- Department of Food Science, Foshan Polytechnic, Foshan, China
Oysters are one of the main aquatic products sold in coastal areas worldwide and are popular among consumers because of their delicious taste and nutritional value. However, the microorganisms present in oysters may pose health risks to consumers. In this study, the microbial communities of Pacific oysters (Crassostrea gigas) collected from aquatic product markets in three cities (Guangzhou, Zhuhai, and Jiangmen) of Guangdong Province, China, where raw oysters are popular, were investigated. The plate counts of viable bacteria in oysters collected in the three cities were all approximately 2 log colony-forming units/g. High-throughput sequencing analysis of the V3–V4 region of the 16Sribosomal DNA gene showed a high level of microbial diversity in oysters, as evidenced by both alpha and beta diversity analysis. Proteobacteria, Bacteroidetes, and Firmicutes were the dominant phyla of the microorganisms present in these samples. A variety of pathogenic bacteria, including the fatal foodborne pathogen Vibrio vulnificus, were found, and Vibrio was the dominant genus. Additionally, the relationship between other microbial species and pathogenic microorganisms may be mostly symbiotic in oysters. These data provide insights into the microbial communities of retail oysters in the Guangdong region and indicate a considerable risk related to the consumption of raw oysters.
Introduction
Oysters are one of the most commonly farmed shellfish in China, with a total output of approximately 5.14 million tons in 2018 (Fisheries administration of Ministry of Agriculture and Rural affairs of the People’s Republic of China, 2019). They are considered a nutrient-rich source of proteins, unsaturated fatty acids, vitamins, and minerals (Ackman, 1990; Cabello et al., 2004; Caglak et al., 2008; Cruz-Romero et al., 2008a; Rey et al., 2012). The Pacific oyster is the main oyster species sold in the coastal areas of southern China. However, similar to most seafood with abundant water and nutrient contents, oysters tend to undergo spoilage during transportation and storage. Microbial proliferation results in spoilage and unacceptable quality during storage (Gram and Dalgaard, 2002; Boziaris and Parlapani, 2017).
In addition to the deterioration of the quality of oysters caused by spoilage, the risks to human health attributed to pathogenic microorganisms present in raw oysters have attracted considerable attention. Oysters are one of the few animal foods that are consumed whole and raw by humans. The consumption of raw oysters has become popular in Asia, including China. Thus, the safety of a product is vital to consumer health (Froelich and Noble, 2016). Pseudomonas and Vibrio were found to be the predominant bacteria in spoiled Pacific oysters bred by conventional cultivation (Cao et al., 2010; Cruz-Romero et al., 2008b). V. vulnificus and V. parahaemolyticus are the most common types of life-threatening foodborne pathogens and have been reported in Vibrio-associated wound infections (Froelich and Noble, 2016). Vibrio infections associated with eating uncooked or undercooked oysters continue to increase (Huss et al., 2014). In the US, V. vulnificus is the most fatal foodborne pathogen, and it is responsible for 95% of all seafood-related deaths. Even with aggressive medical treatment, its fatality rate are about 50%. These bacteria also have a second route of infection. They can enter the body via wounds, either preexisting or related to activities such as seafood handling or oyster harvesting (Jones and Oliver, 2009; Letchumanan et al., 2014; Froelich and Noble, 2016). Additionally, the leading causes of foodborne gastroenteritis in Japan include V. parahaemolyticus infection. This infection affects more than 10,000 individuals in 500–800 outbreaks annually (Food and Agriculture Organization, 2011). This pathogen is estimated to account for half of the foodborne illnesses reported in Asian countries (Chen et al., 2018). In the Guangdong province of China, the current trend suggests that raw oysters are consumed more frequently by an increasing proportion of the population, which results in new food safety problems. Individuals with impaired immune systems, children, pregnant women, and older adults are among the most susceptible to foodborne infections (Lund and O’Brien, 2011). Therefore, the microbial diversity of oysters at the consumer terminal is very important for food hygiene and safety.
To evaluate the quality of retail Pacific oysters in the Guangdong region and to assess their potential health risks to humans, we collected Pacific oysters sold in three coastal cities (Guangzhou, Zhuhai, and Jiangmen) in Guangdong Province. High-throughput sequencing was performed to determine the bacterial profiles of the samples. These data will offer insight into the microbial status of Pacific oysters in this area and may provide a reference for establishing targeted risk reduction strategies for oyster consumption.
Materials and Methods
Sample Collection
Oysters were collected from three cities, namely Zhuhai, Guangzhou, and Jiangmen, of Guangdong Province, China. The fresh oysters were purchased from two markets in Guangzhou, three markets in Jiangmen and one market in Zhuhai, and at least four different stalls in each market were selected (Supplementary Figure 1). The oysters in markets of Guangzhou were transported from other places, and those in markets of Jiangmen and Zhuhai were obtained locally (Supplementary Table 1). Every two oysters randomly purchased from the same stall were mixed into one sample to exclude sampling deviation, and the accumulated 52 samples from different stalls were divided into 3 groups depending on the cities from which they were collected. All oyster samples were rinsed with tap water to remove contaminants, such as dirt, on the surface.
Microbiological Assays
Total viable count (TVC) assays were conducted as per previously described methods with slight modifications (Sanna et al., 2018). Ten grams of each sample was weighed and placed in a sterile homogenizing bag containing 90 mL of normal saline. In the homogenizer, the sample was subjected to flapping for 2.5 min to prepare a 1:10 homogenate (1 mL of homogenate was collected using a pipette and outsourced for DNA extraction and sequencing). A suitable diluent for plate count analysis was selected and cultured for 48–72 h. Clones was counted when the cells were visible.
DNA Extraction
DNA was extracted from the homogenate of the oysters using a DNA extraction kit (Magen Hipure Soil DNA Kit, Angen Biotech, Guangzhou, China) according to the manufacturer’s instructions, and quality control was performed using the Qubit® DSDNA HS Assay Kit (Thermo Fisher Scientific, MA, United States).
16Sribosomal DNA (rDNA) Amplification Sequencing
The Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) was used to control the quality of the DNA, which was subsequently used to perform amplification of the V3–V4 hypervariable regions of prokaryotic16S rDNA. Pairing primers were designed by GENEWIZ (South Plainfield, NJ, United States). The sequence of the forward primer was “CCTACGGRRBGCASCAGKVRVGAAT,” and the reverse primer contained the sequence “GGACTACNVGGGTW TCTAATCC.” Each polymerase chain reaction volume was 25 μL, containing 2.5 μL of TransStart Buffer (TransGen, Beijing, China), 2 μL of dNTPs, 1 μL of each primer, and 20–30 ng of template DNA. Then, the indexed adapters were attached to the ends of the amplicons to generate indexed libraries for subsequent next-generation sequencing using the Illumina platform (San Diego, CA, United States). The libraries were validated using the Qubit3.0 Fluorometer (Thermo Fisher Scientific) and quantified to 10 nmol. The Illumina MiSeq instrument was used to load and sequence the DNA libraries according to the manufacturer’s instructions. Sequencing was performed under the paired-end 250-bp mode. The raw reads were trimmed using the Cutadapt software to generate clean reads.
Microbial Diversity Analysis
Data with clean reads were analyzed by using the nf-core/ampliseqv1.2.0 pipeline, and the microbial diversity within our datasets were determined by setting the optional parameters “- -multiple Sequencing Runs,” “- -trunclenf 220,” and “- -trunclenr 180” (to resemble the truncation values of QIIME2 with DADA2) (Straub et al., 2020). The 16s rRNA gene comparison database, SILVA v132 (Quast et al., 2013), was used to perform clustering at 99% similarity. Based on the species annotation and abundance of effective operational taxonomic units (OTUs), functional annotations were obtained based on Kyoto Encyclopedia of Genes and Genomes pathway analysis using PICRUSt2 (Douglas et al., 2020).
Phylogenetic Analysis
The OTU sequences of potential pathogenic genera and the reference 16S rRNA sequences of corresponding pathogenic species were aligned by using MUSCLE v3.8.1551 and the parameter of “-maxiters 2” (Edgar, 2004). Maximum likelihood phylogenetic trees were constructed with the best-fit substitution model implemented in IQ-TREE v2.0.3 (Nguyen et al., 2015).
Microbiome Interaction Analysis
The interaction network of bacteria within the microbiome was estimated at the genus level according to the Sparcc method (You et al., 2019). Connected retention data were saved with satisfaction of the P-value < 0.05 and correlation value > 0.6 thresholds in both methods. The interaction networks were visualized using the Cytoscape 3.7.2 software (Shannon et al., 2003).
Results
Bacterial Richness and Diversity
According to the TVC assay results, all plate counts of viable bacteria in oysters were approximately 2 log colony-forming units (CFU)/g. The samples from Guangzhou (GZ) have the highest richness of viable bacteria, and it showed no significant difference compared to the samples from Jiangmen (JM) and but a significant difference compared to those from Zhuhai (ZH) (P < 0.05, Figure 1). To further determine the microbial diversity in oysters, we analyzed the diversity of the microbiome within these samples. An average of 65,000 clean sequence reads, ranging from 40,691–105,248, were obtained for each sample. These clean sequences were clustered into 6,025 OTUs whose similarity was >99% (Supplementary Table 2).
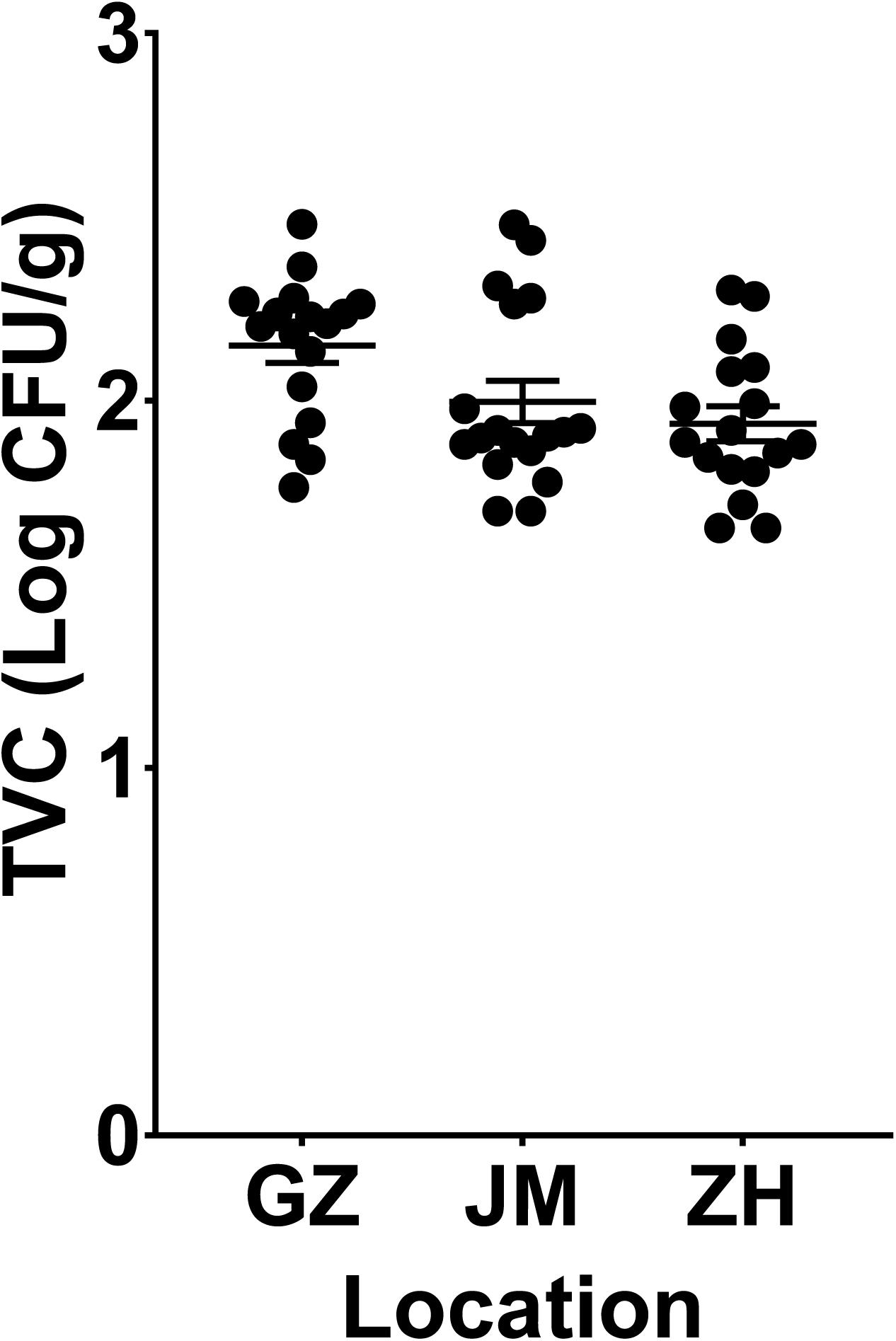
Figure 1. Evaluation of the total viable bacteria count in oyster samples collected from three cities in Guangdong Province, China. GZ, Guangzhou; JM, Jiangmen; ZH, Zhuhai.
We then estimated the alpha diversity of these samples according to the observed OTUs as well as the Chao1, ACE, Shannon, and Simpson indices. The results showed that the oyster samples from Guangzhou had the highest alpha diversity (Figure 2A). Additionally, the samples from Jiangmen had a significantly lower alpha diversity than those from Guangzhou (P = 0.029; Kruskal–Wallis test for the Shannon index), and these findings were different from the TVC assay results. This discrepancy could be easily inferred because the 16S rRNA sequencing revealed the total bacterial species information, whereas the TVC assay results only reflected the viable bacteria (Caporaso et al., 2010). Based on the greater accuracy of OTU estimates, the species abundance of oyster samples from different cities may show a difference, and the transported samples in Guangzhou seemed to be more complicated than those locally obtained samples.
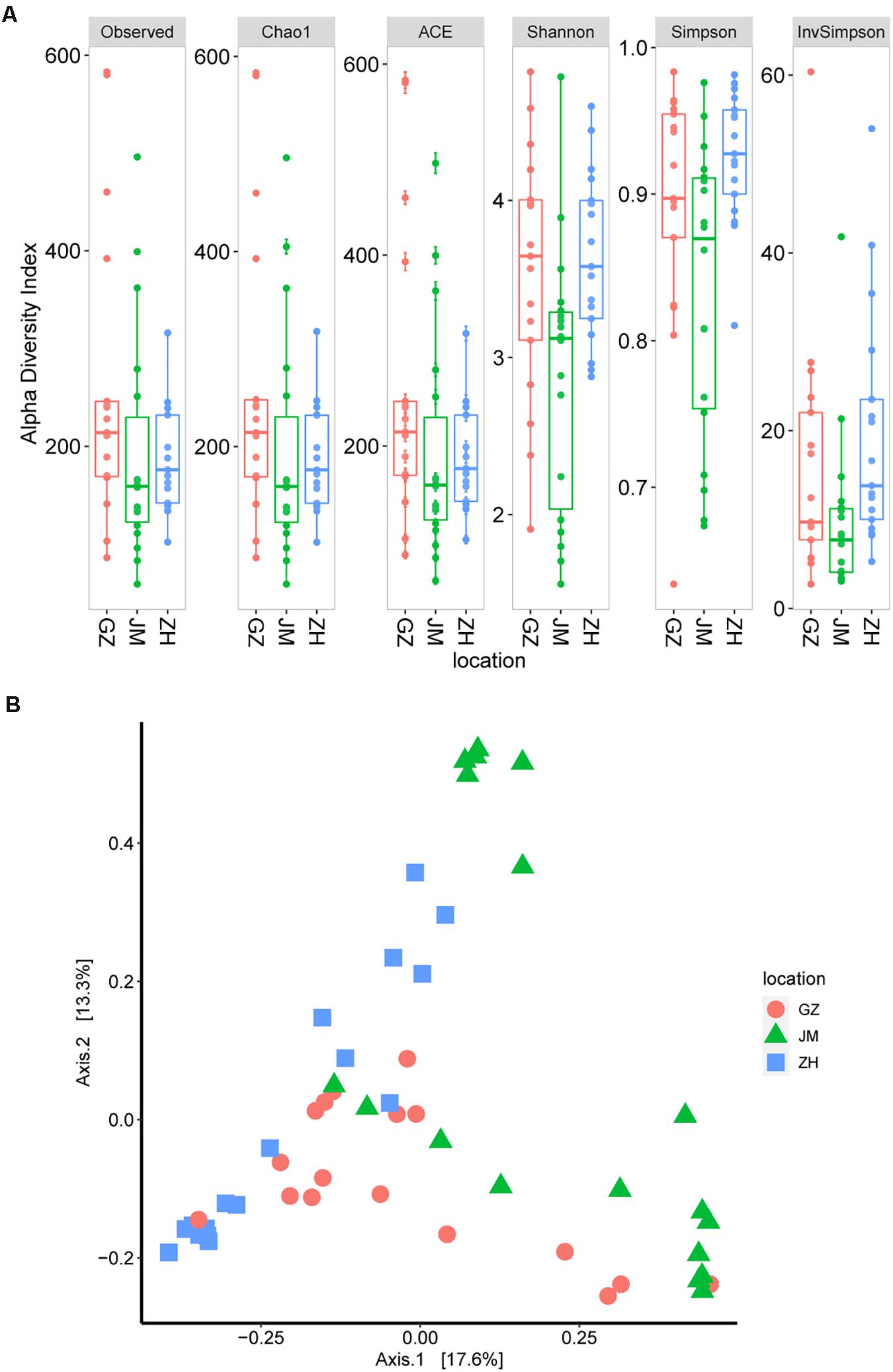
Figure 2. Microbiome diversity in oyster samples. (A) Boxplot of alpha diversity measured by the observed operational taxonomic units as well as theChao1, ACE, Shannon, Simpson, and InvSimpson indices. (B) Principal coordinate analysis plot illustrating the data based on beta-diversity metrics measured by using the Bray–Curtis distance.
The high diversity of microbial communities in oysters could also be inferred from the beta diversity index. According to the principal coordinate analysis of beta diversity, which is generally measured by using the Bray–Curtis distance (Pannaraj et al., 2017), no evident clusters were observed across geographical differences (Figure 2B). These results suggest the existence of a high level of bacterial diversity in oyster samples collected from different cities or even in those within the same city.
Composition of the Bacterial Community
The composition of the bacterial community was studied at the phylum and genus levels of classification. At the phylum level, members of Proteobacteria predominant in oysters collected across different cities; however, Firmicutes, Bacteroidetes, and Tenericutes were the most variable phyla (Figure 3A). The abundance of Firmicutes in the oysters from Zhuhai reached the highest bacterial community composition, up to 43.62%, whereas they accounted for only 4.47% in the oysters collected from Guangzhou. However, this trend showed reverse results when the abundance of Bacteroidetes was investigated. Bacteroidetes constituted the largest proportion of the bacterial community, up to 36.98%, in the oysters collected from Guangzhou; however, they accounted for only 9.57% in the oysters collected from Zhuhai. The variable tendencies of Tenericutes obtained from Guangzhou to Jiangmen to Zhuhai were the same as those of Bacteroidetes; however, Tenericutes represented a lower community abundance than Bacteroidetes. At the genus level, the most variable genera were Lactococcus, Vibrio, and Aeromonas. Lactococcus and Vibrio constituted the largest proportions in Zhuhai (28.90 and 28.44%, respectively), whereas Aeromonas accounted for a higher proportion in Jiangmen (15.68%) (Supplementary Figure 2). Notably, an important undetermined genus accounted for 19.72% bacterial abundance in samples collected from Guangzhou, and the presence of another unidentified genus was found to be consistent among the genera isolated from samples collected from three cities (Supplementary Figure 2). The results showed that the dominant microbes were highly variable. Therefore, we examined the detection rates of various genera in the samples. The heatmap indicated that Vibrio, Shewanella, Aeromonas, Mycoplasma, and Acinetobacter were the most prevalent and chief genera present in the oysters, while Ralstonia, Ochrobactrum, Clostridium, Streptococcus, and Sphaerochaeta displayed relatively high abundance levels but were not present in all samples (Figure 3B).
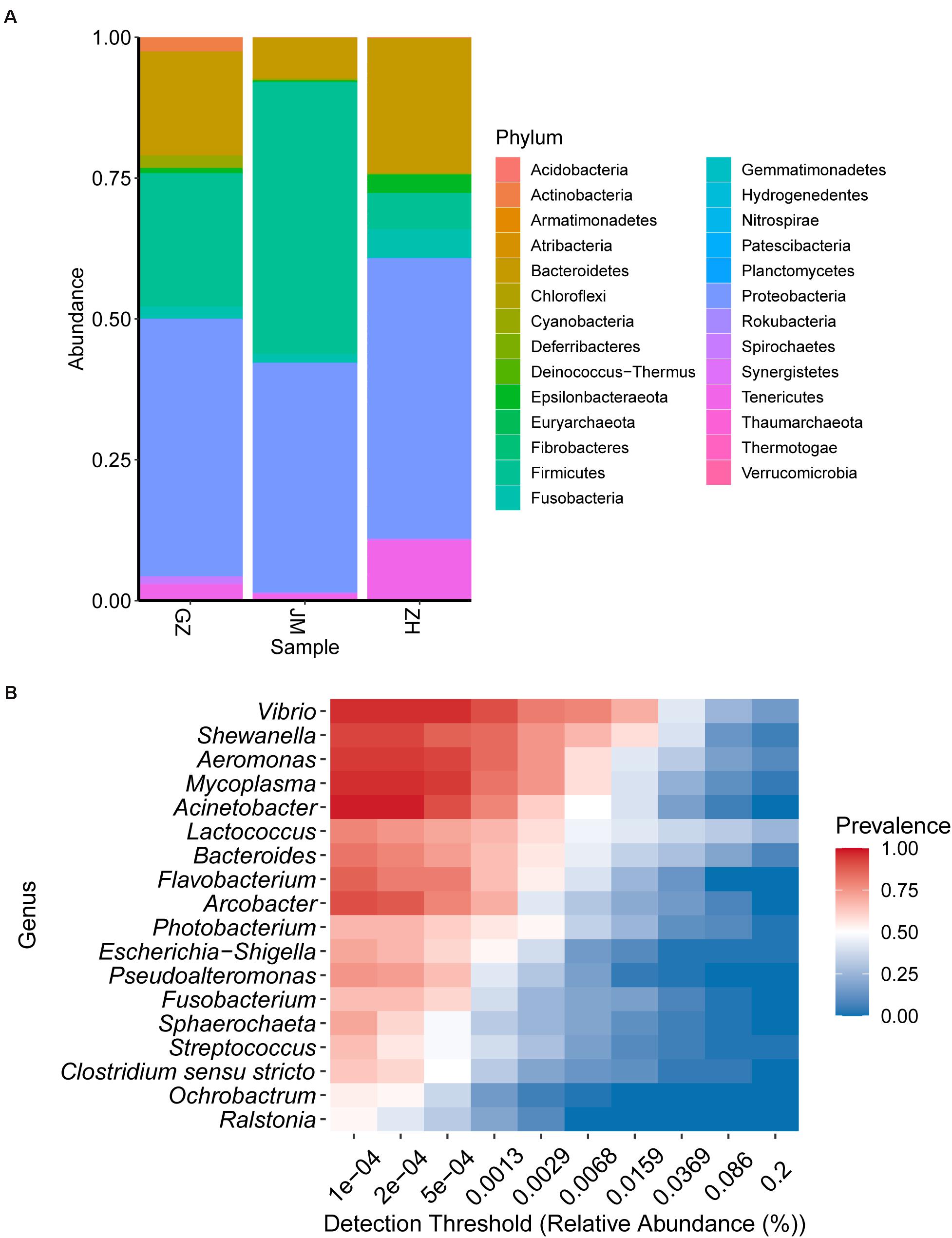
Figure 3. Bacterial community structure in oyster samples. (A) The relative abundance of bacterial species at the phylum level. (B) Heatmap illustrating the distribution of the major genera among all samples.
Functional Properties of the Bacterial Community
According to the composition of the bacterial community investigated herein, the functional properties of the bacterial community were determined across all samples using PICRUSt2 (Supplementary Table 3). We then analyzed the correlation between highly abundant genera and the top annotated metabolic pathways. The results predicted that the core dominant genus, Vibrio, was positively correlated with fatty acid elongation, adenine, and adenosine salvage III pathways; however, Shewanella, the second most abundant genus, showed a negative correlation with most of the top annotated metabolic pathways. Similar negative correlation patterns were also observed for the genera Mycoplasma, Acinetobacter, Flavobacterium, Fusobacterium, and Sphaerochaeta. Additionally, Lactococcus showed a strong positive correlation with the pathway of pyrimidine nucleobase salvage and a negative correlation with aerobic respiration I (cytochrome c). Notably, although the genera Ralstonia and Ochrobactrumhad relatively lower detectable rates, they exhibited remarkable correlations with many metabolic pathways (Figure 4).
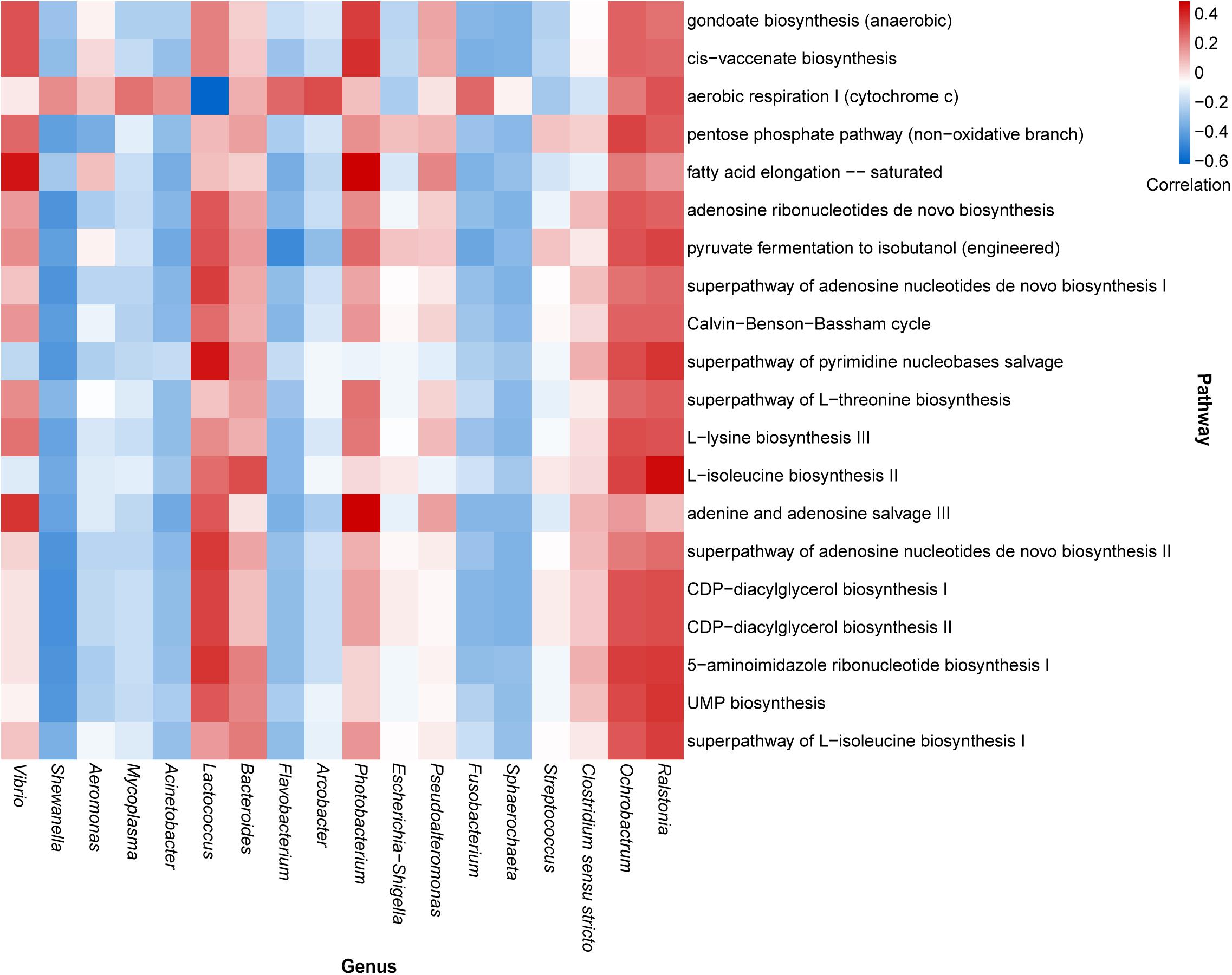
Figure 4. Heatmap illustrating the correlation between the highly abundant genera and top enriched metabolic pathways in oyster samples.
Interaction Network of Potentially Pathogenic Bacteria and Other Bacteria in Oysters
Vibrio, Staphylococcus, Escherichia-Shigella, Cronobacter, Bacillus, Klebsiella, Pseudomonas, and Helicobacter are the genera that are known to include certain pathogenic species. Also, some OTUs belonged to these genera which were indeed highly homologous to the notorious pathogenic species in corresponding genus based on the phylogenetic analysis (Supplementary Figures 3–10). In the samples analyzed herein, pathogenic Vibrio species, such as V. parahaemolyticus, V. fluvialis, V. anguillarum, V. vulnificus, V. cholerae, V. fortis, and V. metschnikovii, were detected (Supplementary Figure 3). Among them, the prevalence of V. anguillarum was the highest (35/52), followed by V. parahaemolyticus (23/52), V. vulnificus (13/52), and V. metschnikovii (3/52). V. fluvialis and V. cholerae were detected in one sample (1/52) (Supplementary Table 2). We then constructed an interaction network between the genera potentially harboring pathogenic species and all other genera. Accordingly, Vibrio was found to establish marked interactions with Photobacterium, followed by Propionigenium, and these interactions were positively correlated (Figure 5). Furthermore, Vibrio was positively correlated with Lactococcus, one of the most abundant genera observed in oyster samples. Bacteroides, the genus with higher relative abundance, was positively correlated with Cronobacter, Bacillus, Escherichia-Shigella, and Klebsiella. Cronobacter and Klebsiella were positively correlated with many genera; however, they were also negatively correlated with a few genera, such as Janthinobacterium and Leisingera. Additionally, Janthinobacterium and Leisingera were negatively correlated with Staphylococcus. Notably, the interactions of Staphylococcus and Helicobacter with other bacteria in oysters showed negative correlation (Figure 5).
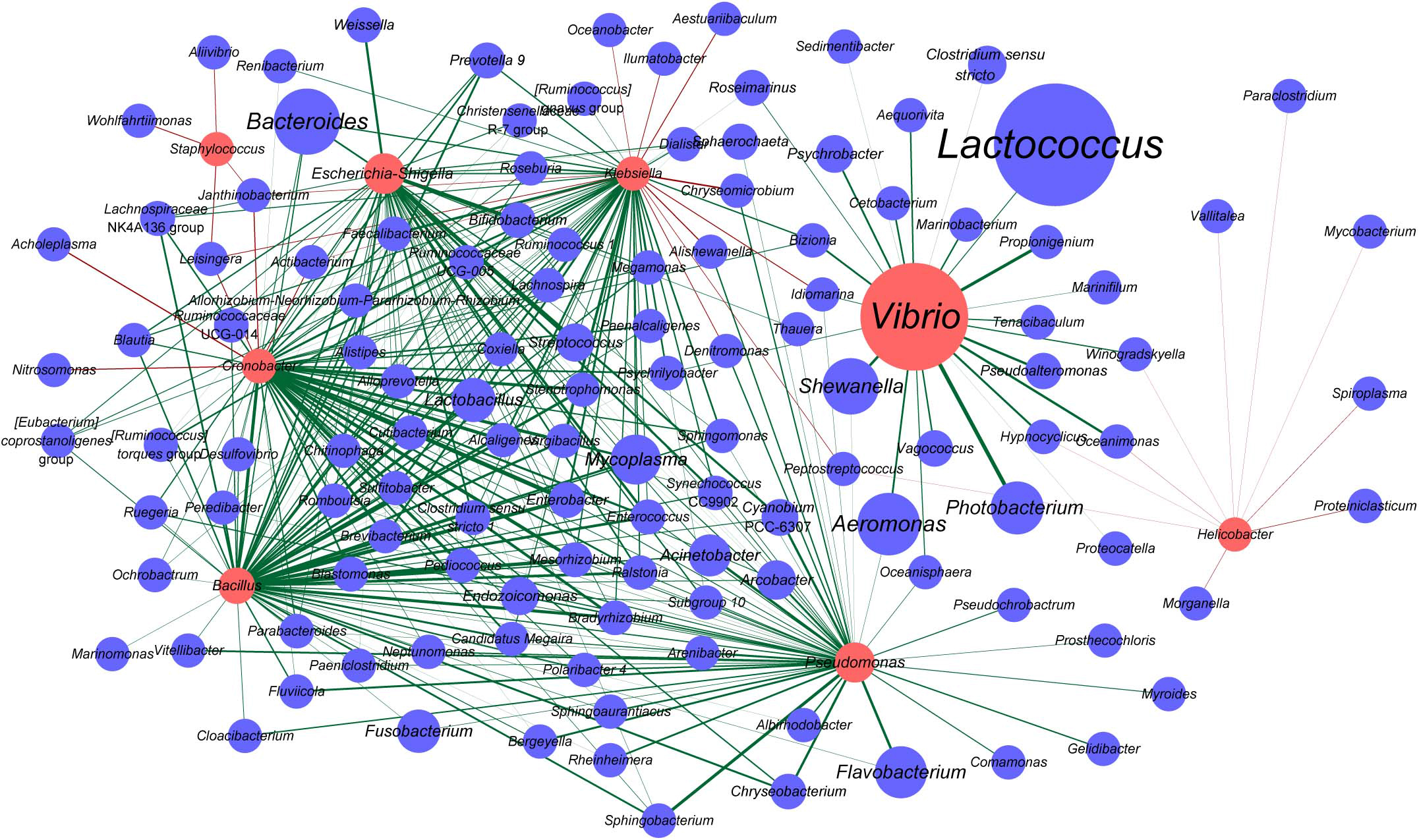
Figure 5. Interaction network of the genera harboring potentially pathogenic species and other bacteria in oysters. The nodes represent taxa at the genus level, and the edges represent correlations between taxa pairs. The nodes in red indicate potentially pathogenic genera. The sizes of the nodes vary according to the total abundance of the corresponding genus in the oyster samples. The edges are colored based on the correlation types, with green indicating positive correlations and red indicating negative correlations.
Discussion
Since oysters are typically reared in uncontrolled, and often dynamic, coastal and estuarine environments, prediction, management, and control of their microbial communities poses challenges (King et al., 2019). Depending on the environment in which oysters are reared, they are subject to different microbial infections, which can also be affected by human activities (Guo and Ford, 2016). The microbial criteria for obtaining satisfactory-quality oysters at wholesale have been set at 5.7–6.2 log CFU/g by Australia and New Zealand and the U.S. Food and Drug Administration (US Food and Drug Administration, 2007; Food Standards Australia New Zealand, 2011; Fernandez-Piquer et al., 2013). In addition, the TVCs in the retail oyster samples collected from the Guangdong area were lower than the reported oysters collected in other places. The TVCs in the Pacific oysters harvested by a commercial grower in Pipeclay Lagoon, Tasmania, Australia, fluctuate between 4 and 6 log CFU/g (Fernandez-Piquer et al., 2011). And in the Pacific oysters harvested in Tasmania, Australia, the mean TVCs predicted in the short supply chain were greater than 6 log CFU/g (Fernandez-Piquer et al., 2013). Therefore, the TVCs in the collected oyster samples in our study were acceptable (Figure 1). All indices presented in Figure 2A jointly indicated that there was a high bacterial diversity of oysters in the Guangdong area. Compared with the oyster samples from JM and ZH, the samples from GZ showed higher microbial richness and diversity. The richness and diversity could be influenced by many factors including growing environment and storage (Trabal et al., 2012; Chen et al., 2017). The raw oysters in Guangzhou seafood market are transported from other places, while the oysters sold in JM and ZH markets are local. The oysters sold in GZ underwent a longer storage, and their growing environment were unknown. These may be the reasons for the higher abundance and diversity of microorganisms in these oysters.
Proteobacteria was the dominant phylum in samples collected from all three cities. Among the five genera with the highest prevalence in oyster samples from the Guangdong region, Vibrio, Shewanella, Aeromonas, and Acinetobacter belong to the phylum Proteobacteria. Notably, Shewanella is spoilage-causing bacteria present in iced fresh seafood (Gram and Huss, 1996), and the higher abundance of this genera in samples of Guangzhou indicated that the transport process might promoter the growth of spoilage bacteria (Supplementary Figure 2). Additionally, the genera of the bacteria identified in the samples, including Escherichia-Shigella, Photobacterium, Arcobacter, Pseudoalteromonas, and Ochrobactrum, also belong to Proteobacteria. Firmicutes was the dominant phylum in oyster samples from Guangzhou and Jiangmen. Lactococcus, Streptococcus, and Clostridium sensustricto were identified in the samples belonging to Firmicutes. Bacteroidetes was the dominant phylum in oysters collected in Guangzhou and Zhuhai. Bacteroides and Flavobacterium present in the samples belong to the phylum Bacteroidetes. These three phyla were also the main phyla present in oysters in previous reports (Math et al., 2010; Adams et al., 2019; Chen et al., 2019).
In fact, most of the bacterial isolates detected in our samples were typically obtained from oysters. Some isolates are human pathogens, and some others accelerate oyster spoilage. The most abundant species in our sample belonged to the genus Vibrio. Members of the genus Vibrio, usually found in oysters, are natural inhabitants of the marine environment (Prapaiwong et al., 2009). Although many Vibrio species are harmless to humans, few species can cause diseases in humans (Froelich and Noble, 2016). Especially, the major pathogenic Vibrio species, V. vulnificus, which can cause sepsis and wound infection, was detected in multiple samples. We also detected several OTUs homologous to V. parahaemolyticus according to the phylogenetic analysis (Supplementary Figure 3). The presence of these two Vibrio sp. in oysters were occasionally reported (Cook et al., 2002; Kaufman et al., 2003; Jones et al., 2014). Although not all of the V. parahaemolyticus and V. vulnificus strains are pathogenic to humans, the presence of important virulence genes in the two Vibrio sp. isolated from oysters could reach a relatively high frequency in some cases (>50%) (DePaola et al., 2003; Turner et al., 2014; Davis et al., 2021). Climate change and rising sea temperatures that favor the spread of Vibrio were also thought to be partly responsible for the rise in vibriosis (Baker-Austin et al., 2018). Particularly, the prevalence of the thermostable direct hemolysin (tdh) and thermostable direct-related hemolysin (trh) genes in V. parahaemolyticus was found to increase with temperature (Turner et al., 2014). Davis et al. also found that the prominent pathogenic V. parahaemolyticus strains in South Puget Sound flourish with exposure to relatively warm temperature (Davis et al., 2021). Our samples were collected in mid-October 2020 (Supplementary Table 1), and the average temperature in Guangdong region during this period was 23.7°C, which was considered to be relatively suitable for the growth of Vibrios (Turner et al., 2014). In addition, although short-term storage at low temperatures can limit bacterial growth in oysters (Spaur et al., 2020), V. parahaemolyticus will not thoroughly death and can recover their viability in appropriate situation (Xie et al., 2019). This may partially explain the observation that the oysters sold in Guangzhou city still showed high abundance of V. parahaemolyticus and V. vulnificus after a period of storage (Supplementary Table 2). Thus, further survey targeting the occurrence of known virulence factors for V. parahaemolyticus or V. Vulnificus and their viable densities is needed to exactly clarify the health risk from these two Vibrio sp.
Besides, V. cholerae, V. anguillarum, V. fluvialis, and V. furnissii, which can cause diarrhea or other human infections (Chowdhury et al., 2013; Ballal et al., 2017; Baker-Austin et al., 2018; Sinatra and Colby, 2018), were also found in a small number of oyster samples. V. cholerae has become the most studied Vibrio due to the serious impact of cholera on human health caused by it (Faruque et al., 1998). Patients with diarrhea due to the infection of V. cholerae often die if they do not receive prompt treatment (Chen et al., 2020). Although V. metschnikoviiis were rarely observed in human infections, it has been associated with a few cases of sepsis, wound infection, cholecystitis, and pneumonia (Jean-Jacques et al., 1981; Hansen et al., 1993; Linde et al., 2004; Wallet et al., 2005). A number of virulence factors make Vibrio infections fatal to humans. Bacterial hemolysins have been identified as important virulence factors of Vibrio due to their contribution to hemorrhagic septicemia (Zhang and Austin, 2005). Cholera toxin is important for the severe diarrhea caused by V. cholerae (Rivera-Chávez and Mekalanos, 2019). Therefore, the assessment of the presence of pathogenic Vibrio sp. was considered necessary due to the increasing importance of some related infections (Koh et al., 1994; Tantillo et al., 2004).
In addition to Vibrio, other microbial genera identified in the samples also include species that are pathogenic to humans and often cause a variety of infections. Acinetobacter species are non-fermentative, gram-negative coccobacilli that are ubiquitous in the environment. A few species of this genus reportedly cause human infections, including A. radioresistens, A. calcoaceticus, and A. lwoffii (Rathinavelu et al., 2003; Nonaka et al., 2014; Wang et al., 2019). Bacteroides fragilis is one of the most prevalent members of the genus Bacteroides has been reported as a common opportunistic pathogen in clinical infections. It may cause a range of diseases involving a permeable intestinal barrier (Sun et al., 2019). Arcobacter species are considered emerging gastrointestinal pathogens (Van den Abeele et al., 2018). A. skirrowii is a gram-negative pathogenic microorganism that is abundant in various aquatic environments (Morita et al., 2004; Ertas et al., 2010). It is responsible for diseases, including watery diarrhea and septicemia in humans (Oliveira et al., 2018). Regarding other genera, although we did not find a specific disease-causing microbial species, there were a few species in the samples that could not be identified, and these unknown microbes could pose a potential health risk to humans. For example, in these samples, abundance of Aeromonas and Escherichia-Shigella was observed, and species of these genera cannot be easily distinguished from one another. Aeromonas species can cause a variety of infections, including sepsis and gastrointestinal diseases (Altwegg and Geiss, 1989). Escherichia-Shigellais the gram-negative bacterium that can cause bacillary dysentery (shigellosis) in humans (Belotserkovsky and Sansonetti, 2018).
Certain core genera may be correlated with metabolic pathways in oysters. For example, certain Vibrio species reportedly exhibit a strong ability to produce long-chain fatty acids (Massengo-Tiassé and Cronan, 2008; Estupiñán et al., 2020), as evidenced by the correlation analysis presented in Figure 4. Vibrio has also been correlated with adenine and adenosine salvage pathways and the recycling of adenine and adenosine into adenosine triphosphates (Schuster and Kenanov, 2005). It has been reported that V. parahaemolyticus can use adenosine produced by the hydrolysis of nucleotides (Sakai et al., 1987). Although Shewanella, Mycoplasma, Acinetobacter, Flavobacterium, and Fusobacterium were negatively correlated with most of the metabolic pathways, they were markedly correlated with aerobic respiration I (cytochrome c). Cytochrome c oxidase plays a central role in aerobic respiration, which is an efficient energy-producing metabolic process (Tosha and Shiro, 2013). This reflects the active energy metabolism of these microorganisms. Lactococcus is a facultative anaerobic bacterium. Although it showed a negative correlation with aerobic respiration I, other energy metabolism pathways might also provide energy for the process. The most studied species of Lactococcus is L. lactis, which can be used to perform fermentation in the food industry and can be used in medical engineering (Liu et al., 2019). L. lactis was not identified in our samples, but there was a considerable number of unknown Lactococcus species in our sample. As Lactococcus exhibited a strong correlation with many metabolic pathways, including the biosynthesis of a variety of substances (Figure 4), and as there was a presence of L. hircilactis that might be used as aromatic cultures in cheesemaking (Tidona et al., 2018), we suggest that identification of new Lactococcus species that synthesize specific metabolites can be achieved using the isolates obtained in the present study and can be used in food fermentation processes.
Interestingly, according to the co-occurrence network analyzed herein (Figure 5), Vibrio and Lactococcus, the two dominant bacteria in oysters, were found to co-occur. Other microorganisms that establish interactions with Vibrio are not mutually exclusive to Vibrio. These results showed that the dominant Vibrio population might be symbiotic with a wide range of bacterial genera in oysters, and microorganisms that were negatively correlated with Vibrio remain to be identified. We suggest that this may also be one of the contributing factors to Vibrio exhibiting predominance in oysters.
Conclusion
In conclusion, a variety of microorganisms were detected in retail Pacific oysters collected in Guangdong, China. The dominant bacteria in oysters belonged to the genus Vibrio, which includes a variety of potentially pathogenic bacteria. Thus, the detection of pathogenic bacteria, in particular the pathogenic Vibrio sp., as well as their viable densities and the presence of their virulence factors, is necessary before oysters entering the consumer market. In addition, it will be useful to establish a certification program in oyster farming areas that classifies oysters based on microbial testing results. These measures can help to ensure food security when consuming raw oysters. In addition, certain microbes in other genera may also cause intestinal diseases in humans, several measures can be implemented to reduce the risk of infection. Most bacteria, including those belonging to the genus Vibrio, cannot survive at high temperatures. Compared to raw seafood, Vibrio species in fully cooked seafood were less frequently detected (Monsreal et al., 2015). Thus, oyster consumers are advised to avoid eating raw oysters to reduce the risk of infection. Oyster processors should also avoid wound contact with oysters.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA736520.
Author Contributions
MY designed and supervised the entire study, helped in microbial diversity analysis and microbiome interaction analysis, and wrote the manuscript. XW carried out sample collection and DNA extraction. AY carried out microbiological assays. All authors read and commented on the manuscript.
Funding
This work was supported by the Foundation of Foshan Polytechnic (KY201902 and KY201903) and the Innovation Projects of Colleges and Universities in Guangdong Province (2019GKTSCX120).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.689520/full#supplementary-material
Supplementary Figure 1 | The sampling sites of oyster samples in this study.
Supplementary Figure 2 | The relative abundance species of bacteria at the genus level.
Supplementary Figure 3 | Maximum likelihood phylogeny of OTUs belonged to genus Vibrio and the notorious pathogenic Vibrio species. The pathogenic species are indicated with orange.
Supplementary Figure 4 | Maximum likelihood phylogeny of OTUs belonged to genus Staphylococcus and the notorious pathogenic Staphylococcus species. The pathogenic species are indicated with orange.
Supplementary Figure 5 | Maximum likelihood phylogeny of OTUs belonged to genus Shigella and the notorious pathogenic Shigella species. The pathogenic species are indicated with orange.
Supplementary Figure 6 | Maximum likelihood phylogeny of OTUs belonged to genus Cronobacter and the notorious pathogenic Cronobacter species. The pathogenic species are indicated with orange.
Supplementary Figure 7 | Maximum likelihood phylogeny of OTUs belonged to genus Bacillus and the notorious pathogenic Bacillus species. The pathogenic species are indicated with orange.
Supplementary Figure 8 | Maximum likelihood phylogeny of OTUs belonged to genus Klebsiella and the notorious pathogenic Klebsiella species. The pathogenic species are indicated with orange.
Supplementary Figure 9 | Maximum likelihood phylogeny of OTUs belonged to genus Pseudomonas and the notorious pathogenic Pseudomonas species. The pathogenic species are indicated with orange.
Supplementary Figure 10 | Maximum likelihood phylogeny of OTUs belonged to genus Helicobacter and the notorious pathogenic Helicobacter species. The pathogenic species are indicated with orange.
Supplementary Table 1 | The information of sample collection in this study.
Supplementary Table 2 | Relative abundance of the observed OTUs among all samples.
Supplementary Table 3 | Functional annotations based on the KEGG pathway using PICRUSt 2.
References
Ackman, R. G. (1990). Seafood lipids and fatty acids. Food Rev. Int. 6, 617–646. doi: 10.1080/87559129009540896
Adams, S., Che, D., Hailong, J., Zhao, B., Rui, H., Danquah, K., et al. (2019). Effects of pulverized oyster mushroom (Pleurotusostreatus) on diarrhea incidence, growth performance, immunity, and microbial composition in piglets. J. Sci. Food Agric. 99, 3616–3627. doi: 10.1002/jsfa.9582
Altwegg, M., and Geiss, H. K. (1989). Aeromonas as a human pathogen. Crit. Rev. Microbiol. 16, 253–286. doi: 10.3109/10408418909105478
Baker-Austin, C., Oliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers 4:8. doi: 10.1038/s41572-018-0005-8
Ballal, M., Shetty, V., Bangera, S. R., Prabhu, M., and Umakanth, S. (2017). Vibrio furnissii, an emerging pathogen causing acute gastroenteritis: a Case Report. JMM Case Rep. 4:e005111. doi: 10.1099/jmmcr.0.005111
Belotserkovsky, I., and Sansonetti, P. J. (2018). Shigella, and enteroinvasive Escherichia coli. Curr. Top. Microbiol. Immunol. 416, 1–26. doi: 10.1007/82_2018_104
Boziaris, I. S., and Parlapani, F. F. (2017). Specific spoilage organisms (SSOs) in fish inThemicrobiologicalquality of food. Amsterdam: Elsevier, 61–98. doi: 10.1016/B978-0-08-100502-6.00006-6
Cabello, A. M., Lezama, R. D. V., Garcia, B. E. F., Marcano, M. C. R., Figueroa, Y. D. M., and Gonzalez, O. M. V. (2004). Freshness parameters of mollusks. Rev. Cient. Fac. Cienc. Vet. 14, 457–466.
Caglak, E., Cakli, S., and Kilinc, B. (2008). Microbial, chemical and sensory assessment of mussels (Mytilus galloprovincialis) stored under modified atmosphere packaging. Eur. Food Res. Technol. 226, 1293–1299.
Cao, R., Liu, Q., Yin, B. Z., and Zhu, L. L. (2010). Combined effect of ozonated water and chitosan on the shelf-life of Pacific oyster (Crassostrea gigas). Innov. Food Sci. Emerg. Technol. 11, 108–112. doi: 10.1016/j.ifset.2009.08.006
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, H., Wang, M., Lin, X., Shi, C., and Liu, Z. (2017). Bacterial microbiota profile in gills of modified atmosphere-packaged oysters stored at 4 °C. Food Microbiol. 61, 58–65. doi: 10.1016/j.fm.2016.08.006
Chen, H., Wang, M., Yang, C., Wan, X., Ding, H. H., Shi, Y., et al. (2019). Bacterial spoilage profiles in the gills of Pacific oysters (Crassostrea gigas) and Eastern oysters (C. virginica) during refrigerated storage. Food Microbiol. 82, 209–217. doi: 10.1016/j.fm.2019.02.008
Chen, J., Qi, L., Zheng, Y., and Gao, L. (2020). Review on pathogenic Vibrio and its phage control. J. Food Safety Qual. 11, 9288–9294. doi: 10.19812/j.cnki.jfsq11-5956/ts.2020.24.039
Chen, X., Zhu, Q., Yu, F., Zhang, W., Wang, R., Ye, X., et al. (2018). Serology, virulence and molecular characteristics of Vibrio parahaemolyticus isolated from seafood in Zhejiang Province. PLoS One 13:e0204892. doi: 10.1371/journal.pone.0204892
Chowdhury, G., Sarkar, A., Pazhani, G. P., Mukhopadhyay, A. K., Bhattacharya, M. K., and Ramamurthy, T. (2013). An outbreak of foodborne gastroenteritis caused by dual pathogens, Salmonella enterica serovar Weltevreden and Vibrio fluvialis in Kolkata, India. Foodborne Pathog. Dis. 10, 904–906. doi: 10.1089/fpd.2013.1491
Cook, D. W., Leary, P. O., Hunsucker, J. C., Sloan, E. M., et al. (2002). Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: A national survey from June 1998 to July 1999. J. Food Prot. 65, 445–445.
Cruz-Romero, M., Kerry, J. P., and Kelly, A. L. (2008a). Fatty acids, volatile compounds and colour changes in high-pressure-treated oysters (Crassostrea gigas). Innov. Food Sci. Emerg. Technol. 9, 54–61. doi: 10.1016/j.ifset.2007.05.003
Cruz-Romero, M., Kelly, A. L., and Kerry, J. P. (2008b). Effects of high-pressure treatment on the microflora of oysters (Crassostrea gigas) during chilled storage. Innov. Food Sci.Emerg. Technol. 9, 441–447. doi: 10.1016/J.IFSET.2008.04.002
Davis, B. J. K., Corrigan, A. E., Sun, Z., Atherly, E., DePaola, A., and Curriero, F. C. (2021). A case-control analysis of traceback investigations for Vibrio parahaemolyticus infections (vibriosis) and pre-harvest environmental conditions in Washington State, 2013-2018. Sci. Total Environ. 752:141650. doi: 10.1016/j.scitotenv.2020.141650
DePaola, A., Nordstrom, J. L., Bowers, J. C., Wells, J. G., and Cook, D. W. (2003). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69, 1521–1526. doi: 10.1128/aem.69.3.1521-1526.2003
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 5, 1792–1797.
Ertas, N., Dogruer, Y., Gonulalan, Z., Guner, A., and Ulger, I. (2010). Prevalence of arcobacter species in drinking water, spring water, and raw milk as determined by multiplex PCR. J. Food Prot. 73, 2099–2102. doi: 10.4315/0362-028x-73.11.2099
Estupiñán, M., Hernández, I., Saitua, E., Bilbao, M. E., Mendibil, I., Ferrer, J., et al. (2020). Novel Vibrio spp. Strains producingomega-3 fatty acids isolated from coastal seawater. Mar. Drugs 18:99. doi: 10.3390/md18020099
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314.
Fernandez-Piquer, J., Bowman, J. P., Ross, T., and Tamplin, M. L. (2011). Predictive models for the effect of storage temperature on Vibrio parahaemolyticus viability and counts of total viable bacteria in Pacific oysters (Crassostrea gigas). Appl. Environ. Microbiol. 77, 8687–8695. doi: 10.1128/AEM.05568-11
Fernandez-Piquer, J., Bowman, J. P., Ross, T., Estrada-Flores, S., and Tamplin, M. L. (2013). Preliminary stochastic model for managing Vibrio parahaemolyticus and total viable bacterial counts in a Pacific oyster (Crassostrea gigas) supply chain. J. Food Prot. 76, 1168–1178. doi: 10.4315/0362-028X.JFP-12-401
Fisheries administration of Ministry of Agriculture and Rural affairs of the People’s Republic of China (2019). China Fishery Statistical Yearbook.23. Beijing: National Aquatic Technology Extension Center.
Food and Agriculture Organization (2011). Risk Assessment of Vibrio Parahaemolyticus in Seafood: Interpretative Summary and Technical Report. Microbiological Risk Assessment Series No. 16. Rome: FAO.
Food Standards Australia New Zealand (2011). Australia New Zealand food standards code. Part 1.6. Microbiological and processing requirements. Standard 1.6.1. Melbourne: Food Standards Australia New Zealand.
Froelich, B. A., and Noble, R. T. (2016). Vibrio bacteria in raw oysters: managing risks to human health. Philos. Trans. R. Soc. Lond. B 371:209. doi: 10.1098/rstb.2015.0209
Gram, L., and Dalgaard, P. (2002). Fish spoilage bacteria–problems and solutions. Curr. Opin. Biotechnol. 13, 262–266. doi: 10.1016/S0958-1669(02)00309-9
Gram, L., and Huss, H. H. (1996). Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 33, 121–137. doi: 10.1016/0168-1605(96)01134-8
Guo, X., and Ford, S. E. (2016). Infectious diseases of marine molluscs and host responses as revealed by genomic tools. Philos. Trans. R. Soc. Lond. B 371:26880838. doi: 10.1098/rstb.2015.0206
Hansen, W., Freney, J., Benyagoub, H., Letouzey, M. N., Gigi, J., and Wauters, G. (1993). Severe human infections caused by Vibrio metschnikovii. J. Clin. Microbiol. 31, 2529–2530. doi: 10.1128/JCM.31.9.2529-2530.1993
Huss, H. H., Ababouch, L., and Gram, L. (2014). “Assessment and management of seafood safety and quality: current practices and issues,” in FAO fisheries and aquaculture technical Paper No. 574, eds J. Ryder, I. Karunasagar, and L. Ababouch (Rome), FAO.
Jean-Jacques, W., Rajashekaraiah, K. R., Farmer, J. J., Hickman, F. W., Morris, J. G., and Kallick, C. A. (1981). Vibrio metschnikovii bacteremia in a patient with cholecystitis. J. Clin. Microbiol. 14, 711–712. doi: 10.1128/JCM.14.6.711-712.1981
Jones, J. L., Lüdeke, C. H. M., Bowers, J. C., DeRosia-Banick, K., Carey, D. H., and Hastback, W. (2014). Abundance of Vibrio cholerae, V. vulnificus, and V. parahaemolyticus in oysters (Crassostrea virginica) and clams (Mercenaria mercenaria) from Long Island sound. Appl. Environ. Microbiol. 80, 7667–7672. doi: 10.1128/AEM.02820-14
Jones, M. K., and Oliver, J. D. (2009). Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77, 1723–1733. doi: 10.1128/IAI.01046-08
Kaufman, G. E., Bej, A. K., Bowers, J., and DePaola, A. (2003). Oyster-to-oyster variability in levels of Vibrio parahaemolyticus. J. Food Prot. 66, 125–129. doi: 10.4315/0362-028x-66.1.125
King, W. L., Jenkins, C., Seymour, J. R., and Labbate, M. (2019). Oyster disease in a changing environment: decrypting the link between pathogen, microbiome and environment. Mar. Environ. Res. 143, 124–140. doi: 10.1016/j.marenvres.2018.11.007
Koh, E. G., Huyn, J. H., and Larock, P. A. (1994). Pertinence of indicator organisms and sampling variables to Vibrio concentrations. Appl. Environ. Microbiol. 60, 3897–3900. doi: 10.1128/AEM.60.10.3897-3900.1994
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Linde, H. J., Kobuch, R., Jayasinghe, S., Reischl, U., Lehn, N., Kaulfuss, S., et al. (2004). Vibrio metschnikovii, a rare cause of wound infection. J. Clin. Microbiol. 42, 4909–4911. doi: 10.1128/JCM.42.10.4909-4911.2004
Liu, Y., Guo, M., Du, R., He, X., Huang, K., and Xu, W. (2019). Advances and prospects of syntheticbiology in lactic acidbacteria [Biotech bulletin]. 325, 199–210.
Lund, B. M., and O’Brien, S. J. (2011). The occurrence and prevention of foodborne disease in vulnerable people. Foodborne Pathog. Dis. 8, 961–973. doi: 10.1089/fpd.2011.0860
Massengo-Tiassé, R. P., and Cronan, J. E. (2008). Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J. Biol. Chem. 283, 1308–1316. doi: 10.1074/jbc.M708171200
Math, R. K., Islam, S. M., Hong, S. J., Cho, K. M., Kim, J. M., Yun, M. G., et al. (2010). Metagenomic characterization of oyster shell dump reveals predominance of Firmicutes bacteria. Mikrobiologiia 79, 532–542. doi: 10.1134/S0026261710040132
Monsreal, J. F., Serralta-Peralta, L., Gómez, J. R. H., Sosa-Castilla, F., and Castillo-Cocom, J. A. (2015). Prevalence of clinically important species of the genus vibrio in catered seafood of city and port of progreso de castro, yucatan, mexico. Medwave 15, e6147–e6147. doi: 10.5867/medwave.2015.05.6147
Morita, Y., Maruyama, S., Kabeya, H., Boonmar, S., Nimsuphan, B., Nagai, A., et al. (2004). Isolation and phylogenetic analysis of Arcobacter spp. in ground chicken meat and environmental water in Japan and Thailand. Microbiol. Immunol. 48, 527–533.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fastand effective stochastic algorithm for estimating maximum-like lihoodphylogenies. Mol. Biol. Evol. 32, 268–274.
Nonaka, Y., Nagae, M., Omae, T., Yamamoto, S., Horitani, R., Maeda, D., et al. (2014). Community-acquired necrotizing fasciitis caused by Acinetobacter calcoaceticus: a case report and literature review. J. Infect. Chemother. 20, 330–335. doi: 10.1016/j.jiac.2013.12.011
Oliveira, M. G. X., Gomes, V. T. M., Cunha, M. P. V., Moreno, L. Z., Moreno, A. M., and Knöbl, T. (2018). Genotypic characterization of Arcobacter spp. Isolated from chicken meat in brazil. Foodborne Pathog. Dis. 15, 293–299. doi: 10.1089/fpd.2017.2368
Pannaraj, P. S., Li, F., Cerini, C., Bender, J. M., Yang, S., Rollie, A., et al. (2017). Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 171, 647–654. doi: 10.1001/jamapediatrics.2017.0378
Prapaiwong, N., Wallace, R. K., and Arias, C. R. (2009). Bacterial loads and microbial composition in high pressure treated oysters during storage. Int. J. Food Microbiol. 131, 145–150. doi: 10.1016/j.ijfoodmicro.2009.02.014
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rathinavelu, S., Zavros, Y., and Merchant, J. L. (2003). Acinetobacter lwoffii infection and gastritis. Microbes Infect. 5, 651–657. doi: 10.1016/s1286-4579(03)00099-6
Rey, M. S., Miranda, J. M., Aubourg, S., and Barros-Velázquez, J. (2012). Improved microbial and sensory quality of clams (Venerupis rhomboideus), oysters (Ostrea edulis) and mussels (Mytilus galloprovincialis) by refrigeration in a slurry ice packaging system. Int. J. Food Sci. Technol. 47, 861–869. doi: 10.1111/j.1365-2621.2011.02919.x
Rivera-Chávez, F., and Mekalanos, J. J. (2019). Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572, 244–248. doi: 10.1038/s41586-019-1453-3
Sakai, Y., Toda, K., Mitani, Y., Tsuda, M., and Tsuchiya, T. (1987). A novel mechanism for utilization of extracellular AMP in Vibrio parahaemolyticus. Biochem. Biophys. Res. Commun. 144, 382–386. doi: 10.1016/S0006-291X(87)80521-1
Sanna, T., Dallolio, L., Raggi, A., Mazzetti, M., Lorusso, G., Zanni, A., et al. (2018). ATP bioluminescence assay for evaluating cleaning practices in operating theatres: applicability and limitations. BMC Infect. Dis. 18:583. doi: 10.1186/s12879-018-3505-y
Schuster, S., and Kenanov, D. (2005). Adenine and adenosine salvage pathways in erythrocytes and the role of S-adenosyl homocysteine hydrolase. A theoretical study using elementary flux modes. FEBS J. 272, 5278–5290. doi: 10.1111/j.1742-4658.2005.04924.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sinatra, J. A., and Colby, K. (2018). Notes from the field: fatal Vibrio anguillarum infection in an immunocompromised patient - Maine, 2017. MMWR Morb. Mortal. Wkly Rep. 67, 962–963. doi: 10.15585/mmwr.mm6734a5
Spaur, M., Davis, B. J. K., Kivitz, S., DePaola, A., Bowers, J. C., Curriero, F. C., et al. (2020). A systematic review of post-harvest interventions for Vibrio parahaemolyticus in raw oysters. Sci. Total Environ. 745:140795. doi: 10.1016/j.scitotenv.2020.140795
Straub, D., Blackwell, N., Langarica-Fuentes, A., Peltzer, A., Nahnsen, S., and Kleindienst, S. (2020). Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (gene) amplicon sequencing pipeline. Front. Microbiol. 11:550420. doi: 10.3389/fmicb.2020.550420
Sun, F., Zhang, Q., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2019). A potential species of next-generation probiotics? The dark and light sides of Bacteroides fragilis in health. Food Res. Int. 126:108590. doi: 10.1016/j.foodres.2019.108590
Tantillo, G. M., Fontanarosa, M., Di Pinto, A., and Musti, M. (2004). Updated perspectives on emerging vibrios associatedwith human infections. Lett. Appl. Microbiol. 39, 117–126. doi: 10.1111/j.1472-765X.2004.01568.x
Tidona, F., Meucci, A., Povolo, M., Pelizzola, V., Zago, M., Contarini, G., et al. (2018). Applicability of Lactococcus hircilactis and Lactococcus laudensis as dairy cultures. Int. J. Food Microbiol. 271, 1–7. doi: 10.1016/j.ijfoodmicro.2018.02.015
Tosha, T., and Shiro, Y. (2013). Crystal structures of nitric oxide reductases provide key insights into functional conversion of respiratory enzymes. IUBMB Life 65, 217–226. doi: 10.1002/iub.1135
Trabal, N., Mazón-Suástegui, J. M., Vázquez-Juárez, R., Asencio-Valle, F., Morales-Bojórquez, E., and Romero, J. (2012). Molecular analysis of bacterial microbiota associated with oysters (Crassostrea gigas and Crassostrea corteziensis) in different growthphases at two cultivation sites. Microb. Ecol. 64, 555–569. doi: 10.1007/s00248-012-0039-5
Turner, J. W., Malayil, L., Guadagnoli, D., Cole, D., and Lipp, E. K. (2014). Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with respect to seasonal fluctuations in temperature and plankton abundance. Environ. Microbiol. 4, 1019–1028. doi: 10.1111/1462-2920.12246
US Food and Drug Administration (2007). Chap. U. Bacteriological examination of shellfish shipments decision tree, Sect. IV in National shellfish Sanitation Program guide for the control of molluscan shellfish. Washington, DC: U. S. Food and Drug Administration.
Van den Abeele, A. M., Vogelaers, D., Vandamme, P., Vanlaere, E., and Houf, K. (2018). Filling the gaps in clinical proteomics: a do-it-yourself guide for the identification of the emerging pathogen Arcobacter by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Microbiol. Methods 152, 92–97. doi: 10.1016/j.mimet.2018.07.007
Wallet, F., Tachon, M., Nseir, S., Courcol, R. J., and Roussel-Delvallez, M. (2005). Vibrio metschnikovii pneumonia. Emerg. Infect. Dis. 11, 1641–1642. doi: 10.3201/eid1110.050177
Wang, T., Costa, V., Jenkins, S. G., Hartman, B. J., and Westblade, L. F. (2019). Acinetobacter radioresistens infection with bacteremia and pneumonia. IDCases 15:e00495. doi: 10.1016/j.idcr.2019.e00495
Xie, T., Pang, R., Wu, Q., Zhang, J., Lei, T., Li, Y., et al. (2019). Cold tolerance regulated by the pyruvate metabolism in vibrio parahaemolyticus. Front. Microbiol. 10:178. doi: 10.3389/fmicb.2019.00178
You, Y., Liang, D., Wei, R., Li, M., Li, Y., Wang, J., et al. (2019). Evaluation of metabolite-microbe correlation detection methods. Anal. Biochem. 567, 106–111. doi: 10.1016/j.ab.2018.12.008
Keywords: Pacific oysters, 16S ribosomal DNA amplification sequencing, microbial diversity, Vibrio vulnificus, Crassostrea gigas
Citation: Yu M, Wang X and Yan A (2021) Microbial Profiles of Retail Pacific Oysters (Crassostrea gigas) From Guangdong Province, China. Front. Microbiol. 12:689520. doi: 10.3389/fmicb.2021.689520
Received: 01 April 2021; Accepted: 31 May 2021;
Published: 07 July 2021.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
Jinxing Ma, University of New South Wales, AustraliaMarcial Leonardo Lizárraga-Partida, Center for Scientific Research and Higher Education in Ensenada (CICESE), Mexico
Copyright © 2021 Yu, Wang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingjia Yu, eXVtaW5namlhQGZzcHQuZWR1LmNu
 Mingjia Yu
Mingjia Yu Xiaobo Wang
Xiaobo Wang