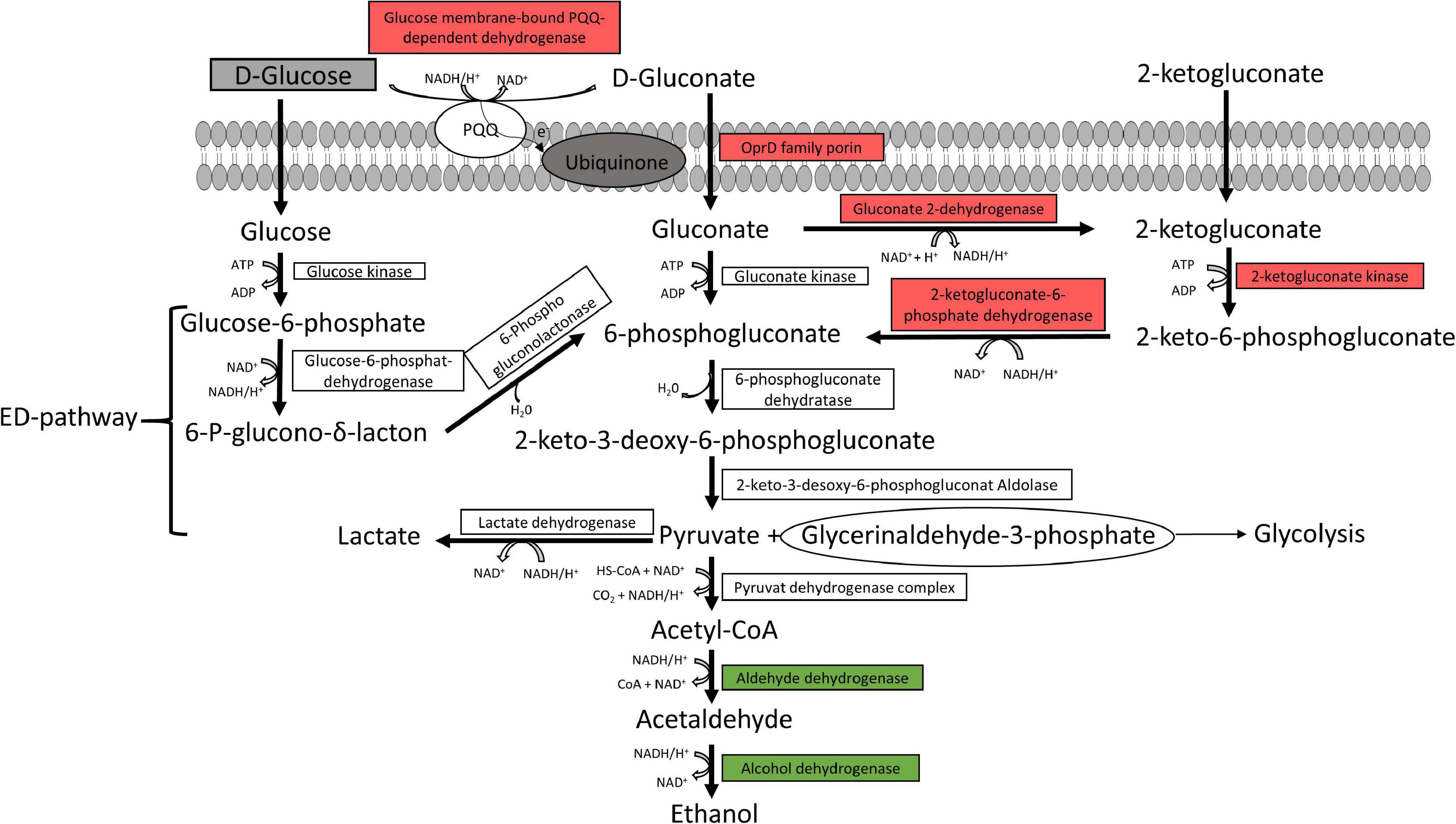
Figure 3. Predicted glucose and pyruvate metabolism. The glucose and pyruvate metabolism were predicted from the genome of the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082, and differentially expressed enzymes based on our proteomic study were marked. ED, Entner-Doudoroff pathway. Enzymes marked in green were upregulated and enzymes marked in red downregulated under anoxic conditions for at least one strain.
Supplementary Figure 3 illustrates the proteins involved in tricarboxylic acid cycle and anaplerotic reactions, which were differentially expressed according to our proteomic data. The aconitase, isocitrate lyase, fumarase, glutaminase A, and glutamate dehydrogenase as well as enzymes of the BCKDH complex, poly-3-hydroxyalkanoate degradation and fatty acid beta oxidation were upregulated under anoxic conditions. Contrary, isocitrate dehydrogenase, succinyl-CoA synthase, asparaginase and aspartate ammonia-lyase were downregulated under anoxic conditions.
Complementary to the listed proteomic regulated enzymes and metabolic pathways in Figures 2, 3 and Supplementary Figure 3, several other differentially expressed enzymes were identified. Those comprise a downregulation of the ferrous metabolism, cellobiose and trehalose degradation, lipopolysaccharide and peptidoglycan synthesis and cell division under anoxic conditions for almost all strains. Contrary, degradation of maltose, sucrose, amylose, benzoate, RNA and murein as well as the glycine cleavage system, fatty acid biosynthesis, urease and oxidative stress proteins were upregulated under anoxic conditions for at least one strain.
In vitro Test for Growth With NaNO3, Arginine, and Glucose
Anoxic growth with or without addition of NaNO3 was tested in yeast extract minimal media for all strains (Supplementary Table 3). P. aeruginosa DSM 1117 was the only strain exhibiting significant growth with sodium nitrate under anoxic conditions. Furthermore, all strains exhibited a negative API 20 NE test regarding reduction of nitrate to nitrite and nitrate to nitrogen (Supplementary Table 4).
All strains were able to grow aerobically and anaerobically on agar plates supplemented with L-arginine (Supplementary Figure 4). This was indicated by a color change from yellow to purple by L-arginine fermentation to ammonia for all strains. Plates incubated oxically were completely purple, while plates incubated anoxically only showed purple color next to the inoculation drops. Growth but no purple color could be observed for all strains cultivated aerobically on plates without arginine. All strains were unable to grow anaerobically on plates without arginine. Furthermore, the arginine deiminase test (ADH) of the API 20 NE was positive for all strains.
No growth and color change could be observed for all our strains on plates containing glucose monohydrate under anoxic conditions (Supplementary Figure 4). Oxically cultivated plates exhibited also no color change but enhanced growth on plates with glucose compared to the growth on plates lacking glucose.
Table 3 summarizes the results of the three physiological assays nitrate respiration, arginine fermentation and glucose metabolism obtained for all analyzed meat strains and P. aeruginosa DSM 1117. Further results of the API 20 NE test can be seen in the Supplementary Table 4 but are not discussed in more detail in this study, as they are not a part of the analyzed topic.
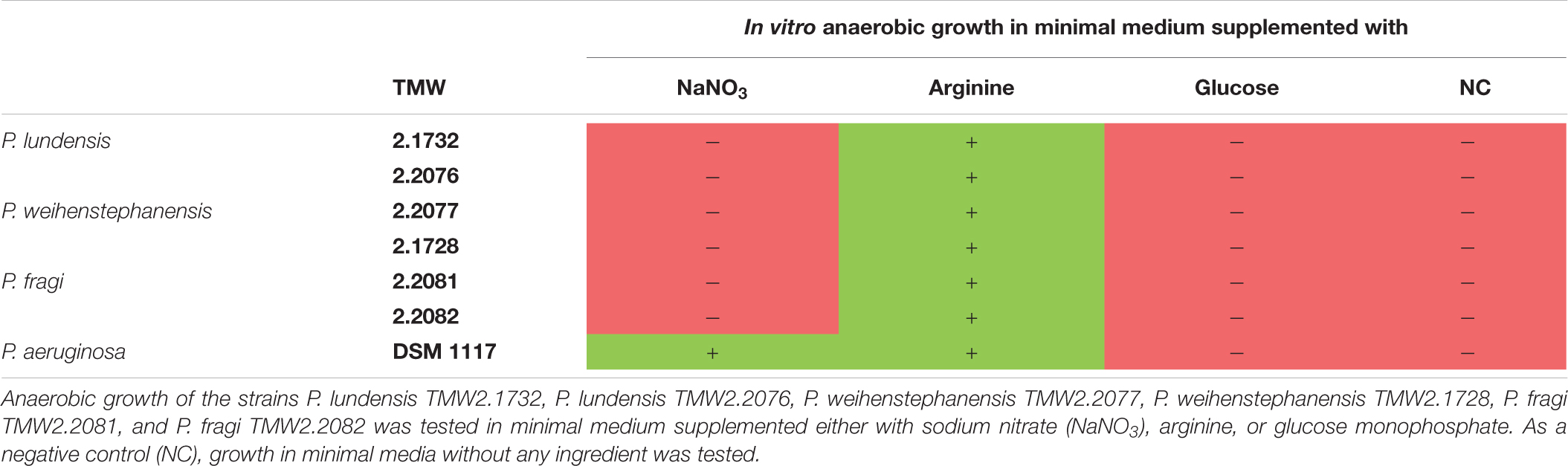
Table 3. Summary of in vitro experiments for identification of the main energy metabolism of Pseudomonas strains on meat.
Discussion
Beside well-studied P. fluorescens, the species P. lundensis, P. weihenstephanensis and P. fragi are known as main aerobic meat spoilers (Erichsen and Molin, 1981; Ercolini et al., 2011; Casaburi et al., 2015; Höll et al., 2016; Hilgarth et al., 2019; Papadopoulou et al., 2020). Recently, strains of those species have been isolated in situ from minced beef stored under high-oxygen MAP atmosphere at a timepoint of oxygen limitation and depletion, and anaerobic growth of the three meat-spoiling species was demonstrated in vitro for the first time (Hilgarth et al., 2019). However, the underlying metabolic response mechanisms remained unknown as well as if selected strains can actually grow on meat in the absence of oxygen or only persist. Consequently, this follow-up study was performed with selected Pseudomonas strains from the study of Hilgarth et al. (2019) in order to study the anaerobic growth of these meat spoilers in detail and identify metabolism regulation to anaerobiosis. Furthermore, this study aimed to provide detailed insight in the hitherto limited research on the anaerobic lifestyle of Pseudomonas species in general, by conducting a comparative full-proteomic approach.
Anaerobic Growth of Pseudomonas Strains on Chicken Breast Filet
We detected restricted growth (cell division rate 0.4–0.8/day) for the analyzed strains of the species P. lundensis (TMW2.1732/TMW2.2076), P. weihenstephanensis (TMW2.2077/TMW2.1728) and P. fragi (TMW2.2081/TMW2.2082) in situ on chicken breast filet packaged with 100% N2 atmosphere. These division rates are unlikely to result in accelerated spoilage if the cell counts are low i.e., average level of initial contamination. However, if levels of Pseudomonas spp. are high e.g., due to previous oxic storage or an unusual high initial contamination, spoilage cannot be delayed by the anoxic packaging as the organisms are still metabolically active. In conclusion, anoxic packaging results in decelerated growth of meat spoiling Pseudomonas spp. compared to oxic conditions, but does not lead to a complete growth repression.
Uncovering the Anaerobic Metabolism and Lifestyle of Pseudomonas Species
We identified several metabolic pathways differentially regulated by the presence and absence of oxygen, which were frequently shared in their regulation between the strains but also between the species. Those metabolic pathways are discussed in detail in the following sections.
Aerobic and Anaerobic Respiration
The respiratory chain is the main energy-producing metabolism of aerobic bacteria, required for cell division and growth. Under oxic conditions, oxygen serves as a terminal electron acceptor for aerobic bacteria including Pseudomonas species. However, under anoxic conditions, some species of this genus e.g., P. aeruginosa and P. stutzeri are capable of restricted growth by denitrification or dissimilatory nitrate reduction using NO3 as a terminal electron acceptor (Fewson and Nicholas, 1961; Carlson and Ingraham, 1983; Zannoni, 1989; Yoon et al., 2002; Wu et al., 2005; Line et al., 2014). This study reveals the meat-spoiling Pseudomonas species P. lundensis, P. weihenstephanensis, and P. fragi to lack corresponding enzymes for dissimilatory nitrate reduction and denitrification in their genomes. This observation was further complemented by the fact that supplementation of NO3 to minimal medium in vitro does not enable anaerobic growth of the bacteria in this study, and nitrate reduction was negative for all analyzed strains in the API 20 NE test. Beside this, anaerobic growth of Pseudomonas strains on meat based on nitrate respiration is rather unlikely, as the amount of nitrate on white and red meat is quite low (max. 0.2 mmol∗kg–1 on red meat) (Lammarino and Di Taranto, 2012). Thus, we conclude that the growth of our strains on chicken meat is not based on energy gained from anaerobic respiration. However, the observed upregulation of several enzymes for different electron donors as well as copper related proteins, which are known to be frequently involved in electron transfer (Dennison, 2005; Farver and Pecht, 2011) under anoxic conditions, can be explained by the attempt of the cells to keep an anaerobic respiratory chain running regardless if substrates/co-factors for their functionality are present or not.
Arginine Fermentation
Arginine conversion via the ADI pathway has been reported to be limited to weak biomass increase of P. aeruginosa under anoxic conditions (Vander Wauven et al., 1984). This is due to the conversion of arginine-derived carbamoyl phosphate to ammonia, carbon dioxide and ATP by the enzymes arginine deiminase, ornithine carbamoyltransferase and carbamate kinase. Regarding these meat-spoiling Pseudomonas strains, we observed an upregulation of the ADI and carbamate kinase enzyme for several of our strains, indicating enhanced arginine fermentation under anoxic conditions. For both P. lundensis strains, respective genes were not differentially but equally strong expressed under both conditions enabling to benefit from arginine degradation under oxic as well as under anoxic conditions.
We further demonstrated that this metabolism provides sufficient energy conservation for anaerobic growth of the strains by performing in vitro plate assays with or without addition of 20 mM arginine. As the amount of arginine in meat is way higher compared to the minimal medium (440 mmol∗kg–1 protein on beef and 344 mmol/kg protein on chicken) (Wladyka and Dawson, 1968; Ahmad et al., 2018) we hypothesize that arginine fermentation is one main source of energy for our Pseudomonas strains, which enables restricted growth also in situ on anoxic packaged meat.
Glucose and Pyruvate Metabolism
There are three possible ways of glucose uptake for Pseudomonas species comprising direct uptake of glucose or indirect uptake of glucose-derived gluconate and 2-ketogluconate, all being ultimately shunted into the Entner-Doudoroff pathway (Chavarría et al., 2013). Enzymes of the indirect uptake system via the glucose membrane-bound PQQ-dependent dehydrogenase, which is known to shuttle electrons into the respiratory chain (Umezawa et al., 2015), and the NADH/H+ producing gluconate 2-dehydrogenase, were down-regulated under anoxic conditions. However, all enzymes for the standard ED-pathway were constitutively expressed without regulation for all strains in our study, conserving one ATP molecule per molecule glucose via production of glyceraldehyde-3-phosphate entering glycolysis pathway. Therefore, the energy yield of this metabolic pathway is equal to arginine fermentation. However, fermentation of arginine is not coupled to the NADH + /NAD + pool, and therefore, no redox equivalents have to be reoxidized to maintain redox balance. This reoxidation can be predictively performed by glucose fermentation to ethanol, indicated by an upregulation of corresponding enzymes (aldehyde and alcohol dehydrogenases) under anoxic conditions. The observed upregulation of enzymes utilizing maltose, sucrose and amylose further support the assumption of glucose utilization under anoxic conditions, especially as glucose is mainly bound in di-, tri- and polysaccharides in meat (Komiyama et al., 2008; Koutsidis et al., 2008). However, our growth experiment under anoxic conditions demonstrated that sole glucose metabolism/fermentation does not conserve enough energy and therefore does not enable true growth of our Pseudomonas strains on meat. Nevertheless, we predict long term survival of our strains by glucose metabolism via the Entner-Doudoroff pathway and NAD+ recycling by ethanolic formation.
Another metabolic pathway enabling long term survival of P. aeruginosa under anoxic conditions is thought to be acetate fermentation via the two enzymes phosphotransacetylase and acetate kinase (Eschbach et al., 2004; Schreiber et al., 2006). However, genomes of our six analyzed strains do not encode those enzymes and therefore employment of acetate fermentation for survival of these Pseudomonas strains on meat can be excluded.
Glyoxylate Shunt
Interestingly, we measured an upregulation of enzymes involved in the glyoxylate shunt as well as a concomitant downregulation of bypassed enzymes of the tricarboxylic acid cycle for our strains under anoxic conditions. The glyoxylate shunt is commonly activated in bacteria under carbon-source limitation. In this bacteria, acetate and fatty acids-derived acetyl-CoA is shuttled into the glyoxylate shunt to obtain C4 compounds which are channeled into gluconeogenesis to synthesize C5 or C6 compounds (Maloy et al., 1980; Son et al., 2007). Indeed, we also observed an upregulation of several proteins involved in anoxic fatty acid beta oxidation, leucine and isoleucine degradation as well as poly-3-hydroxyalkanoate degradation, all yielding acetyl-CoA under anoxic conditions. Gluconeogenetic enzymes replacing irreversible reactions of the glycolysis were not differentially regulated for our strains. However, reduced or disabled respiration has been reported to be associated with an increase in the glyoxylate shunt, preventing NADH/H+ production via the bypassed enzymes isocitrate dehydrogenase, α-ketoglutarate dehydrogenase and succinate dehydrogenase (Tielen et al., 2013; Ahn et al., 2016; Meylan et al., 2017).
Protein Folding
The absence of alternative terminal electron acceptors and impaired respiration imply redox stress within the cell due to an imbalance of redox balance. Thus, cytoplasmatic proteins are kept predominantly in a reduced status and are therefore unable to perform their functionality (Glasser et al., 2014; Lai et al., 2016). Our proteomic data indicated that the DsbA enzyme, which is needed for correct protein folding, was upregulated under anoxic conditions. According to Collet and Bardwell (2002), protein folding takes place by shuttling electrons from the native unfolded peptide chain via DsbA and DsbB proteins to a terminal electron acceptor. As reoxidation of Dsb proteins seems to be impaired in our strains, incorrect folded proteins are generated, which have to be repaired by isomerization of existing S-S bonds. In this context, we detected several corresponding enzymes to be upregulated under anoxic conditions, comprising thioredoxin, peroxiredoxin, cysteine hydrolases, and S-S-oxide reductases. Interestingly, thioredoxin and peroxiredoxin are under control of the transcription factor OxyR in P. aeruginosa, which also regulates the expression of catalase enzymes (Wei et al., 2012). This might also explain the upregulation of the catalase PHII for three of our Pseudomonas stains under anoxic conditions.
Conclusion
To our knowledge this study is the first one demonstrating limited growth of the three meat-spoiling species P. lundensis, P. weihenstephanensis and P. fragi in situ on anoxically packaged chicken meat. Furthermore, we were able to explain the observed anaerobic growth of our strains by uncovering their basic metabolic lifestyle applying a full genomic and comparative proteomic approach. In detail, we predicted arginine fermentation via the ADI pathway to provide the necessary energy for the growth on anoxically packaged meat. We also conclude that glucose fermentation to ethanol via the Entner-Doudoroff pathway contributes to long term survival under those conditions, but alone does not enable true growth. Furthermore, the studied strains are unable to conduct nitrate respiration or fermentation of glucose to acetate due to absence of respective genes, which has previously been described for other Pseudomonas species under anoxic conditions. Under carbon-limited conditions and anoxia, we predict that these strains use the glyoxylate shunt for gluconeogenesis, bypassing enzymes of the TCA cycle producing NADH+. Furthermore, we highlighted the consequences of redox stress triggered by imbalance of the redox equivalents and impaired respiration on protein folding processes, counteracted by overexpression of refolding enzymes. Thus, we also complemented the hitherto limited research on the general anaerobic metabolism of different Pseudomonas species.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD023961.
Author Contributions
SK designed the study, performed the experiments and data analysis, and wrote the first draft of the manuscript. MA performed the mass spectrometric data acquisition and MaxQuant search. MH helped to interpret the data and draft the manuscript, and supervised the work of SK. RV initiated the project and supervised the work of SK. All authors read and approved the final manuscript.
Funding
Part of this work was funded by the German Federal Ministry for Economic Affairs and Energy via the German Federation of Industrial Research Associations (AiF) and the Industry Association for Food Technology and Packaging (IVLV); project number IGF 19993N.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.664061/full#supplementary-material
Supplementary Figure 1 | Growth of our Pseudomonas strains in vitro in MS media. Growth of the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082 in MS medium under oxic (21% O2, 0.03% CO2) and anoxic (100% N2) conditions was monitored for 48 h. Dotted lines: anaerobic growth, solid lines: aerobic growth. Yellow dots represent the timepoint of sample taking for proteomics.
Supplementary Figure 2 | PH development in vitro in MS media. The pH-value of the six strains (a) P. lundensis TMW2.1732, (b) P. lundensis TMW2.2076, (c) P. weihenstephanensis TMW2.2077, (d) P. weihenstephanensis TMW2.1728, (e) P. fragi TMW2.2081, and (f) P. fragi TMW2.2082 in MS medium under oxic (21% O2, 0.03% CO2) and anoxic (100% N2) conditions was recorded for 48 h. Dotted lines: anaerobic growth, solid lines: aerobic growth. All values are based on three independent replicates. Error bars represent standard error values.
Supplementary Figure 3 | Predicted tricarboxylic acid cycle. The tricarboxylic acid cycle and reactions of the anaplerotic metabolism were predicted from the genome of the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082, and differentially expressed enzymes based on our proteomic study were marked. Enzymes marked in green were upregulated and enzymes marked in red downregulated under anoxic conditions for at least on strain.
Supplementary Figure 4 | Anaerobic arginine and glucose fermentation of Pseudomonas strains in vitro in minimal media. Arginine and glucose fermentation was demonstrated by a plate assay were the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, P. fragi TMW2.2082, and the positive control P. aeruginosa DSM 1117 were grown on agar plates containing a minimal medium supplemented with either 20 mM arginine or glucose monohydrate or none of those ingredients. The pH dye phenol red indicates a pH-increase by a color change from yellow to purple red, which is given by catabolism of the amino acid arginine. Plates were either incubated oxically or anoxically.
Supplementary Table 1 | Detailed list of all significantly differential regulated proteins. A list of all significantly differential (p < 0.01, log2 > 2) regulated proteins of the strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081 and P. fragi TMW2.2082 comparing growth under air (21% O2, 0.0% CO2) and anoxic (100% N2) conditions in meat simulation media is provided. Colors indicate the effect of regulation with  log2(diff.) = 2,
log2(diff.) = 2,  log2 (diff.) = 3,
log2 (diff.) = 3,  log2 (diff.) = 4,
log2 (diff.) = 4,  log2 (diff.) = 5,
log2 (diff.) = 5,  log2 (diff.) = 6,
log2 (diff.) = 6,  log2 (diff.) ≥ 6 for downregulated proteins and
log2 (diff.) ≥ 6 for downregulated proteins and  log2 (diff.) = 2,
log2 (diff.) = 2,  log2 (diff.) = 3,
log2 (diff.) = 3,  log2 (diff.) = 4,
log2 (diff.) = 4,  log2 (diff.) = 5,
log2 (diff.) = 5,  log2 (diff.) = 6,
log2 (diff.) = 6,  log2 (diff.) ≥ 6 for upregulated proteins under anoxic conditions. All data are based on three independent replicates. ∗Functional categories are based on NCBI annotation, TIGR annotation, SEED annotation, KEGG annotation and own research.
log2 (diff.) ≥ 6 for upregulated proteins under anoxic conditions. All data are based on three independent replicates. ∗Functional categories are based on NCBI annotation, TIGR annotation, SEED annotation, KEGG annotation and own research.
Supplementary Table 2 | List of the genomic setup of important pathways of the analyzed Pseudomonas strains. All genomically encoded enzymes corresponding to the discussed pathways respiratory chain, Entner-Doudoroff pathway, pyruvate metabolism, arginine metabolism and TCA cycle are listed for the strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082. All data are based on genomic analysis (NCBI annotation and manual blast search). Numbers given are the corresponding locus tags of NCBI annotated genes, ∗ indicates putative genes.
Supplementary Table 3 | Growth with NaNO3 in vitro in minimal media. Anaerobic growth (OD600) of the six strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, P. fragi TMW2.2082, and the positive control P. aeruginosa DSM 1117 was tested in minimal media containing glucose as electron donor supplemented with 20 mM NaNO3 or without NaNO3. Significant differences between growth with or without NaNO3 were defined by a t-test with p < 0.01.
Supplementary Table 4 | API 20 NE test performed for our Pseudomonas strains. Results obtained from the API NE 20 test for the strains P. lundensis TMW2.1732, P. lundensis TMW2.2076, P. weihenstephanensis TMW2.2077, P. weihenstephanensis TMW2.1728, P. fragi TMW2.2081, and P. fragi TMW2.2082 after 48 h.
References
Ahmad, R., Imran, A., and Hussain, M. (2018). “Nutritional composition of meat,”in Meat Science and Nutrition, ed. M.S. Arshad (Faisalabad: Government College University).
Ahn, S., Jung, J., Jang, I.-A., Madsen, E. L., and Park, W. (2016). Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 291, 11928–11938. doi: 10.1074/jbc.m115.708149
Alquati, C., De Gioia, L., Santarossa, G., Alberghina, L., Fantucci, P., and Lotti, M. (2002). The cold-active lipase of Pseudomonas fragi. Eur. J. Biochem. 269, 3321–3328. doi: 10.1046/j.1432-1033.2002.03012.x
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Carlson, C. A., and Ingraham, J. L. (1983). Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans. Appl. Environ. Microbiol. 45, 1247–1253. doi: 10.1128/aem.45.4.1247-1253.1983
Casaburi, A., Piombino, P., Nychas, G.-J., Villani, F., and Ercolini, D. (2015). Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol. 45, 83–102. doi: 10.1016/j.fm.2014.02.002
Chavarría, M., Nikel, P. I., Pérez-Pantoja, D., and De Lorenzo, V. (2013). The Entner–Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 15, 1772–1785. doi: 10.1111/1462-2920.12069
Collet, J.-F., and Bardwell, J. C. A. (2002). Oxidative protein folding in bacteria. Mol. Microbiol. 44, 1–8. doi: 10.1046/j.1365-2958.2002.02851.x
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
De Filippis, F., La Storia, A., Villani, F., and Ercolini, D. (2019). Strain-level diversity analysis of Pseudomonas fragi after in situ pangenome reconstruction shows distinctive spoilage-associated metabolic traits clearly selected by different storage conditions. Appl. Environ. Microbiol. 85, e02212–e02218.
Dennison, C. (2005). Investigating the structure and function of cupredoxins. Coord. Chem. Rev. 249, 3025–3054. doi: 10.1016/j.ccr.2005.04.021
Ercolini, D., Ferrocino, I., Nasi, A., Ndagijimana, M., Vernocchi, P., La Storia, A., et al. (2011). Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl. Environ. Microbiol. 77, 7372–7381. doi: 10.1128/aem.05521-11
Ercolini, D., Russo, F., Blaiotta, G., Pepe, O., Mauriello, G., and Villani, F. (2007). Simultaneous detection of Pseudomonas fragi, P. lundensis, and P. putida from meat by use of a multiplex PCR assay targeting the carA gene. Appl. Environ. Microbiol. 73, 2354–2359. doi: 10.1128/aem.02603-06
Erichsen, I., and Molin, G. (1981). Microbial flora of normal and high pH neef stored at 4°C in different gas environments. J. Food Prot. 44, 866–869. doi: 10.4315/0362-028x-44.11.866
Eschbach, M., Schreiber, K., Trunk, K., Buer, J., Jahn, D., and Schobert, M. (2004). Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186, 4596–4604. doi: 10.1128/jb.186.14.4596-4604.2004
Farver, O., and Pecht, I. (2011). Electron transfer in blue copper proteins. Coord. Chem. Rev. 255, 757–773. doi: 10.1016/j.ccr.2010.08.005
Fewson, C. A., and Nicholas, D. J. D. (1961). Nitrate reductase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 49, 335–349. doi: 10.1016/0006-3002(61)90133-0
Garrity, G. M., Bell, J. A., and Lilburn, T. (2005). “Pseudomonadaceae,” in Bergey’s Manual of Systematic Bacteriology, Volume2: The Proteobacteria, Part B: The Gammaproteobacteria, eds G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (Berlin: Springer), 323–397.
Glasser, N., Kern, S., and Newman, D. (2014). Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 92, 399–412. doi: 10.1111/mmi.12566
Hilgarth, M., Behr, J., and Vogel, R. F. (2018a). Monitoring of spoilage-associated microbiota on modified atmosphere packaged beef and differentiation of psychrophilic and psychrotrophic strains. J. Appl. Microbiol. 124, 740–753. doi: 10.1111/jam.13669
Hilgarth, M., Lehner, E. M., Behr, J., and Vogel, R. F. (2019). Diversity and anaerobic growth of Pseudomonas spp. isolated from modified atmosphere packaged minced beef. J. Appl. Microbiol. 127, 159–174. doi: 10.1111/jam.14249
Hilgarth, M., Nani, M., and Vogel, R. F. (2018b). Assertiveness of meat-borne Lactococcus piscium strains and their potential for competitive exclusion of spoilage bacteria in situ and in vitro. J. Appl. Microbiol. 124, 1243–1253. doi: 10.1111/jam.13710
Höll, L., Behr, J., and Vogel, R. F. (2016). Identification and growth dynamics of meat spoilage microorganisms in modified atmosphere packaged poultry meat by MALDI-TOF MS. Food Microbiol. 60, 84–91. doi: 10.1016/j.fm.2016.07.003
Jääskeläinen, E., Hultman, J., Parshintsev, J., Riekkola, M.-L., and Björkroth, J. (2016). Development of spoilage bacterial community and volatile compounds in chilled beef under vacuum or high oxygen atmospheres. Int. J. Food Microbiol. 223, 25–32. doi: 10.1016/j.ijfoodmicro.2016.01.022
Kolbeck, S., Ludwig, C., Meng, C., Hilgarth, M., and Vogel, R. F. (2020). Comparative proteomics of meat spoilage bacteria predicts drivers for their coexistence on modified atmosphere packaged meat. Front. Microbiol. 11:209. doi: 10.3389/fmicb.2020.00209
Kolbeck, S., Reetz, L., Hilgarth, M., and Vogel, R. F. (2019). Quantitative oxygen consumption and respiratory activity of meat spoiling bacteria upon high oxygen modified atmosphere. Front. Microbiol. 10:2398. doi: 10.3389/fmicb.2019.02398
Komiyama, C., Mendes, A. A., Takahashi, S., Moreira, J., Garcia, R., Sanfelice, C., et al. (2008). Chicken meat quality as a function of fasting period and water spray. Braz. J. Poultry Sci. 10, 179–183. doi: 10.1590/s1516-635x2008000300008
Koutsidis, G., Elmore, J. S., Oruna-Concha, M. J., Campo, M. M., Wood, J. D., and Mottram, D. S. (2008). Water-soluble precursors of beef flavour. Part II: effect of post-mortem conditioning. Meat Sci. 79, 270–277. doi: 10.1016/j.meatsci.2007.09.010
Lai, B., Yu, S., Bernhardt, P. V., Rabaey, K., Virdis, B., and Krömer, J. O. (2016). Anoxic metabolism and biochemical production in Pseudomonas putida F1 driven by a bioelectrochemical system. Biotechnol. Biofuels 9:39.
Lammarino, M., and Di Taranto, A. (2012). Nitrite and nitrate in fresh meats: a contribution to the estimation of admissible maximum limits to introduce in directive 95/2/EC. Int. J. Food Sci. Technol. 47, 1852–1858. doi: 10.1111/j.1365-2621.2012.03041.x
Liang, P., Fang, X., Hu, Y., Yuan, M., Raba, D. A., Ding, J., et al. (2020). The aerobic respiratory chain of Pseudomonas aeruginosa cultured in artificial urine media: role of NQR and terminal oxidases. PLoS One 15:e0231965. doi: 10.1371/journal.pone.0231965
Line, L., Alhede, M., Kolpen, M., Kühl, M., Ciofu, O., Bjarnsholt, T., et al. (2014). Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 5:554. doi: 10.3389/fmicb.2014.00554
Maloy, S. R., Bohlander, M., and Nunn, W. D. (1980). Elevated levels of glyoxylate shunt enzymes in Escherichia coli strains constitutive for fatty acid degradation. J. Bacteriol. 143, 720–725. doi: 10.1128/jb.143.2.720-725.1980
Marchand, S., Heylen, K., Messens, W., Coudijzer, K., De Vos, P., Dewettinck, K., et al. (2009). Seasonal influence on heat-resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environ. Microbiol. 11, 467–482. doi: 10.1111/j.1462-2920.2008.01785.x
Martin, N. H., Murphy, S. C., Ralyea, R. D., Wiedmann, M., and Boor, K. J. (2011). When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. J. Dairy Sci. 94, 3176–3183. doi: 10.3168/jds.2011-4312
Meng, L., Zhang, Y., Liu, H., Zhao, S., Wang, J., and Zheng, N. (2017). Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front. Microbiol. 8:2158. doi: 10.3389/fmicb.2017.02158
Meylan, S., Porter, C. B. M., Yang, J. H., Belenky, P., Gutierrez, A., Lobritz, M. A., et al. (2017). Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 24, 195–206. doi: 10.1016/j.chembiol.2016.12.015
Morales, P., Fernández-García, E., and Nuñez, M. (2005). Production of volatile compounds in cheese by Pseudomonas fragi strains of dairy origin. J. Food Prot. 68, 1399–1407. doi: 10.4315/0362-028x-68.7.1399
Neubeck, M., Huptas, C., Glück, C., Krewinkel, M., Stoeckel, M., Stressler, T., et al. (2016). Pseudomonas helleri sp. nov. and Pseudomonas weihenstephanensis sp. nov., isolated from raw cow‘s milk. Int. J. Syst. Evol. Microbiol. 66, 1163–1173. doi: 10.1099/ijsem.0.000852
Papadopoulou, O. S., Iliopoulos, V., Mallouchos, A., Panagou, E. Z., Chorianopoulos, N., Tassou, C. C., et al. (2020). Spoilage potential of Pseudomonas (P. fragi, P. putida) and LAB (Leuconostoc mesenteroides, Lactobacillus sakei) strains and their volatilome profile during storage of sterile pork meat using GC/MS and data analytics. Foods 9:633. doi: 10.3390/foods9050633
Parte, A. C. (2018). LPSN – List of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 68, 1825–1829. doi: 10.1099/ijsem.0.002786
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450.
Rajmohan, S., Dodd, C. E., and Waites, W. M. (2002). Enzymes from isolates of Pseudomonas fluorescens involved in food spoilage. J. Appl. Microbiol. 93, 205–213.
Scatamburlo, T. M., Yamazi, A. K., Cavicchioli, V. Q., Pieri, F. A., and Nero, L. A. (2015). Spoilage potential of Pseudomonas species isolated from goat milk. J. Dairy Sci. 98, 759–764.
Schreiber, K., Boes, N., Eschbach, M., Jaensch, L., Wehland, J., Bjarnsholt, T., et al. (2006). Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an usp-type stress protein. J. Bacteriol. 188, 659–668.
Son, M. S., Matthews, W. J. Jr., Kang, Y., Nguyen, D. T., and Hoang, T. T. (2007). In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75, 5313–5324.
Stoeckel, M., Lidolt, M., Achberger, V., Glück, C., Krewinkel, M., Stressler, T., et al. (2016). Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. Int. Dairy J. 59, 20–28.
Tielen, P., Rosin, N., Meyer, A. K., Dohnt, K., Haddad, I., Jänsch, L., et al. (2013). Regulatory and metabolic networks for the adaptation of Pseudomonas aeruginosa biofilms to urinary tract-like conditions. PLoS One 8:e71845. doi: 10.1371/journal.pone.0071845
Tyanova, S., Temu, T., and Cox, J. (2016a). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319.
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016b). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740.
Umezawa, K., Takeda, K., Ishida, T., Sunagawa, N., Makabe, A., Isobe, K., et al. (2015). A novel pyrroloquinoline quinone-dependent 2-keto-D-glucose dehydrogenase from Pseudomonas aureofaciens. J. Bacteriol. 197, 1322– 1329.
Vander Wauven, C., Pierard, A., Kley-Raymann, M., and Haas, D. (1984). Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160, 928–934.
Wang, G., Ma, F., Zeng, L., Bai, Y., Wang, H., Xu, X., et al. (2018). Modified atmosphere packaging decreased Pseudomonas fragi cell metabolism and extracellular proteolytic activities on meat. Food Microbiol. 76, 443–449.
Wang, G.-Y., Li, M., Ma, F., Wang, H.-H., Xu, X.-L., and Zhou, G.-H. (2017). Physicochemical properties of Pseudomonas fragi isolates response to modified atmosphere packaging. FEMS Microbiol. Lett. 364:fnx106.
Wei, Q., Minh, P. N. L., Dötsch, A., Hildebrand, F., Panmanee, W., Elfarash, A., et al. (2012). Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 40, 4320–4333.
Wiedmann, M., Weilmeier, D., Dineen, S. S., Ralyea, R., and Boor, K. J. (2000). Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66, 2085–2095.
Wladyka, E. J., and Dawson, L. E. (1968). Essential amino acid composition of chicken meat and drip after 30 and 90 days of frozen storage. J. Food Sci. 33, 453–455.
Wu, M., Guina, T., Brittnacher, M., Nguyen, H., Eng, J., and Miller, S. I. (2005). The Pseudomonas aeruginosa proteome during anaerobic growth. J. Bacteriol. 187, 8185–8190.
Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603.
Keywords: arginine fermentation, anaerobic lifestyle, proteomics, redox stress, modified atmosphere packaging, meat spoilage, Pseudomonas, Entner-Doudoroff pathway
Citation: Kolbeck S, Abele M, Hilgarth M and Vogel RF (2021) Comparative Proteomics Reveals the Anaerobic Lifestyle of Meat-Spoiling Pseudomonas Species. Front. Microbiol. 12:664061. doi: 10.3389/fmicb.2021.664061
Received: 04 February 2021; Accepted: 08 March 2021;
Published: 06 April 2021.
Edited by:
Ivan A. Berg, University of Münster, GermanyReviewed by:
Peter J. Schaap, Wageningen University and Research, NetherlandsJørgen J. Leisner, University of Copenhagen, Denmark
Copyright © 2021 Kolbeck, Abele, Hilgarth and Vogel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maik Hilgarth, bWFpay5oaWxnYXJ0aEB0dW0uZGU=
 Sandra Kolbeck
Sandra Kolbeck Miriam Abele
Miriam Abele Maik Hilgarth
Maik Hilgarth Rudi F. Vogel
Rudi F. Vogel